Abstract
Agents that increase intracellular cAMP inhibit the activation and function of T cells and can lead to cell death. Recently, it has been postulated that cAMP inhibits T cell function in large part by acting as a brake on the T cell receptor and costimulatory receptor pathways. Therefore, for full activation of the T cell to occur, this inhibitory influence must be removed. One likely mechanism for accomplishing this is by up-regulation and/or activation of specific cyclic nucleotide phosphodiesterases (PDEs), and such a mechanism for one phosphodiesterase, PDE7A1, has been reported. In this paper, we extend this mechanism to another isozyme variant of the same PDE family, PDE7A3. We also report the full-length sequence of human PDE8A1 and show that it also is induced in response to a combination of T cell receptor and costimulatory receptor pathway activation. However, the time course for induction of PDE8A1 is slower than that of PDE7A1. The basal level measured and, therefore, the apparent fold induction of PDE7A1 mRNA and protein depend in large part on the method of isolation of the T cells. On the other hand, regardless of the isolation method, the basal levels of PDE7A3 and PDE8A1 are very low and fold activation is much higher. Constitutively expressed PDE8A1 and PDE7A3 also have been isolated from a human T cell line, Hut78.
Full activation of CD4+ T cells requires engagement of both the TCR-CD3 complex and a costimulatory receptor, such as CD28 (1). In vitro, costimulation by antibodies against CD3 and CD28 immobilized on plates can mimic the normal antigen presenting cell stimulation and lead to full activation of CD4+ T cells (2). In some situations, activation of the TCR-CD3 complex alone in the absence of CD28 costimulation can result in T cell anergy or death (3). The detailed mechanism by which this occurs is not thoroughly understood but may occur in part as a consequence of unrestrained activity of the cAMP-dependent protein kinase (PKA) pathway in the absence of costimulation. For example, activation of PKA is known to block the MAP kinase-dependent pathways necessary for IL-2 production and T cell proliferation. Therefore, agents that increase cAMP levels in T cells will diminish the T cell activation response (4) and, conversely, agents that reduce cAMP permit optimal activation of the T cell response (5–7).
The level of cAMP in T cells is controlled in large part by cAMP-specific phosphodiesterases that catalyze the hydrolysis of cAMP to 5′ AMP, thereby lowering cAMP levels in the cell. Therefore, it makes sense that cAMP phosphodiesterase (PDE) protein and activity levels should be regulated in response to appropriate T cell activation signals. Conversely, inhibition of the PDEs that are responsible for controlling the levels of cAMP in the T cell should inhibit IL-2 production and T cell proliferation. We have shown previously that this appears to be the case with PDE7A1. PDE7A1 is up-regulated in CD4+ T cells after CD3 and CD28 stimulation, and inhibition of PDE7A1 leads to inhibition of proliferation and IL-2 production (8).
Currently, 11 major families of PDEs (PDEs 1–11), subdivided on the basis of substrate specificity, kinetic properties, inhibitor profiles, and sequence homology (9–13), have been identified. PDEs 3B, 4A, 4B, 4D, and 7A1 (14–16) are reported in human T cells. Many of these PDEs also are expressed in other inflammatory cell types. As a result, there has been considerable interest in the development of specific phosphodiesterase inhibitors for the treatment of inflammatory diseases. To date, most work has centered on PDE4 inhibitors because PDE4 represents a major isozyme in most T cell preparations (16, 17). However, in vivo, a major drawback has been the significant side effect of emesis seen with all PDE4 inhibitors tested so far (18). To design more efficacious, less toxic inhibitors, it is important to know all of the PDEs that are present in T cells and also which processes they control. In this paper, we identify the major form of PDE8A1 present in human T cells and report its full-length sequence. We also demonstrate that both PDE8A1 and a new PDE7A variant, PDE7A3, are up-regulated in CD4+ T cells after stimulation. Both of these PDEs are cAMP-specific and may provide additional targets for therapeutic intervention.
Materials and Methods
Materials.
Human white blood cell “buffy coats” were obtained from the American Red Cross Blood Bank (Portland, OR). The CD4+ T cell isolation kit (containing antibodies to CD8, CD11b, CD16, CD19, CD36, and CD56), CD69 microbeads, and goat anti-mouse IgG microbeads were from Miltenyi Biotec (Auburn, CA). The anti-CD8 (G10–1), CD16 (FC2), CD20 (2H7), CD25 (7G7), and HLADR (HB10a) mouse mAbs were provided by E. A. Clark (19), and the anti-CD3 and CD28 mAbs were purchased from PharMingen. The human T cell line Hut78 was obtained from American Type Culture Collection. PDE7A1 (P5H7) and PDE8A1 (P4G7) mAbs were obtained from hybridoma cell lines developed by injecting mice with a glutathione S-transferase fusion of the C-terminal 100 residues of PDE7A1 or a thioredoxin fusion of the PAS [Per, ARNT, Sim (10)] domain of PDE8A1, respectively. The PDE8A1 peptide antibody was a gift from N. Robas at Pfizer Central Research (Sandwich, U.K.) and was specific for the N terminus (PIL9: MGCAPSIHTSENRTF) of mouse PDE8A1. The PDE7A3 peptide polyclonal antibody was obtained from Genemed Biotechnologies (South San Francisco, CA) and is specific for the C terminus (6976: QIGNYTYLDIAG) of this enzyme.
