Background: The protein kinase mTOR can negatively regulate transcription factors and stress transcriptional responses.
Results: mTOR and its associated phosphatase PP2A suppress the constitutive nuclear import of STAT1 and apoptosis via karyopherin-α1.
Conclusion: mTOR controls STAT1 activity and apoptosis by regulating STAT1 nuclear import.
Impact: mTOR and karyopherin α1 constitute a molecular link between mitogen or nutrient sensing and apoptosis.
Keywords: Gene Expression, mTOR, Nuclear Transport, Protein Phosphatase, STAT Transcription Factor, Importin, Karyopherin
Abstract
Under conditions of reduced mitogen or nutritional substrate levels, the serine/threonine kinase target of rapamycin can augment the nuclear content of distinct transcription factors and promote the induction of stress response genes. In its latent (i.e., unphosphorylated) form, the transcription factor STAT1 regulates a subset of genes involved in immune modulation and apoptosis. Based on previous work indicating a functional relationship between mammalian target of rapamycin (mTOR) and the nuclear content of latent STAT1, we investigated the mechanism by which mTOR controls STAT1 nuclear import. By fluorescence confocal microscopy, inactivation of mTOR with rapamycin promoted the nuclear translocation of unphosphorylated STAT1, but not that of a STAT1 mutant incapable of binding its nuclear import adaptor karyopherin-α1 (KPNA1). By immunoprecipitation, KPNA1 was physically associated with mTOR and STAT1 in a complex that translocated to the nucleus in response to rapamycin. Although mTOR is not a kinase for KPNA1, the mTOR-associated phosphatase protein phosphatase 2A catalytic interacted directly with KPNA1 and regulated nuclear import of the mTOR-KPNA1 complex. KPNA1, or its interaction with STAT1, was required for the nuclear import of latent STAT1, transcriptional induction of the STAT1 gene, and caspase-3 activation under conditions of reduced mTOR activity (i.e. rapamycin, glucose starvation, serum withdrawal). Therefore, at low mitogen or nutrient levels, mTOR and protein phosphatase 2A catalytically control the constitutive nuclear import of latent STAT1 by KPNA1, which are key modulators of STAT1 expression and apoptosis.
Introduction
Mammalian target of rapamycin (mTOR)2 is a highly conserved serine-threonine kinase that senses intracellular nutrient levels and mitogenic signals and directly modifies anabolic effector proteins. mTOR nucleates two distinct macromolecular protein complexes. mTOR complex 1 (mTORC1) phosphorylates signaling intermediates (e.g. p70 S6 kinase and eIF4E-binding protein 1) that permit protein synthesis, cell growth, and cell proliferation. mTORC1 contains the adaptor protein raptor and is potently inhibited by rapamycin (1–3). mTORC2 contains rictor, is resistant to the acute effects of rapamycin and controls cytokinesis and cell survival (4). Cell stress conditions that reduce mTORC1 activity, such as nutrient starvation, can initiate catabolic responses designed to recycle energy substrates (e.g. autophagy) or reduce energy-consuming processes (e.g. oxidative phosphorylation and ribosomal biogenesis) (5).
In yeast, regulation of gene transcription by TORC1 is an important component of the cellular response to physical or nutrient stress (6). For instance, in a mechanism that requires the TOR-associated phosphatase subunits Tap42 and Pph1, reduced TOR activity led to increased nuclear content of the stress response transcription factors Gln3 or Msn2/4. Nuclear import of Gln3 required the karyopherin Srp1 (7). The mammalian homologues of Tap42 and Pph1 are α4 and PP2Ac; that for Srp1 is karyopherin-α1 (KPNA1; also known as importin-α5). In mammalian cells, the karyopherin-α family contains at least six distinct isoforms; each acts as an adaptor for the importin-β-mediated nuclear import of a different subset of cargo proteins (8, 9). For mammalian stress response proteins (e.g. FoxO3A, NF-κB, and STAT1), nuclear transport in part regulates the transcription of their target genes (10–12).
We recently reported a functional and physical association between mTOR and the transcription factor STAT1 (11). In cells exposed to IFNs, phosphorylation of STAT1 at Tyr-701 permits its homodimerization, translocation to the nucleus, and binding to regulatory regions of interferon-sensitive genes. Phosphorylation of Ser-727 promotes STAT1 transcriptional activity and confers recognition by the nuclear export apparatus (13). However, recent studies indicate that, even in the absence of interferons, “latent” (i.e. unphosphorylated) STAT1 is required for the constitutive expression of apoptosis, cell cycle arrest, and immunomodulatory genes (e.g. caspase-3, p27) (14, 15). Our own studies revealed that inhibition of mTOR kinase activity or depletion of α4 or PP2Ac increased the constitutive nuclear content of latent STAT1 (11). This was associated with enhanced or prolonged expression of STAT1 and other STAT1-dependent genes in cells exposed to IFN-γ. The control of STAT1 nuclear trafficking by mTOR suggested a novel mechanism by which metabolic signals might be coupled to specific stress transcriptional programs.
In the current study, we hypothesized that mTOR regulates KPNA1, a STAT1 karyopherin and mammalian homologue of Srp1. We demonstrate that KPNA1 interacts with mTORC1 in a complex that includes STAT1 and the mTOR-associated phosphatase PP2Ac. KPNA1 was required for the enhancing effect of rapamycin or nutritional stress on constitutive STAT1 nuclear import, the constitutive expression of latent STAT1, and levels of cleaved caspase-3. Our results indicate that mTOR controls an apoptosis transcriptional program via control of its nuclear import.
EXPERIMENTAL PROCEDURES
Cell Culture and Transfection
Human epithelial adenocarcinoma (A549) and HEK 293T cells were cultured as previously described (16, 17). COS7 and mouse embryonic fibroblast (MEF) cells were cultured in DMEM supplemented with 10% fetal bovine serum, penicillin (100 units/ml), and streptomycin (100 μg/ml). MEFs were derived from mice with heterozygous (control) or homozygous genomic deletions of the KPNA1 gene (18) and verified by genotyping. STAT1-deficient (U3A) cells, U3A cells constitutively expressing recombinant STAT1 (U3A-R), and their wild-type parental control (2fTGH) were obtained from Dr. G. Stark (Cleveland Clinic) and propagated as previously described (17, 19). The cells were incubated without or with glucose (DMEM without glucose; Invitrogen), FBS, or rapamycin (EMD; 50 ng/ml), for the indicated times. For the heterologous expression of recombinant proteins, subconfluent A549 or COS7 cells were incubated with serum-free medium and mammalian expression vectors, 0.5–1.0 μg of plasmid DNA/9.6 cm2 of culture surface area and incubated with Lipofectamine 2000 or LTX (Invitrogen) as previously described (11). HEK 293T were transfected using calcium phosphate/DNA precipitates for 24 h in complete medium and incubated with Dulbecco's modified Eagle's medium containing 10% bovine serum albumin for 24 h prior to stimulation. For the siRNA-mediated depletion of KPNA1, A549 cells were transfected with 10 nm siRNA duplexes (siGENOME; Dharmacon) directed against KPNA1 using Dharmafect I, according the manufacturer's protocol. A nontargeting siRNA (siCONTROL) was used as a negative control. After 72 h, experimental protocols were initiated as indicated, and lysates were prepared for detection of protein or mRNA.
Construction of KPNA1 Bacterial and Mammalian Expression Plasmids
The cDNA encoding wild-type KPNA1 (Gene identification number 3836) was obtained in a Gateway pDONR 221 entry vector (ultimate ORF clone IOH3595; Invitrogen) and verified by automated sequencing. For bacterial expression of GST-KPNA1, the KPNA1 cDNA was transferred to Gateway destination vector pDEST15 by recombination, before transformation of Escherichia coli BL21 cells, and induction of protein synthesis with 2% arabinose as per the manufacturer's protocol (Invitrogen). Purification of GST-KPNA1 from crude bacterial lysates was performed by immobilization on glutathione-Sepharose (GE Healthcare), before recovery in elution buffer (50 mm Tris-HCl, pH 8, 40 mm glutathione), and verification by Coomassie Blue staining and Western blot analysis. For mammalian expression, the wild-type KPNA1 cDNA was transferred to the gateway destination vectors pcDNA3.1/nV5/ECFP-DEST or pcDNA3.1/VF2/DEST as described (11).
