Abstract
Candida albicans-associated bloodstream infections are linked to the ability of this yeast to form biofilms. In this study, we used lipidomics to compare the lipid profiles of C. albicans biofilms and planktonic cells, in early and mature developmental phases. Our results showed that significant differences exist in lipid composition in both developmental phases. Biofilms contained higher levels of phospholipid and sphingolipids than planktonic cells (nmol per g biomass, P<0.05 for all comparisons). In the early phase, levels of lipid in most classes were significantly higher in biofilms compared to planktonic cells (P≤0.05). The ratio of phosphatidylcholine to phosphatidylethanolamine was lower in biofilms compared to planktonic cells in both early (1.17 vs 2.52, P≤0.001) and late (2.34 vs 3.81, P≤0.001) developmental phases. The unsaturation index of phospholipids decreased with time, with this effect being particularly strong for biofilms. Inhibition of the biosynthetic pathway for sphingolipid [mannosyl diinositolphosphoryl ceramide, M(IP)2C] by myriocin or aureobasidin A, and disruption of the gene encoding inositolphosphotransferase (Ipt1p), abrogated the ability of C. albicans to form biofilms. The differences in lipid profiles between biofilms and planktonic Candida cells may have important implications for the biology and antifungal resistance of biofilms.
Introduction
Candida albicans-associated bloodstream infections are linked to the ability of this pathogenic yeast to form biofilms (Mukherjee et al., 2005). These biofilms are especially problematic due to their inherent resistance to commonly used antifungal agents. Moreover, Candida biofilms associated with indwelling devices provide a protected niche for the fungal cells where they can evade the host immune system (Chandra et al., 2007). Recent studies have revealed that biofilm formation by C. albicans is associated with differential expression of genes and proteins involved in the synthesis of membrane lipids (Lattif et al., 2008; Yeater et al., 2007), and that alteration in the sterol composition is linked to increased azole resistance of C. albicans biofilms (Mukherjee et al., 2003). Additionally, phosphatidylinositol (PI), a key yeast phospholipid, has been shown to contribute to antifungal resistance (Abu-Elteen & Whittaker, 1997-1998; Hitchcock et al., 1987) and pathogenesis (Reynolds, 2009). PI is a precursor for the biosynthesis of glycosylinositol phosphorylceramides (GIPCs), the yeast-specific sphingolipid constituents of membrane-localized lipid rafts. GIPCs comprise three members: inositolphosphorylceramide (IPC), mannosylinositolphosphorylceramide (MIPC) and mannosyldiinositolphosphorylceramide [M(IP)2C] (Thevissen et al., 2005). The terminal step in M(IP)2C biosynthesis is catalysed by inositolphosphotransferase, encoded by the IPT1 gene. Although several studies have indicated important roles for lipid rafts in modulating fungal growth and pathogenesis (Hallstrom et al., 2001; Martin & Konopka, 2004; Pasrija et al., 2008; Prasad et al., 2005; Siafakas et al., 2006; Thevissen et al., 2000; Toulmay & Schneiter, 2007; Wachtler & Balasubramanian, 2006), their role in biofilm formation and associated drug resistance has not been investigated.
In the current study, we performed lipidomics analysis to characterize the profile of phospholipids and identified the associated molecular classes that are differentially produced in biofilms compared to planktonic cells. Our studies reveal, to our knowledge for the first time, that C. albicans biofilms differ significantly from planktonic cells in their lipid profiles and unsaturation index, in a phase-dependent manner. We also found that PI was overproduced in biofilms. PI is a key precursor for biosynthesis of M(IP)2C, a critical component of lipid rafts; therefore, we investigated the role of lipid rafts in fungal biofilm formation using specific M(IP)2C biosynthesis inhibitors and a C. albicans mutant strain lacking inositolphosphotransferase (Ipt1p), a key enzyme in this biosynthetic pathway. Our results suggest that lipid rafts may be associated with the ability of C. albicans to form biofilms, and may provide a foundation for developing a novel strategy to manage and treat biofilm-associated infections.
Methods
Candida isolates.
The following C. albicans strains were examined in the current study: SC5314, and the isogenic strain set consisting of CAI4 (Δura3 : : imm434/Δura3 : : imm434), Δipt1 mutant (Δura3 : : imm434/Δura3 : : imm434 Δipt1 : : hisG-URA3-hisG/Δipt1 : : hisG), IPT1/Δipt1revertant (Δura3 : : imm434/Δura3 : : imm434 Δipt1 : : hisG/Δipt1 : : hisG : : IPT1-URA3). The isogenic strain set of isolates was a generous gift from R. Prasad (Jawaharlal Nehru University, New Delhi, India) (Prasad et al., 2005). Candida cultures were maintained on Sabouraud dextrose agar (SDA, yeast extract, peptone, glucose, 1 : 2 : 2) or Sabouraud dextrose broth (SDB, Difco), supplemented with uridine (80 µg ml−1) as required, or kept at –80 °C for long-term storage.
Biofilm formation.
C. albicans biofilms were formed on silicone elastomer catheter discs and quantified as described previously (Chandra et al., 2008). Briefly, C. albicans blastospores were grown overnight at 37 °C in a yeast nitrogen base (YNB) medium (Difco) supplemented with 50 mM glucose. A standardized cell suspension was prepared from this culture by adjusting the cell density to 1×107 cells ml−1. Silicone elastomer discs (1.5 cm diameter; Cardiovascular Instrument Corp.) were placed in 12-well tissue culture plates and incubated in fetal bovine serum for 24 h at 37 °C. The disks were immersed in 4 ml of the standardized cell suspension (1×107 cells ml−1) and incubated for 90 min at 37 °C. They were then immersed in YNB medium with 50 mM glucose and incubated for 6 h (early-phase biofilm) or 48 h (mature-phase biofilm) on a rocker at 37 °C. Biofilms were quantified by measuring dry-weight biomass and metabolic activity using a tetrazolium dye reduction assay [2,3-bis (2-methoxy-4-nitro-5-sulfophenyl)-2H-tetrazolium-5-carboxanilide; XTT) (Chandra et al., 2008). For dry-weight determination, biofilms were scraped into 4 ml PBS and transferred to a centrifuge tube. This suspension was filtered through a pre-weighed filter (0.45 µm pore size), washed with PBS, and dried at 35 °C for 24 h. The dried filters were weighed to calculate biofilm biomass from the difference in weight. Planktonic cultures were prepared by the same method as for biofilm formation, except that the silicone elastomer disks were omitted from the tissue-culture wells. Planktonic cultures exhibit similar morphologies to biofilms and consist of both yeast and hyphal forms of Candida.