CD4+ T Cell Preparation.
CD4+ T cells were isolated from a human buffy coat (50 ml) obtained from one donor as described previously (19). CD4+ T cells usually were isolated by using a mixture of mAbs specific for various white cell marker proteins (CD8, CD16, CD20, CD25, and HLADR) and goat anti-mouse IgG conjugated to magnetic beads. In some cases, a CD4+ T cell isolation kit (Miltenyi Biotec) was used in combination with CD69 microbeads (Miltenyi Biotec) to remove activated T cells. The antibody-labeled cells were removed by passage through a CS column (Miltenyi Biotec) placed in a magnetic field. The CD4+ T cells passing through the column were at least 95% pure as determined by FACS analysis. The cells were suspended in RPMI 1640 medium (GIBCO/BRL) containing 10% FBS, 0.1 units/ml penicillin, 0.1 μg/ml streptomycin sulfate, and 0.29 mg/ml l-glutamine. The cells were stimulated as follows. Plates (Corning) were precoated with goat anti-mouse IgG (10 μg/ml) for 2 h at 37°C and then washed with PBS. Cells were added to the plate together with CD3 (0.01 μg/ml) and CD28 (0.1 μg/ml) mAbs and were harvested at various time points.
PDE8A1 and PDE7A3 Sequence Determination.
The complete N terminus of human PDE8A1 was obtained as follows. The previously published truncated human PDE8A1 sequence (20) was extended to residue G47 (Fig. 1A) by performing 5′ rapid amplification of cDNA ends (RACE) on a preparation of mRNA from 16-h-stimulated CD4+ T cells. An expressed sequence tag clone from human kidney (AI474074) was purchased from Genome Systems (St. Louis) and sequenced. This clone contained a complete N terminus that overlapped with both the published sequence and the RACE-determined sequence. Expression in T cells of the C terminus of PDE8A was confirmed by 3′ RACE.
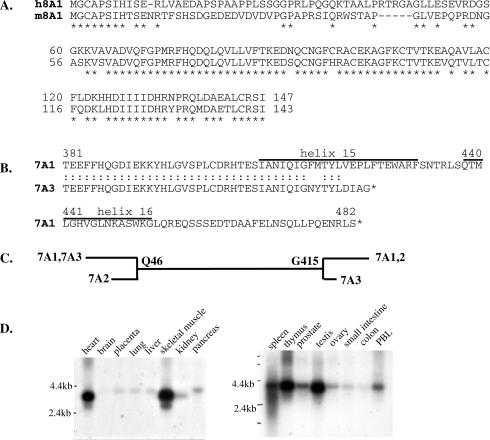
Figure 1.
(A) The complete N-terminal sequence of human PDE8A1 is aligned with the mouse PDE8A1 sequence. (B) Sequence of a new splice variant, PDE7A3. The C termini of PDE7A1 and PDE7A3 are aligned. The numbering refers to PDE7A1 sequence. The positions of helices 15 and 16 are shown. (C) Comparison of the splice variants of PDE7A. (D) Northern blot analysis of PDE7A. The Northern blots (CLONTECH human I and II) were probed with a 1,200-bp PDE7A probe.
The PDE7A3 sequence (Fig. 1B) was obtained by performing RACE on a preparation of mRNA from 16-h-stimulated CD4+ T cells. The new sequence information was obtained with 3′ RACE because PDE7A3 is a C-terminal splice variant. Because we only obtained PDE7A1 N-terminal sequence by using 5′ RACE, we confirmed that this belonged to the PDE7A3 C terminus by reverse transcription–PCR (RT-PCR), which amplified the whole PDE7A3 sequence.
Both 3′ and 5′ RACE were performed by using the SMART RACE cDNA amplification kit (CLONTECH) and a pair of nested gene-specific primers. RACE PCR products were cloned into a pCRII-TOPO vector (Invitrogen) and sequenced.
Northern Blot Analysis.
Northern blots (human I and human II) were purchased from CLONTECH. The probe for PDE7A3 was synthesized by performing RT-PCR on mRNA isolated from a 16-h-stimulated preparation of CD4+ T cells. The 1,200-bp fragment was used as a template for probe synthesis with [α-32P]ATP and exonuclease-free Klenow fragment. The probe was purified on a Centrisep column (Princeton Separations, Adelphia, NJ) and hybridized to the membrane according to the manufacturer's protocol.
RT-PCR Analysis.
RNA was isolated from the cytoplasm of CD4+ T cells by using the Qiagen RNeasy kit, and cDNA was synthesized by using the Promega reverse transcription system. PCR was performed by using 1 μl of undiluted or serially diluted cDNA (1:10 and 1:100 dilutions) and gene-specific primers for 35 cycles (94°C, 1 min; 55°C, 1 min; 72°C, 2 min). The primers used had the following sequences: 7A1, GATATTTGTAACCCATGTCGGACG and GAAAGCTTGGCGGTACTCTACGAT; 7A3, ACGGAGGAATTCTTCCATCAAGGAGAT and AGCTTCCACATGAGCGAATAATGGATT; 8A1, GTAATGCCTTTCAATTCTGCTGGATTTACA and ACGAGTGTCAGACTGAACACATTCGGATAT. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH)-specific primers were used as a control for equivalent cDNA levels.
