Abstract
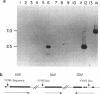
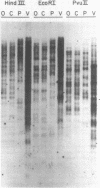
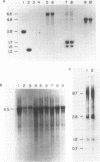
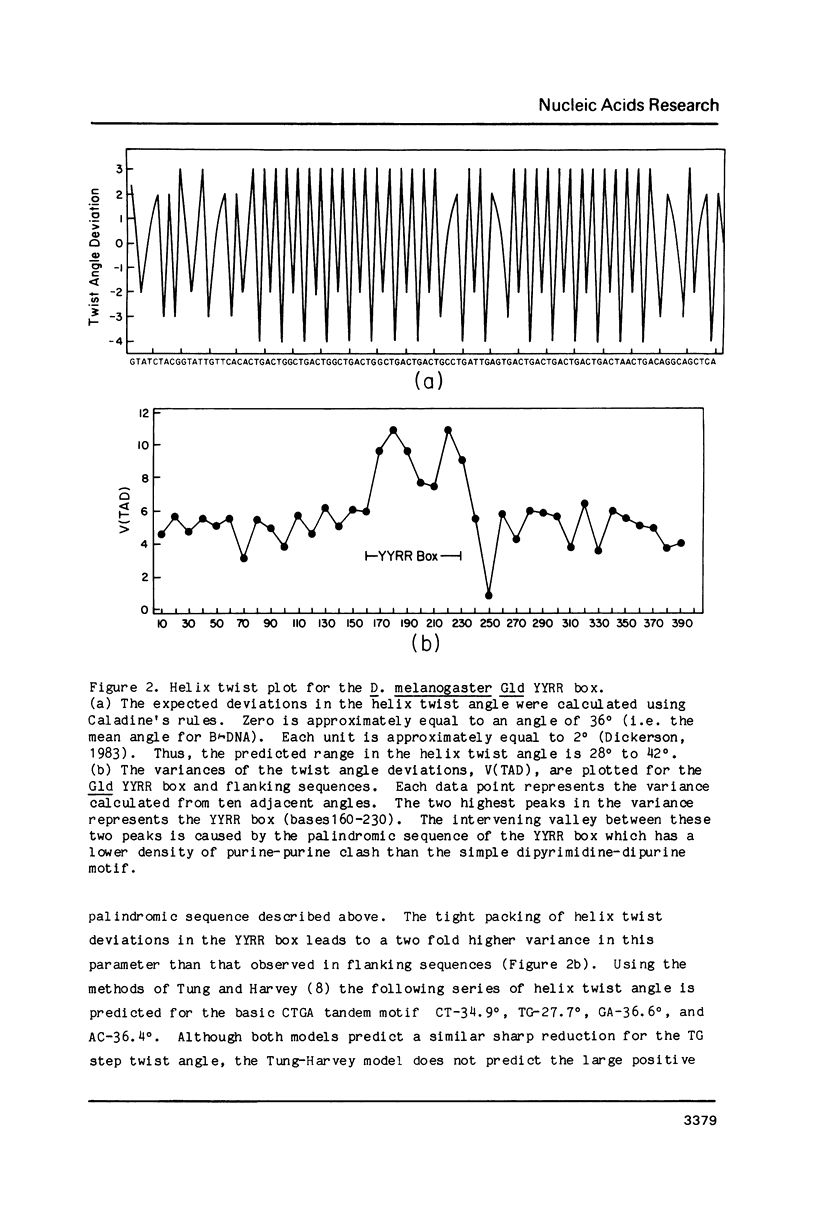
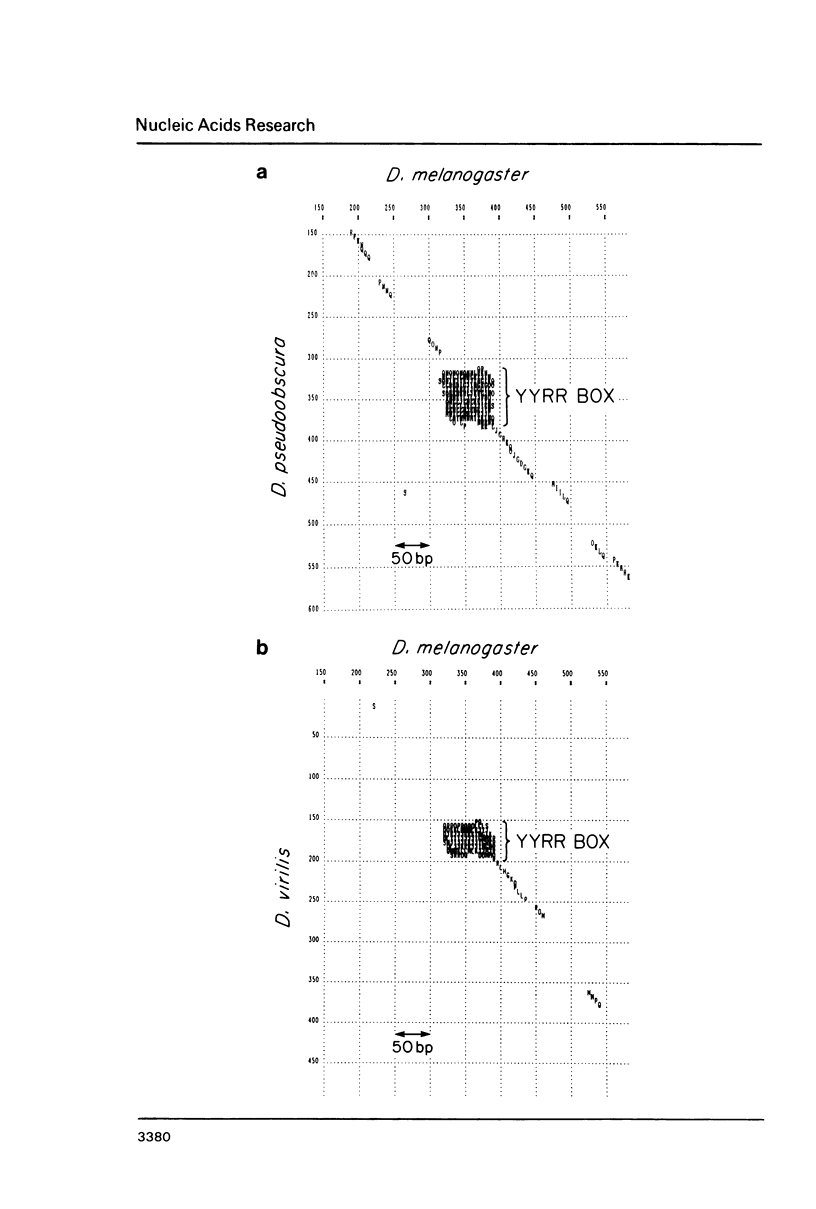
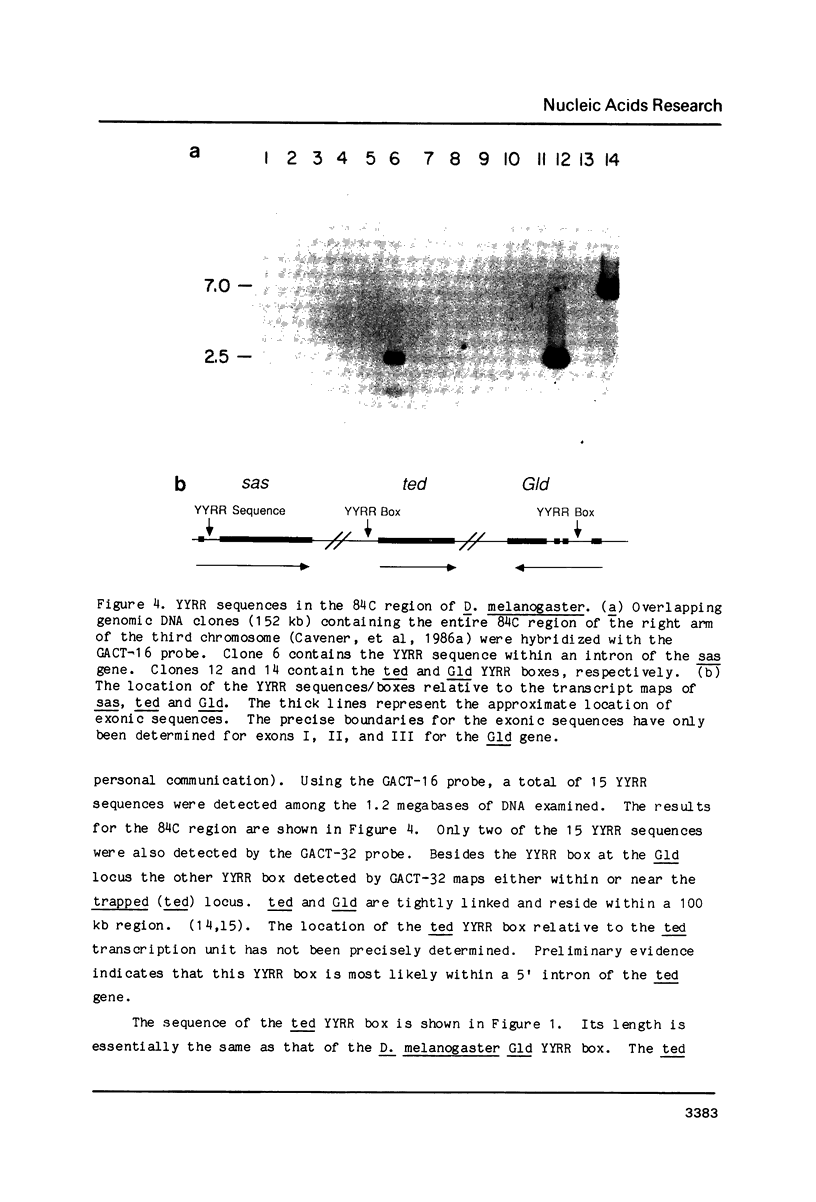
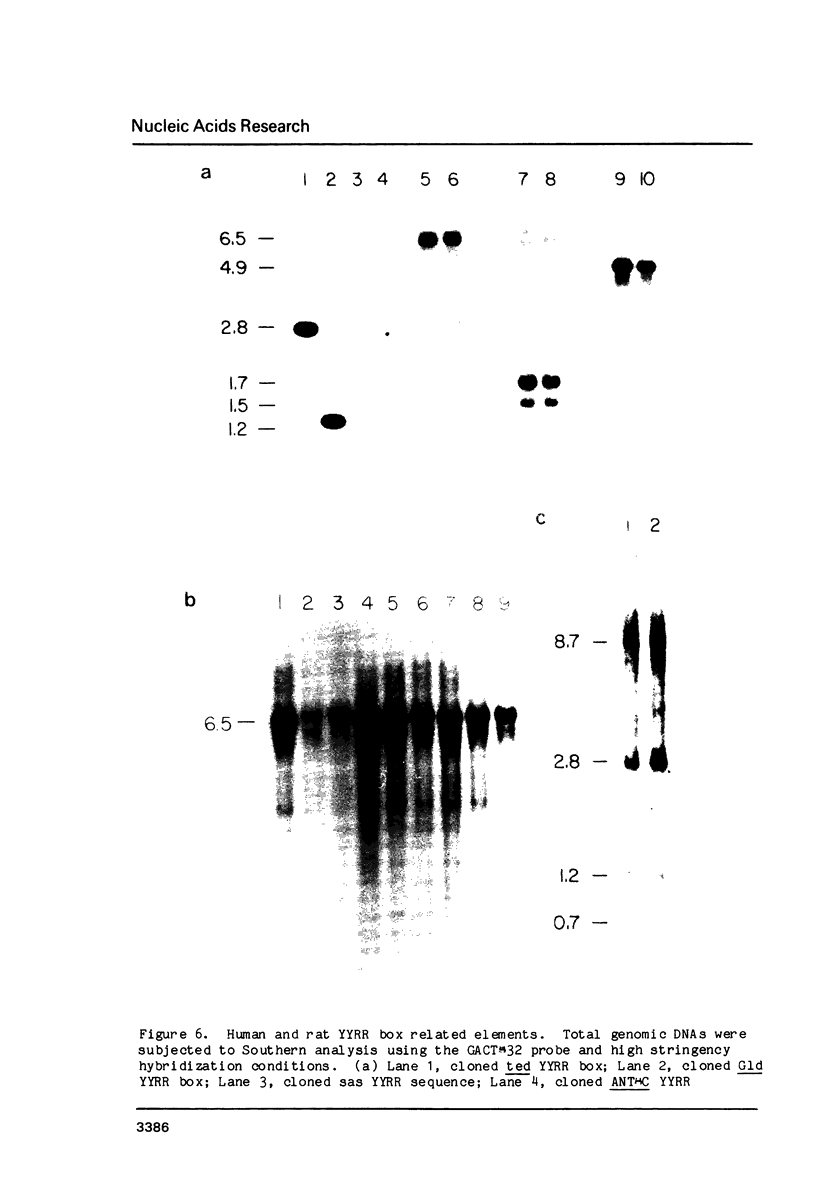
We have discovered a novel DNA sequence element in Drosophila which is based upon a CTGA tandem repeat. This element has been named the YYRR box to emphasize its dipyrimidine-dipurine nature which is predicted to have unusual structural features. Southern hybridization analysis of genomic DNA indicates the presence of 25-30 copies of the YYRR box in each of three Drosophila species (melanogaster, pseudoobscura, and virilis) and conservation of genomic location within species. Similar analysis of human and rat DNA indicates the presence of YYRR related sequences in mammals as well. YYRR boxes have been localized to two genetic loci in Drosophila: Gld and a gene tentative identified as ted. These two genes exhibit correlated patterns of developmental expression and an identical mutant phenotype. Sequence analysis of the Gld YYRR box in three Drosophila species revealed a high degree of conservation despite its intronic location.
