Abstract
Our concept of a stable genome is evolving to one in which genomes are plastic and responsive to environmental changes. Growing evidence shows that a variety of environmental stresses induce genomic instability in bacteria, yeast, and human cancer cells, generating occasional fitter mutants and potentially accelerating adaptive evolution. The emerging molecular mechanisms of stress-induced mutagenesis vary but share telling common components that underscore two common themes. The first is the regulation of mutagenesis in time by cellular stress responses, which promote random mutations specifically when cells are poorly adapted to their environments, i.e., when they are stressed. A second theme is the possible restriction of random mutagenesis in genomic space, achieved via coupling of mutation-generating machinery to local events such as DNA-break repair or transcription. Such localization may minimize accumulation of deleterious mutations in the genomes of rare fitter mutants, and promote local concerted evolution. Although mutagenesis induced by stresses other than direct damage to DNA was previously controversial, evidence for the existence of various stress-induced mutagenesis programs is now overwhelming and widespread. Such mechanisms probably fuel evolution of microbial pathogenesis and antibiotic-resistance, and tumor progression and chemotherapy resistance, all of which occur under stress, driven by mutations. The emerging commonalities in stress-induced-mutation mechanisms provide hope for new therapeutic interventions for all of these processes.
Keywords: adaptive mutation, evolution, mutation rates, error-prone DNA polymerases, stress responses, recombination, DNA repair, SOS response, σS, RpoS, cancer, genome instability, microbial pathogenesis, antibiotic resistance
INTRODUCTION
Evolution results from natural selection acting on variability in populations, which ultimately stems from mutations. In large populations of mostly asexual cells, such as bacteria, and also somatic cells such as in developing cancers, mutation is the most important driving force behind evolution. But how do mutations, and thus how does evolution, occur? Is acquisition of mutations constant and gradual, a ticking clock, independent of selective environments and inexorable, and is selection the panning by the environment for pre-existing genetic gold? Or does mutation, and by extension evolution, occur in bursts stimulated by changing selective environments? The former mutation mode has been recognized since the elegant demonstrations of Luria and Delbrück (1943) and the Lederbergs (Lederberg and Lederberg, 1952) that Escherichia coli virus-resistant and antibiotic-resistant mutants can arise before exposure to virus and antibiotics, respectively. Elegant mathematics describe a constant process dependent on cell generations, and perhaps the result of inevitable errors in DNA replication (Lea and Coulson, 1949; Luria and Delbrück, 1943) (called spontaneous generation-dependent mutagenesis). However, changing environments are often stressful to maladapted cells and organisms, and a growing literature indicates that the same cellular stress responses long appreciated to shore-up damaged cellular hardware (other than DNA) can, surprisingly and importantly, also remodel genomic software (DNA) by increasing rates of random mutagenesis. Stress-inducible mutagenesis mechanisms can potentially accelerate adaptive evolution in populations specifically when organisms are maladapted to their environments, i.e., when they are stressed, and then return genomes to low mutation rates in rare adapted mutants that thrive in the new environment and so are stressed no longer.
Stress-induced genomic instability has been studied in a variety of strains, organisms, stress conditions and circumstances, in various bacteria, yeast, and human cancer cells. Many kinds of genetic changes have been observed, including small (1 to few nucleotide) changes, deletions and insertions, gross chromosomal rearrangements and copy-number variations, and movement of mobile elements, all induced by stresses. Similarly, diversity is seen in the genetic and protein requirements, and other aspects of the molecular mechanisms of the stress-induced mutagenesis pathways. In this review, we will survey several cases/experimental systems in which genomic instability appears to be inducible by stress and focus on evidence bearing on the molecular mechanisms of stress-inducible mutagenesis in each. We will see that unlike, e.g., DNA replication and transcription, there is no single universal molecular mechanism of stress-inducible mutagenesis, but rather, it is a collection of mechanisms with similarities and differences. The good news is that there are several common components and broad themes evident in these mechanisms, which we shall highlight. For previous reviews of the history of ideas and early experiments in this field, see Foster (1999) and Rosenberg (2001).
In our view, the most interesting aspects of the emerging mechanisms of stress-induced mutagenesis are those either demonstrating or implying its regulation. We shall review many instances in which mutagenesis is coupled to cellular stress responses, which regulate mutagenesis in time. Stress responses restrict mutagenesis to times of stress, when, by definition, cells or organisms are poorly adapted to their environments. A second theme is potential restriction/localization of mutagenesis in genomic space via coupling of mutagenesis to localized processes, such as double-strand-break repair or transcription. This may localize mutagenesis within genomes, potentially reducing accumulation of non-adaptive mutations in rare adaptive mutants, and also potentially facilitating concerted evolution within genes or gene clusters.
The general picture across many strains, organisms and stress conditions is of diverse mechanisms with common elements and themes, which suggests multiple independent evolutions of stress-induced mutagenesis, that have probably been selected in changing environments, lost in static ones, and “re-invented” repeatedly.
Is There A Controversy?
It has been argued (Roth et al., 2006) that the existence of stress-inducible mutagenesis mechanisms is controversial. In our view, there is currently no real controversy. We shall touch on a specific mathematical model proposed to account for mutagenesis without stress-inducibility in one E. coli model (the Lac assay), and note several lines of experimental evidence that have superceded the mathematical model, demonstrated control of mutagenesis by stress responses, and provided a clear picture of a molecular mechanism of stress-induced mutagenesis. We compare this with several other strains, stresses, and organisms, illustrating the commonalities.
The Important Questions
In our view, the most important current questions in this field are not whether stress-induced mutagenesis occurs—the evidence for this being substantial and widespread—but rather what are the specific molecular mechanisms of stress-induced mutagenesis in many different experimental and natural systems/circumstances, organisms, and stresses? What are the common themes and components in these mechanisms, and what do they indicate about how evolution works? Can, and how might, such mechanisms be selected? We suggest that understanding of the common components in mechanisms of stress-induced mutagenesis will allow the design of anti-evolution drugs that will short-circuit mutagenesis responses that drive evolution of antibiotic resistance, microbial pathogenesis strategies, tumor progression, radiation- and chemotherapy-resistance mechanisms, all of which are driven by mutation and selection under stress.
MUTAGENESIS AND EVOLVABILITY
Constitutive Mutators Win in Competitive Environments
The concept of evolvability relates to the intrinsic capacity of organisms to evolve, and reflects the extent of genetic variability in populations (Sniegowski and Murphy, 2006). Most mutations in well adapted organisms are deleterious or neutral, and consequently low, constant mutation rates are thought to be advantageous in the long term. Although generation-dependent mutation rates are mostly low and remarkably constant between different organisms (Drake et al., 1998), bacterial mutants with constitutively high mutation rates (constitutive mutators) are fitter in competition experiments with non-mutators. This is true whether the mutation rate is elevated by defects in mismatch repair (MMR) (Table 1) (Gibson et al., 1970), the post-replicational error-correction system (Kunkel and Erie, 2005), or by transposon mobility (Chao et al., 1983), in competition between bacterial strains colonizing mice (Giraud et al., 2001), in repetitive rounds of selection in the lab (Mao et al., 1997), and is predicted by computer modeling (Taddei et al.,1997b) in which mutators increase the fitness of populations during the adaptation period, then burden the adapted cells with deleterious mutations after adaptation. In agreement with this idea, constitutive mutator mutants, most of which are MMR-defective, arise spontaneously and are selected in long-term cultures (Sniegowski et al., 1997), and are found much more often than expected in natural populations of pathogenic (LeClerc et al., 1996) and commensal (Matic et al., 1997) E. coli and Salmonella enterica, constituting up to 1% of individuals, and can be as prevalent as 20% of pathogenic Pseudomonas isolated from the lungs of patients undergoing antibiotic therapy (Blazquez, 2003). The mutator alleles “hitchhike” along with favorable mutations that they generate and so are selected indirectly (“second-order” selection).
TABLE 1.
Definitions of some components in stress-induced mutagenesis pathways
| RpoS/σS (General Stress Response Activator) | A bacterial transcriptional activator protein (sigma factor) that controls a large general stress response. The RpoS (AKA general-, starvation-, or stationary-phase stress) response is induced, upon entry into the stationary phase and in response to starvation, acid pH, osmotic shock, cold shock and oxidative stresses. RpoS upregulates transcription of ~340 genes (Weber et al., 2005), many of which play various roles in stress resistance. Reviewed by Hengge-Aronis (2002). |
| SOS Response | The DNA-damage stress response of bacteria. SOS upregulates ~40 genes that function in DNA repair, cell-cycle check-point control (cell division arrest), DNA damage-tolerance including translesion DNA synthesis via the use of three DNA polymerases Pol II, Pol IV (DinB), and Pol V (UmuD2C). RecA senses DNA damage and then facilitates auto-proteolytic cleavage of LexA transcriptional repressor, thus de-repressing the expression of the SOS regulon. Reviewed by Friedberg et al. (2006). |
| Competence Response of B. subtilis | Competence (for natural transformation) is a differentiated state in which cells take up and incorporate foreign DNA. Competence development occurs in response to nutrient deprivation and high cell density, controlled by the ComP ComA two-component regulatory system. The ComA response regulator activates expression of competence genes when phosphorylated by ComP. When cells become competent, auto-regulated transcriptional activator, ComK, mediates DNA synthesis arrest, inhibition of cell division, and expression of DNA-binding, uptake and recombination genes. Reviewed by Claverys et al. (2006). |
| Stringent Response | Amino-acid-starvation-stress response activated by nutritional deprivation in bacteria that upregulates aminoacid-biosynthesis genes, and is activated by guanosine nucleotides GDP 3′-diphosphate or GTP 3′-diphosphate, commonly referred to as (p)ppGpp. Other effects of ppGpp include facilitation of RpoS-dependent gene expression, and DNA replication arrest. Reviewed by Chatterji and Ojha (2001). |
| Release from Catabolite Repression | A response to carbon starvation that globally regulates carbon utilization genes. When released from catabolite repression, the repression of genes involved in the utilization of some sugars in the presence of better carbon sources, like glucose is released. The CAP/CRP protein mediates this response by binding cAMP and becoming a transcriptional activator for promoters containing a CAP binding site, like the lac, mal, gal, and ara operons. Synthesis of cAMP by adenylate cyclase (product of the cya gene) is turned on when the levels of glucose in cells are low. Reviewed by Snyder and Champness (2002). |
| PhoPQ Response | A two-component transcriptional regulatory system, that responds to several stresses such as acidic pH, low intracellular concentrations of divalent cations and antimicrobial peptides. Several genes involved in stress response and virulence are regulated by this system. Reviewed by Groisman (2001). |
| Mad1/Max and Mnt/Max Complexes | Max is a mammalian transcriptional regulator whose activity is modulated by interaction with several members of the helix-loop-helix leucine zipper family, like c-Myc and the Mad subfamily. c-Myc/Max complexes function in transcriptional activation, whereas complexes with Mad proteins such as Mad1, Mnt and MxiI are repressors. Expression of MMR genes, normally activated by c-Myc/Max complexes, is down-regulated during hypoxia due to the formation of repressive complexes of Max with Mad proteins. See Bindra and Glazer (2007a) and references therein. |
| p53 | Mammalian transcription factor regulating the expression of genes implicated in apoptosis, DNA repair and cell cycle arrest in response to DNA damage. p53 is a very important gate keeper of genome integrity, acting as a tumor suppressor gene. Mutations inactivating p53 are found in at least 50% of all human cancers. Reviewed by Toledo and Wahl (2006). |
| HIF-1alpha | A mammalian transcription factor regulating gene expression in response to hypoxia. Normally degraded by the ubiquitin-proteasome, this protein is stabilized under hypoxia conditions, leading to the transcriptional activation of genes containing HRE (hypoxia responsive element) sequences in their promoters. Reviewed by Ke and Costa (2006). |
| E2F4/p130 | E2Fs compose a family of mammalian transcriptional factors controlling genes involved in cell-cycle regulation, DNA-damage response and apoptosis. Depending on the family member and association with other proteins, E2Fs may function as transcriptional activators or repressors. In particular, E2F4 and p130 associate to form a repressive complex demonstrated to inhibit gene expression under hypoxia conditions. See (Bindra and Glazer, 2007b) and references therein. |
| BRCA1 | Human protein required for DSB repair via homologous recombination. Acts with RAD51. Defects cause breast-cancer predisposition. Reviewed by (Gudmundsdottir and Ashworth, 2006). |
| RAD51 | Eukaryotic RecA homologue required for DSB-repair via homologous recombination. Acts with BRCA1 protein in humans. Reviewed by Friedberg et al. (2006). |
| RecA | Bacterial DNA repair protein that catalyzes homologous genetic recombination (used in single-strand-gap repair and double-strand-break repair) and also induction of the SOS response. RecA forms a nucleoprotein filament on single-stranded DNA that promotes its interaction with homologous sequences in double-stranded DNA, and also activates the SOS response by promoting proteolytic cleavage of the LexA transcriptional repressor of the DNA damage-response genes. The eukaryotic homologue, Rad51, functions similarly in recombinational DNA repair. Reviewed by Cox (2007). |
| RecBCD | The main double-strand DNA end (DSE)-recognition and double-strand-break-repair (DSBR) enzyme complex of E. coli and related bacteria, composed of RecB, RecC, and RecD subunits. An exonuclease and helicase, RecBCD processes only DSEs at which it promotes DSBR by exposing single-strand DNA and loading RecA onto the single-strands. Reviewed by Friedberg et al. (2006). |
| RuvABC | E. coli recombinational DNA repair proteins that function in DSBR and other homologous recombination. This complex catalyzes endonucleolytic cleavage of Holliday-junction intermediates in recombination. Reviewed by Yamada et al. (2004). |
| DinB and Y-family DNA Polymerases | A DNA polymerase superfamily conserved in all three domains of life, with two representatives (of the five total DNA polymerases) in E. coli (DinB or Pol IV and Pol V or UmuD’2C), and four (of nearly 20 total) in humans. These are poorly processive, specialized DNA polymerases that catalyze “translesion” DNA synthesis, which allows replication forks halted at non-instructive DNA lesions to insert one or two bases and continue, allowing cell survival. Most are high-fidelity (not mutagenic) when inserting bases across from their cognate lesions but are highly error-prone/mutagenic when synthesizing on undamaged DNA. The DinB subfamily contains bacterial members required for stress-induced mutagenesis in several different assay systems and organisms, and is the only subfamily conserved in eubacteria, archaea, and eukaryotes (human orthologue of E. coli DinB: DINB1). Other subfamilies: UmuC (eubacterium-specific) Rad30 and Rev1 (eukaryote-specific). Reviewed by Nohmi (2006). |
| DNA Polymerase I | E. coli high-fidelity DNA polymerase used in DNA replication (Okazaki-fragment processing), various DNA repair pathways, and stress-induced gene amplification. Expressed constitutively. Reviewed by Friedberg et al. (2006). |
| DNA Polymerase II | E. coli high-fidelity DNA polymerase used in some DNA repair reactions and some stress-induced point mutagenesis pathways. Expressed constitutively and upregulated by the SOS response. Pol II may also assist the Y-family DNA polymerases IV (DinB) and V in translesion synthesis (Napolitano et al., 2000) and is also considered a specialized DNA polymerase. Reviewed by Friedberg et al. (2006). |
| DNA polymerase III | E. coli major replicative DNA polymerase, a high-fidelity DNA polymerase and an essential protein. Reviewed by Friedberg et al. (2006). |
| DNA polymerase V (UmuD’C) | E. coli Y-family translesion DNA polymerase responsible for “targeted” ultraviolet-light-induced SOS mutagenesis, and some other stress-induced mutagenesis responses. Reviewed by Friedberg et al. (2006); Schlacher and Goodman (2007). |
| Rev3/DNA Pol zeta | Eukaryotic translesion DNA polymerase not in the Y family. Also error-prone on non-lesion template DNAs. Causes mutations associated with DSBR in yeast (Holbeck and Strathern, 1997). |
| Mismatch Repair (MMR) | Highly and widely conserved DNA repair pathway that corrects errors in DNA synthesis post-synthetically by recognizing mispaired bases in DNA, and excising them from the new DNA strand. Perhaps the most important cellular enforcer of genetic and chromosomal stability, MMR—reduces mutations via post-synthesis DNA-polymerase-error correction; suppresses recombination between partially identical DNA sequences, which can lead to chromosome rearrangements; and inhibits transposon mobility. MMR-defective organisms/cells have high mutation rates (“mutator” phenotype) and cancer-predisposition in humans. Reviewed by Kunkel and Erie (2005). In several bacteria and human cells, stress induces transient dysfunction of MMR promoting transiently increased mutagenesis (reviewed here). |
| Non-Homologous DNA End Joining (NHEJ) and Ku Protein | A low-fidelity DNA DSB repair pathway, alternative to high-fidelity homologous recombination, in which DSEs are processed and then religated, mediated by Ku end-binding proteins. More likely to cause genome rearrangements (e.g., translocations, deletions) than DSB repair via homologous recombination. Operative in eukaryotes and some bacteria, not E. coli. Reviewed by Bowater and Doherty (2006). |
Genomic analyses suggest that there has been alternating selection for and against the functions of MMR genes during evolution (“periodic” selection) (Denamur et al., 2000); the MMR gene sequences are highly mosaic, within species, showing sequence characteristics of multiple species, indicating that they have been subject to multiple rounds of loss and reacquisition through horizontal transfer. This fits with the idea that higher mutation rates are selected in some, presumably variable, environments, but eventually become disadvantageous in well-adapted populations, where reacquisition of MMR capability is then selected.
These observations of selective sweeps driven by constitutive mutator mutants illustrate the point that selection favors high mutation rates in competitive environments, and imply that elevated mutation rate is under alternating (periodic) positive and negative selection. Stress-inducible mutagenesis allows the alternation between mutator and stable states to occur controlled by stress responses. Computer modeling and studies of natural isolates described below lead to similar conclusions concerning selection for this, we will suggest, somewhat more refined evolutionary strategy.
STRESS-INDUCED MUTAGENESIS IN NATURAL BACTERIAL POPULATIONS: WIDESPREAD AND SELECTED
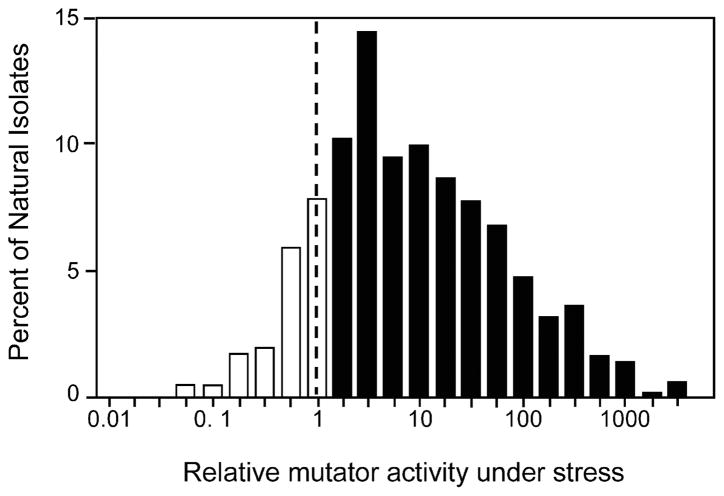
Like constitutive mutators, strains that increase mutation rate transiently during stress are found in natural environments, but appear to be far more abundant than constitutive mutators. Among 787 natural isolates of E. coli from a wide range of habitats worldwide, an astonishing >80% exhibited stress-inducible mutagenesis (Bjedov et al., 2003) (Figure 1). The authors analyzed mutations arising in bacterial colonies aged for 1 or 7 days. Rifampicin-resistant mutants increase in frequency among cells in the colony, which grows little after 1 day. Because the rifampicin-resistant mutants, which carry base-substitution mutations in the gene encoding RNA polymerase, have no growth advantage in these colonies (demonstrated with rigorous control experiments), this indicates an increase in rates of random mutagenesis in the old, stressed colonies. This finding was confirmed with other mutation assays. Several important observations and conclusions come from this study:
FIGURE 1.
Most E. coli natural isolates display stress-inducible mutagenesis. Bjedov et al. (2003) examined 787 natural isolates from various habitats for mutagenesis in aging colonies on solid medium, a starvation stress. The frequencies of rifampicin-resistant (base-substitution) mutants per viable cell after seven days in an aging colony relative to that after only one day are shown for all 787 strains. Numbers greater than one indicate induction of mutagenesis. More than 80% showed stress-inducible mutagenesis, indicating a common feature of many natural isolates. Figure re-drawn from Rosenberg and Hastings’s (2003) review of the Bjedov et al. paper.
First, the capacity for stress-inducible mutagenesis is prevalent in nature.
Second, the magnitude of stress-inducible mutability varied from a few-fold to more than a thousand-fold in the positive strains (Figure 1), revealing variability in the factors modulating and mechanisms of stress-induced mutagenesis in the different strains.
Third, the strengths of the stress-inducible mutagenesis phenotypes are negatively correlated with constitutive, generation-dependent mutation rates. This implies that strains adapt to changing environments via either a constitutive or a stress-inducible mutator pathway (suggested previously, Rosenberg et al., 1998) implying that, like constitutive mutator phenotype, stress-inducible mutability is selected.
Fourth, stress-inducible mutagenesis phenotypes are better correlated with ecological niche than with phylogenetic relatedness of strains, suggesting a recent and active role of the environment in modulating the magnitude of this stress response. This implies that, either directly or indirectly, stress-induced mutagenesis is selected (suggested also by Radman, 1974; Echols, 1981).
Fifth, computer modeling shows that stress-inducible-mutagenesis ability can be selected through the beneficial mutations that are generated, and the mutator pathway or genes hitch-hike along with them. This second-order selection for beneficial mutations and the hitchhiking of the mutator genes is as reviewed above for constitutive mutators, and is also shown by the negative correlation between constitutive and stress-inducible mutability in the study of Bjedov et al. (2003).
The mechanism of mutagenesis in one of the natural isolates from this study is discussed below (Mutagenesis in Aging Colonies) after many of the DNA repair and synthesis components are introduced in the following sections. Table 1 also introduces components discussed.
