Abstract
Genome-wide expression quantitative trait loci (eQTL) studies have emerged as a powerful tool to understand the genetic basis of gene expression and complex traits. In a typical eQTL study, the huge number of genetic markers and expression traits and their complicated correlations present a challenging multiple-testing correction problem. The resampling-based test using permutation or bootstrap procedures is a standard approach to address the multiple-testing problem in eQTL studies. A brute force application of the resampling-based test to large-scale eQTL data sets is often computationally infeasible. Several computationally efficient methods have been proposed to calculate approximate resampling-based P-values. However, these methods rely on certain assumptions about the correlation structure of the genetic markers, which may not be valid for certain studies. We propose a novel algorithm, rapid and exact multiple testing correction by resampling (REM), to address this challenge. REM calculates the exact resampling-based P-values in a computationally efficient manner. The computational advantage of REM lies in its strategy of pruning the search space by skipping genetic markers whose upper bounds on test statistics are small. REM does not rely on any assumption about the correlation structure of the genetic markers. It can be applied to a variety of resampling-based multiple-testing correction methods including permutation and bootstrap methods. We evaluate REM on three eQTL data sets (yeast, inbred mouse, and human rare variants) and show that it achieves accurate resampling-based P-value estimation with much less computational cost than existing methods. The software is available at http://csbio.unc.edu/eQTL.
GENOME-WIDE studies of expression quantitative trait loci (eQTL) have been widely used to dissect the genetic basis of gene expression and molecular mechanisms underlying complex traits (Bochner 2003; Rockman and Kruglyak 2006; Michaelson et al. 2009). In a typical eQTL study, the association between each expression trait and each genetic marker [e.g., single-nucleotide polymorphism (SNP)] is assessed separately, which leads to a huge number of correlated tests. Appropriate multiple-testing correction is crucial for eQTL studies. The resampling-based test using permutation or bootstrap (Good 2005) has been widely used for multiple-testing correction across multiple genetic markers for each phenotype (Barrett et al. 2005; Purcell et al. 2007) by simulating the null distribution using permuted or bootstrapped phenotype values (Westfall and Young 1993; Churchill and Doerge 1994; McClurg et al. 2007). More specifically, the phenotype values are randomly shuffled and reassigned to individuals with or without replacement (i.e., bootstrap and permutation, respectively). For each resampled phenotype a whole-genome scan is performed to find the maximum test statistic among all SNPs. The corrected P-value is the proportion of the resampled phenotypes where the maximum test statistics are greater than the maximum test statistic in the original data. We refer to such a corrected P-value as the resampling-based P-value. The resampling-based test preserves the correlation structure of the SNPs and does not require any distribution assumption on the test statistic.
Another level of multiple-testing problem in eQTL studies is the multiple tests across tens of thousands of gene expression traits (Kendziorski and Wang 2006). One standard solution is first to estimate the resampling-based P-value (of the most significant association) for each expression trait and then to determine a threshold for the corrected P-values across all expression traits by controlling the false discovery rate (FDR) (Benjamini and Hochberg 1995; Storey 2003).
Although this approach is ideal for eQTL studies, its intensive computational burden has greatly limited its practical use. For example, supposing that a corrected P-value threshold of 0.01 is needed to control the FDR, up to 100,000 permutations/bootstraps may be needed to estimate such a resampling-based P-value accurately. Consider a typical scenario where there are 500,000 SNPs and 20,000 expression traits. The total number of tests is 500,000 × 20,000 × 100,000 = 1015. A brute force implementation of the resampling-based test is clearly not practical. Computationally efficient methods are highly desirable.
To tackle the computational challenge, several methods have been proposed to approximate the resampling-based P-values. One approach is to estimate the effective number of independent tests from the eigenvalues of the correlation matrix of the SNPs (Cheverud 2001; Nyholt 2004; Li and Ji 2005; Gao et al. 2008; Moskvina and Schmidt 2008; Pe’er et al. 2008). Previous studies have shown that these methods may yield inaccurate results (Salyakina et al. 2005; Han et al. 2009). Another approach, relying on the assumption that the test statistics over multiple SNPs asymptotically follow a multivariate normal distribution (MVN), calculates the permutation P-values by directly sampling from the MVN distribution (Lin 2005; Seaman and Müller-Myhsok 2005; Conneely and Boehnke 2007; Han et al. 2009). Both the “effective number of independent tests” and MVN methods cannot be directly applied to genome-wide association studies when the number of SNPs is much larger than the sample size. A common strategy is to divide all the SNPs into contiguous blocks, to apply these two methods to each block of SNPs, and then to combine the results across the SNP blocks. This partition–ligation approach implicitly assumes that the SNPs between two blocks are weakly correlated. From a different perspective, an approximation method based on a geometric interpretation of permutation P-values has been proposed in Sun and Wright (2010). This method does not require any asymptotic distribution of the test statistics, but it assumes a Markov type of dependency among the markers. Another method has been developed to correct very significant P-values in disease association studies by importance sampling (Kimmel and Shamir 2006). This method is designed for binary phenotypes and cannot be directly applied to quantitative expression traits.
Despite the success of the aforementioned approximation methods, their accuracies rely on the validity of the following assumption: the nearby SNPs are correlated whereas the distant SNPs are not. This assumption may not be valid. Please refer to supporting information, File S1 for further discussion on the assumptions of the existing approximation methods.
For the resampling-based test, the total computational time is equal to the average time needed to calculate each test statistic multiplied by the total number of tests. Several algorithms have been developed to speed up the exhaustive resampling-based test by calculating each test statistic more efficiently. For example, if the phenotype is binary, computational efficiency can be improved by efficient computation of contingency tables (Browning 2008) or by bit arithmetic operations (Pahl and Schafer 2010). For eQTL studies, a summation tree can be utilized to compute the test statistics incrementally (Gatti et al. 2009). Note that these methods are specifically designed for permutation tests and are not applicable to bootstrapping. In this article we tackle the computational challenge of the exhaustive resampling-based test from another perspective. That is, we try to reduce the number of test statistics that need to be calculated. We present an algorithm, rapid and exact multiple testing correction by resampling (REM), which prunes the search space and performs tests only on a small proportion of the SNPs. This is achieved by constructing a two-layer indexing structure that groups SNPs on the basis of their genotypes. The SNPs in one group share a common upper bound on the test statistics. Actual tests are performed only in the groups whose upper bounds are greater than a certain threshold. REM is guaranteed to find the exact resampling-based P-values. It can be applied to a variety of resampling-based methods, including permutation (Westfall and Young 1993; Churchill and Doerge 1994) and bootstrap (McClurg et al. 2007). We evaluate the performance of REM on yeast segregants, mouse inbred strains, and human rare variants data. The results demonstrate that REM not only returns the exact resampling-based P-values, but also runs orders of magnitude faster than alternative methods.
