The subcellular localization and trafficking of cellular RNAs are essential processes governing plant growth and development. RNAs are targeted to specific regions/organelles mainly by active transport that requires RNA zip codes that are recognized by specific RNA-binding activities, accessory proteins such as adaptors for cytoskeletal movement and, in some instances, components for translation (Okita and Choi, 2002; Michaud et al., 2010). Unlike the progress made in studies on cell-to-cell and long-distance RNA trafficking (Lucas et al., 2001; Gómez and Pallas, 2004; Fernandez-Calvino et al., 2011; Pallas et al., 2011), our understanding of intracellular RNA trafficking in plants remains limited.
Viroids, the smallest noncoding RNAs able to subvert plant cell metabolism to induce pathogenesis in their hosts, have recently emerged as ideal probes for exploring intracellular RNA trafficking (Wang and Ding, 2010). Viroids are single-stranded circular RNAs ranging in size from 250 to 400 nucleotides with a high degree of self-complementarity that promotes compact folding (Tabler and Tsagris, 2004; Flores et al., 2005; Ding, 2009). Lacking mRNA activity, these pathogenic RNAs are completely dependent on functional genomic domains that interact with host factors to complete their life cycle in the infected cell (Diener, 1987). Viroids belong to one of two families: Pospiviroidae, the replication of which takes place in the nucleus, and Avsunviroidae, whose members possess ribozyme activity and replicate in the chloroplasts of infected cells (Tabler and Tsagris, 2004; Flores et al., 2005; Ding, 2009).
Upon mechanical introduction into a susceptible cell, viroid infection comprises a series of sequential events: (1) subcellular compartmentalization for replication; (2) export to neighboring cells; and (3) entry into vascular tissue for long-distance trafficking and to establish systemic infection (Flores et al., 2005; Ding, 2009). In these circumstances, trafficking into cellular organelles (chloroplasts for Avsunviroidae and nucleus for Pospiviroidae) is the first hurdle to be overcome by the viroids after infecting the host. As a consequence, elucidating how these noncoding RNAs are trafficked to the nucleus and chloroplasts to initiate their replication is fundamental to fully understand the biology of these RNAs. Furthermore, such knowledge would yield insights into the poorly understood mechanisms of the intracellular localization of RNAs in plant cells (Ding and Itaya, 2007).
Initial evidence suggested that the subcellular localization of viroids in the Pospiviroidae family is mediated in cis by RNA sequences or structural motifs, which are required for nuclear import by a specific receptor via a cytoskeleton-independent route (Woo et al., 1999; Zhao et al., 2001; Abraitiene et al., 2008). However, how members of the Avsunviroidae are translocated into the chloroplast is an intriguing and challenging question because they are the only known pathogenic RNAs able to traffic into this organelle (Daròs et al., 2006; Wang and Ding, 2010). Although the mechanism of import of RNAs into chloroplasts has previously been suggested for both nucleus-encoded tRNAs (Bungard, 2004) and the mRNA coding for the eukaryotic initiation factor 4E (Nicolaï et al., 2007), the plant cell endogenous mechanisms for RNA import that are exploited by viroids when entering chloroplasts remain unknown.
In recent work, we demonstrated that a chimeric (containing partial length plus- and minus-RNA) sequence derived from Eggplant latent viroid (ELVd; a member of the Avsunviroidae family) acting as the 5′ untranslated region (UTR) end was able to mediate the specific trafficking and accumulation of functional GFP mRNA from the nucleus into chloroplasts (Gómez and Pallas, 2010a, 2010b). This formed the experimental basis for proposing the existence of a novel plant signaling mechanism (mediated by noncoding RNAs) able to regulate the selective import of nuclear transcripts into chloroplasts. Furthermore, we speculated that Avsunviroidae members may subvert this signaling mechanism to mediate their specific trafficking to this organelle. However, this pathway requires the existence of a previous step involving the transport of the viroid from the cytoplasm to the nucleus, such that during the initial phase of their life cycle Avsunviroidae members would be first transported from the cytoplasm into the nucleus and then specifically delivered to the chloroplasts, where they replicate.
To determine whether this RNA import pathway is used by members of the Avsunviroidae family for transit to the chloroplasts, we analyzed the main stages in this route. First, we developed a potato virus X (PVX)-based in vivo assay that used a GFP-intron-ELVd construct as a reporter to determine whether this RNA is able to traffic from the cytoplasm to the nucleus. Next, we used an engineered reporter containing the same full-length ELVd sequence fused as the 5′ UTR end to the GFP cDNA. This construct was used in transient expression assays to confirm the selective trafficking of canonical ELVd monomeric sequence from the nucleus into the chloroplast. The results shown here provide evidence that in members of the Avsunviroidae family, selective intracellular trafficking is a complex process comprising a first nuclear step prior to their delivery to the chloroplast of the infected cell.
