Abstract
Diamond-Blackfan anemia (DBA) is a broad developmental disease characterized by anemia, bone marrow (BM) erythroblastopenia, and an increased incidence of malignancy. Mutations in ribosomal protein gene S19 (RPS19) are found in ~25% of DBA patients; however, the role of RPS19 in the pathogenesis of DBA remains unknown. Using global gene expression analysis, we compared highly purified multipotential, erythroid, and myeloid BM progenitors from RPS19 mutated and control individuals. We found several ribosomal protein genes downregulated in all DBA progenitors. Apoptosis genes, such as TNFRSF10B and FAS, transcriptional control genes, including the erythropoietic transcription factor MYB (encoding c-myb), and translational genes were greatly dysregulated, mostly in diseased erythroid cells. Cancer-related genes, including RAS family oncogenes and tumor suppressor genes, were significantly dysregulated in all diseased progenitors. In addition, our results provide evidence that RPS19 mutations lead to codownregulation of multiple ribosomal protein genes, as well as downregulation of genes involved in translation in DBA cells. In conclusion, the altered expression of cancer-related genes suggests a molecular basis for malignancy in DBA. Downregulation of c-myb expression, which causes complete failure of fetal liver erythropoiesis in knockout mice, suggests a link between RPS19 mutations and reduced erythropoiesis in DBA.
Keywords: Diamond-Blackfan anemia, Bone marrow failure, Global gene expression, Ribosomal protein genes, Apoptosis Cancer
Introduction
Diamond-Blackfan anemia (DBA) (Online Mendelian Inheritance in Man 105650) is a congenital form of red-cell aplasia with marked clinical heterogeneity. The disease is usually characterized by diminished erythroid precursors in the bone marrow (BM). The majority of DBA patients present with macrocytic anemia, reticulocytopenia, and elevated erythrocyte adenosine deaminase activity (eADA) [1, 2]. Growth retardation and variable congenital anomalies, in particular of the head and upper limbs, are present in approximately 30%–40% of patients indicating that DBA is a broad disorder of development. The anemia is often initially responsive to steroids, but more than half of these patients require long-term, low-dose steroid treatment to maintain normal erythropoiesis [1–3]. Analysis of natural history reveals a substantial increase in the incidence of malignancies in DBA patients, particularly acute myelogenous leukemia and solid tumors [4 – 6].
Ribosomal protein S19 (RPS19), on chromosome 19q13.2 [7], is mutated in approximately 25% of both sporadic and familial probands [7–10]. However, its role in the pathogenesis of DBA remains to be determined. Expression studies show that rpS19 protein levels are high in proerythroblasts but decline progressively with maturation of erythroid progenitors, suggesting that high levels may be critical at the earliest stages of erythropoiesis [11]. Our data show that RPS19 mRNA and protein are deficient in DBA cases, with mutations leading to premature stop codons, and suggest that haploinsufficiency is likely the pathogenetic mechanism in DBA patients with RPS19 mutations [10]. Deficiency of rps19 in yeast leads to a block in ribosomal RNA processing [12], but whether that is true in mammalian cells is unknown. Whether DBA is due to a defect in ribosomal biogenesis, an abnormality in translation, and/or disruption of an rps19 extraribosomal function(s) important for erythropoiesis, hematopoiesis, and development, remains an open question.
To investigate the molecular changes secondary to rps19 haploinsufficiency, we performed global gene expression analysis of purified BM subsets from RPS19-mutated DBA patients and unaffected control individuals. By examining erythroid progenitor cells, we expected to identify both primary- and secondary-disease-related changes that would reflect the unique pathophysiology of this highly affected population of cells. Parallel studies of a myeloid population, as well as multipotential progenitor cells, allowed for the determination of erythroid-specific changes, as well as general cell-type independent changes that are more likely to reflect gene-specific proximal responses to rps19 deficiency. Our results provide evidence that rpS19 deficiency leads to codownregulation of multiple ribosomal protein genes, as well as downregulation of the genes involved in transcription and translation in DBA cells. Identification of expression changes for multiple cancer-related genes suggests a molecular basis for the increased risk for malignancy in these patients.
Materials and Methods
Patients
Three adult DBA patients (D1–D3) with RPS19 mutations and six sex- and age-matched control individuals (C1–C6) participated in the study; all nine individuals are of Northern European (Caucasian) origin. The patients were in remission without any treatment of anemia for at least 10 years. We specifically selected patients in remission who have unequivocal evidence of persistent abnormalities in erythropoiesis (see below). Abnormal erythropoiesis was also demonstrated in liquid culture by Ohene-Abuakwa et al. in erythroid progenitor cells from DBA individuals including untreated patients [13]. Furthermore, it would be difficult, if not impossible, to control adequately for steroid or transfusion dependence. At the time of our study, the patients’ red blood cell counts and hemoglobin levels were in the normal or low/borderline range, and their leukocytes and platelets were normal (Table 1). Bone marrow samples were obtained from the patients and control individuals with informed consent under a protocol at Children’s Hospital Boston. For control experiments, replicate BM aspirations were obtained from one diseased individual (D2) and from two control individuals (C4 and C5); the BM sample from C6 was divided into two portions for further analysis.
Table 1.
Diamond-Blackfan anemia patients D1–D3
| D1 | D2 | D3 | |
|---|---|---|---|
| Age (years) | 46 | 30 | 42 |
| Sex | Female | Male | Male |
| HCT (%) | 41.9 (n, 30.0–49.5) | 31.6 (n, 42.0–52.0) | 41.1 (n, 42.0–52.0) |
| HGB (g/dL) (n, 12.2) | 14.09 (n, 11.0–15.6) | 11.0 (n, 13.0–18.0) | 13.9 (n, 14.0–18.0) |
| RBCs (106/μL) | 4.09 (n, 3.5–5.5) | 3.01 (n, 4.7–6.0) | 4.92 (n, 4.7–6.1) |
| MCV (FL) | 102.1 (n, 80.0–94.0) | 105 (n, 80.0–98.0) | 84 (n, 80.0–98.0) |
| WBCs (103/μL) | 5.46 (n, 4.5–11.0) | 5.8 (n, 4.0–10.0) | 6.3 (n, 4.5–11.0) |
| PLTs (103/μL) | 198.0 (n, 130–400) | 223 (n, 140–440) | 210 (n, 130–400) |
| eADA (EU/gm Hgb) | 1.39 (n, 0.33–0.96) | 1.92 (n, 0.33–0.96) | 1.45 (n, 0.33–0.96) |
| RPS19 mutation | ins 250A, ex 4a | 185G→A; Arg62Gln | 392T→G; Leu131Arg |
Frameshift at codon 84; stop codon 7 amino acids behind coding region.
Abbreviations: D1, diseased 1; D2, diseased 2; D3, diseased 3; ex, exon; HCT, hematocrit; HGB, hemoglobin; ins, insertion; MCV, mean corpuscular volume; n, normal range; PLT, platelet; RBC, red blood cell; WBC, white blood cell.
