Background: Subunit NuoI contains two iron-sulfur clusters, N6a and N6b, in the electron transfer pathway of Escherichia coli NDH-1.
Results: Contrary to predictions, cluster N6a was EPR-detectable. Conserved residues presumed to be in electron tunneling are not essential.
Conclusion: EPR signals and key residues were assigned.
Significance: Missing information on the electron flow was provided.
Keywords: Bioenergetics, Dehydrogenase, Electron Paramagnetic Resonance (EPR), Electron Transfer, Escherichia coli, Mutagenesis Site-specific, NDH-1, Complex I
Abstract
Bacterial proton-translocating NADH:quinone oxidoreductase (NDH-1) consists of a peripheral and a membrane domain. The peripheral domain catalyzes the electron transfer from NADH to quinone through a chain of seven iron-sulfur (Fe/S) clusters. Subunit NuoI in the peripheral domain contains two [4Fe-4S] clusters (N6a and N6b) and plays a role in bridging the electron transfer from cluster N5 to the terminal cluster N2. We constructed mutants for eight individual Cys-coordinating Fe/S clusters. With the exception of C63S, all mutants had damaged architecture of NDH-1, suggesting that Cys-coordinating Fe/S clusters help maintain the NDH-1 structure. Studies of three mutants (C63S-coordinating N6a, P110A located near N6a, and P71A in the vicinity of N6b) were carried out using EPR measurement. These three mutations did not affect the EPR signals from [2Fe-2S] clusters and retained electron transfer activities. Signals at gz = 2.09 disappeared in C63S and P110A but not in P71A. Considering our data together with the available information, gz,x = 2.09, 1.88 signals are assigned to cluster N6a. It is of interest that, in terms of gz,x values, cluster N6a is similar to cluster N4. In addition, we investigated the residues (Ile-94 and Ile-100) that are predicted to serve as electron wires between N6a and N6b and between N6b and N2, respectively. Replacement of Ile-100 and Ile-94 with Ala/Gly did not affect the electron transfer activity significantly. It is concluded that conserved Ile-100 and Ile-94 are not essential for the electron transfer.
Introduction
The proton-translocating NADH-quinone oxidoreductase (EC 1.6.5.3) (complex I)3 is the first enzyme in the respiratory chain that couples scalar transfer of electrons from NADH to quinone with vectorial proton translocation across the energy-transducing membrane. Mammalian complex I is composed of 45 different subunits and has the most intricate structure of the membrane-bound mitochondrial enzyme complexes (1–3). Dysfunction of complex I has been implicated in numerous human neurodegenerative diseases, including Parkinson disease, Leber hereditary optic neuropathy, and cancer. Complex I is believed to be a major source of reactive oxygen species in mitochondria causing mitochondrial DNA damage and may be one of the causes of aging (4–6). The bacterial NADH-quinone oxidoreductase enzyme (NDH-1) is simplified version of complex I, which is composed of 13–15 subunits, all of which have homologs in the mitochondrial enzyme (7, 8). We have shown in a series of publications that the Escherichia coli NDH-1 is a very useful model system to elucidate the structure and function of complex I due to its structural simplicity and ease of gene manipulation (7, 9–16). Complex I/NDH-1 has a characteristic L-shaped structure with two distinct domains as follows: a hydrophilic peripheral arm projected into the mitochondrial matrix (or bacterial cytoplasm) and a transmembrane hydrophobic arm (17). Recently, the crystal structures of the peripheral and the hydrophobic domains separately, as well as that of the entire complex, have been determined. The revealed structures suggest unprecedented mechanisms for electron transfer and proton translocation (18–22).
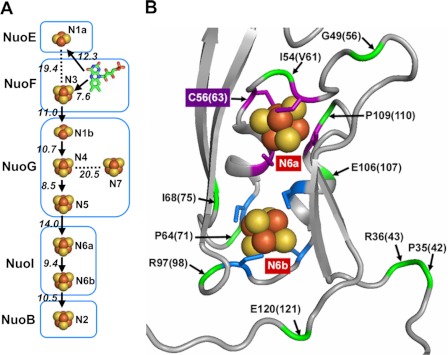
The high resolution three-dimensional structure of the peripheral domain of Thermus thermophilus HB-8 NDH-1 established that the peripheral domain carries one noncovalently bound flavin mononucleotide (FMN) and nine iron-sulfur (Fe/S) clusters (traditional terminology, N1a, N1b, N2, N3, N4, N5, N6a, N6b, and N7) as cofactors (see Fig. 1A) (20, 23). Out of the nine Fe/S centers in NDH-1, N1a and N7 are not directly involved in the electron transfer pathway (see Fig. 1A).
FIGURE 1.
Arrangement of Fe/S clusters in E. coli NDH-1. A, schematic drawing of all Fe/S centers and cofactor FMN displaying their locations in the subunits. Edge to edge distances are given. Postulated flow of electrons through the main pathway is indicated with arrows. B, N6a and N6b clusters and their surroundings in the Nqo9 subunit (E. coli NuoI homolog) from hydrophilic domain of T. thermophilus complex I (Protein Data Bank code 3IAS). The coordinating cysteine residues for the clusters N6a and N6b are highlighted in purple and blue, respectively. The locations of some of the selected residues used for mutations in the vicinity of the two clusters are shown in green and marked by an arrow with their T. thermophilus numbering. In addition, E. coli numbering is displayed in the parentheses. It should be noted that Ile-54 in T. thermophilus is replaced by Val-61 in E. coli.
Electron paramagnetic resonance (EPR) spectroscopy has been one of the most successful and informative methods to study the properties of Fe/S clusters of complex I (24–26). However, not all of them are detectable by EPR spectroscopy. For example, an Fe/S cluster is not paramagnetic under chemical or electronic conditions (25). In addition, when the Fe/S cluster has very fast spin relaxation, extensive broadening of the EPR absorption is observed. Such an effect leads to undetectable EPR signals. Furthermore, considerable overlap of signals exists in the observed EPR spectra for NDH-1/complex I, particularly for [4Fe-4S] clusters (24–27). This makes the actual assignment of observed g values to the individual clusters and their respective subunits an extremely challenging and debatable issue (25–28). The spectra of E. coli NDH-1 has at least six EPR-detectable Fe/S clusters described (N1a, N1b, N2, N3, N4, and N7) (24, 29–36). However, a segment of the electron transfer pathway within subunit NuoI harboring centers N6a or N6b escaped detailed study. Additionally, it had to be clarified whether these two clusters are EPR-detectable. Therefore, extensive studies on the segment of the electron transfer pathway within subunit NuoI harboring centers N6a and N6b is indispensable for the advancement of our knowledge of the entire electron transfer.
The primary sequence of E. coli NuoI contains two sets of conserved cysteine motifs markedly similar to the typical CXXCXXC motif found in the 2× [4Fe-4S] ferredoxin (20, 23). In this study, using an elaborate mutation approach combined with the available three-dimensional structure of the peripheral domain of NDH-1, we were able to analyze features of cluster N6a. We show that cluster N6a is EPR-detectable and exhibits the EPR signals (gz,y,x = 2.09, 1.88, and 1.88). We further report that clusters N6a and N6b display no spin-spin interaction even though these two clusters are located close to each other (Fig. 1B). On the basis of these results, characteristics of clusters N6a and N6b in the electron transfer have been discussed.
Furthermore, it was hypothesized that the electron tunneling pathway in NDH-1 primarily consist of two cysteine ligands and one additional key residue (37). In the case of NuoI, the conserved Ile-100 and Ile-94 are predicted to play the role. However, the mutations of I100G/I100A or I94G/I94A did not significantly affect the electron transfer activities of NDH-1, indicating that these two residues are not indispensable. The contribution of NuoI to the electron transfer mechanism has been discussed.
EXPERIMENTAL PROCEDURES
Materials
The pCRScript cloning kit was from Stratagene (Cedar Creek, TX). The kits for PCR product purification, gel extraction, and plasmid preparation were obtained from Qiagen (Valencia, CA). The QuikChange® II XL site-directed mutagenesis kit and the Herculase®-enhanced DNA polymerase were from Stratagene (Cedar Creek, TX). The endonucleases were from New England Biolabs (Beverly, MA). The pKO3 vector was a generous gift from Dr. George M. Church (Harvard Medical School, Boston). Bis-(3-propyl-5-oxoisoxazol-4-yl) pentamethine oxonol (oxonol VI) and 9-amino-6-chloro-2-methoxyacridine (ACMA) were obtained from Molecular Probes (Eugene, OR). Capsaicin-40 was a generous gift provided by Dr. Hideto Miyoshi (Kyoto University, Kyoto, Japan). The BCA protein assay kit and the SuperSignal West Pico chemiluminescent substrate were from Pierce. p-Nitro blue tetrazolium was from EMD Biosciences (La Jolla, CA). n-Dodecyl β-d-maltoside (DDM) was from Biosynth International Inc. All other chemicals, including dNADH, antibiotics, sodium dithionite, etc., were from Sigma. Q-Sepharose, DEAE-Sepharose, and Superdex columns and goat anti-rabbit IgG horseradish peroxidase conjugate were from GE Healthcare. Oligonucleotides were synthesized by ValueGene (San Diego).
