Abstract
The discovery of the miRNA pathway revealed a new layer of molecular control of biological processes. To uncover new functions of this gene regulatory pathway, we undertook the characterization of the two miRNA-specific Argonaute proteins in Caenorhabditis elegans, ALG-1 and ALG-2. We first observed that the loss-of-function of alg-1 and alg-2 genes resulted in reduced progeny number. An extensive analysis of the germline of these mutants revealed a reduced mitotic region, indicating fewer proliferating germ cells. We also observed an early entry into meiosis in alg-1 and alg-2 mutant animals. We detected ALG-1 and ALG-2 protein expressions in the distal tip cell (DTC), a specialized cell located at the tip of both C. elegans gonadal arms that regulates mitosis-meiosis transition. Re-establishing the expression of alg-1 specifically in the DTC of mutant animals partially rescued the observed germline defects. Further analyses also support the implication of the miRNA pathway in gametogenesis. Interestingly, we observed that disruption of five miRNAs expressed in the DTC led to similar phenotypes. Finally, gene expression analysis of alg-1 mutant gonads suggests that the miRNA pathway is involved in the regulation of different pathways important for germline proliferation and differentiation. Collectively, our data indicate that the miRNA pathway plays a crucial role in the control of germ cell biogenesis in C. elegans.
Keywords: argonaute, miRNA, germline
Introduction
MiRNAs are 21-23 nucleotides long non-coding RNA molecules processed from hairpin-structured RNAs by Drosha and Dicer RNaseIII enzymes. These short RNA molecules induce translational repression and gene silencing of their target mRNAs via interaction with an Argonaute protein. Members of this protein family are classified into three groups: Argonaute-like proteins (AGO); Piwi-like proteins and C. elegans-specific AGOs (WAGOs) (reviewed in1). In C. elegans, AGO-clade proteins ALG-1 and ALG-2 have been identified to be involved exclusively in the miRNA pathway2. MiRNAs regulate a plethora of biological processes including cell proliferation, cell differentiation and apoptosis, processes important to coordinate developmental timing (reviewed in3,4). Since the discovery of the prominent miRNA families of lin-4 and let-7 in determining C. elegans developmental timing (reviewed in4), their mammalian homologs have also been identified to control cell proliferation in human cell lines5,6. Recent studies suggest that mammalian let-7 miRNA could regulate developmental processes via regulation of several cell cycle-related genes7,8. Since the deletion of a single family of miRNA often fails to induce severe defects in vivo9,10, it has been suggested that most biological processes are subjected to regulation of a cumulative effect by various miRNAs.
Among these processes, the studies of animals carrying mutations of important components of the miRNA pathway indicate the contribution of miRNAs in the animal germline. In Drosophila, mutations of dcr-1, ago-1 and loquacious genes resulted in defects in germline maintenance11,12,13,14. In C. elegans, alteration of the dcr-1 gene rendered animals sterile despite retaining a normal gonad due to the strong maternal rescue15. Additionally, deletion of the Drosha-encoding gene drsh-1 also led to sterility in nematode16.
Here, we report that Argonaute proteins ALG-1 and ALG-2 are expressed in the distal tip cells (DTCs) of the C. elegans germline. Mutations in alg-1 and alg-2 result in drastically reduced number of progeny. We observed that this reduction in fertility is caused by defects in multiple processes in germline development, including reduced germ cell proliferation and increase in apoptosis. Our findings suggest that ALG-1/2 function, together with a set of miRNAs expressed in the DTCs to regulate diverse biological pathways important to maintain animal germline proliferation and differentiation.
Results
MicroRNA-specific Argonautes are required for germ cell proliferation
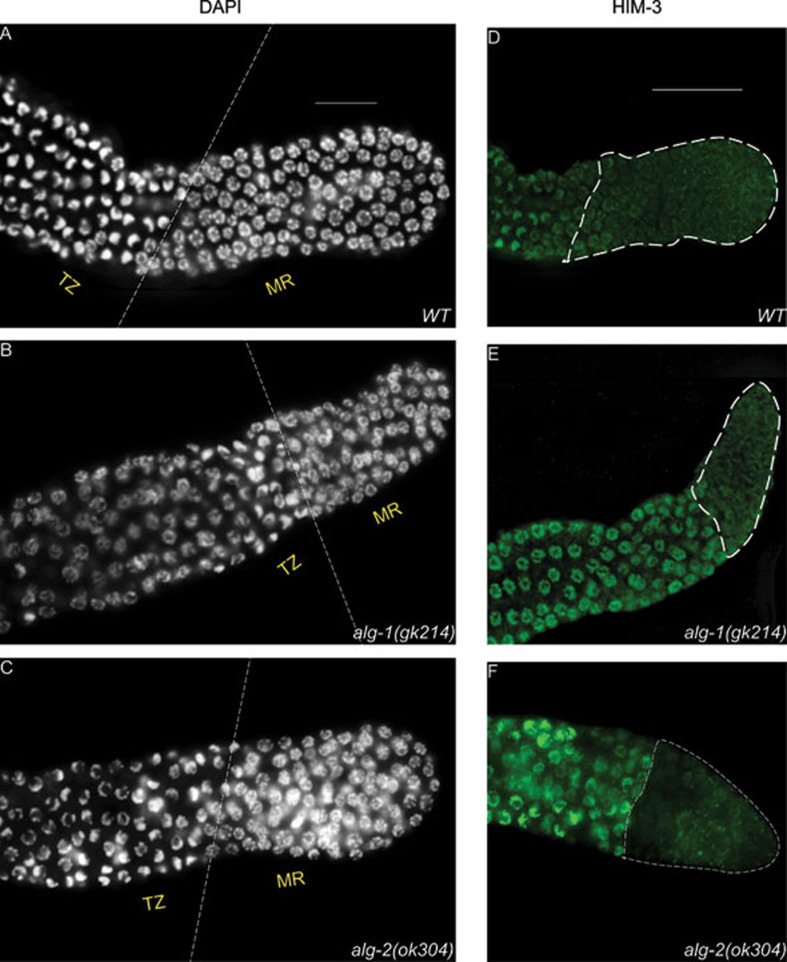
Since defects in the development of the germline will be reflected in the progeny number, we first investigated the brood size of ALG-1 and ALG-2 loss-of-function mutants, named as alg-1(gk214) and alg-2(ok304), respectively. Compared to wild-type animals, we noticed a significantly reduced brood size in alg-1(gk214) and alg-2(ok304), though such reduction was more drastic in alg-1(gk214) (Table 1). As any defects during germline proliferation, meiosis or gamete formation could affect progeny number, we first asked if these mutants have defects in cell proliferation in the germline. We extruded germline of young adults from wild-type, alg-1(gk214) and alg-2(ok304) animals, followed by staining of DNA with DAPI to monitor the spatio-temporal progression of different meiotic phases. Cells with crescent-shaped nuclear morphology mark the transition zone, which represents the leptotene/zygotene stage of meiosis17,18. Compared to the wild-type animals, the mitotic regions of alg-1(gk214) and alg-2(ok304) are shorter as determined by the morphology of the germ cell nuclei (Figure 1A-1C) and by the number of cells in the mitotic region (Table 1). To corroborate our findings from the DAPI staining, we used an antibody specific to HIM-3, a meiosis-specific axis component between sister chromatids19, as a bona fide marker of entry into meiosis. In agreement with our earlier findings, anti-HIM-3 antibody revealed an early entry into meiosis in alg-1(gk214) and alg-2(ok304), compared to the wild type (Figure 1D-1F). Similar defects were observed with another loss-of-function allele of the alg-1 gene (alg-1(tm492)) as well as in alg-1(RNAi) animals (Supplementary information, Figure S1). These results suggest that ALG-1 and ALG-2 are involved in the regulation of germline proliferation.
