Abstract
Mammalian alphaherpesviruses are major causes of human and veterinary disease. During productive infection, these viruses exhibit complex and robust patterns of gene expression. These viruses also form latent infections in neurons of sensory ganglia in which productive cycle gene expression is highly repressed. Both modes of infection provide advantageous opportunities for regulation by microRNAs. Thus far, published data regarding microRNAs are available for six mammalian alphaherpesviruses. No microRNAs have yet been detected from varicella zoster virus. The five other viruses -- herpes simplex viruses-1 and -2, herpes B virus, bovine herpesvirus-1, and pseudorabies virus -- representing both genera of mammalian alphaherpesviruses have been shown to express microRNAs. In this article, we discuss these microRNAs in terms of where they are encoded in the viral genome relative to other viral transcripts; whether they are expressed during productive or latent infection; their potential targets; what little is known about their actual targets and functions during viral infection; and what little is known about the interactions of these viruses with the host microRNA machinery.
1. Introduction: Mammalian Alphaherpesviruses
Mammalian alphaherpesviruses, which include important human and veterinary pathogens, fall within the order Herpesvirales (herpesviruses). Membership in this order is based, largely, on the morphology of the virion, which includes a core containing a linear double-stranded DNA, which is surrounded by an icosahedral capsid. The capsid is surrounded by the tegument, which is an amorphous proteinaceous material, and that in turn is surrounded by a lipid bilayer envelope [1, 2]. The order embraces highly divergent viruses that infect various hosts ranging from bivalves to humans. Members of the order are subclassified into three families: the Herpesviridae (herpesviruses of mammals, birds and reptiles), the Alloherpesviridae (fish and amphibian viruses) and the Malacoherpesviridae (bivalve viruses) [2]. Viruses in the family Herpesviridae share many biological properties including the ability to form latent infections, but have been divided into three subfamilies, Alphaherpesvirinae, Betaherpesvirinae and Gammaherpesvirinae, based on host range, replication properties, transmission routes and sites of latent infection. The members of the subfamily Alphaherpesvirinae (alphaherpesviruses) have broad host ranges (although most alphaherpesviruses have a single natural host, they can infect multiple species in artificial settings such as laboratories, zoos, and farms), and relatively short replication cycles. This subfamily includes two distinct genera that infect mammals --Simplexvirus, whose prototypes are Herpes simplex virus 1 (HSV-1) and 2 (HSV-2), and Varicellovirus, whose prototype is Varicella-zoster virus (VZV). More than 30 mammalian alphaherpesviruses have been identified. (There are also two genera of alphaherpesviruses that infect birds -- Mardivirus and Iltovirus; miRNAs encoded by these avian herpesviruses will be covered in another article in this volume.) All mammalian alphaherpesvirus genomes are highly related, with very similar sets of genes, although there are certain genes in given viruses that are not shared, and viruses in both genera share the property of forming latent infections in neurons [1, 3]. However, while infections by simplexviruses usually involve infection at the peripheral mucosa followed by rather limited dissemination though neurons that innervate the site of infection, infections by varicelloviruses often proceed via other routes (e.g. the respiratory tract) and involve widespread dissemination (e.g. via viremia) to many tissues [1, 4, 5].
2. Advantageous Features of miRNAs for Mammalian Alphaherpesviruses
MicroRNAs (miRNA) are ~22 nucleotide (nt) small RNA molecules that regulate gene expression of plants, animals and viruses [6, 7]. Briefly, canonical miRNAs arise from longer RNA polymerase II transcripts, called pri-miRNAs, that contain one or more characteristic hairpin structures [8]. The hairpin can be recognized by the nuclear microprocessor complex, which includes the RNaseIII enzyme Drosha and its co-factor DGCR8. Drosha cleaves the hairpin within its stem and relatively proximal to the terminal loop, to release a ~60 nt hairpin, called a pre-miRNA [9, 10]. The pre-miRNAs are transported from the nucleus by Exportin 5/RAN-GTP to the cytoplasm where they are cleaved by another RNAseIII enzyme, Dicer in association with TRBP to generate ~22-nt miRNA duplexes that are usually not perfectly complementary and carry 2-nt 3’ overhangs at both ends [11-13]. The miRNA duplexes associate with Argonaute (Ago) proteins and GW182 to form a functional RNA-induced silencing complex (RISC), which leads to separation of strands of the duplex [14, 15]. One strand, designated as the mature miRNA, is retained and serves as a guide to target complementary mRNAs. The other strand (called the star, or passenger strand) usually, but not always, is subsequently degraded. Indeed, certain herpesvirus pre-miRNAs are processed into two strands of similar abundance. Pairing of miRNA nucleotides 2-8 (seed sequence) with target sequences is thought to be crucial for target specificity; however, a lack of perfect seed complementarity can be compensated by downstream complementarity [16]. In animal genes, the majority of miRNA target sites lie within 3’ untranslated regions (UTRs) of mRNAs, but some sites have been found within 5’UTRs and coding sequences [17, 18]. Binding of RISC to targeted mRNAs interferes with their translation and stability, and results in reduced protein levels [19-21].
In principle, miRNAs have advantages over proteins for regulation of gene expression by viruses, particularly mammalian alphaherpesviruses that form latent infections. MiRNAs require relatively little coding capacity and are prone to accelerated evolution, since even single nucleotide changes can dramatically alter miRNA specificity [16, 22, 23]. MiRNAs are also non-immunogenic, which might be particularly important for viruses that can establish life-long latent infections, minimizing exposure to immune-surveillance. In addition, miRNAs can modulate, to varying degrees, the expression of as many as hundreds of different target genes. This could be useful to establish conditions optimally permissive for viral replication, for establishment of latency, or to allow rapid responses to changes in the environment, such as those that trigger reactivation from latency.
3. Discovery of Mammalian Alphaherpesvirus miRNAs
The first hints that mammalian alphaherpesviruses express miRNAs came from the Tuschl laboratory in 2005 [24]. They applied computational analyses of hairpin structures that might represent pre-miRNAs to predict that HSV-1 and HSV-2 encode seven and six miRNAs, respectively. However, they did not validate these predictions experimentally. Interestingly, these authors also predicted that VZV does not encode any miRNAs. In 2006, Cui et al., using a different computational approach, predicted 13 HSV-1 pre-miRNAs, of which only one overlapped with the predictions from the Tuschl lab [25]. These authors experimentally identified the first HSV-1 miRNA, miR-H1. This miRNA was detected in productively infected cells in culture. Subsequently in 2008, Umbach et al. detected five additional virally encoded miRNAs in ganglia from mice latently infected with HSV-1, and Tang et al. detected three related miRNAs in ganglia from guinea pigs latently infected with HSV-2 [26-28]. More recently, advances in massively parallel sequencing technologies have allowed high-throughput miRNA analyses with unprecedented sensitivity. However, to date, of the more than 30 known mammalian alphaherpesviruses, there are published data on analyses of miRNAs from only six -- HSV-1 and -2, VZV, herpes B virus, Bovine herpesvirus 1 (BoHV-1) and Pseudorabies virus (PRV, Suid herpesvirus 1) [26-35]. So far, all of these viruses except VZV have been found to express at least several miRNAs. Thus, it is highly likely that many more mammalian herpesvirus miRNAs will be identified.
4. The Prototype Simplexvirus, HSV-1, and HSV-2
The prototype simplexvirus, HSV-1 and its very close relative, HSV-2, are ubiquitous human pathogens, widely recognized as the causative agents of cold sores and genital herpes, respectively [1]. Although infections with HSV are usually self-limiting, they can result in severe morbidity and life-threatening diseases, particularly in individuals with undeveloped (e.g. newborns) or compromised (e.g. transplant patients) immunity. HSV-1 is the leading infectious cause of blindness, and the most common cause of sporadic encephalitis in the U.S., which fortunately occurs rarely [1]. HSV-2 has also been recognized as an important co-factor for sexually transmitted diseases, including acquired immunodeficiency syndrome (AIDS) [36].
Within the virion, the HSV genome is a linear molecule of ~150 kbp (Fig. 1). The genome consists of two unique segments, unique long (UL) and unique short (US), each bounded by inverted repeat segments (RL and RS, respectively). (There are also repeat segments at the tips of the genome and between the internal copies of RL and RS, but for simplicity, they will be ignored here.)
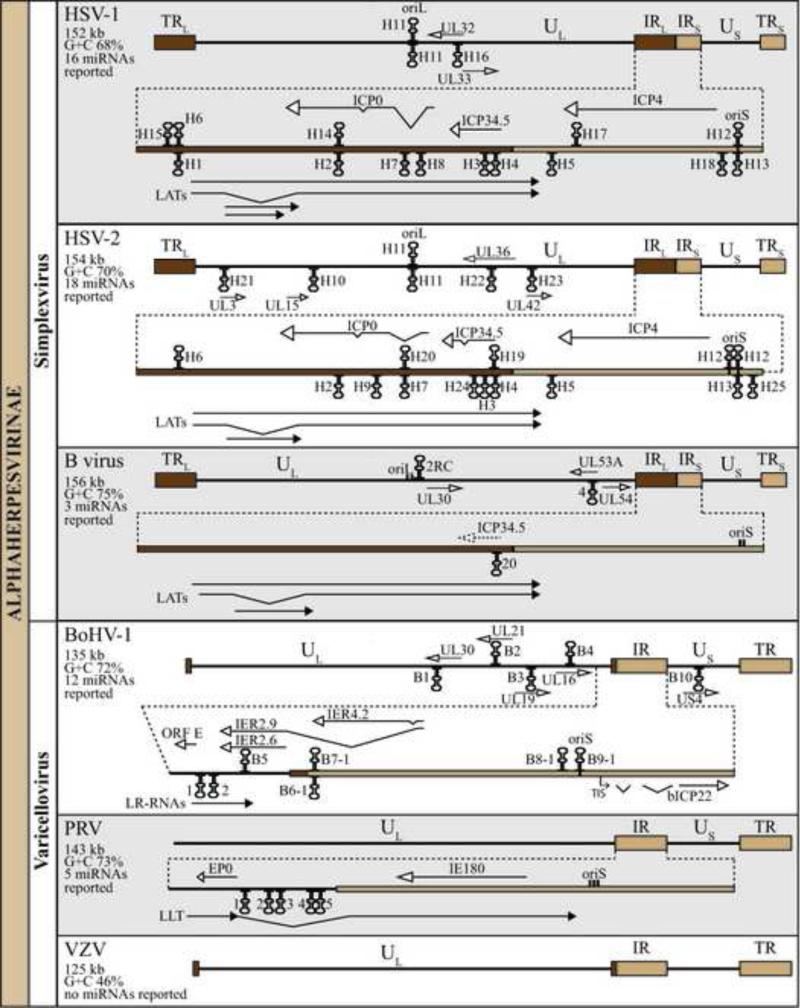
Fig. 1. Genomic organization and location of sequences encoding miRNAs of mammalian alphaherpesviruses.
Schematic of the HSV-1, HSV-2, BV, BoHV-1, PRV and VZV genomes,. The genera (Simplexvirus, Varicellovirus) to which these viruses belong, and the length of each genome and its G+C content are indicated to the left. The locations of sequences encoding miRNAs and viral transcripts (not to scale) proximal to or spanning the miRNA loci are presented [29, 30, 32, 33, 35]). UL and US denote the unique sequences of long (L) and short (S) components of the genome, which are shown as solid lines. Repeat sequences flanking UL (TRL and IRL) and US (IRS or IR, and TRS or TR) are shown as darkly and lightly shaded brown boxes, respectively. One copy of the internal repeat sequences and, for the varicelloviruses whose RL's are very short, the adjacent part of the UL region, is expanded below the schematic for each viral genome. Relative locations and orientations of transcripts in the miRNA regions are denoted by solid arrows, with angled lines denoting portions removed by splicing. Protein coding genes and non-coding RNA transcripts are shown with empty and black-shaded arrows, respectively. A homolog of HSV ICP34.5 is absent in BV, but its homologous position is shown with dotted-lines. OriL and oriS denote viral origins of DNA replication and are shown as small black rectangles (the number of rectangles corresponds to the number of oriL or oriS copies). Locations of miRNA precursors in the viral genomes are shown as hairpins. MiRNA precursors shown below the line are transcribed from left to right, while those above the line are transcribed from right to left. The direction of transcription for the precursor of HSV miR-H11 arising from the oriL palindrome has not been determined and is therefore shown both below and above the line.
