Abstract
The p110 Cut homeobox 1 (CUX1) transcription factor regulates genes involved in DNA replication and chromosome segregation. Using a genome-wide-approach, we now demonstrate that CUX1 also modulates the constitutive expression of DNA damage response genes, including ones encoding ATM and ATR, as well as proteins involved in DNA damage-induced activation of, and signaling through, these kinases. Consistently, RNAi knockdown or genetic inactivation of CUX1 reduced ATM/ATR expression and negatively impacted hallmark protective responses mediated by ATM and ATR following exposure to ionizing radiation (IR) and UV, respectively. Specifically, abrogation of CUX1 strongly reduced ATM autophosphorylation after IR, in turn causing substantial decreases in (i) levels of phospho-Chk2 and p53, (ii) γ-H2AX and Rad51 DNA damage foci and (iii) the efficiency of DNA strand break repair. Similarly remarkable reductions in ATR-dependent responses, including phosphorylation of Chk1 and H2AX, were observed post-UV. Finally, multiple cell cycle checkpoints and clonogenic survival were compromised in CUX1 knockdown cells. Our results indicate that CUX1 regulates a transcriptional program that is necessary to mount an efficient response to mutagenic insult. Thus, CUX1 ensures not only the proper duplication and segregation of the genetic material, but also the preservation of its integrity.
INTRODUCTION
The Cut homeobox gene 1 (CUX1) encodes several transcription factor isoforms with distinct DNA binding and regulatory properties [(1,2), reviewed in (3)]. One isoform, p110 CUX1, functions as a transcriptional regulator of cell cycle progression (4,5). Genetic studies in Drosophila melanogaster showed that Cut is an important determinant of cell-type specificity in several tissues [reviewed in (6)]. Homozygous inactivation of Cux1 in mice causes perinatal lethality in a large proportion of animals due to delayed lung development and associated respiratory failure (7). Surviving mice are usually male and exhibit growth retardation, disrupted hair follicle morphogenesis, purulent rhinitis, infertility, cachexia and reduction of B and T cell content in bone marrow and thymus, respectively (7–9). The basis for some among these multiple phenotypes appears to involve both cell-autonomous and non-autonomous processes. In transgenic mouse models, overexpression of CUX1 generated various cancer-associated disorders depending on the specific isoform and tissue type expression. These include multi-organ organomegaly, glomerulosclerosis and polycystic kidneys, pre-cancerous lesions in the liver, myeloproliferative-disease-like myeloid leukemias and mammary tumors sometimes associated with lung metastasis (10–14). Immunohistochemical analysis of human breast and pancreatic cancer tissues demonstrated that CUX1 protein expression was increased in high histological grade tumors relative to low grade ones (15,16). It has been proposed that the participation of CUX1 in tumor progression involves its role in cell motility. Consistent with this notion, siRNA-mediated knockdown of CUX1 caused a decrease in, whereas overexpression of p110 or p75 CUX1 stimulated, both cell migration and invasion (15,17).
Biochemical activities that implicate CUX1 in tumor initiation likely involve roles for this protein in cell cycle progression [(18–20); reviewed in (3)]. CUX1 expression and activity are tightly regulated in a cell cycle-dependent manner, mostly through phosphorylation/dephosphorylation by cyclin A/Cdk2, cyclin A/Cdk1 cyclin B/Cdk1 and Cdc25A, as well as through proteolytic processing by nuclear cathepsin L and a caspase-like protease (4,21–26). Genome-wide location analysis revealed that p110 CUX1 binds to the promoter of several genes that participate in DNA replication and cell cycle progression from S phase through the end of mitosis (5). In agreement with these findings, G1 was prolonged in mouse embryo fibroblasts derived from Cux1z/z knockout mice, whereas constitutive expression of p110 CUX1 accelerated entry into S phase and stimulated cell proliferation (20). More recently, CUX1 was shown to up-regulate the expression of genes that fulfill important functions in mitosis and the spindle assembly checkpoint. Although these activities of CUX1 in normal cells ensure proper chromosomal segregation, higher CUX1 expression in cancer cells can lead to chromosomal instability following cytokinesis failure (27).
Of major relevance here, another category of genes enriched among transcriptional targets of CUX1 is known to be involved in the processing of DNA damage. Thus, the aim of the present study was to investigate a potential role of CUX1 in the cellular response to mutagenic insult, commonly referred to as the DNA damage response (DDR), which depends on the activity of numerous proteins acting as sensors, mediators, signal transducers and effectors (28). The early DDR is largely mounted in a DNA lesion-specific manner. In the case of DNA double strand breaks (DSBs) generated by clastogens such as ionizing radiation (IR), the Mre11-Rad50-NBS1 (MRN) complex (Mre11-Rad50-NBS1) senses the break and initiates recruitment and activation (i.e. autophosphorylation) of ATM kinase (29). On the other hand, helix-distorting adducts, including UV-induced pyrimidine dimers, strongly block DNA replication which results in formation of large tracts of single-stranded DNA (ssDNA) due to functional uncoupling of DNA synthetic enzymes at stalled replication forks (30,31). The ensuing avid binding of replication protein-A (RPA) to ssDNA tracts specifically activates ATR kinase by facilitating the association of Ataxia-Telangiectasia and Rad3-Related – ATR-Interacting Protein (ATR–ATRIP), TopBP1 and the 911 complex (Rad9-Rad1-Hus1) (30,32). ATR or ATM then rapidly phosphorylate hundreds of downstream targets (many in common) including, very prominently, the transducers Chk1 or Chk2 respectively, and the p53 and BRCA1 tumor suppressors, which regulate cell cycle checkpoints and/or DNA repair (33). Following IR exposure, rapid phosphorylation of H2AX by ATM (to form γ-H2AX) is required for recruitment of the DDR machinery to DSB sites (34). The biological role of a similarly rapid ATR-mediated γ-H2AX induction in response to replication-blocking adducts generated by UV is considerably less clear, but may serve a similar purpose (35).
Genome-wide location analysis suggested that p110 CUX1 localizes to the promoters of several genes involved in the DDR (5). We confirm using independent chromatin immunoprecipitation assays and expression studies that p110 CUX1 indeed binds the promoters, and regulates expression, of DDR genes. Of particular note, CUX1 is shown to be required for full expression of the ATM and ATR checkpoint kinases in the absence of genotoxic stress, as well as of proteins required for activation of these kinases after treatment with DNA damaging agents. Moreover abrogation of CUX1 expression engenders defects in ATM/ATR-mediated DDR signaling, cell cycle checkpoint control and cell survival following exposure to either UV or IR. Our data reveal that the CUX1 transcription factor, in addition to its critical role in regulating normal cell growth, is also required for robust ATM-/ATR-mediated cellular responses to genotoxic stress.
MATERIALS AND METHODS
Cell culture
All cells were maintained in Dulbecco's modified minimum essential medium (DMEM, Wisent) supplemented with 10% Fetal Bovine Serum (Invitrogen) (5% FBS for Hs578T) and penicillin–streptomycin (Invitrogen). The Hs578T and MCF7 cells were grown at 37°C, 5% CO2 and atmospheric O2, whereas mouse embryonic fibroblasts (MEFs) were grown at 37°C, 5% CO2 and 3% O2.
MEF cell isolation
Cux1 mutant mice in the albino OF1 outbred strain were obtained from the laboratory of Meinrad Busslinger and were maintained in the OF1 genetic background (7). Primary MEFs were prepared from 13.5-day-old embryos, and heads were used for genotyping. Limbs and internal organs were removed, and the body was minced and incubated for 10 min in trypsin. Cells were then washed once in complete medium and seeded in a 100-mm dish.
siRNA knockdown
CUX1 knockdown was performed by transfecting cells with a pair of siRNA constructs specific for CUX1 mRNA (5′ GAAUCUUCUCGUUUGAAACUUUGAA and 5′ GCUUCAGAGCGAUAAUACACUAUUA) using Lipofectamine2000 (Invitrogen) according to the manufacturer's instructions. Knockdown was performed 7 days and again 2 days prior to performing experiments. In the case of clonogenic survival assays, knockdown was performed 5 days and again 5 h prior to the experiment.
Retroviral infection
Retroviruses were produced by transfecting 293VSV cells with either the pREV/TRE empty vector (Clontech) or one encoding p110 CUX1 [CUX1 aa 747–1505, Myc tagged at the amino terminus and hemagglutinin (HA) tagged at the carboxyl terminus]. Viruses were applied to cells along with 8 µg/ml polybrene and cells were centrifuged at 300g for 1 h. The mRNA was isolated from infected cells after 24 h.
Reporter assays
PCR amplification was performed to obtain fragments of genomic DNA from DDR gene promoters. These fragments were cloned into the luciferase reporter vector, pGL3 (Promega). Luciferase assays were performed as described previously (36).
Measurement of mRNA levels
RNA was extracted using TRIzol reagent (Invitrogen), and cDNA was prepared using Superscript II RNase H-reverse transcriptase kit (Invitrogen) following the manufacturer's instructions. Real time PCR was performed on a Rotor-Gene instrument (Corbett Life Science) using the QuantiTect SYBR Green PCR Kit (Qiagen) and specific primer pairs for each gene (Supplementary Table S1).
Clonogenic survival assays
The siRNA-treated MCF7 cells or MEF Cux1Z/Z and Cux1 wild-type cells were exposed to either IR at doses of 1, 2 and 4 Gy, or to UV at doses of 2, 5 and 10 J/m2. For MCF7, 500 cells were plated in 60-mm dishes in triplicate. For MEFs, 5000 cells were plated. After 10 days of incubation, cells were washed with phosphate-buffered saline (PBS), fixed with 10% phosphate buffered formalin for 10–20 min then stained with 0.1% crystal violet (Acros Organics) in 20% methanol for 5–10 min. The number of colonies with 50 cells or more was counted.
