Abstract
The discovery that the single p53 gene encodes several different p53 protein isoforms has initiated a flurry of research into the function and regulation of these novel p53 proteins. Full-length p53 protein level is primarily regulated by the E3-ligase Mdm2, which promotes p53 ubiquitination and degradation. Here, we report that all of the novel p53 isoforms are ubiquitinated and degraded to varying degrees in an Mdm2-dependent and -independent manner, and that high-risk human papillomavirus can degrade some but not all of the novel isoforms, demonstrating that full-length p53 and the p53 isoforms are differentially regulated. In addition, we provide the first evidence that Mdm2 promotes the NEDDylation of p53β. Altogether, our data indicates that Mdm2 can distinguish between the p53 isoforms and modify them differently.
Keywords: degradation, isoform, Mdm2, NEDD8, neddylation, nutlin, p53, splice, Ubiquitination
Introduction
Since its discovery, p53 had been considered an “only child” until the discovery of two sibling proteins, p731and p63.2 Both of these proteins have been shown to have important roles during development, and there is interplay between p53 and the other family members in tumor suppression, as when p63 and p73 are knocked out, p53 is not able to induce apoptosis.3
p63 and p73 contain alternative promoters and splice sites, resulting in the expression of several different isoforms. Both proteins can be expressed from a second promoter in intron-3, downstream of their transactivation domain, and can therefore exist as TAp63 and TAp73 [proteins that contain the transactivation (TA) domain] and ΔNp63 and ΔNp73 (proteins that start from the second promoter and lack the transactivation domain). The ΔN isoforms of both p63 and p73 can transactivate target genes through an alternative transactivation domain present in their distinct N-terminal ends and have dominant-negative activities toward TA isoforms of p63 and p73 and toward full-length p53 (FLp53). The C-termini of p63 and p73 introduce further complexity, as multiple splice sites in the last exons lead to the expression of several different splice variants.4
The roles of these different isoforms are not yet fully understood, but the p53 family members and their individual isoforms have been shown to have differential tissue expression and distinct as well as overlapping functions. For reviews, see refs. Five and 6.
Originally, p53 was considered to be a single protein and an evolutionarily late, highly specialized addition to the p53 family. However, an alternative C-terminally-spliced isoform (p53AS) was reported in mouse by Rotter and coworkers,5 and further isoforms were discovered in human p53: p53i9 (also called p53β), which is generated by an alternative splice in intron-9,6,7 and Δ40p53, which is expressed from an alternative initiation of translation at codon 40 and encodes a protein lacking the first 39 amino acids.8,9 A similar Δ40p53 isoform has also been reported to be produced by alternative splicing of intron-2 of human p53 mRNA.10
Our recent data has shown that the TP53 gene encodes at least 12 p53 protein isoforms.11,12 This is due to the presence of an internal promoter in intron-4 (generating Δ133p53) and an additional splice variant of intron-9 (designated γ), which, along with the previously described p53i9 (p53β) and the alternative initiation of translation at codon 40 and at codon 133, collectively leads to the expression of full-length (FL) p53, p53β, p53γ, Δ133p53α, Δ133p53β, Δ133p53γ, Δ160p53α, Δ160p53β, Δ160p53γ, Δ40p53 and the two splice variants of Δ40p53— Δ40p53β and Δ40p53γ (Fig. 1A).7,13 These isoforms have been shown to be expressed at the mRNA level, to varying degrees, in a number of different normal and tumor tissues.11,12 p53β and Δ133p53 proteins have been shown to have important functions in regulating cell senescence and apoptosis.14-17 Furthermore, relative expression levels of the isoforms show a degree of tissue specificity, suggesting the selective regulation of each isoform.7 This presents a picture analogous to p63 and p73 and reveals a previously undiscovered diversity of p53 proteins.
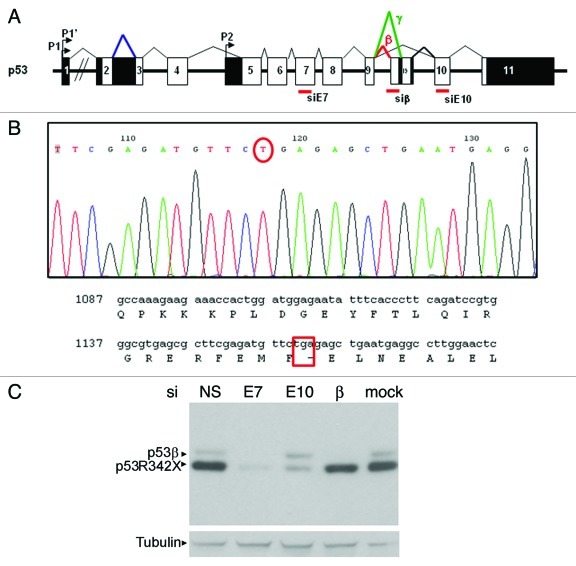
Figure 1. Characterization of p53 protein isoform expression in SK-N-AS cells. (A) Diagram of p53 gene showing coding exons (white boxes), non-coding exons (black boxes), the internal p53 promoter (Δ133 isoforms) and the β and γ splicings. The position of siRNA against Exon 7 (siE7), the β splicing of p53 (siβ) and exon 10 (siE10) are marked. (B) Sequence showing the single nucleotide nonsense mutation that results in the truncated p53R342X mutant present in SK-N-AS cells. (C) Western blot showing both forms of p53 detected in SK-N-AS cells. Cells are transfected with non-specific siRNA (siNS), siE7 (which targets both p53β and p53R342X), siE10 (which only targets p53R342X), siβ (which only targets p53β) and mock transfected, as indicated. Tubulin was used as a loading control.
Because of the diversity of functions attributed to the many isoforms of p63 and p73, it is likely that the p53 isoforms will have important p53-dependent and -independent functions. p53 isoforms are abnormally expressed in a variety of human cancers, suggesting that they play a role in carcinogenesis.18 We were the first to investigate p53 isoform expression in relation to clinical parameters and disease outcome in a cohort of 127 primary breast tumors. Thus, we determined that p53γ expression allows the identification of two distinct subpopulations of mutant p53 breast cancer patients. Mutant p53 breast cancer patients expressing p53γ had as good a disease outcome as patients with WTp53 breast cancer. Conversely, patients expressing only mutant p53, without p53γ expression, had a particularly poor prognosis.19 Therefore, elucidation of the mechanisms that control the function of p53 isoforms will be critical, as they emerge as important players in tumor progression and interfere with cancer treatments.
Mdm2 is a ubiquitin E3-ligase and key regulator of p53 function through promotion of its ubiquitin-dependent degradation.20 Additionally, Mdm2 has been shown to promote the NEDDylation of FLp53, leading to a repression of its transcriptional activity.21 Here, we investigate the role of Mdm2 in the ubiquitination and proteasomal degradation of five of the novel p53 isoforms (p53β, p53γ, Δ133p53α, Δ133p53β and Δ133p53γ). We show that Mdm2 differentially controls the modification of the p53 isoforms, indicating distinct roles for posttranslational modifications in the regulation of the p53 proteins.
