Abstract
PALB2/FANCN is mutated in breast and pancreatic cancers and Fanconi anemia (FA). It controls the intranuclear localization, stability, and DNA repair function of BRCA2 and links BRCA1 and BRCA2 in DNA homologous recombination repair and breast cancer suppression. Here, we show that PALB2 directly interacts with KEAP1, an oxidative stress sensor that binds and represses the master antioxidant transcription factor NRF2. PALB2 shares with NRF2 a highly conserved ETGE-type KEAP1 binding motif and can effectively compete with NRF2 for KEAP1 binding. PALB2 promotes NRF2 accumulation and function in the nucleus and lowers the cellular reactive oxygen species (ROS) level. In addition, PALB2 also regulates the rate of NRF2 export from the nucleus following induction. Our findings identify PALB2 as a regulator of cellular redox homeostasis and provide a new link between oxidative stress and the development of cancer and FA.
INTRODUCTION
The two major high-penetrance breast cancer susceptibility genes BRCA1 and BRCA2 encode very large proteins with a critical function in homologous recombinational repair (HRR) of DNA double-strand breaks (15). PALB2 was discovered as a major BRCA2 binding partner that controls its intranuclear localization, stability, recombinational repair, and DNA damage checkpoint functions (38). Immediately after its discovery, germ line truncating mutations in PALB2 were identified in familial breast cancer (4, 22, 31) and the N subtype of Fanconi anemia (FA-N) (23, 37). Later, PALB2 was also found to be mutated in familial pancreatic cancer, being the second most highly mutated pancreatic susceptibility gene after BRCA2 (7, 25). Furthermore, hypermethylation of the PALB2 promoter occurs in a significant fraction (∼7%) of both sporadic and familial breast/ovarian cancer cases (21). To date, dozens of truncating PALB2 mutations have been identified in cancer families around the world, causing moderate to very high risks of breast cancer (1, 26, 30). Finally, certain single nucleotide polymorphisms (SNPs) in the gene have been suggested to confer an increased risk of breast cancer (2).
The PALB2 protein contains a coiled-coil domain at the N terminus and a series of WD repeats at its C terminus (Fig. 1A). The WD repeats together form a β-propeller structure that directly binds BRCA2 (18), and the N-terminal coiled-coil motif was later found to directly bind BRCA1 (28, 41, 42). Approximately 50% of PALB2 and 50% of BRCA2 are associated with each other in the cell with high affinity (28, 38). The stoichiometry of PALB2-BRCA1 binding appears to be much lower (28, 42). However, the interaction has proved crucial for HRR as several mutations generated in each protein that abrogate the interaction have been shown to greatly compromise repair efficiency (28, 41, 42). Furthermore, multiple naturally occurring, patient-derived missense variants that disrupt PALB2 binding have been identified in both BRCA1 and BRCA2, and all of them have been found to abrogate HRR (28, 38). Thus, PALB2 links BRCA1 and BRCA2 in HRR and breast cancer suppression (30).
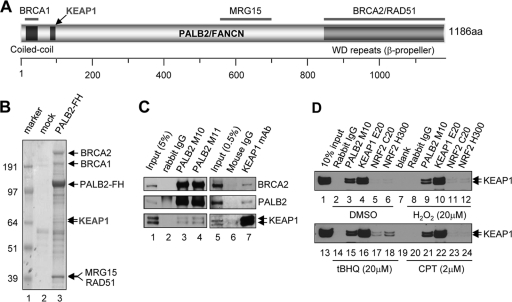
Fig 1.
PALB2 interacts with KEAP1. (A) Domain structure and known binding partners of PALB2. (B) Tandem affinity purification of PALB2 from a HeLa S3 cell line stably expressing a FLAG-HA-double-tagged PALB2 protein (PALB2-FH). Numbers at left are molecular masses in kilodaltons. (C) Reciprocal co-IP of endogenous PALB2 and KEAP1 in U2OS cells. (D) Endogenous KEAP1 association with PALB2 and NRF2 under normal and stress conditions. U2OS cells were treated with DMSO (mock), H2O2, tBHQ, and CPT at the indicated concentrations for 2 h, and the interactions were analyzed by IP-Western blotting.
Oxidative stress occurs when the level of cellular reactive oxygen species (ROS) exceeds the defense capacity of the cell. NRF2 is a master transcription factor that activates the expression of a large battery of antioxidant response element (ARE)-containing genes, which function together to mitigate oxidative damage and maintain cellular redox homeostasis (13, 29). Under normal conditions, KEAP1, a cysteine-rich oxidative stress sensor, binds to NRF2 and functions as an E3 ubiquitin ligase that targets NRF2 for degradation (3, 5, 8, 39, 40). NRF2 has two different KEAP1 binding motifs, a DLG motif and an ETGE motif (9, 14, 32), Interestingly, KEAP1 forms a dimer in the cell, and one NRF2 molecule can bind both monomers—one with the DLG and the other with the ETGE motif (14, 32). Since the two motifs have different binding affinities to KEAP1, the above binding mode is referred to as a hinge-and-latch configuration (33). When both the ETGE motif (hinge) and the DLG motif (latch) bind to a KEAP1 dimer, NRF2 is positioned for ubiquitination and subsequent degradation.
Upon oxidative and certain other electrophilic stresses, KEAP1 is modified on its cysteine residues, leading to a conformational change that disrupts its binding at NRF2's DLG motif, which has a weaker binding affinity to KEAP1, with the ETGE motif still bound. As such, NRF2 is no longer ubiquitinated and instead sequesters the bound KEAP1, allowing other, free or newly synthesized NRF2 to enter and accumulate in the nucleus to activate ARE-containing genes (29). In addition to targeting NRF2 for destruction in the cytoplasm, KEAP1 also represses NRF2 function in the nucleus by blocking its binding to AREs and by exporting NRF2 out of the nucleus during the postinduction phase (27). Moreover, increasing evidence suggests that NRF2 itself may also function as a sensor of oxidative stress through its redox-sensitive nuclear export signal and an internal ribosomal entry site (IRES) in its mRNA (10, 11).
In this study, we discovered a direct interaction between PALB2 and KEAP1. Intriguingly, PALB2 contains an extended ETGE motif (LDEETGE) identical to that of NRF2. Being a nuclear protein, PALB2 prevents KEAP1 from binding to NRF2 in the nucleus, thus promoting its nuclear concentration and ARE association. Therefore, a new mechanism of NRF2 regulation within the nucleus has been identified. Our findings may have important implications for the role of oxidative stress in the development of PALB2-associated diseases as well as cancer and FA in general.
MATERIALS AND METHODS
Plasmids, small interfering RNAs (siRNAs), and antibodies.
The FLAG-hemagglutinin (HA)-double-tagged PALB2 and KEAP1 expression plasmids (pOZ-FH-N-PALB2, pOZ-FH-C1-PALB2, and pOZ-FH-N-KEAP1) were constructed as described before (38). pOZ-FH-C1 is a variant of pOZ-FH-C and contains a base change that terminates an upstream open reading frame whose protein product may potentially interfere with proper interpretation of results. Site-directed mutagenesis was performed according to Stratagene's QuikChange protocol to construct plasmids expressing PALB2 mutations (ΔE4, ΔE4/5, T200, T400, 551X, T800, Δ50–75, Δ76–100, Δ101–125, Δ126–150, Δ151–175, Δ175–200, ΔDEETGE, E91R, G93R, T92E, Y28A, and A1025R) and KEAP1 mutations (NBTB, ΔKC, and ΔC). Myc-green fluorescent protein (GFP)-double-tagged KEAP1- and PALB2-expressing plasmids were constructed by cloning PCR-amplified KEAP1 or PALB2 cDNAs into a modified pEGFP-N3 (Clontech) containing an N-terminal Myc tag.
