Abstract
Variation in the social environment is a fundamental component of many vertebrate societies. In humans and other primates, adverse social environments often translate into lasting physiological costs. The biological mechanisms associated with these effects are therefore of great interest, both for understanding the evolutionary impacts of social behavior and in the context of human health. However, large gaps remain in our understanding of the mechanisms that mediate these effects at the molecular level. Here we addressed these questions by leveraging the power of an experimental system that consisted of 10 social groups of female macaques, in which each individual's social status (i.e., dominance rank) could be experimentally controlled. Using this paradigm, we show that dominance rank results in a widespread, yet plastic, imprint on gene regulation, such that peripheral blood mononuclear cell gene expression data alone predict social status with 80% accuracy. We investigated the mechanistic basis of these effects using cell type-specific gene expression profiling and glucocorticoid resistance assays, which together contributed to rank effects on gene expression levels for 694 (70%) of the 987 rank-related genes. We also explored the possible contribution of DNA methylation levels to these effects, and identified global associations between dominance rank and methylation profiles that suggest epigenetic flexibility in response to status-related behavioral cues. Together, these results illuminate the importance of the molecular response to social conditions, particularly in the immune system, and demonstrate a key role for gene regulation in linking the social environment to individual physiology.
Keywords: inflammation, social gradient, differential gene expression, sociogenomics
The social organization of many group-living mammals, including many primates, is marked by readily recognizable differences in the social environments experienced by individual group members. The causes and consequences of variation in the social environment have been of long-standing interest in behavioral ecology and evolution, and strong evidence indicates the importance of the social environment in human health as well. In particular, a large body of work in humans and nonhuman primates indicates that adverse social conditions can have important consequences, both for disease susceptibility in the short term (1–3) and for evolutionarily important parameters such as fertility and survival over the long term (4–7). Characterizing the mechanisms underlying these effects is therefore a priority not only in the study of evolution and behavior but also for understanding the protective and pathological effects of the social environment in our own lives.
Social status is one of the most important predictors of the quality of an individual's social environment. Consequently, social status has been intensively studied in nonhuman primates, including as a model for the effects of social stress and socioeconomic status (SES) in humans (reviewed in ref. 8). Social status in nonhuman primates is encoded by dominance rank, which defines which individuals yield to other individuals during competitive encounters. In settings in which hierarchies are strongly enforced or subordinates have little social support, low dominance rank can lead to chronic stress, immune compromise, and reproductive dysregulation (3, 9). In socially housed captive female rhesus macaques (Macaca mulatta), for instance, the physiological effects of dominance rank include changes in glucocorticoid (GC) and sex steroid hormone regulation (10), serotonergic and dopaminergic signaling (11), and lymphocyte counts and proliferation rates (12). Interestingly, many of these rank-associated effects are detectable even in the absence of rank-related asymmetries in access to resources, suggesting that the stress of social subordinacy alone can trigger a physiological response. This relationship echoes the effects of social stress and SES in humans, in which observed “social gradients” in disease risk remain in large part unexplained by resource access alone (8, 13, 14).
Much remains unresolved, however, about the physical intermediates that link the social environment with immunological and physiological changes, especially on the molecular level. In particular, we know little about how social status impacts gene regulation, either in primates or in mammals more generally, although several lines of evidence suggest that these effects may be important. First, the potential for social regulation of gene expression is supported by correlations between social integration, early-life SES, and gene expression variation in humans (15–17). Second, gene–environment interactions identified in humans and nonhuman primates frequently involve allelic variants that act via altering gene expression levels (e.g., 18, 19). Finally, social hierarchies are known to influence gene expression in other organisms. Dominance rank ascendancy in social cichlids, for example, is associated with strong induction of the transcription factor egr1 and its downstream target, gonadotropin-releasing hormone (20). Similarly, in honey bees, genome-wide gene expression profiles strongly differentiate queen bees from worker castes, sterile workers from reproductive workers, and active foragers from hive workers (21, 22). Although the nature of social hierarchies differs between insects, fish, and primates, these lines of evidence suggest that social status might also directly influence gene regulation in primates. Further, the strong ties between social status and disease risk in humans suggest that such effects might be particularly pronounced in the primate immune system.
To test this possibility, we turned to social groups of female rhesus macaques, a well-established model for the physiological effects of social stress and social status. Specifically, we focused on experimentally constructed social hierarchies, in which dominance rank assignments were imposed on each female by manipulating her order of introduction into a new social group [earlier introduction predicted higher rank (23); Results]. Using this powerful animal model, we were able to test for an association between rank and gene regulation in social groups in which dominance ranks were experimentally assigned, and in which other known environmental influences were carefully controlled. Together, our findings indicate that dominance rank in female rhesus macaques explains substantial variation in gene expression levels in peripheral blood mononuclear cells, an important component of immune surveillance and defense.
