Abstract
We demonstrate the feasibility of a single-molecule microfluidic approach to both sequence detection and obtaining kinetic information for restriction endonucleases on dsDNA. In this method, a microfluidic stagnation point flow is designed to trap, hold, and linearize double-stranded (ds) genomic DNA to which a restriction endonuclease has been pre-bound sequence-specifically. By introducing the cofactor magnesium, we determine the binding location of the enzyme by the cleavage process of dsDNA as in optical restriction mapping, however here the DNA need not be immobilized on a surface. We note that no special labeling of the enzyme is required, which makes it simpler than our previous scheme using stagnation point flows for sequence detection. Our accuracy in determining the location of the recognition site is comparable to or better than other single molecule techniques due to the fidelity with which we can control the linearization of the DNA molecules. In addition, since the cleavage process can be followed in real time, information about the cleavage kinetics, and subtle differences in binding and cleavage frequencies among the recognition sites, may also be obtained. Data for the five recognition sites for the type II restriction endonuclease EcoRI on λ-DNA are presented as a model system. While the roles of the varying fluid velocity and tension along the chain backbone on the measured kinetics remain to be determined, we believe this new method holds promise for a broad range of studies of DNA-protein interactions, including the kinetics of other DNA cleavage processes, the dissociation of a restriction enzyme from the cleaved substrate, and other macromolecular cleavage processes.
Introduction
Restriction enzymes are routinely used for DNA modification and manipulation due to their ability to cut or cleave DNA at specific nucleotide sequences known as restriction or recognition sites. They are widely used in molecular biology, recombinant DNA technology, 1 gene mapping techniques2–10 and DNA sequencing.11, 12 In optical restriction mapping, 3–7 for example, genomic length dsDNA is stretched and immobilized on a substrate, digested with a restriction enzyme, stained with a fluorescent dye, and the positions at which the enzyme has cut the DNA are used to identify the locations of the restriction sites. Hence, understanding enzyme function, specificity, and kinetics is technologically important for their extensive application. However, some basic questions, such as how to easily determine cleavage mechanisms and kinetics for multiple recognition sites simultaneously, remain challenging due to the limitations of existing approaches.13–24 The type II restriction endonuclease EcoRI has been extensively studied;13–20, 22 in the absence of the cofactor Mg2+ it binds to its target sequence with high affinity and specificity but does not cleave the DNA substrate. We use it here with wild-type bacteriophage λ DNA (which contains five EcoRI restriction sites) as a model system to demonstrate our single-molecule, microfluidic method of determining target sequence location and studying its kinetics.
Traditional bulk studies of EcoRI cleavage at its recognition sites have revealed that the cleavage rate varies from site to site on λ-DNA, possibly due to effects of flanking sequences. 13–15, 17, 25, 26 For some substrates and reaction conditions, bulk studies report the reaction mechanism is altered, 16, 26, 27 complicating the interpretation and comparison of bulk studies on different substrates. In addition, some bulk assays use mutant or short DNA substrates on which there are only one or two recognition sites, 14 making comparison with data obtained for wildtype DNA containing multiple recognitions sites ambiguous.
Single molecule studies offer a spectrum of advantages over bulk measurements, including the direct measurement of the distribution of properties rather than the single, ensemble-average value of a property, and the ability to follow complex, multi-step processes in real-time without the need to synchronize the initial conditions for many molecules. Single molecule approaches to studying the interactions of DNA and enzymes have revealed a host of new insights into properties and function that are obscured by ensemble methods.28–32 However, the vast majority of single molecule studies involve immobilization of either the enzyme or the DNA substrate, typically on a glass slide, in a gel, or on a porous support.31 This immobilization results in modification of the species conformation and may create steric obstacles to interactions of the enzyme with the DNA and/or additional diffusional limitations to these interactions. Typically, a process that includes immobilization also precludes the recovery of the species for downstream processing or analysis.
Here we use recent advances in trapping and controlling the conformation of large DNA molecules in microfluidic stagnation point flows to a) determine the locations of target sequences on genomic-length DNA without specially modified probes or surface immobilization and b) to study enzyme kinetics by recognition site. Stagnation point flows have a point of zero velocity at which a DNA molecule may be trapped and held using flow forces without tethering the DNA to a surface or otherwise modifying it; flow forces may then be used to linearize or alter the conformation of the DNA while holding it at a fixed point.33, 34, 35–37 The steady-state extension of the DNA can be easily controlled through the flow strength, and near complete extension of the DNA molecules has been achieved.33, 34 We have recently demonstrated the use of such a flow to detect target sequences on λ-DNA using specially designed and synthesized probes based on a biotinylated mutant of the restriction enzyme EcoRI attached to Neutravidin-coated fluorescent nanospheres.21 In the present work, we extend this research by demonstrating sequence detection using unmodified, wildtype EcoRI. The target sequence location is determined by introducing the dsDNA with the enzyme pre-bound (as a DNA-enzyme complex), linearizing the DNA-enzyme complex at the stagnation point, and then introducing the cofactor Mg2+ which is required for cleavage of the dsDNA. By observing the cleavage location, the position of the target sequence is determined as in optical restriction mapping, but without surface immobilization or the development of special probes. As in our previous work, we note that this approach allows the detection of widely spaced target sequences across genomic lengths of DNA while preserving information about order. Furthermore, by following the cleavage process in real time at the single-molecule level and by restriction site, we may extract information about cleavage kinetics by site. Ultimately, one may be able to extract subtle differences in binding and cleavage frequencies among multiple recognition sites on a single DNA substrate. This single molecule microfluidic method is suitable for a broad range of studies, including the kinetics of other DNA or macromolecular cleavage processes, and, with a slight modification, the dissociation of enzyme from the cleaved DNA substrate.
