Abstract
Chemotaxis allows bacteria to follow gradients of nutrients and other environmental stimuli. The bacterium Escherichia coli performs chemotaxis via a run-and-tumble strategy in which sensitive temporal comparisons lead to a biased random walk, with longer runs in the preferred gradient direction. The chemotaxis network of E. coli has developed over the years into one of the most thoroughly studied model systems for signal transduction and behaviour, yielding general insights into such properties of cellular networks as signal amplification, signal integration, and robustness. Despite its relative simplicity, the operation of the E. coli chemotaxis network is highly refined and evolutionarily optimized at many levels. For example, recent studies revealed that the network adjusts its signaling properties dependent on the extracellular environment, apparently to optimize chemotaxis under particular conditions. The network can even utilize potentially detrimental stochastic fluctuations in protein levels and reaction rates to maximize the chemotactic performance of the population.
Introduction
The bacterial chemotaxis system is the best-studied biological gradient sensor. The system has remarkable properties – sensitivity to concentrations as low as 3 ligands per cell volume (3 nM), range of response up to five-orders of magnitude of ligand concentration, and integration of multiple signals, including pH, osmolarity, and temperature. Since the core of the chemotaxis network is essentially universal among prokaryotes [1], we limit this review to the workhorse of chemotaxis studies, Escherichia coli. Happily, the E. coli chemotaxis network (Box Figure) is superficially simple with relatively few components and connections (even compared to other chemotaxis networks). Nevertheless, the network has proven to perform sophisticated computation and to be highly organized and evolutionarily optimized at many levels. Since the biochemical and structural aspects of chemotaxis signaling in E. coli have been extensively reviewed recently [2-6], in this review we focus on general principles brought to light by gradient sensing in E. coli.
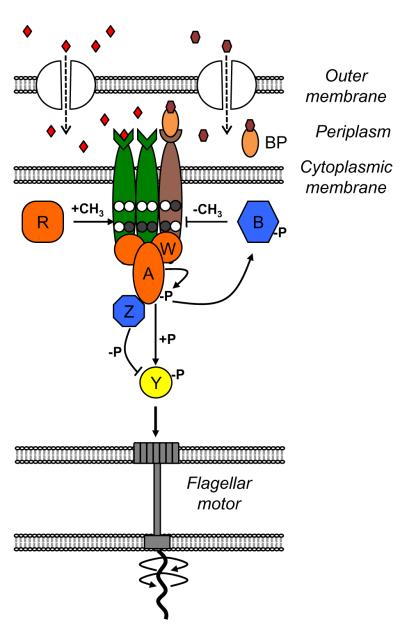
Box Figure.
Fundamental limits of sensitivity
Bacteria like E. coli sense gradients by making temporal comparisons (Fig. 1). Cells set off swimming in one direction, and determine if conditions are getting better or worse. If things are getting better cells tend to keep swimming, if worse (or if the environment remains constant) cells tend to change directions, or “tumble”. The result is a biased random walk leading to progress up an attractant gradient. To make temporal comparisons, cells must both sense their current environment and remember their recent past. Bacteria efficiently combine both these functions in their receptors, which sense ligands and also remember the past via methylation status at specific glutamate residues (Box Figure). Multi-protein receptor complexes, which are organized in the cell in large clusters, also integrate and amplify chemotactic signals (Fig. 2). Ultimately, chemotactic efficiency is limited by how well receptors can sense local ligand concentrations. In their seminal study, Berg & Purcell established biophysical limits on sensing due to the stochasticity both of diffusion and binding/unbinding of ligand molecules [7]. Recently, it was shown that the sensing noise increases due to repetitive rebinding of ligand molecules [8] and can therefore be reduced when the whole cell measures each ligand molecule only once [9]. Additional noise reduction can in principle be achieved by schemes that require expenditure of energy [10] – akin to the energy requirement for kinetic proofreading [11]. However, all of these recent works on the limits of sensing reinforce the central conclusion of Berg & Purcell that the fundamental uncertainty in concentration measurement scales inversely with the number of independently sampled ligand molecules.
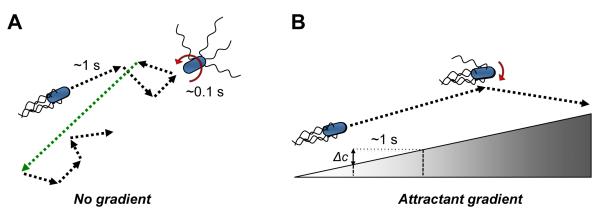
Figure 1. Chemotaxis strategy of Escherichia coli.
