Abstract
G-quadruplex has demonstrated its biological functions in vivo. Although G-quadruplex in single-stranded DNA (ssDNA) has been well characterized, investigation of this species in double-stranded DNA (dsDNA) lags behind. Here we use chemical footprinting and laser-tweezers-based single-molecule approaches to demonstrate that a dsDNA fragment found in the insulin-linked polymorphic region (ILPR), 5′-(ACA GGGG TGT GGGG)2 TGT, can fold into a G-quadruplex at pH 7.4 with 100 mM K+, and an i-motif at pH 5.5 with 100 mM Li+. Surprisingly, under a condition that favors the formation of both G-quadruplex and i-motif (pH 5.5, 100 mM K+), a unique determination of change in the free energy of unfolding (ΔGunfold) by laser-tweezers experiments provides compelling evidence that only one species is present in each dsDNA. Under this condition, molecules containing G-quadruplex are more stable than those with i-motif. These two species have mechanical stabilities (rupture force ≥ 17 pN) comparable to the stall force of RNA polymerases, which, from a mechanical perspective alone, could justify a regulatory mechanism for tetraplex structures in the expression of human insulin.
Introduction
A DNA sequence with tandem G-rich repeats can fold into a tetraplex structure called G-quadruplex (1), which consists of a stack of four guanine residues interconnected by Hoogsteen basepairs (2–4). These G-rich sequences are prevalent in the human genome, particularly in promoter regions (5). Given the demonstrated biological roles of G-quadruplexes in gene-expressing and processing events (6–8), it is imperative to determine the mechanical stabilities of G-quadruplex in the context of naturally occurring double-stranded DNA (dsDNA). Such information can be used to evaluate tetraplexes as regulatory elements for replication and transcription processes in which the mechanical interaction between secondary DNA structures and motor proteins may play a significant role.
In the complementary strand of G-rich repeats, another tetraplex structure, i-motif (9), can form. An i-motif is composed of a stack of hemiprotonated cytosine-cytosine (C:CH+) pairs. Due to the hemiprotonated nature of the C:CH+ pairs, the formation of i-motif often requires acidic conditions. However, depending on particular C-rich sequences, i-motif can fold close to neutral pH (10). Folding at similar pH has also been observed in a DNA template with negative superhelicity (11) or in a molecular crowding environment (12,13), two physiologically relevant conditions. With the recent discovery of proteins that can bind specifically to i-motif-forming DNA fragments, the biological relevance of this structure has begun to emerge (14–16).
The majority of investigations into the DNA tetraplexes have focused on the properties of G-quadruplexes in the context of single-stranded DNA (ssDNA). Recent reports have started to show the formation of G-quadruplexes in the context of dsDNA templates. For example, interconversion between duplex and quadruplex structures has been observed on a DNA hairpin (17) or in a DNA fragment without flanking dsDNA handles (18–22). To address the effect of dsDNA junctions on the DNA tetraplex (23), researchers have investigated the formation of G-quadruplex on dsDNA fragments flanked by long dsDNA handles under specialized conditions, such as in a molecular crowding environment or on a negative supercoiled template (11,24). Interestingly, a single-molecule fluorescence resonance energy transfer analysis on a 96-bp duplex DNA in the c-kit region showed that G-quadruplex can form in the presence of a complementary strand without these specialized settings (25). Likewise, the formation of G-quadruplexes has been demonstrated in dsDNA by dimethyl sulfate (DMS) footprinting and stop assay analyses under similar conditions (24,26). These observations suggest that G-quadruplexes can readily compete with the reannealing of dsDNA in vitro.
With the formation of a G-quadruplex, the complementary C-rich strand becomes single-stranded, which should facilitate the folding of i-motif. However, tetraplex investigations in the dsDNA context are often performed at neutrality, a condition in which it is difficult to form an i-motif. As a result, it is still an open question whether an i-motif can fold in the complementary single-stranded region after a G-quadruplex is formed in the opposite strand. The answer to this question has two important implications. First, the formation of an i-motif is expected to change the formation kinetics or thermodynamic stability of a proximal G-quadruplex in the opposing strand. This may influence the biological functions of the G-quadruplex. Second, it is likely that i-motif has its own biological roles in gene regulation. Together with G-quadruplex, this can introduce a functional versatility for DNA tetraplexes. Compared with the study of G-quadruplex in the dsDNA context, however, relatively few investigations have focused on the formation of i-motif in dsDNA (11,21).
Here, by employing laser-tweezers-based single-molecule methods and DMS/bromine (Br2) footprinting approaches, we obtained compelling evidence that either G-quadruplex or i-motif, but not both, is formed in a long dsDNA construct that contains an insulin-linked polymorphic region (ILPR) fragment, without template superhelicity or molecular crowding environment. We observed that G-quadruplex is more stable than i-motif in the presence of a respective complementary strand.
Materials and Methods
Unless otherwise specified, all DNA fragments were purchased from Integrated DNA Technology (Coralville, IA) and purified on a 10% polyacrylamide gel electrophoresis (PAGE) gel before use. All chemicals (with purity > 95%) were purchased from VWR (West Chester, PA) and used directly without further treatment unless otherwise specified.
DMS and Br2 footprinting
We first labeled a 87-mer ssDNA strand containing an ILPR G-quadruplex forming sequence (underlined; 5′- CTA GAC GGT GTG AAA TAC CGC ACA GAT GCG ACA GGGG TGT GGGG ACA GGGG TGT GGGG ACA GCC AGC AAG ACG TAG CCC AGC GCG TC; G-rich strand) or an ILPR i-motif forming sequence (underlined; 5′- GAC GCG CTG GGC TAC GTC TTG CTG GC TGT CCCC ACA CCCC TGT CCCC ACA CCCC TGT CGC ATC TGT GCG GTA TTT CAC ACC GTC TAG; C-rich strand) at the 5′ end with 32P by incubating the DNA with T4 polynucleotide kinase (New England Biolabs, Ipswich, MA) and [γ-P32] ATP (Perkin-Elmer, Waltham, MA). Labeled DNA was purified using G-25 columns (GE Healthcare, Buckinghamshire, UK).
For the DMS footprinting, the radiolabeled G-rich strand was mixed with its unlabeled complementary C-rich strand in equimolar ratio (1 μM each in 30 μL) in 10 mM Tris (pH 7.4) or 10 mM MES (pH 5.5) buffer with 100 mM KCl, 100 mM LiCl, or without salt. The mixtures were heated at 95°C for 10 min and quickly quenched by submersion in an ice-water bath or in a water bath set at 30°C. Then 0.3 μL DMS was added to the above sample and incubated for 1.5 min at room temperature. The reaction was stopped by addition of 300 μL of a stop buffer (250 μg/mL salmon sperm DNA, 30% β-mercaptoethanol, and 300 mM sodium acetate) followed by addition of 750 μL of absolute ethanol. The detailed procedure for Br2 footprinting has been described elsewhere (27,28). Briefly, the radiolabeled C-rich strand and unlabeled complementary G-rich strand were prepared separately in 10 mM of sodium phosphate buffer (pH 5.5 or 7.0) supplemented with 100 mM KCl or LiCl. The samples were heated at 95°C for 10 min followed by rapid cooling in a water bath set at 30°C. This was followed by mixing of the complementary strand at equimolar concentration (1 μM) in a 30 μL reaction volume. The samples were incubated at the same temperature for a specific time (15 min to 5 days). Cytosine-specific cleavage was performed by incubating the DNA samples with molecular Br2 generated in situ from the reaction between KBr (0.6 μL, 20 mM) and KHSO5 (0.6 μL, 10 mM). The reactions were performed for 10 min at room temperature, terminated by addition of a stop buffer (250 μg/mL sheared salmon sperm DNA, 300 mM NaAc, and 4 mM HEPES) followed by addition of 750 μL absolute ethanol.
