Abstract
The C-terminal domain (CTD) of RNA polymerase II (Pol II) consists of conserved heptapeptide repeats that function as a binding platform for different protein complexes involved in transcription, RNA processing, export, and chromatin remodeling. The CTD repeats are subject to sequential waves of posttranslational modifications during specific stages of the transcription cycle. These patterned modifications have led to the postulation of the “CTD code” hypothesis, where stage-specific patterns define a spatiotemporal code that is recognized by the appropriate interacting partners. Here, we highlight the role of CTD modifications in directing transcription initiation, elongation, and termination. We examine the major readers, writers, and erasers of the CTD code and examine the relevance of describing patterns of posttranslational modifications as a “code.” Finally, we discuss major questions regarding the function of the newly discovered CTD modifications and the fundamental insights into transcription regulation that will necessarily emerge upon addressing those challenges.
1. Introduction
The transcription of DNA to RNA in eukaryotes is catalyzed by three structurally related RNA polymerases, with each acting on a different class of genes [1]. RNA polymerase I synthesizes most of the ribosomal RNA (rRNA) subunits while RNA polymerase III synthesizes tRNAs, 5S rRNA, and other small RNAs [2–4]. These two polymerases account for 75% and 15% of transcription in the cell, respectively [5]. However, the most studied polymerase is RNA Polymerase II (Pol II), which is responsible for the transcription of protein-coding genes, small nuclear RNA (snRNA), and small nucleolar RNA (snoRNA) [6–8]. In higher eukaryotes, Pol II generates long noncoding RNA (lncRNA) and microRNA (miRNA) [9, 10]. Pol II also transcribes cryptic unstable transcripts (CUTs) and stable unannotated transcripts (SUTs), which are degraded after synthesis [11–13]. The suppression of CUTs is important to prevent inappropriate transcription within ORFs, to enhance processivity during transcription elongation, and to prevent gene silencing via histone deacetylation [14–18].
Of the twelve Pol II subunits, five are common between the three polymerases [1, 19–21]. It is believed that the specific functions attributed to each polymerase arise from the combined action of remaining nonidentical subunits and other factors that associate with them. An especially unique feature of Pol II is the carboxy-terminal domain (CTD) of its large subunit Rpb1 (Figure 1(a)). The CTD serves as the primary point of contact for a wide variety of molecular machines involved in RNA biogenesis during the transcription cycle (reviewed in [8, 22–32]). This domain consists of a highly conserved heptapeptide repeat: Y1S2P3T4S5P6S7 [33–36]. The number of times this sequence is repeated varies among eukaryotic organisms, ranging from 15 repeats in amoeba, to 26 repeats in the budding yeast Saccharomyces cerevisiae, to 52 repeats in humans. When fully extended, the yeast CTD can span a distance of up to 650 Å, over 4 times the diameter of the core polymerase (Figure 1(b)) [24, 34, 35]. The ability of this repetitive sequence to interact with a wide range of nuclear factors stems from the dynamic plasticity of its structure and the diversity of binding surfaces generated by the multitude of post-translational modifications it can accommodate. Tyrosine, threonine, and three serines can all be phosphorylated, the threonine and serine can be glycosylated, and the prolines can undergo isomerization (Figure 1(c)) [27, 37, 38]. In humans, CTD repeats further away from core Pol II bear noncanonical repeats that can be methylated [39]. Taken together, at least 1059 unique modification patterns can occur on the CTD. The combinatorial nature of these modifications, which is reminiscent of the histone code, led to the hypothesis of a CTD code, where the patterns of modifications are read by the transcriptional machinery and these patterns dictate the association or disassociation of complexes [40, 41]. To date, much effort has been made towards characterizing these modifications and understanding the interactions between the CTD and components of various protein machines that play a role in RNA biogenesis. Our current knowledge of the integration of these events by Pol II CTD is summarized in Figure 2, and the known yeast CTD-interacting factors are displayed in Table 1. The focus of this paper is to highlight the recent advances in our understanding of the role of CTD in the early stages of the Pol II transcription cycle, expand on the concept of the CTD code hypothesis, and address the current questions and challenges within the field.
Figure 1.
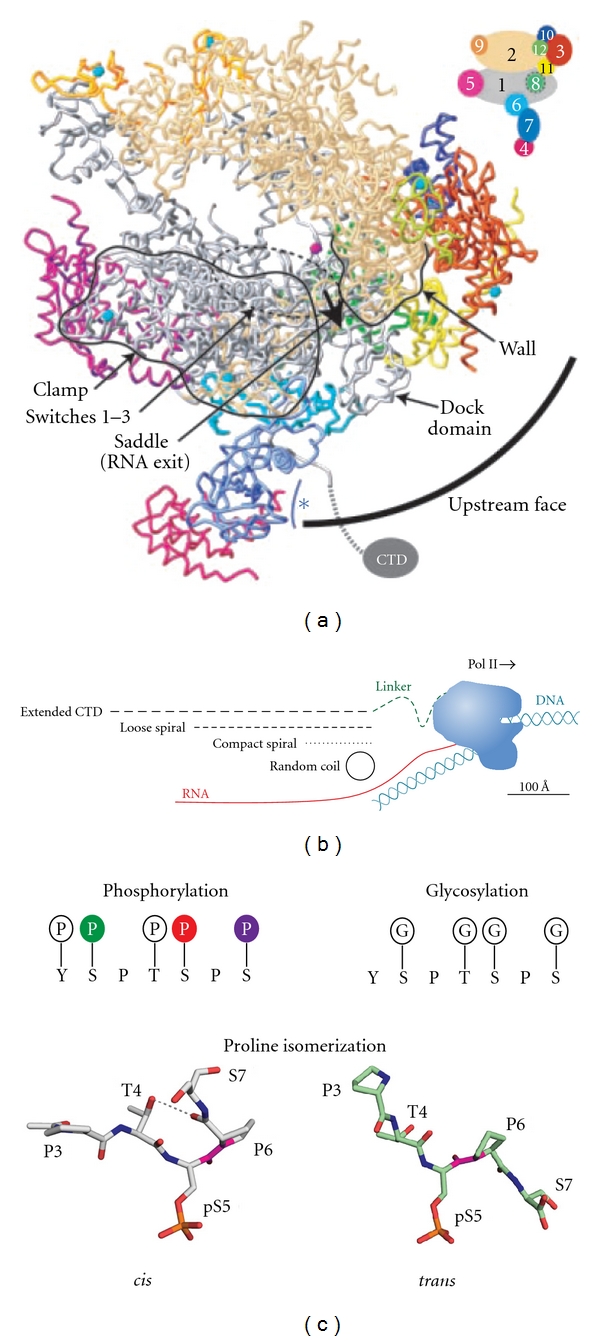
RNA polymerase II structure. (a) Side view of the core Pol II crystal structure containing all twelve subunits and displaying the RNA exit channel (bold arrow) and the positioning of the CTD adapted from Armache et al. [71]. Cartoon in the upper right displays the color coding for the Pol II subunits used in the crystal structure. (b) Illustration of the relative length(s) between the CTD in various conformations and the core Pol II adapted from Meinhart et al. [72]. RNA positioning (red) upon exit of the Pol II and the positioning of the DNA template (blue) upstream and downstream of the core Pol II are also displayed. (c) Known modifications possible on the Pol II CTD are displayed. Glycosylation and phosphorylation are mutually exclusive modifications. Structural images of a heptad repeat in the cis- and trans-conformation are also shown [73–75]. G: β-O-linked N-acetylglucosamine [76]; P: O-linked phosphate.
Figure 2.
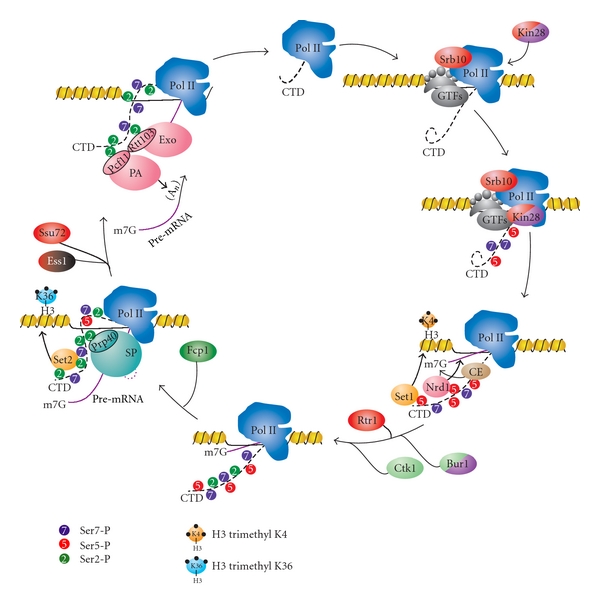
The primary components of the RNA biogenesis machinery and their interactions with the RNA polymerase II C-terminal domain (CTD). Briefly, hypophosphorylated Pol II assembles at the preinitiation complex (PIC) with the Mediator and general transcription factors (GTFs), with TFIIH associating last. The TFIIH-associated kinase Kin28 phosphorylates Ser5 (shown in red) and Ser7 (shown in purple) on the CTD. Mediator-associated kinase Srb10 also contributes to the phosphorylation of Ser5-P. This mark enables promoter release and mediates interactions with the capping enzyme (CE) complex, Nrd1 component of termination machinery, and Set1 histone methyltransferase, which places trimethyl marks on histone H3K4. The Ser5-P mark also facilitates recruitment of Bur1 kinase. Bur1 places initial Ser2-P marks, which facilitate recruitment of Ctk1 kinase, and continues to replenish Ser7-P marks during elongation. Ctk1 is the primary Ser2 kinase, and its phosphorylation recruits splicing machinery (SP) through Prp40, as well as Set2 histone methyltransferase, which places di- and trimethyl marks on histone H3K36. Cleavage and polyadenylation (PA) machinery are recruited through many factors associating with the CTD. One of the factors, Pcf11, binds cooperatively to Ser2-P with Rtt103. The exonuclease complex (Exo) is also recruited through interaction between CTD and Rtt103 and through cooperative interaction between Rtt103 and Pcf11. Finally, the hypophosphorylated CTD is regenerated through three CTD phosphatases. Ser2-P is removed by the phosphatase Fcp1, while two phosphatases, Rtr1 and Ssu72, combine to remove Ser5-P marks during elongation and at termination, respectively. Upon de-phosphorylation, Pol II is released with the assistance of a mechanism involving Pcf11 and can begin another cycle of transcription.
Table 1.
Proteins known to bind RNA polymerase II C-terminal domain in S. cerevisiae.
| Protein/complex | Role in RNA biogenesis | Phospho-CTD bound | References |
|---|---|---|---|
| TFIIE | Preinitiation complex | Hypophosphorylated CTD | [63, 77] |
| TFIIF | Preinitiation complex | Hypophosphorylated CTD | [77] |
| TBP | Preinitiation complex (TFIID) | Hypophosphorylated CTD | [78] |
| Mediator Complex | Transcription activation/repression | Hypophosphorylated CTD | [48, 79] |
| Ceg1 | Capping | Ser5-P | [80–85] |
| Abd1 | Capping | PCTD | [83] |
| Set1 | Histone methylation | Ser5-P | [86] |
| Rpd3C(Rco1) | Histone deacetylation | Ser2-P + Ser5-P | [87, 88] |
| Spt6 | Histone chaperone | Ser2-P | [89] |
| Nrd1 | Transcription termination/processing | Ser5-P | [90] |
| Sen1 | Transcription termination/processing | Unknown | [91] |
| Asr1 | Pol II ubiquitylation (Rpb4/7 Ejection) | Ser5-P | [92] |
| Ess1 | Proline isomerase | Ser2-P | [93, 94] |
| Set2 | Histone methylation | Ser2-P + Ser5-P | [95, 96] |
| Prp40 | Splicing | PCTD | [97] |
| Npl3 | Promotes elongation/prevents polyadenylation | Ser2-P | [98] |
| Pcf11 | Cleavage/polyadenylation (CF1A) | Ser2-P | [99, 100] |
| Rna14 | Cleavage/polyadenylation (CF1A) | PCTD | [101] |
| Rna15 | Cleavage/polyadenylation (CF1A) | PCTD | [101] |
| Ydh1 | Cleavage/polyadenylation (CPF) | PCTD | [102] |
| Yhh1 | Cleavage/polyadenylation (CPF) | PCTD | [103] |
| Pta1 | Cleavage/polyadenylation (CPF) | Ser5-P | [104] |
| Rtt103 | 5′-3′ Exonuclease (Rat1) | Ser2-P | [105] |
| Sus1 | mRNA export | Ser5-P | [106] |
| Yra1 | mRNA export | Hyperphosphorylated CTD | [107] |
| Rsp5 | Pol II ubiquitylation (DNA damage response) | Ser2-P | [108, 109] |
| Hrr25 | DNA damage repair | PCTD | [24, 110] |
CTD-interacting proteins, the processes they are involved in, the phosphorylation state of the CTD with which they associate, and where in the literature the interaction is documented. Ser2-P refers to phosphorylated serine 2, Ser5-P refers to phosphorylated serine 5, and PCTD refers to a mixed phosphorylation state generated by in vitro phosphorylation of a CTD peptide with cell extracts. Additional protein-CTD interactions are described [110] but have not been directly tested.
1.1. RNA Pol II Transcription Cycle
1.1.1. Transcription Initiation
Initiation of transcription begins with the recruitment of gene-specific transcription factors (TFs), general transcription factors (GTFs), the Mediator complex, and Pol II. These factors self-assemble into a pre-initiation complex (PIC) at the promoters of Pol II-transcribed genes [29, 32]. Recognition of the promoter is only partially understood, but it is believed to occur via the recognition of the various cis-elements in the promoter region, such as the TATA box. Binding generally occurs within upstream nucleosome-free regions—the DNA centered over promoters flanked by well-positioned nucleosomes [42–45]. There are two main models for how these factors assemble at this region: the sequential model and the holoenzyme model (Figure 3). In both models, TFs first bind at the upstream activating/repressing sequences (UAS/URS) and recruit the transcriptional machinery. In the sequential model, TBP/TFIID/SAGA assembly at the promoter is accompanied by TFIIA, followed by TFIIB [46, 47]. Then, the Mediator complex arrives, connecting the PIC to transcription factors assembled at the UAS/URS [48–51]. This massive complex consists of three large modules known as the head, middle, and tail and an additional kinase module containing a cyclin-dependent kinase (Srb10 in yeast, Cdk8 in metazoans) [52–57]. The Mediator complex is important for basal transcription and plays a central role in facilitating communication between transcription factors bound to regulatory elements and the PIC [49–51, 56–60]. However, there are studies that suggest the Mediator is not present at most genes, and it only associates with a few UAS/URS in an activator- and stress-specific manner [61, 62]. Pol II is then recruited, followed by the last GTF, TFIIH, which is brought to the PIC by TFIIE [63]. It is possible that several pathways of ordered recruitment exist for GTFs. Other components, including Pol II, TFIIE, and TFIIH, may be recruited via interactions with the Mediator [64]. The holoenzyme model originated from the observation that Srb proteins, which are components of the Mediator, are tightly associated with core Pol II in the absence of DNA [65]. In this model, Pol II is associated with the Mediator and other general transcription factors as a massive holoenzyme supercomplex that is recruited immediately after TBP binds [66–68]. These complexes have been identified in yeast and mammalian systems [69]. Importantly, Pol II is fully able to activate transcription upon arrival in this state [68, 70].