(A) Swimming of E. coli in the absence of a gradient. Movement of E. coli cells in a uniform environment consists of smooth runs that last up to several seconds and are interrupted by short (~0.1 sec) tumbles. Runs result from the counterclockwise (CCW) rotation of flagella, which results in formation of a propelling flagellar bundle behind the cell. Tumbles are caused by the clockwise (CW) rotation of one or several flagella, which destabilizes the bundle. Tumbles randomly reorient the cell body before the next run, with the angle of reorientation (indicated by red arrow) being dependent on the number of CW-rotating flagella [43]. The resulting random walk ensures effective foraging in the environment, and may be further enhanced by occasional long runs (green) resulting from stochastic fluctuation in the pathway activity. (B) Chemotaxis in gradients. The chemotaxis strategy of E. coli and other bacteria is based on a biased random walk, whereby cells make temporal comparisons of chemoeffector concentrations during a run and suppress the onset of the next tumble if the level of positive stimulation increases. As a consequence, runs in the positive direction (i.e., up the chemoattractant gradient) are prolonged. Moreover, since on average fewer flagella participate in tumbles when cells are moving up the gradient, the degree of cell body reorientation during such tumbles is smaller. The magnitude of response to the gradient depends on the change in attractant concentration (Δc) experienced by the swimming cell during a run before the cell’s memory is reset by the adaptation system, with the typical run time ~1 s and the corresponding measurement distance ~20 μm.
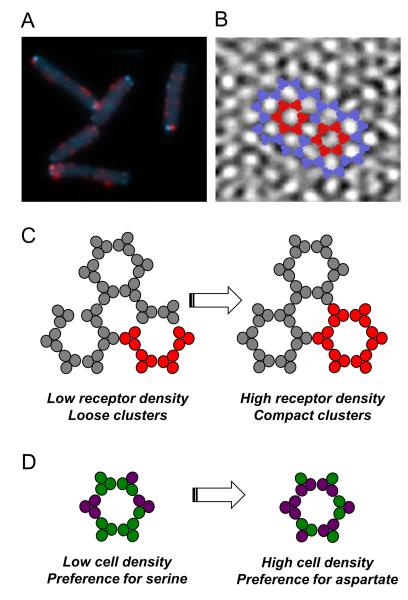
Figure 2. Spatial organization of the chemosensory machinery.
(A) Cellular distribution of chemotaxis and motor proteins. The receptor-kinase sensory complexes and associated chemotaxis proteins are organized into large macromolecular clusters that are visible at the cell poles and along the cell body in fluorescence images. Here receptor clusters are labeled by CheR-CFP (cyan) that directly binds to receptors. Also shown is the cellular distribution of flagellar motors labeled by FliM-YFP (red). (B) Receptor arrangement in clusters. Cryo-electron microscopy image showing a honeycomb lattice of trimers of receptor dimers (courtesy of Ariane Briegel and Grant J Jensen, based on [69] and reproduced from [5]). Individual hexagons of trimers within the lattice are highlighted in red. (C,D) Regulation of allosteric interactions between receptors in clusters. Receptors in clusters are believed to function as allosteric signaling teams of ~2-6 mixed trimers of dimers, with all receptors in a team switching cooperatively between active (i.e. kinase-activating) and inactive (i.e. kinase-inactivating) states, thereby amplifying and integrating chemotactic signals. (C) Regulation of cooperativity among receptors in clusters. High expression levels of receptors and other chemotaxis proteins in nutrient-poor medium results in higher receptor density and higher cooperativity (i.e. signaling team size) (right panel; red indicates one signaling team). (D) Regulation of ligand preference within signaling teams. Receptor dimers of different specificities (green: serine receptor Tsr, magenta: aspartate receptor Tar) are mixed within teams (only one team is shown). Because each team’s response is determined by the total number of bound ligands, higher expression of Tar (as observed at high cell density) changes the ligand preference of the team from serine to aspartate.
The cell’s memory of recent conditions depends on an adaptation system in which the enzymes CheR and CheB reversibly methylate and demethylate receptors (Box Figure). Insofar as the rates of both these enzymes depend only on average receptor activity (and not directly on bound ligand or methylation level), the steady-state requirement that methylation rate equals demethylation rate implies a unique steady-state receptor activity [12]. The result is that receptors adapt to the same fixed activity level for any steady stimulus, which has been shown to be a requirement for optimal chemotactic performance in an unpredictable environment [13]. Control theorists recognize this mechanism for achieving precise adaptation as an example of integral feedback [14,15]. Several recent studies [16-20], have addressed the dynamics and degree of precision of adaptation and what these can teach us about, e.g., receptor organization.
