ABSTRACT
Tropical forest soils decompose litter rapidly with frequent episodes of anoxic conditions, making it likely that bacteria using alternate terminal electron acceptors (TEAs) play a large role in decomposition. This makes these soils useful templates for improving biofuel production. To investigate how TEAs affect decomposition, we cultivated feedstock-adapted consortia (FACs) derived from two tropical forest soils collected from the ends of a rainfall gradient: organic matter-rich tropical cloud forest (CF) soils, which experience sustained low redox, and iron-rich tropical rain forest (RF) soils, which experience rapidly fluctuating redox. Communities were anaerobically passed through three transfers of 10 weeks each with switchgrass as a sole carbon (C) source; FACs were then amended with nitrate, sulfate, or iron oxide. C mineralization and cellulase activities were higher in CF-FACs than in RF-FACs. Pyrosequencing of the small-subunit rRNA revealed members of the Firmicutes, Bacteroidetes, and Alphaproteobacteria as dominant. RF- and CF-FAC communities were not different in microbial diversity or biomass. The RF-FACs, derived from fluctuating redox soils, were the most responsive to the addition of TEAs, while the CF-FACs were overall more efficient and productive, both on a per-gram switchgrass and a per-cell biomass basis. These results suggest that decomposing microbial communities in fluctuating redox environments are adapted to the presence of a diversity of TEAs and ready to take advantage of them. More importantly, these data highlight the role of local environmental conditions in shaping microbial community function that may be separate from phylogenetic structure.
IMPORTANCE
After multiple transfers, we established microbial consortia derived from two tropical forest soils with different native redox conditions. Communities derived from the rapidly fluctuating redox environment maintained a capacity to use added terminal electron acceptors (TEAs) after multiple transfers, though they were not present during the enrichment. Communities derived from lower-redox soils were not responsive to TEA addition but were much more efficient at switchgrass decomposition. Though the communities were different, diversity was not, and both were dominated by many of the same species of clostridia. This reflects the inadequacy of rRNA for determining the function of microbial communities, in this case the retained ability to utilize TEAs that were not part of the selective growth conditions. More importantly, this suggests that microbial community function is shaped by life history, where environmental factors produce heritable traits through natural selection over time, creating variation in the community, a phenomenon not well documented for microbes.
Introduction
Liquid biofuels derived from nonfood crops, such as switchgrass, are a promising replacement for nonrenewable petroleum-based fuels. The U.S. Department of Energy has identified cellulosic biofuels as integral to meeting our current and future energy needs, partly due to a large renewable reservoir of plant biomass (1, 2). Although corn- and sugarcane-derived ethanol production is already under way on a significant commercial scale, cellulosic ethanol production is inefficient and costly; enzymes derived from fungi are expensive to produce and still require feedstocks to be pretreated before fermentation. Alternative biofuels do not compete with food sources, require less energy input, and produce fewer greenhouse gases than corn ethanol (3, 4). Biofuel production from switchgrass and other perennial grasses has the potential to provide more than one-third of the energy for transportation fuels domestically (1, 5) and produce 540% more renewable energy than that consumed for production, with 94% lower greenhouse gas emissions than for gasoline (6). While switchgrass presents a viable solution to improving biofuel cost and energy efficiency, conversion of lignocellulose to sugars stands to be improved through microbial processing.
Tropical forest ecosystems have very high litter and root decomposition rates (7, 8), and these soils have demonstrated the potential to efficiently deconstruct switchgrass into simple compounds (9). High rates of decomposition are fueled by a high microbial biomass and consistent warm and humid conditions (7, 10). It has been hypothesized that iron cycling fuels considerable decomposition; tropical forest soils experience frequent episodes of anoxic conditions (11, 12), so direct or indirect iron reduction could be an avenue of growth, with biotic or abiotic regeneration of this terminal electron acceptor under oxic conditions. Fluctuating redox potential conditions create an environment where anaerobic and facultative bacterial metabolisms play a large role in leaf litter decomposition.
Under anaerobic conditions, a complex microbial community comprised of fermenters, anaerobic respirers using a range of terminal electron acceptors (TEAs), and syntrophic and acetoclastic methanogens likely contributes to decomposition (13, 14). This is distinct from temperate systems where fungi dominate litter decomposition through the release of phenol oxidase and peroxidase, enzymes that require oxygen (O2) to create free radicals (15). Anaerobic bacteria capable of depolymerizing lignin use different enzymes, such as the beta-aryl etherase produced by Sphingomonas paucimobilis (16). There is a large unexplored diversity of bacteria involved in anaerobic decomposition, likely representing novel approaches to lignin depolymerization in particular that address the challenges of biomass conversion. This would be advantageous for large-scale plant biomass conversion, since the production and metabolic engineering of bacteria are easier than those for fungi (17). Understanding the mechanisms of decomposition in these naturally occurring microbial communities will improve development of lignocellulosic biofuels.
We explored patterns in enzymatic activity and microbial diversity between soil types at the extreme ends of an established elevation, rainfall, and soil redox gradient in the Luquillo Experimental Forest, Puerto Rico (12). The two sites were a low-elevation rain forest site with rapidly fluctuating redox (RF) and a high-elevation cloud forest site with low and slowly fluctuating redox (CF). Decomposition is fast at both sites, but degradation of leaf litter in situ was nearly an order of magnitude faster in the lower-elevation sites than in the high-elevation sites (7; W. L. Silver, R. Ostertag and A. Thompson, unpublished observation). Both sites have measurable methane production, though net methane production is considerably higher at the upper-elevation site (18). Both soils are rich in poorly crystalline Fe oxide minerals, which are insoluble but somewhat unstable and available for microbial reduction. In soils that boast up to 1011 cells per g, iron reducers are found at up to 109 cells per g soil, or 1% of total microbial abundance (10). Iron cycling, which itself is driven both by high Fe abundance and frequent fluctuations in soil redox potential (12, 19, 20), can oxidize C equivalent to 44% of annual litter production (7). Dissimilatory Fe(III)-reducing bacteria compete effectively for limited C with methanogens (21), and there is excellent correlation between CO2 production and Fe(II) production (10). These soils also cycle N and S rapidly (22–24), which suggests that the identity and abundance of TEAs may have a significant impact on the microbial community and their associated decomposition enzymes.
To understand the role of different TEAs in decomposition and investigate the possibility for exogenous TEAs to change either the microbial communities or the decomposition rates under controlled laboratory conditions, we set up anaerobic feedstock-adapted consortia (FAC) derived from the rain forest site (RF) and cloud forest site (CF). After three transfers, the richness of consortia was about one-tenth that of the original soil inocula, as previously described (9). Once these three replicate consortia were established on switchgrass under anaerobic conditions, we used them to seed new consortia (each with three biological replicates) and measured switchgrass decomposition under fermentative conditions (switchgrass alone) and also with potential denitrification (nitrate added), Fe reduction [iron(III) added], and sulfate reduction (sulfate added). We then examined how exogenously added TEAs affect switchgrass C mineralization as well as microbial community function and structure.
RESULTS
Microbial activity.