Western Blot Analysis.
CD4+ T cells (5 × 106) were stimulated and harvested at various time points by centrifugation. The samples were run on SDS gels (8% polyacrylamide) and blotted onto poly(vinylidene difluoride) membrane. Membranes were blocked with blocking buffer (10 mM Tris, pH 7.5/100 mM NaCl/0.2% Tween 20/3% nonfat milk). PDE7A and PDE8A primary antibodies and the appropriate secondary antibody each was incubated with the blot for 1 h. To avoid problems in detecting PDE7A1, which migrates in SDS/PAGE very near to the heavy chains of the stimulating CD3 and CD28 antibodies, the mAb to PDE7A1, P5H7, was conjugated to biotin. Bands were visualized with streptavidin-conjugated horseradish peroxidase. An alternative procedure was to use the unconjugated mAb for PDE7A1 and a goat anti-mouse-κ-horseradish peroxidase secondary antibody. PDE8A1 was visualized by the mAb P4G7 or the polyclonal antibody PIL9. PDE7A3 was visualized by the polyclonal antibody 6976. Blots were developed by using SuperSignal Chemiluminescent substrate (Pierce) and by exposure to x-ray film.
Mono Q Chromatography.
Hut78 cells (2 × 108) were resuspended in 20 mM Tris (pH 7.5) buffer and protease inhibitor mixture (Complete, Mini, EDTA-free; Boehringer Mannheim) and sonicated. The supernatant was removed after centrifugation at 15,000 × g for 10 min and applied to a Mono Q column attached to a Rainin Dynamax HPLC system. The column was developed with a NaCl gradient (0–0.8 M at 0.5 ml/min), and 250-μl fractions were collected. The fractions were assayed for activity as described (21) by using either 1 μM or 0.01 μM cAMP as substrate and the indicated concentration of inhibitor. Western blot analysis also was performed (10 μl/well) by using PDE7A1, PDE7A3, or 8A1 antibodies.
Results
PDE8A1 and PDE7A3 Are Present in CD4+ T Cells.
To understand the roles that phosphodiesterases play in CD4+ T cell activation, we set out to determine whether PDEs that previously have not been reported were present in T cells and whether or not their levels were regulated. As an initial approach we performed preliminary RT-PCR experiments with combinations of primers for all known phosphodiesterases. Positive results then were followed up by using RACE techniques. For example, using a preparation of mRNA from highly enriched CD4+ T cells that had been activated by CD3 and CD28 costimulation, we performed RACE by using primers designed from known PDE8A sequence (20). Sequence analysis of the RACE product indicated that a PDE8A similar or identical to known PDE8A sequence (20) indeed was present in CD4+ T cells. As a positive control, we also used RACE techniques to amplify PDE7A1 because this PDE is known to be expressed in CD4+ T cells (16). As expected, we isolated products corresponding to PDE7A1 but also amplified what appears to be a previously unknown splice variant of PDE7A, which we are calling HSPDE7A3.
Sequence analysis of the 3′ RACE product of PDE8A confirmed that the sequence amplified was the same as contained in the previously published truncated sequence for human PDE8A1 isolated from human testis and stomach cDNA libraries (20). Although the 5′ end of this cDNA is very GC-rich, we were able to extend this sequence by 5′ RACE to position G47 (Fig. 1A). However, we were not able to obtain further sequence 5′ of G47 by RACE probably because of the very high GC content of this region. Fortunately, using a database search, we noted an expressed sequence tag clone, AI474074, from human kidney that appeared to contain the complete N terminus of human PDE8A1 (by analogy to the published mouse sequence). Moreover, upon sequencing of this expressed sequence tag clone, we found that the sequence overlapped both the sequence of the T cell PDE8A RACE product and part of the published human PDE8A sequence (i.e., the sequence extends to residue I147; numbering as shown in Fig. 1A). This sequence is highly homologous to the full-length N terminus of the published mouse PDE8A sequence (10) (Fig. 1A) but does contain a stretch of about 50 residues at which the sequence diverges from the mouse homolog. This may be a species difference or may indicate a slightly different splice variant. Because to date no alternate PDE8A N-terminal sequences have appeared in the public database nor have they been identified by RACE reactions, we are tentatively calling the cDNA identified in Fig. 1A HSPDE8A1. In addition, we have confirmed that this N terminus of PDE8A1 is present in commercial cDNA isolated from human testis and human leukocyte cells by sequencing RT-PCR products from these tissues. Also, Western analysis using an antibody (PIL9) made to the highly homologous N terminus of mouse