Abstract
Coordinated, subcellular trafficking of proteins is one of the fundamental properties of the multicellular eukaryotic organisms. Trafficking involves a large diversity of compartments, pathways, cargo molecules, and vesicle-sorting events. It is also crucial in regulating the localization and, thus, the activity of various proteins, but the process is still poorly genetically defined in plants. In the past, forward genetics screens had been used to determine the function of genes by searching for a specific morphological phenotype in the organism population in which mutations had been induced chemically or by irradiation. Unfortunately, these straightforward genetic screens turned out to be limited in identifying new regulators of intracellular protein transport, because mutations affecting essential trafficking pathways often lead to lethality. In addition, the use of these approaches has been restricted by functional redundancy among trafficking regulators. Screens for mutants that rely on the observation of changes in the cellular localization or dynamics of fluorescent subcellular markers enable, at least partially, to circumvent these issues. Hence, such image-based screens provide the possibility to identify either alleles with weak effects or components of the subcellular trafficking machinery that have no strong impact on the plant growth.
Keywords: plant, protein trafficking, screening, forward genetics, fluorescent protein
Introduction
In the last decade, the field of biological imaging had progressed at a remarkable pace. Advances in microscopy and fluorescent reporter protein technology have facilitated the development of imaging-based approaches aimed at unraveling the mechanisms that govern plant growth and development.
The major model plant Arabidopsis thaliana is amenable to genetic manipulation and one can relatively easy generate transgenic lines expressing proteins of interest, genetically fused to fluorescent reporters (Hanson and Köhler, 2001; Nelson et al., 2007). The root meristem of Arabidopsis has a simple and transparent architecture and is widely used as a model system to study the localization and dynamics of fluorescent protein markers in vivo (Haseloff, 1999). The green fluorescent protein (GFP) marker, coupled with either conventional epifluorescence microscopy or confocal laser-scanning microscopy, has become a fundamental tool for plant cell biologists (Haseloff, 1999; Megason and Fraser, 2003; Held et al., 2008).
The genetically and functionally complex endomembrane system of plant cells may compensate the sedentary lifestyle of plants. Therefore, the intracellular compartments of plant cells fulfill a multitude of functions, including, storage of proteins, ions, and metabolites as well as biosynthesis and delivery of cell-wall precursors (Staehelin, 1997; Marty, 1999; Crowell et al., 2010). Thus, the plant vesicular trafficking network is crucial for plant development, signal transduction, and responses to biotic and abiotic stresses (Shimada et al., 1997; Shimada et al., 2002; Surpin and Raikhel, 2004).
Over the last years, various Arabidopsis transgenic lines expressing different fluorescent markers were mutagenized by ethyl methanesulfonate (EMS) and used in forward genetic screens to find novel regulators of protein trafficking or intracellular compartments integrity and function. Effective visualization of the subcellular phenotypes is critical to select mutants and, subsequently, functionally characterize genes responsible for the observed phenotypes. Fortunately, a wealth of molecular and genetic tools are available for Arabidopsis, Consequently, in this model organism, mutated genes can be nowadays efficiently identified by mapping or next-generation sequencing (Lukowitz et al., 2000; Austin et al., 2011).
Screening Strategies
One of the first fluorescence-based screening approaches successfully used Arabidopsis lines that expressed a fluorescent tonoplast marker, GFP:δ-tonoplast-intrinsic protein (TIP; Cutler et al., 2000; Avila et al., 2003). This genetic screen also made use of automated imaging by means of the Atto Pathway high-throughput confocal microscope system (Atto Bioscience, Rockville, MD, USA). The GFP:δ-TIP-mutagenized population was screened for broken or malformed vacuoles as well as mistargeting of the GFP:δ-TIP protein (Avila et al., 2003; Chary et al., 2008; Agee et al., 2010; Table 1).
Table 1.
Summarizes different screening strategies, the markers that have been used and their cellular localization.
| Background | Localization | Screened population | Phenotype | Mutant | Gene | Reference |
|---|---|---|---|---|---|---|
| GFP:δ-TIP | Tonoplast | 9,000 M2 seedlings | Abnormal, subcellular GFP:δ-TIP-labeled membrane structures, general disruption of the intracellular compartments | mvp1/gold36 | Gene encoding putative myrosinase-associated protein MyAP, member of a family of GDSL plant lipases | Agee et al. (2010), Marti et al. (2010) |
| Defects in the leaf epidermis shape and loss of pavement cell lobes | csp-1 | Gene encoding trehalose-6-phosphate synthase | Chary et al. (2008) | |||
| ST-GFP | Golgi apparatus | 10,000 M2 seedlings | Additional to Golgi stacks, large fluorescent globular structures in the cotyledons cells | g92/ermo2 | SEC24a locus, member of the coat protein complex of COPII vesicles responsible for anterograde transport from the ER to Golgi | Faso et al. (2009), Nakano et al. (2009) |
| Partial distribution of the Golgi marker to the ER, defects in the general ER protein export and ER organization. | gold36/mvp1 | Gene encoding putative myrosinase-associated protein MyAP, member of a family of GDSL plant lipases | Marti et al. (2010), Agee et al. (2010) | |||
| Defects in the mobility of Golgi stacks | gom8 | Gene encoding a putative GTPase, distant member of the dynamin superfamily | Stefano et al. (2012) | |||
| SP-GFP-HDEL | Endoplasmic reticulum | 1,746 M2 families | Disorganized ER morphology | ermo1/gnl1 | Guanine-nucleotide exchange factors for ADP-ribosylation factor GTPases (ARF GEF GNL1) | Nakano et al. (2009), Faso et al. (2009) |
| ermo2/g92 | SEC24a locus, member of the coat protein complex of COPII vesicles responsible for anterograde transport from the ER to Golgi | |||||
| SP-GFP-2SC | Endomembrane system | 12,000 M2 seedlings | Abnormal aggregation of the whole endomembrane | kam1/mur3 | Xyloglucan galactosyltransferase | Tamura et al. (2005), Madson et al. (2003) |
| Deformed endosomes and abnormal aggregates of various cellular organelles | kam2/gfs2/grv2 | Receptor-mediated endocytosis8 (RME8) | Silady et al. (2004), Fuji et al. (2007), Tamura et al. (2007) | |||
| secGFP | Secreted protein | 141,000 M2 seedlings | Enhanced intracellular GFP fluorescence | gnl1 | Guanine-nucleotide exchange factors for ADP-ribosylation factor GTPases (ARF GEF GNL1) | Teh and Moore (2007) |
| GFP–CT24 | Protein storage vacuoles in seeds | 3,000,000 M2 seeds | A vacuole-targeted GFP secreted into the extracellular space, causing the green fluorescence of the seeds | gfs2/grv2/kam2 | Receptor-mediated endocytosis8 (RME8) | Silady et al. (2004), Fuji et al. (2007), Tamura et al. (2007) |
| gfs10 | Unknown gene that encodes a novel membrane protein | Fuji et al. (2007) | ||||
| vsr1 | Vacuolar sorting receptor 1 VSR1 | Fuji et al. (2007) | ||||
| PIN1–GFP | Polarly localized, plasma membrane protein | 1,500 M1 families | Ectopic intracellular PM protein agglomeration | pat2 | AP-3 β putative subunits of adaptor complex | Feraru et al. (2010) |
| 1,920 M1 families | pat4 | AP-3 δ putative subunits of adaptor complex β | Zwiewka et al. (2011) | |||
| 25,550 M2 seedling | Resistant to inhibitory effect of BFA to exocytosis | ben1 | Homolog of ARF GEF, also known as MIN7/BIG5, which belongs to the BIG class of the ARF GEF subfamily. | Tanaka et al. (2009) | ||
| 3,500 M1 families | Resistant to inhibitory effect of auxin on endocytosis | doc1 | BIG//TIR3/UMB1/ASA1 encodes a huge member of the Calossin/Pushover family | Paciorek et al. (2005) |
Moreover, it includes genetic populations that have been screened, mutant phenotype, and the function of the gene characterized in the screen. For each characterized mutant, table contains also reference information.
Another, similarly designed forward genetics screen based on the mislocalization of a well-established marker of the Golgi apparatus, a rat sialyl transferase (ST)-GFP that consisted of a cytosolic tail and a transmembrane domain of a glycosylation enzyme fused to GFP (Boevink et al., 1998) allowed the identification of mutants with an altered distribution of the Golgi stacks in leaf epidermal cells. Interestingly, this screen of which the aim was the characterization of genes regulating the Golgi activity and integrity also contributed to the identification of regulators involved in protein exchange mechanisms between the endoplasmic reticulum (ER) and Golgi (Boulaflous et al., 2008; Faso et al., 2009; Marti et al., 2010; Stefano et al., 2012; Table 1).
An Arabidopsis transgenic line SP-GFP-HDEL that expressed the 2S albumin signal peptide (SP) fused to GFP and followed by the ER retention signal HDEL, has proven to be a useful tool for the visualization of the ER morphology within living cells (Mitsuhashi et al., 2000; Hayashi et al., 2001). In plant cells, the ER network is distributed evenly in the cytoplasm between the plasma membrane and the vacuolar membrane. Therefore, plant cells are a good model to observe the fine structure of these organelles by confocal microscopy. Such an Arabidopsis transgenic line with a fluorescently marked ER was chemically mutagenized and used in a screen for mutants with an abnormal ER organization (Nakano et al., 2009; Table 1).
To isolate mutants with an abnormal endomembrane structure within the cells, seeds of the transgenic Arabidopsis line SP-GFP-2SC were used for EMS mutagenesis. This marker line stably expressed a vacuole-targeting signal peptide of the 2S albumin of pumpkin (Cucurbita sp.) genetically fused to GFP (Mitsuhashi et al., 2000). In light-grown SP-GFP-2SC seedlings, the entire endomembrane system remained fluorescent, including the ER network and the dot-like structures of the Golgi complex, but not the vacuoles (Tamura et al., 2003). This screen resulted in the isolation and characterization of two katamari (Japanese word for aggregate) mutants, kam1 and kam2 (Tamura et al., 2005; Tamura et al., 2007; Table 1).
Another strategy in designing fluorescent based forward genetics screen for membrane trafficking regulators was to use secretory GFP (secGFP) line. As the ER retrieval signal had been deleted from the sequence of the ER-localized GFP, this protein was translocated from the lumen of the ER and transported to the apoplast (Wieland et al., 1987; Denecke et al., 1990; Martínez-Menárguez et al., 1999; Vitale and Denecke, 1999), where the GFP signal is largely quenched due to the acidic pH of the extracellular environment (Batoko et al., 2000; Zheng et al., 2004). Selection of seedlings displaying strong intracellular accumulation of secGFP allowed the identification of two allelic mutants in a previously unidentified guanine-nucleotide exchange factor for ADP-ribosylation factor GTPase (ARF GEF), component of secretion (Teh and Moore, 2007; Table 1).
To clarify the sorting machinery for protein storage vacuoles (PSVs) in seeds, an elegant screen has been designed. Putative vacuolar sorting-deficient mutants would produce strongly fluorescent seeds if vacuole-targeted GFP (GFP–CT24) were not degraded in the vacuole, but mislocalized (Nishizawa et al., 2003). For this purpose, the Arabidopsis GFP–CT24 line was EMS-mutagenized and the desired mutant seeds were easily identified by the strong green fluorescence caused by missorting of GFP–CT24 (Fuji et al., 2007; Table 1).
