Abstract
Noscapine is a benzylisoquinoline alkaloid produced in opium poppy (Papaver somniferum) and other members of the Papaveraceae. It has been used as a cough suppressant and more recently was shown to possess anticancer activity. However, the biosynthesis of noscapine in opium poppy has not been established. A proposed pathway leading from (S)-reticuline to noscapine includes (S)-scoulerine, (S)-canadine, and (S)-N-methylcanadine as intermediates. Stem cDNA libraries and latex extracts of eight opium poppy cultivars displaying different alkaloid profiles were subjected to massively parallel pyrosequencing and liquid chromatography-tandem mass spectrometry, respectively. Comparative transcript and metabolite profiling revealed the occurrence of three cDNAs encoding O-methyltransferases designated as SOMT1, SOMT2, and SOMT3 that correlated with the accumulation of noscapine in the eight cultivars. SOMT transcripts were detected in all opium poppy organs but were most abundant in aerial organs, where noscapine primarily accumulates. SOMT2 and SOMT3 showed strict substrate specificity and regiospecificity as 9-O-methyltransferases targeting (S)-scoulerine. In contrast, SOMT1 was able to sequentially 9- and 2-O-methylate (S)-scoulerine, yielding (S)-tetrahydropalmatine. SOMT1 also sequentially 3′- and 7-O-methylated both (S)-norreticuline and (S)-reticuline with relatively high substrate affinity, yielding (S)-tetrahydropapaverine and (S)-laudanosine, respectively. The metabolic functions of SOMT1, SOMT2, and SOMT3 were investigated in planta using virus-induced gene silencing. Reduction of SOMT1 or SOMT2 transcript levels resulted in a significant decrease in noscapine accumulation. Reduced SOMT1 transcript levels also caused a decrease in papaverine accumulation, confirming the selective roles for these enzymes in the biosynthesis of both alkaloids in opium poppy.
Benzylisoquinoline alkaloids (BIAs) are a large and diverse group of approximately 2,500 defined structures found primarily in members of the Papaveraceae, Ranunculaceae, Berberidaceae, and Menispermaceae (Facchini, 2001; Facchini and De Luca, 2008). Among these, opium poppy (Papaver somniferum) remains a valuable source for several pharmaceuticals, including the narcotic analgesics morphine and codeine, the muscle relaxant papaverine, and the cough suppressant and potential anticancer drug noscapine. BIA biosynthesis begins with the formation of (S)-norcoclaurine via the condensation of the Tyr derivatives dopamine and 4-hydroxyphenylacetaldehyde (Samanani and Facchini, 2002; Minami et al., 2007; Lee and Facchini, 2010). (S)-Norcoclaurine is subsequently converted via several steps to the key branch point intermediate (S)-reticuline (Fig. 1), from which numerous BIA structural subgroups are derived (Frenzel and Zenk, 1990; Pauli and Kutchan, 1998; Morishige et al., 2000; Choi et al., 2002). Rearrangement of the benzylisoquinoline backbone is achieved through carbon-carbon or carbon-oxygen coupling catalyzed by a variety of specific cytochromes P450 or by a FAD-linked oxidoreductase. Intermediates within each structural subgroup undergo a variety of functional group additions or substitutions catalyzed by S-adenosyl-l-methionine (SAM)-dependent O-methyltransferases (OMTs) and N-methyltransferases, different NADPH-dependent reductases, 2-oxoglutarate-dependent dioxygenases, and an acetyl-CoA-dependent acyltransferase (Facchini and De Luca, 2008; Hagel and Facchini, 2010).
Figure 1.
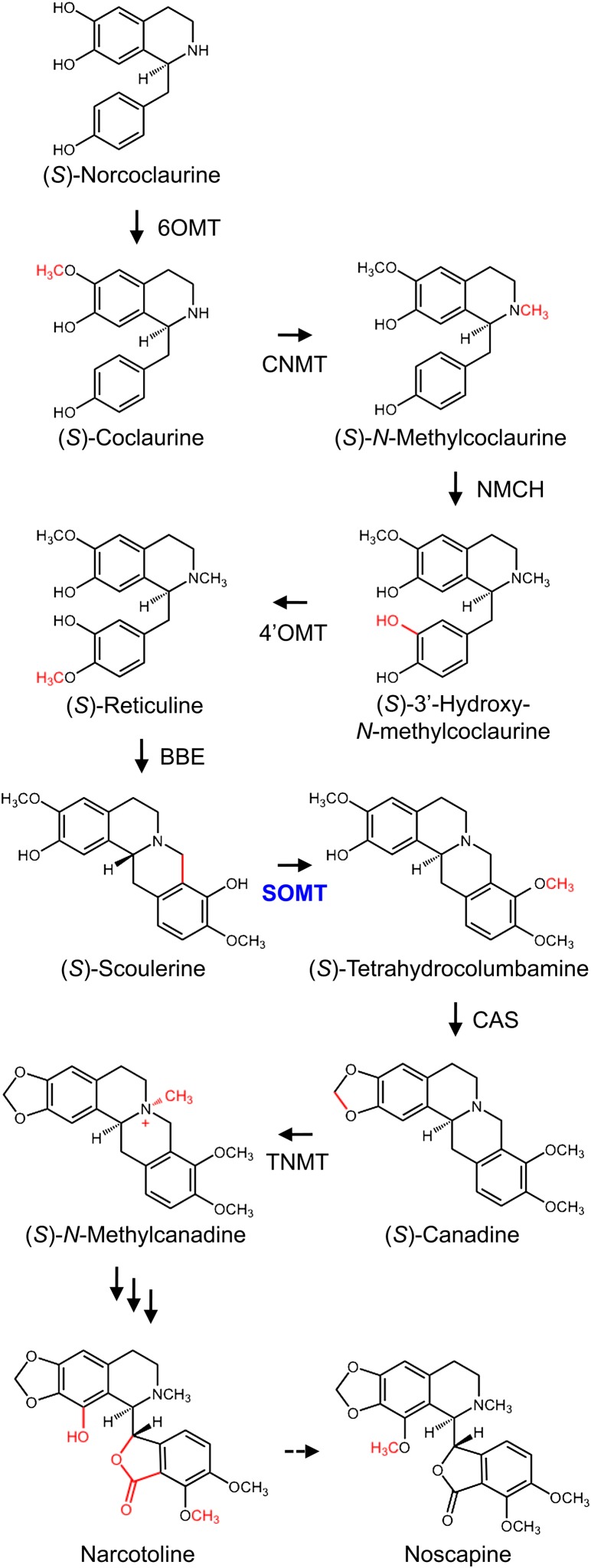
Proposed biosynthetic pathway from (S)-norcoclaurine to noscapine in opium poppy. The proposed central intermediate (S)-reticuline is converted to (S)-scoulerine by the berberine bridge enzyme (BBE). The formation of (S)-canadine from (S)-scoulerine through the intermediate (S)-tetrahydrocolumbamine involves SOMT and canadine synthase (CAS). Tetrahydroprotoberberine cis-N-methyltransferase (TNMT) from opium poppy converts (S)-canadine to (S)-N-methylcanadine. Intermediates between (S)-N-methylcanadine and narcotoline are not known. The 1-O-methylation of narcotoline or an unidentified upstream intermediate yields noscapine. The point of action for a perceived SOMT is shown in blue. Corresponding cDNAs have been isolated for all enzymes shown in black. Multiple arrows indicate several uncharacterized steps, whereas the dashed arrow represents a putative conversion. Abbreviations not defined in the text are as follows: CNMT, coclaurine N-methyltransferase; NMCH, N-methylcoclaurine 3′-hydroxylase.
A common elaboration in BIA metabolism is O-methylation, which involves the transfer of the methyl group of SAM to the hydroxyl group of an acceptor molecule, resulting in the formation of a methyl ether derivative and S-adenosyl-l-homocysteine. SAM-dependent OMTs participate in the formation of the central intermediate (S)-reticuline and are involved in its multistep conversion to a multitude of BIA end products. All known OMTs involved in BIA metabolism exhibit extensive amino acid homology and form a distinctive clade with respect to OMTs involved in other metabolic pathways, such as flavonoid metabolism (Ibrahim et al., 1998). Characterized OMTs involved in BIA biosynthesis include norcoclaurine 6-O-methyltransferase (6OMT; Sato et al., 1994; Morishige et al., 2000; Ounaroon et al., 2003), 3′-hydroxy-N-methylcoclaurine 4′-O-methyltransferase (4′OMT; Morishige et al., 2000; Ziegler et al., 2005), reticuline 7-O-methyltransferase (7OMT; Ounaroon et al., 2003), norreticuline 7-O-methyltransferase (N7OMT; Pienkny et al., 2009), scoulerine-9-O-methyltransferase (SOMT; Takeshita et al., 1995), and columbamine O-methyltransferase (CoOMT; Morishige et al., 2002). Functional homologs for all enzymes except SOMT and CoOMT have been identified in opium poppy (Facchini and De Luca, 2008).
Phthalideisoquinoline alkaloids are a structural subgroup of BIAs that includes noscapine, which has long been used as a cough suppressant (Facchini et al., 2007). More recently, noscapine has been shown to possess anticancer activity (Ye et al., 1998; Barken et al., 2008). The biosynthesis of phthalideisoquinoline alkaloids, including noscapine, has not been well established. Battersby and Hirst (1965) demonstrated that the biosynthetic route to noscapine involved the incorporation of a benzylisoquinoline moiety by in vivo feeding of [14C]Tyr and [14C]norlaudanosoline. Other key early experiments involved feeding labeled reticuline and double-labeled (S)-scoulerine to opium poppy plants, both of which were incorporated into noscapine (Battersby and Hirst, 1965; Battersby et al., 1968). The lactone carbonyl group of noscapine was suggested to derive from the N-methyl group of reticuline (Battersby and Hirst, 1965). Additional support for the enzymatic transformation of (S)-reticuline to noscapine has also been reported (Gözler, 1983; Sariyar, 1986, 2002; Sariyar et al., 1990). Initial steps involved in the conversion of the protoberberine alkaloid (S)-scoulerine to a phthalideisoquinoline alkaloid intermediate ultimately leading to the formation of noscapine are proposed in Figure 1. Because noscapine possesses the same 2,3-methylenedioxy-9,10-dimethoxy substitution pattern as (S)-canadine, also known as (S)-tetrahydroberberine, the first two steps leading to noscapine from (S)-scoulerine are potentially identical to those involved in the formation of berberine (Facchini and De Luca, 2008). Accordingly, (S)-scoulerine would be methylated by SOMT to yield (S)-tetrahydrocolumbamine, which would then be converted to (S)-canadine by canadine synthase, a methylenedioxy bridge-forming P450-dependent monooxygenase (Takeshita et al., 1995; Ikezawa et al., 2003). Subsequently, (S)-canadine would be N-methylated by tetrahydroprotoberberine cis-N-methyltransferase to produce N-methylcanadine (Liscombe and Facchini, 2007), which would be oxidized via several steps and 1-O-methylated at some point to yield noscapine. Previously, cDNAs encoding berberine bridge enzyme and tetrahydroprotoberberine cis-N-methyltransferase have been isolated from opium poppy (Facchini et al., 1996; Liscombe and Facchini, 2007), but their involvement in the noscapine pathway has not been validated. SOMT and canadine synthase cDNAs have only been isolated and characterized from berberine-producing species, including Coptis japonica (Takeshita et al., 1995; Ikezawa et al., 2003, 2007).
In this paper, we report the isolation and characterization of three SOMT homologs in opium poppy based on the integration of deep transcript and metabolite profiles established for cultivars with different alkaloid phenotypes. One SOMT also exhibited 3′OMT activity with (S)-reticuline and (S)-norreticuline as substrates, which represents a previously uncharacterized enzymatic function in BIA metabolism implicated in the formation of the major alkaloid papaverine. The involvement of each SOMT in noscapine and papaverine biosynthesis was determined in opium poppy plants using virus-induced gene silencing (VIGS).