Results
Due to the increasing data emerging on isoform p53β, we initially analyzed the stability of this isoform in neuroblastoma cell line SK-N-AS, which expresses p53β but not FLp53.22,23 The SK-N-AS cell line expresses two p53 forms, p53β and a C-terminally truncated p53. After sequencing, we determined that TP53 gene contains a nonsense mutation at codon 342 that leads to expression of a truncated p53 protein p53R342X (Fig. 1B). In order to identify the two forms of p53 protein by western blotting, SK-N-AS cells were transfected with different siRNAs that specifically target p53β (siβ), exon-10 (siE10) or exon 7 of p53 (siE7). The mRNAs encoding p53β or p53R342X proteins have different 3′UTRs. Indeed, the exon-10 and exon-11 are not coding in the mRNA encoding p53β, whereas they are translated in mRNA encoding p53R342X. This may explain why siRNA siE10 inhibits p53R342X but not p53β protein expression.24,25
Using p53 polyclonal antibody, two bands were detected. Both bands were reduced after transfection of siE7 compared with a non-specific siRNA (siNS) or mock transfected cells, indicating the presence of the expected p53 proteins. In the presence of siβ, only the upper p53 band was downregulated, whereas only the lower band was downregulated in the presence of siE10, indicating that the upper and lower p53 proteins are p53β and p53R342X, respectively (Fig. 1C).
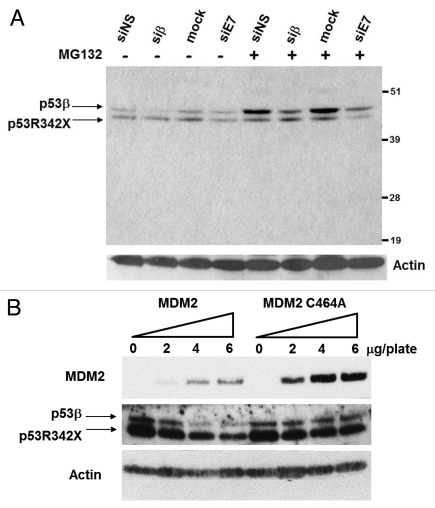
Following transfection with siRNAs targeting p53 isoforms, treatment with proteasome inhibitor MG132 caused a significant accumulation of p53β. This suggests that endogenous p53β is regulated posttranslationally and is subject to proteasomal degradation (Fig. 2A). Only minimal p53R342X accumulation was observed upon MG132 treatment, suggesting that this mutant p53 protein is relatively stable under normal conditions. As the stability of FLp53 is primarily regulated by Mdm2, we analyzed the involvement of Mdm2 in the degradation of p53β. SK-N-AS cells were transfected with increasing amounts of wild-type Mdm2 or mutant Mdm2 C464A, which has no ubiquitin-ligase activity (Fig. 2B). Expression of mutant Mdm2 C464A has no effect on p53β protein level, while expression of Mdm2 promotes degradation of endogenous p53β in a dose-dependent manner.
Figure 2. Endogenous p53β is accumulated upon inhibition of the proteasome by MG132 and Mdm2 can promote degradation of p53β. (A) SK-N-AS cells were transfected with either non-specific siRNA (siNS), siRNAs against p53β (siβ), exon 7 of p53 (siE7) or mock transfected as indicated. Actin was used as a loading control. (B) SK-N-AS cells were transfected with increasing amount of expression vector encoding wild-type Mdm2 or mutant Mdm2 C464A, devoid of ubiquitin-ligase activity. Actin was used as a loading control.
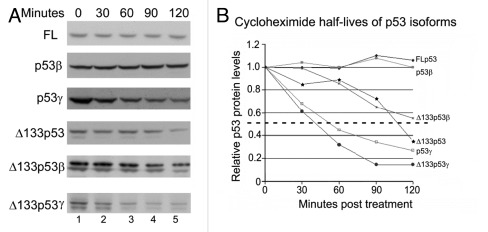
Having established that Mdm2 can induce the degradation of p53β, we wished to analyze whether Mdm2 is able to mediate the degradation or posttranslational modification of other p53 isoforms. As there are no specific antibodies against most of the isoforms, it is difficult to distinguish with pan-tropic p53 antibodies endogenous isoforms from degradation products of FLp53 or other p53 forms (e.g., p53R342X in SK-N-AS cells). Therefore experiments were performed by ectopic expression of the isoforms individually in the p53-negative H1299 lung carcinoma cell line. A cycloheximide pulse-chase analysis was performed to determine the half-life of each isoform. In the absence of an overexpressed E3 ligase, ectopically expressed FLp53 had a relatively long half-life, with protein levels remaining constant over the 120 min period of analysis (Fig. 3A, upper panel). The density of the western blot bands was measured using QuantityOne software, and a graph of the p53 band density at each time point was plotted (Fig. 3B). Ectopically expressed p53β was also stable over the time period analyzed; the other isoforms, however, have varying stabilities: Δ133p53γ and p53γ displayed the shortest half-lives (approximately 40 and 50 min, respectively); Δ133p53α had a half-life of approximately 110 min, and Δ133p53β of over 120 min (approximately 135 min by extrapolation of the curve) (Fig. 3A and B). The lower band detected below the Δ133p53 proteins corresponds to an alternative initiation of translation from a downstream methionine (residue 160).13 The degradation of the Δ160p53 forms was concomitant with the degradation of their Δ133p53 counterparts.
Figure 3. The p53 isoforms have distinct half-lives. (A) H1299 cells were transfected with 1μg of the respective p53 isoform separately (as indicated). At 30, 60, 90 or 120 min prior to harvesting (lanes 2, 3, 4 and 5, respectively) 10μM cycloheximide was added to the medium of each plate. Cells corresponding to the 0 time point were untreated (lane 1). p53 was detected with anti-p53 antibodies (DO-1 for FLp53, p53β and p53γ and DO-12 for Δ133p53, Δ133p53β and Δ133p53γ, throughout). (B) Graph showing the protein level of each isoform as a percentage of the initial (untreated) level over time. The dashed line indicates 50% of the initial protein level (as measured using QuantityOne software).
