Abstract
Mutations in the gene encoding superoxide dismutase 1 (SOD1) account for about 20% of the cases of familial amyotrophic lateral sclerosis (fALS). It is not known how the mutant protein causes disease, or why only a subset of cell types (motor neurons) are targeted. The aggregation and misfolding of mutant SOD1 are implicated in disease pathogenesis in both animal models and humans. We used a monoclonal antibody, C4F6, which specifically reacts with mutant and/or “misfolded” SOD1, to investigate the regional distribution of mutant SOD1 protein in rodent and human tissues. C4F6 reacted only with mutant SOD1 and showed remarkable selectivity for disease-affected tissues and cells. Tissue not affected by disease but containing high levels of mutant protein (sensory neurons) did not stain with C4F6. Additionally, C4F6 intensely stained some motor neurons while leaving adjacent motor neurons unstained. Although C4F6 was generated against the G93A SOD1 mutant, it also recognized other SOD1 mutants. In human autopsy tissues from patients carrying SOD1 mutations, C4F6 identified skein-like intracellular inclusions in motor neurons, similar to those seen in rodents, and again stained only a subset of motor neurons. In spinal cords from patients with sporadic ALS, other neurodegenerative diseases, and normal controls, C4F6-immunoreactive inclusions were not detected, but the antibody did reveal diffuse immunostaining of some spinal motor neurons. The ability of C4F6 to differentiate pathologically affected tissue in mutant SOD1 ALS rodent models and humans, specifically motor neuron populations, suggests that this antibody may recognize a “toxic” form of the mutant SOD1 protein.
Keywords: aggregate, misfold, soluble
Amyotrophic lateral sclerosis (ALS) is a progressive, adult-onset motor neuron disease with an incidence of one to two persons per 100,000 (1, 2). ALS typically causes death within 3–5 y of diagnosis; there is no cure and treatment options are very limited (3–5). Ten percent of ALS cases are familial (fALS), with 20% of fALS being linked to mutations in the homodimeric protein Cu/Zn superoxide dismutase 1 (SOD1) (6–8). In animal models and in humans carrying SOD1 mutations, mutant SOD1 is “toxic” only to motor neurons, and only after it has been present for an extended period (aging), leading to the important questions of how mutant SOD1 exerts its toxicity and why motor neurons are particularly sensitive to the pathogenic process.
Mutant SOD1 has a high propensity to undergo conformational changes and aggregation when stressed in vitro, and SOD1 aggregates and misfolded aggregate precursors have been localized specifically to pathologically affected tissues in animal models of disease, suggesting that SOD1 conformational changes may contribute to the toxicity of the mutant protein (9–14). It has been further demonstrated that conformational changes in wild-type SOD1 can be induced through oxidation and/or metal depletion and that these conformers of the wild-type protein are also toxic in cellular models (15–18). Additionally, misfolded wild-type SOD1 has been identified in both neurons and glia of sporadic ALS (sALS) patients (19, 20), and misfolded wild-type SOD1 derived from sALS tissue inhibits fast axonal transport to a degree reminiscent of that of mutant SOD1 (20). Therefore, misfolded SOD1 may provide a mechanistic link between sporadic and familial forms of ALS.
Several immunological tools have been developed in an attempt to isolate the toxic SOD1 species. Antibodies have been created against the SOD1 dimer interface, against residues in the hydrophobic core, and against specific SOD1 mutant proteins (21–24). We present here further characterization of the C4F6 antibody, which is known to react specifically with mutant human SOD1 (hSOD1) proteins (20, 24). The antibody was generated against the apo (metal-free) form of hSOD1G93A, which exhibits a disordered conformation (25). Immunization with recombinant hSOD1G93A protein increases the life span of hSOD1G37R mice while simultaneously decreasing C4F6 detection of mutant hSOD1 protein, suggesting that C4F6 recognizes toxic mutant SOD1 (24). It has been difficult to identify the precise C4F6 immunoreactive epitope; however, exon 4 of the SOD1 protein is known to be crucial for C4F6 recognition (20). We provide a detailed characterization of this antibody's immunoreactivity both in vitro and in mouse, rat, and human tissues. We demonstrate a unique pattern of C4F6 immunoreactivity that is selective for clinically and pathologically affected tissues in transgenic ALS rodent models and in humans with SOD1 mutations and characterize the biochemical nature of SOD1 protein that is immunoreactive with C4F6. These data indicate that C4F6 recognizes a subset of mutant hSOD1 that is instrumental in the clinical and pathological pattern of motor neuron disease.
Results
The C4F6 antibody selectively stained pathologically affected cell populations (i.e., motor neurons) in both mouse and rat ALS disease models. C4F6 was immunoreactive only with motor neuron populations in these models; no immunoreactivity was observed in secondary sensory neurons in the dorsal horn or in primary sensory neurons in the dorsal root ganglia (Fig. 1 A and I). Immunoreactivity was not uniformly observed across the ventral horn; some motor neurons were intensely stained by C4F6, whereas neighboring neurons with similar morphologies remained unstained (Fig. 1 E and F). Ventral roots exiting the spinal cord were C4F6-immunoreactive, whereas the corresponding dorsal roots did not stain with C4F6 (Fig. 1 G and H). C4F6 staining of hSOD1G93A spinal cord was observed throughout the disease course (presymptomatic through end stage); the distribution of staining expanded with aging, but remained relatively specific for pathologically affected cell populations (Fig. S1). This selectivity for disease-targeted tissues was not due to varying amounts of hSOD1 in these tissues, as staining with a pan-SOD1 antibody demonstrated high levels of SOD1 in clinically unaffected, non-C4F6–immunoreactive tissue (Fig. 1 B and J). Furthermore, C4F6 did not stain in transgenic animals overexpressing wild-type hSOD1, even though hSOD1 was present at high levels in these animals (Fig. 1 C and D).
