Abstract
Ubiquitin (Ub) and the ubiquitin-like proteins (Ubls) comprise a remarkable assortment of polypeptides that are covalently conjugated to target proteins (or other biomolecules) to modulate their intracellular localization, half-life and/or activity. Identification of Ub/Ubl conjugation sites on a protein of interest can thus be extremely important for understanding how it is regulated. While mass spectrometry (MS) has become a powerful tool for the study of many classes of post-translational modifications, the identification of Ub/Ubl conjugation sites presents a number of unique challenges. Here, we present an improved Ub/Ubl conjugation site identification strategy, utilizing SUMmOn analysis and an additional protease (LysC), as a complement to standard approaches. As compared to standard trypsin proteolysis-database search protocols alone, the addition of SUMmOn analysis can; (a) identify Ubl conjugation sites that are not detected by standard database searching methods, (b) better preserve Ub/Ubl conjugate identity, and (c) increase the number of identifications of Ub/Ubl modifications in lysine-rich protein regions. Using this methodology, we characterize for the first time a number of novel Ubl linkages and conjugation sites, including alternative yeast (K54) and mammalian SUMO chain (SUMO-2 K42, SUMO-3 K41) assemblies, as well as previously unreported NEDD8 chain (K27, K33 and K54) topologies.
Keywords: NEDD8, SUMO, SUMmOn, ubiquitin, ubiquitin-like proteins
1 Introduction
Ubiquitin (Ub) and the ubiquitin-like proteins (Ubls; Fig. 1A) are a family of polypeptides that are covalently conjugated to “target” proteins (or other biomolecules) to modulate their intracellular localization, half-life, and/or activity [1-4]. While the Ubls vary significantly in their degree of homology with Ub, and in the populations of molecules that they target, all Ubls appear to be conjugated to their targets in a manner similar to that of Ub, via an E1/E2/E3 activation, conjugation, ligation cascade [1-4]. Covalent conjugation of Ub/Ubls to proteins is an extremely versatile post-translational modification (PTM), implicated in a variety of critical cellular processes [1-5]. A vast number of proteins are modified by Ub and the Ubls [reviewed in 6], and deregulation of the Ub/Ubl systems have been implicated in a number of human diseases [1-5].
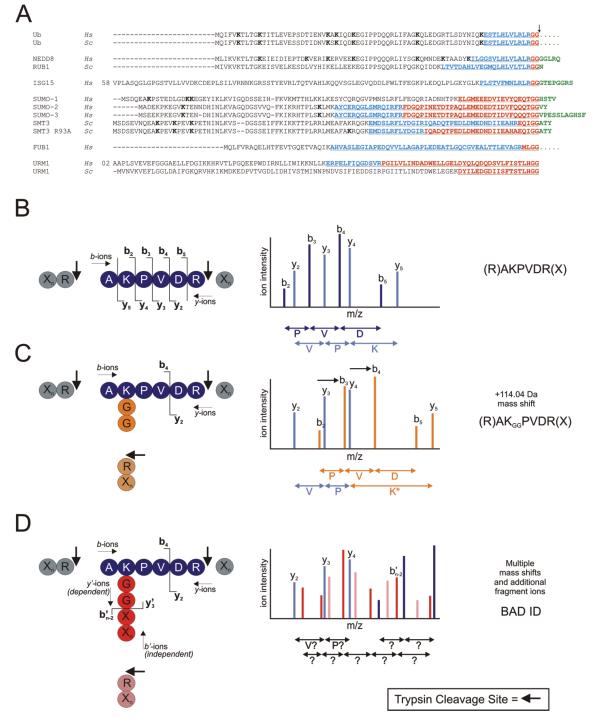
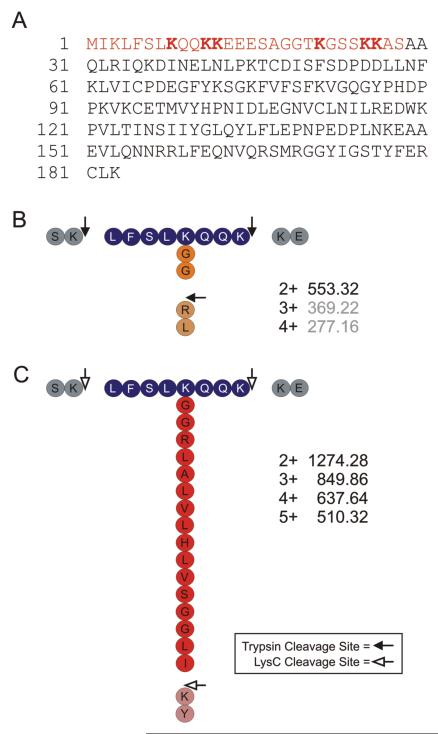
Figure 1.
(A) Amino acid sequence alignment of Ub and Ubls. Gene name and species (Homo sapiens (Hs); and Saccharomyces cerevisiae (Sc)) are indicated at left. C-terminal amino acids present in the pro-protein, but absent in the mature protein, are indicated in green. FUB1 and Ub are synthesized as fusion proteins (as indicated by ..…). The Ubl C-terminal “tail” that remains conjugated to a target molecule following trypsin (red text) or LysC (underlined text) digestion is highlighted. Additional residues resulting from LysC digestion are indicated in blue. Lysine residues implicated in Ub/Ubl chain formation (previously published, or as reported here) are indicated in bold. (B) A theoretical unmodified peptide and its corresponding (simplified) CID spectrum. Trypsin cleavage sites indicated by arrows. b- and y-ion series are indicated by dark and light blue peaks, respectively. (C) A theoretical ubiquitylated tryptic peptide, and its corresponding (simplified) CID spectrum. The entire b-ion series, as well as the y5 ion, undergo a +114.04 Da mass shift (shifted ions highlighted in orange). (D) A theoretical Ubl-modified peptide, and its corresponding (simplified) CID spectrum. b’- and y’-ion series denote the fragmentation ion series of the Ubl (in red and pink, respectively). The additional Ubl fragment ions create a highly complex CID spectrum which is uninterpretable by standard database search software. For example, the y’3 ion includes the entire target peptide (and is therefore considered a dependent ion), and would remain unassigned. The b’n-2 ion, derived only from the Ubl tail, is independent of the target peptide. The y2, y3, and y4 ions are unaffected, but would not be assigned by database search software.
Monoubiquitylation (i.e. conjugation of a single Ub polypeptide to a single target protein lysine residue) is important in processes such as endocytosis and plasma membrane receptor recycling [7]. However, Ub itself contains seven internal lysine residues, all of which can also be ubiquitylated to form poly-Ub multimers, or Ub “chains” [reviewed in 8]. The ability to form chains dramatically increases the functional diversity of Ub, in that multimers of varying lengths and linkage types are structurally distinct, interact with different subsets of binding proteins, and can therefore confer very different biological outcomes to a targeted substrate [1-5, 9]. The best characterized function of Ub chains (in this case, consisting of at least four Ub units linked via K48) is the targeting of a protein substrate for 26S proteasome-dependent degradation [10]. Other chain types (such as those linked via K11 or K29) may also be involved in degradation [11]. By contrast, K63-linked chains play roles in intracellular signaling, the DNA damage response, endocytosis and aggresome formation [12-15].