The DNA samples from DMS or Br2 footprinting reactions were ethanol-precipitated and washed twice with 70% ethanol. The dry DNA pellet was suspended in 70 μL freshly prepared piperidine (10%) for 30 min at 90°C. It was dried by vacuum centrifuge and the resultant DNA fragments were resolved on a 6% denaturing PAGE (19:1 bis/acrylamide) gel. The gel was dried, exposed to a PhosphorImager screen overnight (∼12 h), and scanned with a Typhoon 8600 instrument (GE Healthcare). Kodak digital camera software (Eastman Kodak, Rochester, NY) was used to acquire the line scans (Fig. 1) and measure the band intensities.
Figure 1.
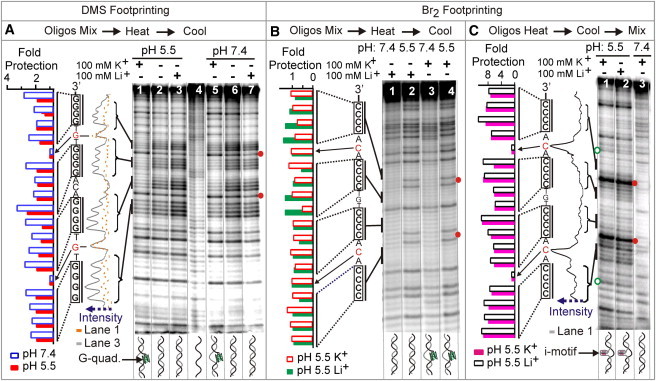
Chemical footprinting of an 87-bp ILPR dsDNA under different pH and salt conditions at 23°C. Black vertical lines depict the G4 and C4 tracts involved in the tetraplex formation. The predominant species is shown at the bottom of each lane. The blue dotted arrows indicate the increasing band intensities. (A) DMS footprinting of the 87-bp dsDNA in 10 mM of MES buffer at pH 5.5 (lanes 1–3) and 10 mM of Tris buffer at pH 7.4 (lanes 5–7). Lane 4 is the GA ladder for the G-rich strand. The orange dotted and gray traces are line scans for lanes 1 and 3, respectively. Fold protections at pH 7.4 (lane 3 versus lane 1, blue open bars) and pH 5.5 (lane 7 versus lane 5, red solid bars) in 100 mM K+ are shown to the left. The red solid dots to the right of the gel show the guanine residues in the TGT loops. (B) Br2 footprinting of 87-bp dsDNA in a pH 7.4 or pH 5.5 sodium phosphate buffer with 100 mM ions. The premixed sample was denatured at 95°C for 10 min and rapidly quenched in a water bath set at 30°C before Br2 treatment. The similar band intensities of all cytosines in the i-motif-forming region (see the quantitation to the left of the gel) indicate that i-motif was not formed under these experimental conditions. The red solid dots to the right of the gel show the cytosine residues in the ACA section. (C) Br2 footprinting of dsDNA at pH 5.5 (lanes 1 and 2) and pH 7.4 (lane 3). G-rich and C-rich strands were heated separately to 95°C and rapidly quenched in a water bath set at 30°C before mixing. The black trace to the left of the gel is the line scan for lane 1. Fold protections at pH 5.5 with 100 mM K+ (lane 3 versus lane 1, pink solid bars) and 100 mM Li+ (lane 3 versus lane 2, black open bars) are shown at the left side. The red solid dots to the right of lane 2 depict cytosines in the ACA loops. The green open circles to the left of lane 1 indicate the cytosines closest to the i-motif-forming region, which are used to quantify the hybridization of flanking DNA strands (see text).
To prepare the GA ladder, a radiolabeled 87-mer G-rich strand was mixed with salmon sperm DNA (0.1 μg/μL) in 10 μL reaction volume; 1 μL of 1 M formic acid (pH 2.0) was added to the above reaction mixture and incubated at 37°C for 30 min, followed by piperidine cleavage (150 μL, 10% piperidine) at 90°C for 30 min. The sample was cooled in an ice-water bath for 5 min and precipitated with n-butanol. The DNA pellet was resuspended in 1% sodium dodecyl sulfate (150 μL), followed by another precipitation with n-butanol. Finally, the DNA pellet was resuspended in a loading buffer for PAGE analysis.
To calculate the fold protection for both DMS and Br2 footprinting experiments (Fig. 1), we measured the net intensity of the bands in the four G4/C4-tracts or the two cytosine bands close to the C4 regions (green open circles in Fig. 1 C) in each lane, normalized against the intensity of the guanine band in the TGT loop in DMS footprinting or the cytosine band in the ACA loop in Br2 footprinting. We determined the fold protection by dividing the band intensity of corresponding G/C in the control lane (Li+ lane for the DMS footprinting at the same pH or the pH 7.4 lane for the Br2 footprinting) by the lane of interest. A similar approach was used to calculate the fold protection of each G/C residue in each G4/C4 tract in the footprinting gel (see Fig. 3 A). The fold protection for a given time (see Fig. 3 B) was averaged from the four G4/C4 tracts with the error bar representing the standard deviation (SD).
Figure 3.
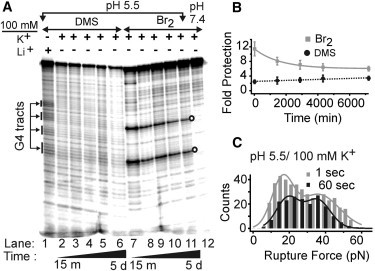
Kinetic analysis of G-quadruplex and i-motif in ILPR dsDNA at pH 5.5. (A) DMS (lanes 1–6) and Br2 (lanes 7–12) footprinting of an equimolar mixture of G-rich and C-rich strands after the heat→cool→mix procedure (see text). All lanes are from experiments performed in 10 mM of MES buffer at pH 5.5 with 100 mM K+, except for lane 1 (with 100 mM Li+) and lane 12 (pH 7.4). Lanes 2–6 and lanes 7–11 represent footprinting on the dsDNA samples taken from five intervals during 5 days. Open circles at the right of lane 11 depict the cytosines in the ACA loops. (B) Fold protection of C4 or G4 tracts over time. DMS (solid circles) and Br2 (squares) footprinting results are fitted with a dotted line and an exponential curve, respectively. (C) Rupture force histograms for structures in the ILPR dsDNA by mechanical unfolding experiments performed in 10 mM of MES buffer at pH 5.5 with 100 mM K+. The DNA molecules were incubated at zero force for 1 s (gray) or 60 s (black) before unfolding.