Table 1. Phenotypes observed in the germline of alg-1(gk214) and alg-2(ok304) animals.
| Strain | Brood size (N=25) | Nb of cells in MR (N=5) | Oocyte / arm day 1 (N=5) | Oocyte / arm day 2 (N=5) | Average corpses / arm (N=5) |
|---|---|---|---|---|---|
| Wild-type | 266.4 ± 3.22 | 240.3 ± 0.63 | 9.06 ± 0.06 | 9.84 ± 0.12 | 2.98 ± 0.19 |
| alg-1(gk214) | 77.0 ± 5.64 | 203.9 ± 1.26 | 5.48 ± 0.16 | 6.08 ± 0.07 | 4.38 ± 0.18 |
| *(5.68E–20) | *(1.93E–05) | *(6.51E–06) | *(3.33E–05) | *(0.01) | |
| alg-2(ok304) | 178.8 ± 4.27 | 213.7 ± 0.43 | 7.0 ± 0.07 | 7.18 ± 0.24 | 4.12 ± 0.05 |
| *(4.24E–14) | *(1.99E–06) | *(1.03E–05) | *(0.0015) | *(0.001) | |
| lag-2p::rfp::alg-1 | 142.12 ± 5.6 | 237.2 ± 1.4 | 6.7 ± 0.17 | 6.4 ± 0.2 | 3.28 ± 0.08 |
| *(3.70E–17) | *(0.134) | *(9.61E–05) | *(8.68E–05) | *(0.184) | |
| **(5.34E–08) | **(0.0001) | **(0.005) | **(0.186) | **(0.004) |
N: number of animals (brood size) or number of replicates (10 animals/replicate); MR: Mitotic region. The brood sizes were defined as the numbers of viable larvae that developed to the L1 stage descended from a single hermaphrodite of its strain. In parenthesis, P-values were calculated with a Student's t-test to compare numbers with wild-type (*) or alg-1(gk214) (**) animals, and represented as ± SEM.
Figure 1.
alg-1 and alg-2 mutant gonads have shorter mitotic region and display early entry into meiosis. (A-C) DAPI-stained germline depicting mitotic region (MR) and transition zone (TZ) in wild-type (WT), alg-1(gk214) and alg-2(ok304) mutant animals. (D-F) Staining with HIM-3 antibody (green) depicting entry into meiosis in wild-type (WT) gonads and alg-1(gk214) and alg-2(ok304). Scale bar measures 20 μm.
Expression of ALG-1 in the DTC controls germ cell proliferation
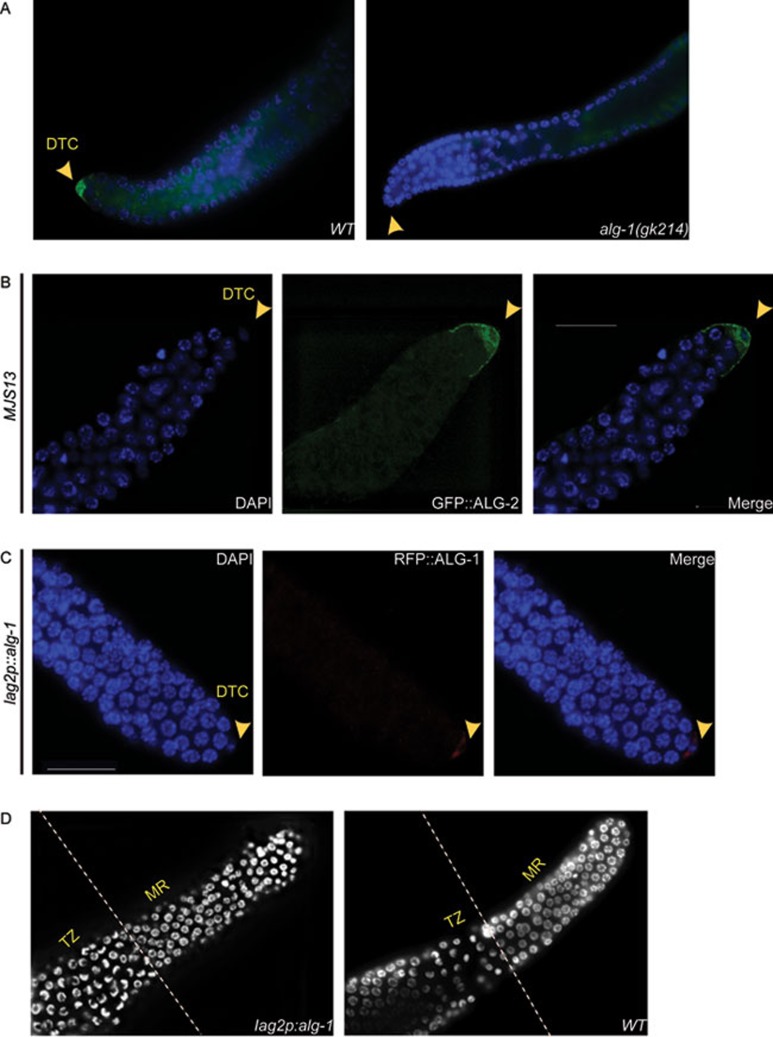
To better decipher how alg-1 and alg-2 control germ cell proliferation and meiosis entry, we next decided to observe the expression pattern of ALG-1 and ALG-2 proteins in animal gonads. By immunostaining of extruded gonads using a newly generated ALG-1-specific antibody (Supplementary information, Figure S2), we observed that ALG-1 is localized to the DTC of the wild-type gonads but not in alg-1(gk214) (Figure 2A). To overcome the non-availability of ALG-2-specific antibody, we generated a transgenic line somatically expressing GFP-tagged ALG-2 (MJS13) and found that ALG-2 is also localized in the DTC of the gonads (Figure 2B). DTC caps the distal end of the germline, and provides the stem cell niche. These specialized cells are also responsible for maintaining proliferation in the distal part of the gonad arm, which is the mitotic region20,21,22,23. When the contact between the DTC and germ cells is breached, cells enter into meiosis. To determine whether the presence of ALG-1 in the DTC is crucial to control germ cell proliferation and differentiation, we generated a transgenic line where ALG-1 expression is under control of the promoter of lag-2, a membrane-bound Delta/Serrate/LAG-2 ligand expressed exclusively in the DTC24. When re-establishing ALG-1 expression in the DTC (Figure 2C), we were able to partially rescue the brood size compared to the alg-1 mutant (Table 1), as well as fully restore the normal length of the mitotic region (Figure 2D and Table 1, compared to wild type). To further decipher the role of alg-1 beyond the DTC, we scored the brood size in transgenic animals expressing RFP-tagged ALG-1 under the control of endogenous alg-1 promoter and 3′UTR. Apart from the DTC, RFP::ALG-1 is also expressed in sheath cells besides other somatic tissues (AVR, unpublished data). We observed that these transgenic animals showed a significant increase in brood size compared to animals with DTC-expressed alg-1 (158 ± 2.7 vs 142.12 ± 5.6 for plag-2::alg-1; P < 0.017). Taken together, our results indicate that ALG-1 plays a role within the DTC as well as in other tissues of the gonads.
