Abstract
Summary: The eukaryotic heat shock response is an ancient and highly conserved transcriptional program that results in the immediate synthesis of a battery of cytoprotective genes in the presence of thermal and other environmental stresses. Many of these genes encode molecular chaperones, powerful protein remodelers with the capacity to shield, fold, or unfold substrates in a context-dependent manner. The budding yeast Saccharomyces cerevisiae continues to be an invaluable model for driving the discovery of regulatory features of this fundamental stress response. In addition, budding yeast has been an outstanding model system to elucidate the cell biology of protein chaperones and their organization into functional networks. In this review, we evaluate our understanding of the multifaceted response to heat shock. In addition, the chaperone complement of the cytosol is compared to those of mitochondria and the endoplasmic reticulum, organelles with their own unique protein homeostasis milieus. Finally, we examine recent advances in the understanding of the roles of protein chaperones and the heat shock response in pathogenic fungi, which is being accelerated by the wealth of information gained for budding yeast.
INTRODUCTION
Cells grow optimally within a relatively narrow temperature range but tolerate moderate deviations, some of which impinge upon cell structure and function, via rapid physiological adaptations. One of the most powerful adaptation mechanisms is the heat shock response (HSR), a highly conserved program of changes in gene expression that result in the repression of the protein biosynthetic capacity and the induction of a battery of cytoprotective genes encoding the heat shock proteins (HSPs). Many HSPs function as molecular chaperones to protect thermally damaged proteins from aggregation, unfold aggregated proteins, and refold damaged proteins or target them for efficient degradation. Physiological changes such as the synthesis of compatible solutes, cell wall restructuring, and the transient interruption of the cell cycle also contribute to cellular survival. Much of what we know regarding the HSR in eukaryotic cells has been elucidated with the model yeast Saccharomyces cerevisiae due to its facile genetics, biochemistry, and cell biology as well as the wealth of genome-level tools made available in the last decade. This review will provide a broad overview of the effects of heat shock on S. cerevisiae and the control of the HSR at multiple regulatory levels. We focus on the cellular biology of the HSPs, defined as operational networks within the major cellular compartments. While the last 30 years or so of research has been a period of intense and fruitful discovery, current efforts are now being targeted to address how the various components of the HSR work together in multiprotein and multicomplex networks. Lessons learned from the budding yeast model may now be applied to intervention therapies to treat human diseases and disorders characterized by defects in protein homeostasis and folding.
PHYSIOLOGICAL EFFECTS OF HEAT SHOCK
The HSR is appropriately considered to be a fundamental cytoprotective pathway conferring resistance to heat shock. However, by its very definition, the response is considered one of repair and adaptation to damage caused by the stress rather than a prophylactic measure. As discussed later in the review, evidence suggests that the HSR may in fact be evolutionarily selected to prevent damage caused by an anticipated future stress rather than to promote recovery from an existing insult. We address the physiological impacts of moderate to severe heat stress, with emphasis on cellular processes sensitive to thermal damage (Fig. 1).
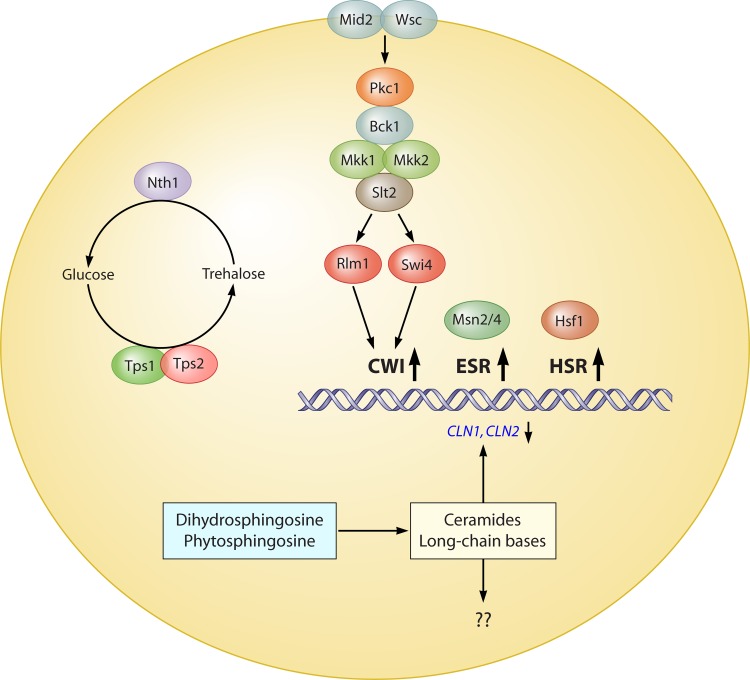
Fig 1.
Physiological effects of heat shock. Immediate consequences of thermal stress are depicted as described in the text. Relevant proteins are depicted as colored balls. Three response pathways are shown to be induced by heat shock: the CWI (cell wall integrity) pathway, the ESR (environmental stress response), and the HSR (heat shock response). The physiological effects of ceramide and long-chain base synthesis and accumulation after heat shock are unknown.
Physiological and Metabolic Adaptation
Cell cycle arrest.
Yeast cells complete a cell cycle in rich medium in approximately 70 to 90 min, and work in the 1980s defined Start as a key regulatory checkpoint in the G1-to-S-phase transition (35). Cells arrested in the G1 phase have unreplicated chromosomes and exist in the unbudded state. Heat shock induces transient arrest at precisely this stage in the cell cycle, likely due to a reduction of transcript levels of the G1/S cyclins CLN1 and CLN2, as the overexpression of CLN2 from the GAL1 promoter is sufficient to prevent heat-induced arrest (Fig. 1) (373). Interestingly, CLN3 transcripts are unaffected, suggesting a posttranscriptional regulation of this cyclin gene product. Consistent with this hypothesis, the Cln3 protein was recently shown to be tethered to the endoplasmic reticulum (ER) membrane in a complex with farnesylated Ydj1, a key J-type molecular chaperone and Hsp70 cofactor (477). Temperature-dependent cell cycle inhibition can be mimicked by treatment with low concentrations of the imino acid analog azetidine-2-carboxylic acid (AZC), a compound that causes the misfolding of nascent polypeptides via a substitution for the amino acid proline. AZC is toxic at high concentrations, but at low concentrations (10 mM or lower), it causes both G1 arrest and the repression of CLN1 and CLN2 (466). These data suggest that G1 arrest is not necessarily a direct physiological phenomenon but rather a signaled event. This idea is supported by the finding that both heat shock and AZC treatments of EXA3-1 cells expressing a dominant negative allele of the heat shock transcription factor HSF1 (see below) do not result in arrest (467). Instead, it is likely that the accumulation of misfolded proteins in both scenarios rapidly activates Hsf1, which in turn induces the expression of one or more proteins that block CLN1/2 expression. What is the competitive advantage of the G1 arrest in response to protein misfolding? Although no data specifically address this question, it is possible that proceeding with DNA synthesis and/or mitosis in the face of proteotoxic damage might be catastrophic and that the G1 delay allows protected time to restore protein homeostasis. This model is consistent with cell cycle checkpoints that halt progression in the presence of DNA-damaging agents or improperly paired chromosomes.
Defects in the HSR also impact cell cycle progression during thermal stress. A temperature-sensitive mitochondrial import mutant, mas3, was found to result in arrest as large-budded cells, indicative of a G2/M transition block (432). This allele was mapped to the HSF1 locus, identifying this transcription factor and its HSR regulon as critical components of chronic heat tolerance. This finding was recapitulated in two subsequent papers that characterized a G2/M arrest phenotype for two additional HSF1 alleles, hsf1-82 and a truncation mutant defective in transcriptional activation, HSF(1–583) (303, 525). Remarkably, in both those reports, the primary defect was found to be a reduced level of expression of the Hsp90 molecular chaperone. Zarzov et al. observed via electron microscopy specific defects in spindle pole body (SPB) duplication which could be corrected by the overexpression of the Hsp90-encoding gene HSP82, implying a role for this chaperone in maintaining SPB function during heat shock (525). SPB defects in these mutants are also consistent with the activation of the spindle pole checkpoint and the G2/M arrest point (136).
Metabolic reprogramming.
Does the position within the cell cycle affect thermotolerance? Early work showed that starving cells (G0 phase) are significantly more thermotolerant than exponentially dividing populations (331). In contrast, cells arrested pharmacologically in the G1, S, or G2 phase of the cell cycle are as sensitive to heat shock as nonsynchronized cultures (14, 347). These findings led to the speculation that quiescent cells that have exited the cell cycle concomitantly acquire substantial heat shock resistance in a process linked to nutrient availability. The nonreducing disaccharide trehalose is an important storage carbohydrate in S. cerevisiae, and the ability of cells to withstand severe heat shock (usually considered to be 45°C or higher) correlates with cellular trehalose levels: the inactivation of the trehalose biosynthetic genes TPS1 and TPS2 results in reduced thermotolerance, and a loss of the trehalose-degrading enzyme NTH1 (neutral trehalase) extends thermotolerance during recovery (Fig. 1) (98). Trehalose levels also rise in response to heat shock and confer thermotolerance in the related fission yeast Schizosaccharomyces pombe at temperatures that preclude HSP synthesis (above 40°C) (365). The intrinsic thermotolerance of stationary-phase cells also correlates well with the activity of the metabolic regulatory enzyme protein kinase A (PKA). As discussed below, PKA is a negative regulator of the environmental stress response (ESR), which includes both the trehalose biosynthetic and catabolic genes, and cells limited or starved for glucose exhibit low levels of PKA activity, leading to the derepression of these enzymes. Importantly, trehalose levels must be tightly regulated, as cells lacking NTH1 exhibit impaired recovery from heat shock (503).
How is trehalose such a powerful contributor to thermotolerance? In vitro studies demonstrated that trehalose is an effective stabilizer of proteins at physiological concentrations (192). The disaccharide is also well known as an antidehydration agent, likely due to its unique propensity to displace the “water shell” around macromolecules, thereby minimizing the effects of desiccation (86). Singer and Lindquist demonstrated that trehalose can suppress the aggregation of misfolded proteins in vivo, effectively preventing one of the most deleterious consequences of severe heat shock (429, 430). In contrast, high levels of trehalose prevent protein refolding, providing a possible molecular explanation for the heat shock recovery defects associated with trehalase mutants (429). The Hsp104 protein chaperone possesses similar properties and works synergistically with trehalose to stabilize the yeast proteome at high temperatures. Indeed, both trehalose and Hsp104 are required for tolerance to heat shock, suggesting that they play complementary but not overlapping roles (119, 383). Interestingly, the protein-stabilizing effects of trehalose can also be observed for the endoplasmic reticulum (ER) lumen, suggesting that the sugar must be transported into the endomembrane system (426). Trehalose may also confer a broader range of protection for proteins, as it was demonstrated to enhance the survival of yeast cells treated with hydrogen peroxide and to reduce the extent of protein carbonylation, a prime indicator of oxidative damage (21). Lastly, Nelson and colleagues reported the surprising finding that trehalose is required for maximal transcriptional activation by Hsf1, which may be tied to its observed ability to stabilize the tertiary structure of the carboxy-terminal activation domain (42, 68).
Altered cell wall and membrane dynamics.
In addition to the effects of heat shock on internal cellular processes, thermal stress appears to impact the cell surface. The fungal cell is limited by the plasma membrane and surrounded by an outer cell wall composed of glucose- and mannose-based polysaccharides and N-acetylglucosamine (44, 45). The intervening periplasmic space contains numerous secreted enzymes and membrane-associated surface proteins. An elaborate signaling pathway has been elucidated, linking outer membrane transmembrane proteins that serve as putative heat and/or pressure sensors, the small GTPase Rho1, and a protein kinase cascade starting with protein kinase C (Pkc1) and terminating in effector transcription factors (Fig. 1) (reviewed in reference 250). This cell wall integrity (CWI) pathway is activated in response to perturbations in the cell ultrastructure, including treatment with compounds that interfere with cell wall synthesis and changes in pH and temperature. The importance of this pathway is made clear by the phenotype of cells lacking the terminal mitogen-activated protein (MAP) kinase (MAPK) Slt2 (also known as Mpk1), which are exquisitely temperature sensitive at 37°C due to autolysis (280). Mutations in nearly every component of the CWI pathway lead to identical phenotypes, consistent with the linear nature of the signaling pathway (220). Importantly, temperature sensitivity can be remediated by the inclusion of a compatible solute such as sorbitol for osmotic support, demonstrating that cell lysis is the primary cause of the observed phenotype. Yeast cells maintain a high internal turgor pressure, making even minor defects in the cell wall structure potentially lethal. The CWI pathway is activated by heat through an unknown mechanism that requires at least one member of the putative sensors Mid2 and Wsc1 to Wsc4. In the absence of these proteins, the HSR is activated normally, but cells are heat shock sensitive, are autolytic, and do not activate the CWI transcription factor Rlm1 (478, 532). RAD6 encodes a ubiquitin-conjugating enzyme required for the resumption of growth after heat shock-induced arrest via an unknown mechanism. Interestingly, the overexpression of WSC2 was found to reverse the rad6 phenotype, implying an intersection between the CWI pathway and the ubiquitin-proteasome machinery to regulate the transient heat-induced arrest (354). However, an absolute requirement for the Wsc proteins to mediate heat shock-induced G1 arrest was not established in that study, leaving open the question of whether these sensors are required for this checkpoint. Another putative plasma membrane pressure sensor, Sho1, is required for the activation of the high-osmolarity glycerol (HOG) pathway in response to heat shock, suggesting that thermal stress may in fact lead to a transient change in the perceived turgor pressure (513).
The role of membranes and lipids in the heat shock response is enigmatic. Little work has been done with S. cerevisiae specifically to assess perturbations in the membrane lipid composition or structure in response to heat shock. A study including budding yeast and the dimorphic pathogenic fungus Histoplasma capsulatum showed that the ratio of saturated to unsaturated fatty acids affects the temperature set point at which the HSR is induced (54). Heat shock is expected to alter membrane fluidity, and the packing constraints of membranes rich in saturated fatty acids likely would differ from those containing a higher proportion of unsaturated bonds. However, this temperature-sensing mechanism is predicated on a membrane-embedded protein component that has not been identified. The Wsc proteins are obvious candidates that must be excluded, since their presence is not required for HSR induction (532). Stress-induced alterations in membrane fluidity can also result in changes in ion transport. The Ca2+-regulated protein phosphatase calcineurin is composed of catalytic (Cna1 or Cna2) and regulatory (Cnb1) subunits and is responsible for the upregulation of genes involved in cell wall biosynthesis, small-molecule transport, and the synthesis of membrane lipids and ergosterol in response to stress via the dephosphorylation of the transcription factor Crz1. Cells deficient in calcineurin activity due to molecular genetic ablation or pharmacological treatment with the inhibitor FK506 or cyclosporine are highly stress sensitive, underscoring the importance of this pathway (87). Calcineurin is also a client of the Hsp90 chaperone system (see below), further integrating the HSR with other cellular stress defense networks (196).
Substantially more is known about post-heat-shock signaling involving lipids and lipid-derived compounds. Sphingolipids are a class of membrane components that include long-chain alkane bases (LCBs) with hydroxyl and/or amine groups at one end of the molecule. S. cerevisiae synthesizes primarily the 18-carbon LCBs dihydrosphingosine and phytosphingosine but transiently accumulates 20-carbon LCBs within 5 to 10 min of heat shock (Fig. 1) (100, 101). These compounds are rapidly converted into other complex sphingolipids and ceramide, the product of a condensation reaction between phytosphingosine and C26-fatty acyl-coenzyme A (CoA) (500). A strain lacking the ability to synthesize LCBs is heat shock sensitive, and this phenotype is reversed by genetic or chemical supplementation with sphingolipids, suggesting that these molecules are required for thermotolerance or signaling (204). This idea was reinforced by the observation that the treatment of cells with dihydrosphingosine activates the transcription of the TPS2 gene and a stress-responsive element (STRE)-lacZ fusion (see below) that reports on the activity of the Msn2/4 stress pathway (101). However, a subsequent study using microarray analysis to evaluate differences in gene expression in response to heat shock in wild-type versus an lcb1-100 mutant strain, which is defective in sphingoid base production, failed to reveal a global defect in STRE-controlled genes (72). This same strain is defective in heat-induced G1 arrest, implying a role for sphingolipid signaling in the cell cycle. The lcb1-100 strain displays an aberrant transcription of cell cycle genes in response to heat shock, supporting this observation (203). Sphingoid bases may also play important posttranscriptional roles in the response to heat stress. The heat-induced increase in the level of phytosphingosine is required for both translation and ubiquitin-dependent proteolysis as well as for the proper organization of the actin cytoskeleton (65, 89, 289). Recently, sphingolipids have also been implicated in the formation of P bodies (discussed below), consistent with translation inhibition in their absence and in aiding cellular recovery from thermal stress (71).
Protein Aggregation and Sequestration
The conventional view of heat shock stress is primarily one of proteotoxicity: an increase in the ambient temperature destabilizes cellular proteins. Lethality could then be predicted to result from misfolding and the subsequent loss of function of one or more essential proteins. Alternatively, the accumulation of a significant number of misfolded polypeptides could have secondary consequences, such as an inhibition of normal protein degradation by the ubiquitin-proteasome system (UPS) or the formation of toxic protein aggregates. While both of these scenarios have been observed at lethal heat shock temperatures (>50°C), little is known about the state of the proteome at the more standard heat shock temperature for mesophilic yeast of 37°C. Little to no protein misfolding, as measured by differential centrifugation to isolate insoluble aggregates, occurs at temperatures between 36°C and 37°C, even in strains defective in the cytoplasmic chaperone Hsp90 (310). Despite this observation, the disruption of Hsp90 cochaperones, including SSE1, STI1, and YDJ1 (see below), and a number of hypomorphic mutations in HSF1 result in a temperature-sensitive growth phenotype at 37°C (59, 197, 303, 409, 468, 516, 525). As described above, temperature-sensitive growth is also a defining characteristic of mutants of the CWI pathway, as is the remediation of this phenotype by external osmotic support. Consistently, the inclusion of 1 M sorbitol in the growth medium suppresses the growth defect of strains deficient in either Hsp90 or HSF1 at 37°C (197, 409, 468, 516). This effect was shown to be a consequence of a requirement for the Hsp90 chaperone system to stabilize the CWI MAP kinase Slt2 (468). Together, these results support the conclusion that the inability of cells deficient in HSP production and function to grow under heat shock conditions is not due to gross protein misfolding but rather to a specific defect in promoting the function of the CWI pathway.
An alternative explanation for the observed lack of significant protein misfolding or phenotypic consequences of heat shock at 37°C is that the protein quality control system efficiently copes with these problems. In addition to HSPs, which are capable of unfolding and refolding aggregated proteins (see below), the UPS identifies such targets and rapidly degrades ubiquitinated substrates (reviewed in reference 223). This idea is supported by the observation that the disruption of proteasome activity genetically or pharmacologically with the inhibitor MG132 results in the accumulation of ubiquitinated proteins (242). What is the fate of these misfolded and ubiquitinated proteins? An analysis of two model misfolded proteins (a temperature-sensitive allele of UBC9 and the E364K actin [ACT1] point mutant) revealed that at 37°C, fluorescently tagged versions of these misfolded proteins accumulate in a juxtanuclear compartment (termed JUNQ) (Fig. 2) (218). Resident proteins in this compartment remain soluble and diffusible and likely interact with colocalized proteasomes, which are also concentrated there. Prolonged exposure to these conditions results in the accumulation of aggregated substrates in a second distinct compartment termed the IPOD (insoluble protein deposit), which exhibits a perivacuolar localization. These aggregates are not ubiquitinated, suggesting that these proteins have escaped recognition by the UPS but are nevertheless sequestered. The Hsp104 chaperone localizes to both compartments, but the total chaperone complement has not been determined, nor is it known whether chaperones contribute to the formation or dissolution of the compartments. Given that the detection of JUNQ and IPOD requires the imposition of stress and, in the case of JUNQ, proteasome inhibition, it is also not clear if these compartments are intermediates in a normal protein quality control pathway or are off-pathway products. Lastly, the identification of these compartments provides an important link between the inclusion bodies of prokaryotes and the aggresomes of mammalian cells, all of which operate in thematically similar if perhaps mechanistically distinct manners (11).
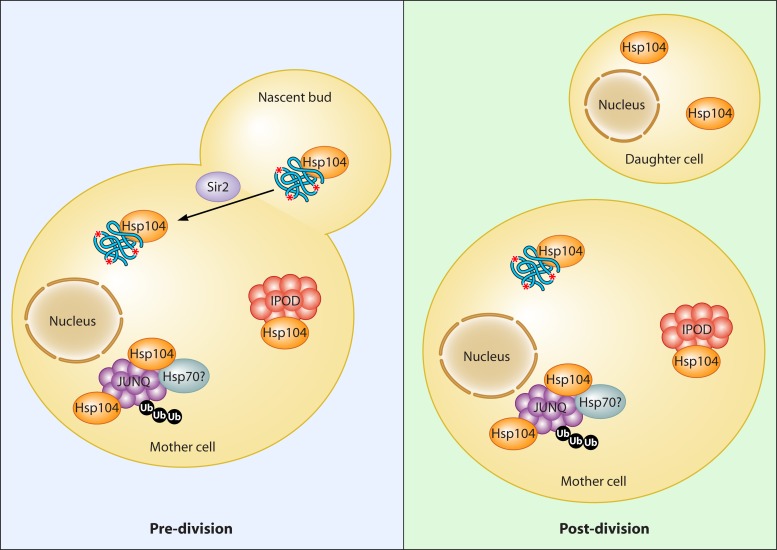
Fig 2.
Asymmetric distribution of damaged proteins during growth. Budding (predivision) and budded (postdivision) cells are depicted, with the net retention of damaged proteins in the mother cell resulting from Sir2-dependent transport. The two recently described “compartments” of protein aggregation, JUNQ and IPOD, are shown with known or suspected associated chaperones. Ub, ubiquitin; red asterisk, carbonylation or other protein damage; blue squiggle, unfolded protein.
Interestingly, long-lived proteins are also damaged over time as cells age or in response to oxidative stress. Heat shock is known to induce oxidative stress in a process linked to the dysfunction of the mitochondrial electron transport chain. Protein oxidation frequently takes the form of the carbonylation of a number of amino acid side groups, resulting in the formation of irreversible semialdehydes (363). Carbonylated proteins can be selectively detected through a technique involving derivatization with 2,4-dinitrophenol hydrazine followed by decoration with an antibody that specifically recognizes the hydrazine moiety (251). Immunofluorescence microscopy has shown that carbonylated proteins tend to form higher-order aggregates in vivo. Fascinatingly, these “clumps” of damaged proteins are selectively retained in the mother cell during the asymmetric division of budding yeast (Fig. 2) (3). This phenomenon likely contributes to the observed replicative senescence of yeast cells after 20 to 30 generations and to the finding that daughter cells are born free of damaged proteins. Indeed, a similar process appears to function during gametogenesis (spore formation) in yeast, with damaged proteins being excluded from the developing spores (472). The segregation of carbonylated proteins away from daughter cells was shown to require Hsp104, cytoskeletal function, and the regulator Sir2, further linking protein sequestration to aging (121). However, the involvement of the actin cytoskeleton in retrograde transport via the formin Bni1 is under dispute, with data supporting both diffusion-based (random) and polarized-transport (targeted) mechanisms to explain the apparent asymmetry of damaged proteins in actively dividing cells (260, 530). Further elucidations of factors governing this clearly important process should help resolve the question, including the possibility that both random and nonrandom events may be in play.
TRANSCRIPTIONAL CONTROL OF THE HEAT SHOCK RESPONSE
The Heat Shock Transcriptome
In addition to the physiological changes described above, cells respond to heat shock by dramatically altering their gene expression programs. For many years, analyses of the heat shock response occurred on a gene-by-gene basis, gradually describing a coordinated response orchestrated by a small number of transcription factors. The invention of DNA microarray technology revolutionized the field by allowing the simultaneous analysis of the entire heat shock transcriptome. Two studies documented the depth and breadth of what is termed the environmental stress response (ESR), including insults such as osmotic stress, salt stress, nutrient starvation, and cold shock in addition to heat shock (58, 147). Indeed, approximately 300 genes are induced in the ESR, and double that number are transcriptionally repressed. The latter category consists largely of protein biosynthesis genes, including ribosomal components, RNA-processing factors, and other progrowth genes. Induced genes include all of the previously known HSPs, a number of metabolic genes, and a significant fraction of genes of unknown function (148). Remarkably, the induction and repression of both gene classes are transient and scale with the magnitude (intensity) of the stress applied, demonstrating a reciprocal relationship (58, 147). The HSR can be considered a subset of the ESR, as essentially all HSR genes are accounted for within the ESR regulon, whereas a number of ESR genes are not necessarily induced by heat shock. As detailed below, the HSR is governed by the action of primarily two transcription factors, Hsf1 and Msn2/4 (Fig. 3). Array studies examining the contributions of each factor revealed a significant overlap in target gene expression, consistent with the presence of the appropriate binding sites within the promoters (147). The Hsf1 regulon was examined in detail and was found to comprise approximately 165 genes (not exclusive of the influence of Msn2/4), which was confirmed by a chromatin immunoprecipitation assay (165). Surprisingly, many of the genes induced by heat shock are not required for heat shock survival; that is, the respective null mutants are not grossly heat shock sensitive (153). This finding suggests a potential disconnect between the regulation and role of a gene product but does not take into account the possibility that gene functions may be redundant or stress specific. A potential explanation is provided by the observation that the induction of the ESR/HSR is required not for survival of the stimulating stress but rather for survival of a subsequent stress. This phenomenon is termed “acquired stress resistance” and was appreciated anecdotally until 2008, when it was formally investigated. Cells unable to mount a protective response after a mild to moderate stress due to protein synthesis inhibition or the deletion of Msn2/4 showed no defect in recovery from that stress but pronounced defects in surviving a secondary insult (24). Moreover, MSN2 or MSN4 appears to play a nonredundant role in mediating acquired tolerance, suggesting some degree of specificity between the two highly related factors. Lastly, because distinct stresses induce a common ESR, the phenomenon of cross-protection, defined as acquired tolerance to a dissimilar stress, is also explained. Therefore, heat shock induces resistance to oxidative stress and vice versa.
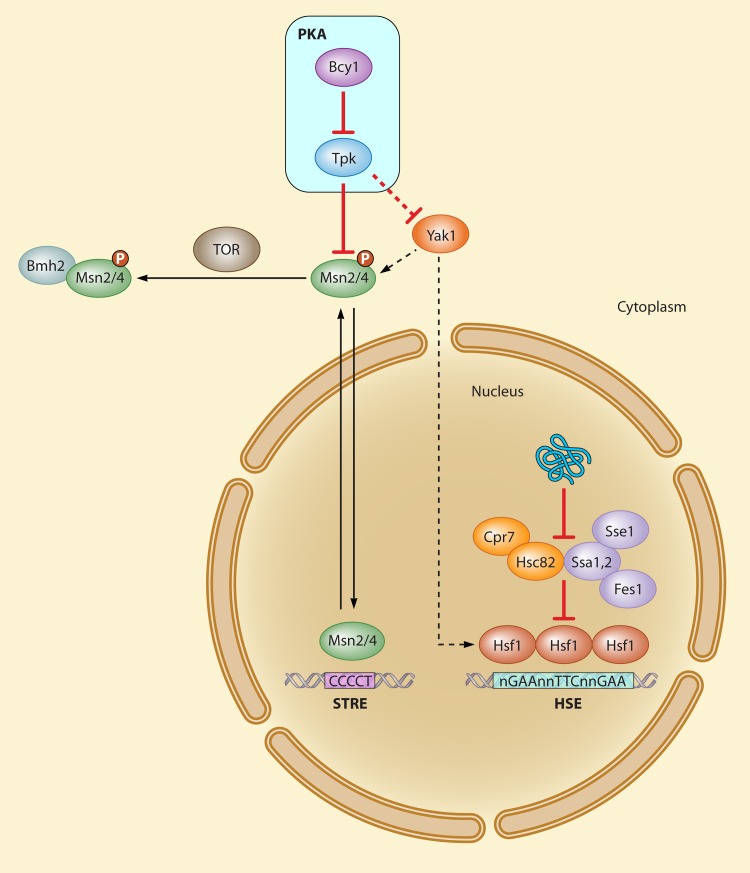
Fig 3.
Hsf1 and Msn2/4, primary modulators of the heat shock response. Dashed lines represent postulated interactions of the Yak1 kinase in the regulation of both Msn2/4 and Hsf1. Red lines indicate regulatory interactions of protein kinase A. P, phosphorylation; STRE, stress response element; HSE, heat shock element.
Hsf1
In most eukaryotes, the increased expression of heat shock proteins (HSPs) in a stressed cell is mediated primarily by so-called heat shock transcription factors (HSFs). Vertebrates and plants have evolved a family of four HSF members, i.e., HSF1 to HSF4. Among these four different HSFs, HSF1 plays a primary role in the transcriptional regulation of HSP expression (see reference 5 for a comparison of yeast and metazoan HSFs). On the contrary, yeast and other invertebrates express a single HSF with functional equivalence to HSF1. Yeast HSF1 is a single-copy, essential gene encoding an 833-amino-acid protein (441, 511). The fundamental architecture of yeast Hsf1 is consistent with all HSF isoforms, including a DNA-binding domain (DBD), three leucine zipper (LZ) repeats responsible for the trimerization of the factor, and a carboxyl-terminal transactivation domain (CTA) (Fig. 4). In addition, budding yeast Hsf1 is unique in having an additional transcriptional activation domain at the amino terminus (N-terminal transactivation domain [NTA]) (320). The DBD is the most conserved region within the HSF family and is the only functional domain of Hsf1 for which detailed structural data are available. The DBD belongs to the “winged” helix-turn-helix family of DNA-binding proteins. Like other members of the family, the Hsf1 DBD recognizes the DNA through helix α3, the second helix of the motif (193, 460). However, a crystallographic study suggested that the recognition helix of the Hsf1 DBD is not buried as deeply in the DNA major groove as those of other winged helix-turn-helix proteins, and the flexible loop or “wing” does not function to contact DNA. Instead, it contributes to interactions with adjacent Hsf1 DBDs bound to the DNA (259). The removal of the wing region decreases Hsf1 DNA affinity and reduces its transcriptional activity under conditions of normal growth and heat-shocked conditions (66).
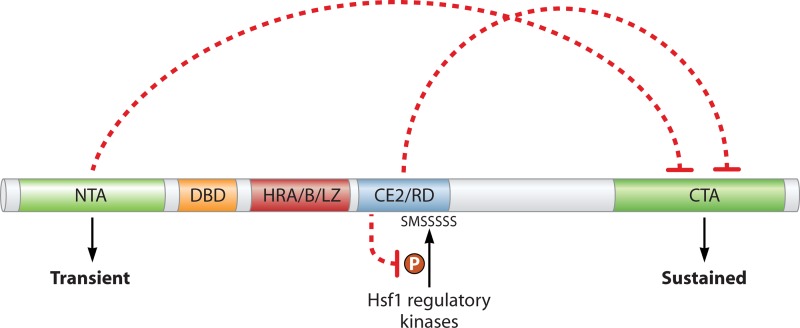
Fig 4.
Architecture and regulation of yeast Hsf1. Relevant domains of the budding yeast transcription factor are indicated. Dashed lines represent regulatory relationships between the NTA (amino-terminal transactivation domain) and the CE2 (control element 2)/RD (regulatory domain) on the CTA (carboxy-terminal transactivation domain). The serine-rich region within the RD is phosphorylated by unknown kinases to promote the repression of the CTA through CE2. As described in the text, the NTA promotes a transient transcriptional response, whereas the CTA is responsible for sustained responses. DBD, DNA-binding domain; HRA/B/LZ, heptad repeats A and B, also called the leucine zipper; P, phosphorylation.
Heat shock elements.
All Hsf1 target genes contain multiples of the pentameric sequence nGAAn, where “n” can be any nucleotide, termed heat shock transcription elements (HSEs), in their promoters (Fig. 3) (440). The architecture and spacing of the pentameric units vary considerably in different promoters, falling into three distinct classes. The “perfect”-type HSE consists of three continuous inverted repeats of the pentameric unit (nTTCnnGAAnnTTCn). The discontinuous or “gap”-type [nTTCnnGAAn(5 bp)nGAAn] and “step”-type [nTTCn(5 bp)nTTCn(5 bp)nTTCn] HSEs contain insertions between the consensus sequences, with a conservation of the 5-bp spacing but not the sequence, presumably to preserve the proper spatial orientation (173, 381, 522). Human HSF1 preferentially binds to continuous HSEs over discontinuous HSEs. Yeast Hsf1, in contrast, recognizes both continuous and discontinuous repeats of the nGAAn unit (381). This observation is consistent with the fact that vertebrates have four HSFs involved in diverse gene expression programs, while yeast cells are limited to a single Hsf1 for the control of the expression of HSPs and other targets under both normal and stress conditions. The active DNA-binding form of yeast and other HSFs is a homotrimer (28). Although each nGAAn unit in an HSE is a recognition site for a single Hsf1 monomer, a minimum of three pentameric units is required for stable binding in vitro (338). Some target genes contain four to six contiguous units that make contact with two neighboring Hsf1 trimers and seven to eight units that may recruit up to three colocalized trimers. Cooperative binding between yeast Hsf1 trimers is not as significant as that of vertebrate HSF1: a single yeast Hsf1 trimer is sufficient to activate transcription (431), and thus, additional nGAAn units in genes with multiple HSEs possibly function to increase the overall stability of the Hsf1-DNA interaction.
Hsf1 functional domains.
The oligomerization domain is another highly conserved functional region among all identified HSF genes (132, 480). The 91-amino-acid domain located carboxy terminally to the DBD mediates the formation of a homotrimer of HSF monomers via a triple-stranded α-helical coiled coil, similar to the trimerization domain found in the influenza virus hemagglutinin protein (Fig. 4) (339). Structure studies of Kluyveromyces lactis revealed that the trimerization domain comprises 7-residue repeating sequences termed heptad repeats in two subdomains: helix A (HR-A), located at the N terminus of the trimerization domain, and helix B (HR-B), located at the C terminus of the domain (340). Both HR-A and HR-B are amphiphilic helices containing hydrophobic residues that occupy the interhelical surface and thus are also known as leucine zipper (LZ) domains. Proteolysis and nuclear magnetic resonance (NMR) studies suggested that the isolated Hsf1 trimerization domain may form an all-parallel, elongated structure (340). Since the activation of gene expression by Hsf1 requires three HSE repeats in vivo, the trimerization of Hsf1 might increase the affinity of DNA binding and stabilize protein-DNA interactions (108). Trimerization is also a key point of regulation of HSF1 activity in higher eukaryotes. In vertebrate and Drosophila melanogaster cells, inactive HSF1 is maintained in the cytoplasm as a monomer (12, 352). Elevated temperatures and other HSF-activating stresses lead to the trimerization of HSF1 to permit DNA binding (510). However, this step of activation is not universal. In yeasts such as S. cerevisiae and K. lactis, Hsf1 appears to bind DNA constitutively as a trimer (28, 439). A distinguishing feature of Hsf1 from S. cerevisiae and the closely related yeast K. lactis is the presence of distinct transactivation regions at both the N and C termini of Hsf1 (Fig. 4). The N-terminal transactivation domain (NTA) is located within the first 170 amino acids (320, 437). The C-terminal transactivation domain (CTA) is found between residues 595 and 783 (62). A structural analysis showed that the NTA is unstructured, as probed by heteronuclear NMR (64). The CTA is predominantly unfolded under physiological conditions but exhibits a certain amount of secondary and tertiary structures, as measured by circular dichroism (CD) and protease resistance. The α-helical content can be significantly increased at high temperatures, at acidic pHs, or by the addition of the disaccharide trehalose, suggesting that the CTA undergoes distinct conformational changes under different conditions (42, 336). Although both transactivation domains are strong constitutive activators when fused to a heterologous DNA-binding domain such as lexA, studies of a synthetic HSE-lacZ reporter suggested that the two transactivation domains respond differently to thermal stress (437). The NTA appears to mediate “transient” increases in levels of Hsf1 activity, while the CTA is required for “sustained” increases. The transient and sustained activities of Hsf1 are regulated over different temperature ranges, and increases in both activities lead to increased levels of Hsf1 phosphorylation (437). The deletion of either transactivation domain does not affect cell growth under normal growth conditions (320, 437). However, the elimination of the CTA, but not the NTA, leads to a temperature-sensitive phenotype and the arrest of the cell cycle in both the G1/S and G2/M phases due to the depletion of Hsp90 at 37°C, as described above in this review (303). The deletion of the NTA between residues 40 and 170 results in the constitutive activation of Hsf1 in the absence of a heat shock (30). High-resolution mapping of the CTA found that point mutations that abolish the activation of the heat shock response result in temperature-sensitive growth (62). These observations suggest that the NTA functions as a negative regulator by masking the CTA and that the CTA is not sufficient for Hsf1 activation during stress (Fig. 4). The presence of two distinct transactivation domains in yeast may provide additional levels of regulation or selectivity in gene activation. For example, the CTA is required for the heat- or glucose starvation-induced activation of the yeast metallothionein gene CUP1 but is dispensable for the transient heat shock induction of the yeast Hsp70 homologs SSA1 and SSA3 (386). In addition, the CUP1 gene differs from typical HSP genes by requiring a temperature of 39°C for robust induction, rather than the standard 37°C for most Hsf1 targets (456).
Regulation of Hsf1 transcriptional activity.
