Abstract
Plastids are the defining organelles of all photosynthetic eukaryotes. They are the site of photosynthesis and of a large number of other essential metabolic pathways, such as fatty acid and amino acid biosyntheses, sulfur and nitrogen assimilation, and aromatic and terpenoid compound production, to mention only a few examples. The metabolism of plastids is heavily intertwined and connected with that of the surrounding cytosol, thus causing massive traffic of metabolic precursors, intermediates, and products. Two layers of biological membranes that are called the inner (IE) and the outer (OE) plastid envelope membranes bound the plastids of Archaeplastida. While the IE is generally accepted as the osmo-regulatory barrier between cytosol and stroma, the OE was considered to represent an unspecific molecular sieve, permeable for molecules of up to 10 kDa. However, after the discovery of small substrate specific pores in the OE, this view has come under scrutiny. In addition to controlling metabolic fluxes between plastid and cytosol, the OE is also crucial for protein import into the chloroplast. It contains the receptors and translocation channel of the TOC complex that is required for the canonical post-translational import of nuclear-encoded, plastid-targeted proteins. Further, the OE is a metabolically active compartment of the chloroplast, being involved in, e.g., fatty acid metabolism and membrane lipid production. Also, recent findings hint on the OE as a defense platform against several biotic and abiotic stress conditions, such as cold acclimation, freezing tolerance, and phosphate deprivation. Moreover, dynamic non-covalent interactions between the OE and the endomembrane system are thought to play important roles in lipid and non-canonical protein trafficking between plastid and endoplasmic reticulum. While proteomics and bioinformatics has provided us with comprehensive but still incomplete information on proteins localized in the plastid IE, the stroma, and the thylakoids, our knowledge of the protein composition of the plastid OE is far from complete. In this article, we report on the recent progress in discovering novel OE proteins to draw a conclusive picture of the OE. A “parts list” of the plastid OE will be presented, using data generated by proteomics of plastids isolated from various plant sources.
Keywords: plastid outer envelope, endoplasmic reticulum, plastid associated membranes, stromules
Introduction
Plastids are the eponymous cellular organelles of the Archaeplastida (i.e., photosynthetic eukaryotes that contain plastids of primary endosymbiotic origin, also known as the Plantae) and they host the majority of anabolic pathways. Archaeplastida that have lost the ability to photosynthesize, such as parasitic plants, still contain plastids. De novo fatty acid synthesis is exclusively localized in plastids. Fatty acid synthesis is based on the production of acetyl-Coenzyme A (acetyl-CoA) by the pyruvate dehydrogenase complex in the plastids (Johnston et al., 1997). The plastidic acetyl-CoA carboxylase drives the first reaction in the fatty acid biosynthesis resulting in malonyl-CoA (Konishi et al., 1996). These fatty acids are used for lipid biosynthesis in the plastid envelopes and in the endoplasmic reticulum (ER).
Of the 20 proteinogenic amino acids, nine are synthesized exclusively in plastids: the aspartate derived amino acids methionine, threonine, and lysine (Mills and Wilson, 1978; Mills, 1980; Wallsgrove and Mazelis, 1980; Curien et al., 2005, 2009), the aromatic amino acids phenylalanine, tryptophan, and tyrosine (Bickel et al., 1978), and the threonine and pyruvate derived branched-chain amino acids leucine, valine, and isoleucine (Singh and Shaner, 1995; Binder et al., 2007).
Purines, the building blocks of RNA and DNA bases and of ATP are also synthesized in plastids, as is the pyrimidine backbone. All 10 enzymatic steps of inosine monophosphate biosynthesis occur in the plastids (Zrenner et al., 2006). Furthermore, five of the six steps of pyrimidine synthesis are catalyzed by the plastidic enzymes carbamoylphosphate synthase (Giermann et al., 2002), aspartate transcarbamoylase, dihydroorotase, and uracil monophosphate synthase (Doremus and Jagendorf, 1985).
The reduction of sulfate to sulfide is also confined to the plastid (summarized in Takahashi et al., 2011) as well as the production of sulfolipids (compare Okanenko, 2002). Chloroplasts fix inorganic carbon in the form of CO2 in the Calvin–Benson cycle and incorporate nitrogen in the form of ammonia into glutamate and glutamine. The chloroplasts provide energy in the form of reducing equivalents through photosynthesis. Further chloroplast products, such as triose phosphates (TP), carbohydrates, and amino acids are exported from the chloroplasts to feed processes in the cell (Weber, 2004; Weber et al., 2004, 2006; Linka and Weber, 2009; Weber and Linka, 2011).
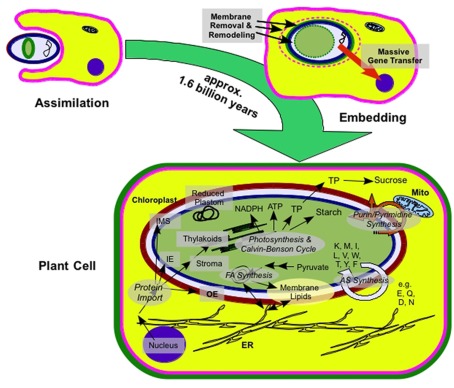
The anabolic versatility of chloroplasts traces back to their prokaryotic ancestor. An ancestral cyanobacterium was engulfed and stably integrated into the eukaryotic host during primary endosymbiosis (for a recent review, see Weber and Osteryoung, 2010). Approximately 1, 6 billion years of co-evolution irreversibly integrated the photoautotrophic prokaryote into the host cell, creating the plant cell as we know it (Figure 1; Yoon et al., 2004; Reyes-Prieto et al., 2007; Tyra et al., 2007).
Figure 1.
From endosymbiont to plastid. Schematic representation of plastid origin and extant anabolic pathways in plastids; ER, endoplasmic reticulum; Mito, mitochondrion; IE, inner envelope; IMS, intermembrane space; OE, outer envelope; FA, fatty acid; AS, amino acid (specific amino acids are denoted in one letter code); TP, triosephosphate
Plastids derived from this initial event, primary plastids, are bound by two surrounding envelope membranes, the inner (IE) and the outer (OE) envelope membrane. It is believed that both envelope membranes are derived from cyanobacterial membranes (Gould et al., 2008). The IE traces back to the plasma membrane (PM) of the ancestral cyanobacteria. The OE traces to the bacterial outer membrane since (i) it contains galactolipids (Jarvis et al., 2000), (ii) β-barrel forming proteins are in both envelope membranes and the bacterial outer envelope (Schleiff et al., 2003a), and (iii) traces of peptidoglucan biosynthesis are present in plastids of glaucophytes (Steiner et al., 2005).
Although the plastid harbors the majority of anabolic pathways and is well separated from the cytosol by two membranes, the extant plastid is only semiautonomous (Gould et al., 2008). During domestication, the organelle lost almost its complete genome to the nucleus. Only approximately 2% of the plastid proteome are encoded on the plastom (Abdallah et al., 2000), and almost all of the proteins the plastid needs to function are imported from the cytosol (Schnell et al., 1994; Hinnah et al., 1997). It also lost the ability to freely replicate, because the complete protein set for division is encoded in the nucleus (Hashimoto and Possingham, 1989; Kuroiwa et al., 1998). Furthermore, cell- and plastid-divisions are synchronized (El-Shami et al., 2002; Raynaud et al., 2005), although it can be uncoupled to a certain degree, as demonstrated by several mutant lines defective in plastid division (Pyke and Leech, 1992; Osteryoung et al., 1998; Asano et al., 2004; Raynaud et al., 2004).
Extant plastids are well integrated into their host cells, exchanging metabolites, proteins, membrane lipids, and information. The ultimate barriers between these compartments are the plastid envelope membranes. For the IE the reader is directed to several recent reviews about the function of the inner envelope membrane (Linka and Weber, 2009; Kovacs-Bogdan et al., 2010; Facchinelli and Weber, 2011; Fischer, 2011; Weber and Linka, 2011). In this review, we will focus on the role of the outer envelope and we provide a parts list of the OE proteome (Tables 1 and 2).
Table 1.
The known solute transport proteins of the outer envelope.
| Name (structure) | Transport | Homologs (mentioned in this review) | Publications |
|---|---|---|---|
| PLASTID OUTER ENVELOPE SOLUTE TRANSPORTER | |||
| OEP16 (α-helix) | Export of amino acids and amines | PsOEP16.1, PsOEP16.2 AtOEP16.1 (At2g28900) AtOEP16.2 (At4g16160) AtOEP16.4 (At3g62880) HvOEP16 (COR TMC-AP3) ZmOEP16 |
Pohlmeyer et al. (1997), Baldi et al. (1999), Rassow et al. (1999), Steinkamp et al. (2000), Linke et al. (2004), Reinbothe et al. (2004), Drea et al. (2006), Duy et al. (2007), Murcha et al. (2007), Philippar et al. (2007), Bräutigam and Weber (2009), Pudelski et al. (2010) |
| OEP21 (β-barrel) | Phosphorylated carboxylates, Pi | PsOEP21 AtOEP21.1 (At1g20816) |
Bolter et al. (1999), Hemmler et al. (2006), Bräutigam et al. (2008) |
| Anion | AtOEP21.2 (At1g76405) | ||
| Rectifying | ZmOEP21 | ||
| OEP24 (β-barrel) | TP, hexose-phosphates, sugar, ATP, Pi, dicarboxylates, charged amino acids | PsOEP24 Putative AtOEP24 (At3g52230, At5g42960, At1g45170) |
Pohlmeyer et al. (1998), Schleiff et al. (2003a), Duy et al. (2007), Sun et al. (2009), Ferro et al. (2010) |
| ZmOEP24 | |||
| OEP37 (β-barrel) | Peptides | PsOEP37 | Schleiff et al. (2003a), Goetze et al. (2006), Bräutigam et al. (2008) |
| Cations | AtOEP37 | ||
| Rectifying | ZmOEP37 | ||
Table 2.
Parts list of the proteome of the OE.
| Name | Function | Isoforms and homologs (locus on Arabidopsis genome) | Publication |
|---|---|---|---|
| PROTEIN TRANSLOCON COMPLEX | |||
| Toc34 family | GTPase, protein import receptor | AtToc33/PPI1 (At1g02280), AtToc34/PPI2 (At5g05000), PsToc34 | Jarvis et al. (1998), Chen et al. (2000), Gutensohn et al. (2000), Schleiff et al. (2003b), Ivanova et al. (2004), Andres et al. (2010), Dhanoa et al. (2010), Huang et al. (2011) |
| Toc64 | Protein import co-receptor/co-chaperone | AtToc64-III (At3g17970), AtToc64-I/Ami1 (At1g08980), AtToc64-like/OEP61 (At5g21990) | Becker et al. (2004), Aronsson et al. (2007), Qbadou et al. (2007), Bae et al. (2008), Barsan et al. (2010), Von Loeffelholz et al. (2011) |
| Toc75 | Protein import translocator pore | AtToc75-III/Mar1 (At3g46740), AtToc75-IV (At4g09080), AtToc75-V/AtOEP80 (At5g19620), PsOEP75 | Perry and Keegstra (1994), Schnell et al. (1994), Tranel et al. (1995), Hinnah et al. (1997), Sveshnikova et al. (2000), Jackson-Constan and Keegstra (2001), Hinnah et al. (2002), Schleiff et al. (2003a), Wallas et al. (2003), Gentle et al. (2004), Baldwin et al. (2005), Patel et al. (2008) |
| Toc159 family (Toc86) | GTPase, protein import receptor | AtToc159 (At4g02510), AtToc132/Mar2 (At2g16640), AtToc120 (At3g16620), AtToc90 (At5g20300) | Bauer et al. (2000), Jackson-Constan and Keegstra (2001), Schleiff et al. (2003b), Hiltbrunner et al. (2004), Ivanova et al. (2004), Kubis et al. (2004), Stanga et al. (2009), Andres et al. (2010), Huang et al. (2011), Infanger et al. (2011) |
| LIPID METABOLISM | |||
| LACS9 | Long-chain acyl-CoA synthase | AtLacs9 (At1g77590) | Schnurr et al. (2002), Zhao et al. (2010) |
| DGD | Digalactosyldiacylglycerol (DGDG) synthase | AtDGD1 (At3g11670), AtDGD2 (At4g00550) | Dörmann et al. (1995), Dörmann et al. (1999), Härtel et al. (2000), Froehlich et al. (2001a), Kelly and Dörmann (2002), Xu et al. (2003) |
| MGD | Monogalactosyldiacylglycerol (MGDG) synthase | AtMGD2 (At5g20410), AtMGD3 (At2g11810) | Miege et al. (1999), Härtel et al. (2000), Awai et al. (2001) |
| GGGT/SFR2 | Galactolipid:galactolipid galactosyltransferase | AtGGGT/AtSFR2 (At3g06510) | Heemskerk et al. (1983), Heemskerk et al. (1986), Kelly and Dörmann (2002), Xu et al. (2003), Thorlby et al. (2004), Fourrier et al. (2008), Moellering et al. (2010) |
| PLASTID MOVEMENT AND DIVISION | |||
| CHUP1 | Anchor protein for plastid movement | AtCHUP1 (At3g25690) | Oikawa et al. (2003), Oikawa et al. (2008), Von Braun and Schleiff (2008), Kadota et al. (2009), Suetsugu et al. (2010), Whippo et al. (2011) |
| PDV | Plastid division | AtPDV1 (At5g53280), AtPDV2 (At2g16070) | Gao et al. (2003), Miyagishima et al. (2006), Glynn et al. (2008), Glynn et al. (2009) |
| CRL1 | Unknown; mutant effect on plastid number and size | AtCRL1 (At5g51020) | Asano et al. (2004), Chen et al. (2009) |
| MOLECULAR AND BIOCHEMICAL TOOLS OF UNKNOWN FUNCTION | |||
| OEP7 | Unknown | AtOEP7 (At3g52420), PsOEP14, SoOEP7 (E6.7) | Salomon et al. (1990), Li et al. (1991), Tu and Li (2000), Lee et al. (2001), Dyall et al. (2004), Bae et al. (2008), Oikawa et al. (2008), Kim et al. (2011) |
| OEP9 | Unknown | OEP9.1 (At1g16000), Putative homolog OEP9.2 (At1g80890) | Dhanoa et al. (2010) |
| OTHERS | |||
| PTM | Plant homeodomain transcription factor with transmembrane domains | PTM (At5g35210) | Sun et al. (2011) |
| HPL | Hydroxide lyase | AtHPL/AtCYP74B (At4g15440), LeHPL, StHPL | Blee and Joyard (1996), Froehlich et al. (2001b), Vancanneyt et al. (2001), Shiojiri et al. (2006), Kishimoto et al. (2008) |
| KO | ent-kaurene oxidase/gibberellin synthesis | AtKO1/GA3 (At5g25900) | Helliwell et al. (2001) |
| OMP24 | Unknown; outer membrane protein of spinach chloroplasts | SoOMP24 | Fischer et al. (1994) |
| WBC7 | Unknown; transport of hydrophobic compounds | AtWBC7 (At2g01320) | Schleiff et al. (2003a), Zybailov et al. (2008) |
| OEP6 | Unknown | AtOEP6 (At3g3160) | Ferro et al. (2010) |
| Putative OE proteins in PPDB database | Unknown | OMP85-family proteins of 35 kDA (At3g48620) and 39 kDa (At3g44160) | Sun et al. (2009) |
| Putative GTPase of 15kDa (At4g02482) | |||
| Putative p-loop containing nucleoside triphosphate hydrolase 100 kDa (At4g15810) | |||
This list contains the proteins discussed in this review with residence in the outer plastid envelope except proteins already listed in Table 1.
Solute Transport Across the Outer Envelope
Since the plastid is the anabolic factory of the cell, substrates that cannot be produced from intermediates of the Calvin–Benson cycle need to be imported while many products need to be exported. All substrates and products cross two envelope membranes, the IE and the OE. The IE contains a diverse set of substrate specific proteins processing and possibly controlling export and import to the organelle (Linka and Weber, 2009; Bräutigam and Weber, 2011; Facchinelli and Weber, 2011; Weber and Linka, 2011). Pores with broad substrate specificity have not yet been described in the inner envelope. For a long time the OE was considered as a molecular sieve for molecules larger than 10 kDa not contributing to the barrier between stroma and cytosol. This view has been disputed (Pohlmeyer et al., 1997; Flügge, 2000; Soll et al., 2000). In the past 15 years, four pore forming proteins of different selectivity have been characterized and were named by their apparent molecular masses as OEP16 (Pohlmeyer et al., 1997), OEP21, OEP24 (Pohlmeyer et al., 1998), and OEP37 (Schleiff et al., 2003a; Table 1). They were initially identified in Pisum sativum (pea) and later pursued in Arabidopsis.
OEP24
OEP24 is a member of β-barrel forming proteins and is proposed to consist of seven β-strands spanning through the membrane (Pohlmeyer et al., 1998). OEP24 shows no similarity in its primary structure to mitochondrial and bacterial porins, and does not show sensitivity to bacterial porin inhibitors (Pohlmeyer et al., 1998). However, the high amounts of hydrophilic amino acids (49%) reflect the properties of other pore forming proteins in bacteria and mitochondria (Röhl et al., 1999).
When reconstituted in proteoliposomes in vitro, the channel is slightly selective for cations and it is highly conductive. The 2.5- to 3-nm wide pore is created by at least two OEP24 proteins. This homodimer facilitates the transport of triose phosphates (TP), hexose-phosphates, sugars, ATP, phosphates (Pi), dicarboxylates like 2-oxoglutarate, and charged amino acids (Table 1; Pohlmeyer et al., 1998). Hence, OEP24 carries the major fluxes across the envelope membrane in the shape of TP, the product of photosynthesis as well as dicarboxylates and amino acids needed for nitrogen assimilation. The importance of OEP24 is underlined by its expression pattern. OEP24 pores exhibit equal distribution in plastids of all kind and each tissue. Moreover, paralogs can be found in monocotyledons and in dicotyledons (Pohlmeyer et al., 1998).
The pea protein PsOEP24 can functionally complement a yeast mutant that lacks the mitochondrial voltage-dependent anion channel (VDAC). Also, like VDAC proteins, PsOEP24 can induce apoptosis in cancer cells (Liguori et al., 2010) indicating a role in programmed cell death for PsOEP24. In both heterologous systems PsOEP24 is targeted to the mitochondrial outer membrane (Röhl et al., 1999).
