Abstract
We have developed a range of vectors for allelic replacements in Staphylococcus aureus to facilitate genetic work in this opportunistic pathogen. The central feature of the vector range is a selection/counterselection system that takes advantage of the 5-fluoroorotic acid (FOA) resistance and pyrimidine prototrophy caused by the loss and gain, respectively, of the pyrF and pyrE genes. This system allows for stringent counterselection of the vectors during the second homologous recombination of a classic allelic replacement. The basic vector pRLY2, which contains the pyrFE genes from Bacillus subtilis, was combined with chloramphenicol, erythromycin, and tetracycline resistance genes and four different versions of nonreplicative or conditionally replicative origins of replication. The choice between these 12 different pRLY vectors allows for high versatility and ensures that the vectors can be used in virtually any genetic background. Finally, as proof of concept, we present six deletions or modifications of components in the S. aureus degradosome as well as the operon containing the cshB DEAD box helicase.
INTRODUCTION
Staphylococcus aureus is an important opportunistic pathogen that can cause various diseases from furuncles to life-threatening osteomyelitis and endocarditis (17). Moreover, acquisition of antibiotic resistance and biofilm formation genes resulting in persistent infections are of major concern. An in-depth genetic analysis of S. aureus virulence genes is therefore highly desirable.
One of the major problems, especially when examining clinical S. aureus strains, is that genetic manipulation is relatively difficult and time-consuming compared to most model organisms. Much work has been carried out using laboratory strains that are easy to work with, such as the mutagenized strains S. aureus RN6390 and RN4220 (14, 19, 22). Furthermore, complementation with genes carried on a plasmid can lead to markedly different phenotypes compared to a single chromosomal copy of the gene, and as a consequence, there is increasing interest in performing allelic replacements on the chromosomes of clinical S. aureus strains.
Several vectors are currently available for generating mutations in S. aureus via double homologous recombination, including pBT2, pMAD, pKOR1, and a range of pCN vectors (1, 2, 5, 7). Common to all of them is that they carry a thermosensitive origin of replication (pE194ts-ORI in pBT2, pMAD, and pKOR1; pT181ts-ORI in pCN39, pCN49, and pCN50), which allows the vectors to replicate in S. aureus at 30°C, but not at 42°C. This enables the passage of the vectors through the restriction-defective, but modification-proficient strain RN4220, by maintaining growth at 30°C, before transforming into the final recipient strain (14, 25). Once colonies have been established after transformation, the temperature is shifted to the nonpermissive temperature of 42°C, while antibiotic selection is kept up, thus selecting for cells where the plasmid has recombined into the chromosome via one of the two regions of homology that correspond to the adjacent regions of the desired mutation. The next step is to find colonies where the vector has recombined a second time via the other region of homology, thereby losing the plasmid and leaving only the desired mutation. This is usually done by growing the culture under nonselective conditions at 42°C and by finally screening individual colonies for loss of the plasmid. This screening can be extremely cumbersome, especially if the desired mutation results in a loss of fitness to the cell, thereby allowing wild-type cells to outgrow the mutant cells during the liquid culture passages. Other potential problems include temperature sensitivity of the desired mutant and the risk of acquiring secondary mutations during the passages at elevated temperature. Although pMAD encodes a beta-galactosidase gene, which allows blue/white screening for the loss of the vector on plates containing 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal), in our hands, white colonies could often be shown to retain pMAD, suggesting that the beta-galactosidase is not stably expressed. Moreover, a reoccurring problem with the thermosensitive origins is that their replication is not completely blocked at the nonpermissive temperature, and therefore, if a mutant divides slower than the replication rate of the vector, then it is extremely difficult to isolate plasmid-free cells. The pKOR1 plasmid goes a step further and incorporates a counterselection system based on a tetracycline-inducible transcript that is antisense to the essential secY secretion gene (2). However, many clinical and lab strains of S. aureus, such as strain COL or our clinical strain S30 (11, 24), carry the pT181 plasmid that carries the gene that encodes a tetracycline efflux pump, which may limit the use of the inducible promoter in pKOR1. A further complication with pKOR1 is the lack of alternatives to the chloramphenicol resistance marker on the backbone vector, which prevents its use in strains that are already resistant to this drug. Additionally, alternative origins would also be desirable, since both pMAD and pKOR1 use the pE194ts origin of replication.
An efficient counterselection system, based on resistance to the drug 5-fluoroorotic acid (FOA) has been used for many years, primarily in yeasts but also in bacteria and archaea (3, 16, 18, 23). FOA is not toxic in itself, but it is converted to the toxic 5-fluoro-UMP by the two genes encoding orotate phosphoribosyltransferase (pyrE) and orotidine 5-phosphate decarboxylase (pyrF). Plating cells on rich medium and FOA will allow extremely efficient selection for the inactivation of pyrE, pyrF, or both. Since pyrE and pyrF catalyze the last two steps in the pyrimidine biosynthesis pathway, they are essential for growth without pyrimidines in the medium. Thus, revertants to the wild type can easily be selected by plating on a pyrimidine-free medium.
Here we present a range of vectors for allelic replacement in S. aureus, incorporating a pyrFE/FOA-based counterselection system and using different origins of replication and different antibiotic markers. As proof of principle, the system was used to delete or modify components of the Gram-positive degradosome: four different RNases, the enolase, and cshA, an RNA DEAD box helicase. Furthermore, a second DEAD box helicase in the S. aureus genome, cshB, and its downstream cistron SA1386 in the operon were deleted individually and together.
MATERIALS AND METHODS
Detailed protocols for allelic replacements, using the pRLY vectors, can be found in Protocols S1, S2, and S3 in the supplemental material.
Media and growth of bacteria.
Escherichia coli was grown in LB medium (Merck, Whitehouse Station, NJ), supplemented as needed with 100 mg/liter ampicillin, 50 mg/liter spectinomycin, and 100 mg/liter X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) (Fermentas Inc., Glen Burnie, MD).
S. aureus was grown in Mueller-Hinton broth (Becton Dickinson and Company, Le Pont de Claix, France), supplemented as needed with 10 mg/liter chloramphenicol (MHC), 10 mg/liter erythromycin (MHE), 2 mg/liter tetracycline (MHT), 200 mg/liter 5-fluoroorotate (MHFOA) (US Biological, Swampscott, MA), or 10 mg/liter uracil (MHU). Agar (13 g/liter) was added to make plates. For pyrimidine-free medium, RH medium, a modified RPMI 1640 medium (catalog no. R7388; Sigma), which contains no NaHCO3 but is buffered with 20 mM HEPES (Sigma-Aldrich Chemie, Steinheim, Germany), was used. When used for plates, 500 ml of RH medium was mixed at 55°C with 7 g of agar (Merck, Whitehouse Station, NJ) that had been autoclaved in 100 ml H2O.