The SUMO (Small Ubiquitin-related MOdifier) proteins, a family of Ubls expressed in all eukaryotes, are also covalently conjugated to the epsilon amine group of lysine residues in a large number of target proteins [16-24]. A consensus sequence for SUMOylation, ΨKxE/D, where Ψ is a hydrophobic residue (usually I, L or V), K is the lysine to which SUMO is conjugated, and x is any amino acid, was first described for the yeast septin proteins [25]. However, as more SUMO conjugation sites have been identified, it has become clear that not every SUMO consensus site is SUMOylated, and that SUMO is often conjugated to target proteins at “non-consensus” sites [see Table 2 in 26]. Like Ub, the yeast and metazoan SUMO proteins can form multimeric chains. The major SUMO branching sites are located at K15 in the yeast SUMO protein (Smt3p); K7 and K17 in human SUMO-1; and at K11 in human SUMOs-2/3 [27-29]. The function of SUMO chains is unknown at present.
NEDD8 (Neural precursor cell Expressed Developmentally Down-regulated 8) is a 9.1 kDa Ubl displaying 58% sequence homology with Ub. The S. cerevisiae NEDD8 ortholog is encoded by the Related to Ubiquitin 1 (RUB1) gene. A number of proteins are NEDDylated [30-32], and NEDDylation appears to activate and/or stabilize multicomponent Ub E3 ligases [33]. NEDD8 was also reported to form chains in vitro and in vivo [31, 32], but the function of these structures is unknown.
Sequence database searching (as performed by programs such as SEQUEST [34], Mascot [35] or X!Tandem [36]) compares MS/MS fragment ion spectra against theoretical fragmentation patterns derived from a user-defined protein database, and assigns scores based on the quality of the match [e.g. 37, 38]. Most PTMs alter one or more peptide fragment ions by a predictable, indivisible mass, and do not undergo a significant degree of fragmentation during the CID process. The theoretical spectra used in the matching process may thus simply be recalculated to reflect an appropriate mass shift on one or more fragment ions.
For example, following trypsin cleavage, a Ub-conjugated peptide is modified at a lysine residue with a diglycine (-GG) remnant (Fig. 1A), which shifts the mass of the peptide fragments containing the modified lysine residue by +114 Da (Figs. 1B and 1C). Many ubiquitin conjugation sites have been successfully identified using this type of approach [e.g. 39]. Importantly, however, Ub is not the only PTM that yields a GG-modified target peptide following trypsin cleavage. NEDD8, Rub1p, and the interferon-stimulated gene 15 (ISG15) proteins also harbor C-terminal –(R)GG sequences (Fig. 1A). Following trypsin cleavage, these Ubl-derived -GG remnants are therefore indistinguishable from a Ub-derived -GG moiety, resulting in a loss of modifier identity.
Trypsin cleavage of many of the other Ubls (e.g. all eukaryotic SUMO proteins, as well as FUB1, URM1, HUB1 and FAT10) results in a much longer C-terminal peptide remnant or “tail” (Fig. 1A). Unlike most PTMs, Ubl tails produce multiple fragment ions during the CID process. If the tail is conjugated to a target peptide, fragment ions derived from the Ubl are combined with fragment ions from the target peptide, and a complex overlapping fragmentation pattern is generated (Fig. 1D). This type of CID spectrum is generally not correctly interpreted by standard search software, and is therefore not correctly identified. Due to the presence of an additional N-terminus and extra charged residues, Ubl-modified peptides are also often highly charged. Standard database search algorithms do not typically search for peptides with charge states higher than 3+ or 4+. Thus, very few Ubl conjugation sites have been unambiguously identified using standard mass spectrometric methods.
To improve our ability to identify Ubl conjugation sites using MS/MS, we developed the SUMmOn pattern recognition software tool [28]. SUMmOn utilizes a novel type of search strategy, scanning MS/MS spectra for the presence of a user-defined fragment ion series, corresponding to the Ubl of interest. We previously demonstrated the utility of SUMmOn by identifying several novel human SUMO-1 conjugation sites.
Here, we describe an integrated strategy for the identification of Ub and Ubl conjugation sites using SUMmOn, standard database search engines, and a combination of proteolytic enzymes. Using this methodology, we: (a) demonstrate that, unlike standard trypsin-database search protocols alone, the use of SUMmOn and alternative proteases can maintain the identity of the original Ub/Ubl conjugate, (b) we identify a number of putative NEDDylation sites in the UbcH12 protein, indicating that our protocol can also improve the identification of Ub/Ubl conjugation sites in lysine-rich protein regions, and finally (c) we identify several previously unreported alternative SUMO and NEDD8 chain topologies.
2 Materials and methods
2.1 LC/MS analysis
Lysyl endopeptidase C (LysC) was purchased from Wako Chemical (Osaka, Japan), and TPCK sequencing grade trypsin was purchased from Promega (Madison, WT). The HeLa S100 protein fraction was from Boston Biochem (Boston, MA). Analytical columns (75μm inner diameter) and pre-columns (100μm) for LC-MS analysis were made in-house from silica capillary tubing from InnovaQuartz (Phoenix, AZ), and packed in-house with 5μM 100Å C18 –coated silica particles (Magic, Michrom Bioresources, Auburn, CA).
Peptides were subjected to LC-ESI-MS/MS, using a 120 min RPLC (95% water-95% acetonitrile, 0.1% formic acid) buffer gradient running at 250nl-400nl/min on a Proxeon EASY-nLC pump in-line with a hybrid LTQ-Orbitrap mass spectrometer (Thermo Fisher Scientific, Waltham, MA). A parent ion scan was performed in the Orbitrap, using a resolving power of 60,000, then the four most intense peaks were selected for MS/MS (minimum ion count of 1000 for activation), using standard CID fragmentation. Fragment ions were detected in the LTQ. Dynamic exclusion was activated such that MS/MS of the same m/z (within a −0.1 and +2.1 Th window; exclusion list size = 500) detected 3 times within 45 seconds were excluded from analysis for 30 seconds.
For protein identification, Thermo .RAW files were converted to the .mzXML format using ReAdW software [40], then searched using X!Tandem [36] against the yeast ORF (Saccharomyces Genome Database, December 2005) or human (Ensembl 44.36F) databases, supplemented with a curated custom database containing the mature Ubl sequences. (Since Ub and most of the Ubls are translated as pro-proteins, then cleaved at a C-terminal diglycine motif to form the active or “mature” Ub/Ubl (e.g. see Fig. 1A), the mature protein sequences are not represented in most protein databases, and therefore must be added manually.) X!Tandem search parameters were: complete modifications, none; cysteine modifications, none; potential modifications, +114.04@K, +16@M and W, +32@M and W, +42@N-terminus, +1@N and Q; −17@N-term Q; −18@N-term E; parent mass error ±10ppm; fragment error 0.4 Da; maximum charge 4+; missed cleavage sites 3; semi-cleavage, no. For determination of conjugation site location, MS/MS data were subjected to SUMmOn analysis [28] using a digestion tolerance of 25 ppm, and a mass tolerance of 1000 ppm. SUMmOn data cutoffs were modification and target scores ≥0.8.