The activation of metazoan HSF1 is a multistep process, including trimerization, nuclear translocation, DNA binding, and posttranslational modifications. However, S. cerevisiae HSF1 is essential for cell viability at all temperatures and consistently is constitutively bound on promoters of HSP genes as a trimer in the absence of stress. Furthermore, the transactivation potency of yeast Hsf1 is negatively regulated for both transactivation domains. These properties strongly suggest that the Hsf1 function is modulated largely posttranslationally. In addition, the loss of two potential control nodes—nuclear translocation and trimerization—suggests that the derepression/activation of the Hsf1 transactivation domains is a plausible regulatory mechanism. The primary amino acid sequence of yeast Hsf1 predicts a molecular mass of 93.2 kDa. However, the protein usually migrates as a spaced doublet of 150 to 160 kDa from nonstressed cells and up to 190 kDa from heat-shocked cells on SDS-polyacrylamide gels, suggesting that significant posttranslational modifications are involved in both the “resting state” and stress-induced levels of activity (438, 441). A major feature of Hsf1 is that its conversion into the active form occurs simultaneously with extensive phosphorylation detectable by a significant retardation of migration on SDS-PAGE gels. Increases in Hsf1 activity levels measured by an analysis of target gene expression or an HSE-lacZ reporter assay correlate with the degree of phosphorylation when cells are shifted between 15°C, 20°C, and 30°C. This observation is further supported by the phosphatase treatment of cell lysates from heat-shocked cells, which significantly decreases the mobility shift (441). Moreover, a detailed kinetic study demonstrated that Hsf1 is rapidly phosphorylated after heat shock, declining to a low degree of phosphorylation coincident with the transient induction of HSP genes (262). Interestingly, the phosphorylation induced by menadione, a pro-oxidant that generates a superoxide anion in vivo, displays a different and sustained kinetic pattern. The two-dimensional resolution of tryptic phosphopeptides also showed that Hsf1 is phosphorylated at different phosphor-acceptor sites in response to heat or menadione, suggesting that Hsf1 undergoes stress-specific phosphorylation (262). Although hyperphosphorylation generally correlates with the transactivation of Hsf1, many phosphorylation events have been established to repress its transcriptional activity. Sorger first reported that yeast Hsf1 remains hyperphosphorylated after the termination of the transient activation of the heat shock response (437). Structural data suggest that a yeast-specific heptapeptide termed CE2 may regulate phosphorylation in the “resting state.” CE2 restrains the activity of the CTA domain, while the sequence adjacent to CE2 is rich in serine residues (SMSSSSS) (Fig. 4) (201, 437). The replacement of all six serines with alanine causes a derepression of Hsf1 activity and the elimination of most but not all electrophoretic mobility shifts on SDS-PAGE gels. On the contrary, when these serines are replaced with negatively charged amino acids to mimic phosphorylation, Hsf1-mediated transcription is repressed upon heat shock (188). These observations suggest that the phosphorylation of the serine-rich domain functions to repress Hsf1 basal activity and/or return Hsf1 to the inactive state in the attenuation phase. Interestingly, the deletion of CE2 leads to constitutively high levels of phosphorylation within the serine-rich domain in nonstressed cells (188). It seems likely that CE2, which represses the CTA, is a major control element for phosphorylation within the serine-rich domain. This finding is consistent with another study demonstrating that an alteration of two arginine residues (residues 826 and 830) in the CTA to glutamate completely abrogated the heat-inducible phosphorylation of Hsf1 (172). Taken together, phosphorylation likely plays both positive and negative roles in the regulation of Hsf1 transcriptional activity. A complete understanding of Hsf1 phosphorylation requires the identification and characterization of the involved kinases and phosphatases. A phospho-amino acid analysis of Hsf1 showed that at 20°C, the majority of the phosphate resides on serine residues, while at 39°C, phosphoserine and phosphothreonine are present at approximately a 3:1 ratio, respectively. Moreover, the absolute levels of both phosphorylated residues increase during heat shock (437). Hsf1 therefore is very likely phosphorylated by one or more serine-threonine protein kinases. Snf1, a homolog of mammalian AMP-activated kinase, is required for the glucose starvation-induced, Hsf1-dependent activation of the CUP1 metallothionein gene (456). However, the activation of Hsf1 by heat shock is Snf1 independent, suggesting the involvement of other kinases (166). Utilizing a Tn7-based insertional mutagenesis approach, Ferguson et al. found that protein kinase A (PKA) represses the Hsf1-dependent expression of the HSP26 gene in nonstressed cells, but regulation was not universally observed among all Hsf1 target genes. In addition, diminished PKA activity paradoxically leads to increased levels of Hsf1 phosphorylation, indicating that PKA indirectly inhibits Hsf1 activity (130). Recently, the dual-specificity, tyrosine phosphorylation-regulated kinase Yak1 was shown to play a key role in mediating the PKA-dependent regulation of Hsf1. Yak1 was first identified as a growth antagonist and is negatively regulated by PKA (146, 244). An in vitro kinase assay established that purified Yak1 phosphorylates truncated Hsf1 between residues 1 and 180. Moreover, Yak1 activates Hsf1 by inducing its DNA-binding activity under conditions of low PKA activity, such as acute glucose depletion. However, the deletion of YAK1 showed no significant effect on Hsf1 activation upon heat shock (243). To date, no definitive support for Hsf1-specific protein phosphatases has been obtained. In sum, despite the abundance of Hsf1 phosphorylation events and significant efforts to understand them, no clear picture has yet emerged to encapsulate the positive and negative effects of this posttranslational modification. Indeed, even phosphorylation events previously thought to be worked out, in this case, the phosphorylation of serines 303 and 307 in mammalian HSF1 by the glycogen synthase kinase 3 (GSK3) family of protein kinases, are now being brought back into question by contrasting data (15).
Chaperone regulation of Hsf1.
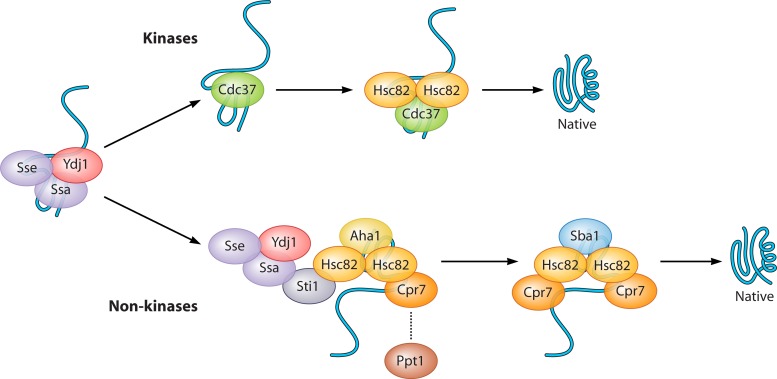
Early studies of cultured Drosophila cells showed that the expression levels of HSP genes increased rapidly after the initiation of a heat shock treatment, followed by a decrease in gene expression levels to slightly above the levels under prestress conditions (257). This observation suggests that the heat shock response is self-regulated via repression under nonstress conditions and attenuation under conditions of moderate thermal stress. As discussed below, substantial genetic and biochemical evidence suggests that two classes of heat shock proteins, Hsp70 and Hsp90, serve as trans-acting Hsf1 repressors that may fulfill both roles (Fig. 3) (reviewed in references 79 and 481). The deletion of SSA1 and SSA2, the two constitutively expressed Hsp70 chaperones in the cytoplasm, leads to the activation of Hsf1 at normal growth temperatures (81, 284). The ATPase activity of Hsp70 seems to play an important role in Hsf1 repression, as mutants lacking the Hsp70 nucleotide exchange factors SSE1 and FES1 result in the constitutive transcriptional competence of Hsf1 (263, 412; Y. Wang and K. A. Morano, unpublished data). The Ssb1 and Ssb2 members of the Hsp70 family, which are ribosome-associated chaperones, were also detected to form a stable and ATP-sensitive complex with Hsf1 (29, 266). Interestingly, the ssb1Δ ssb2Δ double mutant leads to an increase in Hsf1 activity in heat-shocked cells but does not derepress Hsf1 in nonstressed cells (29). These findings suggest that the Ssa and Ssb subclasses of Hsp70 play subtly distinct roles in Hsf1 regulation. Despite these observations, Hsp70 alone is insufficient to suppress HSF1 in mammalian cells (353). Instead, Hsp70 may act in conjunction with the Hsp90 chaperone complex, which is responsible for the maturation and regulation of various client proteins. Hsp90 associates with a number of cochaperones, including Hsp70, Sti1, Cpr6/7, and Sba1, to achieve its cellular functions at different steps of the client-specific folding cycles (see below). Some of the functions attributed to Hsp70 and its nucleotide exchange factors in the regulation of Hsf1 activity may reflect a joint effort with Hsp90. In human cells, Hsp90 is found to be associated with HSF1 in vivo and in vitro (531). In yeast, a double mutant lacking constitutively expressed Hsp90 and a cyclophilin 40 homolog, hsc82Δ cpr7Δ, exhibits high levels of Hsf1 activity in the absence of stress and is thermotolerant (113, 169). Moreover, studies to identify Hsf1 pharmacological modulators showed that some Hsf1 activators also function as Hsp90-specific inhibitors, such as geldanamycin and radicicol (67, 241, 403). Recently, celastrol, an active component of Chinese medicine, was found to promote HSP gene expression through Hsf1 activation and to block the maturation of Hsp90-dependent steroid receptors in yeast and human cells (185, 465, 507). Therefore, the Hsp70/Hsp90 chaperone complex represses the transcriptional activation of Hsf1 under nonstress conditions. During heat shock, the accumulation of unfolded or damaged proteins may titrate the chaperone machinery from Hsf1, allowing the derepression of the transcription factor. An obvious deficiency in this model for the yeast system is the lack of concrete evidence for a physical association between Hsp90 and Hsf1 in nonstressed cells, despite the abundance of genetic support. However, it is possible that the chaperone-transcription factor interaction is not robust enough to survive standard copurification or affinity isolation protocols. This hypothesis is in keeping with the fact that the chaperone repression of Hsf1 appears to operate with a “hair-trigger” mechanism that would not be consistent with high-affinity binding.
Msn2/4
The stress response element.
In addition to heat shock gene transcription mediated by Hsf1, a parallel pathway in S. cerevisiae senses and responds to a remarkable variety of stresses besides heat shock. The regulatory element of this “general” stress pathway was originally identified as an Hsf1-independent sequence in the promoters of the DNA damage-responsive gene DDR2 and the nutrient stress-responsive gene CTT1 (231, 512). This “stress-responsive element” (STRE) is a 5-bp sequence functional in both orientations (CCCCT or AGGGG). Analyses of mutational variants indicated that a sequence alteration within the CCCCT element completely abolishes the regulatory efficacy of the STRE, while base changes in the flanking sequence and a modulation of the spacing between elements only slightly reduce the transcriptional response (462). A single iteration of the STRE is sufficient for the basal and stress-induced expression of a heterologous CYC1-lacZ reporter, but multiple STREs confer a more robust induction of gene expression in a noncooperative manner (231).
Function and regulation of Msn2/4.
Two functionally related transcription factors, Msn2 and Msn4, mediate STRE-mediated gene expression (396). MSN2 was initially identified as the multicopy suppressor of temperature-sensitive protein kinase SNF1 mutant and contains two zinc finger motifs near the C terminus of the Cys2His2 type, which are closely related to those of the yeast Mig1 and Rgm1 repressors (125, 126). A highly related gene, MSN4, bears 41% identity in amino acid sequence to MSN2. The deletion of both MSN2 and MSN4 leads to sensitivity to thermal, oxidative, and osmotic stresses (126, 282). Of the two genes, MSN2 seems to play a more pronounced role, as the overexpression of MSN4 can only partially suppress phenotypes of an msn2Δ mutant (396). Genetic analyses of msn2Δ, msn4Δ, and msn2Δ msn4Δ mutants found that although Msn2 and Msn4 exhibit functional redundancy, they may play distinct roles in the regulation of stress-induced gene expression. For example, the stress-dependent induction of PDE2, a gene encoding a high-affinity cyclic AMP (cAMP) phosphodiesterase, is not affected by Msn2 but is completely eliminated in the double mutant strain. On the contrary, the induction of the yeast phosphoglucomutase isozyme gene PGM2 is dependent exclusively on Msn2 (463). Furthermore, MSN2 and MSN4 display different expression patterns at the diauxic transition (96). Genomic expression studies of yeast cells utilizing DNA microarrays revealed that the expression of MSN2 is constitutive under all conditions, whereas MSN4 gene expression is stress induced, and induction is mediated by itself and Msn2 (147).
The multistress response mediated by Msn2/4 is generally transient, and the intensity and duration of the response are dependent on the strengths of the stresses (147). In vivo footprinting analyses suggested that the occupancy of STREs increases rapidly in an Msn2/4-dependent manner under stress conditions (158). This observation is further supported by fluorescence microscopy analyses of the subcellular localization of myc9- and green fluorescent protein (GFP)-Msn2 fusion proteins. Both fusions are found primarily in the cytoplasm and are largely excluded from the nucleus in nonstressed cells (Fig. 3). Stress treatments, including temperature upshift, ethanol, sorbate, and osmotic stress, lead to the accumulation of Msn2 in the nucleus, suggesting an oscillatory localization (158). Two nutrient-sensing pathways have been described to play important regulatory roles in controlling Msn2/4: the cAMP-protein kinase A (PKA) pathway and the TOR pathway (Fig. 3). PKA activity is regulated by nutrient sufficiency through the modulation of cellular cAMP levels by activating G proteins and adenylate cyclase. Low levels of PKA activity brought about by heat shock or growth under glucose-replete conditions result in the nuclear accumulation of Msn2 and Msn4 in the absence of stress, whereas high levels of PKA activity effectively block the nuclear localization of the transcription factors in stressed cells (145, 158–160). Msn2 and Msn4 each contain a nuclear localization signal (NLS) near the C terminus. Deletion and mutagenesis analyses demonstrated that the substitution of S288 with alanine or aspartate in MSN2 leads to constitutive nuclear accumulation, which is reversible by high exogenous levels of cAMP. Export is completely abolished when S288 is modified in combination with S620, S625, and S633. Thus, the PKA consensus site required for Msn2 nuclear export includes S288, and the cAMP levels are partially redundant with PKA to regulate the trans-localization of the two factors (158). Recent studies showed that Yak1 may contribute to the PKA-dependent inhibition of Msn2/4. Yak1 kinase activates Msn2/4 under conditions of glucose starvation and directly phosphorylates the two factors in vitro (243). Yak1 is also restrained in the cytoplasm under high-glucose conditions by an association with the 14-3-3 protein Bmh1, in a PKA-dependent manner (244). Furthermore, genetic evidence suggests that Bcy1, a regulatory subunit of PKA, mainly affects the phosphorylation status of the Msn2 NLS by the downregulation of PKA (159) Several studies also demonstrated that the protein phosphatase PP1 dephosphorylates Msn2 (99, 248, 285). Taken together, when cells encounter acute glucose starvation, PKA activity is downregulated via Bcy1 or PP1. The decrease in the activity of PKA activates Yak1, which in turn phosphorylates Msn2/4. Hyperphosphorylated Msn2 and Msn4 fail to be exported, and consequently, the accumulation of the two factors leads to an induction of STRE-mediated gene expression. Interestingly, YAK1 gene expression is mediated by Msn2/4, suggesting a potential autoregulatory loop (275).
The TOR signaling pathway also impacts the activities of Msn2 and Msn4. Unlike the cAMP-PKA pathway, which appears to regulate primarily nuclear export, TOR prevents the nuclear import of Msn2 and Msn4 (18). The rapamycin-sensitive TOR signaling pathway is known to control cellular responses to nutrient stress, especially carbon and nitrogen starvation (458). TOR inhibits the expression of STRE-containing genes by stimulating the association of Msn2/4 with the cytoplasmic 14-3-3 protein Bmh2 (Fig. 3) (18). However, the localization of Msn2/4 is not the sole regulatory point in STRE-mediated gene expression. The nuclear localization of Msn2/4 is dependent on the expression of MSN5, encoding a nucleus export receptor (6). The deletion of MSN5 results in the accumulation of the two factors in the nucleus under normal growth conditions but has no effect on the regulation of STRE-dependent gene expression (124). This important observation suggests a functional redundancy within the Msn2/4 regulatory network and the presence of another posttranslocation activating step. A recent systematic study examined the effects of 35 single-deletion mutants of Msn2/4 partners on STRE-dependent gene expression after exposure to heat, oxidative, and osmotic stresses (377). That study suggested that Msn2/4 activity is precisely modulated by multiple partners to provide an optimal stress response. Regulatory inputs included those governing not only nuclear localization but also differential activation, proteasomal degradation, and chromatin remodeling. The combinatorial control of the “general” stress response is critical to effectively manage gene expression induced by multiple different environmental stresses.
Cross-Protection and Acquired Thermotolerance
The ability of cells to survive exposure to a sudden lethal temperature shock is defined as thermotolerance. Pretreatment at sublethal temperatures conditions cells to survive severe heat shock, which would otherwise be lethal. This phenomenon is termed acquired thermotolerance (383). It is commonly assumed that the gain of thermotolerance is due to the induced synthesis of heat shock proteins, and in fact, wild-type yeast cells shifted from 30°C to 37°C before exposure to 50°C showed significant increases in the survival rate compared with cells shifted directly to 50°C (162, 383). A strain lacking both alleles of the constitutive cytosolic Hsp70, ssa1Δ ssa2Δ, renders the heat shock transcription factor Hsf1 constitutively active, and this strain is more tolerant of extreme temperatures than wild-type cells (81). Conversely, the deletion of the inducible chaperone HSP104 dramatically decreased the transient thermotolerance conferred by a sublethal heat shock, suggesting that Hsp104 is one of the major heat shock proteins that contribute to acquired thermotolerance (162). As described above in this review, in addition to Hsp104, trehalose levels appear to be another major determinant of thermotolerance. However, trehalose may contribute more to prolonged stress protection, whereas heat shock proteins are essential for the transient acquisition of thermotolerance (162). Interestingly, the production of Hsp104 is regulated by both the heat shock transcription factor Hsf1 and the general stress transcription factors Msn2 and Msn4, while trehalose levels are modulated primarily by Msn2/4 (282).
Yeast cells exposed to sublethal stress gain tolerance not only to higher doses of the same stress but also to other disparate environmental stresses. A meta-analysis of stress microarray data sets indicated that 21 out of 37 predicted stress-responsive regulators (for example, Hsf1, Msn2/4, and the oxidant response factor Yap1) have overlapping functions under at least half of the eight conditions of environmental stresses surveyed, including oxidative stress, heat/cold shock, and osmotic stress (61). Several studies supported the observation that thermotolerance is tightly linked to aerobic metabolism, likely through the generation of oxidative stress. Mutants deficient in the key antioxidant enzymes catalase, superoxide dismutase (SOD), and cytochrome c peroxidase demonstrate pronounced thermal sensitivity at 50°C, while the overexpression of these enzymes confers thermotolerance (90). The superoxide anion (O2−) not only activates the yeast Yap1 oxidant defense transcription factor but also selectively induces the Hsf1-dependent expression of the copper metallothionein CUP1 (4, 246, 262). Heavy metals and other noxious chemicals are also potent HSF activators in both yeast and human cells. A constitutively active HSF1 allele exhibiting high-level basal transcription activity was shown to result in enhanced cadmium resistance (405). The treatment of cells with the natural product celastrol or diverse chemical electrophiles activates both Hsf1 and Yap1, leading to both thermotolerance and oxidant resistance (465; Wang and Morano, unpublished). Together, these finding suggest that in addition to Yap1, yeast Hsf1 can sense oxidative stress and assist in mounting a defensive transcriptional response. Yap1 has been shown to sense hydrogen peroxide and other oxidants and electrophiles through reactive cysteines in its primary sequence (10, 94, 95). Because yeast Hsf1 lacks cysteines, how this stress factor senses and responds to oxidants remains unclear. It is likely that one or more unidentified cellular factors may act as proxy sensors.
Posttranscriptional Control of the Heat Shock Response
Nuclear mRNA export during heat shock.
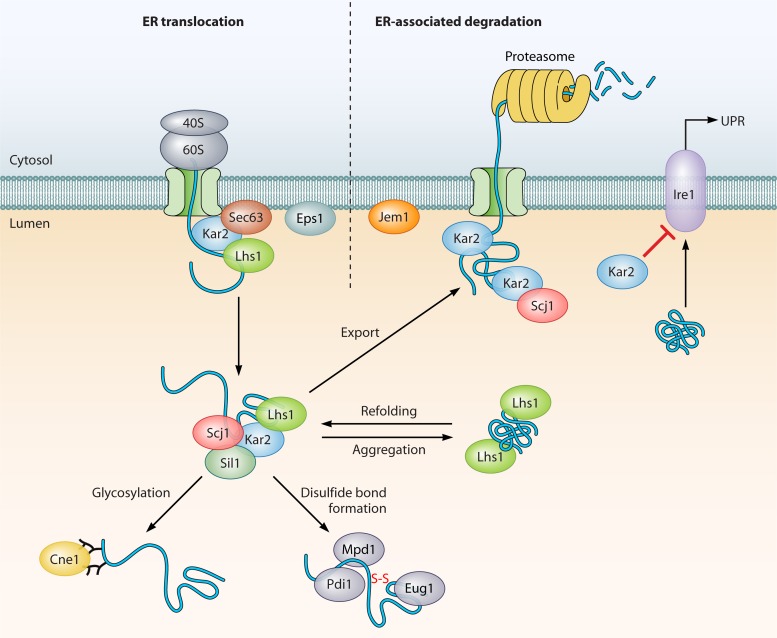
How does heat shock affect gene expression posttranscriptionally? Many nascent transcripts are processed after synthesis in the nucleus but must then be exported to the cytoplasm for translation. In response to heat shock (42°C), bulk poly(A)+ RNA accumulates in the nucleus (375), while HSP mRNAs are translated, and therefore presumably exported, efficiently. Signals in the 5′ and 3′ untranslated regions (UTRs) of the message were found to be required for the export of SSA4 transcripts, encoding an Hsp70 isoform (375). At least one nuclear pore protein, Rip1, is required for the export of heat shock transcripts during thermal stress but not under normal growth conditions, defining a specific transport pathway for these important mRNAs (376). The CWI MAP kinase Slt2 is also required for mRNA retention during heat shock, via the phosphorylation of the mRNA-binding protein Nab2 (52). Interestingly, another pair of RNA-decapping enzymes (Edc1/2) is required for the efficient translation of mRNAs during heat shock but not growth at normal temperatures (311). These results suggest that the processing requirements for efficient mRNA translation may differ under the two conditions, possibly due to an inactivation of other required components. In support of this idea, the major mRNA export factor Gle2 and the DEAD box protein Rat8 dissociate from nuclear pore complexes at 42°C, thereby limiting bulk transport (369). Interestingly, the pretreatment of cells at 37°C prevents the dissociation of both proteins, consistent with the general theme of acquired thermotolerance via preexposure to mild stress (198).
mRNA sequestration in response to stress.
Recent work has defined novel ribonucleoprotein assemblies, termed processing bodies (P bodies) and stress granules (SGs), that appear to concentrate nontranslating mRNAs in exchangeable but sequestered pools in response to a variety of stress conditions, including glucose starvation and osmotic stress. Heat shock preferentially induces the formation of SGs that contain translation initiation factors (eukaryotic initiation factor 2 [eIF2] and eIF3), 40S ribosomal subunits, and non-heat-shock mRNAs (40, 333). These components are capable of redistributing into the cytoplasm and engaging in translation upon a return to normal temperatures (163). Heat-shock-induced SGs also contain a subset of P-body components involved in RNA degradation, including Dcp2 and Dhh1, yet are spatially distinct from other P-body markers (163). The precise roles of these assemblages are still unknown at present, as RNA-processing steps such as decapping and degradation occur in the absence of detectable PB formation. It is tempting to speculate that the formation of SGs may be yet another way for cells to reduce total protein synthesis under unfavorable protein-folding conditions such as heat shock. Moreover, a model wherein SGs act as temporary, protected “storage” compartments for translatable mRNAs is attractive, as it would allow for the rapid reinitiation of the translation of existing transcripts when cells return to proliferative conditions. Such a model predicts that cells incapable of producing SGs in response to heat shock might exhibit reduced survival or delayed reentry into normal growth, but this hypothesis is yet to be tested. Given that both PBs and SGs are sizable multiprotein complexes, it is also conceivable that HSPs may play a role in their assembly or disassembly. Indeed, a number of known PB/SG protein subunits contain glutamine-rich prion-like domains that could serve as potent recruitment regions for the Hsp104 and Hsp70 machinery (91).
MOLECULAR CHAPERONES OF THE CYTOPLASM
At any given time, hundreds of macromolecular processes involving proteins are occurring in the cytosol of a cell. Given the extremely high protein concentration in this environment (over 300 mg/ml), protein-protein interactions must be governed and modulated appropriately (140). In addition, the constant influx of newly synthesized polypeptides provides a significant protein-folding problem, as does the recognition of damaged proteins that must be targeted and shepherded for degradation. One way in which cells maintain the proper homeostatic balance of the proteome is through the deployment of protein molecular chaperones. Molecular chaperones are a ubiquitous group of proteins involved in the folding and remodeling of other proteins (120). Although the term “heat shock protein” is commonly used synonymously with “chaperone,” distinctions must be made, as not all heat shock proteins are chaperones, and not all chaperones are induced by heat shock. A panoply of different classes of chaperones participate in protein biogenesis and quality control, and there is a growing appreciation that these machines cooperate in multichaperone networks (157). In this section, we review the current understanding of the roles that molecular chaperones play in the yeast cytosol and nucleus as a model for an understanding of the protein-folding and remodeling requirements of a eukaryotic cell (Table 1).
Table 1.
Cytosolic chaperones
| Class | Protein(s) | Function(s) |
|---|---|---|
| Hsp100 | Hsp104 | Unfoldase; disaggregase |
| Hsp90 | Hsp82 | Protein maturation, stress inducible |
| Hsc82 | Protein maturation, constitutively expressed | |
| Hsp90 cochaperone | Sti1 | Hsp70/Hsp90-organizing protein homolog, TPR containing |
| Cns1 | Similar to Sti1, TPR containing | |
| Cdc37 | Protein kinase folding | |
| Sba1 | Hsp90 ATPase regulator | |
| Cpr6 | Immunophilin homolog, TPR containing, stress inducible | |
| Cpr7 | Immunophilin homolog, TPR containing, constitutively expressed | |
| Sgt1 | TPR-containing Hsp90 adaptor protein | |
| Aha1 | Hsp90 ATPase regulator | |
| Ppt1 | TPR-containing protein phosphatase | |
| Hsp70 | ||
| GRP170 | None | |
| Hsp110 | Sse1 | Hsp70 nucleotide exchange, substrate binding, constitutively expressed |
| Sse2 | Hsp70 nucleotide exchange, substrate binding, stress inducible | |
| Hsp70 | Ssa1, Ssa2 | Protein folding, translocation, constitutively expressed |
| Ssa3, Ssa4 | Protein folding, translocation, stress inducible | |
| Ssb1, Ssb2 | Nascent-chain folding | |
| Hsp70 NEF | Fes1 | Hsp70 nucleotide exchange |
| Snl1 | Hsp70 nucleotide exchange, ER tethered | |
| Hsp40/J protein | Ydj1 | Hsp70 ATPase activator, substrate binding |
| Sis1 | Hsp70 ATPase activator, substrate binding | |
| Zuo1 | ||
| Caj1 | Hsp70 ATPase activator, substrate binding | |
| Djp1 | Hsp70 ATPase activator, substrate binding, peroxisomal import | |
| Xdj1 | Hsp70 ATPase activator, substrate binding | |
| Apj1 | Hsp70 ATPase activator, substrate binding | |
| Jjj1 | Hsp70 ATPase activator, ribosome biogenesis | |
| Jjj2 | Hsp70 ATPase activator | |
| Jjj3 | Hsp70 ATPase activator | |
| Hlj1 | Hsp70 ATPase activator, ERAD | |
| Cwc23 | Hsp70 ATPase activator, mRNA splicing | |
| Swa2 | Hsp70 ATPase activator, vesicle transport | |
| Chaperonin | TriC/Cct1–Cct8 | Protein folding, cytoskeleton substrates |
| Chaperonin cochaperone | Pfd1–Pfd6 | Protein folding, cytoskeleton substrates |
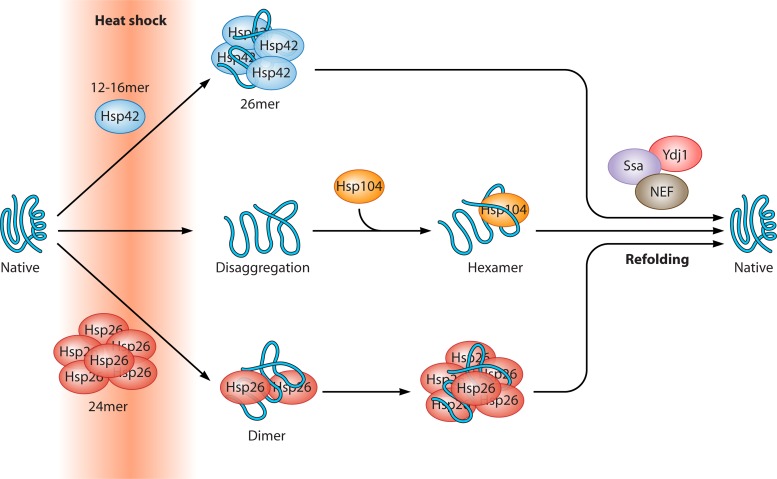
| sHSP | Hsp42 | Antiaggregase |
| Hsp26 | Antiaggregase | |
| Other | Hsp12 | Membrane chaperone? |
Hsp70 and Cofactors
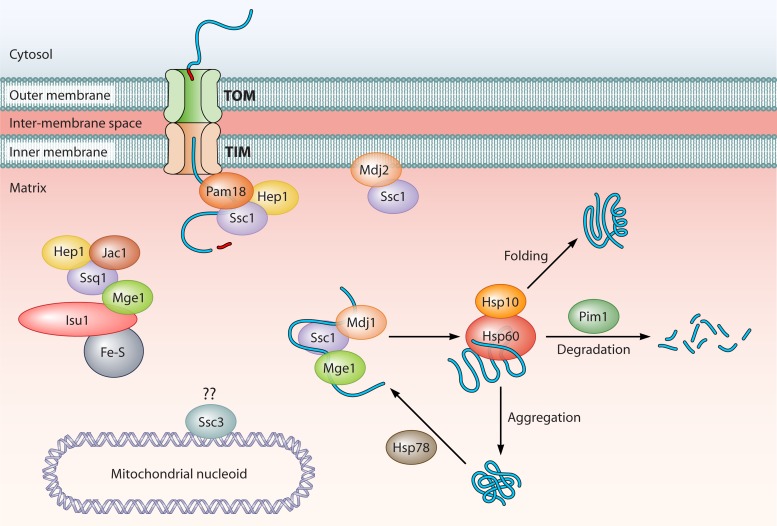
The 70-kDa family of heat shock proteins (Hsp70s) is arguably the most highly conserved family of proteins throughout evolution. In yeast, this ubiquitous family of chaperones is found in many cellular compartments and plays major roles in cell viability (78). Hsp70s function primarily to ensure the proper folding of nascent or misfolded proteins through the recognition of determinants in the tertiary structure, usually the solvent exposure of hydrophobic patches normally buried within a properly folded protein. Hsp70s are functionally divided into two domains: an N-terminal nucleotide-binding domain (NBD) and C-terminal substrate-binding domain (SBD). The NBD is approximately 44 kDa and is related to the hexokinase and actin ATP-binding folds with a bilobular structure (131). Traditionally referred to as the “ATPase” domain, the discovery (see below) that at least some Hsp70-related proteins bind ATP but exhibit weak to nonexistent ATPase activity has prompted a change in terminology. An interdomain linker plays an important role in allosteric communication between the two regions, transmitting conformational information to the SBD based on nucleotide occupancy in the NBD (483, 484). The SBD is itself composed of two subdomains, an 18-kDa substrate interaction domain and a 10-kDa variable domain located at the extreme C terminus. The SBD is made up primarily of β-sheets that form a binding interface for substrates and an α-helical “lid” that assists in conferring high-affinity substrate binding. A short and relatively unstructured intervening sequence connects the β-sandwich and lid domains. While substrate binding requires only the presence of the β-sandwich domains, the active folding of substrate proteins requires allosteric communication between the NBD and SBD (483, 484). The variable domain is important for adaptor protein interactions; for example, some Hsp70s contain an “EEVD” sequence necessary for binding to tetratricopeptide repeat (TPR) domains of cochaperones (139). Classical Hsp70s function through a nucleotide-dependent cycle. When bound to ATP, the SBD of Hsp70 is in a low-affinity substrate-binding conformation. ATP hydrolysis results in conformational changes within the NBD that are transmitted to the SBD, resulting in further conformational changes that increase substrate affinity. Ultimately, the release of the ADP and replacement with ATP confer the release of the folded or partially folded substrate, allowing the cycle to repeat. Two major classes of cochaperones interact with the NBD of Hsp70 to assist protein folding by regulating its cycle: the J-domain proteins and the nucleotide exchange factors (NEFs), which will be discussed below. In S. cerevisiae, the cytosolic Hsp70 superfamily includes the Ssa, Ssb, and Sse families and the atypical Ssz1 (stress seventy A, B, E, Z) family (505). The paralogs Sse1 and Sse2, while clearly related to Hsp70, are best classified as members of the Hsp110 subfamily of Hsp70-like proteins. As described in detail below, Hsp110s were recently shown to act as nucleotide exchange factors for the Hsp70s (106, 360, 410). Based on in vitro and in vivo experiments examining substrate specificity and protein interactions, it was proposed that the Hsp70s can be classified into two broad categories: generalist and specialist. Generalists promiscuously bind to hydrophobic regions of unfolded proteins to promote folding and are responsible for many of the protein quality control functions in the cell; this group includes the Ssa family. The specialists function in specific processes or with specific substrates; this group includes the Ssb family, which is associated with ribosomes and is primarily involved in cotranslational protein folding (77, 202, 343).
The Ssa Hsp70s.
Four genes encode members of the Ssa family. SSA1 is constitutively expressed, but at lower levels than SSA2, and is induced under stress conditions. SSA3 and SSA4 are expressed only under stress conditions and in strains deleted for SSA1/2 due to the derepression of Hsf1 (313). In addition, SSA3 is highly induced upon entering the stationary phase (31, 504). Viability can be conferred by the constitutive expression of any single Ssa isoform, suggesting a high degree of functional redundancy. Ssa1 and Ssa2 are required for diverse protein homeostatic functions in the cell, such as protein folding, translocation, and degradation. While the disruption of either gene results in no detectable phenotypes, the simultaneous deletion of SSA1 and SSA2 results in slow growth and thermosensitivity at 37°C (506). This phenotype is surprising, as SSA3 and SSA4 are induced upon heat shock and are transcriptionally derepressed in the absence of the constitutive Hsp70s, and suggests that Ssa3/4 cannot completely complement the loss of Ssa1/2 despite the high degree of similarity. This distribution of constitutive and heat-inducible Hsp70 isoforms is entirely analogous to mammalian cells that express both Hsc70 (heat shock cognate) and Hsp70 (heat shock protein) and suggests a strong selection pressure for an additional Hsp70 functional capacity in heat shock or other environmental stress situations (80). Due to their semiredundant nature and the existence of four independent genes, genetic analyses of Ssa function have been challenging. To date, nearly all phenotypic consequences for Ssa− cells have been elucidated by using either an ssa1Δ ssa2Δ deletion strain or a temperature-sensitive allele generated in the laboratory of Elizabeth Craig, termed ssa1-45 (19). The latter strain lacks all four endogenous Ssa genes and expresses a temperature-sensitive allele of SSA1 that inactivates within 30 min at 37°C. A shift to the nonpermissive temperature therefore renders cells devoid of all Ssa activity, which ultimately leads to cell death. Experiments using this allele must therefore be carefully monitored, as it is possible that the observed phenotypes could be due to cell morbidity rather than specific defects in Ssa function.
The protein-folding activity of Ssa proteins is one of the most well-known and well-studied functions, and it has been shown that the depletion of the Ssa proteins in vivo leads to folding defects for endogenous enzymes such as ornithine carboxylase and the commonly used model protein firefly luciferase (FFL) (225, 473). In addition, complementing studies showed that the immunodepletion of Ssa1/2 disrupted the refolding of denatured FFL in yeast lysates (225, 252). Initially, it was thought that the Ssa family interacts with proteins only posttranslationally. However, the deletion of the nonessential ribosome-associated Ssb increased the cotranslational interaction of nascent polypeptides with Ssa1, suggesting that Ssa chaperones are competent to interact with chains emerging from the ribosome (521). The inactivation of SSA1 in the ssa1-45 strain resulted in a nearly complete abrogation of protein synthesis within 90 min, strongly supporting a role for this chaperone in bulk translation (225). Ssa has been found to physically interact with two ribosome-associated factors, the Hsp40 Sis1 and the poly(A)-binding protein Pab1, providing a direct link between the chaperone and translating ribosomes (191). The Ssa family is also involved in protein translocation across cellular membranes, including the vacuole, the nucleus, mitochondria, and the endoplasmic reticulum (ER) (97, 286, 388, 420). The depletion of Ssa activity through a GAL-SSA1 shutoff approach provided the first demonstration that a cytosolic Hsp70 is required for organellar translocation (97). The loss of Ssa function resulted in the defective translocation of both the yeast pheromone alpha-factor (αF) and the β-subunit of the mitochondrial F1-ATPase. Another major Ssa-mediated function is protein degradation. The model misfolded protein construct ΔssCG* (a mutated cytosolic form of the vacuolar protease carboxypeptidase Y [CPY] fused to a GFP tag) is a substrate for proteasomal degradation and was found to aggregate and display reduced degradation kinetics in ssa1-45 cells at the nonpermissive temperature (332). In that same study, the deletion of other chaperones, such as HSP104, HSC82, HSP82, the small Hsps (sHsps) HSP26 and HSP42, and SSE1, did not result in degradation defects. In contrast, the finding that Sse1 is not involved in degradation was disputed by a recent report showing that the ubiquitination and, ultimately, the degradation of the same model protein require both Sse1 and Ssa1 (181). A study using the endogenous yeast protein fructose-1,6-bisphosphatase (FBPase), which is expressed in the presence of nonfermentable carbon sources and degraded upon a shift to glucose, likewise uncovered a requirement for Ssa function in degradation (213). Fascinatingly, despite the high degree of sequence conservation between Ssa1 and Ssa2, only Ssa1 is competent to mediate FBPase degradation. A recent study narrowed the cause for this specificity to a single residue in the ATPase domain, alanine 83 (417). Substitution with glycine (the analogous residue in Ssa2) blocked Ssa1 functions in this biological process, and conversely, the replacement of the glycine with alanine in Ssa2 allowed this chaperone to complement the transport defects in the ssa1Δ null strain, demonstrating that a methyl group determines the functional delineation between Ssa1 and Ssa2 in this pathway. Additional reports have implicated Ssa in regulatory roles in RNA degradation and multidrug resistance (115, 407). In these scenarios, the recruitment of Ssa is required to stabilize and promote the activity of substrate proteins. The diverse functional roles that the Ssa family plays reflect its ability to nonspecifically interact with substrates and to be regulated by a large number of structurally distinct cochaperones that are discussed below.
The SSB Hsp70s.