MECHANISMS OF STRESS-INDUCED MUTAGENESIS BEGINNING WITH CLASSICAL SOS MUTAGENESIS
Most of the examples discussed in this review concern mutagenesis induced during growth-limiting stress conditions to which cells are maladapted: selective environments such as starvation, hypoxia, antibiotic or other stresses. Some of the mutations produced can adapt the cell to the stress condition. These mechanisms have also been called “adaptive mutagenesis” and historically have been discussed separately from classical SOS mutagenesis, which is induced when DNA is damaged (but see Cirz and Romesberg, this volume). This historical separation might reflect a bias that DNA damaging stress might not reflect selective environments per se. Both Radman (1974) and Echols (1981) suggested that DNA-damage stress could reflect selective environments, and that SOS mutagenesis might speed evolution specifically during stress producing better adapted variants then. In agreement with their ideas, and because of the similarity between classical SOS mutagenesis and some of the previously-named “adaptive” mutagenesis mechanisms discussed below, we group them together here. The mechanism of stress-induced mutagenesis understood in greatest detail is classical SOS mutagenesis. Two excellent recent reviews cover the SOS DNA-damage response (Friedberg et al., 2006) and present the history and mechanisms of SOS mutagenesis (Schlacher and Goodman, 2007), current understanding of which is outlined in simplified form here.
SOS, the prototypic bacterial DNA-damage stress response, is induced when damage to DNA and/or or stalled replication forks cause exposure of single-strand (ss)DNA, the SOS inducing signal. ssDNA becomes coated with RecA protein which together form a nuceleoprotein filament. This filament of “activated” RecA can facilitate the autoproteolytic cleavage of LexA, a transcriptional repressor, leading to upregulation of roughly 40 genes involved in DNA repair, DNA synthesis past damaged bases, cell-division arrest, and other functions.
SOS mutagenesis is a consequence of the action of specialized, low-fidelity DNA polymerases induced by the SOS response. “Targeted” SOS mutagenesis occurs when an otherwise replication-blocking lesion in DNA is traversed by a special translesion-synthesis (TLS) DNA polymerase which may insert an incorrect base(s). It is targeted to the damaged DNA. In E. coli, DNA polymerase (Pol)V of the Y family of specialized, error-prone DNA polymerases (Table 1), encoded by the umuC and umuD genes, carries out TLS across from pyrimidine dimers and causes targeted SOS mutagenesis in response to UV damage. TLS allows the DNA to be replicated, and so become double-stranded, which is a prerequisite to repair and so is an important survival mechanism.
Targeted mutagenesis is sometimes regarded as an unavoidable consequence of this necessary damage-tolerance pathway. However, this view might be a simplification of the biology underlying SOS mutagenesis, as first appreciated by Radman (1974) and Echols (1981) (before TLS mechanisms were understood). We now know that TLS events can occur in a high-fidelity manner, as exemplified by E. coli DinB (Pol IV) action (Table 1) on guanine adducts, but causes SOS “untargeted” mutagenesis when operating on undamaged template DNA (reviewed by Cirz and Romesberg, 2007; Schlacher and Goodman, 2007). That is, it appears to be possible for TLS to occur without high-frequency mutagenesis, and these TLS polymerases cause mutations when not engaged in TLS, suggesting that mutagenesis might, as Radman and Echols suggested, itself be a selected feature of the SOS response. Regardless of whether or not this is so, both targeted and untargeted mutations are an important biological consequence of the induction of the SOS response.
The SOS response features prominently in many of the experimental systems showing an increase in mutagenesis in response to a variety of stresses, collectively called stress-induced mutagenesis in this essay (see below). Importantly, SOS-controlled inducible mutagenesis is a general phenomenon in bacterial physiology. Several bacterial species have been demonstrated to undergo SOS mutagenesis, and the vast majority of bacterial genomes encode one or more members of the Y-family of specialized DNA polymerases, which includes Pol V and DinB/PolI V discussed here.
THE E. COLI LAC SYSTEM: REGULATION BY TWO STRESS RESPONSES AND THE COUPLING TO DNA BREAK REPAIR
After classical SOS mutagenesis, the most detailed picture of a mechanism of stress-induced mutagenesis has come from studies of the E. coli Lac assay, in which current evidence indicates that stress-induced mutagenesis results from a stress-response-controlled switch in the fidelity of DNA double-strand break repair (DSBR) via homologous recombination (HR), from a high-fidelity mechanism in growing cells, to an error-prone, mutagenic version of that mechanism during starvation, controlled by two stress responses (Ponder et al., 2005).
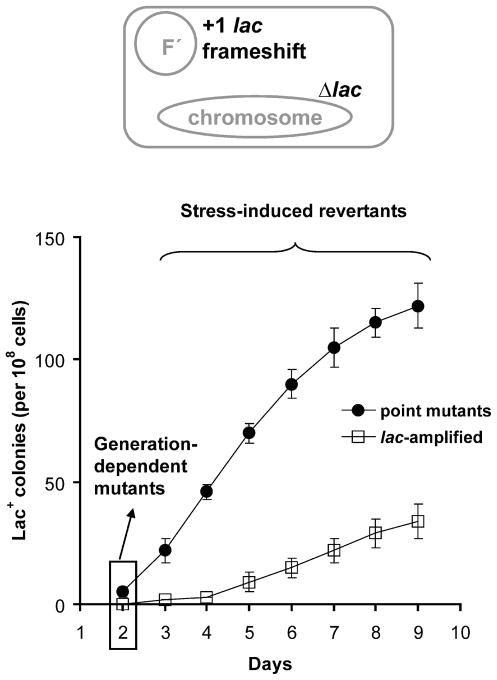
In the Lac assay, an E. coli strain carrying a lacI-lacZ fusion gene with a +1 frameshift mutation in lacI in an F′ conjugative plasmid, and a deletion of the chromosomal lac operon, is grown in a non-lactose carbon source then spread onto lactose plates on which only cells that become Lac+ form colonies (Figure 2) (Cairns and Foster, 1991). Generation-dependent Lac+ mutants that arose prior to starvation on lactose form visible colonies after ~2 days of incubation. Stress-induced Lac+ mutant colonies accumulate continuously from day 3 onward (Figure 2) from a population of stressed starving cells that show no net proliferation (Cairns and Foster, 1991).
FIGURE 2.
The E. coli Lac assay for stress-induced mutagenesis. Above: schematic representation of the strain used in the Lac assay. This strain bears a ~200 kb F′ conjugative plasmid carrying the mutant lacIZ33 allele, a lacI-lacZ fusion gene with a +1 frameshift mutation. Below: generation-dependent mutation events occurring during growth of the culture prior to plating on lactose medium are detected as Lac+ colonies present on about day 2. During subsequent days of incubation, stress-induced Lac+ colonies accumulate, and include both Lac+ point mutants with a compensatory frameshift mutation or lac-amplified cells, with 20 to 50 copies of DNA spanning the lac gene, which provides sufficient beta-galactosidase activity for growth without a frameshift reversion mutation (Hastings et al., 2004).
The stress-induced mutants include two types (Figure 2): Lac+ “point mutants” carry a compensatory frameshift mutation in lac (Foster and Trimarchi, 1994; Rosenberg et al., 1994), whereas lac-amplified cells, carry a tandem array of 20–50 or more repeats of a region of 7–134 kb including the lac gene (Hastings et al., 2000; Kugelberg et al., 2006; Powell and Wartell, 2001; Slack et al., 2006). The lac frameshift allele is leaky, producing ~1% of the beta-galactosidase activity of wild-type (Foster, 1994) such that multiple copies provide sufficient beta-galactosidase for growth on the lactose medium. lac-amplified clones are a minority in earlier days, but constitute up to 40% of colonies appearing on days 8 and 9 (Figure 2). Both the point mutants (Cairns and Foster, 1991; McKenzie et al., 1998) and lac-amplified clones (Hastings et al., 2000) result from genetic changes that occur after exposure to starvation (not slow-growing pre-existing mutants). Similarly, formation of both requires the RpoS-controlled general- or starvation-stress response, which is expressed in stationary-phase (Layton and Foster, 2003; Lombardo et al., 2004) (Table 1).
Because RpoS is required specifically for formation of these mutants (not merely for survival or colony formation under the conditions of the experiment, or for generation-dependent reversion) (Lombardo et al., 2004), we feel that these must now legitimately be called “stress-induced” point mutations and amplifications. Previous names for stress-induced mutagenesis (such as adaptive mutagenesis, and others reviewed by Rosenberg, 2001) suggest an artificial distinction from classical stress-induced SOS mutagenesis, which we think are more usefully grouped together.
The point mutagenesis mechanism will be reviewed here and the mechanism of stress-induced amplification of lac in a separate section (and in detail by Hastings, 2007).
Double-Strand-Break Repair Proteins, Two Stress Responses, the DinB Error-Prone DNA Polymerase and Limiting Mismatch Repair Function
Stress-induced Lac+ point mutations are different from generation-dependent Lac+ reversions and form via a different mechanism controlled by two stress responses. The following are aspects of the mechanism of stress-induced Lac+ point mutagenesis and will be part of common themes in many stress-induced mutagenesis mechanisms reviewed below:
Recombination and DSBR
The E. coli RecA (Cairns and Foster, 1991; Harris et al., 1994), RecBCD (Harris et al., 1994), and RuvABC (Foster et al., 1996; Harris et al., 1996) proteins (Table 1), used in DNA DSBR via HR, are required. When these genes are inactivated, generation-dependent (day-2) Lac+ colonies are present as usual, but late-arising (stress-induced) mutants are reduced ≥10-fold. RecF HR protein is also required for most point mutation (McKenzie et al., 2000). Some of these proteins play roles in induction of the SOS response, in addition to their roles in HR, but all are required in stress-induced point mutation for roles other than or in addition to SOS induction, as seen by the down phenotypes of these mutants in cells with SOS constitutively induced (He et al., 2006).
DinB Error-Prone DNA Polymerase
Formation of most (85%) of Lac+ stress-induced point mutants requires the DinB error-prone DNA polymerase (McKenzie et al., 2001), whereas generation-dependent mutagenesis does not (McKenzie et al., 2001, 2003; Wolff et al., 2004). DinB, a Y-family DNA polymerase (Table 1), has homologues in all three domains of life (Ohmori et al., 2001). Most, Y-family DNA polymerases, including DinB (Jarosz et al., 2006), are poorly processive TLS polymerases that allow the replisome to pass over otherwise replication-blocking lesions, promoting cell survival. Most, including DinB, are relatively high-fidelity (non-mutagenic) and insert the correct base(s) opposite their cognate lesions, but are highly error-prone and mutagenic on undamaged template DNA (Bjedov et al., 2007; Jarosz et al., 2006; Nohmi, 2006). Humans have four DinB homologues including one orthologue, DINB1 (Pol kappa), which is upregulated and promotes genome instability in small-cell lung cancers (Bavoux et al., 2005; Wang et al., 2001). The other three (Pol eta, Pol iota and REV1) all play some role in developmentally programmed somatic hypermutation of immunoglobulin genes (reviewed by Diaz and Lawrence, 2005). E. coli DinB is normally present in growing cells, but is transcriptionally upregulated about 10-fold by the SOS response (Kenyon and Walker, 1980; Kim et al., 1997) and about twofold by the RpoS stress response (Layton and Foster, 2003). DinB is also upregulated independently of the SOS response in response to beta-lactam antibiotics, which target synthesis of cell wall components (Perez-Capilla et al., 2005), though beta-lactam antibiotics induce the SOS response as well (Miller et al., 2004). Thus, DinB is upregulated by a variety of stressors. Cells lacking the GroE chaperone show reduced levels of DinB protein, indicating that it might also be regulated by its rate of degradation (Layton and Foster, 2005). DinB is very likely to be the DNA polymerase whose errors persist to become stress-induced Lac+ point mutations, because the errors spectrum of DinB in vitro (Tang et al., 2000; Wagner et al., 1999) and in vivo (Wagner and Nohmi, 2000) is predominantly −1 deletions in small mononucleotide repeats, which constitute nearly all stress-induced Lac+ point mutations (Foster and Trimarchi, 1994; Rosenberg et al., 1994), and also some base substitutions, predominantly G:C to T:A transversions (which would not be observed when selecting reversion of a frameshift allele).
SOS—the DNA Damage Stress Response
Induction of one or more genes of the SOS/LexA DNA damage response regulon (Table 1) is required for most point mutagenesis, roughly the same fraction as requires DinB (McKenzie et al., 2000, 2001). DinB is a probable candidate for the SOS-induced component required for stress-induced point mutagenesis, but this has not been demonstrated. Whereas point mutagenesis requires SOS and DinB, lac amplification requires neither (McKenzie et al., 2000, 2001).
The RpoS General/Starvation-Stress Response
This response is required for virtually all stress-induced Lac reversion (Lombardo et al., 2004). It is not known which component(s) of this stress response are required, but as we will see below, RpoS in some way licenses the use of DinB in DSBR during stress, making DSBR become mutagenic specifically at that time (Ponder et al., 2005). This might work directly via the observed ~two-fold upregulation of DinB by RpoS (Layton and Foster, 2003), or it might be via some other mechanism. DinB does not contribute to amplification (McKenzie et al., 2001), which requires RpoS (Lombardo et al., 2004), and so clearly RpoS controls at least one other factor that promotes genome instability under stress. At bare minimum, the important role of RpoS in mutagenesis in this system indicates that mutagenesis is a stress response, regulated temporally by RpoS (and SOS).
Mismatch Repair Becomes Limiting
Mismatch repair (MMR) is a highly and widely conserved DNA repair pathway that repairs mispaired bases and 1-few nucleotide insertion/deletion heteroduplexes arising from replication errors or other sources, increasing the fidelity of DNA replication 100 to 1000-fold (Kunkel and Erie, 2005) (Table 1). To begin repair, E. coli MutS and MutL proteins recognize the mispaired bases, and their eukaryotic orthologues (called MSH1-MSH6 for MutS homologues, and MLH1-MLH3 and PMS1 and PMS2, the MutL homologues) do similarly. Cells lacking MMR proteins are constitutive mutators (discussed above). The function of the MMR system becomes limiting transiently for mutation avoidance during stress-induced Lac+ mutagenesis via a limitation in functional MutL, as shown in experiments in which overproduction of MutL specifically reduces stress-induced and not generation-dependent Lac+ point mutagenesis (Harris et al., 1997, 1999) (or homologous recombination, Harris et al., 1999). The mechanism of MutL functional limitation is not understood. MutL protein levels do not decrease in most starving cells (Feng et al., 1996; Harris et al., 1997), though they might possibly do so in a cell subpopulation giving rise to stress-induced point mutants (discussed below). One possibility is that excess errors made by DinB polymerase titrate/saturate MutL, transiently exhausting MMR capacity (Harris et al., 1997). Overproduction of DinB has such a titrating effect on MutL (Wagner and Nohmi, 2000), as do other situations in which cells experience a heavy burden of DNA polymerase errors, including in an error-prone mutant of the replicative DNA Pol III (Schaaper and Radman, 1989). Stationary-phase conditions decrease the levels of MutS and MutH MMR proteins (Feng et al., 1996; Tsui et al., 1997), but MutS is not functionally limiting for mutation avoidance in the Lac system (Harris et al., 1997) and MutH has not been tested. MutS does become limiting during other stress situations discussed below.
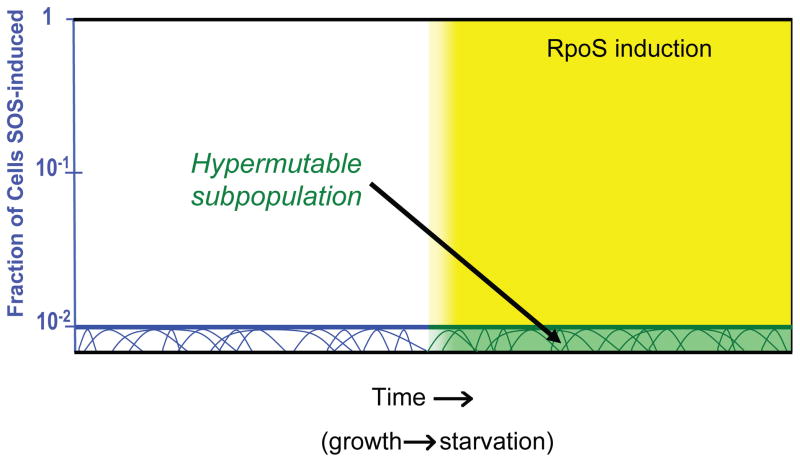
A Hypermutable Cell Subpopulation
Cells that have experienced a stress-induced Lac+ reversion show ~50-fold higher frequencies of unselected secondary mutations affecting many genes throughout their genomes (Godoy et al., 2000; Rosche and Foster, 1999; Torkelson et al., 1997), compared with either unstressed cells or their stressed Lac− neighbor cells from the same selection plates. This implies first, that mutagenesis is not specifically targeted to the lac gene or surrounding DNA (as had been hypothesized in early “directed mutation” models, e.g., Cairns et al., 1988). Mutagenesis of unselected non-lac genes was also shown in the Lac− stressed cells by direct observation of DSBR-protein-dependent reversions of a tetA frameshift allele next to lac in the F′ (Foster, 1997), and of the same allele in the chromosome (Bull et al., 2001). The latter was also shown to be DinB-dependent. Second, the 50-fold higher incidence of chromosomal mutations among Lac+ point mutants compared with the whole population of Lac− stressed cells on the plate demonstrates that a subpopulation of cells experiences increased mutagenesis relative to the main Lac− population. This subpopulation is transiently mutable (not composed of constitutive mutator mutants) as described in the following section. The possible origin of the hypermutable cell subpopulation (HMS) and the importance of its contribution to most Lac+ stress-induced mutagenesis will be considered below. We will suggest a model in which the HMS is differentiated based on coincident induction of the SOS and RpoS stress responses.
Transience of Stress-Induced Mutability
Once cells have acquired a Lac+ mutation that restores their ability to grow, they display normal mutation rates in various mutagenesis assays (Longerich et al., 1995), and this is also true for those with evidence of secondary mutations, demonstrating that the HMS is transient (Godoy et al., 2000; Rosche and Foster, 1999; Torkelson et al., 1997). Moreover, Rosenberg et al. (1998) reintroduced the lac frameshift allele into cells that had become Lac+ via stress-induced mutagenesis and showed that, when recycled though a second round of stress-induced-mutagenesis, these were no better than the original Lac− population at generating stress-induced Lac reversions. Thus, as expected for a process controlled by regulated gene expression of two transient stress responses (RpoS and SOS), stress-induced point mutability is a transient state.
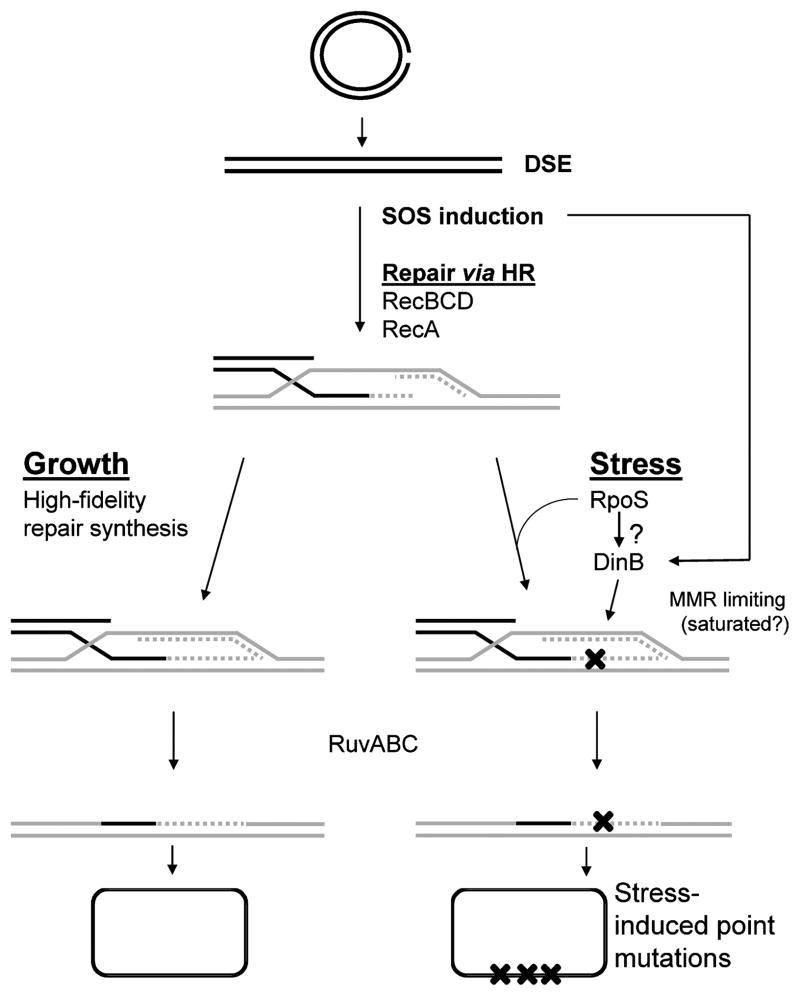
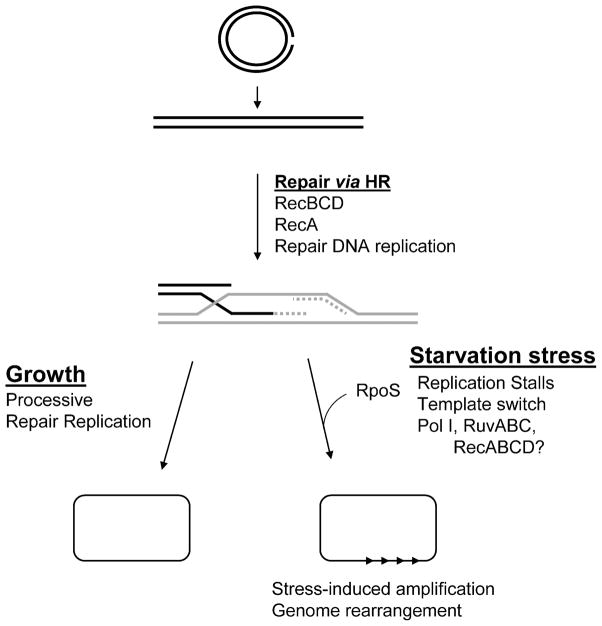
A Stress-Response-Controlled Switch from High-Fidelity to Error-Prone Double-Strand Break Repair
The basic model that stress-induced point mutagenesis results from error-prone-DSBR via HR (Harris et al., 1994) (a version shown in Figure 3) has received strong direct support (Ponder et al., 2005). Additionally, the same study made the important discovery that the RpoS stress response controls a switch from a high-fidelity DSBR mechanism (HR) to an error-prone version of that mechanism leading to mutagenesis specifically under stress (Ponder et al., 2005). In this study, the authors introduced a single genomic DSB with I-SceI endonuclease during stress-induced mutagenesis and found the following (Ponder et al., 2005):
FIGURE 3.