Preparation of Cell Lysates for Detection of Proteins or Protein Complexes
Endogenous or recombinant proteins in whole cell or fractionated lysates were detected by immunoprecipitation or Western blot analysis using the antibodies listed in supplemental Table S1. Whole cell lysates were generated after washing cells once with cold PBS and incubating for 15 min on ice in lysis buffer (20 mm Tris, pH 8.0, 0.3% CHAPS, 1 mm EDTA, 10 mm β-glycerophosphate, aprotinin, 10 μg/ml, leupeptin, 10 μg/ml, 1 mm PMSF, 50 mm NaF, 100 μm sodium orthovanadate). After freezing and thawing, the cells were homogenized and centrifuged at 1000 × g for 5 min. Supernatants were then centrifuged (16,000 × g for 30 min) to generate particulate-free lysates. For immunoprecipitation experiments, the proteins from whole cell lysates were incubated with control IgG, anti-STAT1, mTOR, HA, PP2Ac, or V5 antibody (each 5 μg) overnight at 4 °C before the addition of 20 μl of protein G-Sepharose for 1 h. The pellets were washed three times with PBS containing 0.3% CHAPS before solubilization of bound proteins in SDS sample buffer for 5 min at 95 °C, separation by SDS-PAGE, and detection of bound proteins by Western blot analysis. The cloning of HA-PP2Ac was described previously (11). Proteins from nuclear lysates were immunoprecipitated with 2 μg of IgG, anti-V5, or PP2Ac antibodies. Protein G pellets were washed three times with 1 m LiCl in PBS, three times with 0.3% CHAPS in PBS, and once with PBS, before solubilization and Western blot analysis.
Preparation of Nuclear Lysates
A549 cell nuclear lysates were prepared as described previously (20). Briefly, cell pellets were resuspended in nuclear lysis buffer 1 (20 mm Tris, pH 7.5, 10 mm KCl, 1 mm DTT, aprotinin, 1 μg/ml, leupeptin, 1 μg/ml, 0.5 mm PMSF, 100 μm sodium orthovanadate) before homogenization (Dounce, Pestle B) and centrifugation (1100 × g for 3 min). The supernatants were then centrifuged (16,000 × g for 30 min) and used as cytosolic fractions, whereas pelleted nuclei were resuspended in 2 volumes of nuclear lysis buffer 2 (20 mm Tris, pH 7.5, 500 mm KCl, 1 mm DTT, aprotinin, 1 μg/ml, leupeptin, 1 μg/ml, 0.5 mm PMSF, 100 μm sodium orthovanadate) before freezing, thawing, and centrifugation for 30 min at 16,000 × g. HEK 293T cells were incubated with nuclear lysis buffer (10 mm HEPES, pH 7.9, 10 mm NaCl, 3 mm MgCl2, 1 mm DTT, 1 mm PMSF, aprotinin, 1 μg/ml, leupeptin, 1 μg/ml, 100 μm sodium orthovanadate), and nuclei were extracted by homogenization (Dounce Pestle B, 8 strokes), pelleted by centrifugation (800 × g for 3 min at 4 °C), and washed twice with nuclear lysis buffer. Supernatants were centrifuged at 100,000 × g for 1 h at 4 °C to generate the cytosolic (supernatant) and membrane (pellet) fractions. The pellets were washed twice with nuclear lysis buffer before sonication (three or four short bursts on medium setting). Proteins from the nuclear, cytosolic, and membrane fractions were separated by SDS-PAGE, and the indicated proteins were detected by Western blot analysis.
In Vitro Phosphorylation Assay
HEK 293T cells were transfected with Myc-tagged Raptor and either wild-type or kinase inactive mTOR as previously described (21). mTORC1 was immunoprecipitated using anti-Myc antibodies, and beads were washed three times in CHAPS lysis buffer followed by three washes in CHAPS buffer supplemented with 150 mm NaCl. The assays were performed for 30 min at 30 °C with bacterially purified recombinant GST-eIF4E-binding protein 1 (21) or GST-KPNA1 (each 2 μg/assay), and 100 μm ATP with 10 μCi of [γ-32P]ATP in mTOR kinase buffer (25 mm HEPES, pH 7.4, 50 mm NaCl, 50 mm β-glycerophosphate, 10 mm MnCl2). All of the samples were subjected to SDS-PAGE, and incorporation of radioactive phosphate (32P) was quantified using a Fuji PhosphorImager with ImageQuant software. In separate experiments, recombinant V5-CFP-KPNA1 heterologously expressed in HEK 293T cells was immunoprecipitated using anti-V5 antibody before washing and incubation with mTOR kinase buffer without or with 100 μm ATP or 50 nm okadaic acid for 60 min at 30 °C. After separation by SDS-PAGE, phospho-KPNA1 was detected using an anti-serine/threonine antibody by Western blot analysis.
Real Time PCR
RNA was extracted (Illustra RNAspin kit; GE Healthcare), and cDNA was generated by reverse transcription from 2 μg of RNA (Superscript II, Invitrogen). Sybr green-based real time PCR was performed using 1 μl of cDNA and Power Sybr green universal PCR master mix (ABI). Primers are listed in supplemental Table S2. PCRs were carried out for 45 cycles (ABI 7500 real time PCR system). The results are expressed as fold induction in mRNA levels as calculated by the ΔΔCT method (22).
Fluorescence Imaging of STAT1
For detection of endogenous STAT1, A549, COS7, or mouse embryonic fibroblast cells were fixed with 4% paraformaldehyde for 15 min at room temperature before permeabilization with 0.2% Triton X-100 and incubation with mouse αSTAT1 antibody (Santa Cruz), and then Alexa Fluor 568 was conjugated to anti-mouse antibody (Invitrogen). Construction of the plasmid for mammalian expression of recombinant ECFP-tagged STAT1 (pcDNA3.1/nV5/ECFP-DEST-STAT1) was previously described (11). We used the vector pDONOR221 containing wild-type STAT1 as a template to change serine 727 to alanine (S727A), tyrosine 701 to phenylalanine (Y701F), and leucine 407 to alanine (L407A) by site-directed mutagenesis (Stratagene) using the oligonucleotide primers listed in supplemental Table S3. The mutated STAT1 cDNAs were transferred from pDONR 221 to pcDNA3.1/nV5/ECFP-DEST by recombination (11). After heterologous expression in A549 or COS7 cells, fluorescence was detected by multi-track image acquisition (ECFP: excitation, 458 nm, and emission, 475 nm; Alexa Fluor 568: excitation, 578 nm, and emission, 603 nm; DAPI: excitation, 405 nm, and emission, 475 nm) using a Plan-Neofluar 40×/1.3× oil differential interference contrast objective and Zeiss LSM 510 META scanning confocal microscope. The images were acquired at room temperature using the integrated Zeiss AxioCam HR digital sensor and LSM 510 software. Fluorescence intensity was assessed by measuring pixel density with ImageJ software (version 1.38x; National Institutes of Health), and the images were displayed using Zeiss LSM image browser (version 4.2.0.121; Carl Zeiss MicroImaging). The results are expressed as the ratio of nuclear to cytoplasmic pixel density.
Protein Fragment Complementation Assay
Gateway destination plasmid vectors for protein fragment complementation assays and the cloning of Gateway entry vector pDONR221-PP2Ac wild type and dominant negative (i.e. L199P) are described previously (11). pDONR221-KPNA1 was obtained from Invitrogen (Ultimate ORF Clone IOH3595). cDNAs encoding wild-type PP2Ac, dominant negative PP2Ac, or wild-type KPNA1 were transferred by recombination to pcDNA3.1-based protein fragment complementation assay mammalian expression vectors 3′ to each of the complementary fragments of venus yellow fluorescence protein (VF1 and VF2). Vector plasmids, each 1 μg/9.6 cm2 of dish surface area, were co-transfected in a 1:1 ratio (VF1-KPNA1:VF2-PP2Ac) by liposomal transfection (Lipofectamine LTX reagent; Invitrogen). For each biological replicate, the cells were counted, and fluorescence was measured (excitation, 512 nm, and emission, 529 nm) using a SpectraMax M2 fluorometer (Molecular Devices). The data are expressed as fluorescence units/cell.
