Abstract
Mutations in the him-5 gene in Caenorhabditis elegans strongly reduce the frequency of crossovers on the X chromosome, with lesser effects on the autosomes. him-5 mutants also show a change in crossover distribution on both the X and autosomes. These phenotypes are accompanied by a delayed entry into pachytene and premature desynapsis of the X chromosome. The nondisjunction, progression defects and desynapsis can be rescued by an exogenous source of double strand breaks (DSBs), indicating that the role of HIM-5 is to promote the formation of meiotic DSBs. Molecular cloning of the gene shows that the inferred HIM-5 product is a highly basic protein of 252 amino acids with no clear orthologs in other species, including other Caenorhabditis species. Although him-5 mutants are defective in segregation of the X chromosome, HIM-5 protein localizes preferentially to the autosomes. The mutant phenotypes and localization of him-5 are similar but not identical to the results seen with xnd-1, although unlike xnd-1, him-5 has no apparent effect on the acetylation of histone H2A on lysine 5 (H2AacK5). The localization of HIM-5 to the autosomes depends on the activities of both xnd-1 and him-17 allowing us to begin to establish pathways for the control of crossover distribution and frequency.
CROSSING over between homologous chromosomes during meiosis promotes genetic diversity by creating new combinations of alleles over generations. Crossovers also create physical connections between the homologs that ensure their proper alignment on the meiotic spindle and subsequent apposite segregation. Accordingly, homologous chromosomes require a crossover to prevent nondisjunction, and each of the events of meiosis I functions to promote this exchange.
A necessary early step in crossing over is the SPO11-dependent formation of double strand breaks (DSBs) (Keeney et al. 1997). In Saccharomyces cerevisiae, at least nine other proteins interact with SPO11 to regulate the recruitment and activation of SPO11 (Keeney and Neale 2006). These proteins that regulate the action of SPO11 are not highly conserved at the amino acid level, but recent studies have identified functional homologs of several of these components in mice (Cole et al. 2010; Kumar et al. 2010). Nevertheless, relatively little is known about the regulation of the SPO11 machinery in organisms other than S. cerevisiae.
Meiotic breaks occur preferentially in regions of open chromatin structure known as hotspots (Ohta et al. 1994; Wu and Lichten 1994). The pattern of crossovers and the recombination frequency vary among different chromosomes even within a species. One of the most distinctive patterns is seen in Caenorhabditis elegans, where the recombination rate on autosomes is repressed in the central region of each autosome, which contains a tight central cluster of genes. Instead crossovers occur preferentially on the chromosome arms where genes are widely spaced (Barnes et al. 1995). The X chromosome has a different pattern, in which genes are more uniformly spaced and the recombination frequency is relatively uniform across the chromosome at a rate that is intermediate between that of autosomal clusters and arms (Barnes et al. 1995; Rockman and Kruglyak 2009).
The genetic differences between the X chromosome and the autosomes during meiosis are also correlated with a variety of molecular and cytological differences seen in germline chromosomes. The X chromosome is transcriptionally repressed throughout most of germline development (Kelly et al. 2002) and histone modifications associated with closed and open chromatin configurations are enriched on the X and autosomes, respectively (Schaner and Kelly 2006). Furthermore, a number of mutations differentially affect crossover (CO) frequencies on the X chromosome vs. the autosomes. A subset of these genes, him-1, him-17, xnd-1, and dpy-28, influences DSB formation and also has been implicated in the regulation of germline chromatin architecture (Hodgkin et al. 1979; Reddy and Villeneuve 2004; Tsai et al. 2008; Mets and Meyer 2009). Together, these mutations suggest that X chromosome architecture may predispose this chromosome to defects in crossover formation.
One of the original mutants found to have very strong defects on X chromosome disjunction and recombination was him-5 (Hodgkin et al. 1979; Broverman and Meneely 1994). Mutations in him-5 are similar to the phenotypes seen for these other meiotic mutants, particularly those in xnd-1. Mutations of him-5 exhibit a much stronger effect on the X chromosome than the autosomes, although some effects on autosomal recombination and disjunction have been consistently observed (Hodgkin et al. 1979; Broverman and Meneely 1994). In this article, we present the cloning and characterization of him-5 and show that it is required for the normal distribution of crossovers genome-wide and specifically potentiates crossover formation on the X chromosome. We present evidence that HIM-5 protein is enriched on the autosomes in xnd-1– and him-17–dependent fashions. We also present evidence that xnd-1 and him-5 differentially affect DSB repair kinetics and present models for how these genes may function to control meiotic events.
Materials and Methods
Genetics and worm handling
All strains were grown at 20° on standard media (Brenner 1974). Progeny counts to determine the frequency of males and the rate of hatching were done by placing a single L4 hermaphrodite onto a plate and transferring it daily until no further eggs were laid. him-5(ok1896) was outcrossed >10 times prior to analysis. No differences in embryonic lethality or frequency of males were observed between F1 homozygotes or later generations; therefore, all him-5 stocks were maintained as homozygotes. Mutant strains used in these studies were: LG I, dpy-5(e61); LG II, mes-2(bn27) unc-4(e120)/mnC1dpy-10(e128) unc-52(e444); unc-4(e120); LG III, xnd-1(ok709); unc-25(e156); LG IV, him-6(e1423); unc-24(e138); and LG V, him-5(e1467); him-5(e1490); him-5(ok1896); dpy-11 (e224).
Cosuppression and RNAi of D1086.4
A region of 3.65 kb was amplified by PCR on lysed worms using the primers indicated at the top of Figure 1. The PCR product was purified and co-injected by Verena Plunger Jantsch (University of Vienna) with a plasmid containing rol-6+ into wild-type hermaphrodites by standard methods, and the injected worms were scored by P. M. Meneely. Each injected hermaphrodite was cultured individually and transferred daily, and the percentage of male offspring among the surviving progeny was determined. Lines with a high incidence of males (Him phenotype) were recultured regardless of their rolling phenotype by picking L3 or L4 hermaphrodites; L4 larval hermaphrodites were used to avoid any possibility that the males were arising from cross-fertilization. The lines were transferred at every generation for more than 20 generations, with a consistent Him phenotype observed each generation.
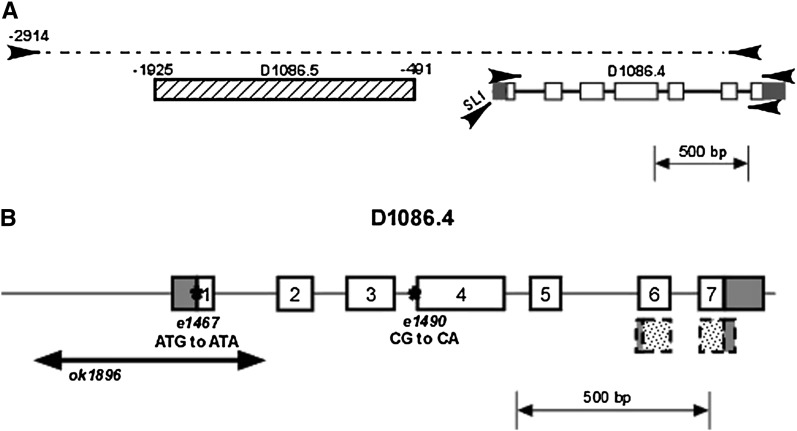
Figure 1 .
D1086.4 corresponds to him-5. (A) The genomic locus on cosmid D1086, showing some of the primers (arrowheads) used for analysis of the D1086.4. The primers at the top were used to amplify a 3.65 kB fragment used for cosuppression. The other pairs of primers where used to find the 3′ and 5′ of the D1086.4 gene using RT–PCR. The gene structure inferred from sequencing agreed with the structure displayed in WormBase. (B) Locations of the three him-5 mutations used in this study. The deletion ok1896 removes 519 bp including the upstream region, the 5′-UTR, all of the first exon, and nearly all of the first intron. Both e1467 and e1490 are G-to-A transitions, affecting the ATG start codon and the splice acceptor site at the start of exon 4 (or at the end of intron 3), respectively. The putative small isoform identified by WormBase is shown in the dashed lines and stippled boxes; this consists of the final 47 amino acids with short 5′- and 3′-UTRs. This isoform would not have been detected in our experiments.
The D1086.4 locus was amplified from wild-type and him-5 mutant worms using primers that flanked the gene, the PCR products were purified, and sequencing was done in both directions from primers located roughly every 400 bp. The sequence found in wild-type worms agreed exactly with what had been reported on WormBase. Each mutant was sequenced more than five times in both directions, with only a single base-pair change observed every time in e1467 and e1490, and the deletion previously found by the C. elegans Knockout Consortium in ok1896. The 5′ end, the 3′ end, and the overall exon–intron structure were determined by RT–PCR using highly purified RNA isolated from a mixed stage population (a gift from Alex Ensminger and Nelson Lau, Bartel lab, MIT, Cambridge, MA). The 5′ end was found using primers corresponding to a region immediately upstream of the putative ATG, to SL1, and SL2; the 3′ end was found using primers corresponding to different regions downstream of the stop codon. The structure determined from our experiments agreed with what had been found by cDNAs and ESTs sequenced by the C. elegans Genome Project.
dsRNAs were made using T7 RNA polymerase on PCR products corresponding to the following regions of the him-5 cDNAs: 5′ end of the gene between 145 and 420 bp; 3′ end from bp 589 to the stop codon. dsRNAs of D1086.5 corresponded to 61–453 bp of the predicted cDNAs. These regions were first subcloned into a double T7 vector (Timmons and Fire 1998), sequence verified, and then reamplified with plasmid-specific primers to generate unique PCR fragments for the upper and lower strands. These were then transcribed according to the manufacturer’s instructions (Ambion/Applied Biosystems, Austin, TX) and annealed by incubating for 10 min at 70° and slow cooling to room temperature. dsRNA was injected at a total RNA concentration of 20 ng/µl into 1-day-old adult worms, which were transferred every 24 hr for 3 days to new plates. The presence of male progeny was assessed 3–5 days later. A subset of F1 progeny was also individually plated to determine whether broods contained males from both the P0 48–72 hr postinjection and from the F1 progeny (Supporting Information, Table S1). We note that the efficacy of the RNAi was low (Table S1): fewer than 10–25% of the injected animals gave male progeny; the rate of 2–10% males was lower than that observed for the known mutations (compare with Table 1), and only progeny from the 48- to 72-hr time point postinjection were Him. The low Him rates could be explained if the dsRNA concentration was lower than anticipated (since the optical density accounts for both single and double stranded RNAs in the mixture), if the amount of dsRNA injected was suboptimal, and/or if dsRNA did not spread well through the animal postinjection (since either the gut or only one germline was injected). Nonetheless, the presence of males in a subset of these populations with two different D1086.4 dsRNAs strongly suggests that D1086.4 is him-5. Furthermore, the presence of males only at late time points after injection raised the possibility, borne out below, that him-5 plays a role in early meiotic nuclei. In this scenario, the numerous meiotic nuclei that were in the pachytene stage at the time of injection needed to be cleared from the animals before an effect could be observed. Our time course is consistent with studies that showed that it takes ∼48 hr to move from premeiotic S phase to fertilization after adult day 1 (Jaramillo-Lambert et al. 2007).