The primary site of infection for HSV is the mucosal epithelium (e.g. the lip, cornea, or vagina), where the virus initiates its productive cycle. As other mammalian alphaherpesviruses have productive cycles highly similar to that of HSV, the HSV cycle will be described in some detail here with emphasis on aspects relevant to viral miRNAs. The cycle begins when the virus attaches to specific cell-surface receptors, which leads to fusion of the viral envelope with cell membranes, and release of tegument and the DNA-containing capsid into the cell. The capsid is then transported to a nuclear pore, where it releases viral DNA into the nucleus. Viral DNA circularizes in the nucleus. HSV expresses its genes in a regulated cascade in which expression of immediate early (IE) genes is followed by the expression of early (E) and then late (L) genes. At the same time, HSV induces extensive nuclear remodeling that provides a framework for efficient virus replication [1]. The expression of IE genes does not require prior viral protein synthesis; however it is strongly simulated by at least one tegument protein, the transcriptional activator VP16 [37-39]. IE proteins promote the expression of subsequent viral gene products, and, at least some have a role in suppressing anti-viral defense mechanisms. One IE protein is infected cell protein (ICP) 4, which interacts with the host transcriptional machinery to upregulate transcription of E and L genes, and binds to the promoters of IE genes to downregulate their expression [40-45]. A second IE protein is ICP0, which acts as promiscuous transcriptional transactivator in transfection experiments [46-50]. ICP0 is a ubiquitin ligase; it promotes the dispersal of cellular nuclear domain 10 (ND-10) structures and degradation of their components (incoming viral genomes initiate transcription and replication near ND-10); interacts with the CoREST/REST HDAC complex to prevent silencing of HSV DNA; and interferes with interferon signaling [51-60].
The E proteins include viral enzymes involved in viral DNA replication such as a DNA polymerase and a thymidine kinase. Viral DNA synthesis requires both specific viral proteins and specific origin (ori) sequences. Following replication of viral DNA, expression of the L genes becomes maximal. There are two kinds of L genes, true L, which are expressed very poorly (e.g. <1%) in the absence of viral DNA synthesis, and leaky L, which are expressed weakly prior to DNA synthesis and exhibit relatively modest reductions in expression when viral DNA synthesis is inhibited. The L genes encode proteins required for the packaging of newly synthesized viral genomes in the nucleus, and egress of infectious virus from the cell [1].
Productive HSV infection is rapid and devastating to the infected cell. Although there are many host mechanisms to combat virus infections, HSV has evolved numerous, sometimes redundant, strategies to circumvent such mechanisms [1]. For example, the leaky L protein ICP34.5 prevents translational arrest mediated by the interferon-induced protein kinase R (PKR) by recruiting protein phosphatase 1 (PP1) to dephosphorylate eIF2α, a PKR substrate [61, 62]. ICP34.5 also targets Toll-like receptor signaling component TANK-binding kinase and blocks maturation of dendritic cells[63, 64]; it binds Beclin 1 protein and thus inhibits autophagy [65, 66]; and it interferes with MHC II expression [67]. The true L protein US11, which is an RNA binding protein, also can act to prevent PKR-mediated translational arrest, in this case by inhibition of PKR, and, if ectopically expressed as an IE protein, can compensate for the loss of ICP34.5 [68-72].
Following productive infection of the mucosal epithelium, HSV can enter sensory neurons that innervate the sites of primary infection and travel through axons to cell bodies in sensory ganglia where virus can initiate an acute productive infection. Eventually, the acute infection clears, yet a fraction of sensory neurons are latently infected for the life of the host (latency). During latency, no infectious virus is detected, but following various stimuli such as stress, virus can reactivate and cause recurrent disease [1]. During latency, productive cycle gene expression is repressed, and the only abundant transcripts arise from the latency-associated transcript (LAT) locus [73]. There are multiple LATs, including an unstable 8.3 kb primary transcript, an unstable spliced 6.3 kb transcript, and two stable introns of 2 kb and 1.5 kb [74, 75]. Despite the fact that LATs have been intensely studied for more than 25 years, the exact roles of LATs remain only vaguely defined. However, depending on the virus strain, animal model, and investigating laboratory, there is evidence for LATs having a role in repression of productive-cycle gene expression and inhibition of apoptosis, and for roles in establishment of latency, maintenance of latency, and/or reactivation from latency (e.g. [76-81]).
5. Herpes simplex virus 1 and 2 miRNAs
5.1. Genome Location and Expression
This review will provide the most detail about HSV miRNAs, because we have more information about them than those from other mammalian alphaherpesviruses, and because they provide comparisons for the miRNAs from the other viruses. To date, sixteen and eighteen miRNAs expressed by HSV-1 and HSV-2, respectively, have been identified (Fig. 1; Table 1). HSV-1 encodes miRNAs named miR-H1 – H8 and miR-H11 – H18, whereas HSV-2 encodes miR-H2 - H7, miR-H9 - H13 and miR-H19 - H25. This nomenclature was based on the order of the discoveries, homologies between HSV-1 and HSV-2 miRNAs, and recommendations from miRBase, the miRNA database [82]. Some HSV-1 and -2 encoded miRNAs have names that reflect their positional and, to varying extents, sequence homology, but do not necessarily imply their functional homology; e.g., HSV-1 and -2 miRNA-H6, which do not share seed sequence homology. It is important to note that HSV-2 miRNA-H2, -H3 and -H4 were originally identified and named as miR-III, -I and –II, respectively, by Tang et al. [27, 28].
Table 1.
miRNAs expressed by Alphaherpesviruses in productive and/or latent infection
| HS V-1 | HSV-2 | |||||
|---|---|---|---|---|---|---|
| miRNA | Productivea | Latenta | Productivea | Latenta | Location in the genome | Target b |
| H1 | + | + | NH | NH | Upstream of LATs; complementary to miR-H6 | Unknown |
| H2 (III)c | + | + | + | + | Within 2nd LAT exon; complementary to ICP0 ORF | ICP0 |
| H3 (I)c | + | + | + | + | Within 2nd LAT exon and L/STs; complementary to ICP34.5 ORF (i.e. in HSV-2 1st exon of ICP34.5) | ICP34.5 |
| H4 (II)c | + | + | + | + | Within 2nd LAT exon and L/STs; complementary to ICP34.5 5′UTR | ICP34.5 |
| H5 | + | + | + | + | Within 2nd LAT exon and L/STs | Unknown |
| H6 | + | + | + | + | Opposite strand and upstream of LATs; complementary to miR-H1 | ICP4e |
| H7 | + | + | + | + | Within 2nd LAT exon, complementary to ICP0 1st intron | Unknown |
| H8 | + | + | NH | NH | Within 2nd LAT exon, complementary to ICP0 1st intron | Unknown |
| H9 | NH | NH | + | + | Within 2nd LAT exon, complementary to ICP0 2nd exon | Unknown |
| H10 | NH | NH | - | + | UL region of the genome; downstream of UL15 | Unknown |
| H11 | + | - | + | - | oriL | Unknown |
| H12 | + | - | + | - | oriS | Unknown |
| H13 | + | - | + | - | oriS | Unknown |
| H14 | + | - | NH | NH | Within 3rd exon of ICP0; complementary to miR-H2 | Unknown |
| H15 | + | - | NH | NH | Downstream of miR-H6 | Unknown |
| H16 | + | - | NH | NH | Upstream of UL33 coding sequence; complementary to UL32 | Unknown |
| H17 | + | - | NH | NH | Within ICP4 coding sequence | Unknown |
| H18 | - | + | NH | NH | Upstream of oriS | Unknown |
| H19 | NH | NH | + | - | 5′UTR of ICP34.5; complementary to miR-H4 | Unknown |
| H20 | NH | NH | + | - | Within 1st LAT intron; complementary to miR-H7 | Unknown |
| H21 | NH | NH | + | - | Within UL3 coding sequence | Unknown |
| H22 | NH | NH | + | - | Complementary to UL36 coding sequence | Unknown |
| H23 | NH | NH | + | - | Within UL42 coding sequence | Unknown |
| H24 | NH | NH | + | + | Within 2nd LAT exon and L/STs; Upstream of miR-H3 and complementary to 1st exon of ICP34.5 | Unknown |
| H25 | NH | NH | + | - | Downstream of miR-H13; proximal to oriS | Unknown |
| B virus | ||||
|---|---|---|---|---|
| miRNA | Productivea | Latent | Location in the genome | Target |
| 2RC | + | NT | Complementary to likely UL30 5′UTR | Unknown |
| 4 | + | NT | Complementary to UL53A coding sequence | Unknown |
| 20 | NT | Within 2nd LAT exon | Unknown | |
| BoHV-1 | BoHV-5 | ||||
|---|---|---|---|---|---|
| miRNA | Productivea | Latenta | Predicted homologg | Location in the genome | Target |
| B1 | + | NT | P | Complementary to UL30 coding sequence | Unknown |
| B2 | + | NT | P | Within UL21 coding sequence | Unknown |
| B3 | + | NT | P | Within UL19 coding sequence | Unknown |
| B4 | + | NT | P | Complementary to UL16 coding sequence | Unknown |
| B5 | + | NT | P | Within bICP0 coding sequence; complementary to LR- RNA 3′UTR | Unknown |
| B6 | + | NT | P | Complementary to 3rd bICP0 intron and miR-B7 | Unknown |
| B7 | + | NT | P | Within 3rd ICP0 intron; complementary to miR-B6 | Unknown |
| B8d | + | NT | P | Upstream of oriS | Unknown |
| B9 | + | NT | P | oriS | Unknown |
| B10 | + | NT | NP | Within US4 coding sequence | Unknown |
| 1f | NT | + | NP | Within LR gene | ICP0 |
| 2f | NT | + | NP | Within LR gene | ICP0 |
| PRV | ||||
|---|---|---|---|---|
| miRNA | Productivea | Latent | Location in the genome | Target |
| 1 | + | NT | Within intron of the LLT transcript | Unknown |
| 2 | + | NT | Within intron of the LLT transcript | Unknown |
| 3 | + | NT | Within intron of the LLT transcript | Unknown |
| 4 | + | NT | Within intron of the LLT transcript | Unknown |
| 5 | + | NT | Within intron of the LLT transcript | Unknown |
miRNA was detected (+) or not detected (-) by Northern Blot, qRT-PCR, or as sequence reads [25-35, 96]; NH, no homolog detected; NT, not tested.
Target identified in transfection studies.
The original HSV-2 nomenclature.
Both 5p and 3p strands were detected and were considered to represent two different miRNAs.
Target identified only for HSV-1.
Species about the size of miRNAs were detected by Northern blot using a large probe (464 nucleotide).
Sequence comparison between BoHV-1 and -5 showed 73-97% sequence conservation; P, predicted; NP, no homolog predicted.
Although many HSV miRNAs have been identified, relatively little is known regarding their kinetics and levels of expression during various stages of infection in vitro and in vivo. For several miRNAs, Northern blot and quantitative stem-loop real-time reverse transcription-PCR (qRT-PCR) analyses have been performed. The latter is the most quantitative, but still suffers from limitations. In particular, the stem-loop assay has a high specificity for a precise miRNA sequence and miRNAs often have heterogeneous 5' and 3' ends, which can affect how accurately a particular stem-loop assay will assess the abundance of the miRNA. Additionally, at least in productively infected cells, the levels of mature miRNAs are substantially lower than the levels of their corresponding pre-miRNAs and possibly their pri-miRNAs (see 5.3 below). These longer species can be detected by the stem-loop procedure, albeit less efficiently ([83]; M.F. Kramer and D.M. Coen, unpublished results). Some investigators have used total RNA rather than RNA <40 nt for their qRT-PCR assays, even in assays of productively infected cells, so those results should be interpreted with caution. For most of the miRNAs, the only information is the number of sequence reads, which can be misleading as methods for library preparation and deep sequencing are biased toward certain miRNAs [84]. Similarly, little is known about the pri-miRNAs from which many of the miRNAs are derived. In general, these pri-miRNAs might be difficult to detect as they are cleaved into pieces during generation of pre-miRNAs and those pieces are usually rapidly degraded. In some cases, previously described transcripts spanning the miRNA locus can help predict potential pri-miRNAs, and also provide some clues regarding expression kinetics.
Most HSV-1 and -2 miRNAs are encoded proximal to or within the LAT locus, i.e. within the repeat sequences RL and RS flanking the UL and US components of the genome, respectively. Thus, the coding sequences of these miRNAs are present in two copies per genome (Fig. 1). HSV miR-H2, H3, H4, H5, H7, and HSV-1 miR-H8, and HSV-2 miR-H9 and -H24 map within sequences encoding the 8.3 kb primary LAT. Indeed, miR-H2 - -H5 can be expressed following transfection from a plasmid encoding the 8.3 kb LAT under the control of a strong promoter [26-28], and the expression of these miRNAs in latently infected ganglia is greatly reduced by LAT promoter mutations [27, 85]. These observations are consistent with the observation that the 8.3kb LAT is very difficult to detect, both in vitro and in vivo [86], which may be due to its processing into miRNAs. They are also consistent with these miRNAs having been detected predominantly in latently infected tissues (human, mouse, or guinea pig sensory ganglia) (Table 1). Interestingly, however, in productively infected cells in culture, these miRNAs might arise from additional transcription units [27, 28, 85].