RESULTS
A STAT1 Mutant (L407A) That Does Not Bind KPNA1 Cannot Undergo Nuclear Import in Cells Exposed to Rapamycin
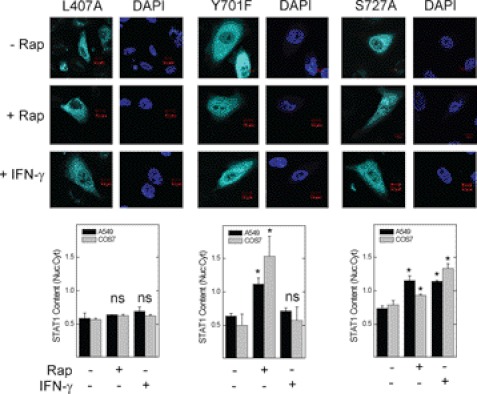
To determine whether inactivation of mTOR initiates the import of STAT1 to the nucleus, we expressed the STAT1 L407A mutant, which cannot bind the STAT1 karyopherin KPNA1 or translocate to the nucleus in response to IFN-γ (23). To simulate unphosphorylated (latent) STAT1, we expressed the STAT1 Y701F mutant, which cannot dimerize or translocate to the nucleus in response to interferons (24). In A549 or COS7 cells, the L407A mutant failed to undergo nuclear translocation in response to rapamycin (Fig. 1, left panel), suggesting a role for KPNA1 in mediating the effect of mTOR on nuclear import. Consistent with previous studies (25), nuclear import of the L407A or Y701F mutant was blocked in cells exposed to IFN-γ (Fig. 1, left and middle panels); however, in cells exposed to rapamycin, nuclear import of the Y701F mutant was retained. Thus, rapamycin induces STAT1 nuclear import independent of Tyr-701 phosphorylation or IFN-γ receptor ligation. Phosphorylation at Ser-727 permits STAT1 nuclear export (11); however, the enhancing effect of rapamycin was unaltered by expression of the STAT1 S727A mutant. Our data suggest that mTOR regulates constitutive nuclear import of unphosphorylated STAT1 via the importin KPNA1.
FIGURE 1.
A KPNA1 binding mutation in STAT1 blocks rapamycin-induced STAT1 nuclear import. A549 or COS7 cells expressing the recombinant ECFP-STAT1 trafficking mutants L407A, Y701F, or S727A were incubated without or with 50 ng/ml rapamycin (Rap) or 100 units/ml IFN-γ for 0 or 1 h. ECFP-STAT1 (cyan) was detected by direct fluorescence confocal microscopy. The slides were mounted with solution containing the nuclear marker DAPI (navy blue). Summarized data (mean nuclear to cytoplasmic pixel density ratio ± S.E., n = 3–5 cells/experiment) are shown below the images and are representative of three independent experiments. *, p < 0.05 versus control by Student's t test; ns, not statistically significant.
STAT1 and KPNA1 Interact with mTORC1
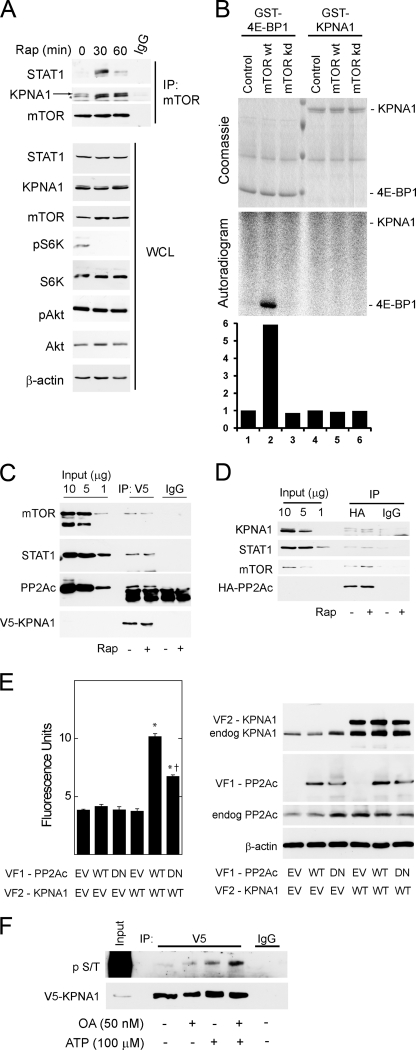
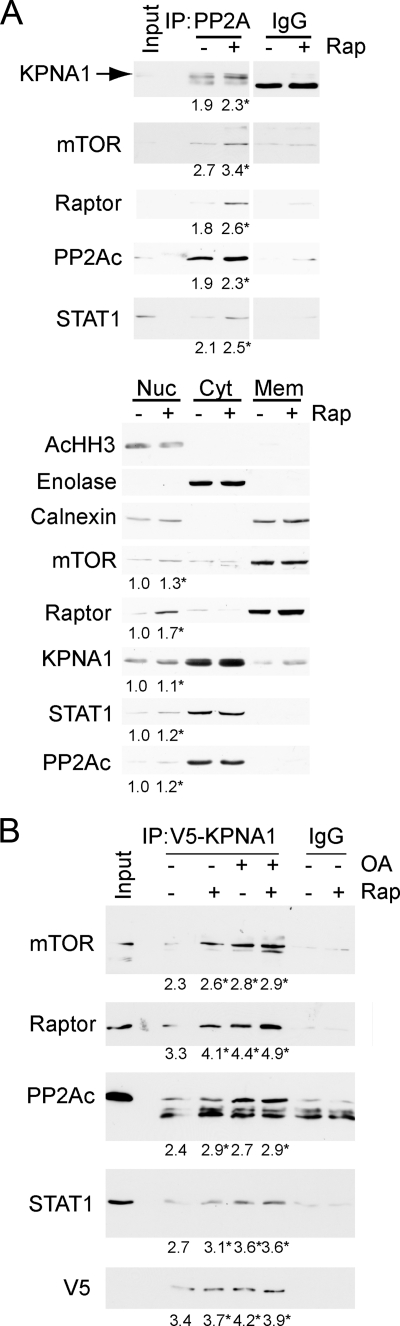
To demonstrate a physical association between mTOR and KPNA1, mTOR-containing complexes were immunoprecipitated in HEK 293T cells exposed to rapamycin for 0, 30, or 60 min (Fig. 2A). Rapamycin led to a time-dependent increase in the association between mTOR and KPNA1 or STAT1. Two distinct bands were detected by anti-KPNA1 antibody, and both were associated with mTOR. Consistent with previous studies, the higher molecular weight band was absent in KPNA1-depleted or knock-out cells (supplemental Fig. S1A; see Figs. 4A and 7B) (26). Rapamycin did not affect levels of mTOR, STAT1, or KPNA1 in whole cell lysates but potently blocked the phosphorylation of p70 S6 kinase at Thr-389, a marker of mTORC1 activity (1, 2). As shown previously, short term exposure to rapamycin failed to inhibit the phosphorylation of Akt at Ser-473, a marker of mTORC2 activity (27, 28). STAT1 immunoprecipitates contained raptor, but not rictor, consistent with its regulation by mTORC1 and its sensitivity to rapamycin (supplemental Fig. S2). Full-length recombinant KPNA1 was not a direct substrate for mTORC1 kinase activity by in vitro kinase assays (Fig. 2B). These results indicate that endogenous mTORC1, STAT1, and KPNA1 can associate in a macromolecular complex, the recovery of which is enhanced by rapamycin. Like STAT1 (11), KPNA1 does not appear to be a direct substrate for the kinase activity of mTOR, suggesting that mTORC1 might regulate the constitutive nuclear trafficking of KPNA1 and STAT1 via its associated phosphatase PP2Ac. In agreement, co-immunoprecipitation experiments revealed that mTOR, raptor, PP2Ac, and STAT1 were detected in V5-tagged KPNA1 immunoprecipitates (Fig. 2C).
FIGURE 2.