The nonessential Ssb family is a fungal-specific class of Hsp70 composed of Ssb1 and Ssb2, which share 99% sequence identity and are considered to be functionally interchangeable (314). The levels of transcription of both genes decrease upon a shift to heat shock temperatures, in sharp contrast to the heat-inducible Ssa family (80, 266). The expression of SSB1/2 is regulated similarly to ribosomal proteins, supporting their role in cotranslational protein folding: they are most highly expressed when cells are growing in the presence of glucose, and expression is diminished at the diauxic shift, along with ribosomal gene synthesis (34, 147). The Ssbs are associated primarily with translating ribosomes and the ribosome-associated complex (RAC), composed of Ssz1 and the J-domain protein zuotin (Zuo1) (149). The RAC binds close to the ribosomal exit tunnel, which is also the predicted region for Ssb binding due to its association with nascent chains and the RAC (149, 343). The interaction of Ssb with the ribosome is not dependent on but is stabilized by nascent chain interactions. Treatment with the aminoacyl-tRNA analog puromycin releases both Ssb and the nascent chain from translating ribosomes. In addition, salt sensitivity assays were used to determine that the binding of Ssb to the ribosome is stabilized by interactions with nascent polypeptides. Ssb also cross-links to nascent chains and can be immunoprecipitated with incompletely synthesized proteins. Taken together, these data support the function of Ssb in cotranslational protein folding. Furthermore, the deletion of SSB1/2, SSZ1, or ZUO1 causes the same phenotypes of slow growth, cold sensitivity, and hypersensitivity to the aminoglycosides hygromycin B and paromomycin, underscoring both their involvement in translation and the shared functions of the RAC and Ssb (149, 194). However, aminoglycoside sensitivity associated with the deletion of SSB1/2 or the RAC appears to be associated at least in part with a general hypersensitivity to cations (226). These phenotypes, while useful proxies for Ssb/RAC functions, must therefore be interpreted with caution with regard to physiological roles in translation. The overexpression of the SSA and SSB genes does not complement each other's deletion phenotypes, suggesting that these two families of Hsp70 have evolved unique functions (202, 342). A study using fluorescence anisotropy to determine Ssb binding to polypeptides revealed that this Hsp70 does not generally bind unfolded substrates, characteristic of the Ssa family. Chimeric constructs of Ssa and Ssb functional domains showed that the Ssb NBD, but not the SBD, is necessary to rescue cold sensitivity, while the Ssb SBD, but not the NBD, is involved in mediating resistance to hygromycin B (342). These results suggest that the two domains play distinct roles in specific Ssb-mediated processes. At the time when these studies were undertaken, few Ssa or Ssb cofactors were known; it would be informative to revisit this analysis by defining cochaperone interactions with the chimeras compared to the endogenous proteins. For example, is the interaction with the RAC maintained in all constructs or only those with the Ssb NBD?
To better understand how Ssb and the RAC affect translation, translation fidelity was monitored by using a luciferase reporter construct in the presence or absence of a functional Ssb1/2 or RAC (355). That study revealed that the translation defects that occurred were due to faulty translation termination rather than misincorporation, which was less compromised in mutant strains. This phenotype was enhanced by paromomycin treatment or reduced levels of the translation termination factor Sup35. Ssz1 is an atypical Hsp70 that was originally thought to act as a scaffold between Ssb and Zuo1 but was recently shown to function independently of the RAC (69). That study also showed that neither its ability to bind substrate nor its ability to hydrolyze ATP was necessary for function, as assayed by growth phenotypes. The Zuo1-binding and nucleotide-binding domains could be eliminated individually, but Ssz1 is nonfunctional if both domains are disrupted in cis, suggesting that at least one intact domain is necessary for function. Independent of its roles in the ribosome, Ssz1 has been shown to activate Pdr1, a transcription factor associated with the induction of genes involved in the efflux of cytotoxic compounds, the stress response, lipid metabolism, and the ER-associated degradation (ERAD) pathway (33). Ssb is thought to be involved primarily in cotranslational protein folding and the promotion of nascent-chain movement through the exit tunnel. This behavior is shared by the structurally unrelated Escherichia coli protein trigger factor (TF). In recent studies, TF was shown to partially suppress the aminoglycoside sensitivity, but not general growth defects, of strains lacking either Ssb or RAC proteins. This finding suggests that the activity of TF-mediated protein folding at the ribosome exit tunnel is the Ssb function associated with aminoglycoside tolerance, but cold sensitivity and other growth defects may reflect distinct Ssb functions (234, 359). Alternatively, TF may not interact with some or all of Ssb's client substrates.
What other roles does Ssb play in vivo? A study in 2009 investigated Ssb's role in the glucose-sensing pathway mediated by Snf1 kinase (487). In the presence of glucose, hyperphosphorylated Snf1 is dephosphorylated by the protein phosphatase Glc7, rendering it inactive. Reg1 is required to target Glc7 to Snf1, and it was shown by using a two-hybrid assay that Ssb interacts with the Reg1 protein (104). In addition, the hyperphosphorylation of Snf1 in the presence or absence of glucose in the reg1Δ strain is suppressed by the overexpression of Ssb. These data suggest that Ssb plays an important role in glucose sensing through the regulation of Snf1, which is also required for the tolerance of toxic cations and the activation of cation transporters. This mechanism could explain the observed pleiotropic cation sensitivity of cells lacking Ssb1/2 (349). Two possible models to account for the observed interaction between Ssb and the SNF pathway have been proposed (337). The first model is that Ssb has a Km for ATP binding approximately 1,000 times higher than that of Ssa; in addition, the ATP hydrolysis rate is 50 times higher, which, taken together, suggest that Ssb might sense ATP levels to influence this pathway. Alternatively, both Ssb and Snf1 interact with and regulate Hsf1, raising the possibility that both proteins converge on a common target during glucose starvation.
The Hsp70 catalytic cycle is regulated at two major nodes: the rate of ATP hydrolysis and the rate of exchange of ADP for ATP. Correspondingly, there are two main groups of Hsp70 cochaperones: the Hsp40s (J-domain-containing proteins, or J proteins), and the nucleotide exchange factors (NEFs). These cofactors provide avenues for both specificity and variability in Hsp70 function and are thus analogous to the well-known cognate regulatory factors for G proteins, which likewise operate by a nucleotide cycle: J proteins mimic the function of GTPase-activating proteins (GAPs), and the Hsp70 NEFs perform the same functions as the well-characterized GEFs.
J proteins.
J-domain proteins (J proteins) (also called Hsp40 due to the apparent molecular masses of the most abundant yeast and mammalian homologs) are so called due to homology with the family archetype DnaJ from E. coli and the ability to accelerate the ATPase activity of Hsp70 ((161; reviewed in reference 60). The J-domain module is a four-helix bundle approximately 70 amino acids in length and contains the Hsp70 interaction site, an invariable histidine-proline-aspartic acid (HPD) motif, between helices II and III (470). Although it is not entirely clear how J proteins activate Hsp70 ATPase activity, at least one cocrystal structure suggests that the HPD motif inserts near the base of the ATPase domain, accelerating the conformational change necessary for ATP hydrolysis and the subsequent closure of the Hsp70 α-helical lid domain (206). Additional domain elements in various J proteins have prompted their classification into distinct structural groups (see reference 221 for an excellent recent review). Thirteen cytosolic J proteins have been identified in S. cerevisiae, which can be reclassified into four major groups based on their known functional characteristics: promiscuous client binding, which includes Ydj1, Sis1, Caj1, Djp1, Xdj1, and Apj1; selective client binding, which includes Jjj1, Jjj3, Cwc23, and Swa2; client binding unclear, which includes Jjj2; and no client binding, which includes Zuo1 and Hlj1 (380). Ydj1 is the most well-studied yeast J protein and consists of four functional modules: the J domain, a Gly-Phe-rich region, two carboxy-terminal domains (CTDs) that incorporate a zinc finger-like module, and a dimerization domain (50). In addition, Ydj1 is farnesylated at the extreme C terminus. The latter modification is essential for full function in vivo and localizes a subpopulation of Ydj1 to the ER membrane (51, 134). The CTDs function in substrate binding, with the Zn finger domains likely playing a modulatory role in substrate transfer to Hsp70 (450). The deletion of YDJ1 results in slow-growing and stress-sensitive cells, while the deletion of another major cytosolic J protein, SIS1, is lethal (50, 529). The Sis1 protein also contains a CTD substrate-binding domain, and while the deletion of the CTDs from either Ydj1 or Sis1 is tolerated, the loss of both is lethal, underscoring the biological relevance of substrate interactions by these Hsp70 cofactors (209). Ydj1 specifically partners with Ssa to promote the activity of Hsp90-dependent clients (see below). Interestingly, ydj1Δ cells paradoxically derepress heterologously expressed steroid hormone receptors, attributed to a negative regulatory function for this class of ligand-activated transcription factors (210). Ydj1 and Sis1 differentially recognize and interact with the various yeast prions described in the last decade, and the interested reader is referred to a recent review for a full discussion (450). A comprehensive analysis of the level of redundancy and functional overlap in the yeast cytosolic J proteins revealed that the J-domain fragments of many of the other cytosolic J proteins were sufficient to replace YDJ1 in vivo, suggesting that this minimal region is sufficient to carry out the “general” roles of this J protein (380). It is important to note that with the exception of Zuo1, described above, Ydj1 and Sis1 are the most highly expressed J proteins, at approximately 105 molecules/cell; the remaining 10 J proteins have expression levels that range from 102 to 104 molecules/cell (152). It is therefore not surprising that ydj1Δ and sis1Δ cells exhibit strong phenotypes, while many of the other J proteins do not.
A subset of cytosolic J proteins appear to play highly specialized cellular roles, and consistently, these mutants cannot be suppressed simply by the overexpression of the generalist YDJ1. Cwc23 was recently shown to be required for pre-mRNA splicing, possibly at the step of spliceosome disassembly (380). Unexpectedly, the Cwc23 J domain is not required for these in vivo functions despite retaining its Hsp70-stimulating capacity. Cells lacking the Djp1 J protein are defective in only one demonstrated function, peroxisomal protein import (183). The ER membrane-localized Hlj1 is involved in the degradation of ER proteins but is functionally redundant with Ydj1 (524). The JJJ1 gene encodes a J protein dedicated to the biogenesis of the large ribosomal subunit, and the null mutant exhibits cold sensitivity in keeping with translation defects (291). Jjj1 operates in conjunction with Zuo1 to assist in the maturation of both ribosomal protein and RNA components, and overexpressed JJJ1 recruits Ssa to the ribosome, effectively duplicating the Zuo1-Ssb chaperone machinery and suppressing mutations in both components. Lastly, Swa2 (also known as Aux1 or auxilin) is a dedicated adaptor for recruiting Ssa to clathrin-coated vesicles, promoting their uncoating (142, 517). In addition to its J domain, Swa2 contains a TPR module that works in tandem to enhance Ssa localization. Little to nothing is known about the remaining J proteins, highlighting the need for further genetic, cell biological, and biochemical analyses to understand the breadth of J-protein functions in the eukaryotic cytosol.
Nucleotide exchange factors.
The NEFs, unlike the J proteins, are a group of structurally unrelated proteins that promote ADP release from the NBD of Hsp70, resetting the cycle for another round of ATP binding and hydrolysis. The canonical NEF for E. coli DnaK is the protein GrpE, homologs of which have not been detected in any eukaryotic cytosol (a mitochondrial GrpE homolog exists [see below]) (170, 238). This prompted the supposition that eukaryotic cytosolic Hsp70s did not utilize a NEF as part of the catalytic cycle until the discovery of three distinct types in the early 2000s with both yeast and human/mammalian counterparts: the Hsp110s (Sse1/2), HspBP1 (Fes1), and Bag domain proteins (Snl1). The Hsp110 family consists of Sse1 and Sse2 in S. cerevisiae. These proteins were first isolated in a biochemical screen to identify calmodulin-binding proteins and were recognized as being Hsp70-related proteins due to a high level of amino acid similarity (309). Sse1 and Sse2 are 76% identical to each other and 70% similar to Ssa1, making them relatively distant Hsp70 relatives. While both are expressed under normal conditions, Sse2 is 10 times less abundant than Sse1 (152). SSE2 gene expression is highly induced in response to a wide range of stresses, while SSE1 levels increase only modestly (147). The deletion of SSE1 results in a slow-growth phenotype exacerbated by temperature stress, while in contrast, the loss of SSE2 is not associated with any phenotypic effects (309). The simultaneous disruption of both genes is lethal, although one group reported the construction of a double deletion that was viable under normal growth conditions (464, 521). An explanation for this discrepancy has not been put forward, but the gene pair is generally considered to be essential for growth. The domain architecture of Sse1/2 is similar to that of all Hsp70s, with the exception of an extended spacer region between the β-sandwich and α-helical bundle domains and variable extensions at the C terminus (116). A number of unusual features of the Sse proteins suggested that they might function differently than canonical Hsp70s. Sse1 was found to bind but not hydrolyze ATP, and mutations expected to abolish ATPase activity in the NBD had no effect on complementation (411). Sse1 was shown to be capable of holdase but not foldase activity, regardless of the nucleotide state (411). Finally, Sse1 and Sse2 were found to exist in stable heterodimeric complexes with both Ssa and Ssb, a behavior unknown for Hsp70s (412, 521). Multiple laboratories subsequently showed that the Sse proteins function as potent NEFs for both cytosolic Hsp70s, culminating in elegant cocrystal structures that depicted a novel binding interface between the NBDs of Sse1 and its partner Hsp70 (106, 348, 361, 402, 410). In addition, the α-helical lid domain of Sse1 exists in an extended conformation that wraps around the distal face of Hsp70 to make additional NBD contacts. This structure and additional genetic experiments support the interpretation that the Sse proteins may bind the substrate when present as a monomer but are incapable of doing so in the heterodimer (410). This has led to the hypothesis that Sse may recruit substrates or at least assist in substrate binding, followed by a “handoff” of the substrate to Hsp70 for folding, although this idea remains untested.
The Fes1 cochaperone was initially identified as a cytosolic homolog of the ER NEF Sls1 that interacts with the ER Hsp70 Kar2 (see below) but is also highly homologous to the mammalian NEF HspBP1 (214, 216). The deletion of FES1 results in a temperature-sensitive growth defect and genetically was shown to act antagonistically with Ydj1. Fes1 was shown to bind to and activate nucleotide exchange for Ssa in vivo and for both Ssa and Ssb in vitro (107). Snl1 is the only Bag-domain-containing protein identified in yeast and was genetically isolated in a multicopy suppressor screen for lethality caused by the expression of a truncation mutant of the nuclear pore protein Nup116 (186). It is unique among all known Hsp70 NEFs in containing an amino-terminal transmembrane region that tethers it to the ER membrane, with the Bag NEF/Hsp70-binding domain facing the cytosol (434). Snl1 also interacts with both Ssa and Ssb. What is the evidence that the NEFs are physiologically relevant for Hsp70 functions in vivo? The deletion of SSE1 results in the accumulation of untranslocated pre-pro-α-factor (ppαF), demonstrating that this NEF partners with Ssa1 to promote posttranslational translocation (412). Sse1 is involved in both cell wall integrity and morphogenesis based on the regulation of the MAP kinase Slt2, which is activated upon heat shock or under other cell wall-perturbing conditions. While the loss of SSE1 does not directly reduce the stability of Slt2 or its ability to be phosphorylated, transcriptional activation by the downstream target Rlm1 is diminished (409). The contribution of Sse1 to Hsp70-mediated folding has been documented by several studies. Sse1 is required for the efficient flux of nascent substrates through the Ssb and Ssa systems (521). Another study used FFL as a model substrate to show that Sse1 is required for efficient de novo folding in vivo and the refolding of denatured protein in vitro (106). Sse1 is also strongly implicated in protein degradation and appears to be required for ubiquitination and the subsequent proteasomal degradation of Hsp70-bound substrates (181, 276). This role is manifest in the somewhat paradoxical stabilization of query proteins in sse1Δ mutants, which would be expected to be deficient in folding by virtue of their pro-Hsp70 cycling activity. In this scenario, it is postulated that Sse1 must assist Hsp70 in transitioning to a low-affinity binding state to allow for substrate recognition by associated ubiquitin ligases. Very little effort has been invested in an understanding of the cellular roles of Fes1 or Snl1, and the elimination of the latter NEF appears to be phenotypically inconsequential. Although dramatic structural differences in the binding mechanisms and mechanics of nucleotide exchange distinguish the three NEFs, they all perform essentially the same biochemical function, making it a puzzle why three distinct families of cytosolic NEFs have been conserved in eukaryote evolution (88). By analogy to the J-protein cofactors, it is possible that the NEFs may confer target or cellular process-based specificity to the promiscuous Hsp70s. At present, the Sse proteins appear to be the dominant NEFs, consistent with their essential nature and cellular abundance. However, since fes1Δ cells exhibit moderate to severe phenotypes, the Sse proteins are clearly unable to perform all the NEF roles in the cell. Sse1 has also been shown to play a role in prion propagation, which cannot be replaced by Fes1, suggesting that the Hsp110 family may also possess Hsp70-independent activities (379). A comprehensive analysis of the shared and unique roles of the cytosolic NEFs is required and is under way (J. Abrams, J. Verghese, and K. A. Morano, unpublished data).
The Hsp90 Chaperone System
Hsp90 is an evolutionarily conserved molecular chaperone unique in both function and client protein profile (see reference 208 for a recent review). In contrast to the Hsp70 chaperones, which recognize unfolded or misfolded proteins indiscriminately, Hsp90 functions primarily in the “final” maturation of proteins and the assembly of complex macromolecular structures. It also functionally interacts with a much more select group of substrates, termed “client” proteins, which include many kinases and transcription factors. Hsp90 is composed of an N-terminal domain comprising the ATP-binding pocket, which is attached to a middle domain (M domain) by a charged linker region (167, 350). The C terminus contains a dimerization region followed by the residues EEVD, which, like the identical sequence in Hsp70, binds tetratricopeptide (TPR) repeats in cochaperone proteins (133, 390). The charged linker region is important for providing flexibility to Hsp90, allowing the conformational changes necessary for cochaperone interactions and ATPase activity. In addition, this linker domain is important for communication between the N, M, and C domains (167). Hsp90 also functions in an ATP-dependent cycle that dictates and is in turn influenced by a complex network of cochaperone proteins. Biophysical studies have highlighted the existence of multiple distinct conformations at different points in the chaperone cycle. Starting with an apo, or open-dimer, conformation, Hsp90 binds ATP, causing the N-terminal lid domain to fold over, locking the nucleotide into a kinked binding pocket. The closely apposed N-terminal domains then dimerize, leading to the twisting and compaction of the M domain, activating the ATPase activity of the protein. Finally, once Hsp90 is in its closed state, ATP hydrolysis is completed, ADP is released, and the protein returns to the open conformation (182). In yeast, two genes, HSC82 and HSP82, which are expressed constitutively and inducibly upon heat shock, respectively, encode Hsp90 (32). Hsp90 is also regulated posttranslationally through modifications, including phosphorylation, acetylation, and S-nitrosylation (see reference 302). Despite this wealth of biochemical and biophysical data, precisely how Hsp90 promotes the maturation of client proteins and the mechanism behind its ability to chaperone a diverse set of substrates while retaining selectivity remain unknown.
Hsp90 cochaperones.
A growing number of cochaperones play important roles in regulating the Hsp90 cycle and providing specificity for client proteins. An intricate and coordinated dance between the cochaperones promotes the transition between functional Hsp90 states required for substrate maturation (Fig. 5). An early-acting cochaperone is Sti1, homologous to the mammalian protein HOP (Hsp90/Hsp70-organizing protein), which plays at least two roles in Hsp90 complex functions (207). First, it is an ATPase regulator that binds to the EEVD sequence in the C terminus and also to either the N or M domain of Hsp90 in the open conformation. Because it is such a strong inhibitor of ATPase activity, only one molecule is necessary per dimer of Hsp90 to completely inhibit N-terminal dimerization; this allows for an asymmetric assembly of Hsp90, Sti1, and other TPR-containing cofactors, which is an important step in cycle progression (253, 366). The second critical activity is the ability to simultaneously bind both Hsp90 and Hsp70 through distinct TPR sites. It is currently thought that Hsp70 and its cofactors bind most Hsp90 client proteins first, assembling a “prefolding” complex. Client-loaded Hsp70 is then brought into close proximity with an Hsp90 dimer through binding to Sti1/HOP, facilitating substrate transfer followed by the release of Hsp70 (497). The ability of Sti1/HOP to act as a strong noncompetitive inhibitor of Hsp90 ATPase activity has also been shown to be important for client transfer (207). Two additional cochaperones modulate Hsp90's weak intrinsic ATPase activity to govern client maturation. Aha1 binds Hsp90 in the M domain, causing a rearrangement of the catalytic loop and allowing it to contact ATP within the N-terminal pocket (292). It is a potent activator of ATPase activity, yielding a 12-fold increase in in vitro ATPase assays with yeast-derived proteins (329). It was suggested that Aha1 may act as a general ATPase activator independent of the stage of the Hsp90 cycle because it exists in both early and late cochaperone complexes. More recent studies support a model wherein Aha1 acts primarily in the earlier stages of the Hsp90 cycle to remodel the protein, favoring N-domain dimerization and ATP hydrolysis (182). The cochaperone Sba1 (p23) both stabilizes ATP binding by Hsp90 and acts as an ATPase inhibitor (127). The interaction of Sba1 with Hsp90 is indirectly dependent on ATP binding because the cochaperone selectively binds the closed, N-terminally dimerized conformation (7, 211). The association of Sba1 with the ATPase domain stabilizes the closed conformation, preventing ATP hydrolysis, and may also play a role in preventing Hsp90 inhibition by natural products such as radicicol and the ansamycins (geldanamycin and macbecin), all of which act through ATP displacement from the nucleotide pocket (135). Sba1 is itself a molecular chaperone and has been shown to regulate telomerase activity independently of Hsp90 (459). A recent high-throughput proteomic study uncovered a wealth of cellular targets and processes for Sba1, many of which do not overlap those of Hsp90, underscoring the idea that this chaperone and cofactor plays a much broader and unappreciated role in cell biology (117). The immunophilin homologs Cpr7 and Cpr6 (heat inducible) are two of many additional TPR-containing proteins and bind the C-terminal EEVD domain of Hsp90 in the closed conformation, thus relieving the ATPase inhibition imparted by Sba1 (112, 211). PPT1, encoding a protein phosphatase, is another TPR-containing cochaperone with a regulatory role that helps promote Hsp90 activity (495).
Fig 5.
The Hsp90 folding cycle. Yeast proteins participating in the Hsp90 folding cycle are indicated. The complexes depicted are from known yeast protein interactions or inferred from in vitro reconstitution experiments with metazoan counterparts, as described in the text. Unfolded client proteins are indicated by the wavy blue line, and the native folded state is labeled. Kinase clients are thought to mature through a Cdc37-specific pathway (kinases), while nearly all other clients proceed through the multichaperone pathway (nonkinases). The cyclophilin homolog Cpr7 (also Cpr6 [see the text]) is a TPR domain-containing protein that competes for binding with other TPR cofactors, including the phosphatase Ppt1, shown by the dashed line.
The kinase chaperone Cdc37.
CDC37 is an essential gene that encodes a protein best described as an adaptor; the N-terminal domain associates with the catalytic domains of protein kinases, while the C terminus binds between the two N-terminal domains of Hsp90, blocking dimerization (Fig. 5) (368, 413, 414, 527). Preventing structural rearrangements is likely how Cdc37 decreases ATP turnover and assists in substrate loading (425, 494). In many cellular pathways, including the high-osmolarity glycerol (HOG) pathway and the cell wall integrity (protein kinase C) MAPK pathway, Cdc37 and Hsp90 collaborate to maintain active levels of Hog1 and Slt2, respectively (180). A mutational analysis showed that in a cdc37-S14A mutant strain in which Cdc37 could not be phosphorylated, the interaction with Hsp90 was severely decreased. In addition, in this genetic background, Hog1 was destabilized during osmotic stress, and the Slt2 activation of downstream targets was decreased. In a screen of the yeast kinome, 75% of kinases were shown to be functionally dependent on Cdc37, demonstrating the breadth and impact of this chaperone (277). Cdc37 is also capable of chaperoning some client protein kinases independently of Hsp90: the kinase-binding domain of Cdc37 is sufficient for cell viability and MAP kinase signaling in sti1Δ and hsc82Δ strains that are severely compromised for Hsp90 function (245). Cdc37 also plays a small role in the function of nonkinase clients, as demonstrated by defects in the activation of the androgen receptor expressed in yeast cells lacking this chaperone (357).
Targets of the Hsp90 chaperone system.
While many studies have exploited S. cerevisiae as a model system to determine the features and players required for the maturation of mammalian Hsp90 client proteins, such as steroid receptors and kinases like v-Src, few endogenous Hsp90 clients have been identified or characterized. Some of the yeast clients of Hsp90 that have been investigated are the kinases Ste11 and Gcn2 and the transcription factors HapI, Mal63, and Hsf1. Ste11 functions in the yeast pheromone signaling pathway as a MAP kinase kinase kinase, analogous to its mammalian counterpart Raf (164). By using a collection of Hsp90 mutants and an Ste11 constitutive mutant, it was revealed that maintaining the levels of Ste11 necessary to elicit cell cycle arrest upon pheromone exposure required both Hsp90 and Cdc37, an Hsp90 cochaperone (2, 268). The relatively low abundance of many client kinases has posed a challenge for studies of chaperone-substrate interactions. Recently, a more tractable version of Ste11, Ste11ΔN-K444R, was constructed. The deletion of the amino-terminal regulatory domain eliminates the pheromone dependence of the kinase, and the substitution of the catalytic lysine renders the kinase all but inactive, allowing overexpression without subsequent cell cycle arrest due to the phosphorylation of the target Far1 (47, 134). By using this allele, Ydj1, and, specifically, the farnesylated population, was shown to be crucial for Ste11 maturation (321). In addition, roles of the Hsp70 system, including the Hsp110 NEFs, in mediating client degradation upon misfolding caused by the pharmacological inhibition of transfer to Hsp90 were revealed by using this key reagent (276). A global analysis of the protein and lipid kinase reliance on the Hsp90/Cdc37 system for function demonstrated that a remarkable 51 of 65 kinases examined were destabilized in a cdc37 mutant strain (277). However, to date, the molecular and/or structural determinants that confer kinase reliance on or independence of chaperones have not been elucidated. This distinction may be subtle, as the mammalian kinase v-src is absolutely dependent on the Hsp90 system when expressed in yeast, yet the close homolog c-src is nearly independent of the chaperone function (520).
HapI is a heme-responsive transcription factor involved in the control of respiratory and oxidative damage genes and is essential under anaerobic growth conditions, regulating over 200 genes (85, 341). Several studies have examined the Hsp90-dependent regulation of HapI, which also involves the Hsp70 Ssa1 and the cochaperones Ydj1 and Sro9. Those studies revealed that in vivo, HapI is always associated with Ssa1 independently of heme interactions, including when it was bound to its own promoter (526). At low heme levels, Ssa1 and Ydj repress HapI, but in the presence of heme, Hsp90 activates HapI (189, 239). In a similar regulatory pathway, Mal63, the maltose-responsive transcription activator, was found to require the Hsp90 system. Mal63 is maintained in its uninduced form by Ssa1 and Sti1; once it is induced in the presence of maltose, it binds to Hsp90, which stabilizes it for transcription activation (13, 356).
The search for additional Hsp90 client proteins continues. In the absence of a clear mechanistic explanation for Hsp90's chaperoning capabilities, a deeper understanding of the range and breadth of endogenous substrates may provide empirical insight. In addition, given the interest in the pharmacological inhibition of Hsp90 as an anticancer therapy, a reliable catalog of known targets in human cells is essential and may be aided by achieving the same goal first in budding yeast. Two genomewide studies have been undertaken to attempt this, in both cases uncovering novel requirements for Hsp90 (cell cycle and vesicular transport [287]) and cofactors (Tah1 and Pih1 [528]). An interpretation of results gleaned from such studies must be carefully done, as Hsp90 interactions inferred from independent two-hybrid, affinity purification, synthetic lethal, and chemical-genetic approaches exhibit remarkably low levels of overlap. However, these functional genomic investigations are likely the only way to generate a comprehensive map of chaperone-client relationships.
Hsp104
Many stress conditions cause protein misfolding, and at high levels, this can lead to aggregation and cell death. The protein chaperone Hsp104 has the unique (in eukaryotes) capability of recognizing misfolded proteins within an aggregate and actively unfolding them, ultimately disassembling the insoluble structure and delivering substrates into refolding pathways (Fig. 6). The so-called Hsp100 family of HSPs includes the bacterial Clp proteins and their yeast homologs Hsp104 and mitochondrial Pim1 protease (393). Although Hsp104 is conserved in fungi and plants, a homolog in metazoans has not been identified. The Hsp104/Clp family is a subgroup of the AAA+ ATPase superfamily, characterized by a ∼200- to 250-amino-acid core composed of an α-helical domain and a Walker-type nucleotide-binding domain (105, 123). AAA+ proteins function as oligomers, wherein ATP hydrolysis allows conformational changes to occur between the AAA+ subdomains in order for the hexameric ring structures to perform processive mechanical work (502). Hsp104 monomers are composed of three functionally distinct regions: the N-terminal, middle (M), and C-terminal domains (335). The N-terminal domain spans residues 1 to 163 and is involved in the initial substrate interaction, possibly limiting substrate access to the inner cavity (269). The M domain, which distinguishes Hsp104 from ClpB, spans residues 412 to 532 and contains coiled-coil structures resembling a two-bladed propeller (501). The C-terminal domain spans residues 871 to 908 and plays roles in substrate binding, oligomerization, and substrate exit; it also contains TPR domains that are likely to play a role in coordination with Hsp70 (57, 274). Both NBDs bind ATP, which stabilizes oligomers and promotes substrate interactions, while ATP hydrolysis allows the restructuring of the domains to allow substrate movement and release (501). When Hsp104 assembles into the homohexameric state, the M domains face the outside of the structure. Recent studies have shown that this domain is required for the interaction with Hsp70, provides substrate-binding sites, and could be involved in allosteric communication between the two NBDs (247, 423).
Fig 6.
The cytosolic disaggregation and refolding machinery. The native protein is shown to be unfolded by heat shock (depicted as a salmon rectangle), which also causes changes in the sHsp oligomerization status. The constitutive Ssa Hsp70 chaperones partner with the J protein Ydj1 and at least one nucleotide exchange factor (NEF) to promote the refolding of disaggregated (Hsp104 pathway) or unfolded but protected (Hsp42 and Hsp26) proteins.
Hsp104 is a highly heat-inducible, nonessential protein in yeast, and deletion under optimal growth conditions does not impact growth (258, 384). However, Hsp104 is required for thermotolerance, and the deletion of HSP104 reduces cell survival 100- to 1,000-fold (384). In fact, increased levels of Hsp104 alone are sufficient to promote survival during lethal heat shock (258). Hsp104 is unique among known protein chaperones in its ability to pull protein aggregates apart, leading to its characterization as a “disaggregase” (Fig. 6). Studies using heat-denatured bacterial luciferase in the presence or absence of Hsp104 revealed that this protein is required for the reactivation of luciferase through resolubilization (335). How are disaggregated proteins restored to functional competency? Hsp104 partners with Hsp70 and Hsp40, which work together to resolubilize and refold substrates. In vitro analyses revealed that both Ssa1 and Ydj1 were necessary, along with Hsp104, to recover 50% of the wild-type activity of FFL after chemical denaturation (156). Hsp104 was also shown to localize within the insoluble cellular fraction after heat shock, along with Hsp26 and Hsp70, where it is likely associated with aggregated substrates (177). In addition, it was shown by using heat-denatured citrate synthase and FFL that both Hsp70 and Hsp104 are necessary to release the substrate from the small heat shock protein Hsp26 (156). This finding suggests that Hsp26 interacts with the protein aggregate, making it accessible to Hsp104. The activity of this small Hsp (sHsp) is described in more detail below. In addition to the Hsp70/Hsp40 chaperone system, the Hsp90 cochaperones Sti1, Cpr7, and Cns1 have been shown to interact with Hsp104 in a manner regulated by growth conditions. These proteins bind Hsp104 when cells are grown on nonfermentable carbon sources, and in a strain where Hsp90 has been truncated to remove its TPR-binding domain, the cochaperones bind to Hsp104 independent of the sugar source (1). This finding suggests competition for cochaperone binding between Hsp90 and Hsp104, which may be physiologically relevant, but to date, no functional significance has been ascribed to this interaction. A genomewide screen of S. cerevisiae to identify genes required for aggregation clearance identified SSD1, a gene known to be involved in cellular integrity pathways. Ssd1 was found to influence Hsp104 hexamerization, interactions with Sti1, and the ability to bind aggregates (296). How Ssd1 exerts control over Hsp104 functions remains an open question. Hsp104 is also a key player in prion inheritance. Hsp70 was also shown to be involved in Hsp104-mediated prion propagation and curing; although the exact role is unclear, it was proposed that the ratio between Hsp70 and Hsp104 could decide whether prions are cured or propagated. This topic has been reviewed extensively, and the interested reader is directed toward these detailed treatments (103, 179, 316).
Small Hsps and Hsp12
sHsps play a vital role in promoting protein solubility when heat or other stresses lead to general cytosolic protein unfolding (200). sHsps, such as Hsp26 and Hsp42, bind unfolded proteins, preventing their aggregation (56). Other sHsps that are less studied, such as Hsp31, have not been well characterized but also appear to play a role in stress tolerance (344, 345, 406). Together, these chaperones provide an additional layer of protection against cellular assaults.
Hsp26.
Hsp26 exists in two major forms in vivo: a large multimer composed of 24 monomers under normal conditions, which dissociates at heat shock temperatures into dimers in a reversible process (Fig. 6) (178). The dimeric form associates with unfolded polypeptides with a stoichiometry of 1 substrate molecule per dimer of Hsp26; these small units ultimately form larger, ordered complexes containing the substrate and chaperone (448). In addition, variability in the size of the oligomers and the stoichiometry of the active form distinguishes these proteins from one another (92). In the case of Hsp26, the oligomeric structure is formed with monomers containing two discrete domains. The N-terminal domain, mostly α-helical in nature, appears to be important for forming the 24-mer and allowing temperature-dependent activation by dissociation at temperatures between 29°C and 43°C. The C-terminal domain is rich in β-sheet structures and is important for the stable formation of the dimeric species, which is inactive in the absence of the N-terminal domain (176). Hsp26 shares significant sequence homology with α-crystallin, a major eye lens protein, and, like this protein and other sHsps, is found as an oligomer in its active state (447). In all organisms studied, sHsps share a conserved C-terminal domain but vary in the N-terminal domain, which ranges from 12 to 40 kDa, making them the most divergent class of chaperones (178, 447). In S. cerevisiae, HSP26 is nonessential and exhibits no phenotypes upon deletion (178). However, the simultaneous deletion of another sHsp, HSP42, results in a 200% increase in levels of insoluble proteins at 30°C compared to levels in wild-type cells (176). The lack of a nucleotide cycle is consistent with a model wherein the sHsps function as energy-independent “holdases” for nonnative proteins until they can be transferred to a chaperone capable of refolding, such as Hsp70. Based on work done with the model protein citrate synthase, binding to Hsp26 results in the stabilization of the client protein at heat shock temperatures and diminishes the thermal inactivation of the protein (178). Consistently, Hsp26 is more effective at maintaining the solubility of proteins at heat shock temperatures than at normal temperature (176). No substrate specificity has been identified for any member of the sHsp family, yet the oligomeric complexes take on a variable size and shape depending on the substrate. Mixed complexes with multiple substrates can be formed, and in this case, the overall shape is determined by the first substrate integrated within the complex (447). It is likely that surface exposure, oligomeric orientation, and shape are optimized to provide maximal protection for the substrate while allowing access to Hsp70 for eventual refolding.
Hsp42.
The Hsp42 monomer is ∼43 kDa and forms 12- to 16-mers at lower concentrations and 24- to 26-mers at higher concentrations. This chaperone has not been extensively studied, and the majority of what is known regarding its function is derived from a few reports (175). Unlike the globular spheres formed by Hsp26, the oligomer is a symmetric assembly of dimers that is ultimately organized into two hexameric rings (Fig. 6). The Hsp42 protein shares homology within the conserved C-terminal α-crystallin domain but possesses an unusually long N-terminal domain bearing no sequence similarity to other sHsps. Both Hsp26 and Hsp42 are poorly expressed during exponential growth, and at heat shock temperatures, there is 10 times more Hsp42 than Hsp26, suggesting that it may be the dominant sHsp in the thermally stressed cell (175). Remarkably, together, these two proteins comprise approximately 1% of cellular proteins at heat shock temperatures. Unlike Hsp26, the enzymatic activity of Hsp42 is not temperature regulated. Strains lacking Hsp42, but not Hsp26, accumulate aggregated proteins in the stationary phase, while heat shock results in aggregates in both strains with single deletions and a strain deleted for both sHsps (175). These results support the notion that Hsp42 is the more potent chaperone of the two. A proteomic study showed that Hsp42 promiscuously binds 30% of yeast cytosolic nonnative proteins, with a remarkable 90% overlap with Hsp26 substrates (174). However, the efficiency with which Hsp42 maintains solubility and activity is substrate dependent. Hsp42 may also be a more effective chaperone than Hsp26, as higher ratios of Hsp26 to substrate are needed to prevent aggregation (515).
Hsp12.
Hsp12 exhibits low sequence homology to the sHsp superfamily and is structurally and functionally distinct, as it appears to exist exclusively as a monomer (499). Like the sHsps, Hsp12 is weakly expressed in exponentially growing cells but highly induced (100-fold) during the stationary phase or heat shock; in the stationary phase, Hsp12 was determined to comprise 2.4% (740,00 molecules) of the total cellular proteins, on par with Hsp90, which comprises 1.3% (420,000 molecules) at the stationary phase (499). Hsp12 is not essential for growth under normal or stress conditions but may play a role in barotolerance (protection against desiccation) (308, 382). Hsp12 has been localized to both the cytosol and cellular membranes. In keeping with this observation, recent studies have revealed that Hsp12 functions in stabilizing membranes under stress conditions by modulating fluidity (499). Interestingly, Hsp12 is unfolded in solution, but in the presence of lipids or lipid-like proteins, it takes on a helical structure essential for membrane interactions (499). These characteristics make it a unique stress factor that may in fact function more as a membrane chaperone than as a protein chaperone.