Four to six individual siRNAs for each gene studied were screened in U2OS cells to determine knockdown efficacy by Western blotting and evident off-target effects which were judged from unusual morphology changes and/or excessive toxicity. Six different control siRNA sequences were screened over the course of this study, and ones that showed noticeable effects on cell growth were excluded. Finally, three control siRNA sequences and three individual sequences for each gene studied were selected and pooled for knockdown analysis. The sense sequences of control siRNAs are as follows: NSC1, UUCGAACGUGUCACGUCAAdTdT; NSCQ, UUCUCCGAACGUGUCACGUdTdT; and LUC1, CGUACGCGGAAUACUCUAUdTdT. The sense sequences of gene-specific siRNAs are as follows: PALB2-718, GGAAAAGACUAAAGGAACAdTdT; PALB2-1493, UCAUUUGGAUGUCAAGAAAdTdT; PALB2-2693, GCAUAAACAUUCCGUCGAAdTdT; KEAP1-1495, ACAACAGUGUGGAGAGGUAdTdT; KEAP1-1803, GAAACAGAGACGUGGACUUdTdT; KEAP1-2529, GAUAAGUAACCCUGUAAUUdTdT; NRF2-786, UGAAGAGACAGGUGAAUUUdTdT; NRF2-1214, UGACAGAAGUUGACAAUUAdTdT; and NRF2-1834, GAGAAAGAAUUGCCUGUAAdTdT. siRNAs were synthesized by Sigma Genosys.
Antibodies used to detect BRCA2 (anti-BRCA2F2 and -F9) were affinity-purified rabbit polyclonal antibodies described previously (38). PALB2 antibodies M10 and M11 were raised in rabbits against residues 1 to 120 and 601 to 880, respectively, and affinity purified. Anti-NRF2 (C20 and H300) and anti-KEAP1 (E20) antibodies were purchased from Santa Cruz. Anti-BRCA1 (catalog no. 07-434) was purchased from Millipore. Monoclonal anti-NRF2 (EP1808Y) and anti-GFP (mAb3E6) were purchased from Abcam and Invitrogen, respectively. Monoclonal anti-KEAP1 (K2769), anti-HA (HA-7), anti-α-tubulin (T9026), anti-FLAG M2 beads, and anti-HA affinity resin were purchased from Sigma.
Cell culture, transfection, extracts, and immunoprecipitation (IP).
293T, T98G, and U2OS cells were all grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS) at 37°C with 5% CO2. HeLa S3 and T98G cells stably expressing PALB2 or harboring an empty vector were generated as previously described (37).
To transiently express PALB2, KEAP1, and NRF2 proteins, 293T cells were transfected in 6-well plates using Fugene 6 (Roche) according to the manufacturer's instructions. At 30 h after transfection, cells were harvested and whole-cell extracts were prepared in 400 μl of NETNG300 (20 mM Tris-HCl, pH 7.4, 300 mM NaCl, 1 mM EDTA, 0.5% NP-40, 10% glycerol). FLAG-tagged and GFP-tagged proteins were precipitated with M2-agarose beads or anti-GFP antibody bound to protein A-agarose beads, respectively.
For siRNA knockdown experiments, U2OS cells were seeded the day before transfection at 200,000 cells per well in 6-well plates, and transfections were performed using Lipofectamine RNAiMax (Invitrogen) according to the manufacturer's instructions. The final concentration of siRNAs was 10 nM. At 48 to 54 h after transfection, cells were harvested and whole-cell extracts were prepared in 100 μl of NETNG420 (20 mM Tris-HCl, pH 7.4, 420 mM NaCl, 1 mM EDTA, 0.5% NP-40, 5% glycerol) for Western blotting analyses. All cell lysis buffers contain the Complete protease inhibitor cocktail (Roche). Nuclear and cytoplasmic extracts were prepared using the NE-PER cell fractionation kit (Pierce).
Purification of PALB2 protein complex.
FLAG-HA-tagged PALB2 protein complex was purified from nuclear extracts (NE) derived from 3 × 108 HeLa S3 suspension cells stably expressing PALB2 by two rounds of affinity purification using anti-FLAG and anti-HA beads, respectively. The final material bound to the HA beads was eluted with 0.1 M glycine (pH 2.5) and neutralized with 0.1 volume of 1 M Tris-HCl (pH 8.5). Purified material was resolved in a 4% to 12% Bis-Tris SDS gel (Invitrogen), and Coomassie blue-stained protein bands were excised from the gel and subsequently analyzed by liquid chromatography-tandem mass spectrometry (LC-MS/MS) at the Biological Mass Spectrometry facility of Rutgers University and Robert Wood Johnson Medical School.
In vitro transcription-translation.
In vitro translation (IVT) was performed using the TNT T7 quick-coupled transcription-translation system (Promega). In brief, T7 promoter and Kozak sequences were added to primers to amplify FLAG-HA-double-tagged or HA-tagged template cDNAs by PCR. Then, the cDNA templates were incubated with the rabbit reticulocyte lysate-based TNT T7 Quick Master mix according to the manufacturer's instructions for 90 min at 30°C.
Immunofluorescence (IF) staining.
Briefly, cells were grown on glass coverslips in 12-well plates to ∼80% confluence. Cells were washed with phosphate-buffered saline (PBS) and fixed with 3% paraformaldehyde in PBS at room temperature (RT) for 10 min. Fixed cells were washed with PBS and permeabilized with PBS containing 0.5% Triton X-100 at RT for 5 min. Cells were then washed and incubated at 37°C for 20 min with primary antibodies in PBS containing 5% goat serum. Coverslips were washed and then incubated with rhodamine (tetramethyl rhodamine isocyanate [TRITC])- or fluorescein (fluorescein isothiocyanate [FITC])-conjugated secondary antibodies (Jackson ImmunoResearch) for 20 min at 37°C. Finally, coverslips were washed 3 times with PBS and mounted on slides with Vectashield mounting medium with DAPI (4′,6-diamidino-2-phenylindole; Vector Labs).
In vitro competitive binding assay.
For assay with extracts only (see Fig. 5A), 293T cells grown in 10-cm dishes were cotransfected with HA-NRF2 and Myc-KEAP1-GFP expression plasmids to produce preformed complex or transfected with either FH-PALB2-WT or FH-PALB2-T92E expression plasmids to generate competitor proteins. At 48 h after transfection, whole-cell extracts were prepared in 1 ml RIPA buffer (10 mM sodium phosphate, pH 8.0, 150 mM NaCl, 1% Triton X-100, 1% sodium deoxycholate, 0.1% SDS) containing 1 mM dithiothreitol (DTT), 1 mM phenylmethylsulfonyl fluoride (PMSF), and protease inhibitor cocktail (Roche). Competitive binding assay was performed by mixing a constant amount of HA-NRF2/Myc-KEAP1-GFP cotransfection extract (containing preformed complex) with increasing amounts of FH-PALB2-WT- or FH-PALB2-T92E transfection extract. Any difference in volume between samples was offset by adding empty-vector-transfected cell extract to make endogenous PALB2 equal. Myc-KEAP1-GFP was immunoprecipitated (IPed) from the mixtures with an anti-GFP antibody and protein A beads. Western blotting was performed to check co-IPed HA-NRF2 and PALB2 using anti-NRF2 (H300) and anti-PALB2, respectively.
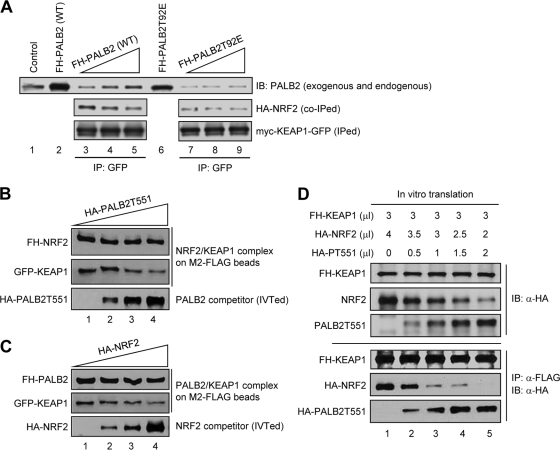
Fig 5.