Fig. 1.
C4F6 is selective for pathologically affected tissue types in ALS rodents. Spinal cord sections from 47-d hSOD1G93A (A and B) and 215-d hSOD1WT (C and D) mice. A and C are stained with the C4F6 antibody and B and D are stained with the pan-SOD1 antibody. (A) Note C4F6 staining of anterior horn cells and ventral root in the ventral spinal cord and the relative lack of staining in the dorsal spinal cord and dorsal root in hSOD1G93A tissue. C4F6 did not stain the spinal cord from hSOD1WT (C). Inset in C further demonstrates this lack of immunoreactivity with a high power image of the hSOD1WT ventral horn stained with C4F6. Staining with the pan-SOD1 antibody demonstrates the diffuse presence of human mutant (B) and wild-type (D) hSOD1 protein. Spinal cord from SOD1 G93A mouse (E) and rat (F) demonstrates that C4F6 stains only a subset of motor neurons within pathologically affected tissue. Furthermore, C4F6 stains rat hSOD1G93A ventral root (G) but not dorsal root (H). Inset in G shows a high power image of ventral root axons showing staining of some, but not all, axons; Inset in H demonstrates minimal staining of dorsal root axons. I–J demonstrate that C4F6 does not recognize mutant SOD1 protein in nonaffected SOD1G93A dorsal root ganglia (I), despite the presence of human mutant SOD1 identified with the pan-SOD1 antibody (J). (Scale bars: E, 50 μm; F–J, 100 μm; all other, 200 μm.) Independent experiments were performed a minimum of three times.
Interestingly, the selectivity of C4F6 for clinically and pathologically affected tissues by immunohistochemistry did not extend to denaturing immunoblot. When the SOD1 protein was denatured through SDS/PAGE, C4F6 recognized mutant hSOD1 protein in dorsal (sensory) spinal cord (Fig. 2A) as well as in ventral spinal cord. C4F6 remained specific for mutant protein in immunoblots and did not stain wild-type hSOD1 (Fig. 2B). However, when SOD1 protein was separated on a native, nondenaturing gel, C4F6 again detected mutant protein only in the ventral horn (Fig. S2).
Fig. 2.
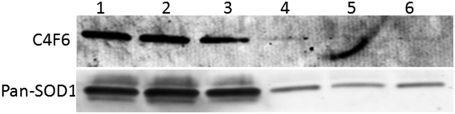
Both disease affected and unaffected hSOD1G93A tissue is C4F6-immunoreactive when denatured through SDS/PAGE. (A) C4F6 is immunoreactive with both denatured dorsal (D) and ventral (V) hSOD1G93A spinal cord. C4F6 does not react with dorsal or ventral cord from either C57/BL6 or hSOD1WT mice. (B) Probing with a pan-SOD1 antibody demonstrates hSOD1 presence; mouse SOD1 (Lower band) runs slightly faster than human SOD1 on SDS/PAGE. Independent experiments were performed a minimum of three times.
Because insoluble aggregates of mutant SOD1 proteins are hypothesized to be toxic (26), and because we suspect that C4F6 reacts with a toxic form of mutant SOD1 protein, we addressed the relative solubility of C4F6-immunoreactive protein using differential solubility extraction. Spinal cords from late-stage hSOD1G93A animals were homogenized in increasingly harsh solvents, and progressive fractions were probed for total SOD1 protein and for C4F6 immunoreactivity. The majority of C4F6-immunoreactive protein was soluble in buffer lacking detergent, with very little requiring harsh detergents for solubility (Fig. 3, Upper). Quantification of the C4F6 immunoreactivity in each solubility condition reveals that ∼59% of total C4F6 immunoreactive protein was soluble in the complete absence of detergent and that ∼27% of total C4F6 immunoreactive protein was soluble with the addition of the relatively mild detergent Nonidet P-40. These results suggest that the protein species preferentially recognized by C4F6 is not insoluble, aggregated protein. Indeed, C4F6 did not recognize the most insoluble hSOD1 species that was detected by the pan-SOD1 antibody (Fig. 3, Lower).
Fig. 3.
C4F6-immunoreactive protein is easily soluble. Dorsal (odd-numbered lanes) and ventral (even-numbered lanes) spinal cord from an end-stage hSOD1G93A mouse extracted in buffer without detergent (lanes 1 and 2), with 1.5% Nonidet P-40 (3, 4), with 1.5% Nonidet P-40 and 2% SDS (5, 6), with 1.5% Nonidet P-40, 2% SDS, and 6 M urea (7, 8), and with the remaining pellet (9, 10). Lane 11 is 215-d hSOD1WT; lane 12 is 70-d hSOD1G93A. Protein was separated through denaturing SDS/PAGE and transferred to PVDF membrane for immunoblot. Upper was probed with C4F6, and Lower was probed with a pan-SOD1 antibody. Note the relatively equal amounts of mutant hSOD1 protein in dorsal vs. ventral spinal cord, in contradistinction to the C4F6 immunoreactivity seen in tissue sections. Independent experiments were performed in duplicate.