2.2 In vitro Ub/Ubl reactions
Wild type and K11R mutant SUMO-2/3 proteins and SUMO-2/3 chains were purchased from Boston Biochem. SUMO-2/3 and yeast SUMO chains were also generated in-house, as in [28]. In vitro NEDDylation reactions were conducted using 50 nM GST-NEDD8 E1, 0.4 μM GST-UbcH12, 10 μM NEDD8 (all from Boston Biochem), 0.45 μM Mdm2 as the E3 (a kind gift from C. Arrowsmith), 2 mM ATP, 5 mM MgCl2, 50 mM NaCl, and 50 mM Tris, pH 7.5. In vitro ubiquitylation reactions were conducted using 20 nM Uba1, 0.6 uM E2D2, and 0.45 uM Mdm2 (all gifts from C. Arrowsmith). All reactions were incubated at 37°C for two hours. Unconjugated Ub/Ubl proteins were removed from the reaction mix, and the reaction buffer exchanged for ammonium bicarbonate (ammbic), using a microcon spin unit with a nominal molecular weight cutoff of 30 kDa (Millipore, Billerica, MA). After two ammbic pH 8.3 washes (400μl), the retentant was resuspended in 250μl ammbic and subjected to protease cleavage with 1μg trypsin or LysC, overnight at 37°C.
3. Results
3.1 Alternative SUMO chain linkages
The mammalian SUMO-2 and SUMO-3 proteins were previously demonstrated to form SUMO-SUMO conjugates (SUMO chains) via K11 [27]. The yeast SUMO protein (Smt3p) forms chains primarily via K15 [29]. Earlier reports have suggested that mutation of these lysine residues dramatically decreases, but does not eliminate, SUMO chain formation [e.g. 27]. To determine whether, like Ub, the SUMO proteins can form additional, alternative chain linkages, we subjected the products of in vitro yeast and mammalian SUMO reactions to MS/SUMmOn analysis.
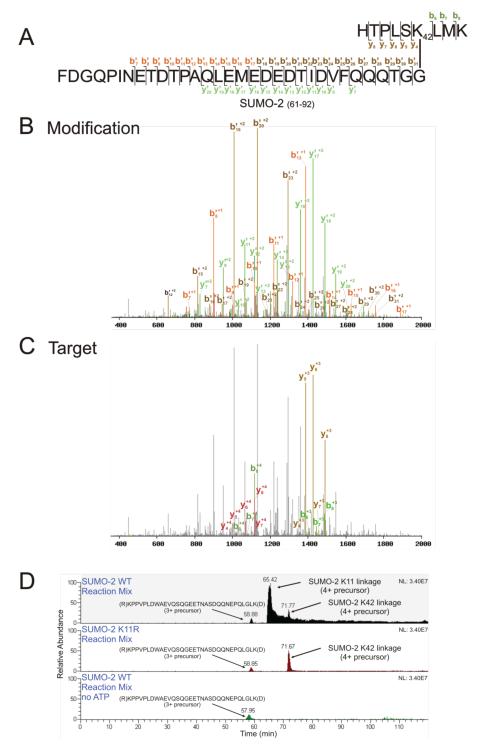
SUMO-2 in vitro autoSUMOylation reactions (as well as purchased, purified SUMO-2 chains; Boston Biochem) were treated with trypsin, and analyzed on an LTQ-Orbitrap. SUMmOn analysis of these data identified SUMO-2 chains formed primarily through K11, but also identified SUMO-2 chains linked via K42 (Figs 2A-C, Table I, and Supporting Information Fig. 1B and 2A. K42-linked SUMO-2 target peptide mass = 1054.61 Da, target peptide sequence (R)HTPLSKLMK(A), aa 37-45). The K42-linked SUMO-2 CIDs accounted for ~3% of all chain spectra detected by SUMmOn (modification score ≥0.8, target score ≥0.8; Table I). Assuming up to one missed trypsin cleavage, and variable modifications at methionine (oxidation) and glutamine (pyroglutamate), no other predicted tryptic peptides in the reaction mix correspond to the m/z of the K42-linked SUMO-2-SUMO-2 peptide assigned by SUMmOn, within a parent mass error of 100 ppm.
Figure 2.
K42-linked SUMO-2 is identified by SUMmOn. (A) Amino acid sequence of the trypsin-digested K42-linked SUMO-2-SUMO-2 peptide, indicating the ion fragments assigned by SUMmOn. (B) SUMmOn-annotated CID spectrum of the K42-linked SUMO-2-SUMO-2 peptide highlighting b’- and y’-ions derived from the modification (i.e. the C-terminal tryptic peptide of SUMO-2, aa 61-92). (C) The same spectrum, highlighting the b- and y-ions derived from the target peptide (aa 37-45 of SUMO-2). (D) Extracted ion chromatograms (EIC) of three different representative SUMO-2 in vitro reaction mixes. The m/z corresponding to the SUMO-2 K11 and K42 linkages elute at ~66 and ~72 min, respectively (top panel). The m/z corresponding to the SUMO-2 K11 linkage is lost when a K11R SUMO-2 mutant protein is used in the in vitro conjugation reaction, while the K42-linked peptide is unaffected (middle panel). The m/z corresponding to the K42 linkage is also lost in a reaction mix lacking ATP (bottom panel). Indicated are (i) the EIC of the SAE2 (SUMO E1) 3+ peptide (R)KPPVPLDWAEVQSQGEETNASDQQNEPQLGLK(D), eluting at ~ 59 min, using an m/z window of 1178.57-1178.59, (ii) an EIC of the 4+ (the most abundant charge state) SUMO-2 K11 linked peptide, using an m/z window of 1291.59-1291.61, and (iii) the EIC of the 4+ SUMO-2 K42 linked peptide (the most abundant charge state), using an m/z window of 1152.29-1152.31. For very large peptides, such as those characterized here, the monoisotopic variant represents only a very small percentage of the peptide population. The m/z windows in Figures 2, 4 and 7 therefore do not bracket the monoisotopic m/z for each linkage, but instead encompass the most abundant isotopic variants (i.e. those that were most often isolated for fragmentation).
Table 1.