Most information on OEP24 is currently based on PsOEP24. Two homologs of OEP24 are found in Arabidopsis (Duy et al., 2007) and proteomic databases suggest that at least three homologs exist (Sun et al., 2009; Ferro et al., 2010). The AtOEP24 encoded by At1g45170 was predicted in silico to contain 12 β-strands (Schleiff et al., 2003a), which may indicate functionality as monomer. Unpublished data of Timper et al. mentioned in Duy et al. (2007) describe defects during pollen germination in a so called AtOEP24.1 mutant. It is hypothesized that this defect is due to a lipid and energy deprivation during early pollen development (Duy et al., 2007). In contrast, the expression of another homolog (AtOEP24.2) is upregulated during late seed development (Duy et al., 2007). At least the findings for AtOEP24.1 indicate that the more selective transport pores of the OE (see below) cannot compensate for the loss of OEP24. The question remains whether, and if so how, OEP24 controls the metabolic flux. Proteomics data indicates that protein abundance in different plastids subtypes does reflect flux (Bräutigam et al., 2008; Bräutigam and Weber, 2009; Bräutigam and Weber, 2011). For example, OEP24 is more abundant in maize mesophyll chloroplasts compared to pea chloroplasts. Since the required flux of TP is at least three times higher in maize mesophyll chloroplasts compared to pea chloroplasts (Bräutigam et al., 2008; Weber and Von Caemmerer, 2010; Bräutigam and Weber, 2011), the increased abundance of OEP24 in maize indicates that an outer envelope porin might be limiting metabolite flux (Bräutigam et al., 2008). Studies with altered levels of AtOEP24 either by knock-outs or knock-downs may shed light on the flux control by OEP24.
OEP16
The first described and best-investigated outer envelope solute channel is OEP16. Like OEP24, OEP16 lacks sequence and structural homology to known porins, which classifies it as a non-classical porin (Pohlmeyer et al., 1997). The OEP16 channel is slightly cation selective and is a high-conductance solute channel (Pohlmeyer et al., 1997) that selectively transports amino acids and amines. Although the pore is principally large enough, OEP16 excludes carbonates such as TP or sugars (Table 1; Pohlmeyer et al., 1997).
The structure of OEP16 was controversially discussed. It was assumed to consist of four β-sheets and three alpha-helices (Pohlmeyer et al., 1997). Based on other porin structures and the transport properties of the channel, it was concluded, that the protein is a β-barrel forming pore (Steinkamp et al., 2000). However, structural analysis and enhanced prediction algorithms later revealed a purely alpha-helical structure (Linke et al., 2004). These findings were supported by the sequence similarity of OEP16 to other alpha-helical transport proteins. The similarity to members of the mitochondrial protein translocon family of the inner membrane (TIM proteins), and to the bacterial amino acid permease LivH led to the classification of OEP16 as preprotein and amino acid transporter (PRAT) relatives (Rassow et al., 1999; Murcha et al., 2007; Pudelski et al., 2010). The similarity to preprotein transporters was also the cause for one group claiming OEP16 to be a protein importer. This claim was recently conclusively disproved (summarized in Pudelski et al., 2010).
The high selectivity for amino acids is achieved via a loop between the pore forming helix1 and helix2 (Linke et al., 2004). Whether this loop is facing the intermembrane space (IMS) or the cytosol is not known (Linke et al., 2004). The model described by Pudelski et al. (2010) puts the loop into the IMS (Pudelski et al., 2010). The location of the regulatory element to the IMS points to a function of OEP16 as amino acid exporter. In addition to this transport regulation, the channel is also redox-regulated by cysteine residues in the first helix (Steinkamp et al., 2000)
Recently a second homolog of PsOEP16 was found and named PsOEP16.2 (Pudelski et al., 2010). Yet, Arabidopsis contains three homologs of OEP16 (Philippar et al., 2007). The protein most similar to PsOEP16.1 is AtOEP16.1 (63%), also called AtOEP16-L (Drea et al., 2006), due to its highest expression levels in leaves. AtOEP16.2/AtOEP16-S is exclusively expressed in mature seeds, cotyledons, and early pollen stages (Drea et al., 2006; Philippar et al., 2007). It contains additional amino acids in the loop responsible for substrate selectivity of the channel (Drea et al., 2006; Philippar et al., 2007). AtOEP16.2 is hypothesized to be involved in seed development, dormancy, and/or desiccation tolerance. Binding elements of the phytohormone abcisic acid in the promoter region of AtOEP16.2 support this hypothesis (Drea et al., 2006).
A third homolog, AtOEP16.4, with a similarity of 20% to PsOEP16 is expressed at low levels throughout all stages of development. It shows higher expression during seed maturation and in pollen, indicating a function as backup for AtOEP16.2 (Pudelski et al., 2010). PsOEP16 also shares slight similarity (comparable with the similarity to AtOEP16.4) to another PRAT protein earlier described as the third homolog of AtOEP16, AtOEP16.3 (Reinbothe et al., 2004; Drea et al., 2006). This protein is localized to the mitochondrial outer membrane and is now labeled PRAT3 (Murcha et al., 2007; Philippar et al., 2007).
A cold regulated protein (COR) TMC-AP3 in barley (Hordeum vulgaris) is also a paralog of OEP16. It was investigated due to its upregulation during cold stress (Baldi et al., 1999). The role in cold acclimation of OEP16 is supported by a recent study on Arabidopsis (Kaplan et al., 2007). Here, increased levels of amino acids were found during cold stress. This indicates that the amino acids are needed as signal substance in cold acclimation and points on OEP16 as necessary transporter in this process. Proteomics also indicates that the OEP16 content is dynamically adjusted between plastid types. Proplastids contain large amounts of OEP16 reflecting the active amino acid synthesis during differentiation (Bräutigam and Weber, 2009).
OEP37
The most recently described member of the transmembrane channels for solutes in the OE is PsOEP37 and its Arabidopsis paralog AtOEP37. It was found in a combined in silico and proteomics study on β-barrel proteins in the OE of pea and Arabidopsis (Schleiff et al., 2003a). Its functional characterization revealed a rectifying, cation selective, high-conductance channel, selective for peptides. It is hypothesized to form a β-barrel with 12 β-strands (Schleiff et al., 2003a). It forms an hourglass shaped pore with a size of 3 nm narrowing to 1.5 nm in the restriction zone (Goetze et al., 2006). A long negatively charged loop responsible for the selectivity is facing the IMS and is regulated by the pH values of the surrounding area (Schleiff et al., 2003a; Goetze et al., 2006). OEP37 is likely regulated by the redox state of the environment due to the oxidation of two neighboring cysteine residues similar to OEP16.
Its expression levels are fairly low but it is ubiquitously distributed in all developmental stages and organs in Arabidopsis. It was observed that during germination the mRNA levels increased indicating a role during early plant development (Goetze et al., 2006).
OEP37 displays binding affinity to the precursor of the inner envelope translocon compound 32 (Tic32), which is imported non-canonically (Nada and Soll, 2004). Consequently, it was assumed to be transported by OEP37 (Goetze et al., 2006). However, reverse genetic analysis of AtOEP37 disproved this hypothesis, since AtOEP37 knock out plants were not lethal while Tic32 mutants are. In addition, the transport of Tic32 into the IE was not impaired in the oep37-1 knock out plants (Goetze et al., 2006).
The in vivo role of OEP37 in plants is unknown. The Arabidopsis oep37-1 mutant has no obvious phenotype although OEP37 is a single copy gene albeit expressed at low levels. OEP37 function may overlap with or is partially redundant in function with OEP16 and/or OEP24. High expression levels during early seedling germination and late embryogenesis indicate a function in early development of the plant (Goetze et al., 2006). It may also become important during stress conditions where higher metabolite fluxes are needed. Similar to OEP24, chloroplast envelopes of the C4 plant maize contain a higher amount of OEP37, which might reflect the higher metabolite flux across this membrane (Bräutigam et al., 2008).
OEP21
The fourth solute pore protein of the OE is OEP21 (Pohlmeyer et al., 1998). Like for the other three OEPs, OEP21 is distributed through all plastid types in varying abundance (Bräutigam et al., 2008). Also, OEP21 is present in both mono- and dicotyledons (Pohlmeyer et al., 1998). It is a rectifying, anion selective channel for phosphorylated carbohydrates and TP (Table 1; Bolter et al., 1999). Like OEP24 and OEP37, OEP21 is a β-barrel forming protein. Its secondary structure displays eight β-strands but just seven are hypothesized to be pore forming. Since this seven β-strands are not enough to form a fully hourglass shaped 2.4 nm pore, OEP21 was proposed to acts at least as dimer. The N- and C-terminus of the protein face the cytosol (Hemmler et al., 2006). The transport through OEP21 is regulated by a substrate gradient and most likely exports TP and phosphorylated carbohydrates during light periods in green tissue. In contrast, import of these compounds would occur during darkness and in non-green tissue (Bolter et al., 1999). Additional regulation OEP21 obtains by two highly affine ATP binding sites (Bolter et al., 1999). The internal ATP binding site is proposed to provide major regulation by blocking the channel. The IMS orientated FX4K motive only provides regulatory function in transport processes, and is 100-fold less affine to ATP. The competitive binding of substrates like TP is initiating the release of ATP from the internal ATP binding side. This leads to the opening of the pore and to reduction of the anion selectivity (Bolter et al., 1999; Hemmler et al., 2006). One of the two Arabidopsis OEP21 homologs, AtOEP21.1, lacks the FX4K motive and has a 50% reduced ability for modulating the ion selectivity of the channel (Hemmler et al., 2006).
In summary, the import and export of cations (e.g., potassium, calcium, iron), anions (e.g., nitrite, sulfate, phosphate), and metabolites across the OE is driven by the set of known solute channels OEP16, OEP21, OEP24, and OEP37. Their ability to shuttle photosynthetic products, amino acids, and nitrogen assimilates likely accounts for the bulk of the metabolic exchange between plastid and cytosol. However, the high and low specificity channel proteins are most likely not the only metabolic shuttling systems of the OE.
Putative Metabolite Shuttles and OE Proteins of Unknown Function
The ABC transporter of unknown transport function WBC7
An ABC transporter of unknown function, white–brown-complex protein 7 (WBC7), was localized to the outer envelope of pea chloroplasts (Schleiff et al., 2003a) and to Arabidopsis chloroplast envelope fractions (Zybailov et al., 2008; Ferro et al., 2010; Joyard et al., 2010) by proteomics. AtWBC7 is a member of the G family of ABC transporters with a single ABC cassette and six transmembrane domains (TMD). The G family contains half size ABC transporters, which form homo- or heterodimers (Kusuhara and Sugiyama, 2007). Two proteins of this family have been investigated in Arabidopsis. AtWBC11 and AtWBC12 reside in the PM and are involved in the export of cuticular lipids in epidermal cells (Mcfarlane et al., 2010). No experimental evidence is available on the function of AtWBC7 although it is tempting to speculate about a role in transport of hydrophobic or partially hydrophobic substances (Figure 2J).
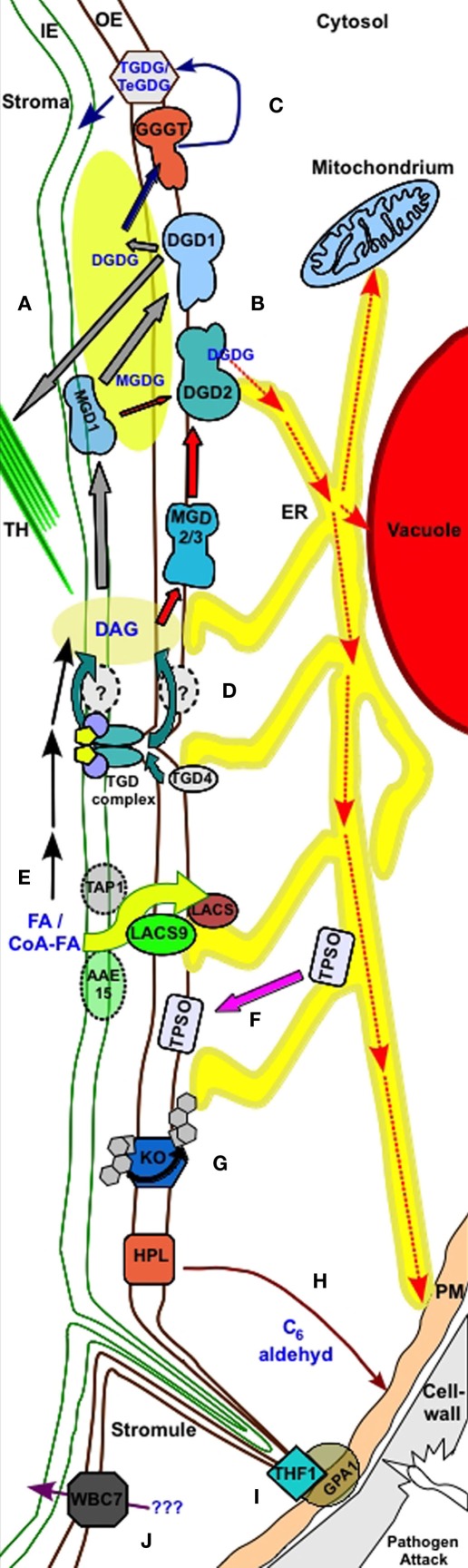
Figure 2.
Processes in the outer plastid envelope. (A) Galactosyl diacylglycerol biosynthesis under normal growth conditions. MGD1 produces MGDG from DAG. MGDG is either used in plastid membranes or is further processed by the OE resident DGD1 to produce DGDG for plastidic membrane use. Gray arrows label transport processes. (B) DGDG production under phosphate deprivation occurs via the OE resident MGD2/3 and DGD2. DGDG is transported to non-plastidic membranes, such as the tonoplast, mitochondrial membranes, and plasma membrane, possibly via the ER. Red arrows label transport processes. (C) GGGT produces TGDG and TeGDG during freezing stress to provide dehydration by thickening hydrophilic parts of the membrane. This process is labeled by blue arrows. (D) TGD1–3 complex disrupts the OE to mediate lipid exchange from ER to the IE. ER resident TGD4 may provide membrane lipid precursors directly to the OE. This process most likely involves further proteins in the OE and/or IE. (E) Free fatty acids are processed by LACS proteins resident in the OE and may IE. An ABC transporter Tap1 putatively involved in esterified fatty acids (CoA-FA) mediates the transport through the IE. At the OE LACS9 takes over the CoA-FA and de-esterifies these during transport across the OE. ER resident LACS further process FA. An alternative pathway involves putative IE resident proteins with LACS activity (AAE15). (F) Transfer of TPSO during salt and osmotic stress from ER to OE. (G) Transfer of ent-kauren across the OE during oxidation to ent-kaurenoic acid by OE resident ent-kauren oxidase. (H) OE resident HPL is integrated into pathogen defense via C6 aldehyde production when wounding occurs. (I) OE resident THF1 interacts with plasma membrane (PM) resident GPA1 in sugar signaling. Stromule bridge the process. (J) The G family ABC transporter WBC7 mediate transport of unknown compounds through the OE.
The putative porphyrin scavenger or transporter TSPO
TSPO is at most a temporary resident in the OE of plastids (Balsemao-Pires et al., 2011). Expression in Arabidopsis thaliana is induced by salt stress (Balsemao-Pires et al., 2011) or ABA treatment (Vanhee et al., 2011). A Physcomitrella TSPO mutant is hypersensitive to oxidative stress (Frank et al., 2007). The protein is conserved throughout eukaryotes including yeast (Vanhee et al., 2011) in which it is degraded by autophagy upon heme binding. AtTSPO co-localizes with autophagy markers (Vanhee et al., 2011). AtTSPO is a membrane protein, which has been localized to the ER and Golgi (Balsemao-Pires et al., 2011; Vanhee et al., 2011) and to the outer envelope of plastids (Balsemao-Pires et al., 2011) where it only appears after salt treatment. The precise function of TSPO is unknown but it is targeted for autophagy after treatments which presumably increase free porphyrins (Guillaumot et al., 2009; Vanhee et al., 2011). TSPO is hypothesized to protect plant cells against oxidative stress by binding and thus detoxifying free porphyrins (Figure 2F; Vanhee et al., 2011).
Transport of gibberellin precursors from the plastid
Gibberellins are produced from geranyl–geranyl-diphosphate generated by the isoprenoid pathway in plastids. The first two biosynthetic enzymes, copalyl diphosphate synthase and ent-kaurene synthase, are soluble stromal proteins (Helliwell et al., 2001). They produce a lipophilic intermediate, ent-kaurene, which likely partitions to the membranes (Helliwell et al., 2001). The next step, the production of the less lipophilic intermediate ent-kaurenoic acid, occurs at the outer envelope of the plastid since ent-kaurene oxidase localizes to the OE of the plastids judged by GFP fusion proteins and in vitro import assays (Helliwell et al., 2001). Whether passage through the IMS requires a dedicated transporter or occurs in conjunction with lipid transfer or spontaneously has not been addressed. Further oxidation of ent-kaurenoic acid occurs in the ER (Figure 2G).
The transfer of the gibberellin precursor presents a model by which lipophilic plastid produced precursors may cross the envelope membranes to their destination in the cytosol without the need of dedicated transporters.
Unusual protein of unknown function
In 1994 a small OE protein from spinach chloroplasts was cloned and biochemically investigated (Fischer et al., 1994). While its calculated weight is approximately 16 kDa, its abnormal amino acid composition led to an apparent molecular weight of approximately 24 kDa on SDS gel for which the protein was named OMP24. The insertion of OMP24 into the membrane is independent of surface receptors and target peptides. ATP has been shown to stimulate the insertion of the protein into the membrane (Fischer et al., 1994). The function of this integral protein is unknown.
The Role of the Outer Envelope in Lipid Synthesis
Unlike small hydrophilic compounds, fatty acids or lipids cannot easily be transported through the aqueous phase. Yet the interplay between the plastid and the cytosol, especially the ER is extensive.
