Abstract
The fundamental mechanisms that control eukaryotic development include extensive regulation at the level of transcription. Gene regulatory networks, composed of transcription factors, their binding sites in DNA, and their target genes, are responsible for executing transcriptional programs. While divergence of these control networks drives species-specific gene expression that contributes to biological diversity, little is known about the mechanisms by which these networks evolve. To investigate how network evolution has occurred in fungi, we used a combination of microarray expression profiling, cis-element identification, and transcription-factor characterization during sexual development of the human fungal pathogen Cryptococcus neoformans. We first defined the major gene expression changes that occur over time throughout sexual development. Through subsequent bioinformatic and molecular genetic analyses, we identified and functionally characterized the C. neoformans pheromone-response element (PRE). We then discovered that transcriptional activation via the PRE requires direct binding of the high-mobility transcription factor Mat2, which we conclude functions as the elusive C. neoformans pheromone-response factor. This function of Mat2 distinguishes the mechanism of regulation through the PRE of C. neoformans from all other fungal systems studied to date and reveals species-specific adaptations of a fungal transcription factor that defies predictions on the basis of sequence alone. Overall, our findings reveal that pheromone-response network rewiring has occurred at the level of transcription factor identity, despite the strong conservation of upstream and downstream components, and serve as a model for how selection pressures act differently on signaling vs. gene regulatory components during eukaryotic evolution.
DURING eukaryotic growth, developmental transitions require the accurate sensing of environmental and cellular signals and subsequent generation of appropriate responses. The fundamental mechanisms that govern these responses include extensive regulation of gene expression. In particular, the accurate timing, location, and extent of the transcription of specific genes are required for normal eukaryotic development. These critical transcriptional regulatory events are governed by gene regulatory networks, composed of DNA-binding proteins, their associated binding sites in DNA, and cohorts of regulated target genes (Davidson EH et al. 2002; Wilczynski and Furlong 2010).
One of the most well characterized eukaryotic gene regulatory networks governs cell type determination in the model yeast Saccharomyces cerevisiae. In S. cerevisiae, two mating types (a and α) are specified by the expression of cell type-specific transcription factors (a1 in a cells and α1 and α2 in α cells) (Herskowitz 1985, 1988). Through coordinate activation and repression of specific gene cohorts, the actions of the transcriptional factors establish three cell types (a, α, and a/α) with distinct properties critical for maintaining the S. cerevisiae sexual cycle (Galgoczy et al. 2004). The more recent elucidation of cell identity circuits in related fungi and subsequent comparative studies with the S. cerevisiae paradigm have been invaluable in revealing features of regulatory network evolution (Tsong et al. 2003, 2006; Baker et al. 2011). For example, the cell identity regulatory network in the related Candida albicans includes an ancient regulator (a2) and diverged cis-regulatory features that indicate the differing mechanisms by which gene regulation evolves and contributes to biological diversity (Tsong et al. 2003, 2006). These studies demonstrate the value of evaluating transcriptional circuits in distinct fungi; such comparative studies have already elucidated nonpredicted regulatory systems governing processes as varied as environmental stress tolerance, carbon source utilization, and dimorphism (Kadosh and Johnson 2001; Martchenko et al. 2007; Homann et al. 2009; Lavoie et al. 2010; Sahni et al. 2010).
A conserved process in fungi that remains unclear in terms of network evolution is sexual development. Sexual development in fungi is the process by which compatible cells fuse with one another (mate), undergo morphological differentiation events, and ultimately generate recombinant progeny (sporulate). In S. cerevisiae, mating is initiated when a and α cells detect one another via chemoattractant pheromones (Dohlman and Thorner 2001). This signal is transduced to the nucleus through a conserved mitogen activated protein kinase (MAPK) cascade to activate the Ste12 transcription factor (Nakayama et al. 1988; Song et al. 1991). Ste12 then induces the expression of target genes whose products execute the cellular fusion event (Fields and Herskowitz 1985; Dolan et al. 1989; Errede and Ammerer 1989). Downstream transcription factors, active in the resulting a/α diploid cell, then control sporulation (Herskowitz 1985, 1988). Like S. cerevisiae, almost all fungal sexual cycles include the processes of mate detection, cell–cell fusion, and recombinant progeny formation (Raper 1966; Ni et al. 2010). However, in the majority of fungi for which sexual cycles have been described, this process has been expanded to include morphological transitions, distinct cell types, multicellular structures, and in some cases fruiting bodies.
A prime example of this more complex development occurs in the meningitis-causing fungus Cryptococcus neoformans (Kwon-Chung 1976; Alspaugh et al. 2000). In C. neoformans, compatible yeast cells mate and then adopt a new morphology in which multicellular filamentous structures are formed. Filamentous growth terminates with the formation of fruiting bodies (basidia) on which four chains of recombinant spores emerge (Idnurm 2010). These features allow the opportunity to investigate how cells adopt new fates and functions in multicellular contexts. Comparisons between regulatory circuits controlling sexual development of C. neoformans and those previously characterized in other fungi are ideal because C. neoformans is phylogenetically distant from many well-characterized systems, allowing the discovery of adaptations that occur over long spans of evolutionary time. Furthermore, C. neoformans sexual development lends itself to comprehensive study because the system is genetically manipulable, and all steps of sexual development occur readily under controlled laboratory conditions in vitro (Kwon-Chung 1976; Hull and Heitman 2002).
As in most fungi, the first step of C. neoformans sexual development is a pheromone-mediated mating event (Davidson et al. 2000; Chung et al. 2002; Shen et al. 2002). In fungi for which molecular data are known, this mating and cellular fusion event is controlled by a DNA-binding protein, known as a pheromone-response factor, that activates target genes via binding to a cis-regulatory sequence known as a pheromone-response element (PRE) (Dolan et al. 1989; Sugimoto et al. 1991; Hartmann et al. 1996; Urban et al. 1996; Sahni et al. 2009). Interestingly, despite the high degree of conservation of the pheromone signaling components in fungi (e.g., G-protein-coupled pheromone receptors and MAP kinase cascade components), the nature and identity of pheromone-response factors vary across species described thus far. The Candida albicans Ste12 homolog Cph1 functions as the pheromone-response factor in the opaque (mating competent) phase. In the fission yeast Schizosaccharomyces pombe and the corn smut Ustilago maydis, there are no detectable Ste12 homologs, and the high-mobility group (HMG) transcription factors Ste11 and Prf1, respectively, function as pheromone-response factors. All identifiable sequence homologs to Ste12 (Cph1), Ste11, and Prf1 in C. neoformans do not exhibit the predicted role in C. neoformans and fail to exhibit the conserved functions of a pheromone-response factor (Wickes et al. 1997; Chang et al. 2001). Thus, the identity of the C. neoformans pheromone-response factor has proven elusive. The lack of bioinformatic predictive power to resolve this question poses a unique opportunity to gain insight into the evolution of the pheromone-response network, because a nonconserved regulator must carry out this conserved function. In other systems, studies have shown that regulatory network evolution is directed by changes in (1) transcription-factor identity and/or behavior, (2) transcription-factor binding sites in DNA, and (3) cohorts of regulated genes (Davidson EH et al. 2002; Gasch et al. 2004; Borneman et al. 2007; Sung et al. 2009; Booth et al. 2010; Wilczynski and Furlong 2010; Baker et al. 2011). A recent discovery by Lin et al. (2010) implicated the more divergent HMG-domain regulator (Mat2) in pheromone signaling. Although the implication of Mat2 suggests a novel regulatory architecture, divergence of the regulatory network cannot be inferred in the absence of functional homology to the pheromone-response factors of other fungi.
To investigate regulatory components in the context of the transcriptional network governing complete sexual development in C. neoformans and how it has evolved in fungi, we first carried out temporal microarray expression analysis of development. Bioinformatic analyses of coexpressed genes revealed the C. neoformans PRE, which we demonstrated to activate transcription in response to pheromone signaling. Our naïve discovery of the PRE as a developmentally important cis-regulatory element led to the search for its binding regulatory factor, which by definition, would be the C. neoformans pheromone-response factor, functionally homologous to those of characterized fungi. Importantly, we showed that Mat2 is required for PRE-mediated activation and interacts directly with PRE sequences in DNA, establishing its role as the C. neoformans pheromone-response factor. These findings demonstrate that rewiring has occurred in this conserved pathway at the level of effector identity, and this particular pathway shows unusual plasticity at the level of regulatory machinery despite the strong conservation of upstream signaling components. These results offer insights into the pheromone-response pathway and sexual-development regulatory network and these novel specialization mechanisms permit further comparative studies to reveal the nature of regulatory network evolution during eukaryotic speciation.
Materials and Methods
Strain manipulations and media
All strains used were of the serotype D background (JEC20 and JEC21) (Kwon-Chung et al. 1992). Yeast strains were maintained on yeast peptone dextrose medium (YPD) or synthetic medium with dextrose (SD) (Sherman 1991). Strains containing telomeric reporter plasmids were maintained on synthetic dextrose media lacking adenine or YPD media containing 100 µg/ml G418. Sexual development assays were conducted on 5% V8 juice agar medium, pH 7.0 (microarray experiment), or 5% V8 juice agar medium with 25 mg/liter uracil, pH 7.0 (reporter assays.)
RNA, cDNA preparation, and microarray hybridization
JEC20 and JEC21 were grown to stationary phase in liquid YPD broth. Five optical density units of each strain were mixed in phosphate buffered saline (PBS) and plated onto V8 juice agar medium. Crosses were incubated at room temperature in the dark for 0.5, 6, 12, 24, 48, or 72 hr. At each time point, the coculture was harvested for RNA extraction using hot acid–phenol as described previously (Ausubel et al. 1997) and purified using the RNEasy Mini Kit (Qiagen, Valencia, CA). cDNA was synthesized using the SuperScript III direct labeling kit (Invitrogen, Carlsbad, CA). The cDNA was purified over a QiaQuick MinElute PCR purification kit (Qiagen, Valencia, CA), both according to manufacturer’s instructions. Fluorescently labeled cDNAs were hybridized in C. neoformans whole-genome spotted microarrays of 70-mer oligonucleotides (Cryptococcus Community Microarray Consortium) containing 7765 open reading frame (ORF) probes. The data presented for each experiment represent eightfold coverage of the genome: quadruplicate experiments (including dye swap), with each slide containing the genome in duplicate. Slides were incubated in prehybridization buffer (5× SSC, 0.1% SDS, 1% BSA) for 1 hr at 42°. cDNAs were then applied to slides in 1× formamide buffer (25% formamide, 5× SSC, 0.1% SDS) containing 10 µg each sheared salmon sperm DNA (Eppendorf, Westbury, NY) and yeast tRNA (Sigma-Aldrich, St. Louis MO). Hybridizations were conducted in a loop design, and each sample served as the reference for the following in the time course. This design enriches the resulting data set for the transcriptional changes that occur as new cell types appear in the population over time. Hybridizations were incubated at 42° for 12 hr. Slides were washed and dried before scanning.