Immunofluorescence
Cells were plated on glass coverslips and fixed in 4% paraformaldehyde. For γ-H2AX staining, the cell membrane was solubilized in PBS containing 5% FBS and 0.5% Triton X-100. The samples were incubated for 1 h in the solubilizing solution containing primary antibodies for γ-H2AX. Secondary detection was done with Alexa Fluor 488-conjugated antibodies (Molecular Probes) and cells were counterstained with 4′,6-diamidino-2-phenylindole (Molecular Probes). Visualization was done using an Axiovert 200 M microscope with an LSM 510 laser module (Zeiss). Images were analyzed using ImageJ64 software.
For Rad51 staining, cells were solubilized in 0.5% Igepal CA-630 and blocked in 10% FBS, 0.1% Igepal for 1 h prior to 3-h incubation with a primary antibody against Rad51 in blocking solution. Secondary detection and visualization was performed as indicated above.
Western blotting
Nuclear extracts were prepared according to the procedure of Lee et al. (37) except that nuclei were obtained by submitting cells to three freeze/thaw cycles in buffer A (10 mM HEPES, pH 7.9, 10 mM KCl, 1.5 mM MgCl2, 1 mM dithiothreitol) along with protease inhibitor and phosphatase inhibitor tablets (Roche). Sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE) was performed, and after electrophoretic transfer to polyvinylidene difluoride, membranes were washed in Tris-buffered saline–0.1% Tween 20 (TBS 0.1%T) and blocked in TBS 0.1%T containing 5% milk and 2% Bovine Serum Albumin. Membranes were probed with antibodies in TBS 0.1%T at room temperature for 1.5 h for primary antibodies and for 45 min for secondary antibodies.
Antibodies
The following antibodies were used for western blotting: anti-Actin (human and mouse), anti-ATR (human) from Santa Cruz Biotechnology (sc-1616, sc-1887); anti-ATM and anti-ATR (mouse) from AbCam (Ab78, Ab2905); anti-ATM (human), anti-p53 from Calbiochem (PC116, OP43); anti-p-ATM from Rockland (200-301-400); Anti-Chk1, p-Chk1, Chk2, p-Chk2 from Cell Signaling (2345, 2344S, 2662, 2661L); anti-53bp1 from BD Biosciences (612522).
For immunofluorescence: anti-γ-H2AX from Cell Signaling (2577); anti-Rad51 from Santa-Cruz (sc-8349).
For flow cytometry experiments: 488-conjugated anti-BrdU from Molecular Probes (A21303)
G1/S and G2/M checkpoint assay
Cells were trypsinized, fixed in 75% ethyl alcohol and stored at −20°C overnight. A quantity of 50 µl of FBS was then added to each sample. The cells were centrifuged, washed in PBS and resuspended in 300 µl of PBS containing 200 µg/ml of RNase (Sigma) and 5 µg/ml of propidium iodide (Sigma). Samples were incubated for 15 min at 37°C and analyzed using a FACScan (Becton Dickinson), with doublet discrimination to gate single cells. Cell cycle profiles were obtained with FlowJo software (Tree Star Software).
BrdU incorporation assay
At the indicated times, 5-Bromo-2′-deoxyuridine (BrdU) was added to the culture media at 100 mM and incubated for 1 h. Cells were then trypsinized and fixed in 4% paraformaldehyde. The cell membrane was solubilized in PBS containing 5% FBS and 0.5% Triton X-100. The samples were incubated for 1 h in the solubilizing solution containing a fluorescent BrdU antibody. The cells were then centrifuged, washed in PBS, stained with propidium iodide and analyzed as for the G1/S and G2/M checkpoint assays, except that BrdU incorporation in S phase cells was scored.
Single cell electrophoresis
To measure strand breaks, single cell electrophoresis (comet assays) using alkaline lysis were carried out as described in (38). Comet tail moments were measured using the CometScore software (TriTeck Corp).
Cytogenetic analyses
Karyotyping, metaphase chromosome counts and breakage studies were carried out at the Quebec Leukemia Cell Bank (http://www.bclq.org/en/index.html).
RESULTS
CUX1 targets a significant number of genes involved in the DDR
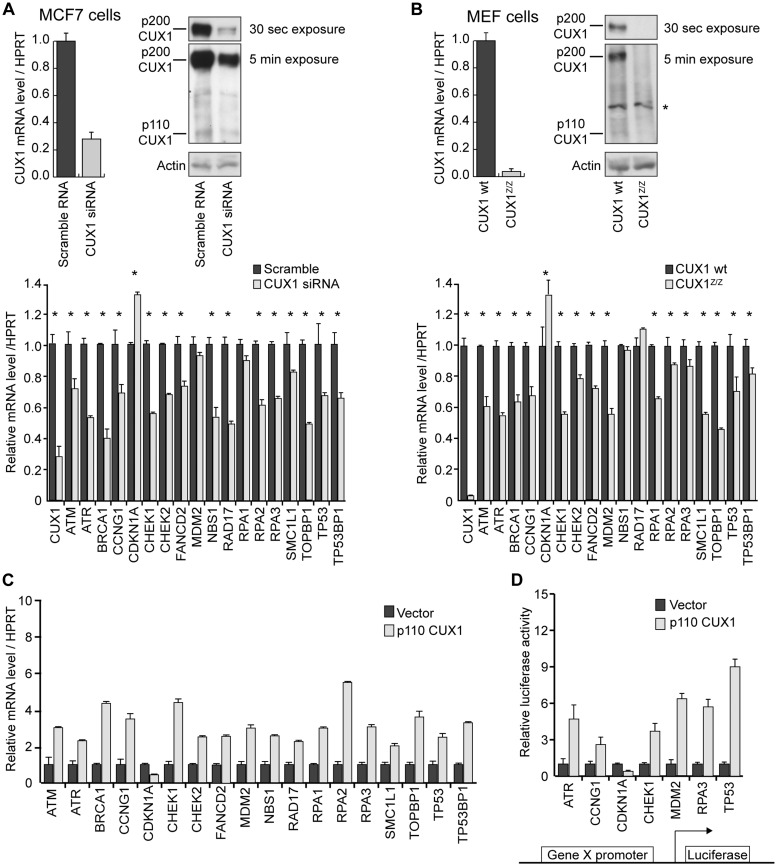
Our laboratory previously carried out genome-wide location analysis on a panel of eight cell lines using promoter microarrays to identify direct transcriptional targets of p110 CUX1 (5). Functional grouping of CUX1 targets using the DAVID software (39) revealed cell cycle checkpoint genes to be among the most enriched categories (Table 1). We used multiple experimental approaches to investigate the role of CUX1 in transcriptional regulation of 18 putative targets known to be involved in DDR checkpoint signaling (Table 2). First, chromatin immunoprecipitation followed by Real-Time PCR (ChIP–qPCR) was employed to measure CUX1 recruitment to each promoter (Table 2, column 2). Second, mRNA expression of DDR target genes was quantified in the context of CUX1 deficiency, either using siRNA-mediated knockdown (Figure 1A) or by comparing MEF cells from wild-type versus Cux1Z/Z mutant mice (Figure 1B) (7). A significant transcriptional effect was observed in both assays for 14 out of 18 genes and in one assay for the remaining 4 genes. All genes manifested decreased expression upon CUX1 knockdown, with the exception of CDKN1A (a.k.a. p21), which was up-regulated (Figure 1 and Table 2, columns 3–6). Third, Hs578t cells were infected with a retroviral vector expressing p110 CUX1 and the expression of target genes measured 24 h later. All genes displayed increased mRNA levels, with the exception of CDKN1A, which was down-regulated (Figure 1C and Table 2, column 7). Fourth, we cloned the proximal promoter region of seven target genes into a reporter construct to verify whether these genomic sequences were sufficient to confer regulation by p110 CUX1 (Figure 1D and Table 2, column 6). Except for CDKN1A which was repressed, all tested genes were activated by p110 CUX1. The regulatory effects of CUX1 on CDKN1A expression observed using these three assays are in agreement with previous studies reporting the role of CUX1 as a transcriptional repressor of this gene (4,22,41–45). Altogether, the above data conclusively show that CUX1 binds to the promoters of many DDR genes and regulates their expression.
Table 1.
Over-represented biological functions of CUX1 targets
| Function | Background (%) | Targets (%) | P-value |
|---|---|---|---|
| Mitotic cell cycle | 2.6 | 4.9 | 2.12E-10 |
| DNA replication and chromosome cycle | 1.5 | 3.0 | 3.46E-08 |
| Cell cycle | 5.6 | 8.3 | 4.18E-08 |
| Cell proliferation | 8.5 | 11.6 | 8.83E-08 |
| M phase of mitotic cell cycle | 1.0 | 2.3 | 9.66E-08 |
| Mitosis | 1.0 | 2.2 | 1.65E-07 |
| M phase | 1.3 | 2.6 | 3.57E-07 |
| DNA replication | 1.1 | 2.3 | 1.44E-06 |
| Nuclear division | 1.3 | 2.5 | 1.90E-06 |
| S phase of mitotic cell cycle | 1.2 | 2.3 | 2.24E-06 |
| DNA metabolism | 4.2 | 6.3 | 2.37E-06 |
| Cell growth and/or maintenance | 30.3 | 34.0 | 7.61E-05 |
| Intracellular transport | 4.5 | 6.1 | 0.00044 |
| Nucleosome assembly | 0.5 | 1.1 | 0.00049 |
| Protein metabolism | 19.9 | 22.6 | 0.00061 |
| DNA-dependent DNA replication | 0.6 | 1.2 | 0.00074 |
| Cell cycle checkpoint | 0.3 | 0.7 | 0.00079 |
| Regulation of cell cycle | 3.1 | 4.3 | 0.00090 |
| Small GTPase mediated signal transduction | 1.9 | 2.8 | 0.00110 |
| Protein folding | 1.0 | 1.7 | 0.00186 |
A single list of putative targets of CUX1 was compiled from eight individual ChIP–chip experiments from cell lines overexpressing p110 CUX1. Genes that were bound by CUX1 (Targets) were compared with all genes present on the microarray (Background) by using a web-based functional annotation tool, DAVID. Overrepresentation of a function depends on the increase in the proportion of genes involved in a given function between CUX1 targets and the background. The P-value is determined using an improved Fisher's exact test from the DAVID software (40). The top 20 significantly over-represented functions are shown.