Transport of fatty acids
Fatty acids are amphipatic: a small hydrophilic head group caps a highly hydrophobic long hydrocarbon tail. Several studies revealed that from 62% (Arabidopsis green tissue, represents “16:3”-plants) up to 90% (non-green tissue and green tissue of “18:3”-plants, see below) of fatty acids are exported from the plastid and transferred to the ER. The major and likely only transport direction is from plastids to the remainder of the cell (Browse et al., 1986, 1993; Somerville and Browse, 1991). The transport of these metabolites across two envelopes has not yet been resolved. However, long-chain-fatty-acid-Coenzyme-A synthetases (LACSs) are expected to play a prominent role in this process since they can catalyze a vectorial reaction in bacteria (Benning, 2009). Nine isoforms of this protein family are distributed to all sub cellular compartments of in Arabidopsis and can be found in peroxisomes, mitochondria, and the plastid (Schnurr et al., 2002; Shockey et al., 2002). Already in the late 1970s LACS activity in spinach chloroplast was shown to localize to the outer envelope membrane (Roughan and Slack, 1977). More than 20 years later, AtLACS9 was located to the envelope membranes by proteomics (Sun et al., 2009; Ferro et al., 2010; Joyard et al., 2010) and is most likely located to the OE (compare Koo et al., 2004; Sun et al., 2009). AtLACS9 is the only known exclusively plastid localized LACS and catalyzes 90% of the acetylation reactions. Its Vmax is higher than needed for complete fatty acid export. However, a knockout mutant shows no apparent phenotype (Schnurr et al., 2002). Possibly, the ER localized AtLACS1 can take over at least part of its function at least in triacylglycerol (TAG) biosynthesis (Zhao et al., 2010), which would require very close contact between the OE and the ER. Proteomics studies suggest three additional proteins with LACS activity in the plastid envelopes, AAE15 and AAE15-like as well as AtLACS8 of which the majority is localized to the ER or peroxisomes (Koo et al., 2004; Ferro et al., 2010; Joyard et al., 2010).
Fatty acid transport via LACS through the OE still leaves the inner envelope to be crossed. Extrapolating from the function of a known fatty acid transporter, peroxisome ABC transporter 1 (PXA1), which is localized to the peroxisomal membrane, the following model is proposed: The inner envelope contains an ABC transporter of the same class as PXA1, transporter associated with antigen processing protein 1 (TAP1), which has been consistently detected in all envelope proteome projects to date (Koo and Ohlrogge, 2002; Garcia et al., 2004; Sugiyama et al., 2006; Bräutigam et al., 2008; Bräutigam and Weber, 2009; Kunz et al., 2009; Ferro et al., 2010; Joyard et al., 2010) and whose function is unknown.
TAP1 or possibly another ABC transporter transports esterified fatty acids across the inner envelope where they are taken over by LACS9 and de-esterified in the process of transport out of the plastid. The free fatty acid is immediately bound by different LACS localized to one of the extraplastidial compartments and esterified again. This modus operandi would limit the exposure of the plastid to free fatty acids, which can act as detergents (Shine et al., 1976; Koo and Ohlrogge, 2002; Koo et al., 2004). However, it requires physically close association of the plastid to the ER to prevent the escape of free fatty acids. 18O labeling indeed showed that the transport involves a free fatty acid stage (Pollard and Ohlrogge, 1999). The reverse pathway of transport is impossible in this model since the ABC transporter is not reversible (Higgins, 1992). Alternatively, if an IE transporter transports free fatty acids, AtLACS9 could transfer the fatty acid by esterification thus leading to the release of CoA-fatty acid esters to the cytosol. Fatty acids are incorporated into lipids in chloroplasts and the ER (Figure 2E; compare Koo et al., 2004).
Oxylipin synthesis at the OE
Oxylipins have prominent roles in plant development and pathogen defense. They are produced from alpha linoleic acid liberated from lipids of the chloroplast. At least one branch of oxylipin synthesis can involve the OE, since the OE localized hydroperoxide lyase (HPL) catalyzes the first step toward C12 omega-keto-fatty acid and C6 aldehydes (Blee and Joyard, 1996; Froehlich et al., 2001b). The C12 omega-keto-fatty acid is the precursor for traumatin while the C6 aldehydes are directly involved in pathogen defense (summarized in Howe and Schilmiller, 2002; Arimura et al., 2009). It has been shown, that during fungal attack AtHPL expression is upregulated and leads to an increase of C6 aldehyde concentration at wound sites (Shiojiri et al., 2006). Defense against the pathogen is directly provided by the toxicity of the HPL metabolized compound (Figure 2H; Kishimoto et al., 2008). Studies on potato HPL also suggest a role for HPL in defense against sucking insects. However, the potato isoforms of the HPL are not localized to the OE (Vancanneyt et al., 2001).
Galactoglycerolipid synthesis in plants
Galactoglycerolipids (GGL) are essential to photosynthetic function (Reifarth et al., 1997; Guo et al., 2005; Hölzl et al., 2006) but can be found throughout the cell depending on the environmental conditions (Härtel et al., 2000; Kelly and Dörmann, 2002). They represent the main membrane lipids in green tissue of land plants and are in majority localized to the thylakoids. Each GGL carries two fatty acids in the sn-1 and sn-2 position which show where the precursor came from before the head groups were attached at the envelopes. Lipid backbones produced by the eukaryotic pathway carry 18:3 fatty acids at positions sn-1 and sn-2 (Browse et al., 1986). In contrast, lipid backbones produced in the plastid carry a 16:3 fatty acid at position sn-2 (Browse et al., 1986).
In the green algae Chlamydomonas reinhardtii the precursors of GGL biosynthesis are exclusively provided by the chloroplast itself since only the sn-2 position carries a 16:3 fatty acid (Giroud et al., 1988). Plants like spinach and Arabidopsis belong to the group of “16:3”-plants. In 16:3 plants the use of ER derived and plastid-derived precursors in GGL biosynthesis is about equal (Browse et al., 1986). Plants like pea use only ER derived precursors for GGL production (Heemskerk et al., 1990) and are called 18:3 plants. It has been shown that “16:3”-plants can be forced to use only ER derived precursors. The knock out of the major enzyme in the prokaryotic diacylglycerol (DAG) production, the plastidic glycerol-3-phosphate acyltransferase (ACT1) leads to a complete shift to ER derived precursor use in Arabidopsis (Kunst et al., 1988). In summary, in all land plants, a high volume of traffic is necessary to supply between 50 and 100% of lipid precursors for GGL synthesis, which is envelope bound, from the ER. The major GGLs are monogalactosyldiacylglycerol (MGDG), synthesized at the IE in Arabidopsis and digalactosyldiacylglycerol (DGDG), synthesized at the OE. MGDG and DGDG represent approximately 50 and 20% of the plastidic membrane lipids, respectively (Block et al., 1983).
The production of MGDG in Arabidopsis is mainly processed by the inner membrane bound protein MGDG synthase 1 (AtMGD1; Figure 2A; Marechal et al., 1994; Jarvis et al., 2000). In pea, the MGD activity is divided equally between the IE and the OE (Cline and Keegstra, 1983; Tietje and Heinz, 1998). This distribution possibly reflects the precursor supply by only the eukaryotic pathway in the ER. Consistent with these findings, OE membranes do contain additional enzymes with MGD activity in Arabidopsis and also soybean and corn. They are classified as type B MGDs, whereas the major MGDs like AtMGD1 and its paralogs are classified as type A proteins (Miege et al., 1999). Arabidopsis contains two type B MGDs, AtMGD2 and AtMGD3 (Figures 2A,B). Compared to MGD1, these enzymes show higher selectivity for eukaryotic pathway derived DAG. In photosynthetic active tissue they are underrepresented compared to AtMGD1, while non-green tissues display equal expression patterns for all three MGDs (Awai et al., 2001). These findings are consistent with the importance of type A MGDs for thylakoid lipid assembly. Knock down mutants of AtMGD1 show that the type B MGDs cannot compensate the complete loss of the type A enzyme (Jarvis et al., 2000).
In contrast to MGDG, DGDG is exclusively synthesized at the OE. The predominant enzyme in DGDG biosynthesis is the OE bound DGDG synthase 1 (AtDGD1; Figure 2A; Froehlich et al., 2001a). This enzyme drives the UDP-galactose dependent glycosylation of MGDG and produces αβDGDG. Knock out mutant analysis in Arabidopsis revealed that AtDGD1 catalyzes 90% of DGDG biosynthesis (Dörmann et al., 1999). The massive decrease in DGDG in mutant plants lead to a strong morphological and developmental defect, which reflects the deficiency in the photosynthetic apparatus (Dörmann et al., 1995). DGD activity in Arabidopsis is also not limited to one enzyme. AtDGD2 is an UDP-galactose dependent galactosyl transferase. It localizes to the OE and similar to alternative MGDs, AtDGD2 also shows far less activity and also lower expression levels than the major enzyme (Figure 2B; Dörmann et al., 1995; Härtel et al., 2000; Kelly and Dörmann, 2002). AtDGD2 can produce trigalactosyldiacylglycerol (TGDG) in vitro (Kelly and Dörmann, 2002), but there is no evidence for the production of TGDG by AtDGD2 in vivo.
An additional enzyme involved in GGL biosynthesis and localized to the OE is the galactolipid:galactolipid galactosyltransferase (GGGT) which uses MGDG as the galactosyl donor (Heemskerk et al., 1983, 1986). Its activity was already described in the 1970s when galactosyltransferase activity was analyzed in spinach (Van Besouw and Wintermans, 1978). GGGT catalyzes the synthesis of TGDG or even tetragalactosyldiacylglycerols (TeGDG), while DAG is released (Benning and Ohta, 2005). All galactosyl groups in GGGT produced GGLs are in β-configuration, while DGDs produce DGDGs with alpha-configuration in the second position (Kelly and Dörmann, 2002; Xu et al., 2003). GGGT is equivalent to sensitive to freezing 2 (SFR2; Moellering et al., 2010), a mutant identified earlier as freezing sensitive (Thorlby et al., 2004; Fourrier et al., 2008). Freezing damage is represented by rupture and fusion of membrane bilayers and non-bilayer structures can provide stability to lamellar membrane structures. The change in the membrane lipid composition affected by oligo GGLs leads to the formation of hexagonal-II-type structures at least in the OE, creating a non-bilayer shape of the membrane. Oligo GGLs mediate protection against the dehydration effect by providing a higher thickness of the hydrophilic part of the OE (Figure 2C; Moellering et al., 2010).
Transport of lipid precursors from the ER to the plastid
Massive lipid traffic occurs between the chloroplast and the ER. Precursors for the galactolipid biosynthesis have to be transported from the ER to the plastid and therefore through the envelopes. The magnitude of lipid flux is increased in “18:3”-plants. While lipid transfer is only partially understood, some proteins involved in ER to plastid transfer have been investigated in detail (summarized in Benning, 2009).
At least four proteins are involved in the transfer of lipids between ER and chloroplast. They are named after the unusual abundance of TGDG in mutant lines, and are called TGD1, 2, 3, and 4 (Xu et al., 2003). TGD1–3 likely form a high molecular weight complex and reside in the IE. While TGD1 is assumed to channel the lipid, TGD3 was demonstrated to be the ATP hydrolyzing component in the super complex, providing the energy for this process (Lu et al., 2007). TGD2 is hypothesized to be anchored in the IE and interact via its C-terminus with the OE. TGD2 is postulated to disrupt or destabilize the OE to form a conduit for lipid transport from the outer envelope across the IMS (Roston et al., 2011). TGD2’s binding affinity to phosphatidic acid (PA) lead to the assumption, that the TGD1–2–3-complex drives the transport of PA from the OE to the IE where it is dephosphorylated to DAG (Figure 2D; Awai et al., 2006; Benning, 2009; Lu and Benning, 2009).
A fourth protein in the lipid shuffling process is TGD4. TGD4 is located to the ER and/or the OE and is assumed to transfer eukaryotic lipid precursors to the plastid together with the TGD1–3 machinery (Xu et al., 2008; Benning, 2009). TGD mutants show that TGD proteins are exclusively involved in the lipid transport to the chloroplast (Xu et al., 2010). It is currently not known which, if any, proteins act between the TDG1–3 complex and TGD4 (Benning, 2009). However, the TGD mutant screens include two more complementation groups with the potential to fill the remaining gaps (Figure 2D; Xu et al., 2003).
Transport of lipids from the plastid to the ER
During phosphate deprivation plant cells reclaim phosphate from phospholipids. Phospholipids in the PM (Andersson et al., 2003, 2005), mitochondrial membranes (Jouhet et al., 2004), and the tonoplast (Andersson et al., 2005) are replaced by GGLs. Under such stress conditions the underrepresented isoforms AtMGD2, AtMGD3, and AtDGD2, all localized to the outer envelope, are highly expressed. The main surrogate for phospholipids during phosphate deprivation is DGDG that can represent more than 30% of extraplastidial membrane lipids in Arabidopsis. The DGDG produced during phosphate limitation is independent of DGD1 since DGD1 knock out plants increase their DGDG content under phosphate starvation (Härtel et al., 2000). These enzymes preferably use precursors of the eukaryotic pathway to provide extraplastidic DGDG. It is currently not known how GGLs produced in the outer envelope reach their destination membranes during phosphate starvation. Although the envelopes can form vesicles toward the stroma (Hatta et al., 1973) as well as to the IMS (Park et al., 1999), no vesicular transfer directed from the plastid to the cytosol has been demonstrated. Possibly, the transport is driven by extensive formation of contact sides by the plastid and the ER (Figure 2B).
Protein Turnover Across and into the Outer Envelope
Protein import through the envelopes
The massive gene transfer from the cyanobacterial endosymbiont to the host nucleus necessitated a protein distribution system for the chloroplast. Most chloroplast-bound preproteins in the cytosol contain an N-terminal amino acid sequence, a transit peptide, to address it for import into the chloroplast through the TIC/TOC Complex (Bionda et al., 2010). This complex is abundant in chloroplasts as well as in non-green plastids, such as proplastids (Bräutigam and Weber, 2009) and etioplasts (Von Zychlinski et al., 2005; Reiland et al., 2011). We briefly summarize the knowledge about import with special focus on the outer envelope. The reader is referred to several excellent recent reviews for more details (Soll and Schleiff, 2004; Inaba and Schnell, 2008; Andres et al., 2010; Schwenkert et al., 2010)
The first recognition of the preprotein occurs by cytosolic chaperones HSP90 and HSP70 as well as by 14-3-3-proteins (Schwenkert et al., 2010). These proteins facilitate the transfer of the preproteins to the different receptors in the OE, proteins of the Toc159- and Toc34-family (Gutensohn et al., 2000; Ivanova et al., 2004). In Arabidopsis the Toc159 family (earlier known as Toc86-family) consists of four known members in Arabidopsis, AtToc90, AtToc120, AtToc132, and AtToc159, named by their different molecular weight. This difference is due to variation in the length of the acidic domain, while they share high sequence similarity in the GTPase-domain and the membrane binding domain (Bauer et al., 2000; Jackson-Constan and Keegstra, 2001; Hiltbrunner et al., 2004; Agne et al., 2010). Since Toc90 can only partially restore the Arabidopsis mutant ppi2, a plant deficient in AtToc159, it can be assumed that the different acidic domains of the proteins lead to specialization of the receptors (Bauer et al., 2000; Hiltbrunner et al., 2004; Ivanova et al., 2004; Kubis et al., 2004; Agne et al., 2010; Infanger et al., 2011).
The smaller GTPases AtToc33 and AtToc34 represent the Toc34-family in Arabidopsis (Jarvis et al., 1998; Chen et al., 2000; Gutensohn et al., 2000). The receptors also partially provide the energy for the protein translocation process by GTP hydrolysis (Figure 3B; Schleiff et al., 2003b; Andres et al., 2010). AtToc159 and AtToc33 facilitate the transport of proteins involved in photosynthesis related processes while AtToc120 and AtToc132 are more associated to Toc34 and seem to process house-keeping proteins (Ivanova et al., 2004; Huang et al., 2011). These roles are supported by proteomic comparisons between proplastids and chloroplasts, since AtToc120 and AtToc132 can be identified from proplastids but not chloroplasts (Bräutigam and Weber, 2009). AtToc159 can be identified from plastids, etioplasts, and chloroplasts (Von Zychlinski et al., 2005; Bräutigam and Weber, 2009; Reiland et al., 2011). Receptors of the TOC complex have not been identified from chromoplasts (Siddique et al., 2006; Barsan et al., 2010), which may either reflect their absence or technical limitations.
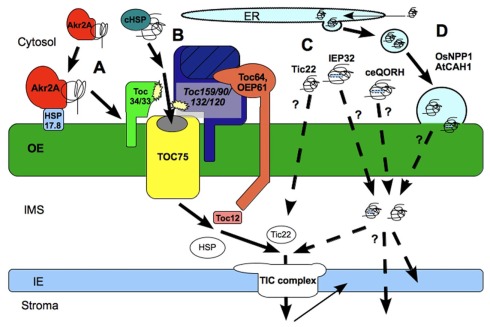
Figure 3.
Protein transfer through and into the outer envelope (OE). (A) Akr2a pathway for the insertion of OE proteins into the membrane, enhanced by HSP17.8; (B) Toc Complex mediated protein translocation via Toc75 and GTPase receptor proteins of the Toc159- and Toc34- family. Translocation is supported by cytosolic HSPs and transmembrane co-chaperones and co-receptors Toc64/OEP61; (C) Non-canonical protein translocation through the OE. Pathways are unclear and diverse. Protein internal signals might support the translocation; (D) Transfer of proteins through the OE via vesicle fusion. Complete proteins are synthesized in the cytosol, transferred to the ER and transported to the OE via vesicle transport; The transport from the inter membrane space (IMS) into the inner Envelope (IE) or stroma is mainly processed by the translocon complex of the IE (TIC) but other mechanisms have been postulated, too.
In addition to their function in protein translocation, some Toc components display also other less investigated features. For example, AtToc159 has binding affinity to actin filaments in vitro (Jouhet and Gray, 2009). The import receptor for house-keeping proteins, AtToc132, and the pore AtToc75-III are involved in root gravitropism (Stanga et al., 2009). Mutations in AtToc132 or in AtToc75-III, called mar2 (modifier of arg1 2) and mar1, modulate the gravitropism defects in altered response to gravity 1 (arg1) mutants. The modulation is not dependent on defects in starch orientation. Since the mutants do not show defects in gravitropism on their own, the molecular connection between the import complex components and gravitropism remains unresolved.
