Abstract
The physiology of ethanologenic Escherichia coli grown anaerobically in alkali-pretreated plant hydrolysates is complex and not well studied. To gain insight into how E. coli responds to such hydrolysates, we studied an E. coli K-12 ethanologen fermenting a hydrolysate prepared from corn stover pretreated by ammonia fiber expansion. Despite the high sugar content (∼6% glucose, 3% xylose) and relatively low toxicity of this hydrolysate, E. coli ceased growth long before glucose was depleted. Nevertheless, the cells remained metabolically active and continued conversion of glucose to ethanol until all glucose was consumed. Gene expression profiling revealed complex and changing patterns of metabolic physiology and cellular stress responses during an exponential growth phase, a transition phase, and the glycolytically active stationary phase. During the exponential and transition phases, high cell maintenance and stress response costs were mitigated, in part, by free amino acids available in the hydrolysate. However, after the majority of amino acids were depleted, the cells entered stationary phase, and ATP derived from glucose fermentation was consumed entirely by the demands of cell maintenance in the hydrolysate. Comparative gene expression profiling and metabolic modeling of the ethanologen suggested that the high energetic cost of mitigating osmotic, lignotoxin, and ethanol stress collectively limits growth, sugar utilization rates, and ethanol yields in alkali-pretreated lignocellulosic hydrolysates.
INTRODUCTION
Escherichia coli is among the best-understood microorganisms and a workhorse for biotechnology, yet its anaerobic physiology in and cellular responses to the complex biomass hydrolysates essential to exploitation of lignocellulosic materials as feedstocks for conversion to chemicals and biofuels remain poorly understood. Conversion of sugars in lignocellulosic hydrolysates to ethanol is a well-developed system with which these questions can be studied (20, 43). Production of lignocellulosic hydrolysates typically requires pretreatment with either an acid or a base, which release different lignin derivatives into hydrolysates (31, 49, 50). We chose to investigate E. coli physiology during ethanologenesis in hydrolysates derived by alkaline pretreatment, specifically, ammonia fiber expansion (AFEX), because it yields a feedstock containing both C5 and C6 sugars and generates fewer lignin-derived inhibitors of microbial growth (19, 81, 82). Furthermore, hydrolysates prepared from AFEX-pretreated biomass, such as AFEX-pretreated corn stover hydrolysate (ACSH), are replete in nutrients and permissive to growth to the extent that engineered strains of E. coli can consistently produce significant amounts of ethanol from glucose (49, 50, 60, 66).
E. coli is a particularly useful organism for design of strains to convert lignocellulosic hydrolysates, because it can use the most abundant hexoses and pentoses present in plant cell walls, and because its extensive study provides a wealth of assisting knowledge (44). The E. coli W-derived strain KO11, which contains a PET cassette, comprised of the Zymomonas mobilis pyruvate decarboxylase (pdc) and alcohol dehydrogenase (adhB) genes, was among the first genetically engineered microbes designed for ethanol production (39, 64). Subsequent improvement of KO11 by placing the PET cassette within an rRNA operon to increase Pdc and AdhB levels, additional pathway engineering, and directed evolution yielded robust E. coli ethanologens with improved ethanol yields, broadened sugar utilization, and increased ethanol tolerance (31, 52, 88–90). Nonetheless, achieving efficient fermentation of concentrated hydrolysates by any microbial ethanologen remains challenging for multiple reasons (59), including the high osmolarity of the medium (57, 67, 83), the presence of lignin derivatives resulting from pretreatment and enzymatic hydrolysis (collectively known as lignotoxins) (93; X. Tang et al., unpublished data), the energetic and regulatory challenges of pentose sugar fermentation (36), and the toxicity of the biofuels themselves (38, 46; D. H. Keating, M. Schwalbach, J. Peters, M. Tremaine, E. Pohlmann, F. Tran, J. Vinokur, A. Higbee, P. Kiley, and R. Landick, submitted for publication). Although many of these inhibitory compounds have been examined individually or in combination in defined media (33, 50, 58, 65, 92), the molecular mechanisms by which lignotoxins act in combination with other stresses induced in fermentations of alkali-pretreated hydrolysates, including osmotic and ethanol stress, have not been examined. To understand the molecular responses of ethanologenic E. coli to alkali-pretreated lignocellulosic hydrolysates, we studied fermentation of ACSH by an E. coli K-12 strain engineered for efficient ethanol production. We compared changes in the composition of the growth medium to changes in the patterns of gene expression during growth in ACSH and during an unusual growth-arrested state during which most ethanol production occurred. To understand the effect of high osmolarity and other stresses associated with ACSH and to relate our findings to earlier studies of E. coli K-12 (GenExpDB [http://genexpdb.ou.edu]), we compared gene expression in ACSH to expression in a synthetic hydrolysate (SynH) and in glucose minimal medium (GMM). These analyses provided insights into the combined demands on cellular energetics caused by stresses in an E. coli ethanologen growing in ACSH.
MATERIALS AND METHODS
Strain construction.
An ethanologenic E. coli K-12 with the same ethanol pathway as the E. coli W ethanologen strain KO11 (64) was constructed by PCR amplification of the Z. mobilis PET cassette genes and flanking pflB sequence from KO11 and subsequent insertion into the pflB locus of MG1655 using lambda Red-mediated recombination (91). This yielded an insertion of pdc, adhB, and cat between nucleotides 396 and 400 of pflB (3 nucleotides [nt] of pflB were eliminated; pdc and adhB were in the same orientation as pflB and thus transcribed from pflB promoters). To enhance ethanol production, the lactate dehydrogenase (ldhA) and acetate kinase (ackA) genes responsible for the production of alternate fermentation end products were deleted as previously described (6, 91), and an additional copy of the PET cassette was constitutively expressed from the low-copy-number plasmid pJGG2 (29). The resulting strain was identified as GLBRCE1. Resequencing of GLBRCE1 by the Joint Genome Institute confirmed the ldhA and ack deletions and the PET insertion in pflB. In addition, variant alleles of crl, ylbE, glpR, and gatC recently reported to be in many “wild-type” E coli strains (28) and single-nucleotide polymorphisms in yodD and gltB were identified. Thus, the genotype of GLBRCE1 relative to GenBank accession number U00096.2 is ldhA(4_969del 3insFRT) ackA(4_1182del 3insFRT) pflB[395ins(pdcZmo adhBZmo cat)] crl(70insIS1) ylbE(253insG) gltB(G3384A) yodD(A85T) glpR(150delG) gatC(916insCC) pJGG2.
The nomenclature used here for the variant alleles is a modification of that described for human sequence variations (23). The first number in the parenthesis specifies the nt position in the orf; ins and del specify insertions or deletions, respectively; the last element specifies the identity of the nt or genetic element (e.g., IS1) inserted or deleted. Substitutions are indicated by, e.g., A85T to indicate that A at position 85 is changed to T. Although the changes to crl, ylbE, glpR, and gatC produced frameshifts and the changes in gltB and yodD produced substitutions (Gly1062Ser and Ile29Phe, respectively) and their impact was not specifically tested, we would not expect that these variant alleles significantly affect the properties of GLBRCE1 reported here.
Production of ACSH.
AFEX pretreatment was adapted from a previously described method (7). Corn stover (Pioneer 36H56) was oven dried at 60°C for ∼2 weeks, after which the material was passed through a Christy hammer mill (Christison Scientific LTD, Gateshead England) equipped with a 5-mm screen. The moisture content of the milled corn stover was raised to 60% by spraying with distilled water, and the material was loaded into a 5-gallon, high-pressure reactor (Parr Instrument Company, Moline, IL). The reactor was charged with nitrogen gas (65 lb/in2). Liquid ammonia was then added (1:1 ammonia-biomass ratio on a dry-weight basis). The reactor was heated to 100°C for 30 min, after which the pressure was rapidly released to allow the majority of the ammonia gas to escape. The resulting AFEX-treated biomass was transferred to plastic trays and allowed to stand overnight, allowing residual ammonia to evaporate. The entire pretreatment process was performed in a walk-in fume hood.
For hydrolysis, AFEX-pretreated corn stover (∼60 g glucan/liter; ∼18% solids, adjusted for moisture content) was suspended in deionized water to a final volume of 10 liters. The mixture was autoclaved for 30 min, cooled to 50°C, and adjusted to 50 mM phosphate using 1 M K-PO4, pH 4.3. Hydrolysis was initiated by adding Genencor Accellerase 1500 to 15 g/liter (cellulase, ∼2,500 U/g; β-galactosidase, ∼650 U/g), Genencor Accellerase XY to 7.5 mg/ml (xylanase, ∼25,000 U/g), and Multifect pectinase (∼155 U/g) to 7.5 g/liter. After 30 to 60 min, the mixture liquefied and was then adjusted to pH 4.9 using 10 N HCl. Hydrolysis proceeded for 7 days at 50°C, after which the remaining solids were removed by centrifugation (8,200 × g, 4°C, 10 to 12 h). The supernatant was filter sterilized in series through 0.5-μm and 0.22-μm filters prior to storage at 4°C. The resulting hydrolysate was used within 2 months. Prior to fermentation, the hydrolysate was adjusted to pH 7.0 using NaOH pellets and again filtered through a 0.22-μm filter to remove precipitates and to ensure sterility.
ACSH and medium composition analysis.
Samples for analysis of culture supernatants were obtained by centrifugation (10,000 × g, 3 min, 4°C) and filter sterilization through a 0.22-μm syringe filter. The elemental composition of ACSH or culture supernatant was determined using an inductively coupled plasma mass spectrometer (PlasmaQuad PQ2 Turbo Plus) at the UW-Madison Soils and Plant Analysis Lab (http://uwlab.soils.wisc.edu/madison). Carbohydrates in hydrolysate were quantified using high performance liquid chromatography and refractive index detection (HPLC-RID), nuclear magnetic resonance (NMR) (see below), and gas chromatography-mass spectrometry (GC-MS) as previously described (2). ACSH osmolality was measured using a Wescor Vapro 5520 osmometer.
Amino acid concentrations were assayed at the Molecular Structure Facility laboratory located at University of California, Davis (http://msf.ucdavis.edu/). Two-milliliter supernatant samples were acidified with sulfosalicylic acid, stored overnight at −20°C, and then centrifuged (16,000 × g, 5 min, room temperature) to remove intact proteins. The supernatant was then mixed with a lithium citrate buffer diluent and spiked with S-(2-aminoethyl)-l-cysteine as an internal standard, and amino acids were assayed on a Hitachi L-8900 amino acid analyzer using commercial standards (Sigma) and the EZ-Chrom software package (Hitachi). Methionine, tryptophan, cysteine, and isoleucine were not reliably detected in ACSH by this method.
Samples were prepared for NMR by addition of D2O (100 μl) containing 6.6 mM 4,4-dimethyl-4-silapentane-1-sulfonic acid (DSS; chemical shift standard) and 500 μM NaN3 (microbial growth inhibitor) to 570 μl of ACSH. The resulting solution was titrated with concentrated DCl or NaOD to pH 7.4. NMR analysis was conducted at the National Magnetic Resonance Facility at Madison on a Bruker Avance III spectrometer operating at 600 MHz for 1H and equipped with a triple-resonance (1H, 13C, 15N, 2H lock) 1.7-mm cryogenic probe and an automated sample changer (SampleJet). The probe temperature was regulated at 298 K. The probe was tuned, matched, and locked to deuterium for the first sample. Sample shimming and the optimal 90° pulse width calibration were determined in an automated fashion; automated shimming was not always sufficient, and manual shimming was carried out when needed. For each sample, data were collected as 1D 1H and 2D 1H-13C HSQC (heteronuclear single-quantum coherence) spectra, processed with NMRPipe (22), and analyzed using rNMR (51). For metabolites quantified by 1D 1H, peak amplitudes were calibrated to DSS. For metabolites quantified by 2D 1H-13C HSQC, concentrations were calculated using calibration curves from metabolite standards.
Synthetic hydrolysate and minimal glucose medium.
A synthetic hydrolysate (SynH) was developed that approximated the composition of ACSH (see Table S1 in the supplemental material). d-Glucose (60 g/liter; 330 mM) and d-xylose (30 g/liter; 200 mM) were added to a minimal salts solution containing 64 mM KPO4 (pH 7.1), 30 mM NH4SO4, micronutrients, and 100 μM (each) adenine, guanine, cytosine, and uracil (see Table S1). Amino acids were added to approximately the concentrations detected in ACSH (see Table S1) plus 100 μM methionine, 250 μM isoleucine, and 50 μM (each) tryptophan and cysteine (62). The resulting SynH was filter sterilized through a 0.22-μm membrane prior to use. Minimal M9 medium containing 10 g glucose/liter (GMM; 56 mM glucose) was prepared as described previously (62).
Fermentation conditions.
