Abstract
The nuclear hormone receptor estrogen receptor α (ERα) mediates the actions of estrogens in target cells and is a master regulator of the gene expression and proliferative programs of breast cancer cells. The presence of ERα in breast cancer cells is crucial for the effectiveness of endocrine therapies, and its loss is a hallmark of endocrine-insensitive breast tumors. However, the molecular mechanisms underlying the regulation of the cellular levels of ERα are not fully understood. Our findings reveal a unique cellular pathway involving the p38 mitogen-activated protein kinase (p38MAPK)-mediated phosphorylation of ERα at Ser-294 that specifies its turnover by the SCFSkp2 proteasome complex. Consistently, we observed an inverse relationship between ERα and Skp2 or active p38MAPK in breast cancer cell lines and human tumors. ERα regulation by Skp2 was cell cycle stage dependent and critical for promoting the mitogenic effects of estradiol via ERα. Interestingly, by the knockdown of Skp2 or the inhibition of p38MAPK, we restored functional ERα protein levels and the control of gene expression and proliferation by estrogen and antiestrogen in ERα-negative breast cancer cells. Our findings highlight a novel pathway with therapeutic potential for restoring ERα and the responsiveness to endocrine therapy in some endocrine-insensitive ERα-negative breast cancers.
INTRODUCTION
The nuclear hormone receptor estrogen receptor α (ERα) is a master regulator of gene expression and the proliferative program of breast cancer cells (18, 29, 36, 38, 50, 54) and, hence, is the main target of endocrine therapies. Approximately 70% of human breast tumors express ERα and depend on estrogens for growth, rendering these tumors amenable to treatment with drugs such as selective estrogen receptor modulators/antiestrogens (such as tamoxifen) and aromatase inhibitors, which are quite effective and have relatively few side effects. These ERα-targeted therapies (7, 27, 28, 40, 41) have resulted in a steady decline in the rate of mortality due to breast cancer but show effectiveness only against ER-positive breast tumors, while ER-negative tumors fail to respond. The regulation of the cellular level of ERα is therefore key to the effectiveness of endocrine therapies in breast cancer, and an understanding of its underlying mechanism is critical for the identification of novel drug targets for the design of combinatorial therapies.
ERα is unusual among nuclear hormone receptors in being a rapidly turning-over protein with a half-life of ca. 4 h in breast cancer cells and in normal target tissues such as the uterus (2, 16, 39), indicating dynamic regulation by modulating factors. The degradation of ERα, and several other nuclear receptors, has been shown to be under the control of the ubiquitin (Ub) proteasome system (2, 31, 32, 48, 51), yet many important aspects of this regulation remain unclear. In view of the importance of ERα in many target tissues and in breast cancer biology, prognosis, and responses to endocrine treatments, we have investigated the underlying mechanism for the cellular turnover of ERα and identify Skp2 (S-phase kinase-associated protein 2), an F-box protein (FBP), and a substrate recognition component of the SCF ubiquitin ligase complex (10) overexpressed in many cancers, including breast cancer (21, 23, 42–44, 46, 47), as a novel E3-ubiquitin ligase that regulates ubiquitination and the turnover of ERα upon specification by the p38 mitogen-activated protein kinase (p38MAPK)-mediated phosphorylation of the receptor while positively regulating the functional activity of this receptor.
We were intrigued to examine the interrelationships between ERα and Skp2, because in our studies of the estrogen receptor and its coregulators, we observed that ERα and the E3 ubiquitin ligase Skp2 appeared to be inversely correlated. The SCFSkp2 complex is under tight bimodal regulation by the concerted actions of various kinases that modulate its activity by phosphorylating either its components (19, 22, 33) or its target proteins (26). Since there is compelling evidence for the requirement of substrate phosphorylation as a signal for SCFSkp2-mediated protein turnover (57, 58), we have investigated the role of such posttranslational modifications in Skp2-mediated ERα turnover and identify the stress-activated kinase p38MAPK as a critical regulator.
Our work highlights the molecular mechanisms governing ERα turnover and the control of receptor proliferative and gene regulatory activities by the coordinated actions of Skp2 and p38MAPK. The findings further reveal a dynamic inverse relationship between ERα and Skp2 and/or active p38MAPK in human breast tumors and breast cancer cell lines and suggest potential new therapeutic strategies for restoring functional ERα protein in some endocrine-insensitive ERα-negative breast cancer cells.
MATERIALS AND METHODS
Cell cultures, antibodies, and other reagents.
Anti-Skp2 (N-19, H-435, and A-2), anti-ERα (HC-20 and F-10), anti-Ub (P4D1), antihemagglutinin (anti-HA) tag (F-7 and Y-11), and anti-glutathione S-transferase (GST) (Z-5) antibodies and horseradish peroxidase (HRP)-conjugated donkey anti-goat, donkey anti-mouse, and donkey anti-rabbit IgG secondary antibodies were obtained from Santa Cruz Biotechnology. Anti-Myc tag (catalog numbers 2272 and 2276), anti-p38MAPK (catalog numbers 9212 and 9228), anti-phospho-Thr-180/Tyr-182 p38MAPK (catalog numbers 9211 and 9216), anti-ATF2, and anti-phospho-ATF2 antibodies were obtained from Cell Signaling; anti-Flag rabbit and mouse antibodies were obtained from Sigma; and antiphosphoserine antibody (ab17465) was obtained from Abcam. Immunoprecipitations (IPs) were performed with antibodies obtained from Santa Cruz (ERα [HC-20], Skp2 [N-19], and RNA polymerase II [N-20]), Abcam (RNA polymerase II phospho-Ser-5 [ab5131]), and Bethyl Laboratories (p38MAPK [A310-212A]). The p38MAPK inhibitor SB203580 was obtained from Calbiochem.
All cell lines were obtained from the American Type Culture Collection (ATCC) and were maintained in culture as suggested by the ATCC, except for MCF-7 cells, which were grown in minimum essential medium (MEM) with phenol red supplemented with 5% heat-inactivated calf serum and were transferred into MEM without phenol red supplemented with 5% charcoal dextran-stripped calf serum prior to treatment, as described previously (18). Plasmid transfections were performed by use of LipofectAMINE 2000 (Invitrogen).
Plasmids and adenoviral constructs.
Skp2 cDNA (Open Biosystems) was cloned into the pcDNA3-Flag expression vector. Various Skp2 deletion mutants were generated from this full-length construct by PCR-based subcloning. ERα was cloned into the pcDNA3-Flag expression vector and the pcDNA3.1-Myc/His expression vector from Invitrogen. Various ERα deletion mutants were generated from the full-length Myc-tagged construct by PCR-based subcloning. A site-directed mutagenesis kit (Stratagene) was used to introduce various Ser-to-Ala or -Glu point mutations in pcDNA3-Flag-ERα or Skp2 (full length). pCMV-ERα/ERβ chimeras were generated previously in our laboratory (37). pCMV-HA-Ubiquitin was generated by subcloning from a GST-ubiquitin expression vector (Addgene plasmid 10861), originally generated by Peter Howley. pMEV-2HA-p38MAPK-WT (wild type [WT]) and the DN (dominant negative) mutant were obtained from Biomyx Technology. Myc-tagged cullins and HA-tagged Rbx1 and Rbx2 were generated by subcloning from cDNA vectors from Addgene. Adenoviral vectors encoding Skp2-WT or the Skp2-ΔFbox mutant were constructed by cloning relevant sequences into the pAdTrack vector from Stratagene. Details on the generation of these expression vectors are available upon request.
Silencing by small interfering RNA.
Cells at a confluence of 40 to 60% were transfected with the small interfering RNA (siRNA) duplexes siSkp2 (Qiagen); siERα; sip38MAPK; siCul1, -2, -3, -4A, -4B, -5, or -7; siRbx1; siRbx2 (Dharmacon); or nontargeting control siGL-3 (Dharmacon or Qiagen), followed by ligand treatment, as indicated. siSkp2 transfections were performed by use of lipitoid reagent, kindly provided by Ronald Zuckermann at the Molecular Foundry, Lawrence Berkeley National Laboratory (25, 55). All other siRNA transfections were performed by use of Dharmafect transfection reagent (Dharmacon).
Expression analysis by quantitative real-time PCR and Western immunoblotting.
RNA was isolated from cells by using TRIzol reagent (Invitrogen) and the mRNA for genes of interest was quantitated by SYBR green-based reverse transcriptase PCR (RT-PCR), using the ABI7600HT sequence detection system (Applied Biosystems). All mRNA quantities were normalized to 36B4. Primer sequences are available upon request. In parallel experiments, total cellular proteins were harvested by using radioimmunoprecipitation assay (RIPA) buffer, followed by Western immunoblotting for proteins of interest.
In vitro kinase assays.
For in vitro kinase assays, 1 μg of recombinant active phospho-GST-p38MAPK or inactive GST-p38MAPK protein (Invitrogen) was incubated with 1 μl of the in vitro-translated ERα-WT or ERα-S294A protein as the substrate in the presence of 1× kinase buffer (25 mM Tris-HCl [pH 7.5], 5 mM beta-glycerophosphate, 2 mM dithiothreitol [DTT], 0.1 mM Na3VO4, 10 mM MgCl2 [Cell Signaling]) supplemented with 200 μM ATP in a total reaction mixture volume of 50 μl. The reaction mixtures were incubated at 30°C for 30 min for enzymatic catalysis to occur; thereafter, reactions were terminated by the addition of 3× SDS sample buffer to the mixtures, and the mixtures were subjected to Western blotting.
Immunohistochemical analysis of primary human breast tumor tissues.
All protocols involving the use of human breast tumor tissues were approved by the University of Illinois and Carle Foundation Institutional Review Boards. Tissues were fixed in 10% neutral buffered formalin and embedded in paraffin, and sections were cut at a 0.2-μm thickness, dried at 37°C overnight, and stained according to standard methods for microwave antigen retrieval. Sections were incubated for 12 h at 4°C with 1:50-diluted anti-ERα, anti-Skp2, or anti-phospho-p38MAPK antibody or a negative control (0.1% bovine serum albumin [BSA] in 1× phosphate-buffered saline [PBS]). After washing in PBS, sections were incubated with a biotinylated secondary antibody for 1 h at room temperature and then incubated with an avidin-biotin complex (Vector Laboratories) for 30 min. Reactivity was visualized by using diaminobenzidine (DAB) chromogen, and sections were counterstained with Mayer's hematoxylin.