Other widely screened EMS population was generated by using the PINFORMED1 (PIN1–GFP) transgenic line expressing PIN1 chimeric protein under the native promoter (Table 1). The auxin efflux carrier PIN1 (Petrášek et al., 2006), is a well-characterized integral membrane protein that undergoes complex and constitutive subcellular dynamics (Geldner et al., 2001; Abas et al., 2006; Dhonukshe et al., 2007; Kleine-Vehn et al., 2008a,b). By using epifluorescence microscopy, the EMS-mutagenized PIN1–GFP population was screened for seedlings displaying an aberrant marker distribution, hinting at a defective intracellular protein trafficking (Feraru et al., 2010; Zwiewka et al., 2011). The PIN1–GFP mutagenized population was also successfully used to better understand the mechanism of constitutive PIN1 cycling. In this approach, the fungal toxin brefeldin A (BFA), an inhibitor of protein trafficking and recycling, was applied to select mutants with changed sensitivity to the drug (Tanaka et al., 2009).
Moreover, this mutagenized marker line was utilized to gain more insights into the pathway by which auxin affects trafficking. A genetic screen had been performed to find mutants resistant to the inhibitory effect of auxin on endocytosis (Paciorek et al., 2005).
The presented examples show that the screens based on changes in the localization or intensity of the fluorescent marker represent a powerful tool to explore the potential of forward genetics in Arabidopsis. This approach allows us to address cell biological questions and to uncover new regulators of protein trafficking and organellar integrity in the endomembrane system.
Technical Considerations for High-Throughput Imaging-Based Screening
Recent developments of high-throughput technology in life sciences provide quantitative profiling data for numerous biomolecules, such as gene transcripts, proteins, and metabolites from single cells, tissues, or whole organs. However, those technologies do not give spatial information about mentioned molecules. On the other hand, confocal laser microscopy and the use of fluorescent tags can reveal dynamics of molecules in vivo at cellular and subcellular resolution (Held et al., 2008; Salomon et al., 2010).
Generally, two main factors seem to be essential for the success of a forward genetics screen. The first factor is the specificity of the screening conditions that enable the identification of mutants involved in the desired process and limit selection of “uninteresting” mutants in the unrelated pathway. The second, and most important, factor is a clear phenotype needed to identify the mutants of interest and later map the mutation.
In forward, reverse, and chemical genetics studies, one is often interested in a specific phenotype based on changes in subcellular morphology, signal intensity, or location. Current standard approaches to these screens include manual microscopy, which is slow (Jorgensen and Mango, 2002), whereas high-throughput microscopy systems (Salomon et al., 2010) have insufficient automations and/or inadequate resolutions (Furlong et al., 2001; Crane et al., 2009). These are frequently a limiting factors in the cell biological screens based on microscopical observation of relatively variable and subtle changes in the subcellular distribution of the marker. Abovementioned aspects can impair the identification of the underlying gene. Therefore, automated systems for handling living plant samples during imaging and development of quantitative image-processing tools are essential to overcome these difficulties. Unfortunately, a reliable and uniformized setup is still missing for plant work.
Traditional strategies for the identification of mutations consist of two discrete steps: recombinant genotyping and candidate gene sequencing. Positional (or map-based) cloning is essentially an indirect approach: mapping narrows down the genetic interval that contains a mutation by successively excluding all other parts of the genome. Once an interval is defined, other criteria have to be employed to find out which of the genes are mutated within the interval. Naturally, the smaller interval or the higher mapping resolution, that is mainly determined by the size of the mapping population, makes this method effortless. Availability of the sequence information and access to basic molecular biology techniques vastly enhance molecular mapping output. As a consequence, it is possible to isolate the mutant and map the mutation within as little as a few months (Lukowitz et al., 2000).
The alternative to traditional strategies are next-generation sequencing technologies by which high-resolution genotypic information can be obtained by rapid and, nowadays, relatively cheap, whole-genome sequencing. Ideally, mutations underlying phenotypes of interest should be identified simply by sequencing the mutant genomes. Unfortunately, this is not possible because numerous unassociated polymorphisms segregate with the causative mutation in a mutagenized population with a very low signal to noise ratio. For this reason, most of the next-generation sequencing approaches overcome the background noise problem either by defining the genomic region of interest through prior genetic analysis or via bulk analysis of a very large number of mutant lines (Austin et al., 2011).
Once a mapping population is available, the short read analysis pipeline (SHOREmap) method allows a single investigator to identify a mutation within a bit more than the week, by the Illumina Genome Analyzer sequencing. Furthermore, the de novo marker prediction that is used in this approach can be an alternative for advanced genetics in many species not yet considered genetically traceable (Schneeberger et al., 2009). Unfortunately SHOREmap might be not robust enough when small mapping populations are used, with occasionally occurring false positives, which is typically the case when an interesting phenotype is difficult to score or when the organism of study is hard to propagate in large number. Thus, mapping of mutations with less obvious and more variable phenotype remains the major challenge for the fluorescent microscopy-based forward genetics approaches.
Signal Versus no Signal Screening Strategies
Clear phenotypes and, thus easy selection of the candidate individuals are crucial aspects, not only for identification of the desirable mutants, but also particularly for the later mapping of the mutation by positional cloning. On the cellular level, the presence or absence of the fluorescent signal is the easiest feature to distinguish, thereby also radically reducing the possibility of selecting false positive individuals.
Secretory GFP screen
Secretory GFP-mutagenized populations have been used to develop membrane trafficking assays based on the differential fluorescence intensity of secreted and retained GFP proteins (Teh and Moore, 2007). The secGFP line was created from the ER-localized GFP version (Haseloff et al., 1997), by replacing its C-terminal ER retention signal with a sequence encoding the C-Myc epitope. As a consequence, the protein is transported to the apoplast where it generates a weak fluorescence signal (Batoko et al., 2000; Zheng et al., 2004; Figure 1A).
Figure 1.
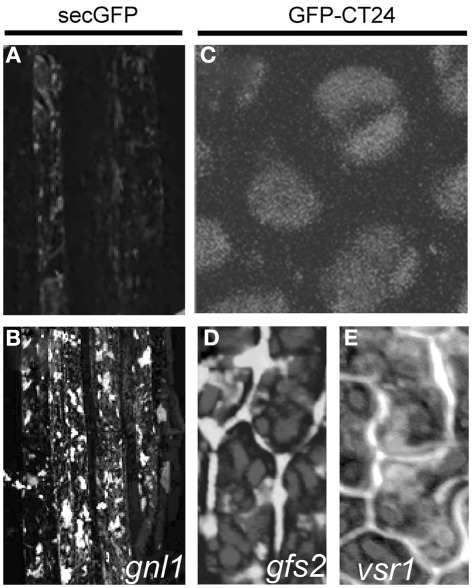
Cellular phenotype of the mutants selected based on the presence of fluorescent signal. Arabidopsis secGFP transgenic line (A) showing weak fluorescence of secreted GFP and (B) pronounced signal accumulation in the root cells of gnl1 mutant. Adapted from Teh and Moore (2007). Wild type GFP–CT24 seeds (C), with the vacuole-targeted marker, signal completely merged with the autofluorescence of PSVs. Mistargeting and secretion into the extracellular space of GFP–CT24 in gfs2 (D) and vsr1 (E) mutant seeds. Adapted from Fuji et al. (2007).
Using a epifluorescence microscope, 141,000 M2 seedlings were screened for enhanced intracellular GFP fluorescence. As a result, two mutants that carried allelic mutations causing the accumulation of secGFP in spheroid bodies were isolated (Figure 1B). Positional cloning and sequencing identified stop codons in the gene encoding an ARF GEF, the nucleotide exchange factor for ARF GTPases, closely related to the Arabidopsis ARF GEF GNOM. Therefore, the mutants were designated Gnom-like1 (Table 1). Both genes, GNOM and GNOM-like1 belong to ancestral GBF1 family of ARF GEFs, known to be involved in the ER to Golgi trafficking in mammals and yeast (Geldner et al., 2003; Gillingham and Munro, 2007). Nevertheless, GNOM has been shown to be essential for recycling of endosomes to the plasma membrane (Steinmann et al., 1999; Geldner et al., 2003) and also to function in the regulation of endocytosis (Naramoto et al., 2010), but not to have a pronounced role in the secretory pathway (Geldner et al., 2003; Richter et al., 2007).
GNL1 has a role in maintaining the Golgi organization, because the gnl1 mutants exhibited a striking increase in the size of the Golgi apparatus stacks and, after BFA treatment, the integral Golgi markers were rapidly transferred to the ER (Teh and Moore, 2007). The GNL1 gene has diverse functions in vesicle coat formation and vesicle–cytoskeleton interactions (Geldner, 2004; D’Souza-Schorey and Chavrier, 2006). Interestingly, the intracellular accumulation of secGFP in gnl1 mutants suggested that this close relative of GNOM is functional on a secretory trafficking pathway.
GFP–CT24 screen
Similar criteria were applied to distinguish mutant phenotypes in the screen for new molecular players that function in vacuolar sorting of storage proteins to PSVs. The Arabidopsis transgenic line used for the screen expressed the GFP–CT24 construct, consisting of a GFP followed by the 24 amino acids from the C-terminal end of the storage protein precursor, β-conglycinin (CT24), under the control of a seed-specific promoter (Nishizawa et al., 2003). In wild type seeds, the GFP–CT24 signal was completely merged with the red autofluorescence of PSVs (Figure 1C), showing that GFP–CT24 was correctly sorted and transported by the vacuolar sorting receptor 1 (VSR1) into PSVs. In planta, only two factors, VSR1 and the homolog of the yeast retromer component VPS29, which is involved in recycling of VSR1 (Shimada et al., 2006), have been shown to function in vacuolar sorting of storage proteins to PSVs. Both the vsr1 and vps29 mutants after crossing with GFP–CT24, secreted a vacuole-targeted marker into the extracellular space, where GFP was stabilized, causing the green fluorescence of the seeds. These results suggested that also other mutants deficient in vacuolar sorting would produce green fluorescent seeds.
Following this idea, transgenic Arabidopsis GFP–CT24 seeds were mutagenized with EMS to obtain populations suitable for screening. By means of a fluorescence stereomicroscope, more than 100 fluorescent seeds were easily selected from 3,000,000 of the mutagenized M2 seeds and 10 green fluorescent seeds (gfs) mutants have been described (Fuji et al., 2007). Molecular characterization revealed that the gfs2 mutant, showing defects in the proper targeting of the GFP marker (Figure 1D), carries the mutation in the same gene as the gravitropism defective 2 (grv2) mutant, previously characterized as deficient in shoot gravitropism and phototropism (Silady et al., 2004), and Arabidopsis kam2 mutant with deformed endosomes and abnormal aggregates of various cellular organelles (Tamura et al., 2007). The GFS2/GRV2/KAM2 gene encodes a homolog of a DnaJ domain-containing the receptor-mediated endocytosis 8 (RME8) that is required for endocytosis and the organization of endosomes in Caenorhabditis elegans (Zhang et al., 2001), Drosophila melanogaster (Chang et al., 2004), and human (Girard et al., 2005; Table 1). It is possible that the DnaJ domain-containing GFS2/GRV2/KAM2 is involved in the folding of protein(s) needed for the proper formation of endosomes that mediate protein trafficking to PSVs and the biogenesis of endosomal compartments, including PSVs (Fuji et al., 2007; Tamura et al., 2007).