Dynamic range of chemotaxis
The bacterial chemotaxis system has apparently evolved a number of features to refine the general strategy of temporal comparisons. One limitation on cells’ ability to follow chemical gradients is imposed by saturation of the sensory system at high ligand concentrations. In the simple case of ligand binding to a receptor with a fixed affinity, the receptor can discriminate about two orders of magnitude of ligand concentration – the range over which receptor occupancy changes from 10% to 90%. Remarkably, the range of concentrations that can be discriminated by the bacterial chemotaxis system (i.e. its dynamic range) can be extended by the adaptation system to over five orders of magnitude. In a nutshell, increased receptor methylation energetically favors the active state of receptors, which both restores kinase activity and reduces the effective ligand binding affinity. A consequence is that the sensitivity of response to an absolute change in ligand concentration, Δc, decreases with the ambient level of ligand, c0. This desensitization has been shown to obey a particular mode of sensing called Weber’s law [21] or fold-change detection [22,23], which is common for biological sensory systems. Weber’s law states that the system detects relative (Δc/c0) rather than absolute changes in ligand concentrations and implies that chemotactic cells will display “logarithmic” tracking or sensing, characterized by a constant amplitude response when moving in a gradient that increases exponentially or nearly exponentially [24,25].
Response sensitivity and its regulation
Recent studies have shown that the sensitivity of chemotactic response in E. coli is further tuned dependent on growth conditions and on ligand availability. In general, the high sensitivity of response is ensured by allosteric interactions between receptors in chemosensory clusters and between switch subunits of the flagellar motor [26-32] (Fig. 2A). Receptors in clusters are organized within a hexagonal lattice of trimers of dimers, with ~10-20 receptors (a “signaling team”, [33]) switching cooperatively between active and inactive states (Fig. 2B,C). The switch complex of the flagellar motor is a ring of ~30 subunits, which all switch cooperatively so that the motor changes the direction of its rotation between CCW and CW. Both transitions can be described using mathematical models of allosteric proteins [31,34-37] and taken together explain the ~100 fold amplification of chemotactic signals by the pathway. While signal amplification by the motor is believed to be fixed, the strength of receptor interactions in the cluster, and hence signaling team size and response sensitivity, was recently observed to increase for cells grown under nutrient limitation, consistent with the greater importance of finding new sources of nutrients under these conditions [38]. Moreover, since receptors of different types are mixed in signaling teams [33], the sensitivity of response to a particular ligand is primarily determined by the fraction of the corresponding receptor in a team; this allows specific regulation of sensitivity via changes in the relative expression of receptors. Indeed, the ratio between the two major receptors, Tar and Tsr, was observed to increase with cell density, leading to an increased sensitivity to the Tar ligand aspartate compared to the Tsr ligand serine [39-41] with a consequent change in the direction of chemotactic drift in opposing chemoeffector gradients of these two ligands [39]. Such adjustment of ligand preference at high cell density is apparently consistent with the order of amino acid consumption by E. coli, with serine being consumed first and aspartate second [42]. Finally, for indirectly binding ligands such as maltose or galactose, response sensitivity is further tuned by expression levels of periplasmic binding proteins [40], which are in typically induced by the presence of their respective ligands.
Fine-tuning of swimming strategy
Another level of fine-tuning in E. coli chemotaxis is apparently achieved by controlling the degree of reorientation during cell tumbling. In the classical model of bacterial chemotaxis, biased cell movement up a gradient is solely achieved by controlling the probability of tumbling. Recent analyses have suggested, however, that the extent of cell reorientation during a tumble is also regulated [43-45]. This implies that cells swimming up an attractant gradient not only tumble less frequently but also make smaller changes in their swimming direction during a tumble (Fig. 1), substantially improving their rate of chemotactic drift up the gradient.
Robustness of signaling in chemotaxis
As for any other cellular function, chemotactic performance is subject to severe perturbations in intracellular and extracellular parameters. Therefore, it is not surprising that the chemotaxis pathway has evolved mechanisms to compensate for the effects of such perturbations. Robustness of chemotaxis against stochastic variations, or noise, in gene expression – the main intracellular source of perturbations – originates both from the chromosomal organization of the chemotaxis genes and from the pathway topology [46-49]. Because all genes for cytoplasmic chemotaxis proteins are organized in E. coli in two adjacent operons with common transcriptional control, their expression is strongly correlated within individual cells. The chemotaxis pathway employs pairs of opposing enzymatic activities (kinase/phosphatase and methyltransferase/methylesterase; Box Figure) such that the steady-state output of the pathway, the level of phosphorylated CheY, is insensitive (robust) to correlated gene-expression noise.