The channel protein AtToc75-III facilitates actual transport of the preprotein across the membrane (Perry and Keegstra, 1994; Schnell et al., 1994; Tranel et al., 1995; Hinnah et al., 1997, 2002; Sveshnikova et al., 2000; Jackson-Constan and Keegstra, 2001). As a member of the Omp85-family it contains a N-terminal POTRA (polypeptide-transport-associated) domain. The 16–18 in silico predicted β-strands at the C-terminus form the β-barrel domain, the pore (Sveshnikova et al., 2000; Hinnah et al., 2002; Schleiff et al., 2003a; Gentle et al., 2004; Baldwin et al., 2005). It has been shown, that AtToc75-III is not selective for a specific form of protein precursors (Baldwin et al., 2005) and this protein can be found in all plastid types investigated by proteomics (e.g., Bräutigam and Weber, 2009; Barsan et al., 2010; Reiland et al., 2011). A protein similar to AtToc75-III, AtToc75-V/OEP80, is required for accumulation of AtToc75-III in the membrane (Wallas et al., 2003). AtToc75-III in turn is required for the TOC-receptor compounds (Wallas et al., 2003).
Several proteins are reportedly associated with the import complex, however their precise function remains elusive. Toc64 and AtOEP61 likely interact with cytosolic chaperones and function as co-chaperone and co-receptor for the TOC complex (Qbadou et al., 2007; Von Loeffelholz et al., 2011). Another protein is the IMS localized Toc12, which interacts with Toc64 (Becker et al., 2004) and the inner membrane space proteins Tic22 and a chaperone (Figure 3B; Becker et al., 2004).
Virtually all known inner envelope, stromal, and thylakoid proteins are imported through the import complex. However, a subset of proteins identified during proteomic analysis of chloroplasts lacks a recognizable transit peptide (Baginsky and Gruissem, 2004). These proteins may either be contaminations or they may enter the plastid through a different pathway. The majority of outer envelope proteins also lacks a recognizable target peptide and enters the outer envelope without the benefit of the TOC Complex.
TOC complex independent protein import
One possible alternative route for proteins into the chloroplast has been shown for the a-type family carbonic anhydrase CAH1 in A. thaliana (Villarejo et al., 2005) and ADP-glucose hydrolytic nucleotide pyrophosphatase/phosphodiesterase (NPP) 1 in rice (Nanjo et al., 2006). CAH1 as well as NPP1 have an N-terminal signal peptide for the secretory pathway, which targets the proteins to the ER. In vivo localization studies with GFP fusion revealed CAH1 and NPP1 localization in the stroma. The localization in both studies was altered by the Golgi vesicle transport inhibitor brefeldin A, and CAH1–GFP as well as NPP1–GFP accumulated in ER and golgi (Villarejo et al., 2005; Nanjo et al., 2006). Based on these results CAH1 and NPP1 are transported from the cytosol into the ER and on to the chloroplast by Golgi-mediated vesicle transport. At the OE it is assumed that Golgi vesicles fuse with the envelope and release the fully folded protein into the IMS between IE and OE (Figure 3D; Villarejo et al., 2005; Nanjo et al., 2006). How the transport from the IMS into the stroma is accomplished is speculative.
An internal signal sequence is important for the transfer of NPP1 to the chloroplast (Kaneko et al., 2010). The requirement of an internal signal sequence was also shown for the TOC-independent insertion of the IE proteins chloroplast envelope quinone oxidoreductase (ceQORH; Miras et al., 2002, 2007), and the inner envelope protein (IEP) 32 (Figure 3C; Nada and Soll, 2004). While the presence of internal sequence motives important for transport appears to be a general theme, ceQORH, IEP32, and the protein Tic22 (Kouranov et al., 1998, 1999) were shown to be transferred to their destinations by different pathways (Figure 3C; Kouranov et al., 1999; Nada and Soll, 2004; Miras et al., 2007).
The study of early branching Archaeplastida provides further insights into the transport independent of the TOC complex. Recent studies on the amoeboid Paulinella chromatophora and its photosynthetic endosymbiont revealed an involvement of the ER in protein sorting to the plastid in earlier evolutionary stages (Mackiewicz and Bodyl, 2010).
Protein insertion into the OE
Unlike TOC based transport, the insertion of OE proteins into the membrane is mostly independent of energy equivalents. Many proteins can insert in thermolysin-treated plastids. This was taken as evidence for protein independent insertion, however, since OEP7 (called OEP14 in pea) insertion is inhibited by trypsin and N-ethylmaleimide but not thermolysin, it is more likely that the proteins involved are not affected by thermolysin treatment. Protein insertion has been studied with several model proteins: OEP7 (Arabidopsis), OEP14 (pea), and E6.7 (spinach) although their function is still not determined, Toc64, (Salomon et al., 1990; Li et al., 1991; Tu and Li, 2000; Lee et al., 2001; Dyall et al., 2004; Nada and Soll, 2004; Bae et al., 2008; Oikawa et al., 2008), the tail-anchored proteins OEP9 and Toc33 and Toc34 (Dhanoa et al., 2010) and the major import pore Toc75-III.
The ankyrin repeat proteins 2A and 2B (Akr2A and Akr2B) are involved in protein trafficking to the OE based on OEP7 and cOEP64/AtToc64 import experiments. The recognition and insertion of OEPs into the membrane requires a single C-terminal TMD and an upstream target signal. This signal is part of the functional protein and not cleaved off after translocation into the membrane (Tu and Li, 2000; Lee et al., 2001; Tu et al., 2004; Bae et al., 2008). The Akr2A protein mediates recognition of the preprotein. Akr2A binds with its N-terminal region to the target signals of the OE protein while the C-terminal region is required for binding to the OE. Akr2A also provides chaperone activity to OEPs (Figure 3A; Bae et al., 2008). Akr2B functions similarly to Akr2A (Bae et al., 2008)
Akr2a mediated import is also required to target tail-anchored proteins to the chloroplast (Dhanoa et al., 2010). Tail-anchored protein import can be divided into delivery of the protein to the appropriate organelle and insertion into the target membrane. While Akr2A mediates targeting to the appropriate organelle, insertion pathways diverge. The insertion of newly synthesized Toc33/34 depends on the presence of Toc33 in the membrane and is thus dependent on proteins and not or not only on the composition of the lipid bilayer. While insertion of OEP9 is also protein dependent, it does not require Toc33 or Toc34 for insertion and may be dependent on Toc75 (Dhanoa et al., 2010).
A second cytosolic factor is required for at least OEP7 insertion. A small cytosolic class 1 heat shock protein, Hsp17.8 which can dimerize or assemble into a temperature induced oligomeric complex, interacts with the C-terminal ankyrin repeat domain of Akr2A and, as a dimer, binds with high affinity to the OE. This mediates an enhanced binding affinity of Akr2A to the OE. Knock down lines of class1 HSPs including HSP17.8 exhibit reduced targeting efficiency for OEP7 and overexpression lines enhance OE targeting (Figure 3A; Kim et al., 2011). At least four other small cytosolic heat shock proteins can also interact with AKR2A to varying degrees (Kim et al., 2011).
Targeting of one of the major outer envelope proteins, AtToc75-III, remains enigmatic. Unlike all other outer envelope proteins tested, AtToc75-III carries a bipartite targeting signal, which consists of a cleavable targeting signal to the chloroplast stroma and a polyglycine stretch required for insertion into the outer envelope (Tranel et al., 1995; Tranel and Keegstra, 1996; Inoue and Keegstra, 2003). The exact mechanism for Toc75 insertion is currently unknown but other OMP85 related proteins like AtTOC75-V/OEP80 (Patel et al., 2008; Huang et al., 2011) and the N-terminal truncated Toc75 homolog, AtToc75-IV, which is inserted into the membrane without any cleavable target peptide (Baldwin et al., 2005), may help to understand the insertion process of AtToc75-III.
The rapid progress in dissecting the import pathways into the outer envelope in recent years (Bae et al., 2008; Dhanoa et al., 2010; Kim et al., 2011) since the focus was shifted from the well understood TIC–TOC Pathway (Soll and Schleiff, 2004; Inaba and Schnell, 2008; Andres et al., 2010; Schwenkert et al., 2010) to other import pathways may indicate that a systematic picture of targeting machinery and signals may soon arise from additional studies.
Protein repair and degradation
The interior of the chloroplast is a hazardously oxidizing environment with multiple protection systems (Baier and Dietz, 2005; Oelze et al., 2008). Yet, no protein repair or turnover mechanism is known in addition to those of cyanobacterial origin affecting the thylakoid membranes themselves (Nixon et al., 2010). Mitochondria can be turned over entirely by autophagy. The complete organelle is engulfed, digested, and its parts recycled (Mijaljica et al., 2007). However, plastids are too big to fit into the autophagy machinery (Ishida and Yoshimoto, 2008; Izumi et al., 2010). There are organisms containing only a single plastid such as C. reinhardtii. Turning over the complete plastid is not a viable route in these organisms. Cytosolic proteins can also be recycled by autophagy or they are targeted for degradation through the proteasome by ubiquitination (Van Doorn and Woltering, 2005; Reape et al., 2008; Uchiyama et al., 2008). Unlike the IE and the proteins within the plastids, the OE is exposed to the cytosol and hence to the protein degradation machinery. However, ubiquitination of any plastid protein has not yet been reported from any of the proteome studies, likely because these studies were not designed to identify such post-translational modifications (e.g., Rolland et al., 2003; Bräutigam et al., 2008; Bräutigam and Weber, 2009; Ferro et al., 2010). Since it is highly unlikely that all plastid proteins survive intact for the duration of plant life (some plants can reach several thousand years in age), one or several methods for protein turnover at the plastid must exist. During plastid differentiation from proplastids to chloroplasts, the protein complement of the stroma (compare Sun et al., 2009; Ferro et al., 2010), the IE, and the OE (e.g., Bräutigam and Weber, 2009) is changed in quality and quantity again necessitating protein degradation. Although it is tempting to speculate about vesicular transport out of the plastids of envelope proteins especially in light of the unresolved lipid transfer from plastids to the remainder of the cell, the question of protein turnover in the plastids envelopes remains completely unresolved.
Interaction of the Outer Envelope with the Cytosol
Sites mediating the contact between the plastid and the ER
Fatty acid and lipid metabolism require close physical contact of the ER and the plastids. Yet, the structural components mediating the interaction have not been identified to date. Plastid associated membranes (PLAMs) are a vesicular structure attached to the plastids from the cytosolic side (Andersson et al., 2007a,b). They were identified by GFP labeling the ER and isolating plastids, which were then decorated with fluorescing vesicles. Optical tweezers needed a force of 400 pN to overcome the attachment, which is a force equivalent to that of a protein–protein interactions (Florin et al., 1994). The vesicles can also be removed by a low pH/low salt wash in MES buffer (pH 6.0) supplemented with sucrose (details in Andersson et al., 2007a,b). Their lipid composition is intermediate between the composition of the outer envelope and the ER.
A second structure has been implicated in mediating interaction between the plastid and the cytosol, especially the ER: the stromules (Köhler and Hanson, 2000). Stromules are defined as stroma-filled tubules jutting out from plastids. They were observed in many tissues and different species, so they are likely a general feature of at least moss and higher plant plastids (Gray et al., 2001; Pyke and Howells, 2002; Waters et al., 2004; Gunning, 2005; Hanson and Sattarzadeh, 2008; Holzinger et al., 2008; Reski, 2009; Shaw and Gray, 2011). Stromules are hypothesized to be built by the joint action of internal pressure and external draw. Filament forming proteins, such as the plastid division protein FtsZ are under debate to be involved in the formation of stromules since they are abundant in tomato chromoplasts where plastid division is unlikely to occur (Reski, 2009; Barsan et al., 2010; Hanson and Sattarzadeh, 2011). The cytosolic compounds in the stromule formation process are most probably the actin cytoskeleton (Kwok and Hanson, 2004) in combination with specific myosin proteins. Stromule formation is decreased by actin inhibitors and by gene silencing of myosin class XI motor proteins (Kwok and Hanson, 2003; Sattarzadeh et al., 2009). Actin inhibitors lead to collapse of existing stromules, which remain tubular but collapse onto the plastid surface (Kwok and Hanson, 2003). Since the material does not spontaneously reinsert, the formation and reinsertion of stromules is probably an active process. In addition to the involvement of actin in stromule movement, motor proteins like myosin XI of the F-class are assumed to drive the chloroplast dynamics (Sattarzadeh et al., 2009). The best evidence for a direct interaction of myosin with the stromule and plastidic OE was described for a myosinXI-2 relative of Arabidopsis in N. benthamiana. This has been shown to interact with its cargo-binding domain at the C-terminus directly with the OE (Natesan et al., 2009). Therefore, one or more myosins of the XI-F-family are proposed to be anchors in the OE, building connection between actin skeleton and stromules (Sattarzadeh et al., 2009).
Similar to PLAMs the functional significance of stromules has not been experimentally tested. Direct substance transport between connected plastids is possible, yet likely slow since dense stroma fills the connecting tubules (Köhler et al., 2000; Hanson and Sattarzadeh, 2011). Plastids have almost perfect lens-shape, which limits the surface to volume ratio. Stromules massively increase the surface area (compare Hanson and Sattarzadeh, 2011) and provide space for proteins such as transporters. If this hypothesis was true, stromules should be present in cells and tissues with increased import or export requirements for metabolites across the envelope membranes. Remarkably, stromules are barely detectable in mature leaf chloroplasts, which actively photosynthesize but frequently observed in other tissues, which rather have sink characteristics (Köhler and Hanson, 2000; Pyke and Howells, 2002; Waters et al., 2004). Apparently, the high flux of photosynthates in leaf chloroplasts does not require surface area increases as is also evident by the absence of stromules in algal chloroplasts. Stromules may also bridge the distance between plastids and other organelles or maintain connections while allowing movement of chloroplasts. Plastids can “hug” or surround completely other organelles via stromules (e.g., Sage and Sage, 2009). Dynamics of stromules and of the cortical ER correlate with each other, mediated either by shared cytoskeletal interaction or by direct interaction (Schattat et al., 2011) possibly through PLAMs. The ER and the plastids use different myosins but a common actin backbone to mediate their individual movements (Sattarzadeh et al., 2009; Ueda et al., 2010). Stromules may mediate signaling between plastids and the PM. The protein thylakoid formation 1 (THF1) is localized to the stroma and to the OE of plastids (Wang et al., 2004; Huang et al., 2006; Joyard et al., 2010). It is also distributed to stromules. While deletion of the THF1 gene leads to defects in thylakoid formation (Wang et al., 2004), it is detectable in chromoplasts (Siddique et al., 2006), a plastid type with frequent stromule sightings (Pyke and Howells, 2002; Waters et al., 2004) but without photosynthesis. Recent findings revealed, that THF1 physically interacts with the plasma membrane G-protein GPA1 (Huang et al., 2006). This interaction requires proximity of the plastid or its stromule with the plasma membrane. Stromules may also play a role in stress response (Holzinger et al., 2007; Gray et al., 2011)
The evidence for stromules interacting with other cellular compartments is circumstantial hence the function of stromules remains as unknown as the mechanism that builds them.
Movement of chloroplast
One of the adaptations of the plant to different light dosage is the dynamic positioning of the chloroplasts inside the cell (Trojan and Gabrys, 1996). On the one hand, the position change increases the light-use-efficiency during low light conditions. On the other hand, it reduces photo-damage during high light conditions. The OE is the interface for the moving devices of the cell. Mutants deficient in chloroplast unusual positioning 1 (CHUP1) display defects in the distribution and positioning of chloroplasts in the cell. CHUP1 is N-terminal anchored in the OE membrane by a single TMD representing also the targeting signal for its insertion. Its C-terminus is facing the cytosol and contains an actin binding motif. CHUP1 directly interacts with the PM via a C-terminal coiled coil domain (Oikawa et al., 2003, 2008; Von Braun and Schleiff, 2008). Two kinesin-like proteins (KAC1 and KAC2) are CHUP1 interaction partners at the PM (Suetsugu et al., 2010). The interaction between CHUP1 and KAC1/2 is mediated by a specific type of chloroplast associated actin filaments (cp-actin). The accumulation of cp-actin at the actin binding domain of CHUP 1 and their connection to the PM are regulated by two blue light receptor phototropins, Phot1 and Phot2 (Kadota et al., 2009). The PM protein THRUMIN1 has also been postulated to be a link for plastidial movement. The study suggests F-actin as the moving devise for plastid movement (Whippo et al., 2011) as described earlier (Oikawa et al., 2003).
While several extra plastidic factors are involved in the chloroplast movement, CHUP1 is the only detected plastidic factor involved in this process so far (Kadota et al., 2009; Whippo et al., 2011).
The role of OE in plastid–nucleus interaction
Retrograde signaling from the plastid to the nucleus is a necessity. However, retrograde signals remain unknown. A recent study describes a protein possibly involved in the retrograde signaling from the plastid outer envelope to the nucleus (Sun et al., 2011). The protein contains a plant homeodomain (PHD) and is bona fide a transcription factor since it can activate transcription. It is bound to the OE via TMDs, hence its name PTM (PHD type transcription factor with TMDs). Immunoblot analysis revealed re-localization of the soluble N-terminal part of PTM to the nucleus upon signal dependent cleavage (Sun et al., 2011). PTM was shown to be involved in the expression of ABI4, which in turn regulates light harvesting complex associated chlorophyll binding proteins (Sun et al., 2011). Ptm knock out plants did not show any phenotype, indicating compensation of the loss of function by other factors. How signals are related from the inside of the plastid to the outer envelope remains unknown. The identification of a chloroplast derived transmitter, the cleavable PTM, holds promise for identification of additional parts in the signaling cascade.