Competition between PALB2 and NRF2 for KEAP1 binding. (A) Disruption of preformed NRF2/KEAP1 complex in cell extracts. Increasing amounts of whole-cell extracts from 293T cells transfected with pOZC1-PALB2WT or pOZC1-PALB2T92E (lanes 2 and 6, respectively) were added to equal aliquots of extract made from cells cotransfected with HA-NRF2 and Myc-KEAP-GFP. Then, KEAP1 was precipitated using a GFP antibody and the amounts of proteins were analyzed by Western blotting. (B and C) Competitive disruption of isolated NRF2/KEAP1 or PALB2/KEAP1 complexes by IVTed PALB2T551 or NRF2, respectively. See Materials and Methods for detailed procedures. (D) In vitro competition between PALB2 and NRF2 for KEAP1 binding. Different amounts of PALB2 and NRF2 were generated together with a fixed amount of KEAP1 in triple IVT reactions by using different combinations of amounts (in μl) of PCR-generated templates for KEAP1, PALB2, and NRF2 (top panels). Then, KEAP1 was IPed and the amounts of KEAP1, PALB2, and NRF2 in the precipitates were analyzed by Western blotting using anti-HA antibody, which recognizes all 3 proteins (bottom panels).
For assays with isolated complexes and in vitro-translated (IVTed) proteins (see Fig. 5B and C), preformed FH-NRF2/KEAP1-GFP complex or FH-PALB2/KEAP1-GFP complex was produced by immunoprecipitation (IP) from cotransfected 293T cells using anti-FLAG M2 beads and washed. The beads were then mixed with increasing amounts of in vitro-translated HA-tagged PALB2T551 or NRF2 and rocked for 3 h at 4°C. Proteins bound to the beads were analyzed by Western blotting.
For assays with in vitro-translated products only (see Fig. 5D), FH-KEAP1 DNA template (3 μl) was mixed with various amounts of HA-NRF2 DNA template (4, 3.5, 3, 2.5, and 2 μl) and HA-PALB2 DNA template (0, 0.5, 1, 1.5, and 2 μl). Then, in vitro transcription-translation reactions were performed to obtain a series of mixtures with a constant amount of FH-KEAP1 and different ratios of HA-NRF2 to HA-PALB2. FH-KEAP1 protein was IPed with M2 beads, and the levels of co-IPed HA-PALB2 and HA-NRF2 were analyzed by Western blotting. All templates were linear PCR products.
ROS measurement.
Cells in 6-well plates were washed twice with PBS and then incubated with fresh phenol red-free DMEM containing 10% fetal bovine serum and 25 μM DCF (2′,7′-dichlorofluorescein diacetate; Sigma; catalog no. D6883) at 37°C for 20 min. Cells were washed twice with PBS and trypsinized. Harvested cells were washed and resuspended in PBS at ∼106 cells/ml. ROS levels were measured by flow cytometry with excitation at 488 nm and emission at 515 to 545 nm.
Luciferase reporter assay.
To determine the effect of overexpression of various PALB2 species on endogenous NRF2 activity (see Fig. 4F), U2OS cells were seeded in 24-well plates at 250,000 cells per well and cotransfected with 200 ng of firefly luciferase reporter plasmid (pARE-LUC), 25 ng of Renilla luciferase expression plasmid (pRL-SV40), and 200 ng of either the pOZ-FH-N vector or pOZ-FH-N-PALB2 for 48 h. To measure the effect of PALB2 depletion (see Fig. 6C), U2OS cells were seeded in 24-well plates at 150,000 cells per well and treated with siRNAs for 48 h first and then cotransfected with 200 ng pARE-LUC and 25 ng of pRL-SV40 for 30 h. Both firefly and Renilla luciferase activities were measured with the dual luciferase reporter assay system (Promega). Firefly luciferase activity was normalized with Renilla luciferase activity to control for sample-to-sample variations in transfection efficiency. All reporter gene assays were repeated independently at least three times.
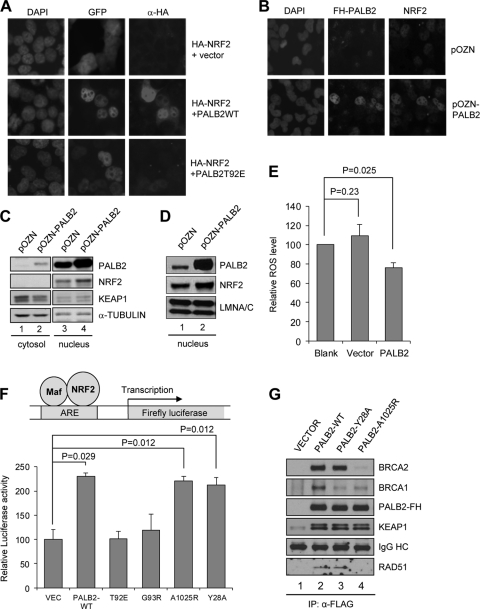
Fig 4.
PALB2 overexpression promotes NRF2 nuclear accumulation and reduces ROS level. (A) An HA-NRF2 plasmid was cotransfected into 293T cells with empty Myc-GFP vector or Myc-PALB2WT-GFP or Myc-PALB2T92E-GFP constructs for 24 h, and their expression and localization were analyzed by fluorescence microscopy. (B) T98G cells overexpressing N-terminally FLAG-HA-double-tagged PALB2 were costained with HA (for PALB2) and NRF2 antibodies and visualized by fluorescence microscopy. (C) T98G cells in panel B were fractionated into cytosol and nuclei, and the amounts of indicated proteins in each compartment were analyzed by Western blotting. (D) Nuclei of HeLa cells harboring the vector or PALB2 overexpression construct were isolated, and protein amounts were analyzed by Western blotting. Lamin A/C, a nuclear protein, was used as a loading control. (E) ROS levels in naïve HeLa cells and the two stable cell lines in panel D were measured using the DCF assay. (F) U2OS cells were cotransfected with the ARE-Luc reporter system and various PALB2 expression vectors, and luciferase activities were measured 48 h after transfection. (G) Cells were transfected with the indicated expression vectors for 30 h, and the tagged PALB2 proteins were IPed with anti-FLAG M2 beads. The IPed PALB2 and co-IPed BRCA2, BRCA1, and KEAP1 proteins were probed with their respective antibodies by Western blotting.
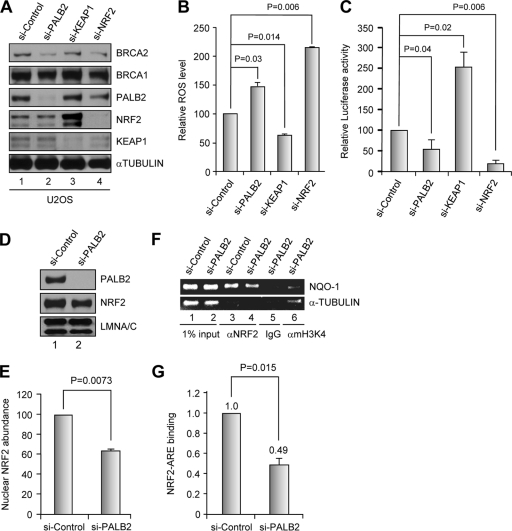
Fig 6.
siRNA-mediated depletion of PALB2 reduces NRF2 abundance and activity in the nucleus and increases cellular ROS level. (A) Western blotting showing the levels of proteins of interest after siRNA treatments. (B) ROS levels in cells depleted of each of the indicated proteins. (C) Endogenous NRF2 activity on an ARE-luciferase reporter following depletion of PALB2 and other proteins. (D and E) Nuclear NRF2 level after depletion of PALB2 in U2OS cells analyzed by Western blotting (D) and quantified by densitometry (E). (F and G) ChIP analysis of endogenous NRF2 binding to NQO1 ARE in control and PALB2-depleted cells. (F) An agarose gel image of the PCR products in a representative experiment. (G) Results quantified by real-time PCR.