A switch from high-fidelity to error-prone DNA double-strand-break repair underlies stress-induced mutagenesis in the Lac system. Double-strand breaks are repaired via high-fidelity HR-DSBR in cells growing in optimal conditions. However, in growth-limited cells expressing the RpoS stress-response regulon, DSBs are repaired mutagenically under the control of RpoS, which somehow licenses the use of the DinB error-prone polymerase, which is upregulated by the SOS and RpoS responses. Mismatch repair (MMR) becomes limiting under this condition, and fails to correct many DNA polymerase errors, we suggest due to saturation/titration of MutL protein by excess DNA polymerase errors (described in the text). Single lines represent strands of DNA except in the two circular molecules at the bottom, where they represent whole bacterial chromosomes composed of double-stranded DNA. X’s represent DNA polymerase errors, and then mutations in the bottom-most molecule.
1. A DSB made near lac more than substitutes for the genetic requirement of stress-induced mutagenesis for TraI. TraI is an F-encoded single-strand endonuclease that nicks constantly at the F origin of transfer. Previous studies showed that some function(s) of the conjugative transfer operon were required for efficient stress-induced mutagenesis of genes in the F′ (Foster and Trimarchi, 1995; Galitski and Roth, 1995) and that these could be substituted by a phage-encoded single-strand endonuclease acting in the F, implying that nicking, not transfer, was the relevant component (Rodriguez et al., 2002). Single-strand nicks were proposed to be precursors to DSBs that promoted stress-induced lac reversion by error-prone DSBR (Kuzminov, 1995; Rosenberg et al., 1995). The results of Ponder et al. (2005) demonstrate that the requirement for TraI can be explained by the need to form DSBs. Lower-frequency spontaneous DSBs are thought to be the basis of the roughly 20-fold lower frequencies of DSBR-protein and DinB-dependent stress-induced mutagenesis in the E. coli chromosome as compared with the F′ (Bull et al., 2001).
2. Single DSBs made near lac stimulated the rate of mutation by ~6000 fold in a traI mutant strain. Both point mutants and lac-amplified clones were stimulated. Thus, DSBs promote both mechanisms. The I-SceI-promoted point mutations require RpoS, DinB, and the same HR-DSBR proteins as normal stress-induced point mutagenesis, and have similar sequences, all indicating that a similar or the same mutation mechanism as usually operates has been elevated 6000-fold (rather than a new mutation pathway activated).
3. I-SceI-induced DSBs promoted mutation ~6000-fold when made locally near lac, but only ~threefold when induced in different molecule in the cell (a plasmid). This shows that their main stimulatory effect is direct and local, suggesting that it occurs via generation of breaks that undergo error-prone repair. The small global stimulation probably occurs via enhanced SOS induction.
4. Compellingly, when DNA near lac carried DNA sequences identical to one end of the linearized plasmid that previously stimulated mutagenesis only weakly, then linearization of that plasmid promoted efficient mutation in the F′ at lac. This shows that homologous interaction between a DSE in one molecule and DNA in another, near lac, promoted mutation. Figure 3 illustrates how this is thought to occur during repair synthesis primed in acts of HR-DSBR.
5. Moreover, delivering DSBs near to a chromosomal lac allele in a different strain background can stimulate mutation, showing that availability of DSBs is the limiting event for chromosomal mutations (Ponder, 2006).
These data provide strong direct support for models in which stress-induced point mutations result from errors made during homologous recombinational DSBR. Additionally, they led to the discovery that a switch in the fidelity of HR-DSBR promotes mutation specifically under stress.
The RpoS-Controlled Switch to Mutagenic DSBR
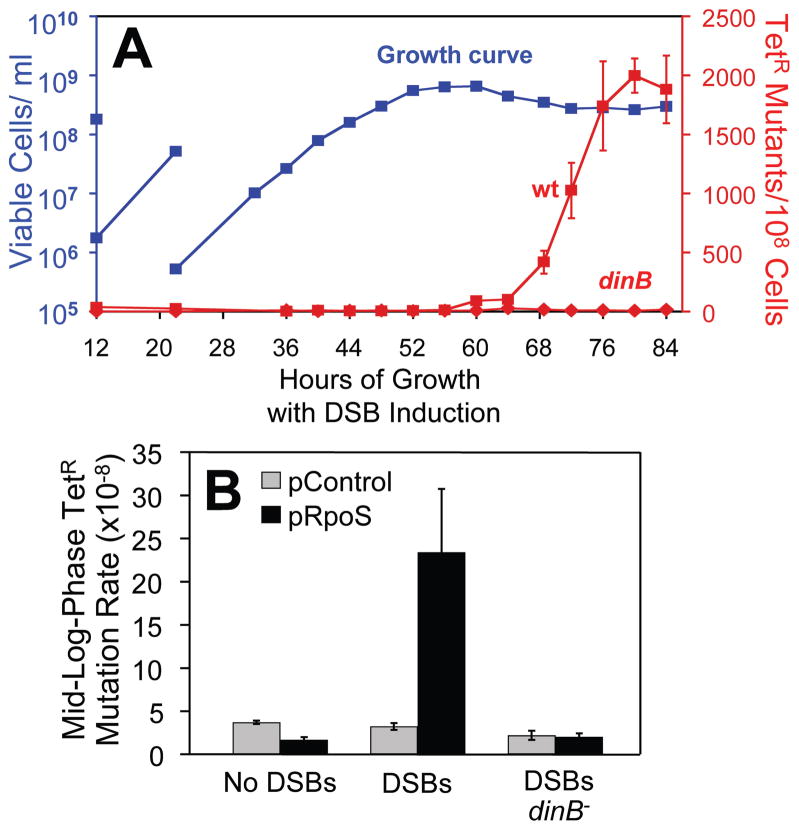
6. Ponder et al. (2005) showed that I-SceI-generated DSBs promote local DinB-dependent mutagenesis only in stationary phase, not during exponential growth (Figure 4A). Liquid cultures were grown and allowed to go stationary with continuous low-level I-SceI induction, and reversions of a tetA gene (near lac and the cutsite in the F′) were assayed at time points throughout. Only after stationary phase were the DSBs mutagenic (Figure 4A), though their formation and repair were measurable throughout. Again, the mutation was wholly DinB-dependent (Figure 4A).
7. However, when RpoS was expressed weakly from a plasmid, then, even during mid-exponential phase, the induced DSBs promoted mutagenesis (Figure 4B), which was both DinB- and enzyme-and-cut-site-dependent. RpoS did not alter the efficiency of repair (Ponder et al., 2005), merely its fidelity. This result demonstrates that RpoS controls a switch that makes the process of DSBR via HR mutagenic during stress, and implies that DSBR is mutagenic in stationary phase because RpoS is expressed then.
FIGURE 4.
The switch to mutagenic double-strand-break repair in stationary phase, or if RpoS is expressed. Data reprinted with permission from Ponder et al. (2005), on stress-induced mutagenesis in the E. coli Lac system. Here, reversion not of lac but of a tetA+1 frameshift allele near to lac is assayed in cells grown in medium without lactose (such that no DNA amplification is selected, reviewed in the text). (A) An I-SceI endonuclease-generated DSB made near tetA is induced at a low level continuously during exponential growth and then stationary phase (blue line represents growth curve), but provokes DinB-dependent tetA reversion (red lines) only in stationary phase. (B) Mutation rate data from a mid-logarithmic-phase time point from experiments as in A, but this time with the RpoS, general stress-response transcriptional activator protein produced weakly from a plasmid. DinB-dependent tetA reversion occurs during mid-log phase if RpoS is expressed and DSBs are induced, showing that stationary phase is not necessary if RpoS is supplied even at low levels.
REGULATION OF MUTAGENESIS AND EVOLVABILITY
Stress Responses Limit Mutagenesis in Time: A Strategy that Enhances Evolvability
Though long appreciated to promote adaptation to stress by protection of cellular hardware, results from the Lac system and many others now indicate that stress responses in addition to SOS also promote changes to genomic software. Most random mutations are neutral or deleterious. Control of stress-induced point mutagenesis by stress responses (SOS and RpoS, and others discussed below) limits the dangerous experiment of random mutagenesis specifically to times when cells are poorly adapted to their environments, by definition, when they are stressed (Lombardo et al., 2004; Ponder et al., 2005). Computer modeling indicates that as for constitutive mutators (reviewed above), the strategy of transient stress-inducible mutator ability should also enhance evolvability and adaptive evolution in competitive environments (Bjedov et al., 2003). In this regard, it is noteworthy that the most widely conserved (or independently evolved) aspect of various stress-induced mutagenesis programs is the requirement for one or more cellular stress responses, in bacteria, RpoS being the most frequent, and SOS less so but also frequent (Table 2, and reviewed below).
TABLE 2.
Common elements between various stress-induced mutagenesis pathways/mechanisms
| Components | Organism | Mutation Assay | Reference |
|---|---|---|---|
| Stress responses | |||
| Requirement for σS/RpoS general stress response | E. coli | Stress-induced point mutation, Lac assay | (Layton and Foster, 2003; Lombardo et al., 2004) |
| E. coli | Stress-induced amplification, Lac assay | (Lombardo et al., 2004) | |
| E. coli | DSBR-associated tetA-frameshift-reversion mutation1 | (Ponder et al., 2005) | |
| E. coli natural isolate | Mutagenesis in aging colonies (MAC) | (Bjedov et al., 2003) | |
| E. coli | Starvation-induced Mu excisions | (Gomez-Gomez et al., 1997; Lamrani et al., 1999) | |
| P. putida | Starvation-induced Phe+ point mutations | (Saumaa et al., 2002) | |
| P. putida | Starvation-induced Tn4652 transposition | (Ilves et al., 2001) | |
| Requirement for decreased function of σS/RpoS general stress response | E. coli | GASP mutations2 | (Zambrano et al., 1993) |
| SOS DNA damage response | E. coli | Stress-induced point mutation, Lac assay | (McKenzie et al., 2000) |
| E. coli | Mutagenesis in aging colonies (MAC) in laboratory strain | (Taddei et al., 1995) | |
| E. coli | Ciprofloxacin-induced resistance mutations in E. coli | (Cirz et al., 2005) | |
| Rad9/Rad17/Rad24 DNA-damage checkpoint | yeast | Telomere-shortening-stress-induced retrotransposition of Ty element | (Scholes et al., 2003) |
| ComAK competence differentiation/starvation-stress response | B. subtilis | Starvation-induced reversion of amino acid auxotrophy | (Sung and Yasbin, 2002) |
| Stringent response to amino acid starvation (ppGpp synthesis) | E. coli | Amino acid-starvation-induced, transcription-associated mutagenesis | (Wright et al., 1999) |
| B. subtilis | Amino acid-starvation-induced mutagenesis | (Rudner et al., 1999) | |
| Cyclic AMP/release from catabolite repression starvation response | E. coli | Mutagenesis in aging colonies (MAC) in laboratory strain | (Taddei et al., 1995) |
| E. coli | Starvation-induced Mu excisions | (Lamrani et al., 1999) | |
| PhoPQ regulon3 | E. coli | Starvation-induced mutations in the ebgR gene | (Hall, 1998) |
| Transcriptional repression by Mad1/Max and Mnt/Max complexes, which down-regulate genes in response to growth arrest | Human | Hypoxia-induced transcriptional down-regulation of MLH1/mismatch-repair function in human cells causing genetic (dinucleotide-repeat) instability | (Bindra and Glazer, 2007a; Mihaylova et al., 2003) |
| HIF-1alpha hypoxia-stress response and p53 DNA-damage response | Human | Hypoxia-induced transcriptional down-regulation of MSH2 and MSH6 mismatch-repair genes in human cells causing genetic (dinucleotide-repeat) instability | (Koshiji et al., 2005) |
| E2F4/p130-mediated transcriptional repression (response to radiation and oxidative damage and hypoxia stress) | Human | Hypoxia-induced down-regulation of RAD51, BRCA1 and DSBR via HR presumably leading to genome instability in human cells via NHEJ substituting in DSBR | (Bindra et al., 2005; Bindra and Glazer, 2007b) |
| Specialized DNA polymerases | |||
| DinB/Pol IV and other Y-family polymerases | E. coli | Stress-induced point mutation, Lac assay (DinB) | (Foster, 2000; McKenzie et al., 2001) |
| E. coli | Ciprofloxacin-induced resistance mutations, requiring three SOS-inducible DNA polymerases, two in the Y-family: DinB/Pol IV and UmuD’C/PolV | (Cirz et al., 2005) | |
| P. putida | Starvation-induced Phe+ point mutations (DinB homologue) | (Tegova et al., 2004) | |
| B. subtilis | Starvation-induced reversion of amino acid auxotrophy (DinB homologue) | (Sung et al., 2003) | |
| Pol II | E. coli natural isolate | Mutagenesis in aging colonies (MAC) | (Bjedov et al., 2003) |
| E. coli | Ciprofloxacin-induced resistance mutations, requiring SOS-inducible DNA Pols II, IV and V | (Cirz et al., 2005) | |
| Pol I | E. coli | Stress-induced amplification, Lac assay | (Hastings et al., 2004) |
| E. coli | Mutagenesis in aging colonies (MAC) in laboratory strain | (Taddei et al., 1997a) | |
| Rev3/DNA Pol zeta | S. cerevisiae | Mutations associated with DNA DSB repair via homologous recombination4 | (Holbeck and Strathern, 1997) |
| Limiting Mismatch Repair Function | |||
| MutL/MLH1 the limiting component | E. coli | Stress-induced point mutation, Lac assay | (Harris et al., 1997, 1999) |
| Human | Mutations induced by hypoxia in human cells, MLH1 transcriptionally down-regulated | (Mihaylova et al., 2003) | |
| MutS/MSH2/MSH6 the limiting component | E. coli natural isolate | Mutagenesis in aging colonies (MAC) | (Bjedov et al., 2003) |
| B. subtilis | Starvation-induced reversion of amino acid auxotrophy | (Pedraza-Reyes and Yasbin, 2004) | |
| E. coli | Decreased mismatch repair of heteroduplexes in phage DNA in stationary-phase cells | (Brégeon et al., 1999) | |
| Human | Dinucleotide instability in human cells, MSH2 and MSH6 transcriptionally down-regulated | (Koshiji et al., 2005; To et al., 2005) | |
| MutH a minor limiting component | E. coli | Decreased mismatch repair of heteroduplexes in phage DNA in stationary-phase cells | (Brégeon et al., 1999) |
| DNA Repair and Recombination | |||
| RecA/Rad51 | E. coli | Stress-induced point mutation, Lac assay | (Cairns and Foster, 1991; Harris et al., 1994) |
| E. coli | Stress-induced amplification, Lac assay | (Slack et al., 2006) | |
| E. coli | Ciprofloxacin-induced resistance mutations | (Cirz et al., 2005) | |
| E. coli | Mutagenesis in aging colonies (MAC) in laboratory strain | (Taddei et al., 1995, 1997a) | |
| S. cerevisiae | Mutations associated with DNA DSB repair via homologous recombination4 | (Strathern et al., 1995) | |
| Down-regulation of RecA homologue RAD51 required | Human | Hypoxia-induced down-regulation of homologous recombination, presumably leading to genome instability | (Bindra et al., 2005) |
| RecBCD | E. coli | Stress-induced point mutation, Lac assay | (Harris et al., 1994) |
| E. coli | Stress-induced amplification, Lac assay | (Slack et al., 2006) | |
| E. coli | Ciprofloxacin-induced resistance mutations | (Cirz et al., 2005) | |
| E. coli | Mutagenesis in aging colonies (MAC) in laboratory strain | (Taddei et al., 1997a) | |
| RuvABC | E. coli | Stress-induced point mutation, Lac assay | (Foster et al., 1996; Harris et al., 1996) |
| E. coli | Stress-induced amplification, Lac assay | (Slack et al., 2006) | |
| E. coli | Ciprofloxacin-induced resistance mutations | (Cirz et al., 2005) | |
| Ku/NHEJ proteins | S. cerevisiae | Stress-induced amino acid-auxotrophy reversion | (Heidenreich et al., 2003) |
| Human | Presumed for hypoxia-induced genome instability in human cells caused by NHEJ substituting for homologous recombination in DSBR due to down-regulation of RAD51 and BRCA1 | (Bindra et al., 2005) | |
| Localized Mutagenesis Processes? | |||
| Mutational clustering of unknown mechanisms | Mouse | Temporally and spatially clustered spontaneous mutations in mouse somatic cells5 | (Wang et al., 2007) |
| Many organisms | Multiple lines of evidence for clustering of mutations in many diverse organisms and in vitro5 | (Drake et al., 2005; Drake 2007) | |
| Mutagenesis associated with DSB repair | E. coli | Stress-induced point mutation, Lac assay | (Ponder et al., 2005) |
| E. coli | Stress-induced amplification, Lac assay | (Ponder et al., 2005; Slack et al., 2006) | |
| E. coli | Presumed for ciprofloxacin-induced resistance mutations | (Cirz et al., 2005) | |
| S. cerevisiae | Mutations associated with DNA double-strand-break repair via homologous recombination4 | (Strathern et al., 1995) | |
| S. cerevisiae | Presumed for stress-induced amino acid auxotrophy reversion dependent on Ku/NHEJ proteins | (Heidenreich et al., 2003) | |
| Human | Presumed for hypoxia-induced genome instability in human cells caused by NHEJ substituting for homologous recombination in DSBR due to down-regulation of RAD51 and BRCA1 | (Bindra et al., 2005; Bindra et al., 2004) | |
| Mutagenesis associated with nucleotide-excision repair (NER) | E. coli | Suggested for mutagenesis in aging colonies (MAC) assay based on requirement for UvrB NER protein | (Taddei et al., 1997a) |
| Mutagenesis associated with transcription | S. cerevisiae | Increased mutation in highly transcribed genes5 | (Datta and Jinks-Robertson, 1995) |
| E. coli | Amino acid-starvation-induced, transcription-associated mutagenesis | (Wright et al., 1999) | |
| E. coli | Transcription-coupled-repair-associated mutations suggested by genome sequence data | (Francino et al., 1996) | |
| B. subtilis | Possible transcription association implied by the requirement for the transcription-coupled-nucleotide-excision-repair factor, Mfd, for these stress-induced mutations. | (Ross et al., 2006). | |
These mutations associated with repair of a restriction-enzyme-produced DSB in vivo can form only if the cells are in stationary phase or if RpoS is expressed inappropriately in the log phase (implying that RpoS expression in stationary phase accounts for the stationary-phase specificity of the mutagenesis) (see Figure 4).
GASP (Growth Advantage in Stationary Phase) mutations are not demonstrated to be stress-induced mutations, but might possibly be. They are mutations that confer increased fitness that allows stationary-phase stressed cells to out-compete neighboring cells. It has not been determined whether the mutations occur before or after the cells enter stationary phase (the stress condition). Mutations that partly diminish function of σS arise early in the GASP process and confer a fitness advantage.
PhoPQ are part of a two-component transcriptional regulatory system involved with scavenging phosphorus and magnesium and are implicated in stress responses partly because they stabilize RpoS (Tu et al., 2006).
These mutations are not known to be starvation- stationary-phase- or stress-associated. Whether stress or stationary phase were involved the activity of this DSBR-associated mutagenesis mechanism was not examined.
Not known to be stress-associated.
Evolvability and the Regulation of Mutagenesis in Genomic Space via Its Coupling with DNA Repair
The coupling of mutagenesis to DSBR reported by Ponder et al. (2005) may have profound implications for the evolvability of organisms that do this, because of its potential to limit stress-induced mutagenesis in genomic space. Unlike housekeeping DNA replication, the replication primed by DSBR is localized to regions near DSBs or between a DSE and the terminus of replication. Ponder et al. (2005) saw that DSBs provoke stress-induced mutations well in the same molecule (in cis, ~6000X) but poorly in trans, in a different molecule (3X), and they suggested that the coupling of mutagenesis to DSBR might also localize mutagenesis within the bacterial chromosome. Whether DSBs do localize mutagenesis within molecules has not yet been tested, but if so, such localization could greatly enhance the adaptive value of the mutagenesis mechanism in two ways. Ponder et al. (2005) suggested that the DSBs that fuel chromosomal mutagenesis might be random spontaneous DSBs that potentially occur anywhere in the genome. In a large population of cells, therefore, the whole genome would be mutagenized, but in any given cell, only localized regions near one or more DSBs in the cell would be mutagenized. Thus, in rare cells that acquire an adaptive mutation, the chance of having acquired deleterious mutations in distant regions in the genome is much reduced. This principle is seen in somatic hypermutation of immunoglobulin genes and several other examples reviewed below.
Potential for Clustering of Mutations
Second, if localized mutated zones are heavily mutagenized, these might acquire multiple mutations, which could potentially promote concerted evolution within genes or gene clusters, such as operons. Multiple mutations are usually required for evolution of new protein functions (e.g. Camps et al., 2003). Ninio (1996, 2000) has discussed the potential evolutionary benefit of clustering mutations in genomic space for concerted evolution within genes, particularly for simultaneous acquisition of compensatory mutations that can ameliorate negative effects of otherwise beneficial function-altering mutations. He suggests that localized clustering of mutations associated with DNA repair processes is a possible selective advantage of constitutively “error-prone” DNA repair mechanisms and meiotic recombination. We suggest that mechanisms that promote clustered mutagenesis could be yet another potential reason for selection of clustering of functionally related genes in genomes, including in operons. In fact, during the course of the stress-induced mutagenesis experiments in the Lac system, mutations accumulating in the codAB genes next to lac were shown not to behave as independent events from Lac+ reversion, whereas mutations at an unlinked site did, suggesting that mutations do indeed occur in clusters (Bull et al., 2000). This phenomenon is echoed in several other mutagenesis mechanisms and circumstances discussed below and in Table 2. Moreover, local clustering of mutations has been argued to be more widespread than expected, and likely to arise from states of transient hypermutation (Drake et al., 2005; Drake 2007). In a study of the sequences of mutations and DNA polymerase errors made in vitro, Drake and colleagues show that the frequencies of multiple events are higher than predicted from the frequencies of singles, demonstrating clustering of mutations in diverse species and circumstances. More recently, evidence for mutational clustering in “showers” of local simultaneous mutations has been obtained in mice (Wang et al., 2007). The Lac system suggests a possible mechanism (coupling of mutagenesis to DNA repair) for mutational clustering.