Figure 3.
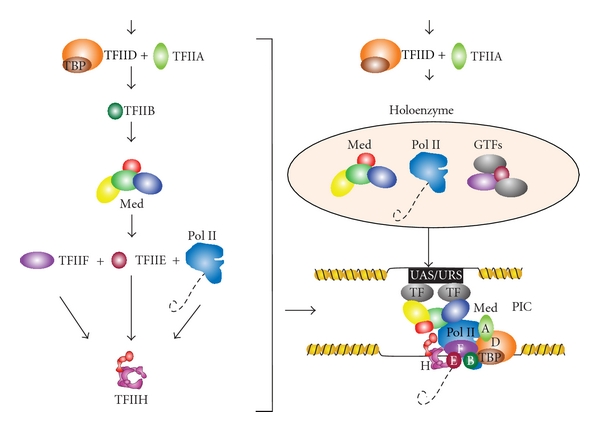
Recruitment and composition of PIC components. Sequential recruitment of the Mediator complex, GTFs, and Pol II (left) or the recruitment of the Pol II holoenzyme (top right), which assembles the pre-initiation complex (PIC) at promoters (bottom right).
Two complexes of the PIC, TFIIH and the Mediator, contain important kinases that phosphorylate the CTD. TFIIH is a ten-subunit complex containing two helicases, an ATPase, a ubiquitin ligase, a neddylation regulator, and a cyclin-dependent kinase (Kin28 in yeast, Cdk7 in metazoans) [111–118]. Both Kin28/Cdk7 and Srb10/Cdk8 have been shown to phosphorylate Ser5 (Ser5-P) in vivo, with Kin28/Cdk7 being the dominant kinase [24, 113, 119–124]. The 5′ enriched Ser5-P mark has been linked to a variety of chromatin-modifying and RNA processing events.
1.1.2. Transcription Elongation
Phosphorylation of Ser5 is involved in coordinating the placement of several key posttranslational modifications on chromatin that constitute the histone code [41] (reviewed in [125–127]). The structural properties of chromatin, such as the +1 nucleosome that resides immediately after gene promoters, are thought to provide a significant physical barrier to transcription. This barrier is weakened or removed through the combined action of posttranslational modifications on the flexible histone tails and chromatin remodeling complexes [127]. In this context, the Ser5-P mark recruits the yeast histone methyltransferase Set1. Trimethylation of histone H3K4 by Set1 and subsequent trimethylation of H3K79 by Dot1 are frequently associated with active transcription and have a reciprocal effect on H3K14 acetylation by SAGA and NuA3 [28, 86, 128, 129]. Ser5-P also recruits the histone deacetylase complexes Set3 and Rpd3C(S) [87], which are important for suppressing CUT initiation at promoters [87, 88].
An especially important role of Ser5-P is the recruitment of the capping enzyme complex. The capping complex places the m7G cap on the nascent transcript as it exits the core polymerase, stabilizing the mRNA by preventing its degradation by 5′-3′ exonucleases. The CTD repeats proximal to the core Pol II are ideally placed near the RNA exit tunnel to facilitate this capping reaction [130, 131]. The guanylyltransferase (Ceg1 in S. cerevisiae) and possibly the methyltransferase (Abd1 in cerevisiae) directly interact with both the Ser5-P and the core polymerase [80–85, 132, 133]. Although the recognition of the CTD is structurally different between yeast and mammalian capping enzymes, both complexes require Ser5-P for binding [81, 131]. A parallel line of experiments showed that inhibition of Kin28 kinase activity using a small-molecule inhibitor leads to a severe reduction in Ser5-P and 5′-capping of transcripts at gene promoters [134, 135]. In agreement with this, tethering the mammalian capping enzyme to the CTD rescues the null Ser5 to alanine mutants in the fission yeast Schizosaccharomyces pombe [136]. Interestingly, inactivation of Kin28 does not eliminate transcription: neither steady-state mRNA levels nor the ability to initiate transcription at the inducible GAL1 gene is significantly compromised by the inhibition [135]. A subsequent study using the same chemical inhibition system confirmed the earlier observations but incorrectly attributed small differences in transcript levels to inappropriate normalization of earlier microarray data [137]. No such global normalization was performed by Kanin et al. [135] and it is unclear why the subsequent study [137] made the unsubstantiated and erroneous claim that the data was treated incorrectly. Kanin et al. were quite cognizant of the consequences of inhibiting an enzyme that could have a role in global transcription. Moreover, quantitative PCR and northern blot assays, experiments that were not reliant on microarray normalization, showed little difference in expression (Hein and Ansari, 2007, unpublished data) [135]. These results strongly support the conclusion that inactivating Kin28 does not significantly impact global transcription. It is important to note that these studies only focused on chemical inhibition of Kin28 and that the inhibition is not an “all or none” phenomenon due to equilibrium binding of the small molecule to the kinase; it is possible that extremely low levels of Ser5 phosphorylation, by either Srb10 or residual Kin28, suffice for transcription initiation. Importantly, chemical inhibition of both Kin28 and Srb10 shows a drop in Pol II across the ORF, supporting the model where Ser5-P may help in promoter clearance [138].
We and others have recently demonstrated that Kin28/Cdk7 is also the primary kinase that phosphorylates Ser7 (Ser7-P) [139–141]. The phosphorylation occurs at protein-coding and noncoding genes and seems to be Mediator dependent [142]. Cyclin-dependent kinases are thought to prefer a substrate bearing Ser-Pro rather than Ser-Tyr dipeptides [143]. Additionally, while Kin28 has been localized to promoters [83], Ser7-P marks were thought to be found only at non-coding genes and at the 3′ end of protein coding genes [144, 145]. The role of Ser7-P at promoters remains an active area of investigation.
Following promoter clearance, transcription initiation factors are exchanged for transcription elongation factors required for RNA processing, passage through chromatin, and suppressing cryptic transcripts. In budding yeast, this exchange occurs immediately after the +1 nucleosome [146]. The association of these elongation factors, which include Paf1, Spt16, Spt4, Spt5, Spt6, Spn1, and Elf1, occurs concurrently on all Pol II genes and is independent of gene length, type, or expression [146]. The recruitment of these factors is essential for transcription processivity (Spt4/5) [147–149], histone regulation (Spt6/16, Spn1, Elf1) [150–156], and gene activation/3′ processing (Paf1) [157]. Similarly, mammalian P-TEFb complex is recruited to Pol II at this stage of transcription [158–161]. This complex contains a cyclin-dependent kinase (Cdk9) that phosphorylates the DRB-sensitivity-inducing factor (DSIF), which allows Pol II to overcome the promoter-proximal pausing induced by the negative elongation factor (NELF) complex [23, 159]. It is unclear if promoter-proximal pausing occurs in yeast, but it is known that Bur1 (the yeast homolog of Cdk9) promotes elongation through post-translational modification of Spt5 (DSIF) (Figure 4(a)) [162]. Bur1 also improves transcription elongation through the recruitment of histone-modifying enzymes and the phosphorylation of CTD. Bur1 activity promotes the ubiquitylation of H2BK123 by the ubiquitin conjugating enzyme Rad6 and Bre1 [129, 163]. H2BK123Ub promotes Set1 trimethylation of histone H3K4 and subsequent trimethylation of H3K79, both of which are important for transcription activation [28, 86, 128, 129]. Bur1 also promotes transcription elongation by coupling promoter-proximal CTD modifications with promoter-distal marks. Bur1 is recruited to the transcription complex by the Ser5-P marks placed at the promoter. It then phosphorylates Ser2 (Ser2-P), priming the CTD for the recruitment of Ctk1 (Cdk12), the major Ser2 kinase [164]. Initial CTD phosphorylation also increases the activity of Ctk1, thereby coupling sequential CTD modifications (Figure 4(b)) [23, 159, 165, 166]. Interestingly, Bur1 travels with Pol II and phosphorylates Ser7-P. Although the exact role of this modification is unclear, it is likely a mark that promotes elongation, as genes with uniformly high levels of Ser7-P are transcribed at significantly higher levels [138].
Figure 4.
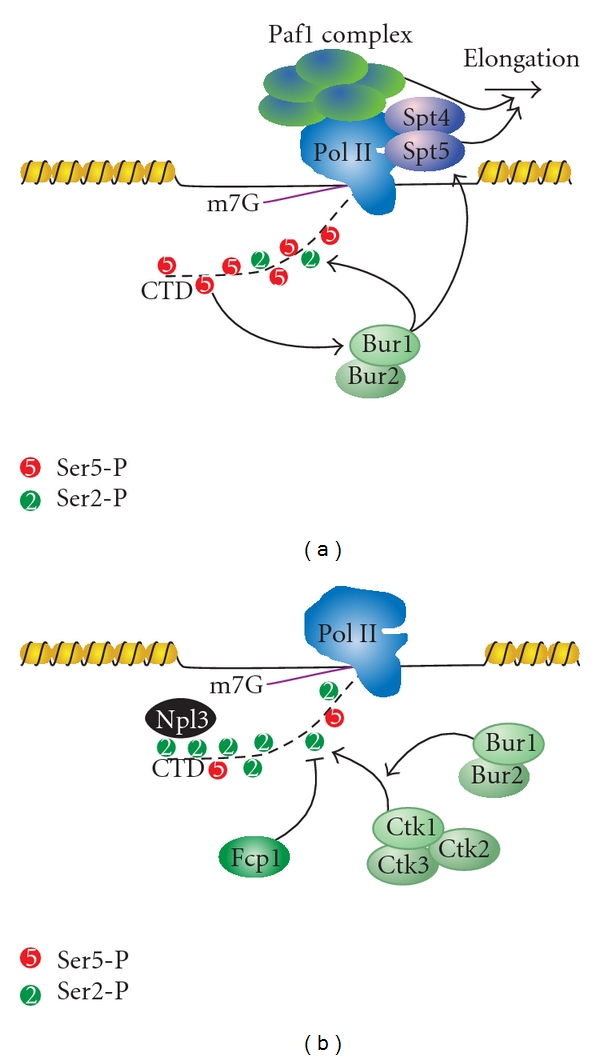
Bur1 phosphorylation of the CTD facilitates the transition from initiation to elongation. (a) Ser5-P enhances recruitment and subsequent phosphorylation of Ser2 by Bur1. Bur1 also phosphorylates Spt5, which acts with the Paf1 complex to promote elongation. (b) CTD phosphorylation by Bur1 enhances the activity of Ctk1 on Ser2. The majority of the Ser2-P is maintained by competition between phosphorylation by Ctk1 and dephosphorylation by Fcp1. This increase in Ser2-P facilitates recruitment of many Ser2-P-binding proteins, such as Npl3.
Most Ser5-P marks are removed near the +1 nucleosome through the action of the newly characterized CTD phosphatase Rtr1 [167]. This phosphatase has been shown to specifically remove Ser5-P marks immediately after promoter clearance. The Ser2-P phosphatase Fcp1 also associates during elongation, but Ser2-P levels remain high across the transcript due to the opposing action of the Ser2-P kinase Ctk1 [168, 169]. It is thought that the Ubp8 component of SAGA travels with Pol II and promotes deubiquitylation of H2BK123Ub [170], which allows the association of Ctk1 and subsequent phosphorylation of Ser2 on the CTD [171].
Ser2-P is critically important for the interaction between the CTD and many histone modifying and RNA processing machines [75, 83, 132, 172–178]. Increasing levels of Ser2-P, in combination with the residual Ser5-P, promote the recruitment of the Set2 methyltransferase, which catalyzes the formation of H3K36me2 and H3K36me3 [95, 96, 179–181]. This leads to the recruitment of the histone deacetylase complex Rpd3C(S) and the removal of acetylation from histones H3 and H4, thereby resetting the transcription state of the nucleosomes and repressing cryptic transcription within ORFs [87, 182, 183]. Ser2-P is involved in the co-transcriptional and posttranscriptional processing of RNA. Cotranscriptional processing of introns via splicing involves the yeast protein Prp40, which preferentially associates with Ser2-P/Ser5-P marked CTD [97]. Ser2-P is also bound by the SR-like (serine/arginine rich) protein Npl3, which functions in elongation, 3′-end processing, hnRNP formation, and mRNA export [184–187]. Finally, increasing levels of Ser2-P, coupled with depletion of Ser5-P, leads to the recruitment of the termination and polyadenylation machinery (discussed below).
1.1.3. Transcription Termination
The role of CTD modifications in orchestrating transcription termination is better described in recent reviews [31, 188]. In essence, two models have been proposed to explain how Pol II termination occurs, with the emerging view being that it is likely a combination of the two models that best describes the mechanism. The first model, known as the “allosteric” or “antiterminator” model, proposes that transcription through the polyadenylation site leads to an exchange of elongation factors for termination factors, resulting in a conformational change of the elongation complex. Indeed, this model is supported by chromatin immunoprecipitation (ChIP) data of elongation factor exchange at the 3′ end of genes [146, 189]. The second model, known as the “torpedo” model, postulates that cleavage of the transcript at the cleavage and polyadenylation site (CPS) creates an entry site for the 5′-3′ exonuclease Rat1 (Xrn2 in mammals), which degrades the 3′ RNA and promotes Pol II release by “torpedoing” the complex [189–191]. In this model, recruitment of Rat1 is likely to be indirect, possibly through its partner Rtt103. Rtt103 has been shown to bind Ser2-P in a cooperative manner with Pcf11 [192], an essential component of the cleavage factor IA (CFIA) complex that also promotes Pol II release [193]. Interestingly, ChIP data shows Pcf11 at both protein-coding and noncoding genes, and mutating Pcf11 results in terminator read-through due to inefficient cleavage at both gene classes [75, 174, 193–196]. Pcf11 may play an important role in both the termination and processing of protein-coding and non-coding genes.