Physical associations between mTOR, its associated phosphatase PP2Ac, and KPNA1 in intact cells. A, HEK 293T cells were exposed to 50 ng/ml rapamycin (Rap) for 0, 30, or 60 min before preparation of whole cell lysates and immunoprecipitation with α-mTOR antibody (IP: mTOR) or normal IgG. Immunoprecipitated proteins or those in whole cell lysates (WCL) were detected by Western blot analysis. The arrow indicates KPNA1, and the asterisk indicates a protein other than KPNA1 that is detected by the anti-KPNA1 antibody. B, HEK 293T cells were transfected with Myc-tagged raptor and either a control vector, wild-type (wt), or kinase inactive (kd) mTOR. After lysis, proteins were immunoprecipitated with α-Myc antibody. mTORC1-containing immunoprecipitates were washed and incubated with [γ-32P]ATP and either recombinant GST-tagged eIF4E-binding protein 1 or KPNA1 in an in vitro kinase reaction. The products were separated by SDS-PAGE before staining with Coomassie Blue or autoradiography. The data in the lower panel show the fold changes in band density (lanes 1–6) versus control = 1 (lane 1). C, HEK 293T cells expressing V5-ECFP-KPNA1 were incubated without or with 50 ng/ml rapamycin (Rap) for 30 min before preparation of whole cell lysates. After immunoprecipitation (IP) with α-V5 antibody, the indicated KPNA1-interacting proteins were detected by Western blot. D, HEK 293T cells expressing HA-PP2Ac were incubated without or with 50 ng/ml rapamycin for 30 min before preparation of whole cell lysates. After immunoprecipitation with anti-HA antibody, the indicated PP2Ac-interacting proteins were detected by Western blot. E, COS7 cells were co-transfected with plasmids for heterologous expression of venus yellow fluorescence protein fragments VF1 or VF2 (empty vector, EV) or fragments linked to wild-type (WT) or dominant negative (DN) PP2Ac or wild-type KPNA1. Direct protein-protein interactions were signaled by increased fluorescence detection upon reconstitution of full-length YFP (excitation, 485 nm; emission, 535 nm; cutoff, 530 nm). Summarized data are the mean fluorescence units/cell ± S.E. in triplicate samples and representative of three individual experiments. *, p < 0.05 versus empty vector control (lane 1); †, p < 0.05 versus lane 5 by Student's t test. Levels of endogenous or recombinant PP2Ac or KPNA1 were detected by Western blot analysis (right panel). F, V5-CFP-KPNA1 heterologously expressed in HEK 293T cells was immunoprecipitated as in C before incubation without or with 100 μm ATP, 50 nm okadaic acid (OA), or both for 1 h at 30 °C. V5 or normal IgG (control) immunoprecipitates were separated by SDS-PAGE before detection of V5-CFP-KPNA1 or phosphoserine/phosphothreonine (p S/T) by Western blot analysis. The images are representative of three individual experiments.
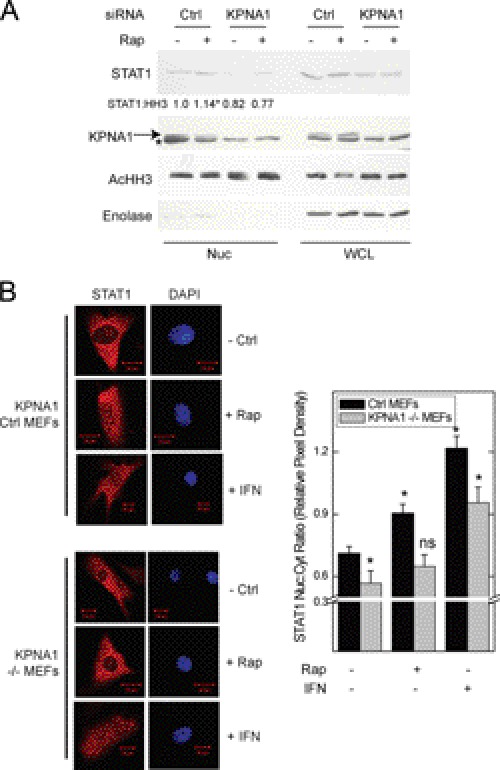
FIGURE 4.
KPNA1 is required for the enhancing effect of rapamycin on STAT1 nuclear content. A, A549 cells were transfected with control (Ctrl) siRNAs or those targeting KPNA1 for 72 h before incubation for 1 h with fresh medium without or with 50 ng/ml rapamycin (Rap) preparation of nuclear (Nuc) or whole cell (WCL) lysates and detection of the indicated proteins by Western blot. The arrow indicates KPNA1, and the asterisk indicates another non-KPNA1 reactive band. Gels are representative of four individual experiments. Nuclear STAT1 protein levels are quantified as fold change in band density relative to control = 1 (means ± S.E.). *, p < 0.05 versus control-transfected cells without rapamycin). B, control or KPNA1-deficient (−/−) MEFs were incubated without or with 50 ng/ml rapamycin or IFN-γ, 100 units/ml, for 0 (− Rap) or 1 h (+ Rap). Endogenous STAT1 (red) was detected by indirect immunofluorescence confocal microscopy. The slides were mounted with solution containing the nuclear marker DAPI (navy blue). Shown to the right of representative images are summarized data (mean nuclear to cytoplasmic pixel density ratio ± S.E., n = 3–5 cells/experiment) representative of five independent experiments. *, p < 0.05 versus control; ns, not significant by Student's t test.
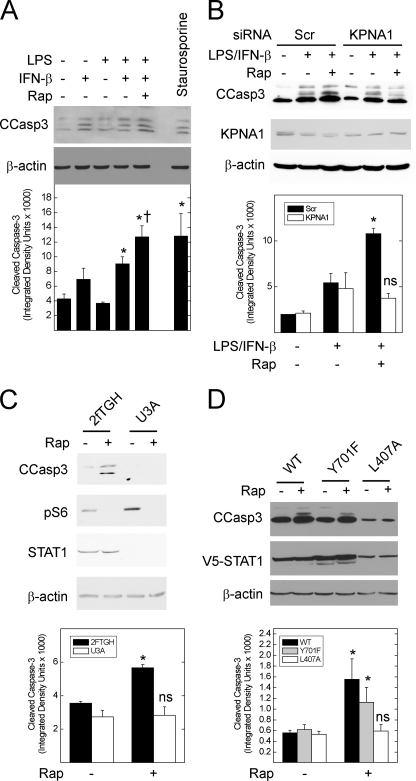
FIGURE 7.
Inhibition of mTOR increases the levels of cleaved caspase-3 in STAT1- and KPNA1-dependent fashion. A, A549 cells were incubated without or with LPS/IFN-β (5 μg/ml/250 units/ml) in the absence or presence of 50 ng/ml rapamycin (Rap) or 20 μm staurosporine for 24 h before generation of whole cell lysates and detection of the indicated proteins by Western blot analysis. Band densitometry for cleaved caspase-3 is shown for each lane (lower panel). B, A549 cell were transfected with scrambled control (Scr) or anti-KPNA1 (KPNA1) siRNA 48 h before the addition of vehicle or rapamycin in the absence or presence of LPS/IFN-β for 24 h and Western blot analysis. C, untransfected 2fTGH or U3A cells were exposed to vehicle or 50 ng/ml rapamycin for 24 h before Western blot analysis. D, U3A cells were transfected with mammalian expression plasmids for wild-type (WT) STAT1 or that containing the Y701F or L407A mutations 24 h before the addition of vehicle or 50 ng/ml rapamycin for 24 h and Western blot analysis. For B and C, mean band densitometry (± S.E.) for cleaved caspase-3 is shown in the lower panels and represents three or four individual experiments. *, p < 0.05 versus control; †, p < 0.05 versus LPS/IFN-β alone; ns, not significant by Student's t test.
PP2Ac Physically and Functionally Associates with KPNA1 and mTORC1 in Intact Cells
In HEK 293T whole cell lysates, heterologously expressed HA-tagged PP2Ac associated with mTOR, KPNA1, and STAT1 (Fig. 2D); the association between HA-PP2Ac and KPNA1 was not significantly altered by rapamycin (Fig. 2, C and D). Because the physical association between PP2Ac and KPNA1 in whole cell lysates appeared weak, we sought additional evidence for a functional interaction. In agreement with immunoprecipitation results, we detected a direct interaction between recombinant PP2Ac and KPNA1 in intact cells by protein fragment complementation assay (Fig. 2E). The interaction was attenuated upon co-expression of a phosphatase-dead mutant (L199P) of PP2Ac. The complementary forms of recombinant KPNA1 and PP2Ac were expressed at similar levels (Fig. 2E, right panel). By in vitro kinase assay, okadaic acid increased the phosphorylation of V5-CFP-KPNA1 in V5 immunoprecipitates (Fig. 2F). Consistent with an associated serine-threonine kinase activity, the addition of exogenous ATP increased phospho-KPNA1 levels. ATP-dependent increases in phospho-KPNA1 levels were further increased by the PP2Ac inhibitor okadaic acid. The enhancing effect of okadaic acid on KPNA1 phosphorylation is consistent with the observed direct interaction between KPNA1 and PP2Ac (Fig. 2E).