Fine-mapping and DNA sequencing of the gfs10 mutant revealed a single base pair mutation in the gene that encodes a novel membrane protein of unknown function with 10 putative transmembrane domains (Table 1). As this protein is specific to higher plants, plants appear to have a novel mechanism for the vacuolar sorting of storage proteins. The phenotype of the gfs10 mutant was very similar to that of the vacuolar sorting mutants vsr1 (Shimada et al., 2003), vps29 (Shimada et al., 2006), and gfs2/grv2/kam2 (Fuji et al., 2007; Tamura et al., 2007). Interestingly, four out of 10 gfs mutants characterized in the screen were mapped as different alleles of vsr1 (Figure 1E). These findings imply that VSR1 might function primarily in vacuolar sorting of storage proteins in maturing seeds, in accordance with VSR1 being one of the seven most abundant VSRs in seeds. Further identification of the responsible genes of the other gfs mutants should provide valuable insights into the complex molecular mechanisms underlying the vacuolar sorting of storage proteins (Fuji et al., 2007).
Screening Strategies Based on Changes in Subcellular Distribution
The fluorescent protein technology applied to live imaging of plant cells helped greatly in providing subcellular markers for the study of the complex spatial and temporal relationships among intracellular endomembrane compartments. The changes in subcellular distribution of various markers have been used as a basis for forward genetics screens.
GFP:δ-TIP screen
One of the first microscopy-based forward genetics screen aimed at identifying mutants with defects in vacuolar biogenesis. Effective visualization of the vacuolar structure was possible by using Arabidopsis transgenic lines constitutively expressing a fluorescent tonoplast marker, GFP:δ-TIP (Cutler et al., 2000; Figure 2A). By means of confocal microscopy, ∼9,000 M2 seedlings were screened for distorted vacuoles based on mistargeting of the GFP:δ-TIP or related phenotypes (Table 1). Originally, from obtained 211 putative mutants with defective vacuole biogenesis, 110 died before setting seed and/or did not produce any seeds, resulting in a population of 101 putative vacuolar mutants (Avila et al., 2003).
Figure 2.
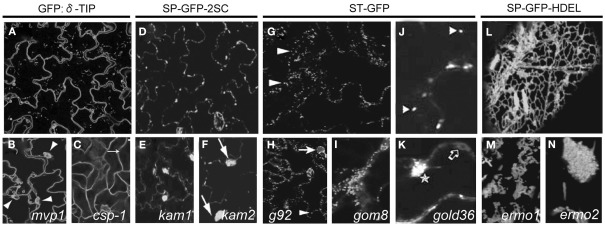
Cellular phenotype of mutants selected based on subcellular distribution of fluorescent signal. Arabidopsis GFP:δ-TIP transgenic line (A) constitutively expressing a fluorescent tonoplast marker. mvp1 mutant (B) accumulating the GFP:δ-TIP fusion protein in static aggregates (arrowheads). Adapted from Agee et al. (2010). csp-1 mutant (C) with defects in the leaf epidermis cell shape and loss of pavement cell lobes (arrow). Adapted from Chary et al. (2008). Arabidopsis line expressing vacuole-targeted GFP-2SC (D), showing GFP fluorescence in entire endomembranes. kam1 (E) and kam2 (F) mutants displaying large aggregates of the endomembranes (arrows). Adapted from Tamura et al. (2005) and Tamura et al. (2007). The ST-GFP Arabidopsis line expressing Golgi marker (G) at the bigger magnification (J). Fluorescent spots correspond to Golgi stacks (arrowheads). Adapted from Faso et al. (2009) and Marti et al. (2010). Mutant g92 (H) showing additional to Golgi stacks (arrowhead), large fluorescent globular structures (arrow). Adapted from Faso et al. (2009). gom8 (I) displaying large static, structures that include aggregates of Golgi stacks and fewer ST-GFP-positive membranes in the cortical region. Adapted from Stefano et al. (2012). ST-GFP marker in the gold36 mutant (K) partially distributed to intracellular network, accumulating in circular structures (empty arrow) and large globular structures (star). Adapted from Marti et al. (2010). Arabidopsis line SP-GFP-HDEL (L) displaying fluorescence of ER network. Mutants ermo1 (M) and ermo2 (N) revealing a number of strongly GFP-labeled spherical structures. Adapted from Nakano et al. (2009).
The modified vacuole phenotype-1 (mvp1) mutant isolated from the screen displayed more static than the wild type, subcellular GFP:δ-TIP-labeled membrane structures and a number of vacuole-related phenotypes (Figure 2B). Studies with other cellular markers indicated a general disruption of the intracellular compartments, including the ER, Golgi, and mislocalization of the plasma membrane cargo. This observation suggested that MVP1 had a more general role early in the endomembrane system and that loss of MVP1 function might detain the proteins destined for other compartments in subcellular aggregates. Positional cloning revealed that the mvp1 mutation was located in a gene coding for a putative myrosinase-associated protein MyAP, which is a member of a large family of GDSL plant lipases. Pull-down assays demonstrated that MVP1 interacted specifically with the Arabidopsis myrosinase protein, thioglucoside glucohydrolase2 (TGG2). MVP1 was proposed to be functional, in part, in the correct localization of the myrosinase TGG2 and the prevention of inappropriate glucosinolate hydrolysis that could generate cytotoxic molecules (Agee et al., 2010).
Because vacuoles occupy most of the volume of plant cells, the tonoplast marker delineates cell shape, allowing the identification of the cell shape phenotype-1 (csp-1) mutant with defects in the leaf epidermis shape and as a consequence, loss of pavement cell lobes (Figure 2C). The gene was annotated in the Arabidopsis genome database as a trehalose-6-phosphate synthase. Enzymatic activities of the CSP-1 protein are necessary for the control of the plant architecture, epidermal pavement cell shape, trichome branching, and other developmental processes by generating a sugar-based signal or, more directly, by participating in transcriptional regulation (Chary et al., 2008).
GFP-2SC screen
Various aspects of plant development and signal transduction rely on the structural and functional maintenance of endomembranes (Surpin and Raikhel, 2004). The transgenic Arabidopsis line expressing vacuole-targeted GFP-2SC, showing GFP fluorescence in entire endomembranes (Tamura et al., 2003), was used as a tool to clarify the molecular mechanisms that regulate endomembrane organization in plants (Figure 2D).
To isolate Arabidopsis mutants showing abnormal endomembrane structure within the cells, GFP-2SC seeds were EMS-mutagenized and about 12,000 M2 seedlings were examined with the fluorescence microscope. This screen resulted in characterization of two mutants, designated katamari (kam1 and kam2) whose endomembrane formed large aggregates (Figures 2E,F). These aggregates, were composed of many small particles indicating fragmentation or deformation of the endomembranes. They were found in most of the cells of cotyledons and hypocotyls as well as in roots, root hairs, and trichomes (Tamura et al., 2005; Tamura et al., 2007).
By using map-based cloning approach, mutation in the kam1 mutant was allocated in the gene encoding a polypeptide sequence with a single putative transmembrane domain and an exostosin-like domain having glycosyltransferases activity (Table 1). Previously described mutant in the same locus, mur3 lacked glycosyltransferase activity (Madson et al., 2003). Interestingly, mur3 mutant exhibited normal endomembrane organization when expressed the vacuole-targeted GFP in the cells which indicates that glycosyltransferase activity is not involved in the organization of the endomembranes.
Isolated kam1 mutant showed abnormal actin distribution and treatment with the actin-depolymerizing reagent, Latrunculin B caused GFP-2SC cells to develop a kam1-like phenotype. This implies that KAM1/MUR3 interacts with actin and regulates the proper distribution of actin filaments which is known to be essential for endomembrane organization and cell elongation in wild type plants (Tamura et al., 2005).
Mapping and DNA sequencing of kam2 mutant showed a single base pair mutation in the RME8 gene (Table 1). This gene was reported to be responsible for the grv2 and gfs2 mutant phenotype (Silady et al., 2004; Fuji et al., 2007). KAM2/GRV2/GFS2 has a single DnaJ domain, known to interact with heat shock protein 70 (Hsp70), a chaperone essential for proper folding of substrate proteins (Miernyk, 2001). These results implied that KAM2/GRV2/GFS2 is involved in the folding of proteins needed for membrane trafficking in Arabidopsis.
Disruption of the formation of endosomal compartments by kam2 mutations caused a depletion of the cytosolic or membrane proteins needed for vesicular trafficking resulting in blocking of vacuolar trafficking. Moreover, in seeds, large amount of precursor form of storage proteins are synthesized on ER and delivered to the PSVs, were the precursors are converted into the mature form (Shimada et al., 2003). kam2 seeds abnormally accumulated the precursors, similarly to seeds of vsr1 that lacks a vacuolar sorting receptor for storage proteins (Shimada et al., 2003). This result suggests that kam2 has a defect in the vacuolar sorting of storage proteins in maturing seeds (Tamura et al., 2007).
Despite the similar cellular phenotype, the kam2 aggregates differ from the aggregates of kam1. The kam2 aggregates contained normal ER bodies, while the kam1 aggregates contained completely deformed ER bodies. kam2 had sheet-like structures of endosomes on the cell surface that was responsible for vesicular trafficking, which have not been seen in kam1. kam2 had normal organization of actin filaments, while kam1 exhibited the disorganization of actin filaments. These differences between the kam1 and kam2 mutants suggest that the maintenance of endomembrane structure is supported by multiple mechanisms (Tamura et al., 2005; Tamura et al., 2007).
ST-GFP screen
Another approach to identify genes that regulate the function and morphology of the Golgi utilized the EMS-mutagenized ST-GFP, the Arabidopsis line expressing the cytosolic tail and transmembrane domain of a rat sialyl transferase fused to GFP (Boevink et al., 1998; Figures 2G,J). The confocal analysis of ∼10,000 individuals from the M2 generation resulted in the discovery of 219 mutants with clear alterations in the Golgi marker distribution in leaf epidermal cells.
Screening of ST-GFP mutagenized line resulted in identification of the mutant, named g92, that showed additional to Golgi stacks, large fluorescent globular structures in most of the cells of the cotyledons (Figure 2H). Interestingly there was no obvious developmental differences between the g92 mutant compared with non-mutagenized ST-GFP (Figures 2G,J).
The affected gene had been identified by map-based cloning and DNA sequencing approach. g92 mutant carried single base pair mutation in one of the three isoforms of the COPII coat component, Sec24a the same as in ermo2 mutant, identified in fluorescence-based screen to characterized new players involved in proper ER organization (Nakano et al., 2009; Table 1). Novel mutation in the COPII subunit Sec24a partially affected the morphology of the ER tubular network as well as the Golgi membrane and soluble secretory protein distribution. The main effect of the Sec24a allele was a partial deformation of the ER network that created a localized membranes cluster containing proteins destined to other compartments. Therefore, those results indicated that in addition to protein export from the ER, COPII machinery is also required for maintenance of the ER tubular network (Faso et al., 2009).
