Abstract
Neurophysiology is increasingly focused on identifying coincident activity among neurons. Strong inferences about neural computation are made from the results of such studies, so it is important that these results be accurate. However, the preliminary step in the analysis of such data, the assignment of spike waveforms to individual neurons (“spike-sorting”), makes a critical assumption which undermines the analysis: that spikes, and hence neurons, are independent. We show that this assumption guarantees that coincident spiking estimates such as correlation coefficients are biased. We also show how to eliminate this bias. Our solution involves sorting spikes jointly, which contrasts with the current practice of sorting spikes independently of other spikes. This new “ensemble sorting” yields unbiased estimates of coincident spiking, and permits more data to be analyzed with confidence, improving the quality and quantity of neurophysiological inferences. These results should be of interest outside the context of neuronal correlations studies. Indeed, simultaneous recording of many neurons has become the rule rather than the exception in experiments, so it is essential to spike sort correctly if we are to make valid inferences about any properties of, and relationships between, neurons.
Keywords: correlated spikes, clustering, point process history, statistical bias, statistical power
The behavior of neural systems is frequently explored by examining the temporal activity of the constituent neuronal cells. Individual cells are characterized by their spike times, descriptive statistics of which are used to characterize responses to stimuli, motor output, and other cognitive states related to memory, emotion, and planning (1). Neuron ensembles are also characterized by temporal relationships between cells, as measured by correlations between cell pairs. Understanding these relationships may be key to deciphering the ensemble neural code of the brain in vivo. For example, cells with correlated activities may be involved in perception (2) and memory formation (3). The correlation structure of cell ensembles also dictates how much information is passed to downstream brain areas (4, 5), and the probability of activating those areas (6). Measurements of correlation structure can also be used to decode subsequent activity (7, 8).
Measuring neuronal correlations requires simultaneously recording multiple cells, and technological developments have made this ensemble approach routine (9). Spike sorting, the assignment of spikes’ waveforms to each cell that emitted them (10), is challenging. When waveforms do not cluster perfectly, some spikes will be incorrectly assigned to the wrong cell. And as the number of cells increases, or the signal-to-noise ratio of the recording decreases, assignments become increasingly ambiguous. Conventionally, a measure of “isolation quality,” the discriminability of a cell’s spike waveforms from those of other cells, determines which cells are worthy of further analysis (11). Cells of inferior isolation quality are typically discarded to preserve the integrity of subsequent analyses.
This results in an unfortunate trade-off. Accepting only the best isolated cells ensures the integrity of analyses but wastes data. For example, if half of recorded cells are excluded then the number of pairs available for computing correlations is reduced by  . Alternatively, we can retain poorly isolated cells at the risk of corrupting subsequent analyses and undermining the biological interpretation of the data. Ref. 12 previously showed that poor isolation yields biased firing rate estimates. Here, we show that neuronal correlation estimates are biased, unless isolation quality is perfect, or cells are Poisson, independent, and have equal firing rates. Hence, the trade-off between wasted data and mistaken inference may seem unavoidable. And as we move to the study of even higher-order relationships in neuronal networks (13), this trade-off worsens exponentially.
. Alternatively, we can retain poorly isolated cells at the risk of corrupting subsequent analyses and undermining the biological interpretation of the data. Ref. 12 previously showed that poor isolation yields biased firing rate estimates. Here, we show that neuronal correlation estimates are biased, unless isolation quality is perfect, or cells are Poisson, independent, and have equal firing rates. Hence, the trade-off between wasted data and mistaken inference may seem unavoidable. And as we move to the study of even higher-order relationships in neuronal networks (13), this trade-off worsens exponentially.
We are at an impasse because the “traditional” analytical framework treats spike sorting and data analysis serially: First, spikes are assigned to cells using waveform information, without regard for correlations between cells; second, these assignments are used to compute those same previously ignored correlations. However, we show that associations between cells contain information about spike identities, and that ignoring this information for spike sorting induces bias in correlation estimates. Refs. 12 and 14 had made a related point: When the firing rates of poorly isolated cells are modulated by covariates, these covariates contain spike identity information, and ignoring it for spike sorting induces bias in firing rate estimates. The solution is to sort spikes using all available information about spike identities, not just waveform information. Refs. 12 and 14 showed how to embed the spiking rates of independent cells into spike sorting. Here we show how to embed the joint spiking probabilities of dependent cells. This results in “ensemble sorting,” whereby spikes are sorted jointly rather than one at a time. We show that ensemble sorting yields unbiased correlation estimates, even for poorly isolated cells. This enables a more efficient use of data, since more cells can be included in correlation analyses, without corrupting scientific interpretations.
Even though the scientific focus of this paper is the estimation of coincidence rates and correlations, it should be of interest to all areas of systems neuroscience. Indeed, ensemble analysis of many cells has become the rule rather than the exception in experimental neuroscience (15), making it essential to sort spikes with these analyses in mind if we are to correctly infer both the individual and the ensemble properties of cells.
Background and Results
Traditional spike-sorting algorithms sort spikes one at a time, so they ignore dependencies between spikes, and therefore between cells. This systematically biases subsequent estimates of neuronal correlation. We propose an “ensemble” approach to sort spikes jointly, which takes into account dependencies between spikes and cells. We show that subsequent correlation analyses are unbiased, and we illustrate our results on simulated data.
Spikes are Sorted One at a Time in Traditional Spike Sorting.
This section presents a synopsis of spike sorting. Additional details can be found in the Supporting Information, Sec. S1. Consider an electrode that records I cells. When its bandpassed voltage exceeds a threshold, we record a snippet of data a around the threshold crossing. All spike-sorting methods assume implicitly or explicitly that suprathreshold waveform measurements a originate from a mixture distribution
 |
[1] |
which simply states that, given a suprathreshold event, the probability that it originated from cell i is πi, and if so, its measurement a arises from a distribution fi centered around the true waveform of cell i. Eq. 1 is often visualized by overlaying the raw waveforms or by plotting their principal components against one another, which aims to reveal clusters that each corresponds to a different cell. Spike sorting consists of separating these clusters.