Figure 2.
ALG-1 and ALG-2 localize to the DTC. (A) Anti-ALG-1 antibody depicts ALG-1 localization to the DTC of wild-type (WT) germline, but not in alg-1(gk214). (B) ALG-2 (green) expression in DTC of gfp::alg-2 (MJS13) transgenic line. Gonads were counter stained with DAPI (blue) to visualize nuclei. (C) ALG-1 localized exclusively in DTC of transgenic animals expressing RFP-tagged ALG-1 protein under control of the DTC-specific lag-2 promoter (lag-2p). Arrows indicate DTC. (D) Restoration of mitotic region in lag-2p::rfp::alg-1-expressing animals shown by DAPI staining compared to wild-type gonad. Scale bar measures 20 μm.
ALG-1 and ALG-2 are important for gamete formation and maintenance
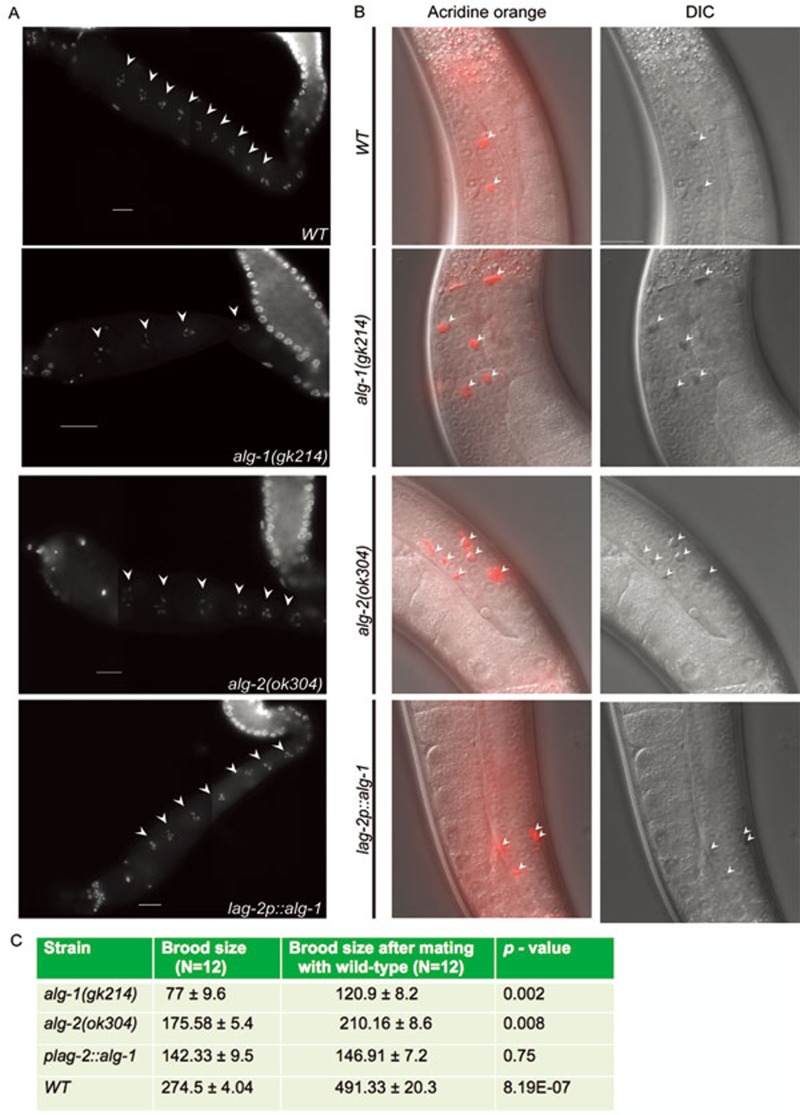
Since germ cells undergo proliferation (mitosis), gametogenesis (meiosis and differentiation) or apoptosis, we examined if the loss of the miRNA-specific Argonautes alg-1 and alg-2 genes can affect the fates of germ cells in animals. Although we did not observe high incidence of males and the dead eggs phenotype in alg-1(gk214) and alg-2(ok304) animals (two notable consequences of meiotic defects in C. elegans, data not shown), we found that both mutants have significantly reduced number of oocytes than the wild-type animals (Figure 3A and Table 1). Using the vital dye acridine orange (AO), which has been used to stain apoptotic cells in live animals25, as well as differential interference contrast (DIC) microscopy, we observed increased germ cell corpses in the proximal region of the gonad arm in alg-1(gk214) and alg-2(ok304) (Figure 3B and Table 1). The fact that both DIC microscopy and AO staining detect the same number of germ cell corpses, suggests that mutants have an increase in apoptosis rather than defective engulfment in the germline. Re-establishing the expression of ALG-1 in the DTC partially restores the number of oocytes and reduces germ cell corpses (Figure 3B and Table 1). We next determined if the decrease in brood size also results from defects in male gametes formation. We observed that sperm nuclei number in alg-1 and alg-2 mutant hermaphrodites is not significantly different from the wild-type animals (data not shown). However, when the two Argonaute mutants were mated with wild-type males, we observed a significant increase in the brood size compared to the unmated mutant hermaphrodites, but brood size is significantly smaller than that of the wild-type hermaphrodites mated with wild-type males (Figure 3C). In addition, the brood size of animals with DTC-expressed alg-1 is not enhanced upon mating with wild-type males (Figure 3C). This observation supports that re-establishing ALG-1 expression in the DTC mainly rescues spermiogenesis, and thus sustains the fact that ALG-1 is required in other gonadal tissues. Together, our data show that ALG-1 and ALG-2 are involved in maintaining C. elegans fertility.
Figure 3.