HSV-1 miR-H1 is encoded ~300 bp upstream of the LAT transcription start site, and it is transcribed in the same direction as LAT. miR-H1 is abundantly expressed as a late gene during productive infection [25, 30, 31]. It has also been detected at low levels in human ganglia by stem-loop RT-PCR and in latently infected mouse ganglia as sequence reads [30, 31]. Thus, it cannot be ruled out that miR-H1 is expressed as part of the latent transcription program. However, it is expressed less abundantly than a number of other miRNAs that are found in latently infected ganglia, and less abundantly during latency than during productive infection [30, 85]. Accordingly, it is possible that this miRNA is expressed less consistently during latency at low levels like various other productive cycle transcripts [76, 87-91]. The pri-miRNA for miR-H1 is not known, but there are a few reports of transcripts that might give rise to this miRNA. One possible candidate is a 1.8kb polyadenylated, low abundance transcript expressed with late kinetics [92, 93], and another is a 0.7 kb relatively abundant polyadenylated transcript that has been reported to encode a protein termed UOL (upstream of LAT) [94, 95]. However, the 0.7kb transcript has been shown to be specific for strain 17 and McKrae, and absent in strain KOS, a strain that expresses miR-H1 (as does strain 17).
Also encoded upstream of the LAT transcription start, but transcribed in the opposite direction are encoded miR-H6 and HSV-1 miR-H15 [26, 30, 31, 96]. miR-H6 is expressed relatively abundantly in productive infection, but is also reproducibly found relatively abundantly in latently infected neurons [26, 30, 31, 96]. Thus, miR-H6 arises from a pri-miRNA other than any previously known LAT that is expressed in latency. Perng et al. had described a transcript named AL-RNA that is expressed antisense to the 5’ end of the primary LAT, and has been reported to encode a protein [97]. However, the reported 3' end of this transcript precludes its containing miR-H6. A possible candidate is a 1.8 kb polyadenylated transcript named TAL (transcript antisense to LAT) detected in latently infected mouse and rabbit neurons (David Bloom, personal communication). This transcript starts within the 5’ exon of the primary LAT, and spans the LAT promoter and miR-H6 locus, suggesting that a promoter other than the LAT promoter is active during latency. Consistent with this hypothesis, Tang et al. reported that expression of HSV-2 miR-H6 is greatly reduced in productively infected cells in culture and in ganglia acutely or latently infected with a virus containing a 624-bp deletion spanning the LAT promoter and a part of exon 1 [96]. Alternatively, the LAT promoter might mediate bi-directional transcription, like many eukaryotic promoters (reviewed in [98]). Consistent with this possibility, a 200 bp deletion that removes the LAT promoter and only a few bases of exon 1 strongly reduces expression of HSV-1 miR-H6 in latently infected mouse ganglia [85]. (A larger LAT deletion does not affect HSV-1 miR-H6 expression in productively infected cells or acutely infected ganglia.) However, the 200 bp deletion might decrease miR-H6 expression by affecting expression of TAL.
HSV-1 miR-H15 is encoded ~800 bp downstream from –H6, and might be derived from the same precursor. However, so far, it has been detected only in productively infected cells.
As mentioned above, certain miRNAs are contained within known HSV transcripts. Three HSV-1 miRNAs, -H14, -H16 and -H17, could be derived from mRNAs for viral proteins ICP0, UL33 (a L protein), and ICP4 respectively. MiR-H14 and -16 have been detected in productive infection only, whereas –H17 has been found in both stages of virus infection [30]. Detection of miR-H17 in latency, in a relatively small number of sequence reads, is consistent with the sporadic detection of ICP4 transcripts at low abundance in latent infections; however, that this miRNA is an authentic latent miRNA cannot be excluded [30, 89]. Two HSV-2 miRNAs, -H19 and –H20, encoded within RL, could derive from the 5’UTR of ICP34.5 mRNA and the first intron of ICP0 primary transcripts, respectively. HSV-2 miR-H21 and –H23 are embedded in coding sequence of the mRNAs for UL23 (thymidine kinase) and UL42 (processivity subunit of DNA polymerase), respectively, and perhaps are derived from these mRNAs. Two other miRNAs encoded within UL, -H10 and –H22, could be processed from the L UL15 and UL33 to -35 read-through transcripts, respectively. HSV-2 miR-H19 – -H23 have been detected only in productively infected cells. HSV-2 miR-H10 has been detected in latently infected human ganglia, but, interestingly, not mouse ganglia, and only as sequence reads [30, 32].
A group of conserved miRNAs, miR-H11, -H12 and H13, which are expressed during productive infection, are encoded within ori sequences in UL (oriL; miR-H11) and in RS (oriS; miR-H12 and -H13) [30]. These miRNAs are notable for several reasons. First, oriL is a long palindromic sequence. A hairpin structure corresponding to HSV-1 oriL would not have any internal bulges (regions with non-complementary bases) while the corresponding HSV-2 hairpin would have only a single nucleotide mismatch, despite Drosha being thought to only recognize pri-miRNAs with bulges [6, 99, 100]. Perhaps Drosha can recognize these hairpins; alternatively, miR-H11 may arise from alternative rather than canonical miRNA biogenesis. Second, the sequences encoding miR-H11 are entirely embedded within the oriL palindrome; i.e. there are two copies of these sequences. The transcriptional direction of the pri-miRNA has not been determined, thus it is possible that both copies of miR-H11 are expressed. In contrast to – H11, miR-H12 and –H13 are derived from the opposite strands of oriS. Like oriL, oriS is palindromic, but the palindrome is shorter. The coding sequences for -H12 and -H13 extend outside of the palindromic region, which distinguishes the two miRNAs from each other. HSV-1 has one and HSV-2 two copies of oriS in each RS, which adds further to the complexity of these miRNAs. A few transcripts that span oriL or oriS have been reported previously [101-103], but these have not yet been investigated in the context of miRNAs. HSV-1 miR-H18, which has been found in a small number of sequence reads from latently infected ganglia, is encoded upstream of miR-H13 [30]. These two miRNAs could share the same precursor, although exclusive detection of –H13 in productive and –H18 in latent infection might suggest otherwise. Similarly, HSV-2 miR-H25 is encoded downstream and close to -H13, and might arise from the same transcript [30]. Detection of miR-H25 only in productive infection, analogous to –H13, is consistent with that hypothesis. Interestingly, another virus, BoHV-1, has been also found to encode one miRNA proximal to and another within its oriS palindrome (see 8.1 below) [33].
5.2. Roles of HSV miRNAs
At this writing, there is relatively little known about the targets of HSV miRNAs (Table 1), and even less about how miRNA-target interactions impact productive and latent infections. In other systems, miRNA target identification has combined computational predictions with experimental analysis, sometimes in a high throughput format, of the effects of miRNAs on the levels of transcripts and proteins and the presence of mRNAs in complexes containing Argonaute when the miRNA is overexpressed. Target prediction algorithms usually rely on the miRNA-target mRNA complementarity (putting highest weight on the seed pairing), features of the surrounding sequence, and evolutionary conservation of the binding site [17, 104-107]. Although evolutionary conservation is a very useful parameter for identifying miRNA targets in metazoans, it might be irrelevant or even contraindicated for identification of targets of viral miRNAs, since most herpesviruses are highly species specific and their miRNAs are poorly conserved, even between closely related viruses [108]. Furthermore, because miRNAs are relatively short and an even shorter sequence engages in base-pairing with target mRNAs, and because virus miRNAs can regulate host and/or viral targets, the large number of possible targets of viral miRNAs is challenging to verify experimentally. Additionally, mammalian herpesvirus transcripts are not always well defined, especially at late times of infection when deregulation of transcription, splicing, and polyadenylation occurs, which further confounds target prediction. Thus far, most work on targets of HSV miRNAs has focused on viral miRNAs. Regardless, it is early days, and it is highly likely that computational and experimental approaches to the identification of both viral and cellular targets will bear fruit soon.
5.2.1 HSV miRNAs that are complementary to or within viral mRNAs
Regardless of these challenges, several HSV miRNAs are encoded antisense to and are thus fully complementary to known viral transcripts, which are likely targets of these miRNAs. HSV miR-H2, H7, HSV-1 miR-H8 and HSV-2 miR-H9 are all complementary to ICP0 transcripts (Fig. 1). MiR-H2 (discussed further below) and HSV-2 miR-H9 are complementary to the coding sequence of ICP0 mRNA, and for these miRNAs, an obvious hypothesis is that they reduce ICP0 expression. It is less straightforward for miR-H7 and HSV-1 miR-H8, which are encoded antisense to an ICP0 intron, as miRNAs function predominantly in the cytoplasm [16, 109]. Unspliced ICP0 transcripts have been detected in latently infected ganglia [87]. Thus, it is possible that miR-H7 and HSV-1 miR-H8 could destabilize and prevent translation of such transcripts. Alternatively these miRNAs might function in the nucleus, which is not without precedent ([110-112]. Similarly, HSV miR-H3 and -H4 (discussed further below) and HSV-2 miR-H24 are complementary to ICP34.5 mRNA; HSV-1 miR-H22 is complementary to UL32 mRNA, which encodes an essential L protein involved in cleavage/packaging of nascent DNA into virions; and HSV-2 miR-H22 is complementary to UL36 mRNA, which encodes an essential L tegument protein with multiple functions.
Interestingly, HSV expresses several complementary pairs of miRNAs (Fig. 1), which are transcribed from palindrome-like sequences [26, 30]. These are HSV-1 miR-H1 and -H6, HSV-1 miR-H2 and -H14, HSV-2 miR-H4 and -H19, and HSV-2 miR-H7 and -H20. Although this phenomenon has been observed for Drosophila, human, and other virus miRNAs [113, 114], the biological significance of such pairs remains largely unknown. It has been suggested that miRNA:miRNA duplexes might have a regulatory role [115] where each miRNA in the pair would have a role in repression of the transcripts encoding the complementary miRNA, or where each miRNA might block the functions of its complementary miRNA.
As has been found with other viruses [116] and discussed above in 5.1, some HSV miRNAs are encoded within mRNAs encoding proteins (Figure 1), and it is possible that processing of these miRNAs would destabilize those mRNAs and reduce their abundance and the abundance of the proteins they encode. Alternatively, the pri-miRNAs for these miRNAs might be distinct from the protein coding mRNAs.
5.2.2 Experimental evidence for targets of HSV miRNAs
To date, there is experimental evidence for roles of only a few HSV miRNAs. Most of the published data derive from studies in which miRNA mimics or plasmids expressing miRNAs or short hairpin (sh) RNAs that mimic miRNAs are co-transfected into cells with plasmids encoding candidate target mRNAs, and the levels of the mRNAs and/or their protein targets are assayed. For several HSV miRNAs, the candidate target mRNAs are completely complementary to the miRNAs, and would thus be expected to act like short interfering (si) RNAs.
As discussed above, miR-H2 is complementary to ICP0 mRNA (see Fig. 1). In transfection studies, HSV-2 miR-H2 (also called miR-III) was reported to reduce levels of both ICP0 mRNA and ICP0 protein, as expected for an siRNA-like mechanism, and this reduction was relieved by co-transfection of an oligonucleotide complementary to miR-H2, but not by an oligonucleotide complementary to miR-H3 [28]. In contrast, it has been reported that HSV-1 miR-H2 interferes only with the expression of ICP0 protein and does not affect the levels of ICP0 mRNA [26]. In this case, a mutant form of miR-H2 did not reduce ICP0 expression, unless the ICP0 gene was mutated to be complementary to the mutant miR-H2. Further investigation of the differences in the two reports, which have interesting mechanistic implications, is warranted.
Regardless, it is not yet clear whether miR-H2 downregulates ICP0 expression in HSV infected cells. In productively infected cells, ICP0 is expressed with IE kinetics, and its expression slightly decreases at later times, which correlates with when expression of miR-H2 peaks [28, 31, 85]. However, ICP0 mRNA and ICP0 protein are both expressed highly abundantly during productive infection, making it less likely that miR-H2, which is relatively non-abundant in productive infection, could have a role in regulation of ICP0 in that setting. On the other hand, in latently infected ganglia, miR-H2 is relatively abundantly expressed and, in that setting, it seems more likely to downregulate the expression of ICP0, which is expressed very weakly during latency [87, 117]. Given the roles of ICP0 in promoting viral gene expression and combating host defenses, miR-H2 seems to be an excellent candidate to tilt the productive-latent balance in sensory neurons towards latency. Indeed, this scenario could help explain how the LAT locus represses productive cycle gene expression in latently infected ganglia [76]. It is also possible that miR-H2 exerts effects during establishment of latency, when the LAT locus also represses productive cycle gene expression [78], or during reactivation from latency. However, this scenario needs to be tested by, for example, the analysis of mutants in which miR-H2 expression is ablated without affecting ICP0 coding integrity.