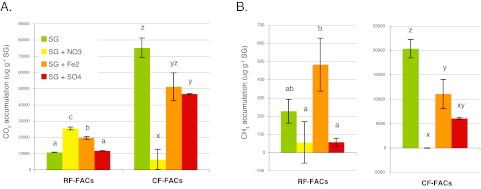
To estimate the C mineralization associated with switchgrass degradation, we measured the amount of CO2 and CH4 emitted; controls with no switchgrass added had no measurable emissions of CO2 or CH4. Carbon mineralization was significantly higher for the cloud forest (CF)-derived FACs than for the rain forest (RF)-derived FACs, regardless of TEAs added, in terms of cumulative CO2 production (P < 0.01) and CH4 production (P < 0.001) per gram switchgrass added. TEA addition had significant effects on CO2 in the RF (P < 0.0001) and CF (P < 0.0001) and on CH4 in the RF (P < 0.05) and CF (P < 0.0001) FACs (Fig. 1). The nitrate amendment had the highest CO2 production in the RF-FACs but the lowest CO2 production in CF-FACs. Iron significantly increased the CO2 above the background level in the RF-FACs. CO2 and CH4 production was suppressed following TEA amendment in the CF-FACs.
FIG 1 .
Net CO2 (A) or CH4 (B), expressed as µg C per gram switchgrass accumulated in the FACs over the course of the incubation. Values shown are means plus or minus standard errors (n = 3). Compared to the RF-FACs, the CF-FACs had significantly more CO2 (P < 0.01) and CH4 emitted (P < 0.001). Within each FAC, the treatment effect was assessed; treatment had a significant effect on FACs in all cases. Lowercase letters denote treatments that are statistically indistinguishable within each set (n = 3), as determined by Tukey’s HSD test at a cutoff of P < 0.05.
Changes in the concentrations of NO3−, NO2−, Fe(II), and SO4− were measured at the beginning and end of the incubation to estimate redox sensitivity of the microbial communities (Table 1). Net reduction of added NO3− to NO2− was detected in the RF-FACs but not the CF-FACs. Net Fe reduction occurred in both the RF- and CF-FACs that received Fe oxide, as evidenced by the generation of high concentrations of Fe(II) at the end of the incubation. A small but significant amount of net sulfate was reduced in the sulfate-amended RF-FACs.
TABLE 1 .
Concentrations of added TEAs at the beginning and end of FAC lab incubations
| Process and consortia | Added TEA | Concn (mM) of TEA |
P valuea | |
|---|---|---|---|---|
| T initial | T final | |||
| Denitrification | ||||
| Rain forest FACs | NO3− | 11.4 ± 0.07 | 0.01 ± 0.01 | *** |
| NO2− | 0.01 ± 0.01 | 3.66 ± 0.44 | ** | |
| Cloud forest FACs | NO3− | 11.6 ± 0.13 | 10.08 ± 1.59 | |
| NO2− | 0.03 ± 0 | 0.02 ± 0 | ||
| Iron reduction | ||||
| Rain forest FACs | Fe(II) | 0.44 ± 0.06 | 6.87 ± 0.07 | *** |
| Cloud forest FACs | Fe(II) | 0.71 ± 0.06 | 6.4 ± 0.26 | *** |
| Sulfate reduction | ||||
| Rain forest FACs | SO4− | 7.89 ± 0.32 | 6.15 ± 0.3 | * |
| Cloud forest FACs | SO4− | 7.85 ± 0.15 | 7.91 ± 0.13 | |
ANOVAs were evaluated at P value cutoffs of <0.05 (*), <0.01 (**), and <0.001 (***) (n = 6).
Beta-galactosidase and cellobiohydrolase activities were significantly higher in CF-derived FACs than in RF-FACs (4-methylumbelliferyl β-d-glucopyranoside (MUB) [BG], P < 0.001; β-d-cellobioside-MUB [CBH], P < 0.05), but there was no difference in activity between sites for N-acetylglucosidase or xylosidase activities. There were significant treatment effects in RF-FACs for BG (P < 0.05), CBH (P < 0. 01), N-acetyl glucosamine-MUB (NAG) (P < 0.0001), and β-d-xylopyranoside-MUB (XYL) (P < 0.0001). The scale of the effect of TEAs on enzyme activities was relatively small, with the largest differences being 31% (BG), 56% (CBH), 234% (NAG), and 61% (XYL). There were weakly significant treatment effects in CF-FACs for NAG (P = 0.081) and XYL (P = 0.078) and no effect for BG or CBH (Fig. 2).
FIG 2 .
Enzyme activities for beta-glucosidase (BG), cellobiohydrolase (CBH), N-acetyl glucosaminidase (NAG), and xylosidase (XYL) are shown for the RF-FACs (left) and CF-FACs (right), expressed as nmol substrate converted per gram of biomass per hour. Values shown are means plus or minus standard errors (n = 3), and ANOVAs were performed to test the null hypothesis of no difference between TEA treatments within an enzyme activity rate, with associated P values shown as “*” (P < 0.05), “**” (P < 0.01) and “***” (P < 0.001).
Microbial community growth and structure.
Higher C mineralization rates observed in CF-FACs than in RF-FACs did not translate to cell abundance or biomass. There was no significant difference between inocula based on total lipids extracted or direct counts, whether comparing final cell counts or fold increase in cell growth (final count divided by initial count) (see Fig. S1 in the supplemental material). There was a significant treatment effect of TEAs on cell growth in RF-FACs (P < 0.0001) but not in CF-FACs. Among the RF-FACs, the sulfate-amended incubations had the most growth, followed by the switchgrass and iron-amended incubations, with the nitrate-amended incubations having the lowest growth (see Fig. S1A). When biomass was measured by lipids extracted, there was no treatment effect for either inoculum type (see Fig. S1B).
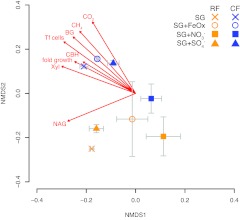
Pyrosequencing resulted in 1,310 unique taxa resolved at 100% nucleotide identity. There were no differences between RF- and CF-FAC inocula in richness or diversity (by Shannon’s H or Simpson’s D index). Treatment significantly affected richness and diversity, where the nitrate treatment was lower in diversity compared to the baseline switchgrass, iron-amended, and sulfate-amended FACs; these data are based on pooled RF- and CF-FAC diversity estimates (Table 2). Nitrate treatments lost taxa primarily from the Proteobacteria and rare species in the RF-FACs (see Table S2 in the supplemental material) and from members of the Proteobacteria, Bacteroidetes, Methanobacteria, and Acidobacteria in the CF-FACs (see Table S3 in the supplemental material). Ordination of microbial communities using phylogenetic distances revealed significant differences in microbial community profiles based on inoculum (A = 0.028 [see Materials and Methods]; P < 0.05) and treatment (A = 0.042; P = 0.054) (Fig. 3). Joint plots for values of microbial community activity showed that C mineralization, enzyme activities, and cell counts were all strongly and significantly correlated to phylogenetic community structure (P < 0.05).
TABLE 2 .
