Abstract
Closely related organisms usually occupy similar ecological niches, leading to intense competition and even extinction. Such competition also can promote rapid phenotypic evolution and ecological divergence. This process may end with the stable occupation of distinct niches or, alternatively, may entail repeated bouts of evolution. Here we examine two Escherichia coli lineages, called L and S, that coexisted for more than 30,000 generations after diverging from a common ancestor. Both lineages underwent sustained phenotypic evolution based on global transcription and resource utilization profiles, with L seeming to encroach over time on the catabolic profile of S. Reciprocal invasion experiments with L and S clones from the same or different generations revealed evolutionary changes in their interaction, including an asymmetry that confirmed the encroachment by L on the niche of the S lineage. In general, L and S clones from the same generation showed negative frequency-dependent effects, consistent with stable coexistence. However, L clones could invade S clones from both earlier and later generations, whereas S clones could invade only L clones from earlier generations. In this system, the long-term coexistence of competing lineages evidently depended on successive rounds of evolution, rather than on initial divergence followed by a static equilibrium.
Keywords: experimental evolution, frequency-dependent selection, gene expression
The competitive exclusion principle precludes the stable coexistence of organisms occupying identical ecological niches (1–4). Closely related organisms tend to retain ancestral traits and thus occupy similar niches (5–9), and so competition is especially intense when they live in sympatry. Natural selection may then reduce their competition by driving apart those traits that determine their ecological niches, thereby allowing their stable coexistence through negative frequency-dependent selection. Examples of such ecological character displacement have been documented in many different organisms (10–14). However, the long-term evolutionary dynamics of competing lineages following such divergence are poorly understood (15). A theoretical model constructed to analyze the coevolution of competitors suggested that this process often led to extinctions (16), although a later study questioned some of the model’s assumptions (17). In an empirical study, the interaction between two bacterial species was observed to evolve during a short-term experiment, leading to increased productivity of the community (18); however, that study examined neither the initial divergence of the bacteria nor the long-term fate of their interaction.
Consider the simple case of two recently diverged lineages that occupy slightly different ecological niches and are able to coexist stably owing to negative frequency-dependent effects. Their subsequent evolution could lead to several distinct scenarios. First, fitness might improve relative to earlier generations within, but not between, the lineages, sustaining their ecological differentiation. Second, competition between the lineages might be further reduced by character displacement, thereby promoting their ecological divergence and stabilizing their interaction. Third, fitness may improve relative to previous generations both within and between lineages, leading to more complex changes in their interaction. For example, one lineage might encroach on the ecological niche of the other, and that encroachment in turn might cause the extinction of the affected lineage, or might lead to further evolution of the affected lineage that enables its persistence.
Here we studied the evolutionary dynamics of two bacterial lineages that diverged from their common ancestor and then coexisted for tens of thousands of generations in a long-term experiment. In that experiment, 12 populations of Escherichia coli were founded from the same strain and propagated in a glucose-limited minimal medium for more than 40,000 generations (19, 20). In one population, designated Ara-2, two lineages, named L and S, had diverged from their ancestor and from one another by 6,500 generations, and they coexisted thereafter (21–23). Their divergence and coexistence involved niche construction, in which organisms modify their environment in ways that promote their own or others’ success (24–27). In particular, the two lineages compete for the limiting glucose, with the L lineage able to grow faster on the glucose but secreting a metabolic byproduct that S is better able to exploit (21, 23).
To characterize and better understand the evolutionary dynamics, we sampled four L and S clones from each of three time points: 6,500, 17,000, and 40,000 generations. We measured many phenotypes by quantifying and comparing their global transcription profiles in the medium where they evolved and their growth abilities in 51 different environments. We also performed reciprocal invasion experiments using the L and S populations from the same or different generations to assess how their ecological interaction had changed and what those changes might indicate about the evolutionary dynamics that allowed their long-term coexistence.
Results and Discussion
Expression Profiles and Growth Abilities of L and S Lineages.