The pathway is also compensated against the main external perturbation that affects chemotaxis – variation in environmental temperature [50]. The major mechanisms of temperature compensation of the steady-state pathway output are similar to those observed for gene expression noise, whereby opposing enzymatic activities (e.g., those of CheR and CheB) show similar temperature dependencies that are mutually compensatory. In contrast, the effect of temperature on the adaptation kinetics is compensated by the pre-programmed adjustment of the expression levels of adaptation enzymes, such that cells perform chemotaxis optimally at their respective growth temperatures.
Noise and bet hedging in chemotaxis
While design for robust function may be a general principle, that does not imply that all cells should behave identically. For example, in an unpredictable environment, variable behavior among closely related cells could be an advantage; by following different individual courses of action these cells may best ensure the survival of the population as a whole. Several reviews have addressed the role of such “bet-hedging” strategies among microbes [51-55]. Could bet hedging also be a characteristic of the chemotaxis network? Substantial cell-to-cell variability in tumble frequency and adaptation times is well known to occur among genetically identical E. coli cells [56,57]. One major source of variability lies in the generally low copy numbers of CheR and CheB, 200-400 copies per cell compared to 15,000 receptors and 8,000 CheY molecules [58], which may lead to large relative fluctuations in their levels – and therefore in adaptation times – across the population [19,59]. Since there is a defined relation between the steepness of a gradient and the adaptation rate that is optimal for chemotaxis in that gradient, such variability in adaptation rates might be evolutionary beneficial by enabling different cells in the population to optimally follow gradients of different steepness [59]. Another consequence of the low levels of CheR and of its stable binding to receptor clusters [60] are stochastic low-frequency fluctuations in the total CheR reaction rate and therefore in the pathway activity over time. This causes the CW bias of flagellar motors in adapted E. coli to fluctuate on the time scale of tens of seconds [61], resulting in periods of long runs and therefore faster spreading of swimming cells (Fig. 1A), which is potentially advantageous for foraging in the absence of gradients [62,63]. However, there are limits of such beneficial variability, because the adaptation system cannot distinguish between spontaneous changes of receptor activity and changes induced by stimulation. As a result, there is an interdependence between fluctuation and response equivalent to the fluctuation-dissipation theorem for equilibrium systems [64]. Cell-to-cell variability is also present in the total number and distribution of receptors [65,66] and at the level of individual motors [67], indicating possible additional mechanisms of bet hedging by a population of chemotactic cells.
Conclusions
The in depth study of the E. coli chemotaxis system in recent years has continued to prove fruitful, revealing new levels of organization as well as general design principles. A number of the recent discoveries, e.g. those regarding robustness to gene-expression noise and temperature and the role of growth conditions in tuning network operation, are likely to apply to a wide range of intracellular networks both in prokaryotes and eukaryotes. Is there anything important left to discover about the bacterial chemotaxis network? At the molecular level, there is still much to learn about how receptor complexes are organized and how receptors cooperate and respond to signals, and much to learn about how the motor switches and generates torque. At the systems level, one can ask how the network responds to additional environmental factors such as pH, osmotic pressure, or the presence of other cells (quorum sensing). Finally, observations of cell-to-cell variability in chemotactic behavior raises conceptual questions about the proper balance between optimization and bet hedging. Overall, the deep study of chemotaxis in E. coli has been richly rewarding, and we anticipate that this mine has not yet played out.
Box. Signaling network in Escherichia coli chemotaxis.
In E. coli, sensing and processing of chemotactic stimuli is performed by complexes that consist of several types of attractant-specific chemoreceptors in the cytoplasmic membrane (different colors indicate different types of receptors), a histidine kinase CheA, and an adaptor protein CheW (Box Figure). The complex is likely to consist of two mixed trimers of receptor homodimers, two monomers of CheW and one dimer of CheA [68], whereas for simplicity only one receptor trimer is shown in Box Figure and dimers of receptors and of CheA are each drawn as single molecules. Chemoeffectors diffuse through channels in the outer membrane and bind periplasmic sensory domains of receptors. Amino acids bind receptors directly whereas sugars and dipeptides bind indirectly through periplasmic binding proteins (BPs). The output of the signaling network is the level of phosphorylated response regulator CheY, which binds to flagellar motors and induces clockwise (CW) rotation. Attractant binding to receptors inhibits CheA autophosphorylation, reducing CheY phosphorylation and thereby promoting smooth swimming. Rapid dephosphorylation of CheY is ensured by the phosphatase CheZ. Initial rapid response to ligand changes is followed by slower adaptation, mediated by methylation or demethylation of receptors on four specific glutamate residues by the methyltransferase CheR or the methylesterase CheB, respectively (unmethylated and methylated glutamates are shown with white and grey circles, respectively). Higher modification of receptors increases the activity of the associated CheA, thereby allowing cells to adapt to a homogeneous chemical environment. Adaptation primarily relies on the feedback provided by substrate specificity of adaptation enzymes, whereby CheR preferentially methylates inactive receptors and CheB preferentially demethylates active receptors. An additional negative feedback is provided by CheA-dependent CheB phosphorylation, which increases CheB activity. Proteins that promote or inhibit CheY phosphorylation are marked in orange or blue, respectively.