Diversity of FACs by TEA treatment, calculated as overall richness, Shannon’s diversity index (H), Simpson’s index (D), and inverse Simpson’s index (invD)
| Statistic | Value for treatmenta |
P valueb | |||
|---|---|---|---|---|---|
| SG | SG + NO3− | SG + FeOx | SG + SO4− | ||
| Richness | 172.2 ± 39.1 b | 54.8 ± 57.1 a | 148.8 ± 105.6 ab | 123.8 ± 29.9 ab | * |
| H | 2.46 ± 0.27 b | 0.75 ± 0.73 a | 2.05 ± 1.04 b | 2.23 ± 0.33 b | ** |
| D | 0.84 ± 0.04 b | 0.33 ± 0.31 a | 0.71 ± 0.35 b | 0.83 ± 0.05 b | ** |
| invD | 6.45 ± 1.51 b | 1.83 ± 0.96 a | 5.94 ± 2.89 b | 6.14 ± 1.71 b | ** |
Lowercase letters denote different levels of significance as determined by Tukey's HSD test at P < 0.05. SG, switchgrass; FeOx, iron oxide.
ANOVAs were evaluated at P value cutoffs of <0.05 (*) and <0.01 (**) (n = 6).
FIG 3 .
Microbial community profile based on unifrac distance calculated for pyrotag-sequenced SSU rRNA genes. A community profile model was made using ordination by nonmetric multidimensional scaling (NMDS) based on a Bray-Curtis distance metric. Community members shown are means of three replicates, with error bars denoting standard errors of the means (n = 3). Joint plots display vectors showing direction and strength (as length) of positive correlation of the community with each of the listed traits; only significant correlations (P < 0.05) are plotted.
Microbial community profile by phospholipid fatty acid analysis (PLFA) showed separation of treatments, though no separation by inoculum (see Fig. S1C in the supplemental material). The 18:0 lipid marker for general biomass was higher in baseline switchgrass and nitrate-amended FACs than in iron and sulfate-amended FACs (see Fig. S2 and Table S1), which may be due to residual lipids from the switchgrass. Of the four lipid markers for Gram-negative bacteria that were significantly affected by TEA treatment, one (17:1w9c) was highest in iron compared to nitrate, while the rest were highest in switchgrass only compared to iron (see Fig. S2 and Table S1). The stress biomarker lipid 16:1w7t was significantly elevated in unamended FACs. Total lipids did not vary significantly by TEA treatment.
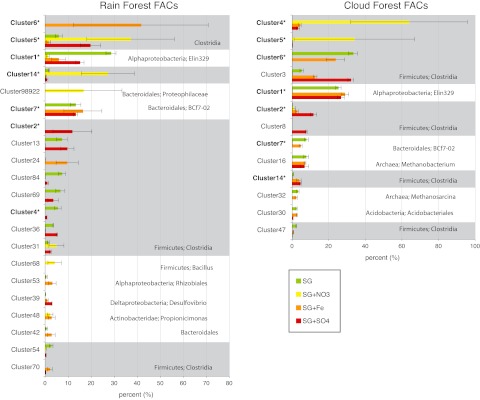
Dominant taxa were defined as taxa with a relative abundance of at least 2% in at least one sample; 23 taxa in RF-FACs and 14 taxa in CF-FACs fit this criterion. Most dominant taxa were members of the Clostridia (phylum Firmicutes); 15 of the RF-FAC dominants and 8 of the CF-FAC dominant taxa were members of the Clostridia (Fig. 4). Taxa in cluster 53 (Rhizobiales), clusters 6 and 24 (Clostridia), and cluster 42 (Bacteroidetes) were all enriched in iron-amended RF-FACs. The switchgrass-only and Fe-amended CF-FACs had the highest relative abundances of the archaeal methanogens Methanosarcina and Methanobacterium and also the highest net methane production. The SO4-amended CF-FACs contained Methanobacterium species but Methanosarcina species as dominant organisms, suggesting that Methanobacterium was primarily responsible for methane production in these microcosms. Among the few eukaryotes grown in these samples, stramenopiles were enriched in sulfate-amended CF-FACs and were dominant (>2% relative abundance) in one replicate (see Table S6 in the supplemental material). Overall, the three replicate microcosms were surprisingly similar considering that they had been separately transferred by 10% serial dilution into fresh media three times.
FIG 4 .
Dominant taxa in the microbial community as determined by pyrosequencing are shown for RF-FACS and CF-FACs based on a relative abundance of 2% or greater. There are 23 taxa in RF-FACs and 14 in CF-FACs that are dominant; taxa are denoted by their cluster number, which was based on 97% SSU rRNA gene sequence identity, and their relative abundance in each TEA treatment is shown as a mean percentage of abundance with standard error (n = 3). Bold cluster numbers with an asterisk “*” indicate taxa that are dominant in both the RF- and the CF-FACs.
DISCUSSION
Through cultivation of feedstock-adapted consortia (FACs) derived from tropical rain forest (RF) and cloud forest (CF) soils, we found that the RF-FACs were able to make use of a broader suite of TEAs than CF-FACs, while the CF-FACs were more productive than RF-FACs. In each case, productivity was indicated by respiration, cell biomass, and enzyme activity during deconstruction of switchgrass under static anoxic conditions. The RF-FACs mostly responded positively to added TEAs. In comparison, the CF-FACs were unresponsive to TEAs and had overall higher CO2 and CH4 accumulation and higher enzyme activities per gram switchgrass. This is particularly surprising given the more rapid decomposition rates of the rain forest site under field conditions (Silver et al., unpublished). The reduction of Fe(III), SO4+, and NO3− by tropical soil communities has the potential to drive considerable C mineralization (10, 22, 24, 25), so these experiments were designed to explore the extent to which anaerobic switchgrass decomposition would be affected by TEA additions in culture. We suggest that a combination of the differential abundances of specific organisms (such as dominance of acetate-utilizing Methanosarcina in the CF-FACs) and the life history of shared microbial populations were responsible for the differences in ability to use different TEAs and in C mineralization efficiency between the two tropical forest soil inocula.
The cloud forest feedstock-adapted consortia (CF-FACs) were more efficient at decomposing switchgrass: higher enzyme activity and rates of C mineralization of CF-FACs were accompanied by similar numbers of cells and amounts of biomass compared to findings for RF-FACs. The greater efficiency of biomass conversion of the CF-FACs could be explained by the different life histories of soil microbes from the tropical cloud forest compared to those from the rain forest. The higher-elevation cloud forest soils experience frequent low-redox conditions, while the lower-elevation rain forest soils experience higher overall redox potential and so are adapted to more-frequent O2 availability. In general, tropical forest soils are likely to be adapted to fluctuating redox; compared to static oxic or anoxic conditions, fluctuating redox resulted in the highest number of active microbes (25, 26). Based on these studies, we would hypothesize that the higher-elevation, lower-O2 soils should have lower field decomposition rates; indeed, this has been shown under field conditions (7). However, under anoxic laboratory conditions the lower-redox CF-derived FACs performed better than the RF-derived FACs. The increased C mineralization of CF-FACs over that of RF-FACs is apparently not due to increased biomass, since these two inocula were indistinguishable by direct counts and by lipid biomass, suggesting that an equivalent number or mass of microbes was able to more efficiently deconstruct the feedstock. Either the planktonic nature of the lab incubations, the static anaerobic conditions, or some property of the plant biomass added was favorable to the CF-FACs compared to the RF-FACs.