One of the characterized mutant, designated Golgi defects 36 (gold36), showed large intracellular accumulation of the Golgi marker ST-GFP, its partial distribution to the ER (Figure 2K) and also partial retention of the soluble marker destined to the apoplast, secRFP (Faso et al., 2009) in the ER, indicating defects in the general ER protein export and ER organization. The mutation causing the gold36 phenotype was located in the same gene as in the mutant mvp1 (Agee et al., 2010; Table 1). Detailed studies indicate that GOLD36 is a protein destined to post-ER compartments. Nevertheless, the mutant phenotype was linked to a reduced amount of GOLD36 or the absence of the protein in the ER, leading to the speculation that GOLD36 is a factor that, while on its way to final destination, might influence the ER integrity. This model proposes that GOLD36 participates in the maintenance of ER integrity either by working as a chaperone for other proteins that are necessary for the ER integrity or by binding and reducing the activity of proteins destined to non-ER compartments that may have deleterious effects in the ER (Marti et al., 2010).
Another mutant isolated in this screen, Golgi movement8 (gom8) displayed defects in the mobility of Golgi stacks (Figure 2I). The mutation responsible for the gom8 phenotype was located in the ROOT HAIR-DEFECTIVE 3 (RHD3) gene, encoding a putative GTPase, distant member of the dynamin superfamily (Wang et al., 1997; Table 1). Detailed studies showed that mutation in the RHD3 hairpin loop domain provoked the accumulation of the mutant protein into large structures and, thus, reduced the availability of putative GTPases over the ER. Proper subcellular distribution of RHD3 is important for maintenance of the ER morphology, but not for tubule fusion in the peripheral ER (Stefano et al., 2012).
This screen was aimed at discovering new factors that regulate biogenesis, organization, and function of a vital organelle, namely the Golgi apparatus in a multicellular context. Some processes that regulate protein traffic along the secretory pathway appear to be conserved among different systems, therefore, this approach may also contribute to a better understanding of how different cell systems have evolved (Boulaflous et al., 2008).
SP-GFP-HDEL screen
Among organelles in eukaryotic cells, the ER has the largest surface area and forms diverse structures. The complex ER morphology supports various cellular functions, including synthesis of proteins and lipids, maintenance of calcium homeostasis, and quality control of proteins (Okita and Rogers, 1996; Staehelin, 1997; Baumann and Walz, 2001).
Because the molecular mechanisms responsible for the organization of these compartments is largely unknown, a forward genetics approach was used to identify novel factors responsible for the organization and maintenance of ER structures. For this purpose, the transgenic Arabidopsis line, designated SP-GFP-HDEL (Figure 2L), expressing the signal peptide of the pumpkin 2S albumin and GFP, followed by an ER retention signal HDEL (Mitsuhashi et al., 2000; Hayashi et al., 2001), was chemically mutagenized. The subcellular phenotypes of seedlings from the 1,746 M2 families were checked with a fluorescence microscope. Two recessive mutants designated ermo1 and ermo2 (endoplasmic reticulum morphology) with a disorganized ER morphology were isolated (Nakano et al., 2009).
Both, ermo1 and ermo2 developed a number of strongly GFP-labeled spherical structures that were not seen in the parental SP-GFP-HDEL (Figures 2M,N). Additionally, both mutants developed a normal polygonal ER network. More detailed studies showed that these compartments were derived from the ER and did not result of an abnormal accumulation of the coat protein COPII vesicles or some other post-ER compartments. Moreover, these compartments did not colocalize with any subcellular marker.
Map-based cloning and DNA sequencing identified a nonsense mutation in the GNL1 locus of ermo1 and in the SEC24a locus of ermo2. Interestingly, also g92 mutant, fund in the screen for regulators of Golgi apparatus distribution was carrying mutation in the same gene locus (Faso et al., 2009; Table 1). Yeast and mammalian homologs of both GNL1 and SEC24a are involved in membrane trafficking between the ER and Golgi (Nakano et al., 2009). The ermo2 phenotype was caused by a point mutation in one of the cargo-binding sites to recruit them into the COPII vesicles. This observation suggests impairment in the packaging of some specific cargos in ermo2/g92, because ERMO1/GNL1 functions in the first step of COPI vesicle budding, hinting at a less specific interaction with cargo proteins. However, defective retrograde transport may also cause partial defects in anterograde transport. As ERMO1/GNL1 contributes to retrograde transport (Teh and Moore, 2007), the protein export from the ER might also be partially defective in the ermo1/gnl1 mutants.
Enhanced defects in the ermo1/ermo2 double mutant suggest that when ERMO1/GNL1 and ERMO2/G92 are both defective, the SEC24a-dependent pathway is blocked and the function of two other SEC24 homologs is partially disrupted because of the defective retrograde transport. Furthermore, ERMO1/GNL1 and ERMO2/G92 might be involved in transport of some specific proteins, which are required for the organization of the ER morphology (Nakano et al., 2009).
PIN1–GFP screen
To identify new regulators of protein trafficking pathways, a fluorescence imaging-based forward genetics screen was carried out to find mutants affecting the subcellular trafficking of the PIN1 auxin efflux carrier (Petrášek et al., 2006) that drives the auxin transport in a polar direction (Wisniewska et al., 2006). The polar localization of PIN proteins (Figure 3A) changes dynamically during plant development (Benková et al., 2003; Friml et al., 2003; Sorefan et al., 2009) and in response to environmental signals (Friml et al., 2002). The PIN proteins are constitutively shuttling between the plasma membrane and endosomes, providing a possible mechanism for rapid relocation of PIN proteins (Geldner et al., 2001). The PIN subcellular movement involves elaborated subcellular dynamics, including secretion (Dhonukshe et al., 2008), clathrin-mediated endocytosis (Dhonukshe et al., 2007), polar recycling (Geldner et al., 2001; Kleine-Vehn et al., 2008a), and trafficking to the vacuole (Abas et al., 2006; Kleine-Vehn et al., 2008b). Due to its complex dynamics, PIN1–GFP is a suitable tool for identification of regulators in multiple trafficking pathways.
Figure 3.
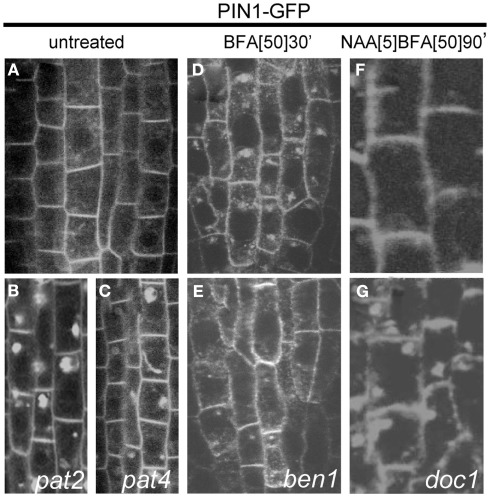
Cellular phenotype of the mutants selected in the screens performed on PIN1–GFP mutagenized population. Plasma membrane, polar localization of PIN1–GFP protein (A) in the steel of Arabidopsis root cells. The mutants pat2 (B) and pat4 (C) displaying ectopic accumulation of PIN1–GFP marker. Adapted from Feraru et al. (2010) and Zwiewka et al. (2011). Wild type line (D) accumulating PIN1–GFP in the BFA compartments. Root cells of ben1 mutant (E) showing smaller BFA-induced internalization. Adapted from Tanaka et al. (2009). In wild type line (F) synthetic auxin NAA blocks PIN1 internalization. doc1 mutant (G) shows resistance to the inhibitory effect of auxin on the endocytosis of PIN1. Adapted from Paciorek et al. (2005).
Using EMS-mutagenized PIN1–GFP population, three independent protein-affected trafficking (pat) mutants were identified from ∼500 M1 families (Feraru et al., 2010). Further screening progenies of 1,920 M1 families, resulted in identifying other additional allele of the pat mutant – pat4 (Zwiewka et al., 2011; Table 1).
pat2 and pat4 are recessive mutants with strong defects in the subcellular trafficking of PIN1–GFP. Besides the normal, basal localization of the PIN1 protein in root stele cells, all these mutants display a strong intracellular accumulation of GFP (Figures 3B,C). Similarly, another polar plasma membrane protein PIN2 (Xu and Scheres, 2005) as well as an apolar aquaporin PIP2 (Cutler et al., 2000), and brassinosteroid receptor BRI1 (Friedrichsen et al., 2000; Geldner et al., 2007) aggregated in intracellular compartments in the pat2 and pat4 mutants. Also the localization of soluble proteins, such as aleurain (Di Sansebastiano et al., 2001), was affected in pat2. Similarly to pat2, pat4 displayed a vacuolar protein accumulation and an abnormal vacuole (lytic and PSV) morphology, but was not defective in protein trafficking to the plasma membrane, recycling, and early endocytosis (Feraru et al., 2010; Zwiewka et al., 2011). Moreover, the pat2 and pat4 mutants grown on medium lacking sucrose showed arrested phenotype correlated with seed age and after long storage, the mutant seeds had a dramatically reduced germination capability, resembling mutants defective in vacuolar function (Feraru et al., 2010; Zwiewka et al., 2011). The molecular characterization of pat2 and pat4 mutants revealed that they were defective in the AP-3 β and AP-3 δ putative subunits of the same adaptor complex. The AP-3 complex have important function in membrane trafficking (Lee et al., 2007; Niihama et al., 2009). Importantly, biochemical analysis confirmed the genetic studies and identified the whole AP-3 complex in Arabidopsis (Zwiewka et al., 2011). Overall, the data indicate that the AP-3 adaptor complex plays a role in the vacuolar biogenesis in Arabidopsis, including mediation of the transition between storage and lytic vacuolar identity.
The pronounced intracellular accumulation of diverse proteins had surprisingly little impact on plant development and physiology. Therefore, usage of the fluorescent microscope as a tool to screen for cellular phenotypes was a successful strategy to reveal new genes involved in trafficking pathways in plants.
Screening Strategies with Trafficking Inhibitors
Some screens for the identification of trafficking mutants combined subcellular localization of fluorescent markers and treatment with inhibitors of different trafficking steps. Specifically, the PIN1–GFP transgenic lines were treated with BFA, which is one of the most useful tool to study vesicle-mediated trafficking in eukaryotic cells (Klausner et al., 1992; Gaynor et al., 1998; Nebenführ et al., 2002; Langhans et al., 2011). BFA interferes with the function of the GEFs that catalyze the activation of the small GTPase ARF on membrane surfaces. The membrane-bound, active form of ARF, ARF–GTP plays a crucial role in vesicle formation (Donaldson and Jackson, 2000; Vernoud et al., 2003; D’Souza-Schorey et al., 2006; Grosshans et al., 2006). BFA rapidly releases the vesicle coat proteins into the cytosol and prevents further vesicle formation (Donaldson et al., 1990; Robinson and Kreis, 1992; Ritzenthaler et al., 2002). As a consequence, the integrity of several subcellular compartments is severely perturbed by this drug (Langhans et al., 2011).
