Abstract
A nitrile labeled amino acid, p-cyanophenylalanine, is introduced near the active site of the semisynthetic enzyme ribonuclease S to serve as a probe of protein dynamics and fluctuations. Ribonuclease S is the limited proteolysis product of subtilisin acting on ribonuclease A, and consists of a small fragment including amino acids 1–20, the S-peptide, and a larger fragment including residues 21–124, the S-protein. A series of two-dimensional vibrational echo experiments performed on the nitrile labeled S-peptide and the RNase S are described. The time-dependent changes in the two-dimensional infrared vibrational echo line shapes are analyzed using the center line slope method to obtain the frequency-frequency correlation function (FFCF). The observations show that the nitrile probe in the S-peptide has dynamics that are similar to, but faster than, those of the single amino acid p-cyanophenylalanine in water. In contrast, the dynamics of the nitrile label when the peptide is bound to form ribonuclease S are dominated by homogeneous dephasing (motionally narrowed) contributions with only a small contribution from very fast inhomogeneous structural dynamics. The results provide insights into the nature of the structural dynamics of the ribonuclease S complex. The equilibrium dynamics of the nitrile labeled S-peptide and the ribonuclease S complex are also investigated by molecular dynamics simulations. The experimentally determined FFCFs are compared to the FFCFs obtained from the molecular dynamics simulations, thereby testing the capacity of simulations to determine the amplitudes and time scales of protein structural fluctuations on fast time scales under thermal equilibrium conditions.
I. Introduction
Two dimensional infrared (2D IR) vibrational echo spectroscopy is employed to probe the dynamics of the semi-synthetic enzyme, ribonuclease S (RNase S).1 RNase S is the limited proteolysis product of subtilisin acting on ribonuclease A (RNase A), and consists of a smaller fragment including amino acids 1–20, the S-peptide, and a larger fragment including residues 21–124, the S-protein. When separated, the S-protein is nonfunctional because the active site is split and the S-peptide is disordered.2 The complex restores nearly native enzymatic activity when the S-peptide is combined non-covalently to form S-protein.1 Because the S-peptide can be readily prepared by solid phase peptide synthesis, it is straightforward to introduce the unnatural amino acid, p-cyanophenylalanine (p-CN-Phe) in the S-peptide, which when non-covalently bound to the S-protein, forms the nitrile (CN) modified RNase S.3 The x-ray crystal structure of this complex shows minimal structural perturbation by p-CN-Phe, and functional assays demonstrate minimal effect on catalytic function, even though the probe is inserted within a few angstroms of key residues in the active site. In this study we report the fast equilibrium structural dynamics of folded RNase S complex as well that of the S-peptide in aqueous buffer using p-CN-Phe as the vibrational dynamics label (VDL). The frequency-frequency correlation function (FFCF), which connects the experimental observables to the underlying protein structural dynamics, is extracted from the time dependent 2D IR spectra using the center line slope (CLS) method.4,5 The results provide insights into the nature of the energy landscape around the free energy minimum of the folded protein structure.
The study of the dynamics of biological molecules undergoing structural changes on a wide range of timescales, from femtoseconds to milliseconds, has been the topic of scientific interest in recent years.6–11 The biological functions of these bio-molecules are related to the ability of the proteins or enzymes to undergo time-dependent structural changes on a variety of timescales.6–8 Understanding the structural dynamics of bio-molecules can provide valuable information regarding the structure-function relationship.12–16
2D IR vibrational echo spectroscopy can measure structural evolution on times scales spanning 100 femtoseconds to 100 picoseconds and has been used to understand the fast protein dynamics.17–21 Applications of 2D IR spectroscopy to biological molecules include studies of protein structure, dynamics, folding and unfolding.18,22–31 2D IR spectroscopy measures protein structural dynamics by determining the time dependent components of the IR absorption band of the vibration under observation. In general, the absorption line has homogeneous and inhomogeneous contributions, both of which are obtained from the 2D IR experiments. Spectral diffusion is the time evolution of the frequency of the VDL within the inhomogeneously broadened vibrational absorption spectrum caused by structural fluctuations. The experimental measurement of spectral diffusion is used to determine the FFCF, which is the connection between the experimental observables and the underlying dynamics of the system.32
The 2D IR experiments show that the nitrile probe in the S-peptide has dynamics that are similar to those of the single amino acid p-CN-Phe in water. Therefore, the dynamics probed by the CN label of the small peptide have substantial contributions from the fluctuations of the solvent although motions of the peptide also contribute. In contrast, the dynamics of the CN label when the peptide is bound to form RNase S are dominated by homogeneous dephasing (motionally narrowed) contributions with only a small contribution from very fast inhomogeneous structural dynamics. The nature of the dynamics of the RNase S is in contrast to many other proteins, enzymes, and peptides that have been elucidated with 2D IR vibrational echo spectroscopy.17,28,29,31 The other systems studied show substantial inhomogeneous broadening and spectral diffusion with the homogeneous component being either relatively minor or large but not the overwhelmingly dominant contribution to the absorption spectrum.28,31 In addition, the single small inhomogeneous component decays very rapidly, ~0.5 ps. This inhomogeneous component is itself close to being motionally narrowed. This single fast almost motionally narrow component is in contrast to other systems where decays of the inhomogeneous dynamics occur from a few to hundreds of picoseconds and longer.25,28,33 Some systems display a very fast decays but these are followed by slower decays. For example, water has a ~0.4 ps decay component followed by a 1.8 ps decay.34,35 A possible, although speculative explanation, for the observed difference of the RNase S enzyme compared to other enzymes28,31 and proteins19,30 is presented.
The equilibrium dynamics of the nitrile labeled S-peptide and the RNase S complex are also investigated by molecular dynamics (MD) simulations. The experimentally determined FFCF is compared to the FFCF obtained from the simulations. While the simulations do yield a very large homogeneous component, the spectral diffusion, (structural fluctuations) is approximately a factor of 10 too slow compared to the experimental results.
II. Materials and Methods
A. Sample Preparation
Type I-A ribonuclease (RNase A) and type VIII Subtilisin Carlsburg were purchased from Sigma Aldrich (St. Louis, MO) and used without further purification. RNase S was prepared using the previously reported procedures.36 In short, 40 µL of 1% subtilisin in 100 mM Tris (pH = 8.0) was added dropwise to 5 mL of 2% RNase A also in 100 mM Tris (pH = 8.0). RNase A was kept on ice and was continuously stirred during the entire process of adding the subtilisin. The mixture was kept for 12–16 hours on ice and the reaction was quenched with few drops of 2M HCl to pH < 4. The mixture was immediately purified by high pressure liquid chromatography on a C-18 reverse phase column using a gradient of 20%–80% acetonitrile in water with 0.1% trifluoroacetic acid (TFA) over 60 min. Liquid chromatography–mass spectrometry (LCMS) was used to identify the fractions containing S-peptide and S-protein.
Fluorenylmethyloxycarbonyl chloride (Fmoc) protected p-cyanophenylalanine was obtained from PepTech Corp. (Burlington, MA). CN modified S-peptide (Phe at position 8 was replaced by p-CNPhe) was synthesized by Elim Biopharmaceuticals Inc. (Hayward, CA). The detailed protocols have been reported earlier.3 S-protein (1.5 mol equiv.) was added to S-peptide in 20mM HEPES (4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid) buffer at pH = 8.0 followed by cation exchange fast protein liquid chromatography employing a gradient in the same buffer, ramping from 50 to 500 mM NaCl over 30 minutes. The fraction containing the RNase S complex showed the presence of both the S-peptide and the S-protein when identified by LCMS. The RNase S complex was lyophilized and the lyophilized product was dissolved in 50:50 glycerol/20 mM HEPES (pH = 8.0) to obtain a final protein concentration of 10 mM. S-peptide was also dissolved in 50:50 glycerol/20 mM HEPES (pH = 8.0). The final concentration of the S-peptide solution used in the experiments was 20 mM.