PDE8A1 (MGCAPSIHTSENRTF) reacts strongly with a band of appropriate size in extracts of the human T cell line, Hut78, and CD4+ T cells. The RACE reactions with PDE7A primers also identified what appeared to be a new splice variant for this enzyme. Fig. 1B shows a C-terminal alignment between PDE7A1 and the new splice variant, PDE7A3. The new sequence diverges at position G415 (PDE7A1 numbering). PDE7A3 has a short C-terminal tail that differs from PDE7A1 sequence and is truncated at residue 424. Fig. 1C shows the relationship of PDE7A3 to the other PDE7A splice variants. To confirm the 5′ end of this isozyme, we amplified the entire sequence by RT-PCR. A product was obtained that has the same 5′ end as PDE7A1, indicating that the PCR product produced in T cells has the 3′ sequence of PDE7A3 and N-terminal sequence common to PDE7A1 and PDE7A3. Fig. 1D shows the results of a Northern blot by using a PDE7A probe. This probe is able to react with all splice variants of PDE7A because of the large amount of common sequence that they share. As expected, using the PDE7A probe, the relatively abundant mRNA for PDE7A1 with a transcript size of 4.2 kb was seen in most tissues. The other previously known form of PDE7A, PDE7A2, is highly expressed in skeletal muscle and heart and has a transcript size of 3.8 kb (22). It is likely that the fainter band at 3.0 kb seen in heart and skeletal muscle mRNA corresponds to the PDE7A3 transcript. Faint bands at 3.0 kb are also seen in spleen, thymus, testis, and peripheral blood leukocytes. We have been unable to do Northern analysis specific only for PDE7A3. Probes specific for the unique C terminus of PDE7A3 gave no signal because of their short length, and probes designed against the 3′ untranslated region gave a very unclean signal. To confirm that PDE7A3 mRNA indeed is present in the cells containing the 3.0-kb band, we sequenced RT-PCR products from testis, skeletal muscle, and leukocytes and obtained PDE7A3 sequence.
Up-Regulation of PDE7A1 and PDE8A1 mRNA and Protein.
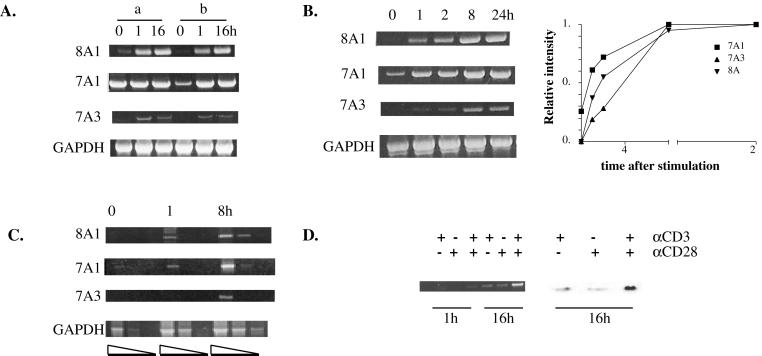
We have reported previously that PDE7A1 is induced when CD4+ T cells are activated with plate-bound CD3 and CD28 antibodies. Activation of T cells causes them to proliferate and produce IL-2. Moreover, inhibition of PDE7A1 expression with an antisense oligonucleotide inhibited proliferation and IL-2 production (8). We now find by using a larger number of different donors that a variable basal level of PDE7A1 can be present in unstimulated CD4+ T cells but that this basal level depends greatly on the donor as well as the method of preparation of CD4+ T cells. Fig. 2A compares two methods used to prepare CD4+ T cells. A commonly used method is negative selection by using a commercial CD4+ T cell isolation kit (Miltenyi Biotec) in combination with CD69 microbeads to remove activated cells (method a, Fig. 2A). Cells isolated in this manner show little induction of PDE7A1, as a high basal level of PDE7A1 is present in the unstimulated cells. This result did not agree with our earlier studies in which we used a different set of mAbs, also in a negative selection protocol. As shown in Fig. 2A, when this earlier method that utilizes a mixture of anti-CD8, CD16, CD20, CD25, and HLADR antibodies is used (method b) along with mouse magnetic beads, a much lower basal value for PDE7A1 is found.
Figure 2.
Up-regulation of PDE7A1, PDE7A3, and PDE8A1 mRNA in CD4+ T cells. CD4+ T cells were stimulated with CD3 and CD28 antibodies and harvested at the indicated time points (hours). RT-PCR analysis was performed as described in Materials and Methods. (A) Comparison of methods of preparation of CD4+ T cells. Cells were prepared as described in Materials and Methods by using either the CD4+ T cell isolation kit in combination with the CD69 microbeads (a) or a mixture of mAbs and goat anti-mouse magnetic beads (b). (B) Time course of the induction of PDE7A1, PDE7A3, and PDE8A1 mRNA compared with a GAPDH control. The bands were scanned and quantified by using nih image. Within each image, the maximum intensity of the band was set to a value of 1 and other bands were calculated as a fraction of the maximum. The values were normalized to the GAPDH signal. The results of the densitometry are graphed. (C) Time course of induction of PDE7A1, PDE7A3, and PDE8A1 by using serially diluted cDNA as described in Materials and Methods. (D) RT-PCR was performed for PDE8A1 from cells harvested at 1 and 16 h after stimulation by using either CD3, CD28, or a combination of the antibodies (Left). Cells were harvested 16 h after stimulation and analyzed by Western blotting by using a PDE8A1 polyclonal antibody (PIL9) (Right).