In Arabidopsis root tissues, recycling of the PIN1 protein to the basal side of vascular cells is highly sensitive to BFA, but its endocytosis is relatively resistant to this compound (Geldner et al., 2001). Consequently, treatment with BFA of Arabidopsis roots results in intracellular accumulation of PIN1 proteins in agglomerated endomembrane compartments, designated “BFA compartments” (Geldner et al., 2003; Figure 2D). The target of BFA in terms of PIN1 recycling is ARF GEF GNOM. GNOM-dependent trafficking has been demonstrated to be required for targeting the PIN1 protein to the basal side of the plasma membrane. Moreover, prolonged treatment with the drug provokes the disappearance of PIN1–GFP from the BFA compartment and its ectopic appearance at the apical site of the cell (Kleine-Vehn et al., 2008a).
To better understand the mechanism of the constitutive cycling of PIN1 and to identify new molecular players regulating this process, forward genetics screen has been designed on BFA-treated PIN1–GFP seedling roots (Tanaka et al., 2009). The screening conditions have been set to select mutants insensitive to BFA.
EMS-mutagenized PIN1–GFP population was screened for mutants that did not efficiently internalize PIN1–GFP and/or accumulate it in the BFA compartments after 2 h of BFA treatment. After 25,550 M2 seedling roots had been screened by epifluorescence microscopy, three mutants were identified, in which BFA did not induce the typical agglomeration of the internalized PIN1–GFP (Figure 3E), in comparison to the wild type seedlings (Figure 3D). These mutant lines were designated BFA-visualized endocytic trafficking defective1 (ben1), ben2, and ben3. The ben1 mutant accumulates less PIN1–GFP proteins in the BFA compartment and the BEN1 gene encodes a homolog of ARF GEF, also known as MIN7/BIG5, which belongs to the BIG class of the ARF GEF subfamily (Table 1). Consistent with its supposed role in early endosomal trafficking, the BEN1 protein localizes to the TGN/EE. This analysis also supports previous findings that early endosomal trafficking is involved in polar localization of PIN proteins, plant growth, and development, because the ben1 knockout mutants show clear, albeit only minor, defects in these aspects (Tanaka et al., 2009). This observation implied that early endosomal trafficking involving an ARF GEF BEN1/MIN7 function is a critical point in establishing and/or maintaining polar PIN localization (Tanaka et al., 2009).
One of the most challenging screens aiming at finding new players involved in intracellular trafficking was done after the breakthrough discovery that the plant hormone auxin can inhibit endocytosis of its own transporters. More detailed studies with the endocytic tracer FM4-64 showed that this phenomenon is more general and concerns the plasma membrane and a number of endogenous and heterologous cargos (Paciorek et al., 2005; Robert et al., 2010).
To visualize the inhibitory effect of auxin on the endocytosis of PIN proteins, PIN1–GFP seedlings from a mutagenized population were first treated with synthetic auxin, naphthalene acetic acid (NAA), and then coincubated with NAA and BFA. From ∼3,500 M1 families, eight candidates were identified. One mutant have been closely characterized that under these conditions, showed resistance to NAA, displaying normal internalization of PIN1–GFP into BFA compartments (Figure 3G), whereas in wild type plants, the BFA-induced PIN1 internalization was inhibited (Figure 3F).
One of the mutants was allelic to transport inhibitor response3 (tir3) that had originally been isolated in a screen for resistance to auxin transport inhibitors (Ruegger et al., 1997). The corresponding gene, designated BIG (formerly DOC1/TIR3/UMB1/ASA1) is known to encode a huge member of the Calossin/Pushover family (Gil et al., 2001; Table 1). In Arabidopsis, BIG is a single-copy gene present also in other plant and animal genomes. Mutations in the BIG protein causes partial resistance to the auxin-inhibitory effect on endocytosis. This protein might be involved in subcellular vesicle trafficking, because it affects the subcellular localization of PIN1 and other plasma membrane proteins, such as PIN2 and plasma membrane-ATPase (Paciorek et al., 2005). There is no experimental support for either a connection between the BIG-dependent auxin signaling pathway inhibiting endocytosis and the previously characterized Skp1/Cullin/F-box protein Transport Inhibitor Response 1-related auxin signaling regulating gene expression (Kepinski and Leyser, 2002; Dharmasiri and Estelle, 2004) or a requirement for gene transcription or protein synthesis (Robert et al., 2010). Unfortunately, this screen did not reveal which molecular pathway auxin uses to exhibit its effect on endocytosis.
Recent studies showed that auxin-mediated inhibition of the PIN protein internalization from the plasma membrane occurs within minutes. These observations imply that non-transcriptional auxin signaling interferes specifically with the general process of clathrin-mediated endocytosis in plant cells. The putative auxin receptor, auxin binding protein 1 (ABP1) seems to be a plant-specific, positive regulatory element of the clathrin-mediated endocytic mechanism. Because ABP1 binds auxin with high affinity (Jones, 1994; Napier et al., 2002), it is proposed that auxin mediates its effect on clathrin-mediated endocytosis via ABP1. Thus, the auxin effect on endocytosis might use a previously unknown and, thus far, molecularly uncharacterized signaling pathway that does not involve the regulation of gene expression.
Conclusion and Outlook
Completion of the Arabidopsis genome sequence allowed the classical genetic screen method to become a powerful tool for function determination of over 25,000 Arabidopsis genes. This ability to map new genes resulted in a rapid exploitation of natural variation and has proven to possess enormous potential in broadening the understanding of plant growth, development, and evolution.
Fluorescent marker-based forward genetics screens performed so far, have demonstrated their efficiency in identifying novel components of subcellular trafficking or in placing the known regulators in another functional context. Previous reports confirmed the robustness of the EMS mutagenesis approach, followed by confocal microscopy of stable Arabidopsis transformants expressing GFP fusions of tonoplast-localized proteins (Avila et al., 2003), soluble vacuolar cargos (Tamura et al., 2005; Tamura et al., 2007), Golgi-localized enzymes (Boulaflous et al., 2008), or secreted proteins (Teh and Moore, 2007). These powerful strategies allowed the isolation of several mutants with an aberrant distribution of vacuolar, secreted, and ER-retained GFP markers that led to the elucidation of secretory pathway components (Table 1).
The recent extensive studies employing molecular tools such as, endosomal markers, dyes, and advanced imaging techniques and genetic studies, enhanced our knowledge about trafficking in plants. Moreover, identification of early and late endosomal compartments as well as molecular dissection of more than one type of vacuole, clearly indicate that the plant endomembrane system is very complex.
Results of fluorescent based forward genetic screens provided evidence that the secretory and the endocytic machinery are tightly connected.
The fact that the different alleles of the same genes were characterized in screens based on different marker lines (Table 1), indicates that the plant endosomal system is not a static set of clearly separable structures but characterized by the dynamic formation and decomposition, of membrane compartments that are derived from each other by maturation.
The screening utilizing the PIN1–GFP marker line and epifluorescence microscopy was aiming to identify mutants showing defects in function of important components of the subcellular trafficking machinery. Nevertheless, so far, all characterized pat mutants from this screen are related rather to vacuolar function or trafficking to the vacuole, but not to the other PIN1 trafficking processes (Feraru et al., 2010; Zwiewka et al., 2011; Table 1). It is possible that other pat mutants will be affected in these processes or that design of the screen was biased toward strong accumulation of GFP due to a defect in lytic degradation.
Combination of the PIN1–GFP-based approach and treatments with the trafficking inhibitors BFA and NAA intended to identify the mutants defective in endocytic step of protein trafficking and the molecular mechanism underlying the auxin effect on endocytosis.
Although, BFA-visualized ben1 carried a mutation in a previously uncharacterized GTP exchange factor on the ARF component of the early endosomal trafficking (Tanaka et al., 2009), unluckily, extensive research revealed that big mutations led to a weak physiological resistance to auxin. So far, experimental data do not support a direct involvement of the BIG protein in the auxin signaling pathway, but rather in more general intracellular trafficking processes (Kanyuka et al., 2003).
From the examples given above, fluorescence imaging-based forward genetics screens can certainly be an excellent tool for the identification of mutants that alter essential trafficking pathways, such as secretion, protein intracellular cycling, sorting of storage proteins as well as vacuole biogenesis and function (Fuji et al., 2007; Teh and Moore, 2007; Tanaka et al., 2009). However, it is important to keep in mind that due to high complexity and dynamicity of the plant endomembrane systems as well as limitations in the imaging techniques, screens based on fluorescent markers localization may bring some difficulties to obtain desired mutants despite their eligible phenotype (Paciorek et al., 2005; Feraru et al., 2010; Zwiewka et al., 2011). Moreover, fluorescent technology combined with genetics screen enabled the more detailed characterization of new molecular regulators of cell shape establishment, maintenance, morphology, and function of cellular compartments, and biogenesis of endosomal compartments (Tamura et al., 2007; Boulaflous et al., 2008; Chary et al., 2008; Nakano et al., 2009; Agee et al., 2010; Marti et al., 2010; Stefano et al., 2012; Table 1).
Classical genetic approaches to study the function of endomembrane system are subject to serious limitations and, specifically, vesicular trafficking concern gametophyte or embryo lethality. In Arabidopsis, it is well known that gene redundancy and loss-of-function mutations frequently leads to failure in producing observable phenotypes. To overcome these limitations, chemical genomics approaches have been initiated to facilitate better understanding of endomembrane trafficking.
Chemical genomics methods are based on the ability of low molecular mass molecules to modify the activity of proteins or pathways and confound only one of several functions of a protein which would be difficult to achieve through gene-based perturbation alone (Kuruvilla et al., 2002; Zouhar et al., 2004; Robert et al., 2008). Furthermore, chemical genomics provides both reversible and tunable plant responses. This gives another possibility to bypass important restrictions indispensable to conventional genetic approaches (Alaimo et al., 2001; Shogren-Knaak et al., 2001). Moreover, chemical genomics that unites the power of chemical screens and genomics strategies, was successfully applied to dissect biological processes such as endomembrane trafficking. This was proven by identification of three compounds (Sortin1, Sortin2, and Sortin3) that resulted in pronounced defects in vacuole biogenesis and inhibition of root growth in Arabidopsis seedlings (Zouhar et al., 2004).
In another screen ES1 compound was identified to affect essential steps in PM/endosome trafficking and disrupt the trafficking of PIN2, AUX1, and BRI1 but not PIN1 and PIN7. ES1 treatment induced the rapid agglomeration of plasma membrane proteins into “endosidin bodies” defined by the SYP61/VHA-a1 as an early endosome compartment. Those results highlights the power of bioactive chemicals to dissect essential endomembrane processes (Robert et al., 2008).
In other approach, visualization of vacuolar morphology in candidate mutants was enabled by using EMS-mutagenized population expressing GFP:δ-TIP (Cutler et al., 2000). This screen revealed that Sortin1-hypersensitive mutants with severe vacuolar morphology phenotypes also showed defects in flavonoid accumulation, suggesting a important role of the flavonoid biosynthesis/transport machinery for vacuolar biogenesis and vacuolar cargo delivery in Arabidopsis (Rosado et al., 2011).
Fluorescence imaging in living cells can directly show whether the tested compounds have functional roles in the cellular process. Straightforward and robust phenotypic screen would allow to test thousands of chemicals in high-throughput manner. Ultimately, bioactive compounds targeted to specific pathways can be useful in plants to understand and dissect molecular and biochemical processes (Norambuena et al., 2009; Robert et al., 2009).