B. FTIR Spectroscopy
All spectra were obtained on a Bruker Vertex 70 FTIR spectrometer with a sample compartment connected to a nitrogen tank to purge atmospheric gasses. 20 µL of the sample solution was loaded into a demountable liquid cell (Bruker) with two sapphire windows (.750” thick, Meller Optics). The windows were separated by using two off-set semicircular mylar spacers (of thickness 75 µm and 100 µm). A liquid N2 cooled InSb detector was employed to detect the signal. For each run, 64 scans were collected at 1 cm−1 resolution. Absorption spectra were calculated from the log-difference of the sample and background transmissions. The absorption spectra were baselined using a polynomial fit.
C. 2D IR Spectroscopy
In the 2D IR vibrational echo experiment, three short IR pulses tuned to the vibrational frequency of the nitrile stretching frequency were crossed in the sample in a noncollinear box-car geometry. The IR excitation pulses (160 fs in duration and 90 cm−1 FWHM) were produced using a Ti:Sapphire oscillator/regenerative amplifier pumped optical parametric amplifier system.37 The three successive IR pulses induced the subsequent generation of the vibrational echo pulse, which propagateed in a unique direction. The vibrational echo pulse was combined with another pulse, the local oscillator (LO), which was fixed in time. Combining the vibrational echo pulse with the LO yielded heterodyne detection that provided both amplitude and phase information necessary for the Fourier transforms from the time domain to the frequency domain that yield the 2D spectra. The combined heterodyned vibrational echo signal was dispersed in a monochromator, and then detected by a 32-element mercury-cadmium-telluride (MCT) IR array, which measures the signal at 32 wavelengths simultaneously. The 2D IR data were obtained as a function of 3 variables: the emitted echo frequency (ωm), the variable time delay between pulses 1 and 2 (τ), and the variable time separation between pulses 2 and 3 (Tw, the waiting time). Scanning τ produceed a temporal interferrogram at each ωm. Numerical Fourier transforms of these temporal interferrogram gave the second variable frequency ωτ. The results were plotted as the 2D spectrum with frequency variables ωτ (horizontal axis) and ωm (vertical axis). A 2D spectrum was produced at each waiting time, where the change in the 2D spectra with Tw provided the dynamical information. The projection slice theorem was employed to obtain the absorptive 2D IR spectra. Pump-probe spectra were simulated using the absorption spectrum, where the absorption spectrum (for 0–1 absorption) and the inverted absorption spectrum (for 1–2 bleach) were separated by the diagonal anharmonicity. The projection of the 2D IR spectrum along ωm at each waiting time was matched with the simulated pump-probe spectrum.
Qualitatively, the experiment works as follows. The first IR pulse creates a coherent superposition of two vibrational states causing the molecules to oscillate in phase with their initial frequencies. The initial phase relationships are lost (free induction decay) because the spectral line is homogeneously and inhomogeneously broadened. The second IR pulse transfers phase relationships of the oscillators over the range of spectral frequencies into populations in the ground vibrational state (0) and the first excited vibrational state (1). The structural evolution of the proteins/peptides in the population period causes the initial frequencies to change. The third pulse ends the population period and creates a second coherence and rephasing that produces the vibrational echo pulse. The vibrational echo pulse reads out the final frequencies of the oscillators following the waiting time, Tw. The structural evolution of the system, which results in spectral diffusion (change in oscillator frequencies) is manifested in the change of 2D line shapes. Detailed information on the dynamics of the molecular system was obtained by analysis the change in the peak shapes as a function of Tw.
The theory, analysis, and interpretation of 2D IR vibrational echo experiments have been described previously.4,5,32,38 With increasing Tw, the structural changes of the proteins and peptides cause the frequencies of the CN stretching mode to change within the distribution of frequencies reflected by the absorption spectrum, resulting in changes in the 2D line shapes. This frequency evolution or spectral diffusion is directly related to the conformational fluctuations of the peptides/proteins. Structural fluctuations of the protein/peptide and the surrounding solvent molecules produce a time dependent electric field at the CN stretching mode, which is the vibrational dynamics label. The structural dynamics induced time evolution of the electric field produce changes in the frequencies through the Stark effect, which gives rise to the spectral diffusion. The Stark mechanism that connects the structural dynamics to spectral diffusion is supported by detailed molecular dynamics (MD) simulation of myoglobin with CO bound at the active side as the VDL.28,39–41 For myoglobin-CO, systematic decomposition of the contributions to dephasing in the MD simulations from concentric shells of the protein show that the vibrational probe not only senses the motions of the atoms in its local vicinity, but also the global dynamics of the protein and the surrounding solvent molecules.40 The MD simulation results on nitrile modified S-peptide and the RNase S complex discussed later in this paper also support the same Stark mechanism for CN as the VDL.
The changes in the 2D IR line shapes were used to determine the timescales and amplitudes that are associated with the structural dynamics of proteins. The FFCF quantifies the connection between the experimental observables and the time scales and amplitudes of the frequency fluctuations caused by protein structural fluctuations. The center line slope (CLS) formalism4,5 was used to obtain the FFCF from the Tw dependent 2D IR spectra and the linear IR absorption spectrum. The CLS is a normalized function closely related to the FFCF with the decay times the same as those in the FFCF. The difference between 1 and the value of CLS at Tw = 0 is related to magnitude of the homogeneous component of the absorption line.4 The CLS analysis combined with the linear absorption spectrum yields the full FFCF, including the Tw independent homogeneous component. The CLS method is based on the diagrammatic perturbation theory description of nonlinear experiments.38 This treatment assumes that the measured structural fluctuations are independent of the position in the inhomogeneous spectroscopic line (Gaussian approximation). This is an excellent assumption for most systems, particularly those with relatively narrow symmetric absorptions lines as those in the current study, and it is supported by simulations that use diagrammatic perturbation theory to compare simulation results to experimental observables.28 For systems such as water, which has very broad asymmetric absorption spectra, the Gaussian approximation still does a reasonable job of reproducing experimental results,34,35 with the magnitude of errors introduced by the Gaussian approximation the same as differences caused by the choice of the water model used a in simulation.34,35,42
The multi-exponential form, C(t), was used to model the FFCF:4,5,23
| (1) |
The Δi and τi terms are the amplitudes and timescales, respectively of the CN vibrational frequency fluctuations induced by protein/peptide and solvent structural dynamics. The static term is the amplitude factor for very slow CN frequency fluctuations arising from structural changes that occur on a time scale longer than the experimental time window, which is limited by the vibrational lifetime. This form of FFCF has been widely used and found applicable to the structural dynamics of proteins.19,28,40,41,43 Δi and τi are obtained from the 2D IR data. If Δiτi < 1, the dynamics are motionally narrowed and contribute to the homogeneous linewidth. The motionally narrowed dynamics are characterized by , where is the pure dephasing time. The total homogeneous dephasing time, T2, is given by . T1 is the vibrational lifetime. The pure dephasing line width is . The total homogeneous linewidth is Γ = 1/πT2.
D. Molecular Dynamics Simulations
Molecular dynamics simulations of both the nitrile modified S-peptide and the nitrile variant of the RNase complex were conducted using GROMACS 3.3.1 using the Amber-99 force field.44 The simulation strategy used here is the same as the approach developed for other systems containing nitrile probes.3,45 The protein was solvated in explicit SPC water. Nonbonded cutoffs of 10 Å were used. Simulations were conducted with periodic boundary conditions, and the calculation of long-range electrostatics was treated with the particle mesh Ewald model. Simulations were equilibrated (100 ps equilibration) and run (20 ns) using the Nose-Hoover thermostat and the Parinello-Raman barostat.