Processing of Pol II transcripts occurs via one of two distinct, gene class-specific pathways in yeast. Many small mRNAs (<550 bp), CUTs, snRNA, and snoRNAs (non-coding genes) are processed via the Nrd1-Nab3 pathway (Figure 5), while longer mRNAs (protein-coding genes) are processed in a polyadenylation-dependent process (Figure 6) [8, 11, 12, 27, 31, 178, 195, 197–199]. The decision to proceed down a certain processing path is modulated by the phosphorylation state of the CTD. Nrd1 preferentially associates with Ser5-P, and its recruitment is also enhanced via histone H3K4 trimethylation by Set1 [90, 200]. Nrd1 and Nab3 scan the nascent RNA for specific sequence elements (GUAA/G or UGGA for Nrd1, and UCUU or CUUG for Nab3) as it exits the core polymerase [90, 199, 201–207]. The helicase Sen1 (senataxin in humans), which exists in complex with Nrd1 and Nab3, resolves the DNA:RNA hybrids known as R-loops that form between the template DNA and the nascent RNA, keeping the specific sequence elements exposed and preserving genomic stability [208–210]. The involvement of Sen1 is dependent on the phosphatase Glc7, which dephosphorylates Sen1 and is essential for the proper termination of snRNA and snoRNA transcripts [211]. Upon detecting its consensus sequence elements, the Nrd1 complex and the Rnt1 endonuclease cleave these short transcripts [195, 212–214], which are then trimmed at the 3′ end by the TRAMP complex and the exosome [6, 215–217]. Nrd1 then disengages from the transcription complex, with help from antagonizing Ser2-P marks [198]. Unlike snRNA/snoRNAs, which have protective structural elements in the RNA, Nrd1-terminated CUTs have no protective elements at their 3′ ends and are thus fully degraded by TRAMP after cleavage [8, 11, 12]. Nrd1 has been mapped to the 5′ end of transcribed regions, but a recent study has demonstrated that Nrd1 occupancy is maintained across the open reading frame of genes [196]. Although no homolog of Nrd1 has been found in mammalian cells, the Integrator complex that is involved in 3′ processing of snRNA transcripts is recruited by Ser7-P [218]. The association of this complex with Ser7-P CTD was demonstrated by the abolishment of this interaction upon mutation of Ser7 to alanine [145]. Subsequent analysis using a panel of CTD peptides determined that the Integrator prefers to bind a diphosphorylated CTD substrate spanning two heptad repeats in the S7-P-S2-P conformation [219]. It is possible that Ser7-P may serve as a similar scaffold for snRNA and snoRNA processing machinery in yeast.
Figure 5.
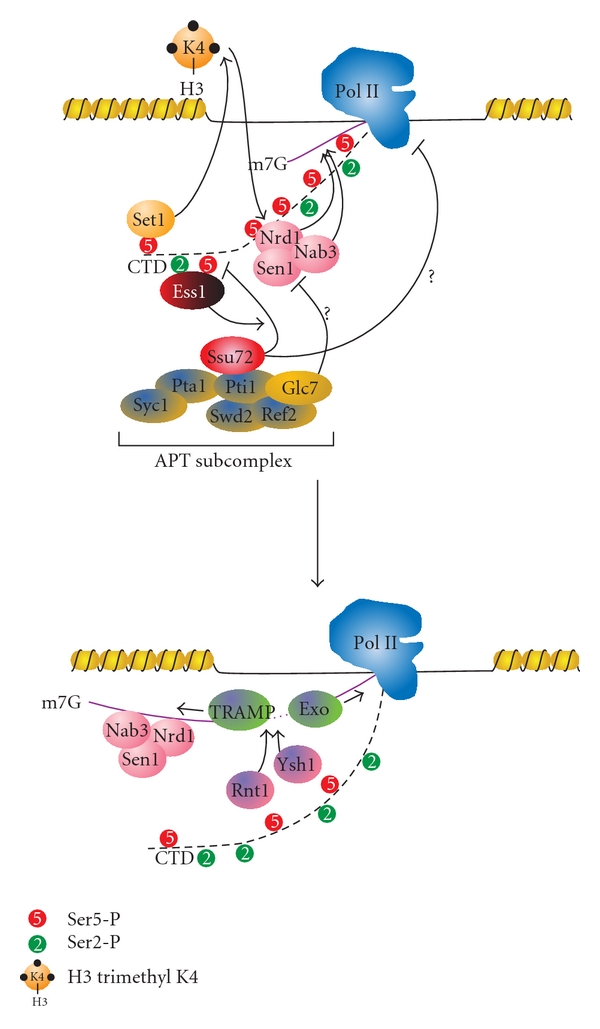
Nrd1-dependent termination pathway. The Nrd1-Nab3-Sen1 complex is recruited via interaction between Nrd1 and Ser5-P. This recruitment is facilitated by H3K4me3, which is placed by the Set1 histone methyltransferase. The mechanisms by which the Ssu72 and Glc7 phosphatases promote termination are still unclear, but it may be that the dephosphorylation of Sen1 by Glc7 and of the CTD by Ssu72 causes the polymerase to pause, and allowing the termination machinery to associate. During elongation, both Nrd1 and Nab3 scan the nascent RNA for their preferred sequences (see text for details). Upon finding their concensus sequences, Nrd1-Nab3-Sen1 complex is able to be associated with the RNA. The endonucleases Rnt1 and Ysh1 may contribute to the cleavage of the RNA, which is followed by 3′-5′ trimming the transcript by the TRAMP complex and by the degradation of the remaining RNA exiting Pol II by the 5′-3′ exonuclease Rat1 (Exo).
Figure 6.
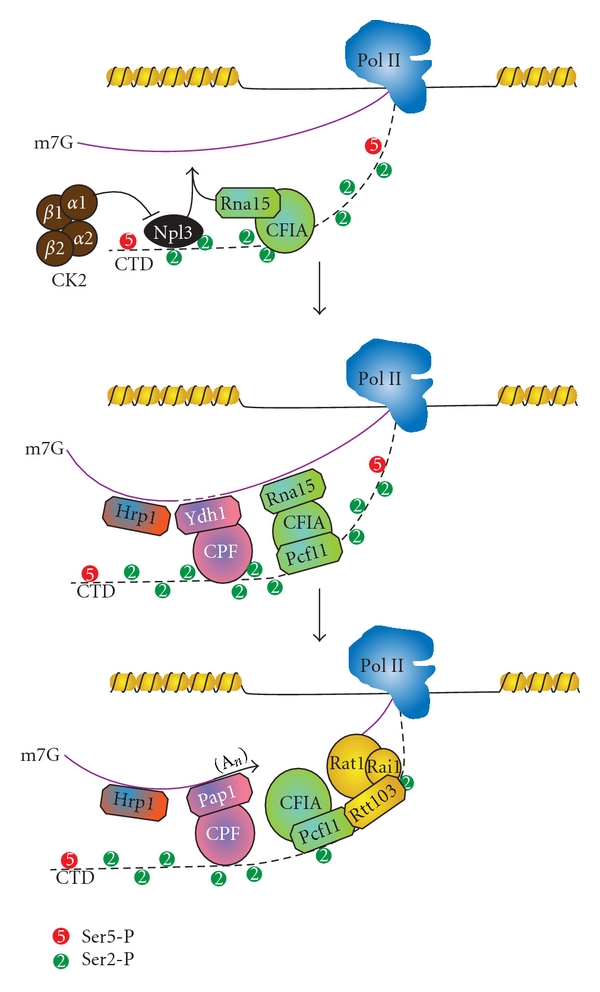
The mRNA termination pathway. Rna15 competes with Npl3 for binding to the nascent RNA. CK2 phosphorylates Npl3, allowing Rna15 to find its preferred binding site (an A/U-rich region) in the RNA. The CPF and CFIA components assemble through interactions with the CTD and the Yth1 component of CPF cleaves the nascent RNA at the polyadenylation site, followed by polyadenylation by Pap1. Then the Rat1 exonuclease complex associates via cooperative interaction between Pcf11 and Rtt103 and leads to termination and dissociation of Pol II.
The second pathway, used for the processing of most mRNA transcripts, involves the cleavage and polyadenylation factor (CPF) complex, cleavage factor IA and IB (CFIA and CFIB) complexes, and the exosome (Figure 6) [31, 195, 197]. Many of the termination and 3′ processing factors involved in this process are known to preferentially associate with Ser2-P or Ser2-P/Ser5-P enriched CTD including: Npl3, Rtt103, Rna14, Rna15, Ydh1, Yhh1, Pta1, and Pcf11. In this pathway, Rna15 competes with Npl3 for recognition of a UA-rich site in the nascent RNA [98, 187]. This competition is removed upon phosphorylation of Npl3 by casein kinase 2 (CK2) [98]. Rna15 can then bind the nascent RNA and promote endonucleolytic cleavage followed by polyadenylation by the polyadenylate polymerase (Pap1) [197, 220]. Polyadenylation-binding proteins (PAB) then protect the mature transcript from exonucleolytic degradation (Figure 6) [221].
In both pathways, the CTD is hypophosphorylated by the combined action of two essential phosphatases at the end of transcription: Ssu72 and Fcp1. Ssu72 is a member of the Associated with Pta1 (APT) complex, which is present at both gene classes and is involved in 3′ processing of non-coding RNAs [222]. As such, Ssu72 is primarily localized at the 3′ end of transcripts [222], although there is one instance in which it has been found at promoters [223]. Temperature-sensitive mutants of Ssu72 exhibit read-through at both protein-coding and non-coding transcripts [224]. Ssu72 is the primary Ser5-P phosphatase [225], and its phosphatase activity is enhanced by the prolyl isomerase Ess1/Pin1 and by interacting with Pta1/symplekin [226–228]. Recently, crystal structures have shed light on the mechanism of Ssu72: the phosphatase binds to Ser5-P only when the adjacent Pro6 is in the cis-conformation [73, 74]. In contrast to Ssu72, Fcp1 associates with TFIIF during transcription and is found across the entire transcribed region [168, 169, 229, 230]. Although it has Ser5-P and Ser2-P phosphatase activity in vitro, Fcp1 is considered a Ser2-P-specific phosphatase in vivo [231, 232]. Fcp1 activity is enhanced upon phosphorylation of Fcp1 by CK2 [233]. Defects in Fcp1 also result in transcription read-through at Nrd1-dependent transcripts [198]. Though it is unclear which phosphatase removes Ser7-P, new data from our lab suggest that Ssu72 may be the phosphatase that removes Ser7-P at both the 5′ and 3′ ends of genes [234]. Removal of this mark may be even more important than its placement as mutation of Ser7 to alanine slows growth while mutating Ser7 to the phosphomimic glutamate is lethal [144].
Global dephosphorylation of the CTD facilitates the release of Pol II from DNA, which can then recycle to promoters for the next cycle of transcription [224, 235, 236]. It has been proposed that transcription termination and subsequent dephosphorylation of the CTD is coupled to transcription reinitiation through gene looping, by which the promoter and terminator regions are brought together, allowing Pol II to associate more rapidly with the PIC [237, 238]. Intriguingly, Ssu72 and the GTF TFIIB have been shown to be essential in gene looping [223, 239]. Taken together, the phosphorylation and dephosphorylation of the CTD is intimately involved in every phase of transcription, from initiation, to elongation, to termination, and possibly reinitiation.
1.1.4. Other Regulatory Roles of the CTD
In addition to its many roles in transcription initiation, elongation, and termination, the CTD has been implicated in a variety of transcription-extrinsic processes, such as mRNA export and stress response. mRNA export (reviewed in [240–242]) requires the packaging of the mRNA into export-competent messenger ribonucleoprotein (mRNP) via association with the Mex67:Mtr2 heterodimer [243]. This heterodimer is brought to the mRNA by Yra1 and Sub2, components of the THO subunit of the TREX1 complex [242]. The process of mRNP export is coordinated by the protein Sus1. This central protein directly interacts with Ser5-P and Ser2-P/Ser5-P of the CTD, Ub8 subunit of the SAGA complex, Yra1 subunit of the TREX1 complex, and Sac3 subunit of the TREX2 complex at the nuclear pore (Figure 7) [106, 244].
Figure 7.
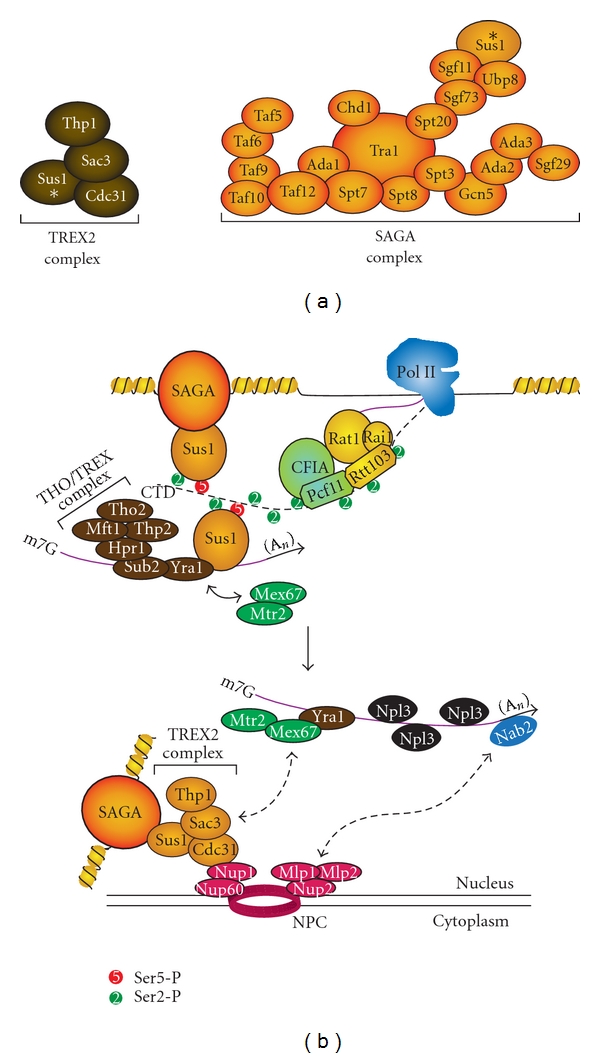
Sus1 in TREX2 and SAGA complexes coordinates mRNA export. (a) Subunit compositions of TREX2 and SAGA complexes are shown, highlighting Sus1 (asterisk). (b) mRNA export coordinated by Sus1. Sus1 binds Ser2-P and Ser2-P/Ser5-P CTD, connecting the CTD to the SAGA histone acetyltransferase complex. Sus1 also interacts with Yra1 component of the THO/TREX complex on the RNA. Mex67-Mtr2 are recruited by interaction with Yra1 and help form the export-competent mRNP. At the nuclear pore complex (NPC), Mlp1-Mlp2 interact with the polyA mRNA-binding protein Nab2 and Mex67 interacts with Sac3 of the TREX2 complex. This interaction brings the export-competent mRNP to the NPC in preparation for export to the cytoplasm. Sus1 is a component of both TREX2 and SAGA and serves to tether actively transcribed gene promoters to the NPC.