Results
Associations Between Dominance Rank and Gene Expression Levels.
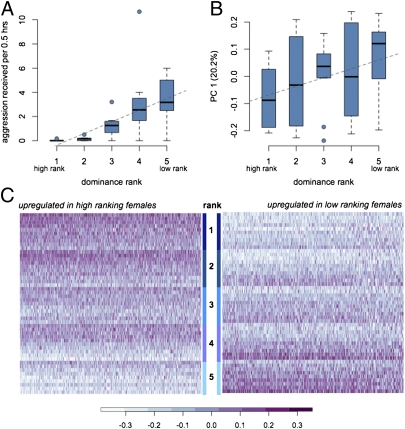
To investigate whether and to what degree dominance rank influences gene regulation, we profiled gene expression levels in peripheral blood mononuclear cells (PBMCs) from 49 female rhesus macaques in 10 replicate social groups, using the Illumina HT-12 BeadChip. Each social group was composed of five females (we sampled only four females in one group), resulting in five dominance rank values for each group. Dominance ranks were strongly correlated with introduction order (Spearman's rho = 0.70, P = 2.62 × 10−8, rho = 0.80 when excluding a group in which multiple individuals recently changed ranks; these individuals were subjects of the plasticity analysis reported below) but not with female age (P = 0.34, rho = −0.14), parity (P = 0.12, rho = −0.23), time since oviarectomy (P = 0.16, rho = 0.20), or time since removal from original breeding colony (P = 0.52, rho = 0.09). Differences in rank were reflected in different social experiences, including rates of received aggression (Fig. 1A). After filtering for probes that mapped uniquely to the macaque genome and for genes that were detectably expressed in our sample, we considered the normalized expression levels of 6,097 genes in each female (listed in Dataset S1). We identified a strong global signature of dominance rank in the gene expression data. In particular, the first principal component of the gene expression data, which accounted for 20.2% of variance in the dataset, was significantly correlated with dominance rank (R2 = 0.10, P < 0.03; Fig. 1B).
Fig. 1.
Parallel effects of dominance rank on social environment and gene expression levels. (A) Low-ranking individuals experience more aggression from group mates than high-ranking individuals (P < 10−6, R2 = 0.42, n = 49). (B) This social experience is mirrored by gene expression profiles that vary according to rank. Principal component (PC)1 explains 20.2% of overall variance in gene expression and is correlated with rank (P = 0.03, R2 = 0.10, n = 49). (C) Heatmaps of log2-transformed gene expression levels for rank-associated genes. Values are shown after controlling for differences in means among social groups; 0 roughly corresponds to mean expression levels.
We then proceeded to analyze the contribution of dominance rank to variation in expression levels for each gene. To do so, we used a linear mixed-effects model in which residual variation in gene expression levels, after taking into account differences across social groups, was treated as the response variable. Dominance rank was incorporated as a fixed effect, and the significance of the dominance rank–gene expression relationship was assessed based on the strength of this effect. We identified an association between interindividual gene expression variation and dominance rank for 987 genes (16.2% of the 6,097 genes we considered; false discovery rate = 10%; Fig. 1C and Dataset S1). Within the set of 987 rank-associated genes, 535 genes (8.8% of all genes we considered) were more highly expressed in high-ranking individuals, and 452 genes (7.4%) were more highly expressed in low-ranking individuals.
To explore the biological functions associated with these genes, we performed categorical enrichment analysis based on publicly available gene set annotations (24) (Dataset S2). We identified little evidence for enrichment of specific functional categories in the rank-associated gene set as a whole, although the most significant category, interleukin signaling (P < 0.02; proportion of false discoveries at this P value threshold, q, = 0.30), suggested a role for immune function. Consistent with this possibility, when conditioning on the direction of the rank effect on gene regulation, we observed enrichments for interleukin signaling (P = 0.01, q = 0.05), T-cell activation (P = 0.01, q = 0.05), and chemokine and cytokine inflammation (P = 0.02, q = 0.07) among genes more highly expressed in low-ranking individuals. Examples of such genes (Fig. 2) include PTGS2 (a proinflammatory signaling molecule that is negatively regulated by immunosuppressive glucocorticoids), IL8RB (a receptor for the strongly proinflammatory cytokine IL8, which is associated with neutrophil migration into injured tissue), and NFATC1 (which is associated with the transcriptional response to T-cell stimulation).
Fig. 2.