Preparation of NuoI Knock-out and Mutagenesis of the nuoI Gene in the E. coli Chromosome
The strategies used for generating the NuoI knock-out (NuoI-KO) mutant and mutagenesis of the E. coli nuoI gene were in principle similar to those we reported previously for the nuoA, nuoH, nuoM, and nuoCD genes (9, 12–15). The NuoI knock-out (NuoI-KO) mutant was generated by employing the pKO3 system according to the method described by Link et al. (38) and Sinha et al. (13) with minor modifications. In brief, an E. coli NuoI-KO was constructed by replacement of the nuoI gene in the NDH-1 operon by spectinomycin (spc) using the pKO3 system. In parallel, a DNA fragment, which includes the nuoI gene, its upstream 1-kb DNA segment, and its downstream 1-kb DNA segment, was amplified from E. coli DH5α by PCR. The amplified DNA fragment containing the nuoI gene was then cloned into the pCRScript vector system, generating pCRScript/nuoI. pCRScript/nuoI was used as a template to obtain the site-specific nuoI mutants. To evaluate possible effects of the entire process of gene manipulation in generation of point mutations on the E. coli cells, we also constructed a control vector pCRScript/(nuoI-Ctrl), using the same procedure except that the wild type nuoI gene, without any mutation with flanking upstream 1-kb DNA segment and its downstream 1-kb DNA segment, was subcloned into the pCRScript vector. The nuoI mutants and control were subcloned into the pKO3 vector generating the pKO3/(nuoI mutants) and pKO3/(nuoI-Ctrl). The pKO3/(nuoI mutants) and pKO3/(nuoI-Ctrl) plasmids were used to replace the spc gene in E. coli nuoI-KO by recombination. The correct introduction of point mutations in the chromosome was finally confirmed by direct DNA sequencing. A list of all the oligonucleotides used for the mutagenesis of E. coli nuoI gene are given in supplemental Table 1.
Bacterial Growth and Membrane Preparation
The E. coli wild type and NuoI mutants were grown, and membrane preparations were done according to the method described previously in our laboratory (10–13). In brief, cells were first grown in 3 ml of LB medium from freshly streaked plates at 37 °C for 10 h. 500 μl of the above preculture was used to inoculate 250 ml of terrific broth (TB), and cells were cultivated at 37 °C with shaking until the A600 was ∼2. The cells were then harvested at 5,800 × g and frozen at −80 °C. After thawing, the cell pellet was resuspended at 10% (w/v) in a buffer containing 10 mm Tris-HCl (pH 7.0), 1 mm EDTA, 1 mm dithiothreitol, 1 mm phenylmethylsulfonyl fluoride, and 15% (w/v) glycerol. The cell suspension was sonicated twice (15 s) and then passed twice through the “French pressure cell press” at 25,000 p.s.i. After cell debris was removed by centrifugation in a Sorvall SS34 rotor at 23,400 × g for 10 min, and the supernatant was further ultracentrifuged at 256,600 × g for 40 min in a Beckman Spinco 60Ti rotor. The collected pellet was resuspended in the same buffer, and the resulting membrane suspension was either used immediately for enzyme assays or stored in small aliquots at −80 °C for later use.
Purification of E. coli NDH-1
The purification process of NDH-1 was done essentially by following the protocol of Sazanov et al. (39) with a few minor modifications in three separate chromatography steps (Q-Sepharose, DEAE-Sepharose, and Superdex; in the same order) using an ÄKTA Explorer chromatography system. In brief, E. coli MC4100 WT and NuoI mutants were grown from a 5-ml overnight culture in a 5-liter TB media at 37 °C until late exponential stage (A600 = 2.0). Cells were harvested by centrifugation at 5,800 × g for 12 min and washed with 50 mm MES buffer (pH 6.0), and then the cell pellets were kept frozen at −80 °C until further use. The cell pellets were resuspended in 250 ml of 50 mm MES buffer (pH 6.0) and 1 mm phenylmethylsulfonyl fluoride. The suspension was passed twice through a French press at 15,000 p.s.i. Cell debris was removed by centrifugation at 18,000 × g for 15 min followed by 23,400 × g for 25 min. The supernatant was ultracentrifuged at 256,600 × g for 1 h. The brown membrane pellet was resuspended in the same buffer as described above, frozen in liquid nitrogen, and stored at −80 °C until their use. DDM was added in a dropwise manner (under constant stirring and on ice) to the membrane suspension to a final concentration of 1% and incubated for 1 h. The nonsolubilized material was removed by centrifugation for 1 h at 150,000 × g. The supernatant was passed through a 0.45-micron filter and adjusted to 150 mm NaCl by adding dropwise a solution of 1 m NaCl. The solubilized membranes were applied onto a HiLoad 26/10 Q-Sepharose column (equilibrated with 20 mm MES (pH 6.0), 0.1% DDM, 10% glycerol, 1 mm phenylmethylsulfonyl fluoride (buffer A)), and the protein was eluted with an 18–35% linear gradient of buffer B (buffer A with 1 m NaCl). Active fractions were pooled, diluted with an equal volume of buffer A, concentrated (Amicon-Ultra, molecular mass cutoff 100 kDa; Millipore) to a volume of about 50 ml, and then applied onto a DEAE column equilibrated with buffer A. The protein was eluted with 15–30% linear gradient of buffer B in buffer A. Fractions containing dNADH-K3Fe(CN)6 reductase activity were pooled and concentrated to a volume of about 2 ml. The concentrated sample was loaded onto a HiLoad Superdex 200 16/60 Prep Grade gel filtration column equilibrated with 20 mm MES (pH 6.0), 200 mm NaCl, 2 mm CaCl2, 0.5% DDM, 10% glycerol, 1 mm phenylmethylsulfonyl fluoride, and the protein was eluted at a flow rate of 0.5 ml/min. Fractions of 1.0 ml were collected and analyzed by SDS-PAGE and dNADH-K3Fe(CN)6 reductase activity assay, and those containing pure NDH-1 were pooled, diluted with buffer A to reduce the NaCl concentration to 50 mm, concentrated using Amicon-Ultra concentrators, and stored in liquid nitrogen for later use. The purification process was repeated several times, and all the pooled fractions were concentrated so that the final concentration of pure NDH-1 was about 3–7 mg/ml for the EPR study. A summary of the purification and procedure is given in supplemental Table 2 and supplemental Fig. 1.
EPR Spectroscopy
EPR samples were prepared anaerobically after their purification. Redox mediators, methyl viologen and benzyl viologen, were added at a concentration of 5 μm each. After the addition of 10 mm neutralized sodium dithionite solution, each sample was incubated for 5–10 min, transferred to an EPR tube, and then frozen immediately. EPR spectra were recorded by a Bruker Elexsys E500 spectrometer at X-band (9.4 GHz) using an Oxford Instrument ESR900 helium flow cryostat.
Gel Electrophoresis and Immunoblotting Analysis
15 μg of protein from each membrane preparation was first subjected to SDS-PAGE using the discontinuous system of Laemmli (40). The expression of the NDH-1 subunits was then determined by using the antibodies specific to NuoB, NuoCD, NuoE, NuoF, NuoG, NuoI, NuoL, and NuoM in Western blot experiments. Blue native-PAGE was performed according to the method of Schägger and von Jagow (41) with some minor modifications as described previously (10, 11, 13). The assembly of NDH-1 was examined by NADH dehydrogenase activity staining and immunoblotting as described previously (10, 11, 13).
Enzymatic Assays
In addition to NDH-1, E. coli also contains an alternative NADH dehydrogenase (NDH-2). Because NDH-2 cannot catalyze dNADH, dNADH was used as a substrate throughout this work to measure the activities derived solely from NDH-1 (42). All measurements of enzymatic activity were carried out at 30 °C using an SLM DW-2000 spectrophotometer according to the methods described previously (13, 43). In brief, dNADH oxidase activity was assayed at 340 nm with 80 μg of protein/ml of membrane samples in 10 mm potassium phosphate buffer (pH 7.0) containing 1 mm EDTA. The reaction was started by the addition of 0.15 mm dNADH. The dNADH-DB reductase activity measurements were conducted in a similar manner, except that 10 mm KCN and 50 μm DB were also included in the assay mixture. The dNADH-K3Fe(CN)6 reductase activity was measured in the presence of 10 mm KCN, 0.15 mm dNADH, and 1 mm K3Fe(CN)6 at 420 nm in the same buffer. Extinction coefficients of ϵ340 = 6.22 mm−1 cm−1 for dNADH and ϵ420 = 1.00 mm−1 cm−1 for K3Fe(CN)6 were used for activity calculations.