Division of the chloroplast
Two systems in concerted action, one on the stromal side of the envelopes and a second one on the cytosolic side of the OE accomplish plastid division. The stromal machinery originated from cyanobacteria (summarized in Yang et al., 2008) while the OE machinery was evolved from host proteins (summarized in Miyagishima, 2005) Both machineries are coordinated through ARC6 (accumulation and replication of chloroplast; Glynn et al., 2008) and PARC6 (paralog of ARC; Glynn et al., 2009) two IEPs that connect the inner machinery to the outer envelope through recruiting PDV1 (plastid division) and PDV2 (Miyagishima et al., 2006; Glynn et al., 2008, 2009). PDV1 and PDV2 in turn recruit ARC5, a dynamin-like protein (Gao et al., 2003), which forms a constriction force on the outside. They can mediate interactions between the plastid and the cytosol since their C-terminus faces the IMS where it can interact with IEPs. Their N-terminus faces the cytosolic site where it can interact with host proteins (Miyagishima et al., 2006). However, PDVs do not control plastid volume per cell; chloroplast number correlates negatively with chloroplast volume, the smaller they are, the more of them are present (Okazaki et al., 2009). The intriguing double mutant in IE mechano-sensitive (MS) ion channels also changes the plastid number per cell but not plastid volume (Haswell and Meyerowitz, 2006; Wilson et al., 2011) as do all known division machinery mutants (summarized in Yang et al., 2008). Pleiotropic effects of the MSL (MS-like) mutant include fewer cells, thicker leaves, and empty leaves. The CRL1 (crumbled leaf) mutants defective in an OE protein of unknown function as well as ARC6 mutants also show developmental abnormalities (Asano et al., 2004; Chen et al., 2009).
All mutants in the division machinery affect the plastid number and size, which are inversely correlated. If the number of plastids is reduced the remaining plastids increase in size and vice versa. Hence it is unlikely that the division machinery, which is localized to the envelopes controls plastid volume. The mechanism by which the cell determines how much of its volume it devotes to plastids is unknown but clearly, different cell types in plants devote different amounts of volume to plastids indicating that plastid volume is developmentally controlled. Recently it was discovered that ER tubules mark the site of mitochondrial division in yeast (Friedman et al., 2011). It is thus tempting to speculate that interactions between chloroplast and ER, such as PLAMs, might be involved in governing or coordinating plastid division.
Conclusion
In this review we described the multitude of functions associated with the outer plastid envelope. The opinions about its relevance for the function of the plant cell have changed over the past decades. Study of the plastids has mostly focused on protein rich, abundant subcompartments, such as the thylakoids. However, a detailed understanding of how the plastids integrate into the cytosol can only be achieved through understanding of the barrier between the compartments, the outer envelope. Metabolite transfer through the outer envelope is reasonably well understood, as are lipid synthesis and the division machinery. Progress has been made in studying ER to chloroplast transfer of lipids and protein targeting. However, transfer of lipids out of the plastids, protein turnover, and signal transduction remain mostly unknown.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by a grant of the Deutsche Forschungsgemeinschaft (WE2231/4-1 to APMW) in the framework of the Arabidopsis Functional Genomics Network (AFGN).
References
- Abdallah F., Salamini F., Leister D. (2000). A prediction of the size and evolutionary origin of the proteome of chloroplasts of Arabidopsis. Trends Plant Sci. 5, 141–142 10.1016/S1360-1385(00)01574-0 [DOI] [PubMed] [Google Scholar]
- Agne B., Andres C., Montandon C., Christ B., Ertan A., Jung F., Infanger S., Bischof S., Baginsky S., Kessler F. (2010). The acidic A-domain of Arabidopsis TOC159 occurs as a hyperphosphorylated protein. Plant Physiol. 153, 1016–1030 10.1104/pp.110.158048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson M. X., Goksor M., Sandelius A. S. (2007a). Membrane contact sites: physical attachment between chloroplasts and endoplasmic reticulum revealed by optical manipulation. Plant Signal. Behav. 2, 185–187 10.4161/psb.2.3.3973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson M. X., Goksor M., Sandelius A. S. (2007b). Optical manipulation reveals strong attracting forces at membrane contact sites between endoplasmic reticulum and chloroplasts. J. Biol. Chem. 282, 1170–1174 10.1074/jbc.M608124200 [DOI] [PubMed] [Google Scholar]
- Andersson M. X., Larsson K. E., Tjellstrom H., Liljenberg C., Sandelius A. S. (2005). Phosphate-limited oat. The plasma membrane and the tonoplast as major targets for phospholipid-to-glycolipid replacement and stimulation of phospholipases in the plasma membrane. J. Biol. Chem. 280, 27578–27586 10.1074/jbc.M503273200 [DOI] [PubMed] [Google Scholar]
- Andersson M. X., Stridh M. H., Larsson K. E., Liljenberg C., Sandelius A. S. (2003). Phosphate-deficient oat replaces a major portion of the plasma membrane phospholipids with the galactolipid digalactosyldiacylglycerol. FEBS Lett. 537, 128–132 10.1016/S0014-5793(03)00109-1 [DOI] [PubMed] [Google Scholar]
- Andres C., Agne B., Kessler F. (2010). The TOC complex: preprotein gateway to the chloroplast. Biochim. Biophys. Acta 1803, 715–723 10.1016/j.bbamcr.2010.03.004 [DOI] [PubMed] [Google Scholar]
- Arimura G.-I., Matsui K., Takabayashi J. (2009). Chemical and molecular ecology of herbivore-induced plant volatiles: proximate factors and their ultimate functions. Plant Cell Physiol. 50, 911–923 10.1093/pcp/pcp030 [DOI] [PubMed] [Google Scholar]
- Aronsson H., Boij P., Patel R., Wardle A., Topel M., Jarvis P. (2007). Toc64/OEP64 is not essential for the efficient import of proteins into chloroplasts in Arabidopsis thaliana. Plant J. 52, 53–68 10.1111/j.1365-313X.2007.03207.x [DOI] [PubMed] [Google Scholar]
- Asano T., Yoshioka Y., Kurei S., Sakamoto W., Machida Y. (2004). A mutation of the CRUMPLED LEAF gene that encodes a protein localized in the outer envelope membrane of plastids affects the pattern of cell division, cell differentiation, and plastid division in Arabidopsis. Plant J. 38, 448–459 10.1111/j.1365-313X.2004.02057.x [DOI] [PubMed] [Google Scholar]
- Awai K., Marechal E., Block M. A., Brun D., Masuda T., Shimada H., Takamiya K., Ohta H., Joyard J. (2001). Two types of MGDG synthase genes, found widely in both 16:3 and 18:3 plants, differentially mediate galactolipid syntheses in photosynthetic and nonphotosynthetic tissues in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 98, 10960–10965 10.1073/pnas.181331498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Awai K., Xu C. C., Tamot B., Benning C. (2006). A phosphatidic acid-binding protein of the chloroplast inner envelope membrane involved in lipid trafficking. Proc. Natl. Acad. Sci. U.S.A. 103, 10817–10822 10.1073/pnas.0602754103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae W., Lee Y. J., Kim D. H., Lee J., Kim S., Sohn E. J., Hwang I. (2008). AKr2A-mediated import of chloroplast outer membrane proteins is essential for chloroplast biogenesis. Nat. Cell Biol. 10, 220–227 10.1038/ncb1683 [DOI] [PubMed] [Google Scholar]
- Baginsky S., Gruissem W. (2004). Chloroplast proteomics: potentials and challenges. J. Exp. Bot. 55, 1213–1220 10.1093/jxb/erh104 [DOI] [PubMed] [Google Scholar]
- Baier M., Dietz K. J. (2005). Chloroplasts as source and target of cellular redox regulation: a discussion on chloroplast redox signals in the context of plant physiology. J. Exp. Bot. 56, 1449–1462 10.1093/jxb/eri161 [DOI] [PubMed] [Google Scholar]
- Baldi P., Grossi M., Pecchioni N., Vale G., Cattivelli L. (1999). High expression level of a gene coding for a chloroplastic amino acid selective channel protein is correlated to cold acclimation in cereals. Plant Mol. Biol. 41, 233–243 10.1023/A:1006375332677 [DOI] [PubMed] [Google Scholar]
- Baldwin A., Wardle A., Patel R., Dudley P., Park S. K., Twell D., Inoue K., Jarvis P. (2005). A molecular-genetic study of the Arabidopsis Toc75 gene family. Plant Physiol. 138, 715–733 10.1104/pp.105.063289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balsemao-Pires E., Jaillais Y., Olson B. J., Andrade L. R., Umen J. G., Chory J., Sachetto-Martins G. (2011). The Arabidopsis translocator protein (AtTSPO) is regulated at multiple levels in response to salt stress and perturbations in tetrapyrrole metabolism. BMC Plant Biol. 11, 108. 10.1186/1471-2229-11-108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barsan C., Sanchez-Bel P., Rombaldi C., Egea I., Rossignol M., Kuntz M., Zouine M., Latche A., Bouzayen M., Pech J. C. (2010). Characteristics of the tomato chromoplast revealed by proteomic analysis. J. Exp. Bot. 61, 2413–2431 10.1093/jxb/erq070 [DOI] [PubMed] [Google Scholar]
- Bauer J., Chen K. H., Hiltbunner A., Wehrli E., Eugster M., Schnell D., Kessler F. (2000). The major protein import receptor of plastids is essential for chloroplast biogenesis. Nature 403, 203–207 10.1038/35003214 [DOI] [PubMed] [Google Scholar]
- Becker T., Hritz J., Vogel M., Caliebe A., Bukau B., Soll J., Schleiff E. (2004). Toc12, a novel subunit of the intermembrane space preprotein translocon of chloroplasts. Mol. Biol. Cell 15, 5130–5144 10.1091/mbc.E04-05-0405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benning C. (2009). Mechanisms of lipid transport involved in organelle biogenesis in plant cells. Annu. Rev. Cell Dev. Biol. 25, 71–91 10.1146/annurev.cellbio.042308.113414 [DOI] [PubMed] [Google Scholar]
- Benning C., Ohta H. (2005). Three enzyme systems for galactoglycerolipid biosynthesis are coordinately regulated in plants. J. Biol. Chem. 280, 2397–2400 10.1074/jbc.R400032200 [DOI] [PubMed] [Google Scholar]
- Bickel H., Palme L., Schultz G. (1978). Incorporation of shikimate and other precursors into aromatic amino acids and prenylquinones of isolated spinach chloroplasts. Phytochemistry 17, 119–124 10.1016/S0031-9422(00)89691-0 [DOI] [Google Scholar]
- Binder S., Knill T., Schuster J. (2007). Branched-chain amino acid metabolism in higher plants. Physiol. Plant 129, 68–78 10.1111/j.1399-3054.2006.00800.x [DOI] [Google Scholar]
- Bionda T., Tillmann B., Simm S., Beilstein K., Ruprecht M., Schleiff E. (2010). Chloroplast import signals: the length requirement for translocation in vitro and in vivo. J. Mol. Biol. 402, 510–523 10.1016/j.jmb.2010.07.052 [DOI] [PubMed] [Google Scholar]
- Blee E., Joyard J. (1996). Envelope membranes from spinach chloroplasts are a site of metabolism of fatty acid hydroperoxides. Plant Physiol. 110, 445–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Block M. A., Dorne A. J., Joyard J., Douce R. (1983). Preparation and characterization of membrane fractions enriched in outer and inner envelope membranes from spinach chloroplasts. II. Biochemical characterization. J. Biol. Chem. 258, 13281–13286 [PubMed] [Google Scholar]
- Bolter B., Soll J., Hill K., Hemmler R., Wagner R. (1999). A rectifying ATP-regulated solute channel in the chloroplastic outer envelope from pea. EMBO J. 18, 5505–5516 10.1093/emboj/18.20.5505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bräutigam A., Hofmann-Benning S., Weber A. P. M. (2008). Comparative proteomics of chloroplast envelopes from C3 and C4 plants reveals specific adaptations of the plastid envelope to C4 photosynthesis and candidate proteins required for maintaining C4 metabolite fluxes. Plant Physiol. 148, 568–579 10.1104/pp.108.121012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bräutigam A., Weber A. P. M. (2009). Proteomic analysis of the proplastid envelope membrane provides novel insights into small molecule and protein transport across proplastid membranes. Mol. Plant 2, 1247–1261 10.1093/mp/ssp070 [DOI] [PubMed] [Google Scholar]
- Bräutigam A., Weber A. P. M. (2011). Do metabolite transport processes limit photosynthesis? Plant Physiol. 155, 43–48 10.1104/pp.110.159442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browse J., Mcconn M., James D., Jr., Miquel M. (1993). Mutants of Arabidopsis deficient in the synthesis of alpha-linoleate. Biochemical and genetic characterization of the endoplasmic reticulum linoleoyl desaturase. J. Biol. Chem. 268, 16345–16351 [PubMed] [Google Scholar]
- Browse J., Warwick N., Somerville C. R., Slack C. R. (1986). Fluxes through the prokaryotic and eukaryotic pathways of lipid synthesis in the “16:3” plant Arabidopsis thaliana. Biochem. J. 235, 25–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K. H., Chen X. J., Schnell D. J. (2000). Initial binding of preproteins involving the Toc159 receptor can be bypassed during protein import into chloroplasts. Plant Physiol. 122, 813–822 10.1104/pp.122.3.813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y. L., Asano T., Fujiwara M. T., Yoshida S., Machida Y., Yoshioka Y. (2009). Plant cells without detectable plastids are generated in the crumpled leaf mutant of Arabidopsis thaliana. Plant Cell Physiol. 50, 956–969 10.1093/pcp/pcp107 [DOI] [PubMed] [Google Scholar]
- Cline K., Keegstra K. (1983). Galactosyltransferases involved in galactolipid biosynthesis are located in the outer membrane of pea chloroplast envelopes. Plant Physiol. 71, 366–372 10.1104/pp.71.2.366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curien G., Bastien O., Robert-Genthon M., Cornish-Bowden A., Cardenas M. L., Dumas R. (2009). Understanding the regulation of aspartate metabolism using a model based on measured kinetic parameters. Mol. Syst. Biol. 5, 271. 10.1038/msb.2009.29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curien G., Ravanel S., Robert M., Dumas R. (2005). Identification of six novel allosteric effectors of Arabidopsis thaliana aspartate kinase-homoserine dehydrogenase isoforms. Physiological context sets the specificity. J. Biol. Chem. 280, 41178–41183 10.1074/jbc.M509324200 [DOI] [PubMed] [Google Scholar]
- Dhanoa P. K., Richardson L. G. L., Smith M. D., Gidda S. K., Henderson M. P. A., Andrews D. W., Mullen R. T. (2010). Distinct pathways mediate the sorting of tail-anchored proteins to the plastid outer envelope. PLoS ONE 5, e10098. 10.1371/journal.pone.0010098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doremus H. D., Jagendorf A. T. (1985). Subcellular localization of the pathway of de novo pyrimidine nucleotide biosynthesis in pea leaves. Plant Physiol. 79, 856–861 10.1104/pp.79.3.856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dörmann P., Balbo I., Benning C. (1999). Arabidopsis galactolipid biosynthesis and lipid trafficking mediated by DGD1. Science 284, 2181–2184 10.1126/science.284.5423.2181 [DOI] [PubMed] [Google Scholar]
- Dörmann P., Hoffmann-Benning S., Balbo I., Benning C. (1995). Isolation and characterization of an Arabidopsis mutant deficient in the thylakoid lipid digalactosyldiacylglycerol. Plant Cell 7, 1801–1810 10.1105/tpc.7.11.1801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drea S. C., Lao N. T., Wolfe K. H., Kavanagh T. A. (2006). Gene duplication, exon gain and neofunctionalization of OEP16-related genes in land plants. Plant J. 46, 723–735 10.1111/j.1365-313X.2006.02741.x [DOI] [PubMed] [Google Scholar]
- Duy D., Soll J., Philippar K. (2007). Solute channels of the outer membrane: from bacteria to chloroplasts. Biol. Chem. 388, 879–889 10.1515/BC.2007.120 [DOI] [PubMed] [Google Scholar]
- Dyall S. D., Brown M. T., Johnson P. J. (2004). Ancient invasions: from endosymbionts to organelles. Science 304, 253–257 10.1126/science.1094884 [DOI] [PubMed] [Google Scholar]
- El-Shami M., El-Kafafi S., Falconet D., Lerbs-Mache S. (2002). Cell cycle-dependent modulation of FtsZ expression in synchronized tobacco BY2 cells. Mol. Genet. Genomics 267, 254–261 10.1007/s00438-002-0660-y [DOI] [PubMed] [Google Scholar]
- Facchinelli F., Weber A. P. M. (2011). The metabolite transporters of the plastid envelope: an update. Front. Plant Sci. 2:50. 10.3389/fpls.2011.00050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferro M., Brugiere S., Salvi D., Seigneurin-Berny D., Court M., Moyet L., Ramus C., Miras S., Mellal M., Le Gall S., Kieffer-Jaquinod S., Bruley C., Garin J., Joyard J., Masselon C., Rolland N. (2010). AT_CHLORO, a comprehensive chloroplast proteome database with subplastidial localization and curated information on envelope proteins. Mol. Cell. Proteomics 9, 1063–1084 10.1074/mcp.M900325-MCP200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer K. (2011). The import and export business in plastids: transport processes across the inner envelope membrane. Plant Physiol. 155, 1511–1519 10.1104/pp.110.170241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer K., Weber A., Arbinger B., Brink S., Eckerskorn C., Flugge U. I. (1994). The 24 kDa outer envelope membrane protein from spinach chloroplasts: molecular cloning, in vivo expression and import pathway of a protein with unusual properties. Plant Mol. Biol. 25, 167–177 10.1007/BF00023235 [DOI] [PubMed] [Google Scholar]
