Abstract
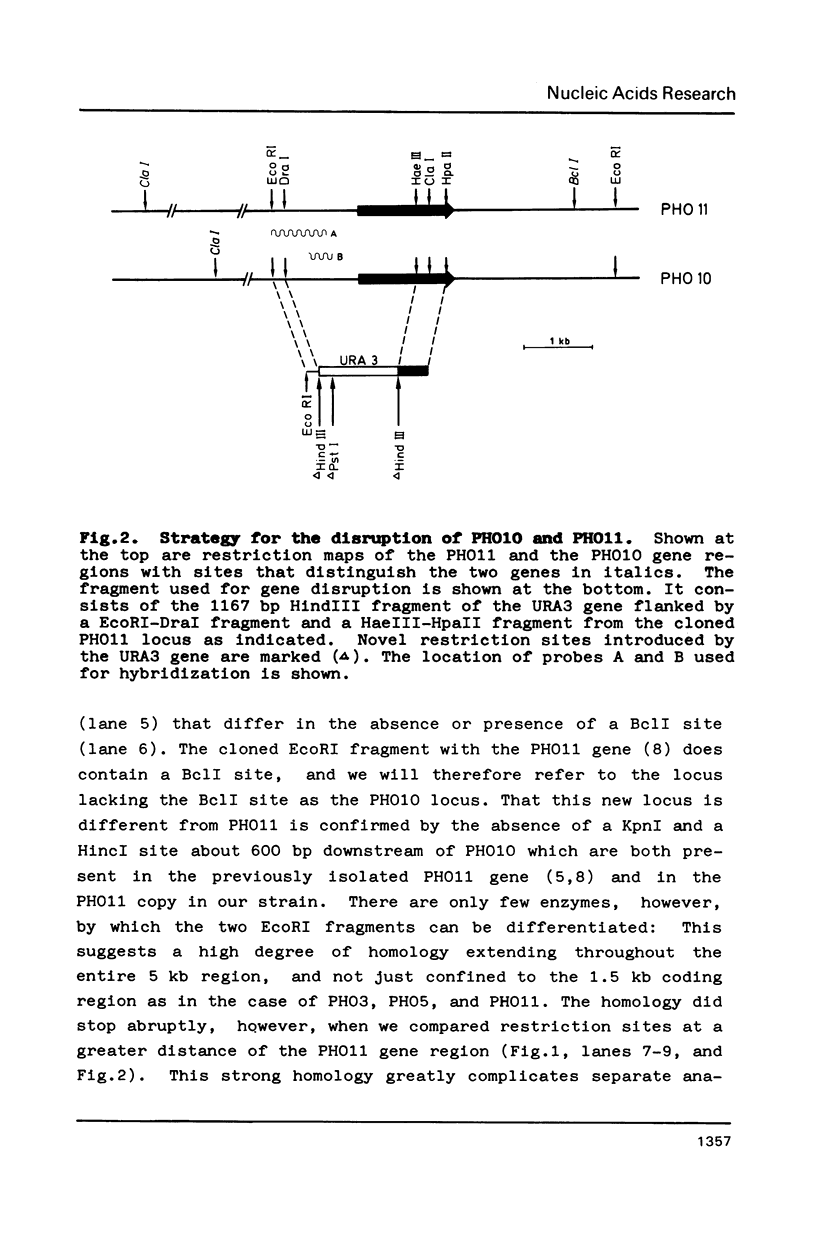
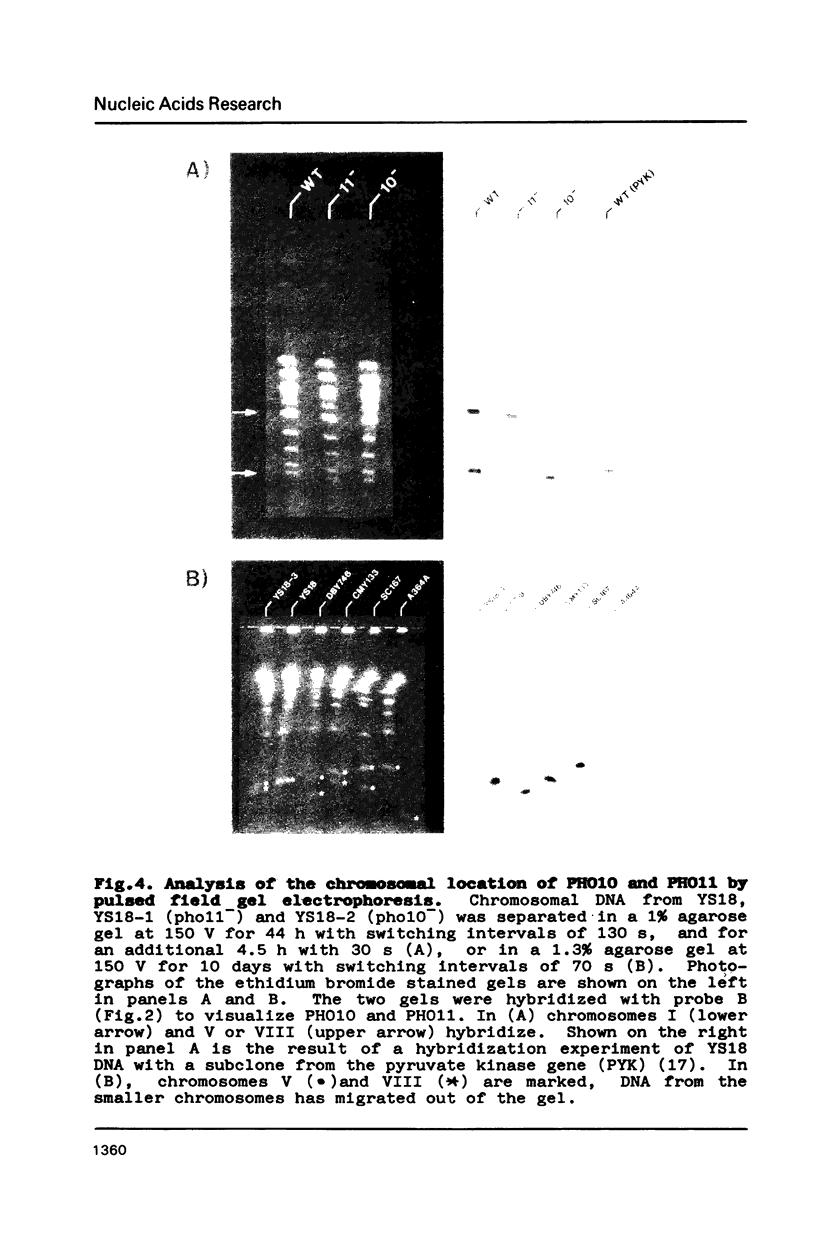
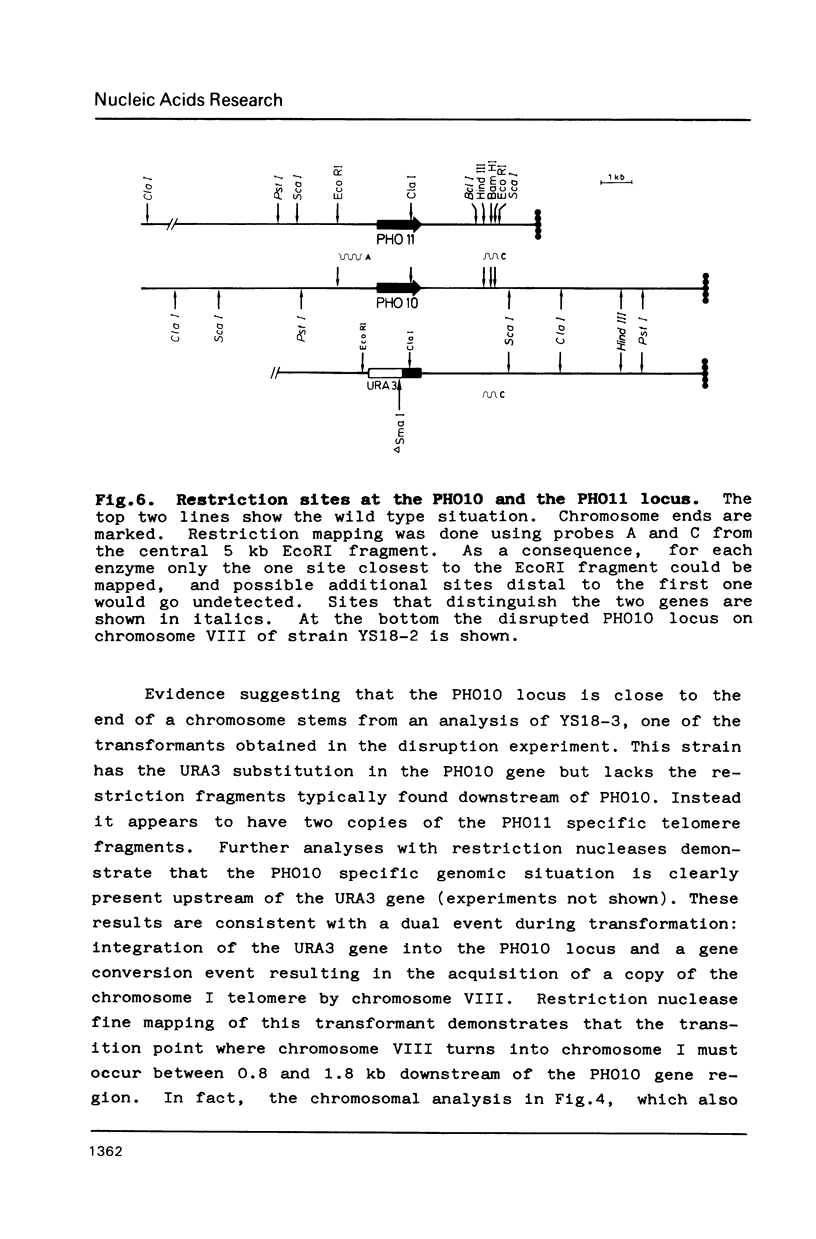
Of the three regulated acid phosphatase genes in S. cerevisiae (PHO5, PHO10 and PHO11) two have previously been cloned (PHO5 and PHO11). We have now identified PHO10 and show by restriction mapping that it is highly homologous to PHO11. This homology includes not only the coding sequence but also a stretch of about 2 kb upstream and 2.2 kb downstream of the genes. Analysis of strains in which either gene had been disrupted shows that the two genes are located at the telomeres of two different chromosomes. PHO10 3.6 kb from the end of a chromosome I. This makes PHO11 the gene closest to the end of a chromosome that has been physically mapped so far in S. cerevisiae. The organization of the two genes varies strongly from strain to strain consistent with a high incidence of telomere rearrangement. In one of twenty transformants examined a conversion event could be directly demonstrated that resulted in a chromosome VIII which had acquired a copy of the telomere from chromosome I.
Full text
PDF













Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Almer A., Hörz W. Nuclease hypersensitive regions with adjacent positioned nucleosomes mark the gene boundaries of the PHO5/PHO3 locus in yeast. EMBO J. 1986 Oct;5(10):2681–2687. doi: 10.1002/j.1460-2075.1986.tb04551.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almer A., Rudolph H., Hinnen A., Hörz W. Removal of positioned nucleosomes from the yeast PHO5 promoter upon PHO5 induction releases additional upstream activating DNA elements. EMBO J. 1986 Oct;5(10):2689–2696. doi: 10.1002/j.1460-2075.1986.tb04552.