Materials and Methods
Problem formulation
We consider binary genotype data. The binary genotype may appear in haploid organisms, such as yeast, inbred diploid organisms, such as inbred mice, or rare variants where the genotype of a homozygous rare allele is unlikely to occur and is collapsed with a heterozygous genotype if it does occur. Without loss of generality, we denote the common and rare genotypes by 0 and 1, respectively.
The permutation test based on the maximum test statistic across all SNPs was originally described in Westfall and Young (1993) and applied to a genome scan in Churchill and Doerge (1994). This approach preserves the correlation structure of the SNPs and requires a much smaller number of permutations than the procedures that first compute pointwise P-values and then apply adjustments (Nettleton and Doerge 2000). The weighted bootstrap approach (McClurg et al. 2007) that accounts for population structure has also been proposed. We collectively refer to the permutation or bootstrap P-values as resampling-based P-values.
Let {X1, … , XN} be the SNPs and {Y1, … , YM} be the gene expression traits. Let (Xn, Ym) denote the test statistic between Xn (1 ≤ n ≤ N) and Ym (1 ≤ m ≤ M). Let be the maximum test statistic for Ym. Suppose that we resample the phenotype K times. For a resampled phenotype , we denote its maximum test statistic as . For k = 1, … , K, let
The resampling-based P-value of Ym is defined as
To calculate the resampling-based P-value of expression trait Ym, the computational problem is to determine, for every , whether its maximum test statistic is larger than . A brute force solution to this problem is that, for every we perform a complete scan of all SNPs to find the maximum test statistic and compare it to . This approach has been adopted in most existing exhaustive methods that calculate the exact resampling-based P-values. Next, we discuss how our algorithm, REM, can efficiently compute the solution without calculating the test statistics for all SNPs.
General idea of the proposed method
The key idea of the proposed algorithm is as follows. We partition SNPs into different groups on the basis of their allele frequencies. For each group of SNPs, we can find an upper bound on the test statistics. If the upper bound is lower than a certain threshold, there is no need to calculate the test statistics for this group of SNPs. The actual tests are performed only for the groups of SNPs whose upper bounds are larger than the threshold.
The groups of SNPs are organized into a two-layer indexing structure. Utilizing indexing structures for efficient computation has been a longstanding research focus of the database management and theoretical computation research communities. The idea of indexing SNPs and exploring the upper bound on test statistics was originally investigated in epistasis detection in a genome-wide association study (Zhang et al. 2009, 2010). In this article we apply this general methodology to the problem of multiple-testing correction in eQTL studies.
Next we first introduce the two-layer indexing structure that groups SNPs on the basis of allele frequencies. Then we discuss how to use this indexing structure to obtain the upper bound on the test statistics for each group of SNPs and thus dramatically reduce the number of test statistics to be calculated.
The indexing structure
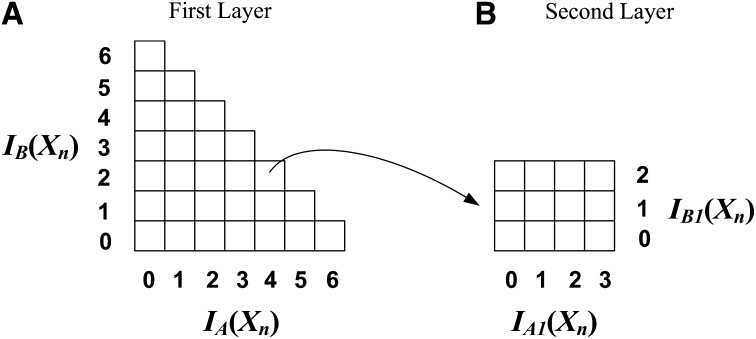
First layer:
Suppose that there are S individuals {I1, I2, … , IS} in the study. We partition the individuals into two subsets: IA = {I1, I2, … , I⌊S/2⌋} and IB = {I⌊S/2⌋+1, I⌊S/2⌋+2, … , IS}. Note that the partition of individuals is random and only done once for the entire data set.
For a SNP Xn, we use |IA(Xn)| and |IB(Xn)| to denote the numbers of rarer genotype (i.e., the number of 1’s) in partitions IA and IB, respectively. We can group the SNPs by their (|IA(Xn)|, |IB(Xn)|) values and index them in a two-dimensional (2D) array. The SNPs with the same (|IA(Xn)|, |IB(Xn)|) values will fall into the same entry in the array.
For example, suppose that there are S = 12 individuals in our study. We can partition them into two subsets, each having 6 individuals. Figure 1A shows the first layer of the 2D indexing structure. The SNPs having the same (|IA(Xn)|, |IB(Xn)|) value fall into the same entry in the 2D array. In this example, the possible values of |IA(Xn)| and |IB(Xn)| are {0, 1, 2, … , 6}. Recall that 1 denotes the rarer genotype, and thus the number of 1’s in any SNP is no greater than ⌊S/2⌋; i.e., |IA(Xn)| + |IB(Xn)| ≤ ⌊S/2⌋. Therefore, the indexing structure has entries only below the diagonal, as shown in Figure 1A.
Figure 1.
The two-layer indexing structure for a study of 12 individuals. (A) The first layer indexing structure. (B) The second layer indexing structure for a particular entry (4, 2) in the first layer indexing structure.
Later we show that the SNPs in the same entry [i.e., with the same (|IA(Xn)|, |IB(Xn)|) value] share a common upper bound on their test statistics.