Using method b, we wanted to compare the regulation of PDE7A1 with the newly discovered PDE8A1. The time course data shown in Fig. 2B indicate that PDE7A1 is up-regulated relatively quickly, with a distinct difference in levels detectable as early as 1 h after stimulation. PDE8A1, conversely, initially is absent and does not reach a maximum until at least 8 h. The amounts of PCR product shown in the time course of Fig. 2B are probably good, quantitative estimates of the mRNA levels because, as shown in Fig. 2C, differing dilutions of cDNA demonstrate that the signal is not saturated under the conditions used. Also, the PCR products in Fig. 2B were quantified and normalized to the GAPDH signal. The results shown in Fig. 2B Right confirm that there is an increase in intensity of the PCR products. Because it has been shown previously that costimulation of CD4+ T cells is required to achieve maximal mRNA and protein levels of PDE7A1 (8), we analyzed the effect of antibodies to CD3 alone and CD28 alone and both in combination on PDE8A1 expression. Fig. 2D shows that PDE8A1 induction also requires both CD3 stimulation and CD28 costimulation for maximal expression of mRNA and protein.
To determine whether the amount of PDE protein expressed also was increased concomitant with the mRNA, Western blot analysis of T cell extracts was performed before and after stimulation. The results of these experiments confirmed the up-regulation of PDE7A1 protein and showed that PDE8A1 protein was increased after stimulation (Fig. 3). Similar to the PCR results, PDE7A1 protein is present in the unstimulated cells and becomes up-regulated, reaching a maximum at about 8 h (Fig. 3, top level). PDE8A1 protein initially is absent and reaches a maximum at about 16 h. The PDE7A1 band migrates at 55 kDa, and the PDE8A1 band migrates at ≈100 kDa, similar to the predicted molecular mass of 93 kDa.
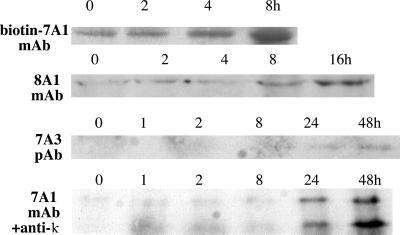
Figure 3.
Western blot analysis of CD4+ T cells shows up-regulation of PDE7A1, PDE7A3, and PDE8A1 proteins. Cells were harvested at various time points and analyzed by Western blotting as described in Materials and Methods. The bottom blot is developed with PDE7A1 mAb and goat anti-mouse-κ-horseradish peroxidase secondary antibody and shows both PDE7A1 (upper band) and PDE7A3 (lower band).
PDE7A3 Is Up-Regulated in CD4+ T Cells.
We also compared the regulation of PDE7A1 expression with the expression of the new splice variant of PDE7A, PDE7A3. Like PDE7A1, both PDE7A3 mRNA (Fig. 2) and protein (Fig. 3) are up-regulated in CD4+ T cells after stimulation, although, like PDE8A1, PDE7A3 initially is absent and only reaches a maximum at about 24–48 h. The band reacting with a PDE7A3-specific polyclonal antibody (Fig. 3) migrates at ≈50 kDa. This is close to the predicted molecular mass of 48.8 kDa. Western blot analysis also was performed with a PDE7A1 mAb directed toward the C-terminal 100 residues of the protein. This antibody was able to react with both splice variants, indicating that they share a common epitope. To avoid interference with the antibody heavy chains originating from the stimulating CD3 and CD28 antibodies, the PDE7A1 mAb was either biotin-conjugated and visualized with streptavidin-conjugated horseradish peroxidase or was left unconjugated and visualized with an anti-κ light chain secondary antibody. Using the latter method, the bands were very faint because of the lower level of amplification of the antibody signal and showed up substantially only in the 24-h lane. The PDE7A1 blot (bottom level of Fig. 3) showed that two bands were up-regulated. The top band migrates at the position of PDE7A1, 55 kDa, whereas the bottom band migrates at the position of PDE7A3, 50 kDa. The biotin-7A1 blot (top level of Fig. 3) shows that at earlier time points there is up-regulation of PDE7A1 protein, although PDE7A3 is absent.
Activity of PDE8A1 and PDE7A3 in T Cells.
