Abstract
The organization of eukaryotic chromosomes into transcriptionally active euchromatin and repressed heterochromatin requires mechanisms that establish, maintain and distinguish these canonical chromatin domains. Post-translational modifications are fundamental in these processes. Monoubiquitylation of histones was discovered more than three decades ago, but its precise function has been enigmatic until recently. It is now appreciated that the spectrum of chromatin ubiquitylation is not restricted to monoubiquitylation of histones, but includes degradatory ubiquitylation of histones, histone-modifying enzymes and non-histone chromatin factors. These occur in a spatially and temporally controlled manner. In this review, we summarize our understanding of these mechanisms with a particular emphasis on how ubiquitylation shapes the physical landscape of chromatin.
Keywords: protein degradation, heterochromatin, histone methylation, monoubiquitylation, ubiquitin ligase
See Glossary for abbreviations used in this article.
Glossary.
- Ams2
CENP-A ts mutant multi-copy suppressor 2
- APC/C
anaphase promoting complex/cyclosome
- BRCA1
breast cancer 1
- Bre1
brefeldin A sensitivity 1
- Cdt1/2
chromatin licensing and DNA replication factor 1/2
- CENP-A
centromere protein A
- chromodomain
chromatin organizer modifier domain
- Clr4
cryptic loci regulator 4
- ClrC
Clr4 complex; H3K9 HMTase complex
- COMPASS
complex proteins associated with Set1
- COP9
constitutive photomorphogenic 9
- CRL
cullin-RING finger ligase
- Cse4
chromosome segregation protein 4
- Ctk1
carboxy-terminal domain kinase 1
- Cul
Cullin, ubiquitin ligase scaffold protein
- DCAF
Ddb1-Cul4-associated factor
- DCDC
Dim-5/7/9, Cul4 Ddb1 complex
- Ddb1
DNA damage-binding protein 1
- Dot1
disruptor of telomeric silencing 1
- DUB
deubiquitylase
- EED
embryonic ectoderm development
- Epe1
enhancer of position effect 1
- FACT
facilitates chromatin transcription, histone chaperone
- HMTase
histone methyltransferase
- Hub1
homologous to ubiquitin
- GAL1
galactose metabolism gene 1
- JmjC
Jumonji domain C
- Msc1
multi-copy suppressor of Chk1
- MSL
male-specific lethal
- N-CoR
nuclear receptor corepressor
- Nedd8
neural precursor cell expressed, developmentally downregulated 8
- PAF
polymerase II association factor
- PcG
Polycomb group
- PCNA
proliferating cell nuclear antigen
- PHD finger
plant homeo domain
- PIP box
PCNA interacting protein box
- Pof1
Pombe F-box 1
- PRC1/2
Polycomb repressor complex 1/2
- PR-DUB
Polycomb repressive deubiquitylating complex
- Psh1
Pob3/Spt16/histone associated protein
- Rad6
radiation-sensitive protein 6
- Raf1/2
Rik1-interacting factor 1/2
- rDNA
ribosomal DNA
- RING
really interesting new gene
- RNAPII
RNA polymerase II
- RNF
RING finger protein
- RYBP
Ring 1 and YY1 binding protein
- SCF
Skp1–Cullin–F-box complex
- SET
Su(var)3-9, Enhancer-of-zeste, Trithorax domain
- Skp
suppressor of kinetochore protein mutan
- Spp1/Cps40
Set1c, PHD finger protein/COMPASS subunit 40
- Spt
suppressor of ty
- SUMO
small ubiquitin-like modifier
- Swd2/Cps35
Set1c, WD40 repeat protein 2/COMPASS subunit 35
- Swi6
Switch 6
- SwrC
Swr1 complex
- TFIIS
transcription factor IIS
- Ubp
ubiquitin protease
- UBR
ubiquitin protein ligase E3 component n-recognin
- USP
ubiquitin-specific protease
- YY1
yin and yang 1, DNA-binding factor
- ZRF1
zuotin-related factor 1
Introduction
In eukaryotic cells, chromatin is compartmentalized into euchromatin—which is associated with active gene expression—and heterochromatin—which is weakly transcribed and characterized by the presence of repetitive DNA, including transposons. These domains are dynamic and respond to DNA, RNA and cell cycle signals. This regulation is achieved by chromatin modification mechanisms that include the exchange of canonical histone proteins with histone variants, the repositioning of nucleosomes and the post-translational modifications of histones. These modifications often control intra- and intermolecular protein–protein and protein–DNA interactions. For instance, histones are acetylated in chromatin regions that are actively transcribed with the addition of a negatively charged acetyl group, altering the net charge of the nucleosome and interfering with chromatin compaction, thereby facilitating access to the transcription machinery. In addition, acetyl groups serve as binding sites for bromodomain and other proteins that recognize this histone mark. By contrast, methyl groups act as docking sites for chromodomain and plant homeodomain finger proteins and are associated with either active or repressed chromatin. Notably, these histone modifications can be removed by specific deacetylases and demethylases. In addition, many of the histone modifications influence each other through trans-histone cross-talk. We focus here on a large post-translational modification of chromatin produced by the conjugation of ubiquitin.
The 76-residue protein ubiquitin is covalently attached to an internal lysine residue of a substrate by an enzymatic cascade, which includes an activating enzyme (E1), a conjugating enzyme (E2) and a ubiquitin ligase (E3) that provides substrate specificity to the ubiquitylation reaction. Whilst the E2 receives the activated ubiquitin from the E1 and mediates the transfer, the E3 provides specificity by binding to the substrate and recruiting it to the conjugation machinery through protein–protein interaction with the E2 enzyme, as in the case of RING domain E3 enzymes. Most organisms have only one E1 enzyme, but dozens of different E2 and hundreds of E3 enzymes, reflecting the need to provide effective substrate specificity [1].
E3 enzymes are often composed of multimeric complexes. Ubiquitin ligases, belonging to the evolutionarily well-conserved family of cullin-RING ligases (CRLs), have a modular architecture in which the individual domains for recruiting the E2 enzyme and substrate are confined to separate subunits, which assemble into large multimeric complexes [2]. Central to this type of E3 ligase is a cullin moiety that acts as a scaffold for recruiting a RING finger protein and, through an adaptor protein, a receptor for recognizing the substrate. Most of the cullin subtypes recruit different adaptor proteins that interact with a series of individual substrate receptors, thereby providing a large spectrum of specific CRL complexes. CRLs themselves are subject to post-translational modification by the ubiquitin-like modifier Nedd8, which controls the activity of the ligase. Conjugation of Nedd8 to the cullin moiety triggers a conformational change within the scaffold, which allows the carboxy-terminal-associated RING–E2 complex to be positioned in close proximity to the substrate receptor, bound at the amino-terminal cullin domain [2]. Conversely, Nedd8 is removed by a multimeric complex, the COP9 signalosome (CSN), thereby adding another layer of regulation that controls CRL activity.
The attachment of ubiquitin alters the function and cellular fate of a substrate either by regulating the interaction with other proteins, by competing with other post-translational modifications involving the same target lysine residue or by targeting it for degradation. As ubiquitin itself can be subject to ubiquitylation, through targeting of one of its seven internal lysine (Lys) residues, several rounds of ubiquitylation result in the formation of a polyubiquitin chain. Importantly, the choice of the individual lysine linkage directly affects the structure of the polyubiquitin chain and therefore has functionally different outcomes [1]. In particular, polyubiquitin containing Lys 48-, Lys 29- or Lys 11-linked chains target substrates for proteasomal degradation, whilst monoubiquitylation or Lys 63 polyubiquitin chains serve as signals in non-proteolytic events. Finally, as the attachment of the relatively bulky ubiquitin molecule can have steric consequences, especially if the substrate is a part of large macromolecules such as the chromatin fibre, ubiquitylation might directly alter the structure.
Despite the fact that histones were identified as the first substrates of ubiquitylation three decades ago, our knowledge of how this modification contributes to the formation of euchromatin and heterochromatin is still fragmentary. Furthermore, several roles for the ubiquitylation of non-histone proteins in defining the chromatin state have emerged recently. Here we discuss these ubiquitylation pathways with particular emphasis on the distinct mechanistic consequences—that is, structural changes, recruitment signals and proteasomal degradation (Fig 1)—through which ubiquitylation shapes the landscape of euchromatin and heterochromatin. For the role of ubiquitylation in other events, such as DNA repair and transcription factor licensing, we refer the reader elsewhere [3,4].
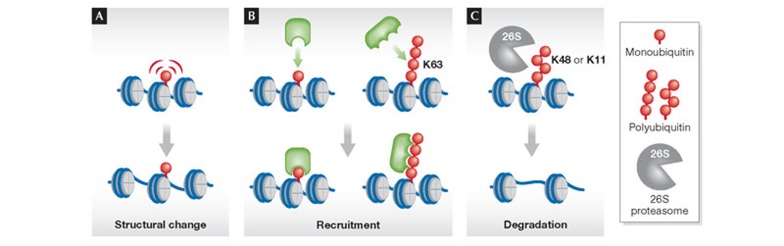
Figure 1. Mechanisms of ubiquitylation on chromatin.
(A) Structural changes: attachment of ubiquitin interferes with chromatin compaction. (B) Recruitment: monoubiquitin or non-degradatory polyubiquitin chains serve as a docking site for recruiting other factors to chromatin. (C) Degradation: polyubiquitin such as Lys 11 or Lys 48-linked chains serve as a recognition signal for proteasomal degradation. Note that the spectrum of ubiquitylated substrates is not restricted to histone proteins but includes also other chromatin-associated factors. K, lysine (Lys).
