Cdc42p is the master regulator of morphogenesis in eukaryotic cells. It has an additional role in cell fusion, acting later in the pathway, after cells have undergone the changes in polarization and growth required for fusion. Cdc42p acts in concert with Fus2p to allow cell fusion.
Abstract
Cell fusion is the key event of fertilization that gives rise to the diploid zygote and is a nearly universal aspect of eukaryotic biology. In the yeast Saccharomyces cerevisiae, several mutants have been identified that are defective for cell fusion, and yet the molecular mechanism of this process remains obscure. One obstacle has been that genetic screens have mainly focused on mating-specific factors, whereas the process likely involves housekeeping proteins as well. Here we implicate Cdc42p, an essential protein with roles in multiple aspects of morphogenesis, as a core component of the yeast cell fusion pathway. We identify a point mutant in the Rho-insert domain of CDC42, called cdc42-138, which is specifically defective in cell fusion. The cell fusion defect is not a secondary consequence of ineffective signaling or polarization. Genetic and morphological data show that Cdc42p acts at a late stage in cell fusion in concert with a key cell fusion regulator, Fus2p, which contains a Dbl-homology domain. We find that Fus2p binds specifically with activated Cdc42p, and binding is blocked by the cdc42-138 mutation. Thus, in addition to signaling and morphogenetic roles in mating, Cdc42p plays a role late in cell fusion via activation of Fus2p.
INTRODUCTION
Cell fusion is widespread in the eukaryotic kingdom. In addition to the familiar fertilization events necessary to restore the diploid state after meiosis (Primakoff and Myles, 2002), fusion occurs throughout the development of many multicellular organisms, including the formation of muscle, bone, and the mammalian placenta (Vignery, 2000; Potgens et al., 2002; Horsley and Pavlath, 2004; Chen and Olson, 2005). In contrast to virus–cell fusion and fusion between intracellular membranes, the molecular mechanism of cell fusion is not well understood.
Cell fusion in budding yeast occurs during mating events between haploid cells (Marsh and Rose, 1997; Ydenberg and Rose, 2008). Yeast haploid cells of the a and α mating types each secrete pheromones sensed by a receptor expressed by the opposite cell type. This induces characteristic gene expression and developmental changes resulting in polarized growth or “shmooing” in the direction of the mating partner, followed by cell fusion and nuclear fusion. Although numerous mutants have been identified that block fusion, many of them are now known to have prior defects in pheromone signaling and polarization, suggesting that the cell fusion defect is a secondary consequence of inefficient completion of earlier mating events (Brizzio et al., 1996; Dorer et al., 1997; Paterson et al., 2008).
In contrast, four proteins have been identified that appear to play a direct role in yeast cell fusion without having detectable defects in earlier steps of mating. Mutations in FUS1, FUS2, and RVS161 block the removal of cell wall material between mating prezygotes (Trueheart et al., 1987; Trueheart and Fink, 1989; Brizzio et al., 1998), whereas PRM1 mutants block fusion after cell wall removal but before plasma membrane fusion (Heiman and Walter, 2000). The molecular function of these proteins is not known, but it may involve the trafficking and/or fusion of mating-specific vesicles that are visualized at the cell fusion zone (Gammie et al., 1998). These vesicles are delocalized in fus1 mutants, as well as in polarization mutants (e.g., bni1 and spa2). The vesicles are properly localized in fus2 and rvs161 mutants, suggesting that these mutations affect later stages of cell fusion than fus1. The behavior of the vesicles in prm1 mutant zygotes has not been reported.
Fus2p and Rvs161p form a complex that is transported to the tips of fusing cells (Brizzio et al., 1998; Paterson et al., 2008; Sheltzer and Rose, 2009). Rvs161p is a BAR domain–containing protein that plays a separate role in endocytosis (Crouzet et al., 1991) by stabilizing membrane curvature (Peter et al., 2004). Fus2p contains a Dbl-homology domain, suggesting that it interacts with a Rho-type G protein. In support of this, a large-scale two-hybrid screen identified an interaction between Fus2p and Cdc42p in the GTP-bound form (Nelson et al., 2004).
Yeast Cdc42p plays multiple essential roles during growth and morphogenesis (Johnson, 1999; Richman et al., 1999; Kozminski et al., 2000; Adamo et al., 2001), including during mating (Simon et al., 1995; Zhao et al., 1995). In late G1, Cdc42p establishes polarized growth toward a specific area of the cell surface to grow a new bud. During mating, the budding axis is overridden by cues that direct Cdc42p activity along the axis of the pheromone gradient (Nern and Arkowitz, 1998, 1999). Cdc42p has been implicated in cell fusion (Barale et al., 2006), but the stage at which it acts and its molecular functions are not clear. Here we present evidence that Cdc42 has multiple roles in cell fusion, that Fus2p is a direct effector of Cdc42p signaling, and that Cdc42p and Fus2p act together late in the cell fusion pathway. Our results have implications for the molecular mechanism of yeast cell fusion, the function of an uncharacterized domain in Cdc42p, and conservation between fungal and metazoan cell fusion mechanisms.
RESULTS
The Fus2p Dbl-homology domain promotes cell fusion
Fus2p contains a Dbl-homology domain (Paterson et al., 2008), suggesting that it may act as a guanine-nucleotide exchange factor for one of the six yeast Rho proteins. Conversely, a two-hybrid study suggested that Fus2p interacted with Cdc42p-Q61L, a GTP-locked allele (Nelson et al., 2004). These results suggested that Fus2p may be an effector for Cdc42p, since it interacted exclusively with the active form of the GTPase.
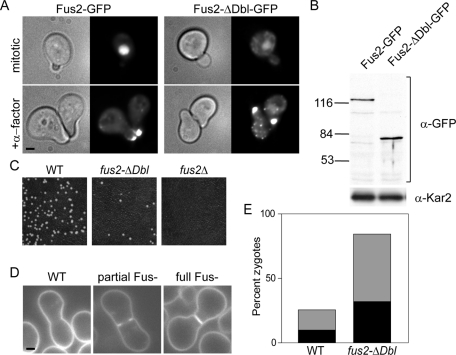
To assess the importance of the Dbl-homology domain in vivo, we examined the phenotype caused by a deletion of this domain. Fus2p-ΔDbl–green fluorescent protein (GFP) localized to the nucleus in mitotic cells and at the shmoo tip in pheromone-arrested cells, similar to wild-type Fus2p-GFP (Patterson et al., 2008). Therefore the Dbl-homology domain is required for neither localization in mitotic and pheromone-arrested cells nor the regulated change in localization (Figure 1A). Both proteins were detected at similar levels (Figure 1B). When this deletion was integrated into the genome to replace the endogenous FUS2 locus, the resulting strain was significantly defective for the formation of diploids, although not quite as defective as a complete deletion mutant (fus2Δ; Figure 1C). To examine the morphology of the zygotes formed by this strain, wild-type and fus2-ΔDbl strains were mated to a fus2Δ tester strain and stained with FM4-64 to visualize the plasma membrane (Figure 1D). We then counted the fully unfused zygotes (in which the septum between the mating partners remains intact) and partially unfused zygotes (in which there is a gap in the septum that fails to open completely). Both types of unfused zygotes are indicative of the defect in Fus2p function. The fus2ΔDbl mutant produced a large number of unfused and partially unfused zygotes (Figure 1E). Again, the phenotype is not as strong as a null fus2Δ strain, in which the majority of zygotes are fully unfused (compare to Figure 2B). These results indicate that the fus2ΔDbl allele is hypomorphic and that this domain is critically important but not absolutely required for Fus2p's cell fusion activity.
FIGURE 1:
The Dbl-homology domain is required for cell fusion. (A) Fus2p-ΔDbl-GFP localizes to the shmoo tip. Cells (MY9181) transformed with a plasmid expressing either Fus2p-GFP (pMR5469) or Fus2p-ΔDbl-GFP (pMR5883) were treated with pheromone as indicated for 2 h and examined. (B) Fus2p-ΔDbl-GFP is expressed at similar levels as the wild-type Fus2p-GFP protein. The same strains as in A were analyzed by Western blot using anti-GFP. (C) Diploid formation is defective in fus2-ΔDbl. Wild type (DDY1300), fus2Δ (MY10016), and fus2-ΔDbl (MY10933) were mated to fus1 fus2 (MY10798) for 4 h at 30°C. (D, E) Cell fusion is defective in fus2-ΔDbl. The same strains as in C were mated to fus2 (MY10797) for 2 h, washed into azide, and stained with FM4-64 to label the plasma membrane. (D) Examples of fusion-defective zygotes. Wild-type zygote (DDY1300xMY10797), partial Fus− zygote (MY10933xMY10797), and full Fus− zygote (MY10933xMY10797). (E) Percentage of zygotes of each morphology produced in matings with the indicated genotype. Gray bars, partial Fus− zygotes; black bars, full Fus− zygotes. n ≥ 133 zygotes imaged in two independent experiments.
