Abstract
Plasma membrane-resident receptor kinases (RKs) initiate signaling pathways important for plant immunity and development. In Arabidopsis (Arabidopsis thaliana), the receptor for the elicitor-active peptide epitope of bacterial flagellin, flg22, is encoded by FLAGELLIN SENSING2 (FLS2), which promotes plant immunity. Despite its relevance, the molecular components regulating FLS2-mediated signaling remain largely unknown. We show that plasma membrane ARABIDOPSIS-AUTOINHIBITED Ca2+-ATPase (ACA8) forms a complex with FLS2 in planta. ACA8 and its closest homolog ACA10 are required for limiting the growth of virulent bacteria. One of the earliest flg22 responses is the transient increase of cytosolic Ca2+ ions, which is crucial for many of the well-described downstream responses (e.g. generation of reactive oxygen species and the transcriptional activation of defense-associated genes). Mutant aca8 aca10 plants show decreased flg22-induced Ca2+ and reactive oxygen species bursts and exhibit altered transcriptional reprogramming. In particular, mitogen-activated protein kinase-dependent flg22-induced gene expression is elevated, whereas calcium-dependent protein kinase-dependent flg22-induced gene expression is reduced. These results demonstrate that the fine regulation of Ca2+ fluxes across the plasma membrane is critical for the coordination of the downstream microbe-associated molecular pattern responses and suggest a mechanistic link between the FLS2 receptor complex and signaling kinases via the secondary messenger Ca2+. ACA8 also interacts with other RKs such as BRI1 and CLV1 known to regulate plant development, and both aca8 and aca10 mutants show morphological phenotypes, suggesting additional roles for ACA8 and ACA10 in developmental processes. Thus, Ca2+ ATPases appear to represent general regulatory components of RK-mediated signaling pathways.
Receptor kinases (RKs) constitute a large gene family in plants, with more than 600 members in Arabidopsis (Arabidopsis thaliana), and are key to ligand-mediated signaling pathways in plant immunity and development (Shiu and Bleecker, 2001). Only a handful of RKs have been studied in detail and matched with their cognate ligand, of which FLAGELLIN SENSING2 (FLS2), EF-TU RECEPTOR (EFR) from Arabidopsis, and rice XA21 encode Leu-rich repeat-RKs, conferring the perception of microbe-associated molecular patterns (MAMPs) from bacteria in these plants (Zipfel, 2009). Perception of the fungal MAMP chitin involves the LysM-RK CHITIN ELICITOR RECEPTOR KINASE1. FLS2 detects a conserved peptide at the N terminus of bacterial flagellin (flg22) and forms an inducible complex with BRI1-ASSOCIATED KINASE1/SOMATIC EMBRYO RECEPTOR KINASE3 (BAK1/SERK3), a Leu-rich repeat-RK initially identified as a coreceptor of BRASSINOSTEROID INSENSITIVE1 (BRI1) regulating brassinosteroid signaling and now reported to also act in various immune pathways and cell death control (Chinchilla et al., 2009; Postel et al., 2010; Schulze et al., 2010; Fradin et al., 2011; Schwessinger et al., 2011). The manifold phenotypes of bak1/serk3 mutant plants suggest that BAK1/SERK3 can potentially interact with multiple RKs to regulate a number of different signaling pathways. This offers the possibility for molecular cross talk between different RK-mediated signaling pathways, as recently demonstrated for brassinosteroid signaling negatively impacting flg22 responses (Albrecht et al., 2012; Belkhadir et al., 2012). At the molecular level, BAK1/SERK3 was shown to transphosphorylate BRI1 and EFR (Wang et al., 2008; Chen et al., 2010; Schwessinger et al., 2011). Moreover, BRI1, FLS2, and EFR can associate with other members of the SERK family, revealing some levels of functional redundancy (Albrecht et al., 2008; Roux et al., 2011).
Interaction between the ligand-binding RKs BRI1, FLS2, EFR, and BAK1/SERK3 is required for proper downstream responses (Chinchilla et al., 2007). Among the earliest responses stimulated by MAMPs are changes in ion fluxes across the plasma membrane, which result in an increased level of calcium ions (Ca2+) in the cytosol (Blume et al., 2000; Wendehenne et al., 2002; Jeworutzki et al., 2010). Ca2+ acts as an important second messenger for a multitude of biotic and abiotic stimuli, whereas different signals trigger unique Ca2+ signatures (Dodd et al., 2010; Kudla et al., 2010). MAMPs typically induce a transient Ca2+ burst, resulting in a rapid (within seconds) increase of free cytosolic Ca2+, which subsequently (within minutes) declines to steady-state Ca2+ levels (Blume et al., 2000; Ranf et al., 2008). The Ca2+ burst occurs upstream of many MAMP-elicited responses, including the rapid production of reactive oxygen species (ROS), the activation of signaling kinases, as well as changes in gene expression (Blume et al., 2000; Boller and Felix, 2009; Ranf et al., 2011; Segonzac et al., 2011). However, genetic studies and the identification of the underlying molecular components of the MAMP-induced Ca2+ burst are largely missing (Ranf et al., 2008; Boller and Felix, 2009). In general, cytosolic Ca2+ levels are regulated by plasma membrane- and endomembrane-bound Ca2+ channels that mediate the influx of Ca2+ and efflux transporters that reestablish Ca2+ homeostasis. A number of ion channels have been identified, some of which have roles in plant immunity, such as DEFENSE NO DEATH1 (DND1; Clough et al., 2000; Lamotte et al., 2004; Kudla et al., 2010). Recently, ionotropic Glu receptor-like proteins were shown to regulate Ca2+ influx at the plasma membrane and were also implicated in MAMP-induced responses (Cho et al., 2009; Kwaaitaal et al., 2011; Michard et al., 2011; Vatsa et al., 2011), and an endoplasmic reticulum-localized P2A-type Ca2+ ATPase was described to contribute to pathogen-induced cell death and to alter the MAMP-triggered Ca2+ burst (Zhu et al., 2010). The relevance of the Ca2+ influx in MAMP-elicited responses is underlined by polysaccharides secreted from bacterial pathogens to chelate Ca2+ in the apoplastic space (Aslam et al., 2008).
Here, we report that the plasma membrane-resident P2B-type Ca2+ ATPase ACA8 interacts with FLS2 in planta. Loss-of-function aca8 plants, the mutant of its closest homolog aca10, and the aca8 aca10 double mutant were more susceptible to bacterial infection. Analyzing individual MAMP responses, aca8 aca10 mutant plants displayed decreased flg22-triggered Ca2+ influx and ROS accumulation. Importantly, flg22-triggered gene expression downstream of mitogen-activated protein kinase (MAPK) signaling was increased, but gene expression downstream of calcium-dependent protein kinase (CDPK) signaling was reduced. This suggests that the MAMP-induced Ca2+ burst is required for proper transcriptional reprogramming upon elicitation. According to their function as Ca2+ pumps, ACA8 and ACA10 are hypothesized to regulate Ca2+ efflux during the flg22-elicited Ca2+ burst, which suggests a molecular link between the FLS2 receptor, Ca2+ signaling, and flg22-triggered downstream responses. In addition, aca10 and aca8 aca10 mutant plants showed developmental phenotypes affecting inflorescence height as well as root length. Together with the finding that ACA8 also interacts with other RKs such as BRI1 and CLV1, these results suggest that plasma membrane Ca2+ ATPases function in multiple RK-mediated signaling pathways.
RESULTS
ACA8 Interacts with FLS2 and Other RKs
In a proteomics approach, we previously isolated proteins colocalizing to FLS2 in plasma membrane microdomains (Keinath et al., 2010). To address whether some of these proteins can associate with FLS2, we focused on Ca2+ ATPases, which have also been identified as differentially phosphorylated and transcriptionally induced in response to flg22 (Zipfel et al., 2004; Benschop et al., 2007). ACA8 and ACA10 belong to the family of type 2B autoinhibited Ca2+ ATPases consisting of 10 members in Arabidopsis (Supplemental Fig. S1A; Boursiac and Harper, 2007). These Ca2+ ATPases comprise 10 transmembrane-spanning domains, harbor a calmodulin-binding domain for autoinhibition of the ATPase active site, and can localize to different membrane compartments (Boursiac and Harper, 2007). ACA8, ACA9, and ACA10 group into a distinct subfamily and accumulate at the plasma membrane (Bonza et al., 2000; Hwang et al., 2000; Lee et al., 2007). Whereas ACA9 expression is restricted to pollen and thereby is critical for pollen tube development, ACA8 and ACA10 are expressed throughout the plant and have not yet been assigned any specific function besides inflorescence growth (Schiøtt et al., 2004).
