Abstract
Despite a wealth of studies examining the toxicity of engineered nanomaterials, current knowledge on their cytotoxic mechanisms (particularly from a physical perspective) remains limited. In this work, we imaged and quantitatively characterized the biomechanical (hardness and elasticity), adhesive, and surface electrical properties of Escherichia coli cells with and without exposure to hematite nanoparticles (NPs) in an effort to advance our understanding of the cytotoxic impacts of nanomaterials. Both scanning electron microscopy (SEM) and atomic force microscopy (AFM) showed that E. coli cells had noticeable deformation with hematite treatment for 45 min with a statistical significance. The hematite-treated cells became significantly harder or stiffer than untreated ones, as evidenced by indentation and spring constant measurements. The average indentation of the hematite-treated E. coli cells was 120 nm, which is significantly lower (P < 0.01) than that of the untreated cells (approximately 400 nm). The spring constant of hematite-treated E. coli cells (0.28 ± 0.11 nN/nm) was about 20 times higher than that of untreated ones (0.01 ± 0.01 nN/nm). The zeta potential of E. coli cells, measured by dynamic light scattering (DLS), was shown to shift from −4 ± 2 mV to −27 ± 8 mV with progressive surface adsorption of hematite NPs, a finding which is consistent with the local surface potential measured by Kelvin probe force microscopy (KPFM). Overall, the reported findings quantitatively revealed the adverse impacts of nanomaterial exposure on physical properties of bacterial cells and should provide insight into the toxicity mechanisms of nanomaterials.
INTRODUCTION
Physicochemical interactions between nanoparticles (NPs) and cell surfaces play a crucial role in the cytotoxicity of engineered NPs (37, 38). For example, the binding of NPs to surface functional groups (e.g., transmembrane proteins) of cells can be reversible or irreversible, resulting in temporary or permanent structural damage (38). Recently, exposure to engineered NPs was reported to cause disorganization and permeability changes in the bacterial cell membrane (6, 24, 29). Particularly, Ag NPs adhered to the Escherichia coli cell surface, altering the membrane properties and affecting the permeability and the respiration of the cell (35). To date, most in vitro and in vivo toxicological studies of engineered nanomaterials on microbial cells employ growth and viability assays (39, 45), proteomic assays, reactive oxygen species (ROS) tests (7, 14), and molecular-level evaluations based on genetic response (55). Relatively fewer studies have focused on the impacts of NP exposure on the mechanical and physical properties of cell systems, although many engineered NPs, including Ag (42), Cu (46), Fe (1, 27), TiO2 (7), CeO2 (50, 59), and ZnO (6, 24), have been shown to compromise the integrity and functions of the bacterial membrane upon exposure to NPs. Clearly, investigations of the impacts of surface interactions between NPs and cells on cellular mechanical and physical properties are important for interpreting toxicity data and predicting the environmental risk associated with engineered NPs.
Potential implications of the changes in biomechanical properties (e.g., hardness and elasticity), adhesiveness, and surface electrical properties of microbial cells are perceivable. For example, hardness or elasticity changes likely influence the surface structural flexibility, the production of mechanical energy for cell division, and cell motility. As for adhesiveness, the cell microenvironment is normally composed of an extracellular matrix (ECM) with specific molecules that allow the cell to adhere to its surroundings (52). Sorption of NPs on cells may alter the adhesion characteristics and affect a variety of microbial processes (e.g., bacterial colonization) (20). Surface charge undoubtedly plays an important role in interactions between cells and their surroundings (18, 25) which, to name a couple, determine the stability of bacterial suspensions (26) and microbial adhesion to solid surfaces (22). In particular, exposure to TiO2 NPs was found to cause aggregation of E. coli cells and suppression of cell division (23, 65). Clearly, characterizing the surface physical properties of cells is essential for the evaluation of the impacts from exposure to engineered NPs.
Although electron microscopy provides direct examination of the cell surface damage, sample fixation and imaging under a vacuum often introduce artifacts. In contrast, atomic force microscopy (AFM) is attractive because microbial cells can be maintained and imaged in aqueous environments at nanometer spatial resolution. Meanwhile, AFM is capable of measuring piconewton forces in liquid, and the generated force-distance curves can be used to determine the hardness and elasticity (48) as well as the adhesiveness (53, 62) of bacterial cells. Moreover, the electrical mode of AFM, known as Kelvin probe force microscopy (KPFM), can be used to map and quantify the local surface potential down to the resolution of the cantilever tip diameter (31, 32). The local surface potential measured from KPFM results from the difference in work function or the contact potential difference (CPD) between the sample surface and the cantilever tip (19). The work function (usually measured in eV) is the minimum energy needed to liberate an electron from the surface of a particular substance (3, 31). In metals, the work function can be the energy difference between the vacuum level and the Fermi energy. In semiconductors or biomolecules (e.g., proteins), the work function may arise from the difference in energy between the vacuum level and the most loosely bound electron inside the sample. The work function difference is affected by the local mechanical and electromagnetic properties, such as surface charge, doping level, or dielectric constant, of the sample (31, 32). As a result, KPFM has been recently demonstrated to quantify the surface electrical properties (e.g., surface heterogeneities and surface charge distribution) of E. coli cells and metallic NPs (40, 49) as well as DNA (63).
Our previous efforts to evaluate the sorption kinetics and adhesion force characteristics of hematite (α-Fe2O3) NPs on E. coli cells and other cell systems, such as the human intestinal cell line (Caco-2), have been made to understand the impacts on cellular viability and integrity (60–62). Hematite NPs were used as a reference nanomaterial with good colloidal stability and a uniform size/morphology distribution (13, 21, 64), characteristics which minimize the potential size and shape effects on the NP-cell interactions. Moreover, hematite itself is relatively chemically inert and does not release toxic metal ions. However, progressive sorption on E. coli cells led to decreased viability (61), which is probably governed by sorption-induced physical injuries. In this study, we further explored the impacts of hematite NP exposure on morphology, biomechanical properties (i.e., hardness and elasticity), adhesiveness, and surface electrical properties of E. coli cells to better understand the cytotoxic effects of engineered NPs. We employed multiple analytical tools, including scanning electron microscopy (SEM), AFM, dynamic light scattering (DLS), and KPFM, to explore the above-mentioned physical properties in individual E. coli cells at the nanometer scale. To the best of our knowledge, this is the first attempt to employ KPFM as a tool in imaging and quantifying the surface potentials of E. coli cells.
MATERIALS AND METHODS
Hematite NP preparation.
Hematite NPs with a mean diameter of 100 nm were synthesized according to the Penners and Koopal method with minor modifications (60, 64). Briefly, 20 mM FeCl3 in 4 mM HCl solution was incubated for 24 h at 100°C in a forced convection oven, and the sediment (hematite NPs) was collected by centrifugation (Eppendorf centrifuge 5430R; Germany) at 5,000 × g for 30 min. The sediment was rinsed three times using 4 mM HCl solution and concentrated using centrifugation. The concentrated hematite NPs were stored in deionized (DI) water (Millipore; >18.2 MΩ) at 4°C before use. The concentration of hematite NPs is expressed in mg of Fe/liter in this study and was determined by inductively coupled plasma-mass spectrometry (ICP-MS) (Elan DRC II; PerkinElmer). The hydrodynamic size distribution and zeta potential were both measured by DLS using a Zetasizer (Nano ZS; Malvern Instruments Ltd.). In addition, the size and morphology of hematite NPs were previously characterized by a Topcon transmission electron microscope (TEM), with detailed results reported elsewhere (60, 61).