Second layer:
Applying a similar idea, we can build a second layer indexing structure for each entry in the first layer. For a group of SNPs in the same entry in the first layer, we further partition individuals in IA into two subsets: IA1 = {I1, I2, … , I⌊S/4⌋} and IA2 = {I⌊S/4⌋+1, I⌊S/4⌋+2, … , I⌊S/2⌋}. Similarly, we partition IB into two subsets IB1 and IB2 of similar sizes. Note that these partitions are random and done only once for the entire data set. For any SNP Xn, we use |IA1(Xn)|, |IA2(Xn)|, |IB1(Xn)|, and |IB2(Xn)| to denote the number of 1’s of Xn in IA1, IA2, IB1, and IB2, respectively. The group of SNPs in the same entry in the first layer is further partitioned by its (|IA1(Xn)|, |IB1(Xn)|) values. The SNPs with the same (|IA1(Xn)|, |IB1(Xn)|) values fall in the same entry in the second layer indexing structure.
Following the previous example where there are 12 individuals, its first layer indexing structure is shown in Figure 1A. For a particular first layer entry (|IA(Xn)|, |IB(Xn)|) = (4, 2), its second layer indexing structure is shown in Figure 1B. It is easy to see that the maximum value that |IA1(Xn)| can take is min{|IA(Xn)|, ⌊S/4⌋}, and the maximum value that |IB1(Xn)| can take is min{|IB(Xn)|, ⌈S/4⌉}. This is reflected in the following example: the maximum value of |IA1(Xn)| is min{4, 3} = 3, and the maximum value of |IB1(Xn)| is min{2, 3} = 2.
Similar to the first layer, the SNPs in the same entry in the second layer also share a common upper bound on their test statistics. We will show that this bound is tighter than (or at least as tight as) the bound derived from the first layer.
Upper bound on the test statistics
In this subsection, we give a common upper bound on the test statistics for the SNPs in the same entry in the indexing structure. An intuitive explanation is as follows. For SNP Xn and a resampled phenotype , let represent the sum of the phenotype values of the individuals with rarer genotypes (i.e., when Xn = 1). It can be shown that many statistics are convex functions of (see File S1 for details). If we can determine the range of , we can easily derive an upper bound of the statistics. It can be shown that the SNPs in the same entry have the same range for and hence share a common upper bound.
Next, we first discuss how to obtain the upper bound for a SNP group in the first layer, and then we show that tighter bounds can be achieved for the groups in the second layer.
First layer:
Recall that when building the first layer indexing structure, we partition the individuals into two subsets, IA and IB. The indexing structure groups SNPs on the basis of their (|IA(Xn)|, |IB(Xn)|) values, i.e., the numbers of rarer genotypes in the two subsets of individuals. For a resampled phenotype vector , we use and to denote the phenotype values of the individuals in IA and IB, respectively.
Let represent the ordered phenotype values in and represent the ordered values in . For any SNP Xn in the first layer entry (|IA(Xn)|, |IB(Xn)|), we have that
| (1) |
where is the sum of the smallest |IA(Xn)| phenotype values of individuals in IA, and is the sum of the smallest |IB(Xn)| phenotype values of individuals in IB. Recall that |IA(Xn)| + |IB(Xn)| is equal to the number of rarer genotypes and is the sum of the phenotype values of individuals with rarer alleles. Clearly, the inequality holds. We use RL to represent the left-hand side of inequality (1).
Similarly we have that
| (2) |
We use RU to represent the right-hand side of inequality (2). For a given resampled phenotype vector , inequalities (1) and (2) give us the range of . That is, for any SNP Xn in the entry (|IA(Xn)|, |IB(Xn)|) of the first layer indexing structure, we have that .
The maximum value of any convex function is attained at the boundary of its convex domain (Boyd and Vandenberghe 2004). Therefore, for the SNPs in the entry (|IA(Xn)|, |IB(Xn)|), the upper bound on test statistics is attained when or .
Second layer:
Similarly, we can obtain an upper bound on the test statistics for the SNPs in an entry of the second layer. We partition the values in the resampled phenotype vector into four groups, each across individuals in IA1, IA2, IB1, and IB2, respectively. Let , , and represent the ordered phenotype values in the four groups. We have the following two inequalities:
| (3) |
| (4) |
Letting be the left (right)-hand side of inequality (3) [inequality (4)], then . The upper bound on the test statistics is attained when or . It is easy to show that and . Therefore, we can get a tighter (or at least an equally tight) upper bound by utilizing the second layer indexing.
The REM algorithm
Given the SNPs and gene expression traits, REM returns the significant traits whose resampling-based P-values are no greater than a user-specified threshold Pt. If Pt = 1, REM returns the resampling-based P-values for all expression traits. The number of permutations/bootstraps is an input parameter provided by the user. The pseudocode of the algorithm is outlined in File S1.
For every phenotype Ym, REM first scans all SNPs to find the maximum statistic . Then REM calculates a variable count, which records the number of resampled phenotypes whose maximum test statistics are greater than , as follows. For each , REM first checks the entries in the first layer of the indexing structure. It goes to the second layer only if the upper bound of a first layer entry is greater than . If the upper bound of the second layer entry is still greater than , REM will perform actual tests on the SNPs in the second layer entry; otherwise this group of SNPs will be skipped because their test statistics are guaranteed to be no greater than .
In addition to applying the upper bound to reduce the number of SNPs to be examined, there are two other strategies to further prune the search space. One is that, for a resampled phenotype vector , as long as we find any SNP whose test statistic is greater than we know its maximum test statistic will also be greater than Therefore, there is no need to scan the remaining SNPs. The other pruning strategy is based on the significance threshold Pt. For an expression trait Ym, once we have already examined enough resampled phenotype vectors such that its corrected P-value is greater than Pt, there is no need to examine the remaining resampled phenotype vectors. The detailed analysis of the time complexity of REM is given in File S1.
REM can be applied to various resampling methods. For example, in addition to the permutation and bootstrap methods (Westfall and Young 1993; Churchill and Doerge 1994; McClurg et al. 2007) that preserve the correlation structure of SNPs, a resampling method that preserves the correlation structure of the expression data was proposed in Breitling et al. (2008). When generating resampled phenotype vectors, this approach permutes the individual labels so that each individual is assigned the genotype of another random individual, while the expression profile of this individual is unchanged. When the mapping is repeated on these permuted data, the correlation structure of gene expression is maintained. These methods differ in how they generate the resampled phenotype vectors, i.e., line 4 in File S1, Algorithm S1. REM will find the exact resampling-based P-values for the resampling method adopted by the user.