Carbon mineralization was inhibited by TEA addition in the CF-FACs. It is possible that the absence of these alternative electron acceptors during the adaptation of microbial consortia to anaerobic switchgrass decomposition could have resulted in a decreased ability to reduce NO3−, Fe(III), and SO4+; further research could address this point. In contrast, we observed considerable plasticity in the metabolism of the RF-FACs, which retained facultative metabolic capacities for coupling NO3− and SO4+ reduction to decomposition. It is possible that the presence of a strong oxidizer, such as NO3−, was inhibitory to the cloud forest-derived communities, which would have been accustomed to low redox. These CF-FAC communities were making 10 times more CH4 than the RF-FACs before the addition of NO3− depressed them; this could be evidence of low-level NO3− utilization below detection of the chemical analysis performed here. It is possible that low levels of nitrite or sulfite were produced in the CF-FACs and that these were enough to inhibit methanogenesis (13, 27). That both FACs retained the ability to utilize Fe(III) as an alternative TEA is a reflection of the prominence of reactive Fe species in these soils and its utility in microbial growth and activity.
Organisms with facultative metabolic capabilities seemed to be preserved throughout the transfers in the RF-FACs but not the CF-FACs, despite the fact that all consortia were adapted without the addition of exogenous TEAs. This was evident in the SO4+-amended FACs, where addition caused a significant increase in cell growth, as well as a significant increase in xylosidase enzyme activity. Compared to findings for other TEA treatments, the SO4+-amended RF-FACs had increased numbers of Desulfovibrio bacteria, which are known SO4+ reducers (28) with innate tolerance for O2 that would aid in survival in fluctuating-redox soils (29). Desulfovibrio was also enriched in the Fe(III)-amended RF-FACs, though to a lesser extent than in the SO4+-amended FACs. This suggests the possibility of Fe(III)-reducing Desulfovibrio, though members of the genera Clostridium and Aeromonas were also present and were likely reducing Fe(III) (30, 31). Siderophores, humics, or other electron shuttles may permit a range of microbes to indirectly reduce Fe(III), producing Fe(II) (32, 33). Like Fe(III), SO4+ can be abiotically oxidized in environments where redox potential fluctuates, creating a sulfur cycle of regenerating terminal electron acceptors to fuel C mineralization (34–36). Both Fe(III) and NO3− increased CO2 production, though both decreased cell abundance. Denitrifiers are facultative aerobes that prefer O2 but have the capacity to switch to NO3− (or NO2−) reduction under O2-limited conditions and are phylogenetically diverse (37). Dissimilatory NO3− reduction to ammonium (DNRA) has also been shown to constitute as much as 75% of the turnover of the NO3− pool in tropical forest soils (22). The organisms performing DNRA in one instance were discovered to consist of spore-forming bacteria (38). The buildup of NO2− in the NO3−-amended FACs could have contributed to the repression of activity and cell growth in the NO3-amended RF-FACs and suggests that while DNRA occurs in the field, it was not likely to be occurring in our FACs.
The addition of different TEAs created changes in C mineralization and populations of methanogens in CF-FACs, where CH4 production was appreciable in the unamended CF-FACs as well as the CF-FACs amended with Fe(III) and SO4+. While CH4 was produced in the Fe(III)-amended CF-FAC microcosms, Fe(III) amendment did not increase CO2 concentrations relative to the background treatment. Iron(III) addition decreased net CH4 production, likely due to competition with dissimilatory Fe reducers for acetate (21). Net CH4 production in Fe(III)-amended CF-FACs was significantly depressed, though not as low as in the other FACs, suggesting that either the Fe(III) reducers were not as efficient as the other community members or that the methanogens competing with the Fe(III) reducers for labile C were not as effective under Fe(III)-reducing conditions. The switchgrass-only, Fe(III)-amended, and SO4+-amended CF-FAC microcosms contained 7% Methanobacterium species, which are hydrogen- and formate-utilizing methanogens, and [in Fe(III) and switchgrass-only treatments] 3% Methanosarcina species, acetate-utilizing methanogens (27, 39, 40). The SO4+-amended CF-FACs did not contain members of Methanosarcina, suggesting that SO4+ was somehow inhibitory. Members of the Clostridia in clusters 8, 3, and 2 were all significantly enriched in SO4+-amended FACs compared to findings for Fe(III)-amended FACs. There is much phenotypic variation within the Clostridia that enables these organisms to live in a variety of environments (41), and apparently even the subtle selective pressures of different additional TEA amendments is enough to select for a specific set of Clostridia bacteria.
The established feedstock-adapted consortia generally contained a few dominant species with long tails of richness extending into the hundreds of rare species, which may play a role in stabilizing the activity of these communities. The dominant species, defined as having a relative abundance of 2% or greater, comprised a substantial portion of the total richness of these consortia, which were mostly comprised of the Clostridia, as well as members of the Rhizobiales, Bacteroidales, methanogenic Archaea, and Desulfovibrio. Taxa in the long tail of the communities were made up of taxa from phyla commonly found in soils but uncultured, with little known about their function. In the RF-FACs these included taxa from the phyla Gemmatimonadetes and Op10. CF-FACs had the same diversity but fewer dominant taxa and different rare taxa from those of the RF-FACs. The rare taxa have been selected for their ability to grow in the anaerobic enrichments but may also contribute to the activity. For example, it is known that anaerobic cellulose degradation by Clostridium straminisolvens CSK1 is stabilized by the presence of noncellulolytic, aerobic bacteria that are thought to reduce the local redox potential (42). Taxa need not be abundant to play important roles in ecosystem function; it has been shown in bog soils that the sulfate reducer Desulfosporosinus was present at low (0.006%) abundance but was responsible for the majority of soil SO4+ reduction (43). Increased richness has been shown to stabilize mixed communities in natural environments, especially when richness is low (below 10 species) (44). Cooperation among disparate taxa in these mixed communities is possible and will become evident with attempts to isolate contributing members under the same conditions.
MATERIALS AND METHODS
Lab incubations.
Soils were collected from the Luquillo Experimental Forest, Puerto Rico, part of the National Science Foundation (NSF)-sponsored Long Term Ecological Research Program. Soils from the Bisley Research Watershed (rain forest site [RF]) and the Pico del Este Short Could Forest (CF) were used as inoculum for three replicate feedstock-adapted consortia (FAC) each. The fieldwork was conducted and samples collected and transported under USDA permit number P526P-08-00634. Site selection was based on redox states of the soil, which change along an elevation and rainfall gradient. The CF site is located in an upper montane tropical cloud forest at approximately 1,050 m above sea level (18°18′N, 65°50′W) and experiences approximately 4 to 5 m rainfall annually and a high frequency of low-redox conditions (45). The RF site is in a lower montane wet tropical forest at approximately 270 m above sea level (18°18′N, 65°50′W) and receives approximately 3.5 m of rainfall annually (46). The soils at this site fluctuate between anoxic and oxic (11, 12). Rainfall at both sites is relatively evenly distributed throughout the year; an average annual temperature is 21°C and 23°C at CF and RF, respectively, with little seasonal variation. RF soils are deep, highly weathered, clayey Ultisols, rich in Fe and Al oxides and hydroxides, while CF soils are isomesic Humic Haplaquox, clay rich, wet, and high in organic matter (45, 47).