ChIP assay.
U2OS cells were seeded the day before transfection at 1.25 × 106/dish in 10-cm dishes and then treated with control siRNA pool or PALB2 siRNA pool. At 48 to 54 h after transfection, cells were treated with 18.5% formaldehyde to cross-link proteins to DNA. Chromatin immunoprecipitation (ChIP) assays were performed using the EZ-ChIP chromatin immunoprecipitation kit (Millipore; catalog no. 17-371) according to the manufacturer's instructions. Briefly, after in vivo cross-linking, cells were lysed and sonications were performed to shear the chromatin to a manageable size (200 to 1,000 bp). Immunoselections of cross-linked protein-DNA were performed with anti-rabbit IgG (negative control), anti-NRF2 (H300), anti-mH3K4 (positive control), and protein G-conjugated agarose beads. Protein-DNA complexes were washed, and then protein-DNA cross-links were reversed to free DNAs. The purified DNAs were analyzed by real-time quantitative PCR as described previously (27).
NRF2 postinduction degradation and export measurement.
U2OS cells were treated with control siRNA pool or PALB2 siRNA pool. At 48 h after transfection, cells were washed twice with PBS and then treated with either dimethyl sulfoxide (DMSO) (as controls) or 100 μM tBHQ in fresh DMEM for 3 h. Cells treated with DMSO were harvested for preparation of whole-cell extracts or nuclear extracts. Cells treated with tert-butylhydroquinone (tBHQ) were further washed twice with PBS to remove tBHQ and then incubated with fresh DMEM for 0 h, 1.5 h, or 3 h. Whole-cell extracts were made and used to check the rate of NRF2 degradation. Nuclear extracts were prepared using the NE-PER nuclear and cytoplasmic extract kit (Pierce) according to the manufacturer's manual to check the rate of NRF2 export.
Statistical analysis.
All pooled data presented are averages of at least 3 independent experiments. The standard deviations and one-sided P values were calculated using the GraphPad Prism software (v.5).
RESULTS
Identification of KEAP1 as a PALB2 binding partner.
In searching for potential new functions of PALB2, we carried out tandem affinity purification to identify its new interacting partners. A C-terminally FLAG-HA-double-tagged PALB2 (PALB2-FH) was purified from HeLa S3 suspension cells stably expressing the protein. As shown in Fig. 1B, known PALB2 binding partners BRCA2, BRCA1, RAD51, and MRG15 were all copurified and identified by mass spectrometry analysis. Interestingly, KEAP1 was found to be a component of the PALB2 complex. To confirm this interaction, we first tested if endogenous PALB2 could be coimmunoprecipitated (co-IPed) with a GFP-tagged KEAP1 protein transiently expressed in 293T cells. As shown in Fig. S1A in the supplemental material, both endogenous PALB2 and BRCA2 were readily co-IPed, although it was unclear if KEAP1 binds PALB2 or BRCA2. Next, we performed reciprocal co-IP of endogenous proteins in U2OS cells with two different affinity-purified PALB2 antibodies and one monoclonal KEAP1 antibody, and co-IP of the endogenous proteins was observed in both cases (Fig. 1C). Since PALB2 is a nuclear protein, we further tested if it indeed interacted with KEAP1 in the nucleus. When U2OS cells were fractionated into cytoplasmic and nuclear components, all PALB2 and approximately a quarter of KEAP1 were found in the nuclear extract (NE), and PALB2-KEAP1 complex formation was clearly detected in this fraction (see Fig. S1B).
Subsequently, we attempted to compare the stoichiometries of PALB2-KEAP1 and NRF2-KEAP1 bindings under different conditions. U2OS cells were treated with DMSO (control), hydrogen peroxide (H2O2), tert-butylhydroquinone (tBHQ; an oxidative stress inducer and NRF2 inducer), or the DNA-damaging agent camptothecin (CPT), and then endogenous PALB2 and NRF2 were IPed. As shown in Fig. 1D, the binding between KEAP1 and PALB2 was not affected by any of the treatments. In contrast, tBHQ treatment clearly enhanced KEAP1-NRF2 complex formation; hydrogen peroxide also elicited a slight increase of the binding, whereas CPT produced no effect. Interestingly, even after tBHQ induction, the amount of KEAP1 co-IPed by either of the two NRF2 antibodies was still much smaller than that in the PALB2 IP (compare lanes 17, 18, and 15 in Fig. 1D). Although we were unable to determine the amounts of NRF2 IPed by the two antibodies due to high background on the Western blot when probed with all available NRF2 antibodies, it appears possible that the stoichiometry of KEAP1-PALB2 binding may be higher than that of KEAP1-NRF2 binding. U2OS cells were mainly used in this study since these cells express relatively low levels of NRF2, which is greatly induced after KEAP1 depletion (see Fig. S1C in the supplemental material and see also Fig. 6A). Also, the expression and behavior of PALB2 as well as its key binding partners BRCA1 and BRCA2 in U2OS cells are well characterized.
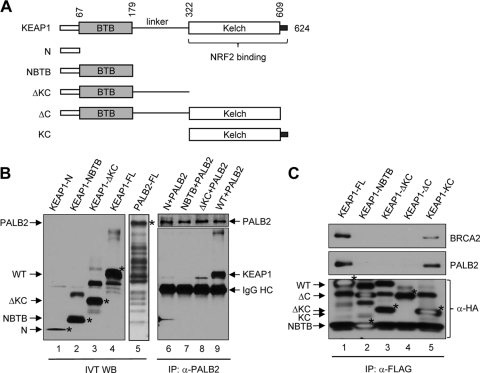
To determine if PALB2 directly binds KEAP1, we generated in vitro-translated (IVTed), epitope-tagged full-length KEAP1 and PALB2 proteins as well as a series of truncated KEAP1 fragments (Fig. 2A and B). We then mixed each KEAP1 species with PALB2 and IPed the latter from the mixture with an anti-PALB2 antibody. As shown in Fig. 2B, full-length KEAP1 was efficiently co-IPed with PALB2 (lane 9) but none of the truncated KEAP1 proteins lacking the KC region was IPed (lanes 6 to 8), indicating that the binding between full-length KEAP1 and PALB2 is specific and direct. Subsequently, we asked if the KC domain is sufficient for PALB2 binding using a transient-transfection and IP-Western blotting experiment. As shown in Fig. 2C, the KC region was able to bind both PALB2 and BRCA2, suggesting that KEAP1 may bind PALB2 with the same structural element as that for NRF2.
Fig 2.
PALB2 directly interacts with the KC domain of KEAP1. (A) Schematic of the KEAP1 constructs. (B) In vitro translations were performed to produce HA-tagged KEAP1 proteins (lanes 1 to 4) and FLAG-HA-PALB2 (lane 5). Subsequently, each of the KEAP1 species was mixed with FLAG-HA-PALB2 and an anti-PALB2 antibody was used to precipitate PALB2. Then, the precipitated materials were analyzed by Western blotting using an anti-HA antibody. (C) Various KEAP1 constructs shown in panel A were transfected into 293T cells, and proteins were IPed by anti-FLAG M2 agarose. The precipitates were analyzed by Western blotting.
Direct interaction between the ETGE motif of PALB2 and the kelch domain of KEAP1.