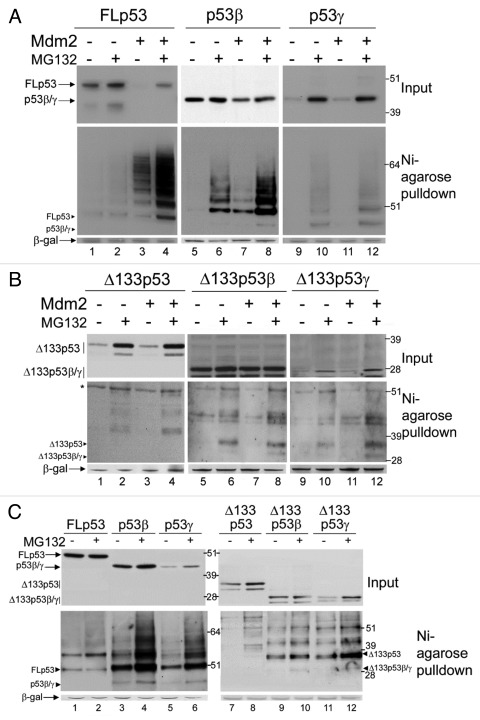
To investigate the role of Mdm2 in the regulation of the protein stabilities, the isoforms were transfected in H1299 cells along with a His6-tagged ubiquitin construct in the absence or presence of co-expressed Mdm2 and treated, or not, with proteasome inhibitor MG132 (Fig. 4). In the absence of Mdm2, the levels of FLp53 protein are slightly increased by MG132 treatment, which was expected, as ectopically expressed FLp53 is stable over this time period in the absence of an ectopic E3 ligase. With co-expression of Mdm2, the levels of FLp53 decrease, consistent with the expected Mdm2-dependent degradation of FLp53, and are restored by MG132 mediated proteasome inhibition (Fig. 4A, upper panel, lanes 1–4). A Nickel-agarose purification of His6-tagged ubiquitin was also performed and ubiquitinated p53 proteins analyzed by western blot (Fig. 4A, lower panel). There is little posttranslational modification of FLp53 in the absence of co-expressed Mdm2. However, in the presence of Mdm2, a ladder of ubiquitinated p53 species is clearly detected and accumulates upon proteasome inhibition by MG132, consistent with many previous reports showing that Mdm2 mediates the degradation of FLp53 in an ubiquitin-dependent manner.20,26,27
Figure 4. Ubiquitination and degradation of p53 isoforms. (A) 1μg FLp53 (lanes 1–4), p53β (lanes 5–8) or p53γ (lanes 9–12) was expressed in H1299 cells in the absence (lanes 1–2, 5–6 and 9–10) or presence (lanes 3–4, 7–8 and 11–12) of 2μg co-expressed Mdm2. Three hours prior to harvesting cells were treated or left untreated, as indicated, with MG132. In all cases 2μg His6-tagged-ubiquitin was co-transfected and DNA concentrations were maintained with empty pcDNA3 control vector. A Ni-agarose purification of His6-tagged ubiquitinated species is shown in the lower panel and corresponding whole cell extract in the upper panel. The molecular weight of each unmodified isoform is indicated with a labeled arrowhead for all ubiquitin-purification assays. (B) As in (A) but Δ133p53, Δ133p53β or Δ133p53γ were expressed instead of FLp53, p53β or p53γ, respectively. A background band detected by DO-12 is indicated with an asterisk *. (C) p53 isoforms are ubiquitinated in Mdm2-negative cells. 1μg FLp53 (lanes1–2), p53β (lanes 3–4), p53γ (lanes 5–6), Δ133p53 (lanes 7–8), Δ133p53β (lanes 9–10) or Δ133p53γ (lanes 11–12) was co-expressed with 2μg His6-tagged-ubiquitin in DKO cells. Cells were treated as indicated with MG132 and His6-tagged ubiquitinated proteins were purified with Ni-agarose. Whole cell extracts are shown in the upper panel and Ni-agarose purification in the lower panel. β-gal was used as a loading and transfection-efficiency control throughout.
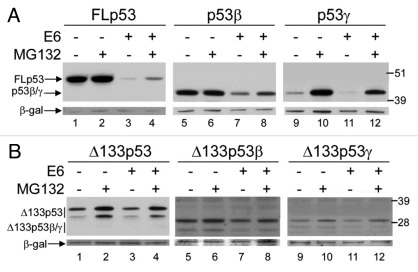
In the absence of ectopically expressed Mdm2, the effect of MG132 treatment on each of the novel p53 isoforms varies. Levels of p53β, which had a relatively long half-life in the absence of an ectopic E3 ligase, do not differ substantially in the presence of MG132 (Fig. 4A, lanes 5–6), whereas levels of p53γ, which was shown to be highly unstable by the cycloheximide pulse-chase analysis, are greatly increased by MG132 treatment (Fig. 4A, lanes 9–10), indicating that p53γ is degraded by the proteasome system. Similarly, levels of Δ133p53α, Δ160p53α, Δ133p53γ and Δ160p53γ are increased by MG132 treatment (Fig. 4B, lanes 1–2 and 9–10) whereas Δ133p53β and Δ160p53β are less affected (Fig. 4B, lanes 5–6), as would be predicted from their long half-life.
Consistent with experiments in SK-N-AS cells, co-expression of Mdm2 promotes a decrease in the level of p53β but not of the other p53 isoforms, suggesting that Mdm2 promotes the degradation of p53β but not of the other isoforms. Similarly, no increase in ubiquitinated species could be detected for any of the p53 isoforms in the presence of ectopic Mdm2 except p53β (Fig. 4A and B). An increase in ubiquitinated p53β proteins that further accumulate in the presence of MG132 can be detected after co-transfection of Mdm2 (Fig. 4A, middle panels), indicating that p53β can be a substrate for Mdm2-mediated ubiquitination. Interestingly, ubiquitinated forms of all of the p53 isoforms accumulated in the presence of MG132 and in the absence of ectopic Mdm2, suggesting that p53 isoforms are ubiquitinated and degraded by the proteasome. A titration of Mdm2 against a constant level of each isoform was also performed to ensure that the lack of degradation was not simply due to insufficient levels of Mdm2 (data not shown). As H1299 cells contain endogenous Mdm2, degradation assays were also performed in a p53 and Mdm2 double knockout (DKO) mouse embryonic fibroblast cell line. Ectopic expression of each isoform in DKO cells confirmed that there are variable levels of degradation (Fig. 4C, upper panel) and accumulation of ubiquitinated species (Fig. 4C, lower panel) of each isoform upon treatment with MG132 in the absence of endogenous Mdm2, consistent with the H1299 data.
Since some p53 isoforms are not degraded by Mdm2, we investigated whether an alternative E3 ligase complex that degrades FLp53, could degrade p53 isoforms. High-risk human papillomavirus E6–16 was co-expressed with each isoform in H1299 cells in the absence or presence of MG132, as indicated. It is clear that Δ133p53, Δ133p53β and Δ133p53γ are not sensitive to degradation by E6–16 (Fig. 5B, compare lanes 1 with 3, 5 with 7 and 9 with 11). However, E6–16 does promote the degradation of FLp53, p53β and p53γ, as indicated by the reduced levels of these proteins in the presence of E6–16 and the inhibition of this reduction by MG132 treatment (Fig. 5A).
Figure 5. High-risk E6 mediates the degradation of some of the p53 isoforms. (A) 1μg FLp53 (lanes 1–4), p53β (lanes 5–8) or p53γ (lanes 9–12) was expressed in H1299 cells in the absence (lanes 1–2, 5–6 and 9–10) or presence (lanes 3–4, 7–8 and 11–12) of 2μg co-expressed E6. Cells were treated or left untreated, as indicated, with MG132. (B) As in (A) but Δ133p53, Δ133p53β or Δ133p53γ isoforms were expressed respectively. β-gal was used as a loading and transfection-efficiency control throughout.