Microarray data extraction and analysis
Arrays were scanned on a GenePix 4000B scanner and the data extracted using GenePix Pro 4.0 (Molecular Devices, Sunnyvale, CA). Extracted data were analyzed using the GeneSpring 10.0 software package (Agilent Technologies, Santa Clara, CA). After initial background correction, data were normalized using the LOWESS (locally weighted scatter plot smoothing) algorithm (Quackenbush 2001) and assessed for statistical significance utilizing ANOVA. Significant genes (P < 0.05) meeting a dynamic expression range greater than 3.5-fold over the course of the experiment or in the top 15th percentile for fold change (up or down) in any single comparison were clustered using Cluster 3.0 (de Hoon et al. 2004) according to a robust K-means 8 algorithm (1000 iterations). The resulting eight clusters were visualized in Java TreeView (http://jtreeview.sourceforge.net/). Clusters were assessed for GO term enrichment using the Genespring 10.0 software package and a likelihood cutoff of P < 0.05 (corrected P value). Likelihood values for the enrichment of unknown genes were determined using the hypergeometric distribution as described previously (Gasch et al. 2004). Upstream of C. neoformans open reading frames (NCBI), 500 or 1000 nucleotides were analyzed for enriched motifs using the MEME algorithm. Motifs were visualized with Weblogo (Crooks et al. 2004). The MAST algorithm was used to identify occurrence of the PRE among other sequence sets (Bailey and Gribskov 1998).
Constructing gpa3mat2 deletion strains
The original gpa3::ADE2 strain WSC75 (Hsueh et al. 2007) was crossed by JEC156 to generate the backcrossed ura5 mutant CHY2226. mat2Δ constructs contained the nourseothricin (NATR) cassette flanked by 1 kb of sequence from upstream and downstream of the MAT2 ORF (Davidson RC et al. 2002). The 5′-flanking region was amplified with primers CHO3473 and CHO3474, the 3′-flanking region was amplified with CHO3475 and CHO3476, and the NATR cassette was amplified with CHO3477 and CHO3478. PCR fusion using CHO3473 and CHO3476 was used to create the final mat2::NATR deletion cassette, which was transformed into CHY2226 by biolistic transformation, grown on medium containing 1 M sorbitol, and selected on medium containing 200 µg/ml nourseothricin (Toffaletti et al. 1993). NATR transformants were screened for the correct integration of the deletion construct by PCR; strain CHY2587 was verified by Southern blot analysis (Ausubel et al. 1997).
Pheromone-response element reporter assays
Sexual development reporter assay:
PRE sequence was used to design oligos CHO2685, CHO2686, CHO2689, and CHO2690, which were phosphorylated, annealed in consecutive pairs, and ligated into the URA5 upstream region at the NheI and BsiWI sites, of pCH703, respectively to generate reporter plasmid pCH871. pCH703 and pCH871 were linearized and transformed into JEC55 (α ade2 ura5). Three independent transformants were assessed alone or in crosses with JEC56 (a ade2 ura5) for URA5 and ADE2 levels by northern blotting.
Reporter assay downstream of constitutive pheromone signaling:
PRE oligos CHO3002 and CHO3003 were ligated into the BsiWI site of pCH1023 to generate pCH1034. pCH1023 and 1034 were linearized and transformed into strains JEC43 (Toffaletti et al. 1993), CHY2226, and CHY2587, on rich media supplemented with G418. The RAM1 (CNF02370) upstream region was amplified with oligos CHO3556 and CHO3559; fusion PCR with oligos CHO3557 and CHO3558 was used to generate a mutant promoter lacking the endogenous 16-bp PRE. PCR products were ligated into pCH1184 to generate an ATG fusion with the genomic URA5 sequence to generate pCH1188 and pCH1185. Constructs were linearized and transformed into C. neoformans strains JEC43, CHY2226, and CHY2587 on rich media supplemented with 100 µg/ml G418. In all cases three independent transformants were assessed on V8 media supplemented with uracil. After 12 hr of incubation, URA5 and GPD1 levels were assessed via Northern blotting.
Northern blot analysis
RNA was prepared from C. neoformans cells as described previously (Ausubel et al. 1997). Northern blots were conducted according to standard protocols using 10 µg total RNA per sample. Probes were generated by PCR amplification utilizing the ExTaq PCR system (TaKaRa Bio) (oligos listed in Supporting information, Table S1). Probes were radiolabeled using Ready-To-GoTM DNA labeling beads (GE Healthcare LifeSciences, Piscataway, NJ) according to manufacturer’s instructions and hybridized to blots at 65° as described previously (Ausubel et al. 1997). Radioactive blots were exposed to a phosphor screen, imaged with a Storm 860 Phosphorimager (Molecular Dymanics, GE Amersham), and quantified using the ImageQuant Software package. Three independently isolated transformants of each reporter construct were assessed in parallel, and URA5 levels were normalized to an internal control gene. ADE2 served as an internal control in Figure 4, expressed on the same episome as the URA5 reporter construct. GPD1 was used for normalization in subsequent experiments. The ADE2 and GPD1 control genes were verified to function similarly (data not shown). The means of sets of biological triplicates ±SE (+PREs vs. –PREs) were compared using Student’s t-test to determine if the difference in URA5 levels was significant (threshold of P < 0.05).
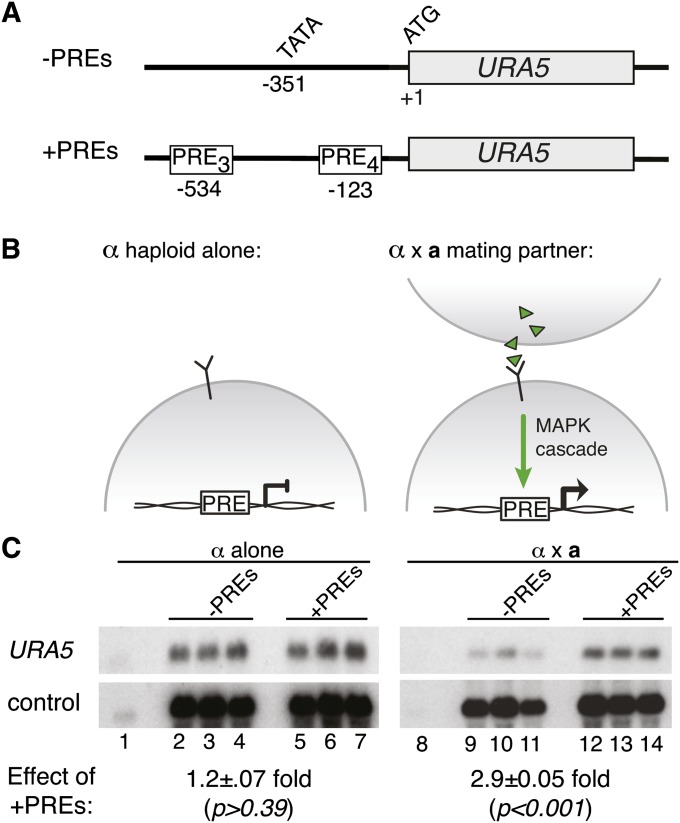
Figure 4 .
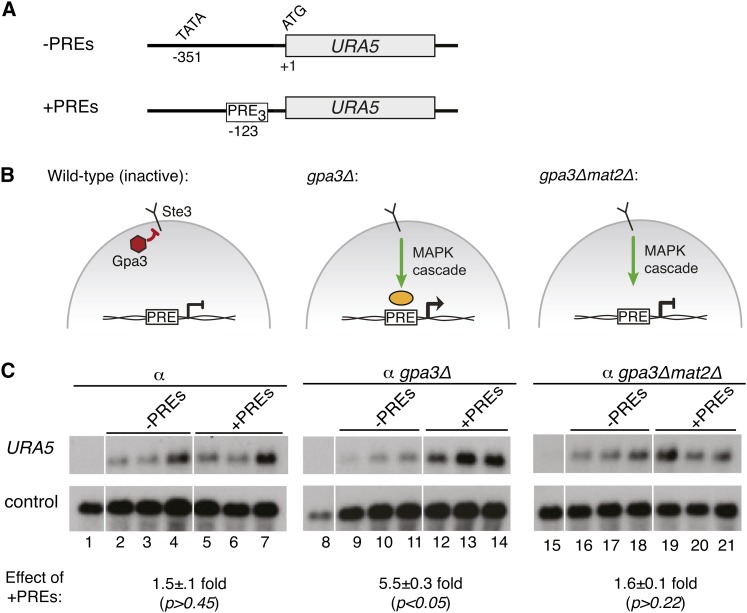
The PRE is sufficient to confer activation of a reporter gene during sexual development. (A) Schematic of constructs used in this assay, showing the reporter gene (URA5), translational start (ATG), and predicted transcriptional start (TATA). The top construct is the control and lacks PREs. The bottom construct includes two PRE insertion sites: three tandem PREs at −123 and four tandem PREs at −534 (PRE sequence = AACAAAAGACA) (B) Schematic of the pheromone response in C. neoformans. The pheromone-response pathway is inactive in the absence of a mating partner (left). In the presence of a mating partner, the pheromone-signaling pathway is activated (right). (C) Northern blot of reporter and control transcript levels. Wild-type and +PRE reporters were expressed in α ura5 strains (three independently isolated strains were assessed for each reporter construct). The resulting reporter strains were incubated alone (lanes 1–7) or in the presence of a mating partner (lanes 8–14). Lanes 1 and 8 show the parental strains lacking reporter plasmid. A probe to URA5 detected reporter readout and a probe to ADE2 served as an internal control. In each case, URA5 levels were normalized to ADE2 levels and mean expression values were compared between sets of biological triplicates to determine fold change with standard error (shown under northern blots). Student’s t-test was used to determine the statistical significance of the differences between mean values among sets of biological triplicates.
Reverse-transcriptase PCR
Single-strand cDNA was synthesized using Superscript III reverse transcriptase (Invitrogen, Carlsbad, CA) and an anchored oligo(dT) primer on 10 µg total RNA harvested from wild-type or mat2Δ crosses (after 12 hr of incubation on solid V8 juice agar medium). Reverse-transcriptase PCR was conducted using diluted cDNAs in a SYBR Green reaction with oligo pairs listed in Table S1. Quantitative real-time PCR (qRT-PCR) was performed using the Bio-Rad CFX96 real-time system with a C1000 thermal cycler (Bio-Rad, Hercules, CA). The normalized expression levels were determined relative to URA5, which was used as the reference gene, and calculated by the Bio-Rad CFX manager software v. 2.0. Reactions were analyzed in triplicate, and mean values were graphed in Microsoft Excel with accompanying standard error. Student’s t-test was used to determine if gene expression differences between wild-type and mat2Δ crosses were statistically significant, applying the commonly accepted standard of significance P < 0.05.
