Abstract
The tumour suppressor gene product Mig-6 acts as an inhibitor of epidermal growth factor (EGF) signalling. However, its posttranslational modifications and regulatory mechanisms have not been elucidated. Here, we investigated the phosphorylation of human Mig-6 and found that Chk1 phosphorylated Mig-6 in vivo as well as in vitro. Moreover, EGF stimulation promoted phosphorylation of Mig-6 without DNA damage and the phosphorylation was inhibited by depletion of Chk1. EGF also increased Ser280-phosphorylated Chk1, a cytoplasmic-tethering form, via PI3K pathway. Mass spectrometric analyses suggested that Ser 251 of Mig-6 was a major phosphorylation site by Chk1 in vitro and in vivo. Substitution of Ser 251 to alanine increased inhibitory activity of Mig-6 against EGF receptor (EGFR) activation. Moreover, EGF-dependent activation of EGFR and cell growth were inhibited by Chk1 depletion, and were rescued by co-depletion of Mig-6. Our results suggest that Chk1 phosphorylates Mig-6 on Ser 251, resulting in the inhibition of Mig-6, and that Chk1 acts as a positive regulator of EGF signalling. This is a novel function of Chk1.
Keywords: Chk1, EGFR, Mig-6, phosphorylation
Introduction
Mig-6 (also called Ralt, Errfi1 and Gene 33) is a negative regulator of epidermal growth factor (EGF) signalling. Mig-6 binds to all EGF receptor (EGFR) family members and inhibits their tyrosine kinase activity (Anastasi et al, 2003). Mig-6 is induced by stimulation with fetal calf serum, insulin, and growth factors, including EGF, as an immediate early-response gene (Zhang and Vande Woude, 2007). Transcription of Mig-6 is also induced by various stresses (Makkinje et al, 2000). Mig-6 functions as a feedback inhibitor of EGFR family signalling via its induction by the Ras/mitogen-activated protein kinase (MAPK) pathway (Fiorentino et al, 2000; Hackel et al, 2001; Anastasi et al, 2003; Xu et al, 2005; Anastasi et al, 2007).
Overexpression of Mig-6 leads to inhibition of EGFR autophosphorylation, and reduces MAPK activity (Anastasi et al, 2003; Xu et al, 2005; Anastasi et al, 2007). Mig-6 binds to EGFR family tyrosine kinases via its EGFR-binding domain (BD) and acts as a specific inhibitor of their signalling, while Mig-6 is suggested to contribute several biochemical functions as a multi-adaptor protein with many interactive domains. It is reported that Mig-6 binds to the GTP-bound form of Cdc42 via the CRIB domain and activates stress-activated protein kinases (Makkinje et al, 2000). The binding of Mig-6 to Cdc42 inhibits the activity of Cdc42, resulting in the inhibition of hepatocyte growth factor-induced cell migration (Pante et al, 2005). It is also reported that the processed CRIB domain of Mig-6, which is derived by limited proteolytic processing, binds to IκB and competes with NFκB, resulting in the activation of NFκB and its signalling (Tsunoda et al, 2002; Mabuchi et al, 2005).
Downregulated expression of the Mig-6 gene is observed in breast carcinomas, in which it correlates with reduced overall survival (Amatschek et al, 2004; Anastasi et al, 2005). Mig-6 is also downregulated in other human cancers such as hepatocellular carcinomas (Reschke et al, 2010) and thyroid cancers. Mig-6 expression correlates with survival and is an independent predictor of recurrence in papillary thyroid cancers (Ruan et al, 2008). Recently, it has been reported that the Mig-6 gene is mutated in the human non-small-cell lung cancer cell lines NCI-H226 and NCI-H 322M, as well as in primary human lung cancer (Zhang et al, 2007). Mig-6-deficient mice show hyperactivation of endogenous EGFR and sustained signalling through the MAPK pathway, resulting in overproliferation and impaired differentiation of epidermal keratinocytes. Furthermore, Mig-6-deficient mice develop spontaneous tumours in various organs and are highly susceptible to chemically induced skin tumours. Inhibition of endogenous EGF signalling by the EGFR inhibitor gefitinib (Iressa), or replacement of wild type (WT) EGFR with a kinase-deficient EGFR, rescues the skin defects in Mig-6-deficient mice (Ferby et al, 2006). Therefore, Mig-6 plays a tumour suppressor role as a specific negative regulator of EGF signalling.
Although Mig-6 is known to be an important tumour suppressor through negative regulation of EGF signalling, its posttranslational modifications such as phosphorylation have not been fully elucidated (Fiorentino et al, 2000; Fiorini et al, 2002). It has been reported that EGF promotes tyrosine phosphorylation of Mig-6 by EGFR tyrosine kinase (Tong et al, 2008), the biological implications of which are unknown. Another report indicated that Mig-6 can bind the phospho-serine binding adaptor protein 14-3-3 via its consensus motif for 14-3-3 binding, but the responsible kinase has not been identified (Makkinje et al, 2000).
One of the kinases that phosphorylates at the 14-3-3 consensus motif is Chk1. If DNA stability is perturbed, Chk1 is activated by ATM/ATR and phosphorylates a Ser residue in the 14-3-3 motifs of human Cdc25A, B, and C (Niida and Nakanishi, 2006). Chk1 is essential not only for checkpoint activation but also for cell viability in the absence of DNA perturbation. Chk1-knockout mice show embryonic lethality at an early stage of development. Chk1-conditional knockout embryonic stem cells and somatic cells also died, even in the absence of DNA damage (Liu et al, 2000; Niida et al, 2005). These phenotypes indicate that Chk1 has an important role during normal cell proliferation. However, the DNA damage-independent functions of Chk1 have not been fully elucidated.
In the present study, we investigated phosphorylation of Mig-6 and found that it is phosphorylated by Chk1 in vitro and in vivo. Moreover, we identified the sites for Chk1 phosphorylation and investigated the functional regulation of Mig-6 via Chk1-mediated phosphorylation in EGF signalling.
Results
Effects of protein kinase inhibitors on Mig-6 phosphorylation
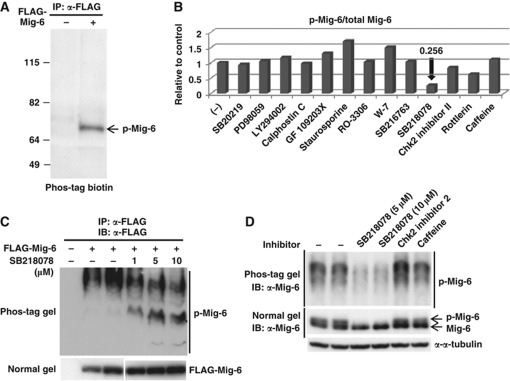
Mig-6 acts as a negative feedback regulator of EGF signalling. However, little is known about its posttranslational modifications or regulatory mechanisms. We first investigated phosphorylation of Mig-6 because many cellular proteins are regulated by phosphorylation. FLAG-Mig-6 was transfected into HEK293 cells and immunoprecipitated (IP) with anti-FLAG antibody, and then the cell lysate were IP and analysed by SDS–PAGE followed by a Phos-tag BTL phosphoprotein detection method (Figure 1A). The result indicated that Mig-6 is phosphorylated in these cells. To identify the responsible kinase, we tested various protein kinase inhibitors (Supplementary Table S1) in the in vivo phosphorylation assay. We found that phosphorylation of Mig-6 was decreased remarkably by the Chk1 inhibitor SB218078 (Figure 1B, Supplementary Figure S1). As shown in Figure 1B, 10 μM SB218078 inhibited phosphorylation of Mig-6 to 25.6% of the control level. To confirm this, we performed Phos-tag SDS–PAGE analysis of Mig-6. Phosphorylation-dependent mobility shifts of Mig-6 were suppressed by SB218078 in a dose-dependent manner (Figure 1C). We next investigated whether the phosphorylation of endogenous Mig-6 (endo Mig-6) was also inhibited by the Chk1 inhibitor. Using MDA-MB-231 cells, in which Mig-6 is endogenously highly expressed, we confirmed that phosphorylation of endo Mig-6 was inhibited by Chk1 inhibitor, whereas Chk2 inhibitor 2 or caffeine (ATM/ATR inhibitor) did not affect it (Figure 1D). This suggests that Chk1 phosphorylates Mig-6 in vivo.
Figure 1.