Ubiquitylation of H2B: docking site or structural change?
Histone H2B is monoubiquitylated within its C-terminus at Lys 123 in budding yeast (Lys 120 in metazoans), which is mediated by the E2 Rad6 and the E3 Bre1 (RNF20/RNF40 in mammals; [5,6,7,8]). The modified form makes up 1–5% of the total H2B pool and is restricted to actively transcribed euchromatin. H2B ubiquitylation seems to take place in a two-step reaction: first, Bre1 is recruited to promoters of active genes by binding to transcriptional activators, as seen, for example, for the human homologue of Bre1, RNF20, and the transcriptional co-activator p53 [9]. Bre1 in turn then mediates the recruitment of Rad6. Second, the activation of the Rad6–Bre1 complex requires a series of interactions with multiple factors implicated in subsequent steps of transcription, which might induce allosteric changes within the E2–E3 complex [10,11]. In addition, in metazoans, modification of Ser 112 of H2B with O-linked N-acetylglucosamine (GlcNAc) has been shown to promote ubiquitylation of H2B and its interaction with Bre1, suggesting that the GlcNAc moiety serves as an anchor for the E3 ligase [12].
Ubiquitylation of H2B (H2Bub) seems to be transient and can be reversed. The removal of this mark is mediated by specific deubiquitylation enzymes (DUBs). In Saccharomyces cerevisiae, this is achieved by Ubp8, a subunit of the SAGA histone acetyltransferase complex, and by Ubp10 [13,14]. Despite the fact that Ubp8 reverses H2B ubiquitylation by Rad6–Bre1, it promotes transcription. Ubp8 traverses with RNAPII through transcribed genes, and deubiquitylation is required for the recruitment of Ctk1, which in turn phosphorylates RNAPII, regulating transcriptional elongation [15]. It has thus been suggested that H2Bub might act as a checkpoint and that perhaps multiple rounds of ubiquitylation and deubiquitylation take place during transcriptional elongation [11]. By contrast, Ubp10 targets a different pool of H2Bub and functions mainly in heterochromatin associated with gene silencing at telomeric regions and the rDNA loci [16]. The fruit-fly homologue of Ubp10, scrawny (scny), functions in gene silencing in stem cells, and prevents untimely expression of key genes involved in differentiation [17]. In addition, other DUBs removing the H2Bub mark have been identified in metazoans (reviewed in [18]).
One main function of monoubiquitylated H2B is the regulation of other histone modifications, that is, the modification of Lys 4 and Lys 79 of histone H3 by the histone methyltransferases (HMTases) COMPASS and Dot1, respectively. This trans-histone cross-talk is unidirectional, that is, defects in the histone methylation of H3 have no reciprocal effect on the upstream ubiquitylation event [19,20]. H2Bub also has other functions in transcription that are independent from the cross-talk to H3 methylation, that is, assisting the histone chaperone FACT in facilitating the passage of transcribing RNAPII through nucleosomes and restoring the chromatin structure in the wake of the polymerase [21,22]. H2Bub also contributes to the repression of a subset of inducible genes that are expressed at low levels under basal conditions [23,24,25].
How does H2Bub mediate these different functions? Given the large size of ubiquitin, it was originally postulated that H2Bub acts as a ‘wedge’ to open chromatin, thereby making it accessible for downstream effectors such as histone HMTases. However, although H2Bub is required for trimethylation of H3K4 and H3K79, it is dispensable for monomethylation and most dimethylation, indicating that COMPASS and Dot1 still have access to chromatin even in the absence of ubiquitin [11]. Moreover, in vivo and in vitro data have demonstrated that other ubiquitin-like modifiers, for example SUMO and Hub1, fused to H2B cannot replace the function of ubiquitin, arguing that it is not merely the addition of steric bulk but rather the intrinsic property of ubiquitin that mediates the cross-talking function of H2Bub [25,26]. Thus, alternative models have been proposed, which we discuss below.
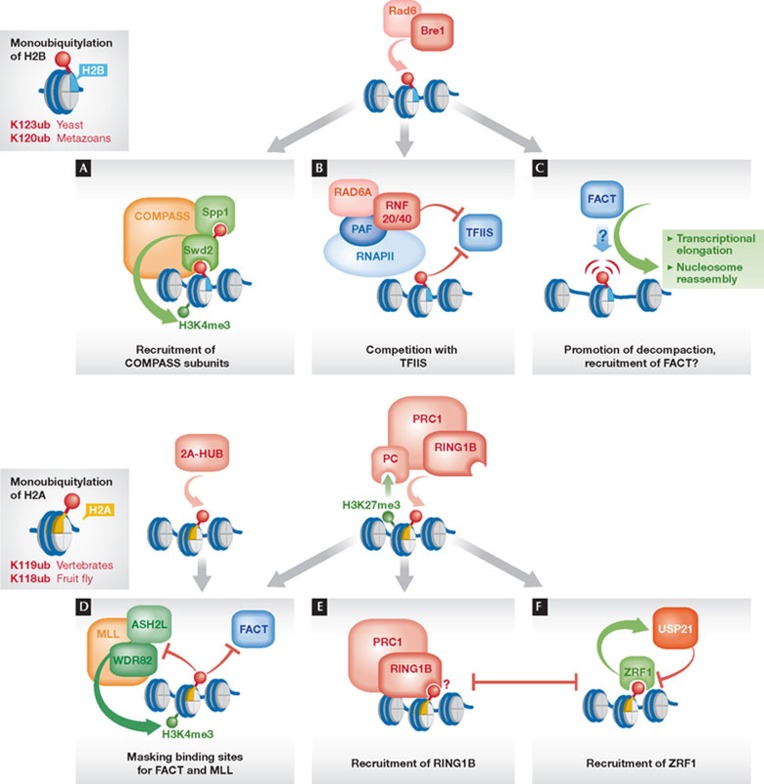
H2Bub might directly recruit subunits of the HMTases, thereby serving as a ‘bridge’ that connects the histone-modifying enzymes with chromatin (Fig 2A). Among the seven subunits of the yeast COMPASS complex, only three subunits (Swd2, Spp1 and Bre2) are essential for H3K4 trimethylation, making them likely candidates for regulating the activity of the HTMase complex [27]. Two studies link H2Bub and the COMPASS subunit Swd2 (Cps35). In the first study, it was shown that H2Bub is essential for the association of Swd2 with the COMPASS complex and mediates the recruitment of this subunit to the GAL1 promoter region. Additionally, Swd2 was shown to interact with Dot1 to control trimethylation of H3K79 in a similar manner, suggesting that the regulation of Swd2 recruitment might represent a universal mechanism by which H2Bub controls histone cross-talk [28]. An independent study revealed another link between Swd2 and the ubiquitylation machinery by demonstrating that Swd2 itself is a target of Rad6–Bre1, and that ubiquitylation of Swd2 also requires H2Bub. The corresponding target lysine residues in Swd2 were found to be required for the chromatin recruitment of another COMPASS subunit that is crucial for trimethylation of H3K4, Spp1 (Cps40) [29]. Although these two studies reported contrary data as to whether the association of Swd2 with COMPASS is regulated through H2B ubiquitylation, the main findings of both reports are consistent with a ‘bridging’ model of H2Bub function. Nevertheless, the structural features that mediate the recognition of the ubiquitin moiety still need to be identified in these chromatin regulators.
Figure 2. Shaping of chromatin through monoubiquitylation of histones H2B and H2A.
(A) H2Bub mediates recruitment of the COMPASS subunit Swd2 required for trimethylation of H3K4. Swd2 itself is ubiquitylated in a manner dependent on Rad6–Bre1, which mediates the association with Spp1, another COMPASS subunit involved in H3K4 trimethylation. (B) RNF20 and H2Bub negatively regulate chromatin recruitment of TFIIS. (C) H2Bub mediates structural changes by interfering with chromatin compaction, thereby facilitating the association of FACT to chromatin. (D) Ubiquitylation of H2A by the E3s 2A-HUB and RING1B negatively regulates the function or recruitment of the mammalian HMTase complex MLL and FACT. (E) H2Aub mediates binding of PRC1 to chromatin, possibly through direct recognition by RING1B. (F) H2Aub mediates the recruitment of ZRF1, which competes with PRC1 for binding to chromatin and facilitates the binding and/or function of the H2A deubiquitylase USP21. COMPASS, complex proteins associated with Set1, H3K4 HTMase complex; FACT, facilitates chromatin transcription, heterodimeric histone chaperone; H2Bub, H2B ubiquitylation; HMTase, histone methyltransferase; K, lysine (Lys); MLL, mixed leukaemia lineage complex, mammalian H3K4 HMTase; PAF, polymerase II association factor; PC, Polycomb; PRC1, Polycomb repressor complex 1; RING1B, really interesting new gene 1B; TFIIS, transcription factor IIS; USP21, ubiquitin-specific protease 21; ZRF1, zuotin-related factor 1.
