Abstract
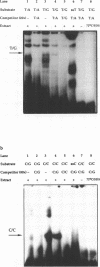
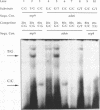
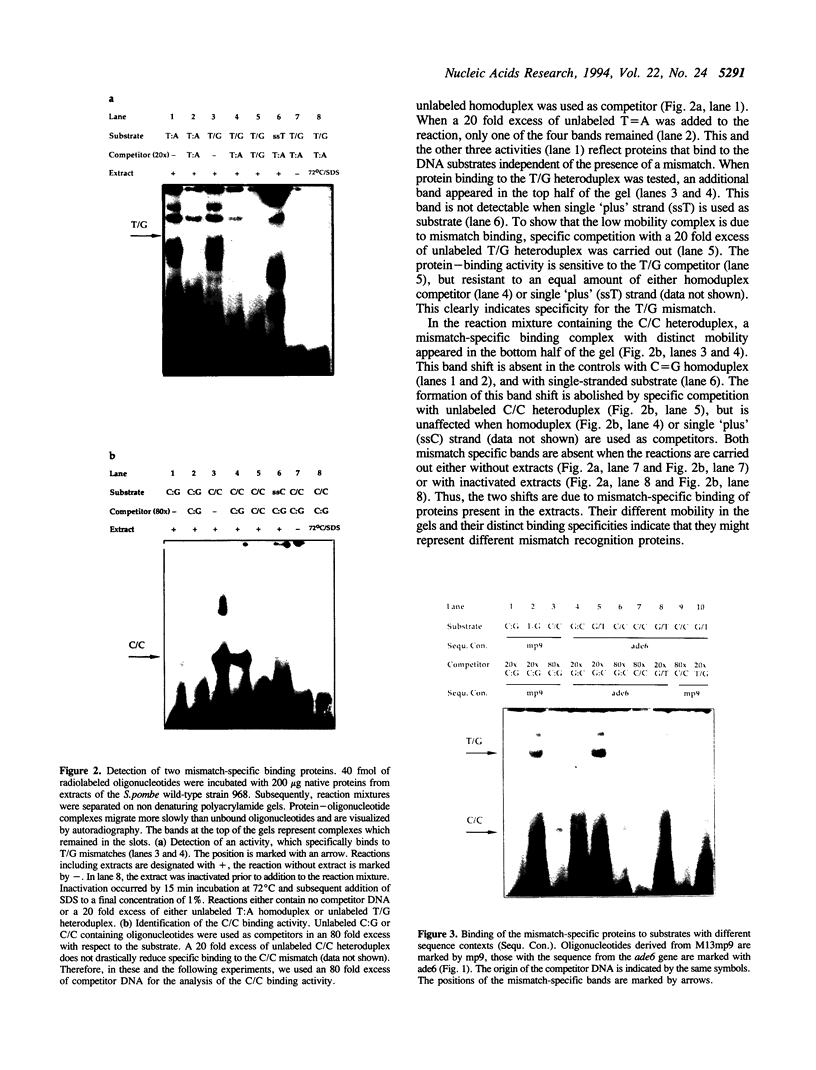
We have performed band-shift assays to identify mismatch-binding proteins in cell extracts of Schizosaccharomyces pombe. By testing heteroduplex DNA containing either a T/G or a C/C mismatch, two distinct band shifts were produced in the gels. A low mobility complex was observed with the T/G substrate, while a high mobility complex was present with C/C. Further analysis of the mismatch-binding specificities revealed that the T/G binding activity also binds to T/C, C/T, T/T, T/-, A/-, C/-, G/-, G/G, A/A, A/C, A/G, G/T, G/A, and C/A substrates with varying efficiencies, but not binds to C/C. The C/C binding activity efficiently binds to C/C, T/C, C/T, C/A, A/C, C/-, and weakly also to T/T, while all other mispairs are not recognized. Protein extracts of a mutant strain, defective in the mutS homologue swi4, displayed both mismatch-binding activities. Thus, swi4 does not encode for either one of the mismatch-binding proteins.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bishop D. K., Andersen J., Kolodner R. D. Specificity of mismatch repair following transformation of Saccharomyces cerevisiae with heteroduplex plasmid DNA. Proc Natl Acad Sci U S A. 1989 May;86(10):3713–3717. doi: 10.1073/pnas.86.10.3713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- Branch P., Aquilina G., Bignami M., Karran P. Defective mismatch binding and a mutator phenotype in cells tolerant to DNA damage. Nature. 1993 Apr 15;362(6421):652–654. doi: 10.1038/362652a0. [DOI] [PubMed] [Google Scholar]
- Claverys J. P., Méjean V., Gasc A. M., Sicard A. M. Mismatch repair in Streptococcus pneumoniae: relationship between base mismatches and transformation efficiencies. Proc Natl Acad Sci U S A. 1983 Oct;80(19):5956–5960. doi: 10.1073/pnas.80.19.5956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper D. L., Lahue R. S., Modrich P. Methyl-directed mismatch repair is bidirectional. J Biol Chem. 1993 Jun 5;268(16):11823–11829. [PubMed] [Google Scholar]
- Dohet C., Wagner R., Radman M. Repair of defined single base-pair mismatches in Escherichia coli. Proc Natl Acad Sci U S A. 1985 Jan;82(2):503–505. doi: 10.1073/pnas.82.2.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egel R., Beach D. H., Klar A. J. Genes required for initiation and resolution steps of mating-type switching in fission yeast. Proc Natl Acad Sci U S A. 1984 Jun;81(11):3481–3485. doi: 10.1073/pnas.81.11.3481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang W. H., Modrich P. Human strand-specific mismatch repair occurs by a bidirectional mechanism similar to that of the bacterial reaction. J Biol Chem. 1993 Jun 5;268(16):11838–11844. [PubMed] [Google Scholar]
- Fishel R., Lescoe M. K., Rao M. R., Copeland N. G., Jenkins N. A., Garber J., Kane M., Kolodner R. The human mutator gene homolog MSH2 and its association with hereditary nonpolyposis colon cancer. Cell. 1993 Dec 3;75(5):1027–1038. doi: 10.1016/0092-8674(93)90546-3. [DOI] [PubMed] [Google Scholar]