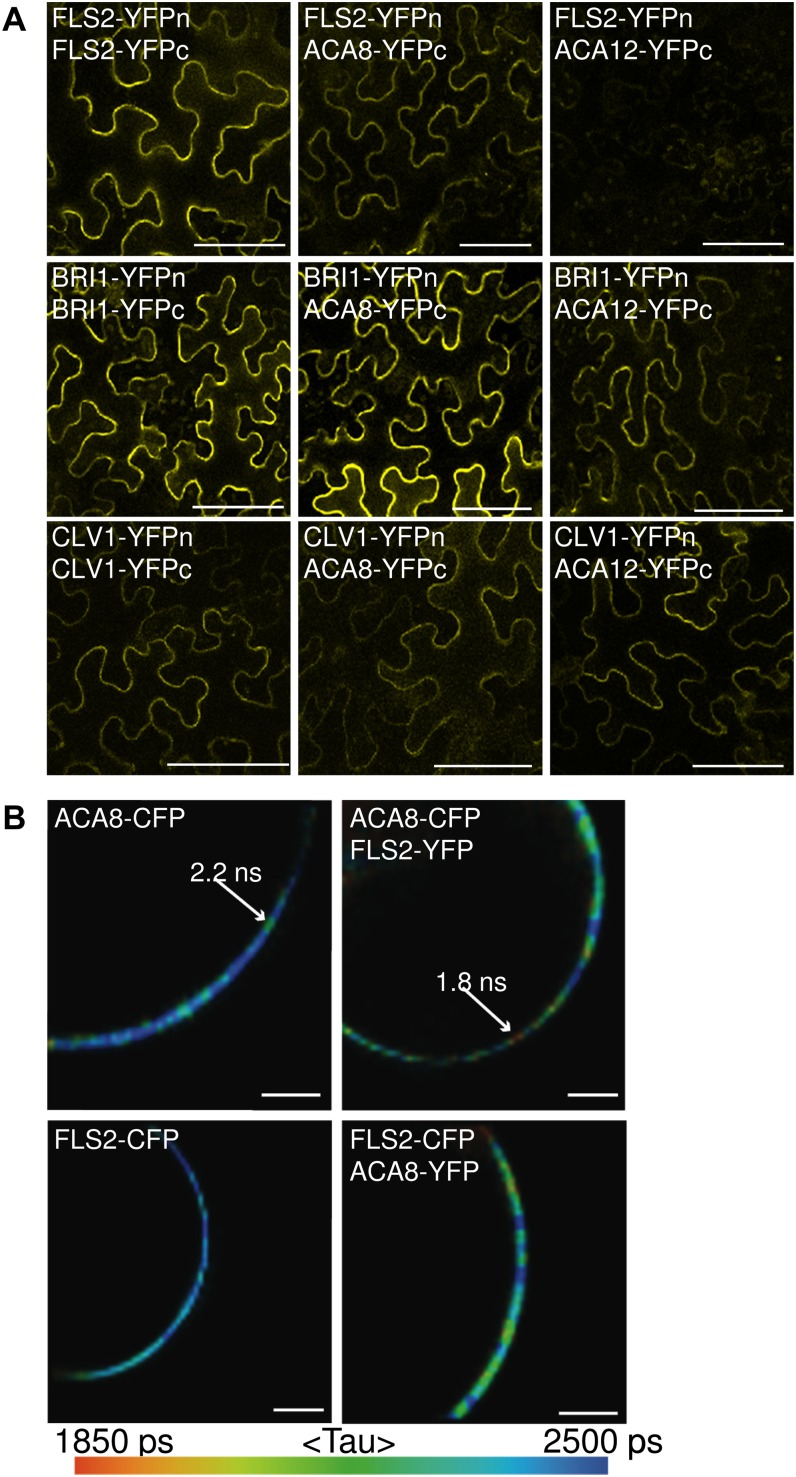
We transiently expressed FLS2 and ACA8 fused to the N- and C-terminal halves of yellow fluorescent protein (YFP), respectively, in Nicotiana benthamiana and examined possible protein-protein interactions by confocal microscopy in a so-called bimolecular fluorescence complementation (BiFC) assay (Bracha-Drori et al., 2004). In this assay, we observed reconstitution of the YFP molecule by the detection of fluorescence when expressing FLS2 fused to both of the YFP halves, indicative of FLS2 homodimerization (Fig. 1A). We also observed BiFC when FLS2 was coexpressed with ACA8. As BiFC assays are based on transient expression in a heterologous system, the tagged proteins can accumulate to high levels, facilitating the reconstitution of a BiFC signal; thus, ACA12, BRI1, and CLV1 were included as controls. No YFP reconstitution could be detected upon coexpressing of FLS2 and ACA12, another plasma membrane-resident Ca2+ ATPase. Notably, ACA8 showed a broader interaction pattern, because BiFC was also observed with other RKs, such as BRI1 and CLV1, of which the latter is functioning in stem cell identity maintenance and is normally not expressed in leaf tissue (Waites and Simon, 2000). Similar to FLS2, both BRI1 and CLV1 formed homodimers in this assay, but unlike FLS2, they also interacted with ACA12 (Fig. 1A). In all cases of BiFC, the YFP signal was recorded at the cell periphery, suggesting complex formation at the plasma membrane.
Figure 1.
FLS2 interacts with ACA8 in planta. A, BiFC. Representative micrographs show YFP signals of epidermal cells from N. benthamiana leaves transformed with plasmids coding for the indicated constructs. YFPc, C-terminal YFP fragment; YFPn, N-terminal YFP fragment. Bars = 100 µm. B, FRET-FLIM measurements. Micrographs show representative false-color-coded fluorescence lifetime images of Arabidopsis protoplasts transfected with plasmids coding for the indicated constructs. Lower fluorescence lifetimes indicate the proximity of the two fluorescence proteins. Similar results were obtained in at least three independent experiments. Bars = 2 µm.
To further overcome the limitations of BiFC assays, we performed Förster resonance energy transfer (FRET) measurements on the basis of fluorescence lifetime imaging microscopy (FLIM) using FLS2 and ACA8 fusions to cyan fluorescent protein (CFP) and YFP, respectively. FRET can be detected using FLIM, where reduction of the fluorescence lifetime of a donor-containing molecule occurs due to the proximity of an acceptor-containing molecule, which is an indication of physical interaction. We examined FLS2-ACA8 association in protoplasts from soil-grown Arabidopsis plants. Under this condition, we observed a significant reduction in fluorescence lifetime when FLS2-CFP and ACA8-YFP were coexpressed as compared with the fluorescence lifetime of ACA8-CFP alone (Fig. 1B; Supplemental Table S1). Similar results were obtained when we used FLS2-CFP and ACA8-YFP. This suggests that both proteins are in close proximity to each other, indicative of a protein-protein interaction. Interaction of fluorophore-tagged FLS2 and ACA8 was detected at the plasma membrane, which is in line with the subcellular localization of the two proteins and substantiates our findings of BiFC in N. benthamiana. However, the interaction of fluorophore-tagged FLS2 and ACA8 was not distributed uniformly across the plasma membrane but seen as patchy areas with strongly reduced fluorescence lifetimes (Fig. 1B), which indicates that the presence of FLS2-ACA8 complexes was restricted to subdomains within the plasma membrane. This observation is in agreement with the notion that FLS2 and ACA8 can localize to flg22-induced plasma membrane microdomains (Keinath et al., 2010). Despite numerous attempts, we failed to clone a full-length ACA10 cDNA, which precluded the analysis of ACA10 by fluorophore-based interaction assays. Despite poor results by coimmunoprecipitation analysis, pull-down experiments of FLS2-GFP followed by mass spectrometric analysis repeatedly revealed the presence of ACA8 and ACA10 peptides, further corroborating the existence of FLS2-ACA8 and FLS2-ACA10 complexes in planta (Supplemental Fig. S2). Taken together, these results indicate that FLS2 forms a complex with ACA8 at the plasma membrane and that ACA8 can interact with multiple RKs, pointing at an important role in the regulation of RK-mediated signaling pathways.
ACA8 and ACA10 Exhibit Partial Overlapping Functions
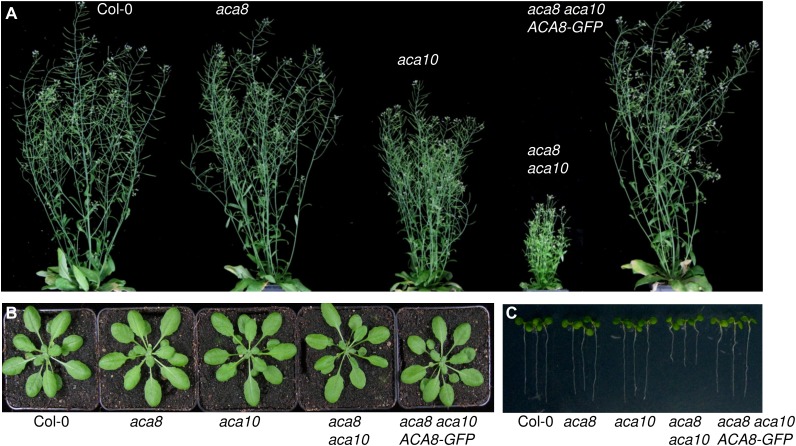
To address ACA8 function, we isolated a T-DNA insertion line and a tilling mutant (both in the ecotype Columbia [Col-0] genetic background) in the ACA8 gene (Supplemental Fig. S1B). Genetic redundancy within members of the ACA family has been documented and could be expected for members of the ACA8, ACA9, and ACA10 subgroup (Boursiac and Harper, 2007). Because ACA9 expression was specific to pollen tubes, we focused on ACA10, isolated a T-DNA insertion line in the ACA10 gene, and generated aca8 aca10 double knockout lines (Supplemental Fig. S1C). In addition, we crossed a 35S::ACA8-GFP-expressing transgenic line into the aca8 aca10 double mutant. Single aca8 mutants displayed no obvious developmental phenotypes (Fig. 2). By contrast, aca10 mutant plants were reduced in inflorescence height and displayed increased axillary stem formation, which was further enhanced in aca8 aca10 plants (Fig. 2A). This phenotype was also present in aca10 plants crossed with the aca8Q70* tilling mutant (Supplemental Fig. S3) and could be rescued by ectopic ACA8-GFP expression, demonstrating functional complementation by the GFP fusion protein (Fig. 2A). Redundant functions of ACA8 and ACA10 in the regulation of inflorescence height were previously reported in the Arabidopsis Wassilewskija background (George et al., 2008). Differences between the single mutants may result from an incomplete overlap of the ACA8 and ACA10 expression patterns. We did not observe any obvious mutant phenotype in rosette leaf development among the genotypes (Fig. 2B), whereas aca8 aca10 mutants showed significantly reduced root growth when cultivated in vitro (Fig. 2C). Reduction in root growth was affecting primary root length and could be correlated with an early differentiation of stem cells compared with wild-type plants (Supplemental Fig. S3).
