Abstract
The vitamin A derivative retinoic acid (RA) is a morphogen that patterns the anterior-posterior axis of the vertebrate hindbrain. Cellular retinoic acid-binding proteins (Crabps) transport RA within cells to both its nuclear receptors (RARs) and degrading enzymes (Cyp26s). However, mice lacking Crabps are viable, suggesting that Crabp functions are redundant with those of other fatty acid-binding proteins. Here we show that Crabps in zebrafish are essential for posterior patterning of the hindbrain and that they provide a key feedback mechanism that makes signaling robust as they are able to compensate for changes in RA production. Of the four zebrafish Crabps, Crabp2a is uniquely RA inducible and depletion or overexpression of Crabp2a makes embryos hypersensitive to exogenous RA. Computational models confirm that Crabp2a improves robustness within a narrow concentration range that optimizes a ‘robustness index’, integrating spatial information along the RA morphogen gradient. Exploration of signaling parameters in our models suggests that the ability of Crabp2a to transport RA to Cyp26 enzymes for degradation is a major factor in promoting robustness. These results demonstrate a previously unrecognized requirement for Crabps in RA signaling and hindbrain development, as well as a novel mechanism for stabilizing morphogen gradients despite genetic or environmental fluctuations in morphogen availability.
Keywords: Danio rerio, Hindbrain, Morphogen
INTRODUCTION
Morphogen gradients induce different cell fates depending on concentration. Gradient shape is determined by the source and rate of ligand production, as well as its transport properties and stability (Ben-Zvi and Barkai, 2010; Sample and Shvartsman, 2010; Umulis et al., 2010). Feedback mechanisms such as self-enhanced receptor-mediated degradation also shape morphogen gradients and make them robust (i.e. able to compensate for changes in morphogen availability), as has been demonstrated for growth factors of the TGFβ, Wingless (Wg) and Hedgehog (Hh) families (Eldar et al., 2003; Lander, 2007; Meinhardt, 2009; Wartlick et al., 2009). The vitamin A derivative retinoic acid (RA) determines the identities of hindbrain rhombomeres along the anterior-posterior (A-P) axis (Dupé and Lumsden, 2001; Maves and Kimmel, 2005; White et al., 2007). However, unlike peptide morphogens, RA is hydrophobic and is typically bound to soluble proteins. How these binding proteins function in modulating RA signaling remains unclear.
Cellular RA-binding proteins (Crabps) bind RA with high affinity and solubilize it intracellularly. Vertebrates have two highly conserved Crabps, both of which transport RA to its receptors (RARs) in vitro (Aström et al., 1991; Delva et al., 1999; Budhu et al., 2001; Budhu and Noy, 2002). In addition, Crabp1 protects cells from excess RA by binding it in the cytosol, away from RARs, and facilitating its degradation by Cyp26s (Boylan and Gudas, 1992; Fiorella and Napoli, 1994; Won et al., 2004). Deletion of both Crabp1 and Crabp2 in mice causes supernumerary digits on the forelimb at low penetrance, but adults are otherwise viable (Lampron et al., 1995), suggesting that other proteins can compensate for Crabps in RA signaling (Romand et al., 2000).
RA signaling is also remarkably robust, as might be expected for a signal derived from a dietary precursor. Hindbrain development occurs normally over a ∼20-fold range of RA concentrations (Hernandez et al., 2007; White and Schilling, 2008). We previously developed a model explaining how robustness critically depends upon negative feedback, and showed that RA induces expression of the RA-degrading enzyme Cyp26a1 (White et al., 2007). However, our models suggest that self-enhanced degradation alone can only account for a small fraction of the robustness. Crabps might provide additional negative feedback by preventing ligand access to receptors and facilitating degradation (Häcker et al., 2005; Lander et al., 2007).
Here we show that, in contrast to mice, zebrafish Crabp2a and Crabp2b are essential for expression of hoxb4 and hoxb5 in the hindbrain. In addition, Crabp2a is uniquely RA inducible and required for robustness. When Crabp2a is depleted or overexpressed, hindbrain patterning becomes hypersensitive to exogenous RA. Computational models with which we simulate the effects of Crabps on gradient robustness indicate that Crabps must be present within a narrow concentration range and must deliver RA both to its receptors and to Cyp26s for degradation.
