Background: A Candida albicans vaccine could prevent life-threatening systemic infections.
Results: A model of a protective antibody developed by NMR, chemical mapping, and computer simulations accounts for strong recognition of the reducing terminal monosaccharide of di- and trimannoside epitopes.
Conclusion: Man(β1–2)Man(β1–2)Man(α- is the optimal oligosaccharide for a conjugate vaccine because internal antigenic determinants dominate recognition of (1→2)-β-mannans.
Significance: Structural information of antibody-carbohydrate interactions identified a candidate conjugate vaccine.
Keywords: Antibodies, Carbohydrate, Epitope Mapping, Nuclear Magnetic Resonance, Vaccine Development, Candida, Glycoconjugate Vaccines, Homology Modeling, STD-NMR, Transferred NOE NMR
Abstract
A self-consistent model of β-mannan oligosaccharides bound to a monoclonal antibody, C3.1, that protects mice against Candida albicans has been developed through chemical mapping, NMR spectroscopic, and computational studies. This antibody optimally binds di- and trisaccharide epitopes, whereas larger oligomers bind with affinities that markedly decrease with increasing chain length. The (1→2)-β-linked di-, tri-, and tetramannosides bind in helical conformations similar to the solution global minimum. Antibody recognition of the di- and trisaccharide is primarily dependent on the mannose unit at the reducing end, with the hydrophobic face of this sugar being tightly bound. Recognition of a tetrasaccharide involves a frameshift in the ligand interaction, shown by strong binding of the sugar adjacent to the reducing end. We show that frameshifting may also be deliberately induced by chemical modifications. Molecular recognition patterns similar to that of mAb C3.1, determined by saturation transfer difference-NMR, were also observed in polyclonal sera from rabbits immunized with a trisaccharide glycoconjugate. The latter observation points to the importance of internal residues as immunodominant epitopes in (1→2)-β-mannans and to the viability of a glycoconjugate vaccine composed of a minimal length oligosaccharide hapten.
Introduction
Candida species are commensal organisms belonging to the human epithelial flora that cause opportunistic infections, with Candida albicans being generally the most common etiologic agent due to its greater prevalence and virulence (1). Vulvovaginal candidiasis is a common syndrome affecting many otherwise healthy women (2), while systemic candidiasis is a life-threatening infection of immunocompromised patients (1). The organism is capable of biofilm formation on the surface of medical device implants, and nosocomial infections are becoming an increasing concern, especially due to the toxicity issues of currently used antifungal treatments (3–5). Additionally, systemic candidiasis is difficult to diagnose, leading to delays in the administration of antifungal drugs that combined with either native or acquired drug resistance of Candida species, contribute to >40% mortality rates associated with systemic infection (1, 6, 7). A vaccine against this organism would, therefore, be highly desirable.
Both humoral and cell-mediated immunity are believed to play major roles in defense against Candida albicans (8). Cutler and co-workers (9–11) have shown that glycoconjugates prepared from native antigen as well as monoclonal antibodies specific for this antigen afford protection in active and passive immunization protocols. Results from our laboratory from immunizations with synthetic di- and trimannoside tetanus toxoid (12) or albumin2 conjugates showed protection in a rabbit model of candidiasis, and work with Cutler and co-workers (13) has demonstrated the promise of trimannoside-peptide conjugates in the design of a Candida vaccine.
The most effective β-mannan-specific mAb in passive protection experiments is the IgG3 C3.1 (14). As described below, this antibody exhibits unique immunochemical characteristics (15), and we have sought to base our design of conjugate vaccines on its specificity and antigen binding profile. To fully appreciate how this antibody binds antigen and what might be the best synthetic construct to elicit antibodies with similar binding profiles, we have delineated the recognition of β-mannan components of the cell-wall phosphomannan.
The β-mannan consists of (1→2)-β-linked mannose oligomers linked to the major α-mannan component of the cell wall either directly or via a phosphodiester (16–20). There is debate in the literature regarding the exact length of the β-mannan chain, with estimates ranging from 1–4 (21) to up to 14 (22) residues. Natural heterogeneity and changes in β-mannan length and abundance depending on growth stages and conditions are also relevant (23).
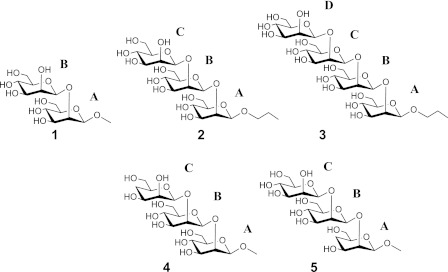
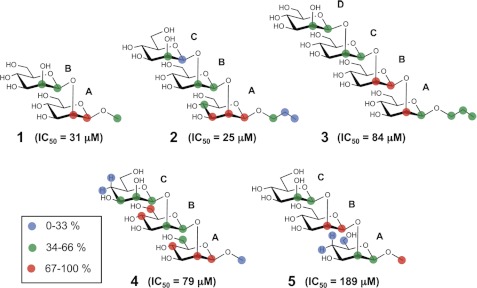
We have shown that the inhibitory power of a series of (1→2)-β-linked mannose oligomers binding to C3.1 decreased sharply for compounds larger than a trisaccharide (15). This immunochemical profile contrasts with a 45-year-old paradigm established by Kabat (24). His work on sera from five individuals showed that the largest proportion of antidextran antibodies had binding site sizes complementary to either a hexasaccharide or to smaller determinants. Although there are examples of exceptions to the Kabat observation (25–27), diminishing inhibitory activity with increasing oligosaccharide length has not been previously reported. To investigate this unusual immunochemistry and our hypothesis that the length of the β-mannan component of a C. albicans conjugate vaccine should correlate with the specificity of the C3.1 antibody (28), we chose to study the molecular recognition of the native oligosaccharides 1–3 and monodeoxy trisaccharides 4 and 5 (Chart 1) by C3.1 using NMR spectroscopic and computational techniques.
CHART 1.
Structures of oligosaccharides 1–5.
EXPERIMENTAL PROCEDURES
NMR Sample Preparation
mAb C3.1 IgG
C3.1 antibody hybridoma cells were grown in bioreactor flasks (Antibody Research), and cell culture supernatant was harvested. IgG was purified by protein A chromatography on a 5-ml column and eluted with 3 m KSCN. Eluted fractions were dialyzed against phosphate-buffered saline (20 mm sodium phosphate, pH 7.2, 140 mm NaCl). A β-mannan disaccharide affinity column was used to estimate the amount of active IgG in the preparation, and ligand:antibody ratios were calculated with respect to active antibody. In preparation for NMR studies, the antibody preparation was dialyzed a second time into 20 mm sodium phosphate, pH 7.2, without NaCl, flash-frozen in a dry ice/ethanol bath, lyophilized, and redissolved in D2O. NaCl was added to 50 mm. The final NMR samples with 1-3 contained 0.4 mm ligand and 13 μm active antibody for a ligand:binding site ratio of 15:1 in 60 mm sodium phosphate with 50 mm NaCl. The final NMR samples of 4 and 5 contained 0.325 mm ligand and 11 μm active antibody with the same ligand:binding site ratio. Antibody-only samples for saturation transfer double difference experiments were prepared at the same concentration and in the same buffer. Control samples of ligands with bovine serum albumin contained an equivalent mass of BSA in the same buffer at the same ligand concentration.
mAb C3.1 scFv
The protein (purified by nickel affinity chromatography and gel filtration, then lyophilized from 10 mm sodium phosphate at pH 7.2) was dissolved in D2O, and its binding affinity to 2 was confirmed by mass spectrometry.3 For the NMR samples, the scFv was used at 29 μm and 2 or 3 at 0.44 mm for a 15:1 ligand:binding site ratio.