Scanning electron microscopy (SEM).
Early- (6 h) and mature- (48 h) phase candidal biofilms were grown as described above. These biofilms were prepared for SEM analysis as described previously (Chandra et al., 2008). Briefly, SEM discs with biofilms were fixed in 2 % glutaraldehyde, and then in osmium tetroxide, tannic acid and uranyl acetate. These fixation steps were followed by a series of ethanol dehydration steps. The dehydrated specimens were placed on microscope stubs and sputter-coated with Au-Pd (60/40 ratio). The prepared samples were examined for their topography using a model XL3C ESEM Philips scanning electron microscope.
Lipid extraction.
Total lipids were extracted from both planktonic cultures and biofilms using chloroform/methanol (2 : 1, v/v) and then purified using established procedures (Ghannoum et al., 1990). Briefly, biofilms and planktonic cells were harvested by centrifugation, and the cell pellet was washed with sterile distilled water; cells were resuspended in 30 ml chloroform/methanol (2 : 1, v/v) and then incubated at room temperature on a shaker overnight. Next, the cell suspension was filtered through 110 mm diameter filter paper (Whatman) to remove denatured protein. Volumes (0.2 ml) of 0.9 % NaCl were added to the cell solution, and the mixture was separated using a separating funnel. The clear lipid-containing phase was carefully collected, and the total lipid mass was estimated gravimetrically after removing the solvent under a gentle stream of N2 gas. Five samples were extracted for each time point, each from an independent replicate.
Lipid characterization
Thin-layer chromatography (TLC).
The extracted lipids contained polar as well as apolar lipids. TLC analysis was carried out using TLC plates (Whatman) with silica gel containing CaSO4 binder (Ibrahim et al., 1996), in rectangular glass tanks saturated with the solvent system. For apolar lipids, the solvent system used was hexane/diethyl ether/acetic acid (90 : 10 : 1, by vol.), while polar lipids were separated using choloroform/methanol/7 M ammonia (65 : 25 : 4, by vol.) as solvent system. Polar lipids were further analysed using two-dimensional TLC using the following solvent systems: first dimension, chloroform/methanol/7 M ammonia (65 : 30 : 4, by vol.); second dimension, chloroform/methanol/acetic acid/water (170 : 25 : 25 : 4, by vol.). The lipids were visualized by exposure to iodine vapour or charring at 220 °C after spraying the plates with 50 % (v/v) H2SO4.
Electrospray ionization-tandem mass spectrometry (ESI-MS/MS).
Extracted lipid samples were subjected to lipidomics analysis of their phospholipids and sphingolipids using an automated ESI-MS/MS approach (Brügger et al., 1997). Briefly, an aliquot of extracted lipids (0.4 mg lipid) was combined with solvents and internal standards, such that the ratio of chloroform/methanol/300 mM ammonium acetate in water was 300 : 665 : 35 and the final volume was 1 ml. Internal standards, obtained and quantified as described previously (Welti et al., 2002), were 0.6 nmol di12 : 0-phosphatidylcholine (PC), 0.6 nmol di24 : 1-PC, 0.6 nmol 13 : 0-lysoPC, 0.6 nmol 19 : 0-lysoPC, 0.3 nmol di12 : 0-phosphatidylethanolamine (PE), 0.3 nmol di23 : 0-PE, 0.3 nmol 14 : 0-lysoPE, 0.3 nmol 18 : 0-lysoPE, 0.3 nmol di14 : 0-phosphatidic acid (PA), 0.3 nmol di20 : 0(phytanoyl)-PA, 0.3 nmol di14 : 0-phosphatidylglycerol (PG), 0.3 nmol di20 : 0(phytanoyl)-PG, 0.2 nmol di14 : 0-phosphatidylserine (PS), 0.2 nmol di20 : 0 (phytanoyl)-PS, 0.23 nmol 16 : 0-18 : 0-phosphatidylinositol (PI) and 0.16 nmol di18 : 0-PI. Lipids were detected with the following scans on a triple quadrupole mass spectrometer (API 4000, Applied Biosystems): PC and lysoPC, [M+H]+ in positive ion mode with precursors of 184.1 (Pre 184.1); PE and lysoPE, [M+H]+ in positive ion mode as neutral loss of 141.0 (NL 141.0); PA, [M+NH4]+ in positive ion mode with NL 115.0; PG, [M+NH4]+ in positive ion mode with NL 189.0; PS, [M+H]+ in positive ion mode with NL 185.0; PI, [M+NH4]+ in positive ion mode with NL 277.0; IPC, [M−H]− in negative ion mode with Pre 259.0; MIPC, [M−H]− in negative ion mode with Pre 421.0; and M(IP)2C, [M−H]− in negative ion mode with Pre 663.1. The assignments of the polar lipid molecular species were in terms of total acyl carbons : total double bonds. Sphingolipid species [IPC, MIPC and M(IP)2C] are indicated by (total carbons in the sphingoid base+fatty amide) : (total carbon–carbon double bonds in sphingoid base+fatty amide)-(number of hydroxyl groups on base+fatty amide chain). It should be noted that in all cases assignments are based on identification of head group and mass/charge ratios only and do not rule out the presence of molecular species with non-normal acyl chains. The phospholipids in each class were quantified in comparison to two internal standards of the class (Welti et al., 2002). The sphingolipids were quantified in comparison to 16 : 0-18 : 0 PI, [M−H]−, in negative ion mode (Pre 241.01). Data acquisition and analysis were carried out as described previously (Devaiah et al., 2006). Lipid levels were normalized to the weight of cells for both biofilms and planktonic cells. Unsaturation index (UI) for each class was calculated as the mean number of double bonds per acyl chain for the species within that class [UI = lipid level (nmol per mg biomass)×number of double bonds/number of acyl chains].
Visualization of lipid rafts.