Many methods exist to separate clusters (10, 16). We focus on model-based clustering because it is optimal when the correct model is used (ref. 17, p. 350], and it can be extended to correct the deficiencies identified herein. Model-based clustering starts with applying Bayes rule to Eq. 1 to obtain the posterior probability that a spike with waveform a was emitted by cell i
 |
[2] |
where the denominator is Eq. 1 and the numerator is one of its summands. The spike is then assigned to the cell with identity x that maximizes P(X = i|a),
 |
[3] |
The most important feature of traditional spike sorting, with respect to our claims, is that spikes are sorted one at a time; i.e., independently of other spikes.
Spikes Should be Sorted Jointly: Ensemble Sorting.
A standard probability result states that only Poisson spike trains are memoryless (18). In that case, spikes are independent, which means that the probability of a spike occurring at a particular time does not depend on the past—i.e., does not depend on the times of previous spikes or on which cells emitted these spikes. Conversely, this implies that if an electrode spike train is not Poisson, then the probability of a spike occurring at a particular time does depend on the times of previous spikes and on which cells emitted them. That is, the history of a non-Poisson spike train contains spike identity information. Current spike-sorting methods ignore that information since they sort spikes independently of other spikes. In Theorem 1.1, we show that this induces bias in estimates of neuronal correlations between cells.
This is of concern because electrode spike trains are seldom Poisson. Indeed, because only Poisson spike trains are memoryless, spikes are independent if and only if all cells are Poisson and are mutually independent. But phenomena such as refractory periods imply that spike timings are not independent; they depend on when and which cells spiked in the past. Refs. 19 and 20 handled this by prohibiting adjacent spikes from being assigned to the same cell. But more crucially in the context of studying neural correlations, the combined spike train of two or more dependent cells cannot be Poisson, even when individual cells are Poisson, since their spikes are dependent. Below, we develop the statistically optimal spike-sorter for non-Poisson electrode spike trains, and in Theorem 1.2, we show that this sorter yields unbiased estimates of neuronal correlations.
Assume that we want to sort jointly n spikes. We denote by  the vector of their unknown identities; Xj can take values in {1,2,…,I}, where Xj = i means that cell i emitted spike j. Spike identity information is contained in the waveform features vector
the vector of their unknown identities; Xj can take values in {1,2,…,I}, where Xj = i means that cell i emitted spike j. Spike identity information is contained in the waveform features vector  , and, as argued above, in the history of the electrode’s spike train. This history is fully summarized by the vector of interspike intervals (ISIs),
, and, as argued above, in the history of the electrode’s spike train. This history is fully summarized by the vector of interspike intervals (ISIs),  , where sj is the time elapsed between spikes (j - 1) and j. Spike sorting aims to estimate X1,…,Xn. The optimal approach relies on calculating the In joint posterior probabilities of possible spike identities, given the variables that contain identity information:
, where sj is the time elapsed between spikes (j - 1) and j. Spike sorting aims to estimate X1,…,Xn. The optimal approach relies on calculating the In joint posterior probabilities of possible spike identities, given the variables that contain identity information:
| [4] |
Then the equivalent of single spike assignments in Eq. 3 consists of assigning the n spikes to the cells with identities
 |
[5] |
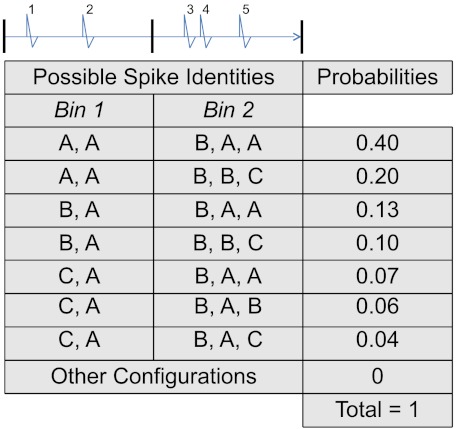
Notice that Eqs. 4 and 2, and 5 and 3, have parallel structures. Eqs. 2 and 3 apply to a single spike, while Eqs. 4 and 5 apply jointly to n spikes, and are further conditioned on the spike train’s history  . To illustrate Eqs. 4 and 5, consider an electrode that records I = 3 cells (A, B, and C), and imagine that n = 5 spikes were observed. The spikes’ joint identities can be one of In = 35 possibilities, with joint posterior probabilities (Eq. 4) listed in Fig. 1. The largest is
. To illustrate Eqs. 4 and 5, consider an electrode that records I = 3 cells (A, B, and C), and imagine that n = 5 spikes were observed. The spikes’ joint identities can be one of In = 35 possibilities, with joint posterior probabilities (Eq. 4) listed in Fig. 1. The largest is  , so joint spike sorting (Eq. 5) assigns the third spike to cell B and all others to cell A.
, so joint spike sorting (Eq. 5) assigns the third spike to cell B and all others to cell A.
Fig. 1.
Hypothetical joint posterior probabilities (Eq. 4) of the identities of five spikes observed on an electrode that records three cells, A, B, and C. For example, the probability that these spikes were emitted successively by cells B, A, B, B, and C is 0.10. Spikes are allotted by time into bins for the computation of coincidence measures.
Ensemble sorting is fully determined by the joint posterior probability in Eq. 4. To evaluate it, we use Bayes rule to rewrite
 |
[6] |
where the denominator is the sum of the numerator over all possible identities  . Eq. 6 is the multivariate equivalent of the spike-sorting rule for a single spike in Eq. 2, but with all quantities now functions of the spike train history
. Eq. 6 is the multivariate equivalent of the spike-sorting rule for a single spike in Eq. 2, but with all quantities now functions of the spike train history  . To use Eq. 6, we need to specify
. To use Eq. 6, we need to specify  and
and  . The first term,
. The first term,  , is the joint distribution of n observed waveforms
, is the joint distribution of n observed waveforms  emitted by cells
emitted by cells  , and with ISIs
, and with ISIs  . If waveforms are stationary, they do not depend on the ISIs, so
. If waveforms are stationary, they do not depend on the ISIs, so  reduces to the product of the marginals,
reduces to the product of the marginals,  , where fi is the waveform distribution of cell i that appeared in Eqs. 1 and 2. Otherwise, a dependent waveform model must be used, e.g., ref. 20 to model waveform attenuations; this is described in Supporting Information, Sec. S2.2. The second term,
, where fi is the waveform distribution of cell i that appeared in Eqs. 1 and 2. Otherwise, a dependent waveform model must be used, e.g., ref. 20 to model waveform attenuations; this is described in Supporting Information, Sec. S2.2. The second term,  , is the probability that the n spikes were emitted by cells
, is the probability that the n spikes were emitted by cells  , given the history
, given the history  of the electrode spike train. If the cells are independent and Poisson, spikes do not depend on the past, so
of the electrode spike train. If the cells are independent and Poisson, spikes do not depend on the past, so  , where πi is the proportion of spikes from cell i that appeared in Eqs. 1 and 2. Otherwise,
, where πi is the proportion of spikes from cell i that appeared in Eqs. 1 and 2. Otherwise,  requires models for the cells firing rates, conditional on the activity of the cell ensemble, and the implied distribution of the ISIs. We discuss this further in the Supporting Information, Sec. S2.2, where we also derive the analytic expression for
requires models for the cells firing rates, conditional on the activity of the cell ensemble, and the implied distribution of the ISIs. We discuss this further in the Supporting Information, Sec. S2.2, where we also derive the analytic expression for  for the example of the illustration section.
