Abstract
Integrating vectors developed on the basis of various retroviruses have demonstrated therapeutic potential following genetic modification of long-lived hematopoietic stem and progenitor cells. Lentiviral vectors (LV) are assumed to circumvent genotoxic events previously observed with γ-retroviral vectors, due to their integration bias to transcription units in comparison to the γ-retroviral preference for promoter regions and CpG islands. However, recently several studies have revealed the potential for gene activation by LV insertions. Here, we report a murine acute B-lymphoblastic leukemia (B-ALL) triggered by insertional gene inactivation. LV integration occurred into the 8th intron of Ebf1, a major regulator of B-lymphopoiesis. Various aberrant splice variants could be detected that involved splice donor and acceptor sites of the lentiviral construct, inducing downregulation of Ebf1 full-length message. The transcriptome signature was compatible with loss of this major determinant of B-cell differentiation, with partial acquisition of myeloid markers, including Csf1r (macrophage colony-stimulating factor (M-CSF) receptor). This was accompanied by receptor phosphorylation and STAT5 activation, both most likely contributing to leukemic progression. Our results highlight the risk of intragenic vector integration to initiate leukemia by inducing haploinsufficiency of a tumor suppressor gene. We propose to address this risk in future vector design.
Introduction
Gene therapy with γ-retroviral vectors has shown its potential to treat inherited immunodeficiencies.1 However, safety concerns arose with the occurrence of severe adverse events, namely leukemias and myelodysplastic syndromes, after insertional activation of proto-oncogenes by the integrated vector.2,3,4 Self-inactivating lentiviral vectors (SIN-LVs), which lack viral enhancer elements and show a potentially safer, albeit more intragenic insertion profile,5,6 are increasingly used in new clinical trials targeting long-term repopulating hematopoietic stem cells.7,8
However, SIN-LVs still contain internal enhancers, promoters, polyadenylation signals, and splice sites. These sequences could potentially interfere with the initiation and processing of cellular messenger RNAs (mRNAs), create truncated proteins with stimulatory or inhibitory functions, or lead to monoallelic inactivation of tumor suppressors. A recent report revealed clonal dominance in a clinical trial to treat β-thalassemia, mediated by deregulation of HMGA2 expression.7 In this case, intronic SIN-LV insertion disturbed correct mRNA splicing and eliminated a target site for a cellular microRNA, thus stabilizing the transcript and potentially leading to a gain-of-function of HMGA2, a known regulator of primitive hematopoiesis.7,9 Aberrant splicing involved a splice site created by genetic rearrangement of a tandem repeat insulator sequence located in the lentiviral SIN long terminal repeat.7 Importantly, to date the dominant clone with activation of HMGA2 did not exceed ~5% of the total hematopoietic cells and has not been reported to trigger a malignant disease.
Furthermore, it was recently shown that intragenic LV insertions can induce expression of the growth-hormone receptor (Ghr)10 or activate proto-oncogenes such as B-Raf or Evi1.11,12 In the case of Ghr and B-Raf, aberrant splicing was involved in gene activation. In both of these cases, the insertions occurred and were selected in cells with pre-existing genetic aberrations, formally questioning their role as potential tumor-inducing events. Lentivirally induced downregulation of gene expression was demonstrated by gene expression analysis in single cell clones.13 However, these analyses did not allow the study of functional consequences of reduced gene expression.
Cell transformation caused by insertional inactivation of tumor suppressor genes is not only a theoretical concern. Proof-of-concept for such events has been obtained in animal studies using replication-competent γ-retroviruses.14,15,16 However, tumors induced by replication-competent retroviruses typically contain several insertions, complicating the distinction of inducer, progression, and bystander events. Although expected to occur with a frequency that still remains to be determined, inactivation of a tumor suppressor gene by a replication-deficient integrating vector may only be functionally relevant if the second allele is lost or if haploinsufficiency disturbs clonal homeostasis.
In the context of the hematopoietic system, the importance of haploinsufficiency has recently been demonstrated in clinical studies and mouse models exploring the role of loss-of-function of Early B-cell factor 1 (EBF1), a master transcription factor responsible for lineage specification of B-cell progenitor cells.17,18,19,20 In human acute B-lymphoblastic leukemia (B-ALL), haploinsufficiency of the transcription factor EBF1 is found as a relatively common genetic lesion.21,22 In a murine model with germline inactivation of one Ebf1 allele, collaboration with STAT5 activation was shown to lead to a high penetrance of B-ALL.23 Here, we report a very similar constellation after LV-induced insertional inactivation of murine Ebf1. This case was observed in the context of our prospective studies addressing the efficiency and safety of SIN-LVs containing human promoter fragments to restrict gene expression to the megakaryocytic lineage.24 We report the leukemia phenotype, genotype, and transcriptome, and the impact of the SIN-LV insertion on Ebf1 transcription and protein expression. Our study highlights the relevance of prospective studies in nontumor-prone murine models to detect rare complications of non-targeted vector insertions.25,26,27,28 Although the incidence of leukemogenic events triggered by haploinsufficiency remains to be established, this case underlines that vectors with a reduced potential of intragenic insertions and/or reduced interference with cellular splicing may increase the safety of untargeted gene addition strategies.
Results
Monoclonal B-ALL originating from SIN-LV–transduced hematopoietic cells
With the aim to characterize vectors designed for expression in the megakaryocytic lineage we transplanted CD45.2+ C57BL/6 mice with syngeneic CD45.1+ bone marrow (BM) cells transduced with a SIN-LV (RRL.PPT.GPIba.eGFP.pre)29 expressing enhanced green fluorescent protein (eGFP) from a human glycoprotein-I-b-α promoter (GPIba) fragment.30 Out of seven mice transplanted, one mouse presented with severe anemia (hematocrit = 26.4%), thrombocytopenia (89 × 103/µl), leukocytosis (179.3 × 103/µl), and splenomegaly (307 mg) 199 days after transplantation. Due to signs of advanced disease, the animal was killed and fresh tissues were collected for detailed investigation.