Materials and Methods
DNA-Enzyme complex preparation
The DNA-Enzyme (DE) complex was prepared outside of the microfluidic device for single-molecule studies as described below. The type II restriction endonuclease EcoRI has been extensively studied,13–20, 22 and here we use commercially available EcoRI as a model system with wild-type bacteriophage λ DNA. EcoRI is a symmetric dimer of two protein subunits17, 38 that binds with extremely high specificity to its recognition site 5′-GAATTC-3′. This sequence appears five times in λ-DNA at 21.226 kb (or 43.7% of the total length from the left end of the DNA, site 1), 26.105 kb (or 53.8%, site 2), 31.747 kb (or 65.5%, site 3), 39.169 kb (or 80.8%, site 4), and 44.973 kb (or 92.7%, site 5). In the absence of the cofactor Mg2+, EcoRI binds sequence-specifically to its target site but cannot cleave the substrate. After the binding of the cofactor Mg2+ (each EcoRI dimer requires two magnesium ions, see Supporting Information Fig. S1), EcoRI will catalyze the cleavage or hydrolysis of dsDNA at each recognition site. 39
First 8 nM wild-type EcoRI (New England Biolabs) and 0.3 nM λ DNA (New England Biolabs) in binding buffer (20 mM sodium phosphate, 100 mM NaCl and 1 M triethylene glycol (TEG), 100μg/mL bovine serum albumin, pH7.3) were incubated in 50 μL reactions at 4 °C for 1 h. For visualization, the DE complexes were fluorescently stained with the intercalating dye YOYO-1; DNA cleavage has been found via bulk experiments to be insensitive to the addition of YOYO-1.40 The above DE complexes were stained by adding 3.6μL of 50 μM YOYO-1 (Invitrogen) and incubating in the dark overnight at room temperature, yielding a staining ratio of 4:1 (bp: YOYO-1). 22
Before single-molecule experiments, the above stained DE complex was diluted to 1.0 pM DNA using H2O (350μL), TEG (150μL), visualization buffer (500μL, 10mM sodium phosphate, 10 mM NaCl, and 1 M TEG, pH 7.3) and viscosified visualization buffer (1mL, 10mM sodium phosphate, 10 mM NaCl, and 25% PEG-8000, pH 7.3). Thirty minutes before starting single molecule experiments, 2μL of 1000x glucose oxidase, 2μL 1000x catalase and 20μL BME were added to the above solution to reduce DNA photocleavage.
Microfluidic stagnation point flow experiments
As in our earlier study,21 the microfluidic stagnation point flow was generated in a cross slot device, consisting of two perpendicular, intersecting channels with two inlets and two outlets. When flow is directed at equal flow rates into the opposing inlets, fluid exits at 90° through the outlets, and a planar extensional flow with a stagnation point centered at the channel intersection is generated. At the stagnation point, a single DE complex may be trapped and held. The DE complex experiences compression along the inflow axis and extension along the outflow axis which linearizes the molecule; the steady-state extension of the DE complex is controlled by the flow rate (see below).
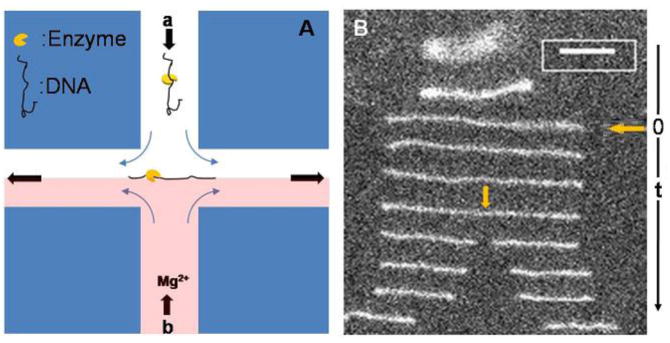
As shown in Figure 1A, the DE complex solution was directed, via a syringe pump (Harvard PHD2000), into one of the two inlet channels (channel a), the cofactor Mg2+ solution, which consists of exactly the same buffer as the DNA solution without DNA or enzyme but with Mg2+, was flowed into the opposing inlet channel (channel b) using the same syringe pump and same flow rates. Movies visualizing the DNA were captured by using a 100x, 1.4-NA oil-immersion objective on a fluorescence microscope (Leica) and a monochrome, image-intensified, cooled CCD camera (Photometrics 512b) and SimplePCI software. Images were captured at 10 frames per second; i.e., the interval between images was 100 ms. Movies were analyzed with ImageJ software to get information about the waiting time for a cleavage event and the location for the first cleavage on each DNA-enzyme complex. Cleavage position is measured on a single frame by taking a 1D-projection of the molecule’s length along the extensional axis and generating an intensity profile using Image J. Fractional cleavage location is determined by normalizing the position of the intensity maximum by the total measured length of the extended DNA molecule. Fractional location is converted to DNA position by multiplying by 48.502 kb, the total length of λ-DNA. Due to the optical indistinguishability of the two DNA ends in this protocol, fractional locations are assigned to yield the best match to expected target locations of EcoRI on λ-DNA.21, 22 This procedure is consistent with that used by others.7–9
Fig. 1.
Experimental setup and images of cleavage event. (A) The scheme for single molecule experiments with stagnation point flow, arrows show the flow directions in the cross-slot device. DNA-Enzyme complexes flow in through the top channel (channel a) and Mg2+ solution flows in from the bottom channel (channel b); DNA-Enzyme complex is trapped at the stagnation point at the intersection of the channels and stretched along the outflow axis. (B) Images of molecular configurations with non-uniform time intervals (smallest time interval, at cleavage, is 100 ms) to show the trapping, stretching and subsequent cleavage of dsDNA. In this experiment, [Mg2+] = 0.25 mM, the steady-state length of the extended DNA is 18.4 μm; the cleavage position is at 53.3% from the left end of the molecule indicated by the yellow arrow. Scale bar: 5 μm.