Filipin staining and fluorescence microscopy were used to compare the distribution of lipid rafts in Candida biofilms and planktonic cells. Filipin is a fluorescent polyene that binds ergosterol (a major component of lipid rafts). This fluorescent dye is commonly used to visualize lipid rafts. Planktonic or biofilm-derived Candida cells were rinsed three times with PBS and fixed with freshly prepared 3 % paraformaldehyde for 1 h at room temperature. Next, cells were washed three times with PBS and incubated with glycine (1.5 g ml−1 in PBS) for 10 min at room temperature. Cells were then stained for 2 h with filipin (0.05 mg ml−1 in PBS/10 % fetal bovine serum). Images were captured with a Leica DMI 6000 B inverted microscope using a Retiga EXI camera (Q-imaging). An excitation filter of 340–380 nm and an emission filter of 425 nm LP were used to visualize the filipin-stained Candida cells. A 63×1.4 NA Plan Apo objective was used with a ×1.6 magnification changer built into the microscope for a final magnification of ×1008. Images were analysed using Metamorph Imaging Software (Molecular Devices).
Analysis of lipid raft formation in C. albicans biofilms in the presence of sphingolipid biosynthesis inhibitors.
Since lipid rafts are enriched in sphingolipids, we determined whether biochemical inhibition of sphingolipid biosynthesis affects C. albicans biofilms. These inhibition studies were performed as described earlier (Martin & Konopka, 2004). Briefly, C. albicans biofilms were treated with sphingolipid biosynthesis inhibitors (myriocin or aureobasidin A, dissolved in methanol) (Martin & Konopka, 2004). The concentration of myriocin ranged from 1.25 to 160 µg ml−1, while that of aureobasidin A ranged from 0.03 to 10 µg ml−1. The methanol concentration in these assays was 0.5 %; this concentration of methanol had no effect on fungal growth. C. albicans biofilms treated with vehicle (methanol) alone served as a control. Changes in lipid rafts following treatment with these inhibitors were monitored using fluorescence microscopy after staining the cells with filipin.
Statistical analysis.
All results represent the mean of at least five independent samples (biological replicates). Comparative results were statistically analysed using independent samples t-test (assuming equal variance) and ANOVA with SPSS software version 19.0. A P-value of <0.05 was considered to be statistically significant.
Results
Biofilms are enriched in specific phospholipid classes
We performed one- and two-dimensional TLC analyses to identify biofilm-specific lipids. These analyses revealed the presence of triacylglycerols, fatty acids, diacylglycerols, sterols, monoacylglycerols and phospholipids (see Supplementary Fig. S1, available with the online version of this paper). These analyses also showed that PC and PE were the two major phospholipids present in biofilms.
Distribution of lipid classes varies between biofilms and planktonic C. albicans and by growth phase in biofilms
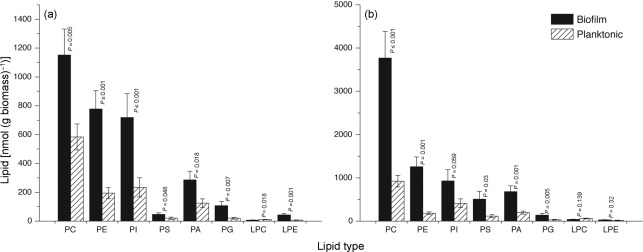
We followed TLC by ESI-MS/MS-based lipidomics, using precursor and neutral loss scans, to identify the classes and molecular species of polar lipids present in biofilms and planktonic cells grown to early and mature phases (Fig. 1). Phospholipid species were quantified in comparison to internal standards of the same class. Since biofilm formation proceeds through distinct developmental phases, we determined whether the profile of the major lipid classes differs between early and mature biofilm phases. Planktonic cells in the early and mature phases were also compared. We found that levels of PC, PE, PI, PS, PA and PG were significantly higher in biofilms compared to planktonic cells in the early phase (Fig. 1a, P≤0.007). A similar trend was observed in biofilms and planktonic C. albicans grown to the mature phase (Fig. 1b). The level of lyso-PE was elevated in early-phase biofilms (Fig. 1a). However, the level of lyso-PC tended to be lower in biofilms than in planktonic cells (Fig. 1a, b). The ratio of PC to PE was lower in biofilms compared to planktonic cells in both early (1.17 vs 2.52, P≤0.001) and late (2.34 vs 3.81, P≤0.001) developmental phases. When comparing the two phases, the PC : PE ratio for biofilms increased from 1.17 to 2.34 (P≤0.001) and for planktonic cells from 2.52 to 3.81 (P = 0.001) in the early and mature developmental phases, respectively. Taken together, these results showed that the distribution of lipid classes in biofilms and planktonic cells is different, particularly in the early phase. In addition, the lipid class profile of early-phase biofilms is significantly different from that of mature-phase biofilms.
Fig. 1.
Distribution of polar lipids in C. albicans biofilms and planktonic cells: lipidomics analysis of phospholipids in biofilms and planktonic cells grown to (a) early phase (6 h) or (b) mature phase (48 h). PC, phosphatidylcholine; PE, phosphatidylethanolamine; PI, phosphatidylinositol; PS, phosphatidylserine; PA, phosphatidic acid; PG, phosphatidylglycerol; LPC, lyso-PC; LPE, lyso-PE. Data represent mean±se (n = 5). P-values represent comparisons between biofilm and planktonic cells.
Molecular species profiles also differ as a function of age and biofilm formation
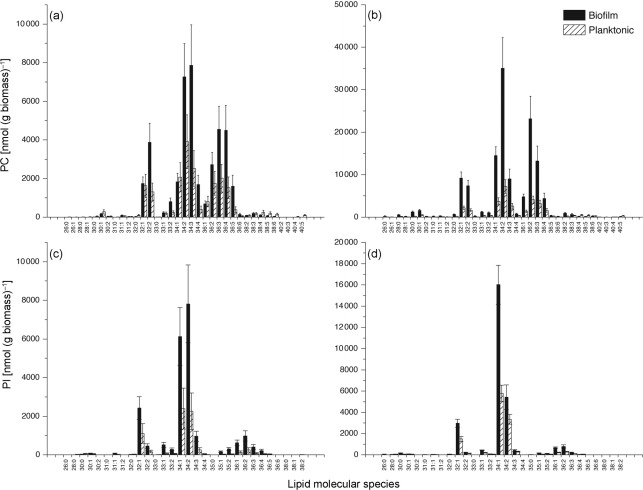
ESI-MS/MS analysis allows identification of individual lipid molecular species. We characterized the lipids present in biofilms and planktonic cells grown for 6 h (early phase) or 48 h (mature phase). Comparison of specific phospholipid species (identified as total acyl carbons in the diacyl species : total acyl carbon–carbon double bonds) showed that the profile of molecular species differed significantly between biofilms and planktonic cells and also between early and mature phases of each growth form. Levels of nearly all detected molecular species for PC and PI were higher in biofilms than in planktonic cells (Fig. 2). Similar trends were observed for PE, PS, PA, PG, and the lysophospholipids (Supplementary Table S1).