Effects of protein kinase inhibitors on Mig-6 phosphorylation in vivo. (A) Mig-6 is phosphorylated in HEK293 cells. pcDNA3-FLAG-Mig-6 was transfected into HEK293 cells. The whole cell lysates were IP with anti-FLAG M2 antibody and resolved by SDS–PAGE. Phosphorylated Mig-6 (p-Mig-6) was detected by a Phos-tag BTL system as described in Materials and methods. (B) Phosphorylation of Mig-6 is inhibited by Chk1 inhibitor (SB218078) in vivo. HEK293 cells were transfected with FLAG-Mig-6 and treated with the indicated kinase inhibitors for 3 h before harvesting. FLAG-Mig-6 was IP with anti-FLAG antibody from the cell lysates and p-Mig-6 was detected by the Phos-tag BTL system. Details of the kinase inhibitors are indicated in Supplementary Table S1. The intensities of the p-Mig-6 bands and total Mig-6 bands are indicated relative to the control (without inhibitor). (C) Phosphorylation of Mig-6 is inhibited by a Chk1 inhibitor in a dose-dependent manner. HEK293 cells were transfected with FLAG-Mig-6 and treated with the Chk1 inhibitor SB218078 at the indicated concentrations for 3 h before harvest. FLAG-Mig-6 was IP with anti-FLAG antibody from the cell lysate and separated by 6% Phos-tag SDS–PAGE (upper panel) or normal SDS–PAGE (lower panel) followed by IB with anti-FLAG antibody. (D) Phosphorylation of endo Mig-6 is inhibited by Chk1 inhibitor. MDA-MB-231 cells were treated without or with 5 or 10 μM SB218078, 10 μM Chk2 inhibitor 2, or 5 mM caffeine for 3 h. The cell lysates were separated by 6% Phos-tag SDS–PAGE (upper panel) or normal SDS–PAGE (lower panel) followed by IB with anti-Mig-6 antibody. Figure source data can be found with the Supplementary data.
Chk1 phosphorylates Mig-6 in vitro
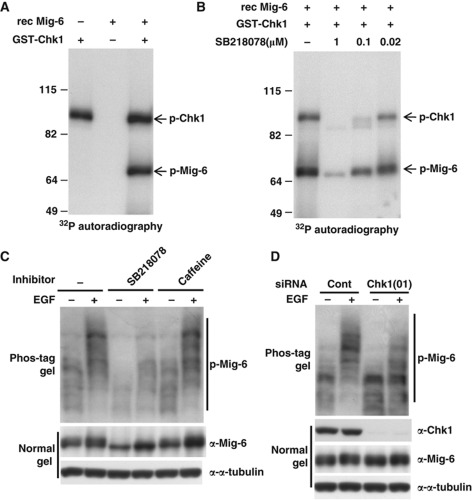
To examine whether Chk1 directly phosphorylates Mig-6, we performed an in vitro kinase assay. Recombinant (rec) Mig-6 protein was incubated with 32P-labelled ATP and rec GST-Chk1 kinase at 30 °C for 30 min. As shown in Figure 2A, phosphorylation of Mig-6 was observed in the presence of Chk1 kinase, and autophosphorylation of Chk1 was also observed in the lane with Chk1. Moreover, both the Chk1-mediated phosphorylation of Mig-6 and autophosphorylation of Chk1 were inhibited by SB218078 in a dose-dependent manner (Figure 2B).
Figure 2.
Chk1 phosphorylates Mig-6 after EGF stimulation. (A) In vitro phosphorylation of Mig-6. rec Mig-6 protein (0.1 μg) was incubated in 20 μl of kinase reaction buffer with 32P-labelled ATP and 0.1 μg of purified rec GST-Chk1 kinase at 30°C for 30 min. The reaction was stopped by the addition of SDS sample buffer, then the proteins were separated by SDS–PAGE. p-Mig-6 was analysed by autoradiography. (B) Phosphorylation of Mig-6 is inhibited by a Chk1 inhibitor in a dose-dependent manner. rec Mig-6 proteins were pre-treated with SB218078 at the indicated concentration for 5 min at room temperature and then subjected to an in vitro kinase assay as described above. (C) EGF stimulation promotes phosphorylation of endo Mig-6. MDA-MB-231 cells were subjected to serum starvation for 16 h. Cells were pretreated with or without 10 μM SB218078 or 5 mM caffeine for 3 h, followed by stimulation with 20 ng/ml EGF for 15 min. Cells were harvested and the cell lysates were separated by 6% Phos-tag SDS–PAGE or normal SDS–PAGE, and subjected to IB with anti-Mig-6 antibody. (D) EGF-promoted phosphorylation of Mig-6 is suppressed by Chk1 depletion. MDA-MB-231 cells were transfected with an siRNA for human Chk1 or a control siRNA (Cont) and serum starved for 16 h, then stimulated with 20 ng/ml EGF for 15 min. Cell lysates were separated by 6% Phos-tag SDS–PAGE or normal SDS–PAGE and analysed by IB with the indicated antibodies. Figure source data can be found with the Supplementary data.
EGF stimulates Chk1-mediated phosphorylation of Mig-6
Previous studies have shown that Mig-6 functions as a feedback inhibitor of EGF signalling. Therefore, we next investigated whether Chk1-mediated phosphorylation of Mig-6 was associated with the EGF signalling pathway. As shown in Figure 2C, we found that phosphorylation of endo Mig-6 was promoted by EGF stimulation in MDA-MB-231 cells, and it was suppressed by the Chk1 inhibitor (Figure 2C, lane 2 versus lane 4), suggesting that Chk1 is involved in EGF-stimulated Mig-6 phosphorylation. Interestingly, caffeine, an ATM/ATR inhibitor, did not affect the phosphorylation of Mig-6 (Figure 2C, lane 2 versus lane 6) even though the same concentration of caffeine could counteract the phosphorylation of Chk1 induced by UV stimulation (Sarkaria et al, 1999; Mailand et al, 2000). Because caffeine did not affect EGF-stimulated Mig-6 phosphorylation, which was observed without genotoxic stress, it is likely that the Mig-6 phosphorylation by Chk1 is induced in a DNA damage-independent manner.
Next, we investigated the effect of Chk1 depletion on Mig-6 phosphorylation. Basal phosphorylation of Mig-6 in the absence of EGF stimulation was suppressed by depletion of Chk1 (Figure 2D, lane 1 versus lane 3). Moreover, EGF-stimulated Mig-6 phosphorylation was severely inhibited by depletion of Chk1 (Figure 2D, lane 2 versus lane 4). Furthermore, we performed a phosphatase-treatment experiment to prove that the smeared Mig-6 band on the Phos-tag gel was because of phosphorylation (Supplementary Figure S2B). We also demonstrated that the smeared Mig-6 band prepared from EGF-stimulated MDA-MB-231 cells on a Phos-tag gel did not indicate ubiquitylation (Supplementary Figure S2C). To confirm the Chk1-mediated Mig-6 phosphorylation, we designed and used another small interfering RNA (siRNA) oligo, Chk1(04). As shown in Figure 2D and Supplementary Figure S2A, depletion of Chk1 by both siRNA(01) and (04) attenuated phosphorylation of Mig-6. These results indicate that EGF stimulation promotes Chk1-mediated Mig-6 phosphorylation.
Analysis of the phosphorylation sites in Mig-6
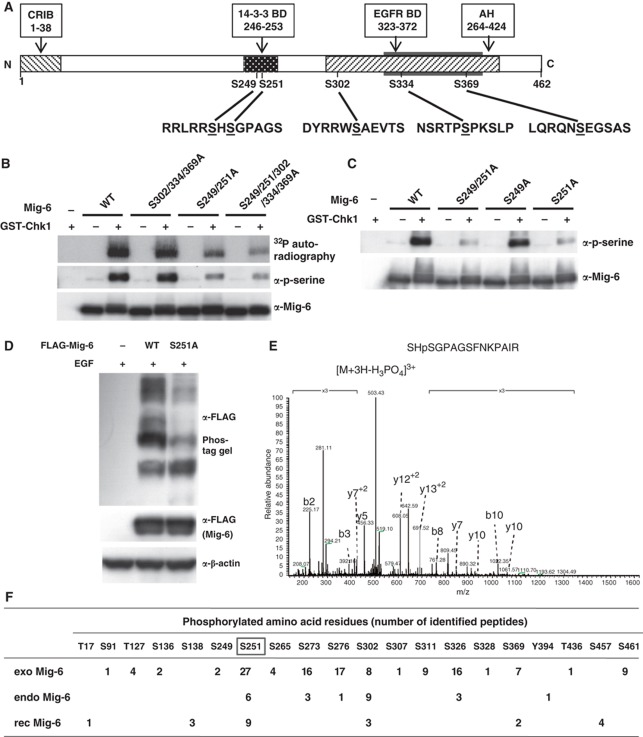
Next, we tried to identify the Chk1-mediated phosphorylation site(s) in Mig-6. Chk1 often phosphorylates serine or threonine residues at an RxxS/T motif (O’Neill et al, 2002). After comparing the amino-acid sequences of Mig-6 between human, mouse, and rat, we selected five conserved serine residues as candidates. S249 and S251 are in the 14-3-3-BD, while S302, S334, and S369 are located in the AH domain, which is involved in binding to EGFR (Figure 3A). We substituted these serine residues with alanine to determine the Chk1 phosphorylation sites using an in vitro kinase assay with rec proteins. We found decreased phosphorylation in the S249/251A and S249/251/302/334/369A mutants but not in the S302/334/369A mutant using 32P autoradiography as well as immunoblotting (IB) with anti-phospho-serine (Figure 3B, Supplementary Figure S3A). Moreover, the Chk1-mediated phosphorylation of S251A but not S249A mutant Mig-6 was apparently decreased compared with that of WT Mig-6 (Figure 3C). To confirm that S251 is a phosphorylation site in vivo, WT or S251A Mig-6 was transfected into HEK293 cells, which were then stimulated with EGF. As shown in Figure 3D, the phosphorylation-dependent mobility shift of the S251A mutant was smaller than that of WT Mig-6, suggesting that S251 is phosphorylated in vivo. At the same time, we found that at least one of S302/334/369 was phosphorylated following EGF stimulation (Supplementary Figure S3B). We tried to generate a Mig-6 phospho-S251-specific antibody to enable further confirmation, but unfortunately were unsuccessful.