Chaperonins
Another relatively less-studied group of chaperones in S. cerevisiae is the chaperonins. Like the above-described chaperones, these proteins are responsible for protection and folding of unfolded or partially unfolded proteins. They are also known to interact with other families of chaperones to fold specific substrates. The chaperonins form double-ring structures and fold proteins within a central cavity in a nucleotide-dependent manner. There are two main classes of chaperonins that have been conserved throughout evolution, groups I and II. Group I includes E. coli GroEL, which is conserved among prokaryotes and endosymbiotic eukaryotic organelles. Group II chaperonins are found in eukaryotes and archaea. The overall structures of the two groups are similar, with two major differences. Group I chaperonins are homomultimers, while there is a hetero-oligomeric assembly in group II. The second difference is in the lid structure, which in both groups closes over the central cavity to encapsulate the substrate (171). In group I, the lid is a separate protein called GroES, while in group II, the lid is an attached flexible extension from each subunit that, once assembled, creates an iris-like structure that can close over the central cavity (53, 474). In yeast, Tcp1 ring complex (TriC) or chaperonin-containing TCP1 (CCT) forms a large cylindrical 900-kDa oligomer composed of a double-ring structure. Each heteromeric ring contains eight orthologous 60-kDa subunits surrounding the cavity where substrates are folded (141, 474). All eight subunits, Cct1 to Cct8, are essential and expressed constitutively under normal conditions. The subunits share 30 to 35% sequence identity, with the highest level of conservation within the ATPase domain. In contrast, the substrate-binding domains are divergent (445). An analysis of a temperature-sensitive mutation in a single subunit (cct4-1) revealed defects in actin filament and tubulin assembly, as indicated by hypersensitivity to the antimicrotubule drug benomyl at the nonpermissive temperature (290, 479). In a more recent study, it was discovered that this mutant displays reduced binding to ATP and that both intra- and inter-ring cooperativity is abolished (419). It was initially thought that these proteins are not stress induced based on results examining transcript levels after heat shock (38°C), but at least one CCT transcript accumulates after cold shock. Additionally, CCT protein levels increase when cells are shifted from 4°C to 10°C, suggesting that CCT may be involved in cold shock recovery (433). This notion is also supported by a suppressor screen that found that the overexpression of ribosomal proteins suppresses CCT defects (217). Initially, it was thought that this chaperone is specialized for folding actin and tubulin, but recently, the interactome of TriC/CCT was analyzed bioinformatically and biochemically to determine how substrate specificity is decided (422). That study revealed that there is a much larger group of substrates, including G-α-transducin and WD repeat proteins, including Cdc20 and Cdh1. Both of these proteins require CCT to function and are involved in cell cycle control: Cdc20 promotes the shift from metaphase to anaphase, and Cdh1 promotes exit from mitosis through the activation of the anaphase-promoting complex or cyclosome (APC/C) (48). Common characteristics of CCT substrates were also uncovered, such as large regions of β-sheets or α-helices, which typically cause proteins to exhibit increased folding kinetics. Additionally, many of the substrates are complex multidomain proteins. Recent work from the laboratory of Dennis Thiele suggested that CCT could have an unexpected role in modulating the activity of the transcription factor Hsf1, but further investigation is necessary to determine the mechanism of this pathway (312).
Additional cofactors modulate CCT activity. Prefoldin/GimC forms a heterohexameric complex composed of six distinct structurally related proteins ranging from 12 to 23 kDa. In addition, prefoldin/GimC is known to act as a cochaperone involved in targeting substrates to CCT. It functions primarily in the binding and stabilizing of unfolded substrate proteins, including α- and β-tubulin, as evidenced by the finding that the deletion of prefoldin/GimC leads to microtubule defects (422). Recently, another cochaperone of CCT was discovered, phosducin-like protein 3 (PhLP3) (yeast homolog, Plp1). This protein binds to CCT, and the deletion of PLP1 rescues benomyl sensitivity in tubulin and prefoldin mutants. Plp1 acts as a negative modulator of CCT ATPase activity and thus may act antagonistically with prefoldin to regulate CCT folding (444).
CHAPERONES OF THE SECRETORY PATHWAY
Protein quality control is a vital aspect of cellular biology that maintains protein homeostasis (proteostasis) under normal and stress conditions (305). Under these conditions, molecular chaperones help to fold proteins and prevent their aggregation, while the cytosolic protein degradation machinery, including the proteasome, degrades damaged or misfolded proteins. Proteins residing in subcellular organelles like the ER and mitochondria are physically separated from the cytosol by phospholipid membranes and are thus shielded from the cytosolic protein quality control machinery. Almost all mitochondrial proteins and all ER and secretory proteins are synthesized in the cytosol and delivered to these compartments by various translocation pathways. The ER lumen has a unique folding environment compared to the cytosol, and proteins in the ER are subject to modifications such as the formation of disulfide bonds and the addition of preassembled oligosaccharides, both of which require their own repertoire of ER-specific molecular chaperones. Mitochondria possess dedicated chaperone machinery in the matrix to assist in the import and folding of protein substrates and specific proteases that degrade misfolded and damaged proteins. In addition, mitochondria and the ER both use the proteasome to degrade damaged proteins and therefore must engage in retrograde transport back to the cytoplasm. The expressions of many of these organellar chaperones are regulated by specific stress-responsive pathways: the unfolded protein response (UPR) for the ER and the so-called retrograde response for mitochondria. Due to space constraints, the reader is referred to recent reviews for detailed overviews of these pathways (23, 265). In eukaryotic cells, almost all secreted proteins enter the ER either during (cotranslational) or soon after (posttranslational) their synthesis. In mammalian cells, proteins are cotranslationally translocated and folded by the chaperone machinery in the ER. In yeast cells, an additional process is present, where select substrates are translocated posttranslationally. A critical step for many proteins that enter the ER is their posttranslational modification. The oxidizing environment in the ER lumen favors the enzyme-assisted formation of disulfide bonds that modify a protein structure. Proteins are also accessorized with N-linked glycans that attract carbohydrate-binding chaperones to increase their capacity to fold into native functional states. The ER of S. cerevisiae harbors three major groups of molecular chaperones and folding enzymes: (i) the heat shock protein (HSP) family of chaperones, which includes Kar2 (Hsp70) and its partners (Hsp40s and NEFs); (ii) the chaperone lectin and calnexin; and (iii) thiol oxidoreductases of the protein disulfide isomerase (PDI) family (Table 2 and Fig. 7).
Table 2.
Endoplasmic reticulum chaperones
| Class | Protein | Function(s) |
|---|---|---|
| Hsp100 | None | |
| Hsp90 | None | |
| Hsp90 cochaperone | None | |
| Hsp70 | ||
| GRP170 | Lhs1 | Kar2 nucleotide exchange, substrate binding |
| Hsp110 | None | |
| Hsp70 | Kar2 | Protein folding, translocation, UPR regulation, karyogamy |
| Hsp70 NEF | Sil1 | Kar2 nucleotide exchange |
| Hsp40/J protein | Sec63 | Kar2 ATPase activator, translocation, ER membrane |
| Scj1 | Kar2 ATPase activator | |
| Jem1 | Kar2 ATPase activator, karyogamy, ER membrane | |
| Chaperonin | None | |
| Chaperonin cochaperone | None | |
| sHSP | None | |
| Other | ||
| Calnexin | Cne1 | Folding of glycosylated proteins |
| Protein disulfide isomerase | Pdi1 | Protein folding, disulfide redox chemistry |
| Mpd1 | Protein folding, disulfide redox chemistry | |
| Mpd2 | Protein folding, disulfide redox chemistry | |
| Eug1 | Protein folding, disulfide redox chemistry? | |
| Eps1 | Protein folding, disulfide redox chemistry?, ER membrane |
Fig 7.
The ER chaperome. ER chaperones and associated cofactors are depicted, along with their respective roles in ER protein biogenesis. Gray 40S and 60S subunits depict docked ribosomes. S-S, disulfide bond; UPR, unfolded protein response.
ER Hsp70
The first Hsp70 family member localized in the ER, termed BiP (for binding protein), was identified in mammalian cells by its binding to immunoglobulin heavy-chain precursors and was later shown to be a member of the Hsp70 family (370). The yeast BiP homolog, termed Kar2 (karyogamy mutant), was unexpectedly discovered in a genetic screen for mutants defective in nuclear fusion during yeast cell mating. Kar2 is essential for yeast cell viability and has 67% identity and 84% similarity to mouse BiP (325). The expression of mouse BiP restored normal karyogamy and complemented the temperature-sensitive growth phenotypes of the kar2-1 yeast strain, suggesting conserved functional homology between the mammalian and yeast proteins (325). Early work established that Kar2 contained structural features similar to those of BiP: an N-terminal secretory sequence, a C-terminal ER retention signal (HDEL) (KDEL in BiP), and the absence of N-linked glycosylation sites. Like other members of the Hsp70 family, Kar2 has an N-terminal nucleotide-binding domain and a C-terminal substrate-binding domain. Three cis-acting elements in the promoter of KAR2 control its expression: (i) a high-GC-rich region that governs constitutive expression, (ii) a 20-bp functional heat shock element (HSE), and (iii) a 22-bp unfolded protein response (UPR) element (UPRE). The latter two elements induce transcription in response to elevated temperatures and the presence of unfolded proteins in the ER, respectively (304). These two elements are functionally independent of each other but additively are responsible for the maximal induction of the KAR2 gene (232). A major difference between the KAR2 and BIP promoters is the presence of the HSE in KAR2, which induces its transcription within 10 min after heat shock, whereas the gene encoding BiP is not heat shock inducible (325). As the only Hsp70 chaperone in the ER lumen, Kar2 plays major roles in a number of different processes, as detailed below.
Protein folding.
The Kar2 function was shown to be required for the folding of the well-characterized vacuolar glycoprotein carboxypeptidase Y (CPY) using three temperature-sensitive strains of KAR2, kar2-113, kar2-159, and kar2-203, that bear inactivating mutations in the N-terminal ATPase domain. These mutants also failed to transport CPY out of the ER to its final destination in the vacuole. This finding suggested that the Kar2 function was necessary for the folding and maturation of CPY. In addition, these mutants accumulated aggregates of CPY in the ER, suggesting that a primary function of Kar2 is to prevent the aggregation of partially folded proteins (428). It is likely that Kar2 interacts with many more substrate proteins as part of their normal biogenesis and certainly upon proteotoxic stress.
Translocation of proteins across the ER membrane.
As mentioned above, translocation into the yeast ER lumen occurs through two pathways. In the cotranslational translocation pathway, nascent polypeptides are targeted to the ER membrane during synthesis, while posttranslational translocation proceeds immediately after the polypeptide has been synthesized and released from the ribosome. Microsomes prepared from the kar2-159 temperature-sensitive mutant strain failed to translocate radiolabeled precursor proteins, ppαF and invertase, which are known substrates for posttranslational and cotranslational translocation, respectively, suggesting that Kar2 is required for both translocation pathways (37). This is also in agreement with previously reported evidence that the kar2-159 strain accumulates both substrate precursors at the nonpermissive temperature and that the depletion of Kar2 results in their cytosolic accumulation (482). Kar2 was shown to function in both the early and late stages of the translocation process and is recruited to the ER membrane by the J-domain protein Sec63 (70). Mutant alleles of KAR2 prevented an ER-targeted preprotein from associating with the Sec61 translocon complex, suggesting a role for Kar2 early in the translocation process. In addition, a decrease in the ability of Sec61 to be cross-linked to a secretory protein that was trapped in translocation in a KAR2 mutant strain is also consistent with a role for Kar2 later in the translocation process (385). How, then, does Kar2 drive the translocation of ER-targeted proteins? Two models have been proposed for this function of Kar2. In the “Brownian ratchet” model, the polypeptide in the translocation channel exhibits Brownian motion, but once enough of the polypeptide moves through the Sec61 translocon and enters the ER lumen, Kar2 binds in a Sec63-dependent manner. Binding will prevent the backward movement of the polypeptide, and since the forward movement into the lumen is thus favored, another Kar2 molecule will bind, and eventually, the entire polypeptide will translocate into the ER. This model fits the notion that Kar2 acts in some cases simply by binding to its substrate (283, 427). In the “translocation motor” model, Kar2, while simultaneously bound to the substrate and Sec63, pulls the polypeptide through the translocon into the ER by hydrolyzing ATP (155). This is proposed to occur by a conformational change in Kar2, which, by replacing ADP with ATP, resets back to its original non-substrate- and non-Sec63-binding state. Multiple iterative rounds of this action would effectively “pull” the polypeptide into the ER (155). While both models require that Kar2 bind the translocating substrate, the ratchet model is well supported by experimental data, while the translocation motor mechanism remains less well validated.
Retrograde transport of aberrant polypeptides from the ER into the cytosol for proteasomal degradation.
Most misfolded proteins are handled in the ER, although some of them escape the quality surveillance system and are transported to and degraded by the lysosome. Results from a number of groups using mutants and inhibitors of the proteasome showed that the degradation of misfolded ER proteins occurred in the cytosol. In order for this to occur, these proteins must first be retrotranslocated from the lumen or ER membrane back into the cytosol (Fig. 7). This pathway is now well established and is known as “ER-associated degradation” (ERAD) and contributes to ER proteostasis (288). Kar2 was shown to be required for the ERAD of a mutated carboxypeptidase Y allele, CPY* (a known substrate for ER degradation). In the kar2-113 mutant strain, a 2-fold increase in the stabilization of lumenal CPY* was seen over the wild type, indicating a defect in the delivery of CPY* to the cytosolic proteasome (346). Kar2 interacts with the lumenal face of Sec61 translocons not engaged in active translocation (those with ribosomes attached to the cytosolic face of the pore) to seal off the cytosol from the ER lumen, and this activity requires that Kar2 be in the nucleotide-bound state (168). How Kar2 maintains this seal while participating in its other ER protein biogenesis roles is yet to be determined.
Regulation of the UPR pathway.
The UPR pathway is activated when unfolded proteins accumulate in the ER, leading to the oligomerization of the transmembrane kinase Ire1 (408). Ire1 initiates the nonconventional splicing of HAC1 mRNA in the cytosol, converting it to its mature form, which is then translated to produce a functional transcription factor, Hac1. Hac1 efficiently induces the transcription of genes that contain one or more unfolded protein response elements (UPREs) in their promoters (76). KAR2 is one such gene and, hence, is induced when unfolded proteins accumulate in the ER lumen. Two important experiments support the notion that Kar2 is a critical modulator of the UPR via the regulation of Ire1. First, mutations in the substrate-binding domain of Kar2 that impaired binding to Ire1 were found to activate the UPR in the absence of ER stress, and second, mutations in the ATPase domain of Kar2 were shown to impair release of Kar2 from Ire1 in the presence of tunicamycin, an ER stressor, resulting in an inability to activate the UPR (227). An alternative model for UPR activation is that unfolded proteins directly stimulate Ire1 independent of its interaction with Kar2. In vivo and in vitro experiments to test this model showed that the core lumenal domain of Ire1 binds directly to the model substrate CPY* (144). In that same study, Ire1 was shown to exhibit peptide-binding preferences similar to those of Kar2, suggesting that these two proteins may compete for binding to unfolded substrate proteins, resulting in UPR activation. Clearly, unfolded polypeptides are competent to bind Ire1 and promote its activation by enhancing oligomerization, relegating Kar2 to a modulatory role. The UPR, along with ERAD, provides a different means of dealing with ER stress caused by protein misfolding in the ER. Two concomitant studies showed that these two pathways are linked. CPY* is stabilized in UPR-deficient cells, suggesting that the UPR is involved in the clearance of at least this model misfolded protein. Additionally, many known ERAD genes are activated by the UPR, and ERAD-deficient mutants constitutively induce the UPR, demonstrating that the inability to export damaged ER cargo causes lumenal proteotoxic stress (317, 461). As the sole Hsp70 of the ER, Kar2 is clearly a multitasking molecular chaperone recruited to perform a number of roles in protein biogenesis (Fig. 7). This diversity of function is possible in part due to the nonselective nature of the Hsp70 class of chaperones and to the action of multiple Kar2 cofactors in the ER that provide both specificity and pathway-specific targeting. As is the case for the cytosolic Hsp70s, these accessory proteins fall into two distinct classes, the Hsp40/J proteins and nucleotide exchange factors.
ER J Proteins
S. cerevisiae expresses three ER-localized J-domain proteins: Sec63, Scj1, and Jem1 (Fig. 7) (322, 378, 394). Sec63, a 73-kDa integral membrane protein, spans the ER membrane three times, with its J domain facing the ER lumen and its C terminus located in the cytosol (129). Sec63 binds and stimulates Kar2, as demonstrated by using a glutathione S-transferase (GST) fusion of the J domain of Sec63 alone (63Jp) (38, 70). Additional genetic evidence supports the in vivo interaction of these two proteins: thermosensitive alleles of SEC63 and KAR2 grow normally at 25°C but are synthetic lethal, and dominant KAR2 alleles were shown to suppress the growth and translocation defects of the sec63-1 strain (404). When expressed in yeast, soluble 63Jp caused defects in the translocation of ppαF, suggesting competition for Kar2 binding and the sequestration of Kar2 from the Sec61 translocon. In another study, translocation defects observed for the sec63-1 strain were narrowed down to a defective interaction between Kar2 and Sec63 (273). These results imply that a major role for Sec63 is to recruit Kar2 to the membrane in the immediate vicinity of the Sec61 translocon for the efficient co- and posttranslational translocation of ER-targeted proteins (273). In addition to its role as a lumenal Hsp70 recruiter, the extreme cytosolic C-terminal region of Sec63 containing a 52-residue acidic domain is required for posttranslational translocation but not for cotranslational translocation (205). Jem1 (DnaJ-like protein of the ER membrane), a nonessential bitopic membrane protein with its J domain in the ER lumen, is required for karyogamy during mating (322). The overexpression of JEM1 from a 2μm plasmid suppressed the karyogamy defect of a kar2-1 mutant, demonstrating a genetic interaction between this Hsp70-Hsp40 pair (36). In contrast, the overexpression of Sec63 in the same background had very little suppressive phenotype, suggesting that Jem1 specifically interacts with Kar2 to promote nuclear fusion. A third soluble ER lumen J protein, Scj1, was identified in a genetic screen for candidates that caused the missorting of a nuclear-targeted protein (25). The swapping of the J domain of Sec63 with that of Scj1 was sufficient to suppress the temperature-sensitive phenotype of the sec63-1 and sec63-101 strains, suggesting that the J domain of Scj1 is competent for Kar2 binding and recruitment (394). However, the replacement of the J domain of Sec63 with those of the non-ER Hsp40s Sis1 (cytosol) and Mdj1 (mitochondria) did not generate functional chimeras, indicating that the Hsp70-Hsp40 interaction is not universally exchangeable (394). The translocation of CPY is unaffected in scj1Δ cells, confirming previous observations that Scj1 does not play a role in protein translocation across the ER (424). The addition of N-linked oligosaccharides to proteins is an important step in protein folding in the ER and is catalyzed by oligosaccharyltransferase (OST). CPY and a nonglycosylated mutant allele were used as substrates to determine if Scj1 is involved in the folding and exit of cargo from the ER. The loss of Scj1 caused a modest delay in the folding of hypoglycosylated forms of CPY but to a lesser extent than that reported for kar2 mutant alleles, suggesting that Scj1 functions with Kar2 to counter the misfolding of proteins due to a lack of carbohydrate modifications (424). Recently, it was shown that yeast cells lacking the Scj1 and Jem1 proteins exhibited defects in the degradation of the heterologously expressed epithelial sodium channel (ENaC), whereas Kar2 function was dispensable. Both Scj1 and Jem1 assisted in the ERAD of ENaC independently of Kar2, indicating that some Hsp40s do not absolutely require Hsp70 to select and process substrates (41). Furthermore, lumenal ERAD substrates were stabilized in either jem1Δ or scj1Δ cells but not in a sec63 temperature-sensitive mutant, whereas a jem1Δ scj1Δ strain lacking both J proteins had no effect on the ERAD of a membrane protein. These findings are consistent with previous observations that the ERAD of membrane proteins is Kar2 independent and that Sec63 is not involved in the recognition of ERAD substrates (323). The latter conjecture is supported by genetic results wherein a single-amino-acid substitution in Kar2 (R217A) compromised its ability to interact with Sec63 but not Jem1, which in turn affected Sec63-dependent protein translocation but not Jem1-dependent ERAD (476). To underscore the separation of duties for the ER J proteins, the ERAD-involved SCJ1 and JEM1 genes, but not the protein biogenesis-based SEC63 gene, contain UPREs in their promoters, with transcription induced by the addition of tunicamycin (322, 394).
ER Nucleotide Exchange Factors
LHS1 (lumenal hsp seventy) encodes a nonessential ER glycoprotein that shares 24% amino acid identity with Kar2. Lhs1 is considered to be an “atypical” member of the Hsp70 superfamily and has been grouped with the Hsp110 family of Hsp70-like proteins that do not function as foldases in vivo. In a landmark paper, Steel and colleagues discovered that Lhs1 acts as a NEF for Kar2, paving the way for similar revelations about the Hsp110 family (443). Consistent with this role, Lhs1 binds preferentially to the apo- and ADP-bound states of Kar2 and not its ATP-bound state (93). Lhs1 and the Hsp110 Sse1 share key conserved Hsp70-binding residues and employ a similar mechanism to trigger nucleotide exchange on their cognate Hsp70s, Ssa1 and Kar2, respectively (8). Unlike Sse1, Kar2 reciprocally stimulates the ATPase activity of Lhs1 in a manner that requires ATP hydrolysis by Kar2. However, ATP binding by Lhs1 is required to promote its nucleotide exchange activity on Kar2 (93). Sse1 possesses holdase activity in vitro—is Lhs1 capable of the same profolding activity? Lhs1 was shown to reduce the thermal aggregation of the model substrate firefly luciferase in a nucleotide-independent manner (93). Likewise, similar experiments with the mammalian Lhs1 homolog Grp170 demonstrated the same properties, indicating that holdase activity is a conserved function of this protein family (498). To investigate the role of Lhs1 in protein translocation across the ER membrane, the biogenesis of a variety of precursor proteins was examined. While the loss of Lhs1 clearly showed defects in the translocation of pre-PDI, ppαF, and pre-Kar2, no defect was observed for preinvertase or pre-dipeptidylaminopeptidase B (DPAPB), suggesting that Lhs1 is required for the efficient translocation of a subset of precursors (84). It was later shown that nucleotide binding to Lhs1, and, hence, its NEF activity, is required for this function (93). This is consistent with the observation that the other ER NEF, SIL1 (suppressor of the ire1Δ lhs1Δ double mutant) (see below) can partially compensate for LHS1 in translocation when overexpressed (471). Lhs1 was found to be required for the refolding, solubilization, and reactivation of the marker protein Hsp150Δ–β-lactamase after heat denaturation but interestingly played no role in its translocation, folding, and secretion under normal conditions (387). Two general conclusions can be drawn from these data. First, while Lhs1 is clearly capable of binding unfolded proteins, this feature may be secondary to its role as a NEF for Kar2, at least with respect to the translocation and biogenesis of ER cargo. Second, the folding repair machinery in the ER may be distinct from the folding of newly synthesized polypeptides. The promoter of LHS1 contains a UPRE similar to KAR2 and is transcriptionally induced both in the presence of tunicamycin and in a kar2-159 mutant strain previously shown to accumulate unfolded proteins in the ER (17, 84). However, unlike KAR2, LHS1 is not a heat shock-inducible gene. Strains lacking LHS1 exhibit elevated KAR2 and PDI1 mRNA levels, consistent with the constitutive induction of the UPR (84). UPR activation is physiologically relevant, as a double mutant strain lacking LHS1 and IRE1 is synthetic lethal, which is indicative of profound folding defects in lhs1Δ cells. Once again, NEF-defective LHS1 mutants phenocopy the null mutant for UPR regulation, highlighting the importance of this biochemical activity (93).
The other NEF of the ER lumen, SIL1, was identified in a screen for suppressors of ire1Δ lhs1Δ lethality. It is a nonessential protein present in the ER lumen and is conserved from yeast to humans. Sil1 is homologous to the Sls1 protein from the yeast Yarrowia lipolytica, which was shown previously to interact with Kar2 (26). Sil1 in S. cerevisiae was subsequently shown to interact specifically with the ATPase domain of Kar2 (215, 471). SIL1 is synthetically lethal when disrupted in combination with the kar2-113 and sec63-1 mutant alleles, suggesting that Sil1 plays a role in the translocation process. In addition, kar2-1 and kar2-133 mutant strains that show defects in folding and ERAD but not translocation are inviable in the absence of SIL1, implicating the NEF in the other functions of Kar2 (215). In a GST pulldown assay, Sil1 promoted the binding of Kar2 with Sec63 in vitro, although the formation of this complex might be only transient in vivo. Since both Sil1 and Lhs1 bind Kar2, it was suggested that they do so in a mutually exclusive manner (443). Like Lhs1, Sil1 stimulates Kar2 ATPase activity and was also found to preferentially bind to the ADP-bound conformation of Kar2 (215). The deletion of SIL1 alone does not result in detectable translocation defects. However, the elimination of both SIL1 and LHS1 results in synthetic lethality (471).
The Glycoprotein Chaperone Calnexin
Calnexin, an ER integral membrane protein, was first identified in mammals as a molecular chaperone that retained incompletely folded glycoproteins in the ER until they were either properly folded or degraded (22, 118). Cne1, the yeast homolog of calnexin, is about 23% identical at the amino acid level to mammalian calnexin and is glycosylated but lacks a cytoplasmic tail and the capacity to bind calcium (Fig. 7) (334). Calreticulin, a calnexin homolog that performs the same function as that performed by calnexin in the mammalian ER lumen, is absent in S. cerevisiae. In addition, UDP-glucose:glycoprotein glucosyltransferase, a key component in the quality control of glycoprotein folding in mammalian cells, is also lacking in yeast (199). Glycoprotein quality control in yeast is therefore potentially less complex than that in higher eukaryotes. However, CNE1 is not essential, suggesting that other uncharacterized proteins may compensate for its absence (436). Cne1 exhibits holdase activity, as demonstrated by its ability to suppress the aggregation of thermally denatured citrate synthase (CS) as well as enhance its reactivation (195, 519). The lectin domain of Cne1 was shown to specifically bind monoglucosylated oligosaccharides, further confirming its function as a component of the glycoprotein quality control system in the ER (519). Another conserved site, called the P (proline-rich) domain, is required for full activity, as the deletion of this region partially decreased the ability of Cne1 to suppress the aggregation of CS and chicken egg yolk immunoglobulin and decreased the refolding of CS (401, 518). Two models have been proposed to describe the roles of calnexin in protein folding. In the “lectin-only” model, calnexin functions only through its lectin domain, with repeated cycles of glycoprotein binding and release through a monoglucosylated oligosaccharide. In this model, calnexin does not act as a classical chaperone to prevent protein aggregation but may recruit chaperones to the unfolded glycoprotein, as demonstrated by its binding to ERp57, a mammalian thiol oxidoreductase that catalyzes disulfide formation in glycoproteins bound to calnexin (195). In the “dual-binding” model, calnexin functions as a lectin and a chaperone. In addition to incorporating the features of the lectin-only model, the model proposes a second site on calnexin that binds directly to polypeptide stretches of unfolded glycoproteins, similar to other chaperones. The latter model is supported by the findings that (i) complexes between calnexin and glycoproteins cannot be dissociated by completely removing oligosaccharides and (ii) calnexin retains interactions with unglycosylated proteins (195).
Protein Disulfide Isomerases
Many ER-resident and secreted proteins contain oxidized disulfide bonds between closely apposed cysteines that maintain their tertiary or quaternary structures. Cargos that form incorrect disulfide bonds or fail to form them are subject to misfolding, the repair of which is impossible without resolving the improper covalent disulfide links. Thus, oxidizing compartments like the ER (and the periplasm in Gram-negative bacteria) contain proteins that catalyze the formation, reduction, and isomerization of disulfide bonds called protein disulfide isomerases (PDIs) (Fig. 7) (138). The oxidase activity of PDI is favored when its active-site cysteines (CxxC) are in the disulfide (oxidized) form, which then catalyzes the oxidation of neighboring sulfhydryl groups in a substrate polypeptide to a disulfide bond. On the contrary, when the active-site cysteines of PDI are reduced (dithiol form), the enzyme catalyzes the reduction or isomerization of disulfides on substrate proteins (272, 493). Yeast Pdi1 is a 522-amino-acid protein that shares 30% identity with mammalian PDIs and is essential for cell viability (128, 454). A significant difference between yeast Pdi1 and its mammalian homolog is the presence of five consensus N-glycosylation sites in Pdi1, all of which are modified in the protein, as seen by a migration shift in SDS-PAGE gels of approximately 10 kDa after endoglycosidase H (EndoH) treatment (297). Mammalian PDI family members contribute both isomerase and chaperone functions to maintain cell growth (46, 351, 435, 523). However, in yeast, only the disulfide isomerase activity of Pdi1 appears to be essential (228).
In addition to PDI1, S. cerevisiae possesses four other PDI homologs, MPD1, MPD2, EUG1, and EPS1, all of which are nonessential (452, 453, 455, 496). All are soluble lumenal proteins, except for Eps1, which is the only membrane-associated PDI1 homolog. Evidence from different groups showed that the overexpression of any of these homologs can partially suppress the lethality of the pdi1Δ strain, suggesting that they have the ability to carry out the minimum Pdi1 function required for cell survival (324). Why, then, is the deletion of PDI1 lethal? A possible explanation lies in the observation that the additional PDI genes are expressed at substantially lower levels in the cell. This notion was tested by placing PDI1 under the control of the weak MPD1 promoter. Interestingly, this construct was able to rescue a pdi1Δ eug1Δ mpd1Δ mpd2Δ eps1Δ strain, suggesting that low PDI levels are sufficient for viability. Moreover, of all the PDI genes, only MPD1 and PDI1 are competent to serve as the sole source of PDI enzyme activity (324). A series of genetic and biochemical experiments suggested that Mpd1 and Mpd2 perform complementary functions. The overexpression of EUG1 was found to complement the lethality of the multiple-PDI-knockout strain only if MPD1 and MPD2 were both present. The overexpression of MPD2 is not able to rescue the pdi1Δ mutation if MPD1 is also absent. A biochemical analysis demonstrated that Mpd1 exhibits very weak isomerase activity and that Mpd2 has a high level of chaperone activity. Both proteins interact with a dissociation constant in the micromolar range, but neither protein increases the isomerase activity or the chaperone activity of the other (228). Unlike all known PDIs, EUG1 lacks one of the two cysteines in the active site (CxxS), cannot form an intramolecular disulfide bond, and, thus, is unable to transfer oxidizing equivalents to substrates. EUG1 expression is induced in the presence of unfolded proteins in the ER, consistent with the presence of a UPRE in its promoter sequence (452). To study the chaperone activity of the PDI homologs, the rate of intracellular folding of proCPY was monitored, since only the correctly folded protein can exit the ER (151). Besides PDI1, none of the PDI homologs were absolutely required for the folding of proCPY. However, they likely recognize it as a substrate in the absence of Pdi1, because proCPY maturation and ER exit proceed but at a much reduced rate. In contrast to a previous observation that Pdi1 played a role in the ERAD of the above-mentioned misfolded substrate CPY* (88), later evidence suggested that neither Pdi1 nor its homologs play a significant role in its degradation (154, 324). The basis for these opposing results is unclear but may be due to physiological differences in the strains. The PDI homolog Mpd1 interacts with Cne1 (calnexin), which increased the reductive activity of Mpd1 but unexpectedly eliminated Cne1 chaperone activity (229). This result suggests that Mpd1 might bind Cne1 near the peptide-binding site of Cne1, competitively inhibiting substrate associations. The membrane-associated Eps1 has strong chaperone activity but no oxidative activity, and similar to Pdi1, it interacts with Kar2 (229). It is possible that Eps1 may function solely as a molecular chaperone in vivo, but its function as a redox enzyme cannot be ruled out, since its reductive activity is increased in the presence of Eug1.
MOLECULAR CHAPERONES OF THE MITOCHONDRION
Mitochondria are essential eukaryotic organelles required for a range of metabolic, signaling, and developmental processes. They also present a unique challenge for protein biosynthesis, targeting, and quality control, given the different milieus within the organelle and its evolutionary history. Mitochondria possess two distinct membrane systems, an inner membrane (IM) and an outer membrane (OM), and two physically separate soluble compartments, the intermembrane space (IMS) and matrix. It is generally accepted that eukaryotic mitochondria arose after a cellular fusion event of a protoeukaryote/archaeon and a Gram-negative eubacterium, and this relationship is supported by the close homology of many bacterial and mitochondrial proteins. While a few mitochondrial proteins are encoded and synthesized within the mitochondrial matrix, the vast majority are synthesized in the cytosol from nuclear genes and must be posttranslationally translocated in an unfolded state into the mitochondria for folding and/or additional targeting. Mitochondria thus possess dedicated chaperone systems to assist in these processes (Table 3 and Fig. 8).
Table 3.
Mitochondrial chaperones
| Class | Protein | Function(s) |
|---|---|---|
| Hsp100 | Hsp78 | Unfoldase, disaggregase |
| Hsp90 | None | |
| Hsp90 cochaperone | None | |
| Hsp70 | ||
| GRP170 | None | |
| Hsp110 | None | |
| Hsp70 | Ssc1 | Protein folding, translocation |
| Ssc3 | Protein folding, translocation | |
| Ssq1 | Folding of FeS proteins | |
| Hsp70 NEF | Mge1 | Hsp70 nucleotide exchange |
| Hsp40/J protein | Mdj1 | Hsp70 ATPase stimulation, translocation |
| Mdj2 | Hsp70 ATPase stimulation, translocation | |
| Jac1 | Ssq1 J-protein partner | |
| Pam16 | Partner with Pam18, Hsp70 ATPase stimulation, translocation | |
| Pam18 | Hsp70 ATPase stimulation, translocation | |
| Chaperonin | Hsp60 | Protein folding, translocation |
| Chaperonin cochaperone | Hsp10 | Partner with Hsp60, protein folding, translocation |
| sHSP | None | |
| Other | Hep1 | Ssc1 partner, stabilization |
| Pim1 | Proteolysis and degradation |
Fig 8.
The mitochondrial chaperome. Chaperones and cofactors of the mitochondrion are shown. OM, outer membrane; IM, inner membrane; IMS, intermembrane space; TOM, transporter outer membrane complex; TIM, transporter inner membrane complex.
Mitochondrial Hsp70s
In S. cerevisiae, three distinct Hsp70s are present within mitochondria, Ssc1, Ssc3, and Ssq1 (16, 83, 391). As discussed below, these chaperones play distinct and occasionally overlapping roles in mitochondrial protein dynamics. Like all members of the Hsp70 family, mitochondrial Hsp70 (mtHsp70) contains an N-terminal ATPase domain (NBD) and a C-terminal substrate-binding domain (SBD). The nuclear gene SSC1 encodes an essential mitochondrial matrix Hsp70 protein that additionally includes a 28-amino-acid mitochondrion-targeting sequence at the amino terminus that is cleaved upon translocation into the matrix (82, 83, 306). The sequence of Ssc1 more closely resembles that of the bacterial Hsp70 DnaK than those of its yeast cytosolic counterparts, Ssa1 and Ssb1, consistent with the hypothesis that mitochondria are of bacterial origin. The two primary functions of Ssc1 are to assist protein translocation and subsequent protein folding. The initial transport of a precursor protein into mitochondria requires an energized inner membrane. The membrane potential (ΔΨ), and not the proton motive force, is required for precursor transport, possibly through the electrophoretic effect on the positively charged presequence required for mitochondrial targeting (281). Pulse radiolabeling of a temperature-sensitive ssc1-2 strain with [35S]methionine showed a substantial accumulation of precursor proteins at the nonpermissive temperature, demonstrating the necessity of Ssc1 for import (222). Cross-linking experiments showed that Ssc1 interacts directly with precursor proteins entering the matrix: this interaction occurs early in the import process, as the cross-linked precursor still contains its cleavable presequence and spans both the inner and outer membranes (222, 389). Does Ssc1 binding to the precursor polypeptide facilitate its unfolding on the cytosolic side? To test this hypothesis, the rate of translocation of a completely unfolded protein into isolated wild-type or ssc1 mutant mitochondria was compared to that of a partially unfolded protein. While transport rates were similar for the completely unfolded substrate, the partially folded protein exhibited slower import into mitochondria defective for Ssc1 (222). This finding supports the idea that the binding of Ssc1 to the precursor protein on the trans side (matrix) facilitates its unfolding on the cytosolic side. Moreover, ATP and a functional ATPase domain of Ssc1 are required to bind the polypeptide and drive its translocation into the matrix (143). However, precursor proteins with an IMS sorting signal do not require Ssc1 function for import, suggesting that these two destinations are functionally distinct in terms of mechanical translocation requirements (488). Mitochondria contain transport machineries in both their outer and inner membranes for the import of nuclear-encoded proteins. The translocase of the outer membrane (Tom), a general import pore, and the translocase of the inner membrane (Tim) are transiently linked via a transiting precursor protein. In S. cerevisiae, Tim44, Tim23, and Tim17 comprise the essential proteins of the Tim complex and were all shown to be in close contact with the precursor protein by in vivo cross-linking approaches (reviewed in reference 315). Interactions between Ssc1 and the Tim complex would support the idea that Ssc1 is recruited to the inner membrane for precursor import into the matrix. Genetic experiments showed that the overexpression of either SSC1 or TIM44 rescues the protein import defects seen in mutant alleles of the other gene. In addition, a severe synthetic growth defect was observed when hypomorphic alleles for both genes were combined (358). Results from biochemical experiments further confirmed this interaction and demonstrated that Ssc1 requires a functional ATPase domain to productively interact with Tim44 to promote substrate translocation (358). ATP hydrolysis by Ssc1 dissociates the Ssc1-Tim44 complex, which can then lead to Tim44 interacting with another Ssc1 molecule. Similar to the ratchet mechanism of Kar2 for precursor import into the ER, the binding and release of Ssc1 from Tim44 at the inner mitochondrial membrane facilitate the movement of the precursor in the forward direction (399). Specifically, Tim44 interacts with the β-stranded portion of the peptide-binding domain of Ssc1, which is postulated to position the substrate-binding domain near the outlet of the import channel to make Ssc1 available immediately for precursor protein binding (307). Genetic and biochemical experiments also showed Ssc1 to interact with the integral membrane protein Tim17 in an ATPase-dependent manner (27). This provides evidence of a second membrane anchor that recruits Ssc1 to the Tim complex for precursor protein import into the matrix. Interestingly, Ssc1-2 binds its substrates efficiently but not Tim17 or Tim44, suggesting that Ssc1 binding to the Tim complex is distinct from its binding to substrates and that both are required for the full function of Ssc1.