The PP2Ac, KPNA1, and mTORC1 Complex Is Enriched in Nuclear Extracts
The amounts of mTOR, KPNA1, and STAT1 recovered with HA-PP2Ac immunoprecipitated from whole cell lysates were small; in fact, they could not be detected using commercially available immunoprecipitating antibodies against endogenous PP2Ac (data not shown). In contrast to whole cell lysates, immunoprecipitation of endogenous PP2Ac from nuclear lysates revealed a robust association with mTORC1 and KPNA1 (Fig. 3A). Rapamycin increased the total and immunoprecipitated levels of nuclear mTORC1, PP2Ac, or STAT1, suggesting that these proteins likely traffic to the nucleus together in a complex (Fig. 3A, top and bottom panels). Similarly, mTOR, raptor, PP2Ac, and STAT1 were recovered from V5-KPNA1 immunoprecipitates from nuclear lysates (Fig. 3B). Rapamycin or the PP2Ac inhibitor okadaic acid enhanced the recovery of mTORC1, PP2Ac, and KPNA1. These data indicate that KPNA1 nuclear cargo includes mTORC1, PP2Ac, and STAT1, and their nuclear import is sensitive to mTORC1 or PP2Ac inhibition.
FIGURE 3.
mTORC1 interacts with endogenous nuclear PP2Ac, KPNA1 and STAT1 in mTORC1- and PP2Ac-dependent fashion. A, HEK 293T cells were exposed to 50 ng/ml rapamycin (Rap) for 0 or 30 min before preparation of nuclear (Nuc), cytosolic (Cyt), or membrane (Mem) homogenates. The nuclear proteins were immunoprecipitated with α-PP2Ac or normal IgG, and associated proteins were detected by Western blot. The images in A are composites from sections of the same gel. Levels of the organelle markers acetylated histone H3 (acHH3, nucleus), calnexin (cytoplasmic membrane), or enolase (cytosol) in each subcellular fraction, as well as the indicated mTORC1-associated proteins, are shown below. B, HEK 293T cells expressing V5-ECFP-KPNA1 were exposed to vehicle or 50 nm okadaic acid before the addition of vehicle or 50 ng/ml rapamycin for 30 min, preparation of nuclear lysates, and immunoprecipitation with α-V5 antibody or normal IgG. Mean band density or mean fold change (untreated control = 1) in band density is indicated below each gel image (standard error of <5% of the mean for three individual experiments). *, p < 0.05 versus untreated control by Student's t test.
KPNA1 Is Required for Rapamycin-induced STAT1 Nuclear Import
The physical and functional interaction between PP2Ac, KPNA1, mTORC1, and STAT1 suggested that KPNA1 might be required for the increased STAT1 nuclear content observed in cells exposed to rapamycin. Nuclear proteins were isolated from A549 cells transfected with control or anti-KPNA1 siRNA and exposed to vehicle or rapamycin. Rapamycin failed to increase STAT1 nuclear import in KPNA1-depleted cells (Fig. 4A). The siRNAs depleted KPNA1 with no apparent off target effects (supplemental Fig. S1C). STAT1 mRNA levels were reduced in cells depleted of KPNA1, consistent with regulation of constitutive STAT1 mRNA levels by latent STAT1 nuclear content (supplemental Fig. S1C) (29). To show that KPNA1 mediates the effect of rapamycin on STAT1 nuclear import independent of STAT1 mRNA levels, we used STAT1-deficient fibroblasts (U3A cells) stably transfected with a lentiviral expression vector for STAT1. Rapamycin increased nuclear levels of STAT1 in U3A-R cells transfected with control siRNA or shRNA, but not in those transfected with anti-KPNA1 siRNA or shRNA (supplemental Fig. S4, A and B). STAT1 protein levels were unchanged in the cytoplasmic fractions of U3A-R cells depleted of KPNA1; STAT1 mRNA levels were also unaffected by RNAi-mediated KPNA1 depletion (supplemental Fig. S4, C and D).
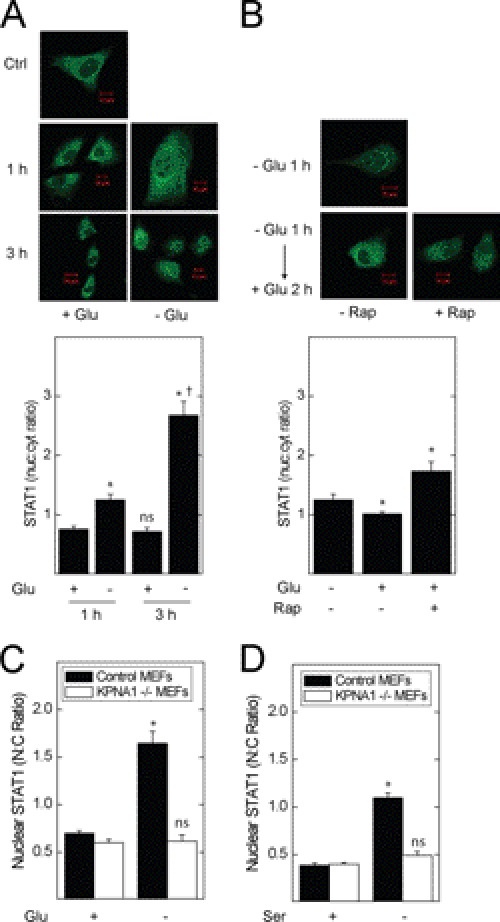
In separate experiments, we showed the requirement for KPNA1 in the rapamycin-induced nuclear translocation of latent STAT1 by immunofluorescence microscopy using MEFs with genetically deleted KPNA1 (KPNA1−/−). We used heterozygous (KPNA1+/−) cells for controls, which permit KPNA1-dependent import and contain the same integrated cassette as their −/− counterparts. Exposure of KPNA1-expressing cells to rapamycin led to the nuclear import of STAT1 (Fig. 4B) (11); this effect of rapamycin was abolished in KPNA1 knock-out cells. In contrast to rapamycin, IFN-γ-induced STAT1 nuclear translocation remained intact, suggesting an alternative mechanism for inducible, but not constitutive, STAT1 nuclear import. The results represent two separately derived clones of KPNA1+/− and KPNA1−/− MEFs. In control experiments, KPNA1 mRNA and protein levels were beyond the limits of detection by real time PCR or Western blot analysis when compared with control cells (supplemental Fig. S1, A and B). These data indicate that the effect of mTOR blockade on STAT1 nuclear import is via KPNA1.
Glucose or Serum Deprivation Increases STAT1 Nuclear Content in mTOR and KPNA1-dependent Fashion
To determine whether physiological modulators of mTOR activity affect STAT1 nuclear import, we exposed A549 cells to nutritional stress. Incubation with glucose-free medium for 1 or 2 h increased STAT1 nuclear content (Fig. 5A). Following starvation for 1 h, repletion of glucose for 2 h partly restored base-line STAT1 nuclear content; rapamycin reversed this effect, indicating a requirement for mTORC1 in the suppression of STAT1 nuclear content by glucose repletion (Fig. 5B). Glucose or serum withdrawal reduced the phosphorylation of p70 S6 kinase, an mTORC1 effector, in A549 cells (supplemental Fig. S3). In KPNA1+/− MEFs, glucose or serum withdrawal increased STAT1 nuclear levels; this effect was absent in KPNA1-deficient cells (Fig. 5, C and D). Therefore, physiological inhibitors of mTORC1 activity increase STAT1 nuclear content in a mTORC1- and KPNA1-dependent fashion.
FIGURE 5.
Glucose or serum levels regulate STAT1 nuclear transport in mTOR- and KPNA1-dependent fashion. A549 cells were incubated with glucose-containing or glucose-free medium for 1 or 3 h (A) or glucose-free medium (B) for 1 h (− Glu 1 h) or glucose-free medium for 1 h followed by glucose repletion for 2 h (− Glu 1 h → + Glu 2 h) in the absence or presence of 50 ng/ml rapamycin. STAT1 (green) was detected by indirect immunofluorescence confocal microscopy. Shown below the representative images are summarized data (mean nuclear to cytoplasmic pixel density ratio ± S.E., n = 3–5 cells/experiment) representative of three independent experiments. In separate experiments, control (Ctrl) or KPNA1-deficient MEFs were exposed to complete, glucose-free (C) or serum-free medium (D) for 1 h before detection of STAT1 by indirect immunofluorescence confocal microscopy. Shown are summarized data (mean nuclear to cytoplasmic pixel density ratio ± S.E., n = 3–5 cells/experiment) representative of three to five independent experiments. *, p < 0.05 versus control; †, p < 0.05 versus − Glu 1 h; ns, not significant versus control by Student's t test.