- Florin E. L., Moy V. T., Gaub H. E. (1994). Adhesion forces between individual ligand-receptor pairs. Science 264, 415–417 10.1126/science.8153628 [DOI] [PubMed] [Google Scholar]
- Flügge U. I. (2000). Transport in and out of plastids: does the outer envelope membrane control the flow? Trends Plant Sci. 5, 135–137 10.1016/S1360-1385(00)01578-8 [DOI] [PubMed] [Google Scholar]
- Fourrier N., Bedard J., Lopez-Juez E., Barbrook A., Bowyer J., Jarvis P., Warren G., Thorlby G. (2008). A role for SENSITIVE TO FREEZING2 in protecting chloroplasts against freeze-induced damage in Arabidopsis. Plant J. 55, 734–745 10.1111/j.1365-313X.2008.03549.x [DOI] [PubMed] [Google Scholar]
- Frank W., Baar K. M., Qudeimat E., Woriedh M., Alawady A., Ratnadewi D., Gremillon L., Grimm B., Reski R. (2007). A mitochondrial protein homologous to the mammalian peripheral-type benzodiazepine receptor is essential for stress adaptation in plants. Plant J. 51, 1004–1018 10.1111/j.1365-313X.2007.03198.x [DOI] [PubMed] [Google Scholar]
- Friedman J. R., Lackner L. L., West M., Dibenedetto J. R., Nunnari J., Voeltz G. K. (2011). ER tubules mark sites of mitochondrial division. Science 334, 358–362 10.1126/science.1207385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Froehlich J. E., Benning C., Dörmann P. (2001a). The digalactosyldiacylglycerol (DGDG) synthase DGD1 is inserted into the outer envelope membrane of chloroplasts in a manner independent of the general import pathway and does not depend on direct interaction with monogalactosyldiacylglycerol synthase for DGDG biosynthesis. J. Biol. Chem. 276, 31806–31812 10.1074/jbc.M104652200 [DOI] [PubMed] [Google Scholar]
- Froehlich J. E., Itoh A., Howe G. A. (2001b). Tomato allene oxide synthase and fatty acid hydroperoxide lyase, two cytochrome P450s involved in oxylipin metabolism, are targeted to different membranes of chloroplast envelope. Plant Physiol. 125, 306–317 10.1104/pp.125.1.306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao H. B., Kadirjan-Kalbach D., Froehlich J. E., Osteryoung K. W. (2003). ARC5, a cytosolic dynamin-like protein from plants, is part of the chloroplast division machinery. Proc. Natl. Acad. Sci. U.S.A. 100, 4328–4333 10.1073/pnas.2334390100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia O., Bouige P., Forestier C., Dassa E. (2004). Inventory and comparative analysis of rice and Arabidopsis ATP-binding cassette (ABC) systems. J. Mol. Biol. 343, 249–265 10.1016/j.jmb.2004.07.093 [DOI] [PubMed] [Google Scholar]
- Gentle I., Gabriel K., Beech P., Waller R., Lithgow T. (2004). The Omp85 family of proteins is essential for outer membrane biogenesis in mitochondria and bacteria. J. Cell Biol. 164, 19–24 10.1083/jcb.200310092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giermann N., Schroder M., Ritter T., Zrenner R. (2002). Molecular analysis of de novo pyrimidine synthesis in solanaceous species. Plant Mol. Biol. 50, 393–403 10.1023/A:1019854531254 [DOI] [PubMed] [Google Scholar]
- Giroud C., Gerber A., Eichenberger W. (1988). Lipids of Chlamydomonas reinhardtii. Analysis of molecular species and intracellular site(s) of biosynthesis. Plant Cell Physiol. 29, 587–595 [Google Scholar]
- Glynn J. M., Froehlich J. E., Osteryoung K. W. (2008). Arabidopsis ARC6 coordinates the division machineries of the inner and outer chloroplast membranes through interaction with pdv2 in the intermembrane space. Plant Cell Online 20, 2460–2470 10.1105/tpc.108.061440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glynn J. M., Yang Y., Vitha S., Schmitz A. J., Hemmes M., Miyagishima S. Y., Osteryoung K. W. (2009). PARC6, a novel chloroplast division factor, influences FtsZ assembly and is required for recruitment of PDV1 during chloroplast division in Arabidopsis. Plant J. 59, 700–711 10.1111/j.1365-313X.2009.03905.x [DOI] [PubMed] [Google Scholar]
- Goetze T. A., Philippar K., Ilkavets I., Soll J., Wagner R. (2006). OEP37 is a new member of the chloroplast outer membrane Ion channels. J. Biol. Chem. 281, 17989–17998 10.1074/jbc.M600700200 [DOI] [PubMed] [Google Scholar]
- Gould S. B., Waller R. F., Mcfadden G. I. (2008). Plastid evolution. Annu. Rev. Plant Biol. 59, 491–517 10.1146/annurev.arplant.59.032607.092915 [DOI] [PubMed] [Google Scholar]
- Gray J. C., Hansen M. R., Shaw D. J., Graham K., Dale R., Smallman P., Natesan S. K. A., Newell C. A. (2011). Plastid stromules are induced by stress treatments acting through abscisic acid. Plant J. 10.1111/j.1365-313X.2011.04800.x [DOI] [PubMed] [Google Scholar]
- Gray J. C., Sullivan J. A., Hibberd J. M., Hansen M. R. (2001). Stromules: mobile protrusions and interconnections between plastids. Plant Biol. 3, 223–233 10.1055/s-2001-15204 [DOI] [Google Scholar]
- Guillaumot D., Guillon S., Morsomme P., Batoko H. (2009). ABA, porphyrins and plant TSPO-related protein. Plant Signal. Behav. 4, 1087–1090 10.4161/psb.4.11.9796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunning B. E. S. (2005). Plastid stromules: video microscopy of their outgrowth, retraction, tensioning, anchoring, branching, bridging, and tip-shedding. Protoplasma 225, 33–42 10.1007/s00709-004-0073-3 [DOI] [PubMed] [Google Scholar]
- Guo J., Zhang Z., Bi Y., Yang W., Xu Y., Zhang L. (2005). Decreased stability of photosystem I in dgd1 mutant of Arabidopsis thaliana. FEBS Lett. 579, 3619–3624 10.1016/j.febslet.2005.05.049 [DOI] [PubMed] [Google Scholar]
- Gutensohn M., Schulz B., Nicolay P., Flugge U. I. (2000). Functional analysis of the two Arabidopsis homologues of Toc34, a component of the chloroplast protein import apparatus. Plant J. 23, 771–783 10.1046/j.1365-313x.2000.00849.x [DOI] [PubMed] [Google Scholar]
- Hanson M. R., Sattarzadeh A. (2008). Dynamic morphology of plastids and stromules in angiosperm plants. Plant Cell Environ. 31, 646–657 10.1111/j.1365-3040.2007.01768.x [DOI] [PubMed] [Google Scholar]
- Hanson M. R., Sattarzadeh A. (2011). Stromules: recent insights into a long neglected feature of plastid morphology and function. Plant Physiol. 155, 1486–1492 10.1104/pp.110.170852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Härtel H., Dörmann P., Benning C. (2000). DGD1-independent biosynthesis of extraplastidic galactolipids after phosphate deprivation in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 97, 10649–10654 10.1073/pnas.180320497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto H., Possingham J. V. (1989). Division and DNA distribution in ribosome-deficient plastids of the barley mutant “albostrians”. Protoplasma 149, 20–23 10.1007/BF01623978 [DOI] [Google Scholar]
- Haswell E. S., Meyerowitz E. M. (2006). MscS-like proteins control plastid size and shape in Arabidopsis thaliana. Curr. Biol. 16, 1–11 10.1016/j.cub.2005.11.044 [DOI] [PubMed] [Google Scholar]
- Hatta T., Bullivan S., Matthews R. E. (1973). Fine-structure of vesicles induced in chloroplasts of chinese cabbage leaves by infection with turnip yellow mosaic-virus. J. Gen. Virol. 20, 37–50 10.1099/0022-1317-20-1-37 [DOI] [PubMed] [Google Scholar]
- Heemskerk J. W., Bogemann G., Wintermans J. (1983). Turnover of galactolipids incorporated into chloroplast envelopes; an assay for galactolipid-galactolipid galactosyltransferase. Biochim. Biophys. Acta 754, 181–189 [Google Scholar]
- Heemskerk J. W., Storz T., Schmidt R. R., Heinz E. (1990). Biosynthesis of digalactosyldiacylglycerol in plastids from 16:3 and 18:3 plants. Plant Physiol. 93, 1286–1294 10.1104/pp.93.4.1286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heemskerk J. W. M., Wintermans J. F. G. M., Joyard J., Block M. A., Dorne A.-J., Douce R. (1986). Localization of galactolipid: galactolipid galactosyltransferase and acyltransferase in outer envelope membrane of spinach chloroplasts. Biochim. Biophys. Acta 877, 281–289 [Google Scholar]
- Helliwell C. A., Sullivan J. A., Mould R. M., Gray J. C., Peacock W. J., Dennis E. S. (2001). A plastid envelope location of Arabidopsis ent-kaurene oxidase links the plastid and endoplasmic reticulum steps of the gibberellin biosynthesis pathway. Plant J. 28, 201–208 10.1046/j.1365-313X.2001.01150.x [DOI] [PubMed] [Google Scholar]
- Hemmler R., Becker T., Schleiff E., Bolter B., Stahl T., Soll J., Gotze T. A., Braams S., Wagner R. (2006). Molecular properties of Oep21, an ATP-regulated anion-selective solute channel from the outer chloroplast membrane. J. Biol. Chem. 281, 12020–12029 10.1074/jbc.M513586200 [DOI] [PubMed] [Google Scholar]
- Higgins C. F. (1992). ABC transporters: from microorganisms to man. Annu. Rev. Cell Biol. 8, 67–113 10.1146/annurev.cellbio.8.1.67 [DOI] [PubMed] [Google Scholar]
- Hiltbrunner A., Grunig K., Alvarez-Huerta M., Infanger S., Bauer J., Kessler F. (2004). AtToc90, a new GTP-binding component of the Arabidopsis chloroplast protein import machinery. Plant Mol. Biol. 54, 427–440 10.1023/B:PLAN.0000036374.92546.51 [DOI] [PubMed] [Google Scholar]
- Hinnah S. C., Hill K., Wagner R., Schlicher T., Soll J. (1997). Reconstitution of a chloroplast protein import channel. EMBO J. 16, 7351–7360 10.1093/emboj/16.24.7351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnah S. C., Wagner R., Sveshnikova N., Harrer R., Soll J. (2002). The chloroplast protein import channel Toc75: pore properties and interaction with transit peptides. Biophys. J. 83, 899–911 10.1016/S0006-3495(02)75216-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holzinger A., Buchner O., Lutz C., Hanson M. R. (2007). Temperature-sensitive formation of chloroplast protrusions and stromules in mesophyll cells of Arabidopsis thaliana. Protoplasma 230, 23–30 10.1007/s00709-006-0222-y [DOI] [PubMed] [Google Scholar]
- Holzinger A., Kwok E. Y., Hanson M. R. (2008). Effects of arc3, arc5 and arc6 mutations on plastid morphology and stromule formation in green and nongreen tissues of Arabidopsis thaliana. Photochem. Photobiol. 84, 1324–1335 10.1111/j.1751-1097.2008.00437.x [DOI] [PubMed] [Google Scholar]
- Hölzl G., Witt S., Kelly A. A., Zahringer U., Warnecke D., Dörmannn P., Heinz E. (2006). Functional differences between galactolipids and glucolipids revealed in photosynthesis of higher plants. Proc. Natl. Acad. Sci. U.S.A. 103, 7512–7517 10.1073/pnas.0600525103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe G. A., Schilmiller A. L. (2002). Oxylipin metabolism in response to stress. Curr. Opin. Plant Biol. 5, 230–236 10.1016/S1369-5266(02)00250-9 [DOI] [PubMed] [Google Scholar]
- Huang J., Taylor J. P., Chen J.-G., Uhrig J. F., Schnell D. J., Nakagawa T., Korth K. L., Jones A. M. (2006). The plastid protein THYLAKOID FORMATION1 and the plasma membrane G-protein GPA1 interact in a novel sugar-signaling mechanism in Arabidopsis. Plant Cell Online 18, 1226–1238 10.1105/tpc.106.045013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W., Ling Q., Bedard J., Lilley K., Jarvis P. (2011). In vivo analyses of the roles of essential Omp85-related proteins in the chloroplast outer envelope membrane. Plant Physiol. 157, 147–159 10.1104/pp.111.181891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inaba T., Schnell D. J. (2008). Protein trafficking to plastids: one theme, many variations. Biochem. J. 413, 15–28 10.1042/BJ20080490 [DOI] [PubMed] [Google Scholar]
- Infanger S., Bischof S., Hiltbrunner A., Agne B., Baginsky S., Kessler F. (2011). The chloroplast import receptor Toc90 partially restores the accumulation of Toc159 client proteins in the Arabidopsis thaliana ppi2 mutant. Mol. Plant 4, 252–263 10.1093/mp/ssq071 [DOI] [PubMed] [Google Scholar]
- Inoue K., Keegstra K. (2003). A polyglycine stretch is necessary for proper targeting of the protein translocation channel precursor to the outer envelope membrane of chloroplasts. Plant J. 34, 661–669 10.1046/j.1365-313X.2003.01755.x [DOI] [PubMed] [Google Scholar]
- Ishida H., Yoshimoto K. (2008). Chloroplasts are partially mobilized to the vacuole by autophagy. Autophagy 4, 961–962 [DOI] [PubMed] [Google Scholar]
- Ivanova Y., Smith M. D., Chen K. H., Schnell D. J. (2004). Members of the Toc159 import receptor family represent distinct pathways for protein targeting to plastids. Mol. Biol. Cell 15, 3379–3392 10.1091/mbc.E03-12-0923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izumi M., Wada S., Makino A., Ishida H. (2010). The autophagic degradation of chloroplasts via rubisco-containing bodies is specifically linked to leaf carbon status but not nitrogen status in Arabidopsis. Plant Physiol. 154, 1196–1209 10.1104/pp.110.158519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson-Constan D., Keegstra K. (2001). Arabidopsis genes encoding components of the chloroplastic protein import apparatus. Plant Physiol. 125, 1567–1576 10.1104/pp.125.4.1567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis P., Chen L. J., Li H. M., Pete C. A., Fankhauser C., Chory J. (1998). An Arabidopsis mutant defective in the plastid general protein import apparatus. Science 282, 100–103 10.1126/science.282.5386.100 [DOI] [PubMed] [Google Scholar]
- Jarvis P., Dörmann P., Peto C. A., Lutes J., Benning C., Chory J. (2000). Galactolipid deficiency and abnormal chloroplast development in the Arabidopsis MGD synthase 1 mutant. Proc. Natl. Acad. Sci. U.S.A. 97, 8175–8179 10.1073/pnas.100132197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston M. L., Luethy M. H., Miernyk J. A., Randall D. D. (1997). Cloning and molecular analyses of the Arabidopsis thaliana plastid pyruvate dehydrogenase subunits. Biochim. Biophys. Acta 1321, 200–206 10.1016/S0005-2728(97)00059-5 [DOI] [PubMed] [Google Scholar]
- Jouhet J., Gray J. C. (2009). Interaction of actin and the chloroplast protein import apparatus. J. Biol. Chem. 284, 19132–19141 10.1074/jbc.M109.012831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jouhet J., Marechal E., Baldan B., Bligny R., Joyard J., Block M. A. (2004). Phosphate deprivation induces transfer of DGDG galactolipid from chloroplast to mitochondria. J. Cell Biol. 167, 863–874 10.1083/jcb.200407022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyard J., Ferro M., Masselon C., Seigneurin-Berny D., Salvi D., Garin J., Rolland N. (2010). Chloroplast proteomics highlights the subcellular compartmentation of lipid metabolism. Prog. Lipid Res. 49, 128–158 10.1016/j.plipres.2009.10.003 [DOI] [PubMed] [Google Scholar]
- Kadota A., Yamada N., Suetsugu N., Hirose M., Saito C., Shoda K., Ichikawa S., Kagawa T., Nakano A., Wada M. (2009). Short actin-based mechanism for light-directed chloroplast movement in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 106, 13106–13111 10.1073/pnas.0906250106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko K., Yamada C., Yanagida A., Koshu T., Umezawa Y., Itoh K., Hori H., Mitsui T. (2010). Differential localizations and functions of rice nucleotide pyrophosphatase/phosphodiesterase isozymes 1 and 3. Plant Biotechnol. 28, 69–76 [Google Scholar]
- Kaplan F., Kopka J., Sung D. Y., Zhao W., Popp M., Porat R., Guy C. L. (2007). Transcript and metabolite profiling during cold acclimation of Arabidopsis reveals an intricate relationship of cold-regulated gene expression with modifications in metabolite content. Plant J. 50, 967–981 10.1111/j.1365-313X.2007.03100.x [DOI] [PubMed] [Google Scholar]
- Kelly A. A., Dörmann P. (2002). DGD2, an Arabidopsis gene encoding a UDP-galactose-dependent digalactosyldiacylglycerol synthase is expressed during growth under phosphate-limiting conditions. J. Biol. Chem. 277, 1166–1173 10.1074/jbc.M110066200 [DOI] [PubMed] [Google Scholar]
- Kim D. H., Xu Z. Y., Na Y. J., Yoo Y. J., Lee J., Sohn E. J., Hwang I. (2011). Small heat shock protein Hsp17.8 functions as an AKR2A cofactor in the targeting of chloroplast outer membrane proteins in Arabidopsis. Plant Physiol. 157, 132–146 10.1104/pp.111.178681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishimoto K., Matsui K., Ozawa R., Takabayashi J. (2008). Direct fungicidal activities of C6-aldehydes are important constituents for defense responses in Arabidopsis against Botrytis cinerea. Phytochemistry 69, 2127–2132 10.1016/j.phytochem.2008.04.023 [DOI] [PubMed] [Google Scholar]
- Köhler R. H., Hanson M. R. (2000). Plastid tubules of higher plants are tissue-specific and developmentally regulated. J. Cell Sci. 113, 81–89 [DOI] [PubMed] [Google Scholar]