As for the Mg2+ concentration range we used, it was found by Pingoud et al.,41 for restriction endonuclease EcoRI, the optimal Mg2+ concentration is 2.5 mM. If [Mg2+] is higher than 2.5 mM, more than two magnesium ions will bind with an EcoRI dimer. The excess binding of Mg2+ will inhibit the enzyme activity. On the other hand, if the [Mg2+] is too low, the longer waiting times for the first cleavage event makes it difficult to rule out photo-cleavage from the enzyme-catalyzed cleavage process. Based on these considerations, we varied the [Mg2+] from 0.25 mM to 2.0 mM.
Microfluidic cross slots with channel widths of 800 μm and inlet lengths of 3 mm were prepared using standard soft lithography techniques. 21, 22 Device masters were fabricated to a 130-μm depth using SU-8 2100 resist (MicroChem). PDMS (Sylgard 184, Dow Corning) molds were cast from the master, and inlets and outlets were created using a 16-gauge blunt-tipped needle. Molds were sealed to glass coverslips after treatment with oxygen plasma. Microbore tubing (Tygon, 0.02”ID) was seated directly into the inlet and outlet holes, providing a water-tight seal. Prior to each experiment, tubing and interior surfaces of the sealed microfluidic device were treated for 10 min at room temperature with isopropyl alcohol and subsequently rinsed with ultrapure (18 MΩ) water. This reduced the incidence of trapped air bubbles during channel loading. Next, the tubing and device were incubated for 10 min with 10 mg/ml BSA in PBS buffer (100 mM sodium phosphate, 1.37 mM NaCl, 27 mM KCl, pH 7.4), which helped to reduce surface adsorption of the fluorescent molecules. BSA was rinsed out of the device using ultrapure water. Immediately before experiments, the tubing and microfluidic device were first primed by loading viscosified visualization buffer into the device. Following experiments, devices were flushed thoroughly with water. The device consists of two channels that intersect at right angles, channel dimensions were 130μm in depth and 800 μm in width.
The flow rate in all studies was held constant at 80μL per hour. For the channel dimensions, flow rate, solution viscosity, and DNA used in all the present experiments, the mean steady-state extension of the DNA is 88% of its contour length. Details of the flow and stagnation point control, velocity field, and distribution of steady-state extensions can be found in reference 21.
Results and Discussion
As shown in Fig. 1A the DNA-enzyme (DE) complexes, formed in the absence of Mg2+, are introduced into one inlet (marked “a” in Fig. 1A) of a microfluidic “cross slot”, a buffer solution containing the cofactor Mg2+ is introduced into the opposite channel (b) at the same flow rate. At the stagnation point, the linearized DE complexes are exposed to the Mg2+ in the opposing stream, which can bind to the EcoRI and catalyze phosphodiester bond cleavage at the recognition site. 42 By fluorescently staining the DE complexes with the intercalating fluorescent dye YOYO-1; such a cleavage event can be directly imaged via fluorescence microscopy, as shown in Fig. 1B. Here, a sequence of images of a single DE complex, separated in time, is shown as the complex undergoes cleavage. In the initial images, the partially coiled DE complex is extended by the flow; time t=0 corresponds to the DE complex reaching its steady-state extension.
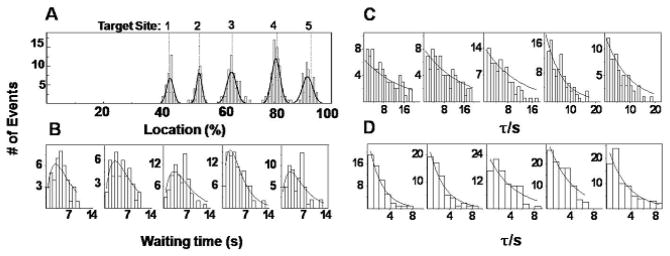
Thus, from movies of cleavage events, we obtained the waiting time for the first DNA cleavage event, defined as the time interval from the trapping of the extended DE complex at the stagnation point to the cleavage or separation of the two fragments after hydrolysis or cleavage of dsDNA. At the flow conditions considered, a large fraction of the DNA molecules enter the stagnation point near their steady-state extension and we focus on only those molecules in measurements of cleavage time. That is, we focus on those molecules that reach steady-state extension within 100 ms (one frame) of entering the stagnation point. We assume that the DNA molecule is exposed to Mg2+ only at the stagnation streamline; that is, we neglect any diffusion of Mg2+ across the stagnation streamline that might result in subtle variations in concentration over the length scale of the DNA molecule. Because the DNA is essentially fully extended prior to cleavage, and because we see no evidence of non-specific binding in bulk studies, 22 the cleavage position corresponds to the location of a recognition sequence. For example, the distributions of the cleavage positions for over 300 DE complexes and the waiting times obtained for an experiment in which stream b contains [Mg2+] =0.5 mM are shown in Fig. 2A and 2B. From Fig. 2A, we see cleavage positions are not randomly distributed along the λ-DNA backbone, but are centered about five distinct positions. These five subpopulations were fitted to Gaussian distributions to obtain the mean cleavage locations and the standard deviations. As expected, these locations coincide with the five target sites of EcoRI on λ DNA as shown in Fig. 2A, indicating these cleavage events should be attributed to the hydrolysis of λ DNA catalyzed by the bound restriction endonuclease EcoRI on recognition sites. The Gaussian best-fit statistics to the five histograms in Fig. 2A, for DNA-enzyme complexes stretched to 88% of the contour length of the DNA, are reported in Table 1. The histograms in Fig. 2B are the distributions of waiting times corresponding to the above five different subpopulations; i.e., the waiting times by recognition site. The average waiting times for the cleavage events are typically a few seconds, and vary from site to site. Data for all recorded cleavage events are included in Fig. 2B.
Fig. 2.
Distribution of cutting location and waiting time. (A) Histogram of measured cleavage positions on 336 λ DNA-Enzyme complexes in an experiment with [Mg2+] =0.5 mM. Solid line is a best fit to the data of a linear combination of Gaussians. The dashed vertical lines indicate the known EcoRI target sites on fully stretched λ DNA numbered from 1 to 5; site 5 is the endmost site. (B) Histograms of waiting times for cleavage events on each of the five target sites of EcoRI on λ DNA; from left to right for target sites from 1 to 5, respectively, with [Mg2+] =0.5 mM, (C) Histograms as in (B), with [Mg2+] =0.25 mM, and (D) [Mg2+] =2.0 mM. Solid lines are the single or double exponential fits.