FIGURE 2:
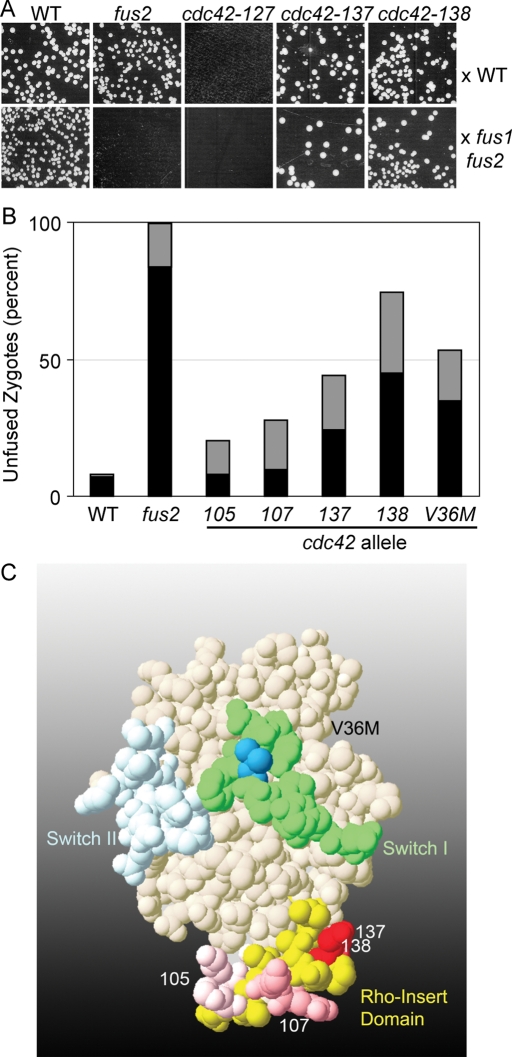
Mutants in the Cdc42p Rho-insert domain are defective for cell fusion. (A, B) A screen for cell fusion–specific alleles of Cdc42p. Wild type (WT; DDY1300), fus2 (MY10016), and 20 alleles from a Cdc42p alanine-scanning set were mated to an anti-WT strain (JY431; A) or an anti– fus1 fus2 strain (JY429; B). Shown are the alleles discussed in the text. Note that the matings against JY431 are more dilute than against JY429, so that colony formation against the two testers cannot be compared with one another. Images are representative of two independent experiments. (B) Microscopic examination of zygotes. Bilateral matings of the indicated genotype were mated for 2 h at 30°C, washed into azide, and stained with FM4-64. Shown is the percentage of zygotes of each morphology in the indicated genotypes (WT, DDY1300 × DDY1301; fus2, MY10016 × MY10303; cdc42-138, DDY1354 × DDY1355; cdc42-V36M, MY10547 × MY10554; see Kozminski et al., 2000, for other strain numbers). Gray bars, partial Fus− zygotes; black bars, full Fus− zygotes. n ≥ 122 zygotes imaged in at least two independent experiments. (C) Structure of Cdc42p indicating mutations affecting cell fusion. Mutations are mapped on a space-filling model of human Cdc42p (Brookhaven Protein Database, accession number 1AN0). The Rho-insert domain is shown in yellow, with mutations indicated in shades of red, correlated with the severity of the defect. The Switch I domain is shown in green, with the V36M mutation in dark blue. The Switch II domain is in light blue. Pseudo–wild-type mutations with no effect on cell fusion are not shown.
Mutations in the Rho-insert domain are defective in cell fusion
The interaction between activated Cdc42p and Fus2p suggested that Cdc42p may play a direct, specific role in cell fusion. Cdc42p has at least two defined roles in the mating pathway already: it is required for response to pheromone by activation of the PAK Ste20p (Moskow et al., 2000) and for defining the site of polarization of the shmoo (Nern and Arkowitz, 2000). One mutant allele of cdc42 that exhibits a defect in cell fusion has been previously characterized, cdc42-V36M (Barale et al., 2006). However, given the many potential roles for Cdc42p in mating, the specific defect of cdc43-V36M remained unclear. To look for additional alleles of CDC42 that might be specifically defective for Fus2p-mediated cell fusion, we used an alanine-scanning collection created previously (Kozminski et al., 2000). Twenty mutant alleles, annotated as wild type for growth, were mated to a wild-type strain or a fusion-defective tester strain and examined for the ability to form diploids by a plate mating assay (Figure 2A and Supplemental Figure S1). Because the fus2Δ single mutant exhibits an obvious defect when mated against a compromised partner (Figure 2A) but not when mated to the wild-type strain, we reasoned that this approach would allow us to identify alleles that were specifically defective in cell fusion. One allele, cdc42-127, did not mate with either the fus1 fus2 or the wild-type partner, indicating that it has a sterile (Ste−) phenotype (Figure 2A). This mutation, V33A, lies within the Switch I region and was not analyzed further. Two alleles, cdc42-137 and -138, exhibited significant mating defects with the fus1 fus2 tester strain but not with the wild-type strain, suggesting that they represent cell fusion–specific (Fus−) alleles (Figure 2B). The two mutations, D121A and D122A, respectively, lie within the Rho-insert domain (Figure 2C) and alter consecutive negatively charged residues.
To confirm that the mating defect against the compromised strain was due to reduced cell fusion efficiency, we quantified zygote morphology in these mutants. For these experiments, we used a bilateral mating configuration in which both partners contain the same mutation of interest. Under these conditions, wild-type zygotes typically have no visible septum between the mating partners, whereas cdc42-137 and 138 zygotes frequently had all or part of the septum intact, indicating that they had not fully fused (Figure 2B; 44% of cdc42-137 zygotes and 75% of cdc42-138 zygotes were fusion defective compared with 8% of wild-type zygotes).
Given the location of these alleles within the Rho-insert domain, we examined two other mutations constructed in the alanine-scanning collection that altered residues in this region but that had not exhibited a defect in the plate mating screen. Both mutants showed a significant cell fusion defect; cdc42-105 (E127A, K128A) produced 20% fusion defective zygotes, and cdc42-107 (R131A, R133A, R135A) produced 28% fusion defective zygotes (Figure 1C). These defects were not as strong as those for cdc42-137 and cdc42-138, explaining why they were missed in the less sensitive plate mating assay. Together the data indicate that the Rho-insert domain of Cdc42p, which had not previously been shown to have a specific function, plays a significant and unexpected role in cell fusion. To further characterize the defect in cell fusion, we focused on the more potent cdc42-138 allele.
The fusion defect in cdc42-138 is not due to defective pheromone signaling
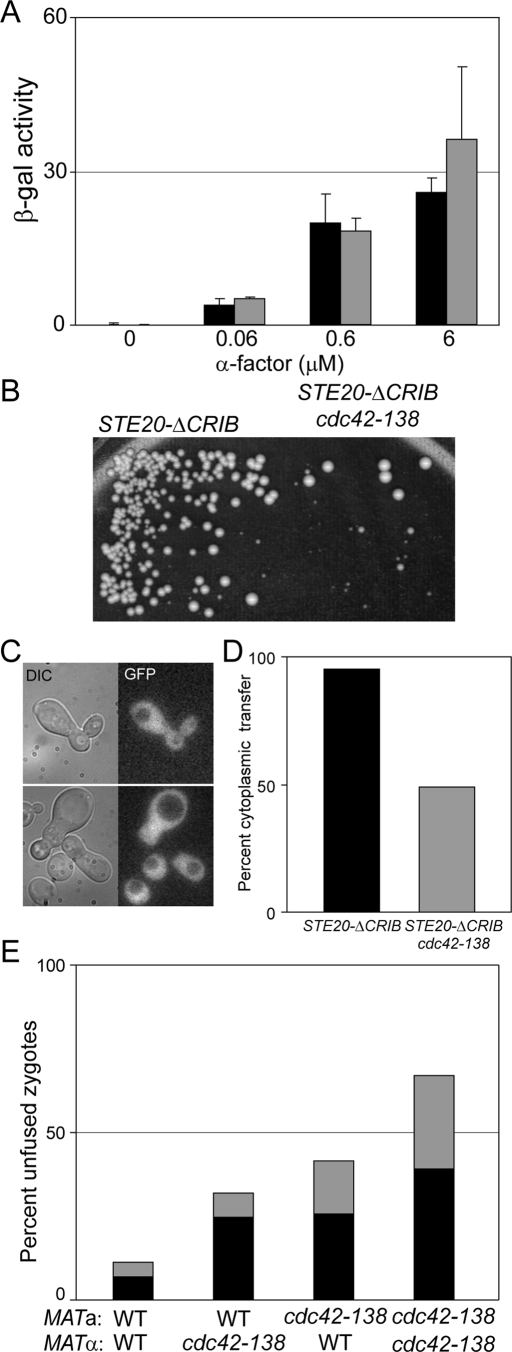
Cdc42p plays a known role in the pheromone-signaling pathway (Simon et al., 1995; Zhao et al., 1995). One possibility was that the defect in cell fusion is simply due to partly compromised signaling (Brizzio et al., 1996). To isolate Cdc42p from any effects on the pheromone pathway, we used a STE20-ΔCRIB mutation (Lamson et al., 2002), which removes the Cdc42p-binding domain in Ste20p, thereby rendering the protein and the pheromone response pathway insensitive to signaling from Cdc42p. In the STE20-ΔCRIB background, cdc42-138 had no effect on pheromone signaling (Figure 3A). In contrast, mating between STE20-ΔCRIB cdc42-138 double mutants was significantly more defective than matings between the STE20-ΔCRIB controls (Figure 3B). Therefore cdc42-138 is defective in an aspect of mating that is independent of signaling to Ste20p. We used cytoplasmic transfer assays to confirm that the mating defect was due to a cell fusion defect (Figure 3, C and D). When a fus2Δ tester strain expressing cytoplasmic GFP was mated to a STE20-ΔCRIB strain, the GFP was transferred to the partner in 95% of the zygotes (corresponding to Fus+ and partial Fus− zygotes). When the STE20-ΔCRIB strain also contained the cdc42-138 mutation, cytoplasmic transfer was observed in only 49% of zygotes, indicating that cdc42-138 had a full cell fusion defect in more than half of the zygotes in this strain (n ≥ 469). We conclude that CDC42 plays a role in cell fusion that is independent of its role in signal transduction.