MATERIALS AND METHODS
Embryo treatments
Wild-type or Tg(RARE-gata2:NTD-eYFP)Id1 transgenic embryos (Perz-Edwards et al., 2001) were treated with all-trans RA (Sigma) or diethylaminobenzaldehyde (DEAB) as described previously (White et al., 2007). Treatments began at 6 hours postfertilization (hpf) and continued until embryos were fixed in 4% paraformaldehyde (PFA).
RT-PCR and cloning
RT-PCR was performed with whole-embryo RNA preparations (for primers see supplementary material Table S1). crabp2a and crabp2b were amplified and cloned into pCS2 (Rupp et al., 1994) for mRNA synthesis.
In situ hybridization
Antisense crabp2a and crabp2b mRNA probes were synthesized from linearized pCS2 clones (Thisse et al., 2004). Bright-field in situ hybridization and fluorescent images were acquired on a Zeiss Axioplan II compound microscope with Improvision software.
Morpholino (MO) and mRNA injections
One-cell stage embryos were injected with 10 ng Crabp2a-MO1 (CGGGAATTTTACGATCCATCTTCCG) or 5 ng Crabp2b-MO1 (TGTTTTCTCCGTGTTAGATTCCATG). Two independent MOs for each gene – Crabp2a-MO2 (TTCTGTCGTTCCTCTGAAAGTAACC) and Crabp2b-MO2 (GTTTCTGCGTGTCTTTCTTTCACTC) – gave similar phenotypes, confirming MO specificity. All MOs were designed to block translation. crabp2a:mCherry and crabp2b:mCherry constructs were generated by fusion PCR of the crabp2a cDNA templates and mCherry from a Gateway vector (Invitrogen) (for primers see supplementary material Table S1). Injections were performed with 200 pg crabp2a:mCherry or crabp2b:mCherry mRNA per embryo, alone or with each MO to confirm MO efficiency. For rescue and overexpression experiments, two full-length crabp2a-pCS2 clones, one with and one without the crabp2a-MO binding sites, were transcribed using SP6 mMessage (Ambion).
Modeling
The model was solved with our newly developed numerical solver (supplementary material Appendices S1, S2), using a scaled domain x∊[0,1] containing the RA gradient between 0≤x≤0.9. A domain of Cyp26a1 posterior to x>0.9 creates a discontinuity in the gradient. Therefore, xref=0.85 was used and the gradient was modeled between x=0 and x=0.9. Established biological ranges of parameter values were used or estimated within a realistic range (supplementary material Appendix S3, Tables S2, S3). Exploration of parameter space was performed on a logarithmic scale to include large orders of magnitude. We measured robustness using a probability density distribution N(E), interpreted as the probability of the system producing robustness values in the range [E1,E2] with the following formula:
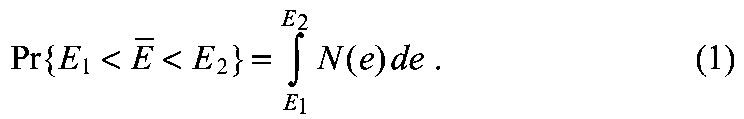 |
To ensure that N(E) was representative of the system, we performed consistency checks using increasing numbers of simulations to find the representative distribution. N(E) typically becomes invariant at ∼10,000 simulations, but we included many more for higher resolution.
RESULTS AND DISCUSSION
A novel requirement for Crabps in Hox gene expression
Crabps facilitate interactions between RA and RARs in vitro (Aström et al., 1991; Delva et al., 1999; Budhu and Noy, 2002), but also bind RA in the cytosol preventing its entry into the nucleus and promoting degradation (Boylan and Gudas, 1991; Boylan and Gudas, 1992; Fiorella and Napoli, 1994; Chen et al., 2003). Consistent with previous studies (Sharma et al., 2005) of the four zebrafish Crabps (crabp1a, crabp1b, crabp2a, crabp2b), all but crabp1a were detected during gastrulation (6-9 hpf; supplementary material Figs S1, S2). crabp2a and crabp2b remained expressed in the presumptive hindbrain at 10-24 hpf. crabp2a was expressed in the posterior hindbrain up to rhombomere (R) 4 (supplementary material Fig. S1) and in the anterior spinal cord. crabp2b was initially expressed throughout the ectoderm during gastrulation and became progressively restricted to the anterior hindbrain (particularly R2) and somites (supplementary material Fig. S1). These expression patterns suggest overlapping but spatially and temporally distinct roles for each Crabp.