Table 2.
Transcriptional targets of CUX1 involved in DDR
| Symbol | ChIP– qPCR | siRNA | MEF CUX1z/z | Overexpression | Reporter |
|---|---|---|---|---|---|
| ATM | 5.1 | 0.74* | 0.61*** | 2.9** | |
| ATR | 3.9 | 0.56*** | 0.56*** | 2.3*** | 4.7* |
| BRCA1 | 2.4 | 0.40** | 0.63*** | 4.5*** | |
| CCNG1 | 4.9 | 0.71* | 0.68*** | 3.5*** | 2.6* |
| CDKN1A | 2.7 | 1.32*** | 1.32* | 0.5* | 0.2*** |
| CHK1 | 3.2 | 0.58*** | 0.57*** | 4.4*** | 3.7** |
| CHK2 | 2.1 | 0.68** | 0.79*** | 2.6*** | |
| FANCD2 | 2 | 0.76** | 0.71*** | 2.6*** | |
| MDM2 | 5.3 | 0.93 | 0.57*** | 3*** | 1.7** |
| NBS1 | 2.2 | 0.56*** | 0.97 | 2.6** | |
| RAD17 | 2.8 | 0.50*** | 1.10 | 2.2** | |
| RPA1 | 2.2 | 0.89 | 0.65*** | 3*** | |
| RPA2 | 3.7 | 0.61*** | 0.87* | 5.5*** | |
| RPA3 | 19.4 | 0.65*** | 0.86** | 3.2*** | 5.7* |
| SMC1L1 | 3.1 | 0.82* | 0.57*** | 2*** | |
| TOPBP1 | 3.3 | 0.52*** | 0.45*** | 3.6** | |
| TP53 | 6 | 0.67* | 0.72* | 2.4* | 9** |
| TP53BP1 | 2.5 | 0.65* | 0.82* | 3.3*** | |
| GAPDH | 0.91 | 1.06 | 0.94 | 1.02 | |
| ACTB | 1.22 | 1.00 | 1.01 | 1.01 | |
| UBC | 0.84 | 0.96 | 1.08 | 1.05 |
Table shows the validation of regulation of DDR genes by CUX1. Column 2 shows enrichment of CUX1 at each gene's promoter. Column 3 shows mRNA levels of DDR genes following siRNA knockdown. Column 4 shows mRNA levels in Cux1Z/Z MEFs relative to wild-type littermates. Column 5 shows mRNA levels following overexpression of p110 CUX1 by retroviral infection. All mRNA levels are shown normalized to HPRT1; GAPDH, Actin Beta (ACTB) and Ubiquitin C (UBC) are shown as additional housekeeping genes that are unaffected by CUX1 levels. Column 6 shows level of activation of reporter constructs by p110 CUX1.
*P < 0.05, **P < 0.01, ***P < 0.001.
Figure 1.
Transcriptional regulation of DDR genes by CUX1. (A) MCF7 cells were transfected with CUX1-specific siRNA. Top panel: mRNA and protein levels of CUX1 are shown following knockdown. Bottom panel: mRNA levels of DDR gene targets are shown. All mRNA levels are normalized to Hypoxanthine-guanine phosphoribosyltransferase (HPRT). The values are the mean of three measurements and error bars represent standard deviation. *P < 0.05 on a student's t-test. (B) Mouse embryonic fibroblasts (MEFs) were obtained from Cux1 knockout embryos (cux1Z/Z) and wild-type littermates. Levels of CUX1 and DDR gene targets are shown as in A; *in top panel corresponds to non-specific band recognized by CUX1 antibody. (C) Hs578T cells were infected with a retrovirus expressing p110 CUX1 or with the empty vector. RNA was prepared 24 h post-infection and levels of DDR target genes was measured by real-time PCR and normalized to HPRT. The values are the mean of three measurements and error bars represent standard deviation. (D) The promoter regions of target genes were cloned into a luciferase reporter plasmid. Hs578T cells were transfected with each reporter plasmid along with a vector expressing p110 CUX1 or with an empty vector. The values are the mean of three measurements and error bars represent standard deviation.
DDR signaling is reduced in CUX1-deficient cells
The decrease in DDR gene expression upon CUX1 knockdown suggests that this transcription factor contributes to the maintenance of a transcriptional program required for cellular responses to mutagenic insult. Among CUX1 targets (Table 2) are critical kinases involved in the transmission of DNA damage signals to downstream effectors, specifically ATM/Chk2 and ATR/Chk1, which are rapidly activated by double strand breaks (e.g. IR-induced) and replication stress (e.g. UV-induced), respectively. We therefore investigated whether DDR signaling through these kinases is impaired in the absence of CUX1.
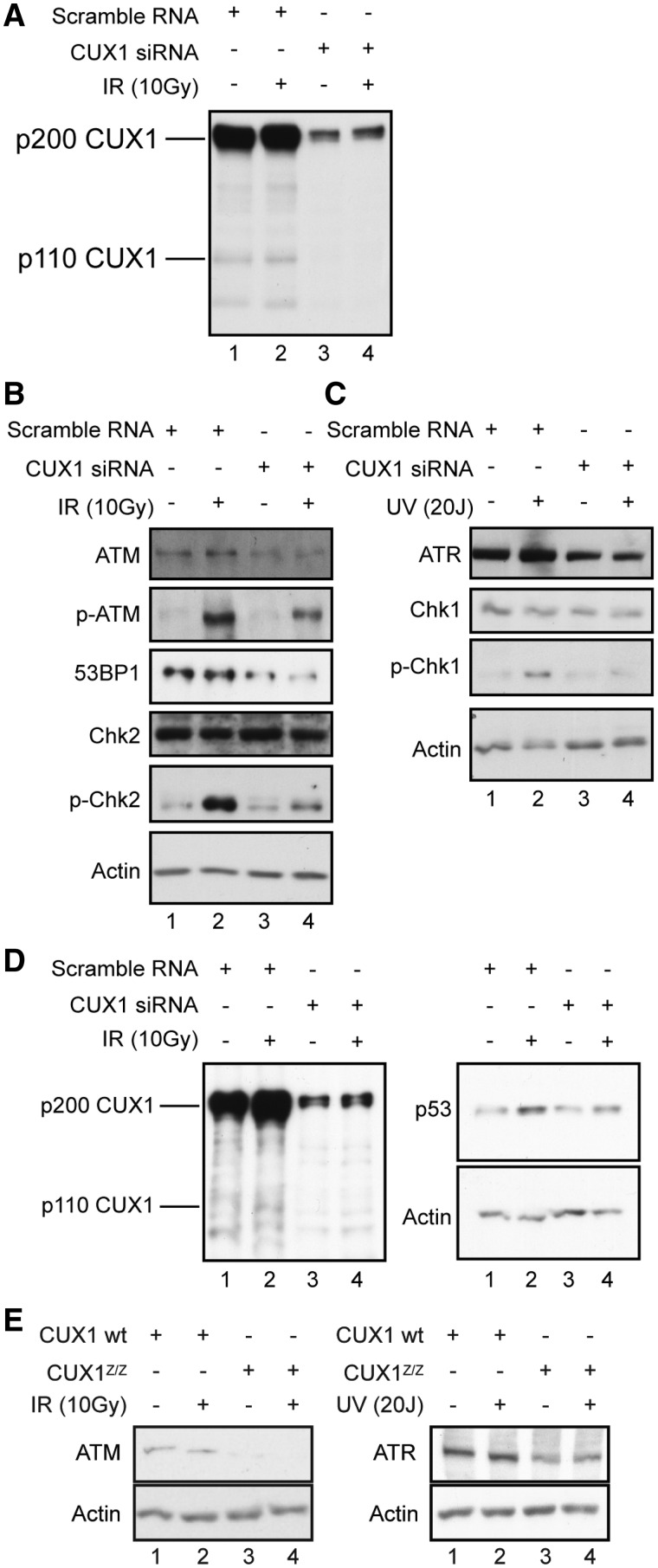
Immunoblotting assays were initially performed to investigate the expression and phosphorylation status of checkpoint kinases following DNA damage. Transfection of MCF7 cells with CUX1-specific siRNA greatly reduced CUX1 expression (Figure 2A, lanes 1 and 3). IR treatment per se did not modulate CUX1 expression in MCF7 cells (Figure 2A, compare lanes 1 and 2, and lanes 3 and 4). Although Chk1 and Chk2 mRNA levels were decreased in CUX1-deficient cells (Figure 1A and Table 2), protein levels were not affected (Figure 2B and C). This suggests that translational or post-translational mechanisms play an important role in regulating Chk1 and Chk2 protein levels. On the other hand, we noted a decrease in steady-state levels of the ATR and ATM kinases, and of the adaptor protein 53BP1 which is critical for ATM-mediated Chk2 activation in response to DSBs (46) (Figure 2B and C, compare lanes 1 and 2 with 3 and 4). Using phospho-specific antibodies, we also detected a strong reduction in phosphorylation of Chk1 Ser317 following UV-irradiation (Figure 2C, compare lanes 2 and 4) and an even more striking decrease in phosphorylation of Chk2 Thr68 and ATM Ser1981 after IR (Figure 2B, compare lanes 2 and 4). These results indicate that both ATM and ATR exhibit reduced activity after DNA damage in CUX1-deficient cells. In addition, p53 accumulation following IR exposure was reduced in siRNA treated cells (Figure 2D, compare lanes 2 and 4), possibly due to impaired ATM/Chk2 signaling (47). Importantly, a decrease in ATM and ATR steady-state levels was also observed in Cux1Z/Z MEFs (Figure 2E).