ELVd RNA MOVES FROM CYTOPLASM TO NUCLEUS
To determine whether ELVd is able to traffic into the nucleus of the host cell, we used an experimental approach similar to that previously employed to demonstrate the selective import of Potato spindle tuber viroid into the nucleus of Nicotiana benthamiana cells (Zhao et al., 2001; Abraitiene et al., 2008). First, the coding region of the GFP was interrupted by the insertion of an intron (IV2) derived from the potato (Solanum tuberosum) ST-LS1 gene (Eckes et al., 1986). The monomeric plus strand of the full-length ELVd sequence (Fadda et al., 2003) was subsequently inserted into the intron to generate an IV2/ELVd-bearing GFP construct (Supplemental Materials and Methods S1). This reporter was cloned in a PVX-based vector (pP2C2S; Chapman et al., 1992; Fig. 1A). These constructs were transcribed in vitro and used to inoculate N. benthamiana leaves. During PVX replication, the different reporters used in this assay were expressed in the cytoplasm of the infected cells.
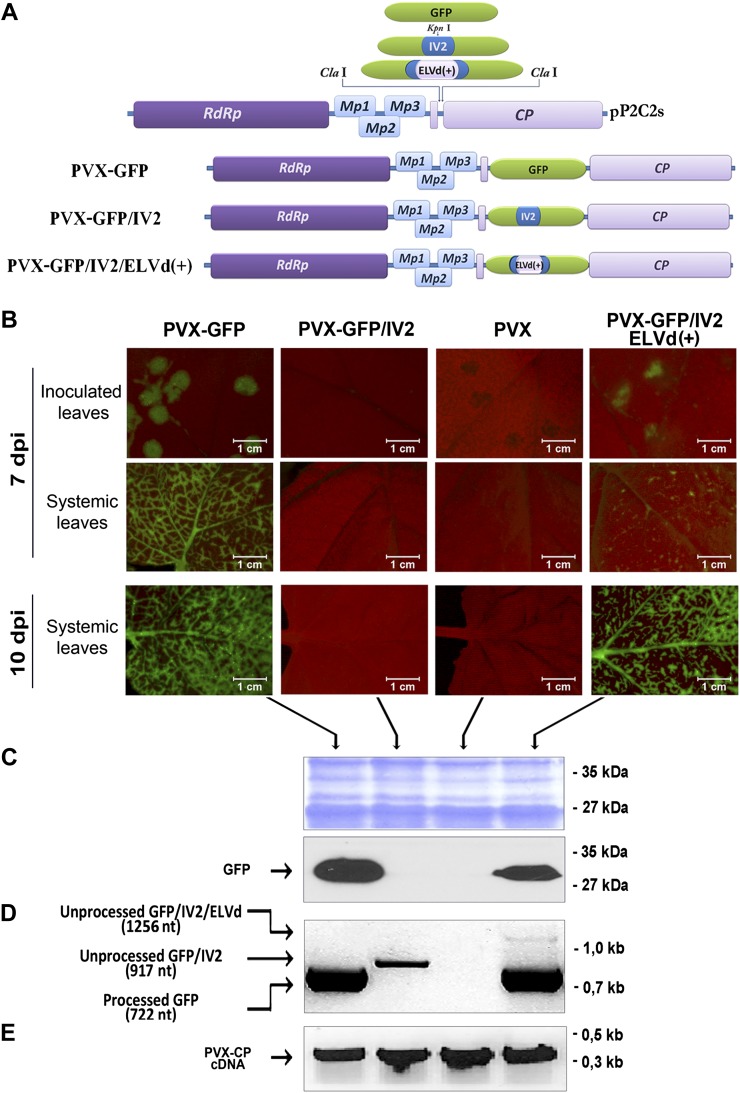
Figure 1.
Analysis of GFP expression in plants infected with PVX-derived transcripts. A, Physical map of the PVX-derived construct used in this work. PVX-based vector P2C2S and cDNAs (GFP, GFP/IV2, and GFP/IV2/ELVd) cloned under the control of the duplicated PVX-CP promoter are shown at top. The bottom shows representations of PVX-GFP, PVX-GFP/IV2, and PVX-GFP/IV2/ELVd(+) chimeric constructs. The constructs are not drawn to scale. B, GFP fluorescence stereomicroscopy images of N. benthamiana leaves inoculated with PVX (unmodified), PVX-GFP, PVX-GFP/IV2, and PVX-GFP/IV2/ELVd(+) transcripts at 7 and 10 d post inoculation. C, Serological detection of GFP. Total proteins were extracted from leaves, electrophoresed by 10% SDS-PAGE (top), and blotted for serological detection (bottom). GFP was clearly detected in both PVX-GFP and PVX-GFP/IV2/ELVd(+) systemically infected plants, confirming the correct processing of the chimeric GFP mRNA carrying the IV2/ELVd(+) insertion. D, RT-PCR amplification of processed and unprocessed GFP mRNAs. E, Confirmation (by RT-PCR) of systemic PVX infection in N. benthamiana plants inoculated with the construct used in this work.
