Abstract
Isoprenoids are a diverse group of molecules found in all organisms, where they perform such important biological functions as hormone signaling (e.g., steroids) in mammals, antioxidation (e.g., carotenoids) in plants, electron transport (e.g., ubiquinone), and cell wall biosynthesis intermediates in bacteria. All isoprenoids are synthesized by the consecutive condensation of the five-carbon monomer isopentenyl diphosphate (IPP) to its isomer, dimethylallyl diphosphate (DMAPP). The biosynthetic pathway for the formation of IPP from acetyl-CoA (i.e., the mevalonate pathway) had been established mainly in mice and the budding yeast Saccharomyces cerevisiae. Curiously, most prokaryotic microorganisms lack homologs of the genes in the mevalonate pathway, even though IPP and DMAPP are essential for isoprenoid biosynthesis in bacteria. This observation provided an impetus to search for an alternative pathway to synthesize IPP and DMAPP, ultimately leading to the discovery of the mevalonate-independent 2-C-methyl-d-erythritol 4-phosphate pathway. This review article focuses on our significant contributions to a comprehensive understanding of the biosynthesis of IPP and DMAPP.
Keywords: biosynthesis, inhibitor, isoprenoid, MEP pathway, mevalonate pathway, terpenoid
Introduction
In 1956, Tamura, who had studied why Japanese rice wine, “sake”, is frequently spoiled by the bacteria Lactobacillus homohiochi or Lactobacillus heterohiochi, discovered “hiochic acid”, a bacterial growth factor, and identified the structure of this chemical to be β-hydroxy-β-methyl-δ-valerolactone.1) In the same year, Wolf et al. discovered an acetate-replacing factor for Lactobacilli and identified it as β-hydroxy-β-methyl-δ-valerolactone.2) This compound was also found to be a precursor of cholesterol.3) Later, the standard name of β-hydroxy-β-methyl-δ-valerolactone was changed to mevalonate (MVA).
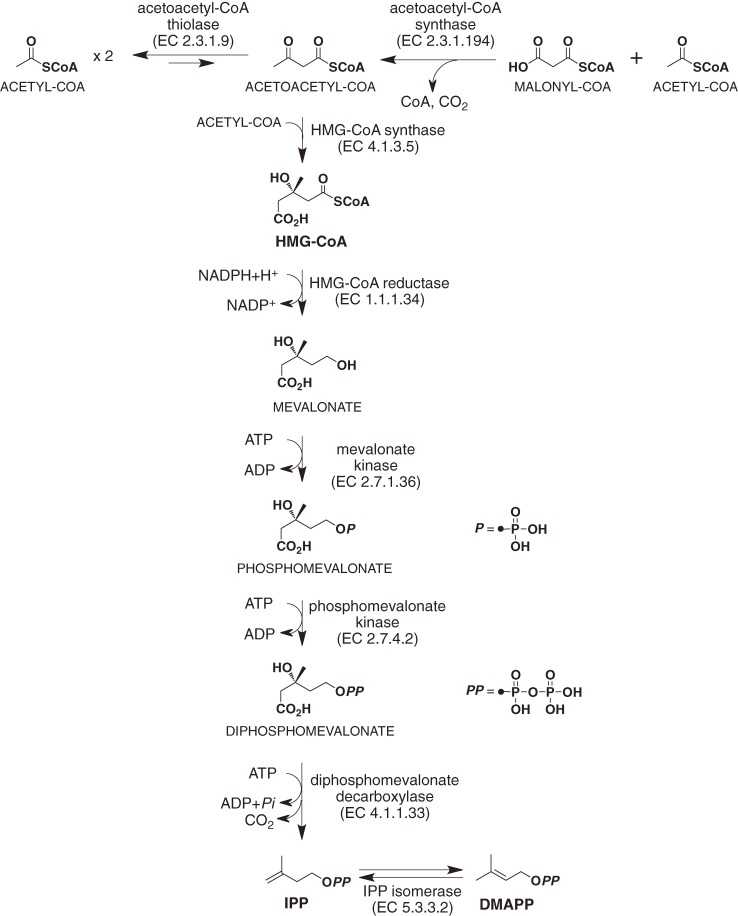
In the 1960s, Bloch and Lynen established the MVA pathway for cholesterol biosynthesis.4,5) In this pathway, three molecules of acetyl-CoA condense successively to form 3-hydroxy-3-methylglutaryl-CoA (HMG-CoA) (Fig. 1). This CoA-derivative is reduced to MVA by HMG-CoA reductase, which is the rate-limiting enzyme of the MVA pathway in humans. Clinically, the specific inhibitors of HMG-CoA reductase, pravastatin and related compounds, are widely used as cholesterol-lowering agents.6) In the next step of the pathway, MVA is phosphorylated twice and finally decarboxylated to form isopentenyl diphosphate (IPP). IPP is then converted to its isomer, dimethylallyl diphosphate (DMAPP), in a reaction that is catalyzed by IPP isomerase. IPP and DMAPP that have been synthesized in the MVA pathway are subsequently used as basic units in the biosynthesis of isoprenoids, such as sterols, carotenoids, and dolichols.
Figure 1.
Mevalonate pathway for IPP and DMAPP biosynthesis.
Since the discovery of the MVA pathway, it had been widely accepted that IPP and DMAPP were formed only through this pathway in all living organisms. However, several results inconsistent with the operation of the MVA pathway in certain bacteria had been reported. For example, [13C]-labeled acetate, a precursor of the MVA pathway, was not incorporated into ubiquinone of Escherichia coli7) or into a sesquiterpene pentalenolactone, which had been produced by Streptomyces chromofuscus (H. Seto, unpublished results). Feeding experiments with [U-13C]-labeled glucose and pentalenolactone-producing Streptomyces exfoliatus UC5319 showed labeling patterns inconsistent with the MVA pathway.8,9) Furthermore, mevinolin, a specific inhibitor of HMG-CoA reductase, did not inhibit the growth of E. coli.7) These results suggested the existence of an alternative pathway for isoprenoid biosynthesis that had been overlooked since the discovery of the MVA pathway.
In 1996, Rohmer identified the first reaction step of an alternative MVA-independent pathway for the formation of IPP and DMAPP in E. coli.10) We were encouraged by this pioneering work and began to study the newly discovered pathway. Initially, this pathway was called the “MVA-independent pathway” or “nonmevalonate pathway”, but at present, the term “2-C-methyl-d-erythritol 4-phosphate (MEP) pathway” is widely accepted and used because MEP is the first biosynthetic intermediate specific for this pathway. This review focuses on our significant contributions to a comprehensive understanding of the MEP and MVA pathways for the essential metabolic precursors for isoprenoid biosynthesis, IPP and DMAPP.