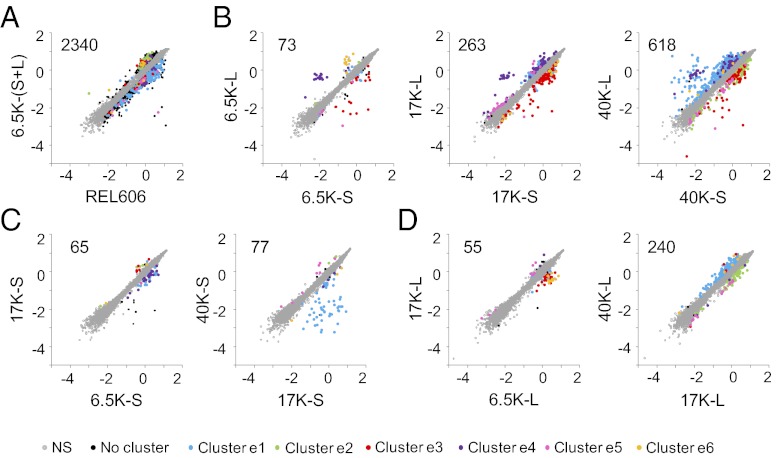
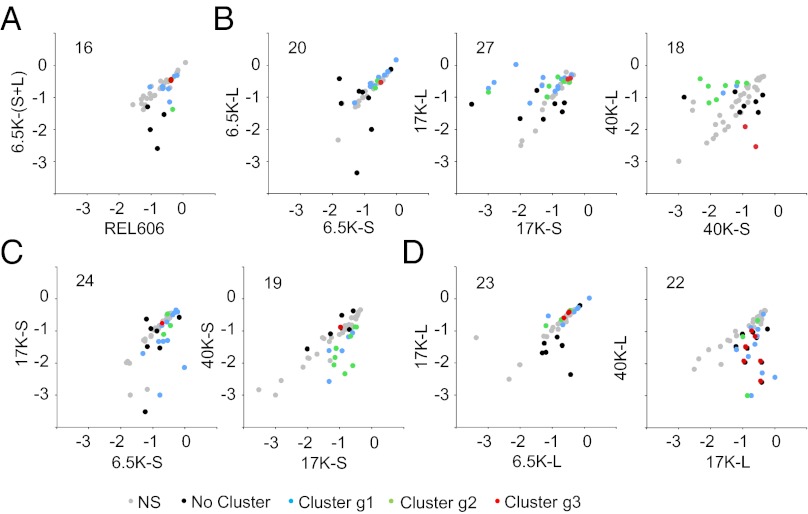
During the initial period through 6,500 generations, the bacteria underwent dramatic changes in global expression profiles relative to their ancestor (Fig. 1A). More than 2,000 genes were differentially expressed between the ancestor and evolved bacteria (including both L and S clones) according to the criteria specified in Materials and Methods. These changes reflect substantial reorganization of gene regulatory networks, and such global changes were observed repeatedly in this long-term experiment (28–30). The growth abilities of the bacteria also changed over this same period in 16 of the 51 environments that yielded informative data (Fig. 2A).
Fig. 1.
Evolutionary dynamics of global transcription profiles for L and S lineages during 40,000 generations. (A) Comparison of the ancestral strain (REL606) with the combined S and L samples at 6,500 generations [6.5K − (S + L)]. (B) Comparisons of S and L samples at 6,500, 17,000, and 40,000 generations. (C) Comparisons of S samples from different generations. (D) Comparisons of L samples from different generations. Each point corresponds to a gene, and the values are log10-transformed expression levels. The number of genes with significantly different expression is shown for each comparison near the upper left corner. Gray symbols indicate genes without significant differences in expression in any of the eight comparisons. Those genes with significant differences in expression in at least one of the seven comparisons between two evolved samples were clustered according to their expression patterns, and the colored symbols indicate genes in expression clusters e1–e6 (Table S1). Black symbols indicate genes that either did not cluster or showed significant differences in expression only between the ancestor and evolved samples.
Fig. 2.
Evolutionary dynamics of growth abilities across 51 environments for L and S lineages during 40,000 generations, as described in Fig. 1. Each point corresponds to a different environment, and the values are log10-transformed growth yields based on optical densities. The number of environments with significant differences in growth is shown for each comparison near the upper left corner. Gray symbols indicate environments without significant differences in growth in any of the eight comparisons. Those environments demonstrating significant differences in growth in at least one comparison between two evolved samples were clustered based on their growth patterns, and the different colored symbols indicate environments in growth clusters g1–g3 (Table S2). Black symbols indicate environments that either did not cluster or showed significant differences only between the ancestor and evolved samples.
We also compared the expression profiles and growth abilities of L and S clones sampled at identical time points. After 6,500 generations, the L and S clones showed significant differences in expression for only 73 genes (Fig. 1B), although the magnitude of the differences (deviations from the 45-degree line) tended to be large compared with their differences relative to the ancestor. The number of genes differentially expressed between the L and S lineages further increased to 263 after 17,000 generations and to 618 after 40,000 generations. The number of test environments in which the growth of the L and S lineages differed showed no clear trend, with 20 differences at 6,500 generations, 27 at 17,000 generations, and 18 at 40,000 generations (Fig. 2B). However, the differences between the two lineages tended to be greater in the later generations, and the environments in which they differed also changed, as described below.
Cluster Analyses.
The differences in gene expression and growth yields demonstrate that many physiological properties of the L and S lineages evolved during the thousands of generations in which they coexisted. We performed cluster analyses of the gene expression and growth datasets to explore the evolution of these multidimensional phenotypes, with a focus on understanding the differences between the L and S lineages and how they evolved over time. Thus, we analyzed those genes and environments (797 and 48, respectively) for which we observed a significant difference in expression or growth in at least one of the seven comparisons between evolved clones, excluding genes and environments that showed no significant difference in any comparison, as well as those for which the only difference was between the ancestral and evolved bacteria. These analyses identified six clusters of genes with differential expression patterns (e1–e6) and three clusters of environments that produced differential growth patterns (g1–g3).