DNA construct preparation
Two DNA oligonucleotides containing ILPR G-quadruplex (5′-CACA GGGG TGT GGGG ACA GGGG TGT GGGG T) and i-motif (5′-CTAGA CCCC ACA CCCC TGT CCCC ACA CCCC TGTGGTAC) forming sequences were annealed with an equimolar ratio to form dsDNA in a 10 mM Tris (pH 8.0) buffer by heating at 97°C for 10 min, followed by slow cooling to 25°C in ∼6 h. At each end of the dsDNA, an XbaI or a KpnI restriction site was introduced to allow the cloning of the DNA into the polycloning site of the plasmid pFOXCAT-362hIns (a gift from Michael German's laboratory) (29). The plasmid with the ILPR G-quadruplex/i-motif insert was then transformed to Escherichia coli (GM2163 strain; Fermentas) for amplification. Bacterial cells were collected through centrifugation after overnight shaking in LB growth medium, and the plasmid was extracted with a Maxiprep kit (Qiagen, Valencia, CA). The ILPR G-quadruplex/i-motif insert was validated by DNA sequencing (DNA Sequencing Facility, University of Maine, Orono, ME). The plasmid containing the ILPR G-quadruplex/i-motif-forming sequence was digested with AflIII (New England Biolabs), labeled with biotin-dUTP (eENZYME, Gaithersburg, MD) using a Klenow enzyme (New England Biolabs) followed by ethanol precipitation. The DNA was then digested by SacI (New England Biolabs), and the larger DNA fragment (∼4 kb) was purified by an agarose gel. The fragment DNA was labeled with digoxigenin at the SacI end with dig-dUTP (Roche, Indianapolis, IN) using terminal transferase (Fermentas, Glen Burnie, MD). Finally, the sample was purified by ethanol precipitation. To prepare a DNA construct without G-quadruplex/i-motif-forming sequences, the pFOXCAT-362hIns plasmid without insert was digested with AflIII and labeled with biotin-dUTP, followed by digestion with SacI and labeled with dig-dUTP as described above.
Single-molecule experiments
The laser-tweezers instrument used for the single-molecule experiments is described elsewhere (30,31). To start the single-molecule experiments, the end-labeled DNA construct was incubated with anti-digoxigenin (Dig) antibody-coated polystyrene particles (2.10 μm diameter; Spherotech, Lake Forest, IL) for ∼1 h to bind the DNA molecules to the bead surface via Dig/anti-Dig antibody linkage. The anti-Dig antibody-coated beads carrying the DNA molecules and the streptavidin-coated beads (0.97 μm diameter; Spherotech) were dispersed in 700 μL of 10 mM buffer (Tris or MES) with 100 mM ions (K+ or Li+) at pH 5.5 or 7.4. They were separately injected in a reaction chamber. The two types of beads were separately trapped by two laser foci in the reaction chamber. The bead carrying the DNA molecules on its surface was moved toward the streptavidin-coated bead so that the free end of the attached DNA molecule could bind to the latter bead via the biotin/streptavidin linkage. Once the DNA was tethered between the beads, the anti-Dig antibody-coated bead was moved away at a load rate of ∼5.5 pN/s to stretch the tethered DNA until a secondary structure in the DNA construct was ruptured. After a specific force was reached (maximum 60 pN), the force was relaxed to zero at the same load rate to allow the structure to refold before subsequent stretching. The single tether was confirmed by a single broken event for the tethered DNA construct. The raw data were recorded at 1000 Hz in LabVIEW (National Instruments, Austin, TX) and Savitzky-Golay filtered to 100 Hz by MATLAB (The MathWorks, Natick, MA), followed by analyses using IGOR (WaveMatrics, Portland, OR) programs. The rupture force was measured directly from the force-extension (F-X) curves, and the change in contour length (ΔL) was calculated from the two data points flanking the rupture event using the worm-like chain model (32) given below:
| (1) |
where x is the end-to-end distance, kB is the Boltzmann constant, T is the absolute temperature, P is the persistent length (51.95 nm (32)), F is the force, and S is the elastic stretch modulus (1226 pN (32)). To obtain the SD in the single-molecule experiments, we performed at least three independent sets of experiments.
Control experiments for single-molecule studies
We performed a number of control experiments at the single-molecule level to validate that the unfolding events observed were due to the unfolding of G-quadruplex or i-motif structures. We conducted these experiments by choosing a buffer condition in which the formation of G-quadruplex or i-motif is discouraged. Alternatively, mechanical unfolding was performed on the DNA construct containing dsDNA handles only (see “DNA construct preparation” above and Table S1 in the Supporting Material).
Calculation of the percentage of formation
For any unfolding events observed in a given buffer, the percentage of unfolding events was calculated as the ratio of the DNA tethers that contain folded secondary structures versus the total number of DNA tethers. To avoid the repetitive counting, each molecule was only counted once. The results for these experiments are summarized in Table 1 and Table S1.
Table 1.
Change in contour length (ΔL), rupture force (Funfold), free energy of unfolding (ΔGunfold), bias of ΔGunfold, and percentage formation (% formation) for fully folded tetraplex structures under different pH and ionic conditions at 23°C
| pH | Salt ions | ΔL (nm) | Funfold (pN) | ΔGunfold (kcal/mol) | Bias of ΔGunfold (kcal/mol) | % Formation |
|---|---|---|---|---|---|---|
| 7.4 | K+ | 6.7 (±0.2) | 24 (±1) | 7.8 (±1.4) | −0.5 | 18 |
| 5.5 | Li+ | 7.2 (±0.1) | 28 (±2) | 8.7 (±1.0) | 1.0 | 34 |
| 5.5 | K+ | 6.7 (±0.2) | 17 (±1) (left peak) | 7.4 (±0.5) (left peak) | 1.2 | 14 |
| 36 (±1) (right peak) | 10.4 (±1.2) (right peak) | 1.1 | 19 |
Values in parentheses are SDs.
Deconvolution of populations
In a histogram with two populations (Fig. 2 C (middle panel) and D (bottom panel), and Fig. S2), the overall population was fit with a two-peak Gaussian function. To account for the stochastic behavior of individual molecules, the population under the intersection region was randomly assigned to one of the populations based on the ratio determined by the Gaussian fitting. The change in contour length (ΔL) and unfolding force histograms were plotted separately for individual populations according to this random assignment. The change in free energy of unfolding (ΔGunfold) was calculated using Eq. 2 (see below) for each population after this deconvolution.
Figure 2.