When leaves infected with PVX carrying the IV2/ELVd(plus)-bearing GFP construct were analyzed at 7 and 10 d post inoculation (Fig. 1B), we observed fluorescence resembling that observed in the N. benthamiana leaves inoculated with the PVX-GFP control. As shown in Figure 1B, although GFP was readily visible throughout the leaves, the fluorescence was more intense in vascular tissues. No GFP expression was observed in the N. benthamiana leaves inoculated with PVX carrying the IV2-bearing GFP construct. GFP expression in the infected leaves was corroborated by western-blot assays. As PVX replication occurs in the cytoplasm of the infected cell, the subgenomic transcripts containing the intron are not processed and translated to produce functional GFP (as observed in the control PVX/IV2-bearing GFP). As a consequence, the fluorescence observed in the leaves infected with PVX carrying the IV2/ELVd(plus)-bearing GFP construct could only be explained by assuming that this cytoplasmatic transcript is directed by the ELVd RNA to the nucleus, where the intron can be efficiently removed to generate a functional GFP mRNA that is later translated.
In order to confirm the correct processing and stability of the constructs in the PVX-infected plants, we analyzed the subgenomic transcripts found in systemically infected N. benthamiana leaves by reverse transcription (RT)-PCR and sequencing. On amplification of the transcripts with GFP-specific primers, we observed a product of 724 bp corresponding to full GFP cDNA in the leaves inoculated with the PVX/GFP and PVX-GFP/IV2/ELVd(+)/constructs, providing evidence that the intron was correctly removed in the ELVd(+)-containing construct. An upper band (approximately 1,250 bp) corresponding to residual nonspliced mRNA was observed in the leaves inoculated with the PVX-GFP/IV2/ELVd(+) transcripts, suggesting that the import of mRNA into the nucleus was not fully efficient. As expected, the amplification product corresponding to nonspliced mRNA was the only band observed in the leaves inoculated with PVX-GFP/IV2 transcripts (Fig. 1D).
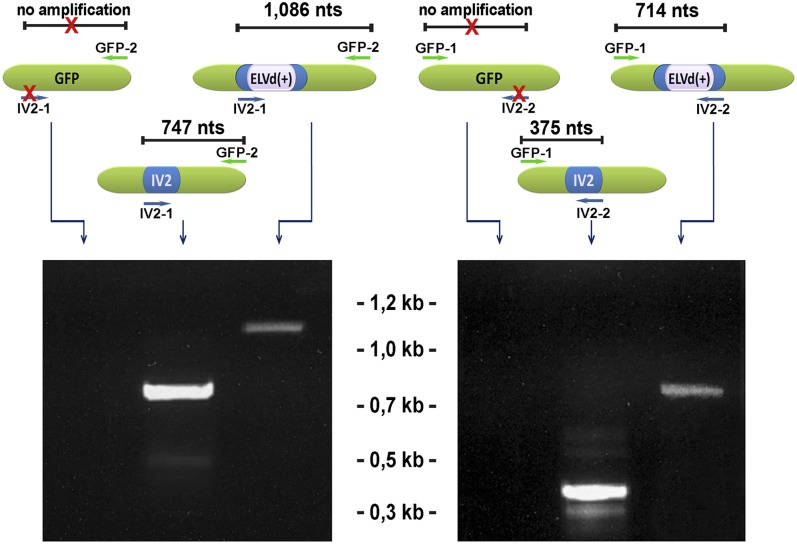
To rule out the possibility that the insertion IV2/ELVd(+) was eliminated from the chimeric PVX during long-distance movement, thereby yielding functional GFP, we assessed the presence of ELVd and IV2 RNA in these leaves at 25 d post inoculation. As shown in Figure 2, both insertions were detected in leaves systemically infected with the constructs PVX-GFP/IV2 and PVX-GFP/IV2/ELVd(+), indicating that the chimeric PVX maintained its stability during their translocation. Sequencing of the amplified cDNAs corroborated the correct processing and stability of the analyzed transcripts (Supplemental Fig. S1).
Figure 2.
Evidence for the stability of chimeric constructs in systemically infected leaves. RT-PCR amplification of IV2 and ELVd(+) cDNAs imbibed in a GFP open reading frame is shown. The sequences and positions of the specific primers (GFP-1 and GFP-2 and IV2-1 and IV2-2) used for the amplification are detailed in Supplemental Figure S2. The sizes in the top panel are not to scale. nt, Nucleotides.