Fermentations were conducted in 2-liter glass vessels (Applikon Biotechnology) containing 1.2 liters of ACSH, GMM, or SynH. ACSH fermentations were conducted in duplicate; GMM and SynH fermentations were in triplicate. Prior to inoculation of anaerobic fermentations, the growth medium was sparged with 95% N2–5% CO2 overnight, and gentamicin (Gent) was then added to 15 μg/ml (to ensure retention of pJGG2). Inocula for ACSH fermentations were prepared from single colonies grown on LB-Gent plates from frozen stocks by initial aerobic growth in LB-Gent to mid-log phase (optical density at 600 nm [OD600], ∼0.3). Cells were diluted 10-fold into ACSH, grown overnight aerobically, transferred to fresh ACSH, grown anaerobically, and then diluted in anaerobic ACSH fermentation vessels to a starting OD600 of 0.02 to 0.06. Inocula for fermentations in SynH or GMM were prepared similarly, except that SynH-Gent or GMM-Gent was used to dilute the initial LB-Gent culture and for subsequent steps. Fermentations were conducted at 37°C with constant stirring (300 rpm) and sparging (150 ml/min; 95% N2, 5% CO2); neutral pH (7.0) was maintained by automated addition of 10 M NaOH. The apparent OD600 of cells diluted 1:10 in water was determined using a Beckman Coulter DU720 spectrophotometer (1.3-mm slit width) in a 1-ml cuvette. Aerobic cultivation of MG1655 in GMM was carried out in sparged Roux bottles (500 ml/min; 69% N2, 30% O2, 1% CO2) inoculated with cultures grown aerobically overnight in GMM.
In amino acid supplementation experiments, amino acids were added at final concentrations approximately three or eight times the amino acid concentrations in ACSH. Supplementation experiments in ACSH were conducted as single fermentations in Sartorius 500-ml fermentation vessels containing 300 ml of growth medium kept anaerobic as described above. Amino acid supplementation assays in SynH medium were conducted in duplicate in covered 2-ml, 96-well plates (Nunc U96) incubated for 48 h in an anaerobic chamber (sparged with 10% CO2, 10% H2, 80% N2) prior to measurement of apparent OD600 with a microtiter plate reader (Tecan Infinite M200).
End product analysis.
Samples (2 ml) for end product analysis were collected throughout the fermentation. Cells were removed by centrifugation (10,000 × g, 4°C, 3 min), and the supernatant was stored at −80°C prior to analysis. For each sample, glucose, xylose, glycerol, xylitol, succinic acid, lactic acid, formic acid, acetic acid, and ethanol were separated by high-performance liquid chromatography (HPLC) and subsequently quantified using refractive index detection (RID). HPLC-RID was conducted with an Agilent 1260 Infinity system (Agilent Technologies, Palo Alto, CA) equipped with an Aminex HPX-87H anion-exchange column, 300 by 7.8 mm (Bio-Rad, Hercules, CA). Samples were diluted with 9 volumes of H2O, injected into the HPLC-RID system (50-μl injection volume), and eluted isocratically with 0.02 N H2SO4 at a flow rate of 0.5 ml/min (RID flow cell, 45°C; column, 50°C). Reference compounds (Fisher Scientific) were diluted in H2O and used to generate a standard curve. Analyte concentrations were calculated using Chem Station software version B.04.03 (Agilent Technologies).
Intracellular ATP concentration.
Cell samples (2 to 10 ml) were rapidly trapped on a 0.44-μm nylon syringe filter (Millipore) using a 15-ml syringe as described previously (4) and then extracted by pushing 2 ml acetonitrile-methanol-water (40:40:20), 0.1% formic acid through the filter into a 15-ml conical tube. The eluent was then pushed through the cells a second time, flash-frozen in a dry ice-ethanol bath, and stored at −80°C.
The intracellular extract was fractionated by anion-exchange chromatography (first mobile phase, 0.5 mM NaOH; second mobile phase, 50 mM NaOH; flow rate, 0.35 ml/min) using a Dionex IonPac AS-11HC column (2 by 250 mm) and ASRS 300 ion suppressor coupled to an Agilent 6460 QQQ mass spectrometer operated in MRM (multiple reaction monitoring) mode with negative-mode electrospray ionization (gas flow, 11 liter/min, 300°C; nebulizer pressure, 45 lb/in2; sheath gas, 11 liters/min, 360°C; capillary at 3,500 V; nozzle at 1,000 V), as described previously (84). The resulting peak area was integrated, and concentrations of ATP in the extracts were determined by comparison to a standard curve. These values were corrected using similarly extracted uninoculated medium and then converted to intracellular molarity assuming 109 cells/ml OD600 and a cell volume of 6.7 × 10−13 ml.
Transcriptomic analysis.
Cells (10 ml) for transcriptomic analysis were collected into tubes containing 1.25 ml ice-cold 5% (vol/vol) unbuffered phenol in ethanol (EP) and pelleted by centrifugation (10,000 × g, 4°C, 3 min), as described previously (70). To remove residual traces of hydrolysate, cell pellets were twice resuspended in ice-cold GMM plus 0.125 volume of EP, repelleted, then flash frozen in dry ice-ethanol, and stored at −80°C. RNA was extracted from cell pellets as described previously (70), analyzed by agarose gel electrophoresis to confirm integrity, quantified using a Nanodrop spectrophotometer (Thermo Scientific), and stored at −80°C.
Transcript levels were measured using custom gene expression microarrays (Roche NimbleGen) that contained two copies of eight 70-nt single-stranded oligonucleotide probes for each E. coli open reading frame and for Z. mobilis pdc and adhB. Random hexamers and SuperScript double-stranded cDNA synthesis reagents (Invitrogen) were used to generate cDNA. Subsequent cDNA labeling and hybridization protocols were conducted according to the manufacturer's instructions (Roche NimbleGen). Hybridized arrays were scanned with a NimbleGen MS 200 microarray scanner, and the probe signal intensities were determined using NimbleScan v 2.6 (Roche NimbleGen). Probe signal intensities were preprocessed using robust multichip averaging (RMA) (15, 40) in the program ArrayStar (DNASTAR), and the resulting gene expression signals were quantile-normalized across all samples using the normalize.quantiles function in the Bioconductor package for R. Bioreplicates in the normalized data set were screened for a high degree of similarity (Pearson coefficient > 0.95) and dissimilar arrays were omitted from subsequent analyses. Significant expression changes were assigned to genes exhibiting a 2-fold or greater change in signal intensity and a Benjamini-Hochberg-adjusted Student's t test P value of <0.05 in a given pairwise comparison. For processed array data, see Table S6 in the supplemental material.
Metabolic modeling.
The iJR904 E. coli metabolic model (67) was used to estimate the minimal production of ATP consistent with the observed rates of growth, uptake, and secretion. Cells were assumed to be at steady-state with no net accumulation of intracellular metabolites and to have a mass of 2.14 g dry weight/liter per OD600 (10). To calculate the ATP production rates, reversible iJR904 reactions were split into forward and reverse reactions, and the rates of growth, carbohydrate uptake, and end product production were fixed to the observed values. The amount of ATP that is produced by the metabolic pathways was then determined using flux balance analysis (68), where the total production of ATP was minimized using the reaction stoichiometry for ATP. Flux balance analysis predicts fluxes through metabolic reactions, where the total production rate minus the consumption rate for each metabolite is set to zero. In flux balance analysis, the values for individual reactions are also constrained using physiological rate measurements (e.g., uptake or growth rates).
Microarray database accession number.
Array information and raw data are available from the Gene Expression Omnibus (accession no. GSE35049).
RESULTS
To study the behavior of an E. coli ethanologen in ACSH, we constructed strain GLBRCE1. GLBRCE1 was derived from the well-studied E. coli K-12 strain MG1655 (12) and was similar in design to the E. coli W ethanologen KO11 (64). Unlike strain KO11, GLBRCE1 contained multiple sources of the Z. mobilis PET cassette: a single chromosomal copy in which expression of Z. mobilis pdc and adhB were driven by the pflB promoters (similar to KO11), and additional copies transcribed from a lac promoter in the low-copy-number plasmid pJGG2 (29). GLBRCE1 also contained deletions of the genes encoding lactate dehydrogenase (ldhA) and acetate kinase (ackA) (Fig. 1A; also, see Materials and Methods). GLBRCE1 was grown in a hydrolysate derived from ammonia-pretreated corn stover (ACSH; see Materials and Methods) that contained ∼60 g glucose/liter (∼330 mM), ∼30 g xylose/liter (∼200 mM), lesser amounts (<35 mM) of arabinose, galactose, mannose, rhamnose, and fucose, and amino acids (from 50 to 700 μM) and other nutrients and minerals (see Tables S1, S2, S4, and S5 in the supplemental material). To understand the metabolic changes that occur when E. coli K-12 uses ACSH as a carbon and energy source anaerobically (and thus inform the design of strains to ferment ACSH efficiently), we correlated changes in global gene expression with the rates of growth, sugar uptake, and production of end products.
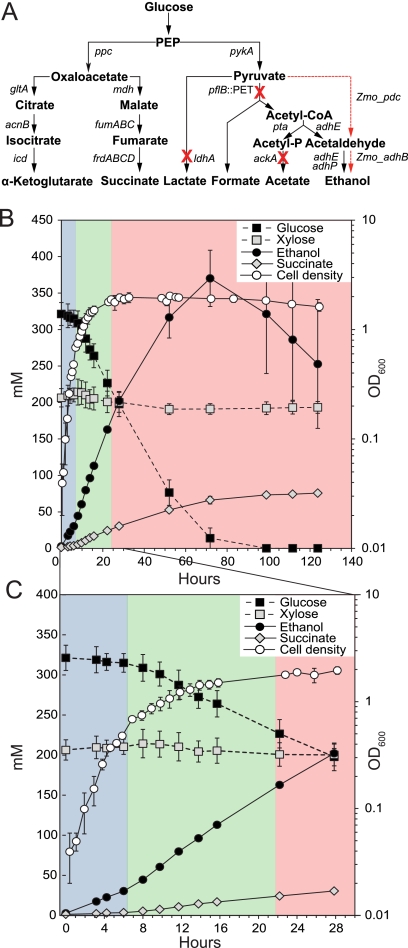
Fig 1.
Genotype and growth of GLBRCE1 in ACSH. (A) Engineered genetic alterations in GLBRCE1 and pathways affected by those changes. A red X indicates a deleted gene. The dashed line indicates the reactions catalyzed by the products of the Z. mobilis pdc and adhB genes. (B) Sugar utilization and metabolic end product accumulation during growth of GLBRCE1 in AFEX-pretreated corn stover hydrolysate (ACSH). GLBRCE1 was cultured in a bioreactor under anaerobic conditions at 37°C, and samples were analyzed for cell density, sugars (glucose and xylose), and metabolic end products (ethanol and succinate) at the time points indicated. (C) Expanded view of sugar utilization and end product accumulation during the first 30 h of growth in ACSH. Blue, green, and red shading shows exponential, transition, and stationary phases, respectively. Error bars represent the ranges of the means of two or three biological replicates.
GLBRCE1 grew robustly under anaerobic conditions in ACSH (μ = 0.37 h−1), 2.5-fold faster than in minimal salt 1% glucose medium (GMM; μ = 0.17 h−1) (Fig. 1B and C; Table 1). Despite the high concentration of available glucose in the hydrolysate, only ∼10% of the glucose was consumed during a relatively short exponential phase (<6 h). An additional ∼15% of glucose was consumed during a protracted transition phase (from 6 to 22 h) that followed and during which growth slowed gradually (Fig. 1B). Complete growth arrest occurred upon entry into stationary phase (22 h), but the cells remained metabolically active and continued assimilating the remaining glucose, albeit at lower rates per cell than observed during the exponential and transition phases (Fig. 1B; Table 1). Disruption of the genes for acetate kinase, lactate dehydrogenase, and pyruvate formate lyase were not responsible for growth arrest, because growth of the parental MG1655 strain containing pJGG2 underwent a similar metabolically active growth arrest in ACSH (data not shown). Furthermore, derivatives of GLBRCE1 containing just the vector or lacking any plasmid also stopped growing in ACSH prior to glucose depletion (data not shown). Growth arrest and the metabolically active stationary phase were of particular interest because they direct carbon to ethanol rather than to cell growth. To investigate these states and the multiphasic behavior of E. coli in ACSH generally, we used gene expression profiling coupled with substrate and end product measurements to examine how the physiology of E. coli changed in the different growth phases.
Table 1.