Note that REM can be easily modified to identify multiple SNPs that are significantly associated with a gene expression trait. One naive approach is to apply REM to each of the top T SNPs that are significantly associated with the gene expression trait. Since we compare the test statistic of the t-th most significant association with the maximum test statistics from the resampled data, the estimate of the resampling-based P-value is conservative. However, as long as T is much smaller than the total number of SNPs, the bias of the estimate is small. A drawback of this naive approach is the computational burden increases linearly with T. An alternative, and more sophisticated approach is to directly calculate the desired percentile of the maximum test statistic. For example, suppose that the desired significance level is α = 0.05, and thus we aim to estimate the 95th percentile of the maximum test statistic from 1000 permutations/bootstraps. In other words, we want to find the 50th largest maximum test statistics among the 1000 maximum test statistics. We can first carry out 50 permutations/bootstraps and record the corresponding 50 maximum test statistics without pruning. Then we carry out the following permutation/bootstraps with pruning by keeping track of the largest 50 maximum test statistics and using the value of the 50th one as the threshold for pruning.
Results
We evaluate the accuracy and computational efficiency of REM and several representative existing methods. For approximation methods, we choose SimpleM (Gao et al. 2008) to represent the approaches of estimating the number of independent tests, and SLIDE (Han et al. 2009) to represent the approaches using the MVN framework. These two methods have been shown to be superior to other alternatives within their classes. We also study the performance of the method in Sun and Wright (2010), which we call GeoP. For exhaustive methods, we study the performance of PLINK (Purcell et al. 2007) and FastMap (Gatti et al. 2009). Note that these two methods are designed to speed up only permutation test but not bootstrapping. Other approaches, such as the approximation method RAT (Kimmel and Shamir 2006) and exhaustive methods PRESTO (Browning 2008) and PERMORY (Pahl and Schafer 2010), are applicable only to binary phenotypes and therefore are not applicable to eQTL mapping. We evaluate the performance of the selected methods on three eQTL data sets: inbred mouse strains, yeast segregants, and human rare variants.
Inbred mouse strains
We use the hypothalamus eQTL data of inbred mice from McClurg et al. (2007). There are 32 mouse strains in the data set. The total number of SNPs is 156,525. The missing values are imputed by the algorithm in Roberts et al. (2007). There are 36,182 probes on the gene expression array. Here we refer to the gene expression captured by each probe as an expression trait. Following the same filtering method as in Gatti et al. (2009), we dropped any expression trait whose expression values are all <200 or whose largest difference in expression across the 32 mouse strains is less than threefold. There are 3,672 expression traits left after filtering.
Accuracy evaluation:
Due to the population structure of these inbred mouse strains, some strains are more similar to each other than other strains, and thus a direct application of a permutation test is not appropriate (Fei et al. 2006; McClurg et al. 2007; Churchill and Doerge 2008). We adopt the weighted bootstrap approach proposed by McClurg et al. (2007) in REM to account for the population structure. We studied 10 expression traits, whose uncorrected P-values range from 1.9 × 10−13 to 2.2 × 10−6. We estimated the corrected P-values by 100 million bootstraps, treated them as the gold standard, and referred to them as the reference P-values. The reference P-values of these 10 expression traits vary from 0.00057 to 0.17. A method is considered accurate if its corrected P-values are close to the reference P-values.
We use 1 million weighted bootstraps to estimate the corrected P-values by REM. Because FastMap and PLINK cannot perform weighted bootstrapping, we apply FastMap and PLINK to these data using 1 million permutations to estimate the corrected P-values. The approximation methods also employ an implicit assumption that all samples are equally exchangeable, for example, in calculating the correlation matrix. For comparison purposes, we apply these approximation methods even though the exchangeability assumption is invalid in these data due to the population structure. We set the window size to be 100 and the number of resamplings to be 1 million for SLIDE. The number of resamplings is also set to be 1 million for GeoP. SimpleM is not a resampling-based method and we use its default setting. Following a similar approach as in Han et al. (2009; Pahl and Schafer 2010), we construct a 95% confidence interval to cover the sampling error of REM.
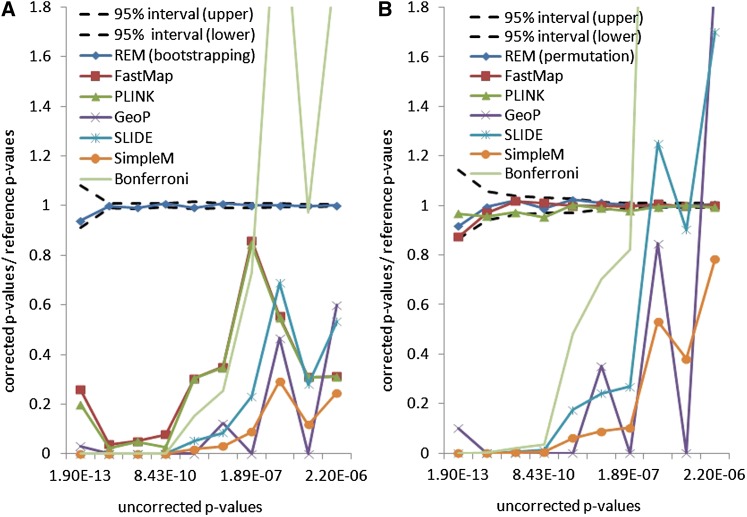
Figure 2A depicts the ratios between the corrected P-values and the reference P-values for the selected methods. A method is accurate if its ratio is close to 1. As shown in Figure 2A, there is discrepancy between the bootstrap and the permutation test (as implemented in PLINK and FastMap). This reflects the effect of the population structure of the inbred mouse strains. The permutation test is anticonservative compared to the weighted bootstrap method. The approximation methods, which are designed to estimate the permutation P-values, are shown to be anticonservative compared to both the bootstrapping and the permutation test. This indicates that these approximation methods are not replacements for exhaustive permutation tests. The corrected P-values from the widely used Bonferroni correction are also shown in Figure 2A. It can be seen that the Bonferroni correction can be either conservative or anticonservative for different uncorrected P-values. Thus it is not an ideal approach for multiple-testing correction in eQTL studies.
Figure 2.