Rank–gene expression associations among inflammation-related immune genes. Low-ranking females tend to overexpress inflammation-related genes: (A) PTGS2 (P = 0.004); (B) IL8RB (P = 0.003); and (C) NFATC1 (P < 10−3).
Finally, we used clustering analysis [modulated modularity clustering (25)] to investigate whether modules of rank-associated genes (>9 genes) with highly correlated expression patterns reflected coherent biological functions. Consistent with our previous results, we found that the largest module (112 genes; mean r = 0.58) was enriched for immune-related processes (P = 0.002, q = 0.08). Additionally, the most highly coregulated module (13 genes; mean r = 0.63) was enriched for specific immune-related gene subsets, including response to IFN-γ (P = 7.50 × 10−6, q = 9.0 × 10−5) and macrophage activation (P = 6.31 × 10−4, q = 3.80 × 10−3). Thus, rank is associated with functionally coordinated changes in immune gene expression.
Predictive Relationships Between Gene Expression and Rank.
The strong effect of dominance rank on gene expression levels suggested that gene expression data alone might be sufficient to predict rank attributes. To test this possibility, we performed 1,000 iterations of a leave-k-out procedure. In each iteration, we fit a support vector machine (SVM)-based model (26) using a training set composed of data from 39 randomly chosen individuals (79.6% of the dataset). We used this model to learn how gene expression data from these females were associated with their relative position in the social hierarchy, specifically to an individual's classification as high-ranking (rank class A, including ranks 1 and 2), middle-ranking (rank class B, including rank 3), or low-ranking (rank class C, including ranks 4 and 5) (SI Appendix). We then used the model to predict rank class in the test set of 10 individuals (20.4% of the dataset) initially removed from the data.
Using this approach, we correctly predicted rank class for a median of 8 of the 10 individuals in a test set (80% predictive accuracy; Fig. 3). We also calculated absolute error in model prediction by summing the absolute value of the true rank class minus the predicted rank class across all test set individuals. The median error rate, three errors per run, was significantly smaller than the same values calculated based on random prediction (P = 0.002; Fig. 3).
Fig. 3.
Gene expression data are sufficient to predict relative position in the social hierarchy. (A) Boxplot of predictive accuracy for 10 training set individuals, across 1,000 cross-validation iterations (dashed line shows expected accuracy under random prediction), and (B) histogram of the sum of absolute error between predicted rank class and true rank class, under random assignment. Black arrow, median sum of absolute error across the 1,000 true leave-k-out iterations (P = 0.002). (C) Shifts in dominance rank experienced by seven females in the dataset. Solid arrows, cases in which females changed rank classes; dashed arrows, cases in which females changed rank within rank classes; check marks, correct predictions; x, incorrect prediction.
Although rank hierarchies in female macaques tend to be stable, female dominance ranks do sometimes change, particularly upon replacement of individuals within a group. As a result of such changes, we were able to opportunistically sample RNA from the same individual, while at different ranks, for seven females, five of whom experienced changes in ordinal rank (1–5) that switched them into a different relative rank class (A–C) as well. These samples allowed us to test the plasticity of the gene expression response to dominance rank. Specifically, we asked whether gene expression data could predict relative rank position for the same individual after her rank had changed. In this case, we used the 49 samples in the original dataset as our training set, and the 7 samples obtained at a different time as our test set (all seven individual females were also represented in the training set, while occupying different ranks). We found that the gene expression data were sufficient to classify six of the seven females (85.7%) in the test set into the correct rank class (Fig. 3).
Regulatory Mechanisms Underlying Rank Effects.
Our data indicated that dominance rank has a strong relationship with gene expression levels. We next evaluated the contribution of two mechanisms that could account for these effects, focusing on the 987 rank-associated genes. Specifically, we investigated whether rank-dependent differences in tissue composition and GC signaling might help explain these associations. In addition, we focused on a smaller dataset of six females (the set of rank 1 and rank 5 individuals from three social groups; it was not feasible to generate whole-genome bisulfite sequencing data for our full set of 49 individuals, so we focused on the extremes of the rank distribution) to explore the possible relationship between dominance rank and another mechanism suggested to mediate social environmental effects on the genome, DNA methylation.
Relationship Between Tissue Composition and Rank-Associated Gene Regulation.