Expression cluster e1 includes 367 genes of diverse functions (Table S1) for which mRNA levels were significantly higher in the L lineage than in the S lineage at generation 40,000 (Fig. 1B). Some of these genes showed substantially reduced expression in the S lineage between 17,000 and 40,000 generations (Fig. 1C), but most exhibited slightly increased expression in the L lineage over the same period (Fig. 1D). Many of the genes with strongly reduced expression in S were found in two chromosomal regions that had been deleted by mutations in that lineage (Table 1). The detection of apparent low expression of the deleted genes indicates background noise (including any cross-hybridization) in the microarrays used to quantify gene expression. Cluster e2 has 133 genes, also with diverse cellular functions, that were expressed at lower levels in the L lineage than in the S lineage at generation 40,000 (Fig. 1B), largely as a consequence of changes in L between 17,000 and 40,000 generations (Fig. 1D).
Table 1.
Large genomic deletions in S lineage at 40,000 generations
| Deletion | Size, kbp | Coordinates* | Deleted genes |
| 1 (ΔkpsD–glcB) | ∼35 | 3015256–3050311 | kpsD, kpsU, kpsC, kpsS, insB, kpsT, kpsM, yghD, yghE, epsF, epsE, epsD, yghF, yghG, pppA, yghJ, yghK, glcB |
| 2 (ΔinsB–ybdK) | ∼41 | 547701–588493 | insB, insA, essD, ybcS, rzpD, borD, ynfO, nohA, appY, ompT, envY, ybcH, nfrA, nfrB, yhhI, cusS, cusR, cusC, cusF, cusB, cusA, pheP, ybdG, nfnB, ybdF, ybdJ, ybdK |
*Coordinates are based on the ancestral strain’s genome sequence (39).
Expression clusters e5 and e6 together include 84 genes that showed more transient changes. The genes in e5 were expressed at slightly higher levels in the L lineage than in the S lineage after 17,000 generations, whereas the reverse pattern was seen at 40,000 generations (Fig. 1B). The genes in cluster e6 had higher expression in L than in S at 6,500 generations, but this pattern was largely eliminated by 40,000 generations (Fig. 1B), primarily as a consequence of reduced expression of these genes in the L lineage between 6,500 and 17,000 generations (Fig. 1D).
Expression clusters e3 and e4 together include another 176 genes for which the mRNA levels differed significantly between the L and S lineages (Fig. 1B). In contrast to the clusters discussed above, the expression of genes in these two clusters remained largely constant between generations 6,500 and 40,000 within each lineage (Fig. 1 C and D). These fixed differences likely include or reflect those physiological traits important for the early ecological specialization of each lineage. Many of the genes that were more highly expressed in the S lineage (cluster e3) are involved in the Entner–Doudoroff pathway (31), an alternative to glycolysis that also catabolizes glucose to pyruvate, and in the glyoxylate cycle (32), a bypass of the tricarboxylic acid cycle that enables growth on two-carbon compounds. These differences may contribute to the lower growth rate of the S cells on glucose and to the ability of the S cells to exploit metabolic byproducts secreted by L cells (21, 23). Some of the genes that were more highly expressed in the L lineage (cluster e4) are involved in glycerol metabolism; these include manXYZ, which encodes a sugar transporter controlled by the glucose-sensing global regulator Mlc (33). The ManXYZ protein has been identified as a secondary glucose transporter (33), and its increased expression may contribute to the faster growth of the L cells on glucose.
Turning to the cluster analyses of differences in growth across environments (Table S2), clusters g2 and g3 together include 18 environments in which the measured yield of the L lineage increased or decreased relative to the S lineage (Fig. 2B). Most of these differences resulted from declines in the yield of one lineage, as opposed to higher yields in the other lineage (Fig. 2 C and D). The finding that many differences reflect diminished growth in the test environments may indicate trade-offs caused by pleiotropic effects of beneficial mutations, accumulation of mutations in genes under relaxed selection in the long-term experiment, or both (34). This population evolved a mutator phenotype even before the L and S lineages diverged (22, 35), and the higher mutation rate in mutators allows for faster accumulation of mutations in those genes that affect nonessential functions (34, 36).
The growth cluster g1 includes 18 other environments in which the growth abilities of the two lineages decreased sequentially, first declining in S between generations 6,500 and 17,000 (Fig. 2C) and then declining in L between generations 17,000 and 40,000 (Fig. 2D). None of these test environments directly reflects the conditions of the medium and metabolic byproducts that were present during the evolution experiment. However, this cluster indicates that some traits that early on were specific to the S lineage later evolved in the L lineage, raising the intriguing possibility that L was encroaching on the ecological niche of S.
Reciprocal Invasion Experiments.
To test the encroachment hypothesis, we ran competition experiments using the same sets of L and S clones. Experiments were performed using a reciprocal-invasion design—in which the initial ratio of the two lineages was either 1:9 or 9:1, to determine whether they could stably coexist or, alternatively, whether one would exclude the other (21, 37).
