Abstract
The ability to track CD4 T cells elicited in response to pathogen infection or vaccination is critical because of the role these cells play in protective immunity. Coupled with advances in genome sequencing of pathogenic organisms, there is considerable appeal for implementation of computer-based algorithms to predict peptides that bind to the class II molecules, forming the complex recognized by CD4 T cells. Despite recent progress in this area, there is a paucity of data regarding their success in identifying actual pathogen-derived epitopes. In this study, we sought to rigorously evaluate the performance of multiple web-available algorithms by comparing their predictions and our results using purely empirical methods for epitope discovery in influenza that utilized overlapping peptides and cytokine Elispots, for three independent class II molecules. We analyzed the data in different ways, trying to anticipate how an investigator might use these computational tools for epitope discovery. We come to the conclusion that currently available algorithms can indeed facilitate epitope discovery, but all shared a high degree of false positive and false negative predictions. Therefore, efficiencies were low. We also found dramatic disparities among algorithms and between predicted IC50 values and true dissociation rates of peptide:MHC class II complexes. We suggest that improved success of predictive algorithms will depend less on changes in computational methods or increased data sets and more on changes in parameters used to “train” the algorithms that factor in elements of T cell repertoire and peptide acquisition by class II molecules.
Introduction
CD4 T cells are known to play a key role in protective immunity to infectious organisms and much current research uses epitope-specific probes to study the role that CD4 T cells play in immunity to complex pathogens. Further success in identification of the peptides that are the focus of an adaptive CD4 T cell response is essential for understanding the mechanisms of protective immunity and the factors that influence the dynamics and specificity of host pathogen interactions. CD4 T cell epitope identification is also needed for vaccine evaluation, tetramer-based studies of T cell phenotype and for development of peptide-based vaccines. With increasing success in genome sequencing of complex bacterial and viral pathogens (reviewed in (1–5)), candidate proteins for vaccines are increasing, but identification of epitopes that are the focus of immune responses remains a bottleneck in this research.
A number of empirical approaches have historically been used for epitope discovery, including biochemical isolation and proteolytic fragmentation of antigenic proteins (6, 7), derivation of genetic constructs that encode all or selected segments of candidate pathogen-derived proteins (8–11), elution and sequencing of peptides from pathogen-infected cells or tumor cells (12–16), and individual epitope mapping, using arrays of synthetic peptides (17–22). These approaches, typically coupled with T cell assays to identify the immunologically active peptide within the candidate antigen, are time consuming and involve significant expenditure of effort and resources to be successful. The labor intensive nature of these approaches is a particularly large obstacle for complex pathogens that express hundreds of proteins, of which only a small fraction may be the target of T cells or B cells or that may serve a protective role as vaccine candidates.
The considerations of time and expense required for empirical approaches have led to the development and refinement of algorithms that use different logic bases and sources of data to predict epitopes that will be presented by particular MHC molecules (reviewed in (23–28)). Because the major selective force in peptide binding to MHC involves side chains of amino acids (“anchors”) in the peptide with depressions (“pockets”) in the MHC molecule, the algorithms focus on scoring these interactions as a means to predict CD4 epitopes. Some methods such as matrix-based algorithms operate with the general model that each amino acid adds or detracts from the binding of the peptide to the MHC protein in a largely predictable, independent and quantifiable manner (29, 30). Large data sets or “training data” are used to construct and refine the algorithms that ultimately search for the highest 9-mer core in a peptide and output the predicted binding affinity of every candidate peptide. Other less rigid algorithms that operate using such methods as neural networks (31, 32) and particle swarm optimization (33) have also been developed and utilized. Finally, Sette and co-workers describe a “Consensus” approach that essentially averages the predicted ranking hierarchy of a given set of peptides scored by what their studies suggest to be the best performing 3–4 web available algorithms (34).
In general, the predictive algorithms developed for MHC class I peptides that activate CD8 T cells significantly outperform those that predict MHC class II-presented peptides that elicit CD4 T cells, in large part because of the nature of their respective peptide binding pockets. MHC class I molecules are closed at their periphery, thus limiting the size of the peptide that binds to 8–10 amino acids (35–37). Therefore, the amino acids that contribute the key anchor positions for pocket interactions are easily identified. In contrast, the binding pocket of MHC class II is open at its periphery. Elution and sequencing studies indicate that peptides bound by class II molecules typically range from 9–25 amino acids in length (38, 39) and long peptides are well presented to CD4 T cells (40, 41). Often, the “register” of these peptides, the amino acids that comprise the 9 amino acid core within the MHC-binding groove and that dictate MHC binding and T cell recognition, is not known. For several peptides analyzed in detail, multiple registers are presented by class II (42–45). Therefore, simple knowledge that a peptide is presented by a class II molecule does not provide insight into what amino acids control its presentation. Because much of the data that trains the algorithms are long peptides, it has been challenging to know how identified antigenic peptides bind to the class II molecule and thus to predict what peptides within an uncharacterized set will be presented by the same or related class II molecules. Databases containing epitopes presented by class II molecules have steadily accumulated (46–48)and been used to refine existing algorithms, but the issue of uncertain binding cores persists.
Because CD4 T cells are known to be critical regulators of both B cell and CD8 T cell responses to pathogens, provide direct function for responses against intracellular pathogens and are critical for vaccine success, it is clear that despite the theoretical challenges, there is considerable appeal in using computation-based algorithms to identify candidate CD4 T cell epitopes, particularly for complex pathogenic organisms that express hundreds of proteins. “Benchmark” studies, that assess the accuracy of the predictions typically measured the ability of the algorithms to predict binding to MHC class II molecules, using existing or new data sets (25, 29, 32–34, 49–51). Evaluation of peptide performance using functional tests of the actual immunogenicity of the predicted peptides have been much more limited. The studies performed are typically restricted to a handful of peptides, are sometimes tested by immunization with free peptides rather than a complex antigen, and often, the presenting class II molecule for CD4 T cell recognition is not unequivocally identified (20, 34, 52–55). In recent years, our laboratory has empirically and comprehensively investigated the peptide specificity of CD4 T cells elicited in response to primary influenza infection using a completely unbiased approach involving overlapping peptide libraries and cytokine EliSpot assays. Multiple strains of mice have been studied, including HLA-DR transgenic mice and common inbred strains of mice (21, 22, 56). We have identified and quantified the responses to more than 500 influenza-derived peptides presented by these different class II molecules that are the focus of CD4 T cell responses.
In the study presented here, we have evaluated the ability of web-available algorithms to predict the specificity CD4 T cells elicited in response to influenza. We had three goals for this study. The first was to evaluate the performance of the algorithms for their efficiency in epitope identification by combining our results in epitope discovery with advances by other groups in developing predictive algorithms. The second goal was to develop useful strategies to implement algorithms for epitope discovery for future investigations on the role that CD4 T cells play in protective immunity to pathogenic organisms. The third goal was to gain the insight needed to improve performance of the algorithms for future efforts to facilitate epitope discovery. To our knowledge, the analyses presented in this study are the first to comprehensively evaluate the performance of algorithms with the results of empirical and non-biased epitope discovery of multiple pathogen-encoded antigens and unrelated class II molecules.
Materials and Methods
Mice
The HLA-DR1 and the HLA-DR4 transgenic mice were obtained from D. Zaller (Merck) through Taconic Laboratories, and were maintained in the specific pathogen-free facility at the University of Rochester according to institutional guidelines. C57Bl/10 mice were purchased from the Jackson Laboratories, Bar Harbor ME. Mice were used at 2–5 months of age.
Ethics Statement
All animal protocols used in this study adhere to the AAALAC, International, the Animal Welfare Act and the PHS Guide. All protocols have been approved by the University of Rochester Committee on Animal Resources; Animal Welfare Assurance Number A3291-01. The protocols under which these studies were conducted were approved on March 4, 2006 (protocol no. 2006-030) and April 10, 2008 (protocol no. 2008-023).
Influenza Infection of mice
A/New Caledonia/20/99 was produced in allantoic fluid of embryonated eggs. Briefly, eggs were purchased from SPAFAS Inc. (North Franklin, CT), and incubated at 70°F and 100% humidity for 9 days, followed by the infection of the allantoic cavity with 100 µl of human (H1N1) influenza virus A/New Caledonia/20/99 (kindly provided by Dr. John Treanor at the University of Rochester). Virus was collected from the allantoic fluid and the IED50 for the produced virus was determined. HLA-DR1, HLA-DR4, transgenic and C57BL/10 (I-Ab) mice were infected intranasally with A/New Caledonia/20/99 at 50,000 EID50 in 30 µl of phosphate-buffered saline (PBS). Groups of mice were 2 to 4 months old, at the time of infection, and were anesthetized by intra-peritoneal injection with tribromoethanol prior to infection. Ten to twelve days post-infection, the mice were euthanized and spleens and mediastinal lymph nodes were isolated and used as sources of CD4 T cells for EliSpot analyses. Lymphocytes were pooled from 4–8 mice and depleted of B cells, CD8 cells and macrophages by negative selection using MACS depletion (Miltenyi Biotech, Gladbach, Germany), according to the manufacturer instructions.
Dissociation Kinetics
The dissociation experiments were performed using the procedure as previously described (57–60). Briefly, 50 µL of 100nM purified soluble DR1 molecules expressed in S2 cells were incubated with 1µL of 250µM fluorescein-labeled binding peptides at a concentration of 100nM and pH 5.3 in McIlvaines buffer, containing 0.025% NaN3. The binding mixture was incubated at 37°C during 2–24 hours and then the class II:peptide complexes were separated from excess labeled peptides at room temperature using micro spin columns (BioRad, Hercules, CA). Unlabeled influenza HA [306–318] (PKYVKQNTLKLAT) peptide was added at final concentration of 5µM in order to prevent possible re-binding of the dissociating peptide. Soluble I-Ad molecules were produced and isolated as previously described (58–60). Briefly, a total of 1 × 1010 I-Ad PI-linked-CHO cells were solubilized with 1.5 L of 50 mM Tris, 150 mM NaCl, 1 mM n-dodecyl maltoside (DDM), and 0.025% NaN3 containing protease inhibitors. Class II molecules were isolated using antibody affinity chromatography. After PI cleavage, the soluble I-Ad was eluted from the column at pH 11.0 and fractions containing class II were pooled, dialyzed against PBS (pH 7.4) containing 0.2 mM DDM and 0.025% NaN3 (DDM/PBS) and concentrated with a Centriprep YM-10 device (Millipore Co., Bedford, MA). 50 µL of 100nM purified soluble I-Ad molecules were incubated with 1µL of 250µM fluorescein-labeled binding peptides at a concentration of 100nM and pH 5.3 in McIlvaines buffer, containing 0.025% NaN3. The complexes were isolated using micro-spin columns as described above for DR1. Dissociation assays were = performed by incubating the purified MHC class II-peptide complexes at 37°C in McIlvaines buffer plus 0.025% NaN3 at a final pH of 5.3 in the presence of either unlabeled influenza HA [306–318] (PKYVKQNTLKLAT), for DR1, or Ea[52–68] (ASFEAQGALANIAVDKK), for I-Ad, in order to prevent possible re-binding of the dissociating peptide. The dissociating peptide was separated from the remaining complex with a BioSep SEC-S 3000 column (Phenomenex, Torrance, CA) using a SHIMADZU chromatograph (Shimadzu Corporation, Japan). The intensity of the remaining class II:peptide complex was determined using a fluorescence detector (at wave lengths of 495 (nm), excitation, and 525 (nm), emission (Shimadzu Corporation, Japan). The intensity of the peak belonging to the complex decreases over time, allowing quantification of the dissociation half time. The graph of the intensity of the MHC class II-peptide complex versus the dissociation time is used to generate the dissociation curve. Baselines values were measured before each dissociation experiment to ascertain that are no traces of previous samples and the program for elution of the complexes by HPLC-SEC includes a wash time that ensures complete removal of all previous materials. Dissociation curves fit to single exponential decay curves with r2>0.9. The half-life is the time at which the initial fluorescence intensity of the complexes decays to half, which is stated as the t1/2 dissociation, proportional to the stability of that complex.
Peptides
17-mer peptides overlapping by 11 amino acids to cover the entire sequences of the HA and NA proteins from the A/New Caledonia/20/99 influenza virus (H1N1), the NS1 sequence from the A/New York/444/2001 influenza virus (H1N1), and the NP and M1 sequences from A/New York/348/2003 influenza virus (H1N1) were used. Peptide arrays were obtained from National Institutes of Health (NIH) Biodefense and Emerging Infections Research Resources Repository, National Institute of Allergy and Infectious Diseases (NIAID). The A/New York/348/2003 amino acid sequences for NP and M1 are 100% conserved, and the A/New York/444/2001 NS1 amino acid sequence is >99% compared to A/New Caledonia/20/99 (H1N1) influenza virus. The peptides were reconstituted to 10mM in PBS, with or without added dimethyl sulfoxide for hydrophobic peptides, and 1mM dithiothreitol for cysteine-containing peptides. Working solutions were prepared in Dulbecco's modified Eagle's medium (Invitrogen Corp., Carlsbad, CA), filter sterilized, and also stored at −20°C. The final concentration of the individual peptides used in the EliSpot assays was 10µM.
