Abstract
Selective sweeps are typically associated with a local reduction of genetic diversity around the adaptive site. However, selective sweeps can also quickly carry neutral mutations to observable population frequencies if they arise early in a sweep and hitchhike with the adaptive allele. We show that the interplay between mutation and exponential amplification through hitchhiking results in a characteristic frequency spectrum of the resulting novel haplotype variation that depends only on the ratio of the mutation rate and the selection coefficient of the sweep. On the basis of this result, we develop an estimator for the selection coefficient driving a sweep. Since this estimator utilizes the novel variation arising from mutations during a sweep, it does not rely on preexisting variation and can also be applied to loci that lack recombination. Compared with standard approaches that infer selection coefficients from the size of dips in genetic diversity around the adaptive site, our estimator requires much shorter sequences but sampled at high population depth to capture low-frequency variants; given such data, it consistently outperforms standard approaches. We investigate analytically and numerically how the accuracy of our estimator is affected by the decay of the sweep pattern over time as a consequence of random genetic drift and discuss potential effects of recombination, soft sweeps, and demography. As an example for its use, we apply our estimator to deep sequencing data from human immunodeficiency virus populations.
Keywords: selective sweep, deep diversity data, selection coefficient
THE frequency and strength of positive selection are key parameters of the evolutionary process, yet reliable estimates are often very difficult to obtain (Nielsen et al. 2005; Eyre-Walker 2006). As it becomes increasingly practicable to analyze the genetic diversity of natural and experimental populations at high depth, we can hope to obtain better estimates from a detailed analysis of the signatures positive selection is expected to leave in such data.
The standard model describing the population genetic signatures of positive selection is the so-called selective sweep (Maynard Smith and Haigh 1974). Selective sweeps produce distinct effects on the patterns of genetic diversity in the vicinity of the adaptive site: (i) Hitchhiking of linked neutral polymorphism with the adaptive allele generates dips in diversity that level off with increasing genetic distance from the selected site (Maynard Smith and Haigh 1974); (ii) because different lineages from a population sample taken after the sweep can coalesce almost instantaneously during the early phase of the sweep, there should be an excess of singletons in the neutral diversity around the selected site (Slatkin and Hudson 1991; Barton 1998; Durrett and Schweinsberg 2004); (iii) derived alleles can be brought to high frequencies (Fay and Wu 2000), which is unlikely under random genetic drift alone; and (iv) the adaptive haplotype should be longer than expected under neutrality since recombination has had only a short time to degrade it (Hudson et al. 1994). These hallmark signatures underlie the majority of approaches to scan for recent adaptation in population genetic data (Hudson et al. 1987; Tajima 1989; Wiehe and Stephan 1993; Fay and Wu 2000; Kim and Stephan 2002; Sabeti et al. 2002, 2007; Przeworski 2003; Nielsen et al. 2005; Voight et al. 2006; Andolfatto 2007).
In addition to detecting selective sweeps, one often wants to know the strength of selection that drove the sweep (Macpherson et al. 2007; Hernandez et al. 2011; Sattath et al. 2011). One common approach is to infer selection coefficients s of the adaptive allele from the size of the dip of approximate length s/r around the adaptive site, where r is the rate of recombination per length in the corresponding region (Maynard Smith and Haigh 1974; Kaplan et al. 1989; Kim and Stephan 2002). Approaches based on this signature rely on the interplay between recombination and ancestral variation and are thus not applicable for scenarios where recombination or ancestral variation is poorly characterized. Furthermore, certain scenarios of adaptation do not always generate clear dips in diversity. Examples are incomplete sweeps, where the adaptive mutation is not fixed in the population, and soft sweeps, where more than one haplotype has swept through the population (Pritchard et al. 2010).
Here, we develop an estimator for selection coefficients at candidate loci where a selective sweep has occurred recently. Our estimator is based on the analysis of the novel haplotype variation that arises from neutral mutations on the sweeping adaptive haplotype. These early variants are very different in kind from the variation due to neutral mutations occurring after a sweep, since they have experienced an initial phase of exponential amplification. Mutations after the sweep will quickly outnumber the few that happened early during the sweep, but they will drift to higher population frequencies comparatively slowly.
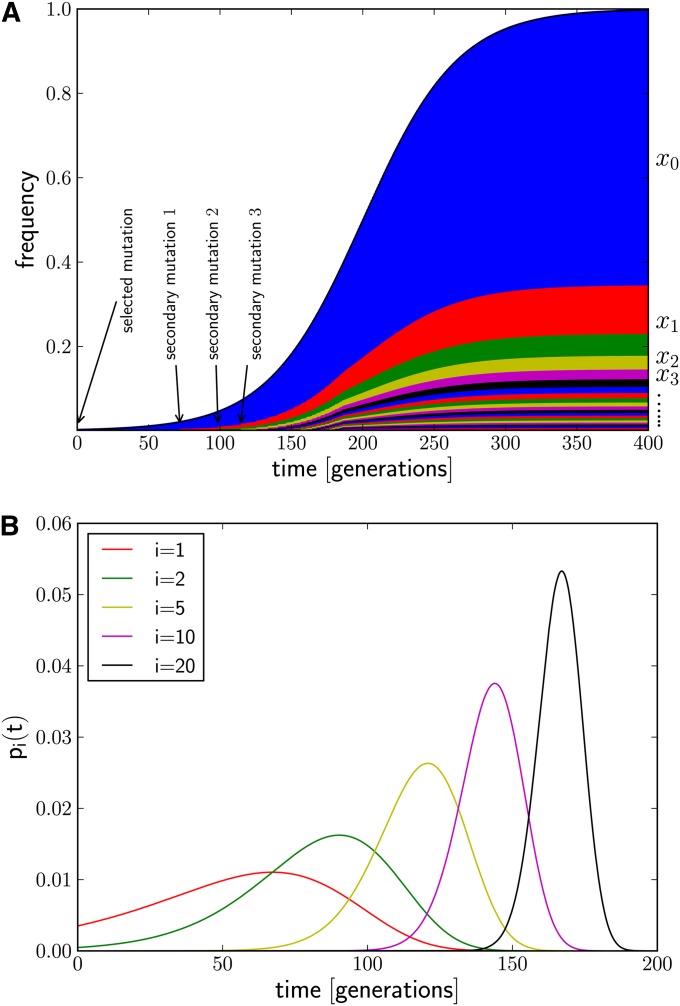
The frequency spectrum of the early haplotype variants is determined by the distribution of their seeding times during the sweep, as illustrated in Figure 1. We show analytically that their rank-frequency spectrum (the frequencies ordered by decreasing abundance) decays according to a simple power law, which differs markedly from the approximately exponential decay expected under neutral evolution. This power law is characterized by only a single parameter: the ratio of the rate at which new haplotypes arise and the selection coefficient s of the sweep.
Figure 1.
(A) Generation of novel haplotype diversity from secondary mutations during a selective sweep. At t = 0 a beneficial mutation establishes and the frequency, x0, of its underlying haplotype (blue) rises. After some time, a neutral mutation occurs in an individual carrying the beneficial mutation, creating a new adaptive haplotype variant (red) that itself increases in frequency, x1. This happens repeatedly, giving rise to a range of low-frequency haplotypes that all descended from the initial founding haplotype. Sweep parameters are s = 0.03 and u = 0.004, and seeding times of new variants are given by Equation 4. (B) The distribution of seeding times for different i according to Equation 3.
We use this result to develop an estimator of s that compares the strength of selection to the rate at which novel haplotypes are produced. Novel haplotypes can be produced by mutation or recombination. In many organisms, recombination is rare and the mutation rate is larger than or similar to the recombination rate (Haag-Liautard et al. 2007; Ossowski et al. 2010; Roach et al. 2010). Hence our estimate is much less sensitive to poorly characterized recombination rates, thereby overcoming several problems of estimators based on dips in diversity. In particular, our estimator should also be applicable to organisms that exchange genomic segments via horizontal transfer (many bacteria) and populations where levels of ancestral variation are very low, for instance in experimental evolution with clonal populations.
Our estimator relies on deep diversity data for accurate measurements of the population frequencies of rare haplotypes. With large-scale sequencing projects such as the 1000 genomes project in humans (1000 Genomes Project Consortium 2010) and similar projects in flies (Drosophila Population Genomics Project 2011) and plants (Cao et al. 2011) currently under way, such data will become available soon.