Spectroscopic observables are linked to molecular dynamics trajectories through the electrostatic model employed in previous studies.39,40,46–48 In this model, the local electric field at the ligand induces an instantaneous frequency shift in the nitrile vibrational frequency (vibrational Stark effect). As the structure fluctuates, the electric field along the CN bond fluctuates. This frequency fluctuation is given by
| (2) |
The electric field E⃗(t) was determined at the midpoint of the CN bond with 2 fs steps for all MD trajectories. In Equation 2, u⃗(t) is a unit vector pointing along the CN bond and the angle brackets denote an equilibrium average. The parameter λ plays the role of a microscopic version of the Stark effect tuning rate. The equilibrium autocorrelation function of the frequency fluctuations (FFCF),
| (3) |
was then computed from Equation 2 by averaging over trajectories. In practice, the electric field along the CN bond is obtained as a function of time. The autocorrelation of the electric field was then determined. The autocorrelation of the electric field multiplied by the Stark coupling constant, λ, is the FFCF. The MD simulations are independent of λ, which is used to convert simulated electric fields to spectroscopic frequencies. As the fluctuation in frequency is linear in λ, the FFCF in Equation 3 is quadratic in λ. Varying λ corresponds only to changing the amplitude factors of C(t) (see Equation 1), leaving the decay times unchanged.
Linear absorption lineshapes were calculated from simulated FFCFs. The simulations yield line shapes, but do not provide absolute frequencies. The bands were normalized, and the center frequencies of the simulated spectra when aligned with the experimental values, facilitate comparison of line shapes. The coupling parameter λ was adjusted to give the best fit to line shapes. This is the single adjustable parameter used to relate simulation results to experimental data. The parameter λ plays the role of the microscopic version of the Stark tuning rate, which can be independently measured experimentally. As discussed below, comparison of λ obtained from the simulations reported here to the value of Stark tuning rate obtained from time independent vibrational Stark spectroscopy provides a direct test of the validity of the fluctuating electric field model.
III. Results and Discussion
A. FTIR Spectroscopy
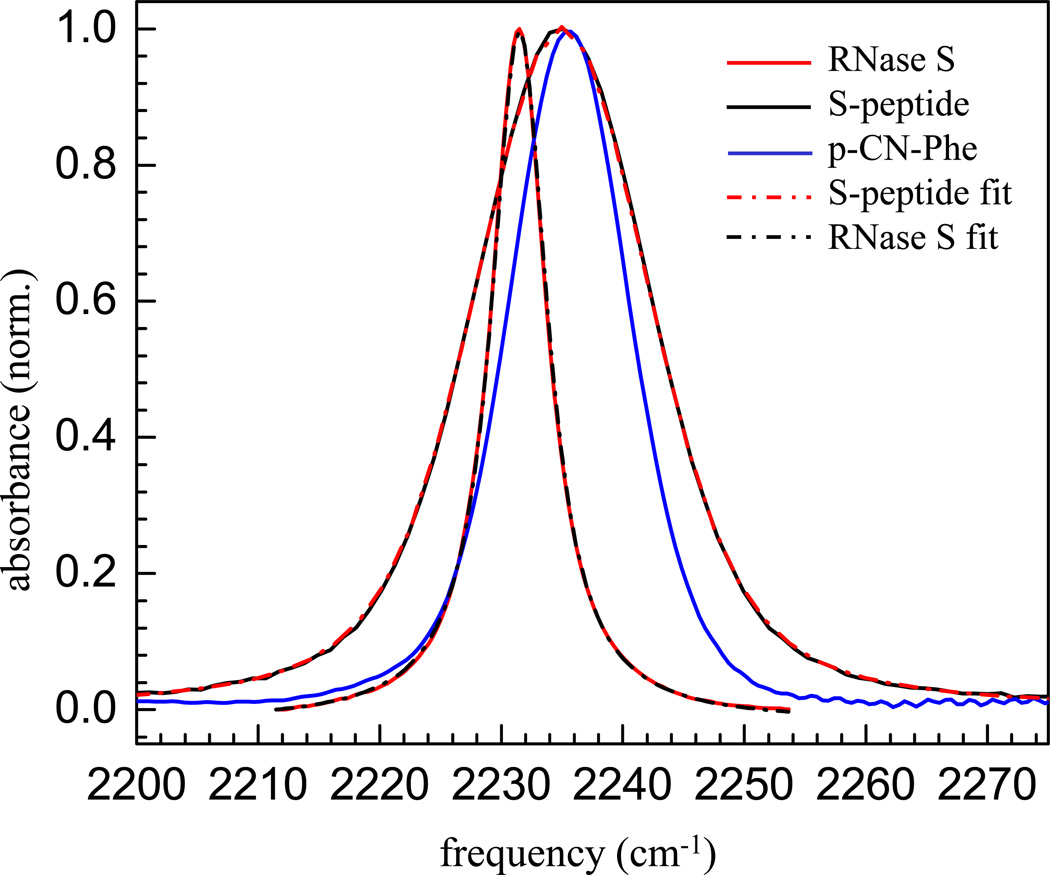
Figure 1 shows the IR absorption spectra of the CN stretching mode of the free S-peptide (black curve), the RNase S protein complex (red curve), and p-CN-Phe (blue curve); each in 50:50 glycerol/aqueous buffer solution at room temperature. The observed peak positions and/or line widths are clearly very different.3 For the free S-peptide there is a single band with its maximum at 2235 cm−1and a full width at half maximum (FWHM) of 17.7 cm−1. Using the Voigt line shape function in the non-linear curve fitting toolbox of OriginPro 8, this spectrum can be fit to a Voigt line shape (red dash-dotted curve in Figure 1)with a dominant Gaussian component of 13 cm−1 (FWHM) and a Lorentzian component of 7.2 cm−1 (FWHM). For the RNase S complex, a single band is observed at 2231 cm−1. This peak is fit with a Lorentzian line shape having FWHM of 5.7 cm−1. This peak can also be fit well with a Voigt profile (black dash-dotted curve in Figure 1) which gives a predominant Lorentzian component of 4.3 cm−1 (FWHM) and a Gaussian component of 2.6 cm−1 (FWHM). As seen from the observed peak positions of the CN mode, the RNase S complex shows a red-shift in the absorption frequency as compared to the free peptide and also shows a substantial narrowing of the absorption line width. The zwitterionic amino acid, p-CN-Phe (in 50/50 glycerol/20 mM HEPES, pH =8.0), has its maximum at 2235.6 cm−1 and has a FWHM of 11.7 cm−1. The S-peptide and the p-CN-Phe have almost the same peak position but the S-peptide spectrum is substantially wider. The spectra will be discussed further below after presentation of the 2D IR spectra.
Figure1.
Normalized FT-IR spectra of the CN label of p-CN-Phe (blue curve), the free S-peptide (black curve), and RNase S protein (red curve); Voigt fit to normalized FT-IR spectra of the CN label of free S-peptide (red dash-dotted curve), and RNase S protein (black dash-dotted curve).