Whereas H2Bub provides a positive function in recruiting subunits of HMTase complexes to chromatin, its cross-talk-independent role in gene repression of certain inducible genes in yeast and mammals seems to rely on preventing chromatin access. A study in human cells revealed that the repression of pro-oncogenic genes relates to the blocking of the chromatin recruitment of the transcription elongation factor TFIIS, which relieves stalled RNAPII [23]. In particular, RNF20—the mammalian homologue of Bre1—interferes with the association of TFIIS to the PAF complex that travels along with RNAPII through chromatin. As RNF20 interacts with PAF, it might directly compete with TFIIS for binding (Fig 2B). Nevertheless, this negative regulation requires the E3 activity of RNF20, suggesting that ubiquitylation of H2B also contributes to this repressive effect. In S. cerevisiae, H2Bub was also found to repress transcription of lowly expressed or quiescent genes, by inhibiting the recruitment of RNAPII to the promoters of these genes [30].
In addition to controlling the recruitment of factors to chromatin by providing or preventing binding sites, several studies have proposed that H2Bub affects the structural properties of chromatin by modulating nucleosome occupancy and chromatin compaction. For instance, loss of H2Bub causes a genome-wide reduction of histone occupancy, particularly at highly expressed genes [30], whilst mutants defective in deubiquitylating H2B show elevated histone levels [25,30]. Moreover, H2Bub cooperates with the FACT subunit Spt16 in maintaining histone occupancy, and both factors influence each other [31]. As changes in nucleosome occupancy can result from diverse mechanisms, that is, nucleosome stability or reassembly, different conclusions have been drawn from these observations. For instance, it has been proposed that H2Bub might promote nucleosome stability and thus contribute to compaction instead of ‘opening’ chromatin [25]. Alternatively, H2Bub has been suggested to promote reassembly of nucleosomes and restore the chromatin structure, along with Spt16, in the wake of elongating RNAPII [30,31]. Interestingly, a recent in vitro study using reconstituted nucleosomes showed that H2Bub causes disruption of local chromatin structure and acts synergistically with histone H4 tail acetylation [26], thus favouring a model by which H2Bub promotes chromatin decompaction. This property is specific to ubiquitin and was not observed for the ubiquitin-like modifier Hub1. Consistent with these in vitro data, H2Bub was found to be present specifically in those chromatin fractions that show increased accessibility and are associated with active transcription, arguing in favour for a similar in vivo role of H2Bub in the disruption of higher-order chromatin structure (Fig 2C). It is not clear how ubiquitin mediates this function in decompaction and why the attachment of similar-sized modifiers is ineffective. It is possible that intrinsic properties of ubiquitin, such as the distribution of surface charges or specific residues, contribute to this function of H2Bub. Hence, whether it is specifically the decompaction of chromatin that promotes the recognition of nucleo-somes by FACT, or whether its subunits directly bind to H2Bub remains to be investigated.
In addition to the canonical Lys 123 target site in yeast (Lys 120 in mammals), other lysine residues are targeted for ubiquitylation in H2B [32,33]. Importantly, in yeast, an H2B mutant that lacks all lysine residues but retains Lys 123 was seen to be severely compromised in GAL gene activation and displayed significant growth defects [32]. Notably, whilst neither Rad6 nor Bre1—nor the Lys 123 residue in H2B—are essential under normal growth conditions, the Lys 123-only mutant becomes inviable in the absence of Bre1. Similarly, mutating all lysine residues in H2B was found to be lethal. Interestingly, other lysine residues were found to be ubiquitylated in vivo in this study. These findings imply two important conclusions. First, ubiquitylation of H2B is essential for viability. Second, this essential function can be compensated by non-canonical lysine residues, and might involve other ubiquitylation reactions and probably other E3 ligases as Rad6–Bre1 is specific to Lys 123 ubiquitylation—although it cannot be excluded that other post-translational modifications might contribute to the crucial functions of these lysine residues. This study further demonstrates that the potential target lysine residues including Lys 123 are modified with more than one ubiquitin moiety, giving rise to specifically Lys 48-linked polyubiquitin chains [32]. This raises the intriguing question as to whether monoubiquitylated Lys 123 is the only active form or whether polyubiquitin chains—or their trimming—might have a physiological function. Notably, direct evidence for the implication of other E3 ligases was provided by a study in mammals that demonstrated that MSL2, a component of the dosage compensation complex (DCC; also known as MSL), has E3 activity towards H2B with a strong preference for the lysine residue Lys 134 [33]. Lys 134ub was found to affect the cross-talk towards methylation of H3K4 and H3K79 in a similar way as seen for Lys 120ub, suggesting that this trans-histone interaction has a certain degree of plasticity. Lys 134ub also had an impact on Lys 120 ubiquitylation, probably by directly affecting the chromatin recruitment of the E3. Whether Lys 120ub and Lys 134ub have identical roles, or instead provide synergistic contributions by complementing each other through similar functions, needs to be examined by future studies.
In conclusion, monoubiquitylation of H2B seems to fulfil a dual function. First, it provides a binding site for chromatin regulators such as HMTases, but also prevents the chromatin association of other factors of the transcriptional machinery through direct competition. Second, H2Bub induces structural changes directly to the chromatin fibre, thereby affecting the three-dimensional organization and compaction of chromatin, although it is possible that it might cause steric effects through the recruitment of other proteins. Nevertheless, it is clear that the underlying mechanisms by which H2B ubiquitylation functions are complex and not fully understood.
Monoubiquitin on H2A—same molecule, opposite function?
In contrast to H2B, monoubiquitylated H2A is associated with inactive chromatin at the pericentromeric regions, the inactive X chromosome and silenced developmental genes. Between 10% and 15% of the cellular pool of H2A is monoubiquitylated on Lys 119 in vertebrates (Lys 118 in Drosophila), but this modification has not been detected in yeast species [10]. Several E3 enzymes contribute to ubiquitylating H2A, showing different specificity with respect to the chromatin environment. Human RING1B was the first E3 identified [34] and is part of the Polycomb repressor complex (PRC1). PRC1 contains a chromodomain protein, Polycomb, and its recruitment to chromatin is believed to occur downstream from H3K27 methylation by PRC2. Two additional RING domain proteins are present in human PRC1, RING1A and BMI1. Although BMI1 only stimulates the activity of RING1B and does not have intrinsic E3 activity, RING1A seems to be able to substitute the function of RING1B in vivo [11]. RING1B is also present in the PRC1-like complexes dRAF (RING-associated factor) and mammalian BCOR (BCL6 co-repressor), and for Drosphilia melanogaster the main H2A ubiquitylation activity was reported to reside in the dRAF complex [35,36]. RING1B also modifies the histone variant H2A.Z. Similarly to H2Aub, monoubiquitylated H2A.Z is found predominantly at the inactivated X chromosome [37]. However, it is not clear whether H2A.Zub has a specific function distinct from H2Aub, or whether RING1B can discriminate between H2A and H2A.Z.
Although RING1B is responsible for the bulk of H2Aub, other E3 ligases seem to have more specific functions in silencing. In macrophages, the E3 2A-HUB, also known as hRUL138, localizes as part of the repressive N-CoR histone deacetylase 1/3 complex to promoter regions of a subset of chemokine genes, and prevents the recruitment of the FACT subunit SPT16 required for RNAPII elongation [38]. In murine spermatocytes, the E3 UBR2 is required for fertility and ubiquitylates H2A at unsynapsed chromosomal regions during the process of meiotic chromatin silencing [39]. The tumour repressor BRCA1 contains a RING domain and ubiquitylates H2A in vitro. Notably, BRCA1-dependent H2Aub is required for transcriptional silencing of pericentromeric tandem repeat satellite-DNA, and de-repression of these repeats was found to phenocopy many of the cellular traits associated with BRCA1 loss [40]. The defect in silencing of satellite-DNA is specific to BRCA1 as neither the loss of RING1B, nor of another H2A E3 involved in DNA damage repair (RNF8), display these phenotypes. Thus, this finding suggests that the tumour suppression function of BRCA1 might originate from heterochromatin-mediated silencing through H2Aub.
Several deubiquitylating enzymes have been identified that reverse the ubiquitylation of H2A and often antagonize the silencing role of H2Aub [11,18]. However, a recent finding revealed a more complex function in the context of H2A ubiquitylation. In Drosophila, the ubiquitin C-terminal hydrolase Calypso is part of the PR-DUB complex that mediates the deubiquitylation of H2Aub [41]. Nevertheless, PR-DUB binds to PcG genes and mutants of this complex, which show surprisingly similar defects in silencing as seen for the loss of H2Aub. Moreover, the combined depletion of PR-DUB and RING1B results in more severe phenotypes than observed for the depletion of the single components. These findings imply that ubiquitylation and deubiquitylation of H2A are not simply reciprocal functions. It has thus been proposed that the E3 complexes and PR-DUB might act either locally on different target genes or together in a temporally controlled manner, similarly to the cycles of adding and removing the H2Bub mark during transcriptional elongation [41].
Considering these multiple roles, raises the question about the underlying mechanism by which H2Aub exerts its silencing function. After the initial step of recruitment to transcriptional start sites, RNAPII is often regulated by transcriptional pausing, and its elongation requires the association of ancillary factors. RING1B-dependent H2Aub seems to specifically cause pausing without affecting the initial recruitment of RNAPII [42]. This idea is further supported by the observations that the E3 2A-HUB also enhances RNAPII pausing by interfering with the recruitment of FACT, and that the FACT subunit Spt16 no longer binds to H2Aub [38]. These findings suggest that at least one function of monoubiquitylation of H2A is to prevent transcriptional elongation by masking specific recruitment sites for FACT (Fig 2D). In addition, H2Aub was found to block in vitro H3K4 methylation and transcriptional initiation, probably also by blocking the access for the HMTase to the substrate [43].