Materials and Methods
Forward simulations of selective sweeps
Simulations were performed with custom-written C++ programs that represent each haplotype in the population by a bit string of fixed length L = 1024 or L = 4096. Populations are initialized with a sample of size 104 individuals obtained from the neutral coalescent, using the program ms (Hudson 2002). Note that this initial variation is needed only to allow diversification by recombination. We constrained ms to return a sample with 1023 (or 4095, respectively) segregating sites, leaving the site in the center of the locus for the beneficial mutation. Each genotype sampled from the coalescent is copied N0/104 times into the initial population. The beneficial mutation is introduced into one randomly chosen individual in generation 0. If the beneficial mutation is lost due to random drift, the population is reset to the neutral sample and the beneficial mutation is introduced again until a successful sweep is observed. Note that our initial condition with each site polymorphic is not supposed to imply that the product of the population size and the per site mutation rate is much larger than one. Instead, those loci correspond to the subset of polymorphic loci scattered along a genomic region. The results do not depend on the number of segregating sites in the initial sample, as long as the pairwise difference between two randomly chosen genomes is almost always much larger than one. We repeated the simulations using only 1/10th of the segregating sites and obtained similar results (see supporting information, File S1).
The Malthusian fitness of a haplotype is given by F = s or 0, depending on whether the haplotype carries the beneficial allele at position L/2. In each generation, a pool of gametes is produced to which each individual contributes a Poisson-distributed number of copies with mean exp(F − 〈F〉 + C), where C = (1 − N/N0) serves as a carrying capacity to keep the population size approximately constant at N0 and 〈F〉 is the mean fitness in the population. A fraction r of the newly produced gametes are paired at random and crossed over at a randomly chosen position (since r ≪ 1, there is no crossover in most gametes). At the L − 1 neutral loci, mutations are introduced at random positions into the gametes with the specified mutation rate u. When simulating the sweeps of different strength, the mutation rate is changed accordingly to keep the ratio of the strength of selection and the total mutation rate in the range specified in Figures 2–4. This is akin to changing the size of the locus simulated and allows more efficient simulations. The code is available on request.
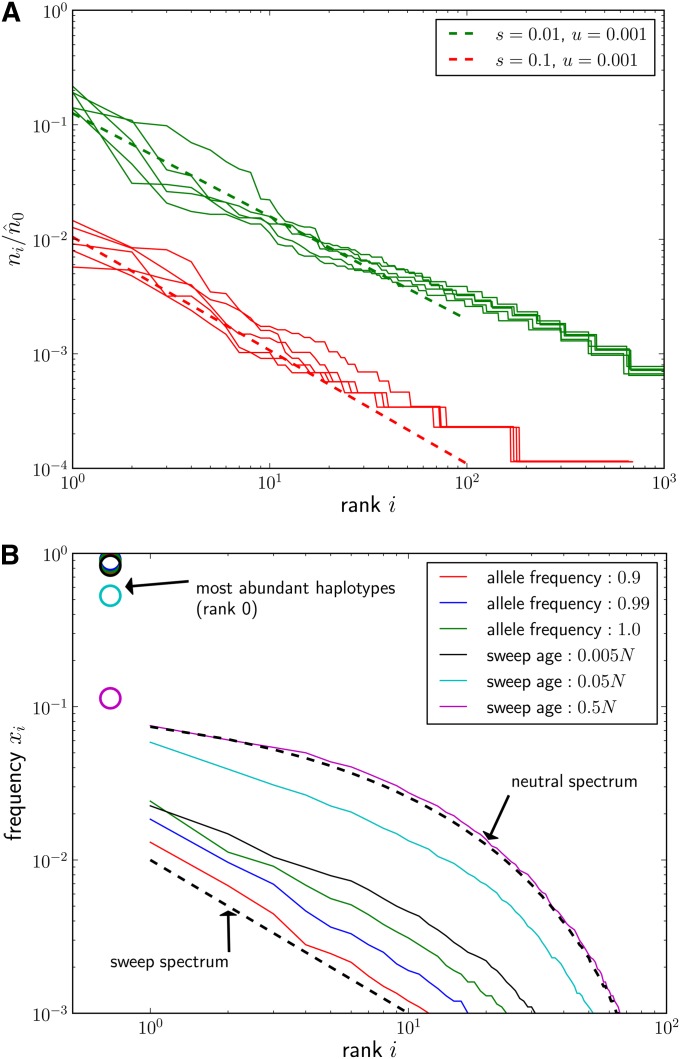
Figure 2.
(A) Ensembles of five haplotype rank-frequency spectra for u = 0.001 and two selection coefficients s = 0.1 and s = 0.01. Spectra were estimated from samples of size n = 104 taken when the allele frequency of the beneficial mutation reached 0.99. The deterministic expectations given in Equation 7 are shown as dashed lines. The normalizer was calculated according to Equation 6 with k = 4. (B) Relaxation of haplotype spectra to the neutral spectrum. Rank-frequency spectra were estimated from samples of size n = 104 when the beneficial allele reached population frequencies 0.9, 0.99, and 1.0 and for several time points after completion of the sweep (time in generations) (s = 0.001, u = 10−5, L = 1024, N = 106, Θ = 2Nu = 20). Each curve is the median of 50 simulated sweeps. The most common haplotypes (rank 0, not represented on log scale) are indicated by circles. Right after the sweep, the population consists of one very common haplotype, x0 ≈ 0.98, and a large number of rare variants whose frequency decays as predicted by Equation 7 (bottom dashed line). Over time of order t ≈ N, the frequency spectrum relaxes to the expected neutral spectrum (top dashed line).
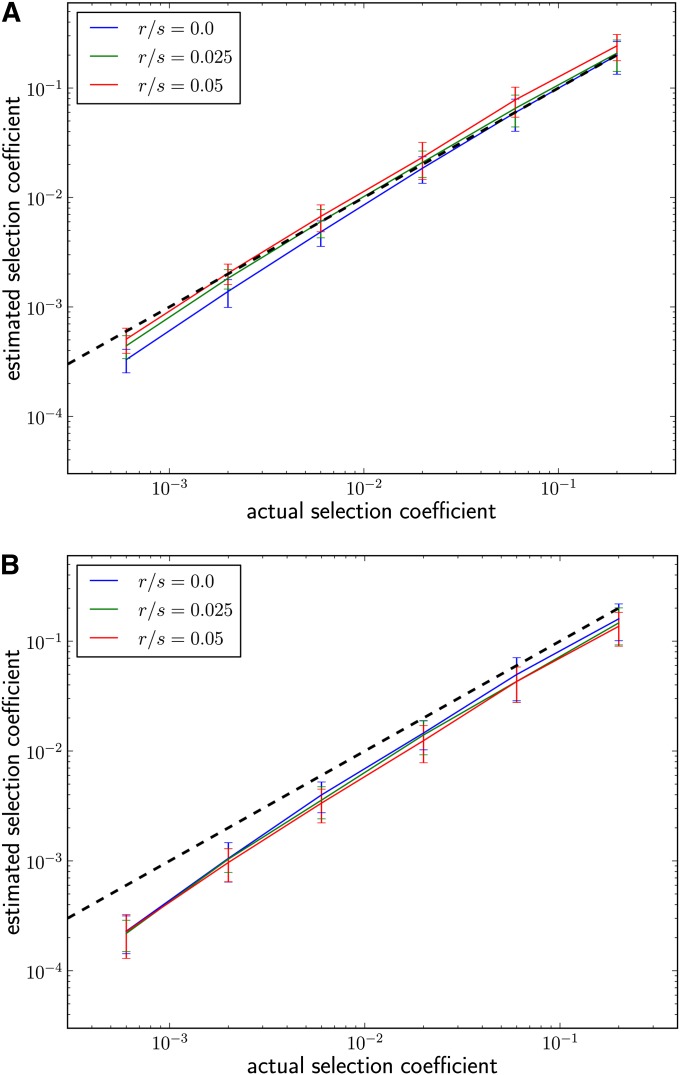
Figure 4.
Estimating selection coefficients in the presence of recombination. (A) Performance of our estimator , using Equation 9 with diversification rate u + r for different r with u/s = 0.05. The slight overestimation at high recombination rates is due to the fact that high- and low-frequency haplotypes degrade at slightly different rates due to the possibility of recombination with an identical haplotype. (B) Estimation of from haplotype spectra restricted to those haplotypes that differ at only one site from the most abundant haplotype. After pruning, there are fewer abundant haplotypes and the estimates are more sensitive to genetic drift.
To compare our method to previously developed methods to estimate selection coefficients (Nielsen et al. 2005), we generated data using the program msms by Ewing and Hermisson (2010); see File S1.