Fluorescence-Activated Cell Sorting Separation of BM Cell Populations
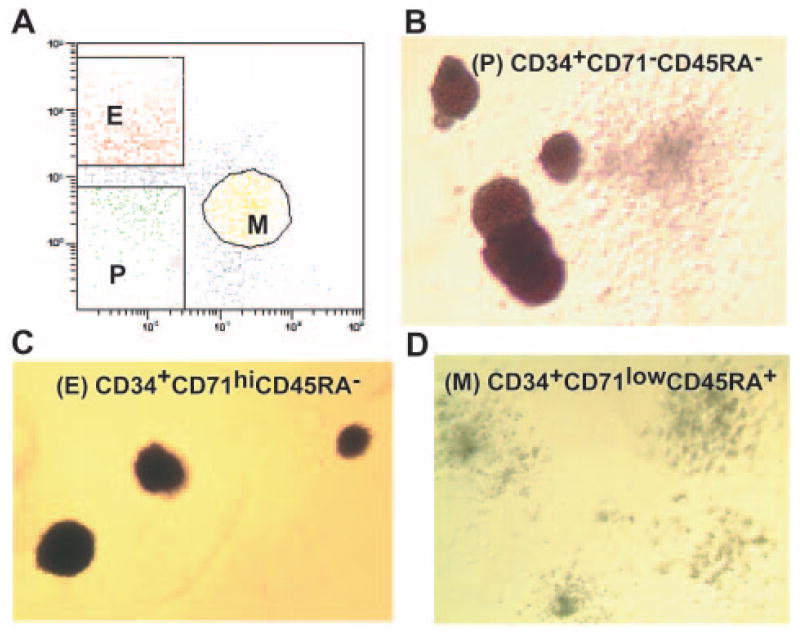
Bone marrow mononuclear cells (1–2 × 108) from DBA patients and control individuals were isolated using Histopaque-107 (Sigma-Aldrich, St. Louis, http://www.sigmaaldrich.com). Isolated cells were stained for 20 minutes on ice with the anti-human antibodies CD34 PE-Cy5, CD71 fluorescein isothiocyanate (BD Biosciences, San Diego, http://www.bdbiosciences.com), and 2H4-RD1 (CD45RA-PE) (Beckman Coulter, Miami, FL, http://www.beckmancoulter.com). The stained cells were separated by fluorescence-activated cell sorting (FACS) using ALTRA HyPeSort System (Beckman Coulter) into three populations: CD34+CD71−CD45RA− (P population), CD34+CD71hiCD45RA− (E population), and CD34+CD71lowCD45RA+ (M population) according to the method of Lansdorp and Dragowska [14].
Methylcellulose Colony Assay
Cells from each sorted population P, E, and M, from all individuals were plated at a concentration of 103 cells per ml in complete methylcellulose medium (MethoCult GF+ H4435; Stem Cell Technologies, Vancouver, BC, Canada, http://www.stemcell.com) containing 30% fetal bovine serum and the human recombinant cytokines stem cell factor, interleukin-3, in-terleukin-6, granulocyte-monocyte colony-stimulating factor, granulocyte colony-stimulating factor, and erythropoietin. The colonies were cultured at 37°C in a water-saturated atmosphere of 5% CO2 and scored after 14 days of culture.
RNA Isolation and Array Hybridization
Total RNA was isolated as previously described [10] from three FACS-separated BM subsets P, E, and M from three patients and six control individuals. The isolated RNA samples were resuspended in diethyl pyrocarbonate-treated water and prepared for hybridization to Affymetrix HG-133A arrays (Affymetrix, Santa Clara, CA, http://www.affymetrix.com) according to the manufacturer’s instructions [15]. Following hybridization, the signal amplification staining option was chosen on the Affymetrix Fluidics station 400, the GeneChips were scanned in an Affymetrix/Hewlett-Packard G2500A Gene Array Scanner (Hewlett-Packard, Palo Alto, CA, http://www.hp.com), and the resulting signals were quantified and stored.
TaqMan Quantitative Real-Time Polymerase Chain Reaction
18S rRNA transcripts from the P, E and M BM populations from diseased and control samples were quantified by real-time polymerase chain reaction (Applied BioSystems, Foster City, CA, http://www.appliedbiosystems.com) with an Assays-On-Demand gene expression kit as previously described [10]. Control reactions with human glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (Assays-On-Demand; Applied BioSystems), as an endogenous reference, were run in together with the 18S rRNA. The outcome of each amplification was calculated with comparative methods according to the manufacturer’s protocol. The 18S rRNA expression level and fold changes between DBA and control samples were normalized to GAPDH in each RNA sample. To validate the microarray data, we quantified the expression of MYB, TNFRSF10B, TNFRSF6, RPL18, and RPS19 RNA in P, E, and M BM populations from diseased and control samples using the Assays-On-Demand gene expression kit and the same conditions as above.
Data Processing
The GeneChip Analysis Suite MAS5.0 (Affymetrix) was used for the initial microarray data processing and noise/quality control [15].
Linear Correlation
Standard linear correlation coefficients were calculated between all pairs of the total 39 samples. Twenty-seven samples were from three diseased individuals (D1–D3) and six control individuals (C1–C6); 12 replicate sample assays were from individuals D2, C4, C5, and C6 as a qualitative assessment of the microarray data.
Normalization
Normalization of the overall original data set was performed prior to differential gene analysis (see below). Each sample expression profile was normalized via linear regression against the expression profile of sample C5.E, which had the highest average correlation against all other samples [16, 17].
Principal Component Analysis
Principal component analysis (PCA) is a standard linear algebraic technique for transforming—typically a high dimensional/feature set— data into a new set of features or principal components (PCs) that correspond with their contribution to the variance structure of the original data [17, 18]. Each PC captures a monotonically decreasing (and “orthogonal”) percentage of variance in the data. PCA was performed using Matlab (Math-Works, Natick, MA, http://www.mathworks.com) on two sets of data. The first set comprises the 27 unique samples (D1–D3 and C1–C6) with 12,593 genes, which have a LocusLink number. The second set of data contains 3,993-gene profiles of all 27 samples. These genes have at least three “Present” calls in all samples and a coefficient of variance between 0.5 and 30 across all 27 sample conditions—i.e., these genes were selected to represent each sample precisely because they had been reliably detected across all 27 sample conditions, and their profile was not static across these conditions.
Hierarchical Clustering
Hierarchical clustering analysis was performed using the Cluster (version 2.0) and Treeview (version 1.6) software (http://rana.lbl.gov/EisenSoftware.htm) [19]. The normalized 27 data sets, containing 22,283 probe sets, and two other sets of 27 samples, already analyzed by PCA, containing 12,593 and 3,993 genes, were analyzed with centered linear correlation as a measure of similarity using average linkage clustering and a SD cut off ≥2,000; 1,000; and 300, respectively. Samples were clustered based on their correlation coefficient without prior knowledge of the disease status.
Statistical Analysis
To evaluate differential gene expression between DBA patients and control individuals for three BM cell populations P, E, and M, we used two separate statistical methods.
Geometric Log (Arithmetic) Fold Analysis
This analysis was applied to identify genes significantly fold changed in diseased versus control subjects [17, 20]. Suppose that the expression levels of a gene G are a1 ≤ a2 ≤ a3 in the three disease cases, and b1 ≤ b2 ≤ … ≤ b6 in the six controls. We placed a threshold for all reported expression levels at a minimum of 50 intensity units. Define Xj = log(aj) − 0.5 · (log(b7 − 2j) + log(b8 − 2j)) for j = 1, 2, 3. The average (AvgLF) and standard deviation (StdLF) of the geometric log fold change of gene G between the diseased and control groups are the average and standard deviation of Xj values, respectively. Define the intra-group log fold change (Noise) as the greater of [log(a3) − log(a1)] or [log(b6) + log(b5) + log(b4) − log(b3) − log(b2) − log(b1)]/3. We say that a gene is significantly differentially fold changed between the two comparison groups if its reported expression levels satisfy the parameters Abs(AvgLF) − StdLF > 0.5 · Noise, Abs(AvgLF) > max(Noise, log(2)), and the gene is called Present in at least three sample conditions. The false discovery rate (FDR) for these parameters is 0.63. The FDR is computed through an iterated series of 10,000 whole data set permutations of the disease labels for each gene.
Significance Analysis of Microarrays
A two-class unpaired data analysis was performed each time, with twofold cutoff and a range of different FDRs [21]. A median FDR of ≤12% was selected for patient to normal data set comparisons, with D values of 1.135, 1.679, and 2.249 in the P, E, and M cell population data sets, respectively. The D parameter, as described [21], enables the user to examine the effect of the false-positive rate in determining significance. Because in the case of the E population analysis over 1,500 probe sets were found to be significantly changed for the 12% FDR cutoff, only the top 390 most significant probe sets were used for further analysis (FDR ≤ 7%). The overlaps of significantly changed probe sets in both analyses in P, E, and M populations were 33%, 14%, and 24%, respectively.