- Fleck O., Michael H., Heim L. The swi4+ gene of Schizosaccharomyces pombe encodes a homologue of mismatch repair enzymes. Nucleic Acids Res. 1992 May 11;20(9):2271–2278. doi: 10.1093/nar/20.9.2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox M. S., Radicella J. P., Yamamoto K. Some features of base pair mismatch repair and its role in the formation of genetic recombinants. Experientia. 1994 Mar 15;50(3):253–260. doi: 10.1007/BF01924008. [DOI] [PubMed] [Google Scholar]
- Fujii H., Shimada T. Isolation and characterization of cDNA clones derived from the divergently transcribed gene in the region upstream from the human dihydrofolate reductase gene. J Biol Chem. 1989 Jun 15;264(17):10057–10064. [PubMed] [Google Scholar]
- Grilley M., Welsh K. M., Su S. S., Modrich P. Isolation and characterization of the Escherichia coli mutL gene product. J Biol Chem. 1989 Jan 15;264(2):1000–1004. [PubMed] [Google Scholar]
- Holmes J., Jr, Clark S., Modrich P. Strand-specific mismatch correction in nuclear extracts of human and Drosophila melanogaster cell lines. Proc Natl Acad Sci U S A. 1990 Aug;87(15):5837–5841. doi: 10.1073/pnas.87.15.5837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes M. J., Jiricny J. The purification of a human mismatch-binding protein and identification of its associated ATPase and helicase activities. J Biol Chem. 1992 Nov 25;267(33):23876–23882. [PubMed] [Google Scholar]
- Jiricny J., Hughes M., Corman N., Rudkin B. B. A human 200-kDa protein binds selectively to DNA fragments containing G.T mismatches. Proc Natl Acad Sci U S A. 1988 Dec;85(23):8860–8864. doi: 10.1073/pnas.85.23.8860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer B., Kramer W., Williamson M. S., Fogel S. Heteroduplex DNA correction in Saccharomyces cerevisiae is mismatch specific and requires functional PMS genes. Mol Cell Biol. 1989 Oct;9(10):4432–4440. doi: 10.1128/mcb.9.10.4432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacks S. A., Dunn J. J., Greenberg B. Identification of base mismatches recognized by the heteroduplex-DNA-repair system of Streptococcus pneumoniae. Cell. 1982 Dec;31(2 Pt 1):327–336. doi: 10.1016/0092-8674(82)90126-x. [DOI] [PubMed] [Google Scholar]
- Lahue R. S., Au K. G., Modrich P. DNA mismatch correction in a defined system. Science. 1989 Jul 14;245(4914):160–164. doi: 10.1126/science.2665076. [DOI] [PubMed] [Google Scholar]
- Lieb M., Allen E., Read D. Very short patch mismatch repair in phage lambda: repair sites and length of repair tracts. Genetics. 1986 Dec;114(4):1041–1060. doi: 10.1093/genetics/114.4.1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieb M. Bacterial genes mutL, mutS, and dcm participate in repair of mismatches at 5-methylcytosine sites. J Bacteriol. 1987 Nov;169(11):5241–5246. doi: 10.1128/jb.169.11.5241-5246.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linton J. P., Yen J. Y., Selby E., Chen Z., Chinsky J. M., Liu K., Kellems R. E., Crouse G. F. Dual bidirectional promoters at the mouse dhfr locus: cloning and characterization of two mRNA classes of the divergently transcribed Rep-1 gene. Mol Cell Biol. 1989 Jul;9(7):3058–3072. doi: 10.1128/mcb.9.7.3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu A. L., Chang D. Y. A novel nucleotide excision repair for the conversion of an A/G mismatch to C/G base pair in E. coli. Cell. 1988 Sep 9;54(6):805–812. doi: 10.1016/s0092-8674(88)91109-9. [DOI] [PubMed] [Google Scholar]