Bacterial strains and plasmids used in this study are listed in Table 1.
Table 1.
Bacterial strains and plasmids used in this study
| Bacterial strain or plasmid | Description, relevant genotype/characteristic, or purposea | Source or reference |
|---|---|---|
| Bacterial strains | ||
| E. coli DH5α | Standard cloning strain | Invitrogen |
| S. aureus | ||
| RN4220 | Restriction deficient, modification proficient | 19 |
| SA564 | Clinical isolate | 9 |
| SA564RD | SA564 hsdR type III mutant (restriction deficient) | 9 |
| SA564RD::pRLBER9 | SA564RD with the pRLBER9 plasmid recombined into the chromosome, thereby disrupting pyrE expression | This work |
| PR01 | SA564RD ΔpyrFE | This work |
| PR02 | RN4220 ΔpyrFE | This work |
| PR03 | RN4220 ΔpyrFE::ORI-pT181ts | This work |
| PR04 | SA564RD ΔpyrFE::ORI-pT181ts | This work |
| PR01-01 | PR01 with the RNase J1 gene deleted (between coordinates 1066843 and 1068378) using the pRLYE1 vector | This work |
| PR01-02 | PR01 with the RNase Y gene deleted (between coordinates 1281956 and 1283492) using the pRLYE1 vector | This work |
| PR01-04 | PR01 with the RNase J2 gene deleted (between coordinates 1268765 and 1270394) using the pRLYE1 vector | This work |
| PR01-05 | PR01 with the enolase gene substituted with the E. coli enolase gene (between coordinates 837567 and 838878) using the pRLYE1 vector | This work |
| PR01-06 | PR01 with the cshA gene substituted with a CAT194 cassette (between coordinates 2136001 and 2137592) using the pRLYE1 vector | This work |
| PR01-07 | PR01 with the PNPase gene deleted (between coordinates 1266484 and 1268492) using the pRLYE1 vector | This work |
| PR01-08 | PR01 with the SA1386 gene deleted (between coordinates 1591115 and 1591987) using the pRLYE1 vector | This work |
| PR01-09 | PR01 with the cshB gene deleted (between coordinates 1592017 and 1593367) using the pRLYE1 vector | This work |
| PR01-10 | PR01 with the cshB and SA1386 genes substituted by an ermC cassette (between coordinates 1591115 and 1593454) using the pRLYC1 vector | This work |
| PR07 | SA564RD with the spa gene deleted (between coordinates 122645 and 124007) using the pRLYC1 vector | This work |
| Plasmids | ||
| pBlueScript II KS+ | Standard E. coli cloning vector | Stratagene |
| pFL | pUC19 derivative with added BglII and ClaI sites | 4 |
| pGB2 | pSC101 origin of replication, spectinomycin resistance marker | 8 |
| pMAD | 1 | |
| pMF35 | Carries the PspacC promoter | 10 |
| pMK4 | 21 | |
| pNL9162 | Contains the targetron homing intron, adapted for use in S. aureus | 28 |
| pT181 | Naturally occurring plasmid, encoding the tetK tetracycline efflux pump | 13 |
| pBS-AB-ERY | Used to generate strains PR01 and PR02 | This work |
| pBS-AB-ERY-TREPrev | Used to generate strains PR03 and PR04 | This work |
Deletions were carried out in strain PR01, which has an S. aureus strain SA564 background, but coordinates are given based on the Staphylococcus aureus n315 genomic sequence (GenBank accession number BA000018.3).
Molecular biology methods.
All methods using standard molecular biology techniques were performed by the methods of Sambrook et al. (20) or according to the recommendations of the manufacturers. Restriction enzymes from New England BioLabs (Ipswich, MA) were used according to the manufacturer's instructions. PCR products used for cloning were amplified using Phusion high-fidelity DNA polymerase enzyme (New England BioLabs), and screening was carried out using REDTaq ReadyMix (Sigma-Aldrich Chemie) according to the manufacturer's instructions.
The primers for sequencing inserts in the pRLY vectors were pRL-MCS-Seq-F1 (MCS stands for multiple cloning site, Seq stands for sequencing, and F stands for forward) (GCGGCATCAGAGCAGATTG) and pRLY-MCS-Seq-R1 (R stands for reverse) (GGAAACGAAATCCCGAGTC). The primers used for sequencing inserts in the pRLB vectors were pRLB-MCS-Seq-F1 (CTAATACGACTCACTATAGGGC) and pRLB-MCS-Seq-R1 (TGCCCCGTTAGTTGAAGAAGGTT). Primers used for vector construction are shown in Table S1 in the supplemental material.
Strain construction.
S. aureus strains PR01 and PR02 were constructed using pBS-AB-ERY (ERY stands for erythromycin), which is a pBluescript II KS+ plasmid (Stratagene, Santa Clara, CA) with ∼1,000 bp upstream of pyrFE cloned between the BamHI and EcoRI sites, ∼1,000 bp downstream of the pyrFE genes cloned between the EcoRI and XhoI sites, and the ermC cassette (see Fig. 2) cloned into the BamHI site. The plasmid was transformed into strains SA564RD (9) and RN4220, respectively, and transformants that grew on RH medium containing erythromycin (ensuring that plasmid integration was downstream of the pyrFE genes) were isolated. Then a round of FOA selection was used to select for a second recombination event that removed the plasmid backbone. FOA-resistant colonies were screened for loss of erythromycin resistance and then sequenced to verify the deletion of pyrFE.
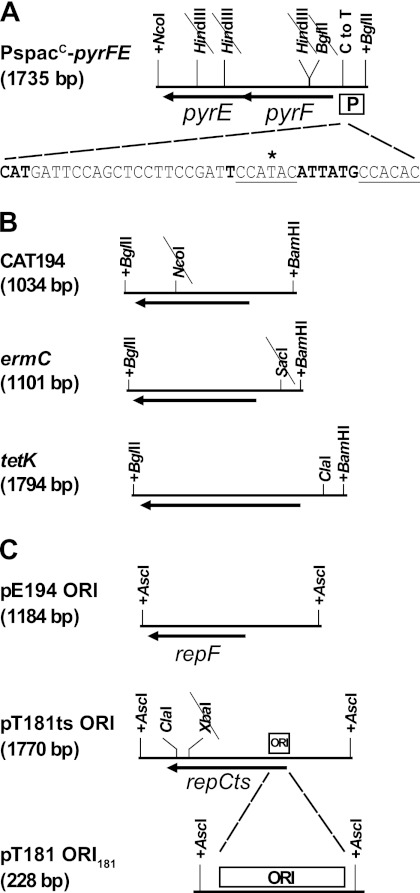
Fig 2.