Table 1 . Hatching rates and male progeny production.
| Genotype | No. eggs | % hatchinga | No. animals | % males |
|---|---|---|---|---|
| Wild type | 3347 | 98.6 | 3300 | 0.06 |
| him-5(e1467) | 4572 | 60 | 2743 | 18 |
| him-5(ok1896) | 4960 | 71 | 3546 | 37 |
| him-5(e1490) | – | – | 873 | 35 |
| e1490/ok1896 | – | – | 1868 | 36 |
| e1467/ok1896 | – | – | 2152 | 27 |
| Cosupp line 1 | – | – | 3786 | 30 |
| Cosupp line 2 | – | – | 3102 | 32 |
| xnd-1(ok709) | 3834 | 60 | 2288 | 23 |
| ok709; e1467 | 1945 | 62 | 1209 | 23 |
| ok709; ok1896 | – | – | 1520 | 39 |
| mes-2(bn27) | – | – | >4000 | 0 |
| mes-2; ok1896 | – | – | 3093 | 24 |
| mes-2/+; ok1896 | – | – | 435 | 46 |
Cosupp, cosuppression.
The total number of viable adult progeny/total no. eggs × 100.
Genetic assays for autosomal nondisjunction
To determine the frequency of autosomal nondisjunction during spermatogenesis in him-5 males and oogenesis in him-5 hermaphrodites, a strategy devised by Haack and Hodgkin (1991) was employed. Mutations in him-6 result in nondisjunction of all chromosomes during both spermatogenesis and oogenesis. A him-6(e1423) male was mated to him-6; dpy; unc hermaphrodites, where dpy and unc correspond to recessive markers found in each autosome. For our experiments, dpy-5(e61)I, unc-4(e120) II, unc-25(e156) III, unc-24(e138) IV, and dpy-11(e224) V were used. Most of the cross-progeny offspring from this mating are non-Dpy non-Unc hermaphrodites and males. However, because him-6 produces nullisomic sperm and disomic ova at detectable frequencies, occasional Dpy non-Unc or non-Dpy Unc offspring are found; these arise when both copies of an autosome are matroclinous. The rate of exceptional progeny with two matroclinous autosomes is ∼0.3% for each autosome.
To test the effect of him-5 on nondisjunction during male spermatogenesis, him-5(e1490) males were used in place of him-6 males and mated to him-6; dpy; unc hermaphrodites; no exceptional offspring were found for any autosome. Similarly, to test the effect on nondisjunction during oogenesis, him-6 males were mated to him-5(e1490); dpy; unc hermaphrodites; exceptional progeny that received both copies of an autosome from the mother were detected at a frequency of ∼0.1%. The hermaphrodites among these exceptional progeny were fertile and segregated offspring consistent with the inferred matroclinous genotype.
Genomic analysis
No statistically significant similarity to any proteins has been observed in other Caenorhabditis species by BLASTP using different scoring matrices and different generations of the genome sequences or by searching against the nonredundant databases found in GenBank. Nonetheless, our attention was drawn to the presence of the sequence KEREKxVxxxDEAD, which is also found in the HSP83 chaperone protein sequence in Drosophila (Figure S1B). This identity is embedded in a region of ∼100 amino acids which, as determined by LALIGN, has an expected similarity value of E <0.05 between HIM-5 and HSP83. The LALIGN program was implemented at the University of Virginia FASTA server. The results are shown in Figure S1. While the similarity is significant at the 5% level, the importance of these amino acids in HSP83 is not known, and they are only very weakly conserved in other HSP82 and HSP83 proteins.
SNP analysis
him-5(ok1896) was introgressed into the Hawaiian CB4856 background by repeated backcrosses. Hawaiian-specific alleles used in these studies were confirmed by PCR genotyping. Spontaneous males from this stock were crossed to dpy-17(e164); him-5(ok1896) and non-Dpy; him-5 trans-heterozygous progeny were crossed to GFP+ males. GFP+ L4 hermaphrodites were individually plated and genotyped as previously described (Lim et al. 2008) using established primer sets for chromosomes X and I (Wagner et al. 2010). Chromosome I was also analyzed in sperm by mating the non-Dpy trans-heterozygous males from the above cross to dpy-18 hermaphrodites and SNP genotyping the individually plated non-Dpy cross-progeny. Wild-type controls for both egg and sperm were previously reported (Wagner et al. 2010) and are included herein for comparison. Chi square tests were performed to test for significant differences in CO frequency and position between wild type and him-5. Map units (m.u.) for the whole chromosome were calculated using the formula: m.u. = (number of single COs (SCOs) + 2 (number of double COs (DCOs))/sample size × 100. For each interval, the map size was calculated using the formula m.u. = (number of CO in interval/number of total COs on the chromosome) × m.u. for the chromosome.
Immunolocalization
Fixation and treatment of gonads were performed according to established protocols (Chan et al. 2003). Briefly, animals were picked onto slides with 2.5 μl of M9, rinsed and dissected in 2.5 μl of 1× sperm salts, and fixed for 5 min in 1% paraformaldehyde with 1% Triton X-100 in a humid chamber. A coverslip was added and slides were placed on a metal block on dry ice for 10–20 min. Samples were freeze cracked and placed in 95% ethanol for 1 min. Prehybridization and hybridization were performed with 1× phosphate buffered saline (PBS) with 0.5% Triton X-100. Except where noted, all stainings were performed on 1-day-old adult animals. Antibodies were used at the following concentrations: rabbit anti–HIM-5 (Novus Biologicals, Littleton, CO) 1:5000; guinea pig anti–XND-1 1:2000 (Wagner et al. 2010); guinea pig anti–SYP-1 1:1000 (Colaiacovo et al. 2003); rabbit anti–RAD-51 1:1000 (Rinaldo et al. 2002); rat anti–HTZ-1 1:2000 (Csankovszki et al. 2009); antinuclear pore monoclonal antibody mAb414 (Abcam, Cambridge, MA) 1:2000; guinea pig anti–HTP-3 1:1000 (Goodyer et al. 2008); guinea pig anti–HIM-8 1:500 (Phillips et al. 2005); anti-H2AacK5 (Cell Signaling, Danvers, MA) 1:1000; secondary antibodies were all Alexa-488, Alexa-568, and Alexa-633 (Invitrogen, Carlsbad, CA), all used at 1:1000–1:2000 dilution. The HIM-5 antibody was made by Strategic Diagnostics (Newark, DE) as part of the modENCODE project using the ∼100 C-terminal amino acids of the protein. DAPI staining bodies were assessed after Carnoy’s fixation of whole animals. Fluorescent in situ hybridization (FISH) was performed using probes to XR and 5S, according to established protocol (Phillips et al. 2005). Analysis of stained nuclei was carried out as previously described (Colaiacovo et al. 2003). All samples were mounted in Prolong Gold with DAPI and imaged on a Nikon A1r confocal microscope (Nikon Instruments, Melville, NY) with 0.2-μm sections. Stacks were reconstructed and analyzed using Volocity 3D imaging software (Perkin-Elmer, Waltham, MA).
Irradiation
Rescue of bivalent formation at diakinesis was performed with 20 Gy irradiation as described previously (Wagner et al. 2010). To determine whether irradiation rescued desynapsis and meiotic progression, 1-day-old adults were exposed to 5 Gy using a Nordion Gamma Cell 1000 Irradiator (Ottawa, ON, Canada). Animals were fixed at t = 0, 2, 4, 6, 8, and 24 hr postirradiation and stained with anti–SYP-1 antibodies and DAPI. Rescue of desynapsis could be seen in a subset of nuclei at t = 4 hr, was apparent in almost all nuclei at t = 8 hr, and persisted until t = 24 hr. Rescue of the clustering phenotypes was apparent in t = 4 and most obvious in t = 6 and t = 8 hr samples.
Analysis of RAD-51 foci
The dynamics of RAD-51 focus formation were assessed by dividing germlines into seven sections between the transition zone (leptotene) and the pachytene/diplotene border (Figure S5). The number of RAD-51 foci in each nucleus was determined using 3D reconstruction and image manipulation software, as described above. RAD-51 foci were plotted as a heat map for each region to rapidly visualize differences in repair dynamics.
To determine whether breaks were made on the X chromosome, wild-type and him-5(ok1896) L1 animals were exposed to rad-54 (RNAi) and dissected as L4 larvae as previously described (Wagner et al. 2010). X chromosomes were distinguished from autosomes on the basis of the lack of anti–HTZ-1 staining (Mets and Meyer 2009). Similar protocols were used to determine the total number of breaks, but germlines were stained with anti–SYP-1 and anti–RAD-51.
Analysis of apoptosis
Day 2 adult wild-type and him-5 mutants were soaked in 0.5–1 ml of either SYTO 12 (33 μM, Molecular Probes S7574) or acridine orange (20 μg/ml, Molecular Probes A3568) directly onto C. elegans growth plates. Plates were incubated at room temperature in the dark for 1 hr, and then worms were transferred to fresh plates without staining solution to destain for another 1 hr in the dark. Destained worms were mounted in 4 μM Levamisole in EN buffer. Fluorescence microscopy was used to count the number of apoptotic nuclei on a Nikon Eclipse Ti wide-field microsocope (Nikon Instruments).
Results
The him-5 gene corresponds to D1086.4
him-5 was mapped using morphological markers and sequence-tagged sites to a region on chromosome V delimited by three cosmids that have 22 predicted protein-coding genes. The predicted gene D1086.4 was considered to be a strong candidate to be him-5 on the basis of its expression in the germline. Three lines of evidence support the conclusion that D1086.4 is the him-5 locus. First, transgenic lines containing a 3.65-kb region beginning 2.9 kb upstream of the predicted start codon on D1086.4 (Figure 1A) produced ∼30% male self-progeny on average, indicating that cosuppression of this locus gives a phenotype that resembles him-5 mutants (Table 1). Second, sequencing him-5 mutant alleles revealed mutations within the D1086.4 region: the molecular lesions for three him-5 mutations are shown in Figure 1B. These include the two previously characterized alleles, e1467 and e1490, as well as a deletion allele created by the C. elegans Knockout Consortium, ok1896. Since the PCR fragment used for cosuppression included most of the neighboring gene, D1086.5, as well as its upstream region, this gene was also sequenced in wild-type and him-5 mutants. No sequence changes in the D1086.5 coding region were found in any him-5 alleles. Third, RNA interference (RNAi) using dsRNA against the 5′ or 3′ regions of the D1086.4 locus, but not against D1086.5, resulted in the production of male progeny.