Figure 2.
ACA8 and ACA10 have a role in plant development. Photographs show growth-related phenotypes of the indicated genotypes in the Col-0 background (Supplemental Fig. S1). A, Inflorescence growth of 8-week-old plants. B, Rosette leaves of 4-week-old plants. C, Root growth of 7-d-old in vitro-grown seedlings.
flg22-Triggered Early Responses Depend on ACA8 and ACA10 Function
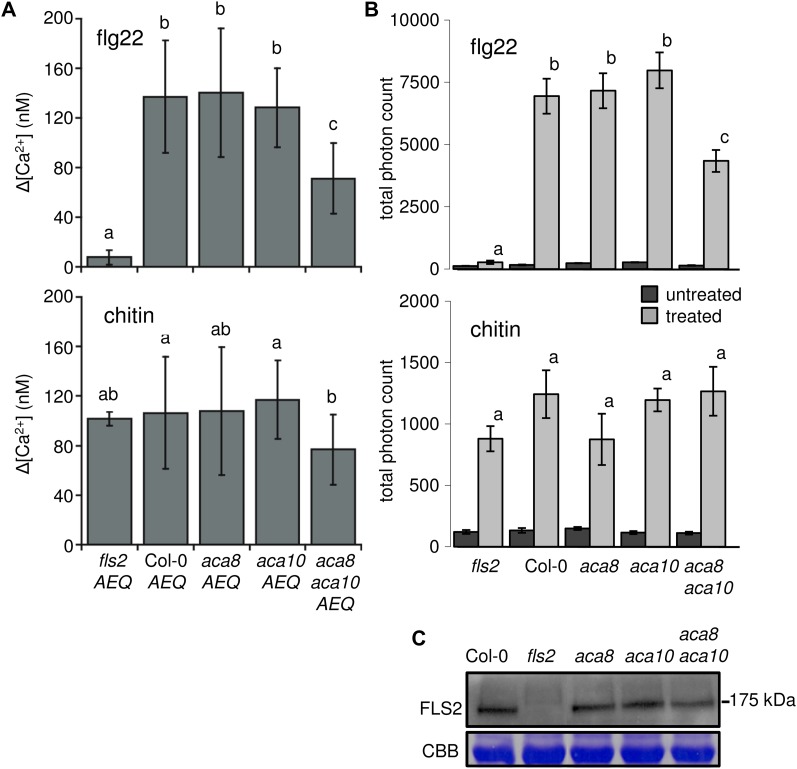
Ca2+ ATPases are responsible for extruding Ca2+ ions from the cytosol into endomembrane compartments or extracellularly into the apoplast (Bonza et al., 2004; Conn et al., 2011). ACA8 has been shown to mediate Ca2+ transport in yeast and is activated by the binding of calmodulin (CaM) to its N terminus (Bonza et al., 2000, 2004; Mersmann et al., 2010). Based on the interaction of ACA8 with FLS2, we addressed whether ACA8 and ACA10 function in the flg22-triggered Ca2+ burst. All genotypes, therefore, were crossed to a transgenic line expressing the aequorin (Aeq) Ca2+ biosensor (Knight et al., 1991). We performed luminescence-based measurements of free cytosolic Ca2+ and revealed slightly elevated constitutive Ca2+ levels in aca8 aca10 Aeq plants (Supplemental Fig. S4). We then monitored the MAMP-induced Ca2+ burst over time. Mutant aca8 Aeq and aca10 Aeq plants responded like wild-type plants upon flg22 treatment. By contrast, the flg22-triggered Ca2+ burst was strongly reduced in the aca8 aca10 Aeq lines and completely abolished in fls2 Aeq plants (Fig. 3A; Supplemental Fig. S5). The overall pattern of the transient increase of Ca2+ remained similar between the wild type and the aca8 Aeq and aca10 Aeq genotypes, but the maximal influx (peak) of the Ca2+ signature was affected in aca8 aca10 Aeq plants (Supplemental Fig. S5). The Ca2+ burst in response to chitin was slightly reduced in aca8 aca10 Aeq lines and for all other genotypes was indistinguishable from the wild type (Fig. 3A). The lower peak in flg22-induced cytosolic Ca2+ influx in the double mutant demonstrates that ACA8 and ACA10 both contribute to the flg22-elicited Ca2+ burst and indicates a role for these proteins in the regulation of FLS2-mediated early responses.
Figure 3.
Early MAMP responses are altered in aca8 aca10 mutants. A, Ca2+ burst in response to flg22 and chitin. The Aeq Ca2+ biosensor was introduced into all indicated genotypes. Data were calculated from curves normalized to steady-state cytosolic [Ca2+]. Shown are average changes (Δ) in [Ca2+] values in the peak between 4 and 5.5 min after elicitation, which was averaged over two independent biological replicates. Error bars indicate sd based on 14 to 16 samples, and letters indicate significant differences at P < 0.05 based on ANOVA with Tukey’s honestly significant difference test. B, ROS burst in response to flg22 and chitin. ROS generation (indicated as total photon counts measured between 2 and 30 min upon elicitation) was monitored over time. Error bars indicate sd based on 28 samples, and letters indicate significant differences at P < 0.05 based on a t test. Similar results were obtained in at least two independent experiments. C, FLS2 protein levels of the indicated genotypes revealed by western blot. Coomassie blue staining (CBB) is included as a loading control. Similar results were obtained in at least two independent experiments. [See online article for color version of this figure.]
The production of ROS upon MAMP treatments is mediated by plasma membrane-resident NADPH oxidases, which depend on Ca2+ signaling for their function (Kobayashi et al., 2007; Mersmann et al., 2010). We examined the flg22-triggered oxidative burst and detected no significant differences between wild-type plants and the aca8 and aca10 single mutants, whereas the aca8 aca10 double mutant displayed an overall decreased ROS production when treated with flg22 (Fig. 3B). ROS production upon chitin treatment remained comparable to the wild type in all tested mutants. The decrease in oxidative burst correlated with the reduced flg22-triggered Ca2+ signature in aca8 aca10 plants, which is in agreement with Ca2+ operating upstream of ROS production. When monitoring these individual MAMP responses, we observed genetic redundancy between ACA8 and ACA10, suggesting that both Ca2+ ATPases exert overlapping functions in these early and transient flg22 responses, which is in contrast to the unequal role of ACA8 and ACA10 in development. Western-blot analysis revealed unaltered FLS2 protein accumulation in the aca mutants compared with wild-type plants (Fig. 3C). Therefore, the observed reduction in flg22-triggered Ca2+ and ROS bursts is likely caused by the loss of ACA8 and ACA10 function rather than altered FLS2 levels.
ACA8 and ACA10 Are Required for Proper flg22-Induced Transcriptional Changes
For more detailed analysis of ACA8/ACA10 functions, we determined the transcriptional changes caused by ACA8 and ACA10 loss of function by microarray analysis. A total of 69 transcripts were identified as showing significantly elevated transcript levels, and 10 had significantly lower transcript abundance in the aca8 aca10 double mutant compared with wild-type seedlings (Supplemental Table S2 and Supplemental Information S1). We validated the differential transcript accumulation of 19 out of 20 tested genes by quantitative reverse transcription-PCR analysis, of which 17 showed wild-type-like expression in the aca8 aca10/35S::ACA8-GFP line, further substantiating the functionality of the ACA8-GFP fusion protein (Supplemental Table S2). Most remarkably, genes belonging to the Gene Ontology categories “calcium ion binding” and “cation binding” were overrepresented among the genes that show higher transcript levels in the aca8 aca10 double mutant (Supplemental Table S2). The first category includes genes coding for CaM-like proteins such as CML35, CML36, CML41, CML45, CML46, and CML47 (McCormack et al., 2005). Increased expression of CaM-like genes could be a compensatory mechanism to counteract the deficiency in extruding Ca2+ ions from the cytosol in aca8 aca10 plants.