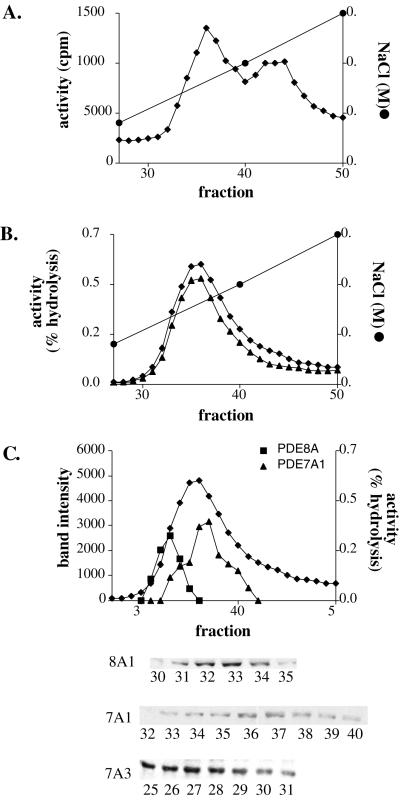
Because we were able to obtain only a limited number of pure CD4+ T cells from a single donor, a human CD4+ T cell line, Hut78, was used to study the activities of PDE8A1 and PDE7A3. Hut78 cells are thought to be analogous to activated CD4+ T cells because they proliferate and produce IL-2. Moreover, PDE7A1 is known to be expressed constitutively in these cells (23). Hut78 cell extract was fractionated by passage through a Mono Q column attached to a Rainin HPLC system. Fig. 4A shows that two main peaks of PDE activity are resolved with 1 μM cAMP as substrate. It has been shown previously that these two peaks contain PDE7 and PDE4 activity, respectively (23, 24). Fig. 4B shows the results from an assay using 0.01 μM cAMP. Under these conditions, only a single peak is seen. At this low concentration, there is negligible contribution to activity by PDE4 and only PDEs having very low Km levels are detected. As expected from the fact that this peak should contain PDE7, the activity of the fractions was resistant to the PDE4-specific inhibitor rolipram. However, Western blot analysis across the peak showed that a PDE8A1-immunoreactive band of the predicted size eluted in the front part of the peak and PDE7A1 eluted in the back part of the peak (Fig. 4C). Therefore, the activity in this peak is contributed by at least two phosphodiesterases, PDE7A1 and PDE8A1.
Figure 4.
Mono Q HPLC profile of the Hut78 cell extract. (A) PDE activity of a Hut78 cell extract by using 1 μM cAMP as a substrate. (B) PDE activity profile by using 0.01 μM cAMP as substrate in the absence (♦) or presence of 10 μM rolipram (▴). (C) The PDE activity profile (0.01 μM cAMP, ♦) overlaid with the band intensities (in arbitrary units) of PDE7A1 (▴) or 8A1(■) from the Western blots in the Inset below. The PDE7A3 band eluted in a region with low activity.
Western blot analysis also showed that PDE7A3 eluted in a part of the HPLC profile that had very low activity (Fig. 4C). A band at about 50 kDa centered at fraction 27 was detected with both the PDE7A mAb and with the PDE7A3 peptide antibody. However, these fractions show very little cAMP PDE activity. We also have expressed large amounts of soluble recombinant PDE7A3 protein in Sf9 cells but still are able to detect little or no activity above basal in extracts from these cells.
Discussion
To understand the role(s) played by cyclic nucleotide phosphodiesterases in T cell regulation, it is clearly essential to know which ones are expressed in the T cells under different physiological conditions. This paper describes the full-length sequence of human PDE8A1 and also the full sequence of a new splice variant of PDE7A, PDE7A3. Importantly, both are shown to be present only in activated T cells. The PDE8A1 sequence reported appears to be full-length because there is an in-frame stop codon upstream from the start methionine (accession number AF332653). Although we have not been able to find an in-frame upstream stop codon for PDE7A3, we believe that the sequence also is likely to be full-length because the protein bands for both PDE7A1 and PDE7A3 detected by Western blot analysis are the same size as predicted by the cDNA sequence. Confirmation of the new sequence information was obtained from the results of a blast search of the genomic database. The gene for human PDE7A is known to be on chromosome 8q13 (25), and the new PDE7A3 C-terminal and 3′ untranslated regions map to sequence AC055822 on chromosome 8. The sequence from residues G388 to G424 and the entire 3′ untranslated sequence are found on a single exon. Currently, there is no genomic sequence available for the 5′ end of PDE7A1 or PDE7A3; therefore, we do not know whether they share a single promoter. It is possible that the apparent difference in time course for the induction of the two transcripts may be a matter of different RNA stabilities or of RNA splicing. Similarly, we found that the N terminus of PDE8A1 maps to sequence AC018732 and is found on chromosome 15. The 5′ untranslated sequence and the sequence from residues M1 to V63 are found on a single exon.
The existence of another splice variant for PDE7A in CD4+ T cells had not been reported previously and was not anticipated because Northern blot analysis of total leukocyte mRNA had shown a signal only at 4.2 kb, the size of PDE7A1 (8). A similar result using a PDE7A3 probe is shown in Fig. 1D. In retrospect, it is likely that the commercial blots used for these experiments were made from RNA that was isolated from unstimulated cells and, therefore, the PDE7A3 mRNA levels were below the limits of detection by Northern analysis. Presumably, PDE8A1 had not been identified previously in CD4+ T cells for similar reasons (20).
We have shown previously that PDE7A1 is up-regulated in human CD4+ T cells (8) and now show that PDE8A1 and PDE7A3 also are up-regulated in a similar manner. However, we also find that the method of isolation of the T cells can affect this result, at least for PDE7A1. Clearly, from the data in Fig. 2A, the second method of preparation (method b) shows a higher fold induction of PDE7A1. However, for either method, a strong induction of PDE7A3 and PDE8A1 is seen. It is possible that the latter method removes a subset of activated T cells that contains PDE7A1. This method utilizes an antibody to HLADR that is thought to remove most cells containing MHCII, a marker of activation. Similarly, the antibody to CD25 removes cells expressing IL-2 receptor, another marker of activation. Presumably, therefore, PDE7A1 is low only in the resulting resting T cell population. In fact, a commercial kit for separation of resting CD4+ T cells has become available recently that contains antibodies to CD25 and HLADR just for the purpose of removing a greater percentage of activated cells (26).