Measurement of Membrane Potential and Proton Pumping Activity
The generation of membrane potential by the NuoI mutants was monitored optically using a reaction mixture containing 0.33 mg/ml E. coli membrane samples in 50 mm MOPS (pH 7.3), 10 mm MgCl2, 50 mm KCl, and 2 μm oxonol VI as described previously (13, 44). The reaction was started by addition of 0.2 mm dNADH, and the absorbance changes at 630 to 603 nm were monitored. The proton ionophore FCCP was added to a final concentration of 2 μm to uncouple the reaction. Proton pumping activity of NDH-1 was estimated from ACMA fluorescence quenching as described by Amarneh and Vik (45). The reaction mixture was the same as that of the membrane potential assay except that the E. coli membrane samples were added at 150 μg of protein/ml, whereas oxonol VI was replaced with 2 μm ACMA. The reaction was initiated by adding 0.2 mm dNADH. SpectraMax M2 fluorescence microplate reader (Molecular Devices Corp.) with an excitation wavelength of 410 nm and an emission wavelength of 480 nm was used to record the fluorescence at room temperature.
Measurements of Production of Superoxide Radicals
Acetylated cytochrome c was used as a reporter for the production of superoxide radicals (46). NuoI membrane samples at 0.24 mg of protein/ml in 10 mm potassium phosphate buffer (pH 7) were mixed with 5 μm acetylated cytochrome c. Reaction was initiated by addition of 0.15 mm dNADH, and the absorbance change was monitored at 550 nm. The rate of reduction of acetylated cytochrome c was calculated using the extinction coefficient of 21 mm−1 cm−1.
Other Analytical Procedures
Protein concentrations were determined by the BCA protein assay kit (Pierce) using bovine serum albumin as the standard according to the manufacturer's instructions. Any variations from the procedures and other details are described in the figure legends.
RESULTS
Sequence Analysis of the NuoI Subunit
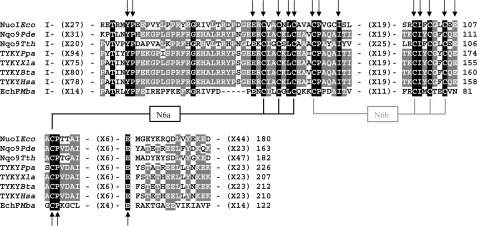
Fig. 2 shows the partial alignment of the deduced amino acid sequences among the E. coli NuoI subunit and its homologs from bacteria to mammals. Subunit NuoI is known to contain two sets of conserved cysteine motifs that are remarkably similar to the typical CXXCXXC motif found in the 2× [4Fe-4S] ferredoxin. As seen in Fig. 2, these cysteine motifs are perfectly conserved among the E. coli NuoI subunit and its homologs. In the crystal structure of the peripheral domain of T. thermophilus NDH-1, cluster N6a is coordinated by Cys-53, Cys-56, Cys-59, and Cys-108 (corresponding to E. coli Cys-60, Cys-63, Cys-66, and Cys-109, respectively). The cluster N6b is coordinated by Cys-63, Cys-98, Cys-101, and Cys-104 (E. coli sequence numbers are Cys-70, Cys-99, Cys-102, and Cys-105, respectively) (23). Therefore, we generated E. coli NDH-1 mutant strains of all the cysteine residues ligating clusters N6a and N6b by replacing them either with alanine, serine, or histidine. Furthermore, we found many residues present in the vicinity of the clusters N6a and N6b that were highly conserved not only in NDH-1/complex I, but also in the EchF subunit of multisubunit membrane-bound energy-converting [Ni-Fe] Ech hydrogenase (Fig. 2 and see also supplemental Fig. 2). The oxidation/reduction of clusters N6a and N6b has been suggested to be dependent on the charge compensation of nearby acidic residue (47). We found two such acidic residues Glu-107 and Glu-121 in NuoI that were highly conserved. Thus, we have made point mutants of highly conserved residues around clusters N6a and N6b, including Pro-71, Ile-94, Ile-100, Glu-107, Pro-110, and Glu-121 (see also Fig. 1B). In this study, a total of 43 point mutations covering 34 different residues in the NuoI subunit of E. coli NDH-1 were generated (Table 1).
FIGURE 2.
Partial alignment of the deduced amino acid sequences among the E. coli NuoI subunit and its homologs from various organisms. The alignment was carried out by the ClustalW program (61). The coordinating cysteine residues for the two tetranuclear clusters N6a and N6b are highlighted. Black boxes with white letters show perfectly conserved residues among the listed organisms. Dark gray boxes with white letters illustrate conserved residues among at least five listed organisms. Amino acids mutated in this study are marked by arrows (also see supplemental Fig. 2). Sequence sources and their Swiss-Prot accession numbers are as follows: NuoIEco, E. coli K-12 NuoI subunit (P0AFD6); Nqo9Pde, Paracoccus denitrificans Nqo9 subunit (A1B486); Nqo9Tth, T. thermophilus HB-8 Nqo9 (Q72GD6); TYKYPpa, Pichia pastoris TYKY subunit (A3LRW9); TYKYXla, Xenopus laevis TYKY subunit (Q66KT2); TYKYBta, Bos taurus TYKY subunit (P42028); TYKYHsa, Homo sapiens TYKY subunit (O00217); EchFMba, M. barkeri EchF subunit (O59657).
TABLE 1.
Enzyme activities of the membrane-bound NDH-1 of E. coli wild-type and various NuoI mutants
| Sample | dNADH-O2a | dNADH-DBa | dNADH-K3Fe(CN)6b |
|---|---|---|---|
| WT | 766 ± 35 (100%) | 786 ± 35 (100%) | 1815 ± 38 (100%) |
| NuoIKO | 16 ± 5 (2%) | 19 ± 5 (2%) | 301 ± 35 (17%) |
| KO-rev | 740 ± 76 (97%) | 1003 ± 59 (128%) | 1920 ± 72 (106%) |
| C60A | 10 ± 8 (1%) | 19 ± 1 (2%) | 312 ± 12 (17%) |
| C60S | 39 ± 4 (5%) | 57 ± 5 (7%) | 561 ± 52 (31%) |
| C60H | 12 ± 3 (2%) | NDc | 365 ± 26 (20%) |
| C63A | 18 ± 2 (2%) | 17 ± 4 (2%) | 409 ± 17 (23%) |
| C63S | 153 ± 32 (20%) | 160 ± 15 (20%) | 905 ± 67 (50%) |
| C66S | 13 ± 2 (2%) | 16 ± 3 (2%) | 323 ± 11 (18%) |
| C66H | 15 ± 1 (2%) | 7 ± 2 (1%) | 352 ± 16 (19%) |
| C70S | 28 ± 4 (4%) | 31 ± 1 (4%) | 343 ± 19 (19%) |
| C70H | 4 ± 1 (1%) | 12 ± 1 (2%) | 265 ± 8 (15%) |
| C99S | 15 ± 2 (2%) | 18 ± 1 (2%) | 308 ± 4 (17%) |
| C99H | 8 ± 1 (1%) | ND | 268 ± 9 (15%) |
| C102S | 14 ± 1 (2%) | 38 ± 2 (5%) | 273 ± 9 (15%) |
| C102H | 9 ± 1 (1%) | 15 ± 1 (2%) | 307 ± 27 (17%) |
| C105S | 11 ± 3 (1%) | 12 ± 1 (2%) | 312 ± 14 (17%) |
| C109S | 16 ± 1 (2%) | 29 ± 8 (4%) | 380 ± 48 (21%) |
| T30A | 470 ± 50 (61%) | 453 ± 43 (58%) | 1169 ± 27 (64%) |
| P34A | 499 ± 20 (65%) | 450 ± 16 (57%) | 1167 ± 79 (64%) |
| P42A | 468 ± 27 (61%) | 510 ± 30 (65%) | 1964 ± 78 (108%) |
| R43A | 350 ± 1 (46%) | 418 ± 13 (53%) | 1520 ± 18 (84%) |
| Y44A | 920 ± 3 (120%) | ND | 2520 ± 12 (139%) |
| G46A | 598 ± 6 (78%) | ND | 1437 ± 33 (79%) |
| R52A | 571 ± 21 (75%) | 504 ± 20 (64%) | 1532 ± 33 (84%) |
| P54A | 563 ± 3 (73%) | 623 ± 42 (79%) | 1717 ± 20 (95%) |
| G56A | 362 ± 11 (47%) | 424 ± 18 (54%) | 1701 ± 46 (94%) |
| E58A | 651 ± 6 (85%) | 503 ± 19 (64%) | 1789 ± 8 (99%) |
| V61A | 238 ± 27 (31%) | 257 ± 18 (33%) | 1225 ± 31 (67%) |
| L65A | 511 ± 49 (67%) | 568 ± 97 (72%) | 1345 ± 17 (74%) |
| P71A | 347 ± 73 (45%) | 437 ± 42 (56%) | 1364 ± 20 (75%) |
| I75A | 218 ± 58 (28%) | 318 ± 83 (40%) | 1191 ± 41 (66%) |
| G85A | 885 ± 57 (116%) | 688 ± 42 (88%) | 1929 ± 23 (106%) |
| F92A | 620 ± 42 (81%) | 599 ± 58 (76%) | 1436 ± 30 (79%) |
| R93A | 777 ± 67 (101%) | 719 ± 55 (92%) | 1837 ± 50 (101%) |
| I94A | 704 ± 3 (92%) | 777 ± 11 (99%) | 1954 ± 26 (108%) |
| I94G | 715 ± 3 (93%) | 935 ± 28 (119%) | 1678 ± 32 (92%) |
| R98A | 151 ± 20 (9%) | 141 ± 18 (18%) | 953 ± 44 (53%) |
| I100A | 649 ± 4 (85%) | 676 ± 14 (86%) | 1827 ± 78 (101%) |
| I100G | 471 ± 10 (61%) | 665 ± 8 (85%) | 1565 ± 5 (86%) |
| G103A | 766 ± 4 (100%) | 631 ± 69 (80%) | 1604 ± 36 (88%) |
| E107A | 441 ± 99 (58%) | 556 ± 86 (71%) | 1211 ± 38 (67%) |
| P110A | 71 ± 8 (9%) | 151 ± 8 (19%) | 1421 ± 22 (78%) |
| I114A | 715 ± 31 (107%) | 954 ± 29 (121%) | 1710 ± 60 (94%) |
| E121A | 569 ± 3 (74%) | 671 ± 32 (85%) | 1798 ± 26 (99%) |
| Y132A | 645 ± 32 (84%) | 829 ± 7 (105%) | 2233 ± 21 (123%) |
a Activity is shown in nanomoles of dNADH/mg of protein/min.