- Köhler R. H., Schwille P., Webb W. W., Hanson M. R. (2000). Active protein transport through plastid tubules: velocity quantified by fluorescence correlation spectroscopy. J. Cell Sci. 113(Pt 22), 3921–3930 [DOI] [PubMed] [Google Scholar]
- Konishi T., Shinohara K., Yamada K., Sasaki Y. (1996). Acetyl-CoA carboxylase in higher plants: most plants other than gramineae have both the prokaryotic and the eukaryotic forms of this enzyme. Plant Cell Physiol. 37, 117–122 [DOI] [PubMed] [Google Scholar]
- Koo A. J., Ohlrogge J. B., Pollard M. (2004). On the export of fatty acids from the chloroplast. J. Biol. Chem. 279, 16101–16110 10.1074/jbc.M311305200 [DOI] [PubMed] [Google Scholar]
- Koo A. J. K., Ohlrogge J. B. (2002). The predicted candidates of Arabidopsis plastid inner envelope membrane proteins and their expression profiles. Plant Physiol. 130, 823–836 10.1104/pp.008052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouranov A., Chen X. J., Fuks B., Schnell D. J. (1998). Tic20 and Tic22 are new components of the protein import apparatus at the chloroplast inner envelope membrane. J. Cell Biol. 143, 991–1002 10.1083/jcb.143.4.991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouranov A., Wang H., Schnell D. J. (1999). Tic22 is targeted to the intermembrane space of chloroplasts by a novel pathway. J. Biol. Chem. 274, 25181–25186 10.1074/jbc.274.35.25181 [DOI] [PubMed] [Google Scholar]
- Kovacs-Bogdan E., Soll J., Bolter B. (2010). Protein import into chloroplasts: the Tic complex and its regulation. Biochim. Biophys. Acta 1803, 740–747 10.1016/j.bbamcr.2010.01.015 [DOI] [PubMed] [Google Scholar]
- Kubis S., Patel R., Combe J., Bedard J., Kovacheva S., Lilley K., Biehl A., Leister D., Rios G., Koncz C., Jarvis P. (2004). Functional specialization amongst the Arabidopsis Toc159 family of chloroplast protein import receptors. Plant Cell 16, 2059–2077 10.1105/tpc.104.023309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunst L., Browse J., Somerville C. (1988). Altered regulation of lipid biosynthesis in a mutant of Arabidopsis deficient in chloroplast glycerol-3-phosphate acyltransferase activity. Proc. Natl. Acad. Sci. U.S.A. 85, 4143–4147 10.1073/pnas.85.12.4143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunz H. H., Scharnewski M., Feussner K., Feussner I., Flugge U. I., Fulda M., Gierth M. (2009). The ABC transporter PXA1 and peroxisomal beta-oxidation are vital for metabolism in mature leaves of Arabidopsis during extended darkness. Plant Cell 21, 2733–2749 10.1105/tpc.108.064857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroiwa T., Kuroiwa H., Sakai A., Takahashi H., Toda K., Itoh R. (1998). The division apparatus of plastids and mitochondria. Int. Rev. Cytol. 181, 1–41 10.1016/S0074-7696(08)60415-5 [DOI] [PubMed] [Google Scholar]
- Kusuhara H., Sugiyama Y. (2007). ATP-binding cassette, subfamily G (ABCG family). Pflugers Arch. 453, 735–744 10.1007/s00424-006-0134-x [DOI] [PubMed] [Google Scholar]
- Kwok E. Y., Hanson M. R. (2003). Microfilaments and microtubules control the morphology and movement of non-green plastids and stromules in Nicotiana tabacum. Plant J. 35, 16–26 10.1046/j.1365-313X.2003.01777.x [DOI] [PubMed] [Google Scholar]
- Kwok E. Y., Hanson M. R. (2004). In vivo analysis of interactions between GFP-labeled microfilaments and plastid stromules. BMC Plant Biol. 4, 2. 10.1186/1471-2229-4-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y. J., Kim D. H., Kim Y. W., Hwang I. (2001). Identification of a signal that distinguishes between the chloroplast outer envelope membrane and the endomembrane system in vivo. Plant Cell 13, 2175–2190 10.1105/tpc.13.10.2175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Moore T., Keegstra K. (1991). Targeting of proteins to the outer envelope membrane uses a different pathway than transport into chloroplasts. Plant Cell Online 3, 709–717 10.1105/tpc.3.11.1167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liguori L., Blesneac I., Madern D., Vivaudou M., Lenormand J.-L. (2010). Single-step production of functional OEP24 proteoliposomes. Protein Expr. Purif. 69, 106–111 10.1016/j.pep.2009.07.004 [DOI] [PubMed] [Google Scholar]
- Linka N., Weber A. P. M. (2009). Intracellular metabolite transporters in plants. Mol. Plant 3, 21–53 10.1093/mp/ssp108 [DOI] [PubMed] [Google Scholar]
- Linke D., Frank J., Pope M. S., Soll J., Ilkavets I., Fromme P., Burstein E. A., Reshetnyak Y. K., Emelyanenko V. I. (2004). Folding kinetics and structure of OEP16. Biophys. J. 86, 1479–1487 10.1016/S0006-3495(04)74216-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu B., Benning C. (2009). A 25-amino acid sequence of the Arabidopsis TGD2 protein is sufficient for specific binding of phosphatidic acid. J. Biol. Chem. 284, 17420–17427 10.1074/jbc.M109.008821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu B. B., Xu C. C., Awai K., Jones A. D., Benning C. (2007). A small ATPase protein of Arabidopsis, TGD3, involved in chloroplast lipid import. J. Biol. Chem. 282, 35945–35953 10.1074/jbc.M611540200 [DOI] [PubMed] [Google Scholar]
- Mackiewicz P., Bodyl A. (2010). A hypothesis for import of the nuclear-encoded PsaE protein of Paulinella chromatophora (Cercozoa, Rhizaria) into its cyanobacterial endosymbonts/plastids via the endomembrane system. J. Phycol. 46, 847–859 10.1111/j.1529-8817.2010.00876.x [DOI] [Google Scholar]
- Marechal E., Block M. A., Joyard J., Douce R. (1994). Comparison of the kinetic properties of MGDG synthase in mixed micelles and in envelope membranes from spinach chloroplast. FEBS Lett. 352, 307–310 10.1016/0014-5793(94)00978-3 [DOI] [PubMed] [Google Scholar]
- Mcfarlane H. E., Shin J. J., Bird D. A., Samuels A. L. (2010). Arabidopsis ABCG transporters, which are required for export of diverse cuticular lipids, dimerize in different combinations. Plant Cell 22, 3066–3075 10.1105/tpc.110.077974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miege C., Marechal E., Shimojima M., Awai K., Block M. A., Ohta H., Takamiya K., Douce R., Joyard J. (1999). Biochemical and topological properties of type A MGDG synthase, a spinach chloroplast envelope enzyme catalyzing the synthesis of both prokaryotic and eukaryotic MGDG. Eur. J. Biochem. 265, 990–1001 10.1046/j.1432-1327.1999.00801.x [DOI] [PubMed] [Google Scholar]
- Mijaljica D., Prescott M., Devenish R. J. (2007). Different fates of mitochondria: alternative ways for degradation? Autophagy 3, 4–9 [DOI] [PubMed] [Google Scholar]
- Mills W. R. (1980). Photosynthetic formation of the aspartate family of amino acids in isolated chloroplasts. Plant Physiol. 65, 1166–1172 10.1104/pp.65.6.1166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills W. R., Wilson K. G. (1978). Amino acid biosynthesis in isolated pea chloroplasts: metabolism of labeled aspartate and sulfate. FEBS Lett. 92, 129–132 10.1016/0014-5793(78)80737-6 [DOI] [Google Scholar]
- Miras S., Salvi D., Ferro M., Grunwald D., Garin J., Joyard J., Rolland N. (2002). Non-canonical transit peptide for import into the chloroplast. J. Biol. Chem. 277, 47770–47778 10.1074/jbc.M207477200 [DOI] [PubMed] [Google Scholar]
- Miras S., Salvi D., Piette L., Seigneurin-Berny D., Grunwald D., Reinbothe C., Joyard J., Reinbothe S., Rolland N. (2007). Toc159- and Toc75-independent import of a transit sequence-less precursor into the inner envelope of chloroplasts. J. Biol. Chem. 282, 29482–29492 10.1074/jbc.M611112200 [DOI] [PubMed] [Google Scholar]
- Miyagishima S. Y. (2005). Origin and evolution of the chloroplast division machinery. J. Plant Res. 118, 295–306 10.1007/s10265-005-0226-2 [DOI] [PubMed] [Google Scholar]
- Miyagishima S. Y., Froehlich J. E., Osteryoung K. W. (2006). PDV1 and PDV2 mediate recruitment of the dynamin-related protein ARC5 to the plastid division site. Plant Cell 18, 2517–2530 10.1105/tpc.106.045484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moellering E. R., Muthan B., Benning C. (2010). Freezing tolerance in plants requires lipid remodeling at the outer chloroplast membrane. Science 330, 226–228 10.1126/science.1191803 [DOI] [PubMed] [Google Scholar]
- Murcha M. W., Elhafez D., Lister R., Tonti-Filippini J., Baumgartner M., Philippar K., Carrie C., Mokranjac D., Soll J., Whelan J. (2007). Characterization of the preprotein and amino acid transporter gene family in Arabidopsis. Plant Physiol. 143, 199–212 10.1104/pp.106.090688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nada A., Soll J. (2004). Inner envelope protein 32 is imported into chloroplasts by a novel pathway. J. Cell Sci. 117, 3975–3982 10.1242/jcs.01265 [DOI] [PubMed] [Google Scholar]
- Nanjo Y., Oka H., Ikarashi N., Kaneko K., Kitajima A., Mitsui T., Munoz F. J., Rodriguez-Lopez M., Baroja-Fernandez E., Pozueta-Romero J. (2006). Rice plastidial N-glycosylated nucleotide pyrophosphatase/phosphodiesterase is transported from the ER-Golgi to the chloroplast through the secretory pathway. Plant Cell 18, 2582–2592 10.1105/tpc.105.039891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natesan S. K. A., Sullivan J. A., Gray J. C. (2009). Myosin XI is required for actin-associated movement of plastid stromules. Mol. Plant 2, 1262–1272 10.1093/mp/ssp078 [DOI] [PubMed] [Google Scholar]
- Nixon P. J., Michoux F., Yu J., Boehm M., Komenda J. (2010). Recent advances in understanding the assembly and repair of photosystem II. Ann. Bot. 106, 1–16 10.1093/aob/mcq059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oelze M. L., Kandlbinder A., Dietz K. J. (2008). Redox regulation and overreduction control in the photosynthesizing cell: complexity in redox regulatory networks. Biochim. Biophys. Acta 1780, 1261–1272 10.1016/j.bbagen.2008.03.015 [DOI] [PubMed] [Google Scholar]
- Oikawa K., Kasahara M., Kiyosue T., Kagawa T., Suetsugu N., Takahashi F., Kanegae T., Niwa Y., Kadota A., Wada M. (2003). CHLOROPLAST UNUSUAL POSITIONING1 is essential for proper chloroplast positioning. Plant Cell Online 15, 2805–2815 10.1105/tpc.016428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oikawa K., Yamasato A., Kong S. G., Kasahara M., Nakai M., Takahashi F., Ogura Y., Kagawa T., Wada M. (2008). Chloroplast outer envelope protein CHUP1 is essential for chloroplast anchorage to the plasma membrane and chloroplast movement. Plant Physiol. 148, 829–842 10.1104/pp.108.123075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okanenko O. A. (2002). Plant sulfolipids. Ukr. Biokhim. Zh. 74, 27–36 [PubMed] [Google Scholar]
- Okazaki K., Kabeya Y., Suzuki K., Mori T., Ichikawa T., Matsui M., Nakanishi H., Miyagishima S. (2009). The PLASTID DIVISION1 and 2 components of the chloroplast division machinery determine the rate of chloroplast division in land plant cell differentiation. Plant Cell 21, 1769–1780 10.1105/tpc.108.063925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osteryoung K. W., Stokes K. D., Rutherford S. M., Percival A. L., Lee W. Y. (1998). Chloroplast division in higher plants requires members of two functionally divergent gene families with homology to bacterial ftsZ. Plant Cell 10, 1991–2004 10.1105/tpc.10.12.1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park H., Eggink L. L., Roberson R. W., Hoober J. K. (1999). Transfer of proteins from the chloroplast to vacuoles in Chlamydomonas reinhardtii (Chlorophyta): a pathway for degradation. J. Phycol. 35, 528–538 10.1046/j.1529-8817.1999.3530528.x [DOI] [Google Scholar]
- Patel R., Hsu S. C., Bedard J., Inoue K., Jarvis P. (2008). The Omp85-related chloroplast outer envelope protein OEP80 is essential for viability in Arabidopsis. Plant Physiol. 148, 235–245 10.1104/pp.108.122754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry S. E., Keegstra K. (1994). Envelope membrane-proteins that interact with chloroplastic precursor proteins. Plant Cell 6, 93–105 10.1105/tpc.6.1.93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippar K., Geis T., Ilkavets I., Oster U., Schwenkert S., Meurer J., Soll J. (2007). Chloroplast biogenesis: the use of mutants to study the etioplast-chloroplast transition. Proc. Natl. Acad. Sci. U.S.A. 104, 678–683 10.1073/pnas.0610062104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pohlmeyer K., Soll J., Grimm R., Hill K., Wagner R. (1998). A high-conductance solute channel in the chloroplastic outer envelope from pea. Plant Cell 10, 1207–1216 10.2307/3870722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pohlmeyer K., Soll J., Steinkamp T., Hinnah S., Wagner R. (1997). Isolation and characterization of an amino acid-selective channel protein present in the chloroplastic outer envelope membrane. Proc. Natl. Acad. Sci. U.S.A. 94, 9504–9509 10.1073/pnas.94.17.9504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard M., Ohlrogge J. (1999). Testing models of fatty acid transfer and lipid synthesis in spinach leaf using in vivo oxygen-18 labeling. Plant Physiol. 121, 1217–1226 10.1104/pp.121.4.1217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pudelski B., Kraus S., Soll J., Philippar K. (2010). The plant PRAT proteins – preprotein and amino acid transport in mitochondria and chloroplasts. Plant Biol. 12, 42–55 10.1111/j.1438-8677.2010.00357.x [DOI] [PubMed] [Google Scholar]
- Pyke K. A., Howells C. A. (2002). Plastid and stromule morphogenesis in tomato. Ann. Bot. 90, 559–566 10.1093/aob/mcf235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyke K. A., Leech R. M. (1992). Chloroplast division and expansion is radically altered by nuclear mutations in Arabidopsis thaliana. Plant Physiol. 99, 1005–1008 10.1104/pp.99.3.1005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qbadou S., Becker T., Bionda T., Reger K., Ruprecht M., Soll J., Schleiff E. (2007). Toc64 – a preprotein-receptor at the outer membrane with bipartide function. J. Mol. Biol. 367, 1330–1346 10.1016/j.jmb.2007.01.047 [DOI] [PubMed] [Google Scholar]
- Rassow J., Dekker P. J. T., Van Wilpe S., Meijer M., Soll J. (1999). The preprotein translocase of the mitochondrial inner membrane: function and evolution. J. Mol. Biol. 286, 105–120 10.1006/jmbi.1998.2455 [DOI] [PubMed] [Google Scholar]
- Raynaud C., Cassier-Chauvat C., Perennes C., Bergounioux C. (2004). An Arabidopsis homolog of the bacterial cell division inhibitor SulA is involved in plastid division. Plant Cell 16, 1801–1811 10.1105/tpc.022335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raynaud C., Perennes C., Reuzeau C., Catrice O., Brown S., Bergounioux C. (2005). Cell and plastid division are coordinated through the prereplication factor AtCDT1. Proc. Natl. Acad. Sci. U.S.A. 102, 8216–8221 10.1073/pnas.0502564102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reape T. J., Molony E. M., Mccabe P. F. (2008). Programmed cell death in plants: distinguishing between different modes. J. Exp. Bot. 59, 435–444 10.1093/jxb/erm258 [DOI] [PubMed] [Google Scholar]
- Reifarth F., Christen G., Seeliger A. G., Dörmann P., Benning C., Renger G. (1997). Modification of the water oxidizing complex in leaves of the dgd1 mutant of Arabidopsis thaliana deficient in the galactolipid digalactosyldiacylglycerol. Biochemistry 36, 11769–11776 10.1021/bi9709654 [DOI] [PubMed] [Google Scholar]
- Reiland S., Grossmann J., Baerenfaller K., Gehrig P., Nunes-Nesi A., Fernie A. R., Gruissem W., Baginsky S. (2011). Integrated proteome and metabolite analysis of the de-etiolation process in plastids from rice (Oryza sativa L.). Proteomics 11, 1751–1763 10.1002/pmic.201000703 [DOI] [PubMed] [Google Scholar]
- Reinbothe S., Quigley F., Springer A., Schemenewitz A., Reinbothe C. (2004). The outer plastid envelope protein Oep16: role as precursor translocase in import of protochlorophyllide oxidoreductase A. Proc. Natl. Acad. Sci. U.S.A. 101, 2203–2208 10.1073/pnas.0307284101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reski R. (2009). Challenges to our current view on chloroplasts. Biol. Chem. 390, 731–738 10.1515/BC.2009.089 [DOI] [PubMed] [Google Scholar]
- Reyes-Prieto A., Weber A. P. M., Bhattacharya D. (2007). The origin and establishment of the plastid in algae and plants. Annu. Rev. Genet. 41, 147–168 10.1146/annurev.genet.41.110306.130134 [DOI] [PubMed] [Google Scholar]
- Röhl T., Motzkus M., Soll J. R. (1999). The outer envelope protein OEP24 from pea chloroplasts can functionally replace the mitochondrial VDAC in yeast. FEBS Lett. 460, 491–494 10.1016/S0014-5793(99)01399-X [DOI] [PubMed] [Google Scholar]
- Rolland N., Ferro M., Seigneurin-Berny D., Garin J., Douce R., Joyard J. (2003). Proteomics of chloroplast envelope membranes. Photosynth. Res. 78, 205–230 10.1023/B:PRES.0000006891.12416.6c [DOI] [PubMed] [Google Scholar]
- Roston R., Gao J. P., Xu C. C., Benning C. (2011). Arabidopsis chloroplast lipid transport protein TGD2 disrupts membranes and is part of a large complex. Plant J. 66, 759–769 10.1111/j.1365-313X.2011.04536.x [DOI] [PubMed] [Google Scholar]
