Abstract
We sequenced the 5′ UTR of the estrogen-related receptor gamma gene (ERR-γ) in ~500 patient and volunteer samples and found that longer alleles of the (AAAG)n microsatellite were statistically and significantly more likely to exist in the germlines of breast cancer patients when compared to healthy volunteers. This microsatellite region contains multiple binding sites for a number of transcription factors, and we hypothesized that the polymorphic AAAG-containing sequence in the 5′ UTR region of ERR-γ might modulate expression of ERR-γ. We found that the 369 bp PCR product containing the AAAG repeat drove expression of a reporter gene in estrogen receptor positive breast cancer cells. Our results support a role for the 5′ UTR region in ERR-γ expression, which is potentially mediated via binding to the variable tandem AAAG repeat, the length of which correlates with breast cancer pre-disposition. Our study indicates that the AAAG tetranucleotide repeat polymorphism in ERR-γ gene 5′ UTR region may be a new biomarker for genetic susceptibility to breast cancer.
Keywords: Microsatellite, Breast cancer, AAAG, Polymorphism, Genetic predisposition
Introduction
Microsatellites are typically defined as tandemly repeated sequences (motifs) of one to six nucleotides that are very widely distributed throughout the genome and are frequently variable in the number of times the motif is repeated. Microsatellite repeats are ubiquitous and frequently polymorphic at rates that far exceed typical single-nucleotide mutation rates [1] in mammalian genomes, and their polymorphism can generate significant phenotype variation [2–4]. Somatic microsatellite length mutations are commonly observed in colorectal, endometrial, breast, gastric carcinomas, and some lung cancers [5–7]. The recurrence of microsatellite mutations in several loci in multiple different cancers, including known tumor suppressor genes (e.g. PTEN), is strong evidence that these microsatellite mutations are indeed important events in the progression of these cancers. Alterations in repeat unit number in and around coding sequences can have important quantitative and qualitative effects on gene expression [8–11] and thus could potentially contribute directly to cancer progression. We hypothesized that microsatellite alterations might play an important etiological role in the development and progression of breast cancer.
We surveyed ~100 individual microsatellite loci in genes suspected to play a role in breast cancer and found a highly polymorphic (AAAG/TTTC)n microsatellite in the 5′ untranslated region (UTR) of the estrogen-related receptor gamma (ERR-γ) gene. The ERR family members, which include ERR-α, ERR-β, and ERR-γ, are orphan nuclear receptors that regulate transcription via estrogen responses elements (EREs) and the closely related ERR response elements (ERREs) but do not bind endogenous estrogen [12]. ERR-α and ERR-γ can both substitute for the estrogen receptor (ER) [12], and all three ERR isoforms have been shown to play a role in HIF-mediated growth of solid tumors [13]. There is evidence to suggest that ERR-γ polymorphisms play a role in breast cancer susceptibility, particularly in post-menopausal women [14], and the chromosomal region containing ERR-γ (1q41) has been linked to breast cancer metastasis [15]. ERR-γ has also been shown to modulate tamoxifen resistance, and the expression of ERR-γ may be a marker of poor tamoxifen response [16].
Our hypothesis was that particular alleles of the (AAAG/TTTC)n microsatellite located in the 5′ untranslated region (UTR) of ERR-γ might affect differential expression of the gene. To examine this, we sequenced the upstream region of ERR-γ in a series of breast cancer patients in cancer-free individuals. We also performed reporter gene expression studies to validate the action of the variable nucleotide repeat (VNTR) as a classical regulatory domain.
Results
Identification of a putative predisposition biomarker for breast cancer
We have been systematically measuring the genotypes of certain 1 to 6-mer repeat motif containing loci within the genomes of cancer patient germline and tumor samples and the genomes of cancer-free individuals. Depending on the exact bioinformatic approach used to identify and count microsatellite containing loci within the reference genome, there are between ~500,000 and ~2,000,000 such loci. Loci were selected and prioritized based on likelihood of polymorphism and proximity to genes known or suspected to be involved cancer development and progression. One of the motifs that we chose to examine (AAAG) represents a relatively small repeat unit size, suggesting a higher likelihood for polymorphism, is prevalent in the genome, and is harbored within a large number of genes that are implicated in cancer. For loci containing this motif, we found 14,311 copies in the entire genome, 4,127 of which are located within genes (exons, introns, UTRs, upstream and downstream areas). When limited to the 7,183 “cancer” genes (defined as those genes found in NCBI's Entrez Gene using the search terms “cancer” and “tumor”), we found 128 in the 5′ UTR and 27 in the potential promoter region, which we defined as 1 kb upstream of those genes. When the upstream region was expanded to include 5 and 10 kb, we found 143 and 266 AAAG repeats that could potentially be located within transcriptional regulatory regions.
