Abstract
Mycolic acids, the major lipid of the Mycobacterium tuberculosis cell wall, are modified by cyclopropane rings, methyl branches, and oxygenation through the action of eight S-adenosylmethionine (SAM)-dependent mycolic acid methyltransferases (MAMTs), encoded at four genetic loci. Mycolic acid modification has been shown to be important for M. tuberculosis pathogenesis, in part through effects on the inflammatory activity of trehalose dimycolate (cord factor). Studies using the MAMT inhibitor dioctylamine have suggested that the MAMT enzyme class is essential for M. tuberculosis viability. However, it is unknown whether a cyclopropane-deficient strain of M. tuberculosis would be viable and what the effect of cyclopropane deficiency on virulence would be. We addressed these questions by creating and characterizing M. tuberculosis strains lacking all functional MAMTs. Our results show that M. tuberculosis is viable either without cyclopropanation or without cyclopropanation and any oxygenated mycolates. Characterization of these strains revealed that MAMTs are required for acid fastness and resistance to detergent stress. Complete lack of cyclopropanation confers severe attenuation during the first week after aerosol infection of the mouse, whereas complete loss of MAMTs confers attenuation in the second week of infection. Characterization of immune responses to the cyclopropane- and MAMT-deficient strains indicated that the net effect of mycolate cyclopropanation is to dampen host immunity. Taken together, our findings establish the immunomodulatory function of the mycolic acid modification pathway in pathogenesis and buttress this enzyme class as an attractive target for antimycobacterial drug development.
INTRODUCTION
Mycobacterium tuberculosis, the causative agent of tuberculosis, remains one of the major causes of infectious morbidity in the world, especially in developing countries. The recent emergence of highly antibiotic-resistant strains (multidrug-resistant [MDR] and extensively drug-resistant [XDR] strains) has complicated the treatment of tuberculosis. New targets for potential new antibiotics are needed, and for this purpose, a better understanding of the importance of specific pathways for bacterial survival in the host is needed.
The outer layer of the mycobacterial cell wall consists mainly of long-chain fatty acids called mycolic acids (4). These mycolic acids comprise approximately 40% of the dry weight of the bacterium and are partially responsible for its acid fastness and the relative impermeability of the cell wall, including impermeability to antibacterial agents (2, 7). In slow-growing pathogenic mycobacteria, mycolic acids are modified with cyclopropane rings, methyl branches, ketones, and methoxy groups to create a series of three major mycolic acids: alpha mycolates (two cis cyclopropane rings), methoxymycolates (a single cis or trans cyclopropane ring and a methoxy group), and ketomycolates (a single cis or trans cyclopropane ring and a ketone group). M. tuberculosis encodes 8 S-adenosylmethionine (SAM)-dependent methyltransferases, 6 of which have been shown to participate in mycolic acid modification (MmaA1 to -4, PcaA, and CmaA2) (3, 5, 9, 13, 14, 16–18). The function of each of these enzymes has been elucidated through a genetic approach in which the gene encoding each mycolic acid methyltransferase (MAMT) was deleted from the M. tuberculosis chromosome and the mycolic acids were analyzed for loss of cyclopropanation. This approach has defined the biosynthetic origin of each cyclopropane ring, methyl branch, and oxygen function in M. tuberculosis mycolic acids. MmaA4 is needed for the synthesis of oxygenated mycolates through its addition of the distal methyl branch and hydroxyl group common to all oxygenated mycolates (9, 13). MmaA3 adds the methyl group to this hydroxyl group to create methoxymycolates (5), and several M. bovis BCG substrains lack methoxymycolates due to mutations in mmaA3 (5, 10, 32). The mmaA1 and cmaA2 gene products both function in the trans cyclopropanation of methoxy- and ketomycolates (3, 17). mmaA2 and pcaA encode the mycolic acid methyltransferases that add the cyclopropane rings to the distal and proximal positions of the alpha mycolate, respectively (16, 18). The mmaA2 gene product also has a redundant role with the cmaA2 gene product in cis cyclopropanation of the oxygenated mycolates (3). No mycolic acid modification function has been elucidated for the umaA1 or cmaA1 gene products (3, 16), despite the cyclopropanating activity of the latter expressed in M. smegmatis (31), a mycobacterial species that does not constitutively produce cyclopropanated mycolates but can produce them at low temperatures (1).
The role of mycolic acid modification in mycobacterial pathogenesis is only partially understood. The fact that many of the responsible genes are conserved among pathogenic slow-growing mycobacteria (such as M. marinum, M. leprae, and the other members of the M. tuberculosis complex) suggests some essentiality for pathogenetic processes. Evidence for the importance of mycolic acid modifications in pathogenesis came from testing of mutants lacking MAMTs in mice. M. tuberculosis lacking mmaA4 (also called hma) has a growth defect during the first 3 weeks of murine infection (13). An M. tuberculosis ΔmmaA3 strain has not been examined in mice, although M. bovis BCG SSI (which lacks a functional MmaA3 protein) is not attenuated compared to mmaA3-complemented BCG SSI, with reconstituted methoxymycolate production (6). An M. tuberculosis ΔpcaA strain (which lacks the proximal cis cyclopropane ring on the alpha mycolates [18]) has multiple phenotypes: defective growth during the first week of mouse infection (24), defective persistence, and attenuated immunopathology (18). An M. tuberculosis ΔcmaA2 strain (which lacks the trans ring on the keto- and methoxymycolates [17]) was shown to be hypervirulent in mice, eliciting an exaggerated immune response leading to more profound tissue damage (25). Thus, there is substantial evidence that specific MAMTs have specific and diverse functions in M. tuberculosis pathogenesis.
Two major hypotheses have been presented for the pathogenic function of mycolic acid modification in M. tuberculosis. The first is that cyclopropanation is important for membrane function, affecting either membrane rigidity or chemical resistance to oxidative stress (13, 15). An alternative model is that these lipid structures affect immune activation. There is substantial evidence that the fine structure of mycolic acids in trehalose dimycolate (TDM) alters its inflammatory properties and can explain many of the phenotypes of MAMT-deficient strains (9, 24, 25). For example, TDM derived from the M. tuberculosis ΔpcaA strain is hypoinflammatory, whereas ΔcmaA2 TDM is hyperinflammatory, and both of these phenotypes correlate with the in vivo phenotypes of the pcaA and cmaA2 mutant strains. In addition, the inflammatory properties of the pcaA and cmaA2 mutant strains can be transferred to delipidated wild-type (WT) cells by petroleum ether extracts, demonstrating that these lipids are direct pathogenesis effectors (24, 25).
All of the data presented above were derived from mutants of M. tuberculosis lacking single MAMTs. The combined effect of complete cyclopropane ring deficiency remains unknown. Whether a strain lacking all cyclopropane rings will be attenuated (and to what extent) or hypervirulent (like the cmaA2 mutant) is also unknown. This question is important in order to understand whether these enzymes are attractive targets for antimicrobial development and to simulate what the in vivo effects of such an inhibitor would be. We and others have recently shown that an inhibitor of multiple mycolic acid methyltransferases is bactericidal, suggesting that deleting genes encoding these enzymes may be synthetically lethal (2, 30).
To investigate the importance of MAMTs as a group to M. tuberculosis pathogenicity, we sought to create strains of M. tuberculosis lacking all cyclopropanation (via deletion of all MAMTs that synthesize cyclopropane rings) or all MAMTs (including those that create methyl branches) and to test these strains in experimental mouse infection.
MATERIALS AND METHODS
Ethics statement.
All mouse procedures performed at the Memorial Sloan Kettering Cancer Center (MSKCC) were conducted following the NIH guidelines for housing and care of laboratory animals and performed in accordance with institutional regulations after protocol review and approval by the Institutional Animal Care and Use Committee of the Sloan Kettering Institute. All animal experiments in Belgium were reviewed and approved by the ethical committee of the Institute of Public Health-Veterinary and Agrochemical Research Institute (IPH-VAR), Brussels, Belgium.
Strains, media, and culture conditions.
The wild-type strain used was M. tuberculosis Erdman. Bacteria were grown at 37°C in Middlebrook 7H9 medium (Difco) containing 0.5% glycerol, 0.05% Tween 80, and 10% oleic acid-albumin-dextrose-catalase (OADC). For growth on plates, 7H10 agar was used, and no Tween was added. For antibiotic selection, we used hygromycin B (50 μg/ml), kanamycin (20 μg/ml), streptomycin (20 μg/ml), and Zeocin (15 μg/ml).
Strain construction.
All gene deletions were performed using homologous recombination via transduction with the temperature-sensitive mycobacteriophage phAE87, as described previously (3). The plasmids and strains used in this study are shown in Table 1. Information regarding exact cloning sites and primers used for PCR amplification is available upon request. To delete the mmaA1-4 locus, the 700-bp region upstream of mmaA1 and the 700-bp region downstream of mmaA4 were cloned from the plasmid pJSC252 on either side of the hygromycin resistance gene in pMSG360. This plasmid was used, via recombineering in Escherichia coli strain EL350, to create the specialized transducing phage phDB1. To delete the pcaA/umaA1 locus, we used the hygromycin-based transducing phage phMSG123. To delete cmaA1 and cmaA2, we used the previously described Zeocin-based transducing phages phDB4 and phDB10, respectively (3).