for the example of the illustration section.
Neuronal Correlation Estimates are Biased.
Several measures of correlated neuronal activity exist: For example, spike count correlations, coincidence rates, joint surprise (21), or information-theoretic measures capturing nonlinear associations; e.g., mutual information. To streamline the presentation, we focus in this section on coincidence rates, where by coincidence we mean co-occurrence within a time bin of width γ, not simultaneity. Results generalize to other measures, as shown in the illustration section, since coincidence rates are related arithmetically to correlations and, under certain assumptions, to other measures.
Let θ denote the true coincidence rate for a pair of cells. The usual estimate for θ is the proportion of bins in which both cells emitted at least 1 spike each; we call it T⊥, where the “independent” subscript ⊥ serves as a reminder that spikes were sorted one at a time. We call T∪ the same estimator calculated from spikes sorted jointly per Eq. 5; the “union” sign ∪ symbolizes that spikes were sorted together. We do not however advocate the use T∪, but that of a related estimator,  , which is shown to have desirable properties in Thm.1.2:
, which is shown to have desirable properties in Thm.1.2:  is the average over bins of the joint posterior probabilities that both cells spiked in these bins;
is the average over bins of the joint posterior probabilities that both cells spiked in these bins;  preserves the uncertainty in determining spike identities when waveform clusters overlap, in contrast to the current practice of considering that the largest posterior probability (Eqs. 3, 5) provides the correct spike assignments. This is discussed further in Supporting Information, Sec. S3.
preserves the uncertainty in determining spike identities when waveform clusters overlap, in contrast to the current practice of considering that the largest posterior probability (Eqs. 3, 5) provides the correct spike assignments. This is discussed further in Supporting Information, Sec. S3.
We illustrate the calculations of T∪ and  using the data in Fig. 1. Imagine that we cut the spike train in two bins of width γ so that the first two spikes belong to the first bin, and the last three to the other bin. We want to estimate the coincidence rate θ of cells A and B in bins of such duration. We determined earlier that joint sorting assigns spike 3 (in bin 2) to cell B and all other spikes to cell A. Only the second bin contains spikes from cells A and B, so the estimate of θ is T∪ = 1/2. To calculate
using the data in Fig. 1. Imagine that we cut the spike train in two bins of width γ so that the first two spikes belong to the first bin, and the last three to the other bin. We want to estimate the coincidence rate θ of cells A and B in bins of such duration. We determined earlier that joint sorting assigns spike 3 (in bin 2) to cell B and all other spikes to cell A. Only the second bin contains spikes from cells A and B, so the estimate of θ is T∪ = 1/2. To calculate  , we need the posterior probabilities that cells A and B spiked in each bin. Standard probability theory obtains these by summing the full joint posterior in Eq. 4 over the identities of all spikes outside of that bin. Hence, the posterior probability that cells A and B both spiked in the first bin is
, we need the posterior probabilities that cells A and B spiked in each bin. Standard probability theory obtains these by summing the full joint posterior in Eq. 4 over the identities of all spikes outside of that bin. Hence, the posterior probability that cells A and B both spiked in the first bin is  . The posterior probability that cells A and B both spiked in the second bin is
. The posterior probability that cells A and B both spiked in the second bin is  . This gives
. This gives  .
.
We now state properties of traditional and ensemble coincidence rate estimates, T⊥ and  .
.
Theorem 1.1.
T⊥ is a biased and inconsistent estimate of θ, i.e., E(T⊥) ≠ θ even in large samples, unless spikes can be classified with perfect confidence, or cells are Poisson, independent, and have the same firing rates.
Proofs are in the Supporting Information, Sec. S4. Although we proved Theorem 1.1 only for coincidence rates, any statistic computed from spike trains of two or more cells will also suffer from estimation bias under traditional spike sorting, if the distribution of that statistic depends on whether the spikes are in fact independent. This applies to all the common measures of association: correlation, coincidence rate, mutual information, and joint surprise (21), since they all depend on (and try to address) whether or not cells are independent. In practice, the bias will be greater the more waveform clusters overlap, the more cells deviate from Poisson, and the greater the cells’ dependence, as illustrated in Figs. 2 and 3. Importantly, the bias does not disappear in large samples—i.e., T⊥ is not consistent for θ—so recording more data does not help.
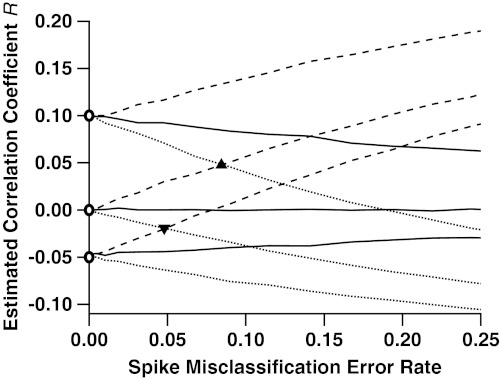
Fig. 2.