In addition to preventing chromatin access to regulators involved in transcription, H2Aub seems to provide a specific binding site for repressive factors (Fig 2E). A study reported that RING1B binds to ubiquitin and ubiquitylated H2A, suggesting that the E3 might recognize the mark that it deposits [44]. This might help to maintain PRC1 complexes bound at silenced chromatin or contribute to the spreading of PRC1 along the chromosomal axis. Indeed, the ability of a ‘writer’ to ‘read’ its own histone mark is a common phenomenon and has been observed for the PRC2 complex that methylates H3K27 and binds to this mark through its EED subunit [45]. Furthermore, this finding might explain why PRC1 and the related dRAF/BCoR complexes can be recruited to chromatin independently of the chromodomain protein Polycomb—which is in fact absent in several RING1 complexes—and the methylation of H3K27 by PRC2. Another interesting link to H2Aub is provided by the finding that the mammalian protein RYBP, involved in chromatin targeting of PRC complexes, contains an N-terminal ubiquitin-binding motif, an N-terminal zinc finger domain, and shows a weak affinity to ubiquitin [46]. RYBP is an adaptor protein that mediates the recruitment of RING1A/RING1B to the DNA-binding factor YY1 [47]. Furthermore, RYBP competes with Polycomb for binding to RING1A and RING1B [48], suggesting that distinct targeting modes—H3K27me compared with YY1 DNA binding sites—are mediated by different complex compositions of PRC1. Thus, an attractive idea would be that chromatin targeting of RYBP might also be influenced by the presence of H2Aub through its ubiquitin-binding domain.
In addition to its repressive role, H2Aub also mediates the de-repression of PcG target genes. This function is mediated through the ubiquitin-binding protein ZRF1, which binds to H2Aub (Fig 2F; [44]). ZRF1 acts here by a two-step mechanism: first, it displaces PRC1 from chromatin through competitive binding to H2Aub; second, it facilitates the removal of the ubiquitin mark by the H2Aub-specific deubiquitylase USP21, which is implicated in transcriptional initiation by relieving the H2Aub-dependent block of H3K4 methylation [43]. Hence, H2Aub mediates its own removal through the recruitment of ZRF1.
Whether H2Aub acts by similar mechanisms in other repressive pathways, such as BRCA1-mediated silencing of pericentromeric heterochromatin, remains to be tested. However, H2A can be polyubiquitylated in vitro by UBR2 [39], an E3 ligase involved in degradation of substrates by the N-end rule proteasome degradation pathways. Thus, it is also possible that degradatory mechanisms might be triggered by H2A ubiquitylation.
In summary, monoubiquitylation of histone H2A seems to have a similar mechanism to that of H2B by providing specific binding sites for downstream factors, or by preventing access to chromatin. However, the functional outcome of these two modifications is opposite. It is not yet understood whether the ‘repelling’ function of H2Aub is of a steric nature or whether it is due to the recruitment of other effectors that mediate its inhibitory effect, nor do we understand what differences between H2Bub and H2Aub might explain the different roles in decompaction and silencing of chromatin.
Shaping chromatin through turnover of histones
Whilst monoubiquitylated H2A and H2B serve as signals for regulating the recruitment to chromatin and its compaction, there is also increasing evidence that histone degradation is fundamental in shaping chromatin. Cells have to prevent the accumulation of large pools of free histone proteins, especially as they are largely positively charged, for instance to prevent unspecific binding to off-targets other than DNA, for example, other nucleic acids and proteins. Therefore, histone levels are tightly controlled throughout all species by regulating both protein synthesis and turnover.
The expression of canonical histone proteins is restricted to S phase, when chromatin is duplicated. In fission yeast, the transcription of histone genes is temporally controlled through the presence of the transcription factor Ams2, which is stable in S phase but degraded by the SCFPof1 E3 ligase complex in other phases of the cell cycle. Notably, mutants defective in Ams2 turnover show defects in chromosome stability and abnormal incorporation of histone H3 into centromeric regions that usually contain the centromere-specific histone H3 variant CENP-A [49]. Thus, the timely controlled expression of canonical histones is required to prevent their ectopic chromosomal distribution and to ensure the proper formation of the centromeres. Although a homologue of Ams2 is not present in budding yeast, excessive levels of histones are prevented, by targeting them directly for degradation with the E3 Tom1 through a surveillance mechanism that relies on the kinase Rad53 [50].
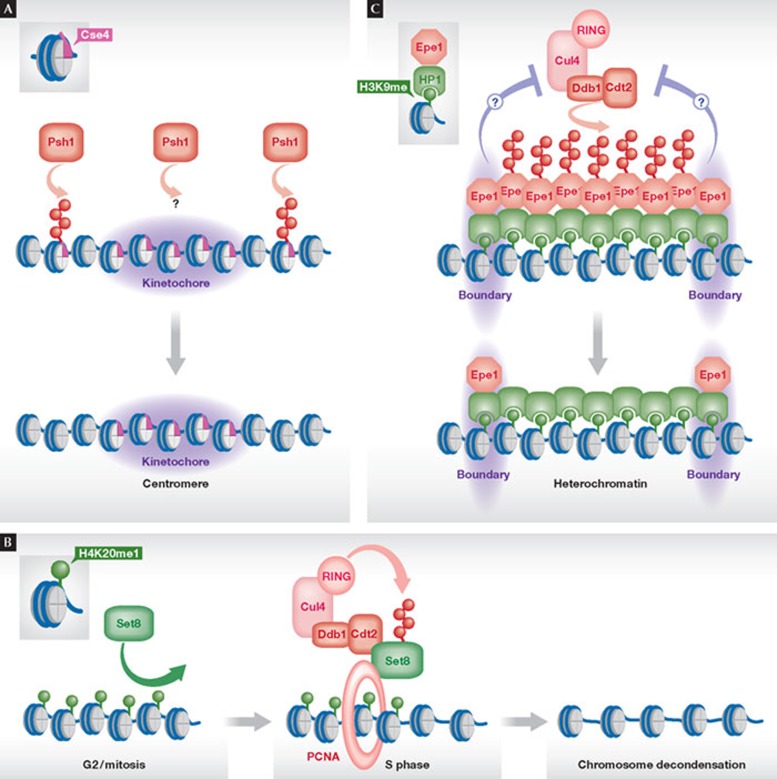
The centromeric H3 variant CENP-A is subject to spatial regulation by ubiquitin-dependent degradation. Centromeric nucleosomes contain the specific H3 CENP-A variant that is recruited to centromeres through its CENP-A-targeting domain (CATD). This is engaged with histone chaperones and kinetochore proteins during the targeting and after its incorporation into centromeres, respectively. Two studies demonstrated that the chromosomal distribution of the CENP-A budding yeast homologue Cse4 is shaped through ubiquitylation by the E3 RING finger ligase Psh1 (Fig 3A; [51,52]). In mutants of Psh1, Cse4 accumulates within euchromatin and overexpression of this histone variant is toxic. Interestingly, the CATD is necessary and sufficient for targeting Cse4 for degradation, indicating that this domain is the key element that provides specificity and promotes degradation of Cse4 when not properly engaged with kinetochore proteins outside centromeres. Similar observations have been made for the Drosophila CENP-A homologue CID [53]. Degradation might require the eviction of Cse4 to expose fully the CATD before its ubiquitylation. Alternatively, degradation might take place directly on chromatin; Psh1 was found to co-localize with Cse4 on chromatin [51]. This finding implies that Psh1 is important not only for preventing ectopic localization of Cse4, but plays a role at the centromeres. Interestingly, Psh1 was originally identified through its interaction with FACT [54], suggesting that additional chromatin-associated factors or substrates might be involved in this regulation.
Figure 3. Shaping of chromatin by ubiquitin-dependent degradation.
(A) Shaping of centromeric chromatin by degradation of the histone H3 variant Cse4 by the E3 Psh1 at ectopic sites outside centromeres. (B) Shaping of S-phase-specific chromatin by degradation of the histone H4K20 mono-HMTase Set8/PR-Set7 by the E3 Cul4–Ddb1Cdt2. Polyubiquitylation requires the formation of a trimeric complex of Set8, Cul4–Ddb1Cdt2 and PCNA. (C) Shaping of heterochromatin boundaries through degradation of the anti-silencing factor Epe1 by Cul4–Ddb1Cdt2. Epe1 is recruited uniformly to heterochromatin by binding to HP1 proteins. Epe1 degradation within the body of heterochromatin results in its local accumulation at the boundaries. The signals preventing degradation of Epe1 at the boundaries are unknown. Cul4, Cullin 4; Cdt2, chromatin licensing and DNA replication factor 2; Ddb1, DNA damage-binding protein 1; Epe1, enhancer of position effect 1; HP1, heterochromatin protein 1; Set7/8, Sul(Var)3–9, Enhancer-of-zeste, Trithorax protein 7/8; PCNA, proliferating cell nuclear antigen processivity factor for DNA polymerase δ.
Shaping chromatin through turnover of histone modifiers
Among the various CRLs, the Cul4-based E3 ligases have a specialized role in regulating chromatin-associated substrates [55]. Cul4 binds to the adaptor protein Ddb1, involved in recruiting various substrate receptors known as DCAFs (Ddb1–Cul4-associated factors). Several studies revealed how CRLs contribute to the shaping of chromatin by degrading the enzymes regulating histone modifications.