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen N., Thill G. P., Kramer R. A. RNA and homology mapping of two DNA fragments with repressible acid phosphatase genes from Saccharomyces cerevisiae. Mol Cell Biol. 1983 Apr;3(4):562–569. doi: 10.1128/mcb.3.4.562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajwa W., Meyhack B., Rudolph H., Schweingruber A. M., Hinnen A. Structural analysis of the two tandemly repeated acid phosphatase genes in yeast. Nucleic Acids Res. 1984 Oct 25;12(20):7721–7739. doi: 10.1093/nar/12.20.7721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackburn E. H. The molecular structure of centromeres and telomeres. Annu Rev Biochem. 1984;53:163–194. doi: 10.1146/annurev.bi.53.070184.001115. [DOI] [PubMed] [Google Scholar]
- Burke R. L., Tekamp-Olson P., Najarian R. The isolation, characterization, and sequence of the pyruvate kinase gene of Saccharomyces cerevisiae. J Biol Chem. 1983 Feb 25;258(4):2193–2201. [PubMed] [Google Scholar]
- Carle G. F., Olson M. V. An electrophoretic karyotype for yeast. Proc Natl Acad Sci U S A. 1985 Jun;82(11):3756–3760. doi: 10.1073/pnas.82.11.3756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson M., Celenza J. L., Eng F. J. Evolution of the dispersed SUC gene family of Saccharomyces by rearrangements of chromosome telomeres. Mol Cell Biol. 1985 Nov;5(11):2894–2902. doi: 10.1128/mcb.5.11.2894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan C. S., Tye B. K. Organization of DNA sequences and replication origins at yeast telomeres. Cell. 1983 Jun;33(2):563–573. doi: 10.1016/0092-8674(83)90437-3. [DOI] [PubMed] [Google Scholar]