Evidence of an Ssc1 function in posttranslocation folding was obtained by using a protease sensitivity assay. A hybrid protein between a native mitochondrial protein and the enzyme dihydrofolate reductase (DHFR) (Su9-DHFR) that localizes to mitochondria was used as a substrate to determine the folding state of the DHFR domain. In wild-type cells, DHFR folds into its mature form after translocation and is resistant to proteolysis, while DHFR in the ssc1-2 mutant is almost completely proteinase K sensitive (222). Similarly, the refolding of chemically denatured luciferase depends on the presence of Ssc1, further supporting its role in substrate folding (261). Ssc1 is also required to stabilize unfolded proteins and maintain them in a soluble state (492). Another SSC1 temperature-sensitive mutant strain, the ssc1-3 strain, carries a mutation in the ATPase domain (G56S) and was defective in recovery after lethal (50°C) heat shock, in contrast to the ssc1-2 strain with a substitution in the peptide-binding domain (P419S) (326). This behavior was correlated with substrate-binding efficacy, as both purified mutant proteins were tested for their abilities to bind a denatured protein, reduced carboxymethylated α-lactalbumin (RCMLA), and while Ssc1-2 and wild-type Ssc1 bound well to the substrate, Ssc1-3 did not. Together, these results suggest that the binding capacity of Ssc1 for unfolded proteins is critical for recovery after stress. Ssc1 therefore plays dual roles as a mitochondrial import motor and a matrix foldase. These duties are coordinated by the participation of Ssc1 in two distinct protein complexes. The import complex associated with the inner membrane contains Ssc1, Tim44, and the NEF Mge1. The folding complex contains Ssc1, the J protein Mdj1, and the NEF Mge1 and is localized in the matrix. Hence, a precursor protein destined for the matrix first binds to the import complex and then is subsequently transferred to the folding complex (190).
Matrix-localized Ssc3 (Ecm10) was first identified in a screen for mutants exhibiting increased sensitivity to the cell wall-perturbing agent calcofluor, but the relationship between this mitochondrion-localized protein and cell wall formation remains unclear (270). The amino acid sequence of Ssc3 is 82% identical to that of Ssc1, and this high degree of similarity implies a functional overlap between both proteins. The overexpression of SSC3 in the temperature-sensitive ssc1-3 strain, which is defective in protein import when grown at the nonpermissive temperature, resulted in the complete restoration of protein import into the matrix (16). In addition, Ssc3 is associated with substrate proteins during or after their import into the matrix, confirming its role as another mitochondrial Hsp70 chaperone with overlapping substrate specificities (16). Cells lacking SSC3 show no obvious growth defects, likely due to functional redundancy with SSC1. However, the disruption of SSC3 in the conditional ssc1-3 strain resulted in heightened cold sensitivity when grown on glycerol compared to the ssc1-3 strain alone, suggesting a role for Ssc3 in protection against cold stress when Ssc1 is partially defective (16). Contrary to the above-described evidence, recent data showed that SSC3, when overexpressed in the ssc1Δ strain complemented with the SSC1 gene on a URA3-marked plasmid, cannot support growth after counterselection for the loss of the complementing plasmid on 5-fluoroorotic acid (5-FOA) (330). These results suggest that SSC3 may not be competent to replace all the biological functions of SSC1 when the latter is absent, as opposed to being compromised by a point mutation. In support of this conjecture, fluorescence anisotropy measurements showed that Ssc3 exhibited a lower affinity with a generic Hsp70-binding peptide than Ssc1. Using chimeras of Ssc1 and Ssc3, the D-helix within the SBD of both proteins was shown to be responsible for their differential affinities for client proteins (330). These functional differences may explain the observation that Ssc3 is present in only a small subset of closely related fungi, including S. cerevisiae, Saccharomyces bayanus, and Candida glabrata. It still remains to be shown what the differences in the phenotypes seen by those two groups represent.
The third Hsp70 family member, Ssq1, is also localized to the mitochondrial matrix but is distantly related to Ssc1, exhibiting 52% amino acid identity (391). Consistently, Ssq1 is capable of binding ATP directly and interacts with the model substrate RCMLA (395). Some functions of Ssq1 and Ssc1 may be related, since the overexpression of SSC1 in ssq1Δ cells partially rescues the cold-sensitive growth defect associated with this strain (391). However, ssc1 mutants that have growth and translocation defects at the nonpermissive temperature cannot be rescued, even when Ssq1 is strongly overexpressed, effectively classifying Ssc1 as a “generalist” Hsp70 and Ssq1 as a “specialist” isoform. The lack of a translocation defect may be due to the inability of Ssq1 to interact with Tim44, thereby making it less likely to be involved in protein import and raising the question of what role a third functional Hsp70 might play in the matrix (271). Strikingly, both a conditional ssq1-2 mutant and ssq1Δ strains accumulate large amounts of iron in the mitochondria (230). A synthetic lethal interaction between ssq1Δ and nfu1Δ, a gene homologous to those involved in iron-sulfur (FeS) center formation in nitrogen-fixing bacteria, further suggested a possible role for Ssq1 in FeS protein biogenesis (392). FeS proteins are present in both prokaryotic and eukaryotic cells and play central roles in a number of cellular processes, including redox reactions, nitrogen fixation, metabolic conversions, and iron and oxygen sensing (212). The biogenesis of FeS-containing proteins takes place in the mitochondria, and increasing evidence shows that Ssq1 is needed for this process. The activity of the FeS cluster-containing enzyme succinate dehydrogenase in the ssq1Δ strain is considerably reduced compared to that in the wild type, providing evidence for its role in FeS biogenesis (392). In addition, mitochondria lacking functional Ssq1 are unable to incorporate FeS clusters into ferredoxin, suggesting a role for Ssq1 in the biogenesis and/or incorporation of FeS clusters rather than the maintenance or protection of FeS-containing proteins (271). While early evidence suggested an interaction between Ssq1 and the FeS cluster scaffolding protein Isu1, this may be indirect, instead requiring the cysteine desulfurase Nfs1 to mediate the differential regulation of Isu1 (9, 114). Defects in iron incorporation into FeS, resulting in Freidreich's ataxia, are linked to mutations in human frataxin (yeast Yfh1), underscoring the relevance of chaperone function to organellar biology (49).
Mitochondrial Hsp70 Cofactors: J Proteins and NEFs
As with the Hsp70s resident in the cytoplasm and ER, mitochondrial Hsp70s are assisted by J proteins and nucleotide exchange factors that play roles in targeting and help confer specificity (Fig. 8). Initial speculation that Tim44 of the Tim translocase machinery was the J-protein partner for Ssc1 for precursor translocation into the mitochondria was shown not to be true, since Tim44 was unable to stimulate the ATPase activity of Ssc1 or bind Ssc1 similarly to the J protein Mdj1, suggesting the involvement of another J protein in this function. An in silico search of the yeast genome database identified the essential PAM18 (presequence translocase-associated motor) gene, whose gene product contains a J domain that is 57% identical to another mitochondrial inner membrane J protein, Mdj2. Substitutions in the conserved HPD sequence of Pam18 rendered the mutant protein unable to rescue the inviability of the pam18Δ strain, demonstrating the importance of the J domain for function (109, 300, 469). The requirement for Pam18 in protein import into the matrix and its ability to stimulate the ATPase activity of Ssc1 confirm its role as the J-domain partner for Ssc1 (109, 300, 469). This stimulation was specific to Ssc1, since Pam18 did not stimulate the ATPase activity of the other mitochondrial Hsp70, Ssq1 (109). Thus, Pam18 localized at the mitochondrial inner membrane with its J domain facing the matrix can stimulate the ATPase activity of Ssc1 that is recruited by Tim44 to the membrane for precursor protein import. Pam16 is a “J-like” protein, as it contains a sequence similar to that of the J domain of Pam18 but lacks the signature HPD motif, having DKE in its place (137, 233). Similar to Pam18, Pam16 is required for precursor translocation into the matrix and forms a stable subcomplex with the Tim23 translocase (137, 233). However, Pam16 efficiently inhibits the Pam18-dependent stimulation of the ATPase activity of Ssc1 (256). The replacement of the DKE motif of Pam16 with HPD did not convert it into a J protein, nor could this mutant protein stimulate the ATPase activity of Ssc1. This may be due to the interaction surface of Pam16, which is mostly neutral or negatively charged, compared to the positively charged surface of Pam18 (299). When carried on a plasmid in the pam18Δ strain, PAM16 failed to rescue the inviability of these cells, suggesting that the main role for Pam16 is to control the activity of Pam18 at the inner mitochondrial membrane (256). To test the hypothesis that Pam16 regulates the interaction of Pam18 with the Tim23 complex, the complex was isolated from a wild-type strain and a mutant strain (pam16-1) and tested for the presence of Pam18. Compared to the wild type, the Tim23 complex isolated from the pam16-1 strain lacked Pam18, which indicates that Pam16 acts as an adaptor protein for Pam18 at the Tim23 complex, thus regulating Pam18's role to stimulate Ssc1 ATPase activity (256). In contrast, a more recent study found that the ability of Pam18 to stimulate Ssc1 was not influenced by Pam16; rather, the primary, if not sole, role for Pam16 is as a tether to recruit Pam18 to the translocon (328). This was confirmed by evidence showing that the J-like domain of Pam16 strongly interacts with the Pam18 J domain and that the formation of the Pam16-Pam18 heterodimer was essential for cell growth and protein import into mitochondria (110). The mitochondrial DnaJ (MDJ1) gene was identified during DNA sequencing of an S. cerevisiae genomic library (374). Mdj1 is a soluble mitochondrial matrix protein and is not required for the Ssc1-dependent import of a number of precursor proteins tested (374). However, Mdj1 binds to precursor proteins entering the matrix in the latter stages of their import, possibly to fold newly imported proteins during their translocation (508). The minimal length of a precursor protein that is in the matrix before Mdj1 can bind is unknown. Mitochondria lacking MDJ1 were found to aggregate approximately 20% of the model fusion substrate Su9-DHFR compared to wild-type mitochondria, and aggregation was more pronounced with heat stress at 37°C (374). This finding suggests a role for Mdj1 in preventing heat-induced protein aggregation in the mitochondria. The enzymatic activity of firefly luciferase targeted to mitochondria was also reduced by 70% in mdj1Δ cells, supporting the conclusion that Mdj1 is involved in protein folding (374). Furthermore, by cross-linking experiments, Mdj1 and Ssc1 were shown to play a role in binding to nascent chains on mitochondrial ribosomes, possibly to prevent unproductive protein folding during translation (508). In addition to folding, Mdj1 also plays a role in protein degradation. A yeast strain containing a disrupted MDJ1 gene was shown to be defective in the degradation of two substrate proteins, indicating a role for Mdj1 in the clearance of misfolded mitochondrial proteins. This process is dependent on its functional interaction with Ssc1, since the release of an unfolded protein from Ssc1 was inhibited in the absence of Mdj1 (492). Mdj2 is an integral membrane protein present in the mitochondrial inner membrane with its J domain facing the matrix (509). As expected of J-domain-containing proteins, Mdj2 simulates the ATPase activity of Ssc1 (301). Although nonessential, the loss of MDJ2 in an mdj1Δ strain is lethal when cells are grown at 35°C compared to strains that lack either of the genes at that temperature (509). This finding suggests that Mdj2 is required for some of the functions of Mdj1 under these conditions. By using various mutations of Mdj1 in the mdj2Δ strain, the complementation of growth at 35°C was tested (509). The J domain of MDJ2 appears to partially complement the J domain of Mdj1, as established by assaying the suppression of the 35°C growth defect of mdj2Δ cells with various MDJ1 mutants (509). On the other hand, the overexpression of MDJ2 cannot suppress the growth defects of mdj1 mutants, suggesting unique functions of Mdj1 (509). Coimmunoprecipitation experiments revealed that Mdj2 associates with the Tim23 translocase, similarly to Pam18. In addition, Mdj2 and Pam18 form two separate complexes with Pam16 (57). What is the functional significance of these distinct complexes? Contrary to its inhibitory effects on Pam18 (see above), Pam16 enhanced the ATPase activity of Ssc1 by Mdj2 (301). Surprisingly, the growth and translocation defects of cells lacking PAM18 can be overcome by overexpressing MDJ2, suggesting that either can function to recruit and activate Ssc1 at the IM to promote import.
Jac1 (J-type accessory chaperone) is an essential member of the J-domain-containing protein family that contains a mitochondrion-targeting sequence and is localized in the matrix (271, 446). The identification in the same genetic screen of ssq1 and jac1 mutants as suppressors of metabolic defects associated with the absence of the copper/zinc superoxide dismutase (Sod1) suggests that Jac1 may play a role as the J-domain partner for Ssq1 (446). Homologs of Ssq1 and Jac1 found in bacteria are closely associated with genes thought to be involved in FeS protein biosynthesis (446). Is Jac1 a dedicated J protein for the mtHsp70 Ssq1? Multiple lines of evidence support this idea: (i) the activity of FeS cluster-containing enzymes, such as aconitase and succinate dehydrogenase, in ssq1Δ and JAC1-depleted strains is significantly reduced (224, 271); (ii) cells which contain a mutation in either SSC1 or JAC1 and which are subjected to increased levels of iron in the growth medium experience a 10-fold increase in iron uptake in the mitochondria (224); and (iii) the incorporation of FeS centers into apo-ferredoxin is compromised in jac1Δ and ssq1Δ mitochondria (271). In line with this function, mitochondria from jac1Δ mutant cells show no defects in general protein import into mitochondria but show a defect in the import of Yfh1, the yeast frataxin homolog required for iron homeostasis (224). Frataxin mutations in humans, associated with the disease Freidreich's ataxia, are characterized by a decrease in the activity of FeS-containing enzymes and an increase in mitochondrial iron levels (49). The phenotype of a yfh1Δ deletion strain is very similar to those of ssq1 and jac1 strains, confirming the role of this chaperone pair in iron homeostasis. The inability of the other mitochondrial matrix J protein, Mdj1, to suppress the growth defect of the jac1Δ strain and the lack of rescue of mdj1Δ growth by JAC1 suggest a functional difference between these two J-domain proteins (485). The basis for this difference remains unresolved, but one possibility may be differential substrate bias.
MGE1 (mitochondrial GrpE homolog) is an essential gene in S. cerevisiae that is related to the E. coli GrpE family, which promotes the release of bound nucleotide on the Hsp70 DnaK. MGE1 is a nuclear gene that encodes a soluble protein of the mitochondrial matrix (238). Coimmunoprecipitation with anti-Ssc1 antibodies revealed that Mge1 associates with Ssc1. Moreover, binding is lost in the Ssc1-3 mutant protein, which is defective in ATP binding and hydrolysis (489). In addition, ATP disrupts the association of Mge1 with Ssc1, suggesting that it binds Ssc1 in the nucleotide-free or ADP state (489). A temperature-sensitive allele, mge1-100, was used to confirm a role in protein import in vivo (238). The association of Ssc1 with Tim44 is required for the import of precursor proteins into the matrix and is dependent on the nucleotide state of Ssc1. Mge1 was shown to modulate the nucleotide-dependent stability of the Ssc1-Tim44 complex in the presence of physiological concentrations of cations, including Na+, K+, and Mg2+ (400). A conserved loop structure on the surface of the ATPase domain of Ssc1 mediates its interaction with Mge1 and for Mge1-induced nucleotide exchange (293). Mge1 stabilizes the complex in the presence of ATP analogs but not the hydrolyzable form of ATP, suggesting that Mge1 assists in the assembly of the ATP-binding form of the complex (400). The overexpression of MGE1 led to a reduced rate of precursor protein import, likely due to the accelerated release of Ssc1 from the precursor as the chaperone was cycled back to the low-affinity ATP state. Mge1 also plays a role in posttranslocational folding, evidenced by the reduced rates of maturation of the Yfh1 protein observed for the mge1-100 strain. A similar defect occurs in strains lacking SSQ1, consistent with the close relationship between the Hsp70 and its accessory factor (395).
As described above, Ssc1 and Ssq1 partner with the dedicated J-domain-containing proteins Mdj1 and Jac1, respectively. In contrast, Mge1 is the only known mitochondrial NEF. The inactivation of Ssc1 in ssc1-3 mitochondria dramatically enhanced the interaction between Ssq1 and Mge1, suggesting that the two Hsp70s might compete for Mge1 associations (395). The relative stoichiometry of the three proteins supports this notion, as the ratio of Ssc1 to Mge1 to Ssq1, is 250:50:1, as determined by immunoprecipitation experiments (395). Taken together, these observations suggest that Ssq1 efficiently interacts with Mge1 but that the large excess of Ssc1 in the matrix may favor the formation of the Ssc1-Mge1 complex. Moreover, the higher relative stability of the Ssc1-Mge1 complex than of the Ssq1-Mge1 complex when treated with high concentrations of salt could factor into the competitive advantage of Ssc1 for Mge1 (395). Correspondingly, MGE1 overexpression increases the activity of Ssq1, indicating that Mge1 is limiting for Ssq1 function in vivo. The mitochondrial matrix thus provides a unique example where the relative activity of multiple Hsp70 chaperones is governed by interactions with a single, limiting NEF.
The Mitochondrial Chaperonin Hsp60
The mitochondrial matrix of S. cerevisiae (and higher eukaryotes) contains another distinct protein-folding machine, Hsp60 (Fig. 8) (362). Mitochondrial Hsp60, bacterial GroEL, and RubisCO-binding protein (in chloroplasts) belong to the type I family of chaperonins. These are differentiated from the type II family, which resides in the cytoplasm of archaea (termed the thermosome) and the eukaryotic cytosol (CCT/TriC [discussed above]). Chaperonins are large, double-ring assemblies that provide an encapsulated cavity to facilitate the folding of newly translated and newly translocated proteins. Type I chaperonins are heat stress inducible, and members of the Hsp60 family form a homo-oligomer of 14 subunits with 7 subunits arranged in a double-stacked ring (490). The inner cavity can accommodate proteins with a maximum mass of 50 kDa and keeps these substrates protected from the environment of the matrix. The importance of Hsp60 for cell function was shown by using temperature-sensitive mutants, as HSP60 null mutants are inviable due to massive mitochondrial folding defects, including the β-subunit of F1-ATPase, cytochrome b2, and the Rieske FeS protein of complex II (63). Imported proteins transiently associate with Hsp60 as incompletely folded intermediates (327). In agreement with this observation, HSP60 conditional mutants accumulate aggregates in the matrix that are unable to assemble into active complexes (63). ATP add-back experiments showed evidence for ATP-dependent folding and release of the substrate protein Su9-DHFR by Hsp60 in apyrase-treated mitochondria. At low levels of ATP, 60% of the substrate was protease sensitive and cofractionated with Hsp60. In contrast, the addition of ATP resulted in the folding of the substrate and its exit from Hsp60, making it protease resistant (327). However, not all proteins that enter the matrix require Hsp60 for folding. The folding of four monomeric proteins (rhodanese, mitochondrial cyclophilin Cpr3, and matrix-targeted variants of dihydrofolate reductase and barnase) after their import into the matrix was monitored in a wild-type strain and an HSP60-inactivated strain. Of these, only rhodanese formed a tight complex with Hsp60 and required the chaperonin for folding (371). To test whether mtHsp70 coordinates with Hsp60 for folding in the matrix, firefly luciferase carrying a mitochondrion-targeting signal (Su9-luciferase) was used as a model substrate to analyze the vectorial coupling of mtHsp70 with Hsp60 with regard to polypeptide transfer (184). Radiolabeled Su9-luciferase that was translated in a rabbit reticulocyte lysate and imported into isolated yeast mitochondria interacted with Ssc1 (by coimmunoprecipitation) at the earliest time points examined. Over a period of a few minutes, the interaction of Su9-luciferase with Ssc1 was reduced, while its interaction with Hsp60 increased. Thus, a vectorial transfer of a substrate from mtHsp70 to Hsp60 occurs in the matrix, similarly to other imported proteins. However, luciferase does not efficiently dissociate from Hsp60 to reach its soluble enzymatically active state, in contrast to other proteins tested previously. A possible explanation for this finding is that luciferase (62 kDa) is too large to fit into the Hsp60 cavity and therefore cannot fold on Hsp60. The same experiment performed in the absence of Hsp60 did not enhance luciferase folding and still resulted in its aggregation, possibly due to the inability of mtHsp70 to efficiently fold this substrate in vivo. This finding is in contrast to evidence that showed that the highly homologous E. coli Hsp70 system (DnaK, DnaJ, and GrpE) efficiently folds luciferase in vitro. These data demonstrate that these two protein-folding machines do not act independently but in an ordered way, where substrate release from Hsp70 precedes its interaction with Hsp60.
Mitochondria also contain a regulator of Hsp60 called Hsp10, the homolog of E. coli GroES, that is essential for cell viability (187). Hsp10 is an important component of various Hsp60-dependent functions, including the folding and assembly of proteins imported into the matrix and the sorting of the Rieske FeS protein en route from the matrix to the intermembrane space. However, consistent with previous in vitro observations, Hsp10 is not required for the folding of the precursor form of dihydrofolate reductase (DHFR). It is possible that small proteins such as DHFR do not require Hsp10 compared to larger proteins. By using an HSP10 mutant (P36S), the release of the substrate from Hsp60 was possible in the absence of Hsp10 function but led to protein aggregation as a result of a defective release from the cavity (187). What features dictate whether a protein becomes a substrate for the chaperonin in vivo is an open question. To address this question, a screen was set up, using temperature-sensitive alleles of HSP60 and HSP10 to test the folding of substrates in the absence or presence of the respective proteins. The identified substrates were classified into three groups: (i) those that require both Hsp60 and Hsp10, (ii) those that require only Hsp60, and (iii) imported Hsp60 itself, which required Hsp10 to be present (111). This finding suggests that Hsp60 does not obligatorily act with Hsp10 to promote matrix protein folding in vivo.
Hsp78, the Mitochondrial Disaggregase
The bacterial chaperone ClpB prevents the terminal accumulation of protein aggregates. In S. cerevisiae, two ClpB homologs are present, Hsp104 in the cytosol and Hsp78 in mitochondria. ClpB and its homologs belong to the AAA+ family, and Hsp78 is 65% similar and 44% identical to Hsp104. Moreover, when expressed in the cytosol, HSP78 can substitute for the loss of HSP104, which is indicative of a highly conserved mode of action (398). Hsp78 binds to misfolded polypeptides in the matrix and stabilizes them, preventing aggregation (397). Hsp78 is a soluble mitochondrial matrix protein whose deletion does not lead to obvious growth defects in cells under normal or heat stress conditions (249). Heat stress inactivates mitochondrial protein synthesis, which is efficiently restored upon a return to normal growth conditions for wild-type, but not hsp78 mutant, cells. Thus, Hsp78 plays a role in the reactivation of damaged proteins, rather than protecting them from heat-induced inactivation (398). Hsp78 was incapable of refolding denatured firefly luciferase in an in vitro system but promoted refolding by Ssc1, suggesting a functional cooperation between an unfoldase (Hsp78) and a foldase (Ssc1), similar to the analogous pairing (Hsp104 and Ssa1) in the cytosol (235). The disruption of HSP78 in ssc1-3 or ssc1-2 mutant cells results in a petite phenotype due to the loss of mitochondrial DNA, suggesting that at least one of the two heat shock proteins is required to maintain genome integrity (298). A molecular rationale for this phenotype is provided by the observation that the Mip1 polymerase is inactivated by heat shock and presumably requires Hsp78 and Ssc1 for a restoration of DNA polymerase activity (150). ssc1-3 hsp78Δ and ssc1-2 hsp78Δ mutant strains also exhibit impaired protein import at the nonpermissive temperature. Conversely, the overexpression of Hsp78 in ssc1-3 cells substantially improves import activity, suggesting that Hsp78 can at least partially complement the functional roles played by Ssc1 (486). Interestingly, Ssc1 itself is subject to misfolding during stress, and Hsp78 is required for its resolubilization (486). Therefore, it is possible that a major role of Hsp78 with regard to thermotolerance is to maintain Ssc1 in a soluble and functional state under stress conditions. This also posits an unusual scenario wherein the “stress” protein is itself structurally labile. In fact, Ssc1 requires the assistance of a novel protein, Hep1 (mtHsp70 escort protein), to maintain solubility and function. Hep1 was identified by the affinity purification of Ssc1 and interacts with Ssc1 when ATP levels are low (421). Fractionation studies of digitonin-digested mitochondria identified approximately 50% of Ssc1 in the insoluble fraction in hep1Δ cells. Hep1 prevents the aggregation of purified Ssc1 but is not capable of resolubilizing the misfolded chaperone and therefore plays a role complementary to Hsp78 in maintaining functional Ssc1 (421).
What happens to proteins not salvageable through the action of the mitochondrial chaperone network? The Pim1 protein complex was shown to degrade misfolded or unfolded reporter proteins (492). Pim1 (proteolysis in mitochondria) is a soluble ring-shaped structure in the mitochondrial matrix that is 30% identical to the E. coli Lon protease. Yeast cells lacking PIM1 are respiratory deficient and lose the integrity of their mitochondrial genome, similarly to hsp78Δ cells. PIM1 mRNA is constitutively expressed, and its levels are increased after heat stress, suggesting that it is required at higher concentrations within the matrix to mediate recovery (475). Ssc1 is also required for efficient proteolysis by Pim1, as demonstrated by a block in protein degradation in ssc1-2 and ssc1-3 mutants (492). Mechanistically, misfolded proteins must first be released from Ssc1 before degradation by Pim1. Mdj1 modulates the release of a substrate from Ssc1 and is thus required for efficient protein degradation in the matrix. The unfoldase Hsp78 is also unexpectedly required for degradation, as mitochondria lacking HSP78 degraded only 40% of imported test substrates compared to the wild type (80%) (372). Therefore, the same chaperones that are responsible for repairing and refolding damaged protein substrates also mediate substrate triage and turnover. How the decision is made to refold or degrade and what determinants influence these outcomes are unknown.
THE HSR IN PATHOGENIC FUNGI
One of the most highly conserved features of all living organisms is the ability to sense and respond to sudden changes in temperature. Interestingly, the heat shock proteins (HSPs) have been frequently shown to be immunodominant in infection by diverse pathogenic fungi, which is against the common rule that the immune response is generally targeted against microbe-specific antigens (55, 102, 219, 236, 294, 295, 364, 418, 442, 457). Candida albicans is a significant human fungal pathogen capable of disseminating through blood and colonizing almost all organs (514). For C. albicans, an elevated temperature of 37°C is critical to undergo the morphological transition between yeast and filamentous growth states, and the capability to sense the temperature upshift and start morphogenesis is tightly linked to its virulence (449). Numerous studies have shown that the heat shock proteins orchestrate temperature-dependent morphogenesis. Together, these observations spurred the push toward molecular genetic analyses of Hsp functions in pathogenic fungi. Two clones encoding Hsp70 were identified in C. albicans by the screening of a cDNA library of the yeast form of the organism with antibodies against heat-activated C. albicans. The deduced amino acid sequences are 79% identical and 84% similar to SSA1 to SSA4 of S. cerevisiae (122, 240). SSB1 has been cloned from C. albicans with 85% similarity to the Ssb subfamily of S. cerevisiae (278). Northern blot analysis revealed that like S. cerevisiae SSB1/2, C. albicans is upregulated after a mild cold shock and is rapidly downregulated after heat shock (279). Interestingly, C. albicans Ssa1 and Ssa2 are expressed on the cell surface of both the yeast and hyphal forms (267). Surface-associated Ssa1 and Ssa2 were identified as receptors for the saliva antimicrobial peptide histatin 5 (254, 255). These two proteins are also essential for the fungicidal activity of human β-defensins 2 and 3 (491). C. albicans expresses a single Hsp90 isoform that is induced at the transition from yeast to filamentous growth, and deletion attenuates the virulence of the fungus in a murine model (43, 415, 451). A series of reports over the last decade have highlighted the role of Hsp90 in promoting drug resistance in C. albicans. Cells lacking Hsp90 activity through pharmacological or genetic inhibition are unable to evolve resistance to antifungal azole drugs (74). Similarly, Hsp90 is required for resistance to the novel class of antifungals called echinocandins, which target cell wall biosynthesis, in Aspergillus spp. (75). At least part of this relationship has been linked to Hsp90's chaperoning of the protein phosphatase calcineurin, providing a molecular mechanism (73). A follow-up study implicated the protein kinase C/cell wall integrity pathway as another component of Hsp90-mediated azole resistance in C. albicans, dovetailing nicely with previous work demonstrating the same interactions in S. cerevisiae (237). The roles of Hsp90 in supporting morphogenesis and drug resistance were recently shown to be relevant to biofilm formation in pathogenic fungi, providing a pharmacological foothold into this clinically intransigent mode of infection (367, 415). The heat shock response has also been found to be highly relevant to pathogenesis, likely in an HSF-dependent context (39, 318, 319). Our understanding of HSPs and the HSR in pathogenic fungi lags far behind that of S. cerevisiae and is clearly at an early stage. However, it is expected that the wealth of knowledge generated from budding yeast should inform and accelerate progress in these diverse systems.
Perspectives
The S. cerevisiae genome sequencing project was completed in 1995, and the first yeast knockout collection was made available in 2002, ushering in the era of yeast genomics and proteomics. Coupled with previous decades' worth of pathway- and gene-specific investigations, our understanding of the heat shock response and the biology of molecular chaperones is rich and detailed. We now have in hand the program and most of the players in multiple cellular compartments. The challenge for the future will be to understand how these distinct systems interact and how they are organized into functional networks to promote the life of a small unicellular yeast. This knowledge can then be applied to an understanding of the same question for human cells, with the key goal being an insight into how we can modulate these powerful machines to further human health. The recent awareness that a wide range of neurodegenerative disorders, including Parkinson's, Alzheimer's, and Lou Gehrig's diseases, are fundamentally pathologies of protein misfolding has dramatically amplified interest in protein quality control systems. Indeed, yeast cells are now being exploited as drug discovery tools for the isolation and rational design of drugs that specifically target the HSR and individual chaperones for induction or repression. For example, a novel compound capable of activating human HSF1 was recently uncovered by using the complementation of yeast Hsf1 as a phenotype in high-throughput screens (312). C. albicans and Plasmodium falciparum Hsp70s have both been expressed in S. cerevisiae to generate safe and tractable experimental systems to utilize for the development of anti-infectives (20, 39). We predict that investigations of the HSR and molecular chaperones in yeast will proceed for some time and will continue to lead the way in discovery and impact.
ACKNOWLEDGMENTS
We thank Kimberly Cope for critical reading of the manuscript. We have endeavored to give credit where appropriate throughout and apologize for any inadvertent omissions.
Work in the laboratory of K.A.M. was supported by NIH grant GM-074696. J.A. was supported by a Robert D. Watkins fellowship from the American Society for Microbiology. J.V. was supported by the Schissler Foundation, and Y.W. was supported by the Cameron Foundation.
Biographies

Jacob Verghese received his undergraduate degree in zoology from Loyola College, Chennai, India. He then obtained an M.S. in biochemistry in the laboratory of Dr. William Widger at the University of Houston, where he investigated the role of the Rho transcription termination factor in plasmid replication control in E. coli. He is currently pursuing a Ph.D. in microbiology and molecular genetics at the University of Texas Medical School and Graduate School of Biomedical Sciences in the laboratory of Dr. Kevin A. Morano, investigating the role of Snl1, an ER membrane-tethered nucleotide exchange factor of the cytosolic Hsp70 family in S. cerevisiae.

Jennifer Abrams obtained a B.S. in biotechnology from the University of Houston—Downtown. She is currently pursuing a Ph.D. in microbiology and molecular genetics at the University of Texas Medical School and Graduate School of Biomedical Sciences in the laboratory of Dr. Kevin A. Morano, where she studies the diversity of three families of cytosolic Hsp70 nucleotide exchange factors in S. cerevisiae.

Yanyu Wang received her B.S. in Environmental Engineering from Shanghai Jiaotong University in China. She then obtained an M.S. in Civil and Environmental Engineering from the University of Houston, investigating high-salt-tolerant bacteria in the treatment of industrial wastewater in the laboratory of Dr. Deborah Roberts. She is currently pursuing a Ph.D. in microbiology and molecular genetics at the University of Texas Medical School and Graduate School of Biomedical Sciences in the laboratory of Dr. Kevin A. Morano, using the model organism Saccharomyces cerevisiae to study the regulation of the Hsf1-mediated heat shock response by Hsp70.

Kevin A. Morano is an associate professor in the Department of Microbiology and Molecular Genetics at the University of Texas Medical School at Houston. He received his B.S. degree in biological sciences from the University of California, Irvine, and his Ph.D. from the University of California, Davis, working on vacuolar protein biogenesis in the laboratory of Dr. Daniel Klionsky. He conducted postdoctoral work with Dr. Dennis Thiele at the University of Michigan, Ann Arbor, studying the heat shock response and Hsp90 in yeast. Dr. Morano's research interests are focused on protein chaperone biology and function in budding yeast.