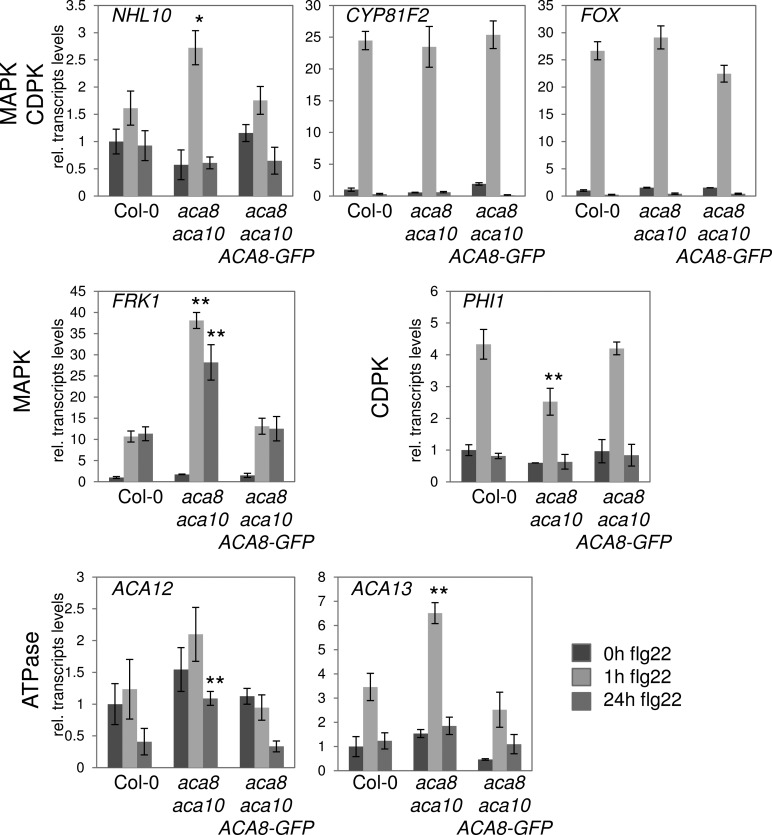
Only a small number of the aca8 aca10 deregulated genes were associated with plant defense (Supplemental Table S2). One of them encodes ACD6, a regulator of salicylic acid (SA)-mediated disease resistance (Lu et al., 2003). Significantly reduced ACD6 transcript levels in aca8 aca10 plants may contribute to the enhanced susceptibility to Pseudomonas syringae pv tomato DC3000 (PtoDC3000). To find out whether any of the other genes that exhibit differential transcript levels in aca8 aca10 has a potential role in MAMP-triggered immunity, we searched publicly available transcriptome databases and identified 27 of the aca8 aca10 up-regulated genes to be induced in response to MAMP treatments (Supplemental Fig. S6A). We then focused on genes downstream of flg22 Ca2+ signaling (Boudsocq et al., 2010). There was little overlap between the aca8 aca10 deregulated and CDPK-dependent genes (Supplemental Fig. S6C), which could be due to the different plant materials used for transcript profiling. Therefore, we studied the flg22-induced expression of selected marker genes specifically downstream of the MAPK and/or CDPK cascade (Boudsocq et al., 2010). flg22-induced expression of MAPK-regulated FLAGELLIN-RESPONSIVE KINASE1 was considerably higher in aca8 aca10 plants compared with the wild type (Fig. 4). This could point at elevated MAPK signaling in aca8 aca10; however, there was no correlation between flg22-induced MAPK activation and the increased expression of MAPK-specific genes (Supplemental Fig. S7). Induction of the downstream genes cytochrome P450 monooxygenase CYP81F2, FAD-LINKED OXIREDUCTASE, and NDR1/HIN-LIKE10, which are controlled by both the MAPK and CDPK pathways, was either wild type like or somewhat enhanced (Fig. 4). By contrast, the flg22-induced transcript accumulation of the CDPK downstream gene PHOSPHATE INDUCED1 was notably reduced compared with wild-type levels (Fig. 4). This indicates that gene induction mediated by CDPK signaling is insufficient, likely due to altered flg22 activation of the CDPK cascade.
Figure 4.
flg22-induced gene expression. Quantitative real-time PCR monitoring is shown for transcript levels of flg22-regulated genes and other Ca2+ ATPases in the indicated genotypes upon flg22 elicitation. Actin was used as a control. Error bars indicate sd based on three biological experiments with three technical replicates each; asterisks indicate significant differences between Col-0 and aca8 aca10 at P < 0.05 (*) and P < 0.01 (**) based on Student’s t test. FOX, FAD-LINKED OXIREDUCTASE; FRK1, FLAGELLIN-RESPONSIVE KINASE1; NHL10, NDR1/HIN-LIKE10; PHI1, PHOSPHATE INDUCED1.
Our data show that knockout of ACA8 and ACA10 function causes pronounced changes in steady-state transcript levels, probably for phenotypic compensation, and also impairs proper flg22-induced transcriptional reprogramming. This was further supported by the differential transcript accumulation of additional marker genes (Supplemental Fig. S8), which either was enhanced (At5g25250, At1g66880, and MYB51/At1g18570) or reduced (At2g47140 and WRKY30/At5g24110) upon flg22 elicitation. We also investigated the flg22-dependent expression of ACA12 and ACA13, which are potential candidates for compensating ACA8 and ACA10 loss of function (Supplemental Fig. S1A). After flg22 treatment, both ACA12 and ACA13 transcripts accumulated to higher levels in the aca8 aca10 double mutant background, whereas no significant differences and a slight up-regulation in ACA12 and ACA13 abundance, respectively, were detected without MAMP stimulus (Fig. 4). Thus, ACA12 and ACA13 may contribute to the control of cytosolic Ca2+ levels during flg22 responses. Intriguingly, ACA12 localizes to the plasma membrane and can interact with the RKs BRI1 and CLV1 (Fig. 1A). ACA12, however, failed to associate with FLS2, which may hamper any possible compensatory effects in flg22 responses.
ACA8 and ACA10 Contribute to Plant Immunity
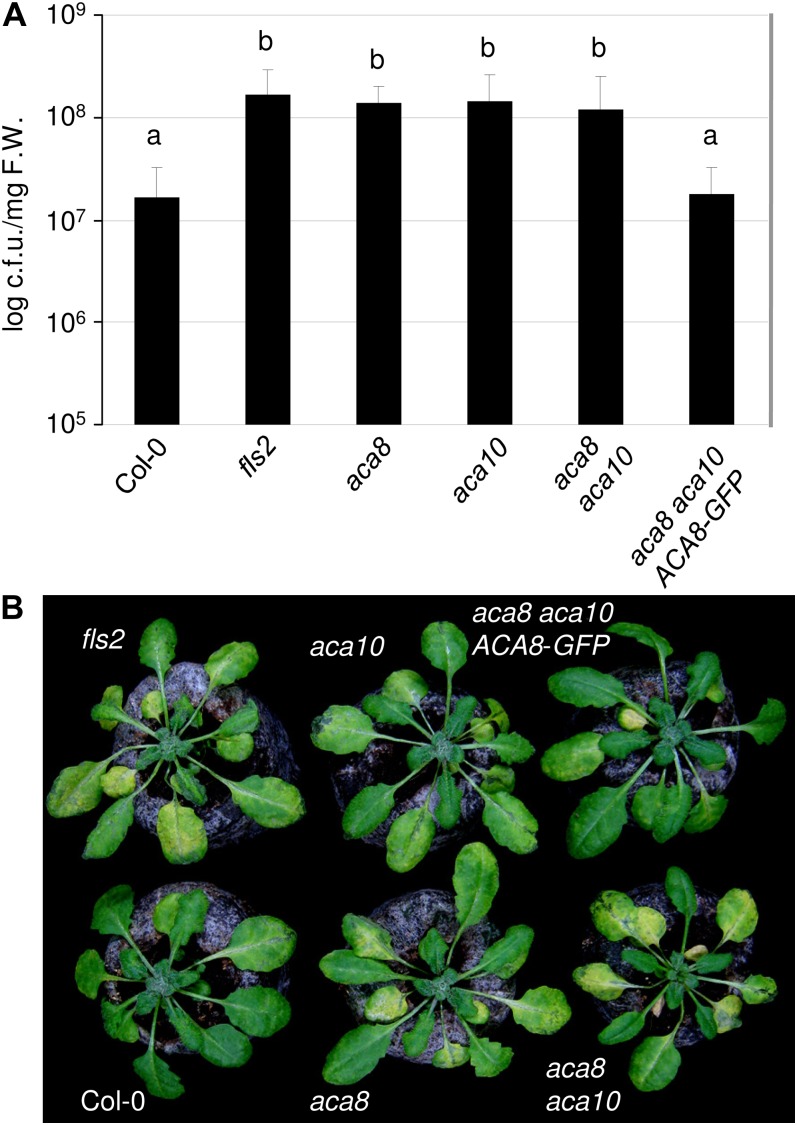
To examine a possible role of ACA8 and ACA10 in plant antibacterial immunity, all genotypes were spray inoculated with virulent PtoDC3000, an infection that is defeated utilizing the FLS2 pathway (Zipfel et al., 2004). Bacterial growth and disease symptom development were monitored 3 and 5 d post inoculation, respectively. PtoDC3000 multiplied to high titers in aca8 and aca10 single mutants as well as in the aca8 aca10 double mutant, which was comparable to the titers detected in immunocompromised fls2 mutants (Fig. 5A). This enhanced susceptibility was reduced to wild-type levels in the transgenic ACA8-GFP complementation line. Moreover, the disease symptom development of aca8 and aca10 single and double mutants was correlated with the enhanced susceptibility phenotype (Fig. 5B). Despite 35S-driven ectopic expression of ACA8-GFP, no increased resistance against PtoDC3000 could be detected in the transgenic line. The aca8 and aca10 single mutants were as affected as the aca8 aca10 double mutant upon PtoDC3000 infection. Thus, ACA8 and ACA10, both individually and equally, contribute to plant immunity in bacterial infections.
Figure 5.
ACA8 and ACA10 contribute to plant immunity. A, Bacterial titers (PtoDC3000) in the indicated genotypes (4-week-old plants) at 3 d post inoculation. Error bars indicate sd of three combined independent biological replicates each with six technical replicates; letters indicate significant differences at P < 0.05 based on a t test. c.f.u., Colony-forming units; F.W., fresh weight. B, Representative photographs showing macroscopic disease symptoms of 4-week-old plants infected with PtoDC3000 at 5 d post inoculation.