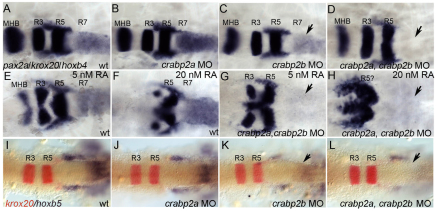
To test functional requirements for Crabps we compared hindbrain patterning in zebrafish embryos injected with antisense morpholino oligonucleotides (MOs) targeting crabp2a and crabp2b. MOs depleted each target effectively (supplementary material Fig. S3) and specifically, and microinjection of crabp2a mRNAs not recognized by MOs targeting their 5′UTRs partially rescued the morphant phenotypes (supplementary material Fig. S4). Embryos injected with either MO alone showed normal patterns of krox20 (egr2 – Zebrafish Information Network) (R3, R5) and reduced hoxb4 (71%, n=14) and hoxb5 (100%, n=25) expression in R7 and spinal cord (Fig. 1A-C,I-K). By contrast, embryos co-injected with Crabp2a-MO and Crabp2b-MO completely lacked hoxb4 (73%, n=15; Fig. 1D) and hoxb5a (90%, n=20; Fig. 1L) expression, suggesting a strong reduction in RA signaling similar to that caused by the loss of aldh1a2 (Begemann et al., 2001). hoxb4 expression was not rescued by treatment with 5-20 nM exogenous RA, despite a strong posteriorization of the hindbrain (n=14; Fig. 1E-H). Thus, Crabps are required for RA-dependent Hox gene expression in the posterior hindbrain.
Fig. 1.
crabp2a and crabp2b are required for zebrafish hindbrain patterning. (A-L) Whole-mount in situ hybridization (ISH) for pax2a at the midbrain-hindbrain boundary (MHB), krox20 in rhombomere (R) 3 and R5, and hoxb4 (A-H) or hoxb5 (I-L) in R7 and spinal cord at 18 hpf; dorsal views, anterior left. Compared with uninjected controls (A), morpholino (MO) depletion of Crabp2a (B,J) or Crabp2b (C,K) slightly reduces and co-injection (D,L) eliminates hoxb4 and hoxb5 expression (arrows). Exogenous RA treatments (5-20 nM) that posteriorize the hindbrain do not rescue hoxb4 expression (E-H). wt, wild type.
Crabp2a is RA inducible and required for signal robustness
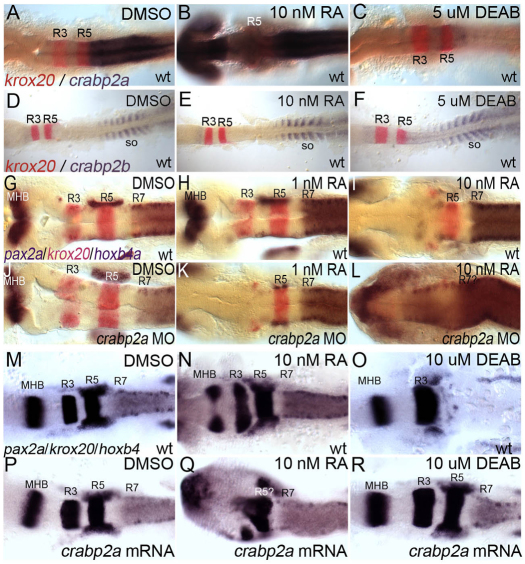
Mammalian Crabp2 contains a retinoic acid response element (RARE) and is RA inducible in vitro (Aström et al., 1994). We examined crabp2a and crabp2b expression after treatment with RA or an Aldh1a2 inhibitor (DEAB) during gastrulation. Whereas 1 nM RA had no effect on the expression of either gene (data not shown), 10 nM RA induced crabp2a expression throughout the CNS (Fig. 2A,B). Conversely, 5 μM DEAB treatments, which phenocopy mutations in aldh1a2 (Begemann et al., 2001), virtually eliminated crabp2a expression (Fig. 2C) but had no effect on crabp2b (Fig. 2D-F; supplementary material Fig. S5). Thus, RA is both necessary and sufficient to induce crabp2a expression.
Fig. 2.
crabp2a is RA inducible and required for signal robustness. (A-F) Whole-mount ISH for crabp2a/b expression at 15 hpf; dorsal views, anterior left. crabp2a (A-C) is expressed posterior to the R5/6 boundary, whereas crabp2b (D-F) expression is restricted to somites. Treatment with 10 nM RA expands crabp2a expression anteriorly throughout the CNS (B) but has no effect on crabp2b (E). Treatment with 5 μM DEAB eliminates crabp2a expression (C) but does not disrupt crabp2b (F). (G-R) pax2a, krox20 and hoxb4 expression at 18 hpf in uninjected controls (G-I), Crabp2a-MO-injected (J-L) and crabp2a mRNA-injected embryos (M-R) treated with 1 nM RA (H,K), 10 nM RA (I,L,N,Q) or 10 μM DEAB (O,R). so, somites.