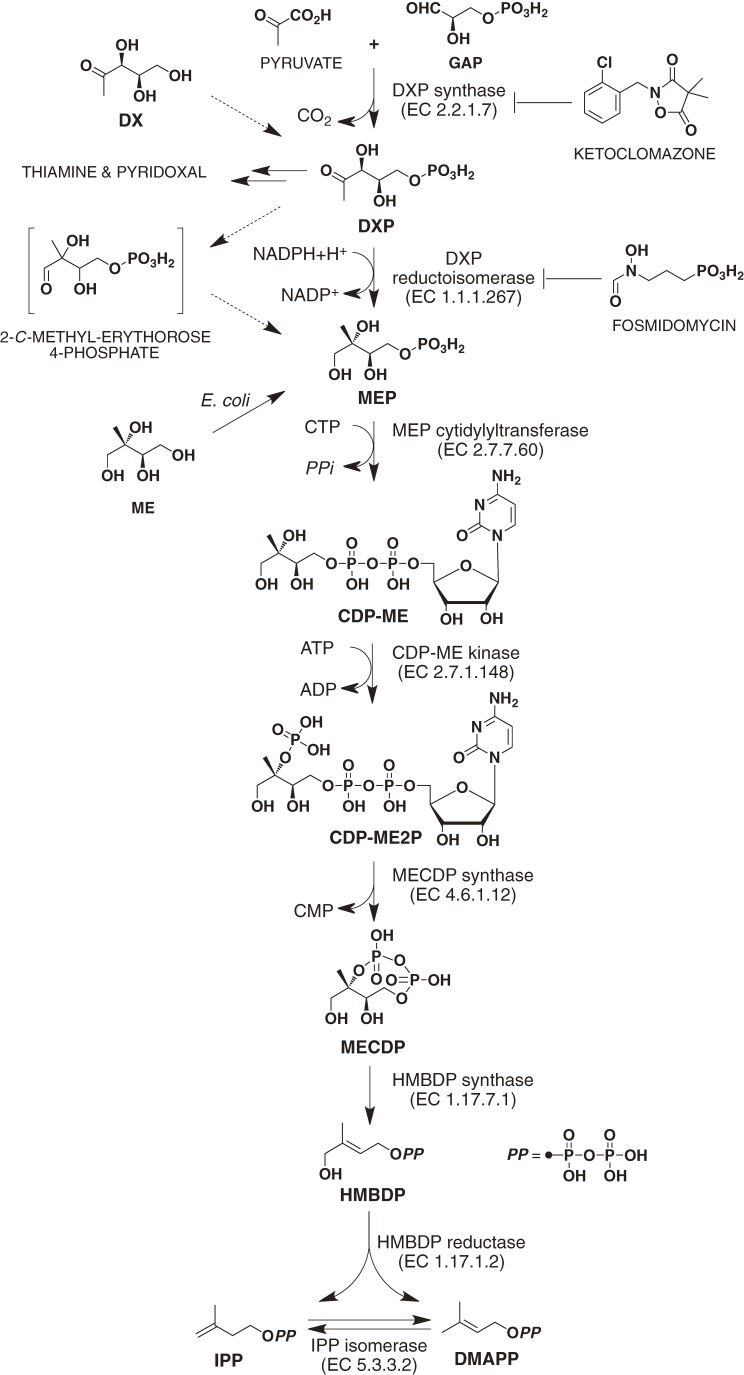
1. First reaction step, catalyzed by DXP synthase (EC 2.2.1.7)
The initial step in the MEP pathway is the formation of 1-deoxy-d-xylulose 5-phosphate (DXP) by the condensation of pyruvate and d-glyceraldehyde 3-phosphate (GAP), and this reaction is catalyzed by DXP synthase (Fig. 2).10) The DXP synthase gene (dxs) was first cloned from E. coli.11,12) This enzyme has a typical thiamine-binding motif and requires both thiamine and a divalent cation, such as Mg2+ or Mn2+, for enzyme activity. The reaction preceding this condensation had been found by Yokota et al., who detected the formation of 1-deoxy-d-xylulose (DX) from pyruvate and d-glyceraldehyde by pyruvate dehydrogenase of E. coli or Bacillus subtilis.13,14)
Figure 2.
MEP pathway for IPP and DMAPP biosynthesis.
We cloned a dxs gene homolog from the Streptomyces sp. strain CL190 by the polymerase chain reaction using oligonucleotide primers based on highly conserved amino acid sequences.15) Next, we compared the enzymatic properties of the protein product of the dxs gene with those of the E. coli DXP synthase. We found that although these two enzymes had different origins, they had nearly identical enzymatic properties.
In E. coli, DXP is a biosynthetic intermediate not only for IPP and DMAPP but also for thiamine and pyridoxal (Fig. 2).16) This finding indicates that DXP synthase is not the only enzyme specific for the MEP pathway.
2. Ketoclomazone, an inhibitor of DXP synthase
Ketoclomazone (2-(2-chlorobenzyl)-4,4-dimethyl-isoxazolidine-3,5-dione) is a derivative of a soil-applied herbicide known as clomazone, dimethazone, or FMC 57020 (Fig. 2).17) Ketoclomazone causes significantly lower chlorophyll and carotenoid levels in barley leaves.18) In addition, 20 µM of ketoclomazone inhibited 47% of the DXP synthase activity in crude extracts from the recombinant E. coli expressing a plant Catharanthus roseus DXP synthase gene,19) suggesting that ketoclomazone is an inhibitor of DXP synthase. Therefore, ketoclomazone and clomazone have been used as chemical tools to suppress the MEP pathway in plants.20,21) In contrast, ketoclomazone has not been reported to have antibacterial activity, although most bacteria, including pathogens, utilize the MEP pathway for isoprenoid biosynthesis.22) However, we recently demonstrated that ketoclomazone inhibits DXP synthases from E. coli and Haemophilus influenzae and exhibits antibacterial activity against these bacteria.23) The kinetics of inhibition by ketoclomazone suggests that this chemical binds to an unidentified inhibitor-binding site that differs from the substrate-binding sites for pyruvate and GAP on DXP synthase.
3. Second reaction step, catalyzed by DXP reductoisomerase (EC 1.1.1.267)
Rohmer and co-workers found that 2-C-methylerythritol (ME) was incorporated into E. coli cells, which used it as the precursor of the ubiquinone side chain.24,25) Because of this finding, the intramolecular rearrangement of DXP had been assumed to yield a hypothetical rearrangement product, 2-C-methylerythrose 4-phosphate, which was subsequently converted to MEP by an unknown reduction process (Fig. 2). For this reason, it was assumed that two enzymes were involved in the formation of MEP from DXP.
To elucidate the details of this mechanism, we decided to clone the gene responsible for MEP synthesis in E. coli. We used a unique strategy of screening for E. coli mutants that were metabolically blocked between the DXP into MEP synthesis steps of the pathway; specifically, these types of mutants would require MEP or ME for growth.26) In the screening, we added ME to the growth medium and were able to select ten mutants with the desired properties; the addition of ME, but not of DX, to the minimal medium facilitated the growth of those mutants. Only the yaeM gene, which had an unknown function, was found to complement these mutations,27) and all ten mutants that needed ME for growth were confirmed by sequencing to be yaeM-gene-deficient mutants.28)
Next, to confirm whether the yaeM gene product (YaeM) was in fact responsible for MEP formation, we constructed a plasmid that would overexpress YaeM in cells. The recombinant protein was purified to apparent homogeneity. Subsequently, the molecular mass of YaeM was estimated to be 42 kDa by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and 165 kDa by gel filtration chromatography, suggesting that YaeM is probably a homotetramer.