Accuracy evaluation of selected methods on real gene expression traits in an inbred mouse data set. (Each line represents the ratio between the corrected P-values and the reference P-values for a method. The reference P-values are obtained using 100 million resamplings. An accurate method should yield a ratio of 1. In A, the reference P-values are estimated by a weighted bootstrap method. In B, the reference P-values are estimated by a permutation test.)
For comparison purposes, in addition to the weighted bootstrap method, we also apply the permutation test using REM. The basic experimental setting is similar to that in the weighted bootstrap method. We estimate the reference permutation P-values of the 10 expression traits by 100 million permutations. The reference permutation P-values range from 0.00018 to 0.053. For the three exact methods, REM, FastMap, and PLINK, we use 1 million permutations to estimate the corrected P-values.
Figure 2B shows the ratio between the corrected P-values and the reference permutation P-values. The corrected P-values generated by the three exact methods fall in the confidence region of the reference P-values. In contrast, the three approximation methods, GeoP, SLIDE, and SimpleM, do not estimate the permutation P-values accurately. Thus these approximation methods cannot replace the exact permutation test. Further comparison between the exhaustive permutation test and the approximation methods will be conducted in yeast segregants and human rare variants data.
Using the genotype data of these 32 mouse inbred strains, we also study the accuracy of the selected methods for three synthetic phenotypes whose values follow standard normal, exponential, and uniform distributions. The approximation methods are shown to be anticonservative. Please see File S1 for further details.
Computational efficiency evaluation:
We evaluate the computational efficiency of the three selected exact resampling methods, REM, FastMap, and PLINK. The approximation methods are usually very fast but do not provide accurate P-value correction. They are not considered in the computational efficiency evaluation. All experiments in this subsection are performed on a single CPU of a 2.6-GHz PC with 8 G memory running the Linux operating system.
Table 1 shows the runtime of the three methods when applied to the entire eQTL data set with 3600 phenotypes. As can be seen, with 100,000 resamplings, REM can finish within 1 day if we set the threshold to be 1. If we find only the expression traits that have significant associations, REM can finish within a few hours. FastMap and PLINK will take a much longer time.
Table 1. Runtime of REM, FastMap, and PLINK on the entire inbred mouse hypothalamus eQTL data.
| No. resamplings | REM (Pt = 0.01) | REM (Pt = 0.05) | REM (Pt = 1) | FastMap | PLINK |
|---|---|---|---|---|---|
| 1,000 | 2.3 min | 4.6 min | 13 min | 1.2 hr | 21 days |
| 10,000 | 0.3 hr | 0.7 hr | 2 hr | 10 hr | 211 days |
| 100,000 | 2.8 hr | 6.7 hr | 0.9 day | 4 days | 5.8 yr |
| 1 million | 1.2 days | 2.8 days | 8.6 days | 40 days | 58 yr |
The data set contains 150,000 SNPs and 3600 gene expression traits over 32 strains. The runtimes >10 days are estimated from smaller-scale experiments.
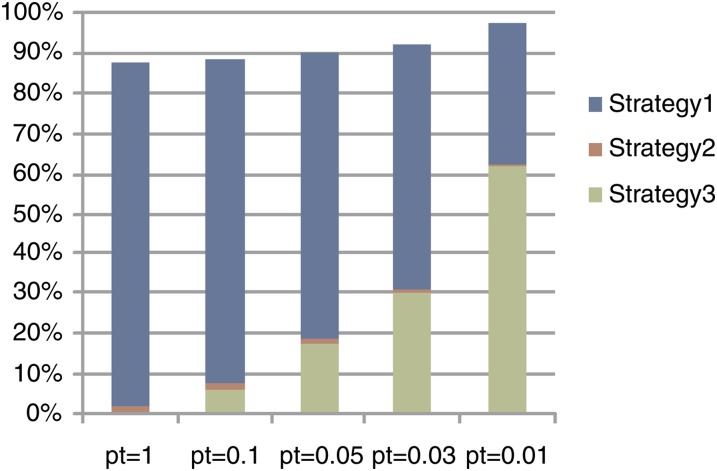
Figure 3 shows the percentage of the SNPs that are pruned by REM under different significance levels. The pruned SNPs are the ones on which we do not perform any test. If we calculate the corrected P-values for all expression traits, >80% of SNPs are pruned. In most cases, we are interested only in the traits whose corrected P-values are less than a certain threshold. At a significance level of 0.01, >97% of SNPs are pruned, which means that we need to calculate test statistics only for <3% of SNPs. This dramatically reduced search space explains the improved computational efficiency of REM. Recall that REM has three pruning strategies to reduce the search space. They are (1) pruning by the upper bound, (2) pruning by the maximum statistic (lines 13–16 in File S1, Algorithm S1), and (3) pruning by the significance threshold (lines 23–25 in File S1, Algorithm S1). Figure 3 also shows the breakdown of which pruning strategy is used to prune the search space. Strategy 2 provides 0.4–2.1% pruning ratios across different significance levels. When the significance level is set to be 1, the pruning strategy 1 alone prunes >80% of the search space. Strategy 3 plays a more important role for smaller significance levels. This is reasonable since, for smaller significance levels, more phenotypes will not be examined once we know they will not become significant. Please refer to File S1 for more results on computational efficiency evaluation.
Figure 3.
The pruning ratio (percentage of the SNPs that are pruned without performing actual tests) of REM for different significance thresholds when using an inbred mouse data set. This also provides the breakdown of the effects of the three pruning strategies used in REM. See text for more details.
Yeast segregants
The original yeast data set consists of 112 yeast segregants generated from two parent strains (Brem and Kruglyak 2005; Brem et al. 2005). Expression levels of 6229 genes and genotypes of 2956 SNPs were measured in each of the segregants. After removing SNPs with >10% missing values and combining SNPs with the same genotype profiles, there are 1017 distinct genotype profiles.