Correlation estimates of true correlations -0.05, 0, and 0.10, as functions of the rate of misclassified spikes. Pairs of spike trains were simulated according to three different gamma interval processes: k = 1 (Poisson, solid lines); k = 0.5 (more bursty, dashed); and k = 2 (more periodic, dotted). These processes mimic the inter-spike-interval (ISI) distributions seen in experimental data (22), classically characterized by the coefficient of variation CV = k-1/2. For each of the three cases, we simulated three different values of true correlation between spike trains: 0.1, 0, and -0.05, marked by the open circles along the y axis. Across the x axis we vary the rate at which a traditional spike-sorting procedure misclassifies spikes, the arithmetic mean of type 1 and type 2 errors. As this error rate increases, the estimation of correlation diverges from its true value. In the Poisson case, the bias is always towards an estimate of zero correlation. In the bursty case, it is always positive, and in the periodic case it is always negative. Also indicated are the rates of misclassification at which an uncorrelated (ρ = 0), bursty (k = 0.5), pair of cells has estimated correlation indistinguishable from a correlated (ρ = 0.1), periodic (k = 2), pair of cells (triangle); and the misclassification rate at which an uncorrelated (ρ = 0), periodic pair of cells becomes indistinguishable from an anticorrelated (ρ = -0.05), bursty pair of cells (inverted triangle).
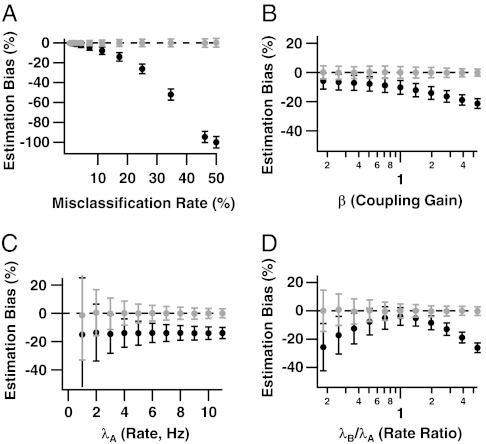
Fig. 3.
Relative biases of coincidence rate estimates from traditional and ensemble sorting, (T⊥ - θ)/θ (black) and  (gray), as functions of (A) the spike misclassification rate due to overlapping waveform clusters, (B) the coupling strength β, (C) the spike rate λA, and (D) the rate ratio λB/λA. The x axes of B and D are on a logarithmic scale. In each panel, parameters held constant are μ = 2 (17.3% misclassified spikes), λA = 4 Hz, λB/λA = 3, and β = 2. Ensemble sorting (gray) always yields unbiased estimates. Traditional sorting (black) never does, except when cells are independent and have equal rates (Theorem 1.1), or when waveform clusters are fully separated (Theorem 1.1 and Fig. 3A, zero error rate). Otherwise, the bias of T⊥ increases as the waveform clusters overlap (Fig. 3A), the coupling strength increases (Fig. 3B), and the firing rates differ (Fig. 3D).
(gray), as functions of (A) the spike misclassification rate due to overlapping waveform clusters, (B) the coupling strength β, (C) the spike rate λA, and (D) the rate ratio λB/λA. The x axes of B and D are on a logarithmic scale. In each panel, parameters held constant are μ = 2 (17.3% misclassified spikes), λA = 4 Hz, λB/λA = 3, and β = 2. Ensemble sorting (gray) always yields unbiased estimates. Traditional sorting (black) never does, except when cells are independent and have equal rates (Theorem 1.1), or when waveform clusters are fully separated (Theorem 1.1 and Fig. 3A, zero error rate). Otherwise, the bias of T⊥ increases as the waveform clusters overlap (Fig. 3A), the coupling strength increases (Fig. 3B), and the firing rates differ (Fig. 3D).
There are two cases when T⊥ is unbiased. When waveform clusters do not overlap, spikes can be classified with no errors and subsequent analyses are valid. Otherwise T⊥ is biased unless (i) cells are independent and Poisson, and their waveforms are independent, and (ii) cells have identical firing rates. Under (i), spikes and waveforms do not depend on the spike train history, so ignoring it for spike sorting, as traditional sorters do, is the correct assumption. But if (ii) is not met, T⊥ suffers from another, unrelated, source of bias: When waveform clusters overlap, one can never be perfectly confident that a spike was emitted by a particular cell; we know that some spikes will be misclassified. Therefore, assigning each spike fully (Eq. 3) to a neuron ignores our uncertainty about spike assignments. Ref. 12 showed that this biases firing rate estimates, unless cells have equal rates. Here, we also show that this biases estimates of neuronal correlations such as T⊥; see Fig. 3. Note that T∪ also suffers from that source of bias, since it is calculated from spikes assigned fully (Eq. 5). But  uses the full posterior distribution of spike identities (Eq. 4), which removes that source of bias, as shown below.
uses the full posterior distribution of spike identities (Eq. 4), which removes that source of bias, as shown below.
Theorem 1.2.
is unbiased; i.e.,
, and under weak conditions stated in the proof, its variability can be reduced arbitrarily by increasing the sample size, so
is consistent for θ.
Theorem 1.2 is valid regardless of how much waveform clusters overlap. In practice, if the overlap is substantial or if the sample is small,  will be relatively variable so it may not match θ closely. However it matches in expectation, i.e.,
will be relatively variable so it may not match θ closely. However it matches in expectation, i.e.,  , and recording more data will help reduce its variability.
, and recording more data will help reduce its variability.
Illustrations: Correlation and Coincidence Rate Estimates for Two Dependent Cells.
To understand the magnitude of the problem for experimental neuroscience, we estimate the spike count correlation ρ and coincidence rate θ between independent and dependent pairs of simulated Poisson and non-Poisson cells.