Table 1.
Gaussian best-fit statistics for histograms shown in Figure 2A. Error is reported as Δ, the distance in percentage or kb from the expected EcoRI target site on λ-DNA; e.g. for an experimentally determined binding location of 43.34% for Site 1, Δ = 43.34% – 43.77% = −0.43%, which multiplies 48.502 kb, the total length of λ-DNA, to be −0.21 kb. Precision is reported as σ, the standard deviation of each Gaussian peak. Both Δ and σ values are shown in two different units: % and kb.
| Δ (%) | Δ(kb) | σ (%) | σ(kb) | |
|---|---|---|---|---|
| Site 1: 43.77% | −0.43 | −0.21 | 2.45 | 1.2 |
| Site 2: 53.83% | −0.26 | −0.13 | 2 | 0.97 |
| Site 3: 65.46% | −0.53 | −0.26 | 2.7 | 1.3 |
| Site 4: 80.76% | −0.28 | −0.14 | 1.1 | 0.53 |
| Site 5: 92.7% | −1.1 | −0.53 | 1.6 | 0.76 |
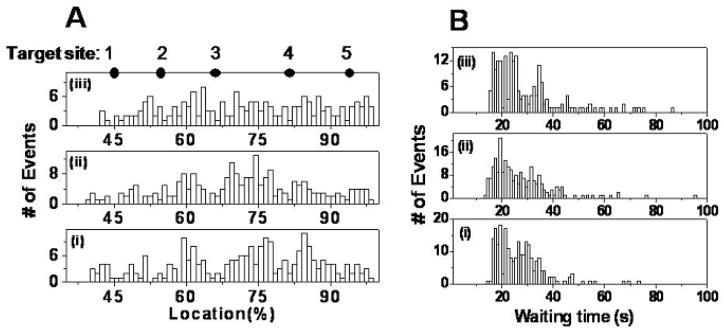
To confirm that these cleavage events are due to the bound EcoRI, we did the following control experiments in the cross slot: (i) DNA, with no enzyme or Mg2+ in channel a, and buffer with no enzyme or Mg2+ in channel b; (ii) DNA with no enzyme or Mg2+ in channel a, and buffer with [Mg2+]= 2.0 mM in channel b; (iii) DNA with bound enzyme, with no Mg2+ in channel a, and buffer with no Mg2+ or enzyme in channel b. As shown in Fig. 3A, the first cleavage events in all three control experiments are randomly distributed over the entire length of the DNA for these events rather than occurring predominantly at the five recognition sites as in Fig. 2A. As shown in Fig. 3B, the average waiting time for the first cleavage of DNA is about half a minute in all these control experiments, and is due to photo-cleavage. The waiting time observed for the first cleavage event in all these control experiments is thus much longer than that observed in experiments with both enzyme and Mg2+ as shown in Fig. 2, and photo-cleavage could be ruled out as the main contributor to the fast cleavage of dsDNA in experiments with both enzyme and Mg2+ binding. Based on these control experiments, then, we conclude that the cleavage reactions in the experiments such as those shown in Fig. 2 are due to the hydrolysis of λ DNA catalyzed by EcoRI.
Fig. 3.
Control experiments. (A) Location distributions of the first cleavage events for three control experiments, ordered, from bottom to top: (i) DNA, with no enzyme or Mg2+ in channel a, and buffer with no enzyme or Mg2+ in channel b; (ii) DNA with no enzyme or Mg2+ in channel a, and buffer with [Mg2+]=2.0 mM in channel b; (iii) DNA with bound enzyme, with no Mg2+ in channel a, and buffer with no Mg2+ or enzyme in channel b. The solid dots indicate expected EcoRI target sites on fully stretched λ DNA numbered from 1 to 5. (B) Distributions of waiting time before the first cleavage event for three control experiments. From bottom to top: (i) <τ>=27.1s; (ii) <τ>=27.2s; (iii) <τ>=29.1s.
Thus, the approach used here is a very simple method for the detection of specific dsDNA sequences since we do not need to label the enzymes with special probes, such as fluorescent beads 18, 21, 22 or otherwise modify the enzyme (for example, by biotinylation). In Table 1, we report target-site prediction to within 1.2 % or 0.6 kb for all five sites with an average precision (reported as the standard deviation) <2.5 % or 1.2 kb. This is comparable to or better than previous single molecule target sequence position determination; for example resolutions of 1–5 kb, based on the full width at half maximum of populations of 100–600 slide-immobilized DNA molecules, have been recently reported by other single molecule techniques.7–9 We previously demonstrated the use of cross slot flows for sequence detection using a biotinylated mutant of EcoRI attached to a NeutrAvidin coated fluorescent nanosphere, 21 however in general the creation of such fluorescent probes is expensive, time-consuming, and the biotinylation or labeling reactions may have low efficiency or interfere with the enzymatic activity. We note that in our earlier study, the error was higher and the precision slightly lower than that reported here; we believe the improved precision in target sequence location in the present case is due to the absence of the nanosphere, which leads to lower fluctuations in the length of the DE complex in this stagnation point flow (see Supplementary information Fig. S1 in reference 21). In addition, we do not need to immobilize the DNA on a surface, so we eliminate the potential steric and diffusion limitations mentioned above; 30, 31, 43 furthermore, in our device the DNA or the fragments following the first cleavage event may be easily recovered for downstream analysis or processing.
Furthermore the magnesium concentration dependence of the cleavage kinetics may also be probed by varying [Mg2+] in the inlet stream (stream b in Fig. 1A). At very low magnesium concentrations, such as [Mg2+] = 0.25mM, the waiting time distribution for each recognition site is a single exponential as shown in Fig. 2C. With increasing magnesium concentration, the distributions of waiting times become double-exponentials (see Fig. 2B with [Mg2+] =0.5 mM). However, when the Mg2+ concentration is sufficiently high, at [Mg2+] = 2.0 mM, the distribution of waiting times becomes a single exponential again (Fig. 2D).