Analysis of HIV data
Sequences from the relevant samples were read by a python script and translated. Sequences corresponding to identical amino acid sequences were grouped together. Within each of these groups, the numbers of transitions and transversions that do not change the amino acid sequence were determined. In the case that the sequence overlapped with the end of the gene and extended into the long terminal repeat (LTR), only the amino acid sequence up to the stop codon was used to group sequences and mutations in the LTR were treated as neutral mutations. Sequences from the study on early immune system escape mutations by Fischer et al. (2010) are available from the NCBI short read archive under accession no. SRA020793. Sequences from the study on drug resistance evolution by Hedskog et al. (2010) were obtained from the authors.
Results
Haplotype frequency spectrum of a selective sweep
Consider a new adaptive mutation with selection coefficient s > 0 in a panmictic haploid population of constant size N. We assume that selection is strong (Ns ≫ 1). Once an adaptive mutation becomes established in the population by reaching a population frequency x ≈ (2Ns)−1 that ensures its escape from future stochastic loss (Maynard Smith 1971), its frequency trajectory can be modeled by logistic growth
| (1) |
Let us first assume that recombination is infrequent compared to mutation (we discuss recombination below). If neutral mutations occur during the early phase of the sweep on the adaptive haplotype, they can generate new variants of the adaptive haplotype that can also rise in frequency (Figure 1A). We adopt an infinite haplotypes model where every such mutation creates a distinct haplotype. When the frequency x(t) of the beneficial allele is still small, novel adaptive haplotypes become established in the population at an approximate rate α(t) = 2s × uNx(t), where u is the rate at which neutral mutations occur on the sweeping haplotype per generation. The factor 2s accounts for the establishment probability of those new haplotypes. After the sweep is completed, a novel haplotype variant will be the more frequent the earlier it arose in the sweep. Thus, to understand the spectrum of haplotype frequencies, we have to study the distribution of times at which these haplotypes are seeded.
Since novel haplotypes are seeded in independent events, the total number of established new haplotypes up to time t is Poisson distributed with parameter
| (2) |
The approximation assumes that the relevant haplotypes are seeded while the sweeping allele is still rare and increases exponentially, which is appropriate as long as t ≪ log(Ns)/s, and that s ≫ u, which can be ensured by reducing the size of the locus under investigation.
The probability density that haplotype i is seeded at time t is given by the rate α(t) of establishing new haplotypes multiplied by the probability of having i − 1 haplotypes at time t,
| (3) |
where we have used the same approximations as in Equation 2 and dropped factors independent of t that ensure normalization. This distribution pi(t) is shown in Figure 1B for different i. The intervals between the peaks of pi(t) and pi+1(t) become smaller with increasing i, while the width of each peak decreases. More precisely, we derive from Equation 3 that the most likely seeding time of the ith haplotype is given by
| (4) |
and that the peak of pi(t) has a width of (if i ≫ 1). The joint distribution of seeding times and the unordered frequency spectrum are derived in File S1, section 2.
Assuming that the ith haplotype establishes at and has same fitness as the founding haplotype, it will also rise in frequency, albeit with a time lag . Together with Equation (4) we obtain an approximation for the expected frequency of the ith haplotype:
| (5) |
The limit corresponds to the asymptotic behavior as the beneficial allele approaches fixation. Note that this expression holds for haplotypes i ≥ 1. The zeroth haplotype, i.e., the haplotype the mutation inititally arose on, grows as x0(t) = (e(s − u)t)/(est + 2Ns) and asymptotes to e−ut. Each of the haplotype variants carrying the adaptive allele thus grows at a slightly smaller rate, s − u, than the overall rate s at which the adaptive allele arises, accounting for the founding of new haplotype variants by mutation.
The interplay between the exponential amplification of the adaptive allele and the generation of new adaptive haplotype variants thus gives rise to a simple power-law decay of the rank-frequency spectrum {x0, x1, …} of adaptive haplotypes: The most abundant adaptive haplotype, on average, is typically s/u times more frequent than the second most abundant adaptive haplotype, 2s/u times more frequent than the third most abundant adaptive haplotype, and so forth. This power-law spectrum with exponent β = 1 − u/s differs markedly from that of neutral evolution, where haplotype frequencies are expected to decay as xi/x0 ∝ exp[−i/(2Nu)] with haplotype rank i (see File S1, section 1).
The distribution of haplotype frequencies after a sweep is related to the distribution of “family sizes” studied by Barton (1998). Barton presented numeric results for the distribution of the size of groups of individuals that share a common ancestor at the beginning of the sweep. Haplotype frequencies, however, refer to the size of groups identical by state, rather than by descent, and characterize diversification that happened after the sweep, rather than the ancestral variation that survived the sweep.
To compare the haplotype counts n0 > n1 > n2 … in a sample of size n to Equation 5, we need an estimate of e−ut. The simplest estimate of e−ut is the sample frequency of the most abundant haplotype, n0/n, whose deterministic value would be precisely e−ut. However, n0 can be quite variable because the variances of the first few seeding times are still large. To circumvent this problem, one can construct a more stable estimate of e−ut on the basis of the first k haplotypes and compare the sum of the sample frequencies to the sum of the deterministic expectation of the frequencies,
| (6) |
where β = 1 − u/s. This effective is insensitive to k as long as the first few haplotypes are included. Figure 2A shows a comparison of the simulated haplotype spectra with
| (7) |
The logarithmic axes of the plot are chosen such that the algebraic decay shows up as a slope −β. The observed slope is correct initially, but the curves become slightly flatter after haplotype rank ≈ 50. This is due to degradation of the sweep pattern by random genetic drift, which we discuss next.
In deriving Equation 7, we have neglected fluctuations in the seeding and establishment times of novel haplotype variants as well as genetic drift after the sweep is complete. Relaxing these assumptions requires a stochastic calculation, which is given in File S1, sections 3A and 3B. Specifically, we calculate the probability of finding ic haplotypes to be present in more than nc copies in the population, assuming the logistic trajectory of the beneficial allele given in Equation 1. These calculations are lengthy, but the resulting effects can be understood by the simple arguments given below.
Variations in the establishment times will cause the haplotype spectra to fluctuate and result in spectra below or above Equation 7 if new haplotypes were seeded fortuitously early or late. As expected from the distribution of seeding times in Figure 1, the frequencies of the first few haplotypes fluctuate substantially, while the later ones do not. Since the number of haplotypes seeded by time t is Poisson distributed, so is the number of haplotypes above a certain frequency after the sweep is complete (neglecting genetic drift, there is a one-to-one mapping between establishment time and eventual frequency). Hence, the fluctuations due to random seeding times are small if the expected number of haplotypes above the chosen frequency is large. In accordance with these arguments, variation of haplotype spectra in Figure 2A decreases with increasing i.
Once the beneficial allele approaches fixation, the strength of selection goes to zero, and exponential amplification ceases. The frequencies of the rare haplotypes thereafter change primarily due to random drift, while the frequencies of the common haplotypes decrease due to additional mutations that produce new rare haplotypes. Both of these processes lead to a homogenization of haplotype frequencies, i.e., common haplotypes becoming rarer and rare haplotypes becoming more common, which ultimately wipes out the sweep signature. This relaxation to the neutral haplotype spectrum is shown in Figure 2B.
Since the time required to drift to frequency xc is approximately Nxc, the haplotype rank-frequency spectrum will soon develop a bulge of drift-dominated haplotypes at low frequency and high rank. This accumulation of rare haplotypes due to genetic drift can be calculated perturbatively (see File S1, section 3). For the expected number of haplotypes ic with frequencies above xc, one obtains
| (8) |
where Δt is the age of haplotype ic, which is of the same order as the age t of the sweep. Note that Equation 8 is essentially Equation 5 solved for i with the “drift contribution” Δt/N subtracted from the frequency xc. Since the age Δt of the haplotype is similar to that of the beneficial allele, t, we conclude that for frequencies less than t/N, the sweep spectrum is eroded, while for frequencies much larger than t/N, it prevails. After a time of order N, the entire spectrum has relaxed to the neutral haplotype frequency spectrum as shown in Figure 2B. Note that the time it takes for the beneficial allele to go from frequency 0.9 to complete fixation can be long, and substantial erosion of the sweep spectrum can happen during this time interval.
In general, the time that has elapsed since the beginning of the sweep is unknown. To obtain an estimate of the age of the sweep, it is useful to reconsider Equation 5, which states that the frequency of the founding haplotype is expected to asymptote to e−ut. This behavior, and in particular the more accurate estimate of e−ut given in Equation 6, allows one to obtain a rough estimate of the sweep’s age.