FIGURE 3:
The cell fusion defect in cdc42-138 is not due to defective pheromone signaling. (A) cdc42-138 has no defect in signaling in the context of STE20-ΔCRIB. STE20Δ−CRIB (MY10925; black bars) and STE20-ΔCRIB cdc42-138 (MY10926; gray bars), transformed with fus1-LacZ (pSB231), were treated with the indicated concentrations of α-factor for 90 min and their response measured via β-galactosidase assay. Error bars represent SD of three independent experiments. (B–D) cdc42-138 still exhibits a cell fusion defect in the STE20-ΔCRIB background. (B) STE20-ΔCRIB (MY10925) and STE20-ΔCRIB cdc42-138 (MY10926) were mated to fus2 (MY10797) for 4 h at 30°C, and diploids were selected by replica plating to selective media (C, D) Cytoplasmic transfer assays were performed using the same strains as in B, in which one partner expresses cytoplasmic GFP. (C) Examples of zygotes showing transfer (top) or in which cell fusion was blocked (bottom). The latter phenotype corresponds to the “Full” Fus− cell fusion defect. (D) Quantification of the data in C. More than 460 zygotes were imaged in two independent experiments. (E) The defect in cdc42-138 is not mating-type specific. WT and cdc42-138 cells were mated in the orientations indicated. Gray bars, partial Fus− zygotes; black bars, full Fus− zygotes. n ≥ 176 zygotes imaged in two independent experiments.
A second possible explanation for the cell fusion defect in cdc42-138 is that the mutants produce reduced levels of a-factor or α-factor, leading to a fusion defect due to reduced signaling in the partner (Brizzio et al., 1996). Because the genes required for production and secretion of a-factor and α-factor are different, cell fusion mutations that affect pheromone levels are mating-type specific (Brizzio et al., 1996). In crosses between cdc42-138 strains and wild-type (WT) strains, the efficiency of cell fusion was independent of whether the a-cell or the α-cell contained the mutation (Figure 3E; a WT × α cdc42-138 produced 32% fusion-defective zygotes, compared with 41% for a cdc42-138 × α WT). Moreover, the defect was significantly stronger when both strains contained the cdc42-138 mutation, showing that Cdc42p function plays a role in cell fusion in both parents. These data indicate that the fusion defect in cdc42-138 is not due to defective pheromone signaling.
CDC42 plays independent roles in cell polarization and fusion
Previously a point mutation within the Switch I domain of Cdc42p (cdc42-V36M) was reported to cause a cell fusion–defective phenotype (Barale et al., 2006). To ensure that any differences observed between cdc42-V36M and cdc42-138 would not be due to strain background, we re-created this allele in our strain background. We confirmed that cdc42-V36M caused a cell fusion defect, producing a significant number of unfused zygotes (Figure 2B; 53% had fusion defects).
Mutations that affect the polarization of cells in response to pheromone (e.g., spa2, bni1, pea2) exhibit corresponding defects in cell fusion (Dorer et al., 1997; Gammie et al., 1998). Because Cdc42p is critically important for polarity during both mating and mitotic growth (Chant, 1999; Pruyne and Bretscher, 2000; Casamayor and Snyder, 2002), we next examined the morphological responses of cdc42-138 and cdc42-V36M to pheromone.
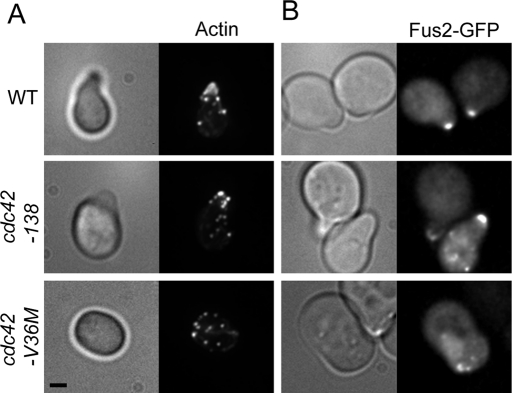
When wild-type or cdc42-138 cells were treated with pheromone, they formed characteristic pointed mating projections (Figure 4A). The actin cytoskeleton became polarized, with actin patches concentrated in the projection and cables oriented toward the shmoo tip (Figure 4A; 85% of wild-type cells and 81% of cdc42-138 cells had polarized actin). In contrast, cdc42-V36M cells were round or oblong, without an obvious projection of any kind (Figure 4A). Actin patches were not tightly localized at any particular point but instead were dispersed throughout the cell (Figure 4A; only 18% had polarized actin).
FIGURE 4:
cdc42-138 is not defective in cell polarization in response to pheromone. (A) Actin localization in cdc42-138 and cdc42-V36M. WT (DDY1300), cdc42-138 (DDY1354), and cdc42-V36M (MY10547) were treated with α-factor for 90 min, fixed, and stained with Texas red–phalloidin. Representative images are shown. n ≥ 241 cells in three independent experiments. (B) Fus2p-GFP localization in cdc42-138 and V36M. The same strains were transformed with FUS2::GFP104 (pMR5482) and imaged after treatment with α-factor for 90 min. Representative images are shown. n ≥ 55 cells in two independent experiments.
We also examined the localization of Fus2p. Fus2p-GFP localizes tightly to the shmoo tip in wild-type cells but is usually dispersed or nuclear in polarization mutants such as bni1 and spa2 (Paterson et al., 2008). Wild-type and cdc42-138 shmoos contained predominantly tip-localized Fus2p-GFP (Figure 4B; Fus2p-GFP was predominantly polarized in 98 and 80% of cells, respectively). The small but significant (p = 0.004, Fisher's exact test) decrease in Fus2-GFP localization is consistent with an interaction with Cdc42p. In contrast, in the majority of cdc42-V36M cells (56%), Fus2p-GFP was dispersed or partially nuclear. In only 44% of cdc42-V36M cells was Fus2p-GFP localized to a point on the cortex (usually at one of the ends of the oval-shaped cells). However, even in these cells, localization was not wild type; time-lapse imaging of Fus2p-GFP in cdc42-V36M showed that the cortical spots were unstable, moving rapidly away from the cortex and showing significant nuclear localization (Supplemental Movie S2). Similar unstable localization was observed in spa2 and bni1 mutants (Paterson et al., 2008).
Together, these data indicate that the cdc42-138 mutant responds to pheromone and polarizes normally. Therefore, the block in cell fusion is not likely to be a secondary consequence of defects in earlier events in mating. In contrast, the cdc42-V36M mutant polarizes poorly in response to pheromone, suggesting that the fusion defect may be largely due to indirect effects on the cytoskeleton during cell polarization.
Cdc42-138 acts late in cell fusion
Several classes of cell fusion mutants are now known that can be distinguished by their morphological characteristics. Mutations in SPA2, FUS1, FUS2, and RVS161 all block prezygotes at the stage of cell wall removal (Gammie et al., 1998). As discussed earlier, SPA2 and BNI1 primarily affect polarization, and mutants form broad adhesion zones between the fusing cells. The respective phenotypes can be distinguished at the level of electron microscopy; fus2 and rvs161 mutants have vesicles tightly clustered at the zone of cell fusion, similar to wild-type zygotes prior to fusion. In contrast, fus1 and spa2 mutants contain broadly dispersed vesicles that do not localize to a particular point. Double-mutant and suppression analysis showed that fus1 and fus2 work in overlapping but distinct pathways, whereas fus2 and rvs161 work together (Trueheart et al., 1987; Brizzio et al., 1998; Gammie et al., 1998).
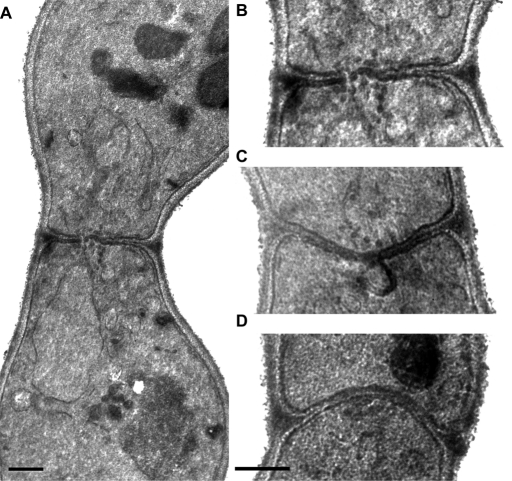
Electron microscopy of cdc42-138 bilateral zygotes revealed that they contain intact cell walls between the mating partners (Figure 5). Vesicles were observed to be tightly clustered in a single region at the zone of cell fusion (Figure 5, B and C; 35% of sections through cdc42-138 prezygotes had vesicles, n = 48; of these, 89% had clustered vesicles, n = 19). This phenotype is very similar to that of fus2 (Figure 5D) and rvs161 but distinct from that of spa2 and fus1 (Gammie et al., 1998). We also observed dark-staining plaques and membrane invaginations in cdc42-138 zygotes (Figure 5C; 31% had plaques and 5% had invaginations, n = 48). These phenotypes were previously shown to be unique to fus2 and rvs161 mutants and were not observed in fus1 and spa2 mutants (Gammie et al., 1998). Furthermore, the formation of these structures was dependent on FUS1 and SPA2 function because they were not observed in fus1 fus2 or spa2 fus2 double mutants. The fact that cdc42-138 formed these structures provides evidence that the SPA2 and FUS1 pathways remain functional.