REFERENCES
- 1. Abbas-Terki T, Donze O, Briand PA, Picard D. 2001. Hsp104 interacts with Hsp90 cochaperones in respiring yeast. Mol. Cell. Biol. 21:7569–7575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Abbas-Terki T, Donze O, Picard D. 2000. The molecular chaperone Cdc37 is required for Ste11 function and pheromone-induced cell cycle arrest. FEBS Lett. 467:111–116 [DOI] [PubMed] [Google Scholar]
- 3. Aguilaniu H, Gustafsson L, Rigoulet M, Nystrom T. 2003. Asymmetric inheritance of oxidatively damaged proteins during cytokinesis. Science 299:1751–1753 [DOI] [PubMed] [Google Scholar]
- 4. Ahn SG, Thiele DJ. 2003. Redox regulation of mammalian heat shock factor 1 is essential for Hsp gene activation and protection from stress. Genes Dev. 17:516–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Akerfelt M, Morimoto RI, Sistonen L. 2010. Heat shock factors: integrators of cell stress, development and lifespan. Nat. Rev. Mol. Cell Biol. 11:545–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Alepuz PM, Matheos D, Cunningham KW, Estruch F. 1999. The Saccharomyces cerevisiae RanGTP-binding protein Msn5p is involved in different signal transduction pathways. Genetics 153:1219–1231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ali MM, et al. 2006. Crystal structure of an Hsp90-nucleotide-p23/Sba1 closed chaperone complex. Nature 440:1013–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Andreasson C, Rampelt H, Fiaux J, Druffel-Augustin S, Bukau B. 2010. The endoplasmic reticulum Grp170 acts as a nucleotide exchange factor of Hsp70 via a mechanism similar to that of the cytosolic Hsp110. J. Biol. Chem. 285:12445–12453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Andrew AJ, Dutkiewicz R, Knieszner H, Craig EA, Marszalek J. 2006. Characterization of the interaction between the J-protein Jac1p and the scaffold for Fe-S cluster biogenesis, Isu1p. J. Biol. Chem. 281:14580–14587 [DOI] [PubMed] [Google Scholar]
- 10. Azevedo D, Tacnet F, Delaunay A, Rodrigues-Pousada C, Toledano MB. 2003. Two redox centers within Yap1 for H2O2 and thiol-reactive chemicals signaling. Free Radic. Biol. Med. 35:889–900 [DOI] [PubMed] [Google Scholar]
- 11. Bagola K, Sommer T. 2008. Protein quality control: on IPODs and other JUNQ. Curr. Biol. 18:R1019–R1021 [DOI] [PubMed] [Google Scholar]
- 12. Baler R, Welch WJ, Voellmy R. 1992. Heat shock gene regulation by nascent polypeptides and denatured proteins: hsp70 as a potential autoregulatory factor. J. Cell Biol. 117:1151–1159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bali M, Zhang B, Morano KA, Michels CA. 2003. The Hsp90 molecular chaperone complex regulates maltose induction and stability of the Saccharomyces MAL gene transcription activator Mal63p. J. Biol. Chem. 278:47441–47448 [DOI] [PubMed] [Google Scholar]
- 14. Barnes CA, Johnston GC, Singer RA. 1990. Thermotolerance is independent of induction of the full spectrum of heat shock proteins and of cell cycle blockage in the yeast Saccharomyces cerevisiae. J. Bacteriol. 172:4352–4358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Batista-Nascimento L, Neef DW, Liu PC, Rodrigues-Pousada C, Thiele DJ. 2011. Deciphering human heat shock transcription factor 1 regulation via post-translational modification in yeast. PLoS One 6:e15976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Baumann F, Milisav I, Neupert W, Herrmann JM. 2000. Ecm10, a novel hsp70 homolog in the mitochondrial matrix of the yeast Saccharomyces cerevisiae. FEBS Lett. 487:307–312 [DOI] [PubMed] [Google Scholar]
- 17. Baxter BK, James P, Evans T, Craig EA. 1996. SSI1 encodes a novel Hsp70 of the Saccharomyces cerevisiae endoplasmic reticulum. Mol. Cell. Biol. 16:6444–6456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Beck T, Hall MN. 1999. The TOR signalling pathway controls nuclear localization of nutrient-regulated transcription factors. Nature 402:689–692 [DOI] [PubMed] [Google Scholar]
- 19. Becker J, Walter W, Yan W, Craig EA. 1996. Functional interaction of cytosolic hsp70 and a DnaJ-related protein, Ydj1p, in protein translocation in vivo. Mol. Cell. Biol. 16:4378–4386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bell SL, Chiang AN, Brodsky JL. 2011. Expression of a malarial Hsp70 improves defects in chaperone-dependent activities in ssa1 mutant yeast. PLoS One 6:e20047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Benaroudj N, Lee DH, Goldberg AL. 2001. Trehalose accumulation during cellular stress protects cells and cellular proteins from damage by oxygen radicals. J. Biol. Chem. 276:24261–24267 [DOI] [PubMed] [Google Scholar]
- 22. Bergeron JJM, Brenner MB, Thomas DY, Williams DB. 1994. Calnexin: a membrane-bound chaperone of the endoplasmic reticulum. Trends Biochem. Sci. 19:124–128 [DOI] [PubMed] [Google Scholar]
- 23. Bernales S, Papa FR, Walter P. 2006. Intracellular signaling by the unfolded protein response. Annu. Rev. Cell Dev. Biol. 22:487–508 [DOI] [PubMed] [Google Scholar]
- 24. Berry DB, Gasch AP. 2008. Stress-activated genomic expression changes serve a preparative role for impending stress in yeast. Mol. Biol. Cell 19:4580–4587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Blumberg H, Silver PA. 1991. A homologue of the bacterial heat-shock gene DnaJ that alters protein sorting in yeast. Nature 349:627–630 [DOI] [PubMed] [Google Scholar]
- 26. Boisrame A, Kabani M, Beckerich JM, Hartmann E, Gaillardin C. 1998. Interaction of Kar2p and Sls1p is required for efficient co-translational translocation of secreted proteins in the yeast Yarrowia lipolytica. J. Biol. Chem. 273:30903–30908 [DOI] [PubMed] [Google Scholar]
- 27. Bomer U, et al. 1997. Multiple interactions of components mediating preprotein translocation across the inner mitochondrial membrane. EMBO J. 16:2205–2216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bonner JJ, Ballou C, Fackenthal DL. 1994. Interactions between DNA-bound trimers of the yeast heat shock factor. Mol. Cell. Biol. 14:501–508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bonner JJ, et al. 2000. Complex regulation of the yeast heat shock transcription factor. Mol. Biol. Cell 11:1739–1751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bonner JJ, Heyward S, Fackenthal DL. 1992. Temperature-dependent regulation of a heterologous transcription activation domain fused to yeast heat shock transcription factor. Mol. Cell. Biol. 12:1021–1030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Boorstein WR, Craig EA. 1990. Transcriptional regulation of SSA3, an HSP70 gene from Saccharomyces cerevisiae. Mol. Cell. Biol. 10:3262–3267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Borkovich KA, Farrelly FW, Finkelstein DB, Taulien J, Lindquist S. 1989. Hsp82 is an essential protein that is required in higher concentrations for growth of cells at higher temperatures. Mol. Cell. Biol. 9:3919–3930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Bosis E, et al. 2009. Ssz1 restores endoplasmic reticulum-associated protein degradation in cells expressing defective Cdc48-Ufd1-Npl4 complex by upregulating Cdc48. Genetics 184:695–706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Brejning J, Arneborg N, Jespersen L. 2005. Identification of genes and proteins induced during the lag and early exponential phase of lager brewing yeasts. J. Appl. Microbiol. 98:261–271 [DOI] [PubMed] [Google Scholar]
- 35. Breter HJ, Ferguson J, Peterson TA, Reed SI. 1983. Isolation and transcriptional characterization of three genes which function at start, the controlling event of the Saccharomyces cerevisiae cell division cycle: CDC36, CDC37, and CDC39. Mol. Cell. Biol. 3:881–891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Brizzio V, et al. 1999. Genetic interactions between KAR7/SEC71, KAR8/JEM1, KAR5, and KAR2 during nuclear fusion in Saccharomyces cerevisiae. Mol. Biol. Cell 10:609–626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Brodsky JL, Goeckeler J, Schekman R. 1995. BiP and Sec63p are required for both co- and posttranslational protein translocation into the yeast endoplasmic reticulum. Proc. Natl. Acad. Sci. U. S. A. 92:9643–9646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Brodsky JL, Schekman R. 1993. A Sec63p-BiP complex from yeast is required for protein translocation in a reconstituted proteoliposome. J. Cell Biol. 123:1355–1363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Brown AJ, Leach MD, Nicholls S. 2010. The relevance of heat shock regulation in fungal pathogens of humans. Virulence 1:330–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Buchan JR, Parker R. 2009. Eukaryotic stress granules: the ins and outs of translation. Mol. Cell 36:932–941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Buck TM, Kolb AR, Boyd CR, Kleyman TR, Brodsky JL. 2010. The endoplasmic reticulum-associated degradation of the epithelial sodium channel requires a unique complement of molecular chaperones. Mol. Biol. Cell 21:1047–1058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Bulman AL, Nelson HC. 2005. Role of trehalose and heat in the structure of the C-terminal activation domain of the heat shock transcription factor. Proteins 58:826–835 [DOI] [PubMed] [Google Scholar]
- 43. Burnie JP, Carter TL, Hodgetts SJ, Matthews RC. 2006. Fungal heat-shock proteins in human disease. FEMS Microbiol. Rev. 30:53–88 [DOI] [PubMed] [Google Scholar]
- 44. Cabib E, Bowers B, Sburlati A, Silverman SJ. 1988. Fungal cell wall synthesis: the construction of a biological structure. Microbiol. Sci. 5:370–375 [PubMed] [Google Scholar]
- 45. Cabib E, Sburlati A, Bowers B, Silverman SJ. 1989. Chitin synthase 1, an auxiliary enzyme for chitin synthesis in Saccharomyces cerevisiae. J. Cell Biol. 108:1665–1672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Cai H, Wang CC, Tsou CL. 1994. Chaperone-like activity of protein disulfide isomerase in the refolding of a protein with no disulfide bonds. J. Biol. Chem. 269:24550–24552 [PubMed] [Google Scholar]
- 47. Cairns BR, Ramer SW, Kornberg RD. 1992. Order of action of components in the yeast pheromone response pathway revealed with a dominant allele of the STE11 kinase and the multiple phosphorylation of the STE7 kinase. Genes Dev. 6:1305–1318 [DOI] [PubMed] [Google Scholar]
- 48. Camasses A, Bogdanova A, Shevchenko A, Zachariae W. 2003. The CCT chaperonin promotes activation of the anaphase-promoting complex through the generation of functional Cdc20. Mol. Cell 12:87–100 [DOI] [PubMed] [Google Scholar]
- 49. Campuzano V, et al. 1996. Friedreich's ataxia: autosomal recessive disease caused by an intronic GAA triplet repeat expansion. Science 271:1423–1427 [DOI] [PubMed] [Google Scholar]
- 50. Caplan AJ, Douglas MG. 1991. Characterization of YDJ1: a yeast homologue of the bacterial dnaJ protein. J. Cell Biol. 114:609–621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Caplan AJ, Tsai J, Casey PJ, Douglas MG. 1992. Farnesylation of YDJ1p is required for function at elevated growth temperatures in Saccharomyces cerevisiae. J. Biol. Chem. 267:18890–18895 [PubMed] [Google Scholar]
- 52. Carmody SR, Tran EJ, Apponi LH, Corbett AH, Wente SR. 2010. The mitogen-activated protein kinase Slt2 regulates nuclear retention of non-heat shock mRNAs during heat shock-induced stress. Mol. Cell. Biol. 30:5168–5179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Carrascosa JL, Llorca O, Valpuesta JM. 2001. Structural comparison of prokaryotic and eukaryotic chaperonins. Micron 32:43–50 [DOI] [PubMed] [Google Scholar]
- 54. Carratu L, et al. 1996. Membrane lipid perturbation modifies the set point of the temperature of heat shock response in yeast. Proc. Natl. Acad. Sci. U. S. A. 93:3870–3875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Caruso M, Sacco M, Medoff G, Maresca B. 1987. Heat shock 70 gene is differentially expressed in Histoplasma capsulatum strains with different levels of thermotolerance and pathogenicity. Mol. Microbiol. 1:151–158 [DOI] [PubMed] [Google Scholar]
- 56. Cashikar AG, Duennwald M, Lindquist SL. 2005. A chaperone pathway in protein disaggregation. Hsp26 alters the nature of protein aggregates to facilitate reactivation by Hsp104. J. Biol. Chem. 280:23869–23875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Cashikar AG, et al. 2002. Defining a pathway of communication from the C-terminal peptide binding domain to the N-terminal ATPase domain in a AAA protein. Mol. Cell 9:751–760 [DOI] [PubMed] [Google Scholar]
- 58. Causton HC, et al. 2001. Remodeling of yeast genome expression in response to environmental changes. Mol. Biol. Cell 12:323–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Chang HC, Nathan DF, Lindquist S. 1997. In vivo analysis of the Hsp90 cochaperone Sti1 (p60). Mol. Cell. Biol. 17:318–325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Cheetham ME, Caplan AJ. 1998. Structure, function and evolution of DnaJ: conservation and adaptation of chaperone function. Cell Stress Chaperones 3:28–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Chen T, Li F, Chen BS. 2009. Cross-talks of sensory transcription networks in response to various environmental stresses. Interdiscip. Sci. 1:46–54 [DOI] [PubMed] [Google Scholar]
- 62. Chen Y, Barlev NA, Westergaard O, Jakobsen BK. 1993. Identification of the C-terminal activator domain in yeast heat shock factor: independent control of transient and sustained transcriptional activity. EMBO J. 12:5007–5018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Cheng MY, et al. 1989. Mitochondrial heat-shock protein hsp60 is essential for assembly of proteins imported into yeast mitochondria. Nature 337:620–625 [DOI] [PubMed] [Google Scholar]
- 64. Cho HS, et al. 1996. Yeast heat shock transcription factor N-terminal activation domains are unstructured as probed by heteronuclear NMR spectroscopy. Protein Sci. 5:262–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Chung N, Jenkins G, Hannun YA, Heitman J, Obeid LM. 2000. Sphingolipids signal heat stress-induced ubiquitin-dependent proteolysis. J. Biol. Chem. 275:17229–17232 [DOI] [PubMed] [Google Scholar]
- 66. Cicero MP, et al. 2001. The wing in yeast heat shock transcription factor (HSF) DNA-binding domain is required for full activity. Nucleic Acids Res. 29:1715–1723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Conde R, Belak ZR, Nair M, O'Carroll RF, Ovsenek N. 2009. Modulation of Hsf1 activity by novobiocin and geldanamycin. Biochem. Cell Biol. 87:845–851 [DOI] [PubMed] [Google Scholar]
- 68. Conlin LK, Nelson HC. 2007. The natural osmolyte trehalose is a positive regulator of the heat-induced activity of yeast heat shock transcription factor. Mol. Cell. Biol. 27:1505–1515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Conz C, et al. 2007. Functional characterization of the atypical Hsp70 subunit of yeast ribosome-associated complex. J. Biol. Chem. 282:33977–33984 [DOI] [PubMed] [Google Scholar]
- 70. Corsi AK, Schekman R. 1997. The lumenal domain of Sec63p stimulates the ATPase activity of BiP and mediates BiP recruitment to the translocon in Saccharomyces cerevisiae. J. Cell Biol. 137:1483–1493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Cowart LA, Gandy JL, Tholanikunnel B, Hannun YA. 2010. Sphingolipids mediate formation of mRNA processing bodies during the heat-stress response of Saccharomyces cerevisiae. Biochem. J. 431:31–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Cowart LA, et al. 2003. Roles for sphingolipid biosynthesis in mediation of specific programs of the heat stress response determined through gene expression profiling. J. Biol. Chem. 278:30328–30338 [DOI] [PubMed] [Google Scholar]
- 73. Cowen LE, Carpenter AE, Matangkasombut O, Fink GR, Lindquist S. 2006. Genetic architecture of Hsp90-dependent drug resistance. Eukaryot. Cell 5:2184–2188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Cowen LE, Lindquist S. 2005. Hsp90 potentiates the rapid evolution of new traits: drug resistance in diverse fungi. Science 309:2185–2189 [DOI] [PubMed] [Google Scholar]
- 75. Cowen LE, et al. 2009. Harnessing Hsp90 function as a powerful, broadly effective therapeutic strategy for fungal infectious disease. Proc. Natl. Acad. Sci. U. S. A. 106:2818–2823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Cox JS, Walter P. 1996. A novel mechanism for regulating activity of a transcription factor that controls the unfolded protein response. Cell 87:391–404 [DOI] [PubMed] [Google Scholar]
- 77. Craig EA, Baxter BK, Becker J, Halladay J, Ziegelhoffer T. 1994. Cytosolic hsp70s of Saccharomyces cerevisiae: roles in protein synthesis, protein translocation, proteolysis, and regulation, p 31–52 In Morimoto RI, Tissieres A, Georgopolous C. (ed), The biology of heat shock proteins and molecular chaperones, vol 26 Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 78. Craig EA, Gambill BD, Nelson RJ. 1993. Heat shock proteins: molecular chaperones of protein biogenesis. Microbiol. Rev. 57:402–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Craig EA, Gross CA. 1991. Is hsp70 the cellular thermometer? Trends Biochem. Sci. 16:135–140 [DOI] [PubMed] [Google Scholar]
- 80. Craig EA, Jacobsen K. 1985. Mutations in cognate genes of Saccharomyces cerevisiae hsp70 result in reduced growth rates at low temperatures. Mol. Cell. Biol. 5:3517–3524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Craig EA, Jacobsen K. 1984. Mutations of the heat inducible 70 kilodalton genes of yeast confer temperature sensitive growth. Cell 38:841–849 [DOI] [PubMed] [Google Scholar]
- 82. Craig EA, Kramer J, Kosic-Smithers J. 1987. SSC1, a member of the 70-kDa heat shock protein multigene family of Saccharomyces cerevisiae, is essential for growth. Proc. Natl. Acad. Sci. U. S. A. 84:4156–4160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Craig EA, et al. 1989. SSC1, an essential member of the yeast HSP70 multigene family, encodes a mitochondrial protein. Mol. Cell. Biol. 9:3000–3008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Craven RA, Egerton M, Stirling CJ. 1996. A novel Hsp70 of the yeast ER lumen is required for the efficient translocation of a number of protein precursors. EMBO J. 15:2640–2650 [PMC free article] [PubMed] [Google Scholar]
- 85. Creusot F, Verdiere J, Gaisne M, Slonimski PP. 1988. CYP1 (HAP1) regulator of oxygen-dependent gene expression in yeast. I. Overall organization of the protein sequence displays several novel structural domains. J. Mol. Biol. 204:263–276 [DOI] [PubMed] [Google Scholar]
- 86. Crowe JH. 2007. Trehalose as a “chemical chaperone”: fact and fantasy. Adv. Exp. Med. Biol. 594:143–158 [DOI] [PubMed] [Google Scholar]
- 87. Cyert MS. 2003. Calcineurin signaling in Saccharomyces cerevisiae: how yeast go crazy in response to stress. Biochem. Biophys. Res. Commun. 311:1143–1150 [DOI] [PubMed] [Google Scholar]
- 88. Cyr DM. 2008. Swapping nucleotides, tuning Hsp70. Cell 133:945–947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Daquinag A, Fadri M, Jung SY, Qin J, Kunz J. 2007. The yeast PH domain proteins Slm1 and Slm2 are targets of sphingolipid signaling during the response to heat stress. Mol. Cell. Biol. 27:633–650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Davidson JF, Whyte B, Bissinger PH, Schiestl RH. 1996. Oxidative stress is involved in heat-induced cell death in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U. S. A. 93:5116–5121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Decker CJ, Teixeira D, Parker R. 2007. Edc3p and a glutamine/asparagine-rich domain of Lsm4p function in processing body assembly in Saccharomyces cerevisiae. J. Cell Biol. 179:437–449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. de Jong WW, Leunissen JA, Voorter CE. 1993. Evolution of the alpha-crystallin/small heat-shock protein family. Mol. Biol. Evol. 10:103–126 [DOI] [PubMed] [Google Scholar]
- 93. de Keyzer J, Steel GJ, Hale SJ, Humphries D, Stirling CJ. 2009. Nucleotide binding by Lhs1p is essential for its nucleotide exchange activity and for function in vivo. J. Biol. Chem. 284:31564–31571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Delaunay A, Isnard AD, Toledano MB. 2000. H2O2 sensing through oxidation of the Yap1 transcription factor. EMBO J. 19:5157–5166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Delaunay A, Pflieger D, Barrault MB, Vinh J, Toledano MB. 2002. A thiol peroxidase is an H2O2 receptor and redox-transducer in gene activation. Cell 111:471–481 [DOI] [PubMed] [Google Scholar]
- 96. DeRisi JL, Iyer VR, Brown PO. 1997. Exploring the metabolic and genetic control of gene expression on a genomic scale. Science 278:680–686 [DOI] [PubMed] [Google Scholar]
- 97. Deshaies RJ, Koch BD, Werner-Washburne M, Craig EA, Schekman R. 1988. A subfamily of stress proteins facilitates translocation of secretory and mitochondrial precursor polypeptides. Nature 332:800–805 [DOI] [PubMed] [Google Scholar]
- 98. De Virgilio C, Hottiger T, Dominguez J, Boller T, Wiemken A. 1994. The role of trehalose synthesis for the acquisition of thermotolerance in yeast. I. Genetic evidence that trehalose is a thermoprotectant. Eur. J. Biochem. 219:179–186 [DOI] [PubMed] [Google Scholar]
- 99. De Wever V, Reiter W, Ballarini A, Ammerer G, Brocard C. 2005. A dual role for PP1 in shaping the Msn2-dependent transcriptional response to glucose starvation. EMBO J. 24:4115–4123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Dickson RC, Lester RL. 2002. Sphingolipid functions in Saccharomyces cerevisiae. Biochim. Biophys. Acta 1583:13–25 [DOI] [PubMed] [Google Scholar]
- 101. Dickson RC, et al. 1997. Sphingolipids are potential heat stress signals in Saccharomyces. J. Biol. Chem. 272:30196–30200 [DOI] [PubMed] [Google Scholar]
- 102. Diez S, Gomez BL, Restrepo A, Hay RJ, Hamilton AJ. 2002. Paracoccidioides brasiliensis 87-kilodalton antigen, a heat shock protein useful in diagnosis: characterization, purification, and detection in biopsy material via immunohistochemistry. J. Clin. Microbiol. 40:359–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. DiSalvo S, Serio TR. 2011. Insights into prion biology: integrating a protein misfolding pathway with its cellular environment. Prion 5:76–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Dombek KM, Kacherovsky N, Young ET. 2004. The Reg1-interacting proteins, Bmh1, Bmh2, Ssb1, and Ssb2, have roles in maintaining glucose repression in Saccharomyces cerevisiae. J. Biol. Chem. 279:39165–39174 [DOI] [PubMed] [Google Scholar]
- 105. Doyle SM, Wickner S. 2009. Hsp104 and ClpB: protein disaggregating machines. Trends Biochem. Sci. 34:40–48 [DOI] [PubMed] [Google Scholar]
- 106. Dragovic Z, Broadley SA, Shomura Y, Bracher A, Hartl FU. 2006. Molecular chaperones of the Hsp110 family act as nucleotide exchange factors of Hsp70s. EMBO J. 25:2519–2528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Dragovic Z, Shomura Y, Tzvetkov N, Hartl FU, Bracher A. 2006. Fes1p acts as a nucleotide exchange factor for the ribosome-associated molecular chaperone Ssb1p. Biol. Chem. 387:1593–1600 [DOI] [PubMed] [Google Scholar]
- 108. Drees BL, Grotkopp EK, Nelson HC. 1997. The GCN4 leucine zipper can functionally substitute for the heat shock transcription factor's trimerization domain. J. Mol. Biol. 273:61–74 [DOI] [PubMed] [Google Scholar]
- 109. D'Silva PD, Schilke B, Walter W, Andrew A, Craig EA. 2003. J protein cochaperone of the mitochondrial inner membrane required for protein import into the mitochondrial matrix. Proc. Natl. Acad. Sci. U. S. A. 100:13839–13844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. D'Silva PR, Schilke B, Walter W, Craig EA. 2005. Role of Pam16's degenerate J domain in protein import across the mitochondrial inner membrane. Proc. Natl. Acad. Sci. U. S. A. 102:12419–12424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Dubaquie Y, Looser R, Funfschilling U, Jeno P, Rospert S. 1998. Identification of in vivo substrates of the yeast mitochondrial chaperonins reveals overlapping but non-identical requirement for hsp60 and hsp10. EMBO J. 17:5868–5876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Duina AA, Chang HC, Marsh JA, Lindquist S, Gaber RF. 1996. A cyclophilin function in Hsp90-dependent signal transduction. Science 274:1713–1715 [DOI] [PubMed] [Google Scholar]
- 113. Duina AA, Kalton HM, Gaber RF. 1998. Requirement for Hsp90 and a CyP-40-type cyclophilin in negative regulation of the heat shock response. J. Biol. Chem. 273:18974–18978 [DOI] [PubMed] [Google Scholar]
- 114. Dutkiewicz R, et al. 2003. Ssq1, a mitochondrial Hsp70 involved in iron-sulfur (Fe/S) center biogenesis. Similarities to and differences from its bacterial counterpart. J. Biol. Chem. 278:29719–29727 [DOI] [PubMed] [Google Scholar]
- 115. Duttagupta R, Vasudevan S, Wilusz CJ, Peltz SW. 2003. A yeast homologue of Hsp70, Ssa1p, regulates turnover of the MFA2 transcript through its AU-rich 3′ untranslated region. Mol. Cell. Biol. 23:2623–2632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Easton DP, Kaneko Y, Subjeck JR. 2000. The hsp110 and Grp170 stress proteins: newly recognized relatives of the Hsp70s. Cell Stress Chaperones 5:276–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Echtenkamp FJ, et al. 2011. Global functional map of the p23 molecular chaperone reveals an extensive cellular network. Mol. Cell 43:229–241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Ellgaard L, Molinari M, Helenius A. 1999. Setting the standards: quality control in the secretory pathway. Science 286:1882–1888 [DOI] [PubMed] [Google Scholar]
- 119. Elliott B, Haltiwanger RS, Futcher B. 1996. Synergy between trehalose and Hsp104 for thermotolerance in Saccharomyces cerevisiae. Genetics 144:923–933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Ellis J. 1987. Proteins as molecular chaperones. Nature 328:378–379 [DOI] [PubMed] [Google Scholar]
- 121. Erjavec N, Larsson L, Grantham J, Nystrom T. 2007. Accelerated aging and failure to segregate damaged proteins in Sir2 mutants can be suppressed by overproducing the protein aggregation-remodeling factor Hsp104p. Genes Dev. 21:2410–2421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Eroles P, Sentandreu M, Elorza MV, Sentandreu R. 1995. Cloning of a DNA fragment encoding part of a 70-kDa heat shock protein of Candida albicans. FEMS Microbiol. Lett. 128:95–100 [DOI] [PubMed] [Google Scholar]
- 123. Erzberger JP, Berger JM. 2006. Evolutionary relationships and structural mechanisms of AAA+ proteins. Annu. Rev. Biophys. Biomol. Struct. 35:93–114 [DOI] [PubMed] [Google Scholar]
- 124. Estruch F. 2000. Stress-controlled transcription factors, stress-induced genes and stress tolerance in budding yeast. FEMS Microbiol. Rev. 24:469–486 [DOI] [PubMed] [Google Scholar]
- 125. Estruch F. 1991. The yeast putative transcriptional repressor RGM1 is a proline-rich zinc finger protein. Nucleic Acids Res. 19:4873–4877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Estruch F, Carlson M. 1993. Two homologous zinc finger genes identified by multicopy suppression in a SNF1 protein kinase mutant of Saccharomyces cerevisiae. Mol. Cell. Biol. 13:3872–3881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Fang Y, Fliss AE, Rao J, Caplan AJ. 1998. SBA1 encodes a yeast hsp90 cochaperone that is homologous to vertebrate p23 proteins. Mol. Cell. Biol. 18:3727–3734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Farquhar R, et al. 1991. Protein disulfide isomerase is essential for viability in Saccharomyces cerevisiae. Gene 108:81–89 [DOI] [PubMed] [Google Scholar]
- 129. Feldheim D, Rothblatt J, Schekman R. 1992. Topology and functional domains of Sec63p, an endoplasmic reticulum membrane protein required for secretory protein translocation. Mol. Cell. Biol. 12:3288–3296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Ferguson SB, et al. 2005. Protein kinase A regulates constitutive expression of small heat-shock genes in an Msn2/4p-independent and Hsf1p-dependent manner in Saccharomyces cerevisiae. Genetics 169:1203–1214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Flaherty KM, DeLuca-Flaherty C, McKay DB. 1990. Three-dimensional structure of the ATPase fragment of a 70K heat-shock cognate protein. Nature 346:623–628 [DOI] [PubMed] [Google Scholar]
- 132. Flick KE, Gonzalez L, Jr, Harrison CJ, Nelson HC. 1994. Yeast heat shock transcription factor contains a flexible linker between the DNA-binding and trimerization domains. Implications for DNA binding by trimeric proteins. J. Biol. Chem. 269:12475–12481 [PubMed] [Google Scholar]
- 133. Flom G, Behal RH, Rosen L, Cole DG, Johnson JL. 2007. Definition of the minimal fragments of Sti1 required for dimerization, interaction with Hsp70 and Hsp90 and in vivo functions. Biochem. J. 404:159–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Flom GA, Lemieszek M, Fortunato EA, Johnson JL. 2008. Farnesylation of Ydj1 is required for in vivo interaction with Hsp90 client proteins. Mol. Biol. Cell 19:5249–5258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Forafonov F, et al. 2008. p23/Sba1p protects against Hsp90 inhibitors independently of its intrinsic chaperone activity. Mol. Cell. Biol. 28:3446–3456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Fraschini R, Venturetti M, Chiroli E, Piatti S. 2008. The spindle position checkpoint: how to deal with spindle misalignment during asymmetric cell division in budding yeast. Biochem. Soc. Trans. 36:416–420 [DOI] [PubMed] [Google Scholar]
- 137. Frazier AE, et al. 2004. Pam16 has an essential role in the mitochondrial protein import motor. Nat. Struct. Mol. Biol. 11:226–233 [DOI] [PubMed] [Google Scholar]
- 138. Freedman RB, Hirst TR, Tuite MF. 1994. Protein disulphide isomerase: building bridges in protein folding. Trends Biochem. Sci. 19:331–336 [DOI] [PubMed] [Google Scholar]
- 139. Freeman BC, Myers MP, Schumacher R, Morimoto RI. 1995. Identification of a regulatory motif in Hsp70 that affects ATPase activity, substrate binding and interaction with HDJ-1. EMBO J. 14:2281–2292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Frydman J. 2001. Folding of newly translated proteins in vivo: the role of molecular chaperones. Annu. Rev. Biochem. 70:603–647 [DOI] [PubMed] [Google Scholar]
- 141. Frydman J, et al. 1992. Function in protein folding of TRiC, a cytosolic ring complex containing TCP-1 and structurally related subunits. EMBO J. 11:4767–4778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Gall WE, et al. 2000. The auxilin-like phosphoprotein Swa2p is required for clathrin function in yeast. Curr. Biol. 10:1349–1358 [DOI] [PubMed] [Google Scholar]
- 143. Gambill BD, et al. 1993. A dual role for mitochondrial heat shock protein 70 in membrane translocation of preproteins. J. Cell Biol. 123:109–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Gardner BM, Walter P. 2011. Unfolded proteins are Ire1-activating ligands that directly induce the unfolded protein response. Science 333:1891–1894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Garreau H, et al. 2000. Hyperphosphorylation of Msn2p and Msn4p in response to heat shock and the diauxic shift is inhibited by cAMP in Saccharomyces cerevisiae. Microbiology 146(Pt 9):2113–2120 [DOI] [PubMed] [Google Scholar]
- 146. Garrett S, Menold MM, Broach JR. 1991. The Saccharomyces cerevisiae YAK1 gene encodes a protein kinase that is induced by arrest early in the cell cycle. Mol. Cell. Biol. 11:4045–4052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Gasch AP, et al. 2000. Genomic expression programs in the response of yeast cells to environmental changes. Mol. Biol. Cell 11:4241–4257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Gasch AP, Werner-Washburne M. 2002. The genomics of yeast responses to environmental stress and starvation. Funct. Integr. Genomics 2:181–192 [DOI] [PubMed] [Google Scholar]
- 149. Gautschi M, et al. 2001. RAC, a stable ribosome-associated complex in yeast formed by the DnaK-DnaJ homologs Ssz1p and zuotin. Proc. Natl. Acad. Sci. U. S. A. 98:3762–3767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Germaniuk A, Liberek K, Marszalek J. 2002. A bichaperone (Hsp70-Hsp78) system restores mitochondrial DNA synthesis following thermal inactivation of Mip1p polymerase. J. Biol. Chem. 277:27801–27808 [DOI] [PubMed] [Google Scholar]
- 151. Gething MJ, McCammon K, Sambrook J. 1986. Expression of wild-type and mutant forms of influenza hemagglutinin: the role of folding in intracellular transport. Cell 46:939–950 [DOI] [PubMed] [Google Scholar]
- 152. Ghaemmaghami S, et al. 2003. Global analysis of protein expression in yeast. Nature 425:737–741 [DOI] [PubMed] [Google Scholar]
- 153. Giaever G, et al. 2002. Functional profiling of the Saccharomyces cerevisiae genome. Nature 418:387–391 [DOI] [PubMed] [Google Scholar]
- 154. Gillece P, Luz JM, Lennarz WJ, de La Cruz FJ, Romisch K. 1999. Export of a cysteine-free misfolded secretory protein from the endoplasmic reticulum for degradation requires interaction with protein disulfide isomerase. J. Cell Biol. 147:1443–1456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Glick BS. 1995. Can Hsp70 proteins act as force-generating motors? Cell 80:11–14 [DOI] [PubMed] [Google Scholar]
- 156. Glover JR, Lindquist S. 1998. Hsp104, Hsp70, and Hsp40: a novel chaperone system that rescues previously aggregated proteins. Cell 94:73–82 [DOI] [PubMed] [Google Scholar]
- 157. Gong Y, et al. 2009. An atlas of chaperone-protein interactions in Saccharomyces cerevisiae: implications to protein folding pathways in the cell. Mol. Syst. Biol. 5:275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. Gorner W, et al. 1998. Nuclear localization of the C2H2 zinc finger protein Msn2p is regulated by stress and protein kinase A activity. Genes Dev. 12:586–597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Gorner W, et al. 2002. Acute glucose starvation activates the nuclear localization signal of a stress-specific yeast transcription factor. EMBO J. 21:135–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160. Gorner W, Schuller C, Ruis H. 1999. Being at the right place at the right time: the role of nuclear transport in dynamic transcriptional regulation in yeast. Biol. Chem. 380:147–150 [DOI] [PubMed] [Google Scholar]
- 161. Greene MK, Maskos K, Landry SJ. 1998. Role of the J-domain in the cooperation of Hsp40 with Hsp70. Proc. Natl. Acad. Sci. U. S. A. 95:6108–6113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. Gross C, Watson K. 1998. Transcriptional and translational regulation of major heat shock proteins and patterns of trehalose mobilization during hyperthermic recovery in repressed and derepressed Saccharomyces cerevisiae. Can. J. Microbiol. 44:341–350 [DOI] [PubMed] [Google Scholar]
- 163. Grousl T, et al. 2009. Robust heat shock induces eIF2alpha-phosphorylation-independent assembly of stress granules containing eIF3 and 40S ribosomal subunits in budding yeast, Saccharomyces cerevisiae. J. Cell Sci. 122:2078–2088 [DOI] [PubMed] [Google Scholar]
- 164. Hagen DC, McCaffrey G, Sprague GF., Jr 1991. Pheromone response elements are necessary and sufficient for basal and pheromone-induced transcription of the FUS1 gene of Saccharomyces cerevisiae. Mol. Cell. Biol. 11:2952–2961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165. Hahn JS, Hu Z, Thiele DJ, Iyer VR. 2004. Genome-wide analysis of the biology of stress responses through heat shock transcription factor. Mol. Cell. Biol. 24:5249–5256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166. Hahn JS, Thiele DJ. 2004. Activation of the Saccharomyces cerevisiae heat shock transcription factor under glucose starvation conditions by Snf1 protein kinase. J. Biol. Chem. 279:5169–5176 [DOI] [PubMed] [Google Scholar]
- 167. Hainzl O, Lapina MC, Buchner J, Richter K. 2009. The charged linker region is an important regulator of Hsp90 function. J. Biol. Chem. 284:22559–22567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168. Hamman BD, Hendershot LM, Johnson AE. 1998. BiP maintains the permeability barrier of the ER membrane by sealing the lumenal end of the translocon pore before and early in translocation. Cell 92:747–758 [DOI] [PubMed] [Google Scholar]
- 169. Harris N, MacLean M, Hatzianthis K, Panaretou B, Piper PW. 2001. Increasing Saccharomyces cerevisiae stress resistance, through the overactivation of the heat shock response resulting from defects in the Hsp90 chaperone, does not extend replicative life span but can be associated with slower chronological ageing of nondividing cells. Mol. Genet. Genomics 265:258–263 [DOI] [PubMed] [Google Scholar]
- 170. Harrison CJ, Hayer-Hartl M, Di Liberto M, Hartl F, Kuriyan J. 1997. Crystal structure of the nucleotide exchange factor GrpE bound to the ATPase domain of the molecular chaperone DnaK. Science 276:431–435 [DOI] [PubMed] [Google Scholar]
- 171. Hartl FU, Hayer-Hartl M. 2002. Molecular chaperones in the cytosol: from nascent chain to folded protein. Science 295:1852–1858 [DOI] [PubMed] [Google Scholar]
- 172. Hashikawa N, Sakurai H. 2004. Phosphorylation of the yeast heat shock transcription factor is implicated in gene-specific activation dependent on the architecture of the heat shock element. Mol. Cell. Biol. 24:3648–3659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173. Hashikawa N, Yamamoto N, Sakurai H. 2007. Different mechanisms are involved in the transcriptional activation by yeast heat shock transcription factor through two different types of heat shock elements. J. Biol. Chem. 282:10333–10340 [DOI] [PubMed] [Google Scholar]
- 174. Haslbeck M. 2006. Recombinant expression and in vitro refolding of the yeast small heat shock protein Hsp42. Int. J. Biol. Macromol. 38:107–114 [DOI] [PubMed] [Google Scholar]
- 175. Haslbeck M, et al. 2004. Hsp42 is the general small heat shock protein in the cytosol of Saccharomyces cerevisiae. EMBO J. 23:638–649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176. Haslbeck M, et al. 2004. A domain in the N-terminal part of Hsp26 is essential for chaperone function and oligomerization. J. Mol. Biol. 343:445–455 [DOI] [PubMed] [Google Scholar]
- 177. Haslbeck M, Miess A, Stromer T, Walter S, Buchner J. 2005. Disassembling protein aggregates in the yeast cytosol. The cooperation of Hsp26 with Ssa1 and Hsp104. J. Biol. Chem. 280:23861–23868 [DOI] [PubMed] [Google Scholar]
- 178. Haslbeck M, et al. 1999. Hsp26: a temperature-regulated chaperone. EMBO J. 18:6744–6751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179. Haslberger T, Bukau B, Mogk A. 2010. Towards a unifying mechanism for ClpB/Hsp104-mediated protein disaggregation and prion propagation. Biochem. Cell Biol. 88:63–75 [DOI] [PubMed] [Google Scholar]