Recombinant Mat2 expression and purification
The full-length MAT2 cDNA was amplified with oligos CHO3843 and CHO3533 and cloned into the BamHI site of pRSET-A (Invitrogen, Carlsbad, CA). The resulting plasmid pCH1226 was transformed into BL21-DE3 pLysS cells (Stratagene, Santa Clara, CA), which were then grown in super-optimal broth (SOB) supplemented with 100 µg/ml ampicillin and 50 µg/ml chloramphenicol. Cells at mid-log phase were induced with 1 mM isopropyl β-d-1-thiogalactopyranoside for 18 hr at 16° with 225 rpm agitation. Cells were harvested, suspended in lysis buffer, and lysed by sonication. The cleared cell lysate was applied to a gravity-flow column packed with Ni-NTA resin (Qiagen, Valencia, CA). The resin was then washed twice with wash buffer (lysis buffer supplemented with 20 mM imidazole), and the Mat2 protein was eluted with lysis buffer supplemented with 0.3 M imidazole.
Electromobility shift assay
Duplex DNA probes corresponding to the MFα1 or RAM1 PRE (44 bp long, containing the PRE and flanking sequence, generated by the annealing of complementary oligos) were end-labeled with [γ-32P]ATP and T4 polynucleotide kinase (oligos listed in Table S1). The resulting probe were used in binding assays containing purified Mat2, 5% glycerol, 37 mM NaCl, 0.5 mM EDTA, 10 mM Tris-Cl pH 7.5, 5 mM MgCl2, 0.3 mg/ml BSA, 25 μg/ml polydeoxyinosinic-deoxycytidylic acid, 25 μg/ml calf thymus DNA, 1 mM dithiothreitol, 0.01% nonidet P-40, and 1 M urea. Unlabeled competitor duplex DNA (40–800× molar excess) was added where indicated. Binding reactions were incubated at room temperature for 30 min and run on a nondenaturing 5% polyacrylamide gel (buffered in 0.5× tris-borate EDTA, pH 7.5) for 2 hr at 200 V at 4°. Radioactive gels were dried and exposed to a phosphor screen for visualization.
Results
Gene expression profiling of C. neoformans during sexual development
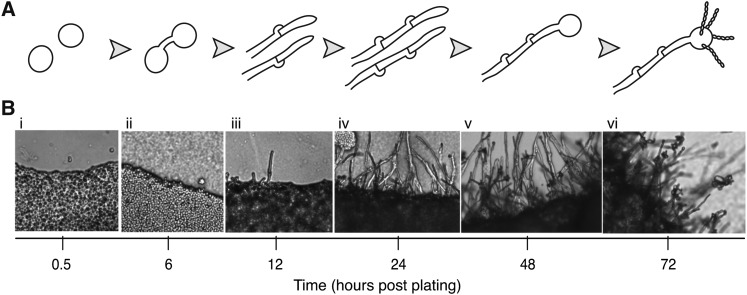
To identify the genome-wide transcriptional changes that occur throughout sexual development, we conducted a time-course microarray experiment spanning six developmental stages to generate a temporal expression pattern for each known gene (Figure 1A). C. neoformans a × α cocultures were grown under sexual development conditions (V8 juice agar), and RNA was harvested at 0.5, 6, 12, 24, 48, and 72 hr postmixing. Time points were chosen on the basis of microscopic identification of cell types of interest: fusants, filaments (early and late), basidia, and spores (Figure 1B). Microarray hybridizations were conducted in a loop design, with each sample serving as a reference for the following time point in the experiment (e.g., 0.5 hr vs. 6 hr, 6 hr vs. 12 hr, etc). The data were filtered for statistical reproducibility, and from these, data were filtered to a final pool of the 3156 (of 7765) most dynamically expressed transcripts over time (Figure 2 and Table S2). Statistically significant, robust changes were detected in the positive and negative directions (exceeding 3.5-fold) for each of the five comparisons. The final pool of statistically significant, highly dynamic genes (3156) was clustered using a robust K-means algorithm to define eight clusters of genes on the basis of similarity of expression patterns over time (Figure 2). The biological relevance of cluster associations was assessed utilizing gene ontology (GO) term-enrichment analyses (Ashburner et al. 2000) (representative enriched GO terms listed in Figure 2; see Table S3 for a comprehensive list).
Figure 1 .
Distinct cell types and multicellular structures form during C. neoformans sexual development. (A) Schematic of sexual development. Haploid yeast of different mating types (a and α) encounter one another and fuse. The fusant then grows filamentously until a basidium forms on the terminal filament cell in which meiosis occurs (nodules on the sides of filaments represent clamp cells). The haploid recombinant progeny are packaged into spores and presented on the basidium surface in long chains. (B) Light microscopy of a sexually developing population on V8 juice agar medium, with images taken at 0.5 (i), 6 (ii), 12 (iii), 24 (iv), 48 (v), and 72 (vi) hours postmixing of the a × α cross (200× magnification), each showing the appearance of a new developmental cell type and corresponding to the schematic in A.
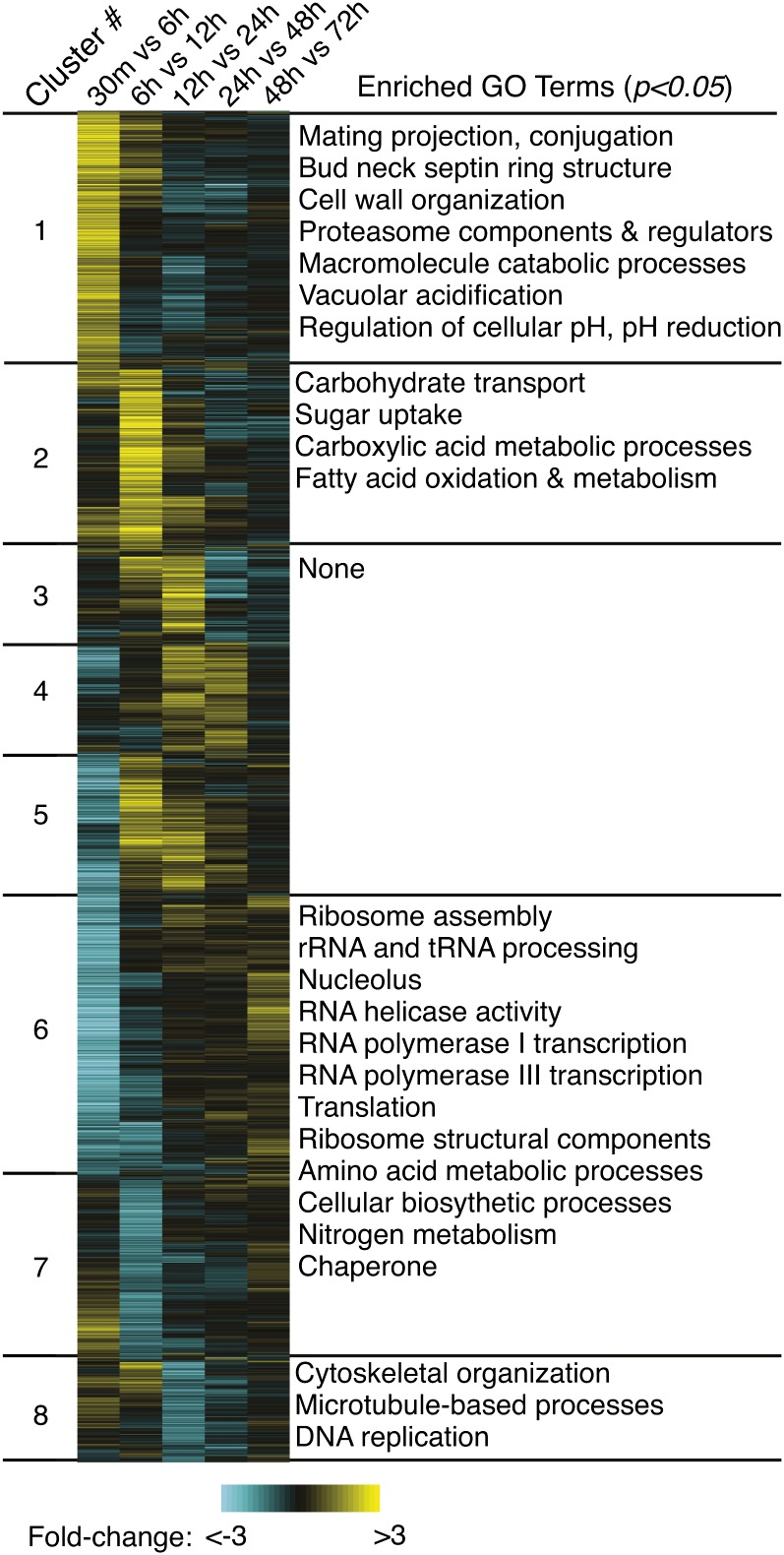
Figure 2 .
Developmental microarray data set defines eight stage-specific gene clusters. The top 3157 genes (as filtered by statistical significance and fold change) are represented as rows, and experiments as columns (labeled above). Yellow indicates an increase over the time period being assessed, blue indicates a decrease, and black indicates that the transcript level was unchanged between time points. The genes are arranged vertically into groups with similar expression profiles, as defined by a robust K-means clustering algorithm (1000 iterations, Pearson correlation similarity metric) with cluster designations on the left (1–8) and representative significantly enriched GO terms (P < 0.05) on the right (Ausubel et al. 1997).
Cluster 1 genes (619 total, 40% unknown) shared a general pattern of strong induction during the first 6 hr of development with levels diminishing thereafter. Active cellular fusion between mating partners occurs during the 0.5- to 6-hr interval, and many genes known to be involved in early sexual development are members of this group. These known “mating genes” included those encoding mating pheromone (MFα1, MFα2, MFα3), the pheromone receptor (STE3α), and other mate recognition and signaling components (CPK1, GPA2, GPA3, RAC1, RAS1, RAS2, STE6, STE7, STE14, and STE12α) (Alspaugh et al. 2000; Chang et al. 2000; Davidson et al. 2003; Hsueh and Shen 2005; Vallim et al. 2005; Hsueh et al. 2007). Structural components known as septins are critical during conjugation tube formation preceding cellular fusion (Kozubowski and Heitman 2010). Known septins CDC3, CDC10, CDC11, and CDC12 and the uncharacterized predicted septin CNB04900 are all members of cluster 1 (average fivefold induction during the 0.5- to 6-hr interval). Additionally, of the 14 genes in our data set identifiable as septins or with septin ring-related functions, 10 are members of cluster 1. This overrepresentation is statistically significant (P = 3 × 10–5, hypergeometric distribution). Other members of cluster 1 are CLP1 and the homeobox gene SXI1α, both of which have been shown to function immediately postfusion to promote dikaryotic growth (Hull et al. 2002; Ekena et al. 2008). GO term-enrichment analysis also identified a number of cellular processes previously unlinked to early sexual development. These included proteasome components and regulators, proton transport, cytoplasmic acidification factors, and genes encoding cell wall and actin cytoskeletal remodeling components.