b Activity is shown in nanomoles of K3Fe(CN)6/mg of protein/min.
c ND means not determined.
Effects of NuoI Mutation on the Electron Transfer Activities of NDH-1
We measured the electron transfer activities of NDH-1 in membrane vesicles obtained from E. coli MC4100 wild type and various NuoI mutants (Table 1). Because the dNADH-K3Fe(CN)6 reductase assay is derived from the NADH dehydrogenase fragment of NDH-1, it was used as a measure for the amount of the peripheral arm of NDH-1. As shown in Table 1, the activity of the NuoI-KO mutant was only about 17% that of the wild type, indicating the absence of a functionally active peripheral arm when the enzyme was devoid of the NuoI subunit. Loss of activity was completely restored in the revertant mutant, KO-rev. The cysteine mutants in the two conserved motifs exhibited greatly suppressed dNADH-K3Fe(CN)6 reductase activities, varying from 15 to 30% of control. Only the mutant C63S, in contrast, showed a significant (∼50%) dNADH-K3Fe(CN)6 reductase activity. Slightly reduced activity ranging from about 70 to 80% of the wild type was observed in mutants V61A, P71A, I75A, and P110A. The R98A mutant, which is present close to N6b, showed about 50% of the dNADH-K3Fe(CN)6 reductase activity as compared with the wild type. The majority of the other point mutants, including I94A, I94G, I100A, I100G, and E121A exhibited dNADH-K3Fe(CN)6 reductase activity more or less similar to the wild type.
We then measured the dNADH oxidase and dNADH-DB reductase activities. As expected, the energy-transducing NDH-1 activities were completely abolished for the cysteine mutants, except for the C63S mutant that showed small but significant activities (about 20%) compared with the wild type. Mutation of the well conserved V61A, I75A, R98A, and P110A showed significantly reduced energy-transducing NDH-1 activity ranging from 10 to 40% of control. Less drastic but still significant diminution of the energy-transducing activity was found in mutants E107A, P71A, P42A, R43A, G56A, and L65A (about 50–70%). The dNADH oxidase and dNADH-DB reductase activities of the other NuoI mutants, including I94A, I94G, and I100A, were found to be mostly comparable with that of wild type. The IC50 values for capsaicin-40 of most of the NuoI mutants were determined to be in the range of 0.1–0.3 μm (wild type; 0.16 μm).
Subunit Contents and Assembly of NDH-1 of NuoI Mutants
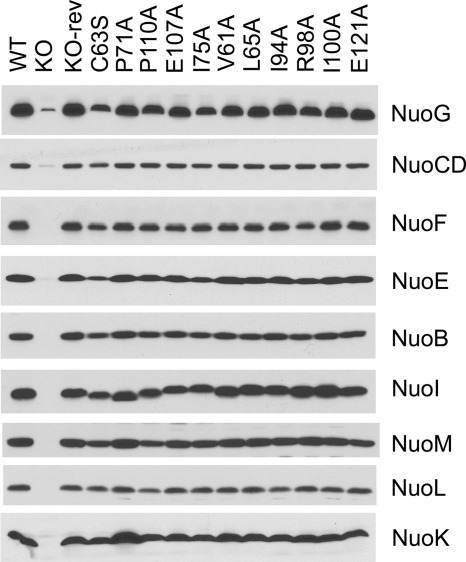
We examined the subunit contents of NDH-1 in the cytoplasmic membranes from NuoI mutants by immunochemical analyses. Peripheral subunits NuoB, NuoCD, NuoE, and NuoF and membrane subunits NuoK, NuoL, and NuoM were almost entirely missing in the membranes of NuoI-KO cells. This confirms that subunit NuoI is indeed required for the efficient assembly of the NDH-1. Cysteine mutants, except for C63S, in the two conserved motifs showed either a very low level or no peripheral subunits (data not shown). The results indicate that clusters N6a and N6b are involved in not only catalytic activity (electron transfer) but also structural stability of E. coli NDH-1. It is conceivable that mutations that affect the structural stability of the two clusters may impact the stability of the NuoI subunit, which in turn affects the stability of the entire complex. Thus, structural stability of the NADH dehydrogenase may be impacted due to the absence of the NuoI subunit. A similar observation was reported for NuoI mutants of Rhodobacter capsulatus (48) and EchF mutants of Methanosarcina barkeri Ech (47). The C63S mutant, which retained about half of the dNADH-K3Fe(CN)6 reductase activity, showed slightly reduced levels of the peripheral subunits NuoG, NuoE, and NuoB (Fig. 3). Subunit contents in point mutants present in the vicinity of the two clusters, including P71A, P110A, and E107A, were found to be almost comparable with the wild type (Fig. 3).
FIGURE 3.
Immunoblotting of membrane preparations from wild type (WT), NuoI knock-out (KO), NuoI knock-out revertant (KO-rev), and site-specific NuoI mutants. E. coli membranes (15 μg of protein per lane) were loaded onto a 15% Laemmli SDS-polyacrylamide gel. After electrophoresis, the proteins were transferred onto nitrocellulose membranes, and Western blotting was carried out using the SuperSignal West Pico system. Antibodies specific to NuoB, NuoCD, NuoE, NuoF, NuoG, NuoI, NuoK, NuoL, and NuoM were used. Goat anti-rabbit IgG horseradish peroxidase conjugate was used as secondary antibody.
We further verified the NDH-1 assembly of NuoI mutants by BN-PAGE with NADH dehydrogenase activity staining and the Western blotting using the NuoB antibody (supplemental Fig. 3). Similar to the results of their subunit content, no assembled NDH-1 was observed in the NuoI-KO mutant and all the cysteine ligand mutants except for C63S. The mutant C63S showed a partial decrease in the amount of assembled NDH-1. The majority of the point mutants, including P71A, I75A, P110A, and E107A, showed either slightly lower or relatively similar amounts of assembled NDH-1 compared with the wild type and KO-rev mutant (supplemental Fig. 3). The R98A mutant lacked proper assembly. This is in contrast to the seemingly normal NDH-1 subunit contents (see Fig. 3) that was seen for this mutant. The R98A mutation thus seems to destabilize the architect of the NADH dehydrogenase complex by the detergent treatment.
Measurements of Proton Pump and Electrochemical Potential of NuoI Mutants
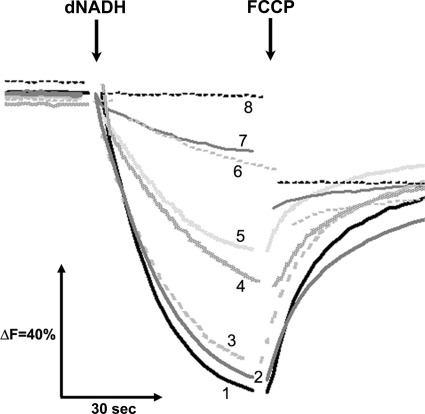
Proton pumping activities in various NuoI mutants were paralleled to their dNADH oxidase activities and the amount of assembled NDH-1. As seen in Fig. 4, membrane vesicles from the wild type showed a maximum proton pump activity, although no or negligible pumping activities were found in the NuoI knock-out and seven cysteine mutants. As expected, the C63S mutant showed small but still significant proton pump activity. A considerable reduction in the pumping ability was observed for mutants P71A, I75A, E107A, and P110A.