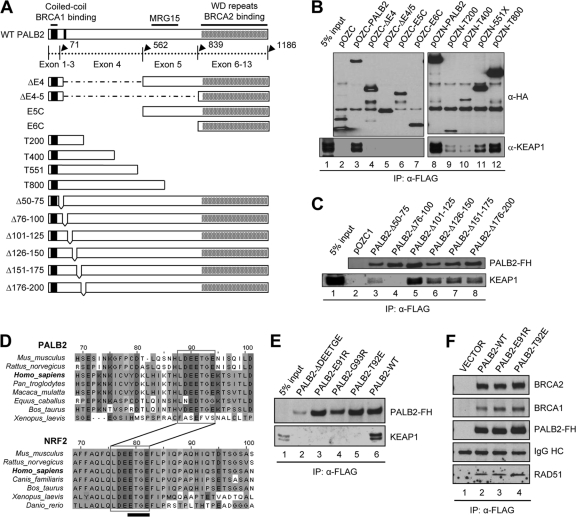
Extensive domain mapping was then carried out to identify the KEAP1 binding site on PALB2. First, deletion of the exon 4-encoded region (residues 71 to 561) was found to abolish the binding (Fig. 3B, lane 3 versus lanes 4 to 7). Then, the binding site was narrowed down to within the N-terminal 200 residues of the protein (Fig. 3B, lane 9). Finally, we generated and tested a series of 25-amino-acid (aa) deletions in the N terminus and found that residues 76 to 100 are essential (Fig. 3C, lane 4). Intriguingly, amino acid sequence alignment identified a 7-aa motif (LDEETGE) between residues 76 and 100 that is highly conserved across mammalian species, and this motif is identical to the ETGE motif of NRF2 that binds KEAP1 (Fig. 3D). Mutations of the critical T residue as well as several other residues within the NRF2 ETGE motif have been shown to abrogate NRF2-KEAP1 interaction, allowing NRF2 to escape KEAP1-mediated repression (12). To test if PALB2 indeed binds KEAP1 with this motif, we generated a deletion mutant and a series of point mutants (E91R, T92E, and G93R). As shown in Fig. 3E, all mutants failed to form complexes with KEAP1. Therefore, we conclude that the ETGE motif of PALB2 directly interacts with the kelch domain of KEAP1 in a fashion similar to the NRF2(ETGE)-KEAP1 interaction (12, 19). As expected, none of the mutations affected the binding of PALB2 to BRCA1 or BRCA2 (Fig. 3F), indicating that the binding events are independent and separable.
Fig 3.
PALB2 binds KEAP1 through a highly conserved ETGE motif. (A) Schematic of PALB2 constructs used in the domain mapping study. (B and C) PALB2 constructs were transiently expressed in 293T cells, proteins were IPed with anti-FLAG M2 agarose, and the precipitates were analyzed by Western blotting. (D) Amino acid sequence alignment of the N-terminal sequences of PALB2 and NRF2 showing the shared LDEETGE KEAP1 binding motif. (E) Deletion or mutations of the ETGE motif in PALB2 all abolish KEAP1 binding. (F) E91R and T92E mutations do not affect the binding of PALB2 to BRCA2, RAD51, and BRCA1.
PALB2 overexpression promotes NRF2 nuclear accumulation and reduces ROS level.
Based on the above finding, we hypothesized that PALB2 may be able to compete with NRF2 for KEAP1 binding and therefore may “protect” NRF2 from KEAP1-mediated repression. To test this hypothesis, we cotransfected HA-tagged NRF2 and Myc-GFP-double-tagged PALB2 into 293T cells and examined their expression and localization. Since high-level overexpression of NRF2 may overwhelm cellular KEAP1 and lead to artifacts, we titrated the amount of HA-NRF2 plasmid used to a level where we could detect weak and mostly cytoplasmic staining of HA-NRF2 when it was cotransfected with Myc-GFP. The largely cytoplasmic localization of the HA-NRF2 observed here may be due to the relatively large amount of KEAP1 in 293T cells (see Fig. S1C in the supplemental material). Under such a condition, overexpression of wild-type (wt) PALB2 strongly increased the level of NRF2 and promoted its accumulation in the nucleus (Fig. 4A). In contrast, overexpression of the T92E mutant showed a much weaker effect, indicating that the effect of wt PALB2 is mediated by its KEAP1 binding.
We also generated T98G glioblastoma cell lines stably expressing PALB2 double tagged with FLAG and HA epitopes (Fig. 4B and C). For reasons so far unknown, T98G cells have a much higher NRF2 level than do U2OS and other cells surveyed in this study (see Fig. S1C in the supplemental material), with the only exception being the A549 lung cancer cell line, which has been shown to harbor a KEAP1 mutation that impairs NRF2 binding (24). The endogenous NRF2 in T98G cells is localized in the nucleus and can be clearly observed using immunofluorescence (IF) (Fig. 4B). While the majority of these stable cells showed a low level of ectopic PALB2 expression, in some cells the tagged PALB2 was overexpressed (Fig. 4B). Interestingly, in all cells expressing ectopic PALB2 at high levels, NRF2 staining signals were clearly stronger than those in cells expressing ectopic PALB2 at low levels or cells harboring the vector alone (Fig. 4B). Consistently, the average abundance of (nuclear) NRF2 in cells expressing tagged PALB2 was also significantly higher than that in control cells (Fig. 4C). A similar observation was also made in HeLa S3 cells overexpressing the same tagged PALB2 (Fig. 4D). Additionally, we observed in PALB2-overexpressing cells a slight decrease of KEAP1 in the cytoplasm and a corresponding increase in the nucleus (Fig. 4C; see also Fig. S2B in the supplemental material), suggesting that PALB2 binding to KEAP1 may promote the nuclear retention of the latter to some degree. However, this effect appears to be small under the setting used and remains to be further investigated under different conditions.
Since a key function of NRF2 is to mitigate reactive oxygen species (ROS) in the cell, we asked if PALB2 overexpression would reduce ROS levels. When ROS levels of the above T98G cell lines were measured using the DCF (2′,7′-dichlorodihydrofluorescein) assay, it was undetectable in both lines, consistent with the high endogenous NRF2 abundance in these cells. In the stable HeLa S3 cell line, we observed a significant decrease in ROS level compared with that in cells harboring the vector (Fig. 4E). Note that PALB2 overexpression was observed in greater than 80% of the cells (see Fig. S2A in the supplemental material). We also generated U2OS stable cell lines, but ROS measurement would not be meaningful since only a minor percentage of cells (∼10 to 15%) showed clear exogenous PALB2 expression even after repeated selections (data not shown).
To directly measure the transcriptional activation activity of endogenous NRF2 in PALB2-overexpressing cells, an ARE-LUC reporter was employed (39). As shown in Fig. 4F, ectopic expression of wt PALB2 increased endogenous NRF2 activity by more than 2-fold, whereas the two KEAP1 binding mutants (T92E and G93R) failed to show any effect. Since PALB2 also directly interacts with BRCA1 and BRCA2, we asked whether these interactions affect its activity to bind KEAP1 and promote NRF2 nuclear localization and function. To this end, we generated PALB2-Y28A and -A1025R mutants, each of which abrogates its BRCA1 or BRCA2 interaction, respectively (Fig. 4G) (18, 28). Interestingly, both mutants were equally active compared with wt PALB2 (Fig. 4F). Like wt PALB2, these two mutants also increased NRF2 accumulation in the nucleus (see Fig. S3 in the supplemental material). These results clearly demonstrate that the basic function of PALB2 to promote NRF2 activity depends on its KEAP1 binding but not its BRCA1/2 interaction, although the possibility that BRCA1/2 may modulate this PALB2 function under certain conditions cannot be ruled out. Surprisingly, the A1025R mutant also impaired PALB2-BRCA1 binding under the setting used (Fig. 4G). However, this does not contradict the conclusion reached above.
PALB2 effectively competes with NRF2 for KEAP1 binding.