It has been well established that nitriles, when exposed to bulk water, can accept a hydrogen bond49–51 causing a blue shift of the nitrile stretching frequency.49,50,52–55 For the nitrile modified free S-peptide, the blue shift in the absorption frequency relative to the complex, in which the nitrile probe is sequestered from water in the protein interior, is consistent with the previous observations. The crystal structure of the nitrile modified RNase S (PDB code: 3OQY)3 shows that the nitrile group of the RNase S complex cannot hydrogen bond to any donor from either the protein or the solvent. This explains the red shift of the CN mode of the RNase S protein upon complex formation, and the narrow line width is consistent with the other nitrile variants of RNase S complex at the same site (m-CN-Phe and SCN variants).3 These other nitrile variants are more complicated, either due to the possibility of multiple orientations evidenced by both the x-ray and the linear IR studies (for m-CN-Phe) or due to much smaller extinction coefficient of the aliphatic nitrile as the vibrational probe (for the SCN variant). So the other nitrile labeled sites of the RNase S complex were not included in the current study.
B. 2D IR Spectroscopy
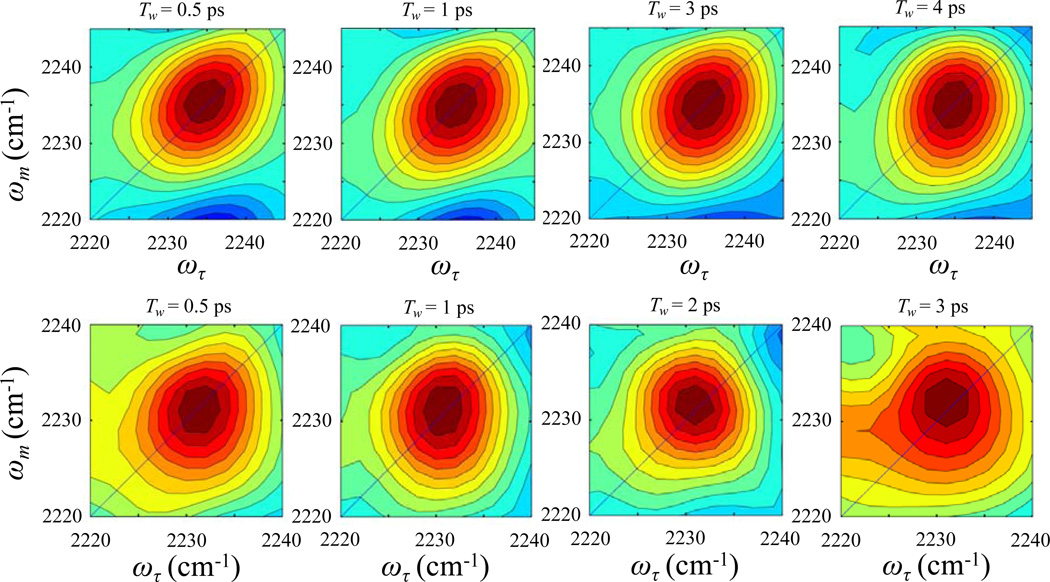
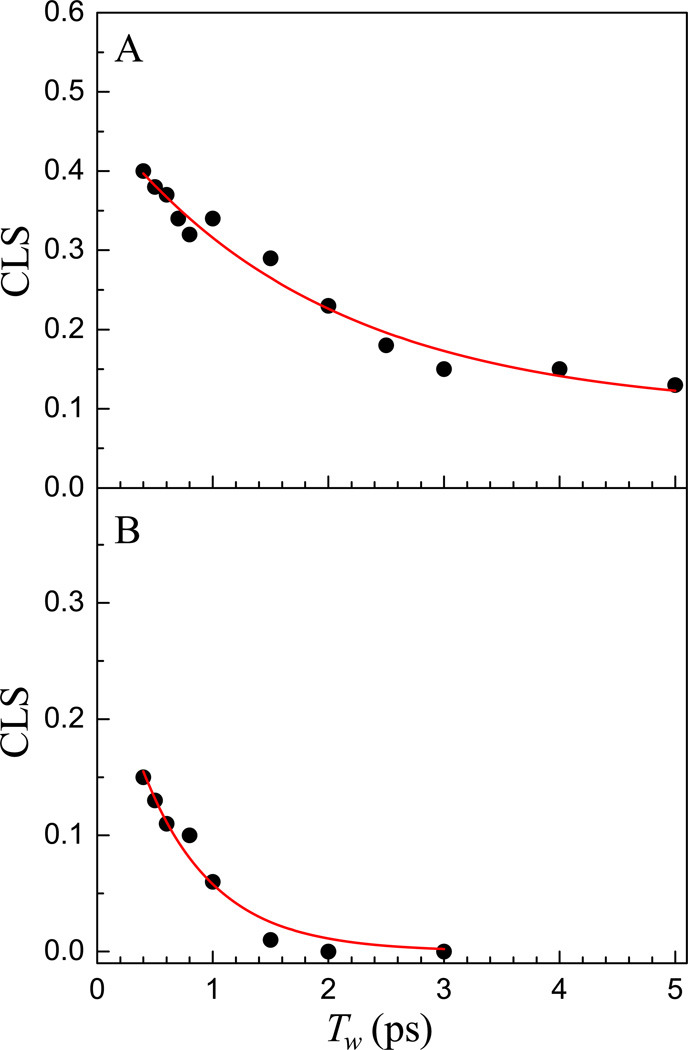
The 2D IR vibrational echo spectra of the CN stretching mode of free S-peptide (top panels) and RNase S complex (bottom panels) are shown in Figure 2 at several waiting times, Tw. A 2D IR spectrum consists of a pair of bands arising from the 0–1 transition on the diagonal (red) and the 1–2 transition (blue) shifted along the vertical (ωm) axis by the anharmonicity of the CN stretching mode.56,57 Here the diagonal 0–1 bands are analyzed, and the figure shows only these bands. A small portion of the 1–2 bands can be seen below the 0–1 bands in the top panels. For the nitrile labeled free S-peptide, the 0–1 band on the diagonal is centered at (ωτ = 2235 cm1, ωm = 2235 cm−1). The 2D IR peak shapes change with increasing Tw. At very short waiting time, the band is elongated along the diagonal. Greater elongation along the diagonal reflects greater inhomogeneity of the molecular system at the particular value of Tw. The bands become less elongated with increasing waiting time, which is the manifestation of spectral diffusion. In both samples, there is a large homogeneous component, which causes the bands, even at the shortest times, to have substantial width along the axis perpendicular to the diagonal. For the nitrile modified RNase S protein, the band is less elongated at the shortest waiting time compared to that of the nitrile modified S-peptide, indicating a larger homogeneous contribution to the spectrum. The 2D IR nitrile stretching band for the RNase S complex becomes almost circular within ~1 ps, which reflects very rapid spectral diffusion. The vibrational lifetime was found to be 4.3 ± 0.2 ps for both the S-peptide and the RNase S complex by fitting the Tw dependent 0–1 peak volumes to single exponential decays. Figure 3A shows the CLS data points and the fit (solid curve) for the CN band in the free S-peptide derived from the 2D IR spectra. The CLS data for the free peptide was fit to a single exponential decay with and without a static term (offset). Detailed statistical analysis using Akaike information criterion (AIC),58 which is a method that compares two models, was used to fit the same data. The method accounts for the number of fitting parameters in selecting between models. The AIC results demonstrated that the fitting model with the exponential decay and a long time offset is significantly superior to the simple single exponential decay. The details of AIC method and its application can be found in reference 55. Figure 3B shows the CLS data and fit for the CN band in the RNase S protein complex, which was fit to a single exponential decay. No static component was required to fit the CLS data of the protein complex as it goes to zero within the time span of the experiment. The absence of an offset shows that there are no dynamics that are so slow as to be outside of the experimental time window, which is set by the vibrational lifetime. The difference between 1 and the CLS value at Tw = 0, found by extrapolation of the curve from 400 fs to Tw = 0, is greater for the RNase S complex than that of the free S peptide. Thus, from inspection it can be seen that the relative contribution of the homogeneous component to their total absorption line shape is larger in the protein complex than in the S-peptide. The 2D IR results also show that the absorption line is inhomogeneously broadened although it is possible to fit the absorption line with a Lorentzian function.