Hypermutable Cell Subpopulation: Significance and Model for Its Origin
A central question is, how is the HMS differentiated? The coupling of stress-induced point mutagenesis to (at least) two different stress responses, SOS and RpoS, means that cells will not experience increased mutation rates until (at least) two different stress inputs are sensed simultaneously.
Model
We suggest that the overlay of the SOS and RpoS stress responses leads to differentiation of the HMS from the main population, and that this occurs as follows (Figure 5). Our laboratory showed recently that the SOS response is induced from spontaneous DNA damage in about 1% (steady-state levels) of cells in growing cultures, some from spontaneous DNA DSBs/DSEs and others from other spontaneous lesions (Pennington and Rosenberg, 2007). Some of the SOS-induced cells will, we suggest, both carry one or more DSB/DSEs and be induced at the right level of this graded response (Friedberg et al., 2006) to differentiate them into cells able to become HMS cells. However, we suggest that they will not become HMS cells unless the RpoS response is activated. This occurs when the growing population enters stationary phase (illustrated Figure 5) but should also occur in response to other RpoS-inducing stresses (reviewed above and Table 1). As discussed above, the SOS-regulated contribution to hypermutability is likely to be its 10-fold upregulation of DinB. The RpoS contribution licenses the use of DinB during DSBR (Ponder et al., 2005, reviewed above), though whether this occurs merely by the RpoS twofold upregulation of DinB levels (Layton and Foster, 2003), or by some other means, remains to be determined.
FIGURE 5.
Model for the origin of the hypermutable cell subpopulation in the Lac system: convergence of two stress responses. We suggest that the hypermutable cell subpopulation (HMS) associated with stress-induced point mutagenesis in the E. coli Lac assay system results from the convergence of cells experiencing both the SOS response (with a DNA double-strand break) and the RpoS response simultaneously. In this version of the model, spontaneous SOS induction and DNA double-strand breakage are constants at the roughly 0.6% level observed in logarithmically growing E. coli cells, with cells cycling into and out of the SOS-induced population at varying rates depicted by curves under the 1% SOS-induced population (Pennington and Rosenberg, 2007). SOS and DSBs are necessary but not sufficient for producing an HMS cell; induction of RpoS, shown as cells enter stationary phase, is also required. This model explains why there is a switch from high-fidelity to error-prone DSBR as cells enter stationary phase or when RpoS is induced (Ponder et al., 2005 and Figure 4).
SOS Induction Properties Can Help Explain HMS Properties
If correct, our model can potentially explain previous perplexing data concerning the HMS. A modern view of the SOS response shows that it is not induced as an all-or-none phenomenon; instead it is heterogeneous in the population, as seen in two studies using SOS promoters fused to gfp. Friedman et al. (2005) saw that SOS-gene expression occurs in waves of induction and repression, and that the cells in a single culture vary, those with greater induction having more cycles of the waves—staying SOS-induced for longer. Pennington and Rosenberg (2007) sorted spontaneously SOS-induced (green) cells by fluorescence-activated cell sorting, and found vastly different times to resumption of proliferation after the response, some forming colonies normally and others in a senescence-like state of viability without proliferation for many hours or days. Also, they found heterogeneity in the SOS response in cells induced due to a single, repairable chromosomal I-SceI-generated DSB: ~27% induced SOS detectably and the rest did not. These results indicate that not all DSB carriers may necessarily induce SOS (relevant to amplification, below), and suggest that those that do may also have a heterogeneous response. All of these results imply that the lengths of time that individual HMS cells spend with SOS induced (and thus in the HMS) is likely to vary considerably.
We suggest that this variability in the length of time that individual cells are induced underlies the observed heterogeneity in accumulation of secondary mutations among Lac+ cells: some of the point mutants arising during the experiments have a much higher frequency of unselected mutations than others (Rosche and Foster, 1999; Torkelson et al., 1997). We suggest (per Bull et al., 2001) that this results from individual cells remaining in the HMS for varying times, as dictated by their variable times remaining SOS induced (illustrated in Figure 5), in addition to their variable time before a Lac+ mutation is encountered, which would end the HMS state when cells grow and turn off RpoS. Also, as predicted by DSBR models, the extent of genome-wide mutagenesis is expected to vary due to uneven distribution of DSBs in cells in the population. In this way, a single differentiated subpopulation (undergoing the SOS and RpoS stress responses) may include cells with widely varying frequencies of secondary mutations, as a consequence of both different lengths of time spent in the transient mutable state and the limiting availability of DSBR events in the genome.
An alternative explanation is that only some stress-induced Lac+ point mutants arise from the HMS (Rosche and Foster, 1999). Those authors suggest that ~90% of Lac+ colonies (including those with no detectable secondary mutation) do not and ~10%, including those with visible secondary mutations, do. In that model, those in the HMS all remain in it for the same length of time and therefore generate about the same number of secondary mutations per cell, but many stress-induced Lac+ clones were never in it and arose via some other means. We find the first model to be simpler and more harmonious with current understanding of the SOS response and the role of DSBR in stress-induced mutagenesis; however, more experimental work is needed to distinguish these models definitively. Our recent work provides support for the first hypothesis by showing first, that Lac+ mutations from demonstrated HMS descendants (cells with a detected secondary mutation) are indistinguishable from most Lac+ mutations implying a common origin; and second, that secondary mutations cannot be uncoupled from most Lac+ mutagenesis when the latter is increased dramatically by I-SceI-induced DSBs. The two processes appear inseparable (Gonzalez C., Ponder R.G., Price M., Hastings P.J., and Rosenberg S.M., unpublished observations).
Other Models
Roth and colleagues (2006) favor a mathematical model for Lac+ point mutagenesis in which there is no transient increase in mutation rates caused by stress responses (Roth et al., 2006). In this model, a small fraction of the cells plated in the experiment have a pre-existing spontaneous duplication of a region containing the leaky lac gene. They propose that the cells carrying duplications can grow on lactose, starting a very slow process of colony formation. Under the strong selection for lactose utilization, eventual recombination-mediated expansion (gene amplification) to many lac copies is favored, improving growth and increasing the chance of acquisition of generation-dependent mutations in the extra copies of lac. This is proposed to be unrelated to stress, and in fact to occur in the least stressed cells on the plate: the ones growing most rapidly (which should not have RpoS induced). They propose that Lac+ point revertants then take over the colony due their optimal growth rate, giving rise to colonies containing a vast majority of point mutants and some remaining lac-amplified cells.
Many aspects of this model are incompatible with previous data, much of it reviewed previously, and we refer readers to the following papers for summary of the literature prior to mid-2004 (Foster, 2004a, 2004b; Rosenberg and Hastings, 2004a, 2004b; Roth and Andersson, 2004a, 2004b, 2004c). At least four important additions to the literature since those reviews have not been reviewed previously. These modern data show that the mathematical model cannot explain most of point mutagenesis in the Lac system.
Evidence that Mutagenesis is Stress Induced
1. Most obviously, mutagenesis in the Lac system has been demonstrated to be a stress response, controlled by RpoS and SOS. The data of Ponder et al. (2005) reviewed above show that DSBR switches to an error-prone mutagenic mode, using DinB, either when cells enter stationary phase or when RpoS is expressed in exponential cells (Figure 4). We know of no argument against these data, or the previous demonstrations that RpoS is required for virtually all stress-induced Lac reversion (Layton and Foster, 2003; Lombardo et al., 2004). Moreover, control by RpoS and other similar stress responses is a widespread bacterial strategy (Table 2 and following sections of this review).
2. Second, DSBR and the stress response or stationary phase are all that is needed to elicit the stress-induced point mutagenesis—amplification is not. In the experiments measuring DSB-stimulation of tet gene reversion in stationary phase (Figure 4A) or with RpoS expression in log phase (Figure 4B) (Ponder et al., 2005), there is no lactose (or tetracycline) in the medium and thus no selection for amplification of DNA including the tet gene near lac. Without selection for multiple copies of some gene, strains carrying amplification do not maintain amplification (Hastings et al., 2000). Thus amplification of lac, tet, or even the dinB gene (proposed to be required in some versions of the model, Roth et al., 2006) cannot be precursors to these stress-induced mutations.
3. In work leading to a similar conclusion, Stumpf et al. (2007) showed that high-level reversion of a tetA allele in the same molecule with lac is not higher if that gene is close to lac and can be co-amplified with lac. This again indicates that amplification is not a precursor to mutagenesis.
4. Some versions of the model of Roth et al. (2006) suggest that dinB, which is near lac, must be located in the F′, where it is expressed more highly (Kim et al., 2001), to cause mutagenesis: a presumed special circumstance, not general to most bacteria. This has been shown not to be true for DinB-dependent chromosomal Lac+ reversions (Ponder, 2006) and is also not the case for the many examples of DinB-dependent stress-induced mutagenesis in E. coli and in other bacterial genera reviewed below (and Table 2).
Finally, previously, the very strongest experimental support for the model of mutations promoted by selection for amplification and growth without stress came from a single experiment reported by (Hendrickson et al., 2002). These authors tested the idea that amplification of lac was a requirement for point mutagenesis by asking whether counter-selection of increased copy number of a tet gene next to lac, which would co-amplify with lac, inhibited stress-induced point mutagenesis, and they reported that it did and that counter-selection of a distant copy of that gene did not.
5. That strongest result has proven to be unrepeatable by another laboratory. Stumpf et al. (2007) find that counter-selection of the gene inhibits lac reversion regardless of its proximity to lac (and ability to affect lac amplification). Thus, this experiment can no longer provide clear support (see also Rosenberg and Hastings, 2004a).
6. Finally, the specific predictions of colony composition and the relationship of amplification to point mutagenesis have not held up to experimental testing (Hastings et al., 2004). In the mathematical model, a microcolony of 105 cells with 100 copies of lac (107total copies) was suggested to mutate at a nearly “normal” generation-dependent rate near 10−7. That means that colonies should contain, on average, only lac-amplified cells until they grow to be ~105 cells big. Hastings et al. (2004) showed that very young colonies are either purely point mutant or amplified, not all purely amplified, as the model demands. Even colonies at the two-cell stage were purely point mutant. This is impossible if 107 lac− copies must precede a Lac+ point mutation. They also separated amplification and point mutagenesis genetically, showing that amplification can be eliminated by polA mutation, inactivating DNA Pol I (Hastings et al., 2004; Slack et al., 2006), without altering point mutagenesis. This is not possible if amplification is a precursor to point mutations. Hastings et al. (2004) report several other critical specific tests, results of which were incompatible with the model.
Thus, we feel that there can be no real controversy regarding whether mutation rates are increased in response to stress in the Lac system. Moreover, the generality of this strategy in many other systems (Table 2 and reviewed below) makes the conclusion widespread.
STRESS-INDUCED AMPLIFICATION: GENOME REARRANGEMENT INDUCED BY DSBS AND CONTROLLED BY A STRESS RESPONSE
Gene amplification is a phenomenon widely observed both in prokaryotic and eukaryotic cells, where it may play important roles in antibiotic resistance and tumorigenesis. For example, amplification of oncogenes is associated with several types of cancer (reviewed by Savelyeva and Schwab, 2001; Volgelstein and Kinzler, 1998); amplification of the DHFR gene induced by the chemotherapeutic agent methotrexate confers tumor resistance to that drug (Huennekens, 1994), and amplification of the beta-lactamase-encoding ampC gene of E. coli promotes resistance to beta-lactam antibiotics such as ampicillin (Edlund and Normark, 1981). As reviewed above, in the E. coli Lac system, amplification of DNA spanning the lac frameshift allele is an adaptive outcome, alternative to point mutation (Hastings et al., 2000, 2004), in which cells acquire 20 to 50 or more tandem repeats of a region containing 7–134 Kb, of the lac-containing DNA, allowing growth on lactose medium (Hastings et al., 2000; Kugelberg et al., 2006; Slack et al., 2006). The lac-amplified colonies appear later than the point mutants (Figure 2) and can be distinguished from point-mutant colonies based on their unstable Lac+ phenotype; upon re-plating on rich medium containing a color indicator for beta-galactosidase activity, such as X-gal, lac-amplified cells give rise to both white (Lac−) and sectored colonies (blue colonies with white sectors), which indicates deamplification of the repeat array by recombination during growth of the colony (discovered and understood by Horiuchi et al., 1963; see also Tlsty et al., 1984). These contrast with the solid blue colonies formed by true lac revertants allowing discrimination between the two classes of event. This has allowed determination of the genetic requirements specific to each (reviewed by Hastings, 2007).
Current understanding of the possible mechanism of stress-induced amplification is reviewed in detail (Hastings, 2007), and here we review only three aspects relevant to the comparison of many mechanisms, though the genetic requirements are summarized in Table 2. Please see that paper for discussion of possible alternative models to the one described here.
The RpoS Response, Coupling to DSBR, and Long-Distance Template-Switching During Repair Replication
Stress-induced lac amplification is a stress response and requires the RpoS general-stress-response master regulator (Lombardo et al., 2004) (Table 1). Amplification requires homologous recombination and DSBR proteins RecA, RecBCD, RuvABC (Slack et al., 2006), and is strongly stimulated by an I-SceI-introduced DSB (Ponder et al., 2005), indicating that DSBs provoke amplification and that HR-DSBR is part of some stage of the stress-induced amplification mechanism. However, the junction sequences for the tandemly repeated DNA segments containing lac indicate that the event that creates a tandem duplication is not homologous recombination, but rather is a “non-homologous” or microhomologous event between 3- and 6-base G-rich imperfectly identical sequences (Kugelberg et al., 2006; Slack et al., 2006). The process also specifically requires the 5′-to-3′ flap endonuclease activity of DNA polymerase I (Pol I, Table 1) (Slack et al., 2006) which is used for removal of RNA primers during lagging-strand replication, and also in some DNA repair reactions, which are not part of amplification (Slack et al., 2006). Those authors propose and support a model in which repeats are formed specifically during stress, by an act of template-switching of the nascent lagging strand from one replication fork to another where it base pairs to a region of microhomology. The switch requires that the initial DNA replication fork should stall. Slack et al. (2006) suggested that the role of DSBs in this processes is that the DNA replication that stalls, allowing the template switch, is replication initiated in acts of DSBR (Figure 6) and thus not under the multiple layers of control of a replication origin, which disallow replication initiation in starving cells. Replication from origins, they suggest, would not pause as frequently. The specific model for how the duplication is generated, and its subsequent amplification by HR between sister chromosomes, is reviewed in detail in another review in this series (Hastings, 2007). Here we wish to highlight three points.
FIGURE 6.
Model for amplification induced by rearrangement-provoking replication fork stalling events, and template switching, during acts of DSBR under stress. Model from Slack et al. (2006), and discussed in the text. Single lines represent strands of DNA except in the two circular molecules at the bottom, where they represent whole bacterial chromosomes composed of double-stranded DNA.
No Hypermutation, SOS or DinB
Unlike Lac+ point mutants, lac-amplified cells show no evidence of genome-wide hypermutation (Hastings et al., 2000), and amplification does not involve the DinB polymerase or the SOS response (McKenzie et al., 2001). The lack of hypermutation in lac-amplified clones probably reflects the lack of requirement for SOS, which we proposed is required, along with RpoS induction, for cells to enter the HMS (Figure 4). Pennington and Rosenberg (2007) find that only ~27% of cells with a reparable chromosomal DSB generated by I-SceI induce SOS, such that only 27% of the amplified clones are predicted to have been hypermutated, a figure which is not ruled out by current data on (lack of) hypermutation among lac-amplified clones (Hastings et al., 2000). Thus, amplification is a process of genome rearrangement controlled by RpoS and not SOS.
Stress-Response Regulation, Genomic Localization of Genome Rearrangements and Evolvability
First, Like Lac+ point mutagenesis, stress-induced amplification is regulated temporally by the RpoS response (Lombardo et al., 2004), such that, in this case, genome rearrangement will occur specifically when cells are stressed and induce the RpoS response. A general model for the switch to rearrangement-provoking replication under stress is shown in Figure 6. Second, the coupling of stress-induced amplification to acts of DSBR (Ponder et al., 2005, reviewed above) should limit amplifications, and other non-homologous rearrangement events, in genomic space to presumably rare sites of spontaneous double-strand breakage in the bacterial chromosome. Could the coupling of genome rearrangement to DSBR be evolutionarily advantageous, e.g., for limiting deleterious rearrangements in the genomes of rare adaptive mutants? Amplifications are transient and unstable, such that those not selected are likely to be lost. However, other genome rearrangements might also be induced via the proposed template-switch model, such as deletions and/or inversions. Limitation of the number of sites per genome that experience rearrangement via its coupling to DSBR could provide an evolutionary advantage similar to that suggested for point mutagenesis: the limitation of deleterious rearrangements in cells that acquire a rare beneficial rearrangement.
ANTIBIOTIC-INDUCED RESISTANCE MUTATIONS
Ciprofloxacin Resistance in E. coli: SOS Response and Error-Prone Double-Strand Break Repair
Stress-induced mutagenesis was predicted to be important for evolution of drug resistance by pathogens (Martinez and Baquero, 2000), and recent work with the antibiotic ciprofloxacin shows a drug-induced mutation pathway nearly identical to stress-induced point mutagenesis in the Lac system described above. Ciprofloxacin is a widely used broad-spectrum antimicrobial agent of the fluoroquinolone family. Quinolones are inhibitors of type II topoisomerases, and known inducers of the SOS response (reviewed by Drlica and Zhao, 1997). In E. coli, point mutations in the gyrase genes gyrA and gyrB, and in the topoisomerase IV genes parC and parE are the main source of resistance to this drug (Drlica and Zhao, 1997). After exposure to this bacteriocidal drug, the ~1% of cells that survive experience no net growth while they generate new resistant mutants for days, mutants that demonstrably did not pre-exist in the cultures before treatment (Riesenfeld et al., 1997). No stepwise improvement of the resistance was detected in the cells on the plate, as determined by measuring the minimum inhibitory concentrations (MICs) of sensitive cells surviving in the plate.
SOS DNA Pols
A recent breakthrough study has provided insight into the mechanism of stress-induced mutagenesis giving rise to ciprofloxacin resistance. Using both laboratory culture experiments and a murine infection model, Romesberg and colleagues showed that E. coli cells are unable to develop resistance either in culture or during infection if they carry a special uncleavable LexA transcriptional repressor, which prevents induction of SOS genes (Cirz et al., 2005; and reviewed in detail by Cirz and Romesberg, this volume). The requirement for the SOS genes in producing ciprofloxacin-resistant mutants probably reflects a need for the induction of the SOS-controlled DNA polymerases Pol II, DinB/Pol IV and Pol V (Tables 1, 2), which they showed are required for mutation to resistance. Mutants defective for one or more of these polymerases were defective in post-exposure mutagenesis, but not in survival during the experiment. Of these DNA polymerases both DinB and Pol V are Y-family error-prone DNA polymerases, and all three are specialized DNA polymerases implicated in translesion DNA synthesis (Table 1). The results reveal a mechanism of SOS-dependent increase in mutation rates after antibiotic stress, that, like point mutagenesis in the Lac system, requires DinB, in addition to the two other SOS and translesion DNA polymerases. They suggest that the Lac system appears to require only DinB because only frameshift reversions are selected (not other alterations of sequence achieved by the other polymerases), i.e., that the two mechanisms are the same in this respect (Cirz and Romesberg, 2007). Moreover, the ciprofloxacin-induced mutagenesis pathway is different from spontaneous mutagenesis leading to ciprofloxacin-resistance in that the latter requires neither SOS nor the SOS DNA polymerases (Cirz et al., 2005; Cirz and Romesberg, 2006). Thus, the mutations arising before and after the antibiotic stress are formed by fundamentally different molecular mechanisms controlled by the SOS response.
DSBR Mechanism
Also like stress-induced Lac point mutagenesis, the ciprofloxacin-induced resistance mutagenesis requires HR-DSBR proteins RecA, RecBC and RuvABC (Cirz et al., 2005). The authors propose a mechanism for the mutagenesis that begins when the antibiotic traps a type II topoisomerase (one that cleaves both strands of DNA prior to strand passage) on DNA that it has cloven but not religated, in a DSB intermediate (see Cirz and Romesberg, this volume). They propose that these DSBs are converted into SOS inducing signals (single-strand DNA), the SOS DNA polymerases are upregulated, and that HR-DSBR then provides an opportunity for error-prone synthesis catalyzed by the SOS DNA polymerases. Their model is similar to the error-prone DSBR model now well supported in the Lac system (e.g., Figure 3). Their data imply a mutagenesis mechanism of error-prone HR-DSBR induced by stress, this time with the stress being caused by the antibiotic rather than starvation. As in the Lac example reviewed above, and the stress is communicated via the SOS stress response.
Antibiotic Therapy
An important implication of this work of interest to the authors is that new drugs that inhibit the mutagenic process itself should be included with the antibiotics to prevent evolution of resistance mutations induced by antibiotics (Cirz et al., 2005, 2006a). In this case, inhibitors of the SOS response, RecA, RecBCD, Ruv proteins or the SOS DNA polymerases might all be appropriate candidates.
Rifampicin Resistance in Mycobacterium tuberculosis
A very similar involvement of the SOS response in the generation of antibiotic resistance during infections was shown in Mycobacterium tuberculosis. In this bacterium, the dnaE2 gene, encoding a second copy of the catalytic subunit of Pol III (Table 1), is regulated by the SOS response (Davis et al., 2002), and is responsible for mutations resulting in rifampicin resistance during infection (Boshoff et al., 2003). Rifampicin inhibits bacterial RNA polymerase. dnaE2 is always accompanied in bacterial genomes by a putative Y-family DNA polymerase (Table 1) encoded by the imuB gene (Galhardo et al., 2005), and these genes seem to be highly conserved components of the SOS response in bacteria (Abella et al., 2004), as recently demonstrated by their LexA-dependent upregulation by ciprofloxacin in Pseudomonas aeruginosa (Cirz et al., 2006b). Thus, upregulation of specialized polymerases by the SOS response is a widely conserved response in bacteria, likely to be relevant in antibiotic-resistance acquisition. The importance and generality of the upregulation of SOS by antibiotics is also indicated by the positive regulation of both the SOS response and error-prone polymerases by various antibiotics (Miller et al., 2004; Perez-Capilla et al., 2005), including, in the latter two studies, beta-lactam antibiotics, which act on cell wall components (not DNA).