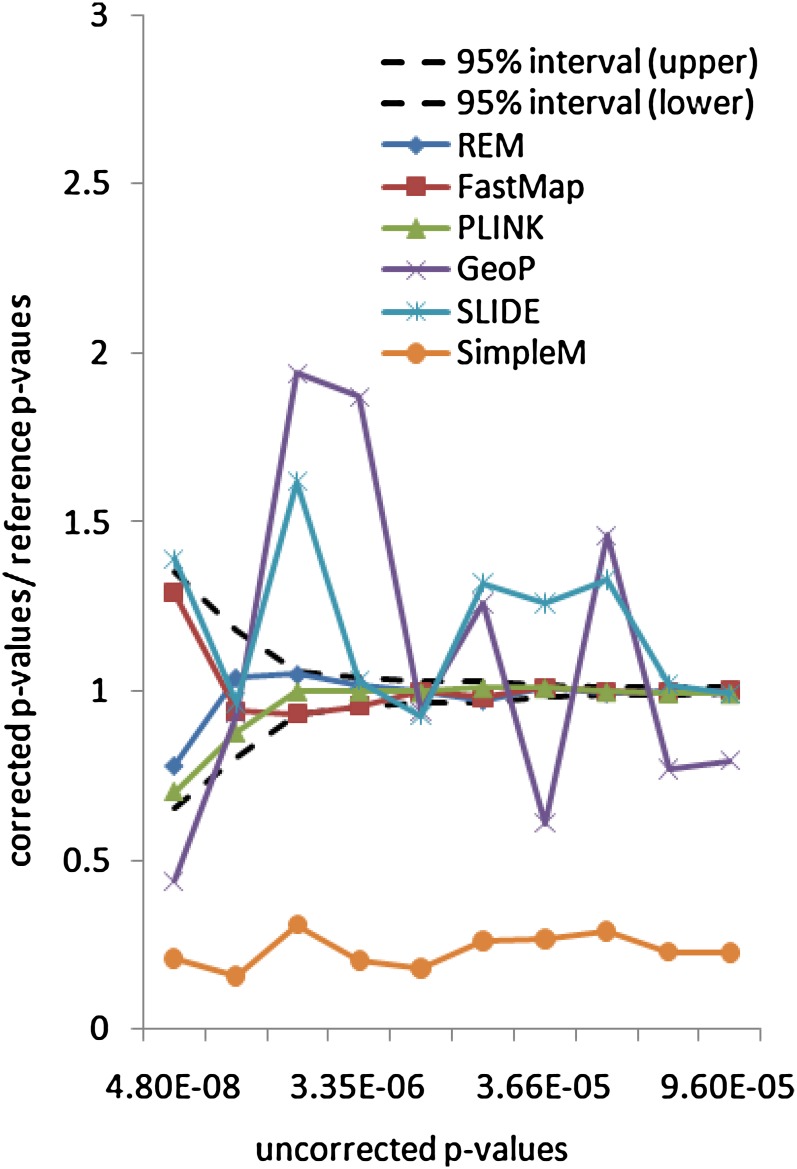
Since this data set only has a very small number of SNPs, the exact methods FastMap and REM can both finish within a few hours for 1 million permutations. To evaluate the accuracy of the selected methods, we use the same ratio measurement as in the inbred mouse data set. Figure 4 shows the ratios between the 10 corrected P-values and the reference P-values (calculated by 100 million permutations) for the selected methods. The uncorrected P-values of the gene expression traits range from 4.8 × 10−8 to 9.6 × 10−5. After correction, the P-values range from 0.000016 to 0.046. As expected, the corrected P-values of the exact methods all fall into the confidence region. Both GeoP and SLIDE provide overall unbiased permutation P-value estimates. This is because the underlying correlation structure assumption of these approximation methods is appropriate in this yeast data set, as illustrated in File S1, Figure S1.
Figure 4.
Accuracy evaluation of selected methods on the yeast data set.
Human rare variants
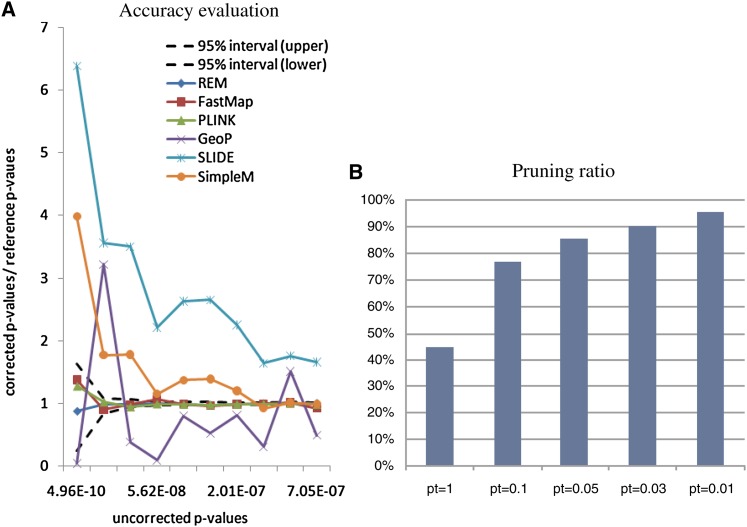
Individual rare variants provide little power for association studies because of their low minor allele frequencies. A commonly used approach is to collapse nearby rare variants and use these collapsed rare variants for association studies (Li and Leal 2008). In this study, we generated a data set of collapsed rare variants on the basis of genotype data from 65 HapMap Yoruba in Ibadan (YRI) samples (Frazer et al. 2007). Specifically, the genotype data of >2 million SNPs were downloaded from the HapMap Web site. We first chose the markers with minor allele frequency <0.05, and no missing values, and then collapsed them within a moving window of 50 kb that shifts 10 kb each time. The final data set includes 143,000 collapsed SNPs. We downloaded expression data of these 65 samples (measured by RNA-seq) from the Pritchard laboratory’s Web site (http://eqtl.uchicago.edu/) (Pickrell et al. 2010).
Figure 5A shows the ratios between the corrected P-values and the reference P-values for the selected methods. The uncorrected P-values of the gene expression traits range from 4.9 × 10−10 to 7.1 × 10−7. After correction, the P-values range from 8 × 10−6 to 0.046. The corrected P-values of the exact methods still fall into the confidence region. For the approximation methods, SLIDE and SimpleM are conservative for small P-values but become more accurate for larger ones. GeoP estimates are overall unbiased but with large fluctuations.
Figure 5.
Accuracy evaluation and pruning ratio on the human rare variant data set.
Figure 5B shows the percentage of the SNPs that are pruned by REM under different significance levels. Note that, at significance level 0.01, >95% SNPs are pruned, and only <5% SNPs need to be examined for their tests values. Compared to FastMap, REM speeds up the process by 20–30 times.
