Abstract
Prolyl-tRNA synthetases (ProRSs) have been shown to activate both cognate and some noncognate amino acids and attach them to specific tRNAPro substrates. For example, alanine, which is smaller than cognate proline, is misactivated by Escherichia coli ProRS. Mischarged Ala-tRNAPro is hydrolyzed by an editing domain (INS) that is distinct from the activation domain. It was previously shown that deletion of the INS greatly reduced cognate proline activation efficiency. In the present study, experimental and computational approaches were used to test the hypothesis that INS deletion alters the internal protein dynamics leading to reduce catalytic function. Kinetic studies with two ProRS variants, G217A and E218A, revealed decreased amino acid activation efficiency. Molecular dynamics studies showed motional coupling between the INS and protein segments containing the catalytically important proline-binding loop (PBL, residues 199–206). In particular, the complete deletion of INS, as well as mutation of G217 or E218 to alanine, exhibited significant effects on the motion of the PBL. The presence of coupled-dynamics between neighboring protein segments was also observed through in silico mutations and essential dynamics analysis. Taken together, the present study demonstrates that structural elements at the editing domain-activation domain interface participate in coupled motions that facilitate amino acid binding and catalysis by bacterial ProRSs, which may explain why truncated or defunct editing domains have been maintained in some systems, despite the lack of catalytic activity.
Prolyl-tRNA synthetases (ProRSs) are class II synthetases that catalyze covalent attachment of proline to the 3´-end of the tRNAPro in a two-step reaction:
| (i) |
| (ii) |
ProRSs from all three kingdoms of life are known to misactivate noncognate alanine and cysteine resulting in mischarged tRNAPro (1–3). To maintain high fidelity in protein synthesis, some ProRSs have acquired editing mechanisms to prevent misaminoacylation of tRNAPro (1, 2, 4). Based on sequence alignments, ProRSs are classified into two broad groups – “eukaryotic-like” and “prokaryotic-like” (5, 6). Escherichia coli (Ec) ProRS, a representative member of the prokaryotic-like group, is a multi-domain protein. The catalytic domain (motifs 1, 2 and 3; residues 64–81, 128–164, and 435–465) catalyzes the activation of proline and the aminoacylation of tRNAPro; the anticodon binding domain (residues 506–570) is critical for recognition of cognate tRNA; the insertion domain (INS; residues 224–407, located between motifs 2 and 3 of the catalytic domain) is the post-transfer editing active site that hydrolyzes mischarged Ala-tRNAPro (7, 8). In contrast, Cys-tRNAPro is hydrolyzed by a free-standing editing domain known as YbaK present in some species (9, 10). Unlike prokaryotic-like ProRSs, eukaryotic-like ProRSs do not possess the INS but in some cases encode free-standing editing domain homologs.
In addition to post-transfer editing, the INS of Ec ProRS was found to have a significant impact on amino acid binding and activation (11). Deletion of the INS (residues 232–394) of Ec ProRS resulted in a 200-fold increase in the KM for proline. The overall proline activation efficiency was reduced by ~1200-fold relative to the wild-type (WT) enzyme (11). Although the specific reason for this drastic effect is not understood, circular dichroism measurements demonstrated that deletion of the INS has no significant effect on the overall folding of the mutant protein (11). Thus, it remains unclear what role the editing domain plays in substrate binding and amino acid activation.
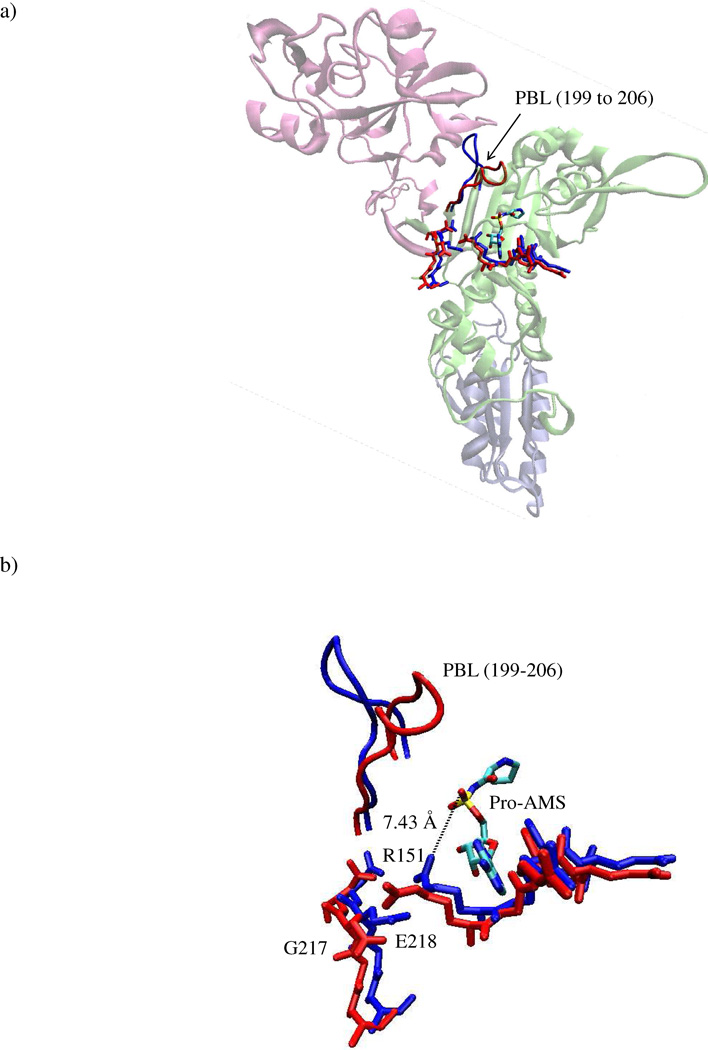
It is known that for multi-domain proteins like ProRS, coupled domain dynamics play an important role in catalytic function (12, 13). Although the relevance of the editing domain to amino acid activation by ProRS is not understood, a substrate-induced conformational change of a neighboring loop, known as the proline-binding loop (PBL, residues 199–206), was revealed by structural studies (14). Three-dimensional structures of two bacterial ProRSs – Rhodopseudomonas palustris ProRS (Rp ProRS) and Enterococcus faecalis ProRS (Ef ProRS, Fig. 1a) showed an induced-fit binding mode with a large displacement (~ 7 Å) of the PBL upon binding of the prolyl-adenylate analog, 5’-O-[N-(prolyl)-sulfamoyl] adenosine (Pro-AMS) (Fig. 1b) (14). Comparison of the substrate bound and unbound structures also showed that the large displacement of the PBL was associated with the re-orientation of several active site moieties, as well as some polypeptide segments that belong to the catalytic domain-editing domain interface (14). These observations together with the observed dramatic change in Ec ProRS function upon deletion of the editing domain, led us to hypothesize that the dynamics of structural elements proximal to the PBL influence substrate binding and catalysis by prokaryotic-like ProRSs.
Figure 1.
a) Cartoon representation of the 3-dimensional structure of the monomeric form of Ef ProRS (residues 1–570, PDB entry: 2J3L, chain B). The structural domains are colored as follows: lime, catalytic domain (residues 1–223 and 408–505); mauve, editing domain (residues 224–407); iceblue, anticodon-binding domain (residues 506–570). The PBL is shown in tube representation. G217, E218, R151, and the prolyl-adenylate analog are shown in licorice representation; “closed” state, red “open” state, blue. b) Closer view of the PBL and the active site residues.
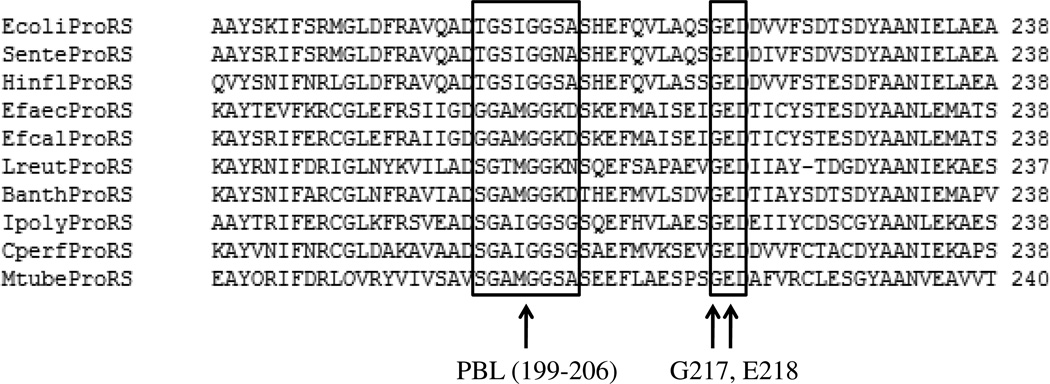
To test the above hypothesis, in the present study the coupling of motions among various structural elements of Ec ProRS was investigated using computational and experimental approaches. In particular, to examine the effect of INS on the PBL dynamics, the motion of the full-length and the truncated enzyme (constructed by deletion of INS, hereafter referred to as ΔINS) was computationally simulated. Also, two highly conserved residues of the prokaryotic-like ProRS family, G217 and E218 (Fig. 2), were mutated. These two residues, located at the junction of the activation domain and the editing domain, are not directly involved in catalysis but undergo substrate-induced conformational changes (14). To evaluate the effect of mutation of these noncatalytic conserved residues on PBL dynamics and enzyme catalysis, enzyme motions were computationally simulated and kinetic parameters were determined experimentally. Taken together, the results of this study shed light on the role of distant domains and noncatalytic residues in producing a catalytically competent state for amino acid binding and activation by prokaryotic-like ProRSs.
Figure 2.
A portion of the multiple sequence alignment of 10 prokaryotic-like ProRSs; the PBL and the highly conserved 217GED219 motif are indicated by rectangular boxes.
MATERIALS and METHODS
All experimental studies were performed using purified Ec ProRS. Since Ec and Ef ProRS possess high sequence identity (48%), computational studies were performed starting with the X-ray crystallographic structure of Ef ProRS [PDB entry codes: 2J3M (“open” state)] (14) and compared with results using a homology model of Ec ProRS developed using Ef ProRS as a template (provided by S. Cusack). All simulations were performed with apoenzymes.