HSV miR-H3 and miR-H4 are complementary to the coding and 5’UTR sequence of ICP34.5 mRNA, respectively [26, 27]. Tang at al. showed that HSV-2 miR-H3 and –H4 (also called miR-I and miR-II, respectively) can each down-regulate ICP34.5 protein in co-transfection experiments, and these reductions were relieved by co-transfection of oligonucleotides complementary to each miRNA [27, 28]. Similar results have been obtained with HSV-1 miR-H3 and -H4 with mutant forms of miRNAs and target providing evidence for selectivity (I Jurak, Mayuri, D.M. Coen unpublished). As is the case for miR-H2 (see above), it remains unclear whether these miRNAs downregulate ICP34.5 expression in productively infected cells. Both miRNAs are found abundantly expressed in HSV latently infected sensory ganglia [26-28, 30, 32]. Thus, it is possible that these miRNAs target ICP34.5 during the establishment, maintenance, and/or reactivation phases of latency, and thus it is possible that suppressing ICP34.5 expression plays a role in one or more phases of latency. However, compared with the effects expected from suppressing an IE regulatory protein such as ICP0, it is less clear how suppressing ICP34.5 would tilt the latency-productive infection balance. One would not expect expression of an L protein like ICP34.5 to be a crucial early step for an miRNA to counter during establishment or reactivation. However, as a leaky L protein, ICP34.5 is expressed relatively early. Moreover, the kinetics of ICP34.5 expression during reactivation are not understood. Perhaps, during acute infection in neurons, miR-H3 and H4 control expression of ICP34.5 to limit virus replication and spread, so that a higher fraction of infected cells would go on to enter latency. Moreover, expression of ICP34.5 in latently infected cells might result in inhibition of antiviral signaling triggered by the adaptive or innate immune system, or by the virus itself, and thus elicit virus reactivation. Viruses deficient for ICP34.5 are highly attenuated for replication and pathogenesis following intracranial inoculation, and severely attenuated for replication in neurons (as well as other cell types), and they reactivate poorly from latency, which would be consistent with the above mechanisms [118-120]. Virus mutants that fail to express functional miR-H3 and/or –H4 while retaining expression of functional ICP34.5 are necessary tools to address the roles of these miRNAs in HSV infection.
HSV miR-H6 is expressed relatively abundantly during productive infection and during latency [26, 30, 96]. HSV-1 miR-H6 has extended seed sequence complementarity to the mRNA for the IE protein ICP4 [26]. An HSV-1 miR-H6 mimic, but not a miR-H2 mimic, repressed ICP4 protein expression in a co-transfection experiment, and this repression depended on the sequence of the predicted target site in the ICP4 mRNA. Given the importance of ICP4 for viral gene expression and replication during productive infection, the repressive activity of HSV-1 miR-H6 could be expected to be important for establishment and maintenance of latency. Also, given that LAT mutations reduce HSV-1 miR-H6 expression [85], this could help explain the repressive effect of the LAT locus on productive cycle gene expression during acute and latent infections of ganglia [76, 78]. No mutant in which HSV-1 miR-H6 has been specifically affected has yet been studied, so this hypothesis remains unproven.
HSV-2 does not express a positional homolog of HSV-1 miR-H1. Instead, it expresses both strands of a positional homolog of miR-H6, but with rather different proportions of the 5' (5p) strand versus the 3' (3p) strand of the miR-H6 duplex when compared to HSV-1. During productive or latent infections with HSV-1, the 3p strand is more abundant [26, 30], whereas in HSV-2, the 5p strand is more abundant [26]. It is possible that small differences in the otherwise well conserved pre-miR-H6 hairpin alter its thermodynamics, which leads to preferential retention of the opposite strands of the duplex [121, 122]. Regardless, the result is that the seed sequence of HSV-2 miR-H6 (on its more abundant 5p strand) does not share sequence homology with the seed sequence of HSV-1 miR-H6 (on its more abundant 3p strand), but rather is identical to the seed sequence of HSV-1 miR-H1. It thus probably regulates the same genes as HSV-1 miR-H1. The targets of HSV-1 miR-H1 and HSV-2 miR-H6 (5p) remain to be determined.
Despite its lack of seed conservation with HSV-1 miR-H6, HSV-2 miR-H6 (3p) does have complementarity with HSV-2 ICP4 mRNA in segments that do not correspond to the HSV-1 miR-H6 target sequence on that mRNA [30]. It is not unusual for such star strand miRNAs to be incorporated into RISC and exert regulatory function [123] . However, despite the complementarity of HSV-2 ICP4 mRNA with the HSV-2 miR-H6 3p, Tang et al. were unable to detect repression of ICP4 expressed from the virus in cells that were transfected with HSV-2 miR-H6 expressing plasmid prior to infection, nor in co-transfection experiments [96]. These authors also studied an HSV-2 mutant with greatly reduced (~100-fold in both cell culture and latently infected ganglia) miR-H6 expression due to insertion of a polyadenylation signal upstream of miR-H6 coding sequences [96]. This mutant replicated similarly to wild type virus in cultured cells, and did not exhibit any differences in several parameters of latent infections of mice and guinea pigs that were examined. However, the mutation did result in attenuation of neurological symptoms such as paralysis during the acute phase of infection of guinea pigs. Whether the attenuation phenotype is due to the effects of the polyadenylation signal on miR-H6 expression remains unclear. Regardless, it seems clear that HSV-2 can establish latency and reactivate from it even when miR-H6 expression is drastically reduced. Thus, the target(s) and role(s) of HSV-2 miR-H6 remain to be established.
Although there remains much work to do to analyze the roles of individual miRNAs and their targets during HSV infection, mutations that ablate LAT expression -- and thus the expression of multiple miRNAs -- have been examined. Such mutations do exert effects on viral gene expression and chromatin structure, as well as biological phenotypes such as reactivation from latency (e.g. [76-81]). Thus, it is possible that one or more miRNAs are important for these phenotypes. Nevertheless, at least in animal models, all LAT mutants examined to date can establish and maintain latency. Thus, either these viral miRNAs are not crucial for latency in these models or the loss of certain miRNAs is compensated by the loss of others; e.g., one miRNA might be important for maintaining latency while another might be important for promoting reactivation.
5.3. Interactions of HSV with Host miRNAs and miRNA Machinery
Based on work with other viruses, including certain herpesviruses [124-139], four related questions arise: 1) Can certain host miRNAs mediate antiviral defense against HSV? 2) Can the virus benefit from expression of other host miRNAs? 3) Does the virus suppress or enhance the host miRNA machinery? 4) Does the virus suppress or enhance the expression or action of particular miRNAs. At present we have only limited information that addresses these questions. Using hybridization of an miRNA array followed by dot blot hybridization, Hill et al. found a ~4-fold increase in signal corresponding to miR-146a, coupled to a decrease in one of its targets, complement factor H, in HSV-1 infected human primary neuronal cells [140]. These authors suggested a role for induction of miR-146a in evasion of HSV-1 from the complement system; i.e., that the virus benefits from expression of a host miRNA. Wu et al. [141] found that RNA interference (RNAi) was less efficient in cells productively infected with HSV-1 and co-transfected with plasmid expressing GFP and shRNAs targeting GFP, than in mock infected cells. These results support the possibility that HSV-1 might suppress the miRNA machinery. Additional evidence that might support this notion is that HSV-1 pre-miRNAs accumulate at high levels relative to mature miRNAs [25, 28, 30, 85]. One possible explanation for this phenomenon is that virus encodes a suppressor, protein or RNA, to inhibit miRNA biogenesis or function. Other explanations would be that the pre-miRNAs are poor substrates for Exportin 5/RAN-GTP or Dicer, that the miRNAs are unstable, or that rapid accumulation of viral hairpin-structured transcripts (pri-, pre-miRNAs, and non-miRNA transcripts) and miRNAs saturate the miRNA machinery. Wu et al. also found that depletion of Argonaute 2 from the cells led to modest burst of virus replication [141]. This might suggest that HSV-1 suppression of the miRNA machinery could benefit the virus, which would be consistent with one or more host miRNAs mediating antiviral defense against HSV (among other possibilities). If true, then HSV might both encode miRNAs and induce host miRNAs that are beneficial to its replication, while simultaneously inhibiting miRNA biogenesis and function. To address this paradox, Wu et al. suggest that a dynamic equilibrium of these processes is required for optimal replication [141]. Clearly, more investigation is required to address these issues in both productive and latent infection.
In summary, although we know more about the miRNAs of HSV than we do about those of other mammalian alphaherpesviruses, we have still just scratched the surface.
6. Herpes B Virus
Herpes B virus (Macacine herpesvirus 1, BV) is a simplexvirus that is enzootic to macaque monkeys. In its natural host BV causes a self-limiting disease similar to HSV disease in humans [142]. BV is highly prevalent in most macaque colonies, including those colonies used for biomedical research. Unique among herpesviruses, BV can zoonotically infect humans to cause a serious disease; typically causing encephalitis that is 80% fatal if untreated [1]. Even with timely antiviral therapy, 20% of those infected die [143]. As a result, working with BV requires Biosafety Level 4 containment. The ability of BV to access the human nervous system, and even establish a latent infection in humans [144, 145], suggests that there may be similarities between the molecular mechanisms used by BV and HSV to interact with the nervous system. The BV genome has been completely sequenced [146]. BV is closely related to HSV and homologous proteins have up to 87% amino acid identity. The overall genome architecture of BV is very similar to that of HSV (Fig. 1), although there are some important differences between the genomes. There are a few genes that are present in HSV, but absent in BV: UL20.5, UL43.5, and UL27.5, and, especially germane here, ICP34.5, which is discussed above. BV does encode a US11 homolog that may compensate for the lack of ICP34.5. BV has one predicted gene that is missing in HSV (UL53A; see below), but there have been no reports demonstrating that a protein is generated from this gene. BV has been reported to have six origins of replication, as it has duplicates of oriS and oriL at each locus. The portions of the BV genome that exhibit the greatest differences with those in HSV are the repeat regions, which is particularly interesting given that most HSV-encoded miRNAs are generated from the repeats. Specifically, BV RL diverges from HSV RL around the flanks of the ICP0 open reading frame, and RS is approximately 1.5 kb longer in BV than in HSV.
6.1. Herpes B Virus miRNAs
Using a combination of computational prediction and experimental validation using Northern blot analysis of short RNAs from infected cells and from cells transfected with plasmids encoding artificial pri-miRNAs, three BV-encoded microRNAs have been reported [29]. None of these miRNAs has homology to any known miRNAs. Two of the BV miRNAs are encoded within the UL region of the genome and one within RL. The two miRNAs encoded by the UL region have interesting potential antisense targets. Hbv-miR-B2RC is fully complementary to a sequence that is ~100 nt upstream of the initiating AUG of the UL30 gene, which encodes the catalytic subunit of the DNA polymerase. Therefore, this miRNA may regulate the expression of BV UL30 in a manner similar to how Epstein Barr virus (EBV) encoded miR-BART2 regulates the EBV DNA polymerase [147]. Hbv-miR-B4 is fully complementary to the putative open reading frame encoded by UL53A, which is a two-exon gene that was identified by statistical analysis of the BV genomic sequence [146]. This is especially interesting as this locus contains one of the few substantial differences between the BV and HSV genomes, and these regions are good candidates to be important for the differences between BV and HSV pathogenesis. Similarly, hbv-miR-B20 is also generated from a region that exhibits a substantial difference between BV and HSV. This miRNA is antisense to a region that based on its position would encode the ICP34.5 protein in HSV, which has been implicated in neurovirulence [119]. Perhaps paradoxically, BV lacks a homolog of ICP34.5 despite exhibiting greater neurovirulence in humans [146]. There are thought to be BV-encoded transcripts antisense to hbv-miR-B20 [29], although more work is required to determine their function and if they are targets of hbv-miR-B20.
Interestingly, the BV miRNAs discovered thus far do not seem to be less abundant than their pre-miRNAs [29], which differs from most of the miRNAs detectable in HSV-infected cells (see 5.3 above).
From studies of HSV it is clear that computational prediction followed by Northern blot hybridization is inadequate to determine the complete complement of virus-encoded miRNAs [25-28, 30-32]. Thus, more sensitive methods are likely required to determine the complete catalog of BV-encoded miRNAs. These studies should include analyses of infected cells in culture, and neuronal tissues from naturally infected macaques.
7. The Prototype Varicellovirus, Varicella-zoster virus
The prototype varicellovirus is varicella-zoster virus (VZV). VZV is a human pathogen that causes chickenpox (varicella) and shingles (herpes zoster). VZV is highly contagious, and typically infects respiratory mucosa and then quickly spreads to lymphocytes, causing viremia. Subsequently, virus is transported to the skin where it induces the well-recognized rash of chickenpox. These aspects of infection differ greatly from those of HSV and the other simplexviruses. Following these stages of infection, VZV then establishes latency in sensory ganglia, much like the simplexviruses. VZV can reactivate from latency to cause shingles, an illness that is very common in the elderly whose immunity to VZV has waned. While HSV exhibits a broad host/cell-type range in experimental settings but limited tissue tropism during natural infections of humans, VZV has a highly restricted host/cell-type range in experimental settings, but a wide range of tissue tropisms during natural infections, including for T lymphocytes [4].
Despite the close relationship of VZV to HSV, there are various molecular differences [5], some of which likely account for the biological differences described above. Germane to miRNAs, the genome arrangement of VZV is similar to that of HSV, however the RL repeats flanking UL are very short (i.e. ~88 bp) compared to those of HSV (~9.2 kb; Fig. 1). Many genes are highly conserved between VZV and HSV, but each virus has a set of genes not present in the other. LAT, ICP34.5, UL45, UL56, US2, US4, US5, US6, US11 and US12 are unique to HSV, whereas ORF1, ORF2, ORF13, ORF32 and ORF57 are unique to VZV. Also, VZV does not have an oriL, but, like HSV, has two copies of oriS, one in each repeat flanking the US region[4, 5]. In addition, VZV has a low G+C content, ~46%, compared to that of HSV, which is ~68%.