- 180. Hawle P, et al. 2007. Cdc37p is required for stress-induced high-osmolarity glycerol and protein kinase C mitogen-activated protein kinase pathway functionality by interaction with Hog1p and Slt2p (Mpk1p). Eukaryot. Cell 6:521–532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181. Heck JW, Cheung SK, Hampton RY. 2010. Cytoplasmic protein quality control degradation mediated by parallel actions of the E3 ubiquitin ligases Ubr1 and San1. Proc. Natl. Acad. Sci. U. S. A. 107:1106–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182. Hessling M, Richter K, Buchner J. 2009. Dissection of the ATP-induced conformational cycle of the molecular chaperone Hsp90. Nat. Struct. Mol. Biol. 16:287–293 [DOI] [PubMed] [Google Scholar]
- 183. Hettema EH, Tabak HF. 2000. Transport of fatty acids and metabolites across the peroxisomal membrane. Biochim. Biophys. Acta 1486:18–27 [DOI] [PubMed] [Google Scholar]
- 184. Heyrovska N, Frydman J, Hohfeld J, Hartl FU. 1998. Directionality of polypeptide transfer in the mitochondrial pathway of chaperone-mediated protein folding. Biol. Chem. 379:301–309 [DOI] [PubMed] [Google Scholar]
- 185. Hieronymus H, et al. 2006. Gene expression signature-based chemical genomic prediction identifies a novel class of HSP90 pathway modulators. Cancer Cell 10:321–330 [DOI] [PubMed] [Google Scholar]
- 186. Ho AK, Raczniak GA, Ives EB, Wente SR. 1998. The integral membrane protein Snl1p is genetically linked to yeast nuclear pore complex function. Mol. Biol. Cell 9:355–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187. Hohfeld J, Hartl FU. 1994. Role of the chaperonin cofactor Hsp10 in protein folding and sorting in yeast mitochondria. J. Cell Biol. 126:305–315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188. Hoj A, Jakobsen BK. 1994. A short element required for turning off heat shock transcription factor: evidence that phosphorylation enhances deactivation. EMBO J. 13:2617–2624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189. Hon T, Lee HC, Hu Z, Iyer VR, Zhang L. 2005. The heme activator protein HapI represses transcription by a heme-independent mechanism in Saccharomyces cerevisiae. Genetics 169:1343–1352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190. Horst M, et al. 1997. Sequential action of two hsp70 complexes during protein import into mitochondria. EMBO J. 16:1842–1849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191. Horton LE, James P, Craig EA, Hensold JO. 2001. The yeast hsp70 homologue Ssa is required for translation and interacts with Sis1 and Pab1 on translating ribosomes. J. Biol. Chem. 276:14426–14433 [DOI] [PubMed] [Google Scholar]
- 192. Hottiger T, De Virgilio C, Hall MN, Boller T, Wiemken A. 1994. The role of trehalose synthesis for the acquisition of thermotolerance in yeast. II. Physiological concentrations of trehalose increase the thermal stability of proteins in vitro. Eur. J. Biochem. 219:187–193 [DOI] [PubMed] [Google Scholar]
- 193. Hubl ST, Owens JC, Nelson HC. 1994. Mutational analysis of the DNA-binding domain of yeast heat shock transcription factor. Nat. Struct. Biol. 1:615–620 [DOI] [PubMed] [Google Scholar]
- 194. Hundley H, et al. 2002. The in vivo function of the ribosome-associated Hsp70, Ssz1, does not require its putative peptide-binding domain. Proc. Natl. Acad. Sci. U. S. A. 99:4203–4208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195. Ihara Y, Cohen-Doyle MF, Saito Y, Williams DB. 1999. Calnexin discriminates between protein conformational states and functions as a molecular chaperone in vitro. Mol. Cell 4:331–341 [DOI] [PubMed] [Google Scholar]
- 196. Imai J, Yahara I. 2000. Role of HSP90 in salt stress tolerance via stabilization and regulation of calcineurin. Mol. Cell. Biol. 20:9262–9270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197. Imazu H, Sakurai H. 2005. Saccharomyces cerevisiae heat shock transcription factor regulates cell wall remodeling in response to heat shock. Eukaryot. Cell 4:1050–1056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198. Izawa S, Takemura R, Inoue Y. 2004. Gle2p is essential to induce adaptation of the export of bulk poly(A)+ mRNA to heat shock in Saccharomyces cerevisiae. J. Biol. Chem. 279:35469–35478 [DOI] [PubMed] [Google Scholar]
- 199. Jakob CA, Burda P, te Heesen S, Aebi M, Roth J. 1998. Genetic tailoring of N-linked oligosaccharides: the role of glucose residues in glycoprotein processing of Saccharomyces cerevisiae in vivo. Glycobiology 8:155–164 [DOI] [PubMed] [Google Scholar]
- 200. Jakob U, Gaestel M, Engel K, Buchner J. 1993. Small heat shock proteins are molecular chaperones. J. Biol. Chem. 268:1517–1520 [PubMed] [Google Scholar]
- 201. Jakobsen BK, Pelham HR. 1991. A conserved heptapeptide restrains the activity of the yeast heat shock transcription factor. EMBO J. 10:369–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202. James P, Pfund C, Craig EA. 1997. Functional specificity among Hsp70 molecular chaperones. Science 275:387–389 [DOI] [PubMed] [Google Scholar]
- 203. Jenkins GM, Hannun YA. 2001. Role for de novo sphingoid base biosynthesis in the heat-induced transient cell cycle arrest of Saccharomyces cerevisiae. J. Biol. Chem. 276:8574–8581 [DOI] [PubMed] [Google Scholar]
- 204. Jenkins GM, et al. 1997. Involvement of yeast sphingolipids in the heat stress response of Saccharomyces cerevisiae. J. Biol. Chem. 272:32566–32572 [DOI] [PubMed] [Google Scholar]
- 205. Jermy AJ, Willer M, Davis E, Wilkinson BM, Stirling CJ. 2006. The Brl domain in Sec63p is required for assembly of functional endoplasmic reticulum translocons. J. Biol. Chem. 281:7899–7906 [DOI] [PubMed] [Google Scholar]
- 206. Jiang J, et al. 2007. Structural basis of J cochaperone binding and regulation of Hsp70. Mol. Cell 28:422–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207. Johnson BD, Schumacher RJ, Ross ED, Toft DO. 1998. Hop modulates Hsp70/Hsp90 interactions in protein folding. J. Biol. Chem. 273:3679–3686 [DOI] [PubMed] [Google Scholar]
- 208. Johnson JL. 2012. Evolution and function of diverse Hsp90 homologs and cochaperone proteins. Biochim. Biophys. Acta 1823:607–613 [DOI] [PubMed] [Google Scholar]
- 209. Johnson JL, Craig EA. 2001. An essential role for the substrate-binding region of Hsp40s in Saccharomyces cerevisiae. J. Cell Biol. 152:851–856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210. Johnson JL, Craig EA. 2000. A role for the Hsp40 Ydj1 in repression of basal steroid receptor activity in yeast. Mol. Cell. Biol. 20:3027–3036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211. Johnson JL, Halas A, Flom G. 2007. Nucleotide-dependent interaction of Saccharomyces cerevisiae Hsp90 with the cochaperone proteins Sti1, Cpr6, and Sba1. Mol. Cell. Biol. 27:768–776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212. Johnson MK. 1998. Iron-sulfur proteins: new roles for old clusters. Curr. Opin. Chem. Biol. 2:173–181 [DOI] [PubMed] [Google Scholar]
- 213. Juretschke J, Menssen R, Sickmann A, Wolf DH. 2010. The Hsp70 chaperone Ssa1 is essential for catabolite induced degradation of the gluconeogenic enzyme fructose-1,6-bisphosphatase. Biochem. Biophys. Res. Commun. 397:447–452 [DOI] [PubMed] [Google Scholar]
- 214. Kabani M, Beckerich JM, Brodsky JL. 2002. Nucleotide exchange factor for the yeast Hsp70 molecular chaperone Ssa1p. Mol. Cell. Biol. 22:4677–4689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215. Kabani M, Beckerich JM, Gaillardin C. 2000. Sls1p stimulates Sec63p-mediated activation of Kar2p in a conformation-dependent manner in the yeast endoplasmic reticulum. Mol. Cell. Biol. 20:6923–6934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216. Kabani M, McLellan C, Raynes DA, Guerriero V, Brodsky JL. 2002. HspBP1, a homologue of the yeast Fes1 and Sls1 proteins, is an Hsc70 nucleotide exchange factor. FEBS Lett. 531:339–342 [DOI] [PubMed] [Google Scholar]
- 217. Kabir MA, Sherman F. 2008. Overexpressed ribosomal proteins suppress defective chaperonins in Saccharomyces cerevisiae. FEMS Yeast Res. 8:1236–1244 [DOI] [PubMed] [Google Scholar]
- 218. Kaganovich D, Kopito R, Frydman J. 2008. Misfolded proteins partition between two distinct quality control compartments. Nature 454:1088–1095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219. Kakeya H, et al. 1997. A 77-kilodalton protein of Cryptococcus neoformans, a member of the heat shock protein 70 family, is a major antigen detected in the sera of mice with pulmonary cryptococcosis. Infect. Immun. 65:1653–1658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220. Kamada Y, Jung US, Piotrowski J, Levin DE. 1995. The protein kinase C-activated MAP kinase pathway of Saccharomyces cerevisiae mediates a novel aspect of the heat shock response. Genes Dev. 9:1559–1571 [DOI] [PubMed] [Google Scholar]
- 221. Kampinga HH, Craig EA. 2010. The HSP70 chaperone machinery: J proteins as drivers of functional specificity. Nat. Rev. Mol. Cell Biol. 11:579–592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222. Kang PJ, et al. 1990. Requirement for hsp70 in the mitochondrial matrix for translocation and folding of precursor proteins. Nature 348:137–143 [DOI] [PubMed] [Google Scholar]
- 223. Kerscher O, Felberbaum R, Hochstrasser M. 2006. Modification of proteins by ubiquitin and ubiquitin-like proteins. Annu. Rev. Cell Dev. Biol. 22:159–180 [DOI] [PubMed] [Google Scholar]
- 224. Kim R, Saxena S, Gordon DM, Pain D, Dancis A. 2001. J-domain protein, Jac1p, of yeast mitochondria required for iron homeostasis and activity of Fe-S cluster proteins. J. Biol. Chem. 276:17524–17532 [DOI] [PubMed] [Google Scholar]
- 225. Kim S, Schilke B, Craig EA, Horwich AL. 1998. Folding in vivo of a newly translated yeast cytosolic enzyme is mediated by the SSA class of cytosolic yeast Hsp70 proteins. Proc. Natl. Acad. Sci. U. S. A. 95:12860–12865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226. Kim SY, Craig EA. 2005. Broad sensitivity of Saccharomyces cerevisiae lacking ribosome-associated chaperone Ssb or Zuo1 to cations, including aminoglycosides. Eukaryot. Cell 4:82–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227. Kimata Y, et al. 2003. Genetic evidence for a role of BiP/Kar2 that regulates Ire1 in response to accumulation of unfolded proteins. Mol. Biol. Cell 14:2559–2569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228. Kimura T, et al. 2004. Functional differences between human and yeast protein disulfide isomerase family proteins. Biochem. Biophys. Res. Commun. 320:359–365 [DOI] [PubMed] [Google Scholar]
- 229. Kimura T, et al. 2005. Interactions among yeast protein-disulfide isomerase proteins and endoplasmic reticulum chaperone proteins influence their activities. J. Biol. Chem. 280:31438–31441 [DOI] [PubMed] [Google Scholar]
- 230. Knight SA, Sepuri NB, Pain D, Dancis A. 1998. Mt-Hsp70 homolog, Ssc2p, required for maturation of yeast frataxin and mitochondrial iron homeostasis. J. Biol. Chem. 273:18389–18393 [DOI] [PubMed] [Google Scholar]
- 231. Kobayashi N, McEntee K. 1990. Evidence for a heat shock transcription factor-independent mechanism for heat shock induction of transcription in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U. S. A. 87:6550–6554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232. Kohno K, Normington K, Sambrook J, Gething MJ, Mori K. 1993. The promoter region of the yeast KAR2 (BiP) gene contains a regulatory domain that responds to the presence of unfolded proteins in the endoplasmic reticulum. Mol. Cell. Biol. 13:877–890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233. Kozany C, Mokranjac D, Sichting M, Neupert W, Hell K. 2004. The J domain-related cochaperone Tim16 is a constituent of the mitochondrial TIM23 preprotein translocase. Nat. Struct. Mol. Biol. 11:234–241 [DOI] [PubMed] [Google Scholar]
- 234. Kramer G, et al. 2004. Functional dissection of Escherichia coli trigger factor: unraveling the function of individual domains. J. Bacteriol. 186:3777–3784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235. Krzewska J, Langer T, Liberek K. 2001. Mitochondrial Hsp78, a member of the Clp/Hsp100 family in Saccharomyces cerevisiae, cooperates with Hsp70 in protein refolding. FEBS Lett. 489:92–96 [DOI] [PubMed] [Google Scholar]
- 236. Kusakabe T, Koga K, Sugimoto Y. 1994. Isolation and characterization of cDNA and genomic promoter region for a heat shock protein 30 from Aspergillus nidulans. Biochim. Biophys. Acta 1219:555–558 [DOI] [PubMed] [Google Scholar]
- 237. LaFayette SL, et al. 2010. PKC signaling regulates drug resistance of the fungal pathogen Candida albicans via circuitry comprised of Mkc1, calcineurin, and Hsp90. PLoS Pathog. 6:e1001069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238. Laloraya S, Gambill BD, Craig EA. 1994. A role for a eukaryotic GrpE-related protein, Mge1p, in protein translocation. Proc. Natl. Acad. Sci. U. S. A. 91:6481–6485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239. Lan C, Lee HC, Tang S, Zhang L. 2004. A novel mode of chaperone action: heme activation of HapI by enhanced association of Hsp90 with the repressed Hsp70-HapI complex. J. Biol. Chem. 279:27607–27612 [DOI] [PubMed] [Google Scholar]
- 240. La Valle R, et al. 1995. Molecular cloning and expression of a 70-kilodalton heat shock protein of Candida albicans. Infect. Immun. 63:4039–4045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241. Lawson B, Brewer JW, Hendershot LM. 1998. Geldanamycin, an hsp90/GRP94-binding drug, induces increased transcription of endoplasmic reticulum (ER) chaperones via the ER stress pathway. J. Cell. Physiol. 174:170–178 [DOI] [PubMed] [Google Scholar]
- 242. Lee DH, Goldberg AL. 1996. Selective inhibitors of the proteasome-dependent and vacuolar pathways of protein degradation in Saccharomyces cerevisiae. J. Biol. Chem. 271:27280–27284 [DOI] [PubMed] [Google Scholar]
- 243. Lee P, Cho BR, Joo HS, Hahn JS. 2008. Yeast Yak1 kinase, a bridge between PKA and stress-responsive transcription factors, Hsf1 and Msn2/Msn4. Mol. Microbiol. 70:882–895 [DOI] [PubMed] [Google Scholar]
- 244. Lee P, Paik SM, Shin CS, Huh WK, Hahn JS. 2011. Regulation of yeast Yak1 kinase by PKA and autophosphorylation-dependent 14-3-3 binding. Mol. Microbiol. 79:633–646 [DOI] [PubMed] [Google Scholar]
- 245. Lee P, et al. 2002. The Cdc37 protein kinase-binding domain is sufficient for protein kinase activity and cell viability. J. Cell Biol. 159:1051–1059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246. Lee S, et al. 2000. The yeast heat shock transcription factor changes conformation in response to superoxide and temperature. Mol. Biol. Cell 11:1753–1764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247. Lee S, Sielaff B, Lee J, Tsai FT. 2010. CryoEM structure of Hsp104 and its mechanistic implication for protein disaggregation. Proc. Natl. Acad. Sci. U. S. A. 107:8135–8140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248. Lenssen E, et al. 2005. The Ccr4-Not complex independently controls both Msn2-dependent transcriptional activation—via a newly identified Glc7/Bud14 type I protein phosphatase module—and TFIID promoter distribution. Mol. Cell. Biol. 25:488–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249. Leonhardt SA, Fearson K, Danese PN, Mason TL. 1993. HSP78 encodes a yeast mitochondrial heat shock protein in the Clp family of ATP-dependent proteases. Mol. Cell. Biol. 13:6304–6313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250. Levin DE. 2005. Cell wall integrity signaling in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 69:262–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251. Levine RL. 2002. Carbonyl modified proteins in cellular regulation, aging, and disease. Free Radic. Biol. Med. 32:790–796 [DOI] [PubMed] [Google Scholar]
- 252. Levy EJ, McCarty J, Bukau B, Chirico WJ. 1995. Conserved ATPase and luciferase refolding activities between bacteria and yeast Hsp70 chaperones and modulators. FEBS Lett. 368:435–440 [DOI] [PubMed] [Google Scholar]
- 253. Li J, Richter K, Buchner J. 2011. Mixed Hsp90-cochaperone complexes are important for the progression of the reaction cycle. Nat. Struct. Mol. Biol. 18:61–66 [DOI] [PubMed] [Google Scholar]
- 254. Li XS, Reddy MS, Baev D, Edgerton M. 2003. Candida albicans Ssa1/2p is the cell envelope binding protein for human salivary histatin 5. J. Biol. Chem. 278:28553–28561 [DOI] [PubMed] [Google Scholar]
- 255. Li XS, Sun JN, Okamoto-Shibayama K, Edgerton M. 2006. Candida albicans cell wall Ssa proteins bind and facilitate import of salivary histatin 5 required for toxicity. J. Biol. Chem. 281:22453–22463 [DOI] [PubMed] [Google Scholar]
- 256. Li Y, et al. 2004. The presequence translocase-associated protein import motor of mitochondria. Pam16 functions in an antagonistic manner to Pam18. J. Biol. Chem. 279:38047–38054 [DOI] [PubMed] [Google Scholar]
- 257. Lindquist S. 1980. Varying patterns of protein synthesis in Drosophila during heat shock: implications for regulation. Dev. Biol. 77:463–479 [DOI] [PubMed] [Google Scholar]
- 258. Lindquist S, Kim G. 1996. Heat-shock protein 104 expression is sufficient for thermotolerance in yeast. Proc. Natl. Acad. Sci. U. S. A. 93:5301–5306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259. Littlefield O, Nelson HC. 1999. A new use for the ‘wing’ of the ‘winged’ helix-turn-helix motif in the HSF-DNA cocrystal. Nat. Struct. Mol. Biol. 6:464–470 [DOI] [PubMed] [Google Scholar]
- 260. Liu B, et al. 2011. Segregation of protein aggregates involves actin and the polarity machinery. Cell 147:959–961 [DOI] [PubMed] [Google Scholar]
- 261. Liu Q, Krzewska J, Liberek K, Craig EA. 2001. Mitochondrial Hsp70 Ssc1: role in protein folding. J. Biol. Chem. 276:6112–6118 [DOI] [PubMed] [Google Scholar]
- 262. Liu XD, Thiele DJ. 1996. Oxidative stress induced heat shock factor phosphorylation and HSF-dependent activation of yeast metallothionein gene transcription. Genes Dev. 10:592–603 [DOI] [PubMed] [Google Scholar]
- 263. Liu XD, Morano KA, Thiele DJ. 1999. The yeast Hsp110 family member, Sse1, is an Hsp90 cochaperone. J. Biol. Chem. 274:26654–26660 [DOI] [PubMed] [Google Scholar]
- 264. Reference deleted.
- 265. Liu Z, Butow RA. 2006. Mitochondrial retrograde signaling. Annu. Rev. Genet. 40:159–185 [DOI] [PubMed] [Google Scholar]
- 266. Lopez N, Halladay J, Walter W, Craig EA. 1999. SSB, encoding a ribosome-associated chaperone, is coordinately regulated with ribosomal protein genes. J. Bacteriol. 181:3136–3143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267. Lopez-Ribot JL, Alloush HM, Masten BJ, Chaffin WL. 1996. Evidence for presence in the cell wall of Candida albicans of a protein related to the hsp70 family. Infect. Immun. 64:3333–3340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268. Louvion JF, Abbas-Terki T, Picard D. 1998. Hsp90 is required for pheromone signaling in yeast. Mol. Biol. Cell 9:3071–3083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269. Lum R, Niggemann M, Glover JR. 2008. Peptide and protein binding in the axial channel of Hsp104. Insights into the mechanism of protein unfolding. J. Biol. Chem. 283:30139–30150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270. Lussier M, et al. 1997. Large scale identification of genes involved in cell surface biosynthesis and architecture in Saccharomyces cerevisiae. Genetics 147:435–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271. Lutz T, Westermann B, Neupert W, Herrmann JM. 2001. The mitochondrial proteins Ssq1 and Jac1 are required for the assembly of iron sulfur clusters in mitochondria. J. Mol. Biol. 307:815–825 [DOI] [PubMed] [Google Scholar]
- 272. Lyles MM, Gilbert HF. 1991. Catalysis of the oxidative folding of ribonuclease A by protein disulfide isomerase: pre-steady-state kinetics and the utilization of the oxidizing equivalents of the isomerase. Biochemistry 30:619–625 [DOI] [PubMed] [Google Scholar]
- 273. Lyman SK, Schekman R. 1995. Interaction between BiP and Sec63p is required for the completion of protein translocation into the ER of Saccharomyces cerevisiae. J. Cell Biol. 131:1163–1171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274. Mackay RG, Helsen CW, Tkach JM, Glover JR. 2008. The C-terminal extension of Saccharomyces cerevisiae Hsp104 plays a role in oligomer assembly. Biochemistry 47:1918–1927 [DOI] [PubMed] [Google Scholar]
- 275. Malcher M, Schladebeck S, Mosch HU. 2011. The Yak1 protein kinase lies at the center of a regulatory cascade affecting adhesive growth and stress resistance in Saccharomyces cerevisiae. Genetics 187:717–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276. Mandal AK, et al. 2010. Hsp110 chaperones control client fate determination in the hsp70-Hsp90 chaperone system. Mol. Biol. Cell 21:1439–1448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277. Mandal AK, et al. 2007. Cdc37 has distinct roles in protein kinase quality control that protect nascent chains from degradation and promote posttranslational maturation. J. Cell Biol. 176:319–328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278. Maneu V, Cervera AM, Martinez JP, Gozalbo D. 1997. Molecular cloning of a Candida albicans gene (SSB1) coding for a protein related to the Hsp70 family. Yeast 13:677–681 [DOI] [PubMed] [Google Scholar]
- 279. Maneu V, Roig P, Gozalbo D. 2000. Complementation of Saccharomyces cerevisiae mutations in genes involved in translation and protein folding (EFB1 and SSB1) with Candida albicans cloned genes. Res. Microbiol. 151:739–746 [DOI] [PubMed] [Google Scholar]
- 280. Martin H, Arroyo J, Sanchez M, Molina M, Nombela C. 1993. Activity of the yeast MAP kinase homologue Slt2 is critically required for cell integrity at 37°C. Mol. Gen. Genet. 241:177–184 [DOI] [PubMed] [Google Scholar]
- 281. Martin J, Mahlke K, Pfanner N. 1991. Role of an energized inner membrane in mitochondrial protein import. Δpsi drives the movement of presequences. J. Biol. Chem. 266:18051–18057 [PubMed] [Google Scholar]
- 282. Martinez-Pastor MT, et al. 1996. The Saccharomyces cerevisiae zinc finger proteins Msn2p and Msn4p are required for transcriptional induction through the stress response element (STRE). EMBO J. 15:2227–2235 [PMC free article] [PubMed] [Google Scholar]
- 283. Matlack KES, Misselwitz B, Plath K, Rapoport TA. 1999. BiP acts as a molecular ratchet during posttranslational transport of prepro-alpha factor across the ER membrane. Cell 97:553–564 [DOI] [PubMed] [Google Scholar]
- 284. Matsumoto R, Akama K, Rakwal R, Iwahashi H. 2005. The stress response against denatured proteins in the deletion of cytosolic chaperones SSA1/2 is different from heat-shock response in Saccharomyces cerevisiae. BMC Genomics 6:141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285. Mayordomo I, Estruch F, Sanz P. 2002. Convergence of the target of rapamycin and the Snf1 protein kinase pathways in the regulation of the subcellular localization of Msn2, a transcriptional activator of STRE (stress response element)-regulated genes. J. Biol. Chem. 277:35650–35656 [DOI] [PubMed] [Google Scholar]
- 286. McClellan AJ, Brodsky JL. 2000. Mutation of the ATP-binding pocket of SSA1 indicates that a functional interaction between Ssa1p and Ydj1p is required for post-translational translocation into the yeast endoplasmic reticulum. Genetics 156:501–512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287. McClellan AJ, et al. 2007. Diverse cellular functions of the Hsp90 molecular chaperone uncovered using systems approaches. Cell 131:121–135 [DOI] [PubMed] [Google Scholar]
- 288. McCracken AA, Brodsky JL. 1996. Assembly of ER-associated protein degradation in vitro: dependence on cytosol, calnexin, and ATP. J. Cell Biol. 132:291–298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289. Meier KD, Deloche O, Kajiwara K, Funato K, Riezman H. 2006. Sphingoid base is required for translation initiation during heat stress in Saccharomyces cerevisiae. Mol. Biol. Cell 17:1164–1175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290. Melville MW, McClellan AJ, Meyer AS, Darveau A, Frydman J. 2003. The Hsp70 and TRiC/CCT chaperone systems cooperate in vivo to assemble the von Hippel-Lindau tumor suppressor complex. Mol. Cell. Biol. 23:3141–3151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291. Meyer AE, Hung NJ, Yang P, Johnson AW, Craig EA. 2007. The specialized cytosolic J-protein, Jjj1, functions in 60S ribosomal subunit biogenesis. Proc. Natl. Acad. Sci. U. S. A. 104:1558–1563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292. Meyer P, et al. 2004. Structural basis for recruitment of the ATPase activator Aha1 to the Hsp90 chaperone machinery. EMBO J. 23:1402–1410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293. Miao B, Davis JE, Craig EA. 1997. Mge1 functions as a nucleotide release factor for Ssc1, a mitochondrial Hsp70 of Saccharomyces cerevisiae. J. Mol. Biol. 265:541–552 [DOI] [PubMed] [Google Scholar]
- 294. Minchiotti G, Gargano S, Maresca B. 1991. The intron-containing hsp82 gene of the dimorphic pathogenic fungus Histoplasma capsulatum is properly spliced in severe heat shock conditions. Mol. Cell. Biol. 11:5624–5630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295. Minchiotti G, Gargano S, Maresca B. 1992. Molecular cloning and expression of hsp82 gene of the dimorphic pathogenic fungus Histoplasma capsulatum. Biochim. Biophys. Acta 1131:103–107 [DOI] [PubMed] [Google Scholar]
- 296. Mir SS, Fiedler D, Cashikar AG. 2009. Ssd1 is required for thermotolerance and Hsp104-mediated protein disaggregation in Saccharomyces cerevisiae. Mol. Cell. Biol. 29:187–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297. Mizunaga T, Katakura Y, Miura T, Maruyama Y. 1990. Purification and characterization of yeast protein disulfide isomerase. J. Biochem. 108:846–851 [DOI] [PubMed] [Google Scholar]
- 298. Moczko M, Schonfisch B, Voos W, Pfanner N, Rassow J. 1995. The mitochondrial ClpB homolog Hsp78 cooperates with matrix Hsp70 in maintenance of mitochondrial function. J. Mol. Biol. 254:538–543 [DOI] [PubMed] [Google Scholar]
- 299. Mokranjac D, Bourenkov G, Hell K, Neupert W, Groll M. 2006. Structure and function of Tim14 and Tim16, the J and J-like components of the mitochondrial protein import motor. EMBO J. 25:4675–4685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300. Mokranjac D, Sichting M, Neupert W, Hell K. 2003. Tim14, a novel key component of the import motor of the TIM23 protein translocase of mitochondria. EMBO J. 22:4945–4956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301. Mokranjac D, et al. 2005. The import motor of the yeast mitochondrial TIM23 preprotein translocase contains two different J proteins, Tim14 and Mdj2. J. Biol. Chem. 280:31608–31614 [DOI] [PubMed] [Google Scholar]
- 302. Mollapour M, Neckers L. 2012. Post-translational modifications of Hsp90 and their contributions to chaperone regulation. Biochim. Biophys. Acta 1823:648–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303. Morano KA, Santoro N, Kock KA, Thiele DJ. 1999. A trans-activation domain in yeast heat shock transcription factor is essential for cell cycle progression during stress. Mol. Cell. Biol. 19:402–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304. Mori K, et al. 1992. A 22 bp cis-acting element is necessary and sufficient for the induction of the yeast KAR2 (BiP) gene by unfolded proteins. EMBO J. 11:2583–2593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305. Morimoto RI. 2008. Proteotoxic stress and inducible chaperone networks in neurodegenerative disease and aging. Genes Dev. 22:1427–1438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306. Morishima N, Nakagawa K, Yamamoto E, Shibata T. 1990. A subunit of yeast site-specific endonuclease SceI is a mitochondrial version of the 70-kDa heat shock protein. J. Biol. Chem. 265:15189–15197 [PubMed] [Google Scholar]
- 307. Moro F, Okamoto K, Donzeau M, Neupert W, Brunner M. 2002. Mitochondrial protein import: molecular basis of the ATP-dependent interaction of MtHsp70 with Tim44. J. Biol. Chem. 277:6874–6880 [DOI] [PubMed] [Google Scholar]
- 308. Motshwene P, Karreman R, Kgari G, Brandt W, Lindsey G. 2004. LEA (late embryonic abundant)-like protein Hsp 12 (heat-shock protein 12) is present in the cell wall and enhances the barotolerance of the yeast Saccharomyces cerevisiae. Biochem. J. 377:769–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309. Mukai H, et al. 1993. Isolation and characterization of SSE1 and SSE2, new members of the yeast HSP70 multigene family. Gene 132:57–66 [DOI] [PubMed] [Google Scholar]
- 310. Nathan DF, Vos MH, Lindquist S. 1997. In vivo functions of the Saccharomyces cerevisiae Hsp90 chaperone. Proc. Natl. Acad. Sci. U. S. A. 94:12949–12956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311. Neef DW, Thiele DJ. 2009. Enhancer of decapping proteins 1 and 2 are important for translation during heat stress in Saccharomyces cerevisiae. Mol. Microbiol. 73:1032–1042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 312. Neef DW, Turski ML, Thiele DJ. 2010. Modulation of heat shock transcription factor 1 as a therapeutic target for small molecule intervention in neurodegenerative disease. PLoS Biol. 8:e1000291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313. Nelson RJ, Heschl MF, Craig EA. 1992. Isolation and characterization of extragenic suppressors of mutations in the SSA hsp70 genes of Saccharomyces cerevisiae. Genetics 131:277–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 314. Nelson RJ, Ziegelhoffer T, Nicolet C, Werner-Washburne M, Craig EA. 1992. The translation machinery and 70 kd heat shock protein cooperate in protein synthesis. Cell 71:97–105 [DOI] [PubMed] [Google Scholar]
- 315. Neupert W. 1997. Protein import into mitochondria. Annu. Rev. Biochem. 66:863–917 [DOI] [PubMed] [Google Scholar]
- 316. Newnam GP, Wegrzyn RD, Lindquist SL, Chernoff YO. 1999. Antagonistic interactions between yeast chaperones Hsp104 and Hsp70 in prion curing. Mol. Cell. Biol. 19:1325–1333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 317. Ng DT, Spear ED, Walter P. 2000. The unfolded protein response regulates multiple aspects of secretory and membrane protein biogenesis and endoplasmic reticulum quality control. J. Cell Biol. 150:77–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 318. Nicholls S, Leach MD, Priest CL, Brown AJ. 2009. Role of the heat shock transcription factor, Hsf1, in a major fungal pathogen that is obligately associated with warm-blooded animals. Mol. Microbiol. 74:844–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 319. Nicholls S, et al. 2011. Activation of the heat shock transcription factor Hsf1 is essential for the full virulence of the fungal pathogen Candida albicans. Fungal Genet. Biol. 48:297–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 320. Nieto-Sotelo J, Wiederrecht G, Okuda A, Parker CS. 1990. The yeast heat shock transcription factor contains a transcriptional activation domain whose activity is repressed under nonshock conditions. Cell 62:807–817 [DOI] [PubMed] [Google Scholar]
- 321. Nillegoda NB, et al. 2010. Ubr1 and Ubr2 function in a quality control pathway for degradation of unfolded cytosolic proteins. Mol. Biol. Cell 21:2102–2116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 322. Nishikawa S, Endo T. 1997. The yeast Jem1p is a DnaJ-like protein of the endoplasmic reticulum membrane required for nuclear fusion. J. Biol. Chem. 272:12889–12892 [DOI] [PubMed] [Google Scholar]
- 323. Nishikawa SI, Fewell SW, Kato Y, Brodsky JL, Endo T. 2001. Molecular chaperones in the yeast endoplasmic reticulum maintain the solubility of proteins for retrotranslocation and degradation. J. Cell Biol. 153:1061–1070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 324. Norgaard P, et al. 2001. Functional differences in yeast protein disulfide isomerases. J. Cell Biol. 152:553–562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 325. Normington K, Kohno K, Kozutsumi Y, Gething MJ, Sambrook J. 1989. S. cerevisiae encodes an essential protein homologous in sequence and function to mammalian BiP. Cell 57:1223–1236 [DOI] [PubMed] [Google Scholar]
- 326. Nwaka S, Mechler B, von Ahsen O, Holzer H. 1996. The heat shock factor and mitochondrial Hsp70 are necessary for survival of heat shock in Saccharomyces cerevisiae. FEBS Lett. 399:259–263 [DOI] [PubMed] [Google Scholar]
- 327. Ostermann J, Horwich AL, Neupert W, Hartl FU. 1989. Protein folding in mitochondria requires complex formation with hsp60 and ATP hydrolysis. Nature 341:125–130 [DOI] [PubMed] [Google Scholar]
- 328. Pais JE, Schilke B, Craig EA. 2011. Reevaluation of the role of the Pam18:Pam16 interaction in translocation of proteins by the mitochondrial Hsp70-based import motor. Mol. Biol. Cell 22:4740–4749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 329. Panaretou B, et al. 2002. Activation of the ATPase activity of hsp90 by the stress-regulated cochaperone Aha1. Mol. Cell 10:1307–1318 [DOI] [PubMed] [Google Scholar]
- 330. Pareek G, Samaddar M, D'Silva P. 2011. Primary sequence that determines the functional overlap between mitochondrial heat shock protein 70 Ssc1 and Ssc3 of Saccharomyces cerevisiae. J. Biol. Chem. 286:19001–19013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 331. Paris S, Pringle JR. 1983. Saccharomyces cerevisiae: heat and gluculase sensitivities of starved cells. Ann. Microbiol. 134B:379–385 [PubMed] [Google Scholar]
- 332. Park SH, et al. 2007. The cytoplasmic Hsp70 chaperone machinery subjects misfolded and endoplasmic reticulum import-incompetent proteins to degradation via the ubiquitin-proteasome system. Mol. Biol. Cell 18:153–165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 333. Parker R, Sheth U. 2007. P bodies and the control of mRNA translation and degradation. Mol. Cell 25:635–646 [DOI] [PubMed] [Google Scholar]
- 334. Parlati F, Dominguez M, Bergeron JJM, Thomas DY. 1995. Saccharomyces cerevisiae CNE1 encodes an endoplasmic reticulum (ER) membrane protein with sequence similarity to calnexin and calreticulin and functions as a constituent of the ER quality control apparatus. J. Biol. Chem. 270:244–253 [DOI] [PubMed] [Google Scholar]
- 335. Parsell DA, Kowal AS, Singer MA, Lindquist S. 1994. Protein disaggregation mediated by heat-shock protein Hsp104. Nature 372:475–478 [DOI] [PubMed] [Google Scholar]
- 336. Pattaramanon N, Sangha N, Gafni A. 2007. The carboxy-terminal domain of heat-shock factor 1 is largely unfolded but can be induced to collapse into a compact, partially structured state. Biochemistry 46:3405–3415 [DOI] [PubMed] [Google Scholar]
- 337. Peisker K, Chiabudini M, Rospert S. 2010. The ribosome-bound Hsp70 homolog Ssb of Saccharomyces cerevisiae. Biochim. Biophys. Acta 1803:662–672 [DOI] [PubMed] [Google Scholar]
- 338. Perisic O, Xiao H, Lis JT. 1989. Stable binding of Drosophila heat shock factor to head-to-head and tail-to-tail repeats of a conserved 5 bp recognition unit. Cell 59:797–806 [DOI] [PubMed] [Google Scholar]
- 339. Peteranderl R, Nelson HC. 1992. Trimerization of the heat shock transcription factor by a triple-stranded alpha-helical coiled-coil. Biochemistry 31:12272–12276 [DOI] [PubMed] [Google Scholar]
- 340. Peteranderl R, et al. 1999. Biochemical and biophysical characterization of the trimerization domain from the heat shock transcription factor. Biochemistry 38:3559–3569 [DOI] [PubMed] [Google Scholar]
- 341. Pfeifer K, Arcangioli B, Guarente L. 1987. Yeast HAP1 activator competes with the factor RC2 for binding to the upstream activation site UAS1 of the CYC1 gene. Cell 49:9–18 [DOI] [PubMed] [Google Scholar]
- 342. Pfund C, Huang P, Lopez-Hoyo N, Craig EA. 2001. Divergent functional properties of the ribosome-associated molecular chaperone Ssb compared with other Hsp70s. Mol. Biol. Cell 12:3773–3782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 343. Pfund C, et al. 1998. The molecular chaperone Ssb from Saccharomyces cerevisiae is a component of the ribosome-nascent chain complex. EMBO J. 17:3981–3989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 344. Piper PW, Ortiz-Calderon C, Holyoak C, Coote P, Cole M. 1997. Hsp30, the integral plasma membrane heat shock protein of Saccharomyces cerevisiae, is a stress-inducible regulator of plasma membrane H(+)-ATPase. Cell Stress Chaperones 2:12–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 345. Piper PW, et al. 1994. Induction of major heat-shock proteins of Saccharomyces cerevisiae, including plasma membrane Hsp30, by ethanol levels above a critical threshold. Microbiology 140(Pt 11):3031–3038 [DOI] [PubMed] [Google Scholar]
- 346. Plemper RK, Bohmler S, Bordallo J, Sommer T, Wolf DH. 1997. Mutant analysis links the translocon and BiP to retrograde protein transport for ER degradation. Nature 388:891–895 [DOI] [PubMed] [Google Scholar]
- 347. Plesset J, Ludwig JR, Cox BS, McLaughlin CS. 1987. Effect of cell cycle position on thermotolerance in Saccharomyces cerevisiae. J. Bacteriol. 169:779–784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 348. Polier S, Dragovic Z, Hartl FU, Bracher A. 2008. Structural basis for the cooperation of Hsp70 and Hsp110 chaperones in protein folding. Cell 133:1068–1079 [DOI] [PubMed] [Google Scholar]
- 349. Portillo F, Mulet JM, Serrano R. 2005. A role for the non-phosphorylated form of yeast Snf1: tolerance to toxic cations and activation of potassium transport. FEBS Lett. 579:512–516 [DOI] [PubMed] [Google Scholar]
- 350. Prodromou C, et al. 1997. Identification and structural characterization of the ATP/ADP-binding site in the Hsp90 molecular chaperone. Cell 90:65–75 [DOI] [PubMed] [Google Scholar]
- 351. Puig A, Lyles MM, Noiva R, Gilbert HF. 1994. The role of the thiol/disulfide centers and peptide binding site in the chaperone and anti-chaperone activities of protein disulfide isomerase. J. Biol. Chem. 269:19128–19135 [PubMed] [Google Scholar]
- 352. Rabindran SK, Haroun RI, Clos J, Wisniewski J, Wu C. 1993. Regulation of heat shock factor trimer formation: role of a conserved leucine zipper. Science 259:230–234 [DOI] [PubMed] [Google Scholar]
- 353. Rabindran SK, Wisniewski J, Li L, Li GC, Wu C. 1994. Interaction between heat shock factor and hsp70 is insufficient to suppress induction of DNA-binding activity in vivo. Mol. Cell. Biol. 14:6552–6560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 354. Raboy B, Marom A, Dor Y, Kulka RG. 1999. Heat-induced cell cycle arrest of Saccharomyces cerevisiae: involvement of the RAD6/UBC2 and WSC2 genes in its reversal. Mol. Microbiol. 32:729–739 [DOI] [PubMed] [Google Scholar]