EXPERIMENTAL METHODS
Materials
All amino acids (Sigma) were of highest quality (>99% pure) and used without further purification. Tritiated proline (83 Ci/mmol) and alanine (75 Ci/mmol) were from Perkin Elmer. Primers for site-directed mutagenesis and PCR were from Integrated DNA Technologies.
Enzyme Preparation
Overexpression and purification of histidine-tagged WT and mutant Ec ProRS was performed as described previously (15, 16). Plasmids encoding G217A and E218A Ec ProRS were generated by QuikChange mutagenesis (Stratagene) of pCS-M1S (16) using the following primers: G217A – top, 5’-GCG CAG AGC GCG GAA GAC GAT GTG G-3’ and bottom, 5’-CCA CAT CGT CTT CCG CGC TCT GCG C-3’; E218A – top, 5’-GCG CAG AGC GGT GCG GAC GAT GTG G-3’ and bottom, 5’-CAA CAT CGT CCG CAC CGC TCT GCG C-3’. Results of mutagenesis were confirmed by DNA sequencing (University of Wisconsin, Biotechnology Center, Madison). Protein expression was induced in Ec SG13009 (pREP4) competent cells with 1 mM isopropyl β-D-thiogalactoside for 4 h at 37°C. Histidine-tagged proteins were purified using a Talon cobalt affinity resin and the desired protein was eluted with 100 mM imidazole. Protein concentrations were determined initially by the Bio-Rad Protein Assay (Bio-Rad Laboratories) followed by active-site titration (17).
RNA Preparation
Ec tRNAPro was transcribed using T7 RNA polymerase from BstN1-linearized plasmid as described (18), and purified by denaturing 12% polyacrylamide gel electrophoresis.
ATP-PPi Exchange Assays
The ATP-PPi exchange assay was performed at 37 °C according to the published method (19). The concentrations of proline and alanine ranged from 0.025 – 50 mM and 1– 850 mM, respectively. The enzyme concentrations used were 10–20 nM for proline and 250–500 nM for alanine activation. Kinetic parameters were determined from Lineweaver-Burk plots and represent the average of at least three determinations.
ATP Hydrolysis Assays
ATP hydrolysis reactions to monitor pre-transfer editing were carried out as described previously (11). Alanine concentration used was 500 mM and proline concentration was 30 mM. The reactions were initiated with a final ProRS concentration of 0.5 µM.
Aminoacylation Assays
Aminoacylation assays were performed under standard conditions (20) with 0.5 µM tRNAPro, 13.3 µM [3H] proline, and 100 nM ProRS.
Aminoacylated tRNA
Aminoacylated tRNA for use in deacylation assays was prepared at room temperature according to published conditions (1). Ec AlaRS (2 µM) was used to acylate G1:C72/U70 tRNAPro (8 µM) in the presence of [3H]Ala (7.3 µM) in buffer containing 50 mM HEPES (pH 7.5), 4 mM ATP, 25 mM MgCl2, 20 mM β-mercaptoethanol, 20 mM KCl and 0.1 mg/ml bovine serum albumin.
Deacylation Assays
Deacylation assays were carried out at room temperature according to published conditions (1). Reactions contained 1 µM G1:C72/U70 [3H] Ala-tRNAPro, 150 mM KPO4 (pH 7.0), 5 mM MgCl2, and 0.1 mg/ml bovine serum albumin. The reactions were initiated with 5 µM ProRS. Negative controls were performed using 150 mM KPO4 (pH 7.0) in place of ProRS.
COMPUTATIONAL METHODS
Molecular Dynamics Simulations
MD simulations were carried out starting with the crystallographic structure of Ef ProRS [chain B; PDB entry code: 2J3M ("open", residues 19–565)]. The ΔINS [constructed by replacing the INS residues 232–394 with a 16-residue Gly12 Ser4 linker (11)] and the three mutants (G217A, E218A, and E218D) were generated with the Mutator plug-in of Visual Molecular Dynamics (VMD) 1.8.6 (21). For all simulations, the all-atom CHARMM22 force field (22) was used within the NAMD (23) package. The three-point charge TIP3P model (24) was used to represent solvent water. Non-bonded interactions were truncated using a switching function between 10 and 12 Å, and the dielectric constant was set to unity. The SHAKE algorithm (25) was used to constrain bond lengths and bond angles of water molecules and bonds involving a hydrogen atom. The MD simulations were performed using isothermal-isobaric (NPT) conditions. Periodic boundary conditions and particle-mesh Ewald methods (26) were used to account for the long-range electrostatic interactions. In all MD simulations, a time step of 2 fs was used. The pressure of the system was controlled by the implementation of the Berendsen pressure bath coupling (27) as the temperature of the system was slowly increased from 100 to 300 K. During the simulations at 300K, the pressure was kept constant by applying the Langevin piston method (28, 29).
The WT and mutant proteins were solvated with water in a periodic rectangular box of dimensions 130 Å × 78 Å × 92 Å with water padding of 12 Å between the walls of the box and the nearest protein atom. The charge neutralization (with sodium ions) of the solvated system was performed with the VMD autoionize extension (21). The resultant systems, containing ~ 84,000 atoms (~74,000 atoms for ΔINS ProRS) including approximately 16,450 water molecules and 33 sodium ions (32 and 14 ions for E218A and ΔINS ProRS, respectively), were equilibrated by slightly modifying previously described procedures (30, 31). Briefly, solvated proteins were further subjected to 1000 steps of conjugate-gradient minimization at 100 K. The temperature of the solvated systems was then increased to 300 K in 3000 steps and was further equilibrated at 300 K for 500 ps. The equilibrated system was then used in 12-ns simulations. The equilibration and stability of the dynamics were checked by calculating the root-mean-square deviations (RMSD) of Cα atoms from their initial coordinates.
Essential Dynamics
The collective dynamics of the protein was studied through essential dynamic analysis (32–34), which involves computation of the principal components of atomic fluctuations. The last 7 ns of the 12 ns MD simulation data was used to extract the principal modes of collective dynamics (called principal components) using the program Carma (35). The mathematical operation behind essential dynamics is called principal component analysis (PCA), which takes a data set from a trajectory of a long time-scale MD simulation as input and extracts the low-frequency (high-amplitude) collective motions of the biomolecule, which are often more relevant for its functions (36). The principal components were computed by performing eigenvalue decomposition of a covariance matrix, and the mathematical formulism is described elsewhere (37). Briefly, the covariance matrix, C is computed with elements Cij for any two points (Cα coordinates) i and j using
| (1) |
where x1, x2 …, x3N are the mass-weighted Cartesian coordinates of an N-particle system and the angular brackets represent an ensemble average calculated over all sampled structures from the simulations. Next, the symmetric 3N×3N matrix C can be diagonalized with an orthonormal transformation matrix R
| (2) |
where c1,c2,…c3N are eigenvalues; columns in the transformation matrix R are eigenvectors, which are also called the principal modes. If X(t) represents the time-evolved coordinates (trajectory) of the water-encapsulated protein active site, it can be projected onto the eigenvectors,
| (3) |
The projection is a measure of the extent to which each conformation is displaced, in the direction of a specific principal mode, and is called principal component (PC). For a trajectory, the projections are obtained as matrix elements qi(t), i = 1, 2, …, M.
PCA was carried out using the following steps: (i) preparing a modified trajectory file by removing the coordinates of the water molecules, selecting only the Cα atoms and removing the overall translational and rotational motions, (ii) calculating the covariance matrix in which the atomic coordinates are the variables, and (iii) diagonalizing the covariance matrix for calculation of the eigenvectors and the corresponding eigenvalues. The first three PCs were used for performing PCA-based cluster analysis as discussed in Carma documentation (35). Briefly, based on contributions of the first three PCs, conformations in the overall trajectory were grouped into several clusters. The cluster with the greatest number of conformations is representative of predominant conformational fluctuations and was used for further analysis of dynamic cross-correlations between Cα atoms. The cross-correlation coefficient between fluctuations of residues i and j (CCij) was calculated using
| (4) |
where σxi and σxj represent the standard deviation of the displacements of the two points (Cα coordinates) i and j, respectively. The correlated motion (CCij > 0) between two Cα atoms occurs when they move in the same direction while the anticorrelated motion is generated when two Cα(CCij < 0) atoms move in opposite direction.
The root-mean-square projections (RMSP) of q were obtained from the last 7 ns of the simulations using the following equation:
| (5) |
To determine if the functional dynamics had undergone significant change due to a single point mutation, a combined essential dynamics analysis was performed following literature methods (32–34). In this procedure, a comparison of dynamics of five protein systems was carried out by concatenating their trajectories to produce a combined covariance matrix. The separate trajectories were then projected onto the resulting eigenvectors and the properties of these projections were compared for these simulations.
RESULTS
The results are presented in the following order. First, the experimental results are reported to show the impact of mutation of the two strongly conserved noncatalytic residues on the enzyme function. Next, the results of the MD simulations are presented to illustrate the flexibility of the ProRS and the overall coupling of various structural elements surrounding its catalytic site. Finally, the molecular-level impact of mutations (deletion and site-directed mutations) on the catalytically-important PBL dynamics was characterized through essential dynamics analysis.
Activation of Proline and Alanine
To investigate the role of the 217GED219 motif in maintaining coupled motions among the protein segments surrounding the synthetic active site, the effect of mutation of G217 and E218 on the function of the enzyme was experimentally tested. The kinetic parameters for proline and alanine activation were determined for both mutants and compared with WT Ec ProRS. We found that E218A ProRS activates proline but with a decreased kcat (3-fold) and elevated KM value (15-fold, Table 1). Overall proline activation efficiency of this mutant was decreased 45-fold compared to the WT enzyme. Reduced catalytic efficiency for proline activation was also observed for the G217A mutant. The kcat/KM of G217A ProRS was reduced 7-fold relative to the WT enzyme (Table 1). In contrast, alanine activation by the G217A mutant was not affected compared to the WT enzyme, and only a 2-fold decrease in alanine activation was observed for the E218A mutant (Table 1).
Table 1.