We prioritized each (AAAG)n locus by copy number, which is positively correlated with a higher likelihood of being polymorphic [17] and subsequently designed and tested 28 PCR primer sets against a panel of 42 samples that included 17 cancer-free volunteers, 17 cancer cell lines, and a variety of controls. We found 11 of these loci to be polymorphic (i.e., 10 that exhibit different sizes and one that is frequently deleted) in the human samples. Of the 11 polymorphic markers, two were of particular interest. One of the two markers containing an AAAG repeat, found in the TBL1Y gene located on the Y chromosome was absent in all female samples. However, this microsatellite was also absent in some lung tumor cell lines but not in their matching B lymphocyte-derived cell lines, consistent with frequent deletion of the entire Y chromosome in some non-small cell carcinomas [18]. The second interesting AAAG tandem repeat locus is located in the 5′ UTR of ERR-γ gene (estrogen-related receptor gamma, ESRRG, located on chromosome 1q41), which has 10 copies of the 4-mer (AAAG) motif, as found in the reference human genome sequence in the UCSC genome browser. ERR-γ is an orphan nuclear receptor and operates independently of estrogen; however, ERR-γ is known to bind to certain estrogen response elements to activate transcription [19]. Also, ERR-γ and its known co-activators have been linked to breast, ovarian, and colon cancer [20] and more recently to tamoxifen resistance in invasive lobular carcinoma of the breast [16].
ERR-γ has two known isoforms, one with an alternative first exon and one with an alternative 5′ UTR. It is possible that the differential AAAG microsatellite length confers alternate regulation of ERR-γ, as is thought to be the case for the gene encoding the parathyroid hormone receptor, which also harbors a polymorphic (AAAG)n repeat sequence in its promoter region that co-varies with adult height [21]. There are 22 candidate transcription factors (data not shown) that could potentially bind to the region of the 5′ UTR of ERR-γ containing the AAAG repeat (the repeat itself plus 100 bp flanking sequences), one of which (paired box gene 2, PAX2) is capable of binding the repeat unit itself.
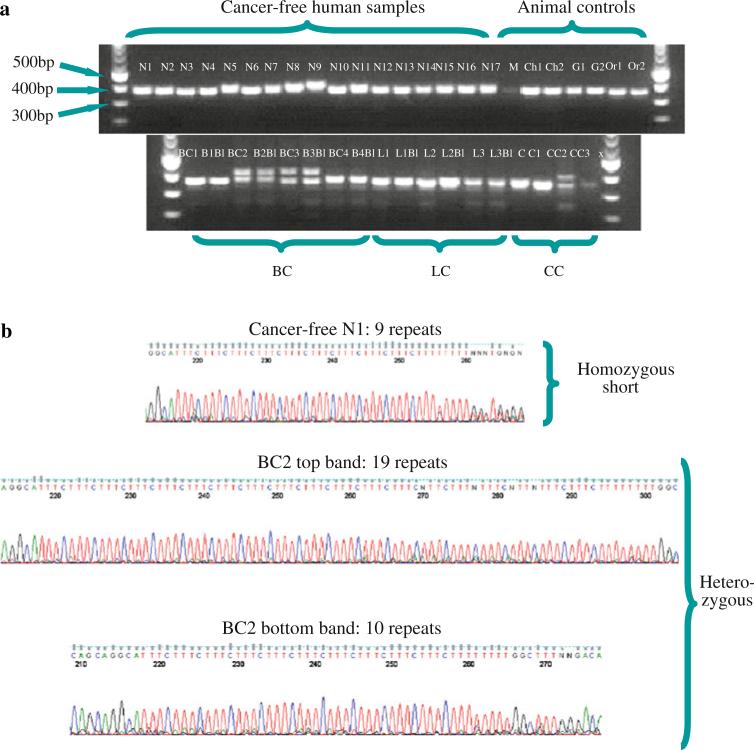
As shown in Fig. 1, two of the four breast cancer cell lines were heterozygous at the ERR-γ (AAAG)n locus, as were the matched blood lines and one of the colon cancer cell lines. Sequencing of the 42 samples indicated that homozygous samples carry a short version of the microsatellite, which ranges between 7 and 12 repeat units, and heterozygous samples carry one short copy and one longer allele ranging from 13–21 repeat units. The frequency of this variation was then measured by sequencing this locus in an expanded set of 528 samples, including 147 breast cancer patients, 104 patients with colorectal neoplasms, 24 prostate cancer patients, 20 cervical cancer patients, 28 ovarian cancer patients, 31 lung cancer cell lines, and 174 cancer-free volunteers with and without a family history of breast cancer.
Fig. 1.