Figure 3.
Analysis of the phosphorylation sites in Mig-6. (A) Primary structure of Mig-6 and schematic representation of its point mutation sites. (B, C). In vitro Chk1 phosphorylation site(s) analysis of Mig-6. WT or mutant rec Mig-6 proteins (0.1 μg) were incubated in 20 μl of kinase buffer with 32P-labelled ATP and 0.1 μg of purified rec GST-Chk1 kinase at 30 °C for 30 min. p-Mig-6 was analysed by autoradiography or IB with anti-phospho-serine antibody. (D) Comparison of phosphorylation status between WT and S251A mutant Mig-6 in HEK293 cells. WT or S251A mutant FLAG-Mig-6 was transfected into HEK293 cells. After 16 h serum starvation, cells were stimulated with 20 ng/ml EGF for 15 min and then harvested. Whole cell lysates were separated by 6% Phos-tag SDS–PAGE (upper panel) or normal SDS–PAGE and analysed by IB with the indicated antibodies. (E) Identification of S251 phosphorylation of endo Mig-6 using MS. Quiescence was induced in MDA-MB-231 cells by 16-h serum starvation; the cells were then treated with 20 ng/ml EGF for 15 min and harvested. Endo Mig-6 was IP with anti-Mig-6 antibody from the lysate and separated by SDS–PAGE. After tryptic digestion of the Mig-6 band, phosphorylated peptides were analysed by LC-MS. The spectrum of the charged ions (m/z 503.38) shows that S251 is phosphorylated in the indicated peptide (top right). b ions, fragmentation ions containing the amino terminus of the peptide; y ions, fragmentation ions containing the carboxy terminus of the peptide. (F) Phosphorylation sites in Mig-6. The number of identified phosphorylated peptides in endo Mig-6 are indicated. exo Mig-6 protein IP from EGF-stimulated HEK293 cells transfected with FLAG-Mig-6 was also analysed by MS. rec Mig-6 protein phosphorylated in vitro by GST-Chk1 (rec Mig-6) was separated by SDS–PAGE and analysed by MS. Figure source data can be found with the Supplementary data.
We next demonstrated the phosphorylation sites of Mig-6 using mass spectrometry (MS). HEK293 cells were transfected with FLAG-Mig-6 and stimulated with EGF. Mig-6 protein was prepared by immunoprecipitation with anti-FLAG antibody from the lysates, separated by SDS–PAGE and analysed by liquid chromatography (LC)-MS. We identified S251-phosphorylated peptides such as SHpSGPAGSFNKPAIR, indicating that Mig-6 was phosphorylated at S251 in vivo (Supplementary Figure S4A). Next, endo Mig-6 prepared from MDA-MB-231 cells treated with EGF was subjected to LC-MS analysis. We identified S251-phosphorylated peptides such as SHpSGPAGSFNKPAIR, indicating that endo Mig-6 was also phosphorylated at S251 (Figure 3E). The numbers of identified phosphorylated peptides in the exogenous FLAG-Mig-6 (exo Mig-6) and the endo Mig-6 protein (endo Mig-6) are indicated in Figure 3F. Because phospho-S251-containing peptides were commonly detected in both exo Mig-6 and endo Mig-6, we believe that S251 is an in vivo phosphorylation site in Mig-6.
As shown in Figure 3F, not only S251, but also S273, S276, S302, and S326 were phosphorylated in both exo and endo Mig-6. These sites might include not only Chkl-mediated phosphorylation sites but also Chk1-independent phosphorylation sites. S251 and S302 are located within a putative consensus motif for Chk1 (R-X-X-S/T), whereas S273, S276, and S326 are not. To determine the Chkl-mediated phosphorylation sites, rec Mig-6 protein was phosphorylated by GST-Chk1 in vitro followed by LC-MS analysis. We found that S251 and S302, but not S273, S276, or S326, were phosphorylated by Chk1 in vitro (Figure 3F, Supplementary Figure S4B). Taken together, Chk1 could directly phosphorylate both S251 and S302 in Mig-6, suggesting that they are major Chk1-mediated phosphorylation sites in Mig-6.
Effects of Chk1-mediated phosphorylation of Mig-6 S251 on EGF signalling
To determine the physiological relevance of Mig-6 phosphorylation, we investigated the effect of substitution at the Chk1-mediated phosphorylation sites on Mig-6 function as a negative feedback inhibitor of EGF signalling. WT, S251A, and S251E (phospho-mimic form) Mig-6 were transfected into HEK293 cells, and the cells were treated with or without EGF. Phosphorylation of EGFR was analysed by IB with anti-phospho-EGFR or anti-phospho-tyrosine antibody. Similar to NIH-EGFR (Anastasi et al, 2007) and Cos-7 cells (Zhang et al, 2007), we confirmed that WT Mig-6 suppressed autophosphorylation of EGFR in EGF-stimulated HEK293 cells (Supplementary Figure S5A, lane 2 versus lane 4). The S251A mutant suppressed this autophosphorylation much more strongly than did WT Mig-6 (Supplementary Figure S5A, lane 4 versus lane 6). In contrast, autophosphorylation of EGFR was partially restored in cells expressing the S251E mutant (Supplementary Figure S5A, lane 6 versus lane 8, and B), suggesting that phosphorylation of S251 in Mig-6 negatively regulates the inhibitory activity of Mig-6 on EGF signalling.
To investigate the effect of Mig-6 S251 phosphorylation on cell growth, we performed proliferation assays. Expression of the S251A mutant markedly inhibited the proliferation of HEK293 cells, whereas the WT and S251E mutant did not affect cell growth (Supplementary Figure S5C). This is presumably because WT Mig-6 is phosphorylated at S251 in HEK293 cells in normal culture medium without further stimulation by EGF.
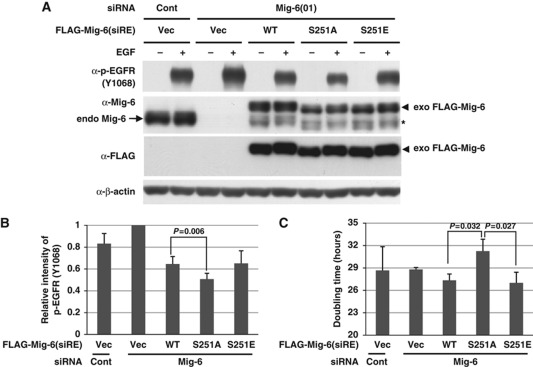
For further information, we performed rescue experiments using siRNA-resistant Mig-6 wild type, S251A, or S251E. MDA-MB-231 cells were infected with retrovirally encoded Mig-6 wild type, S251A, or S251E, and then treated with an siRNA-targeting Mig-6. After EGF stimulation, autophosphorylation of EGFR was analysed by IB. As shown in Figure 4A and B and Supplement Figure S5D, treatment with a Mig-6 siRNA increased the autophosphorylation of EGFR to ∼120% of control levels. Introduction of WT Mig-6 inhibited the autophosphorylation of EGFR to 66% of that for the empty vector control. The S251A mutant suppressed the autophosphorylation of EGFR significantly more strongly than did WT Mig-6. Because Mig-6 is highly phosphorylated in EGF-stimulated MDA-MB-231 cells, the effect of S251E on autophosphorylation was almost the same as that of WT Mig-6. Next, we performed rescue experiments on cell proliferation using siRNA-resistant Mig-6 wild type, S251A, or S251E. As described above, MDA-MB-231 cells were infected with retrovirally encoded Mig-6 wild type, S251A, or S251E, and then treated with siRNA for Mig-6. Then, cell proliferation assays were performed. As shown in Figure 4C, the doubling time of cells expressing S251A was significantly prolonged compared with that of the WT and the S251E mutant. Because Mig-6 is highly phosphorylated in MDA-MB-231 cells in normal culture conditions, it is consistent that the effect of S251E on cell proliferation was almost the same as that of WT Mig-6. Moreover, we obtained almost the same data using HEK293 cells that were transfected with Mig-6 plasmids (Supplementary Figure S5). These results were reproducible in three independent experiments, and the effect of the S251A mutant was always moderate. Therefore, we conclude that phosphorylation of Mig-6 S251 is involved in the regulation of EGF signalling.