Kinetic Parameters for Amino Acid Activation by WT, E218A and G217A Ec ProRSa
| Amino Acid | kcat | KM | kcat/KM | kcat/KM | Fold Change | |
|---|---|---|---|---|---|---|
| sec−1 | mM | sec−1mM−1 | relative | |||
| WT | Proline | 12.7 ± 4.9 | 0.228 ± 0.028 | 55.7 | 1 | - |
| Alanine | 3.52 ± 2.1 | 685 ± 360 | 0.00513 | 1 | - | |
| E218A | Proline | 4.4 ± 2.2 | 3.40 ± 0.68 | 1.29 | 0.0232 | 43 |
| Alanine | 3.26 ± 5.4 | 1360 ± 1300 | 0.0024 | 0.468 | 2 | |
| G217A | Proline | 3.37 ± 1.1 | 0.427 ± 0.077 | 7.89 | 0.142 | 7 |
| Alanine | 2.18 ± 0.23 | 454 ± 78 | 0.0048 | 0.935 | 0 | |
Results are the average of 3 trials with the standard deviation indicated. In each, the kcat/KM of the mutant is relative to the WT kinetics with the corresponding amino acid.
Aminoacylation of tRNAPro
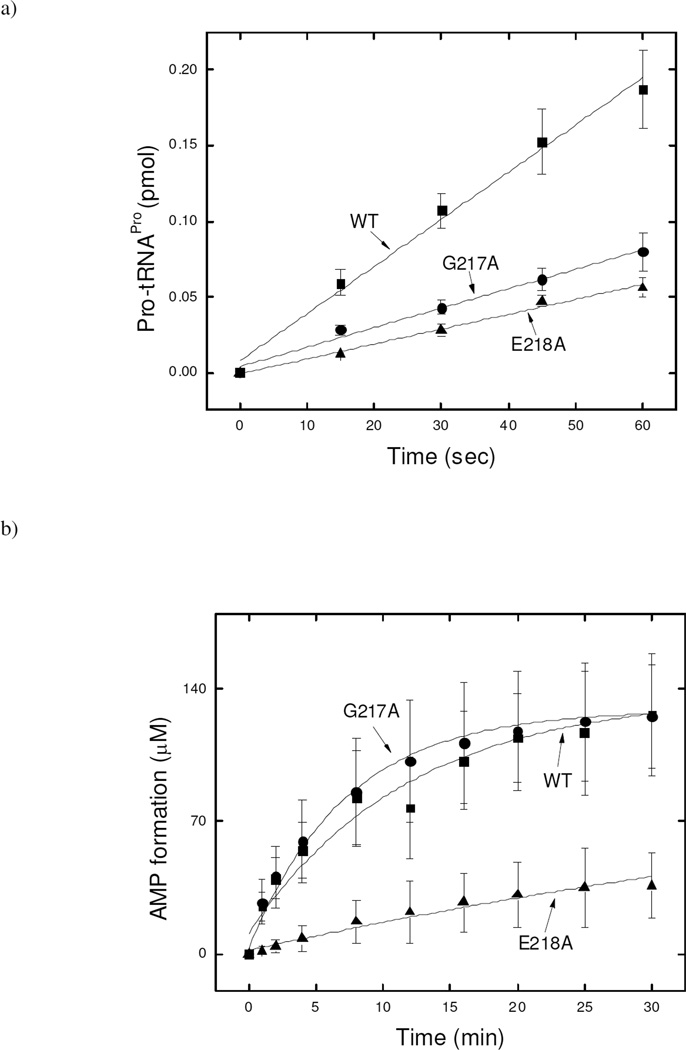
The effect of mutation of G217 and E218 on aminoacylation of proline was also tested. Both G217A and E218A can charge proline onto tRNAPro, albeit with 3-fold reduced efficiency (Fig. 3a).
Figure 3.
a) Aminoacylation of tRNAPro with proline by WT, G217A, and E218A Ec ProRS. The assay was performed at 37 °C with 0.5 µM tRNAPro and 100 nM Ec ProRS. b) Pre-transfer editing in the presence of alanine by WT, G217A, E218A Ec ProRS. The assay was performed at 37 °C using 0.5 µM ProRS and 500 mM alanine. Lines are single exponential fits of the data.
Pre-transfer Editing
Stimulation of ATP hydrolysis is considered indicative of pretransfer editing, presumably because the noncognate amino acids that are hydrolytically edited are repeatedly reactivated by the synthetase, consuming ATP in each cycle (38). In contrast, the cognate amino acid is bound to the synthetase until transferred to the tRNA. Ec ProRS possesses tRNA independent pre-transfer editing against alanine (39). Here, we tested the pre-transfer editing activity of the two mutant proteins and compared them with the WT activity. ATP hydrolysis was stimulated in the presence of alanine for both mutants. However, E218A ProRS exhibited reduced activity (9-fold) compared to the G217A variant, which possessed editing activity that was comparable to the WT enzyme (Fig. 3b). The reduced activity of E218A ProRS may, in part, be due to its poor alanine activation efficiency.
Post-transfer Editing
The post-transfer editing activity of WT and variant ProRSs was also tested by monitoring the hydrolysis of misacylated Ala-tRNAPro. All three enzymes exhibited similar post-transfer editing activity (Fig. S1). Thus, the binding of the mischarged tRNA in the editing active site and the hydrolysis of the ester bond were not affected by mutations at the editing domain-activation domain interface.
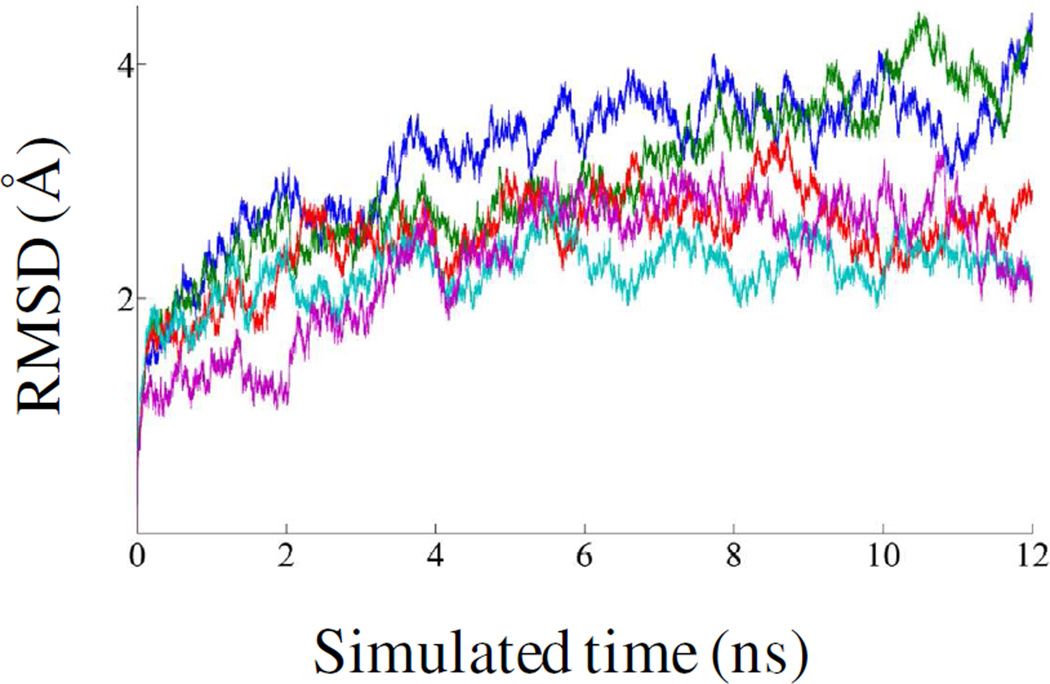
Root-Mean-Square Deviation (RMSD) Profiles
RMSDs were calculated using 12 ns MD simulation data for WT, G217A, E218A, E218D and ΔINS ProRS systems. The plots of RMSD with respect to the initial equilibrated structure are shown in Fig. 4. After ~5 ns simulations, the Cα RMSD values remained within ~1 Å. The last 7 ns simulations data were used for further study.
Figure 4.
RMSD of the Cα atoms from their initial coordinate as a function of time. Calculations of RMSDs for WT (blue), G217A (green), E218A (red), E218D (cyan) and ΔINS (purple) Ef ProRS were performed using 12-ns MD simulation data.
Flexible Regions
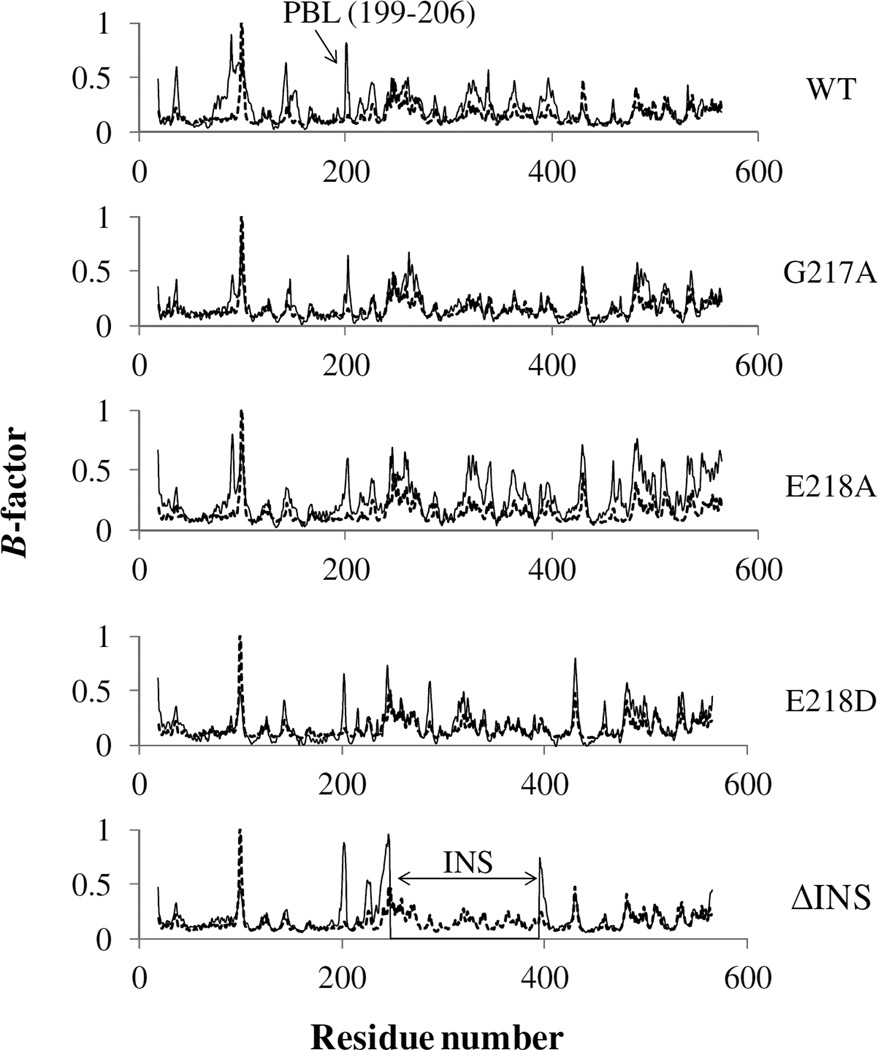
The B-factors analysis revealed several highly flexible regions in Ef ProRS. A plot of normalized experimental B-factors [crystallography, (14)] and calculated B-factors [using Carma (35)] of the Cα atoms of the WT Ef ProRS is shown in Fig. 5. The flexible regions identified by both experimental and computational methods are comparable, except for residues 75–125 and the PBL. It appears that the flexibility of these two regions is experimentally underestimated, possibly due to the crystal packing arrangement of the protein.