Figure 2.
Western blotting of DDR signaling kinases, partners and substrates. (A–D) MCF7 Cells were transfected with CUX1-specific siRNA. Nuclear extract were prepared and analyzed by immunoblotting. (A) Cells were exposed to 10 Gy of IR, incubated for 1 h prior to harvest and the extracts were immunoblotted for CUX1 to assess knockdown. (B) Cells were treated as in (A) and immunoblotted for ATM, p-ATM, 53BP1, Chk2 and p-Chk2. Actin was used to control for equal loading. (C) Cells were exposed to 20 Js of UV, incubated for 2 h prior to harvest and the extracts were immunoblotted for ATR, Chk1 and p-Chk1. (D) Cells were exposed to 10 Gy of IR, incubated for 6 h prior to harvest and the extracts were immunoblotted for p53 and CUX1. Actin was used to control for equal loading. (E) Nuclear extracts from Cux1Z/Z and wild-type MEFs were immunoblotted for ATM or ATR following exposure to 10 Gy of IR (left panel) or 20 Js of UV (right panel), respectively.
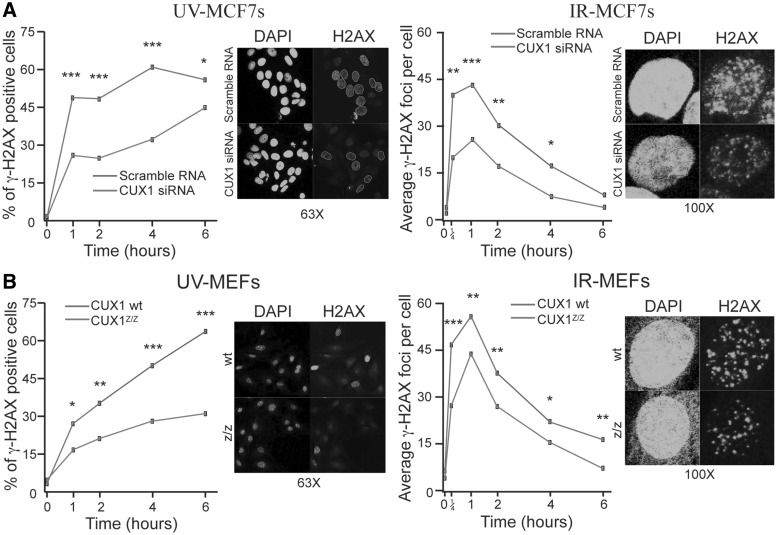
Immunofluorescence microscopy was next employed to evaluate phosphorylation of H2AX (γ-H2AX), well characterized as a very early event mediated by ATR and ATM following exposure to UV and IR, respectively (48). Consistent with previous findings, diffuse γ-H2AX nuclear staining was evident following treatment with 20 J/m2 of UV, whereas distinct foci were discernable in cells treated with 10 Gy of IR (Figure 3) (35). However, in both MEFs and MCF7, knockdown of CUX1 caused a decrease in the proportion of cells showing a positive γ-H2AX signal after UV-irradiation and in the number of γ-H2AX foci per cell after IR (Figure 3A and B).
Figure 3.
γ-H2AX staining in cells after DNA damage. (A) MCF7 cells were transfected with CUX1-specific siRNA. Cells were fixed and stained by immunofluorescence for γ-H2AX. The proportion of γ-H2AX positive cells was counted after treatment with UV. For cells treated with IR, the number of γ-H2AX foci per nuclei was counted. Representative images from experiments at 4 h (UV) and 1 h (IR) are shown next to each graph. *P < 0.05, **P < 0.01, ***P < 0.001; Fisher's exact test for UV treatment or a student's t-test for IR treatment. (B) MEF cells from Cux1Z/Z knockout embryos and wild-type littermates were exposed to DNA damage. Cells were treated and counted as in (A). Representative images from experiments at 4 h (UV) and 1 h (IR) are shown next to each graph.
The above results, taken together, indicate that CUX1 is required for optimal signal transduction downstream of ATM and ATR in response to DNA damage.
CUX1-deficient cells are sensitive to the cytotoxic effects of diverse-acting DNA damaging agents
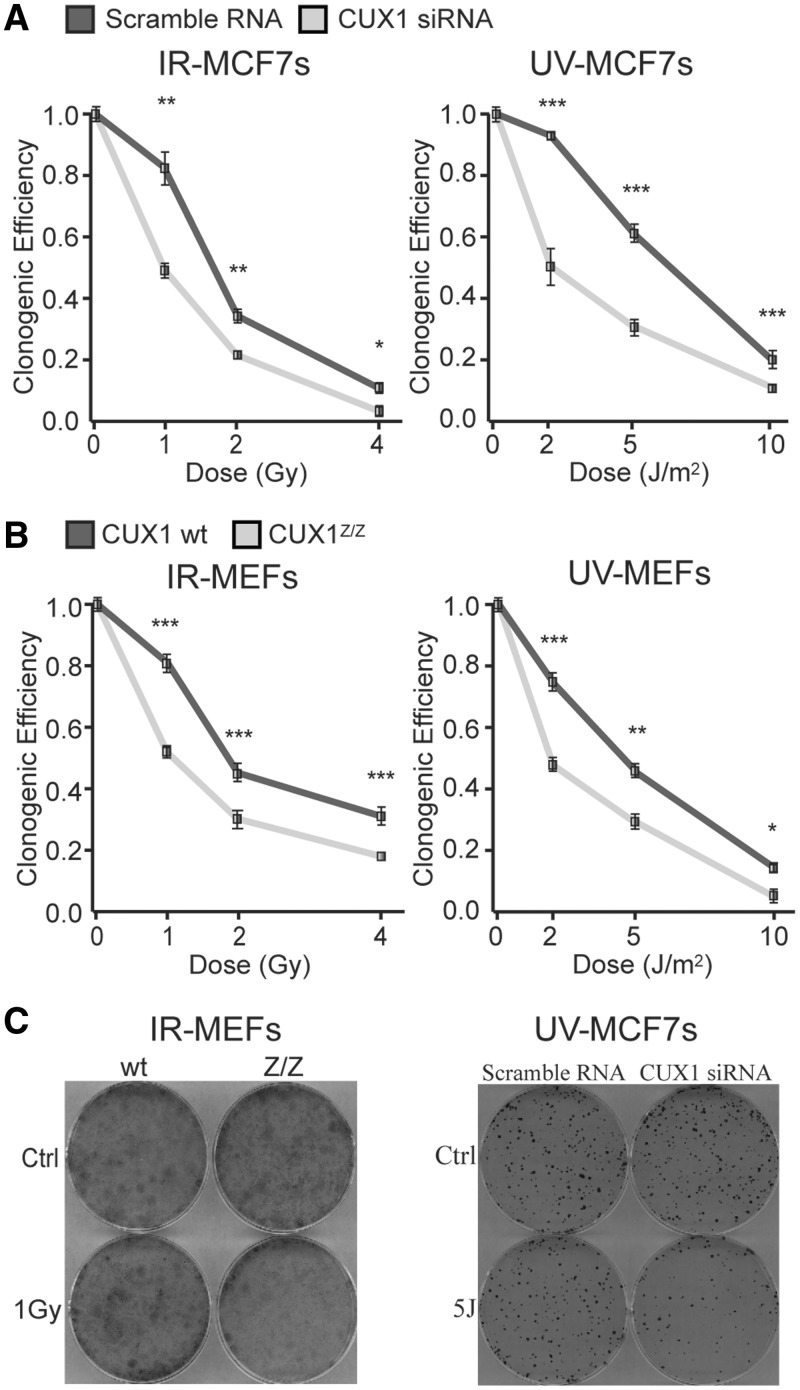
The above results suggest that knockdown of CUX1 would render cells more sensitive to the cytotoxic effects of diverse genotoxic agents. We tested this using either (i) siRNA-mediated knockdown of CUX1 in MCF-7 or (ii) MEFs from wild-type or Cux1Z/Z mutant mice, following exposure to IR or UV. In response to either agent, in both experimental systems, CUX1-deficient cells exhibited significantly decreased clonogenic survival (Figure 4). The well-established role of CUX1 in cell proliferation (20) cannot account for these differences since colony forming ability in mutagen-treated cells is calculated relative to undamaged cells. Furthermore, in the absence of DNA damage, there was no disparity in the absolute number of colonies between cells expressing more versus less CUX1 (Supplementary Figure S1). We conclude that the ability of cells to survive in the face of genotoxic insult is compromised when CUX1 is either inactivated or its expression is reduced.
Figure 4.
Effect of CUX1 knockdown on survival after DNA damage. (A) MCF7 cells were transfected with CUX1-specific siRNA prior to exposure to DNA damage. In total, 500 cells were plated in triplicate and incubated for 10 days. Clones were fixed, stained and counted and the cloning efficiency of unexposed cells was set to 1. *P < 0.05, **P < 0.01, ***P < 0.001; Student's t-test. (B) MEF cells from Cux1Z/Z knockout embryos and wild-type littermates were exposed to DNA damage. 5000 cells were plated in triplicate and incubated for 10 days. Clones were fixed, stained and counted and cloning efficiency of unexposed cells was set to 1. (C) Representative images from experiments with 1 Gy IR in MEF cells (left) and 5 J UV in MCF7 cells (right).