Culture of E. coli cells.
E. coli K-12 cells (strain D21) were purchased from the E. coli Genetic Stock Center (Department of Biology, Yale University, New Haven, CT). E. coli was inoculated in a standard Luria-Bertani (LB) medium at 37°C overnight, and cells were harvested after an incubation time of approximately 13 h (in stationary growth phase), which leads to a cell density of approximately 1.8 × 109 CFU/ml as determined by the optical density (OD) measurement (λ = 600 nm) using a Beckman DU7700 spectrophotometer. The cells were washed with phosphate-buffered saline solution (PBS; 20 mM PO43− and pH 7.2), pelleted by centrifugation at 3,000 × g for 5 min, and finally resuspended in PBS with a final cell density of approximately 109 CFU/ml.
Exposure of E. coli cells to hematite NPs.
Detailed schematics of exposure experiments have been described previously (61). Briefly, the stock suspension of hematite NPs was added to a 15-ml polystyrene test tube at a final total Fe concentration of 100 mg/liter. E. coli cells were dispersed into the tubes at a concentration of approximately 109 CFU/ml; the ratio of the wet mass of E. coli cells to the dry mass of NPs was approximately 1:1 to accelerate cytotoxic effects (47, 61). PBS was added to all tubes to a total volume of 10 ml of the buffered system. The test tubes were stirred gently at a low Reynolds number on a rotational shaker and then incubated at 37°C for 1 h.
The hematite-treated E. coli cells were separated from the mixture to remove loosely bound NPs on the cell surface using the centrifugal method (61). Briefly, the liquid mixture was first rinsed with PBS twice and then centrifuged at 4,000 × g for 10 min on an Eppendorf 5418 centrifuge. Finally, the cell concentrate was resuspended in PBS and used immediately in the following analysis.
Immobilization of E. coli cells on silicon wafer surfaces. (i) Poly-l-lysine-treated silicon wafer surface preparation.
Prior to coating with poly-l-lysine (P8920MW 150 to 300K; Sigma-Aldrich, Saint Louis, MO), silicon wafer disks were cleaned rigorously with 70% ethanol and DI water and dried on a hotplate at 30°C. A 200-μl aliquot of 0.01% poly-l-lysine solution was dropped on the center of the silicon wafer disk, and the disks were placed on the 30°C hotplate for 10 min (15, 48). Finally, DI water was used to gently rinse the silicon wafer surface to remove loosely bound poly-l-lysine molecules.
(ii) Bacteria immobilization.
A 1-ml aliquot of the pelleted E. coli suspension, as prepared previously, was applied to the poly-l-lysine-treated silicon wafer. The sample was allowed to stand for 20 min on the 30°C hotplate before it was rinsed with DI water. The bacterium-immobilized silicon wafer was used immediately in the SEM analysis and the imaging in liquid with AFM.
SEM analysis.
The morphology of E. coli cells with and without hematite treatment was examined by SEM (FEI XL30 EFSEM) with an acceleration voltage of 5 kV. Briefly, the immobilized E. coli cells on the silicon wafer surface were analyzed directly, without a further drying process, to maintain the original morphology.
Imaging and force measurement in liquid with AFM.
An Agilent 5500 AFM (Molecular Imaging, Phoenix, AZ) was used for imaging the immobilized cells in liquid under the acoustic alternating current (AAC) mode and for measuring the interaction forces in PBS between the cantilever tip and the cell surfaces. The hardness, elasticity, and adhesiveness of E. coli cells were evaluated by measuring the indentation in the compliance curve and the adhesion force. Elasticity was evaluated by both the spring constant and Young's modulus, obtained from the Hertz model.
(i) Imaging cells in liquid.
Imaging in PBS was employed in this study to maintain the original morphological and relevant biomechanical properties of E. coli cells in aqueous environments. E. coli cells were immobilized by spreading a 20-μl drop of the bacterial suspension obtained from the previous steps onto the poly-l-lysine-coated silicon wafer surface. The samples were spread to a circular diameter of roughly 5 to 7 mm. After standing for 10 min, the samples were rinsed vigorously with DI water and dried by a focused jet of nitrogen. If the bacteria adhered to the surface, an opaque film was clearly visible. Samples were scanned using a rectangular silicon nitride (Si4N3) cantilever (MLCT model; Bruker AFM Probes) with a nominal spring constant of 0.03 N/m and a nominal resonance frequency of 34 kHz at a scan speed of 1 Hz at 256 pixels per line scan. The initial deflection was set to −1.0 ± 0.2 V with a drive amplitude of 1.8 to 2.0 V. All of the images presented in this article were third-order flattened.
(ii) Determination of the cantilever spring constant.
Prior to the force measurement, deflection amplitude (V)-distance (nm) curves were generated in contact mode by engaging the cantilever with the silicon wafer surface in air. The sweep duration of the force curves was set to 1 s. After each approach, the slope from the force curve in the contact region was recorded and entered into the Thermal K program that interfaces with the imaging software to calculate the spring constant of a cantilever by the thermal method (9). Consequently, the deflection amplitude (V) was converted to force (nN), and the force-distance curve was generated.
(iii) Interaction force measurement.
From the previous imaging step, a single bacterium of interest can be identified within the scanned area. The cursor was placed in the region of the interest on the identified bacterium, and force measurements were performed in PBS. Before engaging the cantilever tip with the sample, the initial deflection of the cantilever was set at −1.0 ± 0.2 V with a set point of 0 V. Depending on the reproducibility, approximately 20 force measurements were recorded on a particular region (62), and then the cursor was relocated to other regions of interest on the same cell to acquire additional force curves. In addition to the force measurement against bacteria, at least six control force curves were similarly obtained between the cantilever tip and the substrate surface. A maximum loading force of approximately 4 nN was consistently applied to effectively engage the tip with the inner cell surface (see more details in the supplemental material).
In this study, the adhesion force measurement between the Si3N4 cantilever tip and the bacterial surface was quantified to evaluate the adhesiveness of the cell surface with and without hematite treatment. The Si3N4 cantilever tip with a small radius of curvature (approximately 20 nm) was used directly to acquire interaction force information between the tip and the bacterial surface at a small scale. In addition, because of their known surface chemical properties, Si3N4 cantilever tips have been frequently used as a model surface to study the interaction force in the bacterial interface (12, 28, 44, 54).
Zeta potential and hydrodynamic size measurements with DLS. (i) Zeta potential.
Hematite NPs and E. coli cells in PBS dispersions were examined using DLS to determine the electrophoretic mobility (EPM), which was then converted to zeta potential using Smoluchowski's approximation. One milliliter of the liquid sample was injected into a folded capillary cell (DTS1060; Malvern Instruments).
(ii) Hydrodynamic sizes of hematite NPs and E. coli cells.
DLS was used to measure the z average intensity-based hydrodynamic size of hematite NPs and E. coli cells on the basis of the Stokes-Einstein equation. Suspensions for the DLS analysis were obtained directly from previous steps as described above. During DLS measurements, refractive indices of 2.94 and 1.40 were used for hematite NPs (13) and E. coli cells (2), respectively, and the instrument temperature was maintained at 25°C. The reported zeta potential and hydrodynamic diameter data show the mean value ± the standard deviation of DLS data collected in three independent experiments.
Local surface potential measurement by KPFM.