Figure 5.
Comparison of the normalized Cα B-factors obtained from the crystal structure (gray, dotted line; PDB entry: 2J3M, chain B) and calculated from MD simulation data (black, solid line). For ΔINS, the calculated B-factors are missing for residues 232–394.
In the case of the two mutants obtained by conservative mutation, G217A and E218D, the overall protein flexibility was reduced compared to the WT enzyme (Fig. 5). However, the substitution of E218 with alanine resulted in increased flexibility of the protein backbone, especially for the Cα atoms of the INS and C-terminal domain. Interestingly, in all the three mutants (G217A, E218A, and E218D) the flexibility of the PBL was reduced compared to the WT protein. On the other hand, the complete deletion of the INS resulted in a less flexible protein with B-factors almost comparable to the experimentally observed results except for the PBL, which becomes more flexible in the absence of the INS (Fig. 5).
Dynamics Cross-Correlations and Essential Dynamics Analyses
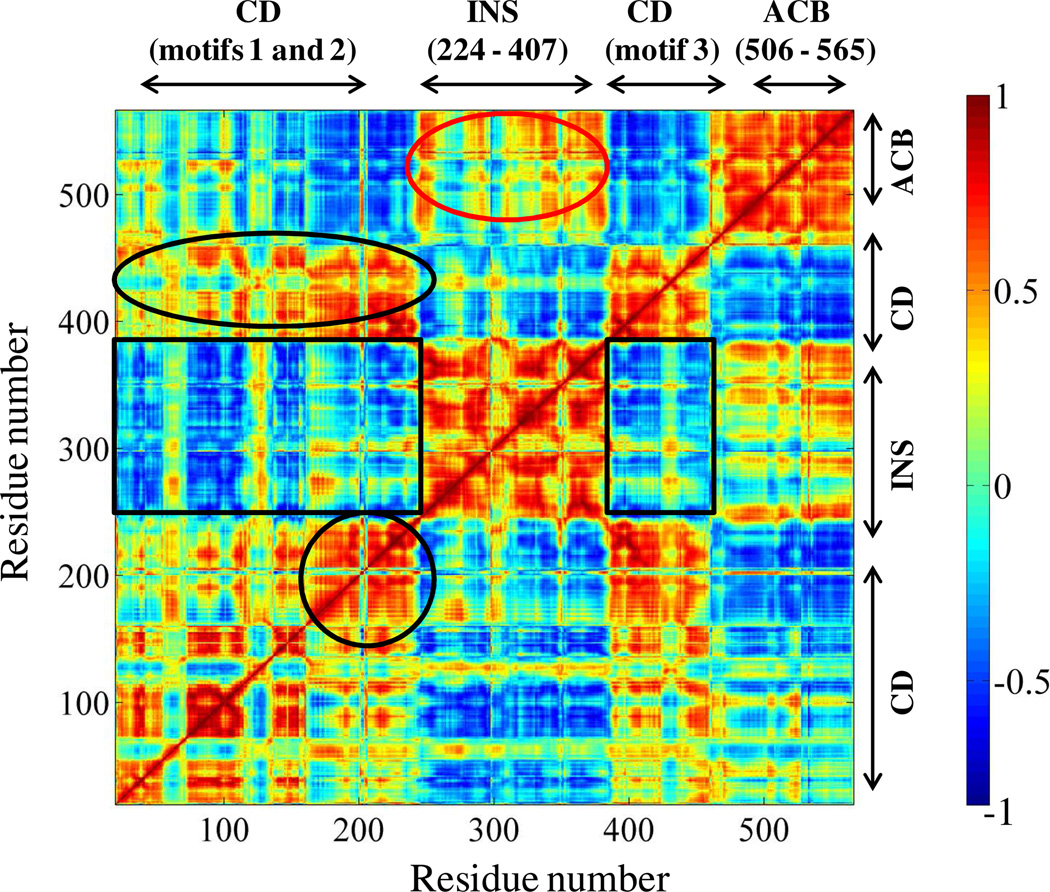
The dynamic cross-correlation map obtained from the MD simulation of Ef ProRS (chain B) is shown in Fig. 6. In the present study, the dynamic cross-correlation matrix was generated using the first 3-PCs. Analysis of the cross-correlation of fluctuations of residues for the first 3 PCs revealed both inter- and intra-domain motional correlation. It was found that the activation domain and the INS are mainly engaged in anticorrelated motions i.e. their displacements are in opposite direction (CCij < 0; Fig. 6, black rectangles). An anticorrelated pattern of motions was also observed between the catalytic domain residues and the anticodon binding elements of Ef ProRS. On the other hand, the motion of the editing domain and the anticodon binding domain is weakly correlated (CCij > 0; Fig. 6, red oval).
Figure 6.
Dynamic cross-correlations between the Cα atoms of Ef ProRS obtained from the cluster analysis and PCA. A value of +1.0 was set for strongly correlated motion and is colored red, whereas −1.0 was used for strongly anticorrelated motions and is colored blue. The boxed and circled regions are discussed in the text. The abbreviations used are: CD, catalytic domain; INS, insertion domain; and ACB, anticodon binding domain.
Various structural elements within the catalytic domain, which are essential for substrate binding and catalysis, are engaged in correlated motions (Fig. 6, black oval). As expected, the adjacent residues of the protein segment (residues 190–220) containing the PBL and the 217GED219 motif are engaged in strong correlated motion (Fig. 6, black circle). Also, the motion of the PBL containing protein segment is mostly correlated in nature with respect to motifs 1–3 of the catalytic domain. However, its motion is anticorrelated with respect to the INS and the anticodon binding domain.
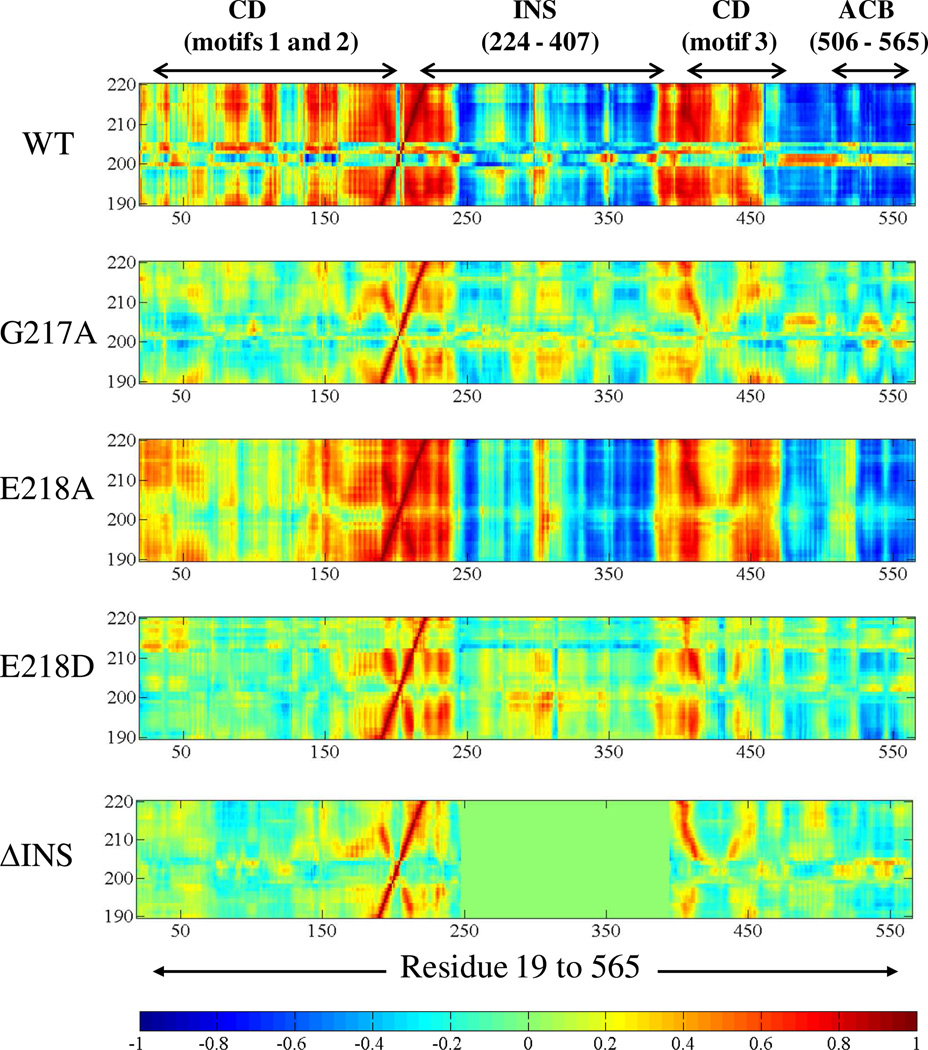
The effects of deletion of INS as well as point mutations of G217 and E218 on Ef ProRS dynamics were also studied. The dynamic-cross-correlation map of the atomic (Cα) fluctuations between the PBL containing protein segments (residues 190–220) and the rest of the molecule for the WT and the mutant variants is shown in Fig. 7. Although, we cannot rule out the change in structure due to these mutations (site-directed/deletion), noticeable alteration in residue fluctuations between the PBL containing protein segment and other structural elements of the protein was observed for all mutant proteins compared to the WT enzyme (Fig. 7). In particular, a significant change in the motional coupling between the PBL containing segment and the editing domain (residues 224–407) was observed for the two ProRS variants, G217A and E218D. In addition, noticeable alteration in dynamic coupling among residues of the entire PBL containing segment (residues 190–235) and residues 150–235 as well as anti-codon binding domain was observed (Fig. 7).