DISCUSSION
It is well known that MAMPs induce a rapid and transient increase of [Ca2+] in the cytosol through the function of plasma membrane-resident Ca2+ channels (Blume et al., 2000; Ranf et al., 2011), but despite its presumed importance in plant immunity, our current understanding of how the MAMP-induced Ca2+ burst is regulated is rather limited (Ranf et al., 2008; Kudla et al., 2010). Although DND1 is important for cytosolic Ca2+ elevation in response to bacterial lipopolysaccharides and endogenous danger peptides (Ma et al., 2009; Qi et al., 2010), it is not required for flg22 and elf18 activation of Ca2+ (Jeworutzki et al., 2010). Similarly, the recently suggested Glu receptor-like-type Ca2+ channels have been implicated in cryptogein- and flg22-triggered responses by pharmacological approaches; however, genetic evidence for their involvement in MAMP signaling is still lacking (Kwaaitaal et al., 2011; Michard et al., 2011; Vatsa et al., 2011). Ca2+ homeostasis is also controlled through the function of Ca2+ ATPases, and our data show that FLS2 forms a complex with ACA8. It is possible that FLS2 transphosphorylates the Ca2+ ATPase to regulate its activity, as ACA10 is differentially phosphorylated upon flg22 treatment (Benschop et al., 2007).
Based on our mutant loss-of-function data, ACA8 and ACA10 cofunction to positively regulate the MAMP-induced Ca2+ burst. Because of their function as Ca2+ pumps, Ca2+ ATPases mediate the efflux of Ca2+ ions out of the cytosol. Therefore, loss of Ca2+ ATPase function should result in an enhanced and prolonged Ca2+ burst (Romani et al., 2004). In line with this assumption, Ca2+ fluxes triggered by the MAMP cryptogein in N. benthamiana were increased in amplitude and duration when endoplasmic reticulum-localized NbCA1 was silenced (Zhu et al., 2010). Our data on ACA8 and ACA10 unexpectedly revealed a reduction in the MAMP-induced Ca2+ burst. We cannot exclude the possibility that other members of the ACA family, such as ACA12 and ACA13, may substitute at least partially for ACA8 and ACA10 function in the mutant backgrounds, as evidenced by the increased ACA12 and ACA13 transcript levels upon flg22 elicitation. This would indicate that the observed phenotypes of aca8 aca10 mutants are rather an indirect effect. However, ACA12 did not associate with FLS2 in our BiFC analysis, ACA12 and ACA13 transcript levels were not generally increased in aca8 aca10 mutants, and our transcriptome data did not point at obvious expression changes of any other member of the ACA family. Alternatively, it is possible that the enhanced transcript levels of CaM-like genes in aca8 aca10 plants reflect a mechanism to compensate for elevated steady-state levels of cytosolic Ca2+. This may in turn lead to the decreased influx of Ca2+ into the cytosol, because CaM-like proteins were shown to regulate cyclic nucleotide-gated channels, a class of cation channels with a documented role in Ca2+ influx (Ali et al., 2007; Boursiac and Harper, 2007). CaM-like proteins can also activate Ca2+ ATPases and are thus key regulators of Ca2+ homeostasis (Boursiac and Harper, 2007). However, we cannot exclude a yet unknown modality of Ca2+ ATPase function implying a direct rather than indirect action. Based on current knowledge, it is possible to speculate that FLS2 may transiently down-regulate ACA8 and ACA10 activities upon flg22 treatments, thereby allowing a cytosolic Ca2+ burst, possibly masked by investigating stable loss-of-function mutants.
The flg22-induced ROS production was decreased in aca8 aca10 mutants, which is in agreement with a reduction of the flg22-triggered Ca2+ burst. Likewise, chemical inhibition of Ca2+ ATPase function resulted in reduced ROS production in response to the fungal MAMP oligogalacturonide, placing ACA proteins upstream of RbohD (Romani et al., 2004). As Rboh proteins contain two EF hand motifs in their N-terminal domains (Ogasawara et al., 2008), an altered Ca2+ signature in aca8 aca10 plants may impair ROS generation catalyzed by the NADPH oxidases. In potato (Solanum tuberosum), CDPK signaling promotes Rboh-mediated ROS production (Kobayashi et al., 2007). This supports the idea of changed CDPK activation in aca8 aca10 plants and ACA8/ACA10 regulating kinase signaling, which is substantiated by altered flg22-induced gene expression caused by ACA8 and ACA10 loss of function. The MAPK/CDPK differential gene expression shows that the flg22-induced Ca2+ burst is required for the concerted activation of the kinase signaling pathways in order to properly reprogram the transcriptome upon MAMP perception.
Ca2+ ATPases have also been shown to regulate defense responses by affecting programmed cell death (Nemchinov et al., 2008). Silencing of NbCA1 causes an enhanced hypersensitive response cell death upon tobacco mosaic virus activation of the tobacco (Nicotiana benthamiana) N immune receptor (Zhu et al., 2010). Knockout plants of ACA4 and ACA11 display cell death-like lesions similar to those triggered by avirulent pathogens, which were dependent on SA accumulation (Boursiac et al., 2010). Cell death-related phenotypes were not observed in aca8 aca10 plants. Instead, they were supersusceptible to infection with PtoDC3000. Unlike the observed genetic redundancy between ACA8 and ACA10 in plant development and the partial phenotype observed when monitoring individual flg22 responses, the two members of the Ca2+ ATPase family are equally required for plant immunity, with single mutants exhibiting a similar level of susceptibility to fls2 mutants. This apparent difference may be due to the different time frames measuring early flg22 responses and the end point of bacterial infections. A sustained increase of cytosolic Ca2+ rather than a transient burst activates downstream defenses (Blume et al., 2000). Additionally, other than MAMP responses, pathogen growth depends on multiple layers of basal immunity (e.g. interference of immunity by effectors from PtoDC3000). Effectors can target MAMP receptors at the plasma membrane (Block and Alfano, 2011), or effectors could directly affect the molecular components controlling Ca2+ fluxes. Alternatively, perception of the complex mixture of different MAMPs present in PtoDC3000 may require independent functions of ACA8 and ACA10 or other members of the ACA family. This is supported by the differential expression pattern of aca8 aca10 deregulated genes in response to flg22 or oligogalacturonides (Supplemental Fig. S6). Moreover, CaM is implicated as a negative regulator in SA-mediated disease resistance, and the CaM-binding protein CBP60g contributes to flg22-elicited SA accumulation and antibacterial defense (Du et al., 2009; Wang et al., 2009), which demonstrates a role for the ACA8/ACA10 deregulated CaM-like genes in plant immunity.
MAMPs are known to trigger a Ca2+ burst, one of the most upstream responses in defense signaling (Boller and Felix, 2009; Segonzac et al., 2011). However, the molecular components underlying the complex regulatory network regulating the Ca2+ fluxes are still poorly described. In this study, we identified two plasma membrane Ca2+ ATPases, ACA8 and ACA10, which, based on mutant loss-of-function data, act as positive regulators of early MAMP responses. Our findings further illustrate the importance of coordinated and fine-tuned MAMP responses, including Ca2+ signaling, for plant immunity. Given the altered MAMP-induced MAPK-/CDPK-dependent transcriptional changes together with the ACA8-FLS2 complex formation at the plasma membrane, our results suggest a mechanistic link between the receptor complex and signaling kinases via the secondary messenger Ca2+. Although root tip growth upon flg22 treatment and in fls2 mutants remains to be inspected in more detail, our data also suggest a broader function of Ca2+ ATPases in RK-mediated signaling. The functional relevance of the interaction of ACA8 and BRI1 is supported by the aca8 aca10 mutant phenotype showing defects in the early differentiation of root stem cells (Clouse and Sasse, 1998; Hacham et al., 2011). BRI1-mediated brassinosteroid signaling has been shown to affect root growth through regulation of the cell cycle (Gonzales-Garcia et al., 2011). Additionally, it is possible that ACA8 and/or ACA10 associate with the Arabidopsis CRINKLY4 RK, known to regulate root stem cells via the CLV3-related peptide CLE40, in particular as root cell type-specific expression data provide evidence for ACA10 transcripts accumulating around the stem cell niche (Brady et al., 2007; Winter et al., 2007; De Smet et al., 2008; Stahl et al., 2009). In analogy to the multiple roles of the coreceptor BAK1/SERK3, this places plasma membrane Ca2+ ATPases as important components of RK signaling pathways, likely through the regulation of Ca2+ fluxes in the cytosol. Dissecting the precise molecular mechanism of the RK-Ca2+ ATPase interaction will further advance our understanding of receptor-mediated signal transduction in the future.
MATERIALS AND METHODS
Plant Lines and Growth Conditions
T-DNA lines for Arabidopsis (Arabidopsis thaliana) ACA8 (GK-688H09) and ACA10 (GK-044H01) were obtained from the European Seed Stock Center Nottingham Arabidopsis Stock Centre (http://arabidopsis.info/), and the tilling ACA8Q70* line was obtained from the Seattle Tilling Project (http://tilling.fhcrc.org/). Homozygous insertions of all aca8, aca10, aca8 aca10, aca8Q70*, and aca8Q70* aca10 mutant plants were validated in the F2 populations by PCR and sequencing. 35S::ACA8-GFP, 35S::Aeq transgenic, and fls2 mutant lines were described previously (Knight et al., 1991; Zipfel et al., 2004; Lee et al., 2007). Homozygous crossed aca8 aca10 ACA8-GFP, aca8 Aeq, aca10 Aeq, aca8 aca10 Aeq, and fls2 Aeq were confirmed by PCR (all oligonucleotides used in this study are summarized in Supplemental Table S3). Arabidopsis plants grown on soil were kept under short-day conditions for 4 to 5 weeks. Arabidopsis seedlings were grown in vitro on plates or in liquid containing Murashige and Skoog (MS) medium and 1% Suc and kept under long-day conditions for 10 to 14 d. Nicotiana benthamiana plants were soil grown under long-day conditions for 4 to 5 weeks.