Soils were collected in January 2009 from the 0-to-10-cm depth and immediately transported fresh to the lab (LBNL, Berkeley CA) under ambient temperature conditions. We weighed out 5 g of fresh soil and added this to deionized water with 0.5 g (dry weight) switchgrass ground to 0.42 mm using a Wiley mill. One control condition consisted of water and soil only, performed in triplicate. Dinitrogen gas was bubbled through all media to remove any dissolved O2, and containers were quickly sealed with airtight stoppers to maintain anaerobic conditions. The original soil slurry was incubated for 8 weeks and then used to inoculate the FACs at a level of 10% final volume into BMM minimal medium (48) with trace minerals (49, 50) and vitamins (51), buffered to pH 5.5 with morpholineethanesulfonic acid (MES) to match the soil pH. The FACs were transferred two more times after 10 weeks each, maintaining separate lines for each of the three individual replicates; cells were not normalized between transfers. The FAC from round three was split into four treatments with three biological replicates each for a total of 24 FAC cultures. The treatments were baseline switchgrass (15 g liter−1) and baseline switchgrass amended with sodium sulfate (10 mM), potassium nitrate (10 mM), or soluble iron (5 mM). A stock solution of soluble iron was obtained by adding ferric chloride hexahydrate [Fe(III)] to a solution of nitrilotriacetic acid disodium salt and sodium bicarbonate. Dinitrogen gas was bubbled through media to remove any dissolved O2, and containers were quickly sealed with airtight stoppers to maintain anaerobic conditions. Containers were autoclaved for 20 min at 121°C. These FACs were incubated with treatments for 8 weeks, and then 10% of these cultures were inoculated into fresh media with treatments (as described above). These FACs were each individually continuously monitored for 19 days for net CO2 and CH4 accumulation using the Micro-Oxymax respirometer (Columbus Instruments, Columbus, OH). At the end of the experiment, we analyzed FACs for biogeochemistry, enzyme activity, and microbial community structure as detailed below.
Iron analysis.
Total iron(II) concentrations were determined by a colorimetric ferrozine assay (52, 53). A 250-µl aliquot was dispensed from each FAC into 5 ml of 0.5 N hydrochloric acid (HCl) and allowed to extract for 20 min. Twenty-five microliters of the HCl solution was then added to 1 ml of ferrozine. Absorbance was immediately recorded at 562 nm on a Beckman DU 640 spectrophotometer (Beckman Coulter, Inc., Brea, CA). A 4-point calibration (0.5, 1, 5, and 15 mM) of iron(II) sulfate was used to quantify sample measurements.
Ion chromatography.
Nitrate, nitrite, and sulfate concentrations were measured by ion chromatography (IC) using a model ICS-2000 IC (Dionex Corporation, Sunnyvale, CA) with an IonPac AG11 guard column (4 by 50 mm), IonPac AS11 analytical column (4 by 250 mm), conductivity suppressor (ASRS Ultra), and eluent generator creating a potassium hydroxide gradient. Samples were filtered with a 0.2-µm nylon membrane and diluted 16-fold in ultrapure Milli-Q water. A 3-point calibration (100, 300, and 600 µM) for each anion was used to determine measured concentrations.
Cell counts.
Direct counts were made of each sample based on an acridine orange staining technique with epifluorescence counting (54). A sample was taken from each lab incubation, preserved using 4% formaldehyde, and stored at 4°C. Direct counts were obtained by diluting samples 10-fold with filter-sterilized 1× phosphate-buffered saline (PBS) (Sigma Aldrich Corp., St. Louis, MO). Ten microliters of each dilution was placed in a 6-mm microscope slide well and allowed to dry. Wells were stained with 25 mg ⋅ ml−1 acridine orange for 2 min in the dark. Unbound acridine orange was rinsed off by dipping slides into deionized water. Cells were imaged using a fluorescein isothiocyanate (FITC) filter on a Zeiss Axioskop microscope (Carl Zeiss, Inc., Germany). Cell counts are expressed as numbers of cells per ml culture, and fold-increase counts are reported as final counts divided by initial counts.
Enzyme assays.
Enzyme activity was determined at the end of the lab incubation by adding 100 µl of culture to wells of a 96-well plate containing 100 µl of the substrates 4-methylumbelliferyl β-d-glucopyranoside (BG) for beta-glucosidase or cellulose activity, β-d-cellobioside-MUB (CBH) for cellobiohydrolase or hemicellulose activity, N-acetyl glucosamine-MUB (NAG) for chitinase activity, and β-d-xylopyranoside-MUB (XYL) for xylosidase activity. The fluorescence of each well was determined after a 4-h incubation at 27°C using a Fluorolog-3 spectrofluorimeter (Horiba Jobin Yvon Inc., Edison, NJ) with 365-nm excitation and 442-nm emission wavelengths. Rates of substrate degradation were calculated as the difference in moles of MUB produced over time based on MUB standards. Enzyme activities for each treatment or replicate were normalized for the initial mass of biomass added and are reported as pmol MUB h−1 g−1 switchgrass.
PLFA.
Samples were extracted by the Bligh-Dyer method (55–57). Briefly, 10 ml of a 10:5:4 mixture of methanol-chloroform (pH 7)-phosphate buffer was added to each sample. Fifty microliters of 500-mg/liter 1,2-dinonadecanoyl-sn-glycero-3-phosphocholine (Avanti Polar Lipids, Alabaster, AL) was added as an internal standard. The mixture was vortexed, sonicated for 2 min, and extracted at room temperature in the dark for 3 h. Phases were separated with the addition of 2 ml of chloroform and 2 ml of water, followed by centrifugation at 2,000 rpm for 15 min to separate the organic and aqueous layers. The lower organic layer was removed to a clean tube, and 2 ml additional chloroform was added to the original extract, which was revortexed and centrifuged and combined with the first layer. This combined organic layer was dried under N2. The dried extracts were separated into neutral, glycerol, and phospholipids on a C18 silica column (Sigma Chemical Company, St. Louis, MO) by sequential elution with chloroform, acetone, and methanol. All collected fractions were dried under N2.
The methanol fractions containing phospholipids were subjected to a mild alkaline hydrolysis to remove the head group and create fatty acid methyl ester (FAME) compounds by resuspending with 1:1 chloroform-methanol and 1 ml of 11.2-mg/liter KOH in methanol. After vortexing for 2 min, they were incubated in a water bath at 37°C for 60 min. The FAME compounds were then neutralized with 200 µl of 0.1 mM acetic acid, extracted three times with 2 ml of hexane, and dried under N2. Fifty microliters of 46.2-mg/liter methyl undecanoate (Sigma Chemical Company, St. Louis, MO) was added to the dried extracts as an external standard. FAME were detected on an Agilent 6890N gas chromatograph/flame ionization detector (GC/FID) on an HP1 60-m column (0.25-mm inside diameter [ID]) and quantified by comparing to known standards. Peak confirmation was accomplished by using an Agilent 6890 GC/mass spectrometer (MS). Lipid classes were grouped into guilds according to configuration (58–62) as outlined in Table S1 in the supplemental material.
Amplicon pyrosequencing of community SSU rRNA.