In addition to genetic drift, limited sampling of the population results in a deviation of the observed from the expected sweep spectra. The detected haplotype counts are a convolution of their true frequencies with the distribution expected from sampling the population. After ranking of haplotype counts, a large number of rare variants leads to a flattening of the spectrum and in particular an excess of singletons.
Estimating selection strength
According to Equation 7, the expected rank-frequency spectrum of haplotypes in a selective sweep is determined by the single parameter u/s—the ratio of the mutation rate over the locus and the strength of selection. Given an independent estimate of u, one can therefore use the haplotype frequency spectrum to estimate the selection coefficient of a sweep, for example by simply counting the number of different haplotypes present above a specified frequency cutoff in the sample. Let ic be the overall number of different haplotypes that are present in at least nc copies each. The estimator for the strength of selection is then
| (9) |
where is either set to the observed n0 or determined via Equation 6.
The estimator allows us to either fix nc and then determine the count ic from the data or fix ic and then measure nc. In either case, the cutoff should be chosen to achieve maximum accuracy. Since the number ic of haplotypes above a frequency threshold is Poisson distributed, fluctuations of decrease with smaller nc and larger ic. Too low ic, however, will include parts of the frequency spectrum that have already been degraded by drift, which predominantly affects the rare haplotypes. In addition, limited sampling depth limits ic. Hence ic should be chosen as large as possible, but such that nc ≫ 1 and that the frequency spectrum is still of power-law form down to haplotype ic. In this case, the relative error of is of order (assuming the model assumptions are met).
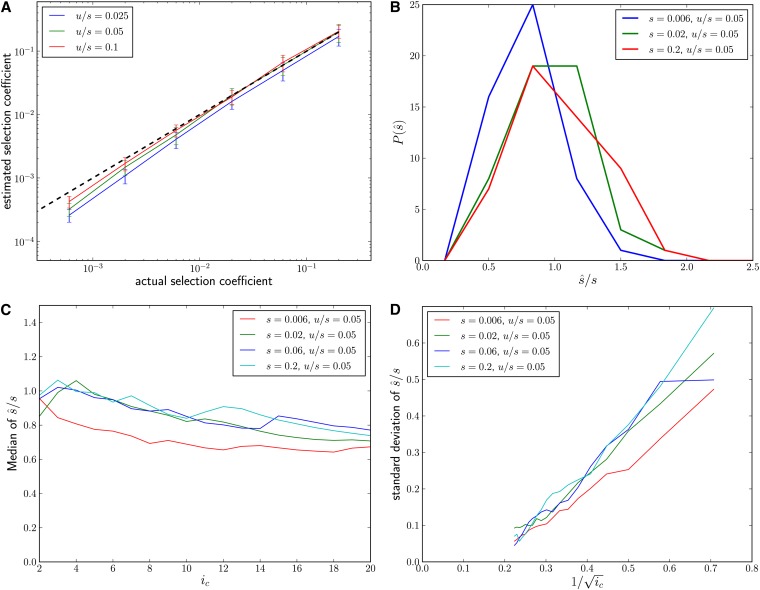
It is generally advisable to fix ic and determine because spectra come flatter than i−1 due to the confounding effect of genetic drift and limited sampling of the population. Figure 3 shows the performance of our estimator when applied to simulated sweeps (see Materials and Methods) for different selection coefficients and mutation rates as well as its dependence on the choice of ic. The simulations confirm that our estimator performs accurately over the range of moderate selection (s ≈ 10−3) to strong selection (s ≈ 0.2) given the parameters used. However, there is a systematic downward bias for small s. This bias is due to genetic drift and limited sampling depth.
Figure 3.
(A) Mean over 50 simulation runs for a wide range of selection strengths and three different ratios of u/s, using Equation 9 with ic = 5. Error bars are ±1 standard deviation. The dashed black line indicates the correct s. Population samples of size n = 103 were taken when the beneficial allele reached fixation. For small s, there is a slight downward bias due to genetic drift. L = 4096 and N = 106. (B) Distribution of at different selection strengths for fixed u/s = 0.05. (C) Performance of as a function of ic. There is a tendency to underestimate the strength of the sweep if ic is large, which arises from the inclusion of rare haplotypes that are affected by genetic drift and may contain haplotypes that arose after the sweep. (D) Standard deviation over 50 estimates as a function of . Variation is suppressed at large ic due the discreteness of low population frequencies.
In File S1, section 3, we derive an estimator that accounts for genetic drift perturbatively; see Equation 24 of File S1. In essence, this estimator contains the same correction present in Equation 8; i.e., it subtracts the drift contribution t/N from the frequency of the rare haplotypes. Importantly, this drift correction sets the limits of applicability of :
-
i.
After the completion of the sweep, we can estimate s only for a limited time. Specifically, for a given sample frequency nc/n, we require that t < Nnc/n.
-
ii.
The range of s that can be reliably estimated by the method is limited: Even if we catch the sweep right at the time of fixation of the adaptive allele, it still needs to have occurred faster than the timescale of its own degradation by drift. Given that the time it takes to complete a sweep is ≈(2/s)log Ns and that nc/n ≈ u/(sic), we require N > (2ic/u)log Ns. Together with s ≫ u, we find that the range of s that can be estimated is bounded from below by Ns ≫ iclog Ns. This estimate is consistent with the deviations in Figure 3 for s < 10−3.
Recombination
Up to now, we have neglected any potential effects of recombination on the frequency distribution of the new adaptive haplotype variants arising in a selective sweep. In fact, one of the advantages of our estimator is that it does not exclusively rely on recombination for its inference of selection coefficients and can thus also be applied to systems that lack recombination or where recombination rates are not well known. However, if recombination occurs on the sweeping haplotype, this can generate new variants of the adaptive haplotype in a manner similar to mutation. This becomes important whenever the recombination rate r over the locus is comparable to its mutation rate u. In this case, the rate at which new haplotype variants are generated during the sweep will actually be higher than assumed on the basis of the mutation rate u, and we thus expect to underestimate the strength of the sweep.
We propose two ways to incorporate recombination: First, one can treat recombination analogously to mutation and assume that every recombination event generates a new variant of the adaptive haplotype that will be different from all other existing variants. Under this assumption, the mutation rate u simply needs to be replaced by the sum u + r. Figure 4A shows how our estimator performs for different ratios of mutation to recombination rate under this approach. The infinite haplotypes assumption is appropriate if ancestral diversity is high and independent recombination events are thus unlikely to yield equal outcomes. If, however, ancestral diversity is low, the infinite haplotypes model will overestimate the rate at which new haplotypes arise.
Alternatively, we can try to effectively exclude most recombined adaptive haplotype variants. Recombined variants are likely to differ from the founding haplotype at more than one site, whereas variants originating from mutation should differ at only one site. By filtering out the former, we can treat the remainder as originating primarily from mutation events. As shown in File S1, section 4, such a set of pruned “mutation-only” haplotypes has a slightly different rank-frequency spectrum: The exponent β is exactly 1, rather than 1 − u/s. Besides that, Equation 9 can be applied with u being the mutation rate and nc being the abundance of the ic haplotype in the pruned set of haplotypes. The estimates obtained by this procedure are shown in Figure 4B. While this modified estimator still seems to underestimate selection coefficients slightly, it performs consistently across different recombination rates.
The difference in the exponent of the rank-frequency spectrum arises from slightly different rates of seeding and amplification of new haplotypes. Without pruning, the rate of establishment of novel haplotypes is proportional to the frequency of the beneficial allele, which follows a logistic growth with rate s. Haplotype frequencies, however, grow only with rate . This slight difference in rates is responsible for the deviation of the exponent β from 1 by u/s or (u + r)/s, where the latter also accounts for recombination. When restricting the analysis to haplotypes that descend directly from the founding haplotype, the rate of establishment of novel haplotypes is proportional to the frequency of the founding haplotype. Hence, establishment and amplification happen with the same rate and the exponent is exactly 1.
We note another minor difference in the dynamics of haplotype frequencies in the presence of recombination. When the beneficial allele reaches fixation, the most abundant haplotype will most likely recombine with itself. Thus it diversifies more slowly after the sweep is complete. Minor haplotypes, however, continue to diversify through recombination, which has the effect of slowly decreasing their frequency and producing novel low-frequency haplotypes. This slow decrease opposes the effects of genetic drift, which has the tendency to increase the frequency of minor alleles. In simulations, estimates of s often become more accurate due to this effect.