A simulation requires (i) a spiking model for individual cells, (ii) a dependence model for cell pairs, and (iii) a model for the waveforms. (i) We consider cells that deviate from Poisson according to parameters found empirically by ref. 22. They observed that the ISIs of cortical cells were well fit by gamma distributions with shape parameters k, and corresponding coefficient of variation  ; k = CV = 1 corresponds to Poisson spike trains, while k > 1 (CV < 1) models periodicity, and k < 1 (CV > 1) “burstiness.” Ref. 22 found that k varied widely within cortical areas, with some cells having k < 1 and others k > 1; and across areas, with median values k = 1.23, 1.95, and 2.70 for MT/MST, LIP, and Area 5, respectively. In Fig. 1, we used k = 0.5, 1 and 2 to investigate a relevant range. (ii) Cells are correlated for various reasons, including common synaptic input, and reciprocal or directed connectivity. Here we consider directed connectivity; results are qualitatively similar under other models. We assume that cell A spikes at constant rate λA and cell B at rate λB, unless cell A spikes, in which case the rate of cell B is multiplied by β for the next 10 msec. The magnitude of β dictates the cells’ degree of association; β = 1 models independence, β > 1 models an excitatory effect, and β < 1 an inhibitory one. For Fig. 2, we selected values of β to generate pairs of uncorrelated cells (ρ = 0, β = 1), and pairs of positively and negatively correlated cells, with true correlations ρ = 0.1 and -0.05. (iii) We assume that waveforms are independent, which is warranted when cells have low firing rates. Without loss of generality, we use one-dimensional waveform features that have Gaussian distributions with means 0 and μ for cells A and B, respectively, and unit variances (f1 and f2 in Eq. 1). The overlap between f1 and f2 depends on μ, which determines the probability of spike misclassifications. (In the limit μ = 0, clusters overlap completely, so waveforms contain no information that can be used to assign spikes.) In the two cell case, a spike misassignment is a type 1 error for one cell and a type 2 error for the other, so we define the spike misclassification rate to be the average of the rates of these two error types. For example, when λA = 4 Hz, λB = 12 Hz, and β = 2, then μ = 2 induces a spike misclassification rate of 17.3%. In Fig. 2, we varied μ so that the spike misclassification rate would range from 0 to 25%.
; k = CV = 1 corresponds to Poisson spike trains, while k > 1 (CV < 1) models periodicity, and k < 1 (CV > 1) “burstiness.” Ref. 22 found that k varied widely within cortical areas, with some cells having k < 1 and others k > 1; and across areas, with median values k = 1.23, 1.95, and 2.70 for MT/MST, LIP, and Area 5, respectively. In Fig. 1, we used k = 0.5, 1 and 2 to investigate a relevant range. (ii) Cells are correlated for various reasons, including common synaptic input, and reciprocal or directed connectivity. Here we consider directed connectivity; results are qualitatively similar under other models. We assume that cell A spikes at constant rate λA and cell B at rate λB, unless cell A spikes, in which case the rate of cell B is multiplied by β for the next 10 msec. The magnitude of β dictates the cells’ degree of association; β = 1 models independence, β > 1 models an excitatory effect, and β < 1 an inhibitory one. For Fig. 2, we selected values of β to generate pairs of uncorrelated cells (ρ = 0, β = 1), and pairs of positively and negatively correlated cells, with true correlations ρ = 0.1 and -0.05. (iii) We assume that waveforms are independent, which is warranted when cells have low firing rates. Without loss of generality, we use one-dimensional waveform features that have Gaussian distributions with means 0 and μ for cells A and B, respectively, and unit variances (f1 and f2 in Eq. 1). The overlap between f1 and f2 depends on μ, which determines the probability of spike misclassifications. (In the limit μ = 0, clusters overlap completely, so waveforms contain no information that can be used to assign spikes.) In the two cell case, a spike misassignment is a type 1 error for one cell and a type 2 error for the other, so we define the spike misclassification rate to be the average of the rates of these two error types. For example, when λA = 4 Hz, λB = 12 Hz, and β = 2, then μ = 2 induces a spike misclassification rate of 17.3%. In Fig. 2, we varied μ so that the spike misclassification rate would range from 0 to 25%.
We used this model to simulate the spike trains and waveform features of cell pairs, which we combined to produce electrode spike trains. We then sorted the spikes using traditional spike sorting (Supporting Information, Sec. S5.2), and estimated the correlation R between pairs of cells’ spike counts in bins of length γ. Fig. 2 shows R against the spike misclassification rate, for λA = λB = 10 Hz and bin size γ = 25 ms; we vary these parameters in Fig. 3. The various curves correspond to various degrees of regularity: Poisson (k = 1), bursty (k = 2), and periodic (k = 0.5), and values of β that correspond to true correlations ρ = 0 (independent cells), 0.1 and -0.05. Fig. 2 shows that R is biased, unless cells are independent and Poisson (ρ = 0, k = 1), or perfectly isolated (zero misclassification rate). Otherwise, the bias increases with the rate of misclassified spikes. When cells are Poisson, R shrinks towards zero regardless of the sign and magnitude of the true correlation ρ; this is consistent with ref. 23; and also with ref. 24, who observed the same with their measure of association, the joint surprise. In the bursty (k < 1) and periodic (k > 1) cases, R has positive and negative bias, respectively. This is consistent with the findings of refs. 25 and 26 who show that misclassification of spikes from non-Poisson spike trains can lead to misestimation of correlations. Hence the sign and magnitude of the bias depend on the ISI distribution, and thus on the brain area the cells are recorded from. It is also possible to identify correlation when it does not exist; i.e., ρ = 0 but R ≠ 0, and fail to identify it when it does exist; i.e., ρ ≠ 0 but R = 0. Importantly, the bias is sufficiently large that sets of cell pairs whose true correlations ρ are very different from one another, may have the same estimated correlations R once the rate of misclassification reaches 5–10% (Fig. 2, triangles). Since a misclassification rate of 5% or more is not rare (11) (also see Supporting Information, Sec. 5.1), our results cast some doubts on the correlation values reported in the neuroscience literature.
Our next simulation serves several purposes. We use the same simulation model, but (i) we consider only Poisson cells to investigate more specifically the bias induced by true neuronal correlation rather than by non-Poisson spiking. (ii) We investigate the effects of more model parameters. (iii) We estimate the coincidence rate θ of cell pairs rather than their correlation ρ, to illustrate that all association measures are biased under traditional spike sorting. (iv) We implement ensemble sorting to confirm that  is indeed unbiased for θ, as proved in Thm.1.2. Detailed descriptions of the traditional and ensemble sorters are in the Supporting Information, Sec. 5.
is indeed unbiased for θ, as proved in Thm.1.2. Detailed descriptions of the traditional and ensemble sorters are in the Supporting Information, Sec. 5.