The structure of the gene (Figure 1B) was confirmed using RT–PCR. him-5 has seven exons encoding a protein of 252 amino acids, which is an exceptionally basic protein, with an inferred isoelectric point (pI) of 10.7 (Figure S1A). WormBase annotations suggest the existence of a short him-5 isoform transcribed from an internal promoter and encoding only the last 47 amino acids (Figure 1B). We would not have observed this transcript in our RT–PCR experiments and cannot rule out the possibility that a very short HIM-5 polypeptide exists. The three him-5 alleles are shown in Figure 1B, the deletion allele ok1896 removes much of the promoter, the translational start and all of exon 1; e1467 mutates the start codon; and e1490 is a splice site mutation, predicted to reduce the efficiency of splicing. The severity of e1490 suggests it may produce a truncated, dominant-negative protein. All three alleles are within the domains unique to the long him-5 isoform and presumably would not affect the structure of the putative shorter isoform. Extensive homology searches and syntenic alignment failed to identify any extended open reading frame or sequence alignments of >35 amino acids (data not shown). In C. briggsae, in which the alignment for this region is most coextensive with C. elegans, there appears to have been an insertion of a sequence of ∼4.3 kb; part of the region also appears to have been inverted, and other base substitutions have occurred. While the precise molecular rearrangements are difficult to reconstruct, it is clear that no orthologous, full-length gene to him-5 could be identified in C. briggsae, C. remaneii, C. japonica, C. brenneri, or in the more distantly related nematodes Brucei malayi and Pristonichus pacificus. Thus, while him-5 plays an important role in meiosis in C. elegans, it may not to be evolutionarily conserved.
Crossover defects in him-5 mutants are X biased
The presence of males in the population is a hallmark of meiotic nondisjunction mutants and indicates underlying defects in the transmission of the X chromosome (Hodgkin et al. 1979). Defects in autosomal segregation are instead manifest as an increase in embryonic lethality because monosomy and trisomy of autosomes is incompatible with life (or subvital) (Zetka and Rose 1992). In mutants defective for CO formation on all chromosomes, e.g., spo-11 (Dernburg et al. 1998), viability of zygotes is <2%, and for strong loss-of-function mutations in other meiotic genes affecting all chromosomes, e.g., him-3(e1256) (Zetka et al. 1999) and him-6(e1423) (Hodgkin et al. 1979), embryonic lethality can be ≥80%. In contrast, in the strongest him-5 alleles, close to 40% of progeny are male, and up to ∼30% of eggs do not hatch (Table 1). Although this rate of embryonic lethality is substantial, the majority of these unhatched eggs can be attributed to X chromosome nondisjuction (Hodgkin et al. 1979). Therefore, compared to the high frequency of males, him-5 may have only a minor effect on segregation of the autosomes, as previously suggested (Hodgkin et al. 1979; Broverman and Meneely 1994).
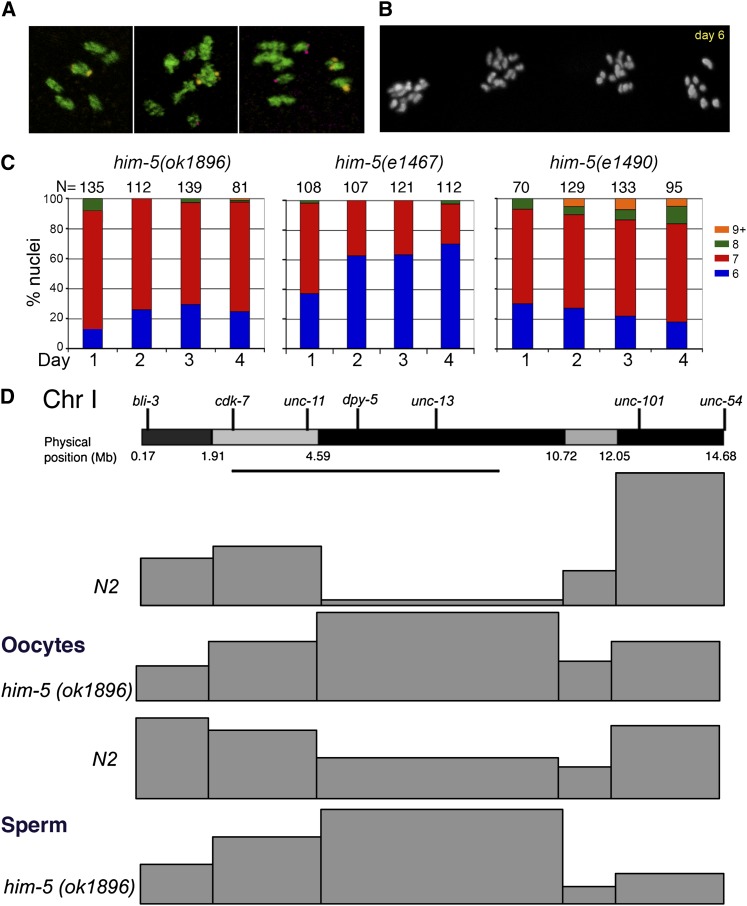
Direct cytology in different him-5 mutant alleles confirmed and extended these findings, as summarized in Table 2 and Figure 2. In wild-type animals, six bivalent chromosomes are observed at diakinesis in almost all nuclei; five bivalents and 2 univalents are seen rarely and only in older animals. Fluorescent in situ hybridization (FISH) confirmed that the large DAPI-staining masses were bivalents as single, large masses were labeled with chromosome V and X probes (Figure 2A). In contrast to wild type, in him-5, only a minority of the ova have the six bivalent chromosomes expected if all homologs have remained paired until the end of prophase I. Numerous ova had five bivalent chromosomes and two univalents, which FISH confirmed were the unpaired X chromosomes in almost all instances (Figure 2A). Ova with more than eight DAPI+ bodies were observed in as many as 10% of nuclei, including a few ova with 12 univalents (Figure 2, B and C). In nuclei with eight or more DAPI-staining bodies, the X was identified as univalent 100% of the time (Table 2), further confirming a more stringent requirement for him-5 activity on the X chromosome.
Table 2 . Fish analysis of bivalent formation in diakinesis oocytes.
| Bivalents | 6 | 5 + 2Xa | 5 + 2Va | 5 + 2Aa | 4+2X+2Va | 4+2X+2Aa | 4 + 4Aa |
|---|---|---|---|---|---|---|---|
| N2 | 100 (63) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| him-5(ok1896) | 24 (20) | 65 (54) | 0 (0) | 1 (1) | 6 (5) | 5 (4) | 0 (0) |
Values are percentage of nuclei (number analyzed).
2X, 2V and 2A or 4A, represent univalents of the X chromosome, chromosome V, or other autosomes, respectively.
Figure 2 .
Bivalent formation and recombination frequency and position. (A and B) Crossover formation is assessed by the number of DAPI-staining bivalents and univalents at diakinesis. (A) FISH of chromosomes X and V reveals defects in bivalent formation in adult day 1 hermaphrodites. (Left) Six bivalents are present in wild-type and a small fraction of him-5(ok1896) nuclei. (Center) Seven foci, five bivalents corresponding to autosomes (V, yellow foci) and two univalent X chromosomes (purple foci) are seen most often in him-5. (Right) Rare nuclei show additional univalents (two purple X foci + 2 yellow chromosome V foci), indicating defects in bivalent formation on autosomes. (B) Chiasma formation is further compromised in aging him-5 germlines as shown by the appearance of multiple nuclei with more than nine DAPI staining bodies in adult day 6 germlines. (C) Quantification of DAPI-staining bodies with age in him-5 alleles. (Day 1 is different from day 4, Mann–Whitney test, e1490, P = 0.04; e1467, P < 0.0001) (D) Recombination landscape is altered on autosomes in him-5 oocytes and sperm. Physical map of chromosome I is shown with the location of physical markers used for polymorphism mapping. Differential staining is used to highlight the different regions assayed by SNP analysis. The line underneath marks the central gene cluster. Map size for each interval is represented by the height of each box for wild type (N2) and him-5(ok1896). Raw data and statistical analyses are provided in Table S2 and Table S3.
One unexplained result is that the frequency of autosomal nondisjunction appears to be age dependent (Figure 2, B and C). For the weakest allele him-5(e1467), the rate of X chromosome univalent formation is highest among the first ova and then declines somewhat with maternal age (Figure 2C). For him-5(e1490), the rate of X chromosome univalent formation shows no age dependence, but the rate of autosomal univalent formation increases with maternal age (Figure 2C). Similarly for the deletion allele, although not statistically different between days 1 and 4, the trend is toward increased numbers of nuclei with autosomal univalents; for example, 12 univalents can be seen in day 4 gonads but in the course of many experiments have never been seen in 1-day-old adults (Figure 2, B and C and data not shown).
To test genetically for the presence of autosomal nondisjunction, we used the assay of Haack and Hodgkin (1991). When him-6 males were mated to hermaphrodites that were mutant for him-6 and two unlinked, recessive autosomal markers [for example, unc-24 (IV) and dpy-11 (V)], 14 Unc and 17 Dpy offspring were found among 6079 cross-progeny, a frequency of 0.25–0.3% of exceptional progeny per chromosome. These exceptional progeny arise from two different nondisjunction events, one in each parent. The experiment was repeated by mating him-5 (e1490) males to him-6unc-24; dpy-11 hermaphrodites to test for the presence of nullisomic sperm arising from him-5 males. No Unc or Dpy animals were found among 9604 cross-progeny, suggesting that autosomal loss during spermatogenesis in him-5 males is extremely low and may not occur at all. To test the effect of him-5 on oogenesis, him-6 males were mated to unc-24; dpy-11him-5(e1490) hermaphrodites. In this case, 12 Unc and 10 Dpy progeny were found among 13,074 cross-progeny. The presence of these progeny that have inherited both copies of an autosome in the ova indicates that him-5 mutations result in autosomal nondisjunction during oogenesis. The rate is somewhat lower in him-5 hermaphrodites than for him-6 hermaphrodites (per chromosome, 0.09% of the progeny for him-5 rather than 0.3% for him-6) but still readily detected above background in which no autosomal nondisjunction has been observed. Similar results were found for other autosomes using dpy-5 I; unc-4 II; him-5 and unc-25 III; dpy-11him-5 hermaphrodites (data not shown). Since these genetic assays require nondisjunction events in both parents as well as the survival of the offspring, it is difficult to compare the rate of autosomal nondisjunction by the genetics and cytology. Nonetheless, these experiments reveal that autosomal nondisjunction occurs in him-5 mutants, albeit at a much lower rate than X chromosome nondisjunction.
him-5 alters recombination distribution and frequency
Prior studies of the weak loss-of-function allele, him-5(e1467), indicated that the genetic map on the X chromosome was decreased to 35% of its wild-type levels (Hodgkin et al. 1979). On the autosomes, the genetic map was increased in some intervals (chromosomes I, II, III, and part of V) and decreased in others (chromosomes IV and part of V). The effect on different genetic intervals suggested the possibility that him-5 would be required not only for crossover frequency, but also for crossover positioning genome-wide. To better understand how the recombination landscape is affected by him-5, we performed single nucleotide polymorphism (SNP) mapping across chromosomes X and I in wild type and him-5(ok1896). SNP mapping of the X chromosome identified only 7/250 recombinants, yielding a map size of 2.8 m.u. for this chromosome. The paucity of recombinant progeny, however, limited our ability to determine whether there was also a change in crossover distribution on the X.