The question whether PALB2 is able to disrupt KEAP1-NRF2 interaction was directly addressed. Since the abundance of endogenous NRF2-KEAP1 complex is low and difficult to obtain by IP with available antibodies (Fig. 1D), we cotransfected HA-NRF2 and Myc-KEAP1-GFP to obtain the complex. When an increasing amount of wt PALB2-FH was added to such preformed HA-NRF2/Myc-KEAP1-GFP complex (in cotransfected cell lysate), a decrease of NRF2 in the Myc-KEAP1-GFP IP along with a steady increase of PALB2 was observed (Fig. 5A), indicating that PALB2 is able to compete with NRF2 for KEAP1 binding and disrupt preformed KEAP1-NRF2 complex. Again, the PALB2T92E mutant protein was unable to elicit any effect, validating the specificity of the competition. Note that lanes 3 to 5 in the PALB2 panel of Fig. 5A contain both endogenous and the tagged PALB2, whereas lanes 7 to 9 contain only endogenous PALB2 since the tagged T92E mutant is defective in KEAP1 binding.
To compare the affinities between NRF2-KEAP1 and PALB2-KEAP1 complex formations, in vitro competition assays were performed. The complexes were obtained by cotransfection followed by IP and washing. Then, increasing amounts of in vitro-translated (IVTed) PALB2 or NRF2 competitors were added to each respective target complex, and the amounts of KEAP1 released were analyzed. Due to the relatively large size of PALB2 (130 kDa), an N-terminal fragment encompassing residues 1 to 550 (PALB2T551) was used for IVT. This fragment has a size similar to those of NRF2 and KEAP1. As shown in Fig. 5B and C, the IVTed PALB2 was able to disrupt the NRF2-KEAP1 complex and released KEAP1 in a dose-dependent manner, and the IVTed NRF2 exhibited similar activity against the PALB2-KEAP1 complex. Furthermore, we also performed a triple IVT experiment in which various amounts of NRF2 and PALB2 were produced along with a fixed amount of KEAP1. As shown in Fig. 5D, when similar amounts of NRF2 and PALB2 were produced in the same reaction (lane 3), significantly more PALB2 was bound to KEAP1 than was NRF2. Taken together, these results indicate that the binding affinity between PALB2 and KEAP1 may be equal to or greater than that between NRF2 and KEAP1 and lend further support to our model that PALB2 promotes NRF2 activity by competitively preventing and/or disrupting KEAP1-NRF2 interaction.
Depletion of PALB2 reduces NRF2 activity and increases cellular ROS level.
Unlike in T98G cells, the ROS level in U2OS cells could be readily detected. Cells were treated with siRNAs against PALB2, KEAP1, and NRF2 to deplete the respective proteins, and the ROS levels were measured. As expected, depletion of NRF2 greatly increased the ROS level, whereas KEAP1 knockdown substantially decreased cellular ROS, presumably through a profound upregulation of NRF2 (Fig. 6A and B). PALB2 depletion increased the ROS level by nearly 50% compared with treatment of cells with control siRNA. Interestingly, the total amount of cellular NRF2 was largely unchanged under the same condition (Fig. 6A). Of note, a moderate reduction of the KEAP1 amount was often observed in NRF2-depleted cells (lane 4).
Next, we directly measured the transcriptional activation activity of endogenous NRF2 in the above siRNA-treated cells using the ARE-LUC reporter. As shown in Fig. 6C, NRF2 depletion reduced the reporter activity by approximately 5-fold and KEAP1 depletion upregulated the activity by ∼2.5-fold, validating the suitability of the assay system. Under this setting, knockdown of PALB2 reduced the luciferase activity by about 2-fold, indicating that endogenous NRF2 transcription activity is impaired in the absence of PALB2.
Subsequently, mRNA amounts of nine NRF2 target genes (NQO1, GCLM, TXN, HMOX1, TXRD1, GSTP1, GCLC, AKR1B10, and AKR1C1) were surveyed following PALB2 knockdown in U2OS cells. To better understand the regulation of these genes by NRF2 under the setting used, their expression levels after KEAP1 and NRF2 depletion were also determined in parallel. As shown in Fig. S4A in the supplemental material, seven out of the nine genes, particularly AKR1C1, showed substantial to dramatic upregulation after KEAP1 depletion. The remaining two, TXRD1 and GSTP1, exhibited weak induction. Upon an acute loss of NRF2, only four genes (NQO1, GCLM, AKR1B10, and AKR1C1) showed significant downregulation, whereas HMOX1 and GCLC expression even increased significantly. These results demonstrate that, under the setting used, most NRF2 target genes positively respond to NRF2 induction but a significant fraction of the genes do not depend on NRF2 for basal expression, with some evidently having alternative, NRF2-independent induction mechanisms. Importantly, mRNA levels of NQO1, GCLM, AKR1B10, and AKR1C1, whose basal expression appeared to depend on NRF2, decreased upon PALB2 depletion. The expression of a similar panel of NRF2 downstream genes was also measured following PALB2 depletion in T98G cells. Although these cells express a high level of NRF2 (see Fig. S1C in the supplemental material), KEAP1 depletion elicited a further and large increase in NRF2 amount (see Fig. S4B), indicating an intact KEAP1 regulation of NRF2. As also shown in Fig. S4B in the supplemental material, the expression of NQO1, GCLC, TXN, and AKR1C1 was reduced after PALB2 depletion, indicating that NRF2 activity on these genes is compromised in the absence of PALB2.
Since loss of PALB2 led to an increased ROS level and reduced NRF2 transcriptional activity without significantly affecting its total abundance in the cell, we suspected that either its concentration in the nucleus or its association with AREs of its target genes may be reduced. In addressing the first possibility, nuclei of U2OS cells treated with control or PALB2 siRNAs were isolated and the NRF2 amount was examined. As shown in Fig. 6D and E, cells depleted of PALB2 exhibited significantly and reproducibly smaller amounts of NRF2 in the nucleus. Next, we asked whether NRF2 binding to the ARE in the NQO1 promoter is impaired by chromatin immunoprecipitation (ChIP) and quantitative PCR analyses and found that depletion of PALB2 reduced NRF2 binding to NQO1 ARE by approximately 2-fold (Fig. 6F and G). Taken together, these results indicate that endogenous PALB2 promotes NRF2 accumulation in the nucleus and binding to ARE and thus its transcription activation activity.
PALB2 impedes NRF2 nuclear export and degradation postinduction.
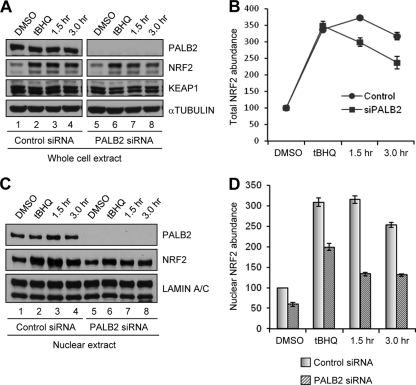
Having analyzed the effect of PALB2 on NRF2 abundance and activity under basal conditions, we further asked whether PALB2 also modulates NRF2 induction as well as its postinduction export and degradation, which are both regulated by KEAP1. Control or PALB2-depleted U2OS cells were subjected to tBHQ treatment (100 μM for 3 h) and NRF2 amounts were examined. As shown in Fig. 7A and B, tBHQ induced NRF2 by approximately 3.5-fold in both control and PALB2-depleted cells, indicating that PALB2 does not play a role in the initial induction of NRF2 by this agent. However, when NRF2 levels were further monitored following tBHQ removal, PALB2-depleted cells showed moderately faster reduction of total NRF2 abundance than did control cells. Then, we analyzed the amounts of NRF2 in cell nuclei under the same conditions. Although the nuclear NRF2 amount was smaller in PALB2-depleted cells before induction, it was induced by a magnitude (∼3-fold) similar to that in control cells (Fig. 7C and D). By 1.5 h after tBHQ removal, whereas nuclear NRF2 in control cells remained unchanged, it had decreased significantly in PALB2-depleted cells. These results indicate that PALB2, by binding KEAP1 and reducing its complex formation with NRF2 in the nucleus, interferes with KEAP1-mediated NRF2 export and degradation in the postinduction phase.
Fig 7.