Use of Web-Based MHC class II:peptide Binding Prediction Servers
The primary sequences for hemagglutinin protein (HA), neuraminidase protein (NA), nucleocapsid protein (NP), non-structural protein 1 (NS1), and matrix protein 1 (M1) proteins from A/NewCaledonia/20/99 (H1N1) were obtained from the protein data bank. The sequences were run using the web-available predictors NetMHCII 2.2, NetMHCII-pan 2.1, and SMM-align at http://www.cbs.dtu.dk/services/ and Consensus, ARB, and TEPITOPE, at http://tools.immuneepitope.org/main/. If only 15-mer peptides were scored by algorithms (Consensus, ARB, and TEPITOPE), the empirically tested peptides were matched to the functionally assayed 17-mer sequences by alignment with their N-termini. The raw scores, the IC50, or the percentile rank values were used to determine the peptides that were selected by the algorithms. For methods that report IC50 values such as NetMHCII-2.2, NetMHCII-pan2.1, SMM-align, and ARB, the predictors qualify a peptide as follows: 1. Non-binders for peptides with IC50 higher than 500nM, 2. Weak binders for peptides with IC50 between 50nM and 500nM, and 3. Strong binders for peptides with IC50 lower than 50nM. These specific cutoffs were used to analyze predictive performance in some experiments based on their use by the developers or users of predictive algorithms (49, 61–64). For Consensus, the percentile rank was used to rank the peptides, while for TEPITOPE, the raw score was used for ranking. When a given functionally evaluated peptide was penalized because its predicted binding core was at either the N- or C- terminus of the scored peptide epitope, we evaluated each 9-amino acid core within the adjacent scored peptides having the same core as the epitope and assigned the score that had the best binding value. When scoring the selected viral “proteome”, the input sequences were the entire amino acids sequences of the five proteins joined together, in the following order: HA, NA, NP, NS1, and M1, with non-native peptides at the junctions of the proteins eliminated from consideration. Only the experimentally tested peptides were scored for analysis.
Derivation of ROC curves and AUC scores
Algorithm prediction accuracy was measured via receiver operating characteristic (ROC) curve and the area under curve (AUC) values were calculated as described by others (23–25, 32, 34). Each algorithm’s binding predictions were compared to the empirically defined set of ‘epitopes’ and categorized under a binary classification system. At each discrimination threshold, peptides were grouped into one of 4 categories. True positive (TP), predicted binders as defined by the threshold that elicited response in T cell assays, false positive (FP), predicted binders as defined by current threshold that did not elicit response in T cell assays, true negative (TN), peptides predicted relative to threshold that did not elicit response in T cell assays, false negative (FN), when peptides predicted as nonbinding relative to threshold elicited response in T cell assays. The true positive rate (TPR), =[TP/(TP+FN)], was plotted against the false positive rate (FPR), = [FP/(FP+TN)], for each threshold to form the ROC curve. The area under each algorithm’s curve was calculated to give the AUC score.
CD4 T cell epitope discovery
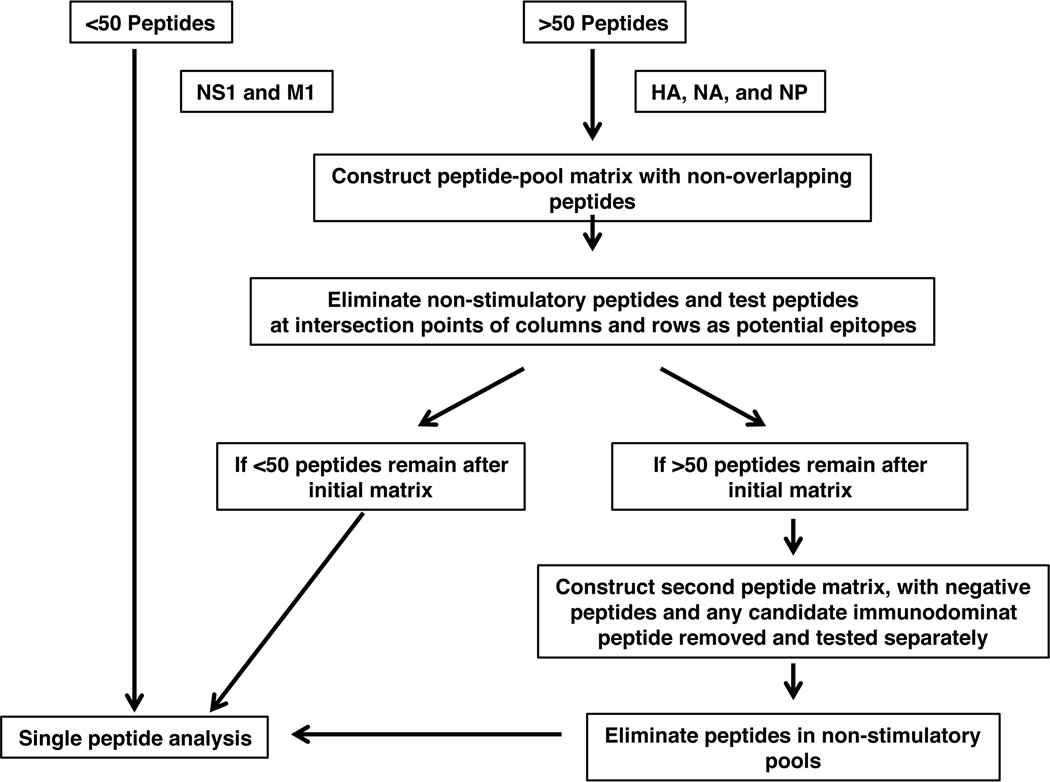
Peptide-specific cytokine EliSpot assays were performed as previously described (65) and as represented schematically in Figure 1. Briefly, 96-well filter plates (Millipore, Billerica, MA) were coated with 2µg/ml purified rat anti-mouse IL-2 (clone JES6-1A12, BD Biosciences, San Jose, CA) in PBS, washed and incubated with media to block non-specific binding. CD4 T cells isolated and purified from previously infected mice (at 100,000 to 300,000 cells per well) were co-cultured with fibroblasts expressing the DR1, or antigen presenting cells (APC) isolated from syngeneic mice and the test peptide at 10µM. After overnight co-culture, plates were processed to visualize IL-2 producing cells as described (21, 22, 56) } and cytokine EliSpots were enumerated using an Immunospot reader series 2A, using Immunospot software version 2.
Figure 1. Schematic of the experimental approach followed for epitope discovery.
To limit the number of peptides tested, a matrix array was used for HA, NA, and NP proteins where peptides were pooled into 8–12 peptides and then arrayed into intersecting rows and columns as described [27,29]. Only peptides within pools that were positive in both columns and rows in the matrix, eliciting a cytokine response equal or greater than 30 spots per 106 cells, were considered positive. All the positive peptides from the matrix were further tested individually. In the case of M1 and NS1 proteins that were comprised of less than 50 peptides all peptides were tested individually.
Results
Identification of CD4 T cell epitopes through an entirely empirical method
Recently, our laboratory has identified the full repertoire of influenza-specific CD4 epitopes from 5 major influenza proteins in several inbred strains of mice that express distinct class II molecules (SJL, C57BL/10, HLA-DR1 (“DR1”) transgenic mice) by using a completely empirical approach (21, 22, 65). HLA-DR4 (“DR4”) epitopes were additionally identified for this study. Mice were infected by intranasal inhalation, representing the natural route of infection for this pathogen in both mice and humans. At 10–14 days post infection, CD4 T cells were isolated from lymphoid tissue, purified and assessed directly for their peptide specificity using synthetic peptides and cytokine ELISPOT assays, allowing direct ex vivo quantification of antigen or peptide-reactive lymphocytes. CD4 T cell specificity was identified from a sequential, iterative method of epitope identification, employing overlapping synthetic 17mer peptides representing the entire translated sequence of these five viral proteins, schematically illustrated in Figure 1. These selected viral proteins that were examined (HA, NA, NP, M1 and NS1) differ with regard to their expression within the influenza virion and in infected cells, allowing us to evaluate whether these variables influenced epitope selection or algorithm performance. HA and NA are expressed as transmembrane proteins in the plasma membrane of infected cells and in the virion envelope, NP is localized within the cytosol and nucleus, while M1 is associated with the inner leaflet of the plasma membrane and is abundant in the virion. NS1 is notable because it is excluded from the virion particle.
For epitope discovery, the approach depended on the number of candidate peptides. For small viral proteins represented by less than 50 different peptides (M1 and NS1), each peptide was tested individually in EliSpot assays. For larger proteins (HA, NA, and NP), we used the peptide pooling matrix strategy outlined on the right in Figure 1, adapted by us (21, 22, 65) from an approach originally described by Tobery and coworkers (19, 66). Three different alleles (HLA-DR1, HLA-DR4 and I-Ab) were the focus of the study here because they have been studied extensively and thus we imagined that the performance of the algorithms would be highest for these. The individual peptide epitopes recognized by the CD4 T cells from infected mice are listed in Supplemental Tables 1A–1C, with the responses adjusted and presented to represent cytokine EliSpots elicited by each peptide per million CD4 T cells tested. Peptides were considered positive if they reproducibly recruited more than 30 cytokine producing cells per million CD4 T cells and data presented represents the average value from at least 3 and typically more than 5 independent assays.
Comparison of algorithm predictions vs. the empirically defined epitopes
To analyze the relationship between predicted MHC class II binding peptide and experimentally identified epitopes, we scored the 17mer peptides used for empirical identification of epitopes using the web-available algorithms. We chose algorithms that integrate much of the available information regarding peptide binding to class II molecules and limited our studies to web-accessible algorithms. In the analyses described here, we evaluated ARB, SMM, Tepitope, NetMHC pan, Net MHCII2.2 and the Consensus method. The “score” of each peptide is provided in different formats for the different algorithms. Some of the algorithms, such as ARB, NN-align and SMM (29, 30, 32), calculate a “predicted IC50” value for each peptide scanned and recommends a cut off for strong, weak and non-binding peptide, typically in the range of 10–500nM. Other algorithms, such as Tepitope (67) and Consensus (34) rank the tested peptides from which an arbitrary cutoff can be chosen, without any estimate of absolute affinity. The Consensus method ranks the peptides based on assimilation of the prediction scores from what their studies have revealed to be the most accurate algorithms. The choice of how to analyze the performance of the algorithms in the current study was driven both by our desire to objectively analyze their accuracy and also by anticipation of the way these tools might be utilized by other investigators.
We began the analyses using the Consensus and the NetMHCII methods, both of which have unique strengths and have been widely used or evaluated by others (25, 29, 49, 50, 62, 68–71). The Consensus method calculates the median percentile rank of each peptide based on the predictions of three independent, high performing algorithms. Because the scoring output is a percentile rank, there is no absolute value of affinity implied and candidate epitopes are identified on inclusion within the top scorers. The recently developed NetMHCII (32) corrects for biases due to replicate binding cores within the training data and is one of the few algorithms that factors in effects of the amino acids that flank the 9 amino acid core of the peptides. It also has added flexibility in that it allows evaluation of peptides of different lengths. This method provides output in the form of predicted IC50 values. Both algorithms are able to predict epitopes for the three alleles of MHC class II molecules studied here and both were found by their originators to outperform other individual state of the art algorithms.
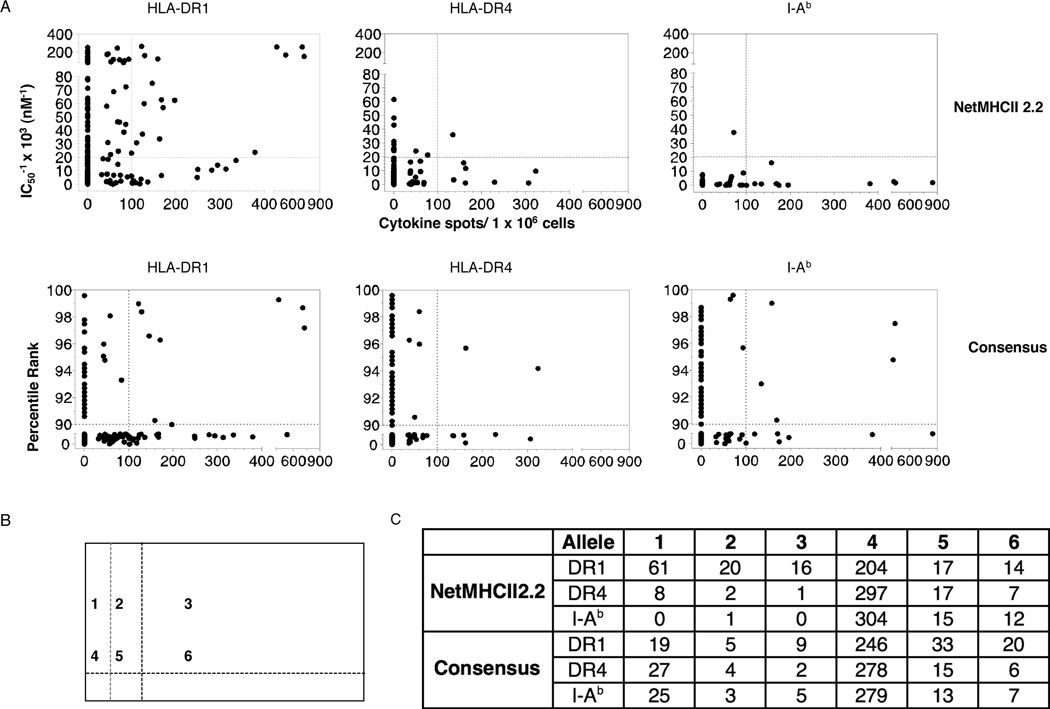
To globally visualize the predicted binding vs. the actual hierarchy of CD4 T cell responses to influenza, the number of cytokine EliSpots elicited by each peptide tested within the 5 viral proteins (the “proteome”) were plotted vs. the predicted affinity (NetMHCII) or percentile rank (Consensus). Predicted values are represented as their reciprocal, so that the height of the epitope on the Y-axis represents a peptide’s actual or predicted “strength”. The CD4 cytokine EliSpot data are represented as spots per million for every peptide analyzed, with an arbitrary cutoff of 100 spots per million CD4 T cells indicated, as this might be a minimal frequency (0.01%) useful for such methods at tetramer-based flow cytometry or intracytoplasmic cytokine staining. These graphs, shown in Figure 2A, are thus divided into 4 quadrants, where the top right quadrant represents the “double positive” peptides-those peptides both predicted by the algorithm and that were also true CD4 T cell epitopes, as detected by cytokine EliSpot assays. The bottom left quadrant represents the double negative peptides, those neither predicted nor observed. The top left quadrant represents the “false positive” peptides, predicted to be above the threshold used and the bottom right quadrant represents the false negatives-peptides found to be epitopes in the influenza-specific CD4 T cell response but not predicted to be high affinity binders. The number of peptides that fell into each quadrant for each allele is tabulated in Figure 2C, that also indicates the fraction of peptides in each category. Also quantified in 2B are minor epitopes, those that recruit low but detectable numbers of cells greater than 30 but less than 100 cells per million. All regions scored are illustrated schematically in Figure 2B.