A polymorphic AAAG repeat-containing sequence in the 5′ UTR of ERR-γ is expanded in some cancer cell lines. A gel survey of the ERR-γ locus was followed by sequencing of each of the PCR products. a The expected product size of the PCR amplicon was 369 bp. PCR amplicons show that all cancer free humans samples (H1-17) possess 7–10 tandem copies of AAAG within the 5′ UTR of the ERR-γ gene (18q21.2), while breast cancer 2 and 3 (BC2 and BC3, HCC2157, and HCC1187 cell lines, respectively) with their matched blood lines (B2Bl, B3Bl), as well as colorectal cancer 3 (CC3, RKO cell line) are heterozygous at the loci, with upper bands ranging from 19–21 repeats. To validate polymorphism specificity in human disease, a series of animal controls were also used: M mouse, Ch chimpanzee, G gorilla and O orangutan. b The band for a cancer-free individual (N1) and upper/lower bands from a heterozygous breast cancer (BC) PCR sample were gel-purified and sequenced, confirming the normal nine copies of the AAAG repeat and products of differing lengths in a heterozygous breast cancer sample. Samples details are provided as Online Resource 3)
Based on genotyping results, the size of the AAAG microsatellite allele ranged between 5 and 21 copies. We chose 13 motif copies as the cut-off length for classification as “long”, as this number was the most rare among samples (only one patient with an allele of this length), and 12 copies was relatively common and equally observed (4–6 incidences) for each class of sample (e.g., cancer and non-cancer). Based on these criteria, carriers and non-carriers of the longer allele for each category of patient are presented in Table 1. As shown, a statistically significant higher incidence of long allele carriers (P value = 0.0195, two tailed Fisher's exact test) was observed for breast cancer patients (14.3%), compared to healthy volunteers (4.8%), which translates to a relative risk ratio of 3 (14.3/4.8). The incidence of carriers in patients without cancer but with a known family history of breast cancer (7.2%), on the other hand, was slightly higher than cancer-free volunteers but lower than breast or colon cancer patients. Likewise, only 7.3% of patients with other cancers (colorectal, lung, prostate, ovarian, or cervical) were carriers of the longer allele. Our results indicate a possible hereditary trend for breast cancer, which does not appear to translate to other potentially heritable cancers, such as ovarian cancer, which is known to be linked to familial (especially BRCA1/2-associated) breast cancer [22]. However, a much larger population is needed to definitively determine the potential contribution of this locus to risk for a spectrum of hereditary cancers.
Table 1.
Summary of the incidence of the ERR-γ repeat in patient samples
| Long allele non-carriers | Long allele carriers | Totals | Incidence (%) | Statistics (P value) |
||
|---|---|---|---|---|---|---|
| Baseline group |
||||||
| Healthy: no BC family hx n = 105 | Healthy: all n = 174 | |||||
| Healthy volunteers | ||||||
| No BC family hx | 100 | 5 | 105 | 4.8 | – | 0.799 |
| BC family hx | 64 | 5 | 69 | 8.2 | 0.520 | 0.514 |
| All healthy volunteers | 164 | 10 | 174 | 5.7 | 0.791 | – |
| Cancer patients | ||||||
| Breast cancer | 126 | 21 | 147 | 14.3* | 0.020 | 0.013 |
| Other cancers | 192 | 15 | 207 | 7.3 | 0.471 | 0.791 |
| Total | 482 | 46 | 528 | 8.7 | – | – |
Note: “BC family hx” refers to first degree (sister, daughter, or mother) or second degree (grandmother, aunt, niece, or granddaughter) family members with breast cancer. “Carriers” refer to persons in which the long allele (defined as at least 13 copies of the AAAG motif) is present. Asterisk indicates a statistically significant difference between the incidence of long allele carriers in breast cancer patients, compared to cancer-free volunteers, based on Fisher's exact test (P value = 0.0134, two-tailed comparison). A detailed list of patients and genotyping information is provided as Online Resource 1
BC breast cancer, hx history
The allele in the human reference sequence genome contains eight copies, though this allele was relatively rare among the patient samples we tested (only 48 alleles were found with eight copies of the motif, compared to 369, 181 and 119 alleles that had 7, 9, and 10 copies, respectively). Observed allelic frequencies of long (n = 13+ copies of AAAG) and short alleles is consistent with Hardy–Weinberg equilibrium. No correlation related to gender (the majority of samples, ~80%, were female) or race/ethnicity was apparent (Online Resource 2), although a much larger patient population would be required to confirm this.
The AAAG-containing promoter region drives luciferase reporter activity
There is evidence to support the involvement of this particular repetitive DNA sequence (i.e., the AAAG motif) in mismatch repair bias, phenotypic variation, and cancer [21, 23, 24]. For instance, a functional (AAAG)5-7 polymorphism in the promoter region of the parathyroid hormone gene was shown to correlate with adult height and bone mineral density in women [21, 23]. The AAAG repeat is located in the most proximal promoter of the parathyroid hormone type I receptor (PTHR1), which mediates skeletal growth. In an osteoblast-like cell line, the AAAG-containing promoter was shown to drive expression of a reporter gene, with the length of the AAAG repeat itself modulating the magnitude of reporter gene expression [23].
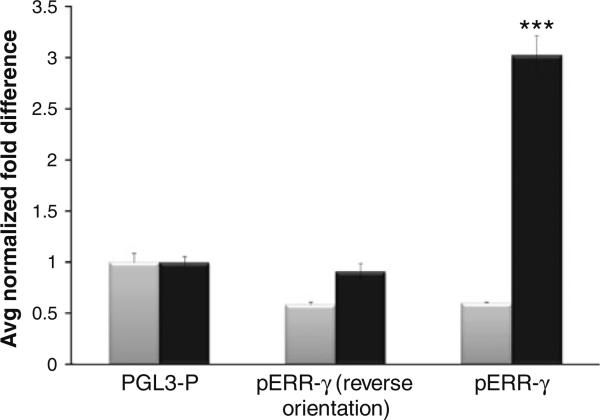
We hypothesized that the AAAG repeat microsatellite might perform a similar function in the ERR-γ promoter. To examine this notion, we performed luciferase assays in ER+ and ER– cells (MCF-7 and MBA-MD-231, respectively), using a plasmid containing a 369 bp PCR fragment of the ERR-γ 5′ UTR amplified from commercial human DNA, including nine copies of the AAAG sequence. This construct drove transcription of luciferase (~threefold, P value < 0.0001, n = 8) in MCF-7 cells, but not in the ER-cell line, MBA-MD-231 (Fig. 2). Our results are consistent with the presence of ER influencing the expression of ERR-γ. Alternatively, disparate results between MCF7 and MBA-MD-231 cells might be due to differences in the transcription factor complement of the two cell lines. Nevertheless, the AAAG domain can support cell-specific reporter gene expression.