Cassettes used in vector construction. Added restriction sites are indicated by a plus sign before the restriction site, whereas a slash through the restriction site indicates that the restriction site was removed by a silent point mutation. The two ClaI sites used for the deletion in pRLYT8 are indicated. (A) The PspacC-pyrFE cassette, with pyrF and pyrE genes from B. subtilis. The C-to-T point mutation used to avoid instability in E. coli is indicated. The −10 box, the transcription start site, and the start codon are indicated in bold type. The 6-bp repeats are underlined, and the asterisk indicates where we introduced the C-to-T point mutation. The three HindIII sites and a BglII site were removed from the pyrFE genes by site-directed mutagenesis to maintain the uniqueness of the corresponding sites. (B) Antibiotic resistance cassettes used for construction of the pRLYx1 vectors. An NcoI site in CAT194 and a SacI site in ermC were removed by site-directed mutagenesis via silent mutations. (C) Origins of replication used for construction of the pRLYx2, pRLYx8, and pRLYx9 vectors. ORI181 (ORI) is indicated, and broken lines indicate the section of the full-length pT181ts origin of replication, which comprises the ORI181, used for the pRLYx2 vectors. The XbaI site was mutated in the pT181ts origin.
For strains PR03 and PR04, the pT181ts origin of replication (see Fig. 2) was cloned into the EcoRI site of pBS-AB-ERY, generating pBS-AB-ERY-TREPrev. The procedure for generating the strains was the same as for PR01 and PR02, but it was carried out at 42°C to avoid plasmid replication via the pT181ts origin.
Vector construction.
All in silico design of vectors and constructs was carried out using pDRAW32 software (www.acaclone.com).
pFL was modified into pRL1 by introducing an XcmI site inside the lacZα gene (generating an unperturbing A53D mutation) and by introducing an NcoI site, an XhoI site, and a second XcmI site. pRL1 was then further modified by introducing an AscI site, an NgoMIV site, and an MluI site near the ClaI site, creating pRL2 (see Fig. 2).
The PspacC-pyrFE cassette was constructed by merging the PspacC promoter from pMF35 with the pyrFE genes from Bacillus subtilis, using fusion PCR. Then the three HindIII sites and a BglII site were removed from the pyrFE genes by site-directed mutagenesis to maintain the uniqueness of the corresponding sites.
The resistance cassettes were all prepared with a BamHI site upstream and a BglII site downstream: the chloramphenicol resistance (Cmr) CAT194 gene, amplified from pMK4 (21); the erythromycin resistance (Eryr) ermC gene, amplified from pMAD (1); and the tetracycline resistance (Tetr) tetK gene from pT181.
The origins of replication were prepared with AscI sites at each end, and the XbaI site in the pT181ts origin was removed by silent mutagenesis.
pRLBC2 was constructed by combining the pSC101-based spectinomycin-resistant vector pGB2 (8) with the multiple cloning site of pBluescript II KS+ and the S. aureus plasmid pC194 (Cmr). In addition, to make plasmid preparation from E. coli easier, the copy number of the pSC101 origin was increased by introducing an G-to-A point mutation at position 1026 of the repA gene (pSC101 numbering) (26). A basic E. coli plasmid, pRLB1, was obtained by removing the pC194 sequence from pRLBC2, while maintaining the MCS. pRLBE2 was generated by introducing a BsrGI site and a BstBI site on each side of the CAT194 gene in pRLBC2, and inserting the ermC cassette between the two sites. pRLBER9 was designed with a relatively short, 652-bp region of homology to the S. aureus chromosome to prevent too frequent plasmid cross-out (from coordinate 201 in the pyrF open reading frame [ORF] to 157 in the pyrE ORF).
Nucleotide sequence accession numbers.
GenBank accession numbers for the vector constructs can be found in Tables 2 and 3.
Table 2.
pRL1-based vectors
| Vector | Selection in S. aureusa | Origin of replication for S. aureus | Length (bp) | GenBank accession no. |
|---|---|---|---|---|
| pRL1 | None | None | 2,702 | JQ410188 |
| pRL2 | None | None | 2,703 | JQ768506 |
| pRLY2 | pyr | None | 4,414 | JQ410189 |
| pRLYC1 | Cm, pyr | None | 5,417 | JQ366074 |
| pRLYC2 | Cm, pyr | ORI181b | 5,627 | JQ768507 |
| pRLYC8 | Cm, pyr | pT181ts | 7,038 | JQ768508 |
| pRLYC9 | Cm, pyr | pE194ts | 6,581 | JQ768509 |
| pRLYE1 | Ery, pyr | None | 5,499 | JQ768510 |
| pRLYE2 | Ery, pyr | ORI181b | 5,709 | JQ768511 |
| pRLYE8 | Ery, pyr | pT181ts | 7,120 | JQ768512 |
| pRLYE9 | Ery, pyr | pE194ts | 6,663 | JQ768513 |
| pRLYT1 | Tet, pyr | None | 6,193 | JQ768514 |
| pRLYT2 | Tet, pyr | ORI181b | 6,403 | JQ768515 |
| pRLYT8 | Tet, pyr | pT181ts | 7,404c | JQ768516 |
| pRLYT9 | Tet, pyr | pE194ts | 7,357 | JQ768517 |
| pRLSAYC9 | Cm | pE194ts | 8,026 | JQ768518 |
| pRLSAYE9 | Ery | pE194ts | 8,108 | JQ768519 |
| pRLSAYT9 | Tet | pE194ts | 8,802 | JQ768520 |
All pRL1-based vectors carry an ampicillin resistance marker for use in E. coli. Abbreviations: pyr, pyrimidine prototrophy; Cm, chloramphenicol; Ery, erythromycin; Tet, tetracycline.
Plasmids with the pT181 origin of replication but missing the repC gene, which is provided in trans in strains PR03 and PR04.
Deletion between the ClaI sites in the Tetr cassette and the pT181ts cassette.
Table 3.
pRLB-based vectors
| Vector | Selection in S. aureusa | Origin of replication for S. aureus | Incompatible vectors | Length (bp) | Description | GenBank accession no. |
|---|---|---|---|---|---|---|
| pRLB1 | None | None | None | 4,105 | Basic E. coli vector, with no similarity to pUC-based plasmids | JQ768521 |
| pRLBC2 | Cm | pC194 | pRLYC vectors | 6,325 | E. coli-S. aureus shuttle vector | JQ768522 |
| pRLBE2 | Ery | pC194 | pRLYE vectors | 6,543 | E. coli-S. aureus shuttle vector | JQ768526 |
| pRLBC3 | Cm | pC194 | pRLYC vectors | 6,575 | pRLBC2 with the Pprot promoter | JQ768523 |
| pRLBE3 | Ery | pC194 | pRLYE vectors | 6,793 | pRLBE2 with the Pprot promoter | JQ768527 |
| pRLBER1 | Ery | None | pRLYE vectors | 5,936 | Carries 652 bp of pyrFE for temporary disruption of pyrE | JQ768528 |
| pRLBCR8 | Cm | pT181ts | pRLYC vectors, pRLYx8 vectors | 7,468 | Carries 652 bp of pyrFE for temporary disruption of pyrE | JQ768524 |
| pRLBCR9 | Cm | pE194ts | pRLYC vectors, pRLYx9 vectors | 7,008 | Carries 652 bp of pyrFE for temporary disruption of pyrE | JQ768525 |
| pRLBER8 | Ery | pT181ts | pRLYE vectors, pRLYx8 vectors | 7,550 | Carries 652 bp of pyrFE for temporary disruption of pyrE | JQ768529 |
| pRLBER9 | Ery | pE194ts | pRLYE vectors, pRLYx9 vectors | 7,090 | Carries 652 bp of pyrFE for temporary disruption of pyrE | JQ768530 |
All pRLB vectors carry a spectinomycin resistance marker for selection in E. coli. Cm, chloramphenicol; Ery, erythromycin.