Figure 7.
Dynamic cross-correlations between the Cα atoms of the PBL containing protein segment (residues 190–220) versus Cα atoms of residues 19–565 of the WT and mutant ProRSs. Color coding is as described in Figure 6. For ΔINS, region for the cross-correlations between residues 190–220 and the INS residues 247–394 is shown in green rectangle. Residues 232–394 are replaced with a 16-residue linker in this plot.
Combined Essential Dynamics
To examine the impact of the deletion of INS or point mutation in the 217GED219 motif on the collective dynamics of the PBL, the essential dynamics of WT Ef ProRS and mutant variants were analyzed using the last 7 ns of the 12 ns MD simulation data. Specifically, we performed a “combined” essential dynamics analysis (32, 33), using the concatenated trajectories (of the Cα atoms) of all the five proteins (WT, ΔINS, G217A, E218A, and E218D).
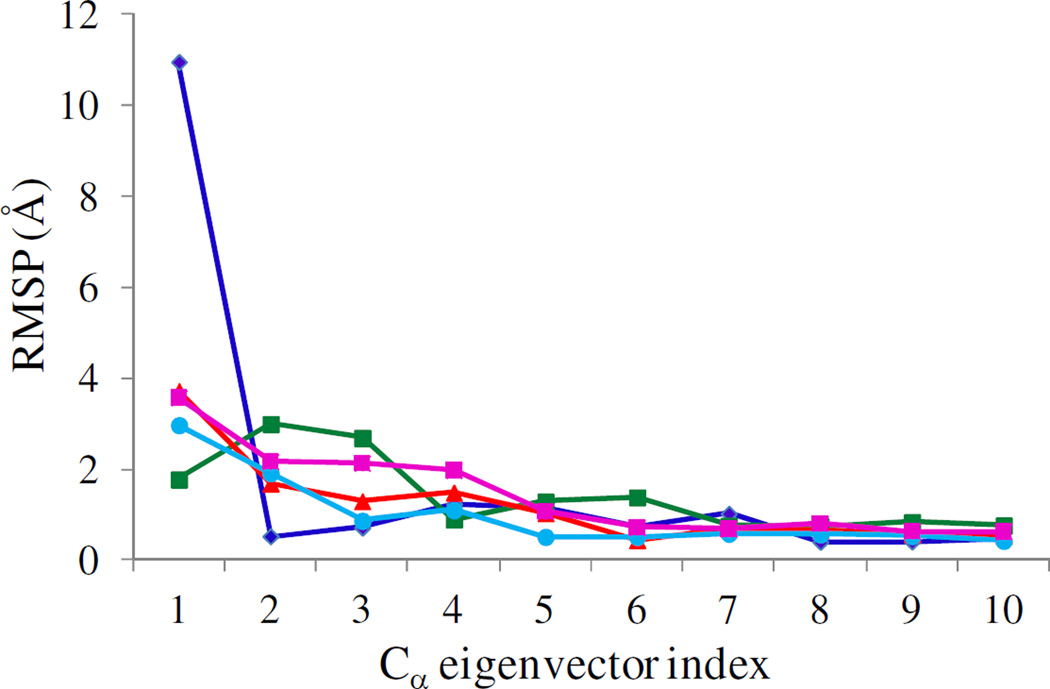
The combined essential dynamics analysis clearly shows that each mutation has impact on the collective PBL (residues 190–210) dynamics. The RMSP’s (eq. 5) as a function of eigenvector indices for the WT and mutant proteins of Ef ProRS are shown in Fig. 8. The fluctuation of the PBL along PC1 is significantly altered for all the mutant proteins compared to the WT enzyme. Noticeable changes were also observed for PC2 and PC3. Therefore, this analysis indicates that the deletion of the INS or mutation at the junction of the INS and activation domain could impact PBL dynamics and potentially alter substrate binding. Similar differences in the slow dynamics of the PBL upon mutation of G217 and E218 to alanine were observed for Ec ProRS (Fig. S2).
Figure 8.
Combined analysis of the computed root-mean-square projections (RMSP, eq. 5) over the last 7 ns of 12-ns simulation data for eigenvectors 1 to 10 for WT (blue), G217A (green), E218A (red), E218D (cyan) and ΔINS (purple) Ef ProRS.
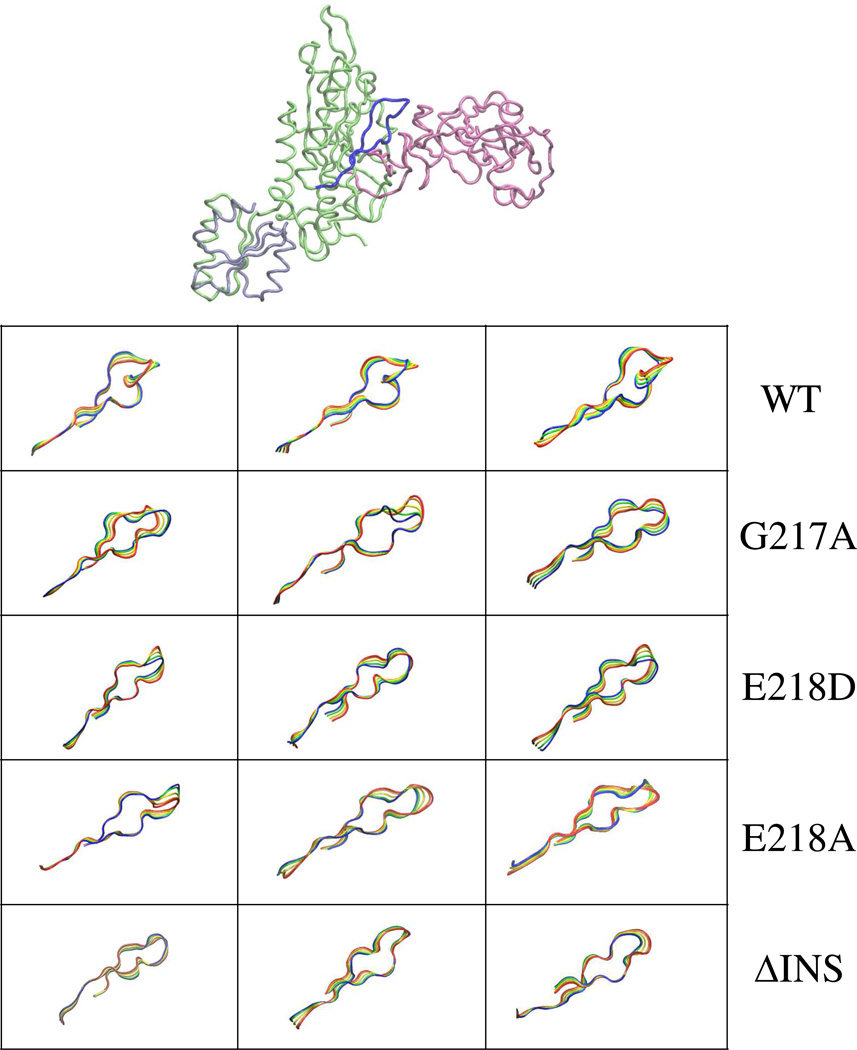
The alteration in the dynamics of the PBL either due to the deletion of the INS or mutations in the 217GED219 motif can be visualized from the superimposition of conformations of the PBL extracted from the essential dynamics analysis. These superimposed conformations correspond to the dynamics of the PBL along the three PCs (i.e. in the direction of collective dynamics) and are displayed in Fig. 9. Only backbone Cα atoms are shown for clarity. In the Cα traces, it is apparent that the pattern of the collective dynamics of the PBL (along the first 3 PCs) was altered by the point mutation at the domain-domain interface as well as by the deletion of the INS. Taken together, combined essential dynamics analysis revealed that deletion of INS or point mutations at the catalytic domain-editing domain junction caused perceptible changes in the collective PBL dynamics.
Figure 9.
Visual representation of the movement of the PBL. Superposition of 4 configurations extracted from the concatenated trajectories by projecting the Cα motion onto eigenvectors 1–3. The four conformations are represented in blue (starting)→green→yellow→red (end).
DISCUSSION
Protein Dynamics and Catalysis
Dynamics is an intrinsic property, encrypted in the three dimensional structure and folding of a protein. Collective dynamics are prevalent in modular proteins and they play an important role in enzyme function. In fact, simulations of mechanochemical properties of enzymes have shown that coupling between catalytic function and collective dynamics is a prerequisite for enzyme activity (40). Several other studies have also revealed that internal motions essentially represent the intrinsic mechanical properties of an enzyme and do not originate because of the presence of a substrate. Nevertheless, these internal protein motions facilitate substrate recognition and binding, and thereby promote catalysis (41–43). In addition, studies have demonstrated that protein motions can modify the catalytic rate by influencing the height of the activation free energy barrier and the transmission coefficient (i.e., the capacity of recrossing the barrier) (44–46). For example, a direct correlation between the frequencies of enzyme motions and catalytic turnover rates was observed in cyclophilin A using NMR relaxation experiments (46).
A number of studies indicate that internal protein motions involve networks of residues extending beyond the catalytic site (41, 44, 45). Enzyme catalysis is found to be augmented by coupled motion through these networks amidst growing evidence that the slower collective protein motions and the faster bond breaking/forming motions are connected. An example of such a synergistic relationship can be found in adenylate kinase (47), where faster (pico-to nanosecond timescale) atomic fluctuations at the hinge regions were found to promote the large-scale displacement of the lid during substrate binding. Also, studies of several enzymes including dihydrofolate reductase and liver alcohol dehydrogenase (42, 45, 48) have demonstrated that mutations of non-catalytic residues alter their catalytic function by modifying internal enzyme motions. Taken together, there is an overwhelming amount of evidence showing the significance of coupled dynamics in enzyme function. The role of coupled dynamics in ProRS structure/function has remained unexplored and constitutes the basis of the present investigation.