FIGURE 5:
EM of cdc42-138 and fus2 zygotes. cdc42-138 bilateral zygotes (DDY1354 × DDY1355) were examined by electron microscopy. (A) A zygote, showing the unfused nuclei and septum, containing cell wall material. Image is representative of 91 zygotes. (B) The waist regions of the same zygotes, showing vesicles clustered at the zone of cell fusion. Images are representative of 32 zygotes in which vesicles were observed. The vesicles were clustered in 91% of this class. (C) Dark plaques and a membrane invagination at the cell fusion zone. These phenotypes were observed in 21 and 10% of all zygotes examined, respectively. (D) A fus2 zygote, for comparison. Scale bar, 0.5 μm.
To further characterize the morphology of cdc42-138, we measured the breadth of the zygote cell fusion zones (referred as the zygote waistlines). In wild-type matings, unfused prezygotes have an average waistline of 1.0 ± 0.3 μm, which expands to 1.6 ± 0.2 μm soon after nuclear fusion (Gammie et al., 1998). When only unfused prezygotes were considered, cdc42-138 bilateral zygotes had waistlines measuring 1.4 ± 0.3 μm (n = 48). It is surprising that this was significantly narrower than for fus2 prezygotes (1.7 ± 0.5 μm, n = 19, p = 0.008, Student's t test). We reasoned that because the cdc42-138 mutant has a weaker phenotype than the fus2 mutant, a significant portion of the zygotes with broad zones may have fused, leading to a shorter average waistline. In support of this, when the entire population of fused and unfused zygotes was considered, there was no difference between the average waistline of the cdc42-138 and fus2 mutant zygotes (1.6 ± 0.4 μm, n = 91, vs. 1.7 ± 0.5 μm, n = 20, respectively; p = 0.22). Most important, the data provide additional evidence that the cdc42-138 mutant is morphologically dissimilar from polarization mutants, which have significantly wider waistlines than the fus2 mutant (Gammie et al., 1998). Together the data show that the cdc42-138 mutant is morphologically most similar to fus2 and rvs161 mutants. Furthermore, they show that CDC42 plays a role in the very latest stages of cell fusion.
FUS2 and CDC42 work together
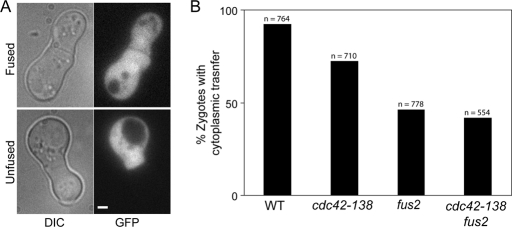
To test whether FUS2 and CDC42 act in the same pathway for cell fusion, we constructed a fus2Δ cdc42-138 double mutant and compared its phenotype to that of the two single mutants. We reasoned that if cdc42-138 specifically compromised Fus2p function in cell fusion (or vice versa), then the double mutants should show no more severe a defect than the most defective single mutant. Such behavior is observed between rvs161 and fus2 mutations, whereas double mutants between either of these mutations and fus1 or spa2 mutations are significantly more severe (Brizzio et al., 1996). For this experiment, we used a cytoplasmic transfer assay (Figure 6A). In this assay, only full Fus− zygotes with no plasma membrane fusion are scored as being defective. The fus2 donor strain was able to transfer cytoplasmic GFP to 92% of wild-type cells (n = 764) but was significantly defective for transfer to fus2 (46% transfer, n = 778, p = 0.001, chi-square) and cdc42-138 (72% transfer, n = 710, p = 0.001, chi-square; Figure 6B). If the two proteins act in two independent pathways, then we predict that the defect of the double mutant should be multiplicative, normalized for the frequency observed for wild type (i.e., [0.72 × 0.46]/0.92). Transfer to the cdc42-138 fus2 double mutant was somewhat more defective than to fus2 alone (42%, n = 554, p = 0.04, chi-square) but significantly different from the multiplicative prediction for independent pathways (36%, p = 0.004, chi-square). Because of the significant difference from the multiplicative model, we conclude that the cdc42-138 and fus2 mutations largely affect the same pathway for cell fusion. However, the smaller difference from the phenotype of the single mutant suggests that one or both proteins may also contribute an independent function to cell fusion.
FIGURE 6:
Fus2p and Cdc42p work in the same pathway. (A) Cytoplasmic transfer assay. Strains of different genotypes were mated to MY10797 (fus2 PGPD1-GFP) for 2 h at 30°C. An example of a zygote displaying cytoplasmic transfer (top) and one not displaying transfer (bottom) are shown. (B) WT (DDY1300), cdc42-138 (DDY1354), fus2 (MY10016), and fus2 cdc42-138 (MY10310) were mated to MY10797, and the percentage of zygotes showing transfer were counted. The total number of zygotes counted in three independent experiments are shown.
Fus2p Interacts with GTP-bound Cdc42p
Our genetic data indicate that Fus2p and Cdc42p work in the same pathway; they do not distinguish which protein acts upstream. Because Fus2p has a Dbl-homology domain required for cell fusion, one possibility is that Fus2p is a guanine nucleotide exchange factor (GEF) for Cdc42p. However, numerous attempts to demonstrate GEF activity have been unsuccessful, even under conditions where Cdc24p (the GEF for Cdc42p) showed strong activity (Ydenberg, Stein, Andrianantoandro, and Rose, unpublished data). We next considered the possibility that Fus2p is a Cdc42p effector, interacting specifically with activated, GTP-bound Cdc42p.
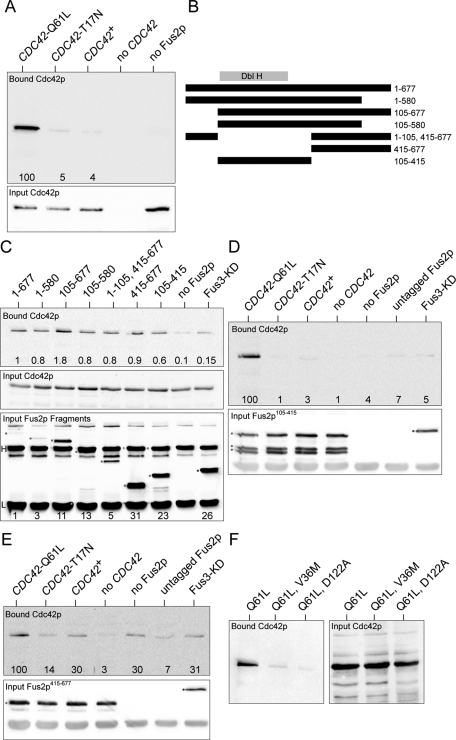
We began by confirming the Fus2p-Cdc42p interaction in an in vitro system. Wild-type Cdc42p, Cdc42p-Q61L, the GTP-locked mutant, and Cdc42p-T17N, the GDP-locked mutant, were expressed as GST fusions in Escherichia coli. Equal amounts of purified GST-Cdc42p were added to yeast extracts containing full-length Fus2p, which was internally FLAG-epitope tagged at residue 104. This protein was shown previously to support wild-type levels of cell fusion, in vivo (Paterson et al., 2008). All three forms of Cdc42p coimmunoprecipitated with Fus2p-FLAG; however roughly 20-times-higher levels of Cdc42p-Q61L (GTP bound) were bound compared with either Cdc42p-T17N (GDP bound) or wild type (Figure 7A). These results confirm that Cdc42p can bind to Fus2p and suggest that Fus2p is not a GEF for Cdc42p. Indeed, because the interaction was strongest with activated Cdc42p, we conclude that that Fus2p is more likely to be a effector or to play a role in localizing activated Cdc42p to the fusion site.
FIGURE 7:
Fus2p binds to Cdc42p in vitro. (A) Fus2p binds most strongly to the activated GTP-bound form of Cdc42p. Wild-type, Cdc42p-Q61L (GTP-locked), and Cdc42p-T17N (GDP-bound) proteins were expressed as GST fusions in E. coli and purified on glutathione Sepharose. The Cdc42p-GST fusions were incubated with yeast cell extracts expressing FLAG epitope–tagged Fus2p for 1 h at 4°C, and interacting proteins were precipitated with anti-FLAG Sepharose. The Cdc42p-GST fusions were detected by Western blot using anti-GST and chemiluminescent imaging. Top, bound Cdc42 proteins. Bottom, input Cdc42 proteins. Numbers indicate the relative level of binding of each Cdc42p species normalized to Cdc42p-Q61L. In the rightmost “no Fus2p” lane, Cdc42p-Q61L was used. (B) Map of Fus2p deletion mutants. The Dbl-homology domain is indicated (Dbl H). The extents of the deletions, in amino acid residues, are indicated. Note that the FLAG epitope is inserted at residue 104. (C) Cdc42p binds to more than one domain of Fus2p. Yeast strains expressing the deletion constructs shown in B were used in binding experiments with GST-Cdc42p-Q61L. The “no Fus2p” control used yeast extracts in which Fus2p was not epitope tagged. FLAG-tagged Fus3-KD (kinase dead) was used as an unrelated protein control. Top, relative levels of bound GST-Cdc42p-Q61L proteins are indicated. Middle, input GST-Cdc42p-Q61L. Bottom, the levels of the input FLAG-tagged Fus2p deletion proteins. Proteins bound to the anti-FLAG Sepharose were eluted and detected by Western blotting with anti-FLAG antibody. The deletion proteins are indicated by a red dot and the Fus3-KD protein with a blue dot, and the heavy and light immunoglobulin chains are indicated (H and L, respectively). The levels of the proteins relative to full-length Fus2p are indicated at the bottom. (D) Nucleotide specificity of binding to the DBH domain. The indicated Cdc42p proteins were incubated with Fus2p105-415. Top, bound Cdc42p proteins; relative levels of binding are indicated. Bottom, input Fus2p105-415. In this experiment some degradation of the Fus2p deletion fragment was observed. In the rightmost three lanes, Cdc42p-Q61L was used. (E) Nucleotide specificity of binding to 415–677. As in D, except that the Cdc42p proteins were incubated with Fus2p415-677. Top, bound Cdc42p proteins; relative levels of binding are indicated. Bottom, input Fus2p415-677. (F) Cdc42p mutations diminished Fus2p binding. The two mutations affecting cell fusion were introduced into the Cdc42p-Q61L construct and used for binding to wild-type Fus2p-FLAG.