Samples were taken of liquid medium plus switchgrass from each microcosm, and DNA was extracted using a modified cetyltrimethylammonium bromide (CTAB) extraction method as previously described (26). Briefly, frozen samples were added to CTAB extraction buffer and phenol in Lysing Matrix E tubes (Qbiogene) and bead beaten in a FastPrep instrument (Bio101), followed by a chloroform extraction, isopropanol precipitation, and use of the AllPrep DNA/RNA extraction kit (Qiagen). Small subunit (SSU) rRNA gene sequences for high-throughput amplicon pyrosequencing were amplified using the universal primers 926F (5′-aaactYaaaKgaattgacgg-3′) and 1392R (5′ acgggcggtgtgtRc 3′), where uppercase letters indicate nucleotide redundancies; these primers target the V8 variable region of the 16S rRNA gene from bacteria and archaea as well as the 18S rRNA gene in eukarya (63). The sequences shown do not include adaptor or bar code sequences, and the reverse primer included a 5-bp bar code for multiplexing of samples during sequencing. Emulsion PCR and sequencing of the PCR amplicons were performed following the manufacturer’s instructions for the Roche 454 GS FLX titanium technology, with the exception that the final dilution was 1e−8. Sequencing tags were analyzed using the software tool PyroTagger (http://pyrotagger.jgi-psf.org/), which filters by removing low-quality sequences from the set based on the qual file, trims using a 225-bp sequence length threshold, dereplicates, clusters at the operational taxonomic unit (OTU) level based on 97% identity, and then classifies. Eighteen chimeras were detected and removed, leaving 1,310 taxa detected. Classification was based on the greengenes database of rRNA genes (64) for bacterial and archaeal amplicons and the Silva database for eukaryotic amplicons (65).
Statistical analysis.
The experimental design included three biological replicates, four treatments, and two soil inoculum types, for a total of 24 samples. Analyses of variance (ANOVAs) were performed to a statistical significance level of 0.05, and Tukey’s honestly significant difference (HSD) test was used to determine treatments with statistically indistinguishable measurements. Ordination of the whole community detected by pyrosequencing was performed using nonmetric multidimensional scaling with the Bray-Curtis distance measure based on unifrac distances (66, 67). Joint plots are vectors indicating the direction and magnitude of correlation to individual samples; joint plots were calculated for all measured variables against the community ordination, and only the factors with significant (P < 0.05) correlations are shown. In all results, means and standard errors of the mean are reported for 3 biological replicates unless otherwise indicated. A multiresponse permutation procedure (MRPP) was used to determine differences in inoculum or TEA amendment treatment within the microbial community distance matrix. This test yields metric A, which describes the within-group homogeneity and can range from 0 if the groups are completely different to 1 if the groups are identical; for ecological data, it is uncommon to observe A values greater than 0.3 (68). We used Benjamini-Hochburg-adjusted P values in PLFA peak analysis (69). To calculate richness or diversity (by Shannon’s H or Simpson’s D index), data from CF- and RF-FACs were pooled after determining that they were not significantly different.
SUPPLEMENTAL MATERIAL
Microbial community abundances were measured by direct counts and PLFA and are shown here as fold increases in counts from the initial to the final time (A) or total lipid biomass as nmol per gram switchgrass (B). Lowercase letters denote differences between TEA treatments within the FAC, as determined by Tukey’s HSD test for FACs that showed a significant treatment effect by ANOVA (P < 0.05). (C) Microbial community profile based on mole fraction of PLFA lipid analysis. A community profile model was made using ordination by nonmetric multidimensional scaling based on a Bray-Curtis distance metric. Community members shown are means of three replicates, with error bars denoting standard errors of the means (n = 3). Download FIG S1, PDF file, 0.1 MB.
Biomass of PLFA lipids (nmol per g sample) that were significantly affected by TEA treatment. Levels of significance are indicated as follows: *, P < 0.05; **, P < 0.01; ***, P < 0.001. All P values are corrected for repeated measures by the Bonferroni method. No lipids were different in abundance between inocula, so lipid measures from BisR- and SCF-FACs are pooled (n = 6). Download FIG S2, PDF file, 0.1 MB.
TEA treatment effect in PLFA lipids.
Abundance of taxa by phylum in rain forest (RF) FACs, mean ± standard deviation.
Abundance of taxa by phylum in cloud forest (CF) FACs, mean ± standard deviation.
Dominant taxa by site.
Dominant taxa in RF-FACs by TEA treatment.
Dominant taxa in CF-FACs by TEA treatment.
ACKNOWLEDGMENTS
This work was conducted by the Joint BioEnergy Institute and was supported by the Office of Science, Office of Biological and Environmental Research of the U.S. Department of Energy under contract no. DE-AC02-05CH11231. The research was also partially supported by DEB-0620910 from NSF to the Institute of Tropical Ecosystem Studies, University of Puerto Rico, and the International Institute of Tropical Forestry (IITF) as part of the Luquillo LTER program.
We thank Ken Vogel of the USDA for switchgrass used in this study.
Footnotes
Citation DeAngelis KM, et al. 2012. Anaerobic decomposition of switchgrass by tropical soil-derived feedstock-adapted consortia. mBio 3(1):e00249-11. doi:10.1128/mBio.00249-11.