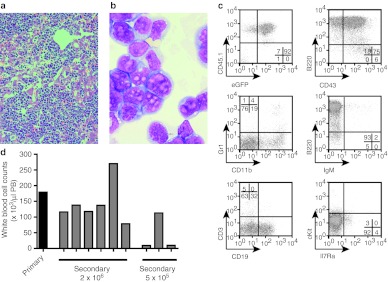
Histopathology revealed strong blast cell infiltrations in BM, spleen, liver, kidney, and lung (Figure 1a,b, Supplementary Figure S1). Flow cytometry specified the cells as B-cell progenitors at the PrePro-B to Pre-B-cell stage expressing B220, CD43, and lacking IgM. CD19 was expressed on ~32% and CD11b on ~19% of the cells, indicating partial conversion to a myeloid phenotype (Figure 1c). The markers CD45.1 and eGFP defined the cells as donor derived and vector transduced (Figure 1c). Secondary transplantation of 2 × 106 or 5 × 105 primary cells induced leukocytosis and organ infiltrations in all (n = 6) or 1/3 (n = 3) of the recipients, respectively (Figure 1d). Compared to the leukemia phenotype in the affected mouse, an increased percentage of cells expressing CD19, cKit, and IL7Ra was observed in secondary recipients (Supplementary Figure S2). Southern blot analyses using three different restriction enzymes identified two independent SIN-LV insertions, with identical pattern and signal strength in the primary and secondary recipients, strongly arguing for monoclonality of leukemic B-lymphoblasts (Figure 2a and Supplementary Figure S3). These data established the diagnosis of a transplantable, monoclonal B-ALL, manifesting 28 weeks after transplantation of SIN-LV–modified BM cells into lethally irradiated syngenic C57BL/6 mice.
Figure 1.
Acute B-lymphoblastic leukemia. (a) Representative histology showing strong infiltration of leukemic cells in the liver. (H&E staining, ×200). (b) Leukemic blasts from the bone marrow of the primary leukemia present with lymphoid and myeloid characteristics. Most blasts cells are larger and have typical myeloid (monocytic) cellular outlines and cytoplasm. The nucleoli are larger and irregular, which are usually seen in lymphoblasts rather than in myeloblasts. (May-Grünwald/Giemsa, ×1,000). (c) Flow cytometry of primary leukemic BM cells indentified the leukemia as B-lymphoblastic with expression of B220 and CD43 while lacking IgM expression. CD19 was expressed on ~32% of the cells while cKit and IL7Ra expression was almost absent. Approximately 19% of the cells expressed the myelo-monocytic marker CD11b. CD45.1 and GFP expression marked cells as donor derived and vector transduced. (d) BM cells of the primary mouse that developed B-lymphoblastic leukemia were transplanted into lethally irradiated secondary recipient mice at two different doses: 2 × 106 and 5 × 105 BM cells. All mice that received the high cell dose succumbed to leukemia 12–17 days after transplantation with high peripheral white blood cell counts, whereas only one of the recipients that were transplanted with the low dose of cells developed leukocytosis. BM, bone marrow; eGFP, enhanced green fluorescent protein; H&E, hematoxylin and eosin; PB, peripheral blood.
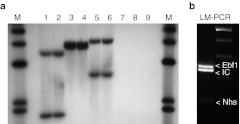
Figure 2.
B-ALL consist of a clone with two insertion sites. (a) Southern blot analysis of BM samples from different secondary recipients (lane 1–6) or control mice (7–9) digested with BsrGI (1, 2, 7), EcoNI (3, 4, 8) or NcoI (5, 6, 9) and probed for the vector-specific post-transcriptional regulatory element (PRE). (b) Ligation-mediated PCR (LM-PCR) on genomic DNA from the BM of a secondary recipient verified two vector integrations which were identified by sequencing to be in the Nhs gene and in the Ebf1 gene. BM, bone marrow.
Lentiviral insertion in Ebf1 as potential inducing mutation
To determine which genes were affected by the lentiviral insertions, we performed ligation-mediated PCR. In addition to the internal control band obtained from the amplification of a vector sequence, we identified two prominent bands. Sequencing of the insertion sites revealed diagnostic junctions of vector long terminal repeat and murine genome sequences. We thus mapped one insertion in the 1st intron of the Nance-Horan Syndrome (Nhs) gene (Figure 2b, Supplementary Table S1). However, expression of Nhs mRNA could neither be detected in wild-type splenocytes nor in the leukemic cells (Supplementary Figure S4). The second integration was located in the 8th intron of the Ebf1 gene (Figure 2b and Supplementary Table S1).
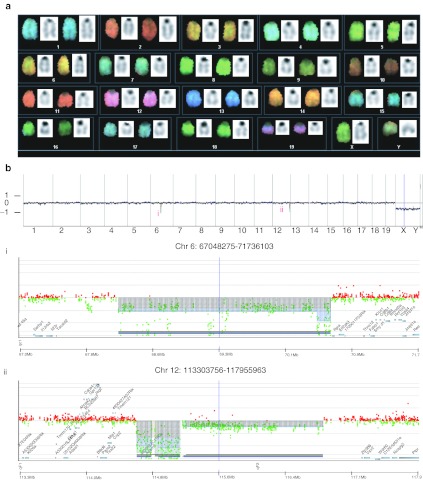
To detect potential additional genetic lesions contributing to leukemogenesis, we searched for chromosomal aberrations (translocations, deletions, amplifications) by spectral karyotyping (Figure 3a) and comparative genome hybridization (array CGH, Figure 3b). While the karyotype was normal, the more sensitive CGH showed microdeletions in two regions located on chromosomes 6 and 12. However, in both cases these regions are not known to contain functional genes. Therefore, we focused on the functional consequences of the SIN-LV insertion in Ebf1.
Figure 3.