MUTAGENESIS IN AGING COLONIES (MAC)
A Natural Isolate: Two Stress Responses, RecA, DNA Pol II, Limiting MMR
The assay system with which 787 E. coli world-wide natural isolates were screened for stress-inducible mutability, described above and in Figure 1, uses E. coli colonies that are allowed to grow old on solid medium, from which the frequencies of unselected rifampicin-resistant (Rifr) mutants are determined after one and 7 days of incubation. The stress-inducible mutagenesis mechanism operating in one natural isolate requires two stress responses: the RpoS-controlled general- or starvation-stress response (reviewed Table 1), and release from catabolite-repression, a carbon-starvation response (Table 1) (Bjedov et al., 2003). Clearly, genesis of the mutants is stress induced. The mechanism also requires oxygen, and DNA Pol II, a relatively high-fidelity DNA polymerase, RecA recombinational DNA repair protein, and a transient limitation/down-regulation of MMR protein MutS, decreasing MMR activity, as shown by the specific inhibition of stress-induced mutagenesis by overproduction of MutS, restoring MMR function (Table 1). The authors suggest a possible mutagenesis mechanism of error-prone repair of oxidative damage to DNA, a known function of Pol II. Components of this mechanism that are common among other stress-induced mutation mechanisms include—regulation by stress responses, particularly RpoS (most common, Table 2); involvement of RecA; use of a specialized (though not an error-prone) DNA polymerase; and transient limitation of MMR, which becomes limiting via limiting MutS (Table 2), not MutL as in the Lac system (discussed above). The proposed mechanism also fits with a theme of mechanisms that potentially localize mutations in genomic space via coupling to a localized process (DNA repair).
A Lab Strain: Three Stress Responses, RecA, DNA Pol I, Limiting MMR
The same assay system used to examine the 787 E. coli natural isolates was developed and used first with a laboratory strain of E. coli. In these experiments (Taddei et al., 1995), cell number increased ~10-fold (only 3 to 4 generations), but Rifr mutant frequencies increased 50-fold over the 7 days, implying mutagenesis induced during growth-limiting stress. In the laboratory strain used initially (Taddei et al., 1995), the increased mutagenesis in ageing colonies requires at least two stress responses: the SOS DNA damage response and cAMP synthesis, part of the release of catabolite repression, a response to carbon starvation (Table 1), indicating that it is a cellular response to starvation stress (Taddei et al., 1995). The authors showed that mutations that block the induction of these responses block the mutagenesis. RecA and RecBCD were required, perhaps for their roles in SOS induction (Taddei et al., 1995, 1997a). Also in this particular laboratory strain, the DNA nucleotide-excision repair protein UvrB and DNA Pol I were required for the stress-induced mutagenesis. The stress-induced mutagenesis was specific to colonies on solid medium, not occurring in liquid cultures, probably because cAMP was used as a necessary communication molecule between cells in close proximity (Taddei et al., 1997a). The requirements for RecA and cAMP were as observed with the natural isolate strain examined by Bjedov et al. (2003) whereas that study found a requirement for Pol II rather than Pol I in the natural isolate. The UvrB requirement suggests possible coupling of mutagenesis to DNA repair, which could cluster mutations, as discussed above.
MutS and MMR Become Limiting
As in stress-induced mutagenesis in the natural isolate of Bjedov et al. (2003) and described above for stress-induced Lac+ point mutagenesis, and several systems discussed below, MMR function becomes limiting during stress-induced mutagenesis of this laboratory strain (Taddei et al., 1997a); mutation in mutS-defective cells did not cause a significant increase in stress-induced mutations, demonstrating that it occurs in cells already experiencing a limitation in MMR activity. Further, overproduction of MutS reduced mutagenesis indicating that MutS was the limiting component.
HOW MMR BECOMES LIMITING
The limitation of MMR activity via limited MutS during MAC is unlike several experimental systems in which MutL becomes limiting, but is like two others in which MutS does: the natural isolate of Bjedov et al. (2003), and an assay measuring MMR itself (not mutagenesis) via repair of heteroduplex DNA molecules introduced into stationary-phase cells (Brégeon et al., 1999). The assays in which MutL becomes limiting are the Lac system (reviewed above, see also Harris et al., 1997, 1999), cells expressing an error-prone mutant DNA polymerase III (Schaaper and Radman, 1989) or overproducing DinB/Pol IV (Wagner and Nohmi, 2000), and cells in which MMR proteins are titrated by overproduction of the interacting repair protein VSR (Doiron et al., 1996). This difference can be understood as follows (Rosenberg and Hastings, 2003): the cases in which MutL becomes limiting appear to be situations in which the number of errors (or protein molecules) potentially titrating MMR are very high, and so might titrate MMR before the significant decline in MutS and MutH levels occurs in stationary phase (Feng et al., 1996; Harris et al., 1997; Tsui et al., 1997). When titrated before the decrease in MutS levels in stationary phase, MutL is proposed to be limiting first. In the studies in which MutS was limiting, the DNA polymerases making the mutations are not “error-prone” DNA polymerases: Pol II (Bjedov et al., 2003) and Pol I (Taddei et al., 1997a), and MMR might not be overwhelmed until later, when MutS levels have dropped significantly in stationary phase. According to this hypothesis, whether MutL or MutS becomes limiting during stress-induced mutagenesis depends on how error-prone the synthesis is and its timing relative to the decline of MutS levels in stationary phase. Regardless of the specific mechanism, work discussed above and below shows that MMR limitation and the resultant increased mutagenesis in response to stress is a general theme in bacteria and human cells.
STARVATION-INDUCED MUTATIONS IN SOME OTHER BACTERIA
Bacillus subtilis: Competence Stress Response, DinB, MMR Limitation, and Possible Transcription Coupling of Mutagenesis
Two other bacterial species are emerging as alternative models for the study of stress-induced mutations, Bacillus subtilis and Pseudomonas putida. Starvation-associated mutations have been studied in B. subtilis using different alleles conferring histidine, methionine or leucine auxotrophy (reviewed in detail by Robleto et al., this volume). All can be reverted by any of several different base substitutions, restoring ability to grow in minimal medium lacking the corresponding amino acid. Using this system, Sung and Yasbin found that stress-induced prototrophic revertants arise after plating of the culture on minimal medium (Sung and Yasbin, 2002). Unlike the E. coli Lac system, mutations form independently of RecA (the SOS response or recombination) but like many of the systems discussed here, a starvation-stress response and a Y-family DNA polymerase are required, and MMR becomes limiting. The competence stress-response genes comA and comK, encoding transcriptional regulators of differentiation and competence development after exponential growth, are required for the generation of stress-induced mutations (Sung and Yasbin, 2002), as is the stringent stress response to aminoacid starvation (Rudner et al., 1999; reviewed Robleto et al., 2007). Competence (for natural transformation with DNA taken up from the environment) is a starvation-stress response, indicating the importance of stationary phase, starvation, and the appropriate stress response to the mutagenesis. The requirement for Com functions is analogous to requirements in other systems discussed above for the RpoS stress response and cAMP (see Table 2). As expected, the competence genes are not involved in mutagenesis in exponentially growing cells. Also paralleling the Lac system, and antibiotic-induced mutations, YqjH, a putative Y-family polymerase homologous to DinB, is responsible for at least half of the adaptive mutations in this system (Sung et al., 2003). Overproduction of MutS and MutL or MutS alone caused a two- to threefold reduction in mutagenesis, indicating that MMR limitation occurs via limitation of MutS (Pedraza-Reyes and Yasbin, 2004).
Possible Transcription Coupling and Limitation in Genomic Space
In addition to its temporal regulation by the Com stress response, the B. subtilis system also shows a hint that there might be spatial restriction of the mutagenesis within the genome. Mfd is a protein that couples DNA nucleotide excision repair to transcription in bacteria, causing preferential repair of actively transcribed genes. Mfd is required for stress-induced mutagenesis in this organism (Ross et al., 2006). This suggests that mutagenesis might be part of transcription-coupled repair, and thus might be localized to genes actively transcribed during the stress, and perhaps further limited to those undergoing a DNA repair event. Like the coupling of mutagenesis to HR-DSBR in the Lac system, this suggests a mechanism with the potential to localize and cluster mutagenesis in genomic space, which could facilitate concerted evolution (per Ninio, 1996; Ponder et al., 2005, discussed above). An important area for future work in this system will be to determine whether or not these mutations are localized and clustered in genomes, and whether this mutation mechanism is in fact coupled to transcription or transcription-coupled repair.
Pseudomonas putida: RpoS Stress Response, DinB, MMR Limitation, and More Than One Mechanism
The P. putida system also involves analysis of mutation arising after imposition of a non-lethal stress. In the first versions of the assay, cells carrying a plasmid containing a promoterless pheAB operon were assayed for post-selection mutagenesis (Kasak et al., 1997). The pheAB genes mediate the degradation of phenol and its use as a carbon source, but cells carrying the promoter-less plasmid version cannot grow on phenol minimal plates. However, mutations occur after plating, creating functional promoters. These mutations are various, and different from those generated during growth prior to selection. They include base substitutions, deletions and insertion of a transposon containing an outward-facing promoter. Intriguingly, mutations do not form in other starvation conditions, but require phenol in the medium. The point mutation sequences differ in the earlier days of starvation from the later days, suggesting that more than one mutagenesis mechanism contributes (Saumaa et al., 2002). Base substitutions prevailed in the first days, and small insertions in the later days.
These mutations are stress-response regulated by RpoS response (Table 1). RpoS is required for 2- to 3-bp insertions occurring in the later days and positively regulates the transposition of Tn4652 by actively regulating the transposase promoter (Ilves et al., 2001; Saumaa et al., 2002, this mechanism discussed below). In another version of the assay for Phe+ mutations, the plasmid-borne pheA gene was altered in the coding sequence by introducing three different nonsense mutations or a +1 frameshift mutations. With this set of alleles, DinB was largely responsible for the generation of −1 frameshift mutations from day 8 onward. These mutations form independently of RecA suggesting that the DinB activity did not require the SOS response in this system (Tegova et al., 2004). Removing the P. putida MMR system by mutations in mutL and mutS caused a marked increase in the base substitutions, but not in the −1 frameshift mutations (Saumaa et al., 2006), suggesting either that the frameshift mutations are generated from a precursor structure not subject to MMR [unlikely given avidity of MMR in other organisms for −1 frameshift mutation intermediates (Kunkel and Erie, 2005)], or that they originate in cells already depleted in MMR activity. The latter possibility would fit with these mutations specifically requiring DinB. Perhaps in this system, as proposed for the E. coli Lac assay (Harris et al., 1997; McKenzie et al., 2001), MMR becomes titrated by high frequency errors generated by DinB, which in this system is not the source of the substitution mutants.
Both the P. putida and B. subtilis examples share many of the key features of E. coli Lac point mutagenesis: temporal regulation by stress responses, DinB-like error-prone polymerases, MMR limitation, and, perhaps for B. subtilis, the possibility of linkage of mutagenesis to local DNA repair events.
CONTROL OF MUTAGENESIS BY PHOPQ: RPOS AGAIN?
PhoPQ is yet another stress response that controls mutagenesis in E. coli. PhoPQ constitute a two-component system of signal transduction that upregulates several genes in response to diminished levels of cellular Mg2+, plays an important role in the expression of virulence genes in pathogenic enterobacteria (Monsieurs et al., 2005), and also stabilizes RpoS (Tu et al., 2006). In a mutagenesis assay used by Hall (1998), the cryptic ebg operon is employed in an elegant selection for forward (gene-inactivating) mutations in the ebgR repressor gene during starvation. Following a screen for mutants with decreased stress-induced ebgR mutagenesis, Hall showed that PhoP and PhoQ are required for the stress-induced mutagenesis specifically, not generation-dependent ebgR mutation, and excluded possible effects of PhoPQ on the growth of ebgR mutants (Hall, 1998). Many of the mutations in ebgR are insertions of mobile elements (IS elements) (Hall, 1999), which left open the possibility that movement of these genetic elements might be regulated by PhoPQ. In light of recent evidence that PhoPQ stabilizes RpoS (Tu et al., 2006), this work now begs the question of whether the RpoS starvation- and general-stress response is required for the ebgR mutagenesis during starvation, and whether the need for RpoS is the reason PhoPQ are required. Whatever its mechanistic basis, these results provide another example of the control of mutagenesis by a stress response(s). Other examples involving movement of mobile elements follow.
MOBILE GENETIC ELEMENTS AND STRESS-INDUCED GENOME REARRANGEMENTS
Mu-Mediated Deletions
The idea that transposons might be a source of genetic variability in response to stress was proposed by Barbara McClintock (1984) working in corn, and the observation of stress-induced movement of mobile genetic elements in bacteria was one of the early instances of stress-inducible genetic change reported. Shapiro used an E. coli strain carrying the transposing prophage Mu inserted between the araB and lacZ genes. The insertion renders the strain Lac−. However, Mu excisions can produce in-frame fusions of araB and lacZ, making cells able to grow on lactose, as long as arabinose is also provided to induce the transcription of the fusion gene. The excision events are very rare in growing cells, but increase by several orders of magnitudeafter prolonged incubation of cells in lactose-arabinose plates (Maenhaut-Michel and Shapiro, 1994; Shapiro, 1984), during long-term incubation of glucose cultures (Mittler and Lenski, 1990), or in response to starvation generally (Foster and Cairns, 1994; Maenhaut-Michel and Shapiro, 1994; Sniegowski, 1995): all starvation stresses. However, the structure of the fusions is different depending on the environment (i.e., selected on lactose-arabinose plates or in liquid with other carbon sources), indicating that a complex regulatory network operates in response to the environment, influencing the genetic rearrangements (Maenhaut-Michel and Shapiro, 1994).
The occurrence of excisions absolutely requires the RpoS stress response (Gomez-Gomez et al., 1997), and proper controls demonstrated that the lack of excisions in the mutant was not due to decreased viability or growth rate of the rpoS strain carrying the araB::lacZ fusion. RpoS mediates de-repression of the lytic promoter of Mu in stationary phase (Lamrani et al., 1999). Similarly, CRP, the transcriptional activator, which upregulates promoters during the release from catabolite repression upon carbon starvation, is also required (Lamrani et al., 1999) (Table 2), indicating positive control of these fusions by two starvation stress responses. Conversely, depletion of the histone-like protein H-NS, which promotes repression of some RpoS-regulated promoters, stimulates excision events (Gomez-Gomez et al., 1997), indicating that several host inputs act to balance the formation of fusions in response to stress.
Stress-Induced Transposition in Old Colonies
More recently, Coros et al. used a papillation assay to monitor transposon movement in single E.coli colonies, and found that transposition of the mobile element IS903 occurs predominantly late in colony development (Coros et al., 2005). Various host factors affect this transposition, including AspA, which implies nutritional regulation of transposition (Twiss et al., 2005). aspA+, encoding aspartase, represses transposition generally and particularly during early colony growth, and also inhibits transposition of the unrelated elements Tn10 and Tn552. Aspartase converts L-aspartate to fumarate in the TCA cycle, suggesting that its inhibitory effect results from avoidance of nutritional deprivation. Demonstration that nutritional stress indeed induces transposition was obtained by supplying extra fumarate in growth medium, which suppressed the early-transposition phenotype of aspA mutants. On the other hand, H-NS, which antagonizes the RpoS stress response, was shown to promote Tn10 transposition directly, by binding to the uncleaved transposasome (Wardle et al., 2005).
Stress-Induced Transposition in Pseudomonas
In P. putida, a similar induction of transposition events by stress was observed using the promoter-creating assay described above (Kasak et al., 1997). Several of the mutations allowing expression of the promoterless pheAB operon were Tn4652 insertions, and these required RpoS, which transcriptionally upregulates the Tn4652 transposase (Ilves et al., 2001). Two other regulatory inputs of Tn4652 transposition specific to stationary phase illustrate a fine-tuned regulation of transposon movement in response to stress: IHF and the ColS-ColR two component system (Horak et al., 2004; Ilves et al., 2004). Upregulation of transposon movement during stress is an interesting observation, because these elements are viewed by some as selfish parasites of the genome. Whatever the possible selective pressure for (or lack of selective pressure against) mechanisms leading to enhanced transposition in times of stress, the result is that transposon movement may be an important source of generation of variability in response to environmental changes.
Telomere Shortening- and DNA Damage-Response-Activated Retrotransposition and Point Mutagenesis in Yeast
Yeast telomeres are stably maintained by the addition of simple DNA repeats at chromosome ends by telomerase. Telomerase uses the reverse transcriptase activity of the Est2 enzyme to synthesize the DNA using an RNA template. In est2 mutants, telomeres shorten progressively, triggering a cell-cycle checkpoint and ultimately provoking cell senescence (Lundblad and Blackburn, 1993). Scholes and colleagues demonstrated that mobility of the yeast LTR-retrotransposon Ty1 is greatly stimulated in response to telomere erosion stress in est2 mutants (Scholes et al., 2003). This enhanced retrotransposon mobility requires the Rad9, Rad17 and Rad24 signal-transduction proteins of a DNA-damage checkpoint pathway, analogous to the SOS DNA-damage response in bacteria.
Additionally, telomere shortening in est2 mutants is associated with increased point mutagenesis in telomere-proximal regions, mediated by the error-prone polymerases Rev1 and Rev3/Rev7 (Meyer and Bailis, 2007), the first of which is a Y-family DNA polymerase homologous with DinB, and all of which are specialized, error-prone translesion polymerases. These authors did not measure mutation of the same gene close to and distant from the telomere, so the differences in mutagenesis upon telomere shortening might be due to either the mutation-target sequences or to telomere proximity per se, in either case, activated by stress. Although the signal transduction pathway(s) of this later phenomenon have not been determined, enhanced movement of the Ty1 retrotransposon and increased mutagenesis might be part of the same global response to telomere shortening. The regulation of retrotransposition by a DNA damage response, and both it and point mutagenesis by stress, make these events analogous to those discussed for bacteria in which the SOS response upregulates genetic instability.
TRANSCRIPTION-ASSOCIATED MUTATIONS IN YEAST AND BACTERIA
A dramatic example of circumscription of mutations in genomic space is the association of mutation and transcription. In Saccharomyces cerevisiae, in experiments in which transcription of a lys2 gene with a frameshift mutation was controlled with an inducible promoter, increased transcription rates were correlated with increased reversion rates (Datta and Jinks-Robertson, 1995). In E. coli, a transcription association with mutagenesis was suggested by genomic analysis (Francino et al., 1996). More directly, auxotrophic mutants carrying mutations in amino acid biosynthetic genes show increased rates of reversion in conditions that stimulate the expression of those specific biosynthetic genes (Wright et al., 1999).
In a particular E. coli example, reversion rates of a leuB allele were perfectly correlated with the expression levels of the gene in several different conditions (Wright et al., 1999). Synthesis of the small starvation-stress signal molecular, ppGpp, and starvation for leucine are necessary for the high levels of expression of leuB, and consequently, elevated reversion rates. Modulation of expression of the biosynthetic gene by a non-native, artificially inducible promoter also demonstrated that increased transcription rates correlate with increased reversion. A similar requirement for ppGpp was observed for the mutation rates in B. subtilis cultures (Rudner et al., 1999). In this assay, reversion rates of two aminoacid auxotrophy markers were positively correlated with the production of ppGpp by cells, unlike mutations giving rise to streptomycin resistance, which were unaltered in ppGpp-synthesis-defective mutant strains.
In one model for a possible mechanism for transcription associated mutagenesis, Wright notes that transcription induces local modifications to supercoiling, which she proposes facilitate the formation of stem-loop structures with unpaired bases, which would be particularly prone to mutation (Wright, 2004). The author observes a correlation between predicted ability of DNA sequences to form stem-loop structures and their acquisition of mutations. This locally facilitated mutagenesis might be an unavoidable consequence of other DNA transactions or a specifically selected mechanism. Either way, it is expected to have broad evolutionary impact, because a link between the need for a gene product in a given situation and mutagenesis could facilitate the evolution of new and better adapted gene functions. The impact of transcription rates on mutagenesis of various genes assayed in stress-induced mutagenesis assays is a very interesting question that still remains to be addressed in most experimental models for stress-induced mutagenesis. Transcription-associated mutagenesis is also well documented in somatic hypermutation of immunoglobulin genes (discussed below).
OTHER BACTERIAL EXAMPLES
What other examples of increased mutation rates in response to stress are available? In an interesting approach, Lowe et al. demonstrated that E. coli cultures held in stationary phase for long periods show a high rate of mutations, as shown by the loss of fitness caused by deleterious mutations (Loewe et al., 2003). Some concerns were raised regarding this study about the possible effects of GASP mutations on the results (de Visser and Rozen, 2004), but the GASP phenomenon itself (see below) might be an example of stress-induced mutagenesis.
GASP: the SOS/Translesion DNA Polymerases and RpoS
GASP (growth advantage in stationary phase) mutants arise in long term stationary-phase populations as a result of new mutations conferring advantageous phenotypes that allow growth in the limiting resources. The cells that acquire these mutations out-compete previous genotypes and overtake the population (Finkel, 2006). While the overall population size stays constant (the definition of stationary phase), the fitter mutant types rise in number, outcompeting their siblings, then later fall as they are overtaken by fitter mutants in successive clonal sweeps.
Early GASP mutations that confer an advantage are diminished function (but not null) alleles of RpoS (Table 2) (Zambrano et al., 1993) in apparent contrast with the several examples given in which RpoS function is required for mutagenesis (Table 2 and above). It appears that in GASP conditions, less activity of the RpoS regulon and more of the housekeeping genes is advantageous, but still some RpoS activity appears to be required in that null alleles are not selected. Ferenci has discussed the inherent problem of the tradeoff between housekeeping genes and the RpoS stress genes in limiting resources (Ferenci, 2003), which this appears to illustrate.
Little is known about the generation of the GASP mutations, including whether they are spontaneous mutations present in cultures before stationary-phase stress, or are stress-induced during starvation. However, the following results imply that they are stress-induced via the SOS response. Intriguingly, all three of the SOS-inducible DNA polymerases (Tables 1, 2) are required for the production of GASP mutants (Yeiser et al., 2002). Strains that lack any of the SOS polymerases are consistently overtaken by wild-type strains, despite the fact that they show no significant alteration in viability in stationary phase. This implies that the three SOS DNA polymerases are required for production of the GASP mutations themselves. These results echo the stories of ciprofloxacin-induced resistant mutagenesis and Lac+ point mutagenesis discussed above, and suggest that the SOS-controlled DNA polymerases may underlie the generation of genetic diversity in stationary phase in this system as they do in others. This is particularly likely, given the very small contribution of these polymerases to spontaneous mutagenesis in growing cells in the absence of DNA damage (reviewed by Nohmi, 2006).