Effects of ALG-1 and ALG-2 on oocytes. (A) DAPI-stained germline. Arrowheads depict oocyte nuclei in diakinesis stage in the proximal gonad arm of wild-type (WT), alg-1(gk214), alg-2(ok304) and plag-2::rfp::alg-1 animals. (B) Merged and nomarski DIC micrographs of AO-stained germline in respective genetic backgrounds. Arrowheads mark apoptotic corpses. (C) Brood size in alg-1 and alg-2 mutant animals as well as in plag-2::alg-1 and wild-type animals after mating with wild-type males represented as average brood size ± SEM, P < 0.01.
miRNAs expressed in the DTC are implicated in germ cell proliferation and differentiation
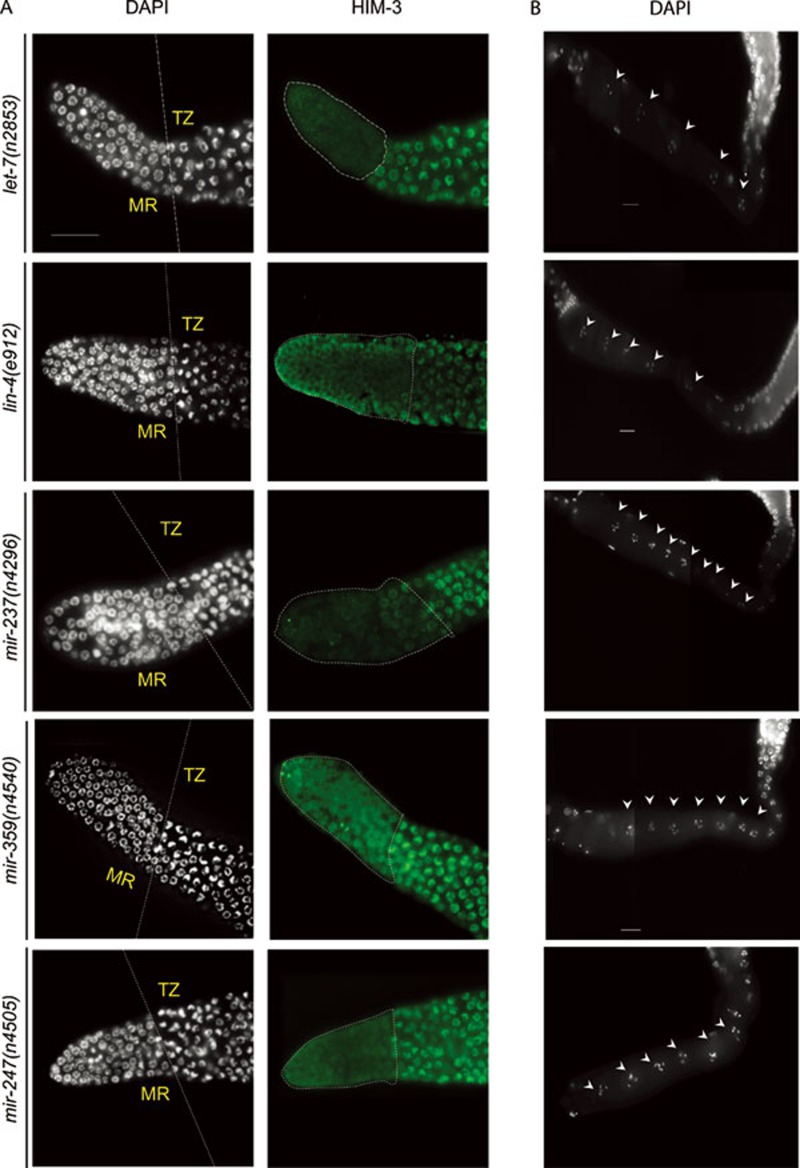
Since ALG-1 and ALG-2 are imperative to miRNA-induced gene silencing, we next decided to identify candidate miRNAs involved in germline maintenance. Recently, the expression pattern of several C. elegans miRNAs has been studied in vivo using miRNA promoter::GFP fusion constructs26. Of the total of 70 transgenic C. elegans strains reported, 8 pmiRNA::gfp strains (plet-7, plin-4, pmir-80, pmir-237, pmir-247-797, pmir-359, pmir-53 and pmir-71) exhibited expression in the DTC. We thus examined animals carrying mutant alleles of the eight miRNA genes, to determine if they display phenotypes similar to alg-1(gk214) and alg-2(ok304). Among them, we found that let-7(n2853), lin-4(e912), miR-237(n4296), miR-359(n4540) and miR-247(n4505) mutants displayed similar phenotypes to alg-1(gk214) and alg-2(ok304). All five mutant strains had significantly smaller brood size and shorter mitotic region with reduced number of cells within the mitotic region compared to wild type (Figure 4 and Table 2). While the let-7(n2853), lin-4(e912), miR-359(n4540) and mir-247(n4505) mutant animals have shorter mitotic region as well as fewer number of oocytes, mir-237(n4296) mutant animals have only shorter mitotic region but normal oocyte number (Figure 4 and Table 2), and miR-80(nDf53), mir-71(n4115) and mir-53(n4113) mutant strains have no apparent germline defect (Supplementary information, Figure S3). These observations suggest that a variety of miRNAs regulate different processes at multiple steps in germline biogenesis.
Figure 4.
A subset of miRNAs expressed in the DTC affects germline proliferation and oocytes. (A) DAPI and HIM-3 staining depicting shorter mitotic region observed in let-7, lin-4, miR-237 and mir-247 in mutant animals. (B) DAPI-stained germline. Arrowheads depict oocyte nuclei in diakinesis stage in the proximal gonad arm of miRNA mutant animals.
Table 2. Different phenotypes observed in miRNA mutants.
| Strain | Brood size (N=20) | Nb of cells in MR (N=5) | Oocyte / arm day 1 (N=5) | Oocyte / arm day 2 (N=5) |
|---|---|---|---|---|
| Wild-type | 266.4 ± 3.7 | 240.3 ± 0.63 | 9.06 ± 0.06 | 9.84 ± 0.12 |
| let-7(n2853) | 43.9 ± 3.1 | 192.02 ± 1.4 | 5.8 ± 0.07 | *ND |
| (6.45E–22) | (3.40E–06) | (1.66E–06) | ||
| lin-4(e912) | 30.0 ± 0.9 | 197.6 ± 1.2 | 5.6 ± 0.12 | *ND |
| (1.07E–23) | (4.11E–06) | (1.81E–06) | ||
| mir-359(n4540) | 235.6 ± 6.8 | 221.3 ± 0.9 | 7.7 ± 0.12 | 8.98 ± 0.09 |
| (0.002) | (5.39E–05) | (0.001) | (0.01) | |
| mir-247(n4505) | 207.7 ± 5.9 | 200.94 ± 2.6 | 7.6 ± 0.12 | 8.84 ± 0.08 |
| (3.10E–07) | (0.0002) | (0.0003) | (0.003) | |
| mir-237(n4296) | 222.1 ± 7.4 | 213.7 ± 1.9 | 8.82 ± 0.05 | 10.12 ± 0.32 |
| (4.37E–05) | (5.45E–05) | **(0.70) | **(0.40) |
N: number of animals (brood size) or number of replicates (10 animals/replicate); MR: Mitotic region; ND: Not determined. In parenthesis, P-values were calculated with a Student's t-test to compare the significance of the numbers with wild-type animals and represented as ± SEM.