Cluster 2 transcripts (428 genes, 67% unknown) accumulated during the 6- to 12-hr interval, when the sexually developing populations were observed to transition from yeast form growth to filamentous growth. CPR2, encoding the mating-type independent pheromone receptor, is a member of this group and exhibits fivefold induction over the 0.5- to 6-hr interval and an additional ninefold induction during the 6- to 12-hr interval. This correlates with the importance of Cpr2 throughout development for proper hyphal morphology (Hsueh et al. 2009). GO term analysis of cluster 2 revealed enrichment of genes with functions related to carbohydrate metabolism such as hexose transporters and those involved in the energy-productive breakdown of fatty acids and large carbohydrates.
Of particular interest were the identities of genes induced during dikaryotic filamentation; clusters 3, 4, and 5 contain genes that are specifically induced during early, middle, and late filamentous growth (Figure 2). These groups contain 148, 263, and 331 genes, respectively, and represent a small fraction of the pool of 3156 dynamically transcribed genes. GO term assessment revealed no enriched GO terms, and clusters 3, 4, and 5 show a statistical overrepresentation of unknown genes (73, 72, and 65%, with corresponding enrichment values of P = 2 × 10–9, 1 × 10–14, and 5 × 10–10, respectively). For reference, the data set of 3156 genes contains 50% unknown genes. This indicates that the molecular and physiological processes underlying dikaryotic growth are distinct from other C. neoformans growth phases and may be undescribed in known biology.
The repressed genes in clusters 6, 7, and 8 (692, 432, and 243 genes respectively) displayed very different functional trends and provide additional insights to the physiological state of developing cells. Those genes repressed earliest in development (clusters 6 and 7) showed the largest number of significantly enriched GO terms, overwhelmingly related to translation and anabolic biosynthetic pathways. Repressed at a later stage were cluster 8 genes, which showed strong downregulation during the 12- to 24-hr interval, the time period with the greatest observed increase in filamentous growth (Figure 1, parts 3 and 4, and Figure 2). This group showed enrichment for genes with functions relating to the cytoskeleton, particularly those components that destabilize microtubules and cytoskeletal components. Cluster 8 also includes BIM1, whose gene product is known to contribute to proper microtubule organization (Staudt et al. 2010).
Overall, these time-course expression data define the changes in gene expression during sexual development and show that they occur in cascades that correlate with distinct morphological transitions, providing the opportunity to investigate regulatory features among coexpressed gene cohorts.
Developmentally important cis-regulatory element identification
To identify the regulatory elements responsible for the expression patterns observed, we conducted motif-finding analyses on the upstream regions of coexpressed gene groups. Many motifs were identified across gene clusters; however, we chose to focus on cluster 1 because of the significant enrichment of mating gene functions among this group and opportunities to compare with regulatory elements founds in other fungi. Within cluster 1, genes associated with a GO process annotation of pheromone-dependent signal transduction, conjugation, or sex determination or with a GO component annotation of mating projection were utilized in subsequent analyses along with any a mating-type counterparts to any α-specific genes (likely components of mate recognition machinery in a cells, but absent from these data because the microarrays used did not contain a-specific probes). This filtering resulted in a total of 31 genes in the putative “pheromone-response group,” whose upstream regions were assessed for enriched motifs using the multiple EM for motif elucidation (MEME) algorithm (Bailey and Elkan 1994; Grundy et al. 1997). The most significant motif identified had an associated likelihood value of e < 10–75 (Figure 3A). Control analyses validated the bioinformatic significance of this motif; 10 independent iterations of 30 random genes from cluster 1 were evaluated with MEME in an identical manner, and no motif was identified with an associated likelihood value e < 0.001 (data not shown). This suggests that the motif identified is not a general transcription element occurring frequently in the C. neoformans genome, but rather is specific to the coexpressed and cofunctional subset of group 1 defined using expression data and functional annotation.
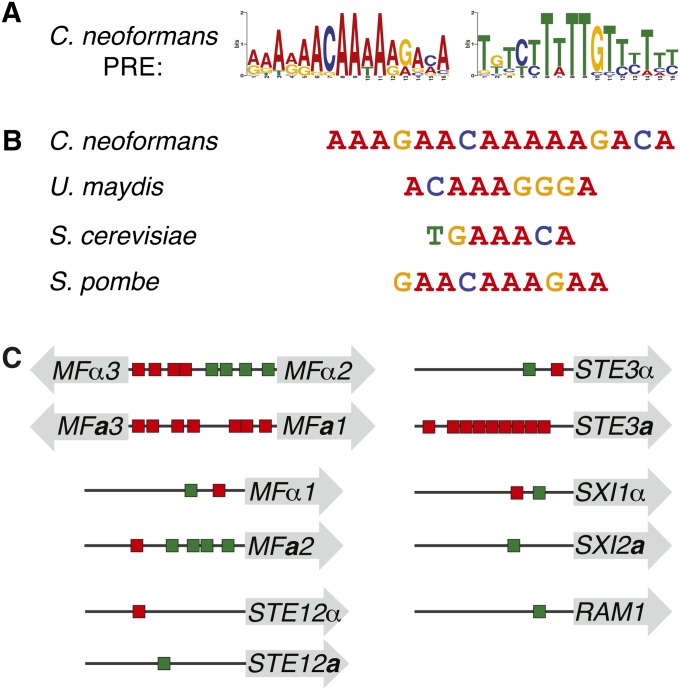
Figure 3 .
The C. neoformans PRE sequence is similar to those of other fungi. (A) Logo of the C. neoformans PRE generated by MEME, shown in forward and reverse complement permutations, with heights of letters corresponding to incidence of a given base at that position (WebLogo assembled from PREs listed in Table 1). The PRE shows no strand bias upstream of target genes and appears in both the A-rich and T-rich permutations on the 5′ strand upstream of target genes. (B) The C. neoformans consensus PRE and those of previously described fungi (Dolan et al. 1989; Sugimoto et al. 1991; Urban et al. 1996; Sahni et al. 2009). (C) Schematic of the PRE content upstream of target genes of particular interest. Shaded arrows represent open reading frames, horizontal lines represent intergenic spaces, and red and green rectangles are representative of PREs, with colors corresponding to direction of PRE as pictured in A. Mating pheromone and receptor genes harbored the most PREs in their upstream regions. Interestingly, orthologous mating-type-specific genes contain differential PRE content, indicating potential regulatory divergence between mating types (e.g., STE3a vs. STE3α).
Because the motif possesses sequence content similar to characterized PREs of other fungi, including those of S. cerevisiae, U. maydis, and S. pombe, it was designated the C. neoformans PRE (Figure 3B). In fungal systems, it has been shown that the PRE controls genes critical for mediating early mating events (Hagen et al. 1991; Sugimoto et al. 1991; Urban et al. 1996; Sahni et al. 2009). In each case, the PRE is a relatively short (6–11 bp) sequence, containing an A/T-rich core, with flanking G/C content (Figure S1). As in other fungi, the C. neoformans PRE appears on both strands of DNA and is present in multiple instances among individual promoters. Numbers of PREs and positions range from a single PRE (STE12a/α, SXI2a, RAM1) to as many as nine (STE3a) within 1000 bp upstream of the ATG, and the PRE content varied upstream of mating-type-specific alleles of numerous genes (Figure 3C and Figure S2). We detected 89 instances of the PRE upstream of the 27 genes in the defined pheromone-response group (Table 1) (Bailey and Gribskov 1998). No significant correlation was detected between number of PREs and the fold induction observed during the first 6 hr of development. The C. neoformans PRE also shows conservation among Cryptoccocus varieties; analyses of the upstream regions from C. neoformans var. grubii (serotype A) orthologs revealed a motif similar to that identified for C. neoformans var. neoformans (serotype D) (Figure S3).
Table 1 . C. neoformans PRE locations among the pheromone-response group.
| Locus | Gene name | PREs within 1 kb of ATG |
|---|---|---|
| AF542530.2 | STE3a | 9 |
| CND05690 | MFα3 | 8 |
| CND05680 | MFα2 | 8 |
| CND05750 | MFα1 | 7 |
| AF542530.2 | MFa1 | 7 |
| AF542530.2 | MFa3 | 7 |
| AF542530.2 | MFa2 | 5 |
| CND05950 | SXI1α | 5 |
| CNE00520 | Mitogen-activated protein kinase | 3 |
| CNF01070 | Plasma membrane fusion-related protein, putative | 3 |
| CNC01050 | Protein-S-isoprenylcysteine O-methyltransferase, putative | 3 |
| CNC02440 | CDC12 (septin) | 3 |
| CND05800 | STE3α | 2 |
| AF542530.2 | STE12a | 2 |
| CND05810 | STE12α | 2 |
| CNE03110 | Pheromone maturation-related protein farnesyltransferase, putative | 2 |
| CNC04960 | Small GTPase RAC1, putative | 2 |
| CNL05710 | Secreted Zn-dependent peptidase (insulinase famly), putative | 2 |
| AF542530.2 | SXI2a | 1 |
| CNB01030 | STE6 | 1 |
| CND04860 | CDC10 (septin ring protein) | 1 |
| CNE03430 | CD11 (septin) | 1 |
| CNE04450 | GPA3 (heterotrimeric G-protein GTPase) | 1 |
| CNF00500 | CDC3 (septin ring protein) | 1 |
| CNA01680 | GPA2 (heterotrimeric G-protein α subunit B) | 1 |
| CND04910 | Prenyl-dependent CAAX protease, putative | 1 |
| CNE00350 | CPK1 | 1 |
The C. neoformans PRE was identified in the upstream regions of 27 genes of the pheromone-response group (within 1000 bp of the predicted translational start).
To validate the significance of the identified PRE, we carried out an additional control analysis. Of the 7765 genes represented on the microarrays used in this study, 79 meet the functional criteria applied to cluster 1 used to define the pheromone-response group. When the upstream regions of these 79 genes were analyzed with MEME for enriched motifs, some weakly conserved motifs were detected, although none resembled PRE identified among the pheromone-response cohort. As such, the expression data were critical in defining only a coexpressed subset to assess for a shared regulatory sequence and mechanism. The initial analyses identified 27 PRE target genes (Table 1). This list, however, is limited to only the functionally related subset of cluster 1. Assessment of the upstream regions from all genes in cluster 1 identified an additional 61 potential targets genes with at least one PRE within 1000 bp of the ATG (Table S4) (MAST algorithm; Bailey and Gribskov 1998).