Incubation of the enzyme with DXP in the presence of NADPH resulted in a decrease in the NADPH level in the solution, thereby indicating that the enzyme converted DXP into an unknown reaction product. We purified the reaction product and identified it as MEP on the basis of NMR and MS data.26) This result demonstrated that MEP is synthesized in the presence of NADPH by rearrangement and reduction of DXP in a single step. We designated this enzyme DXP reductoisomerase and renamed the yaeM gene “dxr”.27)
As mentioned above, DXP is a precursor not only for IPP and DMAPP but also for thiamine and pyridoxal. Therefore, the reaction catalyzed by DXP reductoisomerase is the committed step in the MEP pathway, and MEP is the first biosynthetic intermediate specific for the MEP pathway.
We identified the catalytic residues of DXP reductoisomerase by analyzing the E. coli yaeM-deficient mutants described above. We found that Glu231 of the E. coli enzyme is important for the conversion of DXP to MEP and that His153, His209, and His257 are important for binding of DXP to the enzyme.28) These results were supported by studies of the three-dimensional structure of DXP reductoisomerase.29)
4. Fosmidomycin, an inhibitor of DXP reductoisomerase
DXP reductoisomerase is widely distributed in plants and many bacteria, including pathogens, but not in mammals; therefore, this enzyme could be a target in screens for herbicides and antibacterial drugs.26,27) To screen rapidly for DXP reductoisomerase inhibitors, we constructed an assay system that used a microplate reader.30) Before screening, we performed a literature search to find antibiotics active against E. coli and B. subtilis, both of which use the MEP pathway, but inactive against Staphylococcus aureus, which utilizes the MVA pathway. Our reasoning for doing so was that some inhibitors might have already been reported as antibiotics, and the molecular targets of those drugs may have been unidentified. Fosmidomycin (FR-31564) had the expected antibacterial spectrum and was selected as a candidate for a DXP reductoisomerase inhibitor.
Fosmidomycin has antibacterial activity against most Gram-negative and some Gram-positive bacteria.31,32) Because this antibiotic inhibited menaquinone and carotenoid biosynthesis in Micrococcus luteus, it was proposed that this antibiotic inhibited the biosynthesis of isoprenoids.33) From these studies and the structural similarity between fosmidomycin and 2-C-methylerythrose 4-phosphate, we assumed that the antibiotic might inhibit DXP reductoisomerase.
In our assay, fosmidomycin strongly inhibited DXP reductoisomerase in a dose-dependent manner with an IC50 of 24 nM. The Lineweaver-Burk plot indicated competitive inhibition with a Ki value of 9.4 nM. Therefore, fosmidomycin is a competitive inhibitor of DXP reductoisomerase.30)
Next, we investigated the antibacterial activity of fosmidomycin against E. coli. Fosmidomycin completely inhibited bacterial growth at 6.25 µg/ml; however, growth inhibition of E. coli by the antibiotic was suppressed when ME was added to the growth medium. In addition, fosmidomycin had no effect on the growth of an E. coli DXP reductoisomerase disruptant in the presence of ME.34) The lack of inhibition by fosmidomycin of the disruptant suggested that this mutant lacked a molecular target. From these results, we concluded that fosmidomycin specifically inhibited DXP reductoisomerase in the MEP pathway.30,34)
As mentioned above, we identified the catalytic residues of DXP reductoisomerase28) and determined its X-ray crystal structure.29) Analysis of this structure should provide useful information for the development of antibacterial drugs and herbicides.
5. Reaction steps leading to synthesis of IPP from MEP
To elucidate the later reactions that lead to the synthesis of IPP from MEP, we initiated studies on the cloning of the genes responsible for the conversion of MEP into IPP. To this end, we employed a unique strategy for preparation of mutants of E. coli that were metabolically blocked between the MEP into IPP synthesis steps of the pathway. Because such a mutation would be lethal, we engineered an E. coli transformant that possessed an additional biosynthetic pathway for IPP: the MVA pathway.35) This transformant, which possessed genes encoding MVA kinase, phosphomevalonate kinase, and diphosphomevalonate decarboxylase from Streptomyces sp. strain CL190, could use part of the MVA pathway for IPP biosynthesis but only in the presence of MVA. The MVA added to the growth medium is converted by these three enzymes into IPP, which is later used by the transformant as a precursor for isoprenoids.
With this transformant as the parental strain, we isolated mutants that had an obligatory requirement of MVA for their growth.36) Thirty-three mutants from approximately 60,000 screened colonies showed the expected phenotypes: the addition of MVA, but not ME, to minimal medium facilitated the growth of the mutants. Thus, these phenotypic features unequivocally showed that the mutants had a defect(s) in the pathway leading to IPP from MEP. Using these mutants, we succeeded in cloning four uncharacterized genes, ygbP, ychB, ygbB, and gcpE, that complemented the defects of these mutants in synthesizing IPP from MEP.36–38) It should be noted that these four genes are apparently needed for IPP biosynthesis in E. coli. The distribution patterns of these four genes in bacteria were identical to those of the DXP synthase and DXP reductoisomerase genes of the MEP pathway, thereby supporting the hypothesis that these four genes are essential for this pathway. This bioinformatic analysis also identified the lytB gene as an additional candidate responsible for the MEP pathway.39)
To identify the functions of the gene products of the gcpE, lytB, ygbB, ychB, and ygbP genes, we constructed plasmids that overexpressed the gene products of those genes and were able to prepare sufficiently high concentrations of the enzymes to characterize them. Next, we sought to select appropriate reaction conditions for the conversion of MEP into an unknown product by the action of one of the five enzymes.