Analysis of the genomic stability. (a) No chromosomal aberrations were detected by spectral karyotyping (SKY). Leukemic cell were grown in culture (IL7, Flt3L) for 2 days to induce proliferation and metaphases prepared. (b) Comparative genome hybridization (array-CGH) analysis of leukemic cells. Genomic profiles by means of high resolution array-CGH (180 k): Cye3/Cy5 log2 ratios of fluorescence intensities of probes against their chromosomal localization along chromosomes 1–19 is shown, X: stacked plots of genomic DNA samples of the secondary mice one (brown), two (green), three (black), and the primary mouse (blue); pooled DNA from 10 female C57BL/6 spleen specimen served as reference leading to monosomal X in the male test samples. (i) Microdeletion within chromosomal region 6 in all four probes ranging from 67.847–70.677 Mb containing no genes; (ii) microdeletion within chromosomal region 12 in all four probes ranging from 114.676–115.154 Mb containing no genes.
Truncated transcripts and downregulation of Ebf1
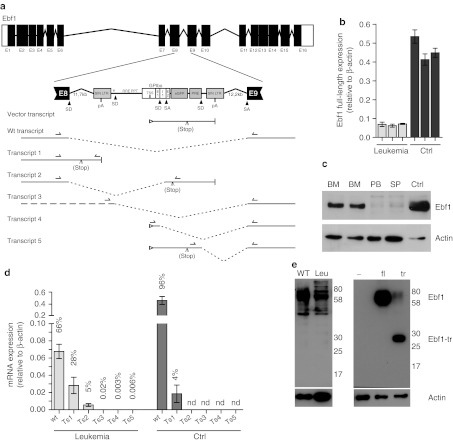
Ebf1 encodes a transcription factor that regulates B-cell development upstream of a whole network of transcriptional regulators and signaling molecules important for B-cell maturation, including Pax5.17 Haploinsufficiency of EBF1 is associated with human B-ALL, which can be recapitulated in a murine model.21,22,23 We thus further investigated a potential deregulation of Ebf1 after the SIN-LV integration and found a downregulation in leukemic cells, both on mRNA and protein level (Figure 4). As the integration event occurred almost in the center of intron 8, and in the same transcriptional orientation as the cellular gene, we examined aberrant splicing events involving the vector's splice sites, including those of an intron contained in the GPIba promoter fragment, with a series of reverse transcription-PCR reactions (Figure 4a). Overall, the transcript was strongly downregulated (~80% reduced, Figure 4b), which was reflected in a strong reduction of protein levels (Figure 4c). Quantitative PCR revealed splicing from Ebf1 exon 8 into the SIN-LV and increased readthrough after Ebf1 exon8 (transcripts 1 and 2, Figure 4d). The detected levels of mis-spliced mRNA of transcript 1 accounted for ~30% of the total Ebf1 mRNA in leukemic cells. Transcripts initiated from the vector that spliced into the downstream Ebf1 exons were detectable (Supplementary Figure S6) although at a very low level. However, nonsense-mediated decay might have caused accelerated mRNA degradation. In support of this hypothesis no truncated EBF1 protein variants could be detected by immunoblot in leukemic samples, while expression of the putative transcript from a LV was well-detectable (Figure 4e). As a putative truncated EBF1 protein may still contain the DNA-binding domain encoded by exon 1–8 but lack the transactivation motif, and considering that the immunoblot may not be sensitive enough despite the choice of reasonable positive controls, we expressed two putative versions of the truncated EBF1 encoded by the transcripts 1 and 2 from SIN-LV containing a strong promoter derived from spleen focus-forming virus.31 Both transcripts will result in a truncated EBF1 containing exons 1–8 (Supplementary Figure S5). Expression of the truncated protein was confirmed by immunoblot (Figure 4e). Lineage-negative BM cells were cultured in conditions that promote B-cell differentiation. However, no evidence was found that a putative truncated protein acts as a dominant-negative variant blocking B-cell differentiation (Figure 5). In support of these findings, the vector expressing truncated EBF1 did not induce leukemia in vivo (10 mice observed for 6 months, data not shown).
Figure 4.
Post-transcriptional deregulation of Ebf1 following lentiviral vector insertion. (a) Schematic overview of the Ebf1 gene locus with the in-sense integrated SIN-LV in intron 8. The SIN-LV harbors the exon (E) and intron (I) containing GPIba promoter, the eGFP reporter gene, the PRE, and splice donors (SD), and splice acceptors (SA) sites as indicated. Splice events can lead to alternative transcripts as indicated in the figure (transcripts 1–5) in addition to the wt and vector transcripts. A stop codon can occur due to frame shifts in transcript 2. (b) Ebf1 mRNA expression levels relative to actin in the BM from three independent leukemic mice (leukemia) and three independently FACS-sorted CD19+CD43+ B-cell progenitor samples from wild-type mice BM (Ctrl). (c) Immunoblot analysis of Ebf1 expression in leukemic samples from bone marrow (BM), spleen (SP), and peripheral blood (PB) of leukemic mice in comparison to wild-type CD19+CD43+ B-cell progenitors (Ctrl). Actin was used as loading control. (d) Quantitative RT-PCR detecting the different splice products of the Ebf1 locus with the inserted LV in comparison to the full-length transcript. Readthrough transcripts into intron 8 (transcript 1) and fusion transcripts from exon 8 to the LV eGFP gene that result in a frame shift and early stop (transcript 2) are well-detectable. Splice products from the LV to the exon 9 (transcripts 3–5) could be amplified and sequences were verified (Supplementary Figure S6) but the overall amount of transcripts was extremely low (percentage of transcript in correlation to the full-length transcript is given). Results from three independent leukemic BM samples (leukemia) in comparison to pooled (n = 3) wild-type BM (Ctrl) (nd = not detected). (e) Immunoblot analysis of wild-type splenocytes (WT) and leukemic cells (Leu) does not show detectable amounts of truncated protein corresponding to the expected protein. Immunoblot analysis of 293T cells transfected with lentiviral constructs expressing dTomato alone (−), full-length Ebf1 (fl) or the truncated Ebf1 (tr) shows that the employed antibody was able to detect the truncated Ebf1 protein. eGFP, enhanced green fluorescent protein; FACS, fluorescence-activated cell sorting; LTR, long terminal repeat; mRNA, messenger RNA; pA, poly-A; PRE, post-transcriptional regulatory element; RT-PCR, reverse transcription-PCR; SIN-LV, self-inactivating lentiviral vector.