Fig. 3 shows plots of the relative biases of T⊥ and  against the spike misclassification rate, the degree of dependence between cells via β, the spike rate of cell A, and the relative spike rate of cell B compared with cell A. A y axis value of 0 implies an unbiased coincidence rate estimate; -1 means that all coincidences are missed; 1 means that the estimate is twofold too large. Fig. 3 shows that the ensemble coincidence rate estimate
against the spike misclassification rate, the degree of dependence between cells via β, the spike rate of cell A, and the relative spike rate of cell B compared with cell A. A y axis value of 0 implies an unbiased coincidence rate estimate; -1 means that all coincidences are missed; 1 means that the estimate is twofold too large. Fig. 3 shows that the ensemble coincidence rate estimate  is unbiased for all configurations of the model parameters. The traditional estimate T⊥ never is, and its bias appears to be a complex function of the model parameters. Fig. 3B displays clearly the two sources of bias—full spike assignments (Eq. 3) and independent sorting – that plague T⊥. The cells are independent when β = 1, yet T⊥ underestimates θ; this is the effect of full spike assignments, which we know (12) becomes more severe as the cells’ firing rates diverge from one another (here λB/λA = 3). As |β - 1| increases, which strengthens the association between cells, the bias arising from independently sorting correlated spikes combines with the first source of bias. Consider also Fig. 3D, which plots relative bias versus λB/λA for a fixed degree of association β = 2. When firing rates are equal (λB/λA = 1), and when, as here, the waveform feature distributions are identical apart from a shift in their mean, it is easy to show that traditional spike sorting misclassifies the same average number of spikes for both cells. In that case, full assignments (Eq. 3) do not bias the coincidence rate estimate, so that the negative bias of T⊥ at λB/λA = 1 must be due to ignoring the dependence between cells when spike sorting. Then as |λB/λA - 1| increases, that bias is compounded with an increasingly large bias from full spike assignments. Considering Fig. 3 overall, we see that the bias of T⊥ is largest when the cells have overlapping waveform clusters, have very different firing rates, and are strongly correlated.
is unbiased for all configurations of the model parameters. The traditional estimate T⊥ never is, and its bias appears to be a complex function of the model parameters. Fig. 3B displays clearly the two sources of bias—full spike assignments (Eq. 3) and independent sorting – that plague T⊥. The cells are independent when β = 1, yet T⊥ underestimates θ; this is the effect of full spike assignments, which we know (12) becomes more severe as the cells’ firing rates diverge from one another (here λB/λA = 3). As |β - 1| increases, which strengthens the association between cells, the bias arising from independently sorting correlated spikes combines with the first source of bias. Consider also Fig. 3D, which plots relative bias versus λB/λA for a fixed degree of association β = 2. When firing rates are equal (λB/λA = 1), and when, as here, the waveform feature distributions are identical apart from a shift in their mean, it is easy to show that traditional spike sorting misclassifies the same average number of spikes for both cells. In that case, full assignments (Eq. 3) do not bias the coincidence rate estimate, so that the negative bias of T⊥ at λB/λA = 1 must be due to ignoring the dependence between cells when spike sorting. Then as |λB/λA - 1| increases, that bias is compounded with an increasingly large bias from full spike assignments. Considering Fig. 3 overall, we see that the bias of T⊥ is largest when the cells have overlapping waveform clusters, have very different firing rates, and are strongly correlated.
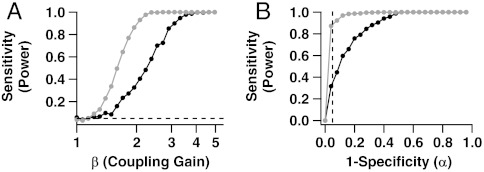
Next, we illustrate that biased estimates not only corrupt scientific inference and interpretation, they also degrade our ability to detect correlations. We consider again the many datasets we simulated to produce Fig. 3B, and for each, we test the null hypothesis that the cells are independent at the 0.05 significance level. This consists of comparing the observed coincidence rate against the rate expected under independence, and calculating a p-value. We then estimate the power of the test—i.e., the probability that it detects correlated activity—by the proportion of times that a significant p-value (< 0.05) is obtained.
Fig. 4A shows the power functions of the two tests based on T⊥ and  as functions of β≥1, the degree of positive association between the cells, for λA = 4 Hz, λB/λA = 3, and μ = 2 (17.3% misclassified spikes). Results are similar for other model settings. We see that ensemble sorting provides uniformly more power. For example, when β = 2, the power to detect that the cells are dependent is approximately 35% using T⊥ and 90% using
as functions of β≥1, the degree of positive association between the cells, for λA = 4 Hz, λB/λA = 3, and μ = 2 (17.3% misclassified spikes). Results are similar for other model settings. We see that ensemble sorting provides uniformly more power. For example, when β = 2, the power to detect that the cells are dependent is approximately 35% using T⊥ and 90% using  .
.
Fig. 4.
Bias in coincident spike rate estimates translates into loss of power to detect correlated cells. (A) Sensitivity (power) of tests for coincident spiking as function of coupling strength β. Other model parameters are held at μ = 2, λA = 4 Hz, and λB/λA = 3. The light and dark curves correspond respectively to the tests based on ensemble and traditional estimates,  and T⊥, of the coincidence rate θ. The test based on ensemble sorting is more powerful. (B) Receiver-operating characteristic (ROC) curves show that for any choice of the significance level α, the sensitivity of the coincident spiking test is higher when using ensemble sorting. The vertical dashed line is at the most commonly used significance level of 5%.
and T⊥, of the coincidence rate θ. The test based on ensemble sorting is more powerful. (B) Receiver-operating characteristic (ROC) curves show that for any choice of the significance level α, the sensitivity of the coincident spiking test is higher when using ensemble sorting. The vertical dashed line is at the most commonly used significance level of 5%.
Note that when the null hypothesis of independence is true (β = 1), both tests reject the null hypothesis approximately 5% of the time. Hence, while the test based on T⊥ lacks power to detect existing associations, it does have the correct rate of false positive (spurious) detections. This should not be surprising here: Traditional spike sorting assumes that spikes are independent, which matches the data in this illustration at β = 1, since we simulated Poisson cells. When cells are not Poisson, we know from Thm.1.1 and Fig. 2 that T⊥ is biased even when cells are independent, which necessarily biases the rate of spurious detections. In contrast, the test based on  retains the correct detection rate since
retains the correct detection rate since  is unbiased.
is unbiased.