To identify the region required for binding to Cdc42p, various deletions of Fus2p were constructed and used in the coimmunoprecipitation assay, using Cdc42p-Q61L (Figure 7, B and C). The most efficient binding was observed with full-length protein and the deletion lacking residues 580–677. Of interest, although the Dbl-homology domain was sufficient to see some binding to Fus2p, it was not required for binding; binding was also observed for protein fragments containing residues 415–677. These results suggest that more than one region of Fus2p interacts with Cdc42p. To analyze this further, we examined the nucleotide specificity of binding to the two regions of Fus2p. Binding to the DBH domain showed strong specificity for binding to the GTP-bound Cdc42p-Q61L but not to the other forms of Cdc42p (Figure 7D). The 415–677 fragment showed lower specific binding, although it still bound preferentially to Cdc42p-Q61L (Figure 7E). We conclude that the region of Fus2p that binds Cdc42 extends beyond the DBH domain.
As a final test, we examined whether the cdc42 mutations affected the interaction with Fus2p. The cdc42-V36M and cdc42-138 alleles were constructed in the plasmid expressing the GTP-locked Q61L mutation and the purified proteins tested for binding to Fus2p-FLAG (Figure 7F). Both proteins showed a significant decrease in binding relative to wild-type Cdc42p (cdc42-V36M and cdc42-138 exhibited 7 and 4% binding, respectively). These results corroborate the genetic data and suggest that cdc42-138, as well as cdc42-V36M, may interfere with cell fusion by disrupting the interaction between Cdc42GTPp and Fus2p.
DISCUSSION
Several lines of evidence previously suggested an interaction between Cdc42p and Fus2p. First, they interact by two-hybrid assay (Nelson et al., 2004). Second, BEM1, encoding a scaffold for Cdc42p and its effectors, is a high-copy suppressor of fus2 (Fitch et al., 2004). Third, high-copy FUS2 suppresses the mating defects of bem1 mutations (Leberer et al., 1996). Finally, Fus2p contains a Dbl-homology domain (Paterson et al., 2008), which is shared with activators of Rho-GTPases. However, Cdc42p has numerous functions and interaction partners and so could potentially have multiple direct and indirect roles during mating.
To implicate CDC42 specifically in cell fusion, we sought mutant alleles defective in cell fusion but not other steps of mating. One mutation, cdc42-138, caused a strong defect in cell fusion but not in mitotic growth, which was not due to impaired pheromone signaling or cell polarization. The mutant phenotype closely resembled fus2 mutations and analysis of double mutants showed that cdc42-138 and fus2 mutations conferred defects in the same step in cell fusion.
The Dbl-homology domain in Fus2p is required for as much as 90% of Fus2p's function in cell fusion. The presence of residual function suggests that Fus2p may have other roles in cell fusion or more than one interaction with Cdc42p. Indeed, we found that Cdc42p interacts with the Dbl-homology domain and additional sequences in Fus2p.
Although Fus2p contains a Dbl-homology domain found in other Cdc42p GEFs, we did not detect GEF activity (unpublished data). Instead, we found that Fus2p interacted more strongly with Cdc42-Q61L, the GTP-bound form of Cdc42p, in vitro, consistent with the two-hybrid data (Nelson et al., 2004). Because GEFs are expected to bind more strongly to the GDP-bound G-protein, these findings suggest that Fus2p is not a GEF but may instead serve either as a mating-specific effector for Cdc42p or to localize the activated Cdc42p to the cell fusion zone. It is notable that another putative GEF, Lte1p, requires its GEF domain for cortical localization and not for activation of Tem1p (Geymonat et al., 2009).
If Fus2p is an effector, what might be the role of the Fus2p–Cdc42p interaction in promoting cell fusion? Fus2p binds the BAR domain–containing protein Rvs161p, and the interaction is essential for cell fusion (Brizzio et al., 1998). BAR-domain proteins preferentially bind to curved membranes (Peter et al., 2004), and Rvs161p, together with Rvs167p, binds to membranes and promotes curvature in vitro (Friesen et al., 2006; Youn et al., 2009). Mutations in Rvs161p that inhibit membrane binding also inhibit mating (Youn et al., 2009), suggesting that Fus2p/Rvs161p's role in mating requires membrane interaction.
Fus2p localizes near clustered vesicles, and both Fus2p and the vesicles are partly delocalized in fus1 mutants (Gammie et al., 1998; Paterson et al., 2008). Deletion of neither Fus2p's Dbl-homology domain nor cdc42-138 affected Fus2p localization. Moreover, vesicles clustered normally in cdc42-138. These data suggest that the Cdc42p interaction might regulate an activity associated with Fus2p/Rvs161p rather than its localization. Recent work showed that membrane bending by the vesicle protein synaptotagmin is required for vesicle fusion (Hui et al., 2009). Perhaps Cdc42p regulates the degree of curvature induced by Fus2p/Rvs161p, deforming vesicle and/or target membranes in aid of membrane fusion. Alternatively, Fus2p/Rvs161p might interact with the curved outer surface of the vesicle fusion pore, stabilizing it to promote fusion. In either case, Fus2p/Rvs161p may play a role in facilitating the exocytosis of secreted proteins required for cell wall dissolution.
A role for the Rho-insert domain
The Rho-insert domain is a ∼20-residue loop containing a 13-residue α-helix unique to the Rho subfamily of small G proteins (Feltham et al., 1997). The residues of Cdc42p required for cell fusion (D121 and D122) are within four charged amino acids at one end of the Rho-insert domain. Previous work in yeast was not informative as to possible function; individual mutations to alanine did not affect growth; mutation of all four was lethal (Kozminski et al., 2000). A role in cell fusion represents the first specific function identified for this region of the protein in yeast.
Although little is known about Cdc42p's Rho-insert domain in yeast, hints may be gleaned from human Cdc42 and Rac1, where it is involved in the interactions with a variety of proteins, including Rho GDP dissociation inhibitors (Wu et al., 1997; Richman et al., 2004), formins (Richman et al., 2004; Lammers et al., 2008), and IQGAPs (McCallum et al., 1996; Li et al., 1999; Owen et al., 2008). It is possible that in yeast other protein interactions may be affected in addition to Fus2p. Although the yeast formin Bni1p is required for cell fusion, it also is required for cell polarization (Evangelista et al., 1997; Matheos et al., 2004; Paterson et al., 2008), likely acting prior to the function identified by cdc42-138. Of interest, the yeast IQGAP, Iqg1p, regulates exocytosis by localizing Sec3p to polarized growth sites (Osman et al., 2002), and Sec3p interacts with activated Cdc42p (Zhang et al., 2001, 2008). Continuous secretion has been shown to be required for the late stages of cell fusion (Grote, 2010). However, a specific requirement for either Iqpg1p or Sec3p in cell fusion has not been tested.
Cdc42 plays roles at multiple stages of mating and cell fusion
Analyzing the many roles of GTPases has required extensive use of point mutations to specifically interfere with individual functions (Mosch et al., 1999; Kozminski et al., 2000; Adamo et al., 2001; Saka et al., 2001; Barale et al., 2006; Heinrich et al., 2007). For example, this approach demonstrated that cdc42-V36M affects cell fusion but not mitotic growth (Barale et al., 2006). We found that cdc42-V36M did not polarize in response to pheromone and showed unstable localization of Fus2p-GFP (Figure 3), closely resembling spa2 and bni1. Fus1p-GFP also failed to localize in cdc42-V36M (Barale et al., 2006). We conclude that cdc42-V36M is primarily defective for polarization and may affect cell fusion indirectly, possibly through Bni1p. The cdc42-V36M mutant may also have a specific defect in cell fusion, which is masked by the earlier polarization defect.
In contrast, cdc42-138 mutant cells polarized normally and were specifically defective in cell fusion. Epistasis analysis showed that Spa2p acts upstream of Fus1p, which in turn acts upstream of Fus2p (Gammie et al., 1998). The morphology of cdc42-138 most closely resembled fus2, arguing that this allele disrupts a function downstream of Fus1p and Spa2p. In confirmation, double-mutant analysis showed that fus2 and cdc42-138 act at the same step.
We conclude that Cdc42p plays multiple roles during cell fusion. Roles in pheromone signaling, via Ste20p (Simon et al., 1995; Zhao et al., 1995), and polarization, via Bni1p (Evangelista et al., 1997), are well established. We propose a separate function in cell fusion mediated by the Rho-insert domain in association with Fus2p/Rvs161p.
A conserved role of Cdc42 homologues in eukaryotic cell fusion?
Among metazoan cell fusion events, Drosophila myoblast fusion may be the best understood (Chen and Olson, 2004). Ultrastructural studies revealed clustered, electron-dense vesicles across the cell fusion zone (Doberstein et al., 1997), similar to yeast cell fusion and vertebrate myoblast fusion (Lipton and Konigsberg, 1972). Recent work showed that, as in yeast, fusion occurs at a single site, coincident with accumulation of actin (Sens et al., 2010). Cdc42p homologues Drac1 and Drac2 are also required for myoblast fusion (Hakeda-Suzuki et al., 2002) and most likely activated by an unconventional GEF (Erickson et al., 1997; Brugnera et al., 2002). Moreover, Cdc42p is required for mouse myoblast fusion (Vasyutina et al., 2009). The conserved requirement for Cdc42p-related proteins in cell fusion suggests that yeast cell fusion will yield important insights into the mechanism in higher cells.