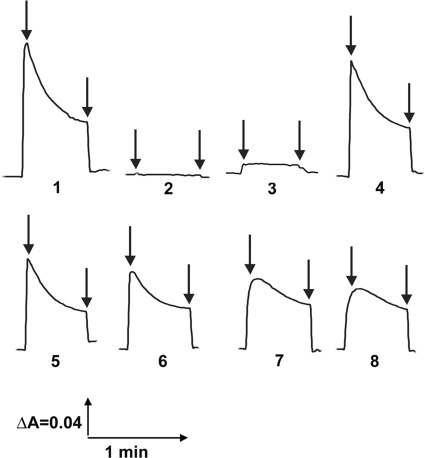
FIGURE 4.
Generation of a pH gradient coupled to dNADH oxidation in E. coli NuoI mutants. Proton translocation was measured by the quenching of the fluorescence of ACMA at room temperature with an excitation wavelength of 410 nm and an emission wavelength of 480 nm. At the time indicated by arrows, 0.2 mm dNADH or 10 μm FCCP, respectively, was added to the assay mixture containing 50 mm MOPS (pH 7.3), 10 mm MgCl2, 50 mm KCl, 2 μm ACMA, and E. coli membrane samples (30 μg of protein/ml). Representative traces from different group of mutants are as follows: 1, WT (or KO-rev, G85A, Y44A, R93A, I94A, G103A, I114A, and Y132A); 2, E107A (or G46A, R52A, P54A, E58A, L65A, F92A, and E121A); 3, I75A (or T30A, P34A, P42A, R52A, and L65A); 4, V61A (or R43A, G56A, and P71A); 5, C63S (or R98A); 6, P110A; 7, C60S (or C70S, C102S, and C109S); and 8, C99H (or KO, C60A, C60H, C63A, C66S, C66H, C70H, C99S, C102H, and C105S)
Similar results were obtained in the membrane potential (ΔΨ) analysis with the membrane vesicles of the NuoI mutants using oxonol VI. Membrane vesicles of the wild type showed a maximum ΔΨ after the addition of dNADH, and FCCP totally abolished the ΔΨ. No ΔΨ was generated in the case of the NuoI-KO mutant. The representative traces from a few key mutants are shown in Fig. 5.
FIGURE 5.
Detection of the membrane potential generated by dNADH oxidation in E. coli NuoI mutants. The membrane potential changes were observed by the absorbance changes of oxonol VI at 630 to 603 nm at 37 °C. The assay mixture typically contained 50 mm MOPS (pH 7.3), 10 mm MgCl2, 50 mm KCl, 2 μm oxonol VI, and E. coli membrane samples (330 μg of protein/ml). The 1st arrow indicates addition of 0.2 mm dNADH, and the 2nd arrow indicates addition of 2 μm FCCP to the assay mixture. Representative traces from different group of mutants are as follows: 1, WT (or KO-rev, G85A, Y44A, R93A, I94A, G103A, I114A, and Y132A); 2, KO (or C60A, C60H, C63A, C66S, C66H, C70H, C99S, C99H, C102H, and C105S); 3, C60S (or C70S, C102S, and C109S); 4, E121A (or G46A, R52A, P54A, E58A, L65A, and F92A); 5, R43A (or G56A and P71A); 6, E107A (or T30A, P34A, P42A, R52A, and L65A); 7, I75A (or V61A); and 8, P110A (or R98A and C63S).
Measurements of Reactive Oxygen Species Generation by NuoI Mutants
The production of superoxide radicals by the NuoI mutant membranes is shown in supplemental Table 3. The V61A, I75A, and P110A mutants produced significant amounts of superoxide radicals induced by dNADH. Furthermore, all the cysteine ligand mutants of NuoI, including C63S, generated considerably high amounts of superoxide radicals. The generation of superoxide radicals was slightly lower for the L65A, P71A, R98A, and E107A mutants; however, it was still much higher than the WT. In general, lesser amount of superoxide radicals were detected in those NuoI mutants that showed NDH-1 activities similar to WT (for example, G85A, I94G, and I100G in supplemental Table 3). Also, addition of capsaicin-40 stimulated the production of superoxide radicals. Thus, the NuoI mutants that showed greater inhibition in the NDH-1 activities also showed an increased production of superoxide radicals. This is in line with the high production of superoxide radicals observed in mammalian mitochondria resulting from complex I inhibition (49, 50)
EPR Analysis of Selected NuoI Mutants in the Vicinity of the Two Clusters
To characterize the properties of bound iron-sulfur clusters, EPR studies are indispensable. However, the physiological expression levels of NDH-1 in E. coli membranes are extremely low. Thus, we decided to purify NDH-1 from NuoI mutants. Because the yield depended on the stability of the assembled NDH-1, we performed EPR analysis only for the following selected NuoI mutants: C63S (ligand for the cluster N6a), P110A (present in the vicinity of cluster N6a), P71A (in the vicinity of cluster N6b, and E107A (relatively distant from both N6a and N6b clusters).
EPR Spectra of the [2Fe-2S] Clusters in Selected NuoI Mutants
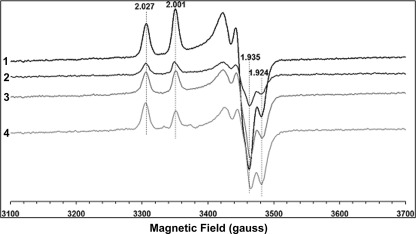
EPR spectra of the binuclear clusters were examined in the dithionite-reduced samples at 40 K and 5 milliwatts (Fig. 6). Two sets of EPR signals, characteristic of N1a (rhombic symmetry (gz,y,x = 2.00, 1.94, and 1.92)) and of N1b (axial symmetry (gz,y,x = 2.02, 1.94, and 1.94)) were observed in the wild type sample (Fig. 6). The EPR line shapes of these clusters were similar to those reported previously for the purified E. coli complex I (29, 51). EPR signals of both binuclear clusters were clearly detected also in NDH-1 from NuoI mutants C63S, P110A, and P71A (Fig. 6). Although signal intensities of these two clusters were varied among the mutants to some extent, the characteristics of [2Fe-2S] clusters were not affected by these NuoI mutations in a significant manner.
FIGURE 6.
EPR spectra of the purified E. coli complex I from wild type and various NuoI mutants at 40K. . Representative traces from different group of mutants are as follows: WT (1), C63S (2), P71A (3), and P110A (4). The samples were reduced with 10 mm sodium dithionite in the presence of 5 μm of benzyl viologen and methyl viologen. EPR spectra were recorded under the following conditions: microwave frequency, 9.45 GHz; microwave power, 5 milliwatts; modulation amplitude, 8.034 G; modulation frequency, 100 kHz; time constant, 82 ms. The gz and gx values are indicated.
EPR Spectra of the [4Fe-4S] Clusters in Selected NuoI Mutants
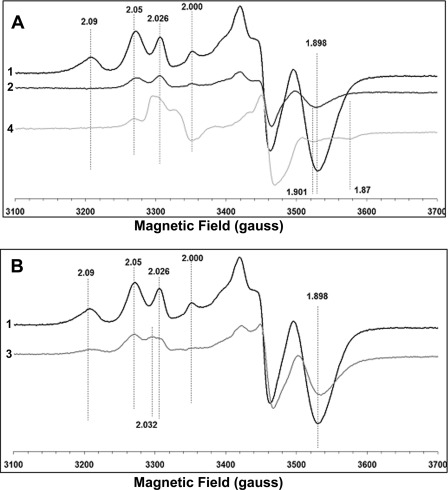
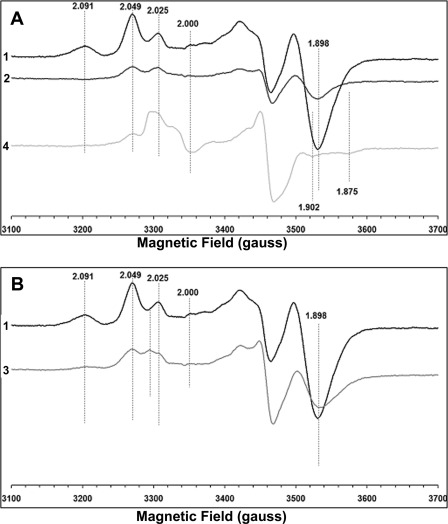
We measured EPR signals arising from [4Fe-4S]+ clusters at two different temperatures 12 K (Fig. 7) and 7.5 K (Fig. 8). As shown previously by other groups (26, 51), the EPR spectra of the wild type showed a substantial overlap of signals from different tetranuclear clusters with distinct g values (Figs. 7A and 8A). The spectra of the wild type clearly showed the EPR signal at g = 2.091, 2.049, and 1.898 at 12 and 7.5 K. However, the EPR spectra of the C63S mutant showed complete absence of the EPR signal at g = 2.091 at both 12 K (Fig. 7A) and 7.5 K (Fig. 8A). A considerable reduction in the signal intensity at g = 1.898 was also observed. The EPR spectra of the P110A mutant also showed a complete loss of the g = 2.091 signal and a significant signal reduction at g = 2.05 and g = 1.898. Cys-63 is a ligand for cluster N6a, and Pro-110 is located in the vicinity of cluster N6a (about 6.7 Å) (Fig. 1B). However, in the EPR spectra of the P71A mutant that is present in the close vicinity (about 3.5 Å) of cluster N6b (Fig. 1B), the g = 2.091 EPR signal clearly was detected, but a significant reduction of the g = 1.898 was observed (Figs. 7B and 8B). The signal intensity of the g = 2.05 was almost comparable with that of the wild type (Fig. 7B). Therefore, our data strongly suggest that the EPR signal at g = 2.091 arises from cluster N6a. In fact, the g = 2.091 signal was also detected in the E107A mutant (supplemental Fig. 4). The polar residue Glu-107 is not present in close vicinity but rather is distant from both N6a and N6b. Interestingly, the reduction in the g = 2.05 and 1.898 signal was also observed in the EPR spectra of this E107A mutant, compared with that of the wild type.