Observed monoisotopic m/z values (mean from four MS scans) for the identified Ub/Ubl linkages
| Chain linkage | Peptide sequencea) | Target peptide (M+H)+ |
Observed conjugate m/z |
Observed spectrab) |
|---|---|---|---|---|
| SUMO-2 K11 (aa 8–21) | (K7)EGVKTENNDHINLK21(V) | 1610.81 | (3+) 1720.79 | 1336 |
| (4+) 1290.85 | ||||
| (5+) 1032.88 | ||||
| (6+) 860.90 | ||||
| SUMO-2 K42 (aa 37–45) | (R36)HTPLSKLMK45(A) | 1054.61 | (3+) 1535.39 | 36 |
| (4+) 1151.79 | ||||
| (5+) 921.64 | ||||
| SUMO-3 K11 (aa 8–20) | (K7)EGVKTENDHINLK20(V) | 1496.77 | (3+) 1682.77 | 356 |
| (4+) 1262.34 | ||||
| (5+) 1010.07 | ||||
| (6+) 841.89 | ||||
| SUMO-3 K41 (aa 36–44) | (R35)HTPLSKLMK44(A) | 1054.61 | (3+) 1535.38 | 23 |
| (4+) 1151.79 | ||||
| ScSUMO K54(aa 48–55) | (R47)LMEAFAKR55(Q) | 965.52 | (3+) 1305.28 | 27 |
| (4+) 979.21 | ||||
| NEDD8 K22 (aa 12–27) | (K11)EIEIDIEPTDKVERIK27(E) | 1927.04 | (4+) 871.50 | 11 |
| (5+) 697.40 | ||||
| NEDD8 K27 (aa 23–33) | (K22)VERIKERVEEK33(E) | 1414.8 | (4+) 743.44 | 31 |
| (5+) 594.95 | ||||
| NEDD8 K33 (aa 28–48) | (K27)ERVEEKEGIPPQQQRLIYSGK48(Q) | 2484.32 | (4+) 1010.82 | 12 |
| (6+) 674.22 | ||||
| NEDD8 K48 (aa 34–54) | (K33)EGIPPQQQRLIYSGKQMNDEK54(T) | 2459.24 | (4+) 1004.55 | 151 |
| (5+) 803.85 | ||||
| (6+) 670.04 | ||||
| NEDD8 K54 (aa 49–60) | (K48)Q(−17.0265)MNDEKTAADYK60(I) | 1396.61 | (3+) 984.86 | 140 |
| (4+) 738.90 |
As expected, SUMmOn did not detect K11 SUMO-2 linkages in an MS analysis of in vitro reactions containing a SUMO-2 K11R mutant protein (which cannot form K11-linked chains). However, the K11R mutation did not affect the m/z corresponding to the K42-linked SUMO-2 peptide assigned by SUMmOn (elution time ~71 min; Fig. 2D, compare middle and top panels). While unmodified peptides derived from components of the reaction mix (e.g. the E1 SAE2 3+ peptide R.KPPVPLDWAEVQSQGEETNASDQQNEPQLGLK.D) were unchanged, SUMmOn did not detect any SUMO linkages in in vitro SUMO-2 reactions lacking ATP (precluding the formation of SUMO conjugates; Fig. 2D, bottom panel) or in in vitro reactions containing a SUMO-2 K11,42R mutant (Supporting Information Fig. 2A). We thus identified a novel, non-consensus SUMO-2 chain linkage.
SUMmOn analysis of in vitro reactions containing recombinant wild type SUMO-3 protein identified SUMO chains formed through both K11 and K41 (Supporting Information Fig. 2B and 3, SUMO-3 K41-linked target peptide mass = 1054.61 Da, target peptide sequence (R)HTPLSKLMK(A), aa 36-44). The SUMO-3 K41 linkage spectra represented ~6% of all chain product spectra assigned by SUMmOn in in vitro SUMOylation reactions (Table I). In reactions containing a SUMO-3 K11R mutant protein, the m/z assigned by SUMmOn as the K11-linked SUMO-3 peptide was not detected. This mutation, however, had no effect on the m/z assigned by SUMmOn as the K41-linked SUMO-3 peptide (Supporting Information Fig. 2B). An in vitro reaction lacking only ATP also lacked the m/z assigned to the K11- and K41-linked SUMO-3 chains (data not shown) as did an in vitro reaction with a SUMO-3 K11,41R mutant (Supporting Information Fig. 2B). The human SUMO-3 protein can thus also form at least two types of SUMO chain linkages in vitro. As previously observed [28], the database search programs X!Tandem and SEQUEST were unable to identify any SUMO-2 or SUMO-3 linkage types in any of the reactions, presumably due to the long C-terminal tryptic tails.
The C-terminal-most tryptic cleavage site in the 98 aa S. cerevisiae SUMO (Smt3p) protein is at R93 (Fig. 1A). Cleavage of yeast SUMO with trypsin thus yields a 5 aa “tail”. SUMmOn can successfully detect this 5aa remnant on covalently modified target proteins [28]. However, a yeast SUMO R93A mutant (with the C-terminal tryptic cleavage site now at R71, tryptic cleavage of this protein results in a 27aa tail; Fig. 1A) is much more efficiently identified by SUMmOn, due to the presence of many more SUMO tail fragment ions. We did not observe any difference in the ability of this point mutant to be conjugated to substrates: e.g. Sc_SUMOR93A was conjugated in vitro to a C-terminal RanGAP1 protein fragment (aa 420-589) as efficiently as the wild type protein (Supporting Information Fig. 4A).
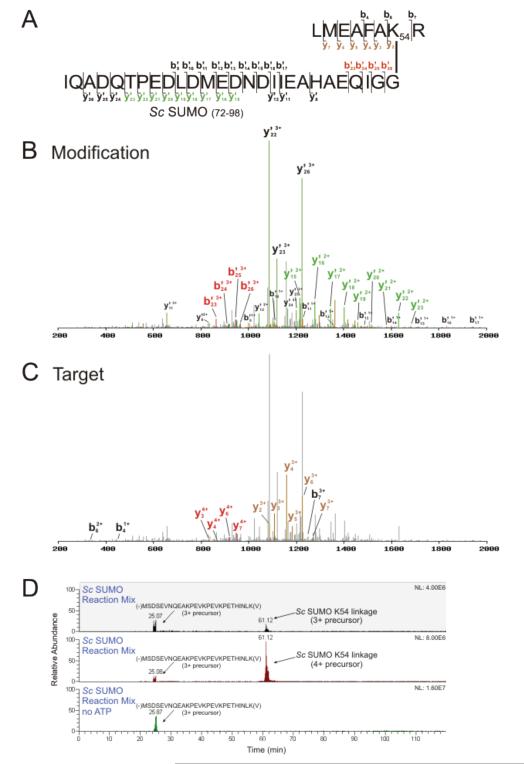
The Sc_SUMOR93A protein was used to determine if, like the mammalian SUMOs, the budding yeast SUMO protein can form alternative chain topologies. SUMmOn analysis of an Sc_SUMOR93A-containing in vitro autoSUMOylation reaction identified a SUMO-SUMO linkage at K54 (Fig. 3A-C, Supporting Information Fig. 4B-C; target peptide mass = 965.52 Da, target peptide sequence (R)LMEAFAKR(Q), aa 48-55). No other predicted tryptic peptide in the reaction mix corresponds to this mass and charge state within 100 ppm. In an identical reaction mix lacking only ATP, the m/z assigned by SUMmOn as the K54-linked peptide was not detected (Fig. 3D).
Figure 3.
A yeast SUMO K54-linkage is identified using SUMmOn. (A) Amino acid sequence of the K54-linked SUMO peptide, and the fragments assigned by SUMmOn. (B) SUMmOn-annotated CID spectrum of a yeast SUMO conjugate highlighting b’- and y’-ions derived from the C-terminal SUMO tryptic peptide, aa 72-98. (C) the same spectrum, highlighting the b- and y-ions derived from the target peptide (yeast SUMO aa 48-55). (D) EIC of representative in vitro yeast SUMO conjugation reactions with and without ATP. The m/z windows correspond to the 3+ (m/z = 1305.94-1305.96, elution at ~61 min) and 4+ (m/z=974.71-974.73) precursor ions of the SUMO K54-linked peptide. An unmodified yeast SUMO 3+ peptide MSDSEVNQEAKPEVKPEVKPETHINLK(V) (m/z window 1026.52-1026.54, eluting at ~25 min) was detected in both reactions.