Estimated rates of growth, uptake, and production by GLBRCE1
| Medium and growth phasea | Growth rate (h−1)b | Rate (mM · unit of OD600−1 · h−1)b |
Aggregate productivity (%)c | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Uptake (observed) |
Production (observed) |
Maximum production of ATPd | Cellular ATP requiremente | ||||||
| Glucose | Xylose | Formate | Ethanol | Succinate | |||||
| ACSH | |||||||||
| Exp | 0.37 ± 0.07 | 13.6 ± 8.2 | 1.8 ± 1.3 | 25.9 ± 8.7 | 7.1 ± 3.7 | 37 ± 22 | 14.4 | 150 ± 80 | |
| Trans | 0.11 ± 0.02f | 3.7 ± 0.9 | 0.6 ± 0.3 | 5.7 ± 1.2 | 1.9 ± 0.6 | 10 ± 2.5 | 130 ± 30 | ||
| Stat | 1.9 ± 0.5 | 0.0 | 1.6 ± 1.0 | 0.9 ± 0.2 | 5.2 ± 0.9 | 80 ± 30 | |||
| SynH | |||||||||
| Exp | 0.29 ± 0.02 | 13.0 ± 3.9 | 17.3 ± 5 | 0.8 ± 0.6 | 36 ± 11 | 12.2 | 70 ± 30 | ||
| Stat | 2.6g | 0.31g | 3.6g | 0.28g | 7.6g | 90 | |||
| SynH-3× AA | |||||||||
| Exp | 0.45 ± 0.03 | 13.8 ± 5.0 | 19.6 ± 6.8 | 1.9 ± 0.6 | 38 ± 14 | 90 ± 40 | |||
| Stat | 3.1g | 0.41g | 4.1g | 0.46g | 9.7g | 80 | |||
| GMM | |||||||||
| Exp | 0.17 ± 0.002 | 8.1 ± 0.8 | 11.9 ± 2.1 | 1.1 ± 0.4 | 22.3 ± 2.2 | 8.4 | 90 ± 20 | ||
Exp, exponential phase; Trans, transition phase; Stat, stationary phase; AA, amino acids.
95% confidence intervals were calculated by sensitivity analysis and the F distribution.
Calculated from the rates of ethanol and succinate production divided by the rate of glucose consumption, assuming 2 ethanol per glucose and 1 succinate per glucose. 95% confidence intervals were propagated from rates of ethanol and succinate production.
Maximum theoretical rate of ATP production possible based on observed sugar uptake rates (2.75 ATP/glucose and 2.04 ATP/xylose).
Combination of growth-dependent ATP requirement and growth-independent maintenance (assumed constant at 3.6 mM · unit of OD−1 · h−1 [86]) calculated with the iJR904 E. coli metabolic model (68); see Materials and Methods.
Growth rates for transition phase are in units of OD · h−1.
Rate estimated from only two time points during which glucose consumption occurred.
The global patterns of gene expression of GLBRCE1 in ACSH were dynamic but highly reproducible during the initial 52 h of the experiment (r2 > 0.95), as illustrated by expression heat maps of the individual replicates (Fig. 2). After ∼70 h, the replicate patterns diverged, with a dramatic change in expression pattern in replicate 1 being evident at 70 h. This change corresponded to the time of glucose depletion, which occurred earlier in replicate 1 than replicate 2 (∼70 h versus ∼100 h). Thus, we restricted our analysis of gene expression changes to times before 70 h and used the growth rates and expression heat maps to group samples into exponential, transition, and stationary phases (Fig. 2). We describe key aspects of gene expression in each growth phase below.
Fig 2.
Global gene expression patterns of GLBRCE1. The heatmap of global gene expression changes in GLBRCE1 during growth in ACSH, SynH, and GMM reveals patterns used to group samples for analysis (38 columns; 4,240 genes per sample). For ACSH bioreplicates 1 and 2, the numbers at the top of heatmap are sample times, in h. Blue, green, and red shading indicate exponential-, transition-, and stationary-phase samples in each replicate. For SynH- and GMM-grown cells, samples were collected from exponential (Exp SynH, 8 h; Exp GMM, 4 h), transition (Tran SynH, 10 h), and stationary (Sta SynH, 14 h; Sta GMM, 16 h) phases as shown in Fig. 5. RNA expression values are shown as fold change relative to expression at the 3-h time point of ACSH bioreplicate 1.
Distribution of carbon and energy sources in hydrolysates.
Despite the unusual growth behavior of the strain, glucose was fermented predominantly to ethanol (Fig. 1B), as expected from the strain design. The rate of glucose uptake relative to cell density was maximal during exponential phase (13.6 mmol · unit of OD600−1 · h−1) (Table 1) and then decreased by a factor of 3.7 in transition phase and by a further factor of 2 in stationary phase (Table 1). The expression of the genes encoding enzymes in the glycolytic pathway and glucose transporters remained relatively high through the transition and stationary phases (see Fig. S3A in the supplemental material). The rate of ethanol production was generally correlated with glucose consumption, showing a high rate of production during exponential phase and a reduced rate during the transition and stationary phases even though transcript profiling indicated that expression of the PET cassette genes pdc and adhB remained high throughout (see Fig. S3A). Overall net ethanol yield was 58 to 67% of the theoretical maximum, but this measure underestimated actual ethanol production, because some ethanol evaporated during fermentation. Ethanol evaporation at ∼2 mM h−1 was evident after glucose depletion (Fig. 1A); this estimate was verified by detection of similar ethanol evaporation from uninoculated, identically sparged bioreactors containing 5% ethanol (data not shown).
Although glucose was ultimately consumed in its entirety, only a small amount of xylose was consumed over the course of the experiment (≤10%) (Fig. 1B; also, see Tables S4 and S5 in the supplemental material). This result is expected with the GLBRCE1 design, which was not optimized for xylose utilization, and was consistent with the observed low expression of xylose utilization genes (xylE, xylFGH, and xylAB) (Table 2; also, see Fig S3B in the supplemental material). Low expression of xylose genes likely reflected repression by arabinose-bound AraC (24), since ACSH contains 33 mM arabinose (see Table S1 in the supplemental material). Further, inducer exclusion by PtsG activated by glucose transport may also limit xylose uptake. Lesser amounts of mannose, fructose, galactose, and arabinose were also present in hydrolysate and represented minor pathways of carbon utilization (see Tables S1 and S5 in the supplemental material). Other potential substrates, such as acetate and glycerol, were not utilized as carbon sources by the ethanologen (see Tables S4 and S5 in the supplemental material).
Table 2.
Genes of interest exhibiting changes in expression in different phases in ACSH
| Function and gene(s) | Fold changea in growth phase |
||
|---|---|---|---|
| Exponential | Transition | Stationary | |
| Acetate stressb | |||
| fliY | −4.2 (1.2e−2) | 1.1 (8.4e−1) | 1.2 (4.9e−1) |
| gatY | −3.0 (6.8e−3) | 1.0 (8.9e−1) | 1.5 (1.2e−1) |
| luxS | 2.1 (1.8e−2) | 1.1 (3.6e−1) | −1.5 (1.9e−2) |
| osmY | 2.1 (5.5e−2) | 1.6 (9.7e−2) | −6.3 (1.8e−4) |
| pfkB | 3.1 (4.5e−3) | 2.6 (3.5e−6) | −1.8 (7.7e−5) |
| slp | 2.9 (8.3e−2) | 1.3 (6.4e−1) | −4.3 (1.1e−2) |
| gadXW | 2.8 (2.6e−3) | 2.3 (3.3e−3) | 1.0 (3.6e−1) |
| hdeAB | 1.4 (7.0e−1) | 2.3 (2.1e−1) | −19.2 (3.3e−4) |
| metA | 2.5 (1.8e−3) | 4.0 (6.5e−2) | −4.8 (4.4e−2) |
| Anaerobic respirationb | |||
| fdhF | 3.0 (1.8e−2) | −1.8 (8.3e−3) | −1.4 (3.5e−2) |
| hycABCDEFGI | 2.5 (1.5e−1) | −2.5 (4.4e−3) | 1.0 (4.0e−1) |
| fdnGHI | 2.4 (3.0e−1) | −3.5 (5.8e−3) | −1.2 (3.8e−1) |
| napFDAGHBC | 2.2 (1.8e−1) | −2.6 (1.2e−3) | −1.3 (3.7e−2) |
| torCAD | 9.6 (4.1e−4) | 1.5 (7.7e−2) | −1.1 (7.2e−1) |
| Amino acid metabolismc | |||
| alaA | 2.5 (4.7e−4) | 1.3 (5.9e−2) | −1.3 (6.0e−2) |
| argA | −19.8 (1.2e−5) | −1.5 (8.8e−2) | 8.6 (8.0e−4) |
| argC | −12.6 (2.0e−4) | −2.3 (4.3e−2) | 14.8 (1.8e−3) |
| asnAB | −2.6 (9.0e−2) | 5.3 (1.0e−2) | −4.6 (2.2e−2) |
| aspC | −2.7 (2.5e−4) | 2.1 (1.6e−5) | 1.2 (2.8e−1) |
| avtA | 2.0 (5.9e−5) | −1.2 (4.1e−1) | −1.6 (3.5e−2) |
| cysCND | 1.0 (4.5e−1) | −2.3 (2.2e−1) | 1.0 (6.2e−1) |
| cysZK | 1.0 (1.3e−1) | −1.7 (1.7e−1) | 1.0 (4.0e−1) |
| cysPUWAM | −1.0 (3.1e−1) | −2.2 (2.5e−1) | 2.0 (2.5e−1) |
| cysHIJ | 1.7 (5.6e−2) | −3.0 (1.6e−1) | 1.9 (4.6e−1) |
| cysG | 3.1 (2.0e−3) | −3.2 (1.4e−5) | −1.2 (9.7e−2) |
| gdhA | −25.7 (2.8e−8) | 4.1 (3.6e−5) | 1.5 (3.6e−1) |
| glnA | −1.9 (2.2e−1) | 1.6 (4.9e−1) | 2.0 (2.8e−1) |
| gltB | −3.9 (2.4e−3) | 2.1 (1.0e−3) | 1.4 (1.4e−1) |
| glnK, amtB | −3.9 (1.8e−3) | 4.6 (1.6e−1) | 1.0 (6.7e−1) |
| ilvE | −1.7 (1.9e−3) | 2.9 (3.5e−4) | −1.2 (4.7e−1) |
| metQIN | −4.3 (3.6e−4) | 1.3 (6.5e−1) | −4.5 (1.4e−2) |
| metFLB | −13.5 (3.4e−6) | 3.1 (1.4e−1) | −4.1 (4.8e−2) |
| metC | −9.8 (6.9e−7) | 1.7 (3.2e−1) | −3.7 (2.4e−2) |
| metRE | −29.9 (8.3e−6) | 5.3 (6.1e−2) | −5.6 (1.4e−2) |
| proAB | −1.6 (2.8e−2) | 1.7 (4.0e−3) | 1.1 (5.7e−1) |
| serAC | −5.7 (8.5e−5) | 3.1 (1.0e−5) | −1.5 (2.3e−2) |
| thrLABC | 1.0 (2.6e−1) | 1.7 (1.4e−1) | 1.0 (1.4e−1) |
| trpEDCBA | −4.6 (1.2e−3) | 3.5 (7.7e−2) | −3.4 (9.8e−2) |
| ansB | 2.4 (3.2e−3) | −3.4 (1.1e−3) | −2.4 (1.1e−2) |
| aspA | 5.9 (3.7e−4) | −4.7 (1.1e−4) | −2.2 (1.5e−2) |
| Osmotic stressc | |||
| proVWX | 1.3 (2.6e−1) | 1.0 (4.1e−2) | 1.7 (2.6e−1) |
| proP | −1.5 (8.8e−2) | 3.0 (3.8e−3) | 2.7 (5.7e−3) |
| otsAB | 2.1 (1.1e−2) | 2.2 (3.6e−4) | −2.9 (3.8e−4) |
| betAB | 2.0 (3.9e−2) | 1.6 (6.5e−2) | 1.4 (2.0e−1) |
| Ethanol stressd | |||
| pspABCDE | 1.2 (4.9e−1) | 2.4 (2.0e−1) | 16.9 (4.0e−3) |
| ibpAB | 1.6 (4.2e−2) | 2.7 (1.2e−1) | 5.8 (8.7e−2) |
| srlABDE gutM | 1.3 (3.5e−1) | 12.9 (2.5e−3) | 11.2 (2.6e−4) |
| Miscellaneousc | |||
| araB | 50.7 (9.2e−6) | −3.8 (1.2e−3) | −4.2 (8.2e−4) |
| xylE | −1.0 (8.7e−1) | 1.0 (8.0e−1) | 1.2 (3.7e−2) |
| xylAB | 2.9 (1.4e−4) | 1.2 (2.2e−1) | −1.2 (3.5e−1) |
| xylFGH | 1.0 (3.4e−1) | −1.1 (3.2e−1) | 1.2 (7.9e−2) |
Changes in expression were calculated as explained in the following footnotes. P values are given in parentheses; e represents exponents of 10 (e.g., 1.1e−4 = 1.1 × 10−4).