Because mutant SOD1 proteins have a tendency to misfold (27–29), we investigated whether the C4F6-immunoreactive hSOD1 showed evidence of being misfolded using hydrophobic interaction chromatography (HIC). HIC operates on the principle that a natively folded protein will shield its hydrophobic core from the hydrophilic environment, whereas misfolded protein will have a greater propensity to expose hydrophobic residues. HIC provides a technical tool to isolate misfolded proteins and has been used to demonstrate that soluble, misfolded forms of hSOD1 are present in fALS mouse spinal cords before the onset of symptoms (14). We hypothesized that, if C4F6 recognizes a misfolded form of SOD1, this form would be selectively enriched in the hydrophobic (misfolded) fraction. However, HIC and subsequent immunoblot of hSOD1G93A spinal cords demonstrated that the majority of C4F6-immunoreactive protein was present in the nonhydrophobic fractions, although some C4F6-immunoreactive protein was identified as hydrophobic (Fig. 4).
Fig. 4.
C4F6 does not robustly detect hydrophobic SOD1. Spinal cord from 135-d hSOD1G93A mouse separated by hydrophobic interaction chromatography and visualized through immunoblot following denaturing SDS/PAGE. C4F6 (Upper) detects the input (lane 1) and flow through not caught on the column (2, 3) but only weakly detects hydrophobic protein eluted off the column (4–6). A pan-SOD1 antibody (Lower) demonstrates that hydrophobic SOD1 is present in the eluate. Blot is representative of duplicate experiments.
Even though the C4F6 antibody was raised against hSOD1G93A mutant protein, it also recognizes linearized hSOD1G37R mutant protein (24), suggesting that the reactive epitope(s) encompass more than the G93A point mutation. To further investigate the reactivity of C4F6 for other SOD1 mutants, CHO cells were transiently transfected with plasmids for A4V, G37R, and G93C human SOD1. C4F6 stained cells expressing hSOD1A4V and hSOD1G37R, but did not stain cells expressing hSOD1G93C (Fig. 5). As in the mice, C4F6 did not stain CHO cells expressing hSOD1WT, whereas the human-specific antibody identified hSOD1 in all transfection conditions. The ability of C4F6 to detect some non-G93A mutant SOD1 proteins confirms previous data showing that the presence of mutant human SOD1 protein, possibly toxic and/or misfolded SOD1, is sufficient for C4F6 recognition (24). However, not all mutants (i.e., hSOD1G93C) are recognized by C4F6. A possible explanation for the inability of C4F6 to recognize G93C is that the 93C mutation is able to form a nonnative disulfide bond that might block the epitope, although we have no evidence to support this hypothesis. As ovary cells are not typically affected in ALS, C4F6 immunoreactivity in transiently transfected CHO cells may at first seem surprising. However, an isolated cell line may not respond to a stressor in the same manner as cells in vivo, and the stress of transient transfection may also increase cell vulnerability.
Fig. 5.
C4F6 detects non-G93A hSOD1 mutants. CHO cells transiently expressing hSOD1G93A, hSOD1A4V, hSOD1G37R, hSOD1G93C, and hSOD1WT were stained with C4F6 (red) or a human SOD1-specific antibody (red) to demonstrate hSOD1 expression. DAPI denotes nuclei (blue). C4F6 detected hSOD1G93A, hSOD1A4V, and hSOD1G37R, but not hSOD1G93C or hSOD1WT. (Scale bar: 50 μm.) Independent experiments were performed three times.
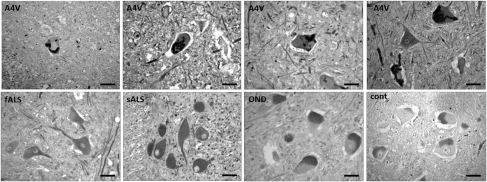
Finally, we investigated the immunoreactivity of C4F6 in human tissues, asking whether the antibody would show similar specificity for motor neurons as was demonstrated in the rodent tissues. In addition, there was a question of whether C4F6 could detect SOD1 even in cases of sporadic ALS, as was demonstrated previously (20). We examined 30 ALS spinal cords and 23 spinal cords from others dying with or without other neurological diseases. Of the 30 ALS patients, three harbored an SOD1 mutation (A4V), and two patients had familial ALS without an SOD1 mutation. Staining with the C4F6 antibody showed distinct, skein-like intracellular inclusions in motor neurons only in the A4V ALS spinal cords (Fig. 6, Upper panels). This pattern of staining was not seen in spinal motor neurons from sALS, non-SOD1 fALS, other neurological disease, or nonneurological controls (Fig. 6, Lower panels). However, we saw diffuse staining of motor neurons in all cases, which was similar in ALS patients and controls (Table 1). The similarity of diffuse neuronal staining in non-SOD1 ALS patients, other neurological disease, and nonneurological controls was not changed by varying the staining methodology. Omitting the antigen-retrieval step from the protocol reduced staining in all cases, but did not change the interpretation of the primary data. In addition, replacement of the C4F6 antibody with normal mouse serum resulted in diffuse staining in some motor neurons, which was similar to that seen with C4F6 in control tissues (Fig. S3). This diffuse staining therefore may be due to a nonspecific background or a reaction with lipofuscin. As is typical in autopsy specimens from adults, many motor neurons showed the granular perikayal staining that is typical of lipofuscin pigment (Fig. S3).
Fig. 6.
Mutant SOD1 human spinal cords show distinct skein-like intracellular inclusions immunoreactive with C4F6. All SOD1A4V cases examined (Upper panels) show skein-like inclusions in motor neurons. Non-SOD1 fALS, sporadic ALS, other neurological diseases, and nondiseased controls (Lower panels) show diffuse staining and do not exhibit skein-like inclusions. (Scale bar: 50 μm.)