On autosomes, the vast majority of crossovers occur toward the chromosome ends in wild type, leaving a region in the central third of the chromosome with a greatly reduced rate of recombination (Figure 2D and Table S2). Similar regions with reduced recombination appear on each autosome and correspond to gene clusters that contain the majority of active genes (Barnes and Hodgkin 1996). In him-5(ok1896) mutants, the crossover landscape is strikingly different: nearly a third of the crossovers occur within the gene cluster on chromosome I (Figure 2D and Table S2). However, the overall length of the genetic map on chromosome I is unchanged (52.5 m.u. in wild type vs. 50.5 m.u. in him-5), and there is no increase in the number of double crossovers (3 DCO/124 CO in wild type vs. 3 DCO/162 CO events in him-5). Therefore, the crossovers that are made within the gene cluster in him-5(ok1896) must occur at the expense of crossovers on the chromosome arms, as reflected by the change in the map size of each of these intervals. These results also suggest the absence of an interchromosomal effect which might have been expected if COs formed on the autosomes at the expense of the X. An interchromosomal effect has been observed for the him-8 mutant that is defective in X chromosome CO formation due to an upstream defect in X chromosome pairing (Phillips et al. 2005). By contrast, no interchromosomal effect was observed in xnd-1 mutants, which functions to promote DSBs on the X.
We also analyzed the effect of him-5 on CO placement in the male germline. The CO landscape of him-5 sperm is similar to that of him-5 oocytes, with a substantial number of COs occurring in the middle of the gene cluster at the expense of COs in more telomere-proximal regions (Figure 2D and Table S3). As in oocytes, the differences in CO position cannot be explained by an increase in crossover formation on this chromosome as the map size of 49.8 m.u. does not differ significantly from wild type, 50.6 m.u. Together the SNP analyses suggest that him-5+ has two different effects on recombination: first, it influences recombination distribution on the autosomes such that crossovers in wild type occur more frequently in the gene-sparse outer regions of the chromosome than in the gene-rich clusters; second, it facilitates crossover formation on the X chromosome.
X chromosome and autosomal nondisjunction in him-5 mutants are rescued by irradiation
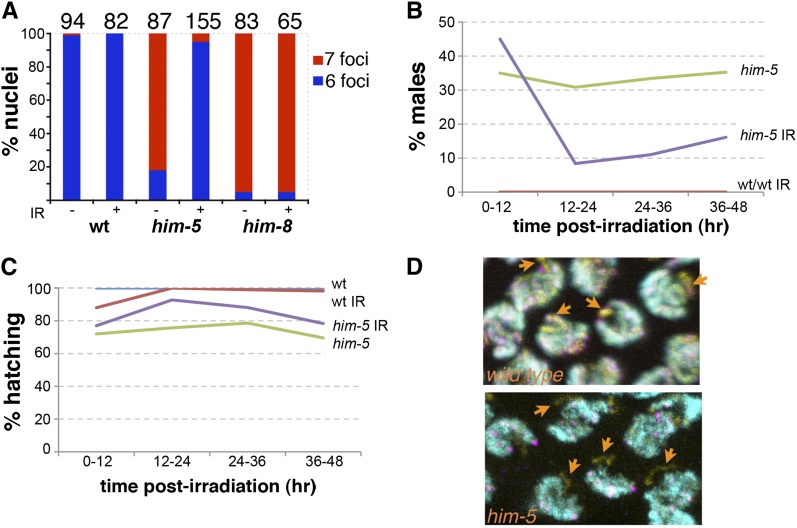
The requirement for him-5+ for normal crossover frequency and distribution suggested the possibility that him-5 may function to regulate the formation of meiotic double strand breaks (DSBs). To determine whether him-5 affects the initiation of meiotic DSBs, we asked whether DSBs introduced by γ-irradiation could rescue the defect in CO formation on the X. This assay has been successfully used to show that a number of meiotic mutants are deficient in the induction of meiotic DSBs (Dernburg et al. 1998; Reddy and Villeneuve 2004; Wagner et al. 2010). As shown in Figure 3, Table S4, and Table S5, γ-irradiation (IR) is an effective suppressor of the him-5 mutant phenotypes. In unirradiated him-5 hermaphrodites, >80% of the ova have five bivalents and two univalents (Figure 3A). Following exposure to a low dose of irradiation, only ∼3% of the ova from a him-5 hermaphrodite have univalent chromosomes, and most ova show six normally paired bivalents. The same dose of irradiation has no effect on wild-type bivalent formation. As expected, irradiation also does not rescue the X chromosome pairing defective mutant, him-8 (Figure 3A), since chromosome pairing is a prerequisite to synapsis and DSB repair off the homologous chromosome in C. elegans. These results indicate that him-5 is competent in processing artificially induced DSBs into crossovers on the X chromosome.
Figure 3 .
him-5 is required for meiotic DSB break formation. (A–C) Ionizing radiation rescues bivalent formation, suppresses male production, and restores full embryonic viability. (A) Number of nuclei with six or seven DAPI-staining bodies is shown for wild type, him-5(ok1896), and him-8 (e1489) hermaphrodites. Irradiation dose has no effect on wild-type animals, but restores bivalent formation in him-5. The X chromosome pairing mutant, him-8, is not rescued by irradiation, as described previously (Wagner et al. 2010). (B and C) Irradiated worms were individually plated and moved every 12 hr to count total eggs and progeny that result. (B) Irradiation suppresses male production in him-5 mutants in the window of time expected for nuclei that were in zygotene or early pachytene when irradiated (see also Table S3). (C) Irradiation also substantially rescues the embryonic lethality of him-5 during this time window. The immediate drop in hatching rates seen in wild type reflects unrepaired damage from nuclei in prophase I at the time of irradiation (see also Table S4). (D) Break formation on the X chromosome is severely reduced in him-5 as revealed by the deficit of RAD-51 foci on the X. Autosomes are labeled with rat anti–HTZ-1 (cyan) to show areas of active transcription; chromosome axes are labeled with guinea pig anti–HTP-3 (yellow), and DSBs with rabbit anti–RAD-51 (magenta). Orange arrows point to the X chromosome. In wild type, foci are observed on the X in 95% of nuclei; in him-5, in only 13% of nuclei (wild type, n = 117; him-5(ok1896), n = 175).
Time course analysis of the irradiated him-5 animals confirmed that the Him phenotype was substantially suppressed from ova that had been exposed to irradiation (Figure 3B). Irradiation also suppressed the low level of embryonic lethality associated with him-5 (Figure 3C). In some of the irradiated animals, hatching rates reached 100%, indicating that complete suppression is possible (data not shown). Since a portion of the lethality must be a consequence of the defect in bivalent formation on autosomes, the ability of irradiation to rescue the hatching defect suggests that autosomal aneuploidy results from a defect in DSB formation. This result also supports our earlier observation that autosomal aneuploidy is minimal in sperm from him-5 males. Since sperm are produced during L3 and early L4 larval stages and stored in the spermathecae, they have already completed meiosis at the time of irradiation. Sperm from irradiated animals were competent to confer full viability to their offspring. Thus, autosomal nondisjunction is minimal or nonexistent during hermaphrodite spermatogenesis.
HIM-5 is required for DSB formation on the X
Our results with irradiation indicate that the aneuploidy defects that arise in him-5 mutants can be effectively bypassed by artificially inducing DSBs. To observe DSB formation more directly, we monitored RAD-51 focus formation in him-5 mutant germlines. RAD-51 is a single-stranded DNA binding protein that is required for strand exchange during DNA damage repair. Since RAD-51 foci overlap SPO-11 induced DSBs with >95% confidence (Mets and Meyer 2009), RAD-51 localization is an accurate and widely used indicator for DSB formation in meiosis. The number and location of loci that have received a DSB can be assessed in situ by looking at RAD-51 foci after rad-54(RNAi) treatment. rad-54(RNAi) prevents repair of the DSBs at a step after the recruitment of RAD-51 to the lesion effectively trapping RAD-51 at the sites of repair (Mets and Meyer 2009). The X chromosome can be unambiguously distinguished from the autosomes on the basis of differential localization of variant histones and histone post-translational modifications, which serve as reporters of transcriptional status (Kelly et al. 2002). In these studies, as previously shown (Wagner et al. 2010), we used antibodies against HTZ-1, a H2A.Z homolog (Csankovszki et al. 2009), to negatively mark the X chromosome. By this combination of methods, recruitment of RAD-51 to the X chromosome was assessed. In agreement with previous results, we could identify a RAD-51 focus on the X chromosome in wild-type worms in almost every nucleus (Figure 3D). In contrast, in him-5(ok1896) mutants close to 90% of the nuclei lacked RAD-51 foci on the X (Figure 3D). These results further support a role for him-5+ in promoting meiotic DSB formation on the X chromosome.
An alternative interpretation of these results is that him-5 is defective in the recruitment of RAD-51 to the break site. We do not favor this interpretation because unlike mutations in rad-51, which lead to fragmented and fused chromosomes at diakinesis, him-5 mutants always show well-formed univalents and bivalents. Nonetheless, we cannot rule out the possibility that him-5 has two roles, one in the formation of DSBs and a second role, immediately downstream, in early repair events.
him-5 mutants are delayed in progression through the meiotic germline
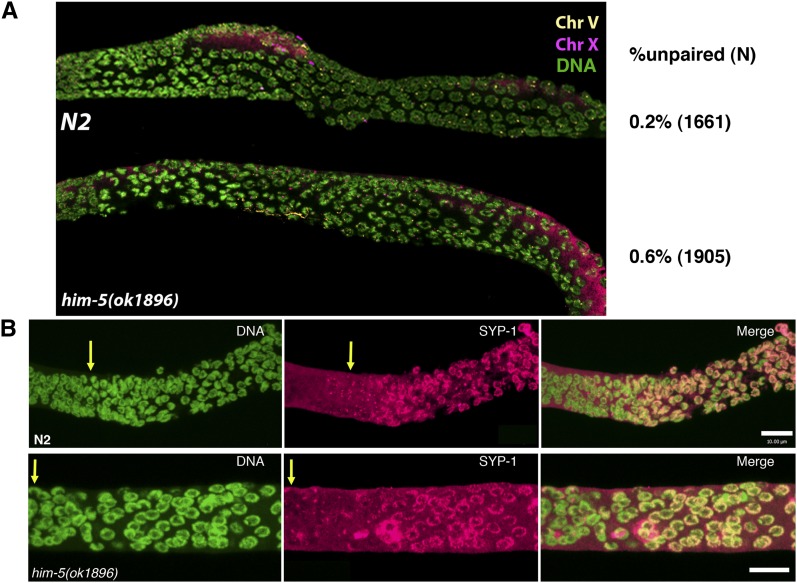
The ability of IR to rescue him-5 but not him-8 (Figure 3A), suggested that chromosome pairing occurs normally in him-5 mutants. In support of this, FISH probes revealed that chromosomes X and V demonstrate pairing with wild-type dynamics and efficiency from zygotene through pachytene (Figure 4 and Figure S2). Quantification of pairing using anti–HIM-8 antibodies confirmed that X chromosome pairing levels are indistinguishable between wild type and mutant from the transition zone through pachytene (Figure 4A). In addition, the chromosome axes and the synaptonemal complex assembled normally in him-5 mutants (Figures 4B, Figure S3A, and data not shown). Thus, the early events of homolog pairing and synapsis do not depend on him-5+ activity.