RESULTS AND DISCUSSION
One of the major challenges in bacterial genetics is to generate mutations that severely limit growth. Mutation in itself is a rare event, but if the mutation additionally confers severe loss of fitness to the cell, then screening for it can be even more difficult due to faster growth of wild-type cells.
To exploit the stringent counterselection offered by FOA to generate an improved vector system for allelic replacement in Staphylococcus aureus, we created a range of plasmids that express pyrE and pyrF, combined with a range of antibiotic resistance genes and different conditionally replicating staphylococcal origins of replication. Moreover, we facilitated cloning into the vectors by retaining the uniqueness of all the restriction sites in the multiple cloning site of pUC19, as well as blue/white screening by the lacZα complementation system.
Counterselection vector construction.
We used the pFL plasmid as the basis. pFL is a pUC19 derivative in which a BglII site and a ClaI site have been introduced upstream of the lacZα gene in a region that is nonessential for plasmid functions (4). pFL was modified into pRL2 by introducing several additional restriction sites (Fig. 1).
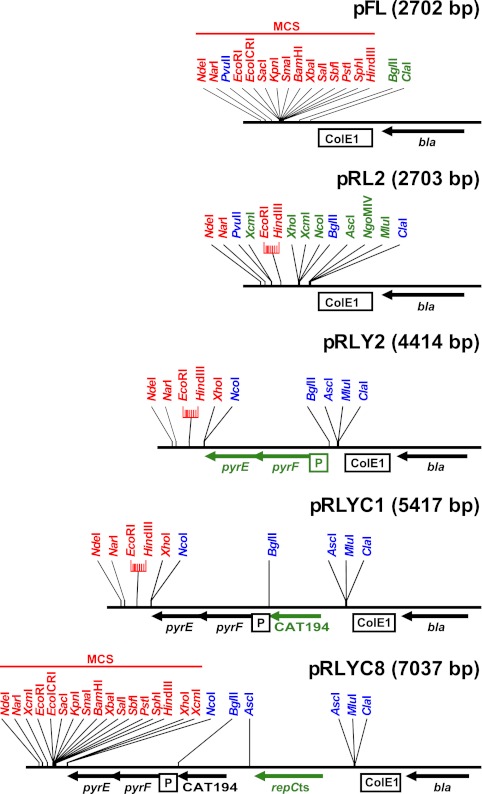
Fig 1.
Different steps in vector construction, exemplified by the pRLYC8 vector. Features that are introduced at each step are shown in green. Unique restriction sites in the multiple cloning site (MCS) are shown in red. Other restriction sites of interest are shown in blue. bla is the beta-lactamase gene. ColE1 is the origin of replication for E. coli. pyrF and pyrE are genes taken from B. subtilis and fused to the PspacC promoter (which is shown as a boxed P). CAT194 is the S. aureus chloramphenicol resistance gene. repCts is the pT181ts replication gene. The full MCS is shown only in pFL and pRLYC8, but the region between the EcoRI and HindIII sites remains unchanged in all vectors.
(i) The pyrFE cassette for counterselection.
To limit unwanted homologous recombination between the vectors and the S. aureus chromosome, endogenous DNA sequence was avoided by fusing the exogenous pyrFE genes from Bacillus subtilis to the artificial constitutive promoter PspacC (10), both of which are significantly different from the S. aureus sequence. Furthermore, while we were testing the vectors, we discovered that the Pspac promoter is slightly unstable in E. coli, due to a 6-bp repeat surrounding the −10 motif, and we therefore introduced a G-to-A mutation that disrupts the repeat but retains promoter activity (Fig. 2A). The resulting construct was introduced into pRL2 between the BglII and NcoI sites, creating pRLY2 (Fig. 1).
Finally, three resistance cassettes, Cmr, Eryr, and Tetr (Fig. 2B), were cloned into the BglII site of pRLY2, creating pRLYC1 (Cmr) (Fig. 1), pRLYE1 (Eryr), and pRLYT1 (Tetr) (see Fig. S1A in the supplemental material).
(ii) Temperature-sensitive origins of replication.
While these three constructs are efficient as suicide integrative vectors, it is often necessary to pass a vector through the restriction-deficient S. aureus strain RN4220, to get it correctly modified, before introducing the vector into a restriction-proficient S. aureus strain. For this to work, the vector will need to replicate in strain RN4220, but not in the wild-type strain.
The pE194ts origin of replication, which is temperature sensitive (12) and the pT181 origin of replication with the cop-634 mutations, which renders it temperature sensitive (6) (Fig. 2C) were therefore cloned into the AscI site of the pRLYx1 vectors (x can stand for C, E, or T), leading to pRLYx8 (pT181ts origin) and pRLYx9 (pE194ts origin) (Fig. 1 and Table 2; also see Fig. S1A in the supplemental material). It should be noted that the tetK cassette and the pT181ts origin of replication have an overlapping sequence, which we removed by deleting the small fragment between the ClaI sites in pRLYT8 indicated in Fig. 2, to avoid potential vector instability.
To ensure that the thermosensitivity of the two origins of replication remained intact when introduced into the pRLYx1 vectors, strain PR01 (Fig. 3) carrying either pRLYx8 or pRLYx9 was grown overnight at 30°C in selective medium, whereupon 10 μl was transferred to MH medium at 42°C for 7 h. Dilution series were then spotted on nonselective (MH medium) and selective (MH medium plus antibiotic) plates, which were incubated at 30°C. Counting the colonies in the spots revealed that during the 7 h of nonselective growth at 42°C, about 90% of the cells had lost pRLYx8 or pRLYx9 (see Fig. S2 in the supplemental material), showing that thermosensitivity remains intact in both vectors.
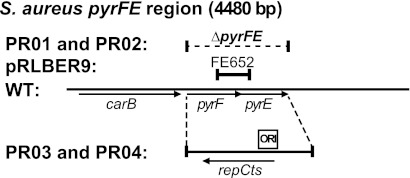
Fig 3.