BiFC
FLS2-Yc, FLS2-Yn, BRI1-Yc, BRI1-Yn, CLV1-Yc, CLV1-Yn, ACA8-Yc, ACA8-Yn, ACA12-Yc, and ACA12-Yn constructs were made by PCR cloning the corresponding full-length cDNAs using Gateway technology in the pAMPAT destination vector series and introduced into Agrobacterium tumefaciens strain GV3101 carrying the p19 silencing suppressor (Voinnet et al., 2003; Lefebvre et al., 2010). Overnight cultures were diluted to an optical density at 600 nm of 0.1 in water supplemented with 100 μm acetosyringone and inoculated into 4-week-old N. benthamiana leaves. Leaf samples were imaged at 1 d post inoculation using a Leica confocal TCS SP5 microscope with the Leica LAS AF system software. YFP emission and chlorophyll autofluorescence were detected at emission spectra of 520 to 600 nm and 680 to 780 nm, respectively, after excitation at 488 nm. All samples were imaged with a 20× objective. Photographs were taken in line averaging of four scans. The same confocal settings were used to image all samples. Representative images of over three biological replicates are shown.
FRET-FLIM Measurements
FLS2-CFP, FLS2-YFP, ACA8-CFP, and ACA8-YFP constructs were PCR cloned as the corresponding full-length cDNAs using Gateway technology in the pCZN575 and pCZN576 vectors and improved sCFP3A and sYFP2 chromophore variants, respectively (Kremers et al., 2006; Karlova et al., 2011). Constructs were transfected into mesophyll protoplasts from soil-grown Arabidopsis Col-0 plants as described before (Russinova et al., 2004), which were prepared using the tape sandwich method (Wu et al., 2009). FRET-FLIM measurements were performed using the Bio-Rad Radiance 2100 MP system combined with a Nikon TE 300 inverted microscope and a Hamamatsu R3809U MCP PMT (Russinova et al., 2004). FRET between CFP and YFP was detected by monitoring donor emission using a 470- to 500-nm band-pass filter. Images with a frame size of 64 × 64 pixels were acquired, and the average count rate was around 0.5 × 104 photons per second for an acquisition time of ±120 s. Donor fluorescence lifetimes (CFP) were analyzed using SPCImage 3.10 software (Becker & Hickl) using a one- and two-component decay model. Average fluorescence lifetimes of different combinations in several cells (n > 10) along the plasma membrane were calculated (Supplemental Table S1). The statistical significance of differences between donor-only and donor-acceptor combinations was determined using Student’s t test.
Ca2+ Measurements
Twelve-day-old sterile-grown seedlings in liquid medium were supplied with 100 µL of MS medium containing 10 µm coelenterazine (Biosynth) and dark incubated overnight. Seedlings were supplied with 100 µL of fresh MS medium and dark incubated for 30 min. Aeq measurements were performed using the Centro LB960 luminometer system (Berthold Technologies). Luminescence from single wells was detected over 0.25 s, and each well was measured every 30 s After 2 min of measurement, flg22 (EZBiolab) and chitin (Sigma) were added to final concentrations of 1 µm or 0.1 mg mL−1, respectively, and luminescence was measured over 40 min. For calculation of Ca2+ concentrations, 100 µL of 2 m CaCl2 in 20% ethanol was added and luminescence was measured over 30 min (0.25 s per well every 63 s). Ca2+ concentrations were calculated according to Rentel and Knight (2004). Differences in Aeq levels due to transgene expression and seedling size were corrected by calculating Ca2+ concentrations and not using luminescence counts. Per treatment, two assays of eight individual wells were averaged. Ca2+ transients were compared between treatments within one experiment unless stated otherwise. Significant differences were evaluated using ANOVA with Tukey’s honestly significant difference test.
ROS Measurements
Leaf discs of 5-week-old plants were used for ROS measurements as described previously (Segonzac et al., 2011). Oxidative burst was elicited with 100 nm flg22 or 100 μg mL−1 chitin oligosaccharide; a negative control without MAMP elicitation was included in all experiments. Luminescence was measured over time using an ICCD photon-counting camera (Photek).
Biochemical Analysis
For western blotting, proteins were separated on 10% SDS-PAGE gels, transferred onto polyvinylidene difluoride membranes using a semidry transfer system, followed by blocking in 5% milk or 3% bovine serum albumin. Antibodies were diluted as follows: anti-p42/44 MAPK (Cell Signaling Technology; 1:1,000), anti-FLS2 (Mersmann et al., 2010; 1:5,000), and alkaline phosphatase-conjugated anti-rabbit (Sigma; 1:20,000–1:30,000). Alkaline phosphatase activity was detected using the CDP-Star (Roche).
For MAPK assay, 14-d-old seedlings grown on MS plates were sprayed with 2 μm flg22 for 0, 5, 15, or 60 min before harvest. A total of 100 mg of plant material was ground and solubilized in 200 μL of buffer (50 mm Tris-HCl, pH 7.5, 150 mm NaCl, 10% glycerol, 1 mm EDTA, 10 mm NaF, 2 mm NaVO3, 25 mm β-glycerophosphate, 1 mm Pefabloc, 1 mm dithiothreitol, 1 mm phenylmethylsulfonyl fluoride, and 0.1% Tween 20) supplied with 3.4 μL per 100 mg fresh weight protease inhibitor cocktail (Sigma). Extracts were centrifuged, solubilized by 5 min of boiling in 2% SDS Laemmli buffer, and equal amounts were loaded onto SDS gels. MAPK activation was detected with anti-p42/44 MAPK antibodies.
Transcript Profiling
For quantitative reverse transcription-PCR analysis, 14-d-old sterile-grown seedlings were untreated or treated with 1 µm flg22 for 1 or 24 h. RNA was extracted and DNA digested using the RNeasy Plant Mini Kit and the RNase-Free DNase Set (Qiagen). A total of 2 µg of RNA was used to synthesize cDNA using the SuperScript II enzyme (Invitrogen). One microliter of a 10× dilution of the cDNA was used for each quantitative PCR, using a Bio-Rad iQ5 apparatus and SYBR Green I detection. All oligonucleotides used in this study are summarized in Supplemental Table S3.
Pathogen Infection Assays
Four-week-old soil-grown (Jiffy pellets) Arabidopsis plants were surface inoculated with Pseudomonas syringae pv tomato DC3000 bacteria at 108 colony-forming units mL−1 and sampled at 3 d post inoculation. Two leaf discs were pooled from six individual plants, and bacterial extraction was done as described before (Zipfel et al., 2004). The results of three independent experiments were combined, and t test analysis was performed.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers FLS2 (AT5G46330), BRI1 (AT4G39400), CLV1 (AT1G75820), ACA8 (AT5G57110), ACA10 (AT4G29900), and ACA12 (AT3G63380).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Characterization of ACA8 and ACA10 loss-of-function lines.
Supplemental Figure S2. Identification of ACA8 and ACA10 peptides by mass spectrometry analysis of immunopurified FLS2-GFP.
Supplemental Figure S3. Developmental phenotypes of aca mutant plants.
Supplemental Figure S4. Steady-state Ca2+ levels before flg22-triggered Ca2+ burst and after the burst.
Supplemental Figure S5. Patterns of the Ca2+ burst induced by flg22 and chitin over time.
Supplemental Figure S6. Altered gene expression in aca8 aca10 mutants.
Supplemental Figure S7. Protein kinase activation in flg22 signaling.
Supplemental Figure S8. Expression analysis of defense marker genes.
Supplemental Table S1. Fluorescence lifetime analysis of the FLS2-ACA8 interaction.
Supplemental Table S2. Microarray expression data of aca8 aca10 deregulated genes.
Supplemental Table S3. List of all oligonucleotides used in this study.
Supplemental Information S1. Supporting information.
Supplementary Material
Acknowledgments
We thank W.S. Chung (Gyeongsang National University) for kindly providing the 35S::ACA8-GFP transgenic line mutant, M. Knight Durham University for the 35S::Aeq line, GABI-KAT for T-DNA insertion lines, and the Seattle Tilling Project for the tilling line. We thank A.M.E. Jones (The Sainsbury Laboratory), J. Sklenar (The Sainsbury Laboratory), and S. Laurent (Max-Planck-Institute Cologne) for technical help, M. Beck (The Sainsbury Laboratory) for constructs, U. Goebel (Max-Planck-Institute Cologne) and E. Ver Loren van Themaat (Max-Planck-Institute Cologne) for data analysis of the Affymetrix Tiling 1.0R array, C. Zipfel (The Sainsbury Laboratory) for providing materials and advice, T. Romeis (Free University Berlin) for fruitful discussions, and G. Oldroyd (John Innes Center Norwich) for reading the manuscript.