Figure 4 .
Pairing and synapsis are normal in him-5 mutants. (A) Wild type and him-5(ok1896) were fixed and hybridized with FISH probes for chromosomes V (yellow) and X (magenta) or anti–HIM-8 antibody (Figure S2) to assess the establishment and maintenance of pairing in zygotene to pachytene nuclei (quantification was done using the HIM-8 images in n = 10 gonads/genotype and total number of nuclei shown). him-5 mutants are indistinguishable from wild type with very few unpaired FISH signals in these regions. (B) Dissected gonads were labeled with DAPI (green) and anti–SYP-1 (magenta) antibodies to assess the kinetics and degree of synapsis. Representative image of him-5(ok1896) is shown compared to wild type (N2). Quantification of synapsis revealed no differences in SC establishment (n = 10 gonads/genotype, data not shown).
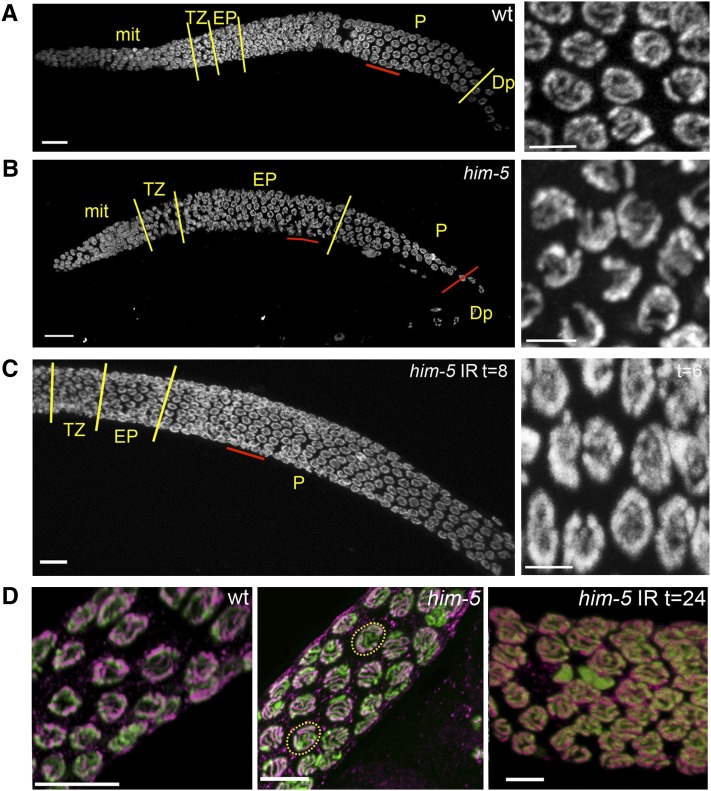
By contrast, the normal progression of meiosis is altered in him-5 mutants. Specifically, him-5 mutant nuclei appear to be delayed in their progression through the early pachytene stage of meiosis, as assessed by comparing the morphology of DAPI-stained nuclei in wild type and him-5 mutants. In wild type, the germline is organized in a spatial and temporal gradient with a stereotypical number of nuclei in each stage of meiosis distinguished by a unique DNA morphology (Figure 5A). Chromosomes in leptotene/zygotene (that is, the transition zone) are tightly clustered at one side of the nuclear periphery and appear as a bright crescent with DAPI staining. As nuclei enter early pachytene, the chromosomes remain attached to the nuclear periphery but are individually visible and thus less tightly packed. By midpachytene, the chromosomes are fully dispersed around the nuclear periphery (Figure 5A, closeup).
Figure 5 .
Deficit in DSBs leads to a delay in pachytene progression and to desynapsis. (A–C) DAPI-stained germline of wild type (A), him-5(ok1896) (B), or irradiated him-5(ok1896) (C) are shown with yellow lines demarcating the different regions of the germline as indicated by chromosome morphology (Bar, 10 μm). The region underlined in red is shown to the right in the higher magnification projection (Bar, 2 μm). Note that him-5 mutants have an extended early pachytene region at the expense of full pachytene (B) and that normal progression is restored after irradiation (C). (D) Desynapsis ensues in the absence of breaks. Full synapsis is seen between all homologs in wild type in late pachytene (left) as shown by the overlap between DNA (green) and anti–SYP-2 (magenta). In him-5(ok1896), the X chromosome desynapses in almost all nuclei (center). In occasional him-5 nuclei, multiple chromosomes without SYP-2 staining are observed (dashed yellow circles). Irradiation suppresses desynapsis (right) and SYP-2 staining persists between homologs in late pachytene nuclei (Bar, 5 μm).
In him-5 mutants, the number of nuclei with the clustered morphology characteristic of early pachytene is dramatically increased, and many fewer nuclei are seen with full pachytene morphology (Figure 5B). The persistence of early pachytene nuclei is not associated with a substantial increase in apoptosis, as expected if DSBs were not repaired or if the synapsis checkpoint were activated (Table 3 and Table S6). To determine whether the delay in progression was instead a consequence of the failure to make a DSB on one or more chromosomes, we asked whether exogenous breaks could promote progression through early pachytene. In wild-type germlines, meiotic progression is unaffected by 20 Gy of irradiation (not shown). The same dose of irradiation completely suppressed the delay in progression observed in the him-5 mutant (Figure 5C). Therefore, we infer that the failure to make DSBs on one or more chromosomes leads to a delay in progression through early pachytene.
Table 3 . Apoptosis analysis with SYTO 12.
| No. apoptotic nuclei/gonad | 0 | 1 | 2 | 3 | 4 | 5 | 6 | N | Mean | SD | SE mean |
|---|---|---|---|---|---|---|---|---|---|---|---|
| N2 | 1 | 10 | 19 | 14 | 4 | 0 | 0 | 48 | 2.208 | 0.9444 | 0.136 |
| him-5(ok1896) | 2 | 1 | 9 | 11 | 4 | 2 | 2 | 31 | 2.903 | 1.4226 | 0.256 |
Student’s t-test, d.f. 47, T-value −2.3999, P-value = 0.0204.
Another cytological feature of him-5 germlines is also noteworthy: the appearance of late pachytene nuclei with desynapsed X chromosomes (Figure 5D). Although full synapsis was observed in early pachytene, him-5(ok1896) nuclei (Figure S3), at slightly later stages of pachytene, a chromosome without SYP-1 labeling is observed in nearly every him-5 mutant nucleus, suggesting that a chromosome pair has desynapsed (Figure 5D). Localization of HIM-8 identified the X as the desynapsed chromosome in these nuclei (Figure S3). In addition, rare nuclei with two unlabeled chromosomes are seen, suggesting that autosomes, as well as the X, are susceptible to desynapsis (Figure 5D). We note that desynapsis at this stage does not immediately cause the homologs to separate from each other since separated X chromosomes were not observed until diplotene (Figure 2A and Figure S2). Induction of exogenous DSBs by IR suppresses desynapsis, as shown by chromosomes in him-5 germlines that are fully labeled with anti–SYP-1 antibodies at 8–24 hr postirradiation (Figure 5D, right). The maintenance of synapsis after irradiation indicates that desynapsis is a secondary consequence of the failure to make a break on the X and autosomes and that him-5 does not directly function in synaptonemal complex (SC) maintenance. These observations are consistent with the results seen in xnd-1 mutants and support the conclusion that DSB formation is coordinated with synapsis and the maintenance of the synaptonemal complex.
him-5 and xnd-1 appear to function in the same genetic pathway
Many of the features of him-5 mutants are strikingly similar to those described for xnd-1 (Wagner et al. 2010). Specifically both mutants appear to have primary defects in making DSBs on the X chromosome and in CO placement on autosomes, as well as defects in pachytene progression and SC maintenance. These similarities prompted us to determine whether him-5+ might function in the same pathway as xnd-1+. To assess genetically whether these genes interact to control crossover formation on the X, we made a double mutant between xnd-1(ok709) and him-5(e1467), both of which independently increase the frequency of males in the population to nearly 20% (Table 1). The xnd-1(ok709); him-5(e1467) double mutant showed no change in male frequency. The lack of suppression or synthetic enhancement between the mutations suggests that him-5 and xnd-1 operate in the same functional pathway.
HIM-5 is enriched on the autosomes
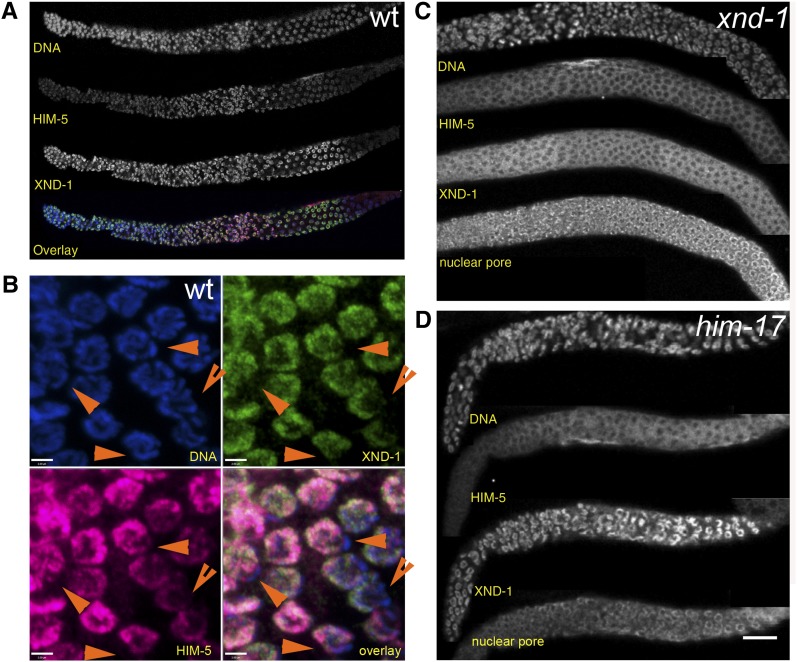
The inferred amino acid sequence of HIM-5 predicts a novel and highly basic protein. To assess the localization of HIM-5 in worms, antibodies generated to the C-terminus were used for immunolocalization (see Materials and Methods). As shown in Figure 6A, HIM-5 protein is found associated with chromosomes from the mitotic region of the germline until late pachytene. No staining is observed outside of the gonad (data not shown). In the germlines of wild-type hermaphrodites, HIM-5 preferentially localizes to most meiotic chromosomes but is clearly excluded from one pair. A number of other proteins have been shown to be enriched on autosomes, including a set that is required for modulating X chromosome gene expression [MES-4 (Bender et al. 2006), MRG-1 (Takasaki et al. 2007), the DRM complex (Tabuchi et al. 2011), and histone modifications associated with transcriptional activation (Schaner and Kelly 2006), as well as XND-1 (Wagner et al. 2010)]. To determine whether HIM-5 is also underrepresented on the X, hermaphrodite germlines were costained with anti–XND-1 and anti–HIM-5 antibodies. As shown in Figure 6B, XND-1 and HIM-5 colocalize to the same subset of chromosomes, indicating that HIM-5 is also enriched on autosomes. We cannot tell from these images whether there are subnuclear differences between the two proteins in their localization on individual autosomes. HIM-5 staining persists in nuclei that are beyond the zone of XND-1 staining. No signal was detected in these regions in the deletion allele him-5(ok1896), confirming the specificity of the antibody (Figure S4).