The CRL Cul4–Ddb1Cdt2 is conserved from fission yeast to human and is pivotal in regulating S phase, by targeting several substrates including the replication licensing factor Cdt1. Most substrates of Cul4–Ddb1Cdt2 have a common recruitment mechanism to chromatin through binding to the DNA polymerase processivity factor PCNA. In most cases, binding to PCNA seems to be crucial for the degradation of substrates, explaining the substrate specificity of this E3 for chromatin-bound proteins. In metazoans, those substrates have a specialized form of the common PIP (PCNA interaction protein) box known as the ‘PIP degron’, which contains an extra basic residue that is crucial for substrate recognition and might make contact with the substrate receptor Cdt2 [56] In fission yeast, the PIP degron is less conserved. A search for potential candidates containing the PIP degron motif led to the identification of the HMTase Set8, also known as PR-Set7, as a new substrate of Cul4–Ddb1Cdt2 [57,58,59,60,61]. Set8 is the only HMTase that mediates monomethylation of histone H4 at Lys 20, a transcriptionally repressive mark that is crucial for chromosome condensation during mitosis. H4K20 methylation levels are low in G1 and early S phase but rise in late S phase and G2, and peak in mitosis. The temporal regulation of this histone modification is reflected by a change in steady state levels of Set8, which is subject to distinct E3s acting at different cell cycle stages: APC/CCdh1 (G1), Cul1–Skp1Skp2 (late G1) and Cul4–Ddb1Cdt2 (S phase and after DNA damage; [56]). Degradation of Set8 by Cul4–Ddb1Cdt2 requires its interaction with PCNA and the presence of a highly conserved PIP degron. Non-degradable mutants of Set8 lacking this motif resulted in elevated H4K20me levels, DNA damage, check-point activation and G2 arrest, and in some cases premature chromosome condensation. Thus, the timely destruction of Set8 ensures the dynamic changes in H4K20 methylation during the passage through the cell cycle (Fig 3B). Further examples underscore the generality of the regulation of histone modifiers, through temporally controlled turnover, by other E3 ligases (see Table 1; [62,63,64]).
Table 1. Roles of E3 ligases in chromatin.
| E3 | Species | Substrate | Process | Mode | Mechanism | Recruitment | Reference | |
|---|---|---|---|---|---|---|---|---|
| Bre1 | Sc Sp Dm Hs | H2B-K123 | Txn initiation | E | Mono | Recruitment | HMTase | 15,16 |
| Sc Hs | H2B-K123 | Txn elongation | E | Mono | Decompact | FACT? | 12,13,18 | |
| Hs | H2B-K123 | Txn repression | E | Mono | Competition | TFIIS | 17 | |
| MSL2 | Hs | H2B-134 | Txn initiation | E | Mono | Recruitment? | HMTase? Bre1? | 20 |
| RING1B | Dm, Hs | H2A (H2A.Z) | Txn repression | H | Mono | Masking | HTMase, FACT | 23,27,28 |
| Hs | H2A | Txn repression | H | Mono | Recruitment | PRC1 | 29 | |
| Hs | H2A | Txn de-repress. | H | Mono | Recruitment | ZRF1 | 29 | |
| Hs | H2A | Txn repression | H | Mono | Recruitment | RYBP? | 31 | |
| 2A-HUB | Hs | H2A | Txn repression | H | Mono | Masking | FACT | 23 |
| UBR2 | Mm | H2A | Meiotic silenc. | H | Poly? | Masking? | FACT? | 24 |
| BRCA1 | Hs | H2A | Txn repression | H | Mono | Masking? | FACT? | 25 |
| SCFPof1 | Sp | Ams2 | Histone levels | S | Poly | Degradation | Proteasome | 34 |
| Tom1 | Sc | histones | Histone levels | S | Poly | Degradation | Proteasome | 35 |
| Psh1 | Sc Sp Dm Hs | Cse4 | Histone incorp. | E | Poly | Degradation | Proteasome | 36, 37 |
| SCFPpa | Ds, Hs | CID | Histone incorp. | E | Poly | Degradation | Proteasome | 38 |
| Cul4–Ddb1Cdt2 | Hs | Set8/PR-Set7 | Condensation | E | Poly | Degradation | Proteasome | 42,43,44,45,46 |
| SCFSkp2 | Hs | MLL | Cell cycle | S | Poly | Degradation | Proteasome | 47 |
| APC/C | Hs | MLL | Cell cycle | S | Poly | Degradation | Proteasome | 47 |
| SCFFbx4 | Hs | JMJD2A | Cell cycle | S | Poly | Degradation | Proteasome | 48 |
| Not4 | Sc (Hs) | Jhd2 (JARID1C) | Txn | S | Poly | Degradation | Proteasome | 49 |
| Cul4–Ddb1Cdt2 | Sp | Epe1 | Boundary | H | ? | ? | ? | 50 |
| Cul4–Rik1Raf1/2 | Sp | ? | H3K9me | H | ? | ? | ? | 51,52,53,54,55 |
| Cul4–Ddb1DCAF26 | Nc | ? | H3K9me | H | ? | ? | ? | 56,57,58,59 |
| Cul2/5–ELC | Cr | ? | H3K9me | H | ? | ? | ? | 60 |
| Cul4–Ddb1Msi1 | At | ? | H3K27me | H | ? | ? | ? | 61 |
| Cul4–Ddb1EED | Hs | ? | H3K27me | H | ? | ? | ? | 62 |
| Msc1 | Sp | ? | HP1 dynamics | H | ? | ? | ? | 63,64,65,66,67,68 |
| Cul8–Mms1Mms22 | Sc | Ctf4? | Silencing | H | ? | ? | ? | 69 |
| SCFPof3 | Sp | Mcl1 (Ctf4)? | Silencing | H | ? | ? | ? | 70 |
| APC/C | Sp | ? | H3K9me | H | ? | ? | ? | 71 |
2A-HUB, 2A-histone ubiquitin ligase; APC/C, anaphase promoting complex/cyclosome; At, Arabidopsis thaliana; BRCA1, breast cancer 1; Bre1, brefeldin A sensitivity; Cr, Chlamydomonas reinhardtii; Ctf4, chromosome transmission fidelity; Cul2/4/5/8, Cullin 2/4/5/8; Ddb1, DNA damage-binding protein 1; de-repress., de-repression; Dm, Drosophila melanogaster; E, euchromatin; ELC, elongin E; Epe1, enhancer of position effect 1; H, heterochromatin; Hs, Homo sapiens; incorp., incorporation; Msc1, multi-copy suppressor of Chk1; MSL2, male-specific lethal; Mm, Mus musculus; Mms1, methyl methane sulfonate sensitivity 1; mono, monoubiquitylation; Nc, Neurospora crassa; Not4, negative on TATA; Psh1, Pob3/Spt16/histone associated protein; poly, polyubiquitylation; RING1B, really interesting new gene 1B; S, soluble; Sc, Saccharomyces cerevisiae; SCF, Skp1-Cullin-F-box; Sp, Schizosaccharomyces pombe; Tom1, temperature-dependent organization in mitotic nucleus 1; txn, transcription; UBR2, ubiquitin protein ligase E3 in component n-recognin 2.
Shaping chromatin by sculpting the boundaries
Our recent study extended the chromatin function of Cul4–Ddb1Cdt2 to a role in controlling the boundaries between euchromatin and heterochromatin [65]. In fission yeast, the JmjC protein Epe1 antagonizes the spreading of heterochromatin and has a potential role in boundary formation. Spreading of heterochromatin is believed to occur by the recruitment of members of the heterochromatin protein 1 (HP1) family that bind to the repressive histone mark H3K9me through their chromodomain, and serve as a docking site for heterochromatic factors including the H3K9 HMTase. HP1 proteins also mediate the recruitment of Epe1 to chromatin. Nonetheless, Epe1 shows a specific strong enrichment at the boundaries and depletion in the body of heterochromatin, which differs substantially from the chromatin landscapes of HP1 proteins and H3K9 methylation. A solution to this paradox was provided by the finding that Epe1 is a substrate of Cul4–Ddb1Cdt2. Importantly, Epe1 is specifically targeted for ubiquitylation within the body of heterochromatin. In the presence of the E3 ligase, Epe1 is specifically removed from the body of heterochromatin, leading to its accumulation at the boundaries. Conversely, in cells lacking Cul4–Ddb1Cdt2, Epe1 invades heterochromatin and adapts a chromatin profile that resembles the chromosomal distribution of H3K9 methylation. Hence, whilst H3K9me and HP1 proteins mediate the initially uniform recruitment of Epe1 to heterochromatin, Cul4–Ddb1Cdt2 removes Epe1 by ‘sculpting’ from selective sites and controls its spatial distribution on chromatin, which specifies the function of this anti-silencer as a boundary factor (Fig 3C). Notably, the inappropriate expansion of Epe1 into heterochromatin in mutants of Cul4–Ddb1Cdt2triggers a silencing defect, which is completely suppressed in mutants that lack both the ubiquitin ligase and the boundary factor, indicating that Epe1 is the sole relevant substrate of Cul4–Ddb1Cdt2 in silencing. Thus far, the signals and factors that direct degradation of Epe1 to the body of heterochromatin or prevent its degradation at the boundaries have not been identified. However, as Epe1 contains two less conserved PCNA-interacting protein box motifs [56], it will be interesting to see whether its spatially controlled degradation also requires binding to PCNA.