Various modifications to the pyrFE locus in S. aureus. The broken horizontal line indicates the chromosomal region of strains SA564RD and RN4220 that was deleted to generate strains PR01 and PR02, respectively. FE652 is the region of the pyrFE locus that allows pRLBER9 to recombine into the chromosome and disrupt pyrE expression. Strains PR03 and PR04 were generated by substituting the pyrFE genes with the pT181ts-ORI cassette from Fig. 2 in strains RN4220 and SA564RD, respectively. The ORI181 region (ORI) is indicated.
(iii) Strain-specific plasmid replication.
Although there are situations where it is desirable to transform with a replicating vector, notably when working with strains for which efficient transformation is not available, the thermosensitive plasmid origins of replication are not always completely inhibited at 42°C. A more stringently conditional system was therefore developed by combining an RN4220 derivative that expresses a plasmid replication protein with vectors that carry the corresponding replication origin without encoding the replication protein.
It has been shown that the replication protein encoded by pT181 (repC) can function in trans and recognizes an ∼168-bp sequence to promote replication (13).
We exploited this relationship by replacing the pyrFE genes of S. aureus RN4220 with the full pT181ts origin of replication, which encompasses both the repC gene and the ∼168-bp origin of replication, thus generating strain PR03 (Fig. 3). We then introduced the 168-bp minimal origin of replication (ORI181 [Fig. 2C]) into the AscI site of the pRLYx1 vectors, thereby obtaining pRLYx2 vectors that can replicate in strain PR03 at 30°C but that will be nonreplicative in any other S. aureus strain (Fig. 1 and Table 2), unless the strain carries another pT181 plasmid. However, it should be noted that the copy number of the pRLYx2 plasmids in strain PR03 is very low, and therefore, we recommend that either the plasmid should be prepared from a large culture volume or phage transduction should be used, when introducing the plasmid into a strain that does not allow plasmid replication.
(iv) Multiple cloning site of the pRLY vectors.
The entire multiple cloning site (MCS) of pUC19 has been kept intact by deleting conflicting sites elsewhere in the vectors by silent mutagenesis (Fig. 2). Additionally, the two XcmI sites were designed to each give a single thymidine 3′ overhang, compatible with the single adenosine 3′ overhang created by many DNA polymerases used for PCR, thus allowing ligation to the vector without the need for enzymatic digestion of the PCR product. Thus, the MCS of the pRLY vectors stretch from the NdeI site at position 184 to the XcmI site at position 624 (the NcoI site can also be used in vectors that does not carry the Tetr cassette), and the lacZα gene is functional for blue/white screening in E. coli (Fig. 1).
Construction of new expression vectors to avoid interplasmid recombination.
Sometimes it is desirable to maintain an expression vector in a strain while performing an allelic replacement. However, in such situations, it is important that the plasmid used for expression of proteins has no similarity to the allelic replacement vector (such as the pRLY vectors) to prevent homologous recombination between the two. We therefore designed a series of E. coli-S. aureus shuttle vectors based on the E. coli vector pGB2, which has no homology to pUC19 (8), and the S. aureus pC194 origin of replication, which has no homology to the pE194 or pT181 origins of replication. Two versions of the vector were made: pRLBC2 with chloramphenicol resistance and pRLBE2 with erythromycin resistance (Table 3 and Fig. 4). Furthermore, to allow continuous strong expression of genes and to allow expression to be uncoupled from native promoters, we introduced the constitutive Pprot promoter from a streptococcal plasmid (15) upstream of the KpnI site in the MCS of pRLBC2 and pRLBE2, generating pRLBC3 and pRLBE3, respectively (Table 3; see also Fig. S3 in the supplemental material).
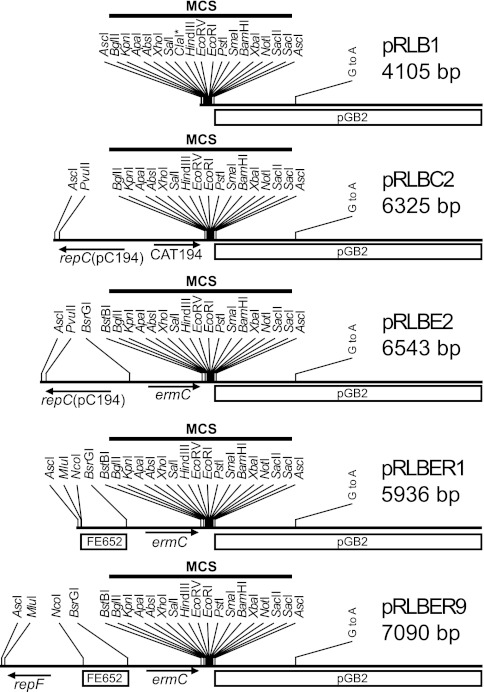
Fig 4.
Layout of selected pRLB vectors. Important restriction sites and genes are indicated. The multiple cloning site (MCS) is indicated. The asterisk on ClaI indicates a ClaI site that is unique in pRLB1 but not in the rest of the vectors. The G-to-A mutation (G to A) that was introduced to increase plasmid copy number in E. coli is shown. pGB2 is the E. coli backbone vector. repC(pC194) is the replication gene from pC194. CAT194 is the chloramphenicol resistance marker. ermC is the erythromycin resistance marker. repF is the pE194ts replication gene. FE652 is the 652-bp sequence from the pyrFE locus in S. aureus.
Generating pyrFE mutants and revertants in S. aureus.
To determine the optimal concentration of FOA for restricting growth of pyrFE+ strains while allowing growth of pyrFE mutant strains, we spotted dilution series of a wild-type and a pyrFE mutant on MH plates with 50, 100, 150, and 200 mg/liter FOA. The 50- and 100-mg/liter concentrations of FOA allow slight growth of wild-type cells, whereas the 150- and 200-mg/liter concentrations abolish growth (not shown). The pyrFE mutant grew on all plates, which shows that the FOA concentrations tested are nontoxic in cells without pyrFE, and also indicates that MH medium contains sufficient pyrimidines to support pyrimidine auxotrophic S. aureus without adding additional uracil, although we find that adding 10 mg/liter uracil is preferable to obtain faster growth and denser cultures. Note that the optimal concentration of FOA might be dependent on the strain.
To use this counterselection system efficiently in almost any S. aureus strain, we developed several strategies to easily generate pyrFE mutations taking in consideration the possibility to revert to a wild-type PyrFE background after having the mutants constructed.