The CTD is also involved in stress response. The ubiquitin ligase Rsp5 binds the CTD and ubiquitylates Pol II in response to DNA damage [245, 246]. Similarly, UV-induced DNA damage in mammalian fibroblasts results in hyperphosphorylation of the CTD by the mammalian positive transcription elongation factor b (P-TEFb), which then regulates Pol II ubiquitylation and subsequent degradation [247]. Under conditions not well understood, Ser5-P can also recruit the Asr1 ubiquitin ligase, which ubiquitylates the Rpb1 and Rpb2 subunits of Pol II. This ubiquitylation promotes ejection of the Rpb4/7 heterodimer from the core polymerase and inactivates Pol II, which may provide a mechanism for stopping polymerases engaged in abortive or cryptic transcription [92].
2. The CTD Code Controversy: Is It a Code?
The concept of the CTD code was first proposed due to the enormous amount of information that can be encoded via post-translational modification of the CTD repeats [40, 248]. The code would coordinate the assembly of complexes that “read, write, and erase” the code during transcription. Historically, the Ser5-P and Ser2-P marks have been the best characterized, with the canonical distribution of Ser5-P being enriched at the 5′ end of genes and Ser2-P enriched towards the 3′ end. Recently, our lab and several others have been able to map the phospho-CTD occupancy profiles across the yeast genome [135, 138, 139, 146, 196]. There are interesting discrepancies between the observations made by various groups. For example, Mayer et al. find the canonical profile to be present at every gene with Ser7-P profiles overlapping with Ser5-P [146], while we find clusters of genes with noncanonical CTD profiles for Ser2-P, Ser5-P, and Ser7-P [138]. We observe gene-specific phosphorylation profiles, with Ser2-P levels being significantly lower at non-coding genes and Ser7-P profiles diverging from Ser5-P profiles only at protein-coding genes. The distinct patterns of CTD marks at these two gene classes reflect the different mechanisms of transcription termination and 3′ end processing machinery that act on these two classes of RNA. Similarly, Kim et al. also observe differences in phospho-CTD profiles at snoRNAs and at introns [196]. However, the positions of the Ser5-P and Ser7-P peaks in Kim et al. are offset from Tietjen et al. and Mayer et al. Importantly, all three genome-wide analyses reveal an unexpected degree of cooccurrence of CTD marks, suggesting a bivalent or even multivalent mode of recognition by docking partners. In support of this idea, the Set2 histone methyltransferase and the Integrator complex have been shown to prefer a bivalent mark rather than a single phosphorylated residue [95, 96, 219].
In addition to the various phosphorylation marks, the isomerization state of the CTD also contributes to the complexity of the code. For example, Pcf11 binds the CTD in the trans-conformation while Ssu72 prefers a cis-CTD as substrate [73–75]. Many in the transcription field have made the argument that the CTD code is not a true code because it does not convey biological information via a rigorous decoding key. However, research in the last several years has demonstrated that specific phosphorylation marks and proline isomerization are important for conveying information from cis-elements encountered by Pol II to the protein complexes necessary for successful progression through the transcription cycle. Further investigation into the mechanism of this information transfer will resolve the controversy over the existence of a CTD code.
3. Future Directions
Extraordinarily rapid progress has been made over the last several years in the field of CTD research; however, many important questions remain unanswered. Although the profiles of Ser7-P have been mapped and several of its kinases discovered, its function at protein coding genes remains unclear. Additionally, most of the kinases identified are established members of the transcription initiation or elongation complexes. One could expect to find new enzymes that could modulate the CTD in response to signals, as post-translational modifications are often used as a mechanism for cells to respond to external stimuli. The recent discovery of Ser7-P at elongating Pol II has also prompted the question of whether Tyr1 and Thr4 phosphorylation (Tyr1-P and Thr4-P) occurs? Tyr1 can be phosphorylated by c-Abl in mammals, but no homolog is present in yeast [249]. In addition, both Tyr1-P and Thr4-P has been detected in S. pombe [250]. Interestingly, Tyr1-P and Thr4-P were found in both the hyperphosphorylated and hypophosphorylated states of Pol II, opening the possibility of CTD function independent of transcription. However, neither the profile nor function of these potential modifications have been extensively characterized.
The role of non-canonical residues and their modification states on mammalian CTD remain to be explored. In mammals, the Ser7 residue is only weakly conserved in polymerase-distal repeats of the CTD, often changed to lysine or arginine [144]. Interestingly, Arg1810 of rpb1 in the human CTD is methylated by the coactivator-associated methyltransferase1 (CARM1) [39]. This methylation occurs prior to both transcription initiation and phosphorylation of Ser2 or Ser5, and mutation of this residue results in the improper expression of a variety of snRNAs and snoRNAs. In addition to methylation, the CTD may also be subject to glycosylation. Recent studies suggest O-GlcNAc are transferred to Ser5 and Ser7 by O-GlcNac transferase and removed by O-GlcNAc aminidase during PIC assembly. This cycling of O-GlcNAc may be important for preventing aberrant CTD phosphorylation by TFIIH [251].
Besides the characterization of novel marks, significant structural challenges remain for understanding the known phosphomarks. One limitation of ChIP is its inability to identify the exact phosphorylation patterns across individual CTD repeats in vivo at different points during the transcription cycle. Recent mutational analysis suggests that the minimal functional unit of the CTD consists of three consecutive Ser-Pro dipeptide residues in a S2-S5-S2 configuration [36], but it is unclear if all three serines can be phosphorylated on one functional unit or if phosphorylation alternates between repeats. The lack of positively charged aminoacids makes the phospho-CTD patterns difficult to decipher via mass spectrometry. Additionally, the highly repetitive nature of the CTD makes it difficult to distinguish between the first repeat and the twenty-first. Consequently, the position along the CTD where interacting partners associate remains a mystery. Mutation of Ser2 to glutamate in the core-distal repeats and mutation of Ser5 to glutamate in the core-proximal repeats are lethal [252]. However, this does not directly demonstrate whether the proteins that bind these phosphorylated residues are located at these repeats. Characterizing the phosphorylation patterns and protein occupancies at individual repeats will help determine the existence of a “CTD recognition” code, and this promises to be one of the most exciting and important challenges in the future of CTD research.
Acknowledgments
The authors gratefully acknowledge the support of the US National Science Foundation (MCB 07147), W. M. Keck, Shaw Scholar and Vilas Associate awards (to A. Z. Ansari). D. W. Zhang was supported by the Burris Pre-Doctoral Fellowship. J. R. Tietjen and J. B. Rodríguez-Molina were supported by a US National Human Genome Research Institute training grant to the Genomic Sciences Training Program (5T32HG002760). C. M. Nemec was supported by Chemistry Biology Interface Training Program (5T32GM008505-18).
References
- 1.Cramer P, Armache KJ, Baumli S, et al. Structure of eukaryotic RNA polymerases. Annual Review of Biophysics. 2008;37:337–352. doi: 10.1146/annurev.biophys.37.032807.130008. [DOI] [PubMed] [Google Scholar]
- 2.Grummt I. Life on a planet of its own: regulation of RNA polymerase I transcription in the nucleolus. Genes & Development. 2003;17(14):1691–1702. doi: 10.1101/gad.1098503R. [DOI] [PubMed] [Google Scholar]
- 3.Russell J, Zomerdijk JCBM. RNA-polymerase-I-directed rDNA transcription, life and works. Trends in Biochemical Sciences. 2005;30(2):87–96. doi: 10.1016/j.tibs.2004.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dieci G, Fiorino G, Castelnuovo M, Teichmann M, Pagano A. The expanding RNA polymerase III transcriptome. Trends in Genetics. 2007;23(12):614–622. doi: 10.1016/j.tig.2007.09.001. [DOI] [PubMed] [Google Scholar]
- 5.Werner M, Thuriaux P, Soutourina J. Structure-function analysis of RNA polymerases I and III. Current Opinion in Structural Biology. 2009;19(6):740–745. doi: 10.1016/j.sbi.2009.10.005. [DOI] [PubMed] [Google Scholar]
- 6.Wyers F, Rougemaille M, Badis G, et al. Cryptic Pol II transcripts are degraded by a nuclear quality control pathway involving a new poly(A) polymerase. Cell. 2005;121(5):725–737. doi: 10.1016/j.cell.2005.04.030. [DOI] [PubMed] [Google Scholar]
- 7.Davis CA, Ares M. Accumulation of unstable promoter-associated transcripts upon loss of the nuclear exosome subunit Rrp6p in Saccharomyces cerevisiae. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(9):3262–3267. doi: 10.1073/pnas.0507783103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lykke-Andersen S, Jensen TH. Overlapping pathways dictate termination of RNA polymerase II transcription. Biochimie. 2007;89(10):1177–1182. doi: 10.1016/j.biochi.2007.05.007. [DOI] [PubMed] [Google Scholar]
- 9.Faller M, Guo F. MicroRNA biogenesis: there’s more than one way to skin a cat. Biochimica et Biophysica Acta. 2008;1779(11):663–667. doi: 10.1016/j.bbagrm.2008.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guttman M, Amit I, Garber M, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458(7235):223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Arigo JT, Eyler DE, Carroll KL, Corden JL. Termination of cryptic unstable transcripts is directed by yeast RNA-binding proteins Nrd1 and Nab3. Molecular Cell. 2006;23(6):841–851. doi: 10.1016/j.molcel.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 12.Thiebaut M, Kisseleva-Romanova E, Rougemaille M, Boulay J, Libri D. Transcription termination and nuclear degradation of cryptic unstable transcripts: a role for the nrd1-nab3 pathway in genome surveillance. Molecular Cell. 2006;23(6):853–864. doi: 10.1016/j.molcel.2006.07.029. [DOI] [PubMed] [Google Scholar]
- 13.Ying SY, Lin SL. Intron-mediated RNA interference and microRNA biogenesis. Methods in Molecular Biology. 2009;487:387–413. doi: 10.1007/978-1-60327-547-7_19. [DOI] [PubMed] [Google Scholar]
- 14.Kaplan CD, Laprade L, Winston F. Transcription elongation factors repress transcription initiation from cryptic sites. Science. 2003;301(5636):1096–1099. doi: 10.1126/science.1087374. [DOI] [PubMed] [Google Scholar]
- 15.Camblong J, Iglesias N, Fickentscher C, Dieppois G, Stutz F. Antisense RNA stabilization induces transcriptional gene silencing via histone deacetylation in S. cerevisiae. Cell. 2007;131(4):706–717. doi: 10.1016/j.cell.2007.09.014. [DOI] [PubMed] [Google Scholar]
- 16.De Santa F, Barozzi I, Mietton F, et al. A large fraction of extragenic RNA Pol II transcription sites overlap enhancers. PLoS Biology. 2010;8(5) doi: 10.1371/journal.pbio.1000384. Article ID e1000384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ørom UA, Derrien T, Beringer M, et al. Long noncoding RNAs with enhancer-like function in human cells. Cell. 2010;143(1):46–58. doi: 10.1016/j.cell.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sato F, Tsuchiya S, Meltzer SJ, Shimizu K. MicroRNAs and epigenetics. FEBS Journal. 2011;278(10):1598–1609. doi: 10.1111/j.1742-4658.2011.08089.x. [DOI] [PubMed] [Google Scholar]
- 19.Young RA. RNA polymerase II. Annual Review of Biochemistry. 1991;60:689–715. doi: 10.1146/annurev.bi.60.070191.003353. [DOI] [PubMed] [Google Scholar]
- 20.Woychik N, Young R. Exploring RNA polymerase II structure and function. In: Conaway RC, Conaway JW, editors. Transcription: Mechanisms and Regulation. New York, NY, USA: Raven Press; 1994. pp. 227–242. [Google Scholar]
- 21.Shpakovski GV, Acker J, Wintzerith M, Lacroix JF, Thuriaux P, Vigneron M. Four subunits that are shared by the three classes of RNA polymerase are functionally interchangeable between Homo sapiens and Saccharomyces cerevisiae. Molecular and Cellular Biology. 1995;15(9):4702–4710. doi: 10.1128/mcb.15.9.4702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hirose Y, Manley JL. RNA polymerase II and the integration of nuclear events. Genes & Development. 2000;14(12):1415–1429. [PubMed] [Google Scholar]
- 23.Sims RJ, Belotserkovskaya R, Reinberg D. Elongation by RNA polymerase II: the short and long of it. Genes & Development. 2004;18(20):2437–2468. doi: 10.1101/gad.1235904. [DOI] [PubMed] [Google Scholar]
- 24.Phatnani HP, Greenleaf AL. Phosphorylation and functions of the RNA polymerase II CTD. Genes & Development. 2006;20(21):2922–2936. doi: 10.1101/gad.1477006. [DOI] [PubMed] [Google Scholar]
- 25.Kornberg RD. The molecular basis of eukaryotic transcription. Proceedings of the National Academy of Sciences of the United States of America. 2007;104(32):12955–12961. doi: 10.1073/pnas.0704138104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hirose Y, Ohkuma Y. Phosphorylation of the C-terminal domain of RNA polymerase II plays central roles in the integrated events of eucaryotic gene expression. Journal of Biochemistry. 2007;141(5):601–608. doi: 10.1093/jb/mvm090. [DOI] [PubMed] [Google Scholar]
- 27.Egloff S, Murphy S. Cracking the RNA polymerase II CTD code. Trends in Genetics. 2008;24(6):280–288. doi: 10.1016/j.tig.2008.03.008. [DOI] [PubMed] [Google Scholar]