KPFM was also conducted on the Agilent 5500 AFM, which is equipped with a MAC III unit and has three lock-in amplifiers (LIAs) that enable multifrequency measurements. Detailed configurations of KPFM have been described previously (63). Briefly, platinum-coated silicon cantilever probes (Olympus AC240TM; Japan) were used as the conductive probes with a force constant of approximately 2 to 5 N/m and a nominal resonance frequency of 70 kHz. The work function of the tips (Φt; 4.91 ± 0.05 eV) was calibrated on freshly cleaved highly oriented pyrolytic graphite (HOPG; grade ZYH, product no. 626; Ted Pella). The reference work function of HOPG in air is 4.65 eV (31, 32). During the operation, the microscope was fully contained in an environmental chamber that controls ambient pressure, temperature (25 ± 2°C), and humidity (approximately 35%), as measured by a VWR humidity/temperature thermometer.
For the NP sample preparation, 0.5 μl of hematite NP stock suspension was deposited on a clean silicon wafer. After air drying for approximately 5 min, the silicon chip was then ready for KPFM. For E. coli cell sample preparation, the air drying time was strictly controlled to avoid severe dehydration of cells. Briefly, 0.5 μl of the bacterial suspension was deposited on a clean silicon wafer and incubated on a 30°C hotplate for approximately 5 min. The fixed bacterial film then was quickly blown under a focused jet of nitrogen to remove residual moisture. The silicon wafer with fixed samples was finally fixed on a small piece (1 cm by 1 cm) of conductive double-sided tape (Ted Pella) which was placed on a grounded microscope stage and ready for KPFM study. To confirm the statistical significance of the observations, local surface potential was measured in at least five different locations for each cell; the images reported here are representative.
Statistical analysis for the force measurements.
Statistical tests were used to determine if the force measurements on hematite-treated E. coli cells were significantly different from those on the untreated cells. Briefly, we randomly selected at least 10 each of untreated and hematite-treated E. coli cells from AFM images. For each selected single cell, approximately 15 to 20 force-distance curves (or compliance curves) were generated for 3 to 5 different regions of the cell surface. We regularly changed cantilever tips to avoid experimental artifacts and tip contamination. Generally, the force measurements within an AFM image were all completed by the same cantilever tip unless the reproducibility in force measurement became worse due to tip damage or contamination. The approach and retraction curves between the cantilever tip and the bare silicon wafer were always reproducible and thus easily averaged, whereas the approach and retraction curves on E. coli cells varied because of the complex heterogeneous bacterial surface; thus, these curves were analyzed individually as described previously (48, 30, 62). Finally, the reported force-distance curves were plotted with averaged data, and the probabilities of occurrence for the indentation and adhesion forces are also discussed below.
RESULTS AND DISCUSSION
Morphology changes in E. coli cells with and without hematite treatment.
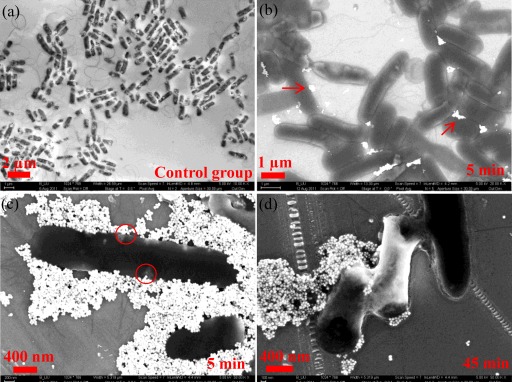
To qualitatively assess the exposure of E. coli cells to hematite NPs, SEM imaging experiments were performed. Figure 1a shows the morphology of representative E. coli cells without exposure to hematite NPs. The cells were rod shaped with some appendages, such as the tail-like flagella, and their sizes were typically 0.5 to 1 μm wide and 2 to 3 μm long, consistent with the common dimensions of E. coli. Figure 1b and c show cells with a few and many hematite NPs (the white dots), respectively, attached to the cell body. However, the cells were still fairly intact and did not exhibit significant deformation; furthermore, flagella or pili could be resolved well. Although some particles appear to be inside or beneath the cells (see the circled region in Fig. 1c), it has been suggested that NPs generally adsorb to but do not penetrate E. coli cells (45). As revealed by additional SEM images (see Fig. S1 in the supplemental material), hematite NPs could merge with or be embedded into the gel-like periplasm or extracellular polymeric substances (EPS), a result which is likely the situation observed in Fig. 1c. With a 45-min exposure to hematite NPs, E. coli cells had become deformed, and the cell size was statistically smaller than that of the untreated cells. Figure 1d shows typical deformed cells with a significant shrinkage in length, approximately 66% ± 15% (within the 95% confidence interval based on comparisons between the measurements of 10 untreated and 35 treated cells).
Fig 1.
SEM images of E. coli cells with and without hematite treatment at different exposure times (indicated in the bottom right of each legend). (a) Untreated E. coli cells. (b) E. coli cells with light attachment of hematite NPs (white dots, as indicated by the arrows). (c) E. coli cells with heavy sorption of hematite NPs. (d) Deformed E. coli cells.
In Fig. 1b and 1c, hematite NPs formed aggregated clusters which may be caused by the surface tension force as the water evaporated during sample preparation. Hematite NPs themselves in PBS should be stabilized by the interparticle electrostatic repulsion, whereas in the presence of E. coli cells, hematite should exhibit high sorption affinity toward E. coli due to the attractive interfacial forces (61).
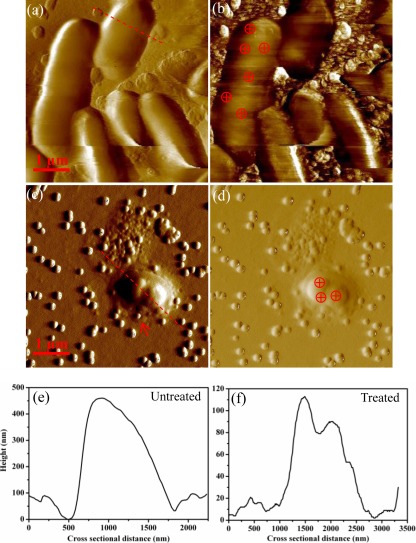
To maintain the physiological properties of the cell under biologically relevant conditions, we also obtained AFM images of E. coli cells in liquid (PBS) to compare the hematite treatment effects on morphology. As shown in Fig. 2a, original untreated E. coli cells maintained an intact appearance, and they appeared to have a smooth outer surface with no apparent surface ultrastructures or flagella resolved. Previous studies attributed the absence of fine surface structures and flagella in liquid AFM imaging to the interference of solvent molecules (water) in interactions between the tip and bacterial surface structures (15, 48, 51). The phase image in Fig. 2b provides better contrast in which some particulates and the rough surfaces on the substrate are clearly resolved. Previous studies showed that the brighter or darker areas of the sample were caused by a positive or negative phase shift (increased or decreased phase angle), respectively, and the phase contrast may reflect changes in surface characteristics (e.g., adhesiveness, hardness, or elasticity) (16, 33).
Fig 2.
(a) Amplitude image of untreated E. coli cells immobilized on the silicon wafer surface. (b) Phase image for the cells shown in panel a. (c) Amplitude image of deformed E. coli cells with exposure to hematite for 45 min. (d) Phase image for the cells shown in panel c. (e and f) The cross-sectional profiles of the heights of the untreated and hematite-treated E. coli cells in panels a and c, respectively, taken along the directions marked with the red dashed lines in panels a and c. The crossed circles indicate examples of force measurement regions.