Unknown substrates—understanding the roles of E3 ligases
Many other E3 ligases have been described that are important in regulating chromatin. However, their functions and underlying mechanism—that is, recruitment or degradation—remain obscure, as the relevant substrates have not yet been identified. However, known homologues of these enzymes or similarities to other pathways can allow us to speculate about possible mechanisms.
An orthologous complex of Cul4–Ddb1Cdt2 exists in fission yeast that comprises the adaptor protein Rik1—44% similarity to Ddb1—and the heterodimeric DCAF Raf1/Raf2 [66,67,68,69,70]. Cul4–Rik1Raf1/2 forms a complex with Clr4, the sole H3K9 HMTase present in Schizosaccharomyces pombe and, importantly, each component of this ClrC complex is essential for H3K9 methylation and heterochromatin formation. However, although Cul4–Rik1Raf1/2 seems to be important for recruiting Clr4 to heterochromatin [69], the identity of the substrate(s) and the role of ubiquitylation remain elusive. Although it has been reported that purified ClrC displays E3 activity towards H2B in vitro, there is no further evidence as to whether H2B is the substrate of this E3 in vivo. Nevertheless, ClrC-associated Cul4 is modified with Nedd8 [66], and lack of Nedd8 phenocopies the silencing defects of ClrC mutants [69], strongly suggesting that CRL activity is indeed required for H3K9 methylation. Thus, it is an attractive idea that Cul4–Rik1Raf1/2 might act similarly to the role of Cul4–Ddb1Cdt2 in ‘sculpting’ Epe1, that is, by removing an inhibitor of H3K9 methylation.
Although no orthologues of Rik1 have been identified outside the Schizosaccharomyces spp., the requirement for coupling ubiquitylation to repressive histone H3 methylation seems to be conserved and involves the canonical adaptor Ddb1. In Neurospora crassa, a CRL consisting of Cul4, DIM-8 (Ddb1), DIM-9 (DCAF26) and DIM-7 (similar to the fission yeast Raf2) assembles with the histone H3K9 HMTase DIM-5 in a complex termed DCDC (DIM-5/-7/-9, Cul4/Ddb1 complex; [71,72,73]). Similarly to the complex in fission yeast, all its components are essential for H3K9 methylation. Analysis of a neddylation-deficient Cul4 mutant revealed that H3K9 methylation requires the activity of the ubiquitin ligase, and that the HMTase DIM-5 preferentially interacts with the neddylated ligase [73]. Furthermore, the recruitment of the HMTase to chromatin requires one of the CRL components, DIM-7 [74]. Further examples for coupling repressive HMTases with CRL complexes can be found in Chlamydomonas [75], Arabidopsis [76] and humans [77] (see Table 1). However, so far, we do not have any knowledge as to whether the E3 activity in these complexes is restricted to mono- or polyubiquitylation, and whether ubiquitylation of the target helps to recruit another factor or has a degradatory function in the course of removing an inhibitor.
Beyond controlling the establishment of repressive histone methylation, other steps in heterochromatin seem to rely on ubiquitylation. The JmjC protein Msc1 is part of the fission yeast SwrC, which deposits the histone variant H2A.Z on chromatin. Lack of Msc1 does not alter the stability of the SwrC but affects the incorporation of H2A.Z to chromatin and results in phenotypes reminiscent of mutants of SwrC and H2A.Z—for example, chromosome instability and synthetic lethality with mutants deficient in kinetochore formation [78,79,80,81]. The role of SwrC in the formation of centromeres is not clear and controversial data have been reported about whether or not H2A.Z is deposited to the centromeric regions in fission yeast, and whether or not the localization of CENP-A is affected in mutants of SwrC [79,80,82]. Nonetheless, Msc1 was found to localize to centromeric regions [83]. In addition, cells lacking Msc1 show an increase in silencing at pericentromeric heterochromatin accompanied by an increase of H3K9 dimethylation and chromatin association of the HP1 protein Swi6 [80,81,83]. Besides the presence of a JmjN and a JmjC domain, which are found in many demethylases, Msc1 contains three individual RING—formerly annotated as PHD—domains that display E3 ligase activity towards artificial substrates in vitro, and Msc1 was found to interact with the E2 Rad6 in vivo [82]. Indeed, the functions of Msc1 in gene silencing and kinetochore formation require the presence of its RING domains [80,81], supporting the idea of a role in ubiquitylation. Intriguingly, cells lacking Msc1 are affected in the dynamics of the chromatin association of Swi6 by displaying a longer residence time of the HP1 protein on chromatin [83], suggesting that Msc1 is a negative regulator of Swi6. Thus, Msc1 might target Swi6 for ubiquitylation leading to its dissociation or degradation from chromatin, which would be consistent with other phenotypes of msc1 mutants [83]. However, ubiquitylated species have yet to be identified for Swi6. Furthermore, in vitro ubiquitylation with artificial substrates revealed a potential role for non-degradatory Lys 63-linked polyubiquitin chains by Msc1 [82]. Thus, alternative substrates and mechanisms other than Lys 48-polyubiquitin-mediated degradation might also be envisaged.
Additional E3 ligases have been described that are implicated in maintaining distinct chromatin domains (see Table 1). Whilst in some cases binding partners have been identified that might represent potential substrates, direct evidence is still lacking, and the genetic phenotypes are not always consistent with a role of a target protein. For instance, the orthologous complex of Cul4–Ddb1Cdt2in budding yeast, Cul8–Mms1Mms22, which affects silencing at telomeres, interacts with the DNA polymerase-α recruitment factor Ctf4 through Mms22; however, protein levels of Ctf4 seem not to be affected in cells lacking Cul8, and the ctf4 mutant is epistatic to mutants of the Cul8–Mms1Mms2 complex [84]. It has thus been suggested that instead of being a substrate, Ctf4 might have regulatory functions or, alternatively, might not be a proteolytic substrate. Similarly, the fission yeast Ctf4 orthologue Mcl1 binds to SCFPof3, but Mcl1 protein levels are relatively stable in wild-type cells. The mcl1 pof3 double mutant shows synthetic phenotypes, implying functions other than regulating the turnover of Mcl1 [85]. Heterochromatin in S. pombe is also compromised in mutants of the SCF-type E3 ligase APC/C, and subunits of this complex were shown to localize to heterochromatin and to interact with Swi6 and Clr4 [85]. Whether this role of APC/C in silencing is mediated by one of its known cell cycle targets or by a new substrate remains to be investigated.
Conclusion and perspectives
Post-translational modifications of chromatin are pivotal in defining the functional states of the genome. Their substrate spectrum is not restricted to histone proteins, which has become particularly evident for ubiquitylation. This modification shapes chromatin by regulating the incorporation of specific histone proteins, the recruitment and stability of other histone-modifying enzymes and the spatio-temporal distribution of other chromatin-associated factors. Many E3 ligases have been identified that contribute to the establishment and maintenance of epigenetic states. However, in numerous cases we have yet to understand the mechanism of regulation as we do not know the consequence of ubiquitylation—recruitment or degradation—the signals that direct ubiquitylation to specific sites on chromatin, or the target proteins of this regulation (see Sidebar A). Identifying substrates of E3 enzymes has proven to be particularly challenging, as the interaction between E3 and substrate is transient, and spatial and temporal regulation is often restricted to specific subpopulations and cell cycle phases. As with other cases of ubiquitin-dependent regulation, finding the relevant E3 targets will be the key to understanding the mode of regulation—mono- compared with polyubiquitylation—and to elucidating the role of this regulation in shaping the chromatin landscape.
Sidebar A | In need of answers.
What are the targets of ubiquitylation? Many E3 ligases have been identified that are involved in defining the chromatin state, however, we still do not know their functions as the relevant substrates have not yet been identified.
What is the mechanism of ubiquitylation? The functions of the individual E3 ligases can only be understood if we know their mode of modification (mono- compared with polyubiquitylation) and targeting (recruitment compared with degradation).
What are the signals that direct ubiquitylation? Ubiquitylation is often subject to spatial and temporal regulation but in numerous cases we do not understand which factors or post-translational modifications define where and when ubiquitylation should take place.
How is ubiquitylation reversed? Most epigenetic changes are reversible and diverse deubiquitylating enzymes are important in chromatin formation, however, we often do not know what defines their substrate specificity.

Sigurd Braun

Hiten D Madhani
Acknowledgments
We thank Andrew Bowman, Maria Hondele, Corey Laverty and Carla Margulies for critical reading of the manuscript. S.B. was a recipient of a postdoctoral fellowship (BR 3511/1-1) from the German Research Foundation and is supported by the EpiGeneSys Network of Excellence from the European Union. Chromatin research in the Madhani laboratory is supported by a National Institutes of Health grant (GM071801).
Footnotes
The authors declare that they have no conflict of interest.