In CD4+ T cells, mRNA levels for PDE7A3 and PDE8A1 become maximally up-regulated by about 8 h after CD3 and CD28 stimulation (Fig. 2B). As predicted from the increased mRNA, the protein levels of PDE7A3 and PDE8A1 also were up-regulated (Fig. 3).
Because of the difficulty in obtaining large numbers of pure CD4+ T cells, the human T cell line, Hut78, was used to characterize the activities of these new PDEs. Because PDE7A1 is known to be constitutively expressed in these cells (23), we assayed Mono-Q-fractionated HPLC fractions with a low concentration of substrate to look for activities that might correspond to PDE7A3 and PDE8A1. PDE7A1 and PDE8A1 have been shown previously to have a Km of 0.2 μM (27) and 55 nM (20). Therefore, by using a very low substrate level, we were able to minimize the contribution of PDE4 activity, which also is known to be expressed in Hut78 cells but has a much higher Km. In confirmation of previous studies (23, 24), two peaks of activity are found when assayed at 1.0 μM cAMP (Fig. 4A). However, the data from assays conducted at 0.01 μM cAMP (Fig. 4B) indicate that only one peak of activity is present that corresponds in position to the first peak as assayed with 1 μM cAMP (Fig. 4A). Further analysis of the HPLC profiles with antibodies to each of the PDEs indicates that this peak actually is composed of a combination of the activities of two PDEs, PDE8A1 and PDE7A1 (Fig. 4C).
Because PDE7A3 has a nearly identical sequence in the catalytic domain, it was expected to have similar activity and affinity for substrate as PDE7A1. Unexpectedly, the PDE7A3-reactive band from Hut78 cells eluted in a part of the HPLC profile that had low activity (Fig. 4C). Similarly, when expressed in Sf9 cells, only PDE7A1 but not PDE7A3 showed measurable activity. Like the Hut78 fractions, the recombinant PDE7A3 had essentially no activity compared with PDE7A1. It is possible that a specific activator for PDE7A3 is missing in our assays. However, it is also possible that a required sequence in that catalytic domain is missing. Because a high-resolution structure for PDE4B2B has been solved (28), we aligned the sequence of PDE4B2B with the PDE7A sequences. We noted that PDE7A1 contains all of the homologous helices found in the catalytic domain crystal structure, whereas PDE7A3 is missing half of helix 15 and all of helix 16 (Fig. 1B). Although this region does not contain residues previously believed to be important for catalysis (based on conserved residues in comparative sequence alignments), this missing sequence may well be important structurally. We currently are investigating the role of these helices in phosphodiesterase function as well as the role of PDE7A3 in T cell function. If this is so and PDE7A3 is intrinsically inactive, then clearly more attention needs to be given to the reason why this polypeptide is induced in response to T cell activation.
Finally, we have shown previously that inhibition of PDE7A1 induction with an antisense oligonucleotide specific for the N terminus of PDE7A inhibits T cell proliferation and IL-2 expression. In these studies, the mRNA and protein levels of PDE7A1 were reduced by the oligonucleotide (8). Because the N termini of PDE7A1 and PDE7A3 are identical, we would expect that the mRNA levels of PDE7A3 also would be reduced after treatment with the antisense oligonucleotide, and, indeed, this appears to be the case (data not shown). Therefore, it is possible that part of the inhibition of proliferation and IL-2 production seen in earlier studies was a result of inhibition of not only PDE7A1 but also PDE7A3 message.
Because there are a number of PDEs known to be expressed in T cells, it is possible that each PDE controls a separate pool of cAMP and, thereby, a separate pathway. An important area of further study will be to inhibit the various phosphodiesterases present in T cells either by specific inhibitors or antisense oligonucleotides and analyze the effect on cellular function.
Acknowledgments
This research is supported by National Institutes of Health Grant DK21723 and grants from the Ono Pharmaceutical Company (Osaka, Japan) and Pfizer Central Research (Sandwich, U.K.).