Another common summary of the power of a test is the receiver-operating characteristic (ROC) curve, which plots power (sensitivity, the probability of rejecting a null hypothesis when it is false) versus significance level (1—specificity, the probability of rejecting a null hypothesis when it is true). A desirable test has high sensitivity for all specificity. Fig. 4B shows the ROC curves of the two tests based on T⊥ and  . The vertical dashed line is at the commonly used significance level of 5%. Again, the test based on
. The vertical dashed line is at the commonly used significance level of 5%. Again, the test based on  has uniformly better power.
has uniformly better power.
Discussion
Several authors have reported that spike misclassifications lead to misestimation of associations between cells. For example, refs. 23 and 24 found that positive correlations were underestimated by both overly liberal and overly conservative spike-sorting procedures, refs. 25 and 26 noted that cells deviating from Poisson also impacted correlation estimates. In this paper, we proved that correlation estimates calculated from spike trains sorted using current methods are biased, unless cells are isolated, or they are Poisson, independent, and have equal rates. Here and previously (12, 14) we argued that this bias occurs because the standard paradigm—spike sorting first, statistical analyses second—exploits the information that spike sorting provides about the quantities of interest (e.g., correlations here, tuning curves in refs. 12 and 14), but ignores the reverse that these quantities also provide information about spike identities. The correct approach is to consider these relationships simultaneously rather than sequentially. Our solution is “ensemble sorting,” which consists of sorting spikes using waveforms as usual, while also accounting for the interdependent spiking of the cell ensemble. We proved that resulting correlation estimates are unbiased, even when spikes cannot be sorted with perfect confidence.
Ensemble sorting involves significant conceptual and practical shifts for spike sorting: Spikes must be sorted jointly, and waveforms must be retained in case some later analysis requires resorting spikes under a different joint spiking model. This is more cumbersome than the current practice of spike sorting once and never considering the waveforms again. But our simulations motivate a serious consideration of ensemble sorting. Indeed, unless the analyst retains only perfectly sorted cells, the size of the biases can be scientifically significant and corrupt inferences about coincident spiking. Bias can also translate into a sizable loss of power to detect existing correlations in data. In contrast, ensemble sorting yields unbiased estimates even for poorly isolated cells, which permits the analysts to retain more cells for analysis. (One might still exclude some poorly isolated cells when the variances of estimates are too high.) Therefore, experimental investigations will be more efficient: Fewer experiments are needed to accurately infer the degree of correlated activity in a neural system; and when the number of experiments is fixed, these inferences are more accurate.
We stress that the bias discussed in this paper is unrelated to the biases that can arise from implementation issues such as choosing poor models for waveform features, having difficulties estimating the spike sorter from data, and determining the number of cells recorded by electrodes. Our results prove that if spikes are sorted independently of other spikes, the bias will not vanish even if an otherwise optimal spike sorter is applied with the true models. But if the correct models are used with ensemble sorting, correlation estimates will be unbiased. The bias discussed in this paper arises because current spike sorters ignore some information about spike identities. Our results should therefore be of interest to all experimentalists, since simultaneous recording of many cells has become the rule rather than the exception in experiments (15). Ref. 12 showed that firing rate estimates are biased. Here we showed that coincident spiking estimates are biased. Estimates of other statistical quantities are also presumably biased, and it would be worth investigating when these biases are scientifically significant.
In this paper, we formulated the concept and framework for ensemble sorting, which relies on the posterior probabilities of spike identities in Eq. 6. To obtain Eq. 6, one must specify a model for potentially nonstationary waveforms, and a model for the cells’ firing rates, conditional on the previous activity of the cell ensemble—i.e., conditional on when past spikes were emitted and on which cells emitted them. Such conditional rate models are routinely used in the analysis of neural data (27). Finally, one must also determine the ISI distribution induced by the conditional rate model. When spikes are independent, this distribution is exponential. Otherwise it must be derived analytically based on the properties of the assumed joint spiking model. The Supporting Information, Sec. S5.3 contains that derivation for the unidirectional coupling two-cell model application in Figs. 3 and 4, and shows that the ISI distribution involves mixtures of truncated and shifted exponential distributions. We have not provided the form of the ISI distribution for a network involving an arbitrary number of cells, but we are confident we can do this in future work using the same framework.
Once all its components are specified, Eq. 6 must also be fitted to data. This requires that models for waveforms and for ensemble spiking be specified together. The traditional approach requires the same components, but they are used separately: The waveform model is used for spike sorting first, the joint spiking model for statistical analyses second. The important point is that ensemble sorting requires no more model assumptions than the traditional approach. Moreover, ensemble sorting makes consistent assumptions across spike sorting and data analysis—the traditional approach does not—which is why the former alone yields unbiased analyses. Since Eq. 6 is a mixture distribution, we are confident that estimation can be done by maximum likelihood via an expectation-maximization (EM) algorithm. We implemented a related algorithm (14), as well as the algorithm needed to estimate the ensemble sorter for the connectivity model of the illustration section; the detailed implementation will be reported elsewhere.
We have not addressed any computational issues in this paper. They include choosing appropriate waveform and spiking models, choosing initial values of the EM algorithm, and determining the number of neurons. The same issues arise in traditional spike sorting and analyses. The methods proposed here are conceptual; they are not designed to resolve implementation issues. When we implement the proposed procedures to data in future work, we will choose currently available models for waveforms and joint spiking, we will fit the joint spike-sorting rule (Eq. 6) using an EM algorithm, we will determine the number of cells using penalized likelihood, and test the goodness of model fits by penalized likelihood or likelihood ratio tests (14).
Finally, the results in this paper concern cells recorded on the same electrode. But if cells recorded on different electrodes do not behave independently, these electrodes may contain information about the spike identities on other electrodes, and ignoring that information will bias correlation estimates. To see this, assume that cells A and B are recorded on an electrode, cell C on another, that A is independent of B and C, but that B and C always spike together. Assume that A and B fire at equal rates and have the same spike waveforms. If we sort the electrodes separately, we will assign spikes to A and B at random, with spike misclassification 50%; then estimates of correlation between A and C, and between B and C, are inflated and deflated, respectively. But if we sort the electrodes jointly, we will (correctly) assign to B all the spikes that occur concurrently with a spike from C, and to A all the other spikes, with spike misclassification rate 0%; then correlation estimates are unbiased. This is related to refs. 28 and 23, who report that for imperfect spike classification, the degree of true correlation of cells recorded on different electrodes will influence estimates of the degree of correlation of cells recorded on the same electrode.