RESULTS
Isolation of OMT Candidates Using Transcript and Metabolite Profiles
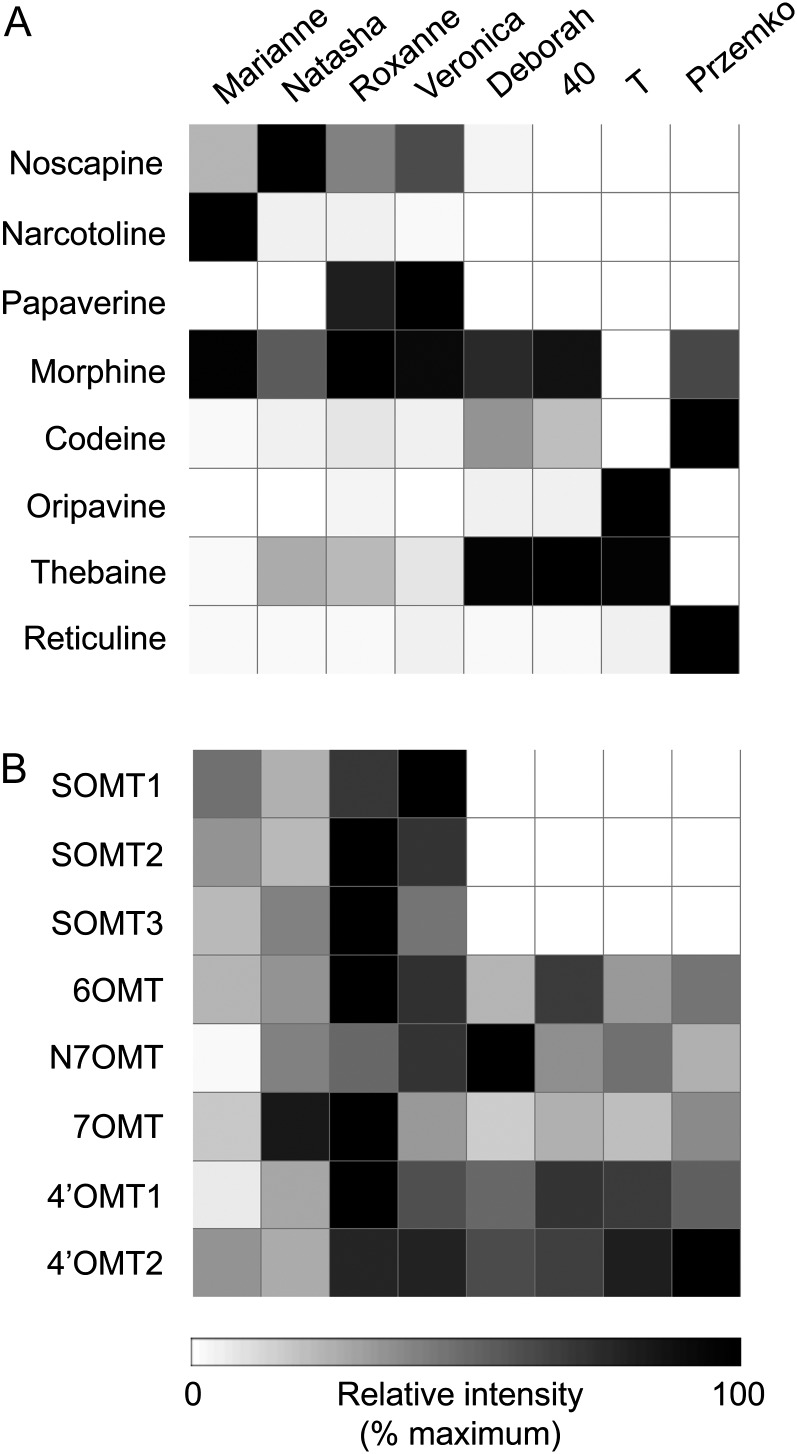
The major BIA content and minor alkaloid profiling of each opium poppy cultivar were determined by HPLC and liquid chromatography-tandem mass spectrometry (LC-MS/MS), respectively (Desgagné-Penix et al., 2012). HPLC analysis revealed substantially different major alkaloid profiles among the cultivars. In the context of this work, cv Marianne, Natasha, Roxanne, and Veronica accumulated high levels of noscapine, whereas cv Deborah, 40, T, and Przemko displayed only trace to undetectable levels of noscapine (Fig. 2A; Supplemental Table S1A). Natasha contained considerably higher levels of noscapine, whereas Marianne accumulated more narcotoline compared with the other cultivars. Roxanne and Veronica were the only two cultivars that accumulated substantial levels of papaverine. The absence of noscapine generally correlated with higher levels of reticuline and the morphinan alkaloids codeine, thebaine, and oripavine in Deborah, 40, and T.
Figure 2.
Heat maps showing the relative abundance of major latex alkaloids determined by HPLC (A) and the relative levels of selected SOMT candidates compared with previously characterized BIA OMTs (B) in eight opium poppy cultivars. GenBank accession numbers for each SOMT candidate are provided in “Materials and Methods.” The highest alkaloid and transcript levels were normalized to 100%.
All eight cultivars were subjected to 454 GS-FLX Titanium pyrosequencing, which provided a deep transcriptome database to search for differentially expressed genes in the noscapine-producing compared with the noscapine-free cultivars. The initial differential expression analysis performed using transcriptome databases for the Marianne and Deborah cultivars revealed more than 500 contigs that were detected only in Marianne. These data were then used to mine SOMT candidates based on (1) predicted amino acid sequence similarity searches for OMT homologs and (2) the correlation of gene expression profiles with the accumulation of noscapine in different cultivars. Five independent assembled contigs were annotated as methyltransferases, including three OMTs, one S-methyltransferase, and one homocysteine methyltransferase. Among the three contigs that were annotated as OMTs, contig1 (designated SOMT1) was annotated as scoulerine 9-O-methyltransferase, whereas contig2 (SOMT2) and contig3 (SOMT3) were annotated as norcoclaurine 6-O-methyltransferase. The relative transcript abundance based on the number of 454 pyrosequencing reads corresponding to SOMT1, SOMT2, and SOMT3 compared with the total number of reads in databases for each of the eight opium poppy cultivars (Fig. 2B; Supplemental Table S1B) showed a strong positive correlation with respect to the accumulation of noscapine (Fig. 2A). In contrast, the previously characterized opium poppy OMT genes 4′OMT1, 4′OMT2, 6OMT, 7OMT, and N7OMT were expressed in stems of all cultivars. SOMT1, SOMT2, and SOMT3 transcripts were relatively higher in Roxanne and Veronica, which were also the only two cultivars that accumulated papaverine (Fig. 2; Supplemental Table S1).
Sequence and Phylogenetic Analysis of OMT Candidates
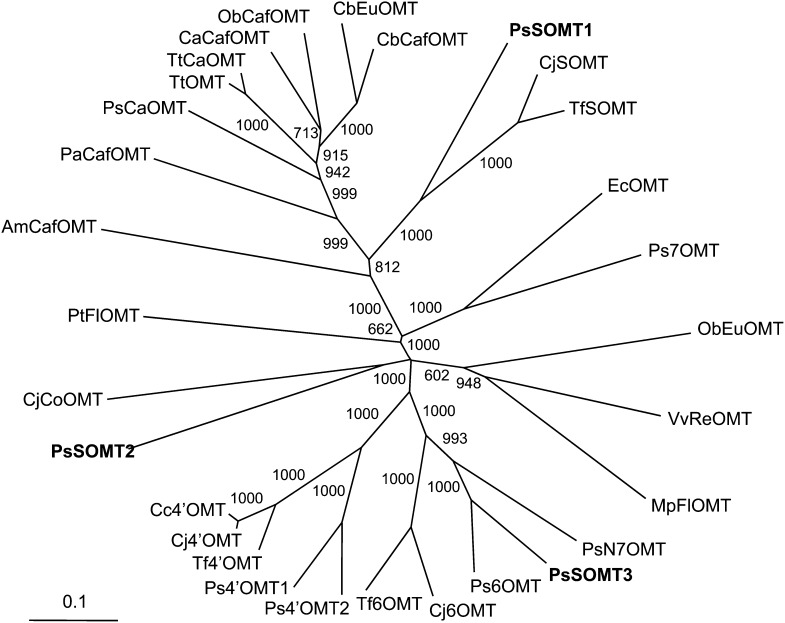
In silico assembly resulted in three full-length SOMT candidates consisting of 1,679 (SOMT1), 1,520 (SOMT2), and 1,255 bp (SOMT3), with corresponding open reading frames of 1,173, 1,071 and 1,020 bp, respectively. SOMT1, SOMT2, and SOMT3 encoded proteins composed of 391, 357, and 340 amino acids, with corresponding molecular mass values of 42.6, 39.0, and 37.4 kD, respectively. With respect to each other, the opium poppy SOMT candidates displayed relatively low amino acid sequence identities of between 19% and 37%. Phylogenetic analysis in the context of several characterized plant OMTs showed that all three opium poppy SOMT candidates formed separate clades with enzymes involved in BIA biosynthesis (Fig. 3). SOMT1 shares 59% and 61% sequence identity with SOMT from C. japonica and Thalictrum flavum, respectively. SOMT2 shows 39% sequence identity with CoOMT from C. japonica and 40% and 43% sequence identity with 6OMT from C. japonica and opium poppy, even though it maps closer to columbamine OMT. SOMT3 displays 79% and 63% sequence identity with 6OMT and N7OMT, respectively, from opium poppy, which accounts for its occurrence in the same clade. The alignment of the opium poppy SOMT candidates with other plant OMTs used to construct the phylogenetic tree is provided in Supplemental Figure S1.
Figure 3.
Unrooted neighbor-joining phylogenetic tree for selected plant OMTs. Bootstrap frequencies for each clade were based on 1,000 iterations. Abbreviations and GenBank accessions numbers for each protein are provided in “Materials and Methods.” PsSOMT1, PsSOMT2, and PsSOMT3 are the three candidate genes from opium poppy.
Purification and in Vitro Characterization of Opium Poppy OMTs
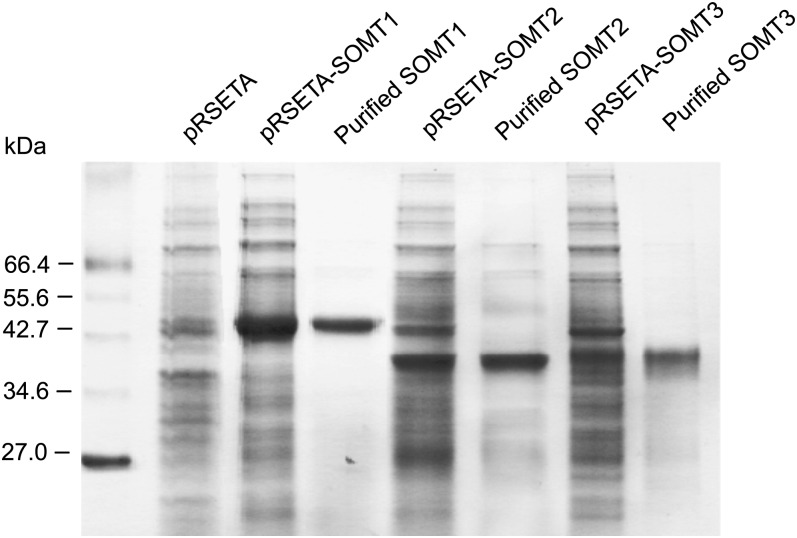
Full-length cDNAs for the three SOMT candidates were cloned into the pRSETA expression vector with an N-terminal His6 tag translational fusion. Recombinant SOMT1, SOMT2, and SOMT3 were purified from total protein extract using a cobalt-affinity resin. The recombinant enzymes displayed molecular mass values of approximately 43, 40, and 39 kD as determined by SDS-PAGE, which were marginally higher than the predicted values owing to the N-terminal peptide fusion (Fig. 4). Enzyme assays were performed on each of the purified, recombinant proteins to screen for O-methylation activity. Fourteen potential BIA substrates containing free hydroxyl groups from six different structural subgroups were tested (Supplemental Fig. S2). Incubation of SOMT1 with (S)-scoulerine, (S)-tetrahydrocolumbamine, (S)-norreticuline, and (S)-reticuline, in the presence of [14C]SAM, yielded radioactivity in the organic phase extracted from the assay reaction mixtures. (S)-Scoulerine was the best substrate for SOMT1 (100%), followed by (S)-tetrahydrocolumbamine (78%), (S)-norreticuline (33%), and (S)-reticuline (20%; Supplemental Fig. S2). In contrast, radioactivity in the extracted organic phase of assay reaction mixtures containing SOMT2 or SOMT3 was only detected when (S)-scoulerine was used as the potential substrate, and in both cases with lower efficiency than reactions containing SOMT1 and (S)-scoulerine. No other BIAs yielded radioactivity in the organic phase of assay reaction mixtures containing SOMT1, SOMT2, or SOMT3.