During VZV latent infection, as with HSV, productive cycle gene expression is restricted. However VZV does not possess a homolog of the LAT locus, and latent virus expresses at least five proteins (ORF4, ORF21, ORF29, ORF62 and ORF63) relatively abundantly. Interestingly, during latency, these proteins localize to the cytoplasm, whereas during reactivation and productive infection, they are recruited to the nucleus [4, 148]. Additionally, whereas HSV is relatively insensitive to interferons and efficiently counteract their action by several mechanisms, including inhibition of eIF2α phosphorylation by US11 and ICP34.5 (as described above in 4), VZV is very sensitive to interferons, which may relate to the absence of homologs of ICP34.5 and US11.
7.1. Does VZV express miRNAs?
Sequencing of trigeminal ganglia from three patients who were latently infected with VZV did not reveal any miRNAs encoded by this virus, although VZV DNA was readily detectable in the same samples, and HSV miRNAs were detected in ganglia from two of the three individuals [31]. These data tend to suggest that VZV does not express any miRNAs during latency. Consistent with this possibility, it was reported as unpublished data that sequencing of trigeminal ganglia from rhesus macaques latently infected with the closely related simian varicella virus (SVV, or Cercopithecine herpesvirus 9) also did not yield any miRNAs encoded by this virus [31]. As mentioned above, VZV lacks the LAT locus, which encodes most HSV miRNAs expressed during latency, but expresses several proteins during latency. Perhaps those proteins serve the regulatory functions that miRNAs otherwise would perform. Interestingly, both VZV and SVV have a very low G+C content, 46% and 40% respectively, which might, in general, influence secondary structures of transcripts, i.e. fewer and less stable pri-miRNA hairpins would be predicted [149].
Nevertheless, it should be noted that there was less VZV DNA than HSV DNA in the human ganglia tested for VZV miRNAs, yet certain HSV miRNAs were detected in only a few sequence reads. Thus, it is possible that VZV does express miRNAs during latency, but that they were below the level of detection in the assay. Moreover, there have been no reports, to our knowledge, addressing whether VZV expresses any miRNAs during productive infection. Indeed, the varicelloviruses bovine herpesvirus-1 (BoHV-1) and Pseudorabies virus (PRV) express miRNAs during productive infection (see below) [33-35]. Thus, whether VZV expresses miRNAs remains an open question.
8. Bovine Herpesvirus 1
BoHV-1 is a varicellovirus that causes economically important diseases in cattle. BoHV-1 is best known for causing a respiratory disease known as infectious rhinotracheitis. BoHV-1 can also cause conjunctivitis, pneumonia, genital disease and abortions, and it is thought to cost the United States cattle industry over $500 million per year [150-152]. BoHV-1 establishes a latent infection in trigeminal ganglia, and latency is characterized by restricted gene expression [153]; the only RNA species expressed abundantly during latency are known as the latency-related RNA (LR-RNA) or latency associated transcripts (LAT). Its genome structure is very similar to that of VZV. The LR-RNAs arise from UL sequences (Fig. 1)
8.1. BoHV-1 miRNAs
There have been two reports regarding BoHV-1 miRNAs [33, 34]. Glazov et al. used deep sequencing of RNA isolated from productively infected cells and validation by qRT-PCR and Northern blot hybridization to detect 11 mature miRNAs and one miRNA star strand. Four of the BoHV-1-encoded miRNAs map to the UL region of the genome and one maps to the unique short (US) region [33]; to date, this is the only report of a miRNA being encoded by the US of a mammalian alphaherpesvirus. All of the miRNAs encoded in the unique regions were either fully complementary to viral transcripts (UL16 and UL30) or derived from known transcripts (UL19 and UL21). Of these, the antisense relationship of BoHV-1 mir-B1 to UL30, which encodes the catalytic subunit of the viral DNA polymerase, is similar to that seen in BV (see 6.1 above). As hypothesized for BV [29], this may suggest viral miRNA-dependent regulation of DNA polymerase expression, as reported for EBV [147]. Bhv1-miR-B6-1 and B7 are, respectively, and encoded antisense to and within intron 3 of the BoHV-1 ICP0 gene, whose product is called bICP0. They are also complementary to each other. This arrangement is akin to the arrangement of HSV-2 miR-H7 and H20 (Fig. 1). Interestingly, BoHV-1 miR-B5 is encoded within transcripts encoding bICP0, and is antisense to the 3' end of BoHV-1 LR-RNA). Whether BoHV-1 miRNAs downregulate the expression of any of these gene products, remains to be seen. BoHV-1 mir-B9 is located within oriS of BoHV-1, while BoHV-1 miR-B8 is very close by; this arrangement is similar to those in HSV-1 and HSV-2 (Fig. 1). Further work will be required to determine the function of these miRNAs, but their locations suggest functions that are conserved with HSV-1 and HSV-2.
In addition, Glazov et al. [33] investigated the evolutionary conservation of the BoHV-1 miRNAs throughout the Varicellovirus genus, searching for highly similar sequences in the genomes of bovine herpesvirus 5 (BoHV-5), equid herpesvirus 1, equid herpesvirus 4, PRV, and VZV. Of the five viruses tested, BoHV-5, which is most closely related to BoHV-1, contained sequences with sufficient identity to the BoHV-1 miRNAs for the authors to report homology. In the BoHV-5 genome, nine loci showed high conservation in sequence (73 to 97% identity) and genomic location with BoHV-1 miR-B1 – B9, leading to the prediction that BoHV-5 encodes orthologs of these miRNAs (Table 1). Interestingly, no homolog of the BoHV-1 miRNA encoded within US, miR-B10, was reported. Experimental validation that these miRNAs are expressed from BoHV-5 is pending.
In their Northern blot analyses, Glazov et al. [33] found that several miRNAs were substantially less abundant than their corresponding pre-miRNAs, similar to what is observed with HSV. However, they also found some miRNAs that were substantially more abundant than their corresponding pre-miRNAs. Thus, there does not seem to be any global suppression of miRNA biogenesis in BoHV-1 infected cells.
Jaber et al. [34] transfected a short region of the viral genome from the LR locus, and found that a number of small non-coding RNAs (as the authors were careful to describe them) were expressed. At least one of these small non-coding RNAs repressed bICP0 expression and, modestly, BoHV-1 production in co-transfection assays. These small non-coding RNAs were larger than miRNAs, but the authors predicted that miRNAs could be derived from them, which they named 1 and 2 (Fig. 1). The predicted miRNAs (delivered as siRNAs) also repressed bICP0 expression and, modestly, BoHV-1 production in co-transfection assays. It is not clear how these predicted miRNAs function. Although they do have some complementarity with ICP0 mRNA, they lack seed complementarity, and no mutant forms of the miRNAs or the target were tested to assess the mechanism of repression. Of note, Northern blot hybridization, using a relatively large fragment that could express the small non-coding RNAs as probe, detected ~miRNA size species in latently infected ganglia whose abundance decreased upon a reactivation stimulus. It will be interesting to learn whether deep sequencing analyses of ganglia latently infected with BoHV-1 detect an authentic miRNA from this region of the genome.
9. Pseudorabies Virus
First described in 1902 by Aujeszky [154], pseudorabies (Aujeszky's disease, mad itch, infectious bulbar paralysis) is an economically important disease of domestic swine, although it has been documented in many other domestic and wildlife species [155]. The causative agent is the varicellovirus, pseudorabies virus (PRV, Suid herpesvirus 1). The virus typically infects via the oro-nasal route and is responsible for a variety of symptoms including abortion and meningoencephalitis. Lethality is related to the strain of the virus and the age of the pig; piglets are more susceptible to a fatal infection than older animals [155]. PRV establishes a latent infection in trigeminal ganglia, and latency is characterized by restricted gene expression; the only RNA species abundantly expressed during latency is the large latency transcript (LLT), which arises from UL (Fig. 1; [156]). The PRV genome has no RL (Fig. 1).
9.1. PRV miRNAs
Using deep sequencing of RNA harvested from PRV-infected pig dendritic cells, five PRV-encoded miRNAs have been reported [35]. Although the miRNAs were detected from productively infected cells, they all mapped to the LLT region of the genome. This would be consistent with expression of these miRNAs during latency, but that has yet to be determined. None of the miRNAs is fully complementary to known viral mRNAs. These miRNAs do not appear to have homologous sequences in VZV, SVV, or BoHV-1 (A.G., unpublished). Using a combination of algorithms, several PRV genes were predicted to be targets of the miRNAs. Interestingly, the predicted targets included the viral genes encoding EP0 and IE180. These two proteins are homologous to HSV ICP0 and ICP4, respectively, which can be repressed by HSV-1 miR-H2 and H6, respectively. Although further work is necessary to confirm action on these PRV genes, these data strengthen the hypothesis that certain miRNAs encoded by different mammalian alphaherpesvirus may share common targets.
10. Concluding Comments
A major conclusion from this review of mammalian alphaherpesvirus miRNAs is that we know very little about them. The miRNAs encoded by most of these viruses remain to be discovered. The targets for most of the miRNAs that have been discovered remain to be identified. The miRNA-target interactions that have been identified in transfection studies remain to be tested for importance during virus infection. We also know little about how these viruses interact with host miRNAs and the miRNA machinery.
Despite these major limitations, several themes emerge. First, in at least four of the mammalian alphaherpesviruses (HSV-1, HSV-2, BV, PRV, and possibly BoHV-1), certain miRNAs are encoded within transcription units active during latency and seem likely to play interesting roles in latent infection. Second, despite the well-deserved focus on latency, many miRNAs are expressed during productive infection and seem likely to play interesting roles there. Third, in at least three of the viruses, miRNAs are encoded within origins of replication. Fourth, among the viruses, a substantial number of miRNAs are encoded antisense to and are thus fully complementary to and likely to target viral mRNAs. In more than one virus, these mRNAs encode ICP0 or the DNA polymerase catalytic subunit. Fifth, in several cases, miRNAs are encoded antisense to each other. These common themes suggest that further comparative analyses of mammalian alphaherpesvirus miRNAs may be helpful in the discovery of the roles of these miRNAs during infection.
Highlights.
> We review the published literature on mammalian alphaherpesviruses and microRNAs.
> So far, five of these viruses are known to express microRNAs.
> The most is known about herpes simplex virus microRNAs.
> Some microRNAs are expressed abundantly during latent infections of sensory neurons.
> Transfection studies suggest some microRNAs might promote latency.
> Overall, we know little about microRNA targets and roles in virus infection.