Figure 2. Accuracy of NetMHC2.2 and Consensus for prediction of influenza-specific CD4 T epitopes.
A. All of peptides analyzed functionally for CD4 T cells isolated from previously infected HLA-DR1, HLA-DR4 or C57Bl/10 mice were compared for their response magnitude (represented as cytokines spots/million on the X axis) vs. their predicted affinity (IC50 values, Net MHCII2.2) or ranking (Consensus), indicated on the Y axis. The inverse value of the affinity is used for NetMHCII2.2 so that greater values represent increasing predicted affinity of the peptide for MHC class II. The threshold indicated by the vertical dotted lines in each panel in A represents major epitopes (eliciting more than 100 spots per million). B. The grid represents the areas scored in Figure 2A. Area 1 includes peptides predicted but not found, area area 3 represents peptides both predicted and found, are 4 are peptides neither predicted or found, area 6 are peptides that were not predicted to bind but that were found. Areas 2 and 5 represents the minor epitopes that recruit more than 30 but less than 100 spots per million CD4 T cells. The values in C quantify the number of peptides in each area shown in B.
These analyses indicated that both the number and percentage of each candidate peptide that in is the “double positive” quadrant varies significantly with the allele and algorithm. For example, DR1 has the highest fraction of double positive peptides chosen by NetMHCII (16 out of 30 total positives or 53%), most notably the four highly dominant peptides, recruiting more than 400 cells per million CD4 T cells and estimated to be of very high affinity. These true positive peptides were offset by a total of 61 false positive peptides. DR4 had only one peptide that was both predicted and found empirically, while I-Ab has none. The range for Consensus ranged from 25%–41% true positives, depending on the allele, and between 19–27 peptides predicted but not found, with DR1 having three peptides that were highly ranked and found to be immunodominant. In all cases, although the algorithms selected some epitopes, there were many epitopes that were not predicted to be MHC binding peptides by either algorithm and many peptides that were predicted to be binders that were not CD4 epitopes.
Because some cellular proteins have preferential access to the MHC class II presentation pathway (39, 72–74), we analyzed the individual peptide epitopes for each viral protein separately to determine if algorithm performance varied among the proteins examined. Supplemental Figures 1 and 2 show the results of these analyses for the three alleles analyzed and the predictions made by NetMHCII and Consensus. Although there were not striking disparities between predicted vs. true epitopes, two rather consistent trends were noted. First, NetMHCII predictions for NS1, M1 and NP appeared to have a somewhat higher “hit rate” across alleles and fewer false positives. For example, for DR1, 11 NS1-derived peptides were predicted to be high affinity binders and of these, 6 of these were true epitopes. Secondly, we noted that epitopes within HA seemed to have an opposite pattern, where 30 peptides were predicted to be high affinity binders to this allele, but only 8 of these were true epitopes. Suppl. Figure 2 shows a similar type of analysis performed using the Consensus method. By the nature of the scoring for this algorithm, all protein/allele combinations will have similar numbers of peptides that fall above and below any cutoff, independently of whether they are truly high affinity binders. Using a 10% cutoff as a “prescreen” would have indeed allowed identification of some of the major epitopes that were recognized by CD4 T cells after influenza infection. In the case of DR1 of the 25 major epitopes, only 6 were identified to be in the top 10% rank, 2 in NS1 and 2 in M1. For DR4, of the 8 major epitopes, only 3 were identified as strong binders and they were all in NP. For I-Ab, there were 11 immunodominant epitopes, and of these, 3 epitopes were within the top 10% predicted and two were in NP.
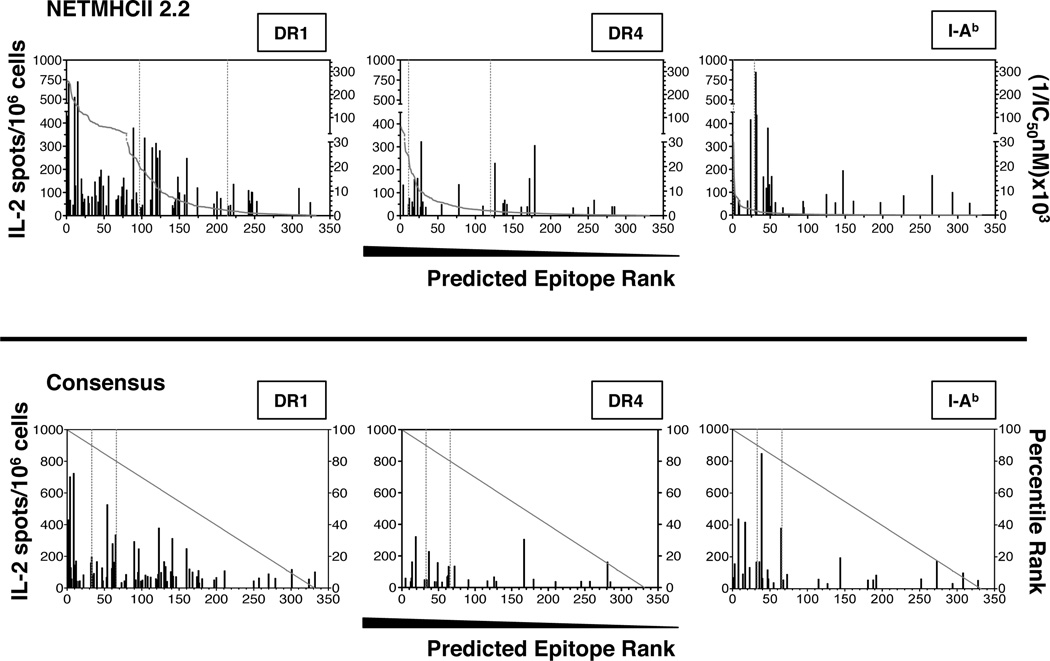
The relationship between cutoff choices for selection of peptides compared to the yield of actual epitopes for both algorithms is most readily seen in Figure 3, that represents each peptide contained in the tested proteome plotted in order of decreasing predicted binding affinity or rank by NetMHCII and Consensus, respectively, with two potential cutoffs shown, corresponding to IC50 values of 50nM or 500nM for NetMHC2.2 and 10% or 20% predicted top binders for Consensus. From this analysis, several conclusions were made. First, the epitopes identified empirically through functional studies are mostly contained within the top 50% of the 332 peptides scored. This indicates that the actual epitopes recognized by CD4 T cells are enriched in the high scoring group of peptides. Second, although there is clustering of epitopes in this top half of the peptides, the most robust CD4 T cell specificities are not clustered within the highest of the predicted affinities. This is particularly apparent for the human DR4 molecule. Thus, although this type of display allows use of algorithms to select peptides, the “cutoff” for selection of peptides for further confirmation needs to be generous if it is hoped to capture the most dominant of epitopes with the candidate peptides. Finally, using NetMHCII, the ranking of affinity vs. response is different for each allele, so selection of a consistently appropriate cutoff may be difficult. For example, for DR1, the use of a 50nM predicted affinity cutoff would select approximately 80 out of 332 peptides and would have successfully identified 30% of the true epitopes, including the three most immunodominant. The same predicted affinity for DR4 would select for less than 15 peptides and only 4 epitopes, none of which are the most immunodominant. For the DR4 allele, the 500nM cutoff would be more effective. For I-Ab, the 50 nM cutoff would select only 2 peptides, of which none of the 11 major epitopes would be found and here the lower threshold of 500nM would be more effective. Thus, the algorithms do not perform equivalently for all alleles, in terms of epitopes “captured” at any given cutoff. The bottom panel of Figure 3 shows Consensus prediction, where the peptides are simply ranked. Consensus was the most successful for I-Ab where most of the peptide epitopes would have been identified with the 20% cutoff and many identified even with the 10% cutoff. Finally, although the identified epitopes are generally within the top third, even the lowest ranked peptides contain some epitopes. In conclusion, both methods facilitated identification of epitopes and if only a few epitopes are needed, a very stringent cutoff will select a subset of peptides that contains at least a few epitopes, but many epitopes, including major immunodominant epitopes will be missed.
Figure 3. Ranking of experimentally identified epitopes using the NetMHCII2.0 and Consensus algorithms.
Representation of the predicted ranking, based on the indicated algorithms, for each of the experimentally identified epitopes. The x-axis shows the relative position of every epitope among the total of 332 synthetic peptides that span the entire HA, NA, NP, NS1, and M1 proteins. Shown for each peptide is the magnitude of the cytokine response elicited in recall experiments. The experimentally tested peptides were placed in the hierarchy for each algorithm calculated either as [(1/IC50)*100] for NetMHCII2.2 or percentage in the ranking of all peptides in Consensus. Vertical dotted lines within each graph indicate the cutoffs of 50nM or 500nM IC50 values for NetMHCII2.2 or 10% or 20% best predicted binding peptides, for Consensus.
Evaluation of the relationship between predicted affinity and epitope dominance
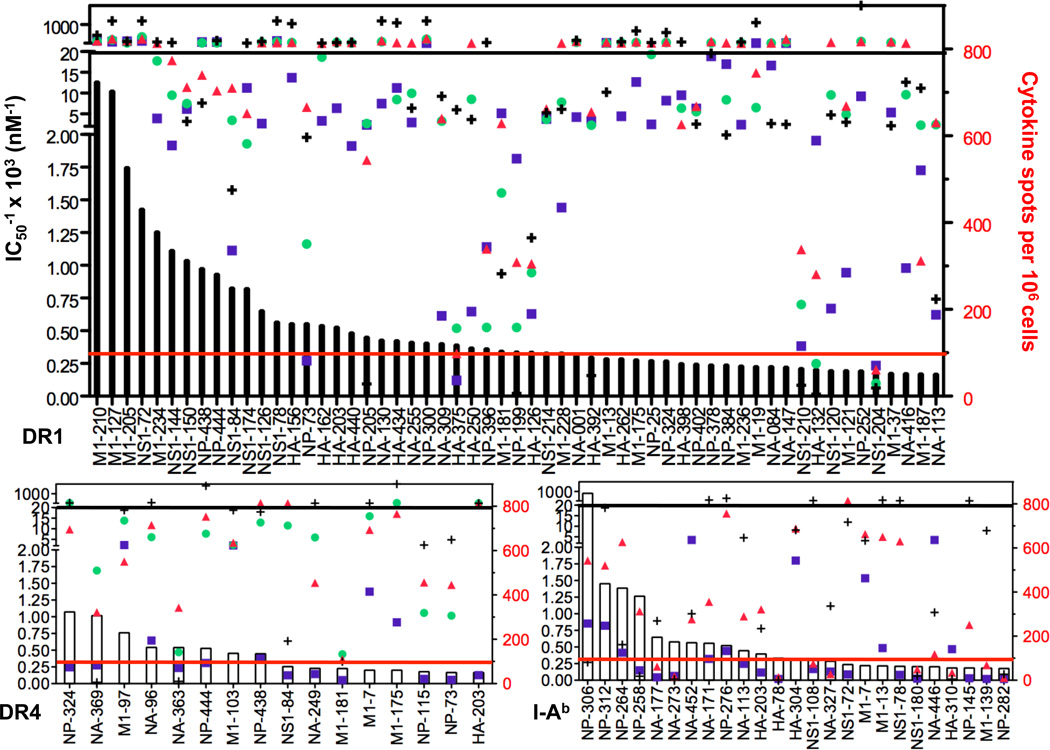
We next extended our analyses to additional algorithms. We had two goals: first to assess the agreement among algorithms with each other and second to examine the relationship between immunodominance and the affinity estimates generated by the algorithms. Shown in Figure 4 is this analysis performed with several algorithms that present estimated affinity in the form of IC50, each of which is represented by a different symbol. Only SMM, NetMHCII and ARB incorporate I-Ab, so the analysis of this MHC class II molecule was restricted to these three algorithms. For simplicity, we only included peptide epitopes that recruited at least 50 spots per million CD4 T cells. For several epitopes, there is good agreement among all of the algorithms for predicted binding and that these estimates could allow selection of epitopes. For example, for DR1, among the 32 dominant peptide epitopes, 20 were predicted by at least three algorithms to possess high affinity binding and three highly dominant M1 peptides and one NS1 peptide were predicted to be high affinity by all four algorithms, suggesting that some peptides have features that are well recognized to promote binding to this allelic form of human class II.
Figure 4. Affinity predictions from different algorithms for influenza-specific CD4 T epitopes.
Shown by bars for each allele is the magnitude of the response recalled by the indicated peptide, represented as cytokine spots per million CD4 T cells in the right Y-axis. Shown by the symbols are the affinity predictions from each algorithm for the peptides relevant to the indicated allele, with the axis shown in the left Y-axis. A: HLA-DR1, B. HLA-DR4 and C. I-Ab. The black line indicates the threshold of 50nM, where all symbols above that line are peptides predicted to bind with high affinity. The solid red line indicates the threshold of 100 spots/million for dominant peptide epitopes.