Bimolecular fluorescence complementation assays.
Bimolecular fluorescence complementation (BiFC) was performed as described previously (24). Cos-1 cells were transfected with the YN and YC fusion constructs alone or in combination and were then incubated for 24 h at 37°C in 5% CO2 and humidified air. Cell nuclei were visualized with CFP-NLS (cyan fluorescent protein [CFP] fused to a nuclear localization signal), which was cotransfected along with the YN and YC fusion constructs. Yellow fluorescent protein (YFP) fluorescence was measured by excitation at 513 nm and emission at 527 nm. Fluorescence emissions were observed in live cells by using an inverted phase-contrast microscope.
GST fusion protein purification, in vitro translation, and GST pulldown assays.
The purification of the GST-fused ERα protein from cultures of bacteria grown overnight and GST pulldown assays thereafter were performed as described previously (20). The in vitro translation of Skp2A and Skp2B and the incorporation of 35S-labeled methionine (Perkin-Elmer) were performed by use of the TNT T7-coupled reticulocyte lysate system kit (Promega). Labeled Skp2A or Skp2B and GST-ERα (or GST alone) were incubated with glutathione-Sepharose beads (Amersham) in binding buffer (20 mM Tris [pH 7.5], 50 nM NaCl, 10% glycerol, 10 mM NaF, 1% Nonidet P-40, 1 mM NaVO4, 1× protease inhibitor cocktail [Roche], and 1 mM phenylmethylsulfonyl fluoride [PMSF]) at 4°C for 4 h with gentle rotation. The beads were washed three times with the same buffer. Bound proteins were eluted with Laemmli buffer containing SDS supplemented with β-mercaptoethanol and subjected to 10% SDS-PAGE. The gels were then dried and visualized by autoradiography.
Coimmunoprecipitation assays.
Cos-1 cells seeded into 6-well plates at a confluence of 80 to 90% were transfected with plasmids of interest, followed by treatment with a vehicle (0.1% ethyl alcohol [EtOH]) or ligands, as indicated. At 24 h posttransfection, cells were harvested, washed with ice-cold PBS, and incubated with 0.5 ml coimmunoprecipitation (CoIP) buffer (50 mM Tris [pH 7.5], 150 mM NaCl, 0.1% Nonidet P-40, 5 mM EDTA, 0.1 mM NaVO3, 1 mM DTT, 1 mM PMSF, and 1× protease inhibitor cocktail from Roche). Immunoprecipitation was carried out by using 2 to 5 μg specific antibody by incubation at 4°C overnight with gentle rotation. The precipitated proteins were collected by incubation with protein G-Sepharose (GE Healthcare) for 2 h at 4°C. After low-speed centrifugation to remove the supernatant, the Sepharose beads were washed with CoIP buffer, boiled in SDS sample buffer, and subjected to SDS-PAGE and Western blotting. In the case of MCF-7 cells, endogenous proteins were immunoprecipitated from lysates obtained from 15-cm2 plates as described above but without the exogenous expression of factors.
Ubiquitination assays.
Cos-1 cells transfected with ERα and other plasmids were pretreated with 10 μM MG-132 (Calbiochem) for 5 h to block proteasome activity, before they were harvested at 24 h posttransfection. Cells were lysed in buffer containing 50 mM Tris (pH 7.5), 150 mM NaCl, 10% glycerol, 5 mM EDTA, 1% Nonidet P-40, 0.1 mM NaVO3, 1 mM DTT, 1 mM PMSF, and 1× protease inhibitor cocktail from Roche. Immunoprecipitation was carried out by using ERα antibody by incubation at 4°C overnight with gentle rotation. The precipitated proteins were collected by incubation with protein G-Sepharose for 2 h at 4°C with gentle rotation. After low-speed centrifugation to remove the supernatant, the Sepharose beads were washed with CoIP buffer, boiled in SDS sample buffer, and subjected to SDS-PAGE and Western blotting with an anti-HA or anti-ERα antibody (Santa Cruz). For MCF-7 cells, endogenous ERα was immunoprecipitated from cell lysates after 48 h of expression of either β-galactosidase (β-gal) (control) or the Skp2 or p38MAPK adenovirus and 5 h of treatment with 10 μM MG-132, and ubiquitinated ERα was detected with an antiubiquitin or anti-ERα antibody.
ChIP and sequential ChIP assays.
Chromatin immunoprecipitation (ChIP) and ChIP-sequential ChIP (reChIP) assays were performed as described previously (54). The DNA isolated was subjected to quantitative real-time PCR with 36B4 used as an internal control, and a recruitment index was calculated (ratio of the specific antibody signal over the IgG signal). In ChIP-reChIP experiments, after the first pulldown, immunoprecipitated material was recovered with 10 mM dithiothreitol in IP buffer at 37°C for 30 min, diluted, and subjected to a second round of immunoprecipitation.
Synchronization of cells and fluorescence-activated cell sorter (FACS) analysis.
MCF-7 cells were synchronized at various stages of the cell cycle (G1 phase, G1/S-phase boundary, S phase, and G2/M-phase boundary), and the DNA content was analyzed by flow cytometry using a BD FACS Canto flow cytometer (BD Biosciences). To synchronize cells in the G1 phase, the cells were serum starved for 48 h, fed with 10% serum-containing medium thereafter, and treated for 24 h with 100 μM indole 3-carbinol, which induces G1-phase cell cycle arrest by inhibiting CDK6 expression. To synchronize cells in the S phase, the cells were serum starved for 48 h to first synchronize them in the G0/G1 phase and then synchronize them at the G1/S boundary by incubation with 1.5 μg of 4-hydroxyurea/ml medium for 24 h in 10% serum-containing medium. The cells were thereafter washed with Hanks' buffered saline (HBS) and released into the S phase with serum-free medium. Synchronization in the G2/M phase was carried out by synchronizing the cells at the G1/S boundary with 1.5 μg of 4-hydroxyurea/ml medium for 48 h in 10% serum-containing medium, washing the cells with HBS, and synchronizing the cells in the G2/M phase by treatment with 1 μg/ml nocodazole in 10% serum-containing medium for 24 h.
FACS analysis.
MCF-7 cells were washed once in cold PBS, scraped from plates, and pelleted by centrifugation at 340 × g at 4°C. The cells were resuspended in 2 ml of 0.9% sodium chloride and fixed for 30 min in 5 ml of 90% ethanol. Ethanol was added dropwise with constant vortexing. After 30 min, the cells were centrifuged out of the fixing solution and resuspended in 1 ml propidium iodide (50 μg/ml) diluted in PBS and supplemented with 100 μg of RNase A (Roche), as described previously (5). The cells were incubated at 37°C for 15 min, and the cell cycle distribution was analyzed by flow cytometry using a BD FACS Canto flow cytometer (BD Biosciences).
Mass spectrometry analysis of ERα.
Semiquantitative tandem mass spectrometry (MS/MS) analysis of posttranslational modifications was performed on the ERα protein in MCF-7 cells in the absence or presence of overexpressed Skp2 or p38MAPK. MCF-7 cells from five 150-mm dishes, 90% confluent, with or without the overexpression of Skp2 or p38MAPK were lysed by using CoIP buffer, sonicated, and then subjected to blocking by use of an excess of salmon sperm DNA and nonspecific IgG for 5 h at 4°C, followed by immunoprecipitation using a specific antibody against ERα or nonspecific IgG (negative control) at 4°C overnight. All ERα-bound protein complexes were then washed three times with CoIP buffer, and the specific ERα-bound protein complexes were extracted by using Laemmli sample buffer. The ERα protein band at 66 kDa in SDS-PAGE gels was visualized by Coomassie blue staining. Gel slices were excised, destained, lyophilized, and digested with trypsin (Promega, Madison, WI) overnight at 37°C. The tryptic peptides were extracted from gel slices by using 70% acetonitrile containing 5% formic acid and were further purified through PepClean C18 Spin columns (Pierce, Rockford, IL). The peptide solution was lyophilized and reconstituted in 0.1% formic acid prior to nanoflow reversed-phase liquid chromatography (nanoRPLC) MS/MS analysis using either an LTQ Orbitrap XL or an LTQ-FT mass spectrometer (ThermoElectron, San Jose, CA). The samples were injected onto a nanoRPLC column, and the peptides were eluted by use of a gradient of mobile phase A (0.1% formic acid in water) and mobile phase B (0.1% formic acid in acetonitrile) under the following conditions: 2% mobile phase B at 500 nl/min for 20 min, linear increases of 2 to 42% mobile phase B at 250 nl/min in 40 min and 42 to 98% mobile phase B at 250 nl/min in 10 min, and 98% mobile phase B at 500 nl/min for 18 min. The mass spectrometer was operated in a data-dependent mode, where the seven most abundant peptide molecular ions in every MS scan were sequentially selected for fragmentation and the acquisition of MS2 spectra. The acquisition of neutral-loss MS3 spectra was triggered when a neutral loss of phosphoric acid (H3PO4) was detected with a loss of m/z 98, 49, or 32.7 Da among the peptide fragment ions in MS2 scans. The normalized collision energy was set at 35% for both MS2 and MS3 collision-induced dissociations (CIDs). Dynamic exclusion was enabled to minimize the repeated selection of peptides previously selected for CID. The ion source capillary voltage and temperature were set at 45 V and 160°C, respectively. The electrospray voltage was set at 1.7 kV.