- 28.Venters BJ, Pugh BF. How eukaryotic genes are transcribed regulation of eukaroytic gene transcription. Critical Reviews in Biochemistry and Molecular Biology. 2009;44(2-3):117–141. doi: 10.1080/10409230902858785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Buratowski S. Progression through the RNA Polymerase II CTD Cycle. Molecular Cell. 2009;36(4):541–546. doi: 10.1016/j.molcel.2009.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Perales R, Bentley D. ‘Cotranscriptionality’: the transcription elongation complex as a nexus for nuclear transactions. Molecular Cell. 2009;36(2):178–191. doi: 10.1016/j.molcel.2009.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Richard P, Manley JL. Transcription termination by nuclear RNA polymerases. Genes & Development. 2009;23(11):1247–1269. doi: 10.1101/gad.1792809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nechaev S, Adelman K. Pol II waiting in the starting gates: regulating the transition from transcription initiation into productive elongation. Biochimica et Biophysica Acta. 2010;1809(1):34–45. doi: 10.1016/j.bbagrm.2010.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Corden JL, Cadena DL, Ahearn JM, Dahmus ME. A unique structure at the carboxyl terminus of the largest subunit of eukaryotic RNA polymerase II. Proceedings of the National Academy of Sciences of the United States of America. 1985;82(23):7934–7938. doi: 10.1073/pnas.82.23.7934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Corden JL. Tails of RNA polymerase II. Trends in Biochemical Sciences. 1990;15(10):383–387. doi: 10.1016/0968-0004(90)90236-5. [DOI] [PubMed] [Google Scholar]
- 35.Chapman RD, Heidemann M, Hintermair C, Eick D. Molecular evolution of the RNA polymerase II CTD. Trends in Genetics. 2008;24(6):289–296. doi: 10.1016/j.tig.2008.03.010. [DOI] [PubMed] [Google Scholar]
- 36.Liu P, Kenney JM, Stiller JW, Greenleaf AL. Genetic organization, length conservation, and evolution of RNA polymerase II carboxyl-terminal domain. Molecular Biology and Evolution. 2010;27(11):2628–2641. doi: 10.1093/molbev/msq151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zeidan Q, Hart GW. The intersections between O-GlcNAcylation and phosphorylation: implications for multiple signaling pathways. Journal of Cell Science. 2010;123(1):13–22. doi: 10.1242/jcs.053678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fuchs SM, Laribee RN, Strahl BD. Protein modifications in transcription elongation. Biochimica et Biophysica Acta. 2009;1789(1):26–36. doi: 10.1016/j.bbagrm.2008.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sims III RJ, Rojas LA, Beck D, et al. The C-terminal domain of RNA polymerase II is modified by site-specific methylation. Science. 2011;332(6025):99–103. doi: 10.1126/science.1202663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Buratowski S. The CTD code. Nature Structural Biology. 2003;10(9):679–680. doi: 10.1038/nsb0903-679. [DOI] [PubMed] [Google Scholar]
- 41.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293(5532):1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 42.Pedersen AG, Baldi P, Chauvin Y, Brunak S. The biology of eukaryotic promoter prediction—a review. Computers and Chemistry. 1999;23(3-4):191–207. doi: 10.1016/s0097-8485(99)00015-7. [DOI] [PubMed] [Google Scholar]
- 43.Basehoar AD, Zanton SJ, Pugh BF. Identification and distinct regulation of yeast TATA box-containing genes. Cell. 2004;116(5):699–709. doi: 10.1016/s0092-8674(04)00205-3. [DOI] [PubMed] [Google Scholar]
- 44.Yuan GC, Liu YJ, Dion MF, et al. Genome-scale identification of nucleosome positions in S. cerevisiae. Science. 2005;309(5734):626–630. doi: 10.1126/science.1112178. [DOI] [PubMed] [Google Scholar]
- 45.Venters BJ, Pugh BF. A canonical promoter organization of the transcription machinery and its regulators in the Saccharomyces genome. Genome Research. 2009;19(3):360–371. doi: 10.1101/gr.084970.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Orphanides G, Lagrange T, Reinberg D. The general transcription factors of RNA polymerase II. Genes & Development. 1996;10(21):2657–2683. doi: 10.1101/gad.10.21.2657. [DOI] [PubMed] [Google Scholar]
- 47.Zanton SJ, Pugh BF. Full and partial genome-wide assembly and disassembly of the yeast transcription machinery in response to heat shock. Genes & Development. 2006;20(16):2250–2265. doi: 10.1101/gad.1437506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Svejstrup JQ, Li Y, Fellows J, Gnatt A, Bjorklund S, Kornberg RD. Evidence for a mediator cycle at the initiation of transcription. Proceedings of the National Academy of Sciences of the United States of America. 1997;94(12):6075–6078. doi: 10.1073/pnas.94.12.6075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Myers LC, Kornberg RD. Mediator of transcriptional regulation. Annual Review of Biochemistry. 2000;69:729–749. doi: 10.1146/annurev.biochem.69.1.729. [DOI] [PubMed] [Google Scholar]
- 50.Kornberg RD. Mediator and the mechanism of transcriptional activation. Trends in Biochemical Sciences. 2005;30(5):235–239. doi: 10.1016/j.tibs.2005.03.011. [DOI] [PubMed] [Google Scholar]
- 51.Malik S, Roeder RG. Dynamic regulation of pol II transcription by the mammalian Mediator complex. Trends in Biochemical Sciences. 2005;30(5):256–263. doi: 10.1016/j.tibs.2005.03.009. [DOI] [PubMed] [Google Scholar]
- 52.Kang JS, Kim SH, Hwang MS, Han SJ, Lee YC, Kim YJ. The structural and functional organization of the yeast mediator complex. Journal of Biological Chemistry. 2001;276(45):42003–42010. doi: 10.1074/jbc.M105961200. [DOI] [PubMed] [Google Scholar]
- 53.Borggrefe T, Davis R, Erdjument-Bromage H, Tempst P, Kornberg RD. A complex of the Srb8, -9, -10, and -11 transcriptional regulatory proteins from yeast. Journal of Biological Chemistry. 2002;277(46):44202–44207. doi: 10.1074/jbc.M207195200. [DOI] [PubMed] [Google Scholar]
- 54.Guglielmi B, van Berkum NL, Klapholz B, et al. A high resolution protein interaction map of the yeast Mediator complex. Nucleic Acids Research. 2004;32(18):5379–5391. doi: 10.1093/nar/gkh878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chadick JZ, Asturias FJ. Structure of eukaryotic Mediator complexes. Trends in Biochemical Sciences. 2005;30(5):264–271. doi: 10.1016/j.tibs.2005.03.001. [DOI] [PubMed] [Google Scholar]
- 56.Casamassimi A, Napoli C. Mediator complexes and eukaryotic transcription regulation: an overview. Biochimie. 2007;89(12):1439–1446. doi: 10.1016/j.biochi.2007.08.002. [DOI] [PubMed] [Google Scholar]
- 57.Toth-Petroczy Á, Oldfield CJ, Simon I, et al. Malleable machines in transcription regulation: the Mediator complex. PLoS Computational Biology. 2008;4(12) doi: 10.1371/journal.pcbi.1000243. Article ID e1000243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Biddick R, Young ET. Yeast Mediator and its role in transcriptional regulation. Comptes Rendus Biologies. 2005;328(9):773–782. doi: 10.1016/j.crvi.2005.03.004. [DOI] [PubMed] [Google Scholar]
- 59.Bjorklund S, Gustafsson CM. The yeast Mediator complex and its regulation. Trends in Biochemical Sciences. 2005;30(5):240–244. doi: 10.1016/j.tibs.2005.03.008. [DOI] [PubMed] [Google Scholar]
- 60.Conaway RC, Sato S, Tomomori-Sato C, Yao T, Conaway JW. The mammalian Mediator complex and its role in transcriptional regulation. Trends in Biochemical Sciences. 2005;30(5):250–255. doi: 10.1016/j.tibs.2005.03.002. [DOI] [PubMed] [Google Scholar]
- 61.Fan X, Chou DM, Struhl K. Activator-specific recruitment of Mediator in vivo. Nature Structural & Molecular Biology. 2006;13(2):117–120. doi: 10.1038/nsmb1049. [DOI] [PubMed] [Google Scholar]
- 62.Fan X, Struhl K. Where does mediator bind in vivo? PLoS One. 2009;4(4) doi: 10.1371/journal.pone.0005029. Article ID e5029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Maxon ME, Goodrich JA, Tjian R. Transcription factor IIE binds preferentially to RNA polymerase IIa and recruits TFIIH: a model for promoter clearance. Genes & Development. 1994;8(5):515–524. doi: 10.1101/gad.8.5.515. [DOI] [PubMed] [Google Scholar]
- 64.Esnault C, Ghavi-Helm Y, Brun S, et al. Mediator-dependent recruitment of TFIIH modules in preinitiation complex. Molecular Cell. 2008;31(3):337–346. doi: 10.1016/j.molcel.2008.06.021. [DOI] [PubMed] [Google Scholar]
- 65.Koleske AJ, Young RA. An RNA polymerase II holoenzyme responsive to activators. Nature. 1994;368(6470):466–469. doi: 10.1038/368466a0. [DOI] [PubMed] [Google Scholar]
- 66.Liao SM, Zhang J, Jeffery DA, et al. A kinase-cyclin pair in the RNA polymerase II holoenzyme. Nature. 1995;374(6518):193–196. doi: 10.1038/374193a0. [DOI] [PubMed] [Google Scholar]
- 67.Hengartner CJ, Thompson CM, Zhang J, et al. Association of an activator with an RNA polymerase II holoenzyme. Genes & Development. 1995;9(8):897–910. doi: 10.1101/gad.9.8.897. [DOI] [PubMed] [Google Scholar]
- 68.Barberis A, Pearlberg J, Simkovich N, et al. Contact with a component of the polymerase II holoenzyme suffices for gene activation. Cell. 1995;81(3):359–368. doi: 10.1016/0092-8674(95)90389-5. [DOI] [PubMed] [Google Scholar]
- 69.Chao DM, Gadbois EL, Murray PJ, et al. A mammalian SRB protein associated with an RNA polymerase II holoenzyme. Nature. 1996;380(6569):82–85. doi: 10.1038/380082a0. [DOI] [PubMed] [Google Scholar]
- 70.Koleske AJ, Young RA. The RNA polymerase II holoenzyme and its implications for gene regulation. Trends in Biochemical Sciences. 1995;20(3):113–116. doi: 10.1016/s0968-0004(00)88977-x. [DOI] [PubMed] [Google Scholar]
- 71.Armache KJ, Kettenberger H, Cramer P. Architecture of initiation-competent 12-subunit RNA polymerase II. Proceedings of the National Academy of Sciences of the United States of America. 2003;100(12):6964–6968. doi: 10.1073/pnas.1030608100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Meinhart A, Kamenski T, Hoeppner S, Baumli S, Cramer P. A structural perspective of CTD function. Genes & Development. 2005;19(12):1401–1415. doi: 10.1101/gad.1318105. [DOI] [PubMed] [Google Scholar]
- 73.Xiang K, Nagaike T, Xiang S, et al. Crystal structure of the human symplekin-Ssu72-CTD phosphopeptide complex. Nature. 2010;467(7316):729–733. doi: 10.1038/nature09391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Werner-Allen JW, Lee C-J, Liu P, et al. cis-proline-mediated ser(P)5 dephosphorylation by the RNA polymerase II C-terminal domain phosphatase Ssu72. Journal of Biological Chemistry. 2011;286(7):5717–5726. doi: 10.1074/jbc.M110.197129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Meinhart A, Cramer P. Recognition of RNA polymerase II carboxy-terminal domain by 3′-RNA-processing factors. Nature. 2004;430(6996):223–226. doi: 10.1038/nature02679. [DOI] [PubMed] [Google Scholar]
- 76.Kelly WG, Dahmus ME, Hart GW. RNA polymerase II is a glycoprotein. Modification of the COOH-terminal domain by O-GlcNAc. Journal of Biological Chemistry. 1993;268(14):10416–10424. [PubMed] [Google Scholar]
- 77.Kang ME, Dahmus ME. The photoactivated cross-linking of recombinant C-terminal domain to proteins in a HeLa cell transcription extract that comigrate with transcription factors IIE and IIF. Journal of Biological Chemistry. 1995;270(40):23390–23397. doi: 10.1074/jbc.270.40.23390. [DOI] [PubMed] [Google Scholar]
- 78.Usheva A, Maldonado E, Goldring A, et al. Specific interaction between the nonphosphorylated form of RNA polymerase II and the TATA-binding protein. Cell. 1992;69(5):871–881. doi: 10.1016/0092-8674(92)90297-p. [DOI] [PubMed] [Google Scholar]
- 79.Myers LC, Gustafsson CM, Bushnell DA, et al. The Med proteins of yeast and their function through the RNA polymerase II carboxy-terminal domain. Genes & Development. 1998;12(1):45–54. doi: 10.1101/gad.12.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Cho EJ, Takagi T, Moore CR, Buratowski S. mRNA capping enzyme is recruited to the transcription complex by phosphorylation of the RNA polymerase II carboxy-terminal domain. Genes & Development. 1997;11(24):3319–3326. doi: 10.1101/gad.11.24.3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Fabrega C, Shen V, Shuman S, Lima CD. Structure of an mRNA capping enzyme bound to the phosphorylated carboxy-terminal domain of RNA polymerase II. Molecular Cell. 2003;11(6):1549–1561. doi: 10.1016/s1097-2765(03)00187-4. [DOI] [PubMed] [Google Scholar]
- 82.Ho CK, Shuman S. Distinct roles for CTD Ser-2 and Ser-5 phosphorylation in the recruitment and allosteric activation of mammalian mRNA capping enzyme. Molecular Cell. 1999;3(3):405–411. doi: 10.1016/s1097-2765(00)80468-2. [DOI] [PubMed] [Google Scholar]
- 83.Komarnitsky P, Cho EJ, Buratowski S. Different phosphorylated forms of RNA polymerase II and associated mRNA processing factors during transcription. Genes & Development. 2000;14(19):2452–2460. doi: 10.1101/gad.824700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.McCracken S, Fong N, Rosonina E, et al. 5-Capping enzymes are targeted to pre-mRNA by binding to the phosphorylated carboxy-terminal domain of RNA polymerase II. Genes & Development. 1997;11(24):3306–3318. doi: 10.1101/gad.11.24.3306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Schroeder SC, Schwer B, Shuman S, Bentley D. Dynamic association of capping enzymes with transcribing RNA polymerase II. Genes & Development. 2000;14(19):2435–2440. doi: 10.1101/gad.836300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ng HH, Robert F, Young RA, Struhl K. Targeted recruitment of Set1 histone methylase by elongating Pol II provides a localized mark and memory of recent transcriptional activity. Molecular Cell. 2003;11(3):709–719. doi: 10.1016/s1097-2765(03)00092-3. [DOI] [PubMed] [Google Scholar]