The PRE is both necessary and sufficient to inducing gene expression in response to pheromone signaling
To assess the biological role of the PRE, we conducted a series of transcriptional reporter assays. Because a standard reporter system had not yet been developed for use in C. neoformans, we tested the effects of the PRE by inserting sequences upstream of the URA5 gene and monitoring levels of URA5 transcript. First, to test the ability of the PRE to confer regulation during sexual development, we inserted tandem repeats of the consensus PRE into the URA5 promoter region (Figure 4A). Strains containing the reporter constructs (−PREs vs. + PREs) were incubated with and without a mating partner (PRE activity modeled in Figure 4B). When the strains were incubated without a mating partner, URA5 levels were not affected by the presence or absence of PRE sequences. (Figure 4C, lanes 2–4 vs. 5–7, 1.2 ± 0.07 fold change, P > 0.39). In contrast, when the reporter strains were incubated with a mating partner under identical conditions, those with the +PREs reporter construct showed transcript levels 2.9 ± 0.05-fold greater than those expressing the control constructs lacking PREs (Figure 4C, lanes 9–11 vs. 12–14, P < 0.001). Because the reporter constructs were expressed in only one-half of the mating population (a cells), seven PREs were necessary to observe this effect. Constructs containing only three PRE repeats also showed regulation, but to a lesser extent (data not shown). We did observe that in the absence of PRE sequences, the levels of URA5 were higher in haploid strains relative to those undergoing sexual development (Figure 4C, lanes 2–4 vs. 9–11). Independent of this effect, however, we conclude that the putative PRE is sufficient to mediate increased gene expression of URA5 during early sexual development.
We next assessed whether the PRE activity detected during early sexual development was in fact mediated by a pheromone signal. We predicted that pheromone-mediated signaling through the PRE would be dependent on the conserved MAPK cascade characterized previously (Alspaugh et al. 1997; Davidson et al. 2003; Hsueh et al. 2007). Thus, we carried out reporter assays using a modified genetic background in which the GPA3 gene was deleted. This mutation has been shown to cause constitutive signaling downstream of the Ste3 pheromone receptor via the pheromone-induced MAPK cascade in the absence of a mating partner or mating pheromone (Hsueh et al. 2007). Reporter constructs with and without repeats of the PRE were expressed in both wild-type and gpa3Δ strains (Figure 5, A and B). In the wild-type background, URA5 levels were unaffected by the presence of PREs in the reporter (Figure 5C, lanes 2–4 vs. 5–7, 1.5 ± 0.1-fold, P > 0.45). In contrast, in the gpa3Δ background, URA5 expression levels were 5.5 ± 0.3-fold greater for constructs containing PREs relative to those without (Figure 5C, lanes 2–4 vs. 5–7, P < 0.05). We conclude from these data that the PRE is a cis-regulatory element that activates transcription downstream of pheromone signaling in vivo.
Figure 5 .
The PRE activates transcription in response to pheromone signaling. (A) Schematic of constructs used in this assay, showing the reporter gene (URA5), translational start (ATG), and predicted transcriptional start (TATA). The top construct is the control, and lacks PREs. The bottom construct includes an insertion of three tandem PREs at −123 (PRE sequence = ATTAAACAAAAAGAAA). (B) Representation of the pheromone response in C. neoformans. Gpa3 represses MAPK signaling in the absence of pheromone ligand (left). gpa3Δ mimics pheromone activation (middle) (Hsueh et al. 2007). gpa3Δmat2Δ is predicted to eliminate signaling through PREs (right). (C) Northern blot of reporter and control transcript levels. Wild-type and +PRE reporters were expressed in ura5 (lanes 2–7), gpa3Δura5 (lanes 9–14), and gpa3Δmat2Δura5 (lanes 16–21) strains (three independently isolated strains were assessed for each reporter construct in each genetic background). Lanes 1, 8, and 15 show the parental strains lacking reporter plasmid. A probe to URA5 detected reporter readout and a probe to GPD1 served as a control. In each case, URA5 levels were normalized to GPD1 levels and mean expression values were compared between sets of biological triplicates to determine fold change with standard error (shown under Northern blots). Student’s t-test was used to determine the statistical significance of the differences between mean values among sets of biological triplicates.
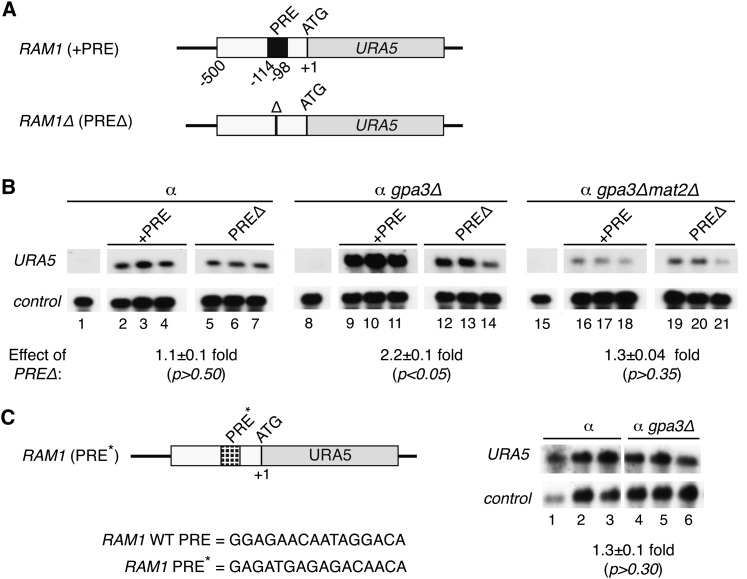
Having established that the PRE was sufficient to activate transcription downstream of pheromone signaling, we then conducted reporter assays to determine whether the PRE was necessary for the induction of endogenous pheromone-responsive promoters. An identified pheromone-responsive gene in C. neoformans, CNF02370, encodes a predicted pheromone modifying enzyme tentatively named RAM1 (based on sequence identity to RAM1 of S. cerevisiae) that shows induction during the first 6 hr of sexual development (Powers et al. 1986; Vallim et al. 2004). The RAM1 promoter (500 bp), with and without its endogenous PRE, was cloned upstream of the ATG of the URA5 reporter gene and expressed in wild-type and gpa3Δ strains (Figure 6A). The native (+PRE) and deletion (PREΔ) versions of the RAM1 reporter construct showed similar URA5 levels in wild-type strains (Figure 6B, lanes 2–4 vs. lanes 5–7, 1.1 ± 0.1-fold change, P > 0.5). As expected, the native RAM1 promoter showed pheromone responsiveness, and the mean URA5 levels were increased in the gpa3Δ background relative to wild type (Figure 6B, lanes 9–11 vs. control lanes 2–4). This induction was dependent on the PRE, as its removal causes a 2.2-fold decrease in URA5 levels (Figure 6B, lanes 9–11 vs. lanes 12–14, 2.2 ± 0.1-fold change, P < 0.05). We also tested a mutant version of the RAM1 promoter in which the PRE sequence was randomized (Figure 6C). This mutant RAM1 promoter failed to respond to pheromone signaling, and the mean URA5 levels showed no significant difference in the gpa3Δ background relative to wild type (Figure 6C, lanes 27–29 vs. lanes 30–32, 1.3 ± 0.1-fold change, P > 0.30). We conclude from these data that the PRE sequence within the RAM1 promoter is critical for its pheromone responsiveness.
Figure 6 .
The induction of the RAM1 promoter in response to pheromone signaling requires the endogenous PRE and Mat2. (A) Schematic of the reporter constructs used in this assay, showing the reporter gene (URA5), translational start (ATG), and location of endogenous PRE within the RAM1 promoter (PRE sequence = GGAGAACAATAGGACA). PREΔ indicates deletion of the 16-bp PRE. (B) Northern blot of reporter and control transcript levels. Reporter constructs with and without the PRE were transformed into ura5 (lanes 2–7), gpa3Δura5 (lanes 9–14), and gpa3Δmat2Δura5 (lanes 16–21) strains (three independently isolated strains were assessed for each reporter construct in each genetic background). Lanes 1, 8, and 15 show the parental strains lacking reporter plasmid. A probe to URA5 detected reporter readout and a probe to GPD1 served as a control. In each case, URA5 levels were normalized to GPD1 levels and mean expression values were compared between sets of biological triplicates to determine fold change with standard error (shown under Northern blots). Student’s t-test was used to determine the statistical significance of the differences between mean values among sets of biological triplicates. (C) Schematic of a mutant reporter construct, showing the reporter gene (URA5), translational start (ATG), and location of the randomized PRE (PRE*) within the RAM1 promoter (PRE* = GAGATGAGAGACAACA). Northern blot of wild-type (lanes 1–3) and gpa3Δ (lanes 4–6) strains expressing the RAM1 PRE* reporter construct. Similar to B, URA5 levels served as the reporter readout and were normalized to control gene GPD1.
Mat2 is required for PRE-mediated transcriptional activation
To determine whether Mat2 functions downstream of the pheromone-activated MAPK cascade and activates transcription via PREs in C. neoformans, we tested the previously used reporter constructs (those in Figure 4A and Figure 6A) in gpa3Δmat2Δ mutant strains. We discovered that MAT2 is required for PRE-mediated activation in response to pheromone signaling. In gpa3Δmat2Δ strains, PREs had no significant effect on URA5 levels (Figure 5C, lanes 16–18 vs. 19–21, 1.6 ± 0.1-fold change, P > 0.22). Similarly, the RAM1 endogenous PRE had no effect on URA5 levels in the gpa3Δmat2Δ background (Figure 6B, lanes 16–18 vs. 19–21, 1.3 ± 0.04-fold change, P > 0.35). Thus, the responsiveness of PREs in the gpa3Δ background requires the activity of Mat2.
Additionally, the gpa3Δmat2Δ mutants are sterile when crossed by a wild-type tester strain (Figure S4), demonstrating the central role of Mat2 downstream of the Ste3 pheromone receptor and the pheromone-induced MAPK cascade during development. The dominance of the mat2Δ sterile phenotype, even in the hyperactive gpa3Δ background, provides genetic evidence for Mat2 function as the central regulator of the pheromone response.