Tandem mass spectra were searched against the human database by using SEQUEST software (ThermoFinnigan, San Jose, CA). Fully tryptic cleavage constraints and two missed cleavage sites were applied for the search. The phosphorylation of serine, threonine, and tyrosine (+79.9663 Da); the acetylation of lysine (+42.0106 Da); and the oxidation of methionine (+15.9949 Da) residues were included as dynamic modifications when searching MS2 spectra, whereas a loss of water at serine and threonine (−18.0106 Da) residues was added as a dynamic modification when searching MS3 spectra. The SEQUEST filtering criteria for the initial identification of peptides were cross correlation (Xcorr) scores of 1.9 for [M + H]1+, 2.2 for [M + 2H]2+, and 3.1 for [M + 3H]3+ ions and a minimum delta correlation (ΔCn) value of 0.08. The MS2 and MS3 spectra of eligible peptides were manually inspected to confirm the identification of the correct peptide sequence and modifications.
Cell proliferation assays.
Assays were performed with ERα-positive MCF-7 cells or ERα-negative MDA-MB-453 and -468 cells in the absence or presence of the overexpressed Skp2-WT, Skp2-S64A mutant, or Skp2-S64E mutant protein; siGL-3 or siSkp2; vehicle (0.1% dimethyl sulfoxide [DMSO]); or the p38MAPK inhibitor (1 μM), as indicated, followed by treatment with the vehicle or ligands indicated and analysis of cell density by using WST-1 reagent (Roche Applied Science; catalog number 11644807001) according to the manufacturer's protocols.
Statistics.
All data were analyzed by using an unpaired, 2-tailed Student t test or analysis of variance (ANOVA), as appropriate. A P value of less than 0.05 was considered significant.
RESULTS
Skp2 regulates ERα protein levels and stability in breast cancer cells: inverse correlation between ERα and Skp2.
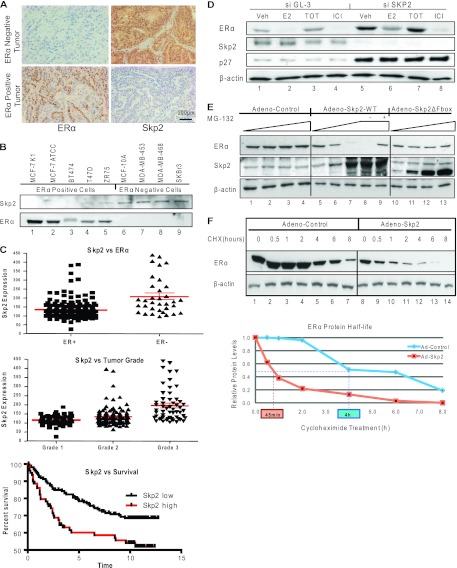
We observed ERα and Skp2 protein levels to be inversely correlated in multiple breast cancer cell lines and also in primary human breast tumors (Fig. 1A and B, and see Fig. S1A in the supplemental material). In addition, data from an analysis of publically available breast tumor database information were consistent with our findings of higher Skp2 levels in ER-negative tumors (Fig. 1C). The Skp2 level was also higher in higher-grade tumors, and elevated Skp2 expression levels were associated with reduced patient survival (Fig. 1C), implying an increased aggressiveness of tumors with higher Skp2 levels, also a hallmark of ER-negative tumors. We thus hypothesized that Skp2, by virtue of its E3-ubiquitin ligase activity, might be regulating the level of ERα and contributing to the increased aggressiveness of ER-negative breast cancers. To investigate this, we performed an siRNA-mediated knockdown of Skp2 in ERα-positive MCF-7 (and ZR-75 [data not shown]) breast cancer cells (Fig. 1D). This siSkp2 exposure reduced Skp2 protein levels to ∼10% of the levels in control siGL-3 (luciferase)-treated cells and produced a 4-fold increase in the cellular level of the ERα protein. This regulation of ERα by Skp2 appeared to be ligand independent (Fig. 1D, and see Fig. S1B in the supplemental material).
Fig 1.
Skp2 regulates ERα protein levels and stability in breast cancer cells. (A) Immunohistochemical staining for Skp2 and ERα proteins in ER-negative and -positive human breast tumors. (B) Western analysis of breast cancer cell lines. (C) Microarray data analysis of 209 ERα-positive and 77 ERα-negative human breast tumors (56) showing the relationship of Skp2 with ERα, tumor grade, and survival. (D) Western analysis of MCF-7 cells transfected with the control or Skp2 siRNA, followed by ligand treatment. Veh, vehicle. (E) Western analysis of MCF-7 cells infected with the control, Skp2-WT, or Skp2-ΔFbox adenovirus with or without MG-132. (F) Western analysis of MCF-7 cells infected with the control or Skp2 adenovirus and treated with cycloheximide (50 μg/ml) for the indicated times.
Notably, with the exogenous expression of the Skp2-WT (wild type) or Skp2-ΔFbox (dominant negative Skp2 lacking the F-box domain) protein, we observed a progressive decrease in ERα protein levels with increasing Skp2-WT expression levels that did not occur with dominant negative Skp2 (Fig. 1E, and see Fig. S1C in the supplemental material) and was blocked by the proteasomal inhibitor MG-132 (Fig. 1E, lane 9). To explore the regulation of the ERα protein half-life by Skp2, we blocked de novo protein synthesis using cycloheximide (CHX) and monitored ERα protein levels in the presence or absence of exogenously expressed Skp2. The half-life of ERα, which was about 4 h in control virus-infected cells, was shortened to about 45 min with overexpressed Skp2 (Fig. 1F, and see Fig. S1D in the supplemental material), implying SCFSkp2 E3-ligase to be a regulator of ERα protein turnover.
Skp2 and ERα interact in the cell nucleus through their N-terminal regions.
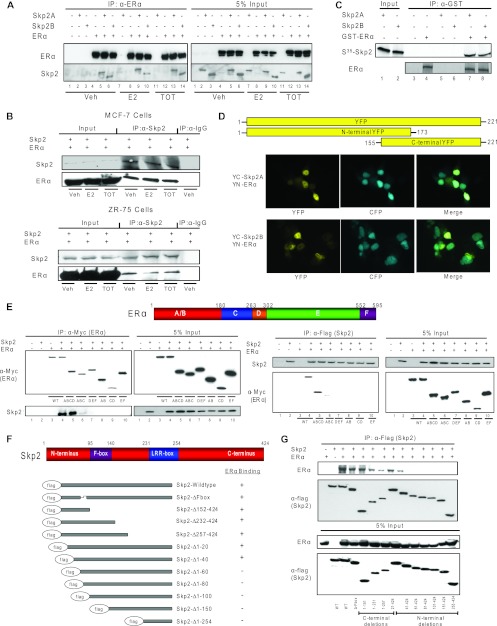
To elucidate the mechanism of ERα degradation by Skp2, we examined the interaction of the two proteins. Coimmunoprecipitation assays of ERα with Skp2A and Skp2B, isoforms that differ in their C-terminal protein sequences, showed a ligand-independent interaction of ERα with both isoforms (Fig. 2A). Data from coimmunoprecipitation studies with endogenous proteins in MCF-7 and ZR-75 cells were consistent with this finding (Fig. 2B), and in vitro GST pulldown assays confirmed a direct interaction between ERα and Skp2 (Fig. 2C).
Fig 2.
Skp2 and ERα proteins interact in the cell nucleus through their N-terminal regions. (A) CoIP of Cos-1 cells treated with ligands for ERα and Skp2A or Skp2B. (B) CoIP of MCF-7 or ZR-75 cells treated with ligands for endogenous Skp2 and ERα. (C) In vitro GST pulldown assay. Shown is an immunoprecipitation of GST-tagged ERα in the presence or absence of in vitro-translated 35S-tagged Skp2. Coimmunoprecipitated Skp2 was analyzed by autoradiography. (D) Bimolecular fluorescence complementation (BiFC) analysis. Cos-1 cells were transfected with YN-ERα along with YC-Skp2A or YC-Skp2B and CFP fused with the nuclear localization signal (to visualize cell nuclei), and YFP fluorescence was monitored by confocal microscopy. (E) CoIP of the ERα deletion mutant with Flag-Skp2 in Cos-1 cells. (F) Schematic of Skp2 deletion mutants showing their relative abilities to interact with ERα. (G) CoIP of the Skp2 deletion mutant with ERα in Cos-1 cells. All data are representative of 3 experiments.
We next used bimolecular fluorescence complementation (BiFC) to visualize the intracellular location of the ERα-Skp2 interaction in living cells. This technique exploits the reconstitution of yellow fluorescent protein (YFP) from nonfluorescent N-terminal (YN) and C-terminal (YC) YFP fragments when brought together by two interacting proteins fused to the YFP fragments. Upon expressing ERα-YN and Skp2-YC fusion constructs in Cos-1 cells, we found YFP fluorescence to be localized almost exclusively to the cell nucleus, as confirmed by the colocalization with CFP-NLS (cyan fluorescent protein fused to a nuclear localization signal) (Fig. 2D). No fluorescence was detected in control cells transfected with c-Jun-YN and c-Fos-mut-YC (c-Fos mutant lacking the c-Jun interaction domain), a control for background fluorescence.
To identify the regions of interaction between ERα and Skp2, we generated a series of Myc epitope-tagged ERα deletion constructs and Flag epitope-tagged Skp2 deletion derivatives. The deletion of the C-terminal region of ERα did not diminish its interaction with Skp2, whereas truncations in the N-terminal region resulted in a loss of interactions, with a dramatic reduction with the ABC region alone and a complete loss with the AB domain only, indicating that ABCD is the smallest portion of ERα that gives an efficient interaction (Fig. 2E). For Skp2, the deletion of either the C-terminal region with or without the leucine-rich-repeat (LRR)-box or the F-box domain had no effect on its interaction with ERα. However, the deletion of the N-terminal regions between amino acids 40 and 60 was very detrimental (Fig. 2F and G). Thus, we conclude that ERα and Skp2 directly interact, primarily in the cell nucleus, and that the N-terminal regions of both proteins are essential for their mutual interaction.
Skp2 mediates proteasomal degradation of ERα with Cul7 and Rbx1 as part of the SCFSkp2 complex.