For DR4 and I-Ab, the relationship between predicted IC50 among algorithms vs. epitope dominance was much weaker. Among the 8 major epitopes restricted by DR4 that recruited at least 100 CD4 T cells per million, two were predicted to bind with high affinity by more than 2 algorithms, while for I-Ab, of the 12 major epitopes, only 2 were selected by at least two algorithms to have high affinity binding. This analysis also revealed that a number of epitopes that were identified empirically were not predicted to be MHC-binding by any of the algorithms. For DR1, 9 of 32 were judged to be non-binders by at least 3 algorithms, for DR4, 4 out of 8 were judged to be non-binders, while for IAb, the vast majority of epitopes were predicted to be poor or non-binders. There was also a significant range in the IC50 values predicted for individual peptides, as evidenced by the high degree of spread among the different symbols in Figure 4.
Analysis of the efficiency of predictive algorithms for response “capture”
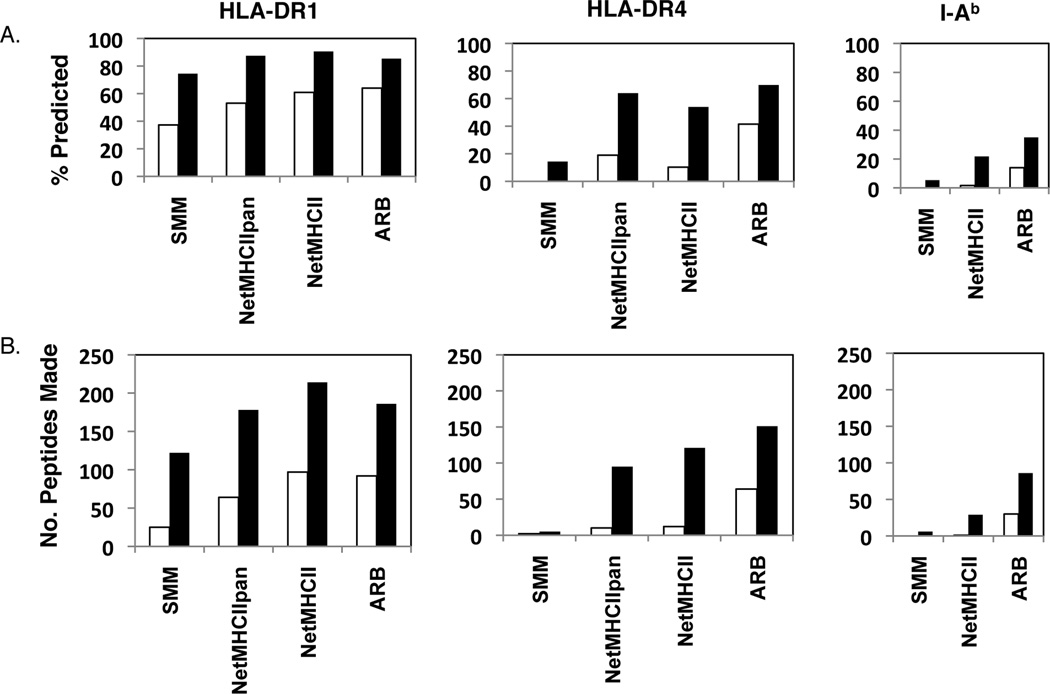
Because there are many false positive peptides predicted by each algorithm, we conclude that secondary screens are important, and next sought to evaluate different thresholds that might be used for selection of candidate peptides. Of particular interest was the cost (how many peptides would be tested in secondary screens) vs. benefit (how many epitopes found) of different thresholds. In this analysis, we selected the subset of algorithms that provides output in the form of predicted affinity (SMM, NetMHCpan, Net MHCII and ARB). Figure 5A shows the fraction of the response predicted or what would have been “captured” by pre-selection of epitopes by the indicated algorithm, at either an IC50 value of 50nM or 500nM. The cutoffs were chosen based on common use in the literature by those who derive or use the predictive algorithms (49, 61–64). Although these two values are somewhat arbitrary and others cutoffs, such as 5nM, for very high affinity or 1000nM, for very low affinity could be used, the two cutoffs selected provide a framework that can be used to compare the predictive efficacy of the individual algorithms. The number of cytokine producing cells elicited by all of the peptides identified by an algorithm were summed and then divided by the total number of cytokine producing CD4 T cell response identified empirically. One can consider this to be the “benefit” of using the algorithms to select peptides for further study. Figure 5B presents the number of peptides that were selected by each algorithm at the two cut-off values, and thus the number that would need to be made if all of the peptides predicted to be in that group of binders were tested in functional studies.
Figure 5. Algorithm performance in predicting CD4 T cell response specificity using different affinity cutoffs.
The cytokine EliSpots for all epitopes recognized by CD4 T cells in each strain were summed to represent the total response. Predictions of MHC class II binding peptides were calculated by each algorithm and the epitopes that were successfully predicted at either a 50nM (open bars) or 500nM (filled bars) IC50 values were identified. Responses to these two subsets of peptides were summed and divided by the total response elicited by all of the peptides to yield “% predicted” in A, the fraction of the response that would have been identified by using only the algorithm-selected peptides at the indicated threshold. B. The total number of peptides predicted by each algorithm for each allele at either the 50 or 500nM threshold is shown. The total number of peptides that would have been made using no selection is 332.
Use of a 50nM IC50 value of predicted affinity did capture at least 30% of the response for DR1, with increasing performance using SMM<NetMHCPan<NetMHC<ARB. However, the apparent greater performance came at the “cost” of increasing numbers of peptides to be tested (panel B, open bars) for these algorithms, based on additional peptides estimated to be better than the 50nM IC50 value. For the less well-studied DR4 molecule, ARB outperformed the other algorithms, and would have allowed identification of 40% of the response using the 50nM cutoff, and would have required testing of only 60 peptides. At the 500nM, Net MHCpan appeared to outperform the other algorithms in terms of cost vs. benefit, with over 60% of the response captured and only requiring testing of 90 peptides. For I-Ab, ARB seemed to be the best performing, with over 30% of the response predicted at 500 nM predicted affinity cutoff, which would have selected approximately 80 peptides to be tested in secondary screens. This comparative retrospective analysis shows that it is difficult to identify algorithms or parameters that will efficiently identify epitopes for untested class II molecules.
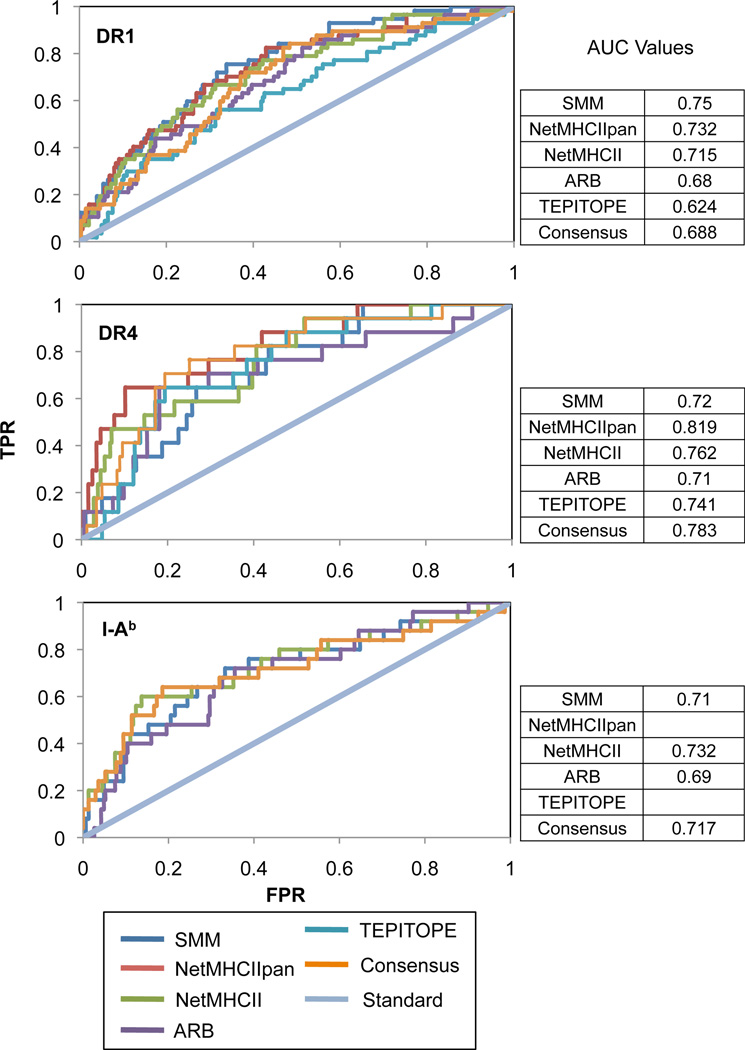
Analysis of algorithm performance by ROC curves
The large discrepancies within the absolute affinities predicted by the different algorithms introduced the rather large caveat of having to pick a threshold to adequately compare predictive ability between algorithms. As observed in the previous section, a good predictive algorithm in a relative sense can hypothetically be too stringent or lenient with its predicted affinities and be eclipsed by an algorithm with lesser predictive ability but more moderate affinity estimate. to circumvent this caveat, the binary classifier system – sorting predictions into true positive, false positive, true negative, and false negative – was imposed at all possible thresholds and for each threshold, a true positive rate and false positive rate were calculated and plotted as ROC curves for each algorithm (Figure 6), a method that is commonly used when multiple algorithms are compared for predictive performance (23–25, 32, 34) and that is generally used to measure the distinguishing capability of a classification or diagnostic system (75). The area under the curve (AUC) values were calculated to score the overall predictive ability of each algorithm without the need for a binding cutoff and compared to what would have been predicted by chance alone. From the given analysis, NetMHCHIIpan and NetMHCII seem to score consistently higher than the mean AUC value. However, aside from TEPITPOE’s relatively poor performance in DR1, there is no algorithm that significantly outperforms or underperforms relative to the other algorithms. Furthermore, the differences between algorithms seem to decrease in relation to how extensively the MHC context is studied. The range and standard deviation between AUC values is largest in DR1 while near negligible in I-Ab.
Figure 6. ROC analysis and AUC values of predictive algorithms.
The predictive ability of SMM, NetMHCIIpan, NetMHCII, ARB, TEPITOPE, and Consensus are shown in terms of ROC curves. The ‘standard’ (y=x) line represents the curve formed had peptides been chosen at random. Predictive ability is gauged by how far the points on the curve fall to the upper left of the standard line. Because better performing algorithms encompass more area under their respective curves, AUC values are a numerical representation of algorithm efficiency, with 1 representing a perfect predictor and 0.5 representing a non-predictive or random algorithm
Potential advantage of percentile ranking vs. predicted affinity for selection of peptides as candidates for further studies
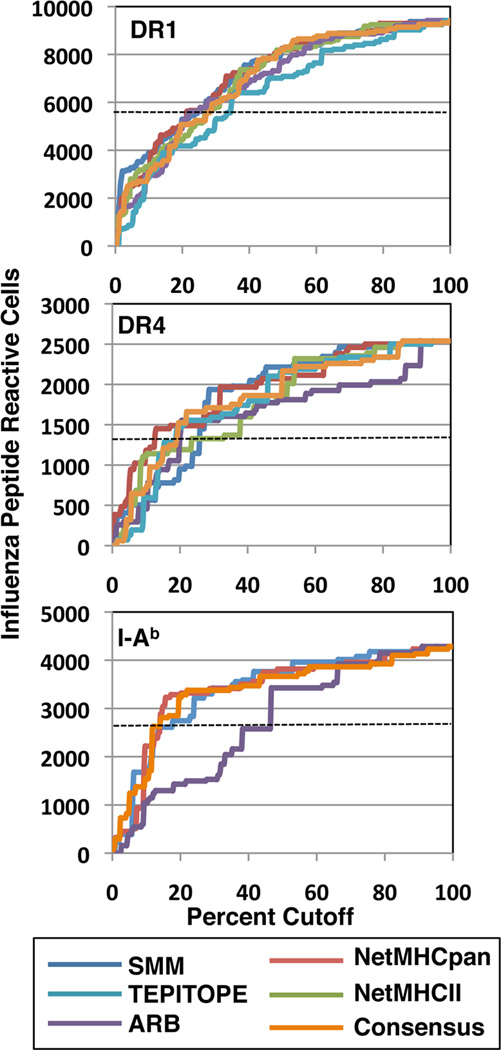
The overall inconsistency in using affinity estimates to predict epitopes, as well as the tendency of some algorithms to estimate high affinity to many peptides (ARB) and other to estimate low affinity to many of the peptides (SMM), prompted us to consider the possibility that the affinity estimates can be inaccurate in an absolute sense but might be useful in a relative sense. We therefore asked if ranking of peptides in a hierarchy of affinity rather than absolute affinity estimates would lead to more consistent results and more efficient use of the algorithms, while allowing us to still estimate the actual fractional response successfully predicted. To evaluate this, we simply ranked all the peptides by their estimated IC50 values and then used a simple percentile cut off, rather than affinity threshold to select peptides (Figure 7). Using this method of ranking, we were also able to extend our studies to Consensus and Tepitope, which simply rank peptides. When this fixed percentile ranking rather than estimated affinity was used to select peptides, a much better “performance” was apparent for all alleles and algorithms, with as much as 40–60% of the response predicted at even the 10% cutoff, depending on the allele. There was a particularly striking agreement when comparing the different algorithms for DR1 using this method. This result indicates that algorithm outputs of IC50 values for peptide:class II interactions are the most useful for simple ranking of candidate peptide epitopes, rather than for use as absolute values for selection. This conclusion from our studies is consistent with recently published approaches that assimilate the rankings of peptides, rather than their estimated affinities, to select peptides for further empirical study (34, 71, 76).
Figure 7. Percentile rank cutoff vs. fraction of CD4 response predicted by individual algorithms.