Mat2 is required for the expression of PRE-containing genes during development
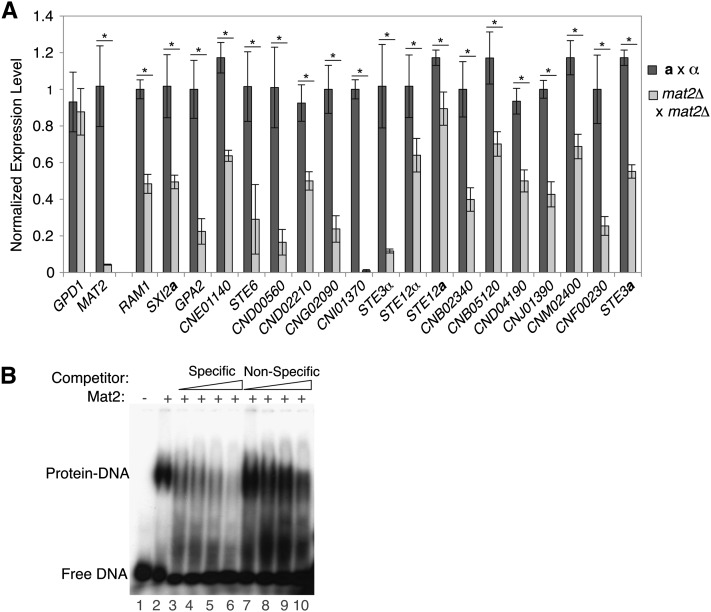
On the basis of strong molecular and genetic evidence mapping Mat2 downstream of the pheromone-activated MAPK cascade and upstream of the PREs tested via reporter assays, we predicted that Mat2 would function globally to activate endogenous PRE-containing genes during early development. To test this hypothesis, we assessed relative transcript levels of numerous target genes in wild-type and mat2Δ crosses via quantitative reverse-transcriptase PCR. GPD1 expression served as a control and did not exhibit MAT2-dependent expression during development (P > 0.50). MAT2 expression served as a positive control for differential expression between wild-type and mat2Δ crosses. Target genes with one or more PREs in their upstream regions overwhelmingly showed a statistically significant decrease in expression levels in the absence of MAT2 (Figure 7A). The +PRE target genes meeting P < 0.05 contained varying numbers of PREs in their upstream regions: one PRE (RAM1, SXI2a, GPA2, CNE01140, STE6, CND00560, CND02210, CNG02090, CNI01370), two PREs (STE3α, STE12a, STE12α, CNB02340, CNB05120, CND04190, CNJ01390, CNM02300), four PREs (CNF00230), or nine PREs (STE3a). The remaining five +PRE targets, while failing to meet our significance threshold of P < 0.05, consistently showed lower expression levels in mat2Δ crosses relative to wild type. A set of 10 control genes (lacking PREs in their upstream regions) was assessed in parallel, and none exhibited a significant difference in expression levels between the wild type and mat2Δ (Figure S5). Importantly, 5 of these are members of cluster 1 and showed no significant change in expression upon the deletion of MAT2 (CNL04900, CNL06590, CND05650, CNE03790, CNB04860). Remaining control target genes included ACT1 and members of other clusters in the data set (cluster 2, CNB03380; cluster 3, CNF01060; and cluster 6, CNB01090). This indicates that the mat2Δ mutation is not affecting global transcript levels.
Figure 7 .
Mat2 is required for the expression of PRE-containing genes and binds specifically to the PRE. (A) qRT-PCR analysis of PRE-containing target genes in wild-type (dark gray bars) and mat2Δ (light gray bars) crosses. Expression levels presented on the y-axis are mean values (of triplicate experiments), normalized to internal reference gene URA5, with associated standard error. Student’s t-test was used to determine if the expression levels were statistically different between the wild-type and mat2Δ crosses (* indicates P < 0.05). (B) Recombinantly expressed Mat2 protein was incubated with radiolabeled probe corresponding to the MFα1 PRE. Lane 1 contains the probe without protein, and lanes 2–10 contain constant amounts of recombinant Mat2. The formation of the Mat2–DNA complex (lane 2) is diminished by the addition of increasing amounts of unlabeled specific competitor DNA corresponding to MFα1 PRE sequence (lanes 3–6). Increasing amounts of unlabeled nonspecific competitor DNA (similarly sized fragment of the pUC18 vector) did not have this effect (lanes 7–10). Competitor DNA was added at 40×, 200×, 400×, and 800× molar excess.
Mat2 interacts directly with the PRE
Given the strong genetic evidence linking Mat2 to PRE-mediated transcriptional induction and the similarity of the PRE to known binding sites of HMG-domain proteins, we tested whether Mat2 binds directly to PRE sequences in DNA using electromobility shift assays (Matys et al. 2003). Recombinantly expressed Mat2 incubated with a functional PRE sequence (that from the MFα1 promoter) caused decreased mobility of the PRE probe (Figure 7B, lane 2). Competing this binding with increasing amounts of unlabeled MFα1 probe eliminated the observed shift (Figure 7, lanes 3–6). Binding was less affected by competition with an unlabeled nonspecific competitor DNA (pUC18 vector sequence of a similar length) (Figure 7B, lanes 7–10). Similar binding patterns were observed for Mat2 and a probe corresponding to the RAM1 PRE (data not shown). These data indicate that functional PRE sequences in vivo are bound directly and specifically by Mat2 in vitro. This connection maps the regulatory network responsible for mating downstream of the pheromone-activated MAPK cascade, via Mat2 binding directly to PREs, resulting in the activation of target genes.
Discussion
Inference of the evolution of regulatory circuits requires three kinds of information: gene expression data, regulatory proteins, and cis regulatory features (Tsong et al. 2003; Galgoczy et al. 2004; Gasch et al. 2004). Here, we present a study in which these three kinds of information were generated and analyzed to assess evolution of a conserved signaling pathway in fungi. Through our investigation, we have connected the Mat2 transcription factor of C. neoformans to the pheromone-activated MAPK cascade and to transcriptional induction via the PRE cis-regulatory element, establishing Mat2 as the C. neoformans pheromone-response factor. In determining the mechanism of Mat2 activity during sexual development of C. neoformans, we demonstrate that the conserved regulatory circuit governing mating and the pheromone response has undergone rewiring over evolutionary time.
Key in this study was a time-course microarray expression experiment spanning complete sexual development of C. neoformans in which we generated a temporal expression pattern for each known gene. While most microarray analyses have assessed gene-expression changes in a binary fashion to compare two conditions (e.g., exposure to environmental stress or wild-type and mutant states), the mechanisms of gene regulation are challenging to infer from this design. In contrast, studies assessing transcriptional dynamics under many conditions or over time often provide the necessary information for the discovery of the molecular mechanisms of gene regulation, including cis-regulatory element identification (Lyons et al. 2000; Galgoczy et al. 2004; Gasch et al. 2004; Campbell et al. 2010).
Using GO terms in concert with the temporal coexpression data we identified the PRE among a functionally related subset of cluster 1 genes. The microarray data were essential in this analysis, because when functionally related genes across the genome were assessed in the absence of expression data, no significant motifs could be identified (data not shown). The identification and characterization of the PRE using both bioinformatic and phylogenetic approaches provided us the opportunity to then test the biological relevance of the site in vivo.
Although the C. neoformans system is highly amenable to many molecular genetic techniques, the use of transcriptional reporter constructs has been limited (Zhang et al. 1999, 2006; Mare et al. 2005; Tommasino et al. 2008). As a consequence, there is no generally established reporter system in which to assay cis-regulatory element function. To test the biological activity of the PRE we developed a reporter assay using the endogenous URA5 gene as the readout of a plasmid-based expression system. These are the first reporter assays of their kind conducted in C. neoformans, as all other transcriptional reporter assays in this system have utilized large, intact promoter sequences of interest rather than inserting small regulatory sequences into an alternate promoter context (Mare et al. 2005; Zhang et al. 2006; Tommasino et al. 2008). By introducing PRE sequences to the URA5 promoter, we were able to confer Mat2-dependent pheromone responsiveness to this normally nonresponsive promoter.
It is clear that the PRE mediates a statistically significant, biologically relevant response in the reporter constructs, but we note that the magnitude of the induction (5.5-fold; Figure 5) is relatively low when compared to that detected in PRE reporter assays conducted in other fungi (ranging from 2.5 to >40-fold (Sengupta and Cochran 1990; Hagen et al. 1991; Urban et al. 1996)). It is possible that the URA5 promoter is not optimal for PRE-mediated activation. The magnitude of the effects of PREs may be suppressed by as-yet-uncharacterized regulatory elements in the URA5 promoter. This is not surprising given that studies in S. cerevisiae and other fungi have demonstrated that the spacing, orientation, and context of PRE repeats influence expression levels of reporter constructs (Urban et al. 1996; Su et al. 2010). Another reason that the promoter context of PREs may be important in this case is the nature of the Mat2 transcription factor. Specifically, Mat2 belongs to the high-mobility group class I (HMG-I) transcription factors, which are known to bind DNA and activate transcription via extreme DNA bending (Urban et al. 1996; Su et al. 2010). The mammalian HMG-I transcription factor SRY has been observed to introduce DNA bends of up to 80°, and the T-cell-specific HMG-I protein LEF-1 causes DNA bend angles exceeding 110° (Su et al. 2010). If the URA5 promoter used in our experiments is not sufficiently flexible, Mat2 may not be able to induce full PRE-mediated activation seen at endogenous locations. Numerous PRE-containing genes in S. cerevisiae have been demonstrated to harbor additional cis-regulatory features in their promoter regions that contribute to overall regulation (van de Wetering et al. 1991). It is very likely that such combinatorial mechanisms contribute to PRE-mediated activation in C. neoformans. Regardless of the magnitude of the fold-induction detected, our reporter assays establish that PREs activate the transcription of target genes downstream of signaling through the pheromone MAPK cascade, requiring the activity of Mat2.
The Mat2-PRE connection was essential in establishing Mat2’s central role in the regulatory cascade governing C. neoformans development. Signaling through the pheromone MAPK cascade activates Mat2, which then binds to PREs in the upstream regions of target genes to induce their expression. The target genes of Mat2 contribute to the proper recognition of and subsequent cellular fusion with a mating partner. Among Mat2-PRE targets are genes encoding the previously described, developmentally important transcriptional regulators STE12a, STE12α, SXI1α, and SXI2a (Wickes et al. 1997; Yue et al. 1999; Chang et al. 2001; Lin et al. 2010). Via these downstream transcription factors, Mat2 initiates a cascade of regulatory events that are required for subsequent developmental transitions.
Although Mat2 had been implicated as the pheromone-response factor on the basis of phenotypic data from a prior genetic screen by Lin et al., additional candidates had also been identified by sequence similarity to those of other fungi. On the basis of pheromone-response mechanisms in other fungal systems, the C. neoformans pheromone-response factor would be predicted to exhibit three conserved behaviors: function downstream of the pheromone-activated MAPK cascade, mediate transcriptional activation via PREs, and bind directly to PRE sequences (Fields and Herskowitz 1985; Sugimoto et al. 1991; Hartmann et al. 1996; Magee et al. 2002). Because bioinformatic predictions were unable to resolve the identity of the C. neoformans pheromone-response factor, a nonconserved regulator was hypothesized to play this conserved role and bind to our newly discovered PRE. The experiments presented here show that Mat2 meets all of the criteria of a pheromone-response factor and is the functional homolog to pheromone-response factors described in other systems.