Similarly, in Pseudomonas putida cultures, the presence of a plasmid encoding a homologue of DinB/Pol V facilitates the acquisition of the GASP phenotype (i.e., population takeover in stationary phase) (Tark et al., 2005). GASP seems to be an important principle in microbial evolution that is wide open for exploration of the mechanisms and regulation of the mutagenesis that drives it.
STRESS-INDUCED MUTATIONS IN YEAST
Yeast cells exposed to starvation also accumulate adaptive mutations that relieve the growth impairment (reviewed in detail by Heidenreich, this volume). Histidine auxotrophic mutants containing a nonsense mutation in the HIS4 gene, show accumulation of HIS+ adaptive mutants detected as papillae in colonies grown in medium with limiting amounts of histidine (Hall, 1992). Cells from the papillae were shown to have formed during starvation, and were not slowly growing mutants present before plating.
A similar observation was made in strains containing a lys2 frameshift mutation, which shows reversion to lysine prototrophy after plating in medium lacking this amino acid, in the absence of detectable cell growth (Steele and Jinks-Robertson, 1992). This allele has been used for studies of stress-induced mutation in two laboratories using different yeast strains. In one strain background housing this allele, lack of functional mitochondria does not affect starvation-stress-induced mutations, strongly suggesting that oxidative DNA lesions are not precursors in the mutagenic process (Heidenreich and Wintersberger, 1998). In a remarkable similarity with stress-induced mutations in the lac system, adaptive reversion of the +4 lys2 allele shows a strong bias for −1 deletions in mononucleotide repeats, in contrast to a variety of different mutations occurring in the same locus in growing cells (Heidenreich and Wintersberger, 2001), although, when assayed in the other strain, both the stress-induced and generation-dependent mutations were mostly deletions in simple repeats (Greene and Jinks-Robertson, 1999). Yeast strains can be quite different genetically, and we will confine the rest of this discussion to the strain used by Heidenreich and colleagues (2003, 2004), only because more information exists about the mechanism of mutagenesis in it. In that strain, the mutation sequences are suggestive of diminished MMR function, but this has not been tested directly.
A Possible Switch To Mutagenic DSBR via NHEJ Under Stress
In the Heidenreich yeast system, two proteins that function in DSBR by non-Homologous End Joining (NHEJ), Ku70 and DNA ligase IV, are required for at least half of the mutations arising specifically under stress (Heidenreich et al., 2003) (Tables 1 and 2).
NHEJ (Table 1) is a mechanism of DSBR that is completely different from homologous recombination (HR), discussed above for the Lac system, and which is inherently mutagenic. In bakers yeast, NHEJ is the less used DSBR pathway and is specific to situations in which there is no homologous DNA available for repair via HR-DSBR (reviewed by Dudasova et al., 2004). In NHEJ, two DSEs are united without any need for homology, involving only recognition of the DNA ends, recruiting of proteins involved in resection, and ligation (Bowater and Doherty, 2006).
In the yeast stress-induced lys2 reversion assay, enhancement of DNA breakage by gamma irradiation increased the rate of adaptive mutagenesis in a NHEJ dependent manner, showing that NHEJ can indeed lead to mutations in starved cells (Heidenreich et al., 2004). The process by which NHEJ produces the frameshift mutations in starved cells is not understood. Although it has not been demonstrated in this yeast system (but see below for human), a plausible mechanism for the stress-induced mutagenesis is that, as in the E. coli Lac system, mutagenesis might result from a switch from high-fidelity DSBR to mutagenic DSBR under stress. This could occur by a switch from HR-DSBR to DSBR via NHEJ. If this hypothesis is correct, the mechanism emerging would appear to be analogous to that in E. coli but evolutionarily unrelated: a common strategy (possible coupling of stress-induced mutagenesis to DSBR) evolved independently and differently, but, we suggest, potentially with similar selective advantages for localization of mutagenesis in genomic space. Below we will see evidence that in human cells, such a switch from high-fidelity HR-DSBR to mutagenic NHEJ is induced by stress, via stress-induced transcriptional down-regulation of key HR-DSBR genes.
Mutagenic HR-DSBR in Yeast
Mutagenesis coupled with HR-DSBR events, just as now appreciated in the Lac system, are also well documented in yeast. Strathern and colleagues showed that the induction of DSBs by an endonuclease specifically provokes both frameshift and base substitution mutations near the break, in the region that has undergone HR-DSBR (Strathern et al., 1995). The error-prone, translesion DNA polymerase REV3 is required for the generation of the base substitutions mutations (Holbeck and Strathern, 1997). Whether these mutations have any relationship to cellular stress or stress responses (including DNA-damage response) has not been investigated, so it is unknown whether there is any temporal regulation of this process in yeast, as there is in E. coli (Ponder et al., 2005). However, the coupling of mutagenesis to HR-DSBR events using an error-prone DNA polymerase is just as observed in the Lac system, and may confer similar potential advantages for evolvability, discussed above.
PROGRAMMED ERROR-PRONE DNA BREAK REPAIR IN THE IMMUNE RESPONSE
An interesting parallel with point mutagenesis in the Lac system, antibiotic-induced mutagenesis, and yeast mutagenesis coupled to HR-DSBR is seen in the diversification of the immunoglobulin genes. Somatic hypermutation of immunoglobulin genes involves Y-family polymerases to generate the mutations giving rise to the antibody repertoire, and apparently concentrates those mutations near to DNA breaks, in this case, programmed DNA breaks in the immunoglobulin genes, also associated with transcription (Diaz and Lawrence, 2005; Neuberger et al., 2003).
STRESS-INDUCED MUTAGENESIS IN HUMAN CELLS AND CANCER
Transcriptional Down-regulation of MMR and High-Fidelity DSBR
Tumor development is a multi-stage process, during which cells progressively acquire a series of characteristics that lead to unrestricted growth. The cascade of events bestowing cancer development requires mutations in multiple genes, and tumor cell lines often display a mutator phenotype (Bielas et al., 2006). The parallels between cancer and evolution, and the role of mutagenesis in promoting both, are discussed in detail in the following review and hypothesis articles (Cairns, 1975; Loeb et al., 1974; Nowell, 1976; Echols, 1981; S.M. Rosenberg, E.C. Cox, R.C. von Borstel and L.A. Loeb, manuscript in preparation). In early tumor microenvironments, cells often experience states of hypoxia, which is counteracted by angiogenesis in later stages of tumor development. Hypoxia plays a very important role in tumor progression, because this condition itself increases genetic instability, as detected in wide variety of cell lines and assay systems (reviewed by Huang et al., 2007).
Although hypoxic conditions can cause DNA damage directly, an interesting set of findings reveals that genetic instability itself is induced by hypoxia, triggered by down-regulation of at least five DNA repair genes by stress responses. This regulated process bears two striking parallels to stress-induced mutagenesis in the Lac system and other microbes.
Down-Regulated MMR
First, mismatch repair is down-regulated by decreased transcription of three genes: MLH1, MSH2, and MSH6, homologues of E. coli MutL and MutS (Table 1). Repression of MLH1 transcription involves histone deacetylation and the Max regulatory network, and is accompanied by an increase in (CA) dinucleotide-repeat instability and increased mutation in a chromosomal marker, demonstrating that MLH1 becomes functionally limiting (Bindra and Glazer, 2007a; Mihaylova et al., 2003). MLH1 down-regulation is p53-independent, and thus likely to fuel mutagenesis and tumor progression in the large numbers of cancers in which the p53 tumor-suppressor gene is mutant. Additionally, MSH2 and MSH6 are transcriptionally down-regulated by the HIF-1alpha repressor protein, which is removed normally by the ubiquitin-proteasome but becomes stable during hypoxia (Koshiji et al., 2005). This decreases MSH2 and MSH6 levels and is also accompanied by genetic instability in a mononucleotide repeat marker. The MSH2/MSH6 down-regulation is p53-dependent and thus potentially relevant to fewer tumors, those with functional p53. Finally, the Max network may also regulate MSH2 in a HIF1-alpha-independent manner (Bindra and Glazer, 2007a). These findings parallel the microbial examples of down-regulation of MutS and functional limitation of MutL and MutS in response to stress. The transcriptional regulatory networks are different, and are understood in greater detail, but the outcomes for MMR function are similar, and the same components (homologues of MutS and MutL) become limiting (Table 2).
Altered DSBR
Second, HR-DSBR is down-regulated via transcriptional repression of BRCA1 (breast cancer tumor suppressor protein) and RAD51 (RecA homologue, see Table 1 for both). RAD51 is transcriptionally down-regulated in response to hypoxia, and this down-regulation is correlated with a substantial decrease in HR in hypoxic conditions (Bindra et al., 2004). The repression is mediated by the formation of repressive E2F4/p130 complexes, triggered by p130 dephosphorylation in response to hypoxia (Bindra and Glazer, 2007b). Similarly, the BRCA1 gene, which functions in HR-DSBR and suppresses use of the more error-prone NHEJ pathway of DSBR (reviewed by Gudmundsdottir and Ashworth, 2006), is also transcriptionally down-regulated by repressive E2F complexes (Bindra et al., 2005). BRCA1 depletion leads to decreased HR, while the NHEJ pathway remains unaffected, potentially leading to increasing genomic instability via a switch to reliance on the inherently mutagenic NHEJ-DSBR pathway during hypoxia. This is highly analogous to, though a different mechanism from, the switch to mutagenic HR-DSBR induced by stress and controlled by RpoS in the Lac system. This resembles the yeast system in which stress-induced mutations require NHEJ proteins (reviewed above), suggesting a switch to error-prone NHEJ from HR-DSBR under stress.
Whatever the selective pressure (or lack of it) that allowed evolution of these responses in multicellular animals, both are expected to drive cancer evolution by promoting mutagenesis under stress. As suggested elsewhere (SM Rosenberg, EC Cox, RC von Borstel and LA Loeb, manuscript in preparation), anti-evolution drugs designed to interfere with the mutator mechanisms themselves would provide powerful accompaniments to standard chemotherapies, most of which block cell growth, induce stress, and have the potential to induce stress-induced resistance mutations.
OVERVIEW AND SIGNIFICANCE FOR EVOLVABILITY
There are similarities and differences in the various molecular mechanisms of mutagenesis discussed above, with several repeating examples of use of common components and/or common strategies that limit when and where mutagenesis occurs. These are summarized in Table 2. The many examples support a picture of mutagenesis as a highly regulated process, regulated both in time, and at least sometimes, in genomic space. The restriction of mutagenesis to times of stress by coupling it to stress responses provokes mutagenesis specifically when cells are maladapted to their environments, which is when mutator phenotypes will drive their adaptation. The limitation of mutagenesis to local genomic regions might, as reviewed above, limit deleterious mutations in genomes of rare adaptive mutants, and promote concerted evolution within genes and linked gene clusters (shown compellingly by Drake et al., 2005; Drake, 2007; Wang et al., 2007). Although these principles have been demonstrated most directly in the Lac system, many other systems share common components—like use of RpoS, the SOS response, RecA, specialized and Y-family and SOS-controlled DNA polymerases, and transiently limiting mismatch repair—and/or common strategies: control of mutagenesis by stress responses, and evidence for localization of mutagenesis in genomic space. We have discussed the probable adaptive value of these strategies in microbial populations. It is less clear why multicellular organisms like humans should share these strategies, but it is clear that they do, and that stress-induced mutagenesis is likely to underlie evolution of tumorigenesis. Elsewhere the case is made that cancer formation, progression and evolution of resistance to treatments are also evolutionary processes driven by mutation and selection, often under stress (SM Rosenberg, EC Cox, RC von Borstel and LA Loeb, manuscript in preparation).
ARE STRESS-INDUCIBLE MUTAGENESIS MECHANISMS SELECTED?
Second-Order Selection
Is stress-inducible mutator character of itself a selectable trait, or is it an incidental byproduct of a stress-inducible function that is selected? There is little evidence to reject or favor either hypothesis (Tenaillon et al., 2004). That some aspect of stress-induced mutagenesis mechanisms is selected is strongly implied by the correlation of stress-inducible mutability with ecological niche rather than phylogeny (Bjedov et al., 2003). As for constitutive mutators, mutator alleles become fixed in populations under strong selective pressure by hitchhiking along with the advantageous mutations that they may induce (Introduction and Tenaillon et al., 1999). This second-order selection could also apply to stress-induced mutation mechanisms, and computer simulations suggest that both constitutive and stress-inducible mutator phenotypes can be selected in this way (Bjedov et al., 2003). The other possibility is that natural selection favors some characteristic(s) that are accompanied by the byproduct of increased mutability during stress: the pleiotropy hypothesis. In this case, evolution favors transient increases in mutation rates via other benefits, such as a strategy for saving energy for other cellular functions by shutting down replication fidelity mechanisms, or upregulating error-prone polymerases to achieve increased resistance to DNA damage encountered during the stress period, etc. Although, as noted, the “error-prone” DNA polymerases are usually error-prone only when not performing translesion synthesis; so random mutagenesis using them would be expected to be avoidable if it were selected against. That is, survival promoted by (high-fidelity) translesion synthesis could occur without concurrent mutagenesis on non-lesion template DNAs. Importantly, stress-inducible mutability will accelerate evolution in competitive environments regardless of why cells possess the ability.
Periodic Selection
We have seen above that there is strong evidence that constitutive mutator alleles are repeatedly lost and regained, revealed by the mosaic phylogeny of the sequence of mutS (Denamur et al., 2000). This tells us that a mutator phenotype is advantageous in some environments/circumstances and not in others, i.e., it is subject to periodic (positive then negative) selection. Could this also apply to the possession of a stress-inducible mutator character? The answer to this question bears profoundly the question of whether stress-inducible mutability is itself a selectable trait.
We see evidence, presented above, that it is indeed likely that stress-inducible mutability is a capacity that is lost and regained as needed. First, there is huge variability in the strength of stress-inducible mutators in wild isolates (Bjedov et al., 2003; Saint-Ruf and Matic, 2006) suggesting that different stress-inducible mutator activities have different mechanisms. Second, as reviewed above, there is in fact wide variation in the molecular mechanisms of stress-inducible mutagenesis among the different systems, as revealed by the differing genetic requirements. The picture appears to be patchwork of mechanisms with common parts and strategies used interchangeably in different stains, species, and stresses, as if stress-inducible mutability has been “invented,” lost and “reinvented” independently many times. Third, the distribution of stress-inducible mutability does not correlate with phylogenetic relationships among natural isolates, but rather, correlates with habitat (Bjedov et al., 2003). This strongly suggests that it is under short-term environment-specific selection. These facts argue strongly that stress-inducible mutability is under periodic selection.
RpoS in Periodic Selection?
Finally, we note that in bacteria, regulation of mutagenesis by RpoS is a common feature even in systems with different mutagenesis mechanisms. We suggest that periodic selection of RpoS could provide a critical master “switch” underlying periodic selection of some and perhaps many bacterial stress-inducible mutagenesis mechanisms. Like mutS, the rpoS gene itself is highly polymorphic in different E. coli populations demonstrating that its function is under periodic selection (Ferenci, 2003). Ferenci has argued that loss of RpoS function is selected due to fitness costs associated with this stress response in decreasing transcription of needed housekeeping genes, as in GASP (Ferenci, 2003). We suggest that that could account for selection against stress-inducible mutagenesis pathways by selecting against their most appropriate master regulator, and that second-order selection for stress-inducible mutability in competitive habitats swings the pendulum back. Stress-inducible mutagenesis mechanisms should be advantageous in competitive environments, and irrelevant in environments to which cells are well adapted, in which the RpoS component is at times demonstrably sometimes costly.
Acknowledgments
We are indebted to Ted Cox and Evelyn Witkin for comments on the manuscript and to the many members of our labs past and present who propelled our work on stress-induced mutagenesis. RSG was supported by a postdoctoral fellowship from the Pew Latin American Fellows Program. Supported by National Institutes of Health grants R01-GM64022 (PJH) and R01-GM53158 (SMR).