The general mechanism of cleavage of dsDNA by a restriction endonuclease in our system, in which the homodimeric enzyme is prebound to the dsDNA to form a DNA-enzyme complex, may be written as the following: 14, 15
| (1) |
In Equation (1), DE represents the DNA-enzyme complex, DEMg and DE2Mg the complex after one and two cofactor Mg2+ ions, respectively, bind with the enzyme, (the prefactor “2” is due to the dimeric structure of EcoRI in the DE complex),41 P’E2Mg is the product in which one of the two DNA strands has been cleaved at the recognition site, and PE2Mg is the final product (or two fragments of dsDNA) after the cleavage of both strands of dsDNA. k1a and k1b are the apparent rate constants for the first and second Mg2+ binding, respectively, and k2a and k2b are apparent first-order rate-constants for the two steps in the DNA cleavage. In this mechanism, in order to simplify the data analysis, we assume the reverse process of magnesium dissociation from the enzyme (which could in principle be represented by additional rate constants k−1a and k−2a) is negligible compared with the binding process in the magnesium concentration range used in our experiments.
At high magnesium concentrations, the overall rate of Mg2+ ion binding will be fast and as a result the waiting time will be dominated by the cleavage steps. If the cleavage of dsDNA in our system is a two-step process as indicated in Eq 1 with rate constants k2a and k2b of the same order of magnitude,15 then the distribution of waiting times should follow a double exponential (see supplementary information). However, as shown in Fig. 2D the distribution of the waiting times at high [Mg2+] is only a single exponential, which suggests cleavage of the first strand of DNA is fast compared with the second strand cleavage (or both strands are cleaved simultaneously) at these conditions.15 Thus, we can only discriminate one step for the cleavage of dsDNA at these conditions. For similar reasons related to the Mg2+ binding process, the single exponential distribution of waiting times we observe at low [Mg2+] (Fig. 2C) suggests the binding of the two Mg2+ ions takes place in essentially one step. That is, at low magnesium concentrations, the waiting time will be dominated by the slow rate of Mg2+ ion binding; if the rate constants k1a and k1b are the same order of magnitude, the distribution of waiting times is expected to follow a double exponential. The measured single exponential distributions at low [Mg2+] in Fig. 2C are consistent with a simpler, one-step binding process. Based on this reasoning, the mechanism for the kinetic study of EcoRI with λ DNA was simplified as follows:
| (2) |
Note that each waiting time starts at the trapping of the DE complex at the stagnation point and ends at the separation of the two DNA fragments. The waiting time thus does not include the dissociation of the enzyme from the substrate following cleavage or the recombination of free enzyme with another recognition site following cleavage since these steps are not visible in our present experiments. Thus, our single molecule experiments, in addition to allowing determination of recognition site location, measure the single-turnover kinetics of dsDNA cleavage. We emphasize that the results reported pertain to the rate of cleavage after binding of the enzyme to the DNA substrate and may not be relevant to which site would be the cleaved most rapidly when DNA, enzyme, and Mg2+ are simultaneously combined in solution. In addition, our model does not account for the varying tension along the DNA backbone due to the extensional flow field (i.e., the tension is highest near the center of the molecule and lowest at the ends), nor does it account for the varying fluid velocity, and hence varying magnesium ion residence time, along the length of the extended DE complex.
From single molecule theory (Supplementary information) for the waiting time (τ) distribution, we can obtain an expression for the probability density function (pdf), f(τ) as shown in the following:
| (3) |
where . Note that at very low [Mg2+] such that k2 ≫ k10, the second term in Eq. (3) dominates and Eq. (3) predicts a single exponential; similarly, at high [Mg2+] such that k2 ≪ k10, the first term dominates and Eq. (3) again simplifies to a single exponential form.
More generally, at arbitrary [Mg2+], from Eq (3) we can get (see Supplementary information):
| (4) |
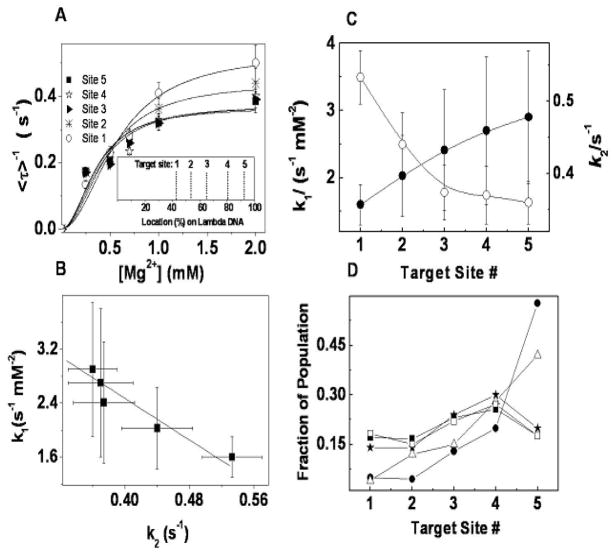
Statistically, <τ>−1, where <τ> denotes averaging, represents the time-averaged single recognition site endonuclease cleavage rate. When averaged over many cleavage events from the same recognition site, Eq. 4 predicts the dependence of <τ>−1 on magnesium concentration [Mg2+]. In our experiments, <τ>−1 for each recognition site indeed shows a dependence on magnesium concentration [Mg2+] consistent with the form of Eq. (4) as shown in Fig. 4A. By fitting the data with Eq. 4, we obtain the kinetic parameters simultaneously for all five recognition sites: k1= 1.6±0.3 s−1mM−2 (site 1), 2.0±0.6 s−1mM−2 (site 2), 2.4±0.9 s−1mM−2(site 3), 2.7±1.1 s−1mM−2(site 4), and, 2.9±1.0 s−1mM−2(site 5) and k2=0.53±0.04 s−1(site 1), 0.44±0.04 s−1(site 2), 0.38±0.04 s−1(site 3), 0.37±0.04 s−1(site 4) and 0.36±0.03 s−1(site 5). The values of k1 and k2 characterize the magnesium binding and cleavage reactivity, respectively, of each recognition site. While these k2 values are much larger than the apparent rate constants obtained by Halford et al. 14 with plasmid or mutant λ DNA on which there was only one recognition site, Halford and co-workers used a different experimental protocol (adding the enzyme to a solution containing DNA and Mg2+) and a different kinetic model which neglected the binding of the enzyme and the Mg2+. We note that our k2 values are consistent in magnitude with values of about 0.5 s−1 obtained for EcoRV with a short oligonucleotide substrate and Mg2+ as cofactor. 24 In the EcoRV study and in the present work, the binding of Mg2+ or enzyme are explicitly included in the kinetic mechanism. Fig. 4A thus supports our kinetic mechanism and the inclusion of the Mg2+ binding step, and we believe one reason for the differences between our k2 values and those of Halford et al.14 relates to the different kinetic models used to analyze the data, although the reasons related to our extensional flow noted above likely also play a role. In addition, we note that in contrast to Halford’s experiments, our measurements were performed on DNA which had been labeled with the intercalating dye YOYO-1, which may affect the cleavage reaction kinetics.