Change for exponential phase was calculated as expression in ACSH exponential phase divided by expression in SynH exponential phase. Change for transition phase was calculated as expression in ACSH transition divided by expression in ACSH exponential phase. Change for stationary phase was calculated as expression in ACSH stationary divided by expression in ACSH transition phase.
Change for exponential phase was calculated as expression in ACSH exponential phase divided by expression in anaerobic GMM exponential phase. Change for transition phase was calculated as expression in ACSH transition divided by expression in ACSH exponential phase. Change for stationary phase was calculated as expression in ACSH stationary divided by expression in ACSH transition phase.
Change for all phases was calculated as expression in ACSH divided by expression in aerobic GMM exponential phase.
Small amounts of the anaerobic respiratory substrate formate (11 mM) and the electron acceptor nitrate (∼270 μM) were also present in ACSH (see Tables S1 and S2 in the supplemental material) but were rapidly consumed in exponential phase; these changes suggested that some anaerobic respiration occurred during initial cell growth. In accordance with the rapid loss of nitrate from the medium and the known nitrate-dependent expression of napFDAGHBC and fdnGHI (encoding the Nap periplasmic nitrate reductase and formate dehydrogenase-N, respectively), expression of these operons decreased 2- to 4-fold as cells entered transition phase (Table 2). It is unlikely that this limited anaerobic respiration contributed significantly to total ATP synthesis during the exponential phase, because glucose uptake rates were ∼7-fold greater than formate uptake rates (Table 1), and there was insufficient nitrate present to respire all of the formate. Rather, we hypothesize that most formate was likely oxidized to CO2 and H2 via formate hydrogenlyase, consistent with maximal expression of the genes (fdhF and hycABCDEFGI) encoding these enzymes early in exponential phase (Table 2; also, see Table S6 in the supplemental material). Expression of genes encoding trimethylamine N-oxide reductase [torCAD; specifically inducible by trimethylamine N-oxide (8)] was highly upregulated compared to GMM-grown cells throughout all growth phases (Table 2; also, see Table S6 in the supplemental material), suggesting that the electron acceptor trimethylamine N-oxide is also present in ACSH.
In addition to ethanol, the other significant and expected end product of ACSH fermentation by GLBRCE1 was succinate (Fig. 1), which accumulated to 62 to 76 mM (see Tables S4 and S5 in the supplemental material). During anaerobic growth, succinate is produced from oxaloacetate (OAA) by reversal of carbon flux through the tricarboxylic acid (TCA) cycle (Fig. 1A) (37, 64). However, not all the succinate appeared to derive from glucose, because the aggregate rates of ethanol and succinate production during exponential and stationary phases exceeded what could be theoretically produced based only on the glucose uptake rate (Table 1). We investigated this question by testing specific gene deletions and found that, even though deletion of frdA eliminated succinate production, deletion of ppc, which encodes the phosphoenolpyruvate (PEP) carboxylase responsible for converting PEP to oxaloacetate (Fig. 1A), eliminated only 50% of succinate production (data not shown). This result confirmed the idea that cells must utilize another succinate precursor in ACSH. Although a possible source would be aspartate or asparagine, which can be deaminated to oxaloacetate or fumarate, neither was present at a high enough concentration to contribute significantly to succinate production (Fig. 3C; also, see Table S1 in the supplemental material). An alternative source of succinate could be malate, which was present at 9 mM in ACSH and consumed (see Table S5 in the supplemental material). Additionally, citrate was a likely succinate precursor, based on the exceptionally high expression of the citrate-inducible citCDEFXG operon, which encodes citrate lyase (Fig. 4A; Table 3). Although difficult to quantify, citrate was detectable in ACSH (data not shown). We conclude that succinate was derived both from glucose and from alternate carbon sources, like malate and citrate, present in ACSH.
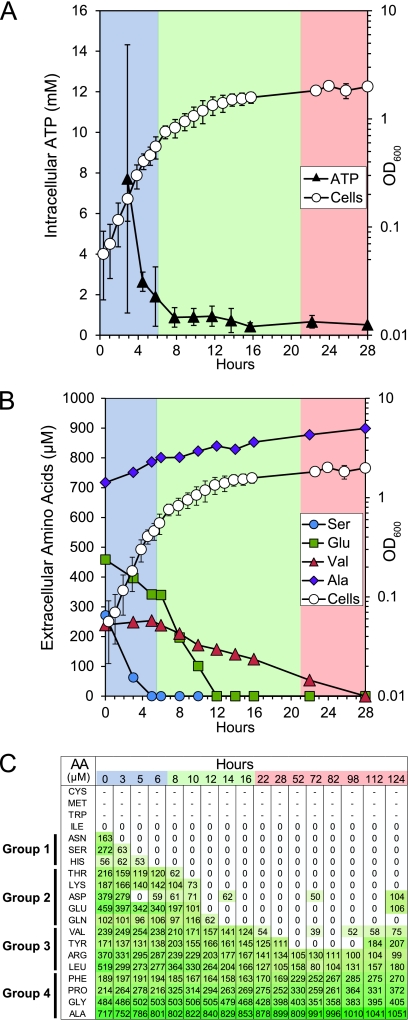
Fig 3.
Intracellular ATP concentrations and extracellular amino acid concentrations during anaerobic growth of GLBRCE1 in ACSH. (A) Mean intracellular ATP concentration measured during growth in ACSH in relation to cell density (same as in Fig. 1). Error bars represent the ranges of measurements. (B) Rates of depletion of individual amino acids that were representative of groups of amino acids with similar depletion patterns during the fermentation. Serine, group 1; glutamate, group 2; valine, group 3; alanine, group 4. (C) Time-dependent changes in concentration of all amino acids detectable in medium during growth in ACSH bioreplicate 1. Group assignments are on the left. Cysteine, methionine, tryptophan, and isoleucine were not detected (see the text). The growth data are from Fig. 1. The shading in panel C reflects the amino acid concentration (with darker shades of green representing higher concentrations).
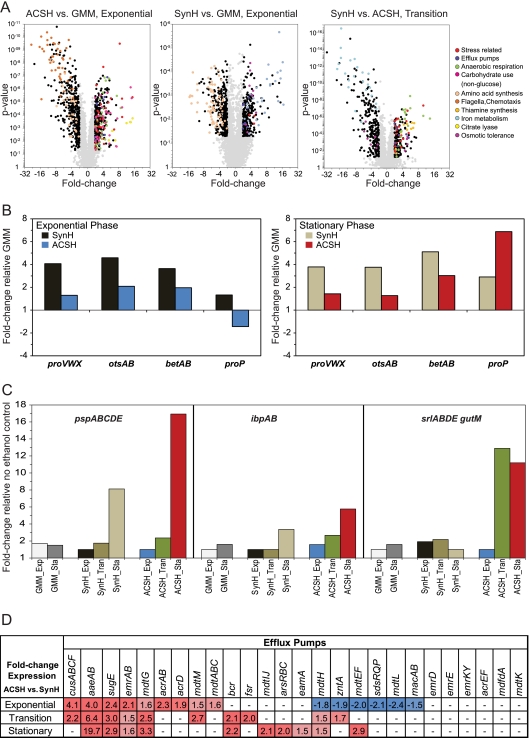
Fig 4.
Expression profiling of strain GLBRCE1 during growth in ACSH, SynH, and GMM. (A) Gene expression levels are plotted as a function of fold change (log2) and P value. Downregulated genes are represented by negative values. Genes with significantly different expression levels (>2-fold) are black or colored according to their functional classification, as defined on the right. (B) Mean expression of genes or operons associated with osmotic stress during growth of GLBRCE1 in ACSH and SynH during exponential and stationary phases. For ACSH in exponential phase, the data from 3, 5, and 6 h of growth were averaged; for ACSH in stationary phase, the data from 22, 28, and 52 h of growth were averaged (see Fig. 2). Expression values are plotted as fold change relative to expression levels during exponential-phase (8 h) or stationary-phase (16 h) growth in GMM (Fig. 5). (C) Mean expression of genes or operons associated with ethanol stress during exponential, transition, and stationary phases during growth of GLBRCE1 in ACSH and SynH. ACSH exponential-phase and stationary-phase data were averaged as described for panel B; for ACSH in transition phase, the data from the 8- to 16-h time points were averaged (see Fig. 2). Expression values are plotted as fold change with respect to expression level of the nonethanologenic E. coli strain MG1655 grown aerobically in GMM. (D) Expression of genes associated with efflux of toxic molecules during exponential, transition, and stationary phases of growth of GLBRCE1 in ACSH. ACSH data were averaged as described for panels B and C. Expression values represent fold change with respect to expression level of GLBRCE1 during the corresponding growth phase in SynH. Dashes represent no change. The P values for genes highlighted in dark red and blue ranged between 0.05 and 1e−5. Values highlighted in light red were marginally significant (elevated expression levels as indicated but P values were >0.05).
Table 3.
Genes exhibiting the greatest changes in expression in different growth phases in ACSH
| Growth phase and gene | Gene product description or functiona | Fold changeb in growth phase |
||
|---|---|---|---|---|
| Exponential | Transition | Stationary | ||
| Exponential | ||||
| yhcN | Stress-induced protein | 62.9 | −1.0 | −1.5 |
| citE | Citrate lyase | 50.2 | −3.9 | −7.6 |
| citD | Citrate lyase | 46.9 | −4.5 | −7.7 |
| citF | Citrate lyase | 36.7 | −4.6 | −8.9 |
| marR | Multiple antibiotic resistance regulator | 29.8 | −1.7 | 1.4 |
| marA | Multiple antibiotic resistance regulator | 28.4 | −1.6 | 1.2 |
| frmB | S-Formylglutathione hydrolase | 19.8 | −11.0 | 2.3 |
| araB | l-Ribulokinase | 18.6 | −3.8 | −4.2 |
| ybjC | Predicted inner membrane protein | 16.9 | −2.6 | 1.3 |
| citC | Citrate lyase | 16.0 | −4.1 | −4.9 |
| entB | Isochorismatase | −87.0 | −1.2 | 1.0 |
| fepA | Ferric enterobactin transport | −87.9 | 1.0 | 1.1 |
| cirA | Ferric dihyroxybenzoylserine transport | −88.7 | 1.1 | 1.6 |
| fliZ | Regulator of σS activity | −103.7 | 1.1 | 1.2 |
| fliD | Flagellar cap protein | −115.9 | 1.1 | 1.2 |
| flgD | Flagellar biosynthesis | −145.0 | 1.0 | −1.1 |
| tap | Chemotaxis | −145.5 | 1.1 | 1.0 |
| fliC | Flagellar biosynthesis | −165.0 | 1.5 | 1.4 |
| tar | Chemotaxis | −180.4 | 1.0 | 1.0 |
| flgC | Flagella | −182.0 | 1.1 | 1.0 |
| Transition | ||||
| yjiH | Conserved inner membrane protein | −30.8 | 34.5 | −1.6 |
| srlE | Glucitol/sorbitol PTS transport | −4.4 | 23.8 | 1.0 |
| yjiG | Conserved inner membrane protein | −18.7 | 22.5 | −1.6 |
| srlA | Glucitol/sorbitol PTS transport | −1.9 | 20.4 | 1.1 |
| srlD | Sorbitol 6-P dehydrogenase | −3.7 | 19.0 | −1.3 |
| srlB | Glucitol/sorbitol PTS transport | −1.4 | 15.1 | −1.4 |
| iadA | Isoaspartyl dipeptidase | −10.1 | 11.1 | −1.8 |
| yeaG | Protein kinase | 1.1 | 8.9 | −2.3 |
| rmf | Ribosome modulation factor | −1.2 | 8.2 | 1.8 |
| metE | Homocysteine transmethylase | −5.1 | 8.1 | −6.7 |
| hmp | Nitric oxide dioxygenase | 6.9 | −9.0 | −1.0 |
| edd | Phosphogluconate dehydratase | 5.0 | −9.8 | −1.5 |
| gntK | d-Gluconate kinase | 12.9 | −10.4 | −1.1 |
| gntU | Gluconate transporter | 8.2 | −10.6 | 1.1 |
| frmB | S-Formylglutathione hydrolase | 19.8 | −11.0 | 2.3 |
| guaB | IMP dehydrogenase | −1.9 | −11.1 | 1.6 |
| mtlA | Mannitol PTS permease | 9.3 | −11.4 | 1.1 |
| frmA | Formaldehyde dehydrogenase | 5.3 | −11.7 | 3.9 |
| frmR | Regulator of frmA | 6.0 | −13.5 | 4.4 |
| hcp | Hybrid cluster protein | 13.5 | −27.9 | −1.1 |
| Stationary | ||||
| artJ | Arginine ABC transport | 2.1 | −2.5 | 15.0 |
| argC | N-Acetylglutamylphosphate reductase | 4.2 | −2.3 | 14.8 |
| argB | Acetylglutamate kinase | 4.2 | −2.1 | 14.2 |
| argD | Acetylornithine transaminase | 1.6 | −3.8 | 12.3 |
| argI | Ornithine carbamoyltransferase | −1.7 | −1.5 | 12.0 |
| argF | Ornithine carbamoyltransferase | 2.6 | −1.7 | 11.1 |
| yhjX | MFS transporter | −3.2 | 2.4 | 10.0 |
| pspD | Phage shock protein D | 1.5 | 2.6 | 8.9 |
| argA | N-Acetylglutamate synthase | 1.4 | −1.5 | 8.6 |
| argG | Argininosuccinate synthase | 1.9 | −1.1 | 8.5 |
| metE | Homocysteine transmethylase | −5.1 | 8.1 | −6.7 |
| ybaS | Glutaminase | −1.0 | 2.0 | −7.1 |
| gadB | Glutamate decarboxylase B | −2.8 | 4.1 | −7.3 |
| citE | Citrate lyase | 50.2 | −3.9 | −7.6 |
| citD | Citrate lyase | 46.9 | −4.5 | −7.7 |
| citF | Citrate lyase | 36.7 | −4.6 | −8.9 |
| hchA | Glyoxalase III | −1.9 | 7.0 | −10.0 |
| gadC | Glutamate/γ-aminobutyrate antiporter | −4.3 | 6.2 | −13.4 |
| hdeA | Acid resistance | 1.4 | 2.3 | −17.9 |
| hdeB | Acid stress chaperone | 1.4 | 2.2 | −20.4 |
A complete description of gene functions is available at www.ecocyc.org (44).