KPNA1 Is Required for Enhancing Effect of Serum Starvation on Constitutive STAT1 Gene Expression
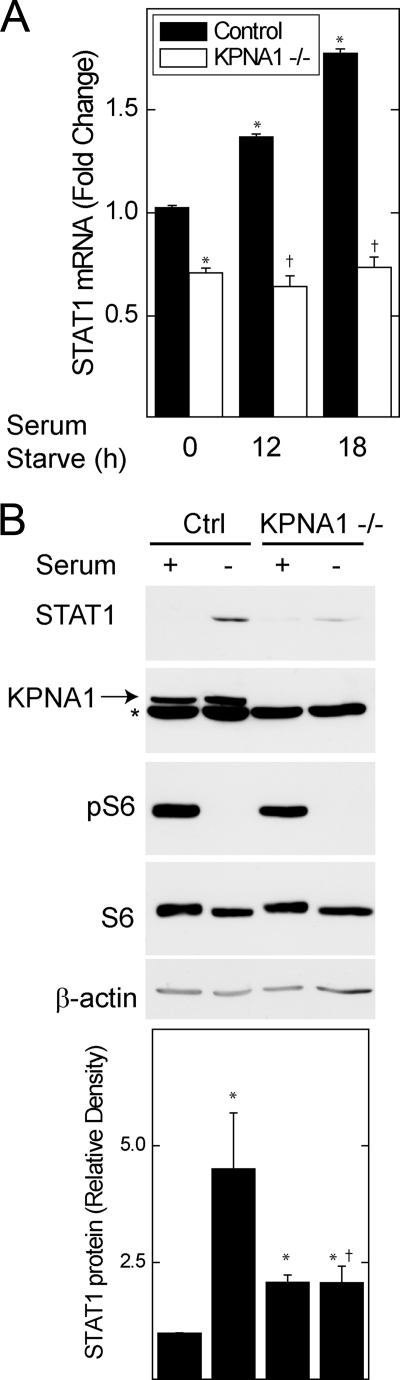
STAT1 is a STAT1-dependent gene (30), and its induction can be enhanced by augmented levels of unphosphorylated, or latent, STAT1 (29). In control MEFs, serum starvation increased STAT1 mRNA levels and protein levels in the absence of cytokine inducers of STAT1 phosphorylation at Tyr-701 (Fig. 6). The starvation-induced increase in STAT1 mRNA and protein levels was abolished in KPNA1 knock-out MEFs. Consistent with inhibition of mTORC1, phosphorylation of S6 at Ser-235/236 was blocked in serum-starved cells. Despite a reduction in STAT1 mRNA levels in KPNA1−/− cells (supplemental Fig. S1A), STAT1 protein levels were slightly increased (Fig. 6B), indicating that the effects of KPNA1 deficiency on STAT1 bioactivity (Figs. 5–7) were not due to a global reduction in STAT1 mRNA or protein levels. Together, our results indicate that mitogen and nutrient levels regulate the nuclear import activity of KPNA1, the nuclear import of latent STAT1, and the constitutive expression of STAT1 via mTORC1.
FIGURE 6.
Growth factor withdrawal increases STAT1 mRNA levels in KPNA1-dependent fashion. A, control or KPNA1-deficient MEFs were exposed to serum-free medium for 0, 12, or 18 h before detection of STAT1 mRNA by real time PCR. The data are shown as the mean fold changes versus control = 1 ± S.E. as determined by the ΔΔCT method. B, control (Ctrl) or KPNA1-deficient MEFs were exposed to serum-free medium for 0 or 18 h before detection of STAT1 protein levels by Western blot. The arrow indicates KPNA1, and the asterisk indicates a protein other than KPNA1 that is detected by the anti-KPNA1 antibody. Shown below are the means of STAT1 normalized to β-actin band densities (± S.E., n = 3). The data represent experiments on two separately derived cultures for each cell type. *, p < 0.05 versus control; †, not significant versus KPNA1−/− control by Student's t test.
Inactivation of mTOR Enhances Cleaved Caspase-3 Levels in STAT1- and KPNA1-dependent Fashion
To determine whether the enhancement of STAT1 bioactivity by rapamycin translates into a biological effect, we measured cleaved caspase-3 levels in A549 cells exposed to bacterial LPS and IFN-β, two important mediators of STAT1-dependent innate immune responses (31), for 24 h. Rapamycin increased cleaved caspase-3 levels in cells exposed to LPS/IFN-β (Fig. 7A). Incubation with LPS/IFN-β and rapamycin achieved a level of cleaved caspase-3 similar to that observed in cells exposed to staurosporine, a potent inducer of apoptosis. Depletion of KPNA1 by RNAi blocked the augmentation of LPS/IFN-β-induced cleavage of caspase-3 (Fig. 7B).
In contrast to A549 cells, rapamycin alone increased cleaved caspase-3 levels in a human fibrosarcoma (2fTGH) cell line; in the STAT1-deficient counterpart (U3A cells), rapamycin failed to increase cleaved caspase-3 levels (Fig. 7C). Expression of wild-type STAT1 or STAT1 Y701F restored rapamycin-induced caspase-3 cleavage in U3A cells, indicating that the effect of rapamycin is via unphosphorylated STAT1 (Fig. 7D). In contrast, rapamycin did not increase cleaved caspase-3 levels in U3A cells expressing the STAT1 L407A mutant, which cannot bind KPNA1. Because U3A cells are STAT1-deficient, rapamycin affects recombinant STAT1 directly via KPNA1 and not by reducing endogenous STAT1 mRNA levels. Taken together, the results support a requirement for KPNA1 and STAT1 in the increased caspase-3 cleavage observed under conditions of reduced mTOR activity.
DISCUSSION
Here, we demonstrate a requirement for the α-importin KPNA1 in the control of constitutive STAT1 nuclear import by mTORC1, a sensor of cellular metabolism and mitogen levels. mTOR associated with raptor, KPNA1, PP2Ac, and STAT1 in a complex that was enriched in nuclear extracts under conditions of reduced mTOR or PP2Ac activity. Consistent with a physiological role for mTOR in the regulation of constitutive STAT1 nuclear import, nutritional deprivation or mitogen withdrawal increased STAT1 nuclear import in KPNA1-dependent fashion. The enhancing effect of mitogen withdrawal on STAT1 nuclear trafficking was followed by increased STAT1 mRNA and protein levels, and both required KPNA1. In addition, both STAT1, its interaction with KPNA1, and KPNA1 were required for rapamycin-induced or enhanced cleavage of caspase-3, a marker of apoptosis. Our results indicate that mTORC1 can sense low cellular nutrient levels or mitogen withdrawal and subsequently associate with KPNA1 and STAT1 in a PP2Ac-regulated mechanism that promotes the nuclear import of latent STAT1 and its bioactivity. The constitutive levels of latent STAT1 regulate STAT1 mRNA levels, as well as anti-microbial, immune surveillance, and apoptotic responses (24, 29, 32).
The current study indicates that mTORC1 controls the nuclear import of STAT1 independent of ligation of the interferon receptor or phosphorylation at its Tyr-701 residue (Fig. 1). Similarly, the expression of STAT1 Y701F in U3A cells restores rapamycin-induced caspase-3 cleavage, confirming that the regulation of latent STAT1 by mTOR is biologically relevant. The phosphorylation of STAT1 Tyr-701 is required for its homodimerization and binding to interferon-γ activation sequence elements in the promoters of anti-microbial genes. However, emerging evidence indicates an important role for unphosphorylated, or latent, STAT1 in the control of immune modulatory or apoptosis genes (15, 32). STAT1 was required for the constitutive expression of apoptosis genes and TNF-α-induced cell death in the absence of interferons (14). STAT1, or its Y701F mutant, constitutively occupied the LMP2 (low molecular mass peptide 2) gene promoter and supported the transcription of LMP2, a gene involved in immune modulation (24). Stable expression of the STAT1 Y701F mutant prolonged the expression of a subset of interferon-sensitive genes involved in anti-viral responses and immune regulation, including STAT1 itself (29). Our data provide evidence for the regulation of latent STAT1 nuclear import by mTOR, a sensor of cellular metabolism, via the STAT1 karyopherin KPNA1. Consistent with a functional role for mTOR regulation of KPNA1 and nuclear trafficking, its inhibition by rapamycin, glucose deprivation, or mitogen withdrawal increased STAT1 mRNA levels in KPNA1-containing, but not deficient, mammalian cells (Figs. 4–6). Also, depletion of KPNA1 by siRNA or expression of KPNA1 binding mutant of STAT1 (L407A) blocked the ability of rapamycin to enhance or induce apoptosis (Fig. 7).