Fig. 2.
The AAAG repeat-containing sequence in the 5′ UTR of ERR-γ drives the expression of firefly luciferase in estrogen receptor positive (ER+) MCF-7 cells but not in ER negative (ER-) MDA-MB-231 cells. The activity of the AAAG repeat-containing ERR-γ promoter-luciferase constructs representing the reference genome polymorphic variant (nine copies of AAAG) was analyzed in transiently transfected human breast cancer cell lines (ER+ MCF-7 and ER– MDA-MB-231 cells). In MCF-7 cells (black bars), the correctly oriented (but not reverse oriented) ERR-γ fragment drove higher levels of luciferase reporter gene activity, compared to the pGL3-P backbone alone. However, this effect was not observed in ER- MDA-MB 231 cells (grey bars). Transfections were carried out in triplicate; each experiment was repeated thrice, and independently prepared plasmid DNAs were used for each promoter construct. Luciferase activity was measured and normalized to renillin, and mean fold changes compared to pGL3-P are given. Statistics were performed using Student's t test, with *** P < 0.0001 (compared to pGL3-P). SE are given as Y-axis error bars (n = 8)
Discussion
We discovered a variable repetitive microsatellite allele in the 5′ UTR of ERR-γ that exhibits a significantly higher incidence in patients with breast cancer. ERR-γ expression has previously been implicated as a potential prognostic marker in breast cancer [12, 16]. We hypothesized that the differential AAAG microsatellite confers alternate regulation of ERR-γ, similar to what has been observed for the polymorphic (AAAG)n repeat sequence located in the promoter of the gene encoding the parathyroid hormone receptor [21]. Our analysis indicated that the AAAG-containing 5′ UTR region of ERR-γ could indeed drive expression of a promoter gene. To identify putative transcription factors, the AAAG-containing region of ERR-γ, including 100 bp flanking sequences, was searched against the Transfac database using BLAST, MATCH, and TFSEARCH tools [25]. Based on this analysis, we found 22 candidate transcription factors that could potentially bind to this region (data not shown). This finding suggests a potential mechanism of action, but further studies would be required to determine if any of these transcription factor binding sites in close proximity to the repeat are affected by (AAAG)n length variations.
Because microsatellites have in many cases been shown to impact expression of associated genes [2, 26], it is interesting to speculate that ERR-γ expression differences related to different AAAG copy numbers may impact breast cancer risk. If the frequency of this potentially predictive marker is sustained in a larger population, and the direct mechanistic link by which it confers the cancer phenotype can be verified, it may contribute substantially to our understanding of this cancer and as a biomarker offering better informed surveillance, prophylactic surgery, and chemoprevention options to patients. Based on our assessment, this allele carries a relative risk of 3 for the development of breast cancer. As a comparison, deleterious germline mutations of the BRCA1 gene have a 3–7% frequency in breast cancer patients (age < 45), which is significantly elevated in those with a family history (up to 33%). Such mutations are associated with a 3–7 times higher risk of breast cancer, compared to non-mutation carriers [27, 28]. The incidence of BRCA1 mutation in the general population is estimated at 0.2 to 0.4% [29].
There are at least five possible explanations for our results: (1) direct transcriptional influence of ERR-γ based on the length of the repeat, (2) linkage of an ancestral “lengthening” mutation with a cancer causing mutation in/ around ERR-γ, (3) the repeat resides in an uncharacterized biologically active RNA which is affected by the length of the repeat, (4) misregulation of splicing due to over-expansion of the polymorphic repeat, or (5) a spurious association due to various sampling errors or population issues (albeit unlikely). Our preliminary results strongly support a role for this repeat—or the immediate surrounding sequence—in the transcription of ERR-γ. To our knowledge, there are no previous descriptions of an association of a polymorphic microsatellite repeat with breast cancer susceptibility with the notable exception of a tandem repeat biomarker located in the first exon of GPX1, the gene encoding glutathione peroxidase 1 [30]. However, there is precedence for the involvement of this particular repetitive DNA sequence (i.e., the AAAG motif) in mismatch repair bias, phenotypic variation, and cancer [21, 23, 24].