Figure 4.
Purification of recombinant proteins in E. coli expressing pRSETA-SOMT1, pRSETA-SOMT2, and pRSETA-SOMT3. The left lane contains protein markers, with corresponding molecular masses indicated on the left. All other lanes show crude extract or purified protein from E. coli strain Rosetta induced with 1.0 mm isopropyl β-d-thiogalactopyranoside. Recombinant proteins were purified using cobalt-affinity chromatography. Bacteria harboring the empty pRSETA vector were included as the negative control. Proteins were separated by SDS-PAGE and visualized by Coomassie Blue staining.
Kinetic analysis showed that SOMT1 displayed greater affinity and substantially higher efficiency toward (S)-scoulerine, which are reflected by lower Km and higher kcat/Km values compared with (S)-reticuline (Table I; Supplemental Fig. S3). SOMT2 and SOMT3 showed similar affinity and efficiency with (S)-scoulerine as the substrate, although both enzymes displayed a kcat/Km value 50-fold lower than that of SOMT1. The affinity of SOMT1 was also higher than that of SOMT2 and SOMT3 toward the cosubstrate SAM (Table I). No significant substrate or product inhibition was detected. The limited availability of norreticuline and tetrahydrocolumbamine precluded the determination of kinetic data for these substrates.
Table I. Kinetic data for SOMT1, SOMT2, and SOMT3.
| Enzyme | Substrate | Km | Vmax | kcat | kcat/Km |
|---|---|---|---|---|---|
| μm | nmol min−1 mg−1 protein | s−1 | s−1m−1 | ||
| SOMT1 | (S)-Scoulerine | 28.5 ± 6.8 | 2,036 ± 160.9 | 1.44 ± 0.27 | 50.52 |
| (S)-Reticuline | 70.3 ± 13.7 | 184.4 ± 38.73 | 0.13 ± 0.07 | 0.19 | |
| SAM | 19 ± 2.66 | 1,290 ± 73.18 | 0.91 ± 0.0006 | 47.89 | |
| SOMT2 | (S)-Scoulerine | 73.28 ± 18.6 | 145.8 ± 28.10 | 0.09 ± 0.013 | 1.23 |
| SAM | 69.62 ± 20.9 | 134.01 ± 20.50 | 0.087 ± 0.037 | 1.25 | |
| SOMT3 | (S)-Scoulerine | 50.83 ± 13.63 | 100.26 ± 11.57 | 0.063 ± 0.014 | 1.24 |
| SAM | 101.2 ± 29.41 | 115.6 ± 11.48 | 0.072 ± 0.016 | 0.71 |
Analysis and Structure Elucidation of Reaction Products
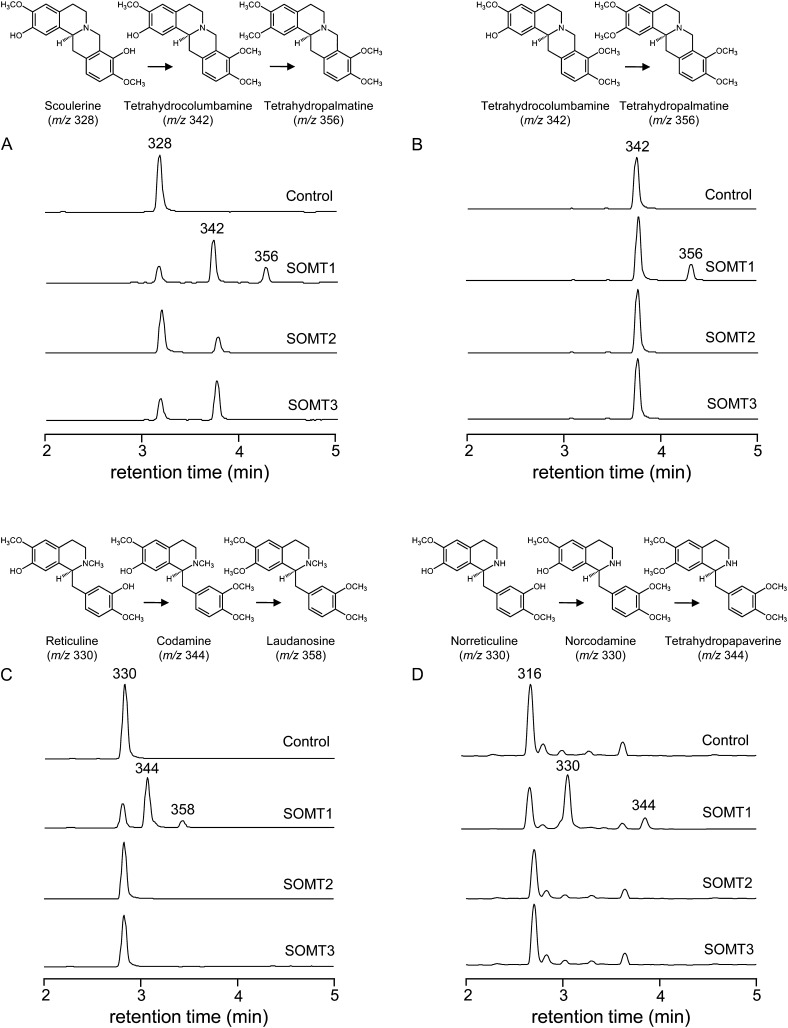
Enzyme assays containing each SOMT enzyme, unlabeled SAM, and individual BIA substrates were subjected to positive-mode LC-MS/MS analysis for reaction product characterization. Incubation of SOMT1 with (S)-scoulerine (mass-to-charge ratio [m/z] 328) yielded two new peaks with m/z 342 at 3.67 min and m/z 356.1 at 4.21 min, suggesting single and double O-methylation events, respectively. Incubation of SOMT2 and SOMT3 with (S)-scoulerine yielded only one peak with m/z 342 (Fig. 5A). Reaction mixtures were injected into an electrospray ionization (ESI) source and detected using a triple-quadruple mass analyzer operating in positive ion mode to obtain collision-induced dissociation (CID) spectra and fragmentation patterns. Typically, cleavage of simple benzylisoquinoline and protoberberine alkaloids at low ionization energy yields their isoquinoline and benzyl moieties. Therefore, the position of new O-methyl groups can often be identified based on the m/z of the resulting fragment ions. Because (S)-scoulerine possesses only one hydroxyl group on each of the isoquinoline and benzyl moieties, SOMT1 was clearly able to O-methylate both moieties. In contrast, SOMT2 and SOMT3 specifically O-methylated only the benzyl moiety.
Figure 5.
Total ion chromatograms (TICs) showing the O-methylation activity of SOMT1, SOMT2, and SOMT3 on (S)-scoulerine (A), (S)-tetrahydrocolumbamine (B), (S)-reticuline (C), and (S)-norreticuline (D). For each substrate, the top (control) TIC corresponds to boiled enzyme as the negative control, whereas the three other TICs show assays performed using native SOMT1, SOMT2, and SOMT3. Incubation of native SOMT1 with (S)-scoulerine (m/z 328) yielded two reaction products identified as tetrahydrocolumbamine (m/z 342) and tetrahydropalmatine (m/z 356) based on their CID spectra. Incubation of native SOMT2 and SOMT3 yielded only tetrahydrocolumbamine. SOMT1 also converted tetrahydrocolumbamine (m/z 342) to tetrahydropalmatine (m/z 356), reticuline (m/z 330) to codamine (m/z 344) and laudanosine (m/z 358), and norreticuline (m/z 316) to norcodamine (m/z 330) and norlaudanosine (m/z 344). Neither SOMT2 nor SOMT3 accepted tetrahydrocolumbamine, reticuline, or norreticuline as substrates.
The first SOMT1-catalyzed O-methylation product of (S)-scoulerine (m/z 328) exhibited a parent ion of m/z 342, which produced a CID spectrum corresponding to authentic (S)-tetrahydrocolumbamine (Supplemental Table S2). The double O-methylation product corresponded to a parent ion of m/z 356, which produced a CID spectrum matching that of (S)-tetrahydropalmatine. This scheme was validated in assays containing SOMT1 and (S)-tetrahydrocolumbamine, which yielded a reaction product of m/z 356 (Fig. 5B) with a CID spectrum identical to that of (S)-tetrahydropalmatine. The single reaction products of SOMT2 and SOMT3 both showed a parent mass of m/z 342 and CID spectra corresponding to that of (S)-tetrahydrocolumbamine (Fig. 5A).
LC-MS/MS analysis of enzyme assays containing SOMT1 and (S)-reticuline (m/z 330) also showed two reaction products, one with m/z 344 and another with m/z 358 (Fig. 5C). Although authentic standards were not available, these compounds could be annotated using published spectra or characterized based on the predictable fragmentation patterns of simple benzylisoquinolines. In CID mode, five ions for m/z 344 and 11 ions for m/z 358 were detected (Supplemental Table S2). Compared with the CID spectrum of (S)-reticuline, which shows the fragment ions m/z 192 (isoquinoline moiety) and m/z 137 (benzyl moiety), the m/z 344 reaction product yielded fragment ions of m/z 192 and m/z 151 (methylated benzyl moiety) corresponding to 3′-O-methylated (S)-reticuline, which is also known as codamine. The CID spectrum of the reaction product with m/z 358 matched that of laudanosine, which displays fragment ions of m/z 206 (methylated isoquinoline moiety) and m/z 151 (Supplemental Table S2). Incubation of SOMT1 with (S)-norreticuline also resulted in two reaction products, the first of which produced a CID spectrum similar to that of codamine except for the replacement of m/z 192 with m/z 178, owing to the absence of an N-methyl group. As such, this reaction product with m/z 330 was characterized as norcodamine. The CID spectrum of the second product matched that of tetrahydropapaverine (Fig. 5D; Supplemental Table S2). SOMT2 and SOMT3 did not accept (S)-tetrahydrocolumbamine, (S)-norreticuline, or (S)-reticuline as substrates (Fig. 5, B–D).
Involvement of SOMT1, SOMT2, and SOMT3 in Noscapine Biosynthesis
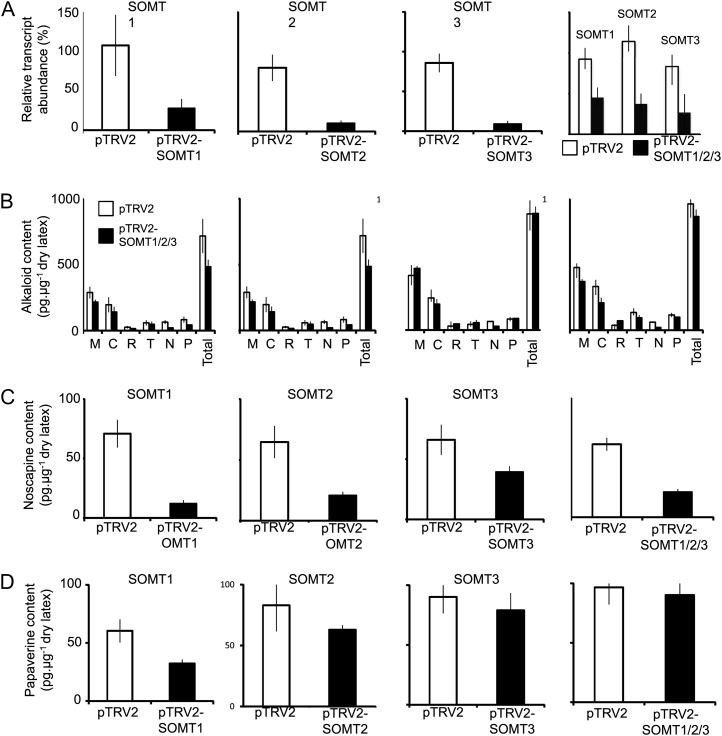
Unique regions of SOMT1, SOMT2, and SOMT3 were deployed to specifically knock down the expression of these genes in opium poppy plants using VIGS (Supplemental Fig. S4). Individual silencing of SOMT1, SOMT2, and SOMT3 and simultaneous cosilencing of all three genes were performed in the Bea’s Choice cultivar of opium poppy, which accumulates noscapine and papaverine in addition to morphinan alkaloids. Tobacco rattle virus (TRV) infection of infiltrated plants was confirmed by reverse transcription-PCR amplification of the viral coat protein (Supplemental Fig. S5). SOMT1, SOMT2, and SOMT3 transcript levels were significantly reduced in plants infiltrated with Agrobacterium tumefaciens harboring the pTRV2-SOMT1, pTRV-SOMT2, and pTRV2-SOMT3 constructs compared with empty pTRV2 vector controls (Fig. 6A). Plants infiltrated with A. tumefaciens harboring pTRV2-SOMT1/2/3 showed reduced expression of all three SOMT genes, although the transcript levels were not as low as those resulting from individually targeted SOMT constructs (Fig. 6A). Silencing SOMT1 or SOMT2 did not affect the relative abundance of transcripts encoding several other tested OMTs (Supplemental Fig. S6). However, silencing SOMT3 was accompanied by a significant reduction in N7OMT transcript levels, which is likely due to its similarity to SOMT3 (Fig. 3). Although 7-O-methylation of the simple benzylisoquinoline backbone is required for papaverine biosynthesis, N7OMT is not involved in the formation of noscapine (Fig. 1).