- Roughan P. G., Slack C. R. (1977). Long-chain acyl-coenzyme A synthetase activity of spinach chloroplasts is concentrated in the envelope. Biochem. J. 162, 457–459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sage T. L., Sage R. F. (2009). The functional anatomy of rice leaves: implications for refixation of photorespiratory CO2 and efforts to engineer C-4 photosynthesis into rice. Plant Cell Physiol. 50, 756–772 10.1093/pcp/pcp033 [DOI] [PubMed] [Google Scholar]
- Salomon M., Fischer K., Flugge U. I., Soll J. (1990). Sequence analysis and protein import studies of an outer chloroplast envelope polypeptide. Proc. Natl. Acad. Sci. U.S.A. 87, 5778–5782 10.1073/pnas.87.15.5778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sattarzadeh A., Krahmer J., Germain A. D., Hanson M. R. (2009). A myosin XI tail domain homologous to the yeast myosin vacuole-binding domain interacts with plastids and stromules in Nicotiana benthamiana. Mol. Plant 2, 1351–1358 10.1093/mp/ssp094 [DOI] [PubMed] [Google Scholar]
- Schattat M., Barton K., Baudisch B., Klosgen R. B., Mathur J. (2011). Plastid stromule branching coincides with contiguous endoplasmic reticulum dynamics. Plant Physiol. 155, 1667–1677 10.1104/pp.110.170480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schleiff E., Eichacker L. A., Eckart K., Becker T., Mirus O., Stahl T., Soll J. (2003a). Prediction of the plant beta-barrel proteome: a case study of the chloroplast outer envelope. Protein Sci. 12, 748–759 10.1110/ps.0237503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schleiff E., Jelic M., Soll J. (2003b). A GTP-driven motor moves proteins across the outer envelope of chloroplasts. Proc. Natl. Acad. Sci. U.S.A. 100, 4604–4609 10.1073/pnas.0730860100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnell D. J., Kessler F., Blobel G. (1994). Isolation of components of the chloroplast protein import machinery. Science 266, 1007–1012 10.1126/science.7973649 [DOI] [PubMed] [Google Scholar]
- Schnurr J. A., Shockey J. M., De Boer G. J., Browse J. A. (2002). Fatty acid export from the chloroplast. Molecular characterization of a major plastidial acyl-coenzyme A synthetase from Arabidopsis. Plant Physiol. 129, 1700–1709 10.1104/pp.003251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwenkert S., Soll J., Bolter B. (2010). Protein import into chloroplasts-how chaperones feature into the game. Biochim. Biophys. Acta 1808, 901–911 [DOI] [PubMed] [Google Scholar]
- Shaw D. J., Gray J. C. (2011). Visualisation of stromules in transgenic wheat expressing a plastid-targeted yellow fluorescent protein. Planta 233, 961–970 10.1007/s00425-011-1351-x [DOI] [PubMed] [Google Scholar]
- Shine W. E., Mancha M., Stumpf P. K. (1976). Fat metabolism in higher plants. The function of acyl thioesterases in the metabolism of acyl-coenzymes A and acyl-acyl carrier proteins. Arch. Biochem. Biophys. 172, 110–116 10.1016/0003-9861(76)90054-0 [DOI] [PubMed] [Google Scholar]
- Shiojiri K., Kishimoto K., Ozawa R., Kugimiya S., Urashimo S., Arimura G., Horiuchi J., Nishioka T., Matsui K., Takabayashi J. (2006). Changing green leaf volatile biosynthesis in plants: an approach for improving plant resistance against both herbivores and pathogens. Proc. Natl. Acad. Sci. U.S.A. 103, 16672–16676 10.1073/pnas.0607780103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shockey J. M., Fulda M. S., Browse J. A. (2002). Arabidopsis contains nine long-chain acyl-coenzyme A synthetase genes that participate in fatty acid and glycerolipid metabolism. Plant Physiol. 129, 1710–1722 10.1104/pp.003269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddique M. A., Grossmann J., Gruissem W., Baginsky S. (2006). Proteome analysis of bell pepper (Capsicum annuum L.) chromoplasts. Plant Cell Physiol. 47, 1663–1673 10.1093/pcp/pcl033 [DOI] [PubMed] [Google Scholar]
- Singh B. K., Shaner D. L. (1995). Biosynthesis of branched chain amino acids: from test tube to field. Plant Cell 7, 935–944 10.1105/tpc.7.7.935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soll J., Bolter B., Wagner R., Hinnah S. C. (2000). … response: the chloroplast outer envelope: a molecular sieve? Trends Plant Sci. 5, 137–138 10.1016/S1360-1385(00)01579-X [DOI] [PubMed] [Google Scholar]
- Soll J., Schleiff E. (2004). Protein import into chloroplasts. Nat. Rev. Mol. Cell Biol. 5, 198–208 10.1038/nrm1333 [DOI] [PubMed] [Google Scholar]
- Somerville C., Browse J. (1991). Plant lipids: metabolism, mutants, and membranes. Science 252, 80–87 10.1126/science.252.5002.80 [DOI] [PubMed] [Google Scholar]
- Stanga J. P., Boonsirichai K., Sedbrook J. C., Otegui M. S., Masson P. H. (2009). A role for the TOC complex in Arabidopsis root gravitropism. Plant Physiol. 149, 1896–1905 10.1104/pp.109.135301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner J. M., Yusa F., Pompe J. A., Loffelhardt W. (2005). Homologous protein import machineries in chloroplasts and cyanelles. Plant J. 44, 646–652 10.1111/j.1365-313X.2005.02559.x [DOI] [PubMed] [Google Scholar]
- Steinkamp T., Hill K., Hinnah S. C., Wagner R., Rohl T., Pohlmeyer K., Soll J. (2000). Identification of the pore-forming region of the outer chloroplast envelope protein OEP16. J. Biol. Chem. 275, 11758–11764 10.1074/jbc.275.16.11758 [DOI] [PubMed] [Google Scholar]
- Suetsugu N., Yamada N., Kagawa T., Yonekura H., Uyeda T. Q. P., Kadota A., Wada M. (2010). Two kinesin-like proteins mediate actin-based chloroplast movement in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 107, 8860–8865 10.1073/pnas.0912773107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiyama A., Shitan N., Sato S., Nakamura Y., Tabata S., Yazaki K. (2006). Genome-wide analysis of ATP-binding cassette (ABC) proteins in a model legume plant, Lotus japonicus: comparison with Arabidopsis ABC protein family. DNA Res. 13, 205–228 10.1093/dnares/dsl013 [DOI] [PubMed] [Google Scholar]
- Sun Q., Zybailov B., Majeran W., Friso G., Olinares P. D., Van Wijk K. J. (2009). PPDB, the plant proteomics database at Cornell. Nucleic Acids Res. 37, D969–D974 10.1093/nar/gkp493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X., Feng P., Xu X., Guo H., Ma J., Chi W., Lin R., Lu C., Zhang L. (2011). A chloroplast envelope-bound PHD transcription factor mediates chloroplast signals to the nucleus. Nat. Commun. 2, 477–487 10.1038/ncomms1532 [DOI] [PubMed] [Google Scholar]
- Sveshnikova N., Grimm R., Soll J., Schleiff E. (2000). Topology studies of the chloroplast protein import channel Toc75. Biol. Chem. 381, 687–693 10.1515/BC.2000.089 [DOI] [PubMed] [Google Scholar]
- Takahashi H., Kopriva S., Giordano M., Saito K., Hell R. (2011). Sulfur assimilation in photosynthetic organisms: molecular functions and regulations of transporters and assimilatory enzymes. Annu. Rev. Plant Biol. 62, 157–184 10.1146/annurev-arplant-042110-103921 [DOI] [PubMed] [Google Scholar]
- Thorlby G., Fourrier N., Warren G. (2004). The sensitive to freezing2 gene, required for freezing tolerance in Arabidopsis thaliana, encodes a beta-glucosidase. Plant Cell 16, 2192–2203 10.1105/tpc.104.024018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tietje C., Heinz E. (1998). Uridine-diphospho-sulfoquinovose: diacylglycerol sulfoquinovosyltransferase activity is concentrated in the inner membrane of chloroplast envelopes. Planta 206, 72–78 10.1007/s004250050375 [DOI] [Google Scholar]
- Tranel P. J., Froehlich J., Goyal A., Keegstra K. (1995). A component of the chloroplastic protein import apparatus is targeted to the outer envelope membrane via a novel pathway. EMBO J. 14, 2436–2446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tranel P. J., Keegstra K. (1996). A novel, bipartite transit peptide targets OEP75 to the outer membrane of the chloroplastic envelope. Plant Cell 8, 2093–2104 10.2307/3870415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trojan A., Gabrys H. (1996). Chloroplast distribution in Arabidopsis thaliana (L.) depends on light conditions during growth. Plant Physiol. 111, 419–425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu S.-L., Chen L.-J., Smith M. D., Su Y.-S., Schnell D. J., Li H.-M. (2004). Import pathways of chloroplast interior proteins and the outer-membrane protein OEP14 converge at Toc75. Plant Cell Online 16, 2078–2088 10.1105/tpc.104.023952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu S.-L., Li H.-M. (2000). Insertion of OEP14 into the outer envelope membrane is mediated by proteinaceous components of chloroplasts. Plant Cell Online 12, 1951–1960 10.1105/tpc.12.10.1951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyra H. M., Linka M., Weber A. P., Bhattacharya D. (2007). Host origin of plastid solute transporters in the first photosynthetic eukaryotes. Genome Biol. 8, R212. 10.1186/gb-2007-8-10-r212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchiyama Y., Shibata M., Koike M., Yoshimura K., Sasaki M. (2008). Autophagy-physiology and pathophysiology. Histochem. Cell Biol. 129, 407–420 10.1007/s00418-008-0406-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda H., Yokota E., Kutsuna N., Shimada T., Tamura K., Shimmen T., Hasezawa S., Dolja V. V., Hara-Nishimura I. (2010). Myosin-dependent endoplasmic reticulum motility and F-actin organization in plant cells. Proc. Natl. Acad. Sci. U.S.A. 107, 6894–6899 10.1073/pnas.1008136107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Besouw A., Wintermans J. F. (1978). Galactolipid formation in chloroplast envelopes. I. Evidence for two mechanisms in galactosylation. Biochim. Biophys. Acta 529, 44–53 [DOI] [PubMed] [Google Scholar]
- Van Doorn W. G., Woltering E. J. (2005). Many ways to exit? Cell death categories in plants. Trends Plant Sci. 10, 117–122 10.1016/j.tplants.2005.08.003 [DOI] [PubMed] [Google Scholar]
- Vancanneyt G., Sanz C., Farmaki T., Paneque M., Ortego F., Castanera P., Sanchez-Serrano J. J. (2001). Hydroperoxide lyase depletion in transgenic potato plants leads to an increase in aphid performance. Proc. Natl. Acad. Sci. U.S.A. 98, 8139–8144 10.1073/pnas.141079498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanhee C., Zapotoczny G., Masquelier D., Ghislain M., Batoko H. (2011). The Arabidopsis multistress regulator TSPO is a heme binding membrane protein and a potential scavenger of porphyrins via an autophagy-dependent degradation mechanism. Plant Cell 23, 785–805 10.1105/tpc.110.081570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villarejo A., Buren S., Larsson S., Dejardin A., Monne M., Rudhe C., Karlsson J., Jansson S., Lerouge P., Rolland N., Von Heijne G., Grebe M., Bako L., Samuelsson G. (2005). Evidence for a protein transported through the secretory pathway en route to the higher plant chloroplast. Nat. Cell Biol. 7, 1224–1231 10.1038/ncb1330 [DOI] [PubMed] [Google Scholar]
- Von Braun S. S., Schleiff E. (2008). The chloroplast outer membrane protein CHUP1 interacts with actin and profilin. Planta 227, 1151–1159 10.1007/s00425-007-0688-7 [DOI] [PubMed] [Google Scholar]
- Von Loeffelholz O., Kriechbaumer V., Ewan R. A., Jonczyk R., Lehmann S., Young J. C., Abell B. M. (2011). OEP61 is a chaperone receptor at the plastid outer envelope. Biochem. J. 438, 143–153 10.1042/BJ20110448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Von Zychlinski A., Kleffmann T., Krishnamurthy N., Sjolander K., Baginsky S., Gruissem W. (2005). Proteome analysis of the rice etioplast – metabolic and regulatory networks and novel protein functions. Mol. Cell. Proteomics 4, 1072–1084 10.1074/mcp.M500018-MCP200 [DOI] [PubMed] [Google Scholar]
- Wallas T. R., Smith M. D., Sanchez-Nieto S., Schnell D. J. (2003). The roles of Toc34 and Toc75 in targeting the Toc159 preprotein receptor to chloroplasts. J. Biol. Chem. 278, 44289–44297 10.1074/jbc.M307873200 [DOI] [PubMed] [Google Scholar]
- Wallsgrove R. M., Mazelis M. (1980). The enzymology of lysine biosynthesis in higher plants: complete localization of the regulatory enzyme dihydrodipicolinate synthase in the chloroplasts of spinach leaves. FEBS Lett. 116, 189–192 10.1016/0014-5793(80)80640-5 [DOI] [PubMed] [Google Scholar]
- Wang Q., Sullivan R. W., Kight A., Henry R. L., Huang J. R., Jones A. M., Korth K. L. (2004). Deletion of the chloroplast-localized thylakoid formation1 gene product in Arabidopsis leads to deficient thylakoid formation and variegated leaves. Plant Physiol. 136, 3594–3604 10.1104/pp.104.042754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters M. T., Fray R. G., Pyke K. A. (2004). Stromule formation is dependent upon plastid size, plastid differentiation status and the density of plastids within the cell. Plant J. 39, 655–667 10.1111/j.1365-313X.2004.02164.x [DOI] [PubMed] [Google Scholar]
- Weber A. P. M. (2004). Solute transporters as connecting elements between cytosol and plastid stroma. Curr. Opin. Plant Biol. 7, 247–253 10.1016/j.pbi.2004.03.008 [DOI] [PubMed] [Google Scholar]
- Weber A. P. M., Linka M., Bhattacharya D. (2006). Single, ancient origin of a plastid metabolite translocator family in Plantae from an endomembrane-derived ancestor. Eukaryotic Cell 5, 609–612 10.1128/EC.5.3.609-612.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber A. P. M., Linka N. (2011). Connecting the plastid: transporters of the plastid envelope and their role in linking plastidial with cytosolic metabolism. Annu. Rev. Plant Biol. 62, 53–77 10.1146/annurev-arplant-042110-103903 [DOI] [PubMed] [Google Scholar]
- Weber A. P. M., Osteryoung K. W. (2010). From endosymbiosis to synthetic photosynthetic life. Plant Physiol. 154, 593–597 10.1104/pp.110.161216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber A. P. M., Schneidereit J., Voll L. M. (2004). Using mutants to probe the in vivo function of plastid envelope membrane metabolite transporters. J. Exp. Bot. 55, 1231–1244 10.1093/jxb/erh091 [DOI] [PubMed] [Google Scholar]
- Weber A. P. M., Von Caemmerer S. (2010). Plastid transport and metabolism of C(3) and C(4) plants – comparative analysis and possible biotechnological exploitation. Curr. Opin. Plant Biol. 13, 257–265 10.1016/j.pbi.2010.01.007 [DOI] [PubMed] [Google Scholar]
- Whippo C. W., Khurana P., Davis P. A., Deblasio S. L., Desloover D., Staiger C. J., Hangarter R. P. (2011). THRUMIN1 is a light-regulated actin-bundling protein involved in chloroplast motility. Curr. Biol. 21, 59–64 10.1016/j.cub.2010.11.059 [DOI] [PubMed] [Google Scholar]
- Wilson M. E., Jensen G. S., Haswell E. S. (2011). Two mechanosensitive channel homologs influence division ring placement in Arabidopsis chloroplasts. Plant Cell 23, 2939–2949 10.1105/tpc.111.088112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C., Fan J., Riekhof W., Froehlich J. E., Benning C. (2003). A permease-like protein involved in ER to thylakoid lipid transfer in Arabidopsis. EMBO J. 22, 2370–2379 10.1093/emboj/cdg021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C., Moellering E. R., Muthan B., Fan J., Benning C. (2010). Lipid transport mediated by Arabidopsis TGD proteins is unidirectional from the endoplasmic reticulum to the plastid. Plant Cell Physiol. 51, 1019–1028 10.1093/pcp/pcq053 [DOI] [PubMed] [Google Scholar]
- Xu C. C., Fan J. L., Cornish A. J., Benning C. (2008). Lipid trafficking between the endoplasmic reticulum and the plastid in Arabidopsis requires the extraplastidic TGD4 protein. Plant Cell 20, 2190–2204 10.1105/tpc.108.061176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Glynn J. M., Olson B. J., Schmitz A. J., Osteryoung K. W. (2008). Plastid division: across time and space. Curr. Opin. Plant Biol. 11, 577–584 10.1016/j.pbi.2008.10.001 [DOI] [PubMed] [Google Scholar]
- Yoon H. S., Hackett J. D., Ciniglia C., Pinto G., Bhattacharya D. (2004). A molecular timeline for the origin of photosynthetic eukaryotes. Mol. Biol. Evol. 21, 809–818 10.1093/molbev/msh075 [DOI] [PubMed] [Google Scholar]
- Zhao L., Katavic V., Li F., Haughn G. W., Kunst L. (2010). Insertional mutant analysis reveals that long-chain acyl-CoA synthetase 1 (LACS1), but not LACS8, functionally overlaps with LACS9 in Arabidopsis seed oil biosynthesis. Plant J. 64, 1048–1058 10.1111/j.1365-313X.2010.04396.x [DOI] [PubMed] [Google Scholar]
- Zrenner R., Stitt M., Sonnewald U., Boldt R. (2006). Pyrimidine and purine biosynthesis and degradation in plants. Annu. Rev. Plant Biol. 57, 805–836 10.1146/annurev.arplant.57.032905.105421 [DOI] [PubMed] [Google Scholar]
- Zybailov B., Rutschow H., Friso G., Rudella A., Emanuelsson O., Sun Q., Van Wijk K. J. (2008). Sorting signals, N-terminal modifications and abundance of the chloroplast proteome. PLoS ONE 3, e1994. 10.1371/journal.pone.0001994 [DOI] [PMC free article] [PubMed] [Google Scholar]