Figure 5.
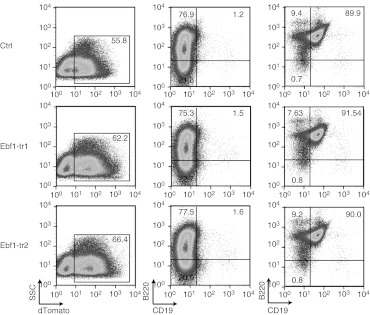
Expression of a truncated Ebf1 protein does not interfere with in vitro B-cell differentiation. Lineage negative BM cells were transduced with a SIN-lentiviral vector expressing the truncated variants of Ebf1, Ebf1-tr1 (RRL.PPT.SFFV.Ebf1-tr1.IRES.dTomato.pre) or Ebf1-tr2 (RRL.PPT.SFFV.Ebf1-tr2.IRES.dTomato.pre) which correspond to the truncations induced by readthrough or splicing to the GPIbaP splice acceptor, respectively. For detection by flow cytometry a dTomato was coexpressed from an IRES. As control, cells were transduced with a vector expressing dTomato alone (RRL.PPT.SFFV.IRES.dTomato.pre). (i) Transgene expression as indicated by dTomato expression in a representative culture on day 7 of coculture on OP9 stromal cells, (ii) expression of B-cell markers B220 and CD19 after 7 days, and (iii) after 18 days was assessed by flow cytometry. Percentages of the distinct populations are indicated in corresponding quadrants. No differences in B-cell differentiation were seen in cells expressing the truncated EBF1 protein in comparison to control vector-transduced cells.
Molecular evidence for Ebf1 loss-of-function connected to activation of STAT5
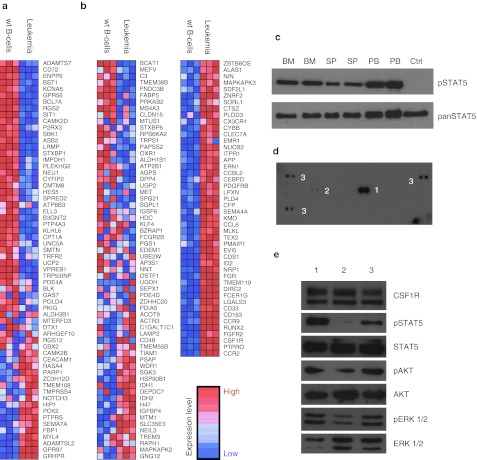
With confirmed downregulation of Ebf1 and potential involvement of aberrant mRNA processing, we next addressed Ebf1 target gene expression in leukemic cells. Gene expression microarrays comparing leukemic blasts to differentiation stage matched B-cell progenitors (CD19+CD43+) from C57BL/6 mice confirmed downregulation of Ebf1 and its target genes (Figure 6a). One of the target genes that was found to be downregulated was Aiolos (Ikzf3), a tumor suppressor which is known to be frequently deleted and coincided with Ebf1 haploinsufficiency in B-ALL patients.21 We did not find Ikzf3 to be genetically deleted according to the CGH analysis (Supplementary Figure S7). As a known downstream target of Ebf120 the downregulation of Ikzf3 may therefore be caused by Ebf1 haploinsufficiency. Gene expression data also provided further evidence for dedifferentiation to the myeloid lineage with upregulation of Csf1r (c-Fms), Runx2 and further genes associated with myeloid differentiation32 (Figure 6b).
Figure 6.
Downregulation of Ebf1 target genes coincides with reactivation of myeloid genes and Csf1r upregulation. (a) cDNA-microarray analysis performed on three independent leukemic BM samples in comparison to three independent samples of CD19+CD43+ wild-type B-cell progenitors (wt B-cells). Gene set enrichment analysis (GSEA) shows downregulation of Ebf1 target genes17 in leukemic cells as compared to wild-type B-cell progenitors (NES = 1.55; P = 0.012, FDR <0.02). (b) GSEA analysis of myeloid lineage-specific genes32 shows strong enrichment of myeloid genes in leukemic cells (NES = −2.49; P < 0.001, FDR <0.001). (c) Phospho-tyrosine immunoblot analysis detected activation of STAT5 (Tyr694) in leukemic cells from bone marrow (BM), spleen (SP), and peripheral blood (PB) in comparison to wild-type CD19+CD43+ B-cell progenitors (Ctrl). (d) Strong phosphorylation of Csf1r [1] (aka. M-Csfr or c-Fms) and some minor activation of Pdgfr-β [2] was found by phospho-receptor-tyrosine-kinase protein arrays. Both genes had been found upregulated in leukemic cells in the cDNA-microarray analysis. Ctrl spots [3]. (e) Immunoblot analysis showing CSFR1, pSTAT5, STAT5, pERK 1/2, ERK 1/2, pAKT, and AKT expression/activation in leukemic cells grown on OP9 stromal cells with 20 ng/ml SCF, 20 ng/ml FLT3-L, and 10 ng/ml IL7 [1], starved for 3 hours [2], and stimulated with 40 ng/ml M-CSF for 10 minutes after 3 hours starvation [3]. Loss of STAT5 phosphorylation upon starvation and regain after stimulation with M-CSF shows ligand-dependency of upregulated CSF1R. FDR, false discovery rate; M-CSF, macrophage colony-stimulating factor; NES, normalized enrichment score.
Gene ontology analysis also identified cytokine receptor signaling target genes to be upregulated in leukemic cells. Considering this transcriptome signature and the known leukemogenic collaboration of Ebf1 haploinsufficiency with STAT5 activation,23 we detected strong STAT5 activation in the murine B-ALL by immunoblot (Figure 6c).