Genetic screens are designed to become more sophisticated by using specific genetic backgrounds that allow more complex processes to be assessed. However, the conceptual framework for genetic screens is still the same as that laid out by the pioneers of Arabidopsis genetics and will remain the basis for future genetic approaches (Page and Grossniklaus, 2002). Rapid progress in imaging, high-throughput automation, and next-generation sequencing technologies is expected and will allow further development of more refined and efficient cell biological screens that will provide promising insights into the complex subcellular trafficking pathways in plants.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Tomasz Nodzyński for critical reading of the manuscript and Martine De Cock for help in preparing it. This work was supported by the Odysseus Program of the Research Foundation-Flanders grant (to Jiří Friml).
References
- Abas L., Benjamins R., Malenica N., Paciorek T., Wiśniewska J., Moulinier-Anzola J. C., Sieberer T., Friml J., Luschnig C. (2006). Intracellular trafficking and proteolysis of the Arabidopsis auxin-efflux facilitator PIN2 are involved in root gravitropism. Nat. Cell Biol. 8, 249–256 10.1038/ncb1369 [DOI] [PubMed] [Google Scholar]
- Agee A. E., Surpin M., Sohn E. J., Girke T., Rosado A., Kram B. W., Carter C., Wentzell A. M., Kliebenstein D. J., Jin H. C., Park O. K., Jin H., Hicks G. R., Raikhel N. V. (2010). MODIFIED VACUOLE PHENOTYPE1 is an Arabidopsis myrosinase-associated protein involved in endomembrane protein trafficking. Plant Physiol. 152, 120–132 10.1104/pp.109.145078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alaimo P. J., Shogren-Knaak M. A., Shokat K. M. (2001). Chemical genetic approaches for the elucidation of signaling pathways. Curr. Opin. Chem. Biol. 5, 360–367 10.1016/S1367-5931(00)00215-5 [DOI] [PubMed] [Google Scholar]
- Austin R. S., Vidaurre D., Stamatiou G., Breit R., Provart N. J., Bonetta D., Zhang J., Fung P., Gong Y., Wang P. W., McCourt P., Guttman D. S. (2011). Next-generation mapping of Arabidopsis genes. Plant J. 67, 715–725 10.1111/j.1365-313X.2011.04619.x [DOI] [PubMed] [Google Scholar]
- Avila E. L., Zouhar J., Agee A. E., Carter D. G., Chary S. N., Raikhel N. V. (2003). Tools to study plant organelle biogenesis. Point mutation lines with disrupted vacuoles and high-speed confocal screening of green fluorescent protein-tagged organelles. Plant Physiol. 133, 1673–1676 10.1104/pp.103.033092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batoko H., Zheng H.-Q., Hawes C., Moore I. (2000). A Rab1 GTPase is required for transport between the endoplasmic reticulum and Golgi apparatus and for normal Golgi movement in plants. Plant Cell 12, 2201–2217 10.2307/3871115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann O., Walz B. (2001). Endoplasmic reticulum of animal cells and its organization into structural and functional domains. Int. Rev. Cytol. 205, 149–214 10.1016/S0074-7696(01)05004-5 [DOI] [PubMed] [Google Scholar]
- Benková E., Michniewicz M., Sauer M., Teichmann T., Seifertová D., Jürgens G., Friml J. (2003). Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell 115, 591–602 10.1016/S0092-8674(03)00924-3 [DOI] [PubMed] [Google Scholar]
- Boevink P., Oparka K., Santa Cruz S., Martin B., Betteridge A., Hawes C. (1998). Stacks on tracks: the plant Golgi apparatus traffics on an actin/ER network. Plant J. 15, 441–447 10.1046/j.1365-313X.1998.00208.x [DOI] [PubMed] [Google Scholar]
- Boulaflous A., Faso C., Brandizzi F. (2008). Deciphering the Golgi apparatus: from imaging to genes. Traffic 9, 1613–1617 10.1111/j.1600-0854.2008.00769.x [DOI] [PubMed] [Google Scholar]
- Chang H. C., Hull M., Mellman I. (2004). The J-domain protein Rme-8 interacts with Hsc70 to control clathrin-dependent endocytosis in Drosophila. J. Cell Biol. 164, 1055–1064 10.1083/jcb.200311084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chary S. N., Hicks G. R., Choi Y. G., Carter D., Raikhel N. V. (2008). Trehalose-6-phosphate synthase/phosphatase regulates cell shape and plant architecture in Arabidopsis. Plant Physiol. 146, 97–107 10.1104/pp.107.107441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crane M. M., Chung K., Lu H. (2009). Computer-enhanced high-throughput genetic screens of C. elegans in a microfluidic system. Lab Chip 9, 38–40 10.1039/b813730g [DOI] [PubMed] [Google Scholar]
- Crowell E. F., Gonneau M., Stierhof Y.-D., Höfte H., Vernhettes S. (2010). Regulated trafficking of cellulose synthases. Curr. Opin. Plant Biol. 13, 700–705 10.1016/j.pbi.2010.07.005 [DOI] [PubMed] [Google Scholar]
- Cutler S. R., Ehrhardt D. W., Griffitts J. S., Somerville C. R. (2000). Random GFP::cDNA fusions enable visualization of subcellular structures in cells of Arabidopsis at a high frequency. Proc. Natl. Acad. Sci. U.S.A. 97, 3718–3723 10.1073/pnas.97.7.3718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denecke J., Botterman J., Deblaere R. (1990). Protein secretion in plant cells can occur via a default pathway. Plant Cell 2, 51–59 10.1105/tpc.2.1.51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dharmasiri N., Estelle M. (2004). Auxin signaling and regulated protein degradation. Trends Plant Sci. 9, 302–308 10.1016/j.tplants.2004.04.003 [DOI] [PubMed] [Google Scholar]
- Dhonukshe P., Aniento F., Hwang I., Robinson D. G., Mravec J., Stierhof Y.-D., Friml J. (2007). Clathrin-mediated constitutive endocytosis of PIN auxin efflux carriers in Arabidopsis. Curr. Biol. 17, 520–527 10.1016/j.cub.2007.01.052 [DOI] [PubMed] [Google Scholar]
- Dhonukshe P., Tanaka H., Goh T., Ebine K., Mähönen A. P., Prasad K., Blilou I., Geldner N., Xu J., Uemura T., Chory J., Ueda T., Nakano A., Scheres B., Friml J. (2008). Generation of cell polarity in plants links endocytosis, auxin distribution and cell fate decisions. Nature 456, 962–966 10.1038/nature07409 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Di Sansebastiano G. P., Paris N., Marc-Martin S., Neuhaus J.-M. (2001). Regeneration of a lytic central vacuole and of neutral peripheral vacuoles can be visualized by green fluorescent proteins targeted to either type of vacuoles. Plant Physiol. 126, 78–86 10.1104/pp.126.1.78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson J. G., Jackson C. L. (2000). Regulators and effectors of the ARF GTPases. Curr. Opin. Cell Biol. 12, 475–482 10.1016/S0955-0674(00)00119-8 [DOI] [PubMed] [Google Scholar]
- Donaldson J. G., Lippincott-Schwartz J., Bloom G. S., Kreis T. E., Klausner R. D. (1990). Dissociation of a 110-kD peripheral membrane protein from the Golgi apparatus is an early event in brefeldin A action. J. Cell Biol. 111, 2295–2306 10.1083/jcb.111.6.2295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Souza-Schorey C., Chavrier P. (2006). ARF proteins: roles in membrane traffic and beyond. Nat. Rev. Mol. Cell Biol. 7, 347–358 10.1038/nrm1910 [DOI] [PubMed] [Google Scholar]
- Faso C., Chen Y.-N., Tamura K., Held M., Zemelis S., Marti L., Saravanan R. S., Hummel E., Kung L., Miller E., Hawes C., Brandizzi F. (2009). A missense mutation in the Arabidopsis COPII coat protein Sec24A induces the formation of clusters of the endoplasmic reticulum and Golgi apparatus. Plant Cell 21, 3655–3671 10.1105/tpc.109.068262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feraru E., Paciorek T., Feraru M. I., Zwiewka M., De Groodt R., De Rycke R., Kleine-Vehn J., Friml J. (2010). The AP-3 β adaptin mediates the biogenesis and function of the lytic vacuoles in Arabidopsis. Plant Cell 22, 2812–2824 10.1105/tpc.110.075424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedrichsen D. M., Joazeiro C. A. P., Li J., Hunter T., Chory J. (2000). Brassinosteroid-insensitive-1 is a ubiquitously expressed leucine-rich repeat receptor serine/threonine kinase. Plant Physiol. 123, 1247–1255 10.1104/pp.123.4.1247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friml J., Vieten A., Sauer M., Weijers D., Schwarz H., Hamann T., Offringa R., Jürgens G. (2003). Efflux-dependent auxin gradients establish the apical-basal axis of Arabidopsis. Nature 426, 147–153 10.1038/nature02085 [DOI] [PubMed] [Google Scholar]
- Friml J., Wiśniewska J., Benková E., Mendgen K., Palme K. (2002). Lateral relocation of auxin efflux regulator PIN3 mediates tropism in Arabidopsis. Nature 415, 806–809 10.1038/415806a [DOI] [PubMed] [Google Scholar]
- Fuji K., Shimada T., Takahashi H., Tamura K., Koumoto Y., Utsumi S., Nishizawa K., Maruyama N., Hara-Nishimura I. (2007). Arabidopsis vacuolar sorting mutants (green fluorescent seed) can be identified efficiently by secretion of vacuole-targeted green fluorescent protein in their seeds. Plant Cell 19, 597–609 10.1105/tpc.106.045997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furlong E. E. M., Profitt D., Scott M. P. (2001). Automated sorting of live transgenic embryos. Nat. Biotechnol. 19, 153–156 10.1038/84422 [DOI] [PubMed] [Google Scholar]
- Gaynor E. C., Graham T. R., Emr S. D. (1998). COPI in ER/Golgi and intra-Golgi transport: do yeast COPI mutants point the way? Biochim. Biophys. Acta 1404, 33–51 10.1016/S0167-4889(98)00045-7 [DOI] [PubMed] [Google Scholar]
- Geldner N. (2004). The plant endosomal system – its structure and role in signal transduction and plant development. Planta 219, 547–560 10.1007/s00425-004-1302-x [DOI] [PubMed] [Google Scholar]
- Geldner N., Anders N., Wolters H., Keicher J., Kornberger W., Muller P., Delbarre A., Ueda T., Nakano A., Jürgens G. (2003). The Arabidopsis GNOM ARF-GEF mediates endosomal recycling, auxin transport, and auxin-dependent plant growth. Cell 112, 219–230 10.1016/S0092-8674(03)00003-5 [DOI] [PubMed] [Google Scholar]
- Geldner N., Friml J., Stierhof Y.-D., Jürgens G., Palme K. (2001). Auxin transport inhibitors block PIN1 cycling and vesicle trafficking. Nature 413, 425–428 10.1038/35096571 [DOI] [PubMed] [Google Scholar]