Fold changes are shown for the ratio of expression levels in ACSH exponential phase compared to SynH exponential phase (Exponential), ACSH transition phase compared to ACSH exponential phase (Transition), and ACSH stationary phase compared to ACSH transition phase (Stationary). The statistical significance of expression changes identified here range from a P value of 1.6 × 10−10 (guaB; −11.1-fold change in exponential phase) to 0.036 (citF; −8.9-fold change in stationary phase). The distribution of P values also is depicted in the volcano plots in Fig. 5A; all but those for metE and citFDE are less than 0.01.
Depletion of amino acids from ACSH during growth.
Free amino acids remain in corn stover after AFEX treatment (47) and are a logical contributor to cell growth. To determine if amino acids contributed to growth of ethanologenic E. coli in ACSH, we quantified the levels of the amino acids in the medium over the course of fermentation (Fig. 3B and C). We detected significant concentrations of alanine, aspartate, glutamate, phenylalanine, glycine, histidine, lysine, leucine, asparagine, proline, glutamine, arginine, serine, threonine, valine, and tyrosine, but isoleucine, methionine, tryptophan, and cysteine were not detected by the assay we used, possibly due to interfering compounds in ACSH. During growth of the ethanologen, the concentrations of most amino acids decreased. However, the patterns of changes in amino acid concentrations were disparate and fell roughly into four groups. The first group (serine, asparagine, and histidine) was depleted from the medium in the first 3 to 6 h of incubation. A second group (glutamate, threonine, lysine, aspartate, and glutamine) was depleted later in the fermentation, after 10 to 14 h of incubation. The third group (valine, tyrosine, arginine, and leucine) remained detectable into stationary phase. Finally, a fourth group either remained nearly constant (glycine) or increased in concentration (phenylalanine, proline and alanine) over the course of the fermentation, suggesting net synthesis of these amino acids by the ethanologen.
Expression levels of amino acid biosynthetic genes were broadly consistent with the patterns of amino acid depletion from ACSH. For example, expression of serAC and asnAB, thrAB, and aspC genes increased after approximately 8 to 12 h of growth, approximately the point at which these amino acids became undetectable (Table 2). In addition, gltB, gdhA, and glnA, which encode enzymes for glutamate and glutamine biosynthesis, were upregulated when the cognate amino acids became depleted (early in transition phase; 8 to 14 h). Coincident with the upregulation of gltB and gdhA, we observed increased expression of glnK and amtB. glnK encodes a PII protein that regulates Ntr gene expression when nitrogen is limiting (13, 76). Upregulation of genes involved in nitrogen utilization likely represented a metabolic switch that occurred when the cells transitioned from use of glutamate and glutamine as a nitrogen source to synthesis of glutamate and glutamine from inorganic nitrogen sources, such as ammonia.
Transcriptional profiling indicated the initial presence of most acids in ACSH, including methionine, tryptophan, and isoleucine that escaped detection by direct assay. However, cysteine appeared to be absent in ACSH, as cysZCANHGWUDIPJ, which encode enzymes for cysteine biosynthesis, were expressed at levels similar to those observed in GMM even during exponential phase in ACSH (Table 2; also, see Table S6 in the supplemental material). In contrast, the trp and met operons were initially repressed in ACSH and reached GMM levels only in transition phase (Table 2; also, see Table S6); this pattern indicated that tryptophan and methionine were present in ACSH but were rapidly consumed. Expression of the biosynthetic genes for two amino acids diverged from this overall trend. Genes for biosynthesis of alanine (alaA, ilvE, and avtA) and proline (proAB) were expressed at high levels throughout the fermentation (Table 2; also, see Table S6), even though extracellular alanine and proline levels were high and increased over time (Fig. 3B).
E. coli K-12 encodes catabolic systems for l-serine, l-aspartate, l-tryptophan, l-glutamate, glycine, l-threonine, l-alanine, l-proline, l-arginine, l-glutamine, l-asparagine, l-cysteine, and l-lysine (72). With the exception of aspA, which encodes aspartate lyase, an enzyme that can degrade aspartate to fumarate, and ansB, which encodes asparaginase II and is involved in the degradation of asparagine to aspartate, expression of genes associated with amino acid degradation were expressed at low levels during growth in ACSH (Table 2; also, see Table S6). Thus, most amino acids present in ACSH were likely used as precursors for protein synthesis, rather than being catabolized for use as carbon sources.
Effect of amino acids on growth of the ethanologen in hydrolysate.
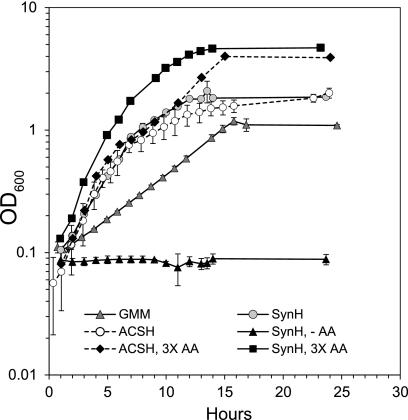
Since the entry of the ethanologen into the transition and stationary phases correlated with the depletion of subsets of amino acids (Fig. 3), we tested whether the slowing of E. coli growth in ACSH resulted from decreases in amino acids. We first tested supplementation of amino acids to three times the concentrations measured in ACSH (see Fig. S1 in the supplemental material), along with methionine (300 μM), isoleucine (700 μM), tryptophan (150 μM), and cysteine (50 μM). Increasing amino acid concentrations delayed entry into stationary phase, increased final cell density, and increased growth rate during transition phase (Fig. 5). We concluded that amino acids significantly impact E. coli growth dynamics in ACSH.
Fig 5.
Growth of GLBRCE1 in ACSH, SynH, and GMM in the presence and absence of amino acid supplements. Growth of GLBRCE1 in GMM, SynH, and ACSH was compared to growth in SynH and ACSH containing amino acids at concentrations three times those measured in ACSH (3× AA) or in SynH lacking amino acids (−AA). Error bars represent the range of values for duplicate samples. 3× AA supplements included methionine, cysteine, tryptophan, and isoleucine (see the text).
We hypothesized two possible ways amino acids could improve growth. First, amino acids could provide osmolytes or osmolyte precursors to mitigate the osmotic stress caused by the concentrated hydrolysate medium. Second, amino acids could reduce the energetic load of amino acid biosynthesis, which could increase the energy available to combat stresses associated with growth in ACSH. To test these hypotheses, we compared gene expression in ACSH to that in synthetic, chemically defined medium.
Comparative transcriptomics suggests physiological roles of amino acids during growth in hydrolysate.
GLBRCE1 grew at a uniform rate in GMM and entered stationary phase precipitously when glucose was exhausted (data not shown), without the apparent transition phase that was evident in ACSH and SynH (Fig. 5). When grown in ACSH or SynH, GLBRCE1 exhibited similar exponential, transition, and stationary phases and reached comparable final cell densities before growth arrest (Fig. 5). In addition, the strain produced similar amounts of ethanol (450 mM, SynH; 370 mM, ACSH), grew at similar exponential rates (0.37 h−1, ACSH; 0.29 h−1, SynH), assimilated glucose similarly (13 mM · unit of OD600 · h−1, SynH; 13.6 mM · unit of OD600 · h−1, ACSH) but exhibited somewhat different patterns of amino acid depletion (Table 1; also, see Tables S3 and S4 and Fig. S1 in the supplemental material). During stationary phase, the glucose uptake rate was slightly higher in SynH than in ACSH (2.6 versus 1.9 mM · unit of OD600 · h−1, respectively) (Table 1), which led to glucose being consumed more quickly in SynH than in ACSH (compare Tables S3 and S4 in the supplemental material). In addition, more xylose was consumed in SynH (Table 1; also, see Tables S3 and S4). Strikingly, proline was rapidly depleted from SynH, in contrast to its accumulation in ACSH (see Fig. S1). Given that proline is a major osmoprotectant in E. coli, this led us to consider how osmotic stress may differ in ACSH and SynH.
Osmotic stress.
Cells growing in concentrated lignocellulose hydrolysates are expected to experience osmotic stress due to the high sugar concentration (∼10%). The sugars and other solutes generate osmolality near one mol/kg, which decreases ∼20 to 30% during fermentation (initial to final osmolality of 1.44 to 0.97 mol/kg for ACSH and 0.97 to 0.75 mol/kg for SynH). Such high solute levels are known to reduce growth rate, reduce ethanol yield, and inhibit xylose utilization by E. coli ethanologens (67, 83).
Despite the slightly lower osmotic strength of SynH, osmotic stress responses were stronger in SynH than in ACSH. In SynH, genes typically induced by osmotic stress were upregulated relative to their expression in GMM. These included proVWX and proP, which encode transporters for the osmoprotectants glycine betaine and proline (9, 54, 55), otsAB, which encode enzymes for synthesis of the osmoprotectant trehalose (32, 67), and betABT, encoding enzymes for glycine betaine synthesis and transport (∼4-, ∼3-, ∼5-, and ∼2-fold upregulation in SynH for proVWX, proP, otsAB, and betABT, respectively [Fig. 4B; Table 2]). To test whether growth in SynH depended on osmoprotectants, we measured final cell ODs for cultures incubated in SynH lacking amino acids and supplemented with different amino acids (Fig. 6). Without supplementation, GLBRCE1 did not grow, but proline, glycine betaine, or high concentrations of some amino acid combinations could restore growth (Fig. 6; also, see Fig. S2 in the supplemental material). We conclude that alleviation of osmotic stress is a crucial contribution of amino acids to growth of GLBRCE1 in SynH.
Fig 6.
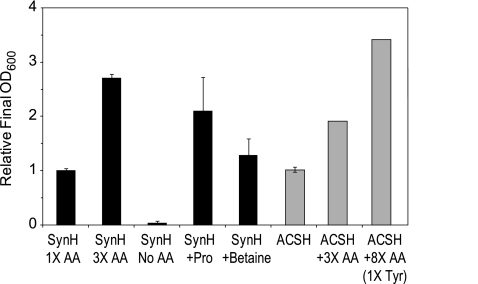
Effect of amino acids on growth of GLBRCE1 in SynH (black) or ACSH (gray). The final OD600 of cells grown anaerobically in SynH with or without amino acids (AA) or glycine betaine was measured, and the values from replicate cultures are plotted relative to the value observed for SynH with 1× AA (0.24 OD600) or relative to ACSH alone (2.06 OD600). Unless noted otherwise, AA were added at 1×, 3×, or 8× the concentrations measured in ACSH (see Table S1 in the supplemental material); Proline and glycine betaine alone were added at 0.67 and 2 mM, respectively, to SynH No AA base medium. Tyrosine was not increased in the ACSH +8× AA culture due to its limited solubility. Error bars represent the ranges of values from duplicate assays.
However, gene expression patterns and supplementation revealed a somewhat different picture in ACSH. During exponential phase in ACSH, proVWX, proP, otsAB, and betABT exhibited less induction or even repression relative to GMM (1.3-, −1.5-, 2.1-, and 2-fold, respectively [Fig. 4B; Table 2]). Lower expression of proVWX and proP was consistent with the lack of proline depletion from ACSH (Fig. 3C). As the cells entered stationary phase in ACSH, expression of proP and betAB increased, suggestive of elevated osmotic stress. Overall, despite the high osmolality of ACSH, the ethanologen exhibited only modest expression of genes associated with osmotic stress.