Pathway Analysis
To study the potential biological significance of the genes changed in DBA, we applied pathway analysis MetaCore (Gene-Go, St. Joseph, MI, http://www.genego.com).
Gene Ontology
Gene Ontology (GO) analysis was performed using GeneSpring (version 6.0) (Agilent Technologies, Inc., Palo Alto, CA, http://www.agilent.com).
Results
Isolation of the Three Highly Purified BM Subsets from DBA Patients and Control Individuals
Bone marrow mononuclear cells from three RPS19 mutated DBA patients (diseased individuals D1–D3) (details are given in Table 1) and from six control individuals (C1–C6) were FACS-separated into three populations (27 samples). We isolated P, E, and M population progenitors, which comprised CD34+CD71−CD45RA−, CD34+CD71hiCD45RA−, and CD34+CD71lowCD45RA+ subsets, respectively (Fig. 1A) [14]. We found that the percentages of CD34+ cells, as well as the relative proportions of the three sorted populations from diseased and normal bone marrow samples, were comparable (data not shown). As control experiments for BM interaspiration variability, we sorted second sets of P, E, and M cells from replicate aspirations from individuals D2, C4, and C5 (an additional nine samples). To further assess intersort, interlabeling, and interchip hybridization reproducibility, we performed another sort of the three populations from the second portion of the BM aspiration from C6, for a total of 39 samples. The purity of each sorted population was more than 97% based on FACS reanalysis. To assess cell sorting accuracy, we performed methylcellulose colony assays and demonstrated that the E populations were highly enriched for mature erythroid burst-forming units (BFU-E) and erythroid colony-forming units (CFU-E) (>90% of all colonies), whereas more than 99% of colonies from M populations were granulocyte colony-forming units (CFU-G), monocyte colony-forming units (CFU-M), and granulocyte-macrophage colony-forming units CFU-GM (Fig. 1B–1D). As expected, the P populations gave rise to granulocyte-erythroid-monocyte-megakaryocyte colony-forming units (CFU-GEMM), primitive BFU-E, and primitive CFU-GM. The numbers of the colonies in all diseased BM populations were comparable with control samples, but the size of DBA BFU-E and CFU-E colonies was strikingly smaller (data not shown).
Figure 1.
Fluorescence-activated cell sorting separation and methyl-cellulose colony assays of bone marrow multipotential, erythroid and myeloid progenitors. (A): Three bone marrow (BM) populations isolated by flow cytometry: P (CD34+CD71−CD45RA− cells), E (CD34+CD71hiCD45RA− cells), and M (CD34+CD71lowCD45RA+ cells). The methylcellulose colony assays demonstrate that three populations of BM progenitors were highly enriched in primitive erythroid burst-forming units (BFU-E) and granulocyte-macrophage colony-forming units (CFU-GM) colonies from multipotential progenitors (average enrichment, 28 of 1,000 and 50 of 1,000 colonies, respectively) (B), more mature BFU-E and erythroid colony-forming units (CFU) colonies from erythroid progenitors (average enrichment for erythroid colonies, 160 of 1,000) (C), and granulocyte CFU, monocyte CFU, and granulocyte-macrophage CFU colonies from myeloid progenitors (average enrichment for myeloid colonies, 45 of 1,000) (D). Abbreviations: E, erythroid progenitors; M, myeloid progenitors; P, multipotential progenitors.
Linear Correlation Revealed High Correlation Between Data Sets
For details of linear correlation, see supplemental online Results.
Three BM Progenitor Populations Are Genomically Distinguished
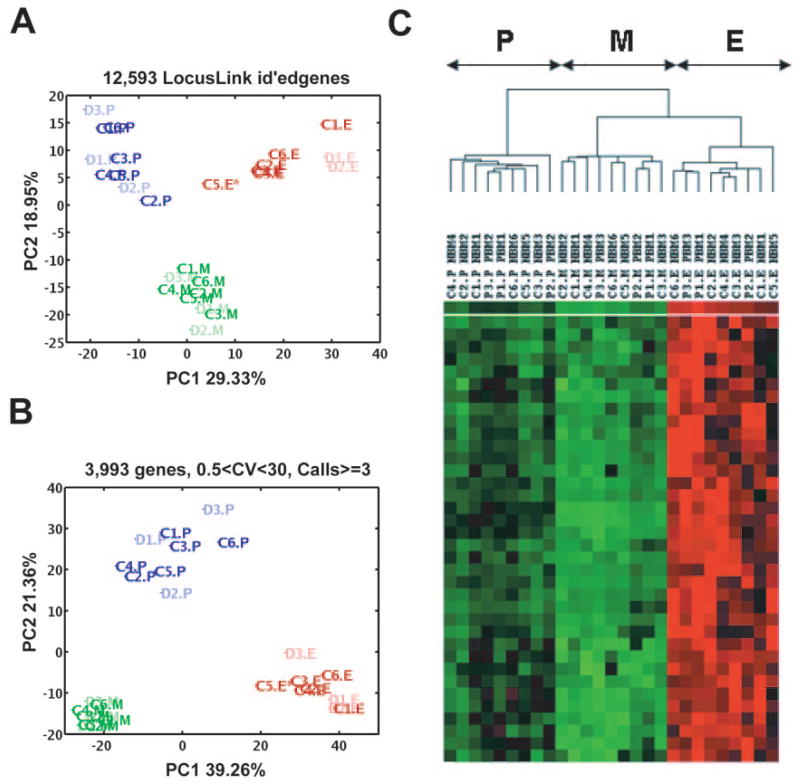
We investigated the question of how the 27 unique samples (i.e. excluding the 12 replicates) relate to one another based solely on their overall RNA (genomic) profiles and ignoring a priori specimen labels P, E, and M. PCA was used to address this question [17, 18]. PCA is an unsupervised linear algebraic technique for decomposing a data set with a high number of dimensions or features (e.g., genes in this case) into an equivalent or lower dimensional representation of the features, called PCs, that most prominently contribute to the inherent variance of the original data. Conceptually, one can think of the samples in the original data set as individual points sitting in a genomic space of n = 12,593 dimensions/microarray probes with LocusLink identification (ID). The first principal component (PC1) represents the direction of the greatest variance in the data, PC2 the direction of the next greatest variance in the data, etc. Within the first two most prominent genomic PCs, the 27 samples separate into three distinct clusters of nine samples each, coinciding with the P, E, and M cell populations, respectively (Fig. 2A). Figure 2B shows results of PCA on 3,993 genes (with at least three “Present” calls in all samples and a coefficient of variance between 0.5 and 30 across all 27 samples). This analysis also separated 27 samples into three clusters, P, E, and M, defined by PC1 and PC2. In fact, these three clusters are cleanly separable along genomic PC1 alone—responsible for 39.3% of the variance—with the E and M populations being the two most genomically dissimilar clusters and with the common progenitor cells (P population) occupying an intermediate space along PC1. When projecting each sample along genomic PC2, the P population is separated from the joint E and M populations, suggesting that genomic PC2 might correspond to a temporal/maturation axis. Finally, PCA also shows the P population to be the most genomically heterogenous of the three, as visually surmised from the greater intracluster scatter of P specimens. When quantified, the relative areas occupied by P:E:M are 13:7:1. The direction of third greatest variance (PC3) corresponds to disease status by distinguishing the diseased and control samples (supplemental online Fig. 1).
Figure 2.
Distinguishing three bone marrow populations by principal component analysis and hierarchical clustering. (A): PC1 and PC2 performed on 12,593 genes (with LocusLink identification) and 27 samples clearly distinguished three cell populations, P, E, and M. Blue, P population (dark blue, control samples [C1–C6]; light blue, diseased samples [D1–D3]); red, E population (dark red, control samples; light red, diseased samples); green, M population (dark green, control samples; light green, diseased samples). (B): PC1 and PC2 performed on 3,993 genes also separate three cell populations (P, E, and M) and show the P population to be the most genomically heterogenous of the three; the areas of P:E:M are 13:7:1. (C): Hierarchical clustering identifies three major clusters, with different expression patterns perfectly overlapped with three bone marrow progenitors (P, E, and M). Erythroid and myeloid populations form related but distinct subsets, whereas the more primitive P population is more distant. Abbreviations: C, control sample; CV, coefficient of variance; D, diseased sample; E, erythroid; id, identification; M, myeloid; P, multipotential; PC, principal component.