Application of to deep sequencing data from human immunodeficiency virus
Deep population diversity data of the kind required for the application of our estimator have recently been obtained for human immunodeficiency virus (HIV) populations by sequencing viral RNA from plasma samples (Tsibris et al. 2009; Fischer et al. 2010; Hedskog et al. 2010). We present three examples from such studies to validate our method and discuss its applicability.
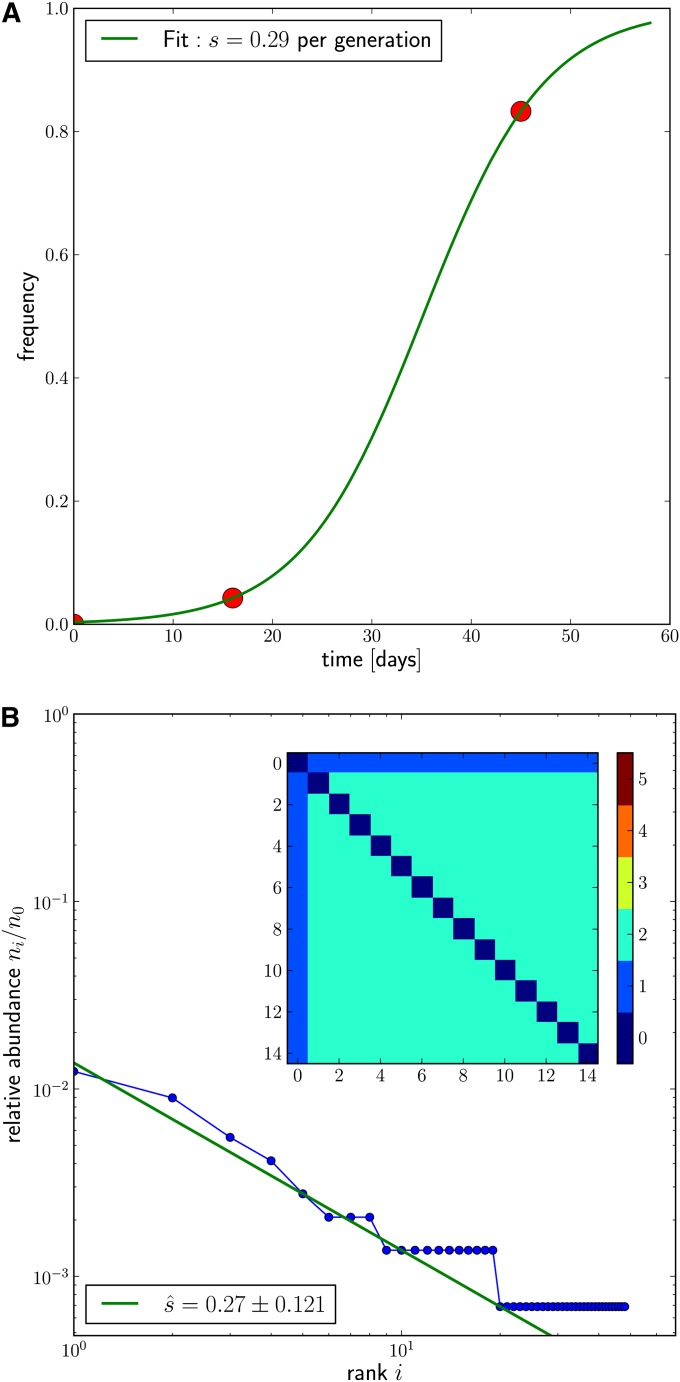
As a first example, we investigated a sample taken shortly after infection when HIV evolution is primarily driven by mutations that allow the virus to escape the immune system. For each of the patients studied in Fischer et al. (2010), samples from several time points were investigated and amplicons sequenced using 454 technology. In one of the epitopes studied (gene nef in patient CH40), a mutation spread sufficiently slowly such that it was possible to observe the mutation rise from low frequency at day 16 to high frequency at day 45 (Figure 5A). From this time series Fischer et al. (2010) estimate a selection coefficient of 0.3, assuming a generation time of HIV between 1.5 and 2 days (Perelson et al. 1996).
Figure 5.
The rise of a mutation selected by the immune system in the gene Nef of patient CH40 from Fischer et al. (2010). (A) Frequency trajectory of the adaptive allele with a fitted curve for s ≈ 0.3. (B) Observed haplotype spectrum of variation due to synonymous transitions from all sequences containing the selected allele at day 45. The straight line indicates the 1/i behavior. The pairwise nucleotide distance between all haplotypes ordered by abundance is shown in the inset. The error of the estimate is obtained assuming Poisson statistics with ic = 5. Note, however, that the uncertainty of the mutation rate is of the same order.
To validate our estimator, we can compare this direct estimate of the selection coefficient obtained from time-course data with that obtained from the frequency spectrum of haplotype diversity at the locus, which is shown in Figure 5B. Figure 5B contains the frequency of haplotypes with the dominant amino acid sequence that differ only by putatively neutral synonymous mutations. The haplotype rank-frequency spectrum according to Equation 9 suggests a ratio of u/s ≈ 0.014. There is considerable uncertainty in the mutation rate estimates for HIV, ranging from 3.4 × 10−5 (Mansky and Temin 1995) to 9 × 10−5 per site and generation (O’Neil et al. 2002 for mutations in the LTR). The majority of these mutations are transitions. In our example the sequenced locus tolerates a total of 93 transitions that do not change the amino acid sequence of Nef or fall into the LTR that overlaps the sequenced region. Using 4 × 10−5 per generation as an averaged estimate for the transition rate, we arrive at
| (10) |
The uncertainty stemming from the random times at which the haplotypes are seeded is expected to result in relative errors , which for ic = 5 used here amounts to ±0.12. Additional uncertainty (probably larger) in the mutation rate needs to be added to these error bars. Given these uncertainties we consider our estimate to be in excellent agreement with the independent estimate from time-course data. Note that recombination is not expected to make a large contribution to haplotype diversification since the effective HIV recombination rate is less than half the mutation rate (Neher and Leitner 2010; Batorsky et al. 2011). Furthermore, the HIV population has undergone several sweeps prior to the date this sample was taken such that the ancestral diversity was very low.
We also checked whether the haplotype variation in our sample is consistent with the characteristic star phylogeny expected for mutations that arose on the background of the sweeping founder haplotype. In particular, under the assumption of an infinite sites model where each such mutation gives rise to a new haplotype, we would expect that descendants should always be one mutation away from the founding haplotype, while any two descendants should be two mutations away from each other. Our example shows precisely this pattern, as can be seen in the inset in Figure 5A. This is further evidence that recombination did not contribute substantially to haplotype diversification.
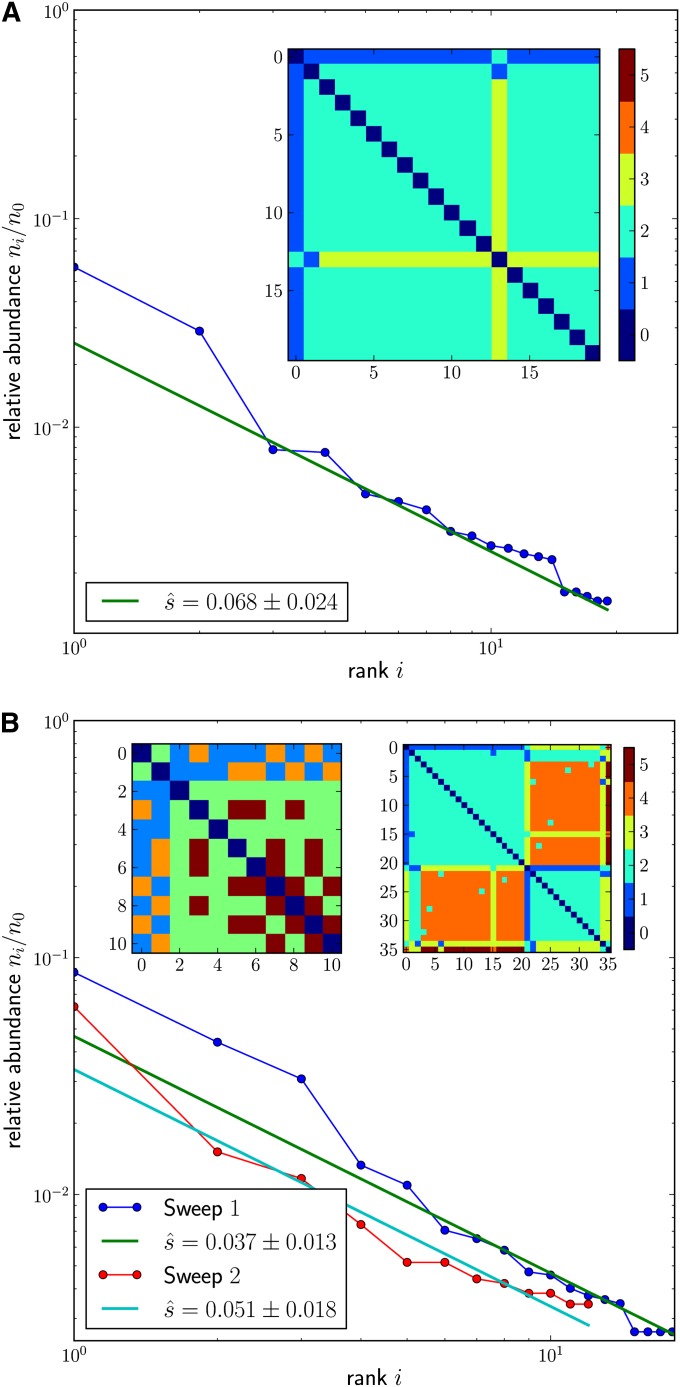
As a second example, we consider deep sequencing data of the reverse transcriptase (RT) locus of HIV (120 bp, amino acids 180–220) (Hedskog et al. 2010). The patients analyzed in this study were under antiretroviral treatment, which implies strong selection pressure for resistance mutations at the RT locus. During the course of treatment resistance has evolved repeatedly in several patients. We therefore expect to see signatures of strong selective sweeps in these viral populations.