In cancer, variations in AAAG are considered to be a novel form of microsatellite instability, termed elevated microsatellite alterations at selected tetranucleotide repeats (EMAST). EMAST has been proposed to account for some sporadic cancers that do not have defects in the most commonly mutated mismatch repair genes, hMLH1 and hMSH2 [31]. As such, AAAG polymorphism in various genes has been used diagnostically to detect a variety of cancers, including non-small cell lung cancer [32, 33], urinary tract tumors [34], skin cancer [35], and ovarian cancer, particularly advanced serous carcinomas [36]. More recently, it was shown that the mismatch repair enzyme human postmeiotic segregation 2 (hPMH2), exhibits repair bias specific to AAAG repeats throughout the genome [24], which could account for mismatch repair deficient colorectal cancers that are not indentified by classical microsatellite instability tests designed to detect only hypervariable mononucleotide and dinucleotide repeats (Bethesda markers).
Methods
Sample acquisition and preparation
Genomic DNA was extracted from blood samples collected from volunteers (Online Resource 1) by the McDermott Center for Human Growth and Development Genetics Clinical Laboratory in accordance with Institutional Review Board (UTSW IRB# 1287-355). Most cell lines were provided by Drs. Girard, Minna, and Boothman. Patient samples were provided by Drs. Perou, Lewis, and the UTSW Tissue Repository, with each institution's review board approval. All other genomic DNA was purchased from Coriell Cell Repositories (Camden, NJ, USA) or American Type Culture Collection (Manassas, VA, USA).
Genotyping
Forward (5′-ACCTAGGAGATAGAGGTTGC-3′) and reverse (5′-CTTCTTCTGCACTATCAGGG-3′) primers were designed to amplify a 369 bp length fragment of the ERR-γ gene including the 5′ UTR AAAG repetitive sequence. PCR was performed using Promega 2× PCR Master Mix (Promega) per manufacturer instructions. Products were gel-purified using Qiagen gel extraction kit (Qiagen, Valencia, California) and sequenced by the McDermott Center Sequencing Core Facility. Hardy– Weinberg equilibrium was tested using χ2 test of goodness of fit, with 1 degree of freedom, checking for long and short allele distribution (where “long” is defined as 13+ copies of the AAAG motif, and “short” is defined as 13 or fewer than 13 copies). To identify putative transcription factors, the AAAG-containing region of ERR-γ, including 100 bp flanking sequences, was searched against the Transfac database using BLAST, MATCH, and TFSEARCH tools [25].
Cloning
The PCR products were subsequently blunt end cloned into the pGL3p (Promega) luciferase reporter gene vector with an SV-40 promoter. The original genotyping forward primer was modified to incorporate a Hind III restriction enzyme site to determine the orientation of the insert once cloned (5′ TAAGCTTACCTAGGAGATAGAGGTTGC 3′). PCR was conducted using a blunt end generating phu polymerase, the original reverse genotyping primer and the modified forward primer. pGL3p was digested with the Sma I restriction endonuclease and incubated with purified PCR products and T4 ligase (New England Biolabs) and competent E. coli were transformed with the resulting construct and plated onto ampicillin-containing agar plates.
Cell transfection
MCF-7 or MDA-MB 134 cells were plated out onto sterile 24-well plates and maintained until 70% confluent. Immediately prior to transfection, cells were washed with PBS and fresh medium was added. Cells were transfected with ExGen 500 (Fermentas) according to manufacturer's guidelines. Briefly, 1 μg reporter gene construct (pGL3-p-ESRRG-native or pGL3p-reverse) and 20 nanograms of TK renillin (Promega) were combined with each construct to normalize and act as an internal control. This was combined with 3.3 μl of ExGen 500/1 μg DNA and incubated for 20 min at room temperature. The resulting ExGen-DNA mix was added to each well of cells and incubated for 48 h before being processed for the luciferase assay.
Luciferase reporter gene assay
Cells were washed twice in sterile PBS, lysed with 100 μl passive lysis buffer (Promega) and incubated with gently rocking for 20 min. Lysates were transferred to 1.5 ml Eppendorf tubes and briefly centrifuge at 1,500 rpm for 1 min to remove cellular debris. Dual luciferase assay was performed by a GlomaxTM 96 Microplate Luminometer (Promega) according to manufacturer's instructions. The ratio of Firefly luciferase to renillin luciferase units were calculated and compared to reporter gene construct alone (no expression constructs transfected).
Supplementary Material
Acknowledgments
This work was funded by the P.O'B. Montgomery Distinguished Chair, the Hudson Foundation and directors funds at the Virginia Bioinformatics Institute. This work was also partially supported by the University of Texas NIH/NCI SPORE in Lung Cancer (P50CA70907). We would like to extend a special thanks to Linda Gunn, Zhaohui Sun, and Tanishia Choice, M.D. for their valuable technical assistance. We also appreciate the assistance of Jennifer Sayne, manager of the UTSW Tissue Repository, who provided the majority of patient samples used in this study, and Jingshen Yan with the Division of Biostatistics, Department of Clinical Sciences at UTSW, who provided statistical assistance. Dr. Adi Gazdar, with the Department of Pathology at UTSW and Dr. Cheryl Lewis, with the Department of Surgery at UTSW, provided additional cell lines and patient samples.
Footnotes
Electronic supplementary material The online version of this article (doi:10.1007/s10549-010-1237-9) contains supplementary material, which is available to authorized users.