Similar to SEM images, AFM images also provided visualization of the hematite NPs adsorbed on E. coli cells in Fig. 2c and d, which show a typical deformed cell after a 45-min exposure. Ten untreated and 25 hematite-treated cells were selected to compare the changes in the bacterial length, which was found statistically to decrease by approximately 59% ± 21% (within the 95% confidence interval) with hematite exposure, consistent with the SEM results. Furthermore, the height profiles for the untreated and hematite-treated cells were also plotted in Fig. 2e and f; these cross-sectional profiles are drawn across the cell body along the red dotted lines in Fig. 2b and d, respectively. The untreated cells had a maximum height of approximately 0.45 μm that is comparable to the range reported previously (51), whereas the hematite-treated cells had an obviously reduced height of ∼0.11 μm, providing more solid evidence of the cell deformation in the vertical or z scale.
Hardness and elasticity analysis of E. coli cells.
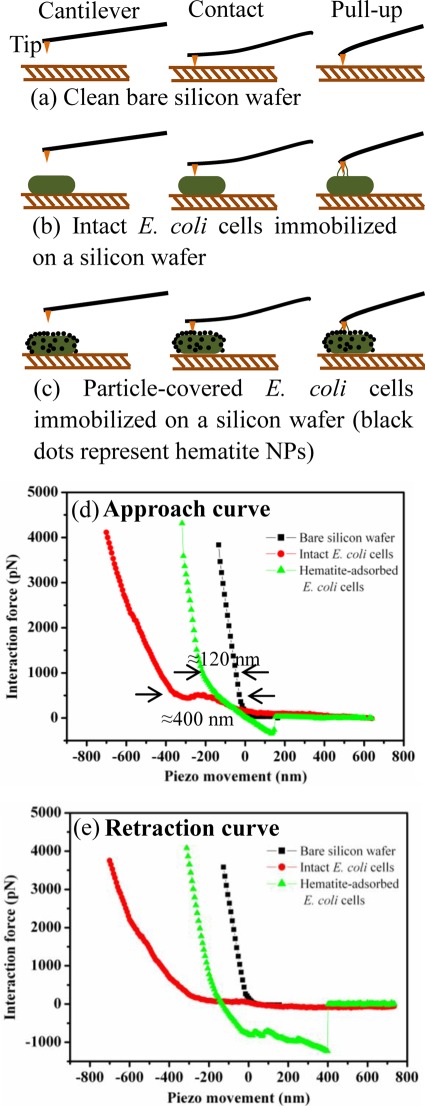
Force-distance curves record the vertical cantilever deflection as the AFM tip is driven by the vertical piezo to approach, contact, and retract from the sample surface. These curves can be used to evaluate the cell surface hardness or elasticity as well as the adhesiveness (48, 28, 53). To evaluate the hardness or elasticity, force-distance curves generated on a relatively rigid surface (i.e., the bare silicon wafer surface) are used as a control for force measurements on bacterial cells, as illustrated in Fig. 3a to c. Force-distance curves were collected on randomly selected regions of the untreated and hematite-treated E. coli cells (Fig. 3b and c). Upon the engagement of the tip with the cell surface, the force-distance curve includes both nonlinear and linear portions, as shown in Fig. 3d and e. The nonlinearity is a consequence of interfacial forces, such as the van der Waals, electrostatic, and steric (electrosteric) forces, and the potential nonlinear deformation of the bacterium, whereas the linear response is due to the elastic deformation of the bacterium (51).
Fig 3.
(a to c) Schematic of the interaction force measurement between the cantilever tip and the sample surface (i.e., the bare silicon wafer surface and untreated and hematite-treated E. coli cells). The illustrations are not drawn to scale. (d to e) Force-distance curves as the tip approached (d) and retracted from (e) contact between the tip and the E. coli cell. Arrows in panel d indicate the measured indentations of untreated (≈400 nm) and treated (≈120 nm) E. coli cells relative to the substrate surface.
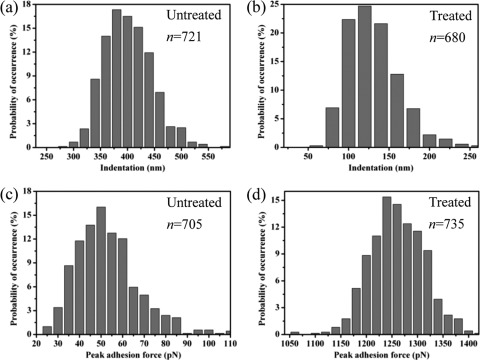
Indentation is the response of a material under a certain compression force and a quick measure of the sample hardness (41). In AFM, the indentation on the local cell surface is measured as the difference between the approach and retraction curves at the start of the linear region (or constant compliance region) because the maximum indentation is clearly proportional to the loading force or compression force from the tip (56, 62). To estimate indentation, approach curves generated on the silicon wafer surface and E. coli cells were plotted together in Fig. 3d. The indentation on the untreated cells was approximately 270 to 550 nm, as shown by the red dots, whereas after exposure to hematite NPs, the indentation was significantly reduced to an average of 120 nm, as shown by the green triangles. The difference in indentation is further illustrated in the distributions of indentation in Fig. 4a and b. As the bacterial surface is heterogeneous, it is not surprising to see a wide distribution of indentation, indicating that some regions are rigid whereas others are soft. Pairwise statistical t tests indicate that the measured indentations of untreated and treated cells were significantly different from one another (P < 0.01; n = 680). The smaller indentation corresponds to a stiffer surface, indicating that the accumulation of hematite NPs stiffened the cell surface, probably owing to the binding or even embedment of hematite NPs into the biopolymer matrix on the E. coli surface, as shown in Fig. 1d and Fig. S1 in the supplemental material.
Fig 4.
Histograms showing the distributions of indentations (a and b) and peak adhesion forces (c and d) measured on untreated and hematite-treated E. coli cells. The probabilities of occurrence were normalized by the total numbers of force measurements, which are indicated by the values of n for each case.
Alternatively, cell hardness or elasticity can be evaluated by measuring the spring constant of the cells (48, 34, 51, 53). The spring constants (kb) were determined from the slope of the linear portion of the constant compliance region, which is due to elastic deformation of the bacterium, as mentioned above (52). When the tip interacts with a cell, two springs are present in series (the cell and the cantilever). In this case, the cantilever deflection may occur over a long distance because soft samples initially exhibit a gradual change in deflection (Fig. 3b and c). The spring constant of the cell (kb) can be determined using the equation (5) kb = kcs/(1 − s), where kc is the spring constant of the cantilever and s is the slope in the linear constant compliance region of the force-distance curve. The spring constant of the cantilever was predetermined by the Thermal K method. Slopes for untreated and hematite-treated E. coli cells were measured and are shown in Table 1. The mean spring constants of untreated and hematite-treated E. coli cells are 0.01 and 0.28 nN/nm, respectively, with a statistically significant difference (P < 0.01; n = 250), also indicating that hematite-treated E. coli cells became stiffer. Previously reported spring constants for E. coli commonly range from 0.02 to 0.26 N/m (30, 34, 58). The deviations may arise from variations in experimental methods or the strain types of the studied bacteria. For example, our AFM force measurement was performed in liquid, whereas some previous work performed it in air (34). The difference in hydration states should vary the spring constant of the cell surface. Moreover, the rigid hematite NPs adsorbed on cells likely affected the physical integrity of the cell membrane and led to the cell deformation and cell hardness changes, as indicated by the different spring constants of untreated and treated cells. We compared the spring constants of the cells rather than Young's modulus because of the considerable deviations between the Young's modulus value calculated using the Hertz model and other literature-reported values (see details in section S3 in the supplemental material).