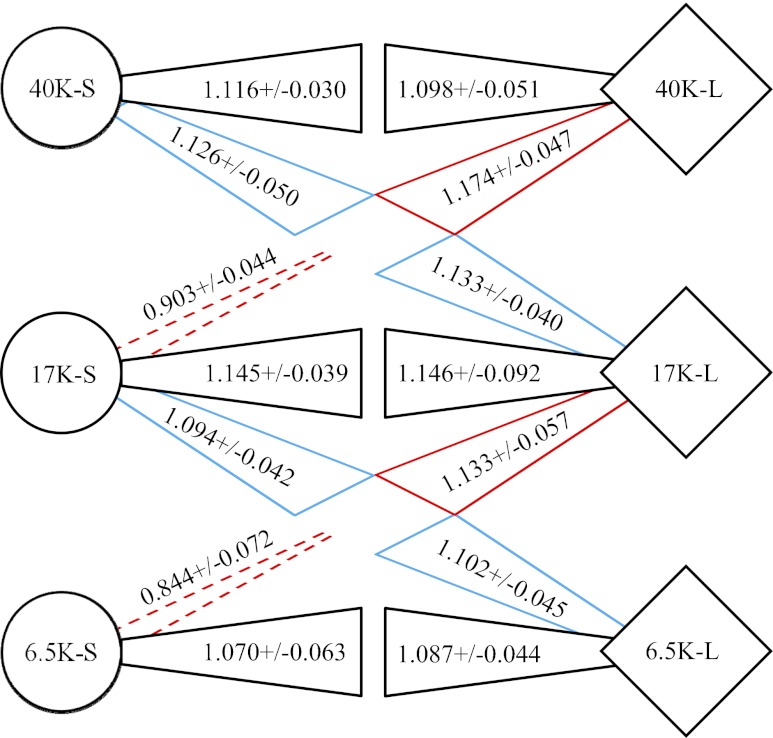
When the competitors were from the same generation, each lineage had a significant advantage when it was initially rare (Fig. 3, side-facing black wedges), which demonstrates a negative frequency-dependent interaction (38). That relationship also confirms that the lineages occupied sufficiently distinct ecological niches to allow them to coexist. When the competitors came from different generations, however, we observed a striking asymmetry in outcomes. In particular, both L and S clones could invade the alternative lineage from the earlier generation when rare (Fig. 3, downward-angled red and blue wedges). Moreover, initially rare L were able to invade S from later generations (Fig. 3, upward-angled blue wedges), but S could not invade L from later generations (Fig. 3, upward-angled dashed red lines); that is, earlier S clones were significantly less fit than later L clones, even when the S clones started at low frequency.
Fig. 3.
Changes in ecological interactions between L and S lineages during 40,000 generations. Sets of clones from the L (diamonds) and S (circles) lineages were sampled at 6,500, 17,000, and 40,000 generations. Each set from each time point competed against two or three other sets from the same or different generations. Black, red, and blue wedges indicate competitions between contemporary lineages, between later L and earlier S, and between earlier L and later S, respectively. Competitions were performed at two starting ratios, with each competitor either initially rare (10%) or common (90%). Wedges that expand (solid lines) or narrow (dashed lines) from each competitor indicate its advantage (fitness >1) or disadvantage (fitness <1) when initially rare. Values represent the mean fitness (with 95% confidence interval) of the rare competitor relative to the common competitor. With 15-fold replication of each competition, all values differ significantly from unity.
This last result demonstrates that the S lineage would have been driven to extinction by the continued evolution of the L lineage had S itself not evolved in response. This result also implies that L encroached on the ecological niche of S, because otherwise S would not have been affected by the further adaptation of L. This encroachment could, in principle, result from the improved performance of L on some shared resource (including glucose), from a newly evolved capacity of L to use a secreted metabolite that was previously used only by S, or from some change in an allelopathic effect whereby the organisms produce substances that harm their competitors.
Synthesis and Perspective.
The coexistence of two competitors may be dynamically stable in a strictly ecological context, but evolutionary changes in one or both populations may affect the stability of the interaction over longer periods. Evolution might enhance stability by promoting character displacement, thereby reducing the intensity of competition between the lineages, or evolution might destabilize the interaction if one lineage encroaches on the other’s niche. In the latter case, the encroached-upon lineage might be driven to extinction, or it might evolve in some way that allows it to persist. Although previous studies have documented character displacement and the resulting coexistence of closely related lineages (10–14), the long-term dynamics of these interactions have not received much attention.
Our ability to analyze these dynamics benefitted from two features of the system under study. First, owing to the short generation times of bacteria, we could observe both the initial divergence of lineages (over several thousand generations) and their subsequent evolution (for tens of thousands of generations). Second, bacteria can be frozen and revived, which allowed us to compete organisms that lived at different times and thereby observe the asymmetry in their evolving interaction. Our results illustrate the rich and complex evolutionary dynamics that can occur between two recently diverged lineages even in a simple environment. The initial diversification involved the process of niche construction (24–27, 37) in which one lineage produced a byproduct that the other lineage was better able to exploit as a resource, allowing the two lineages to occupy distinct niches and thereby coexist (21–23). The two lineages then continued to evolve, with both undergoing substantial changes in gene expression profiles and growth abilities under various conditions. Their competitive interaction also evolved as one lineage encroached on the ecological niche of the other lineage, which in turn responded in ways that allowed it to persist. More generally, the character displacement that enables closely related organisms to coexist need not be an evolutionary endpoint but instead may be only the first step in a long evolutionary dance.