Acknowledgements
We thank Tom Shimizu for critical reading of the manuscript. This work was supported by grant GM082938 from the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Wuichet K, Zhulin IB. Origins and diversification of a complex signal transduction system in prokaryotes. Sci Signal. 2010;3:ra50. doi: 10.1126/scisignal.2000724. *This study provides an excellent overview and classification of bacterial chemotaxis networks.
- 2.Hazelbauer GL, Lai WC. Bacterial chemoreceptors: providing enhanced features to two-component signaling. Curr Opin Microbiol. 2010;13:124–132. doi: 10.1016/j.mib.2009.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hazelbauer GL, Falke JJ, Parkinson JS. Bacterial chemoreceptors: high-performance signaling in networked arrays. Trends Biochem Sci. 2008;33:9–19. doi: 10.1016/j.tibs.2007.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sourjik V. Receptor clustering and signal processing in E. coli chemotaxis. Trends Microbiol. 2004;12:569–576. doi: 10.1016/j.tim.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 5.Sourjik V, Armitage JP. Spatial organization in bacterial chemotaxis. EMBO J. 2010;29:2724–2733. doi: 10.1038/emboj.2010.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wadhams GH, Armitage JP. Making sense of it all: bacterial chemotaxis. Nat Rev Mol Cell Biol. 2004;5:1024–1037. doi: 10.1038/nrm1524. [DOI] [PubMed] [Google Scholar]
- 7.Berg HC, Purcell EM. Physics of chemoreception. Biophys J. 1977;20:193–219. doi: 10.1016/S0006-3495(77)85544-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bialek W, Setayeshgar S. Physical limits to biochemical signaling. Proc Natl Acad Sci U S A. 2005;102:10040–10045. doi: 10.1073/pnas.0504321102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Endres RG, Wingreen NS. Accuracy of direct gradient sensing by single cells. Proc Natl Acad Sci U S A. 2008;105:15749–15754. doi: 10.1073/pnas.0804688105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tu Y. The nonequilibrium mechanism for ultrasensitivity in a biological switch: sensing by Maxwell’s demons. Proc Natl Acad Sci U S A. 2008;105:11737–11741. doi: 10.1073/pnas.0804641105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hopfield JJ. Kinetic proofreading: a new mechanism for reducing errors in biosynthetic processes requiring high specificity. Proc Natl Acad Sci U S A. 1974;71:4135–4139. doi: 10.1073/pnas.71.10.4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barkai N, Leibler S. Robustness in simple biochemical networks. Nature. 1997;387:913–917. doi: 10.1038/43199. [DOI] [PubMed] [Google Scholar]
- 13.Celani A, Vergassola M. Bacterial strategies for chemotaxis response. Proc Natl Acad Sci U S A. 2010;107:1391–1396. doi: 10.1073/pnas.0909673107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yi TM, Andrews BW, Iglesias PA. Control analysis of bacterial chemotaxis signaling. Methods Enzymol. 2007;422:123–140. doi: 10.1016/S0076-6879(06)22006-8. [DOI] [PubMed] [Google Scholar]
- 15.Yi TM, Huang Y, Simon MI, Doyle J. Robust perfect adaptation in bacterial chemotaxis through integral feedback control. Proc Natl Acad Sci U S A. 2000;97:4649–4653. doi: 10.1073/pnas.97.9.4649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Clausznitzer D, Oleksiuk O, Lovdok L, Sourjik V, Endres RG. Chemotactic response and adaptation dynamics in Escherichia coli. PLoS Comput Biol. 2010;6:e1000784. doi: 10.1371/journal.pcbi.1000784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hansen CH, Sourjik V, Wingreen NS. A dynamic-signaling-team model for chemotaxis receptors in Escherichia coli. Proc Natl Acad Sci U S A. 2010 doi: 10.1073/pnas.1005017107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lan G, Schulmeister S, Sourjik V, Tu Y. Adapt locally and act globally: strategy to maintain high chemoreceptor sensitivity in complex environments. Mol Syst Biol. 2011;7:475. doi: 10.1038/msb.2011.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Meir Y, Jakovljevic V, Oleksiuk O, Sourjik V, Wingreen NS. Precision and kinetics of adaptation in bacterial chemotaxis. Biophys J. 2010;99:2766–2774. doi: 10.1016/j.bpj.2010.08.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shimizu TS, Tu Y, Berg HC. A modular gradient-sensing network for chemotaxis in Escherichia coli revealed by responses to time-varying stimuli. Mol Syst Biol. 2010;6:382. doi: 10.1038/msb.2010.37. *This study used FRET measurements of the intracellular pathway response to time-varying stimuli to characterize kinetic properties of the adaptation feedback in chemotaxis.