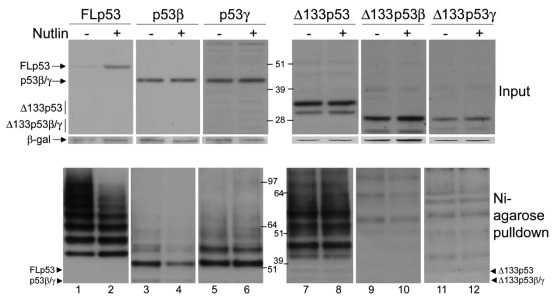
Treatment with Nutlin-3, which specifically disrupts the binding between p53 and Mdm2,28 has no effect on the levels on any p53 isoform other than FLp53 (Fig. 6, upper panels). p53 isoforms were co-transfected with Mdm2 and treated with Nutlin-3, as indicated, and, as expected, levels of FLp53 increased in the presence of Nutlin-3 with a concomitant decrease in accumulated ubiquitinated species (Fig. 6). p53β levels remained unaffected by Nutlin-3 treatment, although, in contrast to the other isoforms, a reduction in ubiquitinated p53β species is observed in the presence of Nutlin-3, confirming that Mdm2 does promote the ubiquitination of p53β (Fig. 6, lower panels). However, inhibition of p53β ubiquitination upon Nutlin treatment does not inhibit p53β degradation, suggesting that Mdm2 promotes degradation of p53β independently of ubiquitination.
Figure 6. Mdm2 promotes the ubiquitination of p53β. 1μg FLp53 (lanes1–2), p53β (lanes 3–4), p53γ (lanes 5–6), Δ133p53 (lanes 7–8), Δ133p53β (lanes 9–10) or Δ133p53γ (lanes 11–12) was expressed individually in H1299 cells along with 2μg Mdm2. Sixteen hours prior to harvesting, the medium was replaced with fresh medium containing 20µM Nutlin-3 or fresh untreated medium, as indicated. The upper panel shows whole-cell extracts, and the lower panel shows Ni-agarose purification of His6-tagged-ubiquitinated p53 proteins. β-gal was used as a loading and transfection-efficiency control throughout.
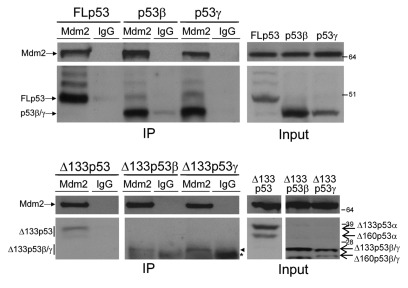
The inability of Mdm2 to degrade Δ133p53 and its splice variants is not surprising, as these isoforms do not contain the N-terminal Mdm2 binding site29,30 that has been shown to be required for p53 degradation.26 However, both p53β and p53γ contain this binding site, and it was therefore unexpected that degradation of p53β and p53γ was not inhibited upon Nutlin treatment. Therefore, co-immunoprecipitation of the isoforms with Mdm2 was performed under the same conditions used for the degradation assays. FLp53, p53β and p53γ all co-immunoprecipitated with Mdm2, confirming that Mdm2 can form a protein complex with p53β and p53γ (Fig. 7, upper panel). In addition, we consistently detected binding of Mdm2 to the Δ133p53 and Δ160p53 isoforms above the background (isotype control) level, suggesting that some interaction between these isoforms and Mdm2 also occurs at a very low level (Fig. 7, lower panel).
Figure 7. Mdm2 interacts with the p53 isoforms. 1μg of relevant p53 isoform (as indicated) was co-expressed with 1μg Mdm2 in H1299 cells. Three hours prior to harvesting, all cells were treated with MG132. Cells were lysed under native conditions and then split for immunoprecipitation with anti-Mdm2 antibody or IgG isotype control antibody, as indicated. Co-immunoprecipitations are shown in the left panel and the inputs in the right panel. *Denotes molecular weight of light chain, and closed arrowhead indicates Δ133p53β and Δ133p53γ isoforms.
Previous reports indicate that Mdm2 requires the C-terminal lysines of p53 for efficient degradation and shows a preference for oligomerized rather than monomeric p53 forms. As the β and γ splice variants both lack the oligomerization and regulatory domains, we utilized three mutant p53 constructs to investigate these properties. Both oligomerization-deficient p53 (p53 I332A)31 and p53 lacking the six C-terminal lysines (p53 6KR)32 show reduced susceptibility to Mdm2-mediated degradation compared with FLp53, and a mutant lacking both properties (p53CΔ60) is almost completely resistant to degradation (Fig. S1A). Co-immunoprecipitation of p53I332A and p53CΔ60 with Mdm2 confirmed that these mutants interact with Mdm2 (Fig. S1B).
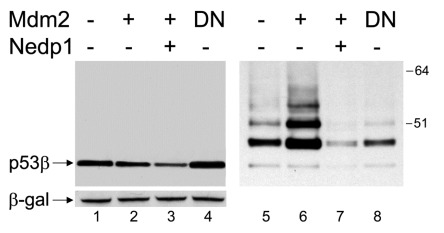
Mdm2 also promotes the conjugation of ubiquitin-like protein NEDD8 to FLp53.21 As Mdm2 interacts with p53β, the ability of Mdm2 to promote the NEDDylation of this isoform was investigated. Surprisingly, Mdm2 promoted the NEDDylation of p53β (Fig. 8, right panels), which was inhibited in the presence of a non-functional Mdm2 that harbors a mutation in the E3-ligase ring domain (Mdm2 C464A), verifying that NEDDylation requires functional Mdm2 (Fig. 8, lanes 4 and 8). Additionally, the de-NEDDylating enzyme Nedp1, which has been shown to be highly specific for deconjugation of NEDDylated species21,33-35 was used to verify the specificity of the NEDDylated p53 bands detected (Fig. 7, lanes 3 and 7). Furthermore, expression of Nedp1 decreases the levels of p53β, indicating that NEDDylation protects p53β from proteasomal degradation.
Figure 8. Mdm2 promotes the NEDDylation of p53β. One μg p53β was expressed in H1299 cells in the absence (lanes 1 and 5) or presence (lanes 2–3 and 6–7) of 2μg co-expressed Mdm2 or Mdm2 C464 (designated DN, lanes 4 and 8). 5μg of de-NEDDylating enzyme, Nedp1, was co-transfected in lanes 3 and 7. In all cases, 2μg His6-tagged-NEDD8 was co-transfected and DNA concentrations were maintained with empty pcDNA3 control vector. A Ni-agarose purification of His6-tagged NEDDylated species is shown in the right panel and corresponding whole-cell extract in the left panel. β-gal was used as a loading and transfection-efficiency control throughout.
Altogether, all p53 protein isoforms are modified by ubiquitination, and their protein expression level is regulated by the proteasome. However, only p53β is ubiquitinated by Mdm2, although Mdm2 can directly interact and form a protein complex with all p53 isoforms. The p53 isoforms are ubiquitinated and degraded in an Mdm2-independent manner. Interestingly, MDM2 NEDDylates p53β inhibiting thus degradation of p53β by the proteasome.