Figure 2.
2D IR vibrational echo spectra of free S-peptide (top panels) and RNase S protein (bottom panels) at various waiting times, Tw.
Figure 3.
CLS data (points) and fits (red curves) of free S-peptide (A) and RNase S protein (B).
The FFCF parameters obtained from the fits to the CLS data and the linear absorption spectra for the CN stretching mode in both the S peptide and the RNase S complex are given in the Table 1. The FFCF parameters for p-CN-Phe in aqueous buffer (pH 5.0) are also given in Table 1.29 These will be used for comparisons to the current data. In addition to the FFCF parameters, the total inhomogeneous contributions (full width at half maximum) to the absorption linewidth (Inhomo) are also given in Table 1. The experiments on p-CN-Phe were conducted in solutions of 25%/75% glycerol/aqueous buffer.29 The experiments conducted here were performed in 50%/50% glycerol/aqueous buffer solutions, which have a higher viscosity (9.5 cP vs. 2.3 cP) than the solutions from the previously reported work. Results from the previously reported work showed that for nitrile modified Villin headpiece (a 35 amino acid peptide modified at 2 positions with p-CN-Phe) in 25%/75% glycerol/aqueous buffer, the viscosity of 2.6 cP of the concentrated sample (16mM) is very similar to that of 25%glycerol/75%water solution (2.3 cP). Thus it is assumed that viscosity of the 10 mM solution of S-peptide (a 20 amino acid peptide) in 50%/50% glycerol/aqueous buffer solutions is similar to that of pure 50%glycerol/50%water mixture.
Table 1.
FFCF parameters of S-peptide and RNase S complex
| Sample | Γ* (cm−1) | T2* (ps) | Δ1 (cm−1) | τ1 (ps) | Δs (cm−1) | Inhomo# (cm−1) |
|---|---|---|---|---|---|---|
| S-peptide (Experiment) | 7 ± 0.3 | 1.5 ± 0.3 | 5 ± 0.1 | 2.0 ± 0.5 | 2.4 ± 0.1 | 13.0 |
| RNase S (Experiment) | 4.5 ± 0.1 | 2.4 ± 0.1 | 1.1 ± 0.03 | 0.6 ± 0.08 | - | 2.6 |
| PheCN† | 4.7 ± 0.1 | 2.3 ± 0.1 | 3.1 ± 0.1 | 2.9 ± 0.5 | 2.1 ± 0.1 | 8.8 |
| S-peptide (Simulation) | 6 ± 0.2 | 1.8 ± 0.3 | 6 ± 0.2 | 2.5 ± 0.4 | 1.1 ± 0.03 | 14.3 |
| RNase S (Simulation) | 4 ± 0.1 | 2.7 ± 0.1 | 0.9 ± 0.02 | 4 ± 0.5 | 0.7 ± 0.1 | 2.7 |
From reference 29. The experiments from reference 29 were conducted in 25%/75% glycerol/water solutions while the experiments conducted here are in 50%/50% glycerol/water solutions.
Inhomo is the full width at half maximum (FWHM) of the inhomogeneous contribution to the absorption lines. The values of Δ are given as standard deviations. Inhomo is obtained by multiplying the convolution of Δ’s with .
The S-peptide CN absorption peak at 2235 cm−1 has a FWHM of 17.7 cm−1, while the p-CN-Phe (taken here in 50%/50% glycerol/water) peak is at 2235.6 cm−1 has a FWFM of 11.7 cm−1 (see Figure 1). Both lines are predominantly Gaussian in shape. These are in contrast to the CN absorption peak for RNase S at 2231.5 cm−1. This band has a FWHM of 5.7 cm−1 with a Lorentzian component of 4.3 cm−1 (FWHM) and a Gaussian component of 2.6 cm−1 (FWHM). The p-CN-Phe is surrounded by solvent, and the CN will accept a solvent hydroxyl hydrogen bond. The similarity in peak positions of the S-peptide and the p-CN-Phe indicates that the CN in the S-peptide is hydrogen bonded to a solvent hydroxyl. However, the substantially broader line of the S-peptide CN absorption compared to the p-CN-Phe demonstrates that the peptide plays a role in the nature and distribution of environments resulting in a broader distribution of conformational states that contribute to the absorption spectrum.
From the 2D IR experiments, the S-peptide has a decay component τ1 = 2.0 ps and a static component of Δs = 2.4 cm−1. These values are similar to those found previously for p-CN-Phe, that is, τ1 = 2.9 ps and a static component Δs = 2.1 cm−1.29 As discussed in connection with Equation 1, the amplitude of the static component is a measure of the extent of the dynamics that are so slow that they are well outside the experimental time window. Although the FFCF parameters are similar, it is important to consider that the S-peptide dynamics measured here are in a 50/50 glycerol/aqueous buffer solution while the previously reported dynamics for the p-CN-Phe are in 25/75 glycerol water solution. The higher viscosity of the 50/50 mixture (9.5 cP) compared to the 25/75 mixture (2.3 cP) should slow the dynamics of the S-peptide considerably if its dynamics arose solely from solvent dynamics. 2D IR experiments on p-CN-Phe in water (1 cP) give τ1 = 2.1 ps and no offset (Δs = 0).29 In fact, the observed S-peptide dynamics are ~30% faster than the p-CN-Phe in 25%75% glycerol/water although the viscosity is much greater. The τ1 value is about the same as for p-CN-Phe in water but the S-peptide has a substantial Δs while p-CN-Phe in water does not. For p-CN-Phe the spectral diffusion reflects solvent dynamics. However the results for the S-peptide indicate that sampling of conformations of the peptide contribute to the spectral diffusion, as sensed by the nitrile VDL, in addition to solvent structural evolution. Therefore, both the substantially wider absorption linewidth and the fast dynamics of the S-peptide compared to p-CN-Phe considering the high viscosity of the solvent show that the peptide is a substantial contributor to the distribution of structural environments as well as the rate at which they are sampled. While the fluctuations of the CN in p-CN-Phe are dominated by the dynamics in the solvent arise from the inhomogeneous environments and dynamics produced by the peptide as well as the solvent.
One might expect that binding the S peptide to the remainder of the protein to form the complete RNase S complex would cause the dynamics to slow as the CN label is no longer solvent exposed and the small 20 amino acid S-peptide is no longer free to undergo structural fluctuations that are only inhibited by the solvent. The crystal structure shows that the CN in the protein complex is sequestered from the solvent.3 In several other proteins studied with 2D IR, the observable dynamics occur on multiple time scales from a few picoseconds to 100 picoseconds or slower.19,23,28,30,31,33,40,41 These proteins frequently have a large static component in the FFCF indicating substantial slow dynamics outside the experimental time window. There are several examples where the inhomogeneous dynamics in proteins occur within a few picoseconds with no static component.25,33 For an inhibitor of HIV reverse transcriptase that has two nitriles, complete spectral diffusion occurs with a single time constant of 7.1 ps.33 In a study on formate dehydrogenase with azide anion located at the active site as the vibrational probe, Cheatum and co-workers fit the CLS decay of the ternary complexes with NAD+ (NADH) to a double exponential, with a fast decay component of 210 fs (150 fs) along with another decay time constant of 3.2 ps (4.6 ps).25 They assign the fastest decay component to the local hydrogen bond fluctuations of the hydrogen bond partners that bind and orient the substrate in the active site. In contrast to these proteins, the spectral diffusion sensed by the nitrile VDL in RNase S complex shows a very large homogeneous component and a single sub-picosecond decay constant.