These observations were further confirmed by subjecting the 27 normalized data sets, containing 22,283 probe sets, to hierarchical clustering analysis, which also identified three major specimen clusters perfectly overlapping with the three different cell populations under study (P, E, and M). Erythroid and myeloid populations formed related but distinct subsets, whereas the P population CD34+CD71−CD45RA− was more distant (Fig. 2C). Since dendrogram patterns may depend on number of data sets, we assessed the robustness of this analysis using a “take one out” strategy whereby the analysis was repeated nine times after random removal of three samples (one sample from each population [P, E, and M]). In all 10 analyses, each population reproducibly segregated into distinct cell-type-specific clusters (data not shown). The hierarchical clustering analysis was also performed on 27 data sets containing: (a) 12,593 probe sets with LocusLink ID, and (b) 3,993 probe sets (representing highly expressed transcripts with the greatest variation) (as for PCA). Again, in both these analyses, the P and M populations formed subsets that were more closely related and the E population was more distant (data not shown), confirming the PCA results. Here, again, we used a “take out strategy,” and the analysis for the each set of data was repeated nine times after random removal of three samples (as above). Neither the PCA nor the hierarchical clustering analysis separated the specimens by disease status, indicating that only a small subset of genes are likely to exhibit DBA-specific changes in expression.
Fold Change Analysis Shows the Highest Number of the Significantly Changed Genes in DBA Erythroid Progenitors
To evaluate differential gene expression between diseased and control individuals for three BM cell populations (P, E, and M), we used two separate statistical methods, geometric fold change analysis [20] and significance analysis of microarrays [21]. These two analyses each revealed the highest number of changed genes in DBA erythroid progenitors. Combined results of both analyses identified 545 genes (565 probe sets) with ≥2-fold expression changes (62 genes overexpressed and 482 underexpressed) in DBA E populations compared with control individuals (supplemental online Table 1). Parallel analysis showed 106 genes (109 probe sets) significantly changed in multipotential progenitors (41 genes overexpressed and 65 underexpressed) and 72 genes (74 probe sets) (34 genes overexpressed and 38 underexpressed) in myeloid progenitors in DBA patients (supplemental online Table 1). A statistical analysis using the χ2 test revealed that the E, P, and M groups of significantly changed genes are statistically different (χ2 = 590; p < .0001). Twenty-nine significantly changed genes in two or more cell types are presented in supplemental online Figure 2 and supplemental online Table 2. Analysis of GO categories revealed no major groups of overrepresented categories among the genes changed in E, M, and P populations (data not shown). However, pathway analysis using MetaCore (GeneGo) showed that ribosomal proteins, transcriptional control, and apoptosis genes figured prominently among the transcripts with the greatest fold changes.
Alteration of Expression of Ribosomal Protein Genes, Genes Involved in Translation, and 18S rRNA in DBA Patients with RPS19 Mutations
Interestingly, among genes with ≥2-fold changed expression in the DBA patients with RPS19 mutations, we found 10 additional ribosomal protein genes (RPS10, RPS14, RPS28, RPL10L, RPL14, RPL15, RPL18, RPL18A, RPL28, RPL36) significantly underexpressed in E, P, and/or M populations (Table 2). Two of these genes were underexpressed in P populations, seven in E populations, and five in M populations. In addition, mitochondrial ribosomal protein gene L23 (MRPL23) was 4-fold and 3.7-fold downregulated in DBA E and M subsets of cells, respectively.
Table 2.
Expression of ribosomal protein, transcription, and translation genes in three Diamond-Blackfan anemia (DBA) cell populations
| Cell population | Affymetrix identification | Gene symbol | Fold change | LocusLink |
|---|---|---|---|---|
| P | 222297_x_at | RPL18 | −2.27 | 604179 |
| 219762_s_at | RPL36 | −2.45 | 25873 | |
| 218128_at | NFYB | −2.25 | 4801 | |
| 202823_at | TCEB1 | −2.16 | 6921 | |
| 202163_s_at | CNOT8 | −2.42 | 9337 | |
| 206118_at | STAT4 | 2.53 | 6775 | |
| 203882_at | ISGF3G | 2.84 | 10379 | |
| E | 217336_at | RPS10 | −2.04 | 6204 |
| 208903_at | RPS28 | −2.08 | 6234 | |
| 217559_at | RPL10L | −6.66 | 140801 | |
| 219138_at | RPL14 | −13.8 | 9045 | |
| 222297_x_at | RPL18 | −2.10 | 6141 | |
| 216383_at | RPL18A | −3.14 | 6142 | |
| 200003_s_ | RPL28 | −2.06 | 6158 | |
| 213897_s_at | MRPL23 | −4.08 | 6150 | |
| 221616_s_at | TAF9L | −4.31 | 51616 | |
| 209463_s_at | TAF12 | −2.08 | 6883 | |
| 209152_s_at | TCF3 | −2.80 | 6929 | |
| 215167_at | CRSP2 | −3.18 | 9282 | |
| 204109_s_at | NFYA | −4.49 | 4800 | |
| 214766_s_at | AHCTF1 | −4.79 | 25909 | |
| 214918_at | HNRPM | −4.42 | 4670 | |
| 215152_at | MYB | −6.20 | 4602 | |
| 209511_at | POLR2F | −2.10 | 5435 | |
| 214144_at | POLR2D | −8.44 | 5433 | |
| 201026_at | EIF5B | −6.68 | 9669 | |
| 222294_s_at | EIF2C2 | −3.07 | 27161 | |
| 213087_s_at | EEF1D | −5.52 | 1936 | |
| 213907_at | EEF1E1 | −38.0 | 9521 | |
| 204906_at | RPS6KA2 | −49.2 | 6196 | |
| M | 208646_at | RPS14 | −2.30 | 6208 |
| 217559_at | RPL10L | −9.18 | 140801 | |
| 217266_at | RPL15 | −2.04 | 6138 | |
| 208903_at | RPS28 | −2.08 | 6234 | |
| 219762_s_at | RPL36 | −2.45 | 25873 | |
| 213897_s_at | MRPL23 | −3.74 | 6150 | |
| 212955_s_at | POLR2I | −2.03 | 5438 |
A negative value indicates downregulation in cells from DBA patients.
Abbreviations: E, bone marrow erythroid progenitor; M, myeloid progenitor; P, multipotential progenitor.
Furthermore, the DBA E populations exhibited significant downregulation of genes encoding proteins important for translation including the eukaryotic translation initiation factors EIF5B and EIF2C2, and two eukaryotic translation elongation factors, factor 1 δ (EEF1D) and factor 1 ε 1 (EEF1E1). In addition, ribosomal protein S6 kinase 90-kDa polypeptide 2 (RPS6KA2) was highly downregulated in the DBA E populations (supplemental online Tables 1, 2). The activity of this protein has been implicated in controlling cell growth and differentiation [22]. The fact that several significantly underexpressed genes (in E and P populations) encode proteins involved in translation suggests that this process is dysregulated in DBA cells.
To explore whether RPS19 mutations in DBA patients result in abnormal rRNA processing, we performed quantitative real-time polymerase chain reaction (qRT-PCR) experiments to measure the amount of 18S rRNA in the three cell populations from diseased and control samples. The amount of the target transcripts of 18S rRNA was normalized to a reference gene, GAPDH. We found that the expression of 18S rRNA is 3.5–7-fold upregulated in the DBA P populations, 1.5– 4-fold upregulated in the E populations, and unchanged in the M populations (Table 3). The upregulation of 18S rRNA by qRT-PCR in DBA may reflect the mechanism described in yeast [12] and indicate lack of processing and abnormal accumulation of pre-18S rRNA in the RPS19 mutated patients with a further defect of small subunit rRNA maturation.
Table 3.