Bylebyl et al. [41] demonstrated that yeast strains lacking one of the two SUMO proteases (Ulp2p) accumulate high molecular weight SUMO conjugates (presumably SUMO chains), and display hypersensitivity to hydroxyurea (HU). HU hypersensitivity is alleviated by replacing the endogenous SUMO (SMT3) gene with a sequence coding for a SUMO protein that is unable to form chains (all lysine codons mutated to arginine codons). Replacing the endogenous gene with a SUMO coding sequence lacking the primary SUMO-SUMO conjugation site (as well as two nearby lysine residues; K11,15,19R) partially alleviated HU sensitivity. Interestingly, a SUMO K54,58R mutant was also able to partially alleviate HU sensitivity. Consistent with our in vitro data, these in vivo data suggest that multiple types of SUMO chains may exist in yeast, and that the K54 linkage may play an important (but unknown) physiological role.
3.2 NEDD8 chain linkages
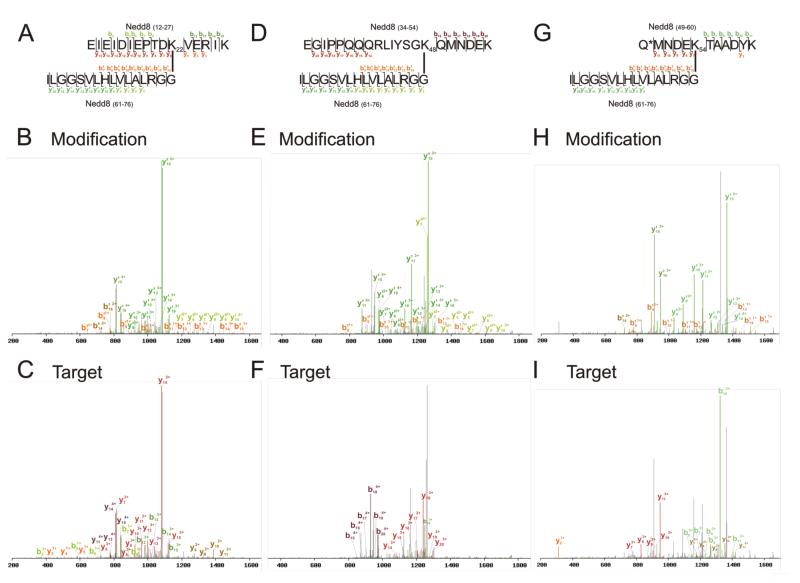
To determine whether SUMmOn could identify other types of Ubl conjugation sites, we also generated in vitro autoNEDDylation reactions (using the NEDD8 E1, APPBP1/Uba3, the E2 UbcH12/UBE2M and Mdm2 as the E3). Half of the sample was subjected to trypsin cleavage, and the other half proteolyzed with LysC. The resulting peptides were analyzed by both SUMmOn and X!Tandem. Previous reports have indicated that in vitro NEDDylation reactions produce K48- and K22-linked NEDD8 chains [31, 32]. SUMmOn also identified both K22- and K48-linked NEDD8 chains in the LysC-treated reaction mix (Fig. 4A-F, Supporting Information Fig. 5A and B). The K48 linkage (target peptide mass = 2459.24 Da, target peptide sequence (K)EGIPPQQQRLIYSGKQMNDEK(T), aa 34-54) accounted for ~44% of observed NEDD8 chain linkage spectra, while K22 linkages (target peptide mass = 1927.04 Da, target peptide sequence (K)EIEIDIEPTDKVERIK(E), aa 12-27) accounted for ~3% of chain spectra assigned by SUMmOn. SUMmOn also identified a previously unreported NEDD8 chain linkage at K54 (Fig. 4G-I and Supporting Information Fig. 5C, target peptide mass = 1396.61 Da, target peptide sequence (K)Q*MNDEKTAADYK(I), *pyroglutamate, aa 49-60), representing ~41% of the NEDD8 chain spectra detected in the analysis. The m/z assigned by SUMmOn as the K54-linked NEDD8 peptide could not be assigned to any other predicted peptide in the reaction mix, within 100ppm. An identical reaction mix lacking ATP also lacked the m/z corresponding to the NEDD8 K54 linkage, and SUMmOn did not assign any NEDD8 conjugates in this reaction (data not shown). X!Tandem was unable to identify any NEDD8 linkages in the LysC-digested samples (data not shown), but successfully identified the GG-moiety at NEDD8 K48 and K54 in the reaction mix digested with trypsin (Supporting Information Fig. 6). Thus, using a combination of SUMmOn and standard database searching, we were able to unambiguously identify two known linkage sites, as well as an abundant novel NEDD8 chain linkage site.
Figure 4.
NEDD8 K22, K48 and K54 linkages identified by SUMmOn. (A) Amino acid sequence of the LysC-digested K22-linked NEDD8-NEDD8 peptide, indicating fragments identified using SUMmOn. (B) SUMmOn-annotated CID spectrum of the LysC-digested K22-linked NEDD8-NEDD8 peptide highlighting the b’- and y’-ions derived from the C-terminal LysC fragment of NEDD8, aa 61-76. (C) Identical CID spectrum, highlighting the b- and y-ions derived from the target peptide (NEDD8 aa 12-27). (D-F) As described for A-C, but for the K48-linked NEDD8 peptide. (G-I) As described for A-C, but for the K54-linked NEDD8 peptide. Asterisk (*) denotes pyroglutamate.
Two less abundant, previously unreported NEDD8 linkages were also detected by SUMmOn, at K27 (Supporting Information Fig. 5D; target peptide mass = 1414.80 Da, target peptide sequence (K)VERIKERFEEK(E), aa 23-33) representing ~9% of the detected chain products, and K33 (Supporting Information Fig. 5E; target peptide mass = 2484.32 Da, target peptide sequence (K)ERFEEKEGIPPQQQRLIYSGK(Q), aa 28-48) representing ~3% of the chain spectra. X!Tandem did not identify these linkages. These data indicate that, like Ub, NEDD8 is capable of forming chains on many different lysine residues in vitro.
3.3 Improved identification of Ubl conjugation sites in lysine-rich protein regions
In the same set of in vitro NEDDylation reactions, SUMmOn also identified several NEDD8 modification sites on the E2 enzyme UbcH12/UBE2M. This set of NEDD8 conjugation sites all occur within a lysine-rich ~28aa N-terminal extension of UbcH12, at K8, K11, K12, K21, K25 and K26 (Fig. 5A, Supporting Information Fig. 7A-F and Table I). None of these conjugation sites have been previously reported, and none were detected in trypsin digests of the same in vitro reactions, as analyzed by X!Tandem, presumably due to the small size of the GG-modified tryptic peptides (Fig. 5B, Supporting Information Table I), which largely precludes their detection in the standard 400-1800 m/z window used in typical ion trap instrument MS parent ion scans. The longer LysC-generated NEDD8 tail results in much larger peptides (Fig. 5C), allowing them to be trapped and analyzed by the mass spectrometer. Thus, another advantage of using LysC digestion and SUMmOn analysis is that it can allow for the identification of Ub/Ubl conjugation sites on small target peptides in lysine rich regions that may otherwise go undetected.