- 21.Mesibov R, Ordal GW, Adler J. The range of attractant concentrations for bacterial chemotaxis and the threshold and size of response over this range. Weber law and related phenomena. J Gen Physiol. 1973;62:203–223. doi: 10.1085/jgp.62.2.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shoval O, Goentoro L, Hart Y, Mayo A, Sontag E, Alon U. Fold-change detection and scalar symmetry of sensory input fields. Proc Natl Acad Sci U S A. 2010;107:15995–16000. doi: 10.1073/pnas.1002352107. *This study developed the concept of fold-change detection and discussed its application to bacterial chemotaxis.
- 23.Lazova MD, Ahmed T, Bellomo D, Stocker R, Shimizu TS. Response rescaling in bacterial chemotaxis. Proc Natl Acad Sci U S A. 2011;108:13870–13875. doi: 10.1073/pnas.1108608108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kalinin YV, Jiang L, Tu Y, Wu M. Logarithmic sensing in Escherichia coli bacterial chemotaxis. Biophys J. 2009;96:2439–2448. doi: 10.1016/j.bpj.2008.10.027. *This study confirmed that chemotactic E. coli cells in a gradient detect relative rather than absolute changes in ligand concentration.
- 25.Tu Y, Shimizu TS, Berg HC. Modeling the chemotactic response of Escherichia coli to time-varying stimuli. Proc Natl Acad Sci U S A. 2008;105:14855–14860. doi: 10.1073/pnas.0807569105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bai F, Branch RW, Nicolau DV, Jr., Pilizota T, Steel BC, Maini PK, Berry RM. Conformational spread as a mechanism for cooperativity in the bacterial flagellar switch. Science. 2010;327:685–689. doi: 10.1126/science.1182105. **This study revealed real-time dynamics of allosteric transitions during switching of individual motors.
- 27.Cluzel P, Surette M, Leibler S. An ultrasensitive bacterial motor revealed by monitoring signaling proteins in single cells. Science. 2000;287:1652–1655. doi: 10.1126/science.287.5458.1652. [DOI] [PubMed] [Google Scholar]
- 28.Gestwicki JE, Kiessling LL. Inter-receptor communication through arrays of bacterial chemoreceptors. Nature. 2002;415:81–84. doi: 10.1038/415081a. [DOI] [PubMed] [Google Scholar]
- 29.Lai RZ, Manson JM, Bormans AF, Draheim RR, Nguyen NT, Manson MD. Cooperative signaling among bacterial chemoreceptors. Biochemistry. 2005;44:14298–14307. doi: 10.1021/bi050567y. [DOI] [PubMed] [Google Scholar]
- 30.Li G, Weis RM. Covalent modification regulates ligand binding to receptor complexes in the chemosensory system of Escherichia coli. Cell. 2000;100:357–365. doi: 10.1016/s0092-8674(00)80671-6. [DOI] [PubMed] [Google Scholar]
- 31.Sourjik V, Berg HC. Functional interactions between receptors in bacterial chemotaxis. Nature. 2004;428:437–441. doi: 10.1038/nature02406. [DOI] [PubMed] [Google Scholar]
- 32.Vaknin A, Berg HC. Physical responses of bacterial chemoreceptors. J Mol Biol. 2007;366:1416–1423. doi: 10.1016/j.jmb.2006.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ames P, Studdert CA, Reiser RH, Parkinson JS. Collaborative signaling by mixed chemoreceptor teams in Escherichia coli. Proc. Natl. Acad. Sci. USA. 2002;99:7060–7065. doi: 10.1073/pnas.092071899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bray D, Levin MD, Morton-Firth CJ. Receptor clustering as a cellular mechanism to control sensitivity. Nature. 1998;393:85–88. doi: 10.1038/30018. [DOI] [PubMed] [Google Scholar]
- 35.Keymer JE, Endres RG, Skoge M, Meir Y, Wingreen NS. Chemosensing in Escherichia coli: Two regimes of two-state receptors. Proc Natl Acad Sci U S A. 2006;103:1786–1791. doi: 10.1073/pnas.0507438103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mello BA, Tu Y. Quantitative modeling of sensitivity in bacterial chemotaxis: the role of coupling among different chemoreceptor species. Proc. Natl. Acad. Sci. USA. 2003;100:8223–8228. doi: 10.1073/pnas.1330839100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Duke TA, Le Novere N, Bray D. Conformational spread in a ring of proteins: a stochastic approach to allostery. J. Mol. Biol. 2001;308:541–553. doi: 10.1006/jmbi.2001.4610. [DOI] [PubMed] [Google Scholar]
- 38.Khursigara CM, Lan G, Neumann S, Wu X, Ravindran S, Borgnia MJ, Sourjik V, Milne J, Tu Y, Subramaniam S. Lateral density of receptor arrays in the membrane plane influences sensitivity of the E. coli chemotaxis response. EMBO J. 2011;30:1719–1729. doi: 10.1038/emboj.2011.77. *This study demonstrated increased cooperativity in receptor clusters of cells grown in minimal medium compared to cells grown in rich medium.