Expression of 18S rRNA related to GAPDH in three BM subsets in diseased and control samples
| BM cell population | Sample identification | 18S rRNA, average CT | GAPDH, average CT | ΔCT 18S rRNA- GAPDH | ΔΔCT (ΔCT − ΔCT,C) | 18S rRNA relative to GAPDH (2−ΔΔCT) |
|---|---|---|---|---|---|---|
| P | (C3, C5, C6) | 20.60 ± 0.48 | 21.83 ± 0.32 | −1.24 ± 0.58 | 0.00 ± 0.58 | 1.0 (0.67–1.49) |
| D2 | 19.54 ± 0.49 | 22.69 ± 0.028 | −3.155 ± 0.49 | −1.91 ± 0.49 | 3.76 (2.68–5.27) | |
| D3 | 19.12 ± 0.07 | 23.20 ± 0.15 | −4.075 ± 0.17 | −2.83 ± 0.17 | 7.11 (6.32–8.00) | |
| E | (C1, C3–C6) | 18.46 ± 0.87 | 21.16 ± 0.37 | −2.70 ± 0.95 | 0.00 ± 0.95 | 1.0 (0.52–1.93) |
| D2 | 16.75 ± 0.00 | 20.03 ± 0.03 | −3.28 ± 0.03 | −0.58 ± 0.03 | 1.49 (1.46–1.52) | |
| D3 | 17.13 ± 0.07 | 21.89 ± 0.02 | −4.76 ± 0.07 | −2.06 ± 0.07 | 4.16 (3.97–4.37) | |
| M | (C1, C3–C6) | 16.38 ± 1.49 | 19.91 ± 0.99 | −3.53 ± 1.78 | 0.00 ± 1.78 | 1.0 (0.29–3.43) |
| D2 | 17.30 ± 0.98 | 21.06 ± 0.63 | −3.76 ± 1.16 | −0.23 ± 1.16 | 1.17 (0.52–2.62) | |
| D3 | 19.36 ± 0.64 | 22.48 ± 0.28 | −3.12 ± 0.69 | 0.39 ± 0.69 | 0.76 (0.47–0.81) |
Abbreviations: BM, bone marrow; C, control sample; CT, number of cycles; CT,C, number of control cycles; D2, diseased 2; D3, diseased 3; E, erythroid progenitor; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; M, myeloid progenitor; P, multipotential progenitor.
Transcriptional Control Genes Are Dysregulated in Diseased BM Cell Populations
We found that BM cells from DBA patients showed significant downregulation of several genes that encode proteins involved in transcription regulation, particularly in erythroid progenitors (supplemental online Table 1). Two genes encoding TATA-binding protein-associated factor (TAF) protein (TAF9L and TAF12, which take part in transcription regulation) and two genes encoding RNA polymerase II polypeptides (POLR2F and POLR20, which are responsible for synthesizing messenger RNA, including RP genes), were downregulated in E populations from DBA patients. Three transcription factor genes, transcription factor 3 (TCF3), nuclear transcription factor Y (NFYA), and AT hook containing transcription factor 1 (AH-CTF1), involved in transcription regulation, as well as the heterogenous nuclear ribonucleoprotein M gene (HNRPM), which influences pre-mRNA processing, metabolism, and transport, were also found to be significantly underexpressed in the DBA E populations (supplemental online Table 1; Table 2). In diseased P populations, two transcription factor genes, nuclear transcription factor Yβ (NFYB) and CCR4-NOT transcription complex subunit 8 (CNOT8), and transcription elongation factor B polypeptide 1 (15 kDa, elongin C) (TCEB1) were significantly underexpressed. Two other transcription factors, signal transducer and activator of transcription 4 (STAT4), which is essential for mediating responses to IL12 in lymphocytes, and interferon-stimulated transcription factor 3 (ISGF3G) were upregulated in P populations (supplemental online Table 1; Table 2). The POL2I gene encoding the other RNA polymerase II polypeptide, POL2I, was downregulated in DBA M populations (supplemental online Table 1; Table 2).
Transcription Factor MYB Is Downregulated in DBA BM Subsets
c-Myb is the cellular homolog of the myeloblastosis viral oncogene v-myb. We found that the gene that encodes c-Myb, MYB, is expressed in all three BM cell populations. Interestingly, it was sixfold downregulated in diseased E populations (supplemental online Table 1; Table 2). In contrast, other transcription factor genes known to be involved in regulation of erythropoiesis, such as TAL1 (SCL) interrupting locus (SIL), LIM domain only 2 (LMO2), GATA binding protein 1 (GATA1), GATA binding protein 2 (GATA2), Kruppel-like factor 1 (erythroid) (KLF1), and signal transducer and activator of transcription 5A (STAT5), were unchanged in DBA subsets.
DBA Erythroid Cells Exhibit Gene Expression Abnormalities Related to Apoptosis and Cancer
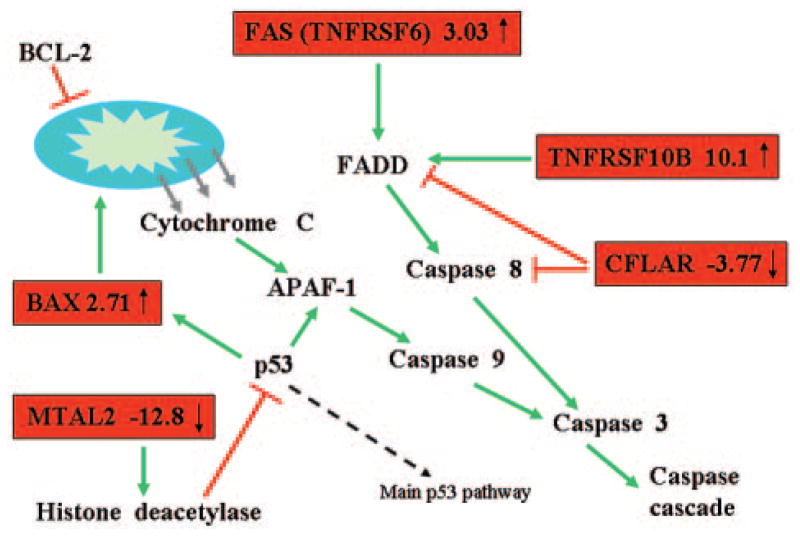
This study also revealed several potentially important groups of genes, such as apoptosis and cancer-related genes, as well as genes involved in DNA repair, that were over- or underexpressed mostly in the E populations (supplemental online Table 1). Among the upregulated transcripts were several proapoptotic genes, including tumor necrosis factor receptor superfamily member 10b TNFRSF10B and tumor necrosis factor receptor superfamily member 6 (TNFRSF6) (FAS); they were upregulated 10- and 3-fold, respectively. Both of these genes encode proteins that stimulate procaspase 8 cleavage and caspase cascade activation [23] through the FAS-associated via death domain (FADD) protein. Other upregulated apoptotic genes in DBA erythroid progenitors included BCL2-associated X protein (BAX) (2.7-fold) and metastasis-associated 1 family member 2 (MTA2) (12.8-fold), whereas the gene encoding apoptosis inhibitory protein CASP8 and FADD-like apoptosis regulator (CFLAR) is downregulated (3.77-fold) in diseased E populations (Fig. 3; Table 4).
Figure 3.
The apoptotic genes changed in Diamond-Blackfan anemia (DBA) are shown here as a truncated apoptosis pathway. The genes on the red background are changed in erythroid population in DBA patients. Green arrows symbolize activation; red lines symbolize inhibition. Abbreviations: APAF-1, apoptotic protease activating factor 1; BAX, BCL-2-associated X protein; BCL-2, anti-apoptotic BCL-2 family members; CFLAR, CASP8 and FADD-like apoptosis regulator; FADD, FAS-associated via death domain protein; FAS (INFRSF6), tumor necrosis factor receptor superfamily, member 6; MTA2, metastasis-associated 1 family, member 2; TNFRSF10b, tumor necrosis factor receptor superfamily, member 10b.
Table 4.