To identify the upstream events leading to STAT5 activation, we used phospho-proteome arrays of freshly harvested leukemic cells. We found that CSF1R was strongly activated in leukemic cells and also obtained a minor phosphorylation signal of PDGFR-β (Figure 6d). We thus tested the response of leukemic cells to macrophage colony-stimulating factor (M-CSF) in vitro, and noted strong activation of STAT5, ERK 1/2, and AKT in leukemic cells. These data revealed that CSF1R activation was nonautonomous and induced a strong signal response in the leukemic clone (Figure 6e). While the microarray supported that M-CSF was not transcriptionally induced in the B-ALL cells, the induction of Csf1r might be a direct consequence of the loss-of-function of the B-cell identity factor EBF1. Indeed, the B-lymphoblastic leukemia showed a partial myeloid expression profile based on both, surface phenotype (Figure 1c) and microarray analysis.
Discussion
This study provides strong support for the potential induction of an acute leukemia by insertional gene inactivation, caused by the integration of an otherwise neutral lentiviral gene marking vector. The analysis of the leukemic phenotype and underlying genetic lesions pointed to a crucial role of transcriptional downregulation of a known tumor suppressor gene, Ebf1, after intronic insertion of the LV.
Indeed, previous studies have shown that murine B-ALL can be induced by monoallelic germline inactivation of Ebf1,23 triggering a block of B-cell differentiation which in our case was associated with upregulation of myeloid growth factor receptors that may mediate growth-promoting signals in response to a supportive cytokine milieu. As karyotype and CGH analyses did not uncover further leukemogenic events, the identification of additional leukemogenic mutations may require genome-wide sequencing. However, as lentiviral integration occurred in BM cells of wild-type C57BL/6 mice, additional leukemogenic mutations likely occurred after the insertion event, connected to the expansion of the leukemic clone. We would thus propose that this study can be interpreted as a proof-of-concept for insertional haploinsufficiency caused by a replication-deficient integrating gene vector as a tumor-initiating event.
One remaining important question is why the second Ebf1 allele was unable to compensate the monoallelic loss of Ebf1 expression. A potential positive feedback loop of EBF1 acting on its own promoter may have contributed to the substantial loss of expression detected at the RNA and protein level.33 Alternatively, antisense transcripts originating from the altered allele may be involved although the transcription unit of the vector was in the transcriptional orientation of the allele, the leukemic clone may have undergone random transcriptional silencing or may even have acquired cryptic mutations of the second allele, confounding the selective advantage of transformed cells.
Furthermore, recent studies of cytogenetically abnormal human B-ALL uncovered heterozygous deletions/mutations of EBF1 as a recurrent event,21,22 which, in line with mechanistic studies in heterozygous knock-out mice,23 provide strong evidence for a role of Ebf1 in B-ALL as a haploinsufficient tumor suppressor. However, the latter study used a transgenic mouse model in which lesions were present in all hematopoietic cells, thus providing multiple opportunities for accumulation of secondary genetic lesions contributing to leukemogenesis. In contrast, our study suggested an insertional event in a single cell as the initiating event, leading to progressive clonal expansion with subsequent transformation.
While haploinsufficiency of a tumor suppressor is a noteworthy initiation mechanism of a leukemia, our observation raises concerns that vectors with a preferred intragenic integration profile may establish a new category of mutagenesis in gene therapy. As transcript truncation by integrating gene vectors may also lead to the upregulation or gain-of-function of proto-oncogenes,10,11 future vector design could tackle this problem by modification of the integration pattern. We and others therefore explore vector systems with a less biased integration pattern.34,35,36 Novel approaches for vector integration into bona fide “safe harbors” may also contribute to the prevention of this risk.37,38,39,40
However, it needs to be stressed that the magnitude and clinical relevance of the risk associated with insertional haploinsufficiency remains to be determined. Depending on the target tissue of the genetic modification, the number and type of potential haploinsufficient tumor suppressors may vary, and some of the safeguarding mechanisms operating in human cells may prevent tumorigenesis on this basis. To understand the frequency of these events and their contribution to clonal skewing in the long-term follow-up of patients receiving gene-modified cells, and to define the most relevant vector sequences involved in alternative splicing with cellular genes, longitudinal “genome-wide” studies will be of great importance.
Materials and Methods
Animals. C57BL/6 (B6.Ly5.2) and C57BL/6 PeP3b (B6 SJL/Ly5.1) mice were obtained from The Jackson Laboratory (Bar Harbor, ME) and kept in the specified pathogen-free animal facilities of the Hannover Medical School, Germany. Animal experiments were approved by the local ethical committee and performed according to their guidelines.
LVs and vector production.
The lentiviral backbone is the RRL SIN-LV with minor modifications.24,29,41 The 596 bp human GPIba promoter has been described30 spanning the region −288 to +308.
BM cell purification and transduction. Briefly, lineage-marker depleted (lin-) cells were isolated from complete BM by magnetic sorting using lineage-specific antibodies (Gr1, CD11b, CD45R/B220, CD3e, TER-119; Miltenyi Biotech, Bergisch Gladbach, Germany). Before lentiviral transduction lin- BM cells were prestimulated for 18 hours in StemSpan SFEM medium (StemCell Technologies, Grenoble, France), 10 ng/ml murine SCF, 20 ng/ml murine THPO, 10 ng/ml human FGF-1, 20 ng/ml murine IGF2, 1% penicillin/streptomycin, 2 mmol/l glutamine, plated in wells precoated with 10 µg/cm2 Retronectin (TaKaRa, Otsu, Japan). For transduction concentrated viral supernatant was added.
PCR. FACS sorted wild-type B-cell progenitors and leukemic cells were lysed in RLT lysis buffer and stored at −80 °C until processing with the RNeasy Micro Kit (Qiagen GmbH, Hilden, Germany). cDNA synthesis was performed using the QuantiTect reverse transcription kit (Qiagen GmbH). Quantitative reverse transcription-PCR was performed using SYBR-Green for detection (Qiagen GmbH). Primers are summarized in Supplementary Table S2. LV integrations in leukemic cells were analyzed by ligation-mediated PCR. See Supplementary Materials and Methods.