Fig. 4.
Kinetic analysis of cleavage. (A) Magnesium concentration dependence for five recognition sites of EcoRI on λ DNA. Each point is an average of > 60 cleavage events on the same recognition site. Solid lines are the fits with equation (4): k1 = 1.6 ± 0.3 s−1mM−2(site 1), 2.0 ± 0.6 s−1mM−2(site 2), 2.4 ± 0.9 s−1mM−2(site 3), 2.7 ± 1.1 s−1mM−2(site 4), 2.9 ± 1.0 s−1mM−2(site 5), and k2=0.53±0.04 s−1(site 1), 0.44±0.04 s−1(site 2), 0.38±0.04 s−1(site 3), 0.37±0.04 s−1(site 4), 0.36±0.03 s−1(site 5), respectively. Inset shows the digestion map of EcoRI on λ DNA. (B) The correlation between the parameters k1 and k2; and (C) the comparison between the parameters k1 and k2 obtained from [Mg2+] titration (● for k1 and ○ for k2) at each recognition site of EcoRI on λ DNA. (D) Average fraction of population for each target site: ■ from our experiments; △ from Ref. 21 with limited mixing; ★ from Ref. 21 with thorough mixing; □ from Ref. 21 with thorough mixing after scaling by k2; ● from Ref. 23. All the errors here are standard deviations.
In order to get more information about the kinetic properties of the five recognition sites, we studied the correlation between k1 and k2 obtained above for different recognition sites as shown in Fig. 4B (■). A correlation coefficient of −0.95 was obtained, indicating a negative linear relationship between these two kinetic parameters. This relationship clearly indicates for each recognition site on the same DNA molecule, that higher cleavage activity on dsDNA is associated with weaker binding of the cofactor Mg2+ with the enzyme. Similar relationships have been observed in other heterogeneous surface systems 44, 45, however we believe this is the first example revealed on the molecular surface at the single molecule level. We also studied the distribution of k1 and k2 among different recognition sites as shown in Fig. 4C; k1, the magnesium binding rate constant, increases and k2 decreases with the recognition site’s proximity to the end of the DNA molecule (i.e., with target site number). Variations in the activity of EcoRI on sites 2, 4, and 5, under certain conditions, were first noted by Halford et al.14, using bulk studies on mutants of λ-DNA containing only one recognition site. Halford suggested the different reactivities were related to variations in the sequences adjacent to each of the recognition sites. In our study, the monotonic variations in k1 and k2 with distance from the end of the molecule (or target site number), suggest that flow forces may also be playing a role. Clearly, additional studies, to tease apart the effects of flanking sequence, the presence of YOYO-1, tension along the backbone, and residence time of magnesium ions near a particular site are needed. These studies are currently underway using different DNA substrates and different restriction enzymes.
Furthermore by fitting the five subpopulations such as those shown in Fig. 2A, we obtained the average fraction of the population cleaved by each target site averaged over all [Mg2+] shown in Fig. 4D (■). The cleavage frequency at the endmost target site is not the maximum one. We note that Thomas and Davis 23 reported similar data from a bulk study (Fig. 4D (●)), but found the lowest cleavage frequency at the innermost target (site 1, located near the center of the fully-extended DNA molecule, cf Fig. 2A) and the highest frequency at the endmost target (site 5). However, their experimental protocol, which involved a very brief pre-incubation of the DNA with the enzyme in the absence of Mg2+, suggests that their results reflect a convolution of the diffusion-limited enzyme binding to the recognition site, the binding of the Mg2+ to the DE complex, and the cleavage reaction. Indeed, the trend observed by Thomas and Davis 23 is similar to the trend reported by Dylla-Spears et al. 21 (Fig. 4D (△)) for the frequency of EcoRI binding on λ DNA in the absence of Mg2+ under conditions that were mass-transfer limited; that is, when either little mixing or short incubation times are used, EcoRI binds with highest frequency to the endmost recognition site. In contrast, Dylla-Spears and co-workers 21 found that with sufficient mixing during incubation (Fig. 4D(★)), the enzyme binding frequency is more uniform across target sites. In our present experiments, the fraction of the population that cleaves a particular site is a function of both the EcoRI binding frequency and the cleavage activity of the DNA-enzyme-Mg2+ complex. As shown in Fig. 4C, the cleavage rates for the last three sites (3, 4, and 5) are lower than for the innermost sites. So, for example, if EcoRI dimers are bound to target sites 5 and 1 on the same DNA molecule, since only the first cleavage event is counted in our experiments then due to the higher cleavage rate of site 1, there is a high probability that only cleavage at site 1 is observed or counted in our statistics. Obviously this will lead to a completely different fraction of the population in our experiments (■ in Fig. 4D) compared with the fractions obtained from a pure binding assay (■ in Fig. 4D). 21 We note that if we scale the pure binding data (Fig. 4D (★)) by multiplying them with corresponding k2’s obtained from [Mg2+] titration, the result (shown in Fig. 4D (□)) is in very good agreement with our present data (■). Thus, these single molecule experiments may allow us to discriminate, by site, subtle differences in binding and cleavage frequency.