Table 1.
Summary of spring constants for the cantilever tip and untreated and hematite-treated E. coli cellsa
| Tip or group of cells | Slope (nm/nm)b | Spring constant (nN/nm) |
|---|---|---|
| Cantilever | 0.03 ± 0.01 | |
| Untreated E. coli cells | 0.31 ± 0.02 | 0.01 ± 0.01 |
| Hematite-treated E. coli cells | 0.90 ± 0.02 | 0.28 ± 0.11 |
The average data and standard deviations are presented. The standard deviations of kb (the spring constants of the cells) were computed by the method of propagation of uncertainty.
The slope in the table (nm/nm) was converted from the slope in the force-distance curve (nN/nm) by dividing by the spring constant of the cantilever (kc). A total of 250 force-distance curves on 3 untreated and 5 hematite-treated cells were selected to determine the slopes in the corresponding linear constant compliance region.
Surface adhesiveness analysis.
Adhesiveness of the cell surface is evaluated using the adhesion events from the retraction curve, in which the tip pulls away from the cell. Figure 3e shows representative adhesion events that were measured on different surfaces. For the bare silicon wafer and untreated E. coli cells, there were almost no noticeable adhesion peaks that could be quantified from the retraction curves. In contrast, hematite-treated E. coli cells produced apparent adhesion peaks (the green triangles). Each adhesion curve may vary in the pattern of adhesion events due to the cell surface heterogeneity. Overall, visual inspection indicates that the adhesion force in the treated cell retraction curves was generally larger than that in the retraction curves for the silicon wafer or untreated cells. The low adhesion between the tip and the silicon wafer might be due to their hydrophilic surfaces, which attract water molecules and produce hydrophilic repulsion. For untreated cells, the cell surface is hydrated and fairly soft; this led to a weak contact between the tip and the cell which resulted in less binding and low adhesion. However, the hematite-treated cell surface was shown to be stiffer, and consequently, the tip might effectively penetrate the slime layer and make contact with more biopolymer molecules, such as lipopolysaccharides (LPSs), resulting in a wide range of adhesion binding between the tip and the cell (17). Thus, the multiple adhesion peaks in Fig. 3e might correspond to the multiple break-up points of the chemical binding of LPSs to the Si3N4 surface (44). Furthermore, for hematite-treated E. coli cells, we observed a “jump to contact” in the approach curve, an event which occurs when the attractive force gradient exceeds the spring constant of the cantilever (11). Clearly, after the sorption of hematite, a stronger attraction which could be due to the increasing electrostatic attraction was present between E. coli cells and the tip. This is probably because E. coli cells had increased surface charge with hematite treatment (to be shown in the following sections) while the surface charge of Si3N4 tips should be close to neutral at pH 7.2 (4). Moreover, attachment of hematite NPs on E. coli cells may also alter other surface characteristics (e.g., hydrophobicity and/or surface tension), a possibility which requires further research to verify.
To determine the statistical significance of the adhesiveness changes, we further compared the peak adhesion forces observed in the retraction curves of untreated and treated cells (Fig. 4c and d). Again, a wide distribution of peak adhesion forces was generally observed due to the surface heterogeneity (44, 62). Clearly, the magnitude of the peak adhesion force of treated cells (∼1 nN) was significantly greater than that of untreated cells, the latter of which was on the order of 20 to 100 pN (P < 0.01; n = 705), which is just above the detection limit (approximately 10 pN) of the AFM force measurement (36). The above-described comparison of peak adhesion forces provides solid evidence of the changes in cell surface adhesiveness with hematite exposure.
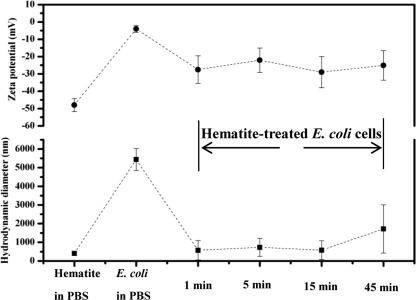
Zeta potential and hydrodynamic size measurements.
To quantify the effect of hematite exposure on the surface electric property of E. coli cells, zeta potentials, as a measure of the average electric potential for suspended colloidal particles in the aqueous phase, were determined. Despite the possible inaccuracy of using the Smoluchowski formula in calculating the zeta potential of bacteria (soft particles) (12), zeta potential is still widely employed as a convenient and approximate measure of the surface electrical potential of bacteria (8, 26). As shown in Fig. 5, the mean zeta potential of E. coli cells in PBS was originally −4 ± 2 mV and immediately shifted to a more negative value (−27 ± 8 mV) upon exposure to hematite NPs; no significant changes were observed over the exposure time. The overall charge of E. coli cells at biological pH values should be negative, owing to the excess carboxylic groups (e.g., LPS) present in the lipoproteins at the bacterial surface (46). The rapid changes in the zeta potential of E. coli cells were likely due to the sorption of hematite NPs, which had a negative surface charge (approximately −48 mV) when dispersed in PBS. Hematite NPs in DI water at neutral pH are positively charged (64). The charge reversal of hematite NPs from positive (in DI water) to negative can be explained by phosphate complexation (10); functional groups ( FeOH,
FeOH,  FeO−, and
FeO−, and  FeOH2+) on hematite surfaces may react with phosphate species to form negatively charged species (e.g., FePO42− and
FeOH2+) on hematite surfaces may react with phosphate species to form negatively charged species (e.g., FePO42− and  FePO4H−). In addition to the zeta potential measurement, the EPM of E. coli cells is presented in Table S1 in the supplemental material (see more details in section S4 in the supplemental material). As the surface potential of E. coli cells became more negative, the electrostatic interactions between E. coli cells and hematite NPs and the aqueous stability (e.g., dispersion state) of E. coli cells were both likely to be affected.
FePO4H−). In addition to the zeta potential measurement, the EPM of E. coli cells is presented in Table S1 in the supplemental material (see more details in section S4 in the supplemental material). As the surface potential of E. coli cells became more negative, the electrostatic interactions between E. coli cells and hematite NPs and the aqueous stability (e.g., dispersion state) of E. coli cells were both likely to be affected.
Fig 5.
Zeta potentials and hydrodynamic sizes of hematite NPs as well as untreated and hematite-treated E. coli cells in PBS with different exposure times (as labeled on the x axis). The dashed lines are a guide for the eye.
To verify the size changes of E. coli cells upon exposure to hematite, the hydrodynamic diameters of E. coli cells were measured. Figure 5 shows that the mean hydrodynamic diameter of E. coli cells before exposure to hematite NPs was approximately 5,400 nm, whereas hematite NPs had a mean hydrodynamic diameter of approximately 400 nm, probably due to some aggregation, as observed in the SEM and AFM images. Interestingly, the hydrodynamic size of E. coli cells seemed to decrease to approximately 1,000 nm immediately after exposure to hematite (e.g., after approximately one min of exposure) and showed no significant changes over the exposure time. This rapid change in size, however, may not correspond to the cell deformation and could be an artifact caused by either the cell damage from the centrifugal separation or potential interference from light scattering on the aggregated clusters of hematite that remained in the bacterial suspension. For instance, the possible interference from hematite is evidenced by the multimodal particle size distribution (PSD) in Fig. S2 in the supplemental material. Upon exposure, the suspension became more polydisperse, and the initially monomodal PSD (time = 0) of the E. coli suspension became bimodal and trimodal over time. Clearly, the multimodal PSD reflects the hydrodynamic size component of hematite, which interferes with the DLS measurement and interpretation of the hydrodynamic size of E. coli cells as also reported previously (23). Thus, it might be less accurate to rely on DLS to characterize the hydrodynamic size changes of E. coli cells in the presence of NPs.