Table 1.
Plasmids and M. tuberculosis strains used in this work
| Plasmid or strain | Antibiotic marker or resistance | Description or genotype | Mycolic acid phenotype |
|---|---|---|---|
| Plasmids | |||
| pMV306kan | Kanamycin | attB-integrating empty vector | |
| pDB22 | Zeocin | attB-integrating vector expressing mmaA1, mmaA2, mmaA3, and mmaA4 | |
| pDB32 | Kanamycin | Multicopy, episomal empty expression vector | |
| pDB46 | Kanamycin | Multicopy, episomal expression vector, based on pDB32, expressing mmaA1, mmaA2, mmaA3, mmaA4, pcaA, and cmaA2 | |
| pDB49 | Kanamycin | attB-integrating vector expressing mmaA3 and mmaA4 | |
| pDB60 | Streptomycin | attB-integrating empty vector | |
| pDB61 | Streptomycin | attB-integrating vector expressing mmaA3 and mmaA4 | |
| pDB90 | Kanamycin | attB-integrating vector, based on pMV306kan, expressing mmaA1, mmaA2, mmaA3, mmaA4, pcaA, and cmaA2 | |
| pDB111 | Kanamycin | attB site integration vector expressing mmaA3 and mmaA4; does not carry an integrase gene; can be inserted into bacteria only with another integrase-containing plasmid at the attB site; cannot be lost spontaneously | |
| Strains | |||
| Erdman | None | Wild type | Wild type |
| MGM1953 | Hygromycin, Zeocin | ΔmmaA1-4::loxP-hyg-loxP attB::mmaA1-4 (zeocin) | Wild type |
| MGM1960 | Hygromycin, Zeocin | ΔmmaA1-4::loxP ΔpcaA/umaA1::hyg attB::mmaA1-4 | Fully defective in proximal alpha cyclopropanation |
| MGM1961 | Hygromycin, streptomycin | ΔmmaA1-4::loxP ΔpcaA/umaA1::hyg attB::mmaA3-4 (strep) | Fully defective in proximal alpha cyclopropanation, partial defect in distal alpha cyclopropanation, fully defective in trans-cyclopropanation of methoxy- and ketomycolates |
| MGM1962 | Hygromycin, streptomycin | ΔmmaA1-4::loxP ΔpcaA/umaA1::hyg attB::(strep) | No oxygenated mycolates; alpha mycolates are fully defective for proximal cyclopropanation and partially defective for distal cyclopropanation |
| MGM1985 | Streptomycin | attB::(strep) | Wild type |
| MGM1990 | Hygromycin, Zeocin, streptomycin | ΔmmaA1-4::loxP attB::mmaA3-4 (strep) ΔpcaA/umaA1::hyg ΔcmaA2::zeo | Has all 3 classes (alpha, methoxy, keto) but no cyclopropane rings |
| MGM1991 | Hygromycin, Zeocin, streptomycin | ΔmmaA1-4::loxP attB::(strep) ΔpcaA/umaA1::hyg ΔcmaA2::zeo | Has no oxygenated mycolates, only alpha mycolates; has no cyclopropane rings |
| MGM1995 | Hygromycin, Zeocin | ΔmmaA1-4::loxP ΔpcaA/umaA1::hyg ΔcmaA2::zeo | Identical to MGM1991 |
| MGM1998 | Hygromycin, Zeocin, kanamycin | ΔmmaA1-4::loxP attB::mmaA3-4 (kan) ΔpcaA/umaA1::hyg ΔcmaA2::zeo | Identical to MGM1990 but no integrase at attB |
Construction of complementation plasmids and other plasmids used in this study.
To precomplement M. tuberculosis with a second copy of the mmaA1-4 locus, we excised the entire locus from pJSC252 by using EcoRI and a partial digestion with NotI and cloned the resulting 4,517-bp piece into the same sites in the Zeocin-marked attB site (L5) integration plasmid pDB19, creating pDB22. To express only mmaA3 and mmaA4 from an attB site integration plasmid, pJSC252 was cut using EcoRI and NaeI, and the resulting 2,442-bp piece was ligated into the EcoRI and EcoRV sites of the kanamycin-marked integration vector pMV306kan, creating pDB49. To make an attB site integration vector conferring streptomycin resistance, the streptomycin resistance gene was first PCR amplified from pTE-2MOX by use of primers with XbaI sites at both ends. The product was digested with XbaI and cloned into the NheI/SpeI sites of pMV306kan and pDB49, creating pDB60 and pDB61, respectively. To make a kanamycin-marked mmaA3- and mmaA4-expressing vector that can integrate into the bacterial chromosome but cannot spontaneously self-excise, we first cloned a 949-bp piece containing the ampicillin resistance gene into the NaeI site of pMV306kan, creating pDB59. This interrupted the open reading frame of the integrase gene after 300 bp (out of 1,021 bp). We then cloned the 2,441-bp EcoRI/NaeI piece from pJSC252 into the EcoRI and EcoRV sites of pDB59, creating pDB111. To create an attB site integration plasmid expressing all 6 functional methyltransferase genes (mmaA1 to -4, cmaA2, and pcaA), we first cloned the cmaA2 gene (as an EcoRI/HindIII fragment) into the same sites in the previously mentioned pDB22 vector. We then changed the selection marker on the plasmid by cutting out the Zeocin resistance marker with NheI and SpeI and instead ligating the kanamycin resistance gene cut from pMV306kan, using the same enzymes. This resulted in pDB83. We then cloned the pcaA gene, after amplification using primers with XbaI sites at both ends, into the NheI site of pDB83, creating pDB90. Finally, to express all 6 functional methyltransferases from an episomal multicopy plasmid, we used the previously described pDB46 vector (2).
Preparation and analysis of MAMEs and phthiocerol dimycocerosate (PDIM).
14C-labeled mycolic acid methyl esters (MAMEs) were prepared from logarithmic-phase cultures of M. tuberculosis with 50 μCi of [1-14C]acetic acid (58.9 mCi/mmol) (PerkinElmer Life Sciences) for 24 h. MAMEs were prepared from whole bacilli and analyzed by two-dimensional argentation thin-layer chromatography (TLC) as described previously (16). Briefly, 50 ml of culture was collected by centrifugation, and the bacterial pellet was resuspended in water, diluted with 40% tetrabutylammonium hydroxide (TBAH) to a final concentration of 20% TBAH, and then heated overnight at 100°C. An equal volume of dichloromethane and 0.15 ml of methyl iodide were added, and the suspension was rotated at room temperature for 1 h. After phase separation, the aqueous phase was discarded and the organic fraction was evaporated under a nitrogen stream. The residue was extracted with ethyl ether and dried. Finally, MAMEs were precipitated from 2:1 toluene-acetonitrile by addition to 2 volumes of acetonitrile at −20°C. The resulting MAMEs were resuspended in ethyl ether. For two-dimensional TLC separation, 90% of the TLC plate was immersed in 10% silver nitrate and then left to dry at 100°C for 15 min. The mycolic acid sample was first developed on the non-silver-immersed strip for 6 developments and then developed 5 times on the silver-immersed area. The solvent for both dimensions was hexanes-ethyl acetate (95:5).
Because of the prolonged in vitro passaging required to construct the MAMT-deficient strains, we confirmed PDIM production in the final strains (19). PDIM analysis was done as described previously (11). Briefly, 100 μl of bacterial culture in logarithmic phase was added to 1 ml of 7H9 broth-OADC containing 0.1 mCi/ml [1-14C]propionic acid in sealed microtubes and labeled for 5 days. Following centrifugation, cells were extracted using 600 ml methanol–0.3% aqueous NaCl (10:1 [vol/vol]) and 300 ml petroleum ether. After shaking, the suspension was centrifuged again, and the upper level was collected into a new tube and allowed to air dry. The extract, containing PDIM, was resuspended in 50 μl petroleum ether, and 10 μl was separated on a TLC plate, using petroleum ether-ethyl acetate (98:2 [vol/vol]) as the solvent. The plates were developed three times and imaged on a phosphorimager.
Animal infections.
For determination of virulence, 8-week-old female C57BL/6 mice were infected with M. tuberculosis by the aerosol route as described previously (28). Bacterial numbers were determined by plating organ homogenates for CFU determination at the indicated times. For immunophenotyping of the cyclopropane-deficient strains, C57BL/6 mice were bred in the animal facilities of the Scientific Institute of Public Health (Brussels, Belgium) from breeding pairs obtained from Harlan Laboratories (The Netherlands). Mice were 6 to 8 weeks of age at the start of experiments. Bacteria were grown on liquid 7H9-OADC medium and harvested after 3 weeks, and CFU numbers were determined by plating on Middlebrook 7H11 agar supplemented with oleic acid-albumin-dextrose-catalase. Aliquots were stored frozen at −70°C until use. Bacteria were instilled into C57BL/6 mice by the intratracheal route (104 CFU) as reported before (21a). At day 21 postinfection, mice were sacrificed and their lungs and spleens removed aseptically for gene expression studies and cytokine analysis.