- Lu A. L., Chang D. Y. Repair of single base-pair transversion mismatches of Escherichia coli in vitro: correction of certain A/G mismatches is independent of dam methylation and host mutHLS gene functions. Genetics. 1988 Apr;118(4):593–600. doi: 10.1093/genetics/118.4.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messing J., Vieira J. A new pair of M13 vectors for selecting either DNA strand of double-digest restriction fragments. Gene. 1982 Oct;19(3):269–276. doi: 10.1016/0378-1119(82)90016-6. [DOI] [PubMed] [Google Scholar]
- Michaels M. L., Cruz C., Grollman A. P., Miller J. H. Evidence that MutY and MutM combine to prevent mutations by an oxidatively damaged form of guanine in DNA. Proc Natl Acad Sci U S A. 1992 Aug 1;89(15):7022–7025. doi: 10.1073/pnas.89.15.7022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miret J. J., Milla M. G., Lahue R. S. Characterization of a DNA mismatch-binding activity in yeast extracts. J Biol Chem. 1993 Feb 15;268(5):3507–3513. [PubMed] [Google Scholar]
- Modrich P. Mechanisms and biological effects of mismatch repair. Annu Rev Genet. 1991;25:229–253. doi: 10.1146/annurev.ge.25.120191.001305. [DOI] [PubMed] [Google Scholar]
- Muster-Nassal C., Kolodner R. Mismatch correction catalyzed by cell-free extracts of Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1986 Oct;83(20):7618–7622. doi: 10.1073/pnas.83.20.7618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- New L., Liu K., Crouse G. F. The yeast gene MSH3 defines a new class of eukaryotic MutS homologues. Mol Gen Genet. 1993 May;239(1-2):97–108. doi: 10.1007/BF00281607. [DOI] [PubMed] [Google Scholar]
- Parsons R., Li G. M., Longley M. J., Fang W. H., Papadopoulos N., Jen J., de la Chapelle A., Kinzler K. W., Vogelstein B., Modrich P. Hypermutability and mismatch repair deficiency in RER+ tumor cells. Cell. 1993 Dec 17;75(6):1227–1236. doi: 10.1016/0092-8674(93)90331-j. [DOI] [PubMed] [Google Scholar]
- Priebe S. D., Hadi S. M., Greenberg B., Lacks S. A. Nucleotide sequence of the hexA gene for DNA mismatch repair in Streptococcus pneumoniae and homology of hexA to mutS of Escherichia coli and Salmonella typhimurium. J Bacteriol. 1988 Jan;170(1):190–196. doi: 10.1128/jb.170.1.190-196.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prudhomme M., Martin B., Mejean V., Claverys J. P. Nucleotide sequence of the Streptococcus pneumoniae hexB mismatch repair gene: homology of HexB to MutL of Salmonella typhimurium and to PMS1 of Saccharomyces cerevisiae. J Bacteriol. 1989 Oct;171(10):5332–5338. doi: 10.1128/jb.171.10.5332-5338.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radicella J. P., Clark E. A., Fox M. S. Some mismatch repair activities in Escherichia coli. Proc Natl Acad Sci U S A. 1988 Dec;85(24):9674–9678. doi: 10.1073/pnas.85.24.9674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reenan R. A., Kolodner R. D. Characterization of insertion mutations in the Saccharomyces cerevisiae MSH1 and MSH2 genes: evidence for separate mitochondrial and nuclear functions. Genetics. 