Measurements of local surface potentials by KPFM.
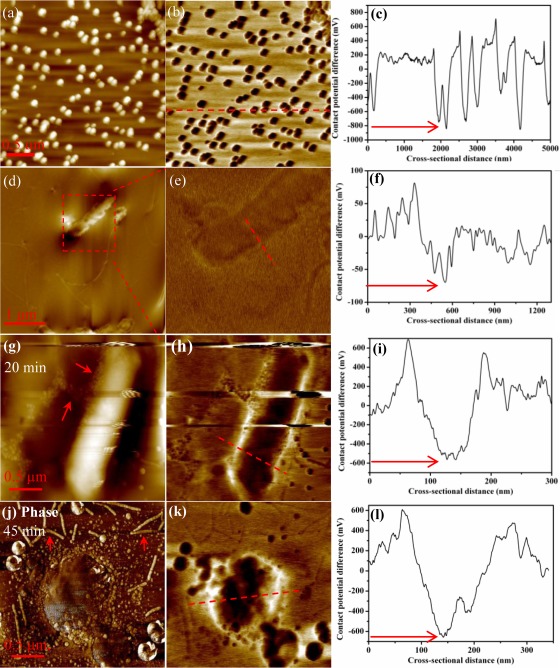
E. coli cells have an uneven or heterogeneous surface charge distribution. However, zeta potential reflects only the overall surface electric property of dispersed colloidal particles. To evaluate the surface charge heterogeneity and quantify the surface potential at the nanoscale, we used KPFM as an alternative method to measure the surface potentials of hematite and the untreated/treated E. coli cells. Figure 6a shows the typical topographical image of hematite NPs deposited on the silicon substrate. Hematite NPs were close to spherical with a diameter of 100 ± 8 nm (n = 30). The morphology and size were consistent with those in our previous studies (60, 61). Figure 6b shows the surface potential image of hematite NPs; the contrast is produced on the basis of the work function difference or CPD between the tip and NPs as introduced previously (49). Hematite NPs appeared darker in color than the bare silicon substrate background (which was grounded to 0 V), indicating that hematite NPs were more negative (40). To determine the surface potentials of individual NPs, a red dashed line was drawn randomly across the surface potential image; the resulting cross-sectional profile is shown in Fig. 6c. The surface potential of hematite NPs varied with a wide distribution, as shown in Fig. S3 in the supplemental material, and showed some dependence on particle size, a finding which was also previously observed for quantum dots (63). The mean surface potential was approximately −800 mV with a standard deviation of 145 mV based on measurements of 155 particles. The relation between the work function of the conductive tip (Φt) and that of the sample (Φs) is given by the equation (43) Φs = Φt − eVCPD, where e is the elementary charge (≈1.6 × 10−19 coulombs) and VCPD is the CPD or surface potential measured by KPFM. With Φt being equal to 4.91 ± 0.05 eV, as indicated above, and the VCPD obtained in Fig. 6c, the work function of hematite NPs can be calculated as 5.71 ± 0.20 eV, a value which agreed well with the literature value (57).
Fig 6.
Topographical (a, d, and g), surface potential (b, e, h, and k), and phase (j) images of hematite NPs and E. coli cells generated from KPFM. Cross-sectional profiles of surface potentials, which were generated along the dashed red lines in the surface potential images in panels b, e, h, and k, are shown in panels c, f, i, and l, respectively.
Similarly, we scanned E. coli cells using KPFM. Compared with the smooth bacterial surface acquired in liquid by AFM, the topographical image of the selected single E. coli cell in Fig. 6d shows slightly better resolution of cell surface details. For example, the flagella attached to the surface of E. coli cells are observed in Fig. 6d (as indicated by the arrow; see also additional images in Fig. S4 in the supplemental material). Figure 6e presents a typical image of the surface potential of the selected cell with the contrast slightly lower than that of the topographical image. The cell region appeared to be slightly darker than the substrate, indicating a negative surface charge for E. coli. The cross-sectional profile shows that the surface potential of E. coli cells varied from −25 to −75 mV (n = 39), as indicated in Fig. 6f.
To compare cell surface potentials with and without hematite treatment, E. coli cells harvested at different exposure times were scanned by KPFM. The topographical image in Fig. 6g shows a typical bacterial cell with some particles (hematite NPs) around it, and the cellular morphology was still intact with an approximately 20-min exposure to hematite. With a longer exposure (i.e., 45 min), hematite-treated E. coli cells exhibited noticeable shrinkage, as marked in Fig. 6j, which shows a typical deformed cell. The flagella appeared to detach from the cell, and small shredded pieces were scattered adjacent to the cell body. In contrast, the control group images in Fig. S4 in the supplemental material show long, intact flagella near or attached to the cell (as indicated by the arrows), thus ruling out possible damage from the sample preparation.
Comparisons of the surface potential images in Fig. 6e, h, and k reveal that as exposure time increased, the E. coli cell surface became darker and darker, indicative of the shift to more-negative surface potentials, a result which agreed with the findings from the zeta potential measurements. This is largely due to the progressive sorption of hematite NPs. For instance, the cross-sectional profiles of surface potentials on randomly selected cell surfaces in Fig. 6f to l clearly show that the original E. coli cell had a surface potential of less than −100 mV that gradually shifted to approximately −600 mV as hematite NPs adsorbed to the cells, as indicated by the red arrows. Fig. S5 in the supplemental material further illustrates the decreased surface potential of E. coli cells as a function of the adsorbed mass of hematite NPs. Clearly, with unique advantages relative to other microscopes or regular AFM, KPFM enables us to quantify and map the surface potential of an E. coli cell at the nanoscale, which, to our knowledge, has not yet been extensively studied (40).
Conclusion.
Currently, there is a lack of knowledge about the potential impacts of NP exposure on the surface mechanical and other physical properties of microbial cells, although some earlier work indicated that the sorption of oxide NPs onto E. coli cells can cause adverse effects (6, 24, 47, 50). In this work, we used multiple analytical approaches to explore the physical impacts on E. coli cells without and with exposure to hematite NPs. First, SEM and AFM or KPFM images illustrated that E. coli cells had deformation with possible disruption of surface appendages (flagella) based on statistical comparisons. Second, this study examined changes in biomechanical properties (i.e., hardness and elasticity), adhesiveness, and surface electrical properties of E. coli cells. With hematite attachment, the cell surface became hardened or stiffer, as indicated by the reduced indentation and the increased spring constant, the cell surface became more adhesive or more attractive toward the cantilever tip, and the cells shifted to a more-negative surface charge, as evaluated by zeta potential and KPFM. These changes in the biomechanical and physical properties of bacterial cells may have implications for cellular motility, adhesion to surfaces, and stability. This work should provide unique insight into the cytotoxic effects of engineered nanomaterials via imaging and measuring physical properties.
Supplementary Material
ACKNOWLEDGMENTS
This study was partially supported by U.S. Environmental Protection Agency Science to Achieve Results Program grant RD-83385601 and Semiconductor Research Corporation (SRC)/ESH grant 425.025.
We thank Baicang Liu from the College of Architecture and Environment at Sichuan University for his kind help on the SEM operation.