Polyclonal Antibodies
Trisaccharide-BSA conjugate prepared according to literature procedures (29) was used to immunize rabbits by the protocol reported for trisaccharide tetanus toxoid conjugates (30). Polyclonal sera thus derived were purified on a disaccharide affinity column prepared by conjugating the disaccharide #11 (29) to epoxy-Sepharose according to the procedure reported (31), yielding 8.4 mg of protein in phosphate-buffered saline. The protein was concentrated to 1 ml by ultracentrifugation and exchanged into 20 mm sodium phosphate at pH 7.35, lyophilized, and resuspended in D2O, and NaCl was added to 50 mm. For the NMR samples, a solution of 10 μm IgG was obtained; 2 was added at oligosaccharide: binding site ratios of 15:1 or 100:1.
Enzyme-linked Immunosorbent Assays
IC50 values were measured by inhibition ELISA with synthetic C. albicans (1→2)-β-mannose trisaccharide-BSA conjugate on the plate and compounds 1-5 as inhibitors, as described in Costello and Bundle (47). Assays were carried out in triplicate, and the IC50 values have errors of 5–7%.
NMR Spectroscopy
All NMR experiments were recorded on a Varian VNMRS 700 MHz spectrometer equipped with a 5 mm 1H{13C/15N} z-gradient cryogenic probe. The measurements were performed at 30.5 °C (1), 35.5 °C (2-4), or 37 °C (5); the temperature was chosen to optimize detection of the anomeric protons.
STD-NMR
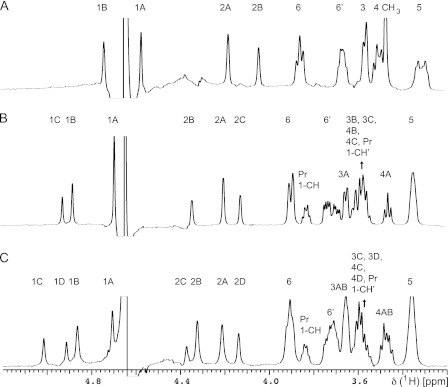
One-dimensional 1H saturation transfer difference (STD)4 experiments (32) were carried out without water suppression with saturation for 2 s at 2.5 ppm using a train of 50 ms E-BURP pulses of 60-Hz rf field strength separated by a 1-ms delay. Off-resonance saturation in alternate scans was applied at 26 ppm, and subtraction was carried out by phase cycling. A spin-lock pulse of 30 ms at 4-kHz rf field strength was applied after the first high power excitation pulse to reduce protein resonances. Because residual protein resonances remained, making quantitation difficult, the experiments were carried out by the saturation transfer double difference method (33); STD-NMR experiments were performed on a ligand/antibody sample and on a sample containing antibody only, and the latter experiment was subtracted from the former. The subtraction was carried out with processed data using the VNMRJ software (Agilent Technologies). Quantitation of the saturation Transfer Double Difference (STDD) enhancements was obtained by comparing the relative intensities of each signal in the STDD-NMR spectra to reference one-dimensional difference spectra (also with antibody signals removed by subtraction) using the VNMRJ graphical interface and using the signal intensity ratios to obtain the STDD enhancement percentages. This procedure was used instead of integration to allow us to correct for the local base line near each signal and avoid errors resulting from residual protein signals in the antibody preparation (e.g. Fig. 1). In the one-dimensional experiments, 10,000–14,000 transients consisting of 16,892 complex points spanning a spectral width of 8446 Hz were recorded. The data were zero-filled to 32,768 complex points and multiplied by an exponential apodization function with a broadening constant of 1 Hz before Fourier transformation.
FIGURE 1.
STDD spectra of oligosaccharides binding to C3.1. A, 1. B, 2. C. 3. Spectra were recorded at 15:1 ligand:binding site ratio, with saturation at 2.5 ppm for 2 s. Individual resonances are labeled; where a residue is not assigned, there are contributions to the signal from all monosaccharide units.
TrNOESY
Two-dimensional transferred NOESY spectra were recorded with double pulsed field gradient spin echo water suppression (34) and with a spin-lock pulse of 30 ms at 3–4 kHz after the first high-power excitation pulse to reduce protein resonances. Control experiments were recorded for each ligand in the presence of an irrelevant protein, BSA, to show that the observed NOEs were either small and positive or near zero in the absence of the antibody. For two-dimensional QUIET-trNOESY spectra (35), double or triple band-selective Q3 inversion pulses were applied to selectively invert H1, H2, and/or H4 resonances of the ligands halfway through the mixing time. 8 or 16 transients consisting of 8192 complex points spanning a spectral width of 6685 Hz in the directly detected dimension and 256 complex points spanning the same spectral width in the indirectly detected dimension were recorded. The data were zero-filled to 16,384 and 2,048 complex points, respectively, and apodized with a squared cosine window function before Fourier transformation. Spectra were analyzed and integrated with VNMRJ, and apparent distances were determined by fitting to a second-order exponential of the form f(t) = p0 × exp(−p2 × t)[1 − exp(−p1 × t)], where p0, p1, and p2 are adjustable parameters, and the initial slope is determined from the first derivative at time 0 [f'(0) = p0 × p1] (36) (other methods for distance determination yielded similar results).
Computational Methods
Molecular Dynamics
Bound conformations of 1-5 were calculated by simulated annealing with AMBER 11 using the GLYCAM06 (revision g) force field (37) and the modified generalized Born solvation model (38). The molecules were heated from 0 to 1200 K and maintained at 1200 K for 10 ps during which time the force constant for NMR restraints was gradually increased to its full value followed by slow cooling to 0 K over 20 ps. NMR restraints were applied as flat-bottomed potential wells with parabolic sides out to ±0.3 Å with respect to the experimentally observed internuclear distances, which were taken as the average of the distances calculated from the trNOESY and QUIET-trNOESY experiments, linear sides beyond those values, and force constants of 100 kcal/mol Å2. 500 separate conformations were generated in this manner and energy-minimized. Twenty conformations with no restraint violations greater than 0.1 Å and lowest AMBER energies were selected to represent the bound conformation of each oligosaccharide. A single representative conformer was selected as that which had the lowest r.m.s.d. to the mean coordinates of the ensemble of 20.
Molecular dynamics simulations of the free oligosaccharides were carried out for 5 ns with AMBER 11 using the GLYCAM06g (37) force field and either the modified generalized Born solvation model (38) or explicit TIP3P water. No separate experimental restraints were included. The molecules were heated from 0 to 298 K over 20 ps and maintained at 298 K for 5 ns. Data were collected every 10 ps. The SHAKE algorithm was used to constrain bonds involving hydrogen.
Antibody Comparative Modeling and Docking
The antibody model was constructed with the Web Antibody Modeling server (39) with the C3.1 variable region sequence as input. As described in “Results and Discussion,” the CDR loops were built by the default procedure of the WAM algorithm, which includes a data base search of the PDB followed by energy- and knowledge-based screens to identify the most likely loop conformations. Before docking, the model was additionally refined with a 5-ps molecular dynamics run at 500 K using the consistent valence force field (40) and a 5 Å layer of explicit water molecules, as implemented in InsightII/Discover (Accelrys, Inc.). The model was evaluated using the programs ANOLEA (41), Verify3D (42), PROCHECK (43), and the Protein Data Bank structure validation server. The model had satisfactory covalent geometry and structural quality. The model exhibited a Verify3D cumulative score of 101.42 and average scores ranging from 0.26 to 0.65, indicating a high quality overall structure and an excellent ANOLEA non-local normalized energy Z-score of −1.24, indicating minimal misfolded or high energy regions (41).
Docking calculations with 1 employed Autodock Vina (44) with a high exhaustiveness value (256) for detailed exploration of the ligand conformational and orientational space. Exocyclic bonds in the ligand were free to rotate, and the protein was rigid. The resulting binding modes were initially selected for agreement with the experimental transferred NOE, STD-NMR, and chemical mapping data. Promising binding modes of 1 by these criteria were further evaluated by superimposing the other oligosaccharides 2-5 and subjecting the modeled complexes to additional molecular dynamics refinement (10 ps in explicit water with AMBER/GLYCAM06g using the AMBER Parm03 force field for the protein (45) with NMR restraints applied to the oligosaccharides and no restraints applied to the protein). The binding mode described above agreed uniquely with experimental data from all ligands.