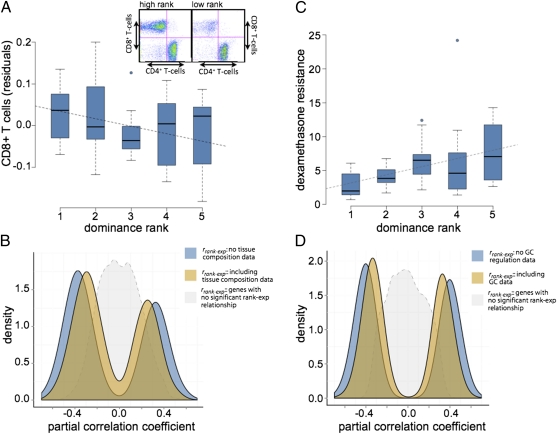
We used fluorescence-activated cell sorting (FACS) analysis to estimate the proportion of the four main PBMC types (CD4+ T cells, CD8+ T cells, B cells, and monocytes) in blood samples from 39 of the individuals included in our study. Consistent with prior reports, we identified a relationship between PBMC composition and dominance rank: low-ranking females had a reduced proportion of CD8+ T cells (a marker associated with cytotoxic T cells) relative to high-ranking individuals (P < 0.05, n = 39; Fig. 4A). Such changes in cellular composition could, in principle, lead to differences in PBMC gene expression levels by rank, even in the absence of other gene regulatory changes.
Fig. 4.
Effects of tissue composition and glucocorticoid-mediated regulation on rank-associated gene expression levels. (A) Low-ranking individuals exhibit lower proportions of CD8+ T cells in PBMCs (P = 0.047, n = 39; y axis shows the residuals of T-cell proportions after controlling for social group). (Inset) Example data for a rank 1 female and a rank 5 female; x axis shows staining for CD4+ (helper) T cells and y axis shows staining for CD8+ (cytotoxic) T cells. (C) Low-ranking individuals exhibit decreased GC negative feedback (P = 0.005, n = 49). The y axis represents levels of circulating cortisol 17 h post-dexamethasone administration. (B and D) Distribution of rank–gene expression partial correlation coefficients for genes in which we inferred a contribution of tissue composition (B, n = 301 genes) or an effect of GC regulation (D, n = 596 genes). Blue density plots show the distribution of partial correlation coefficients without taking into account tissue composition or GC data; tan density plots show the same distribution after considering these data. The magnitude of the rank–gene expression relationship decreases but remains distinct from the background set of all genes (in gray; n = 6,097 genes).
To test this possibility, we assessed whether variation in tissue composition could help explain the relationship between dominance rank and gene expression levels for the 987 rank-associated genes. To do so, we first characterized gene expression levels in pure populations of each of the four main PBMC types, again using Illumina HT-12 arrays. We then asked whether the strength of evidence supporting a direct relationship between rank and gene expression levels (measured by a partial correlation) was significantly reduced after taking into account (i) differences in cell-type composition among individuals and (ii) differences in gene expression levels across cell types.
Specifically, we used our data to calculate the expected gene expression level for each gene in each individual, given tissue composition effects alone. Our estimate was based on the average of the expression levels for the gene across cell types (based on the cell type-specific data), weighted by the proportional representation of that cell type in each sample (based on the FACS analysis). After incorporating these estimates into our analysis, we found that tissue composition effects made a modest but significant contribution to the rank–gene expression relationship for 301 of the 987 rank-associated genes (30.5% at a nominal P value ≤0.05). As expected, genes that exhibited stronger evidence for a tissue composition effect were also more asymmetrically expressed in CD8+ (e.g., cytotoxic) T cells relative to other PBMC types (P < 10−45). Overall, however, the distribution of correlations between rank and expression levels for the 301 genes was still markedly nonzero even after taking tissue composition into account (Fig. 4B).
Relationship Between GC Signaling and Rank-Associated Gene Regulation.
As expected based on previous work, lower-ranking females in our sample exhibited diminished GC negative feedback following experimental administration of dexamethasone (a synthetic GC; P < 0.01, n = 49; Fig. 4C), as well as a more sluggish GC regulatory response to an experimentally induced acute stressor (P < 0.07; SI Appendix, Fig. S4). Such patterns are characteristic of chronic stress exposure and dysregulated hypothalamic-pituitary adrenal axis (HPA) activity.
We therefore asked whether variation in GC-mediated signaling explained, at least in part, the relationship between dominance rank and gene expression for the 987 rank-associated genes. To do so, we used an analysis similar to that for tissue composition effects. Specifically, we asked whether the strength of evidence for a direct relationship between rank and gene expression levels was significantly reduced after taking into account data on GC regulation. We found that the rank–gene expression relationship could be partially explained by dexamethasone suppression data for 596 of the 987 target genes (60.5%, at a nominal P value ≤0.05), including known targets of GC-mediated signaling such as the GC receptor (NR3C1) and the proinflammatory gene PTGS2 (P = 0.015 and P = 0.002, respectively). Similar to our findings for tissue composition, the effects of GC regulation were modest (Fig. 4D and SI Appendix, Fig. S5). However, the contribution of GC-mediated regulation may be twofold, as GC physiology can also impact tissue composition (27). Indeed, for 203 genes, both GC-mediated regulation and tissue composition contributed to the rank–gene expression relationship, a larger overlap than expected by chance (hypergeometric test; P = 0.001). This overlap is also suggested by joint analysis of GC resistance and tissue composition data, which tends to, but does not always, account for more of the rank–gene expression relationship than either dataset alone (SI Appendix, Fig. S5).