Figure 4.
Effect of Mig-6 S251 phosphorylation on EGF signalling. (A) Effect of Mig-6 S251 mutation on EGF signalling. MDA-MB-231 cells were infected with retroviruses encoding Mig-6 wild type, S251A, or S251E, and then treated with an siRNA-targeting Mig-6 or a control siRNA (Cont). After 16 h serum starvation, cells were stimulated with 20 ng/ml EGF, then harvested 15 min later. Cell lysates were separated using SDS–PAGE followed by IB. The asterisk indicates retrovirally expressed Mig-6 protein without the FLAG-tag, which is translated using the original first methionine of the Mig-6 cDNA. (B) The intensity of p-EGFR (Y1068) protein in each condition in (A) was quantified by image analysis. The data from triplicate experiments were evaluated statistically and are shown graphically relative to the data from the vector only. (C) Effect of S251 status on cell growth. MDA-MB-231 cells were infected with retrovirally encoded Mig-6 wild type, S251A, or S251E, and then treated with an siRNA for Mig-6 or a control siRNA (Cont). Cell proliferation assays were performed. The doubling times were calculated and are shown graphically. Figure source data can be found with the Supplementary data.
We tested additional phosphorylation site such as S302 in which phosphorylation as well as S251 was identified by MS in in vitro, in vivo, and endogenous status (Figure 3F). As shown in the Supplementary Figure S6A, S302/334/369A mutant showed the same inhibitory effect on autophosphorylation of EGFR as WT Mig-6 in HEK293 cells. Furthermore, retroviral expression of Mig-6 S302A had no effect on both autophosphorylation of EGFR and proliferation in MDA-MB-231 cells (Supplementary Figure S6, B–D). Taken together, these results suggest that phosphorylation of Mig-6 S251 but not S302 is important for its inhibitory function on EGF signalling and regulation of cell proliferation.
EGF stimulation promotes phosphorylation of Chk1 at S280
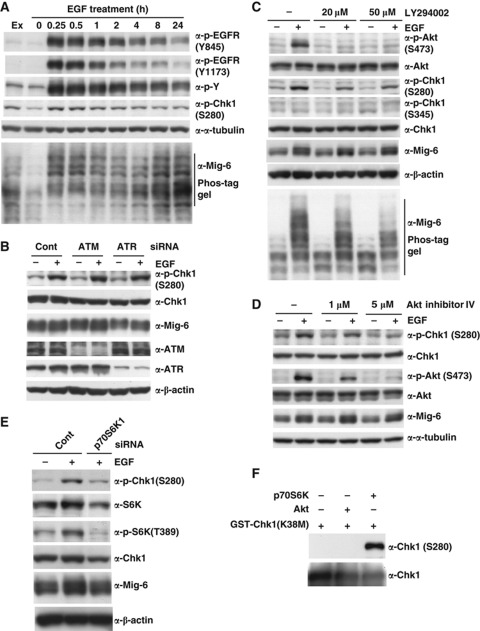
To clarify the Chk1 modification in EGF signalling pathway, we performed quantitative phospho-proteome analysis of EGF-stimulated HeLa cells using LC-MS, a technique termed phospho-iTRAQ (Leitner and Lindner, 2009). Phosphorylation of EGFR (Y1172, Y1192) and PRAS40 (T246) indicating Akt activity peaked at 5 min after stimulation and then oscillated. Chk1 became phosphorylated at S280 from 10 min after stimulation (Supplementary Figure S7, A–C). This suggests that phosphorylation of S280 in Chk1 might be induced via the PI3K/Akt pathway, which is consistent with a previous report that PI3K activation promotes phosphorylation of Chk1 at S280 (Puc et al, 2005). These authors concluded that S280-phosphorylated Chk1 is preferentially tethered in the cytoplasm. We then investigated whether EGF signalling promoted S280 phosphorylation of Chk1 in MDA-MB-231 cells. We found that EGF promoted phosphorylation of Chk1 S280, with a similar duration to Mig-6 phosphorylation, whereas Chk1 S345 was not upregulated (Figure 5A). As shown in Figure 5B, depletion of neither ATM nor ATR affected EGF-dependent S280 phosphorylation of Chk1. This is consistent with the data shown in Figure 2C, in which the ATM/ATR inhibitor caffeine did not affect the phosphorylation of Mig-6.
Figure 5.
Phosphorylation of Chk1 at S280 is promoted by EGF stimulation via the PI3K pathway. (A) Time course of phosphorylated proteins in MDA-MB-231 cells treated with EGF. MDA-MB-231 cells were subjected to serum starvation for 16 h, then treated with 20 ng/ml EGF for the indicated times. The cell lysates were analysed by IB with the indicated antibodies (Ex, without serum starvation and EGF stimulation). (B) Depletion of ATM/ATR had no effect on S280 phosphorylation of Chk1. MDA-MB-231 cells were transfected with an siRNA for human ATM or ATR or a control siRNA (Cont). Quiescence was induced by 16-h serum starvation, and then the cells were stimulated with 20 ng/ml EGF for 15 min. The cell lysates were analysed by IB with the indicated antibodies. (C) Effect of a PI3K inhibitor on Chk1 phosphorylation. MDA-MB-231 cells were subjected to serum starvation for 16 h. Cells were pretreated with or without the PI3K inhibitor LY294002 for 1 h, followed by stimulation with 20 ng/ml EGF for 15 min. Cell lysates were separated by normal SDS–PAGE or 6% Phos-tag SDS–PAGE and immunoblotted. (D) Effect of an Akt inhibitor on Chk1 phosphorylation. MDA-MB-231 cells were made quiescent by 16-h serum starvation, pretreated with or without Akt inhibitor IV for 1 h, then stimulated with 20 ng/ml EGF for 15 min. The cell lysates were analysed by IB with the indicated antibodies. (E) Depletion of p70S6k1 inhibits Chk1 phosphorylation. MDA-MB-231 cells were transfected with an siRNA for human S6K1 or a control siRNA (Cont), then made quiescent by 16-h serum starvation. Cells were harvested after EGF stimulation, and subjected to IB with the indicated antibodies. (F) In vitro phosphorylation of Chk1 S280 by Akt2 and p70S6K. rec GST-Chk1 (kinase-deficient mutant K38M) proteins were incubated in 20 μl of kinase reaction buffer with 50 μM ATP and 1 μl Akt2 (Abcam, ab79798) or p70S6K (Abcam, ab84798) at 30°C for 30 min. The reaction was stopped by the addition of SDS sample buffer, then the proteins were separated using SDS–PAGE followed by IB with indicated antibodies. Figure source data can be found with the Supplementary data.
Next, we investigated the PI3-kinase pathway (Figure 5C–E). The PI3K inhibitor LY294002 inhibited the phosphorylation of not only Akt S473 but also Chk1 S280 and Mig-6 (Figure 5C). Similarly, Akt inhibitor IV inhibited S280 phosphorylation of Chk1 in a dose-dependent manner (Figure 5D). Depletion of p70S6K, a downstream kinase of Akt, inhibited EGF-induced S280 phosphorylation of Chk1 (Figure 5E). To address whether Akt or p70S6K phosphorylates Chk1 at S280 directly, we performed in vitro phosphorylation experiments using recombinant proteins. As shown in Figure 5F, it is p70S6K, but not Akt, phosphorylated Chk1 directly. These results suggest that p70S6K, a downstream kinase of Akt, is involved in phosphorylation of Chk1 at S280 via the EGFR-PI3K/Akt pathway.
Chk1 phosphorylates and inhibits Mig-6, acting as a positive regulator of EGF signalling
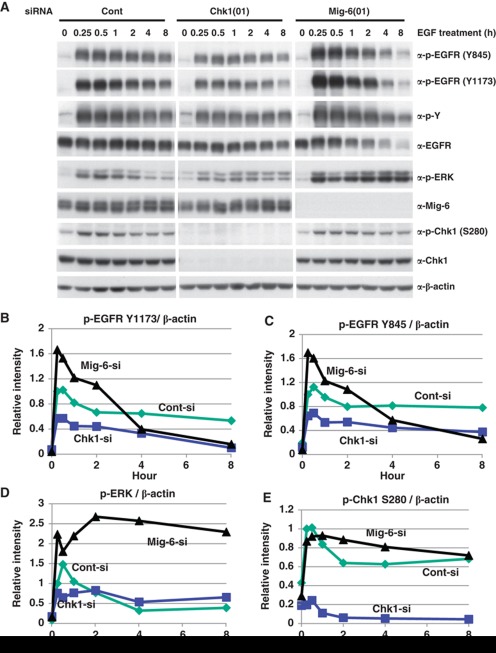
We next investigated the physiological relevance of EGF-promoted phosphorylation of Mig-6 by Chk1. We transfected MDA-MB-231 cells with an siRNA for Chk1(01) or Mig-6(01), or a control siRNA, and then stimulated with EGF. EGF promoted phosphorylation of EGFR, Chk1 S280, and ERK from 5 min after stimulation, peaked at 15–30 min, and reduced from 1 h. Depletion of Chk1 attenuated EGFR phosphorylation and ERK phosphorylation at 5–30 min after EGF treatment. In contrast, depletion of Mig-6 enhanced EGFR phosphorylation and ERK phosphorylation in the same period (Figure 6, Supplementary Figure S9). These tendencies were reproducible in three independent experiments (Figure 6 and Supplementary Figure S8). Moreover, another Chk1 siRNA oligo (04) showed the same result as did Chk1 siRNA oligo (01) (Supplementary Figure S10). These data indicate that depletion of Chk1 results in a decreased phosphorylation of EGFR. Therefore, Chk1 may play a role as a positive regulator of EGF signalling via phosphorylation of Mig-6.