Glossary
- RK
receptor kinase
- ROS
reactive oxygen species
- MAPK
mitogen-activated protein kinase
- CDPK
calcium-dependent protein kinase
- MAMP
microbe-associated molecular pattern
- YFP
yellow fluorescent protein
- BiFC
bimolecular fluorescence complementation
- CFP
cyan fluorescent protein
- FRET
Förster resonance energy transfer
- FLIM
fluorescence lifetime imaging microscopy
- Col-0
ecotype Columbia
- SA
salicylic acid
- PtoDC3000
Pseudomonas syringae pv tomato DC3000
- MS
Murashige and Skoog
References
- Albrecht C, Boutrot F, Segonzac C, Schwessinger B, Gimenez-Ibanez S, Chinchilla D, Rathjen JP, de Vries SC, Zipfel C. (2012) Brassinosteroids inhibit pathogen-associated molecular pattern-triggered immune signaling independent of the receptor kinase BAK1. Proc Natl Acad Sci USA 109: 303–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albrecht C, Russinova E, Kemmerling B, Kwaaitaal M, de Vries SC. (2008) Arabidopsis SOMATIC EMBRYOGENESIS RECEPTOR KINASE proteins serve brassinosteroid-dependent and -independent signaling pathways. Plant Physiol 148: 611–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali R, Ma W, Lemtiri-Chlieh F, Tsaltas D, Leng Q, von Bodman S, Berkowitz GA. (2007) Death don’t have no mercy and neither does calcium: Arabidopsis CYCLIC NUCLEOTIDE GATED CHANNEL2 and innate immunity. Plant Cell 19: 1081–1095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aslam SN, Newman MA, Erbs G, Morrissey KL, Chinchilla D, Boller T, Jensen TT, De Castro C, Ierano T, Molinaro A, et al. (2008) Bacterial polysaccharides suppress induced innate immunity by calcium chelation. Curr Biol 18: 1078–1083 [DOI] [PubMed] [Google Scholar]
- Belkhadir Y, Jaillais Y, Epple P, Balsemão-Pires E, Dangl JL, Chory J. (2012) Brassinosteroids modulate the efficiency of plant immune responses to microbe-associated molecular patterns. Proc Natl Acad Sci USA 109: 297–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benschop JJ, Mohammed S, O’Flaherty M, Heck AJ, Slijper M, Menke FL. (2007) Quantitative phosphoproteomics of early elicitor signaling in Arabidopsis. Mol Cell Proteomics 6: 1198–1214 [DOI] [PubMed] [Google Scholar]
- Block A, Alfano JR. (2011) Plant targets for Pseudomonas syringae type III effectors: virulence targets or guarded decoys? Curr Opin Microbiol 14: 39–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blume B, Nürnberger T, Nass N, Scheel D. (2000) Receptor-mediated increase in cytoplasmic free calcium required for activation of pathogen defense in parsley. Plant Cell 12: 1425–1440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boller T, Felix G. (2009) A renaissance of elicitors: perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu Rev Plant Biol 60: 379–406 [DOI] [PubMed] [Google Scholar]
- Bonza MC, Luoni L, De Michelis MI. (2004) Functional expression in yeast of an N-deleted form of At-ACA8, a plasma membrane Ca(2+)-ATPase of Arabidopsis thaliana, and characterization of a hyperactive mutant. Planta 218: 814–823 [DOI] [PubMed] [Google Scholar]
- Bonza MC, Morandini P, Luoni L, Geisler M, Palmgren MG, De Michelis MI. (2000) At-ACA8 encodes a plasma membrane-localized calcium-ATPase of Arabidopsis with a calmodulin-binding domain at the N terminus. Plant Physiol 123: 1495–1506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudsocq M, Willmann MR, McCormack M, Lee H, Shan L, He P, Bush J, Cheng SH, Sheen J. (2010) Differential innate immune signalling via Ca(2+) sensor protein kinases. Nature 464: 418–422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boursiac Y, Harper JF. (2007) The origin and function of calmodulin regulated Ca2+ pumps in plants. J Bioenerg Biomembr 39: 409–414 [DOI] [PubMed] [Google Scholar]
- Boursiac Y, Lee SM, Romanowsky S, Blank R, Sladek C, Chung WS, Harper JF. (2010) Disruption of the vacuolar calcium-ATPases in Arabidopsis results in the activation of a salicylic acid-dependent programmed cell death pathway. Plant Physiol 154: 1158–1171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bracha-Drori K, Shichrur K, Katz A, Oliva M, Angelovici R, Yalovsky S, Ohad N. (2004) Detection of protein-protein interactions in plants using bimolecular fluorescence complementation. Plant J 40: 419–427 [DOI] [PubMed] [Google Scholar]
- Brady SM, Orlando DA, Lee JY, Wang JY, Koch J, Dinneny JR, Mace D, Ohler U, Benfey PN. (2007) A high-resolution root spatiotemporal map reveals dominant expression patterns. Science 318: 801–806 [DOI] [PubMed] [Google Scholar]
- Chen X, Chern M, Canlas PE, Ruan D, Jiang C, Ronald PC. (2010) An ATPase promotes autophosphorylation of the pattern recognition receptor XA21 and inhibits XA21-mediated immunity. Proc Natl Acad Sci USA 107: 8029–8034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinchilla D, Shan L, He P, de Vries S, Kemmerling B. (2009) One for all: the receptor-associated kinase BAK1. Trends Plant Sci 14: 535–541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinchilla D, Zipfel C, Robatzek S, Kemmerling B, Nürnberger T, Jones JD, Felix G, Boller T. (2007) A flagellin-induced complex of the receptor FLS2 and BAK1 initiates plant defence. Nature 448: 497–500 [DOI] [PubMed] [Google Scholar]
- Cho D, Kim SA, Murata Y, Lee S, Jae SK, Nam HG, Kwak JM. (2009) De-regulated expression of the plant glutamate receptor homolog AtGLR3.1 impairs long-term Ca2+-programmed stomatal closure. Plant J 58: 437–449 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Fengler KA, Yu IC, Lippok B, Smith RK, Jr, Bent AF. (2000) The Arabidopsis dnd1 “defense, no death” gene encodes a mutated cyclic nucleotide-gated ion channel. Proc Natl Acad Sci USA 97: 9323–9328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clouse SD, Sasse JM. (1998) Brassinosteroids: essential regulators of plant growth and development. Annu Rev Plant Physiol Plant Mol Biol 49: 427–451 [DOI] [PubMed] [Google Scholar]
- Conn SJ, Gilliham M, Athman A, Schreiber AW, Baumann U, Moller I, Cheng NH, Stancombe MA, Hirschi KD, Webb AA, et al. (2011) Cell-specific vacuolar calcium storage mediated by CAX1 regulates apoplastic calcium concentration, gas exchange, and plant productivity in Arabidopsis. Plant Cell 23: 240–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Smet I, Vassileva V, De Rybel B, Levesque MP, Grunewald W, Van Damme D, Van Noorden G, Naudts M, Van Isterdael G, De Clercq R, et al. (2008) Receptor-like kinase ACR4 restricts formative cell divisions in the Arabidopsis root. Science 322: 594–597 [DOI] [PubMed] [Google Scholar]
- Dodd AN, Kudla J, Sanders D. (2010) The language of calcium signaling. Annu Rev Plant Biol 61: 593–620 [DOI] [PubMed] [Google Scholar]
- Du J, Xie J, Yue L. (2009) Intracellular calcium activates TRPM2 and its alternative spliced isoforms. Proc Natl Acad Sci USA 106: 7239–7244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fradin EF, Abd-El-Haliem A, Masini L, van den Berg GC, Joosten MH, Thomma BP. (2011) Interfamily transfer of tomato Ve1 mediates Verticillium resistance in Arabidopsis. Plant Physiol 156: 2255–2265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- George L, Romanowsky SM, Harper JF, Sharrock RA. (2008) The ACA10 Ca2+-ATPase regulates adult vegetative development and inflorescence architecture in Arabidopsis. Plant Physiol 146: 716–728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Garcia MP, Vilarrasa-Blasi J, Zhiponova M, Divol F, Mora-Garcia S, Russinova E, Cano-Delgado AI. (2011) Brassinosteroids control meristem size by promoting cell cycle progression in Arabidopsis roots. Development 138: 849–859 [DOI] [PubMed] [Google Scholar]
- Hacham Y, Holland N, Butterfield C, Ubeda-Tomas S, Bennett MJ, Chory J, Savaldi-Goldstein S. (2011) Brassinosteroid perception in the epidermis controls root meristem size. Development 138: 839–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang I, Harper JF, Liang F, Sze H. (2000) Calmodulin activation of an endoplasmic reticulum-located calcium pump involves an interaction with the N-terminal autoinhibitory domain. Plant Physiol 122: 157–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeworutzki E, Roelfsema MR, Anschütz U, Krol E, Elzenga JT, Felix G, Boller T, Hedrich R, Becker D. (2010) Early signaling through the Arabidopsis pattern recognition receptors FLS2 and EFR involves Ca-associated opening of plasma membrane anion channels. Plant J 62: 367–378 [DOI] [PubMed] [Google Scholar]
- Karlova R, Rosin FM, Busscher-Lange J, Parapunova V, Do PT, Fernie AR, Fraser PD, Baxter C, Angenent GC, de Maagd RA. (2011) Transcriptome and metabolite profiling show that APETALA2a is a major regulator of tomato fruit ripening. Plant Cell 23: 923–941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keinath NF, Kierszniowska S, Lorek J, Bourdais G, Kessler SA, Shimosato-Asano H, Grossniklaus U, Schulze WX, Robatzek S, Panstruga R. (2010) PAMP (pathogen-associated molecular pattern)-induced changes in plasma membrane compartmentalization reveal novel components of plant immunity. J Biol Chem 285: 39140–39149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight MR, Campbell AK, Smith SM, Trewavas AJ. (1991) Transgenic plant aequorin reports the effects of touch and cold-shock and elicitors on cytoplasmic calcium. Nature 352: 524–526 [DOI] [PubMed] [Google Scholar]