The cytokine EliSpots for all of the epitopes recognized by CD4 T cells in each strain of mice were summed to represent the total response. Prediction of MHC class II binding peptides were estimated by each algorithm and epitopes were ranked by each algorithm in the order of diminishing predicted affinity. Cytokine EliSpot responses elicited at each incremental rank of the peptides selected by each algorithm were summed and divided by the total response elicited by all of the peptides to yield the number of peptide reactive CD4 T cells successfully predicted at the indicated threshold. Shown by the dotted line is the point at which 50% of the response would have been predicted or “captured” through the use of the indicated algorithm.
An additional and quite striking observation from this analysis is that although irregular in shape, the lines that describe the prediction effectiveness of each algorithm in capturing the response essentially overlap. They all have a fairly sharp slope to the 10% cutoff (successfully predicting approximately 20–30% of the response), then a diminished slope from approximately 10% to 60% cutoff and then approach a plateau after the 60–70% cutoff, representing the low yield of additional epitopes with increasing peptides tested. The conclusion from these analyses is that ranking peptides by different algorithms normalizes their prediction efficiency and when compared in this way, they all perform similarly to each other. The conclusion is in agreement with the ROC analyses discussed earlier. The individual peptides “captured” at each cutoff varies somewhat for each algorithm (not shown), but any given algorithm would apparently allow identification of approximately 25% of the response by selection of 32 peptides out of 332, at least for these three alleles of MHC class II molecules.
Kinetic measure of class II:peptide affinity as a predictor of immunodominance
One issue that is raised by the data in Figures 2 and 4 is why some peptides that are not predicted to bind to the relevant MHC class II at all, do in fact successfully elicit CD4 T cell responses to influenza infection. One possibility to explain these false negatives is that some peptides can bind very weakly to the “presenting” peptide MHC class II molecule but nonetheless have properties that allow elicitation of a CD4 T cell response, for example T cell receptor contacts that recruit a tremendously high number of reactive CD4 T cells, a parameter that is not currently evaluated by available algorithms. The other possibility is that the IC50 values estimated by the algorithms do not accurately assess the true affinity of some MHC class II:peptide complexes. Our laboratory has previously shown that dissociation kinetics of MHC class II:peptide complexes, rather than competition assays are the best predictor of immunogenicity, both in responses to complex proteins and in multi-peptide based vaccines (57, 77–80). In analyses of class II:peptide complexes with dissociation rates that differ by seven orders of magnitude, McConnell’s group (81) showed that the association rates were essentially the same, indicating that dissociation kinetics provide accurate measurements of the affinities of peptide:class II complexes. To evaluate whether this parameter is a better indicator of immunogenicity for influenza-specific responses, we tested the dissociation rates of several DR1 peptide complexes, using purified class II and purified fluoresceinated peptides. Two peptides that elicited greater than 100 CD4 T cells per million but that were not predicted by any of the algorithms to possess high affinity binding to DR1 were chosen for this biochemical study. One peptide was derived from HA (375-SGYAADQKSTQNAIN), and one was from NP (73-ERRNKYLEEHPSAGK). Two control peptides analyzed in earlier work (56) were studied in parallel. Dissociation half times of the peptide:class II complexes (t1/2), cytokine EliSpot and predicted IC50 values for these peptides are shown in Table I. It is clear from this study that all of the immunogenic peptides tested have very high affinity and stable interactions with the presenting class II molecule. The HA (375–389) and NP (73–88) peptides that were not predicted to bind with high affinity to DR1 by any of the algorithms had dissociation half-times of 600 and 500 hr, respectively, indicating that they persist on DR1, even at acidic pH, for more than 20 days. Only the one HA peptide previously examined by our group (ELLVLLENERTLDFHD, t1/2 350 hr) was predicted to be a high affinity binder (low IC50) by most of the algorithms. Strikingly, among the four peptides, there was no correlation between the dissociation rates and predicted IC50 value. The two peptides that had the most rapid dissociation rates (HA-162 and HA-350 with t1/2 of 85 and 350 hr, respectively) had the highest estimated affinity (mean of 106nM, and 148nM IC50 respectively), while the peptides that had the most stable interactions with DR1 (HA-375 and NP-73, t1/2 of 600 hr and 500 hr, respectively) had lowest mean estimated affinity (3354 and 1300nM, respectively). Table II shows a similar type of analyses performed with the murine I-Ad molecule using the algorithms that can be used to score antigenic peptides presented by I-Ad and predict IC50 values (NetMHCII2.2, SMM-align and ARB). In these studies, known immunodominant or cryptic epitopes from model or pathogenic origin, whose binding register and dissociation kinetics were previously determined by us (57, 59, 79, 80, 82) were studied. We also compared peptide variants of these peptides whose binding stability to the class II molecule could be modulated by changes in anchor residues, as we have described (57, 59, 79, 82). Again, as was seen with some of the epitopes presented by HLA-DR1, immunodominant peptides such as LACK (161–173) and an HA peptide 126–138, T128>V) designed to improve its anchor interaction with the P1 pocket (57, 59, 80, 82) were predicted by the algorithms to have variable, but generally weak IC50 values that were not substantially different from peptides that very poorly recruit CD4 T cells in vivo such as HEL (11–25) or HA (126–138) (57, 82). In all cases, however, the measured off-rates of the peptides from the class II molecule correlated with their immunodominance. Table III additionally shows that in many cases incorrect or differing binding registers were predicted by the different algorithms from what our studies have shown to be the correct binding register (57, 59, 82). We conclude from these studies that the ability to bind strongly to the presenting class II molecule is indeed a strong predictor of immunodominance, and that peptide dissociation assays accurately measure this parameter, but that algorithms sometimes fail to recognize the features of high affinity binding.
Table I.
Relationship between the predicted IC50 values, kinetic stability and the immunodominance of influenza-derived peptides presented by HLA-DR1 molecules. Shown are the number of CD4 T cell recruited per million, the t1/2 dissociation values for the DR1:peptide complexes, measured as described in Materials and Methods and affinity (IC50 value) of the peptide for DR1 predicted by the indicated algorithm.
| Peptide | Sequence | Cytokine Spots |
t1/2 (h) pH 5.3 |
SMM IC50 |
NetMHCIIpan IC50 |
NetMHCII IC50 |
ARB IC50 |
Mean IC50 |
|---|---|---|---|---|---|---|---|---|
| NP-73 | ERRNKYLEEHPSAGKDP | 167 | 500 | 3684 | 860 | 151 | 506 | 1300 |
| HA-375 | SGYAADQKSTQNAINGI | 118 | 600 | 8264 | 1930 | 3058 | 166 | 3354 |
| HA-162* | KRNLLWLTGKNGL | 163 | 85 | 302 | 54 | 30 | 40 | 106 |
| HA-440* | ELLVLLENERTLDFHD | 146 | 350 | 523 | 42 | 13 | 15 | 148 |
Peptides with asterisk indicate a sequence extension to improve peptide solubility but that did not change the affinity value predicted by the algorithm.
Table II.
Correlation of predicted IC50 (nM) values with the experimentally determined half-lives of peptide/I-Ad complexes and the immune response.
| Name | Sequence | Immune Response |
t1/2 (h) |
Predicted IC50 (nM) | ||
|---|---|---|---|---|---|---|
| NetMHCII2.2 | SMM-align | ARB | ||||
| Lack[161–173] | SLEHPIVVSGSWD | Dominant | 200 | 5129 | 3376 | 17328 |
| OVA[273–288] | MEERKIKVYLPRMKME | Dominant | 160 | 44 | 1951 | 751 |
| HA[126–138]T128V | HNVNGVTAACSHE | Dominant | 85 | 27215 | 40083 | 474 |
| HA[126–138] | HNTNGVTAACSHE | Cryptic | 26 | 28707 | 50000 | 54 |
| HEL[11–25] | AMKRHGLDNYRGYSL | Cryptic | 6 | 12126 | 50000 | 1906 |
| Lack[161–173]I166A | SLEHPAVVSGSWD | Cryptic | 2 | 5171 | 4406 | 30826 |
Table III.
Accuracy of the peptide binding cores predicted by algorithms for I-Ad compared with cores determined experimentally with anchor-substituted peptides and dissociation kinetics.
| Name | Sequence | Binding Core NetMHCII2.2 |
Binding Core SMM-align |
Binding Core ARB |
Known Binding Core |
|---|---|---|---|---|---|
| Lack[161–173] | SLEHPIVVSGSWD | EHPIVVSGS | HPIVVSGSW | LEHPIVVSG | EHPIVVSGS |
| OVA[273–288] | MEERKIKVYLPRMKME | RKIKVYLPR | RKIKVYLPR | MEERKIKVY | ERKIKVYLP |
| HA[126–138]T128V | HNVNGVTAACSHE | HNVNGVTAA | NGVTAACSH | HNVNGVTAA | VNGVTAACS |
| HA[126–288] | HNTNGVTAACSHE | HNTNGVTAA | NGVTAACSH | HNTNGVTAA | TNGVTAACS |
| HEL[11–25] | AMKRHGLDNYRGYSL | RHGLDNYRG | RHGLDNYRG | MKRHGLDNY | RHGLDNYRG |
| Lack[161–173]I166A | SLEHPAVVSGSWD | EHPAVVSGS | HPAVVSGSW | LEHPAVVSG | EHPAVVSGS |
Discussion
In this study, we have combined the results of an unbiased approach for influenza CD4 T cell epitope discovery with implementation of predictive algorithms to select immunodominant peptide epitopes. Advances in CD4 epitope discovery are becoming increasingly important for the tracking and functional characterization of CD4 T cells during or after infection or vaccination and is also essential for derivation of peptide-based vaccines. Purely empirical strategies for epitope discovery, using either the method we have used or other approaches such as tetramer-based epitope mapping (83–86) are daunting because of their cost and labor intensive nature. Also, for pathogens with large genomes, purely empiric approaches are impractical. Therefore, the potential benefits of computer-based algorithms to facilitate identification of CD4 T cell epitopes are significant. The increasing depth of available databases for peptides presented or bound by class II molecules (46, 87–89), coupled with enhanced sophistication of computer-based algorithms, has led to increasing optimism of using predictive tools as a starting point for epitope discovery. We therefore sought to evaluate the “state of the art” performance of a number of these algorithms to predict peptides that are the focus of the CD4 T cell response to a live influenza infection.
Several important conclusions were possible from the analyses we have performed. First, utilization of predictive algorithms can facilitate epitope discovery. With all of the three allelic forms of class II molecules tested in this study, the identified epitopes were highly represented in the top third to half of the peptides predicted by the different algorithms. Although the degree of enrichment varied with the allele and the algorithm, this result does suggest that use of predictive algorithms can be a good first step toward epitope identification, at least for the alleles analyzed in this study and for pathogens with limited genome sizes or for protein vaccines. Specific strategies to use this enrichment step for these types of applications will depend on the goals of the investigator. For example, if one seeks to identify only a few epitopes for tracking a response to a particular pathogen, fairly stringent selection (i.e. selection of the top 5%–10% of peptides) will likely be successful. If more criteria need to be fulfilled (highly immunodominant peptides or CD4 epitopes within a particular protein), lower thresholds may need to be employed, with the increasing “yield” balanced by increasing “cost” of greater numbers of synthetic peptides to test. Agreement of algorithms and prediction efficiencies were enhanced through use of a ranking system rather than predicted IC50 value to select peptides.
Second and quite striking for each of the algorithms tested, there is a high number of “false positive” peptides identified: peptides that were predicted to be high affinity binders to the host class II molecule but that were not recognized by CD4 T cells elicited in response to influenza infection. The number of peptides in this category varied, depending on the specific allele and algorithm and the cutoff used for selection of peptides. For example, with high stringency (either 50nM for algorithms that predict IC50 values or 10%, for algorithms that rank peptides), the range in false positives for DR1 ranged from 20–65 peptides for the 332-peptide “proteome”, consisting of 5 viral proteins tested. When the threshold included all peptides that are predicted to bind to the relevant MHC class II molecule, there were over 100 false positive peptides. From a practical standpoint, the high false positive rate for many class II/protein combinations indicates in general, that the algorithms should be used only as a first step in epitope discovery. We conclude that secondary screens are essential to identify the true epitopes, after which focused studies can be pursued. For large genome organisms, however, secondary screens of large numbers of candidate peptides may be unfeasible using the currently available computational strategies.
There are several possibilities to explain the high number of false positive peptides. The first, and simplest explanation is that these peptides have been “chosen” correctly by the algorithm to be high affinity binders for the MHC molecules expressed in the host but lack some property needed to be a CD4 T cell epitope. One element that could be lacking in MHC-binding peptides that do not elicit CD4 T cells could be their ability to be processed and presented by the class II molecule, due to the source protein abundance or the location of the peptide within the viral protein. A second feature that might diminish a peptide’s antigenicity despite high affinity binding to class II molecules is that they possess few amino acid residues in solvent-exposed positions that are potent in recruitment of CD4 T cells. The predicted affinity of some peptides may be an overestimate, even for those peptides that have “attractive” TcR contacts and are abundantly released and available for peptide binding during antigen presentation. We do know that the reproducibility in responses among individual animals is reasonably good (Supplemental Figure 3), suggesting that we have not missed epitopes due to pooling of lymphocytes from multiple animals, each of which might display greatly varied responses. Pooling of CD4 T cells from multiple mice is a necessary experimental strategy because of the number of different peptide epitopes tracked in parallel in our studies, and the low yields of CD4 T cells from individual mice. Finally, from a purely technical standpoint, it is possible that poor peptide quality or solubility limits our T cell detection for occasional peptides, thus accounting for false positives. The peptides used in our studies were HPLC-purified, but it is possible that peptide quantity or purity was overestimated by the supplier or alternatively that peptide solubility was limited, thus diminishing the signal in the EliSpot assays.