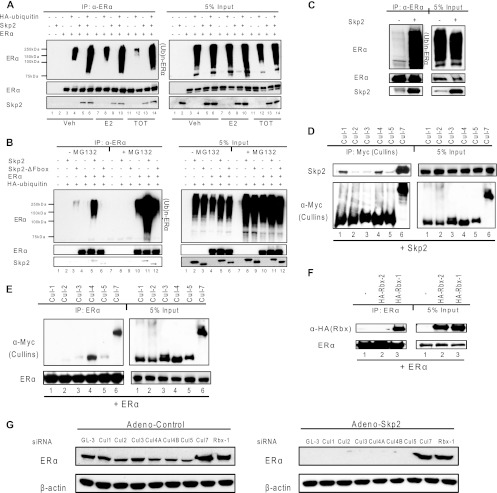
To further understand Skp2-mediated ERα protein turnover, we conducted ubiquitination assays for ERα and observed a significant increase in ubiquitinated ERα levels upon the overexpression of Skp2 (Fig. 3A). Furthermore, the F-box domain in Skp2 was critical for this regulation (Fig. 3B). We also observed an increased ubiquitination of endogenous ERα in MCF-7 cells in the presence of elevated Skp2 levels (Fig. 3C).
Fig 3.
Skp2 mediates ligand-independent proteasomal degradation of ERα with Cul7 and Rbx1 as part of the SCFSkp2 complex. (A) Ubiquitination assay of ERα in Cos-1 cells in the presence of vehicle or ligand (0.1% ethanol vehicle or 10 nM trans-hydroxy-tamoxifen [TOT]) and/or Skp2. (B) Ubiquitination assay of ERα in Cos-1 cells with pCMV-Skp2 or -ΔFbox-Skp2. (C) Ubiquitination assay for endogenous ERα in MCF-7 cells infected with the control or Skp2 adenovirus. (D) CoIP of Flag-Skp2 and various cullin isoforms in Cos-1 cells. (E) CoIP of Flag-ERα and various cullin isoforms in Cos-1 cells. (F) CoIP of Flag-ERα and HA-tagged pCMV-Rbx1 or -Rbx2 in Cos-1 cells. (G) Western analysis of MCF-7 cells transfected with siGL-3 or siRNA against various cullin isoforms or Rbx1, followed by ligand treatment. (H) Western analysis of MCF-7 cells transfected with siGL-3 or siRNA against various cullin isoforms or Rbx1, followed by infection with the control or Skp2 adenovirus. All data are representative of 3 experiments.
We next characterized the SCFSkp2 complex targeting ERα. Immunoprecipitation studies revealed an interaction of Skp2 with Cul1, Cul4, and Cul7 (Fig. 3D) and an interaction of ERα with Cul4 and Cul7 (Fig. 3E). The immunoprecipitation of ERα with HA-tagged derivatives of the two estradiol (E2) (ubiquitin-conjugating enzyme) proteins Rbx1 and Rbx2, known to be a part of the SCFSkp2 complex, revealed that ERα interacted specifically with Rbx1 but not with Rbx2 (Fig. 3F). The siRNA-mediated knockdown of various cullins (each decreased by >80% with specific siRNA) known to be expressed in MCF-7 cells, and of Rbx1, showed that the depletion of Cul7 and Rbx1 prevented the loss of ERα protein upon the overexpression of Skp2 (Fig. 3G, and see Fig. S2C in the supplemental material).
Phosphorylation of ERα at serine 294 is required for its ubiquitination and degradation by Skp2: mutagenesis and mass spectrometry analysis.
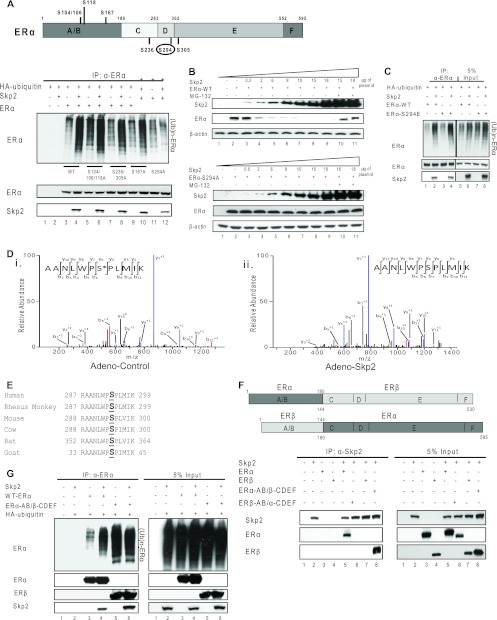
Skp2 is a member of the FBL (L for leucine-rich repeat) subclass of F-box proteins that often require phosphorylation signals on their target proteins to elicit target ubiquitination. We investigated this specific phosphodegron code for the ubiquitination of ERα by Skp2 by generating multiple serine point mutants of ERα at sites previously characterized as being important for ERα functions in breast cancer and found serine 294 in ERα to be critical for its ubiquitination by Skp2 (Fig. 4A). To confirm this, we overexpressed Skp2 in Cos-1 cells in the presence of either the ERα-WT or ERα-S294A protein and observed a dose-dependent decrease in the level of the ERα-WT protein with the overexpression of Skp2 (reversed by the proteasome inhibitor MG-132), whereas the ERα-S294A mutant was resistant to Skp2-mediated degradation (Fig. 4B, and see Fig. S3A in the supplemental material). A similar resistance to the degradation of the ERα-S294A mutant versus the ERα-WT protein was observed over time in experiments conducted with ERα-negative MDA-MB-468 cells (see Fig. S3B in the supplemental material). Of note, the S294E mutant, which acts as a phosphomimic, showed enhanced ubiquitination upon Skp2 overexpression, as did the wild-type receptor (Fig. 4C). Consistent with this, mass spectrometry analysis of ERα with or without the overexpression of Skp2 in MCF-7 cells revealed a loss of ERα peptides phosphorylated at Ser-294 (Fig. 4Di and ii, shown in red) with elevated Skp2 levels, presumably reflecting an enhanced degradation of the receptor by Skp2 (Fig. 4D, and see Fig. S3D in the supplemental material). It is noteworthy that the sequence alignment of the serine 294-containing peptide in ERα indicates that the entire 13-amino-acid sequence, including serine 294, is highly conserved across species (Fig. 4E).
Fig 4.
Phosphorylation of ERα at serine 294 is required for its ubiquitination by Skp2. (A) Ubiquitination assay for ERα-WT or various serine-to-alanine point mutants in Cos-1 cells in the absence or presence of added Skp2. (B) Western analysis of Cos-1 cells transfected with increasing doses of pCMV-Skp2 along with constant levels of pCMV-ERα-WT or pCMV-ERα-S294A. Samples in lanes 10 and 11 were also treated with MG-132. (C) Ubiquitination assay for the ERα-WT or ERα-S294E protein in Cos-1 cells in the absence or presence of added Skp2. (D) Mass spectrometry analysis of ERα in MCF-7 cells. Shown is an MS2 spectrum of an ERα peptide showing modification by phosphorylation at serine 294 (*) in the control (i) and the same but unmodified peptide upon the overexpression of Skp2 (ii). (E) Sequence alignment for ERα across various species showing the highly conserved serine residue 294. (F) CoIP of ERα-WT or the ERα and ERβ chimera with pCMV-Skp2 in Cos-1 cells. (G) Ubiquitination assay of ERα-WT or the ERα and ERβ chimera with pCMV-Skp2 in Cos-1 cells. All data are representative of 3 experiments.
Skp2 targets ERα but not ERβ for degradation.
Having identified Skp2 as a novel E3-ubiquitin ligase for ERα, we next investigated if this regulation was selective for ERα by checking its closely related receptor, estrogen receptor β (ERβ) (Fig. 4F, lane 7), and a few other nuclear receptors and transcription factors, including thyroid receptor, retinoic acid receptor, glucocorticoid receptor, progesterone receptor (PgR), and cyclic AMP (cAMP) response element binding protein, none of which interacted with Skp2 (data not shown), suggesting a great specificity of the interaction of Skp2 with ERα.
ERα and ERβ proteins share considerable sequence homology; however, the most variable domain is the AB domain, which we have shown to be essential for interactions with Skp2 (Fig. 2E). Therefore, we investigated whether the swapping of the AB domains between the two ER proteins would alter their abilities to interact with Skp2. For this, we used two ERα/ERβ chimeras, one with the AB domain of ERα and the CDEF domain of ERβ (ERα-AB/ERβ-CDEF) and the other with the AB domain of ERβ and the CDEF domain of ERα (ERβ-AB/ERα-CDEF). We indeed found that just by interchanging the AB domains, we were able to alter the abilities of the two receptors to interact with Skp2 (Fig. 4F, lanes 6 and 8). Intrigued by this, we investigated whether the transfer of the interaction function to the ERα-AB/ERβ-CDEF chimera would also render it as a target for Skp2-mediated ubiquitination. Whereas Skp2 did increase the ubiquitination of ERα, the ERα-AB/ERβ-CDEF chimera showed high basal ubiquitination levels with no apparent increase with Skp2 (Fig. 4G). This finding suggests that the Skp2-mediated degradation of ERα is a two-step process, the first step being the interaction between the two proteins and the next step being the ubiquitination of ERα, which may require a very precise posttranslational modification(s) of the ERα protein on residues that probably are not conserved between ERα and ERβ. In fact, an examination of the sequences of ERα and ERβ reveals that the important phosphorylation residue, serine 294 in ERα, is not conserved in ERβ, consistent with the lack of increased ubiquitination of the ERα-AB/ERβ-CDEF chimera by Skp2.
p38MAPK-mediated phosphorylation of ERα at serine 294 is required for ERα ubiquitination by Skp2.