- Geldner N., Hyman D. L., Wang X., Schumacher K., Chory J. (2007). Endosomal signaling of plant steroid receptor kinase BRI1. Genes Dev. 21, 1598–1602 10.1101/gad.1561307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gil P., Dewey E., Friml J., Zhao Y., Snowden K. C., Putterill J., Palme K., Estelle M., Chory J. (2001). BIG: a calossin-like protein required for polar auxin transport in Arabidopsis. Genes Dev. 15, 1985–1997 10.1101/gad.905201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillingham A. K., Munro S. (2007). The small G proteins of the Arf family and their regulators. Annu. Rev. Cell Dev. Biol. 23, 579–611 10.1146/annurev.cellbio.23.090506.123209 [DOI] [PubMed] [Google Scholar]
- Girard M., Poupon V., Blondeau F., McPherson P. S. (2005). The DnaJ-domain protein RME–8 functions in endosomal trafficking. J. Biol. Chem. 280, 40135–40143 10.1074/jbc.M505036200 [DOI] [PubMed] [Google Scholar]
- Grosshans B. L., Ortiz D., Novick P. (2006). Rabs and their effectors: achieving specificity in membrane traffic. Proc. Natl. Acad. Sci. U.S.A. 103, 11821–11827 10.1073/pnas.0601617103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson M. R., Köhler R. H. (2001). GFP imaging: methodology and application to investigate cellular compartmentation in plants. J. Exp. Bot. 52, 529–539 10.1093/jexbot/52.356.529 [DOI] [PubMed] [Google Scholar]
- Haseloff J. (1999). GFP variants for multispectral imaging of living cells. Methods Cell Biol. 58, 139–151 10.1016/S0091-679X(08)61953-6 [DOI] [PubMed] [Google Scholar]
- Haseloff J., Siemering K. R., Prasher D. C., Hodge S. (1997). Removal of a cryptic intron and subcellular localization of green fluorescent protein are required to mark transgenic Arabidopsis plants brightly. Proc. Natl. Acad. Sci. U.S.A. 94, 2122–2127 10.1073/pnas.94.6.2122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi Y., Yamada K., Shimada T., Matsushima R., Nishizawa N. K., Nishimura M., Hara-Nishimura I. (2001). A proteinase-storing body that prepares for cell death or stresses in the epidermal cells of Arabidopsis. Plant Cell Physiol. 42, 894–899 10.1093/pcp/pce144 [DOI] [PubMed] [Google Scholar]
- Held M. A., Boulaflous A., Brandizzi F. (2008). Advances in fluorescent protein-based imaging for the analysis of plant endomembranes. Plant Physiol. 147, 1469–1481 10.1104/pp.108.120147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones A. M. (1994). Auxin-binding proteins. Annu. Rev. Plant Physiol. Plant Mol. Biol. 45, 393–420 10.1146/annurev.pp.45.060194.002141 [DOI] [Google Scholar]
- Jorgensen E. M., Mango S. E. (2002). The art and design of genetic screens: Caenorhabditis elegans. Nat. Rev. Genet. 3, 356–369 10.1038/nrg794 [DOI] [PubMed] [Google Scholar]
- Kanyuka K., Praekelt U., Franklin K. A., Billingham O. E., Hooley R., Whitelam G. C., Halliday K. J. (2003). Mutations in the huge Arabidopsis gene BIG affect a range of hormone and light responses. Plant J. 35, 57–70 10.1046/j.1365-313X.2003.01779.x [DOI] [PubMed] [Google Scholar]
- Kepinski S., Leyser O. (2002). Ubiquitination and auxin signaling: a degrading story. Plant Cell 14, S81–S95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klausner R. D., Donaldson J. G., Lippincott-Schwartz J. (1992). Brefeldin A: insights into the control of membrane traffic and organelle structure. J. Cell Biol. 116, 1071–1080 10.1083/jcb.116.5.1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleine-Vehn J., Dhonukshe P., Sauer M., Brewer P., Wiśniewska J., Paciorek T., Benková E., Friml J. (2008a). ARF GEF-dependent transcytosis and polar delivery of PIN auxin carriers in Arabidopsis. Curr. Biol. 18, 526–531 10.1016/j.cub.2008.03.021 [DOI] [PubMed] [Google Scholar]
- Kleine-Vehn J., Leitner J., Zwiewka M., Sauer M., Abas L., Luschnig C., Friml J. (2008b). Differential degradation of PIN2 auxin efflux carrier by retromer-dependent vacuolar targeting. Proc. Natl. Acad. Sci. U.S.A. 105, 17812–17817 10.1073/pnas.0808073105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuruvilla F. G., Shamji A. F., Sternson S. M., Hergenrother P. J., Schreiber S. L. (2002). Dissecting glucose signalling with diversity-oriented synthesis and small-molecule microarrays. Nature 416, 653–657 10.1038/416653a [DOI] [PubMed] [Google Scholar]
- Langhans M., Förster S., Helmchen G., Robinson D. G. (2011). Differential effects of the brefeldin A analogue (6R)-hydroxy-BFA in tobacco and Arabidopsis. J. Exp. Bot. 62, 2949–2957 10.1093/jxb/err007 [DOI] [PubMed] [Google Scholar]
- Lee G.-J., Kim H., Kang H., Jang M., Lee D. W., Lee S., Hwang I. (2007). EpsinR2 interacts with clathrin, adaptor protein–3, AtVTI12, and phosphatidylinositol-3-phosphate. Implications for EpsinR2 function in protein trafficking in plant cells. Plant Physiol. 143, 1561–1575 10.1104/pp.106.095349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukowitz W., Gillmor C. S., Scheible W.-R. (2000). Positional cloning in Arabidopsis. Why it feels good to have a genome initiative working for you. Plant Physiol. 123, 795–805 10.1104/pp.123.3.795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madson M., Dunand C., Li X., Verma R., Vanzin G. F., Caplan J., Shoue D. A., Carpita N. C., Reiter W. D. (2003). The MUR3 gene of Arabidopsis encodes a xyloglucan galactosyltransferase that is evolutionarily related to animal exostosins. Plant Cell 15, 1662–1670 10.1105/tpc.009837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marti L., Stefano G., Tamura K., Hawes C., Renna L., Held M. A., Brandizzi F. (2010). A missense mutation in the vacuolar protein GOLD36 causes organizational defects in the ER and aberrant protein trafficking in the plant secretory pathway. Plant J. 63, 901–913 10.1111/j.1365-313X.2010.04296.x [DOI] [PubMed] [Google Scholar]
- Martínez-Menárguez J. A., Geuze H. J., Slot J. W., Klumperman J. (1999). Vesicular tubular clusters between the ER and Golgi mediate concentration of soluble secretory proteins by exclusion from COPI-coated vesicles. Cell 98, 81–90 10.1016/S0092-8674(00)80608-X [DOI] [PubMed] [Google Scholar]
- Marty F. (1999). Plant vacuoles. Plant Cell 11, 587–599 10.1105/tpc.11.4.587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Megason S. G., Fraser S. E. (2003). Digitizing life at the level of the cell: high-performance laser-scanning microscopy and image analysis for in toto imaging of development. Mech. Dev. 120, 1407–1420 10.1016/j.mod.2003.07.005 [DOI] [PubMed] [Google Scholar]
- Miernyk J. A. (2001). The J-domain proteins of Arabidopsis thaliana: an unexpectedly large and diverse family of chaperones. Cell Stress Chaperones 6, 209–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsuhashi N., Shimada T., Mano S., Nishimura M., Hara-Nishimura I. (2000). Characterization of organelles in the vacuolar-sorting pathway by visualization with GFP in tobacco BY-2 cells. Plant Cell Physiol. 41, 993–1001 10.1093/pcp/pcd040 [DOI] [PubMed] [Google Scholar]
- Nakano R. T., Matsushima R., Ueda H., Tamura K., Shimada T., Li L., Hayashi Y., Kondo M., Nishimura M., Hara-Nishimura I. (2009). GNOM-LIKE1/ERMO1 and SEC24a/ERMO2 are required for maintenance of endoplasmic reticulum morphology in Arabidopsis thaliana. Plant Cell 21, 3672–3685 10.1105/tpc.109.068270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Napier R. M., David K. M., Perrot-Rechenmann C. (2002). A short history of auxin-binding proteins. Plant Mol. Biol. 49, 339–348 10.1023/A:1015259130955 [DOI] [PubMed] [Google Scholar]
- Naramoto S., Kleine-Vehn J., Robert S., Fujimoto M., Dainobu T., Paciorek T., Ueda T., Nakano A., Van Montagu M. C. E., Fukuda H., Friml J. (2010). ADP-ribosylation factor machinery mediates endocytosis of plant cells. Proc. Natl. Acad. Sci. U.S.A. 107, 21890–21895 10.1073/pnas.1016260107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nebenführ A., Ritzenthaler C., Robinson D. G. (2002). Brefeldin A: deciphering an enigmatic inhibitor of secretion. Plant Physiol. 130, 1102–1108 10.1104/pp.011569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson B. K., Cai X., Nebenführ A. (2007). A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J. 51, 1126–1136 10.1111/j.1365-313X.2007.03212.x [DOI] [PubMed] [Google Scholar]
- Niihama M., Takemoto N., Hashiguchi Y., Tasaka M., Terao Morita M. (2009). ZIP genes encode proteins involved in membrane trafficking of the TGN-PVC/vacuoles. Plant Cell Physiol. 50, 2057–2068 10.1093/pcp/pcp137 [DOI] [PubMed] [Google Scholar]
- Nishizawa K., Maruyama N., Satoh R., Fuchikami Y., Higasa T., Utsumi S. (2003). A C-terminal sequence of soybean β-conglycinin α′ subunit acts as a vacuolar sorting determinant in seed cells. Plant J. 34, 647–659 10.1046/j.1365-313X.2003.01754.x [DOI] [PubMed] [Google Scholar]
- Norambuena L., Raikhel N. V., Hicks G. R. (2009). Chemical genomics approaches in plant biology. Methods Mol. Biol. 553, 345–354 10.1007/978-1-60327-563-7_18 [DOI] [PubMed] [Google Scholar]
- Okita T. W., Rogers J. C. (1996). Compartmentation of proteins in the endomembrane system of plant cells. Annu. Rev. Plant Physiol. Plant Mol. Biol. 47, 327–350 10.1146/annurev.arplant.47.1.327 [DOI] [PubMed] [Google Scholar]
- Paciorek T., Zažímalová E., Ruthardt N., Petrášek J., Stierhof Y.-D., Kleine-Vehn J., Morris D. A., Emans N., Jürgens G., Geldner N., Friml J. (2005). Auxin inhibits endocytosis and promotes its own efflux from cells. Nature 435, 1251–1256 10.1038/nature03633 [DOI] [PubMed] [Google Scholar]
- Page D. R., Grossniklaus U. (2002). The art and design of genetic screens: Arabidopsis thaliana. Nat. Rev. Genet. 3, 124–136 10.1038/nrg730 [DOI] [PubMed] [Google Scholar]
- Petrášek J., Mravec J., Bouchard R., Blakeslee J. J., Abas M., Seifertová D., Wiśniewska J., Tadele Z., Kubeš M., Čovanová M., Dhonukshe P., Sk°upa P., Benková E., Perry L., Křeček P., Lee O. R., Fink G. R., Geisler M., Murphy A. S., Luschnig C., Zažímalová E., Friml J. (2006). PIN proteins perform a rate-limiting function in cellular auxin efflux. Science 312, 914–918 10.1126/science.1123542 [DOI] [PubMed] [Google Scholar]