FIGURE 7.
EPR spectra of the purified E. coli complex I from wild type and various NuoI mutants at 12 K. A shows the comparison of EPR spectra of WT (1), C63S (2) and P110A (4), and B shows the EPR spectra from WT (1) and P71A (3). The samples were reduced with 10 mm sodium dithionite in the presence of 5 μm of benzyl viologen and methyl viologen. The EPR conditions used were the same as those in Fig. 6. The gz and gx values are indicated.
FIGURE 8.
EPR spectra of the purified E. coli complex I from wild type and various NuoI mutants at 7.5 K. A shows the comparison of EPR spectra of WT (1), C63S (2), and P110A (4), and B shows the EPR spectra from WT (1) and P71A (3). The EPR conditions used were the same as those in Fig. 6. The gz and gx values are indicated.
DISCUSSION
A thorough understanding of the properties of the individual Fe/S clusters (reduction potentials) and of the clusters as a group is a prerequisite to clearly outline the energy profile for electron transfer through complex I, which in turn is essential in understanding the catalytic mechanism of the enzyme (28, 52). Until recently, we had limited information about clusters N6a and N6b as these two cubane clusters were believed to be EPR-silent (53). In an attempt to clarify the electron pathway around these clusters, we adopted a strategy that involves dissection of the electron transfer pathway at different stages by systematically modifying the residues in the NuoI subunit using site-directed mutagenesis. Point mutations of highly conserved residues in the vicinity of clusters N6a (P110A) and N6b (P71A) and N6a ligand Cys (C63S) provided extremely valuable information. First of all, biochemical data, including the activity and assembly of NDH-1 together with EPR data of the mutants, unequivocally affirmed that clusters N6a and N6b are essential for electron transfer of NDH-1. Second, and more importantly, these three mutants helped in the assignment of the tetranuclear clusters N6a and N6b. EPR data at 12 and 7.5 K showed that the g = 2.091 signal disappeared in the spectra of mutants C63S and P110A; however, it remained clearly visible in the P71A mutant. The data suggest that the g = 2.091 signal belongs to cluster N6a. Because all three mutants retained significant NDH-1 activities, it seems unlikely that the Fe/S cluster (g = 2.091) is lost in mutants C63S and P110A. It is conceivable that these mutants drastically lower the redox potential of the g = 2.091 signal.
One thing that needs clarification is possible contribution of cluster N4 in the EPR signals at g = 2.091. In complex I purified from bovine heart, signals exhibiting different line widths and/or different temperature dependence arising from two Fe/S clusters were detected around gz = 2.11. One EPR signal was observed between 6 and 15 K (27). The second EPR signal at gz = 2.11 became detectable at around 3 K, whose spin relaxation seems to resemble that of cluster N5 (gz = 2.06). In the E. coli subcomplex preparation, NuoCD EFG that lacks NuoB (cluster N2) and Nuo I (clusters N6a and N6b), the EPR signals (gz,x = 2.09, 1.88) were not detected when the temperature was 6 K. However, the gx = 1.88 signal and the gx = 1.90 signal (from cluster N5) were observed at 3 K. From the data, it seemed as if the EPR signals with gz,x = 2.07, 1.88 arose from cluster N4. However, because the Em values of cluster N4 go down easily (it is difficult for the N4 cluster to retain the reduced state) and its line width becomes broader (25), the cluster N4 signal is most likely undetectable under our present EPR measurement conditions. Therefore, contribution of N4 in the EPR signals (gz,x = 2.09, 1.88), if any, would be negligible in our case.
Clusters N4 and N5 (8.5 Å, the shortest distance) are considered to exhibit spin-spin interaction. Clusters N6a and N6b are well within the distance that makes spin-spin interaction possible (9.4 Å, the 2nd shortest distance). However, our results suggest no spin-spin interaction between N6a and N6b. That is because the mutation P71A barely affected the g values of cluster N6a. Similar observations have been reported for EchF (NuoI homolog) (47). Lack of spin-spin interaction between N6a and N6b allows us to determine intrinsic EPR signals of N6b. Thus, in EPR signals gz,x = 2.05, ∼1.90 seem to belong to cluster N6b. Information about assignments of EPR signals to N6a and N6b is useful for discussing the mechanism of electron transfer of NDH-1/complex I as described below.
According to the current consensus for electron transfer pathway proposed by different groups of researchers, the electron transfer starts from the NADH-binding site and FMN, continuing through the “wire” of seven FeS clusters, and terminating with cluster N2 (20, 25, 28). It was assumed that the one-electron potential of cluster N1a is approximately −500 mV and that of cluster N2 is approximately −150 mV, although all the other clusters between FMN and N2 are isopotential (approximately −250 mV) (20, 54). In contrast, an alternating profile of electron transfer chain for complex I has been speculated (54). This study aligns well with the classical (isopotential) pathway as EPR signals for N6a, and possibly the N6b clusters were successfully detected for the first time. This also agrees with the results published previously (34, 53). As far as our present knowledge is concerned, the isopotential electron transfer chain seems to be a unique feature of complex I/NDH-1. It is of interest to investigate how this isopotential electron transfer chain drive the proton pumping machinery of NDH-1/complex I.
The time-dependent excess electron density along the complex I/NDH-1 charge transfer chain was computed by Dutton and co-workers (55). It turned out that the cluster N1b or N4 knock-out mutants are functional (56). To correct these shortcomings, a hopping transfer mechanism involving aromatic amino acids has been proposed to explain the high overall transport rate (56). However, in that study, the internal water has not been taken into account, although it is considered that the intersubunit water is capable of accelerating electron transfer (57–59). Recently, electron tunneling for NDH-1/complex I was proposed wherein specific peptide residues in addition to inter-subunit water were hypothesized to serve as electron wires concerning Fe/S clusters. Individual electron tunneling paths were speculated to involve up to three residues, including two cysteine ligands and one additional key residue (37, 60). A role of Ile-99 of the subunit Nqo9 of T. thermophilus NDH-1 (Ile100 in E. coli) was predicted for electron tunneling between N6b and N2. The same authors proposed that when Fe (N6b) coordinated by Cys-63 mainly accepts the electrons from N6a, the primary pathway is through Cys-59 and Cys-63 with 2.3 Å through-space jump with a significant tunneling flux along Ile-93 of subunit Nqo9 of T. thermophilus NDH-1 (Ile-94 in E. coli) (37). Mutations of these two highly conserved Ile residues to Ala/Gly in the NuoI subunit (I94A/I94G and I100A/I100G) did not bring about any significant changes in the electron transfer or proton pumping activities of NDH-1. These results showed that neither Ile-94 nor Ile-100 is essential, suggesting that the electron tunneling model of NDH-1 does not fit the real electron transfer mechanism. However, one should consider the following points. (i) The three-dimensional structure is available for T. thermophilus but not E. coli. (ii) Mutation of Ile with Ala/Gly can cause conformational changes of NuoI. (iii) The space resulting from mutation is occupied by water, which might construct new electron wire(s), to compensate electron transfer similar to Ile. Therefore, data from various mutations are required to evaluate prediction of theoretical electron tunneling of NDH-1/complex I, and this is currently under investigation in our laboratory. Together, this work has provided a missing piece to the otherwise unresolved section of the entire electron transfer of NDH-1/complex I.
Supplementary Material
Acknowledgments
We thank Drs. Mathieu Marella and Gaurav Patki for discussions, Dr. Jennifer Barber-Singh for critical reading of the manuscript, and Dr. Ronak Y. Patel (Washington University, St. Louis) for help with homology modeling.