5.1. Third reaction step, catalyzed by MEP cytidylyltransferase (EC 2.7.7.60).
Most of our attempts to disclose the next reaction process were unsuccessful because we could not identify the cofactor of the reaction. This problem was finally overcome by an unexpected finding that the ygbP gene product converted MEP into an unknown reaction product in the presence of CTP. We purified the reaction product and identified it by NMR and MS as 4-(cytidine 5′-diphospho)-2-C-methyl-d-erythritol (CDP-ME).36) These results indicated that MEP is converted to CDP-ME in the presence of CTP by the ygbP gene product and that this gene product is essential for IPP biosynthesis. We designated the enzyme MEP cytidylyltransferase.36)
5.2. Fourth reaction step, catalyzed by CDP-ME kinase (EC 2.7.1.148).
Next, we demonstrated that the ychB gene product converted CDP-ME to an unknown reaction product in the presence of ATP. We purified the reaction product and identified it as 2-phospho-4-(cytidine 5′-diphospho)-2-C-methyl-d-erythritol (CDP-ME2P).37) This result showed that CDP-ME is converted to CDP-ME2P in the presence of ATP by the ychB gene product and that this gene product is essential for IPP biosynthesis. We designated the enzyme 4-(cytidine 5′-diphospho)-2-C-methyl-d-erythritol kinase.37)
5.3. Fifth reaction step, catalyzed by MECDP synthase (EC 4.6.1.12).
Finally, we demonstrated that the ygbB gene product converted CDP-ME2P to an unknown reaction product. We purified the reaction product and identified it as 2-C-methyl-d-erythritol 2,4-cyclodiphosphate (MECDP).38) The formation of MECDP was concomitant with the elimination of CMP from CDP-ME2P, suggesting that CDP-ME2P is converted to MECDP by the ygbB gene product and that this gene product is essential for IPP biosynthesis. We designated this enzyme MECDP synthase.38)
It had been reported that certain bacteria, including Corynebacterium ammoniagenes, accumulated MECDP under oxidative stress caused by benzyl viologen,40) but there have been no reports on the role of MECDP in the MEP pathway. We showed that MECDP is an intermediate in the MEP pathway for IPP biosynthesis.
Independent of our studies, German researchers found that MECDP is synthesized from MEP by the consecutive actions of the ygbP, ychB, and ygbB gene products41–43) and that MECDP is efficiently converted into phytoene in the chlomoplasts of Capsicum annuum and Narcissus pseudonarcissus.44)
5.4. Functions of the gcpE and lytB gene products.
Next, we incubated MECDP with the gcpE or lytB gene products in the presence of various possible cofactors, such as NADPH, nucleotide triphosphates, or vitamins, with the expectation that the gene product would convert MECDP to an unidentified reaction product. However, our attempts to detect this reaction product in vitro were unsuccessful. By contrast, German researchers demonstrated that the gcpE and lytB genes encoded iron-sulfur (Fe-S) cluster-containing (E)-4-hydroxy-3-methylbut-2-en-1-yl diphosphate (HMBDP) synthase (EC 1.17.7.1)45) and Fe-S cluster-containing HMBDP reductase (EC 1.17.1.2), respectively.46) E. coli HMBDP synthase converts MECDP into HMBDP in the presence of the flavodoxin/flavodoxin reductase/NADPH system,45) whereas the E. coli HMBDP reductase reduces HMBDP in the presence of ferredoxin and NADPH to yield a 5:1 mixture of IPP and DMAPP.46) Both the reactions require complete anaerobic condition.
6. Identification of the MEP pathway genes in plants
As mentioned above, we isolated E. coli mutants possessing a metabolic block(s) between the steps at which DXP is converted into IPP. These E. coli mutants were also used as tools to confirm the functions of the MEP pathway gene products of Hevea brasiliensis,47) which is a tree that produces natural rubber, and of Ginkgo biloba,48–50) which is a tree that produces a diterpene ginkgolide. For example, the H. brasiliensis ygpP homolog complemented the defect of the E. coli ygbP-deficient mutant that was unable to convert MEP into CDP-ME.47)
7. Two types of IPP isomerases
IPP isomerase (EC 5.3.3.2) catalyzes an essential reaction in the MVA pathway by converting IPP to DMAPP. Many IPP isomerase genes (idi) were cloned from various organisms such as humans, Saccharomyces cerevisiae, E. coli and Rhodobacter capsulatus. Interestingly, database searches using highly conserved amino acid sequences from previously identified IPP isomerases had failed to detect IPP isomerase in archaea and some bacteria that use the MVA pathway. This failure to detect IPP isomerase in these organisms had suggested the existence of an unidentified type of IPP isomerase, the amino acid sequence of which shared no similarity to those of IPP isomerases.
We cloned a gene cluster encoding enzymes of the MVA pathway from Streptomyces sp. strain CL190 and demonstrated that the MVA pathway could function in an E. coli strain transformed with this gene cluster.35,51) In addition to the known genes of the MVA pathway, this gene cluster contains an additional open reading frame, orfD, which encodes a protein with unknown function. We found the orfD homologs in archaea; several Gram-positive bacteria, including Staphylococcus aureus; and in the carotenoid biosynthetic gene cluster of Erwinia herbicola Eho10.52) Homologs of orfD were also detected in the MVA pathway gene cluster in other actinomycetes. These facts led us to assume that orfD encodes an unprecedented type of IPP isomerase.
In 2001, we reported that the orfD product, when expressed in E. coli, catalyzes the isomerization of IPP to produce DMAPP in the presence of both flavin mononucleotide (FMN) and NAD(P)H.52) Based on the enzymatic properties of this enzyme, which are different from those of known IPP isomerases, we proposed that this orfD product is a novel type of IPP isomerase with a sequence that has no similarity to those of known IPP isomerases. In addition, we classified the IPP isomerases into two types: Type 2 for the new FMN- and NAD(P)H-dependent enzymes, and type 1, which was known to be present in many organisms.52) We also identified type 2 IPP isomerase in archaea and certain Gram-positive bacteria, such as S. aureus, Streptococcus pneumoniae, Streptococcus pyrogenes and Enterococcus faecalis.52) This finding suggested that the type 2 IPP isomerase of S. aureus represents an effective target for the screening of anti-staphylococcal drugs.
8. Novel acetoacetyl-CoA synthesizing enzyme found in the Streptomyces MVA pathway gene clusters
As mentioned above, we identified the MVA pathway gene cluster in several actinomycetes strains.35) The MVA pathway begins with the synthesis of acetoacetyl-CoA, which was first reported to be biosynthesized via a thioester-dependent Claisen condensation reaction between two molecules of acetyl-CoA, and is catalyzed by acetoacetyl-CoA thiolase (EC 2.3.1.9). Therefore, acetoacetyl-CoA thiolase was expected to exist in the MVA pathway gene cluster of actinomycetes strains. However, acetoacetyl-CoA thiolase, which is involved in the conversion of acetyl-CoA to acetoacetyl-CoA, is absent. Instead, an open reading frame (for example, nphT7 of Streptomyces sp. strain CL190) that shares homology with β-ketoacyl-(acyl carrier protein) synthase (KAS) III flanks the gene cluster.
In 2010, we reported that NphT7 catalyzes a single condensation of acetyl-CoA and malonyl-CoA to give acetoacetyl-CoA and CoA; however, NphT7 was not able to synthesize acetoacetyl-CoA through the condensation of two molecules of acetyl-CoA.53) Based on the enzymatic properties of NphT7, we proposed that NphT7 was a novel “acetoacetyl-CoA synthase” of the thiolase superfamily. In addition, co-expression of the nphT7 gene with the HMG-CoA synthase and HMG-CoA reductase genes in the heterologous host Streptomyces albus resulted in a 3.5-fold higher production of MVA than when only the HMG-CoA synthase and HMG-CoA reductase genes were expressed.53) This result suggests that nphT7 could be used to significantly increase the production of commodity isoprenoids, such as carotenoids, Taxol, and artemisinin.