- 39.Kalinin Y, Neumann S, Sourjik V, Wu M. Responses of Escherichia coli bacteria to two opposing chemoattractant gradients depend on the chemoreceptor ratio. J Bacteriol. 2010;192:1796–1800. doi: 10.1128/JB.01507-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Neumann S, Hansen CH, Wingreen NS, Sourjik V. Differences in signalling by directly and indirectly binding ligands in bacterial chemotaxis. EMBO J. 2010;29:3484–3495. doi: 10.1038/emboj.2010.224. *This study showed how sensitivity of the chemotaxis response depends on the levels of receptors and periplasmic binding proteins and presented a mathematical model describing this regulation.
- 41.Salman H, Libchaber A. A concentration-dependent switch in the bacterial response to temperature. Nat Cell Biol. 2007;9:1098–1100. doi: 10.1038/ncb1632. [DOI] [PubMed] [Google Scholar]
- 42.Pruss BM, Nelms JM, Park C, Wolfe AJ. Mutations in NADH:ubiquinone oxidoreductase of Escherichia coli affect growth on mixed amino acids. J Bacteriol. 1994;176:2143–2150. doi: 10.1128/jb.176.8.2143-2150.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Turner L, Ryu WS, Berg HC. Real-time imaging of fluorescent flagellar filaments. J Bacteriol. 2000;182:2793–2801. doi: 10.1128/jb.182.10.2793-2801.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Saragosti J, Calvez V, Bournaveas N, Perthame B, Buguin A, Silberzan P. Directional persistence of chemotactic bacteria in a traveling concentration wave. Proc Natl Acad Sci U S A. 2011 doi: 10.1073/pnas.1101996108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vladimirov N, Lebiedz D, Sourjik V. Predicted auxiliary navigation mechanism of peritrichously flagellated chemotactic bacteria. PLoS Comput Biol. 2010;6:e1000717. doi: 10.1371/journal.pcbi.1000717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kollmann M, Lovdok L, Bartholome K, Timmer J, Sourjik V. Design principles of a bacterial signalling network. Nature. 2005;438:504–507. doi: 10.1038/nature04228. [DOI] [PubMed] [Google Scholar]
- 47.Lovdok L, Bentele K, Vladimirov N, Muller A, Pop FS, Lebiedz D, Kollmann M, Sourjik V. Role of translational coupling in robustness of bacterial chemotaxis pathway. PLoS Biol. 2009;7:e1000171. doi: 10.1371/journal.pbio.1000171. **This study demonstrated that selection for robustness against gene expression noise can explain the order of chemotaxis genes in bacterial genomes.
- 48.Lovdok L, Kollmann M, Sourjik V. Co-expression of signaling proteins improves robustness of the bacterial chemotaxis pathway. J Biotechnol. 2007;129:173–180. doi: 10.1016/j.jbiotec.2007.01.024. [DOI] [PubMed] [Google Scholar]
- 49.Steuer R, Waldherr S, Sourjik V, Kollmann M. Robust signal processing in living cells. PLoS Comput Biol. 2011 doi: 10.1371/journal.pcbi.1002218. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Oleksiuk O, Jakovljevic V, Vladimirov N, Carvalho R, Paster E, Ryu WS, Meir Y, Wingreen NS, Kollmann M, Sourjik V. Thermal robustness of signaling in bacterial chemotaxis. Cell. 2011;145:312–321. doi: 10.1016/j.cell.2011.03.013. **This study demonstrated robustness of signaling in chemotaxis against variation in temperature and revealed that E. coli can adjust signaling properties of the chemotaxis network to optimize its chemotaxis at a given growth temperature.