Fig. 2.
Diacylphosphatidylcholine (PC) and diacylphosphatidylinositol (PI) molecular species of C. albicans biofilms and planktonic cells grown to the early or the mature phase. Lipid species are indicated by total acyl carbons : total carbon–carbon double bonds (n = 5 for each). (a) PC early phase, (b) PC mature phase, (c) PI early phase, (d) PI mature phase. Data represent mean±se (n = 5).
There were significant compositional differences in molecular species between planktonic cells and biofilms in each developmental phase and between early and mature phases of each growth form. As shown in Table 1, the unsaturation indices of PC and PS were significantly higher in early-phase biofilms than in early-phase planktonic cells (indicating that biofilm cells had more double bonds per acyl chain than did the planktonic cells). At the mature stage, the unsaturation of PC, PE and PS was significantly lower in biofilms than in planktonic cells. During maturation, the planktonic cells underwent little or no change in composition (i.e. unsaturation) of most phospholipid classes, except for the lysophospholipids, which were reduced in unsaturation. On the other hand, the biofilms underwent a decrease in unsaturation of every lipid class but PA during maturation.
Table 1. Unsaturation index of phospholipids identified in C. albicans biofilms and planktonic cells at early and mature phases.
Unsaturation index for each class was calculated as the mean number of double bonds per fatty acid chain for the species within that class.
| Lipid | Biofilm | Planktonic | ||||||||
| Early phase | Mature phase | P-value* | Early phase | Mature phase | P-value* | |||||
| Mean | sem | Mean | sem | Mean | sem | Mean | sem | |||
| PC | 1.35 | 0.03 | 1.02 | 0.01 | ≤0.001 | 1.21 | 0.01 | 1.14 | 0.01 | ≤0.001 |
| PE | 1.06 | 0.01 | 0.92 | 0.01 | ≤0.001 | 1.04 | 0.00 | 1.00 | 0.01 | ≤0.001 |
| PI | 0.68 | 0.02 | 0.57 | 0.00 | ≤0.001 | 0.65 | 0.01 | 0.59 | 0.01 | 0.01 |
| PS | 0.66 | 0.01 | 0.59 | 0.00 | ≤0.001 | 0.63 | 0.01 | 0.62 | 0.00 | 0.07 |
| PA | 0.69 | 0.02 | 0.70 | 0.02 | 0.81 | 0.68 | 0.08 | 0.68 | 0.03 | 0.96 |
| Lyso-PC | 1.23 | 0.06 | 0.73 | 0.02 | ≤0.001 | 1.04 | 0.07 | 0.67 | 0.02 | 0.00 |
| Lyso-PE | 1.14 | 0.03 | 0.43 | 0.01 | ≤0.001 | 1.03 | 0.04 | 0.43 | 0.01 | ≤0.001 |
P-value for each lipid class represents the comparison of levels in biofilms or planktonic cells grown to early (6 h) vs mature (48 h) phases.
Taken together, the glycerophospholipid molecular species and class results indicate that planktonic cells and biofilms differ in lipid composition, and that biofilms undergo large decreases in unsaturation and an increase in PC to PE ratio during development.
Sphingolipid profile
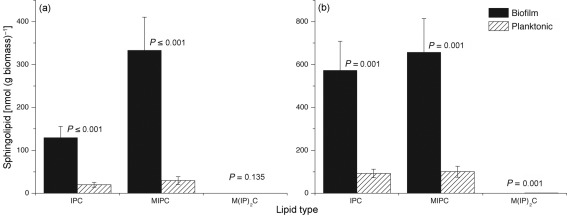
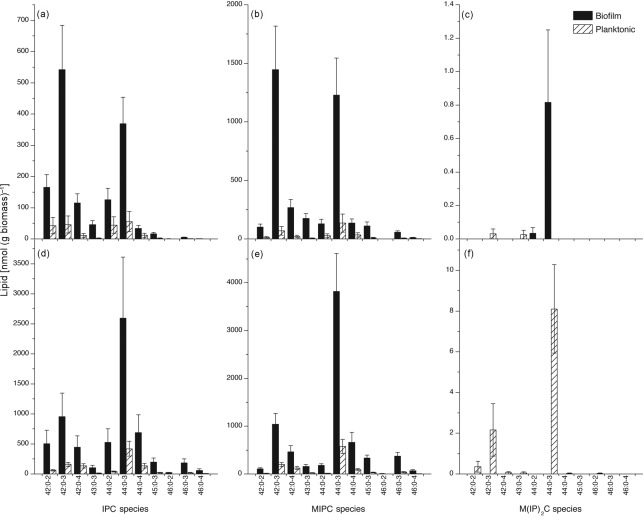
Our results showed that the level of PI was threefold higher in early-phase biofilms compared to early-phase planktonic cells. Since PI is involved in biosynthesis of membrane-localized sphingolipids, we decided to determine the levels of sphingolipids in biofilms and planktonic cells in both the early and mature phases. Among the three known sphingolipids [IPC, MIPC, M(IP)2C], levels of two (IPC, MIPC) were higher in biofilms than in planktonic cells in both early and mature phases (Fig. 3, P≤0.001). Analysis of the molecular species of sphingolipids revealed elevated levels of all species of IPC and MIPC (Fig. 4). Interestingly, there was a shift in the molecular species of both IPC and MIPC in biofilms as they matured. At 48 h, compared to 6 h, the ratio of longer-chain (such as 44 : 0-3) to shorter-chain species (such as 42 : 0-3) was higher (Fig. 4a, b, d, e). The M(IP)2C species (44 : 0-4), which was barely detectable at 6 h in either culture (Fig. 4c; note scale), was still undetectable in biofilms at 48 h, when it was clearly detectable in planktonic cells (Fig. 4f). Overall, the results showed that sphingolipid species were elevated in biofilms compared to planktonic C. albicans in both early and mature phases.