Acknowledgments
We gratefully acknowledges funding support from NCRR (P51 RR013986 and R21 RR026287, AG), and NIAID (RO1 AI26126 and PO1 NS35138, IJ and DMC). We thank Martha Kramer for helpful discussions and David Bloom for communicating results ahead of publication.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Roizman B, Knipe DM, Whitley RJ. Herpes simplex viruses. In: Knipe DM, Howley PM, Griffin DE, Lamb RA, Straus SE, Martin MA, Roizman B, editors. Fields Virology. Fifth Edition Lippincott Williams & Wilkins; Philadelphia: 2007. pp. 2502–2601. [Google Scholar]
- 2.Davison AJ, Eberle R, Ehlers B, Hayward GS, McGeoch DJ, Minson AC, Pellett PE, Roizman B, Studdert MJ, Thiry E. The order Herpesvirales. Arch Virol. 2009;154:171–177. doi: 10.1007/s00705-008-0278-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McGeoch DJ, Dolan A, Ralph AC. Toward a comprehensive phylogeny for mammalian and avian herpesviruses. J Virol. 2000;74:10401–10406. doi: 10.1128/jvi.74.22.10401-10406.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mori I, Nishiyama Y. Herpes simplex virus and varicella-zoster virus: why do these human alphaherpesviruses behave so differently from one another? Rev Med Virol. 2005;15:393–406. doi: 10.1002/rmv.478. [DOI] [PubMed] [Google Scholar]
- 5.Cohen JI, Straus SE, Arvin AM. Varicella-Zoster virus replication, pathogenesis, and management. In: Knipe DM, Howley PM, Griffin DE, Lamb RA, Straus SE, Martin MA, Roizman B, editors. Fields Virology. Fifth Edition Lippincott Williams & Wilkins; Philadelphia: 2007. [Google Scholar]
- 6.Bartel DP. MicroRNAs: genomics, biogenesis mechanisms, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 7.Umbach JL, Cullen BR. The role of RNAi and microRNAs in animal virus replication and antiviral immunity. Genes Dev. 2009;23:1151–1164. doi: 10.1101/gad.1793309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee Y, Jeon K, Lee JT, Kim S, Kim VN. MicroRNA maturation: stepwise processing and subcellular localization. EMBO J. 2002;21:4663–4670. doi: 10.1093/emboj/cdf476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gregory RI, Yan K-P, Amuthan G, Chendrimada T, Doratotaj B, Cooch N, Shiekhattar R. The Microprocessor complex mediates the genesis of microRNAs. Nature. 2004;432:235–240. doi: 10.1038/nature03120. [DOI] [PubMed] [Google Scholar]
- 10.Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Radmark O, Kim S, Kim VN. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- 11.Yi R, Qin Y, Macara IG, Cullen BR. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 2003;17:3011–3016. doi: 10.1101/gad.1158803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hutvagner G, McLachlan J, Pasquinelli AE, Balint E, Tuschl T, Zamore PD. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science. 2001;293:834–838. doi: 10.1126/science.1062961. [DOI] [PubMed] [Google Scholar]
- 13.Elbashir SM, Lendeckel W, Tuschl T. RNA interference is mediated by 21- and 22- nucleotide RNAs. Genes Dev. 2001;15:188–200. doi: 10.1101/gad.862301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu J, Carmell MA, Rivas FV, Marsden CG, Thomson JM, Song JJ, Hammond SM, Joshua-Tor L, Hannon GJ. Argonaute2 is the catalytic engine of mammalian RNAi. Science. 2004;305:1437–1441. doi: 10.1126/science.1102513. [DOI] [PubMed] [Google Scholar]
- 15.Liu J, Rivas FV, Wohlschlegel J, Yates JR, 3rd, Parker R, Hannon GJ. A role for the P-body component GW182 in microRNA function. Nat. Cell Biol. 2005;7:1261–1266. doi: 10.1038/ncb1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 18.Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Selbach M, Schwanhausser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- 20.Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466:835–840. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Baek D, Villen J, Shin C, Camargo FD, Gygi SP, Bartel DP. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brennecke J, Stark A, Russell RB, Cohen SM. Principles of microRNA-target recognition. PLoS Biol. 2005;3:e85. doi: 10.1371/journal.pbio.0030085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McGeoch DJ, Cook S, Dolan A, Jamieson FE, Telford EA. Molecular phylogeny and evolutionary timescale for the family of mammalian herpesviruses. J Mol Biol. 1995;247:443–458. doi: 10.1006/jmbi.1995.0152. [DOI] [PubMed] [Google Scholar]
- 24.Pfeffer S, Sewer A, Lagos-Quintanta M, Sheridan R, Sander C, Grässer FA, van Dyk LF, Ho CK, Shuman S, Chien M, Russo JJ, Ju J, Randall G, Lindenbach BD, Rice CM, Simon V, Ho DD, Zavolan M, Tuschl T. Identification of microRNAs of the herpesvirus family. Nature Methods. 2005;2:269–276. doi: 10.1038/nmeth746. [DOI] [PubMed] [Google Scholar]
- 25.Cui C, Griffiths A, Li G, Silva LM, Kramer MF, Gaasterland T, Wang XJ, Coen DM. Prediction and identification of herpes simplex virus 1-encoded microRNAs. J Virol. 2006;80:5499–5508. doi: 10.1128/JVI.00200-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Umbach JL, Kramer MF, Jurak I, Karnowski HW, Coen DM, Cullen BR. MicroRNAs expressed by herpes simplex virus 1 during latent infection regulate viral mRNAs. Nature. 2008;454:780–783. doi: 10.1038/nature07103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tang S, Bertke AS, Patel A, Wang K, Cohen JI, Krause PR. An acutely and latently expressed herpes simplex virus 2 viral microRNA inhibits expression of ICP34.5, a viral neurovirulence factor. Proc Natl Acad Sci U S A. 2008;105:10931–10936. doi: 10.1073/pnas.0801845105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tang S, Patel A, Krause PR. Novel less-abundant viral microRNAs encoded by herpes simplex virus 2 latency-associated transcript and their roles in regulating ICP34.5 and ICP0 mRNAs. J Virol. 2009;83:1433–1442. doi: 10.1128/JVI.01723-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Besecker MI, Harden ME, Li G, Wang XJ, Griffiths A. Discovery of herpes B virus-encoded microRNAs. J Virol. 2009;83:3413–3416. doi: 10.1128/JVI.02419-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jurak I, Kramer MF, Mellor JC, van Lint AL, Roth FP, Knipe DM, Coen DM. Numerous conserved and divergent microRNAs expressed by herpes simplex viruses 1 and 2. J Virol. 2010;84:4659–4672. doi: 10.1128/JVI.02725-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Umbach JL, Nagel MA, Cohrs RJ, Gilden DH, Cullen BR. Analysis of human alphaherpesvirus microRNA expression in latently infected human trigeminal ganglia. J Virol. 2009;83:10677–10683. doi: 10.1128/JVI.01185-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Umbach JL, Wang K, Tang S, Krause PR, Mont EK, Cohen JI, Cullen BR. Identification of viral microRNAs expressed in human sacral ganglia latently infected with herpes simplex virus 2. J. Virol. 2010;84:1189–1192. doi: 10.1128/JVI.01712-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Glazov EA, Horwood PF, Assavalapsakul W, Kongsuwan K, Mitchell RW, Mitter N, Mahony TJ. Characterization of microRNAs encoded by the bovine herpesvirus 1 genome. J Gen Virol. 2010;91:32–41. doi: 10.1099/vir.0.014290-0. [DOI] [PubMed] [Google Scholar]
- 34.Jaber T, Workman A, Jones C. Small noncoding RNAs encoded within the bovine herpesvirus 1 latency-related gene can reduce steady-state levels of infected cell protein 0 (bICP0) J Virol. 2010;84:6297–6307. doi: 10.1128/JVI.02639-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Anselmo A, Flori L, Jaffrezic F, Rutigliano T, Cecere M, Cortes-Perez N, Lefevre F, Rogel-Gaillard C, Giuffra E. Co-Expression of Host and Viral MicroRNAs in Porcine Dendritic Cells Infected by the Pseudorabies Virus. PLoS One. 2011;6:e17374. doi: 10.1371/journal.pone.0017374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tobian AA, Quinn TC. Herpes simplex virus type 2 and syphilis infections with HIV: an evolving synergy in transmission and prevention. Curr Opin HIV AIDS. 2009;4:294–299. doi: 10.1097/COH.0b013e32832c1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kristie TM, Roizman B. Host cell proteins bind to the cis-acting site required for virion-mediated induction of herpes simplex virus 1 alpha genes. Proc Natl Acad Sci U S A. 1987;84:71–75. doi: 10.1073/pnas.84.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McKnight JL, Kristie TM, Roizman B. Binding of the virion protein mediating alpha gene induction in herpes simplex virus 1-infected cells to its cis site requires cellular proteins. Proc. Natl. Acad. Sci. USA. 1987;84:7061–7065. doi: 10.1073/pnas.84.20.7061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kristie TM, Roizman B. Differentiation and DNA contact points of host proteins binding at the cis site for virion-mediated induction of alpha genes of herpes simplex virus 1. J Virol. 1988;62:1145–1157. doi: 10.1128/jvi.62.4.1145-1157.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Clements JB, Watson RJ, Wilkie NM. Temporal regulation of herpes simplex virus type 1 transcription: location of transcripts on the viral genome. Cell. 1977;12:275–285. doi: 10.1016/0092-8674(77)90205-7. [DOI] [PubMed] [Google Scholar]
- 41.Preston CM. Control of herpes simplex virus type 1 mRNA synthesis in cells infected with wild type virus or the temperature sensitive mutant tsK. J. Virol. 1979;29:275–284. doi: 10.1128/jvi.29.1.275-284.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Watson RJ, Clements JB. A herpes simplex virus type 1 function continuously required for early and late virus RNA synthesis. Nature. 1980;285:329–330. doi: 10.1038/285329a0. [DOI] [PubMed] [Google Scholar]
- 43.Godowski PJ, Knipe DM. Identification of a herpes simplex virus function that represses late gene expression from parental viral genomes. J. Virol. 1985;55:357–365. doi: 10.1128/jvi.55.2.357-365.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Carrozza M, DeLuca N. Interactions of the viral activator protein ICP4 with TFIID through TAF250. Molec. Cell. Biol. 1996;16:3085–3093. doi: 10.1128/mcb.16.6.3085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Faber SW, Wilcox KW. Association of the herpes simplex virus regulatory protein ICP4 with specific nucleotide sequences in DNA. Nuclei Acids Res. 1986;14:6067–6083. doi: 10.1093/nar/14.15.6067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Everett RD. Trans activation of transcription by herpes virus products: requirement for two HSV-1 immediate-early polypeptides for maximum activity. EMBO J. 1984;3:3135–3141. doi: 10.1002/j.1460-2075.1984.tb02270.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nabel GJ, Rice SA, Knipe DM, Baltimore D. Alternative mechanisms for activation of human immunodeficiency virus enhancer in T cells. Science. 1988;239:1299–1302. doi: 10.1126/science.2830675. [DOI] [PubMed] [Google Scholar]
- 48.Gelman IH, Silverstein S. Identification of immediate early genes from herpes simplex virus that transactivate the virus thymidine kinase gene. Proc. Natl. Acad. Sci. USA. 1985;82:256–260. doi: 10.1073/pnas.82.16.5265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.O'Hare P, Hayward GS. Evidence for a direct role for both the 175,000- and 110,000- molecular-weight immediate-early proteins of herpes simplex virus in the transactivation of delayed-early promoters. J Virol. 1985;53:751–760. doi: 10.1128/jvi.53.3.751-760.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Quinlan MP, Knipe DM. Stimulation of expression of a herpes simplex virus DNA-binding protein by two viral functions. Mol. Cell. Biol. 1985;5:957–963. doi: 10.1128/mcb.5.5.957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hagglund R, Roizman B. Role of ICP0 in the strategy of conquest of the host cell by herpes simplex virus 1. J Virol. 2004;78:2169–2178. doi: 10.1128/JVI.78.5.2169-2178.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Roizman B, Gu H, Mandel G. The first 30 minutes in the life of a virus: unREST in the nucleus. Cell Cycle. 2005;4:1019–1021. doi: 10.4161/cc.4.8.1902. [DOI] [PubMed] [Google Scholar]
- 53.Gu H, Liang Y, Mandel G, Roizman B. Components of the REST/coREST/histone deacetylase repressor complex are disrupted, modified, and translocated in HSV-1-infected cells. Proc. Natl. Acad. Sci. USA. 2005;21:7571–7576. doi: 10.1073/pnas.0502658102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Everett RD, Maul GG. HSV-1 IE protein Vmw110 causes redistribution of PML. EMBO J. 1994;13:5062–5069. doi: 10.1002/j.1460-2075.1994.tb06835.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Maul GG, Guldner HH, Spivack JG. Modification of discrete nuclear domains induced by herpes simplex virus type 1 immediate early gene 1 product (ICP0) J Gen Virol. 1993;74(Pt 12):2679–2690. doi: 10.1099/0022-1317-74-12-2679. [DOI] [PubMed] [Google Scholar]