Figure 6.
Effects of silencing SOMT1, SOMT2, SOMT3, or all three genes simultaneously (SOMT1/2/3), on BIA accumulation in opium poppy. A, Relative OMT transcript abundance in plants infiltrated with the empty pTRV2 vector (control), pTRV2-SOMT1, pTRV2-SOMT2, pTRV2-SOMT3, or pTRV2-SOMT1/2/3. For plants infiltrated with pTRV2-SOMT1/2/3, only plants that showed a reduction in the abundance of all target transcripts were analyzed. B, Abundance of major BIAs in control plants and plants infiltrated with pTRV2-SOMT1, pTRV2-SOMT2, pTRV2-SOMT3, and pTRV2-SOMT1/2/3, which silenced SOMT1, SOMT2, SOMT3, and SOMT1/2/3, respectively. C and D, Relative abundance of noscapine (C) and papaverine (D) in plants infiltrated with pTRV2, pTRV2-SOMT1, pTRV2-SOMT2, pTRV2-SOMT3, or pTRV2-SOMT1/2/3. Values represent means ± sd of three technical replicates performed on each of three biological replicates for each of six infiltrated plants. The differences in transcript abundance and alkaloid content of control and silenced plants were analyzed by an unpaired two-tailed Student’s t test. The transcript abundance of all silenced plants is significantly different from that of control plants at P < 0.05. The noscapine abundance of plants silenced with pTRV2-SOMT1, pTRV2-SOMT2, and pTRV2-SOMT1/2/3 was statistically different from that of control plants. Only silencing of SOMT1 resulted in significant reduction of papaverine (D). Plants silenced using pTRV2-SOMT3 showed no significant difference in noscapine levels compared with controls. Silencing of SOMT genes did not affect the total alkaloid content.
Total alkaloid content and the levels of major alkaloids including morphine, codeine, reticuline, and thebaine remained unchanged in all SOMT-silenced plants compared with empty vector controls (Fig. 6B). However, lower SOMT1 and SOMT2 transcript levels were accompanied by a significant reduction in the accumulation of noscapine compared with controls (Fig. 6C; Supplemental Fig. S7). In contrast, noscapine levels were not significantly different in SOMT3-silenced plants compared with controls. Cosilencing of SOMT1, SOMT2, and SOMT3 reduced noscapine accumulation to a level similar to that resulting from the individual silencing of SOMT1. Papaverine accumulation was also significantly reduced in SOMT1-silenced plants compared with controls (Fig. 6D; Supplemental Fig. S7). LC-MS/MS analysis showed that SOMT1-silenced plants also accumulated substantially higher levels of scoulerine and lower levels of narcotoline, whereas SOMT2-silenced plants contained higher levels of tetrahydrocolumbamine compared with controls (Fig. 7; Supplemental Table S3).
Figure 7.
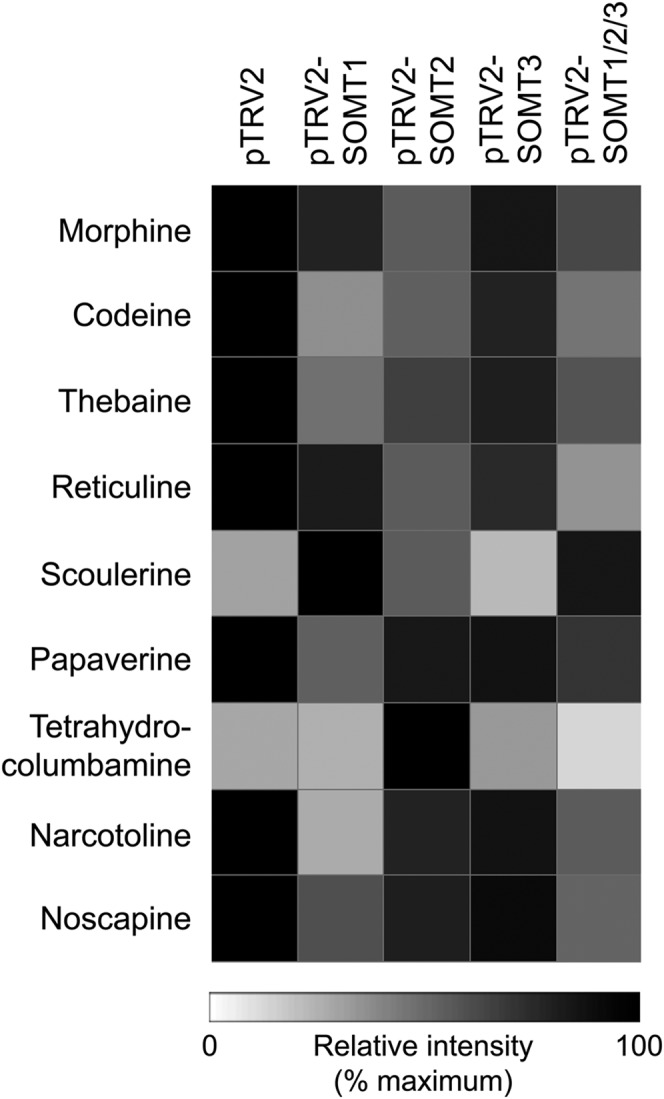
Alkaloid contents of control and SOMT1-, SOMT2-, SOMT3-, and SOMT1/2/3-silenced plants determined using LC-MS/MS. Silencing SOMT1 resulted in increased scoulerine levels and decreased narcotoline levels compared with control plants. A reduction in papaverine accumulation was also observed. Silencing SOMT2 increased tetrahydrocolumbamine levels.
Expression of SOMT1, SOMT2, and SOMT3 in Opium Poppy
Expression analysis by real-time quantitative PCR (RT-qPCR) showed the occurrence of SOMT transcripts in all plant organs, with the highest levels detected in aerial organs (Fig. 8). SOMT1 and SOMT2 transcripts were more abundant in stems, whereas SOMT3 transcript levels were highest in leaves. Transcript levels for all SOMT genes were lowest in roots.
Figure 8.
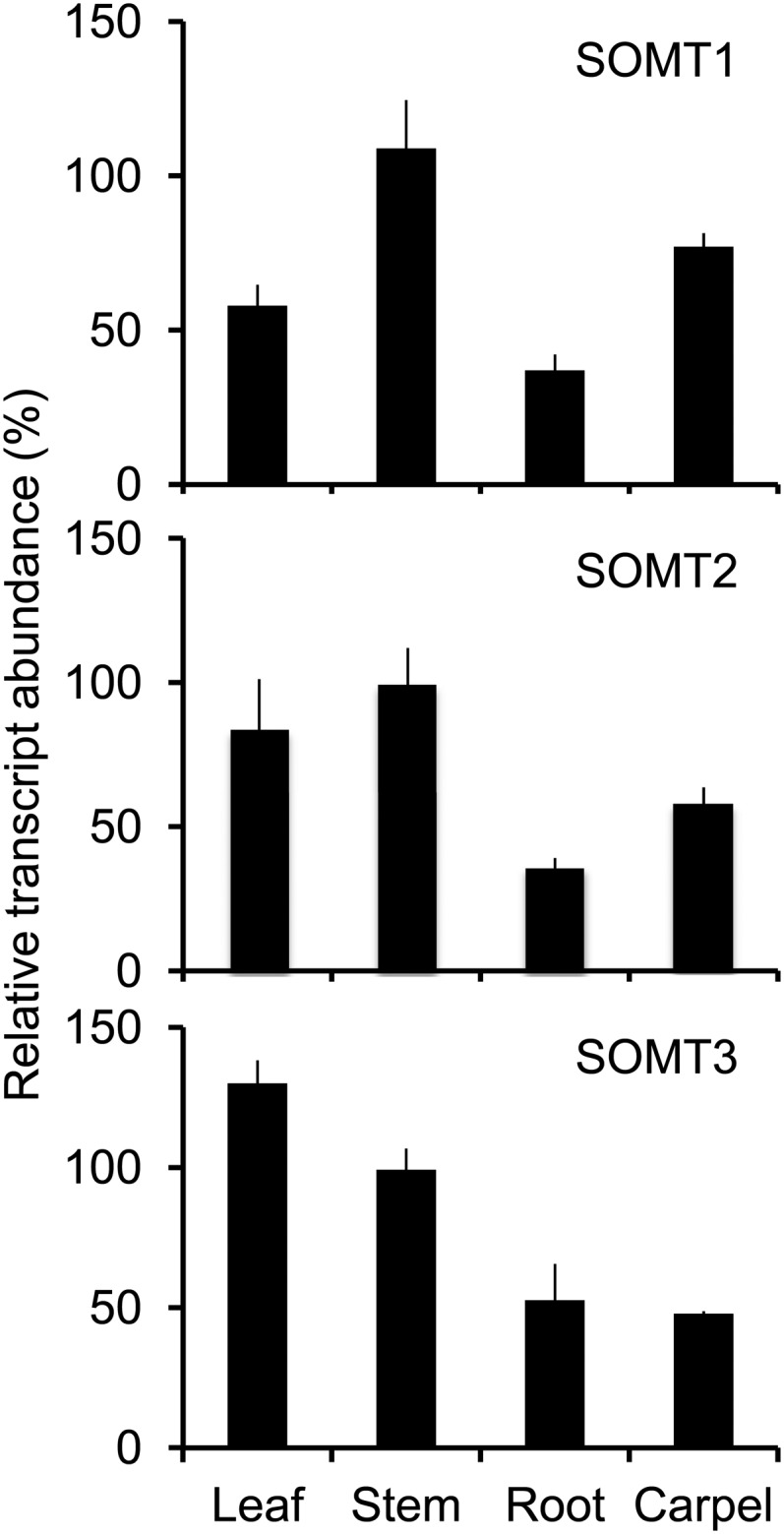
Relative abundance of SOMT1, SOMT2, and SOMT3 transcripts in opium poppy plant organs. RT-qPCR was performed with cDNA synthesized using total RNA from each organ and the primers indicated in Supplemental Table S4.