1992 Dec;132(4):975–985. doi: 10.1093/genetics/132.4.975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reenan R. A., Kolodner R. D. Isolation and characterization of two Saccharomyces cerevisiae genes encoding homologs of the bacterial HexA and MutS mismatch repair proteins. Genetics. 1992 Dec;132(4):963–973. doi: 10.1093/genetics/132.4.963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schär P., Kohli J. Marker effects of G to C transversions on intragenic recombination and mismatch repair in Schizosaccharomyces pombe. Genetics. 1993 Apr;133(4):825–835. doi: 10.1093/genetics/133.4.825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schär P., Munz P., Kohli J. Meiotic mismatch repair quantified on the basis of segregation patterns in Schizosaccharomyces pombe. Genetics. 1993 Apr;133(4):815–824. doi: 10.1093/genetics/133.4.815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohail A., Lieb M., Dar M., Bhagwat A. S. A gene required for very short patch repair in Escherichia coli is adjacent to the DNA cytosine methylase gene. J Bacteriol. 1990 Aug;172(8):4214–4221. doi: 10.1128/jb.172.8.4214-4221.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephenson C., Karran P. Selective binding to DNA base pair mismatches by proteins from human cells. J Biol Chem. 1989 Dec 15;264(35):21177–21182. [PubMed] [Google Scholar]
- Su S. S., Lahue R. S., Au K. G., Modrich P. Mispair specificity of methyl-directed DNA mismatch correction in vitro. J Biol Chem. 1988 May 15;263(14):6829–6835. [PubMed] [Google Scholar]
- Su S. S., Modrich P. Escherichia coli mutS-encoded protein binds to mismatched DNA base pairs. Proc Natl Acad Sci U S A. 1986 Jul;83(14):5057–5061. doi: 10.1073/pnas.83.14.5057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szankasi P., Heyer W. D., Schuchert P., Kohli J. DNA sequence analysis of the ade6 gene of Schizosaccharomyces pombe. Wild-type and mutant alleles including the recombination host spot allele ade6-M26. J Mol Biol. 1988 Dec 20;204(4):917–925. doi: 10.1016/0022-2836(88)90051-4. [DOI] [PubMed] [Google Scholar]
- Valle G., Bergantino E., Lanfranchi G., Carignani G. The sequence of a 6.3 kb segment of yeast chromosome III reveals an open reading frame coding for a putative mismatch binding protein. Yeast. 1991 Dec;7(9):981–988. doi: 10.1002/yea.320070910. [DOI] [PubMed] [Google Scholar]
- Varlet I., Radman M., Brooks P. DNA mismatch repair in Xenopus egg extracts: repair efficiency and DNA repair synthesis for all single base-pair mismatches. Proc Natl Acad Sci U S A. 1990 Oct;87(20):7883–7887. doi: 10.1073/pnas.87.20.7883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsh K. M., Lu A. L., Clark S., Modrich P. Isolation and characterization of the Escherichia coli mutH gene product. J Biol Chem. 1987 Nov 15;262(32):15624–15629. [PubMed] [Google Scholar]
- Williamson M. S., Game J. C., Fogel S. Meiotic gene conversion mutants in Saccharomyces cerevisiae. I. Isolation and characterization of pms1-1 and pms1-2. Genetics. 1985 Aug;110(4):609–646. doi: 10.1093/genetics/110.4.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zell R., Fritz H. J. DNA mismatch-repair in Escherichia coli counteracting the hydrolytic deamination of 5-methyl-cytosine residues. EMBO J. 1987 Jun;6(6):1809–1815. doi: 10.1002/j.1460-2075.1987.tb02435.x. [DOI] [PMC free article] [PubMed] [Google Scholar]