Immunoblot analysis. Cells were collected directly after stimulation and lysed in buffer containing phosphatase inhibitors (50 mmol/l HEPES, 150 mmol/l NaCl, 50 mmol/l NaF, 10 mmol/l Na4P2O7, 10% Glycerin, 1% NP-40); 20–40 µg of protein samples, separated in SDS-polyacrylamide gels, were transferred to nitrocellulose membrane. The rat-anti-Ebf1 antibody has been described42 and was kindly provided by Matthias Kieslinger (Helmholtz Institute, München). Phospho-proteins were detected with antibodies against pSTAT5, pERK1/2, pAKT (all Cell Signaling Technologies, Boston, MA). For loading controls, blots were stripped and reprobed using Csf1r, Actin, pan-STAT5 (Santa Cruz, Biotechnologies, Santa Cruz, CA), pan-ERK1/2, anti-AKT1 antibodies (Cell Signaling Technologies).
Genome analyses. Spectral karyotyping was performed as described previously (see Supplementary Materials and Methods). Array-CGH was performed using the Agilent Mouse Genome Microarray Kit 4x180k (Agilent Technologies, Santa Clara, CA), a high resolution 60-mer oligonucleotide-based microarray with median overall probe spacing of about 10 kb, following the manufacturer's instructions (see Supplementary Materials and Methods).
Antibody RTK-array. Single cell suspensions of spleen and BM of leukemic mice (>90% infiltration) and healthy controls were lysed in NP40 lysis buffer. Mouse Phospho-Receptor Tyrosine Kinase Arrays (ARY014; R&D Systems, Minneapolis, MN) were loaded with lysate containing 1 mg of total protein and processed according to the manufacturer's instructions.
Microarray analysis. Total RNA was prepared from FACS-sorted wild-type B-cell progenitors (CD19+, CD43+) and leukemic cells from bone marrow using the RNeasy Micro Kit (Qiagen GmbH). Quality of RNA was assessed using the Agilent 2100 Bioanalyzer. RNA was amplified by the NuGen WT-Ovation Pico Kit succeeded by biotin labeling of fragmented amplified cDNA using the NuGen Ovation Biotin labeling system (NuGEN Technologies, San Carlos, CA) in triplicates. Fragmented and labeled cDNA was hybridized to Affymetrix (San Jose, CA) Mouse Genome 430 2.0 (MOE430_2) GeneChip arrays (45,101 probe sets). Data was analyzed using R and Bioconductor.43 Data quality was assessed using the ArrayQualityMetrics package.44 Arrays were background corrected, normalized, and summarized employing the RMA algorithm.45 LIMMA46 was used to detect differentially expressed probe sets applying the Benjamini-Hochberg step up multiple testing correction at a false discovery rate of <0.05. We used the Broad Institute GSEA software package47 for testing enrichment of gene sets. The Ebf-1 target gene set was obtained from Treiber et al.17 The myeloid gene set was compiled from the literature, mainly from Ng and Georgopolous et al.32 comprising genes overexpressed in granulo-monocyte progenitors relative to hematopoietic stem cells and common lymphoid progenitors.
SUPPLEMENTARY MATERIAL Figure S1. Histopathological analysis of the B-lymphoblastic leukemia. Figure S2. Leukemia phenotype in secondary recipients. Figure S3. Southern blot analysis of DNA isolated from different tissues and mice. Figure S4. Expression of Nance-Horan-Syndrome (Nhs) mRNA in leukemic cells. Figure S5. Splice products that would result in a truncated EBF1 protein. Figure S6. Verification of transcripts Figure S7. Genomic Ikzf3 locus in the B-ALL is not altered. Table S1. Position of the lentiviral insertion sites. Table S2. Primer. Materials and Methods.
Acknowledgments
The authors thank Rena-Mareike Struß for her excellent technical assistance, Thomas Neumann for performing microarray experiments, Yang Ming for her help with the analysis of cytospins, Matthias Ballmaier from the cell sorting facility, and Joerg Fruehauf from the irradiation facility (all Hannover Medical School, Hannover, Germany). We thank Matthias Kieslinger (Helmholtz Institut, München, Germany) for providing the anti-EBF1 antibody. The research was financed by the grants of Deutsche Forschungsgemeinschaft (SFB 566, SPP1230, and Cluster of Excellence REBIRTH) and the European Union (FP7 integrated project CELL-PID). D.H. was supported by a stipend of the Hannover Biomedical Research School. The authors declared no conflict of interest.
Supplementary Material
Histopathological analysis of the B-lymphoblastic leukemia.
Leukemia phenotype in secondary recipients.
Southern blot analysis of DNA isolated from different tissues and mice.
Expression of Nance-Horan-Syndrome (Nhs) mRNA in leukemic cells.
Splice products that would result in a truncated EBF1 protein.
Verification of transcripts
Genomic Ikzf3 locus in the B-ALL is not altered.
Position of the lentiviral insertion sites.
Primer.