One explanation for the initially modest osmotic stress response in ACSH could be that components of ACSH not present in SynH provide alternative means for cells to mitigate the osmotic stress. Indeed, our NMR analysis of ACSH revealed that glycine betaine is present at 0.7 mM, a concentration previously shown to mitigate osmotic stress (83). Furthermore, carnitine (0.2 mM), a known osmoprotectant transported by the ProP transporter, and choline (0.7 mM), which can be converted to glycine betaine under some conditions (77), were also detected (see Tables S1 and S5 in the supplemental material). We conclude that ACSH contains compounds that can be exploited to mitigate the osmotic stress of the ethanologen.
Acetate and acetamide stress.
Plant cell walls contain many acetyl groups on sugars, which are released as acetamide by AFEX pretreatment and may be converted to acetate during hydrolysate preparation. NMR analysis of ACSH revealed 75 mM acetamide and 33 mM acetate, which increased to 41 mM acetate with concomitant decrease in acetamide by the end of the fermentation (see Table S5 in the supplemental material). Although the effect of acetamide on cell physiology is not well characterized, acetate at concentrations greater than 8 mM is reported to reduce the growth rate of E. coli and to be potentiated by increased osmolality (5, 71, 87). Our analysis of cells grown in ACSH versus SynH identified altered expression for approximately one-third of the genes previously linked to acetate stress (5, 45), including ACSH-dependent upregulation of the hdeA, hdeB, osmY, luxS, fliY, gatY, pfkB, gadW, gadX, metA, and slp genes (Table 2). The limited overlap of our data set of regulated genes with previous studies may result from differences in growth conditions, in that the previous studies were carried out under aerobic conditions and with a nonethanologenic strain of E. coli. However, the similarities in growth rate between cells grown in ACSH and SynH indicate that the acetate and acetamide in ACSH did not greatly impair cell growth.
Ethanol stress.
Growth of the ethanologen in ACSH or SynH yielded 15 to 22 g ethanol/liter (330 to 480 mM). Previous results showed that exogenously supplied ethanol at more than 20 g/liter decreases the growth rate of E. coli (34, 35, 88; Keating et al., submitted). Further, ethanol sensitivity can directly impact ethanol yield during fermentation of E. coli ethanologens (88). To determine if the ethanologen experienced ethanol stress during growth in ACSH, we compared gene expression patterns in exponential-, transition-, and stationary-phase ethanologen cells versus cells of the nonethanologenic strain MG1655 (Fig. 4C). We reasoned that changes in gene expression as ethanol accumulated in transition and stationary phase could identify ethanol stress responses. Indeed, we found that several genes previously associated with ethanol stress were upregulated in the ethanologen (16; Keating et al., submitted). Specifically, expression of pspABCDE, encoding phage shock proteins, and ibpAB, encoding heat shock proteins, increased over the course of the fermentation in both ACSH and SynH media, with sorbitol utilization genes increasing in ACSH as well (Fig. 4C; Table 2). Although differences observed in the osmotic stress response between the two media may account for the differences in the sorbitol response, the similarity of expression in SynH and ACSH of pspABCDE and ibpAB suggests a common cause, the most obvious of which is ethanol stress. Thus, our results support the idea that ethanol stress contributes to growth inhibition in ACSH.
Lignotoxin stress.
Hydrolysates prepared from lignocellulose have previously been reported to contain compounds (collectively referred to as lignotoxins) that affect ethanol productivity, overall growth rate, as well as glucose and xylose consumption (48). These compounds include aromatic carboxylates, like ferulate, coumarate, and salicylate, and aldehydes, like furfural and vanillin. Comparison of gene expression in cells grown in ACSH versus SynH revealed upregulation of diverse stress responses predicted to be associated with lignotoxins, including upregulation of genes associated with small molecule efflux and detoxification of metals and other toxic compounds (Fig. 4D).
Efflux pumps were a prominent class of upregulated genes in comparisons of cells grown in ACSH and SynH. Examples of these pathways include the aaeAB genes, which encode an efflux system associated with tolerance to aromatic hydroxylates, such as p-hydroxybenzoate and cinnamic acid (85), and the emrAB genes, which encode an efflux system reported to confer resistance to hydrophobic aromatic compounds such as carbonyl cyanide, m-chlorophenylhydrazone, tetrachlorosalicylanilide and nalidixic acid (53). Some multidrug resistance (MDR) efflux pumps also were upregulated in ACSH, many of which have been reported to be involved in resistance to hydrophobic compounds and detergents. Upregulated MDR genes include acrB, acrD, and mdtABCGIJM. The acrB and acrD gene products combine with AcrA and TolC to form MDR systems involved in efflux of aminoglycoside antibiotics and polymyxin B, steroid acids such as bile salts, and sodium dodecyl sulfate (SDS), whereas the mdtABCGIJM gene products are involved in resistance to bile salts, aminocoumarins and other aromatic antibiotics, and SDS (41, 61, 63).
Interestingly, expression of the acrAB and tolC genes is regulated by marA gene, which encodes a global regulator that affects tolerance to antibiotics, organic solvents, and oxidative stress agents (3). Expression of the marAB genes is controlled by the MarR repressor, whose function is antagonized by compounds like salicylate, cinnamate, and ferulate (21, 78; Tang et al., unpublished) found in plant cell wall hydrolysates like ACSH. Thus, the MarABR regulatory system appears to be a key point of control for genes associated with lignotoxin stress and a possible point of engineering for generating stress-tolerant organisms.
Finally, stress responses associated with heavy metals and other toxic compounds were found to be upregulated during growth of the ethanologen in ACSH. Examples of these systems include the cusCFBA genes, which encode a permease involved in export of copper and silver ions (27), and arsRBC, which have been reported to be involved in resistance to arsenite, arsenate, and antimonite (17, 25). Although we were unable to determine the concentration of arsenite or related compounds, the concentrations of copper and other heavy metals in hydrolysate were significantly below what has been reported to cause growth inhibition (see Table S2 in the supplemental material). However, these previous experiments were not carried out in concentrated complex media like hydrolysate or anaerobically, and it seems likely that the toxicity of these compounds could synergize with other ACSH-induced stresses to affect cell physiology.
DISCUSSION
The goal of this study was to characterize how E. coli responds metabolically and transcriptionally to concentrated hydrolysates derived from alkali-pretreated lignocellulose, with ACSH as the test case. Rationally engineering microbes to convert lignocellulosic hydrolysates to biofuels requires understanding how microbes respond to these complex mixtures specifically in the anaerobic conditions that optimize retention of hydrolysate energy content in end products. The experiments reported here yielded several results that contribute to this much needed understanding and form a basis for future study. First, robust cell growth that is characteristic of ACSH (47) depends on the presence of amino acids in ACSH, at least for E. coli. Second, even after growth arrest that correlated with depletion of key amino acids, the cells remained metabolically active, and the majority of glucose consumption and ethanol production occurred during this stationary phase. Third, E. coli cells retained metabolic activity in ACSH despite exposure to compound stresses caused by high osmolality, ethanol accumulation, and lignotoxins. By combining knowledge of how E. coli responded to changes in ACSH over exponential, transition, and stationary phases and to stresses caused by ACSH with knowledge of the composition of ACSH over the course of the fermentation, we can propose design improvements to optimize E. coli for anaerobic biofuel synthesis in alkali-pretreated biomass hydrolysates.
Causes of growth arrest in ACSH.
An important question is why cells cease growth in ACSH when ample supplies of glucose remain. An ability to control the timing of this growth arrest is an important goal for bioengineering optimal conversion of ACSH to useful products. Understanding the factors controlling growth cessation will allow optimization of the fraction of ACSH energy content used to accumulate cell biomass versus the rate at which the growth-arrested culture converts ACSH components to desired products like ethanol (i.e., making less biomass consumes less sugar but converts remaining sugar to biofuel more slowly). Our results suggest that a combination of factors underlies growth arrest in ACSH.
First, and most certain, amino acid depletion contributes to growth arrest. The passage of E. coli from exponential to transition to stationary phase was mirrored by successive depletion of amino acids from the growth medium (Fig. 3) and could be delayed by amino acid supplementation (Fig. 5). Cells grown in ACSH or SynH containing amino acids at concentrations three times those measured in ACSH achieved a higher cell density before growth arrest than cells grown in the unsupplemented counterparts, and omission of amino acids from SynH precluded cell growth altogether (Fig. 5). Exactly which amino acids determine growth arrest is less clear, but it is notable that growth arrest in ACSH followed depletion of the major amino acid sources of organic nitrogen (glutamate, glutamine, and aspartate). Consistent with a shift to requiring nitrogen assimilation, we observed a spike in expression of the Ntr regulon during the transition phase (see Table S6 in the supplemental material). Additionally, as the supply of amino acids from ACSH diminished, further growth would have required cells to synthesize them de novo, at a cost ranging from ∼10 to ∼75 ATPs/amino acid (1) plus the cost of synthesizing amino acid biosynthetic enzymes.
Consistent with the idea that increased demands for ATP contributed to growth arrest, we observed a dramatic decrease in intracellular ATP concentration in exponential- and stationary-phase cells: the intracellular ATP concentration in the ethanologen declined from ∼8 mM early in exponential phase (matching values reported for E. coli growing with glucose as a carbon source [11]) to <1 mM upon entry into transition phase and to <0.5 mM in stationary phase (Fig. 3A). These decreases occurred despite continued conversion of glucose to ethanol in the growth-arrested state, which is predicted to generate 5.2 mM ATP · unit of OD600−1 · h−1 (Table 1). Assuming an intracellular volume of 0.67 fl (79), this corresponds to production of ATP at 130 mM min−1. Maintaining cell viability even in optimal growth medium is thought to require significant ATP consumption, ranging from 20 to 90 mM min−1 (80, 86). Cell growth requires significantly more ATP; even a modest growth rate of 0.1 h−1 is predicted to require about twice the maintenance flux of ATP (80, 86). These ATP fluxes required for growth apparently cannot be achieved in ACSH once amino acids are depleted, despite abundant glucose. Given that comparable levels of glucose support anaerobic growth of our strain in less complex media (e.g., GMM), it seems likely that other factors must limit growth either by consuming ATP or by limiting its production.
One possibility is the compounded effect of multiple stresses on E. coli, which could consume ATP, limit ATP production by inhibiting enzymes in the glucose-to-ethanol pathway, or both. Transcriptional profiling identified four possible stresses that might contribute to growth arrest: osmotic stress, acetate stress, ethanol stress, and stresses from components of ACSH, including phenolic compounds derived from lignin and metal ions. The mechanisms by which E. coli responds to several of these stresses are likely to increase the energetic demands for cell maintenance. For instance, the most upregulated efflux pumps (e.g., AaeAB, CusABCF, and the multidrug resistance pumps SugE, EmrAB, and MdtG) are all proton antiporters that would consume ATP to maintain the proton motive force via the F1Fo ATPase proton pump. Thus, these efflux systems may contribute to an ATP demand that limits cell growth in ACSH, as they help E. coli cope with inhibitory molecules present in the hydrolysate. It is also possible that inhibitors in ACSH directly slow growth by binding to cellular targets such as key enzymes. In the case of metal ions, however, none appear to be present in ACSH at levels that, without other stresses, affect growth of E. coli (14, 69).
Another possibility that could contribute to growth arrest is redox imbalance, which is a frequent complication of anaerobic metabolism in E. coli when mixed acid fermentation is blocked (42, 74). The introduction of the Z. mobilis pyruvate decarboxylase pathway corrects the redox imbalance of ethanol synthesis via acetyl coenzyme A in wild-type E. coli (42), but an increase in pyruvate in the growth medium as cell growth slowed (see Table S4 in the supplemental material) was consistent with accumulation of intracellular NADH (possibly limiting NAD+ for glycolysis). Interference from the complex mixture of compounds present in ACSH has so far prevented us from measuring NADH/NAD+ ratios in cells grown in ACSH, but this is an important question for further study of anaerobic metabolism of ethanologenic E. coli in ACSH.
Osmotic stress and ethanol stress.
Two stresses, osmotic and ethanol induced, deserve special mention as potential sources of growth inhibition. Osmotic stress, which is known to affect microbial metabolism at multiple points (67, 83), is an unavoidable consequence of the high sugar concentrations desirable for lignocellulosic conversion. The accumulation of extracellular proline and of alanine during growth in ACSH is likely a consequence of osmotic stress. Although proline accumulation likely reflected proline's known osmoprotective function (56), to our knowledge the use of alanine as a microbial osmoprotectant has not been reported. Alanine has been reported to accumulate during osmotic stress in marine invertebrates and cyclostome fish (75). Furthermore, alanine has been observed to accumulate in plant and mammalian cells under low-oxygen conditions (30, 73), where it has been suggested to function to prevent acidification of the cytoplasm (26). Although additional studies are needed, our results suggest that E. coli maintains high alanine levels for some protective function during growth in ACSH, possibly a direct osmoprotectant, like proline, or a precursor to an unknown protective molecule.