- 87.Govind CK, Qiu H, Ginsburg DS, et al. Phosphorylated Pol II CTD recruits multiple HDACs, including Rpd3C(S), for methylation-dependent deacetylation of ORF nucleosomes. Molecular Cell. 2010;39(2):234–246. doi: 10.1016/j.molcel.2010.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Drouin S, Laramée L, Jacques PE, Forest A, Bergeron M, Robert F. DSIF and RNA polymerase II CTD phosphorylation coordinate the recruitment of Rpd3S to actively transcribed genes. PLoS Genetics. 2010;6(10):1–12. doi: 10.1371/journal.pgen.1001173. Article ID e1001173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Yoh SM, Cho H, Pickle L, Evans RM, Jones KA. The Spt6 SH2 domain binds Ser2-P RNAPII to direct Iws1-dependent mRNA splicing and export. Genes & Development. 2007;21(2):160–174. doi: 10.1101/gad.1503107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Vasiljeva L, Kim M, Mutschler H, Buratowski S, Meinhart A. The Nrd1-Nab3-Sen1 termination complex interacts with the Ser5-phosphorylated RNA polymerase II C-terminal domain. Nature Structural & Molecular Biology. 2008;15(8):795–804. doi: 10.1038/nsmb.1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Finkel JS, Chinchilla K, Ursic D, Culbertson MR. Sen1p performs two genetically separable functions in transcription and processing of U5 small nuclear RNA in Saccharomyces cerevisiae. Genetics. 2010;184(1):107–118. doi: 10.1534/genetics.109.110031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Daulny A, Geng F, Muratani M, Geisinger JM, Salghetti SE, Tansey WP. Modulation of RNA polymerase II subunit composition by ubiquitylation. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(50):19649–19654. doi: 10.1073/pnas.0809372105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Morris DP, Phatnani HP, Greenleaf AL. Phospho-carboxyl-terminal domain binding and the role of a prolyl isomerase in pre-mRNA 3’-end formation. Journal of Biological Chemistry. 1999;274(44):31583–31587. doi: 10.1074/jbc.274.44.31583. [DOI] [PubMed] [Google Scholar]
- 94.Wu X, Wilcox CB, Devasahayam G, et al. The Ess1 prolyl isomerase is linked to chromatin remodeling complexes and the general transcription machinery. The EMBO Journal. 2000;19(14):3727–3738. doi: 10.1093/emboj/19.14.3727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kizer KO, Phatnani HP, Shibata Y, Hall H, Greenleaf AL, Strahl BD. A novel domain in Set2 mediates RNA polymerase II interaction and couples histone H3 K36 methylation with transcript elongation. Molecular and Cellular Biology. 2005;25(8):3305–3316. doi: 10.1128/MCB.25.8.3305-3316.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Vojnic E, Simon B, Strahl BD, Sattler M, Cramer P. Structure and carboxyl-terminal domain (CTD) binding of the Set2 SRI domain that couples histone H3 Lys36 methylation to transcription. Journal of Biological Chemistry. 2006;281(1):13–15. doi: 10.1074/jbc.C500423200. [DOI] [PubMed] [Google Scholar]
- 97.Morris DP, Greenleaf AL. The splicing factor, Prp40, binds the phosphorylated carboxyl-terminal domain of RNA Polymerase II. Journal of Biological Chemistry. 2000;275(51):39935–39943. doi: 10.1074/jbc.M004118200. [DOI] [PubMed] [Google Scholar]
- 98.Dermody JL, Dreyfuss JM, Villén J, et al. Unphosphorylated SR-like protein Npl3 stimulates RNA polymerase II elongation. PLoS One. 2008;3(9) doi: 10.1371/journal.pone.0003273. Article ID e3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Noble CG, Hollingworth D, Martin SR, et al. Key features of the interaction between Pcf11 CID and RNA polymerase II CTD. Nature Structural & Molecular Biology. 2005;12(2):144–151. doi: 10.1038/nsmb887. [DOI] [PubMed] [Google Scholar]
- 100.Hollingworth D, Noble CG, Taylor IA, Ramos A. RNA polymerase II CTD phosphopeptides compete with RNA for the interaction with Pcf11. RNA. 2006;12(4):555–560. doi: 10.1261/rna.2304506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Barillà D, Lee BA, Proudfoot NJ. Cleavage/polyadenylation factor IA associates with the carboxyl-terminal domain of RNA polymerase II in Saccharomyces cerevisae. Proceedings of the National Academy of Sciences of the United States of America. 2001;98(2):445–450. doi: 10.1073/pnas.98.2.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kyburz A, Sadowski M, Dichtl B, Keller W. The role of the yeast cleavage and polyadenylation factor subunit Ydh1p/Cft2p in pre-mRNA 3′-end formation. Nucleic Acids Research. 2003;31(14):3936–3945. doi: 10.1093/nar/gkg478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Dichtl B, Blank D, Sadowski M, Hübner W, Weiser S, Keller W. Yhh1p/Cft1p directly links poly(A) site recognition and RNA polymerase II transcription termination. The EMBO Journal. 2002;21(15):4125–4135. doi: 10.1093/emboj/cdf390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Rodriguez CR, Cho EJ, Keogh MC, Moore CL, Greenleaf AL, Buratowski S. Kin28, the TFIIH-associated carboxy-terminal domain kinase, facilitates the recruitment of mRNA processing machinery to RNA polymerase II. Molecular and Cellular Biology. 2000;20(1):104–112. doi: 10.1128/mcb.20.1.104-112.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kim M, Krogan NJ, Vasiljeva L, et al. The yeast Rat1 exonuclease promotes transcription termination by RNA polymerase II. Nature. 2004;432(7016):517–522. doi: 10.1038/nature03041. [DOI] [PubMed] [Google Scholar]
- 106.Pascual-García P, Govind CK, Queralt E, et al. Sus1 is recruited to coding regions and functions during transcription elongation in association with SAGA and TREX2. Genes & Development. 2008;22(20):2811–2822. doi: 10.1101/gad.483308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.MacKellar AL, Greenleaf AL. Cotranscriptional association of mRNA export factor Yra1 with C-terminal domain of RNA polymerase II. Journal of Biological Chemistry. 2011;286(42):36385–36395. doi: 10.1074/jbc.M111.268144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Chang A, Cheang S, Espanel X, Sudol M. Rsp5 WW domains interact directly with the carboxyl-terminal domain of RNA polymerase II. Journal of Biological Chemistry. 2000;275(27):20562–20571. doi: 10.1074/jbc.M002479200. [DOI] [PubMed] [Google Scholar]
- 109.Somesh BP, Reid J, Liu WF, et al. Multiple mechanisms confining RNA polymerase II ubiquitylation to polymerases undergoing transcriptional arrest. Cell. 2005;121(6):913–923. doi: 10.1016/j.cell.2005.04.010. [DOI] [PubMed] [Google Scholar]
- 110.Phatnani HP, Jones JC, Greenleaf AL. Expanding the functional repertoire of CTD kinase I and RNA polymerase II: novel phosphoCTD-associating proteins in the yeast proteome. Biochemistry. 2004;43(50):15702–15719. doi: 10.1021/bi048364h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Svejstrup JQ, Feaver WJ, LaPointe J, Kornberg RD. RNA polymerase transcription factor IIH holoenzyme from yeast. Journal of Biological Chemistry. 1994;269(45):28044–28048. [PubMed] [Google Scholar]
- 112.Svejstrup JQ, Wang Z, Feaver WJ, et al. Different forms of TFIIH for transcription and DNA repair: holo-TFIIH and a nucleotide excision repairosome. Cell. 1995;80(1):21–28. doi: 10.1016/0092-8674(95)90447-6. [DOI] [PubMed] [Google Scholar]
- 113.Hengartner CJ, Myer VE, Liao SM, Wilson CJ, Koh SS, Young RA. Temporal regulation of RNA polymerase II by Srb10 and Kin28 cyclin-dependent kinases. Molecular Cell. 1998;2(1):43–53. doi: 10.1016/s1097-2765(00)80112-4. [DOI] [PubMed] [Google Scholar]
- 114.Myer VE, Young RA. RNA polymerase II holoenzymes and subcomplexes. Journal of Biological Chemistry. 1998;273(43):27757–27760. doi: 10.1074/jbc.273.43.27757. [DOI] [PubMed] [Google Scholar]
- 115.Tirode F, Busso D, Coin F, Egly JM. Reconstitution of the transcription factor TFIIH: assignment of functions for the three enzymatic subunits, XPB, XPD, and cdk7. Molecular Cell. 1999;3(1):87–95. doi: 10.1016/s1097-2765(00)80177-x. [DOI] [PubMed] [Google Scholar]
- 116.Chang WH, Kornberg RD. Electron crystal structure of the transcription factor and DNA repair complex, core TFIIH. Cell. 2000;102(5):609–613. doi: 10.1016/s0092-8674(00)00083-0. [DOI] [PubMed] [Google Scholar]
- 117.Takagi Y, Masuda CA, Chang WH, et al. Ubiquitin ligase activity of TFIIH and the transcriptional response to DNA damage. Molecular Cell. 2005;18(2):237–243. doi: 10.1016/j.molcel.2005.03.007. [DOI] [PubMed] [Google Scholar]
- 118.Rabut G, Le Dez G, Verma R, et al. The TFIIH subunit Tfb3 regulates cullin neddylation. Molecular Cell. 2011;43(3):488–495. doi: 10.1016/j.molcel.2011.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Dahmus ME. Reversible phosphorylation of the C-terminal domain of RNA polymerase II. Journal of Biological Chemistry. 1996;271(32):19009–19012. doi: 10.1074/jbc.271.32.19009. [DOI] [PubMed] [Google Scholar]
- 120.Bensaude O, Bonnet F, Cassé C, Dubois MF, Nguyen VT, Palancade B. Regulated phosphorylation of the RNA polymerase II C-terminal domain (CTD) Biochemistry and Cell Biology. 1999;77(4):249–255. [PubMed] [Google Scholar]
- 121.Palancade B, Bensaude O. Investigating RNA polymerase II carboxyl-terminal domain (CTD) phosphorylation. European Journal of Biochemistry. 2003;270(19):3859–3870. doi: 10.1046/j.1432-1033.2003.03794.x. [DOI] [PubMed] [Google Scholar]
- 122.Feaver WJ, Svejstrup JQ, Henry NL, Kornberg RD. Relationship of CDK-activating kinase and RNA polymerase II CTD kinase TFIIH/TFIIK. Cell. 1994;79(6):1103–1109. doi: 10.1016/0092-8674(94)90040-x. [DOI] [PubMed] [Google Scholar]
- 123.Rickert P, Corden JL, Lees E. Cyclin C/CDK8 and cyclin H/CDK7/p36 are biochemically distinct CTD kinases. Oncogene. 1999;18(4):1093–1102. doi: 10.1038/sj.onc.1202399. [DOI] [PubMed] [Google Scholar]
- 124.Gebara MM, Sayre MH, Corden JL. Phosphorylation of the carboxy-terminal repeat domain in RNA polymerase II by cyclin-dependent kinases is sufficient to inhibit transcription. Journal of Cellular Biochemistry. 1997;64(3):390–402. [PubMed] [Google Scholar]
- 125.Munshi A, Shafi G, Aliya N, Jyothy A. Histone modifications dictate specific biological readouts. Journal of Genetics and Genomics. 2009;36(2):75–88. doi: 10.1016/S1673-8527(08)60094-6. [DOI] [PubMed] [Google Scholar]
- 126.Lee JS, Smith E, Shilatifard A. The language of histone crosstalk. Cell. 2010;142(5):682–685. doi: 10.1016/j.cell.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Workman JL. Nucleosome displacement in transcription. Genes & Development. 2006;20(15):2009–2017. doi: 10.1101/gad.1435706. [DOI] [PubMed] [Google Scholar]
- 128.Nakanishi S, Sanderson BW, Delventhal KM, Bradford WD, Staehling-Hampton K, Shilatifard A. A comprehensive library of histone mutants identifies nucleosomal residues required for H3K4 methylation. Nature Structural & Molecular Biology. 2008;15(8):881–888. doi: 10.1038/nsmb.1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Wood A, Schneider J, Dover J, Johnston M, Shilatifard A. The Paf1 complex is essential for histone monoubiquitination by the Rad6-Bre1 complex, which signals for histone methylation by COMPASS and Dot1p. Journal of Biological Chemistry. 2003;278(37):34739–34742. doi: 10.1074/jbc.C300269200. [DOI] [PubMed] [Google Scholar]
- 130.Cramer P, Bushnell DA, Kornberg RD. Structural basis of transcription: RNA polymerase II at 2.8 ångstrom resolution. Science. 2001;292(5523):1863–1876. doi: 10.1126/science.1059493. [DOI] [PubMed] [Google Scholar]
- 131.Ghosh A, Shuman S, Lima C. Structural insights to how Mammalian capping enzyme reads the CTD code. Molecular Cell. 2011;43(2):299–310. doi: 10.1016/j.molcel.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Fong N, Bentley DL. Capping, splicing, and 3′ processing are independently stimulated by RNA polymerase II: different functions for different segments of the CTD. Genes & Development. 2001;15(14):1783–1795. doi: 10.1101/gad.889101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Moteki S, Price D. Functional coupling of capping and transcription of mRNA. Molecular Cell. 2002;10(3):599–609. doi: 10.1016/s1097-2765(02)00660-3. [DOI] [PubMed] [Google Scholar]
- 134.Liu Y, Kung C, Fishburn J, Ansari AZ, Shokat KM, Hahn S. Two cyclin-dependent kinases promote RNA polymerase II transcription and formation of the scaffold complex. Molecular and Cellular Biology. 2004;24(4):1721–1735. doi: 10.1128/MCB.24.4.1721-1735.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Kanin EI, Kipp RT, Kung C, et al. Chemical inhibition of the TFIIH-associated kinase Cdk7/Kin28 does not impair global mRNA synthesis. Proceedings of the National Academy of Sciences of the United States of America. 2007;104(14):5812–5817. doi: 10.1073/pnas.0611505104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Schwer B, Shuman S. Deciphering the RNA polymerase II CTD code in fission yeast. Molecular Cell. 2011;43(2):311–318. doi: 10.1016/j.molcel.2011.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Hong SW, Seong MH, Jae WY, et al. Phosphorylation of the RNA polymerase II C-terminal domain by TFIIH kinase is not essential for transcription of Saccharomyces cerevisiae genome. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(34):14276–14280. doi: 10.1073/pnas.0903642106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Tietjen JR, Zhang DW, Rodríguez-Molina JB, et al. Chemical-genomic dissection of the CTD code. Nature Structural & Molecular Biology. 2010;17(9):1154–1161. doi: 10.1038/nsmb.1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Akhtar MS, Heidemann M, Tietjen JR, et al. TFIIH kinase places bivalent marks on the carboxy-terminal domain of RNA polymerase II. Molecular Cell. 2009;34(3):387–393. doi: 10.1016/j.molcel.2009.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Kim M, Suh H, Cho EJ, Buratowski S. Phosphorylation of the yeast Rpb1 C-terminal domain at serines 2,5, and 7. Journal of Biological Chemistry. 2009;284(39):26421–26426. doi: 10.1074/jbc.M109.028993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Glover-Cutter K, Larochelle S, Erickson B, et al. TFIIH-associated Cdk7 kinase functions in phosphorylation of C-terminal domain Ser7 residues, promoter-proximal pausing, and termination by RNA polymerase II. Molecular and Cellular Biology. 2009;29(20):5455–5464. doi: 10.1128/MCB.00637-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Boeing S, Rigault C, Heidemann M, Eick D, Meisterernst M. RNA polymerase II C-terminal heptarepeat domain Ser-7 phosphorylation is established in a mediator-dependent fashion. Journal of Biological Chemistry. 2010;285(1):188–196. doi: 10.1074/jbc.M109.046565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Songyang Z, Lu KP, Kwon YT, et al. A structural basis for substrate specificities of protein Ser/Thr kinases: primary sequence preference of casein kinases I and II, NIMA, phosphorylase kinase, calmodulin-dependent kinase II, CDK5, and Erk1. Molecular and Cellular Biology. 1996;16(11):6486–6493. doi: 10.1128/mcb.16.11.6486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Chapman RD, Heidemann M, Albert TK, et al. Transcribing RNA polymerase II is phosphorylated at CTD residue serine-7. Science. 2007;318(5857):1780–1782. doi: 10.1126/science.1145977. [DOI] [PubMed] [Google Scholar]