RESULTS AND DISCUSSION
Internal Disaccharide Epitope Is Preferred Antigenic Determinant
Chemical mapping studies established that hydroxyl groups at C3, C4, and C-4′ of the disaccharide epitope are involved in crucial hydrogen bonds with the antibody (46). These data suggested that the internal monosaccharide (residue A; Chart 1) made a dominant contribution to specificity and affinity. We elaborated these findings by synthesizing trisaccharides (4 and 5) that incorporate monodeoxy modifications in the terminal C or A residues, respectively (47). The loss of hydrogen bonding capacity in these analogues appears to preclude frameshifting, and the antibody is then obligated to bind either a terminal (5) or internal (4) disaccharide. The binding activities indicate a preference for recognition of internal disaccharide epitopes but also show that frameshifted binding modes are possible for a trisaccharide.2 Here we utilize the native oligomers and deoxy congeners to investigate the fine detail of C3.1 molecular recognition.
STD-NMR Confirms Antibody Binding to Internal Disaccharide Epitopes
The saturation transfer double difference technique (33) was employed for one-dimensional NMR epitope mapping of the native oligosaccharides 1-3 in complex with C3.1. The disaccharide 1 (IC50 = 31 μm) and trisaccharide 2 (IC50 = 25 μm) showed the strongest enhancements in residue A followed by residue B (Fig. 1, Table 1), indicating similar binding modes, although subtle differences were observed, such as the greater enhancement of H2A over H1A in 1. In contrast, the tetrasaccharide 3, which is a 5-fold weaker inhibitor (IC50 = 84 μm) than trisaccharide 2, showed maximal enhancements for residue B followed by the other residues in the order A > C ≈ D. These observations indicate a change in binding mode, which we interpret as a frameshift. A frameshift could arise from the sequence redundancy of the homopolymer and/or steric constraints of the combining site. The latter hypothesis is supported by the fact that the STD enhancement increased in the propyl aglycone of 3 relative to 2, suggesting that the increasing oligosaccharide length of 3 leads to additional contacts with the antibody at the reducing end.
TABLE 1.
STDD enhancements (%) for compounds 1–5 binding to antibody C3.1 at a 15:1 compound:binding site ratio
Enhancements are normalized to the strongest signal in each spectrum.
| Resonance | STD Enhancement |
||||
|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | |
| % | |||||
| Isolated resonances | |||||
| H1A | 73 | 87 | 64 | 100 | 41 |
| H2A | 86 | 76 | 96 | 100 | 64 |
| H3A | 100 | ||||
| H4A | 60 | 86 | 30 | ||
| H4′A | 16 | ||||
| H6A | 18 | ||||
| H6′A | 60 | ||||
| H1B | 55 | 47 | 82 | 60 | 88 |
| H2B | 64 | 44 | 90 | 63 | 88 |
| H4B | 67 | ||||
| H6′B | 92 | ||||
| H1C | 31 | 43 | 38 | 58 | |
| H2C | 49 | 36 | 60 | 54 | |
| H3C | 55 | ||||
| H4C | 20 | ||||
| H4′C | 24 | ||||
| H1D | 40 | ||||
| H2D | 58 | ||||
| Pr 1-CH | 53 | 61 | |||
| Pr 2-CH2 | 29 | 50 | |||
| Pr 3-CH3 | 24 | 50 | |||
| OCH3 | 50 | 67 | 78 | ||
| Overlapping resonances | |||||
| H5AB | 82 | 100 | |||
| H5BC | 100 | ||||
| H5ABC | 64 | ||||
| H5ABCD | 78 | ||||
| H3B,4B,4C | 90 | ||||
| H3A,6B,6C | 78 | ||||
| H5A,6′A,3C | 50 | ||||
| H6AB | 66 | 100 | |||
| H6ABC | 56 | ||||
| H6ABCD | 60 | ||||
| H6′AB | 73 | ||||
| H6′ABC | 53 | ||||
| H6′ABCD | 62 | ||||
| H3AB | 77 | 100 | |||
| H4AB | 100 | 87 | |||
| H6C,6′C,3B,5C | 65 | ||||
| H3B,3C,4B,4C, Pr 1-CH' | 58 | ||||
| H3C,3D,4C,4D, Pr 1-CH' | 55 | ||||
Oligosaccharides 1–3 Have Bound Conformations Similar to Those of Free Ligands in Solution
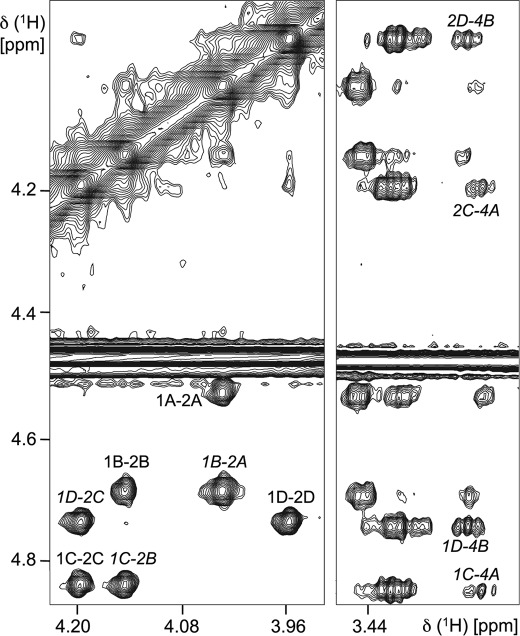
Fig. 2 shows an example of a two-dimensional trNOESY spectrum of 3, with interglycosidic NOE contacts indicated. Medium-range trNOEs such as H1C-H4A were also observed. These (i, i+2) trNOEs were previously observed in β-mannose oligomers in the absence of antibody (15) and established irregular helical conformations as the solution global minima. The (i, i+3) contacts H1D-H4A and H2D-H4A were not observed for 3, except weakly at the longest mixing time used (400 ms), and therefore, we did not attempt to determine this internuclear distance.
FIGURE 2.
Section of a two-dimensional trNOESY spectrum of 3 in the presence of C3.1 (150-ms mixing time). TrNOE contacts used to determine the bound conformation are labeled in italics; other NOEs are labeled in plain text.
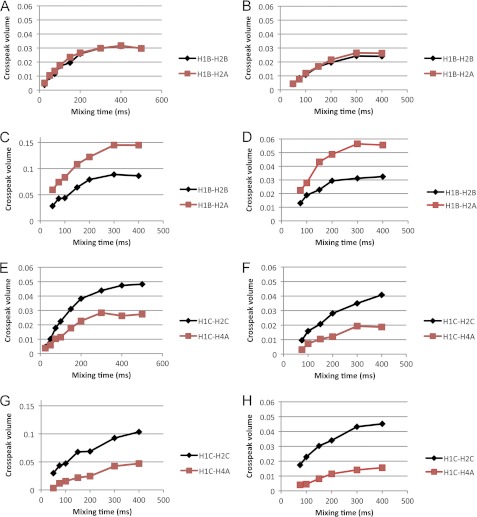
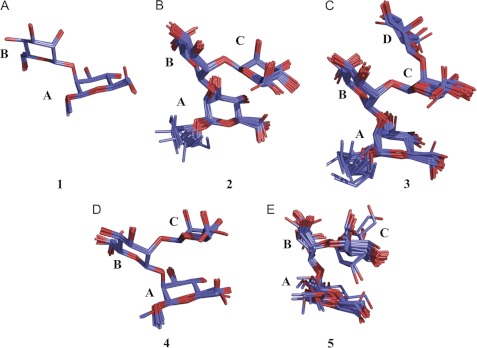
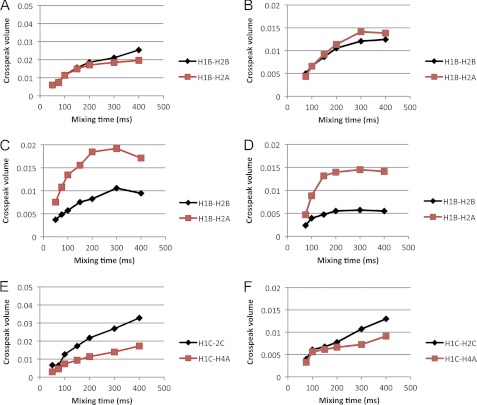
TrNOE buildup curves were collected with mixing times from 50 to 400 ms (Fig. 3) and used to determine internuclear distances with the H1-H2 distance of each monosaccharide as a reference. QUIET-trNOESY experiments were recorded to reduce contributions from spin diffusion. Distances calculated from both sets of data were very similar (Table 2), as were the buildup curves. As expected, there were reductions in the absolute intensities of some signals in the QUIET-trNOESY experiments. There were some observable differences between the oligosaccharides; for example, 3 exhibited a higher intensity for the H1B-H2A trNOE at all mixing times, corresponding to a shorter interglycosidic linkage distance than in 1 or 2 and a lower intensity of the H1C-H4A trNOE. These results indicating minor conformational changes are intriguing taken together with the STD-NMR data which show a change in binding mode for 3. However, these are minor adjustments rather than significant conformational changes (e.g. to another local minimum). Averages of the distances calculated from trNOESY and QUIET-trNOESY experiments were used in simulated annealing calculations to determine bound conformations. Ensembles of 20 conformations that satisfied the distance restraints are shown in Fig. 4, and the dihedral angles characterizing each ensemble are shown in Table 3. The oligosaccharides mainly exhibited similar conformations with dihedral angles in the region (φ, ψ ≈ 40°, 30°), although some variability was observed for the ψ angle of the A-B glycosidic linkage. All of the conformations fell within energetically accessible regions for the free molecules, as assessed by molecular modeling (supplemental Figs. S1 and S2).