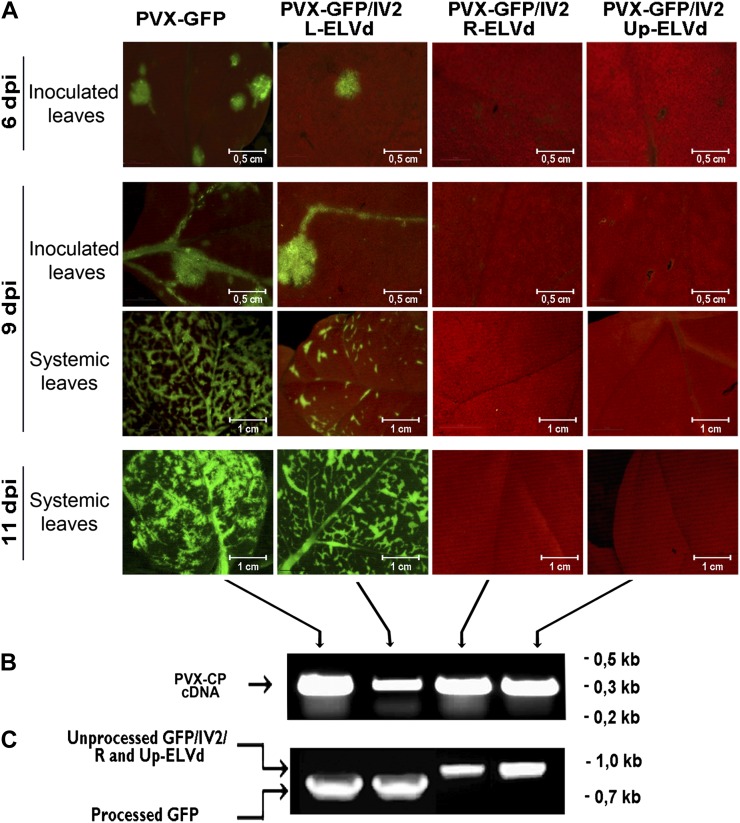
To obtain a more detailed picture, we dissected the ELVd RNA into three arbitrary regions identified as L (left), R (right), and Up (upper; Supplemental Fig. S2). Subsequently, these partial-length sequences were inserted into the PVX carrying the IV2-bearing GFP vector to generate the PVX-GFP/IV2/L-ELVd, PVX-GFP/IV2/R-ELVd, and PVX-GFP/IV2/Up-ELVd constructs. The N. benthamiana plants inoculated with the transcripts derived from these infectious clones were analyzed at 6, 9, and 11 d post inoculation. We observed that the leaves infected with the PVX-GFP/IV2/L-ELVd transcripts exhibit fluorescence resembling that observed in the plants inoculated with the PVX-GFP control (Fig. 3A). However, no GFP expression was observed in the N. benthamiana leaves infected with the PVX-GFP/IV2/R-ELVd and PVX-GFP/IV2/Up-ELVd constructs. This finding reveals that the L region of ELVd is sufficient to mediate the traffic of GFP mRNA into the nucleus, indicating that the nucleus-specific signal is localized in this 168-nucleotide-long ELVd RNA sequence. The correct processing of the chimeric constructs in the infected plants was analyzed by RT-PCR (Fig. 3C).
Figure 3.
Analysis of GFP expression in plants infected with PVX-derived transcripts carrying partial-length ELVd sequences. A, GFP fluorescence stereomicroscopy images of N. benthamiana leaves inoculated with PVX-GFP, PVX-GFP/IV2/L-ELVd, PVX-GFP/IV2/R-ELVd, and PVX-GFP/IV2/Up-ELVd transcripts at 6, 9, and 12 d post inoculation (dpi). B, Confirmation (by RT-PCR) of systemic PVX infection in N. benthamiana plants inoculated with the different constructs used in this assay. C, RT-PCR amplification of processed and unprocessed GFP mRNAs.