- 145.Egloff S, O’Reilly D, Chapman RD, et al. Serine-7 of the RNA polymerase II CTD is specifically required for snRNA gene expression. Science. 2007;318(5857):1777–1779. doi: 10.1126/science.1145989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Mayer A, Lidschreiber M, Siebert M, Leike K, Söding J, Cramer P. Uniform transitions of the general RNA polymerase II transcription complex. Nature Structural & Molecular Biology. 2010;17(10):1272–1278. doi: 10.1038/nsmb.1903. [DOI] [PubMed] [Google Scholar]
- 147.Hartzog GA, Wada T, Handa H, Winston F. Evidence that Spt4, Spt5, and Spt6 control transcription elongation by RNA polymerase II in Saccharomyces cerevisiae. Genes & Development. 1998;12(3):357–369. doi: 10.1101/gad.12.3.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Martinez-Rucobo FW, Sainsbury S, Cheung ACM, Cramer P. Architecture of the RNA polymerase-Spt4/5 complex and basis of universal transcription processivity. The EMBO Journal. 2011;30(7):1302–1310. doi: 10.1038/emboj.2011.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Grohmann D, Nagy J, Chakraborty A, et al. The initiation factor TFE and the elongation factor Spt4/5 compete for the RNAP clamp during transcription initiation and elongation. Molecular Cell. 2011;43(2):263–274. doi: 10.1016/j.molcel.2011.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Adkins MW, Tyler JK. Transcriptional activators are dispensable for transcription in the absence of Spt6-mediated chromatin reassembly of promoter regions. Molecular Cell. 2006;21(3):405–416. doi: 10.1016/j.molcel.2005.12.010. [DOI] [PubMed] [Google Scholar]
- 151.Youdell ML, Kizer KO, Kisseleva-Romanova E, et al. Roles for Ctk1 and Spt6 in regulating the different methylation states of histone H3 lysine 36. Molecular and Cellular Biology. 2008;28(16):4915–4926. doi: 10.1128/MCB.00001-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Orphanides G, Wu WH, Lane WS, Hampsey M, Reinberg D. The chromatin-specific transcription elongation factor FACT comprises human SPT16 and SSRP1 proteins. Nature. 1999;400(6741):284–288. doi: 10.1038/22350. [DOI] [PubMed] [Google Scholar]
- 153.Jamai A, Puglisi A, Strubin M. Histone chaperone Spt16 promotes redeposition of the original H3-H4 histones evicted by elongating RNA polymerase. Molecular Cell. 2009;35(3):377–383. doi: 10.1016/j.molcel.2009.07.001. [DOI] [PubMed] [Google Scholar]
- 154.Zhang L, Fletcher AGL, Cheung V, Winston F, Stargell LA. Spn1 regulates the recruitment of Spt6 and the Swi/Snf complex during transcriptional activation by RNA polymerase II. Molecular and Cellular Biology. 2008;28(4):1393–1403. doi: 10.1128/MCB.01733-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.McDonald SM, Close D, Xin H, Formosa T, Hill CP. Structure and biological importance of the Spn1-Spt6 interaction, and its regulatory role in nucleosome binding. Molecular Cell. 2010;40(5):725–735. doi: 10.1016/j.molcel.2010.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Prather D, Krogan NJ, Emili A, Greenblatt JF, Winston F. Identification and characterization of Elf1, a conserved transcription elongation factor in Saccharomyces cerevisiae. Molecular and Cellular Biology. 2005;25(22):10122–10135. doi: 10.1128/MCB.25.22.10122-10135.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Jaehning JA. The Paf1 complex: platform or player in RNA polymerase II transcription? Biochimica et Biophysica Acta. 2010;1799:379–388. doi: 10.1016/j.bbagrm.2010.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Brès V, Yoh SM, Jones KA. The multi-tasking P-TEFb complex. Current Opinion in Cell Biology. 2008;20(3):334–340. doi: 10.1016/j.ceb.2008.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Peterlin BM, Price DH. Controlling the elongation phase of transcription with P-TEFb. Molecular Cell. 2006;23(3):297–305. doi: 10.1016/j.molcel.2006.06.014. [DOI] [PubMed] [Google Scholar]
- 160.Price DH. P-TEFb, a cyclin-dependent kinase controlling elongation by RNA polymerase II. Molecular and Cellular Biology. 2000;20(8):2629–2634. doi: 10.1128/mcb.20.8.2629-2634.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Viladevall L, Amour CVS, Rosebrock A, et al. TFIIH and P-TEFb coordinate transcription with capping enzyme recruitment at specific genes in fission yeast. Molecular Cell. 2009;33(6):738–751. doi: 10.1016/j.molcel.2009.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Zhou K, Kuo WHW, Fillingham J, Greenblatt JF. Control of transcriptional elongation and cotranscriptional histone modification by the yeast BUR kinase substrate Spt5. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(17):6956–6961. doi: 10.1073/pnas.0806302106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Wood A, Schneider J, Dover J, Johnston M, Shilatifard A. The Bur1/Bur2 complex is required for histone H2B monoubiquitination by Rad6/Bre1 and histone methylation by COMPASS. Molecular Cell. 2005;20(4):589–599. doi: 10.1016/j.molcel.2005.09.010. [DOI] [PubMed] [Google Scholar]
- 164.Jones JC, Phatnani HP, Haystead TA, MacDonald JA, Alam SM, Greenleaf AL. C-terminal repeat domain kinase I phosphorylates Ser2 and Ser5 of RNa polymerase II C-terminal domain repeats. Journal of Biological Chemistry. 2004;279(24):24957–24964. doi: 10.1074/jbc.M402218200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Kim JB, Sharp PA. Positive transcription elongation factor B phosphorylates hSPT5 and RNA polymerase II carboxyl-terminal domain independently of cyclin-dependent kinase-activating kinase. Journal of Biological Chemistry. 2001;276(15):12317–12323. doi: 10.1074/jbc.M010908200. [DOI] [PubMed] [Google Scholar]
- 166.Qiu H, Hu C, Hinnebusch AG. Phosphorylation of the pol II CTD by KIN28 enhances BUR1/BUR2 recruitment and Ser2 CTD phosphorylation near promoters. Molecular Cell. 2009;33(6):752–762. doi: 10.1016/j.molcel.2009.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Mosley AL, Pattenden SG, Carey M, et al. Rtr1 Is a CTD phosphatase that regulates RNA polymerase II during the transition from Serine 5 to Serine 2 phosphorylation. Molecular Cell. 2009;34(2):168–178. doi: 10.1016/j.molcel.2009.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Kobor MS, Archambault J, Lester W, et al. An unusual eukaryotic protein phosphatase required for transcription by RNA polymerase II and CTD dephosphorylation in S. cerevisiae. Molecular Cell. 1999;4(1):55–62. doi: 10.1016/s1097-2765(00)80187-2. [DOI] [PubMed] [Google Scholar]
- 169.Cho EJ, Kobor MS, Kim M, Greenblatt J, Buratowski S. Opposing effects of Ctk1 kinase and Fcp1 phosphatase at Ser 2 of the RNA polymerase II C-terminal domain. Genes & Development. 2001;15(24):3319–3329. doi: 10.1101/gad.935901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Henry KW, Wyce A, Lo WS, et al. Transcriptional activation via sequential histone H2B ubiquitylation and deubiquitylation, mediated by SAGA-associated Ubp8. Genes & Development. 2003;17(21):2648–2663. doi: 10.1101/gad.1144003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Wyce A, Xiao T, Whelan KA, et al. H2B ubiquitylation acts as a barrier to Ctk1 nucleosomal recruitment prior to removal by Ubp8 within a SAGA-related complex. Molecular Cell. 2007;27(2):275–288. doi: 10.1016/j.molcel.2007.01.035. [DOI] [PubMed] [Google Scholar]
- 172.Bentley D. Coupling RNA polymerase II transcription with pre-mRNA processing. Current Opinion in Cell Biology. 1999;11(3):347–351. doi: 10.1016/S0955-0674(99)80048-9. [DOI] [PubMed] [Google Scholar]
- 173.Bentley D. The mRNA assembly line: transcription and processing machines in the same factory. Current Opinion in Cell Biology. 2002;14(3):336–342. doi: 10.1016/s0955-0674(02)00333-2. [DOI] [PubMed] [Google Scholar]
- 174.Licatalosi DD, Geiger G, Minet M, et al. Functional interaction of yeast pre-mRNA 3′ end processing factors with RNA polymerase II. Molecular Cell. 2002;9(5):1101–1111. doi: 10.1016/s1097-2765(02)00518-x. [DOI] [PubMed] [Google Scholar]
- 175.Proudfoot NJ, Furger A, Dye MJ. Integrating mRNA processing with transcription. Cell. 2002;108(4):501–512. doi: 10.1016/s0092-8674(02)00617-7. [DOI] [PubMed] [Google Scholar]
- 176.Ahn SH, Kim M, Buratowski S. Phosphorylation of Serine 2 within the RNA polymerase II C-terminal domain couples transcription and 3′ end processing. Molecular Cell. 2004;13(1):67–76. doi: 10.1016/s1097-2765(03)00492-1. [DOI] [PubMed] [Google Scholar]
- 177.Bentley DL. Rules of engagement: co-transcriptional recruitment of pre-mRNA processing factors. Current Opinion in Cell Biology. 2005;17(3):251–256. doi: 10.1016/j.ceb.2005.04.006. [DOI] [PubMed] [Google Scholar]
- 178.Buratowski S. Connections between mRNA 3′ end processing and transcription termination. Current Opinion in Cell Biology. 2005;17(3):257–261. doi: 10.1016/j.ceb.2005.04.003. [DOI] [PubMed] [Google Scholar]
- 179.Li J, Moazed D, Gygi SP. Association of the histone methyltransferase Set2 with RNA polymerase II plays a role in transcription elongation. Journal of Biological Chemistry. 2002;277(51):49383–49388. doi: 10.1074/jbc.M209294200. [DOI] [PubMed] [Google Scholar]
- 180.Krogan NJ, Kim M, Tong A, et al. Methylation of histone H3 by Set2 in Saccharomyces cerevisiae is linked to transcriptional elongation by RNA polymerase II. Molecular and Cellular Biology. 2003;23(12):4207–4218. doi: 10.1128/MCB.23.12.4207-4218.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Li B, Howe L, Anderson S, Yates JR, Workman JL. The Set2 histone methyltransferase functions through the phosphorylated carboxyl-terminal domain of RNA polymerase II. Journal of Biological Chemistry. 2003;278(11):8897–8903. doi: 10.1074/jbc.M212134200. [DOI] [PubMed] [Google Scholar]
- 182.Carrozza MJ, Li B, Florens L, et al. Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell. 2005;123(4):581–592. doi: 10.1016/j.cell.2005.10.023. [DOI] [PubMed] [Google Scholar]
- 183.Keogh MC, Kurdistani SK, Morris SA, et al. Cotranscriptional set2 methylation of histone H3 lysine 36 recruits a repressive Rpd3 complex. Cell. 2005;123(4):593–605. doi: 10.1016/j.cell.2005.10.025. [DOI] [PubMed] [Google Scholar]
- 184.Gilbert W, Siebel CW, Guthrie C. Phosphorylation by Sky1p promotes Npl3p shuttling and mRNA dissociation. RNA. 2001;7(2):302–313. doi: 10.1017/s1355838201002369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Lei EP, Krebber H, Silver PA. Messenger RNAs are recruited for nuclear export during transcription. Genes & Development. 2001;15(14):1771–1782. doi: 10.1101/gad.892401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Bucheli ME, Buratowski S. Npl3 is an antagonist of mRNA 3′ end formation by RNA polymerase II. The EMBO Journal. 2005;24(12):2150–2160. doi: 10.1038/sj.emboj.7600687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Bucheli ME, He X, Kaplan CD, Moore CL, Buratowski S. Polyadenylation site choice in yeast is affected by competition between Npl3 and polyadenylation factor CFI. RNA. 2007;13(10):1756–1764. doi: 10.1261/rna.607207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Proudfoot NJ. Ending the message: poly(A) signals then and now. Genes & Development. 2011;25(17):1770–1782. doi: 10.1101/gad.17268411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Kim M, Ahn SH, Krogan NJ, Greenblatt JF, Buratowski S. Transitions in RNA polymerase II elongation complexes at the 3′ ends of genes. The EMBO Journal. 2004;23(2):354–364. doi: 10.1038/sj.emboj.7600053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Connelly S, Manley JL. A functional mRNA polyadenylation signal is required for transcription termination by RNA polymerase II. Genes & Development. 1988;2(4):440–452. doi: 10.1101/gad.2.4.440. [DOI] [PubMed] [Google Scholar]
- 191.West S, Gromak N, Proudfoot NJ. Human 5′ → 3′ exonuclease Xm2 promotes transcription termination at co-transcriptional cleavage sites. Nature. 2004;432(7016):522–525. doi: 10.1038/nature03035. [DOI] [PubMed] [Google Scholar]
- 192.Lunde BM, Reichow SL, Kim M, et al. Cooperative interaction of transcription termination factors with the RNA polymerase II C-terminal domain. Nature Structural & Molecular Biology. 2010;17(10):1195–1201. doi: 10.1038/nsmb.1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Zhang Z, Fu J, Gilmour DS. CTD-dependent dismantling of the RNA polymerase II elongation complex by the pre-mRNA 3′-end processing factor, Pcf11. Genes & Development. 2005;19(13):1572–1580. doi: 10.1101/gad.1296305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Sadowski M, Dichtl B, Hübner W, Keller W. Independent functions of yeast Pcf11p in pre-mRNA 3′ end processing and in transcription termination. The EMBO Journal. 2003;22(9):2167–2177. doi: 10.1093/emboj/cdg200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Kim M, Vasiljeva L, Rando OJ, Zhelkovsky A, Moore C, Buratowski S. Distinct pathways for snoRNA and mRNA termination. Molecular Cell. 2006;24(5):723–734. doi: 10.1016/j.molcel.2006.11.011. [DOI] [PubMed] [Google Scholar]
- 196.Kim H, Erickson B, Luo W, et al. Gene-specific RNA polymerase II phosphorylation and the CTD code. Nature Structural & Molecular Biology. 2010;17(10):1279–1286. doi: 10.1038/nsmb.1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Birse CE, Minvielle-Sebastia L, Lee BA, Keller W, Proudfoot NJ. Coupling termination of transcription to messenger RNA maturation in yeast. Science. 1998;280(5361):298–301. doi: 10.1126/science.280.5361.298. [DOI] [PubMed] [Google Scholar]