Fig. 3.
Distribution of sphingolipids in C. albicans biofilms and planktonic cells. Bars represent sphingolipid levels in biofilms and planktonic cells grown to (a) early phase (6 h) or (b) mature phase (48 h). IPC, inositolphosphorylceramide; MIPC, mannosylinositolphosphorylceramide; M(IP)2C, mannosyldiinositolphosphorylceramide. Data represent mean±se (n = 5).
Fig. 4.
Sphingolipid molecular species of C. albicans biofilms and planktonic cells grown to the early (a–c) or mature (d–f) phase. Lipid species are indicated by (total carbons in the sphingoid base+fatty amide) : (total carbon–carbon double bonds in sphingoid base+fatty amide)-(number of hydroxyl groups on base plus fatty amide chain). Data represent mean± se, n = 5 for each panel. (a, d) Inositolphosphorylceramide (IPC); (b, e) mannosylinositolphosphorylceramide (MIPC); (c, f) mannosyldiinositolphosphorylceramide [M(IP)2C].
Lipid rafts are involved in formation of C. albicans biofilms
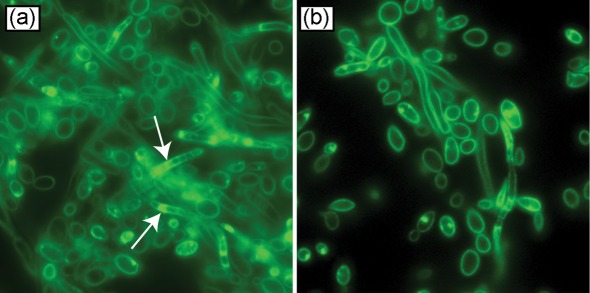
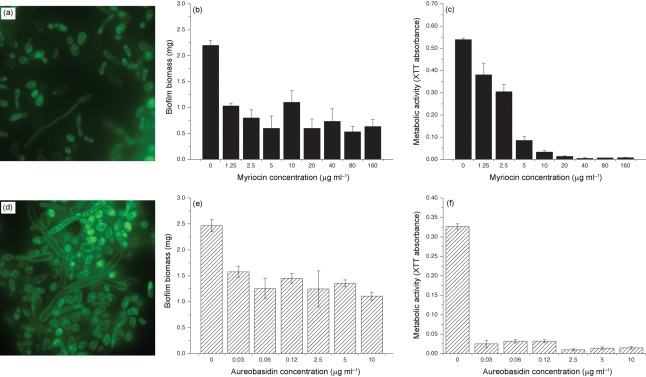
Since sphingolipids are key components of lipid rafts, we hypothesized that levels of lipid rafts are altered in biofilms compared to planktonic cells, and that these rafts are important in biofilm formation. To test this hypothesis, we determined the levels of lipid rafts in C. albicans biofilms using filipin staining. We found that C. albicans biofilms contained more lipid rafts than planktonic cells, especially in the hyphal elements (Fig. 5). Next, we evaluated the effect of myriocin, which inhibits serine palmitoyltransferase (a key enzyme involved in sphingolipid biosynthesis; Martin & Konopka, 2004), on raft formation in biofilms. Biofilms were grown in the absence or presence of different concentrations of myriocin (these concentrations were not toxic for Candida, as determined by cell-viability assay) and quantified by biomass determination. Our results showed that exposure to myriocin resulted in reduction in the raft formation, especially in hyphae (Fig. 6a). Moreover, the ability of C. albicans to form biofilms was significantly reduced in the presence of myriocin, as determined by both dry weight and metabolic activity assays (Fig. 6b, c, P<0.05).
Fig. 5.
Distribution of lipid rafts in C. albicans grown as (a) biofilm or (b) planktonically, visualized by fluorescence microscopy following staining with filipin (image taken at ×200 magnification; arrows, lipid rafts).
Fig. 6.
Effect of the sphingolipid biosynthesis inhibitors myriocin and aureobasidin A on distribution of lipid rafts in C. albicans biofilms. Biofilms exposed to myriocin were characterized for (a) raft distribution, (b) biomass, and (c) metabolic activity; similar assays were performed to determine the effect of aureobasidin A on (d) lipid rafts, (e) biomass, and (f) metabolic activity.
Biosynthesis of IPCs (key sphingolipid constituents) involves transfer of the inositol moiety from PI to sphingosine/dihydrosphingosine, in a reaction catalysed by the enzyme IPC synthase (Aur1p) (Nagiec et al., 1997). To determine whether this transfer reaction is critical for biofilm formation and maintenance of the lipid raft profile in biofilms, we grew biofilms in the presence or absence of aureobasidin A, a known inhibitor of Aur1p (Aeed et al., 2009; Cerantola et al., 2009). Our results showed that exposure to aureobasidin, which also reduced metabolic activity, resulted in reduced biofilm formation, and that these biofilms exhibited fewer lipid rafts in the hyphal membrane and reduced biofilm biomass (Fig. 6d–f). These results suggest that inhibition of the M(IP)2C biosynthesis pathway leads to disruption of lipid rafts and attenuates the ability of C. albicans to form biofilms.
Disruption of M(IP)2C biosynthesis affects biofilm formation by C. albicans
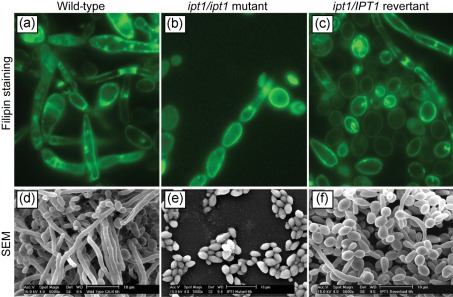
Next, we investigated whether disruption of IPT1, which encodes the enzyme catalysing the terminal step of M(IP)2C biosynthesis (Dickson et al., 1997), influences the ability of C. albicans to form biofilms. Our results showed that the raft formation profile of a Δipt1 mutant was noticeably reduced compared to the isogenic wild-type and Δipt1/IPT1 revertant strains (Fig. 7a–c). SEM analysis showed that biofilms formed by the Δipt1 mutant comprised only clumps of adherent yeast cells, while the revertant and wild-type strains formed robust biofilms with yeasts and hyphae enmeshed in a dense network (Fig. 7d–f). These results confirm that IPT1 plays a critical role in the ability of C. albicans to form biofilms and associated lipid rafts.