Figure 6 .
HIM-5 is enriched on the autosomes. (A and B) Anti–HIM-5 staining in wild-type germlines. (A) HIM-5 can be seen at low levels in the mitotic zone, accumulates in the transition zone and early pachytene, and decreases mildly upon entry into pachytene and more dramatically in late pachytene. Costaining with anti–XND-1 reveals differences in the accumulation patterns of these two proteins. (B) Enlarged region of late pachytene shows the overlap between XND-1 (green), HIM-5 (magenta), and DNA (blue) showing their exclusion from the X chromosome (orange arrowheads). Similar enrichment is observed in mitotic and early pachytene nuclei (not shown). Bar, 2 μm. (C and D) Localization of HIM-5 is impaired in xnd-1 (C) and him-17 (D) mutants. Dissected gonads were colabeled with DAPI, anti–HIM-5, anti–XND-1, and as a positive control for staining, nuclear pore marker anti-mAb414.
The autosomal localization of HIM-5 and the similarities between the xnd-1 and him-5 mutant phenotypes raise the possibility that localization of these proteins could be dependent on one another. Previous studies showed that XND-1 localizes normally in him-5 mutants (Wagner et al. 2010). In contrast, immunolocalization of HIM-5 on autosomes in the mitotic through midpachytene germlines was significantly reduced in the xnd-1 mutant background (Figure 6C). HIM-5 localization was also diminished in him-17(e2806) mutants, another meiotic gene with alleles that preferentially affect X chromosome crossover formation (Figure 6D). Thus, the wild-type activities of both him-17 and xnd-1 appear to be necessary for correct localization of HIM-5 to the autosomes and/or its expression. The stronger Him phenotype in him-5 deletion vs. xnd-1 (40% vs. 25%) suggests that either the unlocalized HIM-5 protein in xnd-1 mutants is functional or that reduced quantities of protein are below the level of detection with our antibodies. Furthermore, the dependency of HIM-5 localization on xnd-1, and not vice versa, suggests that him-5 acts downstream of xnd-1.
him-5 is suppressed by changes in X chromosome architecture
Within the germlines of wild-type worms, the majority of genes on the X chromosome are not expressed, and the X chromosome is replete with histone post-translational modifications associated with heterochromatin and silent chromatin (Kelly and Fire 1998; Kelly et al. 2002). In contrast, modifications associated with transcriptional activation are predominantly found on the autosomes (Schaner and Kelly 2006). We previously showed that xnd-1 could be suppressed by mutations in mes-2 in which X chromosome gene silencing does not occur, raising the possibility that silencing of the X blocks crossover formation on this chromosome (Wagner et al. 2010). To determine whether the requirement for him-5+ activity is diminished when the X chromosome is desilenced, the frequency of males was assessed in the mes-2(bn27); him-5(ok1896) double mutant (Table 1). Loss of mes-2 results in grand-maternal sterility so that the frequency of males can be assessed in the F2 brood. In an otherwise wild-type background, all of the progeny of mes-2 mothers are hermaphrodites, indicating that loss of mes-2 function does not itself cause nondisjunction. him-5(ok1896) broods yield 40–45% males, while mes-2; him-5 broods contained ∼25% males. Therefore, loss of mes-2 function partially suppresses the him-5 X nondisjunction phenotype. These results suggest that mes-2 acts in opposition to him-5 and suggests that him-5+ is sensitive to chromatin configurations on the X.
We showed previously that acetylation of histone H2A lysine 5 (H2AacK5) is increased in xnd-1 mutants, suggesting a role for this mark when crossover frequency is reduced (Wagner et al. 2010). However, we observed no gross changes in H2AacK5 levels in him-5(ok1896) mutant germlines (Figure S5). Therefore, despite the ability of mes-2 to suppress nondisjunction in both xnd-1 and him-5 mutants and the requirement for xnd-1+ function for HIM-5 localization, the two genes appear to function at different steps to control X chromosome crossover formation.
RAD-51 dynamics distinguish him-5 from xnd-1
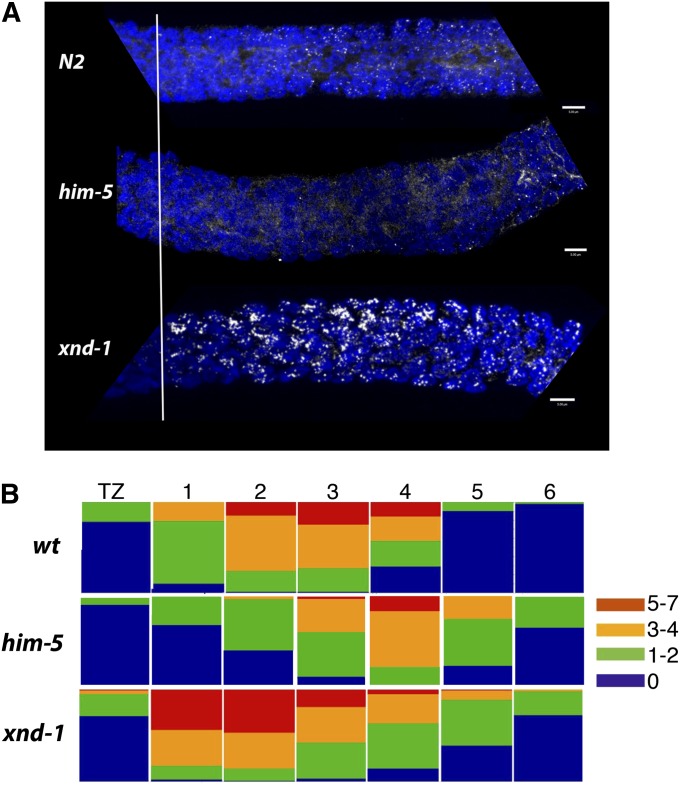
Further evidence that these genes may have distinct functions comes from analysis of RAD-51 accumulation in the mutants. In wild-type nuclei, RAD-51 foci are dynamic, increasing during early pachytene, and peaking with approximately seven RAD-51 foci per nucleus (Figure 7 and Figure S6). RAD-51 foci, and thus DSBs (Mets and Meyer 2009), are initiated within the transition in him-5 mutants and wild type; yet, the number of RAD-51 foci per nucleus in the him-5 mutant is lower at each subsequent stage until late pachytene (Figure 7B). Because initiation of DSBs occurs at the same time in him-5 and wild type, the differences in RAD-51 focus formation could be explained by an overall decrease in breaks, a change in the overall kinetics of DSB formation or both. We observed a decrease in the total number of DSBs by analysis of RAD-51 foci in rad-54(RNAi). Consistent with previous reports, we observed that wild-type animals enjoy ∼14 DSBs (Table S7) (Mets and Meyer 2009). In contrast, in him-5 mutants, we only observed ∼10 breaks/nucleus. This does not rule out an additional role for him-5 in regulating the kinetics of DSB repair, but these results support the hypothesis that him-5 plays a role in controlling CO dynamics through the regulation of meiotic DSB formation.
Figure 7 .
Dynamics of RAD-51 accumulation differ between xnd-1 and him-5 mutants. (A) DNA (blue) and RAD-51 (white) are shown as maximum projections from confocal stacks through wild type (top), him-5(ok1896) (middle), and xnd-1(ok709) (bottom) germlines. Images show the region from the transition zone (marked by horizontal line) through early/mid-pachytene. Full gonad images can be seen in Figure S5. (B) Quantification of RAD-51 foci in the transition zone and six regions of pachytene (as delineated in Figure S5). Each bar represents the percentage of nuclei with the indicated number of foci represented as a heat map. Note the differences in early pachytene accumulation between wild type and each of the mutants. The persistence of foci into late pachytene in both xnd-1 and him-5 is likely the result of the delay in normal pachytene progression (see Figure 4).
We note that RAD-51 foci also persist longer in him-5(ok1896) than in wild type, with foci visible in late pachytene nuclei and disappearing in diplotene (Figure 7B and Figure S6). We attribute the persistence of RAD-51 foci as a consequence of the delay in meiotic progression (described above and Figure 5), although this has not been tested.
The dynamics of break formation in him-5 are strikingly different from that seen in xnd-1 mutants (Figure 7 and Figure S6). In xnd-1 mutants, RAD-51 foci appear simultaneously rather than accumulating over time as they do in wild type and him-5, with the maximum number of foci visible as early as the transition zone/early pachytene nuclei. As nuclei progress into pachytene in xnd-1 mutants, the RAD-51 foci appear to get larger within the extended early pachytene zone and into late pachytene until they disappear upon entry into diplotene (Figure S6). The absence of RAD-51 foci in diplotene indicates that both mutants (and wild type) have repaired all damaged DNA.
Discussion
The him-5 gene in C. elegans was among the original meiotic mutations found in worms, and him-5 mutants are widely used in strain construction and other types of genetic analysis (Hodgkin et al. 1979). Despite its familiarity, the molecular identity of the him-5 gene has not been previously demonstrated. We show that him-5 corresponds to D1086.4, which encodes a novel, small and extremely basic protein. Mutations in him-5 are notable for greatly reduced recombination and elevated nondisjunction of the X chromosome (Hodgkin et al. 1979; Broverman and Meneely 1994), whereas effects on the autosomes appear to be comparatively minor. We find that the overall number of crossovers on the autosomes, as reflected in the genetic map units, is not reduced in him-5 mutants compared to wild type (at least for chromosome I), but crossovers are redistributed such that the recombination map and the physical maps are more congruent in him-5 mutants than in wild type. Defects in crossover formation on other autosomes can be inferred from the presence of multiple pairs of univalents at diakinesis.
Role of HIM-5 in DSB formation and repair
Possibly the most informative results for understanding the mutant phenotype of him-5 are that we do not observe RAD-51 foci on the X in the rad-54(RNAi) and that nearly every aspect of the him-5 mutant phenotype is suppressed by radiation-induced DSBs. These phenotypes include X chromosome and autosome nondisjunction, premature chromosome desynapsis, and a delay in pachytene progression. Two models could explain the ability of IR to rescue: either (1) him-5+ functions upstream of the SPO-11 nuclease to potentiate the X chromosome for recombination or (2) him-5 acts immediately downstream of DSB formation, in a processing step prior to RAD-51 recruitment. The latter model would need to explain how DSBs are ultimately repaired in him-5 mutants since we see no evidence of increased apoptosis (which would occur if there were persistent damage). Additionally, despite passaging for hundreds of generations as homozygotes, no germline transmissible mutations have been identified in these strains, suggesting that the breaks are not repaired by the mutagenic NHEJ pathways nor are they leading to chromosome fusions. For these reasons, we favor a model in which him-5 acts upstream of break formation, although it may also have a role downstream in break repair.