- 56.Everett RD, Freemont P, Saitoh H, Dasso M, Orr A, Kathoria M, Parkinson J. The disruption of ND10 during herpes simplex virus infection correlates with the Vmw110- and proteasome-dependent loss of several PML isoforms. J Virol. 1998;72:6581–6591. doi: 10.1128/jvi.72.8.6581-6591.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Melroe GT, Silva L, Schaffer PA, Knipe DM. Recruitment of activated IRF-3 and CBP/p300 to herpes simplex virus ICP0 nuclear foci: Potential role in blocking IFN-beta induction. Virology. 2007;360:305–321. doi: 10.1016/j.virol.2006.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Melroe GT, DeLuca NA, Knipe DM. Herpes simplex virus 1 has multiple mechanisms for blocking virus-induced interferon production. J Virol. 2004;78:8411–8420. doi: 10.1128/JVI.78.16.8411-8420.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mossman KL, Saffran HA, Smiley JR. Herpes simplex virus ICP0 mutants are hypersensitive to interferon. J Virol. 2000;74:2052–2056. doi: 10.1128/jvi.74.4.2052-2056.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Boutell C, Sadis S, Everett RD. Herpes simplex virus type 1 immediate-early protein ICP0 and its isolated RING finger domain act as ubiquitin E3 ligases in vitro. J. Virol. 2002;76:841–850. doi: 10.1128/JVI.76.2.841-850.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chou J, Roizman B. The gamma 1(34.5) gene of herpes simplex virus 1 precludes neuroblastoma cells from triggering total shutoff of protein synthesis characteristic of programed cell death in neuronal cells. Proc. Natl. Acad. Sci. USA. 1992;89:3266–3270. doi: 10.1073/pnas.89.8.3266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.He B, Gross M, Roizman B. The γ134.5 protein of herpes simplex virus 1 complexes with protein phosphatase 1a to dephosphorylate the a subunit of the eukaryotic initiation factor 2 and preclude the shutoff of protein synthesis by double-stranded RNA-activated protein kinase. Proc. Natl. Acad. Sci. USA. 1997;94:843–848. doi: 10.1073/pnas.94.3.843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jin H, Ma Y, Prabhakar BS, Feng Z, Valyi-Nagy T, Yan Z, Verpooten D, Zhang C, Cao Y, He B. The gamma 1 34.5 protein of herpes simplex virus 1 is required to interfere with dendritic cell maturation during productive infection. J Virol. 2009;83:4984–4994. doi: 10.1128/JVI.02535-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Verpooten D, Ma Y, Hou S, Yan Z, He B. Control of TANK-binding kinase 1-mediated signaling by the γ134.5 protein of herpes simplex virus 1. J. Biol. Chem. 2009;284:1097–1105. doi: 10.1074/jbc.M805905200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Orvedahl A, Alexander D, Talloczy Z, Sun Q, Wei Y, Zhang W, Burns D, Leib DA, Levine B. HSV-1 ICP34.5 confers neurovirulence by targeting the Beclin 1 autophagy protein. Cell Host Microbe. 2007;1:23–35. doi: 10.1016/j.chom.2006.12.001. [DOI] [PubMed] [Google Scholar]
- 66.Leib DA, Alexander DE, Cox D, Yin J, Ferguson TA. Interaction of ICP34.5 with Beclin 1 modulates herpes simplex virus type 1 pathogenesis through control of CD4+ T-cell responses. J Virol. 2009;83:12164–12171. doi: 10.1128/JVI.01676-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Trgovcich J, Johnson D, Roizman B. Cell surface major histocompatibility complex class II proteins are regulated by the products of the gamma(1)34.5 and U(L)41 genes of herpes simplex virus 1. J Virol. 2002;76:6974–6986. doi: 10.1128/JVI.76.14.6974-6986.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bryant KF, Cox JC, Wang H, Hogle JM, Coen DM. Binding of herpes simplex virus-1 US11 to specific RNA sequences. Nucleic Acids Res. 2005;33:6090–6100. doi: 10.1093/nar/gki919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cassady KA, Gross M, Roizman B. The herpes simplex virus US11 protein effectively compensates for the γ134.5 gene if present before activation of protein kinase R by precluding its phosphorylation and that of the α subunit of eukaryotic initation factor 2. J. Virol. 1998;72:8620–8626. doi: 10.1128/jvi.72.11.8620-8626.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cassady KA, Gross M, Roizman B. The second-site mutation in the herpes simplex virus recombinants lacking the γ134.5 genes precludes shutoff of protein synthesis by blocking the phosphorylation of eIF2α. J. Virol. 1998;72:7005–7011. doi: 10.1128/jvi.72.9.7005-7011.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mulvey M, Camarena V, Mohr I. Full resistance of herpes simplex virus type 1-infected primary human cells to alpha interferon requires both the Us11 and gamma(1)34.5 gene products. J Virol. 2004;78:10193–10196. doi: 10.1128/JVI.78.18.10193-10196.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Poppers J, Mulvey M, Khoo D, Mohr I. Inhibition of PKR activation by the proline-rich RNA binding domain of the herpes simplex virus type 1 Us11 protein. J. Virol. 2000;74:11215–11221. doi: 10.1128/jvi.74.23.11215-11221.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Stevens JG, Wagner EK, Devi-Rao GB, Cook ML, Feldman LT. RNA complementary to a herpesvirus alpha gene mRNA is prominent in latently infected neurons. Science. 1987;235:1056–1059. doi: 10.1126/science.2434993. [DOI] [PubMed] [Google Scholar]
- 74.Dobson AT, Sederati F, Devi-Rao G, Flanagan WM, Farrell MJ, Stevens JG, Wagner EK, Feldman LT. Identification of the latency-associated transcript promoter by expression of rabbit beta-globin mRNA in mouse sensory nerve ganglia latently infected with a recombinant herpes simplex virus. J. Virol. 1989;63:3844–3851. doi: 10.1128/jvi.63.9.3844-3851.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Farrell MJ, Dobson AT, Feldman LT. Herpes simplex virus latency-associated transcript is a stable intron. Proc. Natl. Acad. Sci. USA. 1991;88:790–794. doi: 10.1073/pnas.88.3.790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chen S-H, Kramer MF, Schaffer PA, Coen DM. A viral function represses accumulation of transcripts from productive-cycle genes in mouse ganglia latently infected with herpes simplex virus. J. Virol. 1997;71:5878–5884. doi: 10.1128/jvi.71.8.5878-5884.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Cliffe AR, Garber DA, Knipe DM. Transcription of the herpes simplex virus latency-associated transcript promotes the formation of facultative heterochromatin on lytic promoters. J Virol. 2009;83:8182–8190. doi: 10.1128/JVI.00712-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Garber DA, Schaffer PA, Knipe DM. A LAT-associated function reduces productive-cycle gene expression during acute infection of murine sensory neurons with herpes simplex virus type 1. J. Virol. 1997;71:5885–5893. doi: 10.1128/jvi.71.8.5885-5893.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Perng G-C, Jones C, Ciacci-Zanella J, Stone M, Henderson G, Yukht A, Slanina SM, Hofman FM, Ghiasi H, Nesburn AB, Wechsler SL. Virus-induced neuronal apoptosis blocked by the herpes simplex virus latency-associated transcript. Science. 2000;287:1500–1503. doi: 10.1126/science.287.5457.1500. [DOI] [PubMed] [Google Scholar]
- 80.Thompson RL, Sawtell NM. Herpes simplex virus type 1 latency-associated transcript gene promotes neuronal survival. J Virol. 2001;75:6660–6675. doi: 10.1128/JVI.75.14.6660-6675.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wang QY, Zhou C, Johnson KE, Colgrove RC, Coen DM, Knipe DM. Herpesviral latency-associated transcript gene promotes assembly of heterochromatin on viral lytic-gene promoters in latent infection. Proc Natl Acad Sci U S A. 2005;102:16055–16059. doi: 10.1073/pnas.0505850102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Saini HK, Enright AJ, Griffiths-Jones S. Annotation of mammalian primary microRNAs. BMC Genomics. 2008;9:564. doi: 10.1186/1471-2164-9-564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Chen C, Ridzon DA, Broomer AJ, Zhou Z, Lee DH, Nguyen JT, Barbisin M, Xu NL, Mahuvakar VR, Andersen MR, Lao KQ, Livak KJ, Guegler KJ. Realtime quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 2005;33:e179. doi: 10.1093/nar/gni178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Linsen SEV, de Wit E, Jansens G, Heater S, Chapman L, Parkin RF, Fritz B, Wyman SK, de Bruijn E, Voest EE, Kuersten S, Tewarie M, Cuppen E. Limitations and possibilities of small RNA digital gene expression profiling. Nature Methods. 2009;6:474–476. doi: 10.1038/nmeth0709-474. [DOI] [PubMed] [Google Scholar]
- 85.Kramer MF, Jurak I, Pesola JM, Boissel S, Knipe DM, Coen DM. Herpes simplex virus 1 microRNAs expressed abundantly during latent infection are not essential for latency in mouse trigeminal ganglia. Virology accepted for publication. 2011 doi: 10.1016/j.virol.2011.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Spivack JG, Fraser NW. Detection of herpes simplex virus type 1 transcripts during latent infection in mice. J Virol. 1987;61:3841–3847. doi: 10.1128/jvi.61.12.3841-3847.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Chen S-H, Lee LY, Garber DA, Schaffer PA, Knipe DM, Coen DM. Neither LAT nor open reading frame P mutations increase expression of spliced or intron-containing ICP0 transcripts in mouse ganglia latently infected with herpes simplex virus. J. Virol. 2002;76:4764–4772. doi: 10.1128/JVI.76.10.4764-4772.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kramer MF, Chen S-H, Knipe DM, Coen DM. Accumulation of viral transcripts and DNA during establishment of latency by herpes simplex virus. J. Virol. 1998;72:1177–1185. doi: 10.1128/jvi.72.2.1177-1185.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kramer MF, Coen DM. Quantification of transcripts from the ICP4 and thymidine kinase genes in mouse ganglia latently infected with herpes simplex virus. J. Virol. 1995;69:1389–1399. doi: 10.1128/jvi.69.3.1389-1399.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Pesola JM, Zhu J, Knipe DM, Coen DM. Herpes simplex virus 1 immediate-early and early gene expression during reactivation from latency under conditions that prevent infectious virus production. J Virol. 2005;79:14516–14525. doi: 10.1128/JVI.79.23.14516-14525.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Tal-Singer R, Lasner TM, Podrzucki W, Skokotas A, Leary JJ, Berger SL, Fraser NW. Gene expression during reactivation of herpes simplex virus type 1 from latency in the peripheral nervous system is different from that during lytic infection of tissue cultures. J. Virol. 1997;71:5268–5276. doi: 10.1128/jvi.71.7.5268-5276.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ben-Hur T, Moyal M, Rösen-Wolfe A, Darai G, Becker Y. Characterization of RNA transcripts from herpes simplex virus-1 DNA fragment BamHI-B. Virology. 1989;169:1–8. doi: 10.1016/0042-6822(89)90034-2. [DOI] [PubMed] [Google Scholar]
- 93.Singh J, Wagner EK. Transcriptional analysis of the herpes simplex virus type 1 region containing the TRL/UL junction. Virology. 1993;196:220–231. doi: 10.1006/viro.1993.1470. [DOI] [PubMed] [Google Scholar]
- 94.Zhu J, Kang W, Marquart ME, Hill JM, Zheng X, Block TM, Fraser NW. Identification of a novel 0.7-kb polyadenylated transcript in the LAT promoter region of HSV-1 that is strain specific and may contribute to virulence. Virology. 1999;265:296–307. doi: 10.1006/viro.1999.0057. [DOI] [PubMed] [Google Scholar]
- 95.Naito J, Mukerjee R, Mott KR, Kang W, Osorio N, Fraser NW, Perng GC. Identification of a protein encoded in the herpes simplex virus type 1 latency associated transcript promoter region. Virus Res. 2005;108:101–110. doi: 10.1016/j.virusres.2004.08.011. [DOI] [PubMed] [Google Scholar]
- 96.Tang S, Bertke AS, Patel A, Margolis TP, Krause PR. Herpes simplex virus 2 microRNA miR-H6 is a novel latency-associated transcript-associated microRNA, but reduction of its expression does not influence the establishment of viral latency or the recurrence phenotype. J. Virol. 2011;85:4501–4509. doi: 10.1128/JVI.01997-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Perng GC, Maguen B, Jin L, Mott KR, Kurylo J, BenMohamed L, Yukht A, Osorio N, Nesburn AB, Henderson G, Inman M, Jones C, Wechsler SL. A novel herpes simplex virus type 1 transcript (AL-RNA) antisense to the 5' end of the latency-associated transcript produces a protein in infected rabbits. J Virol. 2002;76:8003–8010. doi: 10.1128/JVI.76.16.8003-8010.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Berretta J, Morillon A. Pervasive transcription constitutes a new level of eukaryotic genome regulation. EMBO Reports. 2009;10:973–982. doi: 10.1038/embor.2009.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Han J, Lee Y, Yeom KH, Nam JW, Heo I, Rhee JK, Sohn SY, Cho Y, Zhang BT, Kim VN. Molecular basis for the recognition of primary microRNAs by the Drosha-DGCR8 complex. Cell. 2006;125:887–901. doi: 10.1016/j.cell.2006.03.043. [DOI] [PubMed] [Google Scholar]
- 100.Ambros V, Bartel B, Bartel DP, Burge CB, Carrington JC, Chen X, Dreyfuss G, Eddy SR, Griffiths-Jones S, Marshall M, Matzke M, Ruvkun G, Tuschl T. A uniform system for microRNA annotation. RNA. 2003;9:277–279. doi: 10.1261/rna.2183803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Bludau H, Freese UK. Analysis of the HSV-1 strain 17 DNA polymerase gene reveals the expression of four different classes of pol transcripts. Virology. 1991;183:505–518. doi: 10.1016/0042-6822(91)90980-p. [DOI] [PubMed] [Google Scholar]