Full text
PDF











Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bender W., Akam M., Karch F., Beachy P. A., Peifer M., Spierer P., Lewis E. B., Hogness D. S. Molecular Genetics of the Bithorax Complex in Drosophila melanogaster. Science. 1983 Jul 1;221(4605):23–29. doi: 10.1126/science.221.4605.23. [DOI] [PubMed] [Google Scholar]
- Blackman R. K., Meselson M. Interspecific nucleotide sequence comparisons used to identify regulatory and structural features of the Drosophila hsp82 gene. J Mol Biol. 1986 Apr 20;188(4):499–515. doi: 10.1016/s0022-2836(86)80001-8. [DOI] [PubMed] [Google Scholar]
- Calladine C. R. Mechanics of sequence-dependent stacking of bases in B-DNA. J Mol Biol. 1982 Oct 25;161(2):343–352. doi: 10.1016/0022-2836(82)90157-7. [DOI] [PubMed] [Google Scholar]
- Dickerson R. E. Base sequence and helix structure variation in B and A DNA. J Mol Biol. 1983 May 25;166(3):419–441. doi: 10.1016/s0022-2836(83)80093-x. [DOI] [PubMed] [Google Scholar]
- Drew H. R., Travers A. A. DNA structural variations in the E. coli tyrT promoter. Cell. 1984 Jun;37(2):491–502. doi: 10.1016/0092-8674(84)90379-9. [DOI] [PubMed] [Google Scholar]