Discussion
It has previously been shown that the p53 isoforms show tissue-specific mRNA expression, indicating that the splicing and the activity of the internal promoter of the isoforms can be differentially regulated. However, no previous studies have looked at their posttranslational regulation. Here, we show that proteasome inhibitor, MG132, inhibits endogenous p53 protein isoform degradation and induces accumulation of ubiquitinated forms of all p53 isoforms. p53 isoforms are differentially ubiquitinated and have different stabilities, suggesting that they are also differentially regulated at the protein level. p53β is ubiquitinated and NEDDylated by Mdm2. The other p53 isoforms are not ubiquitinated by Mdm2, although they can form a protein complex with Mdm2. In addition, Mdm2-mediated NEDDylation of p53β protects p53β from degradation by the proteasome.
Analysis of the half-lives and ubiquitination profile of each isoform revealed that there is strikingly little accumulation of ubiquitinated FLp53 in the DKO cell line (i.e., in the absence of endogenous Mdm2) and in H1299 cells in the absence of ectopically expressed Mdm2. This contrasts greatly with p53β and p53γ, which show substantial levels of ubiquitinated forms in the presence of MG132 and in the absence of Mdm2 despite similar or lower total p53 isoform protein levels compared with the level of FLp53. Ubiquitinated forms of the three Δ133p53 isoforms also accumulated in DKO cells in the presence of MG132, albeit to a much lower extent than p53β and p53γ. The inability to detect ubiquitinated FLp53 species in the absence of Mdm2 suggests that Mdm2 acts as the predominant E3 ligase for FLp53 under these conditions, whereas a different, yet unidentified, E3 ligase(s) effectively ubiquitinates the other isoforms in mouse DKO cells.
Comparison of the half-lives and accumulation of ubiquitinated forms of the isoforms also revealed that p53γ is extremely unstable, and that high levels of ubiquitinated p53γ species are accumulated upon proteasome inhibition in Mdm2-negative DKO cells. It is therefore clear that p53γ is efficiently ubiquitinated and degraded, suggesting that this is important for regulating the posttranslational levels of this isoform. The shorter half-life of p53γ compared with FLp53 or p53β, as well as Δ133p53γ compared with Δ133p53 or Δ133p53β, indicates that the alternative γ-ending confers the increased susceptibility to degradation. This may indicate that an E3 ligase interacts with the alternatively spliced ending. However, there is also the interesting possibility that the C-terminal 15 amino-acids that are specific to this isoform can function as a degron, destabilizing the protein by virtue of an aspect that is inherent to its structure.
Conversely, p53β and Δ133p53β are both relatively stable when ectopically expressed in the absence of an ectopic E3 ligase. Endogenous p53β does, however, accumulate upon proteasome inhibition in SK-N-AS cells, suggesting that, under conditions where p53β is subject to endogenous regulatory pathways, the posttranslational regulation of this isoform is important. Interestingly, Mdm2 is able to promote the ubiquitination of p53β although the high level of ubiquitinated p53β forms observed in DKO cells suggests that Mdm2 is neither the only nor the predominant E3 ligase that modifies this isoform.
The ubiquitination and degradation profile of p53CΔ60, which is similar to the p53 splice variants but lacks the β and γ endings, differed from that of both p53β and p53γ, predominantly displaying a low molecular weight ubiquitin species that corresponds to monoubiquitinated p53CΔ60 as well as a strong resistance to degradation. Incidentally, p53R342X, endogenously present in the SK-N-AS cell line, which differs from p53CΔ60 by only eight amino acids, also appears to be resistant to Mdm2 mediated degradation from both the overexpression and siRNA data presented here. The distinct ubiquitination profiles of the p53β and p53γ compared with the p53 C-terminal truncations indicate that these properties are conferred by the additional 10 and 15 amino acids of the β- and γ-endings, respectively.
The inability of Mdm2 to promote the efficient degradation of p53β and p53γ is not due to lack of binding as all of the isoforms co-immunoprecipitated with Mdm2. Previous studies have shown that Mdm2 has lower affinity for monomeric p53 mutants, explaining the resistance of such mutants to Mdm2-mediated degradation.36,37 From our co-immunoprecipitation data, it appears that the monomeric p53 mutants, particularly p53CΔ60, did show reduced binding to Mdm2 compared with WTp53, supporting that the oligomerization status of p53 affects this interaction and could contribute to the reduced susceptibility of the splice variants to Mdm2 mediated degradation. Interestingly, we also observed a weak but consistent interaction between Mdm2 and the Δ133p53 isoforms. This interaction is consistent with data from Ted Hupp and coworkers showing that there is a second Mdm2 binding site in the core domain of p53.38
In addition to oligomerization, the decreased susceptibility of p53β and p53γ to Mdm2-mediated degradation could be due to the loss of critical lysine residues, as indicated by previous reports.32,36,37,39,40 However, we have previously shown that alternative lysine residues can be utilized by Mdm2 when the six C-terminal lysines are mutated.41,42 The importance of this ability was demonstrated in mouse models by Geoffrey Wahl and colleagues, who show that Mdm2-dependent degradation of p53 with the seven C-terminal lysines mutated to arginine is equivalent to that of WTp53 under normal conditions.43 Similar findings were also demonstrated by Feng et al. in murine ES cells that harbor a p53–6KR transgene in the endogenous p53 locus.44 Although both studies showed that Mdm2-dependent ubiquitination was reduced, the half-life of the mutant p53 proteins was not affected compared with WTp53. In both cases, it was considered that the C-terminal lysines are more likely to be involved in the fine-tuning of p53 activity rather than critical for the regulation of p53 stability. This indicates that the reduced degradation of lysine mutants in cell-based assays is probably attributable to a more complex scenario than merely a preference of ubiquitinable lysines, and that more complicated aspects of the C-terminus of FLp53 are likely to render it a good substrate for Mdm2-mediated degradation.
Mdm2 has also been shown to interact with p73 isoforms without targeting them for degradation.45-47 Interestingly, the C terminus of p73 is not conserved between p73 and p53 and is important for p73 degradation.48 As p73 is not degraded by Mdm2, aspects of the C terminus may determine the distinction between FLp53, which is highly susceptible to Mdm2-mediated degradation, and p53 isoforms and sibling proteins that interact with Mdm2 but are not efficient substrates for Mdm2-mediated degradation.
Both p53β and p53γ are substrates for E6–16-mediated degradation, confirming that when ectopically expressed, they can be degraded by an ectopic E3 ligase. However, the efficiency of degradation of both isoforms was lower than that of FLp53. We have previously shown that E6–16 targets FLp53 for degradation through two pathways: an ubiquitin-dependent pathway and an ubiquitin-independent pathway.42 As the C terminus of p53 was shown to be required for the ubiquitin-independent pathway, the reduced efficiency of degradation of p53β and p53γ could be due to the loss of this second route to proteasomal degradation. E6–16 failed to mediate the degradation of the Δ133p53 protein isoform, because it has a mutant conformation due to the loss of conserved box 2 in the DNA binding domain (unpublished data and personal communication, Petr Muller). This is consistent with previous reports showing that high-risk E6 proteins cannot degrade conformational mutant p53 proteins in the DNA binding domain.49
In addition to ubiquitination, we determine that Mdm2 promotes NEDDylation of p53β. As p53β has been shown to interact with p53-responsive promoters and affect FLp53 transcriptional activity,7,22 modifications, such as ubiquitination and NEDDylation, could serve to regulate such activities.