- 102.Hubenthal-Voss J, Starr L, Roizman B. The herpes simplex virus origins of DNA synthesis in the S component are each contained in a transcribed open reading frame. J. Virol. 1987;61:3349–3355. doi: 10.1128/jvi.61.11.3349-3355.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Yager DR, Coen DM. Analysis of the transcript of the herpes simplex virus DNA polymerase gene provides evidence that polymerase expression is inefficient at the level of translation. J. Virol. 1988;62:2007–2015. doi: 10.1128/jvi.62.6.2007-2015.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol. Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Krek A, Grun D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, MacMenamin P, da Piedade I, Gunsalus KC, Stoffel M, Rajewsky N. Combinatorial microRNA target predictions. Nat Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- 106.John B, Enright AJ, Aravin A, Tuschl T, Sander C, Marks DS. Human MicroRNA targets. PLoS Biol. 2004;2:e363. doi: 10.1371/journal.pbio.0020363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Thomas M, Lieberman J, Lal A. Desperately seeking microRNA targets. Nat Struct Mol Biol. 2010;17:1169–1174. doi: 10.1038/nsmb.1921. [DOI] [PubMed] [Google Scholar]
- 108.Skalsky RL, Cullen BR. Viruses, microRNAs, and host interactions. Annu Rev Microbiol. 2010;64:123–141. doi: 10.1146/annurev.micro.112408.134243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Zeng Y, Cullen BR. RNA interference in human cells is restricted to the cytoplasm. RNA. 2002;8:855–860. doi: 10.1017/s1355838202020071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Hwang HW, Wentzel EA, Mendell JT. A hexanucleotide element directs microRNA nuclear import. Science. 2007;315:97–100. doi: 10.1126/science.1136235. [DOI] [PubMed] [Google Scholar]
- 111.Castanotto D, Lingeman R, Riggs AD, Rossi JJ. CRM1 mediates nuclear-cytoplasmic shuttling of mature microRNAs. Proc Natl Acad Sci U S A. 2009;106:21655–21659. doi: 10.1073/pnas.0912384106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Jeffries CD, Fried HM, Perkins DO. Nuclear and cytoplasmic localization of neural stem cell microRNAs. RNA. 2011 doi: 10.1261/rna.2006511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Tyler DM, Okamura K, Chung WJ, Hagen JW, Berezikov E, Hannon GJ, Lai EC. Functionally distinct regulatory RNAs generated by bidirectional transcription and processing of microRNA loci. Genes Dev. 2008;22:26–36. doi: 10.1101/gad.1615208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Buck AH, Santoyo-Lopez J, Robertson KA, Kumar DS, Reczko M, Ghazal P. Discrete clusters of virus-encoded micrornas are associated with complementary strands of the genome and the 7.2-kilobase stable intron in murine cytomegalovirus. J Virol. 2007;81:13761–13770. doi: 10.1128/JVI.01290-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Lai EC, Wiel C, Rubin GM. Complementary miRNA pairs suggest a regulatory role for miRNA:miRNA duplexes. RNA. 2004;10:171–175. doi: 10.1261/rna.5191904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Dolken L, Pfeffer S, Koszinowski UH. Cytomegalovirus microRNAs. Virus Genes. 2009;38:355–364. doi: 10.1007/s11262-009-0347-0. [DOI] [PubMed] [Google Scholar]
- 117.Maillet S, Naas T, Crepin S, Roque-Afonso AM, Lafay F, Efstathiou S, Labetoulle M. Herpes simplex virus type 1 latently infected neurons differentially express latency-associated and ICP0 transcripts. J Virol. 2006;80:9310–9321. doi: 10.1128/JVI.02615-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Whitley RJ, Kern ER, Chatterjee S, Chou J, Roizman B. Replication, establishment of latency, and induced reactivation of herpes simplex virus gamma 1 34.5 deletion mutants in rodent models. J. Clinical Invest. 1993;91:2837–2843. doi: 10.1172/JCI116527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Chou J, Kern ER, Whitley RJ, Roizman B. Mapping of herpes simplex virus-1 neurovirulence to gamma 134.5, a gene nonessential for growth in culture. Science. 1990;250:1262–1266. doi: 10.1126/science.2173860. [DOI] [PubMed] [Google Scholar]
- 120.Bolovan CA, Sawtell NM, Thompson RL. ICP34.5 mutants of herpes simplex virus type 1 strain 17syn+ are attenuated for neurovirulence in mice and for replication in confluent primary mouse embryo cell cultures. J. Virol. 1994;68:48–55. doi: 10.1128/jvi.68.1.48-55.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Schwarz DS, Hutvagner G, Du T, Xu Z, Aronin N, Zamore PD. Asymmetry in the assembly of the RNAi enzyme complex. Cell. 2003;115:199–208. doi: 10.1016/s0092-8674(03)00759-1. [DOI] [PubMed] [Google Scholar]
- 122.Khvorova A, Reynolds A, Jayasena SD. Functional siRNAs and miRNAs exhibit strand bias. Cell. 2003;115:209–216. doi: 10.1016/s0092-8674(03)00801-8. [DOI] [PubMed] [Google Scholar]
- 123.Yang JS, Phillips MD, Betel D, Mu P, Ventura A, Siepel AC, Chen KC, Lai EC. Widespread regulatory activity of vertebrate microRNA* species. RNA. 2011;17:312–326. doi: 10.1261/rna.2537911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Otsuka M, Jing Q, Georgel P, New L, Chen J, Mols J, Kang YJ, Jiang Z, Du X, Cook R, Das SC, Pattnaik AK, Beutler B, Han J. Hypersusceptibility to vesicular stomatitis virus infection in Dicer1-deficient mice is due to impaired miR24 and miR93 expression. Immunity. 2007;27:123–134. doi: 10.1016/j.immuni.2007.05.014. [DOI] [PubMed] [Google Scholar]
- 125.Lecellier CH, Dunoyer P, Arar K, Lehmann-Che J, Eyquem S, Himber C, Saib A, Voinnet O. A cellular microRNA mediates antiviral defense in human cells. Science. 2005;308:557–560. doi: 10.1126/science.1108784. [DOI] [PubMed] [Google Scholar]
- 126.Wang FZ, Weber F, Croce C, Liu CG, Liao X, Pellett PE. Human cytomegalovirus infection alters the expression of cellular microRNA species that affect its replication. J Virol. 2008;82:9065–9074. doi: 10.1128/JVI.00961-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Buck AH, Perot J, Chisholm MA, Kumar DS, Tuddenham L, Cognat V, Marcinowski L, Dolken L, Pfeffer S. Post-transcriptional regulation of miR-27 in murine cytomegalovirus infection. RNA. 2010;16:307–315. doi: 10.1261/rna.1819210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Ramachandran V, Chen X. Degradation of microRNAs by a family of exoribonucleases in Arabidopsis. Science. 2008;321:1490–1492. doi: 10.1126/science.1163728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Chatterjee S, Grosshans H. Active turnover modulates mature microRNA activity in Caenorhabditis elegans. Nature. 2009;461:546–549. doi: 10.1038/nature08349. [DOI] [PubMed] [Google Scholar]
- 130.Ibrahim F, Rymarquis LA, Kim EJ, Becker J, Balassa E, Green PJ, Cerutti H. Uridylation of mature miRNAs and siRNAs by the MUT68 nucleotidyltransferase promotes their degradation in Chlamydomonas. Proc Natl Acad Sci U S A. 2010;107:3906–3911. doi: 10.1073/pnas.0912632107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Ibrahim F, Rohr J, Jeong WJ, Hesson J, Cerutti H. Untemplated oligoadenylation promotes degradation of RISC-cleaved transcripts. Science. 2006;314:1893. doi: 10.1126/science.1135268. [DOI] [PubMed] [Google Scholar]
- 132.Bivalkar-Mehla S, Vakharia J, Mehla R, Abreha M, Kanwar JR, Tikoo A, Chauhan A. Viral RNA silencing suppressors (RSS): novel strategy of viruses to ablate the host RNA interference (RNAi) defense system. Virus Res. 2011;155:1–9. doi: 10.1016/j.virusres.2010.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Lu S, Cullen BR. Adenovirus VA1 noncoding RNA can inhibit small interfering RNA and MicroRNA biogenesis. J Virol. 2004;78:12868–12876. doi: 10.1128/JVI.78.23.12868-12876.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Andersson MG, Haasnoot PC, Xu N, Berenjian S, Berkhout B, Akusjarvi G. Suppression of RNA interference by adenovirus virus-associated RNA. J Virol. 2005;79:9556–9565. doi: 10.1128/JVI.79.15.9556-9565.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Xu N, Segerman B, Zhou X, Akusjarvi G. Adenovirus virus-associated RNAII-derived small RNAs are efficiently incorporated into the rna-induced silencing complex and associate with polyribosomes. J Virol. 2007;81:10540–10549. doi: 10.1128/JVI.00885-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Taddeo B, Roizman B. The virion host shutoff protein (UL41) of herpes simplex virus 1 is an endoribonuclease with a substrate specificity similar to that of RNase A. J Virol. 2006;80:9341–9345. doi: 10.1128/JVI.01008-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Taddeo B, Zhang W, Roizman B. The U(L)41 protein of herpes simplex virus 1 degrades RNA by endonucleolytic cleavage in absence of other cellular or viral proteins. Proc Natl Acad Sci U S A. 2006;103:2827–2832. doi: 10.1073/pnas.0510712103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Wang X, Wang HK, McCoy JP, Banerjee NS, Rader JS, Broker TR, Meyers C, Chow LT, Zheng ZM. Oncogenic HPV infection interrupts the expression of tumor-suppressive miR-34a through viral oncoprotein E6. RNA. 2009;15:637–647. doi: 10.1261/rna.1442309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Kang JG, Pripuzova N, Majerciak V, Kruhlak M, Le SY, Zheng ZM. Kaposi's sarcoma-associated herpesvirus ORF57 promotes escape of viral and human interleukin-6 from microRNA-mediated suppression. J Virol. 2011;85:2620–2630. doi: 10.1128/JVI.02144-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Hill JM, Zhao Y, Clement C, Neumann DM, Lukiw WJ. HSV-1 infection of human brain cells induces miRNA-146a and Alzheimer-type inflammatory signaling. Neuroreport. 2009;20:1500–1505. doi: 10.1097/WNR.0b013e3283329c05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Wu Z, Zhu Y, Bisaro DM, Parris DS. Herpes simplex virus type 1 suppresses RNA-induced gene silencing in mammalian cells. J Virol. 2009;83:6652–6663. doi: 10.1128/JVI.00260-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Whitley RJ, Hilliard J. Cercopithecine Herpes Virus 1 (B virus) In: Knipe DM, Howley PM, editors. Fields Virology. Vol. 2. Lippincott Williams & Wilkins; Philadelphia, PA: 2007. pp. 2889–2903. [Google Scholar]
- 143.Cohen JI, Davenport DS, Stewart JA, Deitchman S, Hilliard JK, Chapman LE. Recommendations for prevention of and therapy for exposure to B virus (cercopithecine herpesvirus 1) Clin Infect Dis. 2002;35:1191–1203. doi: 10.1086/344754. [DOI] [PubMed] [Google Scholar]
- 144.Bryan BL, Espana CD, Emmons RW, Vijayan N, Hoeprich PD. Recovery from encephalomyelitis caused by Herpesvirus simiae. Report of a case. Arch Intern Med. 1975;135:868–870. [PubMed] [Google Scholar]
- 145.Holmes GP, Hilliard JK, Klontz KC, Rupert AH, Schindler CM, Parrish E, Griffin DG, Ward GS, Bernstein ND, Bean TW, et al. B virus (Herpesvirus simiae) infection in humans: epidemiologic investigation of a cluster. Ann Intern Med. 1990;112:833–839. doi: 10.7326/0003-4819-112-11-833. [DOI] [PubMed] [Google Scholar]
- 146.Perelygina L, Zhu L, Zurkuhlen H, Mills R, Borodovsky M, Hilliard JK. Complete sequence and comparative analysis of the genome of herpes B virus (Cercopithecine herpesvirus 1) from a rhesus monkey. J. Virol. 2003;77:6167–6177. doi: 10.1128/JVI.77.11.6167-6177.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Barth S, Pfuhl T, Mamiani A, Ehses C, Roemer K, Kremmer E, Jaker C, Hock J, Meister G, Grasser FA. Epstein-Barr virus-encoded microRNA miR-BART2 down-regulates the viral DNA polymerase BALF5. Nucleic Acids Res. 2008;36:666–675. doi: 10.1093/nar/gkm1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Grinfeld E, Kennedy PG. Translation of varicella-zoster virus genes during human ganglionic latency. Virus Genes. 2004;29:317–319. doi: 10.1007/s11262-004-7434-z. [DOI] [PubMed] [Google Scholar]
- 149.Davis N, Biddlecom N, Hecht D, Fogel GB. On the relationship between GC content and the number of predicted microRNA binding sites by MicroInspector. Comput Biol Chem. 2008;32:222–226. doi: 10.1016/j.compbiolchem.2008.02.004. [DOI] [PubMed] [Google Scholar]
- 150.Bowland SL, Shewen PE. Bovine respiratory disease: commercial vaccines currently available in Canada. Can Vet J. 2000;41:33–48. [PMC free article] [PubMed] [Google Scholar]
- 151.Jones C. Herpes simplex virus type 1 and bovine herpesvirus 1 latency. Clin Microbiol Rev. 2003;16:79–95. doi: 10.1128/CMR.16.1.79-95.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Tikoo SK, Campos M, Babiuk LA. Bovine herpesvirus 1 (BHV-1): biology, pathogenesis, and control. Adv Virus Res. 1995;45:191–223. doi: 10.1016/s0065-3527(08)60061-5. [DOI] [PubMed] [Google Scholar]
- 153.Jones C, Geiser V, Henderson G, Jiang Y, Meyer F, Perez S, Zhang Y. Functional analysis of bovine herpesvirus 1 (BHV-1) genes expressed during latency. Vet Microbiol. 2006;113:199–210. doi: 10.1016/j.vetmic.2005.11.009. [DOI] [PubMed] [Google Scholar]
- 154.Aujeszky A. Ueber eine neue Infektionskrankheit bei Hautieren. Zentralblatt Bakteriologie. 1902;32:353–357. [Google Scholar]
- 155.Stallknecht DE, Howerth EW. Pseudorabies (Aujeszky's Disease) In: Williams ES, Barker IK, editors. Infectious Diseases of Wild Mammals. Blackwell Publishing; Ames: 2001. pp. 164–170. [Google Scholar]
- 156.Pomeranz LE, Reynolds AE, Hengartner CJ. Molecular biology of pseudorabies virus: impact on neurovirology and veterinary medicine. Microbiol Mol Biol Rev. 2005;69:462–500. doi: 10.1128/MMBR.69.3.462-500.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]