Figure 5.
(A) Amino acid sequence of UbcH12/UBE2M. The lysine-rich 28 aa N-terminal extension is highlighted in red, and NEDD8-modified lysine residues identified in this study are in bold. (B and C) Schematic diagram of a NEDD8-modified UbcH12/UBE2M peptide (aa 4-11; K8 modified by NEDD8), cleaved with (B) trypsin (filled arrowhead) or (C) LysC (white arrowhead). Predicted monoisotopic m/z for each peptide at several different charge states are indicated. Grey m/z values fall below those used in typical ion trap mass spectrometer parent ion scans.
3.4 Maintenance of Ub/Ubl identity
Trypsin is the enzyme of choice for most MS-based proteomics analyses. Following trypsin cleavage, Ub-conjugated peptides are modified at a lysine residue with a -GG (diglycine) moiety. Standard database search tools can efficiently identify peptides bearing this modification. However, several of the Ub-like proteins (e.g. human NEDD8 and ISG15, and S. cerevisiae Rub1p) also possess C-terminal –(R)GG amino acid sequences (Fig. 1A, Fig. 5B). Following trypsin proteolysis, these Ubl-derived -GG remnants are therefore indistinguishable from a Ub-derived -GG modification, resulting in a loss of modifier identity. Treatment of the same Ub/Ubl conjugates with LysC yields target peptides conjugated to longer, and therefore more informative, Ub/Ubl tails (Fig. 1A, Fig. 5C). LysC cleavage of Ub results in a 13aa C-terminal peptide, while LysC cleavage of NEDD8, Rub1p or ISG15 yields distinct 16 aa, 23 aa and 14 aa C-terminal peptides, respectively (Fig. 1A). Standard database search tools are generally unable to identify target peptides conjugated to these longer Ubl peptide fragments [21, 28].
We next tested whether SUMmOn could distinguish between peptides modified by Ub, NEDD8 or SUMO-2/3 in the same sample. Ub and NEDD8 in vitro reactions were digested with LysC, and SUMO-2 chains were digested with trypsin. A mixture with an equal ratio of Ub, NEDD8 and SUMO-2 chains was prepared, based on base peak intensities of the m/z corresponding to the predominant linkage (Ub K48, NEDD8 K48 and SUMO-2 K11) in preliminary MS runs. This mixture was subjected to MS, and Ub K48-linked chains (Supporting Information Fig. 8A; target peptide mass 3429.78 Da, target peptide sequence (K)EGIPPDQQRLIFAGKQLEDGRTLSDYNIQK(E), aa 34-63), NEDD8 K48- and K54-linked chains, and SUMO-2 K11-linkages were all identified by SUMmOn. SUMmOn was also able to differentiate between co-eluting Ub/Ubl conjugates: the Ub K48-linked peptide and the SUMO-2 K11-linked peptide both elute at ~65 min (Supporting Information Figs. 8B and C), and both linkage types were successfully identified. Thus, SUMmOn is able to identify multiple Ub/Ubl linkages in a more complex sample, and unlike standard trypsin-database searching approaches, the use of longer, more informative LysC-generated Ub/Ubl tails allows for unambiguous identification of the PTM.
3.5 Enrichment for Ub/Ubl conjugate CIDs using charge state exclusion
Only a fraction of the total number of peptides may be sequenced in highly complex mixtures, using typical shotgun proteomics approaches. Any strategy to enrich for specific subsets of peptides may thus be useful. Our data indicated that a majority of the Ub/Ubl-linked peptides generated by LysC cleavage are highly charged (all are ≥3+, most are ≥4+). We hypothesized that this property could be used to select for fragmentation of Ub/Ubl conjugates, using charge state exclusion on the Orbitrap. To test this approach, 25ng of SUMO-2 chains (corresponding to ~2 pmol of the K11-linked SUMO-2 peptide) was spiked into 3ug of a HeLa S100 protein fraction. This mix was digested with trypsin and subjected to MS analysis using standard settings (+1 charge state exclusion only), or 1+, 2+ and 3+ charge state exclusion. The base peak intensity of the 4+ SUMO-2 K11-linked peptide was low (4E5; 4+ was the most abundant charge state) in comparison to the average base peak intensity of the lysate fraction (7E6; Supporting Information Fig. 9). In standard MS runs (excluding only 1+ ions), this peptide was not efficiently selected for CID. However, when charge-state exclusion (1+, 2+ and 3+) was activated, this peptide was selected for CID fragmentation and successfully identified by SUMmOn (Supporting Information Fig. 10). Thus, SUMmOn is able to identify Ub/Ubl conjugates in more complex samples, and excluding peptides of lower charge from analysis can be used to increase the chances that low abundance Ub/Ubl conjugated peptides will be selected for CID fragmentation.
3.6 HCD for improved fragmentation of some Ub/Ubl conjugates
The LysC-derived K48-linked Ub peptide is highly charged (4+ to 7+; Table I) under standard RPLC buffer conditions. While SUMmOn readily identified the 4+ and 5+ K48-linked Ub ions, standard CID fragmentation settings resulted in poor fragmentation of the 6+ (Supporting Information Fig. 11A-C) and 7+ (data not shown) precursors. The y275+ and y276+ ions account for a majority of the total ion current in these spectra, representing fragmentation at the peptide bond between I36 and P37. Increasing the CID normalized collision energy or activation Q did not result in any additional informative fragmentation events (data not shown). In an attempt to improve the fragmentation of this peptide at higher charge states, in vitro Ub reactions digested with LysC were subjected to Higher-energy C-trap Dissociation (HCD), an alternative fragmentation technique in which the C-trap in the Orbitrap is used as a pseudo-collision cell [42]. HCD was tested using normalized collision energies of 40%, 45% and 50% (data not shown). We found that a normalized collision energy of 45% resulted in a more informative fragmentation of the LysC-derived Ub K48-linked peptide at higher charge states (Supporting Information Fig. 11D-F), and SUMmOn readily identified this linkage site using HCD data. Thus, HCD and SUMmOn analysis can also be used to identify Ubl-linked peptides that do not fragment well under standard CID conditions.