- Fratini A. V., Kopka M. L., Drew H. R., Dickerson R. E. Reversible bending and helix geometry in a B-DNA dodecamer: CGCGAATTBrCGCG. J Biol Chem. 1982 Dec 25;257(24):14686–14707. [PubMed] [Google Scholar]
- Henikoff S. Unidirectional digestion with exonuclease III creates targeted breakpoints for DNA sequencing. Gene. 1984 Jun;28(3):351–359. doi: 10.1016/0378-1119(84)90153-7. [DOI] [PubMed] [Google Scholar]
- Kabsch W., Sander C., Trifonov E. N. The ten helical twist angles of B-DNA. Nucleic Acids Res. 1982 Feb 11;10(3):1097–1104. doi: 10.1093/nar/10.3.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karch F., Weiffenbach B., Peifer M., Bender W., Duncan I., Celniker S., Crosby M., Lewis E. B. The abdominal region of the bithorax complex. Cell. 1985 Nov;43(1):81–96. doi: 10.1016/0092-8674(85)90014-5. [DOI] [PubMed] [Google Scholar]
- Keene M. A., Elgin S. C. Patterns of DNA structural polymorphism and their evolutionary implications. Cell. 1984 Jan;36(1):121–129. doi: 10.1016/0092-8674(84)90080-1. [DOI] [PubMed] [Google Scholar]
- Nussinov R., Lennon G. G. Structural features are as important as sequence homologies in Drosophila heat shock gene upstream regions. J Mol Evol. 1984;20(2):106–110. doi: 10.1007/BF02257370. [DOI] [PubMed] [Google Scholar]