Table 1.
C4F6 immunoreactivity with human autopsy tissues
| None | Sparse diffuse | Majority diffuse | Skein-like | |
| A4V | 0 | 0 | 0 | 3 |
| fALS non-SOD1 | 0 | 0 | 2 | 0 |
| sALS | 0 | 10 | 15 | 0 |
| OND | 1 | 4 | 13 | 0 |
| Nondiseased controls | 1 | 4 | 0 | 0 |
Discussion
The identification of a subset of toxic protein in fALS would be of considerable value to mechanistic research into the pathogenesis of ALS, potentially providing a target for development of therapeutics and screening for disease. Here we show that the C4F6 monoclonal antibody generated against mutant human SOD1 not only can differentiate between mutant and wild-type SOD1 protein, but also specifically reacts with mutant SOD1 protein present in tissues and cells affected by the disease. Although our data do not directly demonstrate that the C4F6-immunoreactive protein is toxic, its propensity to localize only to the pathologically affected neuronal populations in both humans and rodent ALS models strongly suggests that this form of the mutant SOD1 protein plays a role in ALS pathogenesis. The differential reactivity of SOD1 proteins to C4F6 suggests structural variations in the proteins that could provide clues to the difficult question of why mutant SOD1 is preferentially toxic to motor neurons. Additionally, clinicians, pathologists, and patients have long been puzzled by the propensity of ALS to affect some motor neurons while seemingly leaving neighboring neuronal populations intact. Determining why C4F6 reacts only with a subset of neurons harboring mutant SOD1 proteins may help to solve this conundrum.
We approached the characterization of the SOD1 protein that reacts with C4F6 using a number of in vitro and in vivo methods. C4F6 immunohistochemistry and native immunoblot analysis distinguished the presence of mutant SOD1 proteins in diseased tissues (i.e., motor neurons) from clinically and pathologically unaffected tissues (e.g., sensory neurons), suggesting the existence of different conformational epitopes of mutant hSOD1. In SDS/PAGE, C4F6 again reacted with mutant (but not wild-type) SOD1 protein in pathologically affected tissue; however, unaffected mutant tissues, such as sensory neurons in the dorsal spinal cord, were recognized by C4F6 as well. Because proteins separated by SDS/PAGE are denatured and linearized, this finding suggests that the formation of a unique epitope through protein misfolding cannot fully account for C4F6 immunoreactivity. However, the linearized protein also is unlikely to be the sole immunoreactive epitope for C4F6. This conclusion is based on the finding that C4F6 discriminated mutant SOD1 from wild-type SOD1 in the linearized form. The possibility that immunoreactivity is based on the specific G93A point mutation is contradicted by the finding that C4F6 recognizes other linearized mutant SOD1 proteins (24). The precise C4F6 epitope has remained elusive, although epitope mapping has identified exon 4 (20) as necessary to C4F6 immunoreactivity. Regardless, C4F6 is a valuable tool for tracking disease because it is able to discriminate mutant SOD1 protein that is present in pathologically affected versus nonaffected tissues.
To further investigate whether C4F6 immunoreactivity is dependent on misfolding of mutant SOD1 protein, we used hydrophobic interaction chromatography to separate hydrophobic (misfolded) SOD1 from natively folded protein. Surprisingly, although we identified a fraction of mutant SOD1 protein that was hydrophobic, this fraction was only weakly C4F6 immunoreactive, whereas the nonhydrophobic fraction was robustly C4F6 immunoreactive. This does not rule out the possibility that C4F6 recognizes a misfolded hSOD1 species, but it does suggest that hydrophobicity is not a necessary characteristic of this species. It is not clear why C4F6 does not robustly react with hydrophobic SOD1 once it has been denatured. As it is unlikely that the C4F6 epitope is either strictly linear or strictly conformational, there may be an additional modification necessary for C4F6 immunoreactivity that is not present at high levels in the hydrophobic fraction. An alternate explanation is that C4F6 is not sensitive enough to detect the relatively low levels of hydrophobic hSOD1; however, sensitivity assays suggested that this is not the case (Fig. S4). Even so, protein not trapped by the column was C4F6-immunoreactive, demonstrating that hydrophobicity is not exclusively necessary for immunoreactivity.
It is clear that mutant SOD1 proteins are toxic to motor neurons, and there is an ongoing debate about whether this toxicity is due to the presence of SOD1 aggregates (10, 13, 15, 21, 26, 30–33). We found that the subset of mutant SOD1 protein that was recognized by C4F6 also was highly soluble and that the less soluble mutant proteins did not react with the C4F6 antibody. This finding is consistent with previous data supporting the hypothesis that the toxic form of mutant SOD1 may not be wholly contained within protein aggregates (14, 33–35). Indeed, data from models of Alzheimer's and Huntington diseases suggest that, by sequestering soluble, toxic forms of the pathogenic proteins, aggregation into larger polymers may actually serve a protective function (36–38).