REFERENCES
- 1. Perlack RD. 2005. Biomass as feedstock for a bioenergy and bioproducts industry: the technical feasibility of a billion-ton annual supply. DTIC document. Published jointly by the United States Department of Agriculture (USDA) and the U.S. Department of Energy (DOE), Washington, DC. [Google Scholar]
- 2. Simmons BA. 2011. Opportunities and challenges in advanced biofuel production: the importance of synthetic biology and combustion science. Biofuels 2:5–7 [Google Scholar]
- 3. Adler PR, Del Grosso SJ, Parton WJ. 2007. Life-cycle assessment of net greenhouse-gas flux for bioenergy cropping systems. Ecol. Appl. 17:675–691 [DOI] [PubMed] [Google Scholar]
- 4. Charles D. 2009. Biofuels: corn-based ethanol flunks key test. Science 324:587. [DOI] [PubMed] [Google Scholar]
- 5. Blanch HW, et al. 2008. Addressing the need for alternative transportation fuels: the Joint BioEnergy Institute. ACS Chem. Biol. 3:17–20 [DOI] [PubMed] [Google Scholar]
- 6. Schmer MR, Vogel KP, Mitchell RB, Perrin RK. 2008. Net energy of cellulosic ethanol from switchgrass. Proc. Natl. Acad. Sci. U. S. A. 105:464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cusack DF, Chou WW, Yang WH, Harmon ME, Silver WL. 2009. Controls on long-term root and leaf litter decomposition in neotropical forests. Glob. Change Biol. 15:1339–1355 [Google Scholar]
- 8. Parton W, et al. 2007. Global-scale similarities in nitrogen release patterns during long-term decomposition. Science 315:361 [DOI] [PubMed] [Google Scholar]
- 9. DeAngelis KM, et al. 2010. Strategies for enhancing the effectiveness of metagenomic-based enzyme discovery in lignocellulolytic microbial communities. Bioenergy Res. 3:146–158 [Google Scholar]
- 10. Dubinsky EA, Silver WL, Firestone MK. 2010. Tropical forest soil microbial communities couple iron and carbon biogeochemistry. Ecology 91:2604–2612 [DOI] [PubMed] [Google Scholar]
- 11. Liptzin D, Silver WL, Detto M. 2011. Temporal dynamics in soil oxygen and greenhouse gases in two humid tropical forests. Ecosystems 14:171–182 [Google Scholar]
- 12. Silver WL, Lugo A, Keller M. 1999. Soil oxygen availability and biogeochemistry along rainfall and topographic gradients in upland wet tropical forest soils. Biogeochemistry 44:301–328 [Google Scholar]
- 13. Conrad R. 1996. Soil microorganisms as controllers of atmospheric trace gases (H2, CO, CH4, OCS, N2O, and NO). Microbiol. Rev. 60:609–640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Leschine SB. 1995. Cellulose degradation in anaerobic environments. Annu Rev. Microbiol. 49:399–426 [DOI] [PubMed] [Google Scholar]
- 15. Boer W, Folman LB, Summerbell RC, Boddy L. 2005. Living in a fungal world: impact of fungi on soil bacterial niche development. FEMS Microbiol. Rev. 29:795–811 [DOI] [PubMed] [Google Scholar]
- 16. Masai E, Katayama Y, Fukuda M. 2007. Genetic and biochemical investigations on bacterial catabolic pathways for lignin-derived aromatic compounds. Biosci. Biotechnol. Biochem. 71:1–15 [DOI] [PubMed] [Google Scholar]
- 17. Lee SK, Chou H, Ham TS, Lee TS, Keasling JD. 2008. Metabolic engineering of microorganisms for biofuels production: from bugs to synthetic biology to fuels. Curr. Opin. Biotechnol. 19:556–563 [DOI] [PubMed] [Google Scholar]
- 18. Teh YA, Dubinsky EA, Silver WL, Carlson CM. 2008. Suppression of methanogenesis by dissimilatory Fe (III)-reducing bacteria in tropical rain forest soils: implications for ecosystem methane flux. Glob. Change Biol. 14:413–422 [Google Scholar]
- 19. Chacon N, Silver WL, Dubinsky EA, Cusack DF. 2006. Iron reduction and soil phosphorus solubilization in humid tropical forests soils: the roles of labile carbon pools and an electron shuttle compound. Biogeochemistry 78:67–84 [Google Scholar]
- 20. Thompson A, Chadwick OA, Rancourt DG, Chorover J. 2006. Iron-oxide crystallinity increases during soil redox oscillations. Geochim. Cosmochim. Acta 70:1710–1727 [Google Scholar]
- 21. Teh YA, Silver WL, Conrad ME. 2005. Oxygen effects on methane production and oxidation in humid tropical forest soils. Glob. Change Biol. 11:1283–1297 [Google Scholar]
- 22. Silver WL, Herman DJ, Firestone MK. 2001. Dissimilatory nitrate reduction to ammonium in upland tropical forest soils. Ecology 82:2410–2416 [Google Scholar]
- 23. Stanko-Golden K, Fitzgerald J. 1991. Sulfur transformations and pool sizes in tropical forest soils. Soil Biol. Biochem. 23:1053–1058 [Google Scholar]
- 24. Templer PH, Silver WL, Pett-Ridge J, DeAngelis KM, Firestone MK. 2008. Plant and microbial controls on nitrogen retention and loss in a humid tropical forest. Ecology 89:3030–3040 [DOI] [PubMed] [Google Scholar]
- 25. Pett-Ridge J, Silver WL, Firestone MK. 2006. Redox fluctuations frame microbial community impacts on N-cycling rates in a humid tropical forest soil. Biogeochemistry 81:95–110 [Google Scholar]
- 26. DeAngelis KM, Silver WL, Thompson AW, Firestone MK. 2010. Microbial communities acclimate to recurring changes in soil redox potential status. Environ. Microbiol. 12:3137–3149 [DOI] [PubMed] [Google Scholar]
- 27. Fetzer S, Bak F, Conrad R. 1993. Sensitivity of methanogenic bacteria from paddy soil to oxygen and desiccation. FEMS Microbiol. Ecol. 12:107–115 [Google Scholar]
- 28. Boschker H, et al. 1998. Direct linking of microbial populations to specific biogeochemical processes by 13C-labelling of biomarkers. Nature 392:801–805 [Google Scholar]
- 29. Zhou A, et al. 2010. Hydrogen peroxide-induced oxidative stress responses in Desulfovibrio vulgaris Hildenborough. Environ. Microbiol. 12:2645–2657 [DOI] [PubMed] [Google Scholar]
- 30. Bale SJ, et al. 1997. Desulfovibrio profundus sp. nov., a novel barophilic sulfate-reducing bacterium from deep sediment layers in the Japan Sea. Int. J. Syst. Bacteriol. 47:515–521 [DOI] [PubMed] [Google Scholar]
- 31. Lovley DR, Holmes DE, Nevin KP. 2004. Dissimilatory Fe(III) and Mn(IV) reduction. Adv. Microb. Physiol. 49:219–286 [DOI] [PubMed] [Google Scholar]
- 32. Lovley DR, Coates JD, Blunt-Harris EL, Phillips EJP, Woodward JC. 1996. Humic substances as electron acceptors for microbial respiration. Nature 382:445–448 [Google Scholar]
- 33. Nielsen LP, Risgaard-Petersen N, Fossing H, Christensen PB, Sayama M. 2010. Electric currents couple spatially separated biogeochemical processes in marine sediment. Nature 463:1071–1074 [DOI] [PubMed] [Google Scholar]
- 34. Heitmann T, Blodau C. 2006. Oxidation and incorporation of hydrogen sulfide by dissolved organic matter. Chem. Geol. 235:12–20 [Google Scholar]
- 35. Knorr KH, Blodau C. 2009. Impact of experimental drought and rewetting on redox transformations and methanogenesis in mesocosms of a northern fen soil. Soil Biol. Biochem. 41:1187–1198 [Google Scholar]
- 36. Knorr KH, Lischeid G, Blodau C. 2009. Dynamics of redox processes in a minerotrophic fen exposed to a water table manipulation. Geoderma 153:379–392 [Google Scholar]
- 37. Bruns MA, Fries MR, Tiedje JM, Paul EA. 1998. Functional gene hybridization patterns of terrestrial ammonia-oxidizing bacteria. Microb. Ecol. 36:293–302 [DOI] [PubMed] [Google Scholar]
- 38. Yin S, Chen D, Chen L, Edis R. 2002. Dissimilatory nitrate reduction to ammonium and responsible microorganisms in two Chinese and Australian paddy soils. Soil Biol. Biochem. 34:1131–1137 [Google Scholar]
- 39. Grosskopf R, Janssen PH, Liesack W. 1998. Diversity and structure of the methanogenic community in anoxic rice paddy soil microcosms as examined by cultivation and direct 16S rRNA gene sequence retrieval. Appl. Environ. Microbiol. 64:960–969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Rajagopal B, Belay N, Daniels L. 1988. Isolation and characterization of methanogenic bacteria from rice paddies. FEMS Microbiol. Lett. 53:153–158 [Google Scholar]
- 41. Lawson PA, Llop-Perez P, Hutson RA, Hippe H, Collins MD. 1993. Towards a phylogeny of the clostridia based on 16S rRNA sequences. FEMS Microbiol. Lett. 113:87–92 [DOI] [PubMed] [Google Scholar]
- 42. Kato S, Haruta S, Cui ZJ, Ishii M, Igarashi Y. 2004. Effective cellulose degradation by a mixed-culture system composed of a cellulolytic Clostridium and aerobic non-cellulolytic bacteria. FEMS Microbiol. Ecol. 51:133–142 [DOI] [PubMed] [Google Scholar]
- 43. Pester M, Bittner N, Deevong P, Wagner M, Loy A. 2010. A “rare biosphere” microorganism contributes to sulfate reduction in a peatland. ISME J. 4:1591–1602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Nielsen U, Ayres E, Wall D, Bardgett R. 2011. Soil biodiversity and carbon cycling: a review and synthesis of studies examining diversity-function relationships. Eur. J. Soil Sci. 62:105–116 [Google Scholar]
- 45. Silver WL, Liptzin D, Almaraz M. Soil redox dynamics and biogeochemistry along a tropical elevational gradient. Ecol. Bull., in press [Google Scholar]
- 46. Scatena FN. 1989. An introduction to the physiography and history of the Bisley experimental watersheds in the Luquillo mountains of Puerto Rico. General Technical Report SO 72.