Dominance Rank and DNA Methylation Patterns.
Finally, we tested whether DNA methylation, a regulatory mechanism that has been linked to social environmental effects (28), might also play a role in the association between dominance rank and gene expression. In contrast to our analyses of tissue composition and GC resistance, dominance rank effects on DNA methylation have not previously been described. To explore this possibility, we generated whole-genome bisulfite sequencing data (11- to 14-fold CpG coverage) from PBMC DNA from three rank 1 females and three rank 5 females. We then investigated the relationships between DNA methylation, dominance rank, and rank-associated gene expression.
We first attempted to identify individual genomic regions that exhibited a detectable association with rank (rank differentially methylated regions; rankDMRs). We used an empirical approach similar to the one previously used for the analysis of bisulfite sequencing data from cancer and healthy tissues (29). We identified 7,089 putative rankDMRs in the genome, and found that these regions were more likely to be located close to transcription start sites (TSS) than randomly distributed sets of similarly long CpG stretches (P < 0.01). Additionally, rankDMRs that fell close to genes (within 20 kb) were more likely to be associated with TSS for genes whose expression levels were associated with dominance rank (two-tailed Fisher's exact test, P < 0.033).
To assess whether DNA methylation levels might therefore contribute to the rank–gene expression associations, we then collated DNA methylation data in and around the 987 rank-associated genes. We found that methylation data from these regions (within 20 kb of the TSS) clearly discriminated between high- and low-ranking females (Fig. 5). DNA methylation data distinguished rank-associated genes and non-rank-associated genes as well. Specifically, features related to rank-associated differences in methylation (SI Appendix, Table S3) differentiated between the 987 rank-associated genes and a set of 1,000 rank-independent genes (those with the least evidence for rank-associated gene expression) at a prediction accuracy of 58.1% (P < 0.01 based on comparison with permuted data). Thus, information about rank-associated gene expression patterns is embedded within differential DNA methylation data, and epigenetic changes might account, at least in part, for some of the rank–gene expression associations we observed.
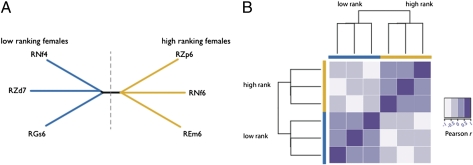
Fig. 5.
DNA methylation levels distinguish high-ranking and low-ranking females. (A) A neighbor-joining tree based on DNA methylation levels within 20 kb around the transcription start sites of rank-associated genes (445,059 CpG sites). (B) A heatmap of the same data reveals positive pairwise correlations between individuals of the same rank but not between individuals of different ranks.
Discussion
Taken together, our findings reveal a strong and widespread association between dominance rank and gene regulation in PBMCs. Our results reinforce the idea that sensitivity to the social environment is reflected in changes in gene expression in the immune system, supporting an increasingly widely recognized link between neural, endocrine, and immune function (30). Moreover, our results demonstrate that these associations also appear to be highly plastic. Not only were gene expression data sufficient to robustly predict relative dominance rank but gene expression profiles also tracked dominance rank shifts closely enough to allow us to predict different rank positions for the same individuals across time. These observations indicate that any causal relationship between dominance rank and gene regulation likely begins with rank, rather than vice versa. Because these study subjects did not experience a completely uniform social history and because unknown variables related to our method of imposing rank (by order of social introduction) might still play a role in the observed rank dynamics, further experiments—for example, imposition of a second controlled rank manipulation on the same individuals—will be necessary to robustly test this hypothesis. However, the current results support the idea that changes in gene regulation help to explain links between the social environment and physiology, potentially supplying an important piece to the puzzle of how social effects “get under the skin” (3, 31).
Changes in gene expression themselves require a mechanistic explanation. A strength of the experimental approach we used here is that it allowed us to move beyond a simple description of gene expression patterns to also investigate the regulatory mechanisms associated with these effects. Our observations suggest that variation in the expression levels of up to 694 (70.3%) of the 987 rank-associated genes may be partially explained by variation in tissue composition or GC regulation. These findings draw a clear connection between gene regulation and prior work on the physiological effects of rank: GCs, for example, have long been implicated in rank effects in the endocrinological literature (9, 32, 33), and also have important downstream effects on immune function.