This work was supported, in whole or in part, by National Institutes of Health Grants R01GM033712 (to T. Y. and A. M.-Y.) and 3R01GM030736-25S1 (to T. O.). This work was also supported by American Heart Association Grant IISDG5560001 (to E. N-O.).

This article contains supplemental Figs. 1–4 and Tables 1–3.
- complex I
- mitochondrial proton-translocating NADH-quinone oxidoreductase
- DB
- 2,3-dimethoxy-5-methyl-6-decyl-1,4-benzoquinone
- dNADH
- reduced nicotinamide hypoxanthine dinucleotide
- Fe/S
- iron-sulfur
- oxonol VI
- bis-(3-propyl-5-oxoisoxazol-4-yl)pentamethine oxonol
- ACMA
- 9-amino-6-chloro-2-methoxyacridine
- FCCP
- carbonyl cyanide-p-trifluoromethoxy-phenylhydrazone
- DDM
- n-dodecyl β-d-maltoside.
REFERENCES
- 1. Carroll J., Fearnley I. M., Shannon R. J., Hirst J., Walker J. E. (2003) Analysis of the subunit composition of complex I from bovine heart mitochondria. Mol. Cell. Proteomics 2, 117–126 [DOI] [PubMed] [Google Scholar]
- 2. Carroll J., Fearnley I. M., Skehel J. M., Shannon R. J., Hirst J., Walker J. E. (2006) Bovine complex I is a complex of 45 different subunits. J. Biol. Chem. 281, 32724–32727 [DOI] [PubMed] [Google Scholar]
- 3. Brandt U. (2006) Energy converting NADH:quinone oxidoreductase (complex I). Annu. Rev. Biochem. 75, 69–92 [DOI] [PubMed] [Google Scholar]
- 4. Lu J., Sharma L. K., Bai Y. (2009) Implications of mitochondrial DNA mutations and mitochondrial dysfunction in tumorigenesis. Cell Res. 19, 802–815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sharma L. K., Lu J., Bai Y. (2009) Mitochondrial respiratory complex I. Structure, function, and implication in human diseases. Curr. Med. Chem. 16, 1266–1277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Janssen R. J., Nijtmans L. G., van den Heuvel L. P., Smeitink J. A. (2006) Mitochondrial complex I. Structure, function, and pathology. J. Inherit. Metab. Dis. 29, 499–515 [DOI] [PubMed] [Google Scholar]
- 7. Sinha P. K., Castro-Guerrero N., Matsuno-Yagi A., Yagi T., Torres-Bacete J. (2009) Bacterial complex I (NDH-1). Functional roles of the hydrophobic domain. Curr. Top. Biochem. Res. 11, 79–90 [Google Scholar]
- 8. Yagi T., Matsuno-Yagi A. (2003) The proton-translocating NADH-quinone oxidoreductase in the respiratory chain. The secret unlocked. Biochemistry 42, 2266–2274 [DOI] [PubMed] [Google Scholar]
- 9. Castro-Guerrero N., Sinha P. K., Torres-Bacete J., Matsuno-Yagi A., Yagi T. (2010) Pivotal roles of three conserved carboxyl residues of the NuoC (30 K) segment in the structural integrity of proton-translocating NADH-quinone oxidoreductase from Escherichia coli. Biochemistry 49, 10072–10080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kao M. C., Di Bernardo S., Nakamaru-Ogiso E., Miyoshi H., Matsuno-Yagi A., Yagi T. (2005) Characterization of the membrane domain subunit NuoJ (ND6) of the NADH-quinone oxidoreductase from Escherichia coli by chromosomal DNA manipulation. Biochemistry 44, 3562–3571 [DOI] [PubMed] [Google Scholar]
- 11. Kao M. C., Nakamaru-Ogiso E., Matsuno-Yagi A., Yagi T. (2005) Characterization of the membrane domain subunit NuoK (ND4L) of the NADH-quinone oxidoreductase from Escherichia coli. Biochemistry 44, 9545–9554 [DOI] [PubMed] [Google Scholar]
- 12. Kao M. C., Di Bernardo S., Perego M., Nakamaru-Ogiso E., Matsuno-Yagi A., Yagi T. (2004) Functional roles of four conserved charged residues in the membrane domain subunit NuoA of the proton-translocating NADH-quinone oxidoreductase from Escherichia coli. J. Biol. Chem. 279, 32360–32366 [DOI] [PubMed] [Google Scholar]
- 13. Sinha P. K., Torres-Bacete J., Nakamaru-Ogiso E., Castro-Guerrero N., Matsuno-Yagi A., Yagi T. (2009) Critical roles of subunit NuoH (ND1) in the assembly of peripheral subunits with the membrane domain of Escherichia coli NDH-1. J. Biol. Chem. 284, 9814–9823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Torres-Bacete J., Nakamaru-Ogiso E., Matsuno-Yagi A., Yagi T. (2007) Characterization of the NuoM (ND4) subunit in Escherichia coli NDH-1. Conserved charged residues essential for energy-coupled activities. J. Biol. Chem. 282, 36914–36922 [DOI] [PubMed] [Google Scholar]
- 15. Torres-Bacete J., Sinha P. K., Castro-Guerrero N., Matsuno-Yagi A., Yagi T. (2009) Features of subunit NuoM (ND4) in Escherichia coli NDH-1. Topology and implication of conserved Glu-144 for coupling site 1. J. Biol. Chem. 284, 33062–33069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Torres-Bacete J., Sinha P. K., Matsuno-Yagi A., Yagi T. (2011) Structural contribution of C-terminal segments of NuoL (ND5) and NuoM (ND4) subunits of complex I from Escherichia coli. J. Biol. Chem. 286, 34007–34014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Guénebaut V., Schlitt A., Weiss H., Leonard K., Friedrich T. (1998) Consistent structure between bacterial and mitochondrial NADH:ubiquinone oxidoreductase (complex I). J. Mol. Biol. 276, 105–112 [DOI] [PubMed] [Google Scholar]
- 18. Berrisford J. M., Sazanov L. A. (2009) Structural basis for the mechanism of respiratory complex I. J. Biol. Chem. 284, 29773–29783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Efremov R. G., Baradaran R., Sazanov L. A. (2010) The architecture of respiratory complex I. Nature 465, 441–445 [DOI] [PubMed] [Google Scholar]
- 20. Sazanov L. A., Hinchliffe P. (2006) Structure of the hydrophilic domain of respiratory complex I from Thermus thermophilus. Science 311, 1430–1436 [DOI] [PubMed] [Google Scholar]
- 21. Baranova E. A., Holt P. J., Sazanov L. A. (2007) Projection structure of the membrane domain of Escherichia coli respiratory complex I at 8 Å resolution. J. Mol. Biol. 366, 140–154 [DOI] [PubMed] [Google Scholar]
- 22. Hunte C., Zickermann V., Brandt U. (2010) Functional modules and structural basis of conformational coupling in mitochondrial complex I. Science 329, 448–451 [DOI] [PubMed] [Google Scholar]
- 23. Hinchliffe P., Sazanov L. A. (2005) Organization of iron-sulfur clusters in respiratory complex I. Science 309, 771–774 [DOI] [PubMed] [Google Scholar]
- 24. Ohnishi T. (1998) Iron-sulfur clusters semiquinones in Complex I. Biochim. Biophys. Acta 1364, 186–206 [DOI] [PubMed] [Google Scholar]
- 25. Ohnishi T., Nakamaru-Ogiso E. (2008) Were there any “misassignments” among iron-sulfur clusters N4, N5, and N6b in NADH-quinone oxidoreductase (complex I)? Biochim. Biophys. Acta 1777, 703–710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Yakovlev G., Reda T., Hirst J. (2007) Re-evaluating the relationship between EPR spectra and enzyme structure for the iron sulfur clusters in NADH:quinone oxidoreductase. Proc. Natl. Acad. Sci. U.S.A. 104, 12720–12725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Nakamaru-Ogiso E., Matsuno-Yagi A., Yoshikawa S., Yagi T., Ohnishi T. (2008) Iron-sulfur cluster N5 is coordinated by an HXXXCXXCXXXXXC motif in the NuoG subunit of E. coli NADH:quinone oxidoreductase (complex I). J. Biol. Chem. 283, 25979–25987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Roessler M. M., King M. S., Robinson A. J., Armstrong F. A., Harmer J., Hirst J. (2010) Direct assignment of EPR spectra to structurally defined iron-sulfur clusters in complex I by double electron-electron resonance. Proc. Natl. Acad. Sci. U.S.A. 107, 1930–1935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Leif H., Sled V. D., Ohnishi T., Weiss H., Friedrich T. (1995) Isolation and characterization of the proton-translocating NADH: ubiquinone oxidoreductase from Escherichia coli. Eur. J. Biochem. 230, 538–548 [DOI] [PubMed] [Google Scholar]