References
- Abella M, Erill I, Jara M, Mazon G, Campoy S, Barbe J. Widespread distribution of a lexA-regulated DNA damage-inducible multiple gene cassette in the Proteobacteria phylum. Mol Microbiol. 2004;54:212–222. doi: 10.1111/j.1365-2958.2004.04260.x. [DOI] [PubMed] [Google Scholar]
- Bavoux C, Leopoldino AM, Bergoglio V, JOW, Ogi T, Bieth A, Judde JG, Pena SD, Poupon MF, Helleday T, et al. Upregulation of the error-prone DNA polymerase kappa promotes pleiotropic genetic alterations and tumorigenesis. Cancer Res. 2005;65:325–330. [PubMed] [Google Scholar]
- Bielas JH, Loeb KR, Rubin BP, True LD, Loeb LA. Human cancers express a mutator phenotype. Proc Natl Acad Sci U S A. 2006;103:18238–18242. doi: 10.1073/pnas.0607057103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bindra RS, Gibson SL, Meng A, Westermark U, Jasin M, Pierce AJ, Bristow RG, Classon MK, Glazer PM. Hypoxia-induced down-regulation of BRCA1 expression by E2Fs. Cancer Res. 2005;65:11597–11604. doi: 10.1158/0008-5472.CAN-05-2119. [DOI] [PubMed] [Google Scholar]
- Bindra RS, Glazer PM. Co-repression of mismatch repair gene expression by hypoxia in cancer cells: Role of the Myc/Max network. Cancer Lett. 2007a;252:93–103. doi: 10.1016/j.canlet.2006.12.011. [DOI] [PubMed] [Google Scholar]
- Bindra RS, Glazer PM. Repression of RAD51 gene expression by E2F4/p130 complexes in hypoxia. Oncogene. 2007b;26:2048–2057. doi: 10.1038/sj.onc.1210001. [DOI] [PubMed] [Google Scholar]
- Bindra RS, Schaffer PJ, Meng A, Woo J, Maseide K, Roth ME, Lizardi P, Hedley DW, Bristow RG, Glazer PM. Down-regulation of Rad51 and decreased homologous recombination in hypoxic cancer cells. Mol Cell Biol. 2004;24:8504–8518. doi: 10.1128/MCB.24.19.8504-8518.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjedov I, Nag Dasgupta C, Slade D, Le Blastier S, Selva M, Matic I. Involvement of Escherichia coli DNA polymerase IV in tolerance of cytotoxic alkylating DNA lesions in vivo. Genetics. 2007;176:1431–1440. doi: 10.1534/genetics.107.072405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjedov I, Tenaillon O, Gerard B, Souza V, Denamur E, Radman M, Taddei F, Matic I. Stress-induced mutagenesis in bacteria. Science. 2003;300:1404–1409. doi: 10.1126/science.1082240. [DOI] [PubMed] [Google Scholar]
- Blazquez J. Hypermutation as a factor contributing to the acquisition of antimicrobial resistance. Clin Infect Dis. 2003;37:1201–1209. doi: 10.1086/378810. [DOI] [PubMed] [Google Scholar]
- Boshoff HI, Reed MB, Barry CE, 3rd, Mizrahi V. DnaE2 polymerase contributes to in vivo survival and the emergence of drug resistance in Mycobacterium tuberculosis. Cell. 2003;113:183–193. doi: 10.1016/s0092-8674(03)00270-8. [DOI] [PubMed] [Google Scholar]
- Bowater R, Doherty AJ. Making ends meet: repairing breaks in bacterial DNA by non-homologous end-joining. PLoS Genet. 2006;2:e8. doi: 10.1371/journal.pgen.0020008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brégeon D, Matic I, Radman M, Taddei F. Inefficient mismatch repair: genetic defects and down regulation. J Genet. 1999;78:21–28. [Google Scholar]
- Bull HJ, Lombardo MJ, Rosenberg SM. Stationary-phase mutation in the bacterial chromosome: recombination protein and DNA polymerase IV dependence. Proc Natl Acad Sci U S A. 2001;98:8334–8341. doi: 10.1073/pnas.151009798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bull HJ, McKenzie GJ, Hastings PJ, Rosenberg SM. Evidence that stationary-phase hypermutation in the Escherichia coli chromosome is promoted by recombination. Genetics. 2000;154:1427–1437. doi: 10.1093/genetics/154.4.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cairns J. Mutation selection and the natural history of cancer. Nature. 1975;255:197–200. doi: 10.1038/255197a0. [DOI] [PubMed] [Google Scholar]
- Cairns J, Foster PL. Adaptive reversion of a frameshift mutation in Escherichia coli. Genetics. 1991;128:695–701. doi: 10.1093/genetics/128.4.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cairns J, Overbaugh J, Miller S. The origin of mutants. Nature. 1988;335:142–145. doi: 10.1038/335142a0. [DOI] [PubMed] [Google Scholar]
- Camps M, Naukkarinen J, Johnson BP, Loeb LA. Targeted gene evolution in Escherichia coli using a highly error-prone DNA polymerase I. Proc Natl Acad Sci U S A. 2003;100:9727–9732. doi: 10.1073/pnas.1333928100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao L, Vargas C, Spear BB, Cox EC. Transposable elements as mutator genes in evolution. Nature. 1983;303:633–635. doi: 10.1038/303633a0. [DOI] [PubMed] [Google Scholar]
- Chatterji D, Ojha AK. Revisiting the stringent response, ppGpp and starvation signaling. Curr Opin Microbiol. 2001;4:160–165. doi: 10.1016/s1369-5274(00)00182-x. [DOI] [PubMed] [Google Scholar]
- Cirz RT, Chin JK, Andes DR, de Crecy-Lagard V, Craig WA, Romesberg FE. Inhibition of mutation and combating the evolution of antibiotic resistance. PLoS Biol. 2005;3:e176. doi: 10.1371/journal.pbio.0030176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirz RT, Gingles N, Romesberg FE. Side effects may include evolution. Nat Med. 2006a;12:890–891. doi: 10.1038/nm0806-890. [DOI] [PubMed] [Google Scholar]
- Cirz RT, O’Neill BM, Hammond JA, Head SR, Romesberg FE. Defining the Pseudomonas aeruginosa SOS response and its role in the global response to the antibiotic ciprofloxacin. J Bacteriol. 2006b;188:7101–7110. doi: 10.1128/JB.00807-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirz RT, Romesberg FE. Induction and inhibition of ciprofloxacin resistance-conferring mutations in hypermutator bacteria. Antimicrob Agents Chemother. 2006;50:220–225. doi: 10.1128/AAC.50.1.220-225.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirz RT, Romesberg FE. Controlling mutation: intervening in evolution as a therapeutic strategy. Crit Rev Bioch Mol Biol. 2007;42:341–352. doi: 10.1080/10409230701597741. [DOI] [PubMed] [Google Scholar]
- Claverys JP, Prudhomme M, Martin B. Induction of competence regulons as a general response to stress in gram-positive bacteria. Annu Rev Microbiol. 2006;60:451–475. doi: 10.1146/annurev.micro.60.080805.142139. [DOI] [PubMed] [Google Scholar]
- Coros AM, Twiss E, Tavakoli NP, Derbyshire KM. Genetic evidence that GTP is required for transposition of IS903 and Tn552 in Escherichia coli. J Bacteriol. 2005;187:4598–4606. doi: 10.1128/JB.187.13.4598-4606.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox MM. Regulation of bacterial RecA protein function. Crit Rev Biochem Mol Biol. 2007;42:41–63. doi: 10.1080/10409230701260258. [DOI] [PubMed] [Google Scholar]
- Datta A, Jinks-Robertson S. Association of increased spontaneous mutation rates with high levels of transcription in yeast. Science. 1995;268:1616–1619. doi: 10.1126/science.7777859. [DOI] [PubMed] [Google Scholar]
- Davis EO, Dullaghan EM, Rand L. Definition of the mycobacterial SOS box and use to identify LexA-regulated genes in Mycobacterium tuberculosis. J Bacteriol. 2002;184:3287–3295. doi: 10.1128/JB.184.12.3287-3295.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Visser JA, Rozen DE. Comment on “High deleterious genomic mutation rate in stationary phase of Escherichia coli”. Science. 2004;304:518. doi: 10.1126/science.1094911. author reply 518. [DOI] [PubMed] [Google Scholar]
- Denamur E, Lecointre G, Darlu P, Tenaillon O, Acquaviva C, Sayada C, Sunjevaric I, Rothstein R, Elion J, Taddei F, et al. Evolutionary implications of the frequent horizontal transfer of mismatch repair genes. Cell. 2000;103:711–721. doi: 10.1016/s0092-8674(00)00175-6. [DOI] [PubMed] [Google Scholar]
- Diaz M, Lawrence C. An update on the role of translesion synthesis DNA polymerases in Ig hypermutation. Trends Immunol. 2005;26:215–220. doi: 10.1016/j.it.2005.02.008. [DOI] [PubMed] [Google Scholar]
- Doiron KM, Viau S, Koutroumanis M, Cupples CG. Overexpression of vsr in Escherichia coli is mutagenic. J Bacteriol. 1996;178:4294–4296. doi: 10.1128/jb.178.14.4294-4296.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drake JW. Too many mutants with multiple mutations. Crit Rev Bioch Mol Biol. 2007;42:249–258. doi: 10.1080/10409230701495631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drake JW, Charlesworth B, Charlesworth D, Crow JF. Rates of spontaneous mutation. Genetics. 1998;148:1667–1686. doi: 10.1093/genetics/148.4.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drake JW, Bebenek A, Kissling GE, Peddada S. Clusters of mutations from transient hypermutability. Proc Natl Acad Sci U S A. 2005;102:12849–12854. doi: 10.1073/pnas.0503009102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drlica K, Zhao X. DNA gyrase, topoisomerase IV, and the 4-quinolones. Microbiol Mol Biol Rev. 1997;61:377–392. doi: 10.1128/mmbr.61.3.377-392.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudasova Z, Dudas A, Chovanec M. Non-homologous end-joining factors of Saccharomyces cerevisiae. FEMS Microbiol Rev. 2004;28:581–601. doi: 10.1016/j.femsre.2004.06.001. [DOI] [PubMed] [Google Scholar]
- Echols H. SOS functions, cancer and inducible evolution. Cell. 1981;25:1–2. doi: 10.1016/0092-8674(81)90223-3. [DOI] [PubMed] [Google Scholar]
- Edlund T, Normark S. Recombination between short DNA homologies causes tandem duplication. Nature. 1981;292:269–271. doi: 10.1038/292269a0. [DOI] [PubMed] [Google Scholar]
- Feng G, Tsui HC, Winkler ME. Depletion of the cellular amounts of the MutS and MutH methyl-directed mismatch repair proteins in stationary-phase Escherichia coli K-12 cells. J Bacteriol. 1996;178:2388–2396. doi: 10.1128/jb.178.8.2388-2396.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferenci T. What is driving the acquisition of mutS and rpoS polymorphisms in Escherichia coli? Trends Microbiol. 2003;11:457–461. doi: 10.1016/j.tim.2003.08.003. [DOI] [PubMed] [Google Scholar]
- Finkel SE. Long-term survival during stationary phase: evolution and the GASP phenotype. Nat Rev Microbiol. 2006;4:113–120. doi: 10.1038/nrmicro1340. [DOI] [PubMed] [Google Scholar]
- Foster PL. Population dynamics of a Lac- strain of Escherichia coli during selection for lactose utilization. Genetics. 1994;138:253–261. doi: 10.1093/genetics/138.2.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster PL. Nonadaptive mutations occur on the F′ episome during adaptive mutation conditions in Escherichia coli. J Bacteriol. 1997;179:1550–1554. doi: 10.1128/jb.179.5.1550-1554.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster PL. Mechanisms of stationary phase mutation: a decade of adaptive mutation. Annu Rev Genet. 1999;33:57–88. doi: 10.1146/annurev.genet.33.1.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster PL. Adaptive mutation in Escherichia coli. Cold Spring Harb Symp Quant Biol. 2000;65:21–29. doi: 10.1101/sqb.2000.65.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster PL. Adaptive mutation in Escherichia coli. J Bacteriol. 2004a;186:4846–4852. doi: 10.1128/JB.186.15.4846-4852.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster PL. Rebuttal: growth under selection stimulates Lac(+) reversion (Roth and Andersson) J Bacteriol. 2004b;186:4861. doi: 10.1128/JB.186.15.4861.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster PL, Cairns J. The occurrence of heritable Mu excisions in starving cells of Escherichia coli. Embo J. 1994;13:5240–5244. doi: 10.1002/j.1460-2075.1994.tb06855.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster PL, Trimarchi JM. Adaptive reversion of a frameshift mutation in Escherichia coli by simple base deletions in homopolymeric runs. Science. 1994;265:407–409. doi: 10.1126/science.8023164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster PL, Trimarchi JM. Adaptive reversion of an episomal frameshift mutation in Escherichia coli requires conjugal functions but not actual conjugation. Proc Natl Acad Sci U S A. 1995;92:5487–5490. doi: 10.1073/pnas.92.12.5487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster PL, Trimarchi JM, Maurer RA. Two enzymes, both of which process recombination intermediates, have opposite effects on adaptive mutation in Escherichia coli. Genetics. 1996;142:25–37. doi: 10.1093/genetics/142.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francino MP, Chao L, Riley MA, Ochman H. Asymmetries generated by transcription-coupled repair in enterobacterial genes. Science. 1996;272:107–109. doi: 10.1126/science.272.5258.107. [DOI] [PubMed] [Google Scholar]
- Friedberg EC, Walker GC, Siede W, Wood RD, Schultz RA, Ellenberger T. DNA repair and mutagenesis. Washington D.C: ASM Press; 2006. [Google Scholar]
- Friedman N, Vardi S, Ronen M, Alon U, Stavans J. Precise temporal modulation in the response of the SOS DNA repair network in individual bacteria. PLoS Biol. 2005;3:e238. doi: 10.1371/journal.pbio.0030238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galhardo RS, Rocha RP, Marques MV, Menck CF. An SOS-regulated operon involved in damage-inducible mutagenesis in Caulobacter crescentus. Nucleic Acids Res. 2005;33:2603–2614. doi: 10.1093/nar/gki551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galitski T, Roth JR. Evidence that F plasmid transfer replication underlies apparent adaptive mutation. Science. 1995;268:421–423. doi: 10.1126/science.7716546. [DOI] [PubMed] [Google Scholar]
- Gibson TC, Scheppe ML, Cox EC. Fitness of an Escherichia coli mutator gene. Science. 1970;169:686–688. doi: 10.1126/science.169.3946.686. [DOI] [PubMed] [Google Scholar]
- Giraud A, Matic I, Tenaillon O, Clara A, Radman M, Fons M, Taddei F. Cost and benefits of high mutation rates: adaptive evolution of bacteria in the mouse gut. Science. 2001;291:2606–2608. doi: 10.1126/science.1056421. [DOI] [PubMed] [Google Scholar]
- Godoy VG, Gizatullin FS, Fox MS. Some features of the mutability of bacteria during nonlethal selection. Genetics. 2000;154:49–59. doi: 10.1093/genetics/154.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Gomez JM, Blazquez J, Baquero F, Martinez JL. H-NS and RpoS regulate emergence of Lac Ara+ mutants of Escherichia coli MCS2. J Bacteriol. 1997;179:4620–4622. doi: 10.1128/jb.179.14.4620-4622.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene CN, Jinks-Robertson S. Comparison of spontaneous and adaptive mutation spectra in yeast. J Genetics. 1999;78:51–55. [Google Scholar]
- Groisman EA. The pleiotropic two-component regulatory system PhoP-PhoQ. J Bacteriol. 2001;183:1835–1842. doi: 10.1128/JB.183.6.1835-1842.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudmundsdottir K, Ashworth A. The roles of BRCA1 and BRCA2 and associated proteins in the maintenance of genomic stability. Oncogene. 2006;25:5864–5874. doi: 10.1038/sj.onc.1209874. [DOI] [PubMed] [Google Scholar]
- Hall BG. Selection-induced mutations occur in yeast. Proc Natl Acad Sci U S A. 1992;89:4300–4303. doi: 10.1073/pnas.89.10.4300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall BG. Adaptive mutagenesis at ebgR is regulated by PhoPQ. J Bacteriol. 1998;180:2862–2865. doi: 10.1128/jb.180.11.2862-2865.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall BG. Spectra of spontaneous growth-dependent and adaptive mutations at ebgR. J Bacteriol. 1999;181:1149–1155. doi: 10.1128/jb.181.4.1149-1155.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris RS, Feng G, Ross KJ, Sidhu R, Thulin C, Longerich S, Szigety SK, Hastings PJ, Winkler ME, Rosenberg SM. Mismatch repair is diminished during stationary-phase mutation. Mutat Res. 1999;437:51–60. [PubMed] [Google Scholar]
- Harris RS, Feng G, Ross KJ, Sidhu R, Thulin C, Longerich S, Szigety SK, Winkler ME, Rosenberg SM. Mismatch repair protein MutL becomes limiting during stationary-phase mutation. Genes Dev. 1997;11:2426–2437. doi: 10.1101/gad.11.18.2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris RS, Longerich S, Rosenberg SM. Recombination in adaptive mutation. Science. 1994;264:258–260. doi: 10.1126/science.8146657. [DOI] [PubMed] [Google Scholar]
- Harris RS, Ross KJ, Rosenberg SM. Opposing roles of the holliday junction processing systems of Escherichia coli in recombination-dependent adaptive mutation. Genetics. 1996;142:681–691. doi: 10.1093/genetics/142.3.681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastings PJ. Adaptive amplification. Crit Rev Biochem Mol Biol. 2007;42:285–311. doi: 10.1080/10409230701507757. [DOI] [PubMed] [Google Scholar]
- Hastings PJ, Bull HJ, Klump JR, Rosenberg SM. Adaptive amplification: an inducible chromosomal instability mechanism. Cell. 2000;103:723–731. doi: 10.1016/s0092-8674(00)00176-8. [DOI] [PubMed] [Google Scholar]
- Hastings PJ, Slack A, Petrosino JF, Rosenberg SM. Adaptive amplification and point mutation are independent mechanisms: evidence for various stress-inducible mutation mechanisms. PLoS Biol. 2004;2:e399. doi: 10.1371/journal.pbio.0020399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He AS, Rohatgi PR, Hersh MN, Rosenberg SM. Roles of E. coli double-strand-break-repair proteins in stress-induced mutation. DNA Repair (Amst) 2006;5:258–273. doi: 10.1016/j.dnarep.2005.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidenreich E. Adaptive mutation in Saccharomyces cerevisiae. Crit Rev Biochem Mol Biol. 2007;42:385–311. doi: 10.1080/10409230701507773. [DOI] [PubMed] [Google Scholar]
- Heidenreich E, Holzmann V, Eisler H. Polymerase zeta dependency of increased adaptive mutation frequencies in nucleotide excision repair-deficient yeast strains. DNA Repair (Amst) 2004;3:395–402. doi: 10.1016/j.dnarep.2003.12.004. [DOI] [PubMed] [Google Scholar]
- Heidenreich E, Novotny R, Kneidinger B, Holzmann V, Wintersberger U. Non-homologous end joining as an important mutagenic process in cell cycle-arrested cells. Embo J. 2003;22:2274–2283. doi: 10.1093/emboj/cdg203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidenreich E, Wintersberger U. Replication-dependent and selection-induced mutations in respiration-competent and respiration-deficient strains of Saccharomyces cerevisiae. Mol Gen Genet. 1998;260:395–400. doi: 10.1007/s004380050909. [DOI] [PubMed] [Google Scholar]
- Heidenreich E, Wintersberger U. Adaptive reversions of a frameshift mutation in arrested Saccharomyces cerevisiae cells by simple deletions in mononucleotide repeats. Mutat Res. 2001;473:101–107. doi: 10.1016/s0027-5107(00)00141-x. [DOI] [PubMed] [Google Scholar]
- Hendrickson H, Slechta ES, Bergthorsson U, Andersson DI, Roth JR. Amplification-mutagenesis: evidence that “directed” adaptive mutation and general hypermutability result from growth with a selected gene amplification. Proc Natl Acad Sci U S A. 2002;99:2164–2169. doi: 10.1073/pnas.032680899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hengge-Aronis R. Signal transduction and regulatory mechanisms involved in control of the sigma(S) (RpoS) subunit of RNA polymerase. Microbiol Mol Biol Rev. 2002;66:373–395. doi: 10.1128/MMBR.66.3.373-395.2002. table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holbeck SL, Strathern JN. A role for REV3 in mutagenesis during double-strand break repair in Saccharomyces cerevisiae. Genetics. 1997;147:1017–1024. doi: 10.1093/genetics/147.3.1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horak R, Ilves H, Pruunsild P, Kuljus M, Kivisaar M. The ColR-ColS two-component signal transduction system is involved in regulation of Tn4652 transposition in Pseudomonas putida under starvation conditions. Mol Microbiol. 2004;54:795–807. doi: 10.1111/j.1365-2958.2004.04311.x. [DOI] [PubMed] [Google Scholar]
- Horiuchi T, Horiuchi S, Novick A. The genetic basis of hypersynthesis of beta-galactosidase. Genetics. 1963;48:157–169. doi: 10.1093/genetics/48.2.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang LE, Bindra RS, Glazer PM, Harris AL. Hypoxia-induced genetic instability-a calculated mechanism underlying tumor progression. J Mol Med. 2007;85:139–148. doi: 10.1007/s00109-006-0133-6. [DOI] [PubMed] [Google Scholar]
- Huennekens FM. The methotrexate story: a paradigm for development of cancer chemotherapeutic agents. Adv Enzyme Regul. 1994;34:397–419. doi: 10.1016/0065-2571(94)90025-6. [DOI] [PubMed] [Google Scholar]
- Ilves H, Horak R, Kivisaar M. Involvement of sigma(S) in starvation-induced transposition of Pseudomonas putida transposon Tn4652. J Bacteriol. 2001;183:5445–5448. doi: 10.1128/JB.183.18.5445-5448.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilves H, Horak R, Teras R, Kivisaar M. IHF is the limiting host factor in transposition of Pseudomonas putida transposon Tn4652 in stationary phase. Mol Microbiol. 2004;51:1773–1785. doi: 10.1111/j.1365-2958.2003.03948.x. [DOI] [PubMed] [Google Scholar]
- Jarosz DF, Godoy VG, Delaney JC, Essigmann JM, Walker GC. A single amino acid governs enhanced activity of DinB DNA polymerases on damaged templates. Nature. 2006;439:225–228. doi: 10.1038/nature04318. [DOI] [PubMed] [Google Scholar]
- Kasak L, Horak R, Kivisaar M. Promoter-creating mutations in Pseudomonas putida: a model system for the study of mutation in starving bacteria. Proc Natl Acad Sci U S A. 1997;94:3134–3139. doi: 10.1073/pnas.94.7.3134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke Q, Costa M. Hypoxia-inducible factor-1 (HIF-1) Mol Pharmacol. 2006;70:1469–1480. doi: 10.1124/mol.106.027029. [DOI] [PubMed] [Google Scholar]
- Kenyon CJ, Walker GC. DNA-damaging agents stimulate gene expression at specific loci in Escherichia coli. Proc Natl Acad Sci U S A. 1980;77:2819–2823. doi: 10.1073/pnas.77.5.2819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SR, Maenhaut-Michel G, Yamada M, Yamamoto Y, Matsui K, Sofuni T, Nohmi T, Ohmori H. Multiple pathways for SOS-induced mutagenesis in Escherichia coli: an overexpression of dinB/dinP results in strongly enhancing mutagenesis in the absence of any exogenous treatment to damage DNA. Proc Natl Acad Sci U S A. 1997;94:13792–13797. doi: 10.1073/pnas.94.25.13792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SR, Matsui K, Yamada M, Gruz P, Nohmi T. Roles of chromosomal and episomal dinB genes encoding DNA pol IV in targeted and untargeted mutagenesis in Escherichia coli. Mol Genet Genomics. 2001;266:207–215. doi: 10.1007/s004380100541. [DOI] [PubMed] [Google Scholar]
- Koshiji M, To KK, Hammer S, Kumamoto K, Harris AL, Modrich P, Huang LE. HIF-1alpha induces genetic instability by transcriptionally downregulating MutSalpha expression. Mol Cell. 2005;17:793–803. doi: 10.1016/j.molcel.2005.02.015. [DOI] [PubMed] [Google Scholar]
- Kugelberg E, Kofoid E, Reams AB, Andersson DI, Roth JR. Multiple pathways of selected gene amplification during adaptive mutation. Proc Natl Acad Sci U S A. 2006;103:17319–17324. doi: 10.1073/pnas.0608309103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel TA, Erie DA. DNA mismatch repair. Annu Rev Biochem. 2005;74:681–710. doi: 10.1146/annurev.biochem.74.082803.133243. [DOI] [PubMed] [Google Scholar]
- Kuzminov A. Collapse and repair of replication forks in Escherichia coli. Mol Microbiol. 1995;16:373–384. doi: 10.1111/j.1365-2958.1995.tb02403.x. [DOI] [PubMed] [Google Scholar]
- Lamrani S, Ranquet C, Gama MJ, Nakai H, Shapiro JA, Toussaint A, Maenhaut-Michel G. Starvation-induced Mucts62-mediated coding sequence fusion: a role for ClpXP, Lon, RpoS and Crp. Mol Microbiol. 1999;32:327–343. doi: 10.1046/j.1365-2958.1999.01352.x. [DOI] [PubMed] [Google Scholar]