Materials and Methods
Strains.
All strains were sampled from population Ara-2 of the E. coli long-term evolution experiment (19, 21–23) or were marked variants thereof. In this population, two lineages, L and S, had diverged from one another by generation 6,500 and then coexisted until at least generation 40,000. Based on derived mutations specific to each lineage (22), we sampled four clones from each lineage from frozen samples obtained at three time points during the evolution experiment: generations 6,500 (6.5K), 17,000 (17K), and 40,000 (40K). The four clones from each lineage and generation were mixed, generating the six evolved samples used in this study, designated 6.5K-S, 6.5K-L, 17K-S, 17K-L, 40K-S, and 40K-L. These samples were used for global transcription profiling, growth assays, and competition experiments.
The Ara-2 population was founded by an ancestral strain, REL606, that cannot use arabinose as a carbon source (19, 39), and all evolved clones were also phenotypically Ara–. To distinguish competitors during the competition experiments, we isolated spontaneous Ara+ mutants of each of the 24 evolved clones, as described elsewhere (21). Again, the four Ara+ clones from each lineage and generation were mixed before the competition experiments. The arabinose utilization marker is effectively neutral under the conditions of the long-term evolution experiment and in the L and S genetic backgrounds (19, 21).
Global Transcription Profiles.
The seven samples, including the six evolved mixtures and the ancestral strain, were grown overnight in 2 mL of Davis minimal (DM) medium (19) containing 1 mg/mL of glucose (DM1000), then diluted 104 fold into 10 mL of DM medium containing 25 μg/mL of glucose (DM25) in 50-mL flasks and incubated for 24 h at 37 °C while shaking at 120 rpm. To obtain sufficient amounts of RNA, these cultures were diluted 1:100 into 200 mL of DM25 in 1-L flasks and incubated until they reached midexponential phase (6 h for the ancestral strain and 4.5 h for the evolved mixtures). Pellets of bacterial cells were produced and total RNAs extracted using the Qiagen RNeasy Kit, following the manufacturer's protocol. We grew six replicate cultures and analyzed six replicate RNA extractions for each sample. Biotinylated cDNA samples were prepared according to the standard Affymetrix GeneChip protocol for bacteria, and the samples were then hybridized to the Affymetrix E. coli Genome 2.0 Microarray for 16 h at 45 °C by Beckman Coulter Genomics. Quality checks and normalization were performed using the Affymetrix GeneChip Command Console and Expression Console according to standard Affymetrix protocols. In brief, the intensity data were subjected to global scaling to compare data from multiple probe arrays.
Identifying Differentially Expressed Genes.
After removing the intergenic regions from the dataset, we identified differentially expressed genes using the optimal discovery procedure (40) implemented in the EDGE v1.1.208 software (41). We performed eight comparisons, including the ancestor vs. the combined S and L samples from generation 6,500 [6.5K − (S + L)], three between-lineage comparisons at different generations (6.5K-S vs. 6.5K-L, 17K-S vs. 17K-L, and 40K-S vs. 40K-L); and four within-lineage comparisons across generations (6.5K-S vs. 17K-S, 17K-S vs. 40K-S, 6.5K-L vs. 17K-L, and 17K-L vs. 40K-L). Each analysis was performed with 200 permutations. Genes were scored as differentially expressed when the false discovery rate (FDR) (42) had a q value <0.05 and the magnitude of the change was greater than twofold. With multiple testing, the FDR estimates the probability, q, of one gene being a false-positive, given the overall distribution of the ordinary P values obtained for genes analyzed one at a time. Those genes identified as differentially expressed in at least one of the seven comparisons between two evolved samples (797 genes in total) were clustered according to their expression patterns (Fig. 1) using the QT CLUST algorithm (43) implemented with TM4 software (44). We used the Pearson correlation metric distance with a maximum cluster diameter of 0.65 and a minimum cluster size of 25; similar clusters were obtained when we varied the parameters. Genes were classified by Gene Ontology (GO) cellular process categories (45) and also by regulons according to RegulonDB (46). We tested the overrepresentation of GO categories and regulons (Table S1) using one-tailed Fisher’s exact tests that compared the number of differentially expressed genes in each of the six clusters with the total number of differentially expressed genes. These tests were performed only when two or more genes within a GO category or regulon were differentially expressed in the focal comparison. The tests were performed multiple times; thus, FDRs were estimated using QVALUE software (42), and categories were identified as overrepresented only for q values <0.05.
Characterization of Deleted Chromosomal Regions in the S Sineage at 40,000 Generations.