Although we observed elevated expression of genes indicative of osmotic stress in SynH (e.g., proVWY, proP, betABT, and otsAB), their expression levels were lower in ACSH. This finding suggests that compounds in ACSH may mitigate osmotic stress. Consistent with this idea, glycine betaine, choline, and carnitine were all identified by NMR in ACSH (see Table S5 in the supplemental material). Although beneficial, import of these osmolytes would place additional energetic demands on the cell. The ProVWX transporter is an ABC transporter that hydrolyzes one ATP per osmolyte imported, whereas ProP, whose gene is expressed at lower levels until stationary phase, is a proton symporter. Thus, engineering increased expression of proP in exponential phase, and possibly lower expression of proVWX, may be a strategy to improve use of osmolytes in ACSH at lower energetic cost. In general, strategies to improve osmotic stress tolerance by E. coli are likely to improve conversion of ACSH to ethanol (57, 83).
An unexpected finding of our study was the elevated expression of genes associated with ethanol stress at low ethanol concentrations. Although expression of pspABCD, ibpB, srlABDE, and gutM was shown previously to be upregulated by ethanol (34, 35, 88; Keating et al., submitted), their induction in ACSH during early stationary phase occurred at lower ethanol concentrations than was typical in the previous studies, which were carried out in laboratory media. It is possible that high osmolality and lignotoxins in ACSH may potentiate the effect of ethanol on E. coli physiology, resulting in stress at lower concentrations of ethanol. Indeed, the growth of the ethanologen in ACSH is slowed by concentrations of ethanol as low as 5 g/liter (0.5%) (data not shown). Although ethanol stress may affect the ability of cells to generate proton-motive force, as reported for Saccharomyces cerevisiae (18), this effect might not be inhibitory at low ethanol concentrations in optimal medium yet become inhibitory in ACSH due to additional sinks for the proton gradient. Collectively, our results suggest that even relatively low concentrations of ethanol may contribute to growth arrest of E. coli in ACSH. Further study will be needed to determine if ethanol also limits the glucose conversion rate after arrest or if the effect of accumulating ethanol can be exploited to help control the point of growth arrest.
Changes observed in E. coli gene expression in ACSH suggest design strategies for improved ethanologens.
A key challenge in metabolic engineering of E. coli for improved conversion of lignocellulosic hydrolysates like ACSH to useful products such as ethanol is the lack of means to control gene expression by design. Conventional strategies for gene regulation in biotechnology use changes in temperature and the addition of compounds like isopropyl β-d-1-thiogalactopyranoside (IPTG), anhydrotetracycline, or arabinose, which are cost-prohibitive in biofuel applications or whose effect will be confounded by compounds present in hydrolysate. Our analysis of gene expression in exponential, transition, and stationary phases of E. coli grown in ACSH suggests an alternative strategy relying on natural changes in gene regulation that occur during these growth phases. In essence, well-characterized promoter/operator sequences for genes that exhibit desired regulation could be used to drive phase-specific expression of transgenes. For example, araB, under the control of PBAD and the AraC regulator, is on in exponential phase but turns off in exponential and stationary phases. The trp operon genes, controlled by the trp promoter and TrpR repressor, are off in exponential and stationary phase but upregulated ∼6-fold in transition phase. Expression of argA and argC, which is controlled by the ArgR repressor and σS, is off in exponential and transition phase but upregulated 50- to 100-fold in stationary phase. These specific patterns of expression will allow selective on- and off-regulation of “programmable gene cassettes” in E. coli cells at different phases of ACSH fermentation without exogenous effectors simply by placing the appropriate promoter regions at the 5′ ends of the gene cassettes. This could, for example allow expression of genes for osmolyte synthesis to be expressed during exponential phase, when energy supplies are more abundant, and repressed in transition phase, stationary phase, or both. Further study of the metabolic and gene-regulatory responses of E. coli to the complex and changing composition of concentrated hydrolysates like ACSH will facilitate implementation of this native-signal gene regulation strategy, will help define the chemical composition of these hydrolysates, and will provide a more complete understanding of how anaerobic physiology adjusts to these important cellular substrates.
Supplementary Material
ACKNOWLEDGMENTS
We thank R. Zinkel for assistance in transcriptomic sample processing, the Michigan Biotechnology Institute for allowing us to use their 5-gallon AFEX batch reactor to pretreat corn stover, T. Record for allowing use of his osmometer, and Genencor for supplying enzymes for hydrolysate production.
This work was funded by the U.S. Department of Energy (DOE) Great Lakes Bioenergy Research Center (DOE BER Office of Science DE-FC02-07ER64494). This study also made use of the National Magnetic Resonance Facility at Madison, which is supported by National Institutes of Health (NIH) grants P41RR02301 (Biomedical Research Technology Program, National Center for Research Resources) and P41GM66326 (National Institute of General Medical Sciences). Equipment in the facility was purchased with funds from the University of Wisconsin, the NIH (P41GM66326, P41RR02301, RR02781, and RR08438), the National Science Foundation (DMB-8415048, OIA-9977486, and BIR-9214394), and the U.S. Department of Agriculture. The work conducted by the U.S. DOE Joint Genome Institute is supported by the Office of Science of the U.S. DOE under contract no. DE-AC02-05CH11231.
Footnotes
Published ahead of print 2 March 2012
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1. Akashi H, Gojobori T. 2002. Metabolic efficiency and amino acid composition in the proteomes of Escherichia coli and Bacillus subtilis. Proc. Natl. Acad. Sci. U. S. A. 99:3695–3700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Albersheim P, Nevins D, English P, Karr A. 1968. A method for the analysis of sugars in plant cell-wall polysaccharides by gas-liquid chromatography. Carbohydr Res. 5:340–345 [Google Scholar]
- 3. Alekshun MN, Levy SB. 1999. Alteration of the repressor activity of MarR, the negative regulator of the Escherichia coli marRAB locus, by multiple chemicals in vitro. J. Bacteriol. 181:4669–4672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Amador-Noguez D, Brasg IA, Feng XJ, Roquet N, Rabinowitz JD. 2011. Metabolome remodeling during the acidogenic-solventogenic transition in Clostridium acetobutylicum. Appl. Environ. Microbiol. 77:7984–7997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Arnold CN, McElhanon J, Lee A, Leonhart R, Siegele DA. 2001. Global analysis of Escherichia coli gene expression during the acetate-induced acid tolerance response. J. Bacteriol. 183:2178–2186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Baba T, et al. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2:2006.0008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Balan V, Bals B, Chundawat SP, Marshall D, Dale BE. 2009. Lignocellulosic biomass pretreatment using AFEX. Methods Mol. Biol. 581:61–77 [DOI] [PubMed] [Google Scholar]
- 8. Baraquet C, et al. 2006. TorT, a member of a new periplasmic binding protein family, triggers induction of the Tor respiratory system upon trimethylamine N-oxide electron-acceptor binding in Escherichia coli. J. Biol. Chem. 281:38189–38199 [DOI] [PubMed] [Google Scholar]
- 9. Barron A, Jung JU, Villarejo M. 1987. Purification and characterization of a glycine betaine binding protein from Escherichia coli. J. Biol. Chem. 262:11841–11846 [PubMed] [Google Scholar]
- 10. Baumler D, Peplinski R, Reen J, Glasner J, Perna N. 2011. The evolution of metabolic networks of E. coli. BMC Syst. Biol. 5:182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bennett BD, et al. 2009. Absolute metabolite concentrations and implied enzyme active site occupancy in Escherichia coli. Nat. Chem. Biol. 5:593–599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Blattner FR, et al. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453–1462 [DOI] [PubMed] [Google Scholar]
- 13. Blauwkamp TA, Ninfa AJ. 2002. Physiological role of the GlnK signal transduction protein of Escherichia coli: survival of nitrogen starvation. Mol. Microbiol. 46:203–214 [DOI] [PubMed] [Google Scholar]
- 14. Bojic A, Purenovic M, Kocic B, Mihailovic D, Bojic D. 2002. The comparison of aluminium effects and its uptake by Escherichia coli in different media. Cent. Eur. J. Public Health 10:66–71 [PubMed] [Google Scholar]
- 15. Bolstad BM, Irizarry RA, Astrand M, Speed TP. 2003. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 19:185–193 [DOI] [PubMed] [Google Scholar]
- 16. Brynildsen MP, Liao JC. 2009. An integrated network approach identifies the isobutanol response network of Escherichia coli. Mol. Syst. Biol. 5:277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Carlin A, Shi W, Dey S, Rosen BP. 1995. The ars operon of Escherichia coli confers arsenical and antimonial resistance. J. Bacteriol. 177:981–986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Cartwright C, et al. 1986. Ethanol dissipates the proton-motive force across the plasma membrane of Saccharomyces cerevisiae. J. Gen. Microbiol. 132:369–377 [Google Scholar]
- 19. Chundawat SP, et al. 2010. Multifaceted characterization of cell wall decomposition products formed during ammonia fiber expansion (AFEX) and dilute acid based pretreatments. Bioresour. Technol. 101:8429–8438 [DOI] [PubMed] [Google Scholar]
- 20. Clomburg JM, Gonzalez R. 2010. Biofuel production in Escherichia coli: the role of metabolic engineering and synthetic biology. Appl. Microbiol. Biotechnol. 86:419–434 [DOI] [PubMed] [Google Scholar]
- 21. Dalrymple BP, Swadling Y. 1997. Expression of a Butyrivibrio fibrisolvens E14 gene (cinB) encoding an enzyme with cinnamoyl ester hydrolase activity is negatively regulated by the product of an adjacent gene (cinR). Microbiology 143:1203–1210 [DOI] [PubMed] [Google Scholar]
- 22. Delaglio F, et al. 1995. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6:277–293 [DOI] [PubMed] [Google Scholar]
- 23. den Dunnen JT, Antonarakis SE. 2001. Nomenclature for the description of human sequence variations. Hum. Genet. 109:121–124 [DOI] [PubMed] [Google Scholar]
- 24. Desai TA, Rao CV. 2010. Regulation of arabinose and xylose metabolism in Escherichia coli. Appl. Environ. Microbiol. 76:1524–1532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Diorio C, Cai J, Marmor J, Shinder R, DuBow MS. 1995. An Escherichia coli chromosomal ars operon homolog is functional in arsenic detoxification and is conserved in gram-negative bacteria. J. Bacteriol. 177:2050–2056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Felle H. 1996. Control of cytoplasmic pH under anoxic conditions and its implication for plasma membrane proton transport in Medicago sativa root hairs. J. Exp. Bot. 47:967–973 [Google Scholar]
- 27. Franke S, Grass G, Rensing C, Nies DH. 2003. Molecular analysis of the copper-transporting efflux system CusCFBA of Escherichia coli. J. Bacteriol. 185:3804–3812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Freddolino PL, Amini S, Tavazoie S. 2012. Newly identified genetic variations in common Escherichia coli MG1655 stock cultures. J. Bacteriol. 194:303–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gardner JG, Keating DH. 2010. Requirement of the type II secretion system for utilization of cellulosic substrates by Cellvibrio japonicus. Appl. Environ. Microbiol. 76:5079–5087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Garza-Quintero R, Ortega-Lopez J, Stein JH, Venkatachalam MA. 1990. Alanine protects rabbit proximal tubules against anoxic injury in vitro. Am. J. Physiol. 258:F1075–F1083 [DOI] [PubMed] [Google Scholar]
- 31. Geddes CC, et al. 2011. Simplified process for ethanol production from sugarcane bagasse using hydrolysate-resistant Escherichia coli strain MM160. Bioresour. Technol. 102:2702–2711 [DOI] [PubMed] [Google Scholar]
- 32. Giaever HM, Styrvold OB, Kaasen I, Strom AR. 1988. Biochemical and genetic characterization of osmoregulatory trehalose synthesis in Escherichia coli. J. Bacteriol. 170:2841–2849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Gonzalez R, et al. 2003. Gene array-based identification of changes that contribute to ethanol tolerance in ethanologenic Escherichia coli: comparison of KO11 (parent) to LY01 (resistant mutant). Biotechnol. Prog. 19:612–623 [DOI] [PubMed] [Google Scholar]