To evaluate the clinical relevance of our rodent and in vitro data, we performed immunohistochemistry with the C4F6 antibody on autopsy tissues from ALS patients and controls. Patients carrying the A4V SOD1 mutation showed C4F6-immunoreactive skein-like intracellular inclusions within spinal motor neurons, similar to those seen in the rodent models. As in the rodent tissues, staining was restricted to the ventral horn and to only a subset of motor neurons. This pattern of staining was not seen in non-SOD1 familial ALS, sporadic ALS, or in control tissue. C4F6 did stain some motor neuron populations in the non-SOD1 tissue, but this staining was more diffuse and was easily distinguished from the patterns seen in the SOD1 cases. Interestingly, this diffuse staining was seen not only in ALS, but also in motor neurons from patients dying with other neurological diseases and in nonneurological controls. Several possibilities could account for this: (i) it may be that other neurological diseases and/or stressors result in SOD1 becoming C4F6-immunoreactive; (ii) C4F6 could be a more general marker of “sick” neurons; or (iii) the diffuse staining noted in sALS and neurological controls could be a result of nonspecific background and/or lipofuscin detection. Methodological issues cannot account for the pathological distinction between the ALS cases with and without SOD1 mutations. Varying antibody concentrations and incubation conditions and the use (or not) of antigen-retrieval techniques did not alter the conclusion that the SOD1 ALS patients show specific staining with C4F6. The finding that C4F6 diffusely stained motor neurons in sporadic ALS is consistent with the results reported by Bosco et al. (20). However, we also saw similar staining in non-ALS tissues; methodological differences between the laboratories may account for this discrepancy.
In summary, these data add to a growing body of literature investigating the specific properties of mutant proteins implicated in neurodegenerative diseases. We demonstrate that C4F6 is specific for pathologically affected tissue in humans and rodents expressing mutant SOD1 and believe C4F6 selectively detects toxic SOD1 protein. The immunoreactive epitope is present primarily in the soluble protein fraction and is specific neither for the G93A mutation nor for the exposed hydrophobic characteristic of a misfolded protein. The ability of C4F6 to detect a ubiquitously expressed protein only in pathologically affected tissue provides a means to investigate how and why motor neurons are uniquely targeted in this disease.
Methods
Animals.
All animal studies were approved by the Emory University Animal Care and Use Committee. Transgenic mice overexpressing hSOD1G93A [B6SJL-Tg(SOD1-G93A)1Gur/J] and hSOD1WT [B6SJL-Tg(SOD1)2Gur/J] were initially purchased from the Jackson Laboratory and were maintained on the C57/BL6 background. In our colony, symptom onset typically occurs at about 90 d of age, and end stage (defined as when the animal can no longer right itself within 30 s when placed on its back) occurs at about 135 d. hSOD1G93A rats were initially obtained from Charles River Laboratory, and the colony was maintained by crossing with Sprague–Dawley rats. All animals were housed in microisolator cages with ad libitum access to food and water.
Cell Culture and Transfections.
CHO cells were maintained in Gibco F-12 Hams media (11765) supplemented with l-glutamine, penicillin/streptomycin, and FBS. Cells were transiently transfected at 95% confluency with Lipofectamine 2000 according to manufacturer protocols.
Immunohistochemistry.
Antibodies used were a pan-SOD1 sheep polyclonal antibody (1:800, Calbiochem) and C4F6 (courtesy of J.-P.J., 1:100 on rodent tissue and 1:50 on human tissue). Paraffin-embedded tissue blocks were cut at 8–10 μm. Slides were rehydrated through a gradient of 2 × 5 min in xylene, 3 × 3 min in 100% (vol/vol) ethanol (EtOH), 3 × 3 min in 95% EtOH, and 3 × 3 min in 70% EtOH and then rinsed in ddH2O, followed by Tris buffer saline (TBS). For human tissue, antigen retrieval was performed at this stage; no antigen retrieval was necessary with rodent tissue. For antigen retrieval, tissue was microwaved 9 × 2 min in sodium citrate buffer (pH 6) with the sample being closely monitored to ensure that the tissue never dried out. The sample was then left at room temperature in buffer for 1 h to allow antigenic sites to reform and then gently rinsed in ddH2O before being rinsed 3 × 3 min in TBS. From this point on, the rodent and human protocols were the same. The samples were incubated for 10 min in 3% H2O2 and then rinsed 5 × 5 min in TBS. Samples were incubated in an avidin preblock solution in TBS composed of 10 μg/μL avidin, 4% normal goat serum, and 0.1% Triton-X. Following 3- × 5-min TBS washes, samples were incubated at 4 °C overnight in primary antibody; primary antibody buffer was made in TBS and contains 50 μg/μL biotin and 2% normal goat serum. Following overnight incubation in primary antibody, tissue was rinsed 4 × 5 min in TBS and incubated with biotinylated secondary antibody in 2% normal goat serum for 1 h at room temperature. Tissue was rinsed 4 × 5 min and incubated for 1 h in a Vector Elite ABC kit, following manufacturer instructions. Slides were then rinsed 5 × 5 min, developed in diaminobenzidine (Sigma; D4293) following manufacturer instructions, rinsed extensively, dehydrated, and coverslipped.
Immunocytochemistry.
Cells were fixed for 20 min in 4% paraformaldehyde and rinsed extensively in TBS. Nonspecific binding was then blocked for 2 h at 4 °C in 10% normal goat serum and 0.4% Triton-X in TBS. Cells were incubated overnight in primary antibody in block buffer at 4 °C; antibody concentration for C4F6 was 1:200 and for the human SOD1-specific antibody (mouse monoclonal, Sigma) was 1:500. The next day, cells were rinsed 3 × 10 min in TBS and incubated with rhodamine goat anti-mouse secondary antibody for 1 h at room temperature. Cells were rinsed extensively, and nuclei were stained with DAPI before coverslipping in Vectashield (Vector Labs) to preserve fluorescence.