*Most of the population died after the first day of fertile adults.
**mir-237(n4296) animals have similar number of oocytes as the wild-type.
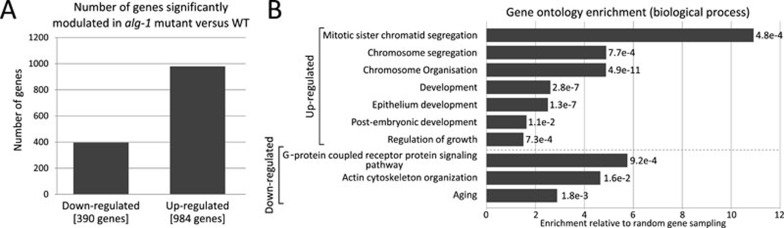
Since the regulation by miRNAs often leads to a decrease in target mRNAs27,28,29,30, we next decided to compare the level of mRNAs found in gonads of wild-type and alg-1 animals to uncover putative targets of let-7, lin-4, miR-237, miR-359 and miR-247 miRNAs in the germline. When we compared microarray data from four independent biological samples, we observed that the level of 1 374 different mRNAs is significantly altered in the absence of alg-1 (with a threshold of > 2-fold change, P ≤ 0.001; Figure 5 and Supplementary information, Table S1). A clustering analysis of the gene expression data revealed a significant alteration in the expression of genes associated with biological pathways important for chromosome organization and segregation (Figure 5). Although they are not significantly enriched, we notably found putative targets predicted by either TargetScan31 or miRWIP32 algorithm for let-7, lin-4/miR-237 (since they are similar in sequence, they are predicted to target the same mRNAs33), miR-359 and miR-247 miRNAs among mRNAs misregulated in the germline of alg-1 mutant (Supplementary information, Table S2). Thus, our results implicate that these miRNAs contribute to the regulation of the process of gamete formation and differentiation in C. elegans by affecting the expression of multiple mRNA targets.
Figure 5.
Comparative microarray analyses of genes expressed in the germline. (A) Number of genes significantly modulated in extruded alg-1 mutant versus wild-type gonads. Selected significant genes have a fold change > 2 and a P-value < 0.001 (N = 4; the list of can be found in Supplementary information, Table S1). (B) Gene ontology biological process enrichment performed using DAVID. The upper part corresponds to enrichment obtained when using the list of downregulated genes and the bottom part of the upregulated genes. Numbers to the right represent Benjamini-Hochberg P-values.
Discussion
Earlier studies performed in Drosophila have highlighted that components of the miRNA pathway are required for germline stem cell self-renewal,11,13,14,34 and that the maternally expressed bantam and miR-184 miRNAs contribute to oogenesis35,36. While our data revealed that the role of the miRNA pathway in germline maintenance is conserved in C. elegans, our results also support for the first time that both miRNA-specific Argonaute proteins ALG-1 and ALG-2 in the stem cell niche are crucial for the proper control of germ cell proliferation and gametes formation.
Previous studies of C. elegans strains carrying mutations in Drosha (drsh-1) and Dicer (dcr-1) genes, two important processors of miRNAs, showed that these mutants were sterile15,16. In our laboratory, we observed that post-embryonic RNAi of ALG-1 on alg-2 background or vice versa could also render animals sterile (unpublished data). These observations indicate that the miRNA pathway is indispensible in animal reproduction. Our current study showed that germline proliferation is reduced in both miRNA-specific Argonaute mutants. The restoration of ALG-1 exclusively in the DTC, rescues the mitotic cell number in the germline, which leads to a partial but significant increase in brood size, suggesting that miRNAs play a role in germline cell division. Additionally, our observation of fewer oocytes and increased apoptotic corpses in both alg-1 and alg-2 suggests that miRNAs are also important regulators in gamete formation. Consistent with our findings, mice with either global deletion of dicer or with oocyte-specific deletion of AGO2 display failures in oogenesis due to arrest at meiosis I37,38,39,40. These animals also have severely reduced number of spermatogonia, which could be due to proliferation defects and an increase in apoptosis39,41,42. However, it has yet to be determined in vertebrates the contribution of miRNAs in gametogenesis, since the loss of Dicer also affects the production of endogenous siRNAs, a type of small RNAs that are also important in this process43,44,45.
In C. elegans, a single DTC in each gonad arm establishes germline stem cell niche. It controls germ cells fate by employing in part the GLP-1/Notch signaling through a network of RNA-binding regulatory proteins, most notably, Pumilio and FBF, to maintain a balance between proliferation and differentiation46. While the RNA-binding proteins GLD-1, GLD-2/GLD-3 promote the meiosis entry of germline stem cells, the activation of the GLP-1/Notch signaling pathway inhibits these signals and retains the cells in mitotic stage47. Interestingly, gld-1 mRNA has been found in ALG-1-immunoprecipitated complex48, and other evidence also indicates that gld-1 is subject to regulation by miRNAs49. These studies suggest the possibility that miRNAs can regulate the mitosis to meiosis decision by controlling key genes involved in the process.
We demonstrate that mutants of five different miRNAs which are known to localize to the DTC display similar phenotypes as the ones observed in alg-1 and alg-2 mutants, suggesting that more than one miRNA participate in stem cell fate regulation. Our extensive microarray analysis of mRNAs expressed in alg-1 mutant gonads, detected the mis-regulation of more than 1 300 genes. Among them, we observed that the expression of the major sperm proteins (MSPs), was upregulated in alg-1 mutant gonads. MSP signaling is known to regulate the oocyte production and development. The proximal MSP signaling works coordinately with the distal GLP-1 signaling to regulate the proper oocyte growth and function50,51. We thus envision that GLP-1 and MSP signaling are both subject to the miRNA regulation, and the loss of the miRNA-specific Argonaute genes leads to alterations in both signaling pathways, likely due to imbalance of these proteins, and thus affecting the fertility of the animals. Therefore, the phenotypes observed in the germline of alg-1 and alg-2 mutants reflect that different miRNAs are involved in germline biogenesis by regulating different pathways. Hence, it may be difficult to recapitulate the alg-1 and alg-2 mutant phenotypes by inactivating specific miRNAs. We therefore propose that the phenotypes of alg-1 and alg-2 mutants result from the combinatorial effect of miRNAs and their targets.