Supplementary Material
ACKNOWLEDGMENTS.
V.V. was supported by National Institutes of Health Grants 2R01MH064537 and 1R01EB005847. R.C.G. was supported by National Institutes of Health Grant F32DC010535.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1115236109/-/DCSupplemental.
References
- 1.Rieke F, Warland D, de Ruyter van Steveninck R, Bialek W. Spikes: Exploring the Neural Code. Cambridge, MA: MIT Press; 1999. [Google Scholar]
- 2.Engel AK, Singer W. Temporal binding and the neural correlates of sensory awareness. Trends Cogn Sci. 2001;5:16–25. doi: 10.1016/s1364-6613(00)01568-0. [DOI] [PubMed] [Google Scholar]
- 3.Bi GQ, Poo MM. Synaptic modification by correlated activity: Hebb’s postulate revisited. Annu Rev Neurosci. 2001;24:139–66. doi: 10.1146/annurev.neuro.24.1.139. [DOI] [PubMed] [Google Scholar]
- 4.Zohary E, Shadlen MN, Newsome WT. Correlated neuronal discharge rate and its implications for psychophysical performance. Nature. 1994;370:140–143. doi: 10.1038/370140a0. [DOI] [PubMed] [Google Scholar]
- 5.Averbeck BB, Latham PE, Pouget A. Neural correlations, population coding and computation. Nat Rev Neurosci. 2006;7:358–366. doi: 10.1038/nrn1888. [DOI] [PubMed] [Google Scholar]
- 6.Salinas E, Sejnowski TJ. Correlated neuronal activity and the flow of neural information. Nat Rev Neurosci. 2001;2:539–550. doi: 10.1038/35086012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pillow JW, et al. Spatio-temporal correlations and visual signalling in a complete neuronal population. Nature. 2008;454:995–999. doi: 10.1038/nature07140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Graf ABA, Kohn A, Jazayeri M, Movshon JA. Decoding the activity of neuronal populations in macaque primary visual cortex. Nat Neurosci. 2011;14:239–245. doi: 10.1038/nn.2733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stevenson IH, Kording KP. How advances in neural recording affect data analysis. Nat Neurosci. 2011;14:139–142. doi: 10.1038/nn.2731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lewicki MS. A review of methods for spike sorting: The detection and classification of neural action potentials. Network. 1998;9:R53–78. [PubMed] [Google Scholar]
- 11.Schmitzer-Torbert N, Jackson J, Henze D, Harris K, Redish AD. Quantitative measures of cluster quality for use in extracellular recordings. Neuroscience. 2005;131(1):1–11. doi: 10.1016/j.neuroscience.2004.09.066. [DOI] [PubMed] [Google Scholar]
- 12.Ventura V. Traditional waveform based spike sorting yields biased rate code estimates. Proc Natl Acad Sci USA. 2009;106:6921–6. doi: 10.1073/pnas.0901771106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Staude B, Grün S, Rotter S. Higher-order correlations in non-stationary parallel spike trains: statistical modeling and inference. Front Comput Neurosci Jul 2. 2010;4. pii:16. doi: 10.3389/fncom.2010.00016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ventura V. Automatic spike sorting using tuning information. Neural Comput. 2009;21:2466–2501. doi: 10.1162/neco.2009.12-07-669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brown EN, Kass RE, Mitra PP. Multiple neural spike trains analysis: state-of-the-art and future challenges. Nat Neurosci. 2004;7:456–461. doi: 10.1038/nn1228. [DOI] [PubMed] [Google Scholar]
- 16.Quiroga R-Q. Spike sorting. Scholarpedia. 2007;2:3583. [Google Scholar]
- 17.Wasserman L. All of Statistics. New York: Springer; 2004. [Google Scholar]
- 18.Daley DJ, Vere-Jones D. Introduction to the Theory of Point Processes. New York: Springer; 1988. [Google Scholar]
- 19.Fee MS, Mitra PP, Kleinfeld D. Variability of extracellular spike waveforms of cortical neurons. J Neurophysiol. 1996;76:3823–3833. doi: 10.1152/jn.1996.76.6.3823. [DOI] [PubMed] [Google Scholar]
- 20.Pouzat C, Delescluse M, Voit P, Diebolt J. Improved spike-sorting by modeling firing statistics and burst-dependent spike amplitude attenuation: a Markov chain Monte Carlo approach. J Neurophysiol. 2004;91:2910–2928. doi: 10.1152/jn.00227.2003. [DOI] [PubMed] [Google Scholar]
- 21.Grün S. Data-driven significance estimation for precise spike correlation. J Neurophysiol. 2009;101:1126–1140. doi: 10.1152/jn.00093.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maimon G, Assad JA. Beyond Poisson: Increased spike-time regularity across primate parietal cortex. Neuron. 2009;62:426–40. doi: 10.1016/j.neuron.2009.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gerstein GL. Cross-correlation measures of unresolved multi-neuron recordings. J Neurosci Methods. 2000;100:41–51. doi: 10.1016/s0165-0270(00)00226-0. [DOI] [PubMed] [Google Scholar]
- 24.Pazienti A, Grün S. Robustness of the significance of spike synchrony with respect to sorting errors. J Comput Neurosci. 2006;21:329–42. doi: 10.1007/s10827-006-8899-7. [DOI] [PubMed] [Google Scholar]
- 25.Ecker AS, et al. Decorrelated neuronal firing in cortical microcircuits. Science. 2010;327:84–587. doi: 10.1126/science.1179867. [DOI] [PubMed] [Google Scholar]
- 26.Cohen MR, Kohn A. Measuring and interpreting neuronal correlations. Nat Neurosci. 2011;14:811–9. doi: 10.1038/nn.2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Truccolo W, Eden UT, Fellows MR, Donoghue JP, Brown EN. A point process framework for relating neural spiking activity to spiking history, neural ensemble and extrinsic covariate effects. J Neurophysiology. 2005 Feb;93:1074–1089. doi: 10.1152/jn.00697.2004. [DOI] [PubMed] [Google Scholar]
- 28.Bedenbaugh P, Gerstein GL. Multiunit normalized cross correlation differs from the average single-unit normalized correlation. Neural Comput. 1997;9:1265–75. doi: 10.1162/neco.1997.9.6.1265. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.