Two sets of physically adjacent genes exhibited large reductions in expression in the S lineage at 40,000 generations (Table 1). These observations suggested that the genes might have been deleted from the genome, with the residual expression reflecting background noise (including any cross-hybridization) in the microarrays. We performed PCR experiments that confirmed the deletion of two large regions, and we identified the precise endpoints of each deletion. We designed primer pairs complementary to the upstream and downstream sequences of each region, which were then used with genomic DNA from a 40,000-generation S clone. For deletion 1 (ΔkpsD-glcB), the pairs were 5′-GCGAAATAAGCGAAAACGAG-3′ and 5′-GTTGCAAAATGGCGATACCT-3′. For deletion 2 (ΔinsB-ybdK), the pairs were 5′-AGTCTCCGACCAGAAGCGTA-3′ and 5′-TTACCATAGGCGACCTGACC-3′.
Growth Assays Using Biolog Microplates.
The same seven biological samples were grown overnight in 2 mL of DM1000, then diluted 104-fold into 10 mL of DM25 in 50-mL flasks and incubated for 24 h at 37 °C with shaking at 120 rpm. For each sample, 2 μL of culture and 100 μL of sterile DM medium were added to each well of a Biolog GENIII microplate. The plates were then incubated at 37 °C in a microplate reader (Infinite M200; Tecan), and the optical density at 600 nm (OD600) was measured periodically for each well. The wells contained 71 carbon sources and 23 other chemicals for assessing substrate use and chemical sensitivity, respectively. Each assay was performed in triplicate. In most cases, we quantified growth yield as the maximum OD600 reached within the first 10 h; however, under four conditions (fusidic acid, d-saccharic acid, l-malic acid, and sodium butyrate), growth began only after a long lag phase, and for these we quantified growth yield as the OD600 reached after 24 h. We excluded from the analysis all conditions for which OD600 did not reach 0.06 in any sample, leaving a total of 51 conditions that were informative.
To analyze the growth data, we performed the same eight comparisons as on the gene expression data. Differences in growth abilities were tested using Wilcoxon’s rank-sum test. Those environments that showed significant growth differences in at least one of the seven comparisons between two evolved samples (48 environments in total) were clustered based on their patterns of growth using the QT CLUST algorithm (43) in TM4 software (44). We used the Pearson correlation metric distance with a maximum cluster diameter of 0.40 and a minimum cluster size of five; similar clusters were obtained when these parameters were varied.
Competition Experiments.
We performed seven sets of pairwise competition experiments to determine whether the L and S lineages from the same or different generations could invade one another when rare. The pairs included 6.5K-S vs. 6.5K-L, 17K-S vs. 17K-L, 40K-S vs. 40K-L, 6.5K-S vs. 17K-L, 6.5K-L vs. 17K-S, 17K-S vs. 40K-L, and 17K-L vs. 40K-S. Each competition experiment ran for 24 h (one complete serial transfer cycle) in the same medium and conditions as used in the long-term evolution experiment (19). The competitors were mixed at two initial ratios (1:9 and 9:1), and 15 replicate experiments were conducted for each pair and each initial ratio. The L and S competitors were distinguished on the basis of an arabinose utilization marker that is neutral (i.e., has no significant effect on fitness) under these conditions (19, 21), with each marker state used for each competitor in approximately half of the replicates. This marker allows the scoring of strains by colony color when plated on an appropriate medium. The fitness of one competitor relative to the other was calculated as the ratio of their net growth rates during the competition experiment (19, 21). The Student t test was used to evaluate whether the mean fitness differed from the null hypothetical expectation of 1 for each pair of competitors at each initial ratio.
Supplementary Material
Acknowledgments
We thank D. Schluter and P. B. Rainey for helpful comments on this paper. This work was supported by Agence Nationale de la Recherche Program “Blanc” ANR-08-BLAN-0283-01, Centre National de la Recherche Scientifique, and Université Joseph Fourier (D.S.); and by National Science Foundation DEB-1019989 and BEACON Center for the Study of Evolution in Action National Science Foundation Cooperative Agreement DBI-0939454 (R.E.L.). M.L.G. was supported by a research fellowship from the Agence Nationale de la Recherche. J.P. was supported by a doctoral fellowship from the French Ministry of Research and the Université Joseph Fourier.
Footnotes
The authors declare no conflict of interest.
Data deposition: The data supporting this paper have been deposited in the DRYAD database, http://datadryad.org (accession: http://dx.doi.org/10.5061/dryad.d7k5r5d2). The microarray data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, http://www.ncbi.nlm.nih.gov/geo (accession no. GSE30639).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1207091109/-/DCSupplemental.