Materials and Methods
Strains
All the strains were maintained according to standard protocols52. The let-7(n2853) and lin-4(e912) mutant strains were maintained at 15 °C and let-7(n2853) (shifted to 20 °C beyond L4 stage). All other strains were maintained at 20 °C. The alg-1(gk214) mutant carries an out-of-frame deletion of 200 bp after the 28th amino acid and terminates by 2 additional amino acids. The alg-2(ok304) allele is an out-of-frame deletion that removes the nucleotides encoding amino acids 34-374 and terminates after encoding 8 additional amino acids from another reading frame. They are therefore likely to be null alleles of alg-1 and alg-2. Further details can be found on the C. elegans Gene Knockout Consortium website. All mutant strains have been outcrossed at least three times before analyses.
Rescue experiments
Transgenic MJS13 line was generated by microinjecting a mix of reporter plasmids (pRF4), MSp59 (alg-1p::RFP::alg-1) and MSp72 (alg-2p::GFP::alg-2) and crossed into alg-1(gk214) strain. Extrachromosomal arrays were UV integrated. The plag-2::alg-1 line (MJS26) was generated by microinjecting a mix of pRF4 and MSp151 plasmids and crossed into alg-1(gk214) strain. A 3 kb promoter of lag-2 was excised by BamHI from pJK59053 and cloned into BamHI-digested pBluescriptII SK+ to generate MSp147. Using a primer set incorporating AflII and NotI sites, the promoter was amplified by PCR, digested and employed to exchange the endogenous alg-1 promoter from AflII/NotI-digested MSp59 plasmid (alg-1p::RFP::alg-1) producing MSp150. Latter, the NotI RFP cassette was reintroduced to generate MSp151 (lag-2p::RFP::alg-1). Both alg-1 and alg-2 constructs contain their respective 3′UTR regions.
Cytological methods
For antibody staining, gonads were dissected from young adults (20-22 h post L4) in PBS. Extruded gonads were immersed in fixing solution (1% PFA+0.1% Tween-20) for 5 min at room temperature, followed by freeze crack in liquid nitrogen and transferred to −20 °C methanol for 1 min. The fixed gonads were washed three times in PBS with 0.1% Tween-20 for 15 min, followed by blocking in PBS-T + 1% BSA for 1 h at room temperature. Gonads were then incubated with primary antibodies (α-ALG-1 (1:500) or α-HIM-3 (1:200)) overnight at 4 °C, and probed with Alexa Fluor 488 anti-rabbit as secondary antibody (1:500). Gonads were counter-stained by 1 μg/ml DAPI in anti-fading agent (Vectashield, Vector Laboratories). Images were captured using Zeiss motorized Axioplan 2 microscope at 630× consisting of 15-20 serial Z sections of 0.5 μm thickness subsuming entire nuclei. Fluorescence images were acquired with an AxioCam HRm camera and AxioVision acquisition software.
Brood size
Single L4 hermaphrodite from wild-type and mutant strains were transferred to seeded NGM plates and maintained at 20 °C. Animals were transferred to fresh plates each day until they stopped laying eggs. The hatched larvae on each plate were counted and total number of viable larvae that developed to the L1 stage descended from a single hermaphrodite was calculated. The average number of viable larvae from 25 hermaphrodites of a strain was plotted as brood size. Significant differences were determined by Student's t-test (P < 0.05) and represented as ± SEM.
Scoring mitotic cell number counts and entry into meiosis
Mitotic region was established previously23. Cell numbers within mitotic region were determined by counting from the row immediately adjacent to the DTC to the row containing multiple crescent-shaped nuclei, which is the early meiotic prophase I (leptotene/zygotene) or to the row where the nuclei stained positive with α-HIM-3 antibody (marker for entry into meiosis). Entry into meiosis was confirmed by looking at the gonad arm within each category for nuclei that stained positive with α-HIM-3 antibody in mitotic region/ transition zone.
Oocyte count and sperm defect
Gonads from L4 animals past 20 h and 40 h (day-1 and day-2, respectively) were dissected and fixed with fixing solution (1% PFA+0.1% Tween-20) for 30 min at room temperature. Gonads were washed stained by 1 μg/ml DAPI in anti-fading agent (Vectashield, Vector Laboratories) and monitored under the microscope at 630× magnification. Oocytes were counted from the loop of the gonad arm in a linear fashion till the most proximal oocyte also called as (−1). To check the sperm defect, L4 animals of mutant background were mated with wild-type males. Progeny was counted from animals, where cross progeny was monitored and compared to progeny from unmated animals.
AO assay
To obtain the number of corpses in worms, 20-22 h past L4 adult animals were stained with AO. Adult animals were incubated in dark for 1.5 h at room temperature on plates containing 1 ml of M9 with 0.08 mg of AO. Stained adults were transferred to fresh NGM plates to incubate for 45 min to clear the stained bacteria. Worms were mounted on agarose pads and monitored under fluorescence microscope. Stained corpses as well as the ones which were clearly visible under DIC as dark spots were counted.
Microarray analysis
Gonads from wild-type N2 and alg-1(gk214) animals from four independent pools (around five hundred gonads for each set) were extruded and immediately placed in cold Tri-Reagent (Sigma) for total RNA extraction. RNA purification was performed using the PicoPure RNA isolation kit (Applied Biosystems), with DNase (Qiagen) treatment on the purification column according to the manufacturer's instructions. Quantity and quality of RNA was verified on a 2100 Bioanalyzer (Agilent Technologies). Samples were stored at −80 °C.
Antisense RNA was produced using the Agilent LowInput QuickAmp Labeling Kit Two Color (Agilent Technologies). A 100 ng of total RNA spiked in with Two-Color RNA Spike-In Kit from Agilent was amplified and labeled as recommended by manufacturer; except for labelled aRNA purification, picopure RNA extraction kit was used. Quantity and labeling of aRNA was determined using a Nanodrop ND-1000 (NanoDrop Technologies).
Samples from four biological replicates of each gonad strain were hybridized on C. elegans Oligo Microarray (Agilent Technologies) using a dye-swap design (technical replicates) for a total of four arrays. The manufacturer's protocol provided with Agilent Gene Expression oligo microarrays Version 6.5 (May 2010) was followed adding the acetonitrile and Stabilization and Drying Solution (Agilent Technologies) to wash steps. Slide was scanned on a G2505 B Agilent Microarray scanner and fluorescence values extract using Feature extraction software (Agilent Technologies).
The raw expression data were imported in FlexArray (http://www.genomequebec.mcgill.ca/FlexArray) and analyzed using LIMMA54. First background was subtracted and the data were normalized using loess normalization. The experimental design was modeled using the function lmFit and the dye-swap arrays were taken into account. Differential expression was assessed using an empirical Bayes' statistics using the eBayes function. Gene enrichment analysis was performed using DAVID on the gene ontology biological processes55.