Strains with spontaneous mutations in either pyrE or pyrF can be obtained by plating on MHFOA medium. Single colonies are restreaked on MHFOA medium and then verified by replica plating on MH, MHFOA, and RH media. Strains growing on MHFOA medium but not on RH medium are strong candidates for pyrFE mutants. Revertants with functional pyrFE genes can be obtained by plating on RH medium. However, if it is important to ensure a perfect reconstitution of the pyrFE genes, then it is preferable to use either phage transduction from a wild-type strain or classical allelic replacement of the pyrFE genes, using pyrimidine prototrophy as a marker. For the latter, we have constructed the pRLSAYC9, pRLSAYE9, and pRLSAYT9 vectors (Table 2), where the pyrFE genes from strain SA564, with an additional ∼1 kb of chromosomal sequence on each side, have been cloned between the XbaI and BglII sites of pRLYC9, pRLYE9, and pRLYT9, respectively. These vectors allow complete reconstruction of the pyrFE locus, regardless of mutation type (see Protocol S2 in the supplemental material for details).
(i) A system for temporary disruptions of pyrFE.
While it is easy to select on FOA to isolate naturally occurring pyrFE mutants, it is practical to be able to disrupt the pyrFE genes in a predictable and completely reversible manner. To accomplish this, the pRLBER9 plasmid was designed to recombine into the pyrFE genes, via a single region of homology (FE652 in Fig. 3 and 4), thereby disrupting expression of the downstream pyrE gene. This disruption can be maintained by selecting on FOA, allowing the pRLY counterselection system to be used for one or more allelic replacements. Once the genetic manipulations have been accomplished, then the pyrFE locus can be reverted back to the wild type by using pyrimidine-free medium to select for cells in which pRLBER9 is removed (see details in Protocol S3 in the supplemental material).
In order to create pRLBER9, the pRLBE2 vector was modified by substituting the pC194 origin of replication with the pE194ts origin, and a 652-bp section of pyrFE was introduced between the origin and the ermC cassette (Fig. 4). pRLBER9 can therefore not be used with pRLY vectors that carry ermC cassettes and/or pE194ts origins, and to remedy this deficiency, versions of pRLBER9 were generated with pT181ts origins and CAT194 cassettes in various combinations (Table 3) to ensure compatibility with any of the pRLY vectors (or any other E. coli-S. aureus shuttle vectors).
In addition to disrupting pyrE, pRLBER9 and its related plasmids can also be used to express genes from a single copy per chromosome by cloning into the MCS and introducing the vector into the pyrFE locus on the chromosome.
Proof of principle. (i) Allelic replacement procedure.
A protocol for allelic replacement, exemplified by using pRLYE1 to exchange your favored gene, yfg, with a kanamycin resistance marker, is briefly summarized here (see Protocol S1 in the supplemental material for details) (Fig. 5 shows a schematic flowchart). An S. aureus pyrFE mutant strain is transformed with 1 μg plasmid DNA and plated on MHE. A liquid culture of each colony to be tested is spotted on plates containing MH, MHE, MHFOA, and RH media (pyrimidine-free defined medium, see Materials and Methods). Strains growing on MHFOA are discarded (the remaining strains can be checked by PCR to verify vector insertion), and one or two strains are chosen for growth in MHU containing kanamycin for 6 h, whereupon they are streaked on MHFOA plus kanamycin. The colonies are picked and restreaked on MHFOA plus kanamycin, then transferred to liquid medium, and spotted onto plates containing MH, MHE, MHFOA, and RH media. Candidates that grow on MHE and/or RH medium are discarded, and remaining candidates are checked by PCR and subsequent sequencing of the genomic region of yfg.
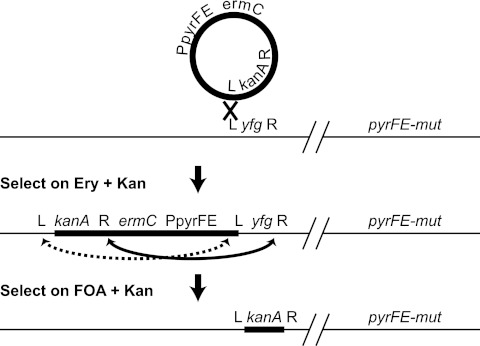
Fig 5.
Allelic replacement flowchart for replacing the yfg allele with a kanamycin resistance cassette (kanA). A nonreplicative vector is forced to recombine (thick X) with the chromosome by selection on erythromycin (Ery) and kanamycin (Kan). The recombination can occur only via L or R, which are homologous to the regions adjacent to yfg in the chromosome (recombination with L is shown). Once the vector has been integrated into the chromosome, then FOA is used to counterselect the presence of the backbone vector, while kanamycin is used to select for the recombination that results in a substitution of yfg by kanA (solid black double-headed arrow). The dotted double-headed arrow indicates an alternative recombination which can happen if there is no selection with kanamycin and which will result in a wild-type strain. Vector DNA (thick black lines) and chromosomal DNA (thin black lines) are indicated. yfg is the gene that is substituted by kanA. kanA is the kanamycin resistance marker. L and R are ∼1 kb of sequence to the left and right of yfg, respectively. ermC is the erythromycin resistance marker. PpyrFE is pyrF and pyrE from B. subtilis with a Pspac constitutive promoter. The chromosomal copies of the pyrF and/or pyrE genes that have been inactivated (pyrFE-mut) (see the text for further information) are indicated.
(ii) Degradosome mutants generated.
The counterselection system worked well in our hands when using spontaneously obtained FOA-resistant (FOAr) mutants. However, for testing purposes, it was decided to use strain PR01, a ΔpyrFE mutant of the clinical strain SA564, because this strain is also mutated in the two restriction systems (9, 27), allowing direct transformation with plasmids isolated from E. coli DH5α.
The S. aureus degradosome consists of five RNases (RNase J1, RNase J2, RNase Y, polynucleotide phosphorylase [PNPase], and RnpA), two metabolic enzymes (enolase and phosphofructokinase), and a DEAD box RNA helicase (CshA) (27).
To test our counterselection system, deletions of SA0940 (RNase J1), SA1118 (RNase J2), SA1117 (PNPase), and SA1129 (RNase Y), and SA1885 (cshA) were performed, generating strains PR01-01, PR01-04, PR01-02, PR01-07, and PR01-06, respectively (Fig. 6). The mutations were checked by sequencing the entire region from ∼1,300 bp downstream to ∼1,300 bp upstream of the deletion. Moreover, since enolase is an essential enzyme in the glyconeogenetic and glycolysis pathways, the S. aureus enolase gene (eno [SA0731]) was substituted with the enolase gene from E. coli, which encodes an enolase that should be sufficiently different to prevent normal assembly of the degradosome, but which still provides the essential enzymatic activity for glycolysis and glyconeogenesis (strain PR01-05) (Fig. 6).