MONOMERIC ELVd RNA IS SPECIFICALLY DELIVERED FROM THE NUCLEUS TO CHLOROPLASTS
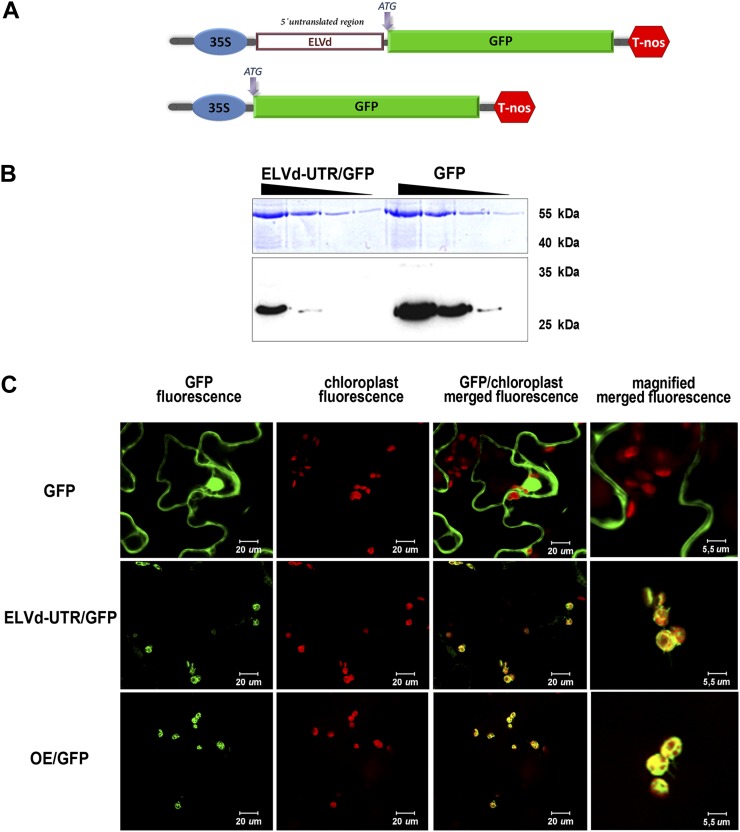
Having established that the monomeric linear form (+) of the ELVd RNA expressed in the cytoplasm is efficiently imported into the nucleus, we investigated whether this biological form of ELVd can be delivered from the nucleus to its specific replication site in the chloroplast. To address this issue, we constructed (Supplemental Materials and Methods S1) a reporter containing a monomeric cDNA derived from the genomic (+)ELVd RNA sequence fused as a UTR to the 5′ end of the cDNA of GFP (Fig. 4A; Supplemental Fig. S3). The fused ELVd-GFP cDNA (ELVd-5′UTR-GFP) was cloned in a binary vector to be transfected into Agrobacterium tumefaciens. This construct was analyzed by transient expression in N. benthamiana plants by means of agroinfiltration assays. First, we evaluated GFP expression in agroinfiltrated leaves by western blot. As shown in Figure 4B, ELVd-5′UTR-GFP exhibited a relative electrophoretic mobility identical to the unmodified GFP used as the control, indicating that the ELVd RNA acts as a true 5′ UTR in this GFP mRNA. Analysis of the infiltrated plants by confocal microscopy revealed that the GFP arising from the ELVd-5′UTR-GFP transcripts was generally concentrated in the chloroplasts of the N. benthamiana cells, resembling the localization of the GFP fused to the chloroplast-specific transit peptide OE23 (Roffey and Theg, 1996) used as a control (Fig. 4C). These results provide direct evidence that the ELVd-5′UTR-GFP mRNA transcribed in the nucleus is specifically targeted to chloroplasts, where it is efficiently retained and translated into functional GFP. This finding confirms that, in a similar way to that observed for a chimeric (plus and minus) ELVd RNA (Gómez and Pallas, 2010a), the biological monomeric form of this Avsunviroidae member contains sufficient information in its genomic RNA to regulate its specific translocation from the nucleus to the chloroplasts of the infected cell.
Figure 4.
The full-length (plus strand) ELVd RNA mediates the trafficking of functional GFP-mRNA from the nucleus to chloroplasts. A, Physical map of the ELVd-5′UTR-GFP and unmodified GFP constructs used in this assay. The construct sizes are not to scale. B, ELVd-5′UTR/GFP and GFP were clearly detected and showed similar relative electrophoretic mobility in the western-blot assay, indicating that the full-length viroid sequence acts as a true untranslated RNA. Serial dilutions(1:5) are shown for each construct. C, Confocal microscopy observation of N. benthamiana leaves expressing unmodified GFP (top panels), ELVd-5′UTR-GFP (middle panels), or OE23/GFP (bottom panels). ELVd-5′UTR/GFP mimics the cellular localization of the OE23/GFP construct, which accumulates specifically in chloroplasts.
DISCUSSION
Recent evidence indicates that it is not only proteins that possess internal localization signals for their targeting to specific destinations, but that RNAs may also harbor appropriate motifs to ensure intracellular trafficking to the appropriate destination (Okita and Choi, 2002; Rymarquis et al., 2008; Nevo-Dinur et al., 2011). Because viroids lack protein-coding capacity, they provide an ideal model system for studying intracellular RNA transport in plants (DiSerio and Flores, 2008; Wang and Ding, 2010). How the members of the Avsunviroidae family traffic into chloroplasts of infected cells to replicate remains a fascinating question that, once elucidated, will advance our understanding of viroid biology and intracellular RNA trafficking in general (Ding, 2009). According to these premises, and employing as a starting point the observation that a chimeric RNA containing partial sequences derived from both (plus and minus) ELVd forms regulates the selective trafficking of a functional mRNA from the nucleus into chloroplasts (Gómez and Pallas, 2010a, 2010b), we used a combined approach involving the cytoplasmic and nuclear expression of the biological monomeric form (plus strand) of the ELVd RNA to study relevant aspects of their intracellular movement in vivo.