Our analyses also revealed a high false negative rate; peptides that were recognized in the CD4 T cell response but not predicted to be MHC binders by most of the algorithms. The analyses here indicated even with well-characterized class II molecules, at least 25% of the peptides that elicit strong CD4 T cell responses were not predicted by most of the algorithms to bind to the MHC molecule in question. Although in the animal studies reported here, we have studied the immunodominance pattern at the peak of the immune response (day 10–14), we have found that the hierarchies in peptide-specific CD4 T cells remain in the memory phase of the immune response (56). These results suggest that epitopes discovered in our studies are representative of those persisting in the memory phase and in human circulating CD4 T cells. In fact, some of the HLA-DR epitopes discovered in our transgenic models predicted to be of very low affinity by all of the algorithms tested, including NP-73 and NP-402 for DR1, and M1–97 for DR4 have been reported in the literature from human subjects (84, 90). Finally, we are confident that the false negative rate is not due to incorrect assignment of the class II restriction element in the mice. Only the HLA-DR1 mice express an additional class II molecule and for these peptide epitopes, MHC restriction was unambiguously assigned through the use of transfected APC in EliSpot assays, as we have described (21, 91).
The false negative score for many epitopes suggests that the algorithms in general fail to recognize certain types of peptides that are presented by MHC class II molecules, even those that have been well studied, such those studied here. In favor of this possibility are results of our studies with the I-Ad class II molecule, which has been extensively analyzed for its peptide binding and cocrystallized with antigenic peptide (92). I-Ad was widely thought to have a “promiscuous” binding pocket with specificity primarily dictated by the P4 and P6 pockets (39, 92–94) but we recently found that the P1 pocket to be a strong determinant of peptide binding particularly with glutamic acid at this position (59, 82). Our studies suggest that this charged reside binds via a novel salt bridge within this pocket. Because of this alternative type of interaction in the P1 pocket, the “value” of glutamic acid in antigenic peptides was previously missed.
One unexpected finding revealed by our analyses is that the efficiency in prediction of epitopes seems to similarly plateau for all the algorithms tested, even if ranking is used to order peptides. It is remarkable that algorithms developed recently do not outperform others developed more than a decade ago when the efficiency of prediction was analyzed in this way. This conclusion prompts us to question whether the approaches used for development and refinement of computation tools have fundamental limitations that will not improve even with more sophisticated computational methods. We suggest that rather than focus on modifications of computational strategies, increased performance of computational aids to epitope prediction may require a serious re-evaluation of the data that is used to “train” the algorithms. One improvement that might significantly enhance performance would be to disproportionately weigh data that provides the most information on the peptide core and stability of binding to the host MHC molecule. Most of the newly developed methods are trained on data derived primarily on competitive binding assays. Because of their relatively low cost and high throughput capacity, these assays are very useful for identification of peptides that do or do not bind to a given class II allele. However, the quantitative ability of peptides to compete for binding may reflect properties that are distinct from those that promote accumulation onto class II molecules, clearly a key event for immunogenicity (57, 80, 82, 95). From the simplest point of view, IC50 values are measures of the ability of a given peptide to diminish accumulation of a labeled “indicator peptide”. Such competitive assays are based on the assumption the data arise from a simple equilibrium between single, homogenous monovalent receptor and ligand and follow the laws of mass action (96–99). It is now clear from the work of many groups that the reactions between peptide and MHC II proteins involve long-lived intermediates, heterogeneous initial states, and a pre-equilibrium between active and inactive forms of the class II protein and that binding reactions compete with inactivation of the MHC protein, an event that is potentiated by formation of unstable MHC:peptide complexes (100–104). The complexities in class II:peptide interactions, coupled with the possibilities of multiple binding cores of antigenic peptides, indicate predictions based solely on competitive binding assays will be limited in their ability to quantitatively assess the ability of the test peptides to bind stably to the class II molecule. Dissociation assays, elution and sequencing or crystallographic studies together provide key insight into the amino acids that promote accumulation of peptides onto the class II molecules. Although these assays are cost and labor prohibitive as routine methods, more investment in generation of these data and more weight given to these data could enhance the predictive capacity of computational strategies. Incorporation of data from these assays might also improve accuracy of the 9 amino acid core predicted to bind to in the MHC class II pocket. We hypothesize that limitations in our understanding of peptide:class II interactions have led to incorrect core assignments, a hypothesis recently supported by discrepancies between predicted cores and co-crystallization of peptide: class II complexes (25, 63) and our own data shown here. Faulty core designation may “contaminate” the prediction accuracy of preferences in binding going forward.
True breakthroughs in algorithm performance for prediction of CD4 T cell epitopes is also likely to be facilitated by considerations of peptide sequences that contribute to recruitment of diverse T cell receptors. Although strength of MHC binding is the key parameter that determines a peptide’s immunogenicity, solvent exposed residues within the peptide likely determine the upper range of the magnitude of a CD4 T cell response that can be recruited by a peptide. Recent theoretical and functional studies, as well as detailed consideration of the molecular events in thymic positive and negative selection events support the view that particular solvent exposed amino acids within MHC-bound peptides dramatically influence TcR repertoire development and elicitation (105–111). Interestingly, recent syntheses of computation and functional studies suggest that the best antigenic epitopes will have TcR contact amino acids that both survive negative selection but also have the ability to recruit diverse TcR and quantifies the amino acids that tend to favor this “equation” (112, 113). Incorporation of these characteristics of TcR recruitment into computational methods of epitope prediction will likely increase the efficiency of computer-based peptide epitope selection. We suggest that more explicit experimental evaluation of these issues, coupled the computational power of many groups of investigators, will dramatically enhance the performance of computation-based predictors, a key step needed for efficient epitope discovery.
Supplementary Material
Acknowledgements
We thank Michael (Rusty) Elliot for comments on the manuscript and faculty in the David H. Smith Center for Vaccine Biology and Immunology for helpful discussions.
Footnotes
This work was supported by grants HHSN266200700008C and R01AI51542 to A. J. Sant from the National Institutes of Health and J. L. Nayak was supported by 1K12HD068373-01 from the National Institutes of Health.
Contributor Information
Francisco A. Chaves, David H. Smith Center for Vaccine Biology and Immunology, Department of Microbiology and Immunology, University of Rochester Medical Center, Rochester, New York 14642
Alvin H. Lee, David H. Smith Center for Vaccine Biology and Immunology, Department of Microbiology and Immunology, University of Rochester Medical Center, Rochester, New York 14642
Jennifer Nayak, Department of Pediatrics, University of Rochester Medical Center.
Katherine A. Richards, David H. Smith Center for Vaccine Biology and Immunology, Department of Microbiology and Immunology, University of Rochester Medical Center, Rochester, New York 14642
Andrea J. Sant, David H. Smith Center for Vaccine Biology and Immunology, Department of Microbiology and Immunology, University of Rochester Medical Center, Rochester, New York 14642
References
- 1.De Groot AS, Rappuoli R. Genome-derived vaccines. Expert Rev Vaccines. 2004;3:59–76. doi: 10.1586/14760584.3.1.59. [DOI] [PubMed] [Google Scholar]
- 2.Herrera-Najera C, Pina-Aguilar R, Xacur-Garcia F, Ramirez-Sierra MJ, Dumonteil E. Mining the Leishmania genome for novel antigens and vaccine candidates. Proteomics. 2009;9:1293–1301. doi: 10.1002/pmic.200800533. [DOI] [PubMed] [Google Scholar]
- 3.Sette A, Rappuoli R. Reverse vaccinology: developing vaccines in the era of genomics. Immunity. 2010;33:530–541. doi: 10.1016/j.immuni.2010.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chaudhuri R, Ahmed S, Ansari FA, Singh HV, Ramachandran S. MalVac: database of malarial vaccine candidates. Malar J. 2008;7:184. doi: 10.1186/1475-2875-7-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dudek NL, Perlmutter P, Aguilar MI, Croft NP, Purcell AW. Epitope discovery and their use in peptide based vaccines. Curr Pharm Des. 2010;16:3149–3157. doi: 10.2174/138161210793292447. [DOI] [PubMed] [Google Scholar]
- 6.Shimonkevitz R, Colon S, Kappler JW, Marrack P, Grey HM. Antigen recognition by H-2-restricted T cells. II. A tryptic ovalbumin peptide that substitutes for processed antigen. J Immunol. 1984;133:2067–2074. [PubMed] [Google Scholar]
- 7.Allen PM, Strydom DJ, Unanue ER. Processing of lysozyme by macrophages: identification of the determinant recognized by two T-cell hybridomas. Proc Natl Acad Sci U S A. 1984;81:2489–2493. doi: 10.1073/pnas.81.8.2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Villarreal-Ramos B, Sanchez-Garcia J, Stoker N, Timms E, Chomer D, Raff K, Mitchison NA. Screening gene expression libraries for epitopes recognized in Mycobacterium leprae by mouse T cells. Eur J Immunol. 1991;21:2621–2624. doi: 10.1002/eji.1830211047. [DOI] [PubMed] [Google Scholar]
- 9.Kurokohchi K, Akatsuka T, Pendleton CD, Takamizawa A, Nishioka M, Battegay M, Feinstone SM, Berzofsky JA. Use of recombinant protein to identify a motif-negative human cytotoxic T-cell epitope presented by HLA-A2 in the hepatitis C virus NS3 region. J Virol. 1996;70:232–240. doi: 10.1128/jvi.70.1.232-240.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Turner MJ, Abdul-Alim CS, Willis RA, Fisher TL, Lord EM, Frelinger JG. T-cell antigen discovery (T-CAD) assay: a novel technique for identifying T cell epitopes. J Immunol Methods. 2001;256:107–119. doi: 10.1016/s0022-1759(01)00436-7. [DOI] [PubMed] [Google Scholar]
- 11.Valentino MD, Maben ZJ, Hensley LL, Woolard MD, Kawula TH, Frelinger JA, Frelinger JG. Identification of T-cell epitopes in Francisella tularensis using an ordered protein array of serological targets. Immunology. 2011;132:348–360. doi: 10.1111/j.1365-2567.2010.03387.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reinhold B, Keskin DB, Reinherz EL. Molecular Detection of Targeted Major Histocompatibility Complex I-Bound Peptides Using a Probabilistic Measure and Nanospray MS(3) on a Hybrid Quadrupole-Linear Ion Trap. Anal Chem. 2010;82:9090–9099. doi: 10.1021/ac102387t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Purcell AW, Gorman JJ. The use of post-source decay in matrix-assisted laser desorption/ionisation mass spectrometry to delineate T cell determinants. J Immunol Methods. 2001;249:17–31. doi: 10.1016/s0022-1759(00)00361-6. [DOI] [PubMed] [Google Scholar]
- 14.Karunakaran KP, Rey-Ladino J, Stoynov N, Berg K, Shen C, Jiang X, Gabel BR, Yu H, Foster LJ, Brunham RC. Immunoproteomic discovery of novel T cell antigens from the obligate intracellular pathogen Chlamydia. J Immunol. 2008;180:2459–2465. doi: 10.4049/jimmunol.180.4.2459. [DOI] [PubMed] [Google Scholar]
- 15.Tan CT, Croft NP, Dudek NL, Williamson NA, Purcell AW. Direct quantitation of MHC-bound peptide epitopes by selected reaction monitoring. Proteomics. 2011;11:2336–2340. doi: 10.1002/pmic.201000531. [DOI] [PubMed] [Google Scholar]
- 16.Strug I, Calvo-Calle JM, Green KM, Cruz J, Ennis FA, Evans JE, Stern LJ. Vaccinia peptides eluted from HLA-DR1 isolated from virus-infected cells are recognized by CD4+ T cells from a vaccinated donor. J Proteome Res. 2008;7:2703–2711. doi: 10.1021/pr700780x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hiemstra HS, Drijfhout JW, Koning F. Synthetic Peptide libraries for T-cell epitope identification. Methods Mol Med. 2000;41:89–96. doi: 10.1385/1-59259-082-9:089. [DOI] [PubMed] [Google Scholar]