- Fogel S., Welch J. W. Tandem gene amplification mediates copper resistance in yeast. Proc Natl Acad Sci U S A. 1982 Sep;79(17):5342–5346. doi: 10.1073/pnas.79.17.5342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horowitz H., Thorburn P., Haber J. E. Rearrangements of highly polymorphic regions near telomeres of Saccharomyces cerevisiae. Mol Cell Biol. 1984 Nov;4(11):2509–2517. doi: 10.1128/mcb.4.11.2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito H., Fukuda Y., Murata K., Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983 Jan;153(1):163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer R. A., Andersen N. Isolation of yeast genes with mRNA levels controlled by phosphate concentration. Proc Natl Acad Sci U S A. 1980 Nov;77(11):6541–6545. doi: 10.1073/pnas.77.11.6541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemire J. M., Willcocks T., Halvorson H. O., Bostian K. A. Regulation of repressible acid phosphatase gene transcription in Saccharomyces cerevisiae. Mol Cell Biol. 1985 Aug;5(8):2131–2141. doi: 10.1128/mcb.5.8.2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyhack B., Bajwa W., Rudolph H., Hinnen A. Two yeast acid phosphatase structural genes are the result of a tandem duplication and show different degrees of homology in their promoter and coding sequences. EMBO J. 1982;1(6):675–680. doi: 10.1002/j.1460-2075.1982.tb01229.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortimer R. K., Schild D. Genetic map of Saccharomyces cerevisiae, edition 9. Microbiol Rev. 1985 Sep;49(3):181–213. doi: 10.1128/mr.49.3.181-213.