- Nussinov R., Shapiro B., Lipkin L. E., Maizel J. V., Jr DNAase I hypersensitive sites may be correlated with genomic regions of large structural variation. J Mol Biol. 1984 Aug 25;177(4):591–607. doi: 10.1016/0022-2836(84)90039-1. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott M. P., Weiner A. J., Hazelrigg T. I., Polisky B. A., Pirrotta V., Scalenghe F., Kaufman T. C. The molecular organization of the Antennapedia locus of Drosophila. Cell. 1983 Dec;35(3 Pt 2):763–776. doi: 10.1016/0092-8674(83)90109-5. [DOI] [PubMed] [Google Scholar]
- Shimizu A., Honjo T. Immunoglobulin class switching. Cell. 1984 Apr;36(4):801–803. doi: 10.1016/0092-8674(84)90029-1. [DOI] [PubMed] [Google Scholar]
- Spradling A. C., de Cicco D. V., Wakimoto B. T., Levine J. F., Kalfayan L. J., Cooley L. Amplification of the X-linked Drosophila chorion gene cluster requires a region upstream from the s38 chorion gene. EMBO J. 1987 Apr;6(4):1045–1053. doi: 10.1002/j.1460-2075.1987.tb04857.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tung C. S., Harvey S. C. A common structural feature in promoter sequences of E. coli. Nucleic Acids Res. 1987 Jun 25;15(12):4973–4985. doi: 10.1093/nar/15.12.4973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tung