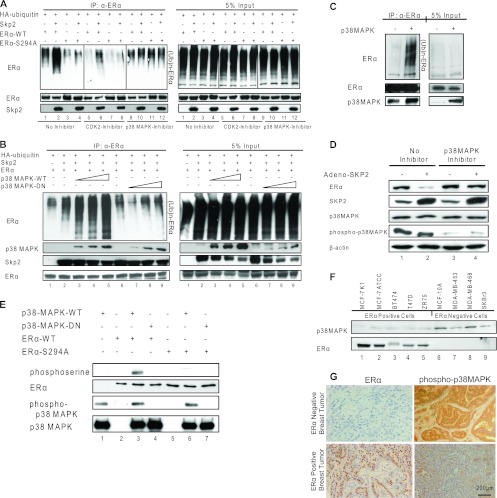
To investigate the kinase regulating ERα phosphorylation at Ser-294, we performed a bioinformatic analysis of the ERα protein sequence, which revealed the Ser-294 residue to be a putative cyclin-dependent kinase 2 (CDK2) or MAPK family target site. We therefore examined ERα ubiquitination by Skp2, this time also with or without specific inhibitors of various CDK family and MAPK family members, including phosphatidylinositol 3-kinase (PI3K); protein kinases A, B, and C; JNK/stress-activated protein kinase (SAPK); and p38MAPK. Data are shown only for the p38MAPK inhibitor (Fig. 5A), since only in the presence of this kinase inhibitor were we able to prevent the Skp2-mediated ubiquitination of ERα. Inhibitors of CDK2 (Fig. 5A) and all other kinases examined were without effect. As expected, Skp2 had no effect on the ERα-S294A mutant (Fig. 5A).
Fig 5.
p38MAPK-mediated phosphorylation of ERα at serine 294 is required for its ubiquitination by Skp2. (A) Ubiquitination assay for the ERα-WT or ERα-S294A protein in Cos-1 cells exposed to the vehicle control, the cyclin-dependent kinase 2 (CDK2) inhibitor (1 μM), or the p38MAPK inhibitor (1 μM) in the absence or presence of Skp2. (B) Ubiquitination assay for ERα in Cos-1 cells in the presence of increasing amounts of either pCMV-p38MAPK-WT or p38MAPK-DN. (C) Ubiquitination assay for endogenous ERα in MCF-7 cells infected with the control or p38MAPK adenovirus. (D) Western analysis of MCF-7 cells infected with the control or Skp2 adenovirus and treated or not with the p38MAPK inhibitor. (E) In vitro kinase assay using an active or inactive recombinant GST-p38MAPK protein with in vitro-translated ERα-WT or ERα-S294A protein as the substrate. (F) Western analysis of p38MAPK and ERα proteins in multiple breast cancer cell lines. (G) Immunohistochemical staining for phospho-p38MAPK and ERα proteins in ER-negative and -positive human breast tumors. All data are representative of 4 experiments.
The dose-dependent increase in Skp2-mediated ERα ubiquitination with the overexpression of wild-type p38MAPK was blocked by the dominant negative p38MAPK mutant (Fig. 5B), consistent with our postulate that the phosphorylation of ERα by p38MAPK is essential for its ubiquitination by Skp2. Since ERα interacted with both wild-type and mutant kinases, the block of Skp2-mediated ERα ubiquitination in the presence of dominant negative p38MAPK appeared to be solely because of the inability of the mutant kinase to phosphorylate ERα. In MCF-7 breast cancer cells, we likewise observed a marked increase in ubiquitinated ERα levels in the presence of elevated p38MAPK levels (Fig. 5C). Furthermore, with the p38MAPK inhibitor, Skp2 was ineffective in eliciting ERα degradation (Fig. 5D, and see Fig. S3D in the supplemental material), and with in vitro kinase assays, we detected the phosphorylation of ERα but not the ERα-S294A mutant in the presence of active p38MAPK (Fig. 5E). Consistent with this, semiquantitative mass spectrometry analysis of MCF-7 cells revealed an increased level of phosphorylation of ERα at serine 294 upon the overexpression of p38MAPK (see Fig. S3B in the supplemental material). Furthermore, the phosphorylation of Skp2 at serine 64 by p38MAPK (S. Bhatt and B. S. Katzenellenbogen, unpublished data) appeared to be critical for the binding of Skp2 to ERα (see Fig. S5A in the supplemental material) and also for the regulation of its function (see Fig. S5B and S5C in the supplemental material). These findings support a critical role for the p38MAPK-dependent phosphorylation of ERα in regulating its turnover by Skp2. Interestingly, as observed for Skp2, levels of active p38MAPK inversely correlated with ERα protein levels in multiple breast cancer cell lines (Fig. 5F) and in primary human breast tumors (Fig. 5G). We also tested the possibility of the regulation of Skp2 and/or p38MAPK by ERα in breast cancer cells (MCF-7 and ZR-75) and also mammary tissue from wild-type versus ERKO (ERα knockout) mice. Interestingly, the loss of the ERα protein was associated with increased expression levels of the Skp2 protein in both breast cancer cells and mammary tissues, whereas no effect on p38MAPK protein levels was observed (see Fig. S5 in the supplemental material).
Skp2 regulates ERα target gene expression, and the two proteins cooperate in driving S-phase entry of MCF-7 cells.
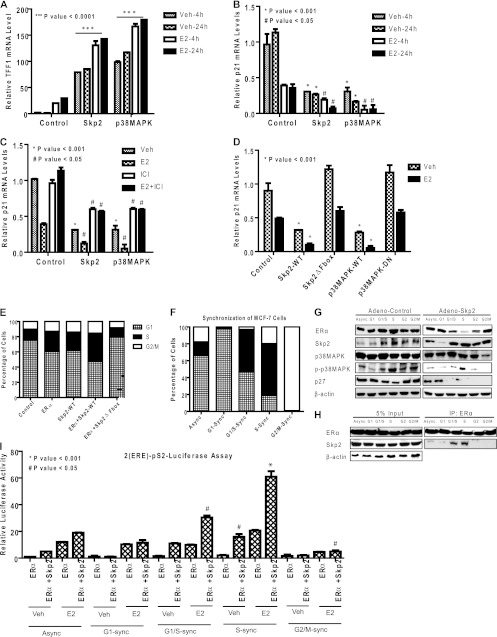
Since Skp2 is an S-phase-regulated E3-ligase, we tested the possibility of Skp2 mediating the ubiquitination-dependent regulation of ERα function driving gene expression and the S-phase entry and proliferation of breast cancer cells. We overexpressed Skp2 or p38MAPK in MCF-7 cells and examined the expressions of several ERα target genes known to have roles in regulating the estrogen response and the proliferation of these cells (TFF1/pS2, p21, and GREB1). We observed greatly upregulated (TFF1 [Fig. 6A] and GREB1 [not shown]) or downregulated (p21) basal expression levels, with a further enhancement of responses in the presence of estradiol, upon the overexpression of either Skp2 or p38MAPK (Fig. 6A and B). This effect of Skp2 on ERα target gene expression was reversed either by cotreatment with the pure antiestrogen ICI 182,780 (shown for p21 in Fig. 6C) or by the depletion of ERα by specific ERα siRNA (see Fig. S4A and S4B in the supplemental material), indicating these gene regulations to be ERα dependent. Also, the Skp2 mutant (Skp2ΔFbox), which was unable to degrade ERα, or the dominant negative p38MAPK mutant (p38MAPK-DN), which was unable to phosphorylate ERα, had no effect on the transcriptional downregulation of the p21 gene by estradiol-bound ERα (Fig. 6D). As expected, in contrast to what was observed with the overexpression of Skp2 or p38MAPK in MCF-7 cells, the depletion of endogenous Skp2 or p38MAPK, which elevates ERα levels, was found to reduce the estrogen stimulation of the pS2/TFF1 and GREB1 genes in these cells (see Fig. S8 in the supplemental material). Also of interest, ERα with the S294A mutation, which is resistant to Skp2-mediated turnover, showed compromised transcriptional activity compared to the activity of the wild-type receptor (see Fig. S3C in the supplemental material). Both of these findings suggest an important role for dynamic ERα turnover in supporting ERα transcriptional activity, as suggested previously by others (48).
Fig 6.
Skp2 regulates ERα target gene expression, and the two proteins cooperate in driving S-phase entry of MCF-7 cells. (A to C) Real-time quantitative PCR analysis of TFF1 mRNA (A) or p21 mRNA (B) in MCF-7 cells infected with the control, Skp2, or p38MAPK adenovirus for 24 h followed by control, vehicle, or estradiol (E2) (10 nM) treatment for 4 h or 24 h and reversal of the E2 effect by the antiestrogen (1 μM) ICI 182,780 (C). (D) Real-time RT-PCR analysis of p21 mRNA in MCF-7 cells infected with control, Skp2-WT, Skp2ΔFbox, p38MAPK-WT, or p38MAPK-DN adenovirus for 24 h, followed by vehicle or ligand treatment. (E) FACS analysis. MCF-7 cells were infected with the control, ERα alone, Skp2 alone, or ERα and Skp2 adenovirus followed by 48 h of E2 (10 nM) treatment and propidium iodide staining to monitor the DNA content. (F) FACS analysis. MCF-7 cells were synchronized at various stages in the cell cycle, as shown in Fig. S5H in the supplemental material, and the percentages of cells in the different cell cycle stages in asynchronous (Async) and synchronized (sync) populations are shown. (G) Western analysis of the indicated proteins in MCF-7 cells synchronized at various stages of the cell cycle and infected with the control or Skp2 adenovirus. (H) Skp2 interacts with ERα preferentially in the G1/S and S phases of the cell cycle, as observed for synchronized MCF-7 cell populations. (I) Luciferase assay. Synchronized MCF-7 cells were transfected with the (ERE)2-pS2-Luciferase expression plasmid along with ERα and β-galactosidase, treated for 24 h with either the vehicle or E2 (10 nM), and monitored for luciferase activity. Data are from 3 experiments and are represented as means ± standard deviations (SD).
By ChIP assays, we observed the recruitment of both Skp2 and p38MAPK to target gene promoters regulated by ERα, such as p21 (see Fig. S4C in the supplemental material), but not to promoters of genes closely related in function that were not regulated by ERα (p27) (see Fig. S4D in the supplemental material). By ChIP-reChIP, we showed the presence of Skp2 and p38MAPK together with ERα at target gene promoters (see Fig. S4E and S4F in the supplemental material). Flow cytometry analysis revealed a cooperativity between ERα and Skp2-WT in promoting the progression of cells through the G1/S-phase checkpoint and their entry into the S phase, while the Skp2 mutant (Skp2ΔFbox) was unable to do so (Fig. 6E, and see Fig. S4G in the supplemental material).