DISCUSSION
Targeted metabolite profiling of eight opium poppy cultivars with different alkaloid profiles revealed four (Marianne, Natasha, Roxanne, and Veronica) containing high levels of narcotoline and/or noscapine (Fig. 2A) and two (Roxanne and Veronica) that also accumulated papaverine (Desgagné-Penix et al., 2012). These three alkaloids were detected only at trace levels, or were undetected, in the other four cultivars. Differential gene expression analysis showed that all three SOMT candidates were expressed only in noscapine-producing cultivars. A cDNA encoding SOMT was originally characterized from C. japonica, where it was implicated in the biosynthesis of berberine (Takeshita et al., 1995). In contrast to C. japonica, in which berberine and other quaternary protoberberine alkaloids are major products, opium poppy accumulates relatively little, if any, berberine (Desgagné-Penix et al., 2012). Nevertheless, the possibility that the phthalideisoquinoline alkaloid biosynthetic pathway involves a three-step enzymatic conversion of (S)-scoulerine to (S)-N-methylcanadine prompted us to consider the role for opium poppy SOMT homologs in the formation of noscapine metabolism through in vitro biochemical characterization and in planta transient gene silencing.
SOMT1, SOMT2, and SOMT3 all showed homology with previously characterized OMTs involved in BIA metabolism, although, interestingly, within three rather distant phylogenetic clades (Fig. 3). The phylogenetic separation of three enzymes with SOMT activity suggests that the common catalytic function with respect to the 9-O-methylation of (S)-scoulerine emerged independently among existing OMTs in opium poppy. From an evolutionary perspective, ancestral enzymes are purported to have exhibited relatively promiscuous activities to provide a starting point for the optimization of new catalytic functions (Khersonsky and Tawfik, 2010). However, the physiological role for an enzyme is not necessarily represented by its in vitro biochemical function. All three opium poppy SOMT homologs accepted (S)-scoulerine as a substrate and yielded (S)-tetrahydrocolumbamine as a reaction product (Fig. 5). Independent or coordinated silencing of SOMT genes in opium poppy plants supported roles for SOMT1 and SOMT2 in the biosynthesis of noscapine and for SOMT1 in the formation of papaverine (Fig. 6; Supplemental Fig. S5). Interestingly, the expression of all three SOMT genes correlated with noscapine accumulation in certain opium poppy cultivars (Fig. 2), suggesting that the cognate enzymes all contribute to the formation of (S)-tetrahydrocolumbamine in the biosynthesis of noscapine and potentially other BIAs.
The three SOMT variants showed differential efficiency for the 9-O-methylation of (S)-scoulerine, with SOMT1 exhibiting the highest affinity and catalytic efficiency (Table I). The higher Km for (S)-reticuline might reflect the larger cellular pool of this alkaloid (0.8% of the total alkaloid content) compared with scoulerine (0.1% of the total alkaloid content; Frick et al., 2005). The lower kcat/Km values of SOMT2 and SOMT3 compared with SOMT1 suggest that SOMT2 and SOMT3 function less efficiently with scoulerine as the substrate. It is notable that none of the SOMT variants converted narcotoline to noscapine (Fig. 1), suggesting that (1) the 1-O-methyl group of noscapine is added to an unavailable intermediate upstream of narcotoline or (2) another OMT is involved. The recent discovery of 2-oxoglutarate/Fe2+-dependent dioxygenases catalyzing the O-demethylation of morphinan alkaloids in opium poppy (Hagel and Facchini, 2010) raises the possibility that narcotoline results from the 1-O-demethylation of noscapine. However, none of the three O-demethylases tested accepted noscapine as a substrate (Hagel and Facchini, 2010).
The molecular mass of SOMT1 was 3.6 and 5.2 kD higher than that of SOMT2 and SOMT3, respectively, partly due to a unique N-terminal extension previously shown to play a role in substrate specificity through an interaction with its partner monomer and the formation of the substrate-binding pocket (Zubieta et al., 2001; Morishige et al., 2010). The 2-O-methylation of (S)-scoulerine by SOMT1 appears dependent on the methylation status of other hydroxyl groups, because the independent formation of 2-O-methylscoulerine was not detected. Moreover, the tetrahydroxylated simple benzylisoquinoline (R,S)-norlaudanosoline was not accepted, whereas (S)-norreticuline with 6- and 4′-O-linked methyl groups is a substrate of SOMT1 (Supplemental Table S2). Other OMTs from opium poppy have been shown to target more than one position on simple benzylisoquinoline, but not protoberberine, substrates. 7OMT was shown to O-methylate (R,S)-orientaline and (R,S)-isoorientaline at C-7 and C-4′, although O-methylation of the C-4′ position represented only a minor fraction (1%) of the reaction products (Ounaroon et al., 2003). In contrast, SOMT1 efficiently catalyzed O-methylation of the C-2 and C-9 hydroxyls on (S)-scoulerine and the C-2 hydroxyl of (S)-tetrahydrocolumbamine (Fig. 5; Supplemental Fig. S2). Similarly, the reaction products of SOMT1 incubated with (S)-reticuline or (S)-norreticuline showed that O-methylation initially occurred at the C-3′ position followed by O-methylation at the C-7 position. Although a native OMT able to 3′-O-methylate a simple benzylisoquinoline substrate has not been reported previously, a chimeric fusion of the N- and C-terminal domains of 6OMT and 4′OMT, respectively, was shown to catalyze the 3′-O-methylation of 3′-hydroxy-N-methylcoclaurine (Morishige et al., 2010). However, the relatively low efficiency of the 3′-O-methylation activity of SOMT1 with respect to simple benzylisoquinoline, compared with protoberberine substrates, suggests the occurrence of a more regiospecific and substrate-specific 3′OMT in opium poppy. The characterization of three enzymes with SOMT activity supports the possibility of similar 3′OMT redundancy.
Plant OMTs show differential selectivity with respect to stereochemistry and the substitution pattern of phenolic hydroxyl groups available on methyl acceptors (Ibrahim et al., 1998). Generally, OMTs involved in BIA biosynthesis display diverse substrate specificities, with enzymes operating early in the pathway being more promiscuous. For example, 6OMT and 4′OMT from C. japonica and opium poppy accept simple benzylisoquinoline and protoberberine substrates (Sato et al., 1994; Morishige et al., 2000; Ziegler et al., 2005). In contrast, N7OMT from opium poppy accepts only norreticuline (Pienkny et al., 2009), whereas 7OMT can catalyze the O-methylation of certain phenolic compounds (e.g. isovanillic acid and guaiacol) in addition to a range of simple benzylisoquinolines (i.e. reticuline, orientaline, protosinomeine, and isoorientaline; Ounaroon et al., 2003). Low activity toward (S)-norreticuline was reported for OMTs from Thalictrum tuberosum plants (Frick and Kutchan, 1999) and Argemone platyceras cell cultures (Rueffer et al., 1983), although the regiospecificity was not determined, and SOMT from C. japonica catalyzed the O-methylation of protoberberine alkaloids (Morishige et al., 2000). Strict specificity for N-desmethyl benzylisoquinoline substrates was also shown by 7OMT (i.e. reticuline) and N7OMT (i.e. norreticuline) from opium poppy (Ounaroon et al., 2003; Pienkny et al., 2009). SOMT1 is the only OMT in BIA biosynthesis shown to function efficiently on both N-methylated and N-desmethyl benzylisoquinolines (Fig. 5; Table I) and to perform sequential regiospecific O-methylations on a protoberberine substrate. Enzymes catalyzing the 2-O-methylation of the isoquinoline moiety on a protoberberine alkaloid, or the 3′-O-methylation of the benzyl moiety on a benzylisoquinoline substrate, have not been reported previously.
Multifunctional OMTs acting on different substrate classes, such as phenylpropanoids, flavonoids, and alkaloids, are well known (Gauthier et al., 1998; Frick and Kutchan, 1999; Chiron et al., 2000), although an OMT capable of the sequential O-methylation of a simple benzylisoquinoline at C-3′ and C-7 has not been reported. A flavonol OMT from Catharanthus roseus was shown to perform two sequential O-methylations (Cacace et al., 2003). Similarly, orcinol OMT was reported to carry out two sequential O-methylations in rose (Rosa × hybrida) scent biosynthesis (Lavid et al., 2002; Scalliet et al., 2002). Sequential methylation has also been reported for certain N-methyltransferases (Nuccio et al., 2000; Charron et al., 2002), but these proteins possess a different structural domain. In comparison, SOMT1 accepted both benzylisoquinoline and protoberberine substrates, showed no discrimination toward N-desmethyl or N-methylated BIAs, and was able to catalyze sequential O-methylations on both the isoquinoline and benzyl moieties. Interestingly, SOMT1 displays equivalent regiospecificity for the hydroxyl functions at the respective C-9 and C-3′ and the C-2 and C-7 positions of the benzylisoquinoline and protoberberine backbones, which differ only by the occurrence of a methylene bridge between C-2′ and the N-methyl carbon in the latter. As such, SOMT should be useful for substrate binding and structural biology investigations.
The abundance of SOMT1, SOMT2, and SOMT3 transcripts in opium poppy aerial organs (Fig. 8) facilitated an in planta assessment of the role for the cognate enzymes in noscapine biosynthesis using VIGS (Lu et al., 2003). The application of VIGS as an effective method to silence specific genes in opium poppy allows for a direct investigation of the physiological roles of putative biosynthetic enzymes (Hagel and Facchini, 2010; Lee and Facchini, 2010, 2011). The substantial reduction in noscapine accumulation in response to the decreased abundance of SOMT1 and SOMT2 transcripts (Supplemental Fig. S5) did not affect total alkaloid content (Fig. 6B) and supports a joint role for these enzymes in phthalideisoquinoline alkaloid metabolism. Compared with SOMT1 and SOMT2, a reduction in SOMT3 transcript levels was accompanied by a modest decrease in total alkaloid accumulation, but SOMT3 transcript levels did not correlate as strongly with the relative abundance of noscapine (Supplemental Fig. S5), suggesting another physiological role for this enzyme. Simultaneously cosilencing all three SOMT genes reduced the relative abundance of noscapine to a level similar to that observed in SOMT2-silenced plants but not as low as in SOMT1-silenced plants, owing most likely to the diminished effectiveness of the tripartite pTRV2-SOMT1/2/3 vector compared with the individual pTRV2-SOMT constructs (Fig. 6). A knockout rather than a knockdown approach would seem necessary to produce the noscapine-free phenotype of the Marianne, Natasha, Roxanne, and Veronica cultivars (Fig. 2A).
SOMT1 silencing also resulted in reduced papaverine levels (Fig. 7), which is in agreement with the activity of the cognate enzyme on the C-3′ hydroxyl group of benzylisoquinoline alkaloid substrates. The biosynthesis of papaverine remains controversial, with suggestions that (S)-reticuline (Han et al., 2010) or (S)-norreticuline (Pienkny et al., 2009) serve as intermediates. The (S)-reticuline pathway requires the terminal N-demethylation of (S)-laudanosine by an unknown enzyme, whereas the (S)-norreticuline pathway proceeds entirely via N-desmethyl intermediates to (S)-tetrahydropapaverine. Conversion of (S)-norlaudanosoline to (S)-tetrahydropapaverine involves the O-methylation of all four hydroxyl groups. Silencing of the cognate gene shows that the 3′OMT activity on benzylisoquinoline substrates of SOMT1 contributes to papaverine biosynthesis (Fig. 7D). However, the acceptance of both (S)-reticuline and (S)-norreticuline as substrates does not discount either the N-desmethyl or the N-methyl benzylisoquinoline routes. The targeted correlation between chemotype and gene expression profile was not focused on the occurrence of papaverine; thus, additional opium poppy variants with 3′OMT activity are expected.
MATERIALS AND METHODS
Plant Material
Eight different opium poppy (Papaver somniferum) cultivars were cultivated in a growth chamber (Conviron; www.conviron.com) at 20°C/18°C (light/dark) with a photoperiod of 16 h and a combination of cool-white fluorescent (Sylvania) and incandescent lights (Facchini and Park, 2003). Stem and latex were harvested 1 to 2 d before anthesis for metabolite and transcript analysis. The opium poppy cv Bea’s Choice was grown under greenhouse conditions for VIGS experiments.