- Layton JC, Foster PL. Error-prone DNA polymerase IV is controlled by the stress-response sigma factor, RpoS, in Escherichia coli. Mol Microbiol. 2003;50:549–561. doi: 10.1046/j.1365-2958.2003.03704.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Layton JC, Foster PL. Error-prone DNA polymerase IV is regulated by the heat shock chaperone GroE in Escherichia coli. J Bacteriol. 2005;187:449–457. doi: 10.1128/JB.187.2.449-457.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lea DE, Coulson CA. The distribution of the numbers of mutants in bacterial populations. J Genet. 1949;49:264–285. doi: 10.1007/BF02986080. [DOI] [PubMed] [Google Scholar]
- LeClerc JE, Li B, Payne WL, Cebula TA. High mutation frequencies among Escherichia coli and Salmonella pathogens. Science. 1996;274:1208–1211. doi: 10.1126/science.274.5290.1208. [DOI] [PubMed] [Google Scholar]
- Lederberg J, Lederberg EM. Replica plating and indirect selection of bacterial mutants. J Bacteriol. 1952;63:399–406. doi: 10.1128/jb.63.3.399-406.1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loeb LA, Springgate CF, Battula N. Errors in DNA replication as a basis of malignant changes. Cancer Res. 1974;34:2311–2321. [PubMed] [Google Scholar]
- Loewe L, Textor V, Scherer S. High deleterious genomic mutation rate in stationary phase of Escherichia coli. Science. 2003;302:1558–1560. doi: 10.1126/science.1087911. [DOI] [PubMed] [Google Scholar]
- Lombardo MJ, Aponyi I, Rosenberg SM. General stress response regulator RpoS in adaptive mutation and amplification in Escherichia coli. Genetics. 2004;166:669–680. doi: 10.1534/genetics.166.2.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longerich S, Galloway AM, Harris RS, Wong C, Rosenberg SM. Adaptive mutation sequences reproduced by mismatch repair deficiency. Proc Natl Acad Sci U S A. 1995;92:12017–12020. doi: 10.1073/pnas.92.26.12017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundblad V, Blackburn EH. An alternative pathway for yeast telomere maintenance rescues est1- senescence. Cell. 1993;73:347–360. doi: 10.1016/0092-8674(93)90234-h. [DOI] [PubMed] [Google Scholar]
- Luria SE, Delbrück M. Mutations on bacteria from virus sensitivity to virus resistance. Genetics. 1943;28:491–511. doi: 10.1093/genetics/28.6.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maenhaut-Michel G, Shapiro JA. The roles of starvation and selective substrates in the emergence of araB-lacZ fusion clones. Embo J. 1994;13:5229–5239. doi: 10.1002/j.1460-2075.1994.tb06854.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao EF, Lane L, Lee J, Miller JH. Proliferation of mutators in a cell population. J Bacteriol. 1997;179:417–422. doi: 10.1128/jb.179.2.417-422.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez JL, Baquero F. Mutation frequencies and antibiotic resistance. Antimicrob Agents Chemother. 2000;44:1771–1777. doi: 10.1128/aac.44.7.1771-1777.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matic I, Radman M, Taddei F, Picard B, Doit C, Bingen E, Denamur E, Elion J. Highly variable mutation rates in commensal and pathogenic Escherichia coli. Science. 1997;277:1833–1834. doi: 10.1126/science.277.5333.1833. [DOI] [PubMed] [Google Scholar]
- McClintock B. The significance of responses of the genome to challenge. Science. 1984;226:792–801. doi: 10.1126/science.15739260. [DOI] [PubMed] [Google Scholar]
- McKenzie GJ, Harris RS, Lee PL, Rosenberg SM. The SOS response regulates adaptive mutation. Proc Natl Acad Sci U S A. 2000;97:6646–6651. doi: 10.1073/pnas.120161797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenzie GJ, Lee PL, Lombardo MJ, Hastings PJ, Rosenberg SM. SOS mutator DNA polymerase IV functions in adaptive mutation and not adaptive amplification. Mol Cell. 2001;7:571–579. doi: 10.1016/s1097-2765(01)00204-0. [DOI] [PubMed] [Google Scholar]
- McKenzie GJ, Lombardo MJ, Rosenberg SM. Recombination-dependent mutation in Escherichia coli occurs in stationary phase. Genetics. 1998;149:1163–1165. doi: 10.1093/genetics/149.2.1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenzie GJ, Magner DB, Lee PL, Rosenberg SM. The dinB operon and spontaneous mutation in Escherichia coli. J Bacteriol. 2003;185:3972–3977. doi: 10.1128/JB.185.13.3972-3977.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer DH, Bailis AM. Telomere dysfunction drives increased mutation by error-prone polymerases Rev1 and zeta in Saccharomyces cerevisiae. Genetics. 2007;175:1533–1537. doi: 10.1534/genetics.106.068130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mihaylova VT, Bindra RS, Yuan J, Campisi D, Narayanan L, Jensen R, Giordano F, Johnson RS, Rockwell S, Glazer PM. Decreased expression of the DNA mismatch repair gene Mlh1 under hypoxic stress in mammalian cells. Mol Cell Biol. 2003;23:3265–3273. doi: 10.1128/MCB.23.9.3265-3273.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller C, Thomsen LE, Gaggero C, Mosseri R, Ingmer H, Cohen SN. SOS response induction by beta-lactams and bacterial defense against antibiotic lethality. Science. 2004;305:1629–1631. doi: 10.1126/science.1101630. [DOI] [PubMed] [Google Scholar]
- Mittler JE, Lenski RE. New data on excisions of Mu from E. coli MCS2 cast doubt on directed mutation hypothesis. Nature. 1990;344:173–175. doi: 10.1038/344173a0. [DOI] [PubMed] [Google Scholar]
- Monsieurs P, De Keersmaecker S, Navarre WW, Bader MW, De Smet F, McClelland M, Fang FC, De Moor B, Vanderleyden J, Marchal K. Comparison of the PhoPQ regulon in Escherichia coli and Salmonella typhimurium. J Mol Evol. 2005;60:462–474. doi: 10.1007/s00239-004-0212-7. [DOI] [PubMed] [Google Scholar]
- Napolitano R, Janel-Bintz R, Wagner J, Fuchs RP. All three SOS-inducible DNA polymerases (Pol II, Pol IV and Pol V) are involved in induced mutagenesis. Embo J. 2000;19:6259–6265. doi: 10.1093/emboj/19.22.6259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuberger MS, Harris RS, Di Noia J, Petersen-Mahrt SK. Immunity through DNA deamination. Trends Biochem Sci. 2003;28:305–312. doi: 10.1016/S0968-0004(03)00111-7. [DOI] [PubMed] [Google Scholar]
- Ninio J. Gene conversion as a focusing mechanism for correlated mutations: a hypothesis. Mol Gen Genet. 1996;251:503–508. doi: 10.1007/BF02173638. [DOI] [PubMed] [Google Scholar]
- Ninio J. Illusory defects and mismatches: why must DNA repair always be (slightly) error prone? Bioessays. 2000;22:396–401. doi: 10.1002/(SICI)1521-1878(200004)22:4<396::AID-BIES10>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- Nohmi T. Environmental stress and lesion-bypass DNA polymerases. Annu Rev Microbiol. 2006;60:231–253. doi: 10.1146/annurev.micro.60.080805.142238. [DOI] [PubMed] [Google Scholar]
- Nowell PC. The clonal evolution of tumor cell populations. Science. 1976;194:23–28. doi: 10.1126/science.959840. [DOI] [PubMed] [Google Scholar]
- O’Wang J, Kawamura K, Tada Y, Ohmori H, Kimura H, Sakiyama S, Tagawa M. DNA polymerase kappa, implicated in spontaneous and DNA damage-induced mutagenesis, is overexpressed in lung cancer. Cancer Res. 2001;61:5366–5369. [PubMed] [Google Scholar]
- Ohmori H, Friedberg EC, Fuchs RP, Goodman MF, Hanaoka F, Hinkle D, Kunkel TA, Lawrence CW, Livneh Z, Nohmi T, et al. The Y-family of DNA polymerases. Mol Cell. 2001;8:7–8. doi: 10.1016/s1097-2765(01)00278-7. [DOI] [PubMed] [Google Scholar]
- Pedraza-Reyes M, Yasbin RE. Contribution of the mismatch DNA repair system to the generation of stationary-phase-induced mutants of Bacillus subtilis. J Bacteriol. 2004;186:6485–6491. doi: 10.1128/JB.186.19.6485-6491.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennington JM, Rosenberg SM. Spontaneous DNA breakage in single living Escherichia coli cells. Nature Genetics. 2007;39:797–802. doi: 10.1038/ng2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Capilla T, Baquero MR, Gomez-Gomez JM, Ionel A, Martin S, Blazquez J. SOS-independent induction of dinB transcription by beta-lactam-mediated inhibition of cell wall synthesis in Escherichia coli. J Bacteriol. 2005;187:1515–1518. doi: 10.1128/JB.187.4.1515-1518.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponder RG. PhD thesis. Baylor College of Medicine; 2006. Error-prone DNA double strand break repair in stress-induced mutation. [DOI] [PubMed] [Google Scholar]
- Ponder RG, Fonville NC, Rosenberg SM. A switch from high-fidelity to error-prone DNA double-strand break repair underlies stress-induced mutation. Mol Cell. 2005;19:791–804. doi: 10.1016/j.molcel.2005.07.025. [DOI] [PubMed] [Google Scholar]
- Powell SC, Wartell RM. Different characteristics distinguish early versus late arising adaptive mutations in Escherichia coli FC40. Mutat Res. 2001;473:219–228. doi: 10.1016/s0027-5107(00)00149-4. [DOI] [PubMed] [Google Scholar]
- Radman M. Phenomenology of an inducible mutagenic DNA repair pathway in Escherichia coli: SOS repair hypothesis. In: Prakash L, FS, Miller M, Lawrence C, Tabor HW, editors. Molecular and environmental aspects of mutagenesis. Springfield; Ill: Rochester, NY: Charles C Thomas; 1974. pp. 128–142. [Google Scholar]
- Riesenfeld C, Everett M, Piddock LJ, Hall BG. Adaptive mutations produce resistance to ciprofloxacin. Antimicrob Agents Chemother. 1997;41:2059–2060. doi: 10.1128/aac.41.9.2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robleto EA, Yasbin R, Pedraza-Reyes M, Ross C. Stationary phase mutagenesis in B. subtilis: a paradigm to study genetic diversity programs in cells under stress. Crit Rev Bioch Mol Biol. 2007 doi: 10.1080/10409230701597717. (this volume) [DOI] [PubMed] [Google Scholar]
- Rodriguez C, Tompkin J, Hazel J, Foster PL. Induction of a DNA nickase in the presence of its target site stimulates adaptive mutation in Escherichia coli. J Bacteriol. 2002;184:5599–5608. doi: 10.1128/JB.184.20.5599-5608.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosche WA, Foster PL. The role of transient hypermutators in adaptive mutation in Escherichia coli. Proc Natl Acad Sci U S A. 1999;96:6862–6867. doi: 10.1073/pnas.96.12.6862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg SM. Evolving responsively: adaptive mutation. Nat Rev Genet. 2001;2:504–515. doi: 10.1038/35080556. [DOI] [PubMed] [Google Scholar]
- Rosenberg SM, Harris RS, Torkelson J. Molecular handles on adaptive mutation. Mol Microbiol. 1995;18:185–189. doi: 10.1111/j.1365-2958.1995.mmi_18020185.x. [DOI] [PubMed] [Google Scholar]
- Rosenberg SM, Hastings PJ. Microbiology and evolution. Modulating mutation rates in the wild. Science. 2003;300:1382–1383. doi: 10.1126/science.1085691. [DOI] [PubMed] [Google Scholar]
- Rosenberg SM, Hastings PJ. Adaptive point mutation and adaptive amplification pathways in the Escherichia coli Lac system: stress responses producing genetic change. J Bacteriol. 2004a;186:4838–4843. doi: 10.1128/JB.186.15.4838-4843.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg SM, Hastings PJ. Rebuttal: growth under selection stimulates Lac(+) reversion (Roth and Andersson) J Bacteriol. 2004b;186:4862–4863. doi: 10.1128/JB.186.15.4862-4863.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg SM, Longerich S, Gee P, Harris RS. Adaptive mutation by deletions in small mononucleotide repeats. Science. 1994;265:405–407. doi: 10.1126/science.8023163. [DOI] [PubMed] [Google Scholar]
- Rosenberg SM, Thulin C, Harris RS. Transient and heritable mutators in adaptive evolution in the lab and in nature. Genetics. 1998;148:1559–1566. doi: 10.1093/genetics/148.4.1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross C, Pybus C, Pedraza-Reyes M, Sung HM, Yasbin RE, Robleto E. Novel role of mfd: effects on stationary-phase mutagenesis in Bacillus subtilis. J Bacteriol. 2006;188:7512–7520. doi: 10.1128/JB.00980-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth JR, Andersson DI. Adaptive mutation: how growth under selection stimulates Lac(+) reversion by increasing target copy number. J Bacteriol. 2004a;186:4855–4860. doi: 10.1128/JB.186.15.4855-4860.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth JR, Andersson DI. Rebuttal: adaptive mutation in Escherichia coli (Foster) J Bacteriol. 2004b;186:4854. doi: 10.1128/JB.186.15.4854.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth JR, Andersson DI. Rebuttal: adaptive point mutation (Rosenberg and Hastings) J Bacteriol. 2004c;186:4844. doi: 10.1128/JB.186.15.4844.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth JR, Kugelberg E, Reams AB, Kofoid E, Andersson DI. Origin of mutations under selection: the adaptive mutation controversy. Annu Rev Microbiol. 2006;60:477–501. doi: 10.1146/annurev.micro.60.080805.142045. [DOI] [PubMed] [Google Scholar]
- Rudner R, Murray A, Huda N. Is there a link between mutation and the stringent response in Bacillus subtilis? Ann N Y Acad Sci. 1999;870:418. doi: 10.1111/j.1749-6632.1999.tb08917.x. [DOI] [PubMed] [Google Scholar]
- Saint-Ruf C, Matic I. Environmental tuning of mutation rates. Environ Microbiol. 2006;8:193–199. doi: 10.1111/j.1462-2920.2005.00968.x. [DOI] [PubMed] [Google Scholar]
- Saumaa S, Tarassova K, Tark M, Tover A, Tegova R, Kivisaar M. Involvement of DNA mismatch repair in stationary-phase mutagenesis during prolonged starvation of Pseudomonas putida. DNA Repair (Amst) 2006;5:505–514. doi: 10.1016/j.dnarep.2005.12.003. [DOI] [PubMed] [Google Scholar]
- Saumaa S, Tover A, Kasak L, Kivisaar M. Different spectra of stationary-phase mutations in early-arising versus late-arising mutants of Pseudomonas putida: involvement of the DNA repair enzyme MutY and the stationary-phase sigma factor RpoS. J Bacteriol. 2002;184:6957–6965. doi: 10.1128/JB.184.24.6957-6965.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savelyeva L, Schwab M. Amplification of oncogenes revisited: from expression profiling to clinical application. Cancer Lett. 2001;167:115–123. doi: 10.1016/s0304-3835(01)00472-4. [DOI] [PubMed] [Google Scholar]
- Schaaper RM, Radman M. The extreme mutator effect of Escherichia coli mutD5 results from saturation of mismatch repair by excessive DNA replication errors. Embo J. 1989;8:3511–3516. doi: 10.1002/j.1460-2075.1989.tb08516.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlacher K, Goodman MF. Lessons from 50 years of SOS DNA-damage-induced mutagenesis. Nat Rev Mol Cell Biol. 2007;8:587–594. doi: 10.1038/nrm2198. [DOI] [PubMed] [Google Scholar]
- Scholes DT, Kenny AE, Gamache ER, Mou Z, Curcio MJ. Activation of a LTR-retrotransposon by telomere erosion. Proc Natl Acad Sci U S A. 2003;100:15736–15741. doi: 10.1073/pnas.2136609100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro JA. Observations on the formation of clones containing araB-lacZ cistron fusions. Mol Gen Genet. 1984;194:79–90. doi: 10.1007/BF00383501. [DOI] [PubMed] [Google Scholar]
- Slack A, Thornton PC, Magner DB, Rosenberg SM, Hastings PJ. On the mechanism of gene amplification induced under stress in Escherichia coli. PLoS Genet. 2006;2:e48. doi: 10.1371/journal.pgen.0020048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sniegowski PD. A test of the directed mutation hypothesis in Escherichia coli MCS2 using replica plating. J Bacteriol. 1995;177:1119–1120. doi: 10.1128/jb.177.4.1119-1120.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sniegowski PD, Gerrish PJ, Lenski RE. Evolution of high mutation rates in experimental populations of E. coli. Nature. 1997;387:703–705. doi: 10.1038/42701. [DOI] [PubMed] [Google Scholar]
- Sniegowski PD, Murphy HA. Evolvability. Curr Biol. 2006;16:R831–834. doi: 10.1016/j.cub.2006.08.080. [DOI] [PubMed] [Google Scholar]
- Snyder L, Champness W. Molecular genetics of bacteria. Washington, DC: ASM Press; 2002. [Google Scholar]
- Steele DF, Jinks-Robertson S. An examination of adaptive reversion in Saccharomyces cerevisiae. Genetics. 1992;132:9–21. doi: 10.1093/genetics/132.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strathern JN, Shafer BK, McGill CB. DNA synthesis errors associated with double-strand-break repair. Genetics. 1995;140:965–972. doi: 10.1093/genetics/140.3.965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stumpf JD, Poteete AR, Foster PL. Amplification of lac cannot account for adaptive mutation to Lac+ in Escherichia coli. J Bacteriol. 2007;189:2291–2299. doi: 10.1128/JB.01706-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung HM, Yasbin RE. Adaptive, or stationary-phase, mutagenesis, a component of bacterial differentiation in Bacillus subtilis. J Bacteriol. 2002;184:5641–5653. doi: 10.1128/JB.184.20.5641-5653.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung HM, Yeamans G, Ross CA, Yasbin RE. Roles of YqjH and YqjW, homologs of the Escherichia coli UmuC/DinB or Y superfamily of DNA polymerases, in stationary-phase mutagenesis and UV-induced mutagenesis of Bacillus subtilis. J Bacteriol. 2003;185:2153–2160. doi: 10.1128/JB.185.7.2153-2160.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taddei F, Halliday JA, Matic I, Radman M. Genetic analysis of mutagenesis in aging Escherichia coli colonies. Mol Gen Genet. 1997a;256:277–281. doi: 10.1007/s004380050570. [DOI] [PubMed] [Google Scholar]
- Taddei F, Radman M, Maynard-Smith J, Toupance B, Gouyon PH, Godelle B. Role of mutator alleles in adaptive evolution. Nature. 1997b;387:700–702. doi: 10.1038/42696. [DOI] [PubMed] [Google Scholar]
- Taddei F, Matic I, Radman M. cAMP-dependent SOS induction and mutagenesis in resting bacterial populations. Proc Natl Acad Sci U S A. 1995;92:11736–11740. doi: 10.1073/pnas.92.25.11736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang M, Pham P, Shen X, Taylor JS, O’Donnell M, Woodgate R, Goodman MF. Roles of E. coli DNA polymerases IV and V in lesion-targeted and untargeted SOS mutagenesis. Nature. 2000;404:1014–1018. doi: 10.1038/35010020. [DOI] [PubMed] [Google Scholar]
- Tark M, Tover A, Tarassova K, Tegova R, Kivi G, Horak R, Kivisaar M. A DNA polymerase V homologue encoded by TOL plasmid pWW0 confers evolutionary fitness on Pseudomonas putida under conditions of environmental stress. J Bacteriol. 2005;187:5203–5213. doi: 10.1128/JB.187.15.5203-5213.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tegova R, Tover A, Tarassova K, Tark M, Kivisaar M. Involvement of error-prone DNA polymerase IV in stationary-phase mutagenesis in Pseudomonas putida. J Bacteriol. 2004;186:2735–2744. doi: 10.1128/JB.186.9.2735-2744.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenaillon O, Denamur E, Matic I. Evolutionary significance of stress-induced mutagenesis in bacteria. Trends Microbiol. 2004;12:264–270. doi: 10.1016/j.tim.2004.04.002. [DOI] [PubMed] [Google Scholar]
- Tenaillon O, Toupance B, Le Nagard H, Taddei F, Godelle B. Mutators, population size, adaptive landscape and the adaptation of asexual populations of bacteria. Genetics. 1999;152:485–493. doi: 10.1093/genetics/152.2.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tlsty TD, Albertini AM, Miller JH. Gene amplification in the lac region of E. coli. Cell. 1984;37:217–224. doi: 10.1016/0092-8674(84)90317-9. [DOI] [PubMed] [Google Scholar]
- To KK, Koshiji M, Hammer S, Huang LE. Genetic instability: the dark side of the hypoxic response. Cell Cycle. 2005;4:881–882. doi: 10.4161/cc.4.7.1839. [DOI] [PubMed] [Google Scholar]
- Toledo F, Wahl GM. Regulating the p53 pathway: in vitro hypotheses, in vivo veritas. Nat Rev Cancer. 2006;6:909–923. doi: 10.1038/nrc2012. [DOI] [PubMed] [Google Scholar]
- Torkelson J, Harris RS, Lombardo MJ, Nagendran J, Thulin C, Rosenberg SM. Genome-wide hypermutation in a subpopulation of stationary-phase cells underlies recombination-dependent adaptive mutation. Embo J. 1997;16:3303–3311. doi: 10.1093/emboj/16.11.3303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsui HC, Feng G, Winkler ME. Negative regulation of mutS and mutH repair gene expression by the Hfq and RpoS global regulators of Escherichia coli K-12. J Bacteriol. 1997;179:7476–7487. doi: 10.1128/jb.179.23.7476-7487.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu X, Latifi T, Bougdour A, Gottesman S, Groisman EA. The PhoP/PhoQ two-component system stabilizes the alternative sigma factor RpoS in Salmonella enterica. Proc Natl Acad Sci U S A. 2006;103:13503–13508. doi: 10.1073/pnas.0606026103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twiss E, Coros AM, Tavakoli NP, Derbyshire KM. Transposition is modulated by a diverse set of host factors in Escherichia coli and is stimulated by nutritional stress. Mol Microbiol. 2005;57:1593–1607. doi: 10.1111/j.1365-2958.2005.04794.x. [DOI] [PubMed] [Google Scholar]
- Volgelstein B, Kinzler KW. The genetic basis of human cancers. New York: McGraw-Hill; 1998. [Google Scholar]
- Wagner J, Gruz P, Kim SR, Yamada M, Matsui K, Fuchs RP, Nohmi T. The dinB gene encodes a novel E. coli DNA polymerase, DNA pol IV, involved in mutagenesis. Mol Cell. 1999;4:281–286. doi: 10.1016/s1097-2765(00)80376-7. [DOI] [PubMed] [Google Scholar]
- Wagner J, Nohmi T. Escherichia coli DNA polymerase IV mutator activity: genetic requirements and mutational specificity. J Bacteriol. 2000;182:4587–4595. doi: 10.1128/jb.182.16.4587-4595.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Gonzalez KD, Scaringe WA, Tsai K, Liu N, Gu D, Li W, Hill KA, Sommer SS. Evidence for mutation showers. Proc Natl Acad Sci U S A. 2007;104:8403–8408. doi: 10.1073/pnas.0610902104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wardle SJ, O’Carroll M, Derbyshire KM, Haniford DB. The global regulator H-NS acts directly on the transpososome to promote Tn10 transposition. Genes Dev. 2005;19:2224–2235. doi: 10.1101/gad.1338905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber H, Polen T, Heuveling J, Wendisch VF, Hengge R. Genomewide analysis of the general stress response network in Escherichia coli: σ S-dependent genes, promoters, and sigma factor selectivity. J Bacteriol. 2005;187:1591–1603. doi: 10.1128/JB.187.5.1591-1603.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff E, Kim M, Hu K, Yang H, Miller JH. Polymerases leave fingerprints: analysis of the mutational spectrum in Escherichia coli rpoB to assess the role of polymerase IV in spontaneous mutation. J Bacteriol. 2004;186:2900–2905. doi: 10.1128/JB.186.9.2900-2905.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright BE. Stress-directed adaptive mutations and evolution. Mol Microbiol. 2004;52:643–650. doi: 10.1111/j.1365-2958.2004.04012.x. [DOI] [PubMed] [Google Scholar]
- Wright BE, Longacre A, Reimers JM. Hypermutation in derepressed operons of Escherichia coli K12. Proc Natl Acad Sci U S A. 1999;96:5089–5094. doi: 10.1073/pnas.96.9.5089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada K, Ariyoshi M, Morikawa K. Three-dimensional structural views of branch migration and resolution in DNA homologous recombination. Curr Opin Struct Biol. 2004;14:130–137. doi: 10.1016/j.sbi.2004.03.005. [DOI] [PubMed] [Google Scholar]
- Yeiser B, Pepper ED, Goodman MF, Finkel SE. SOS-induced DNA polymerases enhance long-term survival and evolutionary fitness. Proc Natl Acad Sci U S A. 2002;99:8737–8741. doi: 10.1073/pnas.092269199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zambrano MM, Siegele DA, Almiron M, Tormo A, Kolter R. Microbial competition: Escherichia coli mutants that take over stationary phase cultures. Science. 1993;259:1757–1760. doi: 10.1126/science.7681219. [DOI] [PubMed] [Google Scholar]