In eukaryotic organisms, intron splicing is a nucleus-specific process catalyzed by the spliceosome, a large ribonucleoprotein complex comprising several small nuclear RNAs, small nuclear ribonucleoprotein particles, and nonsmall nuclear ribonucleoprotein particle protein-splicing factors (Sperling et al., 2008). As a consequence, the observation that the GFP mRNA arising from the PVX-GFP/IV2/ELVd(+) construct is translated into functional fluorescent protein provides strong evidence supporting the nuclear import of the viroid. Because PVX replicates exclusively in the cytoplasm, the mRNA carrying the intron IV2 will not produce a functional GFP unless this RNA can be targeted by the ELVd RNA to the nucleus, where the intron can be efficiently removed. This notion is consistent with previous reports showing that minor fractions of the Avocado sunblotch viroid (Mohamed and Thomas, 1980; Marcos and Flores, 1990) and Peach latent viroid (Bussière et al., 1999; also members of Avsunviroidae family) could be detected in the nuclei of the infected cells. Furthermore, the recent observation that the Avocado sunblotch viroid could be maintained in the nucleus of the yeast Saccharomyces cerevisiae for 25 generations (Delan-Forino et al., 2011) provides additional evidence supporting the potential robustness of chloroplastic viroids to survive in a nuclear environment.
The findings shown suggest that the capacity of a linear monomeric sequence to regulate its nuclear targeting mimics an intrinsic characteristic of the circular mature form of ELVd and that this nuclear stage could occur in the infected cell. It is worth noting that cell-to-cell movement (Ding et al., 1997) and nucleus-specific compartmentalization (Zhao et al., 2001), both natural abilities of the circular form, were retained by linearized Potato spindle tuber viroid RNA fused to reporter sequences. Furthermore, our results indicate that the structural and/or sequence domain involved in this signaling mechanism is localized in a specific region between positions 16 and 182 of the monomeric ELVd cDNA (Supplemental Fig. S4).
Having demonstrated the potential nuclear phase of the viroid RNA, we developed a differential construct to determine whether the monomeric ELVd (plus strand) was able to traffic from the nucleus to the chloroplast. The observation that the GFP arising from the transcript (5′UTR-ELVd-GFP) accumulated specifically in the chloroplasts of the agroinfiltrated plant cells indicates that the ELVd genomic RNA (acting as an untranslated sequence in this transcript) is selectively delivered from the nucleus into chloroplasts, providing biological evidence that, once localized in the nucleus, the monomeric form of ELVd is able to hijack endogenous routes for the export of nuclear RNAs to the chloroplast. This novel and uncharacterized (nucleus-chloroplast) RNA signaling pathway, which was recently proposed as an alternative mechanism to regulate the accumulation of nucleus-encoded proteins lacking canonical transit peptides in chloroplasts (Gómez and Pallas, 2010a), is consistent with previous reports showing that other endogenous RNAs are also targeted to this organelle in Chlamydomonas reinhardtii (Uniacke and Zerges, 2009) and plant cells (Bungard, 2004; Nicolaï et al., 2007). Furthermore, the recent observation that the alternanthera mosaic virus RNA localizes around chloroplast membranes in infected cells (Lim et al., 2010) suggests that association between viral RNA and chloroplasts could have functional implications in the infectious process.
Recent results have shown that the specific trafficking of partial-length ELVd transcripts fused to GFP mRNA viroid from the nucleus to the chloroplast is mediated by a potential structural domain contained in a short fragment localized between positions 52 and 150 of the ELVd cDNA (Gómez and Pallas, 2010b). We note that this fragment involved in the chloroplastic compartmentalization of the viroid overlaps with the region described here as responsible for the nuclear localization of ELVd (between nucleotides 15 and 181), suggesting that an RNA sequence and/or structure, restricted to the left terminal region of ELVd, can mediate both subsequent nuclear and chloroplastic localization. Interestingly, it has previously been shown that an ELVd mutant carrying a deletion in this region (identified as ELVd-D7) failed to infect the natural host eggplant (Solanum melongena), even though it is processed (cleavage and ligation) in vivo in a chloroplastic context using the C. reinhardtii-based experimental system (Martínez et al., 2009). This observation suggests that the deleted region is involved in other essential functions in the ELVd life cycle, such as chloroplastic localization. Interestingly, the possibility that members of the Avsunviroidae family have evolved a specific motif that is recognized by the cell machinery for an as yet undiscovered RNA import mechanism into the chloroplast was previously suggested as a potential way to explain their selective intracellular localization (Ding and Itaya, 2007).