Acknowledgments
We thank members of the Zetka lab for their technical help as well as members of the Simard's group for their critical reading of the manuscript. We also thank Judith Kimble (University of Wisconsin-Madison) and Eric Miska (University of Cambridge) for providing reagents and strains. Some nematode strains were provided by the Caenorhabditis Genetics Center, which is funded by the NIH National Center for Research Resources (NCRR) and by the International C. elegans Gene Knockout Consortium. This research was supported by NSERC grants (C R, J-Y M and M J S). J-Y M is a Chercheur-Boursier Senior from Fonds de la Recherche en Santé du Québec and M J S is a Canadian Institutes of Health Research New Investigator.
Footnotes
(Supplementary information is linked to the online version of the paper on the Cell Research website.)
Supplementary Material
Analyses of alg-1(tm492) and alg-1(RNAi) animals germline.
Specificity of the ALG-1 specific polyclonal antibody.
Set of miRNA mutants with no apparent germline phenotypes.
The list of genes that is mis-regulated in alg-1 (gk214) gonads.
Predictive mRNA targets of microRNAs expressed in gonads distal tip cells that are important for germline proliferation and differentiation.
References
- Hutvagner G, Simard MJ. Argonaute proteins: key players in RNA silencing. Nat Rev Mol Cell Biol. 2008;9:22–32. doi: 10.1038/nrm2321. [DOI] [PubMed] [Google Scholar]
- Grishok A, Pasquinelli AE, Conte D, et al. Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell. 2001;106:23–34. doi: 10.1016/s0092-8674(01)00431-7. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Ambros V. MicroRNAs and developmental timing. Curr Opin Genet Dev. 2011;21:511–517. doi: 10.1016/j.gde.2011.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takamizawa J, Konishi H, Yanagisawa K, et al. Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival. Cancer Res. 2004;64:3753–3756. doi: 10.1158/0008-5472.CAN-04-0637. [DOI] [PubMed] [Google Scholar]
- Lee YS, Kim HK, Chung S, Kim KS, Dutta A. Depletion of human micro-RNA miR-125b reveals that it is critical for the proliferation of differentiated cells but not for the down-regulation of putative targets during differentiation. J Biol Chem. 2005;280:16635–16641. doi: 10.1074/jbc.M412247200. [DOI] [PubMed] [Google Scholar]
- Ramachandran R, Fausett BV, Goldman D. Ascl1a regulates Muller glia dedifferentiation and retinal regeneration through a Lin-28-dependent, let-7 microRNA signalling pathway. Nat Cell Biol. 2010;12:1101–1107. doi: 10.1038/ncb2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson CD, Esquela-Kerscher A, Stefani G, et al. The let-7 microRNA represses cell proliferation pathways in human cells. Cancer Res. 2007;67:7713–7722. doi: 10.1158/0008-5472.CAN-07-1083. [DOI] [PubMed] [Google Scholar]
- Alvarez-Saavedra E, Horvitz HR. Many families of C. elegans microRNAs are not essential for development or viability. Curr Biol. 2010;20:367–373. doi: 10.1016/j.cub.2009.12.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miska EA, Alvarez-Saavedra E, Abbott AL, et al. Most Caenorhabditis elegans microRNAs are individually not essential for development or viability. PLoS Genet. 2007;3:e215. doi: 10.1371/journal.pgen.0030215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Chen D, Duan R, et al. Argonaute 1 regulates the fate of germline stem cells in Drosophila. Development. 2007;134:4265–4272. doi: 10.1242/dev.009159. [DOI] [PubMed] [Google Scholar]
- Forstemann K, Tomari Y, Du T, et al. Normal microRNA maturation and germ-line stem cell maintenance requires Loquacious, a double-stranded RNA-binding domain protein. PLoS Biol. 2005;3:e236. doi: 10.1371/journal.pbio.0030236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Z, Xie T. Dcr-1 maintains Drosophila ovarian stem cells. Curr Biol. 2007;17:539–544. doi: 10.1016/j.cub.2007.01.050. [DOI] [PubMed] [Google Scholar]
- Hatfield SD, Shcherbata HR, Fischer KA, et al. Stem cell division is regulated by the microRNA pathway. Nature. 2005;435:974–978. doi: 10.1038/nature03816. [DOI] [PubMed] [Google Scholar]
- Knight SW, Bass BL. A role for the RNase III enzyme DCR-1 in RNA interference and germ line development in Caenorhabditis elegans. Science. 2001;293:2269–2271. doi: 10.1126/science.1062039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denli AM, Tops BB, Plasterk RH, Ketting RF, Hannon GJ. Processing of primary microRNAs by the Microprocessor complex. Nature. 2004;432:231–235. doi: 10.1038/nature03049. [DOI] [PubMed] [Google Scholar]
- Dernburg AF, McDonald K, Moulder G, et al. Meiotic recombination in C. elegans initiates by a conserved mechanism and is dispensable for homologous chromosome synapsis. Cell. 1998;94:387–398. doi: 10.1016/s0092-8674(00)81481-6. [DOI] [PubMed] [Google Scholar]
- MacQueen AJ, Villeneuve AM. Nuclear reorganization and homologous chromosome pairing during meiotic prophase require C. elegans chk-2. Genes Dev. 2001;15:1674–1687. doi: 10.1101/gad.902601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zetka MC, Kawasaki I, Strome S, Muller F. Synapsis and chiasma formation in Caenorhabditis elegans require HIM-3, a meiotic chromosome core component that functions in chromosome segregation. Genes Dev. 1999;13:2258–2270. doi: 10.1101/gad.13.17.2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen D, Schedl T. The regulatory network controlling the proliferation-meiotic entry decision in the Caenorhabditis elegans germ line. Curr Top Dev Biol. 2006;76:185–215. doi: 10.1016/S0070-2153(06)76006-9. [DOI] [PubMed] [Google Scholar]
- Kimble J, Crittenden SL. Controls of germline stem cells, entry into meiosis, and the sperm/oocyte decision in Caenorhabditis elegans. Annu Rev Cell Dev Biol. 2007;23:405–433. doi: 10.1146/annurev.cellbio.23.090506.123326. [DOI] [PubMed] [Google Scholar]
- Hubbard EJ. Caenorhabditis elegans germ line: a model for stem cell biology. Dev Dyn. 2007;236:3343–3357. doi: 10.1002/dvdy.21335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crittenden SL, Leonhard KA, Byrd DT, Kimble J. Cellular analyses of the mitotic region in the Caenorhabditis elegans adult germ line. Mol Biol Cell. 2006;17:3051–3061. doi: 10.1091/mbc.E06-03-0170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson ST, Gao D, Lambie EJ, Kimble J. lag-2 may encode a signaling ligand for the GLP-1 and LIN-12 receptors of C. elegans. Development. 1994;120:2913–2924. doi: 10.1242/dev.120.10.2913. [DOI] [PubMed] [Google Scholar]