RT-qPCR.
RNA was extracted using TRIzol reagent according to the manufacturer's instructions (Sigma-Aldrich). cDNA was synthesized using SuperScript II reverse transcriptase (Invitrogen). Reverse transcription-quantitative PCR (RT-qPCR) was performed on a CFX96 system (Bio-Rad), using a qPCR core kit for SYBR green I (Eurogentec). Each RT-qPCR amplification was performed in triplicate under the following conditions: 50°C for 2 min and 95°C for 10 min followed by 40 cycles at 95°C for 15 s and 60°C for 1 min. The forward and reverse primers used are shown in Table S1 in the supplemental material. Hydroxymethylbilane synthase mRNA was used as a reference housekeeping gene for normalization. Normalized fold expression of target mRNA was calculated by CFX Manager 2.1 software (Bio-Rad).
Cytokine production.
Mice were sacrificed 3 weeks after infection. Spleens and lungs were removed aseptically and homogenized in a loosely fitting Dounce homogenizer. Lung and spleen cells from five mice per group were tested for cytokine responses to purified protein derivative (PPD; 25 μg/ml) prepared from M. tuberculosis, recombinant E. coli-derived Ag85A (Rv3804c; 5 μg/ml), Ag85A241-260 peptide (immunodominant I-Ab-restricted peptide of Ag85A [Rv3804c] [12]; 10 μg/ml), and ESAT-61-20 peptide (immunodominant I-Ab-restricted peptide of early secretory antigenic target 6 [8]; 10 μg/ml). Supernatants were harvested after 24 h (for tumor necrosis factor alpha [TNF-α]) or 72 h (for gamma interferon [IFN-γ] and interleukin-17A [IL-17A]), when peak values of the respective cytokines can be measured. Spleen cells from individual mice were tested for cytokine responses, whereas lung cells were pooled from multiple mice. Supernatants were stored frozen at −20°C until analysis. IFN-γ was detected using an enzyme-linked immunosorbent assay (ELISA) with purified rat anti-mouse IFN-γ as the capture antibody and biotin-labeled rat anti-mouse IFN-γ as the detection antibody (BD Pharmingen). IL-17A and TNF-α were detected by use of a commercial ELISA kit (eBioscience). The IFN-γ detection limit was 5 pg/ml, that for IL-17A was 4 pg/ml, and that for TNF-α was 8 pg/ml.
IFN-γ and IL-17A ELISPOT assays.
Specific spleen cell IFN-γ secretion was assayed by enzyme-linked immunospot (ELISPOT) assay as described previously (26). Briefly, 96-well surfactant-free mixed cellulose ester membrane plates (Millipore) were incubated overnight at 4°C with 50 μl of purified anti-mouse IFN-γ capture antibody in phosphate-buffered saline (PBS) (15 μg/ml; BD Pharmingen) and then saturated with RPMI 1640 medium (Gibco, Grand Island, NY) supplemented with penicillin, streptomycin, 5 × 10−5 M 2-mercaptoethanol, and 10% fetal calf serum (FCS) for 2 h at 37°C. Lung leukocytes (pooled from five mice per group) were added at a known concentration to the same medium in the presence or absence of peptides (5 or 10 μg/ml), and plates were incubated for 40 h at 37°C and 5% CO2. After extensive washing, plates were incubated overnight at 4°C with 50 μl of biotinylated rat anti-mouse IFN-γ (2 μg/ml) (BD Pharmingen), washed, and incubated for 45 min at 37°C and 5% CO2 with alkaline phosphatase-labeled streptavidin (Sigma). After washing of the plates, spots were revealed with a Bio-Rad (Hercules, CA) alkaline phosphatase conjugate substrate kit following the manufacturer's instructions, and plates were analyzed on a Bioreader 3000 LC plate reader (BioSys, Germany). IL-17A ELISPOT assay was performed in the same manner, with purified rat anti-mouse IL-17A as the capture antibody (BD Pharmingen) and biotin-labeled rat anti-mouse IL-17A as the detection antibody (BD Pharmingen). Results are shown as numbers of mean spot-forming cells (SFC) per million leukocytes.
RESULTS
M. tuberculosis is viable without cyclopropanation or any functional MAMTs.
Previous studies demonstrated that chemical inhibition of mycolic acid methyltransferases is lethal to M. tuberculosis (2, 30) but that individual mycolic acid methyltransferases are nonessential. In addition, although M. tuberculosis strains lacking pcaA or cmaA2 are attenuated or hypervirulent, respectively, in the mouse (18, 24, 25), the in vivo phenotype of an M. tuberculosis strain lacking all mycolic acid modification is not known. To test the importance of the mycolic acid modification system in M. tuberculosis viability and pathogenesis, we attempted to delete all mycolic acid methyltransferase genes from the M. tuberculosis genome. Although there are eight mycolic acid methyltransferase genes, cmaA1 has no defined function (16), and we therefore targeted the seven other genes, which are clustered in three genetic loci (mmaA1-4, pcaA/umaA1, and cmaA2). Because of the possibility of synthetic lethality, we started by integrating a second copy of the mmaA1-4 locus at the attB site of the chromosome of M. tuberculosis, using the Zeocin-marked plasmid pDB22. We then deleted the native mmaA1-4 locus from this strain with the specialized transducing mycobacteriophage phDB1, which carries an mmaA1-4 deletion allele marked by a loxP-flanked hygromycin marker. The ΔmmaA1-4 genotype was confirmed by Southern blotting (Fig. 1A). This mutant was called MGM1953 (ΔmmaA1-4::loxP-Hyg-loxP attB::mmaA1-4), is resistant to hygromycin (and Zeocin), and has a normal mycolic acid profile due to the second copy of the mmaA1-4 locus at the attB site (data not shown). To allow further genetic manipulation, we then removed the hygromycin resistance cassette by electroporating the cells with pMSG381, an episomal plasmid expressing the Cre recombinase and a kanamycin resistance marker. Because the hygromycin cassette on phDB1 is flanked by loxP sites, expression of the Cre recombinase resulted in excision of the resistance marker. We then “cured” the strain of the pMSG381 plasmid by removing kanamycin selection. The loss of the plasmid was confirmed by reversion of the strain to kanamycin sensitivity. We then deleted pcaA/umaA1 in this strain by using the specialized transducing phage phMSG123. The deletion was confirmed by Southern blotting (Fig. 1B), and the resulting mutant was called MGM1960 (ΔmmaA1-4::loxP attB::mmaA1-4 ΔpcaA/umaA1::hyg). Functionally, this mutant lacks only pcaA and umaA1, and mycolic acid analysis confirmed the previously described defect in proximal alpha mycolate cyclopropanation (18) (data not shown).
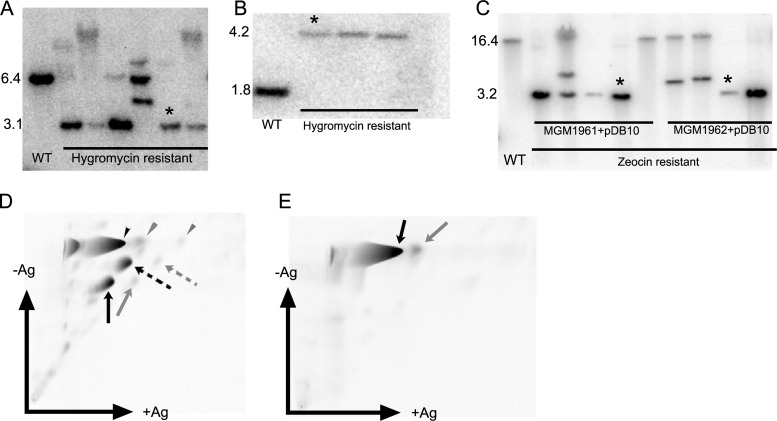
Fig 1.