FIGURE 3.
Examples of transferred NOESY buildup curves for the native oligosaccharides. Panels A–D show the buildup of the interglycosidic distance H1B-H2A (red squares) and the reference distance H1B-H2B (black diamonds). A, 2, trNOESY. B, 2, QUIET-trNOESY. C, 3, trNOESY. D, 3, QUIET-trNOESY. Panels E–H show the buildup of the medium-range distance H1C-H4A (red squares) and the reference distance H1C-H2C (black diamonds). E, 2, trNOESY. F, 2, QUIET-trNOESY. G, 3, trNOESY. H, 3, QUIET-trNOESY.
TABLE 2.
Distances (Å) in the bound conformations of oligosaccharides 1–5 derived by transferred NOE experiments
| 1 |
2 |
3 |
4 |
5 |
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| trNOE | QUIET-trNOE | trNOE | QUIET-trNOE | trNOE | QUIET-trNOE | trNOE | QUIET-trNOE | trNOE | QUIET-trNOE | |
| H1B-H2A | 2.6 | 2.7 | 2.4 | 2.5 | 2.2 | 2.3 | 2.5 | 2.5 | 2.2 | 2.2 |
| H1C-H2B | 2.5 | 2.5 | 2.4 | 2.4 | 2.5 | 2.3 | 2.5 | 2.5 | ||
| H1D-H2C | 2.5 | 2.4 | ||||||||
| H1C-H4A | 2.7 | 2.9 | 3.1 | 3.2 | 2.7 | 2.4 | n.d. | n.d. | ||
| H2C-H4A | 2.7 | 2.8 | 3.1 | 3.1 | 2.6 | 2.8 | n.d. | n.d. | ||
| H1D-H4B | 2.8 | 2.8 | ||||||||
| H2D-H4B | 3.1 | 2.9 | ||||||||
FIGURE 4.
Bound conformations of 1–5. Only heavy atoms are shown. The structures were superimposed using the center and root mean square commands in AMBER 11 and displayed in PyMOL. 20 conformers are shown for each molecule. A, 1, r.m.s.d. = 0.11 ± 0.09 Å. B, 2, r.m.s.d. = 0.47 ± 0.11 Å. C, 3, r.m.s.d. = 0.52 ± 0.10 Å. D, 4, r.m.s.d. = 0.39 ± 0.08 Å. E, 5, r.m.s.d. = 0.65 ± 0.30 Å.
TABLE 3.
Dihedral angles in the bound conformations of the oligosaccharides (mean ± S.D. of the ensembles of Fig. 4)
| 1 |
2 |
3 |
4 |
5 |
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| φ | ψ | φ | ψ | φ | ψ | φ | ψ | φ | ψ | |
| A-B | 46 ± 1 | 26 ± 1 | 48 ± 3 | −1 ± 6 | 41 ± 8 | 13 ± 9 | 44 ± 5 | 20 ± 4 | 14 ± 20 | 1 ± 25 |
| B-C | 44 ± 2 | 28 ± 1 | 38 ± 5 | 26 ± 3 | 41 ± 1 | 27 ± 1 | 36 ± 28 | 18 ± 10 | ||
| C-D | 38 ± 3 | 28 ± 2 | ||||||||
Oligosaccharides Larger Than Trisaccharide Are Forced to Bind in Frameshifted Mode
Since a disaccharide is the minimal unit required for binding affinity to C3.1 and the β-mannan is a homopolymer, any compound consisting of more than two mannose units has the possibility of frameshifting. Modified trisaccharides 4 and 5, deoxygenated either at the 4-position of residue C or A (47), were employed to probe the two possible binding modes. The C-4 hydroxyl group is known from previous work (48) to be essential for antibody recognition, and its removal forces the antibody to bind to the natural disaccharide epitope, preventing frameshifting and, in theory, isolating single binding modes.
The trisaccharide 4 with monodeoxy at the 4-OH of the C residue and IC50 of 79 μm showed similar STD enhancements to 2 (Table 1). Since 4 cannot bind via the B-C disaccharide, this compound provides an essentially pure representation of binding to the internal epitope (the A-B disaccharide). That there are only minimal changes in the STD pattern of 4 relative to 2 shows that contributions from a secondary, frameshifted binding mode where B-C is recognized are not significant even for 2. The similar STD profiles of 2 and 4 are illustrated in Fig. 5.
FIGURE 5.
Graphical representation of STDD enhancements for the oligosaccharides 1–5 binding to the IgG C3.1 (numerical data given in Table 1). The enhancement magnitudes of each resonance (isolated resonances only) are indicated by colored circles; all magnitudes are relative to the most strongly enhanced resonance in each spectrum.
In contrast, the trisaccharide 5, which is deoxygenated at the 4-position of the A residue showed a different STD enhancement pattern (Table 1, Fig. 5). H1 and H2 of B and H5 of B/C were the most strongly enhanced resonances of 5, in contrast to H1 and H2 of A and H5 and H6 of A/B in 4, and H1 and H3 of A in 2. Also, the enhancements of the C residue increased in 5, while that of H1A became weak. Overall, the pattern is consistent with a frameshift induced by the deoxygenation, consisting of the movement of the modified A residue to the edge of the binding site, while the B residue takes the place of the native A residue and the C residue the place of the native B. These observations confirm that a frameshifted binding mode is possible and that it may be deliberately induced by chemical modification. The IC50 value of 5 is 189 μm relative to 25 μm for 2 (47), corresponding to a free energy difference of ∼1.2 kcal/mol and a population of about 11% of the frameshifted conformer. Given that the STD data are a weighted average of the contributing populations, it is not surprising that significant contributions from this frameshifted conformer of the native trisaccharide were not observed.
Fig. 5 also shows the STD profile of 3. The increased enhancements of H1 and H2 of the B residue, the increased enhancements of the aglycone, and the stronger enhancement of H2A over H1A are similar to the STD profile of 5, which we now show represents a frameshifted binding mode. These observations support our hypothesis that the NMR observations of 3, in contrast to 1 and 2, are due to a significant population of a frameshifted binding mode.