MATERIALS AND METHODS
General yeast methods, strains, and plasmid construction
Yeast media, general methods, and transformations were performed as described previously (Adams et al., 1997) with minor modifications. Strains and plasmids are listed in Tables 1 and 2, respectively.
TABLE 1:
Strains used this study.
| Strain | Genotype | Reference |
|---|---|---|
| JY428 | α fus2Δ3 his4-34 trp1Δ1 ura3-52 | G. Fink (Whitehead Institute, Cambridge, MA) |
| JY429 | α fus1Δ1 fus2Δ3 trp1Δ1 ura3-52 | G. Fink |
| JY431 | α his4-34 trp1Δ1 ura3-52 | G. Fink |
| DDY1300a | a ura3-52 leu2-3112 his3Δ200 lys2-801 CDC42::LEU2 | Kozminski et al. (2000) |
| DDY1301 | α ura3-52 leu2-3112 his3Δ200 lys2-801 CDC42::LEU2 | Kozminski et al. (2000) |
| DDY1354 | a ura3-52 leu2-3112 his3Δ200 lys2-801 cdc42-138::LEU2 | Kozminski et al. (2000) |
| DDY1355 | α ura3-52 leu2-3112 his3Δ200 lys2-801 cdc42-138::LEU2 | Kozminski et al. (2000) |
| MY9181 | a ura3Δ0 leu2Δ0 his3Δ1 met15Δ0 fus2Δ::HIS3 | Paterson et al. (2008) |
| MY10016 | a ura3-52 leu2-3112 his3Δ200 lys2-801 CDC42::LEU2 fus2Δ::HIS3 | This study |
| MY10303 | α ura3-52 leu2-3112 his3Δ200 ade2-1 fus2Δ::HIS3 | This study |
| MY10310 | a ura3-52 leu2-3112 his3Δ200 lys2-801 cdc42-138::LEU2 fus2Δ::HIS3 | This study |
| MY10547 | a ura3-52 leu2-3112 his3Δ200 lys2-801 cdc42-V36M::LEU2 | This study |
| MY10554 | α ura3-52 leu2-3112 his3Δ200 ade2-1 cdc42-V36M::LEU2 | This study |
| MY10797 | α fus2Δ3 his4-34 trp1Δ1 ura3-52 SSO1::PGPD1-GFP::URA3 | This study |
| MY10798 | α fus1Δ1 fus2Δ3 trp1Δ1 ura3-52 SSO1::PGPD1-GFP::URA3 | This study |
| MY10925 | a ura3-52 leu2-3112 his3Δ200 lys2-801 CDC42::LEU2 STE20ΔCRIB | This study |
| MY10926 | a ura3-52 leu2-3112 his3Δ200 lys2-801 cdc42-138::LEU2 STE20ΔCRIB | This study |
| MY10933 | a ura3-52 leu2-3112 his3Δ200 lys2-801 CDC42::LEU2 fus2ΔDbl::GFP | This study |
| MY12402 | a ura3-52 leu2-3112 his3Δ200 pep4::HIS3 bar1::kanMX fus2::natMX | This study |
aStrains containing cdc42-105, cdc42-107, and cdc42-137 are isogenic to DDY1300 (or DDY1301). See Kozminski et al. (2000).
TABLE 2:
Plasmids used in this study.
| Plasmid | Markers | Reference |
|---|---|---|
| pSB231 | CEN URA3 fus1-LacZ | Trueheart et al. (1987) |
| pMR5469 | CEN URA3 PGAL1-FUS2::GFP104 | Paterson et al. (2008) |
| pMR5482 | CEN URA3 FUS2::GFP104 | Paterson et al. (2008) |
| pMR5883 | CEN URA3 PGAL1-fus2Δ105-414::GFP | This study |
| pMR5480 | FUS2::FLAG104 URA3 CEN | Paterson et al. 2008 |
| pMR6404 | FUS21-580::FLAG104 URA3 CEN | This study |
| pMR6405 | FUS2105-677::FLAG104 URA3 CEN | This study |
| pMR6406 | FUS2105-580::FLAG104 URA3 CEN | This study |
| pMR6407 | FUS21-104-FLAG104-FUS2415-677URA3 CEN | This study |
| pMR6408 | FUS2415-677::FLAG104 URA3 CEN | This study |
| pMR6409 | FUS2105-415::FLAG104 URA3 CEN | This study |
| pMR6115 | GST-CDC42V36M, Q61L | This study |
| pMR6116 | GST-CDC42Q61L D122A | This study |
| pB2082 | GST-CDC42Q61L | Gladfelter et al. (2002) |
| pB2083 | GST-CDC42T17N | Gladfelter et al. (2002) |
| pB2084 | GST-CDC42 | Gladfelter et al. (2002) |
To create MY10016, the fus2Δ::HIS3 deletion was amplified from MY9181 and transformed into DDY1300. Correct integration was confirmed by PCR across the FUS2 locus. MY10547 and MY10554 were created as described (Kozminski et al., 2000). To create MY10925 and MY10926, pPP1043 (Lamson et al., 2002) was cut with BamHI and integrated in DDY1300 and DDY1354, respectively, by the loop-in/loop-out procedure. Correct loop-outs were confirmed by PCR across the STE20 locus. To create MY10797 and MY10798, pEG746 (Jin et al., 2004) was cut with BglII and transformed into JY428 and JY429, respectively. The integration was confirmed by examining the resulting strains for GFP fluorescence.
pMR5883 was created by a multistep cloning procedure in the vector pRS416 (Sikorski and Hieter, 1989). A similar procedure was used to create pMR5917, which is equivalent but in the integration vector pRS406. pMR5917 was cut with HindIII and fus2Δ105-414::GFP integrated in DDY1300 by the loop-in/loop-out procedure. A loop-out containing GFP fluorescence was denoted MY10933.
To create pMR6115 and pMR6116, mutations were introduced into pB2082 by site-directed PCR mutagenesis (Phusion; Thermo Fisher Scientific, Waltham, MA). Deletion mutants in FUS2 were generated by site-directed PCR mutagenesis using pMR5480 (Paterson et al., 2008) as a template. Deletions were confirmed by sequencing, and pheromone-induced protein expression was confirmed by trichloroacetic acid (TCA) precipitation–Western blot.
β-Galactosidase assays were performed essentially as described (Adams et al., 1997), using cell extracts prepared from ∼1.2 × 108 cells. Actin staining was performed as described with minor modifications (Adams et al., 1997), using cells treated with 6 μM α-factor for 90 min.
Binding assays
GST-Cdc42p and derivatives were expressed and purified from E. coli as described (Gladfelter et al., 2002) with minor modifications. For expression of Fus2p-FLAG in yeast, MY9181, transformed with pMR5480 (or derivative), was grown to early exponential phase (OD600 = 0.2) and treated with 10 μg/ml synthetic mating pheromone (Syn/Seq Facility, Department of Molecular Biology, Princeton University, Princeton, NJ) for 2 h. Cell extracts were prepared as in Brizzio et al. (1998). The cleared lysate was divided into three fractions, and each fraction was incubated with 5 μg of bacterially expressed GST-Cdc42 or derivative as indicated. Binding proceeded for 1 h at 4°C, with gentle rotation. After incubation, 30 μl of equilibrated anti-FLAG Sepharose (Sigma-Aldrich, St. Louis, MO) was added to each reaction and the incubation continued at 4°C for 1 h. The anti-FLAG Sepharose was washed five times and finally resuspended in SDS loading buffer. Experiments using the Fus2p deletion mutants were performed similarly, except that constructs were transformed into MY12402, which harbored deletions of the fus2, pep4, and bar1 genes.
Mating and cell fusion assays
Plate mating assays were performed as described (Gammie and Rose, 2002) by replica plating strains together on yeast extract/peptone/dextrose (YEPD) and selecting diploids by replica plating to minimal media or drop-in media. Microscopic examination of zygotes and cytoplasmic transfer assays were performed as described (Grote, 2008), using ∼1.0 × 106 cells of each mating type. Zygotes were processed for electron microscopy as described (Gammie et al., 1998), using ∼1.2 × 108 cells of each mating type, separated over five filters and mated on YEPD for 2 h.
Cell imaging
Images were acquired using the DeltaVision Microscopy System (Applied Precision, Issaquah, WA) using a TE200 inverted microscope (Nikon, Melville, NY) and a Coolsnap HQ CCD camera (Photometrics, Tucson, AZ) and a 100× objective with a numerical aperture of 1.4. Fus2p-GFP images were acquired as previously described (Ydenberg and Rose, 2009). Figures were prepared for publication using Adobe Photoshop and Adobe Illustrator (Adobe, San Jose, CA). For presentation purposes, pixel density was increased using bicubic resampling, where necessary.
Western blotting
For Western analysis of Fus2p-GFP, protein was prepared from ∼6.0 × 106 exponential phase cells by TCA precipitation, separated by SDS–PAGE, and transferred to a nitrocellulose filter. GFP-tagged Fus2p was detected using mouse anti-GFP (Roche, Indianapolis, IN) at 1:1000. FLAG-tagged Fus2p was detected with anti-FLAG (Sigma-Aldrich) at 1:5000. Rabbit anti-Kar2 (our laboratory) was used at 1:50,000. Glutathione S-transferase (GST)–tagged Cdc42p was detected with anti-GST (Cell Signaling Technologies, Beverly, MA) at 1:1000.