As before, we restricted our analysis to synonymous differences because our model assumes selectively neutral mutations. The 120-bp analysis windows typically contained enough variation such that in most samples more than eight different haplotypes were sampled. Hence we chose ic = 8 for estimating and then measured the cutoff frequency nc of the least abundant haplotype to evaluate Equation 9.
Figure 6A shows the measured haplotype rank-frequency spectrum for one of the patient samples (time point 4, patient 4). The spectrum decays with the characteristic power law of a selective sweep. As discussed above, we determined the number of synonymous transitions in the founding sequence and used 4 × 10−5 as the rate of individual transitions. With this mutation rate estimate, we arrive at per generation. Note that the causative mutation, or the combination of mutations, that drove this sweep does not necessarily have to lie inside the analyzed window but could also be located elsewhere in the genome.
Figure 6.
(A) Exemplary sweep in HIV evolution. The rank-frequency spectrum and pairwise distance matrix are shown for the different haplotypes coding for the same amino acid sequence, amino acids 180–220 of the RT (Hedskog et al. 2010); compare Figure 5. All but haplotype 13 differ from the most abundant haplotype (0) by one mutation. Haplotype 13 is most likely a descendant of haplotype 1. (B) Example of a soft sweep in HIV evolution. In this sample, the two most abundant haplotypes 0 and 1 differ by more than one mutation (top left corner of left inset, showing the distance matrix of the 10 most abundant haplotypes), suggesting that the selected mutation arose independently on different backgrounds. By inspecting the first two columns of the left distance matrix, one finds that most of the other haplotypes differ by one mutation from either haplotype 0 or haplotype 1, consistent with the beneficial mutation having arisen independently on haplotypes 0 and 1. Assigning the rare haplotypes to either of the two abundant “founding” haplotypes and reordering yield the distance matrix shown as the right inset, this time including all 36 haplotypes. The reordered matrix displays two characteristic sweep patterns as blocks on the diagonal. Indeed, the haplotypes of each of these two blocks display the characteristic power-law rank-frequency spectrum. In both panels, ic = 8 and relative errors are .
Figure 6B shows haplotype data from another population sample (time point 4, patient 5). In this third example, the pairwise mutational distances between different haplotypes reveal a more complex pattern (left inset) that does not seem to be compatible with the simple star phylogeny observed in the example from Figure 6A. However, when we focus on the two most abundant haplotypes 0 and 1, we see that these two haplotypes differ at more than one site from each other. The majority of the remaining haplotypes differ by only one mutation from either haplotype 0 or haplotype 1. This pattern can be interpreted as the result of two independent partial sweeps where the two founding haplotypes (0 and 1) differ at two sites.
We can disentangle these two hard sweeps by assigning the rare haplotypes to either of the two founding haplotypes if they differ from the latter by only one mutation. When reordering the distance matrix according to this assignment, we recover the two individual sweep patterns as blocks on the diagonal. This is shown in the right inset of Figure 6B. Both reordered components now exhibit the characteristic power-law decay in their individual rank-frequency spectra with similar selection coefficients, again in the range of a few percent.
A scenario like the one shown in Figure 6B, with several haplotypes carrying the same adaptive mutation rising in frequency simultaneously, is commonly referred to as a “soft sweep” (Hermisson and Pennings 2005). Soft sweeps can occur, for example, if an adaptive mutation arises repeatedly on different haplotypes, which is expected to be common in viruses with large population sizes and high mutation rates such as HIV, where every single point mutation can arise many times each generation. Alternatively, soft sweeps can result when a sudden change in environment renders a previously neutral or deleterious allele selectively beneficial and adaptation involves several different adaptive haplotypes from the standing genetic variation.
An unambiguous decomposition of the haplotype distance matrix into individual sweep components of a soft sweep requires that the founding haplotypes differ by several mutations. This is likely to be the case for a diverse ancestral population. Note that the above approach also suggests how can be applied to incomplete sweeps or sweeps restricted to a subpopulation. In those cases, the reordered distance matrix should display the characteristic sweep pattern only for a subblock on the diagonal that is embedded into the diverse remainder of the population.
Intriguingly, we see very little evidence of degradation of the haplotype spectrum by genetic drift. This might have several causes: (i) a very large population size, (ii) a very recent sweep, or (iii) a scenario where sweeps are so frequent that they start overlapping such that exponential amplification of the most fit variants never ceases. The latter is expected in large continuously adapting populations (Neher and Shraiman 2011). To investigate this matter further, one would need denser time-course data and information on genetic diversity across the genome.
Discussion
We have investigated the pattern of haplotype variation that arises from mutations occurring during the early phase of a selective sweep on the sweeping haplotype. We found that a selective sweep leaves a characteristic signature in the frequency spectrum of such novel haplotypes: When ordering all adaptive haplotype variants by their abundance, the frequency of the ith haplotype variant is ∼u/(is) times the frequency of the first, where u is the mutation rate of the locus and s is the selection coefficient of the sweep. The power-law decay of the rank-frequency spectrum is a consequence of exponential amplification (via selection) competing with diversification (through mutation and recombination), similar to the mechanism of preferential attachment originally proposed by Yule (1925). We applied this finding to construct an estimator for the selection coefficient at a candidate locus where a selective sweep is suspected to have occurred recently. Either such loci could be identified by standard approaches to localize sweeps (Fay and Wu 2000; Kim and Stephan 2002; Sabeti et al. 2002; Nielsen et al. 2005; Voight et al. 2006; Sabeti et al. 2007) or a priori information could suggest that a sweep has occurred, such as in our HIV examples, where the failure of antiretroviral therapy implied an adaptive change at the RT locus.
The classic signature used to infer the strength of a recent sweep has been the size of the dip in diversity around the adaptive site (Maynard Smith and Haigh 1974). Underlying this approach is the rationale that recombination breaks up linkage with increasing genetic distance from the adaptive site: The farther away it is from the sweeping locus, the higher the probability that presweep variation has become unlinked from the adaptive allele and thus remains polymorphic after the sweep. Hence, the width of the dip in genetic diversity scales with the ratio of the selection coefficient and the recombination rate.
While successfully applied in many studies (Kim and Stephan 2002; Sabeti et al. 2002, 2007; Andolfatto 2007; Macpherson et al. 2007; Sattath et al. 2011), this approach suffers from a number of shortcomings that can limit its applicability. First, it relies on recombination and thus cannot be applied to organisms that frequently self, such as many plants or yeast, as well as to organisms that recombine via horizontal gene transfer, such as bacteria. Recombination rates can also fluctuate strongly along the genome and in time (Winckler et al. 2005; Coop and Przeworski 2007), rendering their precise estimation difficult, especially in the regions of reduced genetic diversity around a selective sweep. Furthermore, the approach relies on the presence of preexisting variation at the sweep locus and assumes that we know how much variation was present originally. Ancestral diversity is literally absent in evolution experiments where populations start from a single clone or in populations where adaptation occurs so frequently that genetic diversity is not fully restored between recurrent selective sweeps (Gillespie 1994). Finally, for strong sweeps, the dips in diversity can potentially span very large regions extending up to entire chromosomes (Andersen et al. 2012). In such cases, dip sizes can no longer be assessed accurately.