C. S., Harvey S. C. A molecular mechanical model to predict the helix twist angles of B-DNA. Nucleic Acids Res. 1984 Apr 11;12(7):3343–3356. doi: 10.1093/nar/12.7.3343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tung C. S., Harvey S. C. Base sequence, local helix structure, and macroscopic curvature of A-DNA and B-DNA. J Biol Chem. 1986 Mar 15;261(8):3700–3709. [PubMed] [Google Scholar]
- Tung C. S., Harvey S. C. Computer graphics program to reveal the dependence of the gross three-dimensional structure of the B-DNA double helix on primary structure. Nucleic Acids Res. 1986 Jan 10;14(1):381–387. doi: 10.1093/nar/14.1.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wharton K. A., Yedvobnick B., Finnerty V. G., Artavanis-Tsakonas S. opa: a novel family of transcribed repeats shared by the Notch locus and other developmentally regulated loci in D. melanogaster. Cell. 1985 Jan;40(1):55–62. doi: 10.1016/0092-8674(85)90308-3. [DOI] [PubMed] [Google Scholar]
- Young M. W. Middle repetitive DNA: a fluid component of the Drosophila genome. Proc Natl Acad Sci U S A. 1979 Dec;76(12):6274–6278. doi: 10.1073/pnas.76.12.6274. [DOI] [PMC free article] [PubMed] [Google Scholar]