- 47. Huffaker L. 2002. Soil survey of the Caribbean national forest and Luquillo experimental forest, commonwealth of Puerto Rico (interim publication). U.S: Department of Agriculture, Natural Resource Conservation Service, Washington, DC [Google Scholar]
- 48. Tanner RS. 1997. Cultivation of bacteria and fungi, p 52–60 In Hurst CJ, Manual of environmental microbiology. ASM Press, Washington, DC [Google Scholar]
- 49. Tschech A, Pfennig N. 1984. Growth yield increase linked to caffeate reduction in Acetobacterium woodii. Arch. Microbiol. 137:163–167 [Google Scholar]
- 50. Widdel F, Kohring GW, Mayer F. 1983. Studies on dissimilatory sulfate-reducing bacteria that decompose fatty acids. Arch. Microbiol. 134:286–294 [DOI] [PubMed] [Google Scholar]
- 51. Janssen PH, Schuhmann A, Mörschel E, Rainey FA. 1997. Novel anaerobic ultramicrobacteria belonging to the Verrucomicrobiales lineage of bacterial descent isolated by dilution culture from anoxic rice paddy soil. Appl. Environ. Microbiol. 63:1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Lovley DR, Phillips EJP. 1987. Rapid assay for microbially reducible ferric iron in aquatic sediments. Appl. Environ. Microbiol. 53:1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Stookey LL. 1970. Ferrozine—a new spectrophotometric reagent for iron. Anal. Chem. 42:779–781 [Google Scholar]
- 54. Francisco DE, Mah RA, Rabin AC. 1973. Acridine orange-epifluorescence technique for counting bacteria in natural waters. Trans. Am. Microsc. Soc. 92:416–421 [PubMed] [Google Scholar]
- 55. Guckert JB, Antworth CP, Nichols PD, White DC. 1985. Phospholipid, ester-linked fatty acid profiles as reproducible assays for changes in prokaryotic community structure of estuarine sediments. FEMS Microbiol. Lett. 31:147–158 [Google Scholar]
- 56. Pfiffner SM, et al. 2006. Phospholipid fatty acid profiles and biodensity estimates for water, rock and air samples recovered from Witwatersrand basin mines. Geomicrobiol. J. 23:431–442 [Google Scholar]
- 57. White DC, Ringelberg DB. 1998. Signature lipid biomarker analysis, p. 255–272 In Burlage RS, Atlas R, Stahl D, Geesey G, Sayler G, Techniques in microbial ecology. Oxford University Press, New York, NY. [Google Scholar]
- 58. Cusack DF, Silver WL, Torn MS, Burton SD, Firestone MK. 2011. Changes in microbial community characteristics and soil organic matter with nitrogen additions in two tropical forests. Ecology 92:621–632 [DOI] [PubMed] [Google Scholar]
- 59. Griffiths BS, Ritz K, Ebblewhite N, Dobson G. 1998. Soil microbial community structure: effects of substrate loading rates. Soil Biol. Biochem. 31:145–153 [Google Scholar]
- 60. Moore-Kucera J, Dick RP. 2008. PLFA profiling of microbial community structure and seasonal shifts in soils of a Douglas-fir chronosequence. Microb. Ecol. 55:500–511 [DOI] [PubMed] [Google Scholar]
- 61. Salomonová S, et al. 2003. Determination of phospholipid fatty acids in sediments. Acta Univ. Palacki. Olomuc. Facultas Rerum Nat. Chem. 42:39–49 [Google Scholar]
- 62. Steger K, Jarvis A, Smårs S, Sundh I. 2003. Comparison of signature lipid methods to determine microbial community structure in compost. J. Microbiol. Methods 55:371–382 [DOI] [PubMed] [Google Scholar]
- 63. Engelbrektson A, et al. 2010. Experimental factors affecting PCR-based estimates of microbial species richness and evenness. ISME J. 4:642–647 [DOI] [PubMed] [Google Scholar]
- 64. DeSantis TZ, et al. 2006. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 72:5069–5072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Pruesse E, et al. 2007. Silva: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 35:7188–7196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Hamady M, Lozupone C, Knight R. 2010. Fast UniFrac: facilitating high-throughput phylogenetic analyses of microbial communities including analysis of pyrosequencing and PhyloChip data. ISME J. 4:17–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. McCune B, Grace JB, Urban DL. 2002. Analysis of ecological communities. MjM Software Design, Gleneden; Beach, OR [Google Scholar]
- 68. Legendre P, Legendre L. 1998. Numerical ecology. Elsevier Science, Philadelphia, PA. [Google Scholar]
- 69. Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. B Stat. Methodol. 57:289–300 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Microbial community abundances were measured by direct counts and PLFA and are shown here as fold increases in counts from the initial to the final time (A) or total lipid biomass as nmol per gram switchgrass (B). Lowercase letters denote differences between TEA treatments within the FAC, as determined by Tukey’s HSD test for FACs that showed a significant treatment effect by ANOVA (P < 0.05). (C) Microbial community profile based on mole fraction of PLFA lipid analysis. A community profile model was made using ordination by nonmetric multidimensional scaling based on a Bray-Curtis distance metric. Community members shown are means of three replicates, with error bars denoting standard errors of the means (n = 3). Download FIG S1, PDF file, 0.1 MB.
Biomass of PLFA lipids (nmol per g sample) that were significantly affected by TEA treatment. Levels of significance are indicated as follows: *, P < 0.05; **, P < 0.01; ***, P < 0.001. All P values are corrected for repeated measures by the Bonferroni method. No lipids were different in abundance between inocula, so lipid measures from BisR- and SCF-FACs are pooled (n = 6). Download FIG S2, PDF file, 0.1 MB.
TEA treatment effect in PLFA lipids.
Abundance of taxa by phylum in rain forest (RF) FACs, mean ± standard deviation.
Abundance of taxa by phylum in cloud forest (CF) FACs, mean ± standard deviation.
Dominant taxa by site.
Dominant taxa in RF-FACs by TEA treatment.
Dominant taxa in CF-FACs by TEA treatment.