Abbreviations
- PDE
phosphodiesterase
- RACE
rapid amplification of cDNA ends
- RT-PCR
reverse transcription–PCR
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
Footnotes
References
- 1.Shahinian A, Pfeffer K, Lee K P, Kundig T M, Kishihara K, Wakeham A, Kawai K, Ohashi P S, Thompson C B, Mak T W. Science. 1993;261:609–612. doi: 10.1126/science.7688139. [DOI] [PubMed] [Google Scholar]
- 2.Baroja M L, Lorre K, Van Vaeck F, Ceuppens J L. Cell Immunol. 1989;120:205–217. doi: 10.1016/0008-8749(89)90188-3. [DOI] [PubMed] [Google Scholar]
- 3.Schwartz R H. Science. 1990;248:1349–1356. doi: 10.1126/science.2113314. [DOI] [PubMed] [Google Scholar]
- 4.Kvanta A, Gerwins P, Jondal M, Fredholm B B. Cell Signalling. 1990;2:461–470. doi: 10.1016/0898-6568(90)90042-9. [DOI] [PubMed] [Google Scholar]
- 5.Estes G, Solomon S S, Norton W L. J Immunol. 1971;107:1489–1492. [PubMed] [Google Scholar]
- 6.Skalhegg B S, Landmark B F, Doskeland S O, Hansson V, Lea T, Jahnsen T. J Biol Chem. 1992;267:15707–15714. [PubMed] [Google Scholar]
- 7.Lin J, Gettys T W, Qin L, Chavin K D, Yang Q, Ding Y, Punch J D, Bromberg J S. Pathobiology. 1995;63:175–187. doi: 10.1159/000163949. [DOI] [PubMed] [Google Scholar]
- 8.Li L, Yee C, Beavo J A. Science. 1999;283:848–851. doi: 10.1126/science.283.5403.848. [DOI] [PubMed] [Google Scholar]
- 9.Beavo J A. Physiol Rev. 1995;75:725–748. doi: 10.1152/physrev.1995.75.4.725. [DOI] [PubMed] [Google Scholar]
- 10.Soderling S H, Bayuga S J, Beavo J A. Proc Natl Acad Sci USA. 1998;95:8991–8996. doi: 10.1073/pnas.95.15.8991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Soderling S H, Bayuga S J, Beavo J A. J Biol Chem. 1998;273:15553–15558. doi: 10.1074/jbc.273.25.15553. [DOI] [PubMed] [Google Scholar]
- 12.Soderling S H, Bayuga S J, Beavo J A. Proc Natl Acad Sci USA. 1999;96:7071–7076. doi: 10.1073/pnas.96.12.7071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fawcett L, Baxendale R, Stacey P, McGrouther C, Harrow I, Soderling S, Hetman J, Beavo J A, Phillips S C. Proc Natl Acad Sci USA. 2000;97:3702–3707. doi: 10.1073/pnas.050585197. . (First Published March 21, 2000; 10.1073/pnas.050585197) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ekholm D, Hemmer B, Gao G, Vergelli M, Martin R, Manganiello V. J Immunol. 1997;159:1520–1529. [PubMed] [Google Scholar]
- 15.Erdogan S, Houslay M D. Biochem J. 1997;321:165–175. doi: 10.1042/bj3210165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Giembycz M A, Corrigan C J, Seybold J, Newton R, Barnes P J. Br J Pharmacol. 1996;118:1945–1958. doi: 10.1111/j.1476-5381.1996.tb15629.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Manning C D, Burman M, Christensen S B, Cieslinski L B, Essayan D M, Grous M, Torphy T J, Barnette M S. Br J Pharmacol. 1999;128:1393–1398. doi: 10.1038/sj.bjp.0702911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barnette M S, Bartus J O, Burman M, Christensen S B, Cieslinski L B, Esser K M, Prabhakar U S, Rush J A, Torphy T J. Biochem Pharmacol. 1996;51:949–956. doi: 10.1016/0006-2952(96)00053-6. [DOI] [PubMed] [Google Scholar]
- 19.Sun Y, Pinchuk L M, Agy M B, Clark E A. J Immunol. 1997;158:512–517. [PubMed] [Google Scholar]
- 20.Fisher D A, Smith J F, Pillar J S, St. Denis S H, Cheng J B. Biochem Biophys Res Commun. 1998;246:570–577. doi: 10.1006/bbrc.1998.8684. [DOI] [PubMed] [Google Scholar]
- 21.Hansen R S, Beavo J A. Proc Natl Acad Sci USA. 1982;79:2788–2792. doi: 10.1073/pnas.79.9.2788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Han P, Zhu X Y, Michaeli T. J Biol Chem. 1997;272:16152–16157. doi: 10.1074/jbc.272.26.16152. [DOI] [PubMed] [Google Scholar]
- 23.Bloom T J, Beavo J A. Proc Natl Acad Sci USA. 1996;93:14188–14192. doi: 10.1073/pnas.93.24.14188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ichimura M, Kase H. Biochem Biophys Res Commun. 1993;193:985–990. doi: 10.1006/bbrc.1993.1722. [DOI] [PubMed] [Google Scholar]
- 25.Han P, Fletcher C F, Copeland N G, Jenkins N A, Yaremko L M, Michaeli T. Genomics. 1998;48:275–276. doi: 10.1006/geno.1997.5168. [DOI] [PubMed] [Google Scholar]
- 26.Chun T W, Engel D, Berrey M M, Shea T, Corey L, Fauci A S. Proc Natl Acad Sci USA. 1998;95:8869–8873. doi: 10.1073/pnas.95.15.8869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Michaeli T, Bloom T J, Martins T, Loughney K, Ferguson K, Riggs M, Rodgers L, Beavo J A, Wigler M. J Biol Chem. 1993;268:12925–12932. [PubMed] [Google Scholar]
- 28.Xu R X, Hassell A M, Vanderwall D, Lambert M H, Holmes W D, Luther M A, Rocque W J, Milburn M V, Zhao Y, Ke H, et al. Science. 2000;288:1822–1825. doi: 10.1126/science.288.5472.1822. [DOI] [PubMed] [Google Scholar]