4. Concluding remarks
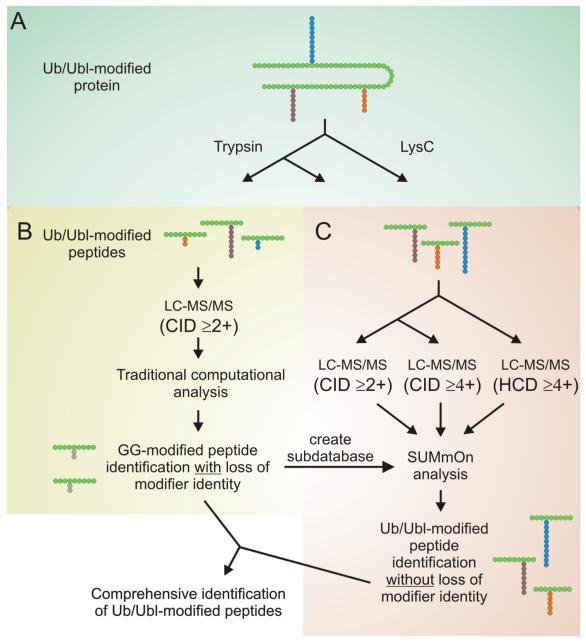
Taking the above observations into account, we propose an improved methodology for the identification of Ub and Ubl conjugates (Fig. 6). A sample containing Ub/Ubl-modified peptides may be digested in two separate reactions using trypsin and LysC. Standard database searching can be used to identify putative GG-modified and unmodified peptides in both reactions, and SUMmOn can be used to identify Ub/Ubl conjugates bearing longer C-terminal tails. The output from both analyses can be combined and used to unambiguously identify Ub/Ubl conjugates and conjugation sites.
Figure 6.
Improved workflow for the identification of Ub/Ubl-conjugation sites. (A) The protein sample of interest is split in two, and digested with trypsin or LysC prior to LC-MS/MS analysis. (B) Standard database searching is used to analyze a portion of the trypsin-digested sample, and to create a subdatabase for SUMmOn. (C) The remainder of the trypsin-digested sample, as well as the LysC-digested sample, are analyzed using SUMmOn. During the initial LC-MS/MS run, CIDs are collected on precursor ions with a charge of 2+ or greater. An additional LC-MS/MS run can be performed in which CIDs are collected on precursor ions with a charge of 4+ or greater to enrich for Ub/Ubl-modified peptides. HCD may also be used to obtain improved fragmentation of highly charged precursor ions. Combining the results of these multiple analyses can generate a comprehensive list of unambiguous Ub/Ubl modification sites.
We have also added spectra from the novel SUMO and NEDD8 chain topologies identified here to our Ub/Ubl spectral library [43], to further facilitate rapid identification of Ub, SUMO and NEDD8 chains in biological samples.
Supplementary Material
Acknowledgements
We thank C. Arrowsmith, Y. Sheng and S. DhePaganon for the generous gifts of recombinant E2 and E3 proteins, and A.-C. Gingras and members of the Raught laboratory for critical reading of the manuscript. SJ was funded by a Canadian Institutes of Health Research (CIHR) CGS Master’s award, and TS was funded by a CIHR CGS Doctoral Award. BR is the Canada Research Chair in Proteomics and Molecular Medicine. This work was supported by funding to BR from CIHR Grant MOP-81268, as well as the Canada Foundation for Innovation, the Ontario Innovation Trust and the McLaughlin Centre for Molecular Medicine.
ABBREVIATIONS
- Ub
ubiquitin
- Ubl
ubiquitin-like protein
- SUMO
small ubiquitin-related modifier
- Smt3p
suppressor of mif two 3 protein
- NEDD8
neural precursor cell expressed developmentally down-regulated protein 8
- LTQ
linear quadrupole ion trap
- HCD
higher-energy C-trap dissociation
- Rub1p
related to ubiquitin 1 protein
- ISG15
interferon-stimulated gene 15 kDa protein
- ammbic
ammonium bicarbonate
5 References
- [1].Schwartz DC, Hochstrasser M. A superfamily of protein tags: ubiquitin, SUMO and related modifiers. Trends Biochem Sci. 2003;28:321–328. doi: 10.1016/S0968-0004(03)00113-0. [DOI] [PubMed] [Google Scholar]
- [2].Hochstrasser M. Origin and function of ubiquitin-like proteins. Nature. 2009;458:422–429. doi: 10.1038/nature07958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Kerscher O, Felberbaum R, Hochstrasser M. Modification of proteins by ubiquitin and ubiquitin-like proteins. Annu Rev Cell Dev Biol. 2006;22:159–180. doi: 10.1146/annurev.cellbio.22.010605.093503. [DOI] [PubMed] [Google Scholar]
- [4].Ciechanover A, Iwai K. The ubiquitin system: from basic mechanisms to the patient bed. IUBMB Life. 2004;56:193–201. doi: 10.1080/1521654042000223616. [DOI] [PubMed] [Google Scholar]
- [5].Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- [6].Kirkpatrick DS, Denison C, Gygi SP. Weighing in on ubiquitin: the expanding role of mass-spectrometry-based proteomics. Nat Cell Biol. 2005;7:750–757. doi: 10.1038/ncb0805-750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Mosesson Y, Yarden Y. Monoubiquitylation: a recurrent theme in membrane protein transport. Isr Med Assoc J. 2006;8:233–237. [PubMed] [Google Scholar]
- [8].Ikeda F, Dikic I. Atypical ubiquitin chains: new molecular signals. ’Protein Modifications: Beyond the Usual Suspects’ review series. EMBO Rep. 2008;9:536–542. doi: 10.1038/embor.2008.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Komander D, Reyes-Turcu F, Licchesi JD, Odenwaelder P, et al. Molecular discrimination of structurally equivalent Lys 63-linked and linear polyubiquitin chains. EMBO Rep. 2009;10:466–473. doi: 10.1038/embor.2009.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Thrower JS, Hoffman L, Rechsteiner M, Pickart CM. Recognition of the polyubiquitin proteolytic signal. Embo J. 2000;19:94–102. doi: 10.1093/emboj/19.1.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Xu P, Duong DM, Seyfried NT, Cheng D, et al. Quantitative proteomics reveals the function of unconventional ubiquitin chains in proteasomal degradation. Cell. 2009;137:133–145. doi: 10.1016/j.cell.2009.01.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Olzmann JA, Li L, Chudaev MV, Chen J, et al. Parkin-mediated K63-linked polyubiquitination targets misfolded DJ-1 to aggresomes bia binding to HDAC6. J Cell Biol. 2007;178:1025–1038. doi: 10.1083/jcb.200611128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Hawryluk MJ, Keyel PA, Mishra SK, Watkins SC, et al. Epsin 1 is a polyubiquitin-selective clathrin-associated sorting protein. Traffic. 2006;7:262–281. doi: 10.1111/j.1600-0854.2006.00383.x. [DOI] [PubMed] [Google Scholar]
- [14].Sobhian B, Shao G, Lilli DR, Culhane AC, et al. RAP80 targets BRCA1 to specific ubiquitin structures at DNA damage sites. Science. 2007;316:1198–1202. doi: 10.1126/science.1139516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Varadan R, Assfalg M, Haririnia A, Raasi S, et al. Solution conformation of Lys63-linked di-ubiquitin chain provides clues to functional diversity of polyubiquitin signaling. J Biol Chem. 2004;279:7055–7063. doi: 10.1074/jbc.M309184200. [DOI] [PubMed] [Google Scholar]