References
- Ye Y, Rape M (2009) Building ubiquitin chains: E2 enzymes at work. Nat Rev Mol Cell Biol 10: 755–764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duda DM et al. (2011) Structural regulation of cullin-RING ubiquitin ligase complexes. Curr Opin Struct Biol 21: 257–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muratani M, Tansey WP (2003) How the ubiquitin-proteasome system controls transcription. Nat Rev Mol Cell Biol 4: 192–201 [DOI] [PubMed] [Google Scholar]
- Moldovan G-L, Pfander B, Jentsch S (2007) PCNA, the maestro of the replication fork. Cell 129: 665–679 [DOI] [PubMed] [Google Scholar]
- Hwang WW, Venkatasubrahmanyam S, Ianculescu AG, Tong A, Boone C, Madhani HD (2003) A conserved RING finger protein required for histone H2B monoubiquitination and cell size control. Mol Cell 11: 261–266 [DOI] [PubMed] [Google Scholar]
- Jentsch S, McGrath JP, Varshavsky A (1987) The yeast DNA repair gene RAD6 encodes a ubiquitin-conjugating enzyme. Nature 329: 131–134 [DOI] [PubMed] [Google Scholar]
- Robzyk K, Recht J, Osley MA (2000) Rad6-dependent ubiquitination of histone H2B in yeast. Science 287: 501–504 [DOI] [PubMed] [Google Scholar]
- Wood A et al. (2003) Bre1, an E3 ubiquitin ligase required for recruitment and substrate selection of Rad6 at a promoter. Mol Cell 11: 267–274 [DOI] [PubMed] [Google Scholar]
- Kim J, Hake SB, Roeder RG (2005) The human homolog of yeast BRE1 functions as a transcriptional coactivator through direct activator interactions. Mol Cell 20: 759–770 [DOI] [PubMed] [Google Scholar]
- Osley MA (2006) Regulation of histone H2A and H2B ubiquitylation. Brief Funct Genomic Proteomic 5: 179–189 [DOI] [PubMed] [Google Scholar]
- Weake VM, Workman JL (2008) Histone ubiquitination: triggering gene activity. Mol Cell 29: 653–663 [DOI] [PubMed] [Google Scholar]
- Fujiki R et al. (2011) GlcNAcylation of histone H2B facilitates its monoubiquitination. Nature 480: 557–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emre NC et al. (2005) Maintenance of low histone ubiquitylation by Ubp10 correlates with telomere-proximal Sir2 association and gene silencing. Mol Cell 17: 585–594 [DOI] [PubMed] [Google Scholar]
- Henry KW et al. (2003) Transcriptional activation via sequential histone H2B ubiquitylation and deubiquitylation, mediated by SAGA-associated Ubp8. Genes Dev 17: 2648–2663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyce A et al. (2007) H2B ubiquitylation acts as a barrier to Ctk1 nucleosomal recruitment prior to removal by Ubp8 within a SAGA-related complex. Mol Cell 27: 275–288 [DOI] [PubMed] [Google Scholar]
- Gardner RG, Nelson ZW, Gottschling DE (2005) Ubp10/Dot4p regulates the persistence of ubiquitinated histone H2B: distinct roles in telomeric silencing and general chromatin. Mol Cell Biol 25: 6123–6139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buszczak M, Paterno S, Spradling AC (2009) Drosophila stem cells share a common requirement for the histone H2B ubiquitin protease scrawny. Science 323: 248–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frappier L, Verrijzer CP (2011) Gene expression control by protein deubiquitinases. Curr Opin Genet Dev 21: 207–213 [DOI] [PubMed] [Google Scholar]
- Briggs SD, Xiao T, Sun ZW, Caldwell JA, Shabanowitz J, Hunt DF, Allis CD, Strahl BD (2002) Gene silencing: trans-histone regulatory pathway in chromatin. Nature 418: 498. [DOI] [PubMed] [Google Scholar]
- Sun Z-W, Allis CD (2002) Ubiquitination of histone H2B regulates H3 methylation and gene silencing in yeast. Nature 418: 104–108 [DOI] [PubMed] [Google Scholar]
- Pavri R, Zhu B, Li G, Trojer P, Mandal S, Shilatifard A, Reinberg D (2006) Histone H2B monoubiquitination functions cooperatively with FACT to regulate elongation by RNA polymerase II. Cell 125: 703–717 [DOI] [PubMed] [Google Scholar]
- Zofall M, Fischer T, Zhang K, Zhou M, Cui B, Veenstra TD, Grewal SI (2009) Histone H2AZ cooperates with RNAi and heterochromatin factors to suppress antisense RNAs. Nature 461: 419–422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shema E, Kim J, Roeder RG, Oren M (2011) RNF20 inhibits TFIIS-facilitated transcriptional elongation to suppress pro-oncogenic gene expression. Mol Cell 42: 477–488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner SD, Ricci AR, Petropoulos H, Genereaux J, Skerjanc IS, Brandl CJ (2002) The E2 ubiquitin conjugase Rad6 is required for the ArgR/Mcm1 repression of ARG1 transcription. Mol Cell Biol 22: 4011–4019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandrasekharan MB, Huang F, Sun ZW (2009) Ubiquitination of histone H2B regulates chromatin dynamics by enhancing nucleosome stability. Proc Natl Acad Sci USA 106: 16686–16691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fierz B, Chatterjee C, McGinty RK, Bar-Dagan M, Raleigh DP, Muir TW (2011) Histone H2B ubiquitylation disrupts local and higher-order chromatin compaction. Nat Chem Biol 7: 113–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider J et al. (2005) Molecular regulation of histone H3 trimethylation by COMPASS and the regulation of gene expression. Mol Cell 19: 849–856 [DOI] [PubMed] [Google Scholar]
- Lee JS, Shukla A, Schneider J, Swanson SK, Washburn MP, Florens L, Bhaumik SR, Shilatifard A (2007) Histone crosstalk between H2B monoubiquitination and H3 methylation mediated by COMPASS. Cell 131: 1084–1096 [DOI] [PubMed] [Google Scholar]
- Vitaliano-Prunier A, Menant A, Hobeika M, Géli V, Dargemont C (2008) Ubiquitylation of the COMPASS component Swd2 links H2B ubiquitylation to H3K4 trimethylation. Nat Cell Biol 10: 1365–1371 [DOI] [PubMed] [Google Scholar]
- Batta K, Zhang Z, Yen K, Goffman DB, Pugh BF (2011) Genome-wide function of H2B ubiquitylation in promoter and genic regions. Genes Dev 25: 2254–2265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming AB, Kao CF, Hillyer C, Pikaart M, Osley MA (2008) H2B ubiquitylation plays a role in nucleosome dynamics during transcription elongation. Mol Cell 31: 57–66 [DOI] [PubMed] [Google Scholar]
- Geng F, Tansey WP (2008) Polyubiquitylation of histone H2B. Mol Biol Cell 19: 3616–3624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, Zee BM, Wang Y, Garcia BA, Dou Y (2011) The RING finger protein MSL2 in the MOF complex is an E3 ubiquitin ligase for H2B K34 and is involved in crosstalk with H3 K4 and K79 methylation. Mol Cell 43: 132–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H et al. (2004) Role of histone H2A ubiquitination in Polycomb silencing. Nature 431: 873–878 [DOI] [PubMed] [Google Scholar]
- Simon JA, Kingston RE (2009) Mechanisms of polycomb gene silencing: knowns and unknowns. Nat Rev Mol Cell Biol 10: 697–708 [DOI] [PubMed] [Google Scholar]
- Lagarou A et al. (2008) dKDM2 couples histone H2A ubiquitylation to histone H3 demethylation during Polycomb group silencing. Genes Dev 22: 2799–2810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarcinella E, Zuzarte PC, Lau PNI, Draker R, Cheung P (2007) Monoubiquitylation of H2A.Z distinguishes its association with euchromatin or facultative heterochromatin. Mol Cell Biol 27: 6457–6468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W, Zhu P, Wang J, Pascual G, Oghi KA, Lozach J, Glass CK, Rosenfeld MG (2008) Histone H2A monoubiquitination represses transcription by inhibiting RNA polymerase II transcriptional elongation. Mol Cell 29: 69–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- An JY, Kim EA, Jiang Y, Zakrzewska A, Kim DE, Lee MJ, Mook-Jung I, Zhang Y, Kwon YT (2010) UBR2 mediates transcriptional silencing during spermatogenesis via histone ubiquitination. Proc Natl Acad Sci USA 107: 1912–1917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Q, Pao GM, Huynh AM, Suh H, Tonnu N, Nederlof PM, Gage FH, Verma IM (2011) BRCA1 tumour suppression occurs via heterochromatin-mediated silencing. Nature 477: 179–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheuermann JC, de Ayala Alonso AG, Oktaba K, Ly-Hartig N, McGinty RK, Fraterman S, Wilm M, Muir TW, Müller J (2010) Histone H2A deubiquitinase activity of the Polycomb repressive complex PR-DUB. Nature 465: 243–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stock JK, Giadrossi S, Casanova M, Brookes E, Vidal M, Koseki H, Brockdorff N, Fisher AG, Pombo A (2007) Ring1-mediated ubiquitination of H2A restrains poised RNA polymerase II at bivalent genes in mouse ES cells. Nat Cell Biol 9: 1428–1435 [DOI] [PubMed] [Google Scholar]
- Nakagawa T et al. (2008) Deubiquitylation of histone H2A activates transcriptional initiation via trans-histone cross-talk with H3K4 di- and trimethylation. Genes Dev 22: 37–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richly H et al. (2010) Transcriptional activation of polycomb-repressed genes by ZRF1. Nature 468: 1124–1128 [DOI] [PubMed] [Google Scholar]
- Margueron R et al. (2009) Role of the polycomb protein EED in the propagation of repressive histone marks. Nature 461: 762–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrigoni R Alam SL, Wamstad JA, Bardwell VJ, Sundquist WI, Schreiber-Agus N (2006) The Polycomb-associated protein Rybp is a ubiquitin binding protein. FEBS Lett 580: 6233–6241 [DOI] [PubMed] [Google Scholar]