CUX1 knockdown impacts cell cycle checkpoints
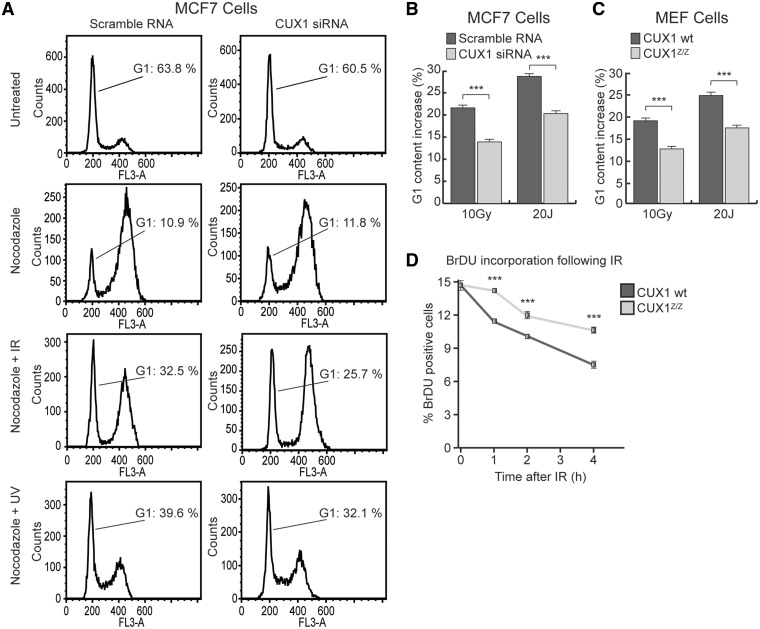
In light of the attenuated ATM/ATR signaling response and reduced viability of CUX1-deficient cells during genotoxic stress, the capacity of such cells to trigger cell cycle checkpoints was investigated. We first evaluated the G1/S checkpoint using flow cytometry to quantify the proportion of cells remaining in G1 24 h post-treatment with IR or UV. The assay was performed on cells synchronized by treatment with the mitotic inhibitor nocodazole prior to irradiation, to exclude the possibility of G1 re-entry of cells that were in G2/M at the time of irradiation. Treatment of MCF7 cells with 10 Gy IR led to a 21.6% increase in the fraction of cells remaining in G1 (Figure 5A, left panels, from 10.9 to 32.5% G1, nocodazole versus nocodazole + IR). This increase reflects cells that were prevented from progressing into S phase as a result of the ATM-mediated G1/S checkpoint. Significantly, siRNA-mediated knockdown of CUX1 attenuated the increase in G1 to 13.9% (Figure 5A, right panels, from 11.8 to 25.7% G1, nocodazole versus nocodazole + IR). Similarly, following treatment with UV, the increase in the G1 fraction was significantly reduced in siRNA-treated MCF7 cells as compared with the control cells (Figure 5A, nocodazole versus nocodazole + UV, results summarized in Figure 5B). A similar trend was observed when the efficiency of the G1/S checkpoint was compared between wild-type and Cux1Z/Z MEF cells following exposure to either IR or UV (Figure 5C). The above data indicate that RNAi knockdown or genetic inactivation of CUX1 compromises the G1/S checkpoint in cells afflicted with either DSBs or increased replication stress.
Figure 5.
Effect of CUX1 knockdown on G1/S and S phase arrest following damage. (A) MCF7 cells were transfected with CUX1-specific siRNA and exposed to either 1 µm Nocodazole, Nocodazole + 10 Gy of IR or Nocodazole + 20 Js of UV. Cells were fixed with ethanol 24 h after exposure, stained with Propidium Iodine (PI) and analyzed for cell cycle distribution by flow cytometry. (B) A histogram of the increase in G1 content after IR and UV in MCF7 cells. *P < 0.05, **P < 0.01, ***P < 0.001; Student's t-test. (C) A histogram of the increase in G1 content after IR and UV in Cux1Z/Z and Cux1 wt MEF cells treated and analyzed as in A. (D) Cux1Z/Z and Cux1 wild-type MEF cells were exposed to 10 Gy IR. 1 to 4 hours post exposure, the cells were labeled with BrdU for 1 h before fixation with 4% Paraformaldehyde. BrdU incorporation was measured by flow cytometry.
The capacity of cells to abrogate DNA replication after irradiation, as controlled by the S phase checkpoint, was also assessed in CUX1 knockdown cells. To this end, we measured incorporation of the deoxyuridine analog bromodeoxyuridine (BrdU) in wild-type versus Cux1Z/Z MEFs. While both cell types showed a reduction in BrdU incorporation following exposure to 10 Gy of IR, wild-type MEFs showed a more rapid and pronounced decrease in the proportion of cells actively synthesizing DNA following irradiation (Figure 5D).
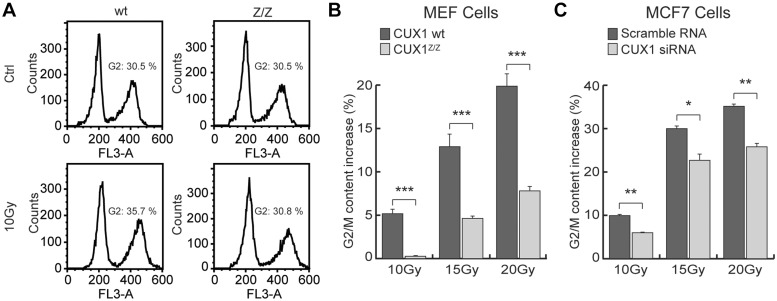
Finally, G2/M arrest was evaluated in wild-type versus Cux1Z/Z MEFs, and in siRNA-treated MCF7 cells. The increase in cellular G2 content was measured by flow cytometry 24 h after exposure to IR. All irradiated cells exhibited an increase in G2 content, although not to the same extent, as illustrated by wild-type versus Cux1Z/Z MEFs (Figure 6A). While the G2 content of wild-type MEFs increased by 5.2, 12.9 and 19.9% after exposure to 10 Gy, 15 Gy and 20 Gy, respectively, the G2 content of MEF CUX1z/z increased by only 0.3, 4.6 and 7.8% under the same conditions (Figure 6B). G2/M arrest was also significantly reduced in siRNA-treated MCF7 cells, although to a less striking extent (Figure 6C).
Figure 6.
Effect of CUX1 knockdown on G2/M arrest following IR. (A) Cux1Z/Z and Cux1 wild-type MEF cells were exposed to 10 Gy IR. Cells were fixed with ethanol 24 h after exposure, stained with Propidium Iodide (PI) and analyzed for cell cycle distribution by flow cytometry. (B) A histogram of the increase in G2 content after IR in MEF cells. *P < 0.05, **P < 0.01, ***P < 0.001; Student's t-test. (C) A histogram of the increase in G2 content after IR in MCF7 cells treated with either CUX1 siRNA or scrambled siRNA.
Taken together, these results indicate that CUX1 is required for cells to mount a complete DNA damage-induced cell cycle checkpoint response, which is fully consistent with the role of this transcription factor in ATM/ATR regulation documented herein.
CUX1 knockdown causes a decrease in Rad51 focus formation and a delay in the repair of DNA strand breaks
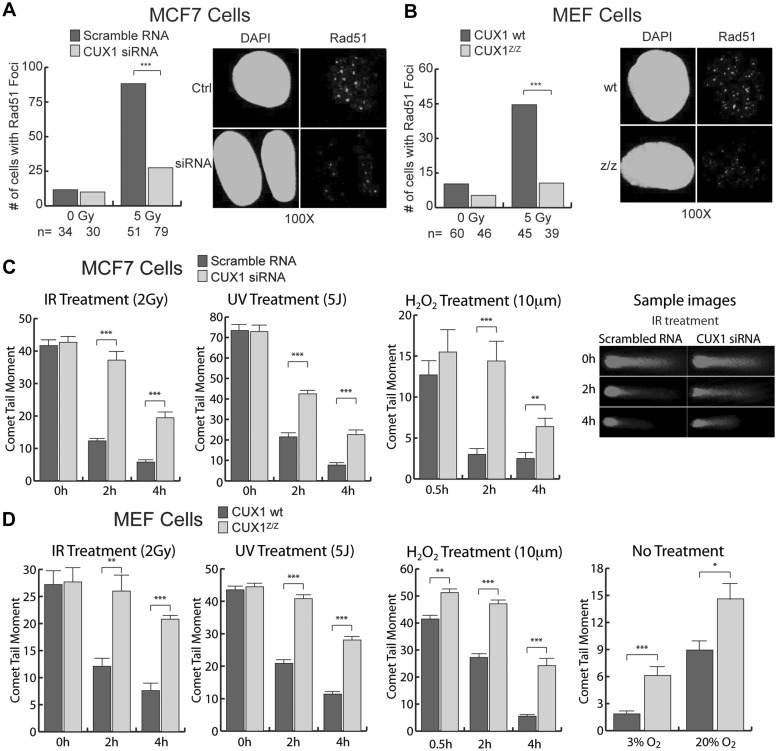
Rad51 focus formation, a well characterized marker of homologous recombination (49), was measured using immunofluorescence. Treatment of MCF7 cells with CUX1 siRNA led to a marked decrease in the proportion of cells displaying five or more Rad51 foci after IR (Figure 7A). Similarly, Cux1Z/Z MEFs displayed a strong reduction in cells displaying Rad51 foci as compared with wild-type counterparts (Figure 7B). These results indicate that CUX1 is required for efficient DNA double strand-break repair by homologous recombination.
Figure 7.