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers D. T., Lemire J. M., Bostian K. A. Acid phosphatase polypeptides in Saccharomyces cerevisiae are encoded by a differentially regulated multigene family. Proc Natl Acad Sci U S A. 1982 Apr;79(7):2157–2161. doi: 10.1073/pnas.79.7.2157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothstein R. J. One-step gene disruption in yeast. Methods Enzymol. 1983;101:202–211. doi: 10.1016/0076-6879(83)01015-0. [DOI] [PubMed] [Google Scholar]
- Schwartz D. C., Cantor C. R. Separation of yeast chromosome-sized DNAs by pulsed field gradient gel electrophoresis. Cell. 1984 May;37(1):67–75. doi: 10.1016/0092-8674(84)90301-5. [DOI] [PubMed] [Google Scholar]
- Sengstag C., Hinnen A. The sequence of the Saccharomyces cerevisiae gene PHO2 codes for a regulatory protein with unusual aminoacid composition. Nucleic Acids Res. 1987 Jan 12;15(1):233–246. doi: 10.1093/nar/15.1.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shampay J., Szostak J. W., Blackburn E. H. DNA sequences of telomeres maintained in yeast. Nature. 1984 Jul 12;310(5973):154–157. doi: 10.1038/310154a0. [DOI] [PubMed] [Google Scholar]
- Struhl K., Stinchcomb D. T., Scherer S., Davis R. W. High-frequency transformation of yeast: autonomous replication of hybrid DNA molecules. Proc Natl Acad Sci U S A. 1979 Mar;76(3):1035–1039. doi: 10.1073/pnas.76.3.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakian V. A., Blanton H. M. Distribution of telomere-associated sequences on natural chromosomes in Saccharomyces cerevisiae. Mol Cell Biol. 1988 May;8(5):2257–2260. doi: 10.1128/mcb.8.5.2257. [DOI] [PMC free article] [PubMed] [Google Scholar]