Expression of apoptosis-related genes, DNA repair, and histone family of genes in DBA BM progenitors
| Affymetrix identification | Gene symbol | LocusLink | Fold change
|
||
|---|---|---|---|---|---|
| P | E | M | |||
| 209295_at | TNFRSF10B | 8795 | — | 10.1 | 3.09 |
| 204780_s | TNFRSF6 | 355 | — | 3.03 | — |
| 209508_x_at | CFLAR | 8837 | — | −3.77 | — |
| 211833_s | BAX | 581 | 3.49 | 2.71 | 2.43 |
| 212969_x_at | MTA2 | 256364 | — | −12.8 | — |
| 208368_s | BRCA2 | 675 | −2.59 | −4.85 | — |
| 203742_s | TDG | 6996 | — | 3.48 | — |
| 213344_s_at | H2AX | 3014 | — | −5.04 | — |
| 208694_at | DNA-PK | 5591 | −2.24 | — | — |
| 203409_at | DDB2 | 1643 | — | 3.73 | 2.08 |
| 214469_at | HIST1H2AE | 3012 | — | −5.49 | — |
| 222067_x_at | HIST1H2BD | 3017 | — | −5.13 | — |
| 208490_x_at | HIST1H2BF | 8343 | — | −6.24 | — |
| 208546_x_at | HIST1H2BH | 8345 | — | −3.98 | — |
| 209398_at | HIST1H1C | 3006 | — | −17.1 | — |
| 208579_x_at | H2BFS | 54145 | — | −7.76 | — |
| 213826_s_at | H3F3A | 3020 | — | −5.85 | — |
| 203582_s_at | RAB4A | 5867 | −2.12 | −5.97 | −2.21 |
| 221960_s_at | RAB2 | 5862 | — | −5.34 | — |
| 205037_at | RABL4 | 11020 | — | −5.09 | — |
| 219622_at | RAB20 | 55647 | — | — | 2.34 |
| 203885_at | RAB21 | 23011 | 2.11 | — | — |
| 212099_at | ARHG | 388 | — | −6.54 | — |
| 203132_at | RB1 | 5925 | −2.3 | — | – |
| 200658_s_at | PHB | 5245 | — | — | −2.48 |
| 202377_at | LEPR | 3953 | 3.07 | — | — |
| 209894_at | LEPR | 3953 | — | — | 4.49 |
A positive value indicates upregulation, and a negative value indicates downregulation in cells from DBA patients. —, not significantly changed.
Abbreviations: DBA, Diamond-Blackfan anemia; E, bone marrow erythroid progenitor; M, myeloid progenitor; P, multipotential progenitor.
Several genes involved in DNA repair (supplemental online Table 1) (including tumor suppressor breast cancer 2, early onset [BRCA2]; thymine-DNA glycosylase [TGD], which is responsible for G/T and G/U mismatch repair; and H2A histone family member X [H2AX], which is critical for facilitating the assembly of specific DNA-repair complexes on damaged DNA [24]) are downregulated in DBA cells, mostly in erythroid progenitors (Table 4). The only gene from this group to be upregulated in E and M diseased populations is damage-specific DNA binding protein 2 (DDB2), the smaller subunit of a heterodimeric protein implicated in the etiology of xeroderma pigmentosum group E. This subunit appears to be required for DNA binding [25] (Table 4). Histones 1, 2A, 2B, and 3 are also significantly downregulated, specifically in erythroid progenitors (Table 4). The downregulation of histones may simply reflect the apoptotic stage of erythroid cells in DBA.
Importantly, we identified 29 cancer-related genes significantly changed in DBA P, E, or M populations, the majority of which were changed in E populations only (Table 4). Several RAB genes, which belong to RAS oncogene superfamily and encode RAB proteins involved in vesicular fusion trafficking, were changed in all three diseased populations of cells. RAB4A was sixfold downregulated in E populations and twofold down-regulated in M populations. RAB2 and RABL4 were fivefold downregulated in E subset of cells, whereas RAB20 and RAB21 were twofold overexpressed in M and P populations, respectively (supplemental online Table 1; Table 4). In addition, member B of the RAS homolog gene family, ARHB, which has a role as a tumor suppressor in lung neoplasms [26] and is essential for DNA damage-induced apoptosis in neoplastically transformed cells [27] is 6.5-fold underexpressed in E subsets of cells. Other downregulated tumor suppressors were BRCA2 in E and P diseased subsets, retinoblastoma 1 (RB1) in P populations, and prohibitin (PHB) in diseased M populations (supplemental online Table 1; Table 4). Interestingly, the leptin receptor, LEPR, which was found to a have promoting effect on carcinogenesis and metastasis of breast cancer [28] and possible involvement in bladder cancer [29], was upregulated 4.5- and 3-fold in E and P diseased populations, respectively (supplemental online Table 1; Table 4). These findings suggest a molecular basis for the increased risk for malignancy in DBA [4 – 6].
Validation of Microarray Gene Expression Data by qRT-PCR
To validate the microarray data, we performed qRT-PCR of several significantly changed and important genes, such as MYB, TNFRSF10B, TNFRSF6, RPL18, and RPS19. We indeed confirmed the downregulation of MYB RNA in the erythroid population from diseased samples (supplemental online Table 3), whereas the overexpressed by microarray DBA erythroid samples of TNFRSF10B and TNFRSF6 were also upregulated by qRT-PCR (supplemental online Tables 4, 5). The twofold downregulation of RPL18 RNA in the P and E populations of the diseased samples were also shown by qRT-PCR (supplemental online Table 6). Since the mutations in the diseased samples are missense mutations or an insertion that does not cause a premature stop codon, we did not expect, and did not find, any expression changes of RPS19 RNA in microarray data in the diseased samples. Supplemental online Table 7 shows results that confirm the microarray gene expression data.
Discussion
Global gene expression profiling has been successfully applied to identify molecular signatures of hematopoietic stem cells [30–33], as well as CD34+ cells, in aplastic anemia [34]. We compared highly purified multipotential, erythroid, and myeloid bone marrow progenitors from diseased and control samples to investigate the molecular changes secondary to rps19 insufficiency in DBA. Our results are representative of at least DBA patients with RPS19 mutations who are in remission. Fold change analysis showed the highest number of significantly changed genes in DBA erythroid progenitors. These results correlate with clinical observation and in vitro studies of the disease, as clinically, the most prominent symptom of DBA is anemia, and bone marrow smears usually show an absence or insufficiency of erythroid precursors with normal myeloid and platelet lineages [1]. Furthermore, in vitro colony assays have revealed deficiencies of BFU-E and CFU-E [35–37], further indicating that erythroid cells are the most affected in DBA. Although our three patients were in remission at the time of BM aspirations, without any history of treatment for anemia for more than 10 years, their complete blood counts revealed evidence of persistent functional abnormalities of erythropoiesis (macrocytosis, low/borderline hemoglobin, and elevated eADA) with normal leukocyte and platelet counts (Table 1). Although, the percentages of both the CD34+ cells and the P, E, and M sorted populations in diseased BMs were very similar to control samples, the size of DBA erythroid colonies were smaller in DBA than in control individuals, as previously reported [13, 38]. In contrast, the CFU-GM and CFU-GEMM colonies appeared to be normal in the diseased samples. These observations and the fact that the most genes were changed in DBA erythroid populations indicate that the molecular defect is mostly expressed in, although not limited to [39], the erythroid lineage. Models of DBA using siRNA-mediated knockdown of RPS19 also show a greater effect on erythroid progenitors, although the myeloid lineage is reduced as well [40, 41].
Interestingly, we found that transcription factor gene MYB is sixfold underexpressed in diseased samples; furthermore, it is the only “erythroid” transcription factor altered in these cells. Yolk sac erythropoiesis in MYB knockout mice is normal, but there is complete failure of erythropoiesis in fetal liver. Progenitors of other lineages, but not megakaryocytes, were also decreased, indicating that c-myb is required for early definitive cellular expansion [42]. Furthermore, a knockdown allele of MYB shows that suboptimal levels of c-myb favor macrophage and megakaryocyte differentiation, whereas higher levels are particularly important for erythropoiesis and lymphopoiesis [43]. Our data suggest a pathway by which rps19 may be involved in erythroid proliferation.