Effect of CUX1 knockdown on Rad51 focus formation and DNA break repair. (A) MCF7 cells were transfected with CUX1-specific siRNA before exposure to 5 Gy of IR. Cells were fixed 1 h after exposure and stained by immunofluorescence for Rad51 foci. The proportion of cells displaying five or more foci is shown. Non-irradiated cells are shown as controls. ***P < 0.001; Student's t-test. (B) Cux1Z/Z and wild-type MEFs were irradiated or mock-irradiated with 5 Gy of IR. Cells were fixed 1 h later and stained by immunofluorescence for Rad51 foci. The proportion of cells displaying five or more foci is shown. (C) MCF7 cells were transfected with CUX1-specific siRNA and then exposed to 2 Gy IR, 5 J UV or 10 mm H2O2 for 30 min. At the indicated times, cells were collected and strand breaks quantified by Alkaline Single Cell Gel Electrophoresis (Comet Assay). Comet tail moments were scored for at least 30 cells per conditions. Error bars represent standard error. *P < 0.05, **P < 0.01, ***P < 0.001; Student's t-test. (D) Cux1Z/Z and wild-type MEFs were exposed to IR, UV or H2O2. and DNA breaks were quantified as in (C). In addition, comet tail moments were measured in MEFs maintained at 3 and 20% oxygen with no further treatment.
Using the single cell gel electrophoresis assay, commonly known as the comet assay, we measured the disappearance of DNA breaks following exposure to IR, UV and H2O2 in MCF7 treated with CUX1 siRNA and in Cux1Z/Z MEFs. Judging from the lengths of comet tails, DNA breaks persisted significantly longer in CUX1-defective cells (Figure 7C and D). Also, Cux1Z/Z MEFs cultured in either a 3 or 20% O2 environment with no additional treatment displayed more damage than the wild-type counterparts, suggesting a higher sensitivity to endogenous DNA damaging agents, e.g. reactive oxygen species produced during oxidative respiration (Figure 7D, rightmost panel)
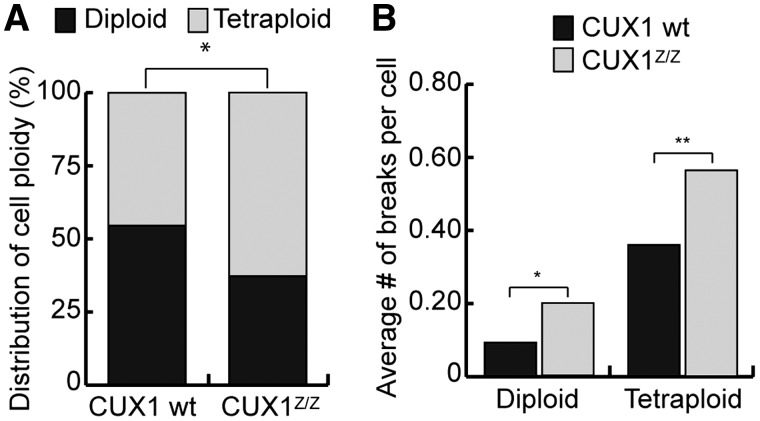
Genomic instability in CUX1z/z MEF cells
Karyotyping analysis was performed on MEFs derived from Cux1Z/Z mutant mice and wild-type littermates. Chromosome counting revealed a greater proportion of tetraploid or near tetraploid cells in CUX1-deficient cells compared with wild-type cells (Figure 8A). In addition, the number of chromosome breaks, as measured following Giemsa staining, was significantly increased in CUX1-deficient cells (Figure 8B). The above results indicate that CUX1-deficient MEFs display increased genomic instability.
Figure 8.
Cux1Z/Z exhibit genomic instability. (A) The number of metaphase chromosomes per cell was counted in Cux1Z/Z and wild-type MEFs. Percentages of diploid/near diploid cells and tetraploid/near tetraploid cells in each population is shown. *P < 0.05; Fisher's exact test. (B) Cells were stained with Giemsa and the number of chromosome breaks per individual cell was counted for Cux1Z/Z and wild-type MEFs. The average number of breaks per cell is shown. *P < 0.05, **P < 0.01; Student's t-test.
DISCUSSION
The present study details the significant role played by CUX1 transcription factor in the cellular response to genotoxic stress. Specifically, in the absence of exogenous DNA damage, CUX1 transcriptionally up-regulates various DDR genes including the ones encoding the critical signaling kinases ATR and ATM. CUX1 similarly stimulates the expression of (i) DNA damage sensors such as NBS1, TopBP1 and RPA, that directly participate in activation of these kinases (29,30,50) and (ii) of the adaptor protein 53BP1 that couples ATM to Chk2 in promoting phosphorylation of the latter (46). It should be noted that relatively modest reductions in steady-state levels of the aforementioned DDR proteins were observed following either partial or complete abrogation of CUX1 in unstressed cells. However, after exposure to DNA damaging agents, the effects of CUX1 depletion on activation of, and signaling through, both ATM and ATR were much more striking. In addition, hallmark ATM/ATR-regulated protective functions including cell cycle checkpoints, survival, homologous recombination and repair of DNA breaks were significantly negatively-impacted following mutagenic insult. This presumably reflects the cooperative effects of CUX1-regulated DDR proteins in triggering a robust ATM/ATR-mediated DDR.
The eukaryotic DDR is classically perceived as being triggered and sustained through post-translational modification of effector proteins imposed after the application of genotoxic stress. However our results provide compelling evidence that prior to sustaining DNA damage, adequate basal DDR protein levels depend on CUX1 transcriptional regulation and moreover, must be in place such that the cells can respond efficiently to mutagenic insult. This function of CUX1 (and quite possibly of other transcription factors; see below), can be distinguished from other, purely DNA damage-inducible mechanisms of DDR gene activation, e.g. the SOS response in bacteria (51–54) and yeast (55), or the p53 tumor suppressor pathway in mammalian cells (56). With this in mind, we speculate that the CUX1-mediated transcriptional response revealed here may be important for full protection against not only exogenous DNA damage, but also highly-genotoxic DNA lesions of endogenous origin such as oxidized bases and strand breaks generated by cellular free radicals. This notion is supported by the comparative analysis of Cux1Z/Z and wild-type MEFs showing a higher number of strand breaks, a greater proportion of tetraploid or near tetraploid cells and an increased number of chromosome breaks in CUX1-deficient MEFs (Figures 7D, 8A and B).
Results from this and other studies reveal a regulatory loop involving CUX1 and key checkpoint kinases. While we have identified CUX1 as an activator of constitutive DDR gene expression, it should be emphasized that this transcription factor itself was shown to be the target of post-translational modification following DNA damage. Specifically, CUX1-derived peptides were identified in two phosphoproteomic studies aimed at identifying proteins phosphorylated after UV or IR (33,57). In total, five phosphorylation sites were identified at positions 322, 734, 1233, 1312 and 1357. Since none of these sites map within one of the four DNA binding domains of CUX1 (CR1, CR2, CR3 and the Cut homeodomain), their phosphorylation would not be expected to inhibit DNA binding. Indeed, CUX1 DNA binding activity is not diminished after DNA damage (Supplementary Figure S2A, B and D), although the manner in which CUX1 regulates transcription may certainly be altered. For example, in agreement with earlier studies from several groups (4,22,41–45), in unstressed cells we observed that CUX1 represses CDKN1A which encodes the p21 cyclin-dependent kinase inhibitor (Figure 1 and Table 2). On the other hand, results using CUX1-deficient cells indicated that CUX1 is required for optimal up-regulation of CDKN1A following DNA damage (Supplementary Figure S2E). We speculate that phosphorylation of CUX1 by checkpoint kinases during periods of genotoxic stress affects its interactions with co-repressors and co-activators, or imparts conformational changes that alter its regulatory properties. The role of CUX1 in the transcriptional program taking place after DNA damage, and the identification of kinases that phosphorylate this transcription factor during genotoxic stress, will be addressed in future studies.
Although a multitude of previous investigations have sought to identify transcription factors that are regulated downstream of DDR signal transduction pathways, only a few attempted to reveal such transcription factors whose activity is required prior to the application of genotoxic stress: (i) expression profiling studies clearly showed a role for E2F family members in the regulation of DNA repair genes involved in homologous recombination, non-homologous end-joining and base excision repair (58–60). Subsequent chromatin immunoprecipitation studies confirmed that E2F transcription factors directly bind the promoters of DDR genes in the absence of DNA damage (59); moreover steady-state changes in the expression of DDR genes in Rb-deficient cells was documented (61,62), (ii) Stat3−/− MEF cells were shown to exhibit a weaker response and decreased survival following irradiation and evidence from reporter assays suggested that MDC1, the gene encoding mediator of DNA damage checkpoint 1, is regulated by STAT3 (63), (iii) NFκB was shown to bind to the promoter of ATM and to be required for optimal expression of this kinase in T cells (64), (iv) the FoxM1 transcription factor regulates baseline levels of p21 and Chk1, and its overexpression is associated with enhanced checkpoint activity (65). Although limited in number, the above studies suggest that preparing cells to cope with DNA damage constitutes a critical function of various transcription factors also known to play roles in cell cycle. Consistent with such a notion, we have shown that CUX1, in addition to regulating cell cycle progression and DNA replication in unstressed cells, also contributes to the maintenance of genomic integrity by ensuring that key players in the DDR are present in stoichiometric amounts prior to DNA damage.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Table 1 and Supplementary Figures 1, 2.
FUNDING
The Canadian Institutes of Health Research (operating grant MOP-98010); Fonds de Recherche en Santé du Québec (FRSQ) (studentship to C.V.). Funding for the open access charge: Operating (grant MOP-98010) from the Canadian Institutes of Health Research.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to thank Ginette Bérubé for preparation of reporter constructs and Maria Drossos for assistance in the preparation of MEF cells. Special thanks go to Ian Hammond-Martel for his expert advice on immunofluorescence procedures.