- [16].Denison C, Rudner AD, Gerber SA, Bakalarski CE, et al. A proteomic strategy for gaining insights into protein sumoylation in yeast. Mol Cell Proteomics. 2005;4:246–254. doi: 10.1074/mcp.M400154-MCP200. [DOI] [PubMed] [Google Scholar]
- [17].Vertegaal AC, Andersen JS, Ogg SC, Hay RT, et al. Distinct and overlapping sets of SUMO-1 and SUMO-2 target proteins revealed by quantitative proteomics. Mol Cell Proteomics. 2006;5:2298–2310. doi: 10.1074/mcp.M600212-MCP200. [DOI] [PubMed] [Google Scholar]
- [18].Vertegaal AC, Ogg SC, Jaffray E, Rodriguez MS, et al. A proteomic study of SUMO-2 target proteins. J Biol Chem. 2004;279:33791–33798. doi: 10.1074/jbc.M404201200. [DOI] [PubMed] [Google Scholar]
- [19].Zhou W, Ryan JJ, Zhou H. Global analyses of sumoylated proteins in Saccharomyces cerevisiae. Induction of protein sumoylation by cellular stresses. J Biol Chem. 2004;279:32262–32268. doi: 10.1074/jbc.M404173200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Wykoff DD, O’Shea EK. Identification of sumoylated proteins by systematic immunoprecipitation of the budding yeast proteome. Mol Cell Proteomics. 2005;4:73–83. doi: 10.1074/mcp.M400166-MCP200. [DOI] [PubMed] [Google Scholar]
- [21].Wohlschlegel JA, Johnson ES, Reed SI, Yates JR., 3rd Global analysis of protein sumoylation in Saccharomyces cerevisiae. J Biol Chem. 2004;279:45662–45668. doi: 10.1074/jbc.M409203200. [DOI] [PubMed] [Google Scholar]
- [22].Panse VG, Hardeland U, Werner T, Kuster B, Hurt E. A proteome-wide approach identifies sumoylated substrate proteins in yeast. J Biol Chem. 2004;279:41346–41351. doi: 10.1074/jbc.M407950200. [DOI] [PubMed] [Google Scholar]
- [23].Hannich JT, Lewis A, Kroetz MB, Li SJ, et al. Defining the SUMO-modified proteome by multiple approaches in Saccharomyces cerevisiae. J Biol Chem. 2005;280:4102–4110. doi: 10.1074/jbc.M413209200. [DOI] [PubMed] [Google Scholar]
- [24].Golebiowski F, Matic I, Tatham MH, Cole C, et al. System-wide changes to SUMO modifications in response to heat shock. Sci Signal. 2009;2:ra24. doi: 10.1126/scisignal.2000282. [DOI] [PubMed] [Google Scholar]
- [25].Johnson ES, Blobel G. Cell cycle-regulated attachment of the ubiquitin-related protein SUMO to the yeast septins. J Cell Biol. 1999;147:981–994. doi: 10.1083/jcb.147.5.981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Jeram SM, Srikumar T, Pedrioli PG, Raught B. Using mass spectrometry to identify ubiquitin and ubiquitin-like protein conjugation sites. Proteomics. 2009;9:922–934. doi: 10.1002/pmic.200800666. [DOI] [PubMed] [Google Scholar]
- [27].Tatham MH, Jaffray E, Vaughan OA, Desterro JM, et al. Polymeric chains of SUMO-2 and SUMO-3 are conjugated to protein substrates by SAE1/SAE2 and Ubc9. J Biol Chem. 2001;276:35368–35374. doi: 10.1074/jbc.M104214200. [DOI] [PubMed] [Google Scholar]
- [28].Pedrioli PG, Raught B, Zhang XD, Rogers R, et al. Automated identification of SUMOylation sites using mass spectrometry and SUMmOn pattern recognition software. Nat Methods. 2006;3:533–539. doi: 10.1038/nmeth891. [DOI] [PubMed] [Google Scholar]
- [29].Bencsath KP, Podgorski MS, Pagala VR, Slaughter CA, Schulman BA. Identification of a multifunctional binding site on Ubc9p required for Smt3p conjugation. J Biol Chem. 2002;277:47938–47945. doi: 10.1074/jbc.M207442200. [DOI] [PubMed] [Google Scholar]
- [30].Xirodimas DP, Saville MK, Bourdon JC, Hay RT, Lane DP. Mdm2-mediated NEDD8 conjugation of p53 inhibits its transcriptional activity. Cell. 2004;118:83–97. doi: 10.1016/j.cell.2004.06.016. [DOI] [PubMed] [Google Scholar]
- [31].Xirodimas DP. Novel substrates and functions for the ubiquitin-like molecule NEDD8. Biochem Soc Trans. 2008;36:802–806. doi: 10.1042/BST0360802. [DOI] [PubMed] [Google Scholar]
- [32].Jones J, Wu K, Yang Y, Guerrero C, et al. A targeted proteomic analysis of the ubiquitin-like modifier nedd8 and associated proteins. J Proteome Res. 2008;7:1274–1287. doi: 10.1021/pr700749v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Pan ZQ, Kentsis A, Dias DC, Yamoah K, Wu K. Nedd8 on cullin: building an expressway to protein destruction. Oncogene. 2004;23:1985–1997. doi: 10.1038/sj.onc.1207414. [DOI] [PubMed] [Google Scholar]
- [34].Eng JK, McCormack AL, Yates JR., 3rd An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J Am Soc Mass Spectrom. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- [35].Perkins DN, Pappin DJ, Creasy DM, Cottrell JS. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis. 1999;20:3551–3567. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- [36].Craig R, Beavis RC. TANDEM: matching proteins with tandem mass spectra. Bioinformatics. 2004;20:1466–1467. doi: 10.1093/bioinformatics/bth092. [DOI] [PubMed] [Google Scholar]
- [37].Aebersold R, Mann M. Mass spectrometry-based proteomics. Nature. 2003;422:198–207. doi: 10.1038/nature01511. [DOI] [PubMed] [Google Scholar]
- [38].Sadygov RG, Cociorva D, Yates JR., 3rd Large-scale database searching using tandem mass spectra: looking up the answer in the back of the book. Nat Methods. 2004;1:195–202. doi: 10.1038/nmeth725. [DOI] [PubMed] [Google Scholar]
- [39].Peng J, Schwartz D, Elias JE, Thoreen CC, et al. A proteomics approach to understanding protein ubiquitination. Nat Biotechnol. 2003;21:921–926. doi: 10.1038/nbt849. [DOI] [PubMed] [Google Scholar]
- [40].Pedrioli PG, Eng JK, Hubley R, Vogelzang M, et al. A common open representation of mass spectrometry data and its application to proteomics research. Nat Biotechnol. 2004;22:1459–1466. doi: 10.1038/nbt1031. [DOI] [PubMed] [Google Scholar]
- [41].Bylebyl GR, Belichenko I, Johnson ES. The SUMO isopeptidase Ulp2 prevents accumulation of SUMO chains in yeast. J Biol Chem. 2003;278:44113–44120. doi: 10.1074/jbc.M308357200. [DOI] [PubMed] [Google Scholar]
- [42].Olsen JV, Macek B, Lange O, Makarov A, et al. Higher-energy C-trap dissociation for peptide modification analysis. Nat Methods. 2007;4:709–712. doi: 10.1038/nmeth1060. [DOI] [PubMed] [Google Scholar]
- [43].Srikumar T, Jeram SM, Lam H, Raught B. A ubiquitin and ubiquitin-like protein spectral library. Proteomics. 2009 doi: 10.1002/pmic.200900627. in press. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.