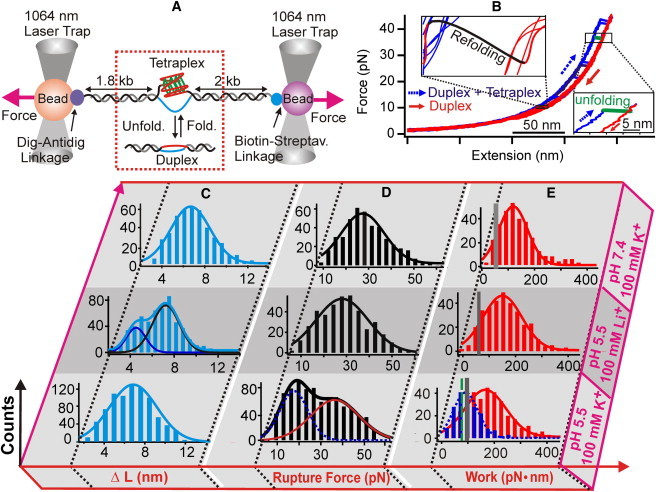
Single-molecule investigation of G-quadruplex/i-motif in ILPR dsDNA. (A) Schematic of the laser-tweezers setup. The interconversion between tetraplex and duplex is highlighted in the red dotted box. (B) An overlay of seven F-X curves from the same molecule at pH 5.5 in a 10 mM MES buffer with 100 mM K+. The stretching and relaxing curves are indicated by blue dotted and red solid arrows, respectively. Top and bottom insets are blowup regions that show the refolding (black) and unfolding (green bold line) events, respectively. (C) Change in contour length (ΔL) histograms. (D) Unfolding force (Funfold) histograms for fully folded species. (E) Unfolding work histograms obtained from the hysteresis area between the stretching and relaxing F-X curves. The vertical shaded strips in E represent the work (mean ± SD) equivalent to the ΔGunfold (dashed green and gray lines in the bottom panel correspond to the blue dotted and red solid histograms, respectively). Experiments were performed in 10 mM of Tris buffer (top panel: pH 7.4, 100 mM K+), or 10 mM of MES buffer (middle panel: pH 5.5, 100 mM Li+; bottom panel: pH 5.5, 100 mM K+). Histograms are fitted with one- or two-peak Gaussian functions.
Results
Chemical footprinting shows that the ILPR G-quadruplex or i-motif is stable in the context of dsDNA
To investigate the effect of the complementary strand on the ILPR tetraplex formation, we first performed DMS footprinting on a 87-bp DNA, 5′-CTA GAC GGT GTG AAA TAC CGC ACA GAT GCG ACA GGGG TGT GGGG ACA GGGG TGT GGGG ACA GCC AGC AAG ACG TAG CCC AGC GCG TC, which consists of a 5′-32P labeled G-rich strand and an unlabeled complementary C-rich strand (Fig. 1 A). This fragment contains an ILPR G-quadruplex-forming sequence (underlined) (24,33,34) sandwiched between a 33-bp sequence and a 29-bp sequence. After these two strands were mixed, incubated at 95°C, and rapidly cooled in an ice-water bath (the mix→heat→cool procedure; Fig. 1 A), they were subjected to DMS footprinting (see Materials and Methods). It is noteworthy that fast temperature quenching was carried out to be consistent with our mechanical unfolding experiment in which the DNA molecules were quickly (∼5.5 pN/s) relaxed to zero force after unfolding (see below). PAGE gel showed less DMS modification for the four G4 tracts, as manifested by the reduced band intensity due to less piperidine cleavage, in a 10 mM MES buffer (lane 1 in Fig. 1 A: pH 5.5 with 100 mM K+) or in a 10 mM Tris buffer (lane 5: pH 7.4 with 100 mM K+; for detailed quantitation, see Materials and Methods). Compared with the control in which G-quadruplex is not expected to form (lanes 2 and 6: without salt; lanes 3 and 7: with 100 mM Li+), the increased cleavage protection in the four G4 tracts suggests the formation of a G-quadruplex employing these G4 tracts. Calculation of the fold protection showed ∼2.5-fold higher protection of these G4 tracts in K+ compared with Li+ at the same pH.
Next, we performed Br2 footprinting to probe the formation of i-motif in the same dsDNA, which contains a 5′-32P labeled i-motif forming C-rich strand (14,27,35), and an unlabeled complementary G-rich strand (see sequence above). However, samples treated with the same procedure (mix→heat→cool) used in the DMS footprinting did not show any formation of i-motif in the pH 5.5 buffer with 100 mM KCl (Fig. 1 B). Whereas lane 2 suggests that the folding of i-motif in the presence of a complementary strand cannot compete with the duplex formation during the mix→heat→cool process, lane 4 implies that with the same process, the coexistence of the i-motif and the G-quadruplex that formed in the complementary strand is not favored. This surprising result indicates that in the single-stranded region complementary to the strand where G-quadruplex has already formed (Fig. 1 A), i-motif does not fold even under favorable pH, possibly due to steric hindrance (see Discussion).
We further evaluated the stability of a preformed i-motif in the presence of a complementary strand. To that end, the G-rich and C-rich strands were heated at 95°C and cooled separately before mixing at 30°C for 15 min (the heat→cool→mix procedure). Notice that this procedure is different from the previous practice in which two complementary strands were mixed before the heating and cooling steps. Compared with the cytosines in the ACA loops, lanes 1 and 2 in Fig. 1 C show significantly reduced (∼9-fold) Br2 cleavage of cytosines closest to the i-motif-formation regions (cytosines highlighted by the green open circles). These observations are in accordance with the previous finding that cytosines in dsDNA are ∼10 times more protected than those in the ssDNA context (28). In the i-motif-forming region, the protection of the C4 tracts (highlighted with vertical black lines) with respect to the loop cytosines was obvious in the pH 5.5 MES buffer with either 100 mM K+ (lane 1, Fig. 1 C) or Li+ (lane 2), which shows that under the heat→cool→mix procedure, the preformed i-motif is stable in the presence of the complementary strand for at least 15 min. Further evidence indicated that such a preformed i-motif was stable in dsDNA even after 5 days (see below). As a control, no protection of cytosines was observed in the pH 7.4 Tris buffer (lane 3, Fig. 1 C) in which ILPR i-motif was not expected to form (27). These experiments demonstrate the strong stability of the ILPR G-quadruplex and the preformed i-motif in the dsDNA context.
Laser-tweezers experiments confirm the presence of ILPR tetraplexes in a dsDNA template
The formation of folded structures in the duplex ILPR DNA was confirmed by laser-tweezers-based single-molecule experiments (30,31) (Fig. 2 A). The dsDNA construct consisted of the ILPR tetraplex-forming sequence, 5′-CACA GGGG TGT GGGG ACA GGGG TGT GGGG T, which was sandwiched between two dsDNA handles of 1800 bp and 2000 bp in length, respectively (see Materials and Methods). This construct was tethered between two optically trapped beads through one of the strands to prevent the accumulation of template superhelicity. The two beads were moved apart to stretch the DNA construct until an unfolding event was observed (Fig. 2 B) in a 10 mM Tris (pH 7.4) or 10 mM MES (pH 5.5) buffer with different ionic conditions (100 mM K+ or Li+). Histograms of the change in contour length (ΔL) due to the unfolding, the rupture force histograms, and the unfolding work histograms are plotted in Fig. 2, C–E.
In these histograms, the ΔL (the ∼7 nm population in Fig. 2 C and Table 1) under all three conditions corresponds to the unfolding of a structure that contains ∼25 nucleotides (see Fig. S1 for calculation). Within experimental error, this number is identical for the nucleotides employed in a fully folded G-quadruplex or i-motif structure in ILPR dsDNA. We also observed a partially folded C-rich species at pH 5.5 with 100 mM Li+ (ΔL ∼4.5 nm population in the middle panel of Fig. 2 C), which is consistent with previous findings (27) under similar conditions.