REFERENCES
- 1.Harada R, Berube G, Tamplin OJ, Denis-Larose C, Nepveu A. DNA-binding specificity of the cut repeats from the human cut-like protein. Mol. Cell Biol. 1995;15:129–140. doi: 10.1128/mcb.15.1.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Neufeld EJ, Skalnik DG, Lievens PM, Orkin SH. Human CCAAT displacement protein is homologous to the Drosophila homeoprotein, cut. Nat. Genet. 1992;1:50–55. doi: 10.1038/ng0492-50. [DOI] [PubMed] [Google Scholar]
- 3.Sansregret L, Nepveu A. The multiple roles of CUX1: Insights from mouse models and cell-based assays. Gene. 2008;412:84–94. doi: 10.1016/j.gene.2008.01.017. [DOI] [PubMed] [Google Scholar]
- 4.Coqueret O, Berube G, Nepveu A. The mammalian Cut homeodomain protein functions as a cell-cycle-dependent transcriptional repressor which downmodulates p21WAF1/CIP1/SDI1 in S phase. EMBO J. 1998;17:4680–4694. doi: 10.1093/emboj/17.16.4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Harada R, Vadnais C, Sansregret L, Leduy L, Berube G, Robert F, Nepveu A. Genome-wide location analysis and expression studies reveal a role for p110 CUX1 in the activation of DNA replication genes. Nucleic Acids Res. 2008;36:189–202. doi: 10.1093/nar/gkm970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nepveu A. Role of the multifunctional CDP/Cut/Cux homeodomain transcription factor in regulating differentiation, cell growth and development. Gene. 2001;270:1–15. doi: 10.1016/s0378-1119(01)00485-1. [DOI] [PubMed] [Google Scholar]
- 7.Ellis T, Gambardella L, Horcher M, Tschanz S, Capol J, Bertram P, Jochum W, Barrandon Y, Busslinger M. The transcriptional repressor CDP (Cutl1) is essential for epithelial cell differentiation of the lung and the hair follicle. Genes Dev. 2001;15:2307–2319. doi: 10.1101/gad.200101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sinclair AM, Lee JA, Goldstein A, Xing D, Liu S, Ju R, Tucker PW, Neufeld EJ, Scheuermann RH. Lymphoid apoptosis and myeloid hyperplasia in CCAAT displacement protein mutant mice. Blood. 2001;98:3658–3667. doi: 10.1182/blood.v98.13.3658. [DOI] [PubMed] [Google Scholar]
- 9.Luong MX, van der Meijden CM, Xing D, Hesselton R, Monuki ES, Jones SN, Lian JB, Stein JL, Stein GS, Neufeld EJ, et al. Genetic ablation of the CDP/Cux protein C terminus results in hair cycle defects and reduced male fertility. Mol. Cell Biol. 2002;22:1424–1437. doi: 10.1128/mcb.22.5.1424-1437.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ledford AW, Brantley JG, Kemeny G, Foreman TL, Quaggin SE, Igarashi P, Oberhaus SM, Rodova M, Calvet JP, Vanden Heuvel GB. Deregulated Expression of the Homeobox Gene Cux-1 in Transgenic Mice Results in Downregulation of p27(kip1) expression during nephrogenesis, glomerular abnormalities, and multiorgan hyperplasia. Dev. Biol. 2002;245:157–171. doi: 10.1006/dbio.2002.0636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brantley JG, Sharma M, Alcalay NI, Heuvel GBV. Cux-1 transgenic mice develop glomerulosclerosis and interstitial fibrosis. Kidney Int. 2003;63:1240–1248. doi: 10.1046/j.1523-1755.2003.00889.x. [DOI] [PubMed] [Google Scholar]
- 12.Cadieux C, Kedinger V, Yao L, Vadnais C, Drossos M, Paquet M, Nepveu A. Mouse mammary tumor virus p75 and p110 CUX1 transgenic mice develop mammary tumors of various histologic types. Cancer Res. 2009;69:7188–7197. doi: 10.1158/0008-5472.CAN-08-4899. [DOI] [PubMed] [Google Scholar]
- 13.Cadieux C, Harada R, Paquet M, Cote O, Trudel M, Nepveu A, Bouchard M. Polycystic kidneys caused by sustained expression of Cux1 isoform p75. J. Biol. Chem. 2008;283:13817–13824. doi: 10.1074/jbc.M709332200. [DOI] [PubMed] [Google Scholar]
- 14.Cadieux C, Fournier S, Peterson AC, Bédard C, Bedell BJ, Nepveu A. Transgenic mice expressing the p75 CCAAT-displacement protein/cut homeobox isoform develop a myeloproliferative disease-like myeloid leukemia. Cancer Res. 2006;66:9492–9501. doi: 10.1158/0008-5472.CAN-05-4230. [DOI] [PubMed] [Google Scholar]
- 15.Michl P, Ramjaun AR, Pardo OE, Warne PH, Wagner M, Poulsom R, D'Arrigo C, Ryder K, Menke A, Gress T, et al. CUTL1 is a target of TGF(beta) signaling that enhances cancer cell motility and invasiveness. Cancer Cell. 2005;7:521–532. doi: 10.1016/j.ccr.2005.05.018. [DOI] [PubMed] [Google Scholar]
- 16.Ripka S, Konig A, Buchholz M, Wagner M, Sipos B, Kloppel G, Downward J, Gress T, Michl P. WNT5A–target of CUTL1 and potent modulator of tumor cell migration and invasion in pancreatic cancer. Carcinogenesis. 2007;28:1178–1187. doi: 10.1093/carcin/bgl255. [DOI] [PubMed] [Google Scholar]
- 17.Kedinger V, Sansregret L, Harada R, Vadnais C, Cadieux C, Fathers K, Park M, Nepveu A. p110 CUX1 homeodomain protein stimulates cell migration and invasion in part through a regulatory cascade culminating in the repression of E-cadherin and occludin. J. Biol. Chem. 2009;284:27701–27711. doi: 10.1074/jbc.M109.031849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Holthuis J, Owen TA, van Wijnen AJ, Wright KL, Ramsey-Ewing A, Kennedy MB, Carter R, Cosenza SC, Soprano KJ, Lian JB, et al. Tumor cells exhibit deregulation of the cell cycle histone gene promoter factor HiNF-D. Science. 1990;247:1454–1457. doi: 10.1126/science.247.4949.1454. [DOI] [PubMed] [Google Scholar]
- 19.van Wijnen AJ, van Gurp MF, de Ridder MC, Tufarelli C, Last TJ, Birnbaum M, Vaughan PS, Giordano A, Krek W, Neufeld EJ, et al. CDP/cut is the DNA-binding subunit of histone gene transcription factor HiNF-D: a mechanism for gene regulation at the G1/S phase cell cycle transition point independent of transcription factor E2F. Proc. Natl Acad. Sci. USA. 1996;93:11516–11521. doi: 10.1073/pnas.93.21.11516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sansregret L, Goulet B, Harada R, Wilson B, Leduy L, Bertoglio J, Nepveu A. The p110 isoform of the CDP/Cux transcription factor accelerates entry into S phase. Mol. Cell Biol. 2006;26:2441–2455. doi: 10.1128/MCB.26.6.2441-2455.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moon NS, Premdas P, Truscott M, Leduy L, Berube G, Nepveu A. S phase-specific proteolytic cleavage is required to activate stable DNA binding by the CDP/Cut homeodomain protein. Mol. Cell Biol. 2001;21:6332–6345. doi: 10.1128/MCB.21.18.6332-6345.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Santaguida M, Ding Q, Berube G, Truscott M, Whyte P, Nepveu A. Phosphorylation of the CCAAT displacement protein (CDP)/Cux transcription factor by cyclin A-Cdk1 modulates its DNA binding activity in G(2) J. Biol. Chem. 2001;276:45780–45790. doi: 10.1074/jbc.M107978200. [DOI] [PubMed] [Google Scholar]
- 23.Goulet B, Baruch A, Moon NS, Poirier M, Sansregret LL, Erickson A, Bogyo M, Nepveu A. A cathepsin l isoform that is devoid of a signal peptide localizes to the nucleus in S Phase and processes the CDP/Cux transcription factor. Mol. Cell. 2004;14:207–219. doi: 10.1016/s1097-2765(04)00209-6. [DOI] [PubMed] [Google Scholar]
- 24.Santaguida M, Nepveu A. Differential regulation of CDP/Cux p110 by cyclin A/Cdk2 and cyclin A/Cdk1. J. Biol. Chem. 2005;280:32712–32721. doi: 10.1074/jbc.M505417200. [DOI] [PubMed] [Google Scholar]
- 25.Sansregret L, Gallo D, Santaguida M, Leduy L, Harada R, Nepveu A. Hyperphosphorylation by cyclin B/CDK1 in mitosis resets CUX1 DNA binding clock at each cell cycle. J. Biol. Chem. 2010;285:32834–32843. doi: 10.1074/jbc.M110.156406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Truscott M, Denault JB, Goulet B, Leduy L, Salvesen GS, Nepveu A. Carboxyl-terminal proteolytic processing of CUX1 by a caspase enables transcriptional activation in proliferating cells. J. Biol. Chem. 2007;282:30216–30226. doi: 10.1074/jbc.M702328200. [DOI] [PubMed] [Google Scholar]
- 27.Sansregret L, Vadnais C, Livingstone J, Kwiatkowski N, Awan A, Cadieux C, Leduy L, Hallett MT, Nepveu A. Cut homeobox 1 causes chromosomal instability by promoting bipolar division after cytokinesis failure. Proc. Natl Acad. Sci. USA. 2011;108:1949–1954. doi: 10.1073/pnas.1008403108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sancar A, Lindsey-Boltz LA, Unsal-Kacmaz K, Linn S. Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu. Rev. Biochem. 2004;73:39–85. doi: 10.1146/annurev.biochem.73.011303.073723. [DOI] [PubMed] [Google Scholar]
- 29.Lee JH, Paull TT. ATM activation by DNA double-strand breaks through the Mre11-Rad50-Nbs1 complex. Science. 2005;308:551–554. doi: 10.1126/science.1108297. [DOI] [PubMed] [Google Scholar]