Our estimator is less dependent on ancestral diversity or accurate recombination rate estimates, since we investigate the new variation that emerges during the sweep and compare selection strength to the rate of haplotype diversification (mutation and recombination) rather than to the recombination rate alone. It is essential for our estimator that one can accurately determine haplotype population frequencies on the order of u/(sic), requiring deep population samples. For example, if we assume a sweep locus with u/s = 0.1 and base our analysis on the five most frequent adaptive haplotype variants (ic = 5), it would be necessary to measure haplotype frequencies of 2% accurately. This calls for a sample size of ≈103. Population genetic data of such depth is already available for HIV, where a large number of sequences can be obtained from plasma samples of infected patients (Tsibris et al. 2009; Fischer et al. 2010; Hedskog et al. 2010). Several efforts are currently being made to achieve a comparable in-depth characterization of the population diversity in several eukaryotes, including humans (1000 Genomes Project Consortium 2010), flies (Drosophila Population Genomics Project 2011), and plants (Cao et al. 2011).
In practice, one typically has a data set (as in the HIV example a sample of size ≈104 sequences of length 120–200 bp) and wants to estimate s. The size of the locus corresponds to a certain mutation rate, which will limit the range of selection coefficients that give rise to an observable sweep spectrum: If u/s is too small (say <10 times the inverse sample size), very little variation will be observed. On the other hand, if u/s is close to one, the founding haplotype diversifies as rapidly as it is amplified and will no longer be the dominant haplotype in the sample. Without a dominant haplotype, the method cannot be applied. Furthermore, our calculations always assume that u ≪ s.
Too large values of u/s can be circumvented by restricting the analysis to only a fraction of the locus, which effectively reduces u. Hence, if a sample contains a large number of rare alleles and haplotypes but no dominant haplotype, one can reduce the window size to see whether a dominant haplotype with a trailing sweep spectrum emerges. Similarly, if one analyzes long genomes using a windowing approach and observes a region almost devoid of diversity (corresponding to u/s ≪ 1), one can increase u by increasing the window size until several low-frequency haplotypes are observed. One can thus tune the sensitivity of the estimator to different ranges of s by adjusting u through the length of the locus. A selection coefficient of s = 10−3 and a mutation rate of 10−8 per site, for example, would require a locus of length 10 kb to achieve u/s = 0.1.
We used the popular program sweepfinder (Nielsen et al. 2005) to compare the performance of our estimator to a traditional approach where the selection coefficient of a sweep is inferred from its signature in the surrounding ancestral diversity. As expected, our estimator consistently outperforms the traditional approach if ancestral diversity is low (Figure 2 in File S1). Interestingly, even when ancestral diversity is rather high (e.g., Θ = 0.01, comparable to the level of neutral diversity observed in Drosophila melanogaster), the estimates from our method still have substantially lower variance than those obtained from sweepfinder. Note, however, that the comparison between the two methods is based on rather different data. While the sweepfinder requires long sequences (r/s ≈ 1) at moderate coverage, our methods works with much shorter regions [(u + r)/s < 0.1] at very deep coverage.
For our estimator to be applicable, haplotype variants that were produced after a sweep and rose in frequency by random genetic drift need to be still at low enough frequency such that they have not yet degraded the sweep signature. The extent of this degradation can be estimated from a simple argument: The lowest population frequency entering our estimator is of the order u/(sic), while drift-dominated haplotypes will typically be at frequencies t/N, where t is the time that has elapsed since the sweep. We thus require u/(sic) ≫ t/N, which translates into the condition that population sizes have to be sufficiently large and that sweeps are not too old. As shown in Figure 2, drift results in a growing bulge at low frequencies where the rank spectrum is exponential rather than a power law. Since the strength of genetic drift is very poorly known, we propose to inspect the ranked haplotype spectrum for deviations from the power law and choose ic small enough such that drift-dominated haplotypes are excluded from the analysis. In addition, one should check whether the sample is compatible with a near star phylogeny. Failure to exhibit these features should indicate either the absence of a selective sweep or a sweep that has already been degraded.
While we have focused on mutation as a source of novel haplotypes, recombination can produce new haplotypes as well. Provided each recombination event results in a unique haplotype, all of the above formulae hold with u + r instead of u alone. If, however, recombination rates are not known, recombinant haplotypes can be filtered out by restricting the analysis to only those haplotypes that differ by a single mutation from the founding haplotype. This removes the majority of recombinant haplotypes since recombination with the diverse ancestral population typically incorporates several polymorphisms at once.
Recent studies suggest that in many selective sweeps the adaptive allele has not actually become fixed in the population (incomplete sweep) or that several different haplotypes, all carrying the adaptive allele, have swept simultaneously through the population (soft sweep) (Hermisson and Pennings 2005; Burke et al. 2010; Karasov et al. 2010; Pritchard et al. 2010). We have demonstrated how incomplete sweeps and soft sweeps can be analyzed by our method (Figure 6B), making use of the fact that the novel haplotype variants our estimator is based on are related to the founding haplotype by a simple star phylogeny, which allows us to easily differentiate them from other haplotypes that are not descendants of the founding haplotype.
Our results on the haplotype pattern of a selective sweep were derived under the assumption of a panmictic population of constant size, raising the question of how they might be affected by past demographic events such as population expansions or bottlenecks. Most current approaches to investigate selective sweeps rely on the specific patterns a sweep leaves in ancestral neutral diversity. These approaches can thus be very sensitive to past demographic events that shape the patterns of ancestral diversity over a timescale of neutral coalescence, which can include quite ancient demographic events. The approach presented here, however, is fundamentally different. In contrast to ancestral neutral variation, which is typically old, we focus on the very recent variation that arises during the early phase of a selective sweep. Demographic events that occurred prior to the onset of the sweep are thus irrelevant. Only very rapid changes in population size that happen while the adaptive allele is sweeping can cause significant deviations in the haplotype frequency spectra.
The key scenario to be discussed in this context is that of a recent population expansion, since our estimator measures specifically the rate at which new haplotype variants were amplified. Indeed, the haplotype frequency spectrum in an expanding population will resemble that of a selective sweep if the expansion lasted long enough for all coalescence to happen during the expansion and if the expansion rate ε is large enough such that the spectrum has not yet been eroded by drift; i.e., Nε ≫ 1. In this case, our estimator turns into an estimator of the expansion rate that might be applicable to scenarios such as the expansion of an HIV population in a newly infected individual or the spread of novel strains of influenza. If, on top of an expansion, a beneficial mutation is spreading through the population, the haplotype carrying the beneficial mutation (and its descendants) is expanding faster than the ancestral haplotypes. The estimate of the selection coefficient will then in fact be an estimate of s + ε.
The spread of a beneficial mutation in the population generally reduces genetic diversity in the vicinity of the adaptive site. That a selective sweep can also amplify new diversity at very low population frequencies is thereby often overlooked. We have shown that the spectrum of this new variation records the exponential amplification of the novel beneficial allele in a clock-like fashion and can thus be used to estimate its selection coefficient. With the recent advances in sequencing technologies, the required information about low-frequency genetic variation is no longer elusive, making our estimator applicable for a wide range of analyses.
Supplementary Material
Acknowledgments
We thank Dmitri Petrov, Nick Barton, Boris Shraiman, Ben Callahan, Taylor Kessinger, and members of the Petrov laboratory for helpful feedback. This research was supported by the European Research Council under grant no. 260686 (to R.A.N.). P.W.M. was an Human Frontier Science Program postdoctoral fellow in the Petrov laboratory at Stanford University. Part of this work was conducted during the program “Microbial and Viral Evolution” at Kavli Institute for Theoretical Physics, supported by the National Science Foundation under grant no. PHY05-51164.