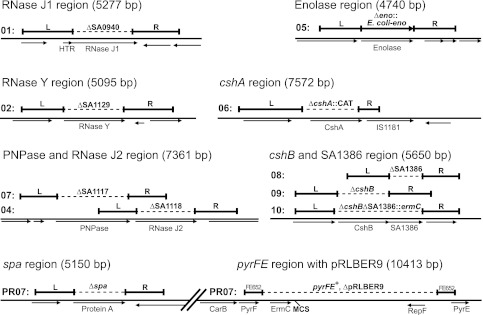
Fig 6.
Overview of deletions and replacements generated in order to test the FOA counterselection system. The deleted regions (broken lines) are indicated. The two regions used for homologous recombination, the regions left and right of the deletion, are indicated by the thick lines with the letter L and R, respectively. Horizontal arrows indicate significant ORFs in the vicinity of the deletions. HTR indicates a hypothetical transcriptional regulator gene, which is 1 bp upstream of the RNase J1 gene. The numbers 01 to 10 indicate the strain name, PR01-01 to PR01-10, respectively. The bottom row shows strain PR07, where a spa deletion (bottom left) was carried out in strain SA564RD that was temporarily a pyrFE mutant, due to a pRLBER9 insertion. Subsequently, pRLBER9 was removed, leaving an intact wild-type pyrFE region (bottom right). FE652 indicates the two 652-bp regions which recombine to cross-out pRLBER9. The multiple cloning site in the chromosomally integrated pRLBER9 (MCS) is indicated.
(iii) Generating a cshB SA1386 double mutant.
To further test the strength of the counterselection, we decided to delete the two genes SA1386 and SA1387 (cshB), coding for a putative DNA endonuclease and an RNA helicase, respectively. First, cshB and SA1386 were deleted individually, giving rise to strains PR01-09 and PR01-08, respectively. PR01-09 is cold sensitive, whereas PR01-08 is able to grow at the same temperatures as the parent strain.
We had attempted several times, unsuccessfully, to obtain an SA1386 cshB double mutant in our laboratory, both via allelic replacement using pMAD and by disruption with a mobile intron, using the pNL9162 targetron system (28), and our continued failure suggested that the double mutation might be synthetically lethal. However, using the pRLYC1 vector, we encountered no problem replacing cshB and SA1386 with an ermC cassette (strain PR01-10 [Fig. 6]), testing three colonies where one had the deletion, thus showing not only that the double mutant is viable (although slowly growing and cold sensitive), but also that the FOA counterselection can be used to obtain mutants that could not be obtained by other means. We speculate that the fitness loss in the double mutant (which grows about 25% slower and has a long lag phase when inoculated) prevented a sufficiently high rate of plasmid loss (using plasmid pMAD or pNL9162) to allow detection of plasmid-free cells via screening, and thus, pyrFE counterselection was needed to overcome this barrier.
(iv) Attempting to delete the essential eno gene.
Finally, we attempted to delete the essential enolase gene (eno) in order to force the appearance of false positives. A construct with pRLYE1 as the backbone and with a Cmr marker situated between the L and R regions (Fig. 6) was recombined into the chromosome. After brief growth in MHC supplemented with uracil, the culture was plated on MHC containing FOA. All six colonies that appeared were restreaked and then tested for erythromycin sensitivity (which would indicate a loss of the backbone vector) and chloramphenicol resistance (which would indicate substitution of the enolase). The six clones were all erythromycin resistant, indicating that the FOA resistance had arisen from a mutation in the pyrFE genes of the integrated vector. No FOAr Erys clones were observed, and we conclude that double mutations, destroying both the vector backbone resistance marker (ermC in this case) and the pyrFE genes, are rare. Furthermore, when performing a parallel experiment where the MHC-plus-uracil culture was plated on MHFOA, many more colonies appeared, and 30 tested colonies had lost both the Cmr marker and the Eryr marker, indicating homologous recombination via the same region as the initial integration into the chromosome (exemplified by the dotted double-headed arrow in Fig. 5).
(v) Generating an spa deletion, using pRLBER9 to temporarily disrupt pyrE.
In order to test the functionality of pRLBER9, the plasmid was transformed into strain SA564RD, the parent strain of PR01, which has an intact and functional pyrFE locus. Plasmid replication was inhibited by shifting the temperature to 42°C, and erythromycin selection was used to select for cells where the plasmid had recombined into the pyrFE locus. It was verified that the resulting strain, SA564RD::pRLBER9, was FOA resistant, and then competent cells were prepared. A pRLYC1-based construct, which was designed to delete the spa gene, encoding protein A, was then transformed into SA564RD::pRLBER9. Transformants were selected on chloramphenicol, and the continued presence of pRLBER9 was confirmed by erythromycin resistance. Following the procedure described above (and in protocol S1 in the supplemental material), we obtained a spa deletion mutant, strain RD::pRLBER9-Δspa. To return to a wild-type pyrFE locus and eliminate pRLBER9, a selection on pyrimidine-free medium was performed (to select for plasmid cross-out) at 42°C (to inhibit plasmid replication). After one restreaking, a screen for loss of erythromycin resistance showed that all 16 colonies had lost pRLBER9. One colony was chosen (PR07; Fig. 6), and the spa and pyrFE loci were PCR amplified and sequenced to confirm the Δspa pyrFE+ genotype (protocol S3 provides details for using pRLBER9).
Conclusion.
We have developed a new and highly efficient system for generating mutants by allelic replacement in S. aureus. The strong FOA counterselection allows for quick and clean selection of mutants, which means that the expensive and time-consuming screening has been reduced to a verification step. While any system for obtaining mutants, including the one presented here, carries the probability of acquiring secondary spontaneous mutations in the genome, a shorter workflow should lessen that risk. Following the detailed protocol presented in supplemental material, we routinely obtain mutants in 8 days after transforming into S. aureus, about 2 to 3 days less than existing systems. Moreover, we have combined our counterselection system with a range of antibiotic resistance genes, and a range of conditional replication origins, which allow the choice of an optimal combination for a given project (Table 2). Therefore, our range of counterselection vectors solves several of the problems inherent in previous vectors, especially for obtaining mutants with significant reduction in fitness.
Furthermore, we have created a range of auxiliary vectors without sequence similarity to the pRLY vectors. These pRLB vectors can be used not only for gene expression during the allelic replacement procedure but can also be used for generating completely reversible pyrFE mutants.
Finally, we have tested our vectors by generating a variety of allelic replacements, including metabolic genes, RNases, and DNA repair enzymes. We have been able to delete or substitute all the alleles we have attempted, with the special exception of a deletion of the essential enolase gene, thus demonstrating the effectiveness and general application of the vector system.
Supplementary Material
ACKNOWLEDGMENTS
We thank W. Kelley for providing the kanamycin and tetracycline cassettes, A. Renzoni for providing the Pprot promoter, D. Belin for providing pGB2, M. Fujita for the PspacC promoter, A. Jousselin for the idea of using RPMI 1640 medium as the base for a defined S. aureus medium, and members of the Linder lab for helpful discussions.