Interestingly a decrease in p53β protein levels was observed in the presence of Nedp1, suggesting that inhibition of p53β NEDDylation lead to an increase in p53β degradation. As NEDD8 and ubiquitin share lysine preference on FLp53,21 it suggests that NEDD8 conjugation interfere with ubiquitination of p53β by Mdm2, protecting p53β from degradation. Similar effects of Nedd8 on protein stability have been observed for ribosomal proteins.50,51
It is interesting that the recognition and structural requirements of p53 that make it susceptible to Mdm2-mediated modification and degradation are subtle, and that the isoforms are differentially modified by Mdm2 compared with each other but also FLp53. The full importance of this differential regulation is likely to only become clear as the intricacies in the crosstalk between FLp53 and the novel isoforms are discerned. In light of this and the known crosstalk between the other p53 family members, the isoforms may also be discovered to affect p63 and p73 activities as well as the activity of FLp53.15
Here, we show that the p53 isoforms are not modified by the same mechanisms as either wild-type or mutant FLp53 protein, and that the modification of p53β is different from that of p53γ, indicating that the specificity of their modification profiles is conferred by the alternative splicing. The differential regulation of the p53 proteins in terms of their modification and stability indicates the importance of their posttranslational regulation.
Materials and Methods
Cells, antibodies and reagents
H1299 cells were cultured in RPMI supplemented with 10% fetal calf serum and gentamycin at 37°C, 5% CO2 in a humidified atmosphere. Neuroblastoma cells (SK-N-AS) and mouse embryonic fibroblast p53/Mdm2 double-knockout (DKO) cells, a kind gift from Guillermina Lozano, were cultured in DMEM supplemented with 10% fetal calf serum and gentamycin. FLp53, p53β and p53γ were detected using the mouse monoclonal antibody DO-1. The Δ133p53 isoforms were detected using mouse monoclonal DO-12. Anti β-galactosidase mouse monoclonal antibody was obtained from Abcam. Anti-tubulin mouse monoclonal antibody (B-7) was obtained from Santa-Cruz Biotechmology. Proteasome inhibitor MG132 and Cycloheximide were obtained from Calbiochem, protein A sepharose beads from Amersham Biosciences, Ni2-NTA agarose beads from Qiagen and N-ethylmaleimide (NEM) from Sigma.
siRNA experiments
Silencing of p53 isoforms was performed in SK-N-AS cells (cell confluence 50%) seeded in antibiotic-free medium and directly transfected with 50 nM of either non-specific (siNS, negative control 0R-0030-Neg05 Eurogentec) or p53-targeted siRNAs using 4 µL of oligofectamine (Life Technologies), following the manufacturer instructions. Three p53 isoform siRNAs were designed: siE7 (ThermoScientific/Dharmacon reference #J-003329–14), targeting all p53 isoforms; siE10 (GUGAGCGCUUCGAGAUGUU), targeting p53α; and siβ (GGACCAGACCAGCUUUCAA), targeting p53β. The efficiency of the siRNA mediated knockdown was evaluated by western blot and/or RTqPCR.
Plasmids
Expression from constructs pCOC-X2mdm2, pcDNA3 β-galactosidase, pcDNA3 His6-tagged ubiquitin, pcDNA3 WTp53, pcDNA3 p53I332A, pcDNA3 p536KR and pcDNA3 p53CΔ60 was under the control of the CMV promoter. Expression of the p53 isoforms was under the control of the SV40 promoter.
Transfection of cells and western blotting
Cells were transfected using Fugene6 transfection agent from Roche according to the manufacturer’s instructions. In all experiments, 1 μg β-gal encoding plasmid was co-transfected as a transfection efficiency control. 48 h post-transfection cells were lysed in Novex loading buffer supplemented with 0.1 M dithiothreitol, and proteins were separated on 4–12% Novex polyacrylamide gels, transferred to nitrocellulose membrane and developed with the relevant antibodies as previously described.52 Band densities were measure using QuantityOne software.
Drug treatments
Cycloheximide was added directly to the medium at a final concentration of 10μM at the times stated prior to harvesting. 20μM MG132 was added in the same way 3 h prior to harvesting. For Nutlin treatments, growth medium was changed to RPMI containing 20μM Nutlin-3, as indicated, 16 h prior to harvesting.
Immunoprecipitations
Mdm2-specific SMP14 antibody or mouse IgG were covalently coupled to protein A sepharose beads; cells were lysed under native conditions in RIPA lysis buffer, and co-immunoprecipitations were performed, as previously described.53
Purification of His6-tagged ubiquitin conjugates
Purification of His6-tagged ubiquitin-conjugated proteins was as described.52,54 Briefly, cells were lysed in 6M guanidinium/HCL pH8 with 5mM NEM, and His-tagged conjugates were isolated by incubation with Ni2-NTA agarose beads. His6-tagged ubiquitin-conjugated proteins were washed, eluted and then analyzed by western blot with the relevant antibody against p53.
Supplementary Material
Supplementary PDF file supplied by authors.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We would like to thank Mark Saville for advice and help with the manuscript preparation.
Funding
This work was supported by the Kitty Cameron Fund; Cancer Research UK; and the Agency of Science, Technology and Research (A*Star), Singapore.