Differential Extraction of SOD1 Protein from Tissue.
Spinal cord tissues from hSOD1G93A mice were homogenized in progressively harsh conditions to determine the relative solubility of the mutant SOD1 protein that reacted with the C4F6 antibody. First the tissue was homogenized in detergent-free buffer composed of 142.5 mM KCl, 5 mM MgCl2, 10 mM Hepes, 1 mM EGTA, 2 mM Na3VO4, 6 mM NaF, and a Roche Mini Complete protease inhibitor mixture (Roche Diagnostics) using a glass-on-glass homogenizer apparatus. Samples were then centrifuged at 16,000 × g for 10 min at 4 °C, and the supernatant (detergent-free, soluble protein) was removed. The remaining pellet was rinsed three times in the homogenization buffer and then resuspended in homogenization buffer containing 1.5% Nonidet P-40 before being spun at 16,000 × g for 10 min to isolate the Nonidet P-40 soluble protein. The pellet was washed three times, resuspended in buffer containing 1.5% Nonidet P-40 and 2% SDS, and spun at 16,000 × g, and the SDS soluble protein was removed. The pellet was again washed three times and resuspended in buffer containing 1.5% Nonidet P-40, 2% SDS, and 6 M urea. Following being spun at 16,000 × g for 10 min at 4 °C, the supernatant (urea soluble protein) was removed, and the pellet was rinsed three times before being resuspended in the same buffer and sonicated to solubilize any remaining protein. Quantification was performed using ImageJ densitometry software, and the percentage C4F6 protein in each solubility condition was calculated as the combined dorsal and ventral C4F6 optical density (OD) value for that solubility condition over total C4F6 OD.
Immunoblotting.
Protein samples in 2× laemmli sample buffer (Bio-Rad catalog #161–0737 + 5% βME) were heated at 85 °C for 5 min, separated by SDS/PAGE in a 4–20% gradient gel, and transferred to PVDF membrane before blocking 1 h in 5% milk at room temperature. Membranes were incubated overnight at 4 °C with primary antibody (Calbiochem pan-SOD1 sheep polyclonal, 1:1,000, or C4F6 mouse monoclonal, 1:500). The following day, membranes were rinsed 3 × 5 min in PBS/Tween and then incubated for 1 h with near-infrared dye-conjugated secondary antibody. The membrane was washed extensively and visualized with an Odyssey scanner (LI-COR Biosciences). Protein was normalized by equivalent volume in the HIC and differential extraction techniques and by equivalent total protein concentration in all other immunoblots. Samples for native immunoblot analysis were prepared as for denaturing immunoblot, except that there were no detergents or reducing agents present in either the homogenization buffer (PBS) or the laemmli sample buffer and the samples were not heated before separation on a nondenaturing, 12% Tris/glycine gel. Human SOD1 purified from erythrocytes was purchased from Sigma (catalog #S9636).
Hydrophobic Interaction Chromatography.
HIC protocol was adapted from Zetterstrom et al. (14). Spinal cords from G93A mice were weighed and ultrasonicated in ice-cold PBS (25 mL/g tissue) containing 20 mM iodoacetamide and Roche Mini Complete protease inhibitor mixture (Roche Diagnostics). Homogenates were spun at 16,000 × g for 25 min at 4 °C, and the supernatants were removed for HIC analysis. HIC columns were packed with 1 mL Octyl-Sepharose CL-4B (GE Healthcare) and were allowed to empty spontaneously with gravity. A sample (250 μL) was loaded onto the column, followed by 500 μL PBS. After 15 min of binding time, columns were eluted two times with 2.5 mL PBS and then twice more with 2.5 mL PBS diluted 10 times. The columns were then eluted with 2.5 mL elution buffer, composed of 4% SDS in 50 mM Tris⋅HCl (pH 6.8), followed by 2 mL PBS. Eluted fractions were separated on an electrophoretic gel and probed for C4F6 and pan-SOD1 immunoreactivity as outlined above.
Classifying C4F6 Immunoreactivity.
Human spinal cords were examined and rated for C4F6 immunoreactivity by three independent examiners blinded to the clinical diagnoses of the patients. Scoring was as follows: no staining, diffuse staining in a minority of neurons, diffuse staining in the majority of neurons, or skein-like intracellular inclusions. Discrepancies in scores between reviewers were resolved by consensus, with all examiners viewing the tissues together. Autopsy tissue examined included 25 sporadic ALS cases, 2 fALS without SOD1 mutations, 3 A4V SOD1 mutants, and 23 non-ALS controls. Controls were 18 cases with other neurological diseases (OND) and 5 cases without neurological disease. Primary diagnoses in OND cases were 10 Alzheimer's disease, 2 dementia with Lewy bodies, 1 Parkinson's disease, 1 tauopathy, 1 progressive supranuclear palsy, 1 neuronal intermediate filament inclusion disease, and 2 cases of frontotemporal lobar degeneration without motor neuron disease.
Supplementary Material
Acknowledgments
We thank the patients, families, and staff of the Emory ALS Center for their continued efforts to make a difference in ALS care and research. We also thank Dr. David R. Borchelt (Department of Neuroscience and McKnight Brain Institute, University of Florida) for providing the hSOD1 plasmids and Dennis Dickson (Neuropathology Laboratory, Department of Neuroscience, Mayo Clinic with funding from Mayo Foundation, Robert E. Jacoby Professorship, National Institute of Neurological Disorders and Stroke R01NS77402) for assistance with autopsy tissue preparation. Dr. Lary Walker (Department of Neuroscience, Emory University) provided helpful feedback and discussion. This project was funded through Emory University Alzheimer's Disease Research Center (P50 AG025688); Emory Neuroscience National Institute of Neurological Disorders and Stroke Core Facilities (P30 NS055077); and the Packard Center for ALS Research.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1115009109/-/DCSupplemental.