Supplementary Material
Acknowledgments
We thank Danny Lew, Keith Kozminski, Peter Pryciak, Eric Grote, and Robert Arkowitz for providing strains and reagents and Satoshi Yoshida and Bruce Goode for critical comments on the manuscript. This work was supported by National Institute of General Medical Sciences Grant GM37739 to M.D.R.
Abbreviations used:
- BAR
Bin1, amphiphysin, Rvs
- GAP
GTPase activating protein
- GEF
GTP exchange factor
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E11-08-0723) on February 9, 2012.
REFERENCES
- Adamo JE, Moskow JJ, Gladfelter AS, Viterbo D, Lew DJ, Brennwald PJ. Yeast Cdc42 functions at a late step in exocytosis, specifically during polarized growth of the emerging bud. J Cell Biol. 2001;155:581–592. doi: 10.1083/jcb.200106065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams A, Gottschling DE, Kaiser CA, Stearns T. Methods in Yeast Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1997. [Google Scholar]
- Barale S, McCusker D, Arkowitz RA. Cdc42p GDP/GTP cycling is necessary for efficient cell fusion during yeast mating. Mol Biol Cell. 2006;17:2824–2838. doi: 10.1091/mbc.E05-11-1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brizzio V, Gammie AE, Nijbroek G, Michaelis S, Rose MD. Cell fusion during yeast mating requires high levels of a-factor mating pheromone. J Cell Biol. 1996;135:1727–1739. doi: 10.1083/jcb.135.6.1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brizzio V, Gammie AE, Rose MD. Rvs161p interacts with Fus2p to promote cell fusion in Saccharomyces cerevisiae. J Cell Biol. 1998;141:567–584. doi: 10.1083/jcb.141.3.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brugnera E, Haney L, Grimsley C, Lu M, Walk SF, Tosello-Trampont AC, Macara IG, Madhani H, Fink GR, Ravichandran KS. Unconventional Rac-GEF activity is mediated through the Dock180-ELMO complex. Nat Cell Biol. 2002;4:574–582. doi: 10.1038/ncb824. [DOI] [PubMed] [Google Scholar]
- Casamayor A, Snyder M. Bud-site selection and cell polarity in budding yeast. Curr Opin Microbiol. 2002;5:179–186. doi: 10.1016/s1369-5274(02)00300-4. [DOI] [PubMed] [Google Scholar]
- Chant J. Cell polarity in yeast. Annu Rev Cell Dev Biol. 1999;15:365–391. doi: 10.1146/annurev.cellbio.15.1.365. [DOI] [PubMed] [Google Scholar]
- Chen EH, Olson EN. Towards a molecular pathway for myoblast fusion in Drosophila. Trends Cell Biol. 2004;14:452–460. doi: 10.1016/j.tcb.2004.07.008. [DOI] [PubMed] [Google Scholar]
- Chen EH, Olson EN. Unveiling the mechanisms of cell-cell fusion. Science. 2005;308:369–373. doi: 10.1126/science.1104799. [DOI] [PubMed] [Google Scholar]
- Crouzet M, Urdaci M, Dulau L, Aigle M. Yeast mutant affected for viability upon nutrient starvation: characterization and cloning of the RVS161 gene. Yeast. 1991;7:727–743. doi: 10.1002/yea.320070708. [DOI] [PubMed] [Google Scholar]
- Doberstein SK, Fetter RD, Mehta AY, Goodman CS. Genetic analysis of myoblast fusion: blown fuse is required for progression beyond the prefusion complex. J Cell Biol. 1997;136:1249–1261. doi: 10.1083/jcb.136.6.1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorer R, Boone C, Kimbrough T, Kim J, Hartwell LH. Genetic analysis of default mating behavior in Saccharomyces cerevisiae. Genetics. 1997;146:39–55. doi: 10.1093/genetics/146.1.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson MR, Galletta BJ, Abmayr SM. Drosophila myoblast city encodes a conserved protein that is essential for myoblast fusion, dorsal closure, and cytoskeletal organization. J Cell Biol. 1997;138:589–603. doi: 10.1083/jcb.138.3.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evangelista M, Blundell K, Longtine MS, Chow CJ, Adames N, Pringle JR, Peter M, Boone C. Bni1p, a yeast formin linking cdc42p and the actin cytoskeleton during polarized morphogenesis. Science. 1997;276:118–122. doi: 10.1126/science.276.5309.118. [DOI] [PubMed] [Google Scholar]
- Feltham JL, Dotsch V, Raza S, Manor D, Cerione RA, Sutcliffe MJ, Wagner G, Oswald RE. Definition of the switch surface in the solution structure of Cdc42Hs. Biochemistry. 1997;36:8755–8766. doi: 10.1021/bi970694x. [DOI] [PubMed] [Google Scholar]
- Fitch PG, Gammie AE, Lee DJ, de Candal VB, Rose MD. Lrg1p Is a Rho1 GTPase-activating protein required for efficient cell fusion in yeast. Genetics. 2004;168:733–746. doi: 10.1534/genetics.104.028027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friesen H, Humphries C, Ho Y, Schub O, Colwill K, Andrews B. Characterization of the yeast amphiphysins Rvs161p and Rvs167p reveals roles for the Rvs heterodimer in vivo. Mol Biol Cell. 2006;17:1306–1321. doi: 10.1091/mbc.E05-06-0476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gammie AE, Brizzio V, Rose MD. Distinct morphological phenotypes of cell fusion mutants. Mol Biol Cell. 1998;9:1395–1410. doi: 10.1091/mbc.9.6.1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gammie AE, Rose MD. Assays of cell and nuclear fusion. Methods Enzymol. 2002;351:477–498. doi: 10.1016/s0076-6879(02)51866-8. [DOI] [PubMed] [Google Scholar]
- Geymonat M, Spanos A, de Bettignies G, Sedgwick SG. Lte1 contributes to Bfa1 localization rather than stimulating nucleotide exchange by Tem1. J Cell Biol. 2009;187:497–511. doi: 10.1083/jcb.200905114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gladfelter AS, Bose I, Zyla TR, Bardes ES, Lew DJ. Septin ring assembly involves cycles of GTP loading and hydrolysis by Cdc42p. J Cell Biol. 2002;156:315–326. doi: 10.1083/jcb.200109062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grote E. Cell fusion assays for yeast mating pairs. Methods Mol Biol. 2008;475:165–196. doi: 10.1007/978-1-59745-250-2_10. [DOI] [PubMed] [Google Scholar]
- Grote E. Secretion is required for late events in the cell-fusion pathway of mating yeast. J Cell Sci. 2010;123:1902–1912. doi: 10.1242/jcs.066662. [DOI] [PubMed] [Google Scholar]
- Hakeda-Suzuki S, Ng J, Tzu J, Dietzl G, Sun Y, Harms M, Nardine T, Luo L, Dickson BJ. Rac function and regulation during Drosophila development. Nature. 2002;416:438–442. doi: 10.1038/416438a. [DOI] [PubMed] [Google Scholar]
- Heiman MG, Walter P. Prm1p, a pheromone-regulated multispanning membrane protein, facilitates plasma membrane fusion during yeast mating. J Cell Biol. 2000;151:719–730. doi: 10.1083/jcb.151.3.719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinrich M, Kohler T, Mosch HU. Role of Cdc42-Cla4 interaction in the pheromone response of Saccharomyces cerevisiae. Eukaryot Cell. 2007;6:317–327. doi: 10.1128/EC.00102-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horsley V, Pavlath GK. Forming a multinucleated cell: molecules that regulate myoblast fusion. Cells Tissues Organs. 2004;176:67–78. doi: 10.1159/000075028. [DOI] [PubMed] [Google Scholar]
- Hui E, Johnson CP, Yao J, Dunning FM, Chapman ER. Synaptotagmin-mediated bending of the target membrane is a critical step in Ca(2+)-regulated fusion. Cell. 2009;138:709–721. doi: 10.1016/j.cell.2009.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin H, Carlile C, Nolan S, Grote E. Prm1 prevents contact-dependent lysis of yeast mating pairs. Eukaryot Cell. 2004;3:1664–1673. doi: 10.1128/EC.3.6.1664-1673.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson DI. Cdc42: An essential Rho-type GTPase controlling eukaryotic cell polarity. Microbiol Mol Biol Rev. 1999;63:54–105. doi: 10.1128/mmbr.63.1.54-105.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozminski KG, Chen AJ, Rodal AA, Drubin DG. Functions and functional domains of the GTPase Cdc42p. Mol Biol Cell. 2000;11:339–354. doi: 10.1091/mbc.11.1.339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lammers M, Meyer S, Kuhlmann D, Wittinghofer A. Specificity of interactions between mDia isoforms and Rho proteins. J Biol Chem. 2008;283:35236–35246. doi: 10.1074/jbc.M805634200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamson RE, Winters MJ, Pryciak PM. Cdc42 regulation of kinase activity and signaling by the yeast p21-activated kinase Ste20. Mol Cell Biol. 2002;22:2939–2951. doi: 10.1128/MCB.22.9.2939-2951.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leberer E, Chenevert J, Leeuw T, Harcus D, Herskowitz I, Thomas DY. Genetic interactions indicate a role for Mdg1p and the SH3 domain protein Bem1p in linking the G-protein mediated yeast pheromone signalling pathway to regulators of cell polarity. Mol Gen Genet. 1996;252:608–621. doi: 10.1007/BF02172407. [DOI] [PubMed] [Google Scholar]