Unfolding force histograms for the fully folded species showed a single population in a pH 7.4 Tris buffer with 100 mM K+ (Funfold = 24 ± 1 pN; Fig. 2 D, top) or in a pH 5.5 MES buffer with 100 mM Li+ (Funfold = 28 ± 2 pN; Fig. 2 D, middle). Because only G-quadruplex can form under the former condition, and only i-motif can fold in the latter buffer (Fig. 1) (27,34), we assigned the folded species at pH 7.4 as the G-quadruplex and those at pH 5.5 as the i-motif. These assignments were confirmed by control experiments that showed little formation of any fully folded structures (3.2% at pH 7.4 with 100 mM Li+; Fig. S2 and Table S1), which was expected for the respective tetraplexes (25,27,34). In comparison, the experiments shown in Fig. 2, C and D, had a percentage of formation in the range of 18–34% (Table 1). It is noteworthy that the unfolding force for these ILPR structures has a broad distribution, in similarity to those of tetraplexes in the ssDNA context (27,34). This may reflect the fact that DNA tetraplex structures are rather dynamic (36,37). The single-molecule experiments described here corroborate the footprinting findings that either G-quadruplex or i-motif can stably exist in the dsDNA context.
G-quadruplex and i-motif are mutually exclusive in ILPR dsDNA fragments
In a pH 5.5 buffer (10 mM MES) with 100 mM K+, two fully folded populations with unfolding forces of 17 (±1) and 36 (±1) pN were observed (Fig. 2 D, bottom panel, and Table 1). A G-quadruplex, an i-motif, or a G-quadruplex with an i-motif can form in the dsDNA in this buffer. To identify the exact species, we compared the change in free energy of unfolding (ΔGunfold) for structures formed in each buffer. The mechanical unfolding allowed us to retrieve ΔGunfold by applying the Jarzynski equation for nonequilibrium systems (38,39):
| (2) |
where N is the number of observations, kB is the Boltzmann constant, T is absolute temperature, and W is the nonequilibrium work done to unfold the structure(s), which is equivalent to the hysteresis area between stretching and relaxing F-X curves (see Fig. 2 B as an example, and Materials and Methods for the detailed calculation). This method recovers ΔGunfold by using an exponentially weighted algorithm to count for the dissipated work during the nonequilibrated unfolding processes. According to this weighting pattern, smaller unfolding work contributes more to the change in free energy. Therefore, ΔGunfold is expected to be smaller than the average unfolding work performed under nonequilibrium conditions. When histograms of this unfolding work were plotted for species in all three buffer conditions, we indeed observed that values of ΔGunfold were significantly below the average work (shaded strips in Fig. 2 E). In previous studies, this method allowed accurate retrieval of the ΔGunfold for hairpins (39) and tetraplexes in ssDNA (27,34).
As shown in Table 1, the ΔGunfold values were similar between a G-quadruplex (7.8 ± 1.4 kcal/mol, pH 7.4 with 100 mM K+) and an i-motif (8.7 ± 1.0 kcal/mol, pH 5.5 with 100 mM Li+). When the two populations at pH 5.5 with 100 mM K+ were deconvoluted (Fig. 2 D, bottom panel, and Materials and Methods), similar values of ΔGunfold were obtained for these species (ΔGunfold for the high-force population: 10.4 ± 1.2 kcal/mol; ΔGunfold for the low-force population: 7.4 ± 0.5 kcal/mol; Fig. 2 E, bottom panel, and Table 1). The accuracy of these ΔGunfold values was reflected by their small biases (within ±1.2 kcal/mol; Table 1), which were calculated according to the literature (40). In fact, these ΔGunfold values are in the same range as that of a stand-alone G-quadruplex or an i-motif in the dsDNA context (see above). They are approximately half the value that would be expected to unfold a G-quadruplex and an i-motif together (16.5 kcal/mol). These results indicate that only one tetraplex structure (either i-motif or G-quadruplex, but not both) is formed in a given dsDNA molecule. Such a scenario is fully consistent with the footprinting results (Fig. 1, A and B) that indicate only G-quadruplex, and not i-motif, is formed in the dsDNA when complementary strands are mixed before the heating and cooling steps (see Discussion below).
Kinetic experiments reveal that G-quadruplex is more stable than i-motif in dsDNA
To identify each species in the pH 5.5 MES buffer with 100 mM K+ (Fig. 2 D, bottom), we performed kinetic footprinting experiments using the heat→cool→mix procedure. First, two complementary strands used in the chemical footprinting were heated separately at 95°C for 10 min and quenched rapidly in a water bath set at 30°C, a temperature significantly below the Tm of the ILPR G-quadruplex (77°C and 88°C for parallel and antiparallel G-quadruplexes in ssDNA, respectively (34)) or the Tm of the ILPR i-motif (37°C (27)) in ssDNA. This facilitates the formation of respective structures in ssDNA. Note that even for the ILPR i-motif, 30°C is well within the plateau of the melting curve that indicates fully folded structures (data not shown) (27). These two ssDNA strands with preformed tetraplex structures were then mixed at equimolar ratio and incubated at 30°C to facilitate the hybridization of flanking dsDNA handles over 5 days. To allow the DMS or Br2 footprinting on G-quadruplex (lanes 2–6, Fig. 3 A) or i-motif (lanes 7–11), respectively, either the G-rich or the C-rich strand in the DNA mixture was 5′-32P labeled. Control experiments were performed at pH 5.5 with 100 mM Li+ for DMS footprinting (lane 1) or at pH 7.4 for Br2 (lane 12) footprinting assays. The quantitation (see Materials and Methods) showed that whereas the G-quadruplex population remained relatively constant over time (dotted line, Fig. 3 B), the i-motif decreased slowly (solid curve), and its presence was still significant after 5 days. In addition, the i-motif population monitored by Br2 footprinting at pH 5.5 with 100 mM Li+, a condition in which the G-quadruplex is not expected to form, showed a similar decrease in the i-motif population (data not shown) over 15 min to 3 days. These results suggest that G-quadruplex is thermodynamically more stable than i-motif in the presence of a corresponding complementary strand.
To correlate the kinetic property of tetraplex species obtained from footprinting experiments, we performed a kinetic analysis on the mechanical unfolding of these species in the pH 5.5 MES buffer with 100 mM K+. After unfolding a specific tetraplex structure, we varied the incubation time at 0 pN to allow the refolding of the structure. We then carried out the next round of the stretching procedure, in which the observation of a rupture event would indicate that a tetraplex had been folded during the incubation (41). When we analyzed the rupture force populations at two incubation times (1 s and 60 s), we found that the high-force species increased its population with respect to the low-force species over time (Fig. 3 C; the ratio of high-force to low-force population decreased from 1.6 to 1.1). Because the footprinting assays suggested that the i-motif population decreases with time, we assigned the high-force species as G-quadruplex and the decreasing low-force population as i-motif. This assignment is consistent with the fact that the ILPR G-quadruplex (ΔGunfold, 10.4 kcal/mol; Table 1) has higher stability than the ILPR i-motif (ΔGunfold, 7.4 kcal/mol; Table 1) in the same buffer (27,34). Compared with the ΔGunfold for tetraplexes in the context of ssDNA, changes in the free energy of unfolding for the tetraplexes in dsDNA were significantly smaller. This could be due to the reannealing of the complementary strands after unfolding of the tetraplexes (Fig. 2 A; due to the force-induced melting discussed below, partial, rather than full, reannealing is more likely), which reduces the free energy of the unfolded state with respect to the ssDNA.