In summary, the findings shown here provide insights into aspects of the intracellular movement of viroids belonging to the Avsunviroidae family. The observation that the monomeric linear form of ELVd possesses the potential to selectively traffic from the cytoplasm into the nucleus and subsequently from this organelle into chloroplasts allows us to envision a novel route that could explain their selective compartmentalization in the chloroplast. In this hypothetical pathway (likely mediated by a RNA domain localized in the left terminal region), after the viroid (mainly the plus-strand circular form) is mechanically introduced into the cell cytoplasm, it is imported into the nucleus by means of an unknown host-dependent mechanism. The viroid then uses this organelle as a subcellular shuttle for delivery into the chloroplast, where replication takes place. Our results reveal that the different steps of this pathway take place in the plant and highlight the fact that viroids can be used to help unravel the trafficking of RNAs to specific destinations in the cell.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Sequences of chimeric transcripts recovered from systemically infected plants.
Supplemental Figure S2. Representation of the constructs containing dissected ELVd regions.
Supplemental Figure S3. Partial sequence of the ELVd-5′UTR/GFP constructs.
Supplemental Figure S4. Partial sequences of the PVX-derived constructs used in this study.
Supplemental Materials and Methods S1. Vector construction and expression.
Supplementary Material
Acknowledgments
We thank J.A. Daròs for providing the pELVd plasmid and for his valuable contribution in the development of this work. We also thank S.F. Elena and M. Fares for critical reading of the manuscript. The valuable suggestions of one of the reviewers is greatly appreciated.
Glossary
- ELVd
Eggplant latent viroid
- UTR
untranslated region
- PVX
potato virus X
- RT
reverse transcription
References
- Abraitiene A, Zhao Y, Hammond R. (2008) Nuclear targeting by fragmentation of the potato spindle tuber viroid genome. Biochem Biophys Res Commun 368: 470–475 [DOI] [PubMed] [Google Scholar]
- Bungard R-A. (2004) Photosynthetic evolution in parasitic plants: insight from the chloroplast genome. Bioessays 26: 235–247 [DOI] [PubMed] [Google Scholar]
- Bussière F, Lehoux J, Thompson D-A, Skrzeczkowski L-J, Perreault J. (1999) Subcellular localization and rolling circle replication of peach latent mosaic viroid: hallmarks of group A viroids. J Virol 73: 6353–6360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman S, Kavanagh T, Baulcombe D. (1992) Potato virus X as a vector for gene expression in plants. Plant J 2: 549–557 [DOI] [PubMed] [Google Scholar]
- Daròs J-A, Elena S-F, Flores R. (2006) Viroids: an Ariadne’s thread into the RNA labyrinth. EMBO Rep 7: 593–598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delan-Forino C, Maurel M-C, Torchet C. (2011) Replication of avocado sunblotch viroid in the yeast Saccharomyces cerevisiae. J Virol 85: 3229–3238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diener T-O (1987) The Viroids. Plenum Press, New York [Google Scholar]
- Ding B. (2009) The biology of viroid-host interactions. Annu Rev Phytopathol 47: 105–131 [DOI] [PubMed] [Google Scholar]
- Ding B, Itaya A. (2007) Viroid: a useful model for studying the basic principles of infection and RNA biology. Mol Plant Microbe Interact 20: 7–20 [DOI] [PubMed] [Google Scholar]
- Ding B, Kwon M-O, Hammond R, Owens R. (1997) Cell-to-cell movement of potato spindle tuber viroid. Plant J 12: 931–936 [DOI] [PubMed] [Google Scholar]
- Di Serio F, Flores R. (2008) Viroids: molecular implements for dissecting RNA trafficking in plants. RNA Biol 5: 128–131 [DOI] [PubMed] [Google Scholar]
- Eckes P, Rosahl S, Schell J, Willmitzer L. (1986) Isolation and characterization of a light-inducible, organ-specific gene from potato and analysis of its expression after tagging and transfer into tobacco and potato shoots. Mol Gen Genet 205: 14–22 [Google Scholar]
- Fadda Z, Daròs J-A, Fagoaga C, Flores R, Duran-Vila N. (2003) Eggplant latent viroid, the candidate type species for a new genus within the family Avsunviroidae (hammerhead viroids). J Virol 77: 6528–6532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez Calvino L, Faulkner C, Maule A. (2011) Plasmodesmata as active conduits for virus cell-to-cell movement. In C Caranta, M-A Aranda, M Tepfer, J-J Lopez Moya, eds, Recent Advances in Plant Virology. Caister Academic Press, Norfolk, UK, pp 47–74 [Google Scholar]