Conclusions
The experiments described here demonstrate the feasibility of a single-molecule microfluidic approach to both sequence detection and obtaining information about the kinetics of restriction endonucleases with dsDNA. In this method we “see” the binding location of the enzyme by the cleavage process of dsDNA as in optical restriction mapping. We note that no special labeling of the enzyme is required, which makes it simpler than our previous scheme using stagnation point flows for sequence detection. 21 Our accuracy in determining the location of the recognition site is comparable to or better than other single molecule techniques7–9 due to the fidelity with which we can control the linearization of the DNA molecules. In addition, since the cleavage process can be followed in real time, we obtained kinetic information for all recognition sites simultaneously on the same DNA substrate to which the enzyme has been pre-bound. Among these different target sites, the data show higher activity of cleavage of dsDNA is associated with weaker binding of Mg2+ with the EcoRI enzyme. The data are consistent with a one-step mechanism of cleavage of λ DNA by EcoRI. The single molecule approach reveals that the closer a target site is to the end of the DNA, the lower the activity for the cleavage of dsDNA and the stronger the Mg2+ binding with pre-bound enzyme. While the roles of the intercalating dye YOYO-1 and the varying fluid velocity (and hence magnesium ion residence time) and tension along the chain backbone on the measured kinetics remain to be determined, we believe this new method holds promise for a broad range of studies of DNA-protein interactions. These include the kinetics of other DNA cleavage processes (involving other types of restriction endonucleases, 41 zinc complexes, 46 biosynthesized cleaving proteins, 47 etc. 48) or other macromolecular cleavage processes. Indeed, we expect that this method can yield insights into enzyme kinetics even for those enzymes that bind non-specifically in the absence of their cofactor as long as the time scale for enzymatic cleavage (on exposure to the cofactor at the stagnation point) can be well separated from the time scale for photocleavage. Note that in principle, one of the cleaved fragments may be captured and held at the stagnation point following the first cleavage event with the technique reported recently by Tanyeri et al.35, thus allowing the study of subsequent cleavage events. Finally, if the enzyme as well as the DNA is fluorescently labeled, this microfluidic stagnation point flow may also be used for studies of the enzyme target site search and binding process, and the dissociation of a restriction enzyme from the cleaved DNA substrate.
Supplementary Material
Acknowledgments
This work was funded by NIH award R21HG004342. The authors would also like to acknowledge Rebecca Dylla-Spears, who fabricated the microfluidic devices. SJM would like to acknowledge support from the Radcliffe Institute for Advanced Studies during the preparation of this manuscript.
References
- 1.Villa-Komaroff L, Efstratiadis A, Broome S, Lomedico P, Tizard R, Naber SP, Chick WL, Gilbert W. Proc Natl Acad Sci USA. 1978;75:3727–3731. doi: 10.1073/pnas.75.8.3727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nathans D, Danna KJ. Nat New Biol. 1973;236:200–202. doi: 10.1038/newbio236200a0. [DOI] [PubMed] [Google Scholar]
- 3.Cai W, Aburatanit H, Stanton VJ, Housmant DE, Wang YK, Schwartz DC. Proc Natl Acad Sci USA. 1995;92:5164–5168. doi: 10.1073/pnas.92.11.5164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jing J, Reed J, Huang J, Hu X, Clarke V, Edington J, Housman D, Anantharamani TS, Huff EJ, Mishrai B, Porter B, Shenker A, Wolfson E, Hiort C, Kantor R, Aston C, Schwartz DC. Proc Natl Acad Sci USA. 1998;95:8046–8051. doi: 10.1073/pnas.95.14.8046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Meng X, Benson K, Chada K, Huff EJ, Schwartz DC. Nature. 1995;9:432–438. doi: 10.1038/ng0495-432. [DOI] [PubMed] [Google Scholar]
- 6.Yokota H, Johnson F, Lu H, Robinson RM, Belu AM, Garrison MD, Ratner BD, Trask BJ, Miller DL. Nucleic Acids Research. 1997;25:1064–1070. doi: 10.1093/nar/25.5.1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yu H, Schwartz DC. Analytical Biochemistry. 2008;380:111–121. doi: 10.1016/j.ab.2008.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xiao M, Gordon MP, Phong A, Ha C, Chan TF, Cai D, Selvin PR, Kwok PY. Human Mutation. 2007;28:913–921. doi: 10.1002/humu.20528. [DOI] [PubMed] [Google Scholar]
- 9.Ebenstein Y, Gassman N, Kim S, Antelman J, Kim Y, Ho S, Samuel R, Michalet X, Weiss S. Nano Lett. 2009;9:1598–1603. doi: 10.1021/nl803820b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schwartz DC, Li X, Hernandez LI, Ramnarain SP, Huff EJ, Wang YK. Science. 1993;262:110–114. doi: 10.1126/science.8211116. [DOI] [PubMed] [Google Scholar]