Footnotes
Published ahead of print 30 March 2012
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1. Auffan M, et al. 2008. Relation between the redox state of iron-based nanoparticles and their cytotoxicity toward Escherichia coli. Environ. Sci. Technol. 42:6730–6735 [DOI] [PubMed] [Google Scholar]
- 2. Balaev AE, Dvoretski KN, Doubrovski VA. 2003. Determination of refractive index of rod-shaped bacteria from spectral extinction measurements. Proc. SPIE 5068:375–380 [Google Scholar]
- 3. Berger R, Butt H-J, Retschke MB, Weber SAL. 2009. Electrical modes in scanning probe microscopy. Macromol. Rapid Commun. 30:1167–1178 [DOI] [PubMed] [Google Scholar]
- 4. Bišćan J, Kallay N, Smol T. 2000. Determination of iso-electric point of silicon nitride by adhesion method. Colloids Surf. A Physiochem. Eng. Asp. 165:115–123 [Google Scholar]
- 5. Boulbitch A, Quinn B, Pink D. 2000. Elasticity of the rod-shaped Gram-negative eubacteria. Phys. Rev. Lett. 85:5246–5249 [DOI] [PubMed] [Google Scholar]
- 6. Brayner R, et al. 2006. Toxicological impact studies based on Escherichia coli bacteria in ultrafine ZnO nanoparticles colloidal medium. Nano Lett. 6:866–870 [DOI] [PubMed] [Google Scholar]
- 7. Brunet L, Lyon DY, Hotze EM, Alvarez PJJ, Wiesner MR. 2009. Comparative photoactivity and antibacterial properties of C60 fullerenes and titanium dioxide nanoparticles. Environ. Sci. Technol. 43:4355–4360 [DOI] [PubMed] [Google Scholar]
- 8. Bundeleva IA, et al. 2011. Zeta potential of anoxygenic phototrophic bacteria and Ca adsorption at the cell surface: possible implications for cell protection from CaCO3 precipitation in alkaline solutions. J. Colloid Interface Sci. 360:100–109 [DOI] [PubMed] [Google Scholar]
- 9. Burnham NA, et al. 2003. Comparison of calibration methods for atomic-force microscopy cantilevers. Nanotechnology 14:1–6 [Google Scholar]
- 10. Butkus MA, Grasso D. 1999. Impact of phosphate sorption on water-treatment residual surface characteristics: prelude to reuse. Environ. Eng. Sci. 16:117–129 [Google Scholar]
- 11. Butt H-J, Cappella B, Kappl M. 2005. Force measurements with the atomic force microscope: technique, interpretation and applications. Surf. Sci. Rep. 59:1–152 [Google Scholar]
- 12. Chandraprabha MN, Somasundaran P, Natarajan KA. 2010. Modeling and analysis of nanoscale interaction forces between Acidithiobacillus ferrooxidans and AFM tip. Colloids Surf. B Biointerfaces 75:310–318 [DOI] [PubMed] [Google Scholar]
- 13. Chen KL, Mylon SE, Elimelech M. 2006. Aggregation kinetics of alginate-coated hematite nanoparticles in monovalent and divalent electrolytes. Environ. Sci. Technol. 40:1516–1523 [DOI] [PubMed] [Google Scholar]
- 14. Choi O, Hu ZQ. 2008. Size dependent and reactive oxygen species related nanosilver toxicity to nitrifying bacteria. Environ. Sci. Technol. 42:4583–4588 [DOI] [PubMed] [Google Scholar]
- 15. Doktycz MJ, et al. 2003. AFM imaging of bacteria in liquid media immobilized on gelatin coated mica surfaces. Ultramicroscopy 97:209–216 [DOI] [PubMed] [Google Scholar]
- 16. Dong R, Yu LYE. 2003. Investigation of surface changes of nanoparticles using TM-AFM phase imaging. Environ. Sci. Technol. 37:2813–2819 [DOI] [PubMed] [Google Scholar]
- 17. Dufrêne YF. 2002. Atomic force microscopy, a powerful tool in microbiology. J. Bacteriol. 184:5205–5213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. El Badawy AM, et al. 2011. Surface charge-dependent toxicity of silver nanoparticles. Environ. Sci. Technol. 45:283–287 [DOI] [PubMed] [Google Scholar]
- 19. Gouveia RF, Galembeck F. 2009. Electrostatic charging of hydrophilic particles due to water adsorption. J. Am. Chem. Soc. 131:11381–11386 [DOI] [PubMed] [Google Scholar]
- 20. Harshey RM. 2003. Bacterial motility on a surface: many ways to a common goal. Annu. Rev. Microbiol. 57:249–273 [DOI] [PubMed] [Google Scholar]
- 21. He YT, Wan JM, Tokunaga T. 2008. Kinetic stability of hematite nanoparticles: the effect of particle sizes. J. Nanopart. Res. 10:321–332 [Google Scholar]
- 22. Hermansson M. 1999. The DLVO theory in microbial adhesion. Colloids Surf. B Biointerfaces 14:105–119 [Google Scholar]
- 23. Horst AM, et al. 2010. Dispersion of TiO2 nanoparticle agglomerates by Pseudomonas aeruginosa. Appl. Environ. Microbiol. 76:7292–7298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Huang Z, et al. 2008. Toxicological effect of ZnO nanoparticles based on bacteria. Langmuir 24:4140–4144 [DOI] [PubMed] [Google Scholar]
- 25. Jiang J, Oberdörster G, Biswas P. 2009. Characterization of size, surface charge, and agglomeration state of nanoparticle dispersions for toxicological studies. J. Nanopart. Res. 11:77–89 [Google Scholar]
- 26. Kłodzińska E, et al. 2010. Effect of zeta potential value on bacterial behavior during electrophoretic separation. Electrophoresis 31:1590–1596 [DOI] [PubMed] [Google Scholar]
- 27. Lee C, et al. 2008. Bactericidal effect of zero-valent iron nanoparticles on Escherichia coli. Environ. Sci. Technol. 42:4927–4933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Leonenko Z, Finot E, Amrein M. 2007. Adhesive interaction measured between AFM probe and lung epithelial type II cells. Ultramicroscopy 107:948–953 [DOI] [PubMed] [Google Scholar]
- 29. Li M, Zhu L, Lin D. 2011. Toxicity of ZnO nanoparticles to Escherichia coli: mechanism and the influence of medium components. Environ. Sci. Technol. 45:1977–1983 [DOI] [PubMed] [Google Scholar]
- 30. Li X, Logan BE. 2004. Analysis of bacterial adhesion using a gradient force analysis method and colloid probe atomic force microscopy. Langmuir 20:8817–8822 [DOI] [PubMed] [Google Scholar]
- 31. Liscio A, Palermo V, Mullen K, Samori P. 2008. Tip-sample interactions in Kelvin probe force microscopy: quantitative measurement of the local surface potential. J. Phys. Chem. C Nanomater. Interfaces 112:17368–17377 [Google Scholar]
- 32. Liscio A, Palermo V, Samori P. 2010. Nanoscale quantitative measurement of the potential of charged nanostructures by electrostatic and Kelvin probe force microscopy: unraveling electronic processes in complex materials. Acc. Chem. Res. 43:541–550 [DOI] [PubMed] [Google Scholar]
- 33. Magonov SN, Elings V, Whangbo MH. 1997. Phase imaging and stiffness in tapping-mode atomic force microscopy. Surf. Sci. 375:L385–L391 [Google Scholar]
- 34. Méndez-Vilas A, Gallardo-Moreno AM, González-Martín ML. 2007. Atomic force microscopy of mechanically trapped bacterial cells. Microsc. Microanal. 13:55–64 [DOI] [PubMed] [Google Scholar]