Among the upregulated transcripts in diseased erythroid progenitors were several proapoptotic genes, including TNFRSF10B, FAS, BAX, and MTA2 (10-, 3-, 2.7-, and 12.8-fold, respectively), whereas the gene encoding apoptosis inhibitory protein, CFLAR, was downregulated (3.77-fold) (Fig. 3; Table 4). FAS has been shown to have an important role in regulation of apoptosis in early erythroid cells, whereas other anti-apoptotic genes are underexpressed [44]. Importantly, in vitro studies previously showed that DBA erythroid progenitors were more susceptible to apoptotic death than normal erythroid progenitors after erythropoietin deprivation [45].
It was recently shown in zebrafish that 11 ribosomal protein genes act as haploinsufficient tumor suppressors; the haploin-sufficiency for any of these genes caused malignant tumors of the peripheral nerve sheath [46]. We found one of these genes, RPL36, significantly underexpressed in DBA patients with RPS19 mutations. None of the studied patients showed any signs of malignancy to date; however, DBA is clearly associated with an increased risk of cancer [4 – 6] in patients with or without RPS19 mutation (unpublished data). Although, it remains to be determined whether insufficiency of rps19 protein and disruption of its ribosomal or potential extraribosomal function contribute to neoplasm in humans, the secondary reduction of other RP genes may be a contributing factor in the increased risk of malignancy in DBA patients.
Our findings also indicate that some RP genes are closely coregulated in humans and that rps19 mutation results in down-regulation of the additional RP genes in both erythroid and nonerythroid cells in DBA patients.
In sum, these data suggest that RPS19 mutation and rps19 protein insufficiency in DBA patients may lead to impairment of ribosome biogenesis by the dysregulated stoichiometry of ribosomal components and subsequent reduction of protein translation capacity. This ribosomal abnormality may be particularly crucial for developing erythroid cells, whose survival and division require large amounts of protein synthesis. At the molecular level, erythroid progenitors seem to be most affected in DBA patients. However, it is also possible that specific targets, such as c-myb, are affected through an extraribosomal role of rps19. Since c-myb level is important for erythropoiesis [42], the regulation and expression of this protein will be the subject of future studies.
Supplementary Material
Acknowledgments
This work was supported by the Diamond-Blackfan Anemia Foundation (H.T.G.), the Dana-Mahoney Center for Neuro-Oncology (A.T.K.), NIH Grant P01-NS40828 (A.T.K., D.S., A.H.B.), NIH Grant R01-NS047527 (A.T.K.), Komitet Badan Naukowych Grant Statutowy (J.M.Z.), NIH P01-HL99021 (H.T.G., C.A.S.), NIH Grant R01-HL64775 (C.A.S.), and NIH Grant R01-AR044345 (A.H.B.). We also thank Dr. David G. Nathan for inspirational discussion, encouragement, and critical comments on the manuscript. We thank Travis Burlson and the Harvard Neuromuscular Disease Project core microarray facility, supported by NIH Grant NS40828, for expert assistance with microarray hybridizations; John Daley and Suzan Lazo of the Dana-Farber Cancer Institute sequencing core facility for outstanding help with BM cell separation; and Karen Backer, Carolyn Wong, and Dr. Bert Glader for eADA determinations.
Footnotes
See www.StemCells.com for supplemental material available online.
Disclosures
The authors indicate no potential conflicts of interest.
References
- 1.Alter BP, Young NS. The bone marrow failure syndromes. In: Nathan DG, Orkin HS, editors. Hematology of Infancy and Childhood. Vol. 1. Philadelphia, PA: Saunders; 1998. pp. 237–335. [Google Scholar]
- 2.Willig TN, Niemeyer CM, Leblanc T, et al. Identification of new prognosis factors from the clinical and epidemiologic analysis of a registry of 229 Diamond-Blackfan anemia patients. DBA group of Societe d’Hematologie et d’Immunologie pediatrique (SHIP), Gesellshaft fur Padiatrische Onkologie und Hamatologie (GPOH), and the European Society for Pediatric Hematology and Immunology (ESPHI) Pediatr Res. 1999;46:553–561. doi: 10.1203/00006450-199911000-00011. [DOI] [PubMed] [Google Scholar]
- 3.Ball SE, McGuckin CP, Jenkins G, et al. Diamond-Blackfan anaemia in the U.K: Analysis of 80 cases from a 20-year birth cohort. Br J Haematol. 1996;94:645–653. doi: 10.1046/j.1365-2141.1996.d01-1839.x. [DOI] [PubMed] [Google Scholar]
- 4.Janov AJ, Leong T, Nathan DG, et al. Diamond-Blackfan anemia. Natural history and sequelae of treatment Medicine (Baltimore) 1996;75:77–78. doi: 10.1097/00005792-199603000-00004. [DOI] [PubMed] [Google Scholar]
- 5.van Dijken PJ, Verwijs W. Diamond-Blackfan anemia and malignancy. A case report and a review of the literature. Cancer. 1995;76:517–520. doi: 10.1002/1097-0142(19950801)76:3<517::aid-cncr2820760324>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- 6.Lipton JM, Federman N, Khabbaze Y, et al. Osteogenic sarcoma associated with Diamond-Blackfan anemia: A report from the Diamond-Blackfan Anemia Registry. J Pediatr Hematol Oncol. 2001;23:39–44. doi: 10.1097/00043426-200101000-00009. [DOI] [PubMed] [Google Scholar]
- 7.Draptchinskaia N, Gustavsson P, Andersson B, et al. The gene encoding ribosomal protein S19 is mutated in Diamond-Blackfan anaemia. Nat Genet. 1999;21:169–175. doi: 10.1038/5951. [DOI] [PubMed] [Google Scholar]
- 8.Willig TN, Draptchinskaia N, Dianzani I, et al. Mutations in ribosomal protein S19 gene and diamond blackfan anemia: Wide variations in phenotypic expression. Blood. 1999;94:4294–4306. [PubMed] [Google Scholar]
- 9.Orfali KA, Ohene-Abuakwa Y, Ball SE. Diamond Blackfan anaemia in the UK: Clinical and genetic heterogeneity. Br J Haematol. 2004;125:243–252. doi: 10.1111/j.1365-2141.2004.04890.x. [DOI] [PubMed] [Google Scholar]
- 10.Gazda HT, Zhong R, Long L, et al. RNA and protein evidence for haplo-insufficiency in Diamond-Blackfan anaemia patients with RPS19 mutations. Br J Haematol. 2004;127:105–113. doi: 10.1111/j.1365-2141.2004.05152.x. [DOI] [PubMed] [Google Scholar]
- 11.Da Costa L, Narla G, Willig TN, et al. Ribosomal protein S19 expression during erythroid differentiation. Blood. 2003;101:318–324. doi: 10.1182/blood-2002-04-1131. [DOI] [PubMed] [Google Scholar]
- 12.Leger-Silvestre I, Caffrey JM, Dawaliby R, et al. Specific role for yeast homologs of the diamond blackfan anemia associated Rps19 protein in ribosome synthesis. J Biol Chem. 2005;12 doi: 10.1074/jbc.M506916200. [DOI] [PubMed] [Google Scholar]
- 13.Ohene-Abuakwa Y, Orfali KA, Marius C, et al. Two-phase culture in Diamond Blackfan anemia: Localization of erythroid defect. Blood. 2005;105:838–846. doi: 10.1182/blood-2004-03-1016. [DOI] [PubMed] [Google Scholar]