One of these things is not like the other one: X/autosome differences in crossover control
A number of studies have revealed that the X and autosomes respond differently to loss of certain meiotic gene functions. A subset of mutations, e.g., htp-1, him-3, and cra-1, more severely affect autosomes (Couteau et al. 2004; Couteau and Zetka 2005; Smolikov et al. 2008); whereas other mutations, e.g., him-17, him-19, xnd-1, and him-5 (Hodgkin et al. 1979; Reddy and Villeneuve 2004; Tang et al. 2010; Wagner et al. 2010), have more severe consequences for the X. The latter class all affect very early events in CO formation, in either DSB formation or processing. The observation that double COs have been documented on the autosomes but never on the X is consistent with this chromosome receiving fewer meiotic breaks, although this has not been directly tested in the nematode system. In the analysis of him-17, Reddy and Villeneuve (2004) proposed a model that they referred to as a “window of opportunity” to explain its different effects on the X and the autosomes. Their hypothesis is that the accessibility of the chromosomes to SPO-11 is limited to a specific time period during prophase I. According to their model, the “window” for the X chromosome closes before the window for the autosomes. Thus, in a mutant like him-17, in which overall accessibility for DSBs is limited even further, the X chromosome is more sensitive because its window is normally smaller. Since its localization is dependent on him-17, we can apply this model to explain the effects of him-5 and hypothesize that it is one of the genes responsible for determining when the X chromosome window is open.
X/autosome differences in DSB formation, however, may alternatively be explained by the gross differences in chromatin states between these chromosomes. Because crossovers occur preferentially in “open” chromatin (Ohta et al. 1994; Wu and Lichten 1994; Nicolas 1998), the heterochromatic-like X chromosome of C. elegans might present an inhospitable environment for crossover formation by creating a kinetic barrier to the SPO-11 complex. Therefore, we postulate that him-5 may have evolved as part of a mechanism to promote crossovers in heterochromatin. Our observation that the HIM phenotype is suppressed by mutations in mes-2 and mes-3, components of a histone H3K27 methyltransferase complex that is required for silencing X-linked genes, supports the model that the chromatin architecture of the X normally limits CO formation.
Models of HIM-5 function
Because the total number of breaks is reduced in him-5 mutants, we favor models in which HIM-5 directly regulates the SPO-11 machinery. The simplest model would have him-5 regulating the expression and/or activity of the SPO-11 machinery. However, while this could account for the increase in X and autosomal univalents at diakinesis and resulting males and lethality, as well as the decreased total number of RAD-51 foci observed (9 vs. 14 in him-5 vs. wild type), it is more difficult to explain how reducing SPO-11 activity could account for the change in CO positioning to go from almost none to nearly half of all COs occurring in the autosomal gene clusters. Simply reducing spo-11 expression would be expected to reduce CO formation throughout the genome, not simply divert COs on the autosome to the central gene cluster.
Instead, we favor the possibility that HIM-5 functions, either directly or indirectly, to target the SPO-11 machinery to the chromosome. In other species, SPO11 is known to interact with accessory proteins that regulate its recruitment and function (Keeney 2001). One possibility is that HIM-5 is a functional homolog of one of these components. Another possibility is that him-5 could function to define the recombination hotspot, either as a landmark itself or by creating a critical chromatin signature. Alternatively, him-5 could function indirectly to influence SPO-11 recruitment by modifying the closed chromatin milieu on the X (and distal autosome regions). This could occur either by directly recruiting histone-modifying complexes (or chromatin-associated proteins) to the X (and distal autosomes) or by preventing these complexes from accessing the autosomes so that modifications are made on the X. Each of these models posits that SPO-11 is functional but cannot be properly targeted to recombination hotspots. We propose that without normal cis or trans-acting localization features, the SPO-11 complex would preferentially catalyze breaks in the regions of most open chromatin structure. This would explain the bias for the autosomal gene clusters. In this case, the overall number of breaks may be reduced in him-5 as compared to in wild type either because crossover interference is so strong in the worm that breaks are redistributed to the center of the chromosome effectively inhibiting additional breaks distally to both ends, or because passive recruitment to the DNA is comparatively inefficient.
These models predict that HIM-5 proteins would be found on the X and/or unequally distributed between the gene clusters and distal autosomal regions. Immunofluorescence does not provide adequate resolution to determine whether HIM-5 is biased in its autosomal distribution. Despite our inability to detect HIM-5 proteins on the X, there may be subthreshold levels sufficient for function.
Pathways for X chromosome breaks
him-5 phenotypes closely resemble those of xnd-1. Double mutant analyses are consistent with these genes working in the same pathway rather than parallel pathways. This interpretation is supported by the observation that xnd-1+ activity is required for proper HIM-5 localization and places HIM-5 downstream in the pathway. However, two lines of evidence indicate that HIM-5 and XND-1 must have distinct roles in crossover control. First, previous studies have revealed pronounced differences in H2AacK5 in the xnd-1 mutant (Wagner et al. 2010), whereas no overt differences in this modification are observed in the him-5 mutant (Figure S5). Second, the early dynamics of RAD-51 accumulation in xnd-1 and him-5 differ dramatically (Figure 7). In xnd-1 mutants, the majority of programmed DSBs, as assessed by RAD-51 foci, appear simultaneously at the onset of break formation. By contrast, in him-5, fewer breaks are made, but the time course of break formation is more similar to wild type. Therefore, although both XND-1 and HIM-5 affect crossover formation on the X as well as crossover distribution genome-wide, and HIM-5 localization is dependent on xnd-1+, it appears that these proteins may have additional, independent roles in regulating DSB formation and repair.
One possible model to explain XND-1 and HIM-5 suggests that XND-1’s primary function is to negatively regulate H2AK5 acetylation, allowing heterochromatic regions of the genome to “compete” effectively with the autosomal gene clusters for crossover formation. HIM-5’s association with chromatin could be reduced in the xnd-1 mutant background either because the high levels of H2AacK5 prevent its association with DNA or because XND-1 protein is directly required for its expression and/or localization.
HIM-5 localization also requires functional him-17, a THAP-domain protein required for meiotic DSB break formation on all chromosomes. Weak mutations of him-17 have more severe effects for the X than for autosomes, which we suggest may now be explained by the defect in HIM-5 localization. Whether the effects of him-17 on HIM-5 localization are direct or not await further analysis of these proteins, however, the observation that HIM-17 protein localizes to all chromosomes (Reddy and Villeneuve 2004) favors a model in which him-17 indirectly controls HIM-5 protein distribution.
Desynapsis and delayed meiotic progression
In addition to effects on recombination and disjunction, him-5 mutants also exhibit a delay in meiotic progression. Since the other mutant phenotypes of him-5 can be explained by a role in DSB formation and the delay is suppressed after exposure to irradiation, we suggest that the delay in meiotic progression is also attributed to the defect in DSB formation. Because DSBs would normally be deleterious, the events of meiosis I are closely monitored to ensure that each chromosome receives a crossover and DNA damage is fully repaired. Checkpoints ensure complete synapsis and the repair of DNA damage. In C. elegans, these checkpoints require the activity of conserved pathways mediated by pch-2 and cep-1, respectively (Gartner et al. 2000; Bhalla and Dernburg 2005). In addition, mechanisms to couple pairing, synapsis, and DSB formation have been elucidated (Couteau and Zetka 2005; Goodyer et al. 2008; Smolikov et al. 2008), suggesting that fail-safe mechanisms ensure the completion of meiotic processes.
The analyses of him-5 presented here and our previous studies on xnd-1 (Wagner et al. 2010) suggest that an additional safeguard exists to maintain the synaptonemal complex association with chromosomes after formation of a DSB. In both mutants, the synaptonemal complex polymerizes along every chromosome with normal kinetics and is fully polymerized by the end of the transition zone region. Thus, him-5+ and xnd-1+ do not appear to be needed to initiate synapsis or SC formation. Moreover, it does not appear that him-5+ and xnd-1+ are simply required for the maintenance of the SC since irradiation can prevent desynapsis in the mutant background. Instead, our data are most consistent with a model in which the underlying defect in these mutants, the failure to make a break on the X chromosome, apparently leads to subsequent desynapsis of this chromosome. The ability of a DSB to stabilize the synaptonemal complex does not appear to be limited to the X chromosome. In him-5 mutants, as many as 10% of nuclei have at least one autosome that does not receive a crossover, as revealed by an increase in the number of univalent chromosomes observed at diakinesis. In a subset of him-5 mutant nuclei, we could observe two chromosomes that failed to label with anti–SYP-1 antibodies (Figure 5D), suggesting that the synaptonemal complex has dissociated from both chromosomes. Since this desynapsis is also rescued by irradiation, we propose that the formation of a DSB (or a subsequent crossover intermediate) must trigger a change in the SC allowing for its stabilization on the homologs.
The destabilization of the SC that is observed in him-5 and xnd-1 mutants is not accompanied by a large increase in apoptosis expected from activation of the synapsis or DNA damage checkpoints (Table 2 and J. Yanowitz, unpublished data). Since the delay in progression observed in him-5 is not suppressed by mutations in pch-2 or cep-1, it also appears to be independent of the known germline checkpoints. The inability of him-5 mutants to activate the pch-2–dependent synapsis checkpoint suggests the possibility that this checkpoint is only active during a short window of time in early pachytene. We noted that the most distal him-5 nuclei in which the X chromosomes desynapsed had not yet progressed into full pachytene, on the basis of nuclear morphology. This suggests that monitoring of break formation and stabilization of the SC occurs during early pachytene. The transition into full pachytene could reflect two inherent changes with regard to the synaptonemal complex, both a closing of the window for activation of the synapsis checkpoint and a DSB-induced stabilization of the synaptonemal complex.
It is unclear as yet what controls these transitions, but it could be mediated through communication with the nuclear periphery. The Jantsch laboratory has shown that distinct phosphorylation states of the nuclear membrane protein SUN-1 are required for association of DNA with the nuclear periphery, its movements there, and its ultimate release (Penkner et al. 2009). It is tempting to speculate that signals emanating from an early crossover intermediate lead to a specific alteration in SUN-1 phospho status and contribute to release from the nuclear periphery. Further analysis of him-5 will help to elucidate the mechanisms that control the formation and monitoring of DSBs and how these are coordinated with meiotic events to ensure the timely and accurate execution of meiotic recombination.
Is break formation sensitive to age?
The nondisjunction phenotype of him-5 appears to worsen with age, suggesting a greater requirement for him-5+ activity in older germlines. Maternal age effects on nondisjunction are a familiar feature of human meiosis, but the time scale over which these effects occur in humans is measured in decades rather than in hours, as they are in worms, so the underlying mechanism could be quite different. For him-5, the differences among the three alleles suggest that these mutations may be revealing an underlying property of meiosis in worms, rather than a specific consequence of the effects on him-5 function. It has been well established that parental age decreases recombination and increases X chromosome nondisjunction rates in C. elegans (Rose and Baillie 1979). him-5 joins him-19 in a class of meiotic genes that increase severity with maternal age. Tang et al. (2010) hypothesized that him-19 might affect the stability of an unknown maternally supplied factor that becomes depleted over time. One postulate is that him-5 mutations have different effects on the stability of this factor, which affects autosomal disjunction more than X disjunction. In both him-5 and him-19, the observed defects can be attributed to defects in early meiotic events. We speculate that early events such as DSB formation might be particularly sensitive to aging, because they are influenced by the packaging of chromatin, which has been shown for somatic tissues to be altered during aging (Greer et al. 2010).