Footnotes
Previously published online: www.landesbioscience.com/journals/cc/article/20119
References
- 1.Kaghad M, Bonnet H, Yang A, Creancier L, Biscan JC, Valent A, et al. Monoallelically expressed gene related to p53 at 1p36, a region frequently deleted in neuroblastoma and other human cancers. Cell. 1997;90:809–19. doi: 10.1016/S0092-8674(00)80540-1. [DOI] [PubMed] [Google Scholar]
- 2.Osada M, Ohba M, Kawahara C, Ishioka C, Kanamaru R, Katoh I, et al. Cloning and functional analysis of human p51, which structurally and functionally resembles p53. Nat Med. 1998;4:839–43. doi: 10.1038/nm0798-839. [DOI] [PubMed] [Google Scholar]
- 3.Flores ER, Tsai KY, Crowley D, Sengupta S, Yang A, McKeon F, et al. p63 and p73 are required for p53-dependent apoptosis in response to DNA damage. Nature. 2002;416:560–4. doi: 10.1038/416560a. [DOI] [PubMed] [Google Scholar]
- 4.Dötsch V, Bernassola F, Coutandin D, Candi E, Melino G. p63 and p73, the ancestors of p53. Cold Spring Harb Perspect Biol. 2010;2:a004887. doi: 10.1101/cshperspect.a004887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arai N, Nomura D, Yokota K, Wolf D, Brill E, Shohat O, et al. Immunologically distinct p53 molecules generated by alternative splicing. Mol Cell Biol. 1986;6:3232–9. doi: 10.1128/mcb.6.9.3232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Flaman J-M, Waridel F, Estreicher A, Vannier A, Limacher J-M, Gilbert D, et al. The human tumour suppressor gene p53 is alternatively spliced in normal cells. Oncogene. 1996;12:813–8. [PubMed] [Google Scholar]
- 7.Bourdon JC, Fernandes K, Murray-Zmijewski F, Liu G, Diot A, Xirodimas DP, et al. p53 isoforms can regulate p53 transcriptional activity. Genes Dev. 2005;19:2122–37. doi: 10.1101/gad.1339905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Courtois S, Verhaegh G, North S, Luciani MG, Lassus P, Hibner U, et al. DeltaN-p53, a natural isoform of p53 lacking the first transactivation domain, counteracts growth suppression by wild-type p53. Oncogene. 2002;21:6722–8. doi: 10.1038/sj.onc.1205874. [DOI] [PubMed] [Google Scholar]
- 9.Yin Y, Stephen CW, Luciani MG, Fåhraeus R. p53 Stability and activity is regulated by Mdm2-mediated induction of alternative p53 translation products. Nat Cell Biol. 2002;4:462–7. doi: 10.1038/ncb801. [DOI] [PubMed] [Google Scholar]
- 10.Matlashewski G, Pim D, Banks L, Crawford L. Alternative splicing of human p53 transcripts. Oncogene Res. 1987;1:77–85. [PubMed] [Google Scholar]
- 11.Khoury MP, Bourdon JC. p53 Isoforms: An Intracellular Microprocessor? Genes Cancer. 2011;2:453–65. doi: 10.1177/1947601911408893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marcel V, Olivier M, Mollereau B, Hainaut P, Bourdon JC. First International p53 Isoforms Meeting: ‘p53 isoforms through evolution: from identification to biological function’. Cell Death Differ. 2011;18:563–4. doi: 10.1038/cdd.2010.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marcel V, Perrier S, Aoubala M, Ageorges S, Groves MJ, Diot A, et al. Δ160p53 is a novel N-terminal p53 isoform encoded by Δ133p53 transcript. FEBS Lett. 2010;584:4463–8. doi: 10.1016/j.febslet.2010.10.005. [DOI] [PubMed] [Google Scholar]
- 14.Aoubala M, Murray-Zmijewski F, Khoury MP, Fernandes K, Perrier S, Bernard H, et al. p53 directly transactivates Δ133p53α, regulating cell fate outcome in response to DNA damage. Cell Death Differ. 2011;18:248–58. doi: 10.1038/cdd.2010.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marcel V, Petit I, Murray-Zmijewski F, Goullet de Rugy T, Fernandes K, Meuray V, et al. Diverse p63 and p73 isoforms regulate Δ133p53 expression through modulation of the internal TP53 promoter activity. Cell Death Differ. 2011 doi: 10.1038/cdd.2011.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fujita K, Mondal AM, Horikawa I, Nguyen GH, Kumamoto K, Sohn JJ, et al. p53 isoforms Delta133p53 and p53beta are endogenous regulators of replicative cellular senescence. Nat Cell Biol. 2009;11:1135–42. doi: 10.1038/ncb1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen J, Ng SM, Chang C, Zhang Z, Bourdon JC, Lane DP, et al. p53 isoform delta113p53 is a p53 target gene that antagonizes p53 apoptotic activity via BclxL activation in zebrafish. Genes Dev. 2009;23:278–90. doi: 10.1101/gad.1761609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Khoury MP, Bourdon JC. The isoforms of the p53 protein. Cold Spring Harb Perspect Biol. 2010;2:a000927. doi: 10.1101/cshperspect.a000927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bourdon JC, Khoury MP, Diot A, Baker L, Fernandes K, Aoubala M, et al. p53 mutant breast cancer patients expressing p53γ have as good a prognosis as wild-type p53 breast cancer patients. Breast Cancer Res. 2011;13:R7. doi: 10.1186/bcr2811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Honda R, Tanaka H, Yasuda H. Oncoprotein MDM2 is a ubiquitin ligase E3 for tumor suppressor p53. FEBS Lett. 1997;420:25–7. doi: 10.1016/S0014-5793(97)01480-4. [DOI] [PubMed] [Google Scholar]
- 21.Xirodimas DP, Saville MK, Bourdon JC, Hay RT, Lane DP. Mdm2-mediated NEDD8 conjugation of p53 inhibits its transcriptional activity. Cell. 2004;118:83–97. doi: 10.1016/j.cell.2004.06.016. [DOI] [PubMed] [Google Scholar]
- 22.Goldschneider D, Horvilleur E, Plassa LF, Guillaud-Bataille M, Million K, Wittmer-Dupret E, et al. Expression of C-terminal deleted p53 isoforms in neuroblastoma. Nucleic Acids Res. 2006;34:5603–12. doi: 10.1093/nar/gkl619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nakamura Y, Ozaki T, Niizuma H, Ohira M, Kamijo T, Nakagawara A. Functional characterization of a new p53 mutant generated by homozygous deletion in a neuroblastoma cell line. Biochem Biophys Res Commun. 2007;354:892–8. doi: 10.1016/j.bbrc.2007.01.057. [DOI] [PubMed] [Google Scholar]
- 24.Overhoff M, Alken M, Far RK, Lemaitre M, Lebleu B, Sczakiel G, et al. Local RNA target structure influences siRNA efficacy: a systematic global analysis. J Mol Biol. 2005;348:871–81. doi: 10.1016/j.jmb.2005.03.012. [DOI] [PubMed] [Google Scholar]
- 25.Schubert S, Grünweller A, Erdmann VA, Kurreck J. Local RNA target structure influences siRNA efficacy: systematic analysis of intentionally designed binding regions. J Mol Biol. 2005;348:883–93. doi: 10.1016/j.jmb.2005.03.011. [DOI] [PubMed] [Google Scholar]
- 26.Haupt Y, Maya R, Kazaz A, Oren M. Mdm2 promotes the rapid degradation of p53. Nature. 1997;387:296–9. doi: 10.1038/387296a0. [DOI] [PubMed] [Google Scholar]