Conflicts of interest None of the authors have any conflicts of interest to declare. Authors also do not have a financial relationship with any of the organizations that sponsored the research. Authors have full control of all primary data and agree to allow the journal to review the data if requested.
Contributor Information
C. L. Galindo, Virginia Bioinformatics Institute, Virginia Polytechnic Institute and State University, Washington Street (0477), Blacksburg, VA 24601-0477, USA
J. F. McCormick, Virginia Bioinformatics Institute, Virginia Polytechnic Institute and State University, Washington Street (0477), Blacksburg, VA 24601-0477, USA
V. J. Bubb, School of Biomedical Sciences, University of Liverpool, Sherrington Buildings, Ashton Street, L69 3GE Liverpool, UK
D. H. Abid Alkadem, School of Biomedical Sciences, University of Liverpool, Sherrington Buildings, Ashton Street, L69 3GE Liverpool, UK.
Long-Shan Li, Simmons Comprehensive Cancer Center, University of Texas Southwestern Medical Center, 5323 Inwood Rd, Dallas, TX 75395, USA.
L. J. McIver, Virginia Bioinformatics Institute, Virginia Polytechnic Institute and State University, Washington Street (0477), Blacksburg, VA 24601-0477, USA
A. C. George, Virginia Bioinformatics Institute, Virginia Polytechnic Institute and State University, Washington Street (0477), Blacksburg, VA 24601-0477, USA
D. A. Boothman, Simmons Comprehensive Cancer Center, University of Texas Southwestern Medical Center, 5323 Inwood Rd, Dallas, TX 75395, USA
J. P. Quinn, School of Biomedical Sciences, University of Liverpool, Sherrington Buildings, Ashton Street, L69 3GE Liverpool, UK
M. A. Skinner, Department of Surgery, University of Texas Southwestern Medical Center, Dallas, TX, USA Children's Medical Center, 5323 Harry Hines Blvd, Dallas, TX 75395, USA.
H. R. Garner, Virginia Bioinformatics Institute, Virginia Polytechnic Institute and State University, Washington Street (0477), Blacksburg, VA 24601-0477, USA
References
- 1.Ellegren H. Microsatellites: simple sequences with complex evolution. Nat Rev Genet. 2004;5:435–445. doi: 10.1038/nrg1348. [DOI] [PubMed] [Google Scholar]
- 2.Fujisawa T, Ikegami H, Kawaguchi Y, Yamato E, Nakagawa Y, Shen GQ, Fukuda M, Ogihara T. Length rather than a specific allele of dinucleotide repeat in the 5′ upstream region of the aldose reductase gene is associated with diabetic retinopathy. Diabet Med. 1999;16:1044–1047. doi: 10.1046/j.1464-5491.1999.00192.x. [DOI] [PubMed] [Google Scholar]
- 3.Laidlaw J, Gelfand Y, Ng KW, Garner HR, Ranganathan R, Benson G, Fondon JW., III Elevated basal slippage mutation rates among the Canidae. J Hered. 2007;98:452–460. doi: 10.1093/jhered/esm017. [DOI] [PubMed] [Google Scholar]
- 4.Rubinsztein DC, Leggo J, Coetzee GA, Irvine RA, Buckley M, Ferguson-Smith MA. Sequence variation and size ranges of CAG repeats in the Machado-Joseph disease, spinocerebellar ataxia type 1 and androgen receptor genes. Hum Mol Genet. 1995;4:1585–1590. doi: 10.1093/hmg/4.9.1585. [DOI] [PubMed] [Google Scholar]
- 5.Forgacs E, Wren JD, Kamibayashi C, Kondo M, Xu XL, Markowitz S, Tomlinson GE, Muller CY, Gazdar AF, Garner HR, Minna JD. Searching for microsatellite mutations in coding regions in lung, breast, ovarian and colorectal cancers. Oncogene. 2001;20:1005–1009. doi: 10.1038/sj.onc.1204211. [DOI] [PubMed] [Google Scholar]
- 6.Girard L, Zochbauer-Muller S, Virmani AK, Gazdar AF, Minna JD. Genome-wide allelotyping of lung cancer identifies new regions of allelic loss, differences between small cell lung cancer and non-small cell lung cancer, and loci clustering. Cancer Res. 2000;60:4894–4906. [PubMed] [Google Scholar]
- 7.Wistuba II, Behrens C, Virmani AK, Mele G, Milchgrub S, Girard L, Fondon JW, 3rd, Garner HR, McKay B, Latif F, Lerman MI, Lam S, Gazdar AF, Minna JD. High resolution chromosome 3p allelotyping of human lung cancer and preneoplastic/preinvasive bronchial epithelium reveals multiple, discontinuous sites of 3p allele loss and three regions of frequent breakpoints. Cancer Res. 2000;60:1949–1960. [PubMed] [Google Scholar]
- 8.Bontekoe CJ, Bakker CE, Nieuwenhuizen IM, van der Linde H, Lans H, de Lange D, Hirst MC, Oostra BA. Instability of a (CGG)98 repeat in the Fmr1 promoter. Hum Mol Genet. 2001;10:1693–1699. doi: 10.1093/hmg/10.16.1693. [DOI] [PubMed] [Google Scholar]