Supplementary Material
Acknowledgments
We thank Verena Jantsch for suggesting the cosuppression assay and doing the injections that facilitate P.M.M.’s analysis of these lines. We also thank Dave Baillie for pointing out the similarity to HSP83 and Alex Ensminger and Nelson Lau for their generosity in providing purified RNAs to P.M.M. We thank Monique Zetka, Anne Villeneuve, Adriana la Volpe, Verena Jantsch, Abby Dernburg, and Gyorgyi Csankovski for antibodies and Barbara Meyer and David Mets for technical advice. P.M.M. thanks the many Haverford College students who worked on different aspects of him-5. J.L.Y. thanks Cynthia Wagner and Mainpal Rana for discussion and reading of the manuscript. This work has been funded by National Science Foundation grants RUI MCB 0131321 and RUI MCB 061470 to P.M.M. and by National Institutes of Health K01AG031206 and MWRI start-up funds to J.L.Y.
Footnotes
Communicating editor: S. Keeney
Literature Cited
- Barnes T. M., Hodgkin J., 1996. The tra-3 sex determination gene of Caenorhabditis elegans encodes a member of the calpain regulatory protease family. EMBO J. 15: 4477–4484 [PMC free article] [PubMed] [Google Scholar]
- Barnes T. M., Kohara Y., Coulson A., Hekimi S., 1995. Meiotic recombination, noncoding DNA and genomic organization in Caenorhabditis elegans. Genetics 141: 159–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender L. B., Suh J., Carroll C. R., Fong Y., Fingerman I. M., et al. , 2006. MES-4: an autosome-associated histone methyltransferase that participates in silencing the X chromosomes in the C. elegans germ line. Development 133: 3907–3917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhalla N., Dernburg A. F., 2005. A conserved checkpoint monitors meiotic chromosome synapsis in Caenorhabditis elegans. Science 310: 1683–1686 [DOI] [PubMed] [Google Scholar]
- Brenner S., 1974. The genetics of Caenorhabditis elegans. Genetics 77: 71–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broverman S. A., Meneely P. M., 1994. Meiotic mutants that cause a polar decrease in recombination on the X chromosome in Caenorhabditis elegans. Genetics 136: 119–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan R. C., Chan A., Jeon M., Wu T. F., Pasqualone D., et al. , 2003. Chromosome cohesion is regulated by a clock gene paralogue TIM-1. Nature 423: 1002–1009 [DOI] [PubMed] [Google Scholar]
- Colaiacovo M. P., MacQueen A. J., Martinez-Perez E., McDonald K., Adamo A., et al. , 2003. Synaptonemal complex assembly in C. elegans is dispensable for loading strand-exchange proteins but critical for proper completion of recombination. Dev. Cell 5: 463–474 [DOI] [PubMed] [Google Scholar]
- Cole F., Keeney S., Jasin M., 2010. Evolutionary conservation of meiotic DSB proteins: more than just Spo11. Genes Dev. 24: 1201–1207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couteau F., Zetka M., 2005. HTP-1 coordinates synaptonemal complex assembly with homolog alignment during meiosis in C. elegans. Genes Dev. 19: 2744–2756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couteau F., Nabeshima K., Villeneuve A., Zetka M., 2004. A component of C. elegans meiotic chromosome axes at the interface of homolog alignment, synapsis, nuclear reorganization, and recombination. Curr. Biol. 14: 585–592 [DOI] [PubMed] [Google Scholar]
- Csankovszki G., Collette K., Spahl K., Carey J., Snyder M., et al. , 2009. Three distinct condensin complexes control C. elegans chromosome dynamics. Curr. Biol. 19: 9–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dernburg A. F., McDonald K., Moulder G., Barstead R., Dresser M., et al. , 1998. Meiotic recombination in C. elegans initiates by a conserved mechanism and is dispensable for homologous chromosome synapsis. Cell 94: 387–398 [DOI] [PubMed] [Google Scholar]
- Gartner A., Milstein S., Ahmed S., Hodgkin J., Hengartner M. O., 2000. A conserved checkpoint pathway mediates DNA damage–induced apoptosis and cell cycle arrest in C. elegans. Mol. Cell 5: 435–443 [DOI] [PubMed] [Google Scholar]
- Goodyer W., Kaitna S., Couteau F., Ward J. D., Boulton S. J., et al. , 2008. HTP-3 links DSB formation with homolog pairing and crossing over during C. elegans meiosis. Dev. Cell 14: 263–274 [DOI] [PubMed] [Google Scholar]
- Greer E. L., Maures T. J., Hauswirth A. G., Green E. M., Leeman D. S., et al. , 2010. Members of the H3K4 trimethylation complex regulate lifespan in a germline-dependent manner in C. elegans. Nature 466: 383–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haack H., Hodgkin J., 1991. Tests for parental imprinting in the nematode Caenorhabditis elegans. Mol. Gen. Genet. 228: 482–485 [DOI] [PubMed] [Google Scholar]
- Hodgkin J., Horvitz H. R., Brenner S., 1979. Nondisjunction mutants of the nematode Caenorhabditis elegans. Genetics 91: 67–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaramillo-Lambert A., Ellefson M., Villeneuve A. M., Engebrecht J., 2007. Differential timing of S phases, X chromosome replication, and meiotic prophase in the C. elegans germ line. Dev. Biol. 308: 206–221 [DOI] [PubMed] [Google Scholar]
- Keeney S., 2001. Mechanism and control of meiotic recombination initiation. Curr. Top. Dev. Biol. 52: 1–53 [DOI] [PubMed] [Google Scholar]
- Keeney S., Neale M. J., 2006. Initiation of meiotic recombination by formation of DNA double-strand breaks: mechanism and regulation. Biochem. Soc. Trans. 34: 523–525 [DOI] [PubMed] [Google Scholar]
- Keeney S., Giroux C. N., Kleckner N., 1997. Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell 88: 375–384 [DOI] [PubMed] [Google Scholar]
- Kelly W. G., Fire A., 1998. Chromatin silencing and the maintenance of a functional germline in Caenorhabditis elegans. Development 125: 2451–2456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly W. G., Schaner C. E., Dernburg A. F., Lee M. H., Kim S. K., et al. , 2002. X-chromosome silencing in the germline of C. elegans. Development 129: 479–492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar R., Bourbon H. M., de Massy B., 2010. Functional conservation of Mei4 for meiotic DNA double-strand break formation from yeasts to mice. Genes Dev. 24: 1266–1280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim J. G., Stine R. R., Yanowitz J. L., 2008. Domain-specific regulation of recombination in Caenorhabditis elegans in response to temperature, age and sex. Genetics 180: 715–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mets D. G., Meyer B. J., 2009. Condensins regulate meiotic DNA break distribution, thus crossover frequency, by controlling chromosome structure. Cell 139: 73–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolas A., 1998. Relationship between transcription and initiation of meiotic recombination: toward chromatin accessibility. Proc. Natl. Acad. Sci. USA 95: 87–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta K., Shibata T., Nicolas A., 1994. Changes in chromatin structure at recombination initiation sites during yeast meiosis. EMBO J. 13: 5754–5763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penkner A. M., Fridkin A., Gloggnitzer J., Baudrimont A., Machacek T., et al. , 2009. Meiotic chromosome homology search involves modifications of the nuclear envelope protein Matefin/SUN-1. Cell 139: 920–933 [DOI] [PubMed] [Google Scholar]
- Phillips C. M., Wong C., Bhalla N., Carlton P. M., Weiser P., et al. , 2005. HIM-8 binds to the X chromosome pairing center and mediates chromosome-specific meiotic synapsis. Cell 123: 1051–1063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy K. C., Villeneuve A. M., 2004. C. elegans HIM-17 links chromatin modification and competence for initiation of meiotic recombination. Cell 118: 439–452 [DOI] [PubMed] [Google Scholar]
- Rinaldo C., Bazzicalupo P., Ederle S., Hilliard M., La Volpe A., 2002. Roles for Caenorhabditis elegans rad-51 in meiosis and in resistance to ionizing radiation during development. Genetics 160: 471–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockman M. V., Kruglyak L., 2009. Recombinational landscape and population genomics of Caenorhabditis elegans. PLoS Genet. 5: e1000419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose A. M., Baillie D. L., 1979. The effect of temperature and parental age on recombination and nondisjunction in Caenorhabditis elegans. Genetics 92: 409–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaner C. E., Kelly W. G., 2006. Germline chromatin. WormBook 1–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smolikov S., Schild-Prufert K., Colaiacovo M. P., 2008. CRA-1 uncovers a double-strand break-dependent pathway promoting the assembly of central region proteins on chromosome axes during C. elegans meiosis. PLoS Genet. 4: e1000088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabuchi T. M., Deplancke B., Osato N., Zhu L. J., Barrasa M. I., et al. , 2011. Chromosome-biased binding and gene regulation by the Caenorhabditis elegans DRM complex. PLoS Genet. 7: e1002074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takasaki T., Liu Z., Habara Y., Nishiwaki K., Nakayama J., et al. , 2007. MRG-1, an autosome-associated protein, silences X-linked genes and protects germline immortality in Caenorhabditis elegans. Development 134: 757–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang L., Machacek T., Mamnun Y. M., Penkner A., Gloggnitzer J., et al. , 2010. Mutations in Caenorhabditis elegans him-19 show meiotic defects that worsen with age. Mol. Biol. Cell 21: 885–896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmons L., Fire A., 1998. Specific interference by ingested dsRNA. Nature 395: 854. [DOI] [PubMed] [Google Scholar]
- Tsai C. J., Mets D. G., Albrecht M. R., Nix P., Chan A., et al. , 2008. Meiotic crossover number and distribution are regulated by a dosage compensation protein that resembles a condensin subunit. Genes Dev. 22: 194–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner C. R., Kuervers L., Baillie D. L., Yanowitz J. L., 2010. xnd-1 regulates the global recombination landscape in Caenorhabditis elegans. Nature 467: 839–843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu T. C., Lichten M., 1994. Meiosis-induced double-strand break sites determined by yeast chromatin structure. Science 263: 515–518 [DOI] [PubMed] [Google Scholar]
- Zetka M. C., Rose A. M., 1992. The meiotic behavior of an inversion in Caenorhabditis elegans. Genetics 131: 321–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zetka M. C., Kawasaki I., Strome S., Muller F., 1999. Synapsis and chiasma formation in Caenorhabditis elegans require HIM-3, a meiotic chromosome core component that functions in chromosome segregation. Genes Dev. 13: 2258–2270 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.