- 27.Kubbutat MH, Jones SN, Vousden KH. Regulation of p53 stability by Mdm2. Nature. 1997;387:299–303. doi: 10.1038/387299a0. [DOI] [PubMed] [Google Scholar]
- 28.Vassilev LT, Vu BT, Graves B, Carvajal D, Podlaski F, Filipovic Z, et al. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science. 2004;303:844–8. doi: 10.1126/science.1092472. [DOI] [PubMed] [Google Scholar]
- 29.Chen J, Marechal V, Levine AJ. Mapping of the p53 and mdm-2 interaction domains. Mol Cell Biol. 1993;13:4107–14. doi: 10.1128/mcb.13.7.4107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Oliner JD, Pietenpol JA, Thiagalingam S, Gyuris J, Kinzler KW, Vogelstein B. Oncoprotein MDM2 conceals the activation domain of tumour suppressor p53. Nature. 1993;362:857–60. doi: 10.1038/362857a0. [DOI] [PubMed] [Google Scholar]
- 31.Mateu MG, Fersht AR. Nine hydrophobic side chains are key determinants of the thermodynamic stability and oligomerization status of tumour suppressor p53 tetramerization domain. EMBO J. 1998;17:2748–58. doi: 10.1093/emboj/17.10.2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rodriguez MS, Desterro JM, Lain S, Lane DP, Hay RT. Multiple C-terminal lysine residues target p53 for ubiquitin-proteasome-mediated degradation. Mol Cell Biol. 2000;20:8458–67. doi: 10.1128/MCB.20.22.8458-8467.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mendoza HM, Shen LN, Botting C, Lewis A, Chen J, Ink B, et al. NEDP1, a highly conserved cysteine protease that deNEDDylates Cullins. J Biol Chem. 2003;278:25637–43. doi: 10.1074/jbc.M212948200. [DOI] [PubMed] [Google Scholar]
- 34.Shen LN, Liu H, Dong C, Xirodimas D, Naismith JH, Hay RT. Structural basis of NEDD8 ubiquitin discrimination by the deNEDDylating enzyme NEDP1. EMBO J. 2005;24:1341–51. doi: 10.1038/sj.emboj.7600628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reverter D, Wu K, Erdene TG, Pan ZQ, Wilkinson KD, Lima CD. Structure of a complex between Nedd8 and the Ulp/Senp protease family member Den1. J Mol Biol. 2005;345:141–51. doi: 10.1016/j.jmb.2004.10.022. [DOI] [PubMed] [Google Scholar]
- 36.Marston NJ, Jenkins JR, Vousden KH. Oligomerisation of full length p53 contributes to the interaction with mdm2 but not HPV E6. Oncogene. 1995;10:1709–15. [PubMed] [Google Scholar]
- 37.Maki CG. Oligomerization is required for p53 to be efficiently ubiquitinated by MDM2. J Biol Chem. 1999;274:16531–5. doi: 10.1074/jbc.274.23.16531. [DOI] [PubMed] [Google Scholar]
- 38.Shimizu H, Burch LR, Smith AJ, Dornan D, Wallace M, Ball KL, et al. The conformationally flexible S9-S10 linker region in the core domain of p53 contains a novel MDM2 binding site whose mutation increases ubiquitination of p53 in vivo. J Biol Chem. 2002;277:28446–58. doi: 10.1074/jbc.M202296200. [DOI] [PubMed] [Google Scholar]
- 39.Kubbutat MHG, Ludwig RL, Ashcroft M, Vousden KH. Regulation of Mdm2-directed degradation by the C terminus of p53. Mol Cell Biol. 1998;18:5690–8. doi: 10.1128/mcb.18.10.5690. [In Process Citation] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nakamura S, Roth JA, Mukhopadhyay T. Multiple lysine mutations in the C-terminus of p53 make it resistant to degradation mediated by MDM2 but not by human papillomavirus E6 and induce growth inhibition in MDM2-overexpressing cells. Oncogene. 2002;21:2605–10. doi: 10.1038/sj.onc.1205343. [DOI] [PubMed] [Google Scholar]
- 41.Camus S, Higgins M, Lane DP, Lain S. Differences in the ubiquitination of p53 by Mdm2 and the HPV protein E6. FEBS Lett. 2003;536:220–4. doi: 10.1016/S0014-5793(03)00054-1. [DOI] [PubMed] [Google Scholar]
- 42.Camus S, Menéndez S, Cheok CF, Stevenson LF, Laín S, Lane DP. Ubiquitin-independent degradation of p53 mediated by high-risk human papillomavirus protein E6. Oncogene. 2007;26:4059–70. doi: 10.1038/sj.onc.1210188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Krummel KA, Lee CJ, Toledo F, Wahl GM. The C-terminal lysines fine-tune P53 stress responses in a mouse model but are not required for stability control or transactivation. Proc Natl Acad Sci U S A. 2005;102:10188–93. doi: 10.1073/pnas.0503068102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Feng L, Lin T, Uranishi H, Gu W, Xu Y. Functional analysis of the roles of posttranslational modifications at the p53 C terminus in regulating p53 stability and activity. Mol Cell Biol. 2005;25:5389–95. doi: 10.1128/MCB.25.13.5389-5395.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ongkeko WM, Wang XQ, Siu WY, Lau AW, Yamashita K, Harris AL, et al. MDM2 and MDMX bind and stabilize the p53-related protein p73. Curr Biol. 1999;9:829–32. doi: 10.1016/S0960-9822(99)80367-4. [DOI] [PubMed] [Google Scholar]
- 46.Bálint E, Bates S, Vousden KH. Mdm2 binds p73 alpha without targeting degradation. Oncogene. 1999;18:3923–9. doi: 10.1038/sj.onc.1202781. [DOI] [PubMed] [Google Scholar]
- 47.Zeng X, Chen L, Jost CA, Maya R, Keller D, Wang X, et al. MDM2 suppresses p73 function without promoting p73 degradation. Mol Cell Biol. 1999;19:3257–66. doi: 10.1128/mcb.19.5.3257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lee CW, La Thangue NB. Promoter specificity and stability control of the p53-related protein p73. Oncogene. 1999;18:4171–81. doi: 10.1038/sj.onc.1202793. [DOI] [PubMed] [Google Scholar]
- 49.Medcalf EA, Milner J. Targeting and degradation of p53 by E6 of human papillomavirus type 16 is preferential for the 1620+ p53 conformation. Oncogene. 1993;8:2847–51. [PubMed] [Google Scholar]
- 50.Xirodimas DP, Sundqvist A, Nakamura A, Shen L, Botting C, Hay RT. Ribosomal proteins are targets for the NEDD8 pathway. EMBO Rep. 2008;9:280–6. doi: 10.1038/embor.2008.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sundqvist A, Liu G, Mirsaliotis A, Xirodimas DP. Regulation of nucleolar signalling to p53 through NEDDylation of L11. EMBO Rep. 2009;10:1132–9. doi: 10.1038/embor.2009.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xirodimas D, Saville MK, Edling C, Lane DP, Laín S. Different effects of p14ARF on the levels of ubiquitinated p53 and Mdm2 in vivo. Oncogene. 2001;20:4972–83. doi: 10.1038/sj.onc.1204656. [DOI] [PubMed] [Google Scholar]
- 53.Menéndez S, Khan Z, Coomber DW, Lane DP, Higgins M, Koufali MM, et al. Oligomerization of the human ARF tumor suppressor and its response to oxidative stress. J Biol Chem. 2003;278:18720–9. doi: 10.1074/jbc.M211007200. [DOI] [PubMed] [Google Scholar]
- 54.Tatham MH, Rodriguez MS, Xirodimas DP, Hay RT. Detection of protein SUMOylation in vivo. Nat Protoc. 2009;4:1363–71. doi: 10.1038/nprot.2009.128. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary PDF file supplied by authors.