Figure 6.
Effect of Chk1 depletion on EGF signalling. (A) EGF-promoted phosphorylation of EGFR is suppressed by Chk1 depletion. MDA-MB-231 cells were transfected with an siRNA for Chk1 or Mig-6, or a control siRNA (Cont), serum starved for 16 h, then stimulated with 20 ng/ml EGF for the indicated times. Cell lysates were separated by SDS–PAGE and analysed by IB with the indicated antibodies. (B–E). The intensities of the indicated phosphorylated proteins were quantified by image analysis and are shown graphically. Three independent experiments were performed as shown in Supplementary Figure S8. Figure source data can be found with the Supplementary data.
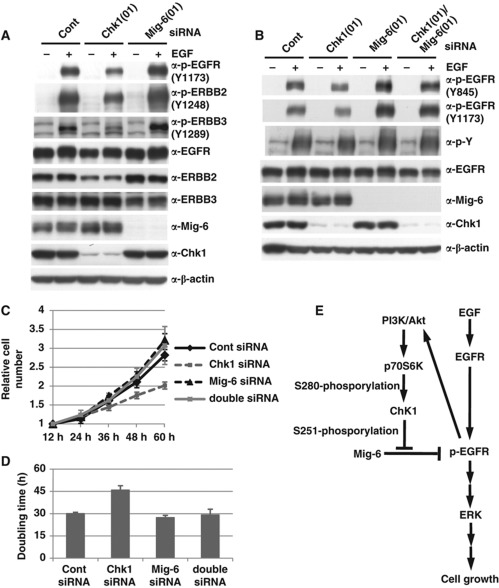
Next, we investigated whether phosphorylation of Mig-6 affects activation of other EGFR family members. As shown in Figure 7A, depletion of Mig-6 facilitated the phosphorylation of not only EGFR but also of ERBB2 and ERBB3 (Figure 7A, lanes 2 versus 6). Moreover, we found that depletion of Chk1 inhibited the phosphorylation of not only EGFR but also of ERBB2 and ERBB3 (Figure 7A, lanes 2 versus 4). These results suggest that Chk1 is also involved in the activation of other EGFR members via phosphorylation of Mig-6.
Figure 7.
Mig-6 is a downstream target for Chk1 in EGF signalling. (A) Effect of Chk1 knockdown on activation of ERBBs. MDA-MB-231 cells were transfected with an siRNA for human Chk1 or Mig-6 or a control siRNA (Cont). Quiescence was induced by 16-h serum starvation, then the cells were stimulated with 20 ng/ml EGF for 15 min. The cell lysates were analysed by IB with the indicated antibodies. (B) Effect of Mig-6 depletion on activation of EGFR promoted by Chk1 depletion. MDA-MB-231 cells were transfected with siRNA for Chk1 and/or Mig-6 or a control siRNA (Cont), serum starved for 16 h, then stimulated with 20 ng/ml EGF for 15 min. Cell lysates were separated by SDS–PAGE and analysed by IB with the indicated antibodies. (C, D) Involvement of Chk1 in cell growth as an upstream regulator of Mig-6. MDA-MB-231 cells were transfected with siRNA for Chk1 and/or Mig-6 or a control siRNA (Cont). Cell numbers were measured at the indicated times (C). The doubling times were calculated from the growth curves (D). Error bars indicate the s.d. of three independent experiments. (E) A model of regulation of EGF signalling by Chk1-mediated phosphorylation of Mig-6. Figure source data can be found with the Supplementary data.
To demonstrate that Mig-6 is a downstream target of Chk1 in EGF signalling, we investigated the effect of Mig-6 depletion on the activation of EGFR promoted by Chk1 depletion. The EGF-dependent activation of EGFR was inhibited by Chk1 depletion, but rescued by co-depletion of Mig-6 (Figure 7B). Next, we investigated the relationship between Mig-6 and Chk1 on cell proliferation and found that cell growth was also suppressed by Chk1 depletion, but was rescued by co-depletion of Mig-6 (Figure 7C and D). Our results suggest that Chk1 phosphorylates Mig-6 on S251, resulting in the inhibition of Mig-6, and is involved in the regulation in EGF signalling as a positive regulator (Figure 7E).
Discussion
Mig-6 as a novel target of Chk1
ATM-Chk2 and ATR-Chk1 play critical roles in the DNA damage stress response pathway (Zhou and Elledge, 2000). In particular, the ATR-Chk1 axis is essential to block the cell cycle through inhibition of the Cdc25 family. Chk1 also regulates Cdc25A (Chen et al, 2003; Lam and Rosen, 2004; Uto et al, 2004) and Cdc25B activities throughout an unperturbed cell cycle (Schmitt et al, 2006). In contrast to the well-characterized role of Chk1 in the checkpoint pathway, little is known about how Chk1 acts in normal cell cycle progression. The kinase activity of Chk1 is maintained during the cell cycle. Recently, Shimada et al (2008) reported that Chk1 bound chromatin and induced cdk1 and cyclin B transcripts by histone H3 Thr11 phosphorylation in the absence of DNA damage. Here, we revealed a novel function of Chk1. We found that the EGFR inhibitor Mig-6 is a novel target of Chk1 under normal cell cycle conditions and in the absence of DNA damage. We noticed that both exo Mig-6 and endo Mig-6 were phosphorylated in living cells. Mig-6 has a PLTP consensus sequence for phosphorylation by ERKs (Gonzalez et al, 1991), a PPLTPI consensus sequence for phosphorylation by cyclin D1-Cdk4 (Kitagawa et al, 1996), PKC (S/T-XR/K) (Woodgett et al, 1986), and several potential sites for phosphorylation by Chk1 (O’Neill et al, 2002). We found that Chk1 is a candidate Mig-6 kinase using various kinase inhibitors and demonstrated this function by depletion of Chk1. We also found that EGF stimulation promoted phosphorylation of Mig-6 in the absence of DNA damage, and that Mig-6 phosphorylation was severely suppressed by a Chk1 inhibitor or by silencing Chk1 by treatment with siRNA. Depletion of ATM or ATR as well as treatment with ATM/ATR inhibitor caffeine did not affect EGF-stimulated Mig-6 phosphorylation. Our results indicated that Chk1 could phosphorylate Mig-6 by EGF stimulation, not via ATR, in a DNA damage-independent manner. It has been reported that S345 and S317 are the main phosphorylation sites of Chk1 in the DNA damage response. We confirmed that phosphorylation of S345 and S317 was upregulated upon DNA damage induced by UV light in MDA-MB-231 cells, but not by EGF stimulation (Figure 5C and unpublished observations). In contrast, phosphorylation of Chk1 at S280 was apparently promoted by EGF signalling. Puc et al (2005) indicated that PI3K activation promotes the phosphorylation of Chk1 at S280 and that S280-phosphorylated Chk1 is preferentially tethered in the cytoplasm. We believe that cytoplasmically localized Chk1 is susceptible to interaction with Mig-6. Moreover, EGFR activity suppressed by Chk1 depletion was rescued by Mig-6 depletion. Therefore, Mig-6 is a downstream target for phosphorylation by Chk1 in a DNA damage-independent manner. The contribution of the Chk1-Mig-6 pathway to EGFR activation is significant and reproducible but moderate, because EGF signalling is regulated by multiple mechanisms. We believe that the Chk1-Mig-6 pathway modulates EGFR activation as one of the regulatory mechanisms of EGF signalling.
Chk1-mediated phosphorylation sites in Mig-6
Tyrosine phosphorylation of Y394 in Mig-6 has been reported by Guha et al (2008), which is consistent with our MS data for endo Mig-6. However, serine/threonine phosphorylation of Mig-6 has not been reported at all. Our results indicated that S251 and S302 are two major phosphorylation sites of Mig-6 by Chk1. However, substitution of serine 302 to alanine did not affect its inhibitory activity against EGFR comparing with WT Mig-6 (Supplementary Figure S6). Therefore, we presume that S251 is the main functional target site for phosphorylation by Chk1 in response to EGF stimulation.