References
- 1.Gause GF. Experimental studies on the struggle for existence, I: Mixed population of two species of yeast. J Exp Biol. 1932;9:389–402. [Google Scholar]
- 2.Hutchinson GE. Concluding remarks. Cold Spring Harb Symp Quant Biol. 1957;22:415–427. [Google Scholar]
- 3.Hardin G. The competitive exclusion principle. Science. 1960;131:1292–1297. doi: 10.1126/science.131.3409.1292. [DOI] [PubMed] [Google Scholar]
- 4.Holt RD. Bringing the Hutchinsonian niche into the 21st century: Ecological and evolutionary perspectives. Proc Natl Acad Sci USA. 2009;106(Suppl 2):19659–19665. doi: 10.1073/pnas.0905137106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Peterson AT, Soberon J, Sanchez-Cordero V. Conservatism of ecological niches in evolutionary time. Science. 1999;285:1265–1267. doi: 10.1126/science.285.5431.1265. [DOI] [PubMed] [Google Scholar]
- 6.Wiens JJ, Graham CH. Niche conservatism: Integrating evolution, ecology, and conservation biology. Annu Rev Ecol Evol Syst. 2005;36:519–539. [Google Scholar]
- 7.Losos JB. Phylogenetic niche conservatism, phylogenetic signal and the relationship between phylogenetic relatedness and ecological similarity among species. Ecol Lett. 2008;11:995–1003. doi: 10.1111/j.1461-0248.2008.01229.x. [DOI] [PubMed] [Google Scholar]
- 8.Gómez JM, Verdú M, Perfectti F. Ecological interactions are evolutionarily conserved across the entire tree of life. Nature. 2010;465:918–921. doi: 10.1038/nature09113. [DOI] [PubMed] [Google Scholar]
- 9.Wiens JJ, et al. Niche conservatism as an emerging principle in ecology and conservation biology. Ecol Lett. 2010;13:1310–1324. doi: 10.1111/j.1461-0248.2010.01515.x. [DOI] [PubMed] [Google Scholar]
- 10.Brown WL, Wilson EO. Character displacement. Syst Zool. 1956;5:49–64. [Google Scholar]
- 11.Schluter D. The Ecology of Adaptive Radiations. Oxford: Oxford Univ Press; 2000. [Google Scholar]
- 12.Grant PR, Grant BR. How and Why Species Multiply. Princeton: Princeton Univ Press; 2008. [Google Scholar]
- 13.Pfennig KS, Pfennig DW. Character displacement: Ecological and reproductive responses to a common evolutionary problem. Q Rev Biol. 2009;84:253–276. doi: 10.1086/605079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brockhurst MA, Hochberg ME, Bell T, Buckling A. Character displacement promotes cooperation in bacterial biofilms. Curr Biol. 2006;16:2030–2034. doi: 10.1016/j.cub.2006.08.068. [DOI] [PubMed] [Google Scholar]
- 15.Ricklefs RE. Evolutionary diversification, coevolution between populations and their antagonists, and the filling of niche space. Proc Natl Acad Sci USA. 2010;107:1265–1272. doi: 10.1073/pnas.0913626107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rummel J, Roughgarden J. A theory of faunal buildup for competition communities. Evolution. 1985;39:1009–1033. doi: 10.1111/j.1558-5646.1985.tb00444.x. [DOI] [PubMed] [Google Scholar]
- 17.Taper ML, Case TJ. Models of character displacement and the theoretical robustness of taxon cycles. Evolution. 1992;46:317–333. doi: 10.1111/j.1558-5646.1992.tb02040.x. [DOI] [PubMed] [Google Scholar]
- 18.Hansen SK, Rainey PB, Haagensen JAJ, Molin S. Evolution of species interactions in a biofilm community. Nature. 2007;445:533–536. doi: 10.1038/nature05514. [DOI] [PubMed] [Google Scholar]
- 19.Lenski RE, Rose MR, Simpson SC, Tadler SC. Long-term experimental evolution in Escherichia coli, I: Adaptation and divergence during 2,000 generations. Am Nat. 1991;138:1315–1341. [Google Scholar]
- 20.Barrick JE, et al. Genome evolution and adaptation in a long-term experiment with Escherichia coli. Nature. 2009;461:1243–1247. doi: 10.1038/nature08480. [DOI] [PubMed] [Google Scholar]
- 21.Rozen DE, Lenski RE. Long-term experimental evolution in Escherichia coli, VIII: Dynamics of a balanced polymorphism. Am Nat. 2000;155:24–35. doi: 10.1086/303299. [DOI] [PubMed] [Google Scholar]
- 22.Rozen DE, Schneider D, Lenski RE. Long-term experimental evolution in Escherichia coli, XIII: Phylogenetic history of a balanced polymorphism. J Mol Evol. 2005;61:171–180. doi: 10.1007/s00239-004-0322-2. [DOI] [PubMed] [Google Scholar]
- 23.Rozen DE, Philippe N, de Visser JA, Lenski RE, Schneider D. Death and cannibalism in a seasonal environment facilitate bacterial coexistence. Ecol Lett. 2009;12:34–44. doi: 10.1111/j.1461-0248.2008.01257.x. [DOI] [PubMed] [Google Scholar]
- 24.Odling-Smee FJ, Laland KN, Feldman MW. Niche construction. Am Nat. 1996;147:641–648. [Google Scholar]