Chemicals
(S)-Reticuline was a gift from Tasmanian Alkaloids (www.tasalk.com). (S)-Scoulerine was converted from (S)-reticuline using opium poppy berberine bridge enzyme produced in Pichia pastoris. Narcotoline was isolated from the latex of the opium poppy cv Marianne. Latex was extracted in methanol, concentrated under reduced pressure, and subjected to thin-layer chromatography on Silica Gel 60 F254 plates (EMD Chemicals; www.emdchemicals.com) developed using toluene:acetone:ammonia ethanol (5:4:1, v/v/v). Silica from regions of the thin-layer chromatography plate corresponding to the RF value of authentic narcotoline was scraped off and extracted with methanol. Aliquots were pooled and concentrated under reduced pressure, and narcotoline purity was confirmed by LC-MS/MS. (S)-Isocorydine, (S)-bulbocapine, (S)-boldine, (S)-corytuberine, and berbamine were purchased from Sequoia Research Products (www.seqchem.com). All other alkaloids were obtained as described previously (Liscombe and Facchini, 2007; Hagel and Facchini, 2010). [14C]SAM was purchased from American Radiolabeled Chemicals (www.arc-inc.com). All other chemicals were purchased from Sigma-Aldrich (www.sigmaaldrich.com).
Phylogenetic Analysis
Amino acid alignment (Supplemental Fig. S1) was performed using ClustalW (Larkin et al., 2007), and a neighbor-joining phylogenetic tree was constructed using TREEVIEW (Page, 1996) with bootstrap values generated using TREECON (Van de Peer and De Wachter, 1994). Abbreviations and GenBank accession numbers for the sequences used are as follows: EcOMT, putative Eschscholzia californica OMT (ACO90220.1); CjCoOMT, Coptis japonica columbamine OMT (Q8H9A8.1); TtOMT, Thalictrum tuberosum catechol OMT (AAD29845.1); TtCaOMT, T. tuberosum catechol OMT (AAD29843.1); PsCaOMT, P. somniferum catechol OMT (AAQ01670.1); PsN7OMT, P. somniferum norreticuline 7OMT (ACN88562.1); Ps6OMT, P. somniferum norcoclaurine 6OMT (AAP45315.1); Ps4′OMT2, P. somniferum 3′-hydroxy-N-methylcoclaurine 4′OMT2 (AAP45314.1); Ps4′OMT1, P. somniferum 3′-hydroxy-N-methylcoclaurine 4′OMT1 (AAP45314.1); Cc4′OMT, Coptis chinensis 3′-hydroxy-N-methylcoclaurine 4′OMT (ABY75613.1); Cj4′OMT, C. japonica 3′-hydroxy-N-methylcoclaurine 4′OMT (Q9LEL5.1); Tf4′OMT, Thalictrum flavum 3′-hydroxy-N-methylcoclaurine 4′OMT (AAU20768.1); Cj6OMT, C. japonica norcoclaurine 6OMT (Q9LEL6.1); Tf6OMT, T. flavum norcoclaurine 6OMT (AAU20765.1); VvReOMT, Vitis vinifera resveratrol OMT (CAQ76879.1); PtFlOMT, Populus trichocarpa flavonoid OMT predicted protein (XP_002312933.1); CjSOMT, C. japonica scoulerine 9OMT (Q39522.1); TfSOMT, T. flavum scoulerine 9OMT (AAU20770.1); PaCafOMT, Picea abies caffeate OMT (CAI30878.1); CaCafOMT, Capsicum annuum caffeate OMT (AAG43822.1); PsOMT1, P. somniferum SOMT1 (JN185323); PsOMT2, P. somniferum SOMT2 (JN185324); PsOMT3, P. somniferum SOMT3 (JN185325); ObEuOMT, Ocimum basilicum eugenol OMT (AAL30424.1); MpFlOMT, Mentha × piperita flavonoid 8OMT (AAR09600.1); ObCafOMT, O. basilicum caffeate OMT (AAD38189.1); CbEuOMT, Clarkia breweri (iso)eugenol OMT (AAC01533.1); CbCafOMT, C. breweri caffeate OMT (O23760.1); and AmCafOMT, Ammi majus caffeate OMT (AAR24095.1).
Transcript and Metabolite Profiling of Opium Poppy Cultivars
Alkaloid extracts of opium poppy latex were subjected to HPLC and LC-MS/MS analysis as described previously (Desgagné-Penix et al., 2012). For the quantification of alkaloids, dextromethorphan was used as an internal standard. Total RNA from stems from which latex was isolated was sequenced using Titanium FLX series reagents on a Genome Sequencer FLX instrument at the McGill University-Genome Quebec Innovation Center (http://gqinnovationcenter.com; Desgagné-Penix et al., 2012). Sequencing data were assembled and annotated on the FIESTA2 platform developed by the National Research Council-Plant Biotechnology Institute (www.pbi.nrc.ca). All raw data from 454 pyrosequencing were uploaded to the sequence read archive in the National Center for Biotechnology Information database (http://www.ncbi.nlm.nih.gov) with the following accession numbers: 40 (SRX012994), Marianne (SRX012995), Natasha (SRX012996), Deborah (SRX012997), Przemko (SRX012998), Roxanne (SRX012999), T (SRX013000), and Veronica (SRX013001; Desgagné-Penix et al., 2012).
Integration of Transcript and Targeted Metabolite Profiles
Stem transcriptomes of the eight opium poppy cultivars were compared to identify differentially expressed genes that potentially correlated with the occurrence of certain alkaloids. A collection of gene candidates was generated based on (1) sequence similarity searches for putative enzymes catalyzing reactions potentially occurring via the same mechanism in hypothetical or empirical pathways using BLAST and (2) correlation of gene expression profiles with specific phenotypes. The sequence database was also searched for genes that annotated as methyltransferases and showed significant similarity with characterized OMTs involved in BIA biosynthesis. The overall result was a collection of OMT candidates expressed only in noscapine-producing cultivars. Full-length SOMT candidates were identified and assembled in silico by screening each database using the tBLASTn algorithm. Relative transcript abundance was determined based on the frequency of representation of a specific sequence in each database (i.e. the number of reads). The number of reads representing a specific gene was normalized over the total number of reads in each database.
Gene Expression Analysis
Total RNA from different organs of the opium poppy cv Bea’s Choice was extracted, and cDNA was synthesized as described previously (Lee and Facchini, 2011). The relative abundance of candidate SOMT transcripts in each organ was determined by RT-qPCR analysis using the primers described in Supplemental Table S4.
Cloning and Expression of OMT Candidates
Full-length open reading frames encoding SOMT candidates were amplified from cDNA templates derived from the opium poppy cv Roxanne using Phusion polymerase (New England Biolabs; www.neb.com) and forward and reverse primers with flanking BamHI and PstI restriction sites (Supplemental Table S4). After A-tailing with Taq DNA polymerase, amplicons were individually inserted into pGEM-T (Promega; www.promega.com) and cloned in Escherichia coli strain XL1BlueMRF. For heterologous expression of His-tagged SOMT proteins, plasmids was digested with BamHI and PstI, ligated into pRSETA (Invitrogen; www.invitrogen.com) precut with BamHI and PstI, and transformed in E. coli strain Rosetta(DE3)pLysS (EMD Chemicals).
For the production of recombinant SOMT proteins, bacteria were cultured in Luria-Bertani medium to an A600 of 0.6 and subsequently induced with 1 mm isopropyl β-d-thiogalactopyranoside. Cultures were incubated at 16°C on a gyratory shaker at 200 rpm. Cells were harvested and sonicated in binding buffer (50 mm potassium phosphate, pH 7.5, 100 mm NaCl, 10% [v/v] glycerol, and 1 mm β-mercaptoethanol). Cleared lysates obtained after centrifugation at 10,000g for 10 min were loaded onto Talon cobalt affinity resin (Clontech; www.clontech.com). The resin was washed three times in binding buffer and eluted using aliquots of binding buffer containing increasing concentrations of imidazole to obtain purified proteins. Purified and His-tagged recombinant SOMT proteins were eluted in binding buffer containing 100 mm imidazole. Desalting was performed on PD10 columns (GE Healthcare; www.gehealthcare.com) in storage buffer (50 mm potassium phosphate, pH 7.5, 100 mm NaCl, and 10% [v/v] glycerol), and protein concentrations were determined using the Bio-Rad Protein Assay (www.bio-rad.com). The purity of recombinant proteins was determined by SDS-PAGE.
Enzyme Assays and Biochemical Characterization of OMTs
The standard enzyme assay for OMT activity was performed using a reaction mixture of 100 mm Gly-NaOH, pH 9.0, 25 mm sodium ascorbate, 100 μm SAM (10% mol/mol [methyl-14C]SAM {specific activity of 55 mCi mmol−1} and 90% mol/mol unlabeled SAM), 10% (v/v) glycerol, 1 mm β-mercaptoethanol, 100 μm of the potential alkaloid substrate, and 50 μg of purified recombinant enzyme. Assays were carried out at 37°C for 1 or 4 h and terminated by adding 200 μL of 1 m NaHCO3. Products were extracted three times with 200 μL of ethyl acetate. The combined organic phases were added to 3 mL of Ultima GOLD MV scintillation cocktail (Perkin-Elmer; www.perkinelmer.com), and radioactivity was quantified with a Beckman LS6000 liquid scintillation counter (Beckman-Coulter; www.beckmancoulter.com). Alkaloids belonging to different structural subgroups were tested as potential enzymatic substrates (Supplemental Fig. S2). For product identification by LC-MS/MS, enzyme assays were conducted using unlabeled SAM. Controls were performed with denatured protein extracts prepared by boiling in water for at least 15 min. Kinetic parameters were determined by varying alkaloid concentrations from 1 to 500 μm at a fixed concentration of 200 μm SAM under optimal temperature and buffer conditions. The kinetic data for SAM were obtained by varying the SAM concentration between 1 and 200 μm at a constant alkaloid concentration of 100 μm. Kinetic constants were determined by fitting initial velocity versus substrate concentration to the Michaelis-Menten equation using GraphPad Prism 5 (www.graphpad.com). Each point represents mean specific activity ± sd monitored as a function of substrate concentration for three independent replicates.
LC-MS/MS
Enzyme assays were diluted 1:10 with 0.2% (v/v) glacial acetic acid and analyzed using a 6410 Triple Quadruple LC-MS/MS system (Agilent Technologies; www.agilent.com). Liquid chromatography was performed using a Zorbax Eclipse Plus C18 column (2.1 × 50 mm, 1.8-μm particle size; Agilent Technologies) at a flow rate of 0.4 mL min−1. The column was equilibrated in solvent A (2% [v/v] acetonitrile, 0.2% [v/v] acetic acid, and 97.8% [v/v] water), and alkaloids were eluted under the following conditions: 0 to 1 min, 0% solvent B (98% [v/v] acetonitrile, 0.2% [v/v] acetic acid, and 2% [v/v] water); 1 to 10 min, to 35% solvent B; and 10 to 11 min, to 100% solvent B. Samples were injected into the mass analyzer by an ESI probe inlet. Ions were generated and focused using an ESI voltage of 4,000 kV, 9 L min−1 gas flow, nebulizing pressure of 40 ψ, and gas temperature of 330°C. MS data were acquired in positive ion mode over 50 to 500 m/z. For product identification and annotation, the CID mass spectra were recorded using collision energies of –25.0 eV (scoulerine, tetrahydrocolumbamine, codamine, laudanosine, norreticuline, reticuline, norcodamine) and –20 eV (tetrahydropalmatine, tetrahydropapaverine). Argon collision gas was set at a pressure of 1.8 × 10−3 torr.