Proposed Role of the Editing Domain
To probe the hypothesis that the collective dynamics involving the editing domain regulates substrate binding and catalysis by ProRS, the motion of ΔINS construct was compared with the full-length WT enzyme. In addition, two non-catalytic but conserved residues (G217 and E218) in the editing domain-activation domain junction were chosen for mutagenesis. If coupled internal dynamics truly exists between structural elements in the vicinity of the PBL, then point mutations in any of these elements should alter the dynamics, as well as the efficiency of catalysis.
Amino Acid Activation and Aminoacylation
Experimental studies show that G217 and E218 are critical for enzyme catalysis. The X-ray crystal structure of bacterial ProRS shows strong interactions between E218 and a conserved arginine residue (R151 of Ef ProRS, see Fig. 1b) that helps to stabilize the phosphate group of the substrate ATP molecule (14). Indeed, a 45-fold decrease in proline activation was measured in the case of E218A ProRS, showing that this residue is critical for cognate amino acid activation. However, only a small decrease (~2-fold) in alanine activation efficiency was observed for this mutant. A 7-fold decrease in proline activation efficiency upon mutation of G217 to alanine was observed, although this residue does not interact directly with any catalytic site residues. Lack of a significant effect on alanine activation for the E218A and G217A variants suggests that these residues might aid in maintaining the internal dynamics of the active-site protein segments and the PBL, which facilitates the binding of the cognate amino acid but plays a more minor role in non-cognate alanine activation. This is also apparent from the fact that the kcat for proline activation by E218A ProRS was only reduced 3-fold, whereas the KM was elevated 15-fold.
The mutation of G217 and E218 to alanine also impacted cognate tRNA aminoacylation (Fig. 3a), although, the impact was less severe (~2- to 3-fold) than for amino acid activation. This observation suggests that the binding of the 3′-acceptor end in the aminoacylation active site was not altered significantly by the alanine substitutions.
Role of PBL in Amino Acid Selection
If the “open” to “closed” conformational transition of the PBL is important for the protection of the cognate aminoacyl-adenylate from spontaneous hydrolysis by the surrounding water, the mutation of G217 and E218 to alanine may be expected to enhance Pro-AMP hydrolysis. However, ATP hydrolysis was only slightly stimulated in the presence of proline for G217A and E218A mutants (Fig. S1(a)), suggesting that the main role of the PBL is to facilitate amino acid selection and binding. Moreover, no noticeable difference in post-transfer editing activity was observed for these mutants relative to the WT enzyme (Fig. S1(b)) demonstrating that mutations in the 217GED219 motif do not affect binding and hydrolysis of misacylated tRNAPro.
Flexibility and Collective Protein Dynamics
The B-factor calculations performed on the Ef ProRS demonstrated that the PBL is quite flexible (Fig. 5). However, the flexibility of this loop was altered by the mutation of G217 and E218. As expected, mutation of G217 to alanine brought some rigidity in the PBL dynamics. On the other hand, mutation of E218 to alanine caused an increase in mobility of the whole protein backbone but had reduced the flexibility of the PBL. The increased mobility of the protein backbone is expected as the substitution of E218 to alanine disrupted the electrostatic interaction between E218 and R151 of the activation domain (Fig. 1b). Interestingly, the mutation of E218 to aspartic acid resulted in overall reduction in protein flexibility. Close scrutiny of the E218D structure revealed existence of some additional H-bond interactions between the surrounding residues and the aspartic acid, which might have brought some extra rigidity into the structure (data not shown). However, the deletion of the INS has the reverse effect on the PBL flexibility. Apparently, the PBL that is essential for substrate binding and catalysis acquired significant flexibility upon deletion of the INS (Fig. 5). This observation suggests that the INS might have a role in maintaining the optimum flexibility of the PBL.
The cross-correlation matrix obtained from the cluster analysis (eq.4) revealed that the editing domain is mainly engaged in anticorrelated motion with the central activation domain (Fig. 6). The existence of anticorrelated motion between these two domains may be critical for providing adequate space for the 3'-end of a tRNA to enter the synthetic active site for aminoacylation. Anticorrelated motion between the editing and activation domains has also been observed in other synthetase systems including isoleucyl- and leucyl-tRNA synthetases (49, 50). Close analysis of the dynamic cross-correlation matrix also revealed the existence of correlated motion among several polypeptide segments within the activation domain. In addition, the adjacent residues of the polypeptide segment that includes both the PBL and the 217GED219 motif (residues 195–225), are found to be engaged in correlated motion among themselves and anticorrelated motion with most of the editing domain elements. Moreover, the simulated collective dynamics analysis of the WT vs. mutant ProRSs revealed that mutation of noncatalytic residues as well as the deletion of INS indeed alter the dynamics of the PBL with respect to the rest of the protein. Analysis of the dynamic cross-correlations between the PBL and other amino acid residues of Ef ProRS (Fig. 7) demonstrated that the extent of correlation/anticorrelation between residue fluctuations depends upon neighboring, as well as distant residues. It also showed that the anticorrelated motion between the editing domain and PBL undergoes perceptible change in the case of the G217A, E218A and E218D variants.
The effect of alanine substitutions at G217 and E218 on the PBL dynamics was also evident from the combined essential dynamics analysis, which showed significant changes in the RMSP of the first 3 major modes (eigenvectors) of collective dynamics of the PBL (Fig. 8). Interestingly, the combined PC analysis shows the impact of deletion of INS or mutation of G217 and E218 has comparable effect on the collective PBL dynamics (Fig. 8 and 9). Although these simulations were carried out in the absence of substrate, the analysis suggests that mutation of residues so close to the PBL has as significant impact on the movement of the PBL as observed for the deletion of the whole INS. Taken together, these observation suggest that coupled dynamics are relevant for PBL movement and, therefore, could impact substrate binding and catalysis.
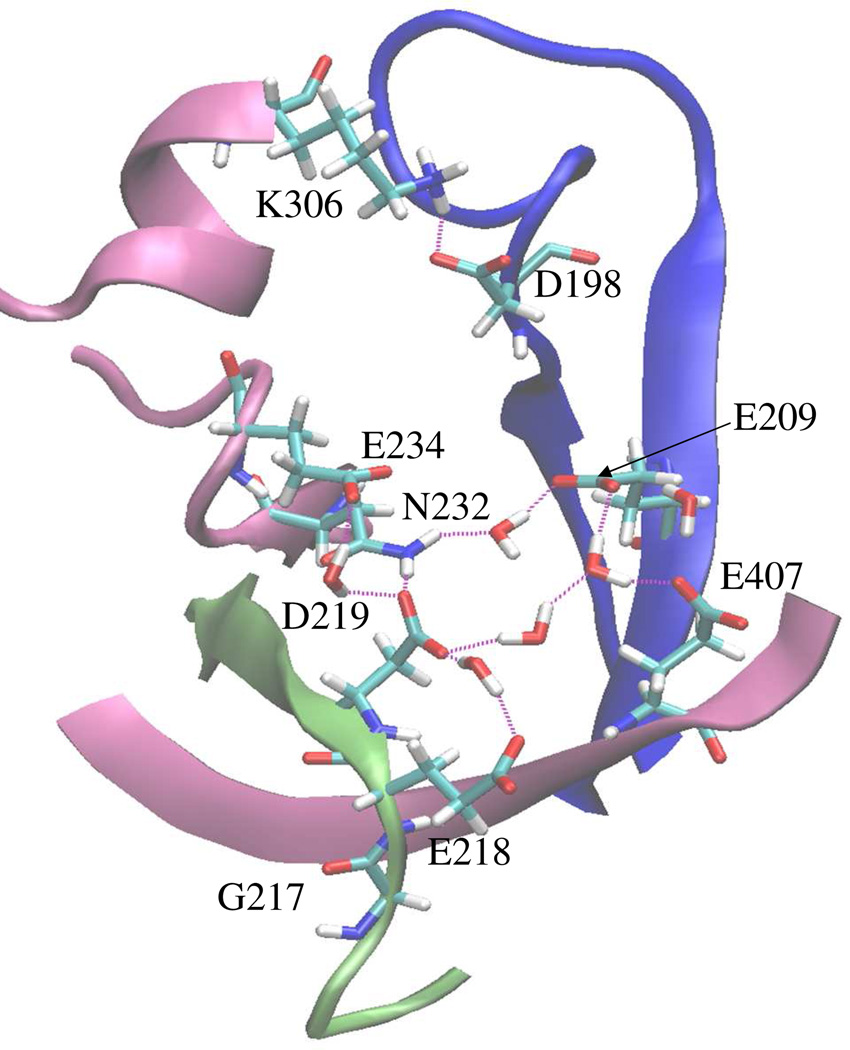
Examination of the polypeptide segment (residues 190–220) at the interface of the activation and editing domains reveals the presence of a number of negatively charged residues, namely E209, E218, D219, E234, and E407 (Fig. 10). These residues, which are conserved in both Ef and Ec ProRSs, are hydrogen-bonded to each other through water molecules and other polar residues like N232 and display significant correlations in the direction of their motions (Table 2). Interestingly, the dynamic correlations among these residues of the INS and the extended part of the PBL were maintained in the E218A variant, whereas correlations between these polar residues were significantly reduced in the case of G218A and E218D mutants (Table 2). On the other hand, analysis of the dynamic coupling between the tip of the PBL (M202 and G203) and several surrounding structural elements (residues 239–244, 345–351, 378–383) of the INS (not shown) revealed that the movements of these editing domain segments are significantly correlated to the tip of the PBL in the WT enzyme. However, these distant correlations are completely abolished in all the three mutants (Table 2). These observations suggest that mutation of either G217 or E218 has a high impact on the collective motion of the PBL despite their varied local impacts. Moreover, structural analysis of the WT and mutant enzymes revealed that INS protein segments are about 2–3 Å closer to the tip of the PBL (residues 201–204) in the WT enzyme compared to the mutant proteins. These neighboring structural elements appear to be critical for maintaining the coupled dynamics between the two functional domains, as well as the optimum flexibility of the PBL. Therefore, the observed dramatic effect on enzyme catalysis in the INS deletion mutant (11) is fully consistent with the present results.