- 25.Laland KN, Odling-Smee FJ, Feldman MW. Evolutionary consequences of niche construction and their implications for ecology. Proc Natl Acad Sci USA. 1999;96:10242–10247. doi: 10.1073/pnas.96.18.10242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Erwin DH. Macroevolution of ecosystem engineering, niche construction and diversity. Trends Ecol Evol. 2008;23:304–310. doi: 10.1016/j.tree.2008.01.013. [DOI] [PubMed] [Google Scholar]
- 27.Kylafis G, Loreau M. Ecological and evolutionary consequences of niche construction for its agent. Ecol Lett. 2008;11:1072–1081. doi: 10.1111/j.1461-0248.2008.01220.x. [DOI] [PubMed] [Google Scholar]
- 28.Cooper TF, Rozen DE, Lenski RE. Parallel changes in gene expression after 20,000 generations of evolution in Escherichia coli. Proc Natl Acad Sci USA. 2003;100:1072–1077. doi: 10.1073/pnas.0334340100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Crozat E, Philippe N, Lenski RE, Geiselmann J, Schneider D. Long-term experimental evolution in Escherichia coli, XII: DNA topology as a key target of selection. Genetics. 2005;169:523–532. doi: 10.1534/genetics.104.035717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Philippe N, Crozat E, Lenski RE, Schneider D. Evolution of global regulatory networks during a long-term experiment with Escherichia coli. Bioessays. 2007;29:846–860. doi: 10.1002/bies.20629. [DOI] [PubMed] [Google Scholar]
- 31.Peekhaus N, Conway T. What’s for dinner?: Entner–Doudoroff metabolism in Escherichia coli. J Bacteriol. 1998;180:3495–3502. doi: 10.1128/jb.180.14.3495-3502.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kornberg H. Krebs and his trinity of cycles. Nat Rev Mol Cell Biol. 2000;1:225–228. doi: 10.1038/35043073. [DOI] [PubMed] [Google Scholar]
- 33.Plumbridge J. Regulation of gene expression in the PTS in Escherichia coli: The role and interactions of Mlc. Curr Opin Microbiol. 2002;5:187–193. doi: 10.1016/s1369-5274(02)00296-5. [DOI] [PubMed] [Google Scholar]
- 34.Cooper VS, Lenski RE. The population genetics of ecological specialization in evolving Escherichia coli populations. Nature. 2000;407:736–739. doi: 10.1038/35037572. [DOI] [PubMed] [Google Scholar]
- 35.Sniegowski PD, Gerrish PJ, Lenski RE. Evolution of high mutation rates in experimental populations of E. coli. Nature. 1997;387:703–705. doi: 10.1038/42701. [DOI] [PubMed] [Google Scholar]
- 36.Funchain P, et al. The consequences of growth of a mutator strain of Escherichia coli as measured by loss of function among multiple gene targets and loss of fitness. Genetics. 2000;154:959–970. doi: 10.1093/genetics/154.3.959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rainey PB, Travisano M. Adaptive radiation in a heterogeneous environment. Nature. 1998;394:69–72. doi: 10.1038/27900. [DOI] [PubMed] [Google Scholar]
- 38.Snyder TP, Ayala FJ. Frequency-dependent selection at the PGM-1 locus of Drosophila pseudoobscura. Genetics. 1979;92:995–1003. doi: 10.1093/genetics/92.3.995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jeong H, et al. Genome sequences of Escherichia coli B strains REL606 and BL21(DE3) J Mol Biol. 2009;394:644–652. doi: 10.1016/j.jmb.2009.09.052. [DOI] [PubMed] [Google Scholar]
- 40.Storey JD, Dai JY, Leek JT. The optimal discovery procedure for large-scale significance testing, with applications to comparative microarray experiments. Biostatistics. 2007;8:414–432. doi: 10.1093/biostatistics/kxl019. [DOI] [PubMed] [Google Scholar]
- 41.Leek JT, Monsen EC, Dabney AR, Storey JD. EDGE: Extraction and analysis of differential gene expression. Bioinformatics. 2006;22:507–508. doi: 10.1093/bioinformatics/btk005. [DOI] [PubMed] [Google Scholar]
- 42.Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci USA. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Heyer LJ, Kruglyak S, Yooseph S. Exploring expression data: Identification and analysis of coexpressed genes. Genome Res. 1999;9:1106–1115. doi: 10.1101/gr.9.11.1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Saeed AI, et al. TM4: A free, open-source system for microarray data management and analysis. Biotechniques. 2003;34:374–378. doi: 10.2144/03342mt01. [DOI] [PubMed] [Google Scholar]
- 45.Riley M, et al. Escherichia coli K-12: A cooperatively developed annotation snapshot—2005. Nucleic Acids Res. 2006;34:1–9. doi: 10.1093/nar/gkj405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gama-Castro S, et al. RegulonDB (version 6.0): Gene regulation model of Escherichia coli K-12 beyond transcription, active (experimental) annotated promoters and Textpresso navigation. Nucleic Acids Res. 2008;36(Database issue):D120–D124. doi: 10.1093/nar/gkm994. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.