Monodeoxy Trisaccharide Congeners Possess Bound Conformations Similar to Native β-Mannans
TrNOESY and QUIET-trNOESY buildup curves for 4 and 5 are shown in Fig. 6, experimentally determined interatomic distances are shown in Table 2, and bound conformations are shown in Fig. 4. 4 showed similar trNOE intensities and buildup rates to 2, indicating a similar bound conformation. The trNOE intensities of 5 were weaker than those of the other compounds. Similarly to 3, 5 showed a rapid buildup rate of the H1B-H2A trNOE. The (i, i+2) trNOEs H1C-H4A and H2C-H4A were not observed except weakly at mixing times >200 ms. This made it difficult to accurately determine the corresponding interatomic distance. The absence of NOEs may result from rapid relaxation, spin diffusion, or other internal mobility differences as well as the inherent low sensitivity of the trNOESY experiment and must be interpreted with caution. We, therefore, did not attempt to interpret this observation as a lower distance limit in the structure calculations. The ensemble of conformers representing the bound conformation of 5 has a higher r.m.s.d. value than those of the other oligosaccharides (Fig. 4E). This is not surprising as there are only two trNOE-derived distance restraints. However, the representative conformer, with minimal r.m.s.d. to the mean coordinates of the ensemble, has dihedral angles of φ, ψ = 34°, −14° and 37°, 27° for the A-B and B-C linkages, respectively. These values are similar to those of the other oligosaccharides, the only significant difference being at the A-B linkage ψ angle, where variation among the other oligosaccharides was previously noted. The bound conformations of 4 and 5 fall within the range of φ, ψ values consistent with the conformations sampled in solution by the free native tetrasaccharide (15) and within the energetically accessible ranges estimated in this study by molecular modeling (supplemental Figs. S1 and S2).
FIGURE 6.
Examples of transferred NOESY buildup curves for the deoxy trisaccharides. Panels A–D show the buildup of the interglycosidic distance H1B-H2A (red squares) and the reference distance H1B-H2B (black diamonds). A, 4, trNOESY. B, 4, QUIET-trNOESY. C, 5, trNOESY. D, 5, QUIET-trNOESY. Panels E and F show the buildup of the medium-range distance H1C-H4A (red squares) and the reference distance H1C-H2C (black diamonds). E, 4, trNOESY. F, 4, QUIET-trNOESY.
Computational Model of Antibody Fv and Docked Epitopes Identifies an Optimal Antigenic Determinant
We modeled the variable VH and VL domains of C3.1. Because antibody Fv fragments exhibit high structural similarity, they are ideal candidates for homology modeling, with the main source of uncertainty being the conformation of CDR H3. C3.1 has an H3 loop consisting of only six residues, which reduces the number of possible conformations significantly. A search of the Protein Data Bank for loops of this length, which is considered to saturate the conformational space of such a short loop (49), followed by energy minimization and knowledge-based ranking based on known structures was used to build the H3 conformation. The model shows a groove-shaped combining site, typical of sites directed against internal carbohydrate epitopes.
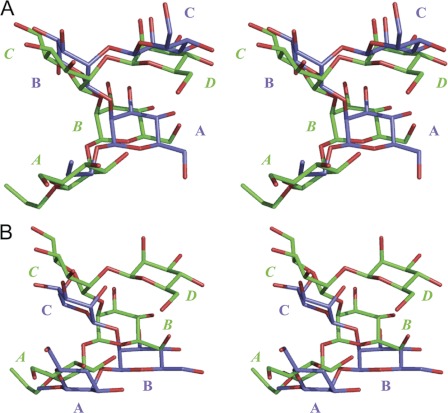
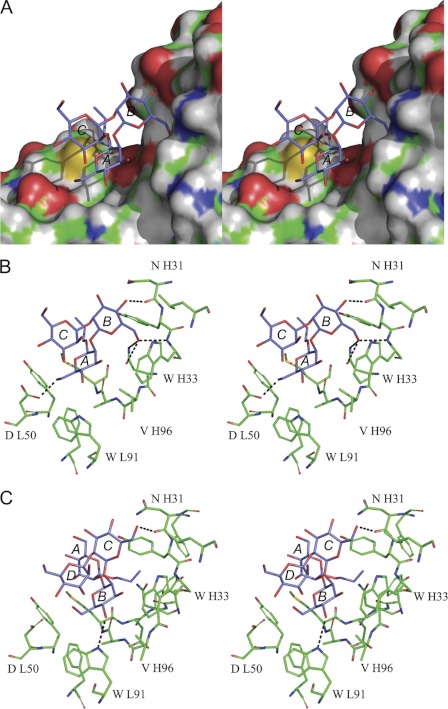
We searched for binding modes by computational docking of 1 followed by interactive evaluation of the resulting possibilities for agreement with the experimental data, molecular dynamics refinement of the representative conformers of 2-5 in the same binding modes, and further evaluation. The most likely binding mode involves residues A and B of 2 (Figs. 7 and 8). In Fig. 7A, the spatial equivalence of the A-B-C residues of 2 with the B-C-D residues of 3 is apparent. The residues participating in binding interactions are the internal disaccharide epitopes A-B of 2 and B-C of 3. In Fig. 7B, the relevant epitopes are B-C of 3 and 5, each with frameshifted binding modes.
FIGURE 7.
Walleye stereo views of the oligosaccharides in their predicted binding modes. A, shown are trisaccharide 2 (blue) and tetrasaccharide 3 (green). B, shown are 4-deoxy trisaccharide 5 (blue) and tetrasaccharide 3 (green). For clarity, only heavy atoms are shown. Oxygen atoms are colored red. Monosaccharide residues are labeled beginning at the reducing end with the labels for 3 in italics.
FIGURE 8.
Walleye stereo views of computational models of oligosaccharides binding to C3.1. A, shown is trisaccharide 2 and all-atom molecular surface representation of the binding pocket. B, shown is the predicted binding mode of trisaccharide 2 (blue) and stick representation of the binding pocket; selected residues are labeled. Hydrogen bonds are indicated by dashed lines. C, shown is the predicted binding mode of the tetrasaccharide 3 (blue).
Further details of the predicted intermolecular interactions are shown in Fig. 8. Fig. 8, A and B, show the predicted complex of 2 with C3.1. In agreement with the STD-NMR data, the hydrophobic face of the A residue is stacked closely against the antibody, interacting with Met H98 and backbone atoms of CDR H3. The 3- and 4-OH hydroxyl groups of A, which are essential for binding, are near the potential hydrogen bond donors Ala H97 NH and Trp L91 NE1, and the 6-OH group hydrogen bonds to the Asp L50 carboxylate side chain. The 6-OH of the B residue is predicted to form hydrogen bonds to Trp H33 N and Asn H95 OD1 and ND1, whereas its 4-OH forms a hydrogen bond to Asn H31 O. This model is broadly consistent with the previously established pattern of essential and non-essential hydroxyl groups determined by chemical mapping (46–47).
The conformation of the trisaccharide then dictates that the hydrophobic faces of the other sugars are exposed to solvent, explaining the reduced STD enhancements of H1 and H2 of the B and C rings. The overlapping multiplet containing H3 and H4 of B and C of 2 has an intermediate STD enhancement of 58%, consistent with a binding mode in which these protons face the protein but are not in as close contact as, for example, the A ring with Met H98. The B ring makes contact mainly with CDR H1, whereas the C ring is completely solvent-exposed, consistent with the observation that this residue does not provide additional binding energy relative to a disaccharide. Its STD enhancements are weaker than those of the A and B residues and may result from indirect transfers/spin diffusion.
Positioning of 3 in the combining site with the A-C residues superimposed on those of 2 shows that there is no room at the nonreducing end for the D residue, because of steric interactions with Trp H33 (Fig. 8C). Our model thus explains why frameshifting is observed for 3 and presumably must occur for any oligomer larger than a trisaccharide to bind. The frameshifted binding mode of 3 was modeled by superimposing the B-D residues on 2 and performing a short molecular dynamics refinement in explicit water. The A residue of 3 is partly solvent-exposed, but its hydrophobic face contacts Met H98 of CDR H3, and the propyl aglycone contacts Asp H99 and CDR H1. As seen in Fig. 7A, the disaccharide epitopes in 2 and 3 adopt similar binding modes and are predicted to make similar intermolecular interactions. Minor changes in the protein conformation and ligand position are apparent; for example, in 3, the 6-OH of C hydrogen bonds to Asn H31 O rather than to Trp H33 and Asn H95 as in 2. The same strategy was employed to generate a model of the antibody with 5.