- 9.Di Marco S, Hel Z, Lachance C, Furneaux H, Radzioch D. Polymorphism in the 3′-untranslated region of TNFalpha mRNA impairs binding of the post-transcriptional regulatory protein HuR to TNFalpha mRNA. Nucl Acids Res. 2001;29:863–871. doi: 10.1093/nar/29.4.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fondon JW, 3rd, Garner HR. Molecular origins of rapid and continuous morphological evolution. Proc Natl Acad Sci USA. 2004;101:18058–18063. doi: 10.1073/pnas.0408118101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tassone F, Hagerman RJ, Chamberlain WD, Hagerman PJ. Transcription of the FMR1 gene in individuals with fragile X syndrome. Am J Med Genet. 2000;97:195–203. doi: 10.1002/1096-8628(200023)97:3<195::AID-AJMG1037>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 12.Ariazi EA, Jordan VC. Estrogen-related receptors as emerging targets in cancer and metabolic disorders. Curr Top Med Chem. 2006;6:203–215. doi: 10.2174/1568026610606030203. [DOI] [PubMed] [Google Scholar]
- 13.Ao A, Wang H, Kamarajugadda S, Lu J. Involvement of estrogen-related receptors in transcriptional response to hypoxia and growth of solid tumors. Proc Natl Acad Sci USA. 2008;105:7821–7826. doi: 10.1073/pnas.0711677105. doi:10.1073/pnas.0711677105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sangrajrang S, Sato Y, Sakamoto H, Ohnami S, Laird NM, Khuhaprema T, Brennan P, Boffetta P, Yoshida T. Genetic polymorphisms of estrogen metabolizing enzyme and breast cancer risk in Thai women. Int J Cancer. 2009;125:837–843. doi: 10.1002/ijc.24434. doi: 10.1002/ijc.24434. [DOI] [PubMed] [Google Scholar]
- 15.Thomassen M, Tan Q, Kruse TA. Gene expression meta-analysis identifies chromosomal regions and candidate genes involved in breast cancer metastasis. Breast Cancer Res Treat. 2009;113:239–249. doi: 10.1007/s10549-008-9927-2. doi:10.1007/s10549-008-9927-2. [DOI] [PubMed] [Google Scholar]
- 16.Riggins RB, Lan JP, Zhu Y, Klimach U, Zwart A, Cavalli LR, Haddad BR, Chen L, Gong T, Xuan J, Ethier SP, Clarke R. ERRgamma mediates tamoxifen resistance in novel models of invasive lobular breast cancer. Cancer Res. 2008;68:8908–8917. doi: 10.1158/0008-5472.CAN-08-2669. doi: 10.1158/0008-5472.CAN-08-2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fondon JW, 3rd, Mele GM, Brezinschek RI, Cummings D, Pande A, Wren J, O'Brien KM, Kupfer KC, Wei MH, Lerman M, Minna JD, Garner HR. Computerized polymorphic marker identification: experimental validation and a predicted human polymorphism catalog. Proc Natl Acad Sci USA. 1998;95:7514–7519. doi: 10.1073/pnas.95.13.7514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Berrieman HK, Ashman JN, Cowen ME, Greenman J, Lind MJ, Cawkwell L. Chromosomal analysis of non-small-cell lung cancer by multicolour fluorescent in situ hybridisation. Br J Cancer. 2004;90:900–905. doi: 10.1038/sj.bjc.6601569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hong H, Yang L, Stallcup MR. Hormone-independent transcriptional activation and coactivator binding by novel orphan nuclear receptor ERR3. J Biol Chem. 1999;274:22618–22626. doi: 10.1074/jbc.274.32.22618. [DOI] [PubMed] [Google Scholar]
- 20.Ariazi EA, Clark GM, Mertz JE. Estrogen-related receptor alpha and estrogen-related receptor gamma associate with unfavorable and favorable biomarkers, respectively, in human breast cancer. Cancer Res. 2002;62:6510–6518. [PubMed] [Google Scholar]
- 21.Scillitani A, Jang C, Wong BY, Hendy GN, Cole DE. A functional polymorphism in the PTHR1 promoter region is associated with adult height and BMD measured at the femoral neck in a large cohort of young Caucasian women. Hum Genet. 2006;119:416–421. doi: 10.1007/s00439-006-0155-8. doi:10.1007/s00439-006-0155-8. [DOI] [PubMed] [Google Scholar]
- 22.Jatoi I, Anderson WF. Management of women who have a genetic predisposition for breast cancer. Surg Clin North Am. 2008;88:845–861. vii–viii. doi: 10.1016/j.suc.2008.04.007. [DOI] [PubMed] [Google Scholar]
- 23.Minagawa M, Yasuda T, Watanabe T, Minamitani K, Takahashi Y, Goltzman D, White JH, Hendy GN, Kohno Y. Association between AAAG repeat polymorphism in the P3 promoter of the human parathyroid hormone (PTH)/PTH-related peptide receptor gene and adult height, urinary pyridinoline excretion, and promoter activity. J Clin Endocrinol Metab. 2002;87:1791–1796. doi: 10.1210/jcem.87.4.8419. [DOI] [PubMed] [Google Scholar]