In addition, our exploratory analysis identified a signal of both rank and rank-associated gene expression in DNA methylation data. The DNA methylation differences that we observed between ranks were modest relative to studies on different tissues or species, or in the context of cancer (e.g., 29). However, changes in epigenetic marks in healthy adults are likely to be muted in comparison, although evidence for such changes is rapidly accumulating (34, 35). Our findings suggest that the timescale for social effects on epigenetic variation also extends to adulthood, and indicate that such effects may include natural components of social structure, such as dominance rank.
Further work will be necessary to more finely evaluate the relative effects of GC regulation, tissue composition, and DNA methylation—among other regulatory mechanisms—on rank-associated gene expression. For instance, NR3C1, the GC receptor gene, plays a key role in linking behavior, HPA axis-mediated stress responses, and gene expression (30). The gene regulatory role of NR3C1 has previously been linked to other social environmental responses via transcription factor motif analysis (16, 17), and could be further investigated using chromatin immunoprecipitation approaches. Tissue composition effects should also be further dissected. Although we identified an important contribution of CD8+ T cells among the four cell types we investigated, additional PBMC subtypes could be relevant (and our estimates of the contribution of tissue composition may therefore be an underestimate). Natural killer cells, for example, are of known importance in the stress response (36), and although they constitute a relatively small proportion of the PBMC pool, their contribution to social stress effects may be disproportionate. Finally, variation across sexes, environmental conditions, species, and genetic backgrounds will be important to take into account, especially as comparative data already indicate that the effects of social status can be highly context-dependent (9), and that high rank can, in some cases, also induce stress (37). Studying the effects of social status in captivity, as we did here, could exacerbate dominance rank effects if females are less able to distance themselves from their group mates. Alternatively, decreased resource competition in captive animals relative to wild populations could produce the opposite effect.
In conclusion, our results emphasize important connections between behavior, social status, and change at the molecular level. Although in one sense these relationships emphasize the potential costs of adverse social environments, the plasticity of these effects is also encouraging, suggesting that mitigating social stress may confer rapidly realized physiological benefits. Our results motivate efforts to develop a nuanced understanding of social effects on gene regulation, with the aim of both exploring its evolutionary and ecological consequences and addressing its effects on human health.
Materials and Methods
We obtained blood samples from 49 adult female rhesus macaques (members of 10 experimentally constructed social groups), and purified PBMCs from each sample (including replicate samples from a separate time period for seven females while they occupied different dominance ranks). In rhesus macaques, dominance ranks are determined by interactions within a social group among the animals themselves. Thus, weighting the directionality of these interactions by experimentally manipulating the order of introduction into groups is the most direct method of experimentally altering rank. Gene expression profiling was performed using the Illumina HT-12 Expression BeadChip. To assess the relative proportions of CD4+ T cells, CD8+ T cells, monocytes, and B cells, we conducted flow cytometry on antibody-stained PBMCs. For cell type-specific gene expression analyses, we physically separated each cell type for five individuals using a BD FACSAria. To identify genes for which gene expression was rank-associated, we used a linear mixed-effects model (38). To test the predictive value of gene expression for relative rank position, we used the program svm-multiclass (26). To investigate the contributions of tissue composition and GC signaling, we tested for a reduction in the strength of the rank–gene expression partial correlation when conditioned on tissue composition-related or GC data. DNA methylation was assessed via bisulfite sequencing in six individuals (three rank 1 and three rank 5, from three different social groups). Reads were mapped and methylation levels were estimated using bismark (39). The predictive value of differential methylation data for differential gene expression was assessed using svm-perf (40). Further details for all experimental and statistical procedures can be found in SI Appendix. Data included in this study were obtained in accordance with Institutional Animal Care and Use Committee protocols approved by Emory University (IACUC 2862008Y).
Supplementary Material
Acknowledgments
We thank S. Mukherjee and R. Irizarry for advice on data analysis; V. Abadie and members of the Y.G. laboratory for helpful discussions; and S. C. Alberts, E. A. Archie, Z. Gauhar, D. Maestripieri, J. Marioni, J. Pritchard, and three anonymous referees for comments on the manuscript. This work was supported by National Institutes of Health (NIH) Grants GM077959 (to Y.G.) and HD046501, MH081816, and RR00165 (to the Yerkes National Primate Research Center). J.T. was supported by a Chicago Fellows fellowship and K.D.H. was supported by NIH Grant R01HG005220.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequence (bisulfite sequencing for DNA methylation) and gene expression datasets reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (SuperSeries no. GSE34129).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1202734109/-/DCSupplemental.