Fig. 4 summarizes our observations. In a pH 7.4 buffer with 100 mM Li+, the duplex DNA was predominant (Fig. 4, top left), as suggested by the few unfolding events (Fig. S2 and Table S1) and lack of G4 or C4 protection in the footprinting experiments (Fig. 1, A and B). At pH 7.4 with 100 mM K+, G-quadruplex formed predominantly (Fig. 4, top right). In a pH 5.5 buffer with 100 mM Li+, i-motif prevailed (Fig. 4, bottom left). Finally, at pH 5.5 buffer with 100 mM K+, either G-quadruplex or i-motif, but not both, formed in each dsDNA (Fig. 4, bottom right). Whereas the G-quadruplex was stable over time, the i-motif reduced its population during the same period.
Figure 4.
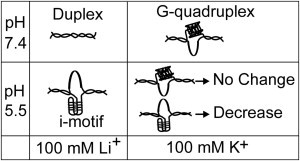
Major species under different pH and ionic conditions.
Discussion
The fact that i-motif did not fold in the dsDNA context after the thermal melting procedure (Fig. 1 B) was probably due to competition from the dsDNA formation during the reannealing. Previously, ILPR i-motif in the single-stranded C-rich sequence showed a melting temperature of 37°C (27), which is much lower than the Tm expected for the dsDNA with the same sequence (80.5°C by nearest-neighbor calculation (42)). Therefore, there is a temperature range (37–80.5°C) in which dsDNA prevails over i-motif. At lower temperatures, the formation of i-motif becomes rather difficult due to the insurmountable energy barrier to separate the two complementary strands that have already been hybridized. On the other hand, ILPR G-quadruplex formed in the single-stranded G-rich sequence has a Tm of ∼77°C and ∼88°C for parallel and antiparallel quadruplexes, respectively (34). Because these Tm values are comparable to that for dsDNA, a mixture of dsDNA and G-quadruplex is expected to form during the cooling. Once the G-quadruplex is formed (Fig. 1 A), it generates a single-stranded complementary region in which i-motif may form. However, Br2 footprinting showed no evidence of i-motif formation (Fig. 1 B). This result strongly supports our laser-tweezers finding that only one tetraplex, in this case G-quadruplex, can form in dsDNA (Fig. 2 E).
During the mechanical folding/unfolding experiments, we observed that either ILPR G-quadruplex or i-motif formed in the dsDNA construct (Fig. 2), which had been stretched above 30 pN in the previous pulling cycle at room temperature (23°C). Such a result was in contrast to the footprinting assays, which showed that only G-quadruplex, but not i-motif, formed in the dsDNA context. We believe it is the different melting processes that led to the different observations. It is known that dsDNA can melt by the force-induced melting process (43,44). As the tension in the dsDNA increases, it generates locally melted regions that can lead to the formation of either a G-quadruplex or an i-motif. In the thermomelting and reannealing processes, however, the competition from the dsDNA hybridization prevents the formation of i-motif but allows G-quadruplex to form (see above). The unprecedented mutual exclusiveness between the G-quadruplex and i-motif may be due to steric hindrance. It is possible that the formation of one tetraplex restricts the degree of freedom for the folding of the other species in the complementary strand. This has interesting biological implications. For example, it provides versatility in the regulation, because either i-motif or G-quadruplex may be involved, and each species has a different set of interacting proteins (14–16,45–47). In addition, the different stability or formation kinetics in the G-quadruplex and i-motif may play an important role in differential regulation of gene expressions.
In contrast to biochemical techniques, the use of the laser-tweezers approach to determine ΔGunfold provides a unique and straightforward way to evaluate the formation of tetraplexes in dsDNA. The dsDNA construct used in the mechanical folding/unfolding experiments closely resembles the physiological situation in which tetraplex-forming sequences in the promoter are always flanked by dsDNA regions. Likewise, the force-induced melting is physiologically more relevant with respect to the thermal melting or denaturant melting. Whereas there exists little possibility for the latter two processes to occur in vivo, numerous events, such as DNA replication, RNA transcription, and cell growth and division, can generate tension in a DNA template, leading to force-induced melting. The other unique aspect of the laser-tweezers approach is that it can evaluate the mechanical stability of the DNA tetraplexes. Here, the ILPR tetraplex structures in the duplex DNA demonstrate a mechanical stability (Frupture ≥ 17 pN) larger or comparable to the stall force of polymerases (14–25 pN) (48–51). From a mechanical perspective alone, this could justify a possible regulatory role played by a DNA tetraplex in the expression of human insulin inside cells in which dsDNA is the predominant form.
Conclusion
By combining two complementary techniques, chemical footprinting and mechanical folding/unfolding, we were able to obtain convincing evidence that either G-quadruplex or i-motif, but not both, forms in the double-stranded ILPR region. Although our experiments were performed at pH 5.5, we anticipate that general aspects of this conclusion can provide insights into the formation of tetraplexes in dsDNA at physiological pH conditions under which recent studies have shown that, in addition to G-quadruplex, i-motif can also fold (11,52). It is interesting to see whether this conclusion holds under in vivo conditions, such as on a DNA template with negative superhelicity or in a molecular crowding environment.
Acknowledgments
This work was supported by the National Institutes of Health (R15DK081191-01), the New Faculty Award Program of the Camille and Henry Dreyfus Foundation, and the Ohio Board of Regents. R.K. thanks the support from National Science Foundation Research Experiences for Undergraduates (grant No. 1004987).