Skp2 interacts with and ubiquitinates ERα specifically in the G1/S and S phases of the cell cycle.
Since Skp2 is known to regulate cell cycle progression through the coordinated control of factors regulating the G1/S checkpoint, we investigated whether the Skp2 interaction with and regulation of ERα were cell cycle stage dependent. We therefore synchronized MCF-7 cells in the early G1, G1/S, S, and G2/M phases of the cell cycle, as shown in Fig. 6F and Fig. S4H in the supplemental material, and monitored the levels of ERα, Skp2, and p38MAPK interactions between Skp2 and ERα at these stages. As shown in Fig. 6G, we found Skp2 levels to be low in the G1 phase, in accordance with data from previous reports (4), and we were unable to detect an Skp2 interaction with ERα at this stage (Fig. 6H). An interaction of ERα with Skp2 could be detected in the G1/S and S phases and was not detectable when cells moved into the G2/M phase (Fig. 6H). Interestingly, we found ERα downregulation by Skp2 to occur primarily in the S phase of the cell cycle, a phase when the phospho-p38MAPK level was high (Fig. 6G).
In our analysis of synchronized breast cancer cell populations, we observed a significant increase in ERα-mediated (ERE)2-pS2-Luciferase activity in the G1/S and S phases of the cell cycle, which appeared to be Skp2 and estradiol dependent (Fig. 6I). These data imply that the regulation of ERα by Skp2, which occurs principally in the G1/S and S phases, is important for the trans-activation functions of this nuclear hormone receptor and for the increased entry of cells into the S phase.
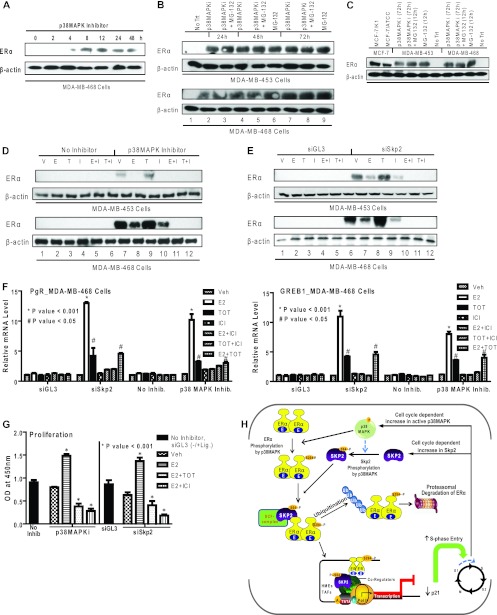
Restoration of functional ERα protein in ER-negative breast cancer cells by treatment with p38MAPK inhibitor or knockdown of Skp2.
In view of our findings for the involvement of p38MAPK and Skp2 in the reduction of ERα protein levels, we investigated whether we might be able to restore ERα levels in some ER-negative breast cancer cell lines by treatment with a p38MAPK inhibitor or the knockdown of Skp2. As shown in Fig. 7A and B, we observed a recovery of ERα protein levels in ER-negative MDA-MB-468 and MDA-MB-453 cells in the presence of either the p38MAPK inhibitor or the proteasomal inhibitor alone or in combination. ERα protein was seen by 4 h (Fig. 7A), and levels remained elevated over the 72 h examined (Fig. 7B). By comparing the level of reexpressed ERα protein in these cells to known amounts of ERα protein in MCF-7 cells, we observed a level of ERα in MDA-MB-453 cells that was ca. one-third of that of MCF-7 cells and nearly half of that in MDA-MB-468 cells after 72 h of p38MAPK inhibitor treatment (Fig. 7C). This receptor showed a normal response to ER ligands with a reduced level in the presence of E2 and ICI 182,780 and stabilization with tamoxifen (Fig. 7D), as we and others have reported previously for ERα-positive breast cancer cell lines (13, 14, 30). siSkp2 treatment also elicited increased ERα protein levels showing the same response to ER ligands (Fig. 7E). Of note, the depletion of Skp2, which increased ERα protein levels, did not change the ERα mRNA level in these cells (see Fig. S7 in the supplemental material). Thus, the regulation of ER by Skp2 is at the protein, and not the mRNA, level. Furthermore, it is of interest that the ER protein downregulates the production of the ER mRNA, and this occurs only when the ER protein is present, i.e., after the downregulation of Skp2 or the inhibition of p38MAPK, as shown in Fig. S7 in the supplemental material. Furthermore, the ERα protein was functional and enabled a marked estradiol stimulation of the well-known ER target genes progesterone receptor (PgR) and GREB1 (Fig. 7F), which was blocked by the antiestrogen trans-hydroxy-tamoxifen (TOT) or ICI 182,780. In addition, p38MAPK inhibition or Skp2 knockdown rendered the cells capable of responding to the E2 stimulation of cell proliferation, which could be suppressed by the antiestrogen tamoxifen or ICI 182,780 (Fig. 7G).
Fig 7.
Restoration of ERα protein and functional activities in ERα-negative breast cancer cells by treatment with a p38MAPK inhibitor or knockdown of Skp2. (A) Western analysis for ERα in MDA-MB-468 cells treated with a p38MAPK inhibitor (SB203580) (1 μM) for the indicated times. (B) Western analysis for ERα in MDA-MB-453 and -468 cells receiving no inhibitor treatment (vehicle only) (No Trt), the p38MAPK inhibitor alone or in combination with MG-132 (10 μM), or MG-132 (10 μM) alone. (C) Western analysis for comparison of ERα in the ERα-positive cell lines MCF-7/K1 and MCF-7/ATCC and ERα-negative cell lines MDA-MB-453 and -468 treated with the vehicle, with the p38MAPK inhibitor for 72 h with or without MG-132 for the last 12 h, or with MG-132 alone. Equal amounts of cell protein were loaded onto each lane. (D) Western analysis of MDA-MB-453 and -468 cells treated with the vehicle (V) or the p38MAPK inhibitor followed by the vehicle or ligand treatments indicated (10 nM E2 [E], 10 nM TOT [T], 1 μM ICI 182,780 [I] alone or E2 and ICI 182,780, or TOT and ICI 182,780). (E) Western analysis of ERα in MDA-MB-453 and -468 cells transfected with siGL-3 or siSkp2 followed by the vehicle or ligand treatments indicated. (F) Real-time RT-PCR analysis to monitor PgR and GREB1 mRNAs in MDA-MB-468 cells after siSkp2 or control siGL-3 treatment for 48 h or treatment with no inhibitor or the p38MAPK inhibitor for 48 h, followed by treatment with the vehicle or the indicated ligands for 24 h. (G) Proliferation of MDA-MB-468 cells in the presence or absence of the p38MAPK inhibitor or after siGL-3 or siSkp2 exposure for 48 h, followed by a 6-day treatment with the indicated ligands. Cell numbers were analyzed by a WST reagent assay. Data are from 3 experiments and are represented as means ± SD. OD, optical density. (H) Model showing the regulation of ERα turnover and functional activity by Skp2 and p38MAPK. The posttranslational modification of ERα by the p38MAPK-dependent phosphorylation of ERα at Ser-294 controls the turnover of ERα by the E3-ubiquitin ligase Skp2 and also the regulation of gene expression and enhanced cell cycle progression. Furthermore, in some ERα-negative breast cancer cells, the inhibition of p38MAPK or the depletion of Skp2 restores cellular ERα protein levels and the responsiveness to estrogen- and antiestrogen-regulated gene expression and cell proliferation.
DISCUSSION
Our study reveals that two proteins important for the regulation of the cell cycle and other cellular activities, Skp2 and p38MAPK, play pivotal roles in controlling the levels and functional activities of ERα in breast cancer cells. Our findings are presented schematically in Fig. 7H and are discussed further below.
Selective regulation of ERα but not ERβ by Skp2 and the ubiquitin-proteasome pathway.
ERα is a rapidly turning-over protein with a half-life of about 4 h in breast cancer cells (16, 39), and in the present study, we have identified Skp2 as a novel E3-ubiquitin ligase that is important in regulating the level of ERα in these cells. Our work is the first evidence for the involvement of an SCFF-box family E3-ubiquitin ligase in regulating the cellular level and functional activity of ERα. The therapeutic importance of this regulation stems from the strong inverse correlation between ERα and Skp2 levels in human breast tumors. Interestingly, Skp2 was found to selectively target ERα and not the closely related receptor ERβ or several other nuclear hormone receptors and transcription factors that we examined, making it an attractive target for pharmacological intervention, because ERβ in breast tumors shows antiproliferative activity, whereas ERα is proproliferative (11, 18).
Insights into the specificity of the interaction of Skp2 with ERα came through our analyses which implicated the N-terminal AB domain, the region least conserved between the two ERs, in the ERα interaction with Skp2. However, the inability of the ERα-AB/ERβ-CDEF chimera to be ubiquitinated by Skp2, despite regained interactions, suggested an additional aspect controlling ER ubiquitination by Skp2 upon their mutual interaction. We showed that this involved the p38MAPK-dependent phosphorylation of ERα at serine 294, a residue not conserved between the two ERs. These data highlight the importance of posttranslational modifications in the unique regulation of these two homologous nuclear receptors. We also analyzed several other ubiquitin ligases of a similar F-box family (data not shown), e.g., βTrCP (beta-transducin repeat-containing protein), which failed to interact with ERα or regulate its level, or CHIP or Stub1, which regulated basal ERα levels, as previously reported (6), but did not regulate ERα through p38MAPK and was insensitive to the S294 phosphorylation of ERα, suggesting the specificity of this regulation toward Skp2.
Involvement of p38MAPK in phosphorylation of ERα and its targeting for degradation by Skp2.