- 11.Sanger F, Coulson AR. J Mol Biol. 1975;94:441–448. doi: 10.1016/0022-2836(75)90213-2. [DOI] [PubMed] [Google Scholar]
- 12.Maxam AM, Gilbert W. Proc Natl Acad Sci USA. 1977;74:560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Halford SE, Johnson NP. Biochem J. 1980;191:593–604. doi: 10.1042/bj1910593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Halford SE, Johnson NP, Grinsted J. Biochem J. 1980;191:581–592. doi: 10.1042/bj1910581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Halford SE, Johnson NP, Grinsted J. Biochem J. 1979;179:353–365. doi: 10.1042/bj1790353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rubin RA, Modrich P. Nucleic Acids Research. 1978;5:2991–2997. doi: 10.1093/nar/5.8.2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Modrich P, Zabel D. J Biol Chem. 1976;251:5866–5874. [PubMed] [Google Scholar]
- 18.Schafer B, Gemeinhardt H, Greulich KO. Angew Chem Int Ed. 2001;40:4663–4666. doi: 10.1002/1521-3773(20011217)40:24<4663::aid-anie4663>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- 19.Berman HM. Science. 1986;234:1482–1483. doi: 10.1126/science.3024319. [DOI] [PubMed] [Google Scholar]
- 20.Taylor J, Fang RMM, Nie S. Anal Chem. 2000;72:1979–1986. doi: 10.1021/ac9913311. [DOI] [PubMed] [Google Scholar]
- 21.Dylla-Spears R, Townsend JE, Jen-Jacobson L, Sohn LL, Muller SJ. Lab-on-a-Chip. 2010;10:1543–1549. doi: 10.1039/b926847b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dylla-Spears R, Townsend JE, Sohn LL, Jen-Jacobson L, Muller SJ. Anal Chem. 2009;81:10049–10054. doi: 10.1021/ac9019895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thomas M, Davis RW. J Mol Biol. 1975;91:315–328. doi: 10.1016/0022-2836(75)90383-6. [DOI] [PubMed] [Google Scholar]
- 24.Sam MD, Perona JJ. J Am Chem Soc. 1999;121:1444–1447. [Google Scholar]
- 25.Li HW, Yeung ES. Anal Chem. 2005;77:4374–4377. doi: 10.1021/ac050408k. [DOI] [PubMed] [Google Scholar]
- 26.Heitman J. BioEssays. 1992:14. doi: 10.1002/bies.950140704. [DOI] [PubMed] [Google Scholar]
- 27.Bennett SP, Halford SE. Curr Top Cell Regul. 1989;30:57–104. doi: 10.1016/b978-0-12-152830-0.50005-0. [DOI] [PubMed] [Google Scholar]
- 28.Deniz AA, Mukhopadhyay S, Lemke EA. J Roy Soc Interface. 2008;5:15–45. doi: 10.1098/rsif.2007.1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xie XS. Single Mol. 2001;2:229–236. [Google Scholar]
- 30.Kim JH, Dukkipati VR, Pang SW, Larson RG. Nanoscale Res Lett. 2007;2:185–201. [Google Scholar]
- 31.Kim JH, Larson RG. Nucleic Acids Research. 2007;35:3848–3858. doi: 10.1093/nar/gkm332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen Q, Groote R, Schonherr H, Vancso GJ. Chem Soc Rev. 2009;38:2671–2683. doi: 10.1039/b903638e. [DOI] [PubMed] [Google Scholar]
- 33.Perkins TT, Smith DE, Chu S. Science. 1997;276:2016–2021. doi: 10.1126/science.276.5321.2016. [DOI] [PubMed] [Google Scholar]
- 34.Schroeder CM, Babcock HP, Shaqfeh ESG, Chu S. Science. 2003;301:1515–1519. doi: 10.1126/science.1086070. [DOI] [PubMed] [Google Scholar]
- 35.Tanyeri M, Johnson-Chavarria EM, Schroeder CM. Appl Phys Lett. 2010;96:224101–224103. doi: 10.1063/1.3431664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schroeder CM, Shaqfeh ESG, Chu S. Macromolecules. 2004;37:9242–9256. [Google Scholar]
- 37.Smith DE, Chu S. Science. 1998;281:1335–1340. doi: 10.1126/science.281.5381.1335. [DOI] [PubMed] [Google Scholar]
- 38.Roulland-Dussoix D, Yoshimori R, Greene P, Betlach M, Goodman HM, Boyer HW. presented in part at the American Society for Microbiology. Conference on Bacterial Plasmids; Washington, D. C. 1974. [Google Scholar]
- 39.Horvath MM, Choi J, Kim Y, Wilkosz P, Rosenberg JM. Rebase Ref. 1999. p. 5897. [Google Scholar]
- 40.Dylla-Spears RJ. Ph D Dissertation. University of California; Berkeley: 2009. [Google Scholar]
- 41.Pingoud V, Wende W, Friedhoff P, Reuter M, Alves J, Jeltsch A, Mones L, Fuxreiter M, Pingoud A. J Mol Biol. 2009;393:140–160. doi: 10.1016/j.jmb.2009.08.011. [DOI] [PubMed] [Google Scholar]
- 42.Pingoud A, Jeltsch A. Nucleic Acids Research. 2001;29:3705–3727. doi: 10.1093/nar/29.18.3705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Namasivayam V, Larson RG, Burke DT, Burns MA. Anal Chem. 2003;75:4188–4194. doi: 10.1021/ac034180h. [DOI] [PubMed] [Google Scholar]
- 44.Xu W, Kong JS, Chen P. Phys Chem Chem Phys. 2009;11:2767–2778. doi: 10.1039/b820052a. [DOI] [PubMed] [Google Scholar]
- 45.Pankratiev YD. React Kinet Catal Lett. 1982;20:3–4. [Google Scholar]
- 46.He J, Sun J, Mao ZW, Ji LN, Sun H. Journal of Inorganic Biochemistry. 2009;103:851–858. doi: 10.1016/j.jinorgbio.2009.02.010. [DOI] [PubMed] [Google Scholar]
- 47.Lee HS, Schultz PG. Journal of the American Chemical Society. 2008;130:13194–13195. doi: 10.1021/ja804653f. [DOI] [PubMed] [Google Scholar]
- 48.Liu J, Saponjic Z, Dimitrijevic NM, Luo S, Rajht PD. Hybrid TiO2 nanoparticles: An approach for developing site specific DNA cleavage. 2006. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.