- García E, Marcos-Gutiérrez C, del Mar Lorente M, Moreno JC, Vidal M (1999) RYBP, a new repressor protein that interacts with components of the mammalian Polycomb complex, and with the transcription factor YY1. EMBO J 18: 3404–3418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R et al. (2010) Polycomb group targeting through different binding partners of RING1B C-terminal domain. Structure 18: 966–975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takayama Y Mamnun YM, Trickey M, Dhut S, Masuda F, Yamano H, Toda T, Saitoh S (2010) Hsk1- and SCF(Pof3)-dependent proteolysis of S. pombe Ams2 ensures histone homeostasis and centromere function. Dev Cell 18: 385–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunjan A, Verreault AA (2003) Rad53 kinase-dependent surveillance mechanism that regulates histone protein levels in S. cerevisiae. Cell 115: 537–549 [DOI] [PubMed] [Google Scholar]
- Hewawasam G, Shivaraju M, Mattingly M, Venkatesh S, Martin-Brown S, Florens L, Workman JL, Gerton JL (2010) Psh1 is an E3 ubiquitin ligase that targets the centromeric histone variant Cse4. Mol Cell 40: 444–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranjitkar P, Press MO, Yi X, Baker R, MacCoss MJ, Biggins S (2010) An E3 ubiquitin ligase prevents ectopic localization of the centromeric histone H3 variant via the centromere targeting domain. Mol Cell 40: 455–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno-Moreno O, Medina-Giró S, Torras-Llort M, Azorín F (2011) The F box protein partner of paired regulates stability of Drosophila centromeric histone H3, CenH3(CID). Curr Biol 21: 1488–1493 [DOI] [PubMed] [Google Scholar]
- Krogan NJ et al. (2002) RNA polymerase II elongation factors of Saccharomyces cerevisiae: a targeted proteomics approach. Mol Cell Biol 22: 6979–6992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connell BC, Harper JW (2007) Ubiquitin proteasome system (UPS): what can chromatin do for you? Curr Opin Cell Biol 19: 206–214 [DOI] [PubMed] [Google Scholar]
- Havens CG, Walter JC (2011) Mechanism of CRL4(Cdt2), a PCNA-dependent E3 ubiquitin ligase. Genes Dev 25: 1568–1582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abbas T, Shibata E, Park J, Jha S, Karnani N, Dutta A (2010) CRL4(Cdt2) regulates cell proliferation and histone gene expression by targeting PR-Set7/Set8 for degradation. Mol Cell 40: 9–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda H, Hübner MR, Beck DB, Vermeulen M, Hurwitz J, Spector DL, Reinberg D (2010) Regulation of the histone H4 monomethylase PR-Set7 by CRL4(Cdt2)-mediated PCNA-dependent degradation during DNA damage. Mol Cell 40: 364–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jørgensen S et al. (2011) SET8 is degraded via PCNA-coupled CRL4(CDT2) ubiquitylation in S phase and after UV irradiation. J Cell Biol 192: 43–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tardat M, Brustel J, Kirsh O, Lefevbre C, Callanan M, Sardet C, Julien E (2010) The histone H4 Lys 20 methyltransferase PR-Set7 regulates replication origins in mammalian cells. Nat Cell Biol 12: 1086–1093 [DOI] [PubMed] [Google Scholar]
- Centore RC et al. (2010) CRL4(Cdt2)-mediated destruction of the histone methyltransferase Set8 prevents premature chromatin compaction in S phase. Mol Cell 40: 22–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Cheng EH, Hsieh JJ (2007) Bimodal degradation of MLL by SCFSkp2 and APCCdc20 assures cell cycle execution: a critical regulatory circuit lost in leukemogenic MLL fusions. Genes Dev 21: 2385–2398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Rechem C et al. (2011) The SKP1-Cul1-F-box and leucine-rich repeat protein 4 (SCF-FbxL4) ubiquitin ligase regulates lysine demethylase 4A (KDM4A)/Jumonji domain-containing 2A (JMJD2A) protein. J Biol Chem 286: 30462–30470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mersman DP, Du H-N, Fingerman IM, South PF, Briggs SD (2009) Polyubiquitination of the demethylase Jhd2 controls histone methylation and gene expression. Genes Dev 23: 951–962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun S, Garcia JF, Rowley M, Rougemaille M, Shankar S, Madhani HD (2011) The Cul4-Ddb1(Cdt)2 ubiquitin ligase inhibits invasion of a boundary-associated antisilencing factor into heterochromatin. Cell 144: 41–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong E-JE, Villén J, Gerace EL, Gygi SP, Moazed D (2005) A cullin E3 ubiquitin ligase complex associates with Rik1 and the Clr4 histone H3-K9 methyltransferase and is required for RNAi-mediated heterochromatin formation. RNA Biol 2: 106–111 [DOI] [PubMed] [Google Scholar]
- Horn PJ, Bastie JN, Peterson CLA (2005) Rik1-associated, cullin-dependent E3 ubiquitin ligase is essential for heterochromatin formation. Genes Dev 19: 1705–1714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Goto DB, Zaratiegui M, Tang X, Martienssen R, Cande WZ (2005) Two novel proteins, dos1 and dos2, interact with rik1 to regulate heterochromatic RNA interference and histone modification. Curr Biol 15: 1448–1457 [DOI] [PubMed] [Google Scholar]
- Jia S, Kobayashi R, Grewal SIS (2005) Ubiquitin ligase component Cul4 associates with Clr4 histone methyltransferase to assemble heterochromatin. Nat Cell Biol 7: 1007–1013 [DOI] [PubMed] [Google Scholar]
- Thon G, Hansen KR, Altes SP, Sidhu D, Singh G, Verhein-Hansen J, Bonaduce MJ, Klar AJ (2005) The Clr7 and Clr8 directionality factors and the Pcu4 cullin mediate heterochromatin formation in the fission yeast Schizosaccharomyces pombe. Genetics 171: 1583–1595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Wang J, Hu Q, Quan Y, Chen H, Cao Y, Li C, Wang Y, He Q (2010) DCAF26, an adaptor protein of Cul4-based E3, is essential for DNA methylation in Neurospora crassa. PLoS Genet 6: e1001132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis ZA, Adhvaryu KK, Honda S, Shiver AL, Knip M, Sack R, Selker EU (2010) DNA methylation and normal chromosome behavior in Neurospora depend on five components of a histone methyltransferase complex, DCDC. PLoS Genet 6: e1001196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Shen Y, Yang S, Wang J, Hu Q, Wang Y, He Q (2010) Ubiquitin ligase components Cullin4 and DDB1 are essential for DNA methylation in Neurospora crassa. J Biol Chem 285: 4355–4365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis ZA, Adhvaryu KK, Honda S, Shiver AL, Selker EU (2010) Identification of DIM-7, a protein required to target the DIM-5 H3 methyltransferase to chromatin. Proc Natl Acad Sci USA 107: 8310–8315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamasaki T, Ohama T (2011) Involvement of Elongin C in the spread of repressive histone modifications. Plant J 65: 51–61 [DOI] [PubMed] [Google Scholar]
- Dumbliauskas E et al. (2011) The Arabidopsis CUL4-DDB1 complex interacts with MSI1 and is required to maintain MEDEA parental imprinting. EMBO J 30: 731–743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higa LA, Wu M, Ye T, Kobayashi R, Sun H, Zhang H (2006) CUL4-DDB1 ubiquitin ligase interacts with multiple WD40-repeat proteins and regulates histone methylation. Nat Cell Biol 8: 1277–1283 [DOI] [PubMed] [Google Scholar]
- Ahmed S, Dul B, Qiu X, Walworth NC (2007) Msc1 acts through histone H2AZ to promote chromosome stability in Schizosaccharomyces pombe. Genetics 177: 1487–1497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchanan L et al. (2009) The Schizosaccharomyces pombe JmjC-protein, Msc1, prevents H2AZ localization in centromeric and subtelomeric chromatin domains. PLoS Genet 5: e1000726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou H, Wang Y, Kallgren SP, Thompson J, Yates JR 3rd, Jia S (2010) Histone variant H2A.Z regulates centromere silencing and chromosome segregation in fission yeast. J Biol Chem 285: 1909–1918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu X, Dul BE, Walworth NC (2010) Activity of a C-terminal plant homeodomain (PHD) of Msc1 is essential for function. J Biol Chem 285: 36828–36835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dul BE, Walworth NC (2007) The plant homeodomain fingers of fission yeast Msc1 exhibit E3 ubiquitin ligase activity. J Biol Chem 282: 18397–18406 [DOI] [PubMed] [Google Scholar]
- Lawrence RJ, Volpe TA (2009) Msc1 links dynamic Swi6/HP1 binding to cell fate determination. Proc Natl Acad Sci USA 106: 1163–1168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimura S, Yamaguchi T, Ishii S, Noro E, Katsura T, Obuse C, Kamura T (2010) Cul8/Rtt101 forms a variety of protein complexes that regulate DNA damage response and transcriptional silencing. J Biol Chem 285: 9858–9867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubey RN, Nakwal N, Bisht KK, Saini A, Haldar S, Singh J (2009) Interaction of APC/C-E3 ligase with Swi6/HP1 and Clr4/Suv39 in heterochromatin assembly in fission yeast. J Biol Chem 284: 7165–7176 [DOI] [PMC free article] [PubMed] [Google Scholar]