- Li R, Debreceni B, Jia B, Gao Y, Tigyi G, Zheng Y. Localization of the PAK1-, WASP-, and IQGAP1-specifying regions of Cdc42. J Biol Chem. 1999;274:29648–29654. doi: 10.1074/jbc.274.42.29648. [DOI] [PubMed] [Google Scholar]
- Lipton BH, Konigsberg IR. A fine-structural analysis of the fusion of myogenic cells. J Cell Biol. 1972;53:348–364. doi: 10.1083/jcb.53.2.348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh L, Rose MD. The pathway of cell and nuclear fusion during mating in S. cerevisiae. In: Pringle JRB, Jones EW, editors. In: The Molecular and Cellular Biology of the Yeast Saccharomyces: Cell Cycle and Cell Biology. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1997. pp. 827–888. [Google Scholar]
- Matheos D, Metodiev M, Muller E, Stone D, Rose MD. Pheromone-induced polarization is dependent on the Fus3p MAPK acting through the formin Bni1p. J Cell Biol. 2004;165:99–109. doi: 10.1083/jcb.200309089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCallum SJ, Wu WJ, Cerione RA. Identification of a putative effector for Cdc42Hs with high sequence similarity to the RasGAP-related protein IQGAP1 and a Cdc42Hs binding partner with similarity to IQGAP2. J Biol Chem. 1996;271:21732–21737. doi: 10.1074/jbc.271.36.21732. [DOI] [PubMed] [Google Scholar]
- Mosch HU, Kubler E, Krappmann S, Fink GR, Braus GH. Crosstalk between the Ras2p-controlled mitogen-activated protein kinase and cAMP pathways during invasive growth of Saccharomyces cerevisiae. Mol Biol Cell. 1999;10:1325–1335. doi: 10.1091/mbc.10.5.1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moskow JJ, Gladfelter AS, Lamson RE, Pryciak PM, Lew DJ. Role of Cdc42p in pheromone-stimulated signal transduction in Saccharomyces cerevisiae. Mol Cell Biol. 2000;20:7559–7571. doi: 10.1128/mcb.20.20.7559-7571.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson B, Parsons AB, Evangelista M, Schaefer K, Kennedy K, Ritchie S, Petryshen TL, Boone C. Fus1p interacts with components of the Hog1p mitogen-activated protein kinase and Cdc42p morphogenesis signaling pathways to control cell fusion during yeast mating. Genetics. 2004;166:67–77. doi: 10.1534/genetics.166.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nern A, Arkowitz RA. A GTP-exchange factor required for cell orientation. Nature. 1998;391:195–198. doi: 10.1038/34458. [DOI] [PubMed] [Google Scholar]
- Nern A, Arkowitz RA. A Cdc24p-Far1p-Gbetagamma protein complex required for yeast orientation during mating. J Cell Biol. 1999;144:1187–1202. doi: 10.1083/jcb.144.6.1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nern A, Arkowitz RA. G proteins mediate changes in cell shape by stabilizing the axis of polarity. Mol Cell. 2000;5:853–864. doi: 10.1016/s1097-2765(00)80325-1. [DOI] [PubMed] [Google Scholar]
- Osman MA, Konopka JB, Cerione RA. Iqg1p links spatial and secretion landmarks to polarity and cytokinesis. J Cell Biol. 2002;159:601–611. doi: 10.1083/jcb.200205084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen D, Campbell LJ, Littlefield K, Evetts KA, Li Z, Sacks DB, Lowe PN, Mott HR. The IQGAP1-Rac1 and IQGAP1-Cdc42 interactions: interfaces differ between the complexes. J Biol Chem. 2008;283:1692–1704. doi: 10.1074/jbc.M707257200. [DOI] [PubMed] [Google Scholar]
- Paterson JM, Ydenberg CA, Rose MD. Dynamic localization of yeast Fus2p to an expanding ring at the cell fusion junction during mating. J Cell Biol. 2008;181:697–709. doi: 10.1083/jcb.200801101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peter BJ, Kent HM, Mills IG, Vallis Y, Butler PJ, Evans PR, McMahon HT. BAR domains as sensors of membrane curvature: the amphiphysin BAR structure. Science. 2004;303:495–499. doi: 10.1126/science.1092586. [DOI] [PubMed] [Google Scholar]
- Potgens AJ, Schmitz U, Bose P, Versmold A, Kaufmann P, Frank HG. Mechanisms of syncytial fusion: a review. Placenta. 2002;23(Supp A):S107–S113. doi: 10.1053/plac.2002.0772. [DOI] [PubMed] [Google Scholar]
- Primakoff P, Myles DG. Penetration, adhesion, and fusion in mammalian sperm-egg interaction. Science. 2002;296:2183–2185. doi: 10.1126/science.1072029. [DOI] [PubMed] [Google Scholar]
- Pruyne D, Bretscher A. Polarization of cell growth in yeast. I. Establishment and maintenance of polarity states. J Cell Sci. 2000;113:365–375. doi: 10.1242/jcs.113.3.365. [DOI] [PubMed] [Google Scholar]
- Richman TJ, Sawyer MM, Johnson DI. The Cdc42p GTPase is involved in a G2/M morphogenetic checkpoint regulating the apical-isotropic switch and nuclear division in yeast. J Biol Chem. 1999;274:16861–16870. doi: 10.1074/jbc.274.24.16861. [DOI] [PubMed] [Google Scholar]
- Richman TJ, Toenjes KA, Morales SE, Cole KC, Wasserman BT, Taylor CM, Koster JA, Whelihan MF, Johnson DI. Analysis of cell-cycle specific localization of the Rdi1p RhoGDI and the structural determinants required for Cdc42p membrane localization and clustering at sites of polarized growth. Curr Genet. 2004;45:339–349. doi: 10.1007/s00294-004-0505-9. [DOI] [PubMed] [Google Scholar]
- Saka A, Abe M, Okano H, Minemura M, Qadota H, Utsugi T, Mino A, Tanaka K, Takai Y, Ohya Y. Complementing yeast rho1 mutation groups with distinct functional defects. J Biol Chem. 2001;276:46165–46171. doi: 10.1074/jbc.M103805200. [DOI] [PubMed] [Google Scholar]
- Sens KL, Zhang S, Jin P, Duan R, Zhang G, Luo F, Parachini L, Chen EH. An invasive podosome-like structure promotes fusion pore formation during myoblast fusion. J Cell Biol. 2010;191:1013–1027. doi: 10.1083/jcb.201006006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheltzer JM, Rose MD. The class V myosin Myo2p is required for Fus2p transport and actin polarization during the yeast mating response. Mol Biol Cell. 2009;20:2909–2919. doi: 10.1091/mbc.E08-09-0923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon MN, De Virgilio C, Souza B, Pringle JR, Abo A, Reed SI. Role for the Rho-family GTPase Cdc42 in yeast mating-pheromone signal pathway. Nature. 1995;376:702–705. doi: 10.1038/376702a0. [DOI] [PubMed] [Google Scholar]
- Trueheart J, Boeke JD, Fink GR. Two genes required for cell fusion during yeast conjugation: evidence for a pheromone-induced surface protein. Mol Cell Biol. 1987;7:2316–2328. doi: 10.1128/mcb.7.7.2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trueheart J, Fink GR. The yeast cell fusion protein FUS1 is O-glycosylated and spans the plasma membrane. Proc Natl Acad Sci USA. 1989;86:9916–9920. doi: 10.1073/pnas.86.24.9916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasyutina E, Martarelli B, Brakebusch C, Wende H, Birchmeier C. The small G-proteins Rac1 and Cdc42 are essential for myoblast fusion in the mouse. Proc Natl Acad Sci USA. 2009;106:8935–8940. doi: 10.1073/pnas.0902501106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vignery A. Osteoclasts and giant cells: macrophage-macrophage fusion mechanism. Int J Exp Pathol. 2000;81:291–304. doi: 10.1111/j.1365-2613.2000.00164.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu WJ, Leonard DA, R AC, Manor D. Interaction between Cdc42Hs and RhoGDI is mediated through the Rho insert region. J Biol Chem. 1997;272:26153–26158. doi: 10.1074/jbc.272.42.26153. [DOI] [PubMed] [Google Scholar]
- Ydenberg CA, Rose MD. Yeast mating: a model system for studying cell and nuclear fusion. Methods Mol Biol. 2008;475:3–20. doi: 10.1007/978-1-59745-250-2_1. [DOI] [PubMed] [Google Scholar]
- Ydenberg CA, Rose MD. Antagonistic regulation of Fus2p nuclear localization by pheromone signaling and the cell cycle. J Cell Biol. 2009;184:409–422. doi: 10.1083/jcb.200809066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youn JY, et al. Dissecting BAR domain function in the yeast amphiphysins Rvs161 and Rvs167 during endocytosis. Mol Biol Cell. 2009;21:3054–3069. doi: 10.1091/mbc.E10-03-0181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Bi E, Novick P, Du L, Kozminski KG, Lipschutz JH, Guo W. Cdc42 interacts with the exocyst and regulates polarized secretion. J Biol Chem. 2001;276:46745–46750. doi: 10.1074/jbc.M107464200. [DOI] [PubMed] [Google Scholar]
- Zhang X, Orlando K, He B, Xi F, Zhang J, Zajac A, Guo W. Membrane association and functional regulation of Sec3 by phospholipids and Cdc42. J Cell Biol. 2008;180:145–158. doi: 10.1083/jcb.200704128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao ZS, Leung T, Manser E, Lim L. Pheromone signalling in Saccharomyces cerevisiae requires the small GTP-binding protein Cdc42p and its activator CDC24. Mol Cell Biol. 1995;15:5246–5257. doi: 10.1128/mcb.15.10.5246. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.