To determine the requirements for Crabps in signal robustness, we treated embryos injected with Crabp2a-MO or Crabp2b-MO, or both, during gastrulation with 1-10 nM RA (Fig. 2G-L). Surprisingly, Crabp2a-deficient embryos were ∼10-fold more sensitive to exogenous RA than wild-type RA-treated embryos; Crabp2b-MO did not affect RA sensitivity alone or when combined with Crabp2a-MO. Treatment of controls with 1 nM RA caused no hindbrain defects (100%, n=22; Fig. 2H), whereas treatment of Crabp2a-MO-injected embryos virtually eliminated the R3 domain of krox20 and pax2a expression at the midbrain-hindbrain boundary (70%, n=10; Fig. 2K). Defects resembled those of controls treated with 10-fold higher amounts of RA (100%, n=18; Fig. 2I). By contrast, treatments of Crabp2a-deficient embryos with 10 nM RA caused complete loss of krox20 and anterior expansion of hoxb4 expression throughout the hindbrain (100%, n=8; Fig. 2L). These patterning defects correlate with an expansion of Tg(RARE-gata2:NTD-eYFP)Id1 transgene expression in Crabp2a-deficient embryos treated with 1 nM exogenous RA (supplementary material Fig. S6) (Linville et al., 2009). Thus, embryos lacking Crabp2a are hypersensitive to small changes in exogenous RA and hindbrain patterning is much less robust.
We also overexpressed Crabp2a and treated embryos with RA or DEAB (Fig. 2M-R). Injection of 50-400 pg crabp2a mRNA caused no hindbrain defects (92%, n=65; Fig. 2M,P). However, treatment of these embryos with 10 nM RA eliminated krox20 expression in R3 and both krox20 and hoxb4 expression in R5 expanded anteriorly (64%, n=11; Fig. 2N,Q). Conversely, Crabp2a-overexpressing embryos were less sensitive to 10 μM DEAB (73%, n=15; Fig. 2R), which in uninjected embryos eliminated krox20 expression in R5 and all hoxb4 expression (89%, n=18; Fig. 2O). Thus, excess Crabp2a also reduces robustness.
A computational model predicts crucial roles for Crabps in robustness
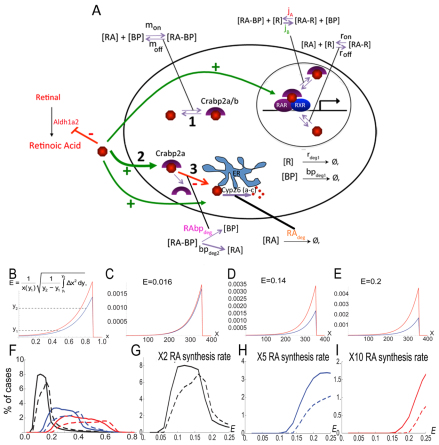
Our previous computational model for RA signaling in the zebrafish hindbrain was based on evidence that RA induces (and Fgf inhibits) Cyp26a1 expression, thereby creating a feedback system of RA degradation (White et al., 2007). This system cannot explain the robustness over a 20-fold concentration range observed experimentally (Hernandez et al., 2007; White and Schilling, 2008), suggesting that reversible binding to receptors and binding proteins is crucial. To test this hypothesis, we expanded our model to include equations that model RA interactions with RARs ([R] in models) and Crabps ([BP]) (supplementary material Appendix S1; Fig. 3A). Our model: (1) represents these in a reaction-diffusion system in one spatial dimension (the A-P axis); (2) assumes that extracellular RA diffuses; and (3) is solved at the steady state. The new model captures receptor (ron/off) and/or binding protein (mon/off) saturation by RA. RA-BP and RA-BP-RAR interactions (jA,B) have been demonstrated (Dong et al., 1999), but there is no direct evidence that RA-BP forms a complex with Cyp26, so we used RAbpdeg to represent Crabp-assisted RA degradation.
Fig. 3.