Our observations of ERα phosphorylation site mutants, kinase inhibitors, and dominant negative kinase and mass spectrometry analyses have highlighted a novel function of the phosphorylation of ERα at serine 294 by p38MAPK in controlling the ubiquitination of the receptor by Skp2. Although this serine residue was recently reported to be phosphorylated in MCF-7 cells (3), no functional aspects of this phosphorylation were explored. Hence, it is of interest that we found that the ERα-S294A mutant was not degraded by Skp2 and was compromised in its ability to upregulate the expression of estrogen-stimulated genes. Since preclinical studies have shown tamoxifen resistance to be associated with elevated levels of p38MAPK (1, 49), the dynamic inverse relationship between ERα, Skp2, and p38MAPK observed in our studies suggests that the degradation of ERα by Skp2, as signaled by its selective phosphorylation by p38MAPK, may be a possible underlying mechanism of this endocrine resistance. Furthermore, our findings imply that the suppression of Skp2 or p38MAPK activity might enable the restoration of an ERα-positive status in some ER-negative breast cancers.
Although our studies have focused on ERα regulation by Skp2 and p38MAPK, there is considerable evidence that other ubiquitin ligases and protein kinases are also involved in ER protein regulation. For instance, the cullin regulation of the ERα protein was previously reported to be mediated through S118 in ERα (8). ERα has also been shown to be ubiquitinated at K302 by BRCA1/BARD1, a ubiquitin ligase (15, 35), although the effect of this on ERα stability was not reported. Nephew and associates also reported previously that lysines 302 and 303 in ERα impact receptor interactions with the E3-ubiquitin ligase CHIP and the proteasome-mediated degradation of the receptor (6, 17). ERα protein turnover is also regulated by p27, a well-established Skp2 substrate, and by Src kinases (12).
Since Skp2 (9, 47, 52) and ERα each have critical roles in regulating cell proliferation and cancer progression, the cooperativity between Skp2 and ERα in driving the progression of breast cancer cells into the S phase of the cell cycle was noteworthy, a phenomenon not elicited by the dominant negative Skp2ΔFbox mutant, which was incapable of degrading ERα. Furthermore, our work revealed that the ectopic expression of Skp2 or p38MAPK greatly increased basal and estradiol-stimulated ERα target gene expression levels. Consistent with this, several previous studies have demonstrated the requirement for the continuous turnover of nuclear receptors, including ERα, on target gene promoters for successful transcription (34, 48, 50, 53). This regulation involved the recruitment of Skp2 and p38MAPK to ERα target gene promoters while modulating the expressions of key cell cycle- and growth-regulating genes, like p21, presumably through the posttranslational modification (phosphorylation and ubiquitination) of the nuclear receptor.
Multilevel regulation of ERα and restoration of ERα protein and hormone responsiveness in ER-negative breast cancer cells by inhibition of p38MAPK or downregulation of Skp2.
It is becoming increasingly understood that ERα, a critical factor in breast cancer prognosis and responsiveness to endocrine therapies, is regulated at multiple levels. These levels include the transcriptional regulation of ER mRNA production, the translational regulation of ERα mRNA involving, at least in part, microRNAs (miRNAs) (59), and the regulation of ERα protein turnover (6, 15, 45). We now report Skp2 to be an important E3-ubiquitin ligase regulating the ERα protein and functional activities in breast cancer cells.
Of note, we were able to restore substantial levels of functional ERα protein in ER-negative breast cancer cells (MDA-MB-453 and MDA-MB-468) with p38MAPK inhibitor treatment or with the knockdown of Skp2, and this ERα was effective in eliciting estrogen- and antiestrogen-regulated cell proliferation and target gene expression. These ERα-negative breast cancer cell lines are ERα protein negative (59) but do express ERα mRNA (our unpublished findings). In contrast, we found p38MAPK inhibitor treatment or Skp2 downregulation to be ineffective in restoring ERα protein levels in SKBr3 cells that are ERα mRNA and protein negative, consistent with our observations that p38MAPK and Skp2 are regulating ERα at the protein level. We also observed that the absence of the ERα protein in MDA-MB-231 breast cancer cells does not appear to be Skp2 dependent, as we found the Skp2 expression level in these cells to be low and comparable to that in the ERα-positive cell line MCF-7, and the further downregulation of Skp2 with siRNA did not restore ERα protein levels in these cells. However, in the other ERα-negative cell lines that we tested (MDA-MB-468, MDA-MB-453, and MCF-10A), our proposed mechanism seems to hold true. This suggests that although Skp2 appears to be an important regulator of the ERα protein in some breast cancer cells, there may be other mechanisms independent of Skp2 that might be responsible for the loss of ERα protein in some breast cancers, including those possibly due to the methylation of the ERα promoter. Our observations may have therapeutic significance and appear to be worthy of further consideration in the clinical setting, where the possible drug targeting of p38MAPK and Skp2 might restore ERα protein levels in a subset of ER-negative breast cancers that express the ERα mRNA but not the protein, in order to render them amenable to endocrine therapy. Collectively, our findings reveal a novel process regulating intracellular ERα protein levels that requires receptor phosphorylation by p38MAPK that is critical for targeting ERα for degradation by Skp2 and for regulating ERα functional activities in breast cancer cells.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by a grant from the NIH (1 P50 AT006268 [B.S.K.] from the National Center for Complementary and Alternative Medicines [NCCAM], the Office of Dietary Supplements [ODS], and the National Cancer Institute [NCI]). This work was also supported by a grant from The Breast Cancer Research Foundation (B.S.K.) and in part with federal funds from the National Cancer Institute, NIH, under contract HHSN261200800001E (Z.X. and Z.M.).
The contents of this work are solely the responsibility of the authors and do not necessarily represent the official views of the NCCAM, ODS, NCI, or National Institutes of Health. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does the mention of trade names, commercial products, or organizations imply endorsement by the U.S. government.
We are grateful to Richard Kollmar and Rex Hess for use of their microscopy facilities and Barbara Pilas for advice with flow cytometry. We thank Fabio Stossi, Daniel Barnett, Anna Bergamaschi, and Maritta Perry Grau for their comments and suggestions.
S.B. conceived of and designed the studies, carried out the experiments and data analysis, interpreted the data, and wrote drafts of the manuscript. Z.X. and Z.M. carried out some of the experimental studies and data analysis and contributed to the writing of the manuscript. B.S.K. conceived of and designed the studies, analyzed and interpreted the data, and wrote the manuscript. All authors read, made suggestions for, and approved the final manuscript.
We declare that we have no conflict of interest.
Footnotes
Published ahead of print 19 March 2012
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1. Aesoy R, et al. 2008. An autocrine VEGF/VEGFR2 and p38 signaling loop confers resistance to 4-hydroxytamoxifen in MCF-7 breast cancer cells. Mol. Cancer Res. 6:1630–1638 [DOI] [PubMed] [Google Scholar]
- 2. Alarid ET. 2006. Lives and times of nuclear receptors. Mol. Endocrinol. 20:1972–1981 [DOI] [PubMed] [Google Scholar]
- 3. Atsriku C, et al. 2009. Systematic mapping of posttranslational modifications in human estrogen receptor-alpha with emphasis on novel phosphorylation sites. Mol. Cell. Proteomics 8:467–480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bashir T, Dorrello NV, Amador V, Guardavaccaro D, Pagano M. 2004. Control of the SCF(Skp2-Cks1) ubiquitin ligase by the APC/C(Cdh1) ubiquitin ligase. Nature 428:190–193 [DOI] [PubMed] [Google Scholar]
- 5. Bergamaschi A, Christensen BL, Katzenellenbogen BS. 2011. Reversal of endocrine resistance in breast cancer: interrelationships among 14-3-3zeta, FOXM1, and a gene signature associated with mitosis. Breast Cancer Res. 13:R70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Berry NB, Fan M, Nephew KP. 2008. Estrogen receptor-alpha hinge-region lysines 302 and 303 regulate receptor degradation by the proteasome. Mol. Endocrinol. 22:1535–1551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Brueggemeier RW, Hackett JC, Diaz-Cruz ES. 2005. Aromatase inhibitors in the treatment of breast cancer. Endocr. Rev. 26:331–345 [DOI] [PubMed] [Google Scholar]
- 8. Byun B, Jung Y. 2008. Repression of transcriptional activity of estrogen receptor alpha by a Cullin3/SPOP ubiquitin E3 ligase complex. Mol. Cells 25:289–293 [PubMed] [Google Scholar]
- 9. Calvisi DF, et al. 2009. SKP2 and CKS1 promote degradation of cell cycle regulators and are associated with hepatocellular carcinoma prognosis. Gastroenterology 137:1816–1826.e10 [DOI] [PubMed] [Google Scholar]
- 10. Cardozo T, Pagano M. 2004. The SCF ubiquitin ligase: insights into a molecular machine. Nat. Rev. Mol. Cell Biol. 5:739–751 [DOI] [PubMed] [Google Scholar]
- 11. Chang EC, et al. 2008. Estrogen receptors alpha and beta as determinants of gene expression: influence of ligand, dose, and chromatin binding. Mol. Endocrinol. 22:1032–1043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chu I, et al. 2007. Src promotes estrogen-dependent estrogen receptor alpha proteolysis in human breast cancer. J. Clin. Invest. 117:2205–2215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Connor CE, et al. 2001. Circumventing tamoxifen resistance in breast cancers using antiestrogens that induce unique conformational changes in the estrogen receptor. Cancer Res. 61:2917–2922 [PubMed] [Google Scholar]