- 51.Veening JW, Smits WK, Kuipers OP. Bistability, epigenetics, and bet-hedging in bacteria. Annu Rev Microbiol. 2008;62:193–210. doi: 10.1146/annurev.micro.62.081307.163002. [DOI] [PubMed] [Google Scholar]
- 52.Davidson CJ, Surette MG. Individuality in bacteria. Annu Rev Genet. 2008;42:253–268. doi: 10.1146/annurev.genet.42.110807.091601. [DOI] [PubMed] [Google Scholar]
- 53.Avery SV. Microbial cell individuality and the underlying sources of heterogeneity. Nat Rev Microbiol. 2006;4:577–587. doi: 10.1038/nrmicro1460. [DOI] [PubMed] [Google Scholar]
- 54.Raj A, van Oudenaarden A. Nature, nurture, or chance: stochastic gene expression and its consequences. Cell. 2008;135:216–226. doi: 10.1016/j.cell.2008.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lidstrom ME, Konopka MC. The role of physiological heterogeneity in microbial population behavior. Nat Chem Biol. 2010;6:705–712. doi: 10.1038/nchembio.436. [DOI] [PubMed] [Google Scholar]
- 56.Berg HC, Tedesco PM. Transient response to chemotactic stimuli in Escherichia coli. Proc Natl Acad Sci U S A. 1975;72:3235–3239. doi: 10.1073/pnas.72.8.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Spudich JL, Koshland DE., Jr. Non-genetic individuality: chance in the single cell. Nature. 1976;262:467–471. doi: 10.1038/262467a0. [DOI] [PubMed] [Google Scholar]
- 58.Li M, Hazelbauer GL. Cellular stoichiometry of the components of the chemotaxis signaling complex. J. Bacteriol. 2004;186:3687–3694. doi: 10.1128/JB.186.12.3687-3694.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vladimirov N, Lovdok L, Lebiedz D, Sourjik V. Dependence of bacterial chemotaxis on gradient shape and adaptation rate. PLoS Comput Biol. 2008;4:e1000242. doi: 10.1371/journal.pcbi.1000242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schulmeister S, Ruttorf M, Thiem S, Kentner D, Lebiedz D, Sourjik V. Protein exchange dynamics at chemoreceptor clusters in Escherichia coli. Proc Natl Acad Sci USA. 2008;105:6403–6408. doi: 10.1073/pnas.0710611105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Korobkova E, Emonet T, Vilar JM, Shimizu TS, Cluzel P. From molecular noise to behavioural variability in a single bacterium. Nature. 2004;428:574–578. doi: 10.1038/nature02404. [DOI] [PubMed] [Google Scholar]
- 62.Emonet T, Cluzel P. Relationship between cellular response and behavioral variability in bacterial chemotaxis. Proc Natl Acad Sci USA. 2008;105:3304–3309. doi: 10.1073/pnas.0705463105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Matthaus F, Jagodic M, Dobnikar J. E. coli superdiffusion and chemotaxis-search strategy, precision, and motility. Biophys J. 2009;97:946–957. doi: 10.1016/j.bpj.2009.04.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Park H, Pontius W, Guet CC, Marko JF, Emonet T, Cluzel P. Interdependence of behavioural variability and response to small stimuli in bacteria. Nature. 2010;468:819–823. doi: 10.1038/nature09551. *This study demonstrated a fixed relation between spontaneous motor switching and motor response to small signals, equivalent to an equilibrium “fluctuation-dissipation” relation.
- 65.Greenfield D, McEvoy AL, Shroff H, Crooks GE, Wingreen NS, Betzig E, Liphardt J. Self-organization of the Escherichia coli chemotaxis network imaged with super-resolution light microscopy. PLoS Biol. 2009;7:e1000137. doi: 10.1371/journal.pbio.1000137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Thiem S, Sourjik V. Stochastic assembly of chemoreceptor clusters in Escherichia coli. Mol Microbiol. 2008;68:1228–1236. doi: 10.1111/j.1365-2958.2008.06227.x. [DOI] [PubMed] [Google Scholar]
- 67.Mora T, Bai F, Che YS, Minamino T, Namba K, Wingreen NS. Non-genetic individuality in Escherichia coli motor switching. Phys Biol. 2011;8:024001. doi: 10.1088/1478-3975/8/2/024001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Li M, Hazelbauer GL. Core unit of chemotaxis signaling complexes. Proc Natl Acad Sci U S A. 2011;108:9390–9395. doi: 10.1073/pnas.1104824108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Briegel A, Ortega DR, Tocheva EI, Wuichet K, Li Z, Chen S, Muller A, Iancu CV, Murphy GE, Dobro MJ, et al. Universal architecture of bacterial chemoreceptor arrays. Proc Natl Acad Sci U S A. 2009;106:17181–17186. doi: 10.1073/pnas.0905181106. **This cryo-EM tomography study revealed that receptor trimers of dimers are themselves organized in a regular honeycomb lattice.