References
- 1.Worms PM. The epidemiology of motor neuron diseases: A review of recent studies. J Neurol Sci. 2001;191(1–2):3–9. doi: 10.1016/s0022-510x(01)00630-x. [DOI] [PubMed] [Google Scholar]
- 2.Logroscino G, et al. EURALS Incidence of amyotrophic lateral sclerosis in Europe. J Neurol Neurosurg Psychiatry. 2010;81:385–390. doi: 10.1136/jnnp.2009.183525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Logroscino G, et al. Descriptive epidemiology of amyotrophic lateral sclerosis: New evidence and unsolved issues. J Neurol Neurosurg Psychiatry. 2008;79(1):6–11. doi: 10.1136/jnnp.2006.104828. [DOI] [PubMed] [Google Scholar]
- 4.Millul A, et al. Survival of patients with amyotrophic lateral sclerosis in a population-based registry. Neuroepidemiology. 2005;25(3):114–119. doi: 10.1159/000086353. [DOI] [PubMed] [Google Scholar]
- 5.Beghi E, Millul A, Logroscino G, Vitelli E, Micheli A. Outcome measures and prognostic indicators in patients with amyotrophic lateral sclerosis. Amyotroph Lateral Scler. 2008;9(3):163–167. doi: 10.1080/17482960801942695. [DOI] [PubMed] [Google Scholar]
- 6.Siddique T, et al. Linkage of a gene causing familial amyotrophic lateral sclerosis to chromosome 21 and evidence of genetic-locus heterogeneity. N Engl J Med. 1991;324:1381–1384. doi: 10.1056/NEJM199105163242001. [DOI] [PubMed] [Google Scholar]
- 7.Rosen DR, et al. Mutations in Cu/Zn superoxide dismutase gene are associated with familial amyotrophic lateral sclerosis. Nature. 1993;362(6415):59–62. doi: 10.1038/362059a0. [DOI] [PubMed] [Google Scholar]
- 8.Chiò A, et al. Prevalence of SOD1 mutations in the Italian ALS population. Neurology. 2008;70:533–537. doi: 10.1212/01.wnl.0000299187.90432.3f. [DOI] [PubMed] [Google Scholar]
- 9.Gaudette M, Hirano M, Siddique T. Current status of SOD1 mutations in familial amyotrophic lateral sclerosis. Amyotroph Lateral Scler Other Motor Neuron Disord. 2000;1(2):83–89. doi: 10.1080/14660820050515377. [DOI] [PubMed] [Google Scholar]
- 10.Johnston JA, Dalton MJ, Gurney ME, Kopito RR. Formation of high molecular weight complexes of mutant Cu, Zn-superoxide dismutase in a mouse model for familial amyotrophic lateral sclerosis. Proc Natl Acad Sci USA. 2000;97:12571–12576. doi: 10.1073/pnas.220417997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Puttaparthi K, Van Kaer L, Elliott JL. Assessing the role of immuno-proteasomes in a mouse model of familial ALS. Exp Neurol. 2007;206(1):53–58. doi: 10.1016/j.expneurol.2007.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Puttaparthi K, Wojcik C, Rajendran B, DeMartino GN, Elliott JL. Aggregate formation in the spinal cord of mutant SOD1 transgenic mice is reversible and mediated by proteasomes. J Neurochem. 2003;87:851–860. doi: 10.1046/j.1471-4159.2003.02028.x. [DOI] [PubMed] [Google Scholar]
- 13.Rakhit R, et al. Monomeric Cu,Zn-superoxide dismutase is a common misfolding intermediate in the oxidation models of sporadic and familial amyotrophic lateral sclerosis. J Biol Chem. 2004;279:15499–15504. doi: 10.1074/jbc.M313295200. [DOI] [PubMed] [Google Scholar]
- 14.Zetterström P, et al. Soluble misfolded subfractions of mutant superoxide dismutase-1s are enriched in spinal cords throughout life in murine ALS models. Proc Natl Acad Sci USA. 2007;104:14157–14162. doi: 10.1073/pnas.0700477104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rakhit R, et al. Oxidation-induced misfolding and aggregation of superoxide dismutase and its implications for amyotrophic lateral sclerosis. J Biol Chem. 2002;277:47551–47556. doi: 10.1074/jbc.M207356200. [DOI] [PubMed] [Google Scholar]
- 16.Ezzi SA, Urushitani M, Julien JP. Wild-type superoxide dismutase acquires binding and toxic properties of ALS-linked mutant forms through oxidation. J Neurochem. 2007;102(1):170–178. doi: 10.1111/j.1471-4159.2007.04531.x. [DOI] [PubMed] [Google Scholar]
- 17.Estévez AG, et al. Induction of nitric oxide-dependent apoptosis in motor neurons by zinc-deficient superoxide dismutase. Science. 1999;286:2498–2500. doi: 10.1126/science.286.5449.2498. [DOI] [PubMed] [Google Scholar]
- 18.Banci L, et al. Metal-free superoxide dismutase forms soluble oligomers under physiological conditions: A possible general mechanism for familial ALS. Proc Natl Acad Sci USA. 2007;104:11263–11267. doi: 10.1073/pnas.0704307104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Forsberg K, Andersen PM, Marklund SL, Brännström T. Glial nuclear aggregates of superoxide dismutase-1 are regularly present in patients with amyotrophic lateral sclerosis. Acta Neuropathol. 2011;121:623–634. doi: 10.1007/s00401-011-0805-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bosco DA, et al. Wild-type and mutant SOD1 share an aberrant conformation and a common pathogenic pathway in ALS. Nat Neurosci. 2010;13:1396–1403. doi: 10.1038/nn.2660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rakhit R, et al. An immunological epitope selective for pathological monomer-misfolded SOD1 in ALS. Nat Med. 2007;13:754–759. doi: 10.1038/nm1559. [DOI] [PubMed] [Google Scholar]