S251 is located in the 14-3-3 binding site (R-S-X-S-X-P) (Muslin et al, 1996) in Mig-6; an interaction between exo Mig-6 and 14-3-3 was reported by Makkinje et al (2000). Our data indicated that WT Mig-6 bound to 14-3-3β, δ, and ξ (Supplementary Figure S11A), whereas S251A mutant showed a low binding affinity to 14-3-3β and ξ but not δ. Moreover, depletion of Chk1 attenuated the binding of Mig-6 to 14-3-3ξ, but not to 14-3-3β and δ (Supplementary Figure S11B). Therefore, phosphorylation of S251, which is located in the putative 14-3-3 binding sequence in Mig-6, may be involved in binding of Mig-6 to 14-3-3ξ.
We noticed that no effect of Mig-6 phosphorylation on its association with EGFR family members was detected using co-immunoprecipitation (data not shown). We speculate that Chk1-mediated phosphorylation of Mig-6 may increase activation of the EGFR family members via some conformational change in the EGFR-Mig-6 complex. Alteration in the binding of Mig-6 to 14-3-3ξ may affect the conformation of Mig-6-EGFR family complex, although further study is required to clarify the mechanism.
Regulation of EGF signalling by the Chk1-Mig-6 pathway
Because Mig-6 is downregulated in many tumour cells, these cells may escape from Mig-6-mediated growth control. EGF signalling is negatively regulated by Mig-6 in cells that express a normal level of Mig-6. Attenuation of Mig-6 activity may be required for efficient activation of EGFR, leading to the accelerated cell growth in these cells. EGF stimulation may promote Chk1-mediated phosphorylation of Mig-6 to negate the EGFR repression. It has been reported that EGF activates the PI3K/Akt pathway (Klein and Levitzki, 2009), and that lack of the PI3K inhibitor PTEN promotes phosphorylation of Chk1 at S280, thereby sequestering S280-phosphorylated Chk1 in the cytoplasm (Puc et al, 2005; Jean et al, 2007). Our results also indicated that EGF stimulation induced phosphorylation of Chk1 at S280, which could be inhibited by a PI3K inhibitor or an Akt inhibitor. Moreover, we found that p70S6K, a downstream kinase of Akt, is involved in the phosphorylation of Chk1 at S280 via EGFR-PI3K/Akt pathway. Therefore, we speculate the following new insight into the regulation of EGF signalling, EGF stimulation activates the PI3K/Akt pathway to phosphorylate Chk1 at S280 in the early stages of EGF signalling. S280-phosphorylated Chk1 is sequestered in the cytoplasm and phosphorylates Mig-6 at S251. Then, the inhibitory activity of Mig-6 on EGFR activation is attenuated by the S251 phosphorylation, and thereby EGFR is fully activated to promote downstream signal transduction and cell growth (Figure 7E). Attenuation of Mig-6 activity may be required for efficient activation of EGFR in the early stage. In the late stages of EGF signalling, transcriptional induction of Mig-6 is increased and Mig-6 is gradually dephosphorylated; EGFR phosphorylation and the downstream signal transduction is therefore attenuated, preventing from overactivity. (Figure 5A). In summary, Chk1 functions as a positive regulator in the early stages of EGF signalling.
Because the EGFR pathway is an important molecular target for cancer therapy, many EGFR tyrosine kinase inhibitors are being tested for an ability to prevent EGF-dependent cancer cell growth. We believe that regulation of the EGFR-Mig-6 pathway is also applicable to cancer therapy. Chk1 kinase inhibitors have been presented as sensitizers against cancer chemotherapy to avoid Chk1-mediated cell cycle arrest (Xiao et al, 2005). In the present study, we found that Chk1 promoted EGF signalling via Mig-6 inhibition. Depletion of Chk1 and Chk1 inhibition suppressed cancer cell growth. Therefore, Chk1 inhibitors may be useful to inhibit EGF-dependent cell growth in Mig-6-positive cancer cells.
Materials and methods
Cell culture
The human cell lines HEK293, HeLa, MDA-MB-231, and retroviral packaging cell line Platinum-A were grown in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum and maintained at 37°C in an atmosphere containing 5% CO2.
Antibodies and inhibitors
The antibodies used in this study were as follows: anti-FLAG antibody M2 (Sigma), anti-Mig-6 antibody PE-16 (Sigma), anti-Mig-6 antibody D-1 (Santa Cruz Biotechnology), anti-phospho-Chk1 (Ser-280) antibody (Cell Signaling), anti-Chk1 antibody G4 (Santa Cruz Biotechnology), anti-phospho-EGFR (Tyr-1068) antibody (Cell Signaling), anti-phospho-EGFR (Tyr-1173) antibody (Cell Signaling), anti-phospho-EGFR (Tyr-845) antibody (UPSTATE), anti-phospho-tyrosine antibody (4G10) (UPSTATE), anti-phospho-p44/p42 MAPK (Thr202/Thr204) antibody (Cell Signaling), anti-phospho-serine antibody (Invitrogen), anti-EGFR antibody (Santa Cruz Biotechnology), anti-ATM antibody (N-19) (Santa Cruz Biotechnology), anti-ATR antibody (2C1) (Santa Cruz Biotechnology), anti-phospho-Akt (Ser-473) antibody (587F11) (Cell Signaling), anti-Akt antibody (Cell Signaling), anti- phospho-p70S6K (Thr-389) antibody (Cell Signaling), anti-p70S6K antibody (Cell Signaling), anti-phospho-ERBB2 (Tyr-1248) antibody (UPSTATE), anti-ERBB2 antibody (Santa Cruz Biotechnology), anti-phospho-ERBB3 (Tyr-1289) antibody (Cell Signaling), anti-ERBB3 antibody (Santa Cruz Biotechnology), anti-β-actin antibody (Sigma), and anti-α-tubulin antibody DM1A (Sigma). The protein kinase inhibitors used in this study are indicated in Supplementary Table 1.
Plasmids and rec proteins
pBabe-EGFR was kindly provided by Wallice Langdon, School of Surgery and Pathology, University of Western Australia. A cDNA encoding WT Mig-6 cloned into pcDNA3.1 was described in a previous report (Tsunoda et al, 2002). All mutants of Mig-6 and Chk1 were constructed using standard rec DNA techniques. WT and mutant cDNA were re-cloned into the pGEX-6P-1 vector (Amersham) and the GST-Mig-6 fusion proteins were prepared as described previously (Frangioni and Neel, 1993). GST-tag was removed by PreScission Protease (GE Healthcare, Little Chalfont, UK) cleavage, and thereby Mig-6 proteins were eluted from Glutathione Sepharose 4B (GE Healthcare).
Immunoprecipitation
Cells were lysed in immunoprecipitation lysis buffer (0.5% Triton X-100, 300 mM NaCl, 25 mM Tris–HCl (pH 7.5)). Cell lysates were incubated with 2 μg of antibodies and protein G+ Sepharose 4FF (GE Healthcare) at 4°C for 2 h. Immunocomplexes were washed four times with lysis buffer, then denatured by treatment with an SDS sample buffer at 100°C for 5 min. IP samples, as well as the original cell lysates (input), were separated by SDS–PAGE and transferred from the gel onto a polyvinylidene difluoride (PVDF) membrane (Millipore, Billerica, MA), followed by IB. The proteins were visualized using an enhanced chemiluminescence system (Perkin Elmer, Waltham, MA).
Immunoblot analysis of phosphorylated proteins
We performed three methods to detect phosphorylation of Mig-6. The first was IB with anti-phospho-serine, anti-phospho-tyrosine, or anti-phospho-EGFR antibodies. The second was chemiluminescent detection using Phos-tag™ biotin (BTL; NARD Institute, Ltd.), which selectively binds to phosphorylated proteins blotted on PVDF membrane. Protein-blotted PVDF membrane was incubated with PB–SH solution (0.2 mM Phos-tag BTL, 0.4 mM Zn(NO3)2, 1 μl streptavidin-conjugated horseradish peroxidase in Tris-buffered saline/0.1% Tween) for 30 min at room temperature. Then, the membrane was washed and the chemiluminescence observed using ECL plus (GE Healthcare) on an X-ray film. The third method was the Phos-tag gel method, which can detect the mobility shifts of phosphorylated proteins using acrylamide-pendant Phos-tag (NARD Institute, Ltd.). The phosphate affinity SDS–PAGE was conducted with 25 μM Phos-tag acrylamide (AAL-107), 25 μM MnCl2, 0.37 M Tris–HCl (pH 8.8), and 0.20% (w/v) SDS. After electrophoresis, the gel was soaked in a general transfer buffer containing 1 mM EDTA and then washed in the same transfer buffer without EDTA. After the treatment, it was transferred from the gel to PVDF membrane and immunoblotted with anti-Mig-6 antibody. The phosphorylated protein forms migrated more slowly than the non-phosphorylated forms.