The linear FT-IR spectrum of the CN label in the RNase S complex shown in Figure 1 can be fit well as a Lorentzian. The 2D IR experiments show the absorption spectrum is actually a Voigt line shape with a dominant Lorentzian contribution. This absorption line shape is very different from the CN line shape of the S peptide or p-CN-Phe, which have dominant Gaussian forms. The absorption line shapes of CO bound to the heme iron in several heme proteins also have dominant Gaussian forms.19,23,28,30,31,40,41 2D IR experiments on these other protein and peptide systems show line shapes that are dominated by inhomogeneous broadening. They all have some homogeneous contribution, but it is not overwhelmingly dominant. However, the RNase S complex has a homogeneous contribution that is the dominant component (~80%) of the total linewidth of the absorption spectrum. In contrast to other proteins, the small inhomogeneous contribution undergoes very fast spectral diffusion, with a single decay time, τ1 = 0.6 ± 0.08 ps (see Table 1). The inhomogeneous dynamics measured by the waiting time dependent 2D IR experiments fall very close to the homogeneous limit, with the product Δτ =1.24. As discussed earlier, if Δτ < 1, the dynamics are motionally narrowed and contribute to the homogeneous linewidth. The spectral diffusion of RNase S complex is just outside of the homogeneous regime. There is no static component in nitrile modified RNase S, which means that all protein structures that contribute to the nitrile absorption line are sampled on a very fast time scale. The RNase S complex is an unusual system in which the protein dynamics, as sensed by a vibrational label using 2D IR vibration echo spectroscopy, are predominantly motionally narrowed, with only a small contribution from exceedingly fast spectral diffusion that lies just outside the homogeneous limit.
The large homogeneous component and the small very fast inhomogeneous sampling demonstrate that the motions of RNase S do not involve large structural conformational changes. Such changes cannot occur on the ultrafast time scale associated with homogeneous dephasing or the 0.6 ps spectral diffusion. These results indicate that RNase S has a very rigid well defined structure that does not undergo substantial conformational evolution.
It is useful to speculate on the reason for the rigid structure of RNase S. It might be possible that the difference in dynamics observed for RNase S compared to other proteins and enzymes is a result of its function. In heme proteins such as myoglobin or hemoglobin, the active site binds small ligands, particularly O2 but also CO and CN. There is no channel to the active site. The small ligands must “diffuse” through the protein to reach the active site, as well as to leave the active site and exit the protein. Such diffusion requires substantial structural fluctuations. Enzymes such as horseradish peroxidase23 and cytochrome C P450cam,31 bind relatively small substrates that are much smaller than the enzyme itself. Such enzymes have channels that allow the substrate to migrate to the pocket next to the active site, but these channels are not clear paths, which in part helps substrate selectivity.28,31 Movement of the substrate to the pocket requires significant structural fluctuations.28,31
RNase S is very different from the heme proteins and enzymes mentioned above. Unlike the heme proteins which bind to smaller substrates, RNase S cleft binds to a long strand of RNA. Furthermore, while the active sites in other proteins have access to solvent (water) molecules, the hydrophobic active site of RNase S is inaccessible to the solvent molecules. The inaccessibility to water molecules might result in a much narrower absorption spectrum due to a narrower conformational distribution, as seen in the case of RNase S. But the motionally narrowed, sub-picosecond dynamics cannot be explained from solvent inaccessibility. The water spectral diffusion timescale is on the order of a few picoseconds.34,35,59,60 The dynamics of HIV reverse transcriptase as probed by a nitrile in the hydrophobic pocket was estimated to be tens of picoseconds.33 Thus the lack of slow large scale structural dynamics in case of RNase S complex is, to date, unique and might be related to its function. It is possible to speculate on how the lack of large scale slow structural fluctuations is related to function. The activity of RNase S requires that two positions on the RNA mate with two histidines (His 12 and His 119) in the cleft. The active site of the RNase complex may need to be relatively rigidly positioned to bind the RNA. Large scale structural fluctuations could result in deformation of the rigidly positioned residues in the active site, thereby inhibiting the binding of the RNA. Such fluctuations would result in the cleft only occasionally having the appropriate shape and not necessarily when the RNase S is appropriately positioned to bind RNA. Thus, the structural rigidity of RNase S could be essential for its function.
C. Molecular Dynamics Simulations
From the linear absorption line shape fits, the values of λ obtained are 0.68 and 0.65 cm−1/(MV/cm) for the nitrile probe in the S-peptide and the RNase complex respectively. The coupling parameter λ is the only adjustable parameter used to relate the MD simulation results to experimental data. These values of λ agree extremely well with the previous experimentally measured Stark tuning rate of an aromatic nitrile61,62 (0.61 cm−1/(MV/cm) for benzonitrile3,61). The fact that the Stark tuning parameters obtained from simulation and fit to the line shapes and the experimentally measured Stark tuning parameters are in good agreement strongly supports the fluctuating electric field (Stark) model that relates the global structural dynamics of the systems as sensed by the nitrile VDL to the 2D IR vibrational echo observables.
For both the S-peptide and the complex, FFCFs are obtained from the respective frequency distributions and the multi-exponential form (Equation 1) is used to fit the FFCFs to 100 ps. The FFCF parameters from MD simulations are given in Table 1. In contrast to an experiment, the simulations will yield the ultrafast dynamics that are not observable in the experiment because they are motionally narrowed (Δτ < 1). The τ and the Δ associated with the ultrafast component are combined as discussed below Equation 1 to give , the homogeneous dephasing time, for comparison to the experiments. For the S-peptide, the FFCF obtained from MD simulations can be fitted to a ~50 fs homogeneous component, that with the Δ, gives a , an inhomogeneous component with τ1 = 2.5 ps, Δ1 = 6 cm−1, and an offset Δs = 1.1 cm−1. At first glance, these values are not in bad agreement with the experiments. However, the simulations were run in water while the experiments were performed in a 50/50 glycerol water mixture. We can get some feel for how much difference the glycerol makes from experiments that have been conducted on the amino acid, p-CN-Phe.29 For HP35 in water, τ1 = 2.1 ps and there is no long time offset. For p-CN-Phe in 25% glycerol/75% water, τ1 = 2.9 ps with an offset, Δs = 2.1 cm−1. So in going from water to 25% glycerol the dynamics slow and a very slow component reflected by Δs is developed. The S-peptide is in 50% glycerol. Thus, the simulations in water should yield dynamics that are significantly faster than the experiment, but they actually give dynamics that are too slow.
For the RNase complex, the FFCF can be fit to a large motionally narrowed component (~50 fs), a dynamical inhomogeneous component, τ1 = 4 ps, and a static component, Δs = 0.7 cm−1. Therefore, the simulations of RNase S produce a τ1 value that is much too slow compared to experiment. The simulation also yields a static component, while the experiment shows that there is no static component. Furthermore, ignoring the numbers, the simulation gets the trend wrong in going from the S-peptide to the RNase S protein. The simulations give a faster τ1 for the S-peptide than for the RNase S protein, while the experiments are just the opposite.
The simulations were successful in predicting the relative proportion of the homogeneous vs. inhomogeneous contributions to the linear absorption line shape. Both the simulated and experimentally measured RNase S protein homogeneous components of the line shape are 80% of the total. For the S peptide, the simulated and experimental homogeneous components are 65% and 60%, respectively. The Δ1 values of both the experimental and simulated S-peptide and RNase S protein agree very well (see Table 1). While the values are off for the static components, at least the Δs values go in the correct direction. In the experiment, Δs values in going from the S-peptide to the RNase protein decrease (2.4 to 0 cm−1), and in the simulation they also decrease (1.1 to 0.7 cm−1). As the simulated linear absorption spectra depend on the relative amplitudes of the different components and the Stark coupling constant, the FWHM of the simulated spectra agree well with experiment given reasonable choices for the Stark coupling constants (see above).