PALB2 regulates NRF2 nuclear export after induction. (A) Western blotting of whole-cell extracts of U2OS cells (transfected with control or PALB2 siRNAs for 48 h) following treatment with DMSO or tBHQ and at indicated time points after tBHQ removal. (B) Results from 3 independent experiments in panel A were quantified by densitometry and plotted. (C) Western blotting of nuclear extracts of U2OS cells following the same treatments as those in panel A. (D) Results from 3 independent experiments in panel C were quantified by densitometry.
DISCUSSION
In this study, we discovered a novel interaction between the PALB2 tumor suppressor and a key oxidative stress sensor, KEAP1. We demonstrated that PALB2 shares a conserved ETGE KEAP1-binding motif with NRF2 and can effectively compete with NRF2 for KEAP1 binding. As such, PALB2 enhances NRF2 function by impeding KEAP1-NRF2 interaction in the nucleus, thus preventing KEAP1-imposed block of NRF2-ARE association and KEAP1-mediated export of NRF2 (Fig. 8). Importantly, depletion of PALB2 led to increased ROS levels and downregulation of a subset of NRF2 target genes whose basal expression relied on NRF2 activity under the setting used. Thus, this study identified PALB2 as a new regulator of NRF2 and redox homeostasis.
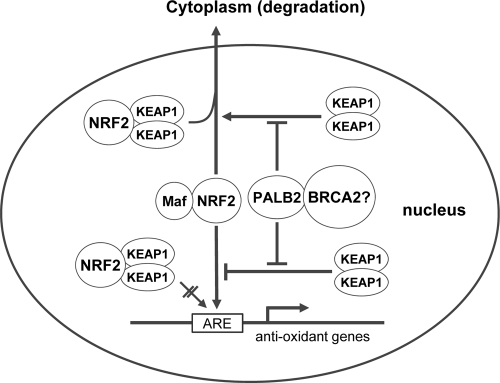
Fig 8.
A proposed model of PALB2 regulation of NRF2 in the nucleus. Nuclear NRF2 forms heterodimers with small Maf proteins and activates antioxidant gene expression through association with the AREs in the promoter region of the genes. KEAP1 represses NRF2 function in the nucleus by blocking NRF2 binding to AREs and exporting it to the cytoplasm for degradation. By competitively binding to KEAP1, PALB2 alleviates the negative impact of KEAP1 on NRF2 function via the two above-noted mechanisms. BRCA2 is also a component of the PALB2-KEAP1 complex, but its role is unclear.
Interestingly, PALB2 depletion reduced NRF2 function without significantly affecting its protein level (Fig. 6A and B). Since PALB2 is a nuclear protein, it is plausible that it may not directly affect KEAP1-mediated NRF2 degradation, which occurs in the cytoplasm (27). KEAP1 was originally thought to be a cytoplasmic protein that sequesters and degrades NRF2 in the cytoplasm, but it has now been made clear that a significant fraction of KEAP1 exists in the nucleus and represses NRF2 therein by blocking NRF2 access to AREs and exporting NRF2 out of the nucleus for degradation (27). In this vein, PALB2-KEAP1 association may reduce the amount of nuclear KEAP1 available to block and export NRF2, thus increasing NRF2 nuclear concentration and activity. This notion is supported by our data showing a smaller NRF2 amount in the nucleus preinduction and a faster decrease of nuclear NRF2 postinduction in PALB2-depleted cells. However, total NRF2 amounts were similar in control and PALB2 knockdown cells, indicating that under the setting used, the exported NRF2 was not completely degraded in a timely manner.
Since KEAP1 is an E3 ubiquitin ligase, it may potentially ubiquitinate PALB2 and earmark it for proteasome-mediated destruction. However, when KEAP1 was depleted in U2OS cells to an extent that NRF2 was greatly upregulated, no meaningful change in PALB2 protein level was observed (Fig. 6A). Similarly, strong induction of NRF2 but lack of change in PALB2 level following KEAP1 depletion was also observed in HeLa cells, MCF-7 and T47D breast cancer cells, and MCF-10A normal breast epithelial cells (data not shown). Therefore, KEAP1 does not appear to be an effective E3 ligase for PALB2 under most conditions, due presumably to the different compartments in which the KEAP1-Cul3 E3 ligase (cytoplasm) and PALB2 (nucleus) are localized. This is further supported by the fact that KEAP1 overexpression in HeLa cells failed to reduce the PALB2 protein level (see Fig. S2B in the supplemental material).
It is reasonable to speculate that the extent of PALB2's influence on NRF2 activity may depend on the relative abundance of PALB2, KEAP1, and NRF2 in the nucleus, which may vary substantially in different tissues or under different conditions. Due to the relatively recent discovery of PALB2, its expression patterns in different tissues have not been well documented. Based on the only information available in www.genecards.org, the PALB2 expression level shows great variation among both normal and cancer tissues/cells. In particular, PALB2 appears to be consistently overexpressed in breast, colon, pancreas, and lung cancers when measured by 3 different approaches—DNA microarray, electronic Northern (eNorthern) blotting, and serial analysis of gene expression (SAGE). Since PALB2 mutations are rare, it is likely that most of the tumors analyzed would have overexpression of the wt protein, which may alter the balance between PALB2 and KEAP1 in the nucleus, leading to reduced NRF2 repression by KEAP1, increased target gene expression, and, ultimately, a favorable redox condition and a growth advantage for the tumor cells. Similarly, certain tissues may express high levels of PALB2 under normal conditions and “enjoy” better protection of NRF2 from KEAP1. However, these tissues may also be more prone to oxidative damage upon PALB2 loss. Further study is needed to address these issues.
Oxidative stress has been implicated in a wide variety of diseases, including cancer and FA (20, 34). Somatic mutations in KEAP1 and NRF2 have each been found in 10 to 15% of lung cancers, and most of these mutations abrogate KEAP1-NRF2 binding (6, 29). In breast cancer, two somatic KEAP1 mutations have been found (36), and one of them has been shown to compromise KEAP1 E3 ligase activity (16). All these mutations presumably lead to constitutive NRF2 overexpression, which appears to confer growth advantage and drug resistance on tumor cells (17, 35). In addition, FA cells are generally hypersensitive to oxidative stress, although the exact mechanism(s) remains poorly defined (20). In this context, our study provides a new and direct link between oxidative stress response and the development of breast cancer, pancreas cancer, and FA.
Supplementary Material
ACKNOWLEDGMENTS
We thank C. Colin (Dana-Farber Cancer Institute) for advice on the quantitative reverse transcription-PCR analysis and helpful comments on the manuscript.
This work was supported by the American Cancer Society (RSG TBG-119822 to B.X. and RSG-07-154 to D.D.Z.), the National Cancer Institute (R01CA138804 to B.X.), the National Institute of Environmental Health Sciences (R01ES015010 to D.D.Z.), and the Cancer Institute of New Jersey (B.X.). J.M. was supported by a postdoctoral fellowship from the New Jersey Commission of Cancer Research (NJCCR).