- 30. Pohl T., Bauer T., Dörner K., Stolpe S., Sell P., Zocher G., Friedrich T. (2007) Iron-sulfur cluster N7 of the NADH:ubiquinone oxidoreductase (complex I) is essential for stability but not involved in electron transfer. Biochemistry 46, 6588–6596 [DOI] [PubMed] [Google Scholar]
- 31. Yano T., Sled V. D., Ohnishi T., Yagi T. (1994) Expression of the 25-kilodalton iron-sulfur subunit of the energy-transducing NADH-ubiquinone oxidoreductase of Paraccocus denitrificans. Biochemistry 33, 494–499 [DOI] [PubMed] [Google Scholar]
- 32. Yano T., Yagi T., Sled V. D., Ohnishi T. (1995) Expression and characterization of the 66-kilodalton (NQO3) iron-sulfur subunit of the proton-translocating NADH-quinone oxidoreductase of Paracoccus denitrificans. J. Biol. Chem. 270, 18264–18270 [DOI] [PubMed] [Google Scholar]
- 33. Yano T., Sled' V. D., Ohnishi T., Yagi T. (1996) Expression and characterization of the flavoprotein subcomplex composed of 50-kDa (NQO1) and 25-kDa (NQO2) subunits of the proton-translocating NADH-quinone oxidoreductase of Paracoccus denitrificans. J. Biol. Chem. 271, 5907–5913 [DOI] [PubMed] [Google Scholar]
- 34. Yano T., Magnitsky S., Sled' V. D., Ohnishi T., Yagi T. (1999) Characterization of the putative 2×[4Fe-4S] binding NQO9 subunit of the proton-translocating NADH-quinone oxidoreductase (NDH-1) of Paracoccus denitrificans. Expression, reconstitution, and EPR characterization. J. Biol. Chem. 274, 28598–28605 [DOI] [PubMed] [Google Scholar]
- 35. Nakamaru-Ogiso E., Yano T., Ohnishi T., Yagi T. (2002) Characterization of the iron-sulfur cluster coordinated by a cysteine cluster motif (CXXCXXXCX27C) in Nqo3 subunit in the proton-translocating NADH-quinone oxidoreductase (NDH-1) of Thermus thermophilus HB-8. J. Biol. Chem. 277, 1680–1688 [DOI] [PubMed] [Google Scholar]
- 36. Yano T., Sklar J., Nakamaru-Ogiso E., Takahashi Y., Yagi T., Ohnishi T. (2003) Characterization of cluster N5 as a fast-relaxing [4Fe-4S] cluster in the Nqo3 subunit of the proton-translocating NADH-ubiquinone oxidoreductase from Paracoccus denitrificans. J. Biol. Chem. 278, 15514–15522 [DOI] [PubMed] [Google Scholar]
- 37. Hayashi T., Stuchebrukhov A. A. (2010) Electron tunneling in respiratory complex I. Proc. Natl. Acad. Sci. U.S.A. 107, 19157–19162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Link A. J., Phillips D., Church G. M. (1997) Methods for generating precise deletions and insertions in the genome of wild type Escherichia coli. Application to open reading frame characterization. J. Bacteriol. 179, 6228–6237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sazanov L. A., Carroll J., Holt P., Toime L., Fearnley I. M. (2003) A role for native lipids in the stabilization and two-dimensional crystallization of the Escherichia coli NADH:ubiquinone oxidoreductase (complex I). J. Biol. Chem. 278, 19483–19491 [DOI] [PubMed] [Google Scholar]
- 40. Laemmli U. K. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685 [DOI] [PubMed] [Google Scholar]
- 41. Schägger H., von Jagow G. (1991) Blue native electrophoresis for isolation of membrane protein complexes in enzymatically active form. Anal. Biochem. 199, 223–231 [DOI] [PubMed] [Google Scholar]
- 42. Matsushita K., Ohnishi T., Kaback H. R. (1987) NADH-ubiquinone oxidoreductases of the Escherichia coli aerobic respiratory chain. Biochemistry 26, 7732–7737 [DOI] [PubMed] [Google Scholar]
- 43. Yagi T. (1986) Purification and characterization of NADH dehydrogenase complex from Paracoccus denitrificans. Arch. Biochem. Biophys. 250, 302–311 [DOI] [PubMed] [Google Scholar]
- 44. Ghelli A., Benelli B., Esposti M. D. (1997) Measurement of the membrane potential generated by complex I in submitochondrial particles. J. Biochem. 121, 746–755 [DOI] [PubMed] [Google Scholar]
- 45. Amarneh B., Vik S. B. (2003) Mutagenesis of subunit N of the Escherichia coli complex I. Identification of the initiation codon and the sensitivity of mutants to decylubiquinone. Biochemistry 42, 4800–4808 [DOI] [PubMed] [Google Scholar]
- 46. Messner K. R., Imlay J. A. (1999) The identification of primary sites of superoxide and hydrogen peroxide formation in the aerobic respiratory chain and sulfite reductase complex of Escherichia coli. J. Biol. Chem. 274, 10119–10128 [DOI] [PubMed] [Google Scholar]
- 47. Forzi L., Koch J., Guss A. M., Radosevich C. G., Metcalf W. W., Hedderich R. (2005) Assignment of the [4Fe-4S] clusters of Ech hydrogenase from Methanosarcina barkeri to individual subunits via the characterization of site-directed mutants. FEBS J. 272, 4741–4753 [DOI] [PubMed] [Google Scholar]
- 48. Dupuis A., Chevallet M., Darrouzet E., Duborjal H., Lunardi J., Issartel J. P. (1998) The complex I from Rhodobacter capsulatus. Biochim. Biophys. Acta 1364, 147–165 [DOI] [PubMed] [Google Scholar]
- 49. Kushnareva Y., Murphy A. N., Andreyev A. (2002) Complex I-mediated reactive oxygen species generation. Modulation by cytochrome c and NAD(P)+ oxidation-reduction state. Biochem. J. 368, 545–553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Marella M., Seo B. B., Matsuno-Yagi A., Yagi T. (2007) Mechanism of cell death caused by complex I defects in a rat dopaminergic cell line. J. Biol. Chem. 282, 24146–24156 [DOI] [PubMed] [Google Scholar]
- 51. Pohl T., Uhlmann M., Kaufenstein M., Friedrich T. (2007) λRed-mediated mutagenesis and efficient large scale affinity purification of the Escherichia coli NADH:ubiquinone oxidoreductase (complex I). Biochemistry 46, 10694–10702 [DOI] [PubMed] [Google Scholar]
- 52. Page C. C., Moser C. C., Chen X., Dutton P. L. (1999) Natural engineering principles of electron tunneling in biological oxidation-reduction. Nature 402, 47–52 [DOI] [PubMed] [Google Scholar]
- 53. Rasmussen T., Scheide D., Brors B., Kintscher L., Weiss H., Friedrich T. (2001) Identification of two tetranuclear FeS clusters on the ferredoxin-type subunit of NADH:ubiquinone oxidoreductase (complex I). Biochemistry 40, 6124–6131 [DOI] [PubMed] [Google Scholar]
- 54. Hirst J. (2010) Toward the molecular mechanism of respiratory complex I. Biochem. J. 425, 327–339 [DOI] [PubMed] [Google Scholar]
- 55. Moser C. C., Farid T. A., Chobot S. E., Dutton P. L. (2006) Electron tunneling chains of mitochondria. Biochim. Biophys. Acta 1757, 1096–1109 [DOI] [PubMed] [Google Scholar]
- 56. Wittekindt C., Schwarz M., Friedrich T., Koslowski T. (2009) Aromatic amino acids as stepping stones in charge transfer in respiratory complex I. An unusual mechanism deduced from atomistic theory and bioinformatics. J. Am. Chem. Soc. 131, 8134–8140 [DOI] [PubMed] [Google Scholar]
- 57. Lin J., Balabin I. A., Beratan D. N. (2005) The nature of aqueous tunneling pathways between electron-transfer proteins. Science 310, 1311–1313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Miyashita O., Okamura M. Y., Onuchic J. N. (2005) Interprotein electron transfer from cytochrome c2 to photosynthetic reaction center. Tunneling across an aqueous interface. Proc. Natl. Acad. Sci. U.S.A. 102, 3558–3563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Tezcan F. A., Crane B. R., Winkler J. R., Gray H. B. (2001) Electron tunneling in protein crystals. Proc. Natl. Acad. Sci. U.S.A. 98, 5002–5006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Hayashi T., Stuchebrukhov A. A. (2011) Quantum electron tunneling in respiratory complex I. J. Phys. Chem. B 115, 19157–19162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Larkin M. A., Blackshields G., Brown N. P., Chenna R., McGettigan P. A., McWilliam H., Valentin F., Wallace I. M., Wilm A., Lopez R., Thompson J. D., Gibson T. J., Higgins D. G. (2007) Clustal W and Clustal X. Version 2.0. Bioinformatics 23, 2947–2948 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.