References
- 1.Cohen S, et al. Chronic social stress, social status, and susceptibility to upper respiratory infections in nonhuman primates. Psychosom Med. 1997;59:213–221. doi: 10.1097/00006842-199705000-00001. [DOI] [PubMed] [Google Scholar]
- 2.Cole SW, Mendoza SP, Capitanio JP. Social stress desensitizes lymphocytes to regulation by endogenous glucocorticoids: Insights from in vivo cell trafficking dynamics in rhesus macaques. Psychosom Med. 2009;71:591–597. doi: 10.1097/PSY.0b013e3181aa95a9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sapolsky RM. The influence of social hierarchy on primate health. Science. 2005;308:648–652. doi: 10.1126/science.1106477. [DOI] [PubMed] [Google Scholar]
- 4.Altmann J, Alberts SC. Variability in reproductive success viewed from a life-history perspective in baboons. Am J Hum Biol. 2003;15:401–409. doi: 10.1002/ajhb.10157. [DOI] [PubMed] [Google Scholar]
- 5.Cowlishaw G, Dunbar RIM. Dominance rank and mating success in male primates. Anim Behav. 1991;41:1045–1056. [Google Scholar]
- 6.Pusey A, Williams J, Goodall J. The influence of dominance rank on the reproductive success of female chimpanzees. Science. 1997;277:828–831. doi: 10.1126/science.277.5327.828. [DOI] [PubMed] [Google Scholar]
- 7.van Noordwijk MA, van Schaik CP. The effects of dominance rank and group size on female lifetime reproductive success in wild long-tailed macaques, Macaca fascicularis. Primates. 1999;40(1):105–130. doi: 10.1007/BF02557705. [DOI] [PubMed] [Google Scholar]
- 8.Sapolsky RM. Social status and health in humans and other animals. Annu Rev Anthropol. 2004;33:393–418. [Google Scholar]
- 9.Abbott DH, et al. Are subordinates always stressed? A comparative analysis of rank differences in cortisol levels among primates. Horm Behav. 2003;43(1):67–82. doi: 10.1016/s0018-506x(02)00037-5. [DOI] [PubMed] [Google Scholar]
- 10.Michopoulos V, Checchi M, Sharpe D, Wilson ME. Estradiol effects on behavior and serum oxytocin are modified by social status and polymorphisms in the serotonin transporter gene in female rhesus monkeys. Horm Behav. 2011;59:528–535. doi: 10.1016/j.yhbeh.2011.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Morgan D, et al. Social dominance in monkeys: Dopamine D2 receptors and cocaine self-administration. Nat Neurosci. 2002;5(2):169–174. doi: 10.1038/nn798. [DOI] [PubMed] [Google Scholar]
- 12.Paiardini M, et al. T-cell phenotypic and functional changes associated with social subordination and gene polymorphisms in the serotonin reuptake transporter in female rhesus monkeys. Brain Behav Immun. 2009;23:286–293. doi: 10.1016/j.bbi.2008.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.House JS, Landis KR, Umberson D. Social relationships and health. Science. 1988;241:540–545. doi: 10.1126/science.3399889. [DOI] [PubMed] [Google Scholar]
- 14.Marmot MG. Status syndrome: A challenge to medicine. JAMA. 2006;295:1304–1307. doi: 10.1001/jama.295.11.1304. [DOI] [PubMed] [Google Scholar]
- 15.Chen E, et al. Genome-wide transcriptional profiling linked to social class in asthma. Thorax. 2009;64(1):38–43. doi: 10.1136/thx.2007.095091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cole SW, et al. Social regulation of gene expression in human leukocytes. Genome Biol. 2007;8:R189. doi: 10.1186/gb-2007-8-9-r189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miller GE, et al. Low early-life social class leaves a biological residue manifested by decreased glucocorticoid and increased proinflammatory signaling. Proc Natl Acad Sci USA. 2009;106:14716–14721. doi: 10.1073/pnas.0902971106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barr CS, et al. Sexual dichotomy of an interaction between early adversity and the serotonin transporter gene promoter variant in rhesus macaques. Proc Natl Acad Sci USA. 2004;101:12358–12363. doi: 10.1073/pnas.0403763101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Newman TK, et al. Monoamine oxidase A gene promoter variation and rearing experience influences aggressive behavior in rhesus monkeys. Biol Psychiatry. 2005;57(2):167–172. doi: 10.1016/j.biopsych.2004.10.012. [DOI] [PubMed] [Google Scholar]