BLAST searches using the NphT7 sequence as a query sequence revealed that NphT7 homologs are widespread in bacteria. We speculate that these NphT7 homologs synthesize CoA derivatives with various acyl groups. Of the homologs, some are located in the biosynthetic gene clusters of unidentified natural products, suggesting that the NphT7-like enzymes are involved in the biosynthesis of acyl-CoA-derived natural products.
9. Diversity of the biosynthesis of IPP and DMAPP
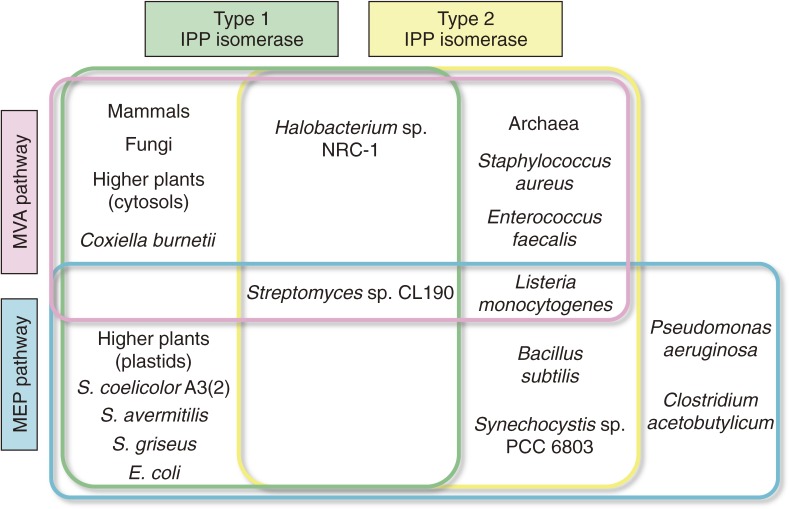
Mammals and fungi synthesize IPP and DMAPP using the MVA pathway (Fig. 3). Nearly all archaea, some Gram-positive bacteria, including Staphylococcus, Streptococcus, Enterococcus, Lactococcus, Lactobacillus, and Leuconostoc, and Myxobacteria (Gram-negative), also use the MVA pathway, whereas other most bacteria including cyanobacteria synthesize IPP and DMAPP using the MEP pathway. Only a few bacteria use both pathways. In higher plants, the MEP pathway functions in plastids, whereas the MVA pathway functions in cytosols. The species distribution of the MVA and MEP pathways are not associated with those of the type 1 and type 2 IPP isomerases. In bacteria that use the MEP pathway, some have a type 1 enzyme, some have a type 2 enzyme, and others have no IPP isomerase. In contrast, bacteria, as well as most archaea, that use only the MVA pathway usually possess a type 2 IPP isomerase. However, the Q-fever pathogen Coxiella burnetii possesses MVA pathway genes and a type 1 IPP isomerase, and the halophilic archaeon Halobacterium sp. NRC-1 possesses both functional type 1 and type 2 IPP isomerases, along with the MVA pathway.
Figure 3.
Diversity of the biosynthesis of IPP and DMAPP.
It may be interesting to note that some actinomycetes utilize the both pathways. For example, Streptomyces sp. strain CL190, a producer of meroterpenoid naphterpin, possesses the genes for both the MVA and MEP pathways. We had proved simultaneous operation of the both pathways for IPP biosynthesis in the strain by detailed tracer experiments using 13C-labeled glucose (for the MEP pathway) and 13C-labeled acetate (for the MVA pathway) on the formation of menaquinone (a primary metabolite) and naphterpin (a secondary metabolite).54) Another actinomycete, Kitasatospora griseola, a producer of diterpene terpentecin, also uses both the MVA and MEP pathways for IPP biosynthesis. We demonstrated that the MEP pathway genes are expressed first and then the MVA pathway genes start to be expressed in the strain.55) Because secondary metabolites such as terpentecin are biosynthesized in the late stage of the growth, the production of terpentecin and the expression of the MVA pathway genes appeared to be synchronized in the producer cells. In addition, the biosynthetic gene clusters of naphterpin and terpentecin flank the MVA pathway gene clusters of each producer.56,57) From these facts, we speculate that the MVA pathway genes in actinomycetes are utilized for the production of the secondary metabolites, such as naphterpin and terpentecin.
Perspective
The MEP pathway is absent in mammals, but this pathway is essential for survival and growth in many pathogenic bacteria, plants, and malaria parasites. Therefore, the enzymes in the MEP pathway could be targets for screening for antibacterial drugs, herbicides, and antimalarial drugs. We expect that our studies will contribute to the development of antimicrobial agents that target the MEP pathway.
Kuzuyama is continuing to study the biosynthesis of some isoprenoids produced by actinomycetes. Yet-undiscovered biosynthetic enzymes for isoprenoids may be buried in the enormous quantity of genetic information, which is accumulating every year. Therefore, we will continue to investigate isoprenoid biosynthesis in microorganisms to illuminate the interplay between functional convergence and divergence in the evolution of metabolic pathways.
Acknowledgments
This review was supported by a Funding Program for Next Generation World-Leading Researchers (LS028 to T.K.) from the Japan Society for the Promotion of Science (JSPS). T.K. sincerely thanks the Japan Academy for the Japan Academy Medal.
Abbreviations
- IPP
isopentenyl diphosphate
- DMAPP
dimethylallyl diphosphate
- MVA
mevalonate
- MEP
2-C-methyl-d-erythritol 4-phosphate
- HMG-CoA
3-hydroxy-3-methylglutaryl coenzyme A
- DXP
1-deoxy-d-xylulose 5-phosphate
- GAP
d-glyceraldehyde 3-phosphate
- DX
1-deoxy-d-xylulose
- ME
2-C-methylerythritol
- CDP-ME
4-(cytidine 5′-diphospho)-2-C-methyl-d-erythritol
- CDP-ME2P
2-phospho-4-(cytidine 5′-diphospho)-2-C-methyl-d-erythritol
- MECDP
2-C-methyl-d-erythritol 2,4-cyclodiphosphate
- HMBDP
(E)-4-hydroxy-3-methylbut-2-en-1-yl diphosphate
Biographies
Profile
Tomohisa Kuzuyama was born in Yokosuka, Japan, in 1966. He completed his Ph.D. for a thesis entitled “Studies on the fosfomycin biosynthesis” under supervision of Professor Haruo Seto in Graduate School of Agricultural and Life Sciences, The University of Tokyo in 1995. In the same year, he was appointed to an assistant professor in the Institute of Molecular and Cellular Biosciences, The University of Tokyo where he began to study the mevalonate pathway and 2-C-methyl-d-erythritol 4-phosphate pathway in bacteria. He joined Professor Joseph P. Noel’s laboratory as a visiting scientist from 2003 through 2004. He was then promoted to an associate professor in Biotechnology Research Center, The University of Tokyo in 2004. His current research interests aim at comprehensive understanding biosynthetic machinery for several natural products. He received The Hamada Award of The Society for Actinomycetes, Japan in 1997, The Japan Bioscience, Biotechnology and Agrochemistry Society Award for the Encouragement of Young Scientists in 2001, The Young Scientists’ Prize in the Commendation for Science and Technology by the Minister of Education, Culture, Sports, Science and Technology in 2006, The Japan Society for the Promotion of Science Prize in 2011, and Japan Academy Medal in 2011.