Identification and annotation of reaction products were achieved by comparing empirical ESI[+]-MS data with those of authentic standards and using published reference spectra, respectively, as indicated in Supplemental Table S2. The putative characterization of BIAs according to compound class was based on the generation of certain fragment ions. The fragmentation patterns and mechanisms of benzylisoquinoline-type alkaloids have been investigated extensively (Gioacchini et al., 2000; Schmidt et al., 2005, 2007) using ESI[+]-CID (Raith et al., 2003; Schmidt et al., 2005, 2007). Generally, fragmentation at the α-carbon linking the isoquinoline and benzyl moieties yields both an isoquinoline and a benzyl ion (Schmidt et al., 2005, 2007). Formation of an ion corresponding to the loss of ammonia or methylamine from the isoquinoline moiety indicated whether the compound was N-methylated or N-desmethyl, respectively (Schmidt et al., 2005, 2007). The benzylisoquinoline moiety is the result of a rearrangement with reversed charge distribution involving the proton on the tetraisoquinoline nitrogen and the aromatic ring of the benzyl substituent (Schmidt et al., 2005). The complementary ion that represents the benzyl group is formed by a secondary fragmentation event. In summary, the fragmentation of simple benzylisoquinolines produces prominent diagnostic ions [M+H]+, [M+H-NH3 or M+H-NCH3]+, [M+H isoquinoline]+, and [M+H benzyl]+ that are useful for characterization. For instance, the [M+H]+ of norcodamine of 330 is the same as (S)-reticuline, but the retention time is different (Supplemental Table S2). The [M+H]+ for norcodamine is 14 mass units higher than norreticuline, corresponding to a methyl group. The diagnostic fragment ion [M+H-NH3]+ at 178 indicates that the compound is not N-methylated. The fragment ion at 151 corresponds to the benzyl moiety [M+H benzyl]+ with an additional 14 mass units compared with (S)-reticuline or (S)-norretriculine. The isoquinoline fragment [M+H isoquinoline]+ at 178 is identical to that of norreticuline. By comparing the CID spectra of norreticuline, reticuline, and codamine, the compound with [M+H]+ of 330 was characterized as 3′-O-methylnorreticuline (norcodamine), the first SOMT1 O-methylation product derived from norreticuline. A similar strategy was used to characterize other simple benzylisoquinolines.
VIGS
To perform VIGS on SOMT candidates in opium poppy, the TRV vector system was used (Dinesh-Kumar et al., 2003). Unique 3′ untranslated region and open reading frame sequences were used to construct VIGS vectors designed to specifically silence genes encoding SOMT1, SOMT2, and SOMT3 and to avoid the suppression of highly homologous genes (Supplemental Fig. S4). For simultaneous silencing of SOMT1, SOMT2, and SOMT3, a construct containing all three SOMT-specific fragments was synthesized. Individual gene fragments were amplified from cDNA using primers flanking XbaI and XhoI restriction endonuclease (Supplemental Table S4). After A-tailing with Taq-DNA polymerase, amplicons were cloned into pGEM-T and subcloned into pTRV2. The pTRV2 constructs (i.e. pTRV2, pTRV2-SOMT1, pTRV2-SOMT2, pTRV2-SOMT3, and pTRV2-SOMT1/2/3) were isolated and introduced independently into Agrobacterium tumefaciens strain GV3101 by electroporation. Overnight cultures of a single colony were used to inoculate 1,000 mL of Luria-Bertani medium supplemented with 10 mm MES, 20 μm acetosyringone, and 50 μg mL−1 kanamycin. Cultures were incubated overnight at 28°C on a gyratory shaker at 200 rpm. Bacteria were collected by centrifugation at 3,000g for 15 min, and the pellet was resuspended in infiltration solution (10 mm MES, 200 μm acetosyringone, and 10 mm MgCl2) to an A600 of 2.5. A. tumefaciens harboring pTRV2, pTRV2-SOMT1, pTRV2-SOMT2, pTRV2-SOMT3, or pTRV2-SOMT1/2/3 were each mixed 1:1 (v/v) with A. tumefaciens containing pTRV1 and incubated at room temperature for 2 h before infiltration. Apical meristems of 2- to 3-week-old seedlings were infiltrated with the mixed cultures using a 5-mL syringe. Seedlings infiltrated with pTRV1 and pTRV2-PsPDS, containing a fragment of the phytoene desaturase gene from opium poppy (Hileman et al., 2005), were used as visual markers of VIGS efficiency based on photobleaching. Infiltrated plants were then grown in a greenhouse for 8 to 10 weeks before collecting stem and latex for analysis.
Metabolite Profiling and Transcript Analysis of VIGS Plants
Three 1-cm stem segments excised immediately below the flower buds and approximately 10 μL of exuding latex were collected and flash frozen in liquid nitrogen. Viral infection of infiltrated plants was confirmed by screening for TRV coat protein transcripts using the primers listed in Supplemental Table S4. The detection of glyceraldehyde 3-phosphate dehydrogenase transcripts was used as a positive PCR control. Latex samples of infected plants were reduced to dryness, resuspended in methanol at a concentration of 0.1 μg μL−1, and extracted overnight. One microliter of each extract was diluted with 300 μL of solvent A (98% [v/v] water, 2% [v/v] acetonitrile, and 0.02% [v/v] phosphoric acid) and analyzed using a System Gold HPLC apparatus and photodiode array detector (Beckman-Coulter). HPLC separations were performed at a flow rate of 1.5 mL min−1 on a LiChrospher RP-Select B 5μ column, 150 × 4.6 mm (Alltech; www.alltech.com) using a gradient of solvent A and solvent B (98% [v/v] acetonitrile, 2% [v/v] water:0.04% [v/v] H3PO4). Chromatography was initiated in 2% solvent B and increased to 10% solvent B after 5 min. The gradient was then increased to 35% solvent B over 40 min and finally to 100% solvent B over 5 min. Peaks corresponding to morphine, codeine, reticuline, thebaine, oripavine, narcotoline, noscapine, and papaverine were monitored at 210 and 280 nm and identified based on their retention times and UV spectra compared with authentic standards. Alkaloid content was calculated as pg alkaloid mg−1 dry weight of latex based on standard quantification curves.
For transcript analysis, cDNA samples derived from stem segments of infected plants were subjected to RT-qPCR analysis using a 7300 Real-Time PCR system (Applied Biosystems; www.appliedbiosystems.com) for 40 cycles. Each 10-μL PCR contained 1 μL of cDNA, 300 nm forward and reverse primers (Supplemental Table S4), and 1× Power SYBR Green PCR Master Mix (Applied Biosystems). The opium poppy Actin gene was used as an endogenous reference, and the plant line showing the highest expression level (empty vector) served as the calibrator for each target gene. Reactions were subjected to 40 cycles of template denaturation, primer annealing, and primer extension. The amplicons of all primer pairs were subjected to dissociation curve analysis using the method suggested by the instrument manufacturer (Applied Biosystems) to evaluate RT-qPCR specificity. Gene expression data for VIGS analysis were analyzed based on 54 independent values per plant line (i.e. three technical replicates performed on each of three stem segments taken from each of six individual plants). To evaluate silencing specificity, plants were subjected to RT-qPCR analysis with primers that targeted 4′OMT2, 6OMT, 7OMT, SOMT1, SOMT2, and SOMT3. Analysis of relative gene expression was performed using the 2−ΔΔCt method (Livak and Schmittgen, 2001). Data were analyzed by paired and unpaired two-way Student’s t-test.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers JN185323 (SOMT1), JN185324 (SOMT2), and JN185325 (SOMT3).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Amino acid sequence alignment of opium poppy SOMT1, SOMT2, and SOMT3 with selected OMTs from different plant species.
Supplemental Figure S2. Substrate specificities of recombinant SOMT1, SOMT2, and SOMT3.
Supplemental Figure S3. Steady-state enzyme kinetics of purified recombinant SOMT1, SOMT2, and SOMT3 using various substrate concentrations.
Supplemental Figure S4. Assembly of VIGS constructs used to silence SOMT1, SOMT2, and SOMT3 and all three genes simultaneously (pTRV2-SOMT1/2/3) in opium poppy.
Supplemental Figure S5. Ethidium bromide-stained agarose gel showing the detection of TRV2 coat protein transcripts amplified by reverse transcription-PCR using total RNA extracted from infiltrated plants.
Supplemental Figure S6. Effects of silencing SOMT1, SOMT2, and SOMT3 or all three genes simultaneously (SOMT1/2/3) on the levels of other OMTs involved in BIA biosynthesis in opium poppy.
Supplemental Figure S7. Correlations in control and gene-silenced plants between the accumulation of noscapine and papaverine and the relative transcript abundance of SOMT1, SOMT2, or SOMT3.
Supplemental Table S1. Relative abundance of major latex alkaloids determined by HPLC in eight opium poppy cultivars and relative abundance of SOMT gene transcripts compared with those of previously characterized OMTs involved in benzylisoquinoline alkaloid biosynthesis in eight opium poppy cultivars.
Supplemental Table S2. Chromatographic and spectral data used for the identification and relative quantification of benzylisoquinoline alkaloids by LC-MS/MS.
Supplemental Table S3. Relative alkaloid contents of control (pTRV2) and SOMT1-, SOMT2-, SOMT3-, and SOMT1/2/3-silenced plants determined using LC-MS/MS.
Supplemental Table S4. Primer sequences used for the assembly of expression vectors, VIGS constructs, confirmation of TRV infection in infiltrated plants, and RT-qPCR analysis.
Supplementary Material
Acknowledgments
We thank Dr. Isabel Desgagné-Penix for constructing the pTRV2-SOMT2 vector and Scott Farrow for assistance with the LC-MS/MS analysis. We are also grateful to the National Research Council-Plant Biotechnology Institute for hosting the sequence databases.