- 355. Rakwalska M, Rospert S. 2004. The ribosome-bound chaperones RAC and Ssb1/2p are required for accurate translation in Saccharomyces cerevisiae. Mol. Cell. Biol. 24:9186–9197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 356. Ran F, Bali M, Michels CA. 2008. Hsp90/Hsp70 chaperone machine regulation of the Saccharomyces MAL-activator as determined in vivo using noninducible and constitutive mutant alleles. Genetics 179:331–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 357. Rao J, et al. 2001. Functional interaction of human Cdc37 with the androgen receptor but not with the glucocorticoid receptor. J. Biol. Chem. 276:5814–5820 [DOI] [PubMed] [Google Scholar]
- 358. Rassow J, et al. 1994. Mitochondrial protein import: biochemical and genetic evidence for interaction of matrix hsp70 and the inner membrane protein MIM44. J. Cell Biol. 127:1547–1556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 359. Rauch T, et al. 2005. Dissecting functional similarities of ribosome-associated chaperones from Saccharomyces cerevisiae and Escherichia coli. Mol. Microbiol. 57:357–365 [DOI] [PubMed] [Google Scholar]
- 360. Raviol H, Bukau B, Mayer MP. 2006. Human and yeast Hsp110 chaperones exhibit functional differences. FEBS Lett. 580:168–174 [DOI] [PubMed] [Google Scholar]
- 361. Raviol H, Sadlish H, Rodriguez F, Mayer MP, Bukau B. 2006. Chaperone network in the yeast cytosol: Hsp110 is revealed as an Hsp70 nucleotide exchange factor. EMBO J. 25:2510–2518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 362. Reading DS, Hallberg RL, Myers AM. 1989. Characterization of the yeast HSP60 gene coding for a mitochondrial assembly factor. Nature 337:655–659 [DOI] [PubMed] [Google Scholar]
- 363. Requena JR, Chao CC, Levine RL, Stadtman ER. 2001. Glutamic and aminoadipic semialdehydes are the main carbonyl products of metal-catalyzed oxidation of proteins. Proc. Natl. Acad. Sci. U. S. A. 98:69–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 364. Rezaie S, et al. 2000. Characterization of a cDNA clone, encoding a 70 kDa heat shock protein from the dermatophyte pathogen Trichophyton rubrum. Gene 241:27–33 [DOI] [PubMed] [Google Scholar]
- 365. Ribeiro MJ, Reinders A, Boller T, Wiemken A, De Virgilio C. 1997. Trehalose synthesis is important for the acquisition of thermotolerance in Schizosaccharomyces pombe. Mol. Microbiol. 25:571–581 [DOI] [PubMed] [Google Scholar]
- 366. Richter K, Muschler P, Hainzl O, Reinstein J, Buchner J. 2003. Sti1 is a non-competitive inhibitor of the Hsp90 ATPase. Binding prevents the N-terminal dimerization reaction during the ATPase cycle. J. Biol. Chem. 278:10328–10333 [DOI] [PubMed] [Google Scholar]
- 367. Robbins N, et al. 2011. Hsp90 governs dispersion and drug resistance of fungal biofilms. PLoS Pathog. 7:e1002257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 368. Roe SM, et al. 2004. The mechanism of Hsp90 regulation by the protein kinase-specific cochaperone p50(cdc37). Cell 116:87–98 [DOI] [PubMed] [Google Scholar]
- 369. Rollenhagen C, Hodge CA, Cole CN. 2004. The nuclear pore complex and the DEAD box protein Rat8p/Dbp5p have nonessential features which appear to facilitate mRNA export following heat shock. Mol. Cell. Biol. 24:4869–4879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 370. Rose MD, Misra LM, Vogel JP. 1989. KAR2, a karyogamy gene, is the yeast homolog of the mammalian BiP/GRP78 gene. Cell 57:1211–1221 [DOI] [PubMed] [Google Scholar]
- 371. Rospert S, et al. 1996. Hsp60-independent protein folding in the matrix of yeast mitochondria. EMBO J. 15:764–774 [PMC free article] [PubMed] [Google Scholar]
- 372. Rottgers K, Zufall N, Guiard B, Voos W. 2002. The ClpB homolog Hsp78 is required for the efficient degradation of proteins in the mitochondrial matrix. J. Biol. Chem. 277:45829–45837 [DOI] [PubMed] [Google Scholar]
- 373. Rowley A, Johnston GC, Butler B, Werner-Washburne M, Singer RA. 1993. Heat shock-mediated cell cycle blockage and G1 cyclin expression in the yeast Saccharomyces cerevisiae. Mol. Cell. Biol. 13:1034–1041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 374. Rowley N, et al. 1994. Mdj1p, a novel chaperone of the DnaJ family, is involved in mitochondrial biogenesis and protein folding. Cell 77:249–259 [DOI] [PubMed] [Google Scholar]
- 375. Saavedra C, Tung KS, Amberg DC, Hopper AK, Cole CN. 1996. Regulation of mRNA export in response to stress in Saccharomyces cerevisiae. Genes Dev. 10:1608–1620 [DOI] [PubMed] [Google Scholar]
- 376. Saavedra CA, Hammell CM, Heath CV, Cole CN. 1997. Yeast heat-shock mRNAs are exported through a distinct pathway defined by Rip1p. Genes Dev. 11:2845–2856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 377. Sadeh A, Movshovich N, Volokh M, Gheber L, Aharoni A. 2011. Fine-tuning of the Msn2/4-mediated yeast stress responses as revealed by systematic deletion of Msn2/4 partners. Mol. Biol. Cell 22:3127–3138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 378. Sadler I, et al. 1989. A yeast gene important for protein assembly into the endoplasmic reticulum and the nucleus has homology to DnaJ, an Escherichia coli heat shock protein. J. Cell Biol. 109:2665–2675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 379. Sadlish H, et al. 2008. Hsp110 chaperones regulate prion formation and propagation in S. cerevisiae by two discrete activities. PLoS One 3:e1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 380. Sahi C, Craig EA. 2007. Network of general and specialty J protein chaperones of the yeast cytosol. Proc. Natl. Acad. Sci. U. S. A. 104:7163–7168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 381. Sakurai H, Takemori Y. 2007. Interaction between heat shock transcription factors (HSFs) and divergent binding sequences: binding specificities of yeast HSFs and human HSF1. J. Biol. Chem. 282:13334–13341 [DOI] [PubMed] [Google Scholar]
- 382. Sales K, Brandt W, Rumbak E, Lindsey G. 2000. The LEA-like protein HSP 12 in Saccharomyces cerevisiae has a plasma membrane location and protects membranes against desiccation and ethanol-induced stress. Biochim. Biophys. Acta 1463:267–278 [DOI] [PubMed] [Google Scholar]
- 383. Sanchez Y, Lindquist SL. 1990. HSP104 required for induced thermotolerance. Science 248:1112–1115 [DOI] [PubMed] [Google Scholar]
- 384. Sanchez Y, Taulien J, Borkovich KA, Lindquist S. 1992. Hsp104 is required for tolerance to many forms of stress. EMBO J. 11:2357–2364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 385. Sanders SL, Whitfield KM, Vogel JP, Rose MD, Schekman RW. 1992. Sec61p and BiP directly facilitate polypeptide translocation into the ER. Cell 69:353–365 [DOI] [PubMed] [Google Scholar]
- 386. Santoro N, Johansson N, Thiele DJ. 1998. Heat shock element architecture is an important determinant in the temperature and transactivation domain requirements for heat shock transcription factor. Mol. Cell. Biol. 18:6340–6352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 387. Saris N, Holkeri H, Craven RA, Stirling CJ, Makarow M. 1997. The Hsp70 homologue Lhs1p is involved in a novel function of the yeast endoplasmic reticulum, refolding and stabilization of heat-denatured protein aggregates. J. Cell Biol. 137:813–824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 388. Satyanarayana C, Schroder-Kohne S, Craig EA, Schu PV, Horst M. 2000. Cytosolic Hsp70s are involved in the transport of aminopeptidase 1 from the cytoplasm into the vacuole. FEBS Lett. 470:232–238 [DOI] [PubMed] [Google Scholar]
- 389. Scherer PE, Krieg UC, Hwang ST, Vestweber D, Schatz G. 1990. A precursor protein partly translocated into yeast mitochondria is bound to a 70 kd mitochondrial stress protein. EMBO J. 9:4315–4322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 390. Scheufler C, et al. 2000. Structure of TPR domain-peptide complexes: critical elements in the assembly of the Hsp70-Hsp90 multichaperone machine. Cell 101:199–210 [DOI] [PubMed] [Google Scholar]
- 391. Schilke B, et al. 1996. The cold sensitivity of a mutant of Saccharomyces cerevisiae lacking a mitochondrial heat shock protein 70 is suppressed by loss of mitochondrial DNA. J. Cell Biol. 134:603–613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 392. Schilke B, Voisine C, Beinert H, Craig E. 1999. Evidence for a conserved system for iron metabolism in the mitochondria of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U. S. A. 96:10206–10211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 393. Schirmer EC, Glover JR, Singer MA, Lindquist S. 1996. HSP100/Clp proteins: a common mechanism explains diverse functions. Trends Biochem. Sci. 21:289–296 [PubMed] [Google Scholar]
- 394. Schlenstedt G, Harris S, Risse B, Lill R, Silver PA. 1995. A yeast DnaJ homologue, Scj1p, can function in the endoplasmic reticulum with BiP/Kar2p via a conserved domain that specifies interactions with Hsp70s. J. Cell Biol. 129:979–988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 395. Schmidt S, Strub A, Rottgers K, Zufall N, Voos W. 2001. The two mitochondrial heat shock proteins 70, Ssc1 and Ssq1, compete for the cochaperone Mge1. J. Mol. Biol. 313:13–26 [DOI] [PubMed] [Google Scholar]
- 396. Schmitt AP, McEntee K. 1996. Msn2p, a zinc finger DNA-binding protein, is the transcriptional activator of the multistress response in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U. S. A. 93:5777–5782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 397. Schmitt M, Neupert W, Langer T. 1995. Hsp78, a Clp homologue within mitochondria, can substitute for chaperone functions of mt-hsp70. EMBO J. 14:3434–3444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 398. Schmitt M, Neupert W, Langer T. 1996. The molecular chaperone Hsp78 confers compartment-specific thermotolerance to mitochondria. J. Cell Biol. 134:1375–1386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 399. Schneider HC, et al. 1994. Mitochondrial Hsp70/MIM44 complex facilitates protein import. Nature 371:768–774 [DOI] [PubMed] [Google Scholar]
- 400. Schneider HC, Westermann B, Neupert W, Brunner M. 1996. The nucleotide exchange factor MGE exerts a key function in the ATP-dependent cycle of mt-Hsp70-Tim44 interaction driving mitochondrial protein import. EMBO J. 15:5796–5803 [PMC free article] [PubMed] [Google Scholar]
- 401. Schrag JD, et al. 2001. The structure of calnexin, an ER chaperone involved in quality control of protein folding. Mol. Cell 8:633–644 [DOI] [PubMed] [Google Scholar]
- 402. Schuermann JP, et al. 2008. Structure of the Hsp110:Hsc70 nucleotide exchange machine. Mol. Cell 31:232–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 403. Schulte TW, et al. 1998. Antibiotic radicicol binds to the N-terminal domain of Hsp90 and shares important biologic activities with geldanamycin. Cell Stress Chaperones 3:100–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 404. Scidmore MA, Okamura HH, Rose MD. 1993. Genetic interactions between KAR2 and SEC63, encoding eukaryotic homologues of DnaK and DnaJ in the endoplasmic reticulum. Mol. Biol. Cell 4:1145–1159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 405. Sewell AK, et al. 1995. Mutated yeast heat shock transcription factor exhibits elevated basal transcriptional activation and confers metal resistance. J. Biol. Chem. 270:25079–25086 [DOI] [PubMed] [Google Scholar]
- 406. Seymour IJ, Piper PW. 1999. Stress induction of HSP30, the plasma membrane heat shock protein gene of Saccharomyces cerevisiae, appears not to use known stress-regulated transcription factors. Microbiology 145(Pt 1):231–239 [DOI] [PubMed] [Google Scholar]
- 407. Shahi P, Gulshan K, Moye-Rowley WS. 2007. Negative transcriptional regulation of multidrug resistance gene expression by an Hsp70 protein. J. Biol. Chem. 282:26822–26831 [DOI] [PubMed] [Google Scholar]
- 408. Shamu CE, Walter P. 1996. Oligomerization and phosphorylation of the Ire1p kinase during intracellular signaling from the endoplasmic reticulum to the nucleus. EMBO J. 15:3028–3039 [PMC free article] [PubMed] [Google Scholar]
- 409. Shaner L, Gibney PA, Morano KA. 2008. The Hsp110 protein chaperone Sse1 is required for yeast cell wall integrity and morphogenesis. Curr. Genet. 54:1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 410. Shaner L, Sousa R, Morano KA. 2006. Characterization of hsp70 binding and nucleotide exchange by the yeast hsp110 chaperone Sse1. Biochemistry 45:15075–15084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 411. Shaner L, Trott A, Goeckeler JL, Brodsky JL, Morano KA. 2004. The function of the yeast molecular chaperone Sse1 is mechanistically distinct from the closely related hsp70 family. J. Biol. Chem. 279:21992–22001 [DOI] [PubMed] [Google Scholar]
- 412. Shaner L, Wegele H, Buchner J, Morano KA. 2005. The yeast Hsp110 Sse1 functionally interacts with the Hsp70 chaperones Ssa and Ssb. J. Biol. Chem. 280:41262–41269 [DOI] [PubMed] [Google Scholar]
- 413. Shao J, et al. 2001. Hsp90 regulates p50(cdc37) function during the biogenesis of the active conformation of the heme-regulated eIF2 alpha kinase. J. Biol. Chem. 276:206–214 [DOI] [PubMed] [Google Scholar]
- 414. Shao J, Irwin A, Hartson SD, Matts RL. 2003. Functional dissection of cdc37: characterization of domain structure and amino acid residues critical for protein kinase binding. Biochemistry 42:12577–12588 [DOI] [PubMed] [Google Scholar]
- 415. Shapiro RS, Cowen L. 2010. Coupling temperature sensing and development: Hsp90 regulates morphogenetic signalling in Candida albicans. Virulence 1:45–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 416. Reference deleted.
- 417. Sharma D, Masison DC. 2011. Single methyl group determines prion propagation and protein degradation activities of yeast heat shock protein (Hsp)-70 chaperones Ssa1p and Ssa2p. Proc. Natl. Acad. Sci. U. S. A. 108:13665–13670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 418. Shearer G, Jr, Birge CH, Yuckenberg PD, Kobayashi GS, Medoff G. 1987. Heat-shock proteins induced during the mycelial-to-yeast transitions of strains of Histoplasma capsulatum. J. Gen. Microbiol. 133:3375–3382 [DOI] [PubMed] [Google Scholar]
- 419. Shimon L, Hynes GM, McCormack EA, Willison KR, Horovitz A. 2008. ATP-induced allostery in the eukaryotic chaperonin CCT is abolished by the mutation G345D in CCT4 that renders yeast temperature-sensitive for growth. J. Mol. Biol. 377:469–477 [DOI] [PubMed] [Google Scholar]
- 420. Shulga N, et al. 1996. In vivo nuclear transport kinetics in Saccharomyces cerevisiae: a role for heat shock protein 70 during targeting and translocation. J. Cell Biol. 135:329–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 421. Sichting M, Mokranjac D, Azem A, Neupert W, Hell K. 2005. Maintenance of structure and function of mitochondrial Hsp70 chaperones requires the chaperone Hep1. EMBO J. 24:1046–1056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 422. Siegers K, et al. 1999. Compartmentation of protein folding in vivo: sequestration of non-native polypeptide by the chaperonin-GimC system. EMBO J. 18:75–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 423. Sielaff B, Tsai FT. 2010. The M-domain controls Hsp104 protein remodeling activity in an Hsp70/Hsp40-dependent manner. J. Mol. Biol. 402:30–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 424. Silberstein S, Schlenstedt G, Silver PA, Gilmore R. 1998. A role for the DnaJ homologue Scj1p in protein folding in the yeast endoplasmic reticulum. J. Cell Biol. 143:921–933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 425. Siligardi G, et al. 2002. Regulation of Hsp90 ATPase activity by the co-chaperone Cdc37p/p50cdc37. J. Biol. Chem. 277:20151–20159 [DOI] [PubMed] [Google Scholar]
- 426. Simola M, Hanninen AL, Stranius SM, Makarow M. 2000. Trehalose is required for conformational repair of heat-denatured proteins in the yeast endoplasmic reticulum but not for maintenance of membrane traffic functions after severe heat stress. Mol. Microbiol. 37:42–53 [DOI] [PubMed] [Google Scholar]
- 427. Simon SM, Peskin CS, Oster GF. 1992. What drives the translocation of proteins? Proc. Natl. Acad. Sci. U. S. A. 89:3770–3774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 428. Simons JF, Ferro-Novick S, Rose MD, Helenius A. 1995. BiP/Kar2p serves as a molecular chaperone during carboxypeptidase Y folding in yeast. J. Cell Biol. 130:41–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 429. Singer MA, Lindquist S. 1998. Multiple effects of trehalose on protein folding in vitro and in vivo. Mol. Cell 1:639–648 [DOI] [PubMed] [Google Scholar]
- 430. Singer MA, Lindquist S. 1998. Thermotolerance in Saccharomyces cerevisiae: the Yin and Yang of trehalose. Trends Biotechnol. 16:460–468 [DOI] [PubMed] [Google Scholar]
- 431. Slater MR, Craig EA. 1987. Transcriptional regulation of an hsp70 heat shock gene in the yeast Saccharomyces cerevisiae. Mol. Cell. Biol. 7:1906–1916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 432. Smith BJ, Yaffe MP. 1991. A mutation in the yeast heat-shock factor gene causes temperature-sensitive defects in both mitochondrial protein import and the cell cycle. Mol. Cell. Biol. 11:2647–2655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 433. Somer L, Shmulman O, Dror T, Hashmueli S, Kashi Y. 2002. The eukaryote chaperonin CCT is a cold shock protein in Saccharomyces cerevisiae. Cell Stress Chaperones 7:47–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 434. Sondermann H, et al. 2002. Prediction of novel Bag-1 homologs based on structure/function analysis identifies Snl1p as an Hsp70 co-chaperone in Saccharomyces cerevisiae. J. Biol. Chem. 277:33220–33227 [DOI] [PubMed] [Google Scholar]
- 435. Song JL, Wang CC. 1995. Chaperone-like activity of protein disulfide-isomerase in the refolding of rhodanese. Eur. J. Biochem. 231:312–316 [DOI] [PubMed] [Google Scholar]
- 436. Song Y, et al. 2001. Effects of calnexin deletion in Saccharomyces cerevisiae on the secretion of glycosylated lysozymes. J. Biochem. 130:757–764 [DOI] [PubMed] [Google Scholar]
- 437. Sorger PK. 1990. Yeast heat shock factor contains separable transient and sustained response transcriptional activators. Cell 62:793–805 [DOI] [PubMed] [Google Scholar]
- 438. Sorger PK, Lewis MJ, Pelham HRB. 1987. Heat shock factor is regulated differently in yeast and HeLa cells. Nature 329:81–84 [DOI] [PubMed] [Google Scholar]
- 439. Sorger PK, Nelson HCM. 1989. Trimerization of a yeast transcriptional activator via a coiled-coil motif. Cell 59:807–813 [DOI] [PubMed] [Google Scholar]
- 440. Sorger PK, Pelham HR. 1987. Purification and characterization of a heat-shock element binding protein from yeast. EMBO J. 6:3035–3041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 441. Sorger PK, Pelham HR. 1988. Yeast heat shock factor is an essential DNA-binding protein that exhibits temperature-dependent phosphorylation. Cell 54:855–864 [DOI] [PubMed] [Google Scholar]
- 442. Stedman TT, Buck GA. 1996. Identification, characterization, and expression of the BiP endoplasmic reticulum resident chaperonins in Pneumocystis carinii. Infect. Immun. 64:4463–4471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 443. Steel GJ, Fullerton DM, Tyson JR, Stirling CJ. 2004. Coordinated activation of Hsp70 chaperones. Science 303:98–101 [DOI] [PubMed] [Google Scholar]
- 444. Stirling PC, et al. 2006. PhLP3 modulates CCT-mediated actin and tubulin folding via ternary complexes with substrates. J. Biol. Chem. 281:7012–7021 [DOI] [PubMed] [Google Scholar]
- 445. Stoldt V, et al. 1996. The Cct eukaryotic chaperonin subunits of Saccharomyces cerevisiae and other yeasts. Yeast 12:523–529 [DOI] [PubMed] [Google Scholar]
- 446. Strain J, et al. 1998. Suppressors of superoxide dismutase (SOD1) deficiency in Saccharomyces cerevisiae. Identification of proteins predicted to mediate iron-sulfur cluster assembly. J. Biol. Chem. 273:31138–31144 [DOI] [PubMed] [Google Scholar]
- 447. Stromer T, Ehrnsperger M, Gaestel M, Buchner J. 2003. Analysis of the interaction of small heat shock proteins with unfolding proteins. J. Biol. Chem. 278:18015–18021 [DOI] [PubMed] [Google Scholar]
- 448. Stromer T, Fischer E, Richter K, Haslbeck M, Buchner J. 2004. Analysis of the regulation of the molecular chaperone Hsp26 by temperature-induced dissociation: the N-terminal domain is important for oligomer assembly and the binding of unfolding proteins. J. Biol. Chem. 279:11222–11228 [DOI] [PubMed] [Google Scholar]
- 449. Sudbery P, Gow N, Berman J. 2004. The distinct morphogenic states of Candida albicans. Trends Microbiol. 12:317–324 [DOI] [PubMed] [Google Scholar]
- 450. Summers DW, Douglas PM, Ramos CH, Cyr DM. 2009. Polypeptide transfer from Hsp40 to Hsp70 molecular chaperones. Trends Biochem. Sci. 34:230–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 451. Swoboda RK, et al. 1995. Structure and regulation of the HSP90 gene from the pathogenic fungus Candida albicans. Infect. Immun. 63:4506–4514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 452. Tachibana C, Stevens TH. 1992. The yeast EUG1 gene encodes an endoplasmic reticulum protein that is functionally related to protein disulfide isomerase. Mol. Cell. Biol. 12:4601–4611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 453. Tachikawa H, et al. 1997. Overproduction of Mpd2p suppresses the lethality of protein disulfide isomerase depletion in a CXXC sequence dependent manner. Biochem. Biophys. Res. Commun. 239:710–714 [DOI] [PubMed] [Google Scholar]
- 454. Tachikawa H, Miura T, Katakura Y, Mizunaga T. 1991. Molecular structure of a yeast gene, PDI1, encoding protein disulfide isomerase that is essential for cell growth. J. Biochem. 110:306–313 [DOI] [PubMed] [Google Scholar]
- 455. Tachikawa H, et al. 1995. Isolation and characterization of a yeast gene, MPD1, the overexpression of which suppresses inviability caused by protein disulfide isomerase depletion. FEBS Lett. 369:212–216 [DOI] [PubMed] [Google Scholar]
- 456. Tamai KT, Liu X, Silar P, Sosinowski T, Thiele DJ. 1994. Heat shock transcription factor activates yeast metallothionein gene expression in response to heat and glucose starvation via distinct signalling pathways. Mol. Cell. Biol. 14:8155–8165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 457. Thanonkeo P, Akiyama K, Jain S, Takata R. 2000. Targeted disruption of sti35, a stress-responsive gene in phytopathogenic fungus Fusarium oxysporum. Curr. Microbiol. 41:284–289 [DOI] [PubMed] [Google Scholar]
- 458. Thomas G, Hall MN. 1997. TOR signalling and control of cell growth. Curr. Opin. Cell Biol. 9:782–787 [DOI] [PubMed] [Google Scholar]
- 459. Toogun OA, Zeiger W, Freeman BC. 2007. The p23 molecular chaperone promotes functional telomerase complexes through DNA dissociation. Proc. Natl. Acad. Sci. U. S. A. 104:5765–5770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 460. Torres FA, Bonner JJ. 1995. Genetic identification of the site of DNA contact in the yeast heat shock transcription factor. Mol. Cell. Biol. 15:5063–5070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 461. Travers KJ, et al. 2000. Functional and genomic analyses reveal an essential coordination between the unfolded protein response and ER-associated degradation. Cell 101:249–258 [DOI] [PubMed] [Google Scholar]
- 462. Treger JM, Magee TR, McEntee K. 1998. Functional analysis of the stress response element and its role in the multistress response of Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 243:13–19 [DOI] [PubMed] [Google Scholar]
- 463. Treger JM, Schmitt AP, Simon JR, McEntee K. 1998. Transcriptional factor mutations reveal regulatory complexities of heat shock and newly identified stress genes in Saccharomyces cerevisiae. J. Biol. Chem. 273:26875–26879 [DOI] [PubMed] [Google Scholar]
- 464. Trott A, Shaner L, Morano KA. 2005. The molecular chaperone Sse1 and the growth control protein kinase Sch9 collaborate to regulate protein kinase A activity in Saccharomyces cerevisiae. Genetics 170:1009–1021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 465. Trott A, et al. 2008. Activation of heat shock and antioxidant responses by the natural product celastrol: transcriptional signatures of a thiol-targeted molecule. Mol. Biol. Cell 19:1104–1112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 466. Trotter EW, Berenfeld L, Krause SA, Petsko GA, Gray JV. 2001. Protein misfolding and temperature up-shift cause G1 arrest via a common mechanism dependent on heat shock factor in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U. S. A. 98:7313–7318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 467. Trotter EW, et al. 2002. Misfolded proteins are competent to mediate a subset of the responses to heat shock in Saccharomyces cerevisiae. J. Biol. Chem. 277:44817–44825 [DOI] [PubMed] [Google Scholar]
- 468. Truman AW, et al. 2007. In the yeast heat shock response, Hsf1-directed induction of Hsp90 facilitates the activation of the Slt2 (Mpk1) mitogen-activated protein kinase required for cell integrity. Eukaryot. Cell 6:744–752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 469. Truscott KN, et al. 2003. A J-protein is an essential subunit of the presequence translocase-associated protein import motor of mitochondria. J. Cell Biol. 163:707–713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 470. Tsai J, Douglas MG. 1996. A conserved HPD sequence of the J-domain is necessary for YDJ1 stimulation of Hsp70 ATPase activity at a site distinct from substrate binding. J. Biol. Chem. 271:9347–9354 [DOI] [PubMed] [Google Scholar]
- 471. Tyson JR, Stirling CJ. 2000. LHS1 and SIL1 provide a lumenal function that is essential for protein translocation into the endoplasmic reticulum. EMBO J. 19:6440–6452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 472. Unal E, Kinde B, Amon A. 2011. Gametogenesis eliminates age-induced cellular damage and resets life span in yeast. Science 332:1554–1557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 473. Unno K, Kishido T, Hosaka M, Okada S. 1997. Role of Hsp70 subfamily, Ssa, in protein folding in yeast cells, seen in luciferase-transformed Ssa mutants. Biol. Pharm. Bull. 20:1240–1244 [DOI] [PubMed] [Google Scholar]
- 474. Valpuesta JM, Martin-Benito J, Gomez-Puertas P, Carrascosa JL, Willison KR. 2002. Structure and function of a protein folding machine: the eukaryotic cytosolic chaperonin CCT. FEBS Lett. 529:11–16 [DOI] [PubMed] [Google Scholar]
- 475. Van Dyck L, Pearce DA, Sherman F. 1994. PIM1 encodes a mitochondrial ATP-dependent protease that is required for mitochondrial function in the yeast Saccharomyces cerevisiae. J. Biol. Chem. 269:238–242 [PubMed] [Google Scholar]
- 476. Vembar SS, Jonikas MC, Hendershot LM, Weissman JS, Brodsky JL. 2010. J domain co-chaperone specificity defines the role of BiP during protein translocation. J. Biol. Chem. 285:22484–22494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 477. Verges E, Colomina N, Gari E, Gallego C, Aldea M. 2007. Cyclin Cln3 is retained at the ER and released by the J chaperone Ydj1 in late G1 to trigger cell cycle entry. Mol. Cell 26:649–662 [DOI] [PubMed] [Google Scholar]
- 478. Verna J, Lodder A, Lee K, Vagts A, Ballester R. 1997. A family of genes required for maintenance of cell wall integrity and for the stress response in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U. S. A. 94:13804–13809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 479. Vinh DB, Drubin DG. 1994. A yeast TCP-1-like protein is required for actin function in vivo. Proc. Natl. Acad. Sci. U. S. A. 91:9116–9120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 480. Voellmy R. 2004. On mechanisms that control heat shock transcription factor activity in metazoan cells. Cell Stress Chaperones 9:122–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 481. Voellmy R, Boellmann F. 2007. Chaperone regulation of the heat shock protein response. Adv. Exp. Med. Biol. 594:89–99 [DOI] [PubMed] [Google Scholar]
- 482. Vogel JP, Misra LM, Rose MD. 1990. Loss of BiP/GRP78 function blocks translocation of secretory proteins in yeast. J. Cell Biol. 110:1885–1895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 483. Vogel M, Bukau B, Mayer MP. 2006. Allosteric regulation of Hsp70 chaperones by a proline switch. Mol. Cell 21:359–367 [DOI] [PubMed] [Google Scholar]
- 484. Vogel M, Mayer MP, Bukau B. 2006. Allosteric regulation of Hsp70 chaperones involves a conserved interdomain linker. J. Biol. Chem. 281:38705–38711 [DOI] [PubMed] [Google Scholar]
- 485. Voisine C, et al. 2001. Jac1, a mitochondrial J-type chaperone, is involved in the biogenesis of Fe/S clusters in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U. S. A. 98:1483–1488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 486. von Janowsky B, Major T, Knapp K, Voos W. 2006. The disaggregation activity of the mitochondrial ClpB homolog Hsp78 maintains Hsp70 function during heat stress. J. Mol. Biol. 357:793–807 [DOI] [PubMed] [Google Scholar]
- 487. von Plehwe U, et al. 2009. The Hsp70 homolog Ssb is essential for glucose sensing via the SNF1 kinase network. Genes Dev. 23:2102–2115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 488. Voos W, Gambill BD, Guiard B, Pfanner N, Craig EA. 1993. Presequence and mature part of preproteins strongly influence the dependence of mitochondrial protein import on heat shock protein 70 in the matrix. J. Cell Biol. 123:119–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 489. Voos W, et al. 1994. Mitochondrial GrpE is present in a complex with hsp70 and preproteins in transit across membranes. Mol. Cell. Biol. 14:6627–6634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 490. Voos W, Rottgers K. 2002. Molecular chaperones as essential mediators of mitochondrial biogenesis. Biochim. Biophys. Acta 1592:51–62 [DOI] [PubMed] [Google Scholar]
- 491. Vylkova S, Li XS, Berner JC, Edgerton M. 2006. Distinct antifungal mechanisms: beta-defensins require Candida albicans Ssa1 protein, while Trk1p mediates activity of cysteine-free cationic peptides. Antimicrob. Agents Chemother. 50:324–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 492. Wagner I, Arlt H, van Dyck L, Langer T, Neupert W. 1994. Molecular chaperones cooperate with PIM1 protease in the degradation of misfolded proteins in mitochondria. EMBO J. 13:5135–5145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 493. Walker KW, Lyles MM, Gilbert HF. 1996. Catalysis of oxidative protein folding by mutants of protein disulfide isomerase with a single active-site cysteine. Biochemistry 35:1972–1980 [DOI] [PubMed] [Google Scholar]
- 494. Wandinger SK, Richter K, Buchner J. 2008. The Hsp90 chaperone machinery. J. Biol. Chem. 283:18473–18477 [DOI] [PubMed] [Google Scholar]
- 495. Wandinger SK, Suhre MH, Wegele H, Buchner J. 2006. The phosphatase Ppt1 is a dedicated regulator of the molecular chaperone Hsp90. EMBO J. 25:367–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 496. Wang Q, Chang A. 1999. Eps1, a novel PDI-related protein involved in ER quality control in yeast. EMBO J. 18:5972–5982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 497. Wegele H, Wandinger SK, Schmid AB, Reinstein J, Buchner J. 2006. Substrate transfer from the chaperone Hsp70 to Hsp90. J. Mol. Biol. 356:802–811 [DOI] [PubMed] [Google Scholar]
- 498. Weitzmann A, Volkmer J, Zimmermann R. 2006. The nucleotide exchange factor activity of Grp170 may explain the non-lethal phenotype of loss of Sil1 function in man and mouse. FEBS Lett. 580:5237–5240 [DOI] [PubMed] [Google Scholar]
- 499. Welker S, et al. 2010. Hsp12 is an intrinsically unstructured stress protein that folds upon membrane association and modulates membrane function. Mol. Cell 39:507–520 [DOI] [PubMed] [Google Scholar]
- 500. Wells GB, Dickson RC, Lester RL. 1998. Heat-induced elevation of ceramide in Saccharomyces cerevisiae via de novo synthesis. J. Biol. Chem. 273:7235–7243 [DOI] [PubMed] [Google Scholar]
- 501. Wendler P, et al. 2007. Atypical AAA+ subunit packing creates an expanded cavity for disaggregation by the protein-remodeling factor Hsp104. Cell 131:1366–1377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 502. Wendler P, et al. 2009. Motor mechanism for protein threading through Hsp104. Mol. Cell 34:81–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 503. Wera S, De Schrijver E, Geyskens I, Nwaka S, Thevelein JM. 1999. Opposite roles of trehalase activity in heat-shock recovery and heat-shock survival in Saccharomyces cerevisiae. Biochem. J. 343(Pt. 3):621–626 [PMC free article] [PubMed] [Google Scholar]
- 504. Werner-Washburne M, Becker J, Kosic-Smithers J, Craig EA. 1989. Yeast Hsp70 RNA levels vary in response to the physiological status of the cell. J. Bacteriol. 171:2680–2688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 505. Werner-Washburne M, Craig EA. 1989. Expression of members of the Saccharomyces cerevisiae hsp70 multigene family. Genome 31:684–689 [DOI] [PubMed] [Google Scholar]
- 506. Werner-Washburne M, Stone DE, Craig EA. 1987. Complex interactions among members of an essential subfamily of hsp70 genes in Saccharomyces cerevisiae. Mol. Cell. Biol. 7:2568–2577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 507. Westerheide SD, et al. 2004. Celastrols as inducers of the heat shock response and cytoprotection. J. Biol. Chem. 279:56053–56060 [DOI] [PubMed] [Google Scholar]
- 508. Westermann B, Gaume B, Herrmann JM, Neupert W, Schwarz E. 1996. Role of the mitochondrial DnaJ homolog Mdj1p as a chaperone for mitochondrially synthesized and imported proteins. Mol. Cell. Biol. 16:7063–7071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 509. Westermann B, Neupert W. 1997. Mdj2p, a novel DnaJ homolog in the mitochondrial inner membrane of the yeast Saccharomyces cerevisiae. J. Mol. Biol. 272:477–483 [DOI] [PubMed] [Google Scholar]
- 510. Westwood JT, Clos J, Wu C. 1991. Stress-induced oligomerization and chromosomal relocalization of heat-shock factor. Nature 353:822–827 [DOI] [PubMed] [Google Scholar]
- 511. Wiederrecht G, Seto D, Parker CS. 1988. Isolation of the gene encoding the S. cerevisiae heat shock transcription factor. Cell 54:841–853 [DOI] [PubMed] [Google Scholar]
- 512. Wieser R, et al. 1991. Heat shock factor-independent heat control of transcription of the CTT1 gene encoding the cytosolic catalase T of Saccharomyces cerevisiae. J. Biol. Chem. 266:12406–12411 [PubMed] [Google Scholar]
- 513. Winkler A, et al. 2002. Heat stress activates the yeast high-osmolarity glycerol mitogen-activated protein kinase pathway, and protein tyrosine phosphatases are essential under heat stress. Eukaryot. Cell 1:163–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 514. Wisplinghoff H, et al. 2004. Nosocomial bloodstream infections in US hospitals: analysis of 24,179 cases from a prospective nationwide surveillance study. Clin. Infect. Dis. 39:309–317 [DOI] [PubMed] [Google Scholar]
- 515. Wotton D, Freeman K, Shore D. 1996. Multimerization of Hsp42p, a novel heat shock protein of Saccharomyces cerevisiae, is dependent on a conserved carboxyl-terminal sequence. J. Biol. Chem. 271:2717–2723 [DOI] [PubMed] [Google Scholar]
- 516. Wright CM, et al. 2007. The hsp40 molecular chaperone Ydj1p, along with the protein kinase C pathway, affects cell-wall integrity in the yeast Saccharomyces cerevisiae. Genetics 175:1649–1664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 517. Xiao J, Kim LS, Graham TR. 2006. Dissection of Swa2p/auxilin domain requirements for cochaperoning Hsp70 clathrin-uncoating activity in vivo. Mol. Biol. Cell 17:3281–3290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 518. Xu X, Azakami H, Kato A. 2004. P-domain and lectin site are involved in the chaperone function of Saccharomyces cerevisiae calnexin homologue. FEBS Lett. 570:155–160 [DOI] [PubMed] [Google Scholar]
- 519. Xu X, Kanbara K, Azakami H, Kato A. 2004. Expression and characterization of Saccharomyces cerevisiae Cne1p, a calnexin homologue. J. Biochem. 135:615–618 [DOI] [PubMed] [Google Scholar]
- 520. Xu Y, Singer MA, Lindquist S. 1999. Maturation of the tyrosine kinase c-src as a kinase and as a substrate depends on the molecular chaperone Hsp90. Proc. Natl. Acad. Sci. U. S. A. 96:109–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 521. Yam AY, Albanese V, Lin HT, Frydman J. 2005. Hsp110 cooperates with different cytosolic HSP70 systems in a pathway for de novo folding. J. Biol. Chem. 280:41252–41261 [DOI] [PubMed] [Google Scholar]
- 522. Yamamoto N, Takemori Y, Sakurai M, Sugiyama K, Sakurai H. 2009. Differential recognition of heat shock elements by members of the heat shock transcription factor family. FEBS J. 276:1962–1974 [DOI] [PubMed] [Google Scholar]
- 523. Yao Y, Zhou Y, Wang C. 1997. Both the isomerase and chaperone activities of protein disulfide isomerase are required for the reactivation of reduced and denatured acidic phospholipase A2. EMBO J. 16:651–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 524. Youker RT, Walsh P, Beilharz T, Lithgow T, Brodsky JL. 2004. Distinct roles for the Hsp40 and Hsp90 molecular chaperones during cystic fibrosis transmembrane conductance regulator degradation in yeast. Mol. Biol. Cell 15:4787–4797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 525. Zarzov P, Boucherie H, Mann C. 1997. A yeast heat shock transcription factor (Hsf1) mutant is defective in both Hsc82/Hsp82 synthesis and spindle pole body duplication. J. Cell Sci. 110:1879–1891 [DOI] [PubMed] [Google Scholar]
- 526. Zhang L, Hach A, Wang C. 1998. Molecular mechanism governing heme signaling in yeast: a higher-order complex mediates heme regulation of the transcriptional activator HAP1. Mol. Cell. Biol. 18:3819–3828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 527. Zhang W, et al. 2004. Biochemical and structural studies of the interaction of Cdc37 with Hsp90. J. Mol. Biol. 340:891–907 [DOI] [PubMed] [Google Scholar]
- 528. Zhao R, et al. 2005. Navigating the chaperone network: an integrative map of physical and genetic interactions mediated by the hsp90 chaperone. Cell 120:715–727 [DOI] [PubMed] [Google Scholar]
- 529. Zhong T, Arndt KT. 1993. The yeast SIS1 protein, a DnaJ homolog, is required for the initiation of translation. Cell 73:1175–1186 [DOI] [PubMed] [Google Scholar]
- 530. Zhou C, et al. 2011. Motility and segregation of hsp104-associated protein aggregates in budding yeast. Cell 147:1186–1196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 531. Zou J, Guo Y, Guettouche T, Smith DF, Voellmy R. 1998. Repression of heat shock transcription factor HSF1 activation by HSP90 (HSP90 complex) that forms a stress-sensitive complex with HSF1. Cell 94:471–480 [DOI] [PubMed] [Google Scholar]
- 532. Zu T, Verna J, Ballester R. 2001. Mutations in WSC genes for putative stress receptors result in sensitivity to multiple stress conditions and impairment of Rlm1-dependent gene expression in Saccharomyces cerevisiae. Mol. Genet. Genomics 266:142–155 [DOI] [PubMed] [Google Scholar]