Supporting Material
References
- 1.Sen D., Gilbert W. Formation of parallel four-stranded complexes by guanine-rich motifs in DNA and its implications for meiosis. Nature. 1988;334:364–366. doi: 10.1038/334364a0. [DOI] [PubMed] [Google Scholar]
- 2.Gellert M., Lipsett M.N., Davies D.R. Helix formation by guanylic acid. Proc. Natl. Acad. Sci. USA. 1962;48:2013–2018. doi: 10.1073/pnas.48.12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Williamson J.R., Raghuraman M.K., Cech T.R. Monovalent cation-induced structure of telomeric DNA: the G-quartet model. Cell. 1989;59:871–880. doi: 10.1016/0092-8674(89)90610-7. [DOI] [PubMed] [Google Scholar]
- 4.Rankin S., Reszka A.P., Neidle S. Putative DNA quadruplex formation within the human c-kit oncogene. J. Am. Chem. Soc. 2005;127:10584–10589. doi: 10.1021/ja050823u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huppert J.L., Balasubramanian S. G-quadruplexes in promoters throughout the human genome. Nucleic Acids Res. 2007;35:406–413. doi: 10.1093/nar/gkl1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Siddiqui-Jain A., Grand C.L., Hurley L.H. Direct evidence for a G-quadruplex in a promoter region and its targeting with a small molecule to repress c-MYC transcription. Proc. Natl. Acad. Sci. USA. 2002;99:11593–11598. doi: 10.1073/pnas.182256799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Paeschke K., Capra J.A., Zakian V.A. DNA replication through G-quadruplex motifs is promoted by the Saccharomyces cerevisiae Pif1 DNA helicase. Cell. 2011;145:678–691. doi: 10.1016/j.cell.2011.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Balasubramanian S., Hurley L.H., Neidle S. Targeting G-quadruplexes in gene promoters: a novel anticancer strategy? Nat. Rev. Drug Discov. 2011;10:261–275. doi: 10.1038/nrd3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gehring K., Leroy J.L., Guéron M. A tetrameric DNA structure with protonated cytosine.cytosine base pairs. Nature. 1993;363:561–565. doi: 10.1038/363561a0. [DOI] [PubMed] [Google Scholar]
- 10.Guo K., Pourpak A., Hurley L.H. Formation of pseudosymmetrical G-quadruplex and i-motif structures in the proximal promoter region of the RET oncogene. J. Am. Chem. Soc. 2007;129:10220–10228. doi: 10.1021/ja072185g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sun D., Hurley L.H. The importance of negative superhelicity in inducing the formation of G-quadruplex and i-motif structures in the c-Myc promoter: implications for drug targeting and control of gene expression. J. Med. Chem. 2009;52:2863–2874. doi: 10.1021/jm900055s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miyoshi D., Matsumura S., Sugimoto N. Duplex dissociation of telomere DNAs induced by molecular crowding. J. Am. Chem. Soc. 2004;126:165–169. doi: 10.1021/ja036721q. [DOI] [PubMed] [Google Scholar]
- 13.Zhao C., Ren J., Qu X. Single-walled carbon nanotubes binding to human telomeric i-motif DNA under molecular-crowding conditions: more water molecules released. Chemistry. 2008;14:5435–5439. doi: 10.1002/chem.200800280. [DOI] [PubMed] [Google Scholar]
- 14.Catasti P., Chen X., Gupta G. Cystosine-rich strands of the insulin minisatellite adopt hairpins with intercalated cytosine+.cytosine pairs. J. Mol. Biol. 1997;272:369–382. doi: 10.1006/jmbi.1997.1248. [DOI] [PubMed] [Google Scholar]
- 15.Lacroix L., Liénard H., Mergny J.L. Identification of two human nuclear proteins that recognise the cytosine-rich strand of human telomeres in vitro. Nucleic Acids Res. 2000;28:1564–1575. doi: 10.1093/nar/28.7.1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marsich E., Piccini A., Manzini G. Evidence for a HeLa nuclear protein that binds specifically to the single-stranded d(CCCTAA)n telomeric motif. Nucleic Acids Res. 1996;24:4029–4033. doi: 10.1093/nar/24.20.4029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hardin C.C., Watson T., Bailey C. Cation-dependent transition between the quadruplex and Watson-Crick hairpin forms of d(CGCG3GCG) Biochemistry. 1992;31:833–841. doi: 10.1021/bi00118a028. [DOI] [PubMed] [Google Scholar]
- 18.Miura T., Thomas G.J., Jr. Structural polymorphism of telomere DNA: interquadruplex and duplex-quadruplex conversions probed by Raman spectroscopy. Biochemistry. 1994;33:7848–7856. doi: 10.1021/bi00191a012. [DOI] [PubMed] [Google Scholar]
- 19.Deng H., Braunlin W.H. Duplex to quadruplex equilibrium of the self-complementary oligonucleotide d(GGGGCCCC) Biopolymers. 1995;35:677–681. doi: 10.1002/bip.360350613. [DOI] [PubMed] [Google Scholar]
- 20.Salazar M., Thompson B.D., Hurley L.H. Thermally induced DNA.RNA hybrid to G-quadruplex transitions: possible implications for telomere synthesis by telomerase. Biochemistry. 1996;35:16110–16115. doi: 10.1021/bi961442j. [DOI] [PubMed] [Google Scholar]
- 21.Phan A.T., Mergny J.-L. Human telomeric DNA: G-quadruplex, i-motif and Watson-Crick double helix. Nucleic Acids Res. 2002;30:4618–4625. doi: 10.1093/nar/gkf597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rangan A., Fedoroff O.Y., Hurley L.H. Induction of duplex to G-quadruplex transition in the c-myc promoter region by a small molecule. J. Biol. Chem. 2001;276:4640–4646. doi: 10.1074/jbc.M005962200. [DOI] [PubMed] [Google Scholar]
- 23.Ren J., Qu X., Chaires J.B. Tiny telomere DNA. Nucleic Acids Res. 2002;30:2307–2315. doi: 10.1093/nar/30.11.2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zheng K.W., Zhang D., Tan Z. Dissecting the strand folding orientation and formation of G-quadruplexes in single- and double-stranded nucleic acids by ligand-induced photocleavage footprinting. J. Am. Chem. Soc. 2011;133:1475–1483. doi: 10.1021/ja108972e. [DOI] [PubMed] [Google Scholar]
- 25.Shirude P.S., Okumus B., Balasubramanian S. Single-molecule conformational analysis of G-quadruplex formation in the promoter DNA duplex of the proto-oncogene c-kit. J. Am. Chem. Soc. 2007;129:7484–7485. doi: 10.1021/ja070497d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Qin Y., Rezler E.M., Hurley L.H. Characterization of the G-quadruplexes in the duplex nuclease hypersensitive element of the PDGF-A promoter and modulation of PDGF-A promoter activity by TMPyP4. Nucleic Acids Res. 2007;35:7698–7713. doi: 10.1093/nar/gkm538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dhakal S., Schonhoft J.D., Mao H. Coexistence of an ILPR i-motif and a partially folded structure with comparable mechanical stability revealed at the single-molecule level. J. Am. Chem. Soc. 2010;132:8991–8997. doi: 10.1021/ja100944j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ross S.A., Burrows C.J. Cytosine-specific chemical probing of DNA using bromide and monoperoxysulfate. Nucleic Acids Res. 1996;24:5062–5063. doi: 10.1093/nar/24.24.5062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Odagiri H., Wang J., German M.S. Function of the human insulin promoter in primary cultured islet cells. J. Biol. Chem. 1996;271:1909–1915. doi: 10.1074/jbc.271.4.1909. [DOI] [PubMed] [Google Scholar]