- Richter S., Geldner N., Schrader J., Wolters H., Stierhof Y.-D., Rios G., Koncz C., Robinson D. G., Jürgens G. (2007). Functional diversification of closely related ARF-GEFs in protein secretion and recycling. Nature 448, 488–492 10.1038/nature05967 [DOI] [PubMed] [Google Scholar]
- Ritzenthaler C., Nebenführ A., Movafeghi A., Stussi-Garaud C., Behnia L., Pimpl P., Staehelin L. A., Robinson D. G. (2002). Reevaluation of the effects of brefeldin A on plant cells using tobacco Bright Yellow 2 cells expressing Golgi-targeted green fluorescent protein and COPI antisera. Plant Cell 14, 237–261 10.1105/tpc.010237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert S., Chary S. N., Drakakaki G., Li S., Yang Z., Raikhel N. V., Hicks G. R. (2008). Endosidin1 defines a compartment involved in endocytosis of the brassinosteroid receptor BRI1 and the auxin transporters PIN2 and AUX1. Proc. Natl. Acad. Sci. U.S.A. 105, 8464–8469 10.1073/pnas.0711650105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert S., Kleine-Vehn J., Barbez E., Sauer M., Paciorek T., Baster P., Vanneste S., Zhang J., Simon S., Čovanová M., Hayashi K., Dhonukshe P., Yang Z., Bednarek S. Y., Jones A. M., Luschnig C., Aniento F., Zažímolová E., Friml J. (2010). ABP1 mediates auxin inhibition of clathrin-dependent endocytosis in Arabidopsis. Cell 143, 111–121 10.1016/j.cell.2010.09.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert S., Raikhel N. V., Hicks G. R. (2009). “Powerful partners: Arabidopsis and chemical genomics,” in The Arabidopsis Book, eds Last R., Chang C., Jander G., Kliebenstein D., McClung R., Millar H., Torii K., Wagner D. (Rockville: The American Society of Plant Biologists; ), e0109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson M. S., Kreis T. E. (1992). Recruitment of coat proteins onto Golgi membranes in intact and permeabilized cells: effects of brefeldin A and G proteins activators. Cell 69, 129–138 10.1016/0092-8674(92)90124-U [DOI] [PubMed] [Google Scholar]
- Rosado A., Hicks G. R., Norambuena L., Rogachev I., Meir S., Pourcel L., Zohar J., Brown M. Q., Boirsdore M. P., Puckrin R. S., Cutler S. R., Rojo E., Aharoni A., Raikhel N. V. (2011). Sortin1-hypersensitive mutants link vacuolar-trafficking defects and flavonoid metabolism in Arabidopsis vegetative tissues. Chem. Biol. 18, 187–197 10.1016/j.chembiol.2010.11.015 [DOI] [PubMed] [Google Scholar]
- Ruegger M., Dewey E., Hobbie L., Brown D., Bernasconi P., Turner J., Muday G., Estelle M. (1997). Reduced naphthylphthalamic acid binding in the tir3 mutant of Arabidopsis is associated with a reduction in polar auxin transport and diverse morphological defects. Plant Cell 9, 745–757 10.1105/tpc.9.5.745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salomon S., Grunewald D., Stüber K., Schaaf S., MacLean D., Schulze-Lefert P., Robatzek S. (2010). High-throughput confocal imaging of intact live tissue enables quantification of membrane trafficking in Arabidopsis. Plant Physiol. 154, 1096–1104 10.1104/pp.110.160325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneeberger K., Ossowski S., Lanz C., Juul T., Høgh Petersen A., Lehmann Nielsen K., Jørgensen J.-E., Weigel D., Uggerjøh Andersen S. (2009). SHOREmap: simultaneous mapping and mutation identification by deep sequencing. Nat. Methods 6, 550–551 10.1038/nmeth0809-550 [DOI] [PubMed] [Google Scholar]
- Shimada T., Fuji K., Tamura K., Kondo M., Nishimura M., Hara-Nishimura I. (2003). Vacuolar sorting receptor for seed storage proteins in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 100, 16095–16100 10.1073/pnas.2530568100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada T., Koumoto Y., Li L., Yamazaki M., Kondo M., Nishimura M., Hara-Nishimura I. (2006). AtVPS29, a putative component of a retromer complex, is required for the efficient sorting of seed storage proteins. Plant Cell Physiol. 47, 1187–1194 10.1093/pcp/pcj103 [DOI] [PubMed] [Google Scholar]
- Shimada T., Kuroyanagi M., Nishimura M., Hara-Nishimura I. (1997). A pumpkin 72-kDa membrane protein of precursor-accumulating vesicles has characteristics of a vacuolar sorting receptor. Plant Cell Physiol. 38, 1414–1420 10.1093/oxfordjournals.pcp.a029138 [DOI] [PubMed] [Google Scholar]
- Shimada T., Watanabe E., Tamura K., Hayashi Y., Nishimura M., Hara-Nishimura I. (2002). A vacuolar sorting receptor PV72 on the membrane of vesicles that accumulate precursors of seed storage proteins (PAC vesicles). Plant Cell Physiol. 43, 1086–1095 10.1093/pcp/pcf152 [DOI] [PubMed] [Google Scholar]
- Shogren-Knaak M. A., Alaimo P. J., Shokat K. M. (2001). Recent advances in chemical approaches to the study of biological systems. Annu. Rev. Cell Dev. Biol. 17, 405–433 10.1146/annurev.cellbio.17.1.405 [DOI] [PubMed] [Google Scholar]
- Silady R. A., Kato T., Lukowitz W., Sieber P., Tasaka M., Somerville C. R. (2004). The gravitropism defective 2 mutants of Arabidopsis are deficient in a protein implicated in endocytosis in Caenorhabditis elegans. Plant Physiol. 136, 3095–3103 10.1104/pp.104.050583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorefan K., Girin T., Liljegren S. J., Ljung K., Robles P., Galván-Ampudia C. S., Offringa R., Friml J., Yanofsky M. F., Østergaard L. (2009). A regulated auxin minimum is required for seed dispersal in Arabidopsis. Nature 459, 583–586 10.1038/nature07875 [DOI] [PubMed] [Google Scholar]
- Staehelin L. A. (1997). The plant ER: a dynamic organelle composed of a large number of discrete functional domains. Plant J. 11, 1151–1165 10.1046/j.1365-313X.1997.11061151.x [DOI] [PubMed] [Google Scholar]
- Stefano G., Renna L., Moss T., McNew J. A., Brandizzi F. (2012). In Arabidopsis, the spatial and dynamic organization of the endoplasmic reticulum and Golgi apparatus is influenced by the integrity of the C-terminal domain of RHD3, a non-essential GTPase. Plant J. 69, 957–966 10.1111/j.1365-313X.2011.04846.x [DOI] [PubMed] [Google Scholar]
- Steinmann T., Geldner N., Grebe M., Mangold S., Jackson C. L., Paris S., Gälweiler L., Palme K., Jürgens G. (1999). Coordinated polar localization of auxin efflux carrier PIN1 by GNOM ARF GEF. Science 286, 316–318 10.1126/science.286.5438.316 [DOI] [PubMed] [Google Scholar]
- Surpin M., Raikhel N. (2004). Traffic jams affect plant development and signal transduction. Nat. Rev. Mol. Cell Biol. 5, 100–109 10.1038/nrm1311 [DOI] [PubMed] [Google Scholar]
- Tamura K., Shimada T., Kondo M., Nishimura M., Hara-Nishimura I. (2005). KATAMARI1/MURUS3 is a novel Golgi membrane protein that is required for endomembrane organization in Arabidopsis. Plant Cell 17, 1764–1776 10.1105/tpc.105.031930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Shimada T., Ono E., Tanaka Y., Nagatani A., Higashi S.-I., Watanabe M., Nishimura M., Hara-Nishimura I. (2003). Why green fluorescent fusion proteins have not been observed in the vacuoles of higher plants. Plant J. 35, 545–555 10.1046/j.1365-313X.2003.01822.x [DOI] [PubMed] [Google Scholar]
- Tamura K., Takahashi H., Kunieda T., Fuji K., Shimada T., Hara-Nishimura I. (2007). Arabidopsis KAM2/GRV2 is required for proper endosome formation and functions in vacuolar sorting and determination of the embryo growth axis. Plant Cell 19, 320–332 10.1105/tpc.106.046631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka H., Kitakura S., De Rycke R., De Groodt R., Friml J. (2009). Fluorescence imaging-based screen identifies ARF GEF component of early endosomal trafficking. Curr. Biol. 19, 391–397 10.1016/j.cub.2009.04.035 [DOI] [PubMed] [Google Scholar]
- Teh O.-K., Moore I. (2007). An ARF-GEF acting at the Golgi and in selective endocytosis in polarized plant cells. Nature 448, 493–496 10.1038/nature06023 [DOI] [PubMed] [Google Scholar]
- Vernoud V., Horton A. C., Yang Z., Nielsen E. (2003). Analysis of the small GTPase gene superfamily of Arabidopsis. Plant Physiol. 131, 1191–1208 10.1104/pp.013052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitale A., Denecke J. (1999). The endoplasmic reticulum – gateway of the secretory pathway. Plant Cell 11, 615–628 10.1105/tpc.11.4.615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Lockwood S. K., Hoeltzel M. F., Schiefelbein J. W. (1997). The ROOT HAIR DEFECTIVE3 gene encodes an evolutionarily conserved protein with GTP-binding motifs and is required for regulated cell enlargement in Arabidopsis. Genes Dev. 11, 799–811 10.1101/gad.11.19.2569 [DOI] [PubMed] [Google Scholar]
- Wieland F. T., Gleason M. L., Serafini T. A., Rothman J. E. (1987). The rate of bulk flow from the endoplasmic reticulum to the cell surface. Cell 50, 289–300 10.1016/0092-8674(87)90224-8 [DOI] [PubMed] [Google Scholar]
- Wisniewska J., Xu J., Seifertová D., Brewer P. B., R°užiČka K., Blilou I., Rouquié D., Benková E., Scheres B., Friml J. (2006). Polar PIN localization directs auxin flow in plants. Science 312, 883–883 10.1126/science.1121356 [DOI] [PubMed] [Google Scholar]
- Xu J., Scheres B. (2005). Cell polarity: ROPing the ends together. Curr. Opin. Plant Biol. 8, 613–618 10.1016/j.pbi.2005.09.003 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Grant B., Hirsh D. (2001). RME-8, a conserved J-domain protein, is required for endocytosis in Caenorhabditis elegans. Mol. Biol. Cell 12, 2011–2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng H., Kunst L., Hawes C., Moore I. (2004). A GFP-based assay reveals a role for RHD3 in transport between the endoplasmic reticulum and Golgi apparatus. Plant J. 37, 398–414 10.1046/j.1365-313X.2003.01969.x [DOI] [PubMed] [Google Scholar]
- Zouhar J., Hicks G. R., Raikhel N. V. (2004). Sorting inhibitors (Sortins): chemical compounds to study vacuolar sorting in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 101, 9497–9501 10.1073/pnas.0402121101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwiewka M., Feraru E., Möller B., Hwang I., Feraru M. I., Kleine-Vehn J., Weijers D., Friml J. (2011). The AP-3 adaptor complex is required for vacuolar function in Arabidopsis. Cell Res. 21, 1711–1722 10.1038/cr.2011.99 [DOI] [PMC free article] [PubMed] [Google Scholar]