Glossary
- BIA
benzylisoquinoline alkaloid
- SAM
S-adenosyl-l-methionine
- OMT
O-methyltransferase
- VIGS
virus-induced gene silencing
- LC-MS/MS
liquid chromatography-tandem mass spectrometry
- m/z
mass-to-charge ratio
- CID
collision-induced dissociation
- RT-qPCR
real-time quantitative PCR
- ESI
electrospray ionization
- TRV
tobacco rattle virus
References
- Barken I, Geller J, Rogosnitzky M. (2008) Noscapine inhibits human prostate cancer progression and metastasis in a mouse model. Anticancer Res 28: 3701–3704 [PubMed] [Google Scholar]
- Battersby AR, Hirst M. (1965) Concerning the biosynthesis of narcotine. Tetrahedron Lett 11: 669–673 [DOI] [PubMed] [Google Scholar]
- Battersby AR, Hirst M, McCaldin DJ, Southgate R, Staunton J. (1968) Alkaloid biosynthesis. XII. The biosynthesis of narcotine. J Chem Soc Perkin 1 17: 2163–2172 [DOI] [PubMed] [Google Scholar]
- Cacace S, Schröder G, Wehinger E, Strack D, Schmidt J, Schröder J. (2003) A flavonol O-methyltransferase from Catharanthus roseus performing two sequential methylations. Phytochemistry 62: 127–137 [DOI] [PubMed] [Google Scholar]
- Charron JBF, Breton G, Danyluk J, Muzac I, Ibrahim RK, Sarhan F. (2002) Molecular and biochemical characterization of a cold-regulated phosphoethanolamine N-methyltransferase from wheat. Plant Physiol 129: 363–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiron H, Drouet A, Claudot AC, Eckerskorn C, Trost M, Heller W, Ernst D, Sandermann H., Jr (2000) Molecular cloning and functional expression of a stress-induced multifunctional O-methyltransferase with pinosylvin methyltransferase activity from Scots pine (Pinus sylvestris L.). Plant Mol Biol 44: 733–745 [DOI] [PubMed] [Google Scholar]
- Choi KB, Morishige T, Shitan N, Yazaki K, Sato F. (2002) Molecular cloning and characterization of coclaurine N-methyltransferase from cultured cells of Coptis japonica. J Biol Chem 277: 830–835 [DOI] [PubMed] [Google Scholar]
- Desgagné-Penix I, Farrow SC, Cram D, Nowak J, Facchini PJ. (2012) Integration of deep transcript and targeted metabolite profiles for eight cultivars of opium poppy. Plant Mol Biol 79: 295–313 [DOI] [PubMed] [Google Scholar]
- Dinesh-Kumar SP, Anandalakshmi R, Marathe R, Schiff M, Liu Y. (2003) Virus-induced gene silencing. Methods Mol Biol 236: 287–294 [DOI] [PubMed] [Google Scholar]
- Facchini PJ. (2001) Alkaloid biosynthesis in plants: biochemistry, cell biology, molecular regulation and metabolic engineering applications. Annu Rev Plant Physiol Plant Mol Biol 52: 29–66 [DOI] [PubMed] [Google Scholar]
- Facchini PJ, De Luca V. (2008) Opium poppy and Madagascar periwinkle: model non-model systems to investigate alkaloid biosynthesis in plants. Plant J 54: 763–784 [DOI] [PubMed] [Google Scholar]
- Facchini PJ, Hagel JM, Liscombe DK, Loukanina N, MacLeod BP, Samanani N, Zulak KG. (2007) Opium poppy: blueprint for an alkaloid factory. Phytochem Rev 6: 97–124 [Google Scholar]
- Facchini PJ, Park S-U. (2003) Developmental and inducible accumulation of gene transcripts involved in alkaloid biosynthesis in opium poppy. Phytochemistry 64: 177–186 [DOI] [PubMed] [Google Scholar]
- Facchini PJ, Penzes C, Johnson AG, Bull D. (1996) Molecular characterization of berberine bridge enzyme genes from opium poppy. Plant Physiol 112: 1669–1677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frenzel T, Zenk MH. (1990) Purification and characterization of three isoforms of S-adenosyl-L-methionine:(R,S)-tetrahydrobenzylisoquinoline-N-methyltransferase from Berberis koetineana cell cultures. Phytochemistry 29: 3491–3497 [Google Scholar]
- Frick S, Kramell R, Schmidt J, Fist AJ, Kutchan TM. (2005) Comparative qualitative and quantitative determination of alkaloids in narcotic and condiment Papaver somniferum cultivars. J Nat Prod 68: 666–673 [DOI] [PubMed] [Google Scholar]
- Frick S, Kutchan TM. (1999) Molecular cloning and functional expression of O-methyltransferases common to isoquinoline alkaloid and phenylpropanoid biosynthesis. Plant J 17: 329–339 [DOI] [PubMed] [Google Scholar]
- Gauthier A, Gulick PJ, Ibrahim RK. (1998) Characterization of two cDNA clones which encode O-methyltransferases for the methylation of both flavonoid and phenylpropanoid compounds. Arch Biochem Biophys 351: 243–249 [DOI] [PubMed] [Google Scholar]
- Gioacchini AM, Czarnocki Z, Arazny Z, Munari I, Traldi P. (2000) Electrospray ionization, accurate mass measurements and multistage mass spectrometry experiments in the characterization of stereoisomeric isoquinoline alkaloids. Rapid Commun Mass Spectrom 14: 1592–1599 [DOI] [PubMed] [Google Scholar]
- Gözler B. (1983) Egenine: a possible intermediate in phthalideisoquinoline biogenesis. Tetrahedron Lett 39: 577–580 [Google Scholar]
- Hagel JM, Facchini PJ. (2010) Dioxygenases catalyze the O-demethylation steps of morphine biosynthesis in opium poppy. Nat Chem Biol 6: 273–275 [DOI] [PubMed] [Google Scholar]
- Han X, Lamshöft M, Grobe N, Ren X, Fist AJ, Kutchan TM, Spiteller M, Zenk MH. (2010) The biosynthesis of papaverine proceeds via (S)-reticuline. Phytochemistry 71: 1305–1312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hileman LC, Drea S, Martino G, Litt A, Irish VF. (2005) Virus-induced gene silencing is an effective tool for assaying gene function in the basal eudicot species Papaver somniferum (opium poppy). Plant J 44: 334–341 [DOI] [PubMed] [Google Scholar]
- Ibrahim RK, Bruneau A, Bantignies B. (1998) Plant O-methyltransferases: molecular analysis, common signature and classification. Plant Mol Biol 36: 1–10 [DOI] [PubMed] [Google Scholar]
- Ikezawa N, Iwasa K, Sato F. (2007) Molecular cloning and characterization of methylenedioxy bridge-forming enzymes involved in stylopine biosynthesis in Eschscholzia californica. FEBS J 274: 1019–1035 [DOI] [PubMed] [Google Scholar]
- Ikezawa N, Tanaka M, Nagayoshi M, Shinkyo R, Sakaki T, Inouye K, Sato F. (2003) Molecular cloning and characterization of CYP719, a methylenedioxy bridge-forming enzyme that belongs to a novel P450 family, from cultured Coptis japonica cells. J Biol Chem 278: 38557–38565 [DOI] [PubMed] [Google Scholar]
- Khersonsky O, Tawfik DS. (2010) Enzyme promiscuity: a mechanistic and evolutionary perspective. Annu Rev Biochem 79: 471–505 [DOI] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23: 2947–2948 [DOI] [PubMed] [Google Scholar]
- Lavid N, Wang J, Shalit M, Guterman I, Bar E, Beuerle T, Menda N, Shafir S, Zamir D, Adam Z, et al. (2002) O-Methyltransferases involved in the biosynthesis of volatile phenolic derivatives in rose petals. Plant Physiol 129: 1899–1907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Lee E-J, Facchini PJ. (2010) Norcoclaurine synthase is a member of the pathogenesis-related 10/Bet v1 protein family. Plant Cell 22: 3489–3503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee E-J, Facchini PJ. (2011) Tyrosine aminotransferase contributes to benzylisoquinoline alkaloid biosynthesis in opium poppy. Plant Physiol 157: 1067–1078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liscombe DK, Facchini PJ. (2007) Molecular cloning and characterization of tetrahydroprotoberberine cis-N-methyltransferase, an enzyme involved in alkaloid biosynthesis in opium poppy. J Biol Chem 282: 14741–14751 [DOI] [PubMed] [Google Scholar]
- Lu R, Martin-Hernandez AM, Peart JR, Malcuit I, Baulcombe DC. (2003) Virus-induced gene silencing in plants. Methods 30: 296–303 [DOI] [PubMed] [Google Scholar]
- Minami H, Dubouzet E, Iwasa K, Sato F. (2007) Functional analysis of norcoclaurine synthase in Coptis japonica. J Biol Chem 282: 6274–6282 [DOI] [PubMed] [Google Scholar]
- Morishige T, Dubouzet E, Choi K-B, Yazaki K, Sato F. (2002) Molecular cloning of columbamine O-methyltransferase from cultured Coptis japonica cells. Eur J Biochem 269: 5659–5667 [DOI] [PubMed] [Google Scholar]
- Morishige T, Tamakoshi M, Takemura T, Sato F. (2010) Molecular characterization of O-methyltransferases involved in isoquinoline alkaloid biosynthesis in Coptis japonica. Proc Jpn Acad Ser B Phys Biol Sci 86: 757–768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morishige T, Tsujita T, Yamada Y, Sato F. (2000) Molecular characterization of the S-adenosyl-L-methionine:3′-hydroxy-N-methylcoclaurine 4′-O-methyltransferase involved in isoquinoline alkaloid biosynthesis in Coptis japonica. J Biol Chem 275: 23398–23405 [DOI] [PubMed] [Google Scholar]
- Nuccio ML, Ziemak MJ, Henry SA, Weretilnyk EA, Hanson AD. (2000) cDNA cloning of phosphoethanolamine N-methyltransferase from spinach by complementation in Schizosaccharomyces pombe and characterization of the recombinant enzyme. J Biol Chem 275: 14095–14101 [DOI] [PubMed] [Google Scholar]
- Ounaroon A, Decker G, Schmidt J, Lottspeich F, Kutchan TM. (2003) (R,S)-Reticuline 7-O-methyltransferase and (R,S)-norcoclaurine 6-O-methyltransferase of Papaver somniferum: cDNA cloning and characterization of methyl transfer enzymes of alkaloid biosynthesis in opium poppy. Plant J 36: 808–819 [DOI] [PubMed] [Google Scholar]
- Page RD. (1996) TreeView: an application to display phylogenetic trees on personal computers. Comput Appl Biosci 12: 357–358 [DOI] [PubMed] [Google Scholar]
- Pauli HH, Kutchan TM. (1998) Molecular cloning and functional heterologous expression of two alleles encoding (S)-N-methylcoclaurine 3′-hydroxylase (CYP80B1), a new methyl jasmonate-inducible cytochrome P-450-dependent mono-oxygenase of benzylisoquinoline alkaloid biosynthesis. Plant J 13: 793–801 [DOI] [PubMed] [Google Scholar]
- Pienkny S, Brandt W, Schmidt J, Kramell R, Ziegler J. (2009) Functional characterization of a novel benzylisoquinoline O-methyltransferase suggests its involvement in papaverine biosynthesis in opium poppy (Papaver somniferum L). Plant J 60: 56–67 [DOI] [PubMed] [Google Scholar]
- Raith K, Neubert R, Poeaknapo C, Boettcher C, Zenk MH, Schmidt J. (2003) Electrospray tandem mass spectrometric investigations of morphinans. J Am Soc Mass Spectrom 14: 1262–1269 [DOI] [PubMed] [Google Scholar]
- Rueffer M, Nagakura N, Zenk MH. (1983) A highly specific O-methyltransferase for nororientaline synthesis isolated from Argemone platyceras cell cultures. Planta Med 49: 196–198 [DOI] [PubMed] [Google Scholar]
- Samanani N, Facchini PJ. (2002) Purification and characterization of norcoclaurine synthase: the first committed enzyme in benzylisoquinoline alkaloid biosynthesis in plants. J Biol Chem 277: 33878–33883 [DOI] [PubMed] [Google Scholar]
- Sariyar G. (1986) Six alkaloids from Papaver species. Phytochemistry 25: 2403–2406 [Google Scholar]
- Sariyar G. (2002) Biodiversity in the alkaloids of Turkish Papaver species. Pure Appl Chem 74: 557–574 [Google Scholar]
- Sariyar G, Sari A, Freyer AJ, Guinaudeau H, Shamma M. (1990) Quaternary isoquinoline alkaloids from Papaver species. J Nat Prod 53: 1302–1306 [Google Scholar]
- Sato F, Tsujita T, Katagiri Y, Yoshida S, Yamada Y. (1994) Purification and characterization of S-adenosyl-L-methionine:norcoclaurine 6-O-methyltransferase from cultured Coptis japonica cells. Eur J Biochem 225: 125–131 [DOI] [PubMed] [Google Scholar]
- Scalliet G, Journot N, Jullien F, Baudino S, Magnard JL, Channelière S, Vergne P, Dumas C, Bendahmane M, Cock JM, et al. (2002) Biosynthesis of the major scent components 3,5-dimethoxytoluene and 1,3,5-trimethoxybenzene by novel rose O-methyltransferases. FEBS Lett 523: 113–118 [DOI] [PubMed] [Google Scholar]
- Schmidt J, Boettcher C, Kuhnt C, Kutchan TM, Zenk MH. (2007) Poppy alkaloid profiling by electrospray tandem mass spectrometry and electrospray FT-ICR mass spectrometry after [ring-13C6]-tyramine feeding. Phytochemistry 68: 189–202 [DOI] [PubMed] [Google Scholar]
- Schmidt J, Raith K, Boettcher C, Zenk MH. (2005) Analysis of benzylisoquinoline-type alkaloids by electrospray tandem mass spectrometry and atmospheric pressure photoionization. Eur J Mass Spectrom (Chichester, Eng) 11: 325–333 [DOI] [PubMed] [Google Scholar]
- Takeshita N, Fujiwara H, Mimura H, Fitchen JH, Yamada Y, Sato F. (1995) Molecular cloning and characterization of S-adenosyl-L-methionine:scoulerine-9-O-methyltransferase from cultured cells of Coptis japonica. Plant Cell Physiol 36: 29–36 [PubMed] [Google Scholar]
- Van de Peer Y, De Wachter R. (1994) TREECON for Windows: a software package for the construction and drawing of evolutionary trees for the Microsoft Windows environment. Comput Appl Biosci 10: 569–570 [DOI] [PubMed] [Google Scholar]
- Ye K, Ke Y, Keshava N, Shanks J, Kapp JA, Tekmal RR, Petros J, Joshi HC. (1998) Opium alkaloid noscapine is an antitumor agent that arrests metaphase and induces apoptosis in dividing cells. Proc Natl Acad Sci USA 95: 1601–1606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegler J, Diaz-Chávez ML, Kramell R, Ammer C, Kutchan TM. (2005) Comparative macroarray analysis of morphine containing Papaver somniferum and eight morphine free Papaver species identifies an O-methyltransferase involved in benzylisoquinoline biosynthesis. Planta 222: 458–471 [DOI] [PubMed] [Google Scholar]
- Zubieta C, He XZ, Dixon RA, Noel JP. (2001) Structures of two natural product methyltransferases reveal the basis for substrate specificity in plant O-methyltransferases. Nat Struct Biol 8: 271–279 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.