- 30.Zou L, Elledge SJ. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science. 2003;300:1542–1548. doi: 10.1126/science.1083430. [DOI] [PubMed] [Google Scholar]
- 31.Byun TS, Pacek M, Yee MC, Walter JC, Cimprich KA. Functional uncoupling of MCM helicase and DNA polymerase activities activates the ATR-dependent checkpoint. Genes Dev. 2005;19:1040–1052. doi: 10.1101/gad.1301205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang XH, Zou L. Checkpoint and coordinated cellular responses to DNA damage. Results Probl. Cell Differ. 2006;42:65–92. [PubMed] [Google Scholar]
- 33.Matsuoka S, Ballif BA, Smogorzewska A, McDonald ER, 3rd, Hurov KE, Luo J, Bakalarski CE, Zhao Z, Solimini N, Lerenthal Y, et al. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science. 2007;316:1160–1166. doi: 10.1126/science.1140321. [DOI] [PubMed] [Google Scholar]
- 34.Stucki M, Jackson SP. gammaH2AX and MDC1: anchoring the DNA-damage-response machinery to broken chromosomes. DNA Repair. 2006;5:534–543. doi: 10.1016/j.dnarep.2006.01.012. [DOI] [PubMed] [Google Scholar]
- 35.Marti TM, Hefner E, Feeney L, Natale V, Cleaver JE. H2AX phosphorylation within the G1 phase after UV irradiation depends on nucleotide excision repair and not DNA double-strand breaks. Proc. Natl Acad. Sci. USA. 2006;103:9891–9896. doi: 10.1073/pnas.0603779103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Truscott M, Raynal L, Premdas P, Goulet B, Leduy L, Berube G, Nepveu A. CDP/Cux stimulates transcription from the DNA polymerase alpha gene promoter. Mol. Cell Biol. 2003;23:3013–3028. doi: 10.1128/MCB.23.8.3013-3028.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee KA, Bindereif A, Green MR. A small-scale procedure for preparation of nuclear extracts that support efficient transcription and pre-mRNA splicing. Gene Anal Tech. 1988;5:22–31. doi: 10.1016/0735-0651(88)90023-4. [DOI] [PubMed] [Google Scholar]
- 38.Olive PL, Banath JP. The comet assay: a method to measure DNA damage in individual cells. Nat. Protoc. 2006;1:23–29. doi: 10.1038/nprot.2006.5. [DOI] [PubMed] [Google Scholar]
- 39.Dennis G, Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4:P3. [PubMed] [Google Scholar]
- 40.Hosack DA, Dennis G, Jr, Sherman BT, Lane HC, Lempicki RA. Identifying biological themes within lists of genes with EASE. Genome Biol. 2003;4:R70. doi: 10.1186/gb-2003-4-10-r70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Alcalay NI, Sharma M, Vassmer D, Chapman B, Paul B, Zhou J, Brantley JG, Wallace DP, Maser RL, Vanden Heuvel GB. Acceleration of polycystic kidney disease progression in cpk mice carrying a deletion in the homeodomain protein Cux1. Am. J. Physiol. Renal Physiol. 2008;295:F1725–F1734. doi: 10.1152/ajprenal.90420.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Goulet B, Watson P, Poirier M, Leduy L, Berube G, Meterissian S, Jolicoeur P, Nepveu A. Characterization of a tissue-specific CDP/Cux isoform, p75, activated in breast tumor cells. Cancer Res. 2002;62:6625–6633. [PubMed] [Google Scholar]
- 43.Nishio H, Walsh MJ. CCAAT displacement protein/cut homolog recruits G9a histone lysine methyltransferase to repress transcription. Proc. Natl Acad. Sci. USA. 2004;101:11257–11262. doi: 10.1073/pnas.0401343101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sharma M, Brantley JG, Alcalay NI, Zhou J, Heystek E, Maser RL, Vanden Heuvel GB. Differential expression of Cux-1 and p21 in polycystic kidneys from Pkd1 null and cpk mice. Kidney Int. 2005;67:432–442. doi: 10.1111/j.1523-1755.2005.67099.x. [DOI] [PubMed] [Google Scholar]
- 45.Vanden Heuvel GB, Brantley JG, Alcalay NI, Sharma M, Kemeny G, Warolin J, Ledford AW, Pinson DM. Hepatomegaly in transgenic mice expressing the homeobox gene Cux-1. Mol. Carcinog. 2005;43:18–30. doi: 10.1002/mc.20091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang B, Matsuoka S, Carpenter PB, Elledge SJ. 53BP1, a mediator of the DNA damage checkpoint. Science. 2002;298:1435–1438. doi: 10.1126/science.1076182. [DOI] [PubMed] [Google Scholar]
- 47.Shiloh Y. ATM and related protein kinases: safeguarding genome integrity. Nat. Rev. Cancer. 2003;3:155–168. doi: 10.1038/nrc1011. [DOI] [PubMed] [Google Scholar]
- 48.Harper JW, Elledge SJ. The DNA damage response: ten years after. Mol. Cell. 2007;28:739–745. doi: 10.1016/j.molcel.2007.11.015. [DOI] [PubMed] [Google Scholar]
- 49.Essers J, Houtsmuller AB, van Veelen L, Paulusma C, Nigg AL, Pastink A, Vermeulen W, Hoeijmakers JH, Kanaar R. Nuclear dynamics of RAD52 group homologous recombination proteins in response to DNA damage. EMBO J. 2002;21:2030–2037. doi: 10.1093/emboj/21.8.2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kumagai A, Lee J, Yoo HY, Dunphy WG. TopBP1 activates the ATR-ATRIP complex. Cell. 2006;124:943–955. doi: 10.1016/j.cell.2005.12.041. [DOI] [PubMed] [Google Scholar]
- 51.Galhardo RS, Hastings PJ, Rosenberg SM. Mutation as a stress response and the regulation of evolvability. Crit. Rev. Biochem. Mol. Biol. 2007;42:399–435. doi: 10.1080/10409230701648502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Friedberg EC, Walker GC, Siede W, Wood RD, Schultz RA, Ellenberger T. DNA Repair and Mutagenesis. 2nd edn. Washington DC: ASM Press; 2006. pp. 461–508. [Google Scholar]
- 53.Walker GC. Mutagenesis and inducible responses to deoxyribonucleic acid damage in Escherichia coli. Microbiol. Rev. 1984;48:60–93. doi: 10.1128/mr.48.1.60-93.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Defais M, Fauquet P, Radman M, Errera M. Ultraviolet reactivation and ultraviolet mutagenesis of lambda in different genetic systems. Virology. 1971;43:495–503. doi: 10.1016/0042-6822(71)90321-7. [DOI] [PubMed] [Google Scholar]
- 55.Fu Y, Zhu Y, Zhang K, Yeung M, Durocher D, Xiao W. Rad6-Rad18 mediates a eukaryotic SOS response by ubiquitinating the 9-1-1 checkpoint clamp. Cell. 2008;133:601–611. doi: 10.1016/j.cell.2008.02.050. [DOI] [PubMed] [Google Scholar]
- 56.Beckerman R, Prives C. Transcriptional regulation by p53. Cold Spring Harbor Perspect Biol. 2010;2:a000935. doi: 10.1101/cshperspect.a000935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Stokes MP, Rush J, Macneill J, Ren JM, Sprott K, Nardone J, Yang V, Beausoleil SA, Gygi SP, Livingstone M, et al. Profiling of UV-induced ATM/ATR signaling pathways. Proc. Natl Acad. Sci. USA. 2007;104:19855–19860. doi: 10.1073/pnas.0707579104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Polager S, Kalma Y, Berkovich E, Ginsberg D. E2Fs up-regulate expression of genes involved in DNA replication, DNA repair and mitosis. Oncogene. 2002;21:437–446. doi: 10.1038/sj.onc.1205102. [DOI] [PubMed] [Google Scholar]
- 59.Ren B, Cam H, Takahashi Y, Volkert T, Terragni J, Young RA, Dynlacht BD. E2F integrates cell cycle progression with DNA repair, replication, and G(2)/M checkpoints. Genes Dev. 2002;16:245–256. doi: 10.1101/gad.949802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.DeGregori J, Johnson DG. Distinct and overlapping roles for E2F family members in transcription, proliferation and apoptosis. Curr. Mol. Med. 2006;6:739–748. doi: 10.2174/1566524010606070739. [DOI] [PubMed] [Google Scholar]
- 61.Liao CC, Tsai CY, Chang WC, Lee WH, Wang JM. RB.E2F1 complex mediates DNA damage responses through transcriptional regulation of ZBRK1. J. Biol. Chem. 2010;285:33134–33143. doi: 10.1074/jbc.M110.143461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ganguly A, Shields CL. Differential gene expression profile of retinoblastoma compared to normal retina. Mol. Vis. 2010;16:1292–1303. [PMC free article] [PubMed] [Google Scholar]
- 63.Barry SP, Townsend PA, Knight RA, Scarabelli TM, Latchman DS, Stephanou A. STAT3 modulates the DNA damage response pathway. Int. J. Exp. Pathol. 2010;91:506–514. doi: 10.1111/j.1365-2613.2010.00734.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.De Siervi A, De Luca P, Moiola C, Gueron G, Tongbai R, Chandramouli GVR, Haggerty C, Dzekunova I, Petersen D, Kawasaki E, et al. Identification of new Rel/NFkappaB regulatory networks by focused genome location analysis. Cell Cycle. 2009;8:2093–2100. doi: 10.4161/cc.8.13.8926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tan Y, Chen Y, Yu L, Zhu H, Meng X, Huang X, Meng L, Ding M, Wang Z, Shan L. Two-fold elevation of expression of FoxM1 transcription factor in mouse embryonic fibroblasts enhances cell cycle checkpoint activity by stimulating p21 and Chk1 transcription. Cell Prolif. 2010;43:494–504. doi: 10.1111/j.1365-2184.2010.00699.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.