To study robustness characteristics, we sampled randomly generated parameter sets (supplementary material Appendix S1), comparing two values for the RA synthesis rate. We quantified the change in the RA gradient with respect to a fold-change in the RA synthesis of a ‘reference’ gradient, to define a robustness index designated ‘E’ (Fig. 3B). E is the normalized mean horizontal shift in two gradients between two thresholds y1 and y2 (representing 20% and 80% of the maximum reference value, respectively) when RA synthesis (or any other parameter) is varied. Smaller E values denote higher similarity between the two gradients and therefore a more robust system (Fig. 3C-E). This integrates spatial information across the entire signaling gradient as opposed to quantifying robustness with respect to a threshold value at one location. Parameter space sampling combined with a robustness index dramatically improves comparisons between systems because a representative distribution of E values is produced for each one.
To quantify influences of Crabps on RA signal robustness, we first compared a model that contains Crabps with one that does not. E value distributions were determined using ∼100,000 simulations of our model for the hindbrain in which the RA synthesis rate was varied up to 10-fold in each case (Fig. 3F-I; supplementary material Figs S7, S8). For both increases (2-, 5-, 10-fold; Fig. 3F) and decreases (2-, 5-, 10-fold; supplementary material Fig. S9) in RA synthesis, E values were consistently lower in models that included Crabps and showed more simulations with extremely low E values (E=0-0.25) (Fig. 3F-I; supplementary material Fig. S9B-D).
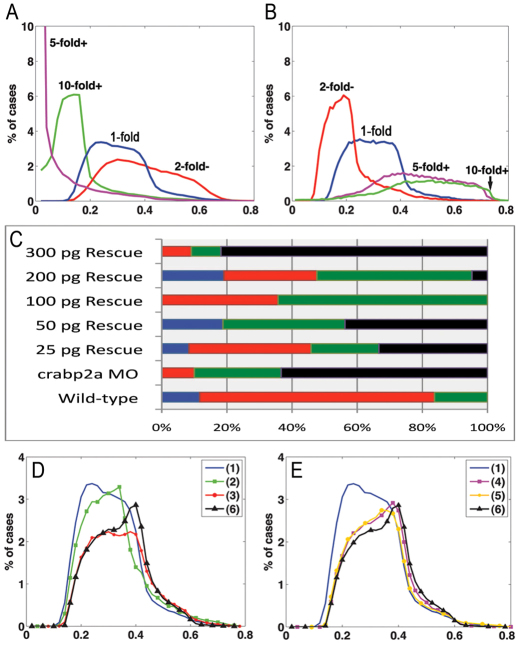
We modeled the effects of varying Crabp levels using randomly generated parameter sets with 5-fold increases or decreases in RA synthesis rate and simulated for 20 different Crabp synthesis rates [Vbp] (supplementary material Fig. S10). Synthesis rates were obtained by logarithmically dividing its parameter range (supplementary material Appendix S3). The lowest E values corresponded to an intermediate level of Vbp in both cases. This suggests an optimal range of Crabp concentration, above or below which robustness is compromised. To examine this in more detail, E value distributions were generated for simulations with a 5-fold increase in RA synthesis rate coupled with either a 5- or 10-fold increase or a 2-fold decrease in the Crabp synthesis rate Vbp (Fig. 4A). E value distributions were lowest with a 5-fold increase in Vbp, with many E values close to zero (Fig. 4A). Similarly, E value distributions for simulations with a 5-fold decrease in RA synthesis rate were lower when Vbp was increased (Fig. 4B; supplementary material Fig. S11). These simulations show that proportional changes in Crabp and RA synthesis produce the best robustness.
Fig. 4.
Robustness gains require Crabp-mediated RA degradation. (A,B) E value distributions for models with either a 5-fold increase (A) or decrease (B) of RA synthesis rate with respect to 10- (green), 5- (magenta), or 2-fold (red) increases in Vbp. (C) Percentages (x-axis) of wild-type (blue bars), mild (red bars), moderate (green bars) and severe (black bars) phenotypes resulting from injection of different amounts of Crabp2a mRNA per embryo (y-axis) treated with 5 nM RA. (D,E) Simulations of 5-fold increases in RA synthesis rate in which binding proteins: (1) have all three mechanisms; (2) do not transport RA to receptors (jB=0); (3) do not sequester RA (jA=0); (4) do not transport RA to Cyp26 degradation (rabpdeg=0); (5) must bind RA to allow degradation (radeg=0); and (6) are absent.