Applying REM to a large sample study through meta-analysis
The computational efficiency of REM comes from the indexing structure. For each entry in the index structure, REM performs a single computation to calculate a bound of the test statistics of all SNPs in this entry. If the number of samples is large, the number of entries in the indexing structure will also be large. This may impair the computational efficiency of the algorithm. This problem may be alleviated by meta-analysis (Munafo and Flint 2004). In contrast to direct analysis of pooled individual-level data, meta-analysis combines the summary statistics from different studies.
We apply REM to a large sample data set (with 1000 samples) through meta-analysis. The samples are partitioned into groups of equal size. For each group, we apply REM to calculate the resampling-based P-values for 1000 simulated phenotypes. For each phenotype, a combined P-value is computed by applying Fisher’s method to the group P-values (Fisher 1925). The combined P-values and original resampling-based P-values (when using all samples) are almost perfectly correlated. Specifically, the correlations are 0.99, 0.98, and 0.96 when we partition the samples into 2, 5, and 10 groups, respectively. Therefore, for large sample studies, we can apply REM to groups and combine the P-values by using meta-analysis. The combined P-values are robust estimations of the original resampling-based P-values. More detailed discussion on applying REM to a large sample study through meta-analysis can be found in File S1.
Discussion
The resampling-based test is widely used to address the multiple-testing correction problem in genetic association studies. Its main disadvantage is the intensive computational burden. In eQTL studies, the computational problem becomes more severe since one needs to correct the P-values for tens of thousands of gene expression traits. In this article, we present a rapid and robust algorithm, REM, that dramatically speeds up the process of the exact resampling-based test. It builds a two-layer indexing structure that groups SNPs by their genotypes. By estimating the upper bound of the test statistics for all SNPs within one group, REM prunes away most of the SNPs. Moreover, since usually we are interested only in the expression traits whose corrected P-values are less than a certain significance level, REM can further improve the computational efficiency by filtering out the insignificant expression traits in early stages. Most importantly, REM guarantees that we find the exact resampling-based P-values even if it performs the actual tests on only a small number of SNPs. REM can be applied to a wide range of resampling procedures. It provides the flexibility to the user to determine the appropriate resampling strategy for the data set under consideration. We use three eQTL data sets to evaluate the performances of several selected algorithms and demonstrate that REM produces accurate estimates of resampling-based P-values with much less computational cost than other alternatives.
We have shown that the performances of approximation methods vary for different data sets with no method being consistently superior to other methods. The approximation methods achieve higher accuracy on the yeast data set than the other two data sets since the correlation structure in the yeast data set matches the assumptions of the approximation methods. However, in inbred mice and human rare variants data sets, nearby markers do not have higher correlations and the heat maps do not show clear banding correlation structure among SNPs (File S1, Figure S1). Therefore, the correlation assumption of the approximation methods is not valid, which leads to their poor performance.
Association studies of the low-frequency/rare variants have recently attracted much research attention (Bodmer and Bonilla 2008; Manolio et al. 2009). With the advance of sequencing techniques, we expect in the near future that rare variants will be used in most association studies. Multiple-testing correction for rare variants association is of great research interest. Since a single rare variant has little power to detect an association signal, a common approach is to collapse the rare variants at a locus so that 1/0 indicates the presence/absence of any rare variant (Li and Leal 2008; Morris and Zegginic 2010). The collapsed rare variants often do not have strong correlations among nearby loci, which violates the assumption underlying most approximation methods for permutation P-value estimation. To the best of our knowledge, this article is the first effort to address the multiple-testing correction problems of rare variant association and our REM algorithm provides an accurate and computationally efficient solution for this problem.
There is room to improve the REM algorithm. In this article, we have focused on the cases where genotype data are binary. The general principle used in REM can also be applied to the situation where one marker may have three possible genotypes, which is among our future research directions.
In summary, the REM algorithm provides an efficient solution to calculate the exact resampling-based P-values for a variety of statistical tests in eQTL studies. It has been demonstrated to be much faster than recently developed methods. The software is implemented in C++ and is publicly available at http://csbio.unc.edu/eQTL. The algorithm can be easily parallelized, for example, by parallelizing the computation for each gene expression trait.
Supplementary Material
Acknowledgments
This work was partially supported by Environmental Protection Agency grant RD-83382501; National Science Foundation awards IIS0448392 and IIS0812464; and National Institutes of Health awards U01CA105417, U01CA134240, and MH090338.
Footnotes
Communicating editor: G. A. Churchill
Literature Cited
- Barrett J., Fry B., Maller J., Daly M., 2005. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 21(2): 263–265 [DOI] [PubMed] [Google Scholar]
- Benjamini Y., Hochberg Y., 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. B 57(1): 289–300 [Google Scholar]
- Bochner B. R., 2003. New technologies to assess genotype–phenotype relationships. Nat. Rev. Genet. 4: 309–314 [DOI] [PubMed] [Google Scholar]
- Bodmer W., Bonilla C., 2008. Common and rare variants in multifactorial susceptibility to common diseases. Nat. Genet. 40(6): 695–701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd S., Vandenberghe L., 2004. Convex Optimization. Cambridge University Press, Cambridge, UK/London/New York [Google Scholar]
- Breitling R., Li Y., Tesson B. M., Fu J., Wu C., et al. , 2008. Genetical genomics: spotlight on QTL hotspots. PLoS Genet. 4(10): e1000232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brem R. B., Kruglyak L., 2005. The landscape of genetic complexity across 5,700 gene expression traits in yeast. Proc. Natl. Acad. Sci. USA 102(5): 1572–1577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brem R. B., Storey J. D., Whittle J., Kruglyak L., 2005. Genetic interactions between polymorphisms that affect gene expression in yeast. Nature 436(7051): 701–703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browning B. L., 2008. PRESTO: rapid calculation of order statistic distributions and multiple-testing adjusted P-values via permutation for one and two-stage genetic association studies. BMC Bioinformatics 9: 309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheverud J. M., 2001. A simple correction for multiple comparisons in interval mapping genome scans. Heredity 87: 52–58 [DOI] [PubMed] [Google Scholar]