- 22.Liu HN, et al. Lack of evidence of monomer/misfolded superoxide dismutase-1 in sporadic amyotrophic lateral sclerosis. Ann Neurol. 2009;66(1):75–80. doi: 10.1002/ana.21704. [DOI] [PubMed] [Google Scholar]
- 23.Gros-Louis F, Soucy G, Larivière R, Julien JP. Intracerebroventricular infusion of monoclonal antibody or its derived Fab fragment against misfolded forms of SOD1 mutant delays mortality in a mouse model of ALS. J Neurochem. 2010;113:1188–1199. doi: 10.1111/j.1471-4159.2010.06683.x. [DOI] [PubMed] [Google Scholar]
- 24.Urushitani M, Ezzi SA, Julien JP. Therapeutic effects of immunization with mutant superoxide dismutase in mice models of amyotrophic lateral sclerosis. Proc Natl Acad Sci USA. 2007;104:2495–2500. doi: 10.1073/pnas.0606201104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Galaleldeen A, et al. Structural and biophysical properties of metal-free pathogenic SOD1 mutants A4V and G93A. Arch Biochem Biophys. 2009;492(1–2):40–47. doi: 10.1016/j.abb.2009.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Matsumoto G, Stojanovic A, Holmberg CI, Kim S, Morimoto RI. Structural properties and neuronal toxicity of amyotrophic lateral sclerosis-associated Cu/Zn superoxide dismutase 1 aggregates. J Cell Biol. 2005;171(1):75–85. doi: 10.1083/jcb.200504050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Durazo A, et al. Metal-free superoxide dismutase-1 and three different amyotrophic lateral sclerosis variants share a similar partially unfolded beta-barrel at physiological temperature. J Biol Chem. 2009;284:34382–34389. doi: 10.1074/jbc.M109.052076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shaw BF, et al. Local unfolding in a destabilized, pathogenic variant of superoxide dismutase 1 observed with H/D exchange and mass spectrometry. J Biol Chem. 2006;281:18167–18176. doi: 10.1074/jbc.M600623200. [DOI] [PubMed] [Google Scholar]
- 29.Tiwari A, et al. Metal deficiency increases aberrant hydrophobicity of mutant superoxide dismutases that cause amyotrophic lateral sclerosis. J Biol Chem. 2009;284:27746–27758. doi: 10.1074/jbc.M109.043729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kopito RR. Aggresomes, inclusion bodies and protein aggregation. Trends Cell Biol. 2000;10:524–530. doi: 10.1016/s0962-8924(00)01852-3. [DOI] [PubMed] [Google Scholar]
- 31.Basso M, et al. Insoluble mutant SOD1 is partly oligoubiquitinated in amyotrophic lateral sclerosis mice. J Biol Chem. 2006;281:33325–33335. doi: 10.1074/jbc.M603489200. [DOI] [PubMed] [Google Scholar]
- 32.Basso M, et al. Characterization of detergent-insoluble proteins in ALS indicates a causal link between nitrative stress and aggregation in pathogenesis. PLoS ONE. 2009;4:e8130. doi: 10.1371/journal.pone.0008130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Witan H, et al. Wild-type Cu/Zn superoxide dismutase (SOD1) does not facilitate, but impedes the formation of protein aggregates of amyotrophic lateral sclerosis causing mutant SOD1. Neurobiol Dis. 2009;36:331–342. doi: 10.1016/j.nbd.2009.07.024. [DOI] [PubMed] [Google Scholar]
- 34.Witan H, et al. Heterodimer formation of wild-type and amyotrophic lateral sclerosis-causing mutant Cu/Zn-superoxide dismutase induces toxicity independent of protein aggregation. Hum Mol Genet. 2008;17:1373–1385. doi: 10.1093/hmg/ddn025. [DOI] [PubMed] [Google Scholar]
- 35.Prudencio M, Durazo A, Whitelegge JP, Borchelt DR. An examination of wild-type SOD1 in modulating the toxicity and aggregation of ALS-associated mutant SOD1. Hum Mol Genet. 2010;19:4774–4789. doi: 10.1093/hmg/ddq408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cohen E, Bieschke J, Perciavalle RM, Kelly JW, Dillin A. Opposing activities protect against age-onset proteotoxicity. Science. 2006;313:1604–1610. doi: 10.1126/science.1124646. [DOI] [PubMed] [Google Scholar]
- 37.Bodner RA, et al. Pharmacological promotion of inclusion formation: a therapeutic approach for Huntington's and Parkinson's diseases. Proc Natl Acad Sci USA. 2006;103:4246–4251. doi: 10.1073/pnas.0511256103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Outeiro TF, et al. Sirtuin 2 inhibitors rescue alpha-synuclein-mediated toxicity in models of Parkinson's disease. Science. 2007;317:516–519. doi: 10.1126/science.1143780. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.