While the simulations to a reasonable job of determining the homogenous components and the amplitudes of the inhomogeneous components, the simulations do a poor job of predicting the structural dynamics beyond the ultrafast contributions to the homogeneous dephasing. The ultrafast motions that contribute to the homogeneous dephasing will involve the very local motions of small groups, such as methyl groups. One possible explanation for the MD simulations failing to predict the experimentally observed inhomogeneous structural dynamics decay timescales is that the potential energy surface determined by the force field is more rugged with higher energy barriers than exist in the real systems. If the barriers are too high, the different conformations will interconvert too slowly. Thus, while the fast motionally narrowed fluctuations are captured in the simulations, the slower conformational interconversions are significantly too slow possibly due to unphysically high energy barriers present in the simulations.
In summary, the MD simulations were able to predict the relative ratios of the homogenous and inhomogeneous contribution and reproduce the linear line shape with a value of the Stark tuning rate in close accord to Stark constant61,62 obtained from time independent vibrational Stark spectroscopy. Furthermore, the MD snapshots successfully show that water does not hydrogen bond with the nitrile probe in RNase S complex. On the other hand, the MD simulations failed to correctly determine the RNase S complex structural dynamics that give rise to the spectral diffusion. The simulated spectral diffusion is almost an order of magnitude too slow compared to the experiments.
IV. Concluding Remarks
In this work, we report a 2D IR vibrational echo study of the dynamics of the RNase S protein and the free S-peptide, a 20 amino acid piece of the total protein. Nitrile (CN) in the form of an artificial amino acid in the S-peptide is used as the vibrational dynamics label to probe the fast dynamics of both the protein and the peptide. In addition, MD simulations of RNase S and the S-peptide were performed and compared to the experimental results.
The important experimental results are that the slower dynamics of the S-peptide lost when the peptide is bound to the rest of the protein to from the complete RNase S complex. The 2D IR experiments determine the homogeneous component of the absorption line and the time dependent spectral diffusion caused by structural evolution, which samples the inhomogeneous distribution of protein structures. In contrast to other proteins and enzymes that have been studied with 2D IR, RNase S dynamics are composed of extremely fast structural fluctuations that can only be caused by the very local motions of small groups. The conclusion is that the protein structure is nearly static, without significant large scale structural evolution. It was speculated that RNase S has a nearly fixed structure to provide a very well defined cleft to bind RNA.
In addition to the experiments, MD simulations were conducted and used to calculate the experimental observables. Both the homogeneous dephasing and spectral diffusion were obtained for the S-peptide and the RNase S. For both molecules, the simulations failed to obtain the correct time scales for the structural evolution that is manifested as spectral diffusion. In both cases, the simulations produce dynamics that are far too slow. However, the simulations do a good job of reproducing the component of the absorption line that is homogeneously broadened.
Acknowledgements
We thank Aaron T. Fafarman for discussions of these experimental and simulation results. This research was supported in part by grants from the National Institutes of Health, 2-R01-GM061137-09 (M.D.F.) and GM27738 (S.G.B.).
References
- 1.Richards FM, Vithayathil PJ. J. Biol. Chem. 1959;234:1459–1465. [PubMed] [Google Scholar]
- 2.Goldberg JM, Baldwin RL. Proc. Nat. Acad. Sci. USA. 1999;96:2019–2024. doi: 10.1073/pnas.96.5.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fafarman AT, Boxer SG. J. Phys. Chem. B. 2010;114:13536–13544. doi: 10.1021/jp106406p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kwak K, Park S, Finkelstein IJ, Fayer MD. J. Chem. Phys. 2007:127. doi: 10.1063/1.2772269. [DOI] [PubMed] [Google Scholar]
- 5.Kwak K, Rosenfeld DE, Fayer MD. J. Chem. Phys. 2008:128. doi: 10.1063/1.2927906. [DOI] [PubMed] [Google Scholar]
- 6.Henzler-Wildman KA, Lei M, Thai V, Kerns SJ, Karplus M, Kern D. Nature. 2007;450:913–916. doi: 10.1038/nature06407. [DOI] [PubMed] [Google Scholar]
- 7.Eisenmesser EZ, Bosco DA, Akke M, Kern D. Science. 2002;295:1520–1523. doi: 10.1126/science.1066176. [DOI] [PubMed] [Google Scholar]
- 8.Hammes-Schiffer S, Benkovic SJ. Annu. Rev. Biochem. Vol. 75. Palo Alto: Annual Reviews; 2006. Relating Protein Motion to Catalysis; pp. 519–541. [DOI] [PubMed] [Google Scholar]
- 9.Erzberger JP, Berger JM. Annu. Rev. Biophys. Biomol. Struct. Vol. 35. Palo Alto: Annual Reviews; 2006. Evolutionary Relationships and Structural Mechanisms of Aaa Plus Proteins; pp. 93–114. [DOI] [PubMed] [Google Scholar]
- 10.Boehr DD, Dyson HJ, Wright PE. Chem. Rev. 2006;106:3055–3079. doi: 10.1021/cr050312q. [DOI] [PubMed] [Google Scholar]
- 11.Hill SE, Bandaria JN, Fox M, Vanderah E, Kohen A, Cheatum CM. J. Phys. Chem. B. 2009;113:11505–11510. doi: 10.1021/jp901321m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Campbell BF, Chance MR, Friedman JM. Science. 1987;238:373–376. doi: 10.1126/science.3659921. [DOI] [PubMed] [Google Scholar]
- 13.Hong MK, Braunstein D, Cowen BR, Frauenfelder H, Iben IET, Mourant JR, Ormos P, Scholl R, Schulte A, Steinbach PJ, Xie AH, Young RD. Biophys. J. 1990;58:429–436. doi: 10.1016/S0006-3495(90)82388-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Frauenfelder H, Sligar SG, Wolynes PG. Science. 1991;254:1598–1603. doi: 10.1126/science.1749933. [DOI] [PubMed] [Google Scholar]
- 15.Frauenfelder H, McMahon BH, Austin RH, Chu K, Groves JT. Proc. Nat. Acad. Sci. USA. 2001;98:2370–2374. doi: 10.1073/pnas.041614298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Andrews BK, Romo T, Clarage JB, Pettitt BM, Phillips GN. Structure. 1998;6:587–594. doi: 10.1016/s0969-2126(98)00060-4. [DOI] [PubMed] [Google Scholar]