Figure 10.
View of region of Ef ProRS (PDB entry 2J3M, chain B) adjacent to the PBL and the ‘GED’ motif showing charged residues at the activation domain-editing domain interface. The color coding is as follows: mauve, editing domain elements; blue, PBL; lime, ‘GED’ motif.
Table 2.
Correlation coefficients (CCij, eq. 4) of fluctuations of residue pairs in Ef ProRS, which were observed to be engaged in hydrogen bonding (Fig. 10).
| Amino Acid pairs | CCij | |||
|---|---|---|---|---|
| WT | G217A | E218A | E218D | |
| D219···E209 | 0.70 | 0.35 | 0.70 | 0.34 |
| E218···E209 | 0.58 | 0.30 | 0.75 | 0.24 |
| D219···N232 | 0.54 | 0.42 | 0.71 | 0.40 |
| E209···N232 | 0.79 | 0.44 | 0.80 | 0.66 |
| E209···E234 | 0.65 | 0.12 | 0.69 | 0.55 |
| D219···E407 | 0.77 | 0.48 | 0.74 | 0.35 |
| E218···E407 | 0.81 | 0.45 | 0.78 | 0.28 |
| ------- | ------- | ------- | ------- | ------- |
| M202···T241 | 0.72 | −0.01 | 0.09 | 0.03 |
| G203···T241 | 0.73 | 0.13 | 0.01 | −0.08 |
| G203···D347 | 0.63 | 0.11 | −0.09 | 0.10 |
| M202···E352 | 0.62 | −0.17 | −0.10 | −0.03 |
| M202···S380 | 0.69 | −0.10 | −0.08 | 0.28 |
| G203···E382 | 0.64 | 0.15 | 0.01 | 0.10 |
| G203···D383 | 0.61 | 0.07 | 0.39 | 0.22 |
| M202···E382 | 0.50 | 0.14 | 0.26 | 0.00 |
| M202···D383 | 0.60 | 0.01 | 0.21 | 0.23 |
CONCLUSIONS
The combined use of computer simulations and mutational analysis has enabled a better understanding of the role of domain dynamics in the enzymatic function of prokaryotic-like ProRSs (Fig. 1). Experimental mutational studies of two conserved residues, G217 and E218 (Fig. 2), revealed significantly reduced catalytic efficiency, while essential dynamics analysis of these mutant proteins showed a reduction in the collective dynamics of the catalytically important proline-binding loop. Overall, the present study provides insights into the interplay of coupled dynamics and enzyme catalysis in prokaryotic-like ProRSs.
The two point mutations, G217A and E218A, were found to significantly impact the proline activation indicating that these noncatalytic residues are crucial for function. The mutation of G217 and E218 to alanine only mildly impacted cognate tRNA aminoacylation. This observation suggests that the binding of the 3′-acceptor end in the aminoacylation active site was not altered significantly by these mutations.
MD simulations of three point mutants (G217A, E218A, and E218D) and the deletion mutant (ΔINS) demonstrated that the overall fluctuations of the backbone were impacted differently among these enzymes. A reduction in backbone fluctuation was evident in the case of G217A and E218D indicating more rigidity in the structure, while for E218A a more flexible backbone was observed. For ΔINS, an overall reduction of flexibility was noted amidst a sharp increase of the fluctuations in the PBL.
The collective motion of PBL was studied by performing dynamic cross-correlation analyses (Fig. 6), which demonstrated that the editing domain in the wild-type enzyme and the three mutants (G217A, E218A, and E218D) is quite flexible and engaged in anticorrelated motion with the activation domain. Although the basic coupling pattern did not change, the extent of correlations and anticorrelations were found to be varying, consistent with the trend observed in the B-factor analysis. In the case of G217A and E218D, the overall correlation among the structural elements surrounding the PBL is decreased, while for E218A it is increased (Fig. 5). The present study indicates the role of E218 is not only limited to stabilizing the substrate, as proposed earlier (14), but also to maintain PBL dynamics through coupled motion.
The present study also provides insights into the severely reduced proline activation efficiency of ΔINS-ProRS (51). In the case of this variant, the analysis of the collective dynamics of the PBL revealed a total abolition of the coupling of motions with surrounding elements. Removal of the editing domain disrupts the hydrogen-bonding network between polar residues at the domain-domain interface, which is important to maintain the coupled protein dynamics (Fig. 10) and optimum flexibility of protein segments surrounding the activation site. Although only the 217GED219 motif was targeted here, the role of other noncatalytic residues, such as N232 and E234 in the editing domain of Ec ProRS can be explored in the future.
Taken together, this work provides an understanding of how noncatalytic residues in a distant site modulate the activity of prokaryotic-like ProRSs by maintaining the coupled protein dynamics essential for catalysis. The present study also reveals a novel role for a synthetase editing domain and may explain why truncated or defunct editing domains have been maintained in some aminoacyl-tRNA synthetases, despite the lack of catalytic activity (51, 52).
Supplementary Material
Acknowledgement
Funding
This work was supported in part by Teragrid [grant number TG-DMR090140 (S.B.)] and National Institute of Health [grant numbers GM049928 (K.M.-F.) and GM085779 (S.H.)] and by the Office of Research and Sponsored Programs of the University of Wisconsin-Eau Claire, Eau Claire, WI.
We gratefully acknowledge the computational support from the LTS, University of Wisconsin-Eau Claire. We thank Dr. Stephen Cusack for providing the coordinates for the model of Ec ProRS used in this work. We are also thankful to the three anonymous reviewers for their constructive comments.
Abbreviations used
- Ec
Escherichia coli
- ED
essential dynamics
- Ef
Enterococcus faecalis
- MD
molecular dynamics
- INS
insertion domain
- PBL
proline-binding loop
- PCA
principal component analysis
- ProRS
prolyl-tRNA synthetase
- RMSD
root-mean-square deviation
- RMSP
root-mean-square projection
- WT
wild-type
Footnotes
SUPPORTING MATERIALS
Kinetic plots of pre- and post-transfer editing reaction, and root-mean-square projections from essential dynamics analysis of WT and two mutants (G217A and E218A) of Ec ProRS are presented as supporting materials. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Beuning PJ, Musier-Forsyth K. Hydrolytic editing by a class II aminoacyl-tRNA synthetase. Proc Natl Acad Sci U S A. 2000;97:8916–8920. doi: 10.1073/pnas.97.16.8916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beuning PJ, Musier-Forsyth K. Species-specific differences in amino acid editing by class II prolyl-tRNA synthetase. J Biol Chem. 2001;276:30779–30785. doi: 10.1074/jbc.M104761200. [DOI] [PubMed] [Google Scholar]
- 3.Ahel I, Stathopoulos C, Ambrogelly A, Sauerwald A, Toogood H, Hartsch T, Soll D. Cysteine activation is an inherent in vitro property of prolyl-tRNA synthetases. J. Biol. Chem. 2002;277:34743–34748. doi: 10.1074/jbc.M206928200. [DOI] [PubMed] [Google Scholar]
- 4.Mascarenhas A, Martinis SA, An S, Rosen AE, Musier-Forsyth K. Protein Engineering. Vol. 22. New York: Springer Verlag; 2009. [Google Scholar]
- 5.Stehlin C, Burke B, Yang F, Liu H, Shiba K, Musier-Forsyth K. Species-specific differences in the operational RNA code for aminoacylation of tRNAPro. Biochemistry. 1998;37:8605–8613. doi: 10.1021/bi980364s. [DOI] [PubMed] [Google Scholar]
- 6.Cusack S, Yaremchuk A, Krikliviy I, Tukalo M. tRNA(Pro) anticodon recognition by Thermus thermophilus prolyl-tRNA synthetase. Structure. 1998;6:101–108. doi: 10.1016/s0969-2126(98)00011-2. [DOI] [PubMed] [Google Scholar]
- 7.Wong FC, Beuning PJ, Nagan M, Shiba K, Musier-Forsyth K. Functional role of the prokaryotic proline-tRNA synthetase insertion domain in amino acid editing. Biochemistry. 2002;41:7108–7115. doi: 10.1021/bi012178j. [DOI] [PubMed] [Google Scholar]
- 8.Wong FC, Beuning PJ, Silvers C, Musier-Forsyth K. An isolated class II aminoacyl-tRNA synthetase insertion domain is functional in amino acid editing. J Biol Chem. 2003;278:52857–52864. doi: 10.1074/jbc.M309627200. [DOI] [PubMed] [Google Scholar]
- 9.An S, Musier-Forsyth K. Trans-editing of Cys-tRNAPro by Haemophilus influenzae YbaK protein. J. Biol. Chem. 2004;279:42359–42362. doi: 10.1074/jbc.C400304200. [DOI] [PubMed] [Google Scholar]
- 10.An S, Musier-Forsyth K. Cys-tRNA(Pro) editing by Haemophilus influenzae YbaK via a novel synthetase. YbaK.tRNA ternary complex. J Biol Chem. 2005;280:34465–34472. doi: 10.1074/jbc.M507550200. [DOI] [PubMed] [Google Scholar]
- 11.Hati S, Ziervogel B, Sternjohn J, Wong FC, Nagan MC, Rosen AE, Siliciano PG, Chihade JW, Musier-Forsyth K. Pre-transfer editing by class II prolyl-tRNA synthetase: role of aminoacylation active site in "selective release" of noncognate amino acids. J. Biol. Chem. 2006;281:27862–27872. doi: 10.1074/jbc.M605856200. [DOI] [PubMed] [Google Scholar]