- Gumienny TL, Lambie E, Hartwieg E, Horvitz HR, Hengartner MO. Genetic control of programmed cell death in the Caenorhabditis elegans hermaphrodite germline. Development. 1999;126:1011–1022. doi: 10.1242/dev.126.5.1011. [DOI] [PubMed] [Google Scholar]
- Martinez NJ, Ow MC, Reece-Hoyes JS, et al. Genome-scale spatiotemporal analysis of Caenorhabditis elegans microRNA promoter activity. Genome Res. 2008;18:2005–2015. doi: 10.1101/gr.083055.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim LP, Lau NC, Garrett-Engele P, et al. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- Bagga S, Bracht J, Hunter S, et al. Regulation by let-7 and lin-4 miRNAs results in target mRNA degradation. Cell. 2005;122:553–563. doi: 10.1016/j.cell.2005.07.031. [DOI] [PubMed] [Google Scholar]
- Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466:835–840. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehwinkel J, Natalin P, Stark A, et al. Genome-wide analysis of mRNAs regulated by Drosha and Argonaute proteins in Drosophila melanogaster. Mol Cell Biol. 2006;26:2965–2975. doi: 10.1128/MCB.26.8.2965-2975.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammell M, Long D, Zhang L, et al. mirWIP: microRNA target prediction based on microRNA-containing ribonucleoprotein-enriched transcripts. Nat Methods. 2008;5:813–819. doi: 10.1038/nmeth.1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambros V, Lee RC, Lavanway A, Williams PT, Jewell D. MicroRNAs and other tiny endogenous RNAs in C. elegans. Curr Biol. 2003;13:807–818. doi: 10.1016/s0960-9822(03)00287-2. [DOI] [PubMed] [Google Scholar]
- Park JK, Liu X, Strauss TJ, McKearin DM, Liu Q. The miRNA pathway intrinsically controls self-renewal of Drosophila germline stem cells. Curr Biol. 2007;17:533–538. doi: 10.1016/j.cub.2007.01.060. [DOI] [PubMed] [Google Scholar]
- Yang Y, Xu S, Xia L, et al. The bantam microRNA is associated with drosophila fragile X mental retardation protein and regulates the fate of germline stem cells. PLoS Genet. 2009;5:e1000444. doi: 10.1371/journal.pgen.1000444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iovino N, Pane A, Gaul U. miR-184 has multiple roles in Drosophila female germline development. Dev Cell. 2009;17:123–133. doi: 10.1016/j.devcel.2009.06.008. [DOI] [PubMed] [Google Scholar]
- Kaneda M, Tang F, O'Carroll D, Lao K, Surani MA. Essential role for Argonaute2 protein in mouse oogenesis. Epigenetics Chromatin. 2009;2:9. doi: 10.1186/1756-8935-2-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh N, Baehner L, Moltzahn F, et al. MicroRNA function is globally suppressed in mouse oocytes and early embryos. Curr Biol. 2010;20:271–277. doi: 10.1016/j.cub.2009.12.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang F, Kaneda M, O'Carroll D, et al. Maternal microRNAs are essential for mouse zygotic development. Genes Dev. 2007;21:644–648. doi: 10.1101/gad.418707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murchison EP, Stein P, Xuan Z, et al. Critical roles for Dicer in the female germline. Genes Dev. 2007;21:682–693. doi: 10.1101/gad.1521307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi K, Chuva de Sousa Lopes SM, Kaneda M, et al. MicroRNA biogenesis is required for mouse primordial germ cell development and spermatogenesis. PLoS One. 2008;3:e1738. doi: 10.1371/journal.pone.0001738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maatouk DM, Loveland KL, McManus MT, Moore K, Harfe BD. Dicer1 is required for differentiation of the mouse male germline. Biol Reprod. 2008;79:696–703. doi: 10.1095/biolreprod.108.067827. [DOI] [PubMed] [Google Scholar]
- Tam OH, Aravin AA, Stein P, et al. Pseudogene-derived small interfering RNAs regulate gene expression in mouse oocytes. Nature. 2008;453:534–538. doi: 10.1038/nature06904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T, Takeda A, Tsukiyama T, et al. Identification and characterization of two novel classes of small RNAs in the mouse germline: retrotransposon-derived siRNAs in oocytes and germline small RNAs in testes. Genes Dev. 2006;20:1732–1743. doi: 10.1101/gad.1425706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T, Totoki Y, Toyoda A, et al. Endogenous siRNAs from naturally formed dsRNAs regulate transcripts in mouse oocytes. Nature. 2008;453:539–543. doi: 10.1038/nature06908. [DOI] [PubMed] [Google Scholar]
- Cinquin O, Crittenden SL, Morgan DE, Kimble J. Progression from a stem cell-like state to early differentiation in the C. elegans germ line. Proc Natl Acad Sci USA. 2010;107:2048–2053. doi: 10.1073/pnas.0912704107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadyk LC, Kimble J. Genetic regulation of entry into meiosis in Caenorhabditis elegans. Development. 1998;125:1803–1813. doi: 10.1242/dev.125.10.1803. [DOI] [PubMed] [Google Scholar]
- Zisoulis DG, Lovci MT, Wilbert ML, et al. Comprehensive discovery of endogenous Argonaute binding sites in Caenorhabditis elegans. Nat Struct Mol Biol. 2010;17:173–179. doi: 10.1038/nsmb.1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M, Liu P, Zhang L, et al. mir-35 is involved in intestine cell G1/S transition and germ cell proliferation in C. elegans. Cell Res. 2011;21:1605–1618. doi: 10.1038/cr.2011.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govindan JA, Nadarajan S, Kim S, Starich TA, Greenstein D. Somatic cAMP signaling regulates MSP-dependent oocyte growth and meiotic maturation in C. elegans. Development. 2009;136:2211–2221. doi: 10.1242/dev.034595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadarajan S, Govindan JA, McGovern M, Hubbard EJ, Greenstein D. MSP and GLP-1/Notch signaling coordinately regulate actomyosin-dependent cytoplasmic streaming and oocyte growth in C. elegans. Development. 2009;136:2223–2234. doi: 10.1242/dev.034603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blelloch R, Anna-Arriola SS, Gao D, et al. The gon-1 gene is required for gonadal morphogenesis in Caenorhabditis elegans. Dev Biol. 1999;216:382–393. doi: 10.1006/dbio.1999.9491. [DOI] [PubMed] [Google Scholar]
- Smyth GK.Linear models and empirical bayes methods for assessing differential expression in microarray experiments Stat Appl Genet Mol Biol 20043Article3. [DOI] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Analyses of alg-1(tm492) and alg-1(RNAi) animals germline.
Specificity of the ALG-1 specific polyclonal antibody.
Set of miRNA mutants with no apparent germline phenotypes.
The list of genes that is mis-regulated in alg-1 (gk214) gonads.
Predictive mRNA targets of microRNAs expressed in gonads distal tip cells that are important for germline proliferation and differentiation.