Footnotes
Published ahead of print 13 February 2012
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1. Casadei S, et al. 2011. Contribution of inherited mutations in the BRCA2-interacting protein PALB2 to familial breast cancer. Cancer Res. 71: 2222– 2229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Chen P, et al. 2008. Association of common PALB2 polymorphisms with breast cancer risk: a case-control study. Clin. Cancer Res. 14: 5931– 5937 [DOI] [PubMed] [Google Scholar]
- 3. Cullinan SB, Gordan JD, Jin J, Harper JW, Diehl JA. 2004. The Keap1-BTB protein is an adaptor that bridges Nrf2 to a Cul3-based E3 ligase: oxidative stress sensing by a Cul3-Keap1 ligase. Mol. Cell. Biol. 24: 8477– 8486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Erkko H, et al. 2007. A recurrent mutation in PALB2 in Finnish cancer families. Nature 446: 316– 319 [DOI] [PubMed] [Google Scholar]
- 5. Furukawa M, Xiong Y. 2005. BTB protein Keap1 targets antioxidant transcription factor Nrf2 for ubiquitination by the Cullin 3-Roc1 ligase. Mol. Cell. Biol. 25: 162– 171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hayes JD, McMahon M. 2009. NRF2 and KEAP1 mutations: permanent activation of an adaptive response in cancer. Trends Biochem. Sci. 34: 176– 188 [DOI] [PubMed] [Google Scholar]
- 7. Jones S, et al. 2009. Exomic sequencing identifies PALB2 as a pancreatic cancer susceptibility gene. Science 324: 217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kobayashi A, et al. 2004. Oxidative stress sensor Keap1 functions as an adaptor for Cul3-based E3 ligase to regulate proteasomal degradation of Nrf2. Mol. Cell. Biol. 24:7130– 7139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kobayashi M, et al. 2002. Identification of the interactive interface and phylogenic conservation of the Nrf2-Keap1 system. Genes Cells 7: 807– 820 [DOI] [PubMed] [Google Scholar]
- 10. Li W, Kong AN. 2009. Molecular mechanisms of Nrf2-mediated antioxidant response. Mol. Carcinog. 48: 91– 104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Li W, et al. 2010. An internal ribosomal entry site mediates redox-sensitive translation of Nrf2. Nucleic Acids Res. 38: 778– 788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lo SC, Li X, Henzl MT, Beamer LJ, Hannink M. 2006. Structure of the Keap1:Nrf2 interface provides mechanistic insight into Nrf2 signaling. EMBO J. 25: 3605– 3617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Malhotra D, et al. 2010. Global mapping of binding sites for Nrf2 identifies novel targets in cell survival response through ChIP-Seq profiling and network analysis. Nucleic Acids Res. 38: 5718– 5734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. McMahon M, Thomas N, Itoh K, Yamamoto M, Hayes JD. 2006. Dimerization of substrate adaptors can facilitate cullin-mediated ubiquitylation of proteins by a “tethering” mechanism: a two-site interaction model for the Nrf2-Keap1 complex. J. Biol. Chem. 281: 24756– 24768 [DOI] [PubMed] [Google Scholar]
- 15. Moynahan ME, Jasin M. 2010. Mitotic homologous recombination maintains genomic stability and suppresses tumorigenesis. Nat. Rev. 11: 196– 207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Nioi P, Nguyen T. 2007. A mutation of Keap1 found in breast cancer impairs its ability to repress Nrf2 activity. Biochem. Biophys. Res. Commun. 362: 816– 821 [DOI] [PubMed] [Google Scholar]
- 17. Ohta T, et al. 2008. Loss of Keap1 function activates Nrf2 and provides advantages for lung cancer cell growth. Cancer Res. 68: 1303– 1309 [DOI] [PubMed] [Google Scholar]
- 18. Oliver AW, Swift S, Lord CJ, Ashworth A, Pearl LH. 2009. Structural basis for recruitment of BRCA2 by PALB2. EMBO Rep. 10: 990– 996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Padmanabhan B, et al. 2006. Structural basis for defects of Keap1 activity provoked by its point mutations in lung cancer. Mol. Cell 21: 689– 700 [DOI] [PubMed] [Google Scholar]
- 20. Pang Q, Andreassen PR. 2009. Fanconi anemia proteins and endogenous stresses. Mutat. Res. 668: 42– 53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Potapova A, Hoffman AM, Godwin AK, Al-Saleem T, Cairns P. 2008. Promoter hypermethylation of the PALB2 susceptibility gene in inherited and sporadic breast and ovarian cancer. Cancer Res. 68: 998– 1002 [DOI] [PubMed] [Google Scholar]
- 22. Rahman N, et al. 2007. PALB2, which encodes a BRCA2-interacting protein, is a breast cancer susceptibility gene. Nat. Genet. 39: 165– 167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Reid S, et al. 2007. Biallelic mutations in PALB2 cause Fanconi anemia subtype FA-N and predispose to childhood cancer. Nat. Genet. 39: 162– 164 [DOI] [PubMed] [Google Scholar]
- 24. Singh A, et al. 2006. Dysfunctional KEAP1-NRF2 interaction in non-small-cell lung cancer. PLoS Med. 3: e420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Slater EP, et al. 2010. PALB2 mutations in European familial pancreatic cancer families. Clin. Genet. 78: 490– 494 [DOI] [PubMed] [Google Scholar]
- 26. Southey MC, et al. 2010. A PALB2 mutation associated with high risk of breast cancer. Breast Cancer Res. 12: R109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sun Z, Zhang S, Chan JY, Zhang DD. 2007. Keap1 controls postinduction repression of the Nrf2-mediated antioxidant response by escorting nuclear export of Nrf2. Mol. Cell. Biol. 27: 6334– 6349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sy SM, Huen MS, Chen J. 2009. PALB2 is an integral component of the BRCA complex required for homologous recombination repair. Proc. Natl. Acad. Sci. U. S. A. 106: 7155– 7160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Taguchi K, Motohashi H, Yamamoto M. 2011. Molecular mechanisms of the Keap1-Nrf2 pathway in stress response and cancer evolution. Genes Cells 16: 123– 140 [DOI] [PubMed] [Google Scholar]
- 30. Tischkowitz M, Xia B. 2010. PALB2/FANCN: recombining cancer and Fanconi anemia. Cancer Res. 70: 7353– 7359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Tischkowitz M, et al. 2007. Analysis of PALB2/FANCN-associated breast cancer families. Proc. Natl. Acad. Sci. U. S. A. 104: 6788– 6793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Tong KI, et al. 2006. Keap1 recruits Neh2 through binding to ETGE and DLG motifs: characterization of the two-site molecular recognition model. Mol. Cell. Biol. 26: 2887– 2900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Tong KI, Kobayashi A, Katsuoka F, Yamamoto M. 2006. Two-site substrate recognition model for the Keap1-Nrf2 system: a hinge and latch mechanism. Biol. Chem. 387: 1311– 1320 [DOI] [PubMed] [Google Scholar]
- 34. Valko M, et al. 2007. Free radicals and antioxidants in normal physiological functions and human disease. Int. J. Biochem. Cell Biol. 39: 44– 84 [DOI] [PubMed] [Google Scholar]
- 35. Wang XJ, et al. 2008. Nrf2 enhances resistance of cancer cells to chemotherapeutic drugs, the dark side of Nrf2. Carcinogenesis 29: 1235– 1243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wood LD, et al. 2007. The genomic landscapes of human breast and colorectal cancers. Science 318: 1108– 1113 [DOI] [PubMed] [Google Scholar]
- 37. Xia B, et al. 2007. Fanconi anemia is associated with a defect in the BRCA2 partner PALB2. Nat. Genet. 39: 159– 161 [DOI] [PubMed] [Google Scholar]
- 38. Xia B, et al. 2006. Control of BRCA2 cellular and clinical functions by a nuclear partner, PALB2. Mol. Cell 22: 719– 729 [DOI] [PubMed] [Google Scholar]
- 39. Zhang DD, Hannink M. 2003. Distinct cysteine residues in Keap1 are required for Keap1-dependent ubiquitination of Nrf2 and for stabilization of Nrf2 by chemopreventive agents and oxidative stress. Mol. Cell. Biol. 23: 8137– 8151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zhang DD, Lo SC, Cross JV, Templeton DJ, Hannink M. 2004. Keap1 is a redox-regulated substrate adaptor protein for a Cul3-dependent ubiquitin ligase complex. Mol. Cell. Biol. 24: 10941– 10953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zhang F, Fan Q, Ren K, Andreassen PR. 2009. PALB2 functionally connects the breast cancer susceptibility proteins BRCA1 and BRCA2. Mol. Cancer Res. 7: 1110– 1118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zhang F, et al. 2009. PALB2 links BRCA1 and BRCA2 in the DNA-damage response. Curr. Biol. 19: 524– 529 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.