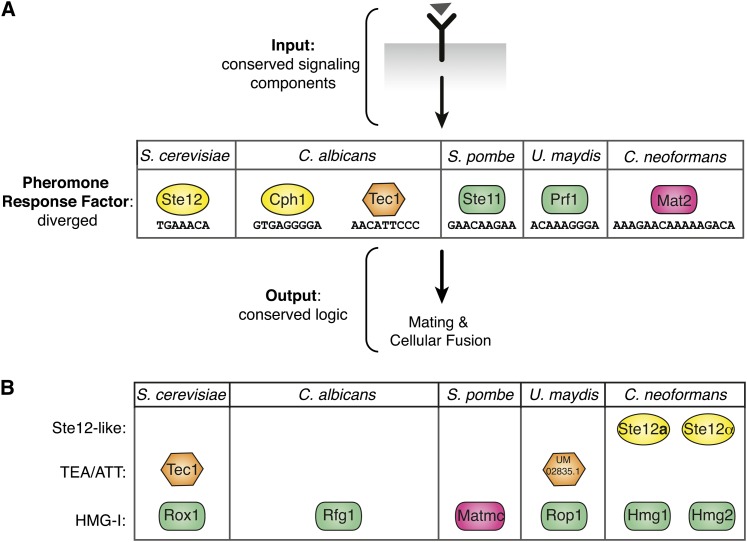
The pheromone-response pathway is an interesting example of evolution because it shows unusual regulatory plasticity despite the strong conservation of upstream signaling components (Figure 8A). It is clear that the use of alternative transcription factors for the same function is a hallmark of this pathway (Figure 8B). For example, in S. cerevisiae in addition to the pheromone-response factor Ste12, its genome contains Tec1 (a global regulator) and Rox1 (a hypoxic gene regulator) (Lowry and Zitomer 1984; Gavrias et al. 1996). The Rox1 sequence homolog in S. pombe is the pheromone-response factor Ste11 (shown in A), and S. pombe does not contain an identifiable Tec1. In contrast, C. albicans contains a Tec1 that functions in a modified pheromone response, but the Ste11 homolog Rfg1 is involved in regulating hyphae formation (not the pheromone response) (Kadosh and Johnson 2001; Sahni et al. 2010; Nobile et al. 2012). The network architecture in C. neoformans is unusual because fungi known to utilize an HMG-domain pheromone-response factor (U. maydis and S. pombe) do not encode detectable Ste12 sequence homologs (Sugimoto et al. 1991; Hartmann et al. 1996). C. neoformans encodes two STE12 sequence homologs, both of which contribute to (but are not required for) sexual development (Wickes et al. 1997; Chang et al. 2001). The intercalation of the noncanonical HMG-domain pheromone-response factor Mat2 between the pheromone MAPK cascade and the Ste12-like factors in C. neoformans is unprecedented.
Figure 8 .
Evolution of the fungal pheromone-response pathway has occurred at the level of the pheromone-response factor. (A) Diverged transcription factors function as pheromone-response factors across fungal systems. The pheromone signal (triangle) is transduced via a conserved receptor and MAPK cascade signaling components, identifiable by sequence homology (conserved input). The transcription factor mediating the conserved response shows divergence among the species characterized (colored shapes representing diverged pheromone-response factors). Responses downstream of the pheromone-response factor are conserved across species (conserved output). Colored shapes (indicating type of DNA-binding domain) with species-specific designations represent the various pheromone-response factors and their binding sites. (B) Rewiring of transcription-factor function. Highest reciprocal BLAST hits were used to identify other predicted DNA-binding proteins with high sequence similarity to the pheromone-response factors in fungi. These factors are present in the genome but have been shown to control different pathways (not pheromone response). C. neoformans is unique because it encodes numerous homologs of the Ste12-like (Ste12a and Ste12α) and HMG-domain (Hmg1 and Hmg2) pheromone-response factors of other fungi, yet uses the diverged HMG-domain protein Mat2 as its pheromone-response factor. Key for DNA binding classes: yellow ovals, Ste12 like; orange hexagon, TEA/ATT; green rectangles, HMG-I; pink rectangles, HMG-I (diverged).
By establishing the role of Mat2 as the C. neoformans pheromone-response factor, we have identified a central adaptive feature of the sexual development regulatory network in this system. Importantly, our findings indicate that rewiring has occurred in this conserved pathway at the level of effector identity and is likely responsible for species-specific and adaptive features that are unique to C. neoformans mating and development in the environment. The discovery of these adaptive regulatory features and their comparisons with other systems are essential to understanding the nature of regulatory network adaptations over long spans of evolutionary time.
Supplementary Material
Acknowledgments
We thank A. Adams, J. L. Ekena, and J. Kusiak for laboratory support. Thanks to C. A. Fox, M. R. Botts, M. E. Mead, M. Huang, and N. Walsh for critical reading of and comments on the manuscript. Microarrays were scanned at the University of Wisconsin Gene Expression Center. This work was supported by National Institutes of Health NIH-R01-AI064287, NIH-R01AI059370, and a Burroughs Wellcome Fund Career Award in Biomedical Sciences, all to C.M.H. E.K.K. was supported by the Molecular Biosciences Training Grant NIH-T32GM0721533, and S.S.G. was supported by the Genomic Sciences Training Grant NHGRI-T32HG002760. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Communicating editor: J. Heitman
Literature Cited
- Alspaugh J., Davidson R., Heitman J., 2000. Morphogenesis of Cryptococcus neoformans. Contrib. Microbiol. 5: 217–238 [DOI] [PubMed] [Google Scholar]
- Alspaugh J. A., Perfect J. R., Heitman J., 1997. Cryptococcus neoformans mating and virulence are regulated by the G-protein α subunit Gpa1 and cAMP. Genes Dev. 11: 3206–3217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M., Ball C. A., Blake J. A., Botstein D., Butler H., et al. , 2000. Gene ontology: tool for the unification of biology: The Gene Ontology Consortium. Nat. Genet. 25: 25–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausubel F., Brent R., Kingston R., Moore D., Seidman J., et al. (Editors), 1997. Current Protocols in Molecular Biology. Wiley, New York [Google Scholar]
- Bailey T. L., Elkan C., 1994. Fitting a mixture model by expectation maximization to discover motifs in biopolymers, 28–36 Proceedings of the Second International Conference on Intelligent Systems for Molecular Biology. AAAI Press, Palo Alto, CA: [PubMed] [Google Scholar]
- Bailey T. L., Gribskov M., 1998. Combining evidence using p-values: application to sequence homology searches. Bioinformatics 14: 48–54 [DOI] [PubMed] [Google Scholar]
- Baker C. R., Tuch B. B., Johnson A. D., 2011. Extensive DNA-binding specificity divergence of a conserved transcription regulator. Proc. Natl. Acad. Sci. USA 108: 7493–7498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booth L. N., Tuch B. B., Johnson A. D., 2010. Intercalation of a new tier of transcription regulation into an ancient circuit. Nature 468: 959–963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borneman A. R., Gianoulis T. A., Zhang Z. D., Yu H., Rozowsky J., et al. , 2007. Divergence of transcription factor binding sites across related yeast species. Science 317: 815–819 [DOI] [PubMed] [Google Scholar]
- Campbell T. L., de Silva E. K., Olszewski K. L., Elemento O., Llinás M., 2010. Identification and genome-wide prediction of DNA binding specificities for the ApiAP2 family of regulators from the malaria parasite. PLoS Pathog. 6: e1001165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Y. C., Wickes B. L., Miller G. F., Penoyer L. A., Kwon-Chung K. J., 2000. Cryptococcus neoformans STE12α regulates virulence but is not essential for mating. J. Exp. Med. 191: 871–882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Y. C., Penoyer L. A., Kwon-Chung K. J., 2001. The second STE12 homologue of Cryptococcus neoformans is MATa-specific and plays an important role in virulence. Proc. Natl. Acad. Sci. USA 98: 3258–3263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung S., Karos M., Chang Y. C., Lukszo J., Wickes B. L., et al. , 2002. Molecular analysis of CPRα, a MATα-specific pheromone receptor gene of Cryptococcus neoformans. Eukaryot. Cell 1: 432–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crooks G. E., Hon G., Chandonia J. M., Brenner S. E., 2004. WebLogo: a sequence logo generator. Genome Res. 14: 1188–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson E. H., Rast J. P., Oliveri P., Ransick A., Calestani C., et al. , 2002. A genomic regulatory network for development. Science 295: 1669–1678 [DOI] [PubMed] [Google Scholar]
- Davidson R. C., Moore T. D., Odom A. R., Heitman J., 2000. Characterization of the MFα pheromone of the human fungal pathogen Cryptococcus neoformans. Mol. Microbiol. 38: 1017–1026 [DOI] [PubMed] [Google Scholar]
- Davidson R. C., Blankenship J. R., Kraus P. R., de Jesus Berrios M., Hull C. M., et al. , 2002. A PCR-based strategy to generate integrative targeting alleles with large regions of homology. Microbiology 148: 2607–2615 [DOI] [PubMed] [Google Scholar]
- Davidson R., Nichols C., Cox G., Perfect J., Heitman J., 2003. A MAP kinase cascade composed of cell type specific and non-specific elements controls mating and differentiation of the fungal pathogen Cryptococcus neoformans. Mol. Microbiol. 49: 469–485 [DOI] [PubMed] [Google Scholar]
- de Hoon M. J., Imoto S., Nolan J., Miyano S., 2004. Open source clustering software. Bioinformatics 20: 1453–1454 [DOI] [PubMed] [Google Scholar]
- Dohlman H. G., Thorner J. W., 2001. Regulation of G protein-initiated signal transduction in yeast: paradigms and principles. Annu. Rev. Biochem. 70: 703–754 [DOI] [PubMed] [Google Scholar]
- Dolan J. W., Kirkman C., Fields S., 1989. The yeast Ste12 protein binds to the DNA sequence mediating pheromone induction. Proc. Natl. Acad. Sci. USA 86: 5703–5707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekena J., Stanton B., Schiebe-Owens J., Hull C., 2008. Sexual development in Cryptococcus neoformans requires CLP1, a target of the homeodomain transcription factors Sxi1α and Sxi2a. Eukaryot. Cell 7: 49–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Errede B., Ammerer G., 1989. Ste12, a protein involved in cell-type-specific transcription and signal transduction in yeast, is part of protein-DNA complexes. Genes Dev. 3: 1349–1361 [DOI] [PubMed] [Google Scholar]
- Fields S., Herskowitz I., 1985. The yeast Ste12 product is required for expression of two sets of cell-type specific genes. Cell 42: 923–930 [DOI] [PubMed] [Google Scholar]
- Galgoczy D., Cassidy-Stone A., Llinás M., O’Rourke S., Herskowitz I., et al. , 2004. Genomic dissection of the cell-type-specification circuit in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 101: 18069–18074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasch A. P., Moses A. M., Chiang D. Y., Fraser H. B., Berardini M., et al. , 2004. Conservation and evolution of cis-regulatory systems in ascomycete fungi. PLoS Biol. 2: e398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavrias V., Andrianopoulos A., Gimeno C. J., Timberlake W. E., 1996. Saccharomyces cerevisiae TEC1 is required for pseudohyphal growth. Mol. Microbiol. 19: 1255–1263 [DOI] [PubMed] [Google Scholar]
- Grundy W. N., Bailey T. L., Elkan C. P., Baker M. E., 1997. Meta-MEME: motif-based hidden Markov models of protein families. Comput. Appl. Biosci. 13: 397–406 [DOI] [PubMed] [Google Scholar]
- Hagen D. C., McCaffrey G., Sprague G. F., 1991. Pheromone response elements are necessary and sufficient for basal and pheromone-induced transcription of the FUS1 gene of Saccharomyces cerevisiae. Mol. Cell. Biol. 11: 2952–2961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann H. A., Kahmann R., Bölker M., 1996. The pheromone response factor coordinates filamentous growth and pathogenicity in Ustilago maydis. EMBO J. 15: 1632–1641 [PMC free article] [PubMed] [Google Scholar]