This work was supported by the Swiss National Science Foundation (to P.L.), the Foundation Ernst et Lucie Schmidheiny, and the Canton of Geneva in Switzerland.
ADDENDUM IN PROOF
While this paper was in proof, a report by Monk and coworkers [mBio 3(2):e00277-11, 2012], describing direct transformation into S. aureus wild-type strains with DNA prepared from their newly developed E. coli strain (DC10B), was published. This should greatly facilitate genetic manipulations of S. aureus in the future, especially in combination with the work presented here.
Footnotes
Published ahead of print 23 March 2012
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1. Arnaud M, Chastanet A, Debarbouille M. 2004. New vector for efficient allelic replacement in naturally nontransformable, low-GC-content, gram-positive bacteria. Appl. Environ. Microbiol. 70:6887–6891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bae T, Schneewind O. 2006. Allelic replacement in Staphylococcus aureus with inducible counter-selection. Plasmid 55:58–63 [DOI] [PubMed] [Google Scholar]
- 3. Boeke JD, Lacroute F, Fink GR. 1984. A positive selection for mutants lacking orotidine-5[prime]-phosphate decarboxylase activity in yeast: 5-fluoro-orotic acid resistance. Mol. Gen. Genet. 197:345–346 [DOI] [PubMed] [Google Scholar]
- 4. Bonneaud N, et al. 1991. A family of low and high copy replicative, integrative and single-stranded S. cerevisiae/E. coli shuttle vectors. Yeast 7:609–615 [DOI] [PubMed] [Google Scholar]
- 5. Bruckner R. 1997. Gene replacement in Staphylococcus carnosus and Staphylococcus xylosus. FEMS Microbiol. Lett. 151:1–8 [DOI] [PubMed] [Google Scholar]
- 6. Carleton S, Projan SJ, Highlander SK, Moghazeh SM, Novick RP. 1984. Control of pT181 replication. II. Mutational analysis. EMBO J. 3:2407–2414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Charpentier E, et al. 2004. Novel cassette-based shuttle vector system for gram-positive bacteria. Appl. Environ. Microbiol. 70:6076–6085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Churchward G, Belin D, Nagamine Y. 1984. A pSC101-derived plasmid which shows no sequence homology to other commonly used cloning vectors. Gene 31:165–171 [DOI] [PubMed] [Google Scholar]
- 9. Corvaglia AR, et al. 2010. A type III-like restriction endonuclease functions as a major barrier to horizontal gene transfer in clinical Staphylococcus aureus strains. Proc. Natl. Acad. Sci. U. S. A. 107:11954–11958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Fujita M, Losick R. 2002. An investigation into the compartmentalization of the sporulation transcription factor sigmaE in Bacillus subtilis. Mol. Microbiol. 43:27–38 [DOI] [PubMed] [Google Scholar]
- 11. Gill SR, et al. 2005. Insights on evolution of virulence and resistance from the complete genome analysis of an early methicillin-resistant Staphylococcus aureus strain and a biofilm-producing methicillin-resistant Staphylococcus epidermidis strain. J. Bacteriol. 187:2426–2438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gryczan TJ, Dubnau D. 1982. Direct selection of recombinant plasmids in Bacillus subtilis. Gene 20:459–469 [DOI] [PubMed] [Google Scholar]
- 13. Khan SA, Adler GK, Novick RP. 1982. Functional origin of replication of pT181 plasmid DNA is contained within a 168-base-pair segment. Proc. Natl. Acad. Sci. U. S. A. 79:4580–4584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kreiswirth BN, et al. 1983. The toxic shock syndrome exotoxin structural gene is not detectably transmitted by a prophage. Nature 305:709–712 [DOI] [PubMed] [Google Scholar]
- 15. Leimeister-Wachter M, Domann E, Chakraborty T. 1992. The expression of virulence genes in Listeria monocytogenes is thermoregulated. J. Bacteriol. 174:947–952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Liu H, Han J, Liu X, Zhou J, Xiang H. 2011. Development of pyrF-based gene knockout systems for genome-wide manipulation of the archaea Haloferax mediterranei and Haloarcula hispanica. J. Genet. Genomics 38:261–269 [DOI] [PubMed] [Google Scholar]
- 17. Lowy FD. 1998. Staphylococcus aureus infections. N. Engl. J. Med. 339:520–532 [DOI] [PubMed] [Google Scholar]
- 18. Meng X, Smith RM, Giesecke AV, Joung JK, Wolfe SA. 2006. Counter-selectable marker for bacterial-based interaction trap systems. Biotechniques 40:179–184 [DOI] [PubMed] [Google Scholar]
- 19. Novick RP. 1990. The Staphylococcus as a molecular genetic system, p 1–40 In Novick RP. (ed), Molecular biology of the staphylococci. VCH Publishers, New York, NY [Google Scholar]
- 20. Sambrook J, Russell DR. 2001. Molecular cloning: a laboratory manual, 3rd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 21. Sullivan MA, Yasbin RE, Young FE. 1984. New shuttle vectors for Bacillus subtilis and Escherichia coli which allow rapid detection of inserted fragments. Gene 29:21–26 [DOI] [PubMed] [Google Scholar]
- 22. Traber K, Novick R. 2006. A slipped-mispairing mutation in AgrA of laboratory strains and clinical isolates results in delayed activation of agr and failure to translate delta- and alpha-haemolysins. Mol. Microbiol. 59:1519–1530 [DOI] [PubMed] [Google Scholar]
- 23. Tripathi SA, et al. 2010. Development of pyrF-based genetic system for targeted gene deletion in Clostridium thermocellum and creation of a pta mutant. Appl. Environ. Microbiol. 76:6591–6599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Tu Quoc PH, et al. 2007. Isolation and characterization of biofilm formation-defective mutants of Staphylococcus aureus. Infect. Immun. 75:1079–1088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Waldron DE, Lindsay JA. 2006. SauI: a novel lineage-specific type I restriction-modification system that blocks horizontal gene transfer into Staphylococcus aureus and between S. aureus isolates of different lineages. J. Bacteriol. 188:5578–5585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Xia GX, et al. 1991. A copy-number mutant of plasmid pSC101. Mol. Microbiol. 5:631–640 [DOI] [PubMed] [Google Scholar]
- 27. Xu SY, Corvaglia AR, Chan SH, Zheng Y, Linder P. 2011. A type IV modification-dependent restriction enzyme SauUSI from Staphylococcus aureus subsp. aureus USA300. Nucleic Acids Res. 39:5597–5610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yao J, et al. 2006. Use of targetrons to disrupt essential and nonessential genes in Staphylococcus aureus reveals temperature sensitivity of Ll.LtrB group II intron splicing. RNA 12:1271–1281 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.