- 24.Shah SN, Eckert KA. Human postmeiotic segregation 2 exhibits biased repair at tetranucleotide microsatellite sequences. Cancer Res. 2009;69:1143–1149. doi: 10.1158/0008-5472.CAN-08-3499. doi:10.1158/0008-5472.CAN-08-3499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Heinemeyer T, Wingender E, Reuter I, Hermjakob H, Kel AE, Kel OV, Ignatieva EV, Ananko EA, Podkolodnaya OA, Kolpakov FA, Podkolodny NL, Kolchanov NA. Databases on transcriptional regulation: TRANSFAC, TRRD and COMPEL. Nucl Acids Res. 1998;26:362–367. doi: 10.1093/nar/26.1.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fondon JW, III, Garner HR. Detection of length-dependent effects of tandem repeat alleles by 3-D geometric decomposition of craniofacial variation. Dev Genes Evol. 2007;217:79–85. doi: 10.1007/s00427-006-0113-4. [DOI] [PubMed] [Google Scholar]
- 27.King MC, Marks JH, Mandell JB. Breast and ovarian cancer risks due to inherited mutations in BRCA1 and BRCA2. Science. 2003;302:643–646. doi: 10.1126/science.1088759. [DOI] [PubMed] [Google Scholar]
- 28.Malone KE, Daling JR, Thompson JD, O'Brien CA, Francisco LV, Ostrander EA. BRCA1 mutations and breast cancer in the general population: analyses in women before age 35 years and in women before age 45 years with first-degree family history. JAMA. 1998;279:922–929. doi: 10.1001/jama.279.12.922. [DOI] [PubMed] [Google Scholar]
- 29.Schwartz GF, Hughes KS, Lynch HT, Fabian CJ, Fentiman IS, Robson ME, Domchek SM, Hartmann LC, Holland R, Winchester DJ, Anderson BO, Arun BK, Bartelink H, Bernard P, Bonanni B, Cady B, Clough KB, Feig SA, Heywang-Kobrunner SH, Howell A, Isaacs C, Kopans DB, Mansel RE, Masood S, Palazzo JP, Pressman PI, Solin LJ, Untch M. Proceedings of the international consensus conference on breast cancer risk, genetics, & risk management, April, 2007. Breast J. 2009;15:4–16. doi: 10.1111/j.1524-4741.2008.00665.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hu YJ, Diamond AM. Role of glutathione peroxidase 1 in breast cancer: loss of heterozygosity and allelic differences in the response to selenium. Cancer Res. 2003;63:3347–3351. [PubMed] [Google Scholar]
- 31.Boland CR, Thibodeau SN, Hamilton SR, Sidransky D, Eshleman JR, Burt RW, Meltzer SJ, Rodriguez-Bigas MA, Fodde R, Ranzani GN, Srivastava S. A National Cancer Institute Workshop on Microsatellite Instability for cancer detection and familial predisposition: development of international criteria for the determination of microsatellite instability in colorectal cancer. Cancer Res. 1998;58:5248–5257. [PubMed] [Google Scholar]
- 32.Ahrendt SA, Decker PA, Doffek K, Wang B, Xu L, Demeure MJ, Jen J, Sidransky D. Microsatellite instability at selected tetranucleotide repeats is associated with p53 mutations in non-small cell lung cancer. Cancer Res. 2000;60:2488–2491. [PubMed] [Google Scholar]
- 33.Xu L, Chow J, Bonacum J, Eisenberger C, Ahrendt SA, Spafford M, Wu L, Lee SM, Piantadosi S, Tockman MS, Sidransky D, Jen J. Microsatellite instability at AAAG repeat sequences in respiratory tract cancers. Int J Cancer. 2001;91:200–204. doi: 10.1002/1097-0215(200002)9999:9999<::aid-ijc1031>3.0.co;2-0. doi:10.1002/1097-0215(20010115)91:2<200::AID-IJC1031>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 34.Catto JW, Azzouzi AR, Amira N, Rehman I, Feeley KM, Cross SS, Fromont G, Sibony M, Hamdy FC, Cussenot O, Meuth M. Distinct patterns of microsatellite instability are seen in tumours of the urinary tract. Oncogene. 2003;22:8699–8706. doi: 10.1038/sj.onc.1206964. doi: 10.1038/sj.onc.1206964. [DOI] [PubMed] [Google Scholar]
- 35.Danaee H, Nelson HH, Karagas MR, Schned AR, Ashok TD, Hirao T, Perry AE, Kelsey KT. Microsatellite instability at tetranucleotide repeats in skin and bladder cancer. Oncogene. 2002;21:4894–4899. doi: 10.1038/sj.onc.1205619. doi:10.1038/sj.onc.1205619. [DOI] [PubMed] [Google Scholar]
- 36.Singer G, Kallinowski T, Hartmann A, Dietmaier W, Wild PJ, Schraml P, Sauter G, Mihatsch MJ, Moch H. Different types of microsatellite instability in ovarian carcinoma. Int J Cancer. 2004;112:643–646. doi: 10.1002/ijc.20455. doi:10.1002/ijc.20455. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.