- 18.Geginat G, Schenk S, Skoberne M, Goebel W, Hof H. A novel approach of direct ex vivo epitope mapping identifies dominant and subdominant CD4 and CD8 T cell epitopes from Listeria monocytogenes. J Immunol. 2001;166:1877–1884. doi: 10.4049/jimmunol.166.3.1877. [DOI] [PubMed] [Google Scholar]
- 19.Tobery TW, Caulfield MJ. Identification of T-cell epitopes using ELISpot and peptide pool arrays. Methods Mol Med. 2004;94:121–132. doi: 10.1385/1-59259-679-7:121. [DOI] [PubMed] [Google Scholar]
- 20.Maciel M, Jr, Kellathur SN, Chikhlikar P, Dhalia R, Sidney J, Sette A, August TJ, Marques ET., Jr Comprehensive analysis of T cell epitope discovery strategies using 17DD yellow fever virus structural proteins and BALB/c (H2d) mice model. Virology. 2008;378:105–117. doi: 10.1016/j.virol.2008.04.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Richards KA, Chaves FA, Sant AJ. Infection of HLA-DR1 transgenic mice with a human isolate of influenza a virus (H1N1) primes a diverse CD4 T-cell repertoire that includes CD4 T cells with heterosubtypic cross-reactivity to avian (H5N1) influenza virus. J Virol. 2009;83:6566–6577. doi: 10.1128/JVI.00302-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nayak JL, Richards KA, Chaves FA, Sant AJ. Analyses of the specificity of CD4 T cells during the primary immune response to influenza virus reveals dramatic MHC-linked asymmetries in reactivity to individual viral proteins. Viral Immunol. 2010;23:169–180. doi: 10.1089/vim.2009.0099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang H, Wang P, Papangelopoulos N, Xu Y, Sette A, Bourne PE, Lund O, Ponomarenko J, Nielsen M, Peters B. Limitations of Ab initio predictions of peptide binding to MHC class II molecules. PLoS One. 2010;5:e9272. doi: 10.1371/journal.pone.0009272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang P, Sidney J, Kim Y, Sette A, Lund O, Nielsen M, Peters B. Peptide binding predictions for HLA DR, DP and DQ molecules. BMC Bioinformatics. 2010;11:568. doi: 10.1186/1471-2105-11-568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lin HH, Zhang GL, Tongchusak S, Reinherz EL, Brusic V. Evaluation of MHC-II peptide binding prediction servers: applications for vaccine research. BMC Bioinformatics. 2008;9(Suppl 12):S22. doi: 10.1186/1471-2105-9-S12-S22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lin HH, Ray S, Tongchusak S, Reinherz EL, Brusic V. Evaluation of MHC class I peptide binding prediction servers: applications for vaccine research. BMC Immunol. 2008;9:8. doi: 10.1186/1471-2172-9-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peters B, Bui HH, Frankild S, Nielson M, Lundegaard C, Kostem E, Basch D, Lamberth K, Harndahl M, Fleri W, Wilson SS, Sidney J, Lund O, Buus S, Sette A. A community resource benchmarking predictions of peptide binding to MHC-I molecules. PLoS Comput Biol. 2006;2:e65. doi: 10.1371/journal.pcbi.0020065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nielsen M, Lund O, Buus S, Lundegaard C. MHC class II epitope predictive algorithms. Immunology. 2010;130:319–328. doi: 10.1111/j.1365-2567.2010.03268.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nielsen M, Lundegaard C, Lund O. Prediction of MHC class II binding affinity using SMM-align, a novel stabilization matrix alignment method. BMC Bioinformatics. 2007;8:238. doi: 10.1186/1471-2105-8-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bui HH, Sidney J, Peters B, Sathiamurthy M, Sinichi A, Purton KA, Mothe BR, Chisari FV, Watkins DI, Sette A. Automated generation and evaluation of specific MHC binding predictive tools: ARB matrix applications. Immunogenetics. 2005;57:304–314. doi: 10.1007/s00251-005-0798-y. [DOI] [PubMed] [Google Scholar]
- 31.Zhang GL, Khan AM, Srinivasan KN, August JT, Brusic V. Neural models for predicting viral vaccine targets. J Bioinform Comput Biol. 2005;3:1207–1225. doi: 10.1142/s0219720005001466. [DOI] [PubMed] [Google Scholar]
- 32.Nielsen M, Lund O. NN-align. An artificial neural network-based alignment algorithm for MHC class II peptide binding prediction. BMC Bioinformatics. 2009;10:296. doi: 10.1186/1471-2105-10-296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang W, Liu J, Niu Y. Quantitative prediction of MHC-II binding affinity using particle swarm optimization. Artif Intell Med. 2010;50:127–132. doi: 10.1016/j.artmed.2010.05.003. [DOI] [PubMed] [Google Scholar]
- 34.Wang P, Sidney J, Dow C, Mothe B, Sette A, Peters B. A systematic assessment of MHC class II peptide binding predictions and evaluation of a consensus approach. PLoS Comput Biol. 2008;4:e1000048. doi: 10.1371/journal.pcbi.1000048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Falk K, Rotzschke O, Stevanovic S, Jung G, Rammensee HG. Allele-specific motifs revealed by sequencing of self-peptides eluted from MHC molecules. Nature. 1991;351:290–296. doi: 10.1038/351290a0. [DOI] [PubMed] [Google Scholar]
- 36.Deres K, Schumacher TN, Wiesmuller KH, Stevanovic S, Greiner G, Jung G, Ploegh HL. Preferred size of peptides that bind to H-2 Kb is sequence dependent. Eur J Immunol. 1992;22:1603–1608. doi: 10.1002/eji.1830220638. [DOI] [PubMed] [Google Scholar]
- 37.Matsumura M, Fremont DH, Peterson PA, Wilson IA. Emerging principles for the recognition of peptide antigens by MHC class I molecules. Science. 1992;257:927–934. doi: 10.1126/science.1323878. [DOI] [PubMed] [Google Scholar]
- 38.Chicz RM, Urban RG, Lane WS, Gorga JC, Stern LJ, Vignali DA, Strominger JL. Predominant naturally processed peptides bound to HLA-DR1 are derived from MHC-related molecules and are heterogeneous in size. Nature. 1992;358:764–768. doi: 10.1038/358764a0. [DOI] [PubMed] [Google Scholar]
- 39.Hunt DF, Michel H, Dickinson TA, Shabanowitz J, Cox AL, Sakaguchi K, Appella E, Grey HM, Sette A. Peptides presented to the immune system by the murine class II major histocompatibility complex molecule I-Ad. Science. 1992;256:1817–1820. doi: 10.1126/science.1319610. [DOI] [PubMed] [Google Scholar]
- 40.Lippolis JD, White FM, Marto JA, Luckey CJ, Bullock TN, Shabanowitz J, Hunt DF, Engelhard VH. Analysis of MHC class II antigen processing by quantitation of peptides that constitute nested sets. J Immunol. 2002;169:5089–5097. doi: 10.4049/jimmunol.169.9.5089. [DOI] [PubMed] [Google Scholar]
- 41.Srinivasan M, Domanico SZ, Kaumaya PT, Pierce SK. Peptides of 23 residues or greater are required to stimulate a high affinity class II-restricted T cell response. Eur J Immunol. 1993;23:1011–1016. doi: 10.1002/eji.1830230504. [DOI] [PubMed] [Google Scholar]
- 42.Landais E, Romagnoli PA, Corper AL, Shires J, Altman JD, Wilson IA, Garcia KC, Teyton L. New design of MHC class II tetramers to accommodate fundamental principles of antigen presentation. J Immunol. 2009;183:7949–7957. doi: 10.4049/jimmunol.0902493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bankovich AJ, Girvin AT, Moesta AK, Garcia KC. Peptide register shifting within the MHC groove: theory becomes reality. Mol Immunol. 2004;40:1033–1039. doi: 10.1016/j.molimm.2003.11.016. [DOI] [PubMed] [Google Scholar]
- 44.Robertson JM, Jensen PE, Evavold BD. DO11.10 and OT-II T cells recognize a C-terminal ovalbumin 323–339 epitope. Journal of Immunology. 2000;164:4706–4712. doi: 10.4049/jimmunol.164.9.4706. [DOI] [PubMed] [Google Scholar]
- 45.McFarland B, Sant A, Beeson C. The Ovalbumin (323–339) peptide binds to the MHC class II I-Ad protein using at least two different registers. Biochem. 1999;38:16663. doi: 10.1021/bi991393l. [DOI] [PubMed] [Google Scholar]
- 46.Peters B, Sidney J, Bourne P, Bui HH, Buus S, Doh G, Fleri W, Kronenberg M, Kubo R, Lund O, Nemazee D, Ponomarenko JV, Sathiamurthy M, Schoenberger S, Stewart S, Surko P, Way S, Wilson S, Sette A. The immune epitope database and analysis resource: from vision to blueprint. PLoS Biol. 2005;3:e91. doi: 10.1371/journal.pbio.0030091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Peters B, Sidney J, Bourne P, Bui HH, Buus S, Doh G, Fleri W, Kronenberg M, Kubo R, Lund O, Nemazee D, Ponomarenko JV, Sathiamurthy M, Schoenberger SP, Stewart S, Surko P, Way S, Wilson S, Sette A. The design and implementation of the immune epitope database and analysis resource. Immunogenetics. 2005;57:326–336. doi: 10.1007/s00251-005-0803-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang GL, Lin HH, Keskin DB, Reinherz EL, Brusic V. Dana-Farber repository for machine learning in immunology. J Immunol Methods. 2011;374:18–25. doi: 10.1016/j.jim.2011.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nielsen M, Justesen S, Lund O, Lundegaard C, Buus S. NetMHCIIpan-2.0 - Improved pan-specific HLA-DR predictions using a novel concurrent alignment and weight optimization training procedure. Immunome Res. 2010;6:9. doi: 10.1186/1745-7580-6-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dimitrov I, Garnev P, Flower DR, Doytchinova I. MHC Class II Binding Prediction-A Little Help from a Friend. J Biomed Biotechnol. 2010;2010 doi: 10.1155/2010/705821. 705821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hu X, Zhou W, Udaka K, Mamitsuka H, Zhu S. MetaMHC: a meta approach to predict peptides binding to MHC molecules. Nucleic Acids Res. 2010;38:W474–W479. doi: 10.1093/nar/gkq407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mustafa AS, Shaban FA, Al-Attiyah R, Abal AT, El-Shamy AM, Andersen P, Oftung F. Human Th1 cell lines recognize the Mycobacterium tuberculosis ESAT-6 antigen and its peptides in association with frequently expressed HLA class II molecules. Scand J Immunol. 2003;57:125–134. doi: 10.1046/j.1365-3083.2003.01204.x. [DOI] [PubMed] [Google Scholar]
- 53.Zhao BP, Chen L, Zhang YL, Yang JM, Jia K, Sui CY, Yuan CX, Lin JJ, Feng XG. In silico prediction of binding of promiscuous peptides to multiple MHC class-II molecules identifies the Th1 cell epitopes from secreted and transmembrane proteins of Schistosoma japonicum in BALB/c mice. Microbes Infect. 2011;13:709–719. doi: 10.1016/j.micinf.2011.03.005. [DOI] [PubMed] [Google Scholar]
- 54.Foo DG, Macary PA, Alonso S, Poh CL. Identification of human CD4 T-cell epitopes on the VP1 capsid protein of enterovirus 71. Viral Immunol. 2008;21:215–224. doi: 10.1089/vim.2007.0089. [DOI] [PubMed] [Google Scholar]
- 55.Fonseca CT, Cunha-Neto E, Goldberg AC, Kalil J, de Jesus AR, Carvalho EM, Correa-Oliveira R, Hammer J, Sidney J, Sette A, Oliveira SC. Identification of paramyosin T cell epitopes associated with human resistance to Schistosoma mansoni reinfection. Clin Exp Immunol. 2005;142:539–547. doi: 10.1111/j.1365-2249.2005.02941.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Richards KA, Chaves FA, Sant AJ. The memory phase of the CD4 T-cell response to influenza virus infection maintains its diverse antigen specificity. Immunology. 2011;133:246–256. doi: 10.1111/j.1365-2567.2011.03435.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lazarski CA, Chaves FA, Jenks SA, Wu S, Richards KA, Weaver JM, Sant AJ. The kinetic stability of MHC class II:peptide complexes is a key parameter that dictates immunodominance. Immunity. 2005;23:29–40. doi: 10.1016/j.immuni.2005.05.009. [DOI] [PubMed] [Google Scholar]
- 58.Chaves FA, Sant AJ. Measurement of peptide dissociation from MHC class II molecules. Curr Protoc Immunol. 2007;Chapter 18(Unit 18):14. doi: 10.1002/0471142735.im1814s77. [DOI] [PubMed] [Google Scholar]
- 59.Chaves FA, Richards KA, Torelli A, Wedekind J, Sant AJ. Peptide-binding motifs for the I-Ad MHC class II molecule: alternate pH-dependent binding behavior. Biochemistry. 2006;45:6426–6433. doi: 10.1021/bi060194g. [DOI] [PubMed] [Google Scholar]
- 60.Chaves FA, Hou P, Wu S, Sant AJ. Replacement of the membrane proximal region of I-A(d) MHC class II molecule with I-E-derived sequences promotes production of an active and stable soluble heterodimer without altering peptide-binding specificity. J Immunol Methods. 2005;300:74–92. doi: 10.1016/j.jim.2005.02.013. [DOI] [PubMed] [Google Scholar]
- 61.Nelson PA, Khodadoust M, Prodhomme T, Spencer C, Patarroyo JC, Varrin-Doyer M, Ho JD, Stroud RM, Zamvil SS. Immunodominant T cell determinants of aquaporin-4, the autoantigen associated with neuromyelitis optica. PLoS One. 2010;5:e15050. doi: 10.1371/journal.pone.0015050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Akhoon BA, Slathia PS, Sharma P, Gupta SK, Verma V. In silico identification of novel protective VSG antigens expressed by Trypanosoma brucei and an effort for designing a highly immunogenic DNA vaccine using IL-12 as adjuvant. Microb Pathog. 2011;51:77–87. doi: 10.1016/j.micpath.2011.01.011. [DOI] [PubMed] [Google Scholar]