The steric requirements of the binding site to accommodate the reducing end as well as the nonreducing end, of the oligomer appear to be quite strict, and when frameshifting, the ligands must adopt conformations that avoid unfavorable interactions with Met H98, Tyr H32, and the protein backbone of CDRs H1 and H3. We do not believe that frameshifting by more than one residue is likely, because the shape of the binding site is not complementary to a longer (1→2)-β polymer. The binding of oligosaccharides larger than 3 would require energetically unfavorable changes to glycosidic torsional angles to compensate for the lack of shape complementarity, explaining the drop in binding affinity observed for penta- and hexasaccharides (15). In contrast, our preliminary modeling studies show that this area of the binding site can efficiently accommodate the α-linkage and phosphate group of the tetrasaccharide Man(β1–2)Man(β1–2)Man(α1–6)Man6P(αOMe), with the phosphate linkage in an energetically accessible conformation (as assessed by molecular dynamics simulations in the presence of explicit water and sodium ions). This suggests that the αMan residue to which the Man(β1–2)Man(β1–2) disaccharide is attached (28) may form part of the epitope recognized by C3.1 and could be important if C3.1-like antibodies are to be raised. Based on these predictions, we recently completed the synthesis of the tetrasaccharide Man(β1–2)Man(β1–2)Man(α1–6)Man6P(αOMe) and showed that its inhibitory power was the same as 2. In parallel we could also show that a disaccharide-protein conjugate afforded protection against live challenge with C. albicans.2
Single-chain Variable Antibody Fragment Exhibits Whole Antibody Binding Profiles
We generated a single-chain variable fragment (scFv) of C3.1 and determined that it binds to oligosaccharides 1-3 with KD values measured by surface plasmon resonance of 26.7 ± 0.2, 18.0 ± 0.3, and 87.7 ± 0.3 μm, respectively (supplemental Fig. S3). These values are very close to those measured by ELISA for the IgG (47). The STDD enhancement pattern for 2 binding to the scFv (supplemental Table S1) showed a similar trend to the IgG, with H1A, H3A, and H1B all being strongly enhanced. The differences between the most strongly and least strongly enhanced resonances were exaggerated relative to the IgG; in the scFv it is clear that H1C, H2C, and the propyl aglycone experienced minimal enhancements, in agreement with the model proposed above. The trend A > B > C was maintained. Observations for 3 were similar, with greatly exaggerated differences between the most strongly and least strongly enhanced resonances and with the trend B > A > C ≈ D maintained. Despite lower overall enhancement strengths, increased enhancements of the aglycone in 3 versus 2, interpreted as additional contacts with the antibody at the reducing end, were also observed with the scFv. The greater spread in enhancement strengths is likely to be due to slower relaxation and spread of saturation in the smaller scFv complexes. Increasing the power level for saturation in the STD experiment increased the enhancements of the weakly enhanced resonances correspondingly but did not bring the enhancement patterns into close similarity with those of the IgG. To our knowledge, this is the first example of a comparison of STD measurements between IgG and scFv forms of the same antibody.
TrNOESY experiments showed similar trends as the IgG. The H1B-H2A buildup rate was more rapid for 3 than 2, whereas the H1C-H4A buildup rate was slower. The model presented in Fig. 8 hypothesizes that the H1C-H4A distance increases in frameshifted binding modes as a result of the steric constraints of the CDR H1/H3 area of the binding site; at the same time, the H1B-H2A distance decreases slightly because of the changed ψ angles. Overall, similar buildup rates and distances, within reasonable error ranges, were observed for the IgG and scFv.
(1→2)-β-Mannan Linkage Exposes Non-terminal Residues to Act as Immunodominant Elements
Our results raise the question of why the antibody appears designed to recognize internal, rather than terminal, (1→2)-β-mannan epitopes, as the internal epitopes are found in the midst of a complex cell wall structure (28). The structural similarity between terminal and internal (1→2)-linked epitopes has been noted by Jann and Westphal (50) based on the fact that the (1→2)-linkage exposes much of the monosaccharide to the solution, as would be the case for a terminal monosaccharide. They further proposed that a sugar in a (1→2)-linkage is the “immunological equivalent” of the same sugar in a terminal position. These conclusions were based on earlier observations of immunological cross-reactivity by Heidelberger (51) and others. In the case of (1→2) linkages, the preference of an antibody for internal over terminal epitopes is consistent with literature precedent.
Optimum Minimal Epitope Is Man(β1–2)Man(β1–2)Man(α-)
An additional consideration for C3.1 is the possible involvement of elements of the αMan-phosphate linkage in cell wall recognition. Initial modeling studies showed that the combining site can accommodate this linkage with the phosphate and αMan making contacts with residues at the edge of the binding site. Our unpublished results show that the tetrasaccharide Man(β1–2)Man(β1–2)Man(α1–6)Man6P(αOMe) has the same inhibitory power as 2 so that these contacts do not provide significant additional binding energy. However, the conformation of the residues that link the β-mannan to the α-mannan side chains allows the (1→2)-β-mannose disaccharide to be positioned properly in the binding site, and this molecule shows greater complementarity to the binding site than would a longer β-mannan polymer. In this sense, the other residues present in the Candida cell wall may also have contributed to the specificity of C3.1.
Polyclonal Sera to Trisaccharide Conjugate Vaccine Exhibit C3.1-like Antigenic Specificity
To assess the prevalence of C3.1-like antibodies elicited by experimental immunization, we carried out STDD-NMR experiments with 2 using sera derived from rabbit immunization experiments with a trisaccharide-BSA conjugate (Table 4). Like C3.1, the polyclonal sera showed strongest enhancements of the A residue, with lesser enhancements of the B and C residues. There were some changes in the enhancement strengths of individual protons. The aglycone enhancement strength remained similar, as did that of the H6/6′ resonances, but there were increases in the enhancement strengths of the H5 and other degenerate multiplets, which is not surprising given that the polyclonal sera contain a mixture of paratopes. Even so, we find the preservation of the C3.1-like pattern with strong recognition of the A residue, indicating a preference for an internal epitope, in experimental immunization trials highly significant and indicative of the biological importance of this monoclonal antibody. A contemporaneous observation with sera directed against the α1,3-galactose epitope was reported recently (52).
TABLE 4.
STDD enhancements (%) for the trisaccharide 2 binding to the mAb C3.1 and to polyclonal sera elicited by experimental immunizations
Enhancements are normalized to the strongest signal in each spectrum.
| Resonance | STDD Enhancement |
||
|---|---|---|---|
| C3.1, 15:1 | pcAb, 15:1 | pcAb, 100:1 | |
| % | |||
| Isolated resonances | |||
| H1A | 87 | 67 | 61 |
| H2A | 76 | 79 | 69 |
| H3A | 100 | 100 | 100 |
| H4A | 60 | 96 | 100 |
| H1B | 47 | 51 | 45 |
| H2B | 44 | 54 | 44 |
| H1C | 31 | 35 | 30 |
| H2C | 49 | 63 | 57 |
| Pr 1-CH | 53 | 73 | 67 |
| Pr 2-CH2 | 29 | 47 | 50 |
| Pr 3-CH3 | 24 | 38 | 35 |
| Overlapping resonances | |||
| H5ABC | 64 | 88 | 95 |
| H6ABC | 56 | 61 | 63 |
| H6′ABC | 53 | 52 | 67 |
| H3B,3C,4B,4C, Pr 1-CH' | 58 | 92 | 91 |
In summary, we report that the di- and trisaccharides 1 and 2 depend heavily on residue A for recognition and share similar binding modes and bound conformations, whereas recognition of a tetrasaccharide 3 involves changes in the binding mode that are probably due to frameshifting of the homo-oligomer. Our model of the bound conformations of 2 and 3 with the binding site of C3.1 demonstrates how the conformation of a β-mannan imposes recognition of the internal monosaccharide. This model clearly explains why a di- or trisaccharide, which share similar binding modes, bound conformations, and binding affinities, would be preferred as haptens over a larger epitope such as 3. This tetrasaccharide has decreased binding affinity and a preference for a frameshifted binding mode, with occupancy of part of the binding site that appears naturally meant to accommodate other parts of the phosphomannan. The model is consistent with the feasibility of antibody binding to the whole phosphomannan, since we show that the α-mannose phosphodiester linkage point is readily accommodated as antigen exits the binding site. These data further suggest that a conjugate vaccine composed of a disaccharide epitope alone or linked to the first residue of the α-mannan should induce C3.1-like binding profiles and may provide protection in the manner afforded by C3.1.