In vitro phosphorylation assay
rec Mig-6 protein (0.1 μg) was incubated in 20 μl of kinase buffer (50 mM Tris–HCl (pH 7.5), 1 mM EGTA, 10 mM MgCl2, 1 mM DTT) and 32P-labelled ATP with or without 0.1 μg of purified rec GST-Chk1 kinase (Carna Biosciences, Kobe, Japan) at 30°C for 30 min with or without pretreatment with SB218078. The reaction was stopped by the addition of SDS sample buffer then analysed by 8% SDS–PAGE followed by autoradiography, or transferred to PVDF membrane and immunoblotted with anti-phospho-serine antibody.
RNA interference
MDA-MB-231 cells were transfected with human Chk1 siRNA, Mig-6 siRNA, or control siRNA oligonucleotides using Lipofectamine™ RNAiMAX (Invitrogen), according to the manufacturer’s protocol. The nucleotide sequence of the Chk1(01) siRNA was 5′-GCGUGCCGUAGACUGUCCA-3′ and Chk1(04) siRNA was 5′-GAAGUUGGGCUAUCAAUGG-3′ with a 3′dTdT overhang. The nucleotide sequence of the human Mig-6(01) siRNA was 5′-CUACACUUUCUGAUUUCAA-3′ and Mig-6(05) siRNA was 5′-GCAGGGUAUCCAUUCUUUA-3′ with a 3′dTdT overhang. The nucleotide sequence of the human ATR siRNA was 5′-CCTCCGTGATGTTGCTTGA-3′ and S6K1 siRNA was 5′-GGACATGGCAGGAGTGTTT-3′ with a 3′dTdT overhang. ATM siRNA used in this study was purchased from Ambion.
Viral infection
Retroviruses were generated by transfection of pMXs-Puro-Mig-6 into Platinum-A retroviral packaging cell line using Fugene 6 Reagent (Roche). Viral particles were collected 48 h after transfection. MDA-MB-231 cells were plated on 10-mm dishes at a density of 4 × 105 cells per dish. On the next day, the medium was replaced with 6 ml of viral supernatant containing 8 μg/ml Polybrene. At 48 h after retroviral infection, cells were selected with 1 μg/ml puromycin for 72 h, then pooled for further use.
Proliferation assay
HEK293 cells were seeded in 10-cm dishes with 60 000 cells per dish. After 24 h, cells were transfected with WT or mutant pcDNA3-FLAG-Mig-6 together with the pEGFP vector. Images were obtained of four fields per dish at 24, 36, 48, and 60 h after transfection. Green cells were counted as transfected cells.
MDA-MB-231 cells were seeded in 6-cm dishes, and transfected simultaneously with an siRNA against human Chk1 or Mig-6, or a control siRNA. After 24 h, cells were reseeded into 96-well plates with 3500 cells per well, and proliferation assays were performed at 12, 24, 36, 48, and 60 h after replating, according to the manufacturer’s instructions (CyQUANT® Cell Proliferation Assay Kit; Invitrogen).
MS analysis
Proteins were subjected to SDS–PAGE followed by in-gel digestion with trypsin. The obtained peptides were dried and then dissolved in 0.1% TFA, 2% ACN prior to LC-MS/MS analysis. Peptides were analysed using a nanoLC-MS/MS system, composed of an LTQ Orbitrap Velos (Thermo Fisher Scientific) coupled with a nanoLC (Advance, Michrom BioResources) and an HTC-PAL autosampler (CTC Analytics). Peptide separation was carried out using an in-house-pulled fused silica capillary (0.1 mm inside diameter, 15 cm length, 0.05 mm inside diameter at the tip), packed with a 3-μm C18 L-column. The mobile phases consisted of 0.1% formic acid (A) and 100% acetonitrile (B). Peptides were eluted by a gradient of 5–35% B for 40 min at a flow rate of 200 nl/min. CID spectra were acquired automatically in the data-dependent scan mode with the dynamic exclusion option. Full MS was obtained by Orbitrap in the range of m/z 300–2000 with resolution 30 000. The nine most intense precursor ions in the full MS spectra were selected for subsequent MS/MS analysis in an ion trap with the automated gain control mode. The lock mass function was activated to minimize mass error during analysis. The peak lists were generated by MSn.exe (Thermo Fisher Scientific) with a minimum scan/group value of 1, and were compared with the Human International Protein Index version 3.1.6 (57366 protein sequences; European Bioinformatics Institute) with the use of the MASCOT algorithm (ver. 2.1.4). Trypsin was selected to be the enzyme used, the permitted number of missed cleavages was set as two, and carbamidomethylation on cysteine was selected as the fixed modification. Oxidized methionine and phosphorylation on serine, threonine, and tyrosine were searched for as variable modifications. The precursor mass tolerances were 10 ppm and the tolerance of the MS/MS ions was 0.8 Da.
Supplementary Material
Acknowledgments
We thank Drs Wallace Langdon and Tsukasa Matsunaga for plasmids, as well as Drs Ping Li and Hidemasa Goto, and Masaki Inagaki, Toshiyuki Tsunoda, Chiharu Uchida, Takayuki Hattori, and Hayato Ihara for their useful discussions. This work was supported in part by grants from the Ministry of Education, Culture, Sports, Science and Technology of Japan (to MK, YK, and KK).
Author contributions: NL and MK wrote the manuscript. NL and MM performed experiments. NL, MM SayuriS and MK analysed the data. NL, KK, YK, SenjiS, KIN, MN, HN, and MK designed the experiments.
Footnotes
The authors declare that they have no conflict of interest.
References
- Amatschek S, Koenig U, Auer H, Steinlein P, Pacher M, Gruenfelder A, Dekan G, Vogl S, Kubista E, Heider KH, Stratowa C, Schreiber M, Sommergruber W (2004) Tissue-wide expression profiling using cDNA subtraction and microarrays to identify tumor-specific genes. Cancer Res 64: 844–856 [DOI] [PubMed] [Google Scholar]
- Anastasi S, Baietti MF, Frosi Y, Alemâ S, Segatto O (2007) The evolutionarily conserved EBR module of RALT/MIG6 mediates suppression of the EGFR catalytic activity. Oncogene 26: 7833–7846 [DOI] [PubMed] [Google Scholar]
- Anastasi S, Fiorentino L, Fiorini M, Fraioli R, Sala G, Castellani L, Alemâ S, Alimandi M, Segatto O (2003) Feedback inhibition by RALT controls signal output by the ErbB network. Oncogene 22: 4221–4234 [DOI] [PubMed] [Google Scholar]
- Anastasi S, Sala G, Huiping C, Caprini E, Russo G, Iacovelli S, Lucini F, Ingvarsson S, Segatto O (2005) Loss of RALT/MIG-6 expression in ERBB2-amplified breast carcinomas enhances ErbB-2 oncogenic potency and favors resistance to Herceptin. Oncogene 24: 4540–4548 [DOI] [PubMed] [Google Scholar]
- Chen MS, Ryan CE, Piwnica-Worms H (2003) Chk1 kinase negatively regulates mitotic function of Cdc25A phosphatase through 14-3-3 binding. Mol Cell Biol 23: 7488–7497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferby I, Reschke M, Kudlacek O, Knyazev P, Pantê G, Amann K, Sommergruber W, Kraut N, Ullrich A, Fässler R, Klein R (2006) Mig6 is a negative regulator of EGF receptor-mediated skin morphogenesis and tumor formation. Nat Med 12: 568–573 [DOI] [PubMed] [Google Scholar]
- Fiorentino L, Pertica C, Fiorini M, Talora C, Crescenzi M, Castellani L, Alemâ S, Benedetti P, Segatto O (2000) Inhibition of ErbB-2 mitogenic and transforming activity by RALT, a mitogen-induced signal transducer which binds to the ErbB-2 kinase domain. Mol Cell Biol 20: 7735–7750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiorini M, Ballaro C, Sala G, Falcone G, Alemâ S, Segatto O (2002) Expression of RALT, a feedback inhibitor of ErbB receptors, is subjected to an integrated transcriptional and post-translational control. Oncogene 21: 6530–6539 [DOI] [PubMed] [Google Scholar]
- Frangioni JV, Neel BG (1993) Solubilization and purification of enzymatically active glutathione S-transferase (pGEX) fusion proteins. Anal Biochem 210: 179–187 [DOI] [PubMed] [Google Scholar]
- Gonzalez FA, Raden DL, Davis RJ (1991) Identification of substrate recognition determinants for human ERK1 and ERK2 protein kinases. J Biol Chem 266: 22159–22163 [PubMed] [Google Scholar]