- Herskowitz I., 1985. A master regulatory locus that determines cell specialization in yeast. Harvey Lect. 81: 67–92 [PubMed] [Google Scholar]
- Herskowitz I., 1988. Life cycle of the budding yeast Saccharomyces cerevisiae. Microbiol. Rev. 52: 536–553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homann O. R., Dea J., Noble S. M., Johnson A. D., 2009. A phenotypic profile of the Candida albicans regulatory network. PLoS Genet. 5: e1000783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsueh Y. P., Shen W. C., 2005. A homolog of Ste6, the a-factor transporter in Saccharomyces cerevisiae, is required for mating but not for monokaryotic fruiting in Cryptococcus neoformans. Eukaryot. Cell 4: 147–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsueh Y. P., Xue C., Heitman J., 2007. G protein signaling governing cell fate decisions involves opposing Gα subunits in Cryptococcus neoformans. Mol. Biol. Cell 18: 3237–3249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsueh Y. P., Xue C., Heitman J., 2009. A constitutively active GPCR governs morphogenic transitions in Cryptococcus neoformans. EMBO J. 28: 1220–1233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hull C., Heitman J., 2002. Genetics of Cryptococcus neoformans. Annu. Rev. Genet. 36: 557–615 [DOI] [PubMed] [Google Scholar]
- Hull C., Davidson R., Heitman J., 2002. Cell identity and sexual development in Cryptococcus neoformans are controlled by the mating-type-specific homeodomain protein Sxi1α. Genes Dev. 16: 3046–3060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Idnurm A., 2010. A tetrad analysis of the basidiomycete fungus Cryptococcus neoformans. Genetics 185: 153–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadosh D., Johnson A. D., 2001. Rfg1, a protein related to the Saccharomyces cerevisiae hypoxic regulator Rox1, controls filamentous growth and virulence in Candida albicans. Mol. Cell. Biol. 21: 2496–2505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozubowski L., Heitman J., 2010. Septins enforce morphogenetic events during sexual reproduction and contribute to virulence of Cryptococcus neoformans. Mol. Microbiol. 75: 658–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon-Chung K. J., 1976. Morphogenesis of Filobasidiella neoformans, the sexual state of Cryptococcus neoformans. Mycologia 68: 821–833 [PubMed] [Google Scholar]
- Kwon-Chung K. J., Edman J. C., Wickes B. L., 1992. Genetic association of mating types and virulence in Cryptococcus neoformans. Infect. Immun. 60: 602–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavoie H., Hogues H., Mallick J., Sellam A., Nantel A., et al. , 2010. Evolutionary tinkering with conserved components of a transcriptional regulatory network. PLoS Biol. 8: e1000329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X., Jackson J. C., Feretzaki M., Xue C., Heitman J., 2010. Transcription factors Mat2 and Znf2 operate cellular circuits orchestrating opposite- and same-sex mating in Cryptococcus neoformans. PLoS Genet. 6: e1000953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowry C. V., Zitomer R. S., 1984. Oxygen regulation of anaerobic and aerobic genes mediated by a common factor in yeast. Proc. Natl. Acad. Sci. USA 81: 6129–6133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyons T. J., Gasch A. P., Gaither L. A., Botstein D., Brown P. O., et al. , 2000. Genome-wide characterization of the Zap1p zinc-responsive regulon in yeast. Proc. Natl. Acad. Sci. USA 97: 7957–7962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magee B. B., Legrand M., Alarco A. M., Raymond M., Magee P. T., 2002. Many of the genes required for mating in Saccharomyces cerevisiae are also required for mating in Candida albicans. Mol. Microbiol. 46: 1345–1351 [DOI] [PubMed] [Google Scholar]
- Mare L., Iatta R., Montagna M. T., Luberto C., Del Poeta M., 2005. APP1 transcription is regulated by inositol-phosphorylceramide synthase 1-diacylglycerol pathway and is controlled by Atf2 transcription factor in Cryptococcus neoformans. J. Biol. Chem. 280: 36055–36064 [DOI] [PubMed] [Google Scholar]
- Martchenko M., Levitin A., Hogues H., Nantel A., Whiteway M., 2007. Transcriptional rewiring of fungal galactose-metabolism circuitry. Curr. Biol. 17: 1007–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matys V., Fricke E., Geffers R., Gössling E., Haubrock M., et al. , 2003. TRANSFAC: transcriptional regulation, from patterns to profiles. Nucleic Acids Res. 31: 374–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama N., Kaziro Y., Arai K., Matsumoto K., 1988. Role of STE genes in the mating factor signaling pathway mediated by GPA1 in Saccharomyces cerevisiae. Mol. Cell. Biol. 8: 3777–3783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni M., Feretzaki M., Sun S., Wang X., Heitman J., 2010. Sex in fungi. Annu. Rev. Genet. 45: 405–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobile C. J., Fox E. P., Nett J. E., Sorrells T. R., Mitrovich Q. M., et al. , 2012. A recently evolved transcriptional network controls biofilm development in Candida albicans. Cell 148: 126–138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers S., Michaelis S., Broek D., Santa Anna S., Field J., et al. , 1986. RAM, a gene of yeast required for a functional modification of RAS proteins and for production of mating pheromone a-factor. Cell 47: 413–422 [DOI] [PubMed] [Google Scholar]
- Quackenbush J., 2001. Computational analysis of microarray data. Nat. Rev. Genet. 2: 418–427 [DOI] [PubMed] [Google Scholar]
- Raper J. R., 1966. Genetics of Sexuality in Higher Fungi. Ronald Press, New York [Google Scholar]
- Sahni N., Yi S., Daniels K. J., Srikantha T., Pujol C., et al. , 2009. Genes selectively up-regulated by pheromone in white cells are involved in biofilm formation in Candida albicans. PLoS Pathog. 5: e1000601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahni N., Yi S., Daniels K. J., Huang G., Srikantha T., et al. , 2010. Tec1 mediates the pheromone response of the white phenotype of Candida albicans: insights into the evolution of new signal transduction pathways. PLoS Biol. 8: e1000363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengupta P., Cochran B. H., 1990. The PRE and PQ box are functionally distinct yeast pheromone response elements. Mol. Cell. Biol. 10: 6809–6812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen W. C., Davidson R. C., Cox G. M., Heitman J., 2002. Pheromones stimulate mating and differentiation via paracrine and autocrine signaling in Cryptococcus neoformans. Eukaryot. Cell 1: 366–377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman F., 1991. Getting started with yeast, 3–21 Methods in Enzymology, edited by Guthrie C., Fink G. Academic Press, San Diego, CA: [DOI] [PubMed] [Google Scholar]
- Song D., Dolan J. W., Yuan Y. L., Fields S., 1991. Pheromone-dependent phosphorylation of the yeast Ste12 protein correlates with transcriptional activation. Genes Dev. 5: 741–750 [DOI] [PubMed] [Google Scholar]
- Staudt M., Kruzel E., Shimizu K., Hull C., 2010. Characterizing the role of the microtubule binding protein Bim1 in Cryptococcus neoformans. Fungal Genet. Biol. 47: 310–317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su T.-C., Tamarkina E., Sadowski I., 2010. Organizational constraints on Ste12 cis-elements for a pheromone response in Saccharomyces cerevisiae. FEBS J. 277: 3235–3248 [DOI] [PubMed] [Google Scholar]
- Sugimoto A., Iino Y., Maeda T., Watanabe Y., Yamamoto M., 1991. Schizosaccharomyces pombe ste11+ encodes a transcription factor with an HMG motif that is a critical regulator of sexual development. Genes Dev. 5: 1990–1999 [DOI] [PubMed] [Google Scholar]
- Sung H.-M., Wang T.-Y., Wang D., Huang Y.-S., Wu J.-P., et al. , 2009. Roles of trans and cis variation in yeast intraspecies evolution of gene expression. Mol. Biol. Evol. 26: 2533–2538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toffaletti D. L., Rude T. H., Johnston S. A., Durack D. T., Perfect J. R., 1993. Gene transfer in Cryptococcus neoformans by use of biolistic delivery of DNA. J. Bacteriol. 175: 1405–1411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tommasino N., Villani M., Qureshi A., Henry J., Luberto C., et al. , 2008. Atf2 transcription factor binds to the APP1 promoter in Cryptococcus neoformans: stimulatory effect of diacylglycerol. Eukaryot. Cell 7: 294–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsong A., Miller M., Raisner R., Johnson A., 2003. Evolution of a combinatorial transcriptional circuit: a case study in yeasts. Cell 115: 389–399 [DOI] [PubMed] [Google Scholar]
- Tsong A., Tuch B., Li H., Johnson A., 2006. Evolution of alternative transcriptional circuits with identical logic. Nature 443: 415–420 [DOI] [PubMed] [Google Scholar]
- Urban M., Kahmann R., Bölker M., 1996. Identification of the pheromone response element in Ustilago maydis. Mol. Gen. Genet. 251: 31–37 [DOI] [PubMed] [Google Scholar]
- Vallim M. A., Fernandes L., Alspaugh J. A., 2004. The RAM1 gene encoding a protein-farnesyltransferase β-subunit homologue is essential in Cryptococcus neoformans. Microbiology 150: 1925–1935 [DOI] [PubMed] [Google Scholar]
- Vallim M. A., Nichols C. B., Fernandes L., Cramer K. L., Alspaugh J. A., 2005. A Rac homolog functions downstream of Ras1 to control hyphal differentiation and high-temperature growth in the pathogenic fungus Cryptococcus neoformans. Eukaryot. Cell 4: 1066–1078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Wetering M., Oosterwegel M., Dooijes D., Clevers H., 1991. Identification and cloning of TCF-1, a T lymphocyte-specific transcription factor containing a sequence-specific HMG box. EMBO J. 10: 123–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickes B. L., Edman U., Edman J. C., 1997. The Cryptococcus neoformans STE12α gene: a putative Saccharomyces cerevisiae STE12 homologue that is mating type specific. Mol. Microbiol. 26: 951–960 [DOI] [PubMed] [Google Scholar]
- Wilczynski B., Furlong E. E., 2010. Challenges for modeling global gene regulatory networks during development: insights from Drosophila. Dev. Biol. 340: 161–169 [DOI] [PubMed] [Google Scholar]
- Yue C., Cavallo L. M., Alspaugh J. A., Wang P., Cox G. M., et al. , 1999. The STE12α homolog is required for haploid filamentation but largely dispensable for mating and virulence in Cryptococcus neoformans. Genetics 153: 1601–1615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S., Varma A., Williamson P. R., 1999. The yeast Cryptococcus neoformans uses “mammalian” enhancer sites in the regulation of the virulence gene, CNLAC1. Gene 227: 231–240 [DOI] [PubMed] [Google Scholar]
- Zhang S., Hacham M., Panepinto J., Hu G., Shin S., et al. , 2006. The Hsp70 member, Ssa1, acts as a DNA-binding transcriptional co-activator of laccase in Cryptococcus neoformans. Mol. Microbiol. 62: 1090–1101 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.