- 35. Morones JR, et al. 2005. The bactericidal effect of silver nanoparticles. Nanotechnology 16:2346–2353 [DOI] [PubMed] [Google Scholar]
- 36. Muller DJ, Engel A. 2007. Atomic force microscopy and spectroscopy of native membrane proteins. Nat. Protoc. 2:2191–2197 [DOI] [PubMed] [Google Scholar]
- 37. Nel A, Xia T, Madler L, Li N. 2006. Toxic potential of materials at the nanolevel. Science 311:622–627 [DOI] [PubMed] [Google Scholar]
- 38. Nel AE, et al. 2009. Understanding biophysicochemical interactions at the nano-bio interface. Nat. Mater. 8:543–557 [DOI] [PubMed] [Google Scholar]
- 39. Oberdörster G, Oberdörster E, Oberdörster J. 2005. Nanotoxicology: an emerging discipline evolving from studies of ultrafine particles. Environ. Health Perspect. 113:823–839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Oh YJ, Jo W, Yang Y, Park S. 2007. Biofilm formation and local electrostatic force characteristics of Escherichia coli O157:H7 observed by electrostatic force microscopy. Appl. Phys. Lett. 90:143901 [Google Scholar]
- 41. Oliver WC, Pharr GM. 2004. Measurement of hardness and elastic modulus by instrumented indentation: advances in understanding and refinements to methodology. J. Mater. Res. 19:3–20 [Google Scholar]
- 42. Pal S, Tak YK, Song JM. 2007. Does the antibacterial activity of silver nanoparticles depend on the shape of the nanoparticle? A study of the Gram-negative bacterium Escherichia coli. Appl. Environ. Microbiol. 73:1712–1720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Palermo V, Palma M, Samori P. 2006. Electronic characterization of organic thin films by Kelvin probe force microscopy. Adv. Mater. 18:145–164 [Google Scholar]
- 44. Park B-J, Haines T, Abu-Lail NI. 2009. A correlation between the virulence and the adhesion of Listeria monocytogenes to silicon nitride: an atomic force microscopy study. Colloids Surf. B Biointerfaces 73:237–243 [DOI] [PubMed] [Google Scholar]
- 45. Pelletier DA, et al. 2010. Effects of engineered cerium oxide nanoparticles on bacterial growth and viability. Appl. Environ. Microbiol. 76:7981–7989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Raffi M, et al. 2010. Investigations into the antibacterial behavior of copper nanoparticles against Escherichia coli. Ann. Microbiol. 60:75–80 [Google Scholar]
- 47. Schwegmann H, Feitz AJ, Frimmel FH. 2010. Influence of the zeta potential on the sorption and toxicity of iron oxide nanoparticles on S. cerevisiae and E. coli. J. Colloid Interface Sci. 347:43–48 [DOI] [PubMed] [Google Scholar]
- 48. Sullivan CJ, Venkataraman S, Retterer ST, Allison DP, Doktycz MJ. 2007. Comparison of the indentation and elasticity of E. coli and its spheroplasts by AFM. Ultramicroscopy 107:934–942 [DOI] [PubMed] [Google Scholar]
- 49. Sun L, Wang JJ, Bonaccurso E. 2010. Nanoelectronic properties of a model system and of a conjugated polymer: a study by Kelvin probe force microscopy and scanning conductive torsion mode microscopy. J. Phys. Chem. C Nanomater. Interfaces 114:7161–7168 [Google Scholar]
- 50. Thill A, et al. 2006. Cytotoxicity of CeO2 nanoparticles for Escherichia coli. Physico-chemical insight of the cytotoxicity mechanism. Environ. Sci. Technol. 40:6151–6156 [DOI] [PubMed] [Google Scholar]
- 51. Velegol SB, Logan BE. 2002. Contributions of bacterial surface polymers, electrostatics, and cell elasticity to the shape of AFM force curves. Langmuir 18:5256–5262 [Google Scholar]
- 52. Volle CB, Ferguson MA, Aidala KE, Spain EM, Núñez ME. 2008. Quantitative changes in the elasticity and adhesive properties of Escherichia coli ZK1056 prey cells during predation by Bdellovibrio bacteriovorus 109J. Langmuir 24:8102–8110 [DOI] [PubMed] [Google Scholar]
- 53. Volle CB, Ferguson MA, Aidala KE, Spain EM, Núñez ME. 2008. Spring constants and adhesive properties of native bacterial biofilm cells measured by atomic force microscopy. Colloids Surf. B Biointerfaces 67:32–40 [DOI] [PubMed] [Google Scholar]
- 54. Webb HK, Truong VK, Hasan J, Crawford RJ, Ivanoya EP. 2011. Physico-mechanical characterisation of cells using atomic force microscopy—current research and methodologies. J. Microbiol. Methods 86:131–139 [DOI] [PubMed] [Google Scholar]
- 55. Xie YP, He YP, Irwin PL, Jin T, Shi XM. 2011. Antibacterial activity and mechanism of action of zinc oxide nanoparticles against Campylobacter jejuni. Appl. Environ. Microbiol. 77:2325–2331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Xu L-C, Vadillo-Rodriguez V, Logan BE. 2005. Residence time, loading force, pH, and ionic strength affect adhesion forces between colloids and biopolymer-coated surfaces. Langmuir 21:7491–7500 [DOI] [PubMed] [Google Scholar]
- 57. Xu Y, Schoonen MAA. 2000. The absolute energy positions of conduction and valence bands of selected semiconducting minerals. Am. Mineral. 85:543–556 [Google Scholar]
- 58. Yao X, et al. 2002. Atomic force microscopy and theoretical considerations of surface properties and turgor pressures of bacteria. Colloids Surf. B Biointerfaces 23:213–230 [Google Scholar]
- 59. Zeyons O, et al. 2009. Direct and indirect CeO2 nanoparticles toxicity for Escherichia coli and Synechocystis. Nanotoxicology 3:284–295 [Google Scholar]
- 60. Zhang W, Kalive M, Capco DG, Chen Y. 2010. Adsorption of hematite nanoparticles onto Caco-2 cells and the cellular impairments: effect of particle size. Nanotechnology 21:355103. [DOI] [PubMed] [Google Scholar]
- 61. Zhang W, Rittmann B, Chen Y. 2011. Size effects on adsorption of hematite nanoparticles on E. coli cells. Environ. Sci. Technol. 45:2172–2178 [DOI] [PubMed] [Google Scholar]
- 62. Zhang W, Stack AG, Chen Y. 2010. Interaction force measurement between E. coli cells and nanoparticles immobilized surfaces by using AFM. Colloids Surf. B Biointerfaces 82:316–324 [DOI] [PubMed] [Google Scholar]
- 63. Zhang W, Yao Y, Chen Y. 2011. Imaging and quantifying the morphology and nanoelectrical properties of quantum dot nanoparticles interacting with DNA. J. Phys. Chem. C Nanomater. Interfaces 115:599–606 [Google Scholar]
- 64. Zhang Y, Chen Y, Westerhoff P, Hristovski K, Crittenden JC. 2008. Stability of commercial metal oxide nanoparticles in water. Water Res. 42:2204–2212 [DOI] [PubMed] [Google Scholar]
- 65. Zhukova L, Kiwi J, Nikandrov V. 2010. Nanoparticles of TiO2 cause aggregation of Escherichia coli cells and suppress their division at pH 4.0-4.5 in the absence of UV irradiation. Dokl. Chem. 435:279–282 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.