Based on these models, we tested the concentration range over which Crabp2a promotes optimal robustness in the hindbrain experimentally, by co-injecting Crabp2a-MO with a range of doses of crabp2a ‘rescue’ mRNA and treating the embryos with 5 nM exogenous RA. This relatively low amount of RA reduces krox20 expression in R3 in wild types (mild), whereas in Crabp2a-depleted embryos R3 expression is lost and R5 is reduced (severe). Co-injection of 100-200 pg/embryo crabp2a mRNA partially rescued these phenotypes, whereas 25-50 pg had little effect and higher amounts (>300 pg) increased the severity of the phenotype (Fig. 4C; supplementary material Fig. S4).
Models suggest that Crabps facilitate RA degradation
Crabp2 in mice has a putative nuclear localization signal and can form a complex with RA and RARs, while Crabp1 enhances the formation of RA degradation products in vitro (Dong et al., 1999). Although our model does not distinguish between different Crabps, it can address questions regarding how positive or negative roles influence gradient robustness. We hypothesize three such roles: (1) Crabps transport RA to receptors, jA in our models (Fig. 3A); (2) Crabps bind RA and prevent interactions with receptors, jA=0 sequestering RA as [RA-BP] and allowing less [RA-R] (supplementary material Figs S12, S13); and (3) Crabps transport RA for degradation by Cyp26s. These roles are not mutually exclusive.
To test each role computationally, we calculated E values for models incorporating different combinations (Fig. 4D,E): (1) all three functions; (2) only sequestration and degradation; (3) transport to receptors and degradation; (4) transport to receptors and sequestration; (5) degradation only when bound to RA (RAdeg=0); and (6) no binding proteins present. Simulations with either 5-fold increases (Fig. 4D,E) or decreases (supplementary material Fig. S14) in RA synthesis rate revealed that combined positive and negative roles were most robust, particularly a model incorporating all three roles. By contrast, a system without sequestration was least robust. The receptor/sequestration model (Fig. 4E) was less robust than the dual sequestration/degradation model, suggesting that transport to Cyp26s is crucial for robustness in the RA system.
These results are consistent with Crabps both facilitating interactions between RA and RARs and promoting RA degradation (Fig. 3A) (Dong et al., 1999). They help explain how hindbrain patterning can be robust to a 20-fold range of RA concentrations (Hernandez et al., 2007). They also reveal an essential role for these proteins that can help explain why Crabps are so highly conserved – zebrafish Crabp2a is 74% similar in protein sequence to human CRABP2 (Sharma et al., 2003). The apparent lack of a requirement for Crabps in mice might reflect differential redundancies among fatty acid-binding proteins or differential regulation in placental mammals at the level of maternal and extra-embryonic tissues (Sapin et al., 1997; Zheng and Ong, 1998).
Supplementary Material
Acknowledgments
We thank Lei Zhang, Tailin Zhang and Ines Gehring for technical assistance.
Footnotes
Funding
This study was supported by The National Institutes of Health [P50 GM-76516 to K.R., A.D.L., Q.N., T.F.S.; R01 GM-67247 to Q.N.; R01 NS-41353 to T.F.S.]; and the National Science Foundation [DMS-0917492 to Q.N.]. Deposited in PMC for release after 12 months.
Competing interests statement
The authors declare no competing financial interests.