Haruo Seto was born in 1939 and started his research career in 1963 with studies on biosynthesis of blasticidin S, an antibiotic effective to rice blast disease, at the Institute of Applied Microbiology, The University of Tokyo, after graduating from the Faculty of Agriculture at The University of Tokyo. In 1968, he moved to Stanford Research Institute, California, where he accomplished the first work on the biosynthesis of microbial metabolites using C-13 labeled precursors and C-13 NMR spectroscopy. In 1970 he became an assistant professor at the Institute of Applied Microbiology, The University of Tokyo. In 1973 he reported the use of C-13/C-13 coupling for labeling experiments; this technique was soon recognized as the very important method for biosynthetic experiments and is now being used worldwide. In 1981 he was promoted to the associate professor of the same laboratory. In 1987 he was promoted to Professor and carried out researches related to biosynthesis of microbial products and screening of bioactive substances of microbial origin and their structural elucidation by NMR spectroscopy. The name of his institute was changed to the Institute of Molecular and Cellular Biosciences in 1993. In 2000 he moved to Tokyo University of Agriculture where he educated many students in the field of natural product chemistry and retired in 2010. He was awarded the Japan Bioscience, Biotechnology and Agrochemistry Society Award for the Encouragement of Young Scientists in 1976, the Sumiki-Umezawa Award, Japan Antibiotic Research Association in 1986, the Award of The Society for Actinomycetes Japan in 1995, the Japan Bioscience, Biotechnology and Agrochemistry Society Award in 1995 and Medal with Purple Ribbon, Japanese Government in 2001.
References
- 1).Tamura G. (1956) Hiochic acid, a new growth factor for Lactobacillus homohiochi and Lactobacillus heterohiochi. J. Gen. Appl. Microbiol. 2, 431–434 [PubMed] [Google Scholar]
- 2).Wolf E.D., Hoffman H.C., Aldrich E.P., Skeggs R.H., Wright D.L., Folkers K. (1956) β-Hydroxy-β-methyl-δ-valerolactone (divalonic acid), a new biological factor. J. Am. Chem. Soc. 78, 4499 [Google Scholar]
- 3).Tavormina A.P., Gibbs H.M., Huff W.J. (1956) Utilization of β-hydroxy-β-methyl-δ-valerolactone in cholesterol biosynthesis. J. Am. Chem. Soc. 78, 4498–4499 [Google Scholar]
- 4).Katsuki H., Bloch K. (1967) Studies on the biosynthesis of ergosterol in yeast: Formation of methylated intermediates. J. Biol. Chem. 242, 222–227 [PubMed] [Google Scholar]
- 5).Lynen F. (1967) Biosynthetic pathways from acetate to natural products. Pure Appl. Chem. 14, 137–167 [DOI] [PubMed] [Google Scholar]
- 6).Watanabe Y., Ito T., Shiomi M., Tsujita Y., Kuroda M., Arai M., Fukami M., Tamura A. (1988) Preventive effect of pravastatin sodium, a potent inhibitor of 3-hydroxy-3-methylglutaryl coenzyme A reductase, on coronary atherosclerosis and xanthoma in WHHL rabbits. Biochim. Biophys. Acta 960, 294–302 [PubMed] [Google Scholar]
- 7).Zhou D., White R.H. (1991) Early steps of isoprenoid biosynthesis in Escherichia coli. Biochem. J. 273, 627–634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8).Cane D.E., Rossi T., Pachlatko J.P. (1979) The biosynthesis of pentalenolactone. Tetrahedron Lett. 20, 3639–3642 [Google Scholar]
- 9).Cane D.E., Rossi T., Tillman A.M., Pachlatko J.P. (1981) Stereochemical studies of isoprenoid biosynthesis: Biosynthesis of pentalenolactone from [U-13C6]glucose and [6-2H2]glucose. J. Am. Chem. Soc. 103, 1838–1843 [Google Scholar]
- 10).Rohmer M., Seemann M., Horbach S., Bringer-Meyer S., Sahm H. (1996) Glyceraldehyde 3-phosphate and pyruvate as precursors of isoprenic units in an alternative non-mevalonate pathway for terpenoid biosynthesis. J. Am. Chem. Soc. 118, 2564–2566 [Google Scholar]
- 11).Lois L.M., Campos N., Putra S.R., Danielsen K., Rohmer M., Boronat A. (1998) Cloning and characterization of a gene from Escherichia coli encoding a transketolase-like enzyme that catalyzes the synthesis of d-1-deoxyxylulose 5-phosphate, a common precursor for isoprenoid, thiamin, and pyridoxol biosynthesis. Proc. Natl. Acad. Sci. U.S.A. 95, 2105–2110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12).Sprenger G.A., Schörken U., Wiegert T., Grolle S., Graaf A.A., Taylor S.V., Begley T.P., Bringer-Meyer S., Sahm H. (1997) Identification of a thiamin-dependent synthase in Escherichia coli required for the formation of the 1-deoxy-d-xylulose 5-phosphate precursor to isoprenoids, thiamine, and pyridoxol. Proc. Natl. Acad. Sci. U.S.A. 94, 12857–12862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13).Yokota A., Sasajima K. (1984) Formation of 1-deoxy-d-threo-pentulose and 1-deoxy-l-threo-pentulose by cell-free extracts of microorganisms. Agric. Biol. Chem. 48, 149–158 [Google Scholar]
- 14).Yokota A., Sasajima K. (1986) Formation of 1-deoxy-ketoses by pyruvate dehydrogenase and acetoin dehydrogenase. Agric. Biol. Chem. 50, 2517–2524 [Google Scholar]
- 15).Kuzuyama T., Takagi M., Takahashi S., Seto H. (2000) Cloning and characterization of 1-deoxy-d-xylulose 5-phosphate synthase from Streptomyces sp. strain CL190, which uses both the mevalonate and nonmevalonate pathways for isopentenyl diphosphate biosynthesis. J. Bacteriol. 182, 891–897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16).Himmeldirk K., Kennedy I.A., Hill R.E., Sayer B.G., Spenser I.D. (1996) Biosynthesis of vitamins B1 and B6 in Escherichia coli: Concurrent incorporation of 1-deoxy-d-xylulose into thiamin (B1) and pyridoxol (B6). Chem. Commun. 10, 1187–1188 [Google Scholar]
- 17).ElNaggar S.F., Creekmore R.W., Schocken M.J., Rosen R.T., Robinson R.A. (1992) Metabolism of clomazone herbicide in soybean. J. Agric. Food Chem. 40, 880–883 [Google Scholar]