References
- Qasim W, Gaspar HB., and, Thrasher AJ. Progress and prospects: gene therapy for inherited immunodeficiencies. Gene Ther. 2009;16:1285–1291. doi: 10.1038/gt.2009.127. [DOI] [PubMed] [Google Scholar]
- Stein S, Ott MG, Schultze-Strasser S, Jauch A, Burwinkel B, Kinner A.et al. (2010Genomic instability and myelodysplasia with monosomy 7 consequent to EVI1 activation after gene therapy for chronic granulomatous disease Nat Med 16198–204. [DOI] [PubMed] [Google Scholar]
- Howe SJ, Mansour MR, Schwarzwaelder K, Bartholomae C, Hubank M, Kempski H.et al. (2008Insertional mutagenesis combined with acquired somatic mutations causes leukemogenesis following gene therapy of SCID-X1 patients J Clin Invest 1183143–3150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hacein-Bey-Abina S, Garrigue A, Wang GP, Soulier J, Lim A, Morillon E.et al. (2008Insertional oncogenesis in 4 patients after retrovirus-mediated gene therapy of SCID-X1 J Clin Invest 1183132–3142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell RS, Beitzel BF, Schroder AR, Shinn P, Chen H, Berry CC.et al. (2004Retroviral DNA integration: ASLV, HIV, and MLV show distinct target site preferences PLoS Biol 2E234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schröder AR, Shinn P, Chen H, Berry C, Ecker JR., and, Bushman F. HIV-1 integration in the human genome favors active genes and local hotspots. Cell. 2002;110:521–529. doi: 10.1016/s0092-8674(02)00864-4. [DOI] [PubMed] [Google Scholar]
- Cavazzana-Calvo M, Payen E, Negre O, Wang G, Hehir K, Fusil F.et al. (2010Transfusion independence and HMGA2 activation after gene therapy of human ß-thalassaemia Nature 467318–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartier N, Hacein-Bey-Abina S, Bartholomae CC, Veres G, Schmidt M, Kutschera I.et al. (2009Hematopoietic stem cell gene therapy with a lentiviral vector in X-linked adrenoleukodystrophy Science 326818–823. [DOI] [PubMed] [Google Scholar]
- Ikeda K, Mason PJ., and, Bessler M. 3'UTR-truncated Hmga2 cDNA causes MPN-like hematopoiesis by conferring a clonal growth advantage at the level of HSC in mice. Blood. 2011;117:5860–5869. doi: 10.1182/blood-2011-02-334425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight S, Bokhoven M, Collins M., and, Takeuchi Y. Effect of the internal promoter on insertional gene activation by lentiviral vectors with an intact HIV long terminal repeat. J Virol. 2010;84:4856–4859. doi: 10.1128/JVI.02476-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montini E, Cesana D, Schmidt M, Sanvito F, Bartholomae CC, Ranzani M.et al. (2009The genotoxic potential of retroviral vectors is strongly modulated by vector design and integration site selection in a mouse model of HSC gene therapy J Clin Invest 119964–975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modlich U, Navarro S, Zychlinski D, Maetzig T, Knoess S, Brugman MH.et al. (2009Insertional transformation of hematopoietic cells by self-inactivating lentiviral and gammaretroviral vectors Mol Ther 171919–1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruggi G, Porcellini S, Facchini G, Perna SK, Cattoglio C, Sartori D.et al. (2009Transcriptional enhancers induce insertional gene deregulation independently from the vector type and design Mol Ther 17851–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beatty J, Terry A, MacDonald J, Gault E, Cevario S, O'Brien SJ.et al. (2002Feline immunodeficiency virus integration in B-cell lymphoma identifies a candidate tumor suppressor gene on human chromosome 15q15 Cancer Res 627175–7180. [PubMed] [Google Scholar]
- Ben David Y, Prideaux VR, Chow V, Benchimol S., and, Bernstein A. Inactivation of the p53 oncogene by internal deletion or retroviral integration in erythroleukemic cell lines induced by Friend leukemia virus. Oncogene. 1988;3:179–185. [PubMed] [Google Scholar]
- Ben-David Y, Lavigueur A, Cheong GY., and, Bernstein A. Insertional inactivation of the p53 gene during friend leukemia: a new strategy for identifying tumor suppressor genes. New Biol. 1990;2:1015–1023. [PubMed] [Google Scholar]
- Treiber T, Mandel EM, Pott S, Györy I, Firner S, Liu ET.et al. (2010Early B cell factor 1 regulates B cell gene networks by activation, repression, and transcription- independent poising of chromatin Immunity 32714–725. [DOI] [PubMed] [Google Scholar]
- Pongubala JM, Northrup DL, Lancki DW, Medina KL, Treiber T, Bertolino E.et al. (2008Transcription factor EBF restricts alternative lineage options and promotes B cell fate commitment independently of Pax5 Nat Immunol 9203–215. [DOI] [PubMed] [Google Scholar]
- Lukin K, Fields S, Guerrettaz L, Straign D, Rodriguez V, Zandi S.et al. (2011A dose-dependent role for EBF1 in repressing non-B-cell-specific genes Eur J Immunol 411787–1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukin K, Fields S, Lopez D, Cherrier M, Ternyak K, Ramírez J.et al. (2010Compound haploinsufficiencies of Ebf1 and Runx1 genes impede B cell lineage progression Proc Natl Acad Sci USA 1077869–7874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullighan CG, Goorha S, Radtke I, Miller CB, Coustan-Smith E, Dalton JD.et al. (2007Genome-wide analysis of genetic alterations in acute lymphoblastic leukaemia Nature 446758–764. [DOI] [PubMed] [Google Scholar]
- Harvey RC, Mullighan CG, Wang X, Dobbin KK, Davidson GS, Bedrick EJ.et al. (2010Identification of novel cluster groups in pediatric high-risk B-precursor acute lymphoblastic leukemia with gene expression profiling: correlation with genome-wide DNA copy number alterations, clinical characteristics, and outcome Blood 1164874–4884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heltemes-Harris LM, Willette MJ, Ramsey LB, Qiu YH, Neeley ES, Zhang N.et al. (2011Ebf1 or Pax5 haploinsufficiency synergizes with STAT5 activation to initiate acute lymphoblastic leukemia J Exp Med 2081135–1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heckl D, Wicke DC, Brugman MH, Meyer J, Schambach A, Büsche G.et al. (2011Lentiviral gene transfer regenerates hematopoietic stem cells in a mouse model for Mpl-deficient aplastic anemia Blood 1173737–3747. [DOI] [PubMed] [Google Scholar]