Fig. 7.
Relationship of IPT1 to membrane-localized lipid rafts in C. albicans biofilms. (a–c) Distribution of lipid rafts in biofilms formed by isogenic (a) wild-type, (b) Δipt1 mutant, and (c) Δipt1/IPT1 revertant. Rafts were visualized by staining with the fluorescent dye filipin. (d–f) SEM analyses of biofilms formed by (d) wild-type, (e) Δipt1 mutant, and (f) Δipt1/IPT1 revertant (scale bars in SEM images, 10 µm).
Discussion
In the current study, we performed lipidomics analysis of C. albicans biofilms and planktonic cells, and demonstrated significant growth-phase-dependent differences in the profile of polar lipid classes, individual molecular lipid species, and unsaturation index. Our results showed that PI was an abundant phospholipid in biofilms, and that sphingolipid levels were elevated in biofilms. In addition, we demonstrated that inositolphosphoryltransferase (Ipt1p), a key enzyme catalysing the biosynthesis of lipid rafts, plays a critical role in the ability of Candida to form biofilms.
In general, the lipid profile observed in planktonic cells in our studies was similar to those reported recently (Singh et al., 2010), but our analysis revealed that the levels of all polar lipids identified were higher in biofilms than in planktonic cells. Differences in lipid levels between biofilms and planktonic cells were observed in both early and mature phases; the difference was more pronounced in the early phase. Since adhesion to a substrate is an important early step in biofilm formation, lipids that were elevated in early-phase biofilms may be important regulators of fungal adherence in biofilms. In this regard, altered lipid composition is reported to affect Candida adherence (Ghannoum et al., 1986) and nutrient uptake (Ansari & Prasad, 1993; Dudani & Prasad, 1985; Singh et al., 1978). In an early study, Ghannoum et al. (1986) showed that lipids from C. albicans and Candida tropicalis, but not from the weakly adherent Candida pseudotropicalis, significantly blocked in vitro adherence of the respective yeast cells to buccal epithelial cells. Moreover, in the same study it was demonstrated that individual phospholipids, sterols and steryl esters blocked adherence significantly, while triacylglycerols and free fatty acids showed no effect.
The unsaturation index of most phospholipids decreased as biofilms matured. Lomb et al. (1975) showed that polyene-resistant Candida cells contained significantly reduced amounts of ergosterol and higher levels of fatty acids containing shorter and more saturated chains. Pesti et al. (1985) reported that sterol mutant strains of C. albicans exhibited a slight increase in saturation and a decrease in the chain length of fatty acids. Furthermore, these investigators showed an increase in the levels of PI and PA, and a decrease in the amount of PC and PS in these mutant strains. Since altered lipid composition (e.g. alteration in sterol levels) is known to be a mechanism of resistance in planktonic Candida cells (Hitchcock et al., 1986; Mago & Khuller, 1989) as well as in Candida biofilms (Mukherjee et al., 2003), it is possible that the increase in unsaturated index in Candida biofilms (of early phase) may contribute to alterations in membrane fluidity, biofilm maturation and resistance against antifungal agents. Our studies also revealed that the PC : PE ratio increased during biofilm maturation, along with a decrease in acyl unsaturation, which may indicate a reduction in the propensity of the membrane to undergo a transition to hexagonal phase (non-bilayer), a feature that may relate to the attachment of the biofilms.
PI has been shown to contribute to antifungal resistance (Abu-Elteen & Whittaker, 1997-1998; Hitchcock et al., 1987), biosynthesis and localization of GPI-anchored proteins, germination (Koul et al., 1995) and host pathogenesis (Reynolds, 2009). Additionally, PI metabolism is intricately tied to biosynthesis of yeast sphingolipids, which are a major constituent of membrane-localized lipid rafts. The major sphingolipids present in yeast lipid rafts are GIPCs: IPC, MIPC and M(IP)2C (Thevissen et al., 2005). We found that levels of sphingolipids, like PI, were elevated in biofilms compared to planktonic cells. Since sphingolipids are linked to formation of lipid rafts, we also analysed lipid rafts present in biofilms. Our studies using specific inhibitors (myriocin and aureobasidin) confirmed that lipid raft formation is correlated with the ability of C. albicans to form biofilms. Further confirmation of the notion that lipid rafts modulate biofilm formation was derived from the experiments showing that disruption of IPT1 resulted in abrogation of the ability of Candida to form biofilms.
The ability of lipid rafts to modulate biofilm formation may be linked to their known association with maintenance of membrane integrity and segregation of membrane-localized proteins. In this regard, lipid rafts have been proposed to play important roles in polarization and compartmentalization of the plasma membrane (Wachtler & Balasubramanian, 2006), surface transport of plasma membrane ATPase (Toulmay & Schneiter, 2007), hyphal growth (Martin & Konopka, 2004), drug resistance (Hallstrom et al., 2001; Pasrija et al., 2008; Prasad et al., 2005; Thevissen et al., 2000) and localization of virulence-associated proteins (Siafakas et al., 2006).
Although our study showed an increase in the levels of all major phospholipid classes, we focused on PI because of its link with biosynthesis of sphingolipids, which are critical components of lipid rafts. Since enzymes involved in the biosynthesis of other phosphoglycolipids (e.g. PS) have also been linked to virulence of C. albicans (Chen et al., 2010), further studies are warranted to investigate the role of these lipids in Candida biofilms.
In conclusion, we have demonstrated significant differences in the distribution of lipid molecular classes between biofilms and planktonically grown Candida. Our results provide support for the notion that lipids play a critical role in modulating microbial biofilms. Additionally, our findings demonstrate that lipid rafts are critical to the ability of Candida to form biofilms.
Supplementary Material
Acknowledgements
This work was supported by funds from the NIH/NIAID (RO1 AI035097) and NIH/NIDCR (R01 DE017486-01A1 and R01DE 13932-4) to M. A. G. and NIH/NIAID (R21AI074077-01A2) to P. K. M. Equipment acquisition and method development at the Kansas Lipidomics Research Center were funded by the National Science Foundation (EPS 0236913, MCB 0455318 and 0920663, DBI 0521587), Kansas Technology Enterprise Corporation, K-IDeA Networks of Biomedical Research Excellence (INBRE) of the National Institutes of Health (P20RR16475) and Kansas State University.