Supplementary Material
Acknowledgments
We thank Dr. Jim E. Cutler for providing the mAb C3.1, Joanna Sadowska, Dr. Adam Szpacenko, and Susmita Sarkar for valuable assistance with protein purification, Casey Costello for the synthesis of compounds 4 and 5, Dr. Thanh Luu for the synthesis of compounds 1-3, and Dr. Tomasz Lipinski for helpful discussions. Funding for the 700-MHz NMR spectrometer was provided by Western Economic Diversification Canada.
This work was supported, in whole or in part, by National Institutes of Health Grants GM094919 and RR005351 (to R. J. W. and N. E. W.). This work was also supported by the Alberta Glycomics Centre and the Natural Sciences and Engineering Research Council of Canada, the Science Foundation of Ireland (08/IN.1/B2070), and the European Union (European Regional Development Fund). D. R. B. is a co-founder and shareholder in a company attempting to advance a vaccine against C. albicans.

This article contains supplemental Table S1 and Figs. S1–S3.
D. R. Bundle, C. Nycholat, C. Costello, T. Lipinski, and R. Rennie, submitted for publication.
E. Kitova, and J. Klassen, unpublished data.
- STD
- saturation transfer difference
- STDD
- saturation transfer double difference
- r.m.s.d.
- root mean square deviation
- trNOE
- transferred NOE
- CDR
- complementarity-determining region
- QUIET
- quenching undesirable indirect external trouble.
REFERENCES
- 1. MacCallum D. M. (2010) In Pathogenic Yeasts, The Yeast Handbook, Ashbee H. R., Bignell E. M., Eds., Springer-Verlag, Berlin, pp. 41–67 [Google Scholar]
- 2. Fidel P. L., Jr., Cutler J. E. (2011) Prospects for development of a vaccine to prevent and control vaginal candidiasis. Curr. Infect. Dis. Rep. 13, 102–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Toscano C. M., Jarvis W. R. (1999) Emerging issues in nosocomial fungal infections. Curr. Infect. Dis. Rep. 1, 347–361 [DOI] [PubMed] [Google Scholar]
- 4. Zhang A. Y., Camp W. L., Elewski B. E. (2007) Advances in topical and systemic antifungals. Dermatol. Clin. 25, 165–183 [DOI] [PubMed] [Google Scholar]
- 5. Stronati M., Decembrino L. (2006) Neonatal invasive candidiasis. Minerva Pediatr. 58, 537–549 [PubMed] [Google Scholar]
- 6. Morrell M., Fraser V. J., Kollef M. H. (2005) Delaying the empiric treatment of Candida bloodstream infection until positive blood culture results are obtained. A potential risk factor for hospital mortality. Antimicrob. Agents Chemother. 49, 3640–3645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Szabo E. K., MacCallum D. M. (2011) The contribution of mouse models to our understanding of systemic candidiasis. FEMS Microbiol. Lett. 320, 1–8 [DOI] [PubMed] [Google Scholar]
- 8. Cutler J. E. (2005) Defining criteria for anti-mannan antibodies to protect against candidiasis. Curr. Mol. Med. 5, 383–392 [DOI] [PubMed] [Google Scholar]
- 9. Han Y., Morrison R. P., Cutler J. E. (1998) A vaccine and monoclonal antibodies that enhance mouse resistance to Candida albicans vaginal infection. Infect. Immun. 66, 5771–5776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Han Y., Cutler J. E. (1995) Antibody response that protects against disseminated candidiasis. Infect. Immun. 63, 2714–2719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Han Y., Ulrich M. A., Cutler J. E. (1999) Candida albicans mannan extract-protein conjugates induce a protective immune response against experimental candidiasis. J. Infect. Dis. 179, 1477–1484 [DOI] [PubMed] [Google Scholar]
- 12. Wu X., Lipinski T., Carrel F. R., Bailey J. J., Bundle D. R. (2007) Synthesis and immunochemical studies on a Candida albicans cluster glycoconjugate vaccine. Org. Biomol. Chem. 5, 3477–3485 [DOI] [PubMed] [Google Scholar]
- 13. Xin H., Dziadek S., Bundle D. R., Cutler J. E. (2008) Synthetic glycopeptide vaccines combining β-mannan and peptide epitopes induce protection against candidiasis. Proc. Natl. Acad. Sci. U.S.A. 105, 13526–13531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Han Y., Riesselman M. H., Cutler J. E. (2000) Protection against candidiasis by an immunoglobulin G3 (IgG3) monoclonal antibody specific for the same mannotriose as an IgM protective antibody. Infect. Immun. 68, 1649–1654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Nitz M., Ling C. C., Otter A., Cutler J. E., Bundle D. R. (2002) The unique solution structure and immunochemistry of the Candida albicans β-1,2-mannopyranan cell wall antigens. J. Biol. Chem. 277, 3440–3446 [DOI] [PubMed] [Google Scholar]
- 16. Kobayashi H., Shibata N., Mitobe H., Ohkubo Y., Suzuki S. (1989) Structural study of phosphomannan of yeast-form cells of Candida albicans J-1012 strain with special reference to application of mild acetolysis. Arch. Biochem. Biophys. 272, 364–375 [DOI] [PubMed] [Google Scholar]
- 17. Kobayashi H., Shibata N., Nakada M., Chaki S., Mizugami K., Ohkubo Y., Suzuki S. (1990) Structural study of cell wall phosphomannan of Candida albicans NIH B-792 (serotype B) strain, with special reference to 1H and 13C NMR analyses of acid-labile oligomannosyl residues. Arch. Biochem. Biophys. 278, 195–204 [DOI] [PubMed] [Google Scholar]
- 18. Shibata N., Kobayashi H., Takahashi S., Okawa Y., Hisamichi K., Suzuki S. (1991) Structural study on a phosphorylated mannotetraose obtained from the phosphomannan of Candida albicans NIH B-792 strain by acetolysis. Arch. Biochem. Biophys. 290, 535–542 [DOI] [PubMed] [Google Scholar]
- 19. Shibata N., Kobayashi H., Tojo M., Suzuki S. (1986) Characterization of phosphomannan-protein complexes isolated from viable cells of yeast and mycelial forms of Candida albicans NIH B-792 strain by the action of Zymolyase-100T. Arch. Biochem. Biophys. 251, 697–708 [DOI] [PubMed] [Google Scholar]
- 20. Tojo M., Shibata N., Ban Y., Suzuki S. (1990) Structure of the d-mannan of Candida stellatoidea IFO 1397 strain. Comparison with that of the phospho-d-mannan of Candida albicans NIH B-792 strain. Carbohydr. Res. 199, 215–226 [DOI] [PubMed] [Google Scholar]
- 21. Shibata N., Suzuki A., Kobayashi H., Okawa Y. (2007) Chemical structure of the cell-wall mannan of Candida albicans serotype A and its difference in yeast and hyphal forms. Biochem. J 404, 365–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Trinel P. A., Lepage G., Jouault T., Strecker G., Poulain D. (1997) Definitive chemical evidence for the constitutive ability of Candida albicans serotype A strains to synthesize β-1,2-linked oligomannosides containing up to 14 mannose residues. FEBS Lett. 416, 203–206 [DOI] [PubMed] [Google Scholar]