- 14.Lansdorp PM, Dragowska W. Long-term erythropoiesis from constant numbers of CD34+ cells in serum-free cultures initiated with highly purified progenitor cells from human bone marrow. J Exp Med. 1992;175:1501–1509. doi: 10.1084/jem.175.6.1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haslett JN, Sanoudou D, Kho AT, et al. Gene expression comparison of biopsies from Duchenne muscular dystrophy (DMD) and normal skeletal muscle. Proc Natl Acad Sci U S A. 2002;99:15000–15005. doi: 10.1073/pnas.192571199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu TD. Analysing gene expression data from DNA microarrays to identify candidate genes. J Pathol. 2001;195:53–65. doi: 10.1002/1096-9896(200109)195:1<53::AID-PATH891>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 17.Kho AT, Zhao Q, Cai Z, et al. Conserved mechanisms across development and tumorigenesis revealed by a mouse development perspective of human cancers. Genes Dev. 2004;18:629–640. doi: 10.1101/gad.1182504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Misra J, Schmitt W, Hwang D, et al. Interactive exploration of microarray gene expression patterns in a reduced dimensional space. Genome Res. 2002;12:1112–1120. doi: 10.1101/gr.225302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eisen MB, Spellman PT, Brown PO, et al. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci U S A. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhao Q, Kho A, Kenney AM, et al. Identification of genes expressed with temporal-spatial restriction to developing cerebellar neuron precursors by a functional genomic approach. Proc Natl Acad Sci U S A. 2002;99:5704–5709. doi: 10.1073/pnas.082092399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhao Y, Bjorbaek C, Weremowicz S, et al. RSK3 encodes a novel pp90rsk isoform with a unique N-terminal sequence: Growth factor-stimulated kinase function and nuclear translocation. Mol Cell Biol. 1995;15:4353–4363. doi: 10.1128/mcb.15.8.4353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kabra NH, Kang C, Hsing LC, et al. T cell-specific FADD-deficient mice: FADD is required for early T cell development. Proc Natl Acad Sci U S A. 2001;98:6307–6312. doi: 10.1073/pnas.111158698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Celeste A, Petersen S, Romanienko PJ, et al. Genomic instability in mice lacking histone H2AX. Science. 2002;296:922–927. doi: 10.1126/science.1069398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Keeney S, Chang GJ, Linn S. Characterization of a human DNA damage binding protein implicated in xeroderma pigmentosum E. J Biol Chem. 1993;268:21293–21300. [PubMed] [Google Scholar]
- 26.Mazieres J, Antonia T, Daste G, et al. Loss of RhoB expression in human lung cancer progression. Clin Cancer Res. 2004;10:2742–2750. doi: 10.1158/1078-0432.ccr-03-0149. [DOI] [PubMed] [Google Scholar]
- 27.Liu A, Cerniglia GJ, Bernhard EJ, et al. RhoB is required to mediate apoptosis in neoplastically transformed cells after DNA damage. Proc Natl Acad Sci U S A. 2001;98:6192–6197. doi: 10.1073/pnas.111137198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ishikawa M, Kitayama J, Nagawa H. Enhanced expression of leptin and leptin receptor (OB-R) in human breast cancer. Clin Cancer Res. 2004;10:4325–4331. doi: 10.1158/1078-0432.CCR-03-0749. [DOI] [PubMed] [Google Scholar]
- 29.Yuan SS, Chung YF, Chen HW, et al. Aberrant expression and possible involvement of the leptin receptor in bladder cancer. Urology. 2004;63:408–413. doi: 10.1016/j.urology.2003.08.038. [DOI] [PubMed] [Google Scholar]
- 30.Ivanova NB, Dimos JT, Schaniel C, et al. A stem cell molecular signature. Science. 2002;298:601–604. doi: 10.1126/science.1073823. [DOI] [PubMed] [Google Scholar]
- 31.Terskikh AV, Miyamoto T, Chang C, et al. Gene expression analysis of purified hematopoietic stem cells and committed progenitors. Blood. 2003;102:94–101. doi: 10.1182/blood-2002-08-2509. [DOI] [PubMed] [Google Scholar]
- 32.Bruno L, Hoffmann R, McBlane F, et al. Molecular signatures of self-renewal, differentiation, and lineage choice in multipotential hemopoietic progenitor cells in vitro. Mol Cell Biol. 2004;24:741–756. doi: 10.1128/MCB.24.2.741-756.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Komor M, Guller S, Baldus CD, et al. Transcriptional profiling of human hematopoiesis during in vitro lineage-specific differentiation. Stem Cells. 2005;23:1154–1169. doi: 10.1634/stemcells.2004-0171. [DOI] [PubMed] [Google Scholar]
- 34.Zeng W, Chen G, Kajigaya S, et al. Gene expression profiling in CD34 cells to identify differences between aplastic anemia patients and healthy volunteers. Blood. 2004;103:325–332. doi: 10.1182/blood-2003-02-0490. [DOI] [PubMed] [Google Scholar]
- 35.Nathan DG, Clarke BJ, Hillman DG, et al. Erythroid precursors in congenital hypoplastic (Diamond-Blackfan) anemia. J Clin Invest. 1978;61:489–498. doi: 10.1172/JCI108960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lipton JM, Kudisch M, Gross R, et al. Defective erythroid progenitor differentiation system in congenital hypoplastic (Diamond-Blackfan) anemia. Blood. 1986;67:962–968. [PubMed] [Google Scholar]
- 37.McGuckin CP, Ball SE, Gordon-Smith EC. Diamond-Blackfan anaemia: Three patterns of in vitro response to haemopoietic growth factors. Br J Haematol. 1995;89:457–464. doi: 10.1111/j.1365-2141.1995.tb08349.x. [DOI] [PubMed] [Google Scholar]
- 38.Sieff CA, Yokoyama CT, Zsebo KM, et al. The production of steel factor mRNA in Diamond-Blackfan anaemia long-term cultures and interactions of steel factor with erythropoietin and interleukin-3. Br J Haematol. 1992;82:640–647. doi: 10.1111/j.1365-2141.1992.tb06938.x. [DOI] [PubMed] [Google Scholar]
- 39.Hamaguchi I, Flygare J, Nishiura H, et al. Proliferation deficiency of multipotent hematopoietic progenitors in ribosomal protein S19 (RPS19)-deficient diamond-Blackfan anemia improves following RPS19 gene transfer. Mol Ther. 2003;7:613–622. doi: 10.1016/s1525-0016(03)00091-1. [DOI] [PubMed] [Google Scholar]
- 40.Ebert BL, Lee MM, Pretz JL, et al. An RNA interference model of RPS19 deficiency in Diamond-Blackfan anemia recapitulates defective hematopoiesis and rescue by dexamethasone: Identification of dexamethasone-responsive genes by microarray. Blood. 2005;105:4620–4626. doi: 10.1182/blood-2004-08-3313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Flygare J, Kiefer T, Miyake K, et al. Deficiency of ribosomal protein S19 in CD34+ cells generated by siRNA blocks erythroid development and mimics defects seen in Diamond-Blackfan anemia. Blood. 2005;105:4627–4634. doi: 10.1182/blood-2004-08-3115. [DOI] [PubMed] [Google Scholar]
- 42.Mucenski ML, McLain K, Kier AB, et al. A functional c-myb gene is required for normal murine fetal hepatic hematopoiesis. Cell. 1991;65:677–689. doi: 10.1016/0092-8674(91)90099-k. [DOI] [PubMed] [Google Scholar]
- 43.Emambokus N, Vegiopoulos A, Harman B, et al. Progression through key stages of haemopoiesis is dependent on distinct threshold levels of c-Myb. EMBO J. 2003;22:4478–4488. doi: 10.1093/emboj/cdg434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.De Maria R, Testa U, Luchetti L, et al. Apoptotic role of Fas/Fas ligand system in the regulation of erythropoiesis. Blood. 1999;93:796–803. [PubMed] [Google Scholar]
- 45.Perdahl EB, Naprstek BL, Wallace WC, et al. Erythroid failure in Diamond-Blackfan anemia is characterized by apoptosis. Blood. 1994;83:645–650. [PubMed] [Google Scholar]
- 46.Amsterdam A, Sadler KC, Lai K, et al. Many ribosomal protein genes are cancer genes in zebrafish. PLoS Biol. 2004;2:E139. doi: 10.1371/journal.pbio.0020139. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.