Supplementary material
Supplementary material available online at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.077065/-/DC1
References
- Allenby G., Janocha R., Kazmer S., Speck J., Grippo J. F., Levin A. A. (1994). Binding of 9-cis-retinoic acid and all-trans-retinoic acid to retinoic acid receptors alpha, beta, and gamma. Retinoic acid receptor gamma binds all-trans-retinoic acid preferentially over 9-cis-retinoic acid. J. Biol. Chem. 269, 16689–16695 [PubMed] [Google Scholar]
- Aström A., Tavakkol A., Pettersson U., Cromie M., Elder J. T., Voorhees J. J. (1991). Molecular cloning of two human cellular retinoic acid-binding proteins (CRABP). Retinoic acid-induced expression of CRABP-II but not CRABP-I in adult human skin in vivo and in skin fibroblasts in vitro. J. Biol. Chem. 266, 17662–17666 [PubMed] [Google Scholar]
- Aström A., Pettersson U., Chambon P., Voorhees J. J. (1994). Retinoic acid induction of human cellular retinoic acid-binding protein-II gene transcription is mediated by retinoic acid receptor-retinoid X receptor heterodimers bound to one far upstream retinoic acid-responsive element with 5-base pair spacing. J. Biol. Chem. 269, 22334–22339 [PubMed] [Google Scholar]
- Begemann G., Schilling T. F., Rauch G. J., Geisler R., Ingham P. W. (2001). The zebrafish neckless mutation reveals a requirement for raldh2 in mesodermal signals that pattern the hindbrain. Development 128, 3081–3094 [DOI] [PubMed] [Google Scholar]
- Ben-Zvi D., Barkai N. (2010). Scaling of morphogen gradients by an expansion-repression integral feedback control. Proc. Natl. Acad. Sci. USA 107, 6924–6929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boylan J. F., Gudas L. J. (1991). Overexpression of the cellular retinoic acid binding protein-I (CRABP-I) results in a reduction in differentiation-specific gene expression in F9 teratocarcinoma cells. J. Cell Biol. 112, 965–979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boylan J. F., Gudas L. J. (1992). The level of CRABP-I expression influences the amounts and types of all-trans-retinoic acid metabolites in F9 teratocarcinoma stem cells. J. Biol. Chem. 267, 21486–21491 [PubMed] [Google Scholar]
- Budhu A., Gillilan R., Noy N. (2001). Localization of the RAR interaction domain of cellular retinoic acid binding protein-II. J. Mol. Biol. 305, 939–949 [DOI] [PubMed] [Google Scholar]
- Budhu A. S., Noy N. (2002). Direct channeling of retinoic acid between cellular retinoic acid-binding protein II and retinoic acid receptor sensitizes mammary carcinoma cells to retinoic acid-induced growth arrest. Mol. Cell. Biol. 22, 2632–2641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen A. C., Yu K., Lane M. A., Gudas L. J. (2003). Homozygous deletion of the CRABPI gene in AB1 embryonic stem cells results in increased CRABPII gene expression and decreased intracellular retinoic acid concentration. Arch. Biochem. Biophys. 411, 159–173 [DOI] [PubMed] [Google Scholar]
- Delva L., Bastie J. N., Rochette-Egly C., Kraïba R., Balitrand N., Despouy G., Chambon P., Chomienne C. (1999). Physical and functional interactions between cellular retinoic acid binding protein II and the retinoic acid-dependent nuclear complex. Mol. Cell. Biol. 19, 7158–7167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong D., Ruuska S. E., Levinthal D. J., Noy N. (1999). Distinct roles for cellular retinoic acid-binding proteins I and II in regulating signaling by retinoic acid. J. Biol. Chem. 274, 23695–23698 [DOI] [PubMed] [Google Scholar]
- Dupé V., Lumsden A. (2001). Hindbrain patterning involves graded responses to retinoic acid signalling. Development 128, 2199–2208 [DOI] [PubMed] [Google Scholar]
- Eldar A., Rosin D., Shilo B. Z., Barkai N. (2003). Self-enhanced ligand degradation underlies robustness of morphogen gradients. Dev. Cell 5, 635–646 [DOI] [PubMed] [Google Scholar]
- Fiorella P. D., Napoli J. L. (1994). Microsomal retinoic acid metabolism. Effects of cellular retinoic acid-binding protein (type I) and C18-hydroxylation as an initial step. J. Biol. Chem. 269, 10538–10544 [PubMed] [Google Scholar]
- Häcker U., Nybakken K., Perrimon N. (2005). Heparan sulphate proteoglycans: the sweet side of development. Nat. Rev. Mol. Cell Biol. 6, 530–541 [DOI] [PubMed] [Google Scholar]
- Hernandez R. E., Putzke A. P., Myers J. P., Margaretha L., Moens C. B. (2007). Cyp26 enzymes generate the retinoic acid response pattern necessary for hindbrain development. Development 134, 177–187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampron C., Rochette-Egly C., Gorry P., Dollé P., Mark M., Lufkin T., LeMeur M., Chambon P. (1995). Mice deficient in cellular retinoic acid binding protein II (CRABPII) or in both CRABPI and CRABPII are essentially normal. Development 121, 539–548 [DOI] [PubMed] [Google Scholar]