Footnotes
Communicating editor: M. A. Beaumont
Literature Cited
- 1000 Genomes Project Consortium, 2010. A map of human genome variation from population-scale sequencing. Nature 467: 1061–1073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen E. C., Gerke J. P., Shapiro J. A., Crissman J. R., Ghosh R., et al. , 2012. Chromosome-scale selective sweeps shape Caenorhabditis elegans genomic diversity. Nat. Genet. 44: 285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andolfatto P., 2007. Hitchhiking effects of recurrent beneficial amino acid substitutions in the Drosophila melanogaster genome. Genome Res. 17: 1755–1762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton N., 1998. The effect of hitch-hiking on neutral genealogies. Genet. Res. 72: 123–133 [Google Scholar]
- Batorsky R., Kearney M. F., Palmer S. E., Maldarelli F., Rouzine I. M., et al. , 2011. Estimate of effective recombination rate and average selection coefficient for HIV in chronic infection. Proc. Natl. Acad. Sci. USA 108: 5661–5666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke M. K., Dunham J. P., Shahrestani P., Thornton K. R., Rose M. R., et al. , 2010. Genome-wide analysis of a long-term evolution experiment with Drosophila. Nature 467: 587–590 [DOI] [PubMed] [Google Scholar]
- Cao J., Schneeberger K., Ossowski S., Günther T., Bender S., et al. , 2011. Whole-genome sequencing of multiple Arabidopsis thaliana populations. Nat. Genet. 43:956–963 [DOI] [PubMed] [Google Scholar]
- Coop G., Przeworski M., 2007. An evolutionary view of human recombination. Nat. Rev. Genet. 8: 23–34 [DOI] [PubMed] [Google Scholar]
- Drosophila Population Genomics Project, 2011 Drosophila Population Genomics Project. Available at www.dpgp.org Accessed April 2012.
- Durrett R., Schweinsberg J., 2004. Approximating selective sweeps. Theor. Popul. Biol. 66: 129–138 [DOI] [PubMed] [Google Scholar]
- Ewing G., Hermisson J., 2010. Msms: a coalescent simulation program including recombination, demographic structure and selection at a single locus. Bioinformatics 26: 2064–2065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyre-Walker A., 2006. The genomic rate of adaptive evolution. Trends Ecol. Evol. (Amst) 21: 569–575 [DOI] [PubMed] [Google Scholar]
- Fay J. C., Wu C. I., 2000. Hitchhiking under positive Darwinian selection. Genetics 155: 1405–1413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer W., Ganusov V. V., Giorgi E. E., Hraber P. T., Keele B. F., et al. , 2010. Transmission of single HIV-1 genomes and dynamics of early immune escape revealed by ultra-deep sequencing. PLoS ONE 5: e12303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillespie J. H., 1994. The Causes of Molecular Evolution. Oxford University Press, Oxford [Google Scholar]
- Haag-Liautard C., Dorris M., Maside X., Macaskill S., Halligan D. L., et al. , 2007. Direct estimation of per nucleotide and genomic deleterious mutation rates in Drosophila. Nature 445: 82–85 [DOI] [PubMed] [Google Scholar]
- Hedskog C., Mild M., Jernberg J., Sherwood E., Bratt G., et al. , 2010. Dynamics of HIV-1 quasispecies during antiviral treatment dissected using ultra-deep pyrosequencing. PLoS ONE 5: e11345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermisson J., Pennings P., 2005. Soft sweeps: molecular population genetics of adaptation from standing genetic variation. Genetics 169: 2335–2352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez R. D., Kelley J. L., Elyashiv E., Melton S. C., Auton A., et al. , 2011. Classic selective sweeps were rare in recent human evolution. Science 331: 920–924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson R. R., 2002. Generating samples under a Wright-Fisher neutral model of genetic variation. Bioinformatics 18: 337–338 [DOI] [PubMed] [Google Scholar]
- Hudson R. R., Kreitman M., Aguadé M., 1987. A test of neutral molecular evolution based on nucleotide data. Genetics 116: 153–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson R. R., Bailey K., Skarecky D., Kwiatowski J., Ayala F. J., 1994. Evidence for positive selection in the superoxide dismutase (sod) region of Drosophila melanogaster. Genetics 136: 1329–1340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan N. L., Hudson R. R., Langley C. H., 1989. The “hitchhiking effect” revisited. Genetics 123: 887–899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karasov T., Messer P. W., Petrov D. A., 2010. Evidence that adaptation in Drosophila is not limited by mutation at single sites. PLoS Genet. 6: 1000924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y., Stephan W., 2002. Detecting a local signature of genetic hitchhiking along a recombining chromosome. Genetics 160: 765–777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macpherson J. M., Sella G., Davis J. C., Petrov D. A., 2007. Genomewide spatial correspondence between nonsynonymous divergence and neutral polymorphism reveals extensive adaptation in Drosophila. Genetics 177: 2083–2099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansky L. M., Temin H. M., 1995. Lower in vivo mutation rate of human immunodeficiency virus type 1 than that predicted from the fidelity of purified reverse transcriptase. J. Virol. 69: 5087–5094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maynard Smith J., 1971. What use is sex? J. Theor. Biol. 30: 319–335 [DOI] [PubMed] [Google Scholar]
- Maynard Smith J., Haigh J., 1974. The hitch-hiking effect of a favourable gene. Genet. Res. 23: 23–35 [PubMed] [Google Scholar]
- Neher R. A., Leitner T., 2010. Recombination rate and selection strength in HIV intra-patient evolution. PLoS Comput. Biol. 6: e1000660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neher R. A., Shraiman B. I., 2011. Genetic draft and quasi-neutrality in large facultatively sexual populations. Genetics 188: 975–996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen R., Williamson S., Kim Y., Hubisz M. J., Clark A. G., et al. , 2005. Genomic scans for selective sweeps using SNP data. Genome Res. 15: 1566–1575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neil P. K., Sun G., Yu H., Ron Y., Dougherty J. P., et al. , 2002. Mutational analysis of HIV-1 long terminal repeats to explore the relative contribution of reverse transcriptase and RNA polymerase II to viral mutagenesis. J. Biol. Chem. 277: 38053–38061 [DOI] [PubMed] [Google Scholar]
- Ossowski S., Schneeberger K., Lucas-Lledó J. I., Warthmann N., Clark R. M., et al. , 2010. The rate and molecular spectrum of spontaneous mutations in Arabidopsis thaliana. Science 327: 92–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perelson A. S., Neumann A. U., Markowitz M., Leonard J. M., Ho D. D., 1996. HIV-1 dynamics in vivo: virion clearance rate, infected cell life-span, and viral generation time. Science 271: 1582–1586 [DOI] [PubMed] [Google Scholar]
- Pritchard J. K., Pickrell J. K., Coop G., 2010. The genetics of human adaptation: hard sweeps, soft sweeps, and polygenic adaptation. Curr. Biol. 20: R208–R215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Przeworski M., 2003. Estimating the time since the fixation of a beneficial allele. Genetics 164: 1667–1676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roach J., Glusman G., Smit A., Huff C., Hubley R., et al. , 2010. Analysis of genetic inheritance in a family quartet by whole-genome sequencing. Science 328: 636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabeti P. C., Reich D. E., Higgins J. M., Levine H. Z. P., Richter D. J., et al. , 2002. Detecting recent positive selection in the human genome from haplotype structure. Nature 419: 832–837 [DOI] [PubMed] [Google Scholar]
- Sabeti P. C., Fry B., Lohmueller J., Hostetter E., Cotsapas C., et al. , 2007. Genome-wide detection and characterization of positive selection in human populations. Nature 449: 913–918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sattath S., Elyashiv E., Kolodny O., Rinott Y., Sella G., 2011. Pervasive adaptive protein evolution apparent in diversity patterns around amino acid substitutions in Drosophila simulans. PLoS Genet. 7: e1001302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slatkin M., Hudson R. R., 1991. Pairwise comparisons of mitochondrial DNA sequences in stable and exponentially growing populations. Genetics 129: 555–562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima F., 1989. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123: 585–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsibris A. M. N., Korber B., Arnaout R., Russ C., Lo C.-C., et al. , 2009. Quantitative deep sequencing reveals dynamic HIV-1 escape and large population shifts during ccr5 antagonist therapy in vivo. PLoS ONE 4: e5683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voight B. F., Kudaravalli S., Wen X., Pritchard J. K., 2006. A map of recent positive selection in the human genome. PLoS Biol. 4: e72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiehe T. H., Stephan W., 1993. Analysis of a genetic hitchhiking model, and its application to DNA polymorphism data from Drosophila melanogaster. Mol. Biol. Evol. 10: 842–854 [DOI] [PubMed] [Google Scholar]
- Winckler W., Myers S. R., Richter D. J., Onofrio R. C., McDonald G. J., et al. , 2005. Comparison of fine-scale recombination rates in humans and chimpanzees. Science 308: 107–111 [DOI] [PubMed] [Google Scholar]
- Yule G. U., 1925. A mathematical theory of evolution, based on the conclusions of Dr. J. C. Willis, F.R.S. Philos. Trans. R. Soc. Lond. B Biol. Sci. 213: 21–87 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.