- 17.Thielges MC, Axup JY, Wong D, Lee HS, Chung JK, Schultz PG, Fayer MD. J. Phys. Chem. B. 2011;115:11294–11304. doi: 10.1021/jp206986v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ishikawa H, Kwak K, Chung JK, Kim S, Fayer MD. Proc. Nat. Acad. Sci. USA. 2008;105:8619–8624. doi: 10.1073/pnas.0803764105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ishikawa H, Finkelstein IJ, Kim S, Kwak K, Chung JK, Wakasugi K, Massari AM, Fayer MD. Proc. Nat. Acad. Sci. USA. 2007;104:16116–16121. doi: 10.1073/pnas.0707718104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mukherjee P, Kass I, Arkin IT, Zanni MT. Proc. Nat. Acad. Sci. USA. 2006;103:3528–3533. doi: 10.1073/pnas.0508833103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ghosh A, Qiu J, DeGrado WF, Hochstrasser RM. Proc. Nat. Acad. Sci. USA. 2011;108:6115–6120. doi: 10.1073/pnas.1103027108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rella CW, Kwok A, Rector K, Hill JR, Schwettman HA, Dlott DD, Fayer MD. Phys. Rev. Lett. 1996;77:1648–1651. doi: 10.1103/PhysRevLett.77.1648. [DOI] [PubMed] [Google Scholar]
- 23.Finkelstein IJ, Ishikawa H, Kim S, Massari AM, Fayer MD. Proc. Nat. Acad. Sci. USA. 2007;104:2637–2642. doi: 10.1073/pnas.0610027104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ganim Z, Chung HS, Smith AW, Deflores LP, Jones KC, Tokmakoff A. Acc. Chem. Res. 2008;41:432–441. doi: 10.1021/ar700188n. [DOI] [PubMed] [Google Scholar]
- 25.Bandaria JN, Dutta S, Nydegger MW, Rock W, Kohen A, Cheatum CM. Proc. Nat. Acad. Sci. USA. 2010;107:17974–17979. doi: 10.1073/pnas.0912190107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Middleton CT, Woys AM, Mukherjee SS, Zanni MT. Methods. 2010;52:12–22. doi: 10.1016/j.ymeth.2010.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim YS, Hochstrasser RM. J. Phys. Chem. B. 2009;113:8231–8251. doi: 10.1021/jp8113978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bagchi S, Nebgen BT, Loring RF, Fayer MD. J. Am. Chem. Soc. 2010;132:18367–18376. doi: 10.1021/ja108491t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chung JK, Thielges MC, Fayer MD. Proc. Nat. Acad. Sci. USA. 2011;108:3578–3583. doi: 10.1073/pnas.1100587108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chung JK, Thielges MC, Bowman SEJ, Bren KL, Fayer MD. J. Am. Chem. Soc. 2011;133:6681–6691. doi: 10.1021/ja111009s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thielges MC, Chung JK, Fayer MD. J. Am. Chem. Soc. 2011;133:3995–4004. doi: 10.1021/ja109168h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhuang W, Hayashi T, Mukamel S. Angewandte Chemie-International Edition. 2009;48:3750–3781. doi: 10.1002/anie.200802644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fang C, Bauman JD, Das K, Remorino A, Arnold E, Hochstrasser RM. Proc. Nat. Acad. Sci. USA. 2008;105:1472–1477. doi: 10.1073/pnas.0709320104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Asbury JB, Steinel T, Stromberg C, Corcelli SA, Lawrence CP, Skinner JL, Fayer MD. J. Phys. Chem. A. 2004;108:1107–1119. doi: 10.1063/1.1803532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Asbury JB, Steinel T, Kwak K, Corcelli SA, Lawrence CP, Skinner JL, Fayer MD. J. Chem. Phys. 2004;121:12431–12446. doi: 10.1063/1.1818107. [DOI] [PubMed] [Google Scholar]
- 36.Doscher MS, Hirs CHW. Biochemistry. 1967;6 doi: 10.1021/bi00853a047. 304-&. [DOI] [PubMed] [Google Scholar]
- 37.Asbury JB, Steinel T, Fayer MD. J. Lumin. 2004;107:271–286. doi: 10.1016/j.jlumin.2003.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mukamel S. Principles of Nonlinear Optical Spectroscopy. New York: Oxford University Press; 1995. [Google Scholar]
- 39.Williams RB, Loring RF, Fayer MD. J. Phys. Chem. B. 2001;105:4068–4071. [Google Scholar]
- 40.Merchant KA, Noid WG, Akiyama R, Finkelstein IJ, Goun A, McClain BL, Loring RF, Fayer MD. J. Am. Chem. Soc. 2003;125:13804–13818. doi: 10.1021/ja035654x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Merchant KA, Noid WG, Thompson DE, Akiyama R, Loring RF, Fayer MD. J. Phys. Chem. B. 2003;107:4–7. [Google Scholar]
- 42.Schmidt JR, Roberts ST, Loparo JJ, Tokmakoff A, Fayer MD, Skinner JL. Chem. Phys. 2007;341:143–157. [Google Scholar]
- 43.Finkelstein IJ, Zheng JR, Ishikawa H, Kim S, Kwak K, Fayer MD. Phys. Chem. Chem. Phys. 2007;9:1533–1549. doi: 10.1039/b618158a. [DOI] [PubMed] [Google Scholar]
- 44.Sorin EJ, Pande VS. Biophys. J. 2005;88:2472–2493. doi: 10.1529/biophysj.104.051938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xu L, Cohen AE, Boxer SG. Biochemistry. 2011;50:8311–8322. doi: 10.1021/bi200930f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Massari AM, Finkelstein IJ, McClain BL, Goj A, Wen X, Bren KL, Loring RF, Fayer MD. J. Am. Chem. Soc. 2005;127:14279–14289. doi: 10.1021/ja053627w. [DOI] [PubMed] [Google Scholar]
- 47.Finkelstein IJ, Goj A, McClain BL, Massari AM, Merchant KA, Loring RF, Fayer MD. J. Phys. Chem. B. 2005;109:16959–16966. doi: 10.1021/jp0517201. [DOI] [PubMed] [Google Scholar]
- 48.Goj A, Loring RF. Chem. Phys. 2007;341:37–44. doi: 10.1021/jp074711w. [DOI] [PubMed] [Google Scholar]
- 49.Eaton G, Penanunez AS, Symons MCR. J. Chem. Soc. Faraday Trans. 1988;84:2181–2193. [Google Scholar]
- 50.Reimers JR, Hall LE. J. Am. Chem. Soc. 1999;121:3730–3744. [Google Scholar]
- 51.Getahun Z, Huang CY, Wang T, De Leon B, DeGrado WF, Gai F. J. Am. Chem. Soc. 2003;125:405–411. doi: 10.1021/ja0285262. [DOI] [PubMed] [Google Scholar]
- 52.Aschaffenburg DJ, Moog RS. J. Phys. Chem. B. 2009;113:12736–12743. doi: 10.1021/jp905802a. [DOI] [PubMed] [Google Scholar]
- 53.Purcell KF, Drago RS. J. Am. Chem. Soc. 1966;88 919-&. [Google Scholar]
- 54.Fawcett WR, Liu GJ, Kessler TE. J. Phys. Chem. 1993;97:9293–9298. [Google Scholar]
- 55.Eaton G, Penanunez AS, Symons MCR, Ferrario M, McDonald IR. Faraday Discuss. 1988;85:237–253. [Google Scholar]
- 56.Rector KD, Kwok AS, Ferrante C, Tokmakoff A, Rella CW, Fayer MD. J. Chem. Phys. 1997;106:10027–10036. [Google Scholar]
- 57.Golonzka O, Khalil M, Demirdoven N, Tokmakoff A. Phys. Rev. Lett. 2001;86:2154–2157. doi: 10.1103/PhysRevLett.86.2154. [DOI] [PubMed] [Google Scholar]
- 58.Akaike H. IEEE Trans. Automat. Contr. 1974;AC19:716–723. [Google Scholar]
- 59.Fayer MD, Levinger NE. Ann. Rev. Analytical Chem. 2010;3:89–107. doi: 10.1146/annurev-anchem-070109-103410. [DOI] [PubMed] [Google Scholar]
- 60.Fayer MD. Physiology. 2011;26:381–392. doi: 10.1152/physiol.00021.2011. [DOI] [PubMed] [Google Scholar]
- 61.Andrews SS, Boxer SG. J. Phys. Chem. A. 2000;104:11853–11863. [Google Scholar]
- 62.Suydam IT, Boxer SG. Biochemistry. 2003;42:12050–12055. doi: 10.1021/bi0352926. [DOI] [PubMed] [Google Scholar]