- 34. Goodarzi H, et al. 2010. Regulatory and metabolic rewiring during laboratory evolution of ethanol tolerance in E. coli. Mol. Syst. Biol. 6:378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Goodarzi H, Hottes AK, Tavazoie S. 2009. Global discovery of adaptive mutations. Nat. Methods 6:581–583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hasona A, Kim Y, Healy FG, Ingram LO, Shanmugam KT. 2004. Pyruvate formate lyase and acetate kinase are essential for anaerobic growth of Escherichia coli on xylose. J. Bacteriol. 186:7593–7600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ingledew WJ, Poole RK. 1984. The respiratory chains of Escherichia coli. Microbiol. Rev. 48:222–271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ingram LO, Buttke TM. 1984. Effects of alcohols on micro-organisms. Adv. Microb. Physiol. 25:253–300 [DOI] [PubMed] [Google Scholar]
- 39. Ingram LO, Conway T, Clark DP, Sewell GW, Preston JF. 1987. Genetic engineering of ethanol production in Escherichia coli. Appl. Environ. Microbiol. 53:2420–2425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Irizarry RA, et al. 2003. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 4:249–264 [DOI] [PubMed] [Google Scholar]
- 41. Jack DL, Storms ML, Tchieu JH, Paulsen IT, Saier MH., Jr 2000. A broad-specificity multidrug efflux pump requiring a pair of homologous SMR-type proteins. J. Bacteriol. 182:2311–2313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Jarboe LR, Grabar TB, Yomano LP, Shanmugan KT, Ingram LO. 2007. Development of ethanologenic bacteria. Adv. Biochem. Eng. Biotechnol. 108:237–261 [DOI] [PubMed] [Google Scholar]
- 43. Jarboe LR, et al. 2010. Metabolic engineering for production of biorenewable fuels and chemicals: contributions of synthetic biology. J. Biomed. Biotechnol. 2010:761042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Keseler IM, et al. 2011. EcoCyc: a comprehensive database of Escherichia coli biology. Nucleic Acids Res. 39:D583–D590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kirkpatrick C, et al. 2001. Acetate and formate stress: opposite responses in the proteome of Escherichia coli. J. Bacteriol. 183:6466–6477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Knoshaug EP, Zhang M. 2009. Butanol tolerance in a selection of microorganisms. Appl. Biochem. Biotechnol. 153:13–20 [DOI] [PubMed] [Google Scholar]
- 47. Lau MW, Dale BE. 2009. Cellulosic ethanol production from AFEX-treated corn stover using Saccharomyces cerevisiae 424A(LNH-ST). Proc. Natl. Acad. Sci. U. S. A. 106:1368–1373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Lau MW, Dale BE. 2010. Effect of primary degradation-reaction products from ammonia fiber expansion (AFEX)-treated corn stover on the growth and fermentation of Escherichia coli KO11. Bioresour. Technol. 101:7849–7855 [DOI] [PubMed] [Google Scholar]
- 49. Lau MW, Dale BE, Balan V. 2008. Ethanolic fermentation of hydrolysates from ammonia fiber expansion (AFEX) treated corn stover and distillers grain without detoxification and external nutrient supplementation. Biotechnol. Bioeng. 99:529–539 [DOI] [PubMed] [Google Scholar]
- 50. Lau MW, Gunawan C, Balan V, Dale BE. 2010. Comparing the fermentation performance of Escherichia coli KO11, Saccharomyces cerevisiae 424A(LNH-ST) and Zymomonas mobilis AX101 for cellulosic ethanol production. Biotechnol. Biofuels 3:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Lewis IA, Schommer SC, Markley JL. 2009. rNMR: open source software for identifying and quantifying metabolites in NMR spectra. Magn. Reson. Chem. 47(Suppl. 1):S123–S126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Lindsay SE, Bothast RJ, Ingram LO. 1995. Improved strains of recombinant Escherichia coli for ethanol production from sugar mixtures. Appl. Microbiol. Biotechnol. 43:70–75 [DOI] [PubMed] [Google Scholar]
- 53. Lomovskaya O, Lewis K, Matin A. 1995. EmrR is a negative regulator of the Escherichia coli multidrug resistance pump EmrAB. J. Bacteriol. 177:2328–2334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Lucht JM, Bremer E. 1994. Adaptation of Escherichia coli to high osmolarity environments: osmoregulation of the high-affinity glycine betaine transport system proU. FEMS Microbiol. Rev. 14:3–20 [DOI] [PubMed] [Google Scholar]
- 55. MacMillan SV, et al. 1999. The ion coupling and organic substrate specificities of osmoregulatory transporter ProP in Escherichia coli. Biochim. Biophys. Acta 1420:30–44 [DOI] [PubMed] [Google Scholar]
- 56. Mahan MJ, Csonka LN. 1983. Genetic analysis of the proBA genes of Salmonella typhimurium: physical and genetic analyses of the cloned proB+A+ genes of Escherichia coli and of a mutant allele that confers proline overproduction and enhanced osmotolerance. J. Bacteriol. 156:1249–1262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Miller EN, Ingram LO. 2007. Combined effect of betaine and trehalose on osmotic tolerance of Escherichia coli in mineral salts medium. Biotechnol. Lett. 29:213–217 [DOI] [PubMed] [Google Scholar]
- 58. Miller EN, et al. 2009. Furfural inhibits growth by limiting sulfur assimilation in ethanologenic Escherichia coli strain LY180. Appl. Environ. Microbiol. 75:6132–6141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Mills TY, Sandoval NR, Gill RT. 2009. Cellulosic hydrolysate toxicity and tolerance mechanisms in Escherichia coli. Biotechnol. Biofuels 2:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Moniruzzaman M, et al. 1996. Ethanol production from AFEX pretreated corn fiber by recombinant bacteria. Biotechnol. Lett. 18:985–990 [Google Scholar]
- 61. Nagakubo S, Nishino K, Hirata T, Yamaguchi A. 2002. The putative response regulator BaeR stimulates multidrug resistance of Escherichia coli via a novel multidrug exporter system, MdtABC. J. Bacteriol. 184:4161–4167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Neidhardt FC, Bloch PL, Smith DF. 1974. Culture medium for enterobacteria. J. Bacteriol. 119:736–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Nishino K, Yamaguchi A. 2001. Analysis of a complete library of putative drug transporter genes in Escherichia coli. J. Bacteriol. 183:5803–5812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Ohta K, Beall DS, Mejia JP, Shanmugam KT, Ingram LO. 1991. Genetic improvement of Escherichia coli for ethanol production: chromosomal integration of Zymomonas mobilis genes encoding pyruvate decarboxylase and alcohol dehydrogenase II. Appl. Environ. Microbiol. 57:893–900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Oliva JM, et al. 2004. Effect of binary combinations of selected toxic compounds on growth and fermentation of Kluyveromyces marxianus. Biotechnol. Prog. 20:715–720 [DOI] [PubMed] [Google Scholar]
- 66. Peterson JD, Ingram LO. 2008. Anaerobic respiration in engineered Escherichia coli with an internal electron acceptor to produce fuel ethanol. Ann. N. Y. Acad. Sci. 1125:363–372 [DOI] [PubMed] [Google Scholar]
- 67. Purvis JE, Yomano LP, Ingram LO. 2005. Enhanced trehalose production improves growth of Escherichia coli under osmotic stress. Appl. Environ. Microbiol. 71:3761–3769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Reed JL, Vo TD, Schilling CH, Palsson BO. 2003. An expanded genome-scale model of Escherichia coli K-12 (iJR904 GSM/GPR). Genome Biol. 4:R54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Rensing C, Mitra B, Rosen BP. 1997. The zntA gene of Escherichia coli encodes a Zn(II)-translocating P-type ATPase. Proc. Natl. Acad. Sci. U. S. A. 94:14326–14331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Rhodius VA, Gross CA. 2011. Using DNA microarrays to assay part function. Methods Enzymol. 497:75–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Roe AJ, McLaggan D, Davidson I, O'Byrne C, Booth IR. 1998. Perturbation of anion balance during inhibition of growth of Escherichia coli by weak acids. J. Bacteriol. 180:767–772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Sezonov G, Joseleau-Petit D, D'Ari R. 2007. Escherichia coli physiology in Luria-Bertani broth. J. Bacteriol. 189:8746–8749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Shingaki-Wells RN, et al. 2011. Differential molecular responses of rice and wheat coleoptiles to anoxia reveal novel metabolic adaptations in amino acid metabolism for tissue tolerance. Plant Physiol. 156:1706–1724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Singh A, Lynch MD, Gill RT. 2009. Genes restoring redox balance in fermentation-deficient E. coli NZN111. Metab. Eng. 11:347–354 [DOI] [PubMed] [Google Scholar]
- 75. Somero GN. 1986. Protons, osmolytes, and fitness of internal milieu for protein function. Am. J. Physiol. 251:R197–213 [DOI] [PubMed] [Google Scholar]
- 76. Soupene E, He L, Yan D, Kustu S. 1998. Ammonia acquisition in enteric bacteria: physiological role of the ammonium/methylammonium transport B (AmtB) protein. Proc. Natl. Acad. Sci. U. S. A. 95:7030–7034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Styrvold OB, et al. 1986. Selection, mapping, and characterization of osmoregulatory mutants of Escherichia coli blocked in the choline-glycine betaine pathway. J. Bacteriol. 165:856–863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Sulavik MC, Gambino LF, Miller PF. 1995. The MarR repressor of the multiple antibiotic resistance (mar) operon in Escherichia coli: prototypic member of a family of bacterial regulatory proteins involved in sensing phenolic compounds. Mol. Med. 1:436–446 [PMC free article] [PubMed] [Google Scholar]
- 79. Sundararaj S, et al. 2004. The CyberCell Database (CCDB): a comprehensive, self-updating, relational database to coordinate and facilitate in silico modeling of Escherichia coli. Nucleic Acids Res. 32:D293–D295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Taymaz-Nikerel H, Borujeni AE, Verheijen PJ, Heijnen JJ, van Gulik WM. 2010. Genome-derived minimal metabolic models for Escherichia coli MG1655 with estimated in vivo respiratory ATP stoichiometry. Biotechnol. Bioeng. 107:369–381 [DOI] [PubMed] [Google Scholar]
- 81. Teymouri F, Laureano-Perez L, Alizadeh H, Dale BE. 2004. Ammonia fiber explosion treatment of corn stover. Appl. Biochem. Biotechnol. 113-116:951–963 [DOI] [PubMed] [Google Scholar]
- 82. Teymouri F, Laureano-Perez L, Alizadeh H, Dale BE. 2005. Optimization of the ammonia fiber explosion (AFEX) treatment parameters for enzymatic hydrolysis of corn stover. Bioresour. Technol. 96:2014–2018 [DOI] [PubMed] [Google Scholar]
- 83. Underwood SA, Buszko ML, Shanmugam KT, Ingram LO. 2004. Lack of protective osmolytes limits final cell density and volumetric productivity of ethanologenic Escherichia coli KO11 during xylose fermentation. Appl. Environ. Microbiol. 70:2734–2740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. van Dam J, et al. 2002. Analysis of glycolytic intermediates in Saccharomyces cerevisiae using anion exchange chromatography and electrospray ionization with tandem mass spectrometric detection. Anal. Chim. Acta 460:209–218 [Google Scholar]
- 85. Van Dyk TK, Templeton LJ, Cantera KA, Sharpe PL, Sariaslani FS. 2004. Characterization of the Escherichia coli AaeAB efflux pump: a metabolic relief valve? J. Bacteriol. 186:7196–7204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Varma A, Palsson BO. 1994. Stoichiometric flux balance models quantitatively predict growth and metabolic by-product secretion in wild-type Escherichia coli W3110. Appl. Environ. Microbiol. 60:3724–3731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Yang S, et al. 2010. Paradigm for industrial strain improvement identifies sodium acetate tolerance loci in Zymomonas mobilis and Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U. S. A. 107:10395–10400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Yomano LP, York SW, Ingram LO. 1998. Isolation and characterization of ethanol-tolerant mutants of Escherichia coli KO11 for fuel ethanol production. J. Ind. Microbiol. Biotechnol. 20:132–138 [DOI] [PubMed] [Google Scholar]
- 89. Yomano LP, York SW, Shanmugam KT, Ingram LO. 2009. Deletion of methylglyoxal synthase gene (mgsA) increased sugar co-metabolism in ethanol-producing Escherichia coli. Biotechnol. Lett. 31:1389–1398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Yomano LP, York SW, Zhou S, Shanmugam KT, Ingram LO. 2008. Re-engineering Escherichia coli for ethanol production. Biotechnol. Lett. 30:2097–2103 [DOI] [PubMed] [Google Scholar]
- 91. Yu D, et al. 2000. An efficient recombination system for chromosome engineering in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 97:5978–5983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Zaldivar J, Martinez A, Ingram LO. 2000. Effect of alcohol compounds found in hemicellulose hydrolysate on the growth and fermentation of ethanologenic Escherichia coli. Biotechnol. Bioeng. 68:524–530 [DOI] [PubMed] [Google Scholar]
- 93. Zaldivar J, Martinez A, Ingram LO. 1999. Effect of selected aldehydes on the growth and fermentation of ethanologenic Escherichia coli. Biotechnol. Bioeng. 65:24–33 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.