- 23. Poulain D., Jouault T. (2004) Candida albicans cell wall glycans, host receptors, and responses. Elements for a decisive crosstalk. Curr. Opin. Microbiol. 7, 342–349 [DOI] [PubMed] [Google Scholar]
- 24. Kabat E. A. (1960) The upper limit for the size of the human antidextran combining site. J. Immunol. 84, 82–85 [PubMed] [Google Scholar]
- 25. Bundle D. R. (1999) in Bioorganic Chemistry (Carbohydrates) (Hecht S. M., ed.) pp. 370–440, Oxford University Press, Oxford [Google Scholar]
- 26. Newman B. A., Kabat E. A. (1985) An immunochemical study of the combining site specificities of C57BL/6J monoclonal antibodies to α (1–6)-linked dextran B512. J. Immunol. 135, 1220–1231 [PubMed] [Google Scholar]
- 27. Jennings H. J. (2012) in Anticarbohydrate Antibodies (Kosma P., Müller-Loennies S., eds) pp. 55–74, Springer-Verlag, Wien, Austria [Google Scholar]
- 28. Bundle D. R., Costello C., Nycholat C., Lipinski T., Rennie R. (2012) in Anticarbohydrate Antibodies (Kosma P., Müller-Loennies S., eds) pp. 121–146, Springer-Verlag, Wien, Austria [Google Scholar]
- 29. Nitz M., Bundle D. R. (2001) Synthesis of di- to hexasaccharide 1,2-linked β-mannopyranan oligomers, a terminal S-linked tetrasaccharide congener, and the corresponding BSA glycoconjugates. J. Org. Chem. 66, 8411–8423 [DOI] [PubMed] [Google Scholar]
- 30. Bundle D. R., Nitz M., Wu X., Sadowska J. M. (2008) A uniquely small, protective carbohydrate epitope may yield a conjugate vaccine for Candida albicans. ACS Symp. Ser. 989, 163–183 [Google Scholar]
- 31. Bundle D. R., Eichler E., Gidney M. A., Meldal M., Ragauskas A., Sigurskjold B. W., Sinnott B., Watson D. C., Yaguchi M., Young N. M. (1994) Molecular recognition of a Salmonella trisaccharide epitope by monoclonal antibody Se155-4. Biochemistry 33, 5172–5182 [DOI] [PubMed] [Google Scholar]
- 32. Mayer M., Meyer B. (1999) Characterization of ligand binding by saturation transfer difference NMR spectroscopy. Angew. Chem. Int. Ed. 38, 1784–1788 [DOI] [PubMed] [Google Scholar]
- 33. Claasen B., Axmann M., Meinecke R., Meyer B. (2005) Direct observation of ligand binding to membrane proteins in living cells by a saturation transfer double difference (STDD) NMR spectroscopy method shows a significantly higher affinity of integrin α(IIb)β3 in native platelets than in liposomes. J. Am. Chem. Soc. 127, 916–919 [DOI] [PubMed] [Google Scholar]
- 34. Hwang T. L., Shaka A. J. (1995) Water suppression that works. Excitation sculpting using arbitrary waveforms and pulsed field gradients. J. Magn. Reson. Ser. A 112, 275–279 [Google Scholar]
- 35. Vincent S. J., Zwahlen C., Bodenhausen G. (1996) Suppression of spin diffusion in selected frequency bands of nuclear Overhauser spectra. J. Biomol. NMR 7, 169–172 [DOI] [PubMed] [Google Scholar]
- 36. Haselhorst T., Espinosa J. F., Jiménez-Barbero J., Sokolowski T., Kosma P., Brade H., Brade L., Peters T. (1999) NMR experiments reveal distinct antibody-bound conformations of a synthetic disaccharide representing a general structural element of bacterial lipopolysaccharide epitopes. Biochemistry 38, 6449–6459 [DOI] [PubMed] [Google Scholar]
- 37. Kirschner K. N., Yongye A. B., Tschampel S. M., González-Outeiriño J., Daniels C. R., Foley B. L., Woods R. J. (2008) GLYCAM06. A generalizable biomolecular force field. Carbohydrates. J. Comput. Chem. 29, 622–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Onufriev A., Case D. A., Bashford D. (2002) Effective born radii in the generalized born approximation. The importance of being perfect. J. Comput. Chem. 23, 1297–1304 [DOI] [PubMed] [Google Scholar]
- 39. Whitelegg N. R., Rees A. R. (2000) WAM. An improved algorithm for modeling antibodies on the WEB. Protein Eng. 13, 819–824 [DOI] [PubMed] [Google Scholar]
- 40. Dauber-Osguthorpe P., Roberts V. A., Osguthorpe D. J., Wolff J., Genest M., Hagler A. T. (1988) Structure and energetics of ligand binding to proteins. Escherichia coli dihydrofolate reductase-trimethoprim, a drug-receptor system. Proteins Struct. Funct. Genet. 4, 31–47 [DOI] [PubMed] [Google Scholar]
- 41. Melo F., Feytmans E. (1998) Assessing protein structures with a non-local atomic interaction energy. J. Mol. Biol. 277, 1141–1152 [DOI] [PubMed] [Google Scholar]
- 42. Eisenberg D., Lüthy R., Bowie J. U. (1997) VERIFY3D. Assessment of protein models with three-dimensional profiles. Methods Enzymol. 277, 396–404 [DOI] [PubMed] [Google Scholar]
- 43. Laskowski R. A., MacArthur M. W., Moss D. S., Thornton J. M. (1993) PROCHECK. A program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26, 283–291 [Google Scholar]
- 44. Trott O., Olson A. J. (2010) AutoDock Vina. Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 31, 455–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Duan Y., Wu C., Chowdhury S., Lee M. C., Xiong G., Zhang W., Yang R., Cieplak P., Luo R., Lee T., Caldwell J., Wang J., Kollman P. (2003) A point-charge force field for molecular mechanics simulations of proteins based on condensed-phase quantum mechanical calculations. J. Comput. Chem. 24, 1999–2012 [DOI] [PubMed] [Google Scholar]
- 46. Nycholat C. (2008) Mapping the Protective Epitope of an anti-Candida albicans Monoclonal Antibody Using Synthetic Di- and Trisaccharide Analogues of (1–2)-β-d-Mannopyranoside. Ph.D. thesis University of Alberta [Google Scholar]
- 47. Costello C., Bundle D. R. (2012) Synthesis of three trisaccharide congeners to investigate frame shifting of β-1,2-mannan homo-oligomers in an antibody binding site. Carbohydr. Res., in press [DOI] [PubMed] [Google Scholar]
- 48. Nycholat C. M., Bundle D. R. (2009) Synthesis of monodeoxy and mono-O-methyl congeners of methyl β-d-mannopyranosyl-(1→2)-β-d-mannopyranoside for epitope mapping of anti-Candida albicans antibodies. Carbohydr. Res. 344, 1397–1411 [DOI] [PubMed] [Google Scholar]
- 49. Martin A. C., Cheetham J. C., Rees A. R. (1989) Modeling antibody hypervariable loops. A combined algorithm. Proc. Natl. Acad. Sci. U.S.A. 86, 9268–9272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Jann K., Westphal O. (1975) in The Antigens (Sela M., ed) Vol. 3, pp. 1–125, Academic Press, Inc., New York [Google Scholar]
- 51. Heidelberger M. (1960) Structure and immunological specificity of polysaccharides. Fortsch. Chem. Org. Naturst. 18, 503–536 [Google Scholar]
- 52. Plum M., Michel Y., Wallach K., Raiber T., Blank S., Bantleon F. I., Diethers A., Greunke K., Braren I., Hackl T., Meyer B., Spillner E. (2011) Close-up of the immunogenic α1,3-galactose epitope as defined by a monoclonal chimeric immunoglobulin E and human serum using STD NMR. J. Biol. Chem. 286, 43103–43111 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.