- 20.Burmeister SS, Jarvis ED, Fernald RD. Rapid behavioral and genomic responses to social opportunity. PLoS Biol. 2005;3:e363. doi: 10.1371/journal.pbio.0030363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grozinger CM, Fan Y, Hoover SE, Winston ML. Genome-wide analysis reveals differences in brain gene expression patterns associated with caste and reproductive status in honey bees (Apis mellifera) Mol Ecol. 2007;16:4837–4848. doi: 10.1111/j.1365-294X.2007.03545.x. [DOI] [PubMed] [Google Scholar]
- 22.Whitfield CW, Cziko AM, Robinson GE. Gene expression profiles in the brain predict behavior in individual honey bees. Science. 2003;302:296–299. doi: 10.1126/science.1086807. [DOI] [PubMed] [Google Scholar]
- 23.Jarrell H, et al. Polymorphisms in the serotonin reuptake transporter gene modify the consequences of social status on metabolic health in female rhesus monkeys. Physiol Behav. 2008;93:807–819. doi: 10.1016/j.physbeh.2007.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thomas PD, et al. PANTHER: A browsable database of gene products organized by biological function, using curated protein family and subfamily classification. Nucleic Acids Res. 2003;31:334–341. doi: 10.1093/nar/gkg115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stone EA, Ayroles JF. Modulated modularity clustering as an exploratory tool for functional genomic inference. PLoS Genet. 2009;5:e1000479. doi: 10.1371/journal.pgen.1000479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Joachims T. Making large-scale SVM learning practical. In: Scholkopf B, Burges C, Smola A, editors. Advances in Kernel Methods—Support Vector Learning. Cambridge, MA: MIT Press; 1999. pp. 169–184. [Google Scholar]
- 27.Fauci AS, Dale DC. The effect of in vivo hydrocortisone on subpopulations of human lymphocytes. J Clin Invest. 1974;53:240–246. doi: 10.1172/JCI107544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Szyf M, McGowan P, Meaney MJ. The social environment and the epigenome. Environ Mol Mutagen. 2008;49(1):46–60. doi: 10.1002/em.20357. [DOI] [PubMed] [Google Scholar]
- 29.Hansen KD, et al. Increased methylation variation in epigenetic domains across cancer types. Nat Genet. 2011;43:768–775. doi: 10.1038/ng.865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Irwin MR, Cole SW. Reciprocal regulation of the neural and innate immune systems. Nat Rev Immunol. 2011;11:625–632. doi: 10.1038/nri3042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Taylor SE, Repetti RL, Seeman T. Health psychology: What is an unhealthy environment and how does it get under the skin? Annu Rev Psychol. 1997;48:411–447. doi: 10.1146/annurev.psych.48.1.411. [DOI] [PubMed] [Google Scholar]
- 32.Creel S. Social dominance and stress hormones. Trends Ecol Evol. 2001;16:491–497. [Google Scholar]
- 33.Sapolsky RM, Alberts SC, Altmann J. Hypercortisolism associated with social subordinance or social isolation among wild baboons. Arch Gen Psychiatry. 1997;54:1137–1143. doi: 10.1001/archpsyc.1997.01830240097014. [DOI] [PubMed] [Google Scholar]
- 34.Guo JU, et al. Neuronal activity modifies the DNA methylation landscape in the adult brain. Nat Neurosci. 2011;14:1345–1351. doi: 10.1038/nn.2900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Miller CA, Sweatt JD. Covalent modification of DNA regulates memory formation. Neuron. 2007;53:857–869. doi: 10.1016/j.neuron.2007.02.022. [DOI] [PubMed] [Google Scholar]
- 36.Esterling BA, Kiecolt-Glaser JK, Bodnar JC, Glaser R. Chronic stress, social support, and persistent alterations in the natural killer cell response to cytokines in older adults. Health Psychol. 1994;13:291–298. doi: 10.1037//0278-6133.13.4.291. [DOI] [PubMed] [Google Scholar]
- 37.Gesquiere LR, et al. Life at the top: Rank and stress in wild male baboons. Science. 2011;333:357–360. doi: 10.1126/science.1207120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kang HM, et al. Efficient control of population structure in model organism association mapping. Genetics. 2008;178:1709–1723. doi: 10.1534/genetics.107.080101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Krueger F, Andrews SR. Bismark: A flexible aligner and methylation caller for bisulfite-Seq applications. Bioinformatics. 2011;27:1571–1572. doi: 10.1093/bioinformatics/btr167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Joachims T. Association for Computing Machinery Conference on Knowledge Discovery and Data Mining (Philadelphia) 2006. Training linear SVMs in linear time. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.