- 30.Luchette P., Abiy N., Mao H. Microanalysis of clouding process at the single droplet level. Sens. Actuators B Chem. 2007;128:154–160. [Google Scholar]
- 31.Mao H., Luchette P. An integrated laser tweezers instrument for microanalysis of individual protein aggregates. Sens. Actuators B Chem. 2008;129:764–771. [Google Scholar]
- 32.Baumann C.G., Smith S.B., Bustamante C. Ionic effects on the elasticity of single DNA molecules. Proc. Natl. Acad. Sci. USA. 1997;94:6185–6190. doi: 10.1073/pnas.94.12.6185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Catasti P., Chen X., Gupta G. Structure-function correlations of the insulin-linked polymorphic region. J. Mol. Biol. 1996;264:534–545. doi: 10.1006/jmbi.1996.0659. [DOI] [PubMed] [Google Scholar]
- 34.Yu Z., Schonhoft J.D., Mao H. ILPR G-quadruplexes formed in seconds demonstrate high mechanical stabilities. J. Am. Chem. Soc. 2009;131:1876–1882. doi: 10.1021/ja806782s. [DOI] [PubMed] [Google Scholar]
- 35.Jolad V.V., Murad F.K., Fisher J. Solution conformation of d(C(4)ACAC(4)TGT)(2); an intramolecularly folded i-motif from the insulin minisatellite. Org. Biomol. Chem. 2005;3:2234–2236. doi: 10.1039/b504606h. [DOI] [PubMed] [Google Scholar]
- 36.Schonhoft J.D., Bajracharya R., Basu S. Direct experimental evidence for quadruplex-quadruplex interaction within the human ILPR. Nucleic Acids Res. 2009;37:3310–3320. doi: 10.1093/nar/gkp181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Töhl J., Eimer W. Interaction energies and dynamics of alkali and alkaline-earth cations in quadruplex-DNA-structures. J. Mol. Model. 1996;2:327–329. [Google Scholar]
- 38.Jarzynski C. Nonequilibrium equality for free energy differences. Phys. Rev. Lett. 1997;78:2690–2693. [Google Scholar]
- 39.Liphardt J., Dumont S., Bustamante C. Equilibrium information from nonequilibrium measurements in an experimental test of Jarzynski's equality. Science. 2002;296:1832–1835. doi: 10.1126/science.1071152. [DOI] [PubMed] [Google Scholar]
- 40.Palassini M., Ritort F. Improving free-energy estimates from unidirectional work measurements: theory and experiment. Phys. Rev. Lett. 2011;107:060601. doi: 10.1103/PhysRevLett.107.060601. [DOI] [PubMed] [Google Scholar]
- 41.Koirala D., Dhakal S., Mao H. A single-molecule platform for investigation of interactions between G-quadruplexes and small-molecule ligands. Nat. Chem. 2011;3:782–787. doi: 10.1038/nchem.1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Allawi H.T., SantaLucia J., Jr. Thermodynamics and NMR of internal G.T mismatches in DNA. Biochemistry. 1997;36:10581–10594. doi: 10.1021/bi962590c. [DOI] [PubMed] [Google Scholar]
- 43.Williams M.C., Wenner J.R., Bloomfield V.A. Entropy and heat capacity of DNA melting from temperature dependence of single molecule stretching. Biophys. J. 2001;80:1932–1939. doi: 10.1016/S0006-3495(01)76163-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Koirala D., Yu Z., Mao H. Detection of single nucleotide polymorphism using tension-dependent stochastic behavior of a single-molecule template. J. Am. Chem. Soc. 2011;133:9988–9991. doi: 10.1021/ja201976r. [DOI] [PubMed] [Google Scholar]
- 45.Paeschke K., Simonsson T., Lipps H.J. Telomere end-binding proteins control the formation of G-quadruplex DNA structures in vivo. Nat. Struct. Mol. Biol. 2005;12:847–854. doi: 10.1038/nsmb982. [DOI] [PubMed] [Google Scholar]
- 46.González V., Guo K., Sun D. Identification and characterization of nucleolin as a c-myc G-quadruplex-binding protein. J. Biol. Chem. 2009;284:23622–23635. doi: 10.1074/jbc.M109.018028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Simonsson T. G-quadruplex DNA structures—variations on a theme. Biol. Chem. 2001;382:621–628. doi: 10.1515/BC.2001.073. [DOI] [PubMed] [Google Scholar]
- 48.Wang M.D., Schnitzer M.J., Block S.M. Force and velocity measured for single molecules of RNA polymerase. Science. 1998;282:902–907. doi: 10.1126/science.282.5390.902. [DOI] [PubMed] [Google Scholar]
- 49.Mejia Y.X., Mao H., Bustamante C. Thermal probing of E. coli RNA polymerase off-pathway mechanisms. J. Mol. Biol. 2008;382:628–637. doi: 10.1016/j.jmb.2008.06.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hodges C., Bintu L., Bustamante C. Nucleosomal fluctuations govern the transcription dynamics of RNA polymerase II. Science. 2009;325:626–628. doi: 10.1126/science.1172926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Galburt E.A., Grill S.W., Bustamante C. Backtracking determines the force sensitivity of RNAP II in a factor-dependent manner. Nature. 2007;446:820–823. doi: 10.1038/nature05701. [DOI] [PubMed] [Google Scholar]
- 52.Zhou J., Wei C., Li C. Formation of i-motif structure at neutral and slightly alkaline pH. Mol. Biosyst. 2010;6:580–586. doi: 10.1039/b919600e. [DOI] [PubMed] [Google Scholar]
- 53.Dietz H., Rief M. Exploring the energy landscape of GFP by single-molecule mechanical experiments. Proc. Natl. Acad. Sci. USA. 2004;101:16192–16197. doi: 10.1073/pnas.0404549101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Greenleaf W.J., Frieda K.L., Block S.M. Direct observation of hierarchical folding in single riboswitch aptamers. Science. 2008;319:630–633. doi: 10.1126/science.1151298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sinden R.R. Academic Press; San Diego, CA: 1995. DNA Structure and Function. [Google Scholar]
- 56.Parkinson G.N., Lee M.P.H., Neidle S. Crystal structure of parallel quadruplexes from human telomeric DNA. Nature. 2002;417:876–880. doi: 10.1038/nature755. [DOI] [PubMed] [Google Scholar]
- 57.Dai J., Carver M., Yang D. Structure of the hybrid-2 type intramolecular human telomeric G-quadruplex in K+ solution: insights into structure polymorphism of the human telomeric sequence. Nucleic Acids Res. 2007;35:4927–4940. doi: 10.1093/nar/gkm522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Luu K.N., Phan A.T., Patel D.J. Structure of the human telomere in K+ solution: an intramolecular (3 + 1) G-quadruplex scaffold. J. Am. Chem. Soc. 2006;128:9963–9970. doi: 10.1021/ja062791w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang Y., Patel D.J. Solution structure of the human telomeric repeat d[AG3(T2AG3)3] G-tetraplex. Structure. 1993;1:263–282. doi: 10.1016/0969-2126(93)90015-9. [DOI] [PubMed] [Google Scholar]
- 60.Phan A.T., Guéron M., Leroy J.L. The solution structure and internal motions of a fragment of the cytidine-rich strand of the human telomere. J. Mol. Biol. 2000;299:123–144. doi: 10.1006/jmbi.2000.3613. [DOI] [PubMed] [Google Scholar]
- 61.Han X., Leroy J.L., Guéron M. An intramolecular i-motif: the solution structure and base-pair opening kinetics of d(5mCCT3CCT3ACCT3CC) J. Mol. Biol. 1998;278:949–965. doi: 10.1006/jmbi.1998.1740. [DOI] [PubMed] [Google Scholar]
- 62.Esmaili N., Leroy J.L. i-motif solution structure and dynamics of the d(AACCCC) and d(CCCCAA) tetrahymena telomeric repeats. Nucleic Acids Res. 2005;33:213–224. doi: 10.1093/nar/gki160. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