- Flores R, Hernández C, Martínez de Alba A-E, Daròs J-A, Di Serio F. (2005) Viroids and viroid-host interactions. Annu Rev Phytopathol 43: 117–139 [DOI] [PubMed] [Google Scholar]
- Gómez G, Pallas V. (2004) A long-distance translocatable phloem protein from cucumber forms a ribonucleoprotein complex in vivo with Hop stunt viroid RNA. J Virol 78: 10104–10110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez G, Pallas V. (2010a) Noncoding RNA mediated traffic of foreign mRNA into chloroplasts reveals a novel signaling mechanism in plants. PLoS ONE 5: e12269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez G, Pallas V. (2010b) Can the import of mRNA into chloroplasts be mediated by a secondary structure of a small non-coding RNA? Plant Signal Behav 5: 1517–1519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim H-S, Vaira A-M, Bae H, Bragg JN, Ruzin S-E, Bauchan GR, Dienelt MM, Owens RA, Hammond J. (2010) Mutation of a chloroplast-targeting signal in Alternanthera mosaic virus TGB3 impairs cell-to-cell movement and eliminates long-distance virus movement. J Gen Virol 91: 2102–2115 [DOI] [PubMed] [Google Scholar]
- Lucas W-J, Yoo B-C, Kragler F. (2001) RNA as a long-distance information macromolecule in plants. Nat Rev Mol Cell Biol 2: 849–857 [DOI] [PubMed] [Google Scholar]
- Marcos J, Flores R. (1990) Subcellular localization of Avocado sunblotch viroid in avocado leaves. Plant Sci 67: 237–244 [Google Scholar]
- Martínez F, Marqués J, Salvador M-L, Daròs J-A. (2009) Mutational analysis of eggplant latent viroid RNA processing in Chlamydomonas reinhardtii chloroplast. J Gen Virol 90: 3057–3065 [DOI] [PubMed] [Google Scholar]
- Michaud M, Maréchal-Drouard L, Duchêne A-M. (2010) RNA trafficking in plant cells: targeting of cytosolic mRNAs to the mitochondrial surface. Plant Mol Biol 73: 697–704 [DOI] [PubMed] [Google Scholar]
- Mohamed N-A, Thomas W. (1980) Viroid-like properties of an RNA species associated with the sunblotch disease of avocado. J Gen Virol 46: 157–167 [Google Scholar]
- Nevo-Dinur K, Nussbaum-Shochat A, Ben-Yehuda S, Amster-Choder O. (2011) Translation-independent localization of mRNA in E. coli. Science 331: 1081–1084 [DOI] [PubMed] [Google Scholar]
- Nicolaï M, Duprat A, Sormani R, Rodriguez C, Roncato M-A, Rolland N, Robaglia C. (2007) Higher plant chloroplasts import the mRNA coding for the eucaryotic translation initiation factor 4E. FEBS Lett 581: 3921–3926 [DOI] [PubMed] [Google Scholar]
- Okita T-W, Choi S-B. (2002) mRNA localization in plants: targeting to the cell’s cortical region and beyond. Curr Opin Plant Biol 5: 553–559 [DOI] [PubMed] [Google Scholar]
- Pallas V, Genoves A, Sanchez-Pina M-A, Navarro J-A. (2011) Systemic movement of viruses via the plant phloem. In C Caranta, M-A Aranda, M Tepfer, J-J Lopez Moya, eds, Recent Advances in Plant Virology. Caister Academic Press, Norfolk, VA, pp 75–102 [Google Scholar]
- Roffey R-A, Theg S-M. (1996) Analysis of the import of carboxyl-terminal truncations of the 23-kilodalton subunit of the oxygen-evolving complex suggests that its structure is an important determinant for thylakoid transport. Plant Physiol 111: 1329–1338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rymarquis L-A, Kastenmayer J-P, Hüttenhofer A-G, Green P-J. (2008) Diamonds in the rough: mRNA-like non-coding RNAs. Trends Plant Sci 13: 329–334 [DOI] [PubMed] [Google Scholar]
- Sperling J, Azubel M, Sperling R. (2008) Structure and function of the pre-mRNA splicing machine. Structure 16: 1605–1615 [DOI] [PubMed] [Google Scholar]
- Tabler M, Tsagris M. (2004) Viroids: petite RNA pathogens with distinguished talents. Trends Plant Sci 9: 339–348 [DOI] [PubMed] [Google Scholar]
- Uniacke J, Zerges W. (2009) Chloroplast protein targeting involves localized translation in Chlamydomonas. Proc Natl Acad Sci USA 106: 1439–1444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Ding B. (2010) Viroids: small probes for exploring the vast universe of RNA trafficking in plants. J Integr Plant Biol 52: 28–39 [DOI] [PubMed] [Google Scholar]
- Woo Y, Itaya A, Owens R-A, Tang L, Hammond R-W, Chou H, Lai M, Ding B. (1999) Characterization of nuclear import of potato spindle tuber viroid RNA in permeabilized protoplasts. Plant J 17: 627–635 [Google Scholar]
- Zhao Y, Owens R-A, Hammond R-W. (2001) Use of a vector based on Potato virus X in a whole plant assay to demonstrate nuclear targeting of potato spindle tuber viroid. J Gen Virol 82: 1491–1497 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.