- 18).Zeidler J., Schwender J., Mueller C., Lichtenthaler H.K. (2000) The non-mevalonate isoprenoid biosynthesis of plants as a test system for drugs against malaria and pathogenic bacteria. Biochem. Soc. Trans. 28, 796–798 [PubMed] [Google Scholar]
- 19).Ferhatoglu Y., Barrett M. (2005) Studies of clomazone mode of action. Pestic. Biochem. Physiol. 85, 7–14 [Google Scholar]
- 20).Sakakibara H., Kasahara H., Ueda N., Kojima M., Takei K., Hishiyama S., Asami T., Okada K., Kamiya Y., Yamaya T., Yamaguchi S. (2005) Agrobacterium tumefaciens increases cytokinin production in plastids by modifying the biosynthetic pathway in the host plant. Proc. Natl. Acad. Sci. U.S.A. 102, 9972–9977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21).TenBrook P.L., Tjeerdema R.S. (2005) Comparative actions of clomazone on β-carotene levels and growth in rice (Oryza sativa) and watergrasses (Echinochloa spp). Pest Manag. Sci. 61, 567–571 [DOI] [PubMed] [Google Scholar]
- 22).Rodriguez-Concepcion M. (2004) The MEP pathway: a new target for the development of herbicides, antibiotics and antimalarial drugs. Curr. Pharm. Des. 10, 2391–2400 [DOI] [PubMed] [Google Scholar]
- 23).Matsue Y., Mizuno H., Tomita T., Asami T., Nishiyama M., Kuzuyama T. (2010) The herbicide ketoclomazone inhibits 1-deoxy-d-xylulose 5-phosphate synthase in the 2-C-methyl-d-erythritol 4-phosphate pathway and shows antibacterial activity against Haemophilus influenzae. J. Antibiot. (Tokyo) 63, 583–588 [DOI] [PubMed] [Google Scholar]
- 24).Duvold T., Bravo J.M., Pale-Grosdemange C., Rohmer M. (1997) Biosynthesis of 2-C-methyl-d-erythritol, a putative C5 intermediate in the mevalonate independent pathway for isoprenoid biosynthesis. Tetrahedron Lett. 38, 4769–4772 [Google Scholar]
- 25).Duvold T., Cali P., Bravo J.M., Rohmer M. (1997) Incorporation of 2-C-methyl-d-erythritol, a putative isoprenoid precursor in the mevalonate-independent pathway, into ubiquinone and menaquinone of Escherichia coli. Tetrahedron Lett. 38, 6181–6184 [Google Scholar]
- 26).Kuzuyama T., Takahashi S., Watanabe H., Seto H. (1998) Direct formation of 2-C-methyl-d-erythritol 4-phosphate from 1-deoxy-d-xylulose 5-phosphate by 1-deoxy-d-xylulose 5-phosphate reductoisomerase, a new enzyme in the non-mevalonate pathway to isopentenyl diphosphate. Tetrahedron Lett. 39, 4509–4512 [Google Scholar]
- 27).Takahashi S., Kuzuyama T., Watanabe H., Seto H. (1998) A 1-deoxy-d-xylulose 5-phosphate reductoisomerase catalyzing the formation of 2-C-methyl-d-erythritol 4-phosphate in an alternative nonmevalonate pathway for terpenoid biosynthesis. Proc. Natl. Acad. Sci. U.S.A. 95, 9879–9884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28).Kuzuyama T., Takahashi S., Takagi M., Seto H. (2000) Characterization of 1-deoxy-d-xylulose 5-phosphate reductoisomerase, an enzyme involved in isopentenyl diphosphate biosynthesis, and identification of its catalytic amino acid residues. J. Biol. Chem. 275, 19928–19932 [DOI] [PubMed] [Google Scholar]
- 29).Yajima S., Nonaka T., Kuzuyama T., Seto H., Ohsawa K. (2002) Crystal structure of 1-deoxy-d-xylulose 5-phosphate reductoisomerase complexed with cofactors: Implications for a flexible loop movement upon substrate binding. J. Biochem. 131, 313–317 [DOI] [PubMed] [Google Scholar]
- 30).Kuzuyama T., Shimizu T., Takahashi S., Seto H. (1998) Fosmidomycin, a specific inhibitor of 1-deoxy-d-xylulose 5-phosphate reductoisomerase in the nonmevalonate pathway for terpenoid biosynthesis. Tetrahedron Lett. 39, 7913–7916 [Google Scholar]
- 31).Okuhara M., Kuroda Y., Goto T., Okamoto M., Terano H., Kohsaka M., Aoki H., Imanaka H. (1980) Studies on new phosphonic acid antibiotics. 3. Isolation and characterization of FR-31564, FR-32863 and FR-33289. J. Antibiot. 33, 24–28 [DOI] [PubMed] [Google Scholar]
- 32).Kuroda Y., Okuhara M., Goto T., Okamoto M., Terano H., Kohsaka M., Aoki H., Imanaka H. (1980) Studies on new phosphonic acid antibiotics. 4. Structure determination of FR-33289, FR-31564 and FR-32863. J. Antibiot. 33, 29–35 [DOI] [PubMed] [Google Scholar]
- 33).Shigi Y. (1989) Inhibition of bacterial isoprenoid synthesis by fosmidomycin, a phosphonic acid-containing antibiotic. J. Antimicrob. Chemother. 24, 131–145 [DOI] [PubMed] [Google Scholar]
- 34).Kuzuyama T., Takahashi S., Seto H. (1999) Construction and characterization of Escherichia coli disruptants defective in the yaeM gene. Biosci. Biotechnol. Biochem. 63, 776–778 [DOI] [PubMed] [Google Scholar]
- 35).Takagi M., Kuzuyama T., Takahashi S., Seto H. (2000) A gene cluster for the mevalonate pathway from Streptomyces sp. strain CL190. J. Bacteriol. 182, 4153–4157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36).Kuzuyama T., Takagi M., Kaneda K., Dairi T., Seto H. (2000) Formation of 4-(cytidine 5′-diphospho)-2-C-methyl-d-erythritol from 2-C-methyl-d-erythritol 4-phosphate by 2-C-methyl-d-erythritol 4-phosphate cytidylyltransferase, a new enzyme in the nonmevalonate pathway. Tetrahedron Lett. 41, 703–706 [Google Scholar]
- 37).Kuzuyama T., Takagi M., Kaneda K., Dairi T., Seto H. (2000) Studies on the nonmevalonate pathway: Conversion of 4-(cytidine 5′-diphospho)-2-C-methyl-d-erythritol to its 2-phospho derivative by 4-(cytidine 5′-diphospho)-2-C-methyl-d-erythritol kinase. Tetrahedron Lett. 41, 2925–2928 [Google Scholar]
- 38).Takagi M., Kuzuyama T., Kaneda K., Dairi T., Seto H. (2000) Studies on the nonmevalonate pathway: formation of 2-C-methyl-d-erythritol 2,4-cyclodiphosphate from 2-phospho-4-(cytidine 5′-diphospho)-2-C-methyl-d-erythritol. Tetrahedron Lett. 41, 3395–3398 [Google Scholar]