Abbreviations:
- GIPC
glycosylinositol phosphorylceramide
- GPI
glycosylphosphatidylinositol
- IPC
inositolphosphorylceramide
- MIPC
mannosylinositolphosphorylceramide
- M(IP)2C
mannosyldiinositolphosphorylceramide
- PA
phosphatidic acid
- PC
phosphatidylcholine
- PE
phosphatidylethanolamine
- PG
phosphatidylglycerol
- PI
phosphatidylinositol
- PS
phosphatidylserine
- SEM
scanning electron microscopy
Footnotes
A supplementary figure and table are available with the online version of this paper.
References
- Abu-Elteen K. H., Whittaker P. A. (1997-1998). Effect of sub-inhibitory concentration of chlorhexidine on lipid and sterol composition of Candida albicans. Mycopathologia 140, 69–76. 10.1023/A:1006852207366 [DOI] [PubMed] [Google Scholar]
- Aeed P. A., Young C. L., Nagiec M. M., Elhammer A. P. (2009). Inhibition of inositol phosphorylceramide synthase by the cyclic peptide aureobasidin A. Antimicrob Agents Chemother 53, 496–504. 10.1128/AAC.00633-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ansari S., Prasad R. (1993). Effect of miconazole on the structure and function of plasma membrane of Candida albicans. FEMS Microbiol Lett 114, 93–98. 10.1111/j.1574-6968.1993.tb06556.x [DOI] [PubMed] [Google Scholar]
- Brügger B., Erben G., Sandhoff R., Wieland F. T., Lehmann W. D. (1997). Quantitative analysis of biological membrane lipids at the low picomole level by nano-electrospray ionization tandem mass spectrometry. Proc Natl Acad Sci U S A 94, 2339–2344. 10.1073/pnas.94.6.2339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerantola V., Guillas I., Roubaty C., Vionnet C., Uldry D., Knudsen J., Conzelmann A. (2009). Aureobasidin A arrests growth of yeast cells through both ceramide intoxication and deprivation of essential inositolphosphorylceramides. Mol Microbiol 71, 1523–1537. 10.1111/j.1365-2958.2009.06628.x [DOI] [PubMed] [Google Scholar]
- Chandra J., McCormick T. S., Imamura Y., Mukherjee P. K., Ghannoum M. A. (2007). Interaction of Candida albicans with adherent human peripheral blood mononuclear cells increases C. albicans biofilm formation and results in differential expression of pro- and anti-inflammatory cytokines. Infect Immun 75, 2612–2620. 10.1128/IAI.01841-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandra J., Mukherjee P. K., Ghannoum M. A. (2008). In vitro growth and analysis of Candida biofilms. Nat Protoc 3, 1909–1924. 10.1038/nprot.2008.192 [DOI] [PubMed] [Google Scholar]
- Chen Y. L., Montedonico A. E., Kauffman S., Dunlap J. R., Menn F. M., Reynolds T. B. (2010). Phosphatidylserine synthase and phosphatidylserine decarboxylase are essential for cell wall integrity and virulence in Candida albicans. Mol Microbiol 75, 1112–1132. 10.1111/j.1365-2958.2009.07018.x [DOI] [PubMed] [Google Scholar]
- Devaiah S. P., Roth M. R., Baughman E., Li M., Tamura P., Jeannotte R., Welti R., Wang X. (2006). Quantitative profiling of polar glycerolipid species from organs of wild-type Arabidopsis and a phospholipase Dalpha1 knockout mutant. Phytochemistry 67, 1907–1924. 10.1016/j.phytochem.2006.06.005 [DOI] [PubMed] [Google Scholar]
- Dickson R. C., Nagiec E. E., Wells G. B., Nagiec M. M., Lester R. L. (1997). Synthesis of mannose-(inositol-P)2-ceramide, the major sphingolipid in Saccharomyces cerevisiae, requires the IPT1 (YDR072c) gene. J Biol Chem 272, 29620–29625. 10.1074/jbc.272.47.29620 [DOI] [PubMed] [Google Scholar]
- Dudani A. K., Prasad R. (1985). Differences in amino acid transport and phospholipid contents during the cell cycle of Candida albicans. Folia Microbiol (Praha) 30, 493–500. 10.1007/BF02927612 [DOI] [PubMed] [Google Scholar]
- Ghannoum M. A., Burns G. R., Elteen K. A., Radwan S. S. (1986). Experimental evidence for the role of lipids in adherence of Candida spp. to human buccal epithelial cells. Infect Immun 54, 189–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghannoum M. A., Swairjo I., Soll D. R. (1990). Variation in lipid and sterol contents in Candida albicans white and opaque phenotypes. J Med Vet Mycol 28, 103–115. 10.1080/02681219080000151 [DOI] [PubMed] [Google Scholar]
- Hallstrom T. C., Lambert L., Schorling S., Balzi E., Goffeau A., Moye-Rowley W. S. (2001). Coordinate control of sphingolipid biosynthesis and multidrug resistance in Saccharomyces cerevisiae. J Biol Chem 276, 23674–23680. 10.1074/jbc.M101568200 [DOI] [PubMed] [Google Scholar]
- Hitchcock C. A., Barrett-Bee K. J., Russell N. J. (1986). The lipid composition of azole-sensitive and azole-resistant strains of Candida albicans. J Gen Microbiol 132, 2421–2431. [DOI] [PubMed] [Google Scholar]
- Hitchcock C. A., Barrett-Bee K. J., Russell N. J. (1987). The lipid composition and permeability to azole of an azole- and polyene-resistant mutant of Candida albicans. J Med Vet Mycol 25, 29–37. 10.1080/02681218780000041 [DOI] [PubMed] [Google Scholar]
- Ibrahim A. S., Ghannoum M. A., Prasad R. (1996). Chromatographic analysis of lipids. In Manual on Membrane Lipids, p. 52 Edited by Prasad R. New York: Springer-Verlag. [Google Scholar]
- Koul A., Chandra J., Prasad R. (1995). Status of membrane lipids and amino acid transport in morphological mutants of Candida albicans. Biochem Mol Biol Int 35, 1215–1222. [PubMed] [Google Scholar]