The KPNA1 nuclear cargo protein STAT1 interacted with raptor, and not rictor; we therefore focused on mTORC1 as a nutrient sensor and regulator of the nuclear import protein KPNA1. The failure of mTORC1 to phosphorylate recombinant KPNA1 in vitro suggested its potential regulation by PP2Ac (Fig. 2B). In agreement, others have shown that mTORC1 can control the phosphorylation of downstream effectors (e.g. p70 S6 kinase, Erk) via PP2Ac (33–35). In fact, by immunoprecipitation, mTORC1 physically associated with KPNA1 in a complex that included its associated phosphatase PP2Ac. The physical associations and their modification by rapamycin appeared weak in whole cell lysates, perhaps because of low affinity or solubility but more likely as a result of their localization to subcellular compartments (Fig. 3). Nonetheless, several observations support their biological significance. We previously showed interactions of similar magnitude between mTOR, α4, PP2Ac, and STAT1, as well as small but significant changes in response to rapamycin (11). In these studies, PP2Ac or its mTOR-associated adaptor protein α4 was required for the basal suppression of STAT1 nuclear content, as well as for the enhancing effect of rapamycin on the induction of STAT1-dependent genes. Consistent with its presence in the complex, KPNA1 interacted directly with PP2Ac (Fig. 2E), and PP2Ac activity regulated its phosphorylation (Fig. 2F), as well as its nuclear trafficking (Fig. 3). Moreover, the mTORC1 inhibitor rapamycin increased the nuclear import of wild-type STAT1, but not that of a STAT1 L407A mutant that cannot bind KPNA1 (Fig. 1). In agreement, mTORC1 control of cleaved caspase-3 levels also required the same physical interaction between KPNA1 and STAT1 (Fig. 7). The current findings are consistent with a functional complex containing mTORC1, KPNA1, and PP2Ac, as well as its involvement in the control of STAT1 bioactivity. This mechanism appears to be evolutionarily conserved, because the yeast homologues of α4 and PP2Ac (i.e. Tap42, Pph, and Sit4) also mediate TOR-dependent suppression of nuclear import via the α-importin Srp1 (6, 7). The exact kinase(s), and their target residue(s) in KPNA1, that control the phosphorylation KPNA1, as well as their roles in the nuclear import process, are the subject of ongoing proteomic studies.
Our results indicate that mTORC1, KPNA1, and STAT1 can traffic to the nucleus as a complex. That is, reduced mTORC1 activity was associated with increased recovery of the complex in nuclear lysates, and this correlated with increased nuclear levels of each of the individual proteins (Fig. 3A). Although several studies have revealed the nuclear localization of TOR, little was known regarding the mechanism of its transport or its nucleus-specific functions (20, 36, 37). Recent studies, however, indicate that TOR can bind chromatin and regulate the transcription of genes involved in ribosomal biogenesis, protein synthesis, or mitochondrial function (38, 39). The finding that mTORC1 regulates its own karyopherin-mediated nuclear import may represent a mechanism for the control of multiple stress response transcription factors and their target genes.
Although mTORC1 interacted with KPNA1, a second associated protein, which is likely another α-importin (26), was detected by Western blot analysis. This raises the possibility that mTOR can associate with and regulate more than one of the six highly homologous mammalian α-importins (8). In this fashion, control of constitutive nuclear import by mTOR may be a mechanism for regulating subsets of proteins related to specific transcriptional programs involved in cell fate (8). Moreover, there is apparent redundancy in the system, because KPNA1 deficiency inhibits rapamycin-induced STAT1 nuclear import but does not abolish that in cells exposed to IFN-γ (Fig. 4B). Identification of the complete set of mTOR-regulated α-importins, as well as their effects on transcriptomic programs or cellular function, will reveal a novel level of transcriptional regulation by nutritional or mitogenic stress. With respect to innate immunity or cancer, the nuclear transport of transcription factors involved in cell death or survival (e.g. FoxO3A, NF-κB, and p53) is likely to represent an important step in the elaboration of their target transcriptional programs, as well as a potential therapeutic target (40). This may be especially relevant in diseases of excessive mTOR activity, such as lymphangioleiomyomatosis or tuberous sclerosis complex (41). Our studies, and those of others, indicate that the nuclear content of latent STAT1 can be regulated by mTOR, PP2Ac, and KPNA1 and can determine the inducibility of genes involved in innate immunity, immune modulation, and oncogenesis (11, 16, 29, 32, 41, 42).
Supplementary Material
Acknowledgments
We thank Dr. J. Moss and S. Levine for critical review of the manuscript and Dr. G. Stark for providing 2fTGH, U3A, and U3A-R cells.
This work was supported, in whole or in part, by National Institutes of Health Grant R01-CA125436 (to A. S. K.). This work was also supported by Canadian Cancer Society Research Institute Grant 018311, a Canada Research Chair grant, and funds from the Human Frontier Science Program Organization (to P. P. R.); funds from Tuberous Sclerosis Alliance and a Canada Research Chair grant (to A. S. K.); and Deutsche Forschungsgemeinschaft Grant BA1374/21-1 (M. B.).

This article contains supplemental Tables S1–S3 and Figs. S1–S5.
- mTOR
- mammalian target of rapamycin
- mTORC
- mTOR complex
- STAT1
- signal transducer and activator of transcription-1
- PP2Ac
- protein phosphatase 2A catalytic subunit
- KPNA1
- karyopherin-α1
- MEF
- mouse embryonic fibroblast
- ECFP
- enhanced cyan fluorescent protein.
REFERENCES
- 1. Hara K., Maruki Y., Long X., Yoshino K., Oshiro N., Hidayat S., Tokunaga C., Avruch J., Yonezawa K. (2002) Raptor, a binding partner of target of rapamycin (TOR), mediates TOR action. Cell 110, 177–189 [DOI] [PubMed] [Google Scholar]
- 2. Kim D. H., Sarbassov D. D., Ali S. M., King J. E., Latek R. R., Erdjument-Bromage H., Tempst P., Sabatini D. M. (2002) mTOR interacts with raptor to form a nutrient-sensitive complex that signals to the cell growth machinery. Cell 110, 163–175 [DOI] [PubMed] [Google Scholar]
- 3. Sarbassov D. D., Ali S. M., Sabatini D. M. (2005) Growing roles for the mTOR pathway. Curr. Opin. Cell Biol. 17, 596–603 [DOI] [PubMed] [Google Scholar]
- 4. Sarbassov D. D., Ali S. M., Kim D. H., Guertin D. A., Latek R. R., Erdjument-Bromage H., Tempst P., Sabatini D. M. (2004) Rictor, a novel binding partner of mTOR, defines a rapamycin-insensitive and raptor-independent pathway that regulates the cytoskeleton. Curr. Biol. 14, 1296–1302 [DOI] [PubMed] [Google Scholar]
- 5. Zoncu R., Efeyan A., Sabatini D. M. (2011) mTOR. From growth signal integration to cancer, diabetes and ageing. Nat. Rev. Mol. Cell Biol. 12, 21–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. De Virgilio C., Loewith R. (2006) Cell growth control. Little eukaryotes make big contributions. Oncogene 25, 6392–6415 [DOI] [PubMed] [Google Scholar]
- 7. Carvalho J., Bertram P. G., Wente S. R., Zheng X. F. (2001) Phosphorylation regulates the interaction between Gln3p and the nuclear import factor Srp1p. J. Biol. Chem. 276, 25359–25365 [DOI] [PubMed] [Google Scholar]
- 8. Yasuhara N., Oka M., Yoneda Y. (2009) The role of the nuclear transport system in cell differentiation. Semin. Cell Dev. Biol. 20, 590–599 [DOI] [PubMed] [Google Scholar]
- 9. Goldfarb D. S., Corbett A. H., Mason D. A., Harreman M. T., Adam S. A. (2004) Importin α. A multipurpose nuclear-transport receptor. Trends Cell Biol. 14, 505–514 [DOI] [PubMed] [Google Scholar]