- Lander A. D. (2007). Morpheus unbound: reimagining the morphogen gradient. Cell 128, 245–256 [DOI] [PubMed] [Google Scholar]
- Lander A. D., Nie Q., Wan F. Y. (2007). Membrane-associated non-receptors and morphogen gradients. Bull. Math. Biol. 69, 33–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linville A., Radtke K., Waxman J. S., Yelon D., Schilling T. F. (2009). Combinatorial roles for zebrafish retinoic acid receptors in the hindbrain and pharyngeal arches. Dev. Biol. 325, 60–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maves L., Kimmel C. B. (2005). Dynamic and sequential patterning of the zebrafish posterior hindbrain by retinoic acid. Dev. Biol. 285, 593–605 [DOI] [PubMed] [Google Scholar]
- Meinhardt H. (2009). Models for the generation and interpretation of gradients. Cold Spring Harb. Perspect. Biol. 1, a001362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Napoli J. L. (1996). Biochemical pathways of retinoid transport, metabolism, and signal transduction. Clin. Immunol. Immunopathol. 80, S52–S62 [DOI] [PubMed] [Google Scholar]
- Perz-Edwards A., Hardison N. L., Linney E. (2001). Retinoic acid-mediated gene expression in transgenic reporter zebrafish. Dev. Biol. 229, 89–101 [DOI] [PubMed] [Google Scholar]
- Romand R., Sapin V., Ghyselinck N. B., Avan P., Le Calvez S., Dollé P., Chambon P., Mark M. (2000). Spatio-temporal distribution of cellular retinoid binding protein gene transcripts in the developing and the adult cochlea. Morphological and functional consequences in CRABP- and CRBPI-null mutant mice. Eur. J. Neurosci. 12, 2793–2804 [DOI] [PubMed] [Google Scholar]
- Rupp R. A., Snider L., Weintraub H. (1994). Xenopus embryos regulate the nuclear localization of XMyoD. Genes Dev. 8, 1311–1323 [DOI] [PubMed] [Google Scholar]
- Sample C., Shvartsman S. Y. (2010). Multiscale modeling of diffusion in the early Drosophila embryo. Proc. Natl. Acad. Sci. USA 107, 10092–10096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sapin V., Ward S. J., Bronner S., Chambon P., Dollé P. (1997). Differential expression of transcripts encoding retinoid binding proteins and retinoic acid receptors during placentation of the mouse. Dev. Dyn. 208, 199–210 [DOI] [PubMed] [Google Scholar]
- Sharma M. K., Denovan-Wright E. M., Boudreau M. E., Wright J. M. (2003). A cellular retinoic acid-binding protein from zebrafish (Danio rerio): cDNA sequence, phylogenetic analysis, mRNA expression, and gene linkage mapping. Gene 311, 119–128 [DOI] [PubMed] [Google Scholar]
- Sharma M. K., Saxena V., Liu R. Z., Thisse C., Thisse B., Denovan-Wright E. M., Wright J. M. (2005). Differential expression of the duplicated cellular retinoic acid-binding protein 2 genes (crabp2a and crabp2b) during zebrafish embryonic development. Gene Expr. Patterns 5, 371–379 [DOI] [PubMed] [Google Scholar]
- Thisse B., Heyer V., Lux A., Alunni V., Degrave A., Seiliez I., Kirchner J., Parkhill J. P., Thisse C. (2004). Spatial and temporal expression of the zebrafish genome by large-scale in situ hybridization screening. Methods Cell Biol. 77, 505–519 [DOI] [PubMed] [Google Scholar]
- Umulis D. M., Shimmi O., O’Connor M. B., Othmer H. G. (2010). Organism-scale modeling of early Drosophila patterning via bone morphogenetic proteins. Dev. Cell 18, 260–274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wartlick O., Kicheva A., González-Gaitán M. (2009). Morphogen gradient formation. Cold Spring Harb. Perspect. Biol. 1, a001255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White R. J., Schilling T. F. (2008). How degrading: Cyp26s in hindbrain development. Dev. Dyn. 237, 2775–2790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White R. J., Nie Q., Lander A. D., Schilling T. F. (2007). Complex regulation of cyp26a1 creates a robust retinoic acid gradient in the zebrafish embryo. PLoS Biol. 5, e304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Won J. Y., Nam E. C., Yoo S. J., Kwon H. J., Um S. J., Han H. S., Kim S. H., Byun Y., Kim S. Y. (2004). The effect of cellular retinoic acid binding protein-I expression on the CYP26-mediated catabolism of all-trans retinoic acid and cell proliferation in head and neck squamous cell carcinoma. Metabolism 53, 1007–1012 [DOI] [PubMed] [Google Scholar]
- Zheng W. L., Ong D. E. (1998). Spatial and temporal patterns of expression of cellular retinol-binding protein and cellular retinoic acid-binding proteins in rat uterus during early pregnancy. Biol. Reprod. 58, 963–970 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.