- 39).Takagi, M. (2001) Studies on the nonmevalonate pathway. Ph.D. thesis in The University of Tokyo. [Google Scholar]
- 40).Ostrovsky D., Diomina G., Lysak E., Matveeva E., Ogrel O., Trutko S. (1998) Effect of oxidative stress on the biosynthesis of 2-C-methyl-d-erythritol-2,4-cyclopyrophosphate and isoprenoids by several bacterial strains. Arch. Microbiol. 171, 69–72 [DOI] [PubMed] [Google Scholar]
- 41).Rohdich F., Wungsintaweekul J., Fellermeier M., Sagner S., Herz S., Kis K., Eisenreich W., Bacher A., Zenk M.H. (1999) Cytidine 5′-triphosphate-dependent biosynthesis of isoprenoids: YgbP protein of Escherichia coli catalyzes the formation of 4-diphosphocytidyl-2-C-methylerythritol. Proc. Natl. Acad. Sci. U.S.A. 96, 11758–11763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42).Lüttgen H., Rohdich F., Herz S., Wungsintaweekul J., Hecht S., Schuhr C.A., Fellermeier M., Sagner S., Zenk M.H., Bacher A., Eisenreich W. (2000) Biosynthesis of terpenoids: YchB protein of Escherichia coli phosphorylates the 2-hydroxy group of 4-diphosphocytidyl-2C-methyl-d-erythritol. Proc. Natl. Acad. Sci. U.S.A. 97, 1062–1067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43).Herz S., Wungsintaweekul J., Schuhr C.A., Hecht S., Lüttgen H., Sagner S., Fellermeier M., Eisenreich W., Zenk M.H., Bacher A., Rohdich F. (2000) Biosynthesis of terpenoids: YgbB protein converts 4-diphosphocytidyl-2C-methyl-d-erythritol 2-phosphate to 2C-methyl-d-erythritol 2,4-cyclodiphosphate. Proc. Natl. Acad. Sci. U.S.A. 97, 2486–2490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44).Fellermeier M., Raschke M., Sagner S., Wungsintaweekul J., Schuhr C.A., Hecht S., Kis K., Radykewicz T., Adam P., Rohdich F., Eisenreich W., Bacher A., Arigoni D., Zenk M.H. (2001) Studies on the nonmevalonate pathway of terpene biosynthesis. the role of 2C-methyl-d-erythritol 2,4-cyclodiphosphate in plants. Eur. J. Biochem. 268, 6302–6310 [DOI] [PubMed] [Google Scholar]
- 45).Hecht S., Eisenreich W., Adam P., Amslinger S., Kis K., Bacher A., Arigoni D., Rohdich F. (2001) Studies on the nonmevalonate pathway to terpenes: The role of the GcpE (IspG) protein. Proc. Natl. Acad. Sci. U.S.A. 98, 14837–14842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46).Rohdich F., Hecht S., Gärtner K., Adam P., Kreiger C., Amslinger S., Arigoni D., Bacher A., Eisenreich W. (2002) Studies on the nonmevalonate terpene biosynthetic pathway: Metabolic role of IspH (LytB) protein. Proc. Natl. Acad. Sci. U.S.A. 99, 1158–1163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47).Sando T., Takeno S., Watanabe N., Okumoto H., Kuzuyama T., Yamashita A., Hattori M., Ogasawara N., Fukusaki E., Kobayashi A. (2008) Cloning and characterization of the 2-C-methyl-d-erythritol 4-phosphate (MEP) pathway genes of a natural rubber producing plant, Hevea brasiliensis. Biosci. Biotechnol. Biochem. 72, 2903–2917 [DOI] [PubMed] [Google Scholar]
- 48).Kim S.M., Kuzuyama T., Chang Y.J., Kim S.U. (2006) Cloning and characterization of 2-C-methyl-d-erythritol 2,4-cyclodiphosphate synthase (MECS) gene from Ginkgo biloba. Plant Cell Rep. 25, 829–835 [DOI] [PubMed] [Google Scholar]
- 49).Kim S.M., Kuzuyama T., Chang Y.J., Song K.S., Kim S.U. (2006) Identification of class 2 1-deoxy-d-xylulose 5-phosphate synthase and 1-deoxy-d-xylulose 5-phosphate reductoisomerase genes from Ginkgo biloba and their transcription in embryo culture with respect to ginkgolide biosynthesis. Planta Med. 72, 234–240 [DOI] [PubMed] [Google Scholar]
- 50).Kim S.M., Kuzuyama T., Chang Y.J., Kwon H.J., Kim S.U. (2006) Cloning and functional characterization of 2-C-methyl-d-erythritol 4-phosphate cytidyltransferase (GbMECT) gene from Ginkgo biloba. Phytochemistry 67, 1435–1441 [DOI] [PubMed] [Google Scholar]
- 51).Takahashi S., Kuzuyama T., Seto H. (1999) Purification, characterization, and cloning of a eubacterial 3-hydroxy-3-methylglutaryl coenzyme A reductase, a key enzyme involved in biosynthesis of terpenoids. J. Bacteriol. 181, 1256–1263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52).Kaneda K., Kuzuyama T., Takagi M., Hayakawa Y., Seto H. (2001) An unusual isopentenyl diphosphate isomerase found in the mevalonate pathway gene cluster from Streptomyces sp. strain CL190. Proc. Natl. Acad. Sci. U.S.A. 98, 932–937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53).Okamura E., Tomita T., Sawa R., Nishiyama M., Kuzuyama T. (2010) Unprecedented acetoacetyl-coenzyme A synthesizing enzyme of the thiolase superfamily involved in the mevalonate pathway. Proc. Natl. Acad. Sci. U.S.A. 107, 11265–11270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54).Seto H., Watanabe H., Furihata K. (1996) Simultaneous operation of the mevalonate and non-mevalonate pathways in the biosynthesis of isopentenyl diphosphate in Streptomyces aeriouvifer. Tetrahedron Lett. 37, 7979–7982 [Google Scholar]
- 55).Hamano Y., Dairi T., Yamamoto M., Kuzuyama T., Itoh N., Seto H. (2002) Growth-phase dependent expression of the mevalonate pathway in a terpenoid antibiotic-producing Streptomyces strain. Biosci. Biotechnol. Biochem. 66, 808–819 [DOI] [PubMed] [Google Scholar]
- 56).Kuzuyama T., Noel J.P., Richard S.B. (2005) Structural basis for the promiscuous biosynthetic prenylation of aromatic natural products. Nature 435, 983–987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57).Hamano Y., Dairi T., Yamamoto M., Kawasaki T., Kaneda K., Kuzuyama T., Itoh N., Seto H. (2001) Cloning of a gene cluster encoding enzymes responsible for the mevalonate pathway from a terpenoid-antibiotic-producing Streptomyces strain. Biosci. Biotechnol. Biochem. 65, 1627–1635 [DOI] [PubMed] [Google Scholar]