- Lattif A. A., Chandra J., Chang J., Liu S., Zhou G., Chance M. R., Ghannoum M. A., Mukherjee P. K. (2008). Proteomic and pathway analyses reveal phase-dependent over-expression of proteins associated with carbohydrate metabolic pathways in Candida albicans biofilms. The Open Proteomics Journal 1, 5–26. 10.2174/1875039700801010005 [DOI] [Google Scholar]
- Lomb M., Fryberg M., Oehlschlager A. C., Unrau A. M. (1975). Sterol and fatty acid composition of polyene macrolide antibiotic resistant Torulopsis glabrata. Can J Biochem 53, 1309–1315. 10.1139/o75-177 [DOI] [PubMed] [Google Scholar]
- Mago N., Khuller G. K. (1989). Influence of lipid composition on the sensitivity of Candida albicans to antifungal agents. Indian J Biochem Biophys 26, 30–33. [PubMed] [Google Scholar]
- Martin S. W., Konopka J. B. (2004). Lipid raft polarization contributes to hyphal growth in Candida albicans. Eukaryot Cell 3, 675–684. 10.1128/EC.3.3.675-684.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee P. K., Chandra J., Kuhn D. M., Ghannoum M. A. (2003). Mechanism of fluconazole resistance in Candida albicans biofilms: phase-specific role of efflux pumps and membrane sterols. Infect Immun 71, 4333–4340. 10.1128/IAI.71.8.4333-4340.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee P. K., Zhou G., Munyon R., Ghannoum M. A. (2005). Candida biofilm: a well-designed protected environment. Med Mycol 43, 191–208. 10.1080/13693780500107554 [DOI] [PubMed] [Google Scholar]
- Nagiec M. M., Nagiec E. E., Baltisberger J. A., Wells G. B., Lester R. L., Dickson R. C. (1997). Sphingolipid synthesis as a target for antifungal drugs. Complementation of the inositol phosphorylceramide synthase defect in a mutant strain of Saccharomyces cerevisiae by the AUR1 gene. J Biol Chem 272, 9809–9817. 10.1074/jbc.272.15.9809 [DOI] [PubMed] [Google Scholar]
- Pasrija R., Panwar S. L., Prasad R. (2008). Multidrug transporters CaCdr1p and CaMdr1p of Candida albicans display different lipid specificities: both ergosterol and sphingolipids are essential for targeting of CaCdr1p to membrane rafts. Antimicrob Agents Chemother 52, 694–704. 10.1128/AAC.00861-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pesti M., Horváth L., Vígh L., Farkas T. (1985). Lipid content and ESR determination of plasma membrane order parameter in Candida albicans sterol mutants. Acta Microbiol Hung 32, 305–313. [PubMed] [Google Scholar]
- Prasad T., Saini P., Gaur N. A., Vishwakarma R. A., Khan L. A., Haq Q. M., Prasad R. (2005). Functional analysis of CaIPT1, a sphingolipid biosynthetic gene involved in multidrug resistance and morphogenesis of Candida albicans. Antimicrob Agents Chemother 49, 3442–3452. 10.1128/AAC.49.8.3442-3452.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds T. B. (2009). Strategies for acquiring the phospholipid metabolite inositol in pathogenic bacteria, fungi and protozoa: making it and taking it. Microbiology 155, 1386–1396. 10.1099/mic.0.025718-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siafakas A. R., Wright L. C., Sorrell T. C., Djordjevic J. T. (2006). Lipid rafts in Cryptococcus neoformans concentrate the virulence determinants phospholipase B1 and Cu/Zn superoxide dismutase. Eukaryot Cell 5, 488–498. 10.1128/EC.5.3.488-498.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh M., Jayakumar A., Prasad R. (1978). The effect of altered lipid composition on the transport of various amino acids in Candida albicans. Arch Biochem Biophys 191, 680–686. 10.1016/0003-9861(78)90407-1 [DOI] [PubMed] [Google Scholar]
- Singh A., Prasad T., Kapoor K., Mandal A., Roth M., Welti R., Prasad R. (2010). Phospholipidome of Candida: each species of Candida has distinctive phospholipid molecular species. OMICS 14, 665–677. 10.1089/omi.2010.0041 [DOI] [PubMed] [Google Scholar]
- Thevissen K., Cammue B. P., Lemaire K., Winderickx J., Dickson R. C., Lester R. L., Ferket K. K., Van Even F., Parret A. H., Broekaert W. F. (2000). A gene encoding a sphingolipid biosynthesis enzyme determines the sensitivity of Saccharomyces cerevisiae to an antifungal plant defensin from dahlia (Dahlia merckii). Proc Natl Acad Sci U S A 97, 9531–9536. 10.1073/pnas.160077797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thevissen K., Francois I. E., Aerts A. M., Cammue B. P. (2005). Fungal sphingolipids as targets for the development of selective antifungal therapeutics. Curr Drug Targets 6, 923–928. 10.2174/138945005774912771 [DOI] [PubMed] [Google Scholar]
- Toulmay A., Schneiter R. (2007). Lipid-dependent surface transport of the proton pumping ATPase: a model to study plasma membrane biogenesis in yeast. Biochimie 89, 249–254. 10.1016/j.biochi.2006.07.020 [DOI] [PubMed] [Google Scholar]
- Wachtler V., Balasubramanian M. K. (2006). Yeast lipid rafts? – an emerging view. Trends Cell Biol 16, 1–4. 10.1016/j.tcb.2005.11.008 [DOI] [PubMed] [Google Scholar]
- Welti R., Li W., Li M., Sang Y., Biesiada H., Zhou H. E., Rajashekar C. B., Williams T. D., Wang X. (2002). Profiling membrane lipids in plant stress responses. Role of phospholipase D alpha in freezing-induced lipid changes in Arabidopsis. J Biol Chem 277, 31994–32002. 10.1074/jbc.M205375200 [DOI] [PubMed] [Google Scholar]
- Yeater K. M., Chandra J., Cheng G., Mukherjee P. K., Zhao X., Rodriguez-Zas S. L., Kwast K. E., Ghannoum M. A., Hoyer L. L. (2007). Temporal analysis of Candida albicans gene expression during biofilm development. Microbiology 153, 2373–2385. 10.1099/mic.0.2007/006163-0 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.