- 14. Dauvois S, Danielian PS, White R, Parker MG. 1992. Antiestrogen ICI 164,384 reduces cellular estrogen receptor content by increasing its turnover. Proc. Natl. Acad. Sci. U. S. A. 89:4037–4041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Eakin CM, Maccoss MJ, Finney GL, Klevit RE. 2007. Estrogen receptor alpha is a putative substrate for the BRCA1 ubiquitin ligase. Proc. Natl. Acad. Sci. U. S. A. 104:5794–5799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Eckert RL, Mullick A, Rorke EA, Katzenellenbogen BS. 1984. Estrogen receptor synthesis and turnover in MCF-7 breast cancer cells measured by a density shift technique. Endocrinology 114:629–637 [DOI] [PubMed] [Google Scholar]
- 17. Fan M, Park A, Nephew KP. 2005. CHIP (carboxyl terminus of Hsc70-interacting protein) promotes basal and geldanamycin-induced degradation of estrogen receptor-alpha. Mol. Endocrinol. 19:2901–2914 [DOI] [PubMed] [Google Scholar]
- 18. Frasor J, et al. 2003. Profiling of estrogen up- and down-regulated gene expression in human breast cancer cells: insights into gene networks and pathways underlying estrogenic control of proliferation and cell phenotype. Endocrinology 144:4562–4574 [DOI] [PubMed] [Google Scholar]
- 19. Gao D, et al. 2009. Phosphorylation by Akt1 promotes cytoplasmic localization of Skp2 and impairs APCCdh1-mediated Skp2 destruction. Nat. Cell Biol. 11:397–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gonzalez F, Delahodde A, Kodadek T, Johnston SA. 2002. Recruitment of a 19S proteasome subcomplex to an activated promoter. Science 296:548–550 [DOI] [PubMed] [Google Scholar]
- 21. Gstaiger M, et al. 2001. Skp2 is oncogenic and overexpressed in human cancers. Proc. Natl. Acad. Sci. U. S. A. 98:5043–5048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Guardavaccaro D, Pagano M. 2006. Stabilizers and destabilizers controlling cell cycle oscillators. Mol. Cell 22:1–4 [DOI] [PubMed] [Google Scholar]
- 23. Hershko DD. 2008. Oncogenic properties and prognostic implications of the ubiquitin ligase Skp2 in cancer. Cancer 112:1415–1424 [DOI] [PubMed] [Google Scholar]
- 24. Hu CD, Chinenov Y, Kerppola TK. 2002. Visualization of interactions among bZIP and Rel family proteins in living cells using bimolecular fluorescence complementation. Mol. Cell 9:789–798 [DOI] [PubMed] [Google Scholar]
- 25. Huang CY, et al. 1998. Lipitoids—novel cationic lipids for cellular delivery of plasmid DNA in vitro. Chem. Biol. 5:345–354 [DOI] [PubMed] [Google Scholar]
- 26. Ji P, et al. 2006. Skp2 contains a novel cyclin A binding domain that directly protects cyclin A from inhibition by p27Kip1. J. Biol. Chem. 281:24058–24069 [DOI] [PubMed] [Google Scholar]
- 27. Johnston SR, Martin LA, Leary A, Head J, Dowsett M. 2007. Clinical strategies for rationale combinations of aromatase inhibitors with novel therapies for breast cancer. J. Steroid Biochem. Mol. Biol. 106:180–186 [DOI] [PubMed] [Google Scholar]
- 28. Jordan VC, O'Malley BW. 2007. Selective estrogen-receptor modulators and antihormonal resistance in breast cancer. J. Clin. Oncol. 25:5815–5824 [DOI] [PubMed] [Google Scholar]
- 29. Katzenellenbogen BS, Frasor J. 2004. Therapeutic targeting in the estrogen receptor hormonal pathway. Semin. Oncol. 31:28–38 [DOI] [PubMed] [Google Scholar]
- 30. Kieser KJ, Kim DW, Carlson KE, Katzenellenbogen BS, Katzenellenbogen JA. 2010. Characterization of the pharmacophore properties of novel selective estrogen receptor downregulators (SERDs). J. Med. Chem. 53:3320–3329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Laios I, et al. 2005. Role of the proteasome in the regulation of estrogen receptor alpha turnover and function in MCF-7 breast carcinoma cells. J. Steroid Biochem. Mol. Biol. 94:347–359 [DOI] [PubMed] [Google Scholar]
- 32. Lange CA, Shen T, Horwitz KB. 2000. Phosphorylation of human progesterone receptors at serine-294 by mitogen-activated protein kinase signals their degradation by the 26S proteasome. Proc. Natl. Acad. Sci. U. S. A. 97:1032–1037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lin HK, et al. 2009. Phosphorylation-dependent regulation of cytosolic localization and oncogenic function of Skp2 by Akt/PKB. Nat. Cell Biol. 11:420–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lonard DM, Nawaz Z, Smith CL, O'Malley BW. 2000. The 26S proteasome is required for estrogen receptor-alpha and coactivator turnover and for efficient estrogen receptor-alpha transactivation. Mol. Cell 5:939–948 [DOI] [PubMed] [Google Scholar]
- 35. Ma Y, et al. 2010. BRCA1 regulates acetylation and ubiquitination of estrogen receptor-alpha. Mol. Endocrinol. 24:76–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Madak-Erdogan Z, Lupien M, Stossi F, Brown M, Katzenellenbogen BS. 2011. Genomic collaboration of estrogen receptor alpha and extracellular signal-regulated kinase 2 in regulating gene and proliferation programs. Mol. Cell. Biol. 31:226–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. McInerney EM, Weis KE, Sun J, Mosselman S, Katzenellenbogen BS. 1998. Transcription activation by the human estrogen receptor subtype beta (ER beta) studied with ER beta and ER alpha receptor chimeras. Endocrinology 139:4513–4522 [DOI] [PubMed] [Google Scholar]
- 38. Metivier R, et al. 2003. Estrogen receptor-alpha directs ordered, cyclical, and combinatorial recruitment of cofactors on a natural target promoter. Cell 115:751–763 [DOI] [PubMed] [Google Scholar]
- 39. Monsma FJ, Jr, Katzenellenbogen BS, Miller MA, Ziegler YS, Katzenellenbogen JA. 1984. Characterization of the estrogen receptor and its dynamics in MCF-7 human breast cancer cells using a covalently-attaching antiestrogen. Endocrinology 115:143–153 [DOI] [PubMed] [Google Scholar]
- 40. Montemurro F, Aglietta M, Del Mastro L. 2009. Aromatase inhibitors as adjuvant therapy for breast cancer. J. Clin. Oncol. 27:2566–2567 [DOI] [PubMed] [Google Scholar]
- 41. Musgrove EA, Sutherland RL. 2009. Biological determinants of endocrine resistance in breast cancer. Nat. Rev. Cancer 9:631–643 [DOI] [PubMed] [Google Scholar]
- 42. Nakayama KI, Nakayama K. 2006. Ubiquitin ligases: cell-cycle control and cancer. Nat. Rev. Cancer 6:369–381 [DOI] [PubMed] [Google Scholar]
- 43. Nalepa G, Rolfe M, Harper JW. 2006. Drug discovery in the ubiquitin-proteasome system. Nat. Rev. Drug Discov. 5:596–613 [DOI] [PubMed] [Google Scholar]
- 44. Radke S, Pirkmaier A, Germain D. 2005. Differential expression of the F-box proteins Skp2 and Skp2B in breast cancer. Oncogene 24:3448–3458 [DOI] [PubMed] [Google Scholar]
- 45. Ramamoorthy S, Nawaz Z. 2008. E6-associated protein (E6-AP) is a dual function coactivator of steroid hormone receptors. Nucl. Recept. Signal. 6:e006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ravaioli A, et al. 2008. p27 and Skp2 immunoreactivity and its clinical significance with endocrine and chemo-endocrine treatments in node-negative early breast cancer. Ann. Oncol. 19:660–668 [DOI] [PubMed] [Google Scholar]
- 47. Reed SI. 2008. Deathproof: new insights on the role of skp2 in tumorigenesis. Cancer Cell 13:88–89 [DOI] [PubMed] [Google Scholar]
- 48. Reid G, et al. 2003. Cyclic, proteasome-mediated turnover of unliganded and liganded ERalpha on responsive promoters is an integral feature of estrogen signaling. Mol. Cell 11:695–707 [DOI] [PubMed] [Google Scholar]
- 49. Riggins RB, Schrecengost RS, Guerrero MS, Bouton AH. 2007. Pathways to tamoxifen resistance. Cancer Lett. 256:1–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Shang Y, Hu X, DiRenzo J, Lazar MA, Brown M. 2000. Cofactor dynamics and sufficiency in estrogen receptor-regulated transcription. Cell 103:843–852 [DOI] [PubMed] [Google Scholar]
- 51. Shen T, Horwitz KB, Lange CA. 2001. Transcriptional hyperactivity of human progesterone receptors is coupled to their ligand-dependent down-regulation by mitogen-activated protein kinase-dependent phosphorylation of serine 294. Mol. Cell. Biol. 21:6122–6131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Signoretti S, et al. 2002. Oncogenic role of the ubiquitin ligase subunit Skp2 in human breast cancer. J. Clin. Invest. 110:633–641 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 53. Stanisic V, Malovannaya A, Qin J, Lonard DM, O'Malley BW. 2009. OTU domain-containing ubiquitin aldehyde-binding protein 1 (OTUB1) deubiquitinates estrogen receptor (ER) alpha and affects ERalpha transcriptional activity. J. Biol. Chem. 284:16135–16145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Stossi F, Madak-Erdogan Z, Katzenellenbogen BS. 2009. Estrogen receptor alpha represses transcription of early target genes via p300 and CtBP1. Mol. Cell. Biol. 29:1749–1759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Utku Y, et al. 2006. A peptidomimetic siRNA transfection reagent for highly effective gene silencing. Mol. Biosyst. 2:312–317 [DOI] [PubMed] [Google Scholar]
- 56. Wang Y, et al. 2005. Gene-expression profiles to predict distant metastasis of lymph-node-negative primary breast cancer. Lancet 365:671–679 [DOI] [PubMed] [Google Scholar]
- 57. Willems AR, et al. 1999. SCF ubiquitin protein ligases and phosphorylation-dependent proteolysis. Philos. Trans. R. Soc. Lond. B Biol. Sci. 354:1533–1550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Willems AR, Schwab M, Tyers M. 2004. A hitchhiker's guide to the cullin ubiquitin ligases: SCF and its kin. Biochim. Biophys. Acta 1695:133–170 [DOI] [PubMed] [Google Scholar]
- 59. Zhao JJ, et al. 2008. MicroRNA-221/222 negatively regulates estrogen receptor alpha and is associated with tamoxifen resistance in breast cancer. J. Biol. Chem. 283:31079–31086 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.