- Kobayashi M, Ohura I, Kawakita K, Yokota N, Fujiwara M, Shimamoto K, Doke N, Yoshioka H. (2007) Calcium-dependent protein kinases regulate the production of reactive oxygen species by potato NADPH oxidase. Plant Cell 19: 1065–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kremers GJ, Goedhart J, van Munster EB, Gadella TW., Jr (2006) Cyan and yellow super fluorescent proteins with improved brightness, protein folding, and FRET Förster radius. Biochemistry 45: 6570–6580 [DOI] [PubMed] [Google Scholar]
- Kudla J, Batistic O, Hashimoto K. (2010) Calcium signals: the lead currency of plant information processing. Plant Cell 22: 541–563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwaaitaal M, Huisman R, Maintz J, Reinstädler A, Panstruga R. (2011) Ionotropic glutamate receptor (iGluR)-like channels mediate MAMP-induced calcium influx in Arabidopsis thaliana. Biochem J 440: 355–365 [DOI] [PubMed] [Google Scholar]
- Lamotte O, Gould K, Lecourieux D, Sequeira-Legrand A, Lebrun-Garcia A, Durner J, Pugin A, Wendehenne D. (2004) Analysis of nitric oxide signaling functions in tobacco cells challenged by the elicitor cryptogein. Plant Physiol 135: 516–529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SM, Kim HS, Han HJ, Moon BC, Kim CY, Harper JF, Chung WS. (2007) Identification of a calmodulin-regulated autoinhibited Ca2+-ATPase (ACA11) that is localized to vacuole membranes in Arabidopsis. FEBS Lett 581: 3943–3949 [DOI] [PubMed] [Google Scholar]
- Lefebvre B, Timmers T, Mbengue M, Moreau S, Hervé C, Tóth K, Bittencourt-Silvestre J, Klaus D, Deslandes L, Godiard L, et al. (2010) A remorin protein interacts with symbiotic receptors and regulates bacterial infection. Proc Natl Acad Sci USA 107: 2343–2348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu H, Rate DN, Song JT, Greenberg JT. (2003) ACD6, a novel ankyrin protein, is a regulator and an effector of salicylic acid signaling in the Arabidopsis defense response. Plant Cell 15: 2408–2420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma W, Smigel A, Verma R, Berkowitz GA. (2009) Cyclic nucleotide gated channels and related signaling components in plant innate immunity. Plant Signal Behav 4: 277–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormack E, Tsai YC, Braam J. (2005) Handling calcium signaling: Arabidopsis CaMs and CMLs. Trends Plant Sci 10: 383–389 [DOI] [PubMed] [Google Scholar]
- Mersmann S, Bourdais G, Rietz S, Robatzek S. (2010) Ethylene signaling regulates accumulation of the FLS2 receptor and is required for the oxidative burst contributing to plant immunity. Plant Physiol 154: 391–400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michard E, Lima PT, Borges F, Silva AC, Portes MT, Carvalho JE, Gilliham M, Liu LH, Obermeyer G, Feijó JA. (2011) Glutamate receptor-like genes form Ca2+ channels in pollen tubes and are regulated by pistil D-serine. Science 332: 434–437 [DOI] [PubMed] [Google Scholar]
- Nemchinov LG, Shabala L, Shabala S. (2008) Calcium efflux as a component of the hypersensitive response of Nicotiana benthamiana to Pseudomonas syringae. Plant Cell Physiol 49: 40–46 [DOI] [PubMed] [Google Scholar]
- Ogasawara Y, Kaya H, Hiraoka G, Yumoto F, Kimura S, Kadota Y, Hishinuma H, Senzaki E, Yamagoe S, Nagata K, et al. (2008) Synergistic activation of the Arabidopsis NADPH oxidase AtrbohD by Ca2+ and phosphorylation. J Biol Chem 283: 8885–8892 [DOI] [PubMed] [Google Scholar]
- Postel S, Küfner I, Beuter C, Mazzotta S, Schwedt A, Borlotti A, Halter T, Kemmerling B, Nürnberger T. (2010) The multifunctional leucine-rich repeat receptor kinase BAK1 is implicated in Arabidopsis development and immunity. Eur J Cell Biol 89: 169–174 [DOI] [PubMed] [Google Scholar]
- Qi Z, Verma R, Gehring C, Yamaguchi Y, Zhao Y, Ryan CA, Berkowitz GA. (2010) Ca2+ signaling by plant Arabidopsis thaliana Pep peptides depends on AtPepR1, a receptor with guanylyl cyclase activity, and cGMP-activated Ca2+ channels. Proc Natl Acad Sci USA 107: 21193–21198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranf S, Eschen-Lippold L, Pecher P, Lee J, Scheel D. (2011) Interplay between calcium signalling and early signalling elements during defence responses to microbe- or damage-associated molecular patterns. Plant J 68: 100–113 [DOI] [PubMed] [Google Scholar]
- Ranf S, Wünnenberg P, Lee J, Becker D, Dunkel M, Hedrich R, Scheel D, Dietrich P. (2008) Loss of the vacuolar cation channel, AtTPC1, does not impair Ca2+ signals induced by abiotic and biotic stresses. Plant J 53: 287–299 [DOI] [PubMed] [Google Scholar]
- Rentel MC, Knight MR. (2004) Oxidative stress-induced calcium signaling in Arabidopsis. Plant Physiol 135: 1471–1479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romani G, Bonza MC, Filippini I, Cerana M, Beffagna N, De Michelis MI. (2004) Involvement of the plasma membrane Ca2+-ATPase in the short-term response of Arabidopsis thaliana cultured cells to oligogalacturonides. Plant Biol (Stuttg) 6: 192–200 [DOI] [PubMed] [Google Scholar]
- Roux M, Schwessinger B, Albrecht C, Chinchilla D, Jones A, Holton N, Malinovsky FG, Tör M, de Vries S, Zipfel C. (2011) The Arabidopsis leucine-rich repeat receptor-like kinases BAK1/SERK3 and BKK1/SERK4 are required for innate immunity to hemibiotrophic and biotrophic pathogens. Plant Cell 23: 2440–2455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russinova E, Borst JW, Kwaaitaal M, Caño-Delgado A, Yin Y, Chory J, de Vries SC. (2004) Heterodimerization and endocytosis of Arabidopsis brassinosteroid receptors BRI1 and AtSERK3 (BAK1). Plant Cell 16: 3216–3229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiøtt M, Romanowsky SM, Baekgaard L, Jakobsen MK, Palmgren MG, Harper JF. (2004) A plant plasma membrane Ca2+ pump is required for normal pollen tube growth and fertilization. Proc Natl Acad Sci USA 101: 9502–9507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze B, Mentzel T, Jehle AK, Mueller K, Beeler S, Boller T, Felix G, Chinchilla D. (2010) Rapid heteromerization and phosphorylation of ligand-activated plant transmembrane receptors and their associated kinase BAK1. J Biol Chem 285: 9444–9451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwessinger B, Roux M, Kadota Y, Ntoukakis V, Sklenar J, Jones A, Zipfel C. (2011) Phosphorylation-dependent differential regulation of plant growth, cell death, and innate immunity by the regulatory receptor-like kinase BAK1. PLoS Genet 7: e1002046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segonzac C, Feike D, Gimenez-Ibanez S, Hann DR, Zipfel C, Rathjen JP. (2011) Hierarchy and roles of pathogen-associated molecular pattern-induced responses in Nicotiana benthamiana. Plant Physiol 156: 687–699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiu SH, Bleecker AB. (2001) Receptor-like kinases from Arabidopsis form a monophyletic gene family related to animal receptor kinases. Proc Natl Acad Sci USA 98: 10763–10768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl Y, Wink RH, Ingram GC, Simon R. (2009) A signaling module controlling the stem cell niche in Arabidopsis root meristems. Curr Biol 19: 909–914 [DOI] [PubMed] [Google Scholar]
- Vatsa P, Chiltz A, Bourque S, Wendehenne D, Garcia-Brugger A, Pugin A. (2011) Involvement of putative glutamate receptors in plant defence signaling and NO production. Biochimie 93: 2095–2101 [DOI] [PubMed] [Google Scholar]
- Voinnet O, Rivas S, Mestre P, Baulcombe D. (2003) An enhanced transient expression system in plants based on suppression of gene silencing by the p19 protein of tomato bushy stunt virus. Plant J 33: 949–956 [DOI] [PubMed] [Google Scholar]
- Waites R, Simon R. (2000) Signaling cell fate in plant meristems: three clubs on one tousle. Cell 103: 835–838 [DOI] [PubMed] [Google Scholar]
- Wang L, Tsuda K, Sato M, Cohen JD, Katagiri F, Glazebrook J. (2009) Arabidopsis CaM binding protein CBP60g contributes to MAMP-induced SA accumulation and is involved in disease resistance against Pseudomonas syringae. PLoS Pathog 5: e1000301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Kota U, He K, Blackburn K, Li J, Goshe MB, Huber SC, Clouse SD. (2008) Sequential transphosphorylation of the BRI1/BAK1 receptor kinase complex impacts early events in brassinosteroid signaling. Dev Cell 15: 220–235 [DOI] [PubMed] [Google Scholar]
- Wendehenne D, Lamotte O, Frachisse JM, Barbier-Brygoo H, Pugin A. (2002) Nitrate efflux is an essential component of the cryptogein signaling pathway leading to defense responses and hypersensitive cell death in tobacco. Plant Cell 14: 1937–1951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter D, Vinegar B, Nahal H, Ammar R, Wilson GV, Provart NJ. (2007) An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS ONE 2: e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu FH, Shen SC, Lee LY, Lee SH, Chan MT, Lin CS. (2009) Tape-Arabidopsis sandwich: a simpler Arabidopsis protoplast isolation method. Plant Methods 5: 16–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X, Caplan J, Mamillapalli P, Czymmek K, Dinesh-Kumar SP. (2010) Function of endoplasmic reticulum calcium ATPase in innate immunity-mediated programmed cell death. EMBO J 29: 1007–1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipfel C. (2009) Early molecular events in PAMP-triggered immunity. Curr Opin Plant Biol 12: 414–420 [DOI] [PubMed] [Google Scholar]
- Zipfel C, Robatzek S, Navarro L, Oakeley EJ, Jones JD, Felix G, Boller T. (2004) Bacterial disease resistance in Arabidopsis through flagellin perception. Nature 428: 764–767 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.