- 198.Gudipati RK, Villa T, Boulay J, Libri D. Phosphorylation of the RNA polymerase II C-terminal domain dictates transcription termination choice. Nature Structural & Molecular Biology. 2008;15(8):786–794. doi: 10.1038/nsmb.1460. [DOI] [PubMed] [Google Scholar]
- 199.Steinmetz EJ, Conrad NK, Brow DA, Corden JL. RNA-binding protein Nrd1 directs poly(A)-independent 3′-end formation of RNA polymerase II transcripts. Nature. 2001;413(6853):327–331. doi: 10.1038/35095090. [DOI] [PubMed] [Google Scholar]
- 200.Terzi N, Churchman LS, Vasiljeva L, Weissman J, Buratowski S. H3K4 trimethylation by Set1 promotes efficient termination by the Nrd1-Nab3-Sen1 pathway. Molecular and Cellular Biology. 2011;31(17):3569–3583. doi: 10.1128/MCB.05590-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Steinmetz EJ, Brow DA. Repression of gene expression by an exogenous sequence element acting in concert with a heterogeneous nuclear ribonucleoprotein-like protein, Nrd1, and the putative helicase Sen1. Molecular and Cellular Biology. 1996;16(12):6993–7003. doi: 10.1128/mcb.16.12.6993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Steinmetz EJ, Brow DA. Control of pre-mRNA accumulation by the essential yeast protein Nrd1 requires high-affinity transcript binding and a domain implicated in RNA polymerase II association. Proceedings of the National Academy of Sciences of the United States of America. 1998;95(12):6699–6704. doi: 10.1073/pnas.95.12.6699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Conrad NK, Wilson SM, Steinmetz EJ, et al. A yeast heterogeneous nuclear ribonucleoprotein complex associated with RNA polymerase II. Genetics. 2000;154(2):557–571. doi: 10.1093/genetics/154.2.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Carroll KL, Pradhan DA, Granek JA, Clarke ND, Corden JL. Identification of cis elements directing termination of yeast nonpolyadenylated snoRNA transcripts. Molecular and Cellular Biology. 2004;24(14):6241–6252. doi: 10.1128/MCB.24.14.6241-6252.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Carroll KL, Ghirlando R, Ames JM, Corden JL. Interaction of yeast RNA-binding proteins Nrd1 and Nab3 with RNA polymerase II terminator elements. RNA. 2007;13(3):361–373. doi: 10.1261/rna.338407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Hobor F, Pergoli R, Kubicek K, et al. Recognition of transcription termination signal by the nuclear polyadenylated RNA-binding (NAB) 3 protein. Journal of Biological Chemistry. 2011;286(5):3645–3657. doi: 10.1074/jbc.M110.158774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Wlotzka W, Kudla G, Granneman S, Tollervey D. The nuclear RNA polymerase II surveillance system targets polymerase III transcripts. The EMBO Journal. 2011;30(9):1790–1803. doi: 10.1038/emboj.2011.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Ursic D, Himmel KL, Gurley KA, Webb F, Culbertson MR. The yeast SEN1 gene is required for the processing of diverse RNA classes. Nucleic Acids Research. 1997;25(23):4778–4785. doi: 10.1093/nar/25.23.4778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Skourti-Stathaki K, Proudfoot N, Gromak N. Human senataxin resolves RNA/DNA hybrids formed at transcriptional pause sites to promote Xrn2-dependent termination. Molecular Cell. 2011;42(6):794–805. doi: 10.1016/j.molcel.2011.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Mischo HE, Gómez-González B, Grzechnik P, et al. Yeast Sen1 helicase protects the genome from transcription-associated instability. Molecular Cell. 2011;41(1):21–32. doi: 10.1016/j.molcel.2010.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Nedea E, Nalbant D, Xia D, et al. The Glc7 phosphatase subunit of the cleavage and polyadenylation factor is essential for transcription termination on snoRNA genes. Molecular Cell. 2008;29(5):577–587. doi: 10.1016/j.molcel.2007.12.031. [DOI] [PubMed] [Google Scholar]
- 212.Chanfreau G, Legrain P, Jacquier A. Yeast RNase III as a key processing enzyme in small nucleolar RNAs metabolism. Journal of Molecular Biology. 1998;284(4):975–988. doi: 10.1006/jmbi.1998.2237. [DOI] [PubMed] [Google Scholar]
- 213.Chanfreau G, Rotondo G, Legrain P, Jacquier A. Processing of a dicistronic small nucleolar RNA precursor by the RNA endonuclease Rnt1. The EMBO Journal. 1998;17(13):3726–3737. doi: 10.1093/emboj/17.13.3726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Allmang C, Kufel J, Chanfreau G, Mitchell P, Petfalski E, Tollervey D. Functions of the exosome in rRNA, snoRNA and snRNA synthesis. The EMBO Journal. 1999;18(19):5399–5410. doi: 10.1093/emboj/18.19.5399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.LaCava J, Houseley J, Saveanu C, et al. RNA degradation by the exosome is promoted by a nuclear polyadenylation complex. Cell. 2005;121(5):713–724. doi: 10.1016/j.cell.2005.04.029. [DOI] [PubMed] [Google Scholar]
- 216.Egecioglu DE, Henras AK, Chanfreau GF. Contributions of Trf4p- And Trf5p-dependent polyadenylation to the processing and degradative functions of the yeast nuclear exosome. RNA. 2006;12(1):26–32. doi: 10.1261/rna.2207206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Vasiljeva L, Buratowski S. Nrd1 interacts with the nuclear exosome for 3′ processing of RNA polymerase II transcripts. Molecular Cell. 2006;21(2):239–248. doi: 10.1016/j.molcel.2005.11.028. [DOI] [PubMed] [Google Scholar]
- 218.Baillat D, Hakimi MA, Näär AM, Shilatifard A, Cooch N, Shiekhattar R. Integrator, a multiprotein mediator of small nuclear RNA processing, associates with the C-terminal repeat of RNA polymerase II. Cell. 2005;123(2):265–276. doi: 10.1016/j.cell.2005.08.019. [DOI] [PubMed] [Google Scholar]
- 219.Egloff S, Szczepaniak SA, Dienstbier M, Taylor A, Knight S, Murphy S. The integrator complex recognizes a new double mark on the RNA polymerase II carboxyl-terminal domain. Journal of Biological Chemistry. 2010;285(27):20564–20569. doi: 10.1074/jbc.M110.132530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Minvielle-Sebastia L, Preker PJ, Keller W. RNA14 and RNA15 proteins as components of a yeast pre-mRNA 3’-end processing factor. Science. 1994;266(5191):1702–1705. doi: 10.1126/science.7992054. [DOI] [PubMed] [Google Scholar]
- 221.Moore MJ. From birth to death: the complex lives of eukaryotic mRNAs. Science. 2005;309(5740):1514–1518. doi: 10.1126/science.1111443. [DOI] [PubMed] [Google Scholar]
- 222.Nedea E, He X, Kim M, et al. Organization and function of APT, a subcomplex of the yeast cleavage and polyadenylation factor involved in the formation of mRNA and small nucleolar RNA 3′-ends. Journal of Biological Chemistry. 2003;278(35):33000–33010. doi: 10.1074/jbc.M304454200. [DOI] [PubMed] [Google Scholar]
- 223.Ansari A, Hampsey M. A role for the CPF 3′ -end processing machinery in RNAP II-dependent gene looping. Genes & Development. 2005;19(24):2969–2978. doi: 10.1101/gad.1362305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Steinmetz EJ, Brow DA. Ssu72 protein mediates both poly(A)-coupled and poly(A)-independent termination of RNA polymerase II transcription. Molecular and Cellular Biology. 2003;23(18):6339–6349. doi: 10.1128/MCB.23.18.6339-6349.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Krishnamurthy S, He X, Reyes-Reyes M, Moore C, Hampsey M. Ssu72 is an RNA polymerase II CTD phosphatase. Molecular Cell. 2004;14(3):387–394. doi: 10.1016/s1097-2765(04)00235-7. [DOI] [PubMed] [Google Scholar]
- 226.Krishnamurthy S, Ghazy MA, Moore C, Hampsey M. Functional interaction of the Ess1 prolyl isomerase with components of the RNA polymerase II initiation and termination machineries. Molecular and Cellular Biology. 2009;29(11):2925–2934. doi: 10.1128/MCB.01655-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Ghazy MA, He X, Singh BN, Hampsey M, Moore C. The essential N terminus of the pta1 scaffold protein is required for snoRNA transcription termination and Ssu72 function but is dispensable for pre-mRNA 3’-end processing. Molecular and Cellular Biology. 2009;29(8):2296–2307. doi: 10.1128/MCB.01514-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Singh N, Ma Z, Gemmill T, et al. The Ess1 prolyl isomerase is required for transcription termination of small noncoding RNAs via the Nrd1 pathway. Molecular Cell. 2009;36(2):255–266. doi: 10.1016/j.molcel.2009.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Archambault J, Chambers RS, Kobor MS, et al. An essential component of a C-terminal domain phosphatase that interacts with transcription factor IIF in Saccharomyces cerevisiae. Proceedings of the National Academy of Sciences of the United States of America. 1997;94(26):14300–14305. doi: 10.1073/pnas.94.26.14300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Kong SE, Kobor MS, Krogan NJ, et al. Interaction of Fcp1 phosphatase with elongating RNA polymerase II holoenzyme, enzymatic mechanism of action, and genetic interaction with elongator. Journal of Biological Chemistry. 2005;280(6):4299–4306. doi: 10.1074/jbc.M411071200. [DOI] [PubMed] [Google Scholar]
- 231.Hausmann S, Erdjument-Bromage H, Shuman S. Schizosaccharomyces pombe carboxyl-terminal domain (CTD) phosphatase Fcp1. Journal of Biological Chemistry. 2004;279(12):10892–10900. doi: 10.1074/jbc.M312513200. [DOI] [PubMed] [Google Scholar]
- 232.Ghosh A, Shuman S, Lima CD. The structure of Fcp1, an essential RNA polymerase II CTD phosphatase. Molecular Cell. 2008;32(4):478–490. doi: 10.1016/j.molcel.2008.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Abbott KL, Renfrow MB, Chalmers MJ, et al. Enhanced binding of RNAP II CTD phosphatase FCP1 to RAP74 following CK2 phosphorylation. Biochemistry. 2005;44(8):2732–2745. doi: 10.1021/bi047958h. [DOI] [PubMed] [Google Scholar]
- 234.Zhang DW, Mosley AL, Ramisetty SR, et al. Ssu72 phosphatase dependent erasure of phospho-Ser7 marks on the RNA Polymerase II C-terminal domain is essential for viability and transcription termination. doi: 10.1074/jbc.M111.335687. Journal of Biological Chemistry. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Cho H, Kim TK, Mancebo H, Lane WS, Flores O, Reinberg D. A protein phosphatase functions to recycle RNA polymerase II. Genes & Development. 1999;13(12):1540–1552. doi: 10.1101/gad.13.12.1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Dichtl B, Blank D, Ohnacker M, et al. A role for SSU72 in balancing RNA polymerase II transcription elongation and termination. Molecular Cell. 2002;10(5):1139–1150. doi: 10.1016/s1097-2765(02)00707-4. [DOI] [PubMed] [Google Scholar]
- 237.O’Sullivan JM, Tan-Wong SM, Morillon A, et al. Gene loops juxtapose promoters and terminators in yeast. Nature Genetics. 2004;36(9):1014–1018. doi: 10.1038/ng1411. [DOI] [PubMed] [Google Scholar]
- 238.Singh BN, Ansari A, Hampsey M. Detection of gene loops by 3C in yeast. Methods. 2009;48(4):361–367. doi: 10.1016/j.ymeth.2009.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Singh BN, Hampsey M. A transcription-independent role for TFIIB in gene looping. Molecular Cell. 2007;27(5):806–816. doi: 10.1016/j.molcel.2007.07.013. [DOI] [PubMed] [Google Scholar]
- 240.Vinciguerra P, Stutz F. mRNA export: an assembly line from genes to nuclear pores. Current Opinion in Cell Biology. 2004;16(3):285–292. doi: 10.1016/j.ceb.2004.03.013. [DOI] [PubMed] [Google Scholar]
- 241.Köhler A, Hurt E. Exporting RNA from the nucleus to the cytoplasm. Nature Reviews Molecular Cell Biology. 2007;8(10):761–773. doi: 10.1038/nrm2255. [DOI] [PubMed] [Google Scholar]
- 242.Stewart M. Nuclear export of mRNA. Trends in Biochemical Sciences. 2010;35(11):609–617. doi: 10.1016/j.tibs.2010.07.001. [DOI] [PubMed] [Google Scholar]
- 243.Segref A, Sharma K, Doye V, et al. Mex67p, a novel factor for nuclear mRNA export, binds to both poly(A)+ RNA and nuclear pores. The EMBO Journal. 1997;16(11):3256–3271. doi: 10.1093/emboj/16.11.3256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Jani D, Lutz S, Marshall NJ, et al. Sus1, Cdc31, and the Sac3 CID region form a conserved interaction platform that promotes nuclear pore association and mRNA export. Molecular Cell. 2009;33(6):727–737. doi: 10.1016/j.molcel.2009.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Huibregtse JM, Yang JC, Beaudenon SL. The large subunit of RNA polymerase II is a substrate of the Rsp5 ubiquitin-protein ligase. Proceedings of the National Academy of Sciences of the United States of America. 1997;94(8):3656–3661. doi: 10.1073/pnas.94.8.3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Beaudenon SL, Huacani MR, Wang G, McDonnell DP, Huibregtse JM. Rsp5 ubiquitin-protein ligase mediates DNA damage-induced degradation of the large subunit of RNA polymerase II in Saccharomyces cerevisiae. Molecular and Cellular Biology. 1999;19(10):6972–6979. doi: 10.1128/mcb.19.10.6972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Heine GF, Horwitz AA, Parvin JD. Multiple mechanisms contribute to inhibit transcription in response to DNA damage. Journal of Biological Chemistry. 2008;283(15):9555–9561. doi: 10.1074/jbc.M707700200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Buratowski S. The CTD code. Protein Science. 1997;6:249–253. [Google Scholar]
- 249.Baskaran R, Escobar SR, Wang JYJ. Nuclear c-Abl is a COOH-terminal repeated domain (CTD)-tyrosine kinase- specific for the mammalian RNA polymerase II: possible role in transcription elongation. Cell Growth & Differentiation. 1999;10(6):387–396. [PubMed] [Google Scholar]
- 250.Sakurai H, Ishihama A. Level of the RNA polymerase II in the fission yeast stays constant but phosphorylation of its carboxyl terminal domain varies depending on the phase and rate of cell growth. Genes to Cells. 2002;7(3):273–284. doi: 10.1046/j.1365-2443.2002.00522.x. [DOI] [PubMed] [Google Scholar]
- 251.Lewis BA. O-GlcNAc modification of the CTD of human RNA Polymerase II is an intermediate step occurring prior to transcription initiation. unpublished data. [Google Scholar]
- 252.West ML, Corden JL. Construction and analysis of yeast RNA polymerase II CTD deletion and substitution mutations. Genetics. 1995;140(4):1223–1233. doi: 10.1093/genetics/140.4.1223. [DOI] [PMC free article] [PubMed] [Google Scholar]