- 63.Bordner AJ, Mittelmann HD. Prediction of the binding affinities of peptides to class II MHC using a regularized thermodynamic model. BMC Bioinformatics. 2010;11:41. doi: 10.1186/1471-2105-11-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nielsen M, Lundegaard C, Blicher T, Peters B, Sette A, Justesen S, Buus S, Lund O. Quantitative predictions of peptide binding to any HLA-DR molecule of known sequence: NetMHCIIpan. PLoS Comput Biol. 2008;4:e1000107. doi: 10.1371/journal.pcbi.1000107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Richards KA, Chaves FA, Krafcik FR, Topham DJ, Lazarski CA, Sant AJ. Direct ex vivo analyses of HLA-DR1 transgenic mice reveal an exceptionally broad pattern of immunodominance in the primary HLA-DR1-restricted CD4 T-cell response to influenza virus hemagglutinin. J Virol. 2007;81:7608–7619. doi: 10.1128/JVI.02834-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tobery TW, Wang S, Wang XM, Neeper MP, Jansen KU, McClements WL, Caulfield MJ. A simple and efficient method for the monitoring of antigen-specific T cell responses using peptide pool arrays in a modified ELISpot assay. J Immunol Methods. 2001;254:59–66. doi: 10.1016/s0022-1759(01)00397-0. [DOI] [PubMed] [Google Scholar]
- 67.Sturniolo T, Bono E, Ding J, Raddrizzani L, Tuereci O, Sahin U, Braxenthaler M, Gallazzi F, Protti MP, Sinigaglia F, Hammer J. Generation of tissue-specific and promiscuous HLA ligand databases using DNA microarrays and virtual HLA class II matrices. Nat Biotechnol. 1999;17:555–561. doi: 10.1038/9858. [DOI] [PubMed] [Google Scholar]
- 68.Cantor JR, Yoo TH, Dixit A, Iverson BL, Forsthuber TG, Georgiou G. Therapeutic enzyme deimmunization by combinatorial T-cell epitope removal using neutral drift. Proc Natl Acad Sci U S A. 2011;108:1272–1277. doi: 10.1073/pnas.1014739108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yauch LE, Prestwood TR, May MM, Morar MM, Zellweger RM, Peters B, Sette A, Shresta S. CD4+ T cells are not required for the induction of dengue virus-specific CD8+ T cell or antibody responses but contribute to protection after vaccination. J Immunol. 2010;185:5405–5416. doi: 10.4049/jimmunol.1001709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chen C, Dow C, Wang P, Sidney J, Read A, Harmsen A, Samuel JE, Peters B. Identification of CD4+ T cell epitopes in C. burnetii antigens targeted by antibody responses. PLoS One. 2011;6:e17712. doi: 10.1371/journal.pone.0017712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Arens R, Wang P, Sidney J, Loewendorf A, Sette A, Schoenberger SP, Peters B, Benedict CA. Cutting edge: murine cytomegalovirus induces a polyfunctional CD4 T cell response. J Immunol. 2008;180:6472–6476. doi: 10.4049/jimmunol.180.10.6472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chicz RM, Urban RG, Gorga JC, Vignali DA, Lane WS, Strominger JL. Specificity and promiscuity among naturally processed peptides bound to HLA-DR alleles. Journal of Experimental Medicine. 1993;178:27–47. doi: 10.1084/jem.178.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sant AJ. Endogenous antigen presentation by MHC class II molecules. Immunol Res. 1994;13:253–267. doi: 10.1007/BF02935617. [DOI] [PubMed] [Google Scholar]
- 74.Loss GE, Jr, Elias CG, Fields PE, Ribaudo RK, McKisic M, Sant AJ. Major histocompatibility complex class II-restricted presentation of an internally synthesized antigen displays cell-type variability and segregates from the exogenous class II and endogenous class I presentation pathways. J Exp Med. 1993;178:73–85. doi: 10.1084/jem.178.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Swets JA. Measuring the accuracy of diagnostic systems. Science. 1988;240:1285–1293. doi: 10.1126/science.3287615. [DOI] [PubMed] [Google Scholar]
- 76.Calvo-Calle JM, Strug I, Nastke MD, Baker SP, Stern LJ. Human CD4+ T cell epitopes from vaccinia virus induced by vaccination or infection. PLoS Pathog. 2007;3:1511–1529. doi: 10.1371/journal.ppat.0030144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sant AJ, Chaves F, Jenks S, Richards K, Menges P, Weaver JM, Lazarski CL. The relationship between immundominance, DM editing, and the kinetic stability of MHC class II:peptide complexes. Immunol. Rev. 2005;207:261–278. doi: 10.1111/j.0105-2896.2005.00307.x. [DOI] [PubMed] [Google Scholar]
- 78.Sant AJ, Chaves FA, Krafcik FR, Lazarski CA, Menges P, Richards K, Weaver JM. Immunodominance in CD4 T-cell responses: implications for immune responses to influenza virus and for vaccine design. Expert Rev Vaccines. 2007;6:357–368. doi: 10.1586/14760584.6.3.357. [DOI] [PubMed] [Google Scholar]
- 79.Weaver JM, Lazarski CA, Richards KA, Chaves FA, Jenks SA, Menges PR, Sant AJ. Immunodominance of CD4 T cells to foreign antigens is peptide intrinsic and independent of molecular context: implications for vaccine design. J Immunol. 2008;181:3039–3048. doi: 10.4049/jimmunol.181.5.3039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Weaver JM, Chaves FA, Sant AJ. Abortive activation of CD4 T cell responses during competitive priming in vivo. Proc Natl Acad Sci U S A. 2009;106:8647–8652. doi: 10.1073/pnas.0811584106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kasson PM, Rabinowitz JD, Schmitt L, Davis MM, McConnell HM. Kinetics of peptide binding to the class II MHC protein I-Ek. Biochemistry. 2000;39:1048–1058. doi: 10.1021/bi9921337. [DOI] [PubMed] [Google Scholar]
- 82.Lazarski CA, Chaves FA, Sant AJ. The impact of DM on MHC class II-restricted antigen presentation can be altered by manipulation of MHC-peptide kinetic stability. J Exp Med. 2006;203:1319–1328. doi: 10.1084/jem.20060058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Grotenbreg GM, Roan NR, Guillen E, Meijers R, Wang JH, Bell GW, Starnbach MN, Ploegh HL. Discovery of CD8+ T cell epitopes in Chlamydia trachomatis infection through use of caged class I MHC tetramers. Proc Natl Acad Sci U S A. 2008;105:3831–3836. doi: 10.1073/pnas.0711504105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Roti M, Yang J, Berger D, Huston L, James EA, Kwok WW. Healthy human subjects have CD4+ T cells directed against H5N1 influenza virus. J Immunol. 2008;180:1758–1768. doi: 10.4049/jimmunol.180.3.1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Yang J, James EA, Huston L, Danke NA, Liu AW, Kwok WW. Multiplex mapping of CD4 T cell epitopes using class II tetramers. Clin Immunol. 2006;120:21–32. doi: 10.1016/j.clim.2006.03.008. [DOI] [PubMed] [Google Scholar]
- 86.Reijonen H, Kwok WW. Use of HLA class II tetramers in tracking antigen-specific T cells and mapping T-cell epitopes. Methods (Duluth) 2003;29:282–288. doi: 10.1016/s1046-2023(02)00350-x. [DOI] [PubMed] [Google Scholar]
- 87.Salimi N, Fleri W, Peters B, Sette A. Design and utilization of epitope-based databases and predictive tools. Immunogenetics. 2010;62:185–196. doi: 10.1007/s00251-010-0435-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zhang Q, Wang P, Kim Y, Haste-Andersen P, Beaver J, Bourne PE, Bui HH, Buus S, Frankild S, Greenbaum J, Lund O, Lundegaard C, Nielsen M, Ponomarenko J, Sette A, Zhu Z, Peters B. Immune epitope database analysis resource (IEDB-AR) Nucleic Acids Res. 2008;36:W513–W518. doi: 10.1093/nar/gkn254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Chung JL, Sun J, Sidney J, Sette A, Peters B. IMMUNOCAT-a data management system for epitope mapping studies. J Biomed Biotechnol. 2010;2010 doi: 10.1155/2010/856842. 856842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ge X, Gebe JA, Bollyky PL, James EA, Yang J, Stern LJ, Kwok WW. Peptide-MHC cellular microarray with innovative data analysis system for simultaneously detecting multiple CD4 T-cell responses. PLoS One. 2010;5:e11355. doi: 10.1371/journal.pone.0011355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Richards K, Chaves F, Sant AJ. The primary response to live influenza infection in HLA-DR1 transgenic mice is diverse and includes more than 100 different CD4 T cell specificities, distributed widely among different influenza proteins. Ms. In preparation. 2008 [Google Scholar]
- 92.Scott CA, Peterson PA, Teyton L, Wilson IA. Crystal structures of two I-Ad-peptide complexes reveal that high affinity can be achieved without large anchor residues.[erratum appears in Immunity 1998 Apr;8(4):531] Immunity. 1998;8:319–329. doi: 10.1016/s1074-7613(00)80537-3. [DOI] [PubMed] [Google Scholar]
- 93.Scott CA, Peterson PA, Teyton L, Wilson IA. Crystal structures of two I-Ad-peptide complexes reveal that high affinity can be achieved without large anchor residues. Immunity. 1998;8:319–329. doi: 10.1016/s1074-7613(00)80537-3. [DOI] [PubMed] [Google Scholar]
- 94.Sette A, Buus S, Colon S, Miles C, Grey HM. I-Ad-binding peptides derived from unrelated protein antigens share a common structural motif. J Immunol. 1988;141:45–48. [PubMed] [Google Scholar]
- 95.Sant AJ, Chaves FA, Jenks SA, Richards KA, Menges P, Weaver JM, Lazarski CA. The relationship between immunodominance, DM editing, and the kinetic stability of MHC class II:peptide complexes. Immunol Rev. 2005;207:261–278. doi: 10.1111/j.0105-2896.2005.00307.x. [DOI] [PubMed] [Google Scholar]
- 96.Hulme EC, Trevethick MA. Ligand binding assays at equilibrium: validation and interpretation. Br J Pharmacol. 2010;161:1219–1237. doi: 10.1111/j.1476-5381.2009.00604.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Zettner A. Some theoretical aspects of competitive binding assays. Hum Pathol. 1974;5:6–8. doi: 10.1016/s0046-8177(74)80096-1. [DOI] [PubMed] [Google Scholar]
- 98.Zettner A. Principles of competitive binding assays (saturation analysis). 1. Equilibrium techniques. Clin Chem. 1973;19:699–705. [PubMed] [Google Scholar]
- 99.Cheng Y, Prusoff WH. Relationship between the inhibition constant (K1) and the concentration of inhibitor which causes 50 per cent inhibition (I50) of an enzymatic reaction. Biochem Pharmacol. 1973;22:3099–3108. doi: 10.1016/0006-2952(73)90196-2. [DOI] [PubMed] [Google Scholar]
- 100.Beeson C, McConnell HM. Kinetic intermediates in the reactions between peptides and proteins of major histocompatibility complex class II. Proceedings of the National Academy of Sciences of the United States of America. 1994;91:8842–8845. doi: 10.1073/pnas.91.19.8842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.de Kroon AI, McConnell HM. Kinetics and specificity of peptide-MHC class II complex displacement reactions. Journal of Immunology. 1994;152:609–619. [PubMed] [Google Scholar]
- 102.Beeson C, Mcconnell HM. Reactions of Peptides with Class-Ii Proteins of the Major Histocompatibility Complex. Journal of the American Chemical Society. 1995;117:10429–10433. [Google Scholar]
- 103.Zarutskie JA, Busch R, Zavala-Ruiz Z, Rushe M, Mellins ED, Stern LJ. The kinetic basis of peptide exchange catalysis by HLA-DM. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:12450–12455. doi: 10.1073/pnas.211439398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Sadegh-Nasseri S, McConnell HM. A kinetic intermediate in the reaction of an antigenic peptide and I-Ek. Nature. 1989;337:274–276. doi: 10.1038/337274a0. [DOI] [PubMed] [Google Scholar]
- 105.Turner SJ, Kedzierska K, Komodromou H, La Gruta NL, Dunstone MA, Webb AI, Webby R, Walden H, Xie W, McCluskey J, Purcell AW, Rossjohn J, Doherty PC. Lack of prominent peptide-major histocompatibility complex features limits repertoire diversity in virus-specific CD8+ T cell populations. Nat Immunol. 2005;6:382–389. doi: 10.1038/ni1175. [DOI] [PubMed] [Google Scholar]
- 106.Hogquist KA, Baldwin TA, Jameson SC. Central tolerance: learning self-control in the thymus. Nat Rev Immunol. 2005;5:772–782. doi: 10.1038/nri1707. [DOI] [PubMed] [Google Scholar]
- 107.Chen JL, Stewart-Jones G, Bossi G, Lissin NM, Wooldridge L, Choi EM, Held G, Dunbar PR, Esnouf RM, Sami M, Boulter JM, Rizkallah P, Renner C, Sewell A, van der Merwe PA, Jakobsen BK, Griffiths G, Jones EY, Cerundolo V. Structural and kinetic basis for heightened immunogenicity of T cell vaccines. J Exp Med. 2005;201:1243–1255. doi: 10.1084/jem.20042323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Stewart-Jones GB, McMichael AJ, Bell JI, Stuart DI, Jones EY. A structural basis for immunodominant human T cell receptor recognition. Nat Immunol. 2003;4:657–663. doi: 10.1038/ni942. [DOI] [PubMed] [Google Scholar]
- 109.Rudolph MG, Stanfield RL, Wilson IA. How TCRs bind MHCs, peptides, and coreceptors. Annu Rev Immunol. 2006;24:419–466. doi: 10.1146/annurev.immunol.23.021704.115658. [DOI] [PubMed] [Google Scholar]
- 110.Chu HH, Moon JJ, Kruse AC, Pepper M, Jenkins MK. Negative selection and peptide chemistry determine the size of naive foreign peptide-MHC class II-specific CD4+ T cell populations. J Immunol. 2010;185:4705–4713. doi: 10.4049/jimmunol.1002276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Obar JJ, Khanna KM, Lefrancois L. Endogenous naive CD8+ T cell precursor frequency regulates primary and memory responses to infection. Immunity. 2008;28:859–869. doi: 10.1016/j.immuni.2008.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Kosmrlj A, Jha AK, Huseby ES, Kardar M, Chakraborty AK. How the thymus designs antigen-specific and self-tolerant T cell receptor sequences. Proc Natl Acad Sci U S A. 2008;105:16671–16676. doi: 10.1073/pnas.0808081105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Kosmrlj A, Read EL, Qi Y, Allen TM, Altfeld M, Deeks SG, Pereyra F, Carrington M, Walker BD, Chakraborty AK. Effects of thymic selection of the T-cell repertoire on HLA class I-associated control of HIV infection. Nature. 2010;465:350–354. doi: 10.1038/nature08997. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.