- 12.Weimer KM, Shane BL, Brunetto M, Bhattacharyya S, Hati S. Evolutionary basis for the coupled-domain motions in Thermus thermophilus leucyl-tRNA synthetase. J. Biol. Chem. 2009;284:10088–10099. doi: 10.1074/jbc.M807361200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bu Z, Biehl R, Monkenbusch M, Richter D, Callaway DJ. Coupled protein domain motion in Taq polymerase revealed by neutron spin-echo spectroscopy. Proc. Natl. Acad. Sci. U. S. A. 2005;102:17646–17651. doi: 10.1073/pnas.0503388102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Crepin T, Yaremchuk A, Tukalo M, Cusack S. Structures of two bacterial prolyl-tRNA synthetases with and without a cis-editing domain. Structure. 2006;14:1511–1525. doi: 10.1016/j.str.2006.08.007. [DOI] [PubMed] [Google Scholar]
- 15.Burke B, Lipman RS, Shiba K, Musier-Forsyth K, Hou YM. Divergent adaptation of tRNA recognition by Methanococcus jannaschii prolyl-tRNA synthetase. J Biol Chem. 2001;276:20286–20291. doi: 10.1074/jbc.m100456200. [DOI] [PubMed] [Google Scholar]
- 16.Stehlin C, Heacock DH, 2nd, Liu H, Musier-Forsyth K. Chemical modification and site-directed mutagenesis of the single cysteine in motif 3 of class II Escherichia coli prolyl-tRNA synthetase. Biochemistry. 1997;36:2932–2938. doi: 10.1021/bi962295s. [DOI] [PubMed] [Google Scholar]
- 17.Fersht AR, Ashford JS, Bruton CJ, Jakes R, Koch GL, Hartley BS. Active site titration and aminoacyl adenylate binding stoichiometry of aminoacyl-tRNA synthetases. Biochemistry. 1975;14:1–4. doi: 10.1021/bi00672a001. [DOI] [PubMed] [Google Scholar]
- 18.Liu H, Musier-Forsyth K. Escherichia coli proline tRNA synthetase is sensitive to changes in the core region of tRNA(Pro) Biochemistry. 1994;33:12708–12714. doi: 10.1021/bi00208a023. [DOI] [PubMed] [Google Scholar]
- 19.Heacock D, Forsyth CJ, Shiba K, Musier-Forsyth K. Bioorganic Chemistry. 1996;24:273–289. [Google Scholar]
- 20.Musier-Forsyth K, Scaringe S, Usman N, Schimmel P. Enzymatic aminoacylation of single-stranded RNA with an RNA cofactor. Proc Natl Acad Sci U S A. 1991;88:209–213. doi: 10.1073/pnas.88.1.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. J. Mol. Graph. 1996;14:33–38. 27–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 22.MacKerell ADJ, Bashford D, Bellott M, Dunbrack RLJ, Evanseck JD, Field MJ, Fischer S, Gao J, Gou J, Ha S, Joseph-McCarthy D, Kuchnir L, Kuczera K, Lau FTK, Mattos C, Michnick S, Ngo T, Nguyen DT, Prodhom B, Reiher WEI, Roux B, Schelenkrich M, Smith JC, Stote R, Straub J, Watanbe M, Wiórkiewicz-Kuczera J, Yin D, Karplus M. Allatom empirical potential for molecular modeling and dynamics studies of proteins. J. Phys. Chem. B. 1998;102:3586. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 23.Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kale L, Schulten K. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983;79:926. [Google Scholar]
- 25.Ryckaert JP, Ciotti G, Berensden HJC. Numerical integration of the Cartesian equations of motion of a system with constraints: molecular dynamics of n-alkanes. J. Comput. Phys. 1977;23:327–341. [Google Scholar]
- 26.Darden T, York D, Pedersen L. Particle Mesh Ewald: An N. Log(N) Method for Ewald Sums in Large Systems. J. Chem. Phys. 1993;98:10089–10092. [Google Scholar]
- 27.Berendsen HJC, Postma JP, van Gunsteren MWF, DiNola A, Haak JR. Molecular dynamics with coupling to an external bath. J. Chem. Phys. 1984;81:3684–3690. [Google Scholar]
- 28.Feller SE, Zhang Y, Pastor RW, Brooks BR. Constant pressure molecular dynamics simulation: The Langevin piston method. J. Chem. Phys. 1995;103:4613–4621. [Google Scholar]
- 29.Martyna GJ, Tobias DJ, Klein ML. Constant pressure molecular dynamics algorithms. J. Chem. Phys. 1994;101:4177–4189. [Google Scholar]
- 30.Bhattacharyya S, Ma S, Stankovich MT, Truhlar DG, Gao J. Potential of mean force calculation for the proton and hydride transfer reactions catalyzed by medium-chain acyl-CoA dehydrogenase: effect of mutations on enzyme catalysis. Biochemistry. 2005;44:16549–16562. doi: 10.1021/bi051630m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rauschnot JC, Yang C, Yang V, Bhattacharyya S. Theoretical determination of the redox potentials of NRH:quinone oxidoreductase 2 using quantum mechanical/molecular mechanical simulations. J. Phys. Chem. B. 2009;113:8149–8157. doi: 10.1021/jp901854a. [DOI] [PubMed] [Google Scholar]
- 32.Peters GH, van Aalten DM, Svendsen A, Bywater R. Essential dynamics of lipase binding sites: the effect of inhibitors of different chain length. Protein Eng. 1997;10:149–158. doi: 10.1093/protein/10.2.149. [DOI] [PubMed] [Google Scholar]
- 33.van Aalten DM, Amadei A, Linssen AB, Eijsink VG, Vriend G, Berendsen HJ. The essential dynamics of thermolysin: confirmation of the hinge-bending motion and comparison of simulations in vacuum and water. Proteins. 1995;22:45–54. doi: 10.1002/prot.340220107. [DOI] [PubMed] [Google Scholar]
- 34.Mueller RM, North MA, Yang C, Hati S, Bhattacharyya S. Interplay of flavin's redox states and protein dynamics: an insight from QM/MM simulations of dihydronicotinamide riboside quinone oxidoreductase 2. J Phys Chem B. 2011;115:3632–3641. doi: 10.1021/jp1107922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Glykos NM. Software news and updates. Carma: a molecular dynamics analysis program. J. Comput. Chem. 2006;27:1765–1768. doi: 10.1002/jcc.20482. [DOI] [PubMed] [Google Scholar]
- 36.Amadei A, Linssen AB, Berendsen HJ. Essential dynamics of proteins. Proteins. 1993;17:412–425. doi: 10.1002/prot.340170408. [DOI] [PubMed] [Google Scholar]
- 37.Kazmierkiewicz R, Czaplewski CC, Lammek B, Ciarkowski J. Essential Dynamics/Factor Analysis for the Interpretation of Molecular Dynamics Trajectories. J. Comp. Aid. Mol. Des. 1999;13:21–33. doi: 10.1023/a:1008052020514. [DOI] [PubMed] [Google Scholar]
- 38.Jabukowski H, Goldman E. Editing of errors in selection of amino acids for protein synthesis. Microbiol. Rev. 1992;56:412–429. doi: 10.1128/mr.56.3.412-429.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Beuning PJ, Musier-Forsyth K. Species-specific differences in amino acid editing by class II prolyl-tRNA synthetase. Journal of Biological Chemistry. 2001 doi: 10.1074/jbc.M104761200. in press. [DOI] [PubMed] [Google Scholar]
- 40.Yang LW, Bahar I. Coupling between catalytic site and collective dynamics: a requirement for mechanochemical activity of enzymes. Structure. 2005;13:893–904. doi: 10.1016/j.str.2005.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tousignant A, Pelletier JN. Protein motions promote catalysis. Chem Biol. 2004;11:1037–1042. doi: 10.1016/j.chembiol.2004.06.007. [DOI] [PubMed] [Google Scholar]
- 42.Hammes-Schiffer S, Benkovic SJ. Relating protein motion to catalysis. Annu Rev Biochem. 2006;75:519–541. doi: 10.1146/annurev.biochem.75.103004.142800. [DOI] [PubMed] [Google Scholar]
- 43.Bakan A, Bahar I. The intrinsic dynamics of enzymes plays a dominant role in determining the structural changes induced upon inhibitor binding. Proc Natl Acad Sci U S A. 2009;106:14349–14354. doi: 10.1073/pnas.0904214106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Agarwal PK. Role of protein dynamics in reaction rate enhancement by enzymes. J Am Chem Soc. 2005;127:15248–15256. doi: 10.1021/ja055251s. [DOI] [PubMed] [Google Scholar]
- 45.Agarwal PK, Billeter SR, Rajagopalan PT, Benkovic SJ, Hammes-Schiffer S. Network of coupled promoting motions in enzyme catalysis. Proc Natl Acad Sci U S A. 2002;99:2794–2799. doi: 10.1073/pnas.052005999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Eisenmesser EZ, Millet O, Labeikovsky W, Korzhnev DM, Wolf-Watz M, Bosco DA, Skalicky JJ, Kay LE, Kern D. Intrinsic dynamics of an enzyme underlies catalysis. Nature. 2005;438:117–121. doi: 10.1038/nature04105. [DOI] [PubMed] [Google Scholar]
- 47.Henzler-Wildman KA, Lei M, Thai V, Kerns SJ, Karplus M, Kern D. A hierarchy of timescales in protein dynamics is linked to enzyme catalysis. Nature. 2007;450:913–916. doi: 10.1038/nature06407. [DOI] [PubMed] [Google Scholar]
- 48.Rajagopalan PT, Lutz S, Benkovic SJ. Coupling interactions of distal residues enhance dihydrofolate reductase catalysis: mutational effects on hydride transfer rates. Biochemistry. 2002;41:12618–12628. doi: 10.1021/bi026369d. [DOI] [PubMed] [Google Scholar]
- 49.Tukalo M, Yaremchuk A, Fukunaga R, Yokoyama S, Cusack S. The crystal structure of leucyl-tRNA synthetase complexed with tRNALeu in the posttransfer-editing conformation. Nat. Struct. Mol. Biol. 2005;12:923–930. doi: 10.1038/nsmb986. [DOI] [PubMed] [Google Scholar]
- 50.Silvian LF, Wang J, Steitz TA. Insights into editing from an ile-tRNA synthetase structure with tRNAile and mupirocin. Science. 1999;285:1074–1077. [PubMed] [Google Scholar]
- 51.SternJohn J, Hati S, Siliciano PG, Musier-Forsyth K. Restoring species-specific posttransfer editing activity to a synthetase with a defunct editing domain. Proc. Natl. Acad. Sci. U. S. A. 2007;104:2127–2132. doi: 10.1073/pnas.0611110104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lue SW, Kelley SO. An aminoacyl-tRNA synthetase with a defunct editing site. Biochemistry. 2005;44:3010–3016. doi: 10.1021/bi047901v. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.