- Churchill G., Doerge R. W., 2008. Naive application of permutation testing leads to inflated type I error rates. Genetics 178: 609–610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchill G. A., Doerge R. W., 1994. Empirical threshold values for quantitative trait mapping. Genetics 138: 963–971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conneely K., Boehnke M., 2007. So many correlated tests, so little time! Rapid adjustment of P values for multiple correlated tests. Am. J. Hum. Genet. 81(6): 1158–1168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fei Z., Xu Z., Vision T., 2006. Assessing the significance of quantitative trait loci in replicable mapping populations. Genetics 174: 1063–1068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher R., 1925. Statistical Methods for Research Worker. Oliver & Boyd, Edinburgh [Google Scholar]
- Frazer K., Ballinger D., Cox D., Hinds D., Stuve L., et al. , 2007. A second generation human haplotype map of over 3.1 million SNPs. Nature 449(7164): 851–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X., Starmer J., Martin E., 2008. A multiple testing correction method for genetic association studies using correlated single nucleotide polymorphisms. Genet. Epidemiol. 32(4): 361–369 [DOI] [PubMed] [Google Scholar]
- Gatti D. M., Shabalin A. A., Lam T.-C., Wright F. A., Rusyn I., et al. , 2009. FastMap: Fast eQTL mapping in homozygous populations. Bioinformatics 25(4): 482–489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Good P., 2005. Permutation, Parametric and Bootstrap Tests of Hypotheses. Springer-Verlag, New York [Google Scholar]
- Han B., Kang H., Eskin E., 2009. Rapid and accurate multiple testing correction and power estimation for millions of correlated markers. PLoS Genet. 5(4): e1000456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendziorski C., Wang P., 2006. A review of statistical methods for expression quantitative trait loci mapping. Mamm. Genome 17(6): 509–517 [DOI] [PubMed] [Google Scholar]
- Kimmel G., Shamir R., 2006. A fast method for computing high-significance disease association in large population-based studies. Am. J. Hum. Genet. 79(3): 481–492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B., Leal S., 2008. Methods for detecting associations with rare variants for common diseases: application to analysis of sequence data. Am. J. Hum. Genet. 83(3): 311–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Ji L., 2005. Adjusting multiple testing in multilocus analyses using the eigenvalues of a correlation matrix. Heredity 95(3): 221–227 [DOI] [PubMed] [Google Scholar]
- Lin D., 2005. An efficient Monte Carlo approach to assessing statistical significance in genomic studies. Bioinformatics 21(6): 781–787 [DOI] [PubMed] [Google Scholar]
- Manolio T., Collins F., Cox N., Goldstein D., Hindorff L., et al. , 2009. Finding the missing heritability of complex diseases. Nat. Genet. 461(7265): 747–753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClurg P., Janes J., Wu C., Delano D., Walker J., et al. , 2007. Genomewide association analysis in diverse inbred mice: power and population structure. Genetics 176: 675–683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaelson J., Loguercio S., Beyer A., 2009. Detection and interpretation of expression quantitative trait loci (eQTL). Methods 48(3): 265–276 [DOI] [PubMed] [Google Scholar]
- Morris A., Zegginic E., 2010. An evaluation of statistical approaches to rare variant analysis in genetic association studies. Genet. Epidemiol. 34(2): 188–193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moskvina V., Schmidt K., 2008. On multiple-testing correction in genome-wide association studies. Genet. Epidemiol. 32(6): 567–573 [DOI] [PubMed] [Google Scholar]
- Munafo M., Flint J., 2004. Meta-analysis of genetic association studies. Trends Genet. 20(9): 439–444 [DOI] [PubMed] [Google Scholar]
- Nettleton D., Doerge R. W., 2000. Accounting for variability in the use of permutation testing to detect quantitative trait loci. Biometrics 56(1): 52–58 [DOI] [PubMed] [Google Scholar]
- Nyholt D. R., 2004. A simple correction for multiple testing for single-nucleotide polymorphisms in linkage disequilibrium with each other. Am. J. Hum. Genet. 74(4): 765–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pahl R., Schafer H., 2010. PERMORY: an LD-exploiting permutation test algorithm for powerful genome-wide association testing. Bioinformatics 26(17): 2093–2100 [DOI] [PubMed] [Google Scholar]
- Pe’er I., Yelensky R., Altshuler D., Daly M., 2008. Estimation of the multiple testing burden for genomewide association studies of nearly all common variants. Genet. Epidemiol. 32(4): 381–385 [DOI] [PubMed] [Google Scholar]
- Pickrell J., Marioni J., Pai A., Degner J., Engelhardt B., et al. , 2010. Understanding mechanisms underlying human gene expression variation with RNA sequencing. Nature 464(7289): 768–772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M., et al. , 2007. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81(3): 559–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts A., McMillan L., Wang W., Parker J., Rusyn I., et al. , 2007. Inferring missing genotypes in large SNP panels using fast nearest-neighbor searches over sliding windows. Bioinformatics 23(13): i401–i407 [DOI] [PubMed] [Google Scholar]
- Rockman M. V., Kruglyak L., 2006. Genetics of global gene expression. Nat. Rev. Genet. 7: 862–872 [DOI] [PubMed] [Google Scholar]
- Salyakina D., Seaman S. R., Browning B. L., Dudbridge F., Müller-Myhsok B., 2005. Evaluation of Nyholt’s procedure for multiple testing correction. Hum. Hered. 60: 19–25 [DOI] [PubMed] [Google Scholar]
- Seaman S. R., Müller-Myhsok B., 2005. Rapid simulation of P values for product methods and multiple-testing adjustment in association studies. Am. J. Hum. Genet. 76(6): 399–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storey J. D., 2003. The positive false discovery rate: a Bayesian interpretation and the q-value. Ann. Stat. 31(6): 2013–2035 [Google Scholar]
- Sun W., Wright F. A., 2010. A geometric interpretation of the permutation p-value and its application in eQTL studies. Ann. Appl. Stat. 4(2): 1014–1033 [Google Scholar]
- Westfall P. H., Young S. S., 1993. Resampling-Based Multiple Testing: Examples and Methods for P-Value Adjustment. Wiley, New York [Google Scholar]
- Zhang X., Pan F., Wang W., 2009. Efficient algorithms for genome-wide association study. ACM Trans Knowl Discov Data 3(4): 19 [Google Scholar]
- Zhang X., Pan F., Xie Y., Zou F., Wang W., 2010. COE: a general approach for efficient genome-wide two-locus epistasis test in disease association study. J. Comput. Biol. 17(3): 401–415 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.