- Guha U, Chaerkady R, Marimuthu A, Patterson AS, Kashyap MK, Harsha HC, Sato M, Bader JS, Lash AE, Minna JD, Pandey A, Varmus HE (2008) Comparisons of tyrosine phosphorylated proteins in cells expressing lung cancer-specific alleles of EGFR and KRAS. Proc Natl Acad Sci USA 105: 14112–14117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackel PO, Gishizky M, Ullrich A (2001) Mig-6 is a negative regulator of the epidermal growth factor receptor signal. Biol Chem 382: 1649–1662 [DOI] [PubMed] [Google Scholar]
- Jean C, Hernandez-Pigeon H, Blanc A, Charveron M, Laurent G (2007) Epidermal growth factor receptor pathway mitigates UVA-induced G2/M arrest in keratinocyte cells. J Invest Dermatol 127: 2418–2424 [DOI] [PubMed] [Google Scholar]
- Kitagawa M, Higashi H, Jung HK, Suzuki-Takahashi I, Ikeda M, Tamai K, Kato J, Segawa K, Yoshida E, Nishimura S, Taya Y (1996) The consensus motif for phosphorylation by cyclin D1-Cdk4 is different from that for phosphorylation by cyclin A/E-Cdk2. EMBO J 15: 7060–7069 [PMC free article] [PubMed] [Google Scholar]
- Klein S, Levitzki A (2009) Targeting the EGFR and the PKB pathway in cancer. Curr Opin Cell Biol 21: 185–193 [DOI] [PubMed] [Google Scholar]
- Lam MH, Rosen JM (2004) Chk1 versus Cdc25: checking one’s levels of cellular proliferation. Cell Cycle 3: 1355–1357 [DOI] [PubMed] [Google Scholar]
- Leitner A, Lindner W (2009) Chemical tagging strategies for mass spectrometry-based phospho-proteomics. Methods Mol Biol 527: 229–243 [DOI] [PubMed] [Google Scholar]
- Liu Q, Guntuku S, Cui XS, Matsuoka S, Cortez D, Tamai K, Luo G, Carattini-Rivera S, DeMayo F, Bradley A, Donehower LA, Elledge SJ (2000) Chk1 is an essential kinase that is regulated by Atr and required for the G(2)/M DNA damage checkpoint. Genes Dev 14: 1448–1459 [PMC free article] [PubMed] [Google Scholar]
- Mabuchi R, Sasazuki T, Shirasawa S (2005) Mapping of the critical region of mitogen-inducible gene-6 for NF-κB activation. Oncol Rep 13: 473–476 [PubMed] [Google Scholar]
- Makkinje A, Quinn DA, Chen A, Cadilla CL, Force T, Bonventre JV, Kyriakis JM (2000) Gene 33/Mig-6, a transcriptionally inducible adapter protein that binds GTP-Cdc42 and activates SAPK/JNK. A potential marker transcript for chronic pathologic conditions, such as diabetic nephropathy. Possible role in the response to persistent stress. J Biol Chem 275: 17838–17847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mailand N, Falck J, Lukas C, Syljuâsen RG, Welcker M, Bartek J, Lukas J (2000) Rapid destruction of human Cdc25A in response to DNA damage. Science 288: 1425–1429 [DOI] [PubMed] [Google Scholar]
- Muslin AJ, Tanner JW, Allen PM, Shaw AS (1996) Interaction of 14-3-3 with signaling proteins is mediated by the recognition of phosphoserine. Cell 84: 889–897 [DOI] [PubMed] [Google Scholar]
- Niida H, Nakanishi M (2006) DNA damage checkpoints in mammals. Mutagenesis 21: 3–9 [DOI] [PubMed] [Google Scholar]
- Niida H, Tsuge S, Katsuno Y, Konishi A, Takeda N, Nakanishi M (2005) Depletion of Chk1 leads to premature activation of Cdc2-cyclin B and mitotic catastrophe. J Biol Chem 280: 39246–39252 [DOI] [PubMed] [Google Scholar]
- O’Neill T, Giarratani L, Chen P, Iyer L, Lee CH, Bobiak M, Kanai F, Zhou BB, Chung JH, Rathbun GA (2002) Determination of substrate motifs for human Chk1 and hCds1/Chk2 by the oriented peptide library approach. J Biol Chem 277: 16102–16115 [DOI] [PubMed] [Google Scholar]
- Pante G, Thompson J, Lamballe F, Iwata T, Ferby I, Barr FA, Davies AM, Maina F, Klein R (2005) Mitogen-inducible gene 6 is an endogenous inhibitor of HGF/Met-induced cell migration and neurite growth. J Cell Biol 171: 337–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puc J, Keniry M, Li HS, Pandita TK, Choudhury AD, Memeo L, Mansukhani M, Murty VV, Gaciong Z, Meek SE, Piwnica-Worms H, Hibshoosh H, Parsons R (2005) Lack of PTEN sequesters CHK1 and initiates genetic instability. Cancer Cell 7: 193–204 [DOI] [PubMed] [Google Scholar]
- Reschke M, Ferby I, Stepniak E, Seitzer N, Horst D, Wagner EF, Ullrich A (2010) Mitogen-inducible gene-6 is a negative regulator of epidermal growth factor receptor signaling in hepatocytes and human hepatocellular carcinoma. Hepatology 51: 1383–1390 [DOI] [PubMed] [Google Scholar]
- Ruan DT, Warren RS, Moalem J, Chung KW, Griffin AC, Shen W, Duh QY, Nakakura E, Donner DB, Khanafshar E, Weng J, Clark OH, Kebebew E (2008) Mitogen-inducible gene-6 expression correlates with survival and is an independent predictor of recurrence in BRAF (V600E) positive papillary thyroid cancers. Surgery 144: 908–913discussion 913-914 [DOI] [PubMed] [Google Scholar]
- Sarkaria JN, Busby EC, Tibbetts RS, Roos P, Taya Y, Karnitz LM, Abraham RT (1999) Inhibition of ATM and ATR kinase activities by the radiosensitizing agent, caffeine. Cancer Res 59: 4375–4382 [PubMed] [Google Scholar]
- Schmitt E, Boutros R, Froment C, Monsarrat B, Ducommun B, Dozier C (2006) CHK1 phosphorylates CDC25B during the cell cycle in the absence of DNA damage. J Cell Sci 119: 4269–4275 [DOI] [PubMed] [Google Scholar]
- Shimada M, Niida H, Zineldeen DH, Tagami H, Tanaka M, Saito H, Nakanishi M (2008) Chk1 is a histone H3 threonine 11 kinase that regulates DNA damage-induced transcriptional repression. Cell 132: 221–232 [DOI] [PubMed] [Google Scholar]
- Tong J, Taylor P, Jovceva E, St-Germain JR, Jin LL, Nikolic A, Gu X, Li ZH, Trudel S, Moran MF (2008) Tandem immunoprecipitation of phosphotyrosine-mass spectrometry (TIPY-MS) indicates C19ORF19 becomes tyrosine-phosphorylated and associated with activated epidermal growth factor receptor. J Proteome Res 7: 1067–1077 [DOI] [PubMed] [Google Scholar]
- Tsunoda T, Inokuchi J, Baba I, Okumura K, Naito S, Sasazuki T, Shirasawa S (2002) A novel mechanism of nuclear factor κB activation through the binding between inhibitor of nuclear factor-κBα and the processed NH2-terminal region of Mig-6. Cancer Res 62: 5668–5671 [PubMed] [Google Scholar]
- Uto K, Inoue D, Shimuta K, Nakajo N, Sagata N (2004) Chk1, but not Chk2, inhibits Cdc25 phosphatases by a novel common mechanism. EMBO J 23: 3386–3396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodgett JR, Gould KL, Hunter T (1986) Substrate specificity of protein kinase C. Use of synthetic peptides corresponding to physiological sites as probes for substrate recognition requirements. Eur J Biochem 161: 177–184 [DOI] [PubMed] [Google Scholar]
- Xiao Z, Xue J, Semizarov D, Sowin TJ, Rosenberg SH, Zhang H (2005) Novel indication for cancer therapy: Chk1 inhibition sensitizes tumor cells to antimitotics. Int J Cancer 115: 528–538 [DOI] [PubMed] [Google Scholar]
- Xu DZ, Makkinje A, Kyriakis JM (2005) Gene 33 is an endogenous inhibitor of epidermal growth factor (EGF) receptor signaling and mediates dexamethasone-induced suppression of EGF function. J Biol Chem 280: 2924–2933 [DOI] [PubMed] [Google Scholar]
- Zhang XW, Pickin KA, Bose R, Jura N, Cole PA, Kuriyan J (2007) Inhibition of the EGF receptor by binding of MIG6 to an activating kinase domain interface. Nature 450: 741–745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YW, Staal B, Su Y, Swiatek P, Zhao P, Cao B, Resau J, Sigler R, Bronson R, Vande Woude GF (2007) Evidence that MIG-6 is a tumour-suppressor gene. Oncogene 26: 269–276 [DOI] [PubMed] [Google Scholar]
- Zhang YW, Vande Woude GF (2007) Mig-6, signal transduction, stress response and cancer. Cell Cycle 6: 507–513 [DOI] [PubMed] [Google Scholar]
- Zhou BB, Elledge SJ (2000) The DNA damage response: putting checkpoints in perspective. Nature 408: 433–439 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.