- 10. Kristof A. S., Fielhaber J., Triantafillopoulos A., Nemoto S., Moss J. (2006) Phosphatidylinositol 3-kinase-dependent suppression of the human inducible nitric-oxide synthase promoter is mediated by FKHRL1. J. Biol. Chem. 281, 23958–23968 [DOI] [PubMed] [Google Scholar]
- 11. Fielhaber J. A., Han Y. S., Tan J., Xing S., Biggs C. M., Joung K. B., Kristof A. S. (2009) Inactivation of mammalian target of rapamycin increases STAT1 nuclear content and transcriptional activity in α4- and protein phosphatase 2A-dependent fashion. J. Biol. Chem. 284, 24341–24353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ziegler E. C., Ghosh S. (2005) Regulating inducible transcription through controlled localization. Sci. STKE. 2005, re6. [DOI] [PubMed] [Google Scholar]
- 13. Meyer T., Vinkemeier U. (2004) Nucleocytoplasmic shuttling of STAT transcription factors. Eur. J Biochem. 271, 4606–4612 [DOI] [PubMed] [Google Scholar]
- 14. Kumar A., Commane M., Flickinger T. W., Horvath C. M., Stark G. R. (1997) Defective TNF-α-induced apoptosis in STAT1-null cells due to low constitutive levels of caspases. Science 278, 1630–1632 [DOI] [PubMed] [Google Scholar]
- 15. Stephanou A., Latchman D. S. (2003) STAT-1. A novel regulator of apoptosis. Int. J. Exp. Pathol. 84, 239–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kristof A. S., Marks-Konczalik J., Billings E., Moss J. (2003) Stimulation of signal transducer and activator of transcription-1 (STAT1)-dependent gene transcription by lipopolysaccharide and interferon-γ is regulated by mammalian target of rapamycin. J. Biol. Chem. 278, 33637–33644 [DOI] [PubMed] [Google Scholar]
- 17. McKendry R., John J., Flavell D., Müller M., Kerr I. M., Stark G. R. (1991) High-frequency mutagenesis of human cells and characterization of a mutant unresponsive to both α and γ interferons. Proc. Natl. Acad. Sci. U.S.A. 88, 11455–11459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Shmidt T., Hampich F., Ridders M., Schultrich S., Hans V. H., Tenner K., Vilianovich L., Qadri F., Alenina N., Hartmann E., Köhler M., Bader M. (2007) Normal brain development in importin-α5 deficient-mice. Nat. Cell Biol. 9, 1337–1339 [DOI] [PubMed] [Google Scholar]
- 19. Müller M., Laxton C., Briscoe J., Schindler C., Improta T., Darnell J. E., Jr., Stark G. R., Kerr I. M. (1993) Complementation of a mutant cell line. Central role of the 91 kDa polypeptide of ISGF3 in the interferon-α and -γ signal transduction pathways. EMBO J. 12, 4221–4228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kim J. E., Chen J. (2000) Cytoplasmic-nuclear shuttling of FKBP12-rapamycin-associated protein is involved in rapamycin-sensitive signaling and translation initiation. Proc. Natl. Acad. Sci. U.S.A. 97, 14340–14345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Carrière A., Cargnello M., Julien L. A., Gao H., Bonneil E., Thibault P., Roux P. P. (2008) Oncogenic MAPK signaling stimulates mTORC1 activity by promoting RSK-mediated raptor phosphorylation. Curr. Biol. 18, 1269–1277 [DOI] [PubMed] [Google Scholar]
- 22. Livak K. J., Schmittgen T. D. (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(−ΔΔCT) method. Methods 25, 402–408 [DOI] [PubMed] [Google Scholar]
- 23. McBride K. M., Banninger G., McDonald C., Reich N. C. (2002) Regulated nuclear import of the STAT1 transcription factor by direct binding of importin-α. EMBO J. 21, 1754–1763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Chatterjee-Kishore M., Wright K. L., Ting J. P., Stark G. R. (2000) How Stat1 mediates constitutive gene expression. A complex of unphosphorylated Stat1 and IRF1 supports transcription of the LMP2 gene. EMBO J. 19, 4111–4122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Shuai K., Schindler C., Prezioso V. R., Darnell J. E., Jr. (1992) Activation of transcription by IFN-γ. Tyrosine phosphorylation of a 91-kD DNA binding protein. Science 258, 1808–1812 [DOI] [PubMed] [Google Scholar]
- 26. Quensel C., Friedrich B., Sommer T., Hartmann E., Kohler M. (2004) In vivo analysis of importin α proteins reveals cellular proliferation inhibition and substrate specificity. Mol. Cell. Biol. 24, 10246–10255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sarbassov D. D., Guertin D. A., Ali S. M., Sabatini D. M. (2005) Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science 307, 1098–1101 [DOI] [PubMed] [Google Scholar]
- 28. Julien L. A., Carriere A., Moreau J., Roux P. P. (2010) mTORC1-activated S6K1 phosphorylates Rictor on threonine 1135 and regulates mTORC2 signaling. Mol. Cell. Biol. 30, 908–921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Cheon H., Stark G. R. (2009) Unphosphorylated STAT1 prolongs the expression of interferon-induced immune regulatory genes. Proc. Natl. Acad. Sci. U.S.A. 106, 9373–9378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wong L. H., Sim H., Chatterjee-Kishore M., Hatzinisiriou I., Devenish R. J., Stark G., Ralph S. J. (2002) Isolation and characterization of a human STAT1 gene regulatory element. Inducibility by interferon (IFN) types I and II and role of IFN regulatory factor-1. J. Biol. Chem. 277, 19408–19417 [DOI] [PubMed] [Google Scholar]
- 31. Akira S., Takeda K. (2004) Toll-like receptor signalling. Nat. Rev. Immunol. 4, 499–511 [DOI] [PubMed] [Google Scholar]
- 32. Yang J., Stark G. R. (2008) Roles of unphosphorylated STATs in signaling. Cell Res. 18, 443–451 [DOI] [PubMed] [Google Scholar]
- 33. Peterson R. T., Desai B. N., Hardwick J. S., Schreiber S. L. (1999) Protein phosphatase 2A interacts with the 70-kDa S6 kinase and is activated by inhibition of FKBP12-rapamycinassociated protein. Proc. Natl. Acad. Sci. U.S.A. 96, 4438–4442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Liu L., Chen L., Luo Y., Chen W., Zhou H., Xu B., Han X., Shen T., Huang S. (2010) Rapamycin inhibits IGF-1 stimulated cell motility through PP2A pathway. PLoS. ONE 5, e10578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Harwood F. C., Shu L., Houghton P. J. (2008) mTORC1 signaling can regulate growth factor activation of p44/42 mitogen-activated protein kinases through protein phosphatase 2A. J. Biol. Chem. 283, 2575–2585 [DOI] [PubMed] [Google Scholar]
- 36. Bachmann R. A., Kim J. H., Wu A. L., Park I. H., Chen J. (2006) A nuclear transport signal in mammalian target of rapamycin is critical for its cytoplasmic signaling to S6 kinase 1. J. Biol. Chem. 281, 7357–7363 [DOI] [PubMed] [Google Scholar]
- 37. Zhang X., Shu L., Hosoi H., Murti K. G., Houghton P. J. (2002) Predominant nuclear localization of mammalian target of rapamycin in normal and malignant cells in culture. J. Biol. Chem. 277, 28127–28134 [DOI] [PubMed] [Google Scholar]
- 38. Cunningham J. T., Rodgers J. T., Arlow D. H., Vazquez F., Mootha V. K., Puigserver P. (2007) mTOR controls mitochondrial oxidative function through a YY1-PGC-1α transcriptional complex. Nature 450, 736–740 [DOI] [PubMed] [Google Scholar]
- 39. Tsang C. K., Liu H., Zheng X. F. (2010) mTOR binds to the promoters of RNA polymerase I- and III-transcribed genes. Cell Cycle 9, 953–957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Faustino R. S., Nelson T. J., Terzic A., Perez-Terzic C. (2007) Nuclear transport. Target for therapy. Clin. Pharmacol. Ther. 81, 880–886 [DOI] [PubMed] [Google Scholar]
- 41. Kristof A. S. (2010) mTOR signaling in lymphangioleiomyomatosis. Lymphat. Res. Biol. 8, 33–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Yarilina A., Park-Min K. H., Antoniv T., Hu X., Ivashkiv L. B. (2008) TNF activates an IRF1-dependent autocrine loop leading to sustained expression of chemokines and STAT1-dependent type I interferon-response genes. Nat. Immunol. 9, 378–387 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.