- Li Z, Düllmann J, Schiedlmeier B, Schmidt M, von Kalle C, Meyer J.et al. (2002Murine leukemia induced by retroviral gene marking Science 296497. [DOI] [PubMed] [Google Scholar]
- Modlich U, Kustikova OS, Schmidt M, Rudolph C, Meyer J, Li Z.et al. (2005Leukemias following retroviral transfer of multidrug resistance 1 (MDR1) are driven by combinatorial insertional mutagenesis Blood 1054235–4246. [DOI] [PubMed] [Google Scholar]
- Kustikova O, Fehse B, Modlich U, Yang M, Düllmann J, Kamino K.et al. (2005Clonal dominance of hematopoietic stem cells triggered by retroviral gene marking Science 3081171–1174. [DOI] [PubMed] [Google Scholar]
- Modlich U, Schambach A, Brugman MH, Wicke DC, Knoess S, Li Z.et al. (2008Leukemia induction after a single retroviral vector insertion in Evi1 or Prdm16 Leukemia 221519–1528. [DOI] [PubMed] [Google Scholar]
- Schambach A, Bohne J, Chandra S, Will E, Margison GP, Williams DA.et al. (2006Equal potency of gammaretroviral and lentiviral SIN vectors for expression of O6-methylguanine-DNA methyltransferase in hematopoietic cells Mol Ther 13391–400. [DOI] [PubMed] [Google Scholar]
- Hashimoto Y., and, Ware J. Identification of essential GATA and Ets binding motifs within the promoter of the platelet glycoprotein Ib alpha gene. J Biol Chem. 1995;270:24532–24539. doi: 10.1074/jbc.270.41.24532. [DOI] [PubMed] [Google Scholar]
- Baum C, Hegewisch-Becker S, Eckert HG, Stocking C., and, Ostertag W. Novel retroviral vectors for efficient expression of the multidrug resistance (mdr-1) gene in early hematopoietic cells. J Virol. 1995;69:7541–7547. doi: 10.1128/jvi.69.12.7541-7547.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng SY, Yoshida T, Zhang J., and, Georgopoulos K. Genome-wide lineage-specific transcriptional networks underscore Ikaros-dependent lymphoid priming in hematopoietic stem cells. Immunity. 2009;30:493–507. doi: 10.1016/j.immuni.2009.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roessler S, Györy I, Imhof S, Spivakov M, Williams RR, Busslinger M.et al. (2007Distinct promoters mediate the regulation of Ebf1 gene expression by interleukin-7 and Pax5 Mol Cell Biol 27579–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, Renaud G, Gomes TJ, Golmes T, Ferris A, Hendrie PC.et al. (2008Reduced genotoxicity of avian sarcoma leukosis virus vectors in rhesus long-term repopulating cells compared to standard murine retrovirus vectors Mol Ther 161617–1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moldt B, Miskey C, Staunstrup NH, Gogol-Döring A, Bak RO, Sharma N.et al. (2011Comparative genomic integration profiling of Sleeping Beauty transposons mobilized with high efficacy from integrase-defective lentiviral vectors in primary human cells Mol Ther 191499–1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suerth JD, Maetzig T, Brugman MH, Heinz N, Appelt JU, Kaufmann KB.et al. (2012Alpharetroviral Self-inactivating Vectors: Long-term Transgene Expression in Murine Hematopoietic Cells and Low Genotoxicity Mol Therepub ahead of print). [DOI] [PMC free article] [PubMed]
- Sadelain M, Papapetrou EP., and, Bushman FD. Safe harbours for the integration of new DNA in the human genome. Nat Rev Cancer. 2012;12:51–58. doi: 10.1038/nrc3179. [DOI] [PubMed] [Google Scholar]
- Mussolino C, Morbitzer R, Lütge F, Dannemann N, Lahaye T., and, Cathomen T. A novel TALE nuclease scaffold enables high genome editing activity in combination with low toxicity. Nucleic Acids Res. 2011;39:9283–9293. doi: 10.1093/nar/gkr597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boch J, Scholze H, Schornack S, Landgraf A, Hahn S, Kay S.et al. (2009Breaking the code of DNA binding specificity of TAL-type III effectors Science 3261509–1512. [DOI] [PubMed] [Google Scholar]
- Moscou MJ., and, Bogdanove AJ. A simple cipher governs DNA recognition by TAL effectors. Science. 2009;326:1501. doi: 10.1126/science.1178817. [DOI] [PubMed] [Google Scholar]
- Dull T, Zufferey R, Kelly M, Mandel RJ, Nguyen M, Trono D.et al. (1998A third-generation lentivirus vector with a conditional packaging system J Virol 728463–8471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieslinger M, Hiechinger S, Dobreva G, Consalez GG., and, Grosschedl R. Early B cell factor 2 regulates hematopoietic stem cell homeostasis in a cell-nonautonomous manner. Cell Stem Cell. 2010;7:496–507. doi: 10.1016/j.stem.2010.07.015. [DOI] [PubMed] [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S.et al. (2004Bioconductor: open software development for computational biology and bioinformatics Genome Biol 5R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauffmann A, Gentleman R., and, Huber W. arrayQualityMetrics–a bioconductor package for quality assessment of microarray data. Bioinformatics. 2009;25:415–416. doi: 10.1093/bioinformatics/btn647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautier L, Cope L, Bolstad BM., and, Irizarry RA. affy–analysis of Affymetrix GeneChip data at the probe level. Bioinformatics. 2004;20:307–315. doi: 10.1093/bioinformatics/btg405. [DOI] [PubMed] [Google Scholar]
- Smyth G.2005Limma: linear models for microarray data Gentleman R, Carey V, Dudoit S, Irizarry S., and, Huber W.eds). Bioinformatics and Computational Biology Solutions Using R and Bioconductor Springer: New York; pp. 397–420. [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA.et al. (2005Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles Proc Natl Acad Sci USA 10215545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Histopathological analysis of the B-lymphoblastic leukemia.
Leukemia phenotype in secondary recipients.
Southern blot analysis of DNA isolated from different tissues and mice.
Expression of Nance-Horan-Syndrome (Nhs) mRNA in leukemic cells.
Splice products that would result in a truncated EBF1 protein.
Verification of transcripts
Genomic Ikzf3 locus in the B-ALL is not altered.
Position of the lentiviral insertion sites.
Primer.