Generation of M. tuberculosis strains lacking all cyclopropanation or all mycolic acid methyltransferases. (A) Deletion of mmaA1-mmaA2-mmaA3-mmaA4 locus from a merodiploid strain of M. tuberculosis Erdman with attB::mmaA1-4. When digested with EcoRI and probed with the 1,000-bp piece immediately upstream of mmaA1, the WT gives a 6.4-kb band, whereas the strain with the mmaA1-4 deletion gives a 3.1-kb band. The strain marked with an asterisk was chosen for further manipulation and was named MGM1953. (B) Deletion of pcaA/umaA1 from Δmma1-4::loxP attB::mmaA1-4 strain. The Southern blot confirms the deletion of pcaA and umaA1 from a strain previously deleted of mmaA1-4 (after removal of the loxP-hyg-loxP marker with Cre recombinase). When digested with SmaI and probed with the 600-bp piece immediately upstream of umaA1, the WT produces a 1.8-kb band, whereas the pcaA/umaA1 deletion strain produces a 4.2-kb band. The strain marked with an asterisk was designated MGM1960 and used for further genetic manipulation. (C) Deletion of cmaA2 from ΔmmaA1-4::loxp ΔpcaA/umaA1::Hyg attB::mmaA3-4 strain (MGM1961) and ΔmmaA1-4::loxp ΔpcaA/umaA1::Hyg attB::Strepr strain (MGM1962) via transduction of ΔcmaA2::zeo (phDB10). The Southern blot confirms the deletion of cmaA2 from MGM1961 or MGM1962. When digested with HindIII and probed with the 600-bp piece immediately upstream of cmaA2, the WT produces a 16.4-kb band, whereas the ΔcmaA2 strain produces a 3.5-kb band. The strains marked with asterisks were designated MGM1990 and MGM1991 and were characterized further. (D) Two-dimensional TLC of mycolic acid methyl esters of the cell wall of MGM1990. All three mycolic acid classes are present (black arrowhead, alpha class; black dashed arrow, methoxy class; and black arrow, keto class), and all are fully unsaturated. Essentially no mature cyclopropanated mycolic acids are made, as shown by the trivial amounts marked by the gray arrows (methoxy and keto classes) or gray arrowhead (alpha class). (E) Two-dimensional TLC of mycolic acid methyl esters of the cell wall of MGM1991. No oxygenated mycolates are present (no methoxy- or ketomycolates), and the remaining alpha mycolate is almost fully unsaturated. Small amounts of alpha mycolate with a single cyclopropane ring are visible (gray arrow).
We then proceeded to remove the attB-integrated copy of the mmaA1-4 locus by marker exchange. We electroporated MGM1960 with either pDB61 (carrying mmaA3 and mmaA4 but not mmaA1 and mmaA2) or pDB60 (not expressing any MAMTs), both of which are attB integration plasmids with a streptomycin resistance marker. Selection for streptomycin resistance allowed for exchange of pDB60 or pDB61 with pDB22. The resulting strains were MGM1961 (ΔmmaA1-4::loxP attB::mmaA3-4 ΔpcaA/umaA1::hyg) and MGM1962 (ΔmmaA1-4::loxP attB::Strepr ΔpcaA/umaA1::hyg). The expected mycolic acid phenotype for both strains was confirmed by TLC of radiolabeled mycolic acids (data not shown). For the last step, we deleted cmaA2 from both MGM1961 and MGM1962 by using the Zeocin-marked specialized transducing phage phDB10. The deletion was confirmed by Southern blotting (Fig. 1C), and the two resulting mutants were named MGM1990 (ΔmmaA1-4::loxP attB::mmaA3-4 ΔpcaA/umaA1::hyg ΔcmaA2::zeo) and MGM1991 (ΔmmaA1-4::loxP attB::Strepr ΔpcaA/umaA1::hyg ΔcmaA2::zeo).
Based on our prior studies, we predicted that the mycolic acids of MGM1990 and MGM1991 would lack all cyclopropanation but produce all major mycolic acid classes (MGM1990) or lack all oxygenated mycolic acids and produce a fully unsaturated alpha mycolate (MGM1991). To examine this prediction, we examined the mycolic acid profiles of MGM1990 and MGM1991 by two-dimensional argentation TLC. The mycolates from MGM1990 showed all three major classes (alpha, methoxy-, and ketomycolates). All three classes were retarded in the silver dimension, indicating double bonds in place of cyclopropane rings (Fig. 1D). MGM1991 produced no methoxy- or ketomycolates, consistent with the absence of MmaA4 activity (9, 13). The alpha mycolate was fully unsaturated, consistent with the loss of both PcaA and MmaA2 (Fig. 1E). We did observe a very small amount of monocyclopropanated alpha mycolate in both MGM1990 and MGM1991, which may have been due to CmaA1, the only MAMT remaining in these strains (Fig. 1D and E). These complex genetic manipulations created strains of M. tuberculosis that lack all cyclopropanation (MGM1990) and all MAMTs (MGM1991).
Sudden changes in mycolic acid methyltransferase function are lethal to M. tuberculosis.
The creation of MGM1990 and MGM1991 indicates that M. tuberculosis is viable without MAMTs. This result contrasts with our prior finding that chemical inhibition of MAMTs with dioctylamine causes severe growth inhibition and cell death (2). There are many examples where chemical inhibition of a protein target does not phenocopy genetic deletion of the gene encoding that target (20). Nevertheless, we sought an explanation for these findings. We hypothesized that the sequential deletion of mycolic acid methyltransferases allowed compensatory changes to accumulate in the membrane, such that the loss of multiple MAMTs could be tolerated. In contrast, the sudden change in membrane fluidity that would accompany dioctylamine inhibition of MAMTs would not allow these compensatory changes to occur. Under this model, we reasoned that reintroduction of mycolic acid methyltransferases into an MAMT-deficient strain might also be lethal, because sudden changes in membrane fluidity would occur with the sudden reacquisition of mycolic acid methyltransferase function. To test this hypothesis, we used strain MGM1995, a derivative of MGM1990 that has lost the attB-integrated mmaA3-mmaA4 cassette and therefore has no methyltransferase genes (except for cmaA1) and has a native, intact attB site. We attempted to introduce either an integrating control vector (pMV306kan), a control episomal plasmid (pDB32), or an integrating (pDB90) or episomal (pDB46) plasmid carrying all six MAMT genes deleted in MGM1990. We found that MGM1995 was easily transformed by both integrating and episomal vectors (pMV306kan and pDB32), indicating that the loss of cyclopropanation does not impair plasmid transformation. In fact, MGM1995 appeared hypertransformable compared to the wild type (Table 2). We were able to transform wild-type M. tuberculosis with pDB90 and pDB46, indicating that these plasmids are tolerated by M. tuberculosis cells with an intact set of MAMTs. In contrast, we could not recover kanamycin-resistant transformants of MGM1995 by using either plasmid (Table 2), indicating that these plasmids are lethal in cells lacking MAMT function. These results are consistent with the model that sudden changes in mycolic acid methyltransferase function are lethal to mycobacteria, presumably through sudden changes in membrane function, and may explain the difference between genetic deletion and chemical inhibition of this enzyme family.
Table 2.
Efficiency of transformation of MAMT-expressing plasmids into M. tuberculosis Erdman or MGM1995a
| Plasmid | Mean no. of transformants/μg of DNA |
MGM1995/WT ratio | |
|---|---|---|---|
| WT (Erdman) | MGM1995 | ||
| pDB32 | 316 | 24,333 | 77 |
| pDB46 | 37 | 0 (2,850) | |
| pMV306Kan | 196 | 2,566 | 13 |
| pDB90 | 88 | 0 (1,150) | |
Reintroduction of the six functional MAMT genes (mmaA1, mmaA2, mmaA3, mmaA4, cmaA2, and pcaA) into a strain lacking all of them (MGM1995) by electroporation is lethal. Results of a representative experiment out of three are shown. The WT Erdman strain or MGM1995 was electroporated with either pDB32 (empty episomal plasmid), pDB46 (similar episomal plasmid expressing the above six methyltransferases), an integrating vector (pMV306Kan), or an integrating plasmid expressing the above six methyltransferases (pDB90). A total of 109 bacteria and 500 ng of DNA were used for each electroporation, and the bacteria were plated in several dilutions, allowing for exact estimation of the number of transformants. Each electroporation was done 3 times (in each of the three experiments mentioned above), and the mean number of transformants per μg of DNA is shown. The last column shows the ratio of the number of transformants of MGM1995 to that of WT Erdman, showing the factor by which MGM1995 is more competent. The data in parentheses show the number of transformants of MGM1995 one would expect by the competency derived from control plasmid transformations.
Methyltransferase-deficient strains lose acid fastness and are more sensitive to detergent stress.
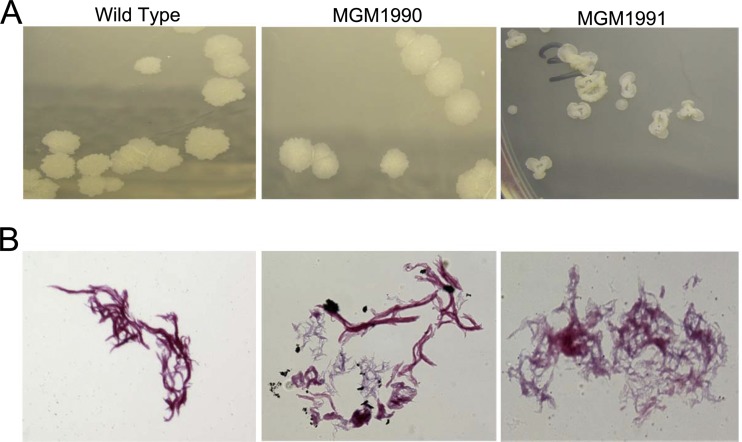
We observed that the colony morphology of MGM1991 was markedly different from that of the wild type or MGM1990, with smaller colonies and a smooth, “donut” morphology (Fig. 2A). This morphology was previously described for the single mmaA4 mutant (9, 13) and is probably related to the lack of oxygenated mycolates. The hallmark of mycobacteria, their acid fastness, has been linked to the mycolic acid layer of the mycobacterial cell wall (7). We previously reported that chemical inhibition of MAMTs by dioctylamine leads to a loss of acid fastness and increased membrane permeability. Accordingly, we tested whether the MAMT- and cyclopropanation-defective mutants were indeed deficient for acid fastness. We found both MGM1990 and MGM1991 to be less acid fast than the wild type, as judged by more intense counterstaining with methylene blue after a decolorization step that left wild-type M. tuberculosis acid fast (Fig. 2B).
Fig 2.
(A) Macroscopic morphology of colonies. The images shown are of representative colonies of the indicated strains on 7H10 agar. The wild type and MGM1990 have similar morphologies, whereas the colonies of MGM1991 are small and smooth and have a “donut” shape. (B) Acid fastness (modified Kinyoun stain) of M. tuberculosis Erdman (WT) compared to MGM1990 and MGM1991.
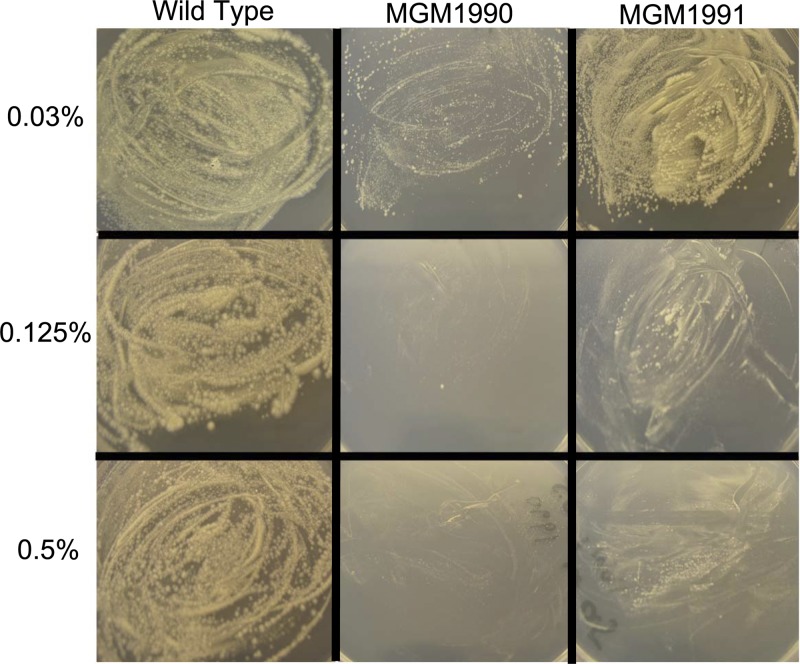
We then tested the relative sensitivities of these strains to the detergent tyloxapol by plating the bacteria on 7H10 plates impregnated with increasing concentrations of tyloxapol. We found that concentrations of tyloxapol of up to 0.5% had no effect on growth of wild-type M. tuberculosis (MGM1985) but that growth of MGM1990 and MGM1991 was completely inhibited at this concentration (Fig. 3). At lower concentrations of tyloxapol, MGM1990 was more susceptible to tyloxapol than MGM1991, with impaired growth at concentrations as low as 0.03%. Similar results were obtained when these strains were grown in liquid medium containing tyloxapol (data not shown). These results implicate mycolic acid modification in detergent resistance but reveal different detergent susceptibility phenotypes for MGM1990 and MGM1991. The presence of oxygenated mycolates in MGM1990 confers severe detergent susceptibility that is partially ameliorated when these oxygenated lipids are lost in MGM1991.
Fig 3.
Loss of cyclopropanation affects detergent susceptibility. Equivalent bacterial numbers of the WT, MGM1990, and MGM1991 strains were plated on 7H10 agar plates supplemented with the detergent tyloxapol and allowed to grow for 4 weeks. The numbers on the left give the concentrations of tyloxapol in the plates. Both mutants were sensitive to tyloxapol, MGM1990 more so than MGM1991.
Cyclopropanation of mycolic acids is essential for pathogenicity in mice.
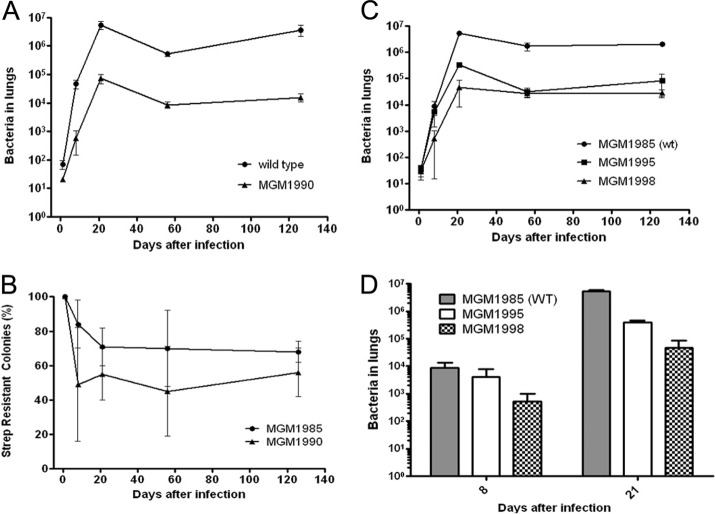
To assess the importance of mycolic acid cyclopropanation for pathogenicity in mice, we infected mice with either wild-type M. tuberculosis or MGM1990. As mentioned above, this mutant lacks all of the functional MAMTs but has mmaA3 and mmaA4 integrated into the chromosome at the attB site and therefore produces all 3 mycolic acid classes (alpha, methoxy, and keto) but no cyclopropane rings, leaving the mycolic acids fully unsaturated (Fig. 1D). This strain therefore affords an opportunity to understand the role of cyclopropanation in M. tuberculosis pathogenicity. After aerosol infection of mice with approximately 100 bacteria, bacterial numbers of MGM1990 were 80-fold lower than those of the wild type at day 8 postinfection (Fig. 4A). This decreased virulence persisted at subsequent time points up to 130 days after infection (Fig. 4A). This phenotype is a more severe attenuation than that previously reported for the pcaA single mutant, which has a transient early attenuation phenotype (24) and a late persistence defect (18). The phenotype of MGM1990 also differs from that of the ΔcmaA2 strain, which does not differ from the wild type in terms of bacterial load but is hypervirulent (25).
Fig 4.
(A) Mice were infected with either WT (circles) or MGM1990 (triangles) bacteria. CFU in both lungs were determined on days 1, 8, 21, 56, and 126 after infection. The combined results of two experiments are shown. (B) In vivo loss of mmaA3-4. The percentage of MGM1990 bacteria retaining the attB-integrated mmaA3-mmaA4 cassette during mouse infection was determined by plating of bacteria on plates with and without streptomycin at the indicated time points. MGM1985 is wild-type M. tuberculosis with an empty attB-integrating streptomycin-selected cassette. (C) Mice were infected with either MGM1985 (WT; circles), MGM1995 (unsaturated alpha mycolates, no oxygenated mycolates; squares), or MGM1998 (all three mycolate classes present, no cyclopropanes; triangles). CFU in both lungs were determined on days 1, 8, 21, 56, and 126 after infection. Each time point represents the mean for 4 or 5 mice, and error bars show standard deviations (SD). (D) Bacterial loads on days 8 and 21 after infection are shown for the experiment in panel C.
During the mouse infection experiments with MGM1990, we noted a mixed-colony morphology of the MGM1990 strain, but not wild-type M. tuberculosis, recovered from mouse organs. We observed colonies that resembled the parental strain and some that resembled the “donut” morphology of MGM1991 (Fig. 2A). On day 8 after infection, almost 50% of the recovered colonies were of the “donut” morphology (Fig. 4B). We hypothesized that these MGM1990 cells had lost the attB-integrated cassette carrying the mmaA3 and mmaA4 genes, whose presence is the only difference between MGM1990 and MGM1991. Such integrated cassettes can be lost because the plasmid encodes an integrase, which can catalyze excision of the cassette (22). We investigated this hypothesis by picking 12 colonies that resembled MGM1991 and testing them for streptomycin sensitivity. All 12 were streptomycin sensitive, indicating loss of the plasmid (data not shown). We further analyzed the mycolic acid profiles of six of these strains and demonstrated that all six had lost keto- and methoxymycolate production, consistent with the loss of MmaA4 function (see Fig. S2 in the supplemental material). One of these clones was designated MGM1995. These experiments confirmed that MGM1990 can lose mmaA3-mmaA4 during mouse infection and imply some selective pressure against retention of oxygenated mycolate production early in infection, at least when the mycolic acids are unsaturated. We further characterized this phenomenon by measuring the in vivo rate of loss of streptomycin-marked, attB-integrated cassettes from M. tuberculosis Erdman attB::Strepr (MGM1985) and MGM1990. The results (Fig. 4B) show that MGM1990 lost the mmaA3-mmaA4 cassette faster than wild-type cells lost the attB::Strepr cassette. This difference was most dramatic in the first 8 days, indicating that there may be a selective advantage for mmaA3-mmaA4 loss during the first week of infection.
The high rate of mmaA3-mmaA4 loss in the MGM1990 strain complicates interpretation of the pathogenesis phenotype, because a mixed genotype develops during infection. To circumvent this problem and more clearly delineate the pathogenesis phenotypes of MGM1990 and MGM1991, we sought to construct a new MGM1990 strain in which the attB-integrated mmaA3-mmaA4 cassette could not be lost. We accomplished this goal by constructing a kanamycin-marked attB integration plasmid bearing mmaA3 and mmaA4 but without a functional integrase gene (pDB111). This plasmid can integrate at the attB site if the integrase is expressed in trans (such as from pDB61 in strain MGM1990), but once it is integrated it cannot be excised. We exchanged pDB111 into MGM1990 and designated this strain MGM1998. This strain has the same mycolic acid profile as MGM1990 (data not shown), is kanamycin resistant, and cannot lose the mmaA3-mmaA4 cassette.
We then infected mice with MGM1985 (to assess for any effect of streptomycin resistance on attenuation), MGM1998 (cyclopropane-deficient strain), and MGM1995 (strain deficient of all MAMTs). The results show that streptomycin resistance has no strong effect on pathogenicity by itself, as the CFU counts of MGM1985 were not different from those usually obtained for the wild type (Fig. 4C and D). We observed that titers of MGM1998 were 15-fold lower than those of the wild type (MGM1985) at 8 days, whereas MGM1995 was not substantially attenuated at this early time point. During the second and third weeks of infection, titers of MGM1995 diverged from those of the wild type such that they were 16-fold lower at 21 days and then declined further, such that the titer 56 days after infection was 50-fold lower than that of the wild type, similar to MGM1998, whose titer was 62-fold lower than that of the wild type at this time point (Fig. 4C and D). These results reveal distinct pathogenesis phenotypes for M. tuberculosis lacking cyclopropanation versus M. tuberculosis lacking all mycolic acid methyltransferases. Cyclopropane deficiency with retention of all three mycolic acid classes causes severe attenuation in the first week of infection. In contrast, loss of all MAMTs confers severe attenuation that begins at week 2 of infection.
Immunologic consequences of complete cyclopropane deficiency in vivo.
Mycolic acids of M. tuberculosis are a well-known trigger of pathogen-associated host innate immune responses (21), and the cyclopropane modification on mycolic acids modulates immune recognition in the context of free mycolic acid (29), trehalose dimycolate (24, 25), or whole bacilli (24, 25). To characterize the immune response to M. tuberculosis completely lacking cyclopropanation, we infected mice intratracheally with 104 CFU of WT M. tuberculosis, MGM1990, or MGM1991 and characterized the mycobacterium-specific immune responses. Both MGM1990 and MGM1991 showed reduced bacterial replication in spleen and lungs compared to WT M. tuberculosis. At day 21 postinfection, the mean log10 CFU in the spleens of mice infected with WT M. tuberculosis was 4.24 ± 0.11, compared to 3.66 ± 0.38 in mice infected with MGM1990 (Δlog10, 0.58; P < 0.05 [Mann-Whitney test]). MGM1991 showed an even more attenuated phenotype, and bacterial numbers in the spleen were below the detection limit (<100 CFU) for mice infected with this mutant. The attenuated phenotype of both mutants was even more pronounced in the lungs. At day 70 after intratracheal infection, pulmonary bacterial numbers were 6.46 ± 0.27 log10 in mice infected with the WT, 5.68 ± 0.17 log10 in mice infected with MGM1990 (Δlog10, 0.78 compared to WT), and 3.78 ± 0.7 log10 in mice infected with MGM1991 (Δlog10, 2.68 compared to WT).
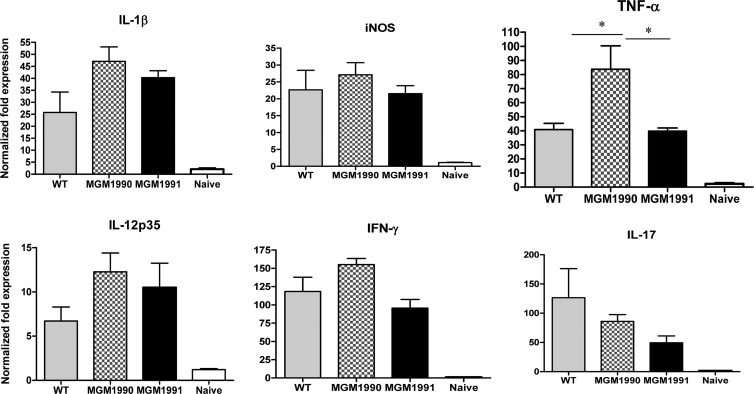
In order to analyze the overall triggering of the immune response by these M. tuberculosis strains, we performed gene expression profiling of different cytokines by RT-qPCR on cDNAs prepared from lung tissues of mice infected with the wild type, the cyclopropane-deficient strain MGM1990, and the MAMT-deficient strain MGM1991. Gene expression was normalized against the hydroxymethylbilane synthase housekeeping gene. We observed that the il-1β, inos, tnf-α, il-12, ifn-γ, and il-17 genes were induced in mice infected with the three M. tuberculosis strains compared to naïve uninfected mice (Fig. 5). Overall, expression levels were comparable for all cytokines, except for the expression of TNF-α, which was significantly higher in lungs from mice infected with MGM1990 than in mice infected with the WT or MGM1991 (Fig. 5). Gene expression profiles of IFN-γ and IL-17A were comparable following infection with the three M. tuberculosis strains (Fig. 5).
Fig 5.
RT-qPCR expression profiling of the lungs of C57BL/6 mice (n = 4/group) infected intratracheally 3 weeks before with 104 CFU of MGM1985 (WT), MGM1990, or MGM1991 or of the lungs of naïve, uninfected mice. Data are expressed as normalized fold expression levels with respect to reference housekeeping genes. Results are representative of 2 separate experiments. *, P < 0.05 (Mann-Whitney test).
Differences in mRNA levels are not necessarily reflected by differences in actual protein levels, and moreover, bulk cytokine measurements in lung homogenates may obscure differences in per cell cytokine secretion because they average cytokine production across multiple cell types present in the lung during infection. Also, analysis of cognate, antigen-specific immune responses cannot be studied in ex vivo transcriptional studies. To measure cognate IFN-γ and IL-17A cytokine production in vitro, we stimulated lung leukocytes, isolated from infected animals 3 weeks after infection, with the mycolyl-transferase Ag85A (Rv3804c), its H-2b-restricted T cell epitope (Ag85A241-260), and the H-2b-restricted epitope of the virulence factor ESAT-61-20. In contrast to the case for naïve mice, strong IFN-γ responses could be detected in culture supernatants by ELISA and ELISPOT assay (see Table S2 in the supplemental material). Lung cells from mice infected with the MGM1990 strain showed a tendency to produce more IFN-γ than lung cells from mice infected with the WT, especially after stimulation by Ag85A and the Ag85A241-260 peptide. The frequency of IFN-γ-producing cells detected by ELISPOT assay was also higher in the lungs of MGM1990-infected mice (see Table S2). In contrast, IFN-γ responses were lower in mice infected with MGM1991 than in mice infected with the WT. Pulmonary IL-17A responses showed a similar trend to that of IFN-γ responses, with the lowest levels in culture supernatants for mice infected with MGM1991 and the highest levels in those for mice infected with MGM1990 (Table 3). Interestingly, cells from mice infected with the latter strain showed significant production of IL-17A in the absence of antigenic stimulation (medium) in the IL-17A ELISPOT assay. Finally, pulmonary TNF-α production was measured after 3 weeks of infection, and confirming the RT-qPCR results, production of this cytokine was highest in cells from mice infected with the MGM1990 mutant (lacking all cyclopropanation) (Table 4). Like the case for IL-17A, cells from mice infected with this mutant released TNF-α in the absence of antigenic stimulation. Mice infected with MGM1991 produced very low levels of TNF-α, particularly following stimulation with ESAT-61-20 (Table 4).
Table 3.
Pulmonary IL-17A response of C57BL/6 mice infected intratracheally with M. tuberculosis WT, MGM1990, or MGM1991a
| Group and assay | IL-17A production (pg/ml or no. of SFC/106 cells) |
|||
|---|---|---|---|---|
| rAg85A | Ag85A241-260 | ESAT-61-20 | Medium | |
| ELISA | ||||
| WT | 787 | 867 | 665 | 665 |
| MGM1990 | 1,319 | 1,342 | 1,258 | 876 |
| MGM1991 | 563 | 446 | 312 | <4 |
| Naïve | 123 | <4 | <4 | <4 |
| ELISPOT assay | ||||
| WT | 168 | 368 | 816 | 24 |
| MGM1990 | 512 | 732 | 832 | 140 |
| MGM1991 | 88 | 596 | 220 | 13 |
| Naïve | ND | 4 | 12 | 12 |
Mice were infected with 104 CFU of M. tuberculosis WT Erdman, MGM1990, or MGM1991 by the intratracheal route. Lungs were recovered 3 weeks after the infection and homogenized, and pulmonary leukocytes were cultured with 5 μg/ml Ag85A, 10 μg/ml Ag85A241-260, or 10 μg/ml ESAT-61-20 or in the absence of antigen (medium). ELISA results are shown in pg/ml, and ELISPOT assay results are shown as the number of SFC/106 cells. ND, no spots detected.
Table 4.
Pulmonary TNF-α response of C57BL/6 mice infected intratracheally with M. tuberculosis WT, MGM1990, or MGM1991a
| Group | TNF-α production (pg/ml) |
|||
|---|---|---|---|---|
| rAg85A | Ag85A241-260 | ESAT-61-20 | Medium | |
| WT | 2,685 ± 167 | 742 ± 144 | 865 ± 118 | 325 ± 78 |
| MGM1990 | 3,625 ± 218 | 2,951 ± 49* | 2,733 ± 256* | 3,722 ± 181* |
| MGM1991 | 1,757 ± 305 | 213 ± 112 | 64 ± 14* | <8 |
| Naïve | 671 ± 260 | <8 | <8 | <8 |
Mice were infected with 104 CFU of M. tuberculosis WT, MGM1990, or MGM1991 by the intratracheal route. Lungs were recovered 3 weeks after the infection and homogenized, and pulmonary leukocytes were cultured with 5 μg/ml Ag85A, 10 μg/ml Ag85A241-260, or 10 μg/ml ESAT-61-20 or in the absence of antigen (medium). Results are expressed as mean ± SD TNF-α levels for ELISAs performed in triplicate.
, P < 0.05 for MGM1990 or MGM1991 versus WT (Mann-Whitney test).
IFN-γ and IL-17A production was also quantified in spleen cell culture supernatants obtained from mice after 3 (see Table S3 in the supplemental material) and 10 (see Table S4) weeks of infection by the intratracheal route. Spleen cells were stimulated with PPD and ESAT-61-20. Overall, 10-fold higher IFN-γ levels were detected in spleen than in lung culture supernatants, whereas IL-17A levels were very low and close to the detection limit. Strong IFN-γ responses were detected in spleen cell cultures from mice infected with the three M. tuberculosis strains in response to PPD of tuberculin. In contrast, at both time points, spleen cell IFN-γ responses to ESAT-61-20 were lower in mice infected with MGM1990 and MGM1991 than in mice infected with the WT Erdman strain, which confirmed the tendency observed in the lungs (for MGM1991 only).
DISCUSSION
The existence of cell wall mycolic acid modifications such as cyclopropanation and their restriction to pathogenic mycobacteria have been known for many years. However, the role of these modifications in the pathogenesis of M. tuberculosis infection has only recently been investigated. The fact that only the slow-growing pathogenic mycobacteria possess significant amounts of cyclopropanated mycolic acids may indicate that these modifications are needed for the survival of these bacteria in macrophages. In recent years, it was shown that the loss of single cyclopropane residues in M. tuberculosis mycolic acids has profound effects on the immune response of the host to this bacterium. Deletion of pcaA alone was enough to elicit a much reduced inflammatory response (that resulted in attenuation), whereas deletion of cmaA2, which abrogates the trans cyclopropanation of keto- and methoxymycolates, created a hyperinflammatory and hypervirulent strain. However, the overall role of mycolic acid modification in the pathogenesis of the bacterium remained unknown.
Role of MAMTs in M. tuberculosis viability.
In this study, we created a series of M. tuberculosis mutants lacking different MAMTs, from a complete absence of cyclopropanation but retention of all three classes of mycolates (MGM1990 and MGM1998) to a complete lack of MAMTs, creating strains with no cyclopropanation and with alpha mycolates only (MGM1991 and MGM1995).
At first, there appears to be a conundrum posed by the creation of these mutants and our prior hypothesis that cyclopropanation is essential for the life of these bacteria (2). There are many examples in the literature of phenotypic differences between chemical inhibition of a protein target and deletion of a gene encoding that target (20). There are many potential mechanisms of these differences, including the difference between complete (genetic) and partial (chemical) inhibition of a target, differential potency of the chemical inhibitor on individual enzymes, disruption of a protein complex, and delayed kinetics of onset of genetic deletion compared to that for chemical inhibition. We present data showing that the nonessentiality of the MAMT gene family is likely due to slow compensatory changes in the membrane that allow the bacterium to adapt to sequential MAMT loss. These compensatory changes are not possible when dioctylamine is used at a concentration that simultaneously inhibits the entire MAMT class. This model is strongly supported by our experiments showing that reintroduction of a full complement of MAMTs was lethal to the MAMT-deficient strain but not to wild-type M. tuberculosis.
Role of cyclopropanation in pathogenicity.
Our results also demonstrate that deletion of these enzymes causes significant attenuation of M. tuberculosis. Prior experiments have shown a variety of phenotypes of M. tuberculosis strains deficient in cyclopropanation. Deletion of pcaA causes a transient growth defect during the first week of infection and a late persistence defect associated with attenuated immunopathology. In contrast, the loss of cmaA2 causes hypervirulence without a difference in bacterial load (24, 25). The severe attenuation of the MGM1990 and MGM1998 strains in the first week of infection is similar to that of the ΔpcaA strain. In contrast to the ΔpcaA strain, in which the early attenuation reverses by day 31 of infection, the completely cyclopropanation-defective strain remains attenuated throughout the infection. This early attenuation is consistent with the sensitivity of the cyclopropane-deficient strain to detergent, as detergent stress has been shown to induce similar gene expression changes to those induced by growth within the phagosome (27). The host factor responsible for this detergent-like stress is not known, but it could be antimicrobial peptides, which kill bacteria while perturbing membranes. Surprisingly, the early attenuation phenotype of the cyclopropane-deficient strain (MGM1990 or MGM1998) is reversed by a loss of oxygenated mycolates, demonstrating that oxygenated mycolates are actually maladaptive lipid modifications in a strain that also lacks cyclopropanation.
Abundant prior data indicate that subclasses of mycolic acids or different cyclopropane modifications within these classes have specific effects on immune activation (9, 21, 23–25, 29). Our characterization of the immune responses to cyclopropane- and MAMT-deficient strains extends these findings in the context of M. tuberculosis infection. We showed that the net effect of cyclopropanation is to suppress the immune response to M. tuberculosis. Lung cells of mice infected with the cyclopropane-deficient MGM1990 strain produced higher levels of IL-17A and TNF-α both in unstimulated cultures and upon antigenic stimulation than lung cells of mice infected with the WT strain. This hyperinflammatory phenotype in the absence of all cyclopropanation is consistent with the results of prior studies using both purified mycolic acids and mutant mycobacterial strains that found trans-cyclopropanated oxygenated mycolates to suppress immune activation. The loss of these trans-cyclopropanated mycolates in the MGM1990 strain may have contributed to the hyperinflammatory phenotype we observed. However, MGM1990 is significantly more attenuated than the previously described ΔcmaA2 strain (25), which also lacks trans-cyclopropanated oxygenated mycolates.
The direct comparison of the MGM1990 strain, which lacks all cyclopropanation, and MGM1991, which has an additional lack of oxygenated mycolates, is informative. Despite its reduced in vivo bacterial replication, MGM1990 was capable of triggering strong mycobacterium-specific pulmonary IFN-γ responses in response to two immunodominant antigens of M. tuberculosis, i.e., the mycolyl-transferase Ag85A and ESAT-6. Pulmonary IFN-γ responses following infection with the MGM1991 strain were lower than those following infection with the WT strain. Finally, strong PPD-specific IFN-γ responses could be detected in spleens from mice infected with both MGM1990 and MGM1991. Whether these mutants (particularly MGM1991) have protective potential as live attenuated vaccines remains to be examined.
Because these two strains differ in the presence of oxygenated mycolates, we concluded that the hyperinflammatory phenotype of MGM1990 depends on the presence of unsaturated oxygenated mycolates. When M. tuberculosis expresses only unsaturated alpha mycolates, without any oxygenated mycolates, virulence is strongly attenuated and antigen-specific immune responses are inhibited in comparison to those against a cyclopropane-deficient strain. Interestingly, the cyclopropane versus MAMT comparison also indicates very specific virulence functions for the two lipids during the first 2 weeks of infection. Cyclopropane deficiency confers attenuation in the first week of lung infection, whereas MAMT deficiency (with consequent loss of oxygenated mycolates) actually reverses this defect (Fig. 4C). In contrast, MAMT deficiency confers attenuation in weeks 2 to 4 of infection in the lung, during which the cyclopropane-deficient strain does not exhibit further attenuation.
Taken together, our findings establish the immunomodulatory function of the mycolic acid modification pathway in pathogenesis and buttress this enzyme class as an attractive target for antimycobacterial drug development.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by NIH grant AI53417 (to M.S.G.) and by the Michael and Ethel L. Cohen Foundation (to D.B.). This work was also partially supported by a grant from the Flemish Scientific Research Foundation (FWO-Vlaanderen G.0063-09N) (to D.H. and K.H.).
Footnotes
Published ahead of print 19 March 2012
Supplemental material for this article may be found at http://iai.asm.org/.
REFERENCES
- 1. Alibaud L, et al. 2010. Temperature-dependent regulation of mycolic acid cyclopropanation in saprophytic mycobacteria: role of the Mycobacterium smegmatis 1351 gene (MSMEG_1351) in cis-cyclopropanation of alpha-mycolates. J. Biol. Chem. 285:21698–21707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Barkan D, Liu Z, Sacchettini JC, Glickman MS. 2009. Mycolic acid cyclopropanation is essential for viability, drug resistance, and cell wall integrity of Mycobacterium tuberculosis. Chem. Biol. 16:499–509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Barkan D, Rao V, Sukenick GD, Glickman MS. 2010. Redundant function of cmaA2 and mmaA2 in Mycobacterium tuberculosis cis cyclopropanation of oxygenated mycolates. J. Bacteriol. 192:3661–3668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Barry CE, III, et al. 1998. Mycolic acids: structure, biosynthesis and physiological functions. Prog. Lipid Res. 37:143–179 [DOI] [PubMed] [Google Scholar]
- 5. Behr MA, Schroeder BG, Brinkman JN, Slayden RA, Barry CE., III 2000. A point mutation in the mma3 gene is responsible for impaired methoxymycolic acid production in Mycobacterium bovis BCG strains obtained after 1927. J. Bacteriol. 182:3394–3399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Belley A, et al. 2004. Impact of methoxymycolic acid production by Mycobacterium bovis BCG vaccines. Infect. Immun. 72:2803–2809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bhatt A, et al. 2007. Deletion of kasB in Mycobacterium tuberculosis causes loss of acid-fastness and subclinical latent tuberculosis in immunocompetent mice. Proc. Natl. Acad. Sci. U. S. A. 104:5157–5162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Brandt L, Oettinger T, Holm A, Andersen AB, Andersen P. 1996. Key epitopes on the ESAT-6 antigen recognized in mice during the recall of protective immunity to Mycobacterium tuberculosis. J. Immunol. 157:3527–3533 [PubMed] [Google Scholar]
- 9. Dao DN, et al. 2008. Mycolic acid modification by the mmaA4 gene of M. tuberculosis modulates IL-12 production. PLoS Pathog. 4:e1000081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dinadayala P, et al. 2003. Tracking the putative biosynthetic precursors of oxygenated mycolates of Mycobacterium tuberculosis. Structural analysis of fatty acids of a mutant strain devoid of methoxy- and ketomycolates. J. Biol. Chem. 278:7310–7319 [DOI] [PubMed] [Google Scholar]
- 11. Domenech P, Reed MB. 2009. Rapid and spontaneous loss of phthiocerol dimycocerosate (PDIM) from Mycobacterium tuberculosis grown in vitro: implications for virulence studies. Microbiology 155:3532–3543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. D'Souza S, et al. 2003. Mapping of murine Th1 helper T-cell epitopes of mycolyl transferases Ag85A, Ag85B, and Ag85C from Mycobacterium tuberculosis. Infect. Immun. 71:483–493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Dubnau E, et al. 2000. Oxygenated mycolic acids are necessary for virulence of Mycobacterium tuberculosis in mice. Mol. Microbiol. 36:630–637 [DOI] [PubMed] [Google Scholar]
- 14. Dubnau E, Marrakchi H, Smith I, Daffe M, Quemard A. 1998. Mutations in the cmaB gene are responsible for the absence of methoxymycolic acid in Mycobacterium bovis BCG Pasteur. Mol. Microbiol. 29:1526–1528 [PubMed] [Google Scholar]
- 15. George KM, Yuan Y, Sherman DR, Barry CE., III 1995. The biosynthesis of cyclopropanated mycolic acids in Mycobacterium tuberculosis. Identification and functional analysis of CMAS-2. J. Biol. Chem. 270:27292–27298 [DOI] [PubMed] [Google Scholar]
- 16. Glickman MS. 2003. The mmaA2 gene of Mycobacterium tuberculosis encodes the distal cyclopropane synthase of the alpha-mycolic acid. J. Biol. Chem. 278:7844–7849 [DOI] [PubMed] [Google Scholar]
- 17. Glickman MS, Cahill SM, Jacobs WR., Jr 2001. The Mycobacterium tuberculosis cmaA2 gene encodes a mycolic acid trans-cyclopropane synthetase. J. Biol. Chem. 276:2228–2233 [DOI] [PubMed] [Google Scholar]
- 18. Glickman MS, Cox JS, Jacobs WR., Jr 2000. A novel mycolic acid cyclopropane synthetase is required for cording, persistence, and virulence of Mycobacterium tuberculosis. Mol. Cell 5:717–727 [DOI] [PubMed] [Google Scholar]
- 19. Kirksey MA, et al. 2011. Spontaneous phthiocerol dimycocerosate-deficient variants of Mycobacterium tuberculosis are susceptible to gamma interferon-mediated immunity. Infect. Immun. 79:2829–2838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Knight ZA, Shokat KM. 2007. Chemical genetics: where genetics and pharmacology meet. Cell 128:425–430 [DOI] [PubMed] [Google Scholar]
- 21. Korf J, Stoltz A, Verschoor J, De Baetselier P, Grooten J. 2005. The Mycobacterium tuberculosis cell wall component mycolic acid elicits pathogen-associated host innate immune responses. Eur. J. Immunol. 35:890–900 [DOI] [PubMed] [Google Scholar]
- 21a. Korf H, et al. 2009. Liver X receptors contribute to the protective immune response against Mycobacterium tuberculosis in mice. J. Clin. Invest. 119:1626–1637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Pashley CA, Parish T. 2003. Efficient switching of mycobacteriophage L5-based integrating plasmids in Mycobacterium tuberculosis. FEMS Microbiol. Lett. 229:211–215 [DOI] [PubMed] [Google Scholar]
- 23. Peyron P, et al. 2008. Foamy macrophages from tuberculous patients' granulomas constitute a nutrient-rich reservoir for M. tuberculosis persistence. PLoS Pathog. 4:e1000204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Rao V, Fujiwara N, Porcelli SA, Glickman MS. 2005. Mycobacterium tuberculosis controls host innate immune activation through cyclopropane modification of a glycolipid effector molecule. J. Exp. Med. 201:535–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Rao V, Gao F, Chen B, Jacobs WR, Jr, Glickman MS. 2006. Trans-cyclopropanation of mycolic acids on trehalose dimycolate suppresses Mycobacterium tuberculosis-induced inflammation and virulence. J. Clin. Invest. 116:1660–1667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Romano M, et al. 2006. Immunogenicity and protective efficacy of tuberculosis DNA vaccines combining mycolyl-transferase Ag85A and phosphate transport receptor PstS-3. Immunology 118:321–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Schnappinger D, et al. 2003. Transcriptional adaptation of Mycobacterium tuberculosis within macrophages: insights into the phagosomal environment. J. Exp. Med. 198:693–704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Stallings CL, et al. 2009. CarD is an essential regulator of rRNA transcription required for Mycobacterium tuberculosis persistence. Cell 138:146–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Vander Beken S, et al. 2011. Molecular structure of the Mycobacterium tuberculosis virulence factor, mycolic acid, determines the elicited inflammatory pattern. Eur. J. Immunol. 41:450–460 [DOI] [PubMed] [Google Scholar]
- 30. Vaubourgeix J, et al. 2009. S-Adenosyl-N-decyl-aminoethyl, a potent bisubstrate inhibitor of Mycobacterium tuberculosis mycolic acid methyltransferases. J. Biol. Chem. 284:19321–19330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Yuan Y, Lee RE, Besra GS, Belisle JT, Barry CE., III 1995. Identification of a gene involved in the biosynthesis of cyclopropanated mycolic acids in Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U. S. A. 92:6630–6634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Yuan Y, Zhu Y, Crane DD, Barry CE., III 1998. The effect of oxygenated mycolic acid